Summary
The interplay between host and pathogen relies heavily on rapid protein synthesis and accurate protein targeting to ensure pathogen destruction. To gain insight into this dynamic interface, we combined click-chemistry with pulsed stable isotope labeling of amino acids in cell culture (pSILAC-AHA) to quantify the host proteome response during macrophage infection with the intracellular bacterial pathogen, Salmonella enterica Typhimurium (STm). We monitored newly synthesised proteins across different host cell compartments and infection stages. Within this rich resource, we detected aberrant trafficking of lysosomal proteases to the extracellular space and the nucleus. We verified active cathepsins re-traffic to the nucleus and are linked to cell death. Pharmacological cathepsin inhibition and nuclear-targeting of a cellular cathepsin inhibitor (Stefin B) suppressed STm-induced cell death. We demonstrate that cathepsin activity is required for pyroptotic cell death via the non-canonical inflammasome, and that LPS transfection into the host cytoplasm is sufficient to trigger active cathepsin accumulation in the host nucleus and cathepsin-dependent cell death. Finally, cathepsin inhibition reduced Gasdermin D expression, thus revealing an unexpected role for cathepsin activity in non-canonical inflammasome regulation. Overall, our study illustrates how resolving host proteome dynamics during infection can drive the discovery of biological mechanisms at the host-microbe interface.
Introduction
A successful intracellular pathogen should avoid detection by host cytoplasmic pattern recognition receptors (PRRs), which constantly survey the host cytoplasm for microbial ligands. Upon detection, activated cytoplasmic PRRs trigger the assembly of a large cytoplasmic protein signaling complex called the inflammasome, which drives an inflammatory form of cell death termed pyroptosis and the release of pro-inflammatory cytokines. Pyroptosis is driven by either the canonical inflammasome cysteine proteases Caspase-1 (mouse and human) or the non-canonical Caspase-11 (mouse) and Caspases-4 and 5 (human)1. Canonical inflammasome activation relies on additional cytoplasmic sensors to detect bacterial ligands, whereas Caspase-11 is both a sensor and activator of the non-canonical inflammasome. Upon sensing lipopolysaccharide (LPS)2, Caspase-11 cleaves Gasdermin D (GSDMD) into an active form. GSDMD then punctures holes in the plasma membrane, ultimately leading to cell death 3,4.
Remarkably, some intracellular pathogens have evolved to survive and replicate within the hostile endo-lysosomal system. For example, Salmonella enterica subsp. enterica serovar Typhimurium (STm) sculpts a specialized endo-lysosomal compartment called the Salmonella-containing vacuole (SCV). The SCV provides a membrane-bound shield that prevents detection of STm’s surface-bound LPS by the non-canonical inflammasome. When SCV integrity is compromised, bacterial LPS is released to the host cytoplasm where it triggers robust activation of the non-canonical inflammasome and cell death5. Thus, host-pathogen interplay within the endo-lysosomal system is critical to both host and pathogen survival, yet the dynamics of this process remain poorly understood.
To shed light onto this complex interface, we leveraged a method recently developed in our lab designed to enrich, identify and quantify newly synthesised host secretory proteins in serum-containing media 6. Here, we extended this approach to include sampling of three host cell compartments from STm infected macrophages spanning three distinct stages of the infection cycle. We detected aberrant subcellular distribution of lysosomal proteases, and demonstrated their active role during STm-induced cell death via the non-canonical inflammasome. Combined, our findings exemplify the treasure trove of functional biology that can be uncovered by spatiotemporally resolving a host-pathogen interface.
Results
Dynamic proteome mapping unravels diverse host responses during STm infection
To model the intracellular STm infection cycle, we infected mouse-derived macrophages with SPI-1 OFF STm constitutively expressing mCherry from a plasmid (not affecting infection progress) 7 (Extended Data Fig. 1a). In order to determine the subcellular distribution of host proteins throughout an infection cycle, we utilised our recently developed proteomic methodology that allows specific enrichment and quantification of the newly synthesised host proteome (Fig. 1a) 6.
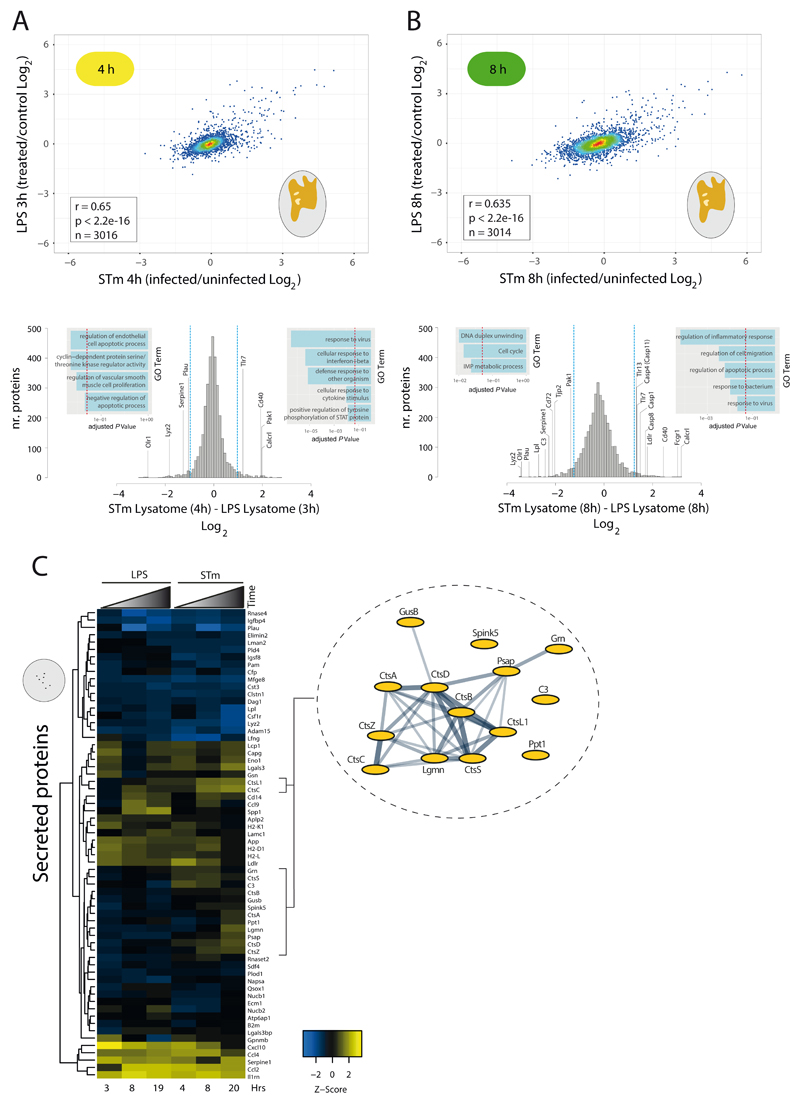
Figure 1. Newly synthesised proteome enrichment detects diverse host responses during STm infection.
a) Experimental design of pSILAC-AHA labelling and subcellular fractionation. (1) RAW264.7 cells were infected with STm 14028s constitutively expressing mCherry (pFCcGi) and grown to stationary phase at an MOI 100:1, followed by centrifugation to facilitate host-pathogen contact. After a 25 min incubation, extracellular STm were killed by moving cells first to media containing 100 μg/ml gentamicin for 1 h, and then to media containing 16 μg/ml gentamicin for the remainder of the experiment. Uninfected samples received a mock inoculum and were otherwise treated identically to infected samples. (2) 3.5 hours prior to harvest, cells were washed and briefly starved for 30 minutes to remove residual amino acids, followed by pSILAC-AHA labelling for 3 hours. A three-hour pulse window is sufficient to allow protein synthesis and subsequent subcellular trafficking to occur 8 (3) Conditioned media was harvested for the secretome samples, whereas cells were washed and partially lysed with a mild detergent (Triton X-100) to extract both the lysatome and nuclear samples. (4) Samples were mixed at 1:1 ratio (infected: uninfected) and then fractionated by centrifugation to separate the secretome, nucleome and lysatome. (5) Newly synthesised proteins containing AHA were covalently linked to alkyne agarose beads via a Click-chemistry reaction, allowing for selective capture of proteins containing incorporated AHA, while serum and other pre-existing background proteins were removed by stringent washing conditions. (6) After on-bead protease digestion, peptides were quantified by LC-MS/MS. (I) and (H) refer to intermediate and heavy isotopic amino acids respectively. Icons distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/) were adapted 32.
b) GO term enrichment of differentially regulated host proteins (4, 8 and 20 hours). Selected enriched GO terms are depicted; all enriched GO terms can be found in Supplementary Table 2. Node size and colours depict significance (p (-Log10) = right-sided hypergeometric test, Bonferroni corrected) and number of proteins (blue shade), respectively. n=2 biologically independent samples.
We quantified the newly synthesised host proteome (4978 proteins) by sampling three distinct subcellular locations from macrophages infected with intracellular STm at 4, 8 and 20 hours post infection (hpi). In total we detected 215 proteins in the secretome (conditioned media), 4640 in the lysatome (Tx-100 soluble; this contains cytoplasmic and organelle content) and 1283 in the nucleome (Tx-100 insoluble) (Supplementary Table 1 and Fig. 1b and Extended Data Fig. 1b). GO term enrichment analysis for host proteins displaying >1.5 ±Log2 fold change revealed a total of 879 enriched GO terms (p = ≤0.05, right-sided hypergeometric test, Bonferroni corrected), with 832 being upregulated and 47 being downregulated (Supplementary Table 2). Consistent with the lysatome containing the majority of quantified proteins, 693 enriched GO terms were detected in the lysatome fraction, whereas 97 and 87 GO terms were enriched in the nucleome and secretome samples respectively. We further validated the secretome data using a custom chemokine and cytokine array for 7 secreted proteins (Extended Data Fig. 2).
In general terms, dynamic changes occurring at distinct time-points of the infection were more frequent in the subcellular compartments, whereas the lysatome was dominated by constant responses, occurring from the first time-point (4 hpi) and remaining stable across time (Fig. 1b). Such early and stable responses included many GO terms related to infection and adaptation to immune stimulation (Supplementary Discussion). For example, in secretome samples, lysosomal proteins displayed enhanced secretion at 20 hpi (GO:0005764, Fig. 1b and Supplementary Table 2). Similarly to the secretome, lysosomal components (GO:0005764, Fig. 1b), consisting of many lysosomal proteases e.g. Cathepsins A (CtsA), B (CtsB), D (CtsD), L (CtsL), S (CtsS), and Z (CtsZ), and Legumain (Lgmn) were more abundant in the nuclear fraction. This nuclear enrichment was specific for cathepsins as other lysosomal lumen proteins, such as aryl-sulfatase (ArsB) and α-glucosidase (Gaa), were abundant in the lysatome, but not detected in the nucleome. Similarly, only a handful of cytosolic proteins increased their abundance in the nucleome during late stages of infection, including peroxiredoxins 1 (Prdx1), 2 (Prdx2) and 4 (Prdx4), a ubiquitous family of antioxidant enzymes (Supplementary Table 3).
STm infection induces distinct host proteome responses compared to LPS
We anticipated that most of the responses in our experimental design would be attributable to the highly immunogenic LPS present on the STm surface. To test this, we directly compared our lysatome samples from STm infected cells with previously published data from RAW264.7 cells stimulated with LPS 8 (Supplementary Table 3). The two time points analysed span two distinct phases of the STm intracellular life cycle, namely i) pre-SPI-2 dependent proliferation (3-4 hours) and ii) active SPI-2 dependent growth (8 hours). Consistent with our expectations, STm infection and LPS stimulation showed strong positive correlations at both 4 h (r = 0.65) and 8 hours (r = 0.635) (Fig. 2a-b). Thus, much of the proteome-response of STm infected cells can be explained by innate immune responses to LPS alone. Beyond the overall similarity, we could identify host responses specific to STm infection by filtering out the LPS response from STm infected samples and performing GO term enrichment (Fig. 2a-b, Supplementary Tables 4 & 5, Supplementary Discussion).
