Abstract
The degradation of misfolded proteins is essential for cellular homeostasis. Misfolded proteins are normally degraded by the ubiquitin-proteasome system (UPS), and selective autophagy serves as backup mechanisms when the UPS is overloaded. Selective autophagy mediates the degradation of harmful material by its sequestration within double membrane organelles called autophagosomes. The selectivity of autophagic processes is mediated by cargo receptors, which link the cargo to the autophagosomal membrane. The p62 cargo receptor has a major function during the degradation of misfolded, ubiquitinated proteins by selective autophagy; here it functions to phase separate these proteins into larger condensates and tether them to the autophagosomal membrane. Recent work has given us crucial insights into the mechanism of action of p62 during selective autophagy and how its activity can be integrated with the UPS. We will discuss these recent insights in the context of protein quality control and the emerging concept of cellular organization mediated by phase transitions.
Keywords: quality control, proteostasis, selective autophagy, cargo receptor, degradation, lysosome, SQSTM1, aggrephagy
Introduction
The degradation of cytoplasmic proteins in eukaryotic cells is regulated by two major pathways: the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal system. Short-lived and misfolded proteins are preferentially degraded by the UPS, while long-lived proteins and proteins that cannot be unfolded, or even entire organelles, can be degraded by autophagy (Dikic, 2017). For a long time, protein degradation accomplished by the UPS and autophagy have been largely viewed independently, but it is becoming increasingly clear that the two pathways are highly interconnected with regard to cellular protein quality control as they use ubiquitination as a common degradation signal (Kirkin et al., 2009b). In this Review, we will discuss the current knowledge about the interplay between the UPS and autophagy in the degradation of cytoplasmic proteins. We will focus on recent insights into the p62/SQSTM-1 protein that have given us clues about how the two pathways may be connected. In particular, we will discuss the most recent findings of how p62 mediates the phase separation of ubiquitinated proteins into larger condensates that can sequester them and perhaps also serve as nucleators for autophagy.
The UPS
As briefly introduced above, two major pathways regulate protein degradation of cytoplasmic proteins in eukaryotic cells: the UPS and the autophagy-lysosomal system. Misfolded proteins are normally degraded by UPS by marking them with the conjugation of ubiquitin, a covalent posttranslational modification (Ciechanover, 2005). The activation of ubiquitin moieties is ATP-dependent and driven by an ubiquitin-activating enzyme (E1). First AMP-ubiquitin is transferred to a cysteine residue in the E1 and from there to a cysteine of an ubiquitin-conjugating enzyme (E2). The E2 then teams up with an ubiquitin-ligase (E3) to ubiquitinate the substrate, which is subsequently degraded by the proteasome (Streich and Lima, 2014, Schulman and Harper, 2009, van Wijk and Timmers, 2010, Ye and Rape, 2009). Substrates can be modified with a single ubiquitin or with ubiquitin chains, where the first substrate-attached ubiquitin serves itself as substrate for further ubiquitin conjugation. Dependent on the lysine of the ubiquitin that is used for chain elongation, diverse ubiquitin chains can be generated (Komander and Rape, 2012). Historically, proteasomal degradation is thought to be driven by the recognition of K48-linked ubiquitin chains, but most chain types can direct proteins for degradation by the proteasome (Komander and Rape, 2012). Certain chain types, including K63-chains have been mainly associated with autophagy and signaling; however, evidence exists that these can also be targeted by the proteasome (Dikic, 2017, Komander and Rape, 2012, Yau and Rape, 2016). At the proteasome, the ubiquitinated substrates are recognized by receptors, and are then deubiquitinated and unfolded in order to funnel them into the catalytic chamber of the protease. Substrates that cannot be unfolded can thus not be degraded by the proteasome (Finley, 2009).
Macroautophagy
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved and highly regulated pathway that plays a key role in quality control by mediating the degradation of cellular material within the lysosomal system. During autophagy, cytoplasmic material is engulfed by double-membrane organelles called autophagosomes that subsequently fuse with lysosomes, wherein their content is degraded. The resulting building blocks are recycled for reuse (Kraft and Martens, 2012). Although the term autophagy was proposed upon detection of double membrane vesicles in rat hepatic cells decades ago (De Duve and Wattiaux, 1966), its molecular understanding is largely based on the discovery of autophagy-related (ATG) genes in yeast genetic screens and the subsequent identification of homologues in complex eukaryotes (Thumm et al., 1994, Tsukada and Ohsumi, 1993, Harding et al., 1995). Autophagy is regulated by more than 40 known ATG genes, but only a restricted subset codes for proteins that are fundamental for all types of autophagy (Xie and Klionsky, 2007). This subset is referred to as the “core” autophagy machinery and can be divided into five subgroups that are thought to act in a hierarchical manner: initiation, nucleation of the isolation membrane (the precursor to the autophagosome), membrane expansion and maturation, fusion with the lysosome and nutrient recycling (Fig. 1). In eukaryotic cells, the initiation of autophagy is controlled by the activation of the unc-51-like kinase (ULK) complex, consisting of ULK1/2, ATG13, FIP200 and ATG101 (Mizushima, 2010, Hosokawa et al., 2009, Mercer et al., 2009, Hara et al., 2008, Young et al., 2006). The nucleation of the isolation membrane is also