Abstract
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) is arguably one of the most abundant proteins in the biosphere and a key enzyme in the global carbon cycle. Although RubisCO has been intensively studied, its evolutionary origins and rise as Nature’s most dominant carbon dioxide (CO2)-fixing enzyme still remain in the dark. In this review we will bring together biochemical, structural, physiological, microbiological, as well as phylogenetic data to speculate on the evolutionary roots of the CO2-fixation reaction of RubisCO, the emergence of RubisCO-based autotrophic CO2-fixation in the context of the Calvin cycle, and the further evolution of RubisCO into the ‘RubisCOsome’, a complex of various proteins assembling and interacting with the enzyme to improve its operational capacity (functionality) under different biological and environmental conditions.
More than 90% of the inorganic carbon that is converted into biomass is fixed by the enzyme RubisCO that catalyzes the carboxylation and cleavage of ribulose-1,5-bisphosphate (RuBP) into two molecules of 3-phosphoglycerate (3PG). RubisCO is found in all three domains of life: bacteria, archaea and eukaryotes. The enzyme makes up 30-50% of the soluble protein in plant leaf and it has been estimated that for every person on earth there is 5 kg of RubisCO [1]. Altogether, this makes RubisCO one of the most abundant enzymes in the global carbon cycle that literally feeds life on earth.
Despite its dominant role in carbon fixation, the enzyme has some peculiarities. First, RubisCO requires a posttranslational activation to perform the carboxylation reaction: A conserved lysine residue in the active site needs to be carbamylated in order to complex an Mg2+ ion that is in turn required for activity [2]. Second, the enzyme is a rather slow catalyst. The turnover frequency of an average RubisCO is only between 1 and 10 s-1 (http://brenda-enzymes.org). which can make it a limiting factor in photosynthetic CO2-fixation under optimal conditions. Finally, RubisCO makes mistakes. Besides the carboxylation reaction RubisCO catalyzes a non-productive oxygenation side-reaction that leads to the formation of 2-phosphoglycolate (2PG). 2PG is a toxic compound that inhibits several enzymes in central carbon metabolism [3–5]. An average C3-plant RubisCO has an error rate of more than 20%. This number can even add up to more than 40% at high temperatures and/or low intracellular CO2, which results in substantial amounts of 2PG that are formed during photosynthesis [6–8]. 2PG is recycled in an energy-demanding process called photorespiration. It has been estimated that approximately 30% of the photosynthetic energy in plants is wasted through photorespiration [8,9].
Why is RubisCO so inefficient? Evidence accumulated that enzyme activity and specificity are reciprocally linked with each other in RubisCO [10–12]. A faster RubisCO has a higher error rate and a more specific RubisCO has a lower catalytic rate. This link has mechanistic and evolutionary reasons. RubisCO evolved before the first great oxygenation event in an atmosphere without oxygen (O2), so that its mechanism was not constrained by O2. However, with the rise of atmospheric O2 concentrations to modern-day levels, as a result of the second great oxygenation event, RubisCO had to learn to discriminate between CO2 and O2. Because discrimination usually comes at the cost of reduced catalytic rate, a more specific enzymes almost inevitably becomes a slower catalyst [13]. As a consequence RubisCO had to evolve along a Pareto front of enzyme activity and specificity, a trade-off in which the modern enzyme apparently became trapped.
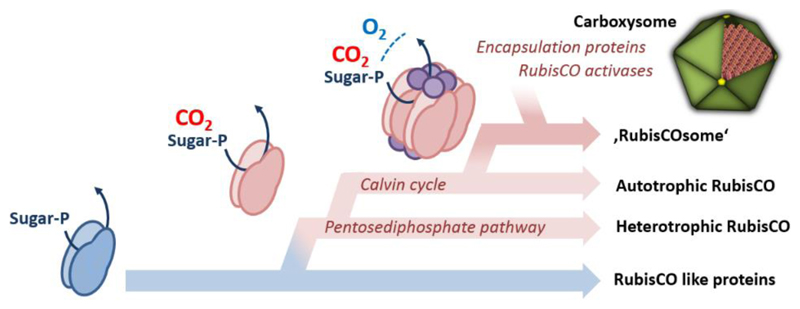
Although this part of RubisCO’s evolutionary history is generally accepted, the answer to the question how RubisCO arose to become Nature’s predominant CO2-fixing enzyme is less well known. In this article we leave firm grounds to illuminate the emergence of ‘proto-RubisCO’, the first RubisCO homolog that catalyzed a carboxylation reaction, its integration into the Calvin cycle and its further evolution into the modern day CO2-fixing enzyme complex, called the ‘RubisCOsome’. The picture we draw is based on works, ideas, discussions and comments of many colleagues and is summarized in three main hypothesis (‘opinions’) that are discussed in the following.