Figure 2. STm infection induces distinct host proteome responses compared to LPS across time and space.
a) Upper panel, scatter plot of pSILAC-AHA lysatome data previously collected in response to LPS stimulation (LPS from Escherichia coli O111:B) 8 versus data collected after STm infection from this study (Supplementary Table 3). N.B. Due to differences in sample collection, the LPS samples contain both the lysatome and nucleome fractions. Lower panel, histogram containing protein expression from lysatome of RAW264.7 cells 4 hpi with STm after subtracting the log2 corresponding LPS signal. Dotted blue lines in histogram indicate ± 2 S.D. from the mean and the dotted red line on inset GO term histograms indicates multiple-test adjusted p value (Bonferroni corrected) cutoff of 0.05. n=2 biologically independent samples.
b) Same as (a) but at a later time point: 8 hpi with STm or LPS stimulation of RAW264.7. Corresponding protein levels for both panels (a) and (b) in Supplementary Table 4 and GO terms in Supplementary Table 5. n=2 biologically independent samples.
c) Heatmap of secreted proteins from pSILAC-AHA labelled RAW264.7 cells after STm infection or LPS stimulation, the latter data obtained previously 8. N.B. Only rows without missing data points were used for analysis. To the right is a STRING network of vacuolar proteins with increased secretion dynamics specific to STm infection. Nodes represent individual proteins and edges reflect experimentally determined interactions and co-expression weighted data from STRING v10.5 33.
We then compared the secretome of LPS and STm treated cells (Fig. 2c). Similar to the lysatome, STm infection and LPS stimulation induced broadly similar protein secretion dynamics over time (Fig. 2c), although clear differences were again apparent. The most profound difference was the enhanced secretion of lysosomal hydrolases, such as CtsC, CtsL, CtsD, CtsZ, CtsA and Lgmn, during STm infection (Fig. 2c). Interestingly, STm was previously shown to enhance the secretion of both β-hexosaminidase and immature CtsD, ultimately depriving lysosomes of hydrolytic enzymes to promote STm proliferation 9. Our findings suggest that STm-induced lysosomal detoxification is broader than previously appreciated.
Lysosomal proteases are enriched in the nuclear fraction during STm infection
Reminiscent of STm-induced secretion of lysosomal proteases observed in the secretome, we also detected a strong and significant increase in several lysosomal hydrolases at 20 hpi in the nucleome (Fig. 3a). As the expression levels of these proteins only marginally increased in the lysatome samples (Fig. 3b, c), their much stronger increase in the nucleus is presumably due to their increased trafficking to this compartment. We excluded the possibility that this may be due to co-fractionation of lysosomal components together with the nucleus in our experiments, as Lamp1 and Lamp2 were only detected in the lystatome (Supplementary Tables 1 & 3). Although some vacuolar membrane proteins were also enriched in the nuclear fraction, this was at a much lower level than their lumen counterparts. This suggested that cathepsin enrichment (lumen proteins) cannot be generally explained by increased levels of lysosomal proteins in the nucleome alone (Fig. 3c). Instead we hypothesized that newly synthesised lysosomal proteins are trafficked to the nucleus during STm infection. Although cathepsins have previously been observed in the nucleus 10–13, this has not been linked to infection. We therefore set out to examine the significance of this organized re-trafficking of lysosomal proteases in more detail.
Figure 3. Lysosomal proteases are enriched in the nuclear fraction upon STm infection.
a) Histogram of nuclear proteins differentially regulated after 20 hours of STm infection, lysosomal hydrolases are annotated in red. Blue and red dotted lines, as well as GO cellular component enrichments (inset) as in Fig. 2a. n=2 biologically independent samples.
b) Scatter plot of lysatome and nucleome data collected from RAW264.7 cells at 20 hours post-infection. Distributions of all proteins found in the lysatome (x-axis) and nuclear fraction (y-axis) (blue), and lysosomal hydrolases (red). Black dotted line represents the line of best fit. p = (two sided unpaired Wilcoxon rank sum test). n=2 biologically independent samples.
c) Boxplots displaying the relative fold change (infected/uninfected) of membrane bound lysosomal versus soluble lysosomal luminal proteins selected from the lysatome and nucleome samples as per Fig 1b from n=2 biologically independent samples. Box boundaries indicate the upper and lower IQR, the median is depicted by the middle boundary and whiskers represent 1.5x IQR.
STm SPI-2 promotes high nuclear cathepsin activity
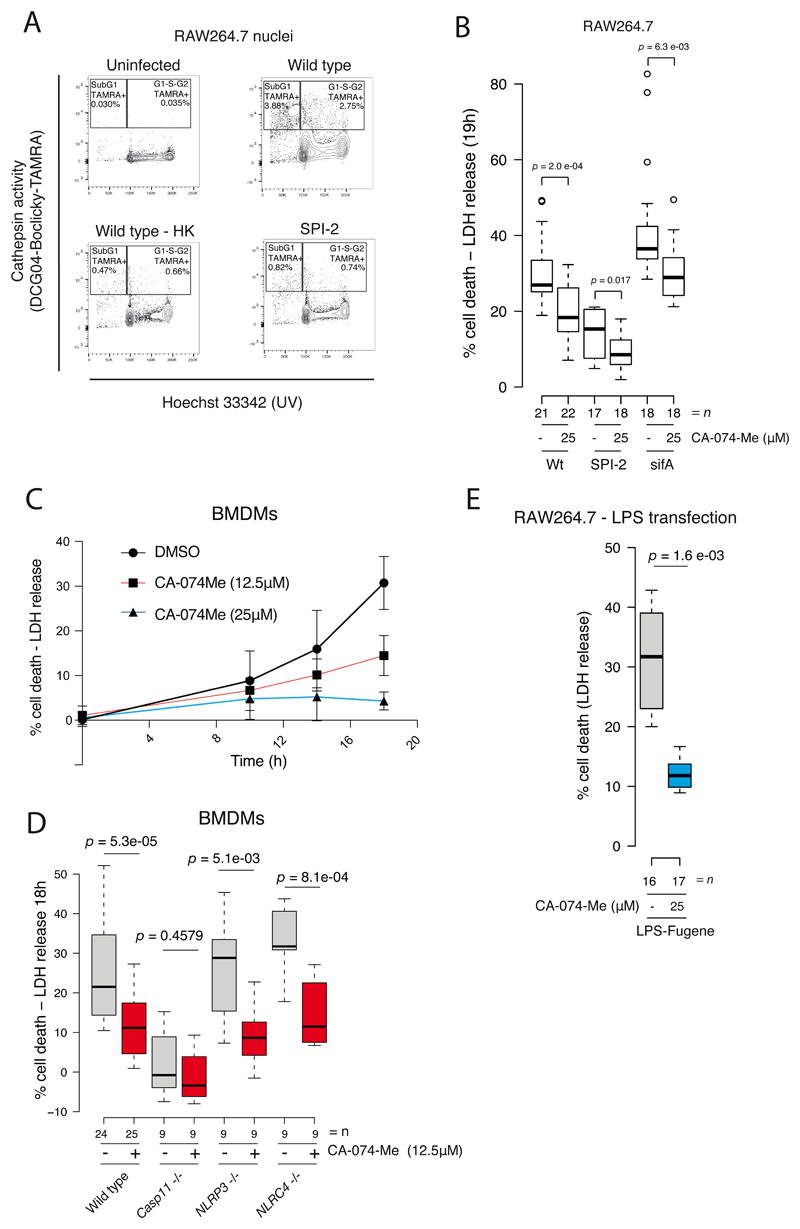
To examine cathepsin localisation and activity, we added a cell permeable cathepsin reactive probe, DCG04-Bodipy-FLike (DCG04-FL), to live cells during STm infection. Analysis of nuclear extracts exhibited a striking increase in nuclear cathepsin activity during infection with wildtype STm compared to the uninfected control (Fig. 4a, right panel). Nuclear cathepsins were also of a higher molecular weight relative to their endolysosomal counterparts (Fig. 4a), which are processed to more active mature forms 14. However, consistent with previous reports describing higher molecular weight nuclear forms of cathepsin CtsB and CtsL 10,11,15, we found these larger forms to be also proteolytically active 10,15 as our probe only binds active enzymes. We verified that DCG04-FL does not cross-react with STm and that increased nuclear cathepsin activity does not lead to increased histone H3 cleavage as previously reported (Extended Data Fig. 3a-b) 16. Thus, newly synthesised active cathepsins are re-trafficked to the nucleus during STm infection.
Figure 4. STm SPI-2 promotes nuclear cathepsin activity.
a) Uninfected and wildtype STm-infected RAW264.7 cells (MOI = 100:1) were treated with the cathepsin probe DCG04-Bodipy-FLike (5 μM), or DMSO vehicle for 3 hours prior to harvesting at 20 hpi. Such inhibitor-based probes covalently link to the reactive cysteine in the catalytic site of endolysosomal proteases, thus the amount of bound probe directly corresponds to cathepsin activity 34. Endosomal organelles containing active lysosomal cysteine proteases were effectively solubilised with the non-ionic detergent Tx-100, as evidenced by the presence of highly active mature cathepsins in cells treated with the cathepsin probe 488nm DCG04-Bodipy-FLike (left panel), and not in DMSO treated cells, thus demonstrating the specificity of the DCG04-Bodipy-FLike probe. Cathepsin activity was elevated in the Tx-100 soluble fraction of infected relative to uninfected cells, particularly for CtsB and CtsZ (left panel). Tx-100 soluble (left) and insoluble (right) extracts were separated by SDS-PAGE and then visualised using a fluorescence scanner (Ex 405 nm/Em 520 nm). Immunoblot with anti-GAPDH and Coomassie staining of histones were used as loading controls for lysatome and nucleome samples respectively. Data is representative of two biological replicates (all blots shown in supplement).
b) RAW264.7 cells infected with wildtype, ΔssaV (SPI-2) or ΔprgK (SPI-1) STm were treated with the cathepsin probe DCG04-Bodipy-FLike (5 μM) and lysatome (left) and nucleome (right) extracts were separated and visualised as described in A. Loading controls as in A. Data is representative of two biological replicates (all blots shown in supplement).
c) The cathepsin activity probe DCG04-Boclicky-TAMRA (5μM) was added to the media 2 hours prior to harvesting at 2 (upper) and 20 (lower) hpi (MOI = 100:1) of RAW264.7 cells with STm constitutively expressing GFP from pDiGc (see Methods). Cells were then fixed and stained with Hoechst 33342 (DNA) and images acquired using a 20x objective. Scale bar represents 50 μm.
d) Violin plots of single-cell nuclear cathepsin activity quantified in RAW264.7 cells infected with wild-type or ΔssaV (SPI-2) STm constitutively expressing GFP from pDiGc (MOI = 100:1) for 20 hours. Infected cells were treated with the cathepsin probe DCG04-Boclicky-TAMRA 2 hours prior to cell fixation. Nuclear cathepsin activity was measured by quantifying DCG04-Boclicky-TAMRA signal that overlapped with Hoechst 33342 stained nuclei. Each data point represents a measurement per cell nuclei, which was further classified as infected or uninfected bystander based on the presence or absence of GFP expressing STm located inside the host cell perimeter, respectively. Nuclear cathepsin activity was normalised to nuclei area. Red bar = median. A two-sided unpaired Wilcoxon rank sum test was used to calculate p. n denotes the total number of single nuclei measurements from 2 biologically independent samples.
e) Cathepsin activity in fixed cells, prepared as in c, were visualised by confocal microscopy. Depicted cells are representative of high and low nuclear cathepsin activity distributions in d. Cells with high nuclear and perinuclear cathepsin activity are more readily observed in cells infected with wildtype-STm compared to those infected with ΔssaV (SPI-2) mutants, uninfected bystanders, and naive cells from control wells not exposed to bacteria. In order to observe clear boundary definition between the nucleus and the non-nuclear area of the cell, single planes from a z-stack are displayed. Scale bars represents 2 μm.
f) Single cell analysis of nuclear and non-nuclear cathepsin activity in RAW264.7 cells infected with wildtype STm for 20 hours as in d. Cells and nuclei were segmented using CellProfiler. Nuclear cathepsin activity was measured by quantifying DCG04-Boclicky-TAMRA signal overlapping with Hoechst 33342 stained nuclei. Non-nuclear cathepsin activity was calculated by subtracting the nuclear cathepsin activity (TAMRA) from the total TAMRA signal per cell. The upper 95th quantile (Q95) representing cells with the highest nuclear cathepsin activity (n = 31) were analysed by directly comparing to the corresponding non-nuclear cathepsin activity. A blue line represents a fitted linear regression model and r denotes the Pearson’s correlation coefficient.
