Abstract
Background
Genome-wide association studies in asthma have repeatedly identified single nucleotide polymorphisms in the ORM (yeast)-like protein isoform 3 (ORMDL3) gene across different populations. Although the ORM-homologues in yeast are well-known inhibitors of sphingolipid (SL) synthesis, it is still unclear whether and how mammalian ORMDL3 regulates SL metabolism and whether altered SL synthesis would be causally related to asthma risk.
Objective
To examine the in vivo role of ORMDL3 in SL metabolism and allergic asthma
Methods
Ormdl3-LacZ reporter mice, gene deficient Ormdl3-/- and overexpressing Ormdl3 Tg/wt mice were exposed to physiologically relevant aeroallergens such as house dust mite or Alternaria alternata to induce experimental asthma. Mass spectrometry based sphingolipidomics was performed, and airway eosinophilia, Th2 cytokine production, Ig synthesis, airway remodeling and bronchial hyperreactivity measured.
Results
HDM challenge significantly increased total sphingolipids in the lung of HDM-sensitized mice compared to controls. In Ormdl3Tg/wt mice, the allergen-induced increase in lung ceramides was significantly reduced, whereas total sphingolipid levels were not affected. Conversely, in liver and serum, total sphingolipids including ceramides were increased in Ormdl3-/- mice, while they were lowered in Ormdl3Tg/wt mice. This difference was independent of allergen exposure. Despite these changes, all features of asthma were identical between wildtype, Ormdl3Tg/wt and Ormdl3-/- mice, across several models of experimental asthma.
Conclusion
ORMDL3 regulates systemic ceramide levels, but genetically interfering with Ormdl3. expression does not result in altered experimental asthma.
Capsule Summary
ORMDL3 regulates systemic ceramide levels, but genetically interfering with Ormdl3 expression does not result in altered experimental asthma.
Keywords: Asthma, ORMDL3, house dust mite, Alternaria alternata, sphingolipids, ceramides
Introduction
Allergic asthma results from a complex gene-by-environment interaction and is currently still on the rise (1). Single nucleotide polymorphisms (SNPs) on chromosome 17q12-21 confer the most significant and replicated genetic susceptibility to childhood-onset asthma and rhinovirus-induced wheezing illness in childhood (2–8). Many of these SNPs fall within the promoter region of the ORM (yeast)-like protein isoform 3 (ORMDL3) gene and induce slightly elevated levels of ORMDL3, particularly in T lymphocytes and following stimulation of PBMCs with allergens (2,9–11).Ormdl3 mRNA is also upregulated in murine asthma models, driven by ovalbumin (OVA), house dust mite (HDM) or Alternaria alternata (12,13). However, studies addressing the functional role of ORMDL3 in asthma generated conflicting conclusions. Both transgenic overexpression as well as genetic deficiency of Ormdl3 can enhance key asthma features, whereas one study showed that Ormdl3 deficiency suppressed only bronchial hyperreactivity (BHR) (14–16). Given the multitude of genetic association studies in humans, the currently prevailing hypothesis is still that ORMDL3 overexpression has a causal role in asthma development or progression.
The molecular mechanism by which ORMDL3 contributes to asthma is still a matter of intense debate (6,8). ORMDL3 is member of an evolutionary conserved family of endoplasmic reticulum (ER)-residing proteins, and has two paralogues in vertebrates, ORMDL1 and ORMDL2, that have not been associated with asthma (17). In yeast, the ORM homologues are described as regulators of de novo sphingolipid synthesis by controlling the activity of the rate limiting enzyme serine palmitoyl transferase (SPT) (18–23). In mammals however, ORMDLs lack the N-terminal phosphorylation site that is crucial for SPT regulation in yeast. Mammalian SPT activity seems to be affected only when all ORMDL paralogues are overexpressed or downregulated simultaneously (17,24–26), making it unlikely that SNPs in only ORMDL3 influence asthma by SPT inhibition. As an ER-resident protein, ORMDL3 has also been described to affect calcium metabolism and the unfolded protein response, influencing cytokine secretion by structural or immune cells (6,12,27–29). However, most molecular studies on ORMDL3 were performed in vitro and have led to contradictory results due to the use of different cell lines and distinct approaches to measure total sphingolipid de novo synthesis and to control ORMDL3 expression. Furthermore, many studies were performed on epithelial cells, macrophages, mast cells and eosinophils (6,12,13,29–31), whereas it has been recently demonstrated that chr17q12-21 SNPs affect ORMDL3 expression most prominently in T-cells (9).
Here, we addressed the role of ORMDL3 in SL metabolism and asthma in newly generated Ormdl3-LacZ reporter mice, full Ormdl3 KO mice (Ormdl3-/-), and mice overexpressing murine Ormdl3 from a Bacterial Artificial Chromosome (BAC)-transgene (Ormdl3 Tg/wt). Using these unique gene-modified mice and their littermate controls, we found that ORMDL3 expression influences ceramide levels in serum and liver and to a lesser extent in lung tissue. At the same time, interfering with Ormdl3 did not impact on key asthma parameters in various allergen driven asthma models. These data do not support the currently prevailing paradigm that ORMDL3 drives asthma by interfering with SPT activity or sphingolipid homeostasis.
Methods
Mice
Ormdl3-LacZ reporter mice were generated by inserting a targeting construct of the European Conditional Mouse Mutagenesis consortium (project 72180) into the first intron of the Ormdl3 gene (Fig. 1A). This construct contains a sequence that consists of an En2 splice acceptor site, an internal ribosome entry site, a LacZ sequence, a polyA-tail, a loxP site, and a neomycin coding sequence driven by a human β-actin promoter that is flanked by 2 Flp recombinase target (FRT) sites. ORMDL3 knockout (Ormdl3-/-) mice and ORMDL3 transgenic (Ormdl3 Tg/wt) mice were previously described (25). All mice were backcrossed for at least 10 crosses onto a C57BL/6 background. 1-DER mice express an MHC class II-restricted TCR specific for Der p 1 on their CD4 T cells and were generated as previously described (32–34). Mice were bred under specific pathogen–free conditions at the animal house of the VIB/UGhent Center for Inflammation Research. Experiments were carried out using age– and gender-matched groups. All mice were used between 6-14 weeks of age. All animal procedures were approved by the Ethical Committee of Ghent University.
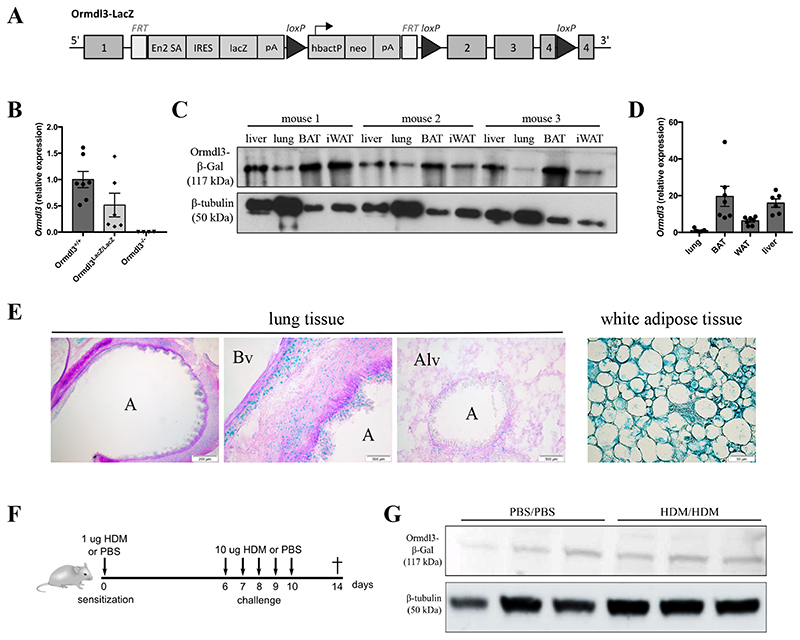
Figure 1. Ormdl3-LacZ reportermice as a useful tool to study ORMDL3 expression.
