Abstract
Inflammatory diseases are frequently treated with Janus kinase (JAK) inhibitors to diminish cytokine signaling. These treatments can lead to inadvertent immune suppression and may increase the risk of viral infection. Tyrosine kinase 2 (TYK2) is a JAK family member required for efficient type I interferon (IFN-α/β) signaling. We report here that selective TYK2 inhibition preferentially blocked potentially detrimental type I IFN signaling whereas IFN-λ-mediated responses were largely preserved. In contrast, the clinically used JAK1/2 inhibitor baricitinib was equally potent in blocking IFN-α/β or IFN-λ-driven responses. Mechanistically, we showed that epithelial cells did not require TYK2 for IFN-λ-mediated signaling or antiviral protection. TYK2 deficiency diminished IFN-α-induced protection against lethal influenza virus infection in mice but did not impair IFN-λ-mediated antiviral protection. Our findings suggest that selective TYK2 inhibitors used in place of broadly acting JAK1/2 inhibitors may represent a superior treatment option for type I interferonopathies to counteract inflammatory responses while preserving antiviral protection mediated by IFN-λ.
Introduction
Janus kinase (JAK) inhibitors are successfully used to treat inflammatory diseases such as type I interferonopathies (1) or psoriasis (2). However, 89 % of type I interferonopathy patients treated with baricitinib, a JAK1/2 inhibitor, suffer from complications including herpes zoster, BK viremia, viral gastroenteritis, and viral infections of the upper respiratory tract (1). JAKs are involved in the signal transduction pathways of several cytokines including interferons (IFNs) (3). The heterodimeric receptor complex for type I IFN (IFN-α/β) consists of IFNAR1 and IFNAR2 (4), whereas the type III IFN (IFN-λ) receptor is formed by IFNLR1 and IL10RB (5).
Tyrosine kinase 2 (TYK2), a member of the JAK family, is physically associated with IFNAR1 (6) and IL10RB (7, 8), whereas JAK1 is associated with IFNAR2 (9) and IFNLR1 (10). Upon ligand binding, IFN receptor-associated JAKs undergo phosphorylation and become activated to phosphorylate receptor-associated signal transducer and activator of transcription 1 (STAT1) and STAT2 which, in complex with interferon regulatory factor 9 (IRF9), bind to IFN-stimulated response elements (ISRE) in the promotor regions of IFN-stimulated genes (ISGs) such as MX1 or ISG15, inducing their transcription (4, 11).
Several patients with TYK2 deficiencies were reported to have surprisingly mild mycobacterial and/or viral infections and were free of inflammatory bowel disease (12, 13). These patients tolerated live attenuated viral vaccines and were seropositive for antibodies against influenza A viruses, indicating that they were able to mount efficient antiviral immune responses (12). This clinical picture was attributed to the leaky phenotype of the TYK2 deficiencies, resulting in residual responses to IFN-α/β, IL-12, IL-23 and IL-10 and seemingly normal responses to IL-21, IL-27, IFN-γ and IFN-λ (13). In agreement with these clinical findings, Tyk2-deficient mice showed only partially impaired responses to TYK2-dependent cytokines (14). At this time, only a single patient with partial JAK1 deficiency has been described (15), and Jak1 deficiency in mice results in postnatal lethality (16).
Since its discovery in 2003 (17, 18), the type III IFN system has been shown to protect mucosal barriers from viral, fungal and protozoal infections (11). Pegylated IFN-λ has been suggested as prophylactic (19–21) and is currently under clinical evaluation for early treatment against COVID-19 (22). IFN-λ has important functions in protecting the intestinal tract (23), the respiratory tract (24, 25) and the reproductive tract (26, 27) against viral infections, and contributes to hepatitis C virus resistance in humans (28). In mucosa-rich tissues, IFN-λ can largely compensate for a defective type I IFN system (24, 29) and induces less tissue damage and inflammation compared with IFN-α/β (25, 30, 31). However, IFN-λ can also impair lung repair after viral infection (32, 33), can increase susceptibility to bacterial infections (11) and promoted immune dysregulation in a murine lupus model (34).
In addition to acting on epithelial cells and protecting barrier surfaces from infection, several recent studies demonstrated that IFN-λ can also modulate the activity of neutrophils (35, 36). By acting directly on neutrophils, IFN-λ was shown to resolve inflammation in a collagen-induced arthritis model (37), counteract fungal pneumonia (38), reduce inflammation in a DSS-induced colitis model (39), and avoid excessive inflammation during influenza A virus infection (25). These observations suggest that IFN-λ is critical to protecting barrier surfaces against a variety of pathogens without inducing excessive inflammation and tissue damage.
TYK2 appears to be required for full-scale IFN-α/β signaling (40, 41), but its role in IFN-λ-mediated gene expression is less clear (11). One study concluded that TYK2 was dispensable for antiviral protection mediated by IFN-λ (12), whereas another study identified TYK2 as an essential positive regulator of IFN-λ-mediated gene expression (42). Other studies have also proposed a role for JAK2 in IFN-λ-mediated signaling (39, 43, 44).
Here we found that the newly developed selective TYK2 inhibitor BMS-986165 (2, 45) preferentially blocked potentially noxious type I IFN signaling but largely preserved IFN-λ-mediated responses. To explain this unexpected observation, we evaluated the responses to IFN-λ treatment in TYK2-deficient primary mouse epithelial cell cultures, mouse neutrophils and two human cell lines. We showed that epithelial cells did not require TYK2 for full-scale IFN-λ-mediated gene expression or antiviral protection, and that mouse neutrophils and human HAP1 cells were only moderately influenced by the complete loss of TYK2. IFN-λ-mediated protection of mice against lethal influenza A virus infection was not diminished in TYK2-deficient mice, whereas IFN-α-mediated protection was abolished. Our findings suggest that specific inhibition of TYK2 might represent a superior treatment option for type I interferonopathies as such drugs provide the possibility to preferentially block IFN-α/β signaling while preserving the antiviral barrier protection mediated by IFN-λ.
Results
IFN-α-but not IFN-λ-mediated gene expression depends on TYK2 in mouse epithelial cells
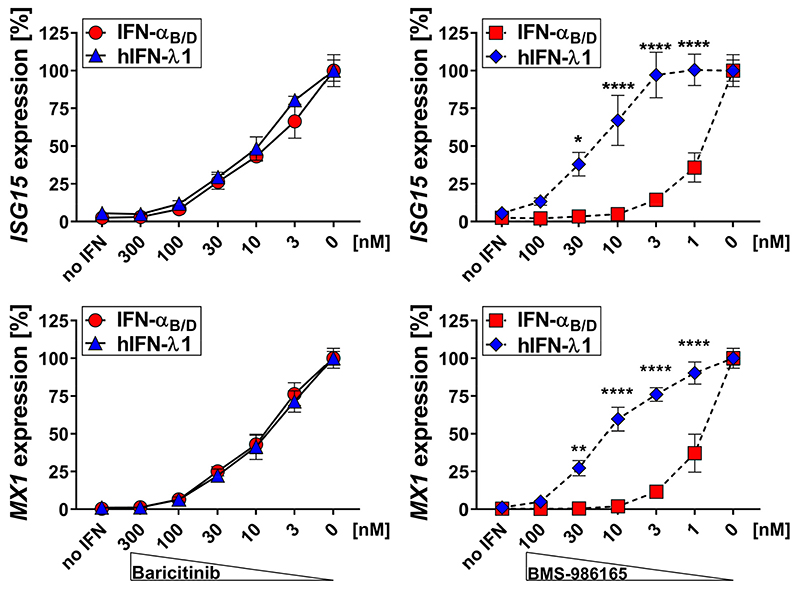
We first determined if inhibition of specific JAK family members would preferentially affect type I versus type III IFN-mediated responses. We treated A549 cells, a human alveolar cell carcinoma-derived cell line (46), with increasing concentrations of either the JAK1/2 inhibitor baricitinib or the selective TYK2 inhibitor BMS-986165 and quantified IFN-mediated gene expression after stimulation with either type I (IFN-αB/D) or type III IFN (hIFN-λ1). Baricitinib inhibited both type I and type III IFN signaling equally well, whereas BMS-986165 preferentially inhibited type I IFN-mediated gene expression. Up to 30-fold higher concentrations of the selective TYK2 inhibitor BMS-986165 were required to reduce type III IFN-mediated gene expression to a similar extent (Fig. 1).
Figure 1. BMS-986165 preferentially inhibits type I compared with type III IFN-mediated gene expression in human A549.
Human A549 cells were treated with the indicated concentrations of the JAK1/2 inhibitor baricitinib (left panels) or the selective TYK2 inhibitor BMS-986165 (right panels) for 1.5 h before 0.03 ng/ml IFN-αB/D or 10 ng/ml hIFN-λ1 was added to the medium. After 4 h, gene expression levels of the representative ISGs ISG15 and MX1 were quantified relative to the housekeeping gene HPRT. Inhibition of IFN-mediated gene expression is indicated as percentages of maximum gene expression levels without inhibitor. Symbols represent means ± SEM, data are pooled from two independent experiments with a total n = 4-7. Statistical analysis: ****P<0.0001, **P<0.01 and * P<0.05, by two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test.
