Abstract
Autophagosomes are double membrane organelles that are formed during a process referred to as macroautophagy. They serve to deliver cytoplasmic material into the lysosome for degradation. Autophagosomes are formed in a de novo manner and are the result of substantial membrane remodeling processes involving numerous protein-lipid interactions. While most studies focus on the proteins involved in autophagosome formation it is obvious that lipids including phospholipids, sphingolipids and sterols play an equally important role. Here we summarize the current knowledge about the role of lipids, especially focusing on phospholipids and their interplay with the autophagic protein machinery during autophagosome formation and fusion.
Keywords: Autophagy, lysosome, conjugation, phosphatidylethanolamine, phosphatidylinositol 3-phosphate, lipid phosphatase, lipid kinase
Introduction
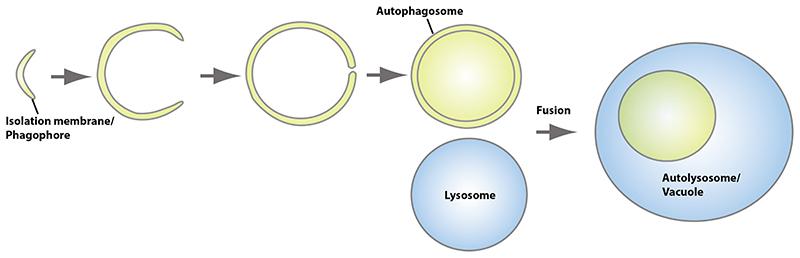
The term autophagy describes processes wherein cytoplasmic material is delivered into the endo-lysosomal compartment for degradation. During macroautophagy (hereafter autophagy) a portion of the cells’ cytoplasm is sequestered within a double membrane organelle referred to as autophagosome. After induction of autophagy by extrinsic or intrinsic stimuli, autophagosomes form in a de novo manner. Initially, small membrane structures called isolation membranes (or phagophores) are assembled. These gradually expand around a portion of the cytoplasm referred to as cargo and eventually close to isolate the cargo from the rest of the cytoplasm. Subsequently, the outer autophagosomal membrane fuses with lysosomes wherein the inner membrane and the cargo are degraded (Figure 1) [1, 2].
Figure 1. The major steps of autophagosome formation.
Upon induction of autophagy, small membrane structures referred to isolation membranes (or phagophores) are generated. These structures expand and gradually enclose cytoplasmic material as they grow. Upon closure of the isolation membranes autophagosomes are formed, which fuse with lysosomes to form autolysosomes, within which the inner membrane and the cargo are degraded.
Autophagy can be massively induced by stress conditions such as starvation but also by cell intrinsic stimuli such as the accumulation of damaged mitochondria, protein aggregates, the presence of damaged endosomes and lysosomes and some cytosolic pathogens [3]. Autophagy thereby serves ensure cellular homeostasis and quality control. Consequently, dysfunctions of autophagy have been implicated in various pathological conditions including neurodegeneration, cancer and uncontrolled infections [4].
Essentially, autophagosome formation is a membrane sculpting process during which cells form a new membrane-bound organelle and as such lipids play a central role. While the lipid composition of autophagosomes is surprisingly still unknown, specific lipids have been shown to be important for the induction and expansion of isolation membranes as well as for their fusion with lysosomes. Here we summarize the recent knowledge of the lipid requirements for autophagosome formation.
