Abstract
Materials patterned with high-aspect-ratio nanostructures have features on similar length-scales to cellular components. These surfaces are an extreme topography on the cellular level and have become useful tools for perturbing and sensing the cellular environment. Motivation comes from the ability of high-aspect-ratio nanostructures to deliver cargoes into cells and tissues, access the intracellular environment, and control cell behavior. These structures directly perturb cells’ ability to sense and respond to external forces, influencing cell fate and enabling new mechanistic studies. Through careful design of their nanoscale structure, these systems act as biological metamaterials, eliciting unusual biological responses. While predominantly used to interface eukaryotic cells, there is growing interest in non-animal and prokaryotic cell interfacing. Both experimental and theoretical studies have attempted to develop a mechanistic understanding for the observed behaviors, predominantly focusing on the cell – nanostructure interface. Here, we consider how high-aspect-ratio nanostructured surfaces are used to both stimulate and sense biological systems and discuss remaining research questions.
Keywords: high-aspect-ratio nanostructure, biological metamaterial, nanoneedle, nanopillar, nanowire
Keywords: Biointerfacing
1. Introduction
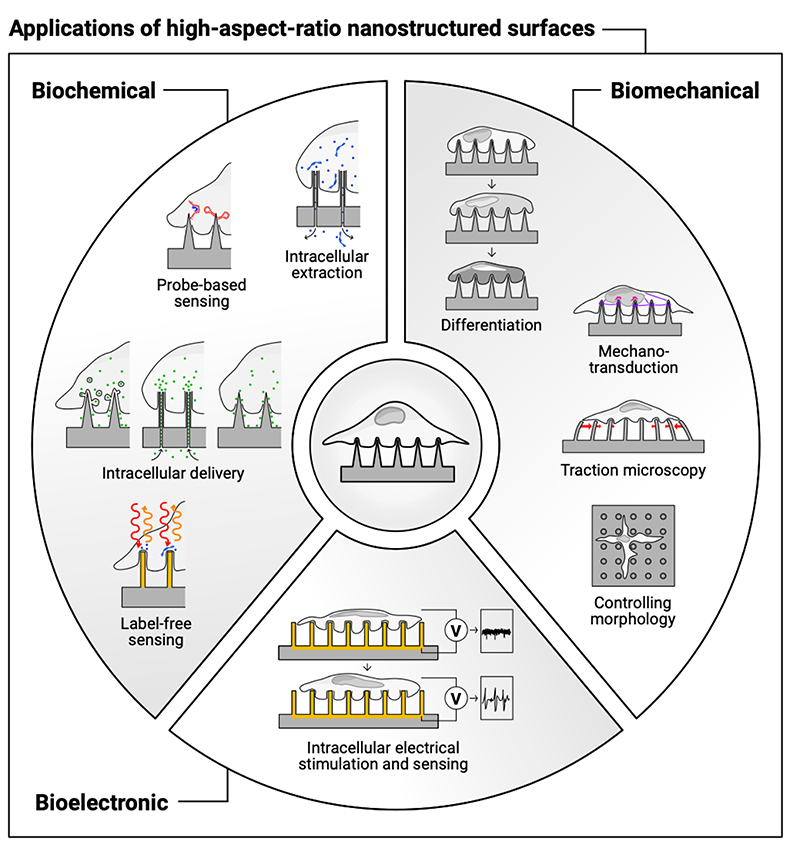
This review summarizes the literature illustrating how high-aspect-ratio nanostructures with defined, cellular-scale dimensions, can both sense and stimulate the extra- and intra-cellular environment (Figure 1). The spacing, sharpness and height of high-aspect-ratio nanostructured surfaces strongly influence the biological response, triggered by membrane – material interactions and subsequent intracellular signaling.
Figure 1. High-aspect-ratio nanostructured surfaces are used to stimulate and sense the biochemical, biomechanical, and bioelectronic environment of cells.
Specifically, we discuss:
different interfacing scenarios, including cellular membrane penetration or engulfment, and the stimulation of endocytosis;
attempts to model the cell membrane – nanostructure interface;
the techniques used to fabricate and characterize high-aspect-ratio nanostructures;
biochemical stimulation and sensing (delivering molecules into tissues and cells, and sensing the intracellular environment);
bioelectronic stimulation and sensing (stimulating electrogenic cells, and recording intra- and extracellular potentials);
biomechanical stimulation and sensing (guiding cell growth, promoting differentiation, studying mechanotransduction, using nanostructures for traction-force microscopy, and to mechanically capture cells);
a brief summary of the emerging use of high-aspect-ratio nanostructures for prokaryotic cell interfacing;
a summary of the fundamental challenges and open questions in the field.
1.1. Why Does This Topic Need a Review?
Much of the literature using high-aspect-ratio nanostructures for biological interfacing tends to exist in discrete silos, related to a given application area (e.g. drug delivery, electronically-stimulating electrogenic cells, promoting cell differentiation).[1] But irrespective of their use, all of these systems are ultimately governed and mediated by the fundamental biological mechanisms occurring at the cell membrane – nanostructure interface. We highlight results that have cross-field importance and where appropriate refer to a number of excellent perspectives and other reviews relevant to each field.[1–11] The wide range of application areas also come with an equally large variety of fabrication and characterization approaches. Hence this review also serves as a practical guide to different techniques, that can be adopted by researchers depending on the application and available resources. We also aim to summarize the questions that we feel are not yet fully satisfactorily answered by the existing body of literature, again to aid the design of future studies.
1.2. High-aspect-ratio nanostructured surfaces as biological metamaterials
Physically patterning a surface with high-aspect-ratio nanostructures can dramatically alter cell interactions with the material. This biological response is strongly influenced by the geometry of the patterned structures and has been harnessed in multiple applications. Underlying all are common biological questions, such as understanding the impact of nanostructures on cellular and nuclear membranes, and cell behavior. While the desired biological response may vary (e.g. how can intracellular access be increased, how can specific differential fates be promoted?), the fundamental consideration remains the same; what is the relationship between material structure and biological response?
We propose a new way of framing high-aspect-ratio nanostructured surfaces as ‘biological metamaterials’. The term metamaterial is borrowed from the physical sciences, where it describes materials whose unusual physical properties arise from their patterned structure, rather than an innate property of the bulk material. For example, electromagnetic metamaterials can have a negative refractive index (which causes incident radiation to refract in the opposite direction to a conventional material).[12,13] Mechanical metamaterials can have a negative Poisson’s ratio (where a material expands laterally as it is stretched longitudinally, rather than contracts).[14–18]
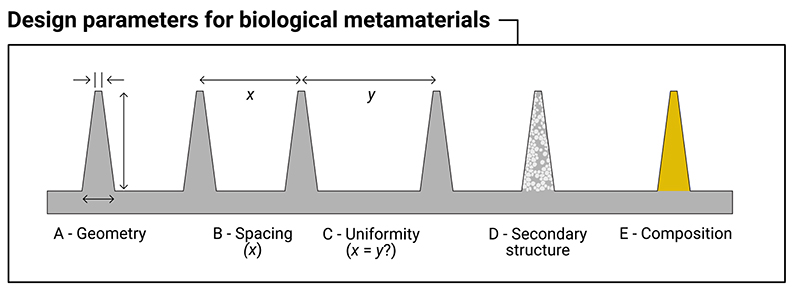
Analogously, a biological metamaterial results in an unnatural biological response. It may cause cells to align in a highly oriented or artificial manner, or promote spontaneous membrane penetration, or result in unnaturally perturbed cellular and nuclear membranes. In some applications, nanostructured materials are proposed as implant coatings, which aim to invisibly integrate biomaterials into the host (and avoid the sequence of biological interactions that result in a foreign body response)[19]. In effect, these nanostructured surfaces attempt to cloak the implant from the rest of the body. We propose that in a biological metamaterial this response is driven primarily by the physical patterning of the material. Changing the constituent material or the surface chemistry may also impact the response (as is true of physical metamaterials), but the main effects derive from the structure. Furthermore, a distinguishing feature of a biological metamaterial is the length scale of these structures. In physical metamaterials, individual structures are smaller than the wavelength of the incident waves. Similarly, in biological metamaterials, individual structures have sub-cellular dimensions (typically in the micron and sub-micron regime for mammalian cells). In both cases, it is this use of structures smaller than the target system which is responsible for the observed properties. Figure 2 summarizes some of the design parameters that can be considered as influencing the behavior of a biological metamaterial.
Figure 2.
Design parameters to consider for high-aspect-ratio nanostructured surfaces that act as biological metamaterials. These are: (A) geometry, including the height, tip-width and base-width of the nanostructure; (B) the spacing between nanostructures; (C) the uniformity of the spacing of the nanostructures (are the nanostructures spaced with a regular periodicity, or stochastically?); (D) the presence of any secondary structure, for example the use of porous materials; (E) the underlying bulk material (e.g. silicon, gold, polymer, etc).
Interestingly, the similar length scales mean biological and electromagnetic metamaterials can overlap. For example, gold nanorod arrays (that would look very familiar in a cell interfacing experiment) have been used as a plasmonic metamaterial biosensor,[20] and Dipalo et al. recently argued for incorporating plasmonic metamaterials as part of an intracellular sensing platform.[21] Careful design could allow the fabrication of hybrid biological/plasmonic metamaterials to simultaneously benefit from desirable biological and physical behaviors.
So why define high-aspect-ratio nanostructured surfaces to be biological metamaterials? Our hope is that it provides an umbrella term to bring together studies from different application areas and highlight common research questions. How can we quantitatively relate cellular and nuclear membrane perturbations to nanostructure geometry? What is the relationship between cell size (or volume), geometry, and biological response (and what can we learn from studies with smaller prokaryotic cells)? What are the geometric thresholds for these biological responses? What are the best metrics to quantify this response? We propose that framing these questions within the scope of biological metamaterials presents a useful approach to help guide research efforts.
1.3. Scope, Terminology and Takeaway Message
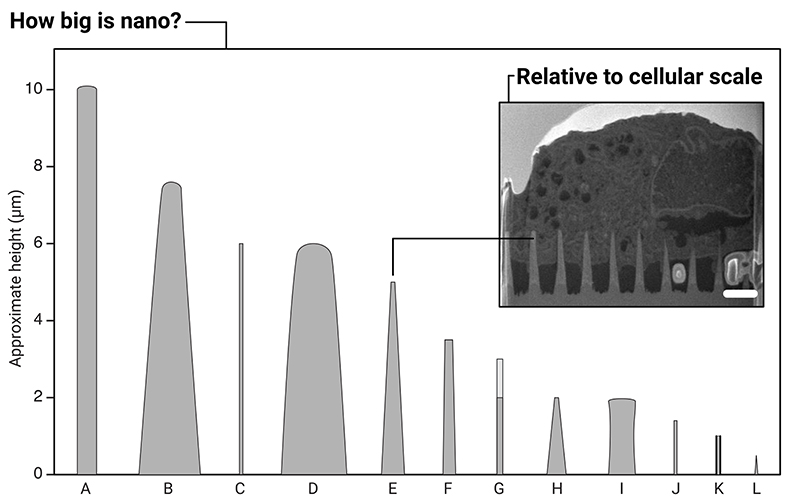
Here we examine surfaces, rather than untethered high-aspect-ratio nanostructures,[35–38] or single-cell probes.[39–47] The description ‘high-aspect-ratio’ is loosely defined in the literature, but is typically applied to structures with an aspect ratio equal to or greater than 10:1.[6,48–51] In this context, this means the majority of nanostructures we review here are less than 10 μm high, with sub-micron tips (with a few exceptions), see Figure 3. We do not consider micropatches (also referred to as microneedles) in this review, which can share similar aspect ratios, but have heights an order-of-magnitude larger.[52,53]
Figure 3.
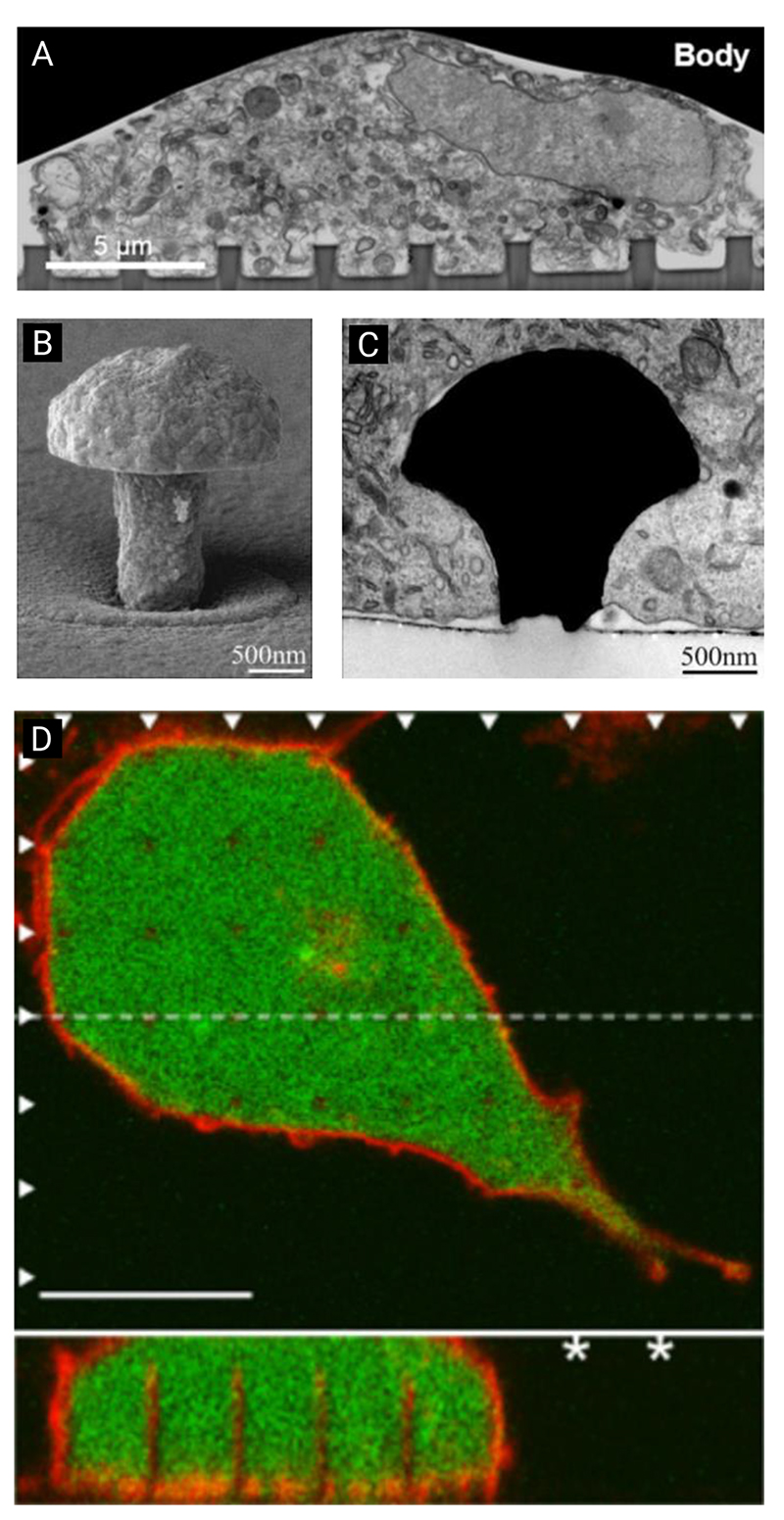
Illustration of the relative sizes of a selection of high-aspect-ratio nanostructures used in biointerfacing studies. A: ordered silicon pillar arrays for cell transfection.[22] B: diamond nanoneedle array for delivering probes and anti-cancer drugs into cells.[23] C: silicon nanowires for gene delivery.[24] D: plasmonic micropillars for cell traction force measurements.[25] E: porous silicon nanoneedles for in vivo growth factor delivery into muscle tissue.[26] F: silicon nanowire arrays for cell transfection.[27] G: vertical nanowire electrode arrays for interfacing neuronal cells.[28] H: diamond nanoneedle arrays for intracellular delivery.[29] I: silicon micropillar arrays for investigating single and collective cell behaviors on structured surfaces.[30] J: vertical nanopillars for studying nuclear deformation.[31] K: hollow nanostraws for intracellular sampling and longitudinal monitoring.[32] L: vertical carbon nanofibre electrodes for electrochemical intracellular communication.[33] (Note: here we use the authors’ original nomenclature for each description, to reflect the variety of terms found within the literature. In some reports, multiple geometries were fabricated, here a representative geometry is shown.) Inset: micrograph of FIB-SEM milled cross-section of a human mesenchymal stem cell interfacing porous silicon nanoneedles, scale bar 2 μm, adapted under the terms of CC BY license.[34] Copyright 2019, The Authors.
Due to the broad range of fields encompassed by this review, there is little consistency in terminology. When referring to the original articles, expect to see high-aspect-ratio nanostructures described as: nanoneedles, nanopillars, nanowires, nanostraws, nanotubes, nanoelectrodes, nanobars, nanoblades, nanospikes, nanoposts, nanowhiskers, vertical nanostructures and more. As each report defines the geometries differently, we use the authors’ original nomenclature wherever practical to aid with follow-up literature searches. But be warned, one scientist’s nanopillar is another’s nanowire...
Our message to engineers reading this review: the most valuable literature not only describes well-engineered nanotopographies, but also presents a clear and relevant application, and/or helps explains the biological mechanisms at the biointerface. For biologists: high-aspect-ratio nanostructures can control and strongly influence the cellular and intracellular microenvironment, and a huge range of materials already exist that can probe fundamental cell behavior, in particular mechanotransduction.[54,55]
2. Understanding the Cell – Nanostructure Interface
2.1. Cell membrane interactions with nanostructured surfaces
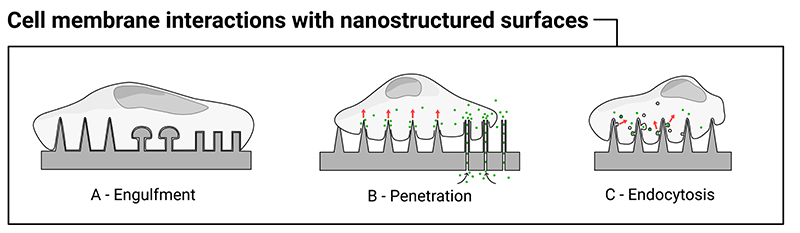
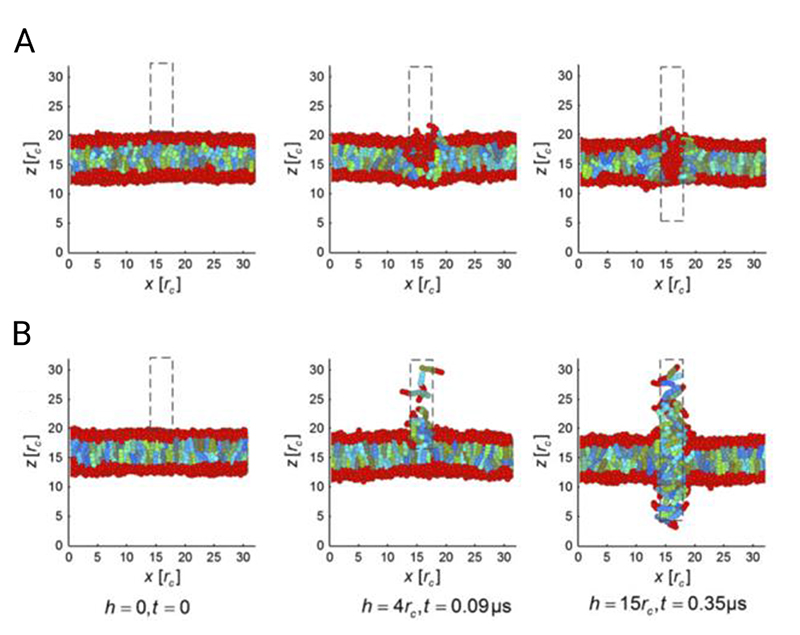
The precise interaction of the cell membrane with high-aspect-ratio nanostructures is critical to the biochemical, bioelectronic and biomechanical effects discussed in the sections below. How high-aspect-ratio nanostructures facilitate intracellular access, particularly when used to deliver biocargoes or electrical stimulate cells is a key question when considering material and experimental design. Throughout the literature interactions are variously described as penetrating, piercing, perturbing, impaling, indenting and mechanically disrupting the cell membrane, which reflects in part the lack of consensus over what is happening. In particular, many reports question whether the cell membrane is spontaneously penetrated by nanostructures,[2,4,11,28,56–65] and this topic has been presented as a source of contention within the field. In this context, spontaneous penetration refers to a high-aspect-ratio nanostructure piercing the membrane of a cell that has been seeded onto a surface (with minimal applied external force). From the literature reviewed here, it is clear that while spontaneous penetration is rare, cells are highly capable of engulfing high-aspect-ratio nanostructured surfaces. This behavior is dependent on a wide-range of factors, including geometry and cell – surface adhesion. Here we consider the evidence for three broadly different interfacing scenarios that are highly relevant when interpreting experimental observations (Figure 4). These are: a) engulfment of the cell membrane around nanostructures, b) penetration of the cell membrane – either spontaneously or under an external driving force, c) and active-uptake by cells seeded on top of nanostructures. These scenarios are not mutually exclusive or exhaustive but are often cited in the literature. (We are not considering the degree of engulfment here – for example when cells perch on top of nanostructures – see the next section for greater discussion.)
Figure 4.
Illustration of the different ways the cell membrane can interact with high-aspect-ratio nanostructures. The cell membrane can engulf nanostructures to varying degrees (A), penetration of the membrane can occur under specific conditions (B), and there is evidence nanostructured surfaces can directly stimulate endocytosis (C). Note: these scenarios are not mutually exclusive.
2.2. Methods for interfacing nanostructured surfaces and cells
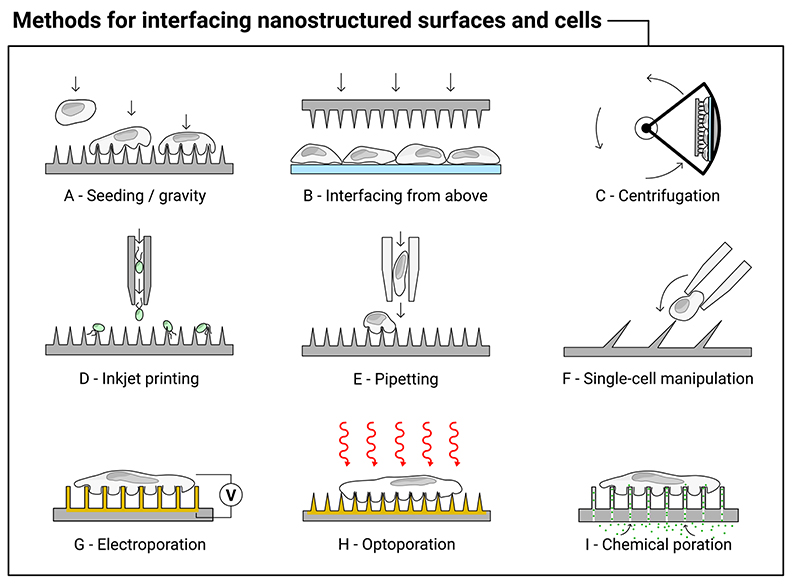
Membrane interactions are highly dependent upon the chosen interfacing method. Figure 5 illustrates a range of interfacing approaches across the literature. Cells can be seeded onto loaded nanostructures (Figure 5A);[66–68] the nanostructured surface can be placed (or mechanically impaled) onto an existing cell layer (Figure 5B);[26,69,70] additional force can be provided by centrifugation (Figure 5C).[26,29,71–73] Cells can also be forcibly impaled by more uncommon techniques, such as inkjet printing (Figure 5D),[74] by repeatedly pipetting cells onto nanostructures (Figure 5E),[23,75] or via single-cell manipulation (Figure 5F).[76]
Figure 5.
Illustration of a range of different methods used to interface high-aspect-ratio nanostructured surfaces with cells, see the main text for corresponding references. The methods include: (A) seeding the cells and allowing them to settle under gravity onto the substrate; (B) either manually or mechanically interfacing the surface from above; (C) sandwiching the nanostructured surface with cells and centrifuging; (D) inkjet printing of cells (algae) directly onto the surface; (E) forcibly and repeatedly pipetting cells onto the surface; and (F) using a micropipette to manually push single cells onto inclined nanostructure. Once on the surface a range of poration methods can be combined to further modify the interfacing behavior, including: (G) electroporation; (H) optoporation; and (I) chemical poration techniques.
Centrifugation increases the penetration force of nanoneedles, ostensibly above the mechanical barrier provided by the membrane.[26,69,77] It has the benefit of rapid interfacing with well-defined force,[78] has been used to penetrate prokaryotic cells with rigid cell walls,[79] but is impractical in most tissue interfacing applications. While one report has suggested centrifugation leads to non-uniform transfection,[80] most report no issues, other than the need to carefully optimize the interfacing parameters. A few studies have systematically studied the impact of increased interfacing force, either by increasing centrifugation speed,[81] or by using a motorized compression test stand to interface nanostructured substrates with cells,[73] and found that increased force correlated with both increased delivery and cell death. To complicate matters, the optimum interfacing force may also vary with cell type.[81]
An unusual interfacing approach has been proposed by Kim et al., who synthesized inclined gallium nitride nanoneedles, with angles as small as ~30° between needle and substrate.[76] Their motivation was predominantly for single-cell interfacing, where the angle of nanoneedle better matches the incoming angle of a cell attached to a glass micropipette (illustrated in Figure 5F), and they successfully delivered dyes and single-stranded DNA in this manner.
2.3. Cells Can Engulf High-Aspect-Ratio Nanostructures
Many electron microscopy studies have illustrated that cell membranes can engulf high-aspect-ratio nanostructures, with no clear evidence of membrane rupture (see Figure 6 for some examples).[26,64,66,82,84–86] Electron microscopy is limited to a fixed moment in time, so may fail to capture the highly dynamic nature of the membrane,[87] which can undergo rapid repair.[11,88] However, engulfment without penetration has also been seen in live cell optical imaging. Berthing et al. imaged intact cell membranes wrapping around indium arsenide nanowire arrays using fluorescence confocal live cell imaging (Figure 6D).[61] Their study revealed intact cell membranes with no penetration in 95% of the nanowires studied (the remaining 5% of nanowires, 29 out of the 542 examined, were ambiguous).
Figure 6.
Examples of cell membranes engulfing nanostructured surfaces. (A) Nanopillar engulfment by a neuronal cell body, imaged by focused-ion-beam scanning-electron microscopy. Reproduced with permission.[82] Copyright 2017, American Chemical Society. (B) Scanning-electron-microscopy micrograph of gold mushroom-shaped electrode, plus (C) engulfment of electrode by a neuroendocrine cell (PC12). Reproduced under terms of CC BY license.[83] Copyright 2018, Spira, Shmoel, Huang and Erez. (D) Fluorescence confocal image of a human embryonic kidney (HEK293) cell cultured on an indium arsenide nanowire array (cell body green and membrane red), showing wrapping of the membrane around each nanowire (scale bar 10 μm). Reproduced with permission.[61] Copyright IOP Publishing Ltd, 2012.
2.4. Spontaneous Penetration of Cells is Rare
Intracellular delivery experiments with hollow nanostraws provide further evidence that spontaneous penetration is rare.[59,89] However, penetration can be enhanced via electroporation,[90,91] optoporation,[92] or by coating nanostraws with strongly cell-adhering coatings.[93,94] Bioelectronic experiments show that nanoelectrodes only measure intracellular potentials after poration techniques have been applied,[62,95,96] and rapidly return to measuring extracellular potentials in the absence further external stimulus, again highlighting the need for an external force to disrupt the membrane. Dipalo et al. explored this behavior explicitly using a range of nanopillar geometries using fluorescent-, and electron-microscopy, and electrophysiological measurements.[64] All three methods consistently showed no spontaneous penetration of the membrane, except in a handful cases.
2.5. The Cell Membrane May Not Be the Only Barrier to Penetration
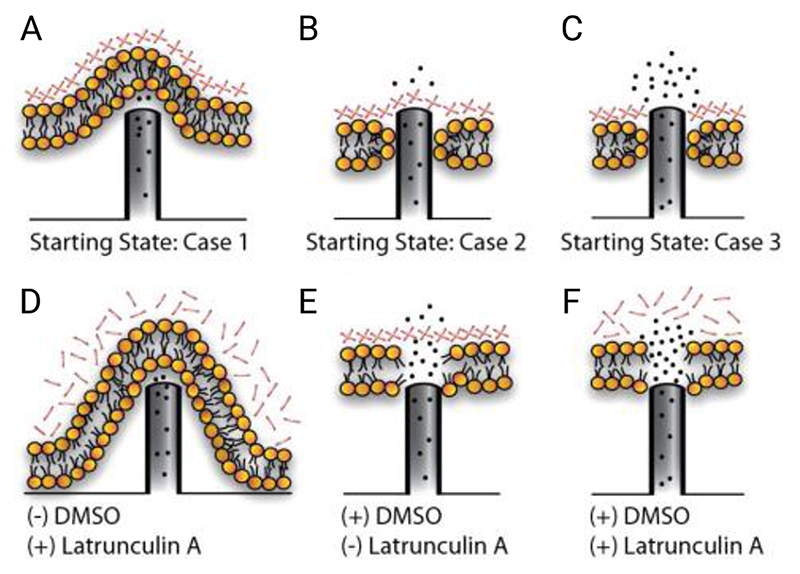
The detection of cytosolic components,[69,71] and delivery of membrane-impermeable cargoes,[29,93,97] are evidence that under the right conditions penetration can occur. From a mechanical perspective, systems such as silicon porous nanoneedles have been found to sustain forces well in excess of those required to penetrate a membrane (as determined using single-cell force microscopy studies).[26,40,98] However, the cell membrane may not be the only barrier to penetration. Dynamic reorganization in response to stimulation by nanostructured surfaces can result in the recruitment of cytoskeletal proteins,[99] creating an actin meshwork in the vicinity of the membrane (the plasmalemmal undercoat).[58,100] The effect of this meshwork depends upon how nanostructures are interfaced with cells. Kagiwada et al. used single-nanoneedle penetration experiments to argue that the actin meshwork reduces membrane fluidity, and is necessary to give the mechanical properties required for penetration.[101] Although this finding was later disputed by Angle et al. who found that they could rupture the cell membrane (of cells without an actin meshwork) by continuing to probe the cell well beyond just the initial indentation.[102] Aalipour et al. found that when cells are seeded onto nanostraws, this same meshwork can instead act as a barrier. They used hollow nanostraws to consecutively deliver a membrane-permeabilizing solvent and actin-depolymerizing toxin (dimethyl sulfoxide and latrunculin A respectively) to cells seeded onto the surface, to separately explore the effects of membrane and cytoskeleton permeability.[58] They found permeabilizing the membrane alone was not sufficient to facilitate intracellular delivery (Figure 7).
Figure 7.
Aalipour et al.’s illustration of nanostraw – cell membrane interfacing behavior. (A – C) In the absence of chemical poration the majority of nanostraws do not penetrate the membrane, (B) a few penetrate the membrane but not the actin meshwork, (C) a few penetrate both the membrane and meshwork. (D – F) Using dimethyl sulfoxide (DMSO) and latrunculin A, the cell membrane and/or actin meshwork can be chemically porated facilitating access. Scenario F provides the greatest degree of intracellular access. Reproduced with permission.[58] Copyright American Chemical Society, 2014.
2.6. High-Aspect-Ratio Nanostructures Can Induce Endocytosis
To further complicate the interpretation of intracellular delivery experiments, nanostructured surfaces have also been observed to induce endocytosis,[103,99,84] causing the cell to actively uptake cargoes without the need for penetration. Generally, eukaryotic cells have a range of mechanisms for the active uptake of molecules surrounding the cell, including: 1) phagocytosis, 2) pinocytosis, and 3) receptor-mediated endocytosis.[104] Curvature-sensitive membrane proteins are well-known to play an active role in these processes,[87,105] and recent reports highlight the interplay between nanostructured surfaces and recruitment of these proteins.
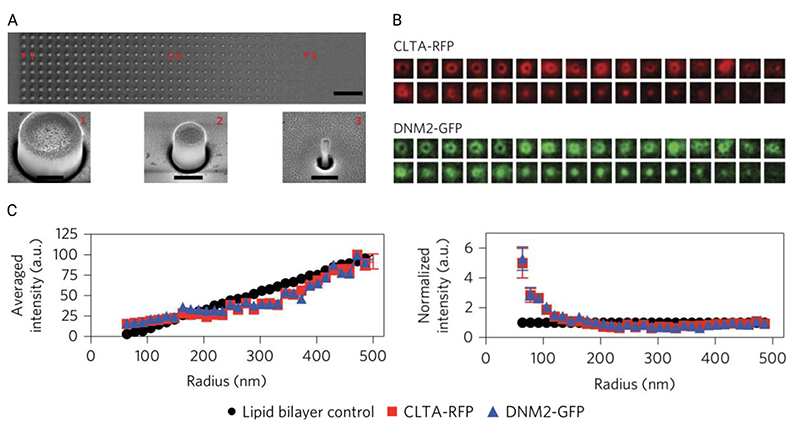
The behavior of cells attempting to phagocytose nano- and microstructured surfaces has previously been reported by a number of groups,[107,108] and other endocytosis pathways appear to be influenced too. Galic et al. showed that tin oxide nanocones cause the recruitment of N-BAR domain proteins to curved regions.[109] Zhao et al. investigated how low-aspect-ratio quartz nanopillars and nanobars can stimulate protein-recruitment during clathrin-mediated endocytosis.[106] (See also their recently reported fabrication and interfacing protocol.[110]) They systematically altered membrane curvature in cancerous skin cells (SK-MEL-2) by seeding them on nanopillars with a range of diameters and observed curvature-dependent clustering of ten endocytosis-related proteins, including clathrin and dynamin2. Critically, they observed that clustering was greatest around features with 200-nm or less radii of curvature, the regime most relevant for high-aspect-ratio nanostructures (Figure 8). They also noted strong actin recruitment to the curved regions, suggesting that the curvature induces further cytoskeletal and mechanotransduction processes.