RubisCO evolved from a non-CO2-fixing ancestor
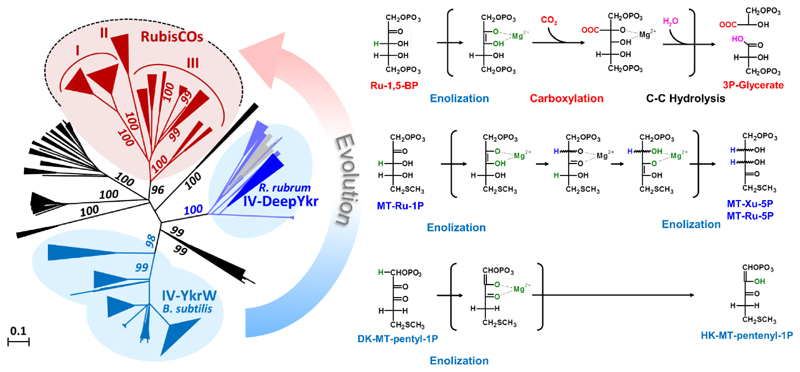
How did RubisCO emerge as Nature’s most dominant CO2-fixing biocatalyst? A key for understanding the evolution of RubisCO are RubisCO-like proteins (RLPs) [14]. RLPs are phylogenetically and evolutionarily related to RubisCOs, with which together they form the RLP/RubisCO enzyme superfamily (Fig. 1). While RLPs share substantial sequence identity with RubisCOs, they lack active site amino acid residues that are known to be essential for the carboxylation reaction of RubisCO.
Figure 1. Evolution of the carboxylation function in the RLP/RubisCO superfamily of enzymes.
The left panel shows a phylogenetic tree of the RLP/RubisCO superfamily, with RubisCOs depicted in red and RLPs of the 1,3-isomerase and tautomerase family shown in blue. The different reaction mechanisms postulated for these enzymes are shown on the right panel. All reactions proceed through a central enolate intermediate that is formed by the initial abstraction of a proton from the respective substrate (highlighted in green). Compared to RubisCO, the reactions of the 1,3-isomerase and tautomerase are mechanistically simpler, indicating that the more complex CO2-fixing reaction of RubisCO emerged in the scaffold of a RLP ancestor. The phylogenetic tree was adapted with changes from Schada et al. [26]. Ribulose-1,5-bisphosphate (Ru-1,5-BP), methylthioribulose-1-phosphate (MT-Ru-1P), methylthioribulose-5-phosphate (MT-Ru-5P), methylthioxylulose-5-phosphate (MT-Xu-5P), 2,3-diketo methylthiopentyl-1-phsphate (DK-MT-pentyl-1P), 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate (HK-MT-pentenyl-1P).
For several RLP subfamilies a biochemical and physiological function has been determined. The tautomerase subfamily of RLPs operates in a variant of the ubiquitous ‘methionine salvage’ pathway that recycles the dead end metabolite methylthioadenosine (MTA) into L-methionine [15]. The 1,3-isomerase subfamily of RLPs function in the so-called MTA-isoprenoid shunt that channels MTA into isoprenoid biosynthesis [16]. Recently, a subfamily of decarboxylase RLPs was identified that serve in a degradation pathway of the four carbon sugar acids erythronate and threonate [17].
Based on sequence diversity, functional evidences, and genomic context, at least four more RLP subfamilies can be distinguished [16,18,19] that are most likely also isomerases and/or epimerases [20]. Thus, the RLP/RubisCO superfamily features at least seven functionally distinct RLP subfamilies besides the subfamily of ‘true RubisCOs’ that can be divided into three distinct phylotypes (RubisCOs Form I, II and III).
How did all these different RLP functions emerge during evolution, and among the different functions, how in particular, did the carboxylation trait evolve that is apparently restricted to the subfamily of true RubisCOs only? Two alternative hypotheses can be formulated that are mutually exclusive.
Hypothesis 1
The carboxylation function in the RLP/RubisCO superfamily is an ancient trait [21]. According to this hypothesis, the carboxylation reaction is an original function and RubisCO-like proteins evolved from a CO2-fixing, ancestral enzyme (a ‘proto-RubisCO’) by loss of the CO2-fixation activity.
Hypothesis 2
The carboxylation function in the RLP/RubisCO superfamily is a novel trait that was acquired during evolution [22,23]. According to this hypothesis, RubisCO emerged from a non-CO2-fixing ancestor. The emergence of the carboxylation function is a secondary event and all RubisCOs are of monophyletic origin.
Although the history of evolutionary events cannot be recapitulated in retrospect, several arguments can be considered to delineate the most plausible evolutionary scenario. Valuable information lies in the various reactions that are catalyzed by the RLP/RubisCO superfamily. The enzymes of the different subfamilies catalyze distinct biochemical reactions. Yet, they share some basic aspects. All reactions in the RLP/RubisCO superfamily center on structurally similar substrates, a C5 or C4 sugar derivative that features a phosphate in the C1 and a keto group in the C2 or C3 position (Fig 1). A general catalytic mechanism can be formulated that is conserved in all members of the RLP/RubisCO superfamily characterized to date. This core mechanism includes the formation of a central enolate intermediate. In most cases this happens through the acid/base catalyzed abstraction of a proton adjacent to the keto group [24,25]. In a subsequent step, the central enolate intermediate then attacks an electrophile that can be a proton, carbon dioxide or oxygen molecule [26].
While the initial steps of catalysis that lead to formation of the central enolate intermediate are mostly conserved between the different subfamilies, they diverge in the later steps of the catalytic cycle, leading to a diverse outcome of reaction products, which explains the different catalytic functions that evolved in the RLP/RubisCO superfamily (Fig. 1). In case of the tautomerase and the decarboxylase RLP subfamilies, the enolate intermediate simply attacks a proton to yield the reaction product. In case of the 1,3-isomerase RLP subfamily two subsequent proton abstraction and attacks take place. Finally, in RubisCOs the central enolate intermediate attacks CO2 (or O2), which is followed by a subsequent water-mediated hydrolysis reaction to generate the two 3-phosphoglycerate molecules.
