Abstract
Spatial metabolomics is an emerging field of omics research that has enabled localizing metabolites, lipids, and drugs in tissue sections, a feat considered impossible just two decades ago. Spatial metabolomics and its enabling technology—imaging mass spectrometry—generate big hyper-spectral imaging data that have motivated the development of tailored computational methods at the intersection of computational metabolomics and image analysis. Experimental and computational developments have recently opened doors to applications of spatial metabolomics in life sciences and biomedicine. At the same time, these advances have coincided with a rapid evolution in machine learning, deep learning, and artificial intelligence, which are transforming our everyday life and promise to revolutionize biology and healthcare. Here, we introduce spatial metabolomics through the eyes of a computational scientist, review the outstanding challenges, provide a look into the future, and discuss opportunities granted by the ongoing convergence of human and artificial intelligence.
Keywords: spatial metabolomics, imaging mass spectrometry, artificial intelligence
Introduction
Spatial metabolomics is a field of omics research focused on the detection and interpretation of metabolites, lipids, drugs, and other small molecules in the spatial context of cells, tissues, organs, and organisms (1). Spatial metabolomics is a rapidly emerging field, fueled by the strong and evergrowing need in biology and medicine to characterize biological phenomena in situ, as well as by the recently revealed key roles of metabolism in health and disease. This field is concerned with a variety of biomedical questions, including the tumor molecular microenvironment (2), functions of immune cells during homeostasis and immunotherapy (3), interactions between host and microbiota (4) and their contribution to inflammation (5), regulation of early development (6), metabolic regulation of epigenetics (7), and metabolic dysregulations during infection (8) and inflammation (9, 10). Over the past decade, this growing interest has stimulated rapid progress in the development of enabling technologies—in particular, imaging mass spectrometry (MS)—that have achieved unprecedented sensitivity, coverage, and robustness as they have become accessible to biologists (11–20).
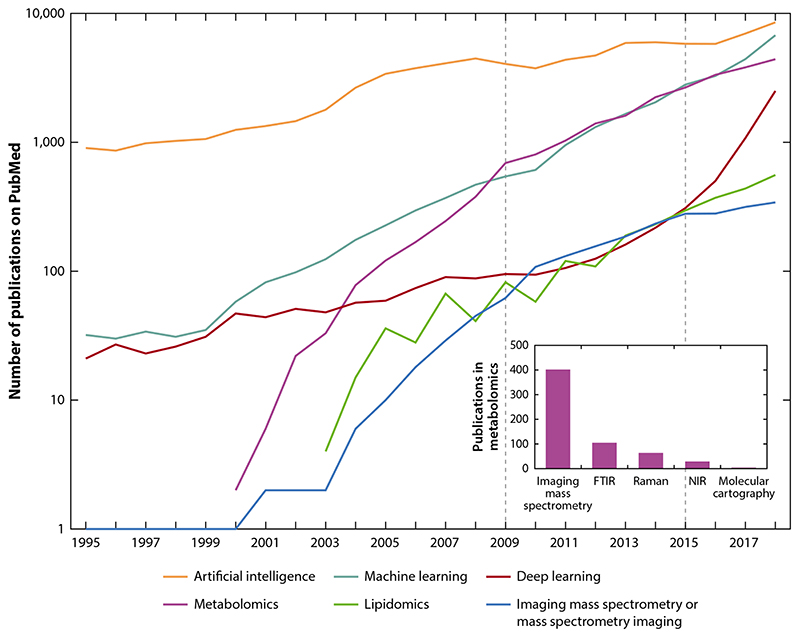
This article provides a review of the challenges in spatial metabolomics and imaging MS, the dominant technology for spatial metabolomics, through the eyes of a computational scientist. In particular, we discuss how artificial intelligence (AI), machine learning, and deep learning are used in spatial metabolomics and what potential they have to transform this field. Our considerations contribute to a broader discussion of an issue recently raised in Nature Metabolism: why the metabolism field risks missing out on the AI revolution (21). Using publication numbers in PubMed, the most comprehensive database of publications in life sciences and biomedicine (Figure 1), one can divide the evolution of technologies for spatial metabolomics in the twenty-first century into three periods: prior to 2009, from 2009 until 2015, and since 2015. Prior to 2009, metabolomics and imaging MS were developing with an over-exponential speed and reached levels comparable to machine learning and deep learning, respectively. From 2009 until 2015, metabolomics and machine learning showed similar exponential growth in the number of publications in the life sciences. Correspondingly, both imaging MS and deep learning were growing at the same pace. However, in 2015 the situation changed, with deep learning exhibiting an over-exponential explosion of popularity, whereas the growth of metabolomics and imaging MS slightly slowed down, despite still growing exponentially. These numbers suggest that, indeed, metabolomics and in particular spatial metabolomics could benefit more from the rise of deep learning. Although this observation is also true for other omics fields such as genomics, proteomics, and transcriptomics that show trends similar to metabolomics (not shown in Figure 1), in this review we focus on challenges and opportunities specific to spatial metabolomics.
Figure 1.
The popularity of different technologies in the life sciences and biomedicine and their evolution over time. The plot shows the numbers of PubMed-indexed publications in a given year containing the keywords shown in the figure key. We highlight three time periods, before 2009, from 2009 until 2015, and after 2015, which we discuss in the main text. The inset shows the popularity of several technologies for metabolomics applications from 1995 until 2018. Abbreviations: FTIR, Fourier-transform infrared spectroscopy; NIR, near-infrared spectroscopy.
We review the principles and unique advantages of spatial metabolomics and outline the state of the art as well as challenges in data analysis in this field. Then, we focus on the outstanding challenges that have received attention but still need solutions and provide an outlook on how novel computational approaches such as machine learning and AI can open up new avenues in spatial metabolomics. We discuss in detail the use of machine learning, big data, and AI approaches in this field, as well as current bottlenecks, in particular, the lack of so-called ground truth data, and how they can be addressed. We provide a list of benchmark datasets and software tools that can serve as entry points for newcomers. Overall, we hope that this review will help beginners in the field of spatial metabolomics orient themselves to the variety of existing computational methods and problems and help experimentalists establish stronger collaborations with computational scientists, in particular by explaining the benefits and prerequisites of modern computational methods.
Bulk Metabolomics and The Need for in Situ Analyses
Bulk metabolomics is performed by extracting metabolites from samples such as in vitro cultured cells, tissues, or biofluids and then subjecting them to further detection and quantification of endogenous and exogenous metabolites, often using MS (22–24). Although this approach is widely accepted and can successfully detect remodeling of metabolism in an organism or its parts, even in bodily fluids (25–27), it cannot pinpoint the localization of metabolites in specific organelles, cells, anatomically or histologically defined parts of tissues, or organs. This makes biological interpretation of metabolomics data challenging and hinders the association of overall metabolic changes with a particular tissue, organ, or spatially localized aberration such as a tumor. This problem can be partially overcome by dissecting tissues (28, 29) or organs (30) before performing bulk metabolomics on individual dissected parts. However, such experiments are hardly scalable, are time consuming, and require expertise and the development of tailored methods for sample handling, normalization, data analysis, and visualization, making them out of reach for most scientists.
Imaging Mass Spectrometry as Enabling Technology
Addressing the need for in situ metabolomics, researchers have developed a variety of spatial metabolomics approaches over the past two decades, enabling the detection of metabolites, lipids, and other small molecules in a broad range of biological samples, with two-dimensional (2D) or 3D spatial resolution on the scales of organisms, organs, tissues, or cells (1). In this review, we focus on spatial metabolomics based on imaging MS (12–20, 31). MS is the most widespread technique for bulk metabolomics (32) and provides an unsurpassed combination of molecular coverage, sensitivity, and specificity. Imaging MS combines the spatially resolved sampling of molecules with their detection in a mass spectrometer. The sampling is systematically performed by dividing the surface of a sample, usually a tissue section, into a virtual grid of pixels. For every pixel in the grid, molecules are desorbed from the area of the pixel using a laser or another ablation method, followed by the generation of a mass spectrum representing relative intensities of molecules within the pixel.
Imaging MS was proposed at the end of the twentieth century, in particular for localizing proteins in cryosections (33, 34); readers are referred to Reference 35 for a historical perspective. Since then, the application’s focus has shifted toward the detection of metabolites, lipids, and small molecules, in particular with the introduction of so-called high-resolving MS (36). Over the past two decades, imaging MS created one of the fastest-growing fields in MS, with most vendors providing commercial solutions. Acknowledging other approaches to spatial metabolomics such as Fourier-transform infrared spectroscopy (37), Raman spectroscopy (38), near-infrared spectroscopy, and molecular cartography (39), here we focus on imaging MS as the enabling and most popular technology for spatial metabolomics (Figure 1).
The various flavors of imaging MS can be categorized by their so-called source, that is, the apparatus by which molecules are ablated from a sample in a spatial manner, and by the analyzer, that is, the mass spectrometer used for detecting and characterizing molecules desorbed from every pixel (35, 40). The most common source type is matrix-assisted laser desorption/ionization (MALDI), where molecules are ablated and simultaneously ionized by a laser (36). Other prominent source types include liquid-based desorption electrospray ionization (DESI) (41); secondary ion mass spectrometry (SIMS), which uses an ion beam (42, 43); infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI), which combines infrared laser and electrospray ionization (44), and the similar laser ablation electrospray ionization (LAESI) (45); nanostructureinitiator mass spectrometry (NIMS) (46); and nanospray desorption electrospray ionization (nanoDESI) (47).
Machine Learning, Deep Learning, and Artificial Intelligence
Machine learning, deep learning, and AI are pushing the boundaries of our understanding of the tasks that can be solved by a computer in everyday life. Examples include autonomous driving, AlphaGo, and IBM Watson. The fields of life sciences and biomedicine are presently embracing these approaches with big hopes that they will transform healthcare and science, in particular when using rich omics or imaging data. In this section, we provide definitions and references introducing and reviewing key concepts and achievements.
Machine learning is an approach to solving a problem that learns from the data rather than encodes decisions based on expert knowledge (48). Deep learning is a subfield of machine learning that uses deep artificial neural networks, a method that has transformed machine learning by outperforming other methods, first for computer vision and later for other problems (49). AI is the broadest field of these three and is concerned with making computers perform intelligent tasks as well as or better than humans (50). In Figure 1, which shows the number of publications in PubMed in various fields, one can clearly see the over-exponential rise of deep learning applications in the life sciences and medicine since 2015 and its impact on the adoption of AI in these fields.
For a comprehensive introduction to machine learning from the medical perspective, including its areas of greatest promise and its challenges, in particular the central challenge of the availability of high-quality data for training (so-called ground truth), readers are referred to Reference 51. To dive deeper into the computational principles of deep learning with applications in biology, see Reference 52, and for applications in biomedicine, see References 53 and 54. Reference 55 reviews the use of machine learning and deep learning in healthcare with a focus on types of machine learning. For a comprehensive review of AI in medicine and its potential to transform the business model in healthcare by capitalizing on massive quantities of data generated in modern medicine, see Reference 56. With the onset of AI in healthcare, there is a hope for positive transformations not only in high-income countries but also in resource-poor healthcare settings (57).