Autophagosome induction
The membrane donor for autophagosomes is still under debate and various sources including the endoplasmic reticulum (ER), the Golgi, mitochondria, the plasma membrane, lipid droplets and endosomes have been suggested to supply lipids to the growing isolation membrane [5–18]. It is possible that multiple sources are able to act as lipid donors, that various cell types or distinct types of autophagy employ different membrane sources or that certain organelles contribute only indirectly to autophagosome formation. However, in mammalian cells it seems well established that when triggered by starvation, isolation membrane formation occurs close to, or at a specialized domain of the ER [5, 6, 9, 10, 16, 19]. Electron microscopy pictures have shown that isolation membranes are connected to the ER [10, 19]. These sites are likely to be identical to structures identified by light microscopy referred to as omegasomes [5]. In yeast, current evidence suggests that isolation membrane growth is driven at least in part by vesicle fusion [20, 21]. Connections of the isolation membranes with the ER as observed in mammalian cells have not yet been reported although ER-derived structured have been seen close to the autophagosome formation site [7, 22]. The distance between the two membranes of the isolation membranes is just a few nanometers and therefore much smaller than for the ER [10, 19]. For this reason, isolation membranes do not simply represent an area of deformed ER.
The molecular mechanisms underlying autophagosome formation are largely unclear. The ULK1 kinase complex (Atg1 in yeast) and the class III PI3K complex 1 are essential for autophagosome formation and act together with the transmembrane protein ATG9 during the induction and nucleation of isolation membranes [23]. The ULK1 kinase complex consists of the ULK1 (Atg1) kinase subunit, FIP200 (Atg17/Atg11 in yeast), ATG13 (Atg13 in yeast) and ATG101 [24–27]. The mechanism of ULK1 recruitment to the autophagosome formation site is not well understood, in particular during starvation induced autophagy but the membrane composition and/or shape of the autophagosome formation initiation site may play an important role. The Atg1 kinase subunit has been shown to contain an amphipathic region termed EAT domain that binds preferentially to highly curved membranes [28]. Amphipathic regions in proteins recognize the curvature as regions with membrane defects as these aid insertion of the protein into the hydrophobic core of membrane and, analogously, lipids with small headgroups will have a similar effect [29, 30].
The class III phosphatidylinositol 3-phosphate kinase (PI3K) complex 1 consists of the VPS34 lipid kinase subunit as well as VPS15, Beclin/ATG6 and ATG14 [31, 32]. VPS34 phosphorylates phosphatidylinositol (PI) to generate phosphatidylinositol 3-phosphate (PI3P) [33]. The enzymatic activity of this complex is the major source of PI3P for autophagosome formation. In addition, other PI3K complexes such as class II PI3K could contribute to PI3P generation [34]. The class III PI3K complex 1 may be targeted to the autophagosome formation site by the ULK1/Atg1 complex and interactions with ER-resident proteins such as syntaxin-17 [9, 35]. In addition, several subunits of the class III PI3K complex 1 have been shown to directly interact with membranes [31, 32]. The VP34 subunit contains a membrane inserting amphipathic helix [36], VPS15 is anchored to the membrane by a N-terminal myristoylation [37], ATG14 contains a C-terminal amphipathic helix [38] and Beclin1 interacts with the membrane via aromatic residues located in its C-terminal evolutionary conserved domain [39]. All these interactions are facilitated on membranes with high degree of defects and thus when they are highly curved and/or contain lipids with small headgroups such as phosphatidylethanolamine (PE). The PI3P generated by the class III PI3K complex 1 subsequently recruits further factors (see below) that mediate the expansion and subsequent closure of the isolation membrane. Downregulation of PI3P levels by the PI3P phosphatase MTMR3 and Jumpy/MTMR14 was shown negatively regulate autophagosome formation [40–42].
