Abstract
The microbiota primes immune defences but the identity of specific commensal microbes which protect against infection is unclear. Conversely, how pathogens compete with the microbiota to establish their host niche is also poorly understood. Here, we investigate the antagonism between the microbiota and Klebsiella pneumoniae during colonization and transmission. We discover that maturation of the microbiota drives the development of distinct immune defence programs in the upper airway and intestine to limit K. pneumoniae colonization within these niches. Immune protection in the intestine depends on the development of Bacteroidetes, IL-36 signalling and macrophages. This effect of Bacteroidetes requires their conserved commensal colonization factor (CCF) polysaccharide utilization locus. Conversely, in the upper airway, Proteobacteria prime immunity through IL-17A, but K. pneumoniae overcomes these defences through encapsulation to effectively colonize this site. Ultimately, we find that host-to-host spread of K. pneumoniae occurs principally from its intestinal reservoir, and that CCF-producing Bacteroidetes are sufficient to prevent transmission between hosts through IL-36. Thus, our study provides mechanistic insight into when, where and how commensal Bacteroidetes protect against K. pneumoniae colonization and contagion, providing insight into how these protective microbes could be harnessed to confer population-level protection against K. pneumoniae infection.
Introduction
The microbiota enhances immune defences to protect against pathogenic microbes1–4. Identifying members of the microbiota that protect against pathogens could therefore provide an alternative means to treat infections resistant to current antimicrobial therapies5. Of these antimicrobial resistant organisms, it is bacteria from the Enterobacteriaceae family which are the greatest clinical problem6–9. Within this family, K. pneumoniae are the most urgent threat to human health because many strains are resistant to multiple antibiotics, are highly virulent, cause disease in both adults and infants, and readily spreading between hosts8,10–14.
Asymptomatic colonization is the pivotal step in K. pneumoniae pathogenesis as this serves as the reservoir of organisms which initiate acute infection and is also the source of organisms which transmit to other hosts10–13,15,16. K. pneumoniae is a versatile colonizer with two principal niches within the human host: the upper airway and intestine11,13,15,17,18. To establish colonization K. pneumoniae must therefore contend with defences established by the microbiota and immune system at both of these sites. Clinical data confirm the importance of the microbiota and immune system in resisting the establishment of K. pneumoniae colonization. Specifically, the association between patients receiving antibiotics and increased incidence of K. pneumoniae colonization10,19; the demonstration that microbiota transplantation, in humans and mice, clears K. pneumoniae from the intestine20–24; and the association between immunosuppression and increased risk of K. pneumoniae colonization25–27 highlight the critical role of the microbiota and immune system in protecting against K. pneumoniae colonization. The mechanistic basis for this protection, and whether there is collaboration between these microbial and immune defences system, is poorly understood hampering the use of microbiota-based approaches to protect against K. pneumoniae.
Once colonization has been established, these hosts serve as reservoirs for the transmission of K. pneumoniae within a population. The process of transmission involves exiting the colonized host, surviving in the environment, acquisition by a new host, and then successful reestablishment of colonization at a new mucosal site. Although the importance of K. pneumoniae transmission is undoubted, and has underpinned its success as a pathogen, the host and bacterial factors dictating the outcome of transmission are poorly defined. Here, by analysing the development of microbiota-mediated defences in the upper airway and intestine, we reveal the commensal taxa that inhibit K. pneumoniae colonization and transmission, the immunological basis for their protection, and the countervailing tactics used by K. pneumoniae to establish colonization in the face of these defences. Through this fundamental analysis, we define a simple consortium of commensal microbes which prevent the host-to-host spread of this contagious pathogen.
Results
The development of the adult microbiota protects against colonization by antibiotic-resistant Klebsiella pneumoniae in the intestine but not upper airway
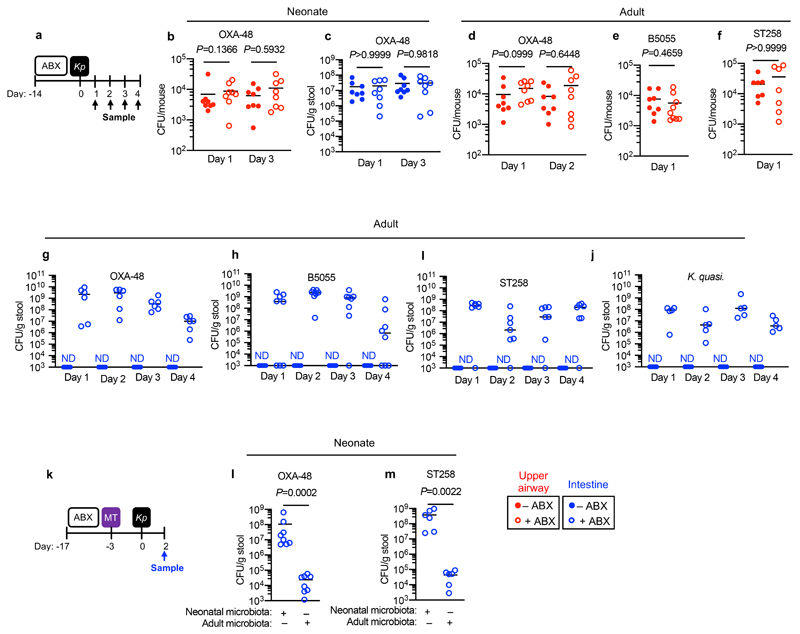
As K. pneumoniae causes disease in infants and adults, we wanted to investigate the development of microbiota-mediated defences against K. pneumoniae colonization across lifecourse. To do this, we depleted the microbiota in neonatal and adult mice (Supplementary Fig. 1a-e). To study upper airway colonization by K. pneumoniae, we used a small inoculation volume and did not anaesthetize mice for intranasal inoculation. This approach means bacteria remain in the nasal cavities and nasopharynx and do not reach the lung28,29 (Supplementary Fig. 2). In neonates, K. pneumoniae was able to colonize both the upper airway and intestine and this unaffected by microbiota-depletion (Fig. 1a-c). Likewise, in the adult upper airway, K. pneumoniae colonization levels were similar between control and microbiota-depleted animals (Fig. 1d-f). In the adult intestine, by contrast, K. pneumoniae could only establish detectable colonization after microbiota-depletion (Fig. 1g-j). Sustained broad spectrum antibiotic treatment (Fig. 1g-j), or a short, clinically-relevant antibiotic regimen (Supplementary Fig. 3a,b) eliminated microbiota-mediated defences. Transferring the adult, but not the neonatal microbiota to neonatal mice protected against intestinal colonization by K. pneumoniae (Fig. 1k-m). The adult intestinal microbiota was broadly protective, inhibiting colonization by clinical isolates of K. pneumoniae producing the OXA-48 carbapenenemase (Fig. 1g), the pandemic K. pneumoniae ST258 (Fig. 1l), and closely related K. quasipneumoniae subsp. similipneumoniae (Fig. 1j). No weight loss was observed during colonization of either niche (Supplementary Fig. 4a,b) supporting the notion that we were modelling asymptomatic colonization. These data show that the development of the adult microbiota creates a barrier sufficient to block K. pneumoniae colonization in the intestine but not the upper airway. This raised three questions: (i) which members of the adult intestinal microbiota prevent K. pneumoniae colonization, and by what mechanism? (ii) What factors allow K. pneumoniae to successfully compete with the upper airway microbiota to colonize this niche? And, most critically, (iii) how does this competition between the microbiota and K. pneumoniae at different mucosal reservoirs impact the host-to-host spread of this contagious pathogen?
Figure 1. The adult microbiota protects against colonization by antibiotic-resistant Klebsiella pneumoniae in the intestine but not upper airway.
(a) Experimental scheme for (b-j): “ABX” = metronidazole, neomycin, vancomycin and ampicillin (MNVA) in drinking water; “Kp” = K. pneumoniae oral or intranasal inoculation; and “Sample” = upper airway or fecal sampling day. Days relative to K. pneumoniae inoculation. (b,c) K. pneumoniae (OXA-48) burden in the (b) upper airway (n=8 animals) or (c) colon of neonatal mice (n=8 animals). (d-f) K. pneumoniae (OXA-48) (n=8 animals) (d), K. pneumoniae (B5055) (n=8,9 animals) (e), and K. pneumoniae (ST258) (n=7 animals) (f) burden in upper airways of adult mice. (g-j) K. pneumoniae (OXA-48) (n=6 animals) (g), K. pneumoniae (B5055) (n=8 animals) (h), K. pneumoniae (ST258) (n=7 animals) (i) and K. quasipneumoniae subsp. similipneumoniae (n=5 animals) (j) fecal burden in adult mice. (k) Experimental scheme for (l,m): “ABX” = MNVA in drinking water; “MT” = microbiota transfer by oral and intranasal inoculation; “Kp” = K. pneumoniae oral inoculation; and “Sample” = fecal sampling day. K. pneumoniae (OXA-48) (n=5 animals) (l) and K. pneumoniae (ST258) (n=6 animals) (m) fecal burden in adult mice. Upper airway colonization data displayed in red and intestinal colonization data in blue. All statistical comparisons were made by Mann-Whitney test (two-tailed), horizontal lines indicate median values, ND is none detected (limit of detection for K. pneumoniae in feces = 103 CFU/g).
