Abstract
In oncology, the holy grail of radiotherapy is specific radiation dose deposition in tumours with minimal healthy tissue toxicity. If used appropriately, injectable, systemic radionuclide therapies could meet these criteria, even for treatment of micrometastases and single circulating tumour cells. The clinical use of α and β− particle-emitting molecular radionuclide therapies is rising, however clinical translation of Auger electron-emitting radionuclides is hampered by uncertainty around their exact subcellular localisation, which in turn affects the accuracy of dosimetry. This review aims to discuss and compare the advantages and disadvantages of various subcellular localisation methods available to localise radiopharmaceuticals and radionuclides for in vitro investigations.
Keywords: Radionuclide, Subcellular localisation, Microautoradiography, Ion beam, Laser ablation, X-ray fluorescence microscopy
1. Introduction
Over the past few decades, molecular imaging tracers and molecular radionuclide therapies (MRT) have emerged as novel tools to specifically deliver radionuclides to biological molecular targets in the tumour or disease, enabling precise diagnosis and treatment of several diseases, as well as disseminated cancer treatment [1–3]. Currently, several radiopharmaceuticals are available on the market, including [177Lu]Lu-DOTA-TATE (LUTATHERA®) for the treatment of somatostatin receptor-positive gastroenteropancreatic neuroendocrine tumours, [223Ra]RaCl2 (Xofigo®) for metastatic castration-resistant prostate cancer and [131I]I-MIBG (AZEDRA®) for neuroblastoma, metastatic pheochromocytoma and paraganglioma [4,5]. MRT development is also quickly expanding to include other exciting α and β− particle emitting radiopharmaceuticals, such as [177Lu]Lu-labelled PSMA-617 for prostate cancer and [225Ac]Ac-labelled antimurine CD38 monoclonal antibody for multiple myeloma, in addition to Auger electron (AE) emitters which are currently extensively studied at the in vitro and preclinical levels [1,6,7].
Radionuclides that emit AEs are of particular interest because these electrons, which can result from radionuclide decay by either electron capture and/or internal conversion, are an appealing tool for the treatment of MRT of small metastases and circulating tumour cells. This potential therapeutic approach is enabled through their low-energy (<25 keV), meaning these AEs deposit their energy over a subcellular range (<1 μm), yielding medium to high linear energy transfer (LET) that can cause high radiotoxicity when located close to radiosensitive targets, such as the DNA [1]. Simultaneously, when using AE-emitters, limited non-specific radiotoxicity is found in healthy, non-targeting neighbouring cells [8,9]. Among several therapeutic AE-emitting radiopharmaceuticals, [111In]In-DTPA-octreotide, [111In]In-DTPA-EFG and 125I-labelled murine anti-EGFR-425 have so far been closest to clinical translation for the treatment of metastatic neuroendocrine malignancies, EGFR-positive breast cancer, and glioblastoma multiforme, respectively [1]. Despite promising results, observed adverse effects, toxicity to healthy tissues, and inadequate tumour control in patients have limited their clinical translation [1,10]. In order to revisit previous AE-emitting radiopharmaceuticals for clinical translation, as well as support the successful implementation of novel AE-emitting radiopharmaceuticals such as [123I]I-MAPi [1,9,11], it is vital to be able to accurately determine radiation dose to these tissues delivered by AE-emitting radiopharmaceuticals. This will consequently allow us to better ascertain how the absorbed dose can predict biological responses and treatment outcomes [10,12,13] and to reassess biological risk [14].
Traditionally, the Medical Internal Radiation Dose (MIRD) formalism of the Society of Nuclear Medicine is the elected method to estimate the absorbed radiation dose at a macroscopic level that is then used to correlate with clinical response. MIRD assumes that radionuclides in the tumour and organs are uniformly distributed [15,16]. The assumption of homogeneous tumour and organ uptake of radionuclides is perceived as adequate for radionuclides that emit γ-rays or β-particles. However, this assumption is not suitable for certain radionuclides traditionally used for diagnostic imaging or being considered for AE-mediated MRT. These radionuclides include technetium-99m, iodine-123, iodine-125, thallium-201, indium-111, gallium-67, cupper-64, iodine-124, and selenium-73 [1,8,11,17–22], where the absorbed radiation dose at the microscopic level will depend highly on the non-uniform distribution of radionuclides at multicellular, cellular, and subcellular levels [17,23], and in particular is reliant on accumulation levels within the cytoplasm, membrane and nucleus of cells [1,8].
Currently, the majority of AEs-emitting radiopharmaceuticals considered for therapy either target cell membrane receptors or the cell nucleus, with the latter still considered the ideal target for maximal therapeutic efficacy [1,9,24]. Alternatively, other therapeutic subcellular targets for AEs-emitters have been identified, e.g. the mitochondria; cytoplasmic targeting of AE-emitters has also been found to be therapeutically effective [1,24–26]. New in silico modelling embracing a non-uniformly absorbed dose approach therefore is being carried out to accurately reflect the therapeutic efficacy AEs can have when localised within range of these targets, including how the effect of irregular cell geometry can affect S-values, hence estimated absorbed dose [9]. However, this also requires a detailed knowledge of where exactly in the cell the radionuclide is localised. Although challenging, the accurate subcellular localisation of AEs and their proximity to cellular organelles must be on a scale comparable to the range of their emissions, i.e. nanometres. γ-ray and β+ particle emissions do not provide the required resolution by single photon emission computed tomography (SPECT) or positron emission tomography (PET), respectively, and thus cannot provide the subcellular localisation information needed to estimate the absorbed dose [26,27]. Thus, other methods are required to provide this information at a microscopic and subcellular level.
This review aims to discuss the various subcellular localisation methods available to localise radiopharmaceuticals and radionuclides in vitro, as well as their advantages and disadvantages. Not all methods described below track radioactivity; instead the localisation of the chemical element itself is observed. The appropriateness of these methods for unchelated, soluble radionuclides is also described.
2. Subcellular fractionation
For the past few decades, subcellular fractionation has been the most common technique to determine the subcellular location of radiopharmaceuticals [26,28,29] by exploiting the physiochemical properties of each cellular compartment, such as density, size, shape or surface charge [26,28–30]. This approach has also been key to studying the intracellular structure and function of organelles and proteins (e.g. mitochondria, lysosomes and exosomes) [30].
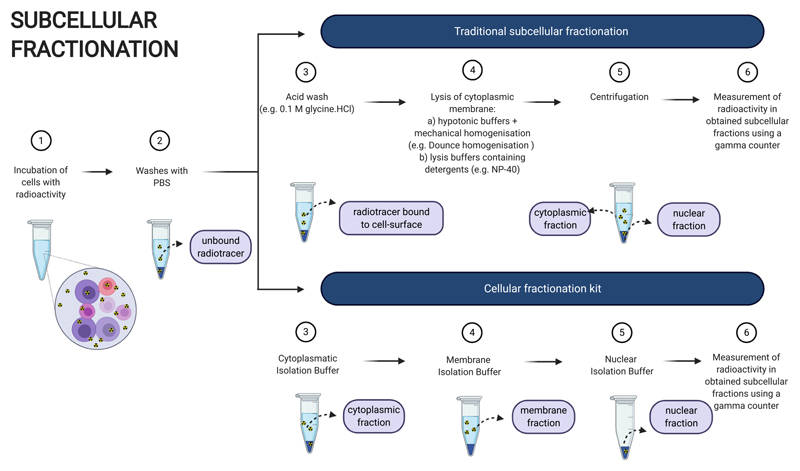
Several methods exist to fractionate cultured cells into nuclear, cytoplasmic and membrane fractions after incubation with a radionuclide or radiopharmaceutical (Fig. 1). Usually, after incubation with a radionuclide or radiopharmaceutical and subsequent washes to remove unbound radioactivity, an acid wash is performed to obtain the percentage of radionuclide or radiopharmaceutical bound to the cell surface (cellular membrane fraction) [31–37]. Then, the cell membrane is disrupted by swelling cells with hypotonic buffers, followed by homogenisation using mechanical methods, such as sonication, Dounce homogenisation and extrusion through a fine needle [36,38–40]. An alternative method to the latter, which takes advantage of the inherent properties and composition of each cellular membrane, is the use of lysis buffers containing mild detergents, such as Triton X-100 and NP-40 [31–34,36]. These nonionic detergents contain uncharged hydrophilic heads that quickly disrupt the cellular membrane, with little damage to the nuclear membrane, if used at optimal incubation times and concentrations [38,41]. Once the cellular membrane is disrupted, the nuclei (nuclear fraction) are separated from other subcellular organelles (cytoplasmic fraction) by low-speed centrifugation [41].
Fig. 1.
Workflow for subcellular fractionation methodologies. Created with BioRender.com.
Alternatively, cellular fractionation kits that contain ready-to-use buffers have also been used to fractionate either cells into cytoplasmic, membrane/organelle and nuclear fractions or to separate nuclear fractions from non-nuclear fractions. These widely available kits are mostly based on differential centrifugation in media of high viscosity, such as sucrose [21,28,30,31] and are designed to fractionate cells in a short period of time (less than 2 h), in an easy and efficient way, usually only requiring a bench top centrifuge [30,42–46].
Regardless of the method through which subcellular components are isolated, the radionuclide content of each subcellular component is subsequently measured with an auxiliary gamma-counter and quantified relative to the total content in the whole cell [1,26,28].
