Abstract
Rituximab (RTX), an antibody targeting CD20, is widely used as a first-line therapeutic strategy in B cell–mediated autoimmune diseases. However, a large proportion of patients either do not respond to the treatment or relapse during B cell reconstitution. Here, we characterize the cellular basis responsible for disease relapse in secondary lymphoid organs in humans, taking advantage of the opportunity offered by therapeutic splenectomy in patients with relapsing immune thrombocytopenia. By analyzing the B and plasma cell immunoglobulin gene repertoire at bulk and antigen-specific single-cell level, we demonstrate that relapses are associated with two responses coexisting in germinal centers and involving preexisting mutated memory B cells that survived RTX treatment and naive B cells generated upon reconstitution of the B cell compartment. To identify distinctive characteristics of the memory B cells that escaped RTX-mediated depletion, we analyzed RTX refractory patients who did not respond to treatment at the time of B cell depletion. We identified, by single-cell RNA sequencing (scRNA-seq) analysis, a population of quiescent splenic memory B cells that present a unique, yet reversible, RTX-shaped phenotype characterized by down-modulation of B cell–specific factors and expression of prosurvival genes. Our results clearly demonstrate that these RTX-resistant autoreactive memory B cells reactivate as RTX is cleared and give rise to plasma cells and further germinal center reactions. Their continued surface expression of CD19 makes them efficient targets for current anti-CD19 therapies. This study thus identifies a pathogenic contributor to autoimmune diseases that can be targeted by available therapeutic agents.
Introduction
B cell depletion with the anti-CD20 chimeric antibody rituximab (RTX) has constituted a major step forward in the treatment of antibody-mediated autoimmune diseases (1, 2). However, a large proportion of these patients eventually relapse during B cell reconstitution (3, 4) as RTX is cleared from the system, starting about 6 months after the last infusion (5, 6). The basic mechanisms underlying this clinical observation are not well understood.
Primary immune thrombocytopenia (ITP) is a prototypic B cell– mediated autoimmune disease in which pathogenic antibodies directed against the platelet membrane glycoprotein IIb-IIIa (GPIIbIIIa) lead to platelet destruction by macrophages in the spleen (7). In ITP, RTX-mediated B cell depletion leads to an immediate clinical response in 50% of patients, but 80% of patients will subsequently relapse in the following months (4, 8). Patients failing to respond or relapsing after RTX treatment are splenectomized when alternative therapies are inefficient or unavailable (9). Therapeutic splenectomy, resulting in a durable platelet response in 60 to 70% of patients with ITP, offers a unique access to study the autoimmune response in the spleen.
The spleen plays a central role in ITP pathogenesis. During the disease, it is the site of an intense autoimmune response with germinal center (GC) expansion and generation of mutated, high-affinity anti-GPIIbIIIa antibody-secreting plasma cells (PC), which play a central role in ITP (10, 11). These GC also produce long-lived autoreactive memory B cells, but their contribution to disease pathogenesis remains elusive, as is the case for many antibody-mediated autoimmune diseases (12). Although PC do not express CD20, RTX affects their generation through depletion of their precursors, including activated and GC B cells (13). Complete B cell depletion is observed in peripheral blood in most patients receiving RTX, but we and others have found evidence for a residual CD19+ B cell population in the spleen of patients with primary failure (that is, during B cell depletion), mainly constituted of nonproliferating PC and memory B cells (10, 14), some of which were autoreactive. We furthermore described that B cell depletion favors the settlement of autoreactive long-lived PC in the spleen and proposed a combined therapy affecting PC survival factors to decrease such cases of primary RTX failure (10, 15).
In contrast, antiplatelet antibodies are scarcely detectable in patients achieving a complete response after RTX (16), suggesting that anti-GPIIbIIIa splenic long-lived PC do not emerge in this situation. A proportion of these patients with fully restored platelet counts eventually relapse at the time of reemergence of B cells (3) after RTX clearance [usually 6 months after the last infusion (5, 6)]. This synchronization between relapse and B cell reconstitution likely results from one of two scenarios that are not mutually exclusive: a de novo breakdown of tolerance involving the recruitment of newly generated naive B cells in the autoimmune response and the reactivation of residual autoreactive memory B cells that survived RTX. Several previous studies have correlated the reemergence of memory B cells in peripheral blood of patients with disease activity in various autoimmune diseases and suggested their role in relapses (17–21). This latter hypothesis nevertheless implies, in the case of ITP, that RTX-resistant memory B cells remain quiescent for months in the splenic environment, despite the constant presence of platelet antigens.
Here, we sought to unravel the cellular mechanisms underlying disease relapse after RTX treatment. To this end, we took advantage of spleen samples obtained from therapeutically splenectomized patients with ITP at various stages after RTX treatment and characterized the onset of disease relapses occurring after an initial complete response to RTX by multiple approaches. We showed that GC giving rise to autoreactive PC are fueled by both anti–GPIIbIIIa-specific RTX-resistant memory B cells and newly generated B cells. To better characterize the phenotype of the memory B cells escaping B cell depletion at early time points after RTX, we studied patients splenectomized for primary RTX failure. This allowed us to identify, at the time of B cell depletion, a memory subset that resisted RTX through the acquisition of a unique phenotype, allowing them to remain quiescent and to contribute to relapses when RTX wanes out. The specific phenotype of this population, targetable by anti-CD19 therapies, provides a therapeutic opportunity in B cell–mediated autoimmune diseases.
Results
Spleens of patients who relapsed after RTX treatment reveal synchronous B cell reconstitution and GC-derived autoimmune response leading to the generation of IgG anti-GPIIbIIIa PC
To decipher the contribution of different B cell subsets in relapses occurring after B cell depletion, we analyzed spleen samples from three types of patients with ITP (table S1): (i) patients who achieved a complete response after a course of RTX and relapsed during B cell reconstitution (RTX relapse), (ii) patients with primary failure of RTX who did not respond to treatment at the time of B cell depletion (RTX failure), and (iii) patients with active ITP that were not treated with RTX (ITP). All were compared when necessary with organ donors with no immune disease who died from stroke or head trauma, hereafter referred to as healthy donors (HD).
We first characterized the presence of the major B cell subpopulations in spleens of patients who relapsed 8.5 months in median (range, 5 to 13) after the last RTX infusion (Fig. 1A). B cell reconstitution had started in all patients, albeit CD19+ cells were present in lower proportions as compared to HD (P = 0.015; Fig. 1B). CD19+ cells were mainly naive B cells (Fig. 1C), with an increased proportion of transitional B cells as compared to HD controls (P = 0.001; Fig. 1D). By contrast, resting CD27+IgD− memory B cells (P < 0.0001; Fig. 1E) and CD27+IgD+ marginal zone B cells (P = 0.007; Fig. 1F) were still durably reduced as compared to HD and patients with ITP. Double-negative CD27−IgD− cells (Fig. 1G) appeared unchanged. This indicated that, although a newly generated wave of transitional and naive B cells had started to repopulate the spleen, the reconstitution of the memory B cell pool had barely begun. Nevertheless, a proportion of memory B cells from RTX relapse patients expressed the Ki67 cell cycle marker, suggesting ongoing or at least recent proliferation events (fig. S1A).
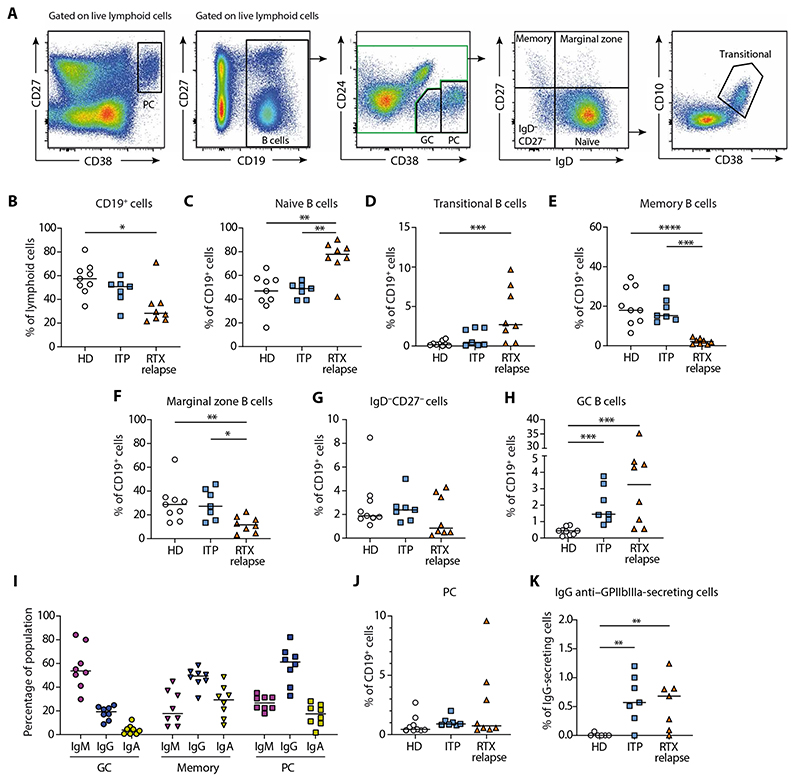
Fig. 1. A GC-derived autoimmune response leads to the generation of IgG anti-GPIIbIIIa plasma cells in the spleen of RTX relapse patients.
(A) Representative dot plots showing gating strategy for flow cytometry analysis of splenic mononuclear cells labeled with antibodies to CD19, CD24, CD27, CD38, and IgD. Plasma cells (PC) were identified as CD27hiCD38hi cells among live lymphoid cells. After gating on CD19+ cells among live lymphoid cells, germinal center (GC) B cells were identified as CD24−CD38int. After exclusion of GC and PC (green gate), B cells were further subdivided into CD27+IgD− resting memory (memory) B cells, CD27−IgD− double-negative B cells, CD27+IgD+ marginal zone B cells, and CD27−IgD+ naive B cells. Transitional B cells were further identified from naive B cells as CD38hiCD10+. (B to H) Proportion of total splenic B cells (B), naive B cells (C), transitional B cells (D), memory B cells (E), marginal zone B cells (F), CD27−IgD− B cells (G), and GC B cells (H), in HD (n = 9), patients with ITP (n = 7), and RTX relapse patients (n = 8). (I) Proportion of IgM, IgG, and IgA expressing splenic GC B cells, memory B cell, and PC assessed by flow cytometry analysis after intracellular staining in RTX relapse patients (n = 8). (J) Proportion of splenic PC and (K) frequency of IgG anti–GPIIbIIIa-secreting cells among IgG-secreting cells assessed by ELISPOT assay in HD (n = 6), patients with ITP (n = 7), and RTX relapse patients (n = 7). Kruskall-Wallis and corrections for multiple comparisons were performed. (***P < 0.001, **P < 0.01, and *P < 0.05). Symbols indicate individual samples; horizontal bars represent median values.
The GC B cell population (identified as CD19+CD24−CD38int) (22, 23) was significantly and largely expanded in RTX relapse patients compared to HD (P < 0.001), suggesting that, as for patients with ITP, these structures were central in the ongoing autoreactive immune response (Fig. 1H). GC B cells were mainly composed of IgM+ cells, IgG+ cells represented a smaller fraction, and the proportion of IgA+ cells was negligible (Fig. 1I). Next, we wondered whether the GC response resulted in the production of autoreactive PC. The proportion of PC was similar between all patients groups (Fig. 1J), including RTX failure patients (fig. S1B), suggesting no expansion or accumulation of this nonproliferating population over time (fig. S1C). Despite this, we observed about 1% of anti-GpIIbIIIa immunoglobulin G (IgG)–secreting cells in RTX relapse patients, a proportion similar to those observed in patients with ITP (Fig. 1K). Using histological analysis, we confirmed the presence of numerous CD20+ GC structures within secondary follicles, PD1+ T follicular helper cells and CD21+ follicular dendritic cells (fig. S2).
Overall, we found that B cell reconstitution in the spleen of RTX relapse patients was characterized by a high proportion of newly generated naive and transitional B cells, whereas the memory B cell pool remained severely reduced, similarly to previous findings in peripheral blood (21). However, we found evidences for an ongoing autoimmune response in the spleen involving GC structures and leading to the generation of IgG anti-GPIIbIIIa PC.
Analysis of splenic B cell IgH repertoires reveals the coexistence of RTX-resistant and newly generated B cells in relapsed patients
Cell proliferation in GC gives rise to clonally related memory B cells and PC. We analyzed the splenic B cell repertoire of three RTX relapse patients and sorted PC (CD19+ CD27hi CD38hi), GC B cells (CD19+ CD24− CD38int IgD− CD20+), memory B cells (CD19+ CD24+ CD27+ CD38− IgD−), and naive B cells (CD19+ CD27− CD38low IgD+) to perform high-throughput sequencing of Ig heavy-chain mRNA sequences (table S2). As expected, variable region of Ig heavy chain (VH) and heavy-chain joining region (JH) usage and CDR3 length showed some differences between naive and antigen-experienced B cells (fig. S3, A to C). Clones of variable sizes were found in GC, memory B cells, and PC populations (fig. S3D). Because IgA expression was minor among GC B cells, we focused our analysis on IgM and IgG sequences.
