Abstract
The two tandem bromodomains of the BET proteins enable chromatin binding to facilitate transcription. Drugs that inhibit both bromodomains equally have shown efficacy in certain malignant and inflammatory conditions. To explore the individual functional contributions of the first (BD1) and second (BD2) bromodomains in biology and therapy, we developed selective BD1 and BD2 inhibitors. We found that steady-state gene expression primarily requires BD1 whereas the rapid increase of gene expression induced by inflammatory stimuli requires both BD1 and BD2 of all BET proteins. BD1 inhibitors phenocopied the effects of pan-BET inhibitors in cancer models whereas BD2 inhibitors were predominantly effective in models of inflammatory and autoimmune disease. These insights into the differential requirement of BD1 and BD2 for the maintenance and induction of gene expression may guide future BET targeted therapies.
The BET (Bromo- and Extra-Terminal domain) family is comprised of the germ cell specific (BRDT) and ubiquitously expressed (BRD2, BRD3, BRD4) epigenetic reader proteins. All family members contain N-terminal tandem bromodomains, a structural feature that enables the recognition and binding to acetylated lysine residues on histones and other cellular proteins that support their function as key transcriptional regulators(1). Following the development of first-in-class BET bromodomain inhibitors(2–4), the BET proteins have become one of the most closely studied protein families in biology. These studies have highlighted the essential role of the BET proteins in coordinating transcription programs necessary for normal development, maintenance of oncogenic gene expression and the physiological response to injury and infection(5, 6). While pre-clinical studies have demonstrated the benefit of BET inhibition in a variety of non-malignant pathologies(3, 7–10), these inhibitors have been most widely studied in the context of cancer(4, 11–14). On the basis of several promising pre-clinical studies in hematological and solid malignancies, these drugs are being evaluated in clinical trials across the world. Early results indicate that specific small molecule inhibitors of the BET bromodomains are safe and capable of inducing complete clinical remissions; however, side-effects and a short duration of clinical response have limited their broad therapeutic application (15, 16).
The first generation of BET inhibitors show equal affinity for the first (BD1) and second (BD2) bromodomains of all the BET proteins and despite enormous investment and interest in the field little is known about the functional commonalities of, and differences between, the two tandem bromodomains. Similarly, although most studies have focused on the role of BRD4 in transcriptional regulation, the individual and redundant roles of BRD2 and BRD3 are yet to be fully determined. To enable this functional dissection, we developed and characterized potent and highly selective bromodomain inhibitors of BD1 and BD2 of the BET proteins.
Development of selective BD1 and BD2 inhibitors
The helical bromodomain modules found within at least 46 human proteins(1) (fig. S1A-B) share a conserved acetyl–lysine (KAc) binding pocket. The divergent ZA and BC loops around the entrance have enabled the development of inhibitors specific to individual bromodomain families. All eight BET-family bromodomains (fig. S1C-D) share structural features, including a narrow ZA channel and a conserved hydrophobic Trp-Pro-Phe (WPF) shelf. First-generation inhibitors exploit these to achieve selectivity for the BET proteins over other bromodomains but show little discrimination within the BET family. The BD1 and BD2 domains form two separate subfamilies, and compounds able to bind preferentially to BD1s or BD2s have been reported (fig. S2), including two in clinical investigation (RVX-208 and ABBV-744)(17, 18). The disparate formats of the published data make direct comparison difficult, but our TR-FRET assays indicate most compounds, except the recently-disclosed ABBV-744, lack the potency or selectivity required to discriminate between BD1 and BD2 inhibition (fig. S3A).
To achieve a high degree of selectivity, we used structure-based design to generate compounds that interact specifically with either BD1 or BD2 of the BET proteins (Fig. 1A). In this manuscript, GSK778 and GSK046 are termed iBET-BD1 and iBET-BD2 respectively. Their affinities for the individual bromodomains of the BET family were initially determined by TR-FRET (Fig. 1B and fig. S1F, table S1). In contrast to other reported domain selective molecules, these compounds showed little binding to bromodomains outside the BET family (fig. S1E and table S2). Surface plasmon resonance (SPR) binding to BRD4 BD1 and BD2 confirmed their selectivity, showing that iBET-BD1 is ≥130-fold selective for BD1 and that iBET-BD2 is >300-fold selective for BD2 (fig. S4). The maintenance of this selectivity within a cellular context was verified using various orthogonal cellular assays employing iBET-BD1 and iBET-BD2 (fig. S6, S7). Together, these data demonstrate the high domain-selectivity of iBET-BD1 and iBET-BD2 and their specificity over other bromodomain proteins.
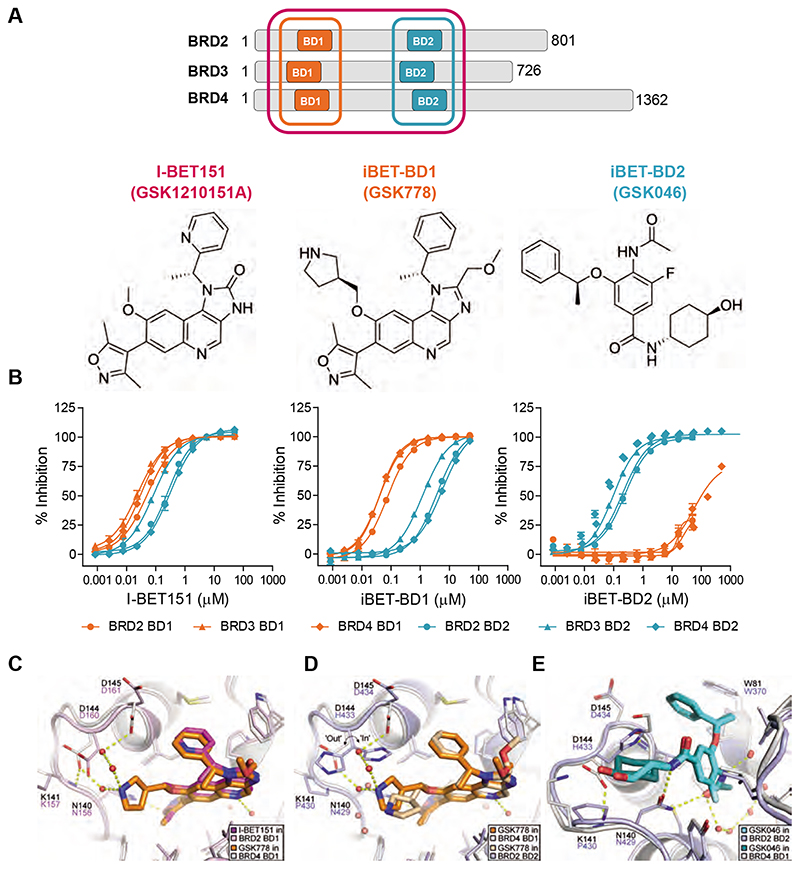
Fig. 1. Selectivity profile of I-BET151, iBET-BD1 (GSK778) and iBET-BD2 (GSK046).
