Summary
Phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is a low abundant membrane lipids essential for plasma membrane function. In plants, mutations in PI4P 5-kinases (PIP5K) suggest that PI(4,5)P2 production is involved in development, immunity and reproduction. However, phospholipid synthesis is highly intricate. It is thus likely that steady-state depletion of PI(4,5)P2 triggers confounding indirect effects. Furthermore, inducible tools available in plants, allow to increase but not decrease PI(4,5)P2, and no PIP5K inhibitors are available. Here, we introduce iDePP (Inducible Depletion of PI(4,5)P2 in Plants), a system for the inducible and tunable depletion of PI(4,5)P2 in plants in less than three hours. Using this strategy, we confirm that PI(4,5)P2 is critical for various aspects of plant development, including root growth, root hair elongation and organ initiation. We show that PI(4,5)P2 is required to recruit various endocytic proteins, including AP2, to the plasma membrane, and thus to regulate clathrin-mediated endocytosis. Finally, we uncover that inducible PI(4,5)P2 perturbation impacts the dynamics of the actin cytoskeleton as well as microtubule anisotropy. Together, we propose that iDePP is a simple and efficient genetic tool to test the importance of PI(4,5)P2 in given cellular or developmental responses, but also to evaluate the importance of this lipid in protein localization.
Phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is a low abundant membrane lipids essential for plasma membrane function1,2. In plants, mutations in PI4P 5-kinases (PIP5K) suggest that PI(4,5)P2 production is involved in development, immunity and reproduction3–5. However, phospholipid synthesis is highly intricate6. It is thus likely that steady-state depletion of PI(4,5)P2 triggers confounding indirect effects. Furthermore, inducible tools available in plants, allow to increase7–9 but not decrease PI(4,5)P2, and no PIP5K inhibitors are available. Here, we introduce iDePP (Inducible Depletion of PI(4,5)P2 in Plants), a system for the inducible and tunable depletion of PI(4,5)P2 in plants in less than three hours. Using this strategy, we confirm that PI(4,5)P2 is critical for various aspects of plant development, including root growth, root hair elongation and organ initiation. We show that PI(4,5)P2 is required to recruit various endocytic proteins, including AP2, to the plasma membrane, and thus to regulate clathrin-mediated endocytosis. Finally, we uncover that inducible PI(4,5)P2 perturbation impacts the dynamics of the actin cytoskeleton as well as microtubule anisotropy. Together, we propose that iDePP is a simple and efficient genetic tool to test the importance of PI(4,5)P2 in given cellular or developmental responses, but also to evaluate the importance of this lipid in protein localization.
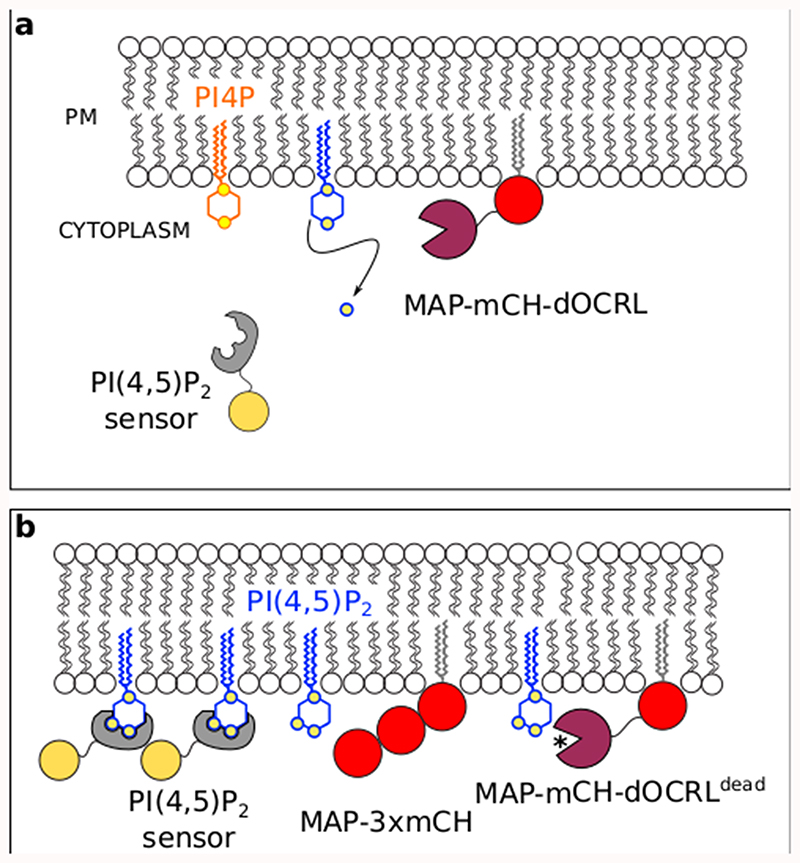
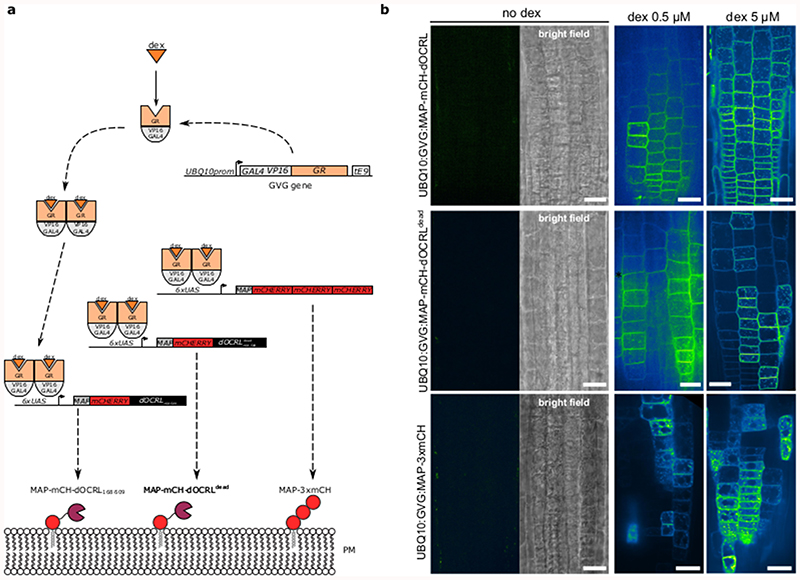
iDePP is a synthetic inducible system designed to specifically dephosphorylate PI(4,5)P2 at the plasma membrane at ~20°C (Extended Data figure 1). It consists of the isolated phosphatase domain of the Drosophila melanogaster OCRL protein (dOCRL), which is artificially targeted to the plasma membrane by a myristoylation and palmitoylation (MAP) sequence (Extended Data figure 2). iDePP is fused with the fluorescent protein mCHERRY for visualization, and its expression is driven by a dexamethasone (dex) inducible promoter (Extended Data figure 3). We included two controls: i) MAP-3xmCH, which serves as a generic control to test for the possible side effects of dex/GVG/MAP, and ii) a catalytic-dead version of dOCRL (MAP-mCH-dOCRLdead), which controls that phenotypes are indeed caused by 5-phosphatase activity and not by other parts of the dOCRL protein (Extended Data figure 2).
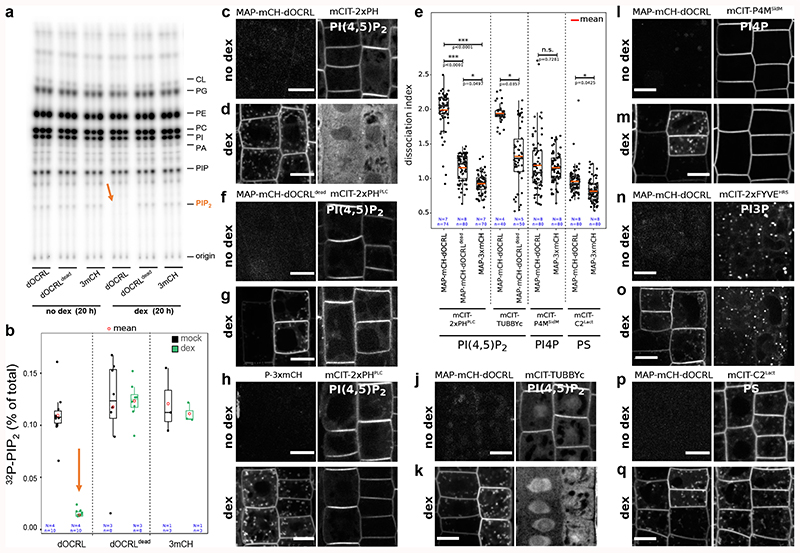
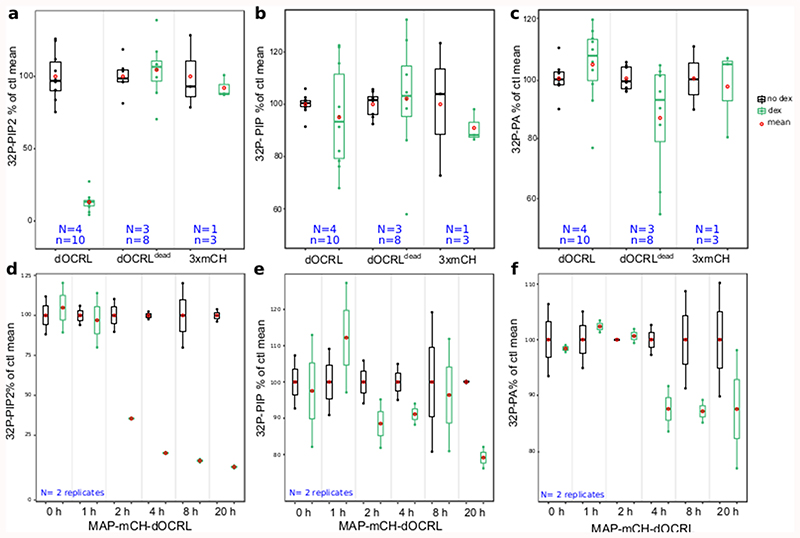
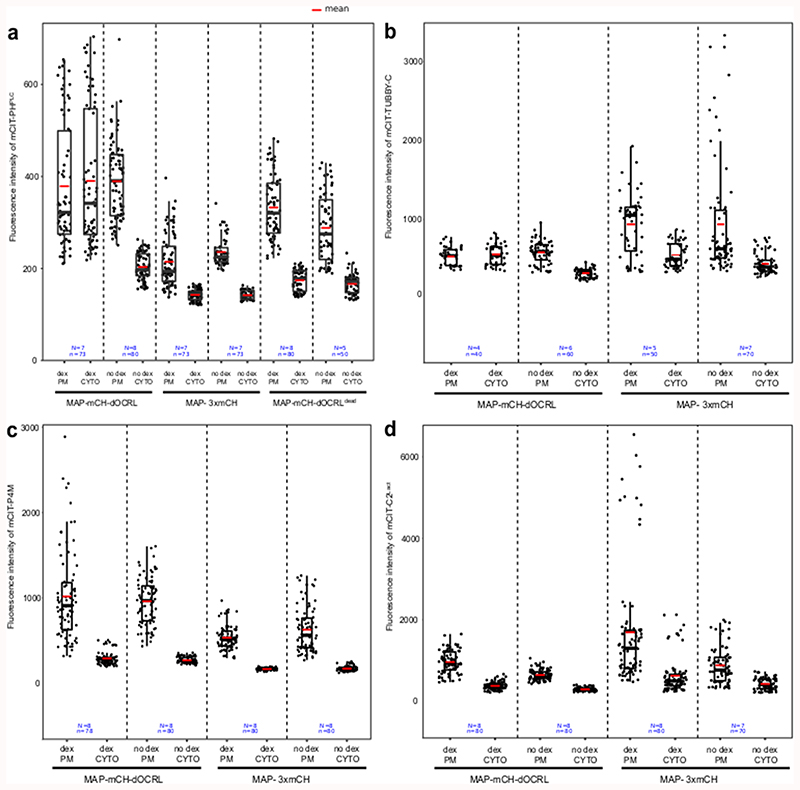
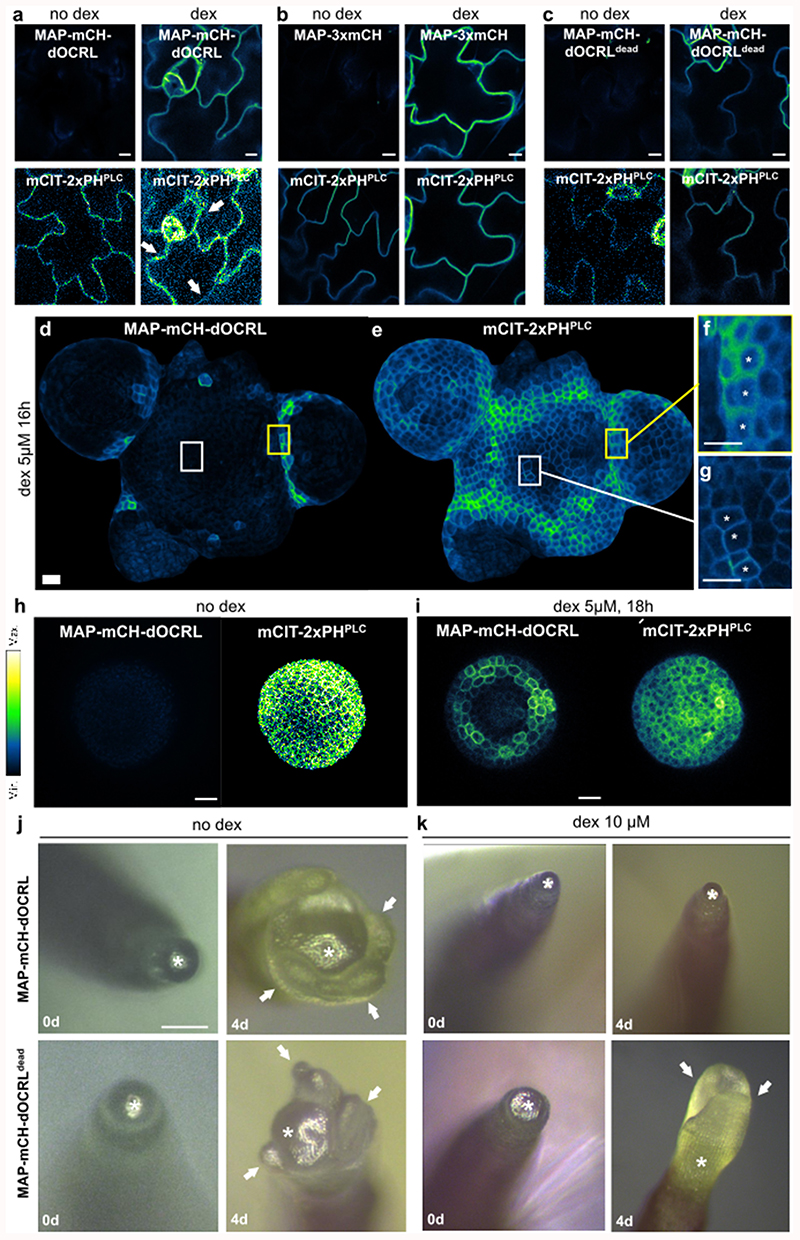
