Abstract
Tissue development and homeostasis are controlled by mechanical cues. Perturbation of the mechanical equilibrium triggers restoration of mechanostasis through changes in cell behavior, while defects in these restorative mechanisms lead to mechanopathologies, for example, osteoporosis, myopathies, fibrosis or cardiovascular disease. Therefore, sensing mechanical cues and integrating them with the biomolecular cell fate machinery is essential for the maintenance of health. The Notch signaling pathway regulates cell and tissue fate in nearly all tissues. Notch activation is directly and indirectly mechanosensitive, and regulation of Notch signaling, and consequently cell fate, is integral to the cellular response to mechanical cues. Fully understanding the dynamic relationship between molecular signaling, tissue mechanics and tissue remodeling is challenging. To address this challenge, engineered microtissues and computational models play an increasingly large role. In this Review, we propose that Notch takes on the role of a ‘mechanostat’, maintaining the mechanical equilibrium of tissues. We discuss the reciprocal role of Notch in the regulation of tissue mechanics, with an emphasis on cardiovascular tissues, and the potential of computational and engineering approaches to unravel the complex dynamic relationship between mechanics and signaling in the maintenance of cell and tissue mechanostasis.
Keywords: Notch signaling, Mechanotransduction, Engineered model systems, Computational modeling, Cardiovascular mechanics
Introduction
Biological tissues constantly respond to their environment to obtain and maintain biological, chemical and mechanical homeostasis. Changes in the mechanical state lead to cellular responses promoting growth and/or remodeling to restore homeostasis. Loss of mechanical homeostasis can trigger the onset of mechanopathologies, including osteoporosis, myopathies, fibrosis and, ultimately, organ failure (Ingber, 2003; Humphrey et al., 2014; Pickup et al., 2014). Cardiovascular development and disease are textbook examples of processes intricately intertwined with changes in tissue mechanics.
Tissues emerge from the collective mechanical properties and behavior of individual cells, and cell fate decisions are at the core of tissue homeostasis. To understand tissue homeostasis, it is essential to understand the processes that integrate the different mechanical and biomolecular cues driving cell fate decisions. The receptors for mechanical cues have been identified as an ensemble of membrane proteins, channels, structural cytoskeletal proteins and mechanosensory organelles, such as the cilium and the cell nucleus (Ferreira et al., 2019; Kirby and Lammerding, 2018). These protein complexes either actively probe the environmental mechanics by exerting force on their surroundings, or sense loads exerted on the cell by changing conformation (Hoffman et al., 2011). In doing so, they convert the mechanical signals into biochemical signals through the process of mechanotransduction (Kindberg et al., 2020; Martino et al., 2018). Understanding how mechanical signals affect cell and tissue functionality can enable the development of medical approaches to restore tissue homeostasis and advance strategies for tissue repair, engineering and regeneration (Drews et al., 2020; Stassen et al., 2017; Wissing et al., 2017). Several components of cellular mechanotransduction cascades have been identified, but a comprehensive picture of the molecular translation from distinct mechanical cues to cell fate decisions remains to be established. Novel mechanosensitive proteins are regularly discovered, such as the recent discovery of plexin D1 as a mechanosensitive receptor (Mehta et al.,2020), with more likely awaiting discovery.
The Notch signaling pathway has an important role in cell fate decisions (Artavanis-Tsakonas et al., 1999) and tissue organization (Sjöqvist and Andersson, 2017). Notch is evolutionary ancient, and has been proposed to have evolved from the fusion of several transmembrane proteins with functions in cell adhesion (Murata and Hayashi, 2016). This evolutionary heritage, combined with the traditional cell–cell signaling function, places Notch at a position to integrate cues coming from the extracellular matrix (ECM), mechanics and direct cellular neighbors (as reviewed by LaFoya et al., 2016). In this Review, we describe recent studies that have uncovered interactions between Notch and mechanical forces, and highlight the tools that might be adopted for the advancement of this field in the future. Given the central role of Notch and mechanics in cardiovascular development and disease, we emphasize cardiovascular tissues, although our conclusions can be generalized to other tissues.
Notch, cell fate and mechanics – a mechanotransductory feedback loop through reciprocal interactions
The canonical Notch signaling pathway has been thoroughly investigated (see Box 1 for an overview). Briefly, Notch ligands interact with Notch receptors on juxtaposed cells. This triggers cleavage of the Notch intracellular domain (NICD), which then translocates to the nucleus and regulates transcription (Bray and Gomez-Lamarca, 2018; Kopan and Ilagan, 2009). Recently, it has been uncovered that mechanical forces regulate Notch signaling via direct or indirect interactions (Gordon et al., 2015; Mack et al., 2017). Conversely, Notch regulates transcriptional programs that are involved in cell fate, including ECM homeostasis, cell and tissue morphogenesis, and cell and tissue mechanics (Lloyd-Lewis et al., 2019; Loerakker et al., 2018). This gives rise to a reciprocal interplay between Notch and mechanics with feedback mechanisms in place, enabling Notch to be an important regulator of mechanical homeostasis.
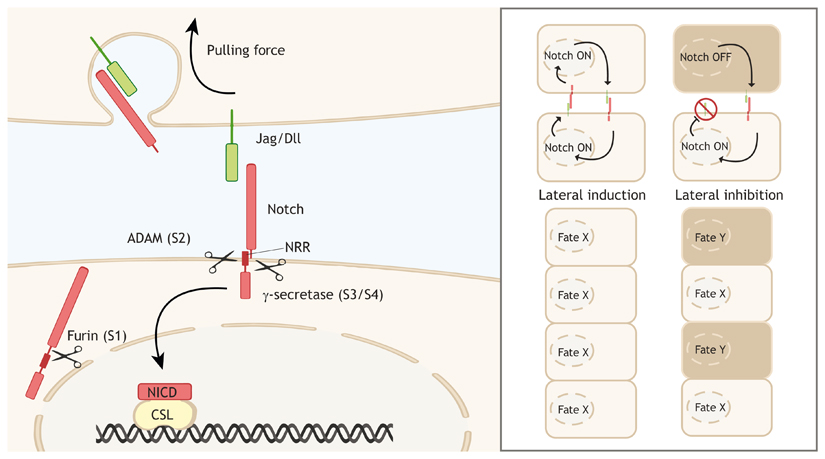
Box 1. Brief overview of Notch signaling regulation.
The canonical signal carried by Notch is linear – a ligand on a cell membrane binds a receptor on the membrane of a neighboring cell, activating a transcription factor complex in the signal-receiving cell (see left-hand panel of the figure). In humans, the pathway consists of the receptor proteins Notch1, Notch2, Notch3 and Notch4, and the ligand proteins delta-like-ligand (Dll)1, Dll3, Dll4, jagged 1 (Jag1) and jagged 2 (Jag2) (Dll/Jag in left-hand panel) (Mašek and Andersson, 2017). Both receptors and ligands consist of an extracellular domain, a transmembrane domain and an intracellular domain. The receptor precursor is cleaved (S1 cleavage) intoa heterodimer by a furin-like-peptidase before insertion into the cell membrane (Logeat et al., 1998). Ligand-receptor binding can activate the receptor by exposing a hidden extracellular site in the negative regulatory region (NRR) to peptidases. ADAM10 or ADAM17 cleaves this site (S2 cleavage) to form the Notch extracellular truncation, which is subsequently recognized by the γ-secretase complex that cleaves inside the transmembrane domain and releases the Notch intracellular domain (NICD) from the membrane (S3 and S4 cleavage) (Kopan and Ilagan, 2009; Shah et al., 2005). The released NICD can then bind transcription factor complexes through CSL [an abbreviation based on the names in different species; CBF1/RBPJκ, Su(H), Lag-1], and either drive or inhibit transcription (Bray and Gomez-Lamarca, 2018), thereby either inducing similar cell fate (lateral induction), or alternating cell fate (lateral inhibition) (right-hand panel of the figure).
The activity and output of Notch signaling is regulated at many levels (Antfolk et al., 2019; Kovall et al., 2017). Availability of ligands and receptors is determined through expression (Chakravarti et al., 2017; Shah et al., 2017), protein stability (Varshney and Stanley, 2018) and trafficking towards the membrane for functional presentation (Itoh et al., 2003; Shao et al., 2017; Takeuchi et al., 2017). The activating cleavage steps are regulated through the control over the peptidases involved in the cleavage and the availability of the S2 target domain, as further discussed in the main text. Apart from its cleavage, the regulation of the NICD also affects signal output, which is influenced by stability of the NICD and available cofactors in the nucleus (Bray andGomez-Lamarca, 2018).
The most direct interaction between mechanical force and Notch signaling is at the stage of receptor activation, that is, a set of cleavage steps to release the NICD from the membrane. During this process, a hidden peptidase-binding site in the extracellular negative regulatory region of Notch is exposed by a conformational unfolding step (Fig. 1) (Chowdhury et al., 2016; Gordon et al.,2015; Morsut et al., 2016). Unfolding is induced by a pulling force acting on the receptor that is mediated by ligand endocytosis after receptor binding. This endocytic pulling force is dependent on dynamin, epsin and actin (Parks et al., 2000; Seugnet et al., 1997). Epsin targets ubiquitylated ligands for clathrin-mediated endocytosis and contributes to membrane bending (Langridge and Struhl, 2017). Dynamin forms a helical multimer that generates force to pinch off vesicles, whereas actin is essential in generating sufficient force to drive endocytosis of the ligand– receptor assembly (Meloty-Kapella et al., 2012). The ability of the ligand–receptor pair to withstand the pulling force depends on their molecular affinity and load-bearing capacity. Interestingly, the Notch ligands jagged 1 (Jag1) and delta-like ligand 4 (Dll4) have distinct mechanical characteristics. The binding of both Jag1 and Dll4 to Notch1 gives rise to a catch bond, a type of bond that strengthens when it is put under tension (often compared to a Chinese finger trap). However, the tension required for Notch activation is different for the two ligands (4 pN for Dll4 and 12 pN for Jag1) and has been suggested to be due to an increased binding rate of Dll4 (Luca et al., 2017). These two ligands are known to have different effects and roles, for example in angiogenesis, inner ear development and airway differentiation (Benedito et al., 2009; Petrovic et al., 2014; Stupnikov et al., 2019). The different force magnitudes required for Notch activation could thereby differentiate between the ligands.
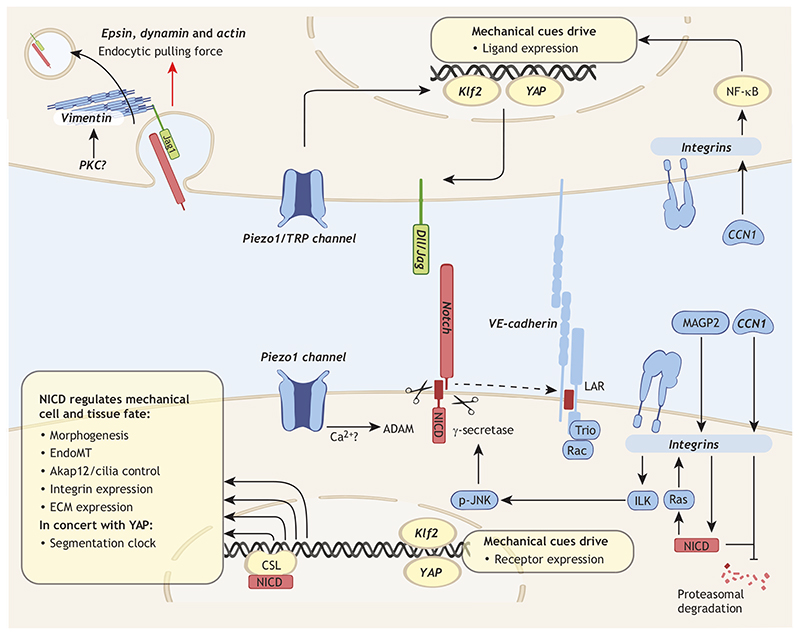
Fig. 1. Direct and indirect links between Notch signaling and mechanical signaling.