Apparently, there is an increasing mechanistic complexity from the simple tautomerase reaction to the multi-step reaction of RubisCO. It is fair to conclude that the increase in mechanistic complexity observed in the RubisCO subfamily reflects an evolutionary development and that the more complex mechanism of RubisCO israther the end of an evolutionary trajectory than its starting point (Fig. 1 & 2).
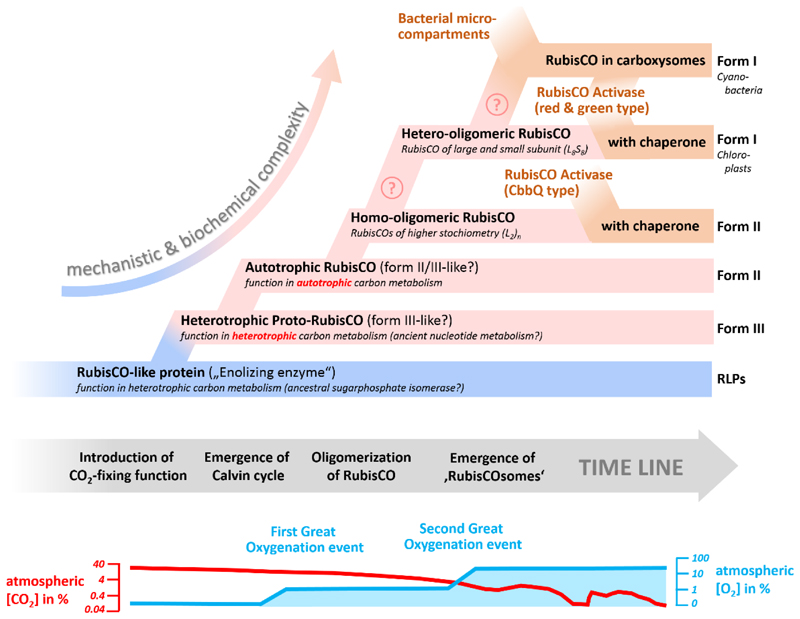
Figure 2. Hypothetical timeline of the emergence and evolution of RubisCO.
The figure summarizes the individual events in the emergence of RubisCO from a non-CO2-fixing ancestor to complex, modern-day RubisCO (Form I) which operates in chloroplasts and cyanobacterial carboxysomes. The postulated events are based on the main steps during evolution that are discussed in the main text. Note that timeline is only relative and that the order of some of the events might differ, which is highlighted by a question mark.
This line of mechanistic evidence is further supported by structural arguments. All members of the RLP/RubisCOs superfamily are eight-stranded α/β-barrels, which is the most frequent and most versatile protein fold used by evolution [27]. However, in contrast to all RLP subfamily members that have been structurally characterized to date, RubisCOs feature an additional β-hairpin in loop number 6 that carries residues essential to the carboxylation reaction. Applying the principle of Occam’s razor, it is rather likely that the subfamily of RubisCOs gained this additional stretch of amino acids during evolution compared to the possibility that all seven other subfamilies lost this additional β-hairpin in loop 6. Current approaches that explore the emergence and evolution of α/β-barrel proteins [28,29] might help to reconstruct and understand the evolutionary history of loop 6 architecture and the evolution of RubisCO in the future.
In summary, physiological, mechanistic and structural data support the hypothesis that the RLP/RubisCO-superfamily is originally a family of enolases and that the carboxylation function is a later invention in this superfamily (Fig. 2).
RubisCO evolved in a non-heterotrophic context
The hypothesis that the emergence of RubisCO is a rather late evolutionary event matches well with the notion that the Calvin cycle is most likely not the first autotrophic CO2-fixation pathway and that RubisCO evolved in a non-autotrophic context [30]. But in what kind of heterotrophic context did RubisCO evolve, what initial function did it serve and what purpose did this CO2-fixing but non-autotrophic pathway have?
Part of the answer to these questions comes from modern day Form III RubisCOs found in archaea and probably represent the most ancient form of the enzyme. Form III RubisCOs are not known to operate in the Calvin cycle for autotrophic CO2-fixation, but in assimilation of ribonucleosides [31–33]. In the so-called pentosebisphosphate pathway in archaea, which lack the ‘canoncial’ pentosephosphate pathway, the ribose moiety of adenosine, guanosine and uridine is metabolized via RuBP and through the action of RubisCO into two molecules of 3PG. Approximately 20% of the cell dry weight is RNA, which makes ribose a fairly abundant carbon sources (up to 8% of the dry weight of biological cellular material). Any pathway like the pentosebisphosphate pathway that channels ribose into central carbon intermediates and, at the same time, allows for the additional fixation of inorganic carbon into biomass would have been of advantage in an early biosphere. This is independent of an early FeS world scenario [34], according to which life arose around primordial metabolic cascades in an organic carbon-limited environment, or an early RNA world scenario that assumes that life began with a prebiotic pool of this biomolecule [35].
Does the function of Form III RubisCO in nucleoside metabolism reflect the ancient function of RubisCO? This hypothesis is further supported by the recent studies on a deep branching class of bacteria referred to as the candidate phyla radiation (CPR). CPR are highly abundant in anaerobic environments and contribute to up to 15% of the bacterial diversity. While CPR have been reluctant to all cultivation efforts until now, metagenomics studies showed that these bacteria are probably fermenters that thrive on organic carbon and hydrogen. For several CPR representatives a RubisCO based nucleotide metabolism similar to the archaeal pentose bisphophate pathway was proposed [36]. This is backed up by functional studies that showed that the CPR enzymes are indeed true RubisCOs. The phylogenetic position of these newly discovered enzymes as Form II/III intermediates and close to archaeal Form III hints to the transfer of RubisCO from archaea to (these) bacteria (Fig. 2).