We validated that induction of either MAP-mCH-dOCRLdead or MAP-3xmCH controls had no effect on PIP2 levels in Arabidopsis seedlings (Figure 1a-b, Extended Data figure 4). By contrast, induction of the active phosphatase MAP-mCH-dOCRL resulted in a dramatic decrease of 32Pi incorporation into PIP2 (32P-PIP2 levels; i.e. ~90%, Figure 1a-b, Extended Data figure 4). In addition, dex induction did not perturb the levels of other phospholipids in any of the lines tested, confirming the specificity of the iDePP system and its controls (Figure 1a, Extended Data figure 4). At the subcellular level, we showed that in the absence of dex, MAP-mCH-dOCRL was not expressed (Figure 1c, left panel), while the PI(4,5)P2 biosensor mCIT-2xPHPLC localized to the plasma membrane (Figure 1c, right panel). Induction of MAP-mCH-dOCRL resulted in the release of the mCIT-2xPHPLC fluorescence from the plasma membrane (Figure 1c-d; Extended Data figure 5), confirming the efficient depletion of PI(4,5)P2 from this membrane. The localization of the mCIT-2xPHPLC biosensor was not affected in the control lines (Figure 1d-f, Extended Data figure 5). We further confirmed the impact of MAP-mCH-dOCRL expression on PI(4,5)P2 using an independent PI(4,5)P2 biosensor (mCIT-TUBBY-C, Figure 1d,g, Extended Data figure 5). In agreement with the results obtained from the biochemical quantification, we did not observe any effect on the localization of the biosensors for PI4P (mCIT-P4MSidM), PI3P (mCIT-2xFYVEHRS) or PS (mCIT-C2Lact) after MAP-mCH-dOCRL induction (Figure 1d, h-j; Extended Data figure 5). Overall, our results show that MAP-mCH-dOCRL efficiently, specifically, and directly hydrolyzes the PI(4,5)P2 pool at the plasma membrane in living plant cells.
Figure 1. Inducible and specific depletion of PI(4,5)P2 in plant cells using iDePP.
(a-b) 32Pi incorporation into PIP2 (32P-PIP2 levels) in iDePP lines and controls ± DEX of 2-5 independent replicates. Seedlings were labelled with 32Pi and incubated ± dex O/N (16-20 hrs). For line MAP-mCH-dOCRL and MAP-mCH-dOCRLdead. Each sample contained the lipid extract of three seedlings, of which 1/5th was analyzed by TLC (a) and quantified by phosphoimaging, of which 32P-PIP2 was calculated as percentage of total 32P-lipids (b). (c) Representative images of the fluorescent signal corresponding to MAP-mCH-dOCRL (left panel) and PI(4,5)P2 sensor mCIT-2xPHPLC (right panel) in the same root cells, without dex treatment. (d) Representative images of the fluorescent signal corresponding to MAP-mCH-dOCRL (left panel) and PI(4,5)P2 sensor mCIT-2xPHPLC (right panel) in the same root cells, after 16h of dex treatment. (e) Dissociation indexes of lipid biosensors upon expression of MAP-mCH-dOCRL or negative controls. Dissociation index is the ratio of (i) plasma membrane to cytosol fluorescence ratio without dex treatment, to (ii) plasma membrane to cytosol fluorescence ratio after dex treatment. Statistical analysis with LMER (Type II Wald χ2 test) and post-hoc. (f-q) Representative fluorescent images of indicated MAP-anchored construct (left panel) and the lipid sensor (right panel) without dex treatment (f, h, j, l, n, and p), or after 16h of dex treatment (g, i, k, m, o, and q). Note that the image on the left and right panels are always the same root cells imaged for mCHERRY (left panel) and mCIT (right panel). Scale bars: 10 μm. In the plots (b, e), middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values. All statistical tests were two-sided.
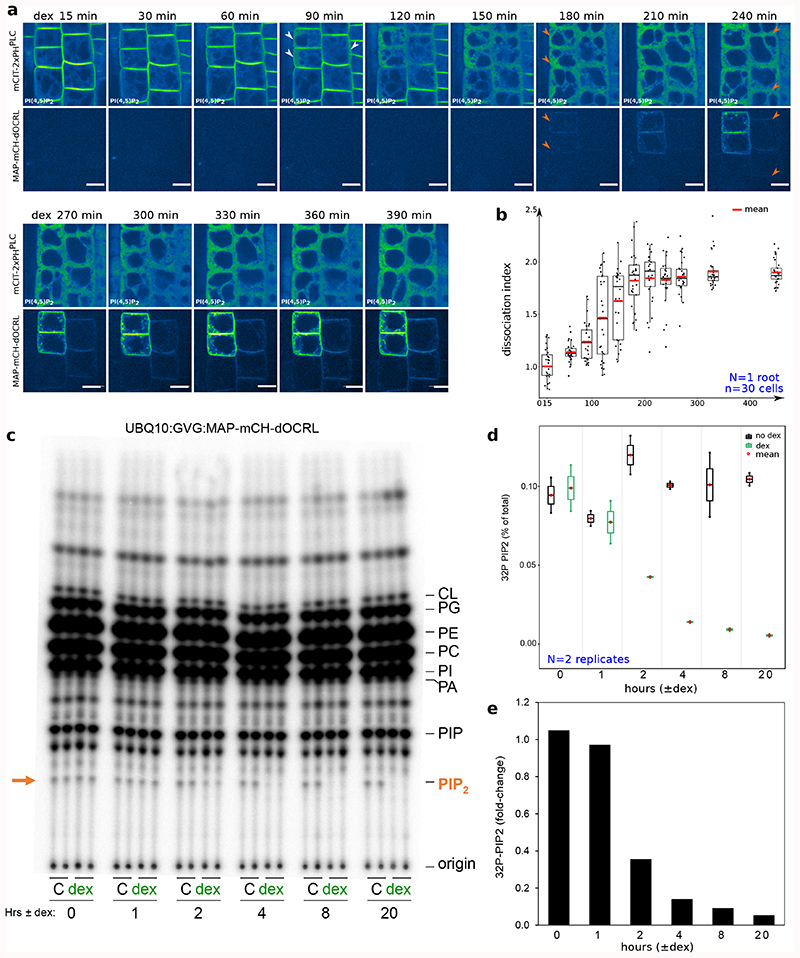
To assess the dynamics of PI(4,5)P2 depletion after iDePP induction, we followed mCIT-2xPHPLC and MAP-mCH-dOCRL localization upon dex treatment. After 15 min of dex treatment (first time-point), mCIT-2xPHPLC labelled the plasma membrane while no signal corresponding to MAP-mCH-dOCRL fluorescence was observed (Figure 2a-b, Supplemental movies 1-2). After 90 min, a fraction of mCIT-2xPHPLC started to be partially released from the plasma membrane in the cytosol of some cells, indicating that PI(4,5)P2 had started to be depleted from the targeted membrane (Figure 2a-b, Supplemental movies 1-2). After 180 min, mCIT-2xPHPLC was cytosolic in most cells, and over the next few hours, mCIT-2xPHPLC remained cytosolic while MAP-mCH-dOCRL fluorescence increased (Figure 2a-b). Biochemical monitoring of the PIP2 levels using 32Pi-prelabelled seedlings ± dex demonstrated that indeed within 2 hours of induction, over 60% of the 32Pi incorporation into PIP2 was gone in MAP-mCH-dOCRL seedlings, while the levels of the other anionic lipids remained stable (Figure 2 c-e, Extended Data figure 4). In addition, controls without dex treatment had no impact on either mCIT-2xPHPLC localization (Supplemental movies 3) or PIP2 level (Figure 2 c-d). Together, these results confirm that iDePP can manipulate PI(4,5)P2 levels within a few hours following induction.
