Abstract
We have identified a proteolysis targeting chimera (PROTAC) of class I HDACs 1, 2 and 3. The most active degrader consists of a benzamide HDAC inhibitor, an alkyl linker, and the von Hippel-Lindau E3 ligand. Our PROTAC increased histone acetylation levels and compromised colon cancer HCT116 cell viability, establishing a degradation strategy as an alternative to class I HDAC inhibition.
Targeting enzymes involved in the regulation of epigenetic modifications have provided therapeutic drugs in treating cancer and have potential in treating other diseases including neurological disorders and cardiovascular disease.1 The Histone Deacetylase (HDAC) family of enzymes, often termed epigenetic erasers, function by removing the acetyl moiety from histone tails thereby modifying chromatin structure and gene expression.2 Currently five HDAC inhibitors have been approved for clinical use to treat T-cell lymphoma and multiple myeloma with other compounds in clinical trials.3 18 HDAC enzymes have been identified in humans, 11 with a divalent zinc cation in the catalytic site and 7 sirtuins whose activity is NAD+ dependent.4 Inhibitors of the Zn2+-dependent enzymes, such as Valproic acid or SAHA (Zolinza), are currently used in the clinic to treat cutaneous T-cell lymphoma (CTCL) and bipolar disorder. However, these drugs exhibit limited selectivity and this has been attributed as a cause of their debilitating side-effects in patients.5
HDAC 1, 2 and 3 are localized in the nucleus, constitute approximately 50% of total cellular deacetylase activity and are the most prominent HDACs in gene expression.6 They do not function as singular entities, but exist in vivo as catalytic subunits in much larger multi-protein corepressor complexes, including Sin3, NuRD, CoREST, MiDAC and NCoR.4 These complexes play an essential role in targeting the HDAC enzyme to specific regions in the genome and demonstrate distinct cell type specific functions.7
The Proteolysis Targeting Chimera (PROTAC) technology has been receiving considerable attention as a novel strategy to target difficult-to-drug proteins of interest (POI). PROTACs are heterobifunctional molecules that couple a ligand for the POI with a ligand for an E3 ligase such that the POI becomes polyubiquitinated and degraded by the proteasome.8 One of the exciting observations regarding PROTACs is their ability to induce selectivity that has otherwise proved very challenging.9 One hypothesis for PROTAC-induced selectivity is the necessity of a protein-protein interaction between the POI and the E3 ligase required for degradation.10 Towards this endeavor we wanted to generate PROTACs specifically for class I HDAC 1, 2 & 3, present in corepressor complexes.
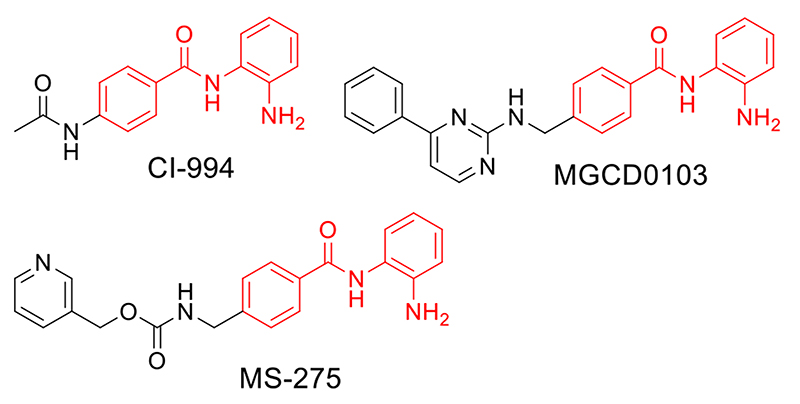
For our PROTAC design we chose benzamide-based HDAC inhibitors that demonstrate selectivity for HDAC 1, 2 & 3. CI-994 has reported Ki values of 0.41 μM for HDAC 1, 0.75μM for HDAC 3, and approximately 100μM for HDAC 8 (Fig 1).11 This inhibitor exhibits phenotypes related to HDAC 1, 2 & 3 activity, inhibition of cell proliferation, induction of apoptosis and broad anti-tumor activity in vitro and in vivo.11,12 CI-994 has been in clinical trials for its anti-tumor properties and analogous benzamides, MS-275 and MGCD0103, are currently in phase III clinical trials for breast cancer and non-small cell lung cancer (Fig 1).3 More recently CI-994 has also been reported for its neuroprotective effects in mice following spinal cord injury, the treatment of cognitive disorders, and reducing atrial fibrillation.13
Figure 1.