(A) Schematic of the BET Bromodomain proteins and chemical structures. (B) Compound binding to the individual bromodomains of BD1 (orange) and BD2 (cyan) of BET tandem bromodomains in TR-FRET assays. (C) X-ray crystal structure of I-BET151 in BRD2 BD1 (magenta, PDB 4A1G)(19) superimposed on the structure of iBET-BD1 in BRD4 BD1 (orange, PDB 6SWN). (D) iBET-BD1 bound to BRD4 BD1 (dark orange/white, PDB 6SWN) and BRD2 BD2 (light orange/blue, PDB 6SWO) highlighting differences in the BC loop. (E) iBET-BD2 bound to BRD2 BD2 (light blue, PDB 6SWP) and BRD4 BD1 (cyan, PDB 6SWQ). In BRD2 BD2, the inhibitor’s benzyl and cyclohexane rings pack against Pro430 and His433, which adopts a single “in” conformation. In BRD4 BD1, iBET-BD2 makes no significant contacts with the corresponding Asp144 and Lys141 sidechains, and the space occupied by the His433 sidechain in BRD2 BD2 is filled by an ethane-1,2-diol molecule from the crystallisation buffer.
Crystal structures of iBET-BD1 and iBET-BD2 with BET BD1 and/or BD2 domains revealed the molecular basis of selectivity and validated our design intent (Fig. 1C-E and fig. S5, table S3). Guided by the BRD2 BD1 crystal structure with I-BET151(19), iBET-BD1 was designed to interact with BRD4 Asp144, an aspartic acid in the BC loop of all BET BD1s, which is replaced by histidine in the BD2s. The X-ray structure of iBET-BD1 in BRD4 BD1 confirms that its binding mode is similar to I-BET151 and that the appended pyrrolidine substituent, which confers BD1 selectivity, is stabilized by a water network involving Asp144 and Asp145 (Fig. 1C). The Asp144 sidechain is constrained by hydrogen-bonding to Lys141. In the BRD2 BD2 domain, replacement of Asp144 by His433 and Lys141 by Pro430 alters the local environment, preventing formation of this water network (Fig. 1D). Asp144 and Lys141 are present in all the BET BD1 domains and substituted by His and Pro in all BD2s, explaining the selectivity of iBET-BD1 for BD1s across the entire BET family.
iBET-BD2 was optimized from a high-throughput BD2-selective screening hit. BRD4 BD1 and BRD2 BD2 crystal structures rationalize its BD2 selectivity (Fig. 1E). In BRD2 BD2, the benzylic WPF-shelf group and cyclohexylamide of iBET-BD2 make extensive hydrophobic contacts with Pro430 and His433. As outlined above, Pro430 and His433 are conserved in BD2 domains and replaced by Lys and Asp in the BD1s. In BRD4 BD1, iBET-BD2 makes no contact with Lys141 or Asp144, accounting for its BD2-selectivity across the family. Additionally, a BRD2 BD2 crystal structure with ABBV-744, a preferential BD2 inhibitor, shows that its ethylamide group contacts His433 and superimposes well on the iBET-BD2 cyclohexylamide moiety, suggesting that similar mechanisms account for the BD2 selectivity of both compounds (fig. S3B)(17).
BD1 inhibition phenocopies the effects of pan-BET inhibitors in cancer
The evolutionarily conserved tandem bromodomain structure of the BET proteins is critical for their binding to acetylated interphase and mitotic chromatin(20). Structural studies have demonstrated that both BD1 and BD2 preferentially engage diacetylated peptides with an optimal spacing of two amino acids (Kac-XX-Kac). While BD1 favors binding to di-acetylated residues on histone H4, particularly H4K5ac/K8ac, BD2 is more permissive and can accommodate a diverse range of di-acetylated peptides(1, 20–22). Subsequent biochemical studies suggested that BD1 of the BET proteins is primarily responsible for chromatin-binding(25, 24) and recent functional studies showed that BD1 is primarily responsible for maintaining the malignant phenotype of acute myeloid leukemia (AML) cells(25). Consistent with these previous studies we found that the iBET-BD1 is efficient at displacing chromatin-bound BRD4, whereas iBET-BD2 was largely ineffective (fig. S6D and S7). These findings raised the prospect that selective BD1 inhibitors may be equally effective to the pan-BET inhibitors in cancer cells.
Treatment of a range of human cancer cell lines showed that iBET-BD1 has a more pronounced effect on the growth and viability of these cells (Fig. 2, A and B, fig. S8A-H). The effects of iBET-BD1 phenocopied the effects of the pan-BET inhibitor I-BET151 in inhibiting proliferation, inducing a cell cycle arrest and apoptosis in these cells (Fig. 2C-D and fig. S8I-K). Moreover, iBET-BD1 reduced the clonogenic capacity of primary human AML cells (fig. S8L). Interestingly, iBET-BD2 was less effective in all these phenotypic assays. These findings contrast with the previously reported BD2 selective compounds, RVX-208 (18) and ABBV-744 (17), and are likely due to the fact that the ability of these compounds to also engage BD1 and other bromodomain proteins in a cellular context has been markedly underappreciated (fig. S6A, S9, S10). We next profiled the in vivo properties of the domain selective inhibitors in mice. Despite the poorer pharmacokinetic properties of iBET-BD1 compared to iBET-BD2 (table S4), iBET-BD1 offered a superior survival advantage to iBET-BD2 in the aggressive MLL-AF9 AML model (Fig. 2E).