Figure 2. Time-lapse analysis of the depletion of PI(4,5)P2 in plant cells using iDePP.
(a) Dual color time-course analysis during dex induction, monitoring mCIT-2xPHPLC PI(4,5)P2 biosensor subcellular localization and MAP-mCH-dOCRL appearance at the root tip in epidermal cells every five minutes for 6h30. White and orange arrowheads indicate mCIT-PHPLC partial- and full release from the plasma membrane into the cytosol, respectively. Images were colored in green fire blue, where yellow showed the max intensity and dark blue the low level of fluorescence. Scale bars: 10 μm. (b) Dissociation indexes of mCIT-2xPHPLC over-time. 30 cells from the displayed root, during the induction of the expression of MAP-mCH-dOCRL, were analyzed. (c-e) Time-course analysis of 32P-PIP2 incorporation (which is related to changed levels of the lipids) in iDePP seedlings ± dex in MAP-mCH-dOCRL line. Seedlings of MAP-mCH-dOCRL line were labelled for 20 hrs with 32Pi and co-incubated with or without dex for the times indicated (0-20 hrs). Each sample contained the lipid extract of three seedlings of which 1/5th was analyzed by TLC (d) and quantified by phosphoimaging, where 32P-PIP2 was calculated as percentage of total 32P-lipids (d) or as fold-change compared to incubation without dex (e). In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values. All statistical tests were two-sided.
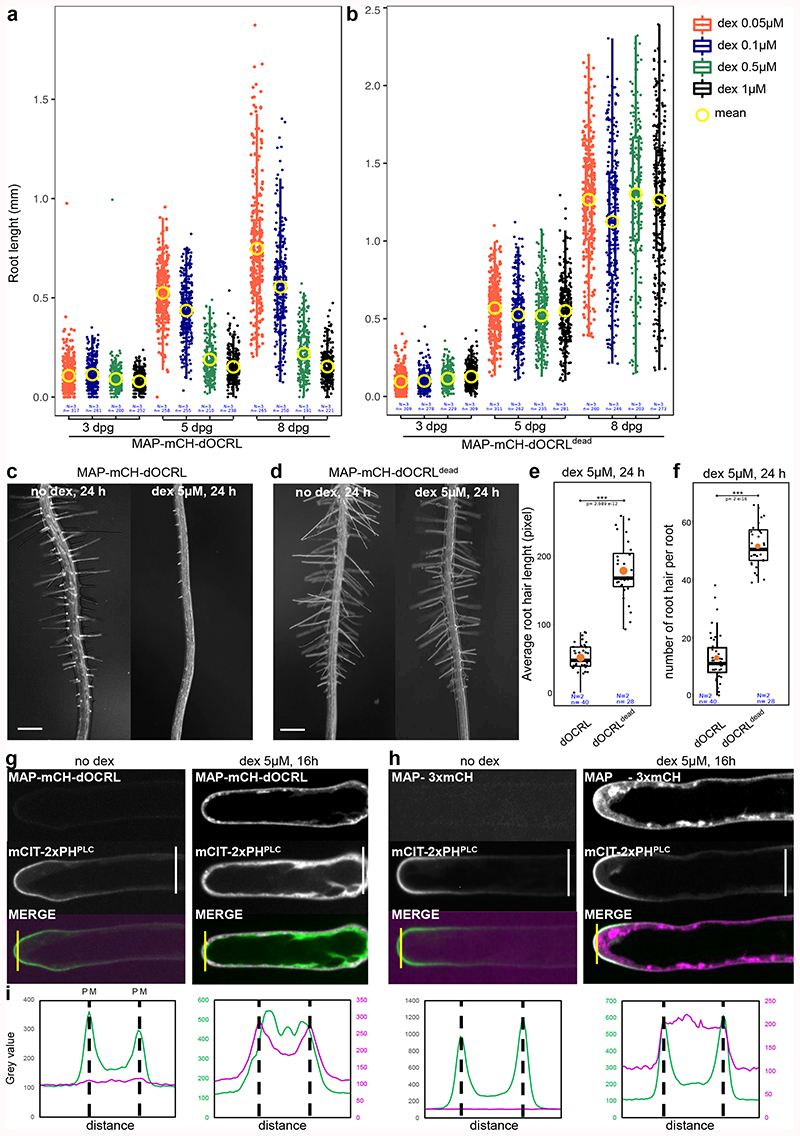
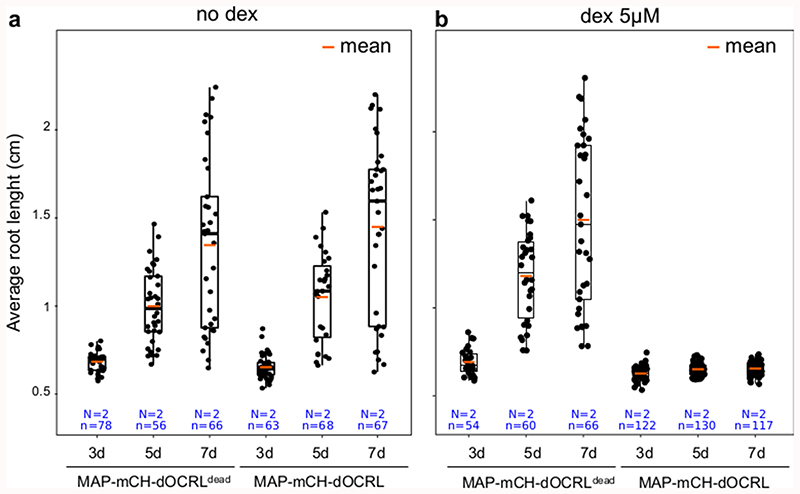
Using the iDePP system, we efficiently depleted PI(4,5)P2 in various cell types and organs, which led to developmental growth arrest in both root and shoot (Figure 3a-b, Extended Data figure 6). Seedlings sawn directly on 5 μM dex (expressing therefore MAP-mCH-dOCRL from day 0) displayed rapid growth arrest, highlighting the important role of PI(4,5)P2 in development (Extended Data figure 7). To by-pass the strong lethality associated with severe PI(4,5)P2 depletion, we first took advantage of the dose-dependent activation of iDePP upon treatment with different concentration of dex. At low dex concentrations (0.05 μM), root growth arrest was mild, while at higher concentrations a complete arrest in root growth was observed (from 0.5 to 5 μM dex, Figure 3a-b). As was performed for the inducible production of PI(4,5)P2 upon human PIP5K induction by estradiol7, we also performed dex treatments later during root development. In 5-day old seedlings transferred to 5 μM dex medium for 22h (+/- 2h), root hair growth was abolished, specifically after induction of MAP-mCH-dOCRL but not MAP-mCH-dOCRdead (Figure 3c-f, Supplemental movie 4). Accordingly, expression of MAP-mCH-dOCRL released mCIT-2xPHPLC fluorescence from the plasma membrane into the cytoplasm of already formed root hair cells, indicating efficient PI(4,5)P2 depletion (Figure 3g-i). These experiments show that dose-response and temporal activation of the iDePP system can be used to perturb plant development at a range of intensities and at fine-tuned temporal resolution. Here, by inducing PI(4,5)P2 perturbation at different time of root epidermis differentiation, we demonstrate that iDePP can be used to dissect the manifold function of this lipid in root hair initiation, i.e. initiation, growth and polarity.
Figure 3. iDePP allows adjusting the timing and level of PI(4,5)P2 depletion to study integrated responses at the organismal level.
(a-b) Quantification over time (at 3, 5 and 8 days) of the root length in transgenic lines MAP-mCH-dOCRL and MAP-mCH-dOCRLdead grown on ½MS plates supplemented with various concentration of dexamethasone. (c-f) Representative image of the phenotype observed at the root tip of MAP-mCH-dOCRL (c) or MAP-mCH-dOCRLdead (d) plantlets transferred for 24h on root hair medium without or with 5 μM dexamethasone, and related quantification of (e) the average root hair length and (f) the number of newly formed root hair. Note that in line expressing MAP-mCH-dOCRL, there is no newly formed root hair after dex induction (and the bulge that we see were formed but are not able to grow in a polar fashion, see supplementary movie 4 for details). (g-h) Subcellular localization of mCIT-2xPHPLC, a PI(4,5)P2 biosensor, and either MAP-mCH-dOCRL (g) or MAP-3xmCH (h) without and with 5 μM dex treatment for 16h. Scale bars: 10 μm. (i) Signal intensity for mCIT-2xPHPLC (green) and MAP-mCH-dOCRL or MAP-3xmCH (magenta) with and without 5 μM dexamethasone treatment for 16h, along the yellow line drawn on panel (g-h). The peaks correspond to plasma membrane (PM) positions, as indicated. Note that for the condition with dex, the signal for mCH channel was weaker that with mCIT and therefore signal intensity was plotted with a different scale, on a secondary axe on the right. In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values. All statistical tests were two-sided.
