Summary
The pathways for ribosomal RNA (rRNA) maturation diverge greatly amongst the domains of life. In the Gram-positive model bacterium, Bacillus subtilis, the final maturation steps of the two large ribosomal subunit (50S) rRNAs, 23S and 5S pre-rRNAs, are catalysed by the double-strand specific ribonucleases (RNases), Mini-RNase III and RNase M5, respectively. Here we present a protocol that allowed us to solve 3.0 and 3.1 Å resolution cryo-electron microscopy structures of these RNases poised to cleave their pre-rRNA substrates within the B. subtilis 50S particle. These data provide the first structural insights into rRNA maturation in bacteria by revealing how these RNases recognise and process double-stranded pre-RNA. Our structures further uncover how specific ribosomal proteins act as chaperones to correctly fold the pre-rRNA substrates and, for Mini-III, additionally anchors the RNase to the ribosome. These r-proteins thereby serve a quality-control function in the process from accurate ribosome assembly to rRNA processing.
Introduction
Ribosome biogenesis dominates the energy consumption and resource utilisation of all living organism. Strict quality control mechanisms accompany the different assembly steps of the small and large ribosome subunits to ensure the fidelity of these energy-costly processes. A crucial step in ribosome biogenesis is the accurate and timely generation of mature rRNAs through processing of their long precursor rRNAs (pre-rRNAs) by RNases (Srivastava, 1990). Failures in these maturation processes can trigger degradation of ribosomes as a safeguard against aberrant assembly (Li et al., 1998). Prokaryotes and Eukaryotes utilise entirely different sets of RNases to perform the maturation reactions, and major differences are also observed within Prokaryotes themselves, e.g. in Archaea and in Proteobacteria and Firmicutes among bacterial phyla (Clouet-D’Orval et al., 2018; Condon, 2009; Henras et al., 2015). In bacteria, the mechanism for the initial maturation of the three rRNAs, i.e. 16S, 23S and 5S, is shared between organisms; the rRNAs are generally synthesised as one long 30S precursor (pre-30S) rRNA, and released by cleavage of the pre-30S transcript by RNase III (French and Miller, 1989) before completion of transcription. During this process, the ribosomal proteins (r-proteins) attach as soon as their binding site has been transcribed (Nomura, 1972). Later maturation steps differ greatly in the three prokaryotic models described thus far. In Firmicutes, such as B. subtilis, the final maturation steps for pre-23S rRNA and pre-5S rRNA is performed by Mini-RNase III (Mini-III) and RNase M5 (M5), respectively. These reactions take place only when ribosomal subunits are accurately assembled and specific r-proteins have joined the premature particles (Redko and Condon, 2009; Stahl et al., 1984).
Mini-III is highly conserved in Firmicutes, a phylum that comprises a major reservoir of human pathogens, but is also found in Cyanobacteria, in unicellular photosynthetic eukaryotes and in higher plants (Redko et al., 2008). Mini-III belongs to the RNase III-like family that also contains the eukaryotic RNA-silencing enzymes Dicer and Drosha. M5 is found mostly in Firmicutes (Condon et al., 2001) and contains a Toprim domain, also present in topoisomerases and reverse gyrases (Allemand et al., 2005). Both M5 and Mini-III cleave double-stranded RNA (dsRNA) by staggered cuts (Figures 1A and 1B) and each requires an r-protein as a cofactor for optimal catalytic activity in vitro. The r-protein uL3, bound to 23S rRNA, enhances the RNase activity of Mini-III by several orders of magnitude in vitro (Redko and Condon, 2009), while the r-protein uL18, bound to 5S rRNA, is strictly required for M5 cleavage activity (Stahl et al., 1984). The necessity for specific r-proteins in rRNA maturation reactions has so far only been demonstrated in B. subtilis, but may also be important in other organisms.
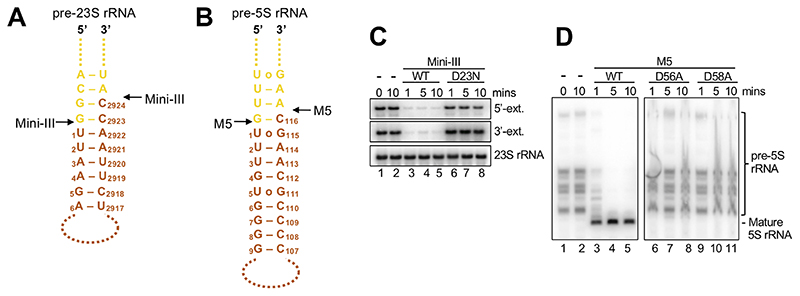
Figure 1. rRNA precursor substrates for Mini-III and M5.