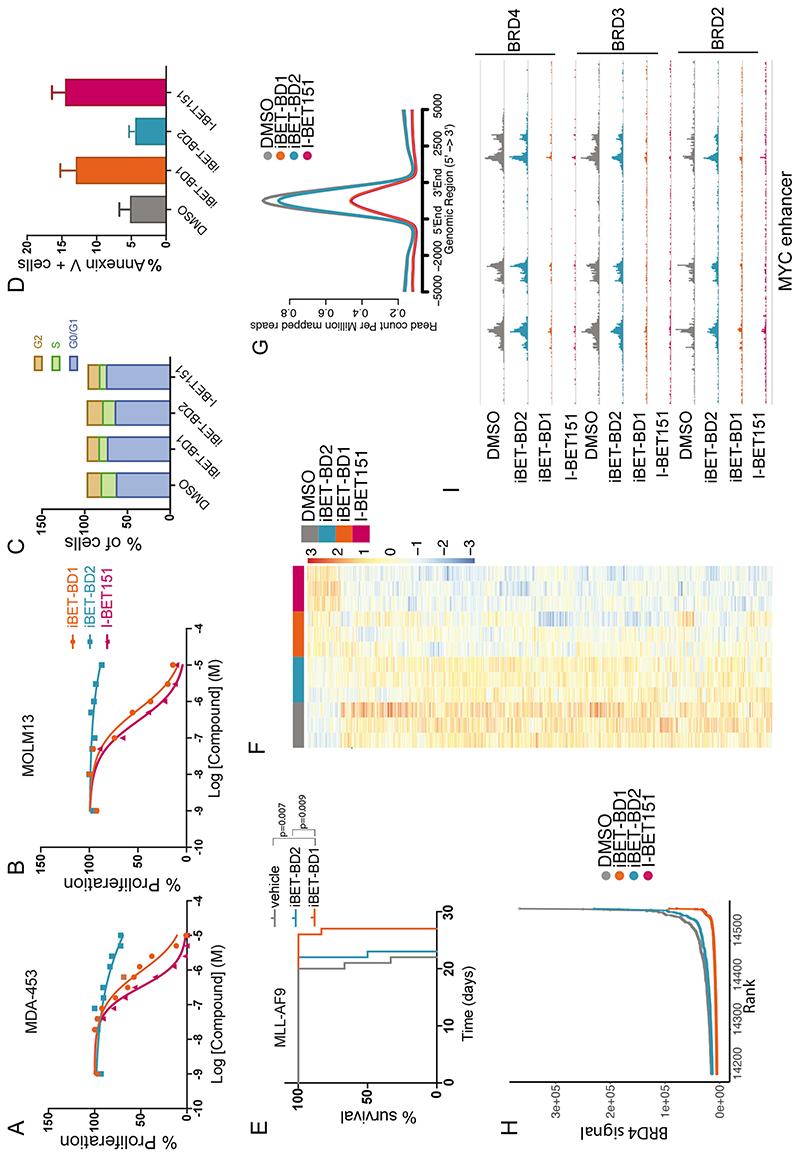
Fig. 2. BET-BD1 inhibition phenocopies pan-BET inhibition.
(A) IC50 assays performed at 72 hrs in MDA-453 and (B) MOLM-13 cells following incubation with a range of doses of iBET-BD1, iBET-BD2 or I-BET151. Data points represent Mean (n= 3 cell culture replicates) plotted as a representative from 3 independent experiments. (C) Cell cycle analysis of MV4;11 cells treated with DMSO, I-BET151 (1000nM), iBET-BD1 (1000nM), or iBET-BD2 (1000nM). (D) Apoptosis assay performed on MOLM13 cells treated with DMSO, I-BET151 (1000nM), iBET-BD1 (1000nM), or iBET-BD2 (1000nM). (E) Kaplan–Meier curve of vehicle-and drug-treated C57BL/6 mice transplanted with 1 ⨯ 106 MLL-AF9 leukemic cells. Treatment commenced at day 9 with bi-daily intraperitoneal injections at 15mg/kg of I-BET151, iBET-BD1, iBET-BD2, or Vehicle. n = 6 mice per group. P values were calculated with the two-tailed log-rank test. (F) Heatmap of differential gene expression from SLAM-seq in THP-1 cells. (G) Average profile of BRD4 ChIP-seq signal at Typical enhancers (TE) and at (H) super-enhancers after treatment with Vehicle (DMSO), I-BET151, iBET-BD1, and iBET-BD2. (I) Genome browser view of the MYC super-enhancer in THP-1 cells showing the occupancy of BRD2, BRD3, and BRD4 after treatment with Vehicle (DMSO), iBET-BD1, iBET-BD2 and I-BET151.
To examine the effects of iBET-BD1 and iBET-BD2 on transcription, we performed global nascent mRNA sequencing with SLAM-Seq (26). We found that the global transcriptome of iBET-BD1-treated cells resembled that of I-BET151-treated cells (Fig. 2F and fig. S11A-D) whereas iBET-BD2 treatment did not induce marked transcriptional changes. Concordant with these findings we found that iBET-BD1 displaced BRD2, BRD3 and BRD4 from chromatin as efficiently as I-BET151; whereas the binding of these proteins was largely unaltered by iBET-BD2 (Fig. 2G and fig. S11G) even at well characterised super-enhancers (27) such as MYC (Fig. 2H, I and fig. S11H).
BD2 facilitates recruitment of BET proteins for the induction of gene expression
While the role of the BET proteins in maintaining oncogenic gene expression programs continues to be an area of intense investigation, therapeutic targeting of this family of proteins has also shown efficacy in a range of non-malignant pathologies, particularly in the context of immuno-inflammation. Here, following inflammatory stimuli such as cytokine stimulation, the BET proteins are specifically recruited to target gene loci to help facilitate the rapid induction of gene expression. To model this scenario, we stimulated K562 cells with the pro-inflammatory cytokine interferon-γ (IFN-γ). K562 cells lack cell surface expression of MHC-I, however, these genes are transcriptionally induced by IFN-γ stimulation leading to the cell surface expression of MHC-I and a functional readout for this pathway(28). Using this established model of cytokine-induced gene expression, we explored the functional effects of the domain selective inhibitors. Here we found that I-BET151 inhibited the IFN-γ induced expression of MHC-I and interestingly, we noted that both iBET-BD1 and iBET-BD2 were also effective in recapitulating this response (Fig. 3A).