A)Ormdl3-LacZ reportermice (Ormdl3 LacZ/LacZ) were generated by inserting a targeting construct of the EUCOMM consortium into the first intron of Ormdl3. Exon 2, exon 3 and part of exon 4 are flanked by two loxP sites, which enable the generation of conditional knockout mice upon crossing with mice expressing Cre-recombinase.
B)Ormdl3 mRNA expression levels in lungs from Ormdl3+/+, Ormdl3 LacZ/LacZ and Ormdl3-/- mice. Expression values are shown relative to means of the wildtype group. Data were pooled from 2 experiments (n=7,6,4; means +/-SEM).
C)Western blot showing β-galactosidase expression in liver, lung, brown adipose tissue (BAT) and white adipose tissue (WAT) in three individual Ormdl3-LacZ reportermice. β-tubulin was used as a loading control.
D)Ormdl3 transcript levels in lung, BAT, WAT and liver in wildtype mice. Expression values are shown relative to means of lung samples (means +/- SEM).
E)Immunohistochemistry analysis of β-galactosidase expression (blue) on lung OCT-inflated cryosections and WAT of Ormdl3-LacZ reportermice. Periodic-acid Schiff staining was used as counterstaining. A = airway; Bv = blood vessel; Alv = alveoli.
F)Scheme representing the acute house dust mite (HDM)-dependent asthma model.
G)Western blot showing β-galactosidase expression in lung tissue from mock- and HDM-challenged Ormdl3-LacZ reportermice.
Models of allergic asthma
The HDM-induced asthma model was performed as described before (35). In brief, mice were sensitized intratracheally (i.t.) on day 0 with 1 μg HDM extract (Greer Laboratories, Lenoir, USA) or saline, followed by 10 μg intranasal (i.n.) challenges from day 6 to 10. On day 14, mice were euthanized by an overdose pentobarbital. In the chronic HDM-induced asthma model, mice were instilled i.n. with 10 ug HDM, or saline as a control, three times a week for 5 weeks. Asthma features were determined 3 days after the last challenge. In the Alternaria-induced asthma model, mice were instilled i.n. with 20ug Alternaria alternata (Greer Laboratories) three times a week for 3 weeks. All i.t. and i.n. treatments were given in 80 and 40 ul PBS, respectively, and under light isoflurane anesthesia.
Bronchoalveolar lavage (BAL) was performed using 3x1ml of EDTA-containing PBS (0,5 mM). Blood was obtained from the iliac vein in non-coated Eppendorf tubes to prepare serum. Single cells suspensions from mediastinal lymph nodes were obtained by homogenizing the organ through a 100 um cell sieve. Cells were restimulated ex vivo with 15 ug/ml HDM for 3 days and supernatants were collected for ELISA (Ready-set-go kits from eBioscience). The left lung was fixed overnight in 4% paraformaldehyde and was used for histology. Right lung tissue was snap frozen in liquid nitrogen and kept at -80°C until used for RNA extraction.
BHR determination
Mice were anesthetized with urethane, paralyzed with D-tubocurarine, tracheostomized, and intubated with a 20-G catheter, followed by mechanical ventilation in a Flexivent apparatus (SCIREQ). Respiratory frequency was set at 150 breaths/min with a tidal volume (Vt) of 10 ml/kg, and a positive–end expiratory pressure (PEEP) of 3 cm H2O was applied. Increasing concentrations of methacholine were nebulized (0–400 mg/ml) for 12 seconds and dynamic resistance, elastance and compliance were recorded by 12 repeat measurements during 2 minutes per dose. Baseline resistance was restored in between the doses.
Histology
After overnight fixation in 4% paraformaldehyde solution, tissues were embedded in paraffin, cut into 5 um slices and stained with either Periodic Acid-Schiff (PAS) or processed for immunochemistry. Periodic Acid-Schiff staining was performed as described in Debeuf et al. (2016). To detect peribronchial α-smooth muscle actin (α-SMA), lung sections were overnight incubated with an anti-α-SMA primary antibody (A2547, Sigma-Aldrich). Species‐and isotype-matched antibodies were used as negative controls. Immunoreactivity was detected by sequential incubations of lung sections with a biotinylated secondary Ab (E0433, Dako, Belgium), Vectastain Elite ABC kit (Vector Laboratories, Burlingame, California, USA), followed by 3,3-diaminobenzidine chromogen (Dako). Sections were mounted by use of Entellan mounting medium (Merck Millipore).
Lipidomics
Individual sphingolipid species were quantified by a novel LC-MS method for a comprehensive analysis of typical and atypical sphingolipid species. A lipidomics profiling was done in 20 μL mouse serum or 20 μg of homogenate from lung tissue were extracted using the MMC protocol as described previously (36). d7-sphinganine, d7-sphingosine, Cer d18:0/12:0, Cer d18:1/12:0, SM d18:1/12:0, GluCer d18:1/18:0 and d7-S1P were used as internal standards. C18-column-based method for chromatographic separation and MS analysis was used, as described previously (36).
Statistical analyses
For all experiments, normality of each group was first checked by Shapiro-Wilk statistical test. If data sets were normally distributed, a One-Way ANOVA test was performed. In case of not normally distributed data, a Kruskal-Wallis test was done. All statistical tests were performed in GraphPad Prism.
Results
Effect of allergen challenge on ORMDL3 expression in asthma
One of the major drawbacks in the ORMDL3 field is the lack of specific antibodies, due to the large sequence homology with ORMDL1 and ORMDL2 (84% and 83%, respectively). Many investigators have therefore relied on ORMDL3 mRNA analysis of isolated tissues or cells, without confirming where exactly ORMDL3 is expressed. To be able to better address ORMDL3 expression, Ormdl3-LacZ reporter mice were generated by inserting a targeting construct into the first intron of Ormdl3 (Fig. 1A)(25). Although these mice still express some basal Ormdl3 (Fig. 1B), β-galactosidase activity can be used as readout for Ormdl3 promoter activation. We first compared ORMDL3 expression levels in different tissues in steady state mice. Despite the fact that ORMDL3 is an asthma susceptibility gene, β-galactosidase expression was relatively low in lung tissue, especially compared to liver, and white and brown adipose tissues where ORMDL3 was abundantly present (Fig. 1C and Suppl. Fig. 1A). Similar results were obtained by RT-qPCR (Fig. 1D), consistent with RNA-seq data derived from the Genevestigator database (Suppl. Fig. 1B)(37). Previous studies suggested that epithelial cells would be the main cell type expressing ORMDL3 in the lung (14). However, in our hands, immunohistochemistry revealed diffuse and low β-galactosidase activity in subepithelial and vascular smooth muscle cells, epithelial cells and fibroblasts (Fig. 1E). On the contrary, high β-galactosidase activity was seen in white adipose tissue (Fig. 1E), consistent with mRNA expression data.
To address whether ORMDL3 expression was induced by allergen challenge in vivo, we employed a previously established model in which inhaled HDM challenge of HDM-sensitized mice leads to airway eosinophilia (Fig. 1F) (35). No increase in lung β-galactosidase activity was seen in allergen-challenged mice compared to control PBS exposed mice (Fig. 1G and Suppl. Fig. 1C). These results showed that ORMDL3 expression in mouse lungs is low and did also not increase after allergen challenge using sensitized mice.