regulated by the class III phosphatidylinositol-3 kinase (PI3K) complex that is composed of VPS34, VPS15, and Beclin and ATG14L (Hurley and Young, 2017, Itakura et al., 2008). Membrane delivery to the expanding isolation membrane is still a largely enigmatic process, but appears to be controlled by the transmembrane protein ATG9A (Young et al., 2006). Vesicle expansion involves two sets of ubiquitin-like (Ubl) conjugation systems, including the ATG12–ATG5-ATG16L1 complex and proteins of the Atg8 family. In humans this family comprises LC3A, LC3B, LC3C, GABARAP, GABARAPL1 and GABARAPL2. The two conjugation systems are highly interconnected and collectively mediate the attachment of the Atg8 C-terminus to phosphatidylethanolamine in the isolation membrane, where it functions as docking site for other factors (Geng and Klionsky, 2008, Mizushima et al., 1998, Ichimura et al., 2000, Ichimura et al., 2000). The closure of the isolation membrane and the recruitment of the SNARE protein syntaxin17 completes autophagosome maturation. Finally, the fusion of completed autophagosomes with lysosomes is mediated by the interaction between syntaxin17, SNAP29 and VAMP8 as well as YKT6, SNAP29 and STX7 (Itakura et al., 2012, Matsui et al., 2018). The homotypic fusion and vacuole protein sorting (HOPS) tethering complex and several other accessory molecules, such as BRUCE and PLEKHM1 promote these fusion events (Hegedus et al., 2013, Takats et al., 2013, Jiang et al., 2014, Takats et al., 2014, Ebner et al., 2018, McEwan et al., 2015). During starvation, the level of autophagy can be drastically increased as a general protective response to compensate for the lack of nutrients (Mortimore and Schworer, 1977, Kopitz et al., 1990, Kuma et al., 2004). For this reason, autophagy has long been considered as a nonselective process that is responsible for the indiscriminate degradation of cytoplasmic components and for the recycling of macromolecules to promote cellular adaptation and survival. However, recent data have revealed that autophagy can selectively direct cargoes to lysosomal degradation in nutrient-rich conditions by encapsulating cytoplasmic material in a selective exclusive manner (reviewed in Zaffagnini and Martens, 2016). Furthermore, the finding that tissue-specific knockout of autophagy genes in mice leads to neurodegeneration or liver cancer (Hara et al., 2006, Komatsu et al., 2006, Takamura et al., 2011) and the fact that cells with defective autophagy are unable to clear certain intracellular pathogens (Randow and Youle, 2014, Deretic et al., 2013, Nakagawa et al., 2004) highlight the essential function of selective autophagy in maintaining cellular homeostasis. Selective autophagy can distinguish between diverse targets and different terms were coined to describe such processes. Among the most studied types of selective autophagy are mitophagy, xenophagy, pexophagy and aggrephagy, in other words the selective disposal of old and/or damaged mitochondria (Rogov et al., 2014, Kanki et al., 2009, Narendra et al., 2008, Novak et al., 2010, Okamoto et al., 2009), intracellular pathogens (Gutierrez et al., 2004, Nakagawa et al., 2004, Thurston et al., 2009, Yoshikawa et al., 2009, Zheng et al., 2009), surplus peroxisomes (Farre et al., 2008, Hutchins et al., 1999, Iwata et al., 2006) and the disposal of aberrant and misfolded cytosolic protein aggregates, respectively (Komatsu et al., 2007, Bjorkoy et al., 2005, Kirkin et al., 2009a, Pankiv et al., 2007, Szeto et al., 2006). In the following sections, we will focus on aggrephagy and, in particular, on the role of the p62/SQSTM1 protein in this process.
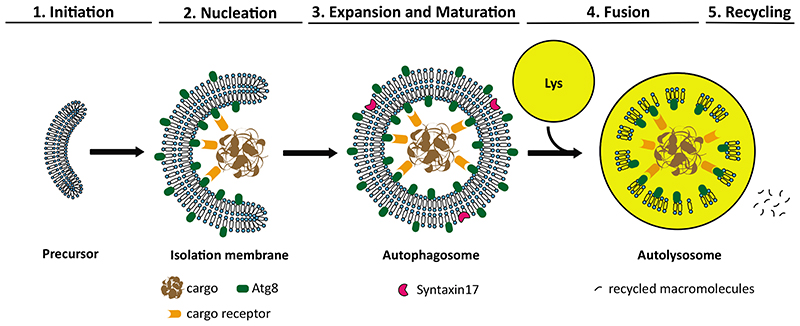
Fig. 1. Brief overview of autophagy.
Autophagy maintains cellular homeostasis by mediating the degradation and recycling of cytoplasmic material. Autophagy can be organized in several hierarchical steps. 1. Initiation, which is controlled by the activation of the ULK complex. 2. Nucleation of an isolation membrane that engulfs the cargo material. 3. Vesicle expansion and maturation, which entails the expansion of the isolation membrane and its closure to form an autophagosome. 4. Fusion of the mature autophagosome with a lysosome, resulting in the degradation of the inner autophagosome membrane and the cargo. 5. Recycling of the macromolecules that result from the breakdown of the cargo and are transported back to the cytosol.
The p62/SQSTM1 cargo receptor in aggrephagy
In contrast to non-selective bulk autophagy, selective autophagy requires i) the specific recognition of the cargo material and ii) its largely specific encapsulation by an isolation membrane (reviewed in Zaffagnini and Martens, 2016). The selective nature of autophagy is mediated by cargo receptors that recognise the cargo destined for degradation and link it to the growing isolation membrane through their interaction with membrane associated Atg8 (Rogov et al., 2014, Zaffagnini and Martens, 2016) (Fig. 2). Many cargo receptors, including p62 and the related NBR1, as well as NDP52 and Optineurin, recognize their cargoes due to its modification with ubiquitin (reviewed in Dikic, 2017).
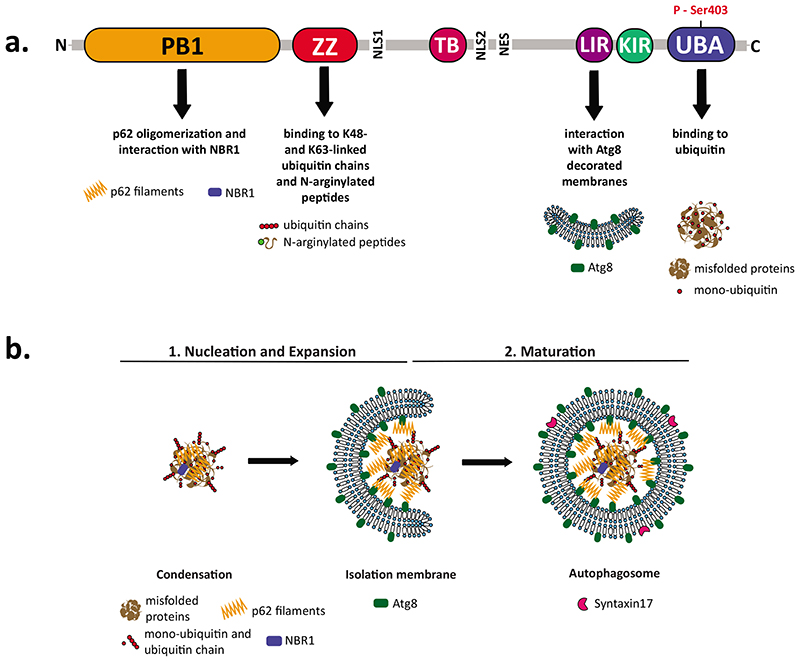
Fig. 2. p62 as a cargo receptor in selective autophagy.
a) Schematic overview of the domain structure of p62, which mediates its functions as indicated. Abbreviations: Phox and Bem1 domain (PB1, 21 – 103 amino acids (aa)); zinc-finger domain (ZZ, 128 - 163 aa); TRAF6 binding domain (TB, 225 - 250 aa); LC3-interacting region (LIR, 338 - 341 aa); Keap1-interacting region (KIR, 346 - 359 aa); ubiquitin-binding domain (UBA, 386 – 440 aa); nuclear localization signal (NLS1 and NLS2); nuclear export signal (NES). b) The mechanism of cargo tethering to the isolation membrane by p62. p62 filaments capture ubiquitinated substrates and link them to the Atg8 decorated isolation membrane to mediate their incorporation into autophagosomes. The NBR1 cargo receptor presumably aids p62 in this process.