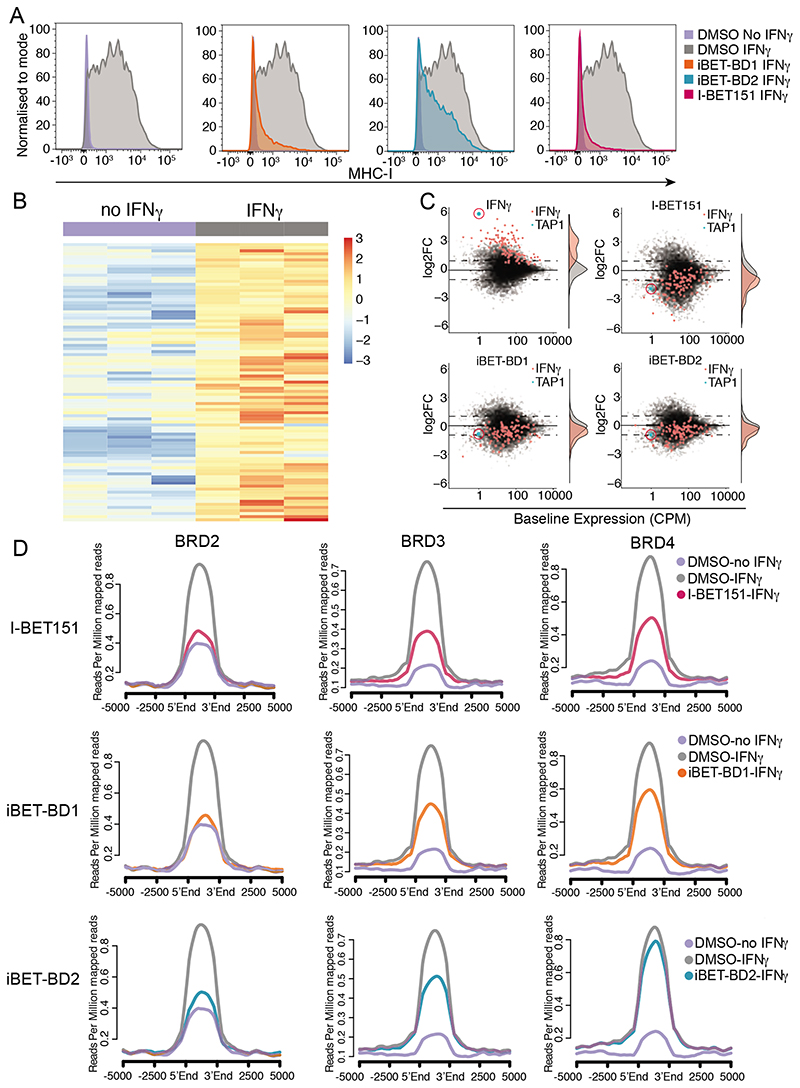
Fig. 3. BET-BD2 is required for activation of IFNg target genes.
(A) FACS analysis of MHC-I expression using antibodies against HLA-A/B/C in K562 cells following stimulation with IFNg (10ng/ml) and treatment with DMSO, iBET-BD1, iBET-BD2 or I-BET151 for 48 hrs. (B) Hierarchical clustering heatmap from SLAM-seq data in K562 cells showing upregulated genes following IFNg treatment for 6 hrs. (C) Scatter plot of differential expression of all genes (grey) and genes significantly upregulated by IFNg (red) in K562 cells, scatter plot and histogram shown for DMSO, iBET-BD1, iBET-BD2 and I-BET151. (D) Average profile plot of BRD2, BRD3 and BRD4 at genomic loci where H3K27ac levels increase following stimulation with IFNg in K562 cells for 6hrs.
To follow the molecular events associated with this phenotype we assessed the global nascent transcriptome of the cells following stimulation with IFN-γ in the presence or absence of the various BET inhibitors (Fig. 3B). We found that I-BET151, iBET-BD1 and iBET-BD2 inhibited the induction of IFN-γ responsive transcripts, which include several components of the MHC-I antigen presentation pathway (Fig. 3C). Notably, the effects of iBET-BD2 at these genes were specific to IFN-γ stimulation, as the inhibitor did not alter the baseline expression of these transcripts (fig. S12A-C). These data suggested that whilst BD2 is not essential for the chromatin binding of the BET proteins to maintain pre-existing transcriptional programs, it plays a more important role in the recruitment of the BET proteins for the induction of gene expression.
In support of this possibility, ChIP-seq analyses showed that in the presence of I-BET151, iBET-BD1 and iBET-BD2, the BET proteins show reduced recruitment to IFN target genes following IFN-γ stimulation (Fig. 3D and fig. S12D). Interestingly, iBET-BD2 appeared to more prominently affect the recruitment of BRD2 and BRD3 compared to BRD4 (Fig. 3D and fig. S12D). To further understand whether the findings associated with cytokine stimulation were also applicable to other cellular contexts and different stimuli that also result in gene induction, we next assessed the global transcriptome and chromatin binding of the BET proteins in THP-1 cells stimulated with phorbol 12-myristate 13-acetate (PMA), a widely used model of macrophage differentiation(29). Here again, we focused on the genes induced by PMA treatment (fig. S13) and although the baseline expression of these genes was not affected by iBET-BD2, treatment with iBET-BD2 prevented the PMA induced gene expression (fig. S13B). Consistent with our data in the context of cytokine stimulation, we find that iBET-BD2, like iBET-BD1 and I-BET151, prevented the PMA induced recruitment of all the BET proteins and the effects of iBET-BD2 were again more pronounced against BRD2 and BRD3 (fig. S13C-E). Moreover, these findings were replicated in a third model using anti-CD3/CD28 mediated stimulation of primary human CD4+ T cells (fig. S14)
BRD4 alone is insufficient for stimulated gene expression
While the majority of the literature assessing the transcriptional function of the BET proteins has focused on the co-activatory effects of BRD4, these studies have mainly examined the requirement of BRD4 to maintain pre-existing transcriptional programs. Whether BRD4 alone is sufficient for rapid induction has not been thoroughly evaluated. Our data raised the prospect that in the context of stimulus-induced gene expression, all of the ubiquitously expressed BET proteins, and not just BRD4, may be required for efficient gene induction. To address this observation, we replicated the effects of a small molecule inhibitor that rapidly and specifically impairs the function of BRD4 without affecting either BRD2 or BRD3 by using an auxin inducible degron tagging strategy to degrade endogenous BRD4 (Fig. 4A and fig. S15A). This method has been employed to highlight the importance of BRD4 in maintaining established malignant gene expression programs(26). Similarly, we also find that specific loss of BRD4 leads to impaired proliferation and survival of cancer cell lines that was comparable in efficacy to I-BET151 and iBET-BD1 (fig. S15B). Moreover, the changes in the established gene expression program following selective BRD4 loss are largely phenocopied by I-BET151 (fig. S15E).