Characterization of Ormdl3Tg/wt and Ormdl3-/- mice
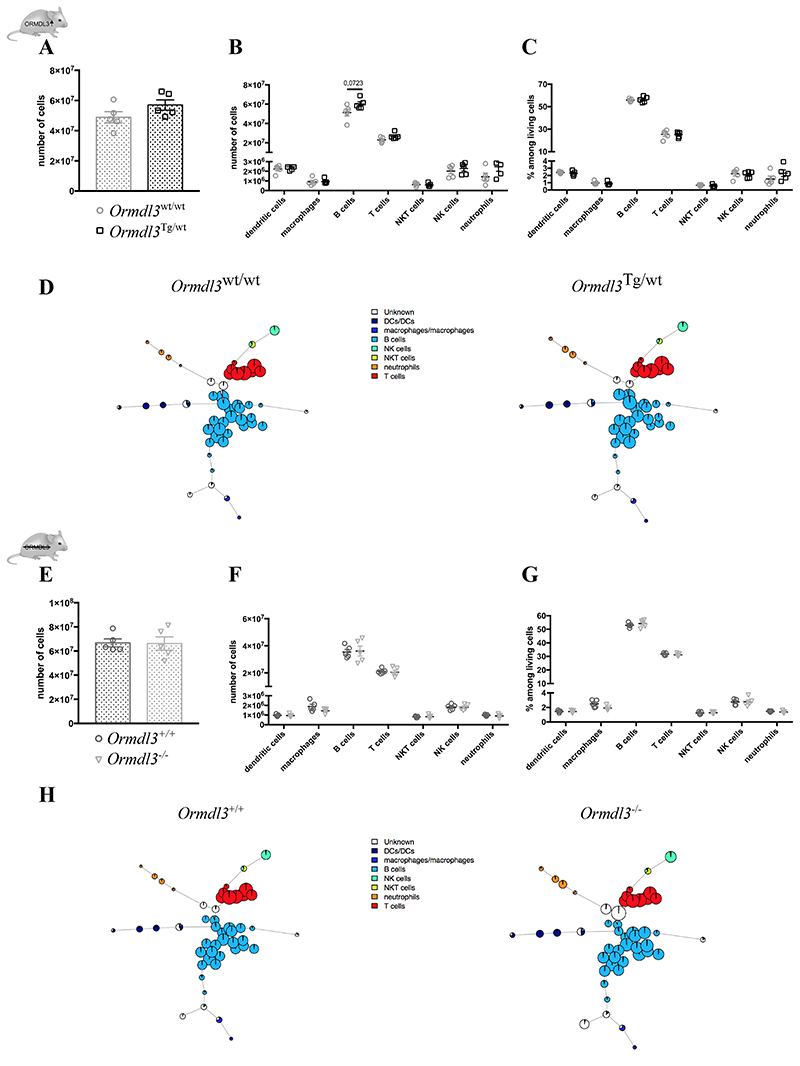
To assess the effects of altered ORMDL3 expression levels in vivo, we have recently generated Ormdl3-overexpressing mice (Ormdl3 Tg/wt) and gene deficient Ormdl3-/- mice (25). Before we used these mice in asthma experiments, we wanted to address if immune cell development was affected by a systemic change in Ormdl3 expression. Mice from both strains were healthy and did not develop a spontaneous phenotype, even with ageing (data not shown). We stained cells with a broad panel of antibodies recognizing various lymphocytes and myeloid cells and used the FlowSOM automatic gating and visualization tool to assess the immune cell landscape of Ormdl3 Tg/wt and Ormdl3-/- mice in the most unbiased manner (38–40). Although there was a trend to an increased number of splenic cells in Ormdl3 Tg/wt mice (Fig. 2A/B), none of the well known or less defined immune cell clusters were significantly altered compared with littermate controls (Fig. 2C/D). Likewise, FlowSOM trees represented a similar immune landscape between Ormdl3 -/- mice and littermate controls (Fig. 2E-H). These data suggest that genetic alteration of ORMDL3 does not affect the immunome in a major manner.
Figure 2. Unbiased analysis of the immune landscape of Ormdl3 Tg/wt and Ormdl3-/- mice.
A)Splenic cell number from 6w old Ormdl3 wt/wt and Ormdl3 Tg/wt mice (n=5,5).
B/C) Numbers (B) and percentages (C) of different immune cell types in spleens from Ormdl3 wt/wt and Ormdl3 Tg/wt mice (n=5,5).
D)FlowSOM trees representing the splenic immune landscape in Ormdl3wt/wt and Ormdl3 Tg/wt mice.
E)Splenic cell number from 6w old Ormdl3+/+ and Ormdl3-/- mice (n=5,5).
F/G) Numbers (F) and percentages (G) of different immune cell types in spleens from Ormdl3+ + and Ormdl3-/- mice (n=5,5).
H)FlowSOM trees representing the splenic immune landscape in Ormdl3+/+ and Ormdl3-/- mice.
Data information: All data were analysed with a Shapiro-Wilk normality test to assess whether the data was normally distributed. Parametric data were analysed with a t-test, whereas non-parametric data were analysed with a Mann-Whitney test. Data are shown as means +/- SEM. *p<0.05.
Effect of altered ORMDL3 expression on sphingolipid concentration
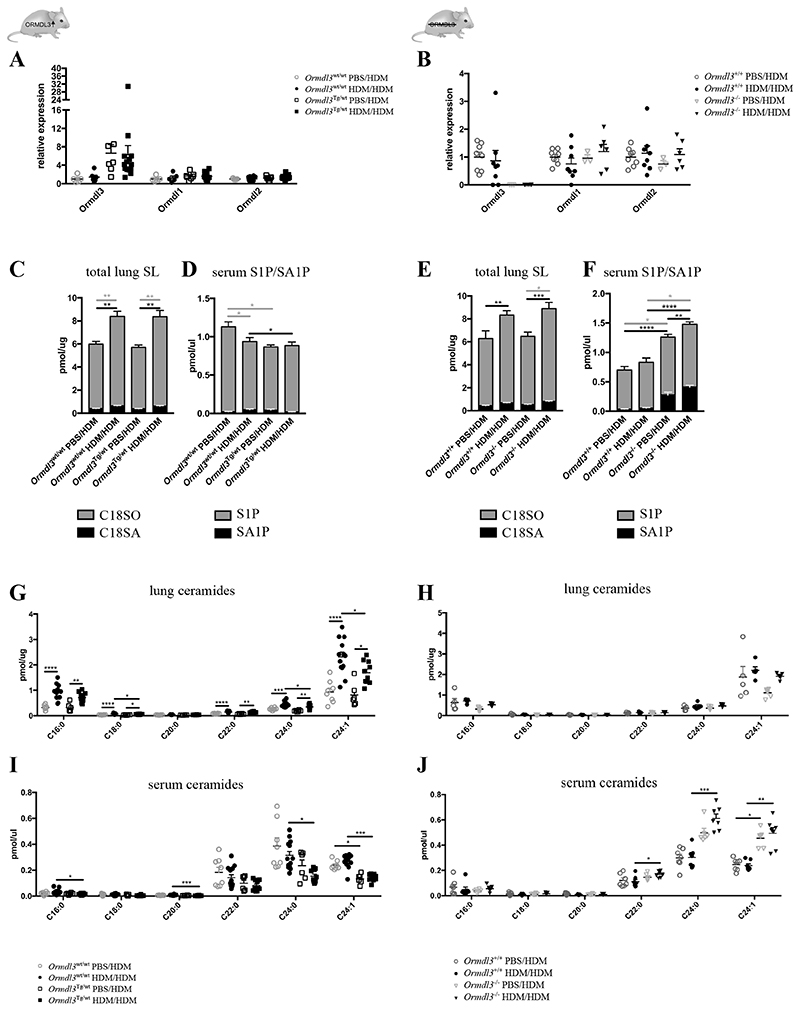
In yeast, it has been conclusively demonstrated that ORM-proteins regulate de novo sphingolipid synthesis via controlling SPT activity. Therefore a dysregulation of sphingolipid de novo synthesis was proposed as a pathophysiological mechanism by which ORMDL3 promotes asthma (for a schematic overview of SL de novo synthesis, see Suppl. Fig. 2A) (8,18–23,41). We therefore assessed whether ORMDL3 affects sphingolipid metabolism in the context of allergic asthma by using our transgenic mice. Since there are three ORM paralogues expressed in mice, it was also important to assess if deficiency or overexpression of Ormdl3 causes a compensatory change in Ormdl1 or Ormdl2 expression. For lung tissue, Ormdl3 transcript levels were 6 fold increased in Ormdl3 Tg/wt and expression levels were comparable for non-asthmatic (PBS/HDM group) and asthmatic (HDM/HDM group) conditions. A compensatory change in Ormdl1 or Ormdl2 expression was not observed (Fig. 3A). In Ormdl3 -/- mice, transcript levels of Ormdl3 were absent, without a sign of compensatory upregulation of Ormdl1 or Ormdl2 (Fig. 3B). Again, we did not observe any induction of Ormdl3 mRNA upon asthmatic challenge (Fig. 3A-B).