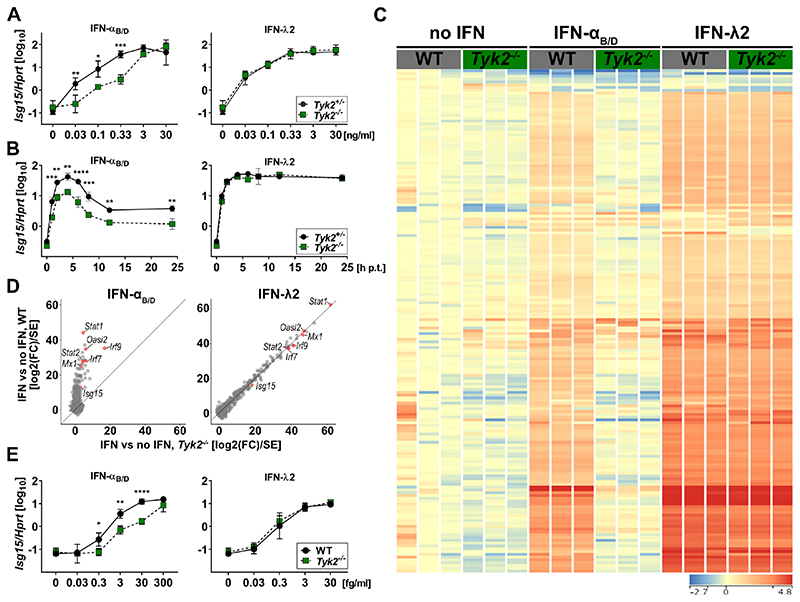
To corroborate the subordinate role of TYK2 in IFN-λ-mediated signaling and to evaluate whether the inhibition of type III IFN-mediated gene expression at high concentrations of BMS-986165 was due to off-target pan-JAK inhibition, we generated primary airway epithelial cells (AECs) from mice lacking TYK2 and performed dose-response experiments with IFN-αB/D and IFN-λ2. Tyk2-deficient AECs showed a dose-dependent defect for IFN-α-mediated gene expression, as expected (Fig. 2A, left panel). In contrast, no defect in IFN-λ-induced gene expression was observed (Fig. 2A, right panel). Next, we investigated the kinetics of gene expression in Tyk2-deficient and control AECs. We stimulated AECs with either IFN-αB/D or IFN-λ2 for 1 h and quantified IFN-mediated expression of Isg15 over time. We observed no defect in IFN-λ-mediated expression of Isg15 in Tyk2-deficient AECs but noted a pronounced deficiency of IFN-αB/D signaling (Fig. 2B).
Figure 2. IFN-α-but not IFN-λ-mediated expression of ISGs depends on TYK2 in primary mouse epithelial cells.
(A). Primary airway epithelial cells (AECs) derived from Tyk2+/− or Tyk2−/− mice were treated with increasing concentrations of IFN-αB/D or IFN-λ2 for 4 h. Expression of Isg15 was quantified relative to Hprt. Symbols represent means ± SD, data representative of two independent experiments with n = 2 per experiment. Statistical analysis: ***P<0.001, **P<0.01 and *P<0.05, by two-way ANOVA with Tukey’s multiple comparisons test.
(B)AECs derived from Tyk2+/− or Tyk2−/− mice were treated with 10 ng/ml of IFN-αB/D or IFN-λ2 for 1 h and analyzed at the indicated time points. Expression of Isg15 was quantified relative to Hprt. Symbols represent means ± SD (n = 2). Statistical analysis: ****P<0.0001, ***P<0.001 and **P<0.01, by two-way ANOVA with Sidak’s multiple comparisons test.
(C)Heat map of genes differentially expressed relative to mock (two-way ANOVA, fold change >2; P<0.01) in WT or Tyk2−/− primary mouse AECs (n = 3) treated with 0.3 ng/ml IFN-αB/D or IFN-λ2 for 4 h. Color range depicted as log2 scale.
(D)Scatter plot of IFN induced genes (fold change >2 over mock in any IFN treatment at any concentration, n = 3). Data used for plot corresponds to the Wald statistic value (log2(fold change) divided by standard error), obtained from DESeq2. Lines indicate the x=y diagonal.
(E) Mini-gut organoids derived from WT or Tyk2−/− mice were treated with increasing concentrations of IFN-αB/D or IFN-λ2 for 4 h. Expression of Isg15 was quantified relative to Hprt. Symbols represent means ± SD, data are pooled from three independent experiments with a total n = 3. Statistical analysis: ****P<0.0001, **P<0.01 and *P<0.05, by two-way ANOVA with Sidak’s multiple comparisons test.
Interferons are known to regulate the transcription of hundreds of genes. We therefore performed RNA-seq analysis of Tyk2-deficient AECs to determine whether a subset of ISGs would require TYK2 for efficient expression upon IFN-λ stimulation. Tyk2 deficiency in AECs led to a global defect in IFN-α-mediated gene expression, whereas IFN-λ-mediated gene expression was not significantly affected (Fig. 2C). We validated this observation by analyzing IFN-induced genes with Wald statistics, which uses log2-transformed fold change values corrected for their variance. Gene induction by IFN-α was nearly absent in Tyk2−/− cells (Fig. 2D, left panel), whereas gene induction by IFN-λ was indistinguishable between Tyk2−/− and WT cells, illustrated by values being aligned with the diagonal (Fig. 2D, right panel). We used a mini-gut organoid culture system (47) to determine whether IFN-λ signaling was equally independent of TYK2 in mouse intestinal epithelial cells. Again, we found that Isg15 expression was independent of TYK2 in IFN-λ-treated cells but not in cells treated with IFN-α (Fig. 2E).
Taken together, these data indicate that, in sharp contrast to IFN-α, IFN-λ is able to induce full-scale global gene expression in primary mouse epithelial cells in the absence of TYK2. Likewise, selective pharmacological inhibition of TYK2 in human epithelial cells preferentially inhibits gene expression induced by IFN-α compared with IFN-λ.
TYK2 moderately influences IFN-λ-mediated gene expression in mouse neutrophils
Several recent studies demonstrated that IFN-λ can modulate the activity of neutrophils in mice (25, 35–39). We performed RNA-seq analyses with purified bone marrow (BM)-derived mouse neutrophils that were treated with either IFN-αB/D or IFN-λ2 to confirm that both treatments induced the expression of an overlapping subset of ISGs (Fig. 3A and Fig. S1A). In line with published data, we found that many pro-inflammatory genes, like Tnf and Il-6 (25), were regulated exclusively by IFN-α (Fig. 3B and Fig. S1B). However, some prominent antiviral genes like Mx1 were not induced by IFN-λ in neutrophils. Of note, Mx1 is known to be regulated by IFN-λ in epithelial cells (24, 48) (Fig. S1C). To test whether the differential regulation of Mx1 by IFN-λ in neutrophils was simply a matter of ligand dose, we treated freshly isolated neutrophils with a high concentration (1 μg/ml) of IFN-λ2. However, IFN-λ still failed to enhance the expression levels of Mx1, in contrast to Isg15 (Fig. 3C), indicating that the integration of the IFN-λ signal may differ between epithelial cells and neutrophils. Of note, IFN-αB/D treatment efficiently induced the expression of Mx1 in neutrophils, indicating that the Mx1 gene locus is not silenced in a cell type-specific manner (Fig. 3C).
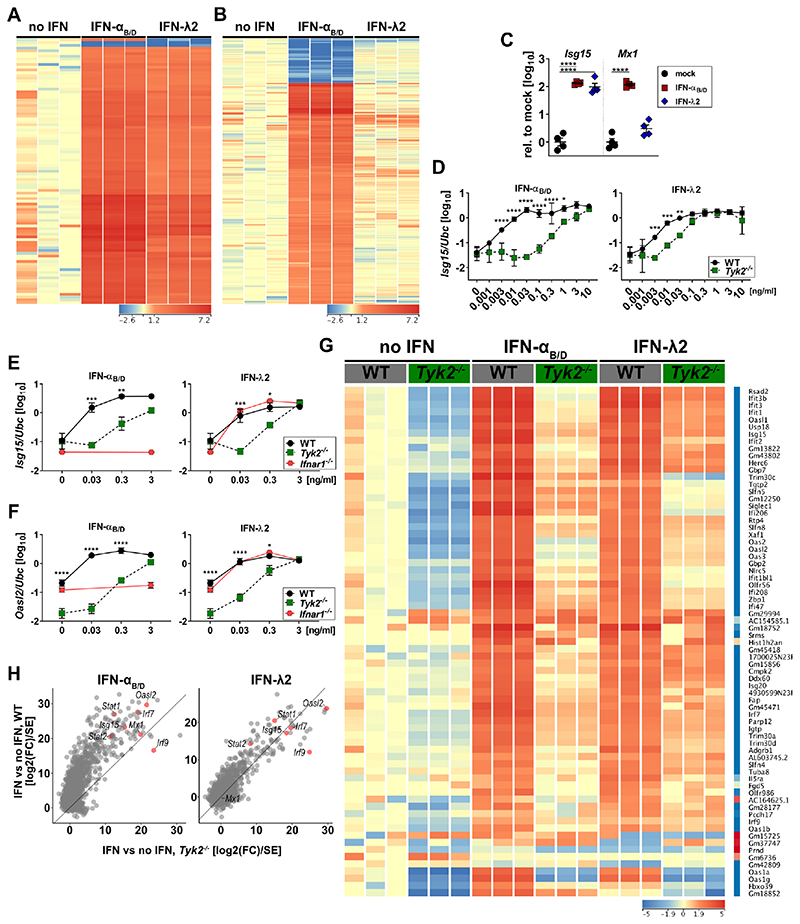
Figure 3. Loss of TYK2 moderately influences IFN-λ-mediated gene expression in mouse neutrophils.
(A-B) BM-derived neutrophils from three individual mice were treated with 3 ng/ml IFN-αB/D or IFN-λ2 for 4 h and then processed for RNA-seq analysis. Heat map shows genes regulated (one-way ANOVA, P<0.01; Fold Change >4 relative to mock) by both types of IFN (A) or exclusively by IFN-αB/D (B). Color range depicted as log2 scale.
(C) WT BM-derived neutrophils from four individual mice were treated with 1 μg/ml of IFN-αB/D or IFN-λ2 for 4 h. Expression levels of Isg15 and Mx1 were quantified relative to Ubc and are shown normalized to untreated controls. Statistical analysis: Ordinary one-way ANOVA with Tukey’s multiple comparisons test with ****p<0.0001.