Figure 8.
Figure adapted from the work of Zhao et al., illustrating the principles of their experimental setup. (A) Scanning-electron-microscopy micrographs of their array of low-aspect-ratio nanopillars, with varying diameters (top row micrograph scale bar 10 μm, bottom row micrographs scale bars 400 nm). (B) They then seeded genome-edited cells (SK-MEL-2) onto these structures, which expressed red-fluorescent-protein-tagged clathrin (CLTA-RFP) and green-fluorescent-protein-tagged dynamin2 (DNM2-GFP). Using immunofluorescence microscopy they averaged multiple cells over multiple geometries to determine differences in intensity. (C) From this analysis they determined that nanopillar radii less than 200 nm resulted in a rapid increase in the quantity of observed proteins. Adapted with permission.[106] Copyright 2017, Springer Nature.
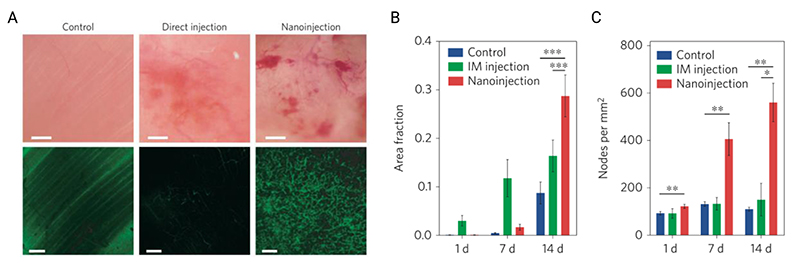
It is unclear whether all high-aspect-ratio nanostructures can induce endocytosis. VanDersarl, Xu et al. saw no significant upregulation of the endocytosis-related gene expression in cells cultured on their nanostraw platform.[89] However, where it does occur, high-aspect-ratio nanostructures can be used to enable endocytosis-mediated intracellular delivery. Gopal et al. investigated the role of silicon nanoneedles in modulating different endocytic pathways in human mesenchymal stem cells.[84] They observed considerable ruffling of the apical membrane (the top surface of the cell), and a strong engulfment of nanoneedles by the basal membrane (the bottom surface, closest to the substrate). Proteins integral to both clathrin- and caveolae-mediated endocytosis (caveolin-1 and clathrin light chain) were found co-localize with nanoneedles in the basal membrane, but not in the apical. At the same time, clathrin pits and caveolae (the membrane invaginations of these processes) were observed at the nanoneedle – basal membrane interface. By using specific cargoes, known to be trafficked by clathrin-, caveolae-mediated endocytosis, and micropinocytosis mechanisms, they were able to show nanoneedles increased uptake by each mechanism (Figure 9). While much of this cargo ends up in the endolysosomal system, a significant proportion (38% of siRNA) is still active in the cytosol, indicating that endocytosis-inducing nanoneedle-mediated delivery still retains biological function. While this efficiency will vary with cargo,[111] it provides insight into non-penetration based mechanisms for intracellular delivery.
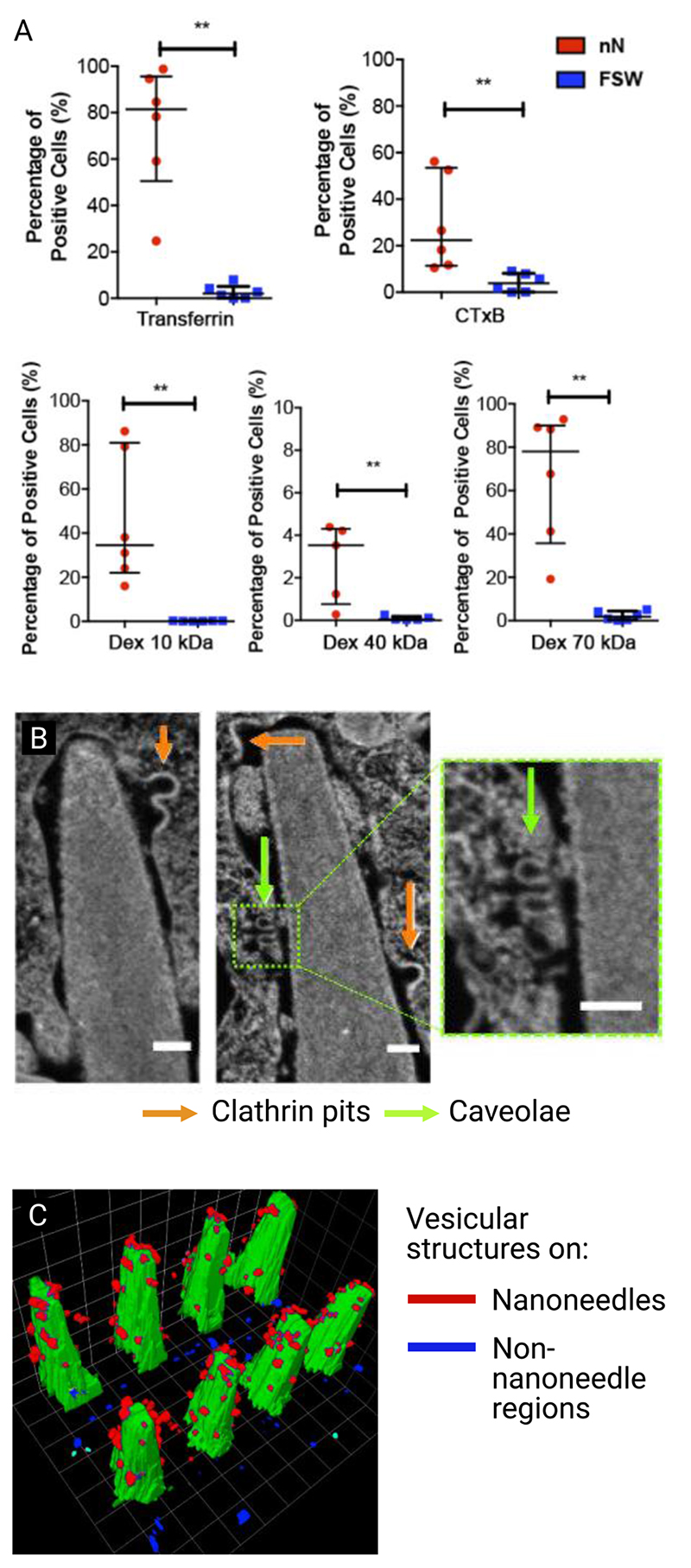
Figure 9.
Gopal et al. nanoinjected cells with different cargoes to explore which uptake mechanisms were stimulated by interfacing with porous silicon nanoneedles. (A) Percentage of positive cells for different mechanism-specific cargoes. Transferrin is a clathrin-mediated endocytosis cargo, cholera toxin B-subunit (CTxB) is a caveolae-specific cargo, and dextran (Dex, tested in different molecular weights) is a micropinocytosis-specific cargo. After confirming that surface area did not affect loading efficacy, they noted that all cargoes were more successfully internalized in cells on nanoneedles compared to flat silicon wafers. (B) Focused-ion-beam scanning-electron-microscopy micrograph of nanoneedle interacting with cell membrane (scale bars 100 nm), showing two different types of vesicular structure (clathrin pits and caveolae). (C) 3D reconstruction of vesicular structures on nanoneedle (red) and non-nanoneedle (blue) regions, nanoneedles shown in green. Reproduced under the terms of CC BY license.[84] Copyright 2019, The Authors.
3. Modelling the Cell – Nanostructure Interface
A number of attempts have been made to model the cell – nanostructure interface to help better understand the range of observed interfacing behaviors. Here, we summarize the design and output from a range of models, categorized by the question they are trying to answer. We focus on animal cell interactions here as the primary interest, but it is worth noting that there have been attempts to model prokaryotic cell interfacing too.[112,113] Similarly, we focus on high-aspect-ratio nanostructures tethered to a surface, for a more general exploration of cell – nanomaterial interactions, see the review of Gao.[114]
Models broadly consist of continuum-type, where the membrane is treated like a continuous sheet that can be characterized by key parameters such as tension, or stiffness; or molecular-based simulations, which attempt to simulate the interactions between constituent molecules directly. Continuum-based models, as first proposed by Helfrich,[115] consider the balance of forces or free-energy at the cell-substrate interface.[56,116,59,117,118] These have the benefit of rapidly showing an ensemble response, at the expense of the role of complex molecular interactions on membrane disruption.[63] Conversely, molecular dynamic simulations can offer greater insight at a smaller scale, but are computationally expensive, limiting the simulation window to very small regions.
3.1. How Likely is Spontaneous Penetration on Nanowires?
Spontaneous membrane penetration by high-aspect-ratio nanostructures is perhaps the largest discussion area in the field of high-aspect-ratio nanostructures and hence multiple attempts have been made to model this interaction. Melosh and colleagues developed a continuum-based model for cell – nanowire interactions.[56,119] Their model is based on balancing the gravitational force acting upon the cell, with the hydrostatic pressure inside (considering the cell as a membrane bound liquid) and the net membrane tension. Given the dependence of the net membrane tension on nanowire geometry, their model suggests that membrane penetration is strongly affected by cell – substrate adhesion, nanowire and array geometry, and cell stiffness.[56] Ultimately, they conclude that spontaneous penetration of the membrane under gravity alone is unlikely for nanowires greater than 20 nm in diameter. They also found that sharper nanowires reduce the required penetration force, while simultaneously reducing contact area and increasing membrane tension. Stiffer cells can be penetrated more efficiently, due to a smaller contact area between the membrane and nanowire, but only for large nanowire spacings. Dense arrays quickly inhibit adhesion between the membrane and substrate, causing cells to sit on top of the nanowire array.
The same authors later elaborated on this model in order to reconcile dynamic effects after the cell has settled on nanostructures.[59] Their revised model considers how the membrane continues to engulf the nanowire after cell settling, due to the adhesive membrane – substrate interaction (Figure 10). They argue that as a greater proportion of membrane contacts the base of the substrate, the effective adhesive force is reduced due to the reduction in remaining contact area. The force required to penetrate the membrane remains broadly constant, so eventually the adhesive force drops below the level required for penetration. Xie et al. use this model to describe a window of time after settling, during which penetration of the membrane is most likely, as validated by experimental results with hollow nanostraws. After this point, further penetration is unlikely, but can occur if the cell provides an additional traction force. This traction force is dependent upon the angle between the membrane and nanowire (pulling straight down on a nanowire requires less force to cause penetration), consistent with a separate study by Santoro et al. who found, after normalization, that cells were twice as likely to engulf nanostructures directly under their center than towards their edge.[66]
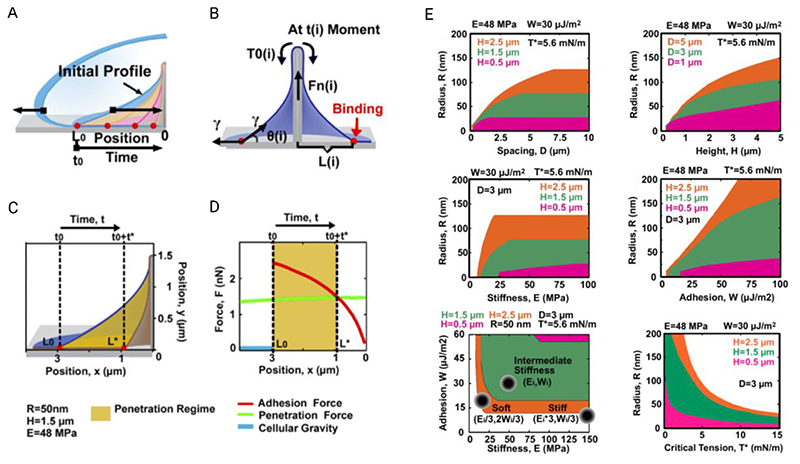
Figure 10.
Illustrations and model outcomes adapted from the report by Xie et al., exploring the dynamic settling behavior of cells on nanostructures. (A) Their adhesion model proposes that, under the appropriate conditions, as a cell settles onto a nanostructure, the membrane will continue to engulf the nanostructure for a short period afterwards. (B) The driving force for engulfment is the relative vertical adhesion force between the membrane and substrate. (C + D) As the remaining contact area decreases with time, so too does the net adhesive force, resulting in a time beyond which the adhesion force becomes less than the penetration force, ultimately making spontaneous penetration increasingly unlikely. (E) The relationship between various geometric, membrane and surface parameters for their adhesion model, where the orange, green and magenta regions indicate the parameter space where penetration is possible, for the corresponding nanowire heights. Adapted with permission.[59] Copyright 2015, American Chemical Society.
The outcome of their modelling again suggests a series of intuitive design rules for influencing the likelihood of spontaneous penetration by altering the spacing and height of nanowire substrates. Broadly speaking, thin, low density nanowires promote penetration, at the expense of limiting the number of cell – nanowire interactions. To overcome this, they propose increasing cell adhesion by modifying the surface chemistry of the nanowires. They found again that cell stiffness plays an important role in whether cells are penetrated, consistent with the wide variety of behavior seen in experimental results. The authors note that their model does not consider dynamic reorganization within the cell, and assumes the force transporting components are infinitely small, when in reality forces are transported by discrete protein units.
Lee et al. adopted this force-based approach to help understand the impact of inkjet printing cells onto silicon nanowires.[74] They were able to model the penetration force as a function on inkjet jetting speed, and used this to optimize the fixation of individual cells on top of each nanowire.
3.2. Will Cells Sink in, or Settle onto Nanostructures?
The result of Xie et al. highlighting the lack of spontaneous penetration,[56] provided an assumption (exploited in later models) that to effectively understand cell behavior, the balance of free energy of the membrane (rather than gravitational force) should be considered. Martinez and colleagues have extensively studied geometry-dependent cell response both experimentally and theoretically.[120,61,116,121–123,117,124,125] Their model considers the cell as a soft-shelled droplet,[126] and defines the free energy of the cell – substrate interaction as: the sum of the cell – substrate adhesion, the change in surface tension caused by an increase in cell surface area, and the change in elastic energy caused by bending the membrane.[116,117] Depending on the combination of these parameters, their model predicts two cell settling states: a ‘top’ state, where cells rest on top of the nanostructures; and ‘bottom’ where cells fully engulf the nanostructures (Figure 11). For realistic cell values, Buch-Månson et al. used this approach to predict that the bending energy term (a function of cell stiffness, nanostructure density, diameter and length) dominates over adhesion and tension effects.[117] The model also suggests that membrane wrapping around the nanowire is not normally energetically favorable, and requires external force. They were able to verify their model against literature and experimental data,[124,125] also observing an intermediate settling between the fully deformed and on-top regimes.
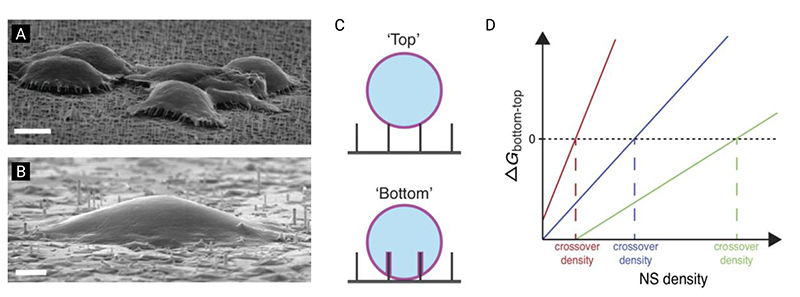
Figure 11.
A free-energy model for cell settling behavior on nanostructured surfaces, as proposed by Buch-Månson et al. (A) Scanning-electron-microscopy micrograph of cells suspended on top of nanostructure arrays in ‘top’ state (scale bar 5 μm). (B) Scanning-electron-microscopy micrograph of cells engulfing nanostructure arrays in ‘bottom’ state (scale bar 2 μm). (C) Illustration of membrane behavior as described by the model. (D) The change in free energy for the membrane – surface interaction, as a function of nanostructure density. In this model, if the overall change in free energy is greater than zero, the system favors cell settling in the ‘top’ state, and vice versa. Depending on the substrate and cell properties, the model predicts that the transition point between states will occur at different nanostructure densities. Adapted with permission.[117] Copyright 2015, John Wiley and Sons.
More recently, Zhou et al. attempted to expand on this approach, to accommodate the impact of nanostructure diameter, by expressing the change in bending energy as three separate terms: membrane unfolding, stretching and edge effects.[118] Their model predicts that for realistic nanostructure densities (25 – 100 nanostructures per 100 μm2), sharper nanostructures tend to favor greater membrane deformation over blunt (Figure 12). In contrast to the previous model, they argue that the limited space between high-density large-diameter nanostructures means the energy required to stretch and unfold the membrane dominates over any adhesion effects. For lower densities and radii, adhesion dominates, resulting in deformed membrane. However, their model also predicts that for very low radii less than approximately 10 nm, the energy required to bend the membrane exceeds that of the adhesion, and cells revert to an on-top state. While this is consistent with the previously discussed model,[117] the result remains true even for very low nanostructure densities, i.e. very sparse, very sharp nanostructures will not deform the membrane, which appears to again be counterintuitive.
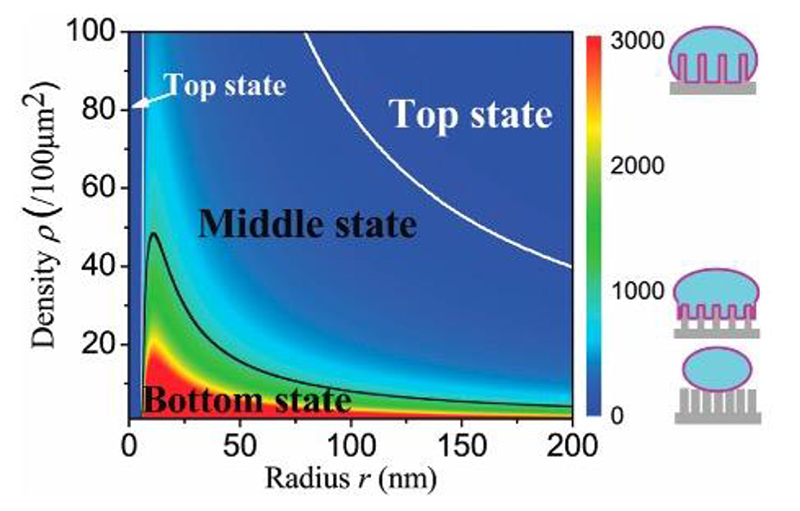
Figure 12.
A phase-diagram illustrating the output from the cell-settling model of Zhou et al. The color scale indicates the degree of adhesion depth (i.e. how far the cell sinks into the nanostructures). The black and white lines indicate the boundary for cells either being a fully-engulfed ‘bottom’ or ‘top’ state. Reproduced with permission.[118] Copyright 2018, Royal Society of Chemistry.
3.3. How Does Surface Chemistry Affect Single Nanopillar Penetration?
In the work discussed above, Buch-Månson et al. also demonstrated both theoretically and experimentally the importance of surface chemistry on cell adhesion behavior.[117] Liu et al. explicitly modelled a single nanopillar penetrating a bilayer membrane, and found that a nanopillar coated with hydrophobic ligands spatially disrupts the membrane far more than hydrophilic ligands (Figure 13). In the latter case the membrane reforms neatly around the nanopillar. Disruption could be further decreased by patterning either axially-orientated, alternating stripes, or randomly-patterned hydrophilic and hydrophobic ligands on the nanopillar.[127] While only applied to a single nanostructure, for those interested in promoting membrane penetration, this result subtly suggests that patterned surface chemistries (achieved perhaps by using self-assembly onto microfabricated layers) are a possible route to improving the likelihood of spontaneous penetration, an idea supported experimentally by the biomimetic probe work of Almquist and Melosh.[128,129]
Figure 13.
Dissipative particle dynamics simulation of either a hydrophilic (A) or hydrophobic (B) probe penetrating a model of a lipid bilayer, for different simulation timepoints. The degree of membrane disruption is strongly influenced by the surface chemistry of the probe. Reproduced with permission.[127] Copyright 2013, Elsevier.
3.4. How Does Nanopillar Curvature Disrupt the Cell Membrane?
Capozza et al. used a coarse-grain molecular dynamics simulation to explore the impact of nanopillar taper on membrane disruption.[63] They argue that the continuum-style models described above fail to account for membrane disruption caused by the sharp transition between nanopillar sidewall and flat top. Their approach considers the interplay between traction forces (pulling on the membrane) and membrane bending, accounting for situations where the membrane is pulled across a sharp edge. They simulated a bilayer membrane, yielding reasonable agreement to realistic membrane properties, given the two-dimensional nature of their model (Figure 14). They found that the spacing between adjacent hydrophilic heads in their membrane increases rapidly with decreasing membrane curvature, causing membrane rupture to occur for even small forces at high curvature. One important outcome of their model is the suggestion that even large diameter nanopillars can induce spontaneous membrane rupturing, provided the taper between the sidewall and top is sharp. They experimentally verified this result by seeding fibroblasts (NIH-3T3) onto silicon nanopillars with either a sharp (~20 nm) or smooth (~250 nm) taper. Using membrane permeable and impermeable dyes, they observed that sharp-tipped nanopillars facilitated about 70% delivery while delivery on blunt nanopillars was negligible. The high delivery was achieved despite using a relatively low-aspect-ratio (~1.25:1) and large tip-diameter (2 μm), a scenario not typically envisaged by other models.
Figure 14.
A two-dimensional coarse-grain molecular dynamics simulation of a strained membrane, rupturing about two different curved edges. (A + B) Sharper edges make membrane rupture more likely for a given traction force. (C + D) Capozza et al. were able to replicate this result experimentally using two different types of nanopillar, with differing sharpness edges. (E + F) Despite being relatively short, wide nanostructures, greater delivery of a membrane-impermeable dye was seen on the sharper-edged structures (compare the greater degree of red staining shown in the fluorescence micrograph E compared to F). Adapted with permission.[63] Copyright 2018, American Chemical Society.
Interestingly, Capozza et al.’s experimental regime is similar to the protein-recruiting studies of Zhao et al. (as discussed above, and part illustrated in Figure 8).[99] In the latter case, quartz flat-topped nanopillars and elongated nanobars of different radii-of-curvature were used to study the recruitment of proteins to the membrane. One might expect that the sharp transition from the vertical sidewall of these structures to the flat top (effectively a tight radius of curvature, in a different plane), might also trigger protein recruitment, however Zhao et al. did not witness this in elongated nanobar structures, where proteins were only recruited around the rounded ends. Further studies exploring the subtlety of in- and out- of plane curvature, as well as whether there is a threshold for membrane-curvature-induced protein recruitment, may help to reconcile these results.
3.5. Can Adding a Cap to the Tip Improve Membrane Engulfment?
Santoro et al. considered the impact of aspect-ratio, and of the addition of a mushroom-shaped cap to the top of nanoelectrodes.[66] They developed a continuum membrane model to support their experimental observation of membrane wrapping around different shaped structures. They found that the presence of a cap substantially increased the degree of membrane engulfment, as did increasing the aspect-ratio of the nanoelectrode. They found that adding a cap to nanowires improves membrane engulfment, preferable for their ultimate application of forming a high-resistance electrical seal between cell and nanoelectrode. Perhaps critically, they also highlighted how this resistance is likely to vary considerably with electrode location under the cell, thanks to the differing cytoskeletal forces acting at the center and periphery.
3.6. Limitations of Current Models
While cell-settling models can provide insight into general cell behaviors, it is also important to note many other types of interaction are also possible. Wierzbicki et al. empirically describe a total of seven discrete cases for fibroblasts (NIH-3T3) cultured on silicon nanowires, including nanowires that had been crushed, underwent cellular uptake, or were being probed by microvilli blebbing from the cell.[130] Similarly, although most theoretical (and indeed experimental) results suggest that spontaneous penetration is highly unlikely, this may not necessarily be true for all cell types. Inspired by the use of black silicon as a bactericidal substrate, Pham et al. studied the interaction of red blood cells with black silicon, an extremely dense and sharp nanowire surface, and observed spontaneous cell lysis.[131] Their modelling suggested that just a handful of nanowire – membrane contacts were enough to cause red blood cells to rupture.
The nature of modelling problems often necessitates careful choice and focus of parameters to yield a computationally tractable problem. The models presented here tend to simplify the nanostructure itself, however the results of Capozza et al. have demonstrated the importance of considering factors such as edge sharpness on local membrane disorder.[63] Future modelling could aim to understand the impact of edge sharpness in three-dimensional membranes, where total membrane rupture (as typically modelled in two-dimensional systems) is unlikely. Given the ability of cell membranes to repair rapidly,[88] modelling either transient or metastable damage may also be required to more accurately model membrane – nanostructure interactions.
4. Fabrication Techniques
4.1. Fabricating High-Aspect-Ratio Nanostructures is a Multi-Step Process
High-aspect-ratio nanostructured surfaces are used not only in the biological applications discussed here, but also to create materials in other research fields, such as optoelectronics and other forms of surface engineering.[6] As a result, there are already a huge range of techniques that can be used to achieve similar structures. In our attempt to codify this, we consider the practical steps needed to fabricate nanostructures (Figure 15). Firstly, a template or pattern is required to define the location of each nanostructure on the surface. This could be a well-defined photopatterned design, or alternatively from randomly deposited metal clusters on the surface of the substrate, or from a naturally occurring material whose pattern specifies the location of each nanostructure. Secondly, this pattern is used as a template to either selectively remove or add material to the substrate. These approaches are often referred to ‘top-down’ and ‘bottom-up’ approaches respectively, in particular with respect to nanofabrication, although the reader may also encounter the related terms ‘subtractive’ or ‘additive’ manufacturing. Once a nanostructured surface has been patterned, some researchers use this substrate as a master pattern, which can be replicated using transfer techniques multiple times. Not all techniques fall neatly into this categorization, sometimes both top-down and bottom-up approaches are combined within one process flow (referred to as a hybrid nanofabrication approach), and more esoteric options are summarized at the end of this section, along with surface chemical functionalization approaches.
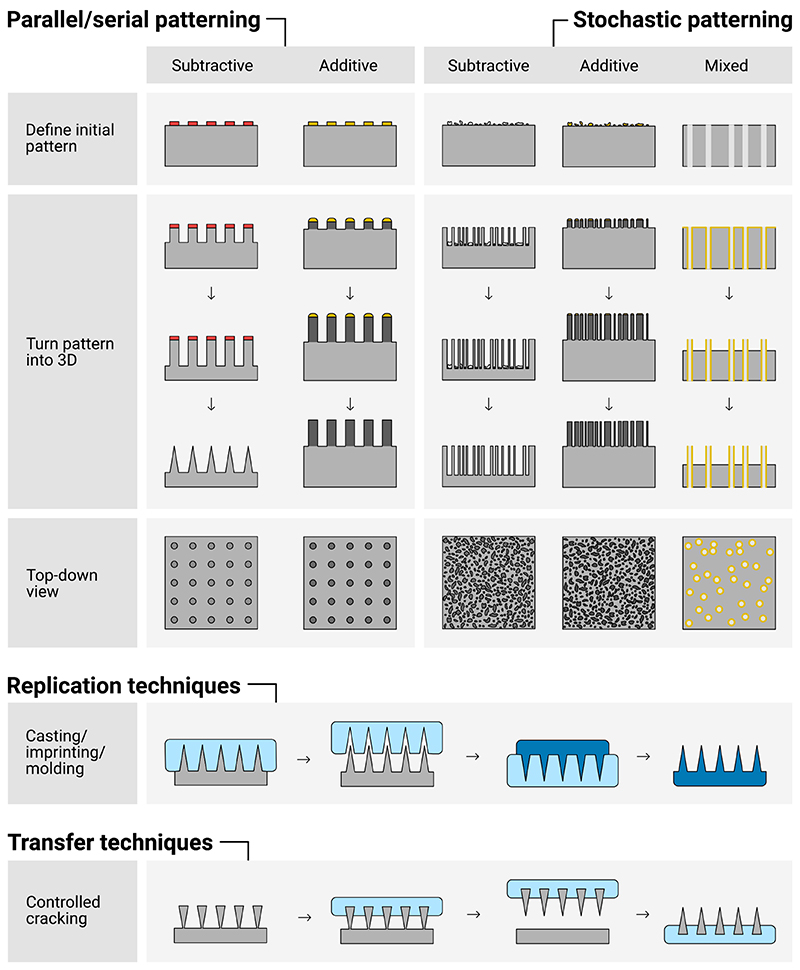
Figure 15.
Multiple fabrication approaches exist for fabricating high-aspect-ratio nanostructures. Patterns can be well-ordered, defined by parallel or serial patterning processes, or stochastically defined by semi-random deposition processes. Subtractive (also called top-down) processes remove material from the substrate, additive (also known as bottom-up) processes deposit material. Once fabricated, a number of techniques exist to replicate and transfer these structures into new materials and substrates.
4.2. Defining an Initial Pattern
Many microfabrication processes require some form of two-dimensional pattern, which is then processed into a three-dimensional structure. The location of individual nanostructures on a surface can be defined using parallel or serial (sometimes referred to as sequential) processes; or by a stochastic patterning techniques, where the pattern is defined pseudo-randomly. Parallel processes simultaneously pattern the entire surface of a substrate and include methods such as photolithography and nanoimprint lithography. Serial processes scan across a surface to define each individual feature, as seen in techniques like electron-beam lithography. In stochastic approaches the absolute position, density and distribution of the pattern is defined loosely by some physically- or chemically driven process, for example, using electroless deposition to deposit metal clusters with a distribution of sizes on the surface of a substrate.
Each approach has pros and cons; parallel processes are generally quick and allow the precise definition of pattern location but require expensive and often unmodifiable tooling. Table 1 gives an overview of the different techniques used for patterning substrates. Serial processes have much greater freedom, as the design can be specified digitally at the time of manufacture, but patterning large areas is often prohibitively slow and expensive. Stochastic processes can be rapid and affordable but lack precise control and can result in a wide distribution of feature sizes. This is potentially problematic where consistent surfaces are required for cell interactions, which some researchers have argued is essential for maximizing the reproducibility of results.[27,132]
Table 1. Microfabrication techniques used for the primary patterning of nanostructures.
Note: relevant exemplars from the literature are cited against each technique. The minimum feature size and length of patternable area are highly equipment and facility dependent, these values are derived from the either manufacturer provided specifications at the time of writing, or from the literature, where available. Techniques are sorted loosely by their prevalence within the field, with the most common listed first. Most fabrication protocols include a combination of techniques, here we are referring to the process used to define the initial pattern.
| Technique | Example applications | Minimum feature size [μm] | Diameter of patternable areaa) [mm] | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Photolithography | Nanoneedles,[26,69,133] hollow nanoneedles/ nanotubes,[134,135] nanowires,[136] | ~0.6 – 3[137] | ≥300 (typically ≥100)[137] | Good resolution | Equipment expensive |
| Parallel patterning | Tooling expensive and unmodifiable | ||||
| Well-established industry process | Complex protocols | ||||
| Sub-micron resolution challenging to achieve in many system | |||||
| Electron-beam lithography | Nanowires,[138,139] nanopillars,[31] nanostructures,[140,141] nanopits / nanopores,[142,143] nanoelectrodes,[28,66,86,144] | ~0.04 – 0.5[145] | ≥300 (in theory, but in reality individual field size ~1 1)[146] | Best resolution | Expensive equipment |
| Flexible design (no fixed tooling required) | Very slow, effectively limiting patternable area | ||||
| Complex protocols | |||||
| Limited resist choices | |||||
| Track-etched membrane / nanopore templates | Nanoelectrodes,[147] spiky microstraws,[94] nanopillar arrays,[148] nanostraws,[90,91] | ~0.1[89] | ≥100[89] | Templates are highly affordable | Limited or no control over location of individual pores |
| Large patternable areas | |||||
| No cleanroom required | |||||
| Nanoimprint lithographyb) | Nanowires,[149] nanopillars,[148,150] nanostructures.[151] | ≥0.04[153] - 25 | ≥150[153] (very large area roll-to-roll patterning reported)[151] | Parallel / quick patterning process | Requires expensive master stamp / shim |
| Good resolution | Care required to optimize resist and surface treatments to ensure good demolding | ||||
| Excellent for reproducing existing designs | |||||
| Very large area patterning possible | |||||
| Nanosphere / colloidal lithography | Nanowires,[27,154,155] nanoelectrodes,[136] nanopillars.[156] | ~0.1 – 2[157,158] | 1×103 (areas of up to 1 m2 reported)[158] | Affordable method | Challenging to align patterns to existing features |
| Achievable with relatively simple equipment | |||||
| Strong interdependence between patterned particle and spacing | |||||
| Very large area patterning possible | |||||
| Ion-beam lithographyc) | Nanoelectrodes,[62] nanoantennas,[159] nanotubes.[92] | ~0.02 – 0.5[160] | ~2.5[160] | High precision | Expensive equipment |
| Best resolution | Very slow, effectively limiting patternable area | ||||
| Interference lithography | Nanostructures;[161] nanoposts.[162] | ~0.05 – 0.5[163] | ≥200[163] | Good resolution | Limited design choices as pattern must be formed by interfering beams |
| Relatively large areas possible | |||||
| Specific tooling required | Requires relatively specialist setup | ||||
| Parallel processing | |||||
| Two-photon / multiphoton lithography | Nanopillars / ridges,[124,164] microneedles.[165] | ~0.15 – 10 (2D patterning, in 3D resolution is lower)[166] | ≥100 (individual field size ≥1)[166] | Good resolution | Expensive equipment |
| Flexible design (no fixed tooling required) | Highest resolution only possible in 2D, 3D structures more typically in micron scale | ||||
| Slow, hence limited write areas | |||||
| Electroless depositiond) | Nanowires.[167,168] | ~0.1 – 0.2[167–169] | ≥100 (limited by wafer handling for acid etching) | Highly affordable | Stochastic – limited control over pattern density and size of features |
| Achievable in chemistry lab, no cleanroom required | Challenging to align to existing features | ||||
| Deposition of particulates from gas phase (e.g. aerosol deposited nanoparticles or sputtering)d) | Nanowires,[170,171] nanoneedles.[29,81] | ~0.04 – 0.1[170,171] | ≥100 (assuming wafer-based system) | Can be performed in-situ with growth mechanisms for efficient processing | Stochastic – limited control over pattern density and size of features |
| Challenging to align to existing features | |||||
| Direct (write) laser lithography | Nanoneedles[172] | 1 – 50[173] | ≥100[173] | Flexible design (no fixed tooling required) | Compromise on resolution due to larger laser beam spot size |
| Typically easier to pattern large areas (e.g. whole wafers) compared to multiphoton approaches | Requires relatively specialist equipment | ||||
This is an estimate of the reasonable diameter over which a given technique can be used to define a pattern, assuming a circular write field;
nanoimprint lithography requires a master stamp (also known as a shim) to define the pattern being imprinted. This stamp is frequently fabricated by other techniques, such as electron-beam lithography;
this refers to using a focused-ion beam microscope to selectively mill (or deposit) a pattern of nanostructures;
these techniques, while mainly used to deposit material and turn 2D structures into 3D, can also be used to define an initial pattern through the stochastic / partial deposition of another catalytic material onto a surface, which is subsequently used as a seed for further growth.