The relative ease, availability and low cost of the cell fractionation techniques mentioned here has led to them being the most ubiquitous in vitro methods for assessing subcellular location of radionuclides and radiopharmaceuticals [28] (Table 1). However, cell fractionation can only ever describe the average intracellular distribution of the radiopharmaceutical in each subcellular fraction for a cell population (Table 1). This thus does not take into account any uptake variation that might be seen between individual cells nor does it inform on the nanometre-range spatial location of AEs in relation to radiosensitive organelles [26,28]. Moreover, the detection of radioactivity in each fraction using a gamma-counter can only be done for γ-ray and β+ particle-emitting radionuclides and, if the method is not optimized and validated by, for instance, western blotting for proteins usually found in each respective cellular component [26,45,47], cross-contamination with other fractions may go undetected.
Table 1. The advantages and disadvantages of the various methods used to determine subcellular localisation of a radionuclide or radiopharmaceutical.
| Characteristics & suitability | Subcellular fractionation | Fluorescence imaging | Micro-autoradiography | Laser ablation – ICP-MS | Ion beam analysis | X-ray fluorescence microscopy |
|---|---|---|---|---|---|---|
| Easy availability | Yes | Yes | No | No | No | No |
| Cheap | Yes | Yes | No | No | No | No |
| Easy sample preparation | Yes | Yes (once dual labelled radiopharmaceutical synthesized) | No | Possibly (depends on sample) | No | No |
| Resolution | N/A | <1 μm | 10–120 μm | >1 μm | 0.2–2 μm | 50 nm |
| Sensitivity | N/A | Depends on fluorophore and microscope | Depends on radionuclide and sample preparation | ppt; pg/mL | ppm; pg/mL | ppb; ng/mL |
| 2D or 3D | N/A | 2D and 3D | 2D and 3D | 2D and 3D (if combinedwith other methods) | 3D | 2D and 3D |
| Unchelated radionuclide | No | No | Possibly | Yes (cold equivalent) | Yes (cold equivalent) | Yes (cold equivalent) |
| Chelated | radiopharmaceutical | Yes | Yes | Yes | Yes (cold equivalent) | |
| Yes (cold equivalent) | Yes (cold equivalent) |
3. Fluorescence imaging
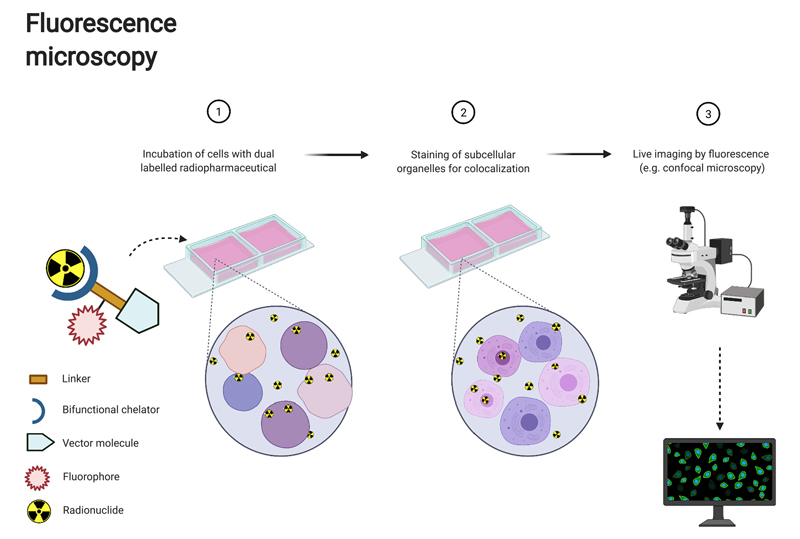
Since 1948, fluorescent dyes, typically in the visible or near-infrared range, have been used during surgery to visualize tumours and verify tumour margins [48,49]. More recently, the conjugation of radiopharmaceuticals with a fluorescent tag has opened up the possibility for multimodal imaging, bringing together the advantages of both techniques [49] (Fig. 2). In the clinic, the radioactive element of the dual labelled compound enables whole body, non-invasive preoperative detection and staging of disease by the highly sensitive modalities of SPECT and PET with subsequent intraoperative localisation of high penetration depth in tissue γ-rays by using a gamma probe. This thus overcomes the major pitfall of fluorescence imaging in general, whose light possesses a limited penetration depth in the tissue of only a few millimetres, thereby restricting detection to superficial tumours [50,51]. Simultaneously, the fluorescent element of the compound enables high cellular spatial resolution during surgery facilitating realtime tumour delineation and improving radical tumour resection accuracy [50–55]. Thus, by assuring complete removal of malignant tissues, this multimodal-imaging could improve surgical outcomes for patients and decrease rates of cancer recurrence [51,52]. Several studies have shown the feasibility of this dual-imaging approach for multimodality image-guided surgery using [111In]In-DOTA-girentuximab-IRDye800CW for multi-guided surgery of clear cell renal cell carcinoma patients, [111In]In-DTPAtrastuzumab-IRDye800CW for HER2-expressing breast cancer, [111In]In-DTPA-D2B-IRDye700DX for PSMA-expressing tumours, [68Ga]Ga-MMC(IR800)-TOC for neuroendocrine tumours and [18F]PARPi-FL for glioblastoma [49,51–53,56]. Similarly, an iodine-124/fluorescein-based dual-modality labelling reagent has enabled a one-step solution to attach a PET radionuclide and a fluo-rescent reporter to any cancer-specific antibody [57].
Fig. 2.
Workflow for fluorescence microscopy where the radiopharmaceutical is directly linked to a fluorescent dye for dual modality imaging. Created with BioRender.com
While the use of this multimodal approach has been explored for in vivo preclinical and clinical applications, only a few studies explore its capacity to inform on the in vitro subcellular localisation of a radiopharmaceutical. And yet, the co-localisation of a fluorescent label with a radionuclide attached to the same tumour targeting antibody or peptide combined with the ability to fluorescently co-stain a cell population for the nucleus with the cell-permeable dye Hoechst 33342, offers an opportunity to study subcellular location (<1 μm) of a radiopharmaceutical in real time in live cells [49,58,59] Fig. 2). For example, it has been shown that the combination of a fluorochrome and radionuclide can be done “low-print”, e.g. without the addition of a linker, by successfully radiolabelling an azide-modified BODIPY-Fl dye with fluorine-18 using an [18F]F/[19F]F exchange reaction of the boron-fluoride core of the BODIPY dye [59]. Using confocal microscopy imaging, this study demonstrated specificity of this compound for cell surface binding [59]. Studies have also been carried out with [111In]In-NLS-trastuzumab where the subcellular localisation of the antibody was then determined by using an AlexaFluor-564-conjugated anti-human IgG secondary antibody and nuclear staining with DAPI [35].
In any approach to determine subcellular localisation of radionuclides using fluorescence, it is key that either the tumour-targeting compound itself is targeted by a secondary fluorescent antibody for staining or that the radiopharmaceutical itself is conjugated with a fluorescent dye. Uncoupling the radionuclide from the fluorescent dye impacts the conclusions that can be made regarding radionuclide localisation from any such studies [47]. One such example is [125I]I-DCIBz, a highly specific small-molecule AE-emitter targeting PSMA, of which the in vitro subcellular localisation within PSMA-positive PC3 PIP tumour cells was inferred from fluorescently-labelled YC-36 that has the same PSMA-binding urea scaffold as the radiolabelled DCIBzL [60]. The use of two different molecules weakens any conclusion made regarding subcellular localisation of the radionuclide.
Fluorescent imaging of either dual labelled radiopharmaceuticals or through immunofluorescent staining of the tumour-targeting compound itself enables subcellular localisation of radionuclides in both 2D and 3D (through performing Z-stacks of the cells) (Table 1). However, fluorescence imaging does not provide the quantitative data required for an accurate estimation of the absorbed radiation dose of radiopharmaceuticals at the microscopic level. To overcome this, previous studies have used this technique in combination with other approaches, such as subcellular fractionation, to quantify the subcellular distribution of radionuclides [37,46,61]. Also, special attention needs to be taken when the subcellular localisation of a radiopharmaceutical is based on fluorescence images, as these images may not be as representative and accurate as described above due to well-known artefacts. For instance, not only can environmental factors such as pH, temperature and solvent polarity affect the fluorescence spectrum or quantum yield, but also the short-range deposition of energy by AEs may result in radiolysis of the fluorescent tag from the radiopharmaceutical [47]. Moreover, although fluorescence microscopy is not an expensive technique itself, the conjugation of a fluorescent tag may not always be favourable when creating a radiopharmaceutical, as it increases synthesis complexity, can be lengthy and costly, and, most importantly, could impact on the in vivo biodistribution and target binding of the radiopharmaceutical [47,59] (Table 1).
4. Microautoradiography
Microautoradiography (MAR) has been used to determine the biological and spatial distribution of drugs at the whole-body, tissue and cellular levels [26,62]. Commonly, the compounds of interest are labelled with low-energy β− emitters carbon-14 and tritium [26,62]. First developed in 1924 by Lacassagne and Lattes, MAR was further refined by Appleton et al. [63] (cryo-fixation and -sectioning to prevent redistribution of diffusible species) and Stumpf and Roth (optimized for receptor-bound species), with the latter two approaches remaining the basis for many studies today [13,64–67]. Despite its long history, the use and prevalence of MAR is not as widespread as cell fractionation and in the last decade, only approximately 18% of all research publications using MAR were applied to nuclear medicine. Instead, MAR has mostly been applied in the fields of microbiology (approximately 49% of publications), with the remaining publications focused on pharmacology and water science. Within nuclear medicine, the vast majority of articles were found to describe ex vivo MAR work, while only three investigations employed MAR with established in vitro cell line samples [20,45,68].