Benzamide based HDAC inhibitors. CI-994 demonstrates selective inhibition for HDAC 1, 2 & 3, inhibits cell proliferation, induces apoptosis and has anti-tumor activity. MS-275 is a HDAC 1 & 3 selective inhibitor with anti-tumor activity currently in phase III clinical trials. MGCD0103 is a HDAC 1, 2, 3 inhibitor in clinical trials.
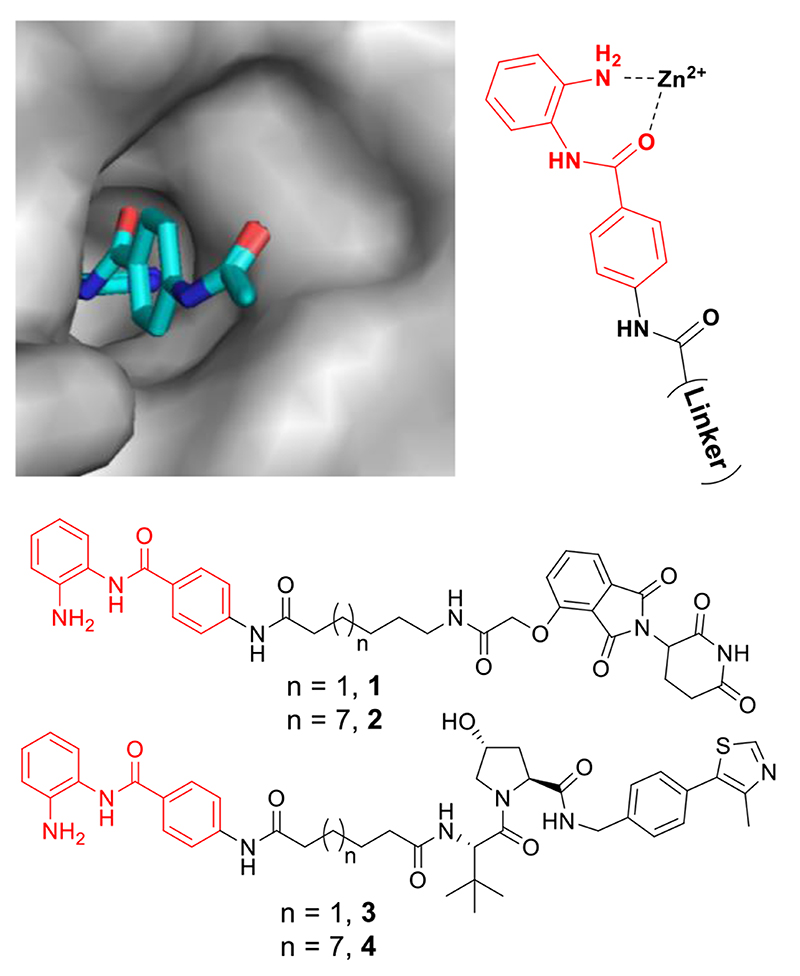
We functionalized CI-994 from the acetyl group of the phenyl moiety (Fig 2, see supplementary information for complete synthesis) as the acetyl group of an analogous benzamide inhibitor is protruding from the catalytic active site and surface exposed in a crystal structure bound to HDAC 2.14 Hence, functionalization at this position should maintain HDAC binding. A short alkyl linker length was prepared (six carbon) and a longer linker length prepared (twelve carbon) as in previous PROTAC studies it has been shown the linker length can play an essential role in inducing degradation.15 Alkyl linkers were chosen to hasten synthesis. Two different E3 ligands were also chosen; the von Hippel-Lindau (VHL) ligand and the cereblon ligand, since successful protein-protein engagement with an E3 ligase is a critical step in promoting degradation.10
Figure 2.
The benzamide acetyl group is surface exposed in a crystal structure bound to the HDAC 2 catalytic active site (PDB: 4LY1). Four preliminary heterobifunctional molecules were synthesised. PROTAC 1 and 3 with a six carbon alkyl linker and either the VHL or Cereblon E3 ligand. PROTAC 2 and 4 with a twelve carbon alkyl linker and either the VHL or Cereblon E3 ligand.