4.2.1. Photolithography
Photolithographic approaches are well suited for patterning flat surfaces, where a large area (greater than a few millimeters squared) of high-density features is required. The surface is coated with a thin-layer of light-sensitive material (a photoresist). Ultraviolet light is projected through a mask onto the surface. The mask is typically a glass or quartz plate, coated in an opaque material such as chrome, patterned to allow light to pass through in specific locations. Exposed photoresist becomes either more or less soluble on exposure, and the soluble material is removed by washing the surface in an appropriate solvent, leaving the pattern in photoresist on the wafer.[174]
The patterning process is carried out using either a mask aligner or a stepper, both operating on similar principles. Mask aligners can achieve resolutions on the order of a few micrometers and are typically found in most research laboratories. Modern stepper systems, using complex optics and deep-ultraviolet light sources, can achieve sub-50 nm resolutions, but are not typically available or economically-feasible in many laboratories. Accordingly, most of the literature uses mask aligners, although there a handful of reports using steppers and other state-of-the-art projection systems.[134,175,176] Nagai et al. have recently illustrated the level of complexity and precision that can be achieved using high-performance lithographic techniques, by fabricating a wide-range of hollow silicon/silicon dioxide nanoneedle arrays (Figure 16).[134]
Figure 16. Scanning-electron-microscopy micrographs illustrating some of the hollow nanoneedle array geometries fabricated by Nagai et al. using i-line stepper lithography. Reproduced with permission.[134] Copyright 2019, Elsevier.
Aside from defining the initial nanostructure pattern, photolithography can also be used to pattern larger order structures, for example to selectively remove nanostructures and nanostraws from unwanted regions,[177,178] to form electrical interconnects with nanopillars,[179] or to create hybrid micro- and nanoscale architectures.[49]
Mask-less variants of photolithography exist, including interference lithography, which uses interfering laser beams to create periodic patterns on the surface of the wafer,[161] and has been used to pattern solid silicon nanoneedles for interfacing fibroblast cells.[162] Interference lithography makes it easy to rapidly pattern small features over large areas, at the expense of design flexibility.
Direct laser lithography is effectively the serial form of photolithography, where an ultraviolet laser beam is scanned over a surface to directly pattern a photoresist.[180] It offers freedom from expensive photomasks, but requires increased patterning time and typically patterns with lower resolution. This technique has been used to successfully prepare molds for casting high-aspect-ratio polymeric structures.[172] A variant is two-photon patterning (sometimes referred to as direct-write or multiphoton lithography), which irradiates the photoresist with a focused infrared laser.[181] The high photon density in the focal point results in upconverted photons with ultraviolet energies, resulting in a smaller patterning region and enhanced resolution. This approach has been used by different groups to directly pattern cell-interfacing polymer nano- and microneedles with tip-diameters in the 500 μm regime.[164,165,124]
4.2.2. Electron-Beam Lithography
Electron-beam lithography is predominantly a serial process, that scans an electron-beam across a resist coated surface. Similar to photolithography, the electron beam changes the solubility of the resist, allowing specific regions to be removed by washing the surface in a solvent. Electron-beam lithography can have significantly higher resolution (sub-50 nm) than photolithographic processes but comes at the cost of long patterning times and limited write areas. As such it provides a useful research tool, for example patterning small regions to study a limited number of cells,[140,142,182] but is not normally feasible where large culture areas are required (for example in high-throughput assays). One potential mitigation is to use electron-beam lithography to define a master stamp, which is then replicated repeatedly using imprint techniques, as discussed below.
Electron-beam lithography has also been used as a direct-write tool, to directly pattern polymeric nanoneedles and nanobars, without the need for further processing.[140] The main limitation in this approach is the penetration depth of electrons, which limits the maximum height (and hence aspect ratio) of nanostructures to around 1 μm.
4.2.3. Nanosphere and Colloidal Lithography
Nanosphere lithography (also referred to as colloidal lithography) uses the self-assembly of polymer or microgel-based nanospheres on a surface.[183,169,184,185] A wide range of variants exist, for a more in depth discussion see the review of Wang et al.[157] Controlling the type and size of particle deposited, the surface chemistry, and deposition conditions allows either well-defined high-density packing,[186] or lower-density stochastic patterns.[125]
Plasma etching and gel-swelling techniques can also be used to vary the pitch and diameter of the pattern,[155,187] which can then be transferred to the surface via multiple methods, such as ion bombardment,[188] or metal-assisted chemical etching.[143] Depositing different sizes of colloidal nanoparticle onto the same substrate allows for more complex pattern formation, a process which has been used to fabricate arrays of periodically-spaced nanowires with two different diameters.[189] Using multiple patterning steps combined allows more complex structures to be created, including hollow silicon nanotubes.[156] Another related technique is block copolymer micelle lithography, which uses the deposition of metal ion core micelles on a substrate, followed by dry etching, to define a pattern.[190,191]
Limitations include the challenge of achieving uniform, regular self-assembly of the spheres without packing defects between different regions, and a lack of design flexibility. The interaction between spheres means parameters such as pitch spacing and nanopillar diameter are interdependent.[27]
4.2.4. Nanoporous Membranes as a Template for Nanostructures
Porous membranes can be used to define an initial template for nanostructures, a technique sometimes referred to as template synthesis. These membranes can be stochastically-patterned, for example using track-etched membranes,[79,90,147] or can originate from self-organized processes such as the anodization of aluminum oxide.[192]
Track-etched membranes (sometimes referred to as ion-track membranes) are formed by irradiating a plastic film with a source of heavy-ions, resulting in the formation of pores. Geometry, orientation and density are influenced by controlling the energy and orientation of the incident ions. Melosh et al. used these membranes as the initial pattern for their nanostraw platform. By depositing 10 nm of alumina inside these pores, and then etching away the surrounding polycarbonate, they were able to form hollow nanostraws.[193] Others have combined track-etched membranes (or photoresist templates) with electrodeposition to fabricate solid nanoelectrodes.[194,195] This approach benefits from being able to readily and rapidly pattern large-areas, with the downside of a relative lack of control over nanostraw placement and distribution uniformity.
Anodized aluminum oxide membranes form a self-organized regularly-spaced pore structure.[196,197] These can be used as both etch masks, or as templates for growing metallic nanowires.[198] This approach has been used to create silicon molds for imprinting and casting nanopillars into polymers,[148,199] and to directly pattern platinum nanowires on an elastomeric substrate.[200] In a similar fashion to nanosphere lithography, this approach has the advantage of large area patterning with uniform distributions, at the expense of limited design and geometry choices.
4.3. Turning Two-Dimensional Patterns into Three-Dimensional Nanostructures
Techniques that define an initial pattern, must then be converted into a three-dimensional nanostructure using either additive (bottom-up) or subtractive (top-down) processes.
4.3.1. Wet Etching
Wet etching immerses the substrate into an acid or other liquid chemical system that attacks unprotected regions on the substrate. The protection may be from a patterned photo- or electron-beam resist, or for a metal or vapor-deposited masking layer. Depending on the etch process, wet etching can be highly isotropic and used to sharpen blunt nanopillars into sharp nanoneedles.[162] However isotropic etching is often undesirable, as it fundamentally limits the maximum achievable aspect ratio.
Where the substrate is a semiconductor such as silicon, metal-assisted chemical etching can be used for anisotropic wet etching. This process uses a patterned metal layer on the substrate surface to selectively catalyze the etch reaction, and has been widely used to create high-aspect-ratio nanostructures for cell and tissue interfacing.[26,27,69,77,125,133,201–203] Metal is deposited in unprotected regions on a surface by evaporation or an electroless-deposition technique.[26,69] When immersed in a solution of hydrofluoric acid and hydrogen peroxide, the metal catalyzes the oxidation of silicon and removes it from the surface.[26] The precise reactions are still under some discussion in the literature,[204] but the etch rate of silicon under the metal catalyst significantly exceeds that of non-metal coated regions, yielding anisotropic etching. By tailoring the etchant composition and silicon doping, the etch behavior can be varied extensively,[169,205,206] allowing the direct formation of porous silicon nanostructures,[26] and inclined silicon nanowires.[207]
If the catalytic metal layer is incomplete, for example a semi-porous layer of silver nanoclusters, then metal-assisted chemical etching can be used to directly fabricate silicon nanowires (sometimes called black silicon or silicon nanograss) where the stochastic deposition of metal clusters acts as the initial patterning step.[167,168,208] Care needs to be taken when fabricating sub-100 nm diameter nanowires as capillary forces and surface tension can cause nanowires to collapse and coalesce during wet processing steps.[169,209,210,168]
Wet etch processes like this have the advantage of generally being cheaper and simpler to implement than dry-etching approaches. Metal-assisted chemical etching can also achieve very high aspect ratios, for example 160:1,[141] which are significantly greater than most other processes. The limitations are: material choice, as metal-assisted chemical etching only works for inorganic semiconductors (silicon, gallium, etc.); the need to carefully control the etchant concentration to achieve uniform and repeatable results; and safety, as the process uses highly hazardous materials such as concentrated hydrofluoric acid.
4.3.2. Dry Etching
Dry etching uses an ionized plasma of reactive molecules to etch surfaces, for example oxygen or fluorine-containing gases such as sulfur hexafluoride. Different types of dry etching process exist, with varying degrees of isotropy. For example, oxygen plasma etching is frequently used in microfabrication processes to clean organic contaminants from surfaces, increase the surface energy to promote adhesion,[65,107,211] or to alter the size of patterned structures.[183] However, the process is highly isotropic, limiting its efficacy for fabricating high-aspect-ratio nanostructures.
Reactive-ion etching is a dry-etching variant that accelerates a plasma towards the substrate using an electric field. Ions bombarding the surface remove material from unprotected regions, and can be used to sharpen nanopillars into nanoneedles.[26,176] Thanks to the electrical bias, reactive-ion etching is more anisotropic than simple oxygen plasma etching, and is used frequently to create cell-interfacing nanostructures.[75,212,213] The process is typically limited to aspect ratios on the order of approximately 10:1.
Deep-reactive-ion etching overcomes the limitations of reactive-ion etching and allows the dry etching of nanostructures with aspect ratios typically in the region of 10:1 to 40:1,[214,48] or in extreme cases up to 100:1,[215] thanks to the use of alternating etch and passivation cycles that increase the overall anisotropy of the process. In the context of biointerfacing, deep-reactive-ion etching is most often used to fabricate solid silicon nanoneedles.[73,97,162] The benefit of the process is the ability to pattern high-aspect-ratio structures in a highly controllable manner (compared to wet-etch techniques). The limitations include the challenge of uniform etching over large areas, and the formation of scalloped vertical edges caused by the cyclical etching process. These can prevent the nanostructured substrate from being used as a master mold for other materials (e.g. polydimethylsiloxane), by preventing clean detachment.[51] Scalloped edges can be reduced by careful tuning of the etch and passivation cycle parameters, or by applying a subsequent isotropic wet or dry etching process to smooth the surface.[216] Cryogenic deep-reactive-ion etching is a variant of the etching process, which allows greater control over the vertical sidewalls of structures by cooling the substrate and modifying the composition of etch gases.[217,218] This approach avoids the scalloped edges formed by non-cryogenic deep-reactive-ion etching, although care is required to ensure the chosen resist material is not damaged by the low temperatures.[215]
Dry etching techniques have also been used to directly pattern high-aspect-ratio nanostructures without an initial patterning layer. This relies on the presence of contaminants during the etching process, which act as a stochastic mask. Diamond nanoneedles have been fabricated in this manner;[23,71,29] where molybdenum from the substrate holder sputters onto the surface during etching. Similarly, silicon nanowires (black silicon/silicon grass) can also be formed from contaminants acting as masking sites on a substrate.[130,209,219] In the case of deep-reactive-ion etching, the source of contaminants can be the incomplete removal of the passivation layer during the etch cycle.[220,221] The difficulty of using contaminants to mask the substrate is the relatively little control over the patterned structures that are formed.
4.3.3. Vapor- and Solution-Based Growth Techniques
Chemical-vapor deposition uses the reaction of chemical components in a gaseous phase to deposit solid material onto a substrate. Depending on the material being deposited, the technique can be used to selectively deposit material in well-defined regions by patterning a catalyst on the surface (for example metallic nanoparticles).[222] The process has been used to fabricate carbon nanofiber substrates for cell interfacing,[223,80,224] and vertically-aligned peptide fibers.[225] By selecting an appropriate substrate and chemical precursors,[226] more abstract geometries, such as inclined gallium nitride nanoneedles,[76] can be grown at a range of angles to the substrate surface.
Another approach is to take advantage of the vapor-liquid-solid growth mechanism.[227,228] This approach also uses the deposition of material from a vapor phase, albeit in a highly controllable manner, and can be used to fabricate high-aspect-ratio nanostructures on semiconductor surfaces.[229,230] A catalyst, typically a layer of gold nanoparticles, is patterned using photolithography, or a stochastic dewetting process.[231] The substrate is then heated under vacuum conditions to a temperature greater than the eutectic point of gold and silicon, enabling the formation of a liquid droplet of gold-semiconductor alloy. A chemical vapor containing the semiconductor (e.g. a silane) is introduced, and preferentially adsorbed by the liquid droplet. Due to a difference in melting points, the semiconductor precipitates out of the alloy at the substrate interface, resulting in the vertical growth of nanowires.[232] Examples include patterning gallium phosphide and indium arsenide nanowires for neuronal cell interfacing,[61,120,233] and silicon nanowires for a range of applications.[47,234,235] The process has also been combined with atomic layer deposition to fabricate hollow nanotubes.[236]
The benefit of vapor-liquid-solid growth is the large parameter space,[3] allowing a variety of complex geometries to be formed,[237,238] including more esoteric structures such as kinked nanowires.[239,240] However the process is generally limited to inorganic semiconductor materials and relatively high process temperatures, and the nature of the growth mechanism means that the orientation of nanowires is dependent upon the crystal orientation of the underlying substrate.
Solution-based growth methods include: electrodeposition, which has been used to deposit iridium oxide nanostructures on microelectrode arrays,[86] along with gold nanoelectrodes;[194,147] the hydrolysis of tetraethyl orthosilicate, which has been used to grow silica nanowires;[241–243] and hydrothermal synthesis.[244,245] The latter uses the combination of high-pressure and/or temperature to trigger the formation of nanostructures, including zinc nanorods,[246,247] and titanium/titanium oxide nanotopographies.[244] Direct thermal oxidation of copper has also been used to form copper oxide nanowires,[248] which have been used to study cell-nanowire interactions.[249]
4.3.4. Ion-Beam Lithography
Ion-beam lithography operates on a similar principle to electron-beam lithography but uses a focused beam of heavier ions (typically a gallium ion source) to either directly ablate or deposit nanoscale features on a surface, without the need for first defining a pattern. De Angelis et al. used a focused beam of gallium ions to mill hollow nanotubes through the back of a silicon membrane,[92,250] as well as to directly pattern nanoantennas.[159] Cui and colleagues used an additive ion-beam process to individually deposit platinum nanopillars on their electrode arrays.[62,251] The benefit is a high level of process control at the expense of throughput and patterning area.
Although rarely reported, interference techniques can also be combined with ion-beam lithography to produce very short periodicity (sub-100 nm pitch) nanostructures on surfaces,[252] which have been used to explore the influence of high-aspect-ratio nanostructures on the basal membrane of corneal epithelial cells.[253]
4.4. Techniques for Transferring Nanostructure Patterns Between Substrates
Over the past few decades multiple pattern-transfer and soft-lithography techniques have been developed.[254–256] Processes, such as nanoimprint lithography, microcontact printing and hot embossing, can be used to transfer patterns of nanostructures between different substrates. This is particularly valuable when a master template has been created using a resource-intensive process such as electron-beam lithography, because it allows the design to be replicated into multiple substrates using a faster and more affordable technique. In some cases, the transfer process itself can be harnessed to tune or taper the formed nanostructures.[199] Another benefit is that nanostructures can be replicated into polymeric or other organic materials, which are typically incompatible with the relatively aggressive microfabrication processes described above.
4.4.1. Casting
Nanostructures can be replicated using casting, where a mold is filled with a liquid that solidifies replicating the underlying pattern.[172,176] Solvent casting typically relies on the evaporation of a volatile solvent from the cast mixture,[148,257,258] or a chemical-crosslinking mechanism, such as the casting of elastomers (e.g. polydimethylsiloxane).[259,260] The simplicity and efficacy of this approach means it has been widely adopted by the research community. Examples include the fabrication of: biomimetic copies of high-aspect-ratio nanostructures found on gecko skins,[261] nanocomposite films for interfacing bacteria,[262] and polymer nanopillars for guided stem cell culture.[50,148] Solvent-cast thin layers can also be used to modify the surface chemistry of nanostructures.[263]
4.4.2. Imprinting
Nanoimprint lithography brings together a thermal- or photo-curable polymer system with a master mold (sometimes referred to as a nanoimprint shim). The polymer is pressed into the mold and cured, forming a negative imprint of the master. The imprint is either used directly or imprinted again to form a copy of the master. Nanoimprint lithography has been used to replicate both artificial and natural nanostructured surfaces,[175,264] to create nanopillars and grooves to study endocytosis,[103] cell dynamics,[265] and mechanotransduction.[67] Nanoimprint lithography can be readily upscaled using roll-to-roll manufacturing techniques, allowing replication on unprecedented (kilometer) scale.[151]
Hot embossing is conceptually similar to nanoimprint lithography and involves pressing a polymer film into a pre-patterned design (referred to as a die). This process transfers the pattern into the film.[266] Hot embossed films have been used to study the influence of nanostructures on stem cell behavior.[142,267] Similarly, injection molding is another widespread manufacturing technique that involves the high-pressure injection of a molten polymer into a cavity.[268] Although not normally considered a microfabrication technique, Stormonth-Darling et al. have shown injection molding can efficiently replicate 100-nm tip diameter polycarbonate nanopillars with very high-aspect-ratios (up to 20:1). Rasmussen et al. showed how injection-molded nanopillars could be used study stem cell differentiation.[268,269] Their work highlights how expensive electron-beam patterned masters can be combined with high-throughput manufacturing processes.
4.4.3. Limitations of Casting and Imprinting
Successful casting and imprinting relies upon good mold filling at the nanoscale (to avoid trapped air bubbles which can cause imperfections), and the careful tailoring of the mold surface chemistry to ensure clean separation of the cast or imprinted material.[184,268,6] This problem is amplified by the large interface area created by high-aspect-ratio nanostructures.[176] (Indeed the high surface area is the same mechanism found in gecko feet to create high adhesion forces.[270]) Improper demolding can lead to a wide-range of defects,[6] although elastomer deformation and swelling can be harnessed to create a wider range of nanostructure geometries.[271,199,272]
4.4.4. Transfer Processes
Vertically-aligned nanostructures can be detached from their original surface by embedding them in an elastomer (typically polydimethylsiloxane).[273,121,274,133] Mumm et al. used this approach to transfer copper oxide nanowires onto a transparent substrate to enable optical imaging of the cell – nanowire interface.[121] Fracture points can be pre-defined in silicon nanowires using multiple wet etch steps, to help ensure even nanowire height.[273] These approaches are particularly interesting as they separate the material properties of the high-aspect-ratio nanostructure from the properties of the supporting bulk material, offering greater control over macro- and nanoscopic properties.
4.5. Esoteric Fabrication Techniques
Many other nanofabrication approaches exist that do not fall neatly into the categorization presented here, including: ultraviolet-assisted capillary force lithography,[275–277] nanodrawing,[278] and indentation lithography,[279]. For an overview of these and more, see the review of Lee et al.[6] However, one final technique we will highlight here is nanoskiving, as originally proposed by Xu et al.[280] The technique involves embedding a pattern in an epoxy resin, and then cutting across the design using an ultramicrotome, before laminating the section onto a new substrate. While this approach only appears to have been applied to horizontally-aligned nanostructures (for cell sensing, guidance and tissue-engineering),[281–283] it presents considerable opportunities for generating arrays of vertically-aligned high-aspect-ratio nanostructures too.[280]
4.6. Surface Chemistry and Functionalization Techniques
4.6.1. Importance of Considering Surface Chemistry
This review considers the impact of surface topography, however it is important to note that in the context of cell interfacing, any biological or biocargo-loading effects cannot be considered in isolation from the substrate surface chemistry. Often in the literature, either substrates are used ‘as-is’ from the fabrication process, or a simple surface functionalization strategy is used to promote cellular adhesion or loading. Relatively few systematic studies of the impact of surface chemistry in combination with high-aspect-ratio topography. Here we summarize the most common surface chemistry modification strategies seen in the nanostructure literature. For those interested in plasmonic biosensing, we recommend the review of Olivero et al.,[7] which explores the wide-range of chemistries available for functionalizing planar surfaces, which could most likely be applied to the nanostructured surfaces discussed here. Similarly, for electrophysiological applications, Blau provides a good overview of microelectrode array functionalization strategies.[284] Stewart et al. also discuss the impact of surface chemistry on intracellular delivery.[11]
The choice of chemistry depends on the target application. Figure 17 gives an overview of different approaches adopted in the literature. Many begin with using oxygen plasma cleaning or acid-based piranha cleaning to introduce hydroxyl (–OH) groups onto the surface to promote physisorption or subsequent bond formation (Figure 17A). Intracellular delivery approaches often involve coating (or ‘loading’) the biocargo onto the surface prior to interfacing. Surface coatings are used to increase the amount of material that binds to the surface and are frequently applied prior to incubating the substrate in media containing the biocargo. Electrostatic bonds are preferable for transient loading, however care must be taken to ensure the biocargo is not immediately released upon contact with cell media. An alternative is to covalently bond the biocargo to the surface, however the strength of this bond can prevent detachment during interfacing.[223,80,288] Hence, covalent bonds are more suited to applications such as tethered intracellular sensing, where it is undesirable for probes to break free in the tissue or cellular environment.[69] Aside from biocargo loading, a handful of reports have also explored phospholipid coatings to promote internalization of individual nanostructures into the cell membrane (Figure 17B).[237,238]
Figure 17.
Illustration of different approaches that have been used to modify the surface-chemistry of high-aspect-ratio nanostructures. Note: surface chemistry mechanisms are often complex and multiple bindings may coexist on the same surface, for example only one mechanism is shown for silane binding in C, but more are possible.[285] Similarly the mechanism of the sulfur-gold bond in D has been of considerable discussion in the literature.[286,287] In F, the blue and bold portions of the line represent the presence of an integrin-recognized peptide-binding sequence within the overall peptide.
4.6.2. Modifying the surface using silane- and thiol-based compounds
Silanes, a collection of compounds based on different substituents of the silane molecule SiH4, are often used promote the physisorption of biocargoes onto surfaces,[289,111,69,76] or to modify the wettability of the surface.[50,290] Silanes can include other reactive groups, making silanes a common coupling agent for joining biomaterials.[291] A common choice is (3-aminopropyl)triethoxysilane (often referred to by the acronym APTES).[69,203,292] This aminosilane can covalently bond with hydroxylated silicon surfaces (those which contain dangling hydroxyl groups), forming –Si–O–Si– bonds.[285] Aminosilanes leave a free amino group (–NH2) on the surface of the silicon (Figure 17C) which can be used as a reactive handle for subsequent reactions. Hence, silanes frequently form the first step in more complex surface modification strategies where secondary components are bound to the surface.[40,71,293] Silanes are not restricted to silicon substrates and have also been used to bind aptamers to diamond nanoneedles.[71]
An alternative to silanes are thiol-based compounds, which have a sulfur-containing thiol group (R–SH, where R is often an alkyl chain), see Figure 17D. Thiols can bind to gold surfaces via a sulfur-gold bond (an approach frequently used with alkanethiols to form self-assembled monolayers),[286] or to materials such as the photoresist SU-8 via thiol-epoxide reactions.[140] Thiol-based self-assembled monolayers were used by Almquist and Melosh in their single probe penetration studies of the cell.[128] Santoro et al. used the selectivity of thiols to readily form bonds with gold (and their poorer binding to a polymeric resist), to selectively pattern regions of mushroom-like microelectrodes, allowing them to promote neuronal cell adhesion along well-defined grids.[294] Cysteine-containing peptides also feature a thiol sidechain, allowing them to bind directly to gold or maleimide functionalized surfaces.[295] This approach was adopted by Spira et al. to promote neuronal cell adhesion to gold microelectrode arrays.[296–298]
4.6.3. Polymer and protein-based coatings
Tailoring the surface chemistry to promote cell adhesion to nanostructured surfaces consists of either creating a favorable electrostatic interaction between the cell and surface, or by replicating naturally occurring binding sites. Electrostatically-charged synthetic polymers such as polylysines and polyornithines have been used to promote the adhesion of cells to diamond nanoneedles,[81] quartz nanopillars,[31,64] alumina nanostraws,[300] gold nanoelectrodes,[147] silicon nanowires,[27] and polymer nanopillars (Figure 17E).[299] Staufer et al. claimed that the cell – substrate adhesion created by coating their gold nanoelectrodes with poly-L-lysine was strong enough to cause widespread spontaneous membrane penetration.[147] Amin et al. used poly-DL-ornithine (a racemic mixture of both D and L forms of polyornithine) in combination with a selectively-patterned nanopillar array to achieve controlled guidance of 90% of primary hippocampal neurons (Figure 18).[299]
Figure 18.
Illustration of the approach used by Amin et al., combining high-aspect-ratio nanostructures with a poly-DL-ornithine coating, to engineer the adhesion and alignment of primary hippocampal neurons on surfaces. Reproduced under the terms of CC BY License.[299] Copyright 2019, The Authors.
An alternative approach is to deposit materials on the surface that mimic in vivo cellular binding sites (Figure 17F). Cells naturally secrete extracellular matrix (a dense network of molecules), which strongly influences cell-specific behavior through complex bidirectional communication.[301,263] Secreted proteins such as fibronectin contain cell-binding motifs (a particular amino acid sequence in a peptide) that strongly influence cell shape and cytoskeletal tension.[260] Artificially patterning these cell-binding motifs has been explored extensively to promote cell response on biomaterials,[302–304] and is another method to increase cell adhesion,[244] and enhance the engulfment of nanostructures by the membrane.[298,305,306] Even without deliberate coating of these materials, proteins in cell culture medium can spontaneously undergo physisorption onto surfaces during cell culture, altering the perceived surface chemistry and binding sites seen by cells.[219] Whether these materials actively or passively promote penetration is an open question; Angle et al. found that coating a range of membrane-related peptides onto single nanoneedle probes did not yield a corresponding change in the force required to manually penetrate the membrane in single-cell experiments.[102]
A few reports have compared the efficacy of both polylysine-based and binding-motif-mimicking coatings. Wrobel et al. examined the distance between the cell basal membrane and planar substrates coated with a range of chemistries. The closest average gap (35 – 40 nm) was observed with coatings of poly-D-lysine, poly-L-lysine and extracellular matrix gel.[307] They later verified this result using surface plasmon microscopy.[308] Interestingly, the authors suggest that coatings of just fibronectin or laminin (some of the main protein components of extracellular matrix) resulted in the formation of focal adhesions that locally perturbed the membrane resulting in a larger gap than the polylysine-based coatings.[307] Given the ability of high-aspect-ratio nanostructures to impact the formation of focal adhesions (discussed later in this review),[34] this further hints at the complexity of the interplay between topography and chemistry, suggesting that care is required when selecting surface coatings.
5. Characterization Approaches
The extreme geometry of high-aspect-ratio nanostructures can make it challenging to characterize cell interactions.[309] While standard optical microscopy is the primary tool, recent reports take advantage of super-resolution imaging techniques to map the distribution of membrane proteins around nanostructures. Specialized electron microscopy protocols have been developed explicitly for imaging the ultrastructure of the cell-nanostructure interface and are also discussed here, along with scanning-conductance-ion microscopy, which allows the cell interface to be imaged in an aqueous environment without fixation.
5.1. Optical Microscopy
Optical microscopy is most commonly used to characterize the cell – nanostructure interface. With appropriate staining, confocal microscopy allows the interface to be imaged in three-dimensions. Super-resolution microscopy techniques are less common, but are becoming increasingly useful to visualize the localization of subcellular components.[27,34] Chien et al. used stochastic optical reconstruction microscopy (STORM,[310,311] which uses the intermittent blinking of fluorophores to exceed normal resolution limits) to show how cells can form mature focal adhesions by deforming polymer nanopillars.[152] Structured-illumination microscopy has also been used to visualize the formation of lamin A (a structural protein) rings in the nuclear membrane, around silicon nanoneedles.[34]
5.2. Atomic-Force Microscopy
Atomic-force microscopy can struggle to map high-aspect-ratio nanostructures due to relative sizes and binding between the microscope cantilever and surfaces.[167] Sharpened high-aspect-ratio cantilevers can help overcome the geometric mismatch,[188,257] but the atomic-force microscope is more often used in this context to understand cell, rather than surface, properties. For single-cell interfacing, custom-machined nanoneedle cantilevers can be used for direct cell delivery,[40,60,98] and to study cell membrane dynamics.[128] These techniques can help verify the results seen in the large-scale parallel interfacing of cells with nanostructured surfaces.[39,312,101,26] In particular, Melosh and colleagues have systematically explored this phenomena in attempt to provide quantitative assessment of the penetration force required by single nanoneedles, in part to understand the relatively low levels of spontaneous penetration by high-aspect-ratio nanostructures.[102,128] They tested both flat-tipped (300 nm diameter) and sharp-tipped (sub-100 nm dimeter) nanoneedles (prepared from AFM-cantilever tips). They found that sharper tips required lower median penetration forces compared to flat. Single-cell force microscopy can also directly probe changes in cell stiffness in the presence of nanotopographies, important for directly measuring changes in cytoskeletal tension.[313]
5.3. Electron Microscopy
Electron microscopy provides the most unambiguous visualization of the cell – nanostructure interface. Critical point drying and other dehydration procedures allow the shape and orientation of cells seeded on nanostructures to be imaged with high resolution, although care needs to be taken to preserve intracellular structures.[130,184,4] Effective protocols facilitate high-contrast imaging while minimizing fixation and vacuum artefacts that might artificially deform structures or alter cell – nanostructure distances.[59,85,130,314] These protocols use multi-stage, heavy-metal and thin plasticization techniques to ensure clear and accurate imaging.[84,85,110,315] When combined with focused-ion-beam milling (a technique sometimes referred to as slice-and-view) the interior ultrastructure of the cell – nanostructure interface can be reconstructed in detail.[188,316,130,26,82,84,260,110] Recently, Gopal et al. have shown how this approach can also be combined with immunogold labelling (whereby antigens are labelled with antibodies conjugated to gold nanoparticles). This allows not only the cell ultrastructure to be mapped, but also the localization of targeted intracellular markers.[317]
Transmission-electron microscopy has also been used to visualize membrane – nanostructure interactions.[65,84,188] This approach facilitates the highest resolutions, with some restrictions on substrate, as inorganic materials such as silicon are challenging to section using ultramicrotomes. This limitation can be mitigated by: using polymeric substrates, which can be sectioned directly;[188] using focused-ion-beam lift-out techniques to first mill thin sections of silicon substrates before imaging;[84] embedding cells in resin, removing the underlying inorganic substrate via acid etching and replacing it with another resin layer, and then sectioning sample for imaging.[65]
A range of non-fixation-based artefacts can occur during electron microscopy, relevant to imaging high-aspect-ratio nanostructures. Wierzbicki et al. highlighted how secondary electron emission from silicon oxide can give the impression of hollow silicon nanowires, despite the structures being solid.[130] Similarly, the resource-intensive preparation protocols ultimately limits the total number of cells that can be imaged, so care must be taken when inferring the characteristics of general populations from a small number of samples. Correlative microscopy approaches, which combine optical, electron and chemical mapping modalities,[318,319] have been proposed as a way to mitigate this issue.[320] Electron tomography is another potential alternative to physical sectioning of the sample.[321,322] Specimens are imaged from multiple angles using a transmission electron microscopy-based technique. These projections are subsequently combined computationally to reconstruct a three-dimensional representation of the sample. This approach can make it easier to interpret features compared to two-dimensional projected electron micrographs, albeit with a number of practical constraints on the sample.[322]
5.4. Scanning-Ion-Conductance Microscopy
Scanning-ion-conductance microscopy attempts to overcome the limitations of fixing and staining cells for imaging, by allowing label-free topographical measurements of cells in solution.[323–326] A hollow-glass micropipette is placed in close proximity to a cell in culture, and an ionic current between pipette and reference electrode measured. The current changes as a function of distance between the pipette tip and cell; hence can be used to map the cell surface topography. Gopal et al. used this technique to demonstrate how human mesenchymal stem cells cultured on porous silicon nanoneedles show increased apical membrane ruffling, consistent with enhanced endocytosis seen on the substrates.[84] Hollow silicon oxide nanoneedle arrays have also been proposed as a multi-tip probe for scanning-ion-conductance microscopy,[327] facilitating both simultaneous imaging and intracellular delivery.[328]
6. Biochemical Delivery
Delivering molecules into cells enables intracellular sensing and control over cell behavior.[11] Delivery efficiency depends on multiple factors, including cargo and cell type, and if done poorly can induce cell death.[4] Exciting new gene-,[329] protein-,[330] and peptide-therapies,[331] have the potential to tackle complex conditions, such as inherited human diseases, but rely upon the ability to simultaneously deliver biomolecules or transfect large numbers of cells in tissues, hence the demand for delivery technologies.
