Abstract
There is an urgent need to improve agricultural productivity to secure future food and biofuel supply. Here, we summarize current approaches that aim at improving photosynthetic CO2-fixation. We critically review, compare and comment on the four major lines of research towards this aim, which focus on (i) improving RubisCO, the CO2-fixing enzyme in photosynthesis, (ii) implementing CO2-concentrating mechanisms, (iii) establishing synthetic photorespiration bypasses, and (iv) engineering synthetic CO2-fixation pathways.
Introduction
The Morrow plots are a landmark of the University of Illinois. They are an experimental corn field that is continuously farmed since 1876 [1]. During the last 140 years, and in particular since the 1950s, crop yield on the Morrow fields (and world-wide) have increased by at least a factor of three [1,2]. Yet, these past achievements in agriculture are challenged by several developments. (i) The current population increase is not matched by the current increase in agricultural productivity, (ii) there is a growing demand to use crops for biofuel and biomass production directly competing with food production, and (iii) global CO2-emissions are continuously rising, accelerating the effects of climate change, including the loss of arable land, increased flooding and droughts. As a consequence, there is an urgent need to further improve agricultural productivity. Because in plants the conversion of light into biomass is the process with the lowest energy conservation (approx. 1%), improving photosynthetic CO2-fixation has been identified as key to increase agricultural productivity [3].
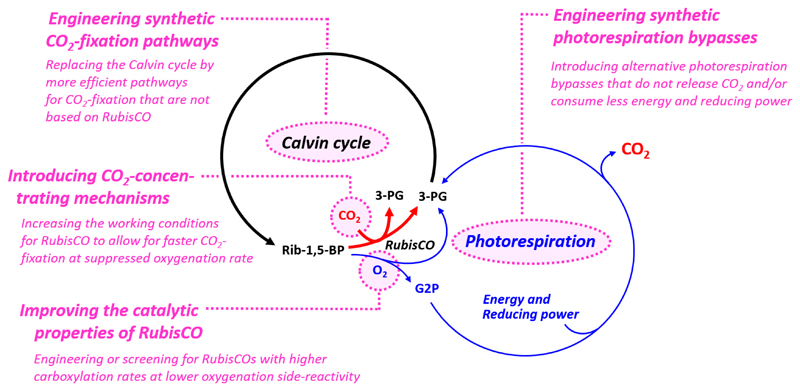
Under optimum conditions, one limiting factor in photosynthetic CO2-fixation is flux through the Calvin cycle, which is often restricted by the activity of the cycle’s CO2-fixing enzyme, ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO). The turnover number of an average RubisCO is between 1 and 10 s-1 (http://brenda-enzymes.org). This is one to two orders of magnitude below the turnover frequencies of other enzymes in central carbon metabolism that lie on average around 50 to 100 s-1 [4]. To allow for sufficient CO2-fixation rates, the low activity of RubisCO is compensated by high expression levels of the enzyme. In a photosynthetic organism, RubisCO can make up to 50% of the soluble protein [5].
Besides showing low specific activity, RubisCO does not discriminate well between O2 and CO2, which results in an oxygenase side reaction of the enzyme. Due to the high O2:CO2 ratio of ambient air (approx. 500:1), an average RubisCO fixes up to two O2 every five CO2-fixation reactions [6]. The products of RubisCO’s side reaction are 3-phosphoglycerate (3-PG) and glycolate-2-phosphate (G2P). The latter is a toxic compound that needs to be removed or recycled. In photosynthetic organisms, this is achieved at the expense of additional energy, reducing power and fixed CO2 in a process called photorespiration (Figure 1, 2). It is estimated that up to 30% of the photosynthetic output is lost through photorespiration [6,7].
Figure 1. Overview of photosynthetic CO2-fixation, photorespiration and current engineering efforts.