Taken together, recent metagenomic, phylogenetic and biochemical data indicate that RubisCO evolved as a CO2-fixing enzyme in an ancient, probable archaeal nucleotide assimilation pathway and that RubisCO-based autotrophy via the Calvin cycle evolved later in bacteria (Fig. 2).
Another interesting point in this respect is the fact that even today the Calvin cycle does not only function in autotrophic CO2 fixation. During anaerobic photoheterotrophic growth of α-proteobacteria, the Calvin cycle is primarily used for the recycling of redox cofactors on growth substrates that are more reduced than the average cell carbon [37]. Furthermore, several non-autotrophic bacteria in the soil harbor RubisCO genes that also serve in redox balancing and carbon by-fixation [38]. It is tempting to speculate that a first Calvin cycle might have evolved from ancient nucleotide metabolism and initially served in redox cofactor balancing and/or mixotrophy, before developing its autotrophic function (Fig. 2).
How fast could a first version of the Calvin cycle have evolved? Surprisingly fast! Recent experiments demonstrate that when RubisCO is heterologously expressed together with phosphoribulokinase in Escherichia coli and selective pressure is applied, a functional Calvin cycle can be established remarkably fast and reproducibly. It takes only a hundred generations and a handful of mutations to convert the heterotrophic organism E. coli into a hemi-autotrophic organism, in which all sugars are formed from CO2 only [39]. From this E. coli laboratory experiment, it still remains unclear when exactly and how many times the Calvin cycle might have been (re-)invented during evolution. Nevertheless, the E. coli evolution experiment revealed a surprising metabolic plasticity of prokaryotic central carbon metabolism and demonstrated impressively how fast new metabolic features, such as the Calvin cycle could have emerged in a heterotrophic background and further evolved to become the dominant autotrophic carbon fixation pathway in the biosphere.
RubisCO evolved from a simple enzyme into a composite enzyme complex
The rise of RubisCO as key enzyme of autotrophic CO2 fixation required that the enzyme operates at high rates and with high efficiency. This is a prerequisite to achieve sufficient flux through central carbon metabolism, because virtually every carbon converted into biomass needs to be fixed via the Calvin cycle. Especially in the aftermath of the emergence of oxygenic photosynthesis and the great oxygenation event, RubisCO underwent strong evolutionary pressure to keep up its CO2 fixation efficiency in the presence of O2. RubisCO ultimately reached the Pareto optimality of enzyme activity and specificity and came to an evolutionary state in which neither of the two catalytic parameters could be further improved without negatively affecting the other one. This forced Nature to develop novel strategies to overcome the catalytic dilemma of RubisCO to further improve the enzymes performance (Fig. 2).
The evolutionary adaptations culminated in the formation of a multifaceted RubisCO complex that can be best viewed as ‘RubisCOsome’. It becomes clearer that modern RubisCO cannot be considered as a stand-alone enzyme, but as an enzyme that functions together with a host of other proteins and enzymes. Formation of the RubisCOsome allowed to improve the working conditions surrounding the enzyme. This in turn enabled organisms that rely on the Calvin cycle to conquer virtually any ecological niche in the biosphere. How can we imagine the evolution of the RubisCOsome and what guided these developments?
Oligomerization of RubisCO
A first step towards evolution of the RubisCOsome was the formation of larger RubisCO complexes. All extant RubisCOs (including the RLPs) share a common feature: their active sites are formed at the interface between two ~50 kDa large subunits that contribute all residues required for catalysis. However, beyond this homodimeric enzyme core (L2), RubisCOs became organized in higher oligomeric states during evolution. Form II & III RubisCOs are known to occur with higher (L2)n stoichiometry (with n up to 5) and Form I RubisCOs are organized in four L2 dimers that assemble together with an additional small subunit (S) of ~15 kDa into a hetero hexadecameric L8S8 RubisCO complex. In Form I RubisCO oligomerization and the presence of the additional small subunits increases catalytic efficiency and specificity. It has been suggested that the additional small subunits function as CO2 reservoir [40]. A subgroup of Form III RubisCOs [41], possess a short (~29 amino acids) insertion domain that apparently mimics the function of the small subunit of Form I RubisCOs [41]. When this domain is truncated, oligomerization is lost and the truncated enzymes shows ~5-fold lower specific activity compared to the wild type enzyme [42]. These findings indicate and underline the general evolutionary trend in the RubisCO subfamily to utilize RubisCO large subunit oligomerization or ‘concentration’ to influence and determine the enzyme kinetic properties (Fig. 2).
Scaffolding RubisCO
A continuation and expansion of this evolutionary theme of aggregating and complexing RubisCO can be found in the emergence of carboxysomes in cyanobacteria [43] as well as the pyrenoids in eukaryotic algae [44]. Both mechanisms are concentrating RubisCO through the interaction with dedicated ‘scaffolding proteins’ in delimited (micro)-compartments together with carbonic anhydrases (CA). CAs convert intracellularly enriched bicarbonate into CO2, thus providing a high local CO2 concentration in the vicinity of RubisCO. Especially the carboxysomes allowed the evolution of cyanobacterial RubisCOs that were able to retain a fairly low substrate specificity and gain much higher catalytic rates [11]. The fact that plants do not have any genes encoding for carboxysome proteins suggests that these microcompartments only arose after the primary endosymbiosis event as a result of the second great oxidation event and drastically decreasing atmospheric CO2 concentrations within the last 400 million years [11]. This forced plants to develop other strategies to cope, such as the invention of C4 metabolism, the fine tuning of the photorespiration pathway, as well as the evolution of RubisCOs with even higher substrate specificities.