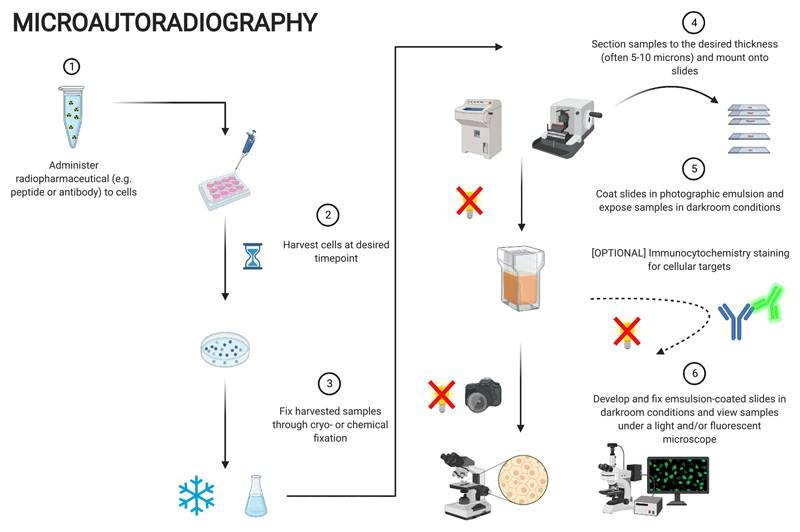
Although there is no unified method, a typical MAR workflow largely follows the same framework, as depicted in Fig. 3. First, the radiopharmaceutical is administered to the sample e.g., monolayer/suspension cell culture, for the desired period of time. The sample is subsequently fixed through cryo- or chemical fixation and sectioned to a thickness of 5–10 μm before being mounted onto microscope slides and placed into contact with photographic emulsion under darkroom conditions (either pre-coated onto microscope slides prior to section mounting or performed post-mounting and coated by slide dipping) for extended periods of time. Slides are then developed and fixed in a fashion similar to photography film procedures and finally imaged under a microscope (light and fluorescence) to visualize the silver grains, indicative of the location of radiolabelled drug species and their co-localisation with cellular structures (if relevant immunocytochemistry was performed) [26,28,62,66,67].
Fig. 3.
Workflow for microautoradiography. Created with BioRender.com
Only a few papers have used MAR to investigate cellular localisation of radionuclides, the most recent example being our study on the therapeutic potential of the AEs emitter gallium-67 for targeted radioimmunotherapy ([67Ga]Ga-trastuzumab) in breast cancer cells [20]. In brief, we exposed cells in suspension to [67Ga]Ga-trastuzumab before subsequent paraffin-embedding, snap-freezing and cryo-sectioning for MAR processing [20]. We noted a diverse range of cell-associated silver grain numbers, on a single-cell basis, despite “a clonally identical population”. This provided evidence for heterogeneous radiopharmaceutical uptake, despite “clonal identity” of the cells. A drawback of this study is the lack of determination of specific subcellular localisation, i.e. the resolution is poor. Also, MAR is usually carried out using cells in suspension. However, utilising a suspension assay with a nominally adherent cell line begs the following question: do these cells in suspension demonstrate normal behaviour when adjacent cell-to-cell contact and communication, usually formed in adherent 2D monolayers, is lacking? Earlier work by Davis et al. [68] showcased a variation of the technique that preserved the 2D-monolayer environment of adherent cultures. Using an in vitro osteoblast cell model to mimic bone mineralisation, the authors reported almost indistinguishable cell uptake behaviours between calcium-45 and strontium-89; the latter is a common calcium mimetic used in palliative cancer treatment [68]. In particular, silver grain localisation, indicative of [89Sr]SrCl2 presence, was mostly observed in regions devoid of cells, which the authors noted as matching with calcium-rich, extracellular mineralisation deposits [68]. The specific, practical elements of their approach could be easily adopted and inserted into investigations as it foregoes sectioning, a technically limiting process and potential source of radioactive loss.
However, a major drawback of both approaches is that the 2D in vitro nature that does not mimic the 3D in vivo environment of tumours. In response to this, Falzone et al. [45] recently showcased the application of MAR with spheroids, 3D cultures of cells for potentiating the mimicry of in vivo tumour geometries in vitro. Utilising a similar protocol to Othman et al. [45], an advantage of this study over others is the generation of similar cellular geometries observed with tissue sectioning and thus allowing for more translatable interpretation of results while continuing basic research.
From these three studies, it is clear that the application of MAR within basic research is possible for answering questions from a range of scenarios. A key aspect of MAR is its potential to visualize the colocalisation of radiopharmaceuticals, or at least their radioactive emissions, with markers of subcellular components through immunohisto-/immunoctyochemical staining [62,66,67]. However, the disadvantages and difficulties associated early on with MAR still permeate to this day owing to a lack of development and improvement since the Appleton and Stumpf era; these have been reiterated in previous reviews many times [26,28,66,67]. As such, these disadvantages will only be touched upon briefly (Table 1).
Firstly, MAR is time consuming, not only in terms of sample preparation, but also because the exposure time of the emulsion to the radioactive samples lasts anywhere between days to weeks and months, depending on the level of radioactivity present and the half-life of the radionuclide. MAR also requires specialised equipment or rooms such as a cryostat and a dark room, the photographic emulsion can be hard to source and tends to be rather expensive, and MAR is, overall, very technically challenging. Finally, resolution, e.g. the distance of silver grains from the actual site of radioactive decay, is highly dependent on and easily influenced by sample preparation, tissue section, emulsion thickness and physical decay properties of the radionuclide being investigated. Ideally, sections and emulsions of <5 μm thickness are used for proximal generation of silver grains at the site of decay itself, especially for AE-emitters where radiation track lengths are particularly short. However, such thin dimensions often result in loss of structural integrity of the sections and more often than not are impossible to obtain with emulsions, especially when performing slide dipping procedures.
5. Laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS)
LA-ICP-MS is a relatively new technique used since the 1980s [69] for the localization of elemental isotopes in biological materials, allowing qualitative and quantitative analysis [70–72]. Apart from the growing interest in biological applications, it has also been used for a chemical analysis of solid samples in a variety of other scientific disciplines such as geology [73], material science [74], and forensics [75].
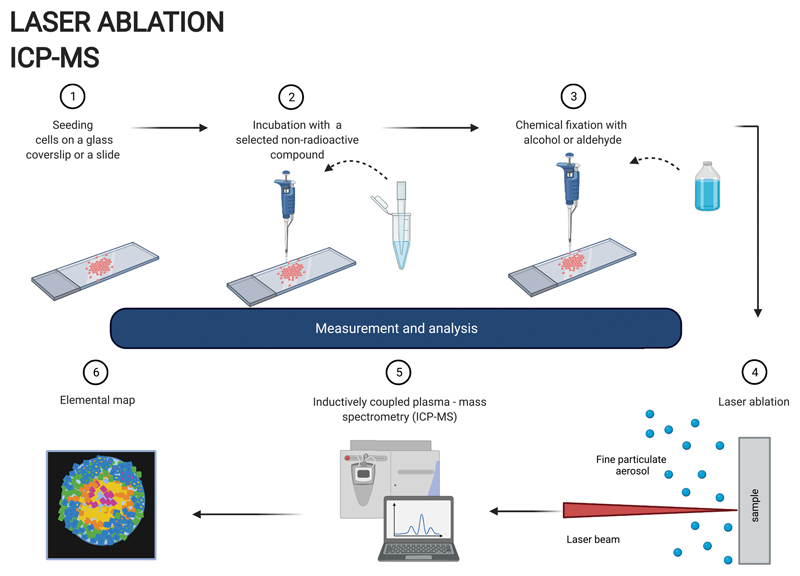
In LA-ICP-MS, samples are ablated by a laser beam (commonly 193 nm or 213 nm) with an adjustable spot size down to 1 μm. The ablated material is directed with a helium flow to a mass spectrometer, where it is atomised and ionised by the inductively coupled plasma and the ions are separated and detected by their mass to charge ratio (m/z) (Fig. 4). LA-ICP-MS allows non-radioactive, multielement mapping providing isotopic information with very high sensitivity. Although this technique has been mostly applied for imaging elemental distribution in tissue sections [72,76], it could also be considered for the purpose of studying localization of radiopharmaceuticals or radionuclides at a single cell level based on the position of the element of interest [77–79].
Fig. 4.
Workflow for LA-ICP-MS. Created with BioRender.com
The high sensitivity (ppt or ng/L) of this technique allows the detection of very low concentrations of an element or a particular isotope thereof, which should more accurately express the real localization of the radionuclide studied, compared to other non-radioactive analytical techniques with higher sensitivity limits. The sample preparation is relatively easy and does not require any additional time-consuming procedures (Table 1). While sectioning of biological tissues is widely described in the literature, e.g. using chemical fixation and paraffin embedding or cryosectioning [73,79], fixation techniques for single cell LA-ICP-MS analysis are much less explored and mostly rely on chemical fixation with alcohol-based solvents, such as ethanol or aldehydes [77,78]. LA-ICP-MS spatial resolution, which is typically between one to a few micrometres, could limit its use in single cells, especially if the cell diameter is less than 20 μm. Improving the spatial resolution to a sub-micron scale, which has recently become possible [80], would greatly enhance the potential of the LA-ICP-MS in the single cell elemental mapping. The depth resolution can be another limitation. As the laser beam penetrates through an entire cell volume or a wider cell section (destructive method), the signal detected is presented as a 2D image and therefore distinguishing from which cell compartment the signal is coming from is difficult. Using additional three dimensional image reconstruction [81] or super-resolution reconstruction [82] techniques can help to overcome these limitations, however at the same time this can make LA-ICP-MS more time consuming, laborious and costly. Some additional information regarding subcellular localization of metals can be achieved by staining cells with an Ir-DNA-intercalator, which binds nuclear DNA and overlaying obtained elemental distribution maps [77,78,81]. Although LA-ICP-MS can be done quantitatively, it can be difficult in some cases to reliably quantify an individual metal concentration in a sample. This is due to the inhomogeneity of the biological samples and the lack of certified matrix matching laboratory standards [76].
