Abstract
Up to 20% of the global population develops gastrointestinal symptoms following a meal1, leading to decreased quality of life, significant morbidity and high medical costs. Although the interest of both the scientific and lay community has increased dramatically with the worldwide introduction of gluten-free and other diets, the underlying mechanisms leading to food-induced abdominal complaints remain largely unknown. Here we show that a bacterial infection and bacterial toxins can trigger an immune response leading to the production of dietary antigen-specific IgE antibodies in mice, a mechanism confined to the intestine. Subsequent oral ingestion of the respective dietary antigen results in increased visceral pain via an IgE- and mast cell-dependent mechanism. This aberrant pain signaling results from histamine receptor H1 (H1R)-mediated sensitization of visceral afferents. Moreover, in patients with irritable bowel syndrome (IBS), we show that injection of food antigens (gluten, wheat, soy and milk) into the rectosigmoid induces local edema and mast cell activation. Hence, we have unveiled and characterized a novel peripheral mechanism underlying food-induced abdominal pain, which creates new opportunities for the treatment of IBS and related abdominal pain disorders.
The mucosal immune system provides a balanced response to pathogens and harmless commensal bacteria or food antigens, thereby limiting unnecessary inflammation and concomitant tissue damage2. This is achieved by an active suppression of cellular and humoral responses to orally administered antigens, a mechanism referred to as oral tolerance3. Viral and bacterial infections can, however, interfere with tolerance to dietary antigens, thereby perturbing intestinal homeostasis4. An infectious gastroenteritis is a significant risk factor to develop IBS, defined as a constellation of abdominal pain and altered bowel patterns. Between 3 and 36% of enteric infections lead to new onset IBS5, while up to 17% of IBS patients report that their symptoms started upon gastrointestinal infection6. Over 10% of the general population experience IBS symptoms, of which food ingestion is one of the most common triggers7. Aberrant pain signaling or visceral hypersensitivity (VHS) is a hallmark symptom of IBS. Increased mast cell (MC) numbers and release of MC-mediators have been proposed to underlie VHS8, although the stimuli responsible for the activation of MC in IBS, especially in response to food intake, remain unclear. IBS is a debilitating and difficult-to-treat condition for which no curative therapies are currently available.
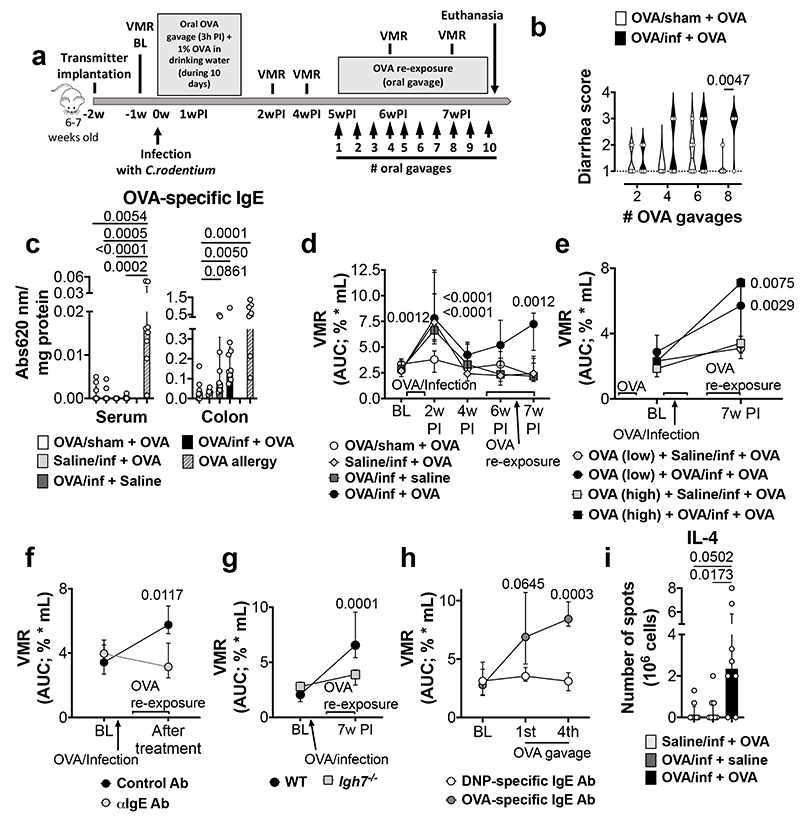
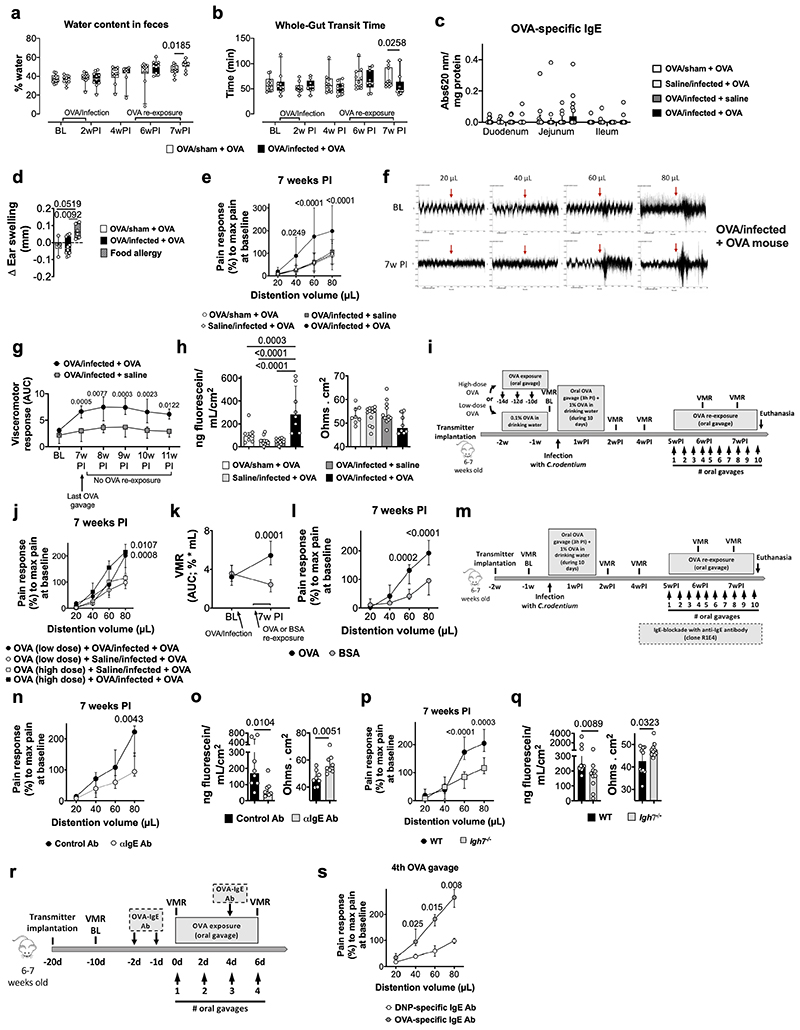
We hypothesized that the break of oral tolerance to food antigens caused by a bacterial infection underlies food-induced VHS. Hence, BALB/c mice were infected with Citrobacter rodentium while exposed to ovalbumin (OVA) in the drinking water (OVA/infected) (Fig. 1a). After clearance of the infection9, repeated oral gavages of OVA led to the development of diarrhea, increased fecal water content and reduced transit time (Fig. 1b, Extended Data Fig. 1a, b) in infected (OVA/infected + OVA) but not in uninfected (OVA/sham + OVA) mice. OVA-specific IgE antibodies were detected in the colon of OVA/infected mice, the site colonized by C. rodentium, but not in the small intestine or serum (Fig. 1c and Extended Data Fig. 1c). Moreover, these mice failed to develop ear-swelling after intradermal injection of OVA, compared to OVA-allergic mice (Extended Data Fig. 1d), suggesting a local rather than a systemic immune response.
Fig. 1. OVA-specific immune response and VHS in post-infectious mice.
a, Experimental protocol. b, Diarrhea score in OVA/sham + OVA and OVA/infected + OVA mice (n = 6 and 8, respectively). c, Quantification of OVA-specific IgE in serum (left) and colon homogenates (right) of OVA/sham + OVA, saline/infected + OVA, OVA/infected + saline, OVA/infected + OVA and OVA-allergy mice (serum: n = 9, 10, 10, 9 and 8, respectively; colon: n = 9, 6, 9, 9 and 7, respectively). d-h, VMR to colorectal distention in (d) OVA/sham + OVA, OVA/infected + saline, saline/infected + OVA and OVA/infected + OVA mice (n = 12, 11, 11 and 13, respectively); (e) mice OVA-tolerized (high-dose) + saline/infected + OVA, OVA-tolerized (high-dose) + OVA/infected + OVA, OVA-tolerized (low-dose) + saline/infected + OVA and OVA-tolerized (low-dose) + OVA/infected + OVA (n = 6, 9, 6 and 8, respectively); (f) OVA/infected + OVA mice treated with anti-IgE antibody or control antibody (n = 8/group); (g); OVA/infected + OVA mice with WT or Igh7-/- background (n = 10/group); (h) and naïve mice treated with monoclonal OVA-specific IgE antibody or monoclonal Dinitrophenyl (DNP) antibody (n = 7 and 6, respectively). i, mIL-4-forming cells in coMLN from saline/infected + OVA, OVA/infected + OVA and OVA/infected + OVA mice (n = 8/group). Two-tailed Mann-Whitney test in b; Kruskal-Wallis test (Dunn’s multiple-comparisons test) in c and i; two-way repeated ANOVA (Sidak’s multiple-comparisons test) in d-h. Data shown as violin plots in b and median ± IQR in c-i. VMR, visceromotor response; BL, baseline; w, week.
To understand the functional implications of this altered immune reaction to a food antigen, we examined if re-exposure to OVA could affect pain perception. As previously described10, mice infected with C. rodentium developed a transient period of increased pain response to colorectal distension, referred to as VHS, which returned to baseline after 4 weeks. Notably, OVA re-exposure 5 weeks post-infection onwards resulted in VHS in OVA/infected, but not in uninfected mice (Fig. 1d and Extended Data Fig. 1e, f), which persisted for at least 4 weeks after the last OVA gavage (Extended Data Fig. 1g). VHS was associated with increased mucosal permeability to sodium fluorescein (Extended Data Fig. 1h), consistent with data obtained in IBS patients11. Similar findings were obtained in mice tolerized to OVA prior to infection, indicating that an infection with C. rodentium can induce a break in oral tolerance once established (Fig. 1e and Extended Data Fig. 1i, j). Moreover, the development of VHS was antigen-specific, as repeated gavages of bovine serum albumin (BSA) failed to affect pain perception in OVA/infected mice (Extended Data Fig. 1k, l). No differences in the volume-pressure relationship (i.e. compliance) were found between groups (Extended Data Table 1), excluding the development of inflammatory-mediated changes in wall stiffness that altered nociceptive signaling.
To further explore the role of the antigen-specific immune response to OVA in VHS, mice were treated with a monoclonal anti-IgE antibody (Extended Data Fig. 1m). This treatment prevented the development of VHS and the increase in colonic permeability upon OVA re-exposure (Fig. 1f and Extended Data Fig. 1n, o). These results were confirmed in IgE-deficient mice (Igh7-/-) (Fig. 1g and Extended Data Fig. 1p, q). Conversely, sensitization of naïve mice with monoclonal OVA-specific IgE antibody resulted in the development of VHS upon repeated OVA administration (Fig. 1h and Extended Data Fig. 1r, s). Notably, the number of lymphocytes producing IL-4, a cytokine required for the production of allergen-specific IgE by B cells12, was significantly increased in colonic draining mesenteric lymph nodes of VHS mice (Fig. 1i). Taken together, these data indicate that a gastrointestinal bacterial infection can break oral tolerance to a dietary antigen and result in an adaptive immune response towards food antigens, leading to increased permeability and abnormal pain signaling upon re-exposure to this antigen.
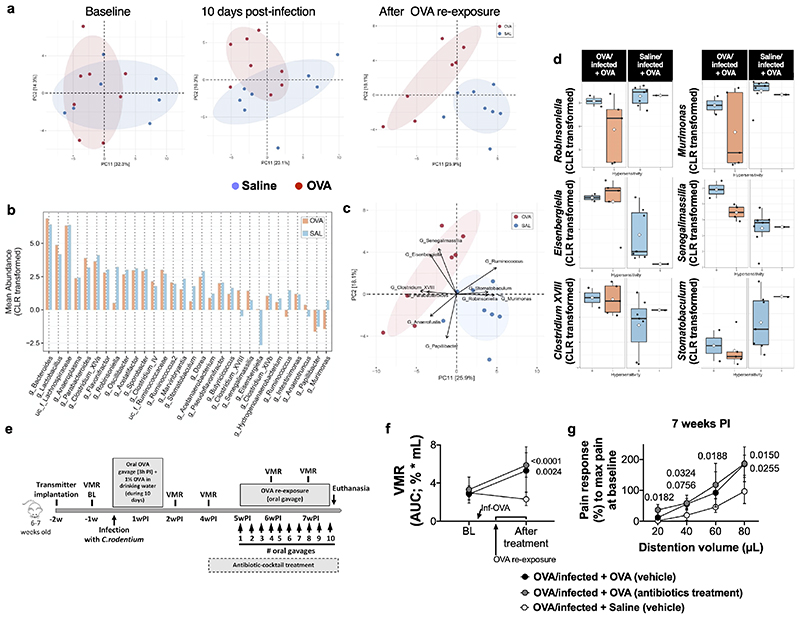
As changes in the gut microbial composition have been associated with IBS13, we investigated if a similar phenomenon is involved in our murine model. Exposure to OVA during C. rodentium infection shifted the community structure of the intestinal microbiome, compared to saline-exposure, 10 days post-infection and after OVA re-exposure (Extended Data Fig. 2a). While Robinsoniella and Eisenbergiella were two of the strongest drivers of this separation, we found no significant correlations with the development of VHS (Extended Data Fig. 2b-d). Furthermore, OVA/infected + OVA mice treated with antibiotics, to deplete the gut microbiota, developed VHS similar to vehicle-treated mice (Extended Data Fig. 2e-g). Despite the shift in microbiota composition observed in VHS mice, we show that this process is likely to be independent from the gut microbiota.