Several mechanotransductory proteins (bold italic in the figure) regulate the expression of Notch receptors or ligands. In addition, Notch reciprocally influences several mechanosensitive pathways, mechanical cell fates and morphogenesis. The top cell represents main interactions between mechanics and Notch ligands, Delta-like ligand (Dll) or Jagged (Jag). Mechanical loading sensed by integrins and mechanosensitive Piezo and transient receptor potential (TRP) channels result in YAP-, Klf2- and NF-κB-mediated ligand transcription, regulating Notch signaling. Proteins responsible for generating endocytic forces (epsin, dynamin and actin) unfold the hidden receptor cleavage site for ADAM10/17. Phosphorylation of vimentin by mechanosensitive kinases, possibly PKC, increases Jag1 endocytosis. The bottom cell represents the main interactions between mechanics and Notch receptors. Integrins are activated by a combination of extracellular matrix (ECM) composition, e.g. cellular CCN1 and MAGP2, and mechanical cues; this affects transcription, cleavage activity and Notch intracellular domain (NICD) stability. Ca2+ influx can regulate ADAM10 activity, possibly coupling Piezo1-mediated mechanosensing to Notch cleavage. The mechanosensitive transcription factors Klf2 and YAP also regulate Notch transcription, depending on cell context, and, in concert, Notch and YAP orchestrate the segmentation clock. Notch activation controls several mechanical fates as indicated. CSL, CBF1/RBPJκ, Su(H), Lag-1; LAR, leukocyte common antigen related.
Recent findings also indicate that there is a direct interaction between cytoskeletal or contractile forces and Notch activation. Inhibition of non-muscle myosin II to reduce contractility in signal-sending cells reduces Notch activation in vivo (Hunter et al., 2019). This reduction is independent of the presentation of ligands or receptors on the cell surface, and it is additive with the effect from dynamin or epsin inhibition, indicating separate contributions of contractile and endocytic forces (Hunter et al., 2019).
Similar to intracellular forces, external forces appear to have an activating effect on Notch. In particular, the timing of flow-induced activation of Notch also points towards a direct role for Notch in shear sensing, as the levels of NICD increase as early as 60 or 30 min after the onset of shear (Fang et al., 2017; Masumura et al.,2009). This rapid onset can however be blocked by inhibition of vascular endothelial growth factor (VEGF) receptor 2 (VEGFR2; also known as KDR), which could indicate both involvement of a mechanosensitive complex or a canonical VEGF-dependent downstream signal (Coon et al., 2015; Lawson et al., 2002; Masumura et al., 2009). However, the quick activation still allows for transcriptional to post-translational regulation of Notch ligands as the point of control (Shimojo et al., 2008). In fact, the mechanoresponsiveness of the Notch transmembrane domain was inhibited by Dll4 knockout or inhibition of endocytosis, suggesting that the regulation of Dll4 could control this mechanoresponse (Polacheck et al., 2017). Recent findings indicate that the mechanosensitive channel Piezo1 might tune the activity of Ca2+- sensitive ADAM10 in mediating the cleavage-dependent activation of Notch through Ca2+ ions, but uncoupling this increased S2-cleavage (see Box 1) from increased ligand production is challenging (Caolo et al., 2020). For these reasons, the evidence for a direct Notch activation by shear stress or stretch is still inconclusive.
Many interactions between Notch and mechanics are indirect. This includes the mechanical regulation of the production of ligands or receptors, and the crosstalk between Notch and mechanosensors (Fig. 1). Yes1-associated transcriptional regulator (YAP; also known as YAP1) and WW domain-containing transcription regulator 1 (TAZ; also known as WWTR1) (collectively YAP/TAZ) are transcriptional regulators that respond to diverse mechanical cues, including extracellular matrix rigidity, cell shape and shear stress (Panciera et al., 2017). Despite recent evidence for a crosstalk between YAP/TAZ and Notch in angiogenesis (Neto et al., 2018), the specific role of this crosstalk is still under investigation. More details have been uncovered in other tissues. For example, during myogenesis, the onset of contractions induces YAP translocation to the nucleus, where it drives expression of Jag2 and subsequent Notch activation (Esteves de Lima et al., 2016), which is associated with the maintenance of the progenitor cell pool required for regeneration (Bröhl et al., 2012), whereas in epidermal stem cells, YAP/TAZ regulates the expression of Notch-inhibiting ligands (Totaro et al., 2017). In the segmentation clock, a molecular oscillator that times the proper segmentation of developing somites, YAP and Notch work in concert to integrate mechanical cues with molecular signals and synchronize cell behavior over the segmenting tissue (Hubaud et al., 2017). The integrins are also mechanosensors that interact with Notch (Fig. 1). Upon activation of integrin linked kinase (ILK), a phosphorylated c-Jun N-terminal kinase family protein (p-JNK) is translocated to the membrane where it activates the γ-secretase complex and increases NICD, whereas direct phosphorylation of NICD by ILK has been shown to decrease NICD half-life, pointing to ILK-mediated regulation of NICD dynamics (Miyagawa et al., 2019; Mo et al., 2007). Cellular communication network factor 1 (CCN1) and microfibril-associated glycoprotein 5 (MAGP2; also known as MFAP5) are matrix proteins that regulate Notch through integrins. CCN1 is a shear-responsive matricellular protein (Hsu et al., 2019). CCN1 acts on integrins, thereby inducing expression of Dll4 in endothelial cells (Chintala et al., 2015) and of Jag1 through nuclear factor κB (NF-κB) in pancreatic ductal carcinoma and the hepatic duct (Haque et al., 2012; Kim et al., 2015). Further substantiating the link between integrin activation and NICD dynamics, is the increase in NICD levels seen upon CCN1 signaling inhibiting proteasomal degradation (Haque et al., 2012). In contrast, the ECM protein MAGP2 binds integrins to phosphorylate NICD through Src, reducing NICD transcriptional activity and half-life (Deford et al.,2016; LaFoya et al., 2018). Vice versa, Notch influences integrin activation through both transcriptional and non-transcriptional mechanisms; in fact NICD can directly activate R-RAS, a small GTPase that activates integrins (Hodkinson et al., 2007). Therefore, Notch is involved in crosstalk with other mechanosensitive signals, which enlarges the range of mechanical cues influencing this important signaling pathway in diverse tissues. In the context of cardiovascular tissues, these interactions might enable a fine-tuning of the Notch response to cell contractility (e.g. via YAP) or blood shear stress (e.g. via CCN1).
The production of Notch receptors and ligands is influenced by mechanics in several tissues. One of the most striking examples is the flow-induced increase or decrease in the levels of Notch receptors and ligands in endothelial cells (ECs) during development (Choi et al., 2017;Samsa et al., 2015; Volz et al., 2015). This is essential for the proper development of the heart, as shown in zebrafish, where heart contraction and subsequent blood flow induce notch1b transcription in the endocardium (Samsa et al.,2015). Similarly, the expression of Jag1 in the coronary artery endothelium depends on the onset of flow (Volz et al., 2015). Flow-induced transcription of Notch signaling components is both cell and tissue dependent. For example, in lymph vessels, flow activates the Ca2+ channel Orai1, which mediates Notch degradation, subsequently allowing lymph vessel sprouting (Choi et al., 2017).
Notch determines cell fate, which in turn strongly influences tissue mechanics. Since an increase or decrease in cell number leads to a change in mechanical stresses within tissues, one way Notch affects mechanics is by driving or inhibiting proliferation and apoptosis. The exact role of Notch in these processes depends on the specific tissue context and the specific Notch receptor and ligand (Dang, 2012; Gao et al., 2013; Liu et al., 2009). Notch influences tissue mechanics by regulating ECM expression in diverse tissues, including cartilage (Mirando et al., 2013), skin (Dees et al., 2011) and heart (Del Monte-Nieto et al., 2018). In the endocardium, this depends on the ligand expressed, as Jag1 regulates the production of ECM, while Dll4 mediates cell adhesion (Torregrosa-Carrión et al., 2019). A bidirectional relationship between Notch and mechanics also is revealed for cilia, as shear induced cilia activation induces Notch1b, Jag1b and Dll4 production in zebrafish, while cilia length and proper mechanosensing are controlled by Notch activation through regulation of A-kinase anchoring protein 12 (Akap12) (Chen et al., 2017; Mukherjee et al., 2020). The fact that Notch is influenced by, and in turn adapts tissue mechanics to attain mechanical homeostasis suggests that Notch could be a key component of the cellular ‘mechanostat’. The term mechanostat was introduced in the context of bone remodeling, describing an equilibrium between the load typically exerted on a bone and the minimal strength of a bone across organisms (Frost, 1987). The concept that any tissue needs to be strong enough to withstand the forces it is typically exposed to, makes the mechanostat a likely ubiquitous mechanism, with Notch as a central candidate.
Notch and mechanics in cardiovascular tissues
Cardiovascular tissues develop and function under mechanical load. From 3 weeks after fertilization until the heart stops beating, there are approximately three billion heartbeats during a human lifetime. The heart and vasculature are constantly exposed to forces derived from blood flow and cellular contractions. Cardiovascular tissues develop to exert and withstand these forces, and they remodel when mechanical homeostasis is perturbed. Mechanically induced remodeling of cardiovascular tissues has an important role in physiology and pathology and is controlled by many different mechanotransduction pathways, as excellently reviewed previously (Baratchi et al., 2017; Hahn and Schwartz, 2009).
The cardiovascular system is exposed to a broad range of dynamic stretches and fluid shears. Small vessels never experience stretch, but the aorta can experience a circumferential stretch of 10%, whereas heart valves can experience up to 30% stretch (Morrison et al., 2009; Oomen et al., 2016; Sacks and Yoganathan,2007). The ECs, the cells lining the inside of the cardiovascular system, are the main cells exposed to wall shear stress (WSS) from blood flow, and experience from 0.1 Pa in the venous system to 1 to 7 Pa in arterial vessels, depending on the location in the vascular tree (Malek et al., 1999). Physiological WSS elicits several physiological responses, including alignment of ECs, formation of the vascular barrier (Polacheck et al., 2017), vascular remodeling (Lucitti et al., 2007) and vasodilation or vasoconstriction to regulate blood flow (Pyke and Tschakovsky, 2005). Disturbed shear stress (DSS) is an important factor in various diseases, including atherosclerosis, aneurysms and collateral vessel formation (Eitenmüller et al., 2006; Hahn and Schwartz, 2009; Humphrey et al., 2015). Cellular forces are also important, as contractility and migration of cells are essential for tissue development and homeostasis, such as in the case of sprouting angiogenesis, the process of blood vessels growing new branches (Vaeyens et al., 2020).
During cardiac development, Notch integrates with other signaling pathways to shape the architecture and functionality of the heart and heart valves (MacGrogan et al., 2018). The role of Notch in cardiovascular development makes it a potential therapeutic target in cardiovascular disease (Box 2) and cardiovascular regeneration (Gálvez-Santisteban et al., 2019; Kefalos et al., 2019; Münch et al., 2017). In vascular development, Notch signaling is crucial for the regulation of both vasculogenesis (i.e. de novo generation of novel blood vessels) and angiogenesis (Alabi et al., 2018; Hellström et al., 2007), as well as hemogenesis, arteriovenous specification and maturation of the vessels (Chen et al., 2017; Gama-Norton et al.,2015; Lawson et al., 2001; Manderfield et al., 2012; Porcheri et al., 2020; Volz et al., 2015). The importance of the interplay between Notch and mechanics is evident at several stages of cardiac development. Early in cardiac development, Notch has an important role in regulating the endothelial–mesenchymal transition (EndoMT) of the endocardium; it initiates the process of trabeculation, the formation of myocardial protrusions into the ventricle that are compacted again later in development, and triggers migration of the endocardium to form heart valves (Grego-Bessa et al., 2007; Timmerman et al., 2004). This process requires the onset of blood flow and endothelial primary cilia, as ablation of either component reduces the expression of notch1b, dll4, jag1, and jag2 and prevents trabeculation in zebrafish (Samsa et al., 2015). Interestingly, the different oscillatory and pulsatile flow profiles caused by the flow disturbance of the trabecula determine Notch activity at different sites of the endocardial lining, leading to different cell fate decisions (Fig. 2B) (Lee et al., 2018). In zebrafish heart valve development, oscillatory flow occurs before heart valves have formed. This activates the mechanosensitive channels Trpv4, Trpp2 and Piezo1 that antagonistically regulate Krüppel-like factor 2 (Klf2), a hallmark shear-responsive factor upstream of Notch expression (Fig. 2B) (Duchemin et al., 2019; Heckel et al., 2015). This flow-induced Notch expression and activation subsequently controls valvular cell EndoMT (Pestel et al., 2016; Timmerman et al., 2004). Damaging valves, and thus disturbing flow patterns, induces Notch reactivation and regeneration of valves (Kefalos et al., 2019). In adulthood, Notch is involved in preventing valve calcification, both in valvular endothelial cells and valvular interstitial cells. Loss of a Notch copy is sufficient to sensitize the cells to mechanical stimulation and calcification (Chen et al., 2015; Theodoris et al., 2015).