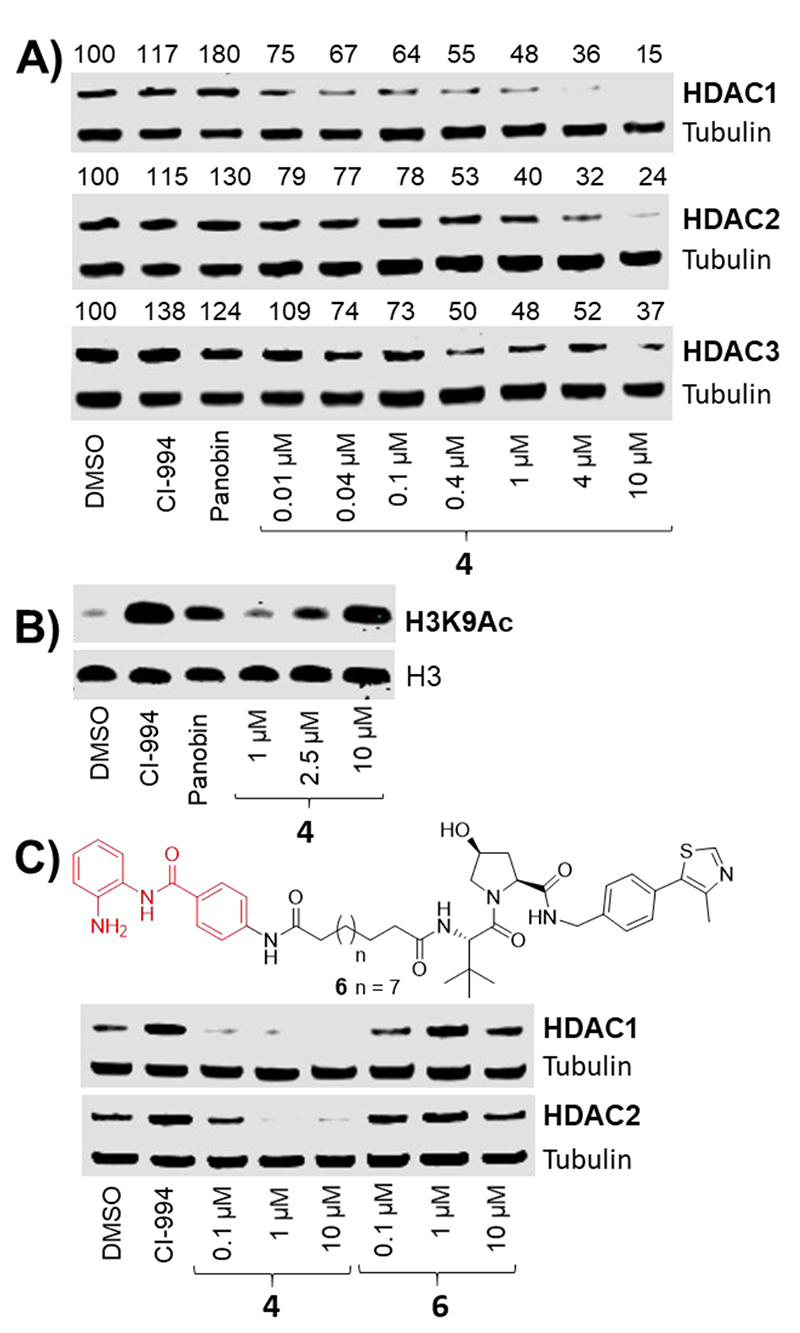
We evaluated compounds 1-4 in vitro using an established fluorescent deacetylase assay with a purified ternary LSD1-CoREST-HDAC1 complex.16 This complex was used as an exemplar of an HDAC multi-protein complex to help determine if heterobifunctional molecules can still engage with the HDAC enzyme when incorporated into a multi-protein entity. The IC50 values of 1-4 were determined side-by-side with CI-994 as a positive control. As a negative control we also synthesized Boc protected CI-994, compound 5. Compound 5 is not capable of chelating zinc in the HDAC active site and should be incapable of HDAC inhibition. We observed that CI-994 had an IC50 value of approx. 0.5 μM consistent with previous reported values,11 and 5 demonstrated no HDAC inhibition (Fig 3A). The putative PROTACs 1 and 3 with shorter linker lengths all engaged HDAC1 in the CoREST complex with IC50 values directly comparable to CI-994, while the longer linker lengths, 2 and 4, still maintained inhibition but at reduced levels compared to CI-994 and compounds 1 and 3 (Fig 3A). We proceeded to assess the effects of these compounds on HDAC activity in cells. In a previous study we demonstrated that acetylated Histone 3 Lysine 56 (H3K56Ac) is a direct substrate of HDAC 1 in embryonic stem (ES) cells.17 To examine the ability of 1-4 to reduce HDAC 1 and 2 activity we began by measuring H3K56 acetylation