The cell membrane provides an effective barrier to molecules such as nucleic acids, making delivery extremely challenging. While many nanoscale delivery methods already exist,[332] researchers are motivated to use high-aspect-ratio nanostructures to: improve transfection efficiencies;[98,103] rapidly transfect many cells in parallel;[73,80] transfect cell lines that are typically hard to modify using other techniques;[333] create a universally-applicable transfection technique, not limited to a particular cell line;[76] minimize the membrane damage seen in techniques such as microinjection,[98] and avoid the off-target effects and safety concerns associated with chemical and viral-based transfection methods.[29,288,289] Nanotopographies significantly increase the surface area available for loading molecules compared to flat substrates,[334] and the basal membrane area, which can also aid delivery.[288]
6.1. Tissue Delivery
6.1.1. Examples of Tissue Delivery
Both silicon nanoneedles and nanowires have been used to deliver plasmids into tissues. Chiappini et al. delivered a growth-factor-coding plasmid into mouse muscle tissue in vivo, resulting in more uniform blood vessel growth compared to direct injection and uncoated nanoneedles (Figure 19).[26] Kubota et al. used silicon nanowires, with sharpened gold tips, to deliver a fluorescent encoding protein in vitro and in vivo into mouse neurons in brain slices.[235] Both these examples used macroscopically rigid silicon substrates, a potential limitation when interfacing with curved tissues. Kim et al. overcame this by using an inverted fabrication process, before embedding an array of porous silicon nanoneedles into an elastomeric substrate (polydimethylsiloxane, PDMS) (Figure 20). Using their conformable nanoneedle patch, they demonstrated both cell interfacing for siRNA delivery, and intradermal interfacing in a mouse model.[133]
Figure 19.
Porous silicon nanoneedles used to nanoinject a growth-factor-encoding plasmid into mouse tissue. (A) intravital bright-field (top row, scale bar 100 μm) and confocal micrographs (bottom row, scale bar 50 μm), showing untreated (left), direct injected (center), and nanoinjected (right) human vascular endothelial growth factor-165 (hVEGF-165). The confocal images show the fluorescent signal from systemically injected fluorescently-tagged dextran, showing a greater degree of neovascularization in the nanoinjected tissue compared to the direct injection. (B + C) quantification of this behavior, both in terms of area of the fluorescent signal and number of nodes observed for different timepoints, averaged over multiple repeats. Adapted with permission.[26] Copyright 2015, Springer Nature.
Figure 20.
Kim et al.’s flexible nanoneedle patch. A: Inverted silicon nanoneedles can be embedded into an elastomer, before a controlled cracking process is used to remove them from the surface. B: Photograph of fabricated patch (scale bar 1.5 cm). C: Scanning-electron-microscopy micrograph of embedded silicon nanoneedles, scale bar 20 μm (inset scale bar 600 nm). D: Confocal laser scanning micrograph, scale bar 30 μm. Reproduced under terms of CC BY-ND license.[133] Copyright 2018, The Authors.
The clinical use of high-aspect-ratio nanostructures is often based around scenarios where the tissue is readily, or already exposed, however some have proposed using high-aspect-ratio nanostructures to facilitate gastrointestinal delivery.[335] Fox et al. incorporated alumina nanostraws onto the surface of a tablet-sized drug reservoir.[336] Their concept is an orally-administered device, which embeds in the intestinal wall to facilitate drug delivery. While still an early proof-of-concept, they found in ex vivo murine studies that the nanostraw surface improved binding to the mucus-covered intestinal wall. The benefit of this approach was that nanostraws help regulate delivery, important for minimizing the side-effects from high doses,[337] and for improving the patient experience.
6.2. Intracellular Delivery
6.2.1. Solid Nanostructure-Mediated Delivery
Cargoes can be coated onto the surface of nanostructures and then directly interfaced with cells. Nucleic acids, such as DNA or RNA, are a particularly desirable cargo, as they can directly modify gene expression.[338] These can either hijack cellular machinery to produce a particular protein, or can interfere with existing nucleic acids to suppress their activity (small interfering RNA). Chiappini et al. used biodegradable silicon nanoneedles to co-deliver enzyme-suppressing small interfering RNA and a green-fluorescent-protein-expressing plasmid into cervical cancer cells (HeLa), with transfection efficiencies of 80% and 90% respectively.[26] Similarly, Harding et al. delivered a similar plasmid into human foreskin fibroblasts (HFF), with transfection efficiencies of 75%.[339] Elnathan et al. demonstrated that the plasmid-transfection efficiency of their silicon nanowires varies as a function of nanowire height, and between four different human cell lines.[27] They observed that thinner (330-nm tip dimeter) nanowires achieved greater transfection efficiencies than larger (600 nm). As the authors note in both this report and their own review,[27,340] a precise understanding of the relationship between tip diameter and delivery efficacy remains unclear. We will discuss models and attempts to understand this behavior later in this review.
DNA can also be engineered to form three-dimensional nanocages, to be used as a delivery vehicle as opposed to transfection. Chan et al. used silicon nanoneedles to deliver peptide-coated DNA nanocages into cancer cells (HeLa).[111] Unlike with a flat control, they found nanocages delivered using nanoneedles did not colocalize with endosomes within the cell, arguing that this provides evidence that the nanoneedles had facilitated direct cytosolic access. As a proof-of-concept, they further added an organelle-targeting peptide sequence to their nanocages, and recorded colocalization with mitochondria.
Immune cells are notoriously harder to transfect than most cell lines, but a desirable target for gene therapies.[4] Shalek et al. used silicon nanowires to deliver siRNA into a range of human and non-human primary immune cells.[333] They found no adverse immune response, and subsequently delivered small interfering RNA for the gene LEF1 into human-sourced B cells from patients with chronic lymphocytic leukemia. This allowed them to identify three different patient groups, depending on the cellular response.
High-aspect-ratio nanostructures can also be used to enhance the delivery of drugs into cells. Diamond nanoneedles have been used to deliver chemotherapy drugs into human lung carcinoma cells.[23,341] Chen et al. found that mechanically impaling and incubating cells onto nanoneedles in the presence of cisplatin (a chemotherapy drug) resulted in a 30-40% drop in cell viability on the timescale of minutes.[23] The same authors also showed that suspended doxorubicin-resistant cells (MCF7/ADR) incubated and centrifuged onto nanoneedles in a doxorubicin containing media saw ~60% reduction in viability.[29] They found that while diamond-nanoneedle treatment did not damage nuclear DNA, they were able to detect an increase in reactive-oxygen-species inside cells, which they attributed in part to nanoneedle-induced depolarization of the mitochondrial membrane.
6.2.2. Nanotube-Mediated Delivery
Hollow nanostraws (or nanotubes) are an alternative to solid nanostructure surface methods. These vertically-aligned arrays of nanoscale tubes allow a suspended or soluble cargo to flow from a microfluidic reservoir directly to the tip of nanostraw – cell interface.[89,92,236,342] Without additional cell poration, nanostraw transfection efficiencies are relatively low compared to nanoneedle and nanowire-mediated approaches. However, nanostraws offer interesting capabilities, such as longitudinal intracellular extraction,[32,91,343] that are challenging to achieve with solid nanostructures. VanDersarl, Xu et al. developed alumina nanostraws, delivering plasmids into an epithelial cell line (CHO) with a transfection efficiency of 5-10% for cells situated on top of the nanostraws.[193] However, by electroporating cells cultured on nanostraws (applying a large oscillating electric field to induce pores in the cell membrane), efficiencies of 60 – 70 % have been reported.[90,300]
In the absence of electroporation, nanostraws have been combined with cell-adhesive surface chemistries to deliver membrane-impermeable azido-functionalized monosaccharides,[93] a type of biorthogonal probe that can be used to study metabolism and other processes inside the cell. Gold nanostraws have also been fabricated into a stamp-style mechanism that can be used to mechanically interface cell cultures, facilitating delivery without electroporation.[342]
Durney at al. showed that a nanostraw-style approach can also be used to fabricate tapered-cone microtube arrays, with tip-diameters of less than 500 nm (Figure 21).[79] Using centrifugation to apply an external force, they were able to deliver 10-nm diameter inorganic quantum dots into microalgae cells (Chlamydomonas reinhardtii), a type of photosynthetic eukaryotic cell with a cell wall. The ability of these nanostructures to deliver through the cell wall illustrates the applicability of high-aspect-ratio nanostructured surfaces to a wide-range of non-animal eukaryotic cells, relevant for industrial biofuel and pharmaceutical applications.[79]
Figure 21.
Scanning-electron-microscopy micrograph of a microalgae cell (Chlamydomonas reinhardtii) impaled on a hollow, tapered microtube, facilitating quantum dot delivery. Reproduced with permission.[79] Copyright 2016, American Chemical Society.
6.3. Variation in Delivery Efficacy
While there is clear evidence for nanostructure-mediated transfection, efficacies are strongly dependent on a wide-range of parameters. The variety of materials and experimental conditions make consistent comparisons across studies challenging at best. As an example, silicon nanoneedle-mediated efficiencies as low as 34% have been reported,[97] compared to the 70-80% efficiencies described above. Similarly, Tao et al. tested a range of cell lines, including human mesenchymal stem cells, monkey fibroblast-like cells (COS7), and human breast cancer cells (MCF7), and found differences in uptake rate and efficiency as a function of cell-line. The uptake of a fluorescently-tagged glucan (FITC-dextrose) was sensitive to variations in substrate topography in human mesenchymal stem cells and COS7 cells, however there was no statistically significant difference between uptake on different geometries, when tested with MCF7 cells.[103] They further noted that the rate of transfection changed as a function of timepoint. In other studies, where endocytosis is the delivery mechanism,[84] temperature is likely to impact the delivery efficiency. And this still fails to take into account possible differences due to changing the surface chemistry, as previously discussed. Given, the complexity of this parameter space, we recommend that researchers designing new experiments do so mindful of these challenges, and design in the appropriate controls (or if possible, consider systematically investigating these parameters).
6.4. Combination with Other Transfection Techniques
One method for directly increasing delivery efficiencies is to combine high-aspect-ratio nanostructured surfaces with other poration techniques. As discussed, electroporation can be used to improve the efficiency of nanostraw delivery,[90,300] however this is not the only approach. Nanostraws have also been combined with laser-based optoporation (optically-induced disruption of the cell membrane), to boost delivery efficiency.[92] The transfection efficiency of diamond nanoneedles was boosted from a few percent to 45%, by coating naked nucleic acids with a cationic liposome (Lipofectamine), a form of lipofection.[81] The coating may have mitigated the degradation of naked nucleic acids on nanostructured surfaces, which were exposed directly to the cell media.[70,81] Magnetic fields have also been used to induce local heating effects in gold-coated micropillars, increasing the uptake of membrane impermeable dyes in colon cancer cells (HCT-116).[344]
Liu et al. combined solid silicon nanoneedles with a biomechanical-energy powered triboelectric generator, which converts body movement into electrical pulses. They claim tip-field enhancement increases the effective electric field at the membrane-nanoneedle interface, increasing the uptake of a membrane-impermeable dye (propidium iodide) from an efficiency of 22% (needles without pulses) to 85%.[345] As well as various molecular weights of a fluorescently-tagged glucan (Dextran-FITC), they also delivered siRNA into human breast cancer cells (MCF-7), with a reported efficiency of 82%.
Solid silicon nanoneedles and nanowires have been combined with mechanoporation techniques, including inkjet printing cells onto nanowires,[74] and by oscillating loaded nanoneedles during interfacing to mechanically tear holes in the cell membrane.[216] Matsumoto et al. used this approach with a genetically-modified reporter cell line (Cre-Lox) to deliver Cre recombinase protein (a bacteria-derived enzyme that facilitates gene editing) into a red-fluorescent-protein expressing cell line (293.RxG, derived from HEK293). They reported Cre delivery efficiencies of up to 42%,[216] although later reported lower transfection efficiencies using a similar approach with a different cargo and cell line.[70]
7. Biochemical Sensing
The intimate interface high-aspect-ratio nanostructures form with the cell membrane enables biochemical sensing as well as delivery. Many reports describe high-aspect-ratio nanostructures as facilitating cytosolic access, as evidenced by their ability to sense known components of the intracellular chemical environment. One key benefit is the ability of each individual nanostructure to effectively act as a sensor, simultaneously probing and spatially-resolving behaviors across cell cultures or tissue sections.[33]
7.1. Intracellular Sensing
7.1.1. Probes Bound to Nanostructured Surfaces
Probes including fluorophores,[69,293] aptamers,[71] molecular beacons,[216] and peptides,[77] have all been chemically bound to the surface of nanostructures. Xie et al. used transparent silicon dioxide nanopillars to localize the fluorescence from fluorophore-tagged nanopillars, demonstrating a technique for localized, sub-diffraction limit sensing using nanopillars.[293]
Wang et al. demonstrated simultaneous stimulation and sensing by binding an aptamer (a short single-stranded DNA or RNA molecule) to diamond nanoneedles.[71] The aptamer was sensitive to a foreign-body-response biomarker (NF-κB), allowing them to simultaneously deliver foreign DNA into cancerous epithelial cells (A549), and later hippocampal neuron tissues slices, and monitor the foreign-body response. They found a reduction in the amount of captured NF-κB with interfacing time, which they concluded was due to the translocation of the transcription factor from the cytoplasm to nucleus in response to the foreign DNA.
Matsumoto et al. bound a molecular beacon (another type of oligonucleotide based probe) to silicon nanoneedles, as a way to verify they had been properly inserted into embryonic kidney cells (HEK293).[216]
Chiappini et al. described two spatially-resolved intracellular sensing techniques. By binding two pH-sensitive fluorophores to silicon nanoneedles and measuring the ratio of their respective fluorescence, the intracellular pH can be mapped across the cell, a potential indicator of cancerous or healthy cells.[69] In a separate report, a fluorescently-tagged peptide was conjugated to porous silicon nanoneedles. This peptide was cleavable by an enzyme (cathepsin B, a cysteine protease) that is normally constrained to the lysosomes of healthy cells but can be found in the cytosol of cancerous cells. This allowed the relative spatial mapping of enzyme activity in healthy and cancerous cells to be determined.[77]
7.1.2. Un-bound Probe Delivery
Most examples in the literature of un-bound probe delivery use simple dyes or fluorophores, to illustrate that a particular nanostructured surface has intracellular access.[89,29] To this end, silicon nanoneedles have also been proposed as a high-efficiency parallel delivery system for imaging probes such as quantum dots both in vitro and in vivo.[69]
7.1.3. Label-Free Raman Sensing
Raman spectroscopy irradiates a surface with laser light, before capturing and analyzing light scattered by the sample. Incident photons interact with different vibrational modes of molecules in the sample, resulting in scattered photons with slightly different energies. The technique is increasingly popular for cell and tissue analysis,[346,347] and is used to identify and classify different tissue and engineered construct regions,[319,348,349] single nanoparticle kinetics,[350] and more.[351,352]
Surface-enhanced Raman spectroscopy (often abbreviated as SERS) is a subset of Raman spectroscopy that uses nanometer-scale metal clusters to locally increase the Raman signal.[353] Light incident on a confined metallic conductor, such as a gold nanoparticle, can excite localized surface plasmons (a coupled oscillation of electrons that resonates about the conductor).[354] The result is a locally-enhanced electric field, which can intensify the Raman signal within a few tens of nanometers of the plasmon, overcoming the limitations of low signal intensity or resolution that can be encountered in non-surface enhanced approaches.[355] Surface-enhanced Raman spectroscopy relies upon roughened or vertically-structured surfaces to induce the appropriate effect,[354] making high-aspect-ratio nanostructured surfaces decorated with metallic nanoparticles well suited to this technique. This approach has been used extensively in general biosensing applications,[356–358,247,359] along with a few examples of direct cell interfacing, as described below.
De Angelis and colleagues have demonstrated a few different approaches to nanostructure enhanced Raman spectroscopy, including using gold nanoantennas to measure the Raman spectra of neuronal cultures,[159] and silver/polymer hollow nanotubes on a silicon nitride/silicon surface.[250] As well as using variants on this approach for electrical sensing (discussed below), Caprettini et al. used a hollow gold/polymer nanotube on quartz to study fibroblast cells (NIH3T3).[355] They first were able to measure extracellular Raman spectra, and after electroporating the cells intracellular spectra too. The authors cite the timescale of 10 – 20 minutes for the signal to revert from intra- to extra-cellular state, consistent with the closing of membrane pores post-electroporation. The Raman spectral intensity shifted between the different states, which they attribute to the orientation of molecular bonds in the lipid membrane. They also saw tentative evidence for nucleic acids in the cytoplasm and suggested nuclear poration may also be occurring.
In a report from the same group, Huang et al. used gold-coated nanostraws, coupled with electroporation to deliver Raman-tagged gold nanorods into fibroblast cells (NIH-3T3) with single-particle precision (Figure 22).[360] Surface-enhanced Raman scattering from the gold nanorods, plus shielding from the delivery reservoir by the gold coated nanostraw layer, means single nanorods could be visualized as a momentary increases in the Raman signal as they passed through the nanostraw. The authors propose this approach as a method for highly-controlled single particle delivery. Interestingly, they observed an absence of intracellular vesicle-related Raman signals, which they suggest means that the delivered particles are directly delivered into the cytosol, and not the endosomal system.
Figure 22.
Illustration of the single-particle intracellular delivery system proposed by Huang et al. A three-electrode system is used to both electroporate the cell surface, while providing electrophoretic control over the flow of charged gold nanorods. Raman Correlation Spectroscopy was used to track surface-enhanced Raman scattering from single gold nanorods passing through the nanostraws. Reproduced with permission.[360] Copyright 2019, American Chemical Society.
7.1.4. Label-free Electrochemical Sensing
The use of electrochemistry for biosensing is well established (see the review of Labib et al.),[8] however there are relatively few reports that have combined electrochemistry with high-aspect-ratio nanostructures for cellular interfacing. Non-planar geometries such as nanostructured electrodes increase the relative surface area available for sensing and can improve the mass transport of electroactive analytes towards the sensor surface.[33,361,362] Rawson et al. have looked at the potential to use high-aspect-ratio nanostructures for intracellular electrochemical communication.[33,363,364] They fabricated arrays of vertically aligned carbon nanofibers using plasma enhanced chemical vapor deposition and used these to interface mouse macrophage cells (RAW 264.7).[33] The cells were preincubated with an electroactive cell stain (methylene blue), and then washed before seeding and voltammetry measurements, to determine whether the carbon nanofibers had intracellular access. While centrifugation was required to facilitate intracellular access, they observed methylene blue oxidation and reduction peaks in cells that had been stained, which they cite as evidence that intracellular electrochemical sensing is possible. More recently, they have also demonstrated a reactive oxygen species sensor capable of sensing an immune response in macrophage cells to a bacterial infection within three seconds.[364] One of their key arguments for electrochemical sensing is this rapid ability to quickly sense changes in the intracellular environment.
7.2. Intracellular Extraction
Physically sampling the intracellular environment avoids the limitation of using discrete probes but comes with the challenge of effectively interfacing cells without killing them. Melosh and colleagues used their nanostraw platform to repeatedly sample cytosol from multiple cell lines for over five days.[32] At regular sampling intervals, they electroporated the cells, causing cytosolic components to diffuse into the adjacent nanostraws. By pooling samples from multiple cells, they were then able to track the expression of over 40 messenger RNA sequences from human-induced pluripotent stem cells over multiple days, with good agreement to lysed controls.
He et al. reported a similar hollow nanoneedle platform, effective at extracting protein from cells. Although not systematically investigated, their results appear to show that extraction efficiency is linked to the diameter of the hollow nanostructure.[91] Their work illustrates neatly how high-aspect-ratio nanostructured surfaces allow parallel cell interfacing, unachievable with single-cell extraction approaches.[343]
7.3. Cell and Virus Capture
The extremely high surface area of high-aspect-ratio nanostructured surfaces makes them well suited for particulate capture.[365,366] Cells can be captured onto a nanostructured surface are then subsequently released for analysis. Antibody-coated silicon nanowires, either grown stochastically on glass slides,[241] or patterned using nanosphere lithography,[367] have been used to capture circulating-tumor cells from blood samples. Porous silicon nanowires, grown inside microfluidic channels, have been used to capture an avian influenza virus (H5N2) with an efficiency of roughly 50%.[368] Others have shown that the capture efficiency of silicon nanopillars is a function of pillar diameter,[369] suggesting that geometry can be used in part to filter which objects are captured.
Kawamura et al. have demonstrated a unique form of cell capture and sorting using antibody-functionalized silicon nanoneedle arrays.[370] They interfaced cocultures of two different cell types using nanoneedle arrays. These arrays were coated with antibodies targeting particular intermediate filaments (a large family of protein-based components of the cell cytoskeleton). Different cell types produce different intermediate filaments, and the authors used this to separate the cell populations. Upon interfacing, a majority of cells with the corresponding filament bound to the needles and were subsequently separated from the other cell type, which remained on the surface. While the authors acknowledge that the efficiency and throughput of the process requires further work, they propose their approach is a feasible alternative technique to fluorescent-activated cell sorting, which often relies upon fluorescently tagging cell surface markers.
He et al. fabricated hollow microstraws with nanoscale spiked coatings and microfluidics to create a combined capture and delivery system.[94] Their microstraw platform is conceptually similar to nanostraws (albeit with a relatively low-aspect-ratio of 1:1.5), and the nanoscale spiked coating an interesting variant on this concept. By coating their structures with a circulating-tumor-cell antibody, they were able to achieve capture efficiencies of ~84%. Post-capture, a microfluidics system incorporated under the hollow microstraws allowed researchers to deliver cell permeable dyes and drugs directly into the captured cancer cells.
While high-aspect-ratio nanostructure-based cell capture systems are relatively unexplored, the recent reports of combined capture – delivery systems,[94] plus the advent of interesting photo-active capture chemistries,[371] suggest this is a growing area of research.
8. Bioelectronic Stimulation and Sensing
Nanomaterials have been proposed as a way of improving the mechanical and electrical properties of neural interfaces.[372–376] While bioelectronic interfaces are already widely used for deep-brain stimulation, pacemakers and cochlear implants,[377] existing electrodes are often physically large, have limited resolution and frequently inducing adverse physiological responses. Similarly, the patch-clamp, the current gold-standard in single-cell electrophysiology, is effective, but complex to implement for any more than a few cells at a time.[378,379] Better neural interfaces are required to understand the role of peripheral nerves on diseases such as diabetes and liver-disease,[380,381] neurological conditions such as epilepsy and Parkinson’s disease,[382] and in the development of brain-machine interfaces.[383,384]
High-aspect-ratio nanostructured electrodes reduce the contact resistance and improve the signal-to-noise ratio of planar electrodes.[62,65,385] Motivations include studying the fundamental cell electrophysiology of cardiac, neural, and skeletal-myotube cells,[66,386] as well as developing platforms for high-throughput drug screening.[95] The spatially-resolved, parallel, intimate interface of high-aspect-ratio nanostructures with cells and tissue is well suited for sensing complex neural network behaviors.[28,95,147,379,387] These benefits have been recognized commercially, with a number of micro- and nanoneedle-coated microelectrode arrays already on the market.[388,389]
8.1. Nanostructured- & High-Density-Electrode Arrays
Cui and colleagues have demonstrated how a range nanostructured electrodes, based on platinum, iridium oxide, and quartz, nanopillars and nanotubes can be used to facilitate intracellular electrical communication.[62,86] Platinum nanopillar electrodes were able to sense both extra- and intracellular potentials generated by cardiac cells (HL-1), in good agreement with patch-clamp measurements.[62] The authors demonstrated their platform by monitoring the impact of ion-channel blocking drugs on the beating behavior of the cardiac cells. Electroporation was required before intracellular signals were detected, as indicated by a significant improvement in the signal-to-noise ratio, and a shift from a bi- to monophasic waveform post-electroporation.[4] By tracking the signal intensity, researchers noted that the signal slowly reverts back to an extracellular waveform over the course of ten minutes, suggesting that the intracellular interface is transient.
Many high-aspect-ratio nanostructure fabrication approaches are based on materials and techniques inherited from the silicon electronics industry, such as complementary metal-oxide semiconductor technology (CMOS). One benefit of this microfabrication legacy, is that stimulating and sensing integrated electronics can be readily incorporated directly under each electrode, to enhance performance.[95,390,391] Very-large-scale integration (the name given to the process of integrating millions of discrete electrical components into a single silicon chip) can also be used to help rapidly upscale the number of electrodes.[392] Park and colleagues have illustrated this approach through the development of an array of 1,024 electrodes, each topped with nine titanium/platinum-coated silicon dioxide nanowires that they used to interface with cells to sense and stimulate electrical activity.[28,383,95,393] They used this platform to record network-level (collective cell behavior) signals and synapse connections in rat cortical neurons,[28] and cardiac cells.[95] Similarly, Braeken et al. fabricated an array of 16,384 individually-addressable tungsten/silicon dioxide/tin nitride sub-cellular electrodes, albeit with relatively low-aspect-ratios (~1.3:1). They used their platform to sense extra- and intracellular potentials of cardiac muscle cells (HL-1) and rat embryonic cardiac cells,[394] as well as to electroporate neuronal cells (NG108-15).[392]
There are a number of different nanoelectrode fabrication approaches that are also worth noting, including carbon-based nanofibers,[224] and carbon-nanotube-coated micropillars,[395] for the stimulation and sensing of hippocampal cells and slices. Electrodeposited gold nanoelectrodes have similarly been used for sensing and stimulating fibroblast, myotube and neuronal assemblies.[147] Gonzales et al. recently demonstrated an interesting alternative to vertically-aligned nanoelectrodes, instead fabricating horizontally-orientated, high-aspect-ratio (25:1), suspended electrodes (named nano-SPEARs), which they used to measure the electrophysiology of roundworms and other animals.[144] The lateral fabrication process is notably different to the majority of other approaches in this field, and has the potential to be laterally-scaled across relatively large distances.
While much progress has been made in this development of electrode arrays, it is important to note that fundamental issues of resolution and scalability do remain. While single nanostructures can address single cells, and single nanostructures can be individually addressed, reconciling both of these behaviors remains a significant fabrication challenge. In particular, when studying network behavior, deconvoluting the complex signal behaviors in confluent cultures of electrogenic cells remains an open area of research.[393]
8.2. Role of Electroporation in Nanoelectrode-Cell Interfacing
The use of electroporation to facilitate intracellular electrical access is common throughout the literature,[88,95,392–394] and is consistent with the use of electroporation to enhance intracellular delivery, as discussed above. Some have argued that this is problematic when studying neuronal networks,[96,393] as electroporation overly perturbs the electroanatomy of the cells under investigation,[396] and can cause damage to the nuclear membrane.[355] Indeed, Hai and Spira have proposed that electroporation on nanostructures can itself be a technique to study membrane repair dynamics.[88]
This problem is driven in part by the continuing development of equivalent circuit models that accurately describe the cell – electrode interface. A complete discussion of this ongoing debate is beyond our scope here, but we recommend the review of McGuire et al.,[4] and the works of Spira et al. for a more complete discussion.[83,385,397]
Suggested alternatives to electroporation include: using mushroom-shaped microelectrodes to promote membrane wrapping,[66] by inducing a phagocytosis-like response;[305] using surface chemistry to facilitate cell membrane penetration;[147] or by using two different physical mechanisms to both stimulate and sense. In the latter case, Dipalo et al. have proposed using their plasmonic-active gold nanopillars to optoporate cells by momentarily irradiating the cell – material interface with infrared light (Figure 23).[96] This approach allows them to continuously monitor the electrical environment via the gold nanopillar, with no interruption from electroporation. Similarly, colloidally-assembled organic semiconducting materials have been proposed as biomimetic high-aspect-ratio nanoscale interfaces with cells, which can be directly photostimulated to study ion- and temperature-gated channels.[398]
Figure 23.
Electrogenic cells are often electroporated in order to allow intracellular potentials to be sensed, however Dipalo et al. have shown plasmonic nanoelectrode-based optoporation also works. (A) Recorded voltage as a function of measurement time, for a cardiac cell seeded on nanoelectrodes, showing two sequential improvements in the signal-to-noise ratio after an increasing number of electrodes are optoporated. (B) The equivalent circuit diagram model of the cell-nanostructure interface, illustrating how sequentially optoporating nanoelectrodes reduces the junction resistance between cell and electrode, while increasing the membrane seal resistance. Adapted with permission.[96] Copyright 2017, American Chemical Society.
9. Biomechanical Cues
The mechanical cell microenvironment has long been understood to strongly influence cell behavior,[399] but continues to remain a much explored area of research.[400] Many empirical and mechanistic studies have illustrated how different cell types preferentially align to regularly-spaced nanostructures,[162,253,401] influencing cell area and spreading.[162,188,199,219,402] However, precise understanding of these mechanisms is lacking, despite their wide-ranging influence on cell behavior, including in some diseases.[403]
Inside the cell, the cytoskeleton (an interconnecting and dynamic network of protein fibers) transfers force from the membrane to nucleus. This outside-in sensing directly influences gene expression through complex biological pathways.[326] Collectively, these mechanisms are referred to as mechanotransduction, the ability of cells to translate mechanical cues into a biological response.[55,404,54] Mechanotransduction influences morphological, differential, apoptotic, and proliferative behaviors,[405] and has been linked to a range of pathologies, including asthma, cardiomyopathies, deafness and cancer.[54,405]
Proponents argue that nanotopographies are ideal for mechanically stimulating cells, because the stimulus can be maintained over long time periods.[277] Physical cues also avoid the use of potential harmful chemicals in vivo,[406] and are highly-localized (unlike chemical cues which can diffuse into surrounding tissue).[148] Nanostructured surfaces have been proposed for: generating specific-cell types in stem-cell-based therapies;[168,269] fabricating better in vitro models; and improving cell integration in tissue engineering.[406,407,244,148]
9.1. Surfaces to Guide Cell Culture and Improve Tissue Integration
In vitro cell cultures of neuronal cells,[154,185,299,408] organoids,[221] and human corneal epithelial cells,[253] can be enhanced using high-aspect-ratio nanostructures that induce more in vivo-like morphologies.[54] Kim et al. showed that varying the height of silicon nanocolumns between 0 – 2 μm influenced both neuron polarization and the length of neurite outgrowth,[154] with more elongated cells on silicon nanocolumns compared to flat control samples. They were able to control the neurite alignment further by patterning a regular grid of nanocolumns, to guide neurites along pre-defined routes (Figure 24). In tissue engineering, a range of nanostructured surfaces can improve the integration of osteochondral hip and dental implants,[301,409] help mitigate fibroblast growth in cochlear implants,[410] or influence wound healing.[411]
Figure 24.
Scanning-electron-microscopy micrographs of unpatterned (A) (scale bar 10 μm) and patterned (C) (scale bar 100 μm) arrays of silicon nanocolumns. The grid pattern provides additional guidance for neurite growth. (B) immunofluorescent micrograph showing neurons on unpatterened nanocolumns after one day in vitro, scale bar 100 μm (inset shows undifferentiated neuron on flat silicon, scale bar 20 μm). (D) Corresponding micrograph for neurons cultured on patterned silicon nanocolumn arrays after seven days in vitro, scale bar 100 μm. Adapted with permission.[154] Copyright 2017, Springer Nature.
9.2. Cellular Mechanotransduction
High-aspect-ratio nanostructured surfaces provide a striking, and often extreme mechanical cue, that can be harnessed to directly stimulate different cellular mechanisms.[263] At the cell membrane, they can modulate the ability of cells to form focal adhesions,[34,147] complex multi-protein assemblies that span the membrane and provide a physical anchor between the cell and the outer environment.[103,412,34] This effect may be particularly pertinent on substrates with nanoscale features (geometry or porosity) that are on a similar length scale to filopodia (nanoscale, environment-sensing cell protrusions) or integrin receptors (transmembrane proteins that facilitate external binding).[413,191,241,414]
Silicon nanoneedles have been shown to directly reduce the formation of focal adhesions in human mesenchymal stem cells, and hence reduce cytoskeletal tension.[34] A similar reduction in focal adhesions has also been observed in human embryonic stem cells cultured on polymer nanotopographies.[184] This relationship between focal adhesion formation and intracellular tension can indirectly alter protein-mediated (small G-proteins) mechanotransduction pathways.[132,142,188,263] Changes in cell adhesion appear to be strongly dependent on nanostructure geometry and/or cell line,[167,191] with Li et al. showing that nanowire density directly impacts the size of focal adhesions in cancer and epithelial cells, with higher densities resulting in smaller, more point-like adhesions.[170]
Correspondingly, nanostructures also strongly influence the behavior of actin (a self-assembling protein that forms cell cytoskeletal fibers). Nanopillars and nanoneedles can cause actin stress fiber formation,[415] induce alignment,[140] and the formation of actin caps and rings.[101,103,416,31] This influence has been linked to multiple mechanisms, including the efficacy of hollow nanostraws to penetrate cells for physical intracellular delivery,[58] and the development of membrane-curvature-influencing endocytosis (as discussed above).[84,99,416]
9.3. Nuclear Mechanotransduction
Experiments stimulating the nucleus through the cell membrane and on isolated nuclei,[417] show the nucleus itself is a potent mechanotransducer, converting mechanical stimuli into changes in cell behavior.[54] The nuclear membrane can be perturbed using sharp nanoneedles;[31,34,69,84,171,355,416] with the degree of perturbation strongly dependent on the density and tip dimeter of the underlying nanostructures.[31,418,416] The cytoskeleton itself couples directly to the nucleus via linking proteins,[406] and mechanical forces can induce conformational changes in nuclear proteins, impacting the organization of chromatin (a complex of DNA and RNA).[417,419]
Sharp-tipped high-aspect-ratio nanostructures can perturb the nuclear membrane directly and have been used to probe the role of different nuclear skeletal proteins. Lamins (not to be confused with laminins) are a family of proteins, sub-divided into two major classes: A-type (comprising two isoforms, lamin A and C), and B-type. They provide structure to the nucleus and are involved in the transcription of different genes. Family-dependent lamin deficiencies have been linked to pathologies such as muscular-dystrophy,[405] and changes in cell viability and mechanotransduction response.[55,420,417,54] Despite this, much about the role of lamins, or indeed nuclear mechanotransduction, is unknown.[54,263] Hansel, Crowder et al. have recently shown that the two lamin types are physically decoupled in human mesenchymal stem cells cultured on silicon nanoneedles,[34] suggesting that lamin A plays a more active role in nuclear mechanotransduction (Figure 25).