Figure 3. Effect of altered ORMDL3 expression on sphingolipid concentration.
A/B) Ormdl3 mRNA expression levels in lung tissue from Ormdl3 wt/wt and Ormdl3 Tg/wt mice (A) or Ormdl3+/+ and Ormdl3-/- mice (B) sensitized with saline (PBS) or house dust mite (HDM) and challenged with HDM. Expression values are shown relative to means of the mock-sensitized wildtype group (A: n=5,10,7,15; B: 8,8,3,6).
C/E) C18-sphingoid base levels in lung tissue from (non)-asthmatic Ormdl3 wt/wt and Ormdl3 Tg/wt mice (C) or Ormdl3+/+ and Ormdl3-/- mice (E). Total SLs were extracted, and sphingosine (C18SO, grey) and sphinganine (C18SA, black) levels were analyzed after acid–base hydrolysis by liquid chromatography-mass spectrometry (C: n=8,13,6,10; E: n=5,5,7,7). D/F) Sphingosine-1-phosphate (S1P, grey) and sphinganine-1-phosphate (SA1P, black) levels in serum from (non)-asthmatic Ormdl3 wt/wt and Ormdl3 Tg/wt mice (D) or Ormdl3 +/+ and Ormdl3-/- mice (F) (D: n=8,13,6,10; F: n=7,7,6,7).
G-J) Ceramide levels in lung tissue (G/H) or serum (I/J) from (non)-asthmatic Ormdl3 wt/wt and Ormdl3 Tg/wt mice (G/I) or Ormdl3+/+ and Ormdl3-/- mice (H/J) (G/I: n=8,13,6,10; H: n=5,5,7,7; J:7,7,6,7).
C/D/G/I) Data were pooled from 3 independent experiments.
Data information: All data were analysed with a Shapiro-Wilk normality test to assess whether the data was normally distributed. Parametric data were analysed with an ordinary one-way ANOVA test with multiple comparison correction. Non-parametric data were analysed with an unpaired Kruskal-Wallis test with multiple correction. Data are shown as means +/- SEM. *p<0.05; **p<0.01, ***p<0.001, ****<0.0001.
Next, we investigated how asthma and ORMDL3 expression influenced the sphingolipid profile in various organs. We observed a mixed picture, depending on the analyzed tissue. In lung, total sphingolipid levels were increased in asthmatic mice compared to non-asthmatic mice, confirming previous observations (13,42), but this increase was clearly independent of Ormdl3 (Fig. 3C/D). In line with this, full sphingolipidomics analysis revealed an increase of ceramides in lungs of asthmatic mice (Fig. 3E/F), which appeared not affected by the absence of ORMDL3 (Fig. 3F). However, upon overexpression of ORMDL3, ceramide levels in the lung were slightly decreased, consistent with a role for ORMLD3 in blocking ceramide synthesis (13,43)(Fig. 3E). This was much more pronounced in the serum (Fig. 3G/H) and several other tissues, such as liver and fat in which ORMDL3 is higher expressed (Fig. 1C, Fig. 3G/H and Suppl. Fig. 2). These effects were not limited to ceramides but could also be observed for other sphingolipid species. For instance, sphingosine-1-phosphate (S1P) and sphinganine-1-phosphate (SA1P) were upregulated in serum from Ormdl3 -/- mice and reduced in Ormdl3Tg/wt mice (Fig. 3C/D). A similar pattern was observed for hexosylceramides and sphingomyelins (Suppl. Fig. 2E-G). In summary, sphingolipid homeostasis seems to be mostly influenced in those tissues where ORMDL3 is abundantly expressed. For lung, we did not see a major influence of ORMDL3 expression on the asthma-induced change in the SL profile.
Genetic alteration of ORMDL3 expression levels does not affect key asthma parameters
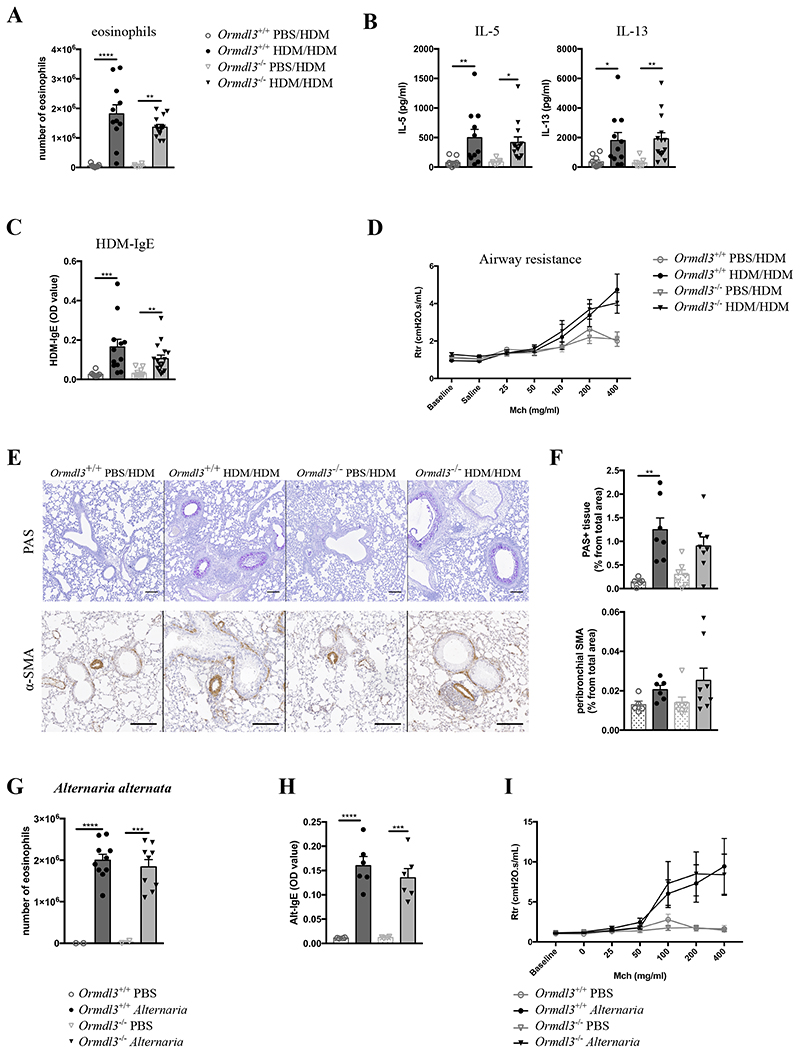
Having found that genetic interference with the expression levels of ORMDL3 led to alterations in ceramide levels, we next addressed if key asthma parameters would be altered in Ormdl3 Tg/wt or Ormdl3 -/- mice, and experiments were each time compared to proper littermate wildtype controls. In HDM-sensitized, but not mock-sensitized, Ormdl3 wt/wt mice, there was eosinophil, T cell and B cell infiltration in bronchoalveolar lavage (BAL) fluid, IL-5 and IL-13 cytokine production in the MLN, and HDM-specific IgE production in the serum, and bronchial hyperreactivity in response to increasing doses of metacholine (Fig. 4A-D and Suppl. Fig. 3A/B). Strikingly, none of these key HDM-induced asthma parameters were altered in Ormdl3 Tg/wt mice. A previous study suggested the involvement of ORMDL3 in airway remodeling, characterized by goblet cell metaplasia (GCM) and hypertrophia of α-smooth muscle actin (α-SMA) expressing myofibroblasts (Miller et al. 2014). In Ormdl3 wt/wt mice, HDM challenge of sensitized mice led to an increase in PAS-positive GCM, and of α-SMA staining around the airways compared to non-sensitized mice (Fig. 4E-F), but again these features were equally induced in Ormdl3 Tg/wt mice. Even in a more chronic 5 week model of HDM exposure, overexpression of ORMDL3 did not significantly influence cardinal features of asthma (Suppl. Fig. 3C-G).