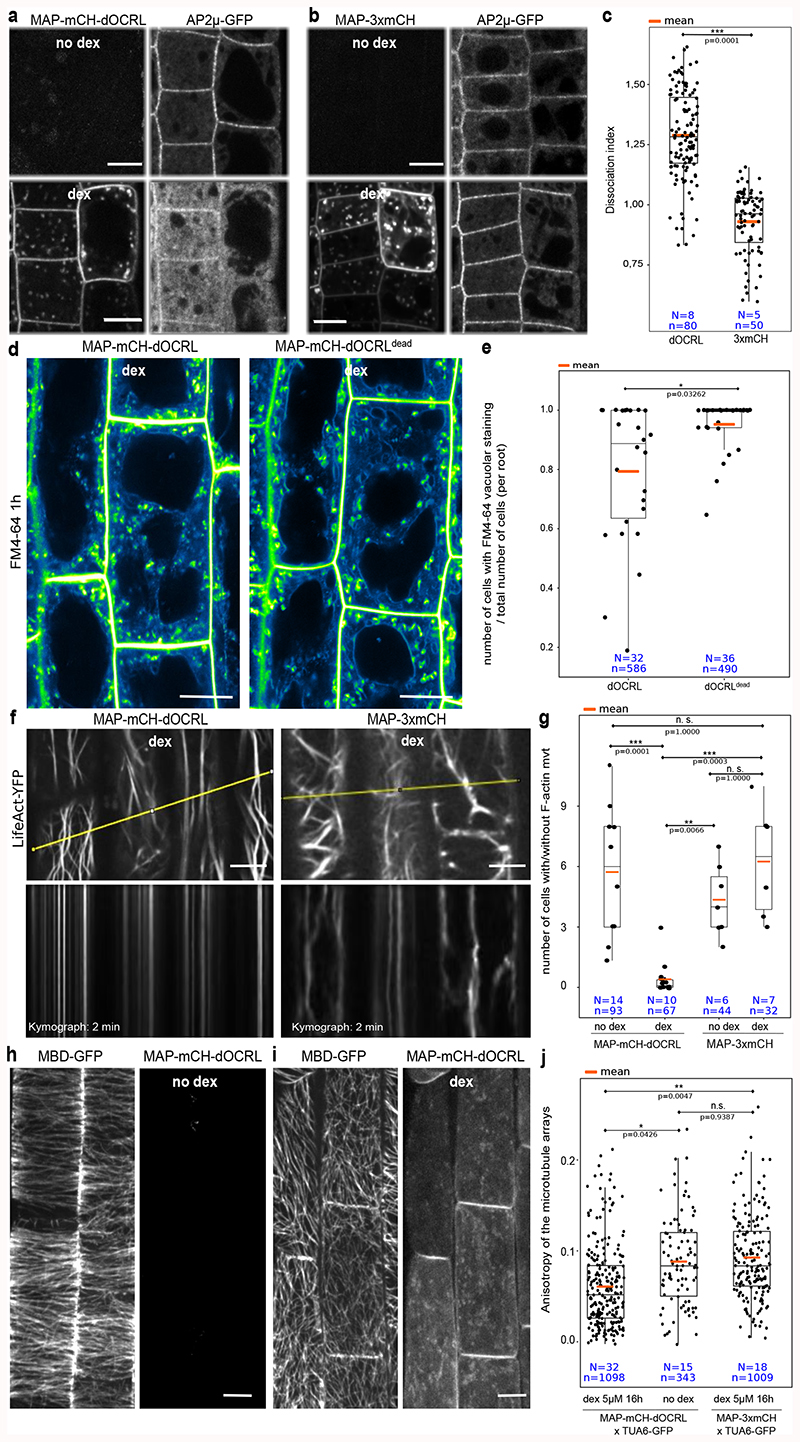
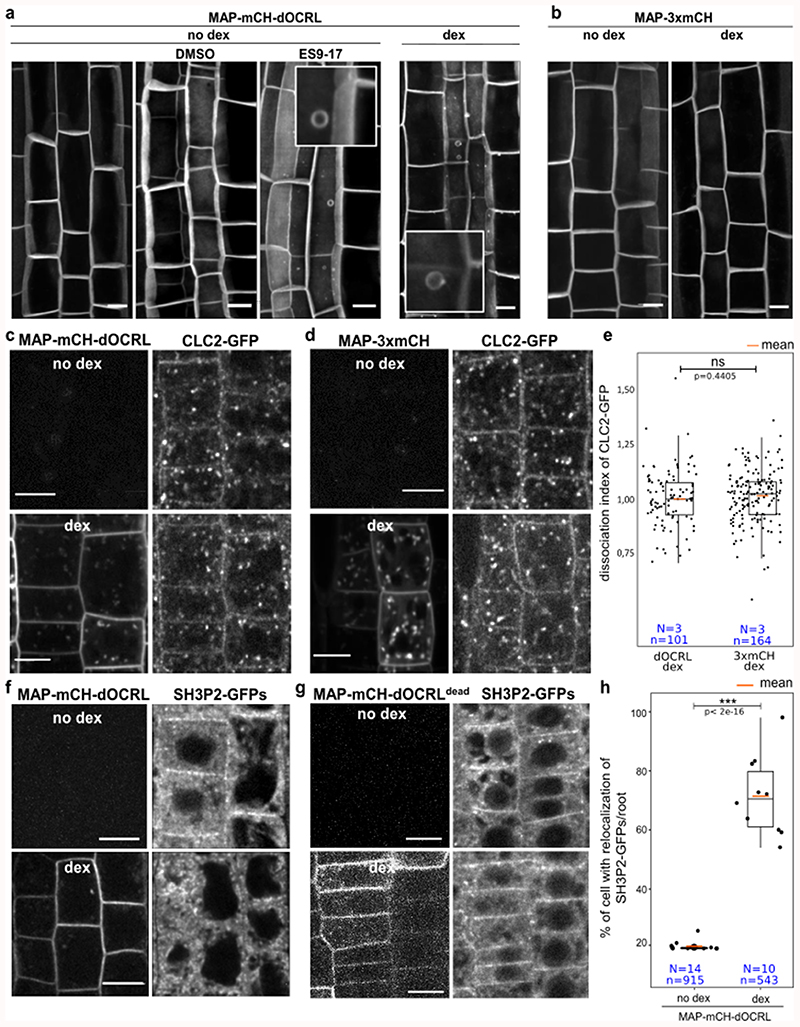
During the course of our experiments, we regularly observed circular plasma membrane protuberances (Extended Data figure 8) that could be the result of altered endocytosis, as observed upon PIP5K6 over-expression in pollen tubes10. Indeed, long-term treatment of non-induced MAP-mCH-dOCRL line with ES9-17, a clathrin-mediated endocytosis inhibitor11, led to similar phenotypes (Extended Data figure 8). We thus assessed the localization of proteins implicated in endocytosis in lines expressing the iDePP system. MAP-mCH-dOCRL induction caused a decrease in both clathrin adaptor protein AP-μ2 (AP-μ2-GFP, Figure 4a-c) and SH3-domain containing protein 2 at the plasma membrane (SH3P2-GFPs; Extended Data figure 8), while the localization of clathrin light chain 2 (CLC2-GFP) was not affected, nor at the trans Golgi network (TGN) (Extended Data figure 8). Since PI(4,5)P2 is not present at the TGN, these results suggest that CLC2 may require other anionic lipids, such as PI4P or phosphatidylserine that are both present at plasma membrane and TGN. CLC2 localization could also require coincidence binding with other proteins that are independent of PI(4,5)P2. FM4-64 uptake experiments confirmed that bulk endocytosis was reduced in line expressing MAP-mCH-dOCRL compared to controls. with a reduction of vacuolar staining after 1 h of FM4-64 incubation (Figure 4d, e). These results are fully in line with the previous findings that clathrin-mediated endocytosis and related trafficking to the vacuole is partially impaired upon PIP5K loss- or gain-of-function 3,7,10,12,13. Thus, our results confirmed the importance of PI(4,5)P2 in regulating endocytosis in plants by further identifying some of the endocytic proteins that rely on PI(4,5)P2 plasma membrane localization. However, unlike animal systems, where PI(4,5)P2 is absolutely required for clathrin-mediated endocytosis14, our results suggest that internalization still occurs upon iDePP induction in plants, albeit at a reduced rate. Because the plant plasma membrane strongly accumulates PI4P15, it is possible that this lipid partially substitutes for PI(4,5)P2 function in endocytosis. It also highlights the fact that clathrin-mediated endocytosis is very different in plant and animal systems 16,17.
Figure 4. Using iDePP to probe the importance of PI(4,5)P2 in protein localization and the dynamics of various cellular processes.
(a-c) Effect of the expression of MAP-mCH-dOCRL (a) or MAP-3xmCh (b) on AP2μ-GFP localization, in absence (top) or presence of dex (16 h), and (c) related quantification of the dissociation index (2 independent experiments). (d-e) Effect of expression of MAP-mCH-dOCRL (left panels) or MAP-mCH-dOCRLdead (right panel) on FM4-64 uptake, 1h after the beginning of the incubation (+/- 10 min). Images were color coded in green fire blue as in Figure 2, and (e) related quantification of the vacuolar staining 1h after FM4-64 incubation. (f) Dynamics of F-actin cytoskeleton labelled by LifeAct-YFPv in root cells expressing MAP-mCH-dOCRL or MAP-3xmCH after 5 μM dex (20 h), and relative kymographs extracted from a time course over 2 min (lower panel). The line drawn to generate the kymograph is shown in yellow. See Supplemental movie 5 for details. (g) Quantification of the number of cells with movement of actin cytoskeleton over 2-min time period relative to cells without movement in MAP-mCH-dOCRL (with or without dex treatment), and MAP-3xmCH (with 5 μM or without dex treatment). (h-j) Representative images of cortical microtubules labelled by MBD-GFP without dex (h) or with dex (16 hours, i) to induce MAP-mCH-dOCRL expression in root cells. All pictures are z-projections and (j) related quantification of the average anisotropy of the microtubule arrays. Scale bars: 10 μm. In the plots, middle horizontal bars represent the median, while the bottom and top of each box represent the 25th and 75th percentiles, respectively. At most, the whiskers extend to 1.5 times the interquartile range, excluding data beyond. For range of value under 1,5 IQR, whiskers represent the range of maximum and minimum values. Details for statistical analysis can be found in the Methods section and supplementary Table 2-9. All statistical tests were two-sided.