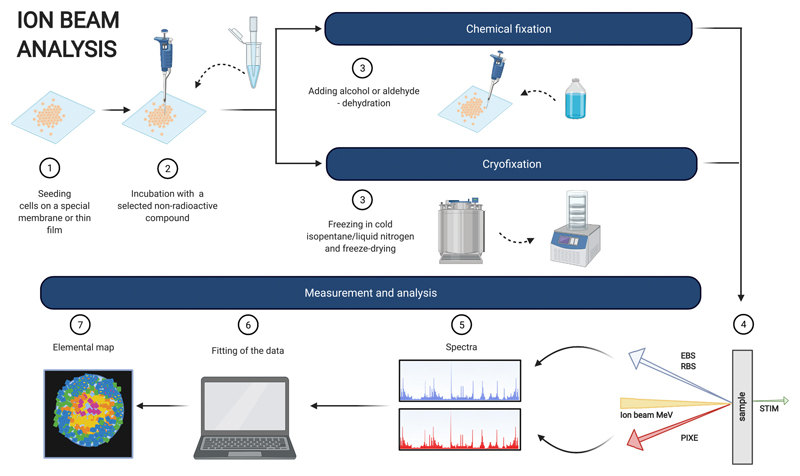
6. Ion beam analysis
Ion Beam Analysis (IBA) is a collective name for a variety of methods using energetic MeV ion beams (protons, α particles or other cations) to probe thin film samples. Although IBA has been applied in analytical chemistry for over half a century, in the last two decades it has gained more popularity among researchers and become a powerful tool used in many different disciplines, such as material science, forensics or biological samples (such as proteins) analysis [83–85].
In order to ascertain the 3D localisation or depth profile of a radionuclide or a stable isotope in a sample, combined IBA methods can be utilised. IBA can be broadly divided into atomic excitation methods (e.g Particle Induced X-ray Emission - PIXE), nuclear excitation methods (e.g Rutherford Back Scattering - RBS, Elastic Back Scattering - EBS, Elastic Recoil Detection - ERD, nuclear reaction analysis - NRA) and Scanning Transmission Ion Microscopy (STIM). After a collision of the incident ions with the target material, IBA is able to detect, depending on the chosen method, back-scattered incident ions (RBS), X-rays resulting from inelastic collisions of the incident ions with the inner shell electrons (PIXE), γ-rays (NRA), forward recoiled particles (ERDA) or energy loss of the primary ions passing through a sample (STIM). However, it is the synergetic approach and ability to combine different, complementary IBA techniques, most commonly PIXE and RBS (called sometimes ‘total IBA’) [86], which enables a comprehensive sample analysis by defining its elemental composition and structural details, as well as depth profiling with good lateral (500 nm) and depth (2 nm) resolution [87].
Recently, the popularity of IBA techniques has grown in biological applications, due to its ability to localise and quantify trace elements in cells at a subcellular level in air or in vacuum. For example, the PIXE methodology has been employed to study platinum accumulation and subcellular localisation in ovarian [88] and lung cancer cells [89] to test the mechanisms of platinum-based anticancer treatment. Although the PIXE method alone does not allow depth profiling, the nuclear localisation of platinum can be confirmed by its co-localisation with bromodeoxyuridine-marked cell nuclei [89]. The distribution of gallium and endogenous trace elements (Mn, Fe, Zn, Cu) has also been analysed in ovarian cancer cells exposed to gallium nitrate by PIXE and RBS methods in order to explain the mechanism behind its antitumor activity and possible interactions with other metals [90]. By combining three different IBA methods, such as PIXE, RBS and STIM, the cellular distribution of iron, suspected to play a crucial role in various neurodegenerative diseases, such as Parkinson’s or Alzheimer’s disease, was identified and quantified by analysing cultured dopaminergic pheochromocytoma cells exposed to an excess of iron. The obtained results suggested that the accumulation of additional iron might influence dopamine storage and release in the distal part of the dopamine-producing cells. [91]. All these studies confirm feasibility and usefulness of IBA methods for the purpose of trace metal analysis, and for the radionuclides’ subcellular localisation purposes, a non-radioactive equivalent could be used.
In terms of sample preparation (Fig. 5), cells are grown on a special membrane, foil or film and fixed with 2% glutaraldehyde, dehydrated with increasing concentrations of ethanol and dried [88] or cryofixed in cold isopentane at liquid nitrogen temperature and freeze-dried [89–91]. Cryofixation, however, is the preferred method as it not only preserves the structure of the cell but also avoids introducing any unwanted chemical changes to the sample and altering elemental distribution [91]. The latter is of the utmost importance, especially for easily diffusible elements.
Fig. 5.
Workflow for ion beam analysis. Created with BioRender.com
IBA requires the use of advanced equipment with high running costs and sophisticated software for data interpretation (Table 1). The detection limit is often described in ppm concentrations (ppm, μg/mL) [87], higher when compared to other techniques, for example, LA-ICP-MS analysis (ppt or pg/mL). However, it offers high quality, multi-elemental analysis with possible quantification and structural information available when multiple methods are combined. It has been also confirmed that the traceable accuracy for RBS is a high as 1% [92] and the method can be regarded as a primary reference method [93] and can provide a model free analysis [86]. Good spatial resolution (0.2–2 μm) [89] allows single-cell analysis, while additional depth profiling might help to localise the trace metal within the cell and eliminates problems described earlier for the LA-ICP-MS technique. Moreover, the total IBA is considered as a matrix effect free method and could be used for producing certified standards for other methods, such as LA-ICP-MS, XRF and SIMS (Secondary Ion Mass Spectroscopy) [94,95].
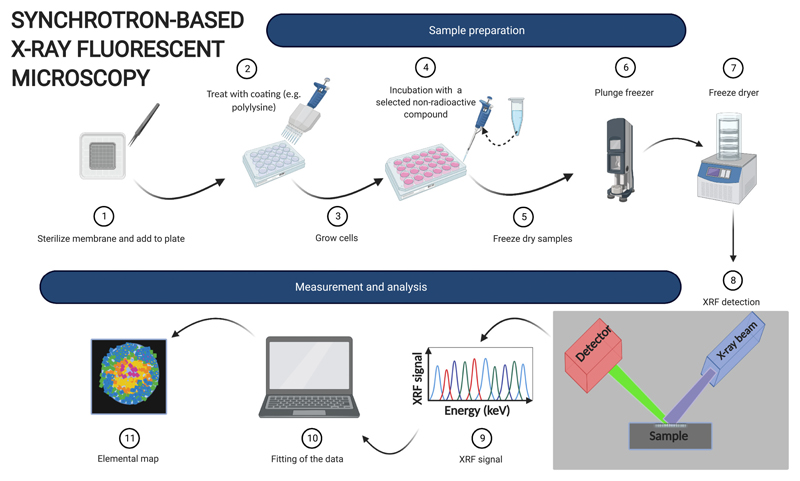
7. Synchrotron-based X-ray fluorescence microscopy
X-ray fluorescence (XRF) microscopy has been used since the late 70s to probe the elemental composition of a variety of materials [96]. However, only in the last two decades has this technique been applied to map elemental composition in biological systems.
XRF is based on the ability of chemical elements to emit X-ray photons when hit by a high-energy X-ray beam. In fact, if the incident photon has enough high energy to cause the emission of an inner-shell electron, an electron from a higher energy orbital will occupy the newly formed hole, resulting in the emission of an X-ray photon, i.e. XRF. The energy of the emitted photon matches that of the electronic transition and is therefore different for different elements. This allows for the simultaneous detection and quantification of multiple elements to produce 2D elemental maps (when the X-rays are used to raster a sample surface) or even 3D tomograms (when cryo-XRF tomography is used) of entire cells or tissue samples [97].
Although XRF has not been applied specifically to mapping of radiopharmaceuticals and radionuclides, studies on “cold” compounds of nuclear medicine-relevant elements such as gallium complexes and iodinated molecules exist demonstrating the feasibility of the approach [98–100].
In principle, different types of X-ray sources can be used for XRF. However, the high flux and brightness of synchrotron radiation allows images to be obtained with excellent resolution (to a subcellular level, 50 nm) and sensitivity (down to ppb, or 10−18 g) [101–103]. In particular, the high sensitivity is extremely well suited for the tracer concentrations generally used for radionuclide imaging [104].
Cells are cultured on Si3N4 membranes or TEM grids (depending on the element of interest for XRF), which are transparent to X-rays. These membrane are often coated (e.g. with poly-l-lysine) prior to cell seeding to promote cell adhesion [105] (Fig. 6). After treatment with the relevant metal, samples are thoroughly washed with and appropriate buffer (e.g. phosphate buffer saline or Hank’s balanced salt solution), then plunge-frozen in liquid ethane and freeze-dried (if cryo-analysis is not available). Sample preparation is key for the success of XRF experiments and special care must be taken in handling the fragile silicon nitride membranes.
Fig. 6.
Workflow for X-ray fluorescence. Created with BioRender.com
Compared to other techniques, synchrotron-based XRF provides superior resolution and sensitivity, which are comparable to those obtained by LA-ICP-MS [106]. However, access to synchrotron nanofocused XRF is still limited, although the number of synchrotron facilities offering this technique is constantly increasing.