Figure 4. ORMDL3 overexpression does not affect allergic asthma driven by HDM or Alternaria alternata .
A)Eosinophil number in BAL fluid from (non)-asthmatic Ormdl3 wt/wt and Ormdl3 Tg/wt mice (n=5,11,7,14).
B)Protein levels of IL-5 and IL-13 secreted by MLN cells during restimulation with HDM (n=5,11,7,14).
C)Serum HDM-specific IgE levels (OD value) (n=4,11,6,13).
A-C) Data were pooled from 2 independent experiments.
D)Bronchial hyperreactivity upon exposure to increasing doses of methacholine (Mch) as measured with flexiVent (n=4,4,6,8).
E)Representative images of lung sections stained with either Periodic Acid-Schiff (PAS) or anti-α-smooth muscle actin (α-SMA) antibody. Scale bar = 100 um.
F)Quantification of PAS+ area and peribronchial α-SMA in lung tissue. Data were pooled from 3 independent experiments (n=1,15,1,9).
G)Eosinophil number in BAL fluid upon exposure to PBS or Alternaria (n=1,8,1,9).
H)Serum Alternaria-specific IgE levels (OD value) (n=1,6,1,4).
I)Bronchial hyperreactivity upon exposure to increasing doses of methacholine (Mch) as measured with flexiVent (SCIREQ) (n=1,7,1,4).
Data information: All data were analysed with a Shapiro-Wilk normality test to assess whether the data was normally distributed. Parametric data were analysed with an ordinary one-way ANOVA test with multiple comparison correction. Non-parametric data were analysed with an unpaired Kruskal-Wallis test with multiple correction. For F-I, a Mann-Whitney statistical test was performed between the HDM–sensitized groups. Data are shown as means +/- SEM. *p<0.05; **p<0.01, ***p<0.001.
Recently, it has been shown that T-cells are the most prominent immune cells affected by chr17q21 SNPs (9). Therefore, we wondered whether T-cell function would be intrinsically affected by ORMDL3 overexpression. Although there were no obvious differences in the outcome of the asthma experiments, it was still possible that T-cell function was slightly affected, but that this only became apparent during competition experiments. Therefore, we crossed Ormdl3 Tg/wt with 1-DER mice, which contain T-cells that specifically react to the Der p 1 peptide of HDM (32).Ormdl3 wt/wt and Ormdl3 Tg/wt 1-DER T cells were fluorescently labelled and transferred into acceptor mice (Suppl. Fig. 4A), which were subsequently exposed to HDM. Similar proportions of Ormdl3 wt/wt and Ormdl3 Tg/wt 1-DER T cells were divided three days after transfer and numbers were equally high (Suppl. Fig. 4B-C). Thus, in line with previous results, ORMDL3 did not affect HDM-specific proliferation by T cells intrinsically.
To exclude allergen-dependent effects, Ormdl3 Tg/wt mice were also exposed to a model driven by the fungal extract Alternaria alternata. Mice were administered 20 μg Alternaria (ALT) or PBS three times a week, for 3 weeks (Suppl. Fig. 5A). ALT-treated mice had elevated expression levels of Ormdl3 mRNA, confirming earlier findings (Miller et al. 2012) (Suppl. Fig. 5B), however this could not be confirmed on protein level with Ormdl3-LacZ reporter mice (Suppl. Fig. 5C). In ALT exposed mice, there was a marked influx of eosinophils in the BAL fluid, an increase in ALT-specific IgE in the serum, Th2 cytokine production in the MLN, and development of BHR to methacholine compared with PBS exposed mice, but none of these features were altered in Ormdl3Tg/wt mice (Fig. 4G-I and Suppl. Fig. 5D-F).
It is most likely that the ORMDL3 SNP at 17q12-21 that confers asthma risk leads to overexpression of ORMDL3, as has been demonstrated in T-cells (9). Since our transgenic mice overexpressing ORMDL3 did not have altered features of asthma, we also decided to perform all key asthma readouts in Ormdl3-/- mice in comparison with Ormdl3+/+ littermates, again sensitizing mice to either HDM or ALT. Whereas all key features of allergic asthma like eosinophilia, Th2 cytokine production, HDM-specific serum IgE, BHR to methacholine, airway remodeling were induced by allergen challenge in Ormdl3+/+ mice, none of these features were altered in Ormdl3 -/- mice (Fig. 5A-F and Suppl. Fig. 6A). There was a small trend towards enhanced IL-17 production by allergen restimulated MLN T cells, but this was not accompanied by airway neutrophilia (Suppl. Fig. 6B). Similar conclusions were reached in the ALT model (Fig. 5G-I and Suppl. Fig. 6C-F). Therefore, neither ORMDL3 overexpression nor deletion affected key asthma parameters in various acute and chronic models driven by two different allergens.
Figure 5. Loss of Ormdl3 expression does not affect allergic asthma driven by HDM or Alternaria alternata .
A)Number of eosinophils in BAL fluid from (non)-asthmatic Ormdl3 +/+ and Ormdl3 -/- mice (n=10,11,7,14).
B)Protein levels of IL-5 and IL-13 secreted by MLN cells during restimulation with HDM (n=10,11,7,14).
C)Serum HDM-specific IgG1, HDM-IgE and total IgE levels (OD value) (n=8,12,12,16). A-C) Data were pooled from 2 experiments.
D)Bronchial hyperreactivity upon exposure to increasing doses of methacholine (Mch) as measured with flexiVent (n=4,4,6,6).
E)Representative images of lung sections stained with either Periodic Acid-Schiff (PAS) or anti-α-SMA antibody. Scale bar = 100um.
F)Quantification of PAS+ area (n=5,7,7,8) and peribronchial α-SMA (n=5,6,7,8) in lung tissue.
G)Number of eosinophils in BAL fluid from Ormdl3+/+ and Ormdl3-/- mice exposed to PBS or Alternaria (n=2,10,2,9).
H)Serum Alternaria-specific IgE levels (OD value) (n=4,6,4,6).
I)Bronchial hyperreactivity upon exposure to increasing doses of methacholine (Mch) as measured with flexiVent (SCIREQ) (n=4,6,4,6).
Data information: All data were analysed with a Shapiro-Wilk normality test to assess whether the data was normally distributed. Parametric data were analysed with an ordinary one-way ANOVA test with multiple comparison correction. Non-parametric data were analysed with an unpaired Kruskal-Wallis test with multiple correction. Data are shown as means +/- SEM. *p<0.05; **p<0.01, ***p<0.001, ****<0.0001.
Discussion
During the last decade, genome-wide association studies have repeatedly and convincingly demonstrated that SNPs in the chr17q12-21 locus are associated with the early life onset of allergic asthma (2,3). Genotypes in the core region defined by the first genome-wide association study correlate with expression of 2 genes, ORMDL3 and gasdermin B (GSDMB), making these prime candidate asthma genes, although recent studies have implicated gasdermin A (GSDMA) distal to and post-GPI attachment to proteins 3 (PGAP3) proximal to the core region as independent loci (8). There is much speculation on how ORMDL3 could regulate asthma. In yeast, ORM proteins control sphingolipid levels by regulating the enzyme SPT, the rate-limiting enzyme of de novo sphingolipid synthesis (18). A mouse study subsequently showed that interfering with SPT using the inhibitor myriocin reduced de novo sphingolipid synthesis and increased bronchial hyperreactivity (44). This led some to conclude that ORMDL3 controls asthma via altered sphingolipid metabolism (41,45). However, despite extensive efforts, the molecular role of ORMDL3 in allergic asthma has not been unraveled yet.