Isolation membrane expansion and closure
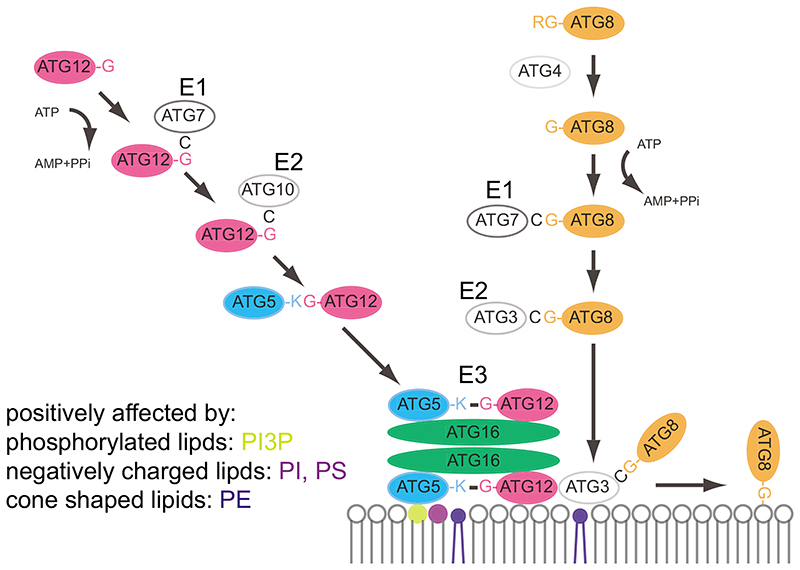
After their nucleation, isolation membranes expand and eventually close. While the lipid composition of isolation membranes and autophagosomes is still unknown, individual lipids are likely to play critical roles. One of the hallmarks of isolation membrane expansion is the attachment of ATG8 proteins to the membrane [43–45]. In yeast there is one Atg8 protein while there are several ATG8 isoforms in mammalian cells, which are subdivided into the LC3s and GABARAPs [46]. ATG8 proteins are ubiquitin-like proteins that become covalently linked to the headgroup of PE via their C-terminal glycine residue (Figure 2)[43–45]. ATG8 proteins are synthesized as precursors with short C-terminal extensions and the C-terminal glycine residues become exposed after proteolytic cleavage by cysteine proteases of the ATG4 family [43]. ATG8 proteins are subsequently activated under consumption of ATP by the E1-like enzyme ATG7. From ATG7, ATG8 is transferred to the E2-like ATG3 and from there to the headgroup of PE [44, 47]. This last step is vastly accelerated by a protein complex composed of the ATG12–ATG5 conjugate and ATG16 [48]. The ATG12–ATG5-ATG16 complex acts analogous to the E3-like enzymes during ubiquitination and is itself a product of a protein conjugation reaction during which the ubiquitin-like ATG12 proteins becomes conjugated via its C-terminal glycine residue to a lysine residue in ATG5 [49]. The localization of the ATG12–ATG5-ATG16 complex is a critical determinant for the localization of ATG8–PE formation [50]. The regulation of selective autophagy and the transport of autophagosomes to lysosomes by ATG8 proteins via interaction with cargo receptors and adaptors is well established [51]. In contrast, the role of ATG8 proteins during the membrane sculpting processes is less well understood. Current evidence points to a contribution of these proteins to isolation membrane expansion and closure [52–55].
Figure 2. The ubiquitin-like protein conjugation machinery in autophagy.
ATG8 proteins, including the LC3 and GABARAP families in mammalian cells, are initially synthesized as precursors with C-terminal extensions. These become cleaved off by the ATG4 cysteine proteases exposing a C-terminal glycine residue. Subsequently, ATG8 proteins are activated by the E1-like ATG7 protein under consumption of ATP. From ATG7, ATG8 proteins are transferred to the E2-like ATG3 and further to the headgroup of the membrane lipid phosphatidylethanolamine (PE). This last step is massively promoted by the ATG12–ATG5-ATG16 complex. This complex is itself the product of an ubiquitin-like conjugation reaction during which the ubiquitin-like ATG12 is transferred via its C-terminal glycine residue to an internal lysine residue of ATG5. The ATG12–ATG5 conjugate subsequently forms a non-covalent complex with ATG16. Lipids that positively influence the recruitment of the ATG12–ATG5-ATG16 complex and ATG3 are highlighted.