(A) Schematic outline of pre-23S rRNA substrate helix highlighting the nts of the 3’-and 5’-extensions in yellow, and mature 23S rRNA nts in orange. Only overhang nts built in the cryoEM models are shown. Mini-III cleavage sites are indicated with black arrows.
(B) As for (A), but for pre-5S rRNA/M5.
(C) Northern blot showing removal of 5’-and 3’-extensions from 50S(pre-23S) particles by wild-type Mini-III (Mini-IIIWT) (lane 3-5), and lack thereof by Mini-IIID23N mutant protein (lane 6-8) at different time points. Lanes 1-2 show pre-23S rRNA at the first and last time point in the absence of Mini-III protein.
(D) As for (C), but for M5WT and M5D56A/M5D58A mutant proteins with 50S(pre-5S) particles.
Here we present high-resolution structures of the final processing steps of the two 50S subunit rRNAs in B. subtilis catalysed by Mini-III and M5. These structures reveal different modes of dsRNA recognition and cleavage for Mini-III and M5, and overlapping but distinct roles for their essential r-protein co-factors in promoting rRNA maturation.
Results
CryoEM structure determination
In B. subtilis, cells lacking the non-essential Mini-III gene (mrnC) can mature pre-23S rRNA via a backup pathway involving the 5’-exoribonuclease RNase J1 (rnjA), and the 3’-exoribonucleases RNase PH (rph) and YhaM (yhaM) (Redko and Condon, 2010). We surmised that knocking out either the 5’- or 3’-exonucleolytic backup pathway would significantly retard the other, due to the double-stranded nature of the processing helix. We therefore made a double ΔmrnC ΔrnjA mutant and used this strain to isolate 50S ribosomal particles containing pre-23S rRNA (50S(pre-23S)) with 5’- and 3’-extensions (Figures 1A and 1C). To produce 50S subunits containing pre-5S rRNA (50S(pre-5S)), we used B. subtilis strains deleted for the gene encoding M5 (ΔrnmV) (Condon et al., 2001). These cells accumulate pre-5S rRNAs of 125 to 165 nucleotides (nts) from the 10 rrn operons, but no mature pre-5S rRNA (116 nts) (Figures 1B and 1D).
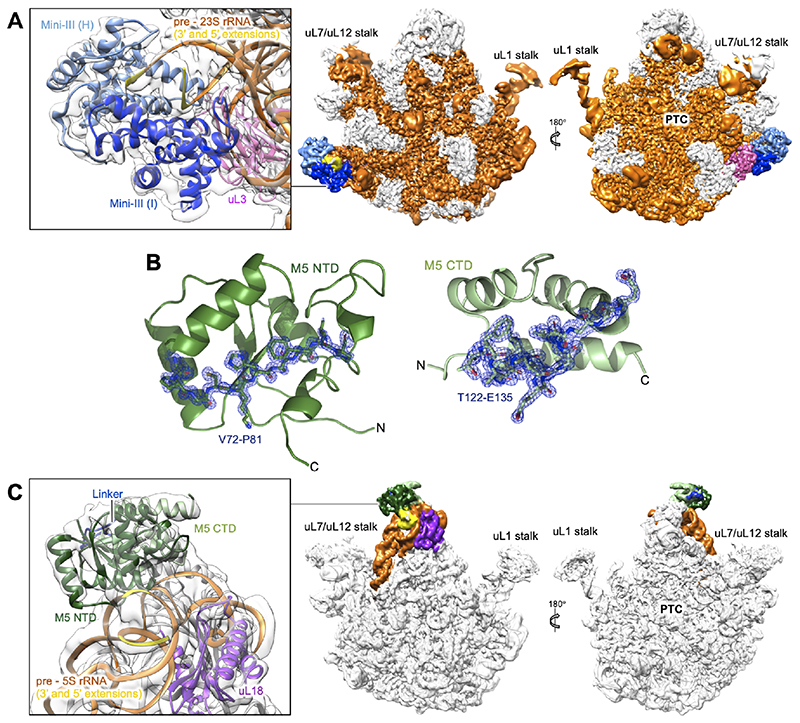
Recombinant purified Mini-III and M5 matured their respective pre-rRNA substrates within the 50S particles in vitro, while catalytically inactive mutant proteins (Mini-IIID23N; M5D56A, M5D58A) did not (Figures 1C and 1D). A dissociation constant (KD) in the micromolar range (7.5±0.7 μM) was measured for M5D58A with in vitro transcribed pre-5S rRNA (Figure S1A) indicating that M5 could have a low affinity for the 50S(pre-5S) particle. Indeed, an initial collection of 1 μM 50S(pre-5S) mixed with 5x excess of M5D58A (i.e, concentration corresponding to less than the KD) did not reveal any density for M5D58A. We therefore subsequently mixed 1 μM 50S(pre-5S) with M5D58A at 50S(pre-5s):M5D58A ratios spanning from 1:10 to 1:1000 to exceed the KD, made cryoEM grids for each mixture, and screened the grids on the microscope. Data were collected on the grid with the highest 50S(pre-5s):M5D58A ratio for which the unbound M5D58A did not disturb