(D) Differentiated WT or Tyk2 Hoxb8 neutrophils were treated (n = 3) with increasing concentrations of IFN-αB/D or IFN-λ2 for 4 h. Expression levels of Isg15 were quantified relative to Ubc. Symbols represent means ± SD, data representative of two independent experiments. Statistical analysis: ****P<0.0001, ***P<0.001, **P<0.01 and *P<0.05, by two-way ANOVA with Sidak’s multiple comparisons test.
(E-F) WT, Tyk2 or Ifnar1 BM-derived neutrophils were treated with increasing concentrations of IFN-αB/D or IFN-λ2 for 4 h (mock, n = 3-4; 0.03 - 0.3 ng/ml, n = 2-3; 3 ng/ml, n = 1-2). Expression levels of Isg15 (e) and Oasl2 (f) were quantified relative to Ubc. Symbols represent means ± SD, data representative of three independent experiments. Statistical analysis: ****P<0.0001, ***P<0.001, **P<0.01 and *P<0.05, by two-way ANOVA with Sidak’s multiple comparisons test. Asterisks indicate significant differences between WT and Tyk2−/−.
(G) BM-derived neutrophils from three individual mice were treated with 0.1 ng/ml IFN-αB/D or IFN-λ2 for 4 h and then processed for RNA-seq analysis. Heat map of IFN-λ regulated genes (two-way ANOVA, P<0.01; Fold Change >4 relative to each mock). Color range depicted as log2 scale. (H) Scatter plot of IFN induced genes (fold change >2 over mock in any IFN treatment at any concentration, n = 3). Data used for plot corresponds to the Wald statistic value (log2(fold change) divided by standard error), obtained from DESeq2. Lines indicate the x=y diagonal
Differentiated mouse neutrophils can be generated ex vivo using the Hoxb8 system (49, 50). Hoxb8 neutrophils started to express the specific IFN-λ receptor chain 1 (Ifnlr1) as soon as they differentiated from their precursors (Fig. S1D) and became readily responsive to IFN-αB/D or IFN-λ2 on day 4 post induction of differentiation (Fig. S1E). To evaluate the role of TYK2 for IFN-λ-mediated gene regulation in neutrophils, we generated Tyk2-deficient Hoxb8 neutrophils, treated them with increasing concentrations of either IFN-αB/D or IFN-λ2 and measured IFN-mediated gene expression by RT-qPCR. Unexpectedly, we observed a clear dose-dependent defect of Tyk2-deficient neutrophils in IFN-λ2-mediated expression of Isg15 (Fig. 3D). We excluded the possibility that the decreased IFN-λ responsiveness of the Tyk2- deficient cells was due to reduced differentiation-induced expression of IFN-λ receptors (Fig. S1F) and used freshly isolated BM-derived neutrophils from WT or Tyk2-deficient mice to confirm the results obtained with the Hoxb8 culture system (Fig. S1G).
Steady-state expression levels of Stat1, Stat2 and Irf9 can be influenced by tonic IFN signaling as they are ISGs themselves (51). Thus, TYK2-dependent defects in tonic type I IFN signaling could indirectly influence gene expression triggered by IFN-λ. To test for such putative indirect negative effects, we measured the IFN-λ response of freshly isolated neutrophils from Tyk2−/−, Ifnar1−/− and WT mice in parallel. Unlike Tyk2−/− neutrophils, cells from Ifnar1−/− mice showed no reduced responsiveness to IFN-λ2 (Fig. 3E-F), arguing against such indirect effects. Interestingly, we found that Oasl2 and Irf9 showed reduced basal expression levels in Tyk2-deficient neutrophils. However, relative to the respective untreated controls, their expression levels were equally well upregulated by IFN-λ in WT and Tyk2−/− cells (Fig. 3F and Fig. S1I). In contrast, Isg15 and Stat2 (Fig. 3E and Fig. S1H) showed normal baseline expression but exhibited some degree of TYK2 dependency for IFN-λ-mediated gene regulation.
To evaluate globally the TYK2 dependency of IFN-λ-mediated signaling in neutrophils, we performed RNA-seq analysis of freshly isolated WT or Tyk2−/− neutrophils treated with either IFN-αB/D or IFN-λ2. We observed reduced basal expression levels for the vast majority of ISGs in Tyk2-deficient neutrophils (Fig. 3G). However, analysis of this dataset by Wald statistics, which are based on relative changes over untreated WT or Tyk2−/− controls, revealed that the response to IFN-λ was only moderately affected in Tyk2-deficient neutrophils (Fig. 3H, right panel), in contrast to type I IFN-mediated gene regulation (Fig. 3H, left panel).
Taken together these data demonstrate that the integration of IFN signals differs between epithelial cells and neutrophils, and that loss of TYK2 leads to a global reduction in basal ISG expression levels in neutrophils. Importantly, we found that type I IFN- but not type III IFN-mediated gene regulation strongly depends on TYK2 in mouse neutrophils.
TYK2 moderately contributes to IFN-λ-induced gene expression in human HAP1 cells
Next, we wanted to investigate whether human neutrophils, similar to mouse neutrophils, might differ from epithelial cells regarding IFN-λ-mediated gene expression. However, naïve human neutrophils isolated from peripheral blood did not upregulate the expression of ISGs upon stimulation with human IFN-λ1 (Fig. S2A).
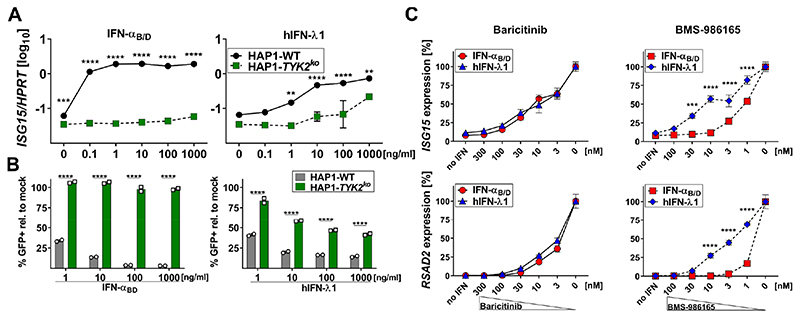
To evaluate the TYK2 dependency of IFN signaling in a human-derived cell line that can respond to IFN-λ (12), we used HAP1 cells, a near-haploid human cell line derived from KBM-7 cells (52) that was originally isolated from a male patient with chronic myeloid leukemia in blast crisis (53). Of note, even though this cell line originated from a myeloid progenitor, it does not represent a surrogate model for neutrophils. Interestingly, one study found that TYK2-deficient HAP1 cells (HAP1-TYK2ko) showed no decrease in IFN-λ-mediated protection against infection with a vesicular stomatitis reporter virus (VSV-GFP) (12), whereas another study identified TYK2 in a genome-wide screening approach to be an essential positive regulator of IFN-λ-mediated gene expression in HAP1 ISRE-GFP reporter cells (42). When we stimulated HAP1-WT and HAP1-TYK2ko cells with increasing concentrations of IFN-αB/D, we found that ISG15 expression (Fig. 4A, left) and antiviral protection against VSV-GFP (Fig. 4B, left) were completely abolished in TYK2-deficient cells. In contrast, IFN-λ-mediated expression of ISG15 (Fig. 4A, right) and antiviral protection against VSV-GFP (Fig. 4B, right) was decreased but not abolished in TYK2-deficient cells. When we measured IFN-induced expression of MX1, we found moderate induction in HAP1-WT cells by IFN-αB/D and, to a lesser extent, by IFN-λ1 (Fig. S2B).
Figure 4. TYK2 inhibition preferentially inhibits type I IFN-mediated gene expression in human HAP1.
(A)HAP1-WT or HAP1-TYK2ko cells were treated with increasing concentrations of IFN-αB/D or hIFN-λ1 for 4 h. Expression levels of ISG15 were quantified relative to HPRT. Symbols represent means ± SD, data representative of two independent experiments with n = 2 per experiment. Statistical analysis: ****P<0.0001, ***P<0.001 and **P<0.01, by two-way ANOVA with Sidak’s multiple comparisons test.
(B)HAP1-WT or HAP1-TYK2ko cells were treated with increasing concentrations of IFN-αB/D or hIFN-λ1 for 16 h before infection with VSV-GFP (MOI~1) for 6 h. Data is shown as percentage of GFP-positive cells normalized to mock-treated controls infected with VSV-GFP. Bars represent means ± SD, data representative of two independent experiments with n = 2 per experiment. Statistical analysis: ****P<0.0001, by two-way ANOVA with Sidak’s multiple comparisons test.
(C) HAP1-WT cells were treated with the indicated concentrations of the JAK1/2 inhibitor baricitinib (left panels) or the selective TYK2 inhibitor BMS-986165 (right panels) for 1.5 h before 0.1 ng/ml IFN-αB/D or 10 ng/ml hIFN-λ1 was added to the medium. After 4 h gene expression levels of the representative ISGs ISG15 and RSAD2 were quantified relative to the housekeeping gene HPRT. JAK-inhibitor-mediated inhibition of IFN-mediated gene expression is indicated as percentages of maximum gene expression levels without inhibitor. Symbols represent means ± SEM, data are pooled from three independent experiments with a total n = 6-10. Statistical analysis: ****P<0.0001 and ***P<0.001, by two-way ANOVA with Sidak’s multiple comparisons test.