As the SPI-2 secretion system of STm has been implicated in re-trafficking of lysosomal contents to the external milieu 9, and nuclear cathepsin activity coincides with SPI-2 dependent proliferation (Extended Data Fig. 3a), we examined whether SPI-2 secretion is also required for nuclear cathepsin activity. Cathepsin activity in Tx-100 soluble lysates was comparable between RAW264.7 cells infected with wildtype, SPI-1 (ΔprgK) or SPI-2 (ΔssaV) secretion system mutants (Fig. 4b left panel). In contrast, nuclear cathepsin activity was reduced in cells infected with the SPI-2 mutant, but not the SPI-1 mutant infected cells (Fig. 4b right panel). To test whether the trafficking of cathepsins to the nucleus is independent of their activity, we probed CtsB localization during infection when cells were treated with the CtsB and CtsL selective inhibitor, CA-074-Me. CtsB was trafficked to the nucleus during infection with wildtype STm independent of the presence of the inhibitor (Extended Data Fig. 3c). Thus, SPI-2 seems to promote active trafficking of cathepsins to the nucleus independently of cathepsin activity.
To ensure our observations of SPI-2 promotion of nuclear cathepsin activity by SDS-PAGE were not artifacts of biochemical fractionation, we used fluorescence microscopy to quantify nuclear cathepsin activity from single cells in situ (Fig. 4c-d). Wildtype infected cells exhibited increased nuclear cathepsin activity relative to uninfected bystanders (Fig. 4d). Furthermore, nuclear cathepsin activity in cells infected with the SPI-2 deficient mutant was reduced compared to wildtype-infected cells (Fig. 4d). This increased cathepsin activity was inside the nucleus (i.e. cathepsin activity within the nuclear boundary defined by Hoechst stained host nuclei) and the perinuclear region of wildtype-infected cells, but not in uninfected bystanders, uninfected naïve or those infected with a SPI-2 deficient mutant (Fig. 4e). Of note, cells displaying elevated nuclear cathepsin activity in some cases exhibited a strong increase in cathepsin activity throughout the cell body (Fig. 4e; 5th image, first row), but in general the two metrics were not strongly correlated for cells with high nuclear cathepsin activity (Fig. 4f). Taken together, STm SPI-2 promotes an accumulation of a molecularly distinct form of cathepsins within the nucleus during infection.
Nuclear cathepsin activity correlates with signatures of cell death
To further examine the consequences of nuclear cathepsin activity, we extracted nuclei from cells treated with the cathepsin activity probe DCG04-Boclicky-TAMRA and analysed by flow cytometry. In cells exposed to wildtype STm, ~5.5 % of nuclei were cathepsin positive compared to 1 - 1.5 % exposed to the ssaV mutant (SPI-2) or heat killed (HK) bacteria (Fig. 5a, Extended Data Fig. 4). A substantial fraction of cathepsin positive nuclei were detected in the sub-G1 region of the FACS plot (Fig. 5a, Extended Data Fig. 4), signifying DNA fragmentation that occurs during pyroptotic cell death induced by STm infection 17,18. STm lacking the SPI-2 secretion system and dead STm (both unable to proliferate intracellularly) elicited a residual amount of cathepsin positive nuclei, in agreement with the previous results (Fig. 4b, d). This residual non-SPI-2 effect is likely due to LPS.
Figure 5. Cathepsin activity promotes STm induced Caspase-11 dependent cell death.
a) Nuclei were extracted from RAW264.7 cells treated with DCG04-Boclicky-TAMRA (5 μM) for 2 hours prior to harvest at 20 hpi with wildtype (Wt), a SPI-2 mutant (ΔssaV) or heat killed wildtype bacteria (Wt HK) (MOI = 100:1). Formaldehyde fixed nuclei were subsequently counterstained with Hoechst 33342 and analysed by flow cytometry. Cells in G1, S or G2 phase of the cell cycle are separated by Hoechst 33342 staining on the x-axis. Cathepsin activity (DCG04-Boclicky-TAMRA) is indicated by the % of total DCG04-Boclicky-TAMRA positive nuclei in either sub-G1 or G1/S/G2 or combined total of sub-G1 and G1/2/G2. Data combining all biological replicates from two independent experiments can be seen in Extended Data Fig. 4.
b) Pyroptotic cell death was assessed by quantifying LDH release into culture supernatants of RAW264.7 cells infected with wildtype, SPI-2 (ΔssaV) and the effector deletion (ΔsifA) mutants in the presence of 25 μM CA-074-Me or DMSO solvent control at 19 hpi. The ΔsifA mutant has a strong replication defect and readily escapes into the cytoplasm, hyperactivating Caspase-11 dependent cell death 5. (n) denotes biologically independent samples combined from three independent experiments (batches); each batch contained a minimum of 4 biological replicate wells per condition. Data represents the % LDH release per condition relative maximum LDH release (see Methods). Box plots are depicted as in Fig. 3C. An unpaired t-test (two-sided) was used to calculate p.
c) BMDMs were infected with wild-type STm (MOI 100:1), followed by incubation in the presence of cathepsin inhibitor CA-074-Me at the indicated concentrations. At the indicated hpi’s, cell death was measured as the % of LDH released into culture supernatants. Data points represent the mean and error bars indicate the 95% CI. (n) denotes biologically independent samples. Time points 0, 10 and 14 hours are derived from three biological replicates per condition (n=3 per condition) from a single batch, whereas the 18-hour time point contains combined data from 3 or more independent experiments (batches), each batch containing 3-4 biological replicates per condition (DMSO n=30; CA-074-Me (12.5 μM) n=28; CA-074-Me (25 μM) n=13).
d) related to a), wildtype, caspase-11/1 -/-, caspase-11 -/-, NLRP3 -/-, NLRC4 -/- BMDMs were infected with wildtype STm (MOI 100:1), followed by incubation in the presence of cathepsin inhibitor CA-074-Me (12.5 μM) for 18 hpi. The % LDH released into culture supernatants was measured 18 hours post-infection. n denotes biologically independent samples combined from >3 (wildtype) and 3 (mutant genotypes) independent experiments (batches), each batch containing 2-4 biological replicates per condition. A two-sided unpaired t-test was used to calculate p.
e) RAW264.7 cell were transfected with LPS with Fugene (Promega) for 20 hours. Pyroptotic cell death was assessed by quantifying LDH release into culture supernatants in the presence of 25 μM CA-074-Me relative to a DMSO solvent control. (n) denotes biologically independent samples combined from three independent experiments (batches), each batch consisting of a minimum of 3 biological replicate wells per condition. Boxplots are depicted as in Fig. 3C. A two-sided unpaired t-test was used to calculate p.
To verify that cathepsins play a role during STm induced cell death, we tested the impact of cathepsin inhibition on host cell death induced by STm by measuring LDH release 19. Both wildtype STm and ΔsifA induced considerable levels of cell death after 19 hours of infection, with the latter exhibiting more cell death, as expected (Fig. 5b). Cell death was largely dependent on the expression of a functional SPI-2 (ΔssaV; Fig. 5b). Importantly, in all cases, cell death was partially inhibited by the addition of CA-074-Me (Fig. 5b). We further confirmed that CA-074-Me does not inhibit STm growth per se (Extended Data Fig. 5), and that it is selective against cathepsins using 2-dimensional Thermal Proteome Profiling (2D-TPP) 20,21. CA-074-Me specifically stabilised cysteine proteases cathepsin B, S, Z, F, K (Extended Data Fig. 6a & Supplementary Table 6), revealing a broader inhibitory spectrum for cathepsins than previously appreciated. Importantly, we found no evidence that CA-074-Me targets any other cysteine protease, including all caspases detected (Extended Data Fig. 6a)22. As proteome coverage for Caspase-11 was incomplete, we used an in vitro assay to verify that CA-074-Me does not affect Caspase-11 proteolytic activity (Extended Data Fig. 6b). Overall, these findings suggest that cathepsin dependent cell death is promoted by STm escape into the cytoplasm and is required in conditions where Caspase-11 is active.
Cathepsin activity is required for STm-induced Caspase-11 dependent cell death
To further understand the host molecules at play, and to ensure our observations go beyond RAW264.7 cells, which lack key inflammasome components 23, we sought to replicate our findings using BMDMs from rodent knockout lines. Consistent with previous reports 24,25, LDH release started ~10 h post-infection (Fig. 5c). This LDH release was dose-dependently suppressed with CA-074-Me, similarly to our observations in RAW264.7 cells.
Delayed lytic cell death of STm infected macrophages depends on the cytoplasmic LPS sensor Caspase-11 2. We therefore tested whether cathepsin dependent cell death requires the presence of Caspase-11. Cathepsin-dependent cell death required Caspase-11 and occurred independently of the canonical inflammasome activators NLRP3 and NLRC4 (Fig. 5d). To test whether cathepsin activity is required for the canonical inflammasome-mediated cell death, we infected BMDMs with SPI-1 (ON) STm, which is known to rapidly trigger the canonical inflammasome 26. In this case, we observed no impact of cathepsin activity on SPI-1 induced cell death (Extended Data Fig. 7). These data demonstrate that cathepsins play a specific functional role in non-canonical inflammasome activation.
Cathepsin activity functions downstream of LPS-triggered Caspase-11 activation
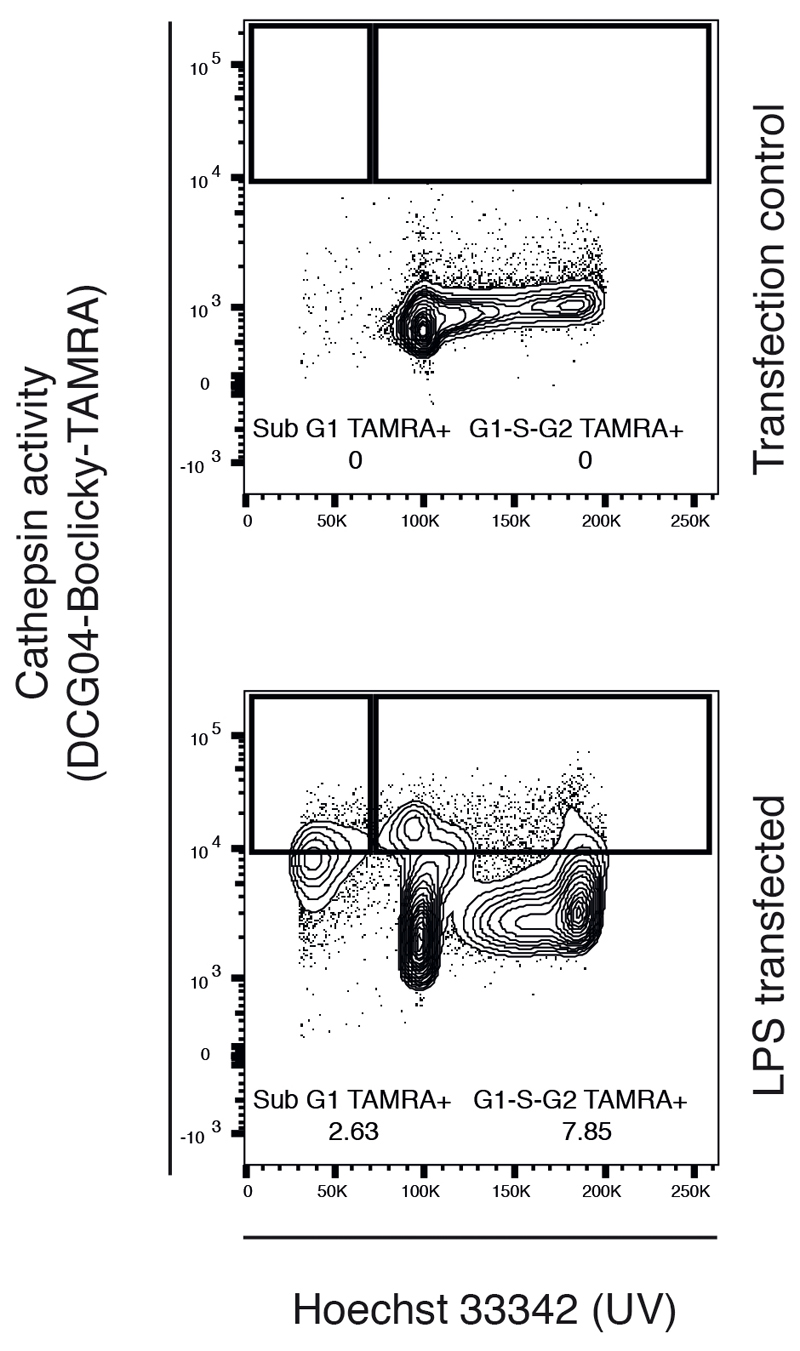
To examine whether cathepsin activity is required upstream or downstream of Caspase-11 activation, we transfected LPS into the host cytoplasm to directly activate the non-canonical inflammasome. Remarkably, direct Caspase-11 activation with cytoplasmic LPS was inhibited by CA-074-Me (Fig. 5e) and on its own promoted nuclear cathepsin activity (Extended Data Fig. 8), demonstrating that the cathepsin activity required for cell death functions downstream of Caspase-11 activation. Consistently, Caspase-11 activation during wildtype STm infection was unaltered by CA-074-Me (Extended Data Fig. 9). Thus, LPS alone can trigger relocation and/or activation of cathepsins in the nucleus, consistent with the basal signal we observed with intracellular non-proliferating STm (Fig. 4b, d & 5). This suggests that cathepsin-dependent cell death is a host-induced process and that cathepsin activity is required for non-canonical inflammasome activation.
