Abstract
Mitochondria are essential for eukaryotic live. These double-membrane organelles often form highly dynamic tubular networks interacting with many cellular structures. Their highly convoluted contiguous inner membrane compartmentalizes the organelle, which is crucial for mitochondrial function. Since the diameter of the mitochondrial tubules is generally close to the diffraction limit of light microcopy, it is often challenging, if not impossible, to visualize sub-mitochondrial structures or protein distributions using conventional light microscopy. This renders super-resolution microscopy particularly valuable, and attractive, to study mitochondria. Super-resolution microscopy encompasses a diverse set of approaches that extend the resolution as well as nanoscopy techniques that even overcome the diffraction limit. In this review, we provide an overview on recent studies using super-resolution microscopy to investigate mitochondria, discuss the strengths and opportunities of the various methods to address specific questions in mitochondrial biology and highlight potential future developments.
Keywords: mitochondria, live-cell microscopy, nanoscopy, super-resolution microscopy
Introduction
Mitochondria, ancient double-membrane organelles, are essential for eukaryotic life (1). They are the ‘powerhouses of the cell’, as they harbor the oxidative phosphorylation (OXPHOS) system that synthesizes the vast majority of the ATP required by cellular processes. Besides, they have a multitude of other cellular functions, including iron-sulfur cluster synthesis, β-oxidation of fatty acids, biosynthesis of haem and of several phospholipids and other metabolites. Mitochondria also play a key role in apoptosis and they participate in developmental processes as well as in ageing. Indeed, there is increasing evidence that numerous devastating human diseases are associated with mitochondrial dysfunctions (2).
In many cell types, mitochondria form loose and dynamic networks of tubules that constantly move, fuse and divide (3–5). These organelles exhibit a smooth outer membrane and a highly folded inner membrane (6–8). The mitochondrial outer membrane primarily harbors the machinery that is required for the communication and physical interactions with other cellular structures. The contiguous inner membrane can be structurally and functionally sub-divided in at least two domains, namely the inner boundary membrane, which parallels the outer membrane, and invaginations of varying shapes, termed cristae (9–13). The inner boundary membrane and crista membranes are connected by crista junctions, narrow tubules or slits, that presumably act as selective barriers for proteins, lipids and metabolites moving in and out of the crista membrane or the crista lumen (14). The intricate architecture of the inner membrane is crucial for proper function of these organelles in cellular metabolism and apoptosis.
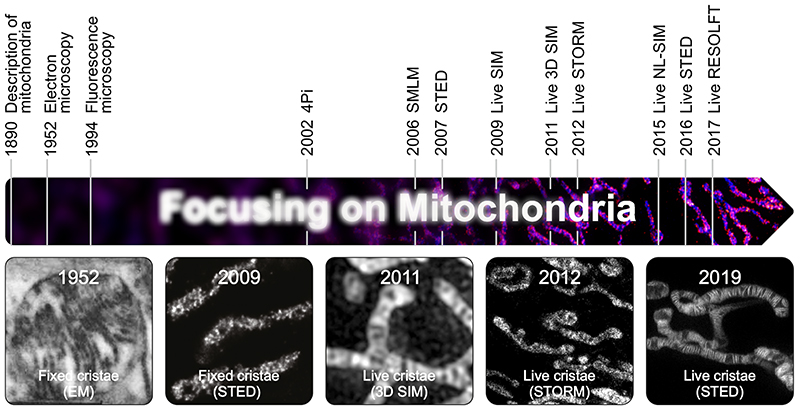
The unique membrane architecture of mitochondria was discovered in the 1950s by the use of transmission electron microscopy, at the time a new enabling technology for cell biology (6; 7) (Fig. 1). Subsequently, it took more than half a century to develop another enabling technology, namely super-resolution microscopy (15–17), facilitating nanoscale resolution also in optical microscopy. As a consequence of this technological advance, researchers can now observe mitochondrial cristae even in living cells (Fig. 1).
Fig. 1. Mitochondria in the focus of microscopy.
Highlighted are important milestones for visualization of mitochondria. Given are the years when a certain imaging method was first used to visualize mitochondria. The images show (from left to right): One of the first images of cristae recorded by EM (6). First light microscopy image of fixed cristae taken by isoSTED nanoscopy (68). First visualization of mitochondrial inner membrane structures in living cells by 3D SIM (53). First live-cell SMLM of the mitochondrial inner membrane (52). One of the first images of individual mitochondrial cristae recorded in living cells using STED nanoscopy (71).
Indeed, mitochondria are challenging objects for optical microscopy, not only because they are very dynamic and sensitive against many stresses in living cells, but also because they are small. Although the mitochondrial tubules can be several μm long, their diameter is typically between 200 and 700 nm, which is around the diffraction limit that restricts the attainable resolution in classical (fluorescence) microscopy (18; 19). Hence, with classical optical microscopy, it is always challenging, often even impossible, to visualize sub-mitochondrial structures. This review aims at providing an overview on recent developments in using super-resolution microscopy to study mitochondria on the nanoscale.
Super-Resolution Microscopy, Nanoscopy and Extended-Resolution Microscopy
The terms ‘super-resolution microscopy’ and ‘nanoscopy’ are sometimes used synonymously, whereas in other contexts distinctions are made. In this review, we will use these terms in the following way: ‘Nanoscopy’ is strictly used for microscopies that fundamentally overcome the diffraction barrier. This includes, amongst others, approaches such as (fluorescence) photo-activated localization microscopy [(f)PALM] (20; 21), stochastic optical reconstruction microscopy [STORM] (22), points accumulation for imaging in nanoscale topography [PAINT] nanoscopy (23), non-linear structured illumination microscopy [NL-SIM] (24), reversible saturable/switchable optical linear (fluorescence) transitions [RESOLFT] nanoscopy (25; 26), and stimulated emission depletion [STED] nanoscopy (27; 28). We use the term ‘extended-resolution microscopy’ (29) to refer to approaches that improve the spatial resolution compared with conventional confocal or widefield fluorescence microscopy, but that do not overcome the diffraction barrier. These methods typically attain a lateral resolution of around 100 nm. Hence, the terms ‘extended-resolution microscopy’ and ‘nanoscopy’ are non-overlapping. Examples for extended-resolution microscopy include linear SIM (30), image scanning microscopy [ISM]/AiryScan microscopy (31; 32), lattice light-sheet microscopy (33), super-resolution optical fluctuation imaging [SOFI] (34), and many others, including further fluctuation-based methods and computer-vision-based methods (for an overview see (35; 36)). In this review, we will use the term ‘super-resolution microscopy’ as a general term for extended-resolution microscopy and nanoscopy.
Next to the methods improving the optical resolution, the recently developed expansion microscopy circumvents the problem of limited spatial resolution by physically increasing the size of the sample (37; 38). With this, it achieves a practical resolution in the nanoscopy regime. Inevitably, this method is restricted to fixed specimen. Indeed, mitochondria were among the first organelles analyzed with expansion microscopy, achieving a detailed imaging of membrane protein distributions and cristae structures (39). Still, to our knowledge, expansion microscopy has not been used to address questions of mitochondrial biology, yet. Consequently, expansion microscopy will not be covered in this review.
Improving the Resolution in Far-Field Light Microscopy
In 1873, Ernst Abbe postulated that the attainable spatial resolution of light microscopy is fundamentally limited (18). He showed that due to the diffraction limit, visible light cannot be focused into an infinitesimal small focal area but only to a spot of about 200 nm width in lateral dimensions und about 500 nm length along the z-axis. As a result, fluorophores within such a diffraction-limited focal spot will all be excited and detected together, and are therefore inseparable in conventional light microscopy. Abbe’s discovery proved to be correct. However, it holds true only under the conditions met by conventional (fluorescence) light microscopy (29). Over the last two decades, several novel microscopy approaches have been implemented to extend the attainable resolution, and even to shatter this resolution limit.