Following its detection in ubiquitin-positive protein aggregates, p62 (also called sequestosome 1; SQSTM1) was the first mammalian cargo receptor identified for selective autophagy (Bjorkoy et al., 2005, Pankiv et al., 2007, Ichimura et al., 2008). Its importance for cellular quality control is demonstrated by mutations in the SQSTM1 gene, which have been associated with several diseases, including amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FD), neurodegeneration with ataxia, distal myopathy with rimmed vacuoles and Padget disease of the bone (Fecto et al., 2011, Goode et al., 2016, Haack et al., 2016, Rubino et al., 2012, Bucelli et al., 2015, Hocking et al., 2002). Studies in cells, mice and Drosophila have shown that p62 is necessary for the formation of the ubiquitin-positive condensates and their subsequent degradation (Komatsu et al., 2007, Nezis et al., 2008). Accordingly, knockout of genes that mediate autophagosome formation and their fusion with lysosomes all result in a marked increase of p62-positive condensates (Bartlett et al., 2011). p62 contains a number of domains and motifs that mediate its function as cargo receptor during aggrephagy (Fig. 2). The N-terminal Phox and Bem1 (PB1) domain drives its oligomerization into long helical oligomers that have the appearance of filaments (Ciuffa et al., 2015, Lamark et al., 2003). The PB1 domain also mediates the interaction with other PB1-containing proteins including the NBR1 cargo receptor (Kirkin et al., 2009a, Lamark et al., 2003). The PB1 domain is followed by a ZZ type zinc finger domain, which binds N-terminally arginylated proteins as well as K48- and K63-linked ubiquitin chains (Zaffagnini et al., 2018, Cha-Molstad et al., 2015). The LC3-interacting region (LIR) is localized in the central part of the molecule within an intrinsically disordered region (Pankiv et al., 2007, Ichimura et al., 2008), and the C-terminal UBA domain mediates its interaction with ubiquitin (Seibenhener et al., 2004).
Both, the UBA domain – ubiquitin and the LIR motif – Atg8 interactions are weak. The low affinity of the UBA domain for ubiquitin is in part due to its homo-dimerization, which is mutually exclusive with ubiquitin binding (Long et al., 2010, Long et al., 2008). Its affinity for ubiquitin can be increased by phosphorylation at Ser403 by the casein kinase 2 (CK2) and TBK1 (Matsumoto et al., 2011, Matsumoto et al., 2015, Pilli et al., 2011). The LIR motif of p62 has the sequence DDDWTHL and binds to LC3B in the low micromolar range (Novak et al., 2010, Pankiv et al., 2007, Ichimura et al., 2008). Oligomerization mediated by its PB1 domain allows p62 to avidly and selectively bind to cargoes, including misfolded proteins, on which ubiquitin is concentrated (Wurzer et al., 2015). A similar effect is seen for the LIR – LC3B interaction, where the PB1-mediated oligomerization results in a very high avidity interaction with membrane-concentrated LC3, such that the off-rate becomes practically zero (Wurzer et al., 2015). This tight interaction enables p62 to bend membranes around cargo material, a property that is conserved in the yeast Atg19 cargo receptor (Wurzer et al., 2015, Sawa-Makarska et al., 2014).
p62 is not merely required for the tethering of ubiquitinated proteins to the Atg8 coated isolation membrane, but also for the preceding condensation of these proteins into larger structures that subsequently become targets for autophagy. How do the different biochemical activities act together to mediate cargo condensation and isolation membrane tethering, and how can this be coordinated with the activity of the UPS and with the autophagy machinery? A number of exciting recent discoveries have given us fascinating clues about these processes and their crosstalk as discussed below.
p62-mediated phase separations as nucleators for autophagosomes
It is becoming increasingly clear that many subcellular structures and compartments are the result of phase-separation reactions that condensate the interacting molecules. These condensates can be the result of low affinity, but multivalent interactions and individual molecules can be highly mobile within them (Box 1) (Banani et al., 2017, Shin and Brangwynne, 2017).
Box 1.
Numerous cellular organelles such as the nucleolus, P granules, stress granules and PML bodies are stable entities even though they are not bound by membranes. It has become clear that many of these membrane-less organelles are the result of the phase separation reactions that mediate the condensation of biomolecules such as proteins, RNA or DNA into larger assemblies (Banani et al., 2017, Shin and Brangwynne, 2017, Brangwynne et al., 2017, Brangwynne et al., 2009). It has emerged that not only orgenelles but also the formation of more transient cellular condensates such as signalling puncta is based on phase separation reactions (Li et al., 2012). Within these condensates biomolecules can display different mobilities giving various condensates distinct physical properties. Condensates resulting from liquid-liquid phase separations show high mobility of the macromolecules that are concentrated within these structures (Banani et al., 2017, Shin and Brangwynne, 2017). While the exact molecular grammar underlying the formation of liquid-liquid phase separation is still not entirely clear, it appears that this phenomenon is often based on the interaction of polymers that undergo multivalent, low affinity interactions with each other (Banani et al., 2016). The presence of unstructured regions containing aromatic site chains also promotes liquid-liquid phase separation (Pak et al., 2016). Intriguingly, the filamentous oligomers of p62 and ubiquitin chains have exactly these properties. On the one hand, the ubiquitin-binding UBA domain of p62 binds ubiquitin with micromolar affinity and is linked to the rest of the p62 protein via a long unstructured region. Ubiquitin chains, on the other hand, harbor multiple interaction sites for p62.