Cathepsin activity promotes Gasdermin D expression
Our previous experiments suggested that cathepsin activity is required downstream of Caspase-11. The only known target of Caspase-11 is Gasdermin D (GSDMD). In our 2D-TPP data, CA-074-Me led to a strong decrease in Gasdermin D expression in a dose dependent manner in RAW 264.7 cells (Fig. 6a). We verified this also occurs in BMDMs by measuring Gasdermin D levels during infection. An infection dependent increase in Gasdermin D abundance could be suppressed by the addition of CA-074-Me (Fig 6b). In addition, CA-704-Me broadly reduced the abundance of several pro-inflammatory proteins e.g. IL1b, IL18, IL1a, GBP1 GBP2, GBP4, GBP5, GBP7, GBP9, IFI202, IFI204, ISG20, ISG15 (Extended Data Fig. 6c & Supplementary Table 6). Taken together, we consider that nuclear cathepsin activity results in increased levels of Gasdermin D and proinflammatory proteins, and hence contributes to non-canonical inflammasome-mediated cell death.
Figure 6. Cathepsin activity promotes Gasdermin D expression and its nuclear activity is required for cell death.
a) Two-dimensional thermal proteome profiling (2D-TPP) of RAW264.7 cells infected with wildtype STm for 20 hours in the presence of increasing concentrations of CA-074-Me post STm uptake. Heatmap of Gasdermin D shows decreased abundance (evident from decrease in protein abundance at lower temperatures, before protein melts) with increasing CA-074-Me concentrations. Key is presented to the left of the heatmap; for each protein and temperature, the signal intensity was normalized to the DMSO vehicle control. 2D-TPP data can be found in Supplementary Table 6 and Extended Data Fig. 6.
b) BMDMs were infected with wildtype Salmonella 14028s (20 hpi) and cell lysates analysed by immunoblot for Gasdermin-D and GAPDH as a loading control. A replicate blot from an independent experiment is located in the Supplementary Information (SI).
c) BMDMs infected with wildtype STm (MOI = 100:1) were treated with CA-074-Me (12.5 or 25 μM) or DMSO control at 1 or 9 hpi. Box plots are depicted as in Fig. 3C. A two-sided unpaired t-test was used to calculate p. n denotes the combined data from 3 independent experiments (batch), each batch containing 3-4 biological replicates per condition.
d) Schematic depicting Stefin B constructs used to generate iMAC cell lines targeting Stefin B to the nucleus (Nuc) with a 3x NLS sequence or expressed in the cytoplasm (Cyto). Stefin B potently inhibits several cathepsins (e.g. CtsL, CtsS, CtsH and to a lesser extent CtsB) 35 and has been previously used to block cathepsin activity and overcome cathepsin redundancy 36. Retrovirally transduced iMACs were FACS sorted for low (Lo) and high (Hi) expression of the Stefin B fusion proteins, which was then verified by immunoblot (Extended Data Fig. 10a) as well as localisation of the Stefin B fusion protein by microscopy (Extended Data Fig. 10b). We tested the sensitivity of Stefin B expressing cells to conditions known to activate the canonical inflammasome (i.e. nigericin + LPS) and the lysosome destablising agent, LLoMe, which triggers a distinct form of cell death. In line with our previous observations that cathepsin activity is not required for canonical inflammasome activation, cells with nuclear-expressed Stefin B were refractory to nigericin+LPS treatment, but also to LLoMe induced cell death (Extended Data Fig. 10c, d).
e) iMACs expressing Stefin B targeted to the nucleus, or expressed in the cytoplasm, were infected with wildtype STm (MOI = 100:1; 19 hpi). LDH release was quantified as in Fig 5b. Sample labels are as described in d. Box plots are depicted as in Fig 3c. A one sided unpaired t-test was used to calculate p. n denotes the combined data from 4 independent experiments (batches), each batch containing >8 biological replicates per condition.
Nuclear cathepsin activity is required for STm induced cell death
To address in which cellular compartment cathepsin activity is required for cell death, we followed a two-pronged approach. First, we hypothesised that if the suppression of STm-induced host cell death by cathepsin inhibition can be explained by cathepsin inhibition in the SCV and thereby SPI-2 activation and STm proliferation, then addition of cathepsin inhibitors subsequent to the initiation of SPI-2-dependent replication should abrogate their ability to inhibit host cell death. However, addition of inhibitor at 8 hpi rescued cathepsin-dependent cell death in BMDMs to a similar degree compared to addition of CA-074-Me at the beginning of infection (Fig. 6c). These data are consistent with a role for late cathepsin activity in promoting Caspase-11-mediated cell death. Since cathepsins relocate to the nucleus at this time point, we reasoned these activities play a role in driving cell death.
Second, we generated iMACs over-expressing the endogenous cathepsin inhibitor Stefin B with or without a nuclear localisation sequence (NLS) (Fig. 6d and Extended Data Fig. 10). We then infected Stefin B expressing cells with wildtype STm and measured LDH release. In this case, the nuclear-targeted Stefin B partially suppressed LDH release in a dose-dependent manner (Fig. 6e). This effect was specific to nuclear-targeted Stefin B and not to cytoplasmic Stefin B (Fig. 6e). Overall, these results are consistent with a specific functional role for nuclear cathepsin activity in promoting non-canonical inflammasome activation.
Discussion
We quantified the host pool of newly synthesised proteins, enabling unprecedented proteome-wide spatiotemporal resolution of a host-pathogen interface. This dataset provides a rich resource for infection biology, and could serve as a basis for new hypotheses. We used it to reveal a role for cathepsins in activating STm-induced pyroptosis via the non-canonical inflammasome. This proof-of-principle illustrates how the selective quantification of the newly synthesised host proteome within different cellular compartments can illuminate mechanisms that would otherwise remain hidden using conventional proteomic approaches.
The host proteome during STm infection has been mapped before: whole proteome analysis during infection of RAW264.7 macrophages (at the time, the authors could identify in total ~1,000 host proteins) 27, and mapping the secretome of human monocytes (THP-1) during early stages of infection - before intracellular SPI-2-dependent proliferation 28. In contrast to prior studies, our approach has three advantages: i) we monitor dynamic host proteome responses, collecting samples at different phases of infection and quantifying the newly synthesized proteins at each stage; ii) we use subcellular fractionation and pSILAC-AHA to assess protein redistribution between cellular compartments, quantifying 4978 newly synthesized host proteins from the cytoplasm/organelles, the nucleus and the extracellular milieu (lysatome, nucleome and secretome); and iii) we deconvolute the STm-specific response by directly comparing our data with similarly acquired data on the macrophage response to LPS 8. To our knowledge, this level of comprehensive proteomic analysis has not been performed before for any host-pathogen interface.
We observed that lysosomal proteases are rewired in their cellular trafficking during STm infection, both to the extracellular space and the nucleus (Supplementary Discussion). Support for nuclear cathepsins playing an active role in cell death comes from prior observations that CtsB is delivered to the nucleus during bile salt-induced apoptosis. In this case, addition of CA-074-Me or silencing ctsB expression abrogated cell death 29. Cathepsin B can also induce nuclear apoptosis in digitonin-permeabilized cells 30. Moreover, deletion of the endogenous cathepsin inhibitor Stefin B led to enhanced Caspase-11 transcription in BMDMs 31. Albeit in the absence of infection, these findings provide strong support that nuclear cathepsins play an active role during cell death. Consistently, we observed an increase in nuclear localization of CtsA, CtsB, CtsL, CtsS, CtsD and CtsZ during STm infection, which correlated with signs of cell death, such as DNA fragmentation. Furthermore, we could: a) inhibit pyroptosis by adding CA-074-Me at 8 hours after infection when cathepsin activity is present in the nucleus and b) partially inhibit cell death by expressing nuclear, but not cytosolic-targeted Stefin B. Both are consistent with nuclear cathepsin activity playing an active role in STm-induced cell death. By directly activating Caspase-11 through transfecting LPS into the host cytoplasm, we demonstrate that cathepsin activity is required downstream of Caspase-11 activation. We observed Gasdermin D expression to be elevated during STm infection in a Cathepsin-dependent manner. As Gasdermin D is the only known downstream substrate of Caspase-11, and is essential for executing cell death via the non-canonical inflammasome, our current working model entails LPS driven translocation of cathepsins to the nucleus to promote Gasdermin D expression and thereby impacting non-canonical inflammasome activation.
In summary, we used a proteomics approach to selectively enrich and quantify the newly synthesised host proteins during STm infection, unmasking a hidden layer of the mammalian innate immune response. This rich tapestry of regulated proteins, resolved throughout time and space offers a resource for new hypotheses governing host-pathogen interactions. We provide a proof-of-principle by further interrogating the re-trafficking of cathepsins outside of the cell and into the nucleus during STm infection. This highlights a role for cathepsins during infection and adds a new layer of complexity to STm-induced pyroptosis.
Methods
No statistical methods were used to predetermine sample size.
Media, chemicals and reagents
The following chemical and reagents used were purchased from Sigma: DMSO (cat. D8418), L-methionine, L-cysteine, L-lysine, L-arginine L-glutamine. Tris(carboxyethyl)phosphine (cat. C4706) and 40 mM 2-Chloroacetamide (cat. 22790), Triton X-100 (x100), heat inactivated Fetal Bovine Serum (FBS) (F9665-500ML), Phalloidin-ATTO 700 (79286-10NMOL), gentamicin (G1914), Gibco; DMEM 4.5 g/L glucose (41965), DMEM 4.5 g/L glucose non GMP formulation ME 100073L1 (without L-lysine HCl and L-arginine HCl), dialysed FBS (26400044), RPMI 1640 without (11835-030) & with (52400-025) phenol red, respectively. DMEM containing high glucose, HEPES buffered and without phenol was purchased from Thermo Fisher (21063029). Cathepsin inhibitor CA-074-Me was purchased from EMD Millipore (205531), L-azidohomoalanine (AHA) from Click chemistry tools (1066-100), cOmplete mini EDTA-free protease inhibitors from Roche (11873580001), recombinant murine M-CSF from PeproTech (315-02), Hoechst 33342 from Life Technologies (H3570), formaldehyde 16% (w/v) from Thermo Scientific Pierce™ (28908) and the CytoTox 96® Non-Radioactive Cytotoxicity kit from Promega (G1780). Antibodies were mostly purchased from Cell Signalling; GAPDH(D16H11) cat. 8884, Histone H3(D1H2) cat. 4499P, Sigma(Merck); anti-rabbit HRP (Sigma/GE - NA934-1ML), M2 antibody (F3165), anti-mouse HRP (Sigma, HVZ-A4416-1ML) and anti-Lamin A (Sigma - SAB4501764). Anti-RNA polymerase Sigma 70 (RpoD) [2G10] cat. GTX12088 was purchased from Acris antibodies. Anti Caspase-11 was purchased from Novus biological 17D9 (cat; NB120-10454SS). Anti Gasdermin D was purchased from Abcam (cat; ab209845). The cathepsin B antibody was purchased from R & D systems AF965-SP. The anti mouse Alexa Flour plus 647 antibody was purchased from Thermo Fischer (cat; A32728). Anti-tubulin (926-42211) was purchased from Li-Cor. Mouse serum was purchased from Abcam (ab7486).
Bacterial culture conditions and strain construction
Salmonella enterica Typhimurium 14028s (STm) was cultured at 37°C in LB Broth (Lennox) with agitation overnight in the presence of antibiotics for plasmid selection when necessary. Strains expressing antibiotic resistance genes were selected and maintained on solid LB agar plates containing citrate and 30μg/mL kanamycin (mutant selection) or 100μg/mL ampicillin (for pDiGc and pFCcGi plasmids). Mutant strains were retrieved from a single-gene mutant collection 37, followed by PCR confirmation and retransduction of the mutated loci into the wildtype STm 14028s background using P22 phage. To visualise bacteria during infection via fluorescence microscopy, a plasmid constitutively expressing GFP - pDiGc - 38 or mCherry - pFCcGi -was introduced into bacteria by electroporation followed by selection on LB agar containing ampicillin 100μg/mL at 37°C.