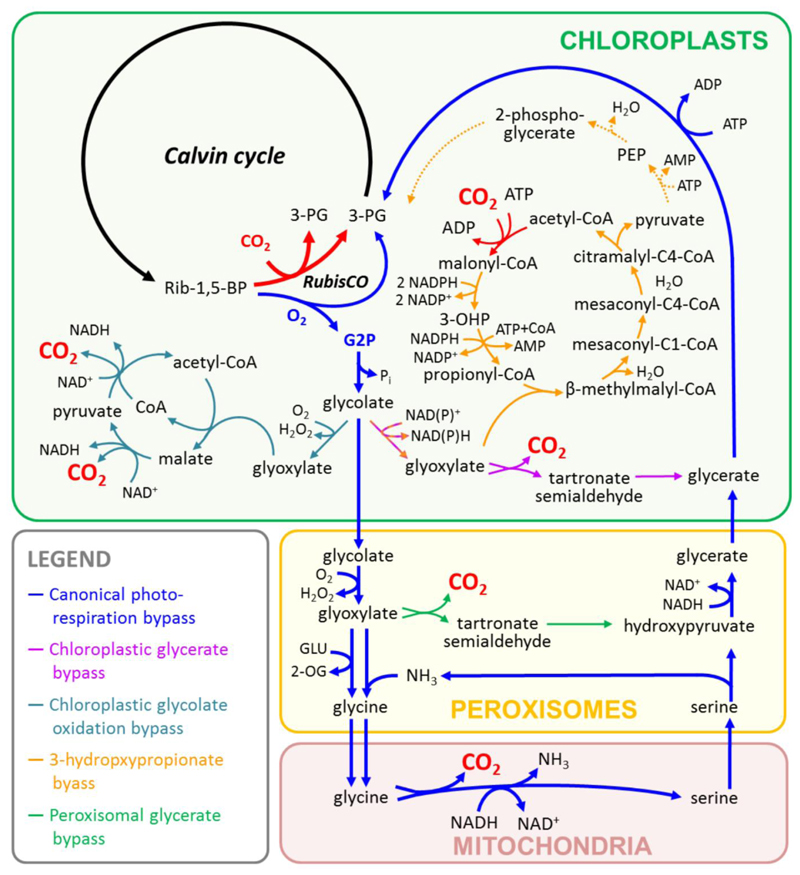
Figure 2. Natural and synthetic photorespiration bypasses.
To increase the yield of photosynthetic CO2-fixation, different strategies were suggested and at least partially pursued (Figure 1). These fall into one of the following four general categories: (i) improving the catalytic properties of RubisCO, (ii) improving the working conditions of RubisCO through CO2-concentrating mechanisms (CCM), (iii) engineering synthetic photorespiration bypasses, and (iv) engineering synthetic CO2-fixation pathways.
Improving the catalytic properties of RubisCO
Initial approaches to improve photosynthetic CO2-fixation focused on identifying [8,9] or engineering [10,11] RubisCOs with higher CO2-specificities and/or higher catalytic rates. These efforts have only met with limited success, because it has become apparent that RubisCO is trapped in an inherent trade-off between activity and specificity. Higher specificity for CO2 usually results in a lower enzyme activity. Vice versa, to engineer a RuBisCO with higher activity its specificity for CO2 has to be sacrificed, resulting in a higher oxygenation rate [12,13].
The reasons for the observed trade-off lie in the evolutionary past of RubisCO, although the emergence of the enzyme’s carboxylation and oxygenation function remain unknown. Recent investigations on the RubisCO superfamily [14,15] suggest that RubisCO was not a CO2-fixing enzyme a priori, but rather that its carboxylation function evolved as a secondary function in the protein scaffold of primordial enolases [16,17]. These findings challenge and extend older theories according to which RubisCO evolved from a primordial carboxylating archaeal enzyme [18,19]. Independent of the true origins of the carboxylation reaction of RubisCO, it is undisputed that the evolutionary roots of RubisCO trace back to a time when the level of O2 in the atmosphere was minimal. Thus, ancient RubisCO was primarily selected for promoting the carboxylation of ribulose-1,5-bisphosphate, but not against suppressing the oxygenation side reaction. This primordial chemistry of the enzyme caused (and still underlies) the inverse coupling of activity and selectivity in RubisCO [12,13]. With the increase of atmospheric O2 during earth’s history, RubisCO evolved along these two parameters, which enabled adaption of a given enzyme towards specificity or activity (depending on its environmental and/or organismic context) [20], but did not allow for an uncoupling of the two opposing catalytic parameters.
Nevertheless, even only slightly improved RubisCOs could have measurable effects. It has been calculated that transplantation of a RubisCO from the red algae Griffithsia monilis into crop could increase carbon gain by 25%, because of a twofold increased CO2-selectivity/activity ratio of the enzyme compared to plant RubisCO [21]. Screening of RubisCOs from wild wheat grasses identified enzyme variants of improved CO2-specificity/activity ratio. These “wild enzymes” were calculated to increase carbon uptake rates by 20% upon substitution of native RubisCO in agriculturally used wheat [22].