using quantitative western blotting in ES cells (Fig 3B). CI-994 and the pan-HDAC inhibitor, Panobinostat, were used as positive controls and H3K56 acetylation increased as expected, 15-20 fold compared to non-treated cells (Fig 3B). Intriguingly, only the compounds 2 and 4 with longer linker lengths, which were less potent in vitro inhibitors against the CoREST complex, caused an increase in histone acetylation (Fig 3B), with 4 resulting in a 10 fold increase at 10 μM (see supporting info Fig S2).
Figure 3.
A) AMC-fluorescence histone deacetylase inhibition assay with the LSD1-CoREST-HDAC complex in vitro. B) Histone 3 Lysine 56 acetylation (H3K56Ac) levels in E14 mouse embryonic stem cells after 24h; Negative = untreated, CI-994 = 40μM, Panobinostat = 30 nM (For fold change in acetylation levels see supplementary info).
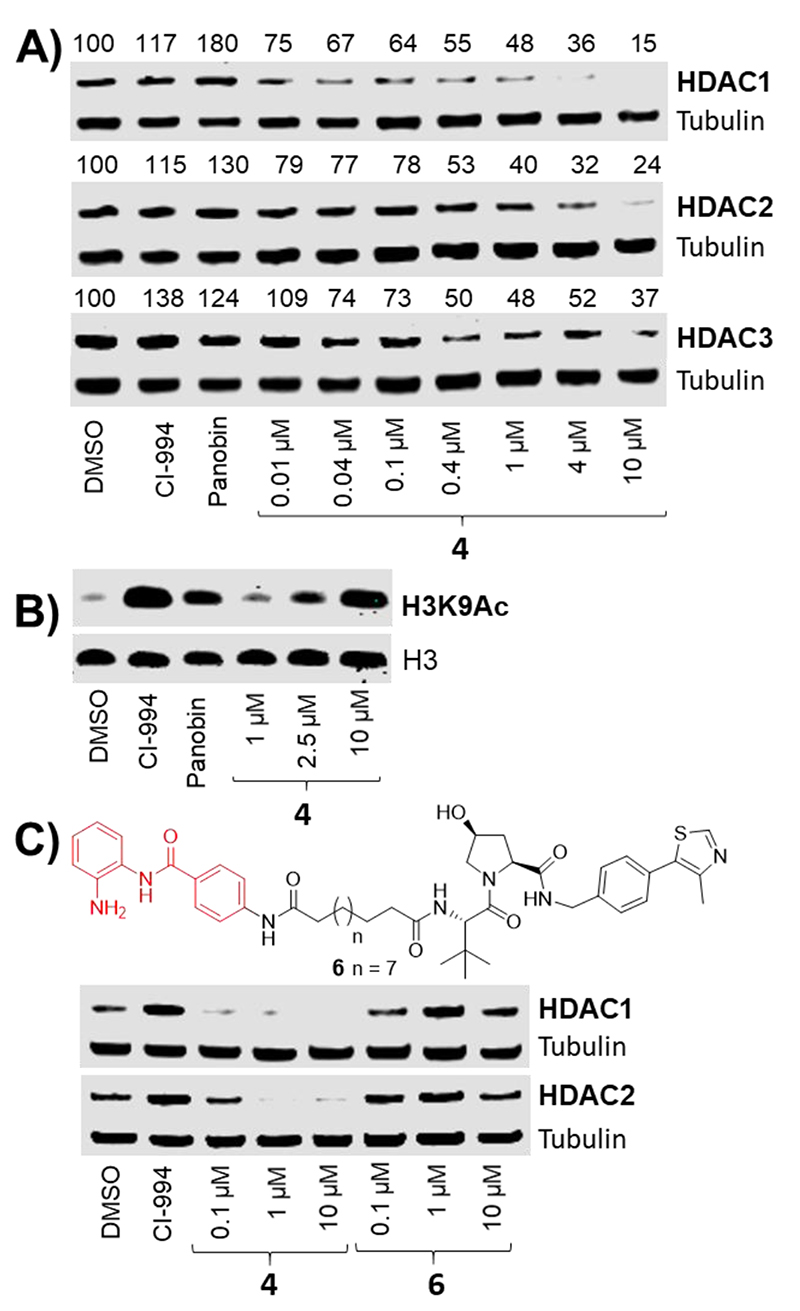
With confirmation that 2 and 4 decrease HDAC activity both in vitro and in cells, we proceeded to quantify HDAC 1, 2 & 3 protein abundance. In ES cells approx. 50% degradation was observed, however, degradation was even more prominent in human colon cancer cell line HCT116. After a 24 hour treatment degradation was observed in a dose-dependent manner with 4 in HCT116 (Fig 4A). PROTAC 4, VHL-based, was a more effective degrader than the cereblon-based PROTAC 2 (see supplementary info Fig S5). HDAC 1 & 2 underwent near complete degradation at 