Imaging Mass Spectrometry Data
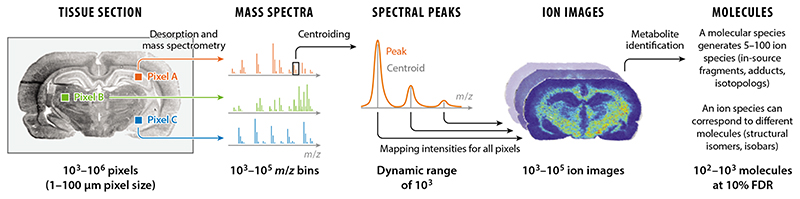
An imaging MS dataset represents a collection of mass spectra, each with its associated x-y pixel position. It can also be viewed as a hyperspectral image consisting of multiple ion images or channels, with every ion channel corresponding to a particular mass-to-charge (m/z) value and representing relative intensities of ions with this m/z value (58). The number of pixels is usually between 10,000 and 100,000 and is often limited by the MS throughput operating at 1–50 Hz (1 Hz representing 1 spectrum/s). The recent progress in high-speed imaging MS, as well as technological solutions supporting the stability of laser focusing (59), enables collecting even bigger datasets.1 This progress will likely enable routine collection of datasets with over one million pixels in the next year or two. The dimensionality of an individual spectrum depends on the analyzer and is usually on the order of 103 for time-of-flight (TOF) analyzers, 104 for quadrupole time-of-flight (QTOF) analyzers, and over 104 for Fourier transformation (FT) analyzers. High-resolution data collected using FT analyzers, such as Fourier transformation ion cyclotron resonance (FTICR) or Orbitrap, are often reduced in the spectral dimension by applying so-called centroiding. Centroiding is a procedure applied to every spectrum that, for every spectral peak, selects one central m/z value (the so-called centroid), and its associated intensity is often calculated as the area under the peak (see Figure 2). Centroiding is essential for FT data as it reduces the dimension and size of multigigabyte data by an order of magnitude.
Figure 2.
An imaging mass spectrometry (MS) dataset represents a collection of spectra acquired from a raster of pixels representing the surface of a tissue section. Peaks are often reduced to centroids, especially for Fourier transform ion cyclotron resonance and Orbitrap high-resolving power analyzers. An ion image represents relative intensities of the ion across all pixels. An imaging MS dataset can represent spatial localization of up to 103 molecules at a false discovery rate (FDR) of 10%.
Key Parameters of Imaging Mass Spectrometry and How They Affect the Data
In Table 1, we summarize key parameters of imaging MS that have the most profound impact on the data. The sample preparation (60), source type, and analyzer polarity predetermine which molecular classes will be desorbed and ionized and thus highly affect the molecular content of the data. These parameters do not directly affect the spectral properties of the data, which makes most of the data analysis methods agnostic to them. The analyzer type, however, has a strong impact on the spectral properties and size of the data and thus affects signal processing, data analysis, and molecular analysis. The mass-resolving power predetermines how detailed the spectra are and whether two molecules with similar m/z values can be resolved from each other; it is a critical parameter affecting data size and molecular ambiguity. Finally, mass accuracy, or how accurately the measured centroids of the spectral peaks for a molecule represent its theoretical m/z value, is a critical property of imaging MS data, as it substantially affects data quality, molecular ambiguity, and overall interpretability. The mass accuracy is different from other parameters, as it cannot be set during the experiment but can be affected by multiple factors, including the calibration and stability of the mass spectrometer and even the thickness of the analyzed sample (for TOF analyzers). Only a few imaging MS software packages (including open-source MSiReader as well as some vendor-provided packages) provide an easy way to estimate mass accuracy for already collected data.
Table 1. Imaging MS parameters and how they affect data properties.
| Parameters of imaging MS | Affected aspects and data properties | |
|---|---|---|
| Sample preparation | MALDI matrix (e.g., DHB, DAN) | Desorption and ionization (and hence detectable molecular classes), spatial resolution (but not critically), sensitivity |
| Solvent (e.g., acetonitrile) | Desorption and ionization, sensitivity | |
| Source | Source type (e.g., MALDI, DESI, SIMS, IR-MALDESI) | Desorption and ionization, sensitivity (and thus the number of ion channels), potentially spatial resolution (and thus the number of pixels) |
| Analyzer | Analyzer type (FTICR, Orbitrap, TOF, QTOF) | Strongly affects spectral properties (dimensionality, noise, background, and, in particular, TOF) and molecular ambiguity; affects spatial properties (noise, resolution) to a lesser degree |
| Polarity (positive, negative) | Which ions are detected (and hence molecular classes) | |
| Mass-resolving power (e.g., 50,000 or 120,000 at 200 m/z) | Critically affects molecular ambiguity (inversely proportional) and data size (proportional) | |
| Mass accuracy | Critically affects data quality, molecular ambiguity, and interpretability | |
Abbreviations: DAN, 1,5-diaminonapthalene; DESI, desorption electrospray ionization; DHB, 2,5-dihydroxybenzoic acid; FTICR, Fourier transformation ion cyclotron resonance; IR-MALDESI, infrared matrix-assisted laser desorption electrospray ionization; MALDI, matrix-assisted laser desorption/ionization; MS, mass spectrometry; QTOF, quadrupole time-of-flight; SIMS, secondary ion mass spectrometry; TOF, time-of-flight.
Common Data Analysis Steps
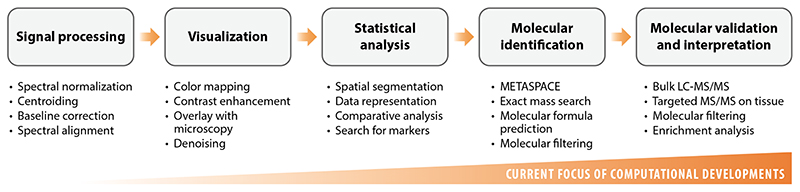
The steps of a typical data analysis workflow in imaging MS–based spatial metabolomics include signal processing, visualization, statistical analysis, molecular identification, molecular validation, and interpretation (Figure 3). Presently, the focus of computational methods development has shifted toward molecular identification as well as molecular validation and interpretation, with low-level signal processing increasingly performed by the vendor software.
Figure 3. Steps of a typical data analysis workflow in imaging mass spectrometry. Abbreviations: LC-MS/MS, liquid chromatography with tandem mass spectrometry; MS/MS, tandem mass spectrometry.
Challenges Compared to Bulk Metabolomics
As discussed above, in situ analysis is often desired and necessary in order to interpret metabolomics data and link them to particular aspects of metabolism compartmentalized within particular organelles, cells, tissues, or organs. Spatial metabolomics using imaging MS provides unique opportunities to investigate tissue sections and overlay metabolite localization with microscopy images. However, these advantages of spatial metabolomics come with some limitations compared to bulk MS-based metabolomics. Here, we discuss two aspects that are critical for the understanding of imaging MS data and that often drive tailored computational developments in this field.
First, in imaging MS the molecular material from a pixel is represented in one spectrum. In contrast, in bulk metabolomics, the molecular material is often separated by another molecular property prior to MS analysis, e.g., by the time of retention through a chromatographic column, as in liquid chromatography–mass spectrometry (LC-MS). This leads to the high complexity of imaging mass spectra and, more specifically, to a high number of peaks in a spectrum, potential overlap of molecular signals, and high noise and variability of spectral intensities. The emerging use of ion mobility separation (IMS) for imaging MS, where the molecules are first separated based on their collisional cross section by flying through a buffer gas, has the potential to overcome this limitation. Computational developments are urgently needed to make use of this additional dimension (61).
Second, imaging MS predominantly exploits the so-called MS1 type of MS where m/z values of ionized analytes are measured in a mass spectrometer. On the contrary, bulk metabolomics largely relies on tandem MS (abbreviated as MS/MS or MS2) where analytes are first fragmented into their structural parts whose m/z values are measured in a mass spectrometer (22, 32, 62). Although MS/MS cannot reconstruct the molecular structure completely, it reduces potential ambiguity, as an MS/MS spectrum encodes additional structural information that is missed in MS1. This leads to a general inability of imaging MS to discern structural isomers, namely, molecules with the same molecular formula. Examples of isomeric molecules include D-glucose, D-galactose, and l-galactose, which all have the molecular formula C6H12O6, or diglycerides DG(16:0_16:0) and DG(18:0/14:0), which all have the molecular formula C35H68O5. In imaging MS, MS/MS spectra can be collected for selected ions only, normally not in a true imaging mode but for a few pixels only, and are generally of low quality due to the overall low amount of molecular material contained in one pixel. Currently, all commercial types of mass spectrometers suffer from this limitation and imaging MS/MS analysis is not supported in vendor-provided software. Below we discuss emerging solutions to this problem.
Recent Computational Advances
The field of spatial metabolomics in its current evolution is enabled by imaging MS technology developed over the past two decades. In particular, in the past decade, the field consolidated a noticeable community of computational methods developers, both in academia and industry, who together addressed several challenges. In this section, we review recent computational advances.
Data Handling and Signal Processing
Data handling was a considerable challenge in imaging MS just a decade ago, particularly when using FTICR or Orbitrap analyzers that can generate millions of m/z bins. Since then, most vendors introduced efficient data formats, often including centroiding or other types of data compression, which reduced data size and facilitated data storage and handling. The crucial success on the way to data interoperability was the development of the imaging MS data format imzML (63). imzML is an open format and is supported by the majority of vendors by allowing users to export their data into this format, and by leading open-source packages where imzML files can be loaded and processed. Thus, imzML is the main means of data exchange between software packages. However, for computational analysis, it is recommended that data be stored in some internal format. imzML is not efficient for data access, particularly when dealing with large data with a large number of pixels, because in imzML every spectrum is stored with its metadata. Similarly, imzML is poorly suited for the emerging use of IMS in imaging MS where the number of spectra is increased by a factor of 102–104. Discussions about a new version of imzML are ongoing but no concrete plans are announced yet. Independently of their support of the imzML format, several vendors have announced that they will either provide access to their data through open interfaces or create APIs for interacting with their software, which should provide computational methods developers fast direct access to data and would help vendors integrate open-source developments.
Signal processing is a foundational step and is essential for any data analysis. Substantial methods development efforts were devoted to this topic in the early years of imaging MS, in particular, to address the problems of spectral normalization and spectral centroiding (58, 64, 65). Various methods of spectral normalization were demonstrated to improve data yet are known to produce artifacts (13). The problem of the choice of normalization is still open, as there are no ground truth data that would contain known distributions of molecules and could be used for objective comparison of existing normalization methods. Currently, every scientist or lab decides based on their own empirical experiences whether to apply spectral normalization at all and which type of normalization to use. This hinders the comparison of the data between labs and can potentially lead to artifacts, particularly when analyzing new types of samples. This problem is an example of how the lack of ground truth data can hamper computational developments; below we discuss this aspect in detail.
Spectral centroiding (previously often called peak picking) is a process of converting a spectral peak corresponding to an ion to a centroid m/z value with an associated intensity (Figure 2). An ion species generates a spectral peak representing a random distribution of m/z measurements of the ions of that species. The shape of a peak is close to a Gaussian function but varies depending on the analyzer type and resolving power. Peaks in FTICR and Orbitrap data have a clean smooth shape whereas peaks in TOF and QTOF data are noisier. Using a higher-resolving power leads to narrower peaks. Working with centroided data is often easier since it simplifies the data and compresses it at approximately a 10x rate. Substantial efforts devoted to spectral centroiding over the past two decades have led to the development of robust methods, and currently, most vendors support the export of centroided data, often into the centroided imzML format.
TOF baseline correction is a process of detecting and removing background baseline in a TOF mass spectrum, which was a considerable challenge in the early days of imaging MS; see Reference 58 for more details. With the introduction of high-resolving power analyzers (FTICR, Orbitrap, QTOF), using TOF in imaging MS became less popular (36), which shifted the interest of methods developers away from TOF-specific signal processing.