Unexpectedly, analysis of mutations in VH genes revealed a bi-modal distribution. A large fraction of sequences had acquired relatively few mutations (Fig. 2A), consistent with the presence of newly activated B cells joining the GC, whereas another peak of highly mutated GC sequences was simultaneously observed (Fig. 2A). Analysis of VH mutation in IgM and IgG sequences confirmed that unmutated or lowly mutated sequences were mostly IgM, whereas there was a predominance of IgG in highly mutated sequences (fig. S4A). This bimodal distribution was also observed in memory and PC populations (Fig. 2, B and C, and fig. S4, B and C), albeit with a stronger bias in highly mutated sequences. By contrast, analysis of VH segment mutations in GC and memory B cells from HD and ITP showed a Gaussian distribution in IgM and IgG sequences (fig. S5, A to D). Because it is unlikely that B cells and PC accumulated up to 30 to 40 mutations during the few weeks after B cell reemergence and relapse (24), this suggested that lowly mutated sequences originated from newly generated B cells, whereas highly mutated sequences were from memory B cells that escaped RTX depletion.
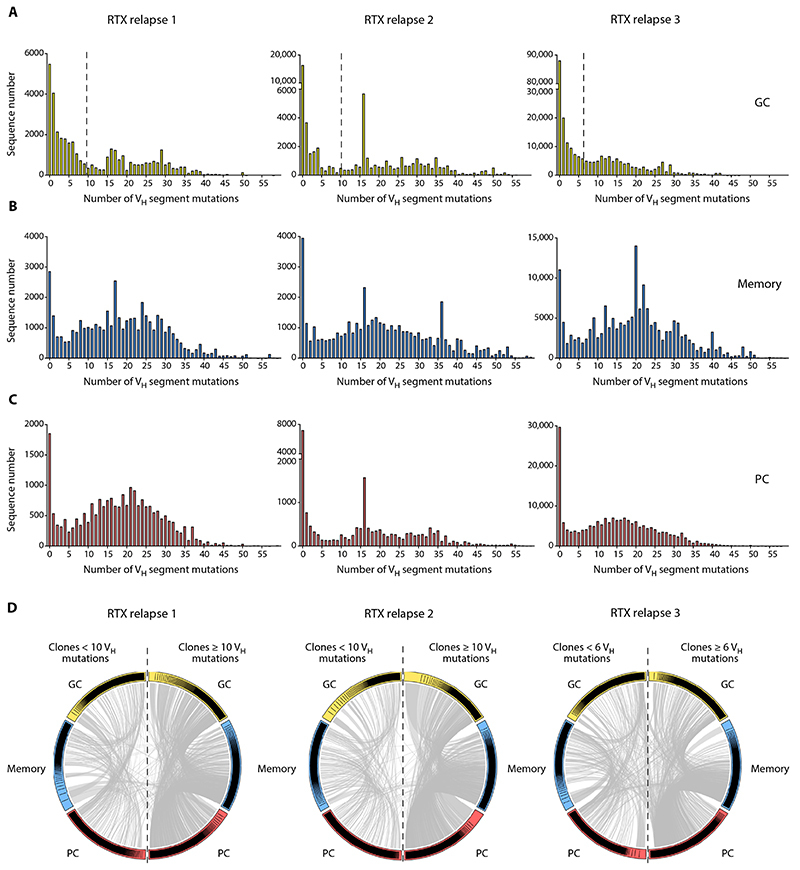
Fig. 2. RTX-resistant and newly generated B cells coexist in the spleen of RTX relapse patients.
(A to C) VH segment mutation distribution in IgM and IgG sequences from (A) splenic GC (yellow), (B) memory (blue), and (C) PC (red) populations from three RTX relapse patients was assessed by high-throughput IgH sequencing. All subpopulations displayed a bimodal distribution with two peaks corresponding to unmutated or lowly mutated cells and highly mutated cells. Dotted vertical lines indicate the threshold used in (D) to discriminate lowly mutated clones and highly mutated clones. (D) Circos plot showing clonal relationships shared between IgM and IgG sequences from GC, memory, and PC splenic populations. According to the mutation distribution in GC shown in (A), clones from each population were classified into “lowly mutated” (left side of the plot) and “highly mutated” (right side of the plot) based on the clone median mutation number. Each colored sector represents one subpopulation and is divided into segments representing individual clones in rank-sized order. The gray internal connections show clones common to multiple subpopulations.
To confirm that lowly mutated and highly mutated sequences indeed originated from distinct populations, we categorized clones in either lowly mutated or highly mutated subsets according to a cutoff value selected from the mutation distribution in GC (≥10 or <10 mutations for patients 1 and 2 and ≥6 or <6 for patient 3). This dichotomy was biologically relevant because mutation distribution inside each clone showed limited dispersion. The SD in mutation numbers for the first 100 clones is shown in table S3.
To address the participation of clones bearing lowly and highly mutated sequences in the B cell response, we then analyzed clonal relationships shared between GC, memory B cells, and PC clones in both populations (Fig. 2D and fig. S6). Numerous clones were shared between GC, memory B cells, and PC subsets in either lowly mutated or highly mutated populations. Among the total number of clonal relationships, only 5.7, 3.5, and 11.5% were shared between lowly mutated and highly mutated populations for patients 1, 2, and 3, respectively. Most of these clones had a median mutation number close to the selected cutoff value, thus accounting for their presence in both mutation categories. Collectively, these results confirmed that lowly mutated and highly mutated clones correspond to distinct populations and suggested that both newly generated naive B cells and RTX-resistant memory B cells were recruited in GC reactions or gave rise to PC in RTX relapse patients.
GPIIbIIIa-specific B cells are found in both RTX-resistant and newly generated B cell populations
IgH repertoire analysis suggests that both RTX-resistant and newly generated B cells participate in new GC reactions in the spleens of RTX relapse patients. To confirm that such GC represent sites of active autoimmune responses, we analyzed whether they contain autoreactive cells. For this, we used a single-cell–based assay adapted from the Nojima culture system (25, 26): Single cells were isolated by sorting from memory and GC B cell subsets, cultured for 15 days, and those harboring detectable IgG secretion were tested by enzyme-linked immune absorbent spot (ELISPOT) to identify GPIIbIIIa-specific clones (Fig. 3A and fig. S7, A to D). To confirm the robustness of the ELISPOT assay, we cloned IgH and IgL genes from two GPIIbIIIa ELISPOT-positive memory B cells and reexpressed them in human embryonic kidney (HEK) cells. Supernatants from both clones reacted with GPIIbIIIa in enzyme-linked immunosorbent assay (ELISA) (fig. S7E) and monoclonal antibody immobilization of platelet antigens (MAIPA) assay (fig. S7F), a standard clinical assay for ITP.
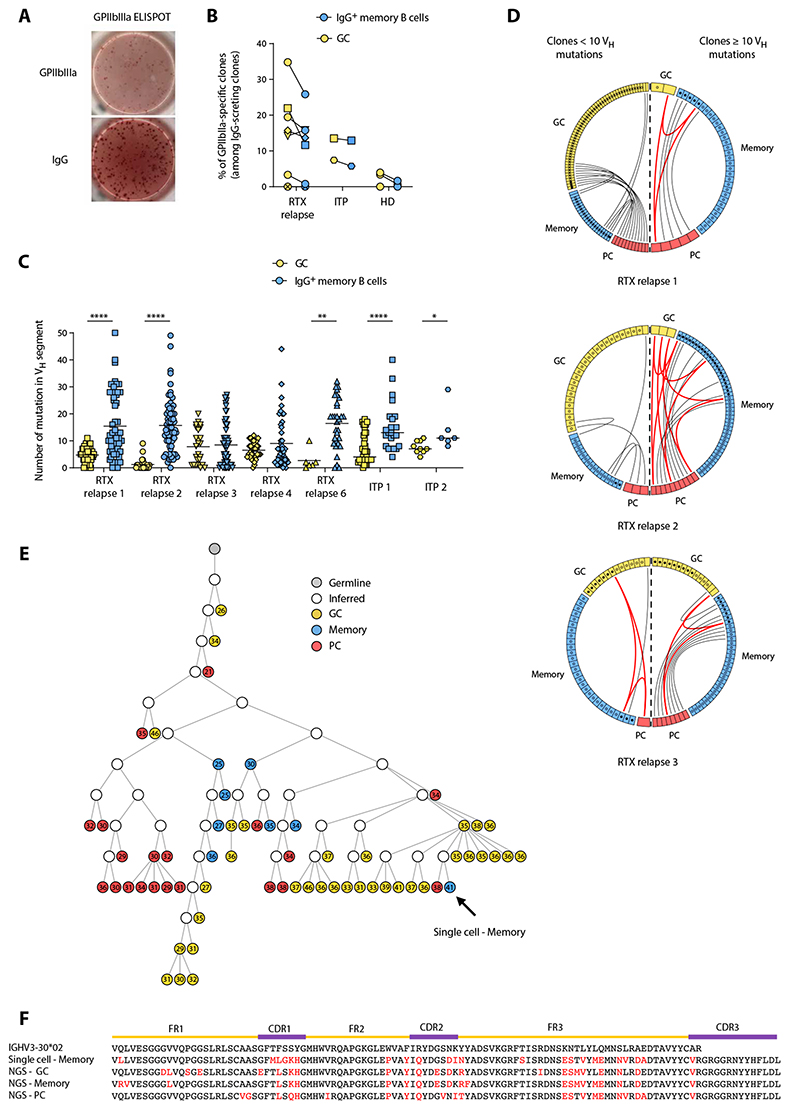
Fig. 3. GPIIbIIIa-specific B cells are found in both RTX-resistant and newly generated B cell populations.
(A) Representative GPIIbIIIa and IgG ELISPOT from a GPIIbIIa-specific clone obtained after single-cell culture of sorted memory B cell from a patient with relapse after RTX. (B) Frequency of GPIIbIIIa-specific clones among GC (yellow) and IgG memory B cell (blue) populations in RTX relapse patients, patients with ITP, and HD. The total number of tested cells is provided in table S2. RTX relapse patient 8 was not tested because of low culture yield. (C) Number of mutations in the VH segment for individual GPIIbIIIa-specific GC (yellow) and IgG memory (blue) B cells from RTX relapse patients #1, #2, #3, #4, and #6 and from two patients with ITP. Two-tailed Mann-Whitney tests (****P < 0.0001, **P < 0.01, and *P < 0.05). Symbols indicate individual cells; horizontal bars represent mean values. (D) Circos plot showing clonal relationships between individual sequences from GPIIbIIIa-specific GC or IgG+ memory B cells and sequences obtained from high-throughput IgH analysis of RTX relapse patients #1, #2, and #3. Each colored sector represents one subpopulation and is divided into segments representing individual clones. Each circle represents a sequence originating from GPIIbIIIa B cells and is filled with black if the sequence was found in high-throughput IgH sequencing data from the same subpopulation. The internal connections show clones shared by two or three subpopulations, depicted in gray or red, respectively. According to the mutation distribution in GC shown in Fig. 2, clones from each population were classified into lowly mutated (left side of the plot) and highly mutated (right side of the plot) based on the clone mutation number (median clone mutation number was used for clones from high-throughput IgH sequencing data). Only clonal relationships shared between individual sequences and high-throughput IgH analysis data are shown. (E) Phylogenetic analysis of a representative clone from the high-throughput IgH sequencing data that includes one of the anti-GPIIbIIIa memory B cell isolated from RTX relapse patient #1. Germline sequence is represented in gray, GC in yellow, memory B cells in blue, and PC in red, and open circles represent inferred precursors. The number of mutations in VH segment is indicated in each circle. All sequences are of the IgG isotype. The arrow indicates the position in the tree of the anti-GPIIbIIIa memory B cell. (F) Alignment of representative sequences from the clone shown in (E); amino acid changes from the germline are shown in red.
Using this single-cell culture followed by ELIPOST, five of seven RTX relapse patients displayed anti–GPIIbIIIa-specific cells. The proportion of GPIIbIIIa-specific cells ranged from 14 to 35% in GC and from 12 to 26% in IgG+ memory B cells in these patients (Fig. 3B and table S4). We then analyzed the IgH sequences from these single anti–GPIIbIIIa-specific cells. The repertoire of VH genes was diverse, VH3-23 and VH3-30 being the most represented ones (fig. S8). Most GPIIbIIIa-specific GC sequences had few VH segment mutations (Fig. 3C), suggesting that a large fraction of these cells originated from newly generated B cells. A low mutation load was also found in some memory B cell sequences, which likely originated from newly generated B cells in GC. In contrast, in all patients analyzed, the major part of anti-GPIIbIIIa memory B cell sequences contained a high number of mutations, indicating that they originated from RTX-resistant memory B cells. Their overall mutation profile was similar to GPIIbIIIa-specific GC and memory B cells from patients with ITP with ongoing disease, in which chronic reactivation and continuous recruitment of B cells is likely to take place (Fig. 3C).