Figure 25.
Illustration from Hansel, Crowder et al., proposing the mechanism for cytoskeletal tension coupling between cellular microenvironment and the nuclear membrane for flat (A) and silicon nanoneedle (B) substrates. Cells on flat surfaces can readily form focal adhesions on strong cytoskeletal tension, activating Yes-associated protein (YAP). Nanoneedles disrupts focal adhesion formation, inhibiting cytoskeletal tension, reducing YAP activity. Simultaneously, direct perturbation of the nucleus results in the formation of lamin protein caps, and a physical decoupling of the two main types of lamin protein in the nucleus. Adapted under the terms of CC BY license.[34] Copyright 2019, The Authors.
In the same study, Hansel, Crowder et al. showed how silicon nanoneedles also influence the activity of transcription factors (such as Yes-associate protein, YAP).[34] Transcription factors control gene expression rates, and those influenced by nanoneedles are important in cellular responses and a number of pathologies.[403,421–423] Their behavior is complex, and has been linked to focal adhesion formation.[424] More generally, SanMartin et al. observed a small but statistically-significant upregulation of cell adhesion and cytoskeletal-related genes in cortical rat neuronal cells cultured on gallium phosphide nanowires,[425] further supporting the idea of a complex interplay between high-aspect-ratio nanostructures and nuclear function.
9.4. Surfaces for Inducing Differentiation
One of the key cell behaviors influenced by mechanotransduction is the differentiation of cells. Nanotopography, along with other biochemical cues, is widely understood to influence stem cell fate.[426,427,326,428,429,402,430] While some high-aspect-ratio surfaces have been used to enhance the delivery of differentiation medium (by effectively acting to permeabilize the membrane),[75] the majority of reports focus on the modulation of focal adhesion formation, which can both directly and indirectly impact differentiation.[67]
Historically in the literature, osteogenesis has been a major focus for nanostructure-driven differentiation. Low-aspect-ratio (~0.8:1) titanium-oxide nanopits have been explored extensively to stimulate osteogenic responses in human mesenchymal stem cells,[142] where differential response has been linked to the impact on cytoskeletal tension,[107,244,267,406,431–433] (for a comprehensive overview of this area, see the reviews of Dalby and colleagues).[9,10] Similar responses have subsequently been demonstrated using high-aspect-ratio titanium nanorods,[407] and polymer-based nano- and micropillars,[148,434] showing that both nanopits and nanostructures can trigger similar behaviors.
Multiple reports link differential fate to nanostructure geometry and density.[168,184,269,277] Kong et al. observed that the spacing of nanostructured surfaces influenced the regulation of differentiation-related protein in human embryonic stem cells.[184] Similarly, Ahn et al. observed that polymer nanopost density influenced the differential fate of human mesenchymal stem cells, with higher densities favoring a fat-cell (adipogenic) lineage, and lower densities favoring bone-cell (osteogenic) lineage.[277] They suggest this behavior may, in part, be due to the direct interaction of the cell mechanosensing machinery with the nanostructured surface. Lin et al. saw similar results, but using smaller, stochastically-patterned silicon nanowires (compared to Ahn et al.’s regularly spaced nanostructures) (Figure 26).[168] In their case, osteogenic fate favored shorter, stiffer bundles of nanowires, whereas longer, less stiff, less bundled nanowires favored an adipogenic fate. It is challenging to compare these results directly, due to the huge variety in material parameters, however they are overall broadly consistent with the existing literature that suggests cytoskeletal tension promotes osteogenesis.[301]
Figure 26.
Illustration from Lin et al. showing the general differentiation fates for human mesenchymal stem cells seeded on silicon nanowires with differing geometries and spring-constants. Group I: ~9 μm-high nanowires, group IV: ~26 μm-high nanowires. Note: geometry and density are convoluted, see the underlying paper for full parameters. Reproduced under the terms of CC BY license.[168] Copyright 2018, The Authors.
While most studies have explored osteogenesis, Wang et al. reported the use of silicon nanowires to promote the differentiation of neural stem cells.[210] They observed that the cells proliferated more readily on nanowires compared to flat silicon wafers, and after seven days of culture saw the formation of elongated neuron-like morphologies and an upregulation in a neural cell biomarker (Tuj-1). Rasmussen et al. explored the differentiation of human embryonic stem cells towards a definitive endoderm cell fate using polymer nanopillar arrays, surrounded by regions of flat polymer.[269] Using chemical factors, they induced differentiation, and saw a higher percentage of cells expressing Sox17 (a definitive endoderm biomarker) on nanopillar arrays compared to flat. However, when trying to differentiate the cells further (towards pancreatic endoderm cells), they found cells subsequently preferred the flat, stiffer polymer substrate, illustrating the complexity of the underlying biology.
9.5. Changes in Cell Viability and Proliferation
Reports of the viability and proliferation of cells on top of high-aspect-ratio nanostructures are mixed. Some report lower,[29,265] complete loss,[246] or no change in viability.[219,435,341,210,147] Similarly cell proliferation is either reduced,[162,415,269,171,436] unchanged,[90,26,34,84] or enhanced on different geometries.[191,265] In the absence of clear trends, it is worth highlighting how experimental design should take into account changes in proliferation and viability, as they are often correlated to the measured experimental outcomes. Changes in proliferation may not occur until sufficient culture time has passed.[437] Reduced proliferation rates have been linked to lower transfection efficiencies,[288] hence will be linked to the efficacy of high-aspect-ratio nanostructures designed for delivery.[27] Similarly, mechanotransduction, intracellular communication, and migration pathways are stimulated by the proportion of cell-to-cell contacts,[438] so studies on nanotopographies are likely to be influenced by proliferation-linked parameters, such as local cell density.[423,439]
9.6. Impact of Nanostructure Stiffness
The stiffness of individual or groups of nanostructures impacts the biomechanical stimulus seen by cells. Lee and colleagues have reported the fabrication of silicon nanowires of different lengths, and hence differing spring constants.[435,168] They have observed that actin fiber expression in the cytoskeleton was considerably lower on longer silicon nanowires, and saw corresponding changes in the regulation of integrin and focal adhesion kinase-related genes.[435] Cells spread more readily, and exhibited greater cytoskeletal tension on shorter, stiffer nanowires. In a later report, they found that the stiffness of nanowire clusters correlated well between promoting osteogenic- or adipogenic-fates, and that even subtle changes in stiffness resulted in differences in differentiation.[168] Similarly, Andolfi et al. concluded that their stochastically patterned sub-100 nm diameter silicon nanowires were too flexible to facilitate actin-stress fiber formation.[437] Polymer micropillars, patterned with a gradient of stiffness have been used to guide cell migration (the process of durotaxis).[440] Much remains unknown about the precise role of high-aspect-ratio nanostructure stiffness as biomechanical cue.
10. Biomechanical Sensing
High-aspect-ratio nanostructures can be used to directly measure biomechanical forces. These are three-dimensional variants of traction force microscopy, which typically uses flat films, embedded with fluorescent markers at regular intervals.[441,442] Deformation of the surface by adherent cells, and knowledge of the material’s mechanical properties, allows the applied force to be determined. Motivations include developing biomechanical sensors that can be used to directly spatially-map the magnitude of forces exerted by cells on their environment,[138] but avoiding the mechanical coupling between sensing sites that convolutes two-dimensional traction-force measurements.[443]
10.1. Traction Force Microscopy Using High-Aspect-Ratio Nanostructures
Individual high-aspect-ratio nanostructures can be treated like mechanical cantilevers, which can be deflected by externally applied forces. Prinz and colleagues have reported on the use of high-aspect-ratio nanostructures as biomechanical sensors,[444,445] and we recommend their recent review in this area.[5] In one of their reports, Hällström et al. fluorescently-labelled regular arrays of gallium phosphide nanowires. Using confocal microscopy they were able to dynamically track nanowire deflection and measure forces as low as 15 pN exerted by growth cones, actin protrusions of neuronal cells (mouse dorsal root ganglia).[445]
Recently, Paulitschke et al. have presented gallium arsenide nanowires to measure the cellular forces exhibited by amoeba (Dictyostelium discoideum).[138] They used inverted conical nanowires which are thinner at the base than the tip, which the authors argue facilitates very small spring-constants and hence high sensitivity, while the large smooth head reflects incident light and enables the nanowire deflection to be readily imaged (Figure 27). Other approaches have incorporated plasmonic-active gold nanoparticles into the tips of polymer micropillars for optical readout,[446] or used atomic-force-microscopy to directly probe nanowire deflection.[231]
Figure 27.
Paulitschke et al. used thin-base, gallium arsenide nanowires to measure the traction forces generated by amoeba (Dictyostelium discoideum). (A) false-colored scanning-electron-microscopy micrograph of a cell interacting with a nanowire. (B) fluorescent micrographs illustrating a top-down view of cells (green) deflecting nanowires (blue), with the degree of deflection indicated by the arrows. (C) individual nanowire deflection as a function of time, with corresponding calculated force (where possible to estimate), illustrating the ability to monitor dynamic changes in force. Adapted with permission.[138] Copyright 2019, American Chemical Society.
10.2. Challenges for Biomechanical Sensing
It is clear that understanding these forces is becoming increasingly important in understanding and modelling the interface of cells with high-aspect-ratio nanostructures.[27,265] One challenge in this area is that structuring the material surface inevitably gives it different properties to the to the bulk (potentially creating, what is in effect, a mechanical metamaterial too), which can be convoluted with the biological response. For example, Viela et al. measured the cell-induced deflection of polymer nanopillars using focused-ion-beam milled scanning-electron microscopy.[265] They observed asymmetric force distributions in migrating cells, and generally lower traction forces for cells on nano- versus microtopography, while simultaneously stimulating the biomechanical environment. As in the durotaxis example discussed above,[440] it may be challenging to deconvolute the biomechanical stimulating and sensing components of high-aspect-ratio nanostructures, but regardless these approaches can provide valuable insight.
11. Prokaryotic Cell Interfacing
While the majority of reports focus on eukaryotic cells, there is growing interest in the interaction between prokaryotic cells and high-aspect-ratio nanostructures. This has primarily emerged from the observation of the antibacterial properties of nanostructured surfaces,[447] where increasing the aspect-ratio can improve bactericidal efficacy.[448] Here, we summarize the handful of reports exploring bacterial cell mechanobiology and transformation on high-aspect-ratio nanostructures. While there are considerable differences between eukaryotic and prokaryotic cell membranes and structures, there is considerable opportunity for understanding in one field to influence the other. Perhaps most exciting of these opportunities, are recent reports that use coated-nanostructured surfaces to simultaneously encourage the osteogenesis of stem cells, while suppressing bacterial growth in coculture experiments.[244] More broadly, there are a number of uniquely prokaryotic cell applications, such as biohybrid carbon capture and photosynthetic energy generation,[449,450] that illustrate the sheer range of application areas for high-aspect-ratio nanostructures.
11.1. Antibacterial Surfaces
Antibiotic resistance is a major public health issue,[451] hence the considerable interest in the antibacterial properties of nanostructured surfaces. In the natural world this effect is wide-spread; cicada wings and gecko skin are composed of vertically-aligned micro- and nanostructures and possess antibacterial properties.[261,447,452–454] The main interest lies in the physical- rather than chemical killing mechanisms.[455–457] This is different to anti-fouling surface, which act by limiting the adhesion of bacteria.[455] Multiple attempts have been made to mimic this physical behavior using a range of nanostructured materials,[458] including polymers,[262] gold,[459] and silicon.
Black silicon (silicon structured into highly light-absorbing, random vertically-aligned nanostructures) can significantly inactivate both gram positive and gram negative bacteria and spores within tens of hours of culture.[460,461] These bactericidal properties are enhanced by coating nanostructured silicon with thin layers of metals,[462] or antibiotics,[203] or an antimicrobial enzyme (lysozyme).[463]
The mechanism for bacterial inactivation is not fully understood and multiple theories have been proposed. Some argue that as bacterial cells settle on the surface of nanostructures, the cell membrane is strained between features and spontaneously ruptures.[112,464,465] Others have suggested that membrane rupture occurs not during settling, but when cells attempt to move about on the surface. The argument is that cells are so strongly adhered to the nanostructures, they lyse themselves while trying to move.[461,466] Researchers investigating titanium-based nanostructures have suggested that rather than membrane rupture, the surface inhibits membrane remodeling after cell division. Improper remodeling leads to cell envelop collapse and hence lack of viability.[467]
In the clinical environment, nanostructured surfaces are being investigated to help reduce the risk of infection.[457] Colonies of microorganisms form biofilms, a complex extracellular matrix of polymers and proteins. Biofilms can prevent the penetration of chemicals, rendering colonies highly resistant to antibiotic treatment.[9,262,458,468] Integrating surface topography and chemical cues, by combining functional peptides with nanostructured surfaces, has been proposed as one solution to this problem.[203] As seen in eukaryotic delivery, high-surface areas also facilitate high loadings of antimicrobial agents. Studies with silicon nanowires coated in a common disinfectant (chlorhexidine digluconate) found cell-dependent effects, due to how different shaped bacteria are able to attach to the surface.[203] Osteogenic implants (such as dental or joint implants) can fail where the interface becomes infected or undergoes aseptic loosening (a lack of integration between the implant and the bone). Fraioli et al. used nanostructured titanium surfaces to encourage osteogenesis in human mesenchymal stem cells, while simultaneously acting as an antibacterial surface to a multidrug-resistant bacteria (Pseudomonas aeruginosa).[244] Similarly, black silicon has been shown to inhibit the growth of pathogenic bacteria (a live coculture of Pseudomonas aeruginosa and Staphylococcus aureus), while simultaneously supporting the proliferation of fibroblast cells (COS-7).[469]
11.2. Prokaryotic Cell Behavior and Transformation
Similar to eukaryotic cell interfacing, prokaryotic cell behavior is geometry dependent. Jeong et al. found that bacteria (Shewanella oneidensis MR-1) cultured on regularly-patterned 10-μm-spaced silicon nanowires were found to vertically-align with the nanowire, despite being significantly smaller than the array spacing (Figure 28).[470] They propose that high-aspect-ratio nanostructured surfaces provide a valuable tool for exploring the single-cell origins of biofilm formation. Similar behavior was seen using different bacterial cells (Sporomusa ovata), with the authors proposing that the local ion concentration could influence orientation.[471] While a number of studies have begun to explore the interaction between nanoscale geometry and bacterial cells,[202,472] much remains unexplored. Similar to eukaryotic techniques, Cotta and colleagues have used indium phosphide nanowire arrays to measure the piconewton adhesion forces exerted by bacterial cells (Xylella fastidiosa).[473,149]
Figure 28.
Bacterial cells (Shewanella oneidensis MR-1) show preferential attachment to silicon nanowire arrays. Cells more frequently aligning parallel to the nanowire (A and B), rather than attaching elsewhere (C and D). A and C: fluorescence micrographs, B and D: scanning-electron-microscopy micrographs, scale bars 500 μm. Adapted with permission.[470] Copyright 2013, American Chemical Society.
Efficient bacterial transformation techniques share the same appeal and motivations as the eukaryotic cell transfection approaches discussed above, albeit with far fewer reports. Yuan et al. coated silicon nanowire arrays with temperature-responsive polymer (poly(N-isopropylacrylamide)). Cycling the substrate through different temperatures changed the substrate from a hydrophobic to superhydrophobic state. The hydrophobic state promoted the adhesion of bacterial cells (Escherichia coli) to the nanowires, the superhydrophobic state caused cell detachment. By switching between the two states, the thermal shock caused the integration of plasmid DNA into the bacteria, resulting in high-efficiency transformation, and an approximately 200-fold increase in efficiency over a flat polymer-coated silicon wafer.[474]
12. Conclusions
12.1. Fundamental Challenges
The main challenge for all investigations of high-aspect-ratio nanostructured surfaces is deconvoluting the influence of geometry, material properties, surface chemistry and differing biological response. Incorporating a range of parameters, for example systematically changing geometry,[99,125,269] or testing multiple cell lines,[103] into the experimental design can help. Approaches such as image-based cell profiling can help to quantitatively analyze large numbers of cells, adding statistical weight to conclusions, as well as in identifying subpopulations and other effects driven by cell heterogeneity.[475,272,175,476,139] For example Reynolds et al. illustrated the potential of these approaches for exploring the impact of topography in endothelial/fibroblast cell cocultures, in this case with low-aspect-ratio nanodot arrays (Figure 29).[272] Likewise, super-resolution microscopy techniques are likely to continue to offer better visualization of transmembrane proteins and interaction sites. In this manner, care should be taken to avoid drawing overly generalized conclusions from results, as changes can arise rapidly from subtle changes in experimental parameters.
Figure 29.
Fluorescent micrographs of a coculture of endothelial (LE2) and fibroblast (hTERT-BJ1) cells seeded on a continuously varying nanopillar array (low-aspect-ratio, maximum height is 250 nm). Both cells are stained red for phalloidin, endothelial cells are also stained green. Cells were segmented using image-based cell profiling and used to quantitatively determine the optimal height favoring endothelial over fibroblast cells, illustrating the benefit of both systematic geometry studies and image-based cell profiling. Adapted with permission.[272] Copyright 2013, American Chemical Society.
Fabrication challenges include: the development of biodegradable and resorbable substrates for tissue interfacing;[148] or soft and conformal substrates,[133] and a particular requirement in chronic neural interfacing.[384,477] Similarly, approaches that apply high-aspect-ratio nanostructures to three-dimensional cell cultures may be useful for in vitro-based tissue and model generation.[412,478] The incorporation of nanostructures into microfluidic or other flow devices is also relatively unexplored.[368] One caveat is to ensure that new fabrication approaches are backed-up by sufficient biology. A relatively large number of orphaned papers exist, detailing a fabrication protocol, followed by an example of cell culture, but lacking any useful insight into underlying mechanisms.
The safety of using high-aspect-ratio nanostructures in vivo also remains a fundamental challenge. In general, while the nanotoxicology of particles is well established,[479] there have been relatively few studies on the safety of high-aspect-ratio nanostructures. It has long been understood that small, micron- and nano-sized particulates have the potential to interfere with proper cell function and growth. Multi-wall carbon nanotubes have been shown to cause inflammatory and fibrotic responses in rat lungs,[480] resulting in comparisons to lung damage caused by the inhalation of asbestos fibers.[481] Eriksson et al. fabricated nanowire arrays on surfaces, before deliberately detaching them and injecting the nanowire suspension into the brain of a mouse model.[482] They observed both degradation and migration of the nanowires, suggesting they could be both removed from the system but more also pass through the blood-brain barrier. In a another study, nanowires were fed to fruit flies (Drosophila melanogaster), and no adverse effects were noted.[483] The challenge is that, analogous to the issues raised above with study design, a large number of parameters (geometry, surface chemistry, surface charge, material) all strongly influence the potential risk of a material.[484,485] This is further complicated by biological effects such as the rapid binding of proteins to nanostructures in vivo that further modify nanoparticle bioactivity.[486] Some have suggested adopting high-throughput screening technologies to rapidly assess the toxicology and safety of different nano-engineered materials.[479] While issues of safety may seem distant (in particular to researchers working on more fundamental applications) practical considerations, such as the ability to insure workers using nanomaterials,[481] or a regulatory environment that permits clinical translation, have the potential to strongly impact future adoption of any technology. As stated by Stewart et al. in their recent review, more research is required.[11]
One potential advantage of the broadly surface-patterned nanostructures patterned here, is that they are typically tethered to a macroscopic surface. While freestanding silicon nanowires readily undergo cellular uptake by cells,[487] SEM studies of cells on nanostructures show their ability to strongly deform or bend nanostructures.[96,152,231] Very few reports discuss detachment of patterned nanostructures from the surface, except where by explicit design.[103] Specifically, studies of nanoneedle-mediated induced endocytosis do not see the phagocytosis of nanoneedle structures,[84] although some authors have described the engulfment of cells of peptide-functionalized-gold-mushroom-shaped nanoelectrodes by neuronal cells as phagocytosis-like.[488]
Fabricating high-aspect-ratio nanostructures from biodegradable materials serves to limit both the interfacing and exposure period. Two main approaches have been reported in the literature: the use of biodegradable porous silicon,[26] and biodegradable polymers.[148] Chiappini et al. studied the in vivo safety of using porous silicon nanoneedles to interface a mouse model, and observing no acute inflammation, blood vessel disruption nor fibrosis, up to fifteen days after interfacing.[26] The main benefit of porous silicon nanoneedles is that the silicon rapidly reacts under physiological conditions to form orthosilicic acid, which is found naturally in the human body and readily excreted.[337] It has also been suggested that porous materials reduce fibrotic response.[69,376] While cytotoxicity may be cell-line dependent,[292,489] biodegradable material systems may ultimately prove better suited to in vivo applications, compared to non-degradable inorganic materials.[78]
12.2. Open Questions
What is relationship between motility and endocytosis for cells on high-aspect-ratio nanostructures? Cell motility relies upon the internalization and redistribution of integrins via clathrin-mediated endocytosis.[490,491] Motility is directly influenced by the presence of nanostructures,[139,219] however the (probably) complex relationship between focal adhesion formation, endocytic-stimulation, and motility remains unclear.
How do membrane proteins associate and act at regions of nanostructure-induced curvature? Recent results have highlighted how high-aspect-ratio nanostructures can result in protein recruitment,[99] alter the dynamics of membrane-embedded proteins,[492] and cause complex interactions with cytoskeletal elements,[34,58] but a precise understanding of these mechanics is still lacking. Both modelling,[63,493] and experimental results,[99,300,94] hint at a complex relationship between the molecular interactions of the cell membrane with the sharp-features and porosity of high-aspect-ratio nanostructures, for example, Dabkowska et al. used supported lipid bilayers on nanowire substrates to experimentally explore this interface, their results suggesting that curvature influences protein localization.[492] Further understanding of the molecular nature of this interface would provide valuable insight.
What is the best way to measure and control the impact of high-aspect-ratio nanostructure stiffness? As discussed above, the stiffness of high-aspect-ratio nanostructures has a clear influence on the observed biological response. Simultaneously, nanopatterning materials can fundamentally alter their mechanical properties (indeed this is one approach to creating mechanical metamaterials).[14] Hence our perception of materials being hard or stiff is often inaccurate at the nanoscopic level, as illustrated by the many examples of cells deforming nanostructures made from macroscopically stiff materials.[494,131,231,152,70,96,133] Even diamond nanoneedles undergo large elastic deformations at the nanoscale.[495] While stiffness-related effects have been studied extensively,[496] and shown to influence biological response, there lacks a clear and consistent approach to measuring the impact of this behavior. It can be a practical challenge to directly measure spring constants, although stroboscopic imaging techniques can be used.[445] Nevertheless, the best approach to characterizing effective stiffness in this context remains an open question. Gadegaard and others have argued for the use of measures such as effective shear stress, to better parameterize high-aspect-ratio nanostructured surfaces.[197,269] Given the extensive impact of stiffness as a biomechanical cue, a clearer approach to deconvoluting this and the effect of geometry is needed.
How do animal eukaryotic cell interfacing behaviors compare to other cell types? High-aspect-ratio nanostructures are increasingly proposed for non-eukaryotic (or non-animal eukaryotic) cell applications in biofuels, agriculture and pharmaceuticals.[449,136,74,497,498] While prokaryotic cells such as bacteria may contain significantly different components, such as a cell wall, bacterial cytoskeletal analogues are present,[499] suggesting that at the very minimum, mutual awareness of both fields may be beneficial. Similarly, application areas focusing on mixed eukaryotic – prokaryotic interfacing would benefit from improved understanding of this interplay.[244]
What happens at the interface between the nuclear membrane and sharp high-aspect-ratio nanostructures? To date, the majority of reports have focused on the cell membrane interface. However, as recent results show,[34] systems such as silicon nanoneedles can strongly perturb the nuclear membrane. Given the close proximity of sharp-tipped nanostructures to the proteins that mediate force between the cytoskeleton and nucleus,[326] do these structures result in a greater influence on mechanotransduction transcription pathways than blunt structures? Can the nucleus, typically an order-of-magnitude stiffer than the cell,[500] also be penetrated? Such insight would help understand how best to influence behaviors such as stem cell differentiation.
Can the functionality of high-aspect-ratio nanostructured surfaces be enhanced through greater material choice and surface chemistries? The majority of articles cited here use a limited palette of materials, predominantly consisting of inorganic systems such as silicon. The casting and molding techniques described above have shown the potential for replicating high-aspect-ratio nanostructured surfaces into polymeric material systems. While these provide clear manufacturing benefits, they also enable a much wider range of organic material systems to be explored. There is considerable scope to incorporate both existing material systems from fields such as tissue engineering,[430] and also new materials from relatively nascent fields such as organic bioelectronics,[501–503] with nanostructured surfaces to create systems that actively modulate the optoelectronic and biochemical environment.
This list is of course not exhaustive but gives an impression for the scope and potential direction of research into high-aspect-ratio nanostructured surfaces.
12.3. Closing remarks
As we have detailed, high-aspect-ratio nanostructured surfaces create complex interfaces with biological systems, facilitating the study and stimulation of important biological mechanisms. With features and geometries on a comparable scale to intracellular machinery, these materials trigger dynamic biological responses far different to the bulk material behavior, which we argue allows these systems to be collectively defined as biological metamaterials, analogous to those found in other disciplines. These materials have been applied in a huge range of areas from enabling drug delivery, studying the intracellular biochemical and biomechanical environment, to enhancing our ability to measure and stimulate the bioelectronic cell environment. The flexibility in fabrication approaches gives researchers ample choice and flexibility when engineering new materials.
While the potential of high-aspect-ratio nanostructured materials has been clearly demonstrated, many exciting challenges remain. For engineers, there is considerable scope for moving beyond predominantly inorganic, semiconductor-based material systems to enable a greater range of material properties to be explored. Additionally, careful pattern control and characterization of materials is critical for enabling more systematic studies. For biologists, much remains unknown about both the membrane – nanostructure interaction, along with the impact of nanostructures on intracellular components. Answering these questions will not only further the understanding of fundamental biological mechanisms, but also power the development of compelling biomedical applications, such as new materials that facilitate high-efficiency cell reprogramming and in vivo interfacing.
Table of Contents Entry.
High-aspect-ratio nanostructured surfaces trigger a wide-range of biological responses and can be thought to act as biological metamaterials. Their nanoscale structuring is comparable with the length-scale of intracellular machinery, interacting directly with the cell membrane and cytoskeleton. They are widely-used for molecular delivery, intracellular sensing and for providing biomechanical cues. This review brings together different fields using these materials to highlight common ideas and open research questions.
Acknowledgements
S.G.H. and M.M.S. acknowledge support from the ERC Seventh Framework Programme Consolidator Grant ‘Naturale CG’ (616417). M.M.S. and M.B. acknowledge support from The Rosetrees Trust. A.B.R. acknowledges a studentship from the EPSRC CDT for the Advanced Characterisation of Materials (EP/L015277/1)”. H.S. acknowledges funding from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A6A3A03007397). S.G.H. acknowledges the many helpful discussions with Spencer Crowder and colleagues within the Stevens Group; the support of Sabrina Skeete and Ben Pierce; and Akemi Nogiwa for extensive proofreading and publishing support. While every attempt was made to locate papers, the authors apologize to any researchers whose work has inadvertently been omitted.
Biographies
 Stuart Higgins received his Ph.D. degree in Experimental Solid-State Physics from Imperial College London in 2014. He worked as a Research Associate in the Optoelectronics Group at the Cavendish Laboratory, University of Cambridge from 2015 – 2016. Since 2016 he has been a Research Associate in the Stevens Group at Imperial College London. His current research interests include the microfabrication of biomaterials for biointerfacing applications, and the development of organic semiconductor-based bioelectronic systems.
Stuart Higgins received his Ph.D. degree in Experimental Solid-State Physics from Imperial College London in 2014. He worked as a Research Associate in the Optoelectronics Group at the Cavendish Laboratory, University of Cambridge from 2015 – 2016. Since 2016 he has been a Research Associate in the Stevens Group at Imperial College London. His current research interests include the microfabrication of biomaterials for biointerfacing applications, and the development of organic semiconductor-based bioelectronic systems.
 Michele Becce is a Ph.D. student in the Stevens Group at Imperial College London and holds a BSc and a MSc in Materials Engineering from Politecnico di Milano. He has previously worked as a research intern at the Nano Carbon Materials group at the Istituto Italiano di Tecnologia (2014-2015) and as a visiting student at the Synthetic Biology Center at the Massachusetts Institute of Technology (2011-2012). His current research activities are in the field of microfabrication of devices for biointerfacing.
Michele Becce is a Ph.D. student in the Stevens Group at Imperial College London and holds a BSc and a MSc in Materials Engineering from Politecnico di Milano. He has previously worked as a research intern at the Nano Carbon Materials group at the Istituto Italiano di Tecnologia (2014-2015) and as a visiting student at the Synthetic Biology Center at the Massachusetts Institute of Technology (2011-2012). His current research activities are in the field of microfabrication of devices for biointerfacing.
 Molly Stevens is Professor of Biomedical Materials and Regenerative Medicine at Imperial College London and Research Director for Biomedical Material Sciences in the Institute of Biomedical Engineering. She is a Foreign Member of the US National Academy of Engineering and a Fellow of six UK academies, including the Royal Society of Chemistry where she is President of the Division of Materials Chemistry. Her group’s research has been recognized by over 20 major awards, including the 2016 Clemson Award for Basic Research from the Society for Biomaterials.
Molly Stevens is Professor of Biomedical Materials and Regenerative Medicine at Imperial College London and Research Director for Biomedical Material Sciences in the Institute of Biomedical Engineering. She is a Foreign Member of the US National Academy of Engineering and a Fellow of six UK academies, including the Royal Society of Chemistry where she is President of the Division of Materials Chemistry. Her group’s research has been recognized by over 20 major awards, including the 2016 Clemson Award for Basic Research from the Society for Biomaterials.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Stuart G. Higgins, Department of Bioengineering, Imperial College London, London, SW7 2AZ, UK.
Hyejeong Seong, Department of Bioengineering, Imperial College London, London, SW7 2AZ, UK.
Julia E. Sero, Department of Bioengineering, Imperial College London, London, SW7 2AZ, UK.
Molly M. Stevens, Email: m.stevens@imperial.ac.uk, Department of Materials, Imperial College London, London, SW7 2AZ, UK; Department of Bioengineering, Imperial College London, London, SW7 2AZ, UK; Institute of Biomedical Engineering, Imperial College London, London, SW7 2AZ, UK.