Cell culture conditions
RAW264.7 cells (ATCC®TIB71™) purchased from ATCC were routinely cultured in DMEM containing 4.5 g/L glucose and passaged by detaching with accutase (StemCell; A1110501) RAW264.7 cells were routinely tested for mycoplasma using the MycoAltert Mycoplasma detection kit (LT07-118, Lonza) which consistently returned a negative result. Only cells below passage number 15 were used for experiments. Bone marrow was isolated from both male and female 8-12 week old mice from wildtype C57Bl/6 mice, Casp1/11-/- 39, Casp11-/- 40, Nlrp3-/- 41 and Nlrc4-/- 42 (C57BL/6 genetic background) from ETH Zurich. Femur and tibia were flushed with PBS. The bone marrow cell suspension (from femur) was filtered through a 70 μm cell strainer (Falcon), washed with 20 ml PBS, spun down at 1200 rpm (4 °C, 15 min) and resuspended in 1 ml of 90 % heat-inactivated FBS (Life Technologies) + 10 % DMSO (Sigma) at a concentration of 1x107, then transferred to liquid nitrogen for storage. For experiments, bone marrow was thawed and washed in 10 mL of pre-warmed RPMI supplemented with 10% FBS (Sigma) (RPMI+FBS). Cells were then resuspended in 20 mL RPMI+FBS without phenol, supplemented with 50 μg/mL gentamicin and 40 ng/mL M-CSF. N.B. M-CSF was reconstituted in 0.1% bovine serum albumin (Carl Roth, cat. 8076.4), then aliquoted and stored at -30°C. Cell suspensions were then split across two 10 cm petri dishes, then incubated at 37°C, 5% CO2 for 6 days to allow bone marrow-derived macrophage (BMDMs) differentiation. BMDMs were washed with 3 mL PBS, and detached by incubating cells in 3 mL cell dissociation buffer (5% FBS, 2.5 mM EDTA in PBS) and incubation on ice for 5 minutes. Resuspended BMDMs were pooled, then pelleted at 500 xg and resuspended in 20 mL RPMI+FBS(5%) without phenol and supplemented with 40 ng/mL M-CSF.
Proteomic sample preparation
RAW264.7 cell infections were performed as previously described 43. Approximately 18-20 hours prior to infection, RAW264.7 cells were seeded in DMEM containing 10% FBS (DMEM+FBS) at a cell density of 0.9x105 per well in 6 well plates. Cell density from overnight cultures of wildtype STm 14028s expressing mCherry from pFCcGi grown in LB Broth (Lennox) at 37°C were measured (OD578) and normalised to OD578. = 1. Cells were then washed in PBS and pelleted at 8,000 xg for 5 min. To opsonize bacteria, pellets were resuspended in DMEM containing 10% mouse serum and incubated at room temperature for 20 min. Opsonized bacteria and mock inoculum, were added directly to wells containing RAW264.7 cells at an MOI 100:1 and centrifuged at 170 g for 5 min to promote bacterial uptake. Infected cells were incubated at 37°C at 5% CO2 for 25 min. Infected cells were then washed once with media and replaced with DMEM+FBS containing 100 μg/mL gentamicin and returned to the incubator for 1 hour. Media was then replaced with DMEM+FBS containing 16 μg/mL gentamicin for the remainder of the experiment; this step denotes t = 0 h. Therefore, for all experiments, t = time since addition of DMEM+FBS containing 16 μg/mL gentamicin.
To quantify the subpopulation of host proteins synthesised within a specific timeframe, we simultaneously pulsed cells with SILAC amino acids and an azide-containing analogue of methionine called azidohomoalanine (AHA). Intermediate (I) or heavy (H) SILAC labels facilitate robust protein quantification relative to uninfected controls, whereas AHA enabled enrichment of the newly synthesised proteome. These amino acid labels can be used as substrates by the hosts’ endogenous translational machinery and are thereby incorporated into newly synthesised proteins (pSILAC-AHA). Cells were pulse-SILAC-AHA labeled as previously described 6 with the following modifications. To deplete the cells of methionine, lysine and arginine, roughly 3.5 hours prior to harvest, infected and corresponding control cells were washed thrice with prewarmed PBS followed by a 30 min incubation in DMEM dropout media; DMEM containing 10% dialysed FBS, 4.5 g/L glucose, 40 mM L-glutamine, 60 μg/mL L-cysteine and 16 μg/mL gentamicin, but lacking L-methionine, L-lysine and L-arginine (GIBCO). This was then replaced with DMEM dropout media media supplemented with 100 μM L-azidohomoalanine and either 84 μg/ml [13C6,15N4] L-arginine and 146 μg/ml [13C6,15N2]L-lysine (heavy) or 84 μg/ml [13C6]L-arginine and 146 μg/ml [4,4,5,5-D4]L-lysine (intermediate) SILAC labels (Cambridge Isotope Laboratories, Inc). Cells were then pulse labeled for 3 hours to allow sufficient time for protein translation and subsequent trafficking throughout the cell.
For cell fractionation, conditioned media containing the “secretome” from pulse SILAC-AHA labeled cells were collected as previously described 6 and stored at -80°C. RAW264.7 cells were then washed three times with prewarmed PBS followed by partial lysis in 1 ml PBS containing 0.1% Tx-100 and protease inhibitors (Roche: cOmplete, mini, EDTA-free) per well for 10 minutes at room temperature. Intermediate and heavy isotopically labeled samples corresponding to either infected or uninfected cells were combined in a 1:1 ratio. To isolate Tx-100 resistant nuclei and bacteria, the lysate was centrifuged at 3,220 xg for 10 min at 4°C. Supernatant containing the “lysatome” was transferred to a separate tube and the pellets containing the “nucleome” were washed with PBS containing 0.1% Tx-100 followed by storage at -80°C. Thus, the lysatome is a nuclear-free cell lysate.
To enrich for the newly synthesised proteome, samples from two biological replicates that were simultaneously pulse labeled with SILAC and AHA labels (biological replicates contained reversed SILAC labels) for 3 h prior to harvest were harvested as previously described 8, with the following modifications. Secretome and lysatome samples were thawed and concentrated to a volume of ~250 ul using a 15 ml Amicon Ultra centrifugal device with a 3 kDa cutoff at 4°C at 3,220 xg. Nucleome pellet was thawed and solubilised in Lysis buffer (ThermoFischer Click-iT: C10416) followed by DNA shearing using a probe sonicator. Newly synthesised proteins were then enriched using 100 ul of beads and according to the manufacturer’s instructions with the following modifications. AHA-containing newly synthesised proteins were reacted with the beads overnight (~16 hours) at room temperature with rotation. Samples were then centrifuged at 2,600 xg. Beads were then washed three times with SDS buffer (1% SDS, 100mM Tris pH8, 5mM EDTA and 500mM NaCl) followed by reduction and alkylation by resuspending the beads in 500 ul SDS buffer containing 10 mM Tris(carboxyethyl)phosphine (Sigma: C4706) and 40 mM 2-Chloroacetamide (Sigma: 22790). Samples were then incubated for 30 minutes at 37 °C with constant agitation at 1,000 rpm. Beads were transferred to retention columns and washed in the following sequence: 7x with 1 ml SDS wash buffer, 10x with 1 ml freshly prepared Urea buffer (8M Urea, Tris-HCl pH 8.0), 10x with 20% 2-propanol and 10x with 20% acetonitrile. Beads were then transferred to low protein binding tubes by resuspending in buffer containing 100 mM Tris pH 8.0, 2 Mm CaCl2 and 4% acetonitrile. Beads were then centrifuged at 2,600 xg for 1 minute and supernatant decanted.
For peptide preparation, on-bead digestion was carried out in 50 ul of digestion buffer (8M Urea, Tris-HCl pH 8.0, 2.5% acetonitrile) by adding 2 μl of a 0.5 μg/μl LysC/Trypsin to each tube and incubation at 37 °C for 4 hours with shaking at 1,000 rpm. Urea was then diluted by adding 150 μl of buffer (100 mM Tris pH 8.0, 2 mM CaCl2 and 4% acetonitrile) and incubated overnight at 37 °C. Supernatants were then transferred to fresh microfuge collection tubes. Beads were then washed with 200 μl of H2O to collect residual peptides and collated. Samples were acidified by adding 8 μl (2% of the sample volume) of a 10% formic acid followed by acidification verification using a pH strip.
Peptides were desalted by binding to a Waters Oasis HLB 96-well μElution Plate (Waters: 186001828BA) using a vacuum manifold. Wells were preconditioned by passing 100 μl of 100% acetonitrile followed by 100 μl of Oasis buffer B (1% formic in 60% MeOH) then 100 μl of Oasis buffer A (1% formic in H2O). Samples were then bound followed by sequential 300 μl, 200 μl and 100 μl washes with Oasis buffer A. To elute peptides, a collection tray was loaded with glass vials over which the Oasis plate was carefully aligned. Peptides were eluted in subsequent 50 μl and 25 μl of Oasis buffer B. Glass vials were then transferred to 2 ml centrifuge tubes, pulse spun then stored at -20°C. Secretome and nucleome fractions were evaporated using a speedVac at 35°C for ~2 hours, then resuspended in 20 μl of injection buffer (96.9% water, 3% ACN and 0.1% Formic acid) for direct analysis by nano LC-MS/MS on Velos orbitrap.
To reduce sample complexity, lysatome samples were subjected to high-pH reversed phase fractionation. In brief, fractionation was performed on an Agilent 1260 HPLC system equipped with a variable wavelength detector (254nm). On the HPLC, fractionation was performed on an XBridge BEH C18 column (1 x 100mm, 3.5 μm, 130Å, Waters). Elution was performed at a flow rate of 0.1mL per minute using a gradient of mobile phase A (20mM ammonium formate, pH 10) and B (acetonitrile), from 1% to 37.5% over 61 minutes. Fractions were collected every 2 minutes across the entire gradient length and concatenated into 8 final samples as discussed previously 44. Fractions were dried in a SpeedVac and reconstituted in 0.1% formic acid prior to desalting on an Oasis uElution plate and analysis by LC-MS/MS. Samples were then analysed on an Orbitrap Velos Pro (Thermo Fisher Scientific) as previously described 8.
For proteomic data analysis, the raw data obtained by Orbitrap mass spectrometers were processed using MaxQuant software (version 1.5.0.0). The MaxQuant implanted search engine Andromeda was used to search the MS/MS spectra, against a mouse database obtained from Uniprot. In the database, the sequences of frequently observed contaminant proteins and the reversed sequences of all entries were included, to indicate the false-positive search hints. Trypsin/P was chosen as the digestion enzyme, and only no more than two miss cleavages were allowed for the peptide identification. Cysteine carbamidomethylation was used as the only fixed modification, while methionine oxidation, N-terminal acetylation and AHA replacement of methionine were chosen as the variable modifications. The minimal peptide length was set to 7 amino acids. The mass tolerance for peptides was 20 ppm in initial search and 6 ppm in main search. The maximal tolerance for fragment ion identification was 0.5 Da. False discovery rates for the identification of proteins and peptides were set to 1%. The minimal unique peptide number for protein identification was set to 1%. At least two ratio counts (quantified peptides for each protein) were set for the protein quantification. The “requantify” and “match between runs” were functionalized in MaxQuant. In the pulse SILAC-AHA samples, only protein groups having a ratio of over 20% identified peptides containing medium or heavy labels were kept for the further analysis. Proteins identified as potential contaminants, with reverse sequences, and from only peptides were removed. Average protein ratios were calculated, only if they were quantified in both biological replicates. The data are available via ProteomeXchange 45 with the identifier (PXD010179). Proteomic data was visualized using R (v 1.0.143) and Cytoscape (v3.3.0). Heatmaps were visualised with Heatmapper 46.
Cytokine and chemokine quantification
RAW264.7 cells were infected with wildtype STm as described above except that DMEM 4.5 g/L glucose containing 0.2% FBS was used for the 20 hour gentamicin 16 μg/L incubation. After 20 hours, conditioned media was collected and centrifuged at 2,000 rpm at 4°C for 10 minutes. Supernatants were then frozen at -80°C until analysis. Samples were analysed using a custom Quantibody® Mouse Cytokine Antibody Array (Protocol number: 061219 Cust-M7 SA14. Test procedure: SOP-TF-QAH-001, SOP-TF-QAH-003) from RayBiotech, Life. Inc.