We tested whether transient TYK2 inhibition would preferentially block IFN-α- mediated gene expression in HAP1 cells. JAK1/2 inhibition by baricitinib blocked signaling by both types of IFN equally well (Fig. 4C, left panels), but TYK2-specific inhibition by BMS-986165 preferentially inhibited type I IFN-mediated gene expression, with ten-fold higher concentrations of the inhibitor required for a comparable reduction of type III IFN-mediated stimulation (Fig. 4C, right panels). These findings indicate that in HAP1 cells, signaling by type I IFN is more strongly dependent on TYK2 than signaling by IFN-λ, although TYK2 is required for full-scale IFN-λ-mediated gene expression and antiviral protection in this cell line.
IFN-λ-responses are TYK2-independent in human epithelial cells
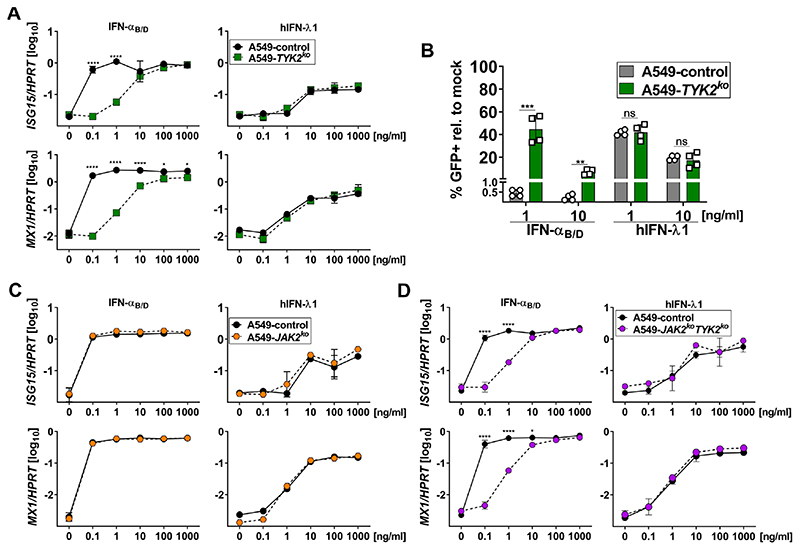
A549 is an epithelial cell line established from a male alveolar cell carcinoma (46). To address whether human epithelia require TYK2 for IFN-λ-responses, we generated TYK2-deficient A549 single cell clones (A549-TYK2ko) using CRISPR/Cas9 (Fig. S3A). A549-TYK2ko cells, in line with mouse-derived cells but in contrast to human HAP1 cells, showed a dose-dependent defect in IFN-αB/D-mediated expression of ISG15 and MX1 (Fig. 5A, left panels), but IFN-λ-mediated expression of ISG15 or MX1 was TYK2-independent (Fig. 5A, right panels), which is similar to our findings in primary mouse epithelial cells (Figs. 2A and 2E). Furthermore, IFN-λ-mediated antiviral protection of A549 cells against VSV-GFP was not affected by TYK2 deficiency, in contrast to IFN-αB/D-mediated protection (Fig. 5B). To investigate the kinetics of IFN-mediated gene expression in TYK2-deficient A549 cells, we pulsed cells with IFN-αB/D or IFN-λ1 for 1 h and quantified IFN-mediated expression of ISG15 and MX1 over time, which is similar to experiments with primary mouse AECs (Fig. 2B). We again observed no defect in IFN-λ-mediated expression of MX1 in TYK2-deficient A549 cells but noted a pronounced reduction of the type I IFN-mediated signaling capacity (Fig. S3B). Of note, in contrast to primary mouse AECs (Fig. 2B), we did not find a similar rapid decline in IFN-αB/D-mediated gene expression for time points later than four hours after treatment, which might indicate that type I IFN-specific negative feedback regulation is not functional in A549 cells.
Figure 5. IFN-λ-mediated gene expression is independent of TYK2 and JAK2 in human A549 cells.
(A)A549 control and TYK2ko cells were treated with increasing concentrations of IFN-αB/D or hIFN-λ1 for 4 h. Expression of ISG15 and MX1 is shown relative to HPRT. Symbols represent means ± SD. Statistical analysis: Two-way ANOVA with Tukey’s multiple comparisons test with ****p<0.0001and *p<0.05. Data representative of two independent experiments with n = 2 per experiment.
(B)A549 control and TYK2ko cells were treated with 1 ng/ml or 10 ng/ml of either IFN-αB/D or hIFN-λ1 for 16 h before infection with VSV-GFP (MOI~1) for 6 h. Data is shown as percentage of GFP-positive cells normalized to mock-treated controls infected with VSV-GFP. Bars represent means ± SD, data pooled from two independent experiments with a total n = 4. Statistical analysis: Unpaired t test with ***p<0.001, **p<0.01, ns = not significant.
(C-D) A549 control and JAK2ko cells (C) or JAK2koTYK2ko cells (D) were treated with increasing concentrations of IFN-αB/D or hIFN-λ1 for 4 h. Expression of ISG15 and MX1 is shown relative to HPRT. Symbols represent means ± SD. Statistical analysis: Two-way ANOVA with Tukey’s multiple comparisons test with ****p<0.0001 and *p<0.05. Data representative of two independent experiments with n = 2 per experiment.
As some studies suggested a role for JAK2 in IFN-λ-mediated gene regulation (39, 43), we tested whether the JAK2 inhibitor HBC (1,2,3,4,5,6,-Hexabromocyclohexane) might influence IFN-λ signaling. However, HBC did not reduce IFN-λ-triggered expression of Isg15 and Mx1 in primary mouse AECs (Fig. S3C) or expression of Isg15 and Oasl2 in BM-derived WT or Tyk2−/− neutrophils (Figs. S3D-E). In contrast, the pan-JAK inhibitor Pyridone 6 (Py6) efficiently reduced IFN-mediated ISG expression in both mouse AECs and neutrophils.
To further investigate the role of JAK2 for IFN-λ-mediated gene regulation, we generated A549-JAK2ko cells (Fig. S3A). Upon treatment with increasing concentrations of IFN-αB/D or IFN-λ1, A549 control and JAK2ko cells upregulated the expression levels for ISG15 and MX1 equally well (Fig. 5C), contrasting previous findings (39, 43) and arguing against an involvement of JAK2 in IFN-λ-mediated expression of ISGs in epithelial cells. To exclude the possibility that JAK2 and TYK2 might compensate for each other in epithelial cells, we generated A549 cells deficient for both kinases (A549-JAK2koTYK2ko). Again, when we treated A549-JAK2koTYK2ko cells with increasing concentrations of IFN-λ1 they upregulated the expression levels of ISG15 and MX1 to the same extent as the control cells (Fig. 5D, right panels).
Taken together these data showed that IFN-λ-mediated gene expression and antiviral protection in human epithelial cells are independent of TYK2 and JAK2.
IFN-λ-mediated protection of mice against influenza A virus-induced disease is TYK2-independent
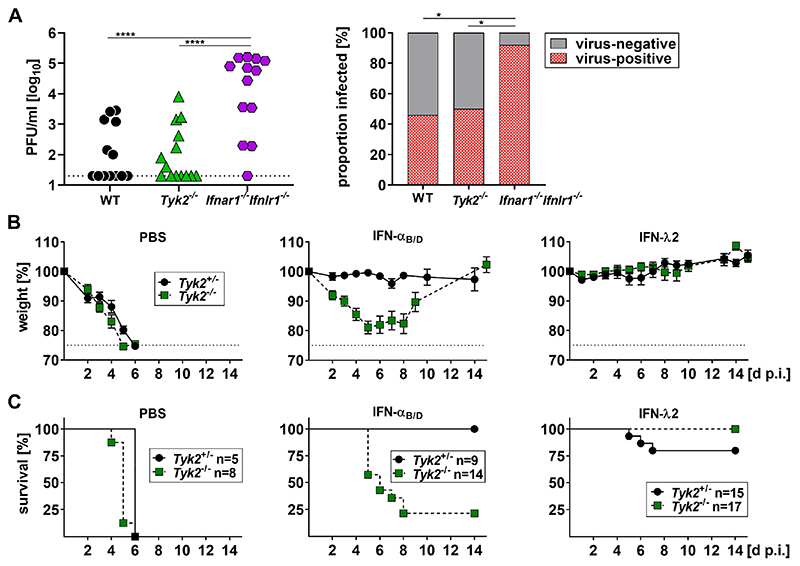
To determine whether Tyk2-deficient mice show increased susceptibility to respiratory viruses, we used an established influenza A virus infection model (24) and infected the upper respiratory tract of WT, Tyk2−/− and Ifnar1−/−Ifnlr1−/− mice with a low dose of influenza A virus strain Udorn. On day 5 post infection, the viral load in upper airways (Fig. 6A, left panel) and the percentage of virus-positive animals (Fig. 6A, right panel) did not differ significantly between Tyk2−/− and WT mice. In contrast, Ifnar1−/−Ifnlr1−/− mice contained significantly more virus in the upper respiratory tract than WT or Tyk2−/− mice and a significantly higher fraction of Ifnar1−/−Ifnlr1−/− mice were productively infected, suggesting that the loss of both type I and type III IFN signaling but not Tyk2 deficiency increases the susceptibility of mice to respiratory viruses.
Figure 6. IFN-λ-mediated protection of mice against influenza A virus-induced disease is not dependent on TYK2.
(A) WT (n=13), Tyk2−/− (n=14) or Ifnar1−IfnlrΓ− (n=13) mice were infected with 200 PFU of the influenza A virus strain Udorn (H3N2) in a 10 μl volume. Viral load in the upper airways was determined 5 d p.i. by plaque-assay. All mice carried two functional alleles of the IFN-regulated influenza virus resistance gene Mx1. Symbols represent individual mice. Statistical analysis left panel: One-way ANOVA with Tukey’s multiple comparisons test with ****p<0.0001. Statistical analysis right panel: Fisher’s exact test with *p<0.05.