It must be noted that, on its own, XRF does not provide detail on cellular structure, although maps of specific elements can provide a rough location of specific organelles (e.g. zinc for cell nucleus) [107]. Combination with techniques such as hard X-ray ptychography (a lens-less method where a sample is scanned with coherent illumination and diffraction patterns are collected) or soft X-ray tomography (based on contrast between low-energy X-ray absorption in water rich vs carbon rich parts of the cell) provide organelle-level detail, helping identification of a specific element’s subcellular location [108]. While combination of XRF with these techniques is becoming more common in recent years, availability at synchrotron facilities is still limited [109,110].
XRF analysis can also be easily combined with X-ray absorption spectroscopy (XAS) and related techniques XANES (X-ray Absorption Near Edge Structure) and Extended X-ray Absorption Fine Structure (EXAFS), to gain information on the oxidation state and coordination environment of a metal. However, this analysis is time consuming and requires higher amount of the investigated compound compared to XRF, thus limiting its potential for trace concentrations.
8. Localisation of unchelated radionuclides
The methods described above can all be used for radionuclides and stable isotopes chelated to compounds targeting particular receptors or proteins overexpressed on cancer cells e.g., for radiopharmaceuticals (Table 1). However, localisation techniques usually used for subcellular localisation studies are not always suitable to accurately determine the intracellular location of non-reactive ionic radionuclides and radiopharmaceuticals that can freely diffuse within the cell, such as [99mTc]TcO4 −, [201Tl]Tl+, [123I]I− and [18F]BF4−. As these radionuclides are not ‘trapped’ within the cell or linked to intercellular proteins, any subcellular location information gained by such methods could be misleading. For instance, subcellular fractionation is disruptive, and hence, the majority of unchelated soluble radionuclide or radiopharmaceutical content will be inherently found in the cytoplasmic fraction [26]. Moreover, when preparing samples for LA-ICP-MS, the commonly used formalin fixation method will not stop radionuclides diffusing throughout the cell. Therefore, sample preparation proves critical, as any additional washes during sample preparation can cause dislocation of the investigated element leading to a false image acquisition. It has been shown before that sample preparation technique can alter metal concentrations and distribution in tissue sections [111] and this effect might be even more prominent on a single cell level.
The MAR method refined by Appleton et al. [63] was developed with the mindset of preventing loss and/or relocation of soluble/diffusible substances such as unchelated radionuclides/radiopharmaceutical fall under [66]. Through the use of cryo-fixation and –sectioning, MAR avoids commonly-used liquid solutions (e.g. paraformaldehyde), thus preserving the spatial localisation of loosely-bound radionuclides/radio-pharmaceuticals is possible [63]. However, as described above, use of MAR for subcellular localisation of tracers is barely applied, due to the lengthy and complicated procedure.
In future, the use of ratiometric fluorescent sensors activated by metal binding might prove a useful technique to study subcellular localisation of endogenous metal ions (most prominently labile pools of Cu+ and Fe2+ as well as Ca2+ and Zn2+) [104]. In principle, these sensors could be used to investigate localisation of unchelated radioactive metals such as [62/63Zn]Zn2+ and [62/64Cu]Cu+ or high concentrations of their stable isotopes. In addition, new advancements in the development of Fe3+ sensors could provide the basis to measure subcellular localisation of unchelated [67/68Ga]Ga3+, owing to the similarity between the two ions [112].
9. Conclusion
Microdosimetry requires accurate subcellular localisation information, especially for radionuclides that emit AEs and are being considered for use in MRT. As highlighted in this review, there is no one method that brings together all the required or desired elements of a successful technique, e.g. that excels on all fronts of easy availability and sample preparation, low cost, resolution and/or sensitivity and at a minimum 2D, but preferably 3D, information. As such, the majority of the work carried out in in vitro nuclear medicine studies has utilised the cell fractionation method, simply because it is fast, easy and low cost. Although a dual labelled radiopharmaceutical could become the ideal method for accurate subcellular localisation, the addition of a fluorophore to a radiopharmaceutical risks decreasing target binding affinity and as such, it is not often explored. Equally, MAR would be a very powerful tool, if only the process of sample preparation and the workflow itself was simpler and less labour- and time-intensive. This is an opportune time to consider methods traditionally explored for chemistry purposes, such as LA-ICP-MS, IBA or XRF, which currently have not yet been regularly applied to nuclear medicine studies. These techniques could be exceptionally powerful in determining the localisation of not only radiopharmaceuticals, but, in some way more importantly, of unchelated radionuclides. Going forward, it is likely that a mix of methodologies is needed to accurately determine subcellular localisation, thus requiring a new interdisciplinary approach utilising different skills of radiobiologists, radiochemists and chemists to further enable accurate dosimetry in collaboration with medical physicists.
Acknowledgment
I.M.C. would like to acknowledge funding from the EPSRC Centre for Doctoral Training in Medical Imaging [EP/L015226/1] and National Physical Laboratory. J.C. and K.O. would like to acknowledge funding from the MRC Doctoral Training Partnership in Biomedical Sciences [MR/N013700/1]. K.O. was also supported by Theragnostics Limited. C.I. was supported by the Wellcome Trust [209173/Z/17/Z]. This work was also supported by the Wellcome/EPSRC Centre for Medical Engineering at King’s College London [WT 203148/Z/16/Z] and by EPSRC Programme Grant [EP/S032789/1].
Finally, authors would like to thank Dr. Melanie Bailey and Dr. Catia Costa from the National Ion Beam Centre [EPSRC funding -NS/A000059/1], Dr. Elizabeth M. Bolitho (University of Warwick), Prof Philip Blower (King’s College London), and Dr. Theodora Stewart from the London Metallomics Facility for their help and support while working on this review.
References
- [1].Ku A, Facca VJ, Cai Z, Reilly RM. Auger electrons for cancer therapy – a review. EJNMMI Radiopharm Chem. 2019;4:27. doi: 10.1186/s41181-019-0075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gudkov S, Shilyagina N, Vodeneev V, Zvyagin A. Targeted radionuclide therapy of human tumors. Int J Mol Sci. 2015;17:33. doi: 10.3390/ijms17010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jadvar H. Targeted radionuclide therapy: an evolution toward precision cancer treatment. Am J Roentgenol. 2017;209:277–88. doi: 10.2214/AJR.17.18264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Malcolm J, Falzone N, Lee B, Vallis K. Targeted radionuclide therapy: new advances for improvement of patient management and response. Cancers (Basel) 2019;11:268. doi: 10.3390/cancers11020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kayano D, Kinuya S. Current consensus on I-131 MIBG therapy. Nucl Med Mol Imaging. 2018;52(2010):254–65. doi: 10.1007/s13139-018-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Agrawal S. The role of 225Ac-PSMA-617 in chemotherapy-naive patients with advanced prostate cancer: is it the new beginning. Indian J Urol. 2020;36:69–70. doi: 10.4103/iju.IJU_266_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sgouros G, Bodei L, McDevitt MR, Nedrow JR. Radiopharmaceutical therapy in cancer: clinical advances and challenges. Nat Rev Drug Discov. 2020;19 doi: 10.1038/s41573-020-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Faraggi M, Gardin I, Stievenart J-L, Bok BD, Le Guludec D. Comparison of cellular and conventional dosimetry in assessing self-dose and cross-dose delivered to the cell nucleus by electron emissions of 99mTc, 123I,111In, 67Ga and 201Tl. Eur J Nucl Med Mol Imaging. 1998;25:205–14. doi: 10.1007/s002590050218. [DOI] [PubMed] [Google Scholar]
- [9].Falzone N, Fernandez-Varea JM, Flux G, Vallis KA. Monte Carlo evaluation of Auger Electron-emitting theranostic radionuclides. J Nucl Med. 2015;56:1441–6. doi: 10.2967/jnumed.114.153502. [DOI] [PubMed] [Google Scholar]