Since PE is the main target for ATG8 conjugation in vivo it is likely that this lipid is present on the isolation membrane. In addition, PE is one of the most abundant lipids in cells and may therefore have a significantly contribution to the lipid mass of the autophagosomal membrane [56]. High levels of PE have been shown to stimulate Atg8-PE conjugation and to facilitate Atg8-mediated hemifusion of small vesicles [47, 48, 53, 54, 57, 58]. In vitro, phosphatidylserine (PS) can also serve as acceptor for ATG8 protein conjugation and it is possible that a small fraction of ATG8 is conjugate to this lipid [59]. Most of the ATG8 proteins are removed from the outer autophagosomal membrane after autophagosome completion by ATG4. The fact that ATG8–PS, as opposed to ATG8–PE, is more resistant to ATG4 cleavage suggests that ATG8–PS is only a minor fraction of the total lipid conjugated ATG8 [60]. PS may have an additional role during ATG8 conjugation since negatively charged lipids aid the recruitment of the ATG12–Atg5-Atg16 complex to the membrane, which stimulates ATG8 conjugation to PE [61].
As discussed above PI3P generated by the class III PI3K complex 1 is critical for the early stages of isolation membrane formation but is also likely to be required during their expansion. Since PI3P is generated from PI, it is reasonable to assume that PI is present in the isolation membrane and that its levels will impact autophagosome formation. The WD40 repeat proteins of the WIPI family directly bind PI3P and PI3,5P2 [31, 62] and become recruited to the expanding isolation membrane, likely in complex with ATG2A/B to facilitate isolation membrane expansion and finally closure [63–65]. WIP2 was also shown to recruit the ATG12–ATG5-ATG16L1 complex to the isolation membrane, thus linking PI3P to the ATG8 conjugation machinery [66, 67].
A further aspect of how lipids may impact autophagosome formation is at the level of their shape. Thus, it was shown that ATG3 preferentially binds highly curved membranes [58]. The S. cerevisiae Atg12–Atg5-Atg16 complex also prefers highly curved membranes [61]. In both cases the curvature is detected due to a higher abundance of membrane defects, aiding the insertion of hydrophobic regions of proteins into the membrane. Lipids with small headgroups such as phosphatidic acid (PA) [68] and/or bulky acyl chains will have similar effects and may promote membrane binding and insertion of ATG3 and the Atg12–Atg5-Atg16 complex and thus autophagosome formation.
Surprisingly, by using quick-freezing and freeze-fracture replica labelling (QF-FRL), it was shown that the distribution of PI3P in autophagosomal membranes is different between yeast and mammals [69]. In yeast, PI3P is more abundant in the luminal than in the cytoplasmic leaflet, while PI3P in mammals is exclusively present in the cytoplasmic leaflet of the autophagosome. In yeast, PI3P is generated equally in both leaflets and the asymmetric distribution is generated by unilateral hydrolysis of PI3P by cytoplasmic phosphatase Ymr1p and Sjl3p. Proper distribution of PI3P in autophagosomes seems to be essential for the subsequent closure and fusion with vacuoles. The difference in PI3P distribution suggests that autophagy could involve different processes in yeast and mammals.
Role of phospholipids in autophagosome-lysosome fusion
In mammals autophagosomes are formed throughout the cytoplasm, while lysosomes are localized at the perinuclear region. Thus autophagosomes have to be transported to the lysosome before they can fuse. The movement of autophagosomes is mediated by the microtubule cytoskeleton [51, 70]. FYCO1 (FYVE and coiled-coil domain containing 1) has been shown to bind to LC3, PI3P as well as the small GTPase Rab7 and is involved in this process. Endogenous FYCO1 is localized on perinuclear cytosolic vesicles, but is also distributed in the cell peripheral region in a microtubule dependent manner upon starvation. FYCO1 functions as an adaptor protein between autophagosomes and microtubule plus end-directed molecular motors, since FYCO1 depleted cells show the accumulation of autophagosomes at perinuclear clusters. [71]. The kinesin proteins interacting with FYCO1 remain to be defined in the future.