Although the studies addressing the effect of inhibition of de novo SL synthesis using myriocin or genetic deficiency of SPT have yielded consistent results (13,24,26,44,46–50), the mouse studies investigating the role of ORMDL3 in asthma have been harder to reconcile. Miller et al. (2014) reported that C57BL/6 mice expressing high levels of human ORMDL3 developed stronger inflammation and BHR compared to wildtype mice in a mouse model driven by intraperitoneal immunization by OVA in alum followed by a series of OVA inhalation challenges (14). However, the same group also reported that BHR (but not inflammation) was increased in C57BL/6 mice lacking ORMDL3 specifically in epithelial cells (16), which makes the results hard to interpret. In contrary, Loser et al. (2016) reported that full Ormdl3 gene deficient C57BL/6 mice were protected from developing BHR to Alternaria alternata, whereas inflammation was only slightly affected. From these conflicting studies, the conclusion would be that the role of ORMDL3 in experimental asthma is highly contextual, and might be explained by subtle differences in animal models used by different investigators, and whether the model employed overexpression of (human) ORMDL3 or genetic deficiency of endogenous mouse Ormdl3. To address this controversy regarding the models used, we applied multiple models (acute versus chronic) and allergens (HDM, Alternaria, and OVA, not shown) in newly generated Ormdl3 BAC transgenic and Ormdl3 -/- mice. We observed no differences in the severity of all key asthma parameters when Ormdl3 was either overexpressed or deleted from the mouse genome. At present, we can only speculate about the differences between our negative results and the positive results reported by others. One major difference is that others studying overexpression have employed the human ORMDL3 gene using a plasmid expression system or adenoviral overexpression of murine Ormdl3 in vivo, which could have led to unphysiological levels of overexpression, or overexpression in cell types that normally do not express ORMDL3. In addition, the reported differences in ORMDL3 expression based on the SNP genotype, is minor and significantly smaller as it was reported for the transgenic mouse models. We have used a BAC transgenic construct in which overexpression of Ormdl3 is still driven by the endogenous Ormdl3 promotor. Modeling asthma in the mouse is difficult. It is possible that other environmental triggers that often occur together with allergen exposure are necessary to unravel the link between ORMDL3 and asthma, such as respiratory viruses or cigarette smoke. In fact, a limitation of this study is the lack of a viral infection model, as recent epidemiological studies suggest that SNPs in the chr17q12-21 locus combined with severe respiratory virus infections do have a driving role in causing childhood-onset asthma (7,51,52). Interestingly, the same SNPs also lead to protection from asthma upon microbial exposure (51), suggesting that this locus is very environment-dependent and might be regulated by epigenetic modifications (53).
None of the studies addressing the relationship between ORMDL3 and asthma have so far studied experimental respiratory viral infections. Efforts are underway in our laboratory addressing the role of ORMDL3 in susceptibility to respiratory mouse pneumovirus infection and subsequent asthma development.
ORM-homologues in yeast are well-known inhibitors of de novo sphingolipid synthesis, whereas the role for the mammalian ORMDLs is still a matter of debate. We therefore examined sphingolipid synthesis by two approaches. First, we analyzed the total SL levels by quantifying the total C18-sphingoid base profile after chemical hydrolysis. If SPT-activity would be inhibited by ORMDL3, total sphingoid bases would be reduced in Ormdl3Tg/wt mice. However, neither in lungs of Ormdl3 Tg/wt nor in Ormdl3 -/- mice, a significant change in total sphingoid base levels could be detected. That corresponds to earlier reports that SPT-activity is not affected by changing ORMDL3 expression levels (25). Next, we performed sphingolipidomic analysis to quantify individual sphingolipid species, including ceramides, sphingomyelins, hexosyl ceramides and phosphorylated sphingoid bases (S1P, SA1P). In particular S1P has been extensively described to be involved in allergic asthma (54–58). Our results demonstrate an increase in serum S1P in Ormdl3 -/- mice (Fig. 3D), which has also been seen in mice lacking ORMDL3 specifically in epithelial cells (16). Furthermore, Ormdl3 Tg/wt mice had reduced levels of S1P (Fig. 3C), which is consistent with previous reports (43). However, the ORMDL3-dependent regulation of S1P did not result in an altered asthma phenotype.
Earlier reports have shown that ceramides are elevated in murine allergic asthma models and asthmatic patients and that inhibition of ceramide synthesis mitigates the allergic response (13,42). This was confirmed in our HDM model, which showed a significant increase in lung ceramides for asthmatic mice compared to non-asthmatic mice. This asthma dependent increase in Cer was not influenced in Ormdl3 -/- mice. On the other hand, it appeared to be hampered in Ormdl3 Tg/wt mice. Of interest, serum ceramides were decreased in Ormdl3 Tg/wt mice and conversely, were increased in Ormdl3 -/- mice. Similar changes were seen in liver but not in BAT or WAT, which also express significant levels of ORMDL3 natively (Fig. 1C). A decrease in long-chain ceramides (C:24) has also been observed in mice overexpressing human ORMDL3 (43). Furthermore, Oyeniran et al. (2015) reported that all ceramide species were reduced upon overexpression of ORMDL3, provided that the degree of overexpression is moderate, which is the case in our Ormdl3 Tg/wt mice. However, although serum (and lung) ceramide levels did change upon changing ORMDL3 expression levels, the allergic response was not affected, suggesting that the association of ORMDL3 with asthma would be hard to explain through alterations in ceramide levels.
In tissues where native ORMDL3 expression was the highest — based on the Ormdl3-LacZ reporter mouse — the effects on sphingolipid homeostasis were also the most pronounced. This may explain the moderate effects observed in lung. However, it is still unclear at which step ORMDL3 interferes in the sphingolipid synthesis pathway. In line with our previous data (25), we did not find a consistent change in total sphingolipid levels, which makes a direct effect of ORMDL3 on SPT activity questionable. Still, especially in liver but also other tissues, levels of several sphingolipid species were altered in mice with deregulated Ormdl3. Interestingly, the largest change was seen for serum SM, which is primarily formed in the liver and then secreted into blood in the form of lipoproteins. Among the different SL classes, we observed the most pronounced changes for Cer and SM whereas HexCer level were not much altered. This shows that mammalian ORMDL3 is somehow involved in the homeostasis of sphingolipids, but likely at a level downstream of SPT.
As an ER-residing protein, in vitro studies also reported a role for ORMDL3 in calcium homeostasis and the unfolded protein response (12,27,28), although there are also reports demonstrating no role for ORMDL3 in the UPR (29). We measured UPR target gene expression in different tissues from Ormdl3 -/- and Ormdl3 Tg/wt mice, but could not find any differences compared to wildtype mice (data not shown), suggesting that the UPR was not affected upon changing ORMDL3 expression levels in vivo. It should be noted that a dramatic overexpression of ER proteins will affect ER homeostasis anyway, so the positive findings described in vitro may not be directly linked to ORMDL3.
Thus, in this study we found no causal relationship between ORMDL3 expression levels and severity of key asthma features in a HDM or Alternaria-driven allergic asthma model, despite clear alterations in sphingolipid levels in mice overexpressing or lacking ORMDL3. We therefore do not support the hypothesis that ORMDL3 controls asthma by altering sphingolipid metabolism. More research is needed on the causal relationship between respiratory virus infection, SNPs at 17q21, ORMDL3 levels and asthma risk, before we can definitively exclude a major role for ORMDL3 in asthma.
Supplementary Material
Key Messages.
-
-
Ormdl3 expression levels did not affect allergic asthma development driven by house dust mite or Alternaria alternata.
-
-
ORMDL3 negatively regulated ceramide levels, but this did not result in altered experimental asthma.
Acknowledgements
We thank all technicians of the Lambrecht-Hammad lab for their technical support, in particular Manon Vanheerswynghels, Sofie De Prijck and Caroline De Wolf.
Funding
ND is supported by a Fonds Wetenschappelijk Onderzoek Vlaanderen (FWO) grant (11Y8417N). BNL is supported by an European Research Council (ERC)-advanced grant (ERC-ADG 789384), a Concerted Research Initiative Grant (GOA) of Ghent University, and an Excellence of Science (EOS) research grant (project 30565447). SJ is a recipient of an ERC Consolidator Grant (ERC-CoG 819314). BNL and SJ are holders of several FWO program grants, a UGent MRP grant (Group-ID) and a Stichting tegen Kanker Grant.