Box 2. Notch mutations and deregulation in cardiovascular disease.
Mutations or misregulation in Notch pathway genes can lead to congenital and acquired cardiovascular malformations (Mašek and Andersson, 2017). Defects in NOTCH1 can cause aortic valve disease (AoVD) (Garg et al., 2005). Mutations in JAG1 or NOTCH2 can give rise to Alagille syndrome, with cardiac symptoms similar to tetralogy of Fallot (McDaniell et al., 2006; Spinner et al., 2001). Mutations in RBPJ (CSL), NOTCH1, DLL4, and EOGT (a Notch-modifying N-acetylglucosamine transferase) can lead to Adams–Oliver syndrome, with 20% of patients having cardiac defects (Chapman et al., 2020; Meester et al., 2015). Mutations in NOTCH3 have been shown to cause cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), but the pathogenesis is under intensive investigation (Joutel et al., 1996; Rajani et al., 2019; Rutten et al., 2019). Mutations in mindbomb1 (MIB1), a regulator of receptor and ligand endocytosis, lead to left ventricular non-compaction cardiomyopathy, where trabeculae are not compacted (Luxán et al., 2013). Among the acquired cardiovascular defects is pulmonary arterial hypertension, during which Notch1 and Notch3 control pathogenic proliferation of ECs and vascular smooth muscle cells (vSMCs), respectively (Dabral et al., 2016; Li et al., 2009; Miyagawa et al., 2019). Owing to vascular wall instability in thoracic/abdominal aortic aneurysm, Notch is likely to have a role, although this is still not clear (Li and Kong, 2020). Dll4 binding to Notch3 and Jag1– Notch1 interactions have been implicated in diabetic vasculopathy and diabetic retinopathy (Miloudi et al., 2019; Wimmer et al., 2019). In cerebral cavernous malformations, Notch also plays a role in the aberrant EndoMT transition and vascular malformations (Kar et al., 2016). Similarly, in arteriovenous malformations, where shunts form between arteries and veins, Notch is upregulated in human patients, and upregulation of Notch1 or Notch4 causes the disease in mouse models (Murphy et al., 2009; Nielsen et al., 2016). In vascular conditions with prominent inflammatory components, Notch ligands also determine cell fate, such as in large vessel vasculitis, vascular graft lesions, or arteriovenous graft remodeling (Guo et al., 2020; Koga et al.,2015; Wen et al., 2017).
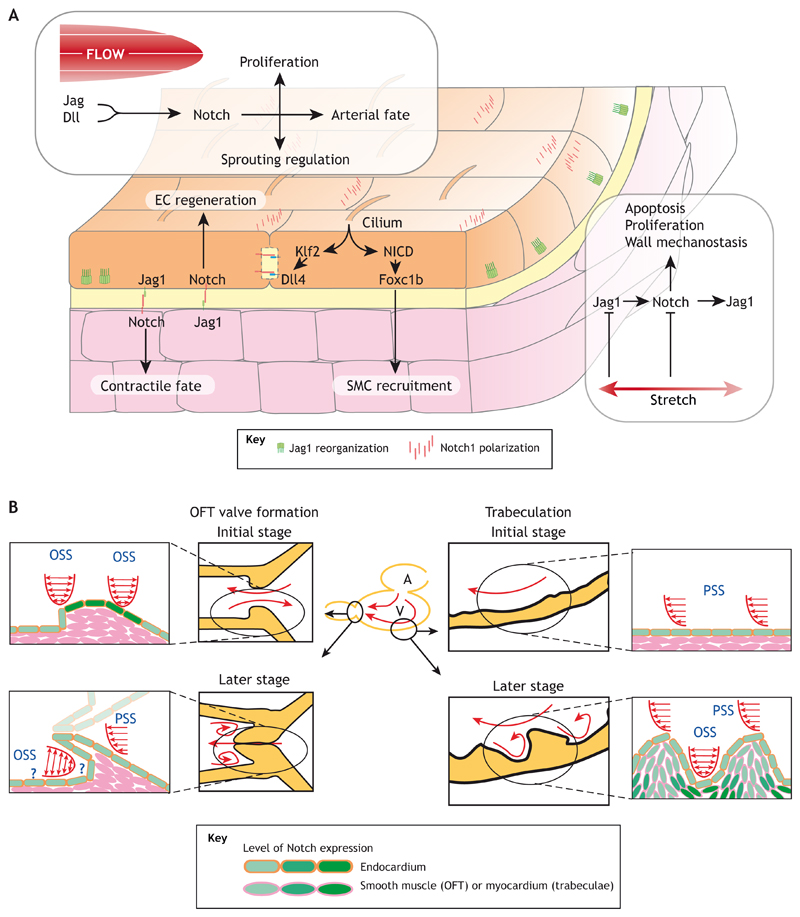
Fig. 2. Interactions of vascular mechanics with Notch signaling and subsequent vascular cell fate effects.
(A) Effect of hemodynamic forces on Notch and vascular cell fate. Blood flow induces ligand transcription, partially through cilia, and Notch activation in endothelial cells (ECs) through Klf2, subsequently leading to Notch activation in neighboring ECs or vascular smooth muscle cells (vSMCs). Notch1 and Jag1 reorganize in the cells, with currently unknown function. In ECs, a balance of Jag1- and Dll4-mediated Notch signaling leads to decisions on sprouting, proliferation, regeneration and arterial fate adoption. Notch activation leads to Foxc1b-mediated vSMC recruitment to the vessel and Jag1-mediated contractile fate adoption in vSMCs. Stretch induces proliferative fate through Jag1 inhibition, possibly as a mechanism for restoring homeostasis upon loss of mechanical balance. (B) Localized hemodynamic forces in zebrafish heart induce Notch and morphogenesis. In zebrafish cardiac development, blood flows from the atrium (‘A’) to the ventricle (‘V’), to the bulbus, or outflow tract (OFT), influencing Notch and morphogenesis. During heart valve formation, pulsatile and oscillating shear stress (PSS and OSS) induce Notch activation, which is required for heart valve morphogenesis (shown on the left). In the ventricle, blood flow initiates Notch mediated trabeculation (shown on the right). The local shear patterns induce a specific Notch response required for morphogenesis. Cells shown in pink are not described to have active Notch in these stages of development. The schemes illustrating trabeculation have been republished with permission of JCI Insight from Lee et al., 2018; permission conveyed through Copyright Clearance Center, Inc.
Notch is a marker for arterial vessels (Swift and Weinstein, 2009). Arterial vessels have a thicker vascular wall, due to a prominent layer of vascular smooth muscle cells (vSMCs), which serve to regulate blood flow and to withstand arterial pressures. Notch signaling occurs both among neighboring ECs and neighboring vSMCS, as well as between ECs and vSMCs (Fig. 2). There are multiple reports about the link between shear stress, Notch activation and arteriovenous fate. This functional link depends both on the organism and organ (Buschmann et al., 2010; Deng et al., 2013; Quillien et al., 2014). In chicken yolk, blood flow determines arteriovenous specification in an interplay with Notch, which is essential for expression of ephrin B2, a hallmark arterial gene (Buschmann et al., 2010; le Noble et al., 2004). In contrast, arteriovenous specification in zebrafish takes place before the onset of flow; here, Notch determines a base pattern of 60% arterial and 40% venous vessels in the zebrafish trunk, which is further balanced to a 50:50 ratio by flow (Geudens et al., 2019; Isogai et al., 2003; Weijts et al., 2018). However, the importance of shear- and strain-regulated expression of Notch for arteriovenous specification and maintenance remains to be further clarified (Gore et al., 2012). Notch signaling is required for Foxc1b-mediated vSMC recruitment (Chen et al., 2017) and differentiation towards a contractile phenotype (Ando et al., 2019; Manderfield et al., 2012). Expression of Notch3 and Jag1 in the wall is reduced by exposure to stretch (Loerakker et al., 2018; Morrow et al., 2005), and this mechanism could play a mechanostatic role by determining the arterial wall thickness in a Notch- and mechanics-driven feedback loop (Loerakker et al., 2018).
Distinct Notch responses, depending on cell type and mechanical cues, occur in the vessel wall. In ECs, different ligands and receptors display distinct responses to shear stress. Jag1 transcription increases under laminar shear of 1 Pa, whereas this downregulates Dll4 (Driessen et al., 2018). Shear stress also induces a reorganization of both Notch1 and Jag1 in the cell, with Notch1 accumulating on the membrane downstream of the shear direction, whereas Jag1 appears in submembranous foci (Fig. 2) (Driessen et al., 2018; Mack et al., 2017). Ligand-mediated Notch activation is also increased by shear stress (Masumura et al., 2009; Polacheck et al., 2017). Dll4 and Jag1 are known to have distinct roles in regulating cell fate in angiogenesis (Benedito et al., 2009), valvular development (MacGrogan et al., 2016), ventricular development (D’Amato et al., 2016) and hemogenesis (Porcheri et al., 2020). Therefore, the distinct effects of shear stress on the expression and localization of different Notch ligand and receptor proteins, in combination with the specific force magnitudes required for Notch activation by these ligands (Luca et al., 2017), are further lines of evidence that mechanical cues are a major regulator of Notch signaling and thus cell fate and function.
Recent discoveries have provided more insight into the molecular mechanisms responsible for Notch mechanosensitivity to blood flow. For example, a novel flow-dependent Notch signal transduction pathway has been shown to be involved in the maintenance of the vascular barrier (Polacheck et al., 2017). Exposure to flow induces ligand-mediated cleavage of the Notch receptor, with S2-cleavage releasing the Notch extracellular truncation and S3-cleavage releasing the NICD, while the short transmembrane domain in between these two cleavages remains in the membrane (Fig. 1). This transmembrane domain forms a complex with the tyrosine phosphatase LAR (PTPRF), the Rac1 guanine exchange factor (GEF) Trio and VE-cadherin, stabilizing cell–cell junctions and the vascular barrier (Fig. 1) (Polacheck et al., 2017).
Recently, we have demonstrated that the mechanosensitive intermediate filament protein vimentin modulates Notch signaling in vascular cells (Antfolk et al., 2017; van Engeland et al., 2019). Vimentin binds the intracellular domain (ICD) of Jag1, but not Dll4, increasing the Jag1 signal sent to neighboring cells. In fact, Jag1 endocytosis and signaling strength are reduced in vimentin-knockout mice, resulting in Dll4 signaling being dominant in the endothelium, consequently reducing sprouting. Vimentin deletion also drives vSMC fate to the proliferative state, indicating that both EC–EC and EC–vSMC signaling is hampered (Antfolk et al., 2017; van Engeland et al., 2019). Vimentin phosphorylation on serine 38 is increased after exposure to shear stress, and phosphorylation increases the signal-sending capacity of Jag1 (van Engeland et al., 2019). Introduction of the mechanoresponsive protein kinase C (PKC), known to target vimentin (Nakayama et al., 2003; Eriksson et al., 2004), or introduction of the vimentin variants phosphomimicking PKC target sites, both enhance Notch transactivation (Antfolk et al., 2017; van Engeland et al., 2019). This raises the interesting possibility that vimentin responds to shear stress by interacting with PKC and, once phosphorylated, increases the Jag1-mediated Notch activation, thereby forming a mechanosensitive cell-fate-controlling axis.
As described above, Notch interaction with mechanics plays a key role in heart development, heart valves and vascular tissues. Despite recent discoveries, a comprehensive understanding of how mechanics and Notch are integrated in cardiovascular tissues at the molecular level still needs to be fully achieved. This will require technologies that allow for analyses of cell signaling and cell fate under controlled hemodynamic environments in real time, as well as methods to translate molecular-level information on mechanosignaling into tissue-level effects on form and function.