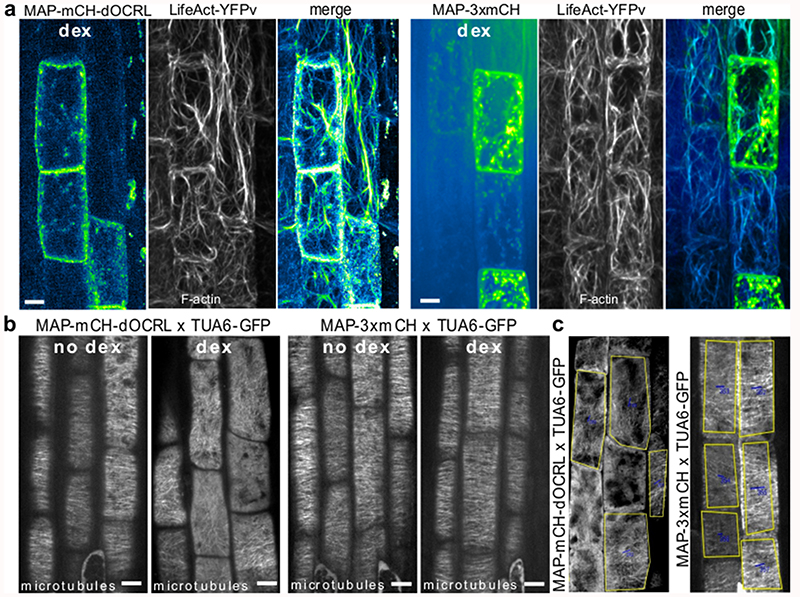
Since phospholipids have been involved in the control of cytoskeleton anchoring and dynamics in plants and animals18–24, we investigated whether PI(4,5)P2 depletion using iDePP might affect the cytoskeleton organization in Arabidopsis. Expression of MAP-mCH-dOCRL in root epidermis did not led to a reduction in polymerized F-actin (Extended Data figure 9) as was reported in animal cells18,25, but, did cause a clear reduction in its dynamics as shown with LifeAct-YFPv (Figure 4f-g, Supplemental movie 5).
To investigate whether PI(4,5)P2 was also affecting the organization of the microtubule cytoskeleton, the cortical microtubules in the root elongation zone were studied, where they are transversely aligned 26, reflecting the unidirectional anisotropic cell expansion (Figure 4h-j, Extended Data figure 8). In the absence of dex in MAP-mCH-dOCRL line, or upon expression of MAP-3xmCH, cortical microtubules of elongating epidermal cells labeled by MBD-GFP or TUA6-GFP were observed to form a network of transversely aligned microtubules, orthogonal to the elongation axis of the cells (Figure 4 h-j, Extended Data figure 8). Depletion of PI(4,5)P2, led to solubilization of the microtubule reporters (Extended Data figure 8), suggesting that PI(4,5)P2 is require for the polymerization of microtubules. Moreover, depletion of PI(4,5)P2, led to a clear loss of microtubule anisotropy (Figure 4h-j, Extended Data figure 8), implying that its organization requires PI(4,5)P2 at the plasma membrane, at least during the elongation process of epidermal root cells.
Here, we describe iDePP, a genetically encoded system that allows to deplete PI(4,5)P2 from the plasma membrane within a few hours in planta. This system by-passes the long term effects of chronic PI(4,5)P2 depletion, for example through a constitutive overexpression of a plasma membrane-targeted 5-phosphatase or genetic elimination of key enzyme responsible for PI(4,5)P2 homeostasis (e.g. PIP5Ks). The use of adequate negative controls demonstrates that the observed effect on PI(4,5)P2 only depends on dOCRL enzymatic activity and is not caused by side effects of the dex-inducible system or the recruitment of an endogenous PI(4,5)P2 phosphatase. We could have used a phospholipase C (PLC) activity to deplete PI(4,5)P2 from the plasma membrane, but we believe a 5-phosphatase produces minimal side effect27. Indeed, PLC activity would increase the production of inositolpolyphosphates, and diacylglycerol (and possibly PA via the subsequent action of diacylglycerol kinases), which act as second messengers in plants20,28. Furthermore, PLCs are often acting on PI4P, which is highly present at plant plasma membranes4,15. By contrast, a 5-phosphatase such as dOCRL specifically dephosphorylates PI(4,5)P2 to produce PI4P. Because PI4P is present in much greater amount at the plasma membrane than PI(4,5)P2 15,20 the additional PI4P produced by the dephosphorylation reaction is unlikely to have a functional impact. This is validated by the absence of any measurable effects on the quantity of various other membrane lipids, including total PIP and PA levels and the localization of PI4P, PS, and PI3P sensors. Thus, iDePP specifically targets PI(4,5)P2 without massively affecting other anionic lipids, although depending on the essays a contribution from the produced PI4P to the observed phenotypes cannot be formally excluded.
Importantly, the release of the fluorescence from mCIT-2xPHPLC PI(4,5)P2 sensor from the plasma membrane was effective in both root and shoot tissues. Here, we mostly focused our analyses on epidermal tissues, because they are easier to image. However, MAP-mCH-dOCRL is induced in the broad expression domain of the UBQ10 promoter and thus iDePP may be used in additional tissues and organs. Indeed, the dex-inducible system that we used was previously shown to be active in many different tissues29. As a testament to the extensive effect of iDePP in seedlings, a reduction of up to 90 % of the total PIP2 levels were observed, indicating the system was active beyond epidermal tissues.
Alternatively, MAP-mCH-dOCRL can easily be expressed in an inducible and tissue specific manner29, as was done with the human PIP5K7. Because the iDePP system is tunable, we predict that the spatial control of PI(4,5)P2 depletion will be of great interest to elucidate additional PI(4,5)P2 functions in cell differentiation and plant development, and will complement the already available tools designed to increase PI(4,5)P2 in an inducible manner7. iDePP can also be used to assess the importance of PI(4,5)P2 to target specific proteins at the plasma membrane, as exemplified for AP-μ2 and SH3P2. While loss-of-function approaches on PIP5Ks previously suggested a general impact of PI(4,5)P2 on clathrin-mediated endocytosis10,12,13, our strategy now allows to specifically pin-point which proteins within this pathway rely on PI(4,5)P2 for localization.
Together, we believe that the iDePP system will have the potential to be useful for numerous research groups working in the fields of plant cell biology, plant development but also immunity30 and reproduction, and will serve as a template to develop additional tools aiming at perturbing membrane lipids in an inducible manner.
Methods
Sequence alignment
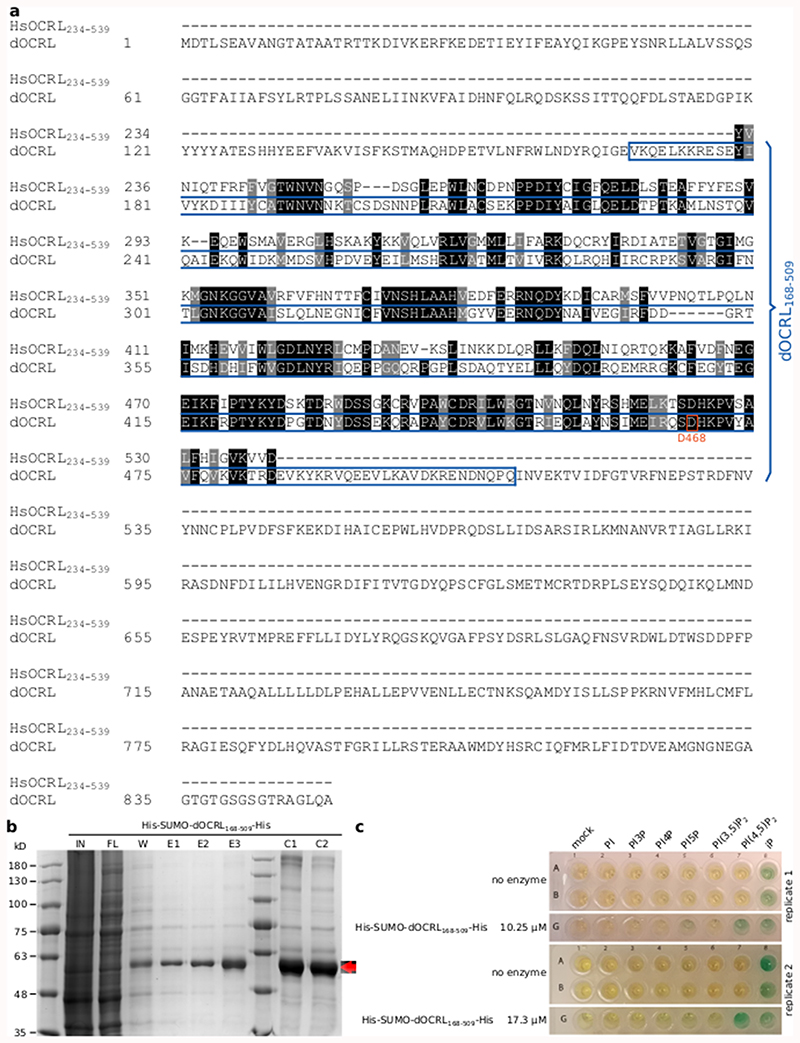
HsOCRL234-539 and dOCRL amino acid sequences were aligned using T-Coffee software (http://tcoffee.crg.cat/apps/tcoffee/do:regular) and the fasta_aln files obtained were then treated with Boxshade (http://www.ch.embnet.org/software/BOX_form.html) and inkscape programs.
Strategy
We initially tested four systems that were previously published in mammalian literature, and were using the following phosphatases: Inp54 (from yeast)31, PSEUDOJANIN (combined yeast and human phosphatases); INPP5E and OCRL (from human) and none of these published phosphatases worked efficiently in plants. We then switched to a new uncharacterized phosphatase from drosophila (dOCRL, which works at the same temperature in plants than in its native organism, see characterization in Extended data file 1). Note that we chose to use an animal enzyme rather than a plant 5-phosphatase because we wanted to be as orthogonal as possible –i.e. to avoid our synthetic enzyme to be regulated by endogenous plant proteins. In addition, we did not simply express a full-length phosphatase, but an engineered chimeric enzyme. To this end, we used only the catalytic domain of the enzyme in order to get rid of all the endogenous targeting sequences from the full-length phosphatase, and then used a plasma membrane targeting sequence to artificially re-target the catalytic activity in a specific subcellular localization of choice. For example, OCRL is known to interact with multiple RABs and with Clathrin. Using the full-length OCRL proteins would have clearly impacted on its localization and would have interfered with our approach. Unfortunately, our structure-function knowledge on plant 5-phosphatases is fragmented compared to animal ones. For example, their catalytic domains have not been well delineated and most importantly the internal sequences responsible for their membrane targeting are largely unknown. This lack of knowledge on plant 5-phosphatase guided our choice toward animal ones. The second module that we had to optimize was the synthetic targeting signal for the plasma membrane localization of the isolated phosphatase domain. In the mammalian field, the most used targeting sequence is the so-called “Lin11” peptide, (and that was used in previous PI(4,5)P2 depletion systems in human cells). We initially used this sequence but we later realized that it destabilized the fusion protein when expressed in stable transgenic plants. We then switched to new targeting sequences, including Lti6b (which did not work efficiently because of rerouting of the chimeric enzyme to the vacuole) and the MAP sequence used in this study.