Visualization
Visualization of imaging MS data in its spatial coordinates can provide an intuitive and easily interpreted view into which metabolites are detected, where they are localized, and what spatial associations exist between molecules. Even more informative can be a visual overlay of ion images with a microscopy image from the same or a consecutive section that can aid in associating metabolites with histopathologically defined regions (e.g., tumor or stroma), cells of a particular type or phenotype (e.g., stem cells, cells exhibiting macrovesicular steatosis or a particular cell cycle phase), or expression of a particular protein or transcript (e.g., a metastasis marker or cell-type marker) imaged using an orthogonal technique such as immunohistochemistry, immunofluorescence, or fluorescence in situ hybridization.
Numerous methods have been developed and implemented in vendor-provided and academic software packages for visualizing ion images, overlaying them with microscopy images and enhancing the contrast (13, 40). Since working with imaging MS data involves visual examination and comparison of numerous ion images, the choice of a colormap for visualization has been a matter of debate at various conferences. Presently, as in many other fields (66), rainbow and jet colormaps are highly discouraged while perceptually uniform colormaps like viridis are recommended. Interestingly, this topic in imaging MS is so important that it stimulated the development of a novel colormap, cividis, which is perceptually uniform and adapted for people with color vision deficiency (67).
Unsupervised Data Analysis
Although visual examination of individual ion images corresponding to specific metabolites is essential and part of everyday work for any imaging mass spectrometrist, this can hardly be done for all 103–105 ion channels contained in a regular imaging MS dataset. Moreover, some compressed visual representation of a whole imaging MS dataset is desired for quality control, expert assessment, and hypothesis generation. This has stimulated the development of unsupervised methods that can represent the molecular and spatial content of an imaging MS dataset with one or a few images. Here, we briefly discuss this topic; for a comprehensive review, see Reference 68, where 40 methods are reviewed and discussed in detail. For an experimental viewpoint and how unsupervised analysis fits into a complete data analysis pipeline, see Reference 13.
Most unsupervised data analysis methods in imaging MS can be categorized into dimensionality reduction (component analysis methods), spatial segmentation (or clustering of pixels), and clustering of ion images. Since the early days, dimensionality reduction has predominantly been performed by using component analysis or factorization methods such as principal component analysis (PCA) and non-negative matrix factorization (NMF). PCA and NMF have obtained substantial popularity, as they can visualize both distinct spatial regions and associated spectral patterns. Other methods have been evaluated and developed; see Reference 68 for a comprehensive list. Spatial segmentation aims at partitioning a sample into distinct regions of molecularly similar pixels and is one of the most popular approaches for data representation in imaging MS. Besides naïve clustering of spectra, segmentation methods tailored for imaging MS have been developed, in particular with the aim of reducing strong pixel-to-pixel variability by employing image processing (69), spatially aware clustering (70), post hoc smoothing of obtained segmentation maps (71), or spatially shrunken centroids (72). Clustering of ion images was proposed to investigate spatial similarities between ion images (73, 74), and its usefulness for supervised analysis was recently demonstrated (75). Among other methods for data representation, self-organizing maps were shown to visualize chemically similar regions of interest (76–78). Manifold learning methods such as t-SNE (t-distributed stochastic neighbor embedding; 79) and UMAP (uniform manifold approximation and projection; 80, 81) are gaining popularity for visualizing spectra or ion images in 2D space, for aiding data interpretation, and for finding clusters of similar pixels or images.
Quantifying the Quality of Ion Images
Even in a high-quality imaging MS dataset, a considerable proportion of ion images either are prohibitively noisy or contain a large number of pixels with zero intensities. This can be due to the images representing molecular concentrations close to the limit of detection. Since they are noisy, such images need special treatment or otherwise can pollute downstream analysis. Expert-performed visual examination of ion images to assess their informativeness is still a part of many workflows in imaging MS. Recently, several methods were developed that can automatically quantify the quality or informativeness of an ion image, including the spatial measure of spatial chaos, gray-level co-occurrence matrices, or the drug homogeneity index (82–85). The developed methods have been evaluated using either a small set of preselected 250 ion images (82) or 634 pairs of crowdsourced expert-ranked images (83). Open-source implementations of these methods are available that have stimulated their integration into academic and commercial software packages. Looking into the future, despite the fact that these methods already provide acceptable performance, they may be further improved by modern computer vision methods such as deep learning, especially if bigger training sets were provided.
Filtering Out Off-Sample Ion Images
Any imaging MS dataset contains ion images corresponding to either solvent (in DESI and nan-oDESI) or MALDI matrices, as well as other background contaminants. These ion images exhibit the so-called off-sample localization with high-intensity pixels outside of the sample area. Representing technical artifacts rather than biological content yet exhibiting high intensities, these images may confound statistical analysis and lead to false positives in molecular identification. Until recently, no automated solutions for filtering off-sample ion images were provided. MSiReader software provides a semisupervised way for filtering off-sample ion images that requires selecting the off-sample area manually (86). We have recently created a high-quality ground truth set of 23,238 ion images where every image has been annotated by an expert as either on-sample, off-sample, or unknown (87). Then, we trained a deep learning model and were able to automatically reproduce expert judgments with high accuracy. Furthermore, we have shared the model and integrated it into the METASPACE cloud software (87), thus making it widely available.
Quantifying Colocalization Between Metabolites
Various applications require calculating the colocalization of molecules in an imaging MS dataset. This is often needed for untargeted spatial metabolomics in order to discover metabolites that are accumulated in particular regions or spatially associated with each other. The most common scenarios include finding metabolites that are spatially associated with another molecule (e.g., either a known oncometabolite or an unknown ion found through statistical analysis) or that exhibit a particular spatial localization (e.g., localized within a region found through spatial segmentation or within a particular histopathological region visible in microscopy).
Various measures for quantifying colocalization between ion images have been proposed, including the Pearson correlation, cosine score, and Euclidean L2-measures (58, 69, 88, 89), applied to either intact or transformed images, e.g., after hotspot removal or log transformation (40). Novel measures adopted from other fields have been proposed, including the structural similarity index and the hypergeometric similarity measure (90–92). For finding molecules colocalized with a region of interest, researchers have developed a non-negative least squares approach (93). We have recently created a gold standard set based on judgments of 42 experts, evaluated several existing measures, and proposed new measures that can reproduce expert judgments, including those based on deep learning (81). This gold standard set is publicly available and can be used for training machine learning and deep learning models to achieve even higher performance.
Spatial Quantitation of Metabolites
Absolute quantitation or estimation of the concentration of a molecule in sampled pixels or a spatial region of interest is essential for modeling in quantitative biology, as well as for pharma-cokinetics/pharmacodynamics studies during drug development. Recently, several protocols for quantitative imaging MS were developed (94–96); see Reference 13 for a review. Until recently, imaging MS was called a semiquantitative technology, meaning that it can be used to compare intensities between pixels but does not provide absolute quantitation of molecular concentrations. However, several studies have demonstrated that imaging MS can achieve quantitation with a high agreement of 70–90% with LC-MS, a gold standard for quantitative bulk metabolomics (97, 98). Similar to quantitative LC-MS, this can be achieved for targeted analysis of a predetermined set of metabolites and requires a careful experimental design, including authentic internal standards. Developing computational methods supporting spatial quantitation is an active area of research and development. The only open-source software that supports quantitation is MSiReader (86), with non-open-source packages also available (99).
Spatial Metabolomics in Three Dimensions
The advance and increasing popularity of 2D spatial metabolomics has led to a growing interest in performing spatial metabolomics in 3D. This would not only provide a richer representation of metabolite localization but would also allow metabolite data to be overlaid with other established and emerging 3D imaging data from magnetic resonance imaging (MRI), microcomputed tomography, or block-face and light-sheet microscopy. Such multimodal imaging would be particularly useful for biological and medical applications involving animal models, e.g., creating a molecular-anatomical atlas or finding metabolites spatially colocalized with xenograft or cancer metastases.
This growing interest has led to the development of methods for 3D spatial metabolomics of tissue specimens, which is normally performed by serial sectioning followed by the 2D analysis of obtained sections and 3D reconstruction; see Reference 100 for a review. Other approaches include SIMS, where an ion beam ablates layers of molecules (101), which can be applied only to small samples such as individual cells. 3D molecular cartography has been proposed for the analysis of not only biological surfaces but also organs like the lung (28, 39). Early reports of serial 3D imaging MS (102–107) revealed associated challenges: long acquisition times, the need for special software for registering serial sections, and the need for special protocols and software for simultaneously coregistering 3D images with other 3D imaging modalities so that hundreds to thousands of ion images can be explored in 3D. Since then, commercial software has been developed that helps create a 3D imaging MS dataset and visualize the localization of ions in 3D; see, e.g., Reference 107. Despite the availability of such software, presently 3D imaging MS is not widespread because it requires substantial manual work for sample sectioning and preparation, takes extensive instrument time, and reduces laser lifetime when used in MALDI imaging MS. In 2015, we argued that 3D imaging MS was at its tipping point (100). Since then, a niche for this technology is still to be found, potentially in clinical applications where 3D imaging methods such as MRI are already in place (108, 109).
Outstanding Computational Challenges
As reviewed in the previous section, over the past two decades many computational challenges have been addressed that have removed barriers for applications of imaging MS in various biomedical areas. However, several challenges are still outstanding and require computational developments. In this section, we review such challenges, what have been the key bottlenecks, and how they can be addressed in the future. This section can be of interest for computational scientists who are interested in contributing to cutting-edge developments. It can also serve as a note of caution for imaging MS practitioners and experimentalists informing them about gaps of knowledge and about areas of active ongoing computational research.
Molecular Identification
Molecular identification is the process of finding molecules represented in imaging MS data and is probably the biggest challenge in imaging MS–based spatial metabolomics. Molecular identification is a key challenge even in bulk metabolomics, where chemical separation (e.g., liquid chromatography, gas chromatography, and, increasingly, IMS) and MS/MS aid finding signals corresponding to known molecules and help de novo reconstruction of the putative structures of unknown molecules (110–112). This challenge was so prominent that several competitions were organized in 2012–2017 to motivate computational developments and aid methods comparison (113). In imaging MS, as discussed above, the problem is exacerbated by both the general inability of collecting spatially resolved MS/MS spectra in an untargeted fashion and the complexity of spectra due to the lack of separation. Since imaging MS cannot resolve isomeric molecules (see discussion above), the problem of metabolite identification in this field is often termed metabolite annotation.
Until recently, a common approach for metabolite annotation in imaging MS was to match spectral signals or peaks of interest against a molecular database by their m/z values (e.g., using the Human Metabolome Database or SwissLipids database), a process often called m/z matching. However, we have shown that m/z matching is unreliable and cannot provide results with a false discovery rate (FDR) lower than 25% even when using a stringent m/z tolerance of ±2.5 ppm (114). This means that among the reported molecules, 25% or more would be false-positive hits. This leads to high ambiguity and increased experimental and data analysis time for follow-up experiments and overall slows down molecular data interpretation.
To provide a reliable solution to this challenge, we have recently developed an FDR-controlled metabolite annotation (114). This approach uses a novel metabolite–signal match score that exploits both spectral and spatial information from imaging MS data and matches it to theoretically predicted properties of a metabolite such as its fine isotopic pattern. Our approach substantially outperforms m/z matching and can annotate 102–103 ions at 10% FDR (115). We have implemented it as the free and open-source cloud software METASPACE, which is currently being used by over 200 users from over 100 imaging MS labs and has already helped annotate more than 5,000 datasets (115).