To confirm that GPIIbIIIa-specific cells identified in our system were implicated in an active immune response, we searched whether they shared clonal relationships with sequences from different effector subsets using our IgH repertoire high-throughput sequencing data (Fig. 3D). We found that some highly mutated sequences from GPIIbIIIa-specific IgG+ memory B cells belonged to clones found in GC or PC populations, suggesting that autoreactive, RTX-resistant memory B cells were able to give rise to autoreactive PC either directly or after reentering GC. In some cases, clonally related sequences could be organized in phylogenetic trees highlighting their hierarchical formation, positioning highly mutated memory B cells both upstream and downstream of GC B cells (Fig. 3, E and F). Nevertheless, clonal relationships shared by GPIIbIIIa-specific highly mutated memory B cells were more frequent with the PC than with the GC B cell subset (77% versus 23% of clonal relationships with PC and GC B cells, respectively). We also found GPIIbIIIa-specific GC clones with few VH mutations shared with memory or PC populations (Fig. 3D), indicating that an ongoing autoimmune response involving newly generated B cells also coexisted in all patients. Overall, these findings revealed that splenic GC from RTX relapse patients display a high frequency of anti-GPIIbIIIa B cells and are composed of a main fraction of newly generated B cells together with highly mutated cells originating from RTX-resistant memory B cells. Both populations gave rise to anti-GPIIbIIIa PC and thus likely contributed to disease relapse.
Patients with ITP treated with RTX harbor a distinct, RTX-resistant memory B cell population
To confirm the existence of RTX-resistant memory B cells in RTX-treated patients with ITP and to perform their characterization, we next analyzed spleen samples from 16 patients who required therapeutic splenectomy between 1 and 6 months after receiving RTX (RTX failure patients) and had not reconstituted their B cell pool yet (table S1), suggesting the continued presence of RTX. In these spleens, CD19+ cells represented a median of 0.5% of lymphoid cells (Fig. 4, A and B), as compared to 50% in patients with ITP and 57% in HD patients (Fig. 1B). As previously reported, PC represented the majority of residual CD19+ cells (median, 73%; range, 39.7 to 97.8%), with memory B cells accounting for most of the remaining cells (25% in median, range of 2.2 to 77.6%) (10). The number of residual memory B cells was highly variable among patients (median, 153 cells per 105 lymphoid cells; range, 2 to 911).
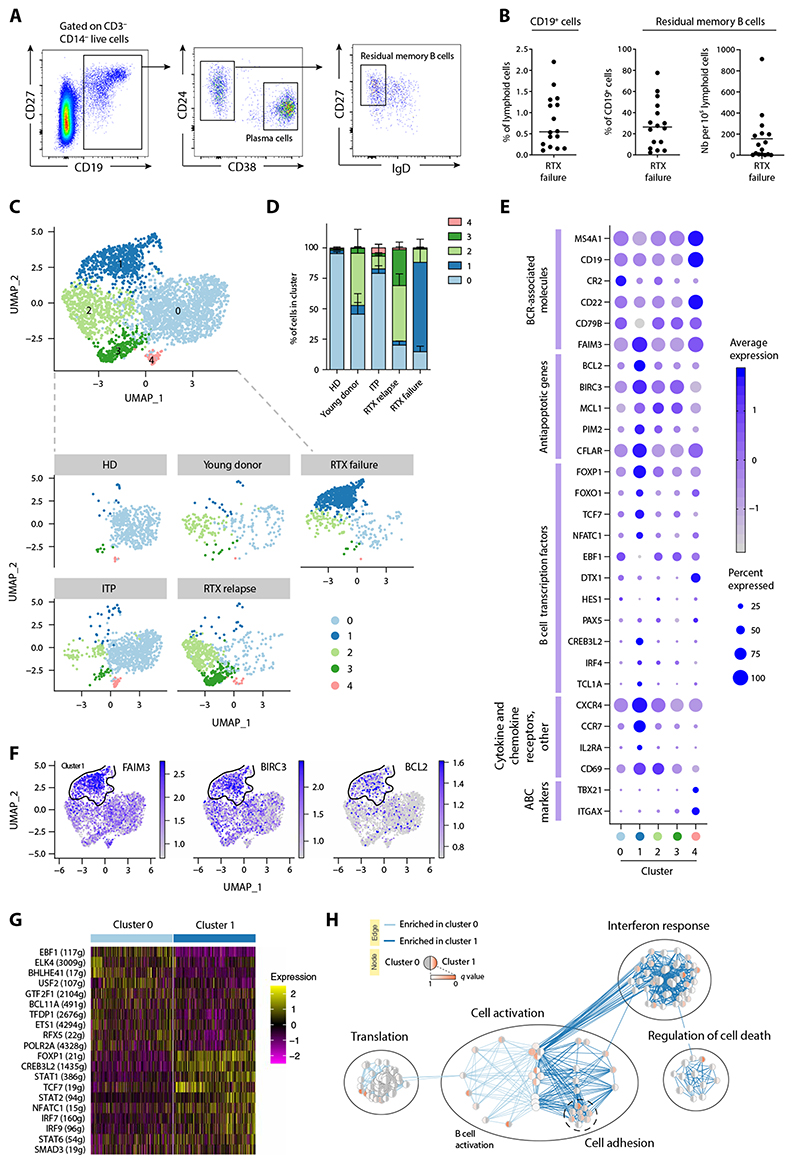
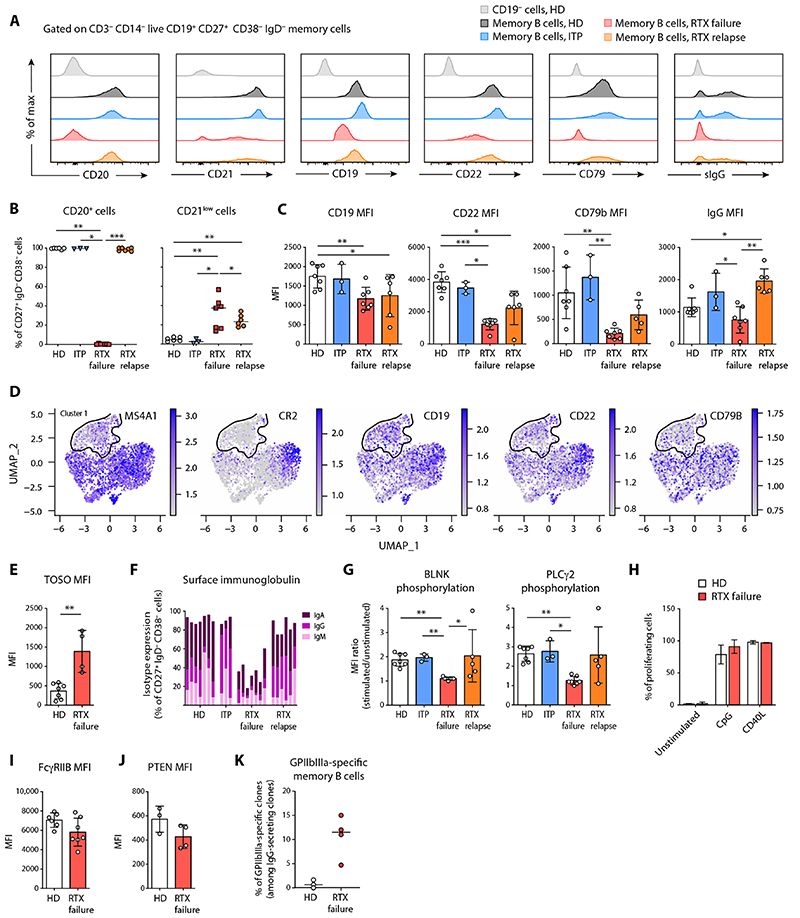
Fig. 4. RTX failure patients harbor a distinct RTX-resistant memory B cell population with a unique transcriptional program.
(A) Representative dot plots showing the gating strategy for flow cytometry analysis of splenic mononuclear cells labeled with antibodies to CD3, CD14, CD19, CD24, CD27, CD38, and IgD. After gating on CD19+ B cells among live CD3−CD14− lymphoid cells, memory B cells were identified as CD24+ CD38−IgD− and PC as CD27+CD24−CD38hi. (B) Proportion of CD19+ B cells (left), residual memory B cells (middle), and number of residual memory B cells (right) in the spleen of RTX failure patients (n = 16). (C to H) Single-cell RNA sequencing was performed after single-cell sorting of memory B cells from healthy donors (HD), young donors, patients with ITP, RTX relapse patients, and RTX failure patients (SORT-seq) and analyzed using the Seurat R pipeline. (C) UMAP and cluster attributions after graph-based clustering (Louvain) of all sorted memory B cells (resolution = 0.6) (top) or memory B cells from each individual donor or patient’s group (bottom). (D) Repartition of single memory B cells in each cluster according to donor type. (E) Dot plots showing a selection of differentially expressed genes between cluster 1 and other clusters. Each dot size is proportional to the number of cells expressing this gene and is colored according to the average expression of that gene in a given cluster. ABC: Atypical B cells (F) Feature plots showing differential gene expression for each cell from all donors for a selection of genes up-regulated in cluster 1. The outline of cluster 1 (highly enriched for RTX-failure memory B cells) is displayed. (G) Heatmap showing the scaled expression (row-normalized area under the curve score) of the top 10 most significantly up-regulated SCENIC regulons in clusters 0 and 1. (H) Pathway enrichment analysis based on gene up- and down-regulated in cluster 1 as compared to cluster 0 resting memory B cells. Each dot represents a unique gene set and is colored on the basis of the P value of its enrichment in cluster 0 (left side) or cluster 1 (right side).
To characterize the phenotype and function of these memory B cells, we next performed single-cell RNA sequencing (scRNA-seq) analysis of splenic memory B cells sorted from three of these RTX failure (RTX failure 1, 2, and 3) as well as RTX relapse patients (RTX relapse 2, 3, and 4), patients with ITP (ITP1, 2, and 3), and HD controls (HD1, 2, and 4). We also included splenic memory B cells from children that underwent splenectomy and are known to have a high frequency of B cell activation secondary to classical childhood infections and vaccinations, reflected by an increased number of GC (27). Unsupervised clustering analysis of all sorted memory B cells revealed that they could be divided in five clusters according to their gene expression profile (Fig. 4, C and D, and table S5). Most memory B cells from RTX failure patients clustered in cluster 1, separately from memory B cells of all other patients and donors. Although cells in this cluster did show expression of all B cell receptor (BCR)–associated genes, confirming their B cell identity, the expression of key members, such as MS4A1 (CD20), CR2 (CD21), or CD79B, appeared reduced as compared to other memory B cell clusters (Fig. 4E). Despite low expression of CD21 and moderate expression of FcRL5, this population clearly differed from atypical memory/ “age-associated B cells” (28, 29), which could be found in cluster 4 and were characterized by high expression of CD19, MS4A1, CD72, ITGAX (CD11c), TBX21, and FcRL5 (Fig. 4, C to E, and fig. S9).
Residual memory B cells from RTX failure patients also clearly differed from memory B cells found in HD as well as both ITP and RTX relapse patients. Memory B cells from HD clustered almost exclusively in cluster 0 (Fig. 4, C and D), along with most memory B cells from patients with ITP and half of memory B cells from young donors. This cluster was characterized by low gene expression without a distinctive transcriptomic signature, and was thus considered as mainly containing quiescent memory B cells (Fig. 4E and fig. S9). Memory B cells from RTX relapse patients segregated in two additional clusters (2 and 3). Cluster 2 was shared with memory B cells from young donors, whereas cluster 3 contained almost exclusively memory B cells from RTX relapse patients.
Overall, gene signatures from clusters 1, 2, and 3 showed very little transcriptional similarity as compared to cluster 0 quiescent memory B cells. We observed a unique down-regulated gene signature specific to cluster 1 that was associated with both translation and the BCR signaling pathways (fig. S10, A and B). Co-clustering with young donor memory B cells, as well as specific up-regulation of pathways linked with response to cytokines or ĴUN/FOS/ATF2 activation (figs. S9 and S10, A and B), suggested that cells from cluster 2 could represent newly generated memory B cells from GC as the result of an active immune response.
Memory B cells from cluster 3 displayed up-regulated pathways linked to the regulation of actin cytoskeleton and chemokine signaling (fig. S10, A and B), with up-regulation of genes implicated in cell migration and calcium signaling (fig. S9). Among up-regulated genes, CD95 and CD29 delineated a particular CD95highCD29high population among CD19+CD27+IgD− memory B cells in RTX relapse patients that was not found in RTX failure or HD and represented less than 1% of memory B cells in patients with ITP (fig. S10C), in line with our scRNA-seq data (Fig. 4D). Single-cell analysis of VH mutations showed that these cells were enriched in highly mutated sequences compared to CD95lowCD29low population, which contained sequences with lower mutation numbers (fig. S10D). These results suggested that cluster 3 memory B cells likely contain recently activated CD95+ effector memory B cells, including some that predate and have resisted RTX treatment. The markedly different transcriptional program of these reactivated, highly mutated memory B cells in relapse patients as compared to RTX-resistant memory B cells found in RTX failure patients further suggests that the unique transcriptional program imposed by RTX is reversible upon RTX clearance from the system.