To our knowledge, such strategy was not used before to manipulate the levels of phosphoinositides in plants. However, overexpression of a human Type I Inositol Polyphosphate 5-Phosphatase was reported in Tabaco cells32. Importantly, this enzyme specifically hydrolyzes inositol 1,4,5-trisphosphate (InsP3) not phosphoinositides. This is thus a fundamentally different type of experiment since InsP3 is soluble, similar to the described enzyme. In our case, we wanted to target PI(4,5)P2 at the plasma membrane, while most known PI(4,5)P2 5-phosphatase actually localize in different membrane compartments.
Cloning
A synthetic gene (https://eu.idtdna.com/pages) corresponding to dOCRL 168-159 codon optimized for expression in Arabidopsis thaliana (see sequence below) was amplified and flanked with NcoI and XhoI restriction sites by PCR and cloned into pMH-HS-SUMO vectors33 by restriction and ligation (see corresponding primers in Supplementary Table 1).
dOCRL168-159
gtaaagcaagagttaaagaagcgagagagtgagtatatcgtctataaggacattatcatttactgcgctacgtggaacgtgaacaataagacatgcagtgatagtaacaatccgctaagagcgtggctcgcatgctccgaaaagccgccagacatatatgcgataggtcttcaggagctagacaccccgaccaaggctatgctcaatagtacccaagtccaagcaattgaaaaacagtggatagataagatgatggacagtgtgcatcctgacgtagagtacgagattttgatgtctcaccgattagttgcgacgatgcttactgtcatcgttagaaaacagcttaggcagcacattatacgttgtcgacccaaatccgtggcccgtggaatattcaacacgctcggaaataagggaggggtggcaatatccctgcagttgaatgagggaaacatatgttttgttaactcccatctggctgcacatatggggtacgtcgaagagaggaatcaagattacaacgctattgtcgaaggcatcaggtttgacgacggtagaactatctccgaccatgaccatatattttgggtaggagacttgaattatcgaatacaggaacctccgggacagcagcgtcctgggccgttgagtgacgcacagacatacgagctcttattacaatacgatcaactccgtcaagagatgcgaaggggaaaatgcttcgaaggttacacggagggggagattaagttcagaccgacatacaaatatgatcctggaacagacaattatgactcttcagagaaacagcgagcacctgcatactgcgatagagtgctatggaaaggtacacgtatcgaacagctagcatacaacagcataatggaaattaggcaaagcgatcataagccagtttatgcggttttccaggtgaaggtaaagacacgagatgaggtgaagtataagcgtgttcaagaggaggtgctaaaggcggttgacaaaagggagaacgataatcagccacagtaa
UBQ10pro:GVG (UBQ10pro:GVG-tE9::6xUAS-35Smini::rigorously)
construct was cloned using Gibson technique and cloned into a pDONR-P4P1R gateway vector (thermofisher cat# 12537023). MAP-mCH was amplified from a mCH/pDONR207 matrix using 5’ phosphorylated primers and ligated into MAP-mCH/pDONR207. The synthetic dOCRL 168-509 sequence (see above) with an N-terminal GAGARS linker (ggggcaggagccagatcc) was flanked with attB2R and attB3 sequences and recombined by BP gateway reaction into pDONR-P2RP3. Site directed mutagenesis of dOCRL was performed by PCR on dOCRL168-509/pDONR-P2RP3. using 5’ phosphorylated primers, and subsequent ligation of the linear plasmid circularized dOCRL168-509 D468G/pDONR-P2RP3. 2xmCHERRY/pDONR-P2RP3 was already available.
UBQ10pro
GVG:MAP-mCH-dOCRL168-509/pH7m34GW, UBQ10pro:GVG:MAP-mCH-dOCRL168-509D468G/pH7m34GW, and UBQ10pro:GVG:MAP-3xmCH//pH7m34GW were obtained by triple LR gateway reaction between UBQ10pro:pDONR-P4P1R, MAP-mCH-pDONR207, the corresponding pDONR-P2RP3 vectors, and pH7m34GW destination vector34. See key resource table for details.
Protein purification
BL21(DE3) pLys competent E. coli (Novagen) were transformed with pMH-HS-Sumo-dOCRL168-509 vector (KanR) and cultured on Luria Broth (LB) liquid or 1 % agar medium supplemented with 50 μM Kanamycin. Two 800 ml 2xTY (10 g/L yeast extract; 16 g/L tryptone; 5 g/L NaCl) liquid cultures supplemented with kanamycin were inoculated with 20 ml from an overnight pre-culture of 150 ml, and left at 37°C with 200 rpm shaking. When OD600nm reached 0.6, (i) protein expression was induced by adding IPTG 0.625 mM final concentration, (ii) 45 ml of glycerol were added, and (iii) temperature was lowered by transferring the liquid culture on ice for 15 min. The induced culture was then grown for 18 h at 18°C with 200 rpm shaking. Bacteria were then centrifuged 30 min at 6000 g and 4 °C, and bacterial pellets scooped, resuspended in lysis buffer (200 mM NaCl; 20 mM Tris-HCl pH 7.5; 1 mM PMSF) with the aid of vortex, flash frozen and stored at -80°C. The day of protein purification, Bacterial pellets corresponding to 33 % (270 ml of the culture) were thawed at room temperature. For then on, all steps were carried on ice. Bacteria were then sonicated in four pulses of 30 sec and 30 W. Sonicated lysates were centrifuge 30 min at 13000 g and 4°C in new falcon tubes.
Supernatants, corresponding to the soluble fraction of the lysate, were then diluted to 50 ml final volume and subjected to affinity chromatography. 2 ml of 50 % HIS-Select® Cobalt affinity resin (Sigma) was washed with milli-Q water and then with equilibration buffer (20 mM imidazole; 200 mM NaCl; 20 mM Tris-HCl pH 7.5; 1 mM). Bacterial lysate was run thought the resin twice. Then, resin was washed with wash buffer (40 mM imidazole; 200 mM NaCl; 20 mM Tris-HCl pH 7.5; 1 mM).
Elution was performed with three fractions of 2 ml of elution buffer (250 mM imidazole; 200 mM NaCl; 20 mM Tris-HCl pH 7.5; 1 mM). Elution fractions were then either directly concentrated (C1 fraction) or kept at 4 °C for a maximum of 16 h before concentration (C2 fraction). Concentration was achieved using 0.5 ml 30 kD molecular-weight cut-off concentration columns (Millipore UFC 501024) centrifuged at 4000 g and 4 °C until 2 ml of elution reached a final volume of 400 μL. Aliquots of 20 μL were sampled from induced bacterial culture (IN), flow thought (FT), wash (W), elution fraction (E1, E2 and E3) and concentrated fractions (C1 and C2). They were mix with SDS loading buffer (0.0625 M Tris HCl pH 6.8; 2.5 % SDS; 2 % DTT (0.13 M); 10 % glycerol; bromophenol blue), vortexed and submitted for 5 min to 95°C treatment, and then stored at -20°C. Protein concentration in concentrated fraction was calculated from absorbance at 280 and 260 nm measured with a nanodrop and calculated extinction coefficient (https://web.expasy.org/protparam/). His-Sumo-dOCRL168-509-His has an expected molecular weight of 54.82 kD.
Malachite green phosphatase assay
Phosphate released was monitored using a Malachite green phosphatase kit (Echelon) and water soluble short chain (di:C8) phosphoinositides (Echelon) stored in glass vials at -20°C as stock solutions of 500 μM in CHCl3/MetOH (9:1; PI, PI3P, PI4P, PI5P), or in CHCL3/MetOH/H2O (5:5:1; PI(3,5)P2, PI(4,5)P2). di:C16 Phosphatidylserine (PS; Echelon) was stored in glass vials at -20°C as a stock solution of 10 mM in CHCl3/MetOH (9:1). Before malachite green assay, inorganic solvent was evaporated by centrifugation in a speed vac at 45 °C for 15 min. Dried lipids (PS and one phosphoinositide) were then resuspended in reaction buffer (200 mM NaCl 50 mM Tris-HCl pH 7.5; 10 mM KCl; 5 mM MgCl2) with a 30 sec of sonication at full power. Reaction were carried out in a 20 μL volume consisting in reaction buffer containing 500 μM of PS, 100 μM of a given short chain phosphoinositide species (except for the “mock” condition), and 5 μL of concentrated protein fractions (C1 or C2; except of “no enzyme” condition) corresponding to the final concentration. In the “iP” condition corresponds to addition of inorganic phosphate. Malachite green assay was carried out at 29 °C for 75 min, with gentle shaking every 15 min, and reaction was eventually quenched by the addition of 90 μL of malachite green reagent. Pictures were taken 25 min after the addition of
Malachite green reagent, and green coloration qualitatively indicated a significant phosphate release. This Experiment was carried out independently three times, with either one or two replicates each time, and gave consistent results.
Growth condition and plant materials
Arabidopsis thaliana Columbia-0 (Col-0) accession was used as wild type (WT) reference genomic background throughout this study. Arabidopsis thaliana seedlings in vitro on half Murashige and Skoog (½ MS) Basal Medium supplemented with 0.8 % plant agar (pH 5.7) in continuous light conditions at 21 °C. Plants were grown in soil under long-day conditions at 21 °C and 70 % humidity 16 h daylight.
Plant transformation and Selection
Plant were transformed and selected as previously described35. Each construct was transformed into Agrobacterium tumefaciens C58 GV3101 strain and selected on YEB media (5g/L beef extract; 1g/L yeast extract; 5g/L peptone; 5g/L sucrose; 15g/L bactoagar; pH 7.2) supplemented with antibiotics (Spectinomycin, Gentamycin, Rifampicin). After two days of growth at 28 °C, bacteria were scooped and re-suspended in roughly 200 mL of transformation buffer (10mM MgCl2; 5 % sucrose; 0.25 % silweet) and Col-0 Arabidopsis were transformed by dipping. Primary transformants (T1) were selected in vitro on the appropriate antibiotic/herbicide (glufosinate for mCIT, hygromycin for mCH-tagged proteins). Approximately 20 independent T1s were selected for each line. In the T2 generation at least two independent transgenic lines were selected using the following criteria when possible: (i) good expression level in the root for detection by confocal microscopy upon dex induction, (ii) relatively uniform expression pattern, (iii) line with no obvious abnormal developmental phenotypes.
Lines were rescreened in T3 using similar criteria as in T2 with the exception that we selected homozygous lines (100 % resistant). At this step, we selected one transgenic line for each marker that was used for further analyses and crosses. NPA treatments were carried out as previously described36. Plants were further kept on NPA-containing medium for all the duration of the experiments.