Extending the Resolution
The super-resolution methods that enhance the resolution compared to the classical diffraction limit, but do not overcome the barrier, are a diverse set of microscopies based on widefield, total internal reflection fluorescence [TIRF] or confocal microscopy (for an overview on the methods see (36)). For imaging mitochondria, SIM and ISM seem to be the most popular microscopy approaches with extended resolution. SIM relies on interference-based periodic light patterns that are projected onto the sample. In SIM, after recording several images for different positions and orientations of the excitation pattern, a final image with about doubled resolution is calculated by applying a reconstruction algorithm (30; 40; 41). SIM is related to ISM (or AiryScan microscopy), which uses a similar physical principle to increase the resolution in a beam scanning microscope (for detailed review see (42)). In ISM, an excitation focus is scanned across the specimen and the fluorescence is recorded by a multi-pixel detector array (for example a camera) in the widefield detection mode. Subsequently, from the many images, each taken at an individual scan position, a final image is calculated. ISM can nearly double the resolution.
Compared to nanoscopy, these extended-resolution microscopies attain only a relative modest spatial resolution, but generally require also only modest light powers and provide fast recoding times, so that they are often considered as rather benign to living cells, and are particularly geared to live-cell imaging approaches. On the other side, most extended-resolution microscopies including SIM require mathematical post processing, which demands significant expertise to detect and counteract reconstruction artifacts (36; 43).
Overcoming the Diffraction Limit
The fact that all fluorophores in a diffraction-limited volume are excited together and therefore fluoresce together ultimately limits the resolution of conventional fluorescence microscopy. The key to fundamentally overcome the diffraction limit is to distinguish fluorophores residing in a diffraction-limited volume by keeping them in separable states (25; 27). To this end, molecules are switched between two states, typically a fluorescent ‘on’- and a non-fluorescent ‘off’-state. Most approaches transfer the majority of the fluorophores in a diffraction-limited volume into the off-state and record few or even a single molecule at a time. This can be achieved by two families of approaches, namely coordinate-targeted nanoscopy and coordinate-stochastic nanoscopy. Coordinate-targeted nanoscopies are scanning approaches with STED nanoscopy (27; 28), RESOLFT nanoscopy (26; 44; 45) and the related NL-SIM (24; 46) being the most prominent methods in this category. Here, the fluorophores are reversibly driven by light between two states. In STED and RESOLFT nanoscopy, a light pattern defining one, several or many minima is used to define sub-diffraction regions at which no off-switching occurs so that the fluorophores can fluoresce only in these regions. In most STED and RESOLFT microscopes a single minimum, or zero, is generated by a focal spot of the shape of a doughnut. This minimum is scanned across the specimen to generate an image with sub-diffraction resolution. The attainable resolution depends on the geometry of the minima and the efficiency of the off-switching process and thereby can be adjusted to the imaging needs. In cellular samples, the attainable resolution is typically between 30 and 50 nm (16). Unlike NL-SIM, which relies on a wide-field detection, beam-scanning STED and RESOLFT nanoscopy require no additional computational efforts to reconstruct the final images.
Coordinate-stochastic nanoscopy methods, often also called single molecule localization microscopy [SMLM], such as (f)PALM (20; 21), STORM (22), PAINT (23), or one of the related approaches (17; 36; 47), are widefield approaches that rely on the localization of individual molecules that have been stochastically turned into a fluorescent on-state. When isolated in space, the positions of single fluorophores can be determined with a precision in the nanometer range. In order to obtain an image reflecting the distribution of all fluorophores, the process of stochastically switching molecules in the on-state and subsequently switching them off (or bleaching them) has to be iterated numerous times, until enough localization events have been determined to reconstruct an image.
Mitochondria as Targets to Evaluate the Resolving Power of Different Super-Resolution Microscopies
Fluorescently labeled cellular structures, notably microtubules and nuclear pores, because of their constant size, geometry and brightness after labeling, are abundantly used targets to evaluate the potential and limitations of various fluorescence microscopies. Likewise, mitochondria have been used frequently as targets to evaluate microscopy techniques; they often served to analyze different characteristics of an imaging scheme, because of the organelle’s complex architecture, structural heterogeneity and dynamics. Due to their sensitivity against photostress, they are also good targets to evaluate the phototoxicity of a given method.
Already in the early 2000s, it was shown that 4Pi-microscopy, which increases the resolution along the axial axis to ~100 nm, enabled a better representation of the complex mitochondrial network in living and fixed cells (48–50). Similarly, early demonstrations of SIM relied on the imaging of mitochondria (51). Introducing 3D SIM, Mats Gustafsson and colleagues were the first to show mitochondrial substructures that were reminiscent of cristae in living HeLa cells (Fig. 1) (52). This approach was later extended to dual-color 3D recordings (53), and with improved data processing, even single cristae could be visualized with an impressive temporal resolution (54). Numerous other studies reporting on developments in SIM and related approaches relied on imaging the dynamics of mitochondria labeled either with dyes or fluorescent proteins (46; 55–57).
The first nanoscopy images of chemically fixed mitochondria were reported in the study introducing PALM (20), relying on the photoconvertible protein mEosFP that was targeted to the mitochondrial matrix. Most subsequent studies using SMLM approaches to visualize mitochondrial proteins also recorded chemically fixed samples (58–60). Notable exceptions relied on the use of several different MitoTracker dyes, which were shown to be live-cell compatible photoswitchable membrane probes (61) (Fig. 1), as well as on a photoactivatable ligand of the Halo-tag (62). Recently, mitochondria, even in thick fixed tissues, have been used as targets for studies relying on PAINT nanoscopy (63; 64).
Also, STED nanoscopy was used early on to visualize mitochondrial proteins. The first dual-color STED study reported on the distribution of the translocase of the outer membrane [TOM] complex (65). Numerous studies followed (66; 67), including the first visualization of (chemically fixed) cristae using isoSTED nanoscopy (Fig. 1) (68), the visualization of mitochondria in bio-banked human tissues (69) and live-cell recordings of mitochondria (70). Recently, the visualization of single cristae in living cells was reported using STED nanoscopy relying on a SNAP-tag fusion protein (Fig. 1), or an inner membrane specific fluorescent probe, respectively (71; 72).
The development of resolution-extended microscopy and diffraction-unlimited nanoscopy in the last two decades allowed addressing questions that were just out of reach before. In the following, we will discuss some recent studies covering topics of mitochondrial biology that benefited from super-resolution microscopy. Given the rapid growth of the field, inescapably, this snapshot represents a selection of studies which is by no means comprehensive.
Mitochondrial Architecture
Protein Distributions Within the Inner Membrane
Early on, the intricate fold of the inner membrane suggested that the crista membranes and the inner boundary membrane have not only different functions, but also different protein compositions. Initially, such different protein localizations were demonstrated by immunogold electron microscopy (11; 73) and by the use of conventional light microscopy on genetically enlarged mitochondria (12). These approaches were also used to demonstrate that proteins move in and out of the crista membrane depending on the cellular conditions (11; 74; 75). By now, several studies using various forms of super-resolution microscopy have confirmed the existence of heterogeneous protein distributions within the inner membrane (60; 76). Accordingly, single molecule tracking of inner membrane proteins suggested that individual cristae represent diffusion-restricting microcompartments (Fig. 2A) (77). A recent study suggested that individual cristae within the same mitochondrion exhibit different membrane potentials, further supporting the view that crista junctions might represent diffusion barriers (78; 79).