The development of Bacteroidetes prevents Klebsiella pneumoniae intestinal colonization
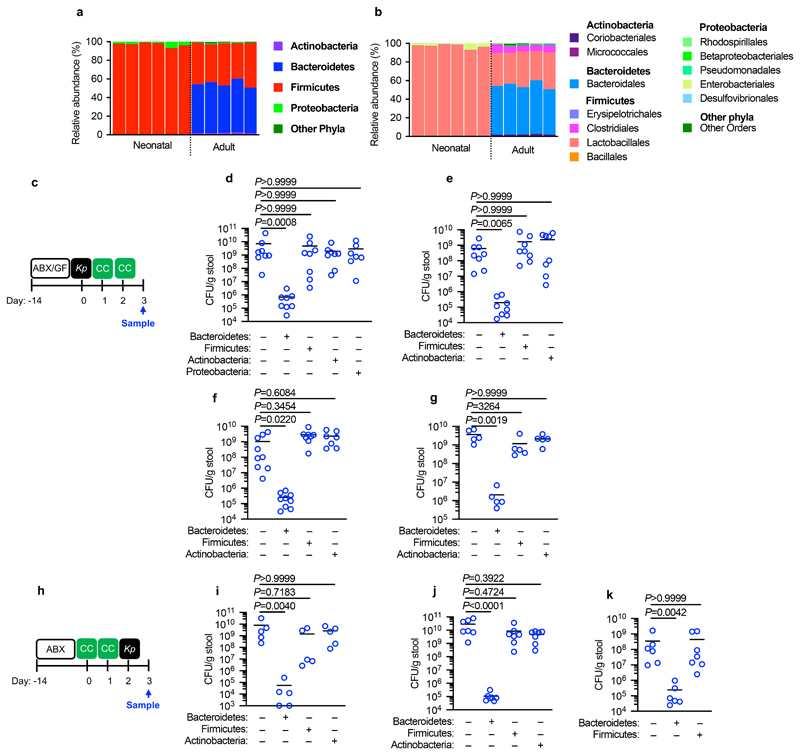
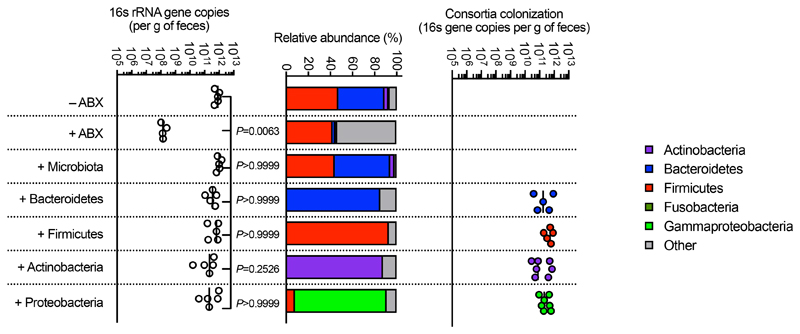
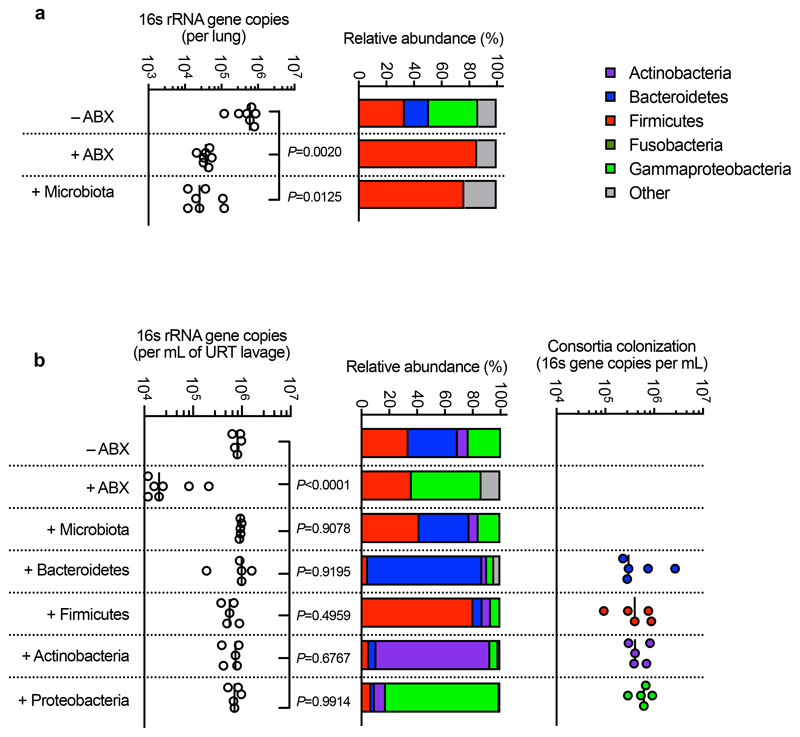
To answer our first question, we sequenced the neonatal and adult mouse intestinal microbiota to determine the respective composition of these permissive and inhibitory microbial communities. The neonatal intestinal microbiota was dominated by bacteria from the Firmicutes phylum, and specifically members of the Lactobacillales order (Fig. 2a,b). The adult intestinal microbiota, by contrast, was composed predominately of commensals from the Bacteroidetes (Bacteroidales) and Firmicutes (Clostridia and Lactobacillales) phyla, with Actinobacteria and Proteobacteria comprising a minor fraction of the commensal taxa present (Fig. 2a,b). We therefore used defined consortia of commensals to determine whether the development of Bacteroidetes, non-Lactobacillales Firmicutes, Actinobacteria, or Proteobacteria drove resistance to intestinal colonization by K. pneumoniae. We found that only Bacteroidetes promoted clearance of K. pneumoniae from the intestine, whether administered before or after the establishment of K. pneumoniae colonization (Fig. 2c-k). Bacteroidetes were protective in both antibiotic treated (Fig. 2d-f,i,j) and germ-free mice (Fig. 2g) and this was consistent using commensal consortia of different compositions (Extended Data Fig. 1). We verified the intestinal microbiota of mice administered our representative commensal consortia was dominated by that consortium and that colonization levels of each consortia were similar (Extended Data Fig. 2). Additionally, we found that Bacteroidetes specifically protected against intestinal colonization by K. pneumoniae in neonates (Fig. 2k), supporting the idea that it is the development of this commensal phylum that protects against K. pneumoniae in adults.
Figure 2. Intestinal Bacteroidetes protect against Klebsiella pneumoniae colonization.
(a,b) Relative abundance of bacterial phyla (a) and orders (b) in (adult feces n=5 animals) and large intestine of neonatal mice (n=6 animals). Each bar represents a single mouse. (c) Experimental scheme for (d-g): “ABX” = MNVA in drinking water; “GF” = germ-free mice; “CC” = commensal consortia inoculation by oral gavage (see Materials and Methods); “Kp” = K. pneumoniae oral inoculation; and “Sample” = fecal sampling day. Days relative to K. pneumoniae inoculation. (d-f) K. pneumoniae (OXA-48) (n=7,8 animals) (d), K. pneumoniae (B5055) (n=8 animals) (e), and K. pneumoniae (ST258) (n=7,8 animals) (f) fecal burden in adult antibiotic treated mice. (g) K. pneumoniae (ST258) fecal burden in germ-free mice (n=5 animals). (h) Experimental scheme for (i-k): acronyms as in (c). K. pneumoniae (OXA-48) (n=5 animals) (i) and K. pneumoniae (ST258) (n=7 animals) (j) fecal burden in adult mice. K. pneumoniae (ST258) (k) fecal burden in neonatal mice (n=6,7 animals). All statistical comparisons were made by Kruskal-Wallis test with Dunn’s correction for multiple comparisons. Horizontal lines indicate median values.