Residual memory B cells in patients who did not respond to RTX treatment express a distinct transcriptional program promoting survival and dormancy
To better understand how RTX-resistant memory B cells survive despite being kept in constant check by RTX, we next focused our analysis on cells from cluster 1, which contained almost exclusively memory B cells from RTX failure patients. As compared to “quiescent” memory B cells (cluster 0), memory B cells from RTX failure patients differentially regulated more than 400 genes, including several transcription factors involved in B cell differentiation or activation. Despite a similar expression of PAX5, the B cell identity transcription factor EBF1 (30) and Notch pathway–associated genes such as DTX1 and HES1 were notably down-regulated in memory B cells from RTX-resistant patients, concomitant with an up-regulation of TCF7 [encoding TCF-1, which is repressed by EBF-1 (30)] as well as associated genes such as IL2RA (Fig. 4E). CREB3L2, a transcription factor induced during PC differentiation, was overexpressed in cluster 1, consistent with an up-regulation of IRF4 and other activation markers such as CD69 (31). However, classical PC transcription factors, such as PRDM-1 and XBP-1, were not expressed in cluster 1, and negative regulators of effector B cell differentiation such as FOXO1 and FOXP1 appeared up-regulated (Fig. 4E) (32, 33).
Memory B cells from RTX failure patients also up-regulated transcriptional regulator TCL1a (Fig. 4E), which has been shown to induce chronic lymphocytic leukemia (CLL) B cells when overexpressed in mice (34, 35). Memory B cells in cluster 1 additionally overexpressed NFATc1 and antiapoptotic genes such as BCL2, BIRC3, or CFLAR (Fig. 4, E and F). NFATc1, low cell surface BCR expression, and overexpression of antiapoptotic genes have all been involved in the survival and anergic phenotype of CLL B cells (36–38).
Regulatory network inference analysis using the single-cell regulatory network inference and clustering (SCENIC) pipeline (39) further confirmed that FOXP1, TCF7, CREB3L2, and NFATc1 were four major regulons in RTX-resistant memory B cells (Fig. 4G). Pathway enrichment analysis on differentially expressed genes confirmed a clear enrichment of pathways linked to the regulation of apoptotic cell death in cluster 1 (Fig. 4H). Pathways linked to cell activation also showed a mixed enrichment profile, correlating with the intermediate differentiation profile of these cells. A clear type 1 interferon signature could also be detected in both our regulatory network inference (STAT1, STAT2, IRF7, and IRF9) and our pathway enrichment analysis (Fig. 4, G and H). This signature, however, appeared mostly driven by cells from one of the RTX failure patient who developed systemic lupus erythematosus several years after splenectomy. Together, our results suggest that memory B cells from RTX failure patients are in a BCR nonresponsive state with a strong up-regulation of several prosurvival factors that may explain their survival despite months of RTX exposure.
RTX-resistant memory B cells down-regulate BCR complex surface expression and contain autoreactive clones
To further validate the suggested phenotype of residual RTX-resistant memory B cells found in RTX failure patients, we next analyzed surface expression of the BCR-associated complex on these cells in a larger set of RTX failure patients (n = 7), selected for harboring the highest number of detectable residual memory B cells. These RTX-resistant cells had undetectable surface expression of CD20 (Fig. 5, A and B) and had decreased surface expression of many BCR-associated molecules such as CD79b, CD19, CD21, and CD22 (Fig. 5, A to C). scRNA-seq data confirmed a decreased expression of these BCR-associated genes (Figs. 4E and 5D), although the absence of CD20 expression could also be explained by epitope masking by RTX or by CD20 internalization upon RTX binding (40–42). The transcript and surface protein of the IgM Fc receptor FAIM3 (TOSO) was overexpressed in residual memory B cells (Fig. 4, E and F, and Fig. 5E). FAIM3 has been shown to be overexpressed on CLL B cells and is considered as a potential therapeutic target (43). Surface and intracellular Ig expression (including IgM, IgA, and IgG) was strongly reduced, resulting in a large fraction of cells without a detectable isotype (Fig. 5F). To test whether this phenotype was induced by RTX, we cultured fresh spleen tissues from HD with and without RTX (fig. S11, A to I) and observed that nondepleted B cells down-regulated CD20, CD79b, and IgM at their surface. This suggested that RTX induces phenotype modifications rather than selecting preexisting CD20-negative B cells. In line with this, we observed a reexpression of CD20 on memory B cells from RTX failure patients after in vitro culture (fig. S11, J and K).
Fig. 5. RTX-resistant memory B cells have a unique surface phenotype and contain autoreactive clones.
(A) Representative overlays of surface markers assessed by flow cytometry expressed on CD19− cells from HD (light gray), memory B cells from HD (dark gray), memory B cells from patients with ITP (blue), memory B cells from RTX relapse patients (orange), and residual memory B cells from RTX failure patients (red). (B) Proportions of CD20+ cells and CD21low cells and (C) median fluorescence intensities (MFIs) ± SD of CD19, CD22, CD79b, and IgG in memory B cells from HD (n = 6), ITP (n = 3), RTX relapse (n = 7), and RTX failure (n = 7) patients. (D) Feature plots showing differential gene expression for each single cell (see Fig. 4) from all donors for CD20 (MS4A1), CD21 (CR2), CD19, CD22, and CD79b. The outline of cluster 1 (highly enriched for RTX-failure memory B cells) is displayed. (E) MFIs ± SD of TOSO in memory B cells from HD (n = 3) and RTX failure (n = 3) patients. (F) Cumulative proportions of surface IgM, IgG, and IgA in memory B cells from HD, ITP, and RTX failure patients. Each bar represents one patient. (G) BLNK and PLCγ2 phosphorylation assessed by stimulating total splenocytes from HD, ITP, and RTX failure patients with anti-IgG/A/M antibodies. Ratio of MFI ± SD from stimulated over nonstimulated cells are indicated for each group. (H) Splenocytes from HD (n = 3) and RTX failure patients (n = 3) were stimulated by CpG or CD40L and interleukin-21 (IL-21), IL-2, IL-4, and B cell activating factor. Percentage of proliferating memory B cells was assessed after 5 days of culture with cell proliferation dye eFluor 450. (I and J) MFIs ± SD of FCγRIIB (I) and PTEN (J) in memory B cells from HD (n = 3) and RTX failure (n = 3) patients. (K) Frequency of GPIIbIIIa-specific clones among memory B cells in RTX failure patients (red) and HD (green). The total number of tested cells is given in table S2. Kruskall-Wallis with corrections for multiple comparisons or two-tailed Mann-Whitney tests were performed. (***P < 0.001, **P < 0.01, and *P < 0.05). Symbols indicate individual samples; horizontal bars represent median values.
To confirm that these cells could not be stimulated through antigen recognition, we assessed the phosphorylation upon BCR ligation of downstream proteins B cell linker (BLNK) and phospholipase Cγ2 (PLCγ2) involved in BCR signaling. Whereas residual memory B cells had similar basal phosphorylation of both proteins, BCR ligation induced BLNK and PLCγ2 phosphorylation only in memory B cells from controls (P < 0.01; Fig. 5G). In vitro stimulation by CpG and CD40L expressing cells, however, induced proliferation and PC differentiation of these residual memory B cells (Fig. 5H). We assessed FcγRIIB expression (Fig. 5I) that has been shown to restrict atypical memory B cell responses to membrane-associated antigens (44) and found it to be similar to HD memory B cells, in line with our scRNA-seq data confirming that residual memory B cells differed from atypical memory B cells (Fig. 4, C to E). Phosphatase and tensin homolog (PTEN), whose expression maintain anergy in human B cells, was not overexpressed in memory B cells from RTX failure patients, suggesting that these cells were not in an anergic state (Fig. 5J).
The potential to reactivate these cells in vitro using CpG- and CD40L-expressing cells further allowed us to make use of the culture assay described in Fig. 3 to search for anti-GPIIbIIIa clones among RTX-resistant residual memory B cells. This analysis showed that 4.7 to 15% of residual memory B cells from patients who did not respond to RTX treatment reacted against GPIIbIIIa (Fig. 5K), confirming that some autoreactive clones do escape RTX depletion. Overall, these data suggest that, during B cell depletion, a residual population of memory B cells containing autoreactive clones persists. However, these cells cannot be activated upon antigen encounter mainly due to down-regulated expression of surface BCR and associated molecules induced by RTX.
RTX-resistant memory B cells can be depleted in vitro by targeting CD19
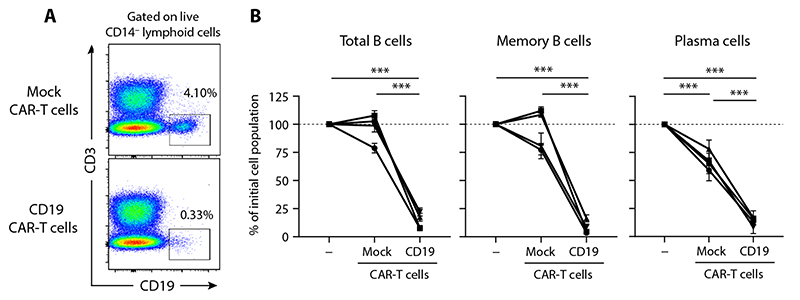
Although its expression was diminished, CD19, unlike CD20, was still present at the surface of RTX-resistant memory B cells (Fig. 5, A to C). We thus asked whether CD19 targeting could efficiently deplete RTX-resistant memory B cells and PC. We incubated in vitro spleno-cytes from RTX failure patients with chimeric antigen receptor (CAR)–T cells transfected either with anti-CD19ScFv-CD28-CD3ζ (anti-CD19 CAR-T cells) or with CD28-CD3ζ without ScFv (mock CAR-T cells). After overnight incubation, there was no reduction of CD19+ B cells cocultured with mock CAR-T cells as compared to splenocytes cultured without CAR-T cells, whereas more than 75% of CD19+ B cells cocultured with anti-CD19 CAR-T cells were depleted (P < 0.001; Fig. 6, A and B). Both CD19+CD38− memory B cells and CD19+CD38+ PC were depleted in this assay (P < 0.001, memory B cells; P < 0.001, PC; Fig. 6B). These results highlight CD19 as a viable option to target RTX-resistant memory B cells.
Fig. 6. RTX-resistant memory B cells can be depleted in vitro by targeting CD19.
(A) Representative flow cytometry analysis of CD3-depleted splenocytes from a RTX failure patient after overnight incubation with either CD19 CAR-T cells or mock CAR-T cells. (B) Percentage of the remaining CD19+ B cells (left), CD19+CD38− memory B cells (middle), and CD19+CD38+ PC (right) after overnight incubation in each condition. n = 4 RTX failure patients analyzed in triplicate, two independent experiments. Data are presented as means ± SD. CD19+ cells without CAR-T cells were used as a reference for calculating percentages. Two-way analysis of variance (ANOVA) tests were performed (***P < 0.001).
Discussion
Splenectomy of patients with ITP relapsing after RTX offers a unique condition to study the precise cause of disease relapse. In a previous study from our group, RTX-resistant PC have been shown to be the main contributors of disease persistence in cases of RTX treatment failure (10). In this study, we focused on a separate clinical situation, where, after an initial complete response to B cell depletion, relapses take place at the time of B cell reemergence. As antiplatelet antibodies are not detected in the serum of patients responding to treatment (16), it is unlikely that RTX-resistant PC make a substantial contribution to relapses. It is more likely that, in patients who relapse after RTX, autoreactive PC arise either from RTX-resistant memory B cells or from a tolerance breakdown event mobilizing naive B cells generated during B cell reconstitution.
In RTX relapse patients, GC were largely expanded, as observed in untreated patients with ITP, suggesting that they were central in disease reinitiation. To better understand the relationships between GC B cells and other B cell populations, we performed high-throughput sequencing of Ig heavy-chain genes of these different subsets. Because mutation accumulation in GC is a time-dependent process (45), we used the number of mutations in VH segment as a surrogate marker to estimate the time elapsed since B cell recruitment in the GC response. Somatic mutations in VH sequences displayed a bimodal distribution in B cells from individuals who relapsed after RTX. In contrast, a Gaussian distribution was observed in HD and in patients with ITP, as expected in an ongoing immune response (46, 47). In the three patients studied, analysis of clonal relationships between GC B cells, memory B cells, and PC showed that lowly mutated and highly mutated cells segregated in mutually exclusive clonal families, therefore confirming that they originated from distinct populations. Repertoire analyses thus revealed the coexistence of two simultaneous responses in the GC of RTX relapse patients, originating from both RTX-resistant and newly generated B cells. Because no RTX-resistant GC could be detected in RTX failure patients, clonal relationships between highly mutated memory B cells, GC B cells, and PC confirmed the contribution of resistant memory B cells to ongoing immune response in patients with ITP.