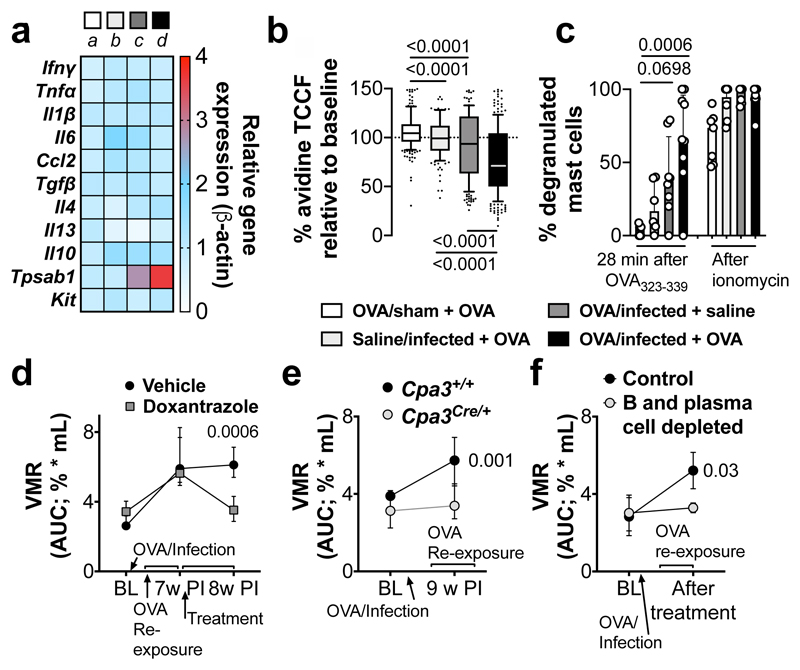
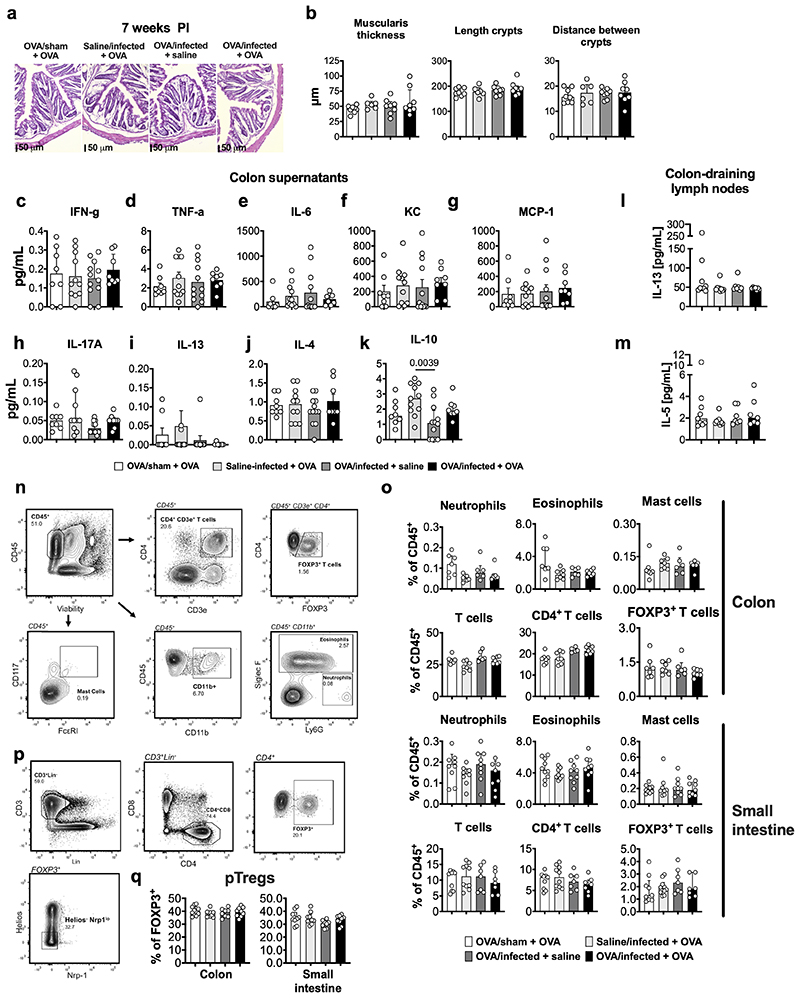
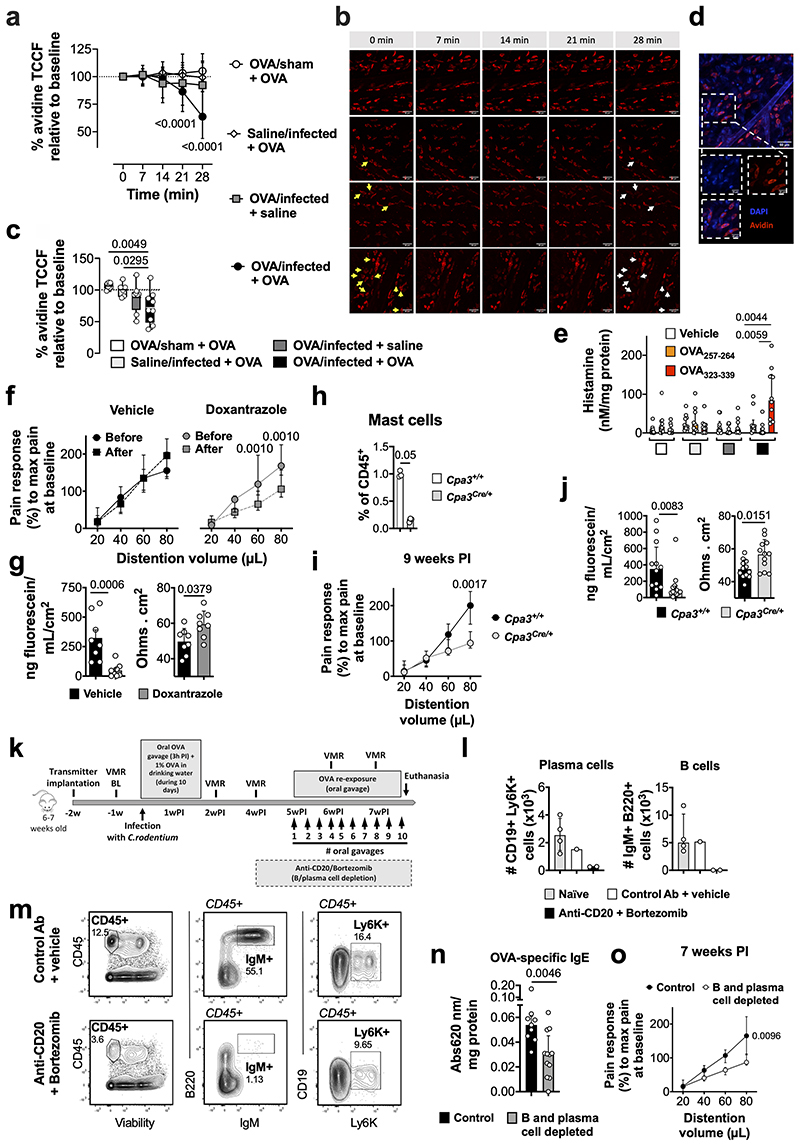
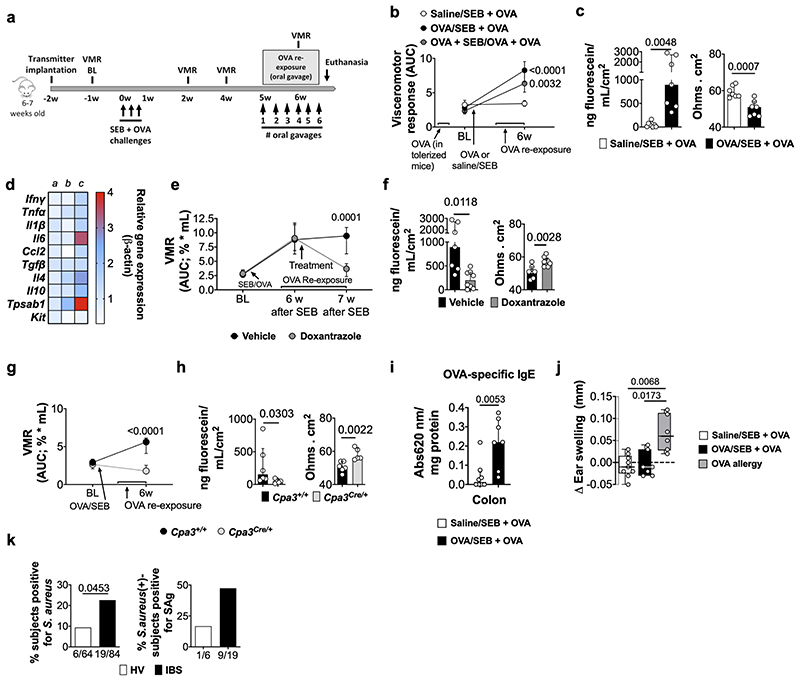
“Low-grade” inflammation and increased MC activation have also been proposed as underlying mechanism in IBS, especially in post-infectious IBS5. While mild colonic inflammation was observed during the acute infectious phase (10-14 days post-infection10), this was resolved in VHS or normosensitive infected mice after 7 weeks (Fig. 2a and Extended Data Fig. 3a-q). Interestingly, however, expression of the MC-restricted tryptase α/β-1 gene (Tpsab1) was upregulated in the colon of OVA/infected mice irrespective of OVA re-exposure (Fig. 2a), suggesting that MC are sensitized or activated as a result of the exposure to the dietary antigen during infection. This was confirmed by increased MC degranulation and increased release of histamine, one of the main MC mediators, in response to OVA-peptide 323-339 incubation in OVA/infected + OVA mice but not in other groups (Fig. 2b, c and Extended Data Fig. 4a-e). Notably, treatment with the MC stabilizer doxantrazole and genetic ablation of MC, normalized and prevented, respectively, VHS and decreased colonic permeability (Fig. 2e, f and Extended Data Fig. 4f-j). Furthermore, elimination of B cells and plasma cells (Extended Data Fig. 4k-m) lowered OVA-specific IgE levels and prevented the development of VHS (Fig. 2g and Extended Data Fig. 4m, o). This indicates that OVA-specific antibodies produced locally by B cells and plasma cells sensitize MC to subsequently mediate MC activation upon re-exposure to OVA.
Fig. 2. Mast cells play a key role in the development of OVA-induced VHS.
a, Relative gene expression in colon samples from OVA/sham + OVA (a), saline/infected + OVA (b), OVA/infected + saline (c) and OVA/infected + OVA (d) (n = 21, 11, 11 and 21, respectively [except for Tpsab1: n = 7, 5, 7 and 8, respectively; and Il13: n = 7, 9, 7 and 8, respectively]). b, c, MC-degranulation in OVA/sham + OVA, saline/infected + OVA, OVA/infected + saline and OVA/infected + OVA mice measured as (b) % of avidin-fluorescence intensity (TCCF) relative to baseline time = 28 min (n = 222 [7 mice], 145 [6 mice], 202 [8 mice] and 239 [9 mice], respectively) and (c) % of total degranulated MC. d-f, VMR to colorectal distention in OVA/infected + OVA mice (d) treated with doxantrazole or vehicle (n = 14 and 11, respectively), (e) with Cpa3Cre/+ background or WT littermates (n = 13 and 10, respectively) and (f) treated with anti-CD20 antibody/bortezomib or control antibody/vehicle (n = 3 and 4, respectively). One-way ANOVA (Sidak’s multiple-comparisons test) in a; Kruskal-Wallis test (Sidak’s multiple-comparisons test) in b and c; two-way repeated ANOVA (Sidak’s multiple-comparisons test) in d-f. Data shown as Box-and-whiskers (center line, median; box, 25th-75th percentiles; whiskers, 10th-90th percentiles; dots depict outliers) in b, and as median ± IQR in c-f. In a, Tpsab1 expression was statistically significant in OVA/infected + saline and OVA/infected + OVA vs. OVA/sham + OVA (adjusted p = 0.0173 and 0.0014, respectively).
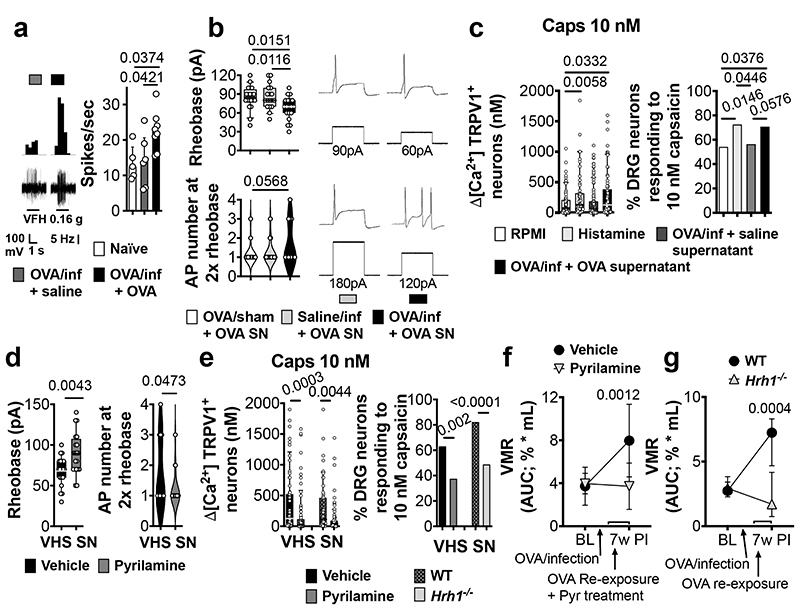
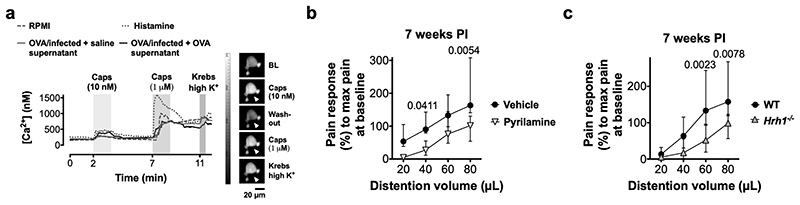
Recently, we and others have demonstrated that both biopsy supernatants from IBS patients and histamine not only activate but also sensitize visceral afferents and dorsal root ganglia (DRG) neurons8,14. Mechanical stimulation of colonic splanchnic nerves, known to transmit visceral pain signals to the spinal cord15, evoked an increased action potential discharge in VHS mice compared to normosensitive mice (Fig. 3a), indicating increased sensitivity of visceral nerves. Moreover, colonic supernatant of VHS mice increased neuronal excitability compared to supernatant of normosensitive mice (Fig. 3b), and significantly increased the sensitivity of DRG neurons to capsaicin, a specific agonist of the transient receptor potential TRPV1, similar to incubation with histamine (Fig. 3c and Extended Fig. 5a). This cation channel on nociceptors is involved in VHS in IBS14. Incubation with the H1R-antagonist pyrilamine prevented the increased excitability and TRPV1-mediated response in DRG neurons treated with VHS colonic supernatant (Fig. 3d, e), similar to our findings in IBS patients14. Furthermore, treatment with pyrilamine reduced the pain response to colorectal distention in VHS mice (Fig. 3f and Extended Fig. 5b). These findings were confirmed in Hrh1-/- mice (Fig. 3g and Extended Fig. 5c), indicating that the MC-mediated increase in visceral pain signaling is mediated via H1R and involves sensitization of TRPV1 and, most likely, other voltage-gated ion channels in sensory neurons.
Fig. 3. H1R-mediated nociceptive neuron sensitization drives OVA-induced VHS.
a, afferent nerve recording in naïve, OVA/infected + saline and OVA/infected + OVA mice (n = 5, 6 and 8, respectively). b, rheobase (up) and action potential (AP) number at 2x the rheobase (down) in DRG neurons incubated with supernatants from OVA/sham + OVA, saline/infected + OVA and OVA/infected + OVA mice (n = 20 [5 supernatants], 18 [6 supernatants] and 19 [5 supernatants], respectively). c, ratiometric Ca2+ response (left) and % cells responding to 10-nM capsaicin (right) in DRG neurons incubated with RPMI, histamine, supernatants from OVA/infected + saline and OVA/infected + OVA mice (n = 85 [5 supernatants], 59 [6 supernatants], 123 [12 supernatants] and 82 [11 supernatants], respectively). d, e, (d) rheobase (left) and action potential number at 2x the rheobase (right) (n = 19 neurons [4 supernatants] and 20 neurons [5 supernatants], respectively) and (e) ratiometric Ca2+ response (left) and % cells responding to 10-nM capsaicin (right) (n = 65 [12 supernatants], 87 [11 supernatants], 51 [3 supernatants] and 100 [8 supernatants], respectively) in DRG neurons incubated with supernatants from VHS mice with vehicle or pyrilamine and (only in e) comparing WT and Hrh1-/-. f, g, VMR to colorectal distention in OVA/infected + OVA mice (f) treated with vehicle or pyrilamine (n = 9 and 8, respectively) and (g) with WT or Hrh1-/- background (n = 13 and 9, respectively). One-way ANOVA (Sidak’s multiple-comparisons test) in a and b; Kruskal-Wallis test (Dunn’s multiple-comparison test) in c (left); two-tailed Fisher’s exact test in c (right) and e (right); two-tailed t-test in d, two-tailed Mann-Whitney test in e (left); two-way repeated measures ANOVA (Sidak’s multiple-comparisons test) in f and g. Data shown as median ± IQR in a, f and g, as Box-and-whiskers (center line, median; box, 25th-75th percentiles; whiskers, 10th-90th percentiles) for b (up), c (left), d (left) and e (left) and as violin plots (center line, median) in b (down), d (right). SN, supernatant.