CCF-producing Bacteroidetes fortifies the intestinal immune barrier via IL-36 and macrophages to prevent Klebsiella pneumoniae intestinal colonization
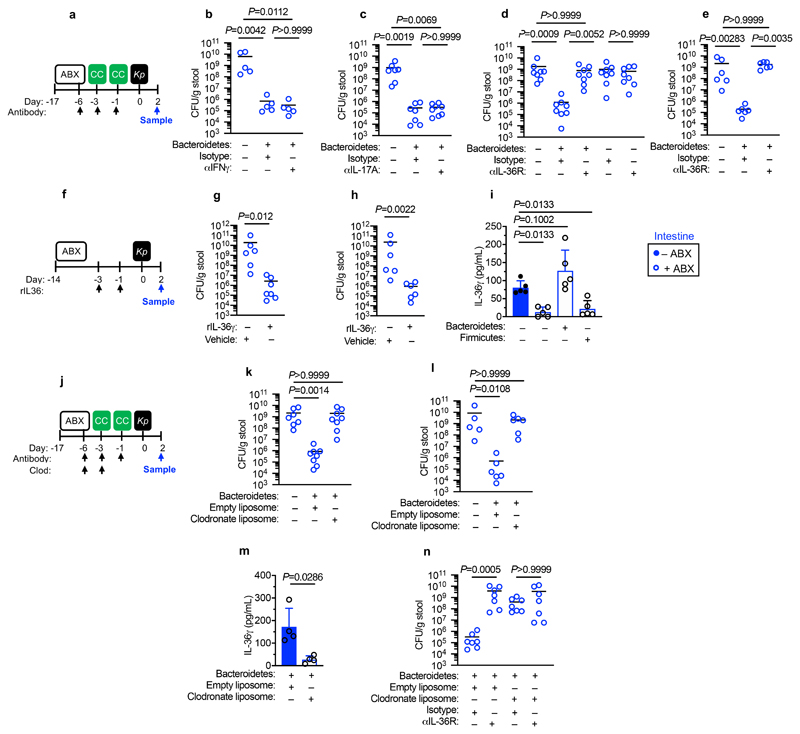
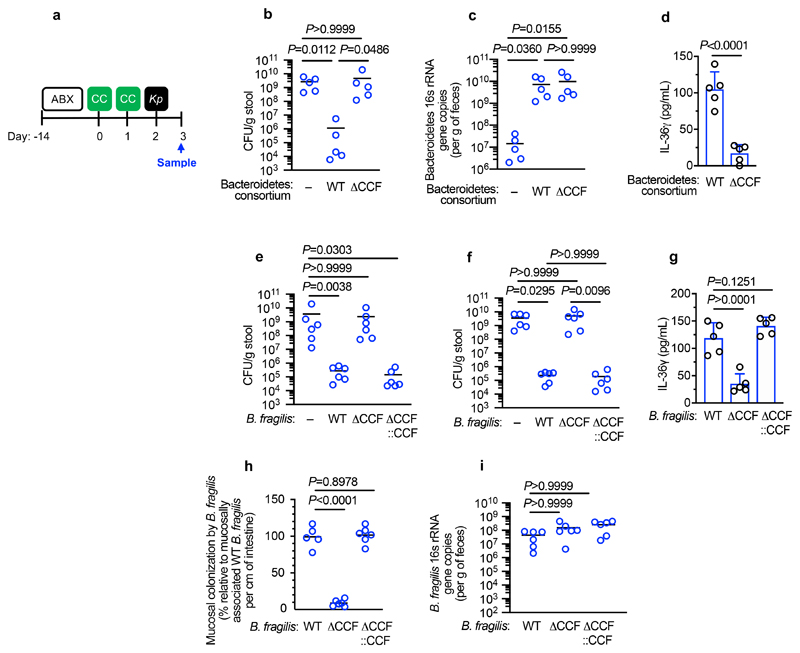
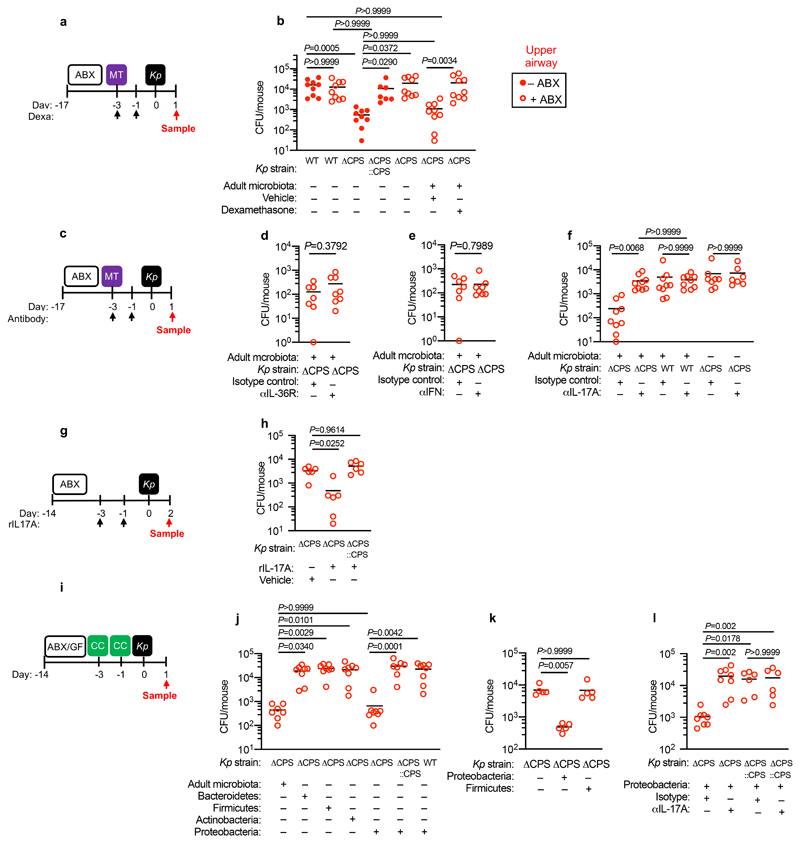
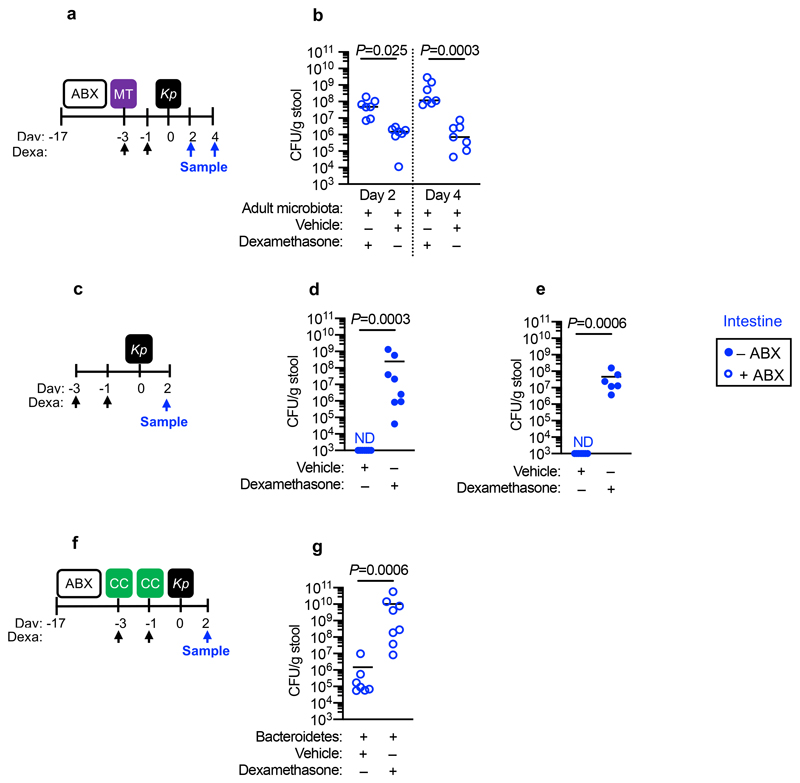
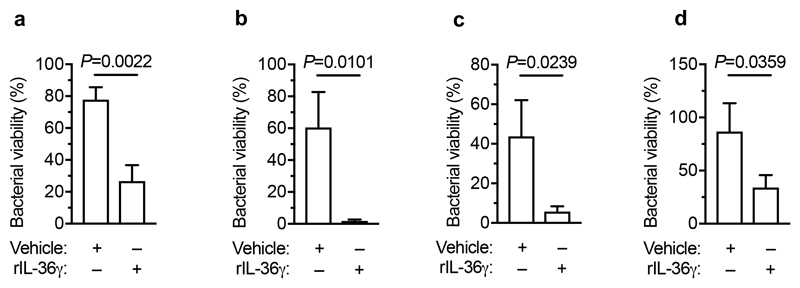
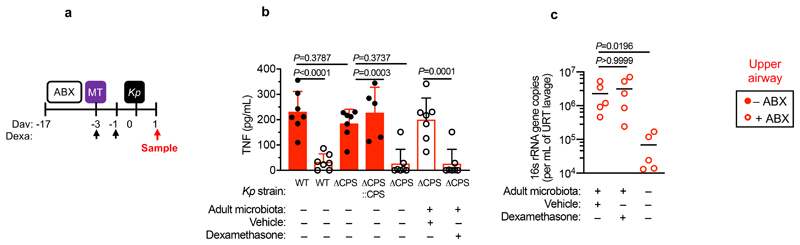
Next, we wanted to determine whether the protective effect of Bacteroidetes required host immunity. We therefore transferred the microbiota to microbiota-depleted mice administered the immunosuppressive dexamethasone, or vehicle control, prior to K. pneumoniae colonization. Dexamethasone eliminated the inhibitory effect of the microbiota on K. pneumoniae colonization (Extended Data Fig. 3a,b), indicating that protection required immune signalling. Furthermore, we found that dexamethasone treatment of non-antibiotic treated mice made these normally resistant mice susceptible to colonization by K. pneumoniae (Extended Data Fig. 3c-e). Similarly, we found that protection conferred by Bacteroidetes required immune signalling (Extended Data Fig. 3f,g). To ensure that dexamethasone was acting on the immune system rather than disrupting the microbiota, we transferred the microbiota from dexamethasone or vehicle control treated mice, to microbiota-depleted mice and then examined the inhibitory effect on K. pneumoniae colonization. Both of these microbiota provided equivalent protection against K. pneumoniae colonization (Supplementary Fig. 5), demonstrating that dexamethasone does not eliminate protective commensals from the microbiota but instead inhibits the stimulatory effect of the microbiota on the immune system. Next, we sought to determine the immune factors which translate the effect of Bacteroidetes into resistance against K. pneumoniae colonization. In the intestine, the transition from neonate to adult was marked by the increased homeostatic expression of a number of cytokines important in maintaining host-microbial homeostasis (Supplementary Fig. 6). To determine whether the protective effect of Bacteroidetes was driven by these cytokine, we abrogated the signalling of cytokines whose homeostatic expression increased from neonates to adults in a microbiota-dependent manner. Protection against intestinal colonization by K. pneumoniae by Bacteroidetes was abrogated after treatment with an antibody targeting the IL-36 receptor signalling, but not IL-17A or IFNγ, (Fig. 3a-e). Supporting the role of IL-36, we found that treatment with recombinant IL-36γ promoted clearance of intestinal K. pneumoniae (Fig. 3f-h) and that intestinal IL-36γ production in microbiota-depleted animals was specifically restored by Bacteroidetes (Fig. 3i). Within the intestine, macrophages act as a critical innate interface with the microbiota, so we therefore investigated their role in protection against K. pneumoniae using liposome clodronate treatment to deplete these cells (Supplementary Fig. 7). After macrophage depletion, Bacteroidetes could not protect against K. pneumoniae, or regulate IL-36γ (Fig. 3j-m). By contrast Bacteroidetes were still protective after neutrophil depletion (Supplementary Fig. 8). Furthermore, in clodronate treated mice, disruption of IL-36R signalling did not increase K. pneumoniae colonization, suggesting that IL-36 and macrophages work along a common pathway (Fig. 3n). To understand how IL-36 controls K. pneumoniae levels in the intestine via macrophages, we investigated if IL-36 promotes the bactericidal activity of these cells. We found that IL-36γ stimulation promoted the killing of multiple strains of K. pneumoniae by macrophages (Extended Data Fig. 4a-d). Next, we wanted to understand the factors in Bacteroidetes required for protection against K. pneumoniae colonization. Common amongst commensals that regulate intestinal immunity is a requirement for intimate contact with the host mucosa to exert their effects30. Highly conserved within Bacteroidetes is a capsular polysaccharide production locus, the commensal colonization factors (CCF), which