Despite the fact that METASPACE provides a viable solution to metabolite annotation in imaging MS, several challenges remain. First, currently we can annotate approximately 1% of imaging MS data (102 molecules out of a dataset with 104 m/z channels). The overwhelming majority of the data is left unannotated, sometimes called dark matter. This gap exists due to our limited understanding of ionization pathways or principles of how an analyte forms a signal in imaging MS. Only empiric and fragmentary knowledge is available about in-source fragmentation, adduct formation, neutral losses, and clusters formation. Second, the lack of MS/MS and additional chemical separation (see discussion of IMS below) does not allow structural isomers or isobars to be resolved. The state-of-the-art approach is to perform MS1-based annotation of imaging MS data, complement it with a bulk LC-MS/MS-based metabolomics, and perform m/z matching between the molecules annotated by imaging MS and identified with LC-MS/MS. This, however, does not necessarily fully resolve the ambiguity. Previously we published an example (114) where two isomeric lipids (a galactosylceramide and a glucosylceramide, both with the molecular formula C48H91NO8) annotated in MALDI imaging MS data of a mouse brain tissue section were both identified by LC-MS/MS. This leaves open the possibilities of both lipids having either the same spatial distribution or different distributions, with the ion image corresponding to C48H91NO8 being a composite of them. Third, our preliminary results (unpublished) show that most of the imaging MS methods are prone to in-source fragmentation. In MS, in-source fragmentation is the process of fragmenting off a structural part of a molecule during ionization or desorption, reported both for metabolites (116) and for lipids (117). An example of in-source fragmentation is the loss of one or two phosphate groups from endogenous ATP, a key energetic metabolite, leading to false detection of ADP or AMP even if they are not present in the sample. The in-source fragmentation is particularly dangerous since it happens in the MS source and no improvements of the analyzer (using MS/MS or additional separation) can help understand whether a detected molecule (e.g., ADP) is an endogenous one or is an in-source fragment of another molecule (e.g., ATP).
Machine learning was successfully employed to perform automated metabolite identification in bulk metabolomics (118, 119) but is yet to be applied to metabolite identification in imaging MS. Using machine learning attracts substantial interest because another approach, developing a comprehensive theoretical model of ionization, is likely unfeasible due to lack of knowledge as well as dependence of ionization on various parameters. The current bottleneck of developing such methods is the lack of a library of authentic molecular standards, similar to MS/MS spectral libraries in LC-MS/MS metabolomics, which should be sufficiently big to represent the complexity of imaging mass spectra. However, once such libraries in imaging MS are created, this should stimulate developments in this field, with some machine learning methods from bulk metabolomics possibly adaptable. This will not only enable detection of currently unannotated—and thus invisible—molecules but also provide more accurate quantitative information by resolving ionization pathways and integrating all signals corresponding to a particular metabolite.
Molecular Digital Pathology
Digital pathology, or using advanced computational methods to assist pathologists in assessing digitalized pathological slides, is an emerging field of medicine facilitated in particular by progress in machine learning and AI (120). In addition to morphometric information captured by microscopy, there is continuing interest in exploiting imaging MS to provide additional molecular layers of spatio-molecular information with the aim of elucidating hidden morphological features, helping precision medicine by visualizing localization of disease- or patient-specific biomarkers, assisting early assessment of treatment, and improving prognosis (14, 20, 121, 122). Developing imaging MS–based molecular digital pathology attracts substantial efforts, in particular, developments aimed at reducing acquisition times (123, 124), protocol developments (125), and computational developments. Presently, imaging MS is not yet employed for digital pathology due to the competitiveness of microscopy-based digital pathology methods and the complexity and high price of imaging mass spectrometers and associated sample preparation. However, the search is open for pathological scenarios and questions where imaging MS would outperform microscopy-based digital pathology. This direction certainly requires the development of advanced and tailored AI-based methods to successfully exploit highly multiplex yet noisy spatial information provided by imaging MS to achieve greater performance than using morphometric information provided by microscopy.
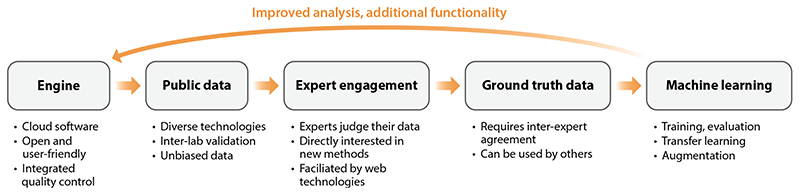
Lack of Ground Truth Data
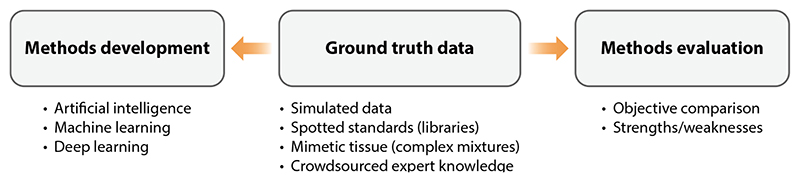
Development of advanced computational methods requires ground truth data or high-quality data with desired labels or outputs (51) (Figure 4). For example, for molecular identification this can be data from known molecules, and for spatial segmentation this can be a dataset comprehensively and manually segmented by experts. Such data are required not only for training machine learning methods but also for evaluating and comparing other computational methods. However, creating such ground truth data is a key challenge since the data should be representative, diverse, big, and either generated in a special manner so that they contain only known outputs (e.g., a mixture of authentic molecular standards) or annotated by experts (51).
Figure 4. Ground truth is needed for methods development (training machine learning and deep learning models) and methods evaluation at the same time, which requires considerable effort.
Progress in addressing many outstanding computational challenges in imaging MS has been hindered by the lack of ground truth data. For example, numerous spatial segmentation methods have been developed (68), but without ground truth data, comparing them is highly subjective because it is not known which spatial regions in a tissue section have distinct molecular compositions. The lack of ground truth for molecular identification is even more critical since it is not known a priori which molecules are contained in the tissue section, which of them are detected by imaging MS, and what are their true localizations, in particular due to various ionization effects affecting the detection and intensities in individual pixels (suppression, in-source fragmentation, and chemical overlap with other isomers, particularly matrix clusters). Overall, this situation leads to arbitrary and empiric choices of methods, slows down computational developments, makes it impossible to train and evaluate machine learning and deep learning models, and leads to overall inertia in adopting novel methods since it is hard to prove objectively that a novel method constitutes an improvement.
Several approaches have been proposed to tackle this formidable challenge, which can be broadly categorized into four classes: data simulation, depositing spots of authentic standards on a glass slide, creating mimetic tissue with authentic standards, and crowdsourcing by engaging experts. Simulating data by composing spectra generated using physical-probabilistic models into an imaging MS dataset with prespecified spatial regions and molecular composition has not received broad recognition. In particular, this approach replicated the complexity of real data only to a limited extent. Depositing spots of authentic standards in a solution onto a glass slide with subsequent drying seems to be the most straightforward approach to obtaining ground truth data that can be used for molecular identification and other problems, similar to spiking authentic standards in bulk metabolomics. However, this approach only works well for a few standards (e.g., drugs in pharmaceutical applications) since the spot formation process depends on the molecular composition and concentration. When not optimized, drying creates inhomogeneous spots with either a so-called coffee ring effect (where most crystals are localized at the perimeter of the spot) or one or a few big crystals forming in the center of the spot. Using mimetic tissue, or a tissue homogenate with authentic standards implanted at different locations or layers (97, 126), was proposed initially for quantitative imaging MS. This approach became the primary approach to estimating the response curve, the relationship between concentration and intensity measured in imaging MS. This method did not become popular in evaluating computational methods, possibly due to a limited number of authentic standards that can be spiked in. However, with considerable experimental effort this can be improved, and hopefully in the future it will provide computer scientists with experimentally obtained ground truth data with sufficiently rich and known molecular composition. Crowdsourcing, an approach successfully used in other areas, was recently exploited by us in three studies (81, 83, 87). In each of these studies, we engaged experts, collected their judgments, curated them, evaluated the agreement between them, and integrated high-quality judgments into high-quality gold standard data. Crowdsourcing has proven useful for problems that are solved by experts during their routine analysis of imaging MS data. We applied it to estimate the quality of ion images (83), recognize off-sample images (87), and quantify colocalization between ion images (81).
Creating ground truth for imaging MS–specific problems is a major effort, but one that can transform this field by employing machine learning and deep learning methods. We expect that the importance of these efforts will become increasingly recognized with more research efforts focused on this topic.
The Future of Spatial Metabolomics
Spatial metabolomics, fueled by biomedical demands and stimulated by recent successes, is attracting growing attention of experimental and computational methods developers. Novel technological solutions are being introduced to address the outstanding challenges and to improve spatial resolution, sensitivity, and specificity. Here, we review the most promising technological directions and provide a look into the future of computational methods—in particular, machine learning, deep learning, and AI—needed to capitalize on these developments.
Integration of Ion Mobility Separation
IMS is a technology to separate ions in gas phase by their structural properties, namely, their collisional cross section (CCS), by flying through a buffer gas (127, 128). IMS can be integrated into an imaging mass spectrometer between the source and analyzer, thus providing another molecular-chemical dimension (61, 129). In bulk proteomics, metabolomics, and lipidomics, IMS was shown to increase specificity (by separating structural isomers by their CCS and by matching signals to either measured or theoretically predicted CCSs) and sensitivity when coupled to a fast analyzer (by analyzing every CCS peak separately, thus avoiding ion suppression) (129). Recently, several vendors released imaging mass spectrometers with IMS. We expect that this will spur computational developments, in particular by enabling novel metabolite identification methods, but also will require novel approaches to data handling and analysis because using IMS increases the data size by a factor of 102–104. Already available libraries of measured CCS values and the high reproducibility of CCS values, even between different platforms, make machine learning a perfect choice for CCS prediction. Early success in machine learning CCS prediction has been achieved for both metabolites (130) and lipids (131, 132). With the increasing popularity of IMS in imaging MS one can expert machine learning to play a greater role in this field.
Using Chemical Derivatization to Detect Invisible Metabolites
Recently, it was repeatedly and convincingly demonstrated that both sensitivity and specificity of imaging MS can be substantially improved when molecules are chemically derivatized in situ by applying a chemical reactant onto the tissue section that modifies particular molecules through a known chemical reaction that increases their ionization (133–136). Chemical derivatization is successfully used in other fields of analytical chemistry (gas chromatography–MS, LC-MS), where protocols and derivatization agents are already available (24, 137). However, using it in imaging MS settings is far from easy. The efficiency of the derivatization can depend on the tissue composition, and partial derivatization can distort quantification and spatial artifacts. We expect that with the mounting interest in applying imaging MS to biological problems focusing on metabolic pathways and classes that are of key importance yet hardly detectable (e.g., glycolysis, tricarboxylic acid cycle, signaling lipids such as phosphatidylinositol phosphates, microbiota-derived molecular signals such as short-chain fatty acids), on-tissue chemical derivatization will be further explored and will require novel computational methods addressing these outlined challenges. One can expect computational chemistry and machine learning methods to play a stronger role in predicting the effects of chemical derivatization in MS and in imaging in particular.