When cells are exposed to proteotoxic stress, such as the inhibition of the proteasome, interference with productive translation or the inhibition of chaperones, ubiquitin-positive proteins are concentrated in μm-sized condensates (Bjorkoy et al., 2005). Interestingly, the formation of these condensates is largely dependent on p62, since its depletion results in a more dispersed distribution of the ubiquitinated proteins in cells (Demishtein et al., 2017, Bjorkoy et al., 2005, Kageyama et al., 2014). The condensation-promoting activity of p62 requires its PB1 and UBA domains, suggesting that the condensates are the result of the interaction of the ubiquitinated substrates with oligomeric, likely filamentous p62 (Bjorkoy et al., 2005, Ciuffa et al., 2015, Seibenhener et al., 2004, Zaffagnini et al., 2018). Two recent studies, including one from our group, demonstrated that p62 is entirely sufficient to phase separate ubiquitinated proteins into μm- sized condensates in vitro (Sun et al., 2018, Zaffagnini et al., 2018). Analogous to the situation in cells, this required the ability of p62 to oligomerize and to bind ubiquitin. The phase separation also required the presence of two or more substrate-attached ubiquitin chains that contain three or more ubiquitins, or very long free ubiquitin chains (Sun et al., 2018, Zaffagnini et al., 2018). Based on our electron microscopy data, we suggested that the formation of condensates is the result of p62 filaments that are crosslinked by the substrate (Zaffagnini et al., 2018).
Potential regulators of p62-ubiquitin phase separation reactions
Substrates modified with M1-linked (where the N-terminus of ubiquitin is linked to the C-terminal glycine of another ubiquitin), K48-linked and K63-linked ubiquitin chains are all able to trigger phase separation in vitro (Sun et al., 2018, Zaffagnini et al., 2018). p62 binds stronger to M1-linked and K63-linked chains compared to K48-linked chains (Long et al., 2008, Wurzer et al., 2015, Seibenhener et al., 2004). Consistently, K48-linked chains were less efficient in triggering phase separation, perhaps preventing p62 from interfering with proteasomal activity under normal conditions. In fact, we found that in vitro the formation of condensates is highly dependent on the concentration of the ubiquitinated substrate (Zaffagnini et al., 2018). Taken together, these recent results suggest that, in principle, the substrates for p62 are not fundamentally different from those of the UPS, but rather that their increased concentration, resulting for example from UPS overload, triggers the p62-mediated phase separation. Indeed, several other lines of evidence suggest that the UPS communicates with the p62 system (Fig. 3). High concentrations of free mono-ubiquitin inhibit condensation, likely by competitively interfering with p62 substrate-binding by interacting with the UBA domain (Zaffagnini et al., 2018). The levels of free monoubiquitin drop substantially when the proteasome is blocked, while at the same time, substrate-attached ubiquitin chains accumulate (Kaiser et al., 2011). Thus, proteasome inhibition decreases the concentration of an inhibitor of the p62 – ubiquitin phase separation, while at the same time, it increases the concentration of substrates triggering the reaction. Furthermore, free K63- and especially K48-chains, but not M1-linked chains inhibit the phase separation (Zaffagnini et al., 2018). Free ubiquitin chains are generated at the proteasome where, at least in yeast, the deubiquitinase Rpn11 cleaves off the ubiquitin chains from the substrate at the base (Lee et al., 2011, Finley, 2009). This activity has been linked to the proteolytic activity of proteasomes, and it is therefore possible that the levels of free ubiquitin chains signal proteasomal activity to the p62 system (Fig. 3). The mechanistic basis for how free K48- and K63-linked ubiquitin chains inhibit phase separation is currently unclear. However, this effect may be related to their ability to disassemble p62 oligomers owing to their interaction with the zinc-finger domain of p62, which is located close to the PB1 domain (Zaffagnini et al., 2018) (Fig. 2). Therefore, they may sterically interfere with oligomerization and consequently with phase separation, as shorter p62 filaments are less efficient to elicit the reaction. In general, little is known about the regulation of p62 filaments length in cells and this will be an important topic for future research.
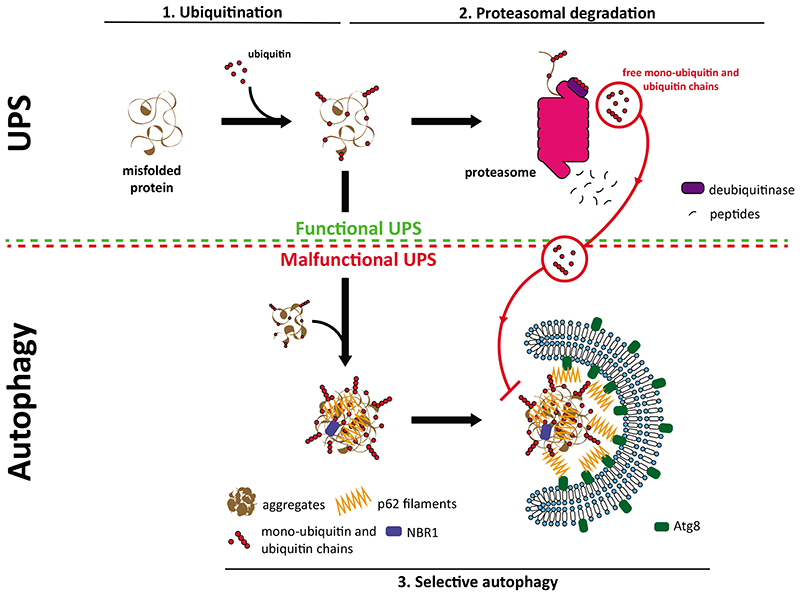
Fig. 3. Crosstalk between the UPS and selective autophagy.
In physiological conditions, misfolded and ubiquitinated proteins (1) are targeted mainly to the proteasome for their degradation (2). In response to proteotoxic stress and as consequence of proteasome overload or malfunction, misfolded proteins can no longer be degraded by the proteasome. In this situation, selective autophagy (3) serves as a backup mechanism and degrades these proteins. p62 filaments act at the intersection between the UPS and autophagy by sensing the accumulation of ubiquitinated substrates and mediate their phase separation into larger condensates. p62 oligomerization is a crucial step in this process and, in vitro, it is inhibited by free K48- and K63-linked ubiquitin chains. Free monoubiquitin also inhibits phase separation, likely by binding to the UBA domain of p62. Since free ubiquitin chains and monoubiquitin are generated by active proteasomes this provides a possible mechanism by which the selective autophagy of proteins and protein aggregates could be inhibited by the UPS.