particle picking and data processing by contributing too much to the background, ultimately corresponding to a 50S(pre-5S):M5D58A ratio of 1:210. This ratio was also used for grid preparation of 50S(pre-23S) particles with Mini-IIID23N. For Mini-IIID23N bound to 50S(pre-23S), single-particle cryoEM data collection and processing (Table S1, Figure S1B) yielded an EM density map at an overall resolution of 3.1 Å with a local resolution of 3.2-4.5 Å for Mini-IIID23N (Figures S1C, S1D and S1E). The crystal structure of Mini-III (PDB 4OUN) (Głów et al., 2015) and the cryoEM structure of the B. subtilis 50S subunit (PDB 3J3V/5NJT) (Beckert et al., 2017; Li et al., 2013) were fitted into the density map (Figure 2A), and pre-23S rRNA extensions were built de novo. For the complex between M5D58A and 50S(pre-5S) particles, the cryoEM map from single-particle analysis (Table S1, Figure S1F) was refined to an overall resolution of 3.0 Å with a local resolution of 4.2-6 Å for M5D58A (Figures S2B, S2C, and S2D). Since no structure was available for M5, we solved the crystal structures of the M5 N-terminal domain (NTD; residues 1-113) to 1.3 Å resolution, and the C-terminal domain (CTD; residues 105-183) to 1.5 Å resolution (Figure 2B, Table S2), and docked these into the cryoEM map (Figure 2C). The linker between the two domains, and the 5’- and 3’-extensions of pre-5S rRNA, was constructed from the cryoEM density map (Figure 2C). The resulting atomic models revealed the high-resolution architecture of the two processing complexes that carry out the final maturation of 50S pre-rRNAs in B. subtilis.
Figure 2. CryoEM density maps for Mini-III/50S(pre-23S) and M5/50S(pre-5S).
(A) CryoEM density map for the Mini-III dimer (light/dark blue) bound to the 50S(pre-23S) ribosomal particle. The pre-23S rRNA substrate (orange) and the uL3 r-protein cofactor (magenta) are highlighted. Ribosomal landmarks including the uL1 and uL7/uL12 stalks and the peptidyl transferase centre (PTC) are labelled for orientation. The insert shows a close up of the Mini-III dimer, its pre-23S rRNA substrate and the r-protein uL3.
(B) X-ray crystal structures of the M5 domains with composite omit 2Fo-Fc difference density map (blue), contoured at 2.0 σ, shown for residues T122-E135 and V72-P81 to emphasise resolution. The N- and C-terminal domains are labelled, with the M5 C-terminal domain (CTD) shown in light green (top) and the N-terminal domain (NTD) in dark green (bottom).
(C) CryoEM density map for the M5 monomer (NTD/CTD/linker in dark green/light green/blue, respectively) bound to the 50S(pre-5S) ribosomal particle. The pre-5S rRNA substrate (orange) and the uL18 r-protein cofactor (purple) are highlighted. Ribosomal landmarks including the uL1 and uL7/uL12 stalks and the peptidyl transferase centre (PTC) are labelled for orientation. The insert shows a close up of the M5 monomer, its pre-5S rRNA substrate and r-protein uL18.
CryoEM structure of the Mini-III/50S(pre-23S) complex