How and when do autophagosomes become competent for fusion with lysosomes? Some hints come from a yeast study. In yeast, dephosphorylation and thus clearance of PI3P by the PI3P phosphatase, Ymr1 on the completed autophagosome are essential for autophagosomevacuole fusion [72]. Clearance of PI3P triggers the dissociation of the ATG machinery from the outer autophagosomal membrane making them competent for fusion with the vacuole. Mammalian forms of PI3P phosphatases such as myotubularin-related protein 3 (MTMR3) and MTMR14/Jumpy appear to have different functions during autophagy [40, 41]. Knockdown of MTMR3 and MTMR14/Jumpy increase autophagosomes formation indicating these phosphatases are negative regulators of autophagy and function at early stage of autophagosome formation.
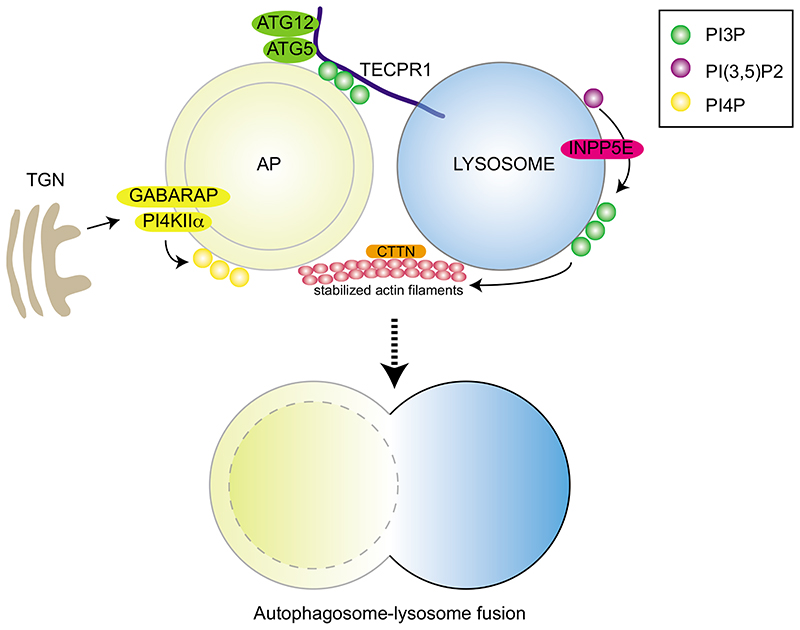
PI3P also participates in fusion process through PI3P binding proteins. In mammals, recent work identified the PI3P binding protein TECPR1 as a tethering factor mediating autophagosome-lysosome fusion [73]. TECPR1 depleted cells show impaired autophagic flux, the accumulation of autophagic vacuoles and autophagic substrates including p62 and lipidated LC3. TECPR1 was originally identified through its interaction with ATG5 [74]. Although the phagophore localization of the ATG12–ATG5-ATG16 complex is dependent on PI3P [50], the authors show that TECPR1 forms a complex with ATG12-ATG5 in a mutually exclusive manner with ATG16. Furthermore, they show that TECPR1 is localized on lysosomes/autolysosomes and recruits the ATG12-ATG5 conjugate to enable binding to PI3P, possibly facilitating autophagosome maturation and autophagosome-lysosome fusion (Figure 3). However, another study showed that TECPR1 also functions in phagophore biogenesis and maturation during selective autophagy [73]. These discrepancies might either reflect the dual roles of TECPR1 or different biological contexts.
Figure 3. Roles of phosphoinositides in autophagosome-lysosome fusion.
TECPR1 on lysosome interacts with ATG12-ATG5 and binds to PI3P on autophagosome, thus functioning as a tethering factor mediating autophagosome-lysosome fusion. GAPARAP recruits palmytoylated PI4KIIα from TGN (Trans Golgi Network) to autophagosomes and the production of PI4P by PI4KIIα is essential for autophagosome-lysosome fusion. Some INPP5E are localized on lysosome and are converting PI(3,5)P2 to PI3P, leading to the activation of cortactin/CTTN and stabilization of actin filaments which are required for autophagosomal fusion. Exact orders of these events need to be clarified in future.