10 μM with 4, while HDAC 3 levels were also decreased, but to a lesser extent. At 1 μM of 4 approximately 50% degradation was observed for HDAC 1, 2 & 3, whilst even at 10 nM of 4 HDAC 1 & 2 levels were still reduced below that of controls. Levels of Histone 3 Lysine 9 acetylation (H3K9Ac), another established residue for HDAC activity,18 were also determined in HCT116 cells after a 24 hour treatment with 4. At 10μM of 4 acetylation levels were highly elevated compared to the DMSO control (Fig 4B), consistent with decreased HDAC 1, 2 & 3 levels at this concentration (Fig 4A).
Figure 4.
HDAC 1, 2 & 3 degradation occurs in a dose dependent manner with 4. A) Western blot showing protein levels of HDAC1, 2 & 3 and α-tubulin in HCT116 cells following 24 hours of the indicated treatments. Numerical values represent percentage of protein compared to DMSO control = 100% B) Histone H3 Lys9 acetylation levels in HCT116 cells following 24 hours of the indicated treatment. C) The inactive diastereoisomer VHL ligand in 6 does not induce degradation. CI-994 = 40μM, Panobinostat = 30 nM
To confirm HDAC degradation was occurring via a VHL mediated proteasome degradation pathway we synthesised 6 with the inactive diastereoisomer VHL ligand (Fig 4C). No degradation of HDAC 1 & 2 was observed with 6 when compared side-by-side with PROTAC 4 confirming degradation is occurring via a VHL-mediated E3-ubiquitin ligase pathway.