- [10].Terry SYA, Nonnekens J, Aerts A, Baatout S, de Jong M, Cornelissen B, et al. Call to arms: need for radiobiology in molecular radionuclide therapy. Eur J Nucl Med Mol Imaging. 2019;46:1588–90. doi: 10.1007/s00259-019-04334-3. [DOI] [PubMed] [Google Scholar]
- [11].Pirovano G, Jannetti SA, Carter LM, Sadique A, Kossatz S, Guru N, et al. Targeted brain tumor radiotherapy using an Auger emitter. Clin Cancer Res. 2020;26:2871–81. doi: 10.1158/1078-0432.CCR-19-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sgouros G, Goldenberg DM. Radiopharmaceutical therapy in the era of precision medicine. Eur J Cancer. 2014;50:2360–3. doi: 10.1016/j.ejca.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stumpf WE. Drug localization and targeting with receptor microscopic autoradiography. J Pharmacol Toxicol Methods. 2005;51:25–40. doi: 10.1016/j.vascn.2004.09.001. [DOI] [PubMed] [Google Scholar]
- [14].Humm JL, Howell RW, Rao DV. Dosimetry of auger-electron-emitting radionuclides: report no. 3 of AAPM nuclear medicine task group no. 6. Med Phys. 1994;21:1901–15. doi: 10.1118/1.597227. [DOI] [PubMed] [Google Scholar]
- [15].Zanzonico P. Cell-level dosimetry and biologic response modeling of heterogeneously distributed radionuclides: a step forward. J Nucl Med. 2011;52:845–7. doi: 10.2967/jnumed.111.087841. [DOI] [PubMed] [Google Scholar]
- [16].Bolch WE, Eckerman KF, Sgouros G, Thomas SR. MIRD pamphlet no. 21 : a generalized schema for radiopharmaceutical dosimetry–standardization of nomenclature. J Nucl Med. 2009;50:477–84. doi: 10.2967/jnumed.108.056036. [DOI] [PubMed] [Google Scholar]
- [17].Vaziri B, Wu H, Dhawan AP, Du P, Howell RW. MIRD pamphlet no. 25: MIRDcell V2.0 software tool for dosimetric analysis of biologic response of multicellular populations. J Nucl Med. 2014;55:1557–64. doi: 10.2967/jnumed.113.131037. [DOI] [PubMed] [Google Scholar]
- [18].Stepanek J, Larsson B, Weinreich R. Auger-electron spectra of radionuclides for therapy and diagnostics. Acta Oncol (Madr) 1996;35:863–8. doi: 10.3109/02841869609104038. [DOI] [PubMed] [Google Scholar]
- [19].McMillan DD, Maeda J, Bell JJ, Genet MD, Phoonswadi G, Mann KA, et al. Validation of 64Cu-ATSM damaging DNA via high-LET Auger electron emission. J Radiat Res. 2015;56:784–91. doi: 10.1093/jrr/rrv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].bin Othman MF, Verger E, Costa I, Tanapirakgul M, Cooper MS, Imberti C, et al. In vitro cytotoxicity of Auger electron-emitting [67Ga]Ga-trastuzumab. Nucl Med Biol. 2020;80–81:57–64. doi: 10.1016/j.nucmedbio.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].van Othman MF, Mitry NR, Lewington VJ, Blower PJ, Terry SYA. Re-assessing gallium-67 as a therapeutic radionuclide. Nucl Med Biol. 2017;46:12–8. doi: 10.1016/j.nucmedbio.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pereira E, do Quental L, Palma E, Oliveira MC, Mendes F, Raposinho P, et al. Evaluation of Acridine Orange derivatives as DNA-targeted radiopharmaceuticals for auger therapy: influence of the radionuclide and distance to DNA. Sci Rep. 2017;7:42544. doi: 10.1038/srep42544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rajon D, Bolch WE, Howell RW. Lognormal distribution of cellular uptake of radioactivity: Monte Carlo simulation of irradiation and cell killing in 3-dimensional populations in carbon scaffolds. J Nucl Med. 2011;52:926–33. doi: 10.2967/jnumed.110.080044. [DOI] [PubMed] [Google Scholar]
- [24].Knapp FF, Dash A. Radiopharm Ther. New Delhi: Springer India; 2016. Auger electron-based radionuclide therapy; pp. 57–67. [DOI] [Google Scholar]
- [25].Paillas S, Ladjohounlou R, Lozza C, Pichard A, Boudousq V, Jarlier M, et al. Localized irradiation of cell membrane by auger electrons is cytotoxic through oxidative stress-mediated nontargeted effects. Antioxid Redox Signal. 2016;25:467–84. doi: 10.1089/ars.2015.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Puncher MRB, Blower PJ. Radionuclide targeting and dosimetry at the microscopic level: the role of microautoradiography. Eur J Nucl Med. 1994;21:1347–65. doi: 10.1007/BF02426701. [DOI] [PubMed] [Google Scholar]
- [27].Hofmann W, Li WB, Friedland W, Miller BW, Madas B, Bardiès M, et al. Internal microdosimetry of alpha-emitting radionuclides. Radiat Environ Biophys. 2020;59:29–62. doi: 10.1007/s00411-019-00826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bavelaar BM, Lee BQ, Gill MR, Falzone N, Vallis KA. Subcellular targeting of theranostic radionuclides. Front Pharmacol. 2018;9:1–17. doi: 10.3389/fphar.2018.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Thakur ML, Segal AW, Louis L, Welch MJ, Hopkins J, Peters TJ. Indium-111-labeled cellular blood components: mechanism of labeling and intracellular location in human neutrophils. J Nucl Med. 1977;18:1022–6. [PubMed] [Google Scholar]
- [30].Nikos P. Subcellular fractionation. Mater Methods. 2013;3:562. doi: 10.13070/mm.en.3.562. [DOI] [Google Scholar]
- [31].O’Neill E, Kersemans V, Allen PD, Terry SYA, Torres JB, Mosley M, et al. Imaging DNA damage repair in vivo after 177Lu-DOTATATE therapy. J Nucl Med. 2020;61:743–50. doi: 10.2967/jnumed.119.232934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Terry SYA, Vallis KA. Relationship between chromatin structure and sensitivity to molecularly targeted Auger electron radiation therapy. Int J Radiat Oncol. 2012;83:1298–305. doi: 10.1016/j.ijrobp.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cornelissen B, Hu M, McLarty K, Costantini D, Reilly RM. Cellular penetration and nuclear importation properties of 111In-labeled and 123I-labeled HIV-1 tat peptide immunoconjugates in BT-474 human breast cancer cells. Nucl Med Biol. 2007;34:37–46. doi: 10.1016/j.nucmedbio.2006.10.008. [DOI] [PubMed] [Google Scholar]
- [34].Reilly RM, Kiarash R, Cameron RG, Porlier N, Sandhu J, Hill RP, et al. 111In-labeled EGF is selectively radiotoxic to human breast cancer cells overexpressing EGFR. J Nucl Med. 2000;41:429–38. [PubMed] [Google Scholar]
- [35].Costantini DL, Chan C, Cai Z, Vallis KA, Reilly RM. 111In-labeled trastuzumab (Herceptin) modified with nuclear localization sequences (NLS): an Auger electron-emitting radiotherapeutic agent for HER2/neu-amplified breast cancer. J Nucl Med. 2007;48:1357–68. doi: 10.2967/jnumed.106.037937. [DOI] [PubMed] [Google Scholar]
- [36].Wang M, Caruano AL, Lewis MR, Meyer LA, VanderWaal RP, Anderson CJ. Subcellular localization of radiolabeled somatostatin analogues. Cancer Res. 2003;63:6864–9. [PubMed] [Google Scholar]
- [37].Cornelissen B, Darbar S, Hernandez R, Kersemans V, Tullis I, Barber PR, et al. ErbB-2 blockade and prenyltransferase inhibition alter epidermal growth factor and epidermal growth factor receptor trafficking and enhance 111In-DTPA-hEGF Auger electron radiation therapy. J Nucl Med. 2011;52:776–83. doi: 10.2967/jnumed.110.084392. [DOI] [PubMed] [Google Scholar]
- [38].Jamur MC, Oliver C. Methods Mol Biol. Humana Press; 2010. Permeabilization of cell membranes; pp. 63–6. [DOI] [PubMed] [Google Scholar]
- [39].Slastnikova TA, Koumarianou E, Rosenkranz AA, Vaidyanathan G, Lupanova TN, Sobolev AS, et al. Modular nanotransporters: a versatile approach for enhancing nuclear delivery and cytotoxicity of Auger electron-emitting 125I. EJNMMI Res. 2012;2:59. doi: 10.1186/2191-219X-2-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Karyagina TS, Ulasov AV, Slastnikova TA, Rosenkranz AA, Lupanova TN, Khramtsov YV, et al. Targeted delivery of 111In into the nuclei of EGFR overexpressing cells via modular Nanotransporters with anti-EGFR affibody. Front Pharmacol. 2020;11:1–15. doi: 10.3389/fphar.2020.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nabbi A, Riabowol K. Isolation of nuclei. Cold Spring Harb Protoc. 2015 doi: 10.1101/pdb.top074583. (2015):pdb.top074583. [DOI] [PubMed] [Google Scholar]
- [42].Maucksch U, Runge R, Wunderlich G, Freudenberg R, Naumann A, Kotzerke J. Comparison of the radiotoxicity of the 99m Tc-labeled compounds 99mTc-pertechnetate, 99mTc-HMPAO and 99mTc-MIBI. Int J Radiat Biol. 2016;92:698–706. doi: 10.3109/09553002.2016.1168533. [DOI] [PubMed] [Google Scholar]
- [43].Freudenberg R, Runge R, Maucksch U, Berger V, Kotzerke J. On the dose calculation at the cellular level and its implications for the RBE of 99mTc and 123I. Med Phys. 2014;41:062503. doi: 10.1118/1.4876296. [DOI] [PubMed] [Google Scholar]
- [44].Cai Z, Kwon YL, Reilly RM. Monte Carlo N-particle (MCNP) modeling of the cellular dosimetry of 64Cu: comparison with MIRDcell S values and implications for studies of its cytotoxic effects. J Nucl Med. 2017;58:339–45. doi: 10.2967/jnumed.116.175695. [DOI] [PubMed] [Google Scholar]