In addition to PI3P, PI4P generation on autophagosomes is also critical for autophagosome-lysosome fusion in mammals [75]. Phosphatidylinositol 4-phosphate kinase type II α (PI4KIIα) is normally localized in the perinuclear region and trans-Golgi network (TGN). Upon starvation, PI4KIIα exits from the TGN and is distributed in the cytoplasm. Some of the PI4KIIα is localized on autophagosomes in a palmytoylation-dependent manner. Consistent with this, PI4P undergoes a similar redistribution and depletion of PI4KIIα reduces the concentration of PI4P in autophagosomes. Interestingly, PI4KIIα interacts with GABARAP and GABARAPL2, but not with LC3. Depletion of GABARAP inhibits PI4KIIα translocation to autophagosomes, while depletion of PI4KIIα does not affect GABARAP distribution, indicating that GABARAP functions upstream of PI4KIIα in this context. Knockdown of either GABARAP or PI4KIIα shows defective large autophagosomes and impaired degradation of LC3 and p62 due to defective autophagosomal fusion with lysosome. Importantly, the fusion defect is rescued by introduction of PI4P, but not by PI(4,5)P2, suggesting that PI4P generation, but not downstream metabolites, is essential for autophagosome-lysosome fusion (Figure 3). The exact role of PI4P in the fusion needs to be clarified in future studies.
Contrary to the roles of phospholipids in the autophagosomal membrane, it was unclear whether the specific composition of phospholipids in the lysosomal membrane is essential for autophagosome-lysosome fusion. Recently a phosphoinositide phosphatases, inositol polyphosphate-5 phosphatase E (INPP5E) has been identified as a novel regulator of autophagy, promoting the fusion step [76]. Inhibition of INPP5E causes the accumulation of autophagosomes as defined by LC3 puncta due to the impairment of autophagosomelysosome fusion. Importantly, knockdown of INPP5E does not affect the endocytic pathway and the integrity of lysosomes, further highlighting the specific role of INPP5E during the autophagosome-lysosome fusion step. Notably, some of the INPP5E protein is localized at lysosomes where it mediates the conversion of PI(3,5)P2 to PI3P in the lysosomal membrane, which in turn is crucial for the fusion with autophagosomes. Previously, it was shown that knockdown of the phosphoinositide kinase PIKfyve decreases the PI(3,5)P2 levels on lysosomes and causes severe autophagy defects due to the loss of lysosomal function [77]. Therefore, proper levels of PI(3,5)P2 in the lysosomal membrane seems to be required for its function and fusion with autophagosomes during autophagy. INPP5E activity on lysosomes is ultimately required for phosphorylation of cortactin/CTTN, which leads to actin polymerization followed by autophagosome-lysosome fusion (Figure 3). Mutations in INPP5E are known to be linked to Joubert syndrome, a rare brain abnormality [78] and these mutant forms of INPP5E could not rescue the autophagy defects caused by INPP5E knockdown in neuronal cells, strongly suggesting that the impairment of autophagy could be causal for this disease [76]. Since INPP5E is predominantly expressed in neuronal cells, future studies need to clarify if a similar mechanism functions during the autophagosome-lysosome fusion process in other cell types as well. Taken together, although several specific PIs both in autophagosome and lysosome have been shown to participate in autophagosome-lysosome fusion, one should address the exact role of each PIs, the timeline of the function of these PIs and how these different phospholipids cooperatively function during the fusion process in upcoming research.