promote the association of Bacteroidetes with the intestinal mucosa31,32. We tested the hypothesis that CCFs are required for Bacteroidetes to inhibit K. pneumoniae colonization. In agreement with this, a consortium of Bacteroidetes, or a single Bacteroidetes species, was able to protect against K. pneumoniae colonization and prime intestinal IL-36γ production with an intact CCF system, but this was lost in the absence of CCFs, and could be reestablished by complementation of the CCFs (Fig. 4a-g). In line with their proposed role in mucosal colonization, we found that the CCFs promoted the association of Bacteroidetes with the mucosa (Fig. 4h), but fecal levels of Bacteroidetes were equivalent in the presence or absence of CCFs (Fig. 4i). Together, our data support a model whereby CCF-producing Bacteroidetes associate with the mucosal barrier to create an immune barrier within the intestine preventing colonization by K. pneumoniae through IL-36 and macrophages.
Figure 3. Bacteroidetes protect against Klebsiella pneumoniae colonization in the intestine through IL-36 signalling and macrophages.
(a) Experimental scheme for (b-e): “ABX” = MNVA in drinking water; “CC” = commensal consortia oral inoculation; “Kp” = K. pneumoniae oral inoculation; and “Sample” = fecal sampling day; “Antibody treatment” = antibody, or isotype control, treatment. Days relative to K. pneumoniae inoculation. (b-e) K. pneumoniae (OXA-48) burden in the feces of adult mice (n=5,7,8 animals). (e) K. pneumoniae (ST258) burden in the feces of adult mice (n=6 animals). (f) Experimental scheme for (g-i): acronyms as in (a), days on which recombinant IL-36γ was administered by intraperitoneal injection are indicated. (g,h) K. pneumoniae (OXA-48) burden (n=6,7 animals) (g), and K. pneumoniae (ST258) burden (n=6 animals) (h) in the feces of adult mice. (i) Intestinal cytokine levels 3 days after oral inoculation with indicated commensal consortia (n=5 animals). (j) Experimental plan for (k-n), acronyms in experimental plan as in (a), “Clodronate” = day of clodronate liposome, or empty liposome treatment. (k,l) K. pneumoniae (OXA-48) burden (n=7,8 animals) (k), and K. pneumoniae (ST258) burden (n=5,6 animals) (l) in the feces of adult mice. (m) Intestinal cytokine levels 3 days after oral inoculation with indicated commensal consortia (n=5). (n) K. pneumoniae (OXA-48) fecal burden in adult mice (n=6,7 animals). Statistical comparisons were made by Kruskal–Wallis test with Dunn’s correction for multiple comparisons (b-e, i-l and n), and by Mann-Whitney test (two-tailed) (g,h,m). Horizontal lines indicate median values, error bars are standard deviation.
Figure 4. Bacteroidetes require their commensal colonization factors to protect against Klebsiella pneumoniae colonization in the intestine.
(a) Experimental scheme: “ABX” = MNVA in drinking water; “CC” = commensal consortia oral inoculation; “Kp” = K. pneumoniae oral inoculation; and “Sample” = fecal sampling day. Days relative to K. pneumoniae inoculation. (b) K. pneumoniae (OXA-48) fecal burden in adult mice (n=5 animals). Mice were orally inoculated with either a consortium of wild-type or isogenic ΔCCF Bacteroidetes (B. fragilis and B. vulgatus). (d) Bacteroidetes levels in feces of mice from (b). (d) Intestinal cytokine levels 3 days after oral inoculation of Bacteroidetes consortia (n=5 animals). (e,f) K. pneumoniae (OXA-48) (n=6 animals) (e) and K. pneumoniae (ST258) (n=6 animals) (f) fecal burden in adult mice. Indicated mice were orally inoculated with either WT B. fragilis, ΔCCF B. fragilis or ΔCCF::CCF B. fragilis. (g) Intestinal cytokine levels 3 days after oral inoculation with either WT B. fragilis, ΔCCF B. fragilis or ΔCCF::CCF B. fragilis (n=5). (h) Mucosal association of WT B. fragilis, ΔCCF B. fragilis and ΔCCF::CCF B. fragilis (n=5,6 animals). (i) B. fragilis levels in feces (n=5,6 animals). Statistical comparisons were made by Kruskal–Wallis test with Dunn’s correction for multiple comparisons (b-c,e-i), and by Student’s t-test (two-tailed) (d). Horizontal lines indicate median values, error bars are standard deviation.