(B-C) Tyk2+/− or Tyk2−/− mice were treated intranasally with PBS or 0.66 μg of either IFN-αB/D or IFN-λ2 in 30 μl. After 18 h, mice were infected with 160 PFU of a highly virulent variant of the influenza A virus strain PR8 (hvPR8; H1N1) in a 40 μl volume. All mice used for these experiments carried one functional allele of the IFN-regulated influenza virus resistance gene Mx1. Weight loss (B) and survival (C) was monitored for 14 days. Symbols in (B) represent means ± SEM; Group sizes are indicated in (C). Pooled data from three experiments are shown.
Next, we tested whether IFN-λ2 treatment would still protect against a lethal influenza A virus challenge if the mice lacked TYK2. All mice used for these experiments carried one functional allele of the IFN-regulated influenza virus resistance gene Mx1 and were either heterozygous (Tyk2+/−) or homozygous (Tyk2−/−) for the defective Tyk2 allele. The animals were treated either with PBS, IFN-αB/D or IFN-λ2 at 18 hours prior to infection with a highly virulent variant of the influenza A virus strain PR8. As shown in Figure 6B (left panels), PBS-treated animals quickly succumbed to the infection regardless of whether they carried a functional Tyk2 allele. IFN-αB/D-treated animals were protected against disease and survived the infection only if they had a functional copy of Tyk2 (Fig. 6B, middle panels). In contrast, Tyk2-deficient mice pretreated with IFN-λ2 were all protected against disease and survived the infection (Fig. 6B, right panels).
Taken together these data indicate that TYK2 deficiency does not increase the susceptibility to influenza A virus infection and does not reduce the protective potential of IFN-λ during a lethal viral challenge.
Discussion
Here we demonstrate that TYK2 deficiency effectively blocks type I IFN signaling but does not negatively influence IFN-λ-mediated gene induction or antiviral protection. Selective inhibition of TYK2 with BMS-986165 preferentially affected type I IFN signaling, while largely preserving type III IFN signaling. This observation highlights fundamental differences in IFN type I and type III signaling and is relevant for various clinical conditions including autoinflammatory type I interferonopathy in which JAK inhibitors are used to limit excessive immune responses. Such disease can be caused by monogenetic defects that result in constitutive activation of the type I IFN signaling pathway (1, 54–56). Although the currently used pan-JAK inhibitors are effective at reducing type I IFN-driven inflammation, they predispose treated patients to viral infections (1). Our data suggests that TYK2-specific inhibitors may represent an improvement of the current treatment.
The signaling pathways downstream of the type I and type III IFN receptors show a high degree of similarity and involve activation of several protein kinases, including JAK1 and TYK2 (4, 11, 57). Type I IFN signaling is strongly impaired in TYK2-deficient mice and humans, but some residual signaling does occur in response to high concentrations of type I IFN (13, 14). Whether type III IFN also requires TYK2 for signaling has to date been unclear. Our work with primary epithelial cells from the airways and the intestinal tract of wild-type and Tyk2-deficient mice clearly demonstrated that TYK2 is dispensable for full-scale IFN-λ signaling. Furthermore, experiments with A549 cells in which the TYK2 gene had been deleted showed that IFN-λ signaling in human epithelial cells is equally independent of TYK2. Finally, IFN-λ retained its protective activity if applied prophylactically to Tyk2−/− mice before infection with highly pathogenic influenza A virus. Under identical experimental conditions, IFN-α was only active in wild-type but not Tyk2−/− mice, unequivocally demonstrating that type I and type III IFN differ with regard to requiring TYK2 for signal transduction.
A previous study showed that IFN-λ protected human HAP1 cells comparably well against infection with VSV whether or not they carried a functional TYK2 gene (12). However, a genome wide trap mutagenesis screen using the same parental cell line identified TYK2 as an essential positive regulator for IFN-λ-mediated gene induction (42). Our work can explain this apparent inconsistency and revealed that IFN-λ responses in HAP1 cells are partially dependent on TYK2. Thus, depending on the experimental setup, the importance of TYK2 in IFN-λ signaling can easily be overestimated when this particular cell line is employed.
While IFN-λ mainly acts on epithelial cells, recent studies showed that mouse neutrophils can also respond to IFN-λ (reviewed in (11)). In line with other studies (34), we found that mouse and human neutrophils differ in their responsiveness to IFN-λ. Neutrophils isolated from the bone marrow of mice readily responded to IFN-λ stimulation, whereas human neutrophils isolated from peripheral blood did not. Previous work suggests that regulation of IFN-λ receptor expression in human neutrophils could be dynamic, as the IFNLR1 was up-regulated in human neutrophils upon co-cultivation with Aspergillus fumigatus (38). The mouse Hoxb8 neutrophil culture system may represent a useful tool to study the up-regulation of the Ifnlr1 gene during differentiation.
Interestingly, IFN-λ induced a substantially smaller subset of ISGs in mouse neutrophils than IFN-α, confirming a previous study which showed that IFN-λ induces fewer pro-inflammatory genes than type I IFN in this cell type (25). Besides pro-inflammatory genes, we found that IFN-λ does not upregulate some well-characterized antiviral ISGs, like Mx1, in neutrophils. To date it is unclear how and why IFN-λ has evolved to induce distinct gene expression profiles in different cell types. Most likely, IFN-λ evolved to serve unique functions in neutrophils that differ from those in epithelial cells.
Comparisons of the IFN-λ responses in neutrophils from WT and Tyk2-deficient mice revealed a reduced baseline expression of most ISGs in knockout cells. Importantly, however, IFN-λ increased the relative expression levels of the vast majority of ISGs in Tyk2-deficient neutrophils to a similar extent as in WT cells. These results indicate that the IFN-λ responsiveness of mouse neutrophils is largely independent of TYK2, as in epithelial cells. We currently do not understand the molecular basis of the reduced baseline expression of ISGs nor the differential induction of ISGs observed in Tyk2-deficient mouse neutrophils. However, we excluded the possibility that the TYK2-dependent defect in type I IFN signaling would negatively affect base-line ISG levels or IFN-λ-mediated responses by demonstrating that Ifnar1−/− neutrophils have similar base-lines and respond equally well to IFN-λ as WT cells. Neutrophils likely differ qualitatively and quantitatively from epithelial cells with regard to the expression of cytokine receptors. This might change the composition of available JAKs as the various receptors might compete for JAK family members (3). Therefore, the availability of JAK1 could influence the degree of the TYK2 dependency of the IFN-λ signaling pathway. Abundant JAK1, associated with IFNLR1, might suffice for maximum IFN-λ-mediated signaling, whereas TYK2, associated with IL10RB, might be needed as an amplifier if JAK1 availability is limited.
Previous studies using inhibitors suggested a role for JAK2 in IFN-λ-mediated gene regulation (39, 43, 44). Contrasting these findings we did not observe reduced IFN-λ-mediated gene expression in JAK2-deficient A549 cells. We can only speculate that cell type specific differences or limited specificity of the JAK2 inhibitors could explain this unexpected discrepancy. We could rule out the possibility that JAK2 and TYK2 compensate for each other in human epithelial cells by showing that IFN-λ-induced ISG expression is the same in A549-JAK2koTYK2ko and control cells. However, we cannot exclude the possibility that JAK2 might serve other functions in the IFN-λ pathway beyond regulating ISG expression.
A limitation of our study is that we currently lack animal models which could be used to demonstrate the potency of selective TYK2 inhibitors for the treatment of type I IFN-driven autoimmune diseases. Nevertheless, our results strongly suggest that specific TYK2 inhibition may be a superior treatment option in clinical situations of excessive type I IFN signaling. The selective TYK2-inhibitor BMS-986165 employed here was already tested in clinical studies for the treatment of psoriasis (2), where no increased risk for infections was observed. We envisage that patients with interferonopathies would benefit from the suppression of unwanted type I IFN-driven inflammation, while barrier protection by IFN-λ and its potential therapeutic use (22) would be preserved. Similarly, in viral infections such as influenza, where type I IFN-driven inflammation and pathology can contribute to severity, selective TYK2 inhibition may alleviate this immunopathology without affecting the antiviral effects of IFN-λ in infected epithelia.
Material and Methods
Study Design
The aim of this study was to clarify the role of TYK2 in type I and type III IFN mediated signaling. The IFN specificity of a selective TYK2 inhibitor was compared to a broadly acting JAK inhibitor in human cell lines. Dose- and time-dependent transcriptional responses to type I or type III IFN stimulation were quantified in control and TYK2-deficient primary mouse epithelial cells, BM-derived mouse neutrophils, a mouse neutrophil culture model, and in two human cell lines, one derived from a myeloid and one from an epithelial progenitor. IFN-mediated antiviral protection was assessed in control and TYK2-deficient human cell lines and in mice. Control and experimental treatments were administered in a randomized fashion to sex- and age-matched mice.
Mice
B6.A2G-Mx1 mice are C57BL/6 mice carrying functional Mx1 alleles (58). B6.A2G-Mx1 or C57BL/6 mice were designated WT in this study. B6.A2G-Mx1-Tyk2−/− and C57BL/6-Tyk2−/− mice (designated Tyk2−/−) lack functional Tyk2 (14). B6.A2G-Mx1-Ifnar1−/− mice (designated Ifnar1−/− Ifnar1−/−) lack functional type I IFN receptors (58). B6.A2G-Mx1-Ifnar1−/−Ifnlr1−/− mice (designated Ifnar1−/−Ifnlr1−/−) lack functional type I and type III IFN receptors (58). All mice used in this study were bred and kept under specific-pathogen-free conditions in the local animal facility. All experiments with mice were carried out in accordance with the guidelines of the Federation for Laboratory Animal Science Associations and the national animal welfare body. Experiments were in compliance with the German animal protection law and were approved by the animal welfare committee of the Regierungspräsidium Freiburg (permit G-18/34). Mice used for experiments or for isolation of primary cells were 6 to 13 weeks old.