Abbreviations
- ALT
Alternaria alternata
- BAL
Bronchoalveolar lavage
- BHR
Bronchial hyperreactivity
- ER
Endoplasmic reticulum
- GCM
Goblet cell metaplasia
- GWAS
Genome-wide association study
- HDM
House dust mite
- Ig
Immunoglobulin
- IL
Interleukin
- MLN
Mediastinal lymph node
- OVA
Ovalbumin
- ORMDL3
ORM (yeast)-like protein isoform 3
- α-SMA
alpha-smooth muscle actin
- SNPs
Single nucleotide polymorphisms
- SPT
Serine palmitoyl transferase
References
- 1.Lambrecht BN, Hammad H. Nat Immunol. 10. Vol. 18. Nature Publishing Group; 2017. Sep 19, The immunology of the allergy epidemic and the hygiene hypothesis; pp. 1076–83. [DOI] [PubMed] [Google Scholar]
- 2.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Nature. 7152. Vol. 448. Nature Publishing Group; 2007. Jul 26, Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma; pp. 470–3. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y-F, Luo Y-M, Xiong W, Wu X-L. Genetic variation in ORMDL3 gene may contribute to the risk of asthma: a meta-analysis. Hum Immunol. 2014 Sep;75(9):960–7. doi: 10.1016/j.humimm.2014.08.202. [DOI] [PubMed] [Google Scholar]
- 4.Bouzigon E, Corda E, Aschard H, Dizier M-H, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008 Nov 6;359(19):1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 5.Andiappan AK, Sio YY, Lee B, Suri BK, Matta SA, Lum J, et al. Functional variants of 17q12-21 are associated with allergic asthma but not allergic rhinitis. J Allergy Clin Immunol. 2016 Mar;137(3):758–66.:e3. doi: 10.1016/j.jaci.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Willis-Owen SAG, Spiegel S, Lloyd CM, Moffatt MF, Cookson WOCM. The ORMDL3 Asthma Gene Regulates ICAM1 and has Multiple Effects on Cellular Inflammation. Am J Respir Crit Care Med. 2018 Oct 19;:rccm.201803-04380C. doi: 10.1164/rccm.201803-0438OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calişkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, et al. N Engl J Med. 15. Vol. 368. Massachusetts Medical Society; 2013. Apr 11, Rhinovirus wheezing illness and genetic risk of childhood-onset asthma; pp. 1398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, et al. A decade of research on the 17q12-21 asthma locus: Piecing together the puzzle. J Allergy Clin Immunol. 2018 Sep;142(3):749–764.:e3. doi: 10.1016/j.jaci.2017.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmiedel BJ, Seumois G, Samaniego-Castruita D, Cayford J, Schulten V, Chavez L, et al. Nat Commun. 1. Vol. 7. Nature Publishing Group; 2016. Nov 16, 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells; p. 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lluis A, Schedel M, Liu J, Illi S, Depner M, von Mutius E, et al. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J Allergy Clin Immunol. 2011 Jun;127(6):1587–94.:e6. doi: 10.1016/j.jaci.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Schedel M, Michel S, Gaertner VD, Toncheva AA, Depner M, Binia A, et al. Polymorphisms related to ORMDL3 are associated with asthma susceptibility, alterations in transcriptional regulation of ORMDL3, and changes in TH2 cytokine levels. J Allergy Clin Immunol. 2015 Oct;136(4):893–903.:e14. doi: 10.1016/j.jaci.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA. 2012 Oct 9;109(41):16648–53. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oyeniran C, Sturgill JL, Hait NC, Huang W-C, Avni D, Maceyka M, et al. Aberrant ORM (yeast)-like protein isoform 3 (ORMDL3) expression dysregulates ceramide homeostasis in cells and ceramide exacerbates allergic asthma in mice. J Allergy Clin Immunol. 2015 Oct;136(4):1035–6. doi: 10.1016/j.jaci.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, et al. J Immunol. 8. Vol. 192. American Association of Immunologists; 2014. Apr 15, ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma; pp. 3475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Löser S, Gregory LG, Zhang Y, Schaefer K, Walker SA, Buckley J, et al. Pulmonary ORMDL3 is critical for induction of Alternaria-induced allergic airways disease. J Allergy Clin Immunol. 2017 May;139(5):1496–1507.:e3. doi: 10.1016/j.jaci.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller M, Tam AB, Mueller JL, Rosenthal P, Beppu A, Gordillo R, et al. J Immunol. 8. Vol. 198. American Association of Immunologists; 2017. Apr 15, Cutting Edge: Targeting Epithelial ORMDL3 Increases, Rather than Reduces, Airway Responsiveness and Is Associated with Increased Sphingosine-1-Phosphate; pp. 3017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzàlez-Duarte R. Genome Biol. 6. Vol. 3. BioMed Central; 2002. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. RESEARCH0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, et al. Nature. 7284. Vol. 463. Nature Publishing Group; 2010. Feb 25, Orm family proteins mediate sphingolipid homeostasis; pp. 1048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proceedings of the National Academy of Sciences; 2010. Mar 30, pp. 5851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, et al. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Riezman H, editor. Mol Biol Cell. 2012 Jun;23(12):2388–98. doi: 10.1091/mbc.E12-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walther TC. Keeping sphingolipid levels nORMal. Proc Natl Acad Sci USA. 2010 Mar 30;107(13):5701–2. doi: 10.1073/pnas.1001503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimobayashi M, Oppliger W, Moes S, Jenö P, Hall MN. TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Riezman H, editor. Mol Biol Cell. 2013 Mar;24(6):870–81. doi: 10.1091/mbc.E12-10-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences; 2011. Nov 29, pp. 19222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiefer K, Carreras-Sureda A, García-López R, Rubio-Moscardó F, Casas J, Fabriàs G, et al. J Biol Chem. 5. Vol. 290. American Society for Biochemistry and Molecular Biology; 2015. Jan 30, Coordinated regulation of the orosomucoid-like gene family expression controls de novo ceramide synthesis in mammalian cells; pp. 2822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhakupova A, Debeuf N, Krols M, Toussaint W, Vanhoutte L, Alecu I, et al. FASEB J. 12. Vol. 30. Federation of American Societies for Experimental BiologyBethesda; MD, USA: 2016. Dec, ORMDL3 expression levels have no influence on the activity of serine palmitoyltransferase; pp. 4289–300. [DOI] [PubMed] [Google Scholar]
- 26.Siow DL, Wattenberg BW. Mammalian ORMDL Proteins Mediate the Feedback Response in Ceramide Biosynthesis. J Biol Chem. 2012 Nov 23;287(48):40198–204. doi: 10.1074/jbc.C112.404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantero-Recasens G, Fandos C, Rubio-Moscardó F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010 Jan 1;19(1):111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 28.Carreras-Sureda A, Cantero-Recasens G, Rubio-Moscardó F, Kiefer K, Peinelt C, Niemeyer BA, et al. ORMDL3 modulates store-operated calcium entry and lymphocyte activation. Hum Mol Genet. 2013 Feb 1;22(3):519–30. doi: 10.1093/hmg/dds450. [DOI] [PubMed] [Google Scholar]