In the upper airways, superantigens (SAgs), microbial antigens known to cause non-specific activation of T cells, have been implicated in the pathogenesis of asthma and atopic rhinitis16 via a bystander Th2-polarized immune response against innocent antigens17. We reasoned that intestinal exposure to SAgs, for instance after swallowing of SAgs during an episode of acute rhinitis or produced by the microbiome, could trigger a similar immune response to dietary antigens as we observed with a bacterial gastroenteritis. Similar to OVA/infected mice, mice receiving OVA in the presence of SEB developed VHS when re-exposed to OVA, even if these were tolerized to OVA prior to SEB administration, and displayed increased mucosal permeability compared to saline/SEB mice (Extended Data Fig. 6a-c). SEB-VHS mice showed an upregulation of Tpsab1, Il4, Il6 and Il10 genes (Extended Data Fig. 6d). MC activation mediated these changes as they were blocked by doxantrazole and were absent in Cpa3Cre/+ mice (Extended Data Fig. 6e-h). Colonic OVA-specific IgE was higher in SEB-VHS mice compared to saline/SEB mice (Extended Data Fig. 6i) and SEB-VHS mice failed to develop ear-swelling after intradermal injection of OVA (Extended Data Fig. 6j), indicating a local but not a systemic immune response to OVA. To evaluate whether SAgs could also play a role in the pathogenesis of IBS, we compared the presence of the main SAg-producing bacteria (i.e. Staphylococcus aureus and Streptococcus pyogenes18) in fecal samples from patients and healthy volunteers (HV). Of interest, 23% of the fecal samples from IBS patients tested positive for S. aureus, compared to 9% in HV (Extended Data Fig. 6k). Moreover, 47% of the positive IBS samples were positive for one or more SAgs compared to 17% in HV. Previous studies have linked S. aureus colonization to conditions associated with antigen-specific sensitization such as allergic rhinitis, asthma and food allergy19. Based on these and our findings, we speculate that SAgs might be involved in initiating VHS in subsets of IBS patients.
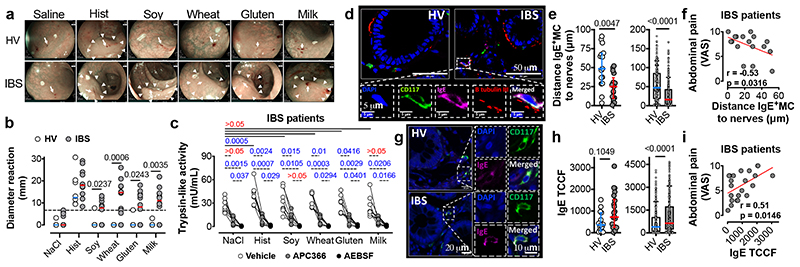
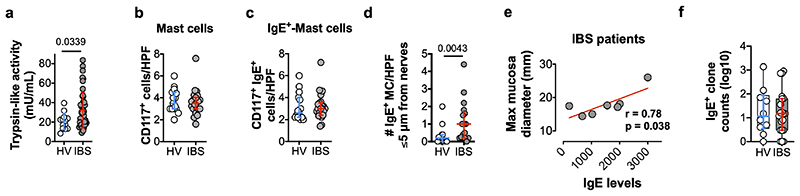
After characterizing the mechanisms underlying food-mediated VHS in two pre-clinical models, we investigated whether a similar process is present in IBS patients. To evaluate if food antigens can indeed evoke a localized immune response in the colon, solutions of soy, wheat, gluten and milk were injected in the rectosigmoid mucosa of 12 IBS patients and 8 HV (Supplementary Information Video 1). Food allergy to these dietary antigens was excluded in all participants (Extended Data Table 2). Notably, all 12 IBS patients showed mucosal reactions to at least one of the food antigens tested (Fig. 4a, b and Supplementary Information Video 2–4), whereas only two HV showed single positive reactions to soy and gluten, respectively. Next, we assessed tryptase activity as a proxy for mast cell degranulation. In basal conditions, total trypsin-like-activity was increased in IBS patients compared to HV (Extended Fig. 7a), as previously described20, and tryptase-activity, discriminated from overall activity by using the specific inhibitor APC-366, was higher after injection of histamine, soy, wheat and gluten (Fig. 4c). AEBSF (a broad-spectrum serine protease inhibitor) completely inhibited trypsin-like activity. Finally, while no differences were found in the total number of MC or IgE+ MC (Extended Data Fig. 7b, c), IBS patients showed an increased number of IgE+ MC in close proximity to nerve fibers (≤5 μm) (Extended Data Fig. 7d), in keeping with previous findings21. The distance between IgE+ MC and nerve fibers was smaller in IBS compared to HV (Fig. 4d, e), and inversely correlated with abdominal pain severity (Fig. 4f). Of interest, MC IgE immunofluorescent intensity was increased in IBS patients (Fig. 4g, h) and positively correlated with both abdominal pain severity (Fig. 4i) and the mucosal-edema diameter (Extended Data Fig. 7e). Finally, using deep sequencing of immunoglobulin genes from tissue cDNA, we found comparable numbers of IgE+ clones in the biopsies from IBS patients and HV (Extended Data Fig. 7f). Evaluation of allergen-specific clonal populations will be needed to further explore potential local sources of pathogenic IgE. Taken together, these results suggest that similar to our pre-clinical models, food antigens trigger MC activation in IBS patients, most likely, via a local IgE-mediated mechanism.
Fig. 4. Intramucosal injection of food antigens induces an immediate mucosal response in IBS patients.
a, representative images (arrows, antigen injection site; arrow-heads, reaction area) and (b) diameter reaction to food antigen injection in HV and IBS patients (n = 8 and 12, respectively) (dotted line represents the threshold for positive reactions). c, trypsin-like activity in rectal biopsies supernatants after antigen injection in IBS patients (n = 8). d, micrographs used to measure (e) IgE+ MC-nerve fibers distance in HV (n = 15 subjects [left], 206 cells [right]) and IBS patients (n = 17 subjects [left], 296 cells [right]). f, correlation between the IgE+ MC-nerve fibers distance and abdominal pain severity in IBS patients (n = 17). g, micrographs used to quantify (h) IgE intensity (TCCF) in MC in mucosal rectal biopsies from HV (n = 15 subjects [left], 261 cells [right]) and IBS patients (n = 22 subjects [left], 315 cells [right]). i, correlation between the IgE TCCF and abdominal pain severity in IBS patients (n = 22). Two-tailed Mann-Whitney test in b, e and h; two-way ANOVA (Sidak’s multiple-comparisons test) in c; two-tailed Spearman’s and Pearson’s correlation in f and i, respectively. Data represented as individual dots and median in b and median ± IQR in e and h left panels. e and h right panes show Box-and-whiskers (center line, median; box, 25th-75th percentiles; whiskers, 10th-90th percentiles).
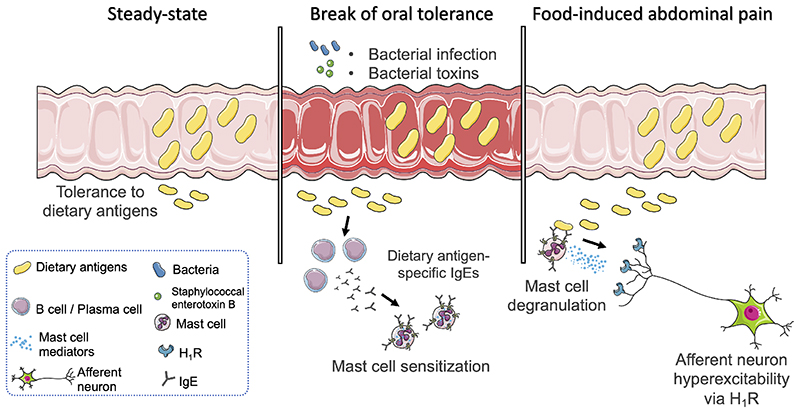
A key breakthrough proposed here is the understanding of how oral tolerance to food antigens, a pivotal regulatory process in homeostasis, can be lost in IBS and lead to aberrant pain signaling. We provide evidence for a role of local IgE antibodies in food-induced abdominal pain (Extended Data Fig. 8). A fundamental difference with food allergy is that OVA-specific IgE antibodies were only detectable in colonic tissue, indicating a local rather than systemic immune response against dietary antigens. A role for local IgE antibodies has previously been demonstrated in the airways, in particular in patients with allergic rhinitis and chronic rhinosinusitis with nasal polyps22, and associated with increased airway reactivity. Based on our data, we propose that IBS, at least in an immunogenetically susceptible subgroup of patients, is part of a spectrum of food-induced disorders mediated by MC activation, with systemic food allergy at the extreme end of the spectrum. The final phenotype most likely depends on the genetic make-up, favoring an atopic immune response. This hypothesis is further supported by the association of IBS with atopic diseases such as allergic rhinitis, allergic eczema or asthma23,24. Whether this concept also explains IBS-like symptoms reported by patients with inflammatory bowel disease in clinical remission25 remains speculative but is certainly of great interest.
The concept of food-mediated VHS proposed herein has major implications with respect to exploring novel treatments for IBS and related disorders. Blocking the effect of MC mediators, as we previously showed in a proof-of-concept study using the H1R-antagonist ebastine14, but especially using compounds targeting upstream mechanisms leading to MC sensitization and activation may be more effective. Thus, IgE-mediated activation of MC could be of special interest as a prognostic biomarker as well as a therapeutic target in these patients. Along the same line, improvement of IBS symptoms has been described in patients with severe asthma treated with omalizumab, a monoclonal antibody to IgE26. Therefore, our findings set the stage for further studies exploring the potential of therapies targeting upstream mechanisms of MC sensitization and/or activation, such as IgE neutralization or spleen tyrosine kinase inhibitors, as the latter is a pivotal signaling transducer coupling antigen-IgE/FcεRI complexes regulating MC degranulation27.
(online-only) Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Corresponding Author, Guy Boeckxstaens (guy.boeckxstaens@kuleuven.be).
Experimental Model and Subject Details
Study patients
All experiments involving humans and human material were approved by the Medical Ethics Committee of the University Hospitals Leuven and all patients provided informed consent (reference approval numbers: S51973, S55484 and S55485).
Food antigen injection: Eight healthy volunteers (HV, 8 female, 50 years, IQR [32-54]) free of abdominal symptoms, with no history of gastrointestinal disease, no previous gastrointestinal surgery or taking gastrointestinal medication, were recruited by public advertisement. Twelve IBS patients meeting the ROME III criteria (8 females, 29 years, IQR [21-39]) were recruited from the outpatient clinic of the University Hospitals Leuven. Allergy to soy, wheat, gluten and milk was excluded in all participants (anamnesis, skin-prick test and total IgE, tryptase and antigen-specific IgE in serum performed by a trained allergologist).
Trypsin-like-activity: 13 HV (10 females, 29 years IQR [23-47]) were recruited by public advertisement and were free of abdominal symptoms, had no gastrointestinal disease or surgery and were not on gastrointestinal medication. 48 IBS patients meeting the ROME III criteria (33 females, 31 years IQR [19-40]) were recruited from the outpatient clinic of the University Hospitals Leuven.
Bacterial identification in fecal samples: 64 HV (35 females, 49 years IQR [32-58]) were recruited by public advertisement and were free of abdominal symptoms, had no gastrointestinal disease or surgery and were not on gastrointestinal medication. 84 IBS patients meeting the ROME III criteria (66 females, 38 years IQR [25-50]) were recruited from the outpatient clinic of the University Hospitals Leuven.
Biopsy collection for immunofluorescence staining: 15 HV (13 females, 41 years IQR [32-52]) were recruited by public advertisement and were free of abdominal symptoms, had no gastrointestinal disease or surgery and were not on gastrointestinal medication. 22 IBS patients meeting the ROME III criteria (20 females, 31 years IQR [27-43]) were recruited from the outpatient clinic of the University Hospitals Leuven. All participants were invited to report their gastro-intestinal symptoms by means of an adapted version of the visual analogue scale (VAS) for IBS28, assessing the intensity of their last episode of abdominal pain, abdominal discomfort and bloating (scored from 0 to 10).
Mice
All animal experiments were performed in accordance with the European Community Council or Canadian Council of Animal Care guidelines and approved by the Animal Care and Animal Experiments Committee of the Medical Faculty of the KU Leuven or Queen’s University Animal Care Committee. Six- to seven-week-old wild type (WT) male BALB/c mice were purchased from Janvier. Six- to seven-week-old male Cpa3Cre/+ and WT male littermates on BALB/c background29, were used to study the role of mast cells. Six- to ten-week-old female Igh7-/- (mice lacking IgE)30 on BALB/c background, were used to study the role of IgE, which were compared to WT female BALB/c mice. Six- to ten-week-old male Hrh1-/- KO mice on BALB/c background (Oriental Bioservice, INC, Kyoto, Japan) were used to study the role of the histamine 1 receptor. Mice were maintained under a 14h/10h dark/light cycle, at a temperature of 20-22 °C (45-70% humidity), provided with food and water ad libitum.
Method Details
Food antigen intramucosal injection
IBS patients and HV were invited to undergo a sigmoidoscopy. During the procedure, food antigens were dissolved in 0.9% NaCl and 200 μL were injected in the rectal mucosa (soy, 3 mg/mL; wheat, 3 mg/mL; gluten, 3 mg/mL; and milk, 30 mg/mL). 0.9% NaCl and histamine (30 μg/mL) were used as negative and positive controls, respectively. Sigmoidoscopy was video-recorded and images of the injection sites were taken prior volume injection, immediately after injection, and 12-min after injection. Mucosal edema-diameter was measured upon blinded evaluation of images taken 12 min after injection, performed independently by 2 researchers (JAL and DB). The 95th percentile of the diameter reactions of the negative control (saline) was used as threshold for determining a positive reaction. Moreover, biopsies of the injection site were collected. Next, to obtain biopsy supernatant for protease activity assays, biopsies from 8 IBS patients injected with different food antigens or controls were incubated in Ad-DMEM/F12 supplemented with GlutaMAX (2 mM), Penicillin, Streptomycin, Gentamycin and Amphotericin B (100 U, 100 μg, 10 μg and 0.25 μg per mL; Invitrogen) for 1 h at 37°C and 5% CO2.
Citrobacter rodentium (C. rodentium) infection
C. rodentium (DBS100, ATCC® 51459™) was cultured overnight in Luria-Bertani (LB) broth medium (MP Biomedicals) at 37°C and 180-200 rpm. After 16 h, the bacteria were centrifuged (4°C, 2000 rpm, 10 min), the supernatant was discarded, and the pellet was resuspended in fresh LB medium. Eight- to nine-week-old mice were infected by oral gavage with 1 x 109 colony forming units (CFU) of C. rodentium or sterile 0.9% NaCl (B. Braun Medical NV/SA).
Telemetric Visceromotor response (VMR) recordings
Six to seven-week-old mice of at least 20 grams were implanted with Physiotel ETA-F10 telemetric transmitters (Data Sciences International). In anaesthetized mice (20 mg/kg ketamine, Nimatek, Eyurovet Animal Health B.V.; and 1 mg/kg xylazine, Rompun 2%, Bayer), transmitters were inserted in the abdominal cavity and electrodes were tunneled through the abdominal wall and sutured into the left external abdominal oblique muscle10.
As previously described10, colorectal distensions were performed to evoke abdominal cramping as read-out of visceral pain, using a distension catheter (volumes from 20 to 80 μL, Fogarty catheter for arterial embolectomy, 4F; Edwards Lifesciences; Extended Data Fig. 1h). Data were calculated as % visceromotor response relative to maximum distention response before infection (i.e. 80 μL distension is set at 100%) or as area under the curve (AUC) of the 4 distention volumes. Acknowledge 3.2.6 software was used for analysis.