IFN treatment and infection of mice
B6.A2G-Mx1, B6.A2G-Mx1-Tyk2−/− and B6.A2G-Mx1-Ifnar1−/− Ifn1−/− were intranasally infected with 200 PFU of the influenza virus strain A/Udorn/72 (H3N2), designated Udorn (24), in a 10 μl volume. MDCK cells were used for the preparation of virus stocks and for virus titration by plaque assay. B6.A2G-Mx1+/−-Tyk2+/− and B6.A2G-Mx1+/−-Tyk2−/− mice were treated intranasally with 30 μl PBS containing 0.66 μg of either IFN-αB/D (59), recombinant mouse IFN-λ2 (24) or PBS only. After 18 h, mice were infected intranasally with 160 PFU of a highly virulent variant of the influenza A virus strain A/PR8/34 (H1N1) (designated hvPR8) (60) in a 40 μl volume. Mice were monitored for weight loss during the following 14 days and were euthanized when body weight loss exceeded 25 %.
Primary mouse tracheal epithelial cell culture
The preparation of primary mouse airway epithelial cells (AECs) was performed as previously described (24). Briefly, cells were isolated from the tracheae of mice with the indicated genotypes by enzymatic treatment and seeded onto a 0.4-μm pore size clear polyester membrane (Corning) coated with a collagen solution. At confluency, the medium was removed from the upper chamber to establish an air-liquid interface. Fully differentiated, 7-10-day-old post-air-liquid interface cultures were used routinely for experiments. For IFN treatment, cells were treated for 4 h from the basolateral side with indicated concentrations of either IFN-αB/D or recombinant mouse IFN-λ2 and then processed for RNA isolation and subsequent RT-qPCR analysis. For time course analysis, AECs were treated for 1 h with 10 ng/ml of either IFN-αB/D or recombinant mouse IFN-λ2 and processed for RNA isolation and subsequent RT-qPCR analysis.
Primary mouse mini-gut organoids
Small intestines of 10-12 week-old C57BL/6 and C57BL/6-Tyk2−/− were used to generate mini-gut organoid cultures (47). Briefly, small intestines were thoroughly cleaned and stem cell containing crypts were isolated following dissociation of tissue samples by Gentle Cell Dissociation Solution (STEMCELL technologies) for 30 min at room temperature. Crypts were collected by centrifugation and washed with PBS. After re-suspending in PBS, crypts were enriched in the supernatant by sedimentation. Resuspension followed by sedimentation and collection of supernatant was repeated to generate crypt enriched fractions which were then filtered using a 70 μm cell strainer and evaluated under the microscope. Fractions containing high numbers of crypts were pooled and collected by centrifugation. Crypts were then re-suspended in Matrigel® for cultivation. Crypts were passaged weekly using Gentle Cell Dissociation Solution and maintained in basal culture medium. For IFN treatment, mouse mini-gut organoids were isolated by dissolving the Matrigel® with PBS two days post-splitting. About 50 organoids per condition were mechanically disrupted and maintained in culture medium with the indicated concentrations of either IFN-αB/D or recombinant mouse IFN-λ2 for 4 h and then processed for RT-qPCR.
Hoxb8 neutrophils
Eight week-old male B6.A2G-Mx1 and B6.A2G-Mx1-Tyk2−/− were used to generate Hoxb8 neutrophil cultures as described (49). Briefly, myeloid progenitor cells were derived from bone marrow, retrovirally transduced with an estrogen-regulated Hoxb8 construct (MSCV-ERHBD-Hoxb8 (50) and selected for 4 weeks in the presence of stem cell factor (SCF) to generate neutrophil progenitor lines. Polyclonal progenitor cell lines were cultured in OptiMEM + GlutaMAX medium (Life Technologies) supplemented with 10 % FCS, 30 μM β-mercaptoethanol (Life Technologies), 1 μM β-estradiol (Sigma-Aldrich) and 1 % supernatant from SCF-producing CHO cells. Differentiation was induced by β-estradiol removal in the presence of 1% SCF supernatant and 20 ng/ml murine recombinant G-CSF (Peprotech). After 3.5-4 days of differentiation cells were used for experiments. For IFN treatment, cells were treated for 4 h with the indicated concentrations of either IFN-αB/D or recombinant mouse IFN-λ2 and then processed for RNA isolation and subsequent RT-qPCR analysis.
Isolation of human and mouse neutrophils
Human neutrophils were isolated by negative selection from peripheral blood samples of healthy donors using EasySep™ Direct Human Neutrophil Isolation Kit (STEMCELL Technologies) according to the manufacturer’s instructions. Mouse neutrophils were isolated by negative selection from bone marrow of mice with the indicated genotypes using EasySep™ Mouse Neutrophil Enrichment Kit (STEMCELL Technologies) according to the manufacturer’s instructions. Human and mouse neutrophils were directly used for experiments and cultured in OptiMEM + GlutaMAX medium (Life Technologies).
Cell lines
HAP1 cells (Horizon Biotec, C631) and HAP1 cells deficient for TYK2 (Horizon Biotec, HZGHC002840c01) were kindly provided by Stephan Ehl (Medical Center University of Freiburg) and cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) with 10 % FCS and 1 % Pen/Strep.
Human lung adenocarcinoma cell lines A549 were originally purchased from the American Type Culture Collection (ATCC) and maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with fetal calf serum (FCS 10% v/v, ThermoFisher Scientific) and penicillin-streptomycin (100 U/ml, ThermoFisher Scientific).
Generation of knockout cell lines
Generation of TYK2 knockout A549 cells: We first generated A549 cell clones stably expressing Cas9 (A549-Cas9) as described previously (61). Guide RNAs targeting TYK2 (NCBI reference sequence; NM_003331.5) and control sgRNA were individually cloned into lentiGuide-Puro (Addgene 52963, kindly gifted by Feng Zhang). Lentiviruses expressing sgRNA targeting TYK2 and control sgRNA were produced as described previously (62). A549-Cas9 cells were transduced with the respective lentiviruses and selected with 1 μg/ml puromycin 2 days post transduction. Single cell clones were seeded by limiting dilution into 96 well-plates and screened by western blot for expression of TYK2 using a monoclonal rabbit anti-TYK2 antibody (Cell signalling, #14193, 1:1000 dilution). Two of those single cell clones were used for experiments. TYK2-deficiency was further confirmed by Sanger sequencing.
Generation of JAK2 and JAK2/TYK2 knockout A549 cells: Two distinct single guide RNAs targeting JAK2 (NCBI reference sequence; NM_004972.3) were cloned into pSpCas9(BB)-2A-GFP (63). The plasmid was transfected into A549-Cas9 control or A549-Cas9-TYK2ko cells using Lipofectamin3000 according to manufactures protocol (ThermoFisher Scientific). GFP expressing single cell clones were sorted into 96-well plates using a BD FACSAria cell sorting system two days after transfection. Single cell clones were screened by western blot for expression of JAK2 using a monoclonal rabbit anti-JAK2 antibody (Cell signalling, #3230, 1:1000 dilution). Two single cell clones per gRNA targeting JAK2 were used for experiments. JAK2-deficiency was further confirmed by Sanger sequencing. Actin was detected using a rabbit serum (SIGMA-ALDRICH®, #A5060).
The following sgRNA target sequences were used:control:5’-AAAAAGCTTCCGCCTGATGG-3’; TYK2: 5’-TGTGCTGCCGGATATGCCGG-3’; JAK2 (KO1):5’-CGTTGGTATTGCAGTGGCAG-3’ and JAK2 (KO2):5’-AGAAAACGATCAAACCCCAC-3’. The following primer sequences for genotyping were used:TYK2:forward 5’-ACTTTGCATATTGCCGTGCC-3’, reverse 5’-TGACCATCTGCTGGGAGAGT-3’;JAK2(KO1):forward5’-TTTGTGCACTGAAGGAGGTAGT-3’, reverse 5’-ACTTCTCTGCACCTGCTGT-3; JAK2 (KO2):forward 5’-AGTGGCGGCATGATTTTGTG-3’, reverse 5’-TCAAAAGGCATGGGTAAACACAG-3.