Outlook
At its core, autophagosome formation is a membrane sculpting process and it therefore not surprising that there is an extensive cross talk between lipids and the autophagic protein machinery. Consequently, individual lipids, mostly phosphorylated forms of PI, have been shown to be required for multiple steps during the life of an autophagosome. However, in order to fully comprehend the role of lipids in autophagy the lipid composition of autophagosome precursors and of completed autophagosomes will have to be known, which is currently not the case. To this end, methods to purify autophagosomes to a high degree and subsequent lipidomics are essential. Improvement of imaging techniques allowing the visualization of lipids using electron microscopy and super resolution microscopy will also help to study the distribution and dynamics of lipids during autophagosome formation. In our view this is one of the major challenges in autophagy research that must be overcome in order to fully appreciate the role of lipids during this process.
Acknowledgements
S.M. is supported by ERC grant (No.646653), by the FWF (No. P25546-B20) and by the EMBO Young Investigator Program. Y.T. is supported by The Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Agency for Medical Research and Development (AMED).
References
- [1].Kraft C, Martens S. Mechanisms and regulation of autophagosome formation. Current opinion in cell biology. 2012;24(4):496–501. doi: 10.1016/j.ceb.2012.05.001. [DOI] [PubMed] [Google Scholar]
- [2].Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Current opinion in cell biology. 2013;25(4):455–60. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- [3].Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. Journal of Molecular Biology. 2016;428(9):1714–1724. doi: 10.1016/j.jmb.2016.02.004. Part A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of cell biology. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24(18):2918–31. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria Supply Membranes for Autophagosome Biogenesis during Starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- [10].Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature cell biology. 2009;11(12):1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- [11].Longatti A, Tooze SA. Recycling endosomes contribute to autophagosome formation. Autophagy. 2012;8(11):1682–3. doi: 10.4161/auto.21486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature cell biology. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Velazquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. The Journal of cell biology. 2016;212(6):621–31. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. The EMBO journal. 2015;34(16):2117–31. doi: 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgiendosomal system. Mol Biol Cell. 2010;21(22):3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. doi: 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yamaguchi H, Arakawa S, Kanaseki T, Miyatsuka T, Fujitani Y, Watada H, Tsujimoto Y, Shimizu S. Golgi membrane-associated degradation pathway in yeast and mammals. The EMBO journal. 2016 doi: 10.15252/embj.201593191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fernandez-Fernandez MR, Ruiz-Garcia D, Martin-Solana E, Chichon FJ, Carrascosa JL, Fernandez JJ. 3D electron tomography of brain tissue unveils distinct Golgi structures that sequester cytoplasmic contents in neurons. Journal of cell science. 2016 doi: 10.1242/jcs.188060. [DOI] [PubMed] [Google Scholar]
- [19].Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5(8):1180–5. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- [20].Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. The Journal of cell biology. 2010;190(6):1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. The Journal of cell biology. 2012;198(2):219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. Journal of cell science. 2013;126(Pt 11):2534–44. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- [23].Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nature reviews Molecular cell biology. 2013;14(12):759–74. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- [24].Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Current opinion in cell biology. 2016;39:61–8. doi: 10.1016/j.ceb.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Current opinion in cell biology. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- [26].Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. The Journal of cell biology. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5(7):973–9. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- [28].Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151(7):1501–12. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–23. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- [30].Vanni S, Hirose H, Barelli H, Antonny B, Gautier R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun. 2014;5:4916. doi: 10.1038/ncomms5916. [DOI] [PubMed] [Google Scholar]
- [31].Baskaran S, Carlson LA, Stjepanovic G, Young LN, do Kim J, Grob P, Stanley RE, Nogales E, Hurley JH. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife. 2014;3 doi: 10.7554/eLife.05115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350(6257):aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10(12):6742–54. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Devereaux K, Dall’Armi C, Alcazar-Roman A, Ogasawara Y, Zhou X, Wang F, Yamamoto A, De Camilli P, Di Paolo G. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS One. 2013;8(10):e76405. doi: 10.1371/journal.pone.0076405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jao CC, Ragusa MJ, Stanley RE, Hurley JH. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. PNAS. 2013;110(14):5486–5491. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, Williams RL. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327(5973):1638–42. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Herman PK, Stack JH, DeModena JA, Emr SD. A novel protein kinase homolog essential for protein sorting to the yeast lysosome-like vacuole. Cell. 1991;64(2):425–37. doi: 10.1016/0092-8674(91)90650-n. [DOI] [PubMed] [Google Scholar]