A first step into this direction was the replacement of native RubisCO of Nicotiana tabaccum through faster homologs from the alphaproteobacterium Rhodospirillum rubrum [23] and the cyanobacterium Synechoccocus elongatus [24], although the transgenic plants were only able to grow under highly elevated CO2-concentrations and showed severe growth deficits compared to the corresponding wild-types. Redesign of the S. elongatus transgene recently restored wild-type-like growth, still under elevated CO2 atmosphere, but notably at tenfold lower RubisCO levels compared to the wild-type [25]. This shows that it is in principle possible to transplant exogenous RubisCOs with improved catalytic properties into plants. Yet, it remains to be demonstrated, whether improved RubisCOs alone would be actually able to substantially increase photosynthetic yield under field conditions.
Improving the working conditions of RubisCO through CCMs
Instead of improving the catalytic parameters of RubisCO, an alternative approach to increase photosynthetic productivity centers on changing the working conditions of the enzyme. By increasing the CO2 concentration around RubisCO, the oxygenation side reaction of the enzyme can be effectively suppressed, which in turn enables faster CO2-fixation rates, resulting in an increased CO2-fixation efficiency.
Different CO2-concentrating mechanisms (CCMs) emerged naturally during evolution [26]. In C4-plants, CO2 is pre-fixed in special cells or dedicated compartments into a C4-acid, which is transported to the place of RubisCO, where it is decarboxylated again, thereby increasing the local CO2:O2 ratio around the enzyme [27]. Cyanobacteria on the other hand evolved several HCO3 - transporters and CO2-uptake systems that enable them to concentrate up to 40 mM HCO3 - intracellularly [28]. In addition, cyanobacteria feature carboxysomes [29], proteinaceous compartments that are filled with RubisCO [30] and carbonic anhydrase [31,32]. These compartments allow the selective influx of HCO3- and ribulose-1,5-bisphosphate through pores. HCO3 - is converted into CO2 and retained within the carboxysome, so that CO2-fixation takes place in a dedicated compartment under a highly enriched CO2 environment.
Current crop production relies mainly on plants that do not possess any of the known natural CCMs. More than 80% of the agricultural land used in crop production is covered by plants that lack CCMs, such as rice, potato, wheat, and barley [33]. First efforts that focused on transplanting CCMs into rice demonstrated that it is not sufficient to simply import the enzyme machinery of C4-plants [34]. Thus, current strategies that are pursued by different consortia (http://C4rice.irri.org; http://www.3to4.org/) aim at mimicking cell-specific expression patterns of C4-CCM genes [35] and introducing the structural and anatomical characteristics of C4-plants into CCM-free crops [36], which apparently is a long-term challenge.
Another line of research aims at introducing carboxysomes into chloroplasts of CCM-free crops, which is predicted to improve yield by up to 60% under hot and dry conditions [37]. Functional carboxysomes were already reconstituted in Escherichia coli [38] demonstrating the potential for robust self-assembly in foreign hosts. The transient expression of several carboxysome subunits in Nicotiana benthaniama at least resulted in the formation of organized structures that resembled empty microcompartments [39]. Together with the fact that carboxysomal RubisCO from S. elongatus is known to be functionally expressed in tobacco [24,25] (see above), these studies might pave the way to produce functional chloroplastic carboxysomes in the future. If the introduction of such complex CCM will actually be beneficial or rather a burden in planta remains to be seen, given the fact that the number of carboxysomes per chloroplast required is still unclear [40]. To reduce the genes necessary for a functional chloroplastic carboxysome, it might become necessary to streamline the assembly process by protein domain fusions [41]. Finally, the supply of these “compartments within compartments” with sufficient CO2 will be crucial [37]. The expression of HCO3 - transporters and carbonic anhydrase in chloroplasts [42,43] could provide a solution to this problem.
Engineering synthetic photorespiration bypasses
The possibility to increase photosynthetic CO2-fixation yield by improving photorespiration has gained considerable interest in recent years. Natural photorespiration is a costly process that involves multiple enzyme reactions, which are located in different organelles in plants. Canonical photorespiration recycles two G2P molecules into one 3-PG molecule (Figure 2), while releasing one molecule of CO2 and NH3 during this process. The recycling of G2P and in particular the re-fixation of the lost CO2 requires input of a considerable amount of energy and reducing power. Several alternative photorespiration bypasses, based on existing routes have been suggested that are advantageous compared to the natural process in terms of either ATP requirement, reducing potential, carbon stoichiometry, or the number of cellular compartments involved (Table 1).