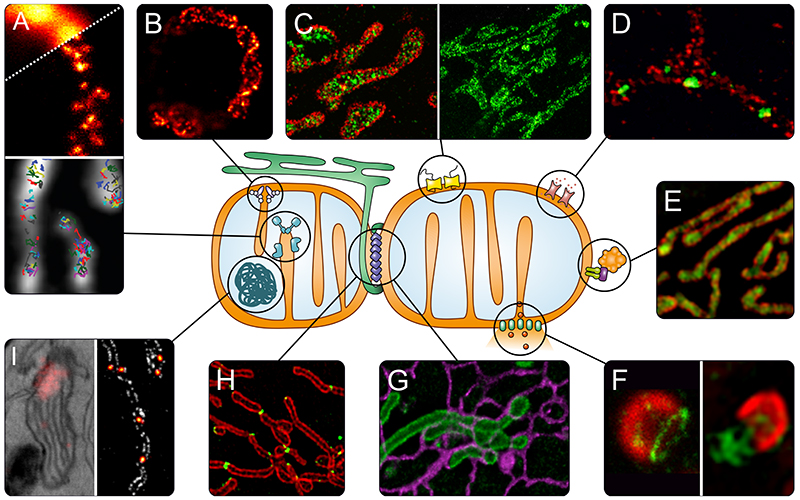
Fig. 2. Super-resolution microscopy to address questions of mitochondrial biology.
A) OXPHOS. Upper panel: Distribution of assembly factors recorded with STED nanoscopy (80). Lower panel: Tracking of single OXPHOS subunits using SMLM (77). B) Localization of the MICOS subunit Mic60 in a yeast cell, revealed by STED nanoscopy (89). C) TOM complex in the outer membrane recorded with DNA-PAINT nanoscopy (left, TOM complex in red) (63) and STED nanoscopy (right) (67). D) Differential distributions of the three human hVDAC (mitochondrial porin) isoforms in the outer membrane of human mitochondria (STED nanoscopy, hVDAC in green) (66). E) Sub-mitochondrial localization of PINK1 (green) shown by 3D-SIM (76). F) Apoptosis. Left: Several pro-apoptotic BAX proteins (green) form ring like structures in the outer membrane that may act as pores (STED nanoscopy) (100). Right: During later steps of apoptosis, these large BAX assemblies facilitate the herniation of the inner membrane and release of mtDNA (green) as shown by SIM (102). G) Interactions between mitochondria (green) and the ER (purple) in living cells recorded by STED nanoscopy (70). H) Spatial dynamics of the dynamin-like GTPase DRP1 (green), which is essential for mitochondrial fission, visualized by SIM (109). I) Nucleoids, analyzed with correlative PALM and EM (left panel) (131), and with STED nanoscopy (right panel, nucleoids in fire) (unpublished).
An example for the spatial segregation of protein distributions and functions within the inner membrane are the Uncoupling Protein 4 (UCP4) and the F1FO-ATP synthase, which both can dissipate the proton gradient across the inner membrane. dSTORM revealed that UCP4 and the F1FO-ATP synthase are spatially separated to the inner boundary membrane and to the crista membranes, respectively (60). Likely, this spatial separation to different domains prevents UCP4 from uncoupling oxidative phosphorylation (OXPHOS) from F1FO-ATP synthase mediated proton pumping under normal cellular conditions; UCP4 might only lower the proton motive force if it exceeds a potentially damaging threshold.
OXPHOS is vital for the regeneration of the vast majority of ATP in eukaryotic cells. In higher eukaryotes, it is carried out by five large multi-subunit protein complexes. The F1FO-ATP synthase is the terminal complex V of the OXPHOS system. In mammals, four of the five OXPHOS complexes are hybrids of subunits encoded in both the nuclear and mitochondrial genomes, and, as such, the assembly of these dual-origin complexes is an enormous logistical challenge for the cell. The fact that the OXPHOS complexes are primarily localized in the crista membranes renders this logistic challenge even more difficult. The assembly of the OXPHOS complexes in yeast mitochondria seems to be rather uniformly distributed along the mitochondrial tubules, as shown by STED nanoscopy (Fig. 2A) (80), suggesting that here OXPHOS assembly occurs in young as well as in old mitochondria.
Still, our understanding of functional domains within the mitochondrial inner membrane is rather rudimentary. Many aspects, such as smaller sub-domains which might be created by proteins such as prohibitins (81), or the role of lipids in the function of this membrane, are hardly explored.
MICOS
The mitochondrial contact site and cristae organizing system (MICOS) is a large multi-subunit protein complex that is essential for the maintenance of the mitochondrial inner membrane architecture and the formation of crista junctions (82; 83). In one of the three studies that first described MICOS (84–86), it had been suggested that MICOS may form a filamentous structure within the inner membrane of budding yeast mitochondria (85). Later, several studies have investigated the sub-mitochondrial distributions of various MICOS subunits and interacting proteins using nanoscopy, showing that individual MICOS subunits are generally found in well-coordinated distinct clusters (87–89).
A core subunit of MICOS is the inner membrane spanning protein Mic60. Initially, it had been reported that in mammalian cells, Mic60 clusters are distributed along two opposing sides (or distribution bands) of the mitochondria (87). In a subsequent study it was found that these distribution bands can also be twisted, resulting in a helical arrangement of Mic60 clusters (Fig. 2B) (89). Since Mic60 is enriched at the crista junctions (84; 87; 90), these findings suggested that also the cristae adapt a helical arrangement. Indeed, focused ion beam scanning electron microscopy (FIB-SEM) revealed that in yeast the twisting of the opposing distribution bands is echoed by the folding of the inner membrane such that the cristae seemed to often adopt a propeller-like arrangement (89). Intriguingly, such spiral-like cristae have also been reported in mitochondria from Drosophila (91). Several other Mic60 interacting proteins in the mitochondrial inner and outer membrane seemed to be arranged in a similar patterned manner (88; 89). Based on these observations, it has been speculated that Mic60 is part of a multi-protein interaction network that exhibits a heterogeneous composition (85; 89) and that is vital for the architecture of the mitochondria as it might scaffold the organelle. Nanoscopy will undoubtedly facilitate the further investigation of this proposed structure.
Outer Membrane Proteins
The mitochondrial outer membrane contains numerous transporters and channels, most of which are so abundant that their distributions cannot be resolved with diffraction-limited microscopy. The TOM complex is the primary entry gate for nuclear encoded proteins into the mitochondria. Several studies reported on the clustering of the TOM complexes in the membrane (Fig. 2C) (39; 59; 63; 65; 92; 93). STED nanoscopy revealed that in multiple human cell lines, the antibody-decorated TOM clusters have a diameter of ~70 nm, suggesting an actual diameter of the clusters of around 40 nm (Fig. 2C) (67). As this is substantially larger than a single TOM pore, which has a diameter of around 15 nm (94), it had been concluded that every TOM cluster represents several interacting import pores. An early high-throughput STED study that analyzed more than 1000 cells revealed that the nanoscale distribution of these TOM clusters is finely adjusted to the energetic demands of the cell so that cell growth conditions or the microenvironment of a cell in a microcolony influence the density of the TOM cluster distribution. Remarkably, even the localization of a mitochondrion in a cell impacts the cluster density. The TOM cluster distribution seems to correlate strongly to the mitochondrial membrane potential, suggesting that the metabolic load of an individual mitochondrion and the number of its TOM import pores is tightly linked (67).
Another abundant outer membrane protein is the voltage-dependent anion channel (VDAC, mitochondrial porin). In human cells, three VDAC isoforms exist (95). A dual-color STED study reported that hVDAC1 and hVDAC2 are localized in distinct domains, whereas hVDAC3 was more uniformly distributed, suggesting also functional differences between these three isoforms (Fig. 2D) (66).
An example for a protein that exhibits different activities depending on its localization is the mitochondrial kinase PINK1. Using 3D SIM, it has been suggested that in healthy HeLa mitochondria, a substantial amount of PINK1 is associated with the crista membrane or localized in the crista lumen (Fig. 2E) (76). Upon mitochondrial depolarization, PINK1 translocates to the outer membrane. This is presumably a requirement for mitophagy, and the different submitochondrial localizations of PINK1 may act as a molecular switch mediating different functions of this protein.