Flow Cytometry of extracted nuclei
Infections were prepared as described above. Heat killed STm inoculum was prepared by heating bacteria to 65°C for 15 minutes prior to infection. Tissue culture media from infected cells were replaced with fresh DMEM containing 5 μM DCG04-Boclicky-TAMRA, followed by a 1 hour incubation at 37°C, 5% CO2. Cells were washed thrice in PBS followed by solubilization in 0.1% Triton X-100. Nuclei were pelleted (500 xg, 3 min), washed and resuspended in PBS containing 0.1% Tx-100. Nuclei were then fixed by adding formaldehyde to 4% wt/vol (Thermo Scientific; 28908) and incubated for 15 minutes at room temperature. Prior to flow cytometry, nuclei were stained with 2 μg/mL Hoechst. Nuclei samples were acquired on a BD LSRFortessa™ (BD Biosciences) equipped with 5 lasers. Hoechst was excited by the 355 nm laser line and fluorescence signal was collected using a 450/50 bandpass filter and TAMRA excited by the 561 nm laser line and collected using a 585/15 bandpass filter. Doublets were carefully excluded by plotting Hoechst-area versus Hoechst-width, nuclei with increased width signal were not considered. Post-acquisition analysis was done in FlowJo software (BD Biosciences).
SDS-PAGE and Immunoblot
RAW264.7 cells and BMDMs were infected in 6 well plates tissue culture plates at an MOI 100:1 as described above. For BMDMs immunoblots, cells lysates were harvested by washing twice with PBS and then partially lysing in 200 μL PBS lysis buffer (PBS containing 0.1% Tx-100 and cOmplete mini EDTA-free protease inhibitors) on ice for 10 min. Nuclei were removed by centrifugation at 500 xg for 5 minutes. Supernatants were added to Laemmli buffer loading dye and heated to 95°C for 3 minutes and analysed by immunoblot. To visualise cathepsin activity, infected and mock treated cells were labeled with 5 μm DCG04-Bodipy-FLike (λex 488 λem520) 3-4 hours prior to harvest. Cells were then washed three times with PBS, then partially lysed in 200 μL PBS lysis buffer (PBS containing 0.1% Tx-100 and cOmplete mini EDTA-free protease inhibitors) on ice for 10 min. Cell lysates were then collected and nuclei pelleted at 50 xg for 3 min, followed by a subsequent spin at 500 xg to pellet bacterial cells. Nuclei and bacterial enriched pellets were washed once and then resuspended in 200 μL PBS lysis buffer. Cell fractions were then combined with 4x Laemmli buffer loading dye, heated to 95°C for 3 min. Samples were then separated by SDS-PAGE and cathepsin activity measured using 488nm laser on Typhoon scanner (FLA 9500). The bottom sections of SDS-PAGE gels were subjected to Coomassie staining to visualise histones followed by western transfer to a 0.45μm PVDF membrane (Millipore Immobilin®-P, IPVH00010). Infected whole cell lysates for analysis by SDS-PAGE were generated by washing cells 3 times in PBS at the indicated time point and then directly lysing cells in 1x Laemmli buffer loading dye. Lysates were then heated to 95°C for 3 min and analysed by immunoblot. Prior to SDS-PAGE, nuclear DNA was mechanically sheared using a Hamilton syringe.
To examine cathepsin B trafficking to the nucleus in the presence of the CA-074-Me inhibitor, RAW264.7 cells were infected with wildtype and heat-killed wildtype (65°C for 15 minutes) in a 6 well format as described above. After cells were treated for 1 hour gentamicin, cells were then incubated in the presence of absence of indicated CA-074-Me concentrations or DMSO solvent control for the remainder of the experiment. 20 hours later, cells were washed twice and lysed in 200 μL PBS lysis buffer (PBS containing 0.1% Tx-100 and cOmplete mini EDTA-free protease inhibitors) and allowed to lyse on ice for 10 min. Nuclei were then resuspended in 1x Laemmli buffer loading dye, sonicated in a bath sonicator and heated to 95°C for 3 min. Samples were then separated by SDS-PAGE and the bottom the gel was subjected to coomassie staining to visualize histones. The rest of the gel was transferred to a PVDF membrane and probed as described below.
Membranes were blocked with 5% skim milk in TBS-T and probed with the following antibodies 5% skim milk in TBS-T overnight; RpoD 1:1000, GAPDH 1:10,000, Lamin A 1:5,000, Histone H3 1:,1,000 and H3.cs1 1:1,000, Cathepsin B (CtsB) 1:2,000; Caspase-11 1:1,000. Secondary antibodies were used at 1:5,000 and incubated with washed membranes for 1 h at room temperature. After washing, chemiluminescence substrate (GE Healthcare, RPN2106) was used for signal development and detected using x-ray film (Advantsta, L-07013-100) or a BioRad ChemoDoc Touch. X-ray film was converted to digital from by scanning at 300x300 dpi. Digital images were then cropped and contrast adjusted in Photoshop and incorporated into figures using Adobe Illustrator. Biological replicate samples were analysed on separate SDS-PAGE gels, unless otherwise stated.
LDH release assay
RAW264.7 and iMACs were seeded in DMEM containing 10% FBS (DMEM+FBS) at a cell density of 3x104 per well in 96 well plates and infected as described above with the following deviations. Unless otherwise stated, inhibitors and DMSO solvent was added at t = 0 h (see definition above). BMDMs were seeded in RPMI+FBS(5%) without phenol and supplemented with 40 ng/mL M-CSF at 5x104 cells per well into a 96 well plate (Thermo, cat.167008) ~18-20 hours prior to infection. The bacterial inoculum was prepared as described above. To opsonize bacteria, pellets were resuspended in RPMI containing 10% mouse serum and incubated at room temperature for 20 minutes. Opsonized bacteria were added directly to wells containing BMDMs, iMACs or RAW264.7 cells at an MOI 100:1 and centrifuged at 170 xg for 5 minutes to promote bacterial uptake. Infected cells were incubated at 37°C at 5% CO2 for 25 min. Cells were then washed once, media was replaced with RPMI + FBS(5%) without phenol, or DMEM+FBS(5%) without phenol for RAW264.7 and iMAC cells, containing 100 μg/mL gentamicin and returned to the incubator for 1 hour. Media was then replaced with RPMI+FBS(5%) (BMDMs) or DMEM+FBS(5%) (RAW264.7 and iMACs) without phenol and with 16 μg/mL gentamicin for the remainder of the experiment: this step denotes t = 0 h. LPS transfection of RAW264.7 cells was performed as previously described 19 using LPS (E. coli 0111:B4; Sigma L2630) except cells were seeded at 3x104 per well of a 96 well plates. LDH release as a measure of cell death was quantified according by measuring the % LDH release as previously described using the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega:G1780) kit 17 according to manufacturer’s instructions. Percentage cell death = 100 x (experimental LDH - spontaneous LDH) / (maximum LDH release - spontaneous LDH). Data was visualised in R and GraphPad Prism. Each experimental batch consisted of 3-8 (RAW264.7 & BMDMs) or 8-29 (iMACs) biological replicate wells per condition, whereby single measurements (490nm) were acquired per well. This measurement was then normalised to the spontaneous LDH release from uninfected wells that were otherwise identically treated, using the aforementioned formula and treated as a separate data point. In our hands, the degree of overall LDH release during STm infection displayed considerable variance across batches of BMDM cells. To reduce this variance, batches whereby wildtype STm induced <10% and >50% LDH release in RAW264.7 cells and BMDMs were removed (except for experiments where cells were infected with exponentially growing SPI-1 ON STm).
Microscopy
RAW264.7 cells were seeded in DMEM containing 10% FBS (DMEM+FBS) at a density of 4x105 cells per well in 24 well glass bottom tissue culture plates (Greiner, 662892). Cells were infected with pDiGc expressing bacteria as described above (MOI 100:1). At the indicated times, cells were then treated with the cell permeable cathepsin probe DCG04-Boclicky-TAMRA for 2 hours prior to harvest. Cells were washed three times with PBS and then fixed with in PBS solution containing 4% formaldehyde. Cells were then stained overnight with 2 μg/mL Hoechst 33342 and in PBS containing 0.05% Tx-100 to visualize host cell nuclei and host cytoskeleton, respectively. Images of stained cells were acquired on a Zeiss Cell Observer microscope using a 20x objective. Images were processed using CellProfiler software 2.1.1 as previously described 47. Nuclear cathepsin activity was quantified by measuring the integrated DCG04-Boclicky-TAMRA signal intensity that overlapped with the host nucleus defined by the Hoechst 33342 channel. Similarly, non non-nuclear cathepsin activity was calculated by subtracting the nuclear cathepsin activity from the total cathepsin activity per cell. Cathepsin activity was then normalised to the nuclear size (area) and non-nuclear cathepsin activity was normalised to the total cell area of the cell minus the nuclear area.
Confocal images were acquired on an Olympus confocal laser scanning microscope (FV3000) using an 60x oil immersion objective. Samples were prepared as described above in a glass bottomed 96 well plate. After three washes in PBS, cells were fixed with 4% formaldehyde and 0.4% Tx-100 in PBS for 1 hour at room temperature. Pictures were captured using the 405 nm laser for excitation for Hoechst 33342, 488 nm for GFP expressing (pDiGc) STm and 561 nm for DCG04-Boclicky-TAMRA. Image overlays and grey scale conversions were done in FIJI, then images were cropped in Photoshop before figure construction with Adobe Illustrator. Single planes from a z-stack are represented to provide a sharp boundary definition between the nucleus (Hoechst overlap) and the non-nuclear cathepsin activity.
Bacterial growth curves
Overnight bacterial cultures grown at 37°C in LB (Lennox) were pelleted at 8000 xg and washed 3 times in fresh media, then back-diluted to OD578 = 0.005 in the presence of inhibitors and solvent at the indicated concentrations. Cells were dispensed, 100 μL per well, into a round bottom 96 well plate in triplicate (Thermo, Nunclon Delta Surface, cat. 168136), then covered with a sealable transparent breathable membrane (Breathe-Easy by Divbio, cat. BEM-1). Cell growth was quantified by measuring the absorbance at 578 nm at 20 min intervals with constant shaking at 37°C.
Generation of Stefin B expressing iMACs
Immortalised wildtype macrophages (iMACs), and the retroviral transduction thereof were previously described 48, and HEK cells were cultured in DMEM containing 10% heat inactivated FCS, 100U/ml penicillin/streptomycin and 2mM GlutaMAX. iMACs were routinely tested for mycoplasma using a direct growth assay on conditioned media. Immortalised wildtype iMACs were retrovirally transduced with the listed Stefin B constructs or a control construct containing mCitrine only. Following transduction the cells were sorted for comparable expression of both constructs at either high or low expression of mCitrine by flow cytometry.
iMAC sample preparation and immunoblot
Samples for analysis of the Stefin B expressing iMACs were prepared and the immunoblot performed as described previously 49. Briefly cells were harvested, washed once in PBS and then lysed in RIPA lysis buffer (cOmplete protease inhibitors (Roche)), then sonicated to disrupt DNA to facilitate gel loading. The samples were then denatured and reduced by adding NuPAGE sample buffer (25% sample volume; Life technologies) and NuPAGE sample reducing agent (10% sample volume; Life technologies) and heating the samples to 95°C for 5 min. Proteins were separated by electrophoresis using a precast 4-12% SDS-PAGE gel (Novex, Invitrogen) and MES buffer (Novex, Invitrogen). The proteins were transferred to an Immobilon-FL PVDF membrane (Millipore) and blocked using 3% BSA in Tris-buffered saline pH7.4 (BSA-TBS) for 1h. The membrane was then probed with an anti-Flag antibody (Clone M2, Sigma) at a 1:1000 dilution and an anti-tubulin antibody (CST) at a 1:1000 dilution in BSA-TBS with 0.1% Tween overnight at 4C. Secondary antibodies (coupled to IRDye 680RD ot IRDye 800CW, Li-Cor Biosciences) were used 1:25,000 dilution.