References
- [1].Chiappini C. ACS Sens. 2017;2:1086. doi: 10.1021/acssensors.7b00350. [DOI] [PubMed] [Google Scholar]
- [2].Prinz CN. J Phys Condens Matter. 2015;27:233103. doi: 10.1088/0953-8984/27/23/233103. [DOI] [PubMed] [Google Scholar]
- [3].Parameswaran R, Tian B. Acc Chem Res. 2018;51:1014. doi: 10.1021/acs.accounts.7b00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].McGuire AF, Santoro F, Cui B. Annu Rev Anal Chem. 2018;11:101. doi: 10.1146/annurev-anchem-061417-125705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lard M, Linke H, Prinz CN. Nanotechnology. 2019;30:214003. doi: 10.1088/1361-6528/ab0326. [DOI] [PubMed] [Google Scholar]
- [6].Lee S, Park B, Kim JS, Kim T. Nanotechnology. 2016;27:474001. doi: 10.1088/0957-4484/27/47/474001. [DOI] [PubMed] [Google Scholar]
- [7].Oliverio M, Perotto S, Messina GC, Lovato L, De Angelis F. ACS Appl Mater Interfaces. 2017;9:29394. doi: 10.1021/acsami.7b01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Labib M, Sargent EH, Kelley SO. Chem Rev. 2016;116:9001. doi: 10.1021/acs.chemrev.6b00220. [DOI] [PubMed] [Google Scholar]
- [9].Mas‐Moruno C, Su B, Dalby MJ. Adv Healthc Mater. 2019;8:1801103. doi: 10.1002/adhm.201801103. [DOI] [PubMed] [Google Scholar]
- [10].Dalby MJ, García AJ, Salmeron-Sanchez M. Nat Rev Mater. 2018;3:17091 [Google Scholar]
- [11].Stewart MP, Langer R, Jensen KF. Chem Rev. 2018;118:7409. doi: 10.1021/acs.chemrev.7b00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu Y, Zhang X. Chem Soc Rev. 2011;40:2494. doi: 10.1039/c0cs00184h. [DOI] [PubMed] [Google Scholar]
- [13].Zheludev NI, Kivshar YS. Nat Mater. 2012;11:917. doi: 10.1038/nmat3431. [DOI] [PubMed] [Google Scholar]
- [14].Lee J-H, Singer JP, Thomas EL. Adv Mater. 2012;24:4782. doi: 10.1002/adma.201201644. [DOI] [PubMed] [Google Scholar]
- [15].Zheng X, Lee H, Weisgraber TH, Shusteff M, DeOtte J, Duoss EB, Kuntz JD, Biener MM, Ge Q, Jackson JA, Kucheyev SO, et al. Science. 2014;344:1373. doi: 10.1126/science.1252291. [DOI] [PubMed] [Google Scholar]
- [16].Bertoldi K, Vitelli V, Christensen J, van Hecke M. Nat Rev Mater. 2017;2:17066 [Google Scholar]
- [17].Kolken HMA, Janbaz S, Leeflang SMA, Lietaert K, Weinans HH, Zadpoor AA. Mater Horiz. 2018;5:28. [Google Scholar]
- [18].Kapnisi M, Mansfield C, Marijon C, Guex AG, Perbellini F, Bardi I, Humphrey EJ, Puetzer JL, Mawad D, Koutsogeorgis DC, Stuckey DJ, et al. Adv Funct Mater. 2018;28:1800618. doi: 10.1002/adfm.201800618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Anderson JM, Rodriguez A, Chang DT. Semin Immunol. 2008;20:86. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kabashin AV, Evans P, Pastkovsky S, Hendren W, Wurtz GA, Atkinson R, Pollard R, Podolskiy VA, Zayats AV. Nat Mater. 2009;8:867. doi: 10.1038/nmat2546. [DOI] [PubMed] [Google Scholar]
- [21].Dipalo M, Melle G, Lovato L, Jacassi A, Santoro F, Caprettini V, Schirato A, Alabastri A, Garoli D, Bruno G, Tantussi F, et al. Nat Nanotechnol. 2018;13:965. doi: 10.1038/s41565-018-0222-z. [DOI] [PubMed] [Google Scholar]
- [22].Harding FJ, Surdo S, Delalat B, Cozzi C, Elnathan R, Gronthos S, Voelcker NH, Barillaro G. ACS Appl Mater Interfaces. 2016;8:29197. doi: 10.1021/acsami.6b07850. [DOI] [PubMed] [Google Scholar]
- [23].Chen X, Zhu G, Yang Y, Wang B, Yan L, Zhang KY, Lo KKW, Zhang W. Adv Healthc Mater. 2013;2:1103. doi: 10.1002/adhm.201200362. [DOI] [PubMed] [Google Scholar]
- [24].Kim W, Ng JK, Kunitake ME, Conklin BR, Yang P. J Am Chem Soc. 2007;129:7228. doi: 10.1021/ja071456k. [DOI] [PubMed] [Google Scholar]
- [25].Xiao F, Wen X, Chiou P-Y. 2017 IEEE 30th Int Conf Micro Electro Mech Syst MEMS; IEEE; Las Vegas, NV, USA. 2017. pp. 243–246. [Google Scholar]
- [26].Chiappini C, De Rosa E, Martinez JO, Liu X, Steele J, Stevens MM, Tasciotti E. Nat Mater. 2015;14:6. doi: 10.1038/nmat4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Elnathan R, Delalat B, Brodoceanu D, Alhmoud H, Harding FJ, Buehler K, Nelson A, Isa L, Kraus T, Voelcker NH. Adv Funct Mater. 2015;25:7215. [Google Scholar]
- [28].Robinson JT, Jorgolli M, Shalek AK, Yoon M-H, Gertner RS, Park H. Nat Nanotechnol. 2012;7:180. doi: 10.1038/nnano.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhu X, Yuen MF, Yan L, Zhang Z, Ai F, Yang Y, Yu PKN, Zhu G, Zhang W, Chen X. Adv Healthc Mater. 2016;5:1157. doi: 10.1002/adhm.201500990. [DOI] [PubMed] [Google Scholar]
- [30].Jahed Z, Zareian R, Chau YY, Seo BB, West M, Tsui TY, Wen W, Mofrad MRK. ACS Appl Mater Interfaces. 2016;8:23604. doi: 10.1021/acsami.6b08668. [DOI] [PubMed] [Google Scholar]
- [31].Hanson L, Zhao W, Lou H-Y, Lin ZC, Lee SW, Chowdary P, Cui Y, Cui B. Nat Nanotechnol. 2015;10:554. doi: 10.1038/nnano.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cao Y, Hjort M, Chen H, Birey F, Leal-Ortiz SA, Han CM, Santiago JG, Paşca SP, Wu JC, Melosh NA. Proc Natl Acad Sci. 2017;114:E1866. doi: 10.1073/pnas.1615375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rawson FJ, Cole MT, Hicks JM, Aylott JW, Milne WI, Collins CM, Jackson SK, Silman NJ, Mendes PM. Sci Rep. 2016;6:37672. doi: 10.1038/srep37672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hansel CS, Crowder SW, Cooper S, Gopal S, João Pardelha da Cruz M, de Oliveira Martins L, Keller D, Rothery S, Becce M, Cass AEG, Bakal C, et al. ACS Nano. 2019 doi: 10.1021/acsnano.8b06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee JH, Zhang A, You SS, Lieber CM. Nano Lett. 2016;16:1509. doi: 10.1021/acs.nanolett.6b00020. [DOI] [PubMed] [Google Scholar]
- [36].Zimmerman JF, Parameswaran R, Murray G, Wang Y, Burke M, Tian B. Sci Adv. 2016;2:1. doi: 10.1126/sciadv.1601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Parameswaran R, Carvalho-de-Souza JL, Jiang Y, Burke MJ, Zimmerman JF, Koehler K, Phillips AW, Yi J, Adams EJ, Bezanilla F, Tian B. Nat Nanotechnol. 2018;13:260. doi: 10.1038/s41565-017-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Safi M, Yan M, Guedeau-Boudeville M-A, Conjeaud H, Garnier-Thibaud V, Boggetto N, Baeza-Squiban A, Niedergang F, Averbeck D, Berret J-F. ACS Nano. 2011;5:5354. doi: 10.1021/nn201121e. [DOI] [PubMed] [Google Scholar]
- [39].Obataya I, Nakamura C, Han S, Nakamura N, Miyake J. Biosens Bioelectron. 2005;20:1652. doi: 10.1016/j.bios.2004.07.020. [DOI] [PubMed] [Google Scholar]
- [40].Han SW, Nakamura C, Obataya I, Nakamura N, Miyake J. Biosens Bioelectron. 2005;20:2120. doi: 10.1016/j.bios.2004.08.023. [DOI] [PubMed] [Google Scholar]
- [41].Zhang Y, Clausmeyer J, Babakinejad B, López Córdoba A, Ali T, Shevchuk A, Takahashi Y, Novak P, Edwards C, Lab M, Gopal S, et al. ACS Nano. 2016;10:3214. doi: 10.1021/acsnano.5b05211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Guillaume-Gentil O, Grindberg RVV, Kooger R, Dorwling-Carter L, Martinez V, Ossola D, Pilhofer M, Zambelli T, Vorholt JAA. Cell. 2016;166:506. doi: 10.1016/j.cell.2016.06.025. [DOI] [PubMed] [Google Scholar]
- [43].Ivanov AP, Actis P, Jönsson P, Klenerman D, Korchev Y, Edel JB. ACS Nano. 2015;9:3587. doi: 10.1021/acsnano.5b00911. [DOI] [PubMed] [Google Scholar]
- [44].Singhal R, Orynbayeva Z, Kalyana Sundaram RV, Niu JJ, Bhattacharyya S, Vitol Ea, Schrlau MG, Papazoglou ES, Friedman G, Gogotsi Y. Nat Nanotechnol. 2011;6:57. doi: 10.1038/nnano.2010.241. [DOI] [PubMed] [Google Scholar]
- [45].Ivanov AP, Actis P, Jönsson P, Klenerman D, Korchev Y, Edel JB. ACS Nano. 2015;9:3587. doi: 10.1021/acsnano.5b00911. [DOI] [PubMed] [Google Scholar]
- [46].Actis P, Maalouf MM, Kim HJ, Lohith A, Vilozny B, Seger RA, Pourmand N. ACS Nano. 2014;8:546. doi: 10.1021/nn405097u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kubota Y, Oi H, Sawahata H, Goryu A, Ando Y, Numano R, Ishida M, Kawano T. Small. 2016;12:2846. doi: 10.1002/smll.201600172. [DOI] [PubMed] [Google Scholar]
- [48].Tang Y, Sandoughsaz A, Owen KJ, Najafi K. J Microelectromechanical Syst. 2018;27:686. [Google Scholar]
- [49].Ma Z, Jiang C, Li X, Ye F, Yuan W. J Micromechanics Microengineering. 2013;23:095027 [Google Scholar]
- [50].Bucaro MA, Vasquez Y, Hatton BD, Aizenberg J. ACS Nano. 2012;6:6222. doi: 10.1021/nn301654e. [DOI] [PubMed] [Google Scholar]
- [51].Addae-Mensah KA, Retterer S, Opalenik SR, Thomas D, Lavrik NV, Wikswo JP. J Microelectromechanical Syst. 2010;19:64. doi: 10.1109/JMEMS.2009.2037440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ma G, Wu C. J Controlled Release. 2017;251:11. doi: 10.1016/j.jconrel.2017.02.011. [DOI] [PubMed] [Google Scholar]
- [53].Shende P, Sardesai M, Gaud RS. Artif Cells Nanomedicine Biotechnol. 2018;46:19. doi: 10.1080/21691401.2017.1304409. [DOI] [PubMed] [Google Scholar]
- [54].Fedorchak GR, Kaminski A, Lammerding J. Prog Biophys Mol Biol. 2014;115:76. doi: 10.1016/j.pbiomolbio.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang N, Tytell JD, Ingber DE. Nat Rev Mol Cell Biol. 2009;10:75. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- [56].Xie X, Xu AM, Angle MR, Tayebi N, Verma P, Melosh NA. Nano Lett. 2013;13:6002. doi: 10.1021/nl403201a. [DOI] [PubMed] [Google Scholar]
- [57].Xu AM, Aalipour A, Leal-Ortiz S, Mekhdjian AH, Xie X, Dunn AR, Garner CC, Melosh NA. Nat Commun. 2014;5:8. doi: 10.1038/ncomms4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Aalipour A, Xu AM, Leal-Ortiz S, Garner CC, Melosh NA. Langmuir. 2014;30:12362. doi: 10.1021/la502273f. [DOI] [PubMed] [Google Scholar]
- [59].Xie X, Aalipour A, Gupta SV, Melosh NA. ACS Nano. 2015;9:11667. doi: 10.1021/acsnano.5b05498. [DOI] [PubMed] [Google Scholar]
- [60].Kawamura R, Shimizu K, Matsumoto Y, Yamagishi A, Silberberg YR, Iijima M, Kuroda S, Fukazawa K, Ishihara K, Nakamura C. J Nanobiotechnology. 2016;14:74. doi: 10.1186/s12951-016-0226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Berthing T, Bonde S, Rostgaard KR, Madsen MH, Sørensen CB, Nygård J, Martinez KL. Nanotechnology. 2012;23:415102. doi: 10.1088/0957-4484/23/41/415102. [DOI] [PubMed] [Google Scholar]
- [62].Xie C, Lin Z, Hanson L, Cui Y, Cui B. Nat Nanotechnol. 2012;7:185. doi: 10.1038/nnano.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Capozza R, Caprettini V, Gonano CA, Bosca A, Moia F, Santoro F, De Angelis F. ACS Appl Mater Interfaces. 2018;10:29107. doi: 10.1021/acsami.8b08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dipalo M, McGuire AF, Lou H-Y, Caprettini V, Melle G, Bruno G, Lubrano C, Matino L, Li X, De Angelis F, Cui B, et al. Nano Lett. 2018;18:6100. doi: 10.1021/acs.nanolett.8b03163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hanson L, Lin ZC, Xie C, Cui Y, Cui B. Nano Lett. 2012;12:5815. doi: 10.1021/nl303163y. [DOI] [PubMed] [Google Scholar]
- [66].Santoro F, Dasgupta S, Schnitker J, Auth T, Neumann E, Panaitov G, Gompper G, Offenhäusser A. ACS Nano. 2014;8:6713. doi: 10.1021/nn500393p. [DOI] [PubMed] [Google Scholar]
- [67].Teo BKK, Wong ST, Lim CK, Kung TYS, Yap CH, Ramagopal Y, Romer LH, Yim EKF. ACS Nano. 2013;7:4785. doi: 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]
- [68].Liu D, Yi C, Wang K, Fong CC, Wang Z, Lo PK, Sun D, Yang M. ACS Appl Mater Interfaces. 2013;5:13295. doi: 10.1021/am404276r. [DOI] [PubMed] [Google Scholar]
- [69].Chiappini C, Martinez JO, De Rosa E, Almeida CS, Tasciotti E, Stevens MM. ACS Nano. 2015;9:5500. doi: 10.1021/acsnano.5b01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Matsumoto D, Yamagishi A, Saito M, Sathuluri RR, Silberberg YR, Iwata F, Kobayashi T, Nakamura C. J Biosci Bioeng. 2016;122:748. doi: 10.1016/j.jbiosc.2016.05.006. [DOI] [PubMed] [Google Scholar]
- [71].Wang Z, Yang Y, Xu Z, Wang Y, Zhang W, Shi P. Nano Lett. 2015;15:7058. doi: 10.1021/acs.nanolett.5b03126. [DOI] [PubMed] [Google Scholar]
- [72].Rawson FJ, Hicks J, Dodd N, Abate W, Garrett DJ, Yip N, Fejer G, Downard AJ, Baronian KHR, Jackson SK, Mendes PM. ACS Appl Mater Interfaces. 2015;7:23527. doi: 10.1021/acsami.5b06493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Park S, Choi S-O, Paik S, Choi S, Allen M, Prausnitz M. Biomed Microdevices. 2016;18:10. doi: 10.1007/s10544-016-0038-2. [DOI] [PubMed] [Google Scholar]
- [74].Lee D, Lee D, Won Y, Hong H, Kim Y, Song H, Pyun J-C, Cho YS, Ryu W, Moon J. Small. 2016;12:1446. doi: 10.1002/smll.201502510. [DOI] [PubMed] [Google Scholar]
- [75].Chong EYW, Ng CYP, Choi VWY, Yan L, Yang Y, Zhang WJ, Yeung KWK, Chen XF, Yu KN. J Mater Chem B. 2013;1:3390. doi: 10.1039/c3tb20114g. [DOI] [PubMed] [Google Scholar]
- [76].Kim K-H, Lee K, Hong H, Yang D, Ryu W, Nam O, Kim Y-C. J Ind Eng Chem. 2018;59:184. [Google Scholar]
- [77].Chiappini C, Campagnolo P, Almeida CS, Abbassi-Ghadi N, Chow LW, Hanna GB, Stevens MM. Adv Mater. 2015;27:5147. doi: 10.1002/adma.201501304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Chen X, Zhang W. Chem Soc Rev. 2017;46:734. doi: 10.1039/c6cs00109b. [DOI] [PubMed] [Google Scholar]
- [79].Durney AR, Frenette LC, Hodvedt EC, Krauss TD, Mukaibo H. ACS Appl Mater Interfaces. 2016;8:34198. doi: 10.1021/acsami.6b11062. [DOI] [PubMed] [Google Scholar]
- [80].McKnight TE, Melechko AV, Hensley DK, Mann DGJ, Griffin GD, Simpson ML. Nano Lett. 2004;4:1213. [Google Scholar]
- [81].Wang Y, Yang Y, Yan L, Kwok SY, Li W, Wang Z, Zhu X, Zhu G, Zhang W, Chen X, Shi P. Nat Commun. 2014;5:4466. doi: 10.1038/ncomms5466. [DOI] [PubMed] [Google Scholar]
- [82].Santoro F, Zhao W, Joubert L-M, Duan L, Schnitker J, van de Burgt Y, Lou H-Y, Liu B, Salleo A, Cui L, Cui Y, et al. ACS Nano. 2017;11:8320. doi: 10.1021/acsnano.7b03494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Spira ME, Shmoel N, Huang S-HM, Erez H. Front Neurosci. 2018;12:212. doi: 10.3389/fnins.2018.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Gopal S, Chiappini C, Penders J, Leonardo V, Seong H, Rothery S, Korchev Y, Shevchuk A, Stevens MM. Adv Mater. 2019;31:1806788. doi: 10.1002/adma.201806788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Belu A, Schnitker J, Bertazzo S, Neumann E, Mayer D, Offenhäusser A, Santoro F. J Microsc. 2016;263:78. doi: 10.1111/jmi.12378. [DOI] [PubMed] [Google Scholar]
- [86].Lin ZC, Xie C, Osakada Y, Cui Y, Cui B. Nat Commun. 2014;5:1. doi: 10.1038/ncomms4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].McMahon HT, Gallop JL. Nature. 2005;438:590. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- [88].Hai A, Spira ME. Lab Chip. 2012;12:2865. doi: 10.1039/c2lc40091j. [DOI] [PubMed] [Google Scholar]
- [89].VanDersarl JJ, Xu AM, Melosh NA. Nano Lett. 2012;12:3881. doi: 10.1021/nl204051v. [DOI] [PubMed] [Google Scholar]
- [90].Xie X, Xu AM, Leal-Ortiz S, Cao Y, Garner CC, Melosh NA. ACS Nano. 2013;7:4351. doi: 10.1021/nn400874a. [DOI] [PubMed] [Google Scholar]
- [91].He G, Yang C, Hang T, Liu D, Chen H-J, Zhang A, Lin D, Wu J, Yang B, Xie X. ACS Sens. 2018;3:1675. doi: 10.1021/acssensors.8b00367. [DOI] [PubMed] [Google Scholar]
- [92].Messina GC, Dipalo M, Rocca RL, Zilio P, Caprettini V, Zaccaria RP, Toma A, Tantussi F, Berdondini L, Angelis FD. Adv Mater. 2015;27:7145. doi: 10.1002/adma.201503252. [DOI] [PubMed] [Google Scholar]
- [93].Xu AM, Wang DS, Shieh P, Cao Y, Melosh NA. ChemBioChem. 2017;18:623. doi: 10.1002/cbic.201600689. [DOI] [PubMed] [Google Scholar]
- [94].He G, Yang C, Feng J, Wu J, Zhou L, Wen R, Huang S, Wu Q, Liu F, Chen H-J, Hang T, et al. Adv Funct Mater. 2019;29:1806484 [Google Scholar]
- [95].Abbott J, Ye T, Qin L, Jorgolli M, Gertner RS, Ham D, Park H. Nat Nanotechnol. 2017:1. doi: 10.1038/nnano.2017.3. [DOI] [PubMed] [Google Scholar]
- [96].Dipalo M, Amin H, Lovato L, Moia F, Caprettini V, Messina GC, Tantussi F, Berdondini L, De Angelis F. Nano Lett. 2017;17:3932. doi: 10.1021/acs.nanolett.7b01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Paik S-J, Park S, Zarnitsyn V, Choi S, Dong Guo X, Prausnitz MR, Allen MG. Solid-State Sens Actuators Microsyst Workshop. 2012:149. [Google Scholar]
- [98].Han S-W, Nakamura C, Kotobuki N, Obataya I, Ohgushi H, Nagamune T, Miyake J. Nanomedicine Nanotechnol Biol Med. 2008;4:215. doi: 10.1016/j.nano.2008.03.005. [DOI] [PubMed] [Google Scholar]
- [99].Zhao W, Hanson L, Lou H-Y, Akamatsu M, Chowdary PD, Santoro F, Marks JR, Grassart A, Drubin DG, Cui Y, Cui B. Nat Nanotechnol. 2017;12:750. doi: 10.1038/nnano.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kagiwada H, Nakamura C, Kihara T, Kamiishi H, Kawano K, Nakamura N, Miyake J. Cytoskeleton. 2010;67:496. doi: 10.1002/cm.20460. [DOI] [PubMed] [Google Scholar]
- [101].Kagiwada H, Nakamura C, Kihara T, Kamiishi H, Kawano K, Nakamura N, Miyake J. Cytoskeleton. 2010;67:496. doi: 10.1002/cm.20460. [DOI] [PubMed] [Google Scholar]
- [102].Angle MR, Wang A, Thomas A, Schaefer AT, Melosh NA. Biophys J. 2014;107:2091. doi: 10.1016/j.bpj.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Teo BKK, Goh S-H, Kustandi TS, Loh WW, Low HY, Yim EKF. Biomaterials. 2011;32:9866. doi: 10.1016/j.biomaterials.2011.08.088. [DOI] [PubMed] [Google Scholar]
- [104].Cooper GM. The Cell : A Molecular Approach. Sunderland, Mass. : ASM Press ; Sinauer Associates, C2000, United States; Washington, D.C: 2000. [Google Scholar]
- [105].Jarsch IK, Daste F, Gallop JL. J Cell Biol. 2016;214:375. doi: 10.1083/jcb.201604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhao W, Hanson L, Lou H-Y, Akamatsu M, Chowdary PD, Santoro F, Marks JR, Grassart A, Drubin DG, Cui Y, Cui B. Nat Nanotechnol. 2017;12:750. doi: 10.1038/nnano.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dalby MJ, Berry CC, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Exp Cell Res. 2004;295:387. doi: 10.1016/j.yexcr.2004.02.004. [DOI] [PubMed] [Google Scholar]
- [108].Moerke C, Mueller P, Nebe B. Biomaterials. 2016;76:102. doi: 10.1016/j.biomaterials.2015.10.030. [DOI] [PubMed] [Google Scholar]
- [109].Galic M, Jeong S, Tsai F-C, Joubert L-M, Wu YI, Hahn KM, Cui Y, Meyer T. Nat Cell Biol. 2012;14:874. doi: 10.1038/ncb2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Li X, Matino L, Zhang W, Klausen L, McGuire AF, Lubrano C, Zhao W, Santoro F, Cui B. Nat Protoc. 2019;14:1772. doi: 10.1038/s41596-019-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chan MS, Lo PK. Small. 2014;10:1255. doi: 10.1002/smll.201302993. [DOI] [PubMed] [Google Scholar]
- [112].Pogodin S, Hasan J, Baulin VA, Webb HK, Truong VK, Phong Nguyen TH, Boshkovikj V, Fluke CJ, Watson GS, Watson JA, Crawford RJ, et al. Biophys J. 2013;104:835. doi: 10.1016/j.bpj.2012.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Elbourne A, Chapman J, Gelmi A, Cozzolino D, Crawford RJ, Truong VK. J Colloid Interface Sci. 2019;546:192. doi: 10.1016/j.jcis.2019.03.050. [DOI] [PubMed] [Google Scholar]
- [114].Gao H. J Mech Phys Solids. 2014;62:312. [Google Scholar]
- [115].Helfrich W. Z Für Naturforschung C. 2014;28:693. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- [116].Bonde S, Berthing T, Madsen MH, Andersen TK, Buch-Månson N, Guo L, Li X, Badique F, Anselme K, Nygård J, Martinez KL. ACS Appl Mater Interfaces. 2013;5:10510. doi: 10.1021/am402070k. [DOI] [PubMed] [Google Scholar]
- [117].Buch-Månson N, Bonde S, Bolinsson J, Berthing T, Nygård J, Martinez KL. Adv Funct Mater. 2015;25:3246. [Google Scholar]
- [118].Zhou J, Zhang X, Sun J, Dang Z, Li J, Li X, Chen T. Phys Chem Chem Phys. 2018;20:22946. doi: 10.1039/c8cp03538e. [DOI] [PubMed] [Google Scholar]
- [119].Verma P, Wong IY, Melosh NA. Biointerphases. 2010;5:37. doi: 10.1116/1.3431960. [DOI] [PubMed] [Google Scholar]
- [120].Berthing T, Bonde S, Sørensen CB, Utko P, Nygård J, Martinez KL. Small. 2011;7:640. doi: 10.1002/smll.201001642. [DOI] [PubMed] [Google Scholar]
- [121].Mumm F, Beckwith KM, Bonde S, Martinez KL, Sikorski P. Small. 2013;9:263. doi: 10.1002/smll.201201314. [DOI] [PubMed] [Google Scholar]
- [122].Rostgaard KR, Frederiksen RS, Liu Y-CC, Berthing T, Madsen MH, Holm J, Nygård J, Martinez KL. Nanoscale. 2013;5:10226. doi: 10.1039/c3nr03113f. [DOI] [PubMed] [Google Scholar]
- [123].Bonde S, Buch-Månson N, Rostgaard KR, Andersen TK, Berthing T, Martinez KL. Nanotechnology. 2014;25:362001. doi: 10.1088/0957-4484/25/36/362001. [DOI] [PubMed] [Google Scholar]
- [124].Buch-Månson N, Spangenberg A, Gomez LPC, Malval J-P, Soppera O, Martinez KL. Sci Rep. 2017;7:9247. doi: 10.1038/s41598-017-09208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Buch-Månson N, Kang D-H, Kim D, Lee KE, Yoon M-H, Martinez KL. Nanoscale. 2017;9:5517. doi: 10.1039/c6nr09700f. [DOI] [PubMed] [Google Scholar]
- [126].Sackmann E, Bruinsma RF. ChemPhysChem. 2002;3:262. doi: 10.1002/1439-7641(20020315)3:3<262::AID-CPHC262>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- [127].Liu F, Wu D, Kamm RD, Chen K. Biochim Biophys Acta BBA - Biomembr. 2013;1828:1667. doi: 10.1016/j.bbamem.2013.03.011. [DOI] [PubMed] [Google Scholar]
- [128].Almquist BD, Melosh NA. Proc Natl Acad Sci. 2010;107:5815. doi: 10.1073/pnas.0909250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Almquist BD, Melosh NA. Nano Lett. 2011;11:2066. doi: 10.1021/nl200542m. [DOI] [PubMed] [Google Scholar]
- [130].Wierzbicki R, Købler C, Jensen MRB, Łopacińska J, Schmidt MS, Skolimowski M, Abeille F, Qvortrup K, Mølhave K. PLOS ONE. 2013;8:e53307. doi: 10.1371/journal.pone.0053307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Pham VTH, Truong VK, Mainwaring DE, Guo Y, Baulin VA, Kobaisi MA, Gervinskas G, Juodkazis S, Zeng WR, Doran PP, Crawford RJ, et al. J Mater Chem B. 2014;2:2819. doi: 10.1039/c4tb00239c. [DOI] [PubMed] [Google Scholar]
- [132].Dalby MJ, Gadegaard N, Oreffo ROC. Nat Mater. 2014;13:558. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- [133].Kim H, Jang H, Kim B, Kim MK, Wie DS, Lee HS, Kim DR, Lee CH. Sci Adv. 2018;4:eaau6972. doi: 10.1126/sciadv.aau6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Nagai M, Miyamoto T, Hizawa T, Shibata T. Precis Eng. 2019;55:439. [Google Scholar]
- [135].Kwon SS, Shin JH, Choi J, Nam S, Park WI. Sens Actuators B Chem. 2018;254:16. [Google Scholar]
- [136].Kim LH, Kim YJ, Hong H, Yang D, Han M, Yoo G, Song HW, Chae Y, Pyun J-C, Grossman AR, Ryu W. Adv Funct Mater. 2016;26:7679. [Google Scholar]
- [137].Süss MicroTec. 2019 [Google Scholar]
- [138].Paulitschke P, Keber F, Lebedev A, Stephan J, Lorenz H, Hasselmann S, Heinrich D, Weig EM. Nano Lett. 2019;19:2207. doi: 10.1021/acs.nanolett.8b02568. [DOI] [PubMed] [Google Scholar]
- [139].Li Z, Kamlund S, Ryser T, Lard M, Oredsson S, Prinz CN. J Mater Chem B. 2018;6:7042. doi: 10.1039/c8tb02049c. [DOI] [PubMed] [Google Scholar]
- [140].Beckwith KS, Cooil SP, Wells JW, Sikorski P. Nanoscale. 2015;7:8438. doi: 10.1039/c5nr00674k. [DOI] [PubMed] [Google Scholar]
- [141].Li H, Ye T, Shi L, Xie C. J Micromechanics Microengineering. 2017:0. [Google Scholar]
- [142].Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. Nat Mater. 2007;6:997. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- [143].Brodoceanu D, Elnathan R, Prieto-Simón B, Delalat B, Guinan T, Kroner E, Voelcker NH, Kraus T. ACS Appl Mater Interfaces. 2015;7:1160. doi: 10.1021/am506891d. [DOI] [PubMed] [Google Scholar]
- [144].Gonzales DL, Badhiwala KN, Vercosa DG, Avants BW, Liu Z, Zhong W, Robinson JT. Nat Nanotechnol. 2017;12:684. doi: 10.1038/nnano.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Vieu C, Carcenac F, Pépin A, Chen Y, Mejias M, Lebib A, Manin-Ferlazzo L, Couraud L, Launois H. Appl Surf Sci. 2000;164:111. [Google Scholar]
- [146].JEOL. JBX-9500FS Electron Beam Lithography System | Products | JEOL Ltd. can be found under https://www.jeol.co.jp/en/products/detail/JBX-9500FS.html n.d.
- [147].Staufer O, Weber S, Bengtson CP, Bading H, Rustom A, Spatz JP. Nano Lett. 2019;19:3244. doi: 10.1021/acs.nanolett.9b00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zhang S, Ma B, Liu F, Duan J, Wang S, Qiu J, Li D, Sang Y, Liu C, Liu D, Liu H. Nano Lett. 2018;18:2243. doi: 10.1021/acs.nanolett.7b04747. [DOI] [PubMed] [Google Scholar]
- [149].da Silva AM, Sahoo PK, Cavalli A, de Souza AA, Bakkers EPAM, Cesar CL, Janissen R, Cotta MA. Small Methods. 2018;2:1700411 [Google Scholar]
- [150].Ankam S, Teo BKK, Pohan G, Ho SWL, Lim CK, Yim EKF. Front Bioeng Biotechnol. 2018;6 doi: 10.3389/fbioe.2018.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Leitgeb M, Nees D, Ruttloff S, Palfinger U, Götz J, Liska R, Belegratis MR, Stadlober B. ACS Nano. 2016;10:4926. doi: 10.1021/acsnano.5b07411. [DOI] [PubMed] [Google Scholar]
- [152].Chien F-C, Dai Y-H, Kuo CW, Chen P. Nanotechnology. 2016;27:475101. doi: 10.1088/0957-4484/27/47/475101. [DOI] [PubMed] [Google Scholar]
- [153].EV Group. EVG®610 UV Nanoimprint Lithography System. can be found under https://www.evgroup.com/products/nanoimprint-lithography/uv-nil-smartnil/evg-610/ n.d.
- [154].Kim S-M, Lee S, Kim D, Kang D-H, Yang K, Cho S-W, Lee JS, Choi IS, Kang K, Yoon M-H. Nano Res. 2018;11:2532. [Google Scholar]
- [155].Rey BM, Elnathan R, Ditcovski R, Geisel K, Zanini M, Fernandez-Rodriguez MA, Naik VV, Frutiger A, Richtering W, Ellenbogen T, Voelcker NH, et al. Nano Lett. 2016;16:157. doi: 10.1021/acs.nanolett.5b03414. [DOI] [PubMed] [Google Scholar]
- [156].Xu X, Yang Q, Wattanatorn N, Zhao C, Chiang N, Jonas SJ, Weiss PS. ACS Nano. 2017;11:10384. doi: 10.1021/acsnano.7b05472. [DOI] [PubMed] [Google Scholar]
- [157].Wang Y, Zhang M, Lai Y, Chi L. Nano Today. 2018;22:36. [Google Scholar]
- [158].Gao P, He J, Zhou S, Yang X, Li S, Sheng J, Wang D, Yu T, Ye J, Cui Y. Nano Lett. 2015;15:4591. doi: 10.1021/acs.nanolett.5b01202. [DOI] [PubMed] [Google Scholar]
- [159].Dipalo M, Messina GC, Amin H, Rocca RL, Shalabaeva V, Simi A, Maccione A, Zilio P, Berdondini L, Angelis FD. Nanoscale. 2015;7:3703. doi: 10.1039/c4nr05578k. [DOI] [PubMed] [Google Scholar]
- [160].Carnegie Mellon University. FEI Nova Nanolab 600 Dual Beam FIB. can be found under https://www.cmu.edu/engineering/materials/facilities/roberts_suite/electron_microscopy_facilities/microscope/fei-nov-nanolab-600-dual-beam-fib.html n.d.
- [161].Zhu M, Zhou L, Li B, Dawood MK, Wan G, Lai CQ, Cheng H, Leong KC, Rajagopalan R, Too HP, Choi WK. Nanoscale. 2011;3:2723. doi: 10.1039/c1nr00015b. [DOI] [PubMed] [Google Scholar]
- [162].Choi C-H, Hagvall SH, Wu BM, Dunn JCY, Beygui RE, CJ Kim C-J. Biomaterials. 2007;28:1672. doi: 10.1016/j.biomaterials.2006.11.031. [DOI] [PubMed] [Google Scholar]
- [163].AMO Automatisierung Messtechnik Optik GmbH. Interference Lithography Gratings. can be found under https://www.amo.de/products-services/interference-lithography-gratings/ n.d.
- [164].Marino A, Ciofani G, Filippeschi C, Pellegrino M, Pellegrini M, Orsini P, Pasqualetti M, Mattoli V, Mazzolai B. ACS Appl Mater Interfaces. 2013;5:13012. doi: 10.1021/am403895k. [DOI] [PubMed] [Google Scholar]
- [165].Kavaldzhiev M, Perez JE, Ivanov Y, Bertoncini A, Liberale C, Kosel J. Biomed Phys Eng Express. 2017;3:025005 [Google Scholar]
- [166].Nanoscribe GmbH. Photonic Professional GT2: World’s highest resolution 3D printer. can be found under https://www.nanoscribe.com/en/solutions/photonic-professional-gt2 n.d.