Mitochondrial Outer Membrane Permeabilization in Apoptosis
Mitochondria-mediated apoptosis is a genetically encoded program leading to cell death (96). It can be elicited by a number of stimuli, and throughout the animal kingdom, apoptosis is essential for normal development and tissue homeostasis. Under normal conditions, a cocktail of pro-apoptotic proteins localized in the mitochondrial intermembrane space is sequestered from the cytosol. Upon induction of apoptosis, a cascade of events cumulates in mitochondrial outer membrane permeabilization (MOMP), resulting in the release of apoptotic proteins, including cytochrome c and Smac/DIABLO (97). In the cytosol, these proteins initiate the activation of caspases, thereby triggering the subsequent apoptotic program. This event is considered as the “point of no return”, and cells undergoing MOMP are destined to die.
The pro-apoptotic Bcl-2 family proteins BAX (Bcl-2-associated X protein) and BAK (Bcl-2-antagonistic killer) are key players in MOMP. Upon reception of apoptotic stress, BAX and BAK undergo conformational changes and mediate the rupture of the mitochondrial outer membrane. It has been known for decades that at later stages of the apoptotic process, BAX and BAK form large clusters on the mitochondrial surface (98; 99). However, the actual mechanism of BAX and BAK mediated membrane rupture has been controversially discussed. Recently, several studies using various forms of super-resolution microscopy have made important contributions to the understanding of the process (100–103). Two studies, utilizing dual-color STED and single color dSTORM/GSDIM of chemically fixed human cells, respectively, demonstrated that BAX proteins form next to large clusters also much lighter extended oligomers (100; 101). Often, these oligomers formed ring-like assemblies in the outer membrane whose interiors were devoid of outer membrane proteins, strongly suggesting that these ring-like structures represent pores (Fig. 2F) (100). Subsequent studies, relying on, amongst others ISM and SIM, revealed that these BAX/BAK ring-like structures can widen into macropores that even allow the herniation of the inner mitochondrial membrane into the cytosol (102; 103). It is suggested that this process is associated with the release of mitochondrial DNA into the cytosol, triggering the innate immune cGAS/STING pathway, resulting in an inflammatory response (Fig. 2F) (102). Remarkably, such inner membrane herniations have been reported previously using electron microscopy (104), but their importance has only been widely acknowledged after their visualization with fluorescence microscopy (105).
Mitochondrial Dynamics and Interactions with Other Cellular Compartments
The highly dynamic mitochondrial networks that constantly change their appearance due to fusion and fission processes entertain numerous, often transient, physical interactions with other cellular components including the actin and the microtubule cytoskeleton, but also with other organelles such as the ER, endo- or lysosomes (Fig. 2G) (70; 106). The sizes of the mitochondria as well as those of the interacting structures are rather in the hundreds of nanometer range than in the sub-50 nm size regime. Hence, for many questions on mitochondrial dynamics, a large field of view and a high temporal resolution is pivotal rather than the ultimate optical resolution. Indeed, a number of studies used SIM-variants and other extended-resolution microscopies to obtain insights into mitochondrial dynamics (36; 107), mitochondria-microtubule (108), mitochondria-actin (109), and mitochondria-purinosome interactions (110).
The dynamin-related GTPase Drp1 is essential for mitochondrial fission (111; 112). Relying on ISM/Airyscan and SIM imaging, Drp1 clusters were visualized showing that some loosely attached Drp1 clusters move along the mitochondrial tubules, whereas other Drp1 clusters are attached to mitochondria and merge, divide and occasionally move along the organelle with a speed of about 50 nm/sec (109). 3D SIM imaging suggested that through a series of merging and reshaping events, mitochondrially bound Drp1 oligomers develop into rings that encircle the mitochondrion (Fig. 2H) (109). The findings led to the conclusion that Drp1 is constantly in an equilibrium between the cytosol and the mitochondria, and that several fission factors, including actin filaments, target Drp1 to fission sites (109).
Several studies described that mitochondrial fission preferentially occurs at sites where ER tubules encircle mitochondria (113–115). In addition, contact sites between the ER and the mitochondria are also important for calcium signaling and lipid transfer (116). In many cultured cell lines, the majority of the mitochondria, the ER and other organelles are found at the basal cell cortex. Grazing incidence SIM (GI-SIM) has been developed to specifically record sub-cellular dynamics in this region with ~100 nm lateral resolution at very high speed (266 frames/s over thousands of time points) (56). A study using GI-SIM showed that ER-mitochondrion contact sites frequently marked mitochondrial constrictions, and fission events usually occurred at these constriction sites. It was also shown that the majority of mitochondrial fusion events (~60%) is taking place at ER-mitochondrion contact sites. Remarkably, mitochondrial fusions at ER-mito interaction sites were nearly twice as fast (12.2 s) as fusions without ER involvement (21.9 s), suggesting an even tighter entanglement of mitochondrial and ER dynamics than anticipated previously. The functional connection between the ER, actin filaments and mitochondrial fission mediated by Drp1 was also investigated by a study that relied on 3D SIM and conventional live-cell imaging (117). The authors demonstrated that a novel isoform of the actin-nucleating protein Spire1 localizes to the outer mitochondrial membrane. This protein interacts with the ER-bound actin polymerase INF2 (Inverted formin 2) and thereby links mitochondria to the ER and the actin cytoskeleton that pre-constricts mitochondrial tubules before Drp1 facilitates fission. These recordings, together with numerous other studies (109; 118; 119), further underscore the view that mitochondria are part of a dynamic multi-organelle network within cells and that they need to be investigated in this context (120). Fast and rather gentle extended-resolution microscopies, even if they do not provide the very highest optical resolution, are a very suitable approach here. Nanoscopy further uncovered the interaction between purinosomes, mitochondria and microtubules (110; 121). Purinosomes are multi-enzyme complexes mediating the de novo synthesis of purine. These are dynamic structures that were shown to partially colocalize with mitochondria in studies relying on 3D STORM. Interestingly, the dysregulation of mitochondrial function and metabolism influenced the number of purinosome-containing cells, suggesting a functional link between mitochondrial function and purinosome formation (121).
Mitochondrial Nucleoids
Mitochondria maintain their own genome with its own genetic code, which is a remnant of their proteobacterial origin (122). Already in the early 1960s, with electron microscopy of chicken tissue sections, an extra-nuclear DNA molecule with a contour length of 5 μm could be identified, which later proved to be mitochondrial DNA [mtDNA] (123; 124). The mtDNA is tightly packed into nucleoprotein complexes, termed nucleoids, from which up to several thousand copies are found in human cells.
Earlier studies based on conventional light microscopy lead to inconsistent estimates regarding the size of nucleoids (125; 126). Likewise, the number of mtDNA molecules within a single nucleoid was unclear, with data suggesting between two and ten mtDNA molecules per single nucleoid (127; 128). When nucleoids came into the focus of super-resolution microscopy in 2011, two independent studies reported their sizes to range between 70 and 110 nm in mammalian cells using STED nanoscopy and iPALM (129; 130). Several studies, including correlative 3D super-resolution fluorescence and electron microscopy to investigate the relationship of mitochondrial nucleoids to membranes followed (Fig. 2I) (131). STED imaging of hundreds of cells revealed a frequent clustering of nucleoids (Fig. 2I). This clustering depends on the cell type and is altered when outer membrane fusion is disrupted (129; 132). As with nanoscopy, individual nucleoids instead of clusters could be resolved, a substantially higher number of nucleoids per cell than previously reported was recorded. By combining the number of nucleoids per cell with biochemical data, it was shown that many nucleoids in human cells contain only a single copy of mtDNA (129). Based on this conclusion, it has been estimated that a single human mtDNA is decorated with approximately 1000 mitochondrial transcription factor A (TFAM) molecules. The insight that many nucleoids contain only one mtDNA refutes the hypothesis of dynamic nucleoids that contain several DNA molecules which can be transferred from one nucleoid to another.
Different nanoscopy studies have come to diverse conclusions on the shape of single nucleoids (129–131; 133). The reported differences on the shape of nucleoids might well be explained by the cell type and the functional status of the cell. In fact, variations and adaptations of nucleoid shape might be functionally important, as the it might reflect the compaction level and consequently gene expression activity (122).