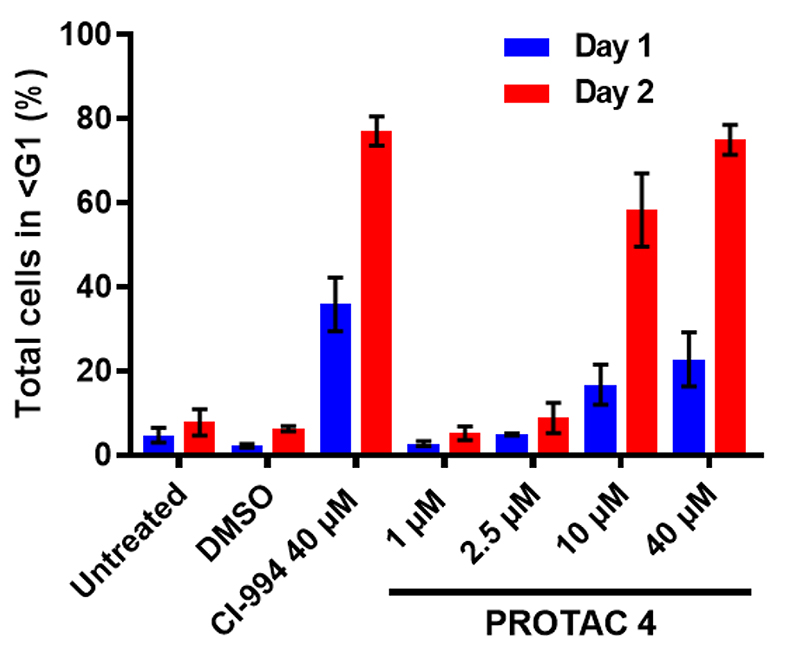
As the most effective of our four PROTAC compounds, we tested the effects of 4 on cell cycle and cell viability of HCT116 cell lines using flow cytometry. After a 48 hour treatment we observed significant cell death at 10μM of 4 (Fig 5). At 40μM cell death after 48 hours was at similar levels to CI-994 (78% vs 73% respectively). Although the effect of 4 on cell viability was comparable to CI-994 it is important to note that the IC50 of 4 against the LSD1-CoREST-HDAC1 complex in vitro was 16.8 μM compared to 0.5 μM for CI-994 (Fig 3A), with near complete HDAC 1, 2 & 3 degradation being observed with 4 in this cell line at 10 μM. Hence, the effects of 4 at 10 μM are most likely due to the absence of HDAC 1, 2 & 3 by a degradation mechanism rather than enzyme inhibition.
Figure 5.
Flow cytometry was used to determine the viability of HCT116 cells following treatment with compound 4 and CI-994 for the indicated times and concentrations. Error bars represent standard deviation of n=3 biological replicates
The PROTAC approach has been applied to a number of protein targets, yet, importantly, not all proteins are as amenable to degradation. This has been observed with PROTACs based on non-selective pan-kinase inhibitors.19 Non-complex forming, cytoplasmic localized HDAC 6 has also recently been identified as a preferential target for degradation with hydroxamic acid based PROTACs.20 In this study, with the synthesis of four heterobifunctional molecules, we have demonstrated the feasibility of targeted degradation of HDAC 1, 2 & 3. These enzymes are localized in the nucleus and present in large multi-protein corepressor complexes. PROTAC 4 produces near complete degradation of HDAC 1, 2 & 3, with an associated increase in global acetylation levels and a loss of cell viability in colon cancer cells. Compounds 1 and 3 despite being a lower molecular weight and more potent HDAC inhibitors in vitro than 2 and 4 against the CoREST complex, failed to alter histone acetylation levels in cells by inhibition or degradation. It is tempting to speculate that the increased hydrophobicity of PROTACs 2 and 4 enhances their cell permeability or nucleus localisation in cells. This may account for the discrepancy in HDAC activities in vitro compared to in cells between 1 & 3 and PROTACs 2 & 4. It seems likely that the linker length and physiochemical properties of Class I HDAC PROTAC linkers will have a profound effect on their activity in cells. Class I HDAC degraders, such as reported in this manuscript, offer an important alternative strategy to inhibition in targeting HDAC corepressor complexes. Such degraders have potential in the development of novel therapeutics in cancer and other diseases related to Class I HDACs.
Supplementary Material
Footnotes
Conflicts of interest
There are no conflicts to declare.
Contributor Information
John W.R. Schwabe, Email: john.schwabe@le.ac.uk.
Shaun M. Cowley, Email: smc57@leicester.ac.uk.
James T. Hodgkinson, Email: JTHodgkinson@le.ac.uk.