RNA isolation and RT-qPCR
RNA from AECs, HAP1 and A549 cells was isolated using the RNeasy Plus Mini Kit (Quiagen) according to the manufacturer’s instructions. RNA from mini-gut organoids was isolated using NucleoSpin RNA extraction kit (Machery-Nagel) according to the manufacturer’s instructions. RNA from 2-5x106 cells per sample Hoxb8 or BM-derived neutrophils was isolated by pelleting cells in FastPrep tubes and subsequent lysis in 300 μl of peqGOLD TriFast™ using a homogenizer (MP Biomedicals). Three cycles of homogenization at 6.5 m/s for 18 s were performed, with samples resting on ice in between. After lysis, RNA was isolated using the Direct-zol RNA Miniprep (Zymo Research) according to the manufacturer’s instructions. For all samples, except mini-gut organoids, cDNA was generated from 850 ng total RNA using LunaScript™ RT SuperMix Kit (New England Biolabs) following the manufacturer’s instructions. In the case of mini-gut organoids, 250 ng of total RNA was reverse transcribed using iSCRIPT reverse transcriptase (BioRad). The cDNA served as template for the amplification of genes of interest (Isg15: forward: 5’-GAGCTAGAGCCTGCAGCAAT-3’; reverse: 5’-TTCTGGGCAATCTGCTTCTT-3’, Mx1: forward:5’-TCTGAGGAGAGCCAGACGAT-3’;reverse:5’-ACTCTGGTCCCCAATGACAG-3’, Ifnar1: forward: 5’-CATGTGTGCTTCCCACCACT-3’; reverse: 5’-TGGAATAGTTGCCCGAGTCC-3’, Ifnar2: forward: 5’-GACCTTCGGATAGCTGGTGG-3’; reverse: 5’-CTCATGATGTAGCCGTCCCC-3’, Ifnlr1: forward:5’-GGAACTGAAGTACCAGGTGGA-3’;reverse:5’-GCCATAGGGAGTGTCAGGAA-3’, Il1Orb: forward: 5’-TCTCTTCCACAGCACCTGAA-3’; reverse: 5’-GAACACCTCGCCCTCCTC-3’, Ubc (QT00245189, QuantiTect Primer Assay, Qiagen), Hprt (mm00446968_m1, Applied Biosystems), GAPD: forward: 5’-TGCACCACCAACTGCTTAGC-3’; reverse: 5’-GGCATGGACTGTGGTCATGAG-3’, HPRT: forward: 5’-TGACACTGGCAAAACAATGCA -3’; reverse: 5’-GGTCCTTTTCACCAGCAAGCT -3’, ISG15: forward: 5’-TCCTGCTGGTGGTGGACAA-3’; reverse: 5’-TTGTTATTCCTCACCAGGATGCT-3’, RSAD2: forward: 5’-CTTTGTGCTGCCCCTTGAG-3’; reverse: 5’-TCCATACCAGCTTCCTTAAGCAA-3’, MX1:forward:5’-GAAAAATCCAGGCTCGGTGG-3’; reverse:5’-TCAATGAGGTCGATGCAGGG -3’) by real-time PCR, using TaqMan Gene Expression Assays (Applied Biosystems), Universal PCR Master Mix (Applied Biosystems) and the QuantStudio 5 Real-Time PCR system (Applied Biosystems by Thermo Fisher Scientific). The increase in mRNA expression was determined by the 2-ΔCt method relative to the expression of the indicated housekeeping gene or by the 2-ΔΔCt method relative to mock.
RNA-seq
RNA-sequencing was carried out on the Illumina HiSeq 4000 platform and typically generated ~30 million 101-bp stranded single-end reads per sample. Adaptor trimming was performed with Trimmomatic/0.36-Java-1.7.0_80 with parameters “LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36” (64). The RSEM package (v.1.2.31) (65) in conjunction with the STAR alignment algorithm (v.2.5.2a) (66) was used for the mapping and subsequent gene-level counting of the sequenced reads with respect to mouse Ensembl GRCm38 - release 89. The parameters used were: -star-output-genome-bam-forward-prob 1. Differential expression analysis was performed with the DESeq2 package (v.1.20.0) (67) within the R programming environment (v.3.5.1) (https://www.r-project.org/). The significance threshold for the identification of differentially expressed genes was set as an adjusted P value ≤0.05.
Ingenuity Pathway Analysis (IPA) was used to annotate and visualize genes by function and pathway.
JAK inhibitors
Cells were pre-treated with 50 nM Pyridone 6 (BioVision) or 10 μM 1,2,3,4,5,6-Hexabromocyclohexane (HBC, Tocris) for 1 h or with the indicated concentrations of baricitinib (Hycultec) or BMS-986165 (Hycultec) for 1.5 h before the indicated amounts of either IFN-αB/D or recombinant mouse IFN-λ2 or recombinant human IFN-λ1 (68) was added to the respective culture medium containing the inhibitors.
VSV-GFP inhibition assay
HAP1 or A549 cells were pre-treated with the indicated concentrations of either IFN-αB/D or recombinant human IFN-λ1 at 18 h prior to infection with green-fluorescent protein expressing vesicular stomatitis virus (designated VSV-GFP) (69). Cells plated in 12-well plates (Corning) were washed once with PBS and subsequently incubated with 500 μl of virus suspension in OptiMEM 0.1 % BSA containing 2.5*106 TCID50 VSV-GFP for 1 h at 37 °C. After inoculum was removed, cells were incubated for 6 h at 37 °C. Hereafter, cells were harvested by trypsinization and fixed with 4 % formaldehyde at 4 °C for 10 min. Cells were analyzed for GFP expression using LSRFortessa™ cell analyzer (BD Biosciences) and data analyzed using FlowJo software. Percentage of GFP-positive cells of IFN-treated samples was normalized to untreated controls.
Statistical analysis
Data shown as the mean ± SD if not indicated otherwise in the figure legend. Sample sizes were designed to give statistical power, while minimizing animal use. All statistical comparisons were performed using GraphPad Prism 8 with the exception of sequencing data. Wald statistic values were plotted (log2(fold change) divided by standard error) instead of log2(fold change) without statistical filtering as these encapsulate the consistency of the replicates. The largest fold changes have a tendency to be the noisiest which distorts the ability to visually interpret log2(fold change) without statistical filtering.
Figure legends denote the specific statistical tests used for each experiment. Unless otherwise indicated, statistical significance was determined as P > 0.05.
Supplementary Material
One sentence summary:
TYK2 is required for efficient IFN-α/β signaling but dispensable for IFN-λ-mediated antiviral protection in epithelial cells.
Acknowledgements
We are grateful to Liang Ye, Jan Becker, Stefanie Koßmann and Annika Kellersohn for their technical support, to Stephan Ehl for providing the HAP1 cell lines, and to Otto Haller for helpful comments. We thank the Advanced Sequencing facility of the Francis Crick Institute for excellent support and expertise, and Gavin Kelly for advice on statistical analysis.
Funding
This work was supported by grant STA 338/15-1 from the German Research Foundation (DFG) to P.S.. S.B. was supported by DFG grants BO 4340/1-1, SFB1129 and TRR186. M.S. was supported by the Brigitte-Schlieben Lange Program from the state of Baden Württemberg, and DFG grant STA 1536/2-1. S.C., M.L. and A.W. were supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001206), the UK Medical Research Council (FC001206) and the Wellcome Trust (FC001206). M.Sch. was supported by DFG grant SFB 1160. T.T was supported by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School). B.S. was supported by grants DOC 32-B28 and SFB-F6101 from the Austrian Science Fund.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author contributions
D.S. designed and performed most of the experiments, analyzed and interpreted the data and performed statistical analyses. S.C., M.S., L.P. performed experiments, analyzed and interpreted the data and performed statistical analyses. A.O. and J.V. performed experiments and analyzed the data. M.L. analyzed and interpreted data and performed statistical analyses. T.T., H.H.G., R.H. B.S. and M.Sch. provided essential materials. R.H., B.S., S.K., S.B., M.Sch. and A.W interpreted data and gave advice. P.S. conceived the project, acquired funding for the study, designed and performed experiments and interpreted the data. D.S. and P.S. wrote the manuscript with input from all authors.
Data availability
Sequencing data are available in GEO under accession code GSE146104.
References
- 1.Sanchez GAM, Reinhardt A, Ramsey S, Wittkowski H, Hashkes PJ, Berkun Y, Schalm S, Murias S, Dare JA, Brown D, Stone DL, et al. JAK1/2 inhibition with baricitinib in the treatment of autoinflammatory interferonopathies. J Clin Invest. 2018;128:3041–3052. doi: 10.1172/JCI98814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papp K, Gordon K, Thaci D, Morita A, Gooderham M, Foley P, Girgis IG, Kundu S, Banerjee S. Phase 2 Trial of Selective Tyrosine Kinase 2 Inhibition in Psoriasis. N Engl J Med. 2018;379:1313–1321. doi: 10.1056/NEJMoa1806382. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov. 2017;16:843–862. doi: 10.1038/nrd.2017.201. [DOI] [PubMed] [Google Scholar]
- 4.Lazear HM, Schoggins JW, Diamond MS. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendoza JL, Schneider WM, Hoffmann HH, Vercauteren K, Jude KM, Xiong A, Moraga I, Horton TM, Glenn JS, de Jong YP, Rice CM, et al. The IFN-lambda-IFN-lambdaR1-IL-10Rbeta Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity. Immunity. 2017;46:379–392. doi: 10.1016/j.immuni.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol. 2014;21:443–448. doi: 10.1038/nsmb.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finbloom DS, Winestock KD. IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol. 1995;155:1079–1090. [PubMed] [Google Scholar]
- 8.Ho AS, Wei SH, Mui AL, Miyajima A, Moore KW. Functional regions of the mouse interleukin-10 receptor cytoplasmic domain. Mol Cell Biol. 1995;15:5043–5053. doi: 10.1128/mcb.15.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domanski P, Fish E, Nadeau OW, Witte M, Platanias LC, Yan H, Krolewski J, Pitha P, Colamonici OR. A region of the beta subunit of the interferon alpha receptor different from box 1 interacts with Jak1 and is sufficient to activate the Jak-Stat pathway and induce an antiviral state. J Biol Chem. 1997;272:26388–26393. doi: 10.1074/jbc.272.42.26388. [DOI] [PubMed] [Google Scholar]
- 10.Ferrao R, Wallweber HJ, Ho H, Tam C, Franke Y, Quinn J, Lupardus PJ. The Structural Basis for Class II Cytokine Receptor Recognition by JAK1. Structure. 2016;24:897–905. doi: 10.1016/j.str.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye L, Schnepf D, Staeheli P. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat Rev Immunol. 2019;19:614–625. doi: 10.1038/s41577-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs S, Kaiser-Labusch P, Bank J, Ammann S, Kolb-Kokocinski A, Edelbusch C, Omran H, Ehl S. Tyrosine kinase 2 is not limiting human antiviral type III interferon responses. Eur J Immunol. 2016;46:2639–2649. doi: 10.1002/eji.201646519. [DOI] [PubMed] [Google Scholar]