The adult microbiota primes upper airway defences through IL-17A, but encapsulation allows Klebsiella pneumoniae to overcome this barrier to establish colonization
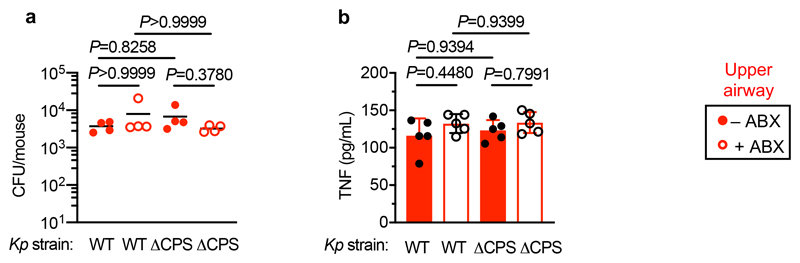
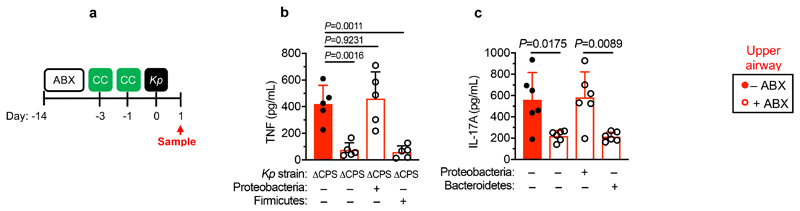
We then wanted to answer our second question: what factors allow K. pneumoniae to successfully compete with the upper airway microbiota to colonize this niche? To do this, we examined the role of K. pneumoniae encapsulation. Traditionally, encapsulation has been viewed as a virulence determinant, however, capsular polysaccharide production is common to both pathogens and non-pathogens, especially in the upper airway33. In adult mice with a microbiota, we found that encapsulated K. pneumoniae colonized the upper airway higher levels than an isogenic unencapsulated K. pneumoniae mutant34 and the ability to withstand microbiota-mediated defences was restored by complementation of the unencapsulated mutant (Fig. 5a,b). This advantage of encapsulation was lost in microbiota-depleted animals, where encapsulated and unencapsulated K. pneumoniae colonized to similar levels (Fig. 5a,b). Next, therefore, we determined whether encapsulation allows K. pneumoniae to withstand direct microbial competition within the upper airway, or indirect effects of the microbiota via the immune system. To address this, we transferred the microbiota back to microbiota-depleted animals treated with dexamethasone or vehicle control, prior to intranasal administration of K. pneumoniae. Microbiota transfer reduced upper airway colonization by unencapsulated K. pneumoniae in vehicle control (Fig. 5a,b), but not dexamethasone treated animals (Fig. 5a,b), supporting the notion that the microbiota enhances defences in the upper airway via immune signalling. In agreement with this, innate cytokine production (TNFα), which is induced by K. pneumoniae during upper airway colonization, was reduced in microbiota-depleted animals, and restored by microbiota transfer through immune signalling (Extended Data Fig. 5a,b). We confirmed that levels of upper airway commensal bacteria after microbiota transfer were equivalent between vehicle and dexamethasone treated mice (Extended Data Fig. 5c) and that microbiota transfer did not result in commensal inoculation into the lung (Extended Data Fig. 6a). Analogously to the intestine, microbiota priming of immune defences in the upper airway was specific to adult animals, as encapsulated and unencapsulated K. pneumoniae colonized the neonatal upper airway to similar levels and induced an equivalent innate cytokine response in microbiota-depleted and control animals (Extended Data Fig. 7a,b). The transition from neonate to adult was accompanied by the increased homeostatic expression of a number of cytokines in the upper airway which were dependent on the microbiota (Supplementary Fig. 9). We therefore abrogated signalling by these cytokines to understand if they were required for the adult microbiota to exert its immunomodulatory effects in the upper airway. Disruption of IL-17A, but not IFNγ or IL-36R signalling, inhibited microbiota-mediated clearance of unencapsulated, but not encapsulated, K. pneumoniae from the upper airway (Fig. 5c-f). Furthermore, treatment with recombinant IL-17A was sufficient to promote clearance of unencapsulated K. pneumoniae (Fig. 5g,h). The upper airway microbiota of mice comprises bacteria from the Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria phyla (Extended Data Fig. 6b)35–41. Of these commensals, we found that Proteobacteria specifically enhanced clearance of unencapsulated K. pneumoniae from the upper airway in antibiotic treated and germ-free mice by IL-17A signalling (Fig. 5i-l). Proteobacteria also restored homeostatic IL-17A production in the upper airway and cytokine production induced by K. pneumoniae colonization (Extended Data Fig. 8a-c). We verified the upper airway microbiota of mice administered our representative commensal consortia was dominated by that consortium and colonization by each consortia were similar (Extended Data Fig. 6b). These data support a model whereby Proteobacteria prime an IL-17A-dependent immune defence program in the adult upper airway, however, encapsulation allows K. pneumoniae to withstand these defences to successfully establish colonization.
Figure 5. Proteobacteria primes upper airway defences through IL-17A but encapsulation allows Klebsiella pneumoniae to overcome these defences.
(a) Experimental scheme for (b): “ABX” = MNVA in drinking water; “MT” = microbiota transfer; “Kp” = K. pneumoniae intranasal inoculation; and “Sample” = upper airway lavage sampling; “Dexa” = dexamethasone treatment. Days relative to K. pneumoniae inoculation. (b) Upper airway K. pneumoniae burden (WT B5055), unencapsulated K. pneumoniae (ΔCPS B5055), and complemented K. pneumoniae (ΔCPS::CPS B5055) in adult mice (n=7,9 animals). (c) Experimental plan for (d-f), acronyms as in (a), “Antibody treatment” = antibody, or isotype control, treatment. Days relative to K. pneumoniae inoculation. (d-f) K. pneumoniae (WT and ΔCPS B5055) upper airway burden in adult mice (n=5,7,8,9 animals). (g) Experimental scheme for (h): acronyms as in (a), days of rIL-17A, or vehicle control, administration are indicated. (h) Upper airway burden of unencapsulated K. pneumoniae (ΔCPS B5055), and complemented K. pneumoniae (ΔCPS::CPS B5055) in adult mice (n=6 animals). (i) Experimental plan for (j-m), acronyms in experimental plan as in (a,c), “CC” = indicated commensal consortia inoculation by intranasal inoculation (see Materials and Methods). (j-m) Upper airway K. pneumoniae burden (WT B5055), unencapsulated K. pneumoniae (ΔCPS B5055), and complemented K. pneumoniae (ΔCPS::CPS B5055) in adult mice (n=5,7,8 animals). Mice were antibiotic treated in (j) and (l) and germ-free in (k). Statistical comparisons were made by Kruskal–Wallis test with Dunn’s correction for multiple comparisons (b,f,h,j-m) and by Mann-Whitney test (two-tailed) (d,e). Horizontal lines are median values
Intestinal Bacteroidetes protect against Klebsiella pneumoniae transmission via IL-36 and this requires the Bacteroidetes commensal colonization factors
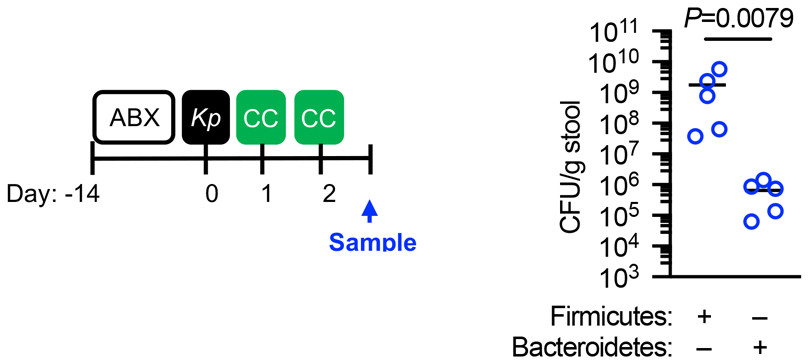
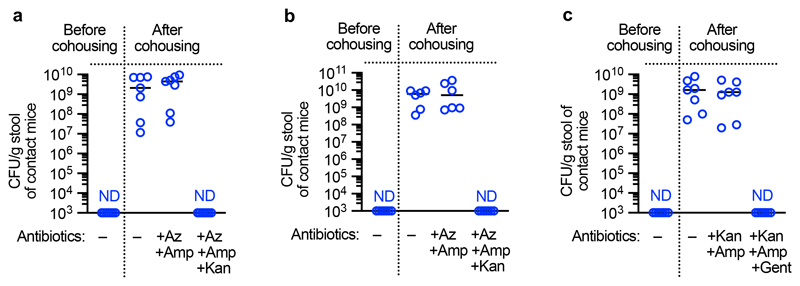
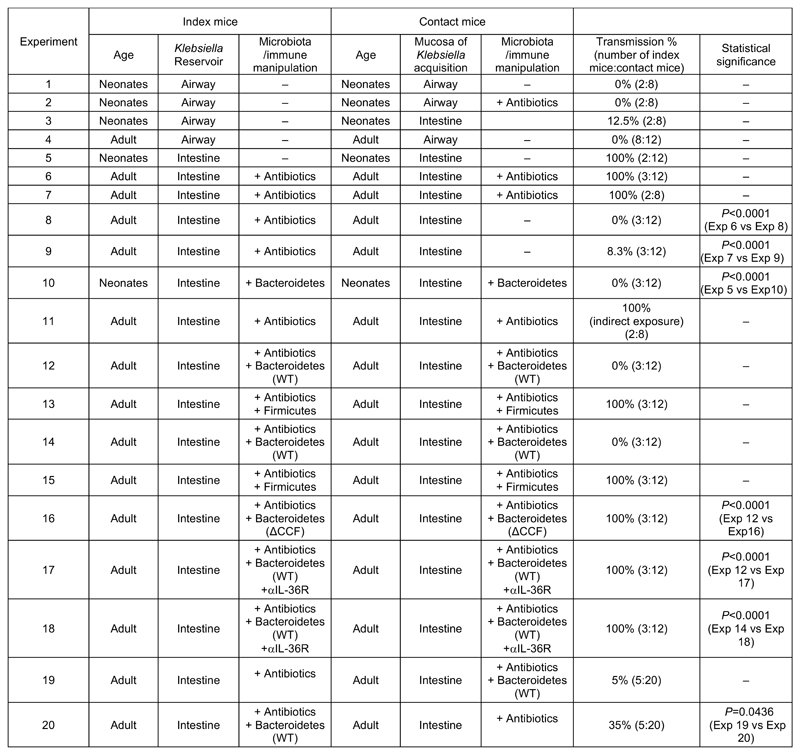
Individuals colonized by K. pneumoniae act as reservoirs for its spread to other hosts within a population15,16,42,43 which led to our third question: how does the battle between K. pneumoniae and the microbiota during colonization of different mucosal niches impact its host-to-host spread? To answer this, first, we investigated the contribution of the upper airway and intestinal reservoirs to K. pneumoniae transmission. To investigate the role of the upper airway reservoir, neonatal mice (7 – 11 days old) with established upper airway K. pneumoniae colonization (referred to as “index” mice) were cohoused with naïve (non-K. pneumoniae colonized) neonatal mice (referred to as “contact” mice) and then to determine transmission, K. pneumoniae colonization levels in contact animals were quantified. After 4 days of cohousing there was no detectable K. pneumoniae in the upper airway of the contacts (Extended Data Figure 10, experiment 1). No transmission was observed if the contact mice were antibiotic treated or non-antibiotic treated before cohousing (Extended Data Figure 10, experiment 2). There was, however, limited transmission from the upper airway of neonatal index mice to the intestine of neonatal contacts (Extended Data Figure 10, experiment 3). This suggests that despite mice not sneezing there is limited shedding of K. pneumoniae from the upper airway. Similarly to neonates, no transmission was observed from the upper airway of adult index to the same niche in adult contact animals (Extended Data Figure 10, experiment 4). We then assessed transmission between intestinal reservoirs in neonates aged between 7 and 11 days as these animals are not coprophagic44. Cohousing neonatal index mice with established intestinal colonization with naïve neonatal contacts resulted in transmission of K. pneumoniae to the intestine of all contacts (Extended Data Figure 10, experiment 5), indicating that in the absence of microbiota-mediated defences, K. pneumoniae transmission occurs primarily from the intestinal reservoir. We confirmed that the antibiotic resistance profiles of K. pneumoniae isolated from neonatal contact mice matched the antibiotic resistance profiles of K. pneumoniae inoculated into index mice (Extended Data Fig. 9a), confirming that the K. pneumoniae isolated from contact mice originated from the index animals. To understand how the development of the adult microbiota, which protects against K. pneumoniae colonization in the intestine, impacts transmission, we used three approaches. Firstly, adult index mice with established intestinal K. pneumoniae colonization were cohoused with either antibiotic treated, or non-antibiotic treated, contact mice. Antibiotic treated adult contact animals all acquired K. pneumoniae (Extended Data Figure 10, experiment 6 and 7), whereas there was limited transmission to non-antibiotic treated adult contact animals (Extended Data Figure 10, experiments 8 and 9). Again, we confirmed that the antibiotic resistance profiles of K. pneumoniae isolated from adult contact mice matched the antibiotic resistance profiles of K. pneumoniae inoculated into index mice (Extended Data Fig. 9b,c), confirming that the K. pneumoniae isolated from contact mice originated from the index animals. Secondly, we orally inoculated the protective Bacteroidetes commensals into K. pneumoniae colonized neonatal index mice, and also naïve neonatal contact mice, prior to cohousing. This was sufficient to block transmission of K. pneumoniae between hosts (Extended Data Figure 10, experiment 10 and Supplementary Fig. 10a). To examine whether transmission required direct contact of animals, adult contacts were housed in a cage that had previously housed intestinally colonized index mice. Despite all index mice and fecal material being removed from the cage prior to housing, all contact mice became colonized by K. pneumoniae (Extended Data Figure 10, experiment 11), demonstrating that transmission does not require direct contact between animals or coprophagy. Thirdly, we orally inoculated the protective Bacteroidetes commensals into both K. pneumoniae colonized adult index mice, and naïve antibiotic treated adult contact mice, prior to cohousing. We found that Bacteroidetes specifically were sufficient to block transmission of K. pneumoniae between hosts (Extended Data Figure 10, experiments 12-15, and Supplementary Fig. 10b). The inhibitory effect of Bacteroidetes was lost upon disruption of IL-36 signaling or in the absence of the commensal colonization factors (Extended Data Figure 10, experiments 16-18). Taken together, these data suggest that the protective effect of Bacteroidetes on K. pneumoniae colonization limits its transmission within a population. We questioned whether the inhibitory effect of Bacteroidetes on K. pneumoniae transmission was due to Bacteroidetes reducing K. pneumoniae shedding from the index animals, or by establishing a protective barrier against K. pneumoniae acquisition in contact animals. To answer this, Bacteroidetes were given to either the K. pneumoniae colonized index mice only, or the naïve contacts only. We found that both approaches reduced K. pneumoniae transmission but that introduction of Bacteroidetes to contacts was more effective than the effect of Bacteroidetes in index mice (Extended Data Figure 10, experiments 19 and 20), demonstrating that the critical point in transmission controlled by Bacteroidetes is fortification of the immune barrier to prevent K. pneumoniae establishment in a new host. Collectively, our data show that CCF-producing Bacteroidetes inhibit the spread of K. pneumoniae within a population.
Discussion
Whilst it has been assumed that colonization resistance is a property common to resident microbial communities colonizing different host mucosae, whether the rules and mechanisms of colonization resistance for a given pathogen are shared between different niches is poorly defined45. We reveal that microbiota development drives separate host defence programs in the adult intestine and upper airway to inhibit colonization by the K. pneumoniae (Supplementary Fig. 11). This major antibiotic-resistant pathogen can overcome these defences in the upper airway but cannot compete with the microbiota in the intestine. Because of this, microbiota dysbiosis within an individual might not only promote K. pneumoniae colonization in that host but also promotes its spread to others. Thus, the defined consortium of Bacteroidetes we identify here could provide population level protection against K. pneumoniae by preventing its transmission between hosts – the ultimate means of infectious disease control.
Materials and Methods
Bacterial Strains
Bacteria used in this study and their growth conditions are outlined in Supplementary Table 1.
PCR amplification and sequencing of wzi gene
To identify the capsule type of the K. pneumoniae strains used in this study, genomic DNA was isolated from specified K. pneumoniae strains using the Wizard Genomic DNA purification kit (Promega) as per the manufacturer’s instructions. A 580 bp segment of the wzi gene was amplified from the genomic DNA by PCR using the wzi primers outlined in Supplementary Table 2 46. The 25 µL PCR reaction contained 200 µM dNTPs, 0.5 µM of each primer, 5 µL genomic DNA template, 1 × GC buffer and 0.25 µL Phusion polymerase (New England Biolabs). The PCR reaction was performed, following denaturation at 94°C for 2 minutes, by running 30 cycles of: 94°C for 30 seconds; 55°C for 40 seconds; and 72°C for 30 seconds. A final elongation step of 72°C for 5 minutes ended the reaction. The PCR was run on a 2% TAE agarose gel and the 580 bp band excised and purified using the Monarch DNA gel extraction kit (New England Biolabs). Sanger sequencing was performed on both strands using the PCR primers. The resulting consensus sequence was used to identify its corresponding wzi gene using the capsular typing database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Mice
The use of mice was performed under the authority of the UK Home Office outlined in the Animals [Scientific Procedures] Act 1986 after ethical review by Imperial College London Animal Welfare and Ethical Review Body (PPL 70/7969). Wild-type C57BL/6 mice were purchased from Charles River (UK). Germ-free Swiss Webster mice were purchased from Taconic (Denmark). All adult mice were female and between 6 – 14 weeks old, except the germ-free mice which were both male and female. Adult mice were housed no more than five per cage with Aspen chip 2 bedding with a 12 hour light and 12 hour dark cycle at 20°C – 22°C. Neonatal mice were 7 – 11 days old. Mice were randomly assigned to experimental groups, water was provided ad libitum and mice were fed RM1 (Special Diet Services).