The active participation of memory B cells to the reemerging autoimmune response was further demonstrated using a single-cell culture assay, allowing us to identify B cell clones reactive against GPIIbIIIa, the main target antigen in ITP, and to determine their Ig sequence (48). Cross-reactive or polyreactive clones were detected at a low frequency in some HD, but there was a clear enrichment in anti–GPIIbIIIa-specific GC and memory B cells in splenic samples from RTX relapse patients. Analysis of VH sequences from anti-GpIIbIIIa clones indicated that a major fraction of anti-GpIIbIIIa GC B cells harbored less than 10 mutations. This reflected the predominance of newly generated B cells in GC, in accordance with the high proportion of B cells expressing an IgM isotype. Highly mutated clones were predominant among memory B cells, indicating that they corresponded to RTX-resistant cells. Such cells were present in five of the five patients studied with detectable anti-GPIIbIIIa B cells, emphasizing the generality of the observation. Clonal relationships between sequences from GPIIbIIIa-specific cells and high-throughput repertoire data in the three patients studied by both approaches indicated that anti-GPIIbIIIa clones were also shared between PC, GC B cell, and memory B cell populations, supporting the notion that RTX-resistant memory B cells directly participated in ITP relapses by joining GC reaction or differentiating into autoreactive PC. Clonal tree reconstruction further confirmed the position of memory B cells as ancestors of either PC or GC B cells. RTX-resistant memory B cells, however, are clearly not the only contributors to these autoimmune GC reactions. We also observed lowly mutated anti-GPIIbIIIa clones shared between GC and PC populations. This observation is consistent with recent work demonstrating that both memory and naive B cells are recruited in GC reaction after influenza virus vaccination in humans (24).
The simultaneous activation of autoreactive B cells from newly arising cells and residual memory B cells raises the question of the respective role and interdependency of both phenomena. Although ranking the absolute contribution of each B cell subpopulation to this process is a difficult task, it is nonetheless notable that we found splenic anti-GPIIbIIIa RTX-resistant memory B cells with clonal relationships with GC or PC in all RTX relapse patients studied, something that one would not expect if their role was fully dispensable. An attractive hypothesis is that responding autoreactive memory B cells may create a favorable microenvironment for a subsequent tolerance breakdown and the recruitment of some platelet-reactive naive B cells to the GC, restarting the autoimmune response (49).
Several studies have documented the presence of residual B cells in secondary lymphoid organs after RTX, with variable proportions of memory B cells reported (10, 14, 50–53). The reasons why some memory B cells resist anti-CD20 remain unclear. To this end, we studied splenectomized patients with ITP with primary RTX failure to decipher the specific features that allow residual splenic memory B cells to persist during B cell depletion. In such patients, residual memory B cells did not express CD20, contrasting with the presence of MS4A1 transcripts. Several mechanisms accounting for the lack of CD20 detection have been described in vitro and in vivo, such as epitope masking by RTX, antigen modulation through CD20 internalization upon RTX binding (40–42), or trogocytosis, mainly in the context of CLL (54–57). RTX can induce a colocalization of CD20 molecules with BCR in lipid rafts on target cells (58). We found a strong reduction of BCR expression, including surface and intracellular Ig and a down-modulation of BCR-associated molecules. Accordingly, residual memory B cells did not respond to BCR stimulation. This phenotype could explain why residual memory B cells remained dormant despite the constant presence of their target antigen.
scRNA-seq analysis revealed that RTX-resistant memory cells displayed a unique transcriptional signature, characterized by the down-modulation of B cell–specific factors and the expression of prosurvival genes. Despite their low or absent CD21 expression, they clearly differed from age-associated B cells described in several infectious or autoimmune diseases (28) by the absence of expression of TBX21 (T-BET) and ITGAX (CD11c). Transcription factors involved in B cell identity such as EBF-1 were down-regulated in RTX-resistant memory cells. This is in line with the up-regulation of TCF7, a transcription factor repressed by EBF-1 whose expression also correlates with the capacity of some exhausted CD8+ T cells to maintain stem-like potential in the presence of chronic stimulation (59). The overexpression of genes involved in B cell activation with repressors of PC differentiation suggests that these memory cells are locked in an intermediate state. In this regard, RTX-resistant memory B cells shared some transcriptional similarities with CLL (such as expression of TCL1a or FAIM3), which are anergic because of constant BCR stimulation. Together, this unique signature with strong expression of antiapoptotic genes, down-regulation of BCR-associated signaling molecules, and intermediate differentiation state highlights the pressure faced by these cells under constant anti-CD20 exposure as well as some of their in vivo survival strategies. This signature was not shared with memory B cells from RTX relapse patients. Accordingly, this dormant or “frozen” state appears to be reversible and shaped by RTX, as suggested by in vitro assays.
Although we cannot exclude the selection of a preexisting CD20− or apoptosis-resistant memory B cell population by RTX, the phenotype and gene expression profile of RTX-resistant memory B cells appears, nevertheless, reversible because no such profile is observed among memory B cells from RTX relapse patients. All memory B cells in disease relapses expressed CD20 as well as classical B cell markers and segregated into two distinct clusters. One corresponded to the major subset observed in young donors, suggesting that they may correspond to newly generated memory B cells. The second one (cluster 3) was only observed in this setting. It displayed several hallmarks of immune activation, as well as surface markers such as FAS and CD29 (β1-integrin) that allowed us to identify them as enriched in highly mutated VH sequences. This strengthens the conclusion that these cells are likely derived from RTX-resistant memory B cells. In addition, up to 40% of memory B cells in ITP relapse patients stain positive for Ki67 (which marks cells in G1 as well) yet did not show a transcriptional signature for active cell division (G2/M or S). This suggests a recent history of division, as one would expect for memory B cells generated in the context of GC or extrafollicular activation. Overall, this suggested that some memory B cells survived and remained quiescent for months during RTX treatment in some patients, constituting a reservoir of pathogenic cells that became reactivated when RTX was cleared.
This study has several limitations. We could not access splenic samples at the time of RTX depletion from patients who initially responded and then relapsed. Therefore, the exact number of residual memory B cells before B cell reconstitution in RTX relapse patients remains unknown. Moreover, although GPIIbIIIa is the main antigen in ITP, other platelet autoantigens such as GPIbIX have been described and were not studied in the present work (48).
From a clinical perspective, targeting residual memory B cells at the time of B cell depletion could represent an attractive therapeutic option to avoid disease relapses. Increased B cell depletion with the use of next-generation anti-CD20 such as obinutuzumab could limit residual memory B cell survival. Profound B cell depletion has also been achieved with anti–CD19-CAR-T cells, which were recently shown to be effective in treating murine lupus (60). A clinical trial is currently conducted to assess anti–CD19-CAR-T cells safety and efficacy in patients with systemic lupus erythematosus (NCT03030976). We showed that residual splenic memory B cells and PC expressed CD19 in sufficient amounts for efficient depletion with anti–CD19-CAR-T cells in vitro. Anti-CD19 antibodies are also under development in autoimmune diseases and could provide an easier way to treat RTX failure patients and to prevent disease relapses in responders.
Because the analysis of secondary lymphoid organs after RTX administration is limited in humans, it remains to be demonstrated whether our findings can be generalized to other autoimmune diseases where RTX is only transiently effective. As mentioned previously, RTX-resistant B cell populations, mainly composed of memory B cells, have been described in several autoimmune diseases including rheumatoid arthritis (52), autoimmune hemolytic anemia (23), and myasthenia gravis (61) as well as after preventive treatments of humoral graft rejection (50, 51, 53), suggesting that persistence of autoreactive memory clones is not limited to ITP.
Collectively, our results strongly suggest that splenic autoreactive residual memory B cells survived for months during RTX-mediated B cell depletion by acquiring a unique transcriptional program. These cells actively participated in disease relapses during B cell reconstitution through differentiation into autoantibody secreting cells and through reinitiation of the autoreactive GC response, which also involved newly generated B cells. Our study shows that this reservoir is targetable by anti-CD19 therapies, which paves the way for the development of strategies optimizing B cell depletion to increase the rate of long-term remissions in antibody-mediated diseases.
Materials and Methods
Study design
Spleen samples from patients with ITP were obtained through the French Network for Adult’s Cytopenia. All patients with ITP included in this study were adults and had ITP diagnosis according to the international guidelines definitions (9). Patients with underlying immunodeficiency, hepatitis C virus infection, lymphoproliferative disorders, thyroid or liver disease, and defined systemic lupus erythematosus (≥4 American Rheumatism Association criteria) were excluded. All patients had persistent (≥3 months) or chronic (≥12 months) ITP. Complete response was defined as platelet count >100 × 109/liter and no bleeding, and failure was defined as platelet count <30 × 109/liter (62). All relapse cases described in this study correspond to patients who first showed a complete response to RTX treatment. Cases of partial restoration of platelet counts (<100 × 109/liter) were excluded because this is likely linked to the persistence of small amount of autoantibody secretion. All healthy controls (HD) were organ donors who died from stroke or head trauma and were obtained from the Agence de la Biomédecine. None of them had lymphoma or autoimmune disease. All young donors included in the scRNA-seq analysis were patients with sickle cell disease who were splenectomized between the age of 4 and 5 years. Throughout the manuscript, all patients are referred to using the nomenclature presented in table S1. No sample size calculation, randomization, or blinding was performed for this study.
This study was conducted in compliance with the Declaration of Helsinki principles and was approved by the Agence de la Biomédecine and the Institutional Review Boards Comité de Protection des Personnes (CPP) Ile-de-France IX (for patients with ITP) and II (for control patients). All patients with ITP or sickle cell disease provided written informed consent before the collection of splenic samples.
Spleen sample processing
Immediately after splenectomy, splenic tissues were maintained at 4°C and crushed using gentleMACS dissociator (Miltenyi). After filtration and 1:2 dilution by RPMI 1640 (Invitrogen), mononuclear cells were isolated by Ficoll density gradient (Ficoll Paque Plus, GE Healthcare) centrifugation at 1300 rpm for 20 min with the brakes off and stored in liquid nitrogen.
Histology
Formol-fixed paraffin-embedded (FFPE) tissue sections of spleen of patients with relapse after RTX were studied after hematein eosin safran (HES) staining using an automated stainer PRISMA (Sakura). Immunostainings were performed on FFPE tissue sections on a BenchMark ULTRA automated immunostainer (Roche Diagnostics). The antibodies used were CD20 (1:800; M755-01, Dako), CD79a (1:100; M705001, Agilent), CD3 (1:200; A0452, Dako), Ki67 (1:50; M7240, Dako), CD10 (1:20; CD10-270-L-CE, Leica), CD21 (1:20; CD21-2G9-L-CE, Leica), and PD1 (1:50; ab52587, Abcam).
Multicolor flow cytometry and cell sorting
After thawing, mononuclear cells were counted and stained with antibodies directed against surface markers (listed in table S6). Intracellular staining was performed after using BD Cytofix/Cytoperm solution (BD Biosciences). Samples were collected using a FACS LSRFortessa flow cytometer and analyzed using DIVA (BD Biosciences) and FlowJo (Treestar Inc.) softwares. Cell sorting was performed with a FACSAria III cell sorter (BD Biosciences).
Single-cell culture
Single-cell culture was performed as previously described (25). Single B cells were sorted in 96-well plates containing MS40 cells expressing CD40L (gift from G. Kelsoe). Cells were cocultured at 37°C with 5% CO2 during 15 days in B cell medium consisting of RPMI 1640 (Invitrogen) supplemented with 10% HyClone fetal bovine serum (Thermo Fisher Scientific), 55 μM 2-mercaptoethanol, 10 mM Hepes, 1 mM sodium pyruvate, penicillin (100 U/ml), streptomycin (100 μg/ml), and Eagle’s minimum essential medium (MEM) non-essential amino acid solution (all Invitrogen), with the addition of recombinant human B cell activating factor (BAFF; 10 ng/ml), interleukin-2 (IL-2; 50 ng/ml), IL-4 (10 ng/ml), and IL-21 (10 ng/ml; all PeproTech). Half of supernatant was carefully removed at days 4, 8, and 12, and the same amount of fresh B cell medium with cytokines was added to the cultures. IgG ELISA was performed at day 15 for all wells to screen for IgG-secreting clones.
ELISPOT assays and identification of anti–GPIIbIIIa-specific clones
Anti-GPIIbIIIa ELISPOTS were performed as previously described (10). Briefly, keyhole limpet hemocyanin (2.5 μg/ml), goat anti-human Ig polyvalent antibody (10 μg/ml) (Invitrogen), or purified GpIIbIIIa (15 μg/ml) (Stago) were coated in phosphate-buffered saline (PBS) and 0.05% CaCl2 in multiscreen 96-well filter plates (MSIPS4510, Millipore) with overnight incubation at 4°C.