- [167].Qi S, Yi C, Ji S, Fong CC, Yang M. ACS Appl Mater Interfaces. 2009;1:30. doi: 10.1021/am800027d. [DOI] [PubMed] [Google Scholar]
- [168].Lin H-I, Kuo S-W, Yen T-J, Lee OK. Sci Rep. 2018;8:12913. doi: 10.1038/s41598-018-30854-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Chiappini C, Liu X, Fakhoury JR, Ferrari M. Adv Funct Mater. 2010;20:2231. doi: 10.1002/adfm.201000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Li Z, Persson H, Adolfsson K, Oredsson S, Prinz CN. Sci China Life Sci. 2018;61:427. doi: 10.1007/s11427-017-9264-2. [DOI] [PubMed] [Google Scholar]
- [171].Persson H, Købler C, Mølhave K, Samuelson L, Tegenfeldt JO, Oredsson S, Prinz CN. Small. 2013;9:4006. doi: 10.1002/smll.201300644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Rajput D, Costa L, Lansford K, Terekhov A, Hofmeister W. ACS Appl Mater Interfaces. 2013;5:1. doi: 10.1021/am302009e. [DOI] [PubMed] [Google Scholar]
- [173].KLOE SA. Photolithography system TableTop : DILASE 250. 2018 can be found under https://www.kloe-france.com/en/laser-lithography/photolithography-systems/direct-laser-writing/dilase-250.
- [174].Mack C. Fundamental Principles of Optical Photolithography: The Science of Microfabrication. John Wiley & Sons Ltd; Chichester: 2007. [Google Scholar]
- [175].Hulshof FFB, Zhao Y, Vasilevich A, Beijer NRM, de Boer M, Papenburg BJ, van Blitterswijk C, Stamatialis D, de Boer J. Acta Biomater. 2017;62:188. doi: 10.1016/j.actbio.2017.08.023. [DOI] [PubMed] [Google Scholar]
- [176].Lee KG, Choi BG, Kim BI, Shyu T, Oh MS, Im SG, Chang S-J, Lee TJ, Kotov NA, Lee SJ. Adv Mater. 2014;26:6119. doi: 10.1002/adma.201401246. [DOI] [PubMed] [Google Scholar]
- [177].Fox CB, Cao Y, Nemeth CL, Chirra HD, Chevalier RW, Xu AM, Melosh NA, Desai TA. ACS Nano. 2016;10:5873. doi: 10.1021/acsnano.6b00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Hulshof FFB, Zhao Y, Vasilevich A, Beijer NRM, de Boer M, Papenburg BJ, van Blitterswijk C, Stamatialis D, de Boer J. Acta Biomater. 2017;62:188. doi: 10.1016/j.actbio.2017.08.023. [DOI] [PubMed] [Google Scholar]
- [179].Xie C, Lin Z, Hanson L, Cui Y, Cui B. Nat Nanotechnol. 2012;7:185. doi: 10.1038/nnano.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Blanchet P, Aubert C, Brillouet N, Coudray P. Adv Fabr Technol MicroNano Opt Photonics II. International Society For Optics And Photonics; 2009. 720513 [Google Scholar]
- [181].Selimis A, Mironov V, Farsari M. Microelectron Eng. 2015;132:83. [Google Scholar]
- [182].Biggs MJP, Fernandez M, Thomas D, Cooper R, Palma M, Liao J, Fazio T, Dahlberg C, Wheadon H, Pallipurath A, Pandit A, et al. Adv Mater. 2017:1.:1702119. doi: 10.1002/adma.201702119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Cheung CL, Nikolić RJ, Reinhardt CE, Wang TF. Nanotechnology. 2006;17:1339. [Google Scholar]
- [184].Kong YP, Tu CH, Donovan PJ, Yee AF. Acta Biomater. 2013;9:6369. doi: 10.1016/j.actbio.2013.01.036. [DOI] [PubMed] [Google Scholar]
- [185].Toma M, Belu A, Mayer D, Offenhäusser A. Small. 2017;13:1700629. doi: 10.1002/smll.201700629. [DOI] [PubMed] [Google Scholar]
- [186].Elnathan R, Delalat B, Brodoceanu D, Alhmoud H, Harding FJ, Buehler K, Nelson A, Isa L, Kraus T, Voelcker NH. Adv Funct Mater. 2015;25:7215. [Google Scholar]
- [187].Lee D, Bae J, Hong S, Yang H, Kim Y-B. Nanotechnology. 2016;27:215302. doi: 10.1088/0957-4484/27/21/215302. [DOI] [PubMed] [Google Scholar]
- [188].Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Biomaterials. 2004;25:5415. doi: 10.1016/j.biomaterials.2003.12.049. [DOI] [PubMed] [Google Scholar]
- [189].Fernández-Rodríguez MÁ, Elnathan R, Ditcovski R, Grillo F, Conley GM, Timpu F, Rauh A, Geisel K, Ellenbogen T, Grange R, Scheffold F, et al. Nanoscale. 2018;10:22189. doi: 10.1039/c8nr07059h. [DOI] [PubMed] [Google Scholar]
- [190].Lohmüller T, Aydin D, Schwieder M, Morhard C, Louban I, Pacholski C, Spatz JP. Biointerphases. 2011;6:MR1. doi: 10.1116/1.3536839. [DOI] [PubMed] [Google Scholar]
- [191].Fiedler J, Özdemir B, Bartholomä J, Plettl A, Brenner RE, Ziemann P. Biomaterials. 2013;34:8851. doi: 10.1016/j.biomaterials.2013.08.010. [DOI] [PubMed] [Google Scholar]
- [192].Roslyakov IV, Gordeeva EO, Napolskii KS. Electrochimica Acta. 2017;241:362. [Google Scholar]
- [193].Vandersarl JJ, Xu AM, Melosh NA. Nano Lett. 2012;12:3881. doi: 10.1021/nl204051v. [DOI] [PubMed] [Google Scholar]
- [194].Staufer O, Weber S, Bengtson CP, Bading H, Spatz JP, Rustom A. Beilstein J Nanotechnol. 2016;7:296. doi: 10.3762/bjnano.7.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Weidlich S, Krause KJ, Schnitker J, Wolfrum B, Offenhäusser A. Nanotechnology. 2017;28:095302. doi: 10.1088/1361-6528/aa57b5. [DOI] [PubMed] [Google Scholar]
- [196].Es-Souni M, Habouti S. Front Mater. 2014;1 doi: 10.3389/fmats.2014.00019. [DOI] [Google Scholar]
- [197].Cho Y, Minsky HK, Jiang Y, Yin K, Turner KT, Yang S. ACS Appl Mater Interfaces. 2018;10:11391. doi: 10.1021/acsami.8b02303. [DOI] [PubMed] [Google Scholar]
- [198].Fan Z, Razavi H, Do J, Moriwaki A, Ergen O, Chueh Y-L, Leu PW, Ho JC, Takahashi T, Reichertz LA, Neale S, et al. Nat Mater. 2009;8:648. doi: 10.1038/nmat2493. [DOI] [PubMed] [Google Scholar]
- [199].Hu W, Crouch AS, Miller D, Aryal M, Luebke KJ. Nanotechnology. 2010;21:385301. doi: 10.1088/0957-4484/21/38/385301. [DOI] [PubMed] [Google Scholar]
- [200].Mátéfi‐Tempfli S, Mátéfi‐Tempfli M. Adv Mater. 2009;21:4005. [Google Scholar]
- [201].Brodoceanu D, Alhmoud HZ, Elnathan R, Delalat B, Voelcker NH, Kraus T. Nanotechnology. 2016;27:075301. doi: 10.1088/0957-4484/27/7/075301. [DOI] [PubMed] [Google Scholar]
- [202].Hu H, Siu VS, Gifford SM, Kim S, Lu M, Meyer P, Stolovitzky GA. Appl Phys Lett. 2017;111:253701 [Google Scholar]
- [203].Susarrey-Arce A, Sorzabal-Bellido I, Oknianska A, McBride F, Beckett AJ, Gardeniers JGE, Raval R, Tiggelaar RM, Fernandez YAD. J Mater Chem B. 2016;4:3104. doi: 10.1039/c6tb00460a. [DOI] [PubMed] [Google Scholar]
- [204].Huang Z, Geyer N, Werner P, De Boor J, Gösele U. Adv Mater. 2011;23:285. doi: 10.1002/adma.201001784. [DOI] [PubMed] [Google Scholar]
- [205].Lin H, Cheung H-Y, Xiu F, Wang F, Yip S, Han N, Hung T, Zhou J, Ho JC, Wong C-Y. J Mater Chem A. 2013;1:9942. [Google Scholar]
- [206].Singh N, Sahoo MK, Kale PG. J Cryst Growth. 2018;496-497:10. [Google Scholar]
- [207].Chen C-Y, Wu C-S, Chou C-J, Yen T-J. Adv Mater. 2008;20:3811. [Google Scholar]
- [208].Fang H, Wu Y, Zhao J, Zhu J. Nanotechnology. 2006;17:3768. [Google Scholar]
- [209].Schmidt MS, Hübner J, Boisen A. Adv Mater. 2012;24:OP11. doi: 10.1002/adma.201103496. [DOI] [PubMed] [Google Scholar]
- [210].Yan Q, Fang L, Wei J, Xiao G, Lv M, Ma Q, Liu C, Wang W. J Biomater Sci Polym Ed. 2017;28:1394. doi: 10.1080/09205063.2017.1329888. [DOI] [PubMed] [Google Scholar]
- [211].Velcheti V, Pennell NA. Ann Transl Med. 2017;5:378. doi: 10.21037/atm.2017.08.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Hanson L, Zhao W, Lou H-Y, Lin ZC, Lee SW, Chowdary P, Cui Y, Cui B. Nat Nanotechnol. 2015;10:554. doi: 10.1038/nnano.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Ji J, Tay FEH, Miao J, Iliescu C. J Micromechanics Microengineering. 2006;16:958. [Google Scholar]
- [214].Liu C, Tang J, Chen HM, Liu B, Yang P. Nano Lett. 2013;13:2989. doi: 10.1021/nl401615t. [DOI] [PubMed] [Google Scholar]
- [215].Zeniou A, Ellinas K, Olziersky A, Gogolides E. Nanotechnology. 2013;25:035302. doi: 10.1088/0957-4484/25/3/035302. [DOI] [PubMed] [Google Scholar]
- [216].Matsumoto D, Rao Sathuluri R, Kato Y, Silberberg YR, Kawamura R, Iwata F, Kobayashi T, Nakamura C. Sci Rep. 2015;5:15325. doi: 10.1038/srep15325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Tachi S, Tsujimoto K, Okudaira S. Appl Phys Lett. 1988;52:616. [Google Scholar]
- [218].de Boer MJ, Gardeniers JGE, Jansen HV, Smulders E, Gilde M, Roelofs G, Sasserath JN, Elwenspoek M. J Microelectromechanical Syst. 2002;11:385. [Google Scholar]
- [219].Łopacińska JM, Grădinaru C, Wierzbicki R, Købler C, Schmidt MS, Madsen MT, Skolimowski M, Dufva M, Flyvbjerg H, Mølhave K. Nanoscale. 2012;4:3739. doi: 10.1039/c2nr11455k. [DOI] [PubMed] [Google Scholar]
- [220].Jansen H, de Boer M, Legtenberg R, Elwenspoek M. J Micromechanics Microengineering. 1995;5:115. [Google Scholar]
- [221].Shen Y, Hou Y, Yao S, Huang P, Yobas L. Sci Rep. 2015;5:9293. doi: 10.1038/srep09293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Hu J, Odom TW, Lieber CM. Acc Chem Res. 1999;32:435. [Google Scholar]
- [223].McKnight TE, Melechko AV, Griffin GD, Guillorn MA, Merkulov VI, Serna F, Hensley DK, Doktycz MJ, Lowndes DH, Simpson ML. Nanotechnology. 2003;14:551. [Google Scholar]
- [224].Yu Z, McKnight TE, Ericson MN, Melechko AV, Simpson ML, Morrison B. Nano Lett. 2007;7:2188. doi: 10.1021/nl070291a. [DOI] [PubMed] [Google Scholar]
- [225].Vasudev MC, Koerner H, Singh KM, Partlow BP, Kaplan DL, Gazit E, Bunning TJ, Naik RR. Biomacromolecules. 2014;15:533. doi: 10.1021/bm401491k. [DOI] [PubMed] [Google Scholar]
- [226].Lee K, Chae S, Jang J, Min D, Kim J, Eom D, Yoo Y-S, Cho Y-H, Nam O. Nanotechnology. 2015;26:335601. doi: 10.1088/0957-4484/26/33/335601. [DOI] [PubMed] [Google Scholar]
- [227].Wagner RS, Ellis WC. Appl Phys Lett. 2004:89. APLCLASS2019. [Google Scholar]
- [228].Morales AM, Lieber CM. Science. 1998;279:208. doi: 10.1126/science.279.5348.208. [DOI] [PubMed] [Google Scholar]
- [229].Hochbaum AI, Fan R, He R, Yang P. Nano Lett. 2005;5:457. doi: 10.1021/nl047990x. [DOI] [PubMed] [Google Scholar]
- [230].Pinion CW, Christesen JD, Cahoon JF. J Mater Chem C. 2016;4:3890. [Google Scholar]
- [231].Park Y-S, Yoon SY, Park JS, Lee JS. NPG Asia Mater. 2016;8:e249 [Google Scholar]
- [232].Schmidt V, Wittemann JV, Gösele U. Chem Rev. 2010;110:361. doi: 10.1021/cr900141g. [DOI] [PubMed] [Google Scholar]
- [233].Hällström W, Mårtensson T, Prinz C, Gustavsson P, Montelius L, Samuelson L, Kanje M. Nano Lett. 2007;7:2960. doi: 10.1021/nl070728e. [DOI] [PubMed] [Google Scholar]
- [234].Kim W, Ng JK, Kunitake ME, Conklin BR, Yang P. J Am Chem Soc. 2007;129:7228. doi: 10.1021/ja071456k. [DOI] [PubMed] [Google Scholar]
- [235].Kubota Y, Numano R, Goryu A, Sawahata H, Yamagiwa S, Matsuo M, Ishida M, Kawano T. 2018 IEEE Micro Electro Mech Syst MEMS. IEEE, Belfast; 2018. pp. 436–439. [Google Scholar]
- [236].Persson H, Beech JP, Samuelson L, Oredsson S, Prinz CN, Tegenfeldt JO. Nano Res. 2012;5:190. [Google Scholar]
- [237].Duan X, Gao R, Xie P, Cohen-Karni T, Qing Q, Choe HS, Tian B, Jiang X, Lieber CM. Nat Nanotechnol. 2012;7:174. doi: 10.1038/nnano.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Tian B, Cohen-Karni T, Qing Q, Duan X, Xie P, Lieber CM. Science. 2010;329:830. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Zimmerman JF, Murray GF, Wang Y, Jumper JM, Austin JR, Tian B. Nano Lett. 2015;15:5492. doi: 10.1021/acs.nanolett.5b01963. [DOI] [PubMed] [Google Scholar]
- [240].Hibst N, Strehle S. Phys Status Solidi A. 2016;213:2901. [Google Scholar]
- [241].Cui H, Wang B, Wang W, Hao Y, Liu C, Song K, Zhang S, Wang S. ACS Appl Mater Interfaces. 2018;10:19545. doi: 10.1021/acsami.8b06072. [DOI] [PubMed] [Google Scholar]
- [242].Kuijk A, van Blaaderen A, Imhof A. J Am Chem Soc. 2011;133:2346. doi: 10.1021/ja109524h. [DOI] [PubMed] [Google Scholar]
- [243].Zhao B, Li D, Long Y, Yang G, Tung C-H, Song K. RSC Adv. 2016;6:32956 [Google Scholar]
- [244].Fraioli R, Tsimbouri PM, Fisher LE, Nobbs AH, Su B, Neubauer S, Rechenmacher F, Kessler H, Ginebra M-P, Dalby MJ, Manero JM, et al. Sci Rep. 2017;7:16363. doi: 10.1038/s41598-017-16385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Hang T, Chen H-J, Xiao S, Yang C, Chen M, Tao J, Shieh H, Yang B, Liu C, Xie X. R Soc Open Sci. 2017;4:171431. doi: 10.1098/rsos.171431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Lee J, Kang BS, Hicks B, Chancellor TF, Chu BH, Wang H-T, Keselowsky BG, Ren F, Lele TP. Biomaterials. 2008;29:3743. doi: 10.1016/j.biomaterials.2008.05.029. [DOI] [PubMed] [Google Scholar]
- [247].Kim W, Lee SH, Kim JH, Ahn YJ, Kim Y-H, Yu JS, Choi S. ACS Nano. 2018;12:7100. doi: 10.1021/acsnano.8b02917. [DOI] [PubMed] [Google Scholar]
- [248].Jiang X, Herricks T, Xia Y. Nano Lett. 2002;2:1333. [Google Scholar]
- [249].Mumm F, Beckwith KM, Bonde S, Martinez KL, Sikorski P. Small. 2013;9:263. doi: 10.1002/smll.201201314. [DOI] [PubMed] [Google Scholar]
- [250].De Angelis F, Malerba M, Patrini M, Miele E, Das G, Toma A, Zaccaria RP, Di Fabrizio E. Nano Lett. 2013;13:3553. doi: 10.1021/nl401100x. [DOI] [PubMed] [Google Scholar]
- [251].Lin ZC, McGuire AF, Burridge PW, Matsa E, Lou H-Y, Wu JC, Cui B. Microsyst Nanoeng. 2017;3:16080. doi: 10.1038/micronano.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Smirnov VK, Kibalov DS, Krivelevich SA, Lepshin PA, Potapov EV, Yankov RA, Skorupa W, Makarov VV, Danilin AB. Nucl Instrum Methods Phys Res Sect B Beam Interact Mater At. 1999;147:310. [Google Scholar]
- [253].Tocce EJ, Smirnov VK, Kibalov DS, Liliensiek SJ, Murphy CJ, Nealey PF. Biomaterials. 2010;31:4064. doi: 10.1016/j.biomaterials.2010.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Xia Y, Whitesides GM. Angew Chem Int Ed. 1998;37:550. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- [255].Xia Y, Whitesides GM. Angew Chem. 1998;110:568. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- [256].Rogers JA, Nuzzo RG. Mater Today. 2005;8:50. [Google Scholar]
- [257].Loesberg WA, te Riet J, van Delft FCMJM, Schön P, Figdor CG, Speller S, van Loon JJWA, Walboomers XF, Jansen JA. Biomaterials. 2007;28:3944. doi: 10.1016/j.biomaterials.2007.05.030. [DOI] [PubMed] [Google Scholar]
- [258].Skoog SA, Kumar G, Narayan RJ, Goering PL. Pharmacol Ther. 2018;182:33. doi: 10.1016/j.pharmthera.2017.07.009. [DOI] [PubMed] [Google Scholar]
- [259].Qin D, Xia Y, Whitesides GM. Nat Protoc. 2010;5:491. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- [260].von Erlach TC, Bertazzo S, Wozniak MA, Horejs C-M, Maynard SA, Attwood S, Robinson BK, Autefage H, Kallepitis C, del A, Hernández R, et al. Nat Mater. 2018;17:237. doi: 10.1038/s41563-017-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Green DW, Lee KK-H, Watson JA, Kim H-Y, Yoon K-S, Kim E-J, Lee J-M, Watson GS, Jung H-S. Sci Rep. 2017;7:41023. doi: 10.1038/srep41023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Armentano I, Arciola CR, Fortunati E, Ferrari D, Mattioli S, Amoroso CF, Rizzo J, Kenny JM, Imbriani M, Visai L. Sci World J. 2014;2014:1. doi: 10.1155/2014/410423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Rizwan M, Tse JW, Nori A, Leong KW, Yim EKF. In: Princ Regen Med Third Ed. Atala A, Lanza R, Mikos AG, Nerem R, editors. Academic Press; Boston: 2019. pp. 437–468. [Google Scholar]
- [264].Hong S-H, Hwang J, Lee H. Nanotechnology. 2009;20:385303. doi: 10.1088/0957-4484/20/38/385303. [DOI] [PubMed] [Google Scholar]
- [265].Viela F, Granados D, Ayuso‐Sacido A, Rodríguez I. Adv Funct Mater. 2016;26:5599. [Google Scholar]
- [266].Peng L, Deng Y, Yi P, Lai X. J Micromechanics Microengineering. 2013;24:013001 [Google Scholar]
- [267].Dalby MJ, Biggs MJP, Gadegaard N, Kalna G, Wilkinson CDW, Curtis ASG. J Cell Biochem. 2007;100:326. doi: 10.1002/jcb.21058. [DOI] [PubMed] [Google Scholar]
- [268].Stormonth-Darling JM, Pedersen RH, How C, Gadegaard N. J Micromechanics Microengineering. 2014;24:075019 [Google Scholar]
- [269].Rasmussen CH, Reynolds PM, Petersen DR, Hansson M, McMeeking RM, Dufva M, Gadegaard N. Adv Funct Mater. 2016;26:815. [Google Scholar]
- [270].Lee DY, Lee DH, Lee SG, Cho K. Soft Matter. 2012;8:4905. [Google Scholar]
- [271].Zhang Y, Reed JC, Yang S. ACS Nano. 2009;3:2412. doi: 10.1021/nn900650n. [DOI] [PubMed] [Google Scholar]
- [272].Reynolds PM, Pedersen RH, Stormonth-Darling J, Dalby MJ, Riehle MO, Gadegaard N. Nano Lett. 2013;13:570. doi: 10.1021/nl304097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Weisse JM, Kim DR, Lee CH, Zheng X. Nano Lett. 2011;11:1300. doi: 10.1021/nl104362e. [DOI] [PubMed] [Google Scholar]
- [274].Guan N, Dai X, Babichev AV, Julien FH, Tchernycheva M. Chem Sci. 2017;8:7904. doi: 10.1039/c7sc02573d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Jeong HE, Kwak R, Kim JK, Suh KY. Small. 2008;4:1913. doi: 10.1002/smll.200800151. [DOI] [PubMed] [Google Scholar]
- [276].Eui Jeong H, Kwak R, Khademhosseini A, Suh KY. Nanoscale. 2009;1:331. doi: 10.1039/b9nr00106a. [DOI] [PubMed] [Google Scholar]
- [277].Ahn EH, Kim Y, Kshitiz, An SS, Afzal J, Lee S, Kwak M, Suh K-Y, Kim D-H, Levchenko A. Biomaterials. 2014;35:2401. doi: 10.1016/j.biomaterials.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Yang D, Hong H, Seo YH, Kim LH, Ryu W. ACS Appl Mater Interfaces. 2015;7:16873. doi: 10.1021/acsami.5b05254. [DOI] [PubMed] [Google Scholar]
- [279].Gong J, Lipomi DJ, Deng J, Nie Z, Chen X, Randall NX, Nair R, Whitesides GM. Nano Lett. 2010;10:2702. doi: 10.1021/nl101675s. [DOI] [PubMed] [Google Scholar]
- [280].Xu Q, Rioux RM, Dickey MD, Whitesides GM. Acc Chem Res. 2008;41:1566. doi: 10.1021/ar700194y. [DOI] [PubMed] [Google Scholar]
- [281].Dai X, Xu Q. Nanotechnology. 2011;22:494008. doi: 10.1088/0957-4484/22/49/494008. [DOI] [PubMed] [Google Scholar]
- [282].Qiu W, Xu M, Li R, Liu X, Zhang M. Anal Chem. 2016;88:1117. doi: 10.1021/acs.analchem.5b04055. [DOI] [PubMed] [Google Scholar]
- [283].Zhou Q, Zhao Z, Zhou Z, Zhang G, Chiechi RC, van Rijn P. Adv Mater Interfaces. 2018;5:1800334 [Google Scholar]
- [284].Blau A. Curr Opin Colloid Interface Sci. 2013;18:481. doi: 10.1016/j.cocis.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Acres RG, Ellis AV, Alvino J, Lenahan CE, Khodakov DA, Metha GF, Andersson GG. J Phys Chem C. 2012;116:6289. [Google Scholar]
- [286].Pensa E, Cortés E, Corthey G, Carro P, Vericat C, Fonticelli MH, Benítez G, Rubert AA, Salvarezza RC. Acc Chem Res. 2012;45:1183. doi: 10.1021/ar200260p. [DOI] [PubMed] [Google Scholar]
- [287].Bürgi T. Nanoscale. 2015;7:15553. doi: 10.1039/c5nr03497c. [DOI] [PubMed] [Google Scholar]
- [288].Adler AF, Leong KW. Nano Today. 2010;5:553. doi: 10.1016/j.nantod.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn D-R, Yoon M-H, Sutton A, Jorgolli M, Gertner RS, Gujral TS, MacBeath G, et al. Proc Natl Acad Sci. 2010;107:1870. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Casanova A, Blatche M-C, Ferre CA, Martin H, Gonzalez-Dunia D, Nicu L, Larrieu G. Langmuir. 2018;34:6612. doi: 10.1021/acs.langmuir.8b00529. [DOI] [PubMed] [Google Scholar]
- [291].Cass T, Ligler FS, editors. Immobilized Biomolecules in Analysis: A Practical Approach. Oxford University Press, Oxford; New York: 1998. [Google Scholar]
- [292].Tieu T, Alba M, Elnathan R, Cifuentes‐Rius A, Voelcker NH. Adv Ther. 2019;2:1800095 [Google Scholar]
- [293].Xie C, Hanson L, Cui Y, Cui B. Proc Natl Acad Sci. 2011;108:3894. doi: 10.1073/pnas.1015589108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Santoro F, Panaitov G, Offenhäusser A. Nano Lett. 2014;14:6906. doi: 10.1021/nl502922b. [DOI] [PubMed] [Google Scholar]
- [295].Spira ME, Kamber D, Dormann A, Cohen A, Bartic C, Borghs G, Langedijk JPM, Yitzchaik S, Shabthai K, Shappir Josef. TRANSDUCERS 2007 - 2007 Int. Solid-State Sens. Actuators Microsyst. Conf; Lyon, France. 2007. pp. 1247–1250. [Google Scholar]
- [296].van Meerbergen B, Raemaekers T, Winters K, Braeken D, Bartic C, Spira M, Engelborghs Y, Annaert WG, Borghs G. J Exp Nanosci. 2007;2:101. [Google Scholar]
- [297].Hai A, Shappir J, Spira ME. J Neurophysiol. 2010;104:559. doi: 10.1152/jn.00265.2010. [DOI] [PubMed] [Google Scholar]
- [298].Hai A, Kamber D, Malkinson G, Erez H, Mazurski N, Shappir J, Spira ME. J Neural Eng. 2009;6:066009. doi: 10.1088/1741-2560/6/6/066009. [DOI] [PubMed] [Google Scholar]
- [299].Amin H, Dipalo M, De Angelis F, Berdondini L. ACS Appl Mater Interfaces. 2018;10:15207. doi: 10.1021/acsami.8b00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].He G, Chen H-J, Liu D, Feng Y, Yang C, Hang T, Wu J, Cao Y, Xie X. Adv Mater Interfaces. 2018;5:1701535 [Google Scholar]
- [301].Donnelly H, Dalby MJ, Salmeron-Sanchez M, Sweeten PE. Nanomedicine Nanotechnol Biol Med. 2018;14:2455. doi: 10.1016/j.nano.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Cavalcanti‐Adam EA, Aydin D, Hirschfeld‐Warneken VC, Spatz JP. HFSP J. 2008;2:276. doi: 10.2976/1.2976662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Huang J, Gräter SV, Corbellini F, Rinck S, Bock E, Kemkemer R, Kessler H, Ding J, Spatz JP. Nano Lett. 2009;9:1111. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Wang X, Yan C, Ye K, He Y, Li Z, Ding J. Biomaterials. 2013;34:2865. doi: 10.1016/j.biomaterials.2013.01.021. [DOI] [PubMed] [Google Scholar]
- [305].Hai Aviad, Ada Dormann, Shappir Joseph, Yitzchaik Shlomo, Bartic Carmen, Borghs Gustaaf, Langedijk JPM, Spira Micha E. J R Soc Interface. 2009;6:1153. doi: 10.1098/rsif.2009.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Santoro F, Schnitker J, Panaitov G, Offenhäusser A. Nano Lett. 2013;13:5379. doi: 10.1021/nl402901y. [DOI] [PubMed] [Google Scholar]
- [307].Günter Wrobel, Matthias Höller, Sven Ingebrandt, Sabine Dieluweit, Frank Sommerhage, Peter Bochem Hans, Andreas Offenhäusser. J R Soc Interface. 2008;5:213. [Google Scholar]
- [308].Toma K, Kano H, Offenhäusser A. ACS Nano. 2014;8:12612. doi: 10.1021/nn505521e. [DOI] [PubMed] [Google Scholar]
- [309].Ting Zheng X, Ming Li C. Chem Soc Rev. 2012;41:2061. doi: 10.1039/c1cs15265c. [DOI] [PubMed] [Google Scholar]
- [310].Rust MJ, Bates M, Zhuang X. Nat Methods. 2006;3:793. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. Nat Protoc. 2011;6:991. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- [312].Vakarelski IU, Brown SC, Higashitani K, Moudgil BM. Langmuir. 2007;23:10893. doi: 10.1021/la701878n. [DOI] [PubMed] [Google Scholar]
- [313].Yim EKF, Darling EM, Kulangara K, Guilak F, Leong KW. Biomaterials. 2010;31:1299. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Lamers E, Walboomers XF, Domanski M, McKerr G, O’Hagan BM, Barnes CA, Peto L, Luttge R, Winnubst LAJA, Gardeniers HJGE, Jansen JA. Tissue Eng Part C Methods. 2010;17:1. doi: 10.1089/ten.TEC.2010.0251. [DOI] [PubMed] [Google Scholar]
- [315].Donselaar EGV, Dorresteijn B, Popov‐Čeleketić D, Wetering WJVD, Verrips TC, Boekhout T, Schneijdenberg CTWM, Xenaki AT, Krift TPVD, Müller WH. J Microsc. 2018;270:359. doi: 10.1111/jmi.12694. [DOI] [PubMed] [Google Scholar]
- [316].Gnauck P, Burkhardt C, Wolburg H, Nisch W. Microsc Microanal. 2005;11:800. [Google Scholar]
- [317].Gopal S, Chiappini C, Armstrong JPK, Chen Q, Serio A, Hsu C-C, Meinert C, Klein TJ, Hutmacher DW, Rothery S, Stevens MM. Adv Mater. 2019:1900488. doi: 10.1002/adma.201900488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Swain RJ, Stevens MM. Biochem Soc Trans. 2007;35:544. doi: 10.1042/BST0350544. [DOI] [PubMed] [Google Scholar]
- [319].Bergholt MS, Serio A, McKenzie JS, Boyd A, Soares RF, Tillner J, Chiappini C, Wu V, Dannhorn A, Takats Z, Williams A, et al. ACS Cent Sci. 2018;4:39. doi: 10.1021/acscentsci.7b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Cheng A, Chen H, Schwartz Z, Boyan BD. J Microsc. 2018;270:41. doi: 10.1111/jmi.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Song H, Yang Y, Geng J, Gu Z, Zou J, Yu C. Adv Mater. 2018:1801564. doi: 10.1002/adma.201801564. [DOI] [PubMed] [Google Scholar]
- [322].Ercius P, Alaidi O, Rames MJ, Ren G. Adv Mater. 2015;27:5638. doi: 10.1002/adma.201501015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Hansma PK, Drake B, Marti O, Gould SA, Prater CB. Science. 1989;243:641. doi: 10.1126/science.2464851. [DOI] [PubMed] [Google Scholar]
- [324].Korchev YE, Bashford CL, Milovanovic M, Vodyanoy I, Lab MJ. Biophys J. 1997;73:653. doi: 10.1016/S0006-3495(97)78100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [325].Novak P, Li C, Shevchuk AI, Stepanyan R, Caldwell M, Hughes S, Smart TG, Gorelik J, Ostanin VP, Lab MJ, Moss GWJ, et al. Nat Methods. 2009;6:279. doi: 10.1038/nmeth.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Crowder SW, Leonardo V, Whittaker T, Papathanasiou P, Stevens MM. Cell Stem Cell. 2016;18:39. doi: 10.1016/j.stem.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Kawashima T, Matsugase T, Tanaka K, Nagai M, Shibata T, Mineta T, Makino E. Microelectron Eng. 2012;98:663. [Google Scholar]
- [328].Shibata T, Ozawa T, Ito Y, Yamamoto K, Nagai M. Jpn J Appl Phys. 2017;56:017001 [Google Scholar]
- [329].Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. Science. 2018;359:eaan4672. doi: 10.1126/science.aan4672. [DOI] [PubMed] [Google Scholar]
- [330].Lagassé HAD, Alexaki A, Simhadri VL, Katagiri NH, Jankowski W, Sauna ZE, Kimchi-Sarfaty C. F1000Research. 2017;6:113. doi: 10.12688/f1000research.9970.1. F1000 Faculty Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Lau JL, Dunn MK. Bioorg Med Chem. 2018;26:2700. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- [332].Yan L, Zhang J, Lee C-S, Chen X. Small. 2014;10:4487. doi: 10.1002/smll.201401532. [DOI] [PubMed] [Google Scholar]
- [333].Shalek AK, Gaublomme JT, Wang L, Yosef N, Chevrier N, Andersen MS, Robinson JT, Pochet N, Neuberg D, Gertner RS, Amit I, et al. Nano Lett. 2012;12:6498. doi: 10.1021/nl3042917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Hammarin G, Persson H, Dabkowska AP, Prinz CN. Colloids Surf B Biointerfaces. 2014;122:85. doi: 10.1016/j.colsurfb.2014.06.048. [DOI] [PubMed] [Google Scholar]
- [335].Fox CB, Kim J, Le LV, Nemeth CL, Chirra HD, Desai TA. J Controlled Release. 2015;219:431. doi: 10.1016/j.jconrel.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [336].Fox CB, Cao Y, Nemeth CL, Chirra HD, Chevalier RW, Xu AM, Melosh NA, Desai TA. ACS Nano. 2016;10:5873. doi: 10.1021/acsnano.6b00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Fine D, Grattoni A, Goodall R, Bansal SS, Chiappini C, Hosali S, van de Ven AL, Srinivasan S, Liu X, Godin B, Brousseau L, et al. Adv Healthc Mater. 2013;2:632. doi: 10.1002/adhm.201200214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn D-R, Yoon M-H, Sutton A, Jorgolli M, Gertner RS, Gujral TS, MacBeath G, et al. Proc Natl Acad Sci. 2010;107:1870. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Harding FJ, Surdo S, Delalat B, Cozzi C, Elnathan R, Gronthos S, Voelcker NH, Barillaro G. ACS Appl Mater Interfaces. 2016;8:29197. doi: 10.1021/acsami.6b07850. [DOI] [PubMed] [Google Scholar]
- [340].Elnathan R, Kwiat M, Patolsky F, Voelcker NH. Nano Today. 2014;9:172. [Google Scholar]
- [341].Zhu X, Kwok SY, Yuen MF, Yan L, Chen W, Yang Y, Wang Z, Yu KN, Zhu G, Zhang W, Chen X. J Mater Sci. 2015;50:7800. [Google Scholar]
- [342].Zhang B, Shi Y, Miyamoto D, Nakazawa K, Miyake T. Sci Rep. 2019;9:6806. doi: 10.1038/s41598-019-43340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Higgins SG, Stevens MM. Science. 2017;356:379. doi: 10.1126/science.aan0228. [DOI] [PubMed] [Google Scholar]
- [344].Kavaldzhiev MN, Perez JE, Sougrat R, Bergam P, Ravasi T, Kosel J. Sci Rep. 2018;8:9918. doi: 10.1038/s41598-018-28194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [345].Liu Z, Nie J, Miao B, Li J, Cui Y, Wang S, Zhang X, Zhao G, Deng Y, Wu Y, Li Z, et al. Adv Mater. 2019;31:1807795. doi: 10.1002/adma.201807795. [DOI] [PubMed] [Google Scholar]
- [346].Ember KJI, Hoeve MA, McAughtrie SL, Bergholt MS, Dwyer BJ, Stevens MM, Faulds K, Forbes SJ, Campbell CJ. Npj Regen Med. 2017;2:12. doi: 10.1038/s41536-017-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Talari ACS, Movasaghi Z, Rehman S, ur Rehman I. Appl Spectrosc Rev. 2015;50:46. [Google Scholar]
- [348].Bergholt MS, Albro MB, Stevens MM. Biomaterials. 2017;140:128. doi: 10.1016/j.biomaterials.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].You AYF, Bergholt MS, St-Pierre J-P, Kit-Anan W, Pence IJ, Chester AH, Yacoub MH, Bertazzo S, Stevens MM. Sci Adv. 2017;3:e1701156. doi: 10.1126/sciadv.1701156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Penders J, Pence IJ, Horgan C, Bergholt M, Wood C, Najer A, Kauscher U, Nagelkerke A, Stevens MM. Nat Commun. 2018:1. doi: 10.1038/s41467-018-06397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [351].Kallepitis C, Bergholt MS, Mazo MM, Leonardo V, Skaalure SC, Maynard SA, Stevens MM. Nat Commun. 2017;8:14843. doi: 10.1038/ncomms14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Zhao Y, Zhang Y-L, Huang J-A, Zhang Z, Chen X, Zhang W. J Mater Chem A. 2015;3:6408. [Google Scholar]
- [353].Yang Y, Tanemura M, Huang Z, Jiang D, Li Z-Y, Huang Y, Kawamura G, Yamaguchi K, Nogami M. Nanotechnology. 2010;21:325701. doi: 10.1088/0957-4484/21/32/325701. [DOI] [PubMed] [Google Scholar]
- [354].Smith E, Dent G. Mod Raman Spectrosc – Pract Approach. John Wiley & Sons, Ltd; Chichester: 2005. pp. 113–133. [Google Scholar]
- [355].Caprettini V, Huang J-A, Moia F, Jacassi A, Gonano CA, Maccaferri N, Capozza R, Dipalo M, De Angelis F. Adv Sci. 2018;5:1800560. doi: 10.1002/advs.201800560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [356].Goldberg-Oppenheimer P, Hutter T, Chen B, Robertson J, Hofmann S, Mahajan S. J Phys Chem Lett. 2012;3:3486. doi: 10.1021/jz301333r. [DOI] [PubMed] [Google Scholar]
- [357].Lin D, Wu Z, Li S, Zhao W, Ma C, Wang J, Jiang Z, Zhong Z, Zheng Y, Yang X. ACS Nano. 2017;11:1478. doi: 10.1021/acsnano.6b06778. [DOI] [PubMed] [Google Scholar]
- [358].Zhang L, Li X, Wang Y, Sun K, Chen X, Chen H, Zhou J. ACS Omega. 2018;3:14181. doi: 10.1021/acsomega.7b02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [359].Cao W, Jiang L, Hu J, Wang A, Li X, Lu Y. ACS Appl Mater Interfaces. 2018;10:1297. doi: 10.1021/acsami.7b13241. [DOI] [PubMed] [Google Scholar]
- [360].Huang J-A, Caprettini V, Zhao Y, Melle G, Maccaferri N, Deleye L, Zambrana-Puyalto X, Ardini M, Tantussi F, Dipalo M, De Angelis F. Nano Lett. 2019;19:722. doi: 10.1021/acs.nanolett.8b03764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [361].Park YM, Lim SY, Jeong SW, Song Y, Bae NH, Hong SB, Choi BG, Lee SJ, Lee KG. Nano Converg. 2018;5:15. doi: 10.1186/s40580-018-0147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Soleymani L, Fang Z, Sargent EH, Kelley SO. Nat Nanotechnol. 2009;4:844. doi: 10.1038/nnano.2009.276. [DOI] [PubMed] [Google Scholar]
- [363].Rawson FJ, Hicks J, Dodd N, Abate W, Garrett DJ, Yip N, Fejer G, Downard AJ, Baronian KHR, Jackson SK, Mendes PM. ACS Appl Mater Interfaces. 2015;7:23527. doi: 10.1021/acsami.5b06493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [364].Hicks JM, Halkerston R, Silman N, Jackson SK, Aylott JW, Rawson FJ. Biosens Bioelectron. 2019;141:111430. doi: 10.1016/j.bios.2019.111430. [DOI] [PubMed] [Google Scholar]
- [365].Zhang F, Jiang Y, Liu X, Meng J, Zhang P, Liu H, Yang G, Li G, Jiang L, Wan LJ, Hu JS, et al. Nano Lett. 2016;16:766. doi: 10.1021/acs.nanolett.5b04731. [DOI] [PubMed] [Google Scholar]
- [366].Wang S, Wang H, Jiao J, Chen K-J, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng H-R. Angew Chem Int Ed. 2009;48:8970. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Lee SK, Kim GS, Wu Y, Kim DJ, Lu Y, Kwak M, Han L, Hyung JH, Seol JK, Sander C, Gonzalez A, et al. Nano Lett. 2012;12:2697. doi: 10.1021/nl2041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Xia Y, Tang Y, Yu X, Wan Y, Chen Y, Lu H, Zheng S-Y. Small. 2016;13:1603135 [Google Scholar]
- [369].Wang S, Wan Y, Liu Y. Nanoscale. 2014;6:12482. doi: 10.1039/c4nr02854f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Kawamura R, Miyazaki M, Shimizu K, Matsumoto Y, Silberberg YR, Sathuluri RR, Iijima M, Kuroda S, Iwata F, Kobayashi T, Nakamura C. Nano Lett. 2017;17:7117. doi: 10.1021/acs.nanolett.7b03918. [DOI] [PubMed] [Google Scholar]
- [371].Parker SG, Yang Y, Ciampi S, Gupta B, Kimpton K, Mansfeld FM, Kavallaris M, Gaus K, Justin Gooding J. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [372].Avants BW, Park H, Robinson JT. In: Micro-Nanosyst Biotechnol. Love JC, editor. Wiley-VCH Verlag; Weinheim, Germany: 2016. pp. 127–142. [Google Scholar]
- [373].Zeck G, Jetter F, Channappa L, Bertotti G, Thewes R. Adv Biosyst. 2017;1:1700107. doi: 10.1002/adbi.201700107. [DOI] [PubMed] [Google Scholar]
- [374].Marchesan S, Ballerini L, Prato M. Science. 2017;356:1010. doi: 10.1126/science.aan1227. [DOI] [PubMed] [Google Scholar]
- [375].Wang Y, Zhu H, Yang H, Argall AD, Luan L, Xie C, Guo L. Nano Res. 2018;11:5065. [Google Scholar]
- [376].Scaini D, Ballerini L. Curr Opin Neurobiol. 2018;50:50. doi: 10.1016/j.conb.2017.12.009. [DOI] [PubMed] [Google Scholar]
- [377].Feiner R, Dvir T. Nat Rev Mater. 2017;3:17076 [Google Scholar]
- [378].Wang G, Wyskiel DR, Yang W, Wang Y, Milbern LC, Lalanne T, Jiang X, Shen Y, Sun Q-Q, Zhu JJ. Nat Protoc. 2015;10:397. doi: 10.1038/nprot.2015.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [379].Jäckel D, Bakkum DJ, Russell TL, Müller J, Radivojevic M, Frey U, Franke F, Hierlemann A. Sci Rep. 2017;7:978. doi: 10.1038/s41598-017-00981-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [380].National Institutes of Health. Stimulating Peripheral Activity to Relieve Conditions (SPARC) 2019 can be found under https://commonfund.nih.gov/sparc.