- [45].Falzone N, Lee BQ, Able S, Malcolm J, Terry S, Alayed Y, et al. Targeting micrometastases: the effect of heterogeneous radionuclide distribution on tumor control probability. J Nucl Med. 2019;60:250–8. doi: 10.2967/jnumed.117.207308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Panosa C, Fonge H, Ferrer-Batallé M, Menéndez JA, Massaguer A, De Llorens R, et al. A comparison of non-biologically active truncated EGF (EGFt) and full-length hEGF for delivery of auger electron-emitting 111In to EGFR-positive breast cancer cells and tumor xenografts in athymic mice. Nucl Med Biol. 2015;42:931–8. doi: 10.1016/j.nucmedbio.2015.08.003. [DOI] [PubMed] [Google Scholar]
- [47].Zheng N, Tsai HN, Zhang X, Rosania GR. The subcellular distribution of small molecules: from pharmacokinetics to synthetic biology. Mol Pharm. 2011;8:1619–28. doi: 10.1021/mp200092v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen Y, Dhara S, Banerjee SR, Byun Y, Pullambhatla M, Mease RC, et al. A low molecular weight PSMA-based fluorescent imaging agent for cancer. Biochem Biophys Res Commun. 2009;390:624–9. doi: 10.1016/j.bbrc.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Carlucci G, Carney B, Brand C, Kossatz S, Irwin CP, Carlin SD, et al. Dual-modality optical/PET imaging of PARP1 in glioblastoma. Mol Imaging Biol. 2015;17:848–55. doi: 10.1007/s11307-015-0858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rijpkema M, Oyen WJ, Bos D, Franssen GM, Goldenberg DM, Boerman OC. SPECT- and fluorescence image-guided surgery using a dual-labeled Carcinoembryonic antigen-targeting antibody. J Nucl Med. 2014;55:1519–24. doi: 10.2967/jnumed.114.142141. [DOI] [PubMed] [Google Scholar]
- [51].Hekman MC, Rijpkema M, Muselaers CH, Oosterwijk E, Hulsbergen-Van de Kaa CA, Boerman OC, et al. Tumor-targeted dual-modality imaging to improve intraoperative visualization of clear cell renal cell carcinoma: a first in man study. Theranostics. 2018;8:2161–70. doi: 10.7150/thno.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Deken MM, Bos DL, Tummers WSFJ, March TL, van de Velde CJH, Rijpkema M, et al. Multimodal image-guided surgery of HER2-positive breast cancer using [111In]in-DTPA-trastuzumab-IRDye800CW in an orthotopic breast tumor model. EJNMMI Res. 2019;9:98. doi: 10.1186/s13550-019-0564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lütje S, Heskamp S, Franssen GM, Frielink C, Kip A, Hekman M, et al. Development and characterization of a theranostic multimodal anti-PSMA targeting agent for imaging, surgical guidance, and targeted photodynamic therapy of PSMA-expressing tumors. Theranostics. 2019;9:2924–38. doi: 10.7150/thno.35274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Perez-Medina C, Abdel-Atti D, Zhang Y, Longo VA, Irwin CP, Binderup T, et al. A modular labeling strategy for in vivo PET and near-infrared fluorescence imaging of nanoparticle tumor targeting. J Nucl Med. 2014;55:1706–11. doi: 10.2967/jnumed.114.141861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhao J, Chen J, Ma S, Liu Q, Huang L, Chen X, et al. Recent developments in multimodality fluorescence imaging probes. Acta Pharm Sin B. 2018;8:320–38. doi: 10.1016/j.apsb.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hernandez Vargas S, Kossatz S, Voss J, Ghosh SC, Tran Cao HS, Simien J, et al. Specific targeting of somatostatin receptor Subtype-2 for fluorescence-guided surgery. Clin Cancer Res. 2019;25:4332–42. doi: 10.1158/1078-0432.CCR-18-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lu Z, Pham TT, Rajkumar V, Yu Z, Pedley RB, Årstad E, et al. A dual reporter iodinated labeling reagent for cancer positron emission tomography imaging and fluorescence-guided surgery. J Med Chem. 2018;61:1636–45. doi: 10.1021/acs.jmedchem.7b01746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sarparanta M, Pourat J, Carnazza KE, Tang J, Paknejad N, Reiner T, et al. Multimodality labeling strategies for the investigation of nanocrystalline cellulose biodistribution in a mouse model of breast cancer. Nucl Med Biol. 2020;80–81:1–12. doi: 10.1016/j.nucmedbio.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Paulus A, Desai P, Carney B, Carlucci G, Reiner T, Brand C, et al. Development of a clickable bimodal fluorescent/PET probe for in vivo imaging. EJNMMI Res. 2015;5:43. doi: 10.1186/s13550-015-0120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kiess AP, Minn I, Chen Y, Hobbs R, Sgouros G, Mease RC, et al. Auger radiopharmaceutical therapy targeting prostate-specific membrane antigen. J Nucl Med. 2015;56:1401–7. doi: 10.2967/jnumed.115.155929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Donaldson JG. Immunofluorescence staining. Curr Protoc Cell Biol. 2015;69:4.3.1–7. doi: 10.1002/0471143030.cb0403s69. [DOI] [PubMed] [Google Scholar]
- [62].Puncher MRB, Blower PJ. Frozen section microautoradiography in the study of radionuclide targeting: application to indium-111-oxine-labeled leukocytes. J Nucl Med. 1995;36:499–505. [PubMed] [Google Scholar]
- [63].Appleton TC. Autoradiography of soluble labelled compounds. J R Microsc Soc. 1964;83:277–81. doi: 10.1111/j.1365-2818.1964.tb00541.x. [DOI] [PubMed] [Google Scholar]
- [64].Stumpf WE, Roth LJ. Vacuum freeze drying of frozen sections for dry-mounting, high-resolution autoradiography. Stain Technol. 1964;39:219–23. doi: 10.3109/10520296409061233. [DOI] [PubMed] [Google Scholar]
- [65].Stumpf WE. Drug Targeting, Delivery, and Prediction. 1DDC-Press; 2003. Drug localization in tissues and cells: receptor microscopic autoradiography: a basis for tissue and cellular pharmacokinetics. [Google Scholar]
- [66].Solon EG, Schweitzer A, Stoeckli M, Prideaux B. Autoradiography, MALDI-MS, and SIMS-MS imaging in pharmaceutical discovery and development. AAPS J. 2010;12:11–26. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Solon EG. Autoradiography techniques and quantification of drug distribution. Cell Tissue Res. 2015;360:87–107. doi: 10.1007/s00441-014-2093-4. [DOI] [PubMed] [Google Scholar]
- [68].Davis J, Cook ND, Pither RJ. Biologic mechanisms of 89SrCl2 incorporation into type I collagen during bone mineralization. J Nucl Med. 2000;41:183–8. [PubMed] [Google Scholar]
- [69].Lin J, Liu Y, Yang Y, Hu Z. Calibration and correction of LA-ICP-MS and LA-MC-ICP-MS analyses for element contents and isotopic ratios. Solid Earth Sci. 2016;1:5–27. doi: 10.1016/j.sesci.2016.04.002. [DOI] [Google Scholar]
- [70].Shariatgorji M, Nilsson A, Bonta M, Gan J, Marklund N, Clausen F, et al. Direct imaging of elemental distributions in tissue sections by laser ablation mass spectrometry. Methods. 2016;104:86–92. doi: 10.1016/j.ymeth.2016.05.021. [DOI] [PubMed] [Google Scholar]
- [71].P M-M, Weiskirchen R, Gassler N, Bosserhoff AK, Becker JS. Novel bioimaging techniques of metals by laser ablation inductively coupled plasma mass spectrometry for diagnosis of fibrotic and cirrhotic liver disorders. PLoS One. 2013;8:e58702. doi: 10.1371/journal.pone.0058702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sabine Becker J. Imaging of metals in biological tissue by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS): state of the art and future developments. J Mass Spectrom. 2013;48:255–68. doi: 10.1002/jms.3168. [DOI] [PubMed] [Google Scholar]
- [73].Liu YS, Hu ZC, Li M, Gao S. Applications of LA-ICP-MS in the elemental analyses of geological samples. Chin Sci Bull. 2013;58:3863–78. doi: 10.1007/s11434-013-5901-4. [DOI] [Google Scholar]
- [74].Becker JS. Applications of inductively coupled plasma mass spectrometry and laser ablation inductively coupled plasma mass spectrometry in materials science. Spectrochim Acta - Part B At Spectrosc. 2002;57:1805–20. doi: 10.1016/S0584-8547(02)00213-6. [DOI] [Google Scholar]
- [75].Orellana FA, Gálvez CG, Orellana FA, Gálvez CG, Roldán MT, García-Ruiz C, et al. Applications of laser-ablation-inductively-coupled plasma-mass spectrometry in chemical analysis of forensic evidence. TrAC - Trends Anal Chem. 2013;42:1–34. doi: 10.1016/j.trac.2012.09.015. [DOI] [Google Scholar]
- [76].Pozebon D, Scheffler GL, Dressler VL, Nunes MAG. Review of the applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) to the analysis of biological samples. J Anal At Spectrom. 2014;29:2204–28. doi: 10.1039/c4ja00250d. [DOI] [Google Scholar]
- [77].Van Acker T, Buckle T, Van Malderen SJM, van Willigen DM, van Unen V, van Leeuwen FWB, et al. High-resolution imaging and single-cell analysis via laser ablation-inductively coupled plasma-mass spectrometry for the determination of membranous receptor expression levels in breast cancer cell lines using receptorspecific hybrid tracers. Anal Chim Acta. 2019;1074:43–53. doi: 10.1016/j.aca.2019.04.064. [DOI] [PubMed] [Google Scholar]
- [78].Löhr K, Traub H, Wanka AJ, Panne U, Jakubowski N. Quantification of metals in single cells by LA-1CP-MS: comparison of single spot analysis and imaging. J Anal At Spectrom. 2018;33:1579–87. doi: 10.1039/c8ja00191j. [DOI] [Google Scholar]