Microbiota-depletion, antibody, inhibitor and recombinant protein treatment
Mice were given broad-spectrum antibiotics (metronidazole 1 g/L, neomycin sulfate 1 g/L, ampicillin 1 g/L, and vancomycin 0.5 g/L (MNVA)) in drinking water for 7 – 14 days as described previously29. For treatment with a β-lactam and macrolide antibiotic combination, mice were given a total of four doses of azithromycin (10 mg/kg/dose) and cefotaxime (40 mg/kg/dose) by intraperitoneal injection 1 and 2 days prior to K. pneumoniae inoculation. Infant mice were indirectly exposed to antibiotics through their dams. Dams were given the MNVA antibiotic cocktail in their drinking water as in47. Indicated groups of mice were treated with dexamethasone (5 mg/kg/dose) (Sigma). Anti-IL-17A (R&D Systems), anti-IL-36R (Invitrogen), anti-IFNγ (Abcam), and isotype control were administered at timepoints indicated in figures prior to oral inoculation of K. pneumoniae via the intraperitoneal route at 75 μg/mouse to disrupt cytokine signalling. Neutrophils were depleted using the anti-mouse Ly-6G antibody (1A8) (Biolegend). Recombinant IL-17A (R&D Systems) and recombinant IL-36γ (R&D Systems) (10 μg/mouse) were administered at timepoints indicated in figures prior to oral inoculation of K. pneumoniae with inoculation via the intraperitoneal route. Macrophages were depleted using clodronate containing liposomes as described previously48.
Models of bacterial colonization, quantification of bacterial load in the upper airway and intestine
For upper airway colonization of K. pneumoniae in adult mice, unanaesthetized animals were intranasally administered indicated bacteria in 20 μL. In neonates, unanaesthetized animals were intranasally administered K. pneumoniae in 5 μL. To establish intestinal colonization in adults, unanaesthetized animals were orally inoculated with K. pneumoniae (5×105 CFU) in approximately 200 μL. For all experiments K. pneumoniae was grown to mid-log phase and resuspended in sterile PBS for inoculation. Microbiota transfer was carried out as described in29, 3 days prior to inoculation with K. pneumoniae. Quantification of commensal bacterial load in the upper airway and intestine was carried out essentially as described in29. For 16s rRNA analysis of consortia engraftment, we used primers outlined in Supplementary Table 2 (see Microbiome sequencing). To quantify Klebsiella in the upper airway and intestine, Klebsiella ChromoSelect Selective Agar Base (Sigma) and for β-lactamase-producing Klebsiella, ESBL ChromoSelect Agar Base (Sigma) were used as per the manufacturer’s instructions. We confirmed that the coloured colonies on these chromogenic plates were Klebsiella by 16s rRNA sequencing. For consortia inoculation mice were orally inoculated 5×108 CFU of either of the following: the intestinal Bacteroidetes consortium consisted of: B. fragilis, B. caccae, B. thetaiotaomicron, B. uniformis, B.ovatus; the intestinal Firmicutes consortium: C. ramosum, C. symbiosum, C. orbiscindens, C. histolyticum, E. faecalis; the intestinal Actinobacteria consortium: B. adolescentis, E. lenta, B. breve; and the intestinal Proteobacteria consortium: E. coli, C. koseri, A. radioresistens. For all experiments using consortia of intestinal commensals, bacteria were grown to mid-log phase. For anaerobic consortia (Firmicutes, Bacteroidetes, and Actinobacteria) bacteria were resuspended in pre-reduced sterile PBS prior to inoculation, and for aerobic consortia (Proteobacteria) bacteria were resuspended in sterile PBS prior to inoculation. For upper airway consortia inoculation unanaesthetized animals mice were intranasally inoculated 1×106 CFU of either of the following in 20 μL: the upper airway Bacteroidetes consortium consisted of: P. nigrescens and P. melaninogenica; the upper airway Firmicutes consortium: S. epidermidis and L. crispatus; the upper airway Actinobacteria consortium: P. acnes and C. propinquum; and the upper airway Proteobacteria consortium: H. influenzae and M. catarrhalis. For all experiments using consortia of upper airway commensals, bacteria were grown to mid-log phase and consortia were resuspended in sterile PBS prior to inoculation. Because of the small inoculation volume and lack of anesthesia these bacteria remain in the upper airway29.
Cytokine measurements
Cytokine levels were determined using ELISA kits for TNFα, IL-17A, IFNγ (Biolegend) and IL-36γ (antibodies-online.com) according to the manufacturer’s instructions.
Macrophage culture and killing assay
Authenticated J774A.1 macrophages (Sigma-Aldrich) were cultured in DMEM supplemented with 10% v/v FBS, penicillin (100 units/mL) and streptomycin (100 mg/mL) and maintained at 37°C and 5% (v/v) CO2. This cell line was free of mycoplasma contamination. For bone marrow-derived macrophages, bone marrow was harvested from the femurs and tibiae of C57BL/6 mice and differentiated into macrophages by culturing in R10 medium containing 15% L-cell conditioned media and maintained at 37°C and 5% (v/v) CO2. Macrophage killing assays were carried out essentially as described before29,49.
Microbiome sequencing
For DNA extraction from feces, lung and upper airway lavage fluid DNA extracted using the E.Z.N.A Stool DNA kit (Omega Bio-Tek) as per the manufacturer’s instructions. Homogenization of the stool and lung was accomplished by bead disruption with approximately 200 mg beads using a FastPrep-24 instrument (MP Biomedicals) by 15 repeated pulses of 20 seconds vortexing and a subsequent 2 minute recovery. All samples were vortexed for 30 seconds to ensure complete disruption before proceeding with the protocol. Processing included the optional extra steps of incubating at 95°C to ensure sufficient lysis of Gram-positive bacteria, and treatment with 100 µg of RNase A for 3 minutes at 37°C to remove unwanted RNA. For quantification of mucosa-associated bacteria, mucus was scraped from intestine that had been washed with sterile PBS to remove all fecal matter, as described in31 and DNA extracted by bead beating as above. Extracted DNA was then used for 16s rRNA gene qPCR and deep sequencing. For qPCR, in a total 10 μL reaction there was 1 × SYBR Green PCR Master Mix (Thermo Fisher Scientific), 200 nM of each forward and reverse primer (Supplementary Table 2), and 2 μL template DNA. qPCR was performed on an Applied Biosystems StepOnePlus instrument and cycled 40 times with 15 seconds at 95 °C and 1 min at 61°C. To calculate 16s rRNA gene copy numbers, a standard curve was generated using plasmids containing a cloned 16s rRNA gene from a representative bacterial species of the indicated phylum (Supplementary Table 2). Plasmid copy numbers were calculated using the formula: (amount of plasmid in ng × 6.0221 × 1023 molecules/mol)/((4500 × 660 g/mol) × 1 × 109 ng/g), where 4500 is the estimated length of the plasmid with the 16s rRNA gene insert and 660 g/mol is the average mass of 1 base of dsDNA. Using this standard curve, 16s rRNA gene copy number per gram of stool was determined. 16s rRNA gene sequencing sample libraries were generated using Illumina’s 16s Metagenomic Sequencing Library Preparation Protocol with some modifications. The V1-V2 16s rRNA gene regions were amplified from stool isolated DNA using primers at a ratio as described previously 50. Index PCR reactions were cleaned and subsequently normalized with the SequalPrep Normalization Plate Kit (Life Technologies). These libraries were quantified by the NEBNext Library Quant Kit for Illumina (New England Biolabs). Sequencing data was generated using the MiSeq Reagent Kit v3 (Illumina) and paired-end 300 bp chemistry on an Illumina MiSeq platform (Illumina Inc). Sequencing data was processed following the DADA2 pipeline as previously described51. The SILVA bacterial database version 132 was used to classify the sequence variants (www.arb-silva.de/).
Models of bacterial transmission
For transmission studies in adult mice, “index” mice colonized by K. pneumoniae either in the upper airway or intestine were cohoused with naïve “contact” mice at indicated “index”:”contact” ratios. “Contact” mice were either treated with azithromycin and cefotaxime as above, or non-antibiotic treated as indicated. To study K. pneumoniae transmission in infant mice, a single infant “index” mouse was colonized by K. pneumoniae in the intestine and returned to its litter of naïve “contact” littermates. To determine whether direct contact between animals was required for transmission, indicated naïve contact mice were housed in a cage which had previously housed K. pneumoniae colonized “index” mice. Before the addition of the “contact” animals, all feces was removed from the cage.