Engineered systems to addressing open questions of Notch signaling
Many biophysical tools to mechanically or spatially control cells or proteins, including optical and magnetic tweezers, microfluidics and microcontact printing, have been key in attaining novel discoveries in mechanobiology (Monteiro et al., 2018; Qin et al., 2010). Some of these tools have already been implemented to investigate Notch signaling as previously reviewed (Lovendahl et al., 2018), while other techniques have the potential to lead to breakthroughs in some of the long-standing questions in the Notch field (highlighted in Box 3). In particular, engineered tools can help to control mechanical, molecular and cellular variables, thereby enabling the investigation of the effects of these variables on Notch, while keeping other variables approximately unchanged. For example, identifying the contribution of the different Notch components to the mechanosensitivity of this pathway will require mechanical stimulation combined with cellular engineering or synthetic biology to separate the mechanosensitivity of different molecular components. The importance of dynamics in Notch signaling is emerging, as evidenced by the role of Notch oscillations in stem cell maintenance, the dose dependence of cell fate decisions, and specific receptor–ligand combinations resulting in different temporal activation curves (Nandagopal et al., 2018; Shimojo et al.,2008; Theodoris et al., 2015). To couple dynamic molecular activity to tissue morphogenesis requires real-time following of systems instead of end-point analyses, and engineered culture systems enable these studies.
Box 3. Main questions pertaining to Notch in mechanotransduction.
-
How sensitive is Notch to the dosage and dynamics of mechanical cues?
This is essential for the proposed role as a mechanostat.If Notch is sensitive to mechanical load dosage and its temporal dynamics, then it can trigger compensating responses in the cell.
-
What is the systemic result of several mechanical cues acting on different Notch components and cell types interacting together?
Answering this question will increase our understanding of the integration of mechanics and cell fate signaling for tissue morphogenesis and homeostasis.
-
Do the different Notch components have distinct mechanotransductory properties?
Dll4 and Jag1 require different force magnitudes to activate Notch (Luca et al., 2017). Additional differences among Notch components might fine-tune Notch mechanosensitivity.
-
Are the peptidases that are involved in Notch cleavage mechanosensitive?
Piezo-mediated Ca2+ influx might influence ADAM10 activity, while membrane tension may affect γ-secretase activity, and these mechanisms might be important in physiology and pathology.
-
Which mechanosensitive proteins interact with Notch?
Further enlarging the set of mechanosensitive pathways crosstalking with Notch and identifying the specific crossroads will potentiate pharmaceutical targeting.
-
Are there mechanosensitive processes influencing Notch protein trafficking and polarization and, if so, what is their role in cell fate?
In ECs, Notch1 and Jag1 reorganize under shear stress (Driessen et al., 2018; Mack et al., 2017). Similar mechanisms might be present in other cell types and for Notch components involving different molecular machineries, and might have consequences in cell fate and tissue morphology.
-
Are the dynamics of Notch signaling integrated with mechanical dynamics?
Notch temporal dynamics, signal strength and oscillations are crucial for cell fate decisions and morphogenesis. Mechanical cues are inherently dynamic, and therefore they might play a role in establishing Notch dynamics.
-
Is the mechanotransductory role of Notch specific to different cell types, and what conveys this specificity?
Cell type specificities with respect to Notch mechanosensitivity might inform new therapeutic options that are more tailored to the target tissue.
Elucidating the molecular machinery around Notch activation that contributes to its mechanosensitivity (Caolo et al., 2020; Mo et al., 2007) will require more screening approaches for molecular interactors. To achieve this, cells must be exposed to forces, either at the macroscopic or microscopic scale, with various methods available (Box 4). Identification of mechanosensitive interactors of Notch signaling might be possible through high cellular yield shear approaches, for example, using an orbital shaker (Driessen et al., 2020). To separate the effect of mechanical loading on the molecular receptor–ligand interaction from the cellular regulation of these components, techniques with higher temporal and spatial resolution of Notch readout, such as luminescent constructs (Ilagan et al., 2011), are required in conjunction with mechanical stimulation.
Box 4. Engineered approaches to study mechanics in cell biology.
Many of the questions on Notch mechanotransduction can be addressed using engineered approaches. These allow the study of diverse aspects, such as cellular transcriptional responses, cell–cell interactions, morphogenesis, and cell–matrix interactions under mechanical cues. Mechanical cues that can be presented to cells include fluid-flow-induced shear stress, stretch, compression or viscoelasticity of the environment. Shear stress is typically imposed on cells by using parallel plate geometries (Driessen et al., 2018; Mack et al., 2017), cone-plate shear systems (Kunnen et al., 2017) or orbital shakers (Driessen et al.,2020), depending on the requirements (e.g. amount of cells for analysis and level of control over the fluid flow). Variations of the orbital shaker, using annular shaped dishes or seeding patterns, have recently opened novel possibilities by enhancing control over flow patterns (Driessen et al., 2020; Ghim et al., 2018). Cells can be stretched by culturing them on cyclically stretched flexible membranes (Loerakker et al., 2018; Morrow et al., 2005) or in deformable 3D microtissues (Foolen et al.,2012; Gould et al., 2012). Compression is applied by culturing cells in a gel or in a well and compressing the gel or culture medium (Di Federicoet al., 2017; Manokawinchoke et al., 2017). Viscoelasticity can be controlled by culturing cells on hydrogels or lipid bilayers (Chaudhuri et al., 2020).
There is also a rapid development of bioengineered systems that more closely resemble (micro)physiological conditions, although they typically offer less control over mechanical variables. Many of these systems provide a 3D environment, resulting in different behavior of signaling pathways compared to that seen in 2D systems (Cukierman et al., 2001; Duval et al., 2017). Spatial constraints in 3D systems perturb mechanical cues, thereby possibly affecting cell behavior (Chen et al., 2018), although the effects of these constraints are difficult to control and quantify. Different types of bioengineered systems include organoids (Sato et al., 2009) and predefined or self-organizing vascular systems in gels (Polacheck et al., 2017; Wimmer et al., 2019). The microfluidic vessel-wall-on-a-chip offers control over independent cellular and mechanical variables in cell–cell contact situations (Huh et al., 2010; van Engeland et al., 2018).
Verifying that Notch functions as a mechanostat will require engineered systems with optimal control over mechanical loading, or with force gradients built into the system, for example, a Y-shaped fluidic channel (Mack et al., 2017). In fact, to function as a mechanostat, Notch should either have a mechanical set point encoded in the pathway and its interactors, or a very high sensitivity to signaling dosage. To understand the effect and regulation of signaling dynamics (Nandagopal et al., 2018; Shimojo et al., 2008) and the interactions of mechanical forces with temporal dynamics, it is necessary to develop systems with temporal control over mechanical loading, as well as Notch readout. Microfluidic chips have been implemented before to pace the rate of oscillations in Notch (Sonnen et al., 2018), and could form a way to investigate the effect of periodicity in mechanical loading, as imposed by the heart beat (Ubezio et al., 2016).
Studying mechanics at the cellular and molecular level is challenging as there is overlap with topographical, or spatial, variables, as occurs for example in integrin and ephrin signaling, where specific spatial organization is essential for the signaling output (Westerfield and Barrera, 2020). Topographical control of ligand presentation can enable a more detailed investigation of Notch activation by mechanical forces. An early study showed that surface-immobilized Dll1 could activate Notch, whereas soluble ligands could not (Varnum-Finney et al., 2000). In contrast, Jag1-derived peptides did not activate Notch in a surface-immobilized form, possibly due to being sterically unavailable to the receptor, but activated Notch when they were self-assembled in solution (Putti et al., 2019). Making use of microcontact printing techniques, which allow the deposition of proteins in predefined patterns, has proven useful to spatially control cell fate (Tiemeijer et al., 2018), and if applied at subcellular resolution could help in resolving the effect of polarized Notch activation.
The distinct Notch responses to flow and stretch and the different cell types making up mechanically activated tissues, can result in interactions that are challenging to understand, but which are still fundamental for the physiology of cardiovascular tissues. To characterize these system-level outcomes it is necessary to allow separate control of flow and stretch and to separately manipulate and monitor different cell populations, to obtain mechanistic insight. A device that enables this is the recently developed vessel-wall-on-a-chip (Box 4), based on a lung-on-a-chip design (Huh et al., 2010), as it allows two cell populations to communicate via Notch through pores, mimicking the myoendothelial projections that are sites of EC– vSMC Notch signaling in vivo (McCallinhart et al., 2020), while separating interacting ECs and vSMCs, which can thus be subjected to different levels of flow and stretch (van Engeland et al., 2018).
While microfluidic devices can be employed to separate variables influencing Notch and mechanics, tissue-engineered constructs are a step closer towards mimicking the complexity of multicellular tissues in vitro. These constructs might be employed both on a level of fundamental understanding and for more translational purposes, such as towards the development of regenerative medicine approaches. These engineered tissues can be used to study how the long-term interplay between Notch and mechanics regulates cell fate and morphogenesis. For example, a bioreactor with a controlled pressurized tissue system has been recently developed to study the long-term effect of constant strain versus constant stress on tissue growth (van Kelle et al., 2017). We expect that these controlled engineered systems will have a great impact on the field, especially when coupled with computational models, as discussed next.
Computational models coupling Notch and mechanics
Despite recent developments, it is unlikely that engineered systems will ever fully control all variables influencing Notch and mechanics. Computational models can help in overcoming this limitation, owing to their potential to predict and isolate the effects of single parameters, and to carefully analyze cell and tissue mechanics. Several computational models of Notch have been proposed and have demonstrated their impact in testing hypotheses and generating new theories based on model predictions (Binshtok and Sprinzak, 2018). For example, computational simulations of sprouting angiogenesis accounting for the crosstalk between Notch and VEGF were fundamental to suggest that ECs shuffle at the top of angiogenic sprouts (Bentley et al., 2008), a computational prediction that was later verified in vivo (Jakobsson et al., 2010). In the context of Notch mechanosensitivity, computational models can enable the investigation of the effects of this phenomenon at a systemic level, as they can be adopted to analyze cell and tissue mechanics, growth, and remodeling, while accounting for Notch signaling among different cell types.
A direct relationship between Notch and mechanics was first included in a computational model of collective epithelial cell migration (Riahi et al., 2015). Because the authors observed in vitro that cells leading the migration exhibit high Dll4 levels and that perturbation of cell contractility affects the number of leading cells, they hypothesized that stress downregulates Notch1 and Dll4 and that this feature is crucial for the regulation of the spatial distribution of leading cells. To test this hypothesis, building on previous computational models of Notch–Delta lateral inhibition (Cohen et al., 2010; Collier et al., 1996; Sprinzak et al.,2010), they developed a computational model accounting for Notch downregulation in response to stress (Riahi et al., 2015). In particular, they assumed that Notch1 and Dll4 production decreases with the cell distance from the migration leading edge, where lower stress levels had been experimentally observed. The experimental finding that Dll4-expressing cells appear at the leading edge could be computationally replicated only when Notch mechanosensitivity was considered, thereby supporting the notion that this phenomenon contributes to regulating the distribution of leading cells during epithelial cell migration (Riahi et al., 2015). Therefore, computational modeling was used here to verify the hypothesis that Notch mechanosensitivity played a role in the observed biological phenomenon.
Although hypothesis verification via modeling is useful, the added value of computational models is more evident when a full cycle of interplay between experiments and simulations is implemented, with simulations suggesting new theories and experiments. Recently, we have shown the potential of coupling experiments and simulations to study the systemic effects of the interplay between Notch and mechanics in arterial walls (van Engeland et al., 2019; Loerakker et al., 2018; Ristori et al.,2020). Using in vitro experiments, we first showed that cyclic strain downregulates Notch3 and Jag1 expression in vSMCs (Loerakker et al., 2018). To understand the implication for arterial remodeling, we extended a previous model of Notch that accounted for Notch– Jagged lateral induction (Boareto et al., 2015), present in vSMCs (Manderfield et al., 2012), by assuming that the production of Jag1 and Notch3 decreases with increasing strain (Loerakker et al.,2018). Coupled with mechanical analysis of arteries with different thickness, the new model predicted that vSMCs in relatively thin arteries experience high strain and thus have low Notch activation, corresponding to proliferative vSMCs and growing arteries. The strain decreased with arterial thickness and, when the arterial wall thickness reached a specific value, the model predicted that Notch signaling induces a collective switch of vSMCs from proliferative to contractile, corresponding to homeostatic non-growing arteries. The thickness for which the model predicted cell fate switching for arteries at different locations of the arterial tree corresponded well with their in vivo homeostatic thickness (Loerakker et al., 2018). Therefore, the model identified a new mechanism by which Notch mechanosensitivity might regulate arterial thickness and homeostasis. In a subsequent study, the same model was crucial for screening of the possible molecular mechanisms causing in vivo arterial thickening after vimentin knockout (van Engeland et al., 2019). In particular, the new simulations suggested that vimentin depletion disrupts Jag1-mediated Notch activation in vSMCs, which was later confirmed by additional in vivo experiments (van Engeland et al., 2019). More recent simulations of the interplay between Notch and mechanics in arteries suggest that Notch lateral induction is key in ensuring the collective switch of vSMCs from proliferative to contractile in growing arteries (Ristori et al., 2020), a computational prediction that hopefully will be verified in future experimental studies. Taken together, these studies therefore demonstrate the potential of computational models in translating findings from in vitro experiments to a higher level of complexity, such as in vivo cell behavior, and in generating new hypotheses about the underlying molecular mechanisms.