- [79].Theiner S, Loehr K, Koellensperger G, Mueller L, Jakubowski N. Single-cell analysis by use of 1CP-MS. J Anal At Spectrom. 2020;35:1784–813. doi: 10.1039/d0ja00194e. [DOI] [Google Scholar]
- [80].Van Malderen SJM, Van Acker T, Vanhaecke F. Sub-micrometer nanosecond LA-1CP-MS imaging at pixel acquisition rates above 250 Hz via a low-dispersion setup. Anal Chem. 2020;92:5756–64. doi: 10.1021/acs.analchem.9b05056. [DOI] [PubMed] [Google Scholar]
- [81].Van Malderen SJM, Van Acker T, Laforce B, De Bruyne M, de Rycke R, Asaoka T, et al. Three-dimensional reconstruction of the distribution of elemental tags in single cells using laser ablation ICP-mass spectrometry via registration approaches. Anal Bioanal Chem. 2019;411:4849–59. doi: 10.1007/s00216-019-01677-6. [DOI] [PubMed] [Google Scholar]
- [82].Westerhausen MT, Bishop DP, Dowd A, Wanagat J, Cole N, Doble PA. Superresolution reconstruction for two- and three-dimensional LA-ICP-MS bioimaging. Anal Chem. 2019;91:14879–86. doi: 10.1021/acs.analchem.9b02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jeynes C, Webb RP, Lohstroh A. Ion beam analysis: a century of exploiting the electronic and nuclear structure of the atom for materials characterisation. Rev Accel Sci Technol Accel Appl Ind Environ. 2011;4:41–82. doi: 10.1142/S1793626811000483. [DOI] [Google Scholar]
- [84].Romolo FS, Christopher ME, Donghi M, Ripani L, Jeynes C, Webb RP, et al. Integrated ion beam analysis (IBA) in gunshot residue (GSR) characterisation. Forensic Sci Int. 2013;231:219–28. doi: 10.1016/j.forsciint.2013.05.006. [DOI] [PubMed] [Google Scholar]
- [85].Garman EF, Grime GW. Elemental analysis of proteins by microPIXE. Prog Biophys Mol Biol. 2005;89:173–205. doi: 10.1016/j.pbiomolbio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- [86].Jeynes C, Bailey MJ, Bright NJ, Christopher ME, Grime GW, Jones BN, et al. “Total IBA” - Where are we? Nucl Instruments Methods Phys Res Sect B Beam Interact with Mater Atoms. 2012;271:107–18. doi: 10.1016/j.nimb.2011.09.020. [DOI] [Google Scholar]
- [87].Jeynes C, Colaux JL. Thin film depth profiling by ion beam analysis. Analyst. 2016;141:5944–85. doi: 10.1039/c6an01167e. [DOI] [PubMed] [Google Scholar]
- [88].Jeynes JCG, Bailey MJ, Coley H, Kirkby KJ, Jeynes C. Microbeam PIXE analysis of platinum resistant and sensitive ovarian cancer cells. Nucl Instruments Methods Phys Res Sect B Beam Interact with Mater Atoms. 2010;268:2168–71. doi: 10.1016/j.nimb.2010.02.042. [DOI] [Google Scholar]
- [89].Sakurai H, Okamoto M, Hasegawa M, Satoh T, Oikawa M, Kamiya T, et al. Direct visualization and quantification of the anticancer agent, cis-diamminedichloro-platinum(II), in human lung cancer cells using in-air microparticle-induced X-ray emission analysis. Cancer Sci. 2008;99:901–4. doi: 10.1111/j.1349-7006.2008.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ortega R, Suda A, Devès G. Nuclear microprobe imaging of gallium nitrate in cancer cells. Nucl Instruments Methods Phys Res Sect B Beam Interact with Mater Atoms. 2003;210:364–7. doi: 10.1016/S0168-583X(03)01052-8. [DOI] [Google Scholar]
- [91].Carmona A, Devès G, Ortega R. Quantitative micro-analysis of metal ions in subcellular compartments of cultured dopaminergic cells by combination of three ion beam techniques. Anal Bioanal Chem. 2008;390:1585–94. doi: 10.1007/s00216-008-1866-6. [DOI] [PubMed] [Google Scholar]
- [92].Jeynes C, Barradas NP, Szilágyi E. Accurate determination of quantity of material in thin films by Rutherford backscattering spectrometry. Anal Chem. 2012;84:6061–9. doi: 10.1021/ac300904c. [DOI] [PubMed] [Google Scholar]
- [93].Colaux JL, Jeynes C, Heasman KC, Gwilliam RM. Certified ion implantation fluence by high accuracy RBS. Analyst. 2015;140:3251–61. doi: 10.1039/C4AN02316A. [DOI] [PubMed] [Google Scholar]
- [94].Greenhalgh CJ, Karekla E, Miles GJ, Powley IR, Costa C, de Jesus J, et al. Exploration of matrix effects in laser ablation inductively coupled plasma mass spectrometry imaging of cisplatin-treated tumors. Anal Chem. 2020;92:9847–55. doi: 10.1021/acs.analchem.0c01347. [DOI] [PubMed] [Google Scholar]
- [95].Colaux JL, Jeynes C, Heasman KC, Gwilliam RM. Certified ion implantation fluence by high accuracy RBS. Analyst. 2015;140:3251–61. doi: 10.1039/c4an02316a. [DOI] [PubMed] [Google Scholar]
- [96].Kirz J, Jacobsen C. The history and future of X-ray microscopy. J Phys Conf Ser. 2009;186:012001. doi: 10.1088/1742-6596/186/1/012001. [DOI] [Google Scholar]
- [97].Conesa JJ, Carrasco AC, Rodríguez-Fanjul V, Yang Y, Carrascosa JL, Cloetens P, et al. Unambiguous intracellular localization and quantification of a potent iridium anticancer compound by correlative 3D Cryo X-ray imaging. Angew Chem Int Ed. 2020;59:1270–8. doi: 10.1002/anie.201911510. [DOI] [PubMed] [Google Scholar]
- [98].Bockman RS, Repo MA, Warrell RP, Pounds JG, Schidlovsky G, Gordon BM, et al. Distribution of trace levels of therapeutic gallium in bone as mapped by synchrotron X-ray microscopy. Proc Natl Acad Sci. 1990;87:4149–53. doi: 10.1073/pnas.87.11.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hummer AA, Bartel C, Arion VB, Jakupec MA, Meyer-Klaucke W, Geraki T, et al. X-ray absorption spectroscopy of an investigational anticancer gallium(III) drug: interaction with serum proteins, elemental distribution pattern, and coordination of the compound in tissue. J Med Chem. 2012;55:5601–13. doi: 10.1021/jm3005459. [DOI] [PubMed] [Google Scholar]
- [100].Konkankit CC, Lovett J, Harris HH, Wilson JJ. X-ray fluorescence microscopy reveals that rhenium(i) tricarbonyl isonitrile complexes remain intact in vitro . Chem Commun. 2020;56:6515–8. doi: 10.1039/D0CC02451A. [DOI] [PubMed] [Google Scholar]
- [101].De Samber B, Niemiec MJ, Laforce B, Garrevoet J, Vergucht E, De Rycke R, et al. Probing intracellular element concentration changes during neutrophil extracellular trap formation using synchrotron radiation based X-ray fluorescence. PLoS One. 2016;11:e0165604. doi: 10.1371/journal.pone.0165604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hummer AA, Rompel A. The use of X-ray absorption and synchrotron based microX-ray fluorescence spectroscopy to investigate anti-cancer metal compounds invivo and in vitro. Metallomics. 2013;5:597. doi: 10.1039/c3mt20261e. [DOI] [PubMed] [Google Scholar]
- [103].Stewart TJ. Across the spectrum: integrating multidimensional metal analytics for in situ metallomic imaging. Metallomics. 2019;11:29–49. doi: 10.1039/C8MT00235E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ackerman CM, Lee S, Chang CJ. Analytical methods for imaging metals in biology: from transition metal metabolism to transition metal signaling. Anal Chem. 2017;89:22–41. doi: 10.1021/acs.analchem.6b04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bissardon C, Reymond S, Salomé M, André L, Bayat S, Cloetens P, et al. Cell culture on silicon nitride membranes and cryopreparation for synchrotron x-ray fluorescence nano-analysis. J Vis Exp. 2019;2019 doi: 10.3791/60461. [DOI] [PubMed] [Google Scholar]
- [106].Davies KM, Hare DJ, Bohic S, James SA, Billings JL, Finkelstein DI, et al. Comparative study of metal quantification in neurological tissue using laser ablation-inductively coupled plasma-mass spectrometry imaging and X-ray fluorescence microscopy. Anal Chem. 2015;87:6639–45. doi: 10.1021/acs.analchem.5b01454. [DOI] [PubMed] [Google Scholar]
- [107].Bafaro E, Liu Y, Xu Y, Dempski RE. The emerging role of zinc transporters in cellular homeostasis and cancer. Signal Transduct Target Ther. 2017;2:17029. doi: 10.1038/sigtrans.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Harkiolaki M, Darrow MC, Spink MC, Kosior E, Dent K, Duke E. Cryo-soft X-ray tomography: using soft X-rays to explore the ultrastructure of whole cells. Emerg Top Life Sci. 2018;2:81–92. doi: 10.1042/ETLS20170086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Deng J, Vine DJ, Chen S, Jin Q, Nashed YSG, Peterka T, et al. X-ray ptychographic and fluorescence microscopy of frozen-hydrated cells using continuous scanning. Sci Rep. 2017;7:445. doi: 10.1038/s41598-017-00569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Guo J, Larabell CA. Soft X-ray tomography: virtual sculptures from cell cultures. Curr Opin Struct Biol. 2019;58:324–32. doi: 10.1016/j.sbi.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bonta M, Török S, Hegedus B, Döme B, Limbeck A. A comparison of sample preparation strategies for biological tissues and subsequent trace element analysis using LA-ICP-MS. Anal Bioanal Chem. 2017;409:1805–14. doi: 10.1007/s00216-016-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Gao J, He Y, Chen Y, Song D, Zhang Y, Qi F, et al. Reversible FRET fluorescent probe for ratiometric tracking of endogenous Fe3+ in ferroptosis. Inorg Chem. 2020;59:10920–7. doi: 10.1021/acs.inorgchem.0c01412. [DOI] [PubMed] [Google Scholar]