- 29.Hsu KJ, Turvey SE. Allergy Asthma Clin Immunol. 1. Vol. 9. BioMed Central; 2013. Feb 1, Functional analysis of the impact of ORMDL3 expression on inflammation and activation of the unfolded protein response in human airway epithelial cells; p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bugajev V, Halova I, Draberova L, Bambouskova M, Potuckova L, Draberova H, et al. Cellular and Molecular Life Sciences. 6. Vol. 73. Springer International Publishing; 2015. Sep 25, Negative regulatory roles of ORMDL3 in the FcεRI-triggered expression of proinflammatory mediators and chemotactic response in murine mast cells; pp. 1265–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha SG, Ge XN, Bahaie NS, Kang BN, Rao A, Rao SP, et al. Nat Commun. 1. Vol. 4. Nature Publishing Group; 2013. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48; p. 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dullaers M, Schuijs MJ, Willart M, Fierens K, van Moorleghem J, Hammad H, et al. House dust mite-driven asthma and allergen-specific T cells depend on B cells when the amount of inhaled allergen is limiting. J Allergy Clin Immunol. 2017 Jul;140(1):76–7. doi: 10.1016/j.jaci.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 33.Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015 Sep 4;349(6252):1106–10. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- 34.Vanheerswynghels M, Toussaint W, Schuijs M, Vanhoutte L, Killeen N, Hammad H, et al. Methods Mol Biol. 2. Vol. 1799. Springer New York; New York, NY: 2018. The Generation and Use of Allergen-Specific TCR Transgenic Animals; pp. 183–210. [DOI] [PubMed] [Google Scholar]
- 35.Debeuf N, Haspeslagh E, van Helden M, Hammad H, Lambrecht BN. Curr Protoc Mouse Biol. 2. Vol. 6. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2016. Jun 1, Mouse Models of Asthma; pp. 169–84. [DOI] [PubMed] [Google Scholar]
- 36.Atkinson D, Nikodinovic Glumac J, Asselbergh B, Ermanoska B, Blocquel D, Steiner R, et al. Sphingosine 1-phosphate lyase deficiency causes Charcot-Marie-Tooth neuropathy. Neurology. 2017 Feb 6;88(6):533–42. doi: 10.1212/WNL.0000000000003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Adv Bioinformatics. Vol. 2008. Hindawi; 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes; 420747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. In: Brinkman RR, Aghaeepour N, Finak G, Gottardo R, Mosmann T, Scheuermann RH, editors. Cytometry A. 7. Vol. 87. Wiley-Blackwell; 2015. Jul, pp. 636–45. [DOI] [PubMed] [Google Scholar]
- 39.Guilliams M, Dutertre C-A, Scott CL, McGovern N, Sichien D, Chakarov S, et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity. 2016 Sep 20;45(3):669–84. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeys Y, Van Gassen S, Lambrecht BN. Nat Rev Immunol. 7. Vol. 16. Nature Publishing Group; 2016. Jul, Computational flow cytometry: helping to make sense of high-dimensional immunology data; pp. 449–62. [DOI] [PubMed] [Google Scholar]
- 41.Levy BD. Sphingolipids and susceptibility to asthma. Phimister EG, editor. N Engl J Med. 2013 Sep 5;369(10):976–8. doi: 10.1056/NEJMcibr1306864. [DOI] [PubMed] [Google Scholar]
- 42.Masini E, Giannini L, Nistri S, Cinci L, Mastroianni R, Xu W, et al. J Pharmacol Exp Ther. 2. Vol. 324. American Society for Pharmacology and Experimental Therapeutics; 2008. Feb, Ceramide: a key signaling molecule in a Guinea pig model of allergic asthmatic response and airway inflammation; pp. 548–57. [DOI] [PubMed] [Google Scholar]
- 43.Miller M, Rosenthal P, Beppu A, Gordillo R, Broide DH. Oroscomucoid like protein 3 (ORMDL3) transgenic mice have reduced levels of sphingolipids including sphingosine-1-phosphate and ceramide. J Allergy Clin Immunol. 2017 Apr;139(4):1373–4. doi: 10.1016/j.jaci.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, et al. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med. 2013 May 22;5(186):186ra67-7. doi: 10.1126/scitranslmed.3005765. [DOI] [PubMed] [Google Scholar]
- 45.Worgall TS. Sphingolipids, ORMDL3 and asthma: what is the evidence? Curr Opin Clin Nutr Metab Care. 2017 Mar;20(2):99–103. doi: 10.1097/MCO.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 46.Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochemical and Biophysical Research Communications. 1995 Jun 15;211(2):396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 47.Hojjati MR, Li Z, Jiang X-C. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim Biophys Acta. 2005 Oct 15;1737(1):44–51. doi: 10.1016/j.bbalip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Park T-S, Li Y, Pan X, Iqbal J, Lu D, et al. Serine palmitoyltransferase (SPT) deficient mice absorb less cholesterol. Biochim Biophys Acta. 2009 Apr;1791(4):297–306. doi: 10.1016/j.bbalip.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakraborty M, Lou C, Huan C, Kuo M-S, Park T-S, Cao G, et al. J Clin Invest. 4. Vol. 123. American Society for Clinical Investigation; 2013. Apr, Myeloid cell-specific serine palmitoyltransferase subunit 2 haploinsufficiency reduces murine atherosclerosis; pp. 1784–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strettoi E, Gargini C, Novelli E, Sala G, Piano I, Gasco P, et al. Inhibition of ceramide biosynthesis preserves photoreceptor structure and function in a mouse model of retinitis pigmentosa. Proc Natl Acad Sci USA. 2010 Oct 26;107(43):18706–11. doi: 10.1073/pnas.1007644107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loss GJ, Depner M, Hose AJ, Genuneit J, Karvonen AM, Hyvärinen A, et al. Am J Respir Crit Care Med. 8. Vol. 193. American Thoracic Society; 2016. Apr 15, The Early Development of Wheeze. Environmental Determinants and Genetic Susceptibility at 17q21; pp. 889–97. [DOI] [PubMed] [Google Scholar]
- 52.Wang X-H, Shu J, Jiang C-M, Zhuang L-L, Yang W-X, Zhang H-W, et al. Mechanisms and roles by which IRF-3 mediates the regulation of ORMDL3 transcription in respiratory syncytial virus infection. Int J Biochem Cell Biol. 2017 Jun;87:8–17. doi: 10.1016/j.biocel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 53.Kothari PH, Qiu W, Croteau-Chonka DC, Martinez FD, Liu AH, Lemanske RF, et al. Role of local CpG DNA methylation in mediating the 17q21 asthma susceptibility gasdermin B (GSDMB)/ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) expression quantitative trait locus. J Allergy Clin Immunol. 2018 Jun;141(6):2282–6. doi: 10.1016/j.jaci.2017.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolly PS, Rosenfeldt HM, Milstien S, Spiegel S. The roles of sphingosine-1-phosphate in asthma. Mol Immunol. 2002 Sep;38(16-18):1239–45. doi: 10.1016/s0161-5890(02)00070-6. [DOI] [PubMed] [Google Scholar]
- 55.Kleinjan A, van Nimwegen M, Leman K, Hoogsteden HC, Lambrecht BN. Topical treatment targeting sphingosine-1-phosphate and sphingosine lyase abrogates experimental allergic rhinitis in a murine model. Allergy. 2013 Feb;68(2):204–12. doi: 10.1111/all.12082. [DOI] [PubMed] [Google Scholar]
- 56.Price MM, Oskeritzian CA, Falanga YT, Harikumar KB, Allegood JC, Alvarez SE, et al. A specific sphingosine kinase 1 inhibitor attenuates airway hyperresponsiveness and inflammation in a mast cell–dependent murine model of allergic asthma. Journal of Allergy and Clinical Immunology. 2013 Feb;131(2):501–1. doi: 10.1016/j.jaci.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karmouty-Quintana H, Siddiqui S, Hassan M, Tsuchiya K, Risse P-A, Xicota-Vila L, et al. Treatment with a sphingosine-1-phosphate analog inhibits airway remodeling following repeated allergen exposure. Am J Physiol Lung Cell Mol Physiol. 2012 Apr 15;302(8):L736-45. doi: 10.1152/ajplung.00050.2011. [DOI] [PubMed] [Google Scholar]
- 58.Oskeritzian CA, Hait NC, Wedman P, Chumanevich A, Kolawole EM, Price MM, et al. The sphingosine-1-phosphate/sphingosine-1-phosphate receptor 2 axis regulates early airway T-cell infiltration in murine mast cell-dependent acute allergic responses. J Allergy Clin Immunol. 2015 Apr;135(4):1008–18.:e1. doi: 10.1016/j.jaci.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.