RubisCO-specific Chaperones
The evolution of complex RubisCOs with higher specificities not only comes at a cost of catalytic rates but also with another problem. Many RubisCOs, and especially Form I RubisCOs suffer from inhibition by several naturally occurring sugar phosphates such as 2-carboxyarabitinol 1-phosphate (CA1P), xylulose 1,5-bisphosphate (XuBP), and even by the native substrate RuBP [45,46]. If RuBP (or XuBP) is bound to an active site that lacks the carbamylated lysine, carboxylation cannot occur and re-carbamylation of the lysine is prevented as well. This leads to a dead-end (inactive) enzyme-substrate complex. Other sugar phosphates like CA1P can be bound to a carbamylated active site and act as a non-reactive RuBP analog. The binding of these compounds to RubisCO results in a very tightly closed conformation, whereby it is observed that the higher the specificity for CO2 over O2 the tighter the binding of inhibitors [46]. Evolution has solved this problem by providing another set of proteins, so called RubisCO activases (RCAs) that became part of the RubisCOsome (Fig. 2). RCAs are dedicated ATP dependent chaperones that can open the active site of RubisCO and release the inhibitory compounds [47–50]. Three classes of RCAs have been identified to-date (green type, red-type, and CbbQO) that all belong to the ATPases superfamily of enzymes [51]. Genes encoding RCAs can often be found up- or downst ream of carboxysomal gene clusters [52]. Moreover, some of these RCAs possess an additional C-terminal domain mimicking the small subunit of RubisCO [53]. Both these findings hint at a probable encapsulation of the RCAs in the carboxysomes. The co-evolution of RCAs with their corresponding RubisCO has resulted in some level of incompatibility [46]. Thus, when transplantation of foreign RubisCOs into crop plants or biotechnologically used microbes are to be attempted one may have to consider bringing along the specific RCA to reconstitute the functional RubisCOsome.
All in all it is thus more appropriate to regard RubisCO as the center (heart) of a multi enzyme complex (Fig. 2), which has to be acknowledged/addressed in future attempts of re-engineering and transplanting these enzymes into hosts of agricultural and biotechnological interest.
The fall of RubisCO? Synthetic approaches to overcome the evolutionary constraints of RubisCO
Even though evolution is generally considered to be a highly creative force, its nature is actually rather conservative. Once a biological solution is found by evolution, the space to explore new evolutionary paths dramatically narrows, as evolution rather tends to work in a tinkering fashion by improving and recombining existing parts and pieces. This pattern is also reflected in RubisCO’s history: The evolution of proto-RubisCO in a scaffold of enolizing enzymes without carboxylation activity, its initial function in a heterotrophic context, which only evolved later to become the Calvin cycle, and the subsequent addition of more proteins to the catalytic core of RubisCO to improve its functionality that cumulated in the formation of the RubisCOsome.
With the advent of synthetic biology, however, it becomes possible to realize completely novel biological solutions that are not bound by historical constraints in a rational fashion. Currently, several ideas are followed to overcome RubisCO’s inefficiency with synthetic biological methods: Some approaches aim at replaying the evolution of RubisCO with RubisCO variants [54] or resurrected ancestors [11], which are placed under strong O2 selection to create alternate RubisCOs that could break the Pareto front of modern-day RubisCOs. Other ideas focus on extending the Pareto front by creating a ‘neo-RubisCO’, an enzyme that catalyzes the same chemistry as RubisCO but in a different protein scaffold that would allow to discriminate better against O2.
The most radical approaches, however, aim at completely redesigning photosynthetic central carbon metabolism. Some efforts aim at realizing a synthetic photorespiration that would even allow for additional CO2 fixation and could cover up for RubisCO’s side reaction with O2 [55]. Other efforts focus on creating novel CO2 fixation pathways that are independent of RubisCO, such as the MOG [56] or the CETCH cycle [57]. The latter is based on the highly efficient and versatile class of enoyl-CoA carboxylases/reductases [58,59] that are notably also not affected by O2, because of their unique CO2-fixation mechanism [60,61]. The CETCH cycle was recently realized with 17 different enzymes from nine different organisms, including three redesigned enzymes and shown to turn at a rate comparable to the Calvin cycle in vitro [57].
However, despite impressive progress in all these efforts, it should be noted that modern cells are highly integrated biological systems, which needs to be considered in any approaches that focus on substituting or bypassing RubisCO. For instance the fact that linear electron flow matches almost perfectly the ATP and NADPH stoichiometries of the Calvin cycle might hint that it will not be sufficient to implement synthetic photorespiration and/or CO2-fixation into photosynthetic organisms without further adaptions. Only the future will tell whether carbon fixation through an alternatively evolved RubisCO, a neo-RubisCO, or RubisCO-independent synthetic pathways will allow us to rewrite or even overwrite the chapter of RubisCO’s evolution.
Bulletpoints.