In addition, it was shown that the zinc-finger domain binds N-terminally arginylated proteins (Cha-Molstad et al., 2015, Cha-Molstad et al., 2017, Yoo et al., 2018). N-terminal arginylation is a N-degron that triggers rapid substrate ubiquitination and degradation by the proteasome (Bachmair et al., 1986, Tasaki et al., 2012). Therefore, accumulation of N-terminally arginylated proteins may signal proteasomal overload to p62. Binding of N-terminally arginylated proteins to the zinc finger enhances oligomerization of p62 (Cha-Molstad et al., 2015, Cha-Molstad et al., 2017, Yoo et al., 2018), which, in turn, should facilitate phase separation of p62 and its ubiquitinated substrates. An interesting but unanswered question is whether p62 is able to simultaneously bind arginylated N-termini and ubiquitinated substrates.
Ubiquitin-binding and oligomerization of p62 are also the subject of regulation by posttranslational modification. Phosphorylation of serine 403 in the UBA domain by TBK1 or casein kinase 2 increases its affinity for ubiquitin and consequently the formation of p62-ubiquitin condensates in vitro and in cells (Matsumoto et al., 2011, Zaffagnini et al., 2018, Matsumoto et al., 2015, Pilli et al., 2011). Phosphorylation of serine 409 in the UBA domain by ULK1 occurs upon proteotoxic stress and increases the binding to ubiquitin by destabilizing the UBA-dimer interface (Lim et al., 2015). Additionally, the UBA domain is subject to ubiquitination at lysine 420, and this modification increases its ability to form condensates, likely because it interferes with the inhibitory homodimerization of the UBA domain, thereby activating ubiquitin binding (Peng et al., 2017, Lee et al., 2017). In contrast, ubiquitination of lysine 7 in the PB1 domain negatively regulates oligomerization and cargo sequestration (Pan et al., 2016). The ubiquitination of p62 may be affected by the overall levels of free ubiquitin in the cell, which decreases upon proteasome inhibition. It has also been shown that upon oxidative stress, oxidization of cysteine 105 and 113, which are located in the linker region between the PB1 domain and the zinc-finger domain, enhances oligomerization of p62 and the formation of condensates in cells (Carroll et al., 2018).
In addition to the role of p62 in substrate condensation and isolation membrane tethering in autophagy, p62 has been suggested to be a direct adaptor for the recruitment of substrates to the proteasome in the cytoplasm (Seibenhener et al., 2004). Furthermore, p62 contains a nuclear localization signal (NLS) and a nuclear export signal (NES) and shuttles between the nucleus and the cytoplasm. p62 has not only been suggested to export polyubiquitinated substrates from the nucleus for their degradation by autophagy in the cytoplasm, but also to attach ubiquitinated proteins to the proteasome in the nucleus (Hewitt et al., 2016, Pankiv et al., 2010). Moreover, there are indications that p62 mediates the degradation of proteasomes by autophagy upon starvation (Cohen-Kaplan et al., 2016).
Although these p62-ubiquitin condensates were for a long time considered rather passive aggregates that become linked to the autophagosomal membrane and subsequently degraded, this view has changed substantially by now. When the in vitro reconstituted p62-ubiquitin condensates were analyzed by fluorescence recovery after photobleaching (FRAP), surprisingly, it turned out that the ubiquitinated substrates showed fast recovery and by implication high mobility within the condensates and considerable exchange with the material in solution (Sun et al., 2018, Zaffagnini et al., 2018). In contrast, p62 displayed very low recovery, demonstrating that it is stably associated with the condensates (Sun et al., 2018, Zaffagnini et al., 2018). Thus, although the condensates form in a manner that is dependent on p62 and ubiquitin, the two interaction partners show a strikingly different behavior within the structures. The reason for the different mobility might be that owing to their size, the p62 filaments have a much lower diffusion coefficient. In addition, while the individual UBA – ubiquitin interactions may be highly transient, allowing the ubiquitinated proteins to diffuse, the p62 filaments have a number of binding sites and thus may be fixed because they are engaged in multiple interactions between UBA domains and ubiquitin at any given time. In cells, the situation might be even more complex. Endogenously-tagged p62 showed a higher exchange in condensates in cells compared to the in vitro formed condensates, but the FRAP recovery was still relatively low (Zaffagnini et al., 2018). The situation for the ubiquitinated substrates is far less clear, because until now, no imaging experiments with endogenous ubiquitinated substrates have been conducted. It is therefore possible that their mobility is lower in vivo than in vitro as some of them could be larger and more aggregated than the proteins used for the in vitro experiments. However, the condensates in vivo may undergo constant ATP-dependent remodeling of the ubiquitin chains by ubiquitin ligases and deubiquitinases (DUBs), chaperones or disaggregases, thereby increasing the mobility of the components in the condensates. The same factors may also regulate the stability and lifetime of the condensates in the cytoplasm. In cells, the p62 condensates have been observed by live cell imaging to undergo fusion followed by relaxation into a spherical shape (Sun et al., 2018). This observation is consistent with a liquid-like behavior of the condensates and thus with high mobility of the components within them.
A further aspect is that in vivo, ubiquitin-chain remodelling processes, or other ubiquitin-chain types, such as K11- and K33-linked ubiquitin chains, branched ubiquitin chains or heterotypic ubiquitin chains composed of multiple ubiquitin chain linkage types, may be additionally required or regulate efficient phase separation (Nibe et al., 2018, Yau et al., 2017, Meyer and Rape, 2015, Yau et al, 2017). As many ubiquitin-chain types have been found to be enriched in insoluble inclusions of autophagy-deficient mice, the relative contribution of specific ubiquitin chains to substrate condensation in cells has still to be elucidated (Riley et al., 2010). Because certain ubiquitin-chain types have a higher affinity for p62, it would, for example, be possible that substrates are specifically targeted to p62 by their modification with K63-linked chains, whereas substrates modified with K48-linked chains are only accepted when they accumulate owing to proteasomal overload.