Molecular Interpretation of the Detected Metabolites
Molecular annotation or identification reduces tens of thousands of signals associated with m/z values to a list of molecules and associated ion images. Statistical analysis can help further reduce the list to those molecules that are associated with some known or discovered spatial pattern or that represent a difference between conditions. Molecular interpretation of the obtained molecular information is currently one of the key challenges, with hardly any tools available. In other omics fields, identification and statistical analysis are often followed by enrichment analysis to find associated pathways or other molecular, cellular, or functional properties (138). Enrichment analysis helps estimate the statistical significance of the representation of a metabolic pathway, molecular class, or other property (e.g., compartmentalization with a particular organelle) in the results. Although in conventional metabolomics and lipidomics, enrichment analysis is becoming commonplace, with open-source software available (139, 140), currently there are no solutions for imaging MS that take into account the imaging specifics (ambiguity, spatial localization, and molecular coverage). We are not aware of machine learning or deep learning being used for these purposes. However, recent advances of Semantic Web approaches such as the resource description framework (RDF) in chemoinformatics, with various SPARQL (SPARQL Protocol and RDF Query Language) endpoints available for many chemical resources including metabolomics [ChEBI (Chemical Entities of Biological Interest)] and lipidomics (SwissLipids), promise automated integration of different omics fields (141–143).
The Emergence of Spatial Single-Cell Metabolomics
Single-cell metabolomics and lipidomics are experiencing a boom in growth (15, 144–151). These advances are motivated by discoveries of the single-cell extent of biology with the help of other single-cell omics, particularly single-cell RNA-seq, and enabled by the rapidly increasing sensitivity of MS–based metabolomics and lipidomics. Along with other approaches such as microextraction and Raman spectroscopy, imaging MS is playing a key role in this field since it ensures high sensitivity and requires minimal sample preparation, thus minimizing analyte loss. At the same time, imaging MS can be used to profile cells in situ, thus not only minimizing the cell perturbation required in conventional metabolomics but also allowing the association of molecular information with the spatial context, such as cell morphometric information or cell–cell interactions. Various single-cell metabolomics and lipidomics imaging MS approaches have been proposed and have demonstrated the potential of obtaining rich metabolic and lipidomics profiles that can predict cell types and associated molecular changes with perturbations (152–155). We have recently demonstrated that single-cell metabolomics can discover molecularly distinct cell subpopulations even in cell cultures (155). Moreover, these subpopulations were associated with nonalcoholic steatohepatitis, a critical pathological condition and a key factor in the progression toward liver cancer (156). Computational methods for analysis and interpretation of the obtained single-cell metabolomics data are yet to be developed, in particular for addressing batch effects, reducing confounding factors, visualizing highly multiplex data from thousands of cells, discovering molecular features and subpopulations hidden behind noise, and ultimately discovering links between cell types, phenotypes, and states and their metabolism. AI and machine learning were demonstrated to be indispensable in the field of single-cell transcriptomics (157). We expect this to happen in the field of single-cell metabolomics as well where such approaches are already needed to analyze, visualize, and interpret big and often spatial data generated from single cells.
Metaspace as A Community Artificial Intelligence Platform
AI, machine learning, and deep learning are revolutionizing the life sciences and biomedicine (51, 52, 55, 56). Do the metabolomics and spatial metabolomics fields risk missing out on the AI revolution (21)? On one hand, the complexity of metabolomics, with hundreds of thousands of possible molecules in the metabolome, lipidome, and exosome (exogenous molecules that come from the environment), poses challenges that can hardly be addressed using theoretical methods and that need AI solutions. Moreover, spatial metabolomics, combining metabolomics questions and spatial imaging data, can be a perfect field for deep learning methods, which perform well at image analysis. On the other hand, presently machine learning and AI approaches are applied in spatial metabolomics only to a limited degree and have not yet demonstrated their full potential.
The key bottleneck in exploiting these modern approaches is the lack of ground truth data, as discussed above. Moreover, engaging machine learners requires clearly formulated and high-impact but yet unsolved problems. The fast development of machine learning–based identification approaches in LC-MS/MS–based metabolomics (158, 159), in particular fueled by competitions on manual and automated identification (113), suggests that molecular identification in imaging MS will be the high-priority challenge for machine learning as soon as sufficient ground truth data are provided. We recently developed several AI approaches that combined formulating a problem, selecting data, collecting expert judgments, creating a gold standard set, and developing deep learning methods. Our developments used public data from METASPACE and a knowledge base of spatial metabolomes and led to the implementation of novel features for METASPACE users. METASPACE already includes over 3,500 public imaging MS datasets and is continuously populated by users from more than 100 labs across the world. Using METASPACE helped us engage experts and select representative data from various imaging MS technologies. Hence, METASPACE already serves as a community AI platform (Figure 5). We aim at further facilitating AI developments on this platform by following open-source and open-access principles, demonstrating the benefits of making data public, providing high-quality data, connecting computer scientists with experimentalists, and overall removing barriers for methods developers.
Figure 5.
METASPACE as an example of an open community platform for artificial intelligence developments in the field of spatial metabolomics. METASPACE provides a free cloud engine for metabolite annotation and encourages users to make their data public, thus creating an open knowledge base of spatial metabolomes. The open public data help engage experts to create ground truth necessary for the development of machine learning methods. Implementing these methods in METASPACE improves the platform and adds new functionality that, in turn, attracts more users, thus creating a sustainable open platform for both imaging mass spectrometry practitioners and computational method developers.
Software
In this section, we review open-source software packages for imaging MS. Our aim is to help experimentalists navigate existing solutions and to provide examples for methods developers. The software packages were selected based on our subjective knowledge of their popularity among users and the existence of training options such as regular workshops, in addition to a short survey we conducted on Twitter.
METASPACE is a cloud software for metabolite and lipid identification as well as a public knowledge base of spatial metabolomes (115). METASPACE requires data in the centroided imzML format and can be used for online browsing of ion images for annotated metabolites and lipids, sharing the results, and organizing them into projects. Since recently, METASPACE also includes methods for visualization and data analysis.
MSiReader is a stand-alone software for visualization, signal processing, and unsupervised analysis of imaging MS data (86). MSiReader is equipped with a rich graphical user interface and since recently supports quantitation. MSiReader supports the imzML format and can be used either in Windows or on any operating system if the Matlab environment is provided.
Cardinal is an R package implementing statistical analysis methods including spatial segmentation, classification, and class comparison (160). Using Cardinal requires basic knowledge of R as no graphical interface is provided.
A growing number of open-source software packages help working with imaging MS data. imzMLConverter (https://github.com/AlanRace/imzMLConverter) helps convert data into the imzML format. pyImzML parser (https://github.com/alexandrovteam/pyimzML) helps with reading and writing imzML files using Python. rMSI (161) and RmsiGUI (162) are R packages with a graphical user interface for data visualization and analysis that provide alternatives to MSiReader. OpenMSI is a cloud data hosting and visualization platform (163). Recently, a Galaxy workflow for imaging MS was developed that represents an attractive alternative to those already using Galaxy (164). MetaboLights is a general repository for metabolomics data deposition that can also be used for imaging MS data (165). For a longer list of software with a focus on exploratory analysis, see Reference 68.
Conclusions and Outlook
Spatial metabolomics is a rapidly emerging field that promises—and has already delivered—discoveries of how metabolism is regulated in the spatial context of tissues. At the intersection of metabolomics, mass spectrometry, and imaging, this technology not only opens novel avenues to understanding biology in its spatial context in situ but also creates novel challenges in the analysis and interpretation of generated high-dimensional data. The revolution of AI, machine learning, and deep learning offers a unique opportunity for addressing these challenges using these modern computational approaches. However, grasping this opportunity requires concerted efforts of experimentalists and computer scientists to create high-quality ground truth data such as libraries for metabolite annotation in imaging MS in order to train machine learning models and evaluate their performance. We hope that this review will help build bridges between experimentalists and computer scientists by introducing key concepts, reviewing outstanding challenges that need computational solutions, and highlighting the need for creating ground truth data, and thus ultimately will benefit this fascinating, challenging, and interdisciplinary field.
Acknowledgments
Work on this paper was supported by the European Research Council (agreement 773089), the European Horizon2020 ICT project CloudButton (agreement 825184), the public–private partnership OpenTargets, and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Kidney Precision Medicine Project (KPMP).
Footnotes
For example, see this public METASPACE dataset from a mouse brain tissue provided by Dhaka Bhandari (University of Giessen), which contains over 500,000 pixels: https://metaspace2020.eu/annotations?db=SwissLipids-2018-02-02&ds=2019-08-19_11h28m42s&offs=0.
Disclosure Statement
Until 2020, T.A. was on the Scientific Advisory Board of SCiLS, a company developing software for imaging mass spectrometry.
Errata
An online log of corrections to Annual Review of Biomedical Data Science articles may be found at http://www.annualreviews.org/errata/biodatasci
Data Availability
In this section, we briefly overview the available data that can be used for methods development, training machine and deep learning models, and methods evaluation. In 2015, we published benchmark datasets from different biological systems from several labs (166). These datasets included serial 3D imaging MS data, but individual serial sections can also be used as 2D datasets. The datasets include MALDI-TOF and DESI-Orbitrap data, and although they still can be used for methods development, they do not represent the state of the art in terms of spatial and spectral resolution. MetaboLights hosts several imaging MS studies where both data and metadata can be downloaded; the data can be found by searching “imaging mass spectrometry.” METASPACE provides the largest collection of public imaging MS datasets. Currently, it provides access to annotated data only; we are planning to implement user-approved access to associated imzML data for public datasets. We have also published ground truth data from three crowdsourcing studies aimed at estimating the quality of ion images (https://github.com/alexandrovteam/IMS_quality), recognizing off-sample ion images (https://github.com/metaspace2020/offsample), and quantifying colocalization between ion images (https://github.com/metaspace2020/coloc).