Induction of gene expression with dexamethasone
In order to study the effect of iDePP system in root, seedlings of each genotype were grown vertically for five to seven-day in vitro on half Murashige and Skoog (½ MS) Basal Medium supplemented with 0.8 % plant agar (pH 5.7) in continuous light conditions at 21 °C. Part of the seedlings were next carefully transferred to ½ MS medium plates, 0.8 % agar (pH 5.7), containing 5 μM dexamethasone. Both non-treated seedlings and dex-treated seedlings were, all together, grown back in continuous daylight (see before). 16 h after transfer on dexamethasone plates, (more or less 90 min) seedlings were mounted in ½ MS medium and imaged by confocal microscopy. For growth experiments, seedlings were directly sawn on ½ MS medium on line and let to grow vertically up to 7 days. For root hair phenotyping, five days old plantlets grown on root hair medium, ½ MS medium, 2 % sucrose, 0.6 % phytagel, 0.01 % myo-inositol and vitamins (0.5mg/L of nicotinic acid and vitamin B6, 0.1mg/L vitamin B1 and 2mg/L glycine), were transferred in 5 μM dexamethasone containing root hair medium. Root were imaged with a manual stereomicroscope Nikon SMZ18 (mag.: x7.5 to x135, zoom ratio 18:1, LED diascopic illumination base, LED fluorescence light source (CoolLed PE300 Lite)), equipped with a camera Hamamatsu Orca Flash9.0LT (Nikon MIS acquisition software) and control by a DELL precision 3630 computer.
For cotyledon analysis, seeds were sterilized and sown on Arabidopsis medium (MS medium without sugars and vitamins) for 7 days. Seedling were then carefully transferred on MS medium (without sugars and vitamins) supplemented with 5μM of Dexamethasone. Cotyledons were placed at the surface of the medium, as flat as possible, while the rest of the plant was immerged into the medium.
Shoot apex were dissected as described by 37 and then placed in a shoot apex medium as described by 36 supplemented with 5μM of Dexamethasone. Upon dex treatment, MAP-mCH-dOCRL was mainly induced in the boundary region of the shoot apical meristem and had a weak or no expression in the central zone. This result was consistent with our previous observations, since MAP-mCH-dOCRL is induced in the UBQ10 promoter expression domain, which is low at the center of the shoot apical meristem in our conditions. We observed a strong correlation between MAP-mCH-dOCRL expression and its impact on mCIT-2xPHPLC localization. Indeed, in the center of the shoot meristem, MAP-mCH-dOCRL was not expressed and mCIT-2xPHPLC labelling clearly accumulated at the cell periphery, likely representing the plasma membrane. By contrast, in the boundary zone, where MAP-mCH-dOCRL was expressed, mCIT-2xPHPLC was not localized sharply at the cell edge and instead filled the entire volume of the cell except the nucleus. This suggested that, similar to root epidermis, expression of MAP-mCH-dOCRL in the shoot epidermis displaced mCIT-2xPHPLC from the plasma membrane into the cytosol, and thus efficiently erased PI(4,5)P2 from the plasma membrane.
Because UBQ10-drivenMAP-mCH-dOCRL induction was not uniform at the shoot apical meristem, we decided to test the iDePP system on N-1-Naphthylphthalamic Acid (NPA) grown shoot meristems. NPA is a polar auxin transport inhibitor, which induces naked shoot apical meristem without any organ. In this condition, the UBQ10 promoter is more uniformly expressed throughout the meristem, including the central and peripheral zones, as exemplified by the homogenous expression of mCIT-2xPHPLC. To generate naked meristems in vitro, seeds were first directly sown on Arabidopsis medium, supplemented with 10 μM NPA. Seedlings with naked meristems were then selected and transferred to a shoot apex medium without NPA and supplemented with 5μM of Dexamethasone. Images were taken at 18 h after induction. To test organ initiation on NPA-treated meristems, seedlings with naked meristems were selected and then transferred on shoot apex medium without NPA and supplemented with 10μM of Dexamethasone. These seedlings were then placed for 4days in long day conditions (16h light, 8h dark, 21°C).
Note that we recommend the following when using iDePP
i) if researchers want to assess fast effects of PI(4,5)P2 depletion, they can do so starting from 90–120 min after dex induction but they should double-check the induction of MAP-mCH-dOCRL in these cells (and if possible the plasma membrane release of mCIT-PHPLC), and ii) if researchers want to quantify the effects on a large number of cells and if rapid depletion is not critical, they may use a 12–16 hours dex treatments in order to obtain more cells with PI(4,5)P2 depletion.
Time-lapse imaging
Time lapse imaging of cell division and root hair growth were performed mostly as described previously 38, with few amendments. In brief, five days old Arabidopsis seedlings were transferred in a chambered cover glass (Lab-Tek II, www.thermoscientific.com), which contained (i) 3 ml of ½ MS medium (pH 5.7) containing 0.8 % plant agar (Duchefa, http://www.duchefa-biochemie.nl/) and 5 μM dexamethasone and (ii) 800 μL of ½ MS medium (pH 5.7) containing 5 μM dexamethasone. Epidermal cells in the meristematic region of the root tip were subjected to time-lapse imaging with spinning disk confocal microscope. Up to three roots were observed simultaneously and images were collected at different Z-positions every 5 min for 8 h. Tracking of growing roots was automatically performed 38.
Microscopy setup
Images of cotyledons, shoot apical meristem and NPA meristems were acquired with a Leica SP8 upright scanning confocal microscope equipped with a water immersion objective (HCX IRAPO L 25x/ 0.95 W). Fluorophores were excited using Led laser (Leica Microsystems, Wetzlar, Germany) emitting at wavelengths of 514 nm for mCIT fluorochrome and 552 nm for mCH fluorochrome. Emission fluorescence was recovered between 519nm - 548nm for mCIT and between 604 nm - 662 nm for mCH. The following scanning settings were used: pinhole size 1 AE, scanning speed of 8000 Hz (resonant scanner), frame averaging between 8 and 10, Z intervals of 0.45 μm.
NPA-treated meristems used to test organ initiation with reduced PI(4,5)P2 content were imaged with a Leica MZ12, equipped with a camera AxioCam ICc 5 (Zeiss). Images were acquired with Zen software, right after and 4 days after the transfers of NPA-treated meristems on the shoot apex medium.
For FM4-64 imaging, root elongation zones were acquired with a confocal laser scanning Zeiss LSM 800 spectral microscope. Acquisitions were performed using the same settings (PMT voltage, laser power and detection wavelengths) with an excitation wavelength: 488 nm, and emission: 515 /640 nm.
Imaging was performed with the following spinning disk confocal microscope set up: inverted Zeiss microscope (AxioObserver Z1, Carl Zeiss Group, http://www.zeiss.com/) equipped with a spinning disk module (CSU-W1-T3, Yokogawa, www.yokogawa.com) and a ProEM+ 1024B camera (Princeton Instrument, http://www.princetoninstruments.com/) or Camera Prime 95B (www.photometrics.com) using a 63x Plan-Apochromat objective (numerical aperture 1.4, oil immersion). GFP and mCIT were excited with a 488 nm laser (150 mW) and fluorescence emission was filtered by a 525/50 nm BrightLine! single-band bandpass filter (Semrock, http://www.semrock.com/), mCH was excited with a 561 nm laser (80 mW) and fluorescence emission was filtered by a 609/54 nm BrightLine! single-band bandpass filter (Semrock). For quantitative imaging, pictures of epidermal root meristem cells were taken with detector settings optimized for low background and no pixel saturation. Care was taken to use similar confocal settings when comparing fluorescence intensity or for quantification. Signal intensity was color-coded (green fire blue scale, https://fiji.sc/).
Drug treatments
ES9-17 was stored at -20°C as a 30 mM stock solution in DMSO. Seedlings were incubated in dark without shaking for the indicated duration (± 7 min) in liquid ¼ MS (pH 5.7) in wells containing 1 % DMSO to help solubilization of ES9-17, and either 1 μM ES9-17 or the corresponding additional volume of DMSO (mock treatment).
Biochemical quantification of PIP2 levels in iDePP lines
Five-days-old seedlings were transferred to 2 ml Safe-lock Eppendorf tubes containing 190 μL labelling buffer (2.5 mM MES (2-[N-Morpholino]ethane sulfonic acid) (pH 5.7 with KOH), 1 mM KCl) and 5 μM dex, with each tube containing three seedlings. To quantify their PIP2 levels, seedlings were metabolically labelled with radioactive phosphate by incubating them overnight for ~16-20 hrs with 10 μL (5-10 μCi) carrier-free32P-PO4 3- (32Pi; PerkinElmer, The Netherlands) in labelling buffer. Incubations were stopped by adding 50 μL of 50 % (w/v) perchloric acid and lipids extracted 39. PIP2 was separated from the rest of the phospholipids by thin-layer chromatography (TLC) using K-oxalate-impregnated and heat-activated silica gel 60 plates (20x20 cm; Merck) and an alkaline TLC solvent, containing chloroform/methanol/25 % ammonia/water (90:70:4:16) 40. Each lane contained 1/5th of the extract. Radioactivity was visualized by autoradiography and quantified by phosphoimaging (Typhoon FLA 7000, GE Healthcare). PIP2 levels were quantified as percentage of total 32P-labelled phospholipids. Experiments were performed in duplo or triplo and repeated 2-5 times, depending on the genotype.
3D projections, dissociation indexes and anisotropy
The fluorescence intensity in root hairs was obtained using Fiji tool. A line of 66 pixels was drawn at the proximity of the root hair tip and the intensity of grey was plot using the Plot Profil tool. The values were transferred in Excel to obtain the graph. Shoot apical meristem and NPA-induced meristem projected images were obtained by using the MorphoGraphX software (https://www.mpipz.mpg.de/MorphoGraphX). The signal projections were generated by extracting the fluorescent signal at the surface of the meristem (between 2- and 5 μm from the meristem surface) and by projecting it on the cellular mesh. Projection of root epidermal cells were obtained using Fiji “Max intensity projection” tool on Z-stacks of 21 slices distant of 0.5 μm from each other.
Dissociation indexes of membrane lipid fluorescent biosensors were measured and calculated as previously described 35. Briefly, we calculated “indexNoDex” in the mock condition, defined as the ratio between the fluorescence intensity (Mean Grey Value function of Fiji software) measured in two elliptical regions of interest (ROIs) from the plasma membrane region (one at the apical/basal plasma membrane region and one in the lateral plasma membrane region) and two elliptical ROIs in the cytosol. Next, we measured a similar ratio after dexamethasone treatments (“indexDex”). The dissociation index is the ratio of (indexNoDex)/(indexDex). This dissociation index reveals the degree of relocalization of the fluorescent reporters from the plasma membrane to the cytosol, between the non-treated and perturbed conditions (pharmacological treatment or mutant). Dissociation indexes of mCIT-2xPHPLC during time-lapse induction of MAP-mCH-dOCRL were measured in 30 cells of the same root at each time point.