Currently, there is little insight on the regulation of replication and gene expression on the single nucleoid level, although several reports demonstrated nucleoid-to-nucleoid variations (129; 134; 135). The recently described mitochondrial organization of gene expression (MIOREX) complexes contain mitochondrial ribosomes as well as proteins required for mRNA maturation, translation and decay and thus the MIOREX complexes might represent large expressosome-like assemblies (136). In yeast, STED nanoscopy revealed that a fraction of the MIOREX complexes co-localize with nucleoids, whereas other nucleoids are apparently not interacting with the MIOREX complexes, further supporting the concept of nucleoid heterogeneity.
About 25% of the mitochondrial proteome is involved in gene expression. Currently, the spatial organization of these factors within mitochondria is largely unknown, rendering this a large and still mostly unexplored field.
Different Methods for Different Questions
As super-resolution microscopies are used to address questions ranging from mitochondrial protein distributions on the nanoscale in fixed cells to network dynamics with very high temporal resolution, it is evident that no single method can serve all purposes (Fig. 3). The choice of the imaging method will depend on the required temporal and spatial distribution, the specimen, the preferred labeling approach, and if live or chemically fixed cells are to be recorded (for detailed reviews comparing different super-resolution approaches see (16; 35; 36)).
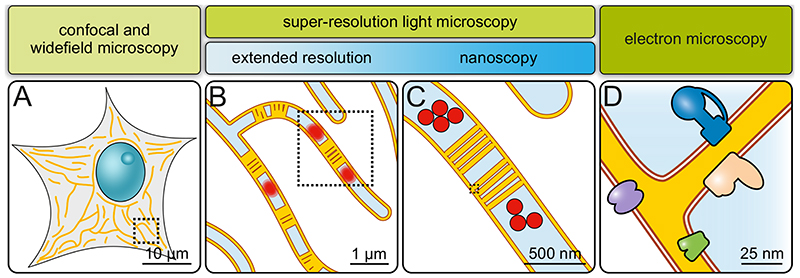
Fig. 3. Imaging mitochondria across scales.
A) Diffraction-limited microscopy (confocal or widefield microscopy). Overall mitochondrial morphology and network dynamics can be visualized. Only limited information on sub-mitochondrial protein distributions. B) Extended-resolution microscopy (diffraction-limited super-resolution microscopy). Network dynamics, but also inner mitochondrial dynamics. Groups of cristae and, under some conditions, single cristae are visible. Mitochondrial sub-compartments can be analyzed. C) Nanoscopy (diffraction-unlimited super-resolution microscopy). Detailed sub-mitochondrial protein distributions and individual cristae can be resolved. D) Electron microscopy. Precise membrane architecture and lipid bilayers are resolved. With specific approaches, protein distributions and even protein structures can be determined.
In the published studies reporting on sub-mitochondrial structures in living cells, STED nanoscopy excels in terms of spatial resolution, whereas SIM shows its full potential when high temporal resolution in combination with a large field of view is required. For example, the SIM images (shown here are only cut-outs) of COS-7 cells labeled with MitoTracker Green (Fig. 4A) (54) or MCC13 cells expressing the outer-membrane marker mEmerald-TOM20 (Fig. 4D) (93), respectively, demonstrate the strength of the approach to record large fields of view at an extended resolution. In comparison, the STED image of a living HeLa cell expressing a SNAP-tag fusion protein targeted to the crista membranes (Fig. 4C) (71), was recorded on a rather small field of view (to ensure a sufficiently high temporal resolution) with a spatial resolution of about 50 nm, facilitating a significantly clearer view on individual cristae. Although SMLM has mostly been performed on fixed mitochondria, Shim et al. demonstrated time-lapse STORM images of BS-C-1 cells labeled with MitoTracker Red, which highlights the inner membrane (61). Their recordings revealed thin, extended tubular intermediates connecting neighboring mitochondria both prior to fission and after fusion (Fig. 4B). Interestingly, the authors report that tightly packed cristae were better resolved after chemical fixation, suggestion that in the living cells the cristae were either motion-blurred during the recording time of two seconds, or because unbound dye molecules were washed away by the fixation process (61).
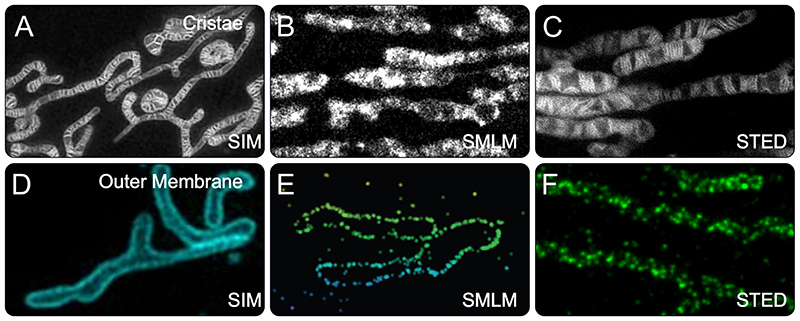
Fig. 4. Cristae and TOM-complexes recorded with different super-resolution microscopies.
A-C) Mitochondrial cristae recorded with Hessian SIM (A) (54), SMLM (STORM) (B) (61) or STED nanoscopy (C) (71) in living human cells. For SIM and STORM, the cristae were labeled with different MitoTracker dyes. For STED nanoscopy, cells expressing a COX8A-Snap-tag fusion protein were labeled with a silicone rhodamine dye. D-F) The mitochondrial outer membrane proteins TOM20 or TOM22 were imaged with SIM (D) (93), SMLM (4PiSMSN) (E) (142) and STED nanoscopy (F) (89) in human cell lines. For 4PiSMSN and STED nanoscopy, fixed cells were decorated with antibodies against TOM20 (4PiSMSN) or TOM22 (STED nanoscopy). SIM was performed on living cells expressing a TOM20-mEmerald fusion protein.
A range of different stresses can elicit excessive mitochondrial fission or, occasionally, mitochondrial fusion, resulting in aberrant network morphologies. Therefore, live-cell recordings of mitochondria require care in avoiding perturbations of the environmental conditions, notably of the growth medium and the temperature. Likewise, protein tagging and/or protein overexpression can interfere with mitochondrial function or architecture, requiring rigorous control experiments (137; 138). In particular, in super-resolution microscopy, the elevated light intensities imposed on the specimen are a notorious concern (139; 140). Nonetheless, SIM has been used to record thousands of images of mitochondria without obvious phototoxic effects (56). Recently, a detailed study on the phototoxic effects of STED nanoscopy came to the conclusion that at least for short term imaging (~10 minutes), living cells can also be imaged by STED nanoscopy without substantial photodamage, although negative long-term effects could not be excluded (141). In this regard, the sensitivity of mitochondria to stresses is simultaneously a blessing and a curse, as damages are readily observable.
In fixed cells, all nanoscopies can exploit their full resolution potential as no constrains with respect to phototoxicity or recording times have to be considered. Unlike extended-resolution methods, nanoscopies like SMLM or STED nanoscopy routinely allow for the visualization of distinct protein clusters. For example, the 4PiSMSN image of a fixed COS-7 cell decorated with antibodies against TOM20 shows a single optical section of a large 3D data set encompassing the entire cell recorded with a 3D resolution of 10 to 20 nm (Fig. 4E) (142). Because of the 3D resolution, the image shows the TOM20 clusters at the rim of the mitochondrial section, whereas the 2D STED image (Fig. 4F) (89) shows a z-projection of TOM22 clusters of an antibody decorated mitochondrion from a human dermal fibroblast.
Preparation of cells for fixation, as well as the actual fixation process and the subsequent sample treatment procedures may damage subcellular structures (143–146). Likewise, the choice of inadequate binders such as poor antibodies may lead to incomplete decoration of the target structure (147; 148). These challenges are obviously also present for conventional microscopy, but are considerably more relevant when cells are imaged at an improved resolution.