- [38].Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proceedings of the National Academy of Sciences of the United States of America; 2011. pp. 7769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, Deng H, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22(3):473–89. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. The EMBO journal. 2009;28(15):2244–58. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11(4):468–78. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- [42].Noda T, Matsunaga K, Taguchi-Atarashi N, Yoshimori T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin Cell Dev Biol. 2010;21(7):671–6. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- [43].Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The Reversible Modification Regulates the Membrane-Binding State of Apg8/Aut7 Essential for Autophagy and the Cytoplasm to Vacuole Targeting Pathway. The Journal of cell biology. 2000;151(2):263–275. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- [45].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal. 2000;19(21):5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12(7):226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. The Journal of biological chemistry. 2004;279(39):40584–92. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- [48].Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. The Journal of biological chemistry. 2007;282(52):37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- [49].Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- [50].Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nature cell biology. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- [52].Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156(3):469–81. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- [53].Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- [54].Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20(4):444–54. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- [55].Wu F, Watanabe Y, Guo XY, Qi X, Wang P, Zhao HY, Wang Z, Fujioka Y, Zhang H, Ren JQ, Fang TC, et al. Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy. Molecular cell. 2015;60(6):914–29. doi: 10.1016/j.molcel.2015.11.019. [DOI] [PubMed] [Google Scholar]
- [56].van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nature reviews Molecular cell biology. 2008;9(2):112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146(2):290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nature cell biology. 2014;16(5):415–24. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. The Journal of biological chemistry. 2006;281(6):3017–24. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- [60].Oh-oka K, Nakatogawa H, Ohsumi Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of Atg8. The Journal of biological chemistry. 2008;283(32):21847–52. doi: 10.1074/jbc.M801836200. [DOI] [PubMed] [Google Scholar]
- [61].Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. The EMBO journal. 2012;31(22):4304–17. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kuhnel K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a betapropeller protein family. Proceedings of the National Academy of Sciences of the United States of America; 2012. pp. E2042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23(5):896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. The Journal of biological chemistry. 2008;283(35):23972–80. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kishi-Itakura C, Koyama-Honda I, Itakura E, Mizushima N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. Journal of cell science. 2014;127(Pt 18):4089–102. doi: 10.1242/jcs.156034. [DOI] [PubMed] [Google Scholar]
- [66].Juris L, Montino M, Rube P, Schlotterhose P, Thumm M, Krick R. PI3P binding by Atg21 organises Atg8 lipidation. The EMBO journal. 2015;34(7):955–73. doi: 10.15252/embj.201488957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Molecular cell. 2014;55(2):238–52. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dall’Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, Yu WH, Robinson KS, Yeku O, Small SA, Duff K, et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun. 2010;1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cheng J, Fujita A, Yamamoto H, Tatematsu T, Kakuta S, Obara K, Ohsumi Y, Fujimoto T. Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat Commun. 2014;5:3207. doi: 10.1038/ncomms4207. [DOI] [PubMed] [Google Scholar]
- [70].Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33(1):109–22. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- [71].Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. The Journal of cell biology. 2010;188(2):253–69. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, Reggiori F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012;22(17):1545–53. doi: 10.1016/j.cub.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S, Koyama T, et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011;9(5):376–89. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- [74].Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang H, Sun HQ, Zhu X, Zhang L, Albanesi J, Levine B, Yin H. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proceedings of the National Academy of Sciences of the United States of America; 2015. pp. 7015–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hasegawa J, Iwamoto R, Otomo T, Nezu A, Hamasaki M, Yoshimori T. Autophagosomelysosome fusion in neurons requires INPP5E, a protein associated with Joubert syndrome. The EMBO journal. 2016 doi: 10.15252/embj.201593148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10(7):883–93. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, Kayserili H, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009;41(9):1032–6. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]