Literature Cited
- 1.Petras D, Jarmusch AK, Dorrestein PC. From single cells to our planet—recent advances in using mass spectrometry for spatially resolved metabolomics. Curr Opin Chem Biol. 2017;36:24–31. doi: 10.1016/j.cbpa.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Lau AN, Vander Heiden MG. Metabolism in the tumor microenvironment. Annu Rev Cancer Biol. 2020;4:17–40. [Google Scholar]
- 3.Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic instruction of immunity. Cell. 2017;169(4):570–86. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014;20(5):719–30. doi: 10.1016/j.cmet.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol. 2019;20:40–54. doi: 10.1038/s41577-019-0198-4. [DOI] [PubMed] [Google Scholar]
- 6.Miyazawa H, Aulehla A. Revisiting the role of metabolism during development. Development. 2018;145(19):dev131110. doi: 10.1242/dev.131110. [DOI] [PubMed] [Google Scholar]
- 7.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16(1):9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escoll P, Buchrieser C. Metabolic reprogramming of host cells upon bacterial infection: Why shift to a Warburg-like metabolism? FEBS J. 2018;285(12):2146–60. doi: 10.1111/febs.14446. [DOI] [PubMed] [Google Scholar]
- 9.Gaber T, Strehl C, Buttgereit F. Metabolic regulation of inflammation. Nat Rev Rheumatol. 2017;13(5):267–79. doi: 10.1038/nrrheum.2017.37. [DOI] [PubMed] [Google Scholar]
- 10.Eming SA, Wynn TA, Martin P. Inflammation and metabolism in tissue repair and regeneration. Science. 2017;356(6342):1026–30. doi: 10.1126/science.aam7928. [DOI] [PubMed] [Google Scholar]
- 11.Baker M. Metabolomics: from small molecules to big ideas. Nat Methods. 2011;8(2):117–21. [Google Scholar]
- 12.Doerr A. Mass spectrometry imaging takes off. Nat Methods. 2018;15:32. [Google Scholar]
- 13.Buchberger AR, DeLaney K, Johnson J, Li L. Mass spectrometry imaging: a review of emerging advancements and future insights. Anal Chem. 2018;90(1):240–65. doi: 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung F, Eberlin LS, Schwamborn K, Heeren RMA, Winograd N, Cooks RG. Mass spectrometrybased tissue imaging: the next frontier in clinical diagnostics? Clin Chem. 2019;65(4):510–13. doi: 10.1373/clinchem.2018.289694. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore IS, Heiles S, Pieterse CL. Metabolic imaging at the single-cell scale: recent advances in mass spectrometry imaging. Annu Rev Anal Chem. 2019;12:201–24. doi: 10.1146/annurev-anchem-061318-115516. [DOI] [PubMed] [Google Scholar]
- 16.Luberto C, Haley JD, Del Poeta M. Imaging with mass spectrometry, the next frontier in sphingolipid research? A discussion on where we stand and the possibilities ahead. Chem Phys Lipids. 2019;219:1–14. doi: 10.1016/j.chemphyslip.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz S, Becker M, Groseclose MR, Schadt S, Hopf C. Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Curr Opin Biotechnol. 2018;55:51–59. doi: 10.1016/j.copbio.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Ryan DJ, Spraggins JM, Caprioli RM. Protein identification strategies in MALDI imaging mass spectrometry: a brief review. Curr Opin Chem Biol. 2018;48:64–72. doi: 10.1016/j.cbpa.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spraker JE, Luu GT, Sanchez LM. Imaging mass spectrometry for natural products discovery: a review of ionization methods. Nat Prod Rep. 2020;37:150–62. doi: 10.1039/c9np00038k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaysse P-M, Heeren RMA, Porta T, Balluff B. Mass spectrometry imaging for clinical research— latest developments, applications, and current limitations. Analyst. 2017;142(15):2690–712. doi: 10.1039/c7an00565b. [DOI] [PubMed] [Google Scholar]
- 21.Editorial. Why the metabolism field risks missing out on the AI revolution. Nat Metab. 2019;1(10):929–30. doi: 10.1038/s42255-019-0133-9. [DOI] [PubMed] [Google Scholar]
- 22.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 23.Patti GJ, Yanes O, Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–69. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu W, Su X, Klein MS, Lewis IA, Fiehn O, Rabinowitz JD. Metabolite measurement: pitfalls to avoid and practices to follow. Annu Rev Biochem. 2017;86:277–304. doi: 10.1146/annurev-biochem-061516-044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinges SS, Hohm A, Vandergrift LA, Nowak J, Habbel P, et al. Cancer metabolomic markers in urine: evidence, techniques and recommendations. Nat Rev Urol. 2019;16(6):339–62. doi: 10.1038/s41585-019-0185-3. [DOI] [PubMed] [Google Scholar]
- 26.Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, et al. Blood metabolome predicts gut microbiome a-diversity in humans. Nat Biotechnol. 2019;37(10):1217–28. doi: 10.1038/s41587-019-0233-9. [DOI] [PubMed] [Google Scholar]
- 27.Weiner J, 3rd, Maertzdorf J, Sutherland JS, Duffy FJ, Thompson E, et al. Metabolite changes in blood predict the onset of tuberculosis. Nat Commun. 2018;9(1):5208. doi: 10.1038/s41467-018-07635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg N, Wang M, Hyde E, da Silva RR, Melnik AV, et al. Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe. 2017;22(5):705–16.:e4. doi: 10.1016/j.chom.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hudry B, de Goeij E, Mineo A, Gaspar P, Hadjieconomou D, et al. Sex differences in intestinal carbohydrate metabolism promote food intake and sperm maturation. Cell. 2019;178(4):901–18.:e16. doi: 10.1016/j.cell.2019.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–18. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodzon-Kulakowska A, Suder P. Imaging mass spectrometry: instrumentation, applications, and combination with other visualization techniques. Mass Spectrom Rev. 2016;35(1):147–69. doi: 10.1002/mas.21468. [DOI] [PubMed] [Google Scholar]
- 32.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 34.Pacholski ML, Winograd N. Imaging with mass spectrometry. Chem Rev. 1999;99(10):2977–3006. doi: 10.1021/cr980137w. [DOI] [PubMed] [Google Scholar]
- 35.Reyzer ML, Caprioli RM. The development of imaging mass spectrometry. In: Gross ML, Caprioli RM, editors. The Encyclopedia of Mass Spectrometry. Elsevier; Boston: 2016. pp. 285–304. [Google Scholar]
- 36.Palmer A, Trede D, Alexandrov T. Where imaging mass spectrometry stands: Here are the numbers. Metabolomics. 2016;12(6):107. [Google Scholar]
- 37.Movasaghi Z, Rehman S, ur Rehman I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl Spectrosc Rev. 2008;43(2):134–79. [Google Scholar]
- 38.Langer J, Jimenez de Aberasturi D, Aizpurua J, Alvarez-Puebla RA, Auguié B, et al. Present and future of surface-enhanced Raman scattering. ACS Nano. 2020;14:28–117. doi: 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Protsyuk I, Melnik AV, Nothias L-F, Rappez L, Phapale P, et al. 3D molecular cartography using LC-MS facilitated by Optimus and ‘ili software. Nat Protoc. 2018;13(1):134–54. doi: 10.1038/nprot.2017.122. [DOI] [PubMed] [Google Scholar]
- 40.Watrous JD, Alexandrov T, Dorrestein PC. The evolving field of imaging mass spectrometry and its impact on future biological research. J Mass Spectrom. 2011;46(2):209–22. doi: 10.1002/jms.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Detection technologies. ambient mass spectrometry. Science. 2006;311(5767):1566–70. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 42.Fletcher JS, Kotze HL, Armitage EG, Lockyer NP, Vickerman JC. Evaluating the challenges associated with time-of-fight secondary ion mass spectrometry for metabolomics using pure and mixed metabolites. Metabolomics. 2013;9(3):535–44. [Google Scholar]
- 43.Passarelli MK, Pirkl A, Moellers R, Grinfeld D, Kollmer F, et al. The 3D OrbiSIMS-label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power. Nat Methods. 2017;14(12):1175–83. doi: 10.1038/nmeth.4504. [DOI] [PubMed] [Google Scholar]
- 44.Robichaud G, Barry JA, Muddiman DC. IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J Am Soc Mass Spectrom. 2014;25(3):319–28. doi: 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79(21):8098–106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 46.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449(7165):1033–36. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 47.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2012;84(1):141–48. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer; New York: 2013. [Google Scholar]
- 49.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 50.Russell SJ, Norvig P. Artificial Intelligence: A Modern Approach. 3rd. Prentice Hall; Upper Saddle River, NJ: 2009. [Google Scholar]
- 51.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347–58. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 52.Angermueller C, Pârnamaa T, Parts L, Stegle O. Deep learning for computational biology. Mol SystBiol. 2016;12(7):878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–48. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baldi P. Deep learning in biomedical data science. Annu Rev Biomed Data Sci. 2018;1:181–205. [Google Scholar]
- 55.Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 56.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 57.Hosny A, Aerts HJWL. Artificial intelligence for global health. Science. 2019;366(6468):955–56. doi: 10.1126/science.aay5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexandrov T. MALDI imaging mass spectrometry: statistical data analysis and current computational challenges. BMC Bioinform. 2012;13(16):S11. doi: 10.1186/1471-2105-13-S16-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kompauer M, Heiles S, Spengler B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat Methods. 2016;14:90–96. doi: 10.1038/nmeth.4071. [DOI] [PubMed] [Google Scholar]
- 60.Shimma S, Sugiura Y. Effective sample preparations in imaging mass spectrometry. Mass Spectrom. 2014;3:S0029. doi: 10.5702/massspectrometry.S0029. Spec. Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sans M, Feider CL, Eberlin LS. Advances in mass spectrometry imaging coupled to ion mobility spectrometry for enhanced imaging of biological tissues. Curr Opin Chem Biol. 2018;42:138–46. doi: 10.1016/j.cbpa.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451–59. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schramm T, Hester A, Klinkert I, Both J-P, Heeren RMA, et al. imzML—a common data format for the flexible exchange and processing of mass spectrometry imaging data. J Proteom. 2012;75(16):5106–10. doi: 10.1016/j.jprot.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 64.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, et al. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int J Mass Spectrom. 2007;260(2-3):212–21. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor AJ, Dexter A, Bunch J. Exploring ion suppression in mass spectrometry imaging of a heterogeneous tissue. Anal Chem. 2018;90(9):5637–45. doi: 10.1021/acs.analchem.7b05005. [DOI] [PubMed] [Google Scholar]
- 66.O’Donoghue SI, Baldi BF, Clark SJ, Darling AE, Hogan JM, et al. Visualization of biomedical data. Annu Rev Biomed Data Sci. 2018;1:275–304. [Google Scholar]
- 67.Nuñez JR, Anderton CR, Renslow RS. Optimizing colormaps with consideration for color vision deficiency to enable accurate interpretation of scientific data. PLOS ONE. 2018;13(7):e0199239. doi: 10.1371/journal.pone.0199239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verbeeck N, Caprioli RM, Van de Plas R. Unsupervised machine learning for exploratory data analysis in imaging mass spectrometry. Mass Spectrom Rev. 2020;39(3):245–91. doi: 10.1002/mas.21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexandrov T, Becker M, Deininger S-O, Ernst G, Wehder L, et al. Spatial segmentation of imaging mass spectrometry data with edge-preserving image denoising and clustering. J Proteom Res. 2010;9(12):6535–46. doi: 10.1021/pr100734z. [DOI] [PubMed] [Google Scholar]