The development of photostable dyes that either accumulate in the inner membrane (61; 72) or that can be used in combination with self-labeling protein-tags such as the SNAP-tag (62; 71; 149), open up new possibilities for live-cell nanoscopy. However, the choice of usable fluorophores is restricted, as all nanoscopies require special dyes to unlock the full potential of the respective method. In contrast, extended-resolution microscopies work principally with any fluorescent marker and therefore provide the flexibility to use established dyes or cell lines.
Although with nanoscopy the sub-50 nm regime can be routinely addressed, the ultimate resolution to visualize the mitochondrial membranes or to localize proteins within the organelle is still provided by electron microscopy (EM) (Fig. 3). However, EM is inescapably limited to fixed cells. MINFLUX nanoscopy (150) provides the resolution required to localize proteins at the single-digit nanometer scale, even in living cells. To our knowledge, optical methods that address this resolution regime, have not been used to address biological questions yet, but we propose that they will become an important part of the method portfolio required to dissect mitochondrial biology on the nanoscale. With increasing resolution it becomes more difficult to assign a specific fluorescence signal to the overall mitochondrial membrane architecture. Therefore, we also predict that correlative approaches that combine (super-resolution) light microscopy with electron microscopy and others on the same specimen will become increasingly important to investigate the nanoscale architecture of mitochondria.
Explanations
- Extended-resolution microscopy
Super-resolution microscopy techniques that can achieve an optical resolution significantly higher (typically ~two-fold) than attainable in conventional confocal or widefield microscopy, but still diffraction limited.
- Nanoscopy
Super-resolution microscopy techniques that overcome the diffraction limit.
- ISM
Image Scanning Microscopy, also referred to as AiryScan microscopy.
- STED
Stimulated Emission Depletion.
- SMLM
Single Molecule Localization Microscopy.
- SIM
Structured Illumination Microscopy.
- MINFLUX
Nanoscopy with minimal photon fluxes; this method achieves the highest levels of localization precision of all nanoscopy approaches.
- RESOLFT
Reversible Saturable/Switchable Optical Linear Fluorescence Transitions.
- TOM
Translocase of the mitochondrial outer membrane.
- MICOS
Mitochondrial contact site and cristae organizing system.
- OXPHOS
Oxidative Phosphorylation.
- MOMP
Mitochondrial Outer Membrane Permeabilization.
Acknowledgements
We apologize to the authors of the numerous papers that could not be cited due to space constraints. We thank Jaydev Jethwa for carefully proof reading the manuscript. Part of the work described in this review was supported by Germany’s Excellence Strategy - EXC 2067/1- 390729940, ERCAdG No. 835102, by the German Research Foundation-funded SFB1190 (project P01) and FOR2848 (project P04).
Literature Cited
- 1.Scheffler IE. John Wiley & Sons, Inc; Hoboken, New Jersey, USA: 2008. [Google Scholar]
- 2.Nunnari J, Suomalainen A. Cell. 2012;148:1145–59. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bereiter-Hahn J, Voth M. Microscopy Research & Technique. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 4.Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Molecular Biology of the Cell. 1997;8:1233–42. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westermann B. Nat Rev Mol Cell Biol. 2010;11:872–84. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 6.Palade GE. Anat Rec. 1952;114:427–51. doi: 10.1002/ar.1091140304. [DOI] [PubMed] [Google Scholar]
- 7.Sjöstrand FS. Nature. 1953;171:30–2. doi: 10.1038/171030a0. [DOI] [PubMed] [Google Scholar]
- 8.Hackenbrock CR. Proc Natl Acad Sci U S A. 1968;61:598–605. doi: 10.1073/pnas.61.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werner S, Neupert W. Eur J Biochem. 1972;25:379–96. doi: 10.1111/j.1432-1033.1972.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 10.Frey TG, Mannella CA. Trends Biochem Sci. 2000;25:319–24. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 11.Vogel F, Bornhovd C, Neupert W, Reichert AS. J Cell Biol. 2006;175:237–47. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wurm CA, Jakobs S. Febs Letters. 2006;580:5628–34. doi: 10.1016/j.febslet.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Zick M, Rabl R, Reichert AS. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Mannella CA, Marko M, Penczek P, Barnard D, Frank J. Microsc Res Tech. 1994;27:278–83. doi: 10.1002/jemt.1070270403. [DOI] [PubMed] [Google Scholar]
- 15.Ji N, Shroff H, Zhong H, Betzig E. Curr Opin Neurobiol. 2008;18:605–16. doi: 10.1016/j.conb.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Sahl SJ, Hell SW, Jakobs S. Nat Rev Mol Cell Biol. 2017;18:685–701. doi: 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- 17.Sigal YM, Zhou R, Zhuang X. Science. 2018;361:880–7. doi: 10.1126/science.aau1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbe E. Arch f Mikroskop Anat. 1873;9:413–20. [Google Scholar]
- 19.Born M, Wolf E. 2002 [Google Scholar]
- 20.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, et al. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 21.Hess ST, Girirajan TP, Mason MD. Biophys J. 2006;91:4258–72. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rust M, Bates M, Zhuang X. Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharonov A, Hochstrasser RM. Proc Natl Acad Sci U S A. 2006;103:18911–6. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafsson MG. Proc Natl Acad Sci U S A. 2005;102:13081–6. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hell SW, Dyba M, Jakobs S. Current Opinion in Neurobiology. 2004;14:599–609. doi: 10.1016/j.conb.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann M, Eggeling C, Jakobs S, Hell SW. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17565–9. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hell SW, Wichmann J. Opt Lett. 1994;19:780–2. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 28.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8206–10. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gustafsson MGL. Current Opinion in Structural Biology. 1999;9:627–34. doi: 10.1016/s0959-440x(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 30.Gustafsson MG. J Microsc. 2000;198:82–7. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 31.Sheppard CJR. Optik. 1988;80:53–4. [Google Scholar]
- 32.Müller CB, Enderlein J. Phys Rev Lett. 2010;104:198101. doi: 10.1103/PhysRevLett.104.198101. [DOI] [PubMed] [Google Scholar]
- 33.Chen BC, Legant WR, Wang K, Shao L, Milkie DE, et al. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. Proc Natl Acad Sci U S A. 2009;106:22287–92. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vangindertael J, Camacho R, Sempels W, Mizuno H, Dedecker P, Janssen KPF. Methods Appl Fluoresc. 2018;6:022003. doi: 10.1088/2050-6120/aaae0c. [DOI] [PubMed] [Google Scholar]
- 36.Schermelleh L, Ferrand A, Huser T, Eggeling C, Sauer M, et al. Nat Cell Biol. 2019;21:72–84. doi: 10.1038/s41556-018-0251-8. [DOI] [PubMed] [Google Scholar]
- 37.Chen F, Tillberg PW, Boyden ES. Science. 2015;347:543–8. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wassie AT, Zhao Y, Boyden ES. Nat Methods. 2019;16:33–41. doi: 10.1038/s41592-018-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gambarotto D, Zwettler FU, Le Guennec M, Schmidt-Cernohorska M, Fortun D, et al. Nat Methods. 2019;16:71–4. doi: 10.1038/s41592-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, et al. Biophys J. 2008;94:4957–70. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraus F, Miron E, Demmerle J, Chitiashvili T, Budco A, et al. Nat Protoc. 2017;12:1011–28. doi: 10.1038/nprot.2017.020. [DOI] [PubMed] [Google Scholar]