Compliance
Colonic compliance was studied in order to exclude inflammatory- or pharmacologically-mediated changes in wall stiffness that can influence in the nociception of the animals. Thus, the volume-pressure relationship of the colon was evaluated in animals subjected to visceromotor response recordings (Extended Data Table 1).
Post-infectious protocol
Two weeks after telemetric transmitter implantation (i.e. 8-9 weeks old), mice were infected with C. rodentium as abovementioned. Three hours after infection, mice were orally gavaged with 50 mg of ovalbumin (OVA) (Grade II, Sigma-Aldrich®) dissolved in 0.9% NaCl. Next, OVA was dissolved in drinking water (1% v/v) and changed daily throughout 10 days following infection. Five weeks after infection, mice were re-exposed to OVA (50 mg) by oral gavage every other day until sacrifice. Visceromotor responses were recorded before the infection (baseline), 2 and 4 weeks after infection, and 1 h after the 4th and the 8th OVA gavage (Fig. 1a). Oral OVA gavage continued every other day until the day of euthanasia (normally after an extra 1 or 2 oral OVA challenges, unless otherwise specified (i.e. VMR in long-term VHS following final OVA re-exposure, ex-vivo re-exposure, etc.). Development of VHS at 7 weeks after infection was routinely confirmed in OVA/infected + OVA mice throughout all experiments in the manuscript. Mice were sacrificed by CO2 intoxication. In the experiment to evaluate the antigen-specificity of the immune response to OVA, mice received repeated gavages with 50 mg bovine serum albumin (BSA, Enzo Life sciences) dissolved in 0.9% NaCl. In the experiment using Cpa3Cre/+ mice, OVA re-exposure started 7 weeks after infection, after normalization of infection-induced VHS (i.e. 6 weeks PI).
Post-infectious protocol with prior OVA tolerization
Two weeks before the infection with Citrobacter rodentium, mice were exposed to 1 mg/mL of OVA in the drinking water (low dose oral tolerance) during 7 days or were orally gavaged 3 times, every other day, with 50 mg of OVA (high dose oral tolerance). Following tolerization, mice underwent the same post-infectious protocol described above.
Staphylococcal enterotoxin B (SEB) protocol
Mice were exposed by oral gavage to 10 μg of SEB from Staphylococcus aureus (Sigma-Aldrich®) in the presence of 50 mg of OVA during 3 consecutive days. Similar to the post-infectious protocol, 5 weeks after OVA/SEB administration mice were re-exposed to OVA (50 mg). Visceromotor responses were measured at different time-points: before the SEB administration (baseline) and after OVA re-exposure (Extended Data Fig. 5a). Mice were sacrificed by CO2 intoxication.
Ovalbumin allergy protocol
Six- to eight-week-old BALB/c mice were sensitized at day 0 and day 14 with 100 μg OVA (grade V) in the presence of 1 mg Alum (ThermoFisher Scientific Inc.) by intraperitoneal (i.p.) injection. From day 28, mice were challenged every other day with OVA (grade II, 50 mg) by oral gavage.
Treatments of mice
Antibiotics treatment
To avoid that mice refrain from drinking due to the foul taste of some antibiotics, we used an adapted protocol from Reikvam DH et al.31. All antibiotics were simultaneously administered by gavage. Ampicillin (25 mg/mL, Sigma-Aldrich®), vancomycin (12.5 mg/mL, Sigma-Aldrich®), neomycin (25 mg/mL, Sigma-Aldrich®) and metronidazole (25 mg/mL, Sigma-Aldrich®) were dissolved in 0.9% NaCl and a volume of 10 mL/kg body weight was orally gavaged every day from week 4 after the infection with C. rodentium until the sacrifice (Extended Data Fig. 2e). Antibiotic concoction was prepared fresh every day. When administered with OVA (250 mg/mL) during the re-exposure, OVA was dissolved and gavaged together with the antibiotic-cocktail. NaCl (NaCl%) was used as vehicle treatment. Absence of cultivable bacteria in antibiotics-treated mice was confirmed by culturing of fecal pellets in LB agar incubated for 24h at 37°C.
Mast cell-stabilizer
Doxantrazole (Sigma-Aldrich®) was dissolved in NaHCO3 (0.5% w/v) by sonication for 25 min. Mice were treated i.p. with doxantrazole (16.5 mg/kg) daily 1h before the OVA re-exposure during 7 consecutive days. NaCHO3 (0.5% w/v) was used as vehicle treatment.
B cell depletion
The B cell depleting antibody, IgG2a anti-CD20 (clone 5D2, isotype IgG2a) and the control antibody (clone 3E5, isotype IgG2a) were kindly provided by Genentech. Mice were treated i.p with anti-CD20 or control antibody (8 mg/kg) dissolved in PBS i.p. every other day32 from 4 weeks after the infection with C. rodentium until the sacrifice (Extended Data Fig. 4h).
Plasma cell depletion
Bortezomib (Selleckchem) was dissolved in PBS (DMSO 0.1% v/v). Mice were treated i.p. with bortezomib (0.65 mg/kg)33 every day from 4 weeks after the infection with C. rodentium until the sacrifice. PBS (DMSO 0.1% v/v) was used as vehicle treatment (Extended Data Fig. 4h).
Anti-IgE antibody
Anti-IgE antibody (clone R1E4, Kerafast) was dissolved in NaCl (0.9%). Mice were treated i.p. with anti-IgE antibody (1 mg/kg) once a week starting before the re-exposure until the sacrifice. NaCl (0.9 %) was used as vehicle treatment.
Pyrilamine
Pyrilamine maleate salt (Sigma-Aldrich®) was dissolved in 0.9% NaCl. Mice were treated i.p. with pyrilamine (5 mg/kg)34 every other day 1h before the OVA re-exposure until the end of the experiment. NaCl (0.9 %) was used as vehicle treatment.
Monoclonal OVA-specific IgE antibody
Mouse anti-OVA monoclonal IgE antibody (clone E-C1, Chondrex) was diluted in PBS. Mice were treated i.p. with OVA-specific IgE antibody (10 μg/mouse) 2 and 1 days before the 1st OVA exposure, and another time 1 day before the 4th OVA exposure (Extended Data Fig. 1n). After OVA-specific IgE antibody injection, mice were orally gavaged with OVA 4 times. Monoclonal Anti-Dinitrophenyl (DNP) antibody (clone SPE-7, Sigma) was used as non-specific IgE control antibody.
Diarrhea development
Diarrhea score
Mice were placed in a metabolic cage provided with water for 1 h after OVA gavage. Diarrhea was scored as follows: 1 = normal and dry pellet; 2 = normal and wetter pellet; 3 = wet and less-shaped pellet; 4 = complete diarrhea.
Whole-gut Transit time
Carmine red dye (6%) was dissolved in methylcellulose (0.5% w/v) and orally gavaged to overnight (12-hours) fasted mice. Then, mice were individually placed in cages and time was monitored until the presence of the first fecal pellet containing carmine red dye.
Water content assessment
Fecal pellets were collected after whole-gut transit time experiment (2 hours after carmine dye gavage) and weighed. Next, samples were again weighed after 24-hour-incubation at 50°C (with the Eppendorf tube lid open), and water content was determined (calculated as weight %).
ELISA OVA-specific IgE
Micro-titer plates were coated with 40 μg/mL OVA (grade V) in carbonate buffer and incubated at 4°C overnight. Next, plates were blocked for 2h at room temperature with PBS containing 3% BSA. Next, plates were loaded with 50 μL of sample and incubated overnight at 4°C. Then, plates were incubated for 1 h at room temperature with detection antibody (Biotin Rat anti-Mouse IgE, 4 μg/mL, BD Biosciences), followed by an incubation at room temperature for 30 min with Strep-AP (R&D systems, bipp) (diluted 1:500 in 1% BSA, 0.05% Tween 20, 0.025 M Tris, 0.5M NaCl; pH 7.4). Finally, BluePhos substrate (KPL) was added to the reaction and incubated for 30 min at room temperature. The reaction was stopped with H2SO4 (2 M) and the absorbance was measured at a wavelength of 620 nm. Absorbance values were normalized by the amount of protein (mg), that were measured using BCA Protein Assay Kit (ThermoFisher Scientific) following manufacturers’ instructions.
Ear prick allergy test
Mice were subjected to intradermal injections of 1 μg of OVA (Grade III, Sigma-Aldrich®) dissolved in sterile saline in one ear and 10 μL of saline in the other. Ear swelling was compared with a caliper 30 min after injection.
Colonic permeability
Freshly isolated colonic tissue was mounted in Ussing chambers (Mussler Scientific Instruments) with an opening of 0.017 cm2 and without removal of the seromuscular layer as previously described35. Transepithelial electrical resistance was continuously measured (open circuit conditions, bipolar constant-current pulses of 16 μA during 200 ms) and passage of fluorescein sodium salt were assessed every 30 min over 2 h. The concentration of fluorescein was measured using a FLUOstar Omega fluorescence reader (BMG Labtech) and the transepithelial electrical resistance with Clamp software (v9.00) (Mussler Scientific Instruments).
DNA extraction and 16S rRNA gene sequencing
Mouse fecal samples were collected at three-time points from the same cohort of mice: before C. rodentium infection (baseline), 10 days after infection and after OVA re-exposure. DNA was extracted from ~150 mg of fecal samples using MagAttract PowerMicrobiome DNA/RNA KF kit. The V4 region of 16S rRNA genes was amplified using the 515F/806R primer pair and purified using the QIAquick PCR Purification Kit. Sequencing was performed using the Illumina MiSeq platform (MiSeq Reagent Kit v2).
Microbiome Analysis
After demultiplexing with the LotuS 1.56536, fastq sequences were further processed following the DADA2 microbiome pipeline37. Briefly, sequence reads were first filtered and trimmed with the parameters: truncQ=11, truncLen=c (150,200), and trimLeft=c (30, 30). Filtered reads were denoised using the DADA2 algorithm, which infers the sequencing errors. After removing chimeras, amplicon sequence variants (ASVs) table was constructed and taxonomy was assigned using the Ribosomal Database Project (RDP) classifier implemented in DADA2. Prior to the downstream analysis, bacterial abundances were transformed using centered log-ratio (CLR) transformation of Aitchinson to control the compositionality of the sequencing data (CoDaSeq R package function codaSeq.clr38). Zero counts in the AVS table were replaced by multiplicative zero imputation using the zCompositions package function cmultRepl39. Ordination analysis (PCA) with contributing bacteria was performed at genus-level using the vegan R package (functions: prcomp and envfit; R version 3.4.1)40. Association of hypersensitivity with genus Robinsoniella and Eisenbergiella was assessed using the Mann–Whitney U test. All analyses were performed in R (version 3.4.1).
Histopathology assessment of murine samples
Samples were fixed in 4% paraformaldehyde (PFA) at 4°C overnight, transferred to sucrose (0.3 g/mL) and stored at 4°C. Samples were embedded using conventional techniques. 5-μm-sections were cut and stained by the heamatoxylin & eosin (H&E) method. Slides were reviewed using an Olympus BX41 light microscope and scored blinded. Crypt length, crypt space and muscle thickness were measured.
Mast cell live imaging in mouse colonic submucosa
The colon was dissected and placed into Krebs solution oxygenated with 95% O2/5% CO2. The colon was cut opened parallel to the mesenteric line, cleaned from fecal pellets and pinned flat in a Sylgard plate. Upon removal of the mucosa, the submucosal layer was dissected, placed on a metal ring to stabilize the tissue and loaded with the dye Sulforhodamine 101–coupled avidin (8 μg/mL in Krebs solution) (Sigma-Aldrich®) for 20 min and washed for 5 min with Krebs solution41,42. For image acquisition, the field of interest was selected randomly based on Avidin staining and incubated with OVA peptide 323-339 (10 μg/mL; InvivoGen). Five Z stacks per section (5 microns depth) were acquired just upon incubation with OVA (0 min) and every 7-minute interval, during 28 minutes for Texas red channels using a 25X water immersion lens under a confocal microscope (Zeiss LSM 780, Belgium). All images were acquired using the same settings using Zen (v14.0.20.201) software. Ionomycin (10 μM, Sigma-Aldich®) was used as positive control for MC degranulation. For fluorescence intensity analysis, Z stacks were processed (ImageJ 2.0.0-rc-54/1.52d) as sum projection used to quantify total corrected cell fluorescence (TCCF) as previously described43 and expressed as % compared to TCCF in baseline (time = 0 min). For calculation of the percentage of MC degranulated, the 95th percentile of TCCF % loss in MC from OVA/sham + OVA group at time = 28 min was used as a threshold for MC to degranulate. Representative images were acquired using a 25X water immersion lens and processed as sum projection. Images were assigned with red color and adjusted similarly for brightness and contrast.
Quantification of histamine release
The release of histamine was assessed by incubating 5-mm-thick colonic tissue rings (3 rings/well in a 24-well plate) for 60 min (37°C 5% CO2) in 2 mL of Krebs buffer supplemented with histamine catabolism inhibitors (10 μM diminazene aceturate, 1 μM R-(-)-deprenyl hydrochloride and 10 μM SKF 91488 dihydrochloride) in the presence of 10 μg/mL OVA323-339. Control samples were incubated with vehicle (supplemented Krebs buffer) or 10 μg/mL OVA257-264. Tissue supernatants were collected, cleared by centrifugation (5 min at 12,000 x g at 4°C) and stored at -80°C until use. Histamine concentration was measured by enzyme immunoassay (Immunotech #IM2015), and normalized to protein content (nM histamine/mg of protein) upon quantification by the BCA method, both as per supplier’s instructions.
RNA extraction and cDNA preparation
RNeasy Mini Kit (Qiagen) was used to isolate total RNA following manufacturers’ instructions (Qiagen). cDNA was then transcribed (Quanta Biosciences, Gaithersburg, Maryland, USA) and subjected to RT-qPCR analysis according to manufacturers’ instructions.
Tissue harvest for single cell suspensions
Colon and small intestine were removed from mesenteric fat tissue, opened longitudinally and subsequently cleaned in cold phosphate-buffered saline (PBS) and Hank’s Balanced Salt Solution (HBSS) (Gibco) supplemented with fetal calf serum (FCS, 1%, Lonza) and Penicillin/Streptomycin (Gibco) (100 μg/mL) (= wash medium). Epithelial cells were removed by an 8-minute incubation on a magnetic stirrer in warm (37°C) wash medium supplemented with DTT (1 mM) and EDTA (1 mM). After a washing step, tissue was cut in pieces smaller than 3mm and digested by a 25- to 30-min incubation on a magnetic stirrer in warm (37°C) MEM α containing Penicillin/Streptomycin (100 μg/mL), β-mercaptoethanol (50 μM, Sigma-Aldrich®), FCS (5%), DNase (5 U/mL, Roche), Collagenase V (0.85 mg/mL, Sigma-Aldrich®), Collagenase D (1.25 mg/mL, Roche), Dispase (1 mg/mL, Gibco). After digestion, cells were filtered through a 70 μm cell strainer and lamina propria cells were purified using a Percoll (VWR) gradient (44 to 67%). Purified cells were washed and re-suspended in PBS supplemented with FCS (1%) and Penicillin/Streptomycin (100 μg/mL) and stained for Flow Cytometry.
Flow Cytometry
Single cell suspensions were labelled for flow cytometry analysis using fluorophore-conjugated anti-mouse antibodies for 30 min at 4°C. Living cells were identified using a viability dye (Fixable Viability Dye eFluor®506, eBioscience). For intracellular staining, cells were fixed for 30 minutes, washed and then permeabilized for 20 minutes using a Foxp3 / Transcription Factor Staining Buffer Set (eBioscience). Cells were incubated with intracellular antibodies for 1 hour at 4°C. Samples were acquired using BD FACSCanto II (BD Biosciences) or BD FACS Symphony A5 (BD Biosciences), and analysis was carried out using FlowJo software (V.10.0.8, BD).