- 70.Alexandrov T, Kobarg JH. Efficient spatial segmentation of large imaging mass spectrometry datasets with spatially aware clustering. Bioinformatics. 2011;27(13):i230–38. doi: 10.1093/bioinformatics/btr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanselmann M, Köthe U, Kirchner M, Renard BY, Amstalden ER, et al. Toward digital staining using imaging mass spectrometry and random forests. J Proteom Res. 2009;8(7):3558–67. doi: 10.1021/pr900253y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bemis KD, Harry A, Eberlin LS, Ferreira CR, van de Ven SM, et al. Probabilistic segmentation of mass spectrometry (MS) images helps select important ions and characterize confidence in the resulting segments. Mol Cell Proteom. 2016;15(5):1761–72. doi: 10.1074/mcp.O115.053918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alexandrov T, Chernyavsky I, Becker M, von Eggeling F, Nikolenko S. Analysis and interpretation of imaging mass spectrometry data by clustering mass-to-charge images according to their spatial similarity. Anal Chem. 2013;85(23):11189–95. doi: 10.1021/ac401420z. [DOI] [PubMed] [Google Scholar]
- 74.Konicek AR, Lefman J, Szakal C. Automated correlation and classification of secondary ion mass spectrometry images using a k-means cluster method. Analyst. 2012;137(15):3479–87. doi: 10.1039/c2an16122b. [DOI] [PubMed] [Google Scholar]
- 75.Inglese P, Correia G, Pruski P, Glen RC, Takats Z. Colocalization features for classification of tumors using desorption electrospray ionization mass spectrometry imaging. Anal Chem. 2019;91(10):6530–40. doi: 10.1021/acs.analchem.8b05598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fonville JM, Carter CL, Pizarro L, Steven RT, Palmer AD, et al. Hyperspectral visualization of mass spectrometry imaging data. Anal Chem. 2013;85(3):1415–23. doi: 10.1021/ac302330a. [DOI] [PubMed] [Google Scholar]
- 77.Franceschi P, Wehrens R. Self-organizing maps: a versatile tool for the automatic analysis of untargeted imaging datasets. Proteomics. 2014;14(7-8):853–61. doi: 10.1002/pmic.201300308. [DOI] [PubMed] [Google Scholar]
- 78.Gardner W, Cutts SM, Muir BW, Jones RT, Pigram PJ. Visualizing ToF-SIMS hyperspectral imaging data using color-tagged toroidal self-organizing maps. Anal Chem. 2019;91(21):13855–65. doi: 10.1021/acs.analchem.9b03322. [DOI] [PubMed] [Google Scholar]
- 79.Abdelmoula WM, Škrášková K, Balluff B, Carreira RJ, Tolner EA, et al. Automatic generic registration of mass spectrometry imaging data to histology using nonlinear stochastic embedding. Anal Chem. 2014;86(18):9204–11. doi: 10.1021/ac502170f. [DOI] [PubMed] [Google Scholar]
- 80.Smets T, Verbeeck N, Claesen M, Asperger A, Griffioen G, et al. Evaluation of distance metrics and spatial autocorrelation in uniform manifold approximation and projection applied to mass spectrometry imaging data. Anal Chem. 2019;91(9):5706–14. doi: 10.1021/acs.analchem.8b05827. [DOI] [PubMed] [Google Scholar]
- 81.Ovchinnikova K, Stuart L, Rakhlin A, Nikolenko S, Alexandrov T. ColocML: Machine learning quantifies co-localization between mass spectrometry images. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa085. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alexandrov T, Bartels A. Testing for presence of known and unknown molecules in imaging mass spectrometry. Bioinformatics. 2013;29(18):2335–42. doi: 10.1093/bioinformatics/btt388. [DOI] [PubMed] [Google Scholar]
- 83.Palmer A, Ovchinnikova E, Thuné M, Lavigne R, Guével B, et al. Using collective expert judgements to evaluate quality measures of mass spectrometry images. Bioinformatics. 2015;31(12):i375–84. doi: 10.1093/bioinformatics/btv266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wijetunge CD, Saeed I, Boughton BA, Spraggins JM, Caprioli RM, et al. EXIMS: an improved data analysis pipeline based on a new peak picking method for exploring imaging mass spectrometry data. Bioinformatics. 2015;31(19):3198–206. doi: 10.1093/bioinformatics/btv356. [DOI] [PubMed] [Google Scholar]
- 85.Prasad M, Postma G, Morosi L, Giordano S, Giavazzi R, et al. Drug-homogeneity index in massspectrometry imaging. Anal Chem. 2018;90(22):13257–64. doi: 10.1021/acs.analchem.8b01870. [DOI] [PubMed] [Google Scholar]
- 86.Bokhart MT, Nazari M, Garrard KP, Muddiman DC. MSiReader v1.0: evolving open-source mass spectrometry imaging software for targeted and untargeted analyses. J Am Soc Mass Spectrom. 2018;29(1):8–16. doi: 10.1007/s13361-017-1809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ovchinnikova K, Kovalev V, Stuart L, Alexandrov T. OffsampleAI: artificial intelligence approach to recognize off-sample mass spectrometry images. BMC Bioinform. 2020;21:129. doi: 10.1186/s12859-020-3425-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCombie G, Staab D, Stoeckli M, Knochenmuss R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal Chem. 2005;77(19):6118–24. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- 89.McDonnell LA, van Remoortere A, van Zeijl RJM, Deelder AM. Mass spectrometry image correlation: quantifying colocalization. J Proteom Res. 2008;7(8):3619–27. doi: 10.1021/pr800214d. [DOI] [PubMed] [Google Scholar]
- 90.Kaddi C, Parry RM, Wang MD. Hypergeometric similarity measure for spatial analysis in tissue imaging mass spectrometry. In: Wu F-X, Zaki M, Morishita S, Pan Y, Wong S, et al., editors. Proceedings of the 2011 IEEE International Conference on Bioinformatics and Biomedicine. IEEE Comput. Sci; Los Alamos, CA: 2011. pp. 604–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ekelöf M, Garrard KP, Judd R, Rosen EP, Xie D-Y, et al. Evaluation of digital image recognition methods for mass spectrometry imaging data analysis. J Am Soc Mass Spectrom. 2018;29(12):2467–70. doi: 10.1007/s13361-018-2073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aaron JS, Taylor AB, Chew T-L. Image co-localization—co-occurrence versus correlation. J Cell Sci. 2018;131(3):jcs211847. doi: 10.1242/jcs.211847. [DOI] [PubMed] [Google Scholar]
- 93.Van de Plas R, Pelckmans K, De Moor B, Waelkens E. Spatial querying of imaging mass spectrometry data: a nonnegative least squares approach; Paper presented at the Machine Learning in Computational Biology workshop at the Conference on Neural Information Processing Systems (NIPS 2007); Vancouver, B.C. Dec. 7; 2007. [Google Scholar]
- 94.Shariatgorji M, Nilsson A, Goodwin RJA, Källback P, Schintu N, et al. Direct targeted quantitative molecular imaging of neurotransmitters in brain tissue sections. Neuron. 2014;84(4):697–707. doi: 10.1016/j.neuron.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 95.Rzagalinski I, Volmer DA. Quantification of low molecular weight compounds by MALDI imaging mass spectrometry—a tutorial review. Biochim Biophys Acta. 2017;1865(7):726–39. doi: 10.1016/j.bbapap.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 96.Swales JG, Dexter A, Hamm G, Nilsson A, Strittmatter N, et al. Quantitation of endogenous metabolites in mouse tumors using mass-spectrometry imaging. Anal Chem. 2018;90(10):6051–58. doi: 10.1021/acs.analchem.7b05239. [DOI] [PubMed] [Google Scholar]
- 97.Groseclose MR, Castellino S. A mimetic tissue model for the quantification of drug distributions by MALDI imaging mass spectrometry. Anal Chem. 2013;85(21):10099–106. doi: 10.1021/ac400892z. [DOI] [PubMed] [Google Scholar]
- 98.Chumbley CW, Reyzer ML, Allen JL, Marriner GA, Via LE, et al. Absolute quantitative MALDI imaging mass spectrometry: a case of rifampicin in liver tissues. Anal Chem. 2016;88(4):2392–98. doi: 10.1021/acs.analchem.5b04409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Källback P, Nilsson A, Shariatgorji M, Andrén PE. msIQuant—quantitation software for mass spectrometry imaging enabling fast access, visualization, and analysis of large data sets. Anal Chem. 2016;88(8):4346–53. doi: 10.1021/acs.analchem.5b04603. [DOI] [PubMed] [Google Scholar]
- 100.Palmer AD, Alexandrov T. Serial 3D imaging mass spectrometry at its tipping point. Anal Chem. 2015;87(8):4055–62. doi: 10.1021/ac504604g. [DOI] [PubMed] [Google Scholar]
- 101.Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev. 2011;30(1):142–74. doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- 102.Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. PNAS. 2008;105(47):18126–31. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trede D, Schiffler S, Becker M, Wirtz S, Steinhorst K, et al. Exploring three-dimensional matrix-assisted laser desorption/ionization imaging mass spectrometry data: three-dimensional spatial segmentation of mouse kidney. Anal Chem. 2012;84(14):6079–87. doi: 10.1021/ac300673y. [DOI] [PubMed] [Google Scholar]
- 104.Oetjen J, Aichler M, Trede D, Strehlow J, Berger J, et al. MRI-compatible pipeline for threedimensional MALDI imaging mass spectrometry using PAXgene fixation. J Proteom. 2013;90:52–60. doi: 10.1016/j.jprot.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 105.Watrous JD, Phelan VV, Hsu C-C, Moree WJ, Duggan BM, et al. Microbial metabolic exchange in 3D. ISME J. 2013;7(4):770–80. doi: 10.1038/ismej.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seeley EH, Wilson KJ, Yankeelov TE, Johnson RW, Gore JC, et al. Co-registration of multi-modalityimaging allows for comprehensive analysis of tumor-induced bone disease. Bone. 2014;61:208–16. doi: 10.1016/j.bone.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Giordano S, Morosi L, Veglianese P, Licandro SA, Frapolli R, et al. 3D mass spectrometry imaging reveals a very heterogeneous drug distribution in tumors. Sci Rep. 2016;6:37027. doi: 10.1038/srep37027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Abdelmoula WM, Regan MS, Lopez BGC, Randall EC, Lawler S, et al. Automatic 3D nonlinear registration of mass spectrometry imaging and magnetic resonance imaging data. Anal Chem. 2019;91(9):6206–16. doi: 10.1021/acs.analchem.9b00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vos DRN, Jansen I, Lucas M, Paine MRL, de Boer OJ, et al. Strategies for managing multi-patient 3D mass spectrometry imaging data. J Proteom. 2019;193:184–91. doi: 10.1016/j.jprot.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 110.Wishart DS. Advances in metabolite identification. Bioanalysis. 2011;3(15):1769–82. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- 111.Blaženović I, Kind T, Ji J, Fiehn O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites. 2018;8(2):31. doi: 10.3390/metabo8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nash WJ, Dunn WB. From mass to metabolite in human untargeted metabolomics: recent advances in annotation of metabolites applying liquid chromatography-mass spectrometry data. Trends Anal Chem. 2019;120:115324 [Google Scholar]
- 113.Schymanski EL, Ruttkies C, Krauss M, Brouard C, Kind T, et al. Critical Assessment of Small Molecule Identification 2016: automated methods. J Cheminform. 2017;9(1):22. doi: 10.1186/s13321-017-0207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Palmer A, Phapale P, Chernyavsky I, Lavigne R, Fay D, et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14(1):57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
- 115.Alexandrov T, Ovchinnikova K, Palmer A, Kovalev V, Tarasov A, et al. METASPACE: A community-populated knowledge base of spatial metabolomes in health and disease. bioRxiv. 2019:539478. doi: 10.1101/539478. [DOI] [Google Scholar]
- 116.Xu Y-F, Lu W, Rabinowitz JD. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal Chem. 2015;87(4):2273–81. doi: 10.1021/ac504118y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gathungu RM, Larrea P, SniatynskI MJ, Marur VR, Bowden JA, et al. Optimization of ESI-source parameters for lipidomics reduces misannotation of in-source fragments as precursor ions. Anal Chem. 2018;90(22):13523–32. doi: 10.1021/acs.analchem.8b03436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. PNAS. 2015;112(41):12549–50. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Monge ME, Dodds JN, Baker ES, Edison AS, Fernández FM. Challenges in identifying the dark molecules of life. Annu Rev Anal Chem. 2019;12:177–99. doi: 10.1146/annurev-anchem-061318-114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry—looking beyond classical histology. Nat Rev Cancer. 2010;10(9):639–46. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 122.Aichler M, Walch A. MALDI imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Lab Investig. 2015;95(4):422–31. doi: 10.1038/labinvest.2014.156. [DOI] [PubMed] [Google Scholar]