For the identification of anti–GPIIbIIIa-specific cultured clones from memory B cells or GC B cells, IgG-secreting clones from single-cell cultures were transferred in individual wells of an ELISPOT plate and incubated overnight in B cell medium at 37°C with 5% CO2. Cells were then carefully removed and stored at 37°C, and the ELISPOT plate was incubated for 4 hours at 4°C with biotinylated goat anti-human IgG Fc (Invitrogen), followed by incubation for 1 hour at room temperature with horseradish peroxidase (HRP) – conjugated avidin (Vector Laboratories). HRP activity was further revealed using 3-amino-9-ethylcarbazole (BD Biosciences) for 8 min at room temperature in the dark. Spots were enumerated in each well with an ELISPOT reader using the AID software (version 3.5; AutoImmun Diagnostika). Clones were considered GPIIbIIIa reactive if more than three dense and specific spots were detected in the well. AP3 hybridoma cells, producing an anti-GPIIbIIIA mouse IgG, were purchased from the American Type Culture Collection.
Alternatively, to identify anti–GPIIbIIIa-specific splenic PC, 106 splenocytes were serially diluted in B cell medium in triplicate before transferring to ELISPOT plates and incubated overnight at 37°C with 5% CO2. IgG-secreting anti-GpIIbIIIa spots were enumerated in each well as above and reported to IgG-positive spots.
IgH sequencing of GPIIbIIIa-specific clones
GPIIbIIIa-specific clones identified in the ELISPOT assay were washed in PBS, and RNA was extracted using an RNeasy micro kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription was performed using SuperScript III enzyme (Thermo Fisher Scientific) in 12-μl volume (42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 5 min) with 6 μl of RNA and random hexamers (GE Healthcare). A polymerase chain reaction (PCR) was then performed on the basis of an established protocol (63). Briefly, 3.5 μl of cDNA was used as a template and amplified in a total volume of 40 μl with forward L-VH primer mix and reverse Cγ primer (sequences in table S7), using HotStarTaq DNA polymerase (Qiagen) for 30 cycles (94°C for 30 s, 58°C for 30 s, and 72°C for 55 s). PCR products were sequenced with the reverse primer CHG-D1 and read on ABI PRISM 3130XL genetic analyzer (Applied Biosystems). Sequence quality was verified with the CodonCode Aligner software (CodonCode Corporation), and data were analyzed with the IMGT/HighV-QUEST web portal (from the International Immunogenetics Information System).
Recombinant antibody production and purification
IgH and IgL chains from clones of interest were cloned as previously described (63). HEK293T cells were transfected with plasmid DNA encoding paired IgH and IgL chains using jetPRIME kit (Polyplus) as previously described (63). Recombinant antibodies were purified from supernatants using protein G beads according to the manufacturer’s instructions (GE Healthcare).
ELISA and MAIPA
Total IgG and anti-GPIIbIIIa IgG from culture supernatants were detected by an in-house ELISA. Briefly, 96-well ELISA plates (Thermo Fisher Scientific) were coated with either goat anti-human Ig (10 μg/ml; Invitrogen) or purified GPIIbIIIa (5 μg/ml; Stago) in sodium carbonate. Cell culture supernatants were added for 1 hour, and then ELISAs were developed using HRP-goat anti-human IgG (1 μg/ml; ImmunoTech) and 3,3’,5,5’-tetramethylbenzidine substrate (Eurobio). Optical densities were measured at 450nm and 620nm, and antibody reactivity was calculated after subtraction of blank wells. Indirect MAIPA assays (apDia) were performed following the manufacturer’s instructions.
Proliferation assay
Cell proliferation was assessed with cell proliferation dye eFluor 450 (Thermo Fisher Scientific). Briefly, splenocytes were resuspended at 2.5 × 106 cell/ml in B cell medium and stained according to the manufacturer’s instructions. Stained cells were cultured for 5 days in B cell medium supplemented or not with CpG (2.5 μg/ml; ODN2006, Invivogen) or cocultured with MS40 cells expressing CD40L and cytokine cocktail [recombinant human BAFF (10 ng/ml), IL-2 (50 ng/ml), IL-4 (10 ng/ml), and IL-21 (10 ng/ml); all PeproTech]. Cell division was assessed by measuring the decrease in eFluor 450 fluorescence via flow cytometry on resting memory B cells (CD3−CD14−CD19+CD38−CD24+CD27+IgD−).
In vitro RTX cultures
Fresh spleen samples from HD were obtained and immediately crushed as described above. Total splenocytes were seeded at 5 × 106 cells per well in 24-well plates with complete B cell medium. RTX (1 or 10 μg/ml) or PBS was added to the culture. After overnight incubation at 37°C with 5% CO2, cells were washed and stained for surface markers for flow cytometry analysis as described above.
CD20 reexpression assay
Thawed spleen samples from RTX failure patients and HD were seeded at 4 × 106 cells per well in 24-well plates in B cell medium with BAFF (10 ng/ml). CD20 expression was assessed by flow cytometry analysis at days 0, 1, 2, and 5 of culture.
Library preparation and Illumina sequencing
Splenic B cell subsets from three RTX relapse patients, including PC (CD19+CD27highCD38highCD24−), GC (CD19+CD38intCD24−IgD−CD20+), resting memory (CD19+CD38−CD24+CD27+IgD−), and naive (CD19+CD38lowCD27−IgD+) B cells, were sorted in PBS and 2% fetal calf serum. Cells (104 to 2 × 105) were obtained for each subset. Cells were washed with PBS, centrifuged, and resuspended in the appropriate volume of lysis/binding buffer from the Dynabeads mRNA DIRECT Micro Kit (Thermo Fisher Scientific). mRNA were isolated directly with Dynabeads oligo (dT) and immediately used in their entirety for reverse transcription in solid phase, according to a protocol adapted from Vergani et al. (64), with modified primers (table S7). Briefly, cDNA was synthetized in 10 μl (50°C for 1 hour and 72°C for 10 min) using SuperScript III Enzyme (Thermo Fisher Scientific). After ribonuclease H treatment, second-strand synthesis was performed in solid phase in 10 μl using Q5 Polymerase (New I England Biolabs) and a mix of 13 primers covering IGHV leader sequences, containing 13 to 16 random nucleotide and partial Illumina adaptor sequences (37°C for 20 min, 98°C for 30 s, 62°C for 2 min, and 72°C for 10 min). IGHV-D-J-CH double-stranded cDNA attached to the magnetic Dynabeads was washed three times in : 10 mM tris-HCl (pH 8.0) to remove the remaining primers, and the I entire sample was used as a template for the PCR1 amplification in 10 μl using Q5 Polymerase with a forward primer that is specific for the partial Illumina adaptor sequence and allowing its extension, : and a mix of reverse isotype specific (μ, α, and γ) primers (98°C for 30 s; 10 cycles of 98°C for 10 s, 58°C for 15 s, and 72°C for 1 min; 72°C for 10 min). Depending on the initial cell fraction, individual semi-nested PCR2 were performed with inner reverse primers specific for the mu, alpha, and gamma constant region and allowing the introduction of partial Illumina adaptors. Two microliters of the PCR1 product was used for the semi-nested PCR2 reactions carried in 20 μl (98°C for 30 s; 15 cycles of 98°C for 10 s, 58°C for 15 s, and 72°C for >1 min; 72°C for 10 min). The PCR2 products were purified with AMPure I XP beads at a ratio of 1:1, and 1 to 10 ng was used to add Illumina Index with the Nextera XT kit (Illumina). After a final purification with AMPure XP beads (ratio 1:1), the library obtained for each sample was diluted at a concentration of 2 nM and pooled equimolarly into a final pool. Sequencing was performed with the Illumina MiSeq sequencer using the V3 kit (2 × 300) with 12 pmol of the final pool and 10% PhiX control to avoid low-complexity library issues.
Bioinformatics analysis of IgH sequencing data
Unique molecular identifier (UMI)–barcoded Illumina MiSeq 2 × 300 reads were analyzed with the packages of the Immcantation framework as well as custom Python, Bash, and R scripts. Raw read preprocessing was done with pRESTO (REpertoire Sequencing TOolkit) (65). Sequences with average Phred quality scores of less than 20 were excluded from further analysis. PCR primers were identified with a mismatch rate of 0.11 for C primer and 0.2 for V primer. UMI read groups having high error statistics were removed using a 0.1 threshold; gap positions were removed if present in more than 50% of the sequences.
After consensus build, the paired reads were assembled, deduplicated, and submitted to IMGT/HighV-QUEST web portal (the International ImMunoGeneTics database) for annotation, mutation analysis, and functionality assessment (66). Annotated sequences with a V region length of less than 250 or with a number of detected somatic hypermutation exceeding 60 were filtered out.
Clonal cluster assignment and germline reconstruction were performed with Change-O toolkit (67) on the sequences having at least two representative reads. The sequences that had the same V-gene, same J-gene, including ambiguous assignments, and same : CDR3 length with maximal nucleotide hamming distance of 0.16 (the threshold determined with SHazaM R package) were considered belonging to the same clonal group.
Further clonal relationship and statistical analysis was implemented in R. The graphics were obtained with packages ggplot2, circlize, as well as with GraphPad tool. Phylogenetic trees were made using Alakazam package and Phylip tool.
Single-cell RNA sequencing
Single-cell mRNA sequencing was performed according to an adapted version of the SOrting and Robot-assisted Transcriptome SEQuencing (SORT-seq) protocol (68) with primers described by van den Brink et al. (69). Briefly, resting memory B cells (CD19+CD38−CD24+CD27+IgD−) from HD, RTX failure, RTX relapse, and young donors were single-cell FACS (fluorescence-activated cell sorting) sorted (Aria III) into 384-well plates containing 384 primers and Mineral oil (Sigma-Aldrich). After sorting, plates were snap-frozen on dry ice and stored at -80°C. Each plate was designed to contain cells from each donor group. Further processing of cells, cDNA library generation, sequencing, and reads alignment were performed at Single Cell Discoveries. For amplification, cells were heat-lysed at 65°C followed by cDNA synthesis using the CEL-Seq2 protocol (70) and robotic liquid handling platforms. After second-strand cDNA synthesis, the barcoded material was pooled into libraries of 384 cells and amplified using in vitro transcription. After amplification, the rest of the CEL-Seq2 protocol was followed for preparation of the amplified cDNA library, using TruSeq small RNA primers (Illumina). The DNA library was paired-end sequenced on an Illumina NextSeq 500, high output, with a 2 × 75 bp kit (Illumina). Read 1 was used for identification of the Illumina library barcode, 8 bp of cell barcode and 6 bp of UMI, and read 2 was used to map to the reference transcriptome of Hg19 with BWA (71). Data were demultiplexed, and transcript counts were adjusted to the expected number of molecules based on counts, 4096 possible UMI’s and Poissonian counting statistics as described in Grün et al. (72).
In total, 4041 memory B cells were sequenced. Initial quality control and cell filtering were performed using the R Scater package (73). After removing wells where more than 50% of transcripts mapped to external RNA control consortium (ERCC) spike-ins, cells with less than 2000 UMIs or 500 genes detected, and keeping only genes that were detected in at least 2% of any given sorted cell population, 3309 cells and 12,166 genes were left for further analysis. Normalization, integration, clustering, and differential expression analysis were then performed through the R Seurat v3 (74) pipeline. Transcript counts were first normalized using the scTransform algorithm (75), additionally correcting for potential bias related to the detected percentage of mitochondrial genes and selecting for the top 3000 variable features for downstream visualization and clustering analysis. After principal components analysis, potential sort-specific batch effects were removed using the Harmony algorithm (76). The first 30 corrected principal components analysis dimensions were then used to construct a k nearest neighbor graph (k = 20 neighbors) and perform graph-based clustering (Louvain) with a resolution parameter of 0.6 as well as compute the uniform manifold approximation and projection (UMAP) coordinates for each cell. For all initial steps, 2159 PC from the same donors, and sequenced using the same protocol, were added to the analysis. This allowed us to remove potential PC contaminants from sorted memory B cell populations and to identify a small mix cluster of apoptotic cells (1% of all cells) that were removed from further analysis. Final clustering steps as presented in the manuscript were focused solely on sorted memory B cells. Differential expression analyses between clusters of interest were then performed using the model-based analysis of single-cell transcriptomics (MAST) algorithm (77) using raw transcript counts and applying a threshold of 0.25 for log fold change and 0.05 for the adjusted P value. Regulon identification and module activity scoring in cells were done using the SCENIC workflow (39) using version 9 of cis-Target motif collection (mc9nr).
Pathway enrichment analysis and visualization (Fig. 5) were performed as described in (78), using up- and down-regulated gene lists generated via MAST as input. Statistically enriched pathways from both the Gene Ontology Biological Process and Reactome curated gene sets were determined using the g:Profiler web interface, ordering input gene lists based on P values, filtering out gene sets containing less than 5 or more than 2000 terms and applying a threshold of 0.05 for P value. Visualization and interpretation of enriched pathways were then performed in Cytoscape v.3.7.2 using the EnrichmentMap and AutoAnnotate plugins and node and edge cutoffs of 0.05 (Q values) and 0.5 (similarity), respectively.