- [381].National Institutes of Health. Brain Initiative. 2019 can be found under https://www.braininitiative.nih.gov/
- [382].Alivisatos AP, Andrews AM, Boyden ES, Chun M, Church GM, Deisseroth K, Donoghue JP, Fraser SE, Lippincott-Schwartz J, Looger LL, Masmanidis S, et al. ACS Nano. 2013;7:1850. doi: 10.1021/nn4012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [383].Robinson JT, Jorgolli M, Park H. Front Neural Circuits. 2013;7:1. doi: 10.3389/fncir.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [384].Lacour SP, Courtine G, Guck J. Nat Rev Mater. 2016;1:16063 [Google Scholar]
- [385].Fendyur A, Mazurski N, Shappir J, Spira ME. Front Neuroengineering. 2011;4 doi: 10.3389/fneng.2011.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [386].Rabieh N, Ojovan SM, Shmoel N, Erez H, Maydan E, Spira ME. Sci Rep. 2016;6:36498. doi: 10.1038/srep36498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [387].Petreanu L, Mao T, Sternson SM, Svoboda K. Nature. 2009;457:1142. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [388].Multi Channel Systems MCS GmbH. Innovations in Electrophysiology. 2019 can be found under www.multichannelsystems.com, https://www.multichannelsystems.com/
- [389].Heuschkel M. “Qwane Biosciences - Technology,”. 2019 can be found under http://www.qwane.com/Technology.html. [Google Scholar]
- [390].Park JS, Grijalva SI, Aziz MK, Chi T, Li S, Sayegh MN, Wang A, Cho HC, Wang H. Lab Chip. 2018;18:3037. doi: 10.1039/c8lc00156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [391].Lopez CM, Andrei A, Mitra S, Welkenhuysen M, Eberle W, Bartic C, Puers R, Yazicioglu RF, Gielen GGE. IEEE J Solid-State Circuits. 2014;49:248. [Google Scholar]
- [392].Braeken D, Huys R, Loo J, Bartic C, Borghs G, Callewaert G, Eberle W. 2010 Annu Int Conf IEEE Eng Med Biol. 2010:6473–6476. doi: 10.1109/IEMBS.2010.5627352. [DOI] [PubMed] [Google Scholar]
- [393].Abbott J, Ye T, Ham D, Park H. Acc Chem Res. 2018;51:600. doi: 10.1021/acs.accounts.7b00519. [DOI] [PubMed] [Google Scholar]
- [394].Braeken D, Jans D, Huys R, Stassen A, Collaert N, Hoffman L, Eberle W, Peumans P, Callewaert G. Lab Chip. 2012;12:4397. doi: 10.1039/c2lc40656j. [DOI] [PubMed] [Google Scholar]
- [395].Wang K, Fishman HA, Dai H, Harris JS. Nano Lett. 2006;6:2043. doi: 10.1021/nl061241t. [DOI] [PubMed] [Google Scholar]
- [396].Spira ME, Hai A. Nat Nanotechnol. 2013;8:83. doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- [397].Shmoel N, Rabieh N, Ojovan SM, Erez H, Maydan E, Spira ME. Sci Rep. 2016;6:27110. doi: 10.1038/srep27110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [398].Sytnyk M, Jakešová M, Litviňuková M, Mashkov O, Kriegner D, Stangl J, Nebesářová J, Fecher FW, Schöfberger W, Sariciftci NS, Schindl R, et al. Nat Commun. 2017;8:91. doi: 10.1038/s41467-017-00135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [399].Huang G, Li F, Zhao X, Ma Y, Li Y, Lin M, Jin G, Lu TJ, Genin GM, Xu F. Chem Rev. 2017;117:12764. doi: 10.1021/acs.chemrev.7b00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [400].Goetzke R, Sechi A, De Laporte L, Neuss S, Wagner W. Cell Mol Life Sci. 2018;75:3297. doi: 10.1007/s00018-018-2830-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Beckwith KS, Cooil SP, Wells JW, Sikorski P. Nanoscale. 2015;7:8438. doi: 10.1039/c5nr00674k. [DOI] [PubMed] [Google Scholar]
- [402].Stevens MM, George JH. Science. 2005;310:1135. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- [403].Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Nat Rev Mol Cell Biol. 2017;18:758. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [404].Uhler C, Shivashankar GV. Nat Rev Mol Cell Biol. 2017;18:717. doi: 10.1038/nrm.2017.101. [DOI] [PubMed] [Google Scholar]
- [405].Jaalouk DE, Lammerding J. Nat Rev Mol Cell Biol. 2009;10:63. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [406].McNamara LE, McMurray RJ, Biggs MJP, Kantawong F, Oreffo ROC, Dalby MJ. J Tissue Eng. 2010;1:120623. doi: 10.4061/2010/120623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [407].Qiu J, Li J, Wang S, Ma B, Zhang S, Guo W, Zhang X, Tang W, Sang Y, Liu H. Small. 2016;12:1770. doi: 10.1002/smll.201503946. [DOI] [PubMed] [Google Scholar]
- [408].Xie C, Hanson L, Xie W, Lin Z, Cui B, Cui Y. Nano Lett. 2010;10:4020. doi: 10.1021/nl101950x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [409].He P, Wang X, Ning C, Liu X, Li M, Xu H, Guo G, Mao G, Liu G, Xu B, Zhang Y, et al. Sci China Technol Sci. 2019;62:559. [Google Scholar]
- [410].Tan F, Walshe P, Viani L, Al-Rubeai M. Trends Biotechnol. 2013;31:678. doi: 10.1016/j.tibtech.2013.09.001. [DOI] [PubMed] [Google Scholar]
- [411].Xu X, Ma L, Wu Y, Tang L. J Biomed Mater Res A. 2018;106:3165. doi: 10.1002/jbm.a.36511. [DOI] [PubMed] [Google Scholar]
- [412].Han J, Lin K-H, Chew LY. J Phys Condens Matter. 2017;29:455101. doi: 10.1088/1361-648X/aa8d49. [DOI] [PubMed] [Google Scholar]
- [413].O’Brien LE, Jou T-S, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, Mostov KE. Nat Cell Biol. 2001;3:831. doi: 10.1038/ncb0901-831. [DOI] [PubMed] [Google Scholar]
- [414].Mas‐Moruno C, Su B, Dalby MJ. Adv Healthc Mater. 2019;8:1801103. doi: 10.1002/adhm.201801103. [DOI] [PubMed] [Google Scholar]
- [415].Minev IR, Moshayedi P, Fawcett JW, Lacour SP. Acta Biomater. 2013;9:6936. doi: 10.1016/j.actbio.2013.02.048. [DOI] [PubMed] [Google Scholar]
- [416].Lou H-Y, Zhao W, Zeng Y, Cui B. Acc Chem Res. 2018;51:1046. doi: 10.1021/acs.accounts.7b00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [417].Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Nat Cell Biol. 2014;16:376. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [418].Liu X, Liu R, Gu Y, Ding J. ACS Appl Mater Interfaces. 2017;9:18521. doi: 10.1021/acsami.7b04027. [DOI] [PubMed] [Google Scholar]
- [419].Makhija E, Jokhun DS, Shivashankar GV. Proc Natl Acad Sci U S A. 2015;113:E32. doi: 10.1073/pnas.1513189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [420].Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin J-W, Tewari M, Rehfeldt F, et al. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [421].Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, et al. Nature. 2011;474:179. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- [422].Piccolo S, Dupont S, Cordenonsi M. Physiol Rev. 2014;94:1287. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- [423].Sero JE, Bakal C. Cell Syst. 2017;4:84. doi: 10.1016/j.cels.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [424].Nardone G, Oliver-De La Cruz J, Vrbsky J, Martini C, Pribyl J, Skládal P, Pešl M, Caluori G, Pagliari S, Martino F, Maceckova Z, et al. Nat Commun. 2017;8:15321. doi: 10.1038/ncomms15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [425].SanMartin A, Johansson F, Samuelson L, Prinz CN. J Nanosci Nanotechnol. 2014;14:4880. doi: 10.1166/jnn.2014.8669. [DOI] [PubMed] [Google Scholar]
- [426].Murphy WL, McDevitt TC, Engler AJ. Nat Mater. 2014;13:547. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [427].Pan H, Xie Y, Li K, Hu D, Zhao J, Zheng X, Tang T. RSC Adv. 2015;5:101834 [Google Scholar]
- [428].Poudineh M, Wang Z, Labib M, Ahmadi M, Zhang L, Das J, Ahmed S, Angers S, Kelley SO. Nano Lett. 2018;18:7188. doi: 10.1021/acs.nanolett.8b03313. [DOI] [PubMed] [Google Scholar]
- [429].Kumari S, Vermeulen S, van der Veer B, Carlier A, de Boer J, Subramanyam D. Tissue Eng Part B Rev. 2018;24:255. doi: 10.1089/ten.TEB.2017.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [430].Place ES, Evans ND, Stevens MM. Nat Mater. 2009;8:457. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- [431].Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Eur J Cell Biol. 2004;83:159. doi: 10.1078/0171-9335-00369. [DOI] [PubMed] [Google Scholar]
- [432].Dalby MJ, Gadegaard N, Riehle MO, Wilkinson CDW, Curtis ASG. Int J Biochem Cell Biol. 2004;36:2005. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- [433].Biggs MJP, Richards RG, Dalby MJ. Nanomedicine Nanotechnol Biol Med. 2010;6:619. doi: 10.1016/j.nano.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [434].Liu X, Liu R, Cao B, Ye K, Li S, Gu Y, Pan Z, Ding J. Biomaterials. 2016;111:27. doi: 10.1016/j.biomaterials.2016.09.023. [DOI] [PubMed] [Google Scholar]
- [435].Kuo S-W, Lin H-I, Hui-Chun Ho J, Shih Y-RV, Chen H-F, Yen T-J, Lee OK. Biomaterials. 2012;33:5013. doi: 10.1016/j.biomaterials.2012.03.080. [DOI] [PubMed] [Google Scholar]
- [436].Persson H, Li Z, Tegenfeldt JO, Oredsson S, Prinz CN. Sci Rep. 2015;5:18535. doi: 10.1038/srep18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [437].Andolfi L, Murello A, Cassese D, Ban J, Zilio SD, Lazzarino M. Nanotechnology. 2017;28:155102. doi: 10.1088/1361-6528/aa5f3a. [DOI] [PubMed] [Google Scholar]
- [438].Hughes BR, Mirbagheri M, Waldman SD, Hwang DK. Acta Biomater. 2018;78:89. doi: 10.1016/j.actbio.2018.07.053. [DOI] [PubMed] [Google Scholar]
- [439].Ladoux B, Mège RM. Nat Rev Mol Cell Biol. 2017;18:743. doi: 10.1038/nrm.2017.98. [DOI] [PubMed] [Google Scholar]
- [440].Sochol RD, Higa AT, Janairo RRR, Li S, Lin L. Soft Matter. 2011;7:4606. [Google Scholar]
- [441].Chan CE, Odde DJ. Science. 2008;322:1687. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- [442].Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, Wilen LA, Xu Y, Dufresne ER. Soft Matter. 2014;10:4047. doi: 10.1039/c4sm00264d. [DOI] [PubMed] [Google Scholar]
- [443].Tymchenko N, Wallentin J, Petronis S, Bjursten LM, Kasemo B, Gold J. Biophys J. 2007;93:335. doi: 10.1529/biophysj.106.093302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [444].Suyatin DB, Hällström W, Samuelson L, Montelius L, Prinz CN, Kanje M. J Vac Sci Technol B Microelectron Nanometer Struct Process Meas Phenom. 2009;27:3092. [Google Scholar]
- [445].Hällström W, Lexholm M, Suyatin DB, Hammarin G, Hessman D, Samuelson L, Montelius L, Kanje M, Prinz CN. Nano Lett. 2010;10:782. doi: 10.1021/nl902675h. [DOI] [PubMed] [Google Scholar]
- [446].Xiao F, Wen X, Chiou P. Micro Electro Mech Syst; MEMS 2017 IEEE 30th Int. Conf. On; 2017. p. 243. [Google Scholar]
- [447].Tripathy A, Sen P, Su B, Briscoe WH. Adv Colloid Interface Sci. 2017;248:85. doi: 10.1016/j.cis.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [448].Lin N, Berton P, Moraes C, Rogers RD, Tufenkji N. Adv Colloid Interface Sci. 2018;252:55. doi: 10.1016/j.cis.2017.12.007. [DOI] [PubMed] [Google Scholar]
- [449].Liu C, Gallagher JJ, Sakimoto KK, Nichols EM, Chang CJ, Chang MCY, Yang P. Nano Lett. 2015;15:3634. doi: 10.1021/acs.nanolett.5b01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [450].Xu L, Zhao Y, Owusu KA, Zhuang Z, Liu Q, Wang Z, Li Z, Mai L. Chem. 2018;4:1538. [Google Scholar]
- [451].Ventola CL. Pharm Ther. 2015;40:277. [Google Scholar]
- [452].Watson GS, Green DW, Schwarzkopf L, Li X, Cribb BW, Myhra S, Watson JA. Acta Biomater. 2015;21:109. doi: 10.1016/j.actbio.2015.03.007. [DOI] [PubMed] [Google Scholar]
- [453].Kelleher SM, Habimana O, Lawler J, O’ Reilly B, Daniels S, Casey E, Cowley A. ACS Appl Mater Interfaces. 2016;8:14966. doi: 10.1021/acsami.5b08309. [DOI] [PubMed] [Google Scholar]
- [454].Hasan J, Webb HK, Truong VK, Pogodin S, Baulin VA, Watson GS, Watson JA, Crawford RJ, Ivanova EP. Appl Microbiol Biotechnol. 2013;97:9257. doi: 10.1007/s00253-012-4628-5. [DOI] [PubMed] [Google Scholar]
- [455].Ivanova EP, Hasan J, Webb HK, Truong VK, Watson GS, Watson JA, Baulin VA, Pogodin S, Wang JY, Tobin MJ, Löbbe C, et al. Small. 2012;8:2489. doi: 10.1002/smll.201200528. [DOI] [PubMed] [Google Scholar]
- [456].Hasan J, Crawford RJ, Ivanova EP. Trends Biotechnol. 2013;31:295. doi: 10.1016/j.tibtech.2013.01.017. [DOI] [PubMed] [Google Scholar]
- [457].Elbourne A, Crawford RJ, Ivanova EP. J Colloid Interface Sci. 2017;508:603. doi: 10.1016/j.jcis.2017.07.021. [DOI] [PubMed] [Google Scholar]
- [458].Hasan J, Chatterjee K. Nanoscale. 2015;7:15568. doi: 10.1039/c5nr04156b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [459].Svensson S, Forsberg M, Hulander M, Vazirisani F, Thomsen P, Trobos M, Palmquist A, Lausmaa J. Int J Nanomedicine. 2014:775. doi: 10.2147/IJN.S51465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [460].Ivanova EP, Hasan J, Webb HK, Gervinskas G, Juodkazis S, Truong VK, Wu AHF, Lamb RN, Baulin VA, Watson GS, Watson JA, et al. Nat Commun. 2013;4:2838. doi: 10.1038/ncomms3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [461].Hasan J, Raj S, Yadav L, Chatterjee K. RSC Adv. 2015;5:44953. doi: 10.1039/C5RA05206H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [462].Tripathy A, Sreedharan S, Bhaskarla C, Majumdar S, Peneti SK, Nandi D, Sen P. Langmuir. 2017;33:12569. doi: 10.1021/acs.langmuir.7b02291. [DOI] [PubMed] [Google Scholar]
- [463].Li Y-Q, Zhu B, Li Y, Leow WR, Goh R, Ma B, Fong E, Tang M, Chen X. Angew Chem. 2014;126:5947. doi: 10.1002/anie.201310135. [DOI] [PubMed] [Google Scholar]
- [464].Xue F, Liu J, Guo L, Zhang L, Li Q. J Theor Biol. 2015;385:1. doi: 10.1016/j.jtbi.2015.08.011. [DOI] [PubMed] [Google Scholar]
- [465].Li X. Phys Chem Chem Phys. 2016;18:1311. doi: 10.1039/c5cp05646b. [DOI] [PubMed] [Google Scholar]
- [466].Bandara CD, Singh S, Afara IO, Wolff A, Tesfamichael T, Ostrikov K, Oloyede A. ACS Appl Mater Interfaces. 2017;9:6746. doi: 10.1021/acsami.6b13666. [DOI] [PubMed] [Google Scholar]
- [467].Köller M, Ziegler N, Sengstock C, Schildhauer TA, Ludwig A. Biomed Phys Eng Express. 2018;4:055002 [Google Scholar]
- [468].Bertoglio F, Bloise N, Oriano M, Petrini P, Sprio S, Imbriani M, Tampieri A, Visai L. Appl Sci. 2018;8:845. [Google Scholar]
- [469].Pham VTH, Truong VK, Orlowska A, Ghanaati S, Barbeck M, Booms P, Fulcher AJ, Bhadra CM, Buividas R, Baulin V, Kirkpatrick CJ, et al. ACS Appl Mater Interfaces. 2016;8:22025. doi: 10.1021/acsami.6b06415. [DOI] [PubMed] [Google Scholar]
- [470].Jeong HE, Kim I, Karam P, Choi H-J, Yang P. Nano Lett. 2013;13:2864. doi: 10.1021/nl401205b. [DOI] [PubMed] [Google Scholar]
- [471].Sakimoto KK, Liu C, Lim J, Yang P. Nano Lett. 2014;14:5471. doi: 10.1021/nl502946j. [DOI] [PubMed] [Google Scholar]
- [472].Michalska M, Gambacorta F, Divan R, Aranson IS, Sokolov A, Noirot P, Laible PD. Nanoscale. 2018;10:6639. doi: 10.1039/c8nr00439k. [DOI] [PubMed] [Google Scholar]
- [473].Sahoo PK, Janissen R, Monteiro MP, Cavalli A, Murillo DM, Merfa MV, Cesar CL, Carvalho HF, de Souza AA, Bakkers EPAM, Cotta MA. Nano Lett. 2016;16:4656. doi: 10.1021/acs.nanolett.6b01998. [DOI] [PubMed] [Google Scholar]
- [474].Yuan L, Wang H, Yu Q, Wu Z, Brash JL, Chen H. J Mater Chem. 2011;21:6148. [Google Scholar]
- [475].Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, et al. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [476].Cutiongco MFA, Jensen BS, Reynolds PM, Gadegaard N. bioRxiv. 2018:495879 [Google Scholar]
- [477].Bensmaia SJ, Miller LE. Nat Rev Neurosci. 2014;15:313. doi: 10.1038/nrn3724. [DOI] [PubMed] [Google Scholar]
- [478].Paşca SP. Nature. 2018;553:437. doi: 10.1038/nature25032. [DOI] [PubMed] [Google Scholar]
- [479].Fadeel B, Farcal L, Hardy B, Vázquez-Campos S, Hristozov D, Marcomini A, Lynch I, Valsami-Jones E, Alenius H, Savolainen K. Nat Nanotechnol. 2018;13:537. doi: 10.1038/s41565-018-0185-0. [DOI] [PubMed] [Google Scholar]
- [480].Muller J, Huaux F, Moreau N, Misson P, Heilier J-F, Delos M, Arras M, Fonseca A, Nagy JB, Lison D. Toxicol Appl Pharmacol. 2005;207:221. doi: 10.1016/j.taap.2005.01.008. [DOI] [PubMed] [Google Scholar]
- [481].Murphy F, Mullins M, Hester K, Gelwick A, Scott-Fordsmand JJ, Maynard T. Nat Nanotechnol. 2017;12:717. doi: 10.1038/nnano.2017.162. [DOI] [PubMed] [Google Scholar]
- [482].Eriksson Linsmeier C, Prinz CN, Pettersson LME, Caroff P, Samuelson L, Schouenborg J, Montelius L, Danielsen N. Nano Lett. 2009;9:4184. doi: 10.1021/nl902413x. [DOI] [PubMed] [Google Scholar]
- [483].Adolfsson K, Schneider M, Hammarin G, Häcker U, Prinz CN. Nanotechnology. 2013;24:285101. doi: 10.1088/0957-4484/24/28/285101. [DOI] [PubMed] [Google Scholar]
- [484].Maynard AD, Aitken RJ, Butz T, Colvin V, Donaldson K, Oberdörster G, Philbert MA, Ryan J, Seaton A, Stone V, Tinkle SS, et al. Nature. 2006;444:267. doi: 10.1038/444267a. [DOI] [PubMed] [Google Scholar]
- [485].Ganguly P, Breen A, Pillai SC. ACS Biomater Sci Eng. 2018;4:2237. doi: 10.1021/acsbiomaterials.8b00068. [DOI] [PubMed] [Google Scholar]
- [486].Walkey CD, Chan WCW. Chem Soc Rev. 2012;41:2780. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- [487].Zimmerman JF, Parameswaran R, Murray G, Wang Y, Burke M, Tian B. Sci Adv. 2016;2:1. doi: 10.1126/sciadv.1601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [488].Meerbergen BV, Jans K, Loo J, Reekmans G, Braeken D, Seon-Ah C, Bonroy K, Maes G, Borghs G, Engelborghs Y, Annaert W, et al. 2008 30th Annu Int Conf IEEE Eng Med Biol Soc. 2008:1833–1836. doi: 10.1109/IEMBS.2008.4649536. [DOI] [PubMed] [Google Scholar]
- [489].Keshavarz M, Tan B, Venkatakrishnan K. ACS Appl Mater Interfaces. 2017;9:6292. doi: 10.1021/acsami.6b14836. [DOI] [PubMed] [Google Scholar]
- [490].Chao W-T, Kunz J. FEBS Lett. 2009;583:1337. doi: 10.1016/j.febslet.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [491].Adler AF, Speidel AT, Christoforou N, Kolind K, Foss M, Leong KW. Biomaterials. 2011;32:3611. doi: 10.1016/j.biomaterials.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [492].Dabkowska AP, Niman CS, Piret G, Persson H, Wacklin HP, Linke H, Prinz CN, Nylander T. Nano Lett. 2014;14:4286. doi: 10.1021/nl500926y. [DOI] [PubMed] [Google Scholar]
- [493].Belessiotis-Richard A, Butterworth A, Higgins SG, Stevens MM, Alexander-Katz A. Nano Lett. 2019 doi: 10.1021/acs.nanolett.9b01990. in revision. [DOI] [PubMed] [Google Scholar]
- [494].Jahed Z, Molladavoodi S, Seo BB, Gorbet M, Tsui TY, Mofrad MRK. Biomaterials. 2014;35:9363. doi: 10.1016/j.biomaterials.2014.07.022. [DOI] [PubMed] [Google Scholar]
- [495].Banerjee A, Bernoulli D, Zhang H, Yuen M-F, Liu J, Dong J, Ding F, Lu J, Dao M, Zhang W, Lu Y, et al. Science. 2018;360:300. doi: 10.1126/science.aar4165. [DOI] [PubMed] [Google Scholar]
- [496].Yang Y, Wang K, Gu X, Leong KW. Engineering. 2017;3:36. doi: 10.1016/J.ENG.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [497].Ha J-G, Song YS, Jung S, Jang S, Kim Y-K, Bai SJ, Park J-H, Lee S-K. Biotechnol Lett. 2017;39:849. doi: 10.1007/s10529-017-2307-4. [DOI] [PubMed] [Google Scholar]
- [498].Mukaibo H. Chem Rec. 2019;19:859. doi: 10.1002/tcr.201800099. [DOI] [PubMed] [Google Scholar]
- [499].Carballido-López R. Microbiol Mol Biol Rev. 2006;70:888. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [500].Caille N, Thoumine O, Tardy Y, Meister J-J. J Biomech. 2002;35:177. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- [501].Rivnay J, Owens RM, Malliaras GG. Chem Mater. 2014;26:679. [Google Scholar]
- [502].Simon DT, Gabrielsson EO, Tybrandt K, Berggren M. Chem Rev. 2016;116:13009. doi: 10.1021/acs.chemrev.6b00146. [DOI] [PubMed] [Google Scholar]
- [503].Tullii G, Giona F, Lodola F, Bonfadini S, Bossio C, Varo S, Desii A, Criante L, Sala C, Pasini M, Verpelli C, et al. ACS Appl Mater Interfaces. 2019;11:28125. doi: 10.1021/acsami.9b08822. [DOI] [PMC free article] [PubMed] [Google Scholar]