Thus, multiple connections between aggrephagy and the UPS exists. These include the use of ubiquitin for marking the substrate, the recognition of N-terminal arginylation and the fact that p62 can act as adaptor for the proteasome and as autophagic cargo receptor (Kirkin et al., 2009b, Cha-Molstad et al., 2015, Seibenhener et al., 2004, Babu et al., 2005, Bjorkoy et al., 2005). Furthermore, at least in plants and yeast proteasomes can themselves become substrates for autophagy (Marshall et al., 2015, Marshall et al., 2016, Waite at al., 2016). Additionally, the level autophagy proteins including p62 can be regulated by the UPS (Platta et al., 2012, Myeku and Figueiredo-Pereira, 2011).
p62-ubiquitin condensates as substrates for autophagy
At least some of the p62-ubiquitin condensates become substrates for autophagy. Indeed, it was demonstrated that p62 is required to trigger autophagosome formation upon proteotoxic stress, proteasome inhibition and oxidative stress (Carroll et al., 2018, Demishtein et al., 2017, Bjorkoy et al., 2005, Pankiv et al., 2007, Babu et al., 2005). However, it was suggested that the condensates function to sequester ubiquitinated proteins in neuroblastoma cells (Sha et al., 2018). How substrate sequestration and autophagosome formation are coordinated is an important open question (Fig. 4). In cells, these activities are modulated by additional factors including ALFY, WDR81, Huntingtin, and the cargo receptor NBR1 (Clausen et al., 2010, Filimonenko et al., 2010, Rui et al., 2015, Liu et al., 2017), which promote p62-mediated substrate condensation and also interact with the autophagy machinery. Among these, NBR1 directly interacts with p62 via its PB1 domain, and in vitro, NBR1 has a direct stimulatory effect on substrate condensation by p62 (Zaffagnini et al., 2018). p62-ubiquitin condensates formed in vitro have also been shown to directly recruit LC3B showing that p62 is able to interact with LC3 when engaged in the phase separation reaction (Sun et al., 2018, Zaffagnini et al., 2018. Interestingly, the addition of LC3B to phase separation reactions slowed down condensation (Zaffagnini et al., 2018). This effect is likely mediated by a LC3B-mediated masking of the LIR motif of p62, which is required for efficient phase separation (Zaffagnini et al., 2018). The mechanistic basis for the role of the LIR motif in cargo condensation is currently unclear, but this observation suggests that substrate condensation and autophagosome formation are coordinated, such that the recruitment of the autophagy machinery slows down cargo condensation. It is further possible that some structural rearrangement of the p62 filaments occurs when the Atg8-decorated isolation membrane forms at the condensates such that p62 can efficiently link the condensates to it.
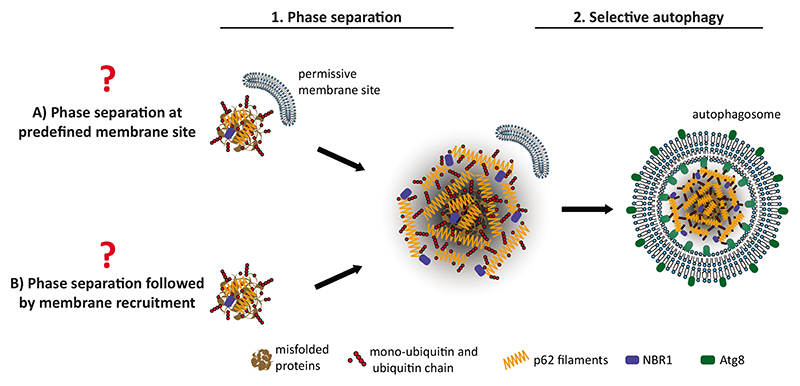
Fig. 4. p62 phase separation in selective autophagy.
Degradation of p62-ubiquitin condensates that form as a result of phase separation reactions. It is currently unclear if the phase separation reaction occurs at predefined, membrane proximal sites that are later on transformed into autophagosomes (A), or if the phase separation of p62 and ubiquitinated substrates can occur anywhere in the cells followed by their recruitment to a membrane site that is permissive for autophagosome formation (B).
p62 has also been demonstrated to mediate the autophagic degradation of stress granules, which are condensates originating from phase separation reactions (Buchan et al., 2013, Molliex et al., 2015). In contrast to aggrephagy, these structures form in a p62 independent manner and it is therefore unclear if the coordination of phase separation and autophagy machinery recruitment follows similar principles.
It is becoming increasingly clear that the cargo material has an active role in the induction and formation of autophagosomes in selective, starvation-independent autophagy (Zaffagnini and Martens, 2016). This activity is conferred by the cargo receptors upon recognition of their cargo. For example, in S. cerevisiae, the Atg19 cargo receptor recruits the scaffold protein Atg11 and the Atg12–Atg5-Atg16 complex to the prApe1 cargo in the cytoplasm-to-vacuole targeting (Cvt) pathway (Fracchiolla et al., 2016, Kamber et al., 2015, Shintani et al., 2002, Torggler et al., 2016). Optineurin and NDP52 recruit the autophagy machinery to damaged mitochondria in mammalian cells (Lazarou et al., 2015). Furthermore, ER proteins have been shown to recruit the autophagy machinery through the Atg11/FIP200 scaffold proteins (Khaminets et al., 2015, Mochida et al., 2015, Smith et al., 2018). In aggrephagy, the ALFY protein was shown to bind ATG5 and phosphatidylinositol 3-phosphate (PI3P) (Filimonenko et al., 2010) and may therefore promote isolation membrane elongation, whereas Huntingtin interacts with ULK1 that is required autophagosome initiation (Rui et al., 2015).
However, with regard to p62 as a cargo receptor, important questions remain. For instance, it is unclear whether p62 itself as a major component of the condensates is also able to interact with the upstream autophagy machinery. It is also unknown if the p62– ubiquitin condensates are formed at predefined sites that provide membranes or lipids that are permissive for autophagosome formation (Fig. 4A), or if the membrane source is recruited to the condensates after their formation (Fig. 4B).
Future Perspectives
It has become clear that the UPS and autophagy are the main systems for the degradation of misfolded proteins in the cytoplasm. Moreover, it is now generally appreciated that the two systems are highly interlinked and that they use the same signals on their targets. While it appears that there is no single, specific signal that specifically targets substrates to either the UPS or autophagy, we still do not fully understand how the decision about the fate of the substrate is made. A key factor in this regard is p62 owing to its ability to phase separate ubiquitinated proteins into larger condensates, which subsequently become targets for autophagy. It remains unclear how cargo condensation and the recruitment of the autophagy machinery are mechanistically and temporally linked. Furthermore, if p62-ubiquitin condensates additionally function to sequester misfolded proteins in the cytoplasm rather than to mediate their immediate degradation as has been suggested, this raises the question of how the switch from sequestration to recruitment to the autophagy machinery is regulated and how this can be coupled to the activity of the UPS? With regard to p62 mediated phase transition, it is possible that it also plays a role in other forms of selective autophagy apart from aggrephagy such as xenophagy or mitophagy, perhaps by aiding the concentration of autophagy proteins at the cargo. Given these important questions, exciting times are ahead for the field.