Table 1.
Comparison of natural and synthetic photorespiration bypasses. Due to the different topologies, the synthetic photorespiration bypasses cannot be simply compared side-by-side. The expected advantages are of multifactorial nature and more than a simple sum of redox equivalents and ATPs comsumed. However, when normalized onto total carbon stochiometry (i.e., the total requirements to regenerate a C3 intermediate and the net fixation of one CO2) the individual photorespiration bypasses can be balanced as followed. Advantages compared to the canonical (i.e., natural) photorespiration bypass are highlighted in blue, disadvantages are marked in red.
| Canonical photorespiration bypass | Chloroplastic glycerate bypass | Chlorplastic glycolate oxidation bypass | Peroxisomal glycerate bypass | 3-hydroxyx-propionate bypass |
|---|---|---|---|---|
| 12 | 5 | 5 | 7 | 13 |
| 8 | 7 | 7 | 9 | 7 |
| 4 NAD(P)H + 2 Fd (2,200 mV) | 5 NAD(P)H (1,700 mV) | 4 NAD(P)H (1,360 mV) | 5 NAD(P)H (1,700 mV) | 3 NADPH (1,020 mV) |
| Yes (25%) | Yes (25%) | Yes (100%) | Yes (25%) | No (0%) |
| mitochondria (far RubisCO) | chloroplast (near RubisCO) | chloroplast (near RubisCO) | peroxisome (far RubisCO) | no CO2 released |
| 4 | 0 | 0 | 2 | 0 |
| Yes | No | No | No | No |
| Calvin cycle | Calvin cycle | Calvin cycle | Calvin cycle | Included in bypass |
| 2 turns | 2 turns | 3 turns | 2 turns | none |
In the chloroplastic glycerate bypass [44], two molecules of G2P are converted under loss of one CO2 into one molecule of glycerate, which is fed back into the Calvin cycle (Figure 2). The entire process circumvents the release of NH3, consumes less ATP, and conserves reducing power. Since the whole process was designed to take place in the chloroplast, the CO2 is released in vicinity of RubisCO, reducing its oxygenation side reaction. When the glycerate pathway from E. coli was introduced into Arabidopsis thaliana [44] or Camelina sativa [45] chloroplasts, transgenic plants showed an enhanced photosynthesis, faster growth and higher biomass generation. However, transgenic lines only expressing glycolate dehydrogenase in the chloroplasts, showed similar results [44,45]. Thus, the role and the fate of the glyoxylate that is produced in the chloroplasts of these transgenic plants is not quite clear.
The peroxisomal glycerate bypass [46] is based on the conversion of glycolate into glycerate in peroxisomes (Figure 2). It bypasses NH3 release, and conserves reducing power. The pathway originally from E. coli could be implemented only partially in Nicotiana tabacum. Transgenic tobacco lacked expression of one of the key enzymes, and plants stunted growth under ambient air [46].
In the chloroplastic glycolate oxidation bypass [47], G2P is converted into glycolate, which is subsequently completely oxidized into CO2 within the chloroplast (Figure 2). This pathway bypasses the release of NH3 and conserves reducing power. A huge disadvantage is that all of the carbon is lost instead of “only” one out of four, as in the other pathways. When experimentally realized by redirecting peroxisomal glycolate oxidase and catalase to the chloroplast, transgenic A. thaliana were demonstrated to support higher dry weight and photosynthetic rates. This effect was significant under energy-limiting growth conditions [47].
In contrast to above strategies that all release CO2, the proposed 3-hydroxypropionate bypass [48] leads to a net fixation of CO2 during synthetic photorespiration by converting G2P into pyruvate (Figure 2). The 3-hydroxypropionate bypass was successfully realized in the cyanobacterium S. elongatus through hetereologous expression of seven enzymes from the filamentous anoxygenic phototroph Chloroflexus aurantiacus and the betaproteobacterium Accumulibacter phosphatis. All enzyme activities were successfully demonstrated. A growth phenotype, however, was not observed, most probably because S. elongatus possesses already very efficient CCMs [48]. Transplantation of the 3-hydroxyproionate bypass into CCM-deficient strains of S. elongatus may provide a growth advantage and proof this concept.
Even though the engineering of synthetic photorespiration bypasses has already shown promising results, all projects so far were based on grafting naturally occurring pathways into photosynthetic hosts. To overcome this restriction onto natural solutions, a new European research initiative aims at systematically exploring and engineering completely artificial routes of higher efficiency in a true synthetic biology effort by combining enzyme engineering and metabolic retrosynthesis (http://FutureAgriculture.eu).