In these previous computational studies accounting for Notch mechanosensitivity, the local tissue mechanics was only approximated. Future studies should aim at overcoming this common limitation by adopting concepts from computational biomechanics to increase the predictive and analytical potential of computational models. Recently, a few computational modeling approaches have been proposed to analyze the local tissue mechanics and the consequential mechanosensitive behavior of interacting cells (Nolan and Lally, 2018a,b; Rouillard and Holmes, 2014; Thorne et al., 2011; Zahedmanesh and Lally, 2012). Although these approaches have not been applied to Notch signaling yet, we anticipate that they might be promising in further elucidating the fundamental role of Notch mechanosensitivity in cardiovascular tissue development and disease.
Conclusions
Despite the increasing amount of studies linking mechanical forces to Notch signaling, mechanistic detail is lacking. One pressing question in the Notch field is the distinct role of the different ligands and receptors, and how the combinatorial communication through distinct receptors and ligands gives rise to different outcomes in different tissues. Knowing which components of the Notch pathway are specifically sensitive to mechanical cues and determining which mechanosensitive proteins interact specifically to regulate Notch, will help researchers to understand this combinatorial code. This potentiates the development of directed therapies for Notch-related diseases and mechanopathologies. Examples of avenues where further mechanistic detail might emerge are the mechanosensitivity of regulatory microRNAs targeting Notch, as these are closely involved in tuning Notch activity and generating its oscillatory dynamics (Roese-Koerner et al., 2017), or the effect of mechanics on NICD stability and transcriptional activity through post-translational modifications (Antila et al., 2018; Wiedermann et al., 2015). The axis involving the mechanosensor PECAM, VE-cadherin, VEGFR, vimentin, Jag1 and Notch proteins is likely of importance in vascular biology (Conway et al., 2013; Coon et al.,2015; van Engeland et al., 2019; Polacheck et al., 2017), but the hierarchy of the signals remains to be clarified. In general, the role of the cell membrane, membrane transport and endo- and/or exo-cytotic processes in response to mechanical forces is a relevant field of study (Boulant et al., 2011; Shi et al., 2018), since membrane dynamics are intrinsically important for both mechanotransduction and Notch signaling, with a recently identified lipid-interacting domain on Notch ligands with as of yet unknown function of particular interest (Suckling et al., 2017). The direct and indirect reciprocal signaling between Notch and mechanics, both at the cellular and the tissue level, places Notch as an integrating hub candidate that is essential for the maintenance of mechanostasis. Systemic approaches, including computational modeling, in vivo experimental designs and advanced engineered culture systems, will prove essential to further unraveling this mechanosensitive signaling hub and establishing its role as a mechanostat.
Acknowledgements
The authors thank Kai-Lan Lin for contributions to graphical design. Fig. 1 and the illustration in Box 1 was created with biorender.com.
Funding
Our work in this area has been supported by the Academy of Finland project numbers 218062 and 33041, and the European Research Council Consolidator Grant 771168-ForceMorph to C.S., and by the research programme NWO Rubicon, which is (partly) financed by the Dutch Research Council (NOW; Nederlandse Organisatie voor Wetenschappelijk Onderzoek), with project number 019.183EN.025 to T.R.
Footnotes
Competing interests
The authors declare no competing or financial interests.
References
- Alabi RO, Farber G, Blobel CP. Intriguing roles for endothelial ADAM10/Notch signaling in the development of organ-specific vascular beds. Physiol Rev. 2018;98:2025–2061. doi: 10.1152/physrev.00029.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Wang W, Peng D, Chiba A, Lagendijk AK, Barske L, Crump JG, Stainier DYR, Lendahl U, Koltowska K, et al. Peri-arterial specification of vascular mural cells from naïve mesenchyme requires Notch signaling. Development. 2019;146:dev165589. doi: 10.1242/dev.165589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antfolk D, Sjoqvist M, Cheng F, Isoniemi K, Duran CL, Rivero-Muller A, Antila C, Niemi R, Landor S, Bouten CVC, et al. Selective regulation of Notch ligands during angiogenesis is mediated by vimentin. Proc Natl Acad Sci USA. 2017;114:E4574–E4581. doi: 10.1073/pnas.1703057114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antfolk D, Antila C, Kemppainen K, Landor SK-J, Sahlgren C. Decoding the PTM-switchboard of Notch. Biochim Biophys Acta Mol Cell Res. 2019;1866:118507. doi: 10.1016/j.bbamcr.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antila CJM, Rraklli V, Blomster HA, Dahlstrom KM, Salminen TA, Holmberg J, Sistonen L, Sahlgren C. Sumoylation of Notch1 represses its target gene expression during cell stress. Cell Death Differ. 2018;25:600–615. doi: 10.1038/s41418-017-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cellfate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Baratchi S, Khoshmanesh K, Woodman OL, Potocnik S, Peter K, McIntyre P. Molecular sensors of blood flow in endothelial cells. Trends Mol Med. 2017;23:850–868. doi: 10.1016/j.molmed.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and jagged1 have opposing effectson angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- Bentley K, Gerhardt H, Bates PA. Agent-based simulation of notch-mediated tip cell selection in angiogenic sprout initialisation. J Theor Biol. 2008;250:25–36. doi: 10.1016/j.jtbi.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Binshtok U, Sprinzak D. Modeling the notch response. Adv Exp Med Biol. 2018;1066:79–98. doi: 10.1007/978-3-319-89512-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boareto M, Jolly MK, Lu M, Onuchic JN, Clementi C, Ben-Jacob E. Jagged-Delta asymmetry in Notch signaling can give rise to a Sender/Receiver hybrid phenotype. Proc Natl Acad Sci USA. 2015;112:E402–E409. doi: 10.1073/pnas.1416287112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh J-C, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ, Gomez-Lamarca M. Notch after cleavage. Curr Opin Cell Biol. 2018;51:103–109. doi: 10.1016/j.ceb.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Brohl D, Vasyutina E, Czajkowski MT, Griger J, Rassek C, Rahn H-P, Purfürst B, Wende H, Birchmeier C. Colonization of the satellite cell niche by skeletal muscle progenitor cells depends on notch signals. Dev Cell. 2012;23:469–481. doi: 10.1016/j.devcel.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Buschmann I, Pries A, Styp-Rekowska B, Hillmeister P, Loufrani L, Henrion D, Shi Y, Duelsner A, Hoefer I, Gatzke N, et al. Pulsatile shear and Gja5 modulate arterial identity and remodeling events during flow-driven arteriogenesis. Development. 2010;137:2187–2196. doi: 10.1242/dev.045351. [DOI] [PubMed] [Google Scholar]
- Caolo V, Debant M, Endesh N, Futers TS, Lichtenstein L, Bartoli F, Parsonage G, Jones EA, Beech DJ. Shear stress activates ADAM10 sheddase to regulate Notch1 via the Piezo1 force sensor in endothelial cells. Elife. 2020;9:e50684. doi: 10.7554/eLife.50684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti B, Yang J, Ahlers–Dannen Katelin E, Luo Z, Flaherty Heather A, Meyerholz David K, Anderson Mark E, Fisher Rory A. Essentiality of regulator of G protein signaling 6 and oxidized Ca2+/calmodulin–dependent protein kinase II in notch signaling and cardiovascular development. J Am Heart Assoc. 2017;6:e007038. doi: 10.1161/JAHA.117.007038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman G, Moreau JLM, Eddie IP, Szot JO, Iyer KR, Shi H, Yam MX, O’Reilly VC, Enriquez A, Greasby JA, et al. Functional genomics and gene-environment interaction highlight the complexity of congenital heart disease caused by Notch pathway variants. Hum Mol Genet. 2020;29:566–579. doi: 10.1093/hmg/ddz270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Cooper-White J, Janmey PA, Mooney DJ, Shenoy VB. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature. 2020;584:535–546. doi: 10.1038/s41586-020-2612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ryzhova LM, Sewell-Loftin MK, Brown CB, Huppert SS, Baldwin HS, Merryman WD. Notch1 mutation leads to valvular calcification through enhanced myofibroblast mechanotransduction. Arterioscler Thromb Vasc Biol. 2015;35:1597–1605. doi: 10.1161/ATVBAHA.114.305095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Gays D, Milia C, Santoro MM. Cilia control vascular muralcell recruitment in vertebrates. Cell Reports. 2017;18:1033–1047. doi: 10.1016/j.celrep.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Vigliotti A, Bacca M, McMeeking RM, Deshpande VS, Holmes JW. Role of boundary conditions in determining cell alignmentin response to stretch. Proc Natl Acad Sci USA. 2018;115:986–991. doi: 10.1073/pnas.1715059115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintala H, Krupska I, Yan L, Lau L, Grant M, Chaqour B. The matricellular protein CCN1 controls retinal angiogenesis by targeting VEGF, Srchomology 2 domain phosphatase-1 and Notch signaling. Development. 2015;142:2364–2374. doi: 10.1242/dev.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Park E, Jung E, Seong YJ, Yoo J, Lee E, Hong M, Lee S, Ishida H, Burford J, et al. Laminar flow downregulates Notch activity topromote lymphatic sprouting. J Clin Invest. 2017;127:1225–1240. doi: 10.1172/JCI87442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury F, Li ITS, Ngo TTM, Leslie BJ, Kim BC, Sokoloski JE, Weiland E, Wang X, Chemla YR, Lohman TM, et al. Defining single molecular forces required for notch activation using nano yoyo. Nano Lett. 2016;16:3892–3897. doi: 10.1021/acs.nanolett.6b01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19:78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Collier JR, Monk NAM, Maini PK, Lewis JH. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J Theor Biol. 1996;183:429–446. doi: 10.1006/jtbi.1996.0233. [DOI] [PubMed] [Google Scholar]
- Conway DE, Breckenridge MT, Hinde E, Gratton E, Chen CS, Schwartz MA. Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr Biol. 2013;23:1024–1030. doi: 10.1016/j.cub.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon BG, Baeyens N, Han J, Budatha M, Ross TD, Fang JS, Yun S, Thomas J-L, Schwartz MA. Intramembrane binding of VE-cadherin to VEGFR2 and VEGFR3 assembles the endothelial mechanosensory complex. J Cell Biol. 2015;208:975–986. doi: 10.1083/jcb.201408103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Dabral S, Tian X, Kojonazarov B, Savai R, Ghofrani HA, Weissmann N, Florio M, Sun J, Jonigk D, Maegel L, et al. Notch1 signalling regulates endothelial proliferation and apoptosis in pulmonary arterial hypertension. Eur Respir J. 2016;48:1137–1149. doi: 10.1183/13993003.00773-2015. [DOI] [PubMed] [Google Scholar]
- D’Amato G, Luxan G, del Monte-Nieto G, Martlnez-Poveda B, Torroja C, Walter W, Bochter MS, Benedito R, Cole S, Martinez F, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18:7–20. doi: 10.1038/ncb3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TP. Notch,apoptosis and cancer. Adv Exp Med Biol. 2012;727:199–209. doi: 10.1007/978-1-4614-0899-4_15. [DOI] [PubMed] [Google Scholar]
- Dees C, Tomcik M, Zerr P, Akhmetshina A, Horn A, Palumbo K, Beyer C, Zwerina J, Distler O, Schett G, et al. Notch signalling regulates fibroblast activation and collagen release in systemic sclerosis. Ann Rheum Dis. 2011;70:1304–1310. doi: 10.1136/ard.2010.134742. [DOI] [PubMed] [Google Scholar]