Gene set enrichment analysis (GSEA) (fig. S9) was performed using the online tool of the GSEA/Molecular Signatures Database MSigDB of the Broad Institute (https://www.gsea-msigdb.org/), computing overlaps between the Gene Ontology biological process gene sets (79) and up- and down-regulated gene lists generated via MAST. All downstream analyses were performed with R v.3.5.2 on a macOS 10.13.6 (64 bit) system, using the following R packages and libraries: scater v.1.10.1, scTransform v. 0.2.1, harmony v0.99.9, Seurat v.3.1.2, reticulate v1.13, umap v0.2.4.1, MAST v.1.8.2, ggplot2 v.3.2.1, SCENIC v 1.1.1-8, GENIE3 v.1.4.3, RcisTarget v1.2.1, and AUCell v1.4.1.
CAR-T cell assay
Anti-CD19ScFv-CD28-CD3ζ (PM CAR 1001) and CD28-CD3ζ without ScFv (PM CAR 1008) CAR-T cells were purchased from ProMab Biotechnologies. Cells were thawed using CAR-T cell medium (ProMab Biotechnologies) with IL-2 (300 UI/ml; PeproTech) and plated at 105 cells per well in 96-well plates. Splenic mononuclear cells from RTX failure patients were thawed and submitted to negative selection using CD3+ microbeads (Miltenyi Biotech) according to the manufacturer’s instructions. Cells (5 × 104) were added to each well containing CAR-T cells, as well as to wells without CAR-T cells as a negative control, and incubated for 24 hours before flow cytometry analysis as described above. All experiments were performed in triplicate for each condition for each patient.
IgH sequencing of single-cell memory B cell
IgH sequencing was performed after single-cell sorting, as described above for GPIIbIIIa-specific cells, but with 2 cycles of PCR (63): the first one, for 50 cycles, with 5’ L-VH primer mix and 3’ Cγ primer, and the second one, for 50 cycles, with a 5’ Age I VH primer mix and 3’ Sal I JH primer mix (primer sequences in table S7).
Statistics
Kruskall-Wallis and corrections for multiple comparisons using Benjamini-Hochberg-Yekutieli method (experiments with three groups or more) or Mann-Whitney test (experiments with two groups) were used to compare continuous variables as appropriate. A P value ≤ 0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism 8.1.
Supplementary Material
One-sentence summary.
Rituximab-resistant memory B cells and newly engaged naive B cells join to fuel subsequent autoimmune responses in patients with relapsing ITP.
Editor’s Summary.
B cells behaving badly
Patients diagnosed with B cell–mediated autoimmune diseases, such as immune thrombocytopenia (ITP), can benefit from treatment with the B cell–depleting antibody, rituximab. Unfortunately, a large proportion of patients relapse after treatment. To understand why, Crickx et al. evaluated B cell phenotypes and specificity, finding two populations of B cells that may contribute to such relapses. The first consisted of a population of rituximab-resistant memory B cells, whereas the second consisted of a new population of autoreactive B cells never exposed to rituximab. Rituximab-resistant B cell populations maintained expression of the B cell marker CD19, suggesting that CD19-targeting therapies could provide another line of treatment for patients who do not respond to rituximab.
Acknowledgments
We thank L. Da Silva and E. Letoriellec for expert technical assistance and J. Mégret (Cell Sorting Facility of the SFR Necker) for cell sorting. We thank D. Bagnara for sharing expertise on high-throughput sequencing, P. Villarese and the McIntyre/Asnafi team for access to an Illumina MiSeq sequencer, L. Languille for logistic coordination, and G. Kelsoe for providing the human cell culture system, together with invaluable advices. We also thank the other physicians, L. Galicier, C. Fieschi, M. Malphettes, K. Sacré, A. Mekinian, and N. Abisror, whose patients were included in this study.
Funding
This work was supported by an ANR Grant (Auto-Immuni-B, ANR-18-CE15-0001), by the Fondation Princesse Grace, and, in part, by an ERC Advanced Grant (B-response). E.C. was supported by a Poste d’Accueil from INSERM, A.S. and G.R. by an Année Recherche from APHP, and G.B. by an SNFMI fellowship.
Footnotes
Author contributions: Conceptualization: E.C., J.-C.W., C.-A.R., and M. Mahévas Data curation: P.C. and T.F. Formal analysis: E.C., P.C., A.S., I.A., A.V., G.B., G.R., and T.F. Funding acquisition: E.C., J.-C.W., C.-A.R., and M. Mahévas Investigation: E.C., P.C., A.S., I.A., A.V., G.B., G.R., and S.W. Methodology: E.C., J.-C.W., C.-A.R., P.C., and M. Mahévas Project administration: J.-C.W., C.-A.R., and M. Mahévas Resources: J.F., V.M., E.R., B.G., M. Michel, and S.S. Software: P.C. and T.F. Supervision: J.-C.W., C.-A.R, M. Mahévas, B.G., and M. Michel Validation: A.S. and A.V. Visualization: E.C., P.C., A.S., M. Mahévas, I.A., and T.F. Writing (original draft): E.C., P.C., A.S., J.-C.W., C.-A.R., and M. Mahévas Writing (review and editing): All authors.
Competing interests: B.G. served as an expert for AMGEN, Novartis, LFB, and Roche. He received funds for research from AMGEN and Roche. M. Michel received consultancy fees from Amgen, Novartis, and Argenx. M. Mahévas received research funds from GSK. J.-C.W. is a scientific consultant of Institut Merieux. The other authors declare that they have no competing interests.
Data and materials availability
All data associated with this study are present in the paper or the Supplementary Materials. All high-throughput VDJ sequencing and scRNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress) and are available under the following accession number: E-MTAB-9953 and E-MTAB-9955.
References and Notes
- 1.Dörner T, Lipsky PE. B cells: Depletion or functional modulation in rheumatic diseases. Curr Opin Rheumatol. 2014;26:228–236. doi: 10.1097/BOR.0000000000000000. [DOI] [PubMed] [Google Scholar]
- 2.Huang H, Benoist C, Mathis D. Rituximab specifically depletes short-lived autoreactive plasma cells in a mouse model of inflammatory arthritis. Proc Natl Acad Sci USA. 2010;107:4658–4663. doi: 10.1073/pnas.1001074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khellaf M, Charles-Nelson A, Fain O, Terriou L, Viallard J-F, Cheze S, Graveleau J, Slama B, Audia S, Ebbo M, Le Guenno G, et al. Safety and efficacy of rituximab in adult immune thrombocytopenia: Results from a prospective registry including 248 patients. Blood. 2014;124:3228–3236. doi: 10.1182/blood-2014-06-582346. [DOI] [PubMed] [Google Scholar]
- 4.Patel VL, Mahévas M, Lee SY, Stasi R, Cunningham-Rundles S, Godeau B, Kanter J, Neufeld E, Taube T, Ramenghi U, Shenoy S, et al. Outcomes 5 years after response to rituximab therapy in children and adults with immune thrombocytopenia. Blood. 2012;119:5989–5995. doi: 10.1182/blood-2011-11-393975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lioger B, Edupuganti SR, Mulleman D, Passot C, Desvignes C, Bejan-Angoulvant T, Thibault G, Gouilleux-Gruart V, Mélet J, Paintaud G, Ternant D. Antigenic burden and serum IgG concentrations influence rituximab pharmacokinetics in rheumatoid arthritis patients. Br J Clin Pharmacol. 2017;83:1773–1781. doi: 10.1111/bcp.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng CM, Bruno R, Combs D, Davies B. Population pharmacokinetics of rituximab (anti-CD20 monoclonal antibody) in rheumatoid arthritis patients during a phase II clinical trial. J Clin Pharmacol. 2005;45:792–801. doi: 10.1177/0091270005277075. [DOI] [PubMed] [Google Scholar]
- 7.Kuwana M, Okazaki Y, Kaburaki J, Kawakami Y, Ikeda Y. Spleen is a primary site for activation of platelet-reactive T and B cells in patients with immune thrombocytopenic purpura. J Immunol. 2002;168:3675–3682. doi: 10.4049/jimmunol.168.7.3675. [DOI] [PubMed] [Google Scholar]
- 8.Deshayes S, Khellaf M, Zarour A, Layese R, Fain O, Terriou L, Viallard J-F, Cheze S, Graveleau J, Slama B, Audia S, et al. Long-term safety and efficacy of rituximab in 248 adults with immune thrombocytopenia: Results at 5 years from the French prospective registry ITP-ritux. Am J Hematol. 2019;94:1314–1324. doi: 10.1002/ajh.25632. [DOI] [PubMed] [Google Scholar]
- 9.Neunert C, Terrell DR, Arnold DM, Buchanan G, Cines DB, Cooper N, Cuker A, Despotovic JM, George JN, Grace RF, Kühne T, et al. American Society of Hematology 2019 guidelines for immune thrombocytopenia. Blood Adv. 2019;3:3829–3866. doi: 10.1182/bloodadvances.2019000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahévas M, Patin P, Huetz F, Descatoire M, Cagnard N, Bole-Feysot C, Le Gallou S, Khellaf M, Fain O, Boutboul D, Galicier L, et al. B cell depletion in immune thrombocytopenia reveals splenic long-lived plasma cells. J Clin Invest. 2013;123:432–442. doi: 10.1172/JCI65689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audia S, Rossato M, Santegoets K, Spijkers S, Wichers C, Bekker C, Bloem A, Boon L, Flinsenberg T, Compeer E, van den Broek T, et al. Splenic TFH expansion participates in B-cell differentiation and antiplatelet-antibody production during immune thrombocytopenia. Blood. 2014;124:2858–2866. doi: 10.1182/blood-2014-03-563445. [DOI] [PubMed] [Google Scholar]
- 12.Vinuesa CG, Sanz I, Cook MC. Dysregulation of germinal centres in autoimmune disease. Nat Rev Immunol. 2009;9:845–857. doi: 10.1038/nri2637. [DOI] [PubMed] [Google Scholar]
- 13.Hiepe F, Radbruch A. Plasma cells as an innovative target in autoimmune disease with renal manifestations. Nat Rev Nephrol. 2016;12:232–240. doi: 10.1038/nrneph.2016.20. [DOI] [PubMed] [Google Scholar]
- 14.Audia S, Samson M, Guy J, Janikashvili N, Fraszczak J, Trad M, Ciudad M, Leguy V, Berthier S, Petrella T, Aho-Glélé S, et al. Immunologic effects of rituximab on the human spleen in immune thrombocytopenia. Blood. 2011;118:4394–4400. doi: 10.1182/blood-2011-03-344051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thai L-H, Le Gallou S, Robbins A, Crickx E, Fadeev T, Zhou Z, Cagnard N, Mégret J, Bole C, Weill J-C, Reynaud C-A, et al. BAFF and CD4+ T cells are major survival factors for long-lived splenic plasma cells in a B-cell-depletion context. Blood. 2018;131:1545–1555. doi: 10.1182/blood-2017-06-789578. [DOI] [PubMed] [Google Scholar]
- 16.Porcelijn L, Huiskes E, Schipperus M, van der Holt B, de Haas M, Zwaginga JJ. Dutch HOVON 64 Study Group, Lack of detectable platelet autoantibodies is correlated with nonresponsiveness to rituximab treatment in ITP patients. Blood. 2017;129:3389–3391. doi: 10.1182/blood-2016-11-751719. [DOI] [PubMed] [Google Scholar]
- 17.Leandro MJ. B-cell subpopulations in humans and their differential susceptibility to depletion with anti-CD20 monoclonal antibodies. Arthritis Res Ther. 2013;15(Suppl. 1):S3. doi: 10.1186/ar3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roll P, Palanichamy A, Kneitz C, Dorner T, Tony H-P. Regeneration of B cell subsets after transient B cell depletion using anti-CD20 antibodies in rheumatoid arthritis. Arthritis Rheum. 2006;54:2377–2386. doi: 10.1002/art.22019. [DOI] [PubMed] [Google Scholar]
- 19.Anolik JH, Barnard J, Owen T, Zheng B, Kemshetti S, Looney RJ, Sanz I. Delayed memory B cell recovery in peripheral blood and lymphoid tissue in systemic lupus erythematosus after B cell depletion therapy. Arthritis Rheum. 2007;56:3044–3056. doi: 10.1002/art.22810. [DOI] [PubMed] [Google Scholar]
- 20.Anolik JH, Friedberg JW, Zheng B, Barnard J, Owen T, Cushing E, Kelly J, Milner ECB, Fisher RI, Sanz I. B cell reconstitution after rituximab treatment of lymphoma recapitulates B cell ontogeny. Clin Immunol. 2007;122:139–145. doi: 10.1016/j.clim.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Leandro MJ, Cambridge G, Ehrenstein MR, Edwards JCW. Reconstitution of peripheral blood B cells after depletion with rituximab in patients with rheumatoid arthritis. Arthritis Rheum. 2006;54:613–620. doi: 10.1002/art.21617. [DOI] [PubMed] [Google Scholar]
- 22.Weill J-C, Weller S, Reynaud C-A. Human marginal zone B cells. Annu Rev Immunol. 2009;27:267–285. doi: 10.1146/annurev.immunol.021908.132607. [DOI] [PubMed] [Google Scholar]