Acknowledgments
We thank Andreas Bachmair for comments on the manuscript. S.M. is supported by an ERC grant (No.646653) and by the Austrian Science Fund (FWF, No. P30401-B21 and W1261).
References
- Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. Compositional Control of Phase-Separated Cellular Bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett BJ, Isakson P, Lewerenz J, Sanchez H, Kotzebue RW, Cumming RC, Harris GL, Nezis IP, Schubert DR, Simonsen A, Finley KD. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy. 2011;7:572–83. doi: 10.4161/auto.7.6.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, JüLicher F, Hyman AA. Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science. 2009;324:1729. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences. 2011;108:4334. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucelli RC, Arhzaouy K, Pestronk A, Pittman SK, Rojas L, Sue CM, Evila A, Hackman P, Udd B, Harms MB, Weihl CC. SQSTM1 splice site mutation in distal myopathy with rimmed vacuoles. Neurology. 2015;85:665–74. doi: 10.1212/WNL.0000000000001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B, Otten EG, Manni D, Stefanatos R, Menzies FM, Smith GR, Jurk D, Kenneth N, Wilkinson S, Passos JF, Attems J, et al. Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis. Nat Commun. 2018;9:256. doi: 10.1038/s41467-017-02746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Sung KS, Hwang J, Kim KA, Yu JE, Yoo YD, Jang JM, Han DH, Molstad M, Kim JG, Lee YJ, et al. Aminoterminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. 2015;17:917–29. doi: 10.1038/ncb3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Yu JE, Feng Z, Lee SH, Kim JG, Yang P, Han B, Sung KW, Yoo YD, Hwang J, Mcguire T, et al. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat Commun. 2017;8:102. doi: 10.1038/s41467-017-00085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Ciuffa R, Lamark T, Tarafder AK, Guesdon A, Rybina S, Hagen WJ, Johansen T, Sachse C. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. 2015;11:748–58. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- Clark SL., JR Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J Biophys Biochem Cytol. 1957;3:349–62. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–44. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- Cohen-Kaplan V, Livneh I, Avni N, Fabre B, Ziv T, Kwon YT, Ciechanover A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci U S A. 2016;113:E7490–E7499. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–92. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Demishtein A, Fraiberg M, Berko D, Tirosh B, Elazar Z, Navon A. SQSTM1/p62-mediated autophagy compensates for loss of proteasome polyubiquitin recruiting capacity. Autophagy. 2017;13:1697–1708. doi: 10.1080/15548627.2017.1356549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–37. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. Proteasomal and Autophagic Degradation Systems. Annu Rev Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- Ebner P, Poetsch I, Deszcz L, Hoffmann T, Zuber J, Ikeda F. The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat Commun. 2018;9:599. doi: 10.1038/s41467-018-02823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, Donkervoort S, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–79. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracchiolla D, Sawa-Makarska J, Zens B, Ruiter A, Zaffagnini G, Brezovich A, Romanov J, Runggatscher K, Kraft C, Zagrovic B, Martens S. Mechanism of cargo-directed Atg8 conjugation during selective autophagy. Elife. 2016;5 doi: 10.7554/eLife.18544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–64. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode A, Butler K, Long J, Cavey J, Scott D, Shaw B, Sollenberger J, Gell C, Johansen T, Oldham NJ, Searle MS, et al. Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy. 2016;12:1094–104. doi: 10.1080/15548627.2016.1170257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Haack TB, Ignatius E, Calvo-Garrido J, Iuso A, Isohanni P, Maffezzini C, Lonnqvist T, Suomalainen A, Gorza M, Kremer LS, Graf E, et al. Absence of the Autophagy Adaptor SQSTM1/p62 Causes Childhood-Onset Neurodegeneration with Ataxia, Dystonia, and Gaze Palsy. Am J Hum Genet. 2016;99:735–743. doi: 10.1016/j.ajhg.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus K, Takats S, Kovacs AL, Juhasz G. Evolutionarily conserved role and physiological relevance of a STX17/Syx17 (syntaxin 17)-containing SNARE complex in autophagosome fusion with endosomes and lysosomes. Autophagy. 2013;9:1642–6. doi: 10.4161/auto.25684. [DOI] [PubMed] [Google Scholar]
- Hewitt G, Carroll B, Sarallah R, Correia-Melo C, Ogrodnik M, Nelson G, Otten EG, Manni D, Antrobus R, Morgan BA, Von Zglinicki T, et al. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy. 2016;12:1917–1930. doi: 10.1080/15548627.2016.1210368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, Nicholson GC, Ward L, Bennett ST, Wuyts W, Van Hul W, et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s disease. Hum Mol Genet. 2002;11:2735–9. doi: 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–9. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Young LN. Mechanisms of Autophagy Initiation. Annu Rev Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112(Pt 22):4079–87. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–57. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–69. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–41. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–37. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R, Kouno T, Bedford L, Mayer RJ, Lee MS, Yamamoto M, et al. Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J Biol Chem. 2014;289:24944–55. doi: 10.1074/jbc.M114.580357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods. 2011;8:691–6. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber RA, Shoemaker CJ, Denic V. Receptor-Bound Targets of Selective Autophagy Use a Scaffold Protein to Activate the Atg1 Kinase. Mol Cell. 2015;59:372–81. doi: 10.1016/j.molcel.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, Mauthe M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–8. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, Mcewan DG, Clausen TH, Wild P, Bilusic I, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009a;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Mcewan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009b;34:259–69. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990;111:941–53. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Martens S. Mechanisms and regulation of autophagosome formation. Curr Opin Cell Biol. 2012;24:496–501. doi: 10.1016/j.ceb.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–81. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.R110.003871. R110 003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Chou TF, Pittman SK, Keith AL, Razani B, Weihl CC. Keap1/Cullin3 Modulates p62/SQSTM1 Activity via UBA Domain Ubiquitination. Cell Rep. 2017;19:188–202. doi: 10.1016/j.celrep.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, Komatsu M, Oh YJ, Zhao Y, Yue Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015;11:e1004987. doi: 10.1371/journal.pgen.1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li Y, Wang X, Xing R, Liu K, Gan Q, Tang C, Gao Z, Jian Y, Luo S, Guo W, et al. The BEACH-containing protein WDR81 coordinates p62 and LC3C to promote aggrephagy. J Cell Biol. 2017;216:1301–1320. doi: 10.1083/jcb.201608039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–40. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- Long J, Garner TP, Pandya MJ, Craven CJ, Chen P, Shaw B, Williamson MP, Layfield R, Searle MS. Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-kappaB signalling. J Mol Biol. 2010;396:178–94. doi: 10.1016/j.jmb.2009.11.032. [DOI] [PubMed] [Google Scholar]