- Deford P, Brown K, Richards RL, King A, Newburn K, Westover K, Albig AR. MAGP2 controls Notch via interactions with RGD binding integrins: identification of a novel ECM-integrin-Notch signaling axis. Exp Cell Res. 2016;341:84–91. doi: 10.1016/Jyexcr.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Monte-Nieto G, Ramialison M, Adam AAS, Wu B, Aharonov A, D’Uva G, Bourke LM, Pitulescu ME, Chen H, de la Pompa JL, et al. Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature. 2018;557:439–445. doi: 10.1038/s41586-018-0110-6. [DOI] [PubMed] [Google Scholar]
- Deng Y, Larrivee B, Zhuang ZW, Atri D, Moraes F, Prahst C, Eichmann A, Simons M. Endothelial RAF1/ERK activation regulates arterial morphogenesis. Blood. 2013;121:3988–3996. doi: 10.1182/blood-2012-12-474601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Federico E, Shelton JC, Bader DL. Complex mechanical conditioning of cell-seeded agarose constructs can influence chondrocyte biosynthetic activity. Biotechnol Bioeng. 2017;114:1614–1625. doi: 10.1002/bit.26273. [DOI] [PubMed] [Google Scholar]
- Drews JD, Pepper VK, Best CA, Szafron JM, Cheatham JP, Yates AR, Hor KN, Zbinden JC, Chang Y-C, Mirhaidari GJM, et al. Spontaneous reversal of stenosis in tissue-engineered vascular grafts. Sci Transl Med. 2020;12:eaax6919. doi: 10.1126/scitranslmed.aax6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen RCH, Stassen OMJA, Sjoqvist M, Suarez Rodriguez F, Grolleman J, Bouten CVC, Sahlgren CM. Shear stress induces expression, intracellular reorganization and enhanced Notch activation potential of Jagged1. Integr Biol. 2018;10:719–726. doi: 10.1039/C8IB00036K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen R, Zhao F, Hofmann S, Bouten C, Sahlgren C, Stassen O. Computational characterization of the dish-in-a-dish, a high yield culture platform for endothelial shear stress studies on the orbital shaker. Micromachines(Basel) 2020;11:552. doi: 10.3390/mi11060552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchemin A-L, Vignes H, Vermot J. Mechanically activated piezo channels modulate outflow tract valve development through the Yap1 and Klf2-Notch signaling axis. Elife. 2019;8:e44706. doi: 10.7554/eLife.44706.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z. Modeling physiological events in 2D vs. 3D cell culture Physiology (Bethesda) 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Eitenmüller I, Volger O, Kluge A, Troidl K, Barancik M, Cai W-J, Heil M, Pipp F, Fischer S, Horrevoets Anton JG, et al. The range of adaptation by collateral vessels after femoral artery occlusion. Circ Res. 2006;99:656–662. doi: 10.1161/01.RES.0000242560.77512.dd. [DOI] [PubMed] [Google Scholar]
- Eriksson JE, He T, Trejo-Skalli AV, Harmala-Brasken A-S, Hellman J, Chou Y-H, Goldman RD. Specific in vivo phosphorylation sites determine the assembly dynamics of vimentin intermediate filaments. J Cell Sci. 2004;117:919–932. doi: 10.1242/jcs.00906. [DOI] [PubMed] [Google Scholar]
- Esteves de Lima J, Bonnin M-A, Birchmeier C, Duprez D. Muscle contraction is required to maintain thepool ofmuscle progenitors via YAPand NOTCH during fetal myogenesis. Elife. 2016;5:e15593. doi: 10.7554/eLife.15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang JS, Coon BG, Gillis N, Chen Z, Qiu J, Chittenden TW, Burt JM, Schwartz MA, Hirschi KK. Shear-induced Notch-Cx37-p27 axisarrests endothelial cell cycle to enable arterial specification. Nat Commun. 2017;8:1–14. doi: 10.1038/s41467-016-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RR, Fukui H, Chow R, Vilfan A, Vermot J. The cilium as a force sensor-myth versus reality. J Cell Sci. 2019;132:jcs213496. doi: 10.1242/jcs.213496. [DOI] [PubMed] [Google Scholar]
- Foolen J, Deshpande VS, Kanters FMW, Baaijens FPT. The influence of matrix integrity on stress-fiber remodeling in 3D. Biomaterials. 2012;33:7508–7518. doi: 10.1016/Jbiomaterials.2012.06.103. [DOI] [PubMed] [Google Scholar]
- Frost HM. Bone “mass” and the “mechanostat”: a proposal. Anat Rec. 1987;219:1–9. doi: 10.1002/ar.1092190104. [DOI] [PubMed] [Google Scholar]
- Galvez-Santisteban M, Chen D, Zhang R, Serrano R, Nguyen C, Zhao L, Nerb L, Masutani EM, Vermot J, Burns CG, et al. Hemodynamic-mediated endocardial signaling controls in vivo myocardial reprogramming. eLife. 2019;8:e44816. doi: 10.7554/eLife.44816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Norton L, Ferrando E, Ruiz-Herguido C, Liu Z, Liu Z, Guiu J, Islam ABMMK, Lee S-U, Yan M, Guidos CJ, et al. Notch signal strength controls cell fate in the haemogenic endothelium. Nat Commun. 2015;6:8510. doi: 10.1038/ncomms9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Yao M, Shi Y, Hao J, Ren Y, Liu Q, Wang X, Duan H. Notch pathway is involved in high glucose-induced apoptosis in podocytes via Bcl-2 and p53 pathways. J Cell Biochem. 2013;114:1029–1038. doi: 10.1002/jcb.24442. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aorticvalve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Geudens I, Coxam B, Alt S, Gebala V, Vion A-C, Meier K, Rosa A, Gerhardt H. Artery-vein specification in the zebrafish trunk is prepatterned by heterogeneous Notch activity and balanced by flow-mediated finetuning. Development. 2019;146:dev181024. doi: 10.1242/dev.181024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghim M, Pang KT, Arshad M, Wang X, Weinberg PD. A novel method for segmenting growth of cells in sheared endothelial culture reveals the secretion of an anti-inflammatory mediator. J Biol Eng. 2018;12:15. doi: 10.1186/s13036-018-0107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, et al. Mechanical allostery: evidence for a force requirement in the proteolytic activation of notch. Dev Cell. 2015;33:729–736. doi: 10.1016/Jdevcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore AV, Monzo K, Cha YR, Pan W, Weinstein BM. Vascular development in the Zebrafish. Cold Spring Harb Perspect Med. 2012;2:a006684. doi: 10.1101/cshperspect.a006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RA, Chin K, Santisakultarm TP, Dropkin A, Richards JM, Schaffer CB, Butcher JT. Cyclic strain anisotropy regulates valvular interstitial cell phenotype and tissue remodeling in three-dimensional culture. Acta Biomater. 2012;8:1710–1719. doi: 10.1016/Jactbio.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama Y-S, Chen H, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/Jdevcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Huang F, Qing Y, Feng S, Xiao X, Wang Y, Liang M, Wang T, Mitch WE, Cheng J. Decreased Jagged1 expression in vascular smooth muscle cells delays endothelial regeneration in arteriovenous graft. Cardiovasc Res. 2020;116:2142–2155. doi: 10.1093/cvr/cvz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C, Schwartz MA. Mechanotransduction in vascular physiologyand atherogenesis. Nat Rev Mol Cell Biol. 2009;10:53–62. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque I, De A, Majumder M, Mehta S, McGregor D, Banerjee SK, VanVeldhuizen P, Banerjee S. The matricellular protein CCN1/Cyr61 isa critical regulator of Sonic Hedgehog in pancreatic carcinogenesis. J Biol Chem. 2012;287:38569–38579. doi: 10.1074/jbc.M112.389064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel E, Boselli F, Roth S, Krudewig A, Belting H-G, Charvin G, Vermot J. Oscillatory flow modulates mechanosensitive klf2a expression through trpv4 and trpp2 during heart valve development. Curr Biol. 2015;25:1354–1361. doi: 10.1016/Jcub.2015.03.038. [DOI] [PubMed] [Google Scholar]
- Hellstrom M, Phng L-K, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson A-K, Karlsson L, Gaiano N, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- Hodkinson PS, Elliott PA, Lad Y, McHugh BJ, MacKinnon AC, Haslett C, Sethi T. Mammalian NOTCH-1 activates β1 integrins viathe small GTPase R-Ras. J Biol Chem. 2007;282:28991–29001. doi: 10.1074/jbc.M703601200. [DOI] [PubMed] [Google Scholar]
- Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P-L, Chen J-S, Wang C-Y, Wu H-L, Mo F-E. Shear-induced CCN1 promotes atheroprone endothelial phenotypes and atherosclerosis. Circulation. 2019;139:2877–2891. doi: 10.1161/CIRCULATIONAHA.118.033895. [DOI] [PubMed] [Google Scholar]
- Hubaud A, Regev I, Mahadevan L, Pourquie O. Excitable dynamics and yap-dependent mechanical cues drive the segmentation clock. Cell. 2017;171:668–682.:e11. doi: 10.1016/Jcell.2017.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JD, Dufresne ER, Schwartz MA. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JD, Schwartz MA, Tellides G, Milewicz DM. Role of mechanotransduction in vascular biology: focus on thoracic aortic aneurysms and dissections. Circ Res. 2015;116:1448–1461. doi: 10.1161/CIRCRESAHA.114.304936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GL, He L, Perrimon N, Charras G, Giniger E, Baum B. A role for actomyosin contractility in Notch signaling. BMC Biol. 2019;17:12. doi: 10.1186/s12915-019-0625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan MXG, Lim S, Fulbright M, Piwnica-Worms D, Kopan R. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci Signal. 2011;4:rs7. doi: 10.1126/scisignal.2001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- Isogai S, Lawson ND, Torrealday S, Horiguchi M, Weinstein BM. Angiogenic network formation in the developing vertebrate trunk. Development. 2003;130:5281–5290. doi: 10.1242/dev.00733. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim C-H, Palardy G, Oda T, Jiang Y-J, Maust D, Yeo S-Y, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is aubiquitin ligase that is essential for efficient activation of notch signaling by delta. Dev Cell. 2003;4:67–82. doi: 10.1016/S1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- Kar S, Baisantry A, Nabavi A, Bertalanffy H. Role of Delta-Notch signaling in cerebral cavernous malformations. Neurosurg Rev. 2016;39:581–589. doi: 10.1007/s10143-015-0699-y. [DOI] [PubMed] [Google Scholar]
- Kefalos P, Agalou A, Kawakami K, Beis D. Reactivation of Notch signaling is required for cardiac valve regeneration. Sci Rep. 2019;9:16059. doi: 10.1038/s41598-019-52558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-H, Chen C-C, Alpini G, Lau LF. CCN1 induces hepatic ductular reaction through integrin αvβ 5–mediated activation of NF-κB. J Clin Invest. 2015;125:1886–1900. doi: 10.1172/JCI79327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindberg A, Hu JK, Bush JO. Forced to communicate: Integration of mechanical and biochemical signaling in morphogenesis. Curr Opin Cell Biol. 2020;66:59–68. doi: 10.1016/Jceb.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol. 2018;20:373–381. doi: 10.1038/s41556-018-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga J, Nakano T, Dahlman JE, Figueiredo J-L, Zhang H, Decano J, Khan OF, Niida T, Iwata H, Aster JC, et al. Macrophage notch ligand delta-like 4 promotes vein graft lesion development: implications for the treatment of vein graft failure. Arterioscler Thromb Vasc Biol. 2015;35:2343–2353. doi: 10.1161/ATVBAHA.115.305516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MaXG. The canonical notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/Jcell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Gebelein B, Sprinzak D, Kopan R. The canonical notch signaling pathway: structural and biochemical insights into shape, sugar,and force. Dev Cell. 2017;41:228–241. doi: 10.1016/Jdevcel.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnen SJ, Leonhard WN, Semeins C, Hawinkels LJAC, Poelma C, Ten Dijke P, Bakker A, Hierck BP, Peters DJM. Fluid shear stress-induced TGF-β/ALK5 signaling in renal epithelial cells is modulated byMEK1/2. Cell Mol Life Sci. 2017;74:2283–2298. doi: 10.1007/s00018-017-2460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFoya B, Munroe JA, Mia MM, Detweiler MA, Crow JJ, Wood T, Roth S, Sharma B, Albig AR. Notch: A multi-functional integrating system of microenvironmental signals. Dev Biol. 2016;418:227–241. doi: 10.1016/Jydbio.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFoya B, Munroe JA, Pu X, Albig AR. Src kinase phosphorylates Notch1 to inhibit MAML binding. Sci Rep. 2018;8:15515. doi: 10.1038/s41598-018-33920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge PD, Struhl G. Epsin-dependent ligand endocytosis activates notch by force. Cell. 2017;171:1383–1396.:e12. doi: 10.1016/Jcell.2017.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, Kim C-H, Chitnis AB, Campos-Ortega JA, Weinstein BM. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–136. doi: 10.1016/S1534-5807(02)00198-3. [DOI] [PubMed] [Google Scholar]