Engineering synthetic CO2-fixation
The most ambitious approach to improve photosynthetic yield is to completely rewire CO2-fixation in plants, algae and cyanobacteria. This research is inspired by the discovery that during the course of evolution nature itself has invented five alternative CO2-fixation pathways to the Calvin cycle, which operate in different bacteria and archaea [49–54]. These “alternative” microbial CO2-fixation pathways are not based on RubisCO [55] and several of them show advantages in respect to energy requirement and efficiency compared to the Calvin cycle [56]. The reconstitution of natural existing CO2-fixation pathways in model organisms, however, has not proven successful so far [57], probably due to the complex interplay and interference with the host’s native carbon and energy metabolism.
Even more progressive are synthetic biology approaches that are based on the principle of metabolic retrosynthesis. Here, completely novel CO2-fixation pathways of high efficiency are supposed to be designed through the free recombination of known enzyme reactions [55,58]. These efforts are further fueled by the discovery [59,60] and rational engineering [61] of highly efficient carboxylases, and the general progress in computational enzyme design [62]. The degree of freedom in these synthetic pathways allows tailoring the conversion of CO2 into virtually any desired product, and their synthetic nature could be advantageous for in vivo transplantations due to a limited interference with natural metabolism. The realization of such synthetic CO2-fixation pathways and their integration into living organisms still poses several challenges, but will be indispensable for freeing natural photosynthetic CO2-fixation from its inherent disadvantages, and transforming biology from a tinkering science into a truly synthetic discipline. Compared to all other strategies discussed here, this approach holds the most promise to substantially improve photosynthetic productivity on a long-term perspective.
Acknowledgments
This work was supported by the European Research Council (ERC 637675 ‘SYBORG’ & ERC 686330 ‘FutureAgriculture’) and the Max-Planck-Society. The authors thank I. Berg and A. Bar-Even for critical comments.
References
• of special interest
•• of outstanding interest
- 1.Odell RT, Walker WM, Boone LV, Oldham MG. The Morrow Plots - a Century of Learning. Illinois Agricultural Experiment Station Bulletin. 1982:1–22. [Google Scholar]
- 2.Fuglie KO, Wang SL, Ball VE. Introduction to Productivity Growth in Agriculture. Productivity Growth in Agriculture: An International Perspective. 2012:1–11. [Google Scholar]
- 3.Long SP, Marshall-Colon A, Zhu XG. Meeting the Global Food Demand of the Future by Engineering Crop Photosynthesis and Yield Potential. Cell. 2015;161:56–66. doi: 10.1016/j.cell.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. The Moderately Efficient Enzyme: Evolutionary and Physicochemical Trends Shaping Enzyme Parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 5.Feller U, Anders I, Mae T. Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J Exp Bot. 2008;59:1615–1624. doi: 10.1093/jxb/erm242. [DOI] [PubMed] [Google Scholar]
- 6.Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR. The Costs of Photorespiration to Food Production Now and in the Future. Annu Rev Plant Biol. 2016;67:107–129. doi: 10.1146/annurev-arplant-043015-111709. [DOI] [PubMed] [Google Scholar]
- 7.Zhu XG, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
- 8.Varaljay VA, Satagopan S, North JA, Witte B, Dourado MN, Anantharaman K, Arbing MA, McCann SH, Oremland RS, Banfield JF, et al. Functional metagenomic selection of ribulose 1,5-bisphosphate carboxylase/oxygenase from uncultivated bacteria. Environ Microbiol. 2016;18:1187–1199. doi: 10.1111/1462-2920.13138. [• A growth based bacterial complementation system was used to screen for active RubisCO from environmental DNA sources. Seventeen functional RubisCO from uncultivated and largely unknown members of natural microbial communities could be identified.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witte B, John D, Wawrik B, Paul JH, Dayan D, Tabita FR. Functional Prokaryotic RubisCO from an Oceanic Metagenomic Library. Appl Environ Microbiol. 