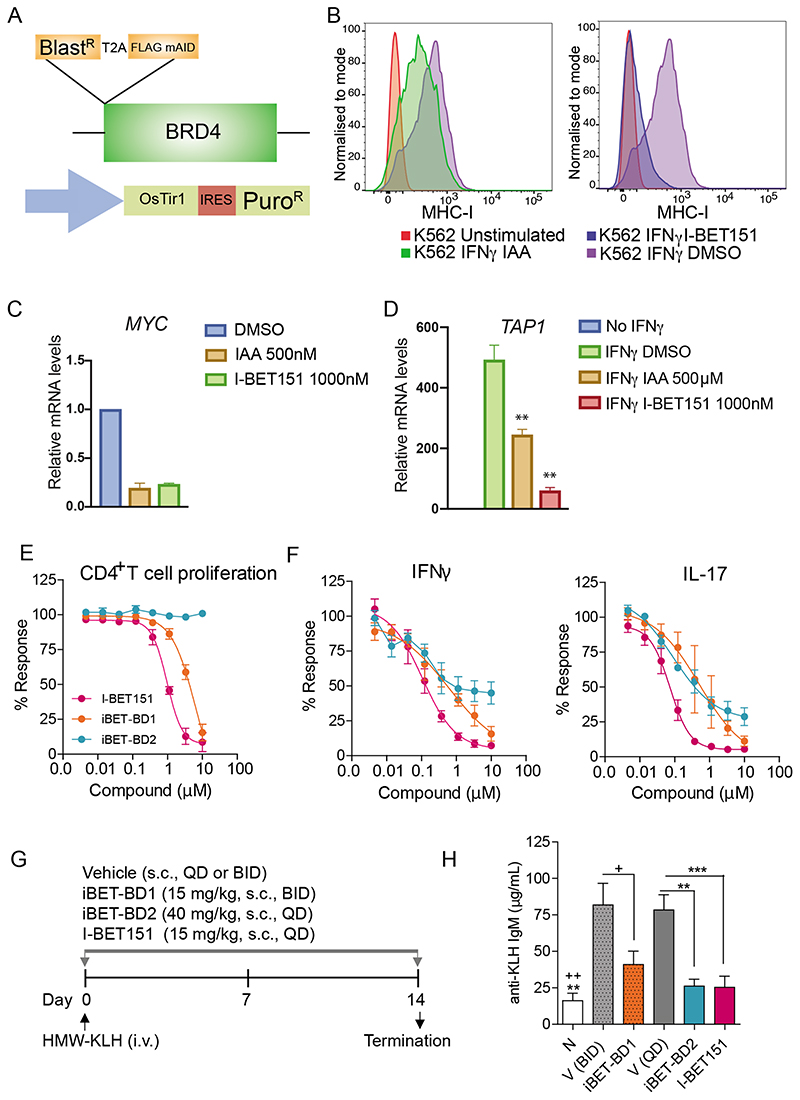
Fig. 4. iBET-BD2 has immunomodulatory activity.
(A) schematic of auxin-inducible degradation strategy for BRD4. BlastR (Blasticidin resistance) linked in-frame with T2A self-cleaving peptide to FLAG-mAID-BRD4. A separate viral vector was used to express CMV promoter driving expression of OsTIR1-IRES-Puro resistance. (B) FACS analysis of HLA/B/C expression in K562 cells pretreated with DMSO, Auxin (IAA) or I-BET151 for 6hrs followed by stimulation with IFNg (10ng/ml) for 48 hrs and 48 hr incubation with compounds. (C) qRT-PCR analysis of MYC expression in K562 cells following treatment with I-BET151 or IAA (Auxin) for 6hrs. Data shown represent the mean ± SD (n=3) (D) qRT-PCR analysis of TAP1 expression in K562 cells before and after IFNg treatment (6 hours) and/or either IAA (Auxin), IBET151, or DMSO (Vehicle control) for 7 hours (1hr pretreatment), Data shown represent the mean (n=3) +/- SD, ** p>0.01. (E) Compound effects on cellular proliferation and (F) cytokine production in anti-CD3/CD28-stimulated human primary CD4+ T cells. Data represent the mean ± SEM (n = 4) (G-H) Efficacy of compounds reducing KLH-induced antibody responses (IgM) in mice. N, naїve (n = 4); V, vehicle; iBET-BD1 (15 mg/kg, s.c., BID), iBET-BD2 (40 mg/kg, s.c., QD), I-BET151 (15 mg/kg, s.c., QD). Data shown as the mean ± SEM (n = 10). One-way ANOVA followed by Bonferroni’s multiple comparison test was used to determine statistical significance compared with respective vehicle controls (***P<0.001, **P<0.005 vs V(QD); ++ P<0.005, + P<0.01 vs V(BID)).
Although these data confirm that BRD4 is required to maintain established oncogenic transcriptional programs, we found that selective BRD4 loss was not as effective as a pan-BET inhibitor at impairing the induction of MHC-I following IFN-γ stimulation (Fig. 4B). Notably, while selective BRD4 loss, but not iBET-BD2 treatment, resulted in profound repression of MYC that was equivalent to the effects seen with I-BET151 or iBET-BD1 (Fig. 4C fig. S15 C, D), it was not as effective as I-BET151 in negating IFN-γ induced gene expression (Fig. 4D). Together, these findings suggest that while BRD4 may be the primary BET protein required to maintain steady state gene expression, cooperation amongst all of the BET proteins is required to efficiently induce gene expression. Moreover, while BD1 of the BET proteins is sufficient to tether the BET family to chromatin and maintain gene expression, both BD1 and BD2 of all BET family members are required for the recruitment and binding to chromatin for the rapid induction of gene expression.
iBET-BD1 and iBET-BD2 have immunomodulatory activity
To further investigate the functional properties of iBET-BD1 and iBET-BD2 we used the BioMap Diversity plus panel to assess the phenotypic responses of cytokine, growth factor and hormonal stimulation in a broad range of primary human cell-based co-culture systems. These data showed that both I-BET151 and iBET-BD1 treatment resulted in a broad phenotypic profile across the entire BioMap panel; whereas iBET-BD2 displayed a more selective phenotypic fingerprint, particularly inhibiting the production of key pro-inflammatory mediators including Th17 cytokines in the B and T cell co-culture system (fig. S16A, B). These effects were subsequently confirmed in anti-CD3/CD28-stimulated human primary CD4+ T cell cultures where unlike I-BET151 and iBET-BD1 treatment, iBET-BD2 did not affect the proliferative activity of the cells but still inhibited the production of effector cytokines including IFNy, IL-17A and IL-22 (Fig. 4E-F, fig. S15F and fig. S16C). Moreover, in line with our molecular data in THP1 cells, we again found that iBET-BD2 impaired macrophage activation following PMA stimulation, similar to iBET-BD1 and I-BET151, without impacting cellular viability (fig. S15G-H).