- 42.Gregor I, Enderlein J. Curr Opin Chem Biol. 2019;51:74–83. doi: 10.1016/j.cbpa.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Demmerle J, Innocent C, North AJ, Ball G, Muller M, et al. Nat Protoc. 2017;12:988–1010. doi: 10.1038/nprot.2017.019. [DOI] [PubMed] [Google Scholar]
- 44.Grotjohann T, Testa I, Leutenegger M, Bock H, Urban NT, et al. Nature. 2011;478:204–8. doi: 10.1038/nature10497. [DOI] [PubMed] [Google Scholar]
- 45.Brakemann T, Stiel AC, Weber G, Andresen M, Testa I, et al. Nat Biotechnol. 2011;29:942–7. doi: 10.1038/nbt.1952. [DOI] [PubMed] [Google Scholar]
- 46.Li D, Shao L, Chen BC, Zhang X, Zhang M, et al. Science. 2015;349:aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumgart F, Arnold AM, Rossboth BK, Brameshuber M, Schutz GJ. Methods Appl Fluoresc. 2018;7:013001. doi: 10.1088/2050-6120/aaed0f. [DOI] [PubMed] [Google Scholar]
- 48.Egner A, Jakobs S, Hell SW. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3370–5. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gugel H, Bewersdorf J, Jakobs S, Engelhardt J, Storz R, Hell SW. Biophysical Journal. 2004;87:4146–52. doi: 10.1529/biophysj.104.045815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dlaskova A, Spacek T, Santorova J, Plecita-Hlavata L, Berkova Z, et al. Biochim Biophys Acta. 2010;1797:1327–41. doi: 10.1016/j.bbabio.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Hirvonen LM, Wicker K, Mandula O, Heintzmann R. Eur Biophys J. 2009;38:807–12. doi: 10.1007/s00249-009-0501-6. [DOI] [PubMed] [Google Scholar]
- 52.Shao L, Kner P, Rego EH, Gustafsson MG. Nat Methods. 2011;8:1044–6. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- 53.Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MG. Proc Natl Acad Sci U S A. 2012;109:5311–5. doi: 10.1073/pnas.1119262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X, Fan J, Li L, Liu H, Wu R, et al. Nat Biotechnol. 2018;36:451–9. doi: 10.1038/nbt.4115. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Liu W, Zhang Z, Zheng C, Huang Y, et al. Nat Commun. 2018;9:4818. doi: 10.1038/s41467-018-07244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y, Li D, Zhang S, Yang Y, Liu JJ, et al. Cell. 2018;175:1430–42.:e17. doi: 10.1016/j.cell.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 57.Liu W, Liu Q, Zhang Z, Han Y, Kuang C, et al. Opt Express. 2019;27:7237–48. doi: 10.1364/OE.27.007237. [DOI] [PubMed] [Google Scholar]
- 58.van de Linde S, Sauer M, Heilemann M. J Struct Biol. 2008;164:250–4. doi: 10.1016/j.jsb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Huang B, Jones SA, Brandenburg B, Zhuang X. Nat Methods. 2008;5:1047–52. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klotzsch E, Smorodchenko A, Lofler L, Moldzio R, Parkinson E, et al. Proc Natl Acad Sci U S A. 2015;112:130–5. doi: 10.1073/pnas.1415261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shim SH, Xia C, Zhong G, Babcock HP, Vaughan JC, et al. Proc Natl Acad Sci U S A. 2012;109:13978–83. doi: 10.1073/pnas.1201882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frei MS, Hoess P, Lampe M, Nijmeijer B, Kueblbeck M, et al. Nat Commun. 2019;10:4580. doi: 10.1038/s41467-019-12480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schueder F, Lara-Gutierrez J, Beliveau BJ, Saka SK, Sasaki HM, et al. Nat Commun. 2017;8:2090. doi: 10.1038/s41467-017-02028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park S, Kang W, Kwon YD, Shim J, Kim S, et al. Mol Brain. 2018;11:17. doi: 10.1186/s13041-018-0361-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donnert G, Keller J, Wurm CA, Rizzoli SO, Westphal V, et al. Biophysical Journal. 2007;92:L67–L9. doi: 10.1529/biophysj.107.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neumann D, Buckers J, Kastrup L, Hell SW, Jakobs S. PMC Biophysics. 2010;3:15. doi: 10.1186/1757-5036-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wurm CA, Neumann D, Lauterbach MA, Harke B, Egner A, et al. Proc Natl Acad Sci U S A. 2011;108:13546–51. doi: 10.1073/pnas.1107553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt R, Wurm CA, Punge A, Egner A, Jakobs S, Hell SW. Nano Letters. 2009;9:2508–10. doi: 10.1021/nl901398t. [DOI] [PubMed] [Google Scholar]
- 69.Ilgen P, Stoldt S, Conradi LC, Wurm CA, Ruschoff J, et al. PLoS One. 2014;9:e101563. doi: 10.1371/journal.pone.0101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bottanelli F, Kromann EB, Allgeyer ES, Erdmann RS, Wood Baguley S, et al. Nat Commun. 2016;7:10778. doi: 10.1038/ncomms10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stephan T, Roesch A, Riedel D, Jakobs S. Sci Rep. 2019 doi: 10.1038/s41598-019-48838-2. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang C, Taki M, Sato Y, Tamura Y, Yaginuma H, et al. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1905924116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilkerson RW, Selker JML, Capaldi RA. FEBS Lett. 2003;546:355–8. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- 74.Suppanz IE, Wurm CA, Wenzel D, Jakobs S. Molecular Biology of the Cell. 2009;20:572–80. doi: 10.1091/mbc.E07-11-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stoldt S, Wenzel D, Hildenbeutel M, Wurm CA, Herrmann JM, Jakobs S. Molecular Biology of the Cell. 2012;23:2292–301. doi: 10.1091/mbc.E11-06-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fallaize D, Chin LS, Li L. Cell Signal. 2015;27:2543–54. doi: 10.1016/j.cellsig.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Appelhans T, Richter CP, Wilkens V, Hess ST, Piehler J, Busch KB. Nano Lett. 2012;12:610–6. doi: 10.1021/nl203343a. [DOI] [PubMed] [Google Scholar]
- 78.Mannella CA, Pfeiffer DR, Bradshaw PC, Moraru II, Slepchenko B, et al. Iubmb Life. 2001;52:93–100. doi: 10.1080/15216540152845885. [DOI] [PubMed] [Google Scholar]
- 79.Wolf DM, Segawa M, Kondadi AK, Anand R, Bailey ST, et al. EMBO J. 2019:e101056. doi: 10.15252/embj.2018101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stoldt S, Wenzel D, Kehrein K, Riedel D, Ott M, Jakobs S. Nat Cell Biol. 2018;20:528–34. doi: 10.1038/s41556-018-0090-7. [DOI] [PubMed] [Google Scholar]
- 81.Tatsuta T, Model K, Langer T. Mol Biol Cell. 2005;16:248–59. doi: 10.1091/mbc.E04-09-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, et al. J Cell Biol. 2014;204:1083–6. doi: 10.1083/jcb.201401006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rampelt H, Zerbes RM, van der Laan M, Pfanner N. Biochim Biophys Acta. 2017;1864:737–46. doi: 10.1016/j.bbamcr.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 84.Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, et al. EMBO J. 2011;30:4356–70. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, et al. J Cell Biol. 2011;195:323–40. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, et al. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 87.Jans DC, Wurm CA, Riedel D, Wenzel D, Stagge F, et al. Proc Natl Acad Sci U S A. 2013;110:8936–41. doi: 10.1073/pnas.1301820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou W, Ma D, Sun AX, Tran HD, Ma DL, et al. Hum Mol Genet. 2019;28:1100–16. doi: 10.1093/hmg/ddy413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stoldt S, Stephan T, Jans DC, Bruser C, Lange F, et al. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1820364116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, et al. J Cell Biol. 2009;185:1047–63. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang YF, Lin SS, Chen JM, Tsai HZ, Hsieh TS, Fu CY. Sci Rep. 2017;7:45474. doi: 10.1038/srep45474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmidt R, Wurm CA, Jakobs S, Engelhardt J, Egner A, Hell SW. Nature Methods. 2008;5:539–44. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 93.Opstad IS, Wolfson DL, Oie CI, Ahluwalia BS. Nanophotonics. 2018;7:935–47. [Google Scholar]