Statistical and reproducibility
Statistical comparisons were performed using GraphPad Prism 8 software. To compare differences between two groups, the Student’s t-test or the Mann–Whitney test were used as appropriate (two-tailed). For statistical analysis of transmission, Fisher’s exact test was used (two-sided). For multiple comparisons, a Kruskal–Wallis test with Dunn’s correction for multiple comparisons was used. All error bars indicated standard deviation. All experiments were from a minimum of 4 biological replicates, with data from all replicates included in statistical analysis and no data excluded. Animals were randomly assigned to groups (cages) prior to any experimentation. Differences between litters of mouse pups were controlled for by mixing pups from different litters into each treatment condition. We used power analysis from published studies and our preliminary experiments to determine the minimum required sample size required for the groups used in our experiments. Microbiota sample preparation and analysis performed in a blinded manner.
Extended Data
Extended Data Figure 6.
Extended Data Figure 7.
Extended Data Figure 7.
Extended Data Figure 7.
Extended Data Figure 7.
Extended Data Figure 7.
Extended Data Figure 7.
Extended Data Figure 7.
Extended Data Figure 7.
Extended Data Figure 7.
Supplementary Material
Acknowledgements
T.B.C. is a Sir Henry Dale Fellow jointly funded by the Wellcome Trust and Royal Society (Grant Number 107660/Z/15Z). For J.A.K.M. and J.R.M., the Division of Integrative Systems Medicine and Digestive Disease at Imperial College London receives financial support from the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre (BRC) based at Imperial College Healthcare NHS Trust and Imperial College London. We would like to thank Dr Abigail Clements (Imperial College London) for providing K. pneumoniae strains, Gregory Donaldson and Sarkis Mazmanian (California Institute of Technology) for providing wild-type and ΔCCF B. fragilis and B. vulgatus strains and Rebecca Brown and Max Larkinson (Imperial College London) for providing macrophages.
Footnotes
Data availability. Microbiome sequencing data have been deposited at the NCBI SRA (BioProject PRJNA579139). The data supporting the findings of the study are available in this article (see Source Data) and its Supplementary Information files, or from the corresponding author upon request.
Competing Interests
The authors declare no competing interests.
Author Contributions
T.B.C. and R.P.S. conceived and performed all colonization, transmission and immunological experiments, analysed data, and wrote the manuscript. J.A.K.M. and J.R.M. performed sequencing experiments, analysed sequencing data and contributed to the manuscript. J.R.M. and T.B.C. acquired funding.
References
- 1.Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown RL, Clarke TB. The regulation of host defences to infection by the microbiota. Immunology. 2017;150:1–6. doi: 10.1111/imm.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke TB. Microbial programming of systemic innate immunity and resistance to infection. PLoS Pathog. 2014;10:e1004506. doi: 10.1371/journal.ppat.1004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keith JW, Pamer EG. Enlisting commensal microbes to resist antibiotic-resistant pathogens. J Exp Med. 2019;216:10–19. doi: 10.1084/jem.20180399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 7.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 8.Munoz-Price LS, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin RM, Bachman MA. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montgomerie JZ. Epidemiology of Klebsiella and hospital-associated infections. Rev Infect Dis. 1979;1:736–753. doi: 10.1093/clinids/1.5.736. [DOI] [PubMed] [Google Scholar]
- 12.Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Antimicrobial resistance : global report on surveillance. World Health Organization; 2014. [Google Scholar]
- 15.Bagley ST. Habitat association of Klebsiella species. Infect Control. 1985;6:52–58. doi: 10.1017/s0195941700062603. [DOI] [PubMed] [Google Scholar]
- 16.Donskey CJ. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis. 2004;39:219–226. doi: 10.1086/422002. [DOI] [PubMed] [Google Scholar]
- 17.Martin RM, et al. Molecular Epidemiology of Colonizing and Infecting Isolates of Klebsiella pneumoniae. mSphere. 2016;1 doi: 10.1128/mSphere.00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose HD, Schreier J. The effect of hospitalization and antibiotic therapy on the gram-negative fecal flora. Am J Med Sci. 1968;255:228–236. doi: 10.1097/00000441-196804000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Bilinski J, et al. Fecal Microbiota Transplantation in Patients With Blood Disorders Inhibits Gut Colonization With Antibiotic-Resistant Bacteria: Results of a Prospective, Single-Center Study. Clin Infect Dis. 2017;65:364–370. doi: 10.1093/cid/cix252. [DOI] [PubMed] [Google Scholar]
- 21.Caballero S, et al. Distinct but Spatially Overlapping Intestinal Niches for Vancomycin-Resistant Enterococcus faecium and Carbapenem-Resistant Klebsiella pneumoniae. PLoS Pathog. 2015;11:e1005132. doi: 10.1371/journal.ppat.1005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh R, et al. Fecal microbiota transplantation against intestinal colonization by extended spectrum beta-lactamase producing Enterobacteriaceae: a proof of principle study. BMC Res Notes. 2018;11:190. doi: 10.1186/s13104-018-3293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorbara MT, et al. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med. 2019;216:84–98. doi: 10.1084/jem.20181639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau HY, Huffnagle GB, Moore TA. Host and microbiota factors that control Klebsiella pneumoniae mucosal colonization in mice. Microbes Infect. 2008;10:1283–1290. doi: 10.1016/j.micinf.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madueno A, et al. Risk factors associated with carbapenemase-producing Klebsiella pneumoniae fecal carriage: A case-control study in a Spanish tertiary care hospital. Am J Infect Control. 2017;45:77–79. doi: 10.1016/j.ajic.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 26.Mills JP, Talati NJ, Alby K, Han JH. The Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae Colonization and Infection among Long-Term Acute Care Hospital Residents. Infect Control Hosp Epidemiol. 2016;37:55–60. doi: 10.1017/ice.2015.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhargava A, et al. Risk factors for colonization due to carbapenem-resistant Enterobacteriaceae among patients exposed to long-term acute care and acute care facilities. Infect Control Hosp Epidemiol. 2014;35:398–405. doi: 10.1086/675614. [DOI] [PubMed] [Google Scholar]
- 28.Nelson AL, Barasch JM, Bunte RM, Weiser JN. Bacterial colonization of nasal mucosa induces expression of siderocalin, an iron-sequestering component of innate immunity. Cell Microbiol. 2005;7:1404–1417. doi: 10.1111/j.1462-5822.2005.00566.x. [DOI] [PubMed] [Google Scholar]
- 29.Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01803-x. 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atarashi K, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donaldson GP, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360:795–800. doi: 10.1126/science.aaq0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rendueles O, Garcia-Garcera M, Neron B, Touchon M, Rocha EPC. Abundance and co-occurrence of extracellular capsules increase environmental breadth: Implications for the emergence of pathogens. PLoS Pathog. 2017;13:e1006525. doi: 10.1371/journal.ppat.1006525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clements A, et al. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J Biol Chem. 2007;282:15569–15577. doi: 10.1074/jbc.M701454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Steenhuijsen Piters WA, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. 2016;10:97–108. doi: 10.1038/ismej.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi H, Yong D, Lee K, Cho YJ, Chun J. Profiling bacterial community in upper respiratory tracts. BMC Infect Dis. 2014;14:583. doi: 10.1186/s12879-014-0583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaspar U, et al. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol. 2016;18:2130–2142. doi: 10.1111/1462-2920.12891. [DOI] [PubMed] [Google Scholar]
- 38.Pettigrew MM, et al. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012;78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassis CM, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Steenhuijsen Piters WA, Sanders EA, Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14:R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris AD, et al. Patient-to-patient transmission is important in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae acquisition. Clin Infect Dis. 2007;45:1347–1350. doi: 10.1086/522657. [DOI] [PubMed] [Google Scholar]
- 43.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004129. 148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebino KY. Studies on Coprophagy in Experimental-Animals. Exp Anim Tokyo. 1993;42:1–9. doi: 10.1538/expanim1978.42.1_1. [DOI] [PubMed] [Google Scholar]
- 45.Schenck LP, Surette MG, Bowdish DM. Composition and immunological significance of the upper respiratory tract microbiota. FEBS Lett. 2016;590:3705–3720. doi: 10.1002/1873-3468.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brisse S, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshmukh HS, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redhu NS, et al. Macrophage dysfunction initiates colitis during weaning of infant mice lacking the interleukin-10 receptor. Elife. 2017;6 doi: 10.7554/eLife.27652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun. 2014;82:4596–4606. doi: 10.1128/IAI.02212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullish BH, et al. Functional microbiomics: Evaluation of gut microbiota-bile acid metabolism interactions in health and disease. Methods. 2018;149:49–58. doi: 10.1016/j.ymeth.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.