These functional assays in vitro raised the intriguing possibility that although iBET-BD2 was largely ineffective at impairing proliferation and survival of cancer cells (Fig. 2 and fig. S8), it may have greater in vivo activity in an immunomodulatory context. To explore this further, we assessed the efficacy of iBET-BD1 and iBET-BD2 in a T cell dependent immunisation model(30). Here mice are immunised with the exogenous antigen keyhole limpet hemocyanin (KLH) resulting in efficient T cell activation, the production of cytokines (IFNγ) and T cell dependent primary antibodies (IgM). All compounds were well tolerated (fig. S16D) and, in agreement with our molecular data and the functional in vitro assays, we found that iBET-BD2 was as effective as iBET-BD1 and I-BET151 in reducing the production of anti-KLH IgM (Fig. 4G-H). Taken together, these data show that BD2 selective inhibition specifically affects the induction of gene expression whilst leaving the maintenance of established transcription programs largely unaltered. This distinction may have important clinical implications as the repression of established gene expression may be essential for the maintenance of cell identity and viability. For instance, as described previously even low doses of pan-BET inhibitors result in the undesired effect of testicular atrophy (31, 32); however these effects are much less pronounced even with high doses of iBET-BD2 (fig. S16E).
BD2-selective inhibition ameliorates inflammatory disease in preclinical models
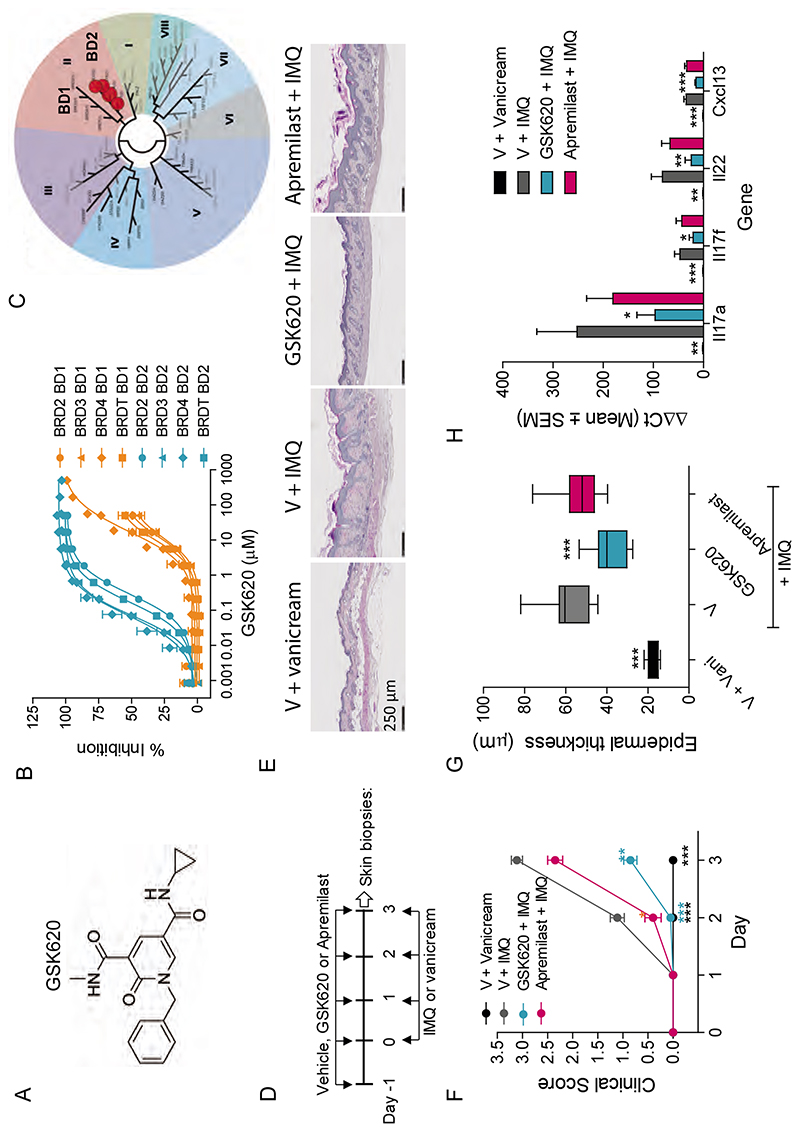
The ability to selectively alter induced gene expression whilst leaving established gene expression programs intact with iBET-BD2 raised the exciting possibility of specifically targeting pathologies initiated by an inflammatory gene expression program. To address this further we optimized the pharmacokinetic properties of iBET-BD2 to develop GSK620 (Fig. 5A-C and tables S4 and S5). This compound retains the marked specificity for binding BD2 in all of the BET family of proteins (Fig. 5C and tables S1 and S2) but has significantly improved oral bioavailability compared to iBET-BD2 (table S5). We initially tested GSK620 in a collagen-induced arthritis model in rats, a commonly used model of pathologies such as rheumatoid arthritis where an immunological response manifests as inflammatory arthritis associated with progressive cartilaginous destruction, joint swelling, and ultimately ankylosis. In this model, GSK620 led to a significant dose-dependent inhibition of both the arthritic score and IgG1 production in response to the immunisation (fig. S17A-E). Importantly, at the well tolerated highest dose, GSK620 showed similar level of efficacy to the pan-BET inhibitor I-BET151 and also dramatically reduced joint swelling, synovitis, cartilage damage, pannus formation and bone resorption (fig. S17C, F-G).
Fig. 5. GSK620 is efficacious in preclinical models of immuno-inflammation.
(A) Chemical structure of the BD2-selective BET inhibitor in vivo tool GSK620. (B) Compound binding to the individual tandem bromodomains (BD1 (orange) and BD2(cyan)) of BRD2, 3, 4 and T determined using TR-FRET. Data are the mean ± SEM (n = 12-21). (C) Phylogenetic tree of bromodomain family demonstrating preferential compound binding for the BD2 domains of the BET family of proteins using the BROMOscan bromodomain competition binding assay. Red dots represent KD values. (D) Mouse imiquimod (IMQ)-induced psoriasis study design. (E) Histology H&P (Hematoxylin-Phloxin) staining of skin sections (F) GSK620 (20mg/kg, p.o., QD) reduces the psoriasis score, (G) the epidermal thickness and (H) the expression levels of inflammatory genes in skin biopsies. V, vehicle; Vani,vanicream; Apremilast (20 mg/kg, p.o., BID). Data represent the mean ± SEM (n = 10). One-way ANOVA followed by Dunnett’s multiple comparison test (***P<0.0001 vs V + IMQ).