- Marshall Richard S, Li F, Gemperline David C, Book Adam J, Vierstra Richard D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Molecular Cell. 2015;58:1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Mcloughlin F, Vierstra RD. Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone. Cell Reports. 2016;16:1717–1732. doi: 10.1016/j.celrep.2016.07.015. [DOI] [PubMed] [Google Scholar]
- Matsui T, Jiang P, Nakano S, Sakamaki Y, Yamamoto H, Mizushima N. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. The Journal of Cell Biology. 2018;217:2633. doi: 10.1083/jcb.201712058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–42. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–89. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Mcewan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda De Stegmann D, Bhogaraju S, Maddi K, Kirchof A, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–62. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Meyer H-J, Rape M. Enhanced Protein Degradation by Branched Ubiquitin Chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–62. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj Anderson P, Kim Hong J, Mittag T, Taylor JP. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–6. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- Myeku N, Figueiredo-Pereira ME. Dynamics of the Degradation of Ubiquitinated Proteins by Proteasomes and Autophagy: ASSOCIATION WITH SEQUESTOSOME 1/p62. Journal of Biological Chemistry. 2011;286:22426–22440. doi: 10.1074/jbc.M110.149252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–71. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibe Y, Oshima S, Kobayashi M, Maeyashiki C, Matsuzawa Y, Otsubo K, Matsuda H, Aonuma E, Nemoto Y, Nagaishi T, Okamoto R, et al. Novel polyubiquitin imaging system, PolyUb-FC, reveals that K33-linked polyubiquitin is recruited by SQSTM1/p62. Autophagy. 2018;14:347–358. doi: 10.1080/15548627.2017.1407889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Kirkin V, Mcewan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Pak Chi W, Kosno M, Holehouse Alex S, Padrick Shae B, Mittal A, Ali R, Yunus Ali A, Liu David R, Pappu Rohit V, Rosen Michael K. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Molecular Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JA, Sun Y, Jiang YP, Bott AJ, Jaber N, Dou Z, Yang B, Chen JS, Catanzaro JM, Du C, Ding WX, et al. TRIM21 Ubiquitylates SQSTM1/p62 and Suppresses Protein Sequestration to Regulate Redox Homeostasis. Mol Cell. 2016;61:720–733. doi: 10.1016/j.molcel.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285:5941–53. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Yang J, Li G, You Q, Han W, Li T, Gao D, Xie X, Lee BH, Du J, Hou J, et al. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27:657–674. doi: 10.1038/cr.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun J-A, et al. TBK-1 Promotes Autophagy-Mediated Antimicrobial Defense by Controlling Autophagosome Maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta Harald W, Abrahamsen H, Thoresen Sigrid B, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochemical Journal. 2012;441:399. doi: 10.1042/BJ20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–11. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, Yamamoto M, et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol. 2010;191:537–52. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–78. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- GROUP, T. S. Rubino E, Rainero I, Chio A, Rogaeva E, Galimberti D, Fenoglio P, Grinberg Y, Isaia G, Calvo A, Gentile S, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–62. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, Cuervo AM, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17:262–75. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa-Makarska J, Abert C, Romanov J, Zens B, Ibiricu I, Martens S. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat Cell Biol. 2014;16:425–433. doi: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–31. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–68. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z, Schnell HM, Ruoff K, Goldberg A. Rapid induction of p62 and GABARAPL1 upon proteasome inhibition promotes survival before autophagy activation. J Cell Biol. 2018;217:1757–1776. doi: 10.1083/jcb.201708168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, Von Kriegsheim A, Behrends C, Wilkinson S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell. 2018;44:217–232.:e11. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC, JR, Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu Rev Biophys. 2014;43:357–79. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Wu R, Zheng J, Li P, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–415. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, Bazett-Jones DP, Brumell JH. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–99. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K, Kovacs AL, Hegedus K, Juhasz G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531–9. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–54. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–89. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–80. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, Von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–21. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Torggler R, Papinski D, Brach T, Bas L, Schuschnig M, Pfaffenwimmer T, Rohringer S, Matzhold T, Schweida D, Brezovich A, Kraft C. Two Independent Pathways within Selective Autophagy Converge to Activate Atg1 Kinase at the Vacuole. Mol Cell. 2016;64:221–235. doi: 10.1016/j.molcel.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Van Wijk SJ, Timmers HT. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24:981–93. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- Waite KA, Mota-Peynado AD-L, Vontz G, Roelofs J. Starvation Induces Proteasome Autophagy with Different Pathways for Core and Regulatory Particles. Journal of Biological Chemistry. 2016;291:3239–3253. doi: 10.1074/jbc.M115.699124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzer B, Zaffagnini G, Fracchiolla D, Turco E, Abert C, Romanov J, Martens S. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife. 2015;4:e08941. doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–86. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, Rape M. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell. 2017;171:918–933.:e20. doi: 10.1016/j.cell.2017.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YD, Mun SR, Ji CH, Sung KW, Kang KY, Heo AJ, Lee SH, An JY, Hwang J, Xie XQ, Ciechanover A, et al. N-terminal arginylation generates a bimodal degron that modulates autophagic proteolysis. Proc Natl Acad Sci U S A. 2018;115:E2716–E2724. doi: 10.1073/pnas.1719110115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. J Mol Biol. 2016;428:1714–24. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G, Savova A, Danieli A, Romanov J, Tremel S, Ebner M, Peterbauer T, Sztacho M, Trapannone R, Tarafder AK, Sachse C, et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018;37 doi: 10.15252/embj.201798308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–16. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]