For quantification of the anisotropy of microtubule arrays in the different transgenic lines, Maximal z projection of z-stack of epidermal root cell in the elongation zone were obtained, using Fiji. In fibriltool, the value for ‘Multiply line length by’ was set up at 1 and ROI using the built-in Polygon tool were generated for individual cells (>100 cells per conditions). The data from the log file was used to extract the average anisotropy of microtubule arrays (a score between 0 and 1) and to run statistical analyses.
Statistical analysis
For dissociation index and anisotropy, we performed all our statistical analyses in R (v. 3.6.1, (R Core Team, 2019), using R studio interface and the packages ggplot2 41, lme4 42, car 43, multcomp 44 and lsmeans 45. For each model assuming normality, we plotted residuals values against normal quantiles to confirm their normal distribution. Graphs were obtained with R and R-studio software, and customized with Inkscape (https://inkscape.org). Details can be found in Supplementary Tables 2-9.
Extended Data
Extended Data Fig. 1.
Extended Data Fig. 2.
Extended Data Fig. 3.
Extended Data Fig. 4.
Extended Data Fig. 5.
Extended Data Fig. 6.
Extended Data Fig. 7.
Extended Data Fig. 8.
Extended Data Fig. 9.
Supplementary Material
Acknowledgments
We are grateful to the SiCE group (RDP, Lyon, France) in particular T. Gaude and V. Bayle (RDP, Lyon), and to Y. Boutté (LBM, Bordeaux, France), D. Van Damme (VIB, Ghent, Belgium), T. Stanislas (ZMBP, Tübingen, Germany), F. Besnard and N. Doll (RDP, Lyon, France), for comments and discussions. We as well thank former interns L. Courgeon (ENS de Lyon) and A. Bauer (ENS de Lyon), who performed preliminary experiments during early steps of this project and helped with plant labor, and K. Grünwald. We thank J. Berger, P. Bolland, and A. Lacroix from our plant facility. We acknowledge the contribution of SFR Biosciences (UMS3444/CNRS, US8/Inserm, ENS de Lyon, UCBL) facilities: C. Lionet, E. Chatre and J. Brocard at the LBI-PLATIM-MICROSCOPY for assistance with imaging. We are also grateful to E. Russinova (VIB, Ghent, Belgium) for kindly providing us ES9-17; S. Bednarek for sharing markers and for discussions, as well as M. Uyttewaal (INRAE, Versailles, France) for the MBD-GFP marker. We would like to thanks E. Isono (University of Konstanz, Konstanz, Germany) for sharing SH3P2-GFPs transgenic line with us. We also acknowledge precious help from R. Boisseau (OBEE department, University of Montana, USA) regarding the statistical analysis. This work was supported by the ERC (no. 3363360-APPL under FP/2007–2013, YJ; EU-FET 828753, TM), the Netherlands Organisation for Scientific Research (NWO 867.15.020; 711.017.005, TM), Austrian Academy of Science through the Gregor Mendel Institute (Y.B), French National Research Agency ANR (caLIPSO; ANR-18-CE13-0025-02; YJ), ANR JC/JC JUNIOR INVESTIGATOR GRANT (INTERPLAY; ANR-16-CE13-0021; MCC, AF) and a SEED FUND ENS LYON-2016 (MCC).
Footnotes
Author Contributions Statement
MD, performed the experiments, analyzed the data, (including statistics), and wrote the paper; AL, performed the experiments and analyzed the data, (including statistics); LC, performed the experiments and analyzed the data; AF, performed the experiment; FDS, performed the biochemical quantification of the PIP2 and analyzed the data; JB, performed the experiment of biochemistry; OH, supervised the work and help with the writing of the paper; YB, supervised the work of biochemistry and analyzed the data; TM, supervised the work of the biochemical quantification of PIP2 and analyzed the data; YJ, supervised the work and wrote the paper; MCC, performed the experiments and analyzed the data, supervised the work and wrote the paper.
Competing Interests Statement
There is no competing interest
Data Availability
The Arabidopsis lines generated in this study are available from the corresponding author on reasonable request.
References
- 1.Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Developmental cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noack LC, Jaillais Y. Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Current opinion in plant biology. 2017;40:22–33. doi: 10.1016/j.pbi.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Noack LC, Jaillais Y. Functions of Anionic Lipids in Plants. Annual review of plant biology. 2020;71:71–102. doi: 10.1146/annurev-arplant-081519-035910. [DOI] [PubMed] [Google Scholar]
- 5.Heilmann I. Phosphoinositide signaling in plant development. Development. 2016;143:2044–2055. doi: 10.1242/dev.136432. [DOI] [PubMed] [Google Scholar]
- 6.Colin LA, Jaillais Y. Phospholipids across scales: lipid patterns and plant development. Current opinion in plant biology. 2019;53:1–9. doi: 10.1016/j.pbi.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Gujas B, et al. Perturbing phosphoinositide homeostasis oppositely affects vascular differentiation in Arabidopsis thaliana roots. Development. 2017;144:3578–3589. doi: 10.1242/dev.155788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ischebeck T, Stenzel I, Heilmann I. Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. The Plant cell. 2008;20:3312–3330. doi: 10.1105/tpc.108.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbosa IC, et al. Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development. 2016;143:4687–4700. doi: 10.1242/dev.137117. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, et al. Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. The Plant cell. 2010;22:4031–4044. doi: 10.1105/tpc.110.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dejonghe W, et al. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nature chemical biology. 2019 doi: 10.1038/s41589-019-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mei Y, Jia WJ, Chu YJ, Xue HW. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell research. 2012;22:581–597. doi: 10.1038/cr.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ischebeck T, et al. Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. The Plant cell. 2013;25:4894–4911. doi: 10.1105/tpc.113.116582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He K, et al. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature. 2017;552:410–414. doi: 10.1038/nature25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon ML, et al. A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants. 2016;2:16089. doi: 10.1038/nplants.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadeyne A, et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell. 2014;156:691–704. doi: 10.1016/j.cell.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 17.Narasimhan M, et al. Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. Elife. 2020;9 doi: 10.7554/eLife.52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raucher D, et al. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, et al. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. The Plant cell. 2012;24:4555–4576. doi: 10.1105/tpc.112.104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Current opinion in plant biology. 2011;14:489–497. doi: 10.1016/j.pbi.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Gungabissoon RA, Jiang C-J, Drøbak BK, Maciver SK, Hussey PJ. Interaction of maize actin-depolymerising factor with actin and phosphoinositides and its inhibition of plant phospholipase C. The Plant Journal. 1998;16:689–696. doi: 10.1046/j.1365-313x.1998.00339.x. [DOI] [Google Scholar]
- 22.Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. The Journal of biological chemistry. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 23.Huang S, Blanchoin L, Kovar DR, Staiger CJ. Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. Journal of Biological Chemistry. 2003;278:44832–44842. doi: 10.1074/jbc.M306670200. [DOI] [PubMed] [Google Scholar]
- 24.Staiger CJ, Gibbon BC, Kovar DR, Zonia LE. Profilin and actin-depolymerizing factor: modulators of actin organization in plants. Trends in plant science. 1997;2:275–281. [Google Scholar]
- 25.Shibasaki Y, et al. Massive actin polymerization induced by phosphatidylinositol-4-phosphate 5-kinase in vivo. The Journal of biological chemistry. 1997;272:7578–7581. doi: 10.1074/jbc.272.12.7578. [DOI] [PubMed] [Google Scholar]
- 26.Vaughn L, et al. 2011. pp. 307–326.
- 27.Balla T. Imaging and manipulating phosphoinositides in living cells. J Physiol. 2007;582:927–937. doi: 10.1113/jphysiol.2007.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenzo-Orts L, Couto D, Hothorn M. Identity and functions of inorganic and inositol polyphosphates in plants. The New phytologist. 2020;225:637–652. doi: 10.1111/nph.16129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marques-Bueno MM, et al. A versatile Multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. The Plant journal : for cell and molecular biology. 2016;85:320–333. doi: 10.1111/tpj.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saile SC, et al. Coiled-coil and RPW8-type immune receptors function at the plasma membrane in a phospholipid dependent manner. bioRxiv. 2020:2020.2011.2018.388520. doi: 10.1101/2020.11.18.388520. [DOI] [Google Scholar]
- 31.Johnson CM, Chichili GR, Rodgers W. Compartmentalization of phosphatidylinositol 4, 5-bisphosphate signaling evidenced using targeted phosphatases. Journal of Biological Chemistry. 2008;283:29920–29928. doi: 10.1074/jbc.M805921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perera IY, Love J, Heilmann I, Thompson WF, Boss WF. Up-regulation of phosphoinositide metabolism in tobacco cells constitutively expressing the human type I inositol polyphosphate 5-phosphatase. Plant physiology. 2002;129:1795–1806. doi: 10.1104/pp.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karimi M, Depicker A, Hilson P. Recombinational cloning with plant gateway vectors. Plant physiology. 2007;145:1144–1154. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platre MP, et al. A Combinatorial Lipid Code Shapes the Electrostatic Landscape of Plant Endomembranes. Developmental cell. 2018;45:465–480 e411. doi: 10.1016/j.devcel.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Grandjean O, et al. In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. The Plant cell. 2004;16:74–87. doi: 10.1105/tpc.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanislas T, Hamant O, Traas J. In-vivo analysis of morphogenesis in plants. Methods Cell Biol. 2017;139:203–223. doi: 10.1016/bs.mcb.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Doumane M, Lionnet C, Bayle V, Jaillais Y, Caillaud MC. Automated Tracking of Root for Confocal Time-lapse Imaging of Cellular Processes. Bio Protoc. 2017;7 doi: 10.21769/BioProtoc.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zarza X, et al. Lipid kinases PIP5K7 and PIP5K9 are required for polyamine-triggered K(+) efflux in Arabidopsis roots. Plant J. 2020;104:416–432. doi: 10.1111/tpj.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munnik T, Zarza X. Plant Lipid Signaling Protocols. Springer; 2013. pp. 3–15. [Google Scholar]
- 41.Wickham H. ggplot2: elegant graphics for data analysis. springer; 2016. [Google Scholar]
- 42.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. 2014 arXiv preprint arXiv:1406.5823. [Google Scholar]
- 43.Fox J, Weisberg S. 2011.
- 44.Hothorn T, Bretz F, Westfall P, Heiberger RM. Multcomp: simultaneous inference for general linear hypotheses. R package version. 2008 1.0-3. [Google Scholar]
- 45.Lenth R, Lenth MR. Package ‘lsmeans’. The American Statistician. 2018;34:216–221. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Arabidopsis lines generated in this study are available from the corresponding author on reasonable request.