- 123.Prentice BM, Chumbley CW, Caprioli RM. High-speed MALDI MS/MS imaging mass spectrometryusing continuous raster sampling. J Mass Spectrom. 2015;50(4):703–10. doi: 10.1002/jms.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tillner J, Wu V, Jones EA, Pringle SD, Karancsi T, et al. Faster, more reproducible DESI-MS for biological tissue imaging. J Am Soc Mass Spectrom. 2017;28(10):2090–98. doi: 10.1007/s13361-017-1714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Basu SS, Regan MS, Randall EC, Abdelmoula WM, Clark AR, et al. Rapid MALDI mass spectrometry imaging for surgical pathology. NPJ Precis Oncol. 2019;3:17. doi: 10.1038/s41698-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barry JA, Groseclose MR, Fraser DD, Castellino S. Revised preparation of a mimetic tissue model for quantitative imaging mass spectrometry. Methods Protoc, Protoc Exch. 2018 doi: 10.1038/protex.2018.104. [DOI] [Google Scholar]
- 127.Burnum-Johnson KE, Zheng X, Dodds JN, Ash J, Fourches D, et al. Ion mobility spectrometry and the omics: distinguishing isomers, molecular classes and contaminant ions in complex samples. Trends Anal Chem. 2019;116:292–99. doi: 10.1016/j.trac.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Levy AJ, Oranzi NR, Ahmadireskety A, Kemperman RHJ, Wei MS, Yost RA. Recent progress in metabolomics using ion mobility-mass spectrometry. Trends Anal Chem. 2019;116:274–81. [Google Scholar]
- 129.Hinz C, Liggi S, Griffin JL. The potential of ion mobility mass spectrometry for high-throughput and high-resolution lipidomics. Curr Opin Chem Biol. 2018;42:42–50. doi: 10.1016/j.cbpa.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 130.Zhou Z, Shen X, Tu J, Zhu Z-J. Large-scale prediction of collision cross-section values for metabolites in ion mobility-mass spectrometry. Anal Chem. 2016;88(22):11084–91. doi: 10.1021/acs.analchem.6b03091. [DOI] [PubMed] [Google Scholar]
- 131.Zhou Z, Tu J, Xiong X, Shen X, Zhu Z-J. LipidCCS: prediction of collision cross-section values for lipids with high precision to support ion mobility-mass spectrometry-based lipidomics. Anal Chem. 2017;89(17):9559–66. doi: 10.1021/acs.analchem.7b02625. [DOI] [PubMed] [Google Scholar]
- 132.Harris RA, Leaptrot KL, May JC, McLean JA. New frontiers in lipidomics analyses using structurally selective ion mobility-mass spectrometry. Trends Anal Chem. 2019;116:316–23. doi: 10.1016/j.trac.2019.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chacon A, Zagol-Ikapitte I, Amarnath V, Reyzer ML, Oates JA, et al. On-tissue chemical derivatization of 3 -methoxysalicylamine for MALDI-imaging mass spectrometry. J Mass Spectrom. 2011;46(8):840–46. doi: 10.1002/jms.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Toue S, Sugiura Y, Kubo A, Ohmura M, Karakawa S, et al. Microscopic imaging mass spectrometry assisted by on-tissue chemical derivatization for visualizing multiple amino acids in human colon cancer xenografts. Proteomics. 2014;14(7-8):810–19. doi: 10.1002/pmic.201300041. [DOI] [PubMed] [Google Scholar]
- 135.Dueñas ME, Larson EA, Lee YJ. Toward mass spectrometry imaging in the metabolomics scale: increasing metabolic coverage through multiple on-tissue chemical modifications. Front Plant Sci. 2019;10:860. doi: 10.3389/fpls.2019.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shariatgorji M, Nilsson A, Fridjonsdottir E, Vallianatou T, Källback P, et al. Comprehensive mapping of neurotransmitter networks by MALDI-MS imaging. Nat Methods. 2019;16(10):1021–28. doi: 10.1038/s41592-019-0551-3. [DOI] [PubMed] [Google Scholar]
- 137.Xu F, Zou L, Liu Y, Zhang Z, Ong CN. Enhancement of the capabilities of liquid chromatographymass spectrometry with derivatization: general principles and applications. Mass Spectrom Rev. 2011;30(6):1143–72. doi: 10.1002/mas.20316. [DOI] [PubMed] [Google Scholar]
- 138.Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14(2):482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marco-Ramell A, Palau-Rodriguez M, Alay A, Tulipani S, Urpi-Sarda M, et al. Evaluation and comparison of bioinformatic tools for the enrichment analysis of metabolomics data. BMC Bioinform. 2018;19(1):1. doi: 10.1186/s12859-017-2006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chong J, Soufan O, Li C, Caraus I, Li S, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–94. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Willighagen EL, Alvarsson J, Andersson A, Eklund M, Lampa S, et al. Linking the Resource Description Framework to cheminformatics and proteochemometrics. J Biomed Semant. 2011;2(1):S6. doi: 10.1186/2041-1480-2-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Galgonek J, Hurt T, Michlíková V, Onderka P, Schwarz J, Vondrášek J. Advanced SPARQL querying in small molecule databases. J Cheminform. 2016;8:31. doi: 10.1186/s13321-016-0144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lombardot T, Morgat A, Axelsen KB, Aimo L, Hyka-Nouspikel N, et al. Updates in Rhea: SPARQLing biochemical reaction data. Nucleic Acids Res. 2019;47(D1):D596–600. doi: 10.1093/nar/gky876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rubakhin SS, Lanni EJ, Sweedler JV. Progress toward single cell metabolomics. Curr Opin Biotechnol. 2013;24(1):95–104. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342(6163):1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 146.Comi TJ, Do TD, Rubakhin SS, Sweedler JV. Categorizing cells on the basis of their chemical profiles: progress in single-cell mass spectrometry. J Am Chem Soc. 2017;139(11):3920–29. doi: 10.1021/jacs.6b12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Evers TMJ, Hochane M, Tans SJ, Heeren RMA, Semrau S, et al. Deciphering metabolic heterogeneity by single-cell analysis. Anal Chem. 2019;91(21):13314–23. doi: 10.1021/acs.analchem.9b02410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fessenden M. Metabolomics: small molecules, single cells. Nature. 2016;540(7631):153–55. doi: 10.1038/540153a. [DOI] [PubMed] [Google Scholar]
- 149.Zhang L, Vertes A. Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew Chem. 2017;57(17):4466–77. doi: 10.1002/anie.201709719. [DOI] [PubMed] [Google Scholar]
- 150.Duncan KD, Fyrestam J, Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2019;144(3):782–93. doi: 10.1039/c8an01581c. [DOI] [PubMed] [Google Scholar]
- 151.Ali A, Abouleila Y, Shimizu Y, Hiyama E, Emara S, et al. Single-cell metabolomics by mass spectrometry: advances, challenges, and future applications. Trends Anal Chem. 2019;20:115436 [Google Scholar]
- 152.Krismer J, Sobek J, Steinhoff RF, Fagerer SR, Pabst M, Zenobi R. Screening of Chlamydomonas reinhardtii populations with single-cell resolution by using a high-throughput microscale sample preparation for matrix-assisted laser desorption ionization mass spectrometry. Appl Environ Microbiol. 2015;81(16):5546–51. doi: 10.1128/AEM.01201-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Do TD, Comi TJ, Dunham SJB, Rubakhin SS, Sweedler JV. Single cell profiling using ionic liquid matrix-enhanced secondary ion mass spectrometry for neuronal cell type differentiation. Anal Chem. 2017;89(5):3078–86. doi: 10.1021/acs.analchem.6b04819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Neumann EK, Comi TJ, Rubakhin SS, Sweedler JV. Lipid heterogeneity between astrocytes and neurons revealed by single-cell MALDI-MS combined with immunocytochemical classification. Angew Chem. 2019;58(18):5910–14. doi: 10.1002/anie.201812892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rappez L, Stadler M, Sierra SHT, Phapale P, Heikenwalder M, Alexandrov T. Spatial single-cell profiling of intracellular metabolomes in situ. bioRxiv. 2019:510222. doi: 10.1101/510222. [DOI] [Google Scholar]
- 156.Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16(7):411–28. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 157.Angerer P, Simon L, Tritschler S, Wolf FA, Fischer D, Theis FJ. Single cells make big data: new challenges and opportunities in transcriptomics. Curr Opin Syst Biol. 2017;4:85–91. [Google Scholar]
- 158.Neumann S, Böcker S. Computational mass spectrometry for metabolomics: identification of metabolites and small molecules. Anal Bioanal Chem. 2010;398(7-8):2779–88. doi: 10.1007/s00216-010-4142-5. [DOI] [PubMed] [Google Scholar]
- 159.Nguyen DH, Nguyen CH, Mamitsuka H. Recent advances and prospects of computational methods for metabolite identification: a review with emphasis on machine learning approaches. Brief Bioinform. 2018;20:2028–43. doi: 10.1093/bib/bby066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bemis KD, Harry A, Eberlin LS, Ferreira C, van de Ven SM, et al. Cardinal: an R package for statistical analysis of mass spectrometry-based imaging experiments. Bioinformatics. 2015;31(14):2418–20. doi: 10.1093/bioinformatics/btv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ràfols P, Torres S, Ramírez N, Del Castillo E, Yanes O, et al. rMSI: an R package for MS imaging data handling and visualization. Bioinformatics. 2017;33(15):2427–28. doi: 10.1093/bioinformatics/btx182. [DOI] [PubMed] [Google Scholar]
- 162.Gamboa-Becerra R, Ramírez-Châvez E, Molina-Torres J, Winkler R. MSI.R scripts reveal volatile and semi-volatile features in low-temperature plasma mass spectrometry imaging (LTP-MSI) of chilli (Capsicum annuum) Anal Bioanal Chem. 2015;407(19):5673–84. doi: 10.1007/s00216-015-8744-9. [DOI] [PubMed] [Google Scholar]
- 163.Rübel O, Greiner A, Cholia S, Louie K, Bethel EW, et al. OpenMSI: a high-performance webbased platform for mass spectrometry imaging. Anal Chem. 2013;85(21):10354–61. doi: 10.1021/ac402540a. [DOI] [PubMed] [Google Scholar]
- 164.Föll MC, Moritz L, Wollmann T, Stillger MN, Vockert N, et al. Accessible and reproducible mass spectrometry imaging data analysis in Galaxy. bioRxiv. 2019:628719. doi: 10.1101/628719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kale NS, Haug K, Conesa P, Jayseelan K, Moreno P, et al. MetaboLights: an open-access database repository for metabolomics data. Curr Protoc Bioinform. 2016;53 doi: 10.1002/0471250953.bi1413s53. 14.13.1-18. [DOI] [PubMed] [Google Scholar]
- 166.Oetjen J, Veselkov K, Watrous J, McKenzie JS, Becker M, et al. Benchmark datasets for 3D MALDI- and DESI-imaging mass spectrometry. GigaScience. 2015;4:20. doi: 10.1186/s13742-015-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
In this section, we briefly overview the available data that can be used for methods development, training machine and deep learning models, and methods evaluation. In 2015, we published benchmark datasets from different biological systems from several labs (166). These datasets included serial 3D imaging MS data, but individual serial sections can also be used as 2D datasets. The datasets include MALDI-TOF and DESI-Orbitrap data, and although they still can be used for methods development, they do not represent the state of the art in terms of spatial and spectral resolution. MetaboLights hosts several imaging MS studies where both data and metadata can be downloaded; the data can be found by searching “imaging mass spectrometry.” METASPACE provides the largest collection of public imaging MS datasets. Currently, it provides access to annotated data only; we are planning to implement user-approved access to associated imzML data for public datasets. We have also published ground truth data from three crowdsourcing studies aimed at estimating the quality of ion images (https://github.com/alexandrovteam/IMS_quality), recognizing off-sample ion images (https://github.com/metaspace2020/offsample), and quantifying colocalization between ion images (https://github.com/metaspace2020/coloc).