2010;76:2997–3003. doi: 10.1128/AEM.02661-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene DN, Whitney SM, Matsumura I. Artificially evolved Synechococcus PCC6301 Rubisco variants exhibit improvements in folding and catalytic efficiency. Biochem J. 2007;404:517–524. doi: 10.1042/BJ20070071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kreel NE, Tabita FR. Serine 363 of a Hydrophobic Region of Archaeal Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase from Archaeoglobus fulgidus and Thermococcus kodakaraensis Affects CO2/O2 Substrate Specificity and Oxygen Sensitivity. PLoS One. 2015;10:e0138351. doi: 10.1371/journal.pone.0138351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tcherkez GG, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA. 2006;103:7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savir Y, Noor E, Milo R, Tlusty T. Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc Natl Acad Sci USA. 2010;107:3475–3480. doi: 10.1073/pnas.0911663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashida H, Saito Y, Kojima C, Kobayashi K, Ogasawara N, Yokota A. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science. 2003;302:286–290. doi: 10.1126/science.1086997. [DOI] [PubMed] [Google Scholar]
- 15.Erb TJ, Evans BS, Cho K, Warlick BP, Sriram J, Wood BM, Imker HJ, Sweedler JV, Tabita FR, Gerlt JA. A RubisCO-like protein links SAM metabolism with isoprenoid biosynthesis. Nat Chem Biol. 2012;8:926–932. doi: 10.1038/nchembio.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashida H, Saito Y, Nakano T, Tandeau de Marsac N, Sekowska A, Danchin A, Yokota A. RuBisCO-like proteins as the enolase enzyme in the methionine salvage pathway: functional and evolutionary relationships between RuBisCO-like proteins and photosynthetic RuBisCO. J Exp Bot. 2008;59:1543–1554. doi: 10.1093/jxb/ern104. [DOI] [PubMed] [Google Scholar]
- 17.Schada von Borzyskowski L, Rosenthal RG, Erb TJ. Evolutionary history and biotechnological future of carboxylases. J Biotechnol. 2013;168:243–251. doi: 10.1016/j.jbiotec.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 2008;59:1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 19.Tabita FR, Hanson TE, Li HY, Satagopan S, Singh J, Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71:576. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih PM, Occhialini A, Cameron JC, Andralojc PJ, Parry MA, Kerfeld CA. Biochemical characterization of predicted Precambrian RuBisCO. Nat Commun. 2016;7:10382. doi: 10.1038/ncomms10382. [• Phylogenetic analysis was used to reconstruct potentially ancestral RubisCO forms. The reconstructed enzymes possesed low carboxylation activities as well as a low CO2- specificities. This suggests that “ancestral RubisCO” evolved into two diverging directions along the two trade-offs, i.e. faster, but more unspecific enzymes (carboxysomal RubisCOs in algae) and more specific, but slower enzymes (plant RubisCOs).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu XG, Portis AR, Long SP. Would transformation of C-3 crop plants with foreign Rubisco increase productivity? A computational analysis extrapolating from kinetic properties to canopy photosynthesis. Plant Cell Environ. 2004;27:155–165. [Google Scholar]
- 22.Prins A, Orr DJ, Andralojc PJ, Reynolds MP, Carmo-Silva E, Parry MA. Rubisco catalytic properties of wild and domesticated relatives provide scope for improving wheat photosynthesis. J Exp Bot. 2016;67:1827–1838. doi: 10.1093/jxb/erv574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitney SM, Andrews TJ. Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco. Proc Natl Acad Sci USA. 2001;98:14738–14743. doi: 10.1073/pnas.261417298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin MT, Occhialini A, Andralojc PJ, Parry MAJ, Hanson MR. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014;513:547–550. doi: 10.1038/nature13776. [•• The RubisCO from the cyanobacterium Synechoccus elongatus was used to replace native RubisCO of tobacco. The transgenic plants showed RubisCO expression levels of about a sixth compared to native RubisCO. Although this study demonstrated for the first time that a cyanobacterial RubisCO can sustain autotrophic growth in planta, the plants required an atmosphere enriched with 9% CO2 and showed a severe growth defect.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Occhialini A, Lin MT, Andralojc PJ, Hanson MR, Parry MA. Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2 . Plant J. 2016;85:148–160. doi: 10.1111/tpj.13098. [• This study aimed at improving the transgenic tobacco line carrying RubisCO from Synechococcus elongatus (see above). Altering the gene regulatory sequences of the RubisCO transgenes enhanced RubisCO expression. The transgenic plants still required elevated CO2-concentrations, but grew at nearly wild-type growth rates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raven JA, Cockell CS, De La Rocha CL. The evolution of inorganic carbon concentrating mechanisms in photosynthesis. Philos Trans R Soc Lond B Biol Sci. 2008;363:2641–2650. doi: 10.1098/rstb.2008.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sage RF, Sage TL, Kocacinar F. Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Biol. 2012;63:19–47. doi: 10.1146/annurev-arplant-042811-105511. [DOI] [PubMed] [Google Scholar]