- 94.Model K, Meisinger C, Kuhlbrandt W. J Mol Biol. 2008;383:1049–57. doi: 10.1016/j.jmb.2008.07.087. [DOI] [PubMed] [Google Scholar]
- 95.Messina A, Reina S, Guarino F, De Pinto V. Biochim Biophys Acta. 2012;1818:1466–76. doi: 10.1016/j.bbamem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 96.Tait SW, Green DR. Nat Rev Mol Cell Biol. 2010;11:621–32. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 97.Kalkavan H, Green DR. Cell Death Differ. 2018;25:46–55. doi: 10.1038/cdd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. J Cell Biol. 1997;139:1281–92. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nechushtan A, Smith CL, Lamensdorf I, Yoon SH, Youle RJ. J Cell Biol. 2001;153:1265–76. doi: 10.1083/jcb.153.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grosse L, Wurm CA, Bruser C, Neumann D, Jans DC, Jakobs S. EMBO J. 2016;35:402–13. doi: 10.15252/embj.201592789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salvador-Gallego R, Mund M, Cosentino K, Schneider J, Unsay J, et al. EMBO J. 2016;35:389–401. doi: 10.15252/embj.201593384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, et al. Science. 2018:359. doi: 10.1126/science.aao6047. [DOI] [PubMed] [Google Scholar]
- 103.Riley JS, Quarato G, Cloix C, Lopez J, O’Prey J, et al. EMBO J. 2018:37. doi: 10.15252/embj.201899238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sesso A, Belizario JE, Marques MM, Higuchi ML, Schumacher RI, et al. Anat Rec (Hoboken) 2012;295:1647–59. doi: 10.1002/ar.22553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, et al. Elife. 2019:8. doi: 10.7554/eLife.40712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lackner LL. Trends Cell Biol. 2019;29:580–90. doi: 10.1016/j.tcb.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heintzmann R, Huser T. Chem Rev. 2017;117:13890–908. doi: 10.1021/acs.chemrev.7b00218. [DOI] [PubMed] [Google Scholar]
- 108.Lawrence EJ, Boucher E, Mandato CA. Cell Div. 2016;11:3. doi: 10.1186/s13008-016-0015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ji WK, Hatch AL, Merrill RA, Strack S, Higgs HN. Elife. 2015;4:e11553. doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chan CY, Pedley AM, Kim D, Xia C, Zhuang X, Benkovic SJ. Proc Natl Acad Sci U S A. 2018;115:13009–14. doi: 10.1073/pnas.1814042115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Mol Biol Cell. 2001;12:2245–56. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fonseca TB, Sanchez-Guerrero A, Milosevic I, Raimundo N. Nature. 2019;570:E34–E42. doi: 10.1038/s41586-019-1296-y. [DOI] [PubMed] [Google Scholar]
- 113.Korobova F, Ramabhadran V, Higgs HN. Science. 2013;339:464–7. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murley A, Lackner LL, Osman C, West M, Voeltz GK, et al. Elife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Korobova F, Gauvin TJ, Higgs HN. Curr Biol. 2014;24:409–14. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. Science. 2011;334:358–62. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, et al. Elife. 2015:4. doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Han Y, Li M, Qiu F, Zhang M, Zhang YH. Nat Commun. 2017;8:1307. doi: 10.1038/s41467-017-01503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wong YC, Ysselstein D, Krainc D. Nature. 2018;554:382–6. doi: 10.1038/nature25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Murley A, Nunnari J. Mol Cell. 2016;61:648–53. doi: 10.1016/j.molcel.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.French JB, Jones SA, Deng H, Pedley AM, Kim D, et al. Science. 2016;351:733–7. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gustafsson CM, Falkenberg M, Larsson NG. Annu Rev Biochem. 2016;85:133–60. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 123.Nass MM, Nass S. J Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nass MM. J Mol Biol. 1969;42:521–8. doi: 10.1016/0022-2836(69)90240-x. [DOI] [PubMed] [Google Scholar]
- 125.Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, et al. Mitochondrion. 2007;7:311–21. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 126.Park CB, Larsson NG. J Cell Biol. 2011;193:809–18. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iborra FJ, Kimura H, Cook PR. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Legros F, Malka F, Frachon P, Lombes A, Rojo M. J Cell Sci. 2004;117:2653–62. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 129.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Proc Natl Acad Sci U S A. 2011;108:13534–9. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, et al. Mol Cell Biol. 2011;31:4994–5010. doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. Proc Natl Acad Sci U S A. 2012;109:6136–41. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Silva Ramos E, Motori E, Bruser C, Kuhl I, Yeroslaviz A, et al. PLoS Genet. 2019;15:e1008085. doi: 10.1371/journal.pgen.1008085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kukat C, Davies KM, Wurm CA, Spahr H, Bonekamp NA, et al. Proc Natl Acad Sci U S A. 2015;112:11288–93. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chatre L, Ricchetti M. J Cell Sci. 2013;126:914–26. doi: 10.1242/jcs.114322. [DOI] [PubMed] [Google Scholar]
- 135.Rajala N, Gerhold JM, Martinsson P, Klymov A, Spelbrink JN. NuCleic Acids Res. 2014;42:952–67. doi: 10.1093/nar/gkt988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kehrein K, Schilling R, Moller-Hergt BV, Wurm CA, Jakobs S, et al. Cell Rep. 2015;10:843–53. doi: 10.1016/j.celrep.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 137.Gibson TJ, Seiler M, Veitia RA. Nat Methods. 2013;10:715–21. doi: 10.1038/nmeth.2534. [DOI] [PubMed] [Google Scholar]
- 138.Ratz M, Testa I, Hell SW, Jakobs S. Sci Rep. 2015;5:9592. doi: 10.1038/srep09592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wäldchen S, Lehmann J, Klein T, van de Linde S, Sauer M. Sci Rep. 2015;5:15348. doi: 10.1038/srep15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Laissue PP, Alghamdi RA, Tomancak P, Reynaud EG, Shroff H. Nat Methods. 2017;14:657–61. doi: 10.1038/nmeth.4344. [DOI] [PubMed] [Google Scholar]
- 141.Kilian N, Goryaynov A, Lessard MD, Hooker G, Toomre D, et al. Nat Methods. 2018;15:755–6. doi: 10.1038/s41592-018-0145-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang F, Sirinakis G, Allgeyer ES, Schroeder LK, Duim WC, et al. Cell. 2016;166:1028–40. doi: 10.1016/j.cell.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wurm CA, Neumann D, Schmidt R, Egner A, Jakobs S. Papkovsky DB, editor. Live Cell Imaging: Methods and Protocols. 2010;591:185–99. doi: 10.1007/978-1-60761-404-3_11. Number of 185-99. [DOI] [PubMed] [Google Scholar]
- 144.Whelan DR, Bell TD. Sci Rep. 2015;5:7924. doi: 10.1038/srep07924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li Y, Almassalha LM, Chandler JE, Zhou X, Stypula-Cyrus YE, et al. Exp Cell Res. 2017;358:253–9. doi: 10.1016/j.yexcr.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Richter KN, Revelo NH, Seitz KJ, Helm MS, Sarkar D, et al. EMBO J. 2018;37:139–59. doi: 10.15252/embj.201695709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lau L, Lee YL, Sahl SJ, Stearns T, Moerner WE. Biophys J. 2012;102:2926–35. doi: 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bachmann M, Fiederling F, Bastmeyer M. J Microsc. 2016;262:306–15. doi: 10.1111/jmi.12365. [DOI] [PubMed] [Google Scholar]
- 149.Lukinavicius G, Umezawa K, Olivier N, Honigmann A, Yang G, et al. Nat Chem. 2013;5:132–9. doi: 10.1038/nchem.1546. [DOI] [PubMed] [Google Scholar]
- 150.Balzarotti F, Eilers Y, Gwosch KC, Gynna AH, Westphal V, et al. Science. 2017;355:606–12. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]