Immunofluorescence of iMAC cells
Cells were fixed in 4% paraformaldehyde (Sigma) for 20 min, washed 5x in PBS, then permeabilised by incubation with 0.3% Triton X-100 diluted in PBS for 5 min. Cells were then incubated in immunofluorescence staining solution (PBS containing 1% heat inactivated FCS, 10% goat serum) for 1h at ambient temperature to block non-specific antibody binding. Primary antibody (anti-Flag 1:1000, (Clone M2, Sigma)) diluted in immunofluorescence staining solution was added to the cells and these incubated overnight at 4C. Cells were washed thrice in staining solution, then incubated with Alexa 647 conjugated mouse IgG antibody (1:800 dilution in staining solution, Life technologies) at RT for 1 h. Cells were then washed 3x, incubated with DAPI for 5 min to visualize nucleic acid (1ug/ml, Life technologies), then washed once. Cells were imaged using an Observer.ZI epifluorescence microscope, 20x objective (dry, PlanApochromat, NA 0.8; Zeiss), Axiocam 506 mono and ZEN blue software.
iMAC stimulation conditions
Stefin B expressing and control iMACs were harvested and plated at 0.8x104 cells/well in a 96 well plate and left to adhere for 3 hours. To assess LDH release after activation of the NLRP3 inflammasome, cells were washed once and primed with 200 ng/ml Ultra-pure LPS (Invivogen; tlrl-3pelps) for 3 hours in DMEM containing 1% heat inactivated FCS. 10 μM nigericin (Invitrogen; N1495) was then added and the cells incubated for a further 1.5 h. To assess LDH release following Leu-Leu-o-Methyl ester (LLoMe - Santa Cruz; 16889-14-8) cells were washed once and treated with 1mM LLoMe for 2 hours in DMEM containing 1% heat inactivated FCS. For both assays the 96 well plate was centrifuged at 400 xg for 5 minutes and supernatants were then harvested and LDH release was measured from the supernatant using the LDH cytotoxicity kit (Pierce; 10008882).
Two-dimensional thermal proteome profiling (2D-TPP)
Two-dimensional thermal proteome profiling was performed as previously described 21. Briefly, RAW264.7 macrophages were infected overnight cultures of wildtype STm as described above and CA-074-Me was added to a final concentration of 1 μM, 4 μM, 20 μM, 100 μM, and a DMSO together with the 16 μg/mL gentamicin treatment. Two wells of a 6 well plate were pooled for each condition (i.e. ~1.8x 106 cells per condition, times 5 conditions). Drug treatment was retained throughout the infection. After 20 hours, cells were washed twice with PBS, aliquoted to a PCR plate, and heated for 3 minutes to ten different temperatures (37 °C – 67 °C) in a PCR machine (Agilent SureCycler 8800). Lysis buffer (final concentration: 0.8% NP40, 1.5 mM MgCl2, 1× protease inhibitor (Roche), 1× phosphatase inhibitor (Roche), 250 U/mL benzonase in PBS) was added and cells were lysed for 1 hour at 4°C. Protein aggregates were then removed as previously described 50, and the remaining soluble proteins were digested according to a modified SP3 protocol 51,52. Peptides were labeled with TMT10plex (ThermoFisher Scientific), fractionated into twelve fractions under high pH conditions and analyzed with liquid chromatography coupled to tandem mass spectrometry, as previously described 50. Protein identification and quantification was performed using IsobarQuant 53 and Mascot 2.4 (Matrix Science) against a concatenated FASTA file with Salmonella Typhimurium (Proteome ID: UP000001014) and Mus musculus (Proteome ID: UP000000589) proteins. Data was analyzed with the TPP package for R 53.
Caspase-11 activity assay
Caspase-11 activity was measured as previously described 54. In brief, 10 U/100 μL of recombinant Caspase-11 (mouse, Enzo life sciences; BML-SE155-5000) in caspase activity buffer (200 mM NaCl, 50 mM HEPES pH 8.0, 50 mM KCl, 10 mM DTT) supplemented with 100 μMAcLEHD-afc (Santa Cruz; sc-311277) was incubated in the presence of CA-074-Me, E64d (Sigma; E8640) and the Caspase-9/11 inhibitor Z-LEHD-FMK (Abcam; ab142026) or a DMSO solvent control as the indicated concentrations on the figure legend. Hydrolysis of the Caspase-11 substrate was measured at regular time intervals at 37°C using an Infinite M1000Pro TECAN spectroflourometer in a 96 well flat bottom transparent plate (Thermo Scientific; 167008) (excitation 400nm, emission 505 nm).
Statistical analyses
For cell death (LDH release) assays, significance testing was performed in R using the unpaired t-test (Figure 5b, 6b & c, 7c & e). To avoid assuming any shape of the distribution for single cell nuclei analysis of cathepsin activity, statistical testing was performed in R using the unpaired Wilcoxon rank sum test (non-parametric) (Figure 4d & S4). For proteomics data, statistical tests were performed using the Limma package in R/Bioconductor as previously described 8. Briefly, after fitting a linear model to the data, an empirical Bayes moderated t test was used and p values were adjusted for multiple testing with Benjamini and Hochberg’s method. Gene Ontology (GO) enrichment was calculated with the ClueGO plugin v2.1.7 of Cytoscape v3.3.0. For global analyses described (Figure 1b), GO enrichment was performed on proteins displaying ±1.5 Log2 fold change (infected/uninfected). For comparison between STm and LPS stimulated cells in (Figure 2a & b), GO enrichment was performed on proteins ±2 s.d. of the Log2 fold change (infected/uninfected). A custom reference set was used for each test and contained all quantified proteins within the same fraction. GO enrichment was performed separately on up- and down-regulated proteins using the right-sided hypergeometric test, and corrected for multiple testing using the Bonferroni step-down (Holm) method. This test was chosen for stringent error control.
Extended Data
Extended Data Fig. 1. Increasing intracellular STm load over time co-occurs with dynamic proteome changes.
Increasing intracellular STm load over time co-occurs with dynamic proteome changes.
a) RAW264.7 cells infected with STm constitutively expressing mCherry (pFCcGi) at MOI 100:1 and incubated for the indicated times were analysed by CellProfiler. SPI-1 OFF bacteria were chosen to mimic the pathogen-host interaction at systemic sites. After bacterial internalization, cells were treated with gentamicin to kill non-internalised bacteria, synchronise the infection and avoid re-infection cycles. The intracellular bacterial load was quantified at the single-cell level to ensure sampling times spanned distinct phases of intracellular STm proliferation states i.e. pre-proliferation (4h), initial (8h) and extensive (20h) proliferation post uptake. Images were captured at 20x objective and analysis was conducted with CellProfiler to segment infected from uninfected cells and quantify bacterial load per cell based on mCherry fluorescence per infected cell. CellProfiler quantification of bacterial count per infected cell (only infected cells) are displayed as a histogram and shows increasing bacterial load with time. Data contains combined counts from two biological replicate experiments, whereby each replicate received reverse SILAC labels (see Methods). 2-5 fields of view (technical replicates) were acquired per biological replicate and per time point. The combined total of infected host cells quantified is indicated (n=). The dotted red line indicates distribution median. Representative images of quantified cells are displayed to the right of histograms. Scale bars represent 50 μm. The % of infected cells per field of view (FOV) at the beginning of the experiment (i.e. immediately post gentamicin 100 μg/ml treatment, t=0) is displayed as a boxplot on the far right. Box plots are depicted as in Fig. 3C. n denotes FOV, amounting to 1224 cells in total (infected & non-infected).
b) Replicate correlation of infected versus uninfected samples for the indicated time points and different subcellular fractions - cytoplasmic and solubilised organelles within the Tx-100 soluble fraction (lysatome; upper), nuclear enriched fraction in the Tx-100 insoluble fraction (nucleome-middle) and proteins secreted into the culture supernatant (secretome-lower). Two biological replicates containing reversed SILAC labels were obtained for each cell fraction and each time point (See Supplementary Table 1). We observed concordant biological replicate correlation (r = Pearson’s correlation) across all time points, and within each subcellular fraction (mean r = 0.886). Data is from n=2 biologically independent samples. A red dotted line represents a linear model by robust regression and r refers to the Pearson’s correlation coefficient.
Extended Data Fig. 2. Quantification of cytokine and chemokine release from RAW264.7 cells during STm infection.
Quantification of cytokine and chemokine release from RAW264.7 cells during STm infection. After RAW264.7 cells were infected with STm for 20 hours, the conditioned media was removed and indicated cytokines and chemokines were quantified using a custom array (Biocat, GmbH). n denotes the combined data from 2 independent experiments (batches), each batch containing 2 biological replicates per condition. Box boundaries indicate the upper and lower IQR, the median is depicted by the middle boundary and whiskers represent 1.5x IQR. Relative infected/uninfected Δ (Log2) from the cytokine array are depicted within the plot for each protein. Corresponding pSILAC-AHA infected/uninfected (Log2) ratios are presented below the corresponding protein for the 20hr post-infection time-point (with the exception of TNFa, which we could only detect at 8 hpi). Non-hits and hits match perfectly between the 2 methods. Fold effect differences between pSILAC-AHA and array-based quantification are most likely due to the fact that the cytokine assay measures steady-state protein levels and pSILAC-AHA newly synthesized proteins. As expected, for more abundant proteins, measuring steady-state levels masks changes in expression dynamics. A two-sided unpaired t-test was used to calculate p.
Extended Data Fig. 3. Nuclear cathepsin translocation occurs already from 8 hpi, is independent of cathepsin activity and does not result in increased Histone 3 cleavage.
Nuclear cathepsin translocation occurs already from 8 hpi, is independent of cathepsin activity and does not result in increased Histone 3 cleavage. a) Time course of nuclear cathepsin activity. Nuclei and STm were enriched by sequential 50 x g and 8,000 x g centrifugation steps, respectively. RAW264.7 cells were infected with wildtype STm 14028s and treated with DCG04-Bodipy-FLike (5μM) for 4 hours prior to harvesting. Samples were separated by SDS-PAGE and visualised using a fluorescent scanner (Ex 405 nm/Em 520 nm), followed by immunoblotting for the soluble cytoplasmic protein GAPDH, the bacterial protein RpoD and Coomassie staining for histones as a loading control for nuclear extracts. L = lysatome, N = nucleome, STm = STm enriched fraction. Experiment was performed once. b) RAW264.7 cells were infected with wildtype STm at MOI 100:1 and harvested at 20 hpi. Whole cell lysates were immunoblotted with the CtsL specific cleavage product of H3 (H3.cs1)16 and histone 3 (H3) as loading control. The experiment was performed in biological duplicate per condition and were analysed in adjacent lanes. c) Nuclear extracts from RAW264.7 cells either mock infected, infected with wildtype STm or heat killed wildtype (STm-HK) for 20 hours. CA-074-Me was present throughout the infection experiment at the indicated concentrations. Nuclear extracts were analysed by immunoblot for CtsB (1:2,000), Lamin A (1:5,000) and histones by Coommassie stain (see Supplementary Information (SI) for second replicate data).
Extended Data Fig. 4. Nuclear cathepsin activity is enhanced by wildtype STm and mildly by an ssaV mutant and heat killed bacteria.
Nuclear cathepsin activity is enhanced by wildtype STm and mildly by an ssaV mutant and heat killed bacteria. Nuclei were extracted from RAW264.7 cells treated with DCG04-Boclicky-TAMRA (5 μM) for 2 hours prior to harvest at 20 hpi with wildtype (Wt), a SPI-2 mutant (ΔssaV) or heat killed wildtype bacteria (Wt HK) (MOI = 100:1). Samples were processed as described in Fig. 5a. Data is combined from two independent experiments (batches). n denotes the number of biologically independent samples. A two-sided unpaired t-test was used to calculate p. Boxplots are depicted as in Fig. 3C.
Extended Data Fig. 5. STm growth is unaffected by the cathepsin inhibitor CA-074-Me in batch culture conditions.
STm growth is unaffected by the cathepsin inhibitor CA-074-Me in batch culture conditions.
STm 14028s growth was measured in the presence of the selective cathepsin inhibitor CA-074-Me in LB 6, 12.5 and 25 μM and DMSO solvent controls. Drug concentrations used are indicated. Experiment was performed in biological triplicate.
Extended Data Fig. 6. CA-074-Me inhibits cathepsins and reduces expression of proinflammatory proteins.