- 28.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 29.Rae BD, Long BM, Badger MR, Price GD. Functions, Compositions, and Evolution of the Two Types of Carboxysomes: Polyhedral Microcompartments That Facilitate CO2 Fixation in Cyanobacteria and Some Proteobacteria. Microbiol Mol Biol Rev. 2013;77:357–379. doi: 10.1128/MMBR.00061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus . Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 31.Cannon GC, Heinhorst S, Kerfeld CA. Carboxysomal carbonic anhydrases: Structure and role in microbial CO2 fixation. Biochim Biophys Acta. 2010;1804:382–392. doi: 10.1016/j.bbapap.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 32.So AKC, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J Bacteriol. 2004;186:623–630. doi: 10.1128/JB.186.3.623-630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leff B, Ramankutty N, Foley JA. Geographic distribution of major crops across the world. Global Biogeochem Cy. 2004;18 [Google Scholar]
- 34.Miyao M, Masumoto C, Miyazawa S, Fukayama H. Lessons from engineering a single-cell C4 photosynthetic pathway into rice. J Exp Bot. 2011;62:3021–3029. doi: 10.1093/jxb/err023. [DOI] [PubMed] [Google Scholar]
- 35.Kajala K, Covshoff S, Karki S, Woodfield H, Tolley BJ, Dionora MJ, Mogul RT, Mabilangan AE, Danila FR, Hibberd JM, et al. Strategies for engineering a two-celled C4 photosynthetic pathway into rice. J Exp Bot. 2011;62:3001–3010. doi: 10.1093/jxb/err022. [DOI] [PubMed] [Google Scholar]
- 36.Feldman AB, Murchie EH, Leung H, Baraoidan M, Coe R, Yu SM, Lo SF, Quick WP. Increasing leaf vein density by mutagenesis: laying the foundations for C4 rice. PLoS One. 2014;9:e94947. doi: 10.1371/journal.pone.0094947. [• A high throughput method with handheld microscopes was developed to screen more than 23,000 rice lines. Eight lines were identified that show significantly increased leaf vein densities. These morphologically changed lines provide an important step towards establishing a functional C4 trait in rice.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath JM, Long SP. Can the Cyanobacterial Carbon-Concentrating Mechanism Increase Photosynthesis in Crop Species? A Theoretical Analysis. Plant Physiol. 2014;164:2247–2261. doi: 10.1104/pp.113.232611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci USA. 2012;109:478–483. doi: 10.1073/pnas.1108557109. [•• The hetereologous expression of the Halothiobacillus neapolitanus carboxysome encoded by 10 genes in Escherichia coli resulted in the production of icosahedral complexes similar to those of native carboxysomes. The complexes were capable of assembling with carboxysomal proteins and fixing CO2 in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MAJ, Hanson MR. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 2014;79:1–12. doi: 10.1111/tpj.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanson MR, Lin MT, Carmo-Silva AE, Parry MA. Towards engineering carboxysomes into C3 plants. Plant J. 2016 doi: 10.1111/tpj.13139. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Esquera CR, Shubitowskia TB, Kerfeld CA. Streamlined Construction of the Cyanobacterial CO2-Fixing Organelle via Protein Domain Fusions for Use in Plant Synthetic Biology. Plant Cell. 2015;27:2637–2644. doi: 10.1105/tpc.15.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pengelly JJL, Förster B, von Caemmerer S, Badger MR, Price GD, Whitney SM. Transplastomic integration of a cyanobacterial bicarbonate transporter into tobacco chloroplasts. J Exp Bot. 2014;65:3071–3080. doi: 10.1093/jxb/eru156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atkinson N, Feike D, Mackinder LC, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ. Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components. Plant Biotechnol J. 2016;14:1302–1315. doi: 10.1111/pbi.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kebeish R, Niessen M, Thiruveedhi K, Bari R, Hirsch HJ, Rosenkranz R, Stabler N, Schonfeld B, Kreuzaler F, Peterhansel C. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol. 2007;25:593–599. doi: 10.1038/nbt1299. [•• A glycolate catabolic pathway from Escherichia coli consisting of three enzyme reactions was introduced into Arabidopsis thaliana chloroplasts, representing the first artifical photorespiration bypass realized in planta. The transgenic plants grew faster, produced more shoot and root biomass, and contained more soluble sugars.] [DOI] [PubMed] [Google Scholar]