- le Noble F, Moyon D, Pardanaud L, Yuan L, Djonov V, Matthijsen R, Breant C, Fleury V, Eichmann A. Flow regulates arterial-venous differentiation in the chick embryo yolk sac. Development. 2004;131:361–375. doi: 10.1242/dev.00929. [DOI] [PubMed] [Google Scholar]
- Lee J, Vedula V, Baek KI, Chen J, Hsu JJ, Ding Y, Chang C-C, Kang H, Small A, Fei P, et al. Spatial and temporal variations in hemodynamic forces initiate cardiac trabeculation. JCI Insight. 2018;3:e96672. doi: 10.1172/jci.insight.96672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kong W. Cellular signaling in abdominal aortic aneurysm. Cell Signal. 2020;70:109575. doi: 10.1016/Jcellsig.2020.109575. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang X, Leathers R, Makino A, Huang C, Parsa P, Macias J, Yuan JX-J, Jamieson SW, Thistlethwaite PA. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat Med. 2009;15:1289–1297. doi: 10.1038/nm.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Breit S, Danckwardt S, Muckenthaler MU, Kulozik AE. Down regulation of Notch signaling by gamma-secretase inhibition can abrogate chemotherapy-induced apoptosis in T-ALL cell lines. Ann Hematol. 2009;88:613–621. doi: 10.1007/s00277-008-0646-x. [DOI] [PubMed] [Google Scholar]
- Lloyd-Lewis B, Mourikis P, Fre S. Notch signalling: sensor and instructor of the microenvironment to coordinate cell fate and organ morphogenesis. Curr Opin Cell Biol. 2019;61:16–23. doi: 10.1016/Jceb.2019.06.003. [DOI] [PubMed] [Google Scholar]
- Loerakker S, Stassen OMJA, ter Huurne FM, Boareto M, Bouten CVC, Sahlgren CM. Mechanosensitivity of Jagged-Notch signaling can induce a switch-type behavior in vascular homeostasis. Proc Natl Acad Sci USA. 2018;115:E3682–E3691. doi: 10.1073/pnas.1715277115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovendahl KN, Blacklow SC, Gordon WR. The molecular mechanism of notch activation. Adv Exp Med Biol. 2018;1066:47–58. doi: 10.1007/978-3-319-89512-3_3. [DOI] [PubMed] [Google Scholar]
- Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, Haltiwanger RS, Zhu C, Ha T, Garcia KC. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science. 2017;355:1320–1324. doi: 10.1126/science.aaf9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucitti JL, Jones EAV, Huang C, Chen J, Fraser SE, Dickinson ME. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development. 2007;134:3317–3326. doi: 10.1242/dev.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxan G, Casanova JC, Martinez-Poveda B, Prados B, D’Amato G, MacGrogan D, Gonzalez-Rajal A, Dobarro D, Torroja C, Martinez F, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- MacGrogan D, D’Amato G, Travisano S, Martinez-Poveda B, Luxan G, Del Monte-Nieto G, Papoutsi T, Sbroggio M, Bou V, Gomez-Del Arco P, et al. Sequential ligand-dependent notch signaling activation regulates valve primordium formation and morphogenesis. Circ Res. 2016;118:1480–1497. doi: 10.1161/CIRCRESAHA.115.308077. [DOI] [PubMed] [Google Scholar]
- MacGrogan D, Münch J, de la Pompa JL. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat Rev Cardiol. 2018;15:685–704. doi: 10.1038/s41569-018-0100-2. [DOI] [PubMed] [Google Scholar]
- Mack JJ, Mosqueiro TS, Archer BJ, Jones WM, Sunshine H, Faas GC, Briot A, Aragon RL, Su T, Romay MC, et al. NOTCH1 is a mechanosensor in adult arteries. Nat Commun. 2017;8:1620. doi: 10.1038/s41467-017-01741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek AM, Alper SL, Izumo S. hemodynamic shear stress and its role in atherosclerosis. JAMA. 1999;282:2035–2042. doi: 10.1001/jama.282.21.2035. [DOI] [PubMed] [Google Scholar]
- Manderfield LJ, High FA, Engleka KA, Liu F, Li L, Rentschler S, Epstein JA. Notch activation of Jagged1 contributes to the assembly of the arterial wall. Circulation. 2012;125:314–323. doi: 10.1161/CIRCULATIONAHA.111.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manokawinchoke J, Pavasant P, Osathanon T. Intermittent compressive stress regulates Notch target gene expression via transforming growth factor-β signaling in murine pre-osteoblast cell line. Arch Oral Biol. 2017;82:47–54. doi: 10.1016/Jarchoralbio.2017.05.020. [DOI] [PubMed] [Google Scholar]
- Martino F, Perestrelo AR, Vinarský V, Pagliari S, Forte G. Cellular mechanotransduction: from tension to function. Front Physiol. 2018;9:824. doi: 10.3389/fphys.2018.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek J, Andersson ER. The developmental biology of genetic Notch disorders. Development. 2017;144:1743–1763. doi: 10.1242/dev.148007. [DOI] [PubMed] [Google Scholar]
- Masumura T, Yamamoto K, Shimizu N, Obi S, Ando J. Shear stress increases expression of the arterial endothelial marker EphrinB2 in murine ES cells via the VEGF-notch signaling pathways. Arterioscler Thromb Vasc Biol. 2009;29:2125–2131. doi: 10.1161/ATVBAHA.109.193185. [DOI] [PubMed] [Google Scholar]
- McCallinhart PE, Biwer LA, Clark OE, Isakson BE, Lilly B, Trask AJ. Myoendothelial junctions of mature coronary vessels express notch signaling proteins. Front Physiol. 2020;11:29. doi: 10.3389/fphys.2020.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meester JAN, Southgate L, Stittrich A-B, Venselaar H, Beekmans SJA, den Hollander N, Bijlsma EK, Helderman-van den Enden A, Verheij JBGM, Glusman G, et al. Heterozygous loss-of-function mutations in DLL4 cause adams-oliver syndrome. Am J Hum Genet. 2015;97:475–482. doi: 10.1016/Jajhg.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta V, Pang K-L, Rozbesky D, Nather K, Keen A, Lachowski D, Kong Y, Karia D, Ameismeier M, Huang J, et al. The guidance receptor plexin D1 is a mechanosensor in endothelial cells. Nature. 2020;578:290–295. doi: 10.1038/s41586-020-1979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloty-Kapella L, Shergill B, Kuon J, Botvinick E, Weinmaster G. Notch ligand endocytosis generates mechanical pulling force dependenton dynamin, epsins, and actin. Dev Cell. 2012;22:1299–1312. doi: 10.1016/Jdevcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miloudi K, Oubaha M, Menard C, Dejda A, Guber V, Cagnone G, Wilson AM, Tetreault N, Mawambo G, Binet F, et al. NOTCH1 signaling induces pathological vascular permeability in diabetic retinopathy. Proc Natl Acad Sci USA. 2019;116:4538–4547. doi: 10.1073/pnas.1814711116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirando AJ, Liu Z, Moore T, Lang A, Kohn A, Osinski AM, O’Keefe RJ, Mooney RA, Zuscik MJ, Hilton MJ. RBP-Jκ–dependent notch signaling is required for murine articular cartilage and joint maintenance. Arthritis Rheum. 2013;65:2623–2633. doi: 10.1002/art.38076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa K, Shi M, Chen P-I, Hennigs JK, Zhao Z, Wang M, Li CG, Saito T, Taylor S, Sa S, et al. Smooth muscle contact drive sendothelial regeneration by BMPR2-Notch1-mediated metabolic and epigenetic changes. Circ Res. 2019;124:211–224. doi: 10.1161/CIRCRESAHA.118.313374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J-S, Kim M-Y, Han S-O, Kim I-S, Ann E-J, Lee KS, Seo M-S, Kim J-Y, Lee S-C, Park J-W, et al. Integrin-linked kinase controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. Mol Cell Biol. 2007;27:5565–5574. doi: 10.1128/MCB.02372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro I, Kollmetz T, Malmstrom J. Engineered systems to study the synergistic signaling between integrin-mediated mechanotransduction and growth factors (Review) Biointerphases. 2018;13:06D302. doi: 10.1116/1.5045231. [DOI] [PubMed] [Google Scholar]
- Morrison TM, Choi G, Zarins CK, Taylor CA. Circumferential and longitudinal cyclic strain of the human thoracic aorta: age-related changes. J Vasc Surg. 2009;49:1029–1036. doi: 10.1016/Jjvs.2008.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow D, Sweeney C, Birney YA, Cummins PM, Walls D, Redmond EM, Cahill PA. Cyclic strain inhibits notch receptor signaling in vascular smooth muscle cells in vitro. Circ Res. 2005;96:567–575. doi: 10.1161/01.RES.0000159182.98874.43. [DOI] [PubMed] [Google Scholar]
- Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, Lim WA. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell. 2016;164:780–791. doi: 10.1016/JCell2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Ratnayake I, Janga M, Fogarty E, Scheidt S, Grassmeyer J, deRiso J, Chandrasekar I, Ahrenkiel P, Kopan R, et al. Notch signaling regulates Akap12 expression and primary cilia length during renal tubule morphogenesis. FASEB J. 2020;34:9512–9530. doi: 10.1096/fJ201902358RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch J, Grivas D, Gonzalez-Rajal Á, Torregrosa-Carrion R, de la Pompa JL. Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development. 2017;144:1425–1440. doi: 10.1242/dev.143362. [DOI] [PubMed] [Google Scholar]
- Murata A, Hayashi S-I. Notch-mediated cell adhesion. Biology. 2016;5:5. doi: 10.3390/biology5010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy PA, Lu G, Shiah S, Bollen AW, Wang RA. Endothelial Notch signaling is upregulated in human brain arteriovenous malformations and amouse model of the disease. Lab Investig. 2009;89:971–982. doi: 10.1038/labinvest.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Obara K, Tanabe Y, Saito M, Ishikawa T, Nishizawa S. Interactive role of tyrosine kinase, protein kinase C, and Rho/Rho kinase systems in the mechanotransduction of vascular smooth muscles. Biorheology. 2003;40:307–314. [PubMed] [Google Scholar]
- Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, Elowitz MB. Dynamic ligand discrimination in the notch signaling pathway. Cell. 2018;172:869–880.:e19. doi: 10.1016/JCell2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto F, Klaus-Bergmann A, Ong YT, Alt S, Vion A-C, Szymborska A, Carvalho JR, Hollfinger I, Bartels-Klein E, Franco CA, et al. YAP and TAZ regulate adherens junction dynamics and endothelial cell distribution during vascular development. eLife. 2018;7:e31037. doi: 10.7554/eLife.31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CM, Huang L, Murphy PA, Lawton MT, Wang RA. Mouse models of cerebral arteriovenous malformation. Stroke. 2016;47:293–300. doi: 10.1161/STROKEAHA.115.002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan DR, Lally C. An investigation of damage mechanisms in mechanobiological models of in-stent restenosis. J Comput Sci. 2018a;24:132–142. doi: 10.1016/Jjocs.2017.04.009. [DOI] [Google Scholar]
- Nolan D, Lally C. Chapter 16-coupled finite element–agent-based models for the simulation of vascular growth and remodeling. In: Cerrolaza M, Shefelbine SJ, Garzon-Alvarado D, editors. In Numerical Methods and Advanced Simulation in Biomechanics and Biological Processes. Academic Press; 2018b. pp. 283–300. [Google Scholar]
- Oomen PJA, Loerakker S, van Geemen D, Neggers J, Goumans M-JTH, van den Bogaerdt AJ, Bogers AJJC, Bouten CVC, Baaijens FPT. Age-dependent changes of stress and strain in the human heart valveand theirrelation with collagen remodeling. Acta Biomater. 2016;29:161–169. doi: 10.1016/Jactbio.2015.10.044. [DOI] [PubMed] [Google Scholar]
- Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Pestel J, Ramadass R, Gauvrit S, Helker C, Herzog W, Stainier DYR. Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development. 2016;143:2217–2227. doi: 10.1242/dev.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic J, Formosa-Jordan P, Luna-Escalante JC, Abello G, Ibanes M, Neves J, Giraldez F. Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development. 2014;141:2313–2324. doi: 10.1242/dev.108100. [DOI] [PubMed] [Google Scholar]
- Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck WJ, Kutys ML, Yang J, Eyckmans J, Wu Y, Vasavada H, Hirschi KK, Chen CS. A non-canonical Notch complex regulate sadherens junctions and vascular barrier function. Nature. 2017;552:258–262. doi: 10.1038/nature24998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcheri C, Golan O, Calero-Nieto FJ, Thambyrajah R, Ruiz-Herguido C, Wang X, Catto F, Guillen Y, Sinha R, Gonzalez J, et al. Notch ligand Dll4 impairs cell recruitment to aortic clusters and limits blood stem cell generation. EMBO J. 2020;39:e104270. doi: 10.15252/embJ2019104270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putti M, Stassen OMJA, Schotman MJG, Sahlgren CM, Dankers PYW. Influence of the assembly state on the functionality of a supramolecular jagged1-mimicking peptide additive. ACS Omega. 2019;4:8178–8187. doi: 10.1021/acsomega.9b00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KE, Tschakovsky ME. The relationship between shear stressand flow-mediated dilatation: implications for the assessment of endothelial function. J Physiol (Lond) 2005;568:357–369. doi: 10.1113/jphysiol.2005.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nat Protoc. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- Quillien A, Moore JC, Shin M, Siekmann AF, Smith T, Pan L, Moens CB, Parsons MJ, Lawson ND. Distinct Notch signaling outputs pattern the developing arterial system. Development. 2014;141:1544–1552. doi: 10.1242/dev.099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani RM, Ratelade J, Domenga-Denier V, Hase Y, Kalimo H, Kalaria RN, Joutel A. Blood brai nbarrier leakage is not a consistent feature of white matter lesions in CADASIL. Acta Neuropathol Commun. 2019;7:187. doi: 10.1186/s40478-019-0844-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riahi R, Sun J, Wang S, Long M, Zhang DD, Wong PK. Notch1-Dll4 signalling and mechanical force regulate leader cell formation during collective cell migration. Nat Commun. 2015;6:6556. doi: 10.1038/ncomms7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristori T, Stassen OMJA, Sahlgren CM, Loerakker S. Lateral induction limits the impact of cell connectivity on Notch signaling in arterial walls. Int J Numer Method Biomed Eng. 2020;36:e3323. doi: 10.1002/cnm.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roese-Koerner B, Stappert L, Brustle O. Notch/Hes signaling and miR-9 engage in complex feedback interactions controlling neural progenitor cell proliferation and differentiation. Neurogenesis (Austin) 2017;4:e1313647. doi: 10.1080/23262133.2017.1313647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard AD, Holmes JW. Coupled agent-based and finite-element models for predicting scar structure following myocardial infarction. Prog Biophys Mol Biol. 2014;115:235–243. doi: 10.1016/Jpbiomolbio.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Rutten JW, Van Eijsden BJ, Duering M, Jouvent E, Opherk C, Pantoni L, Federico A, Dichgans M, Markus HS, Chabriat H, et al. The effect of NOTCH3 pathogenic variant position on CADASIL disease severity: NOTCH3 EGFr 1-6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7-34 pathogenic variant. Genet Med. 2019;21:676–682. doi: 10.1038/s41436-018-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks MS, Yoganathan AP. Heart valve function: a biomechanical perspective. Phil Trans R Soc B. 2007;362:1369–1391. doi: 10.1098/rstb.2007.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa LA, Givens C, Tzima E, Stainier DYR, Qian L, Liu J. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development. 2015;142:4080–4091. doi: 10.1242/dev.125724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- Shah S, Lee S-F, Tabuchi K, Hao Y-H, Yu C, LaPlant Q, Ball H, Dann CE, Südhof T, Yu G. Nicastrin functions as a γ-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/JCell2005.05.022. [DOI] [PubMed] [Google Scholar]
- Shah AV, Birdsey GM, Peghaire C, Pitulescu ME, Dufton NP, Yang Y, Weinberg I, Almagro LO, Payne L, Mason JC, et al. The endothelial transcription factor ERG mediates Angiopoietin-1-dependent controlof Notch signalling and vascular stability. Nat Commun. 2017;8:16002. doi: 10.1038/ncomms16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Ding Z, Zhao M, Liu K, Sun H, Chen J, Liu X, Zhang Y, Hong Y, Li H, et al. Mammalian Numb protein antagonizes Notch by controlling postendocytic trafficking of the Notch ligand Delta-like 4. J Biol Chem. 2017;292:20628–20643. doi: 10.1074/jbc.M117.800946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Graber ZT, Baumgart T, Stone HA, Cohen AE. Cell membranes resist flow. Cell. 2018;175:1769–1779.:e13. doi: 10.1016/JCell2018.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/Jneuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Sjoqvist M, Andersson ER. Do as I say, Not(ch) as I do: lateral control of cell fate. Dev Biol. 2017;447:58–70. doi: 10.1016/Jydbio.2017.09.032. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Lauschke VM, Uraji J, Falk HJ, Petersen Y, Funk MC, Beaupeux M, François P, Merten CA, Aulehla A. Modulation of phase shift between wnt and notch signaling oscillations controls mesoderm segmentation. Cell. 2018;172:1079–1090.:e12. doi: 10.1016/JCell2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner NB, Colliton RP, Crosnier C, Krantz ID, Hadchouel M, Meunier-Rotival M. Jagged1 mutations in Alagille syndrome. Hum Mutat. 2001;17:18–33. doi: 10.1002/1098-1004(2001)17:1<18::AID-HUMU3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen OMJA, Muylaert DEP, Bouten CVC, Hjortnaes J. Current challenges in translating tissue-engineered heart valves. Curr Treat Options Cardiovasc Med. 2017;19:71. doi: 10.1007/s11936-017-0566-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupnikov MR, Yang Y, Mori M, Lu J, Cardoso WV. Jagged and Delta-like ligands control distinct events during airway progenitor cell differentiation. Elife. 2019;8:e50487. doi: 10.7554/eLife.50487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckling RJ, Korona B, Whiteman P, Chillakuri C, Holt L, Handford PA, Lea SM. Structural and functional dissection of the inter play between lipid and Notch binding by human Notch ligands. EMBO J. 2017;36:2204–2215. doi: 10.15252/embJ201796632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift MR, Weinstein BM. Arterial–venous specification during development. Circ Res. 2009;104:576–588. doi: 10.1161/CIRCRESAHA.108.188805. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Yu H, Hao H, Takeuchi M, Ito A, Li H, Haltiwanger RS. O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking. J Biol Chem. 2017;292:15964–15973. doi: 10.1074/jbc.M117.800102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoris CV, Li M, White MP, Liu L, He D, Pollard KS, Bruneau BG, Srivastava D. Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell. 2015;160:1072–1086. doi: 10.1016/JCell2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne BC, Hayenga HN, Humphrey JD, Peirce SM. Toward a multi-scale computational model of arterial adaptation in hypertension:verification of a multi-cell agent based model. Front Physiol. 2011;2:20. doi: 10.3389/fphys.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeijer LA, Frimat J-P, Stassen OMJA, Bouten CVC, Sahlgren CM. Spatial patterning of the Notch ligand Dll4 controls endothelial sprouting in vitro. Sci Rep. 2018;8:6392. doi: 10.1038/s41598-018-24646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregrosa-Carrion R, Luna-Zurita L, Garcia-Marques F, D’Amato G, Pineiro-Sabaris R, Bonzon-Kulichenko E, Vazquez J, de la Pompa JL. NOTCH activation promotes valve formation by regulating the endocardial secretome. Mol Cell Proteomics. 2019;18:1782–1795. doi: 10.1074/mcp.RA119.001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Castellan M, Battilana G, Zanconato F, Azzolin L, Giulitti S, Cordenonsi M, Piccolo S. YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat Commun. 2017;8:15206. doi: 10.1038/ncomms15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubezio B, Blanco RA, Geudens I, Stanchi F, Mathivet T, Jones ML, Ragab A, Bentley K, Gerhardt H. Synchronization of endothelial Dll4-Notch dynamics switch blood vessels from branching to expansion. Elife. 2016;5:e12167. doi: 10.7554/eLife.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaeyens M-M, Jorge-Penas A, Barrasa-Fano J, Steuwe C, Heck T, Carmeliet P, Roeffaers M, Van Oosterwyck H. Matrix deformations around angiogenic sprouts correlate to sprout dynamics and suggest pulling activity. Angiogenesis. 2020;23:315–324. doi: 10.1007/s10456-020-09708-y. [DOI] [PubMed] [Google Scholar]
- van Engeland NCA, Rodriguez FS, Rivero-Müller A, Ristori T, Duran CL, Stassen OMJA, Antfolk D, Driessen RCH, Ruohonen S, et al. Vimentin regulates Notch signaling strength and arterial remodeling inresponse to hemodynamic stress. Sci Rep. 2019;9:12415. doi: 10.1038/s41598-019-48218-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engeland NCA, Pollet AMAO, den Toonder JMJ, Bouten CVC, Stassen OMJA, Sahlgren CM. A biomimetic microfluidic model to study signalling between endothelial and vascular smooth muscle cells under hemodynamic conditions. Lab Chip. 2018;18:1607–1620. doi: 10.1039/C8LC00286J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kelle MAJ, Oomen PJA, Bulsink JA, Janssen-van den Broek MWJT, Lopata RGP, Rutten MCM, Loerakker S, Bouten CVC. A bioreactor to identify the driving mechanical stimuli of tissue growth and remodeling. Tissue Eng Part C Methods. 2017;23:377–387. doi: 10.1089/ten.tec.2017.0141. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;23:4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- Varshney S, Stanley P. Multiple roles for O-glycans in Notch signalling. FEBS Lett. 2018;592:3819–3834. doi: 10.1002/1873-3468.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz KS, Jacobs AH, Chen HI, Poduri A, McKay AS, Riordan DP, Kofler N, Kitajewski J, Weissman I, Red-Horse K. Pericytes are progenitors for coronary artery smooth muscle. Elife. 2015;4:e10036. doi: 10.7554/eLife.10036.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijts B, Gutierrez E, Saikin SK, Ablooglu AJ, Traver D, Groisman A, Tkachenko E. Blood flow-induced Notch activation and endothelial migration enable vascular remodeling in zebrafish embryos. Nat Commun. 2018;9:5314. doi: 10.1038/s41467-018-07732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Shen Y, Berry G, Shahram F, Li Y, Watanabe R, Liao YJ, Goronzy JJ, Weyand CM. The microvascular niche instructs T cells in large vessel vasculitis via the VEGF-Jagged1-Notch pathway. Sci Transl Med. 2017;9:eaa13322. doi: 10.1126/scitranslmed.aa13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield JM, Barrera FN. Membrane receptor activation mechanisms and transmembrane peptide tools to elucidate them. J Biol Chem. 2020;295:1792–1814. doi: 10.1074/jbc.REV119.009457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedermann G, Bone RA, Silva JC, Bjorklund M, Murray PJ, Dale JK. A balance of positive and negative regulators determines the pace of the segmentation clock. eLife. 2015;4:e05842. doi: 10.7554/eLife.05842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, Taubenschmid J, Hammerle M, Esk C, Bagley JA, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing TB, Bonito V, Bouten CVC, Smits AIPM. Biomaterial-driven in situ cardiovascular tissue engineering —a multi-disciplinary perspective. npj Regenerative Medicine. 2017;2:18. doi: 10.1038/s41536-017-0023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedmanesh H, Lally C. A multiscale mechanobiological modelling framework using agent-based models and finite element analysis: application to vascular tissue engineering. Biomech Model Mechanobiol. 2012;11:363–377. doi: 10.1007/s10237-011-0316-0. [DOI] [PubMed] [Google Scholar]