- 13.Kreins AY, Ciancanelli MJ, Okada S, Kong XF, Ramirez-Alejo N, Kilic SS, El Baghdadi J, Nonoyama S, Mahdaviani SA, Ailal F, Bousfiha A, et al. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J Exp Med. 2015;212:1641–1662. doi: 10.1084/jem.20140280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, Pfeffer K, et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–560. doi: 10.1016/s1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 15.Eletto D, Burns SO, Angulo I, Plagnol V, Gilmour KC, Henriquez F, Curtis J, Gaspar M, Nowak K, Daza-Cajigal V, Kumararatne D, et al. Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat Commun. 2016;7:13992. doi: 10.1038/ncomms13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto K, Wehde BL, Radler PD, Triplett AA, Wagner KU. Generation of Janus kinase 1 (JAK1) conditional knockout mice. Genesis. 2016;54:582–588. doi: 10.1002/dvg.22982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nature immunology. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nature immunology. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 19.Prokunina-Olsson L, Alphonse N, Dickenson RE, Durbin JE, Glenn JS, Hartmann R, Kotenko SV, Lazear HM, O’Brien TR, Odendall C, Onabajo OO, et al. COVID-19 and emerging viral infections: The case for interferon lambda. J Exp Med. 2020;217 doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Brien TR, Thomas DL, Jackson SS, Prokunina-Olsson L, Donnelly RP, Hartmann R. Weak Induction of Interferon Expression by SARS-CoV-2 Supports Clinical Trials of Interferon Lambda to Treat Early COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreakos E, Tsiodras S. COVID-19: lambda interferon against viral load and hyperinflammation. EMBO Mol Med. 2020 doi: 10.15252/emmm.202012465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feld JJ, Kandel C, Biondi MJ, Kozak RA, Zahoor MA, Lemieux C, Borgia SM, Boggild AK, Powis J, McCready J, Tan DHS, et al. Peginterferon-lambda for the treatment of COVID-19 in outpatients. medRxiv. 2020:2020.2011.2009.20228098 [Google Scholar]
- 23.Lee S, Baldridge MT. Interferon-Lambda: A Potent Regulator of Intestinal Viral Infections. Front Immunol. 2017;8:749. doi: 10.3389/fimmu.2017.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, Garcin D, Mahlakoiv T, Staeheli P. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 2018;7:e33354. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, Thanos D, Doyle SE, Kotenko SV, Thanopoulou K, Andreakos E. Interferon-lambda Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity. 2017;46:875–890.:e876. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Wells AI, Coyne CB. Type III Interferons in Antiviral Defenses at Barrier Surfaces. Trends Immunol. 2018;39:848–858. doi: 10.1016/j.it.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caine EA, Scheaffer SM, Arora N, Zaitsev K, Artyomov MN, Coyne CB, Moley KH, Diamond MS. Interferon lambda protects the female reproductive tract against Zika virus infection. Nat Commun. 2019;10:280. doi: 10.1038/s41467-018-07993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruening J, Weigel B, Gerold G. The Role of Type III Interferons in Hepatitis C Virus Infection and Therapy. J Immunol Res. 2017;2017:7232361. doi: 10.1155/2017/7232361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLOS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson S, Mc Cabe TM, Crotta S, Gad HH, Hessel EM, Beinke S, Hartmann R, Wack A. IFNlambda is a potent anti-influenza therapeutic without the inflammatory side effects of IFNalpha treatment. EMBO Mol Med. 2016;8:1099–1112. doi: 10.15252/emmm.201606413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson S, Crotta S, Mc Cabe TM, Wack A. Pathogenic potential of interferon alphabeta in acute influenza infection. Nat Commun. 2014;5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Major J, Crotta S, Llorian M, McCabe TM, Gad HH, Priestnall SL, Hartmann R, Wack A. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Lo Cascio A, Clementi N, De Santis M, Mancini N, Granucci F, Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goel RR, Wang X, O’Neil LJ, Nakabo S, Hasneen K, Gupta S, Wigerblad G, Blanco LP, Kopp JB, Morasso MI, Kotenko SV, et al. Interferon lambda promotes immune dysregulation and tissue inflammation in TLR7-induced lupus. Proc Natl Acad Sci U S A. 2020;117:5409–5419. doi: 10.1073/pnas.1916897117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnepf D, Staeheli P. License to kill: IFN-lambda regulates antifungal activity of neutrophils. Sci Immunol. 2017;2:eaap9614. doi: 10.1126/sciimmunol.aap9614. [DOI] [PubMed] [Google Scholar]
- 36.Rivera A. Interferon Lambda’s New Role as Regulator of Neutrophil Function. J Interferon Cytokine Res. 2019 doi: 10.1089/jir.2019.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blazek K, Eames HL, Weiss M, Byrne AJ, Perocheau D, Pease JE, Doyle S, McCann F, Williams RO, Udalova IA. IFN-lambda resolves inflammation via suppression of neutrophil infiltration and IL-1beta production. J Exp Med. 2015;212:845–853. doi: 10.1084/jem.20140995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinosa V, Dutta O, McElrath C, Du P, Chang YJ, Cicciarelli B, Pitler A, Whitehead I, Obar JJ, Durbin JE, Kotenko SV, et al. Type III interferon is a critical regulator of innate antifungal immunity. Sci Immunol. 2017;2:eaan5357. doi: 10.1126/sciimmunol.aan5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broggi A, Tan Y, Granucci F, Zanoni I. IFN-lambda suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nature immunology. 2017;18:1084–1093. doi: 10.1038/ni.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velazquez L, Fellous M, Stark GR, Pellegrini S. A Protein Tyrosine Kinase in the Interferon-Alpha/Beta Signaling Pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 41.Strobl B, Stoiber D, Sexl V, Mueller M. Tyrosine kinase 2 (TYK2) in cytokine signalling and host immunity. Front Biosci-Landmrk. 2011;16:3224–U3248. doi: 10.2741/3908. [DOI] [PubMed] [Google Scholar]
- 42.Lumb JH, Li Q, Popov LM, Ding S, Keith MT, Merrill BD, Greenberg HB, Li JB, Carette JE. DDX6 Represses Aberrant Activation of Interferon-Stimulated Genes. Cell Rep. 2017;20:819–831. doi: 10.1016/j.celrep.2017.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nature immunology. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SJ, Kim WJ, Moon SK. Role of the p38 MAPK signaling pathway in mediating interleukin-28A-induced migration of UMUC-3 cells. Int J Mol Med. 2012;30:945–952. doi: 10.3892/ijmm.2012.1064. [DOI] [PubMed] [Google Scholar]
- 45.Burke JR, Cheng L, Gillooly KM, Strnad J, Zupa-Fernandez A, Catlett IM, Zhang Y, Heimrich EM, McIntyre KW, Cunningham MD, Carman JA, et al. Autoimmune pathways in mice and humans are blocked by pharmacological stabilization of the TYK2 pseudokinase domain. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aaw1736. [DOI] [PubMed] [Google Scholar]
- 46.Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;17:6270. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- 47.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 48.Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLOS Pathog. 2015;11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- 50.Redecke V, Wu R, Zhou J, Finkelstein D, Chaturvedi V, High AA, Hacker H. Hematopoietic progenitor cell lines with myeloid and lymphoid potential. Nat Methods. 2013;10:795–803. doi: 10.1038/nmeth.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andersson BS, Collins VP, Kurzrock R, Larkin DW, Childs C, Ost A, Cork A, Trujillo JM, Freireich EJ, Siciliano MJ, et al. KBM-7, a human myeloid leukemia cell line with double Philadelphia chromosomes lacking normal c-ABL and BCR transcripts. Leukemia. 1995;9:2100–2108. [PubMed] [Google Scholar]
- 54.Alsohime F, Martin-Fernandez M, Temsah MH, Alabdulhafid M, Le Voyer T, Alghamdi M, Qiu X, Alotaibi N, Alkahtani A, Buta S, Jouanguy E, et al. JAK Inhibitor Therapy in a Child with Inherited USP18 Deficiency. N Engl J Med. 2020;382:256–265. doi: 10.1056/NEJMoa1905633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duncan CJA, Thompson BJ, Chen R, Rice GI, Gothe F, Young DF, Lovell SC, Shuttleworth VG, Brocklebank V, Corner B, Skelton AJ, et al. Severe type I interferonopathy and unrestrained interferon signaling due to a homozygous germline mutation in STAT2. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aav7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidson S, Steiner A, Harapas CR, Masters SL. An Update on Autoinflammatory Diseases: Interferonopathies. Curr Rheumatol Rep. 2018;20:38. doi: 10.1007/s11926-018-0748-y. [DOI] [PubMed] [Google Scholar]
- 57.Stanifer ML, Pervolaraki K, Boulant S. Differential Regulation of Type I and Type III Interferon Signaling. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, Staeheli P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLOS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horisberger MA, de Staritzky K. A recombinant human interferon-alpha B/D hybrid with a broad host-range. J Gen Virol. 1987;68(Pt 3):945–948. doi: 10.1099/0022-1317-68-3-945. [DOI] [PubMed] [Google Scholar]
- 60.Grimm D, Staeheli P, Hufbauer M, Koerner I, Martinez-Sobrido L, Solorzano A, Garcia-Sastre A, Haller O, Kochs G. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc Natl Acad Sci U S A. 2007;104:6806–6811. doi: 10.1073/pnas.0701849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karakus U, Thamamongood T, Ciminski K, Ran W, Gunther SC, Pohl MO, Eletto D, Jeney C, Hoffmann D, Reiche S, Schinkothe J, et al. MHC class II proteins mediate cross-species entry of bat influenza viruses. Nature. 2019;567:109–112. doi: 10.1038/s41586-019-0955-3. [DOI] [PubMed] [Google Scholar]
- 62.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. Human interferon-lambda 3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 69.Boritz E, Gerlach J, Johnson JE, Rose JK. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J Virol. 1999;73:6937–6945. doi: 10.1128/jvi.73.8.6937-6945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available in GEO under accession code GSE146104.