RubisCO evolved to become Nature’s predominant CO2-fixing enzyme through three distinct steps
RubisCO evolved from a non-CO2-fixing ancestral enzyme into a true carboxylase
RubisCO emerged in a non-autotrophic context before the Calvin cycle evolved
RubisCO evolved from a simple enzyme into a composite enzyme complex, the RubisCOsome that helped to improve the enzyme’s functionality in an O2-rich atmosphere
Multiple synthetic biological approaches aim at overcoming RubisCO and its catalytic imperfection in the future
Acknowledgments
This work was supported by the European Research Council Grant 637675 (‘SYBORG’), FET-Open Grant 686330 (‘FutureAgriculture’), and the Max-Planck-Society. The authors thank Birgit Alber, Ivan Berg, John Gerlt, Cheryl Kerfeld, Oliver Mueller-Cajar, Justin North, Robert Tabita, Dan Tawfik, Rolf Thauer, Kelly Wrighton and many more colleagues of the ‘C1-community’ for the countless discussions that formed the basis for this article.
References
• of special interest
•• of outstanding interest
- 1.Phillips R, Milo R. A feeling for the numbers in biology. Proc Natl Acad Sci U S A. 2009;106:21465–21471. doi: 10.1073/pnas.0907732106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorimer GH, Miziorko HM. Carbamate formation on the epsilon-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by CO2 and Mg2+ . Biochemistry. 1980;19:5321–5328. doi: 10.1021/bi00564a027. [DOI] [PubMed] [Google Scholar]
- 3.Anderson LE. Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim Biophys Acta. 1971;235:237–244. doi: 10.1016/0005-2744(71)90051-9. [DOI] [PubMed] [Google Scholar]
- 4.Kelly GJ, Latzko E. Chloroplast Phosphofructokinase: II. Partial Purification, Kinetic and Regulatory Properties. Plant Physiol. 1977;60:295–299. doi: 10.1104/pp.60.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norman EG, Colman B. Purification and Characterization of Phosphoglycolate Phosphatase from the Cyanobacterium Coccochloris peniocystis. Plant Physiol. 1991;95:693–698. doi: 10.1104/pp.95.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharkey TD. Estimating the Rate of Photorespiration in Leaves. Physiol Plant. 1988;73:147–152. [Google Scholar]
- 7.Sage RF, Sage TL, Kocacinar F. Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Biol. 2012;63:19–47. doi: 10.1146/annurev-arplant-042811-105511. [DOI] [PubMed] [Google Scholar]
- 8.Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR. The Costs of Photorespiration to Food Production Now and in the Future. Annu Rev Plant Biol. 2016;67:107–129. doi: 10.1146/annurev-arplant-043015-111709. [DOI] [PubMed] [Google Scholar]
- 9.Zhu XG, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
- 10.Savir Y, Noor E, Milo R, Tlusty T. Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc Natl Acad Sci U S A. 2010;107:3475–3480. doi: 10.1073/pnas.0911663107. [• This study analyses and underscores the apparent trade-off between reaction velocity and substrate specificity in extant RubisCOs from various organisms.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih PM, Occhialini A, Cameron JC, Andralojc PJ, Parry MA, Kerfeld CA. Biochemical characterization of predicted Precambrian RuBisCO. Nat Commun. 2016;7:10382. doi: 10.1038/ncomms10382. [• Here the authors infer the primary structures of the most recent common ancestors of RubisCO Form IA & B. The biochemical characterization of these ancestral RubisCOs revealed intermediary substrate specificities as well as reaction velocities. Interestingly, the enzymes could readily be encapsulated into carboxysomes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studer RA, Christin PA, Williams MA, Orengo CA. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proc Natl Acad Sci U S A. 2014;111:2223–2228. doi: 10.1073/pnas.1310811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tawfik DS. Accuracy-rate tradeoffs: how do enzymes meet demands of selectivity and catalytic efficiency? Curr Opin Chem Biol. 2014;21:73–80. doi: 10.1016/j.cbpa.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Hanson TE, Tabita FR. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc Natl Acad Sci U S A. 2001;98:4397–4402. doi: 10.1073/pnas.081610398. [• The first description and definition of RubisCO-like proteins] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashida H, Saito Y, Kojima C, Kobayashi K, Ogasawara N, Yokota A. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science. 2003;302:286–290. doi: 10.1126/science.1086997. [• The first biochemical and physiological characterization of a RubisCO-like protein.] [DOI] [PubMed] [Google Scholar]
- 16.Erb TJ, Evans BS, Cho K, Warlick BP, Sriram J, Wood BM, Imker HJ, Sweedler JV, Tabita FR, Gerlt JA. A RubisCO-like protein links SAM metabolism with isoprenoid biosynthesis. Nat Chem Biol. 2012;8:926–932. doi: 10.1038/nchembio.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Carter MS, Vetting MW, San Francisco B, Zhao S, Al-Obaidi NF, Solbiati JO, Thiaville JJ, de Crecy-Lagard V, Jacobson MP, et al. Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars. Proc Natl Acad Sci U S A. 2016;113:E4161–4169. doi: 10.1073/pnas.1605546113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71:576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc Lond B Biol Sci. 2008;363:2629–2640. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.San Francisco B, Gerlt JA. In: Beach Pete., St, editor. 25 the Enzyme Mechanisms Conference; 2017. Jan, [Google Scholar]