The efficacy of BD2 selective inhibition in these immuno–inflammatory models contrasts sharply with the modest effects seen in cancer models and raised the prospect that BD2 selective BET inhibition may be a valuable therapeutic strategy to specifically counter various immuno-inflammatory diseases. Therefore, we next evaluated the efficacy of GSK620 in a mouse model of imiquimod (IMQ)-induced psoriasis(33). Whilst topical therapies are often used for mild psoriasis, more severe forms of the disease require systemic therapy with anti-inflammatory and immunomodulatory agents including the phosphodiesterase 4 (PDE4) inhibitor, apremilast, which has recently been FDA approved for moderate to severe psoriasis and other autoimmune diseases such as Behcet’s disease(34). In our study, we found that GSK620 was superior to apremilast in reducing the clinical score (erythema and plaque formation) and the epidermal hyperplasia associated with the imiquimod treatment (Fig. 5D-G, fig. S17H). Moreover, the gene expression analyses from the skin of treated mice showed that GSK620 significantly reduced the expression of disease relevant pro-inflammatory genes, including IL-17A, IL17F and IL-22 (Fig. 5H).
Finally, we also tested GSK620 in a mouse model of non–alcoholic fatty liver disease (NAFLD), a widespread problem in western society associated with obesity and insulin resistance that ultimately manifests in liver fibrosis and predisposes to the development of hepatocellular carcinoma. We have recently demonstrated the promising effects of non-selective BET inhibition in the STAM mouse model, which recapitulates many of the features of human NAFLD (8) and wanted to further evaluate if these effects could be phenocopied with BD2 selective inhibition. We assessed the effects of treatment at two distinct time windows to evaluate its efficacy on early non-alcoholic steatohepatitis (NASH) (“NASH phase”, week 6-9) (fig. S18A-C) and during fibrosis onset (“fibrosis phase”, week 9-12) (fig. S18 D). We found that mice treated with GSK620 showed reduced steatosis, lobular inflammation and hepatocyte ballooning when compared to the vehicle-treated group (fig. S18C). Importantly, in comparison to telmisartan, which has shown activity in human NASH, we found that GSK620 showed comparable activity in reducing hepatic fibrosis (fig. S18E). Moreover, GSK620 was highly effective in reducing the expression of pro-inflammatory and pro-fibrotic genes in liver biopsies taken from treated animals (fig. S18F). Taken together, these data in separate models of inflammatory arthritis, psoriasis and hepatitis demonstrate the pre-clinical value of specifically modulating inducible gene-expression by targeting BD2 of the BET family in immuno-inflammatory pathologies.
Discussion
Despite marked progress in studying the role of the BET proteins as key transcriptional regulators several fundamental questions about this family of proteins remain. For instance, it is largely unknown why most cells ubiquitously express three BET proteins. It is also not clear why both bromodomains in all the BET proteins share such high degree of structural conservation and, show a similar preference for substrate binding (particularly di-acetylated histone tails). Our data assessing the specific requirements of BD1 and BD2 in the context of maintenance or induction of gene expression has highlighted that BD1 is the primary module required to bind chromatin to maintain established gene expression programs and phenocopies the effects of pan-BET inhibitors in cancer cells. While first-generation pan-BET inhibitors can induce a complete clinical remission in some patients(15, 16, 35, 36), these remissions are often short-lived suggesting that these drugs will need to be combined with other conventional and targeted therapies to provide meaningful clinical outcomes. As every cancer therapy has side effects, refining the target specificity of current BET inhibitors to focus on BD1 may maintain the observed efficacy but limit the side effects particularly as part of combination regimes.
In contrast to the effects seen with BD1 selective inhibitors, BD2 selective inhibitors have minimal effects in altering pre-existing gene expression programs as they are largely ineffective at displacing chromatin bound BET proteins from established cis-regulatory elements including super-enhancers. Although BD2 inhibition is not critical to maintain gene expression, we found that BD2 of all the BET proteins is necessary following a stimulus that drives rapid gene induction. The requirement for a structurally-related chromatin binding module to facilitate the rapid recruitment and stability of the BET proteins to provide a swift and coordinated transcriptional response helps explain why the two tandem bromodomains are not only structurally similar but also highly conserved through evolution. Moreover, we also found that efficient gene induction demands the functional integrity of all three ubiquitously expressed BET proteins, and BRD4 by itself was insufficient for optimal gene induction. These findings help reconcile a number of previous observations. For instance, global genetic dependency analyses have clearly demonstrated that BRD4 is the only BET protein that is a pan-essential requirement in mammalian cells (37). BRD4 also appears to be the main BET protein required to maintain established gene expression programs(26) making it difficult to understand why its paralogues BRD2 and BRD3 are also ubiquitously expressed. Interestingly, the BET proteins particularly BRD2, have consistently been shown to play a critical role in mediating inflammatory responses(38) and mice that express a hypomorphic BRD2 allele are protected from inflammatory diseases(39). Notably, the first-generation BET inhibitors have been effective in a number of seemingly disparate pathologies including heart failure(40), sepsis(3), and drug induced nephrotoxicity(41). Whilst these pathologies appear to be unrelated, all of them are linked by the common requirement to mount an immediate adaptive transcriptional response following a defined insult or injury. Consistent with this, our data show that BD2 selective inhibition is efficacious in a broad range of inflammatory pathologies and raise the possibility that BD2 inhibition may be a useful and novel therapeutic strategy to counter the immunoinflammatory damage that results in end organ damage following infective, autoimmune or toxic insult. Taken together, our data provide new insights to refine our therapeutic strategy and achieve greater efficacy with few side-effects when targeting BET proteins in distinct pathologies such as cancer and immuno-inflammation.
Supplementary Material
Acknowledgements
We thank members of the Dawson Laboratory, the molecular genomics and flow cytometry core facility at Peter Mac for helpful discussions and technical assistance. We would also like to thank Dr Carleen Cullinane and Susan Jackson for their assistance with the animal experiments.
Funding
We thank the following funders for fellowship and grant support: CCV Dunlop Fellowship and HHMI international research scholarship (M.A.D); Victoria Cancer Agency (O.G) (E.Y.N.L) fellowships), CRUK (M.L.B), CSL Centenary fellowship (S.J.D). Work from the Dawson Lab is supported by grant funding from NHMRC (1106444, 1144649, 1146192), University of Melbourne and Peter Mac foundation.
Footnotes
Author contributions: O.G, I.R, R.K.P, M.A.D designed and analysed experiments and wrote the manuscript with helpful contributions from all authors. M.Y.M, E.Y.N.L, and D.V performed computational analyses that was supervised by S.J.D and M.A.D. O.G, K.K, M.L.B performed experiments and analysed data supervised by M.A.D. M.J.B, N.R.H, A.K.B, E.J.R, A.K.B, P.E.S, D.F.T performed in vitro biology experiments, analysed and interpreted data supervised by I.R and R.K.P. S.J.A, A.G.S.P, C.W, M.L, E.H.D designed and synthesised the compounds and generated and analysed data. J.R.G, S.T, T.G, V.K and A.H designed and generated data from DMPK and in vivo efficacy studies. C.W.C and P.B generated crystallography and data analysis. M.P, T.W, A.M.M, J.V, M.B designed, generated data and analysed omics experiments supervised by G.D and P.G.