- 45.Dalal J, Lopez H, Vasani NB, Hu Z, Swift JE, Yalamanchili R, Dvora M, Lin X, Xie D, Qu R, et al. A photorespiratory bypass increases plant growth and seed yield in biofuel crop Camelina sativa . Biotechnol Biofuels. 2015;8:175. doi: 10.1186/s13068-015-0357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalho JDC, Madgwick PJ, Powers SJ, Keys AJ, Lea PJ, Parry MAJ. An engineered pathway for glyoxylate metabolism in tobacco plants aimed to avoid the release of ammonia in photorespiration. BMC Biotechnol. 2011;11 doi: 10.1186/1472-6750-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier A, Fahnenstich H, von Caemmerer S, Engqvist MK, Weber AP, Flugge UI, Maurino VG. Transgenic Introduction of a Glycolate Oxidative Cycle into A. thaliana Chloroplasts Leads to Growth Improvement. Front Plant Sci. 2012;3:38. doi: 10.3389/fpls.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shih PM, Zarzycki J, Niyogi KK, Kerfeld CA. Introduction of a synthetic CO2-fixing photorespiratory bypass into a cyanobacterium. J Biol Chem. 2014;289:9493–9500. doi: 10.1074/jbc.C113.543132. [• This study describes the design and introduction of a synthetic photorespiration bypass into Synecchococcus elongatus. This is the first synthetic photorespiration bypass that allows to fix CO2 instead of releasing CO2. The pathway consists of six reactions that are based on the autotrophic 3-hydroxypropionate bi-cycle originally identified in Chloroflexus aurantiacus.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Könneke M, Schubert DM, Brown PC, Hugler M, Standfest S, Schwander T, von Borzyskowski LS, Erb TJ, Stahl DA, Berg IA. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA. 2014;111:8239–8244. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans MCW, Buchanan BB, Arnon DI. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc Natl Acad Sci USA. 1966;55:928. doi: 10.1073/pnas.55.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarzycki J, Brecht V, Müller M, Fuchs G. Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus . Proc Natl Acad Sci USA. 2009;106:21317–21322. doi: 10.1073/pnas.0908356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 53.Huber H, Gallenberger M, Jahn U, Eylert E, Berg IA, Kockelkorn D, Eisenreich W, Fuchs G. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis . Proc Natl Acad Sci USA. 2008;105:7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erb TJ. Carboxylases in Natural and Synthetic Microbial Pathways. Appl Environ Microbiol. 2011;77:8466–8477. doi: 10.1128/AEM.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mattozzi M, Ziesack M, Voges MJ, Silver PA, Way JC. Expression of the sub-pathways of the Chloroflexus aurantiacus 3-hydroxypropionate carbon fixation bicycle in E. coli: Toward horizontal transfer of autotrophic growth: Chloroflexus carbon fixation in E. coli . Metab Eng. 2013:130–139. doi: 10.1016/j.ymben.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Bar-Even A, Noor E, Lewis NE, Milo R. Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci USA. 2010;107:8889–8894. doi: 10.1073/pnas.0907176107. [• In this theoretical study a repertoire of more than 5,000 enzymes was used to identify potential alternative CO2-fixation pathways that are predicted to be superior in terms of kinetics, energetics and/or topology compared to natural existing ones.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci USA. 2007;104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erb TJ, Brecht V, Fuchs G, Müller M, Alber BE. Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoylthioester reductase. Proc Natl Acad Sci USA. 2009;106:8871–8876. doi: 10.1073/pnas.0903939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peter DM, Schada von Borzyskowski L, Kiefer P, Christen P, Vorholt JA, Erb TJ. Screening and Engineering the Synthetic Potential of Carboxylating Reductases from Central Metabolism and Polyketide Biosynthesis. Angew Chem Int Ed Engl. 2015;54:13457–13461. doi: 10.1002/anie.201505282. [• Carboxylating enoyl-thioester reductases (ECRs) are a recently discovered class of highly efficient CO2-fixing enzymes that are 10-100 times more efficient than RubisCO. This study identified three active site residues that define substrate speficity in ECRs, which allowed to produce a variety of different α-carboxyl-acyl-thioesters from CO2.] [DOI] [PubMed] [Google Scholar]
- 62.Kries H, Blomberg R, Hilvert D. De novo enzymes by computational design. Curr Opin Chem Biol. 2013;17:221–228. doi: 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]