- 21.Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 2008;59:1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 22.Ashida H, Danchin A, Yokota A. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res Microbiol. 2005;156:611–618. doi: 10.1016/j.resmic.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Ashida H, Saito Y, Nakano T, Tandeau de Marsac N, Sekowska A, Danchin A, Yokota A. RuBisCO-like proteins as the enolase enzyme in the methionine salvage pathway: functional and evolutionary relationships between RuBisCO-like proteins and photosynthetic RuBisCO. J Exp Bot. 2008;59:1543–1554. doi: 10.1093/jxb/ern104. [DOI] [PubMed] [Google Scholar]
- 24.Imker HJ, Fedorov AA, Fedorov EV, Almo SC, Gerlt JA. Mechanistic diversity in the RuBisCO superfamily: the “enolase” in the methionine salvage pathway in Geobacillus kaustophilus . Biochemistry. 2007;46:4077–4089. doi: 10.1021/bi7000483. [DOI] [PubMed] [Google Scholar]
- 25.Imker HJ, Singh J, Warlick BP, Tabita FR, Gerlt JA. Mechanistic diversity in the RuBisCO superfamily: a novel isomerization reaction catalyzed by the RuBisCO-like protein from Rhodospirillum rubrum . Biochemistry. 2008;47:11171–11173. doi: 10.1021/bi801685f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schada von Borzyskowski L, Rosenthal RG, Erb TJ. Evolutionary history and biotechnological future of carboxylases. J Biotechnol. 2013;168:243–251. doi: 10.1016/j.jbiotec.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Hocker B, Jurgens C, Wilmanns M, Sterner R. Stability, catalytic versatility and evolution of the (beta alpha)(8)-barrel fold. Curr Opin Biotechnol. 2001;12:376–381. doi: 10.1016/s0958-1669(00)00230-5. [DOI] [PubMed] [Google Scholar]
- 28.Farias-Rico JA, Schmidt S, Hocker B. Evolutionary relationship of two ancient protein superfolds. Nat Chem Biol. 2014;10:710–715. doi: 10.1038/nchembio.1579. [DOI] [PubMed] [Google Scholar]
- 29.Sperl JM, Rohweder B, Rajendran C, Sterner R. Establishing catalytic activity on an artificial (betaalpha)8-barrel protein designed from identical half-barrels. FEBS Lett. 2013;587:2798–2805. doi: 10.1016/j.febslet.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 30.Schönheit P, Buckel W, Martin WF. On the Origin of Heterotrophy. Trends Microbiol. 2016;24:12–25. doi: 10.1016/j.tim.2015.10.003. [• This theroretical paper hyposizes that amino acid and purine fermentations may have been the basis of ancient heterotrophy and that ribose likely was (and still is) the first abundant sugar source in the biosphere. Archaeal purine fermentation is linked to the emergence of Form III RubisCO and the nucleoside monophosphate pathway.] [DOI] [PubMed] [Google Scholar]
- 31.Aono R, Sato T, Imanaka T, Atomi H. A pentose bisphosphate pathway for nucleoside degradation in Archaea. Nat Chem Biol. 2015;11:355–360. doi: 10.1038/nchembio.1786. [•• This work elucidated the complete metabolic network that serves in nucleoside degradation in Thermococcus kokaradensis to bring the function of archaeal type III RubisCOs and nucleoside metabolism together.] [DOI] [PubMed] [Google Scholar]
- 32.Aono R, Sato T, Yano A, Yoshida S, Nishitani Y, Miki K, Imanaka T, Atomi H. Enzymatic characterization of AMP phosphorylase and ribose-1,5-bisphosphate isomerase functioning in an archaeal AMP metabolic pathway. J Bacteriol. 2012;194:6847–6855. doi: 10.1128/JB.01335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T, Atomi H, Imanaka T. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science. 2007;315:1003–1006. doi: 10.1126/science.1135999. [DOI] [PubMed] [Google Scholar]
- 34.Huber C, Wachtershauser G. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science. 1997;276:245–247. doi: 10.1126/science.276.5310.245. [DOI] [PubMed] [Google Scholar]
- 35.Robertson MP, Joyce GF. The Origins of the RNA World. Cold Spring Harbor Perspectives in Biology. 2012:4. doi: 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrighton KC, Castelle CJ, Varaljay VA, Satagopan S, Brown CT, Wilkins MJ, Thomas BC, Sharon I, Williams KH, Tabita FR, et al. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 2016;10:2702–2714. doi: 10.1038/ismej.2016.53. [•• A multidisciplinary approach of different labs and experts that describes the functional characterization of a novel class of RubisCO that are found in the recently discovered deep-branching phylum of CPR bacteria. This work suggests that these novel RubisCOs operate in nucleoside assimilation, which provides a new perspective onto the evolutionary history of RubisCO.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laguna R, Tabita FR, Alber BE. Acetate-dependent photoheterotrophic growth and the differential requirement for the Calvin-Benson-Bassham reductive pentose phosphate cycle in Rhodobacter sphaeroides and Rhodopseudomonas palustris . Arch Microbiol. 2011;193:151–154. doi: 10.1007/s00203-010-0652-y. [DOI] [PubMed] [Google Scholar]
- 38.Selesi D, Schmid M, Hartmann A. Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl Environ Microbiol. 2005;71:175–184. doi: 10.1128/AEM.71.1.175-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antonovsky N, Gleizer S, Noor E, Zohar Y, Herz E, Barenholz U, Zelcbuch L, Amram S, Wides A, Tepper N, et al. Sugar Synthesis from CO2 in Escherichia coli . Cell. 2016;166:115–125. doi: 10.1016/j.cell.2016.05.064. [•• This study demonstrates the possibility to introduce a Calvin cycle into Escherichia coli to create a hemi-autotrophic strain that synthesizes all sugars from CO2. This was achieved by use of specifically designed selection strains and fine tuned selection pressures in an laboratory evolution approach.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Lun M, Hub JS, van der Spoel D, Andersson I. CO2 and O2 distribution in Rubisco suggests the small subunit functions as a CO2 reservoir. J Am Chem Soc. 2014;136:3165–3171. doi: 10.1021/ja411579b. [DOI] [PubMed] [Google Scholar]