Gene expression quantification
Gene expression in mouse colonic samples by RT-qPCR
All steps were performed according to manufacturers’ instructions and as previously described 10 using LightCycler® 480 multiwell plate 96 (Roche GmBH). Gene expression was calculated as relative quantification of the gene expression using the Pfaffl method 44 and normalized to the control group. The primer pairs used are described in the Supplementary Information Table 1. mRNA expression of each gene was compared to the expression of Actb (reference gene).
Staphylococcus aureus and SAgs qPCR identification in fecal samples
The presence of Staphylococcus aureus and Streptococcus pyogenes were evaluated by identifying the presence of the nuc and spy1258 genes, respectively. The presence of the following SAg of Staphylococcus aureus were evaluated: sea, seb, sec, sed, seg, she, selk, sell, selm, selo, selp, selq and selu. The primer pairs used are described in the Supplementary Information Table 1.
mIL-4 Enzyme-Linked Immunospot Assay
IL-4 producing cells were quantified by means of enzyme-linked immunospot (ELISpot) assay, according to manufacturer’s instructions (CTL-Europe, Bonn, Germany). Total cells were isolated from colon-draining lymph nodes (cMLN). Cells were seeded (5 x 105/well in duplicate) on plates pre-coated with capture antibody for mIL-4 in medium (R10) and incubated at 37° C with 5% CO2 for 4 days. Medium contained 1% penicillin/streptomycin, 1% L-glutamin and 10% fetal bovine serum. Additionally, R10 contained 50 μM β-mercaptoethanol and 20 mM HEPES. As positive control, cells were activated with 50 ng/mL PMA and 500 ng/mL of ionomycin during 4 hours prior to the first washing step. Cytokine-producing cells were resolved by a 25-min-incubation with the Blue Developer Solution provided. Spot-forming cells (SFC) were quantified with ImmunoSpot software (v7.0; CTL-Europe, Bonn, Germany).
Preparation of colonic supernatants
Supernatants (colonic tissue 2% w/v, collected at 7 weeks post-infection, or 7 weeks post-WAS) were incubated for 4 or 24 h at 37°C, 5% CO2 in RPMI (Lonza) supplemented with fetal calf serum (10%), Penicillin, Streptomycin, Gentamycin and Amphotericin B (100 U, 100 μg, 10 μg and 0.25 μg per mL; Invitrogen). Supernatants were collected and stored at -80°C until use.
Quantification of inflammatory cytokines
Colonic supernatants were analyzed for secretion of cytokines (IL-4, IL-6, KC (murine IL-8 homologue), IL-10, IL-13, IL-17A, IFN-γ, MCP-1, TNF-α) by flow cytometry using BD™ Cytometric Bead Array (CBA) (BD Biosciences) according to the manufacturers’ instructions. Cytokine concentrations were calculated using FCAP Array Software (v3.0; BD Biosciences).
Afferent nerve recording
The colon from BALB/c mice, with the associated lumbar splanchnic nerves, was dissected out. Tissues were placed in a bath perfused with carbogenated Krebs buffer (7 mL/min; 32°C–34°C) and lumbar splanchnic nerves were isolated as previously described45. Lumbar splanchnic nerve fibers were identified based on their anatomic location, their receptive field and response to mechanical stimuli such as brush stroking and von Frey hair stroking45. Basal mechanosensitivity was characterized on response of receptive fields to von Frey hair probing (0.16 grams applied 3 times for 3 s) applied to the colonic mucosa. For analysis, individual single-unit discharge was discriminated offline using template-matching wavemark software. For the mechanosensitivity to von Frey hair probing, peak action potential discharge (Hz) over a 2-second period within the 3-second period of probing was determined for each probe and a mean response was calculated from each set of 3. Spike2 software (V.607) was used for the analysis.
Patch-camp recording
DRG neurons (T9-T13) from 8-12 weeks old mice were bilaterally excised under a dissection microscope as previously described 46. Following overnight incubation with colonic supernatants, DRG neuronal activity was then assessed using perforated-patch clamp with amphotericin B (0.24 mg/mL). The composition of the extracellular solution was as follows (in mM): 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 10 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 10 D-glucose; pH 7.4. For the pipette solution (in mM): 110 K-gluconate, 30 KCl, 10 HEPES, 1 MgCl2, 2 CaCl2; pH 7.25. The liquid junction potential was calculated to be 12 mV and corrected. The recording chamber was continuously perfused with external solution at approximately 1-2 mL/min. Final pipette resistance was 2–5 MΩ in bath solution. Signals were amplified using Multiclamp 700B, digitized by Digidata 1440A converters, and recorded using pClamp 10.5 software (Molecular Devices). Only neurons with resting membrane potentials more negative than -40 mV were analyzed. Only neurons with <30 pF capacitance were used because these neurons have been shown to display properties associated with nociceptors 47,48. Neuronal excitability was measured by recording the rheobase (minimum amount of current required to elicit an action potential) and numbers of action potentials discharged at twice the rheobase, as previously described 49. The role of H1R receptor was evaluated using pyrilamine (1 μM), applied 1 h prior to application of colonic supernatant.
Ca2+ imaging
DRG neurons (T9-T13) from 8-12 weeks old mice were bilaterally excised under a dissection microscope and intracellular Ca2+ concentration was measured as previously described 14 Following overnight incubation with colonic supernatants, neurons were loaded with 2 μM Fura-2AM (Invitrogen) for 20 min at 37 °C. The baseline intracellular Ca2+ concentration was monitored for 120 s and neurons were perfused with 10 nM of the TRPV1 channel-agonist capsaicin (Sigma-Aldrich®) to evaluate neuron sensitization. Capsaicin (1 μM) was used to identify TRPV1-expressing neurons. Neurons were identified upon perfusion with of 45 mM K+Cl-supplemented Krebs. We previously performed (Wouters MM, et al., Gastroenterology. 2016) dose-response curves using 10, 30, and 100 nM capsaicin, in the presence or absence of 10 μM histamine. Neurons responding to 10 nM capsaicin were more sensitized by histamine than neurons responding to 30 and 100 nM capsaicin, therefore, the first concentration was chosen for our experiments. The role of H1R receptor was evaluated using pyrilamine (1 μM). Pyrilamine was applied to neurons 1 h prior to application of colonic supernatant and was present throughout the incubation with colonic supernatants. The percentage of neurons responding to 10 nM of capsaicin was calculated among the total amount of neurons recorded. TILLVision software (TILL Photonics) (v4.5.66 build 10002) was used for the analysis.
Colonic tissue homogenates
Immediately after sacrifice, mouse colonic and small intestine tissues were snap frozen upon collection and kept frozen while being mechanically disrupted using a TissueLyser II (Qiagen). Disrupted tissues were suspended (10% w/v) in 0.9% NaCl solution containing Protease Inhibitor Tablets (Compete, Roche Diagnostics). After homogenization by vortex, samples were centrifuged and supernatants were collected and stored at -80°C until use.
DNA extraction from fecal samples
Fecal samples (200 mg) were incubated in Brain Heart Infusion broth (Sigma-Aldrich®) for 24 h shaking at 37°C and DNA was extracted using a commercial kit. Thereafter, bacterial DNA was extracted following manufacturers’ instructions (Genomic DNA extraction from tissue kit, Macherey-Nagel) adapted to the Support Protocol for hard-to-hard bacteria.
SAgs-positive Staphylococcus aureus strains, used as positive controls, were kindly provided by the National Reference Centre (NRC) for Staphylococcus aureus of Université Libre de Bruxelles (ULB) (Belgium).
Immunofluorescence staining, total corrected cell fluorescence and cell distance analyses in human samples
For immunostaining, biopsies were fixed in 4% PFA, then transferred to sucrose (0.3 g/mL) and subsequently embedded in OCT-compound and stored at -80 °C. Cryosections (5 μm) were incubated with 1% BSA in PBS for 1h at room temperature, followed by overnight incubation with the primary antibody at 4 °C. Primary antibodies were prepared in PBS containing 0.3% Triton-X and used to stain mast cells (1:500 anti-CD117 K963, IBL, USA), mast cells (1:500 rabbit anti-CD117 K963, IBL, USA), IgE (1:250 mouse anti-human MHE-18, BioLegend, Fell, Germany) and nerve fibers (1:500 chicken anti-beta III tubulin, Abcam, UK). After washing, sections were incubated with fluorescent secondary antibodies (2h in the dark at room temperature). Secondary antibodies (Jackson ImmunoResearch) included Cy3 donkey anti-rabbit (1:500), Cy5 donkey anti-mouse (1:500) [for immunofluorescence intensity evaluation] and Cy3 donkey anti-chicken (1:300), Cy5 donkey anti-rabbit (1:500), A488 donkey anti mouse (1:500) [for distance evaluation between IgE+ MC and nerve fibers]. Sections were mounted in SlowFade® Gold (Life Technologies Europe B.V., Gent, Belgium) with DAPI. A negative control, absence of primary antibody, was carried out for every subject to ensure antibody specificity. For image acquisition, the field of interest was selected randomly based on DAPI staining and 5-6 Z stacks per section with a step-size of 1.07 μm were acquired for three channels (DAPI, Cy3 and Cy5) using a 40X oil immersion lens under a confocal microscope (Zeiss LSM 880 – Airyscan, Belgium). All images were acquired using the same settings using Zen Black 2.3 SP1 software. For fluorescence intensity analysis, Z stacks were processed (ImageJ 2.0.0-rc-54/1.52d) as sum projection used to quantify total corrected cell fluorescence (TCCF) as previously described 43. Cells were identified using c-kit immunoreactivity and only nucleated profiles were included for analysis. For distance measurements, Z stacks were processed (ImageJ 2.0.0-rc-54/1.52d) as sum projection and used to quantify distance from the periphery of nerve fibers (based on beta tubulin staining) to mast cells periphery (identified based on CD117 staining) by using straight line tool. Cells were identified using CD117 and only nucleated profiles were included for analysis. Immunostaining, image acquisition and mean fluorescence intensity analysis were performed in a blinded manner. Representative images were acquired using a 63X or 40X oil immersion lens and processes as described above. Images were assigned with pseudo colors (immunofluorescence analysis: green for Cy3 channel and magenta for Cy5 channels; distance analysis: green for Cy5 channel, magenta for A488 channels, red for Cy3 channel) and adjusted for brightness and contrast.
Trypsin-like activity assay
Trypsin-like activity was measured as previously described with some modifications 50,51. Briefly, colonic tissue supernatant samples were incubated with 0.1 mM Tos-GPR-AMC.HCl as substrate in 50 mM Tris, 10 mM CaCl2, pH=8 (Sigma-Aldrich). Substrate cleavage was calculated over 30 min at 37°C on a FLUOstar Omega microplate reader (BMG Labtech, GmbH). Sample values were interpolated into a linear regression generated with a standard curve of AMC.HCl (0.05–0.5 nmol; Bachem). Mast cell tryptase activity was characterized by pre-incubating supernatants with the specific inhibitor APC-366 (100 μM; Tocris) for 60 min at 37°C. As controls, samples were pre-incubated with vehicle (0.2% DMSO) or the broad-spectrum serine protease inhibitor AEBSF (1000 μM; Roche). Data were expressed as mU/mL (1 U = conversion of 1 nmol of substrate/min).
Library preparation for high-throughput sequencing of immunoglobulin heavy chain genes
cDNA was synthesized from 1 μg of RNA using SuperScript III (Thermo Fisher Scientific) and primed with random hexamers (Promega) according to manufacturer’s protocols. IgH genes were amplified by multiplexed RT-PCR using Biomed2 IGHV primers in the FR1 framework region (van Dongen et al, 2003) and isotype-specific primers for IgM, IgD, IgG, IgA and IgE located in the CH1 constant region. Each isotype was amplified separately to prevent the formation of cross-isotype chimeric PCR products, and 1/10th of the total pool of cDNA generated from 1 μg of RNA from each sample was used for each RT-PCR reaction. The primer sequences have been published previously 52. Briefly, primers contained 8-mer barcodes used to identify samples as well as half of the Illumina adapter sequence needed for cluster generation and sequencing on the MiSeq instrument. PCR was performed with gene-specific primers for 35 cycles, and then an additional 12 cycles with primers to complete the Illumina adapters. For Illumina cluster recognition, four randomized nucleotides were included in the primers immediately after the Illumina adapter sequence in the constant region primers. The PCR programs were as follows: first PCR: 94°C for 7 min.; 35 cycles of (94°C 30 s, 58°C 45 s, 72°C 120 s); and final extension at 72°C for 10 min.; second PCR: 0.4 μL of first PCR product was used as template for this 30 μL reaction. Conditions: 94°C for 15 min.; 12 cycles of (94°C 30 s, 60°C 45 s, 72°C 90 s); and final extension at 72°C for 10 min. Amplification was confirmed by electrophoresis on agarose gels. To adjust for differences in amplification efficiencies arising from sample quality and biological factors such as low numbers of IgE transcripts relative to IgM, IgD, IgG and IgA transcripts, PCR reactions were pooled in roughly equal amounts based on the visual inspection of band intensity. The pool of products was then separated again by electrophoresis and purified with QIAquick Gel Extraction Kits (Qiagen). IgH libraries were sequenced on an Illumina MiSeq using 600-cycle sequencing kits.
IgH sequence analysis
Paired-end reads were merged by FLASH and sequences with perfect or single mismatch alignment to full-length VH and CH primers were demultiplexed according to the primer-encoded barcodes. Isotypes were called by perfect or single mismatch alignment to known constant region gene sequences, to remove artefactual sequences generated by cross-priming in the CH1 region and sequences with low-certainty subisotype calls. Out-of-frame rearrangements, rearrangements containing stop codons, and reads with no isotype call were not analyzed further. Reads were aligned using a local installation of IgBLAST to identify features including germline IGHV, IGHD and IGHJ genes, framework and heavy chain complementarity determining regions 1, 2 and 3 (CDR-H1, CDR-H2 and CDR-H3). After quality filtering, library sequencing yielded 11,115,260 productively rearranged sequences, which were subsequently clustered into inferred clonal lineages via single linkage clustering, with the restrictions that the sequences had to be from the same individual, use the same IGHV gene and the same IGHJ gene (disregarding allele call), have the same CDR-H3 length, and have at least 90% identity in the nucleotides encoding the CDR-H3 region. IgE+ clones were defined as those that contained at least one member expressing IgE.
Quantification and Statistical Analysis
Sample size and statistical methods
For animal studies, the number of mice was selected based on power calculations performed based on previous or present studies carried out in our laboratory and in the field. Normal (Gaussian) distribution was determined for all data sets using Shapiro-Wilk normality test. Data sets where one (or more) group did not pass the normality test (assuming α = 0.05), were analyzed using non-parametric tests. Otherwise, parametric tests were used. For the assessment of the diarrhea-score, whole total transit time and water content in fecal water we used between 6 and 10 mice per experimental group. For measurement visceromotor response we used between 3 and 13 mice per experimental group/genotype/pharmacological treatment. For ELISpot assay, we used 4 mice per experimental group. For recording of afferent nerves experiment we used between 6 and 8 mice per experimental group. For measurement of colonic permeability, we used between 5 and 12 mice per group/genotype/pharmacological treatment. For analysis of gut microbiota, we used 8 mice per experimental group. For histology studies we used between 6 and 8 mice per experimental group. For measurement of cytokine levels, we used between 8 and 12 mice per experimental group. For measurement of gene expression in colonic murine samples we used between 5 and 21 mice per experimental group. For FACS analysis we used between 6 and 10 mice per experimental group. For measurement of OVA-specific immunoglobulins we used between 6 and 10 mice per experimental group in serum samples; and between 4 and 19 mice per experimental group in colonic samples. For evaluation of live MC degranulation, 145-239 MC were analyzed per group, from tissue isolated from 6-9 different mice/group. For evaluation of ear-swelling upon OVA intradermal injection we used between 6 and 10 mice per experimental group. For patch-clamp experiments in DRG neurons we used between 18 and 24 neurons per experimental group/pharmacological treatment. For measurement of Ca2+ responses we used between 51 and 126 neurons per group/genotype/pharmacological treatment.