- 23.Mahévas M, Michel M, Vingert B, Moroch J, Boutboul D, Audia S, Cagnard N, Ripa J, Menard C, Tarte K, Mégret J, et al. Emergence of long-lived autoreactive plasma cells in the spleen of primary warm auto-immune hemolytic anemia patients treated with rituximab. J Autoimmun. 2015;62:22–30. doi: 10.1016/j.jaut.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Turner JS, Zhou JQ, Han J, Schmitz AJ, Rizk AA, Alsoussi WB, Lei T, Amor M, McIntire KM, Meade P, Strohmeier S, et al. Human germinal centres engage memory and naive B cells after influenza vaccination. Nature. 2020;586:127–132. doi: 10.1038/s41586-020-2711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, Harrison SC. Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity. 2018;48:174–184.:e9. doi: 10.1016/j.immuni.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, Azuma T, Kitamura D. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 27.Weller S, Mamani-Matsuda M, Picard C, Cordier C, Lecoeuche D, Gauthier F, Weill J-C, Reynaud C-A. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+IgD+CD27+ B cell repertoire in infants. J Exp Med. 2008;205:1331–1342. doi: 10.1084/jem.20071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phalke S, Marrack P. Age (autoimmunity) associated B cells (ABCs) and their relatives. Curr Opin Immunol. 2018;55:75–80. doi: 10.1016/j.coi.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Portugal S, Tipton CM, Sohn H, Kone Y, Wang J, Li S, Skinner J, Virtaneva K, Sturdevant DE, Porcella SF, Doumbo OK, et al. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. eLife. 2015;4:e07218. doi: 10.7554/eLife.07218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nechanitzky R, Akbas D, Scherer S, Györy I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 31.Al-Maskari M, Care MA, Robinson E, Cocco M, Tooze RM, Doody GM. Site-1 protease function is essential for the generation of antibody secreting cells and reprogramming for secretory activity. Sci Rep. 2018;8:14338. doi: 10.1038/s41598-018-32705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song S, Matthias PD. The transcriptional regulation of germinal center formation. Front Immunol. 2018;9:2026. doi: 10.3389/fimmu.2018.02026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Keimpema M, Grüneberg LJ, Mokry M, van Boxtel R, van Zelm MC, Coffer P, Pals ST, Spaargaren M. The forkhead transcription factor FOXP1 represses human plasma cell differentiation. Blood. 2015;126:2098–2109. doi: 10.1182/blood-2015-02-626176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pekarsky Y, Palamarchuk A, Maximov V, Efanov A, Nazaryan N, Santanam U, Rassenti L, Kipps T, Croce CM. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci USA. 2008;105:19643–19648. doi: 10.1073/pnas.0810965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, Russo G, Hardy RR, Croce CM. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf C, Garding A, Filarsky K, Bahlo J, Robrecht S, Becker N, Zucknick M, Rouhi A, Weigel A, Claus R, Weichenhan D, et al. NFATC1 activation by DNA hypomethylation in chronic lymphocytic leukemia correlates with clinical staging and can be inhibited by ibrutinib. Int J Cancer. 2018;142:322–333. doi: 10.1002/ijc.31057. [DOI] [PubMed] [Google Scholar]
- 37.Märklin M, Heitmann JS, Fuchs AR, Truckenmüller FM, Gutknecht M, Bugl S, Saur SJ, Lazarus J, Kohlhofer U, Quintanilla-Martinez L, Rammensee H-G, et al. NFAT2 is a critical regulator of the anergic phenotype in chronic lymphocytic leukaemia. Nat Commun. 2017;8:755. doi: 10.1038/s41467-017-00830-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vuillier F, Dumas G, Magnac C, Prevost M-C, Lalanne AI, Oppezzo P, Melanitou E, Dighiero G, Payelle-Brogard B. Lower levels of surface B-cell-receptor expression in chronic lymphocytic leukemia are associated with glycosylation and folding defects of the μ and CD79a chains. Blood. 2005;105:2933–2940. doi: 10.1182/blood-2004-09-3643. [DOI] [PubMed] [Google Scholar]
- 39.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, van den Oord J, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim SH, Vaughan AT, Ashton-Key M, Williams EL, Dixon SV, Chan HTC, Beers SA, French RR, Cox KL, Davies AJ, Potter KN, et al. Fc gamma receptor IIb on target B cells promotes rituximab internalization and reduces clinical efficacy. Blood. 2011;118:2530–2540. doi: 10.1182/blood-2011-01-330357. [DOI] [PubMed] [Google Scholar]
- 41.Reddy V, Cambridge G, Isenberg DA, Glennie MJ, Cragg MS, Leandro M. Internalization of rituximab and the efficiency of B cell depletion in rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheumatol. 2015;67:2046–2055. doi: 10.1002/art.39167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaughan AT, Iriyama C, Beers SA, Chan CHT, Lim SH, Williams EL, Shah V, Roghanian A, Frendéus B, Glennie MJ, Cragg MS. Inhibitory FcγRIIb (CD32b) becomes activated by therapeutic mAb in both cis and trans and drives internalization according to antibody specificity. Blood. 2014;123:669–677. doi: 10.1182/blood-2013-04-490821. [DOI] [PubMed] [Google Scholar]
- 43.Faitschuk E, Hombach AA, Frenzel LP, Wendtner C-M, Abken H. Chimeric antigen receptor T cells targeting Fc μ receptor selectively eliminate CLL cells while sparing healthy B cells. Blood. 2016;128:1711–1722. doi: 10.1182/blood-2016-01-692046. [DOI] [PubMed] [Google Scholar]
- 44.Ambegaonkar AA, Kwak K, Sohn H, Manzella-Lapeira J, Brzostowski J, Pierce SK. Expression of inhibitory receptors by B cells in chronic human infectious diseases restricts responses to membrane-associated antigens. Sci Adv. 2020;6:eaba6493. doi: 10.1126/sciadv.aba6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynaud C-A, Descatoire M, Dogan I, Huetz F, Weller S, Weill J-C. IgM memory B cells: A mouse/human paradox. Cell Mol Life Sci. 2012;69:1625–1634. doi: 10.1007/s00018-012-0971-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagnara D, Squillario M, Kipling D, Mora T, Walczak AM, Da Silva L, Weller S, Dunn-Walters DK, Weill J-C, Reynaud C-A. A reassessment of IgM memory subsets in humans. J Immunol. 2015;195:3716–3724. doi: 10.4049/jimmunol.1500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, Gregoretti I, Schieferl S, Hom J, Jenks S, Feldman RJ, Mehr R, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16:755–765. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He R, Reid DM, Jones CE, Shulman NR. Spectrum of Ig classes, specificities, and titers of serum antiglycoproteins in chronic idiopathic thrombocytopenic purpura. Blood. 1994;83:1024–1032. [PubMed] [Google Scholar]
- 49.Degn SE, van der Poel CE, Firl DJ, Ayoglu B, Al Qureshah FA, Bajic G, Mesin L, Reynaud C-A, Weill J-C, Utz PJ, Victora GD, et al. Clonal evolution of autoreactive germinal centers. Cell. 2017;170:913–926.:e19. doi: 10.1016/j.cell.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamburova EG, Koenen HJPM, Borgman KJE, ten Berge IJ, Joosten I, Hilbrands LB. A single dose of rituximab does not deplete B cells in secondary lymphoid organs but alters phenotype and function. Am J Transplant. 2013;13:1503–1511. doi: 10.1111/ajt.12220. [DOI] [PubMed] [Google Scholar]
- 51.Ramos EJ, Pollinger HS, Stegall MD, Gloor JM, Dogan A, Grande JP. The effect of desensitization protocols on human splenic B-cell populations in vivo. Am J Transplant. 2007;7:402–407. doi: 10.1111/j.1600-6143.2006.01632.x. [DOI] [PubMed] [Google Scholar]
- 52.Ramwadhdoebe TH, van Baarsen LGM, Boumans MJH, Bruijnen STG, Safy M, Berger FH, Semmelink JF, van der Laken CJ, Gerlag DM, Thurlings RM, Tak PP. Effect of rituximab treatment on T and B cell subsets in lymph node biopsies of patients with rheumatoid arthritis. Rheumatology. 2019;58:1075–1085. doi: 10.1093/rheumatology/key428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallin EF, Jolly EC, Suchánek O, Bradley JA, Espéli M, Jayne DRW, Linterman MA, Smith KGC. Human T-follicular helper and T-follicular regulatory cell maintenance is independent of germinal centers. Blood. 2014;124:2666–2674. doi: 10.1182/blood-2014-07-585976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beum PV, Peek EM, Lindorfer MA, Beurskens FJ, Engelberts PJ, Parren PWHI, van de Winkel JGJ, Taylor RP. Loss of CD20 and bound CD20 antibody from opsonized B cells occurs more rapidly because of trogocytosis mediated by Fc receptor-expressing effector cells than direct internalization by the B cells. J Immunol. 2011;187:3438–3447. doi: 10.4049/jimmunol.1101189. [DOI] [PubMed] [Google Scholar]
- 55.Boross P, Jansen JHM, Pastula A, van der Poel CE, Leusen JHW. Both activating and inhibitory Fc gamma receptors mediate rituximab-induced trogocytosis of CD20 in mice. Immunol Lett. 2012;143:44–52. doi: 10.1016/j.imlet.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Rossi EA, Goldenberg DM, Michel R, Rossi DL, Wallace DJ, Chang C-H. Trogocytosis of multiple B-cell surface markers by CD22 targeting with epratuzumab. Blood. 2013;122:3020–3029. doi: 10.1182/blood-2012-12-473744. [DOI] [PubMed] [Google Scholar]
- 57.Taylor RP, Lindorfer MA. Fcγ-receptor-mediated trogocytosis impacts mAb-based therapies: Historical precedence and recent developments. Blood. 2015;125:762–766. doi: 10.1182/blood-2014-10-569244. [DOI] [PubMed] [Google Scholar]
- 58.Walshe CA, Beers SA, French RR, Chan CHT, Johnson PW, Packham GK, Glennie MJ, Cragg MS. Induction of cytosolic calcium flux by CD20 is dependent upon B Cell antigen receptor signaling. J Biol Chem. 2008;283:16971–16984. doi: 10.1074/jbc.M708459200. [DOI] [PubMed] [Google Scholar]
- 59.Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, Yu S, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;51:840–855.:e5. doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kansal R, Richardson N, Neeli I, Khawaja S, Chamberlain D, Ghani M, Ghani Q-U-A, Balazs L, Beranova-Giorgianni S, Giorgianni F, Kochenderfer JN, et al. Sustained B cell depletion by CD19-targeted CAR T cells is a highly effective treatment for murine lupus. Sci Transl Med. 2019;11:eaav1648. doi: 10.1126/scitranslmed.aav1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang R, Fichtner ML, Hoehn KB, Pham MC, Stathopoulos P, Nowak RJ, Kleinstein SH, O’Connor KC. Single-cell repertoire tracing identifies rituximab-resistant B cells during myasthenia gravis relapses. JCI Insight. 2020;5:e136471. doi: 10.1172/jci.insight.136471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodeghiero F, Stasi R, Gernsheimer T, Michel M, Provan D, Arnold DM, Bussel JB, Cines DB, Chong BH, Cooper N, Godeau B, et al. Standardization of terminology, definitions and outcome criteria in immune thrombocytopenic purpura of adults and children: Report from an international working group. Blood. 2009;113:2386–2393. doi: 10.1182/blood-2008-07-162503. [DOI] [PubMed] [Google Scholar]
- 63.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vergani S, Korsunsky I, Mazzarello AN, Ferrer G, Chiorazzi N, Bagnara D. Novel method for high-throughput full-length IGHV-D-J sequencing of the immune repertoire from bulk B-cells with single-cell resolution. Front Immunol. 2017;8:1157. doi: 10.3389/fimmu.2017.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vander Heiden JA, Yaari G, Uduman M, Stern JNH, O’Connor KC, Hafler DA, Vigneault F, Kleinstein SH. pRESTO: A toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alamyar E, Duroux P, Lefranc M-P, Giudicelli V. IMGT® tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 67.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH. Change-O: A toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJP, van Oudenaarden A. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394.:e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, Robin C, van Oudenaarden A. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 70.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, et al. CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 73.McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, Merico D, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. All high-throughput VDJ sequencing and scRNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress) and are available under the following accession number: E-MTAB-9953 and E-MTAB-9955.