- 41.Gunn LH, Valegard K, Andersson I. A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic. J Biol Chem. 2017;292:6838–6850. doi: 10.1074/jbc.M116.767145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witte BH. Ph.D. Thesis. The Ohio State University; Columbus: 2012. Taming the Wild RubisCO: Explorations in Functional Metagenomics. [Google Scholar]
- 43.Kerfeld CA, Melnicki MR. Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol. 2016;31:66–75. doi: 10.1016/j.pbi.2016.03.009. [• A recent review on structural and functional properties of cyanobacterial carboxysomes] [DOI] [PubMed] [Google Scholar]
- 44.Mackinder LC, Meyer MT, Mettler-Altmann T, Chen VK, Mitchell MC, Caspari O, Freeman Rosenzweig ES, Pallesen L, Reeves G, Itakura A, et al. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc Natl Acad Sci U S A. 2016;113:5958–5963. doi: 10.1073/pnas.1522866113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhat JY, Thieulin-Pardo G, Hartl FU, Hayer-Hartl M. Rubisco Activases: AAA+ Chaperones Adapted to Enzyme Repair. Front Mol Biosci. 2017;4:20. doi: 10.3389/fmolb.2017.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueller-Cajar O. The Diverse AAA+ Machines that Repair Inhibited Rubisco Active Sites. Front Mol Biosci. 2017;4:31. doi: 10.3389/fmolb.2017.00031. [• Another review on RubisCO activases highlighting their evolution and the conditions under which they may be required. The author also points out the impact of the activases on bioengineering approaches.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature. 2011;479:194–199. doi: 10.1038/nature10568. [DOI] [PubMed] [Google Scholar]
- 48.Tsai YC, Lapina MC, Bhushan S, Mueller-Cajar O. Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat Commun. 2015;6:8883. doi: 10.1038/ncomms9883. [• The manuscript desribes a third type of RubisCO activase that evovled indepently from the known CbbX and AAA+ type activases. This work shows that the evolution of an activating protein is a conserved pattern in the evolution of modern day RubisCO and the ‘RubisCOsome’.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loganathan N, Tsai YC, Mueller-Cajar O. Characterization of the heterooligomeric red-type rubisco activase from red algae. Proc Natl Acad Sci U S A. 2016;113:14019–14024. doi: 10.1073/pnas.1610758113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salvucci ME, Werneke JM, Ogren WL, Portis AR. Purification and species distribution of rubisco activase. Plant Physiol. 1987;84:930–936. doi: 10.1104/pp.84.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 52.Roberts EW, Cai F, Kerfeld CA, Cannon GC, Heinhorst S. Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J Bacteriol. 2012;194:787–795. doi: 10.1128/JB.06444-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. Cyanobacterial-based approaches to improving photosynthesis in plants. J Exp Bot. 2013;64:787–798. doi: 10.1093/jxb/ers294. [DOI] [PubMed] [Google Scholar]
- 54.Mueller-Cajar O, Whitney SM. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem J. 2008;414:205–214. doi: 10.1042/BJ20080668. [DOI] [PubMed] [Google Scholar]
- 55.Shih PM, Zarzycki J, Niyogi KK, Kerfeld CA. Introduction of a synthetic CO2-fixing photorespiratory bypass into a cyanobacterium. J Biol Chem. 2014;289:9493–9500. doi: 10.1074/jbc.C113.543132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bar-Even A, Noor E, Lewis NE, Milo R. Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci U S A. 2010;107:8889–8894. doi: 10.1073/pnas.0907176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwander T, Schada von Borzyskowski L, Burgener S, Cortina NS, Erb TJ. A synthetic pathway for the fixation of carbon dioxide in vitro . Science. 2016;354:900–904. doi: 10.1126/science.aah5237. [•• This paper describes the design and realization of a RubisCO-independent, artificial CO2-fixation cycle in a test tube. The so-called CETCH cycle was built with 17 different enzymes from nine different organisms and optimized in several round using enzyme engineering and metabolite proof reading.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erb TJ, Berg IA, Brecht V, Muller M, Fuchs G, Alber BE. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci U S A. 2007;104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peter DM, Schada von Borzyskowski L, Kiefer P, Christen P, Vorholt JA, Erb TJ. Screening and Engineering the Synthetic Potential of Carboxylating Reductases from Central Metabolism and Polyketide Biosynthesis. Angew Chem Int Ed Engl. 2015;54:13457–13461. doi: 10.1002/anie.201505282. [• This paper reports on the abundance and substrate specificity of reductive enoyl-CoA carboxylases/reductases, which are catalytically superior to RubisCO, as well as the rational mutagenesis, to create new CO2-fixation reactions that can be used in synthetic CO2-fixation patwhays and biotechnology.] [DOI] [PubMed] [Google Scholar]
- 60.Rosenthal RG, Ebert MO, Kiefer P, Peter DM, Vorholt JA, Erb TJ. Direct evidence for a covalent ene adduct intermediate in NAD(P)H-dependent enzymes. Nat Chem Biol. 2014;10:50–55. doi: 10.1038/nchembio.1385. [DOI] [PubMed] [Google Scholar]
- 61.Rosenthal RG, Vogeli B, Wagner T, Shima S, Erb TJ. A conserved threonine prevents self-intoxication of enoyl-thioester reductases. Nat Chem Biol. 2017;13:745–749. doi: 10.1038/nchembio.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]