References
- 1.Falkenberg KJ, Johnstone RW. Nat Rev Drug Discov. 2014;13:673. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]; Ganesan A, Arimondo PB, Rots MG, Jeronimo C, Berdasco M. Clin Epigenetics. 2019;11:174. doi: 10.1186/s13148-019-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seto E, Yoshida M. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stengel KR, Hiebert SW. Antioxid Redox Signal. 2015;23:51. doi: 10.1089/ars.2014.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon S, Eom GH. Chonnam Med J. 2016;52:1. doi: 10.4068/cmj.2016.52.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millard CJ, Watson PJ, Fairall L, Schwabe JWR. Trends Pharmacol Sci. 2017;38:363. doi: 10.1016/j.tips.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Guha M. Nat Rev Drug Discov. 2015;14:225. doi: 10.1038/nrd4583. [DOI] [PubMed] [Google Scholar]
- 6.Jamaladdin S, Kelly RDW, O’Regan L, Dovey OM, Hodson GE, Millard CJ, Portolano N, Fry AM, Schwabe JWR, Cowley SM. Proc Natl Acad Sci USA. 2014;111:9840. doi: 10.1073/pnas.1321330111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Weichert W. Caner Lett. 2009;280:168. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 7.a) Hayakawa T, Nakayma J. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kelly RD, Cowley SM. Biochem Soc Trans. 2013;41:741. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 8.Pettersson M, Crews CM. Drug Discov Today Technol. 2019;31:15. doi: 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hughes SJ, Ciulli A. Esssays Biochem. 2017;61:505. doi: 10.1042/EBC20170041. [DOI] [PMC free article] [PubMed] [Google Scholar]; Burslem GM, Crews CM. Cell. 2020 doi: 10.1016/j.cell.2019.11.031. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zengerle M, Chan K-H, Ciulli A. ACS Chem Biol. 2015;10:81770. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadd MS, Testa A, Lucas X, Chan KH, Chen W, Lamont DJ, Zengerle M, Ciulli A. Nat Chem Biol. 2017;13:514. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]; Smith BE, Wang SL, Jaime-Figueroa S, Harbin A, Wang J, Hamman BD, Crews CM. Nat Commun. 2019;10:131. doi: 10.1038/s41467-018-08027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckers T, Burkhardt C, Wieland H, Gimmnich P, Ciossek T, Maier T, Sanders K. Int J Cancer. 2007;121:1138. doi: 10.1002/ijc.22751. [DOI] [PubMed] [Google Scholar]
- 12.Lorusso PM, Demchik L, Foster B, Knight J, Bissery MC, Polin LM, Leopold WR, Corbett TH. Invest New Drug. 1996;14:349. doi: 10.1007/BF00180810. [DOI] [PubMed] [Google Scholar]
- 13.Zhang S, Fujita Y, Matsuzaki R, Yamashita T. Cell Death Dis. 2018;9:460. doi: 10.1038/s41419-018-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gräff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, Rei D, Bero AW, Phan TX, Wagner F, Holson E, et al. Cell. 2014;156:261. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Seki M, LaCanna R, Powers JC, Vrakas C, Liu F, Berretta R, Chacko G, Holten J, Jadiya P, Wang T, Arkles JS, et al. J Pharmacol Exp Ther. 2016;358:441. doi: 10.1124/jpet.116.234591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauffer BEL, Mintzer R, Fong R, Mukund S, Tam C, Zilberleyb I, Flicke B, Ritscher A, Fedorowicz G, Vallero R, Ortwine DF, et al. J Biol Chem. 2013;288:26926. doi: 10.1074/jbc.M113.490706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai AC, Toure M, Hellerschmied D, Salami J, Jamie-Figueroa S, Ko E, Hines J, Crews CM. Angew Chem Int Ed. 2016;55:807. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cyrus K, Wehenkel M, Choi EY, Han HJ, Lee H, Swanson H, Kim KB. Mol Biosyst. 2011;7:359. doi: 10.1039/c0mb00074d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y, Dagil I, Fairall L, Robertson N, Wu M, Ragan TJ, Savva C, Saleh A, Morone N, Kunze MBA, Jamieson AG, et al. Cell rep. 2020 doi: 10.1016/j.celrep.2020.01.091. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dovey OM, Foster CT, Cowley SM. Proc Natl Acad Sci USA. 2010;107:8242. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai Q, Du W, Wu J, Wang X, Li X, Qu X, Wu X, Dong F, Yao R, Fan H. Front Mol Neurosci. 2017;10:376. doi: 10.3389/fnmol.2017.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jamie-Figueroa S, Wang J, Hamman BD, Ishchenko A, Crews CM. Cell Chem Biol. 2018;25:78. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An Z, Lv W, Su S, Wu W, Rao Y. Protein Cell. 2019;10:606. doi: 10.1007/s13238-018-0602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu H, Yang K, Zhang Z, Leisten ED, Li Z, Xie H, Liu J, Smith KA, Novakova Z, Barinka C, Tang W. J Med Chem. 2019;62:7042. doi: 10.1021/acs.jmedchem.9b00516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.