Competing interests: M.A.D. has been a member of advisory boards for CTX CRC, Storm Therapeutics, Celgene and Cambridge Epigenetix. The Dawson lab receives research funding from CTX CRC. The following authors: R.K.P, I.R, M.J.B, N.R.H, A.K.B, E.J.R, P.E.S, D.F.T, J.R.G, T.G, V.K, A.H, S.J.A, A.G.S.P, C.W, E.H.D, C.W.C, P.B, M.P, T.W, A.M.M, J.V, M.B, G.D and P.G are employees and shareholders of GSK. M.L is an employee at Biopharmaceuticals R&D, AstraZeneca. S.T is an employee at Pharmaron UK and hold shares at Glaxo Smith Kline (GSK). GSK and the authors S.J.A, A.G.S.P, M.L and E.H.D have a patent/patent application WO 2017037116 A1 related to the invention of certain compounds which are bromodomain inhibitors including GSK620. M.U and D.L.D are employees of Promega. The remaining authors declare no competing financial interests.
Data and materials availability
I-BET151, GSK778, GSK046 and GSK620 are available from R.K.P (rabinder.prinjha@gsk.com) and I.R (inma.5.rioja@gsk.com) under a material transfer agreement with GSK. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with identification code 6SWN, 6SWO, 6SWP and 6SWQ. The RNA-seq and ChIP-seq data have been deposited to the NCBI Sequence Archive under the accession number GSE138210.
References and Notes
- 1.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson MA. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- 6.Tough DF, Tak PP, Tarakhovsky A, Prinjha RK. Epigenetic drug discovery: breaking through the immune barrier. Nat Rev Drug Discov. 2016;15:835–853. doi: 10.1038/nrd.2016.185. [DOI] [PubMed] [Google Scholar]
- 7.Klein K, et al. The bromodomain protein inhibitor I-BET151 suppresses expression of inflammatory genes and matrix degrading enzymes in rheumatoid arthritis synovial fibroblasts. Ann Rheum Dis. 2016;75:422–429. doi: 10.1136/annrheumdis-2014-205809. [DOI] [PubMed] [Google Scholar]
- 8.Middleton SA, et al. BET Inhibition Improves NASH and Liver Fibrosis. Sci Rep. 2018;8:17257. doi: 10.1038/s41598-018-35653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadeem A, et al. Imiquimod-induced psoriasis-like skin inflammation is suppressed by BET bromodomain inhibitor in mice through RORC/IL-17A pathway modulation. Pharmacol Res. 2015;99:248–257. doi: 10.1016/j.phrs.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Duan Q, et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci Transl Med. 2017;9:eaah5084. doi: 10.1126/scitranslmed.aah5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson M, et al. A Phase I Study of GSK525762, a Selective Bromodomain (BRD) and Extra Terminal Protein (BET) Inhibitor: Results from Part 1 of Phase I/II Open Label Single Agent Study in Patients with Acute Myeloid Leukemia (AML) Blood. 2017;130:1377. [Google Scholar]
- 16.Berthon C, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 17.Faivre EJ, et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature. 2020;578:306–310. doi: 10.1038/s41586-020-1930-8. [DOI] [PubMed] [Google Scholar]
- 18.McLure KG, et al. RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist. PLOS ONE. 2014;8:e83190. doi: 10.1371/journal.pone.0083190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seal J, et al. Identification of a novel series of BET family bromodomain inhibitors: binding mode and profile of I-BET151 (GSK1210151A) Bioorg Med Chem Lett. 2012;22:2968–2972. doi: 10.1016/j.bmcl.2012.02.041. [DOI] [PubMed] [Google Scholar]
- 20.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamsjaeger R, et al. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol. 2011;31:2632–2640. doi: 10.1128/MCB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriniere J, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 23.Baud MG, et al. Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science. 2014;346:638–641. doi: 10.1126/science.1249830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller TC, et al. A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat Commun. 2016;7:13855. doi: 10.1038/ncomms13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyler DS, et al. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science. 2017;356:1397–1401. doi: 10.1126/science.aal2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muhar M, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360:800–805. doi: 10.1126/science.aao2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burr ML, et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell. 2019;36:385–401.:e388. doi: 10.1016/j.ccell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One. 2010;5:e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelso A, Groves P, Troutt AB, Pech MH. Rapid establishment of a stable IL-4/IFN-gamma production profile in the antigen-specific CD4+ T cell response to protein immunization. Int Immunol. 1994;6:1515–1523. doi: 10.1093/intimm/6.10.1515. [DOI] [PubMed] [Google Scholar]
- 31.Matzuk MM, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang E, Nickerson HD, Wen D, Wang X, Wolgemuth DJ. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development. 2007;134:3507–3515. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- 33.van der Fits L, et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 34.Hatemi G, et al. Apremilast for Behcet’s syndrome--a phase 2, placebo-controlled study. N Engl J Med. 2015;372:1510–1518. doi: 10.1056/NEJMoa1408684. [DOI] [PubMed] [Google Scholar]
- 35.Amorim S, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3:e196–204. doi: 10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 36.Lewin J, et al. Phase Ib Trial With Birabresib, a Small-Molecule Inhibitor of Bromodomain and Extraterminal Proteins, in Patients With Selected Advanced Solid Tumors. J Clin Oncol. 2018;36:3007–3014. doi: 10.1200/JCO.2018.78.2292. [DOI] [PubMed] [Google Scholar]
- 37.Tsherniak A, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576.:e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F, et al. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J. 2009;425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan Q, et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aah5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun L, Liu J, Yuan Y, Zhang X, Dong Z. Protective effect of the BET protein inhibitor JQ1 in cisplatin-induced nephrotoxicity. Am J Physiol Renal Physiol. 2018;315:F469–F478. doi: 10.1152/ajprenal.00527.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becher I, et al. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat Chem Biol. 2016;12:908–910. doi: 10.1038/nchembio.2185. [DOI] [PubMed] [Google Scholar]
- 43.Riching KM, et al. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem. 2018;13:2758–2770. doi: 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- 44.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
I-BET151, GSK778, GSK046 and GSK620 are available from R.K.P (rabinder.prinjha@gsk.com) and I.R (inma.5.rioja@gsk.com) under a material transfer agreement with GSK. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with identification code 6SWN, 6SWO, 6SWP and 6SWQ. The RNA-seq and ChIP-seq data have been deposited to the NCBI Sequence Archive under the accession number GSE138210.