CA-074-Me inhibits cathepsins and reduces expression of proinflammatory proteins.
a) 2D-TPP of RAW264.7 cells infected with wildtype STm 14028s for 20 hours in the presence of increasing concentrations of CA-074-Me (1-100 μM). To assess CA-074-Me specificity, we applied 2-dimensional Thermal Proteome Profiling (2D-TPP). 2D-TPP enables proteome assessment of protein ligand binding based on the principle that ligand-bound proteins are more thermally stable than proteins not bound to a ligand 20,21, and in parallel provides information of proteome-wide protein abundance. We subjected RAW264.7 cells infected with wildtype STm to increasing doses of CA-074-Me and compared proteome-wide thermostabilisation relative to solvent controlled cells (see methods). Increased (blue) or decreased fold changes are calculated by normalising the abundance of each protein relative to the abundance in the DMSO vehicle per temperature. Protein stabilization (red framed) is demonstrated by increasing (blue) fold changes with respect to increasing temperature (top to bottom) as well as with increasing drug concentrations (left to right). Changes in protein expression (green framed) can be detected if abundance changes are already evident from low temperatures i.e. before proteins melt. All cathepsins detected by 2D-TPP are displayed. CtsA and CtsD are serine and aspartic-acid proteases, respectively, and are therefore not targeted by CA-074-Me and serve as negative controls. Contrary to evidence demonstrating CA-074-Me targets CtsL 35, we failed to detect its stabilisation in our experiment. Cathepsins are arranged in the top row, followed by caspases and other cellular proteases in descending order. Note Caspase-1 stability was not affected by CA-074-Me, in line with previous reports 22. Additionally, CA-074-Me reduced IL-1b expression as previously shown 35,36. 2D-TPP data can be found in Supplementary Table 6.
b) Caspase-11-like proteolysis activity was assessed using purified active Caspase-11 and measuring cleavage of the fluorescent substrate AcLEHD-afc as previously described 37. Caspase-11-like proteolytic activity was assessed in the presence of indicated concentrations of CA-074-Me and the Caspase-11 inhibitor Z-LEHD-FMK as a positive control. Activity is expressed as the % of AcLEHD-afc cleavage relative to the DMSO solvent control. Data is combined from n = 3 biologically independent experiments; error bars depict standard deviation and the center value denotes the mean.
c) 2D-TPP profiles of inflammation related proteins reduced in abundance upon CA-074-Me treatment. Data presented as in (a).
Extended Data Fig. 7. Rapid cell death induced by SPI-1 ON STm is independent of cathepsin activity.
Rapid cell death induced by SPI-1 ON STm is independent of cathepsin activity.
Wildtype and Caspase-1/11 -/- BMDMs were infected with wildtype STm and a SPI-1 mutant (ΔprgK), late-exponential growing STm (SPI-1 ON) in the presence of CA-074-Me at the indicated concentrations. LDH release was measured 1.5 hpi. Boxplots are depicted as in Fig. 3C. n denotes the number of biologically independent samples.
Extended Data Fig. 8.
LPS transfection promotes nuclear cathepsin activity. RAW264.7 cells were transfected with purified LPS for 20 hours. 2 hours prior to harvest cells were treated with DCG04-Boclicky-TAMRA (5 μM) to quantify nuclear cathepsin activity. Nuclei were extracted and analysed by flow cytometry as per Fig. 5A. The percentage of cathepsin positive (TAMRA+) nuclei is indicated and a second biological replicate is located in the supplementary material replicate data section.
Extended Data Fig. 9.
CA-074-Me does not affect Caspase-11 expression or processing. Wildtype BMDMs were infected with wildtype STm for 20 hours in the presence of CA-074-Me (25 μM) or DMSO control. Lysates were probed for Caspase-11 by immunoblot and using GAPDH as a loading control. Experiment was performed once.
Extended Data Fig. 10.
Characterisation of iMACs expressing Stefin B targeted to the nucleus or expressed in the cytoplasm. a) Expression of Stefin B fusions targeted to either the nucleus (Nuc) or expressed in the cytoplasm (Cyto) were verified in iMAC lysates by immunoblot using anti-FLAG (M2) and anti-tubulin as a loading control. Cells were FACS sorted into high (Hi) and low (Lo) Stefin B expressing populations prior to use. Cells treated in parallel expressing mCitrine alone were used as negative controls. Experiment was repeated twice with similar results (see Supplementary Information for replicate data). b) Expression and localisation of Stefin B was assessed by microscopy in cells expressing nuclear-targeted (Nuc-Lo and Nuc-Hi) or cytoplasmic (Cyto-Lo and Cyto-Hi) Stefin B or mCitrine alone as a control. Scale bar denotes 25 μm. Experiment was repeated twice with similar results (see Supplementary Information for replicate data). c) iMACs expressing nuclear or cytoplasmic Stefin B were treated with 1 mM LLoMe or untreated and LDH release was quantified and plotted as described previously in Fig. 5b. Each data point is the combined average of biological replicate wells from three independent experiments. Boxplots are depicted as in Fig. 3c. d) iMACs expressing nuclear or cytoplasmic Stefin B were untreated, LPS treated or LPS and nigericin treated and LDH release was quantified and plotted as described previously in Fig. 5b. Each data point is the combined average of biological replicate wells from three independent experiments. Boxplots are depicted as in Fig. 3c.
Supplementary Material
Acknowledgements
We thank the EMBL Flow Cytometry, Proteomics and ALMF Core Facilities; Nils Kurzawa for 2D TPP data analysis; Dr. Sophie Helaine (Imperial College) for the pDiGc plasmid; Omar Wagih (EBI) for R scripts; Kyung-Min Noh (EMBL) for the H3.cs1 antibody; and members of the Typas and Krijgsveld laboratories for discussions. We acknowledge funding from EMBL for this research. JS, NL and HI were supported by Marie Skłodowska-Curie Actions COFUND grant number 291772 and AM by grant number 664726. EL was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC2151 – 390873048 and the ERC consolidator grant InflammAct. NL is now supported by the National Key Research and Development Program of China (2018YFA0902703), the Shenzhen Science and Technology Innovation Committee (JCYJ20170818164014753) and the National Natural Science Foundation of China (31800694 and 31971354). NKJ and BT were supported by the Slovene Research Agency (P1-0140).
Footnotes
Author contributions
Conceptualization: JS, JK and AT; Investigation: JS, NL, AM, GS, MS, JK (proteomics); JS, JB, AS, MM, MZ (molecular biology, biochemistry and cell biology); Resources: BIF, HSO (cathepsin reactive probes), MM, EL (iMACs), NK-J, BT (Stefin B construct) and AH, W-DH (BMDMs); Data analysis: JS, MM, AS, BED, NL, HI, AM; Writing: JS and AT with input from NL, MM, JB, AH, AS, HI, GS, PB, WDH, PB, and JK; Figures: JS, except for TPP (AM), bacterial growth curves (AM) and iMAC data (MM) - with input from JK and AT; Supervision: AT, JK, MS, EL and PB; Funding Acquisition: AT and JK.
Declaration of interest
The authors declare no competing interests.
Data Availability
Proteomic pSILAC-AHA data has been deposited at ProteomeXchange (http://www.proteomexchange.org/) under the identifier PXD010179 (pSILAC-AHA) and PXD016086 (2D-TPP).
Code Availability
The code and pipelines used for data analysis are available upon request.
References
- 1.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 2.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 3.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 4.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 5.Aachoui Y, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichelbaum K, Winter M, BerrielDiaz M, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol. 2012;30:984–990. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 7.Figueira R, Watson KG, Holden DW, Helaine S. Identification of salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar typhimurium: implications for rational vaccine design. MBio. 2013;4:e00065. doi: 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichelbaum K, Krijgsveld J. Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation. Mol Cell Proteomics. 2014;13:792–810. doi: 10.1074/mcp.M113.030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGourty K, et al. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science. 2012;338:963–967. doi: 10.1126/science.1227037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goulet B, et al. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 11.Tamhane T, et al. Nuclear cathepsin L activity is required for cell cycle progression of colorectal carcinoma cells. Biochimie. 2016;122:208–218. doi: 10.1016/j.biochi.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Goulet B, et al. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol Cancer Res. 2007;5:899–907. doi: 10.1158/1541-7786.MCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 13.Konjar S, Yin F, Bogyo M, Turk B, Kopitar-Jerala N. Increased nucleolar localization of SpiA3G in classically but not alternatively activated macrophages. FEBS Lett. 2010;584:2201–2206. doi: 10.1016/j.febslet.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 14.Turk V, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tedelind S, et al. Nuclear cysteine cathepsin variants in thyroid carcinoma cells. Biol Chem. 2010;391:923–935. doi: 10.1515/BC.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan EM, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol. 2000;38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- 18.Fink SL, Cookson BT. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006;8:1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- 19.Napier BA, et al. Complement pathway amplifies caspase-11-dependent cell death and endotoxin-induced sepsis severity. J Exp Med. 2016;213:2365–2382. doi: 10.1084/jem.20160027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savitski MM, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014;346:1255784. doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 21.Becher I, et al. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat Chem Biol. 2016;12:908–910. doi: 10.1038/nchembio.2185. [DOI] [PubMed] [Google Scholar]
- 22.Hentze H, Lin XY, Choi MSK, Porter AG. Critical role for cathepsin B in mediating caspase-1-dependent interleukin-18 maturation and caspase-1-independent necrosis triggered by the microbial toxin nigericin. Cell Death Differ. 2003;10:956–968. doi: 10.1038/sj.cdd.4401264. [DOI] [PubMed] [Google Scholar]
- 23.Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J Immunol. 2008;180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- 24.van der Velden AW, Lindgren SW, Worley MJ, Heffron F. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype typhimurium. Infect Immun. 2000;68:5702–5709. doi: 10.1128/iai.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broz P, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monack DM, Detweiler CS, Falkow S. Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1. Cell Microbiol. 2001;3:825–837. doi: 10.1046/j.1462-5822.2001.00162.x. [DOI] [PubMed] [Google Scholar]
- 27.Shi L, et al. Proteomic investigation of the time course responses of RAW 264.7 macrophages to infection with Salmonella enterica. Infect Immun. 2009;77:3227–3233. doi: 10.1128/IAI.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hui WW, et al. Salmonella enterica Serovar Typhimurium Alters the Extracellular Proteome of Macrophages and Leads to the Production of Proinflammatory Exosomes. Infect Immun. 2018;86 doi: 10.1128/IAI.00386-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts LR, et al. Cathepsin B contributes to bile salt-induced apoptosis of rat hepatocytes. Gastroenterology. 1997;113:1714–1726. doi: 10.1053/gast.1997.v113.pm9352877. [DOI] [PubMed] [Google Scholar]
- 30.Vancompernolle K, et al. Atractyloside-induced release of cathepsin B, a protease with caspase-processing activity. FEBS Lett. 1998;438:150–158. doi: 10.1016/s0014-5793(98)01275-7. [DOI] [PubMed] [Google Scholar]
- 31.Maher K, et al. A role for stefin B (cystatin B) in inflammation and endotoxemia. J Biol Chem. 2014;289:31736–31750. doi: 10.1074/jbc.M114.609396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateus A, Määtä TA, Savitski MM. Thermal proteome profiling: unbiased assessment of protein state through heat-induced stability changes. Proteome Sci. 2016;15:13. doi: 10.1186/s12953-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenbaum D, et al. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- 35.Turk B, Turk D, Salvesen GS. Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Curr Pharm Des. 2002;8:1623–1637. doi: 10.2174/1381612023394124. [DOI] [PubMed] [Google Scholar]
- 36.Orlowski GM, et al. Multiple Cathepsins Promote Pro-IL-1β Synthesis and NLRP3-Mediated IL-1β Activation. J Immunol. 2015;195:1685–1697. doi: 10.4049/jimmunol.1500509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porwollik S, et al. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One. 2014;9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helaine S, et al. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci U S A. 2010;107:3746–3751. doi: 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 40.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 41.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 42.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 43.Beuzón CR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang F, Shen Y, Camp DG, 2nd, Smith RD. High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev Proteomics. 2012;9:129–134. doi: 10.1586/epr.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vizcaíno JA, et al. The PRoteomicsIDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063-9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Babicki S, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–53. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochoa D, et al. An atlas of human kinase regulation. Mol Syst Biol. 2016;12:888. doi: 10.15252/msb.20167295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stutz A, et al. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med. 2017;214:1725–1736. doi: 10.1084/jem.20160933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mateus A, et al. Thermal proteome profiling in bacteria: probing protein state in vivo. Mol Syst Biol. 2018;14:e8242. doi: 10.15252/msb.20188242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes CS, et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 52.Hughes CS, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franken H, et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- 54.Ross C, Chan AH, VonPein J, Boucher D, Schroder K. Dimerization and auto-processing induce caspase-11 protease activation within the non-canonical inflammasome. Life Sci Alliance. 2018;1:e201800237. doi: 10.26508/lsa.201800237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic pSILAC-AHA data has been deposited at ProteomeXchange (http://www.proteomexchange.org/) under the identifier PXD010179 (pSILAC-AHA) and PXD016086 (2D-TPP).
The code and pipelines used for data analysis are available upon request.