In human studies, no power calculations were performed for the food-antigens injection study and the amount of participant was chosen based on a previous similar study performed in patients with eosinophilic esophagitis53: 8 HV subjects and 12 IBS patients were used. For the measurement of trypsin-like-activity, samples from the food antigens-injection sites of 8 IBS patients were processed, and 13 HV vs 48 IBS patients were compared at basal conditions. For assessment of IgE immunofluorescence in mucosal biopsies, 15 HV and 22 IBS patients were used. For the bacteria and SAg-producing bacteria, the number of samples was chosen based on power calculations of past studies in the field: 64 HV subjects and 84 IBS patients were used.
Diarrhea score and colonic compliance was analyzed with two-way repeated measures ANOVA with Sidak’s multiple-comparisons post-test, each p-value was therefore adjusted to account for multiple-comparison. Whole-gut transient time and fecal water content, visceromotor response, afferent nerve recording, measurement of fluorescein sodium and TEER, FACS analysis, OVA-specific IgE in mouse tissue, association between VHS and bacterial abundance, rheobase and action potential number, measurement of Ca2+ responses in DRG neurons and total corrected cell fluorescence (TCCF) of IgE in human biopsies were compared with two-tailed unpaired t-test for parametric analyses, or Mann-Whitney test for nonparametrical analyses (as indicated in the legend of each figure). ELISpot mIL-4 were analyzed using paired t-test of dependent samples. Visceromotor responses were also analyzed with two-way repeated measures ANOVA with Sidak’s multiple-comparisons post-test, each p-value was therefore adjusted to account for multiple-comparison. Measurement of fluorescein sodium and TEER, thickness of muscularis, length crypts, distance between crypts, cytokines levels, gene expression, ear swelling, rheobase, action potential number and measurement of Ca2+ responses in DRG neurons were analyzed with one-way ANOVA with Sidak’s multiple-comparisons post-test for parametrical analyses or Kruskal-Wallis test with Dunn’s multiple-comparisons post-test for nonparametrical analyses (each p-value was therefore adjusted to account for multiple-comparison). Trypsin-like activity was analyzed by two-way ANOVA with Sidak’s multiple-comparisons post-test. Associations between IgE immunofluorescence levels and symptoms severity scores were assessed by Pearson’s correlation and distance between MC IgE+-nerve fibers and symptom severity scores by Spearman’s correlation. Bacteria and SAgs-producing bacteria proportions and proportion of responding DRG neurons to 10 nM capsaicin were evaluated by two-sided Fisher’s exact test. Data were plotted and statistical analysis were performed with Prism (GraphPad Software version 9.0.0 [86], La Jolla, California).
Replication
For visceromotor response recording (except mice receiving repeated BSA by oral gavage, B and plasma cell depletion, treatment with anti-IgE antibody and IgE-deficient mice and treatment with OVA-specific IgE), experiments were performed twice or more times and data from individual mice were pooled from all experiments. Diarrhea score, WGTT and water content where performed in different batches of animals. Similar number of mice were used per experiment duplicated for control group/treatment and experimental group. For afferent nerve recording and Ussing chambers, experiments were performed twice and data from individual mice were pooled from all experiments. Similar number of mice were used per experiment duplicated for control group/treatment and experimental group. All in vivo and ex vivo experiments were routinely assessed on different days, including every group tested in each day. For FACS analysis, at least 2 experiments were performed for each tissue. For experiments with cultures of DRG neurons, multiple biological replicates were used for each group. For Ca2+ experiments (except for Hrh1-/- DRG neurons), experiments were performed twice and neuron Ca2+ responses were pooled. For experiments with live MC degranulation and histamine release, experiments were performed twice and data were pooled.
Randomization and blinding
For experiments involving transgenic mice and pharmacological treatments, genetically modified mice and WT littermates or drug-treated- and vehicle/control-treated-mice were placed together in the same cages. For pharmacological treatments, the online tool of GraphPad software (QuickCalcs) was used to randomly assign subjects to groups for treatment or vehicle/control.
For pharmacological treatments and experiments with KO mice, the experimenter recording visceromotor responses was blinded for the treatment and control/vehicle or the mouse genotype and was unblinded for the analysis. For the afferent nerve recording experiment, the experimenter was blinded for the animal groups and was unblinded for the analysis. For the experiments involving cultures of DRG neuron, the experimenters were blinded for the analysis of the neurons and were unblinded for pooling the data in groups and analyze statistical differences. For experiments involving microbiota analysis the experimenter was blinded for the analysis. For experiments involving live MC degranulation, the experimenter was blinded during the performance of the experiment and for the analysis of images.
For human experiments, the experimenters analyzing the reactions to food-antigens were blinded for the images/videos analyses and were unblinded once all the reactions were analyzed.
For histopathology assessment murine samples and immunostainings in human samples, five to ten images per sections were randomly captured and analyzed by the experimenter in a blinded manner. Experimenter was unblinded for analysis of statistical differences.
For the analysis of immunoglobulin genes from tissue cDNA (IgE+ clones), the experimenter analyzing the number of clones was only unblinded for analysis of statistical differences.
Extended Data
Extended Data Fig. 1. Extended analysis of the OVA-specific immune response and VHS in post-infectious mice.
a, b, diarrhea development quantification by (a) water content in feces and (b) whole-gut transit time upon gavage of carmine red dye in OVA/sham + OVA, OVA/infected + OVA (n = 10/group) mice. c, quantification of OVA-specific IgE in intestinal homogenates of OVA/sham + OVA, saline/infected + OVA, OVA/infected + saline and OVA/infected + OVA mice (n = 9, 10, 10 and 9, respectively) at 7 weeks post-infection. d, ear-swelling after intradermal injection of OVA in OVA/sham + OVA (n = 6), OVA/infected + OVA (n = 10), at 7 weeks post-infection, and OVA-allergy mice (n = 6, 10 and 6, respectively). e, f, VMR to colorectal distention in (e) OVA/sham + OVA, OVA/infected + saline, saline/infected + OVA and OVA/infected + OVA mice (n = 12, 11, 11 and 13, respectively) and (f) tracing of a electromyographic response to 20-μL-, 40-μL-, 60-μL- and 80-μL-volume colorectal distention in a OVA/infected + OVA mouse at baseline and 7 weeks post-infection. g, VMR to colorectal distention in OVA/infected + OVA (n = 7) and OVA/infected = Saline (n = 7 and 5, respectively) mice at baseline (BL), 7 weeks post-infection (PI) and after 1, 2, 3 and 4 weeks after stopping oral OVA or saline re-exposure, respectively. h, colonic permeability in BALB/c mice expressed as passage of fluorescein sodium (left) and transepithelial resistance (right) in OVA/sham + OVA, saline/infected + OVA, OVA/infected + saline and OVA/infected + OVA mice (n = 8, 11, 12 and 8, respectively) at 7 weeks post-infection. i, scheme illustrating the post-infectious protocol with prior OVA tolerization. j, VMR to colorectal distention in mice OVA-tolerized (high-dose) + saline/infected + OVA, OVA-tolerized (high-dose) + OVA/infected + OVA, OVA-tolerized (low-dose) + saline/infected + OVA and OVA-tolerized (low-dose) + OVA/infected + OVA (n = 6, 9, 6 and 8, respectively). k, l, VMR to colorectal distention in mice OVA/infected repeatedly gavaged with BSA compared to OVA (n = 6 and 10, respectively). m, scheme illustrating the post-infectious protocol in mice treated with anti-IgE antibody. n, VMR to colorectal distention in OVA/infected + OVA mice treated with anti-IgE antibody or control antibody (n = 8/group). o, colonic permeability expressed as passage of fluorescein sodium (left) and transepithelial resistance (right) of (l) VHS mice treated with anti-IgE antibody or control antibody (n = 8/group). p, VMR to colorectal distention in OVA/infected + OVA mice with WT or Igh7-/- background (n = 10/group). q, colonic permeability expressed as passage of fluorescein sodium (left) and transepithelial resistance (right) of OVA/infected + OVA mice with Igh7-/- background or WT mice (n = 10/group) at 7 weeks post-infection. r, scheme illustrating the protocol in mice that received monoclonal OVA-specific IgE antibody. s, VMR to colorectal distention in naïve mice treated with monoclonal OVA-specific IgE antibody or monoclonal Dinitrophenyl (DNP) antibody (n = 7 and 6, respectively). Two-tailed Mann–Whitney test in a and b for every time point, and in o and q (left); two-tailed t-test for q (right). Kruskal-Wallis test (Dunn’s multiple-comparisons test) in c and d. One-way ANOVA (Sidak’s multiple-comparisons test) in h. Two-way repeated ANOVA (Sidak’s multiple-comparisons test) in e, g, j-l, n, p and s. Data shown as Box-and-whiskers (center line, median; box, 25th-75th percentiles; whiskers, 10th-90th percentiles) in a, b and d and median ± IQR in c, e, g, h, j-l, n-q and s. VMR, visceromotor response; BL, baseline; w, week; d; day.
Extended Data Fig. 2. Changes in microbiota composition are not likely to be associated with the development in VHS.
a, changes in the gut microbiome composition over time studied by ordination (PCA) plot before the infection, 10 days after infection (R2 = 0.11, p-value = 0.02), and after OVA gavage (R2 = 0.18, p-value = 0.002) between OVA/infected and saline/infected (n = 8/group) mice. The numbers in brackets indicate the variation explained by each axis (PCA: principal component analysis). b-d, drivers of variation in the murine intestinal community after OVA re-exposure. b, Comparison of mean abundance of taxa between OVA- and saline-infected mice. Bacterial abundances were computed using centered log-ratio transformation (CLR; two-sided Mann–Whitney test, FDR < 0.2). c, ordination biplot of PCA analysis showing the significant contributors in the community variation. Arrows indicate the relative importance of each genus (permutation-based p < 0.05). d, abundance of genus Robinsoniella, Eisenbergiella, Senegalimassilia, Murimonas, Clostridium_XVIII and Stomatobaculum by group and VHS (two-sided Mann-Whitney test, FDR > 0.1). e, scheme illustrating the post-infectious protocol in mice treated with antibiotics. f, g, VMR to colorectal distention in VHS (OVA/infected + OVA) mice treated with antibiotics or control, and compared to OVA/infected + saline mice (n = 10, 7 and 6, respectively) (g depicts the VMM response at 7 weeks post-infection). In d, VHS threshold was established based on the 95th percentile of visceromotor responses at baseline (AUC > 4.8). There was no significant association between VHS and bacterial abundance in any group (two-tailed Mann-Whitney test). X-axis indicates absence (0) and presence (1) of VHS. Two-way ANOVA 9(Sidak’s multiple-comparisons test) in f and g. Data shown as box-and-whiskers ± 1.5 IQR in d and as median ± IQR in f and g. VMR, visceromotor response; BL, baseline; w, week.
Extended Data Fig. 3. Analysis of ‘low grade’ inflammation in colonic samples of post-infectious mice.
a, Representative H&E-stained colonic sections from OVA/sham + OVA, OVA/infected + saline, saline/infected + OVA and OVA/infected + OVA at 7 weeks post-infection. b, thickness of the colonic muscularis (left), length of the crypts (middle) and distance between crypts (right), measured in OVA/sham + OVA, saline/infected + OVA (n = 6), OVA/infected + saline and OVA/infected + OVA mice (n = 8, 6, 8 and 8, respectively) at 7 weeks post-infection. c-k, concentration of (c) IFN-γ, (d) TNF-α, (e) IL-6, (f) KC (murine IL-8 homologue), (g) MCP-1, (h) IL-17A, (i) IL-13, (j) IL-4 and (k) IL-10 assessed in colonic supernatant and (l) IL-13 and (m) IL-5 in colon-draining lymph nodes (coMLN) of OVA/sham + OVA, saline/infected + OVA, OVA/infected + saline and OVA/infected + OVA mice (n = 8, 11, 12 and 8, respectively, for c-k; n = 7, 7, 9 and 9, respectively for l and m). n-q, immune cell quantification along the intestinal tract: (n and p) gating strategy; (o) quantification of population of CD11b+Ly6G+SiglecF- neutrophils, CD11b+SiglecF+ eosinophils, CD117+FcRεI+ mast cells, CD3+ T cells, CD3+CD4+ T cells, CD3+CD4+Foxp3+ T cells and (q) Helios- Nrp1loFOXP3+ pTregs in the colon and small intestine in OVA/sham + OVA, saline/infected + OVA, OVA/infected + saline and OVA/infected + OVA mice (n = 7, 6, 7 and 7, respectively, for o (colon); 9, 8, 9 and 10, respectively, for o (small intestine); 9, 9, 8 and 10, respectively, for q (Helios-Nrp1loFOXP3+ pTregs in colon); 9, 9, 8 and 10, respectively, for q (Helios-Nrp1loFOXP3+ pTregs in small intestine);. One-way ANOVA (Sidak’s multiple-comparisons test) in b-m, o and q. Data shown median ± IQR in b-m, o and q.
Extended Data Fig. 4. Extended analysis of mast cell activation as key players in the development of OVA-induced VHS in post-infectious mice.
a-c, avidin-fluorescence intensity over time (expressed as TCCF) in MC from OVA/sham + OVA, saline/infected + OVA OVA/infected + saline and OVA/infected + OVA mice (n = 222 [7 mice], 145 [6 mice], 202 [8 mice] and 239 [9 mice], respectively), (a) shown as percentage at times = 0, 7, 14, 21 and 28 min, (b) micrographs and (c) shown at time = 28 min as median per mouse. In b, arrows point to representative MC showing degranulation (yellow, 28 min) compared to baseline (white, 0 min). d, representative image of double-staining using DAPI and avidin in fixed colonic tissue from healthy mice (n = 3). e, Histamine quantification in supernatant collected from OVA/sham + OVA, saline/infected + OVA, OVA/infected + saline and OVA/infected + OVA (n = 10/group) at 7 weeks post-infection. f, VMR to colorectal distention in OVA/infected + OVA mice (d) treated with doxantrazole or vehicle (n = 14 and 11, respectively). g, colonic permeability expressed as passage of fluorescein sodium (left) and transepithelial electrical resistance (right) of OVA/infected + OVA mice treated with doxantrazole or vehicle (n = 8/group) at 8 weeks post-infection. h, quantification of population of CD117+FcRεI+ mast cells in intestinal lamina propria from Cpa3Cre/+ and WT mice (n = 3/group). i, VMR to colorectal distention in OVA/infected + OVA mice with Cpa3Cre/+ background or WT littermates (n = 13 and 12, respectively). j, colonic permeability expressed as passage of fluorescein sodium (left) and transepithelial electrical resistance (right) in OVA/infected + OVA mice with Cpa3Cre /+ background or WT littermates (n = 12/group) at 9 weeks post-infection. k, scheme illustrating the post-infectious protocol in mice treated with anti-CD20 antibody and bortezomib. l, anti-CD20 (5D2 clone) antibody and bortezomib depleted CD19+ Ly6K+ plasma cells (left) and B220+ IgM+ B cells (right) from the colon of mice compared to control antibody/vehicle and naïve mice (n = 2, 1 and 4, respectively) and (m) gating strategy used to validate B cell and plasma cell depletion. n, quantification of OVA-specific IgE in colon samples homogenates from OVA/infected + OVA mice treated with anti-CD20 antibody and bortezomib or control antibody and vehicle (n = 12 and 9, respectively). o, VMR to colorectal distention in OVA/infected + OVA mice treated with anti-CD20 antibody/bortezomib or control antibody/vehicle (n = 3 and 4, respectively). Mixed-effects model (Dunnett’s multiple-comparisons test) in a and e. One-way ANOVA (Sidak’s multiple-comparisons test) in c. two-way repeated measures ANOVA (Sidak’s multiple-comparisons test) in f, I and o. Two-tailed Mann-Whitney in g, h, j and n. Data are shown as median ± IQR in a, e-j, l, n and o, and box-and-whiskers (center line, median; box, 25th and 75th percentiles; whiskers, 10th and 90th percentiles) in c. VMR, visceromotor response; BL, baseline; w, week.
Extended Data Fig. 5. H1R mediates OVA-induced VHS in post-infectious mice.
a, representative tracing and Fura-2 ratiometric fields of DRG neurons (TRPV1+ neuron [right] indicated by arrowhead). b, c, VMR to colorectal distention in OVA/infected + OVA mice (a) treated with vehicle and pyrilamine (n = 9 and 8, respectively) and (b) with WT and Hrh1-/- background (n = 13 and 9, respectively). Two-way repeated measures ANOVA (Sidak’s multiple-comparisons test) in b and c. Data are shown as median ± IQR.
Extended Data Fig. 6. SEB induces MC-mediated OVA-induced VHS upon comparable to C. rodentium infection.
a, scheme illustrating the SEB protocol. b, VMR to colorectal distention in saline/SEB + OVA, OVA/SEB + OVA and OVA (low dose) + OVA/SEB + OVA mice (n = 10, 14 and 4, respectively). c, colonic permeability expressed as passage of fluorescein sodium (left) and transepithelial electrical resistance (right) of saline/SEB + OVA and OVA/SEB + OVA (n = 8 and 7, respectively) mice. d, heat-map of the gene expression of inflammatory genes and mast-cell-related genes in OVA/saline + OVA (a), saline/SEB + OVA (b) and OVA/SEB + OVA mice (c) (n = 21, 7 and 7, respectively; except for Tryptase α/β-1: n = 7/group). e, VMR to colorectal distention in SEB-VHS mice treated with doxantrazole or vehicle (n = 8 and 9, respectively). f, colonic permeability expressed as passage of fluorescein sodium (left) and transepithelial electrical resistance (right) of SEB-VHS mice treated with doxantrazole or vehicle (n = 8 and 7, respectively). g, VMR to colorectal distention in OVA/SEB + OVA mice with Cpa3Cre/+ background or WT littermates (n = 5 and 6, respectively). h, colonic permeability expressed as passage of fluorescein sodium (left) and transepithelial electrical resistance (right) of OVA/SEB + OVA mice with Cpa3Cre/+ background or WT littermates (n = 5 and 6, respectively). i, quantification of OVA-specific IgE in colon homogenates of saline/SEB + OVA and OVA/SEB + OVA mice (n = 8 and 7, respectively). j, ear-swelling after intradermal injection of OVA in saline/SEB + OVA, OVA/SEB + OVA and OVA-allergy mice (n = 9, 7 and 6, respectively) mice. k, % of HV and IBS patients (n = 64 and 84, respectively) positive for S. aureus (left) and SAg-encoding S. aureus (right) in fecal samples. Numbers below the bars represent the ratio of positive and total HV and patients. Two-way repeated ANOVA (Sidak’s multiple-comparisons test) in b, e and g. Two-tailed unpaired t-test in c and f. Two-tailed Mann-Whitney test in h and i. Kruskal-Wallis test (Dunn’s multiple-comparisons test) in d and j. Two-sided Fisher’s exact test in k. Data shown as median ± IQR in b, e-i and box-and-whiskers (center line, median; box, 25th-75th percentiles; whiskers, 10th-90th percentiles) in j. VMR, visceromotor response; BL, baseline; w, week. In d, Tpsab1, Il4, Il6 and Il10 gene expression differences were statistically significant in OVA/SEB + OVA vs. OVA/saline + OVA (adjusted p = 0.0005, 0.0004, 0.0163 and 0.0039, respectively).
Extended Data Fig. 7. Extended analysis of mast cells and IgE in human samples.
a, measurement of trypsin-like activity in supernatants of rectal biopsies from HV and IBS patients (n = 13 and 48, respectively) in basal conditions. b, c, number of (a) MC (CD117+) and (b) IgE+-MC (CD117+ IgE+) quantified in mucosal rectal biopsies from HV and IBS patients (n = 15 and 22, respectively). d, quantification of CD117+ IgE+ cells (MC) at ≤5 μm from nerve fibers (β tubulin III+ cells) in HV and IBS patients (n = 15 and 17, respectively). e, correlation between the IgE TCCF and max mucosal diameter after food-antigen injection in IBS patients (n = 7). f, number of IgE+ clones was quantified in mucosal rectal biopsies cDNA from HV and IBS patients (n = 10 and 20, respectively) using deep sequencing. Two-tailed Mann-Whitney test in a-d and f. two-tailed Pearson’s correlation in e. Data are shown as median ± IQR in a-d, and as box-and-whiskers (center line, median; box, 25th and 75th percentiles; whiskers, 10th and 90th percentiles) in f. HPF = high power field.
Extended Data Fig. 8. Graphical representation of the mechanism proposed: Local immune response to dietary antigens triggered by bacterial infection leads to food-induced abdominal pain.
Bacterial infection (or bacterial toxins, SEB) can trigger break of oral tolerance to food antigens leading to food-induced VHS upon food-antigen re-exposure. OVA-specific IgE antibodies bind to and sensitizes tissue-resident mast cells, which are activated upon re-exposure to OVA during feeding and release mediators that sensitize afferent neurons via H1R-mediated pathway.
Extended Data Table 1.
Colonic compliance (volume-pressure relationship) evaluated in animals subjected to visceromotor response recordings. ns = non-significant.
| Group | Genotype / Treatment | Strain | Volume | Weeks post-infection / post SEB-administration / post-WAS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 μL | 40 μL | 60 μL | 80 μL | ||||||||||||
| Mean (mmHg) | SD | Significance compared to control | Mean (mmHg) | SD | Significance compared to control | Mean (mmHg) | SD | Significance compared to control | Mean (mmHg) | SD | Significance compared to control | ||||
| OVA/control + OVA | WT | BALB/c | 24.23 | 5.36 | - | 40.74 | 8.35 | - | 55.02 | 9.54 | - | 71.74 | 10.72 | - | 7 |
| OVA/infected + OVA | WT | BALB/c | 22.74 | 3.85 | ns | 38.35 | 5.92 | ns | 53.52 | 8.47 | ns | 70.03 | 8.80 | ns | 7 |
| OVA/infected + saline | WT | BALB/c | 21.14 | 3.70 | ns | 37.69 | 7.87 | ns | 48.39 | 12.72 | ns | 63.16 | 15.70 | ns | 7 |
| Saline/infected + OVA | WT | BALB/c | 23.49 | 3.26 | ns | 39.76 | 4.91 | ns | 53.54 | 5.66 | ns | 71.63 | 9.03 | ns | 7 |
| OVA/infected + BSA | WT | BALB/c | 22.05 | 4.61 | ns | 40.42 | 7.64 | ns | 56.99 | 9.15 | ns | 76.48 | 10.32 | ns | 7 |
| OVA (low) + saline/infected + OVA | WT | BALB/c | 26.59 | 7.02 | - | 47.35 | 10.31 | - | 60.60 | 11.11 | - | 79.23 | 13.13 | - | 7 |
| OVA (low) + OVA/infected + OVA | WT | BALB/c | 22.71 | 4.29 | ns | 44.34 | 5.89 | ns | 59.29 | 7.00 | ns | 75.31 | 7.40 | ns | 7 |
| OVA (high) + saline/infected + OVA | WT | BALB/c | 27.12 | 6.45 | - | 47.88 | 9.26 | - | 61.13 | 10.56 | - | 79.77 | 12.57 | - | 7 |
| OVA (high) + OVA/infected + OVA | WT | BALB/c | 23.56 | 4.95 | ns | 45.19 | 6.11 | ns | 60.14 | 7.62 | ns | 76.16 | 8.16 | ns | 7 |
| OVA/infected + OVA | WT / Vehicle | BALB/c | 23.77 | 4.29 | - | 45.94 | 5.61 | - | 61.47 | 7.09 | - | 79.49 | 10.81 | - | 7 |
| OVA/infected + OVA | WT / antibiotic-cocktail | BALB/c | 23.24 | 4.57 | ns | 43.87 | 4.93 | ns | 55.99 | 5.59 | ns | 74.75 | 2.88 | ns | 7 |
| OVA/infected + OVA | WT / Vehicle | BALB/c | 25.11 | 1.61 | - | 40.35 | 1.76 | - | 54.97 | 4.84 | - | 70.63 | 7.73 | - | 8 |
| OVA/infected + OVA | WT / Doxantrazole | BALB/c | 17.19 | 1.12 | ns | 32.28 | 2.21 | ns | 49.26 | 4.55 | ns | 66.88 | 3.35 | ns | 8 |
| OVA/infected + OVA | Cpa3 +/+ | BALB/c | 26.86 | 5.79 | - | 43.89 | 12.02 | - | 60.70 | 15.08 | - | 78.63 | 16.41 | - | 9 |
| OVA/infected + OVA | Cpa3 Cre/+ | BALB/c | 21.18 | 5.25 | ns | 39.95 | 7.10 | ns | 51.67 | 6.24 | ns | 64.92 | 5.69 | ns | 9 |
| OVA/infected + OVA | WT / Vehicle | BALB/c | 22.73 | 1.51 | - | 40.32 | 3.36 | - | 56.51 | 7.38 | - | 67.76 | 9.45 | - | 7 |
| OVA/infected + OVA | WT / antiCD-20 + Bortezomib | BALB/c | 23.95 | 4.15 | ns | 40.34 | 13.09 | ns | 53.73 | 19.77 | ns | 67.48 | 23.61 | ns | 7 |
| OVA/infected + OVA | WT / isotype control antibody | BALB/c | 21.22 | 4.64 | - | 40.06 | 9.06 | - | 50.82 | 11.35 | - | 64.83 | 14.84 | - | 7 |
| OVA/infected + OVA | WT / αIgE antibody | BALB/c | 24.78 | 5.03 | ns | 40.69 | 4.62 | ns | 54.58 | 8.33 | ns | 67.02 | 13.27 | ns | 7 |
| OVA/infected + OVA | WT | BALB/c | 16.06 | 3.74 | - | 42.49 | 12.39 | - | 58.23 | 9.98 | - | 74.62 | 10.23 | - | 7 |
| OVA/infected + OVA | Igh KO | BALB/c | 19.62 | 6.11 | ns | 36.95 | 5.33 | ns | 54.50 | 6.86 | ns | 74.47 | 5.45 | ns | 7 |
| Naïve | DNP-specific IgE antibody | BALB/c | 19.71 | 3.52 | - | 35.87 | 7.18 | - | 53.58 | 6.48 | - | 72.98 | 8.25 | - | *After last OVA challenge |
| Naïve | OVA-specific IgE antibody | BALB/c | 17.78 | 5.75 | ns | 38.69 | 6.80 | ns | 56.06 | 6.69 | ns | 78.13 | 5.53 | ns | *After last OVA challenge |
| OVA/infected + OVA | WT / Vehicle | BALB/c | 25.25 | 1.67 | - | 43.77 | 4.10 | - | 56.32 | 10.54 | - | 70.37 | 10.68 | - | 7 |
| OVA/infected + OVA | WT / Pyrilamine | BALB/c | 21.97 | 3.00 | ns | 39.95 | 5.81 | ns | 58.44 | 7.26 | ns | 70.88 | 4.49 | ns | 7 |
| OVA/infected + OVA | WT | BALB/c | 26.41 | 5.08 | - | 42.67 | 6.71 | - | 56.46 | 7.26 | - | 74.55 | 9.70 | - | 7 |
| OVA/infected + OVA | Hrh1 KO | BALB/c | 23.58 | 6.71 | ns | 41.94 | 9.55 | ns | 58.51 | 11.71 | ns | 78.01 | 12.91 | ns | 7 |
| Saline/SEB + OVA | WT | BALB/c | 26.88 | 3.35 | - | 43.39 | 6.12 | - | 57.66 | 7.78 | - | 74.39 | 8.93 | - | 7 |
| OVA/SEB + OVA | WT | BALB/c | 23.14 | 3.83 | ns | 39.70 | 7.41 | ns | 50.40 | 12.14 | ns | 72.17 | 14.31 | ns | 7 |
| OVA/SEB + OVA | WT / Doxantrazole | BALB/c | 23.86 | 5.80 | ns | 38.73 | 9.35 | ns | 53.17 | 12.86 | ns | 68.93 | 15.71 | ns | 8 |
| OVA/SEB + OVA | Cpa3 +/+ | BALB/c | 25.10 | 3.57 | - | 44.37 | 7.07 | - | 53.78 | 5.10 | - | 64.63 | 10.33 | - | 7 |
| OVA/SEB + OVA | Cpa3Cre/+ | BALB/c | 24.75 | 7.69 | ns | 39.70 | 12.39 | ns | 54.87 | 11.99 | ns | 68.16 | 16.28 | ns | 7 |
Extended Data Table 2.
Demographic data and characteristics of IBS patients participating in food-antigen injection protocol. M, male; F, female; IBS-D, IBS diarrhea-predominant; IBS-U, IBS unsubtyped; IBS-M, IBS mixed bowel patter; n.d. = non-detected.
| Patient | IBS subtype | Post-infectious IBS | Gender | Age (years) | Symptom severity (VAS pain score) | Associated psychological distress | Food antigen-specific IgE | Symptom development (time food intake) | Reported relationship between food intake and symptom generation | Reported food items associated with positive mu co sal ed ema reaction | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Soy | Wheat | Gluten | Milk | ||||||||||
| IBS 1 | IBS-D | No | M | 27 | 5 | Yes | n.d. | n.d. | n.d. | n.d. | 5-6 h | Diarrhea, abdominal pain, bloating | - |
| IBS 2 | IBS-U | No | F | 48 | 4 | No | n.d. | n.d. | n.d. | n.d. | 30 min - 1 day | Diarrhea, abdominal pain, abdominal cramps, belching | - |
| IBS 3 | IBS-M | No | F | 25 | 6 | Yes | n.d. | n.d. | n.d. | n.d. | 1-3 h | Diarrhea, abdominal cramps, bloating | Gluten |
| IBS 4 | IBS-D | No | F | 19 | 6 | Yes | n.d. | n.d. | n.d. | n.d. | Immediately | Diarrhea, abdominal pain, abdominal cramps | Soy, milk |
| IBS 5 | IBS-D | No | M | 41 | 2 | No | n.d. | n.d. | n.d. | n.d. | 3h | Abdominal cramps | Wheat, gluten |
| IBS 6 | IBS-D | No | F | 19 | 5 | Yes | n.d. | n.d. | n.d. | n.d. | Immediately - 6 h | Diarrhea, abdominal pain, abdominal cramps, flatulence | Wheat, gluten, milk |
| IBS 7 | IBS-D | No | F | 20 | 5 | Yes | n.d. | n.d. | n.d. | n.d. | 30 min - 3 h | Abdominal pain, abdominal cramps, bloating | Wheat, gluten, milk |
| IBS 8 | IBS-D | No | F | 50 | 6 | Yes | n.d. | n.d. | n.d. | n.d. | 30 min - 4 h | Abdominal pain | Milk |
| IBS 9 | IBS-D | Yes | M | 33 | 4 | Yes | n.d. | n.d. | n.d. | n.d. | 3h | Diarrhea, abdominal cramps, flatulence, bloating | - |
| IBS 10 | IBS-D | Yes | M | 31 | 10 | No | n.d. | n.d. | n.d. | n.d. | 10 min | Abdominal pain, flatulence | - |
| IBS 11 | IBS-D | No | F | 28 | 9 | Yes | n.d. | n.d. | n.d. | n.d. | 30 min | Diarrhea, abdominal pain | - |
| IBS 12 | IBS-D | No | F | 34 | 6 | Yes | n.d. | n.d. | n.d. | n.d. | 30 min - 1 h | Diarrhea, abdominal cramps, bloating, flatulence | Soy, wheat, milk |
| Median | - | - | 8F / 4M | 29, IQR [21-39] | 5.5, IQR [4.25-6] | - | - | - | - | - | 30min - 3h | - | - |
Supplementary Material
Acknowledgments
We thank I. Croux and R. De Keyser for technical assistance. Also, L. Beullens, A.W. Essilfie, H. Willekens and H. Volkaerts for nursing assistance in the studies involving human patients. We also thank M. Rasulova (Rega Institute, KU Leuven, Belgium) for assistance with ELISpot plate read-out. Moreover, we thank the Cell and Tissue Imaging Cluster for the use of the NIKON inverted TillVision microscope, epifluorescence microscope BX 41 Olympus and Zeiss LSM880 – Airyscan – and Zeiss LSM 780 – SP Mai Tai HP DS – confocal microscopes (Cell and Tissue Imaging Cluster, CIC, Supported by Hercules AKUL/15/37_GOH1816N and FWO G.0929.15 to Prof. Pieter Vanden Berghe, KU Leuven). Flow cytometry was done at the FACS Core facility (KU Leuven, Belgium) and we would like to thank Pier Andrée Penttila and Pradeep Kumar for their excellent technical assistance. Also, we thank National Reference Centre (NRC) for Staphylococcus aureus of Université Libre de Bruxelles (ULB) (Belgium) for kindly providing SAgs-positive Staphylococcus aureus strains; and Genentech for providing IgG2a anti-CD20 antibody and its isotype control. MVF, MFV and DB were supported by an FWO PhD fellowship 1110019N, 11C2219N and 1127415N, respectively). CB is supported by the Clinical Research Fund, University Hospitals, Leuven. JAL, MMW, PJ, NS and YAA were supported by an FWO postdoctoral fellowship (12X9820N, 1248513N, 12R5219N, 12V3619N and 12H8220N, respectively). During this project JAL was also previously supported by an FWO PhD fellowship (11Y2116N). TBF and H-RR were supported by SFB-TRR156-A07. SDB is supported by the NIH grant NIAID R01 AI125567 and an endowment from the Crown Family Foundation. GEB was funded by a KU Leuven university grant (Global Opportunities for Associations GOA 14.011, C14/18/086) and FWO grant (G0A9516N).
Footnotes
Author Contributions:
JAL, MVF, ADS, MMW and GEB planned and designed the experiments. JAL and MVF performed mice infections, pharmacological treatments and carrying out of in vivo animal protocols. JAL, MVF, IA and MCE performed immunoglobulin ELISAs. JAL, MCE and SUM performed diarrhea-development experiments. JAL recorded visceromotor responses and compliance, except for pyrilamine treatment and in Hrh1-/-, performed by MVF. EP performed afferent nerve recordings. JAL performed colonic permeability assessments assisted by RF. JS and JR performed analysis of murine microbiota. MVF and ST performed histopathology assessments in murine samples. NS performed cytokine quantification by Cytometric Bead Array. JAL, IA and NF performed RNA extraction and quantitative PCR experiments. MFV, MVF, IA, NF and GB performed FACS experiments assisted by GM, SIM, CB and BNL. ELISPOT assay was performed by LD and ADS. CLL and JJP performed patch-clamp experiments assisted by DER. JAL performed Ca2+ experiments assisted by YAA and KT. JAL performed bacterial DNA extraction from fecal samples. PJ and MFV performed immunofluorescence staining of human samples and total corrected cell fluorescence analysis. PJ and MCE performed and analyzed immunofluorescence staining of murine samples and mast cell live imaging analysis. RB, GEB, KVB and DB performed food-antigens injections and collection of biopsies. JAL and DB performed the analysis of the reactions to food-antigens. CB and RS performed the allergy work-up in humans. ADS designed, performed and analyzed protease activity assays. ADS and MN evaluated histamine presence in mouse and human samples assisted by DC and PA. IgH sequence analysis of IgE+ clones was performed by RAH assisted by SDB. TBF and HRR provided Cpa3Cre/+ mice and JS provided Igh7-/- mice. SH and FAR provided experimental support. JAL, MVF, MFV, ADS, DER, SJV, MMW and GEB reviewed data. JAL, ADS and GEB wrote and revised the manuscript. All other authors corrected and approved the final version of the manuscript. MFV and PJ contributed equally to this manuscript.
Competing Declaration of Interests:
The authors declare no competing interests
Data availability
The data for 16S rRNA gene sequences are available from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/) at accession number PRJEB41204. All other relevant data generated and analyzed during the current study are available upon reasonable request to the corresponding author (GEB). Source data are provided with this paper.
References
- 1.Schäfer T, et al. Epidemiology of food allergy/food intolerance in adults: Associations with other manifestations of atopy. Allergy Eur J Allergy Clin Immunol. 2001;56:1172–1179. doi: 10.1034/j.1398-9995.2001.00196.x. [DOI] [PubMed] [Google Scholar]
- 2.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 3.Mowat AMI. Anatomical basis of tolerance and immunity to intestinal antigens. Nature Reviews Immunology. 2003;3:331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 4.Bouziat R, et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science (80-) 2017;356:44–50. doi: 10.1126/science.aah5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009;136:1979–1988. doi: 10.1053/j.gastro.2009.02.074. [DOI] [PubMed] [Google Scholar]
- 6.Card T, et al. Post-infectious IBS: Defining its clinical features and prognosis using an internet-based survey. United Eur Gastroenterol J. 2018;6:1245–1253. doi: 10.1177/2050640618779923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford AC, Lacy BE, Talley NJ. Irritable Bowel Syndrome. N Engl J Med. 2017;376:2566–2578. doi: 10.1056/NEJMra1607547. [DOI] [PubMed] [Google Scholar]
- 8.Barbara G, et al. Mast Cell-Dependent Excitation of Visceral-Nociceptive Sensory Neurons in Irritable Bowel Syndrome. Gastroenterology. 2007;132:26–37. doi: 10.1053/j.gastro.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Seiffart V, et al. IL10-Deficiency in CD4+T Cells Exacerbates the IFNγ and IL17 Response During Bacteria Induced Colitis. Cell Physiol Biochem. 2015;36:1259–73. doi: 10.1159/000430295. [DOI] [PubMed] [Google Scholar]
- 10.Mondelaers SU, et al. Effect of genetic background and postinfectious stress on visceral sensitivity in Citrobacter rodentium-infected mice. Neurogastroenterol Motil. 2016;28:647–658. doi: 10.1111/nmo.12759. [DOI] [PubMed] [Google Scholar]
- 11.Lee JW, et al. Subjects with diarrhea-predominant IBS have increased rectal permeability responsive to tryptase. Dig Dis Sci. 2010;55:2922–2928. doi: 10.1007/s10620-009-1094-8. [DOI] [PubMed] [Google Scholar]
- 12.Wu YL, Stubbington MJT, Daly M, Teichmann SA, Rada C. Intrinsic transcriptional heterogeneity in B cells controls early class switching to IgE. J Exp Med. 2017;214:183–196. doi: 10.1084/jem.20161056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kassinen A, et al. The Fecal Microbiota of Irritable Bowel Syndrome Patients Differs Significantly From That of Healthy Subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Wouters MM, et al. Histamine Receptor H1-Mediated Sensitization of TRPV1 Mediates Visceral Hypersensitivity and Symptoms in Patients with Irritable Bowel Syndrome. Gastroenterology. 2016;150:875–887.:e9. doi: 10.1053/j.gastro.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 15.Hughes PA, et al. Post-inflammatory colonic afferent sensitisation: Different subtypes, different pathways and different time courses. Gut. 2009;58:1333–1341. doi: 10.1136/gut.2008.170811. [DOI] [PubMed] [Google Scholar]
- 16.Liu JN, et al. The prevalence of serum specific IgE to superantigens in asthma and allergic rhinitis patients. Allergy, Asthma Immunol Res. 2014;6:263–266. doi: 10.4168/aair.2014.6.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes-Blom E, Camberis M, Prout M, Tang SC, Le Gros G. Staphylococcal-derived superantigen enhances peanut induced Th2 responses in the skin. Clin Exp Allergy. 2012;42:305–314. doi: 10.1111/j.1365-2222.2011.03861.x. [DOI] [PubMed] [Google Scholar]
- 18.Proft T, F J. Bacterial Superantigens. Clin Exp Immunol. 2003;133:299–306. doi: 10.1046/j.1365-2249.2003.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones AL, Curran-Everett D, Leung DYM. Food allergy is associated with Staphylococcus aureus colonization in children with atopic dermatitis. J Allergy Clin Immunol. 2016;137:1247–1248. doi: 10.1016/j.jaci.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Rolland-Fourcade C, et al. Epithelial expression and function of trypsin-3 in irritable bowel syndrome. Gut. 2017 doi: 10.1136/gutjnl-2016-312094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbara G, et al. Activated Mast Cells in Proximity to Colonic Nerves Correlate with Abdominal Pain in Irritable Bowel Syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 22.De Schryver E, et al. Local immunoglobulin E in the nasal mucosa: Clinical implications. Allergy, Asthma Immunol Res. 2015;7:321–331. doi: 10.4168/aair.2015.7.4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobin MC, et al. Atopic irritable bowel syndrome: A novel subgroup of irritable bowel syndrome with allergic manifestations. AnN Allergy, Asthma Immunol. 2008;100:49–53. doi: 10.1016/S1081-1206(10)60404-8. [DOI] [PubMed] [Google Scholar]
- 24.Jones MP, Walker MM, Ford AC, Talley NJ. The overlap of atopy and functional gastrointestinal disorders among 23,471 patients in primary care. Aliment Pharmacol Ther. 2014;40:382–391. doi: 10.1111/apt.12846. [DOI] [PubMed] [Google Scholar]
- 25.Halpin SJ, Ford AC. Prevalence of symptoms meeting criteria for irritable bowel syndrome in inflammatory bowel disease: Systematic review and meta-analysis. American Journal of Gastroenterology. 2012:1474–82. doi: 10.1038/ajg.2012.260. [DOI] [PubMed] [Google Scholar]
- 26.Pearson JS, Niven RM, Meng J, Atarodi S, Whorwell PJ. Immunoglobulin E in irritable bowel syndrome: Another target for treatment? A case report and literature review. Therapeutic Advances in Gastroenterology. 2015;8:270–277. doi: 10.1177/1756283X15588875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilfillan AM, Rivera J. The tyrosine kinase network regulating mast cell activation. Immunol Rev. 2009;228:149–169. doi: 10.1111/j.1600-065X.2008.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengtsson M, Persson J, Sjölund K, Ohlsson B. Further validation of the visual analogue scale for irritable bowel syndrome after use in clinical practice. Gastroenterol Nurs. 2013;36:188–98. doi: 10.1097/SGA.0b013e3182945881. [DOI] [PubMed] [Google Scholar]
- 29.Feyerabend TB, et al. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity. 2011;35:832–844. doi: 10.1016/j.immuni.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Oettgen HC, et al. Active anaphylaxis in IgE-deficient mice. Nature. 1994;370:367–70. doi: 10.1038/370367a0. [DOI] [PubMed] [Google Scholar]
- 31.Reikvam DH, et al. Depletion of murine intestinal microbiota: Effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moritoki Y, et al. B-cell depletion with anti-CD20 ameliorates autoimmune cholangitis but exacerbates colitis in transforming growth factor-β receptor II dominant negative mice. Hepatology. 2009;50:1893–1903. doi: 10.1002/hep.23238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MS, Kim TS. IgA+ plasma cells in murine intestinal lamina propria as a positive regulator of Treg differentiation. J Leukoc Biol. 2014;95:461–469. doi: 10.1189/jlb.0613310. [DOI] [PubMed] [Google Scholar]
- 34.Huang Z-L, et al. Altered sleep-wake characteristics and lack of arousal response to H3 receptor antagonist in histamine H1 receptor knockout mice. Proc Natl Acad Sci. 2006;103:4687–4692. doi: 10.1073/pnas.0600451103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauffer A, et al. Subacute stress and chronic stress interact to decrease intestinal barrier function in rats. Stress. 2016;19:225–234. doi: 10.3109/10253890.2016.1154527. [DOI] [PubMed] [Google Scholar]
- 36.Hildebrand F, Tadeo R, Voigt AY, Bork P, Raes J. LotuS: An efficient and user-friendly OTU processing pipeline. Microbiome. 2014;2:30. doi: 10.1186/2049-2618-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gloor GB, Wu JR, Pawlowsky-Glahn V, Egozcue JJ. It’s all relative: analyzing microbiome data as compositions. Annals of Epidemiology. 2016:322–9. doi: 10.1016/Jannepidem.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Palarea-Albaladejo J, Martín-Fernández JA. ZCompositions - R package for multivariate imputation of left-censored data under a compositional approach. Chemom Intell Lab Syst. 2015;143:85–96. [Google Scholar]
- 40.Oksanen J, et al. The vegan package. Community ecology package. 2008 doi: 10.4135/9781412971874.n145. [DOI] [Google Scholar]
- 41.Reber LL, et al. Imaging protective mast cells in living mice during severe contact hypersensitivity. JCIInsight. 2017;2 doi: 10.1172/jci.insight.92900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joulia R, et al. Mast cells form antibody-dependent degranulatory synapse for dedicated secretion and defence. Nat Commun. 2015;25:6174. doi: 10.1038/ncomms7174. [DOI] [PubMed] [Google Scholar]
- 43.McCloy RA, et al. Partial inhibition of Cdk1 in G2phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–12. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffl MW. A new mathematical model for relative quantification in RT-PCR. Nucleic Acids Res. 2001;29:16–21. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brierley SM, Jones RCW, Gebhart GF, Blackshaw LA. Splanchnic and pelvic mechanosensory afferents signal different qualities of colonic stimuli in mice. Gastroenterology. 2004;127:166–178. doi: 10.1053/j.gastro.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Ibeakanma C, Vanner S. TNFα is a key mediator of the pronociceptive effects of mucosal supernatant from human ulcerative colitis on colonic DRG neurons. Gut. 2010;59:612–621. doi: 10.1136/gut.2009.190439. [DOI] [PubMed] [Google Scholar]
- 47.Stewart T, Beyak MJ, Vanner S. Ileitis modulates potassium and sodium currents in guinea pig dorsal root ganglia sensory neurons. J Physiol. 2003;552:797–807. doi: 10.1113/jphysiol.2003.046409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore BA, Stewart TMR, Hill C, Vanner SJ. TNBS ileitis evokes hyperexcitability and changes in ionic membrane properties of nociceptive DRG neurons. Am J Physiol Gastrointest liver Physiol. 2002;282:G1045–51. doi: 10.1152/ajpgi.00406.2001. [DOI] [PubMed] [Google Scholar]
- 49.Ibeakanma C, et al. Brain-Gut interactions increase peripheral nociceptive signaling in mice with postinfectious irritable bowel syndrome. Gastroenterology. 2011;141:2098–2108. doi: 10.1053/j.gastro.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Denadai-Souza A, et al. Functional Proteomic Profiling of Secreted Serine Proteases in Health and Inflammatory Bowel Disease. Sci Rep. 2018;8:7834. doi: 10.1038/s41598-018-26282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denadai-Souza A, et al. Effect of tryptase inhibition on joint inflammation: A pharmacological and lentivirus-mediated gene transfer study. Arthritis Res Ther. 2017;19:124. doi: 10.1186/s13075-017-1326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roskin KM, et al. IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci Transl Med. 2015;7:302ra135. doi: 10.1126/scitranslmed.aab1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warners MJ, et al. Abnormal Responses to Local Esophageal Food Allergen Injections in Adult Patients With Eosinophilic Esophagitis. Gastroenterology. 2018;154:57–60.:e2. doi: 10.1053/j.gastro.2017.08.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for 16S rRNA gene sequences are available from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/) at accession number PRJEB41204. All other relevant data generated and analyzed during the current study are available upon reasonable request to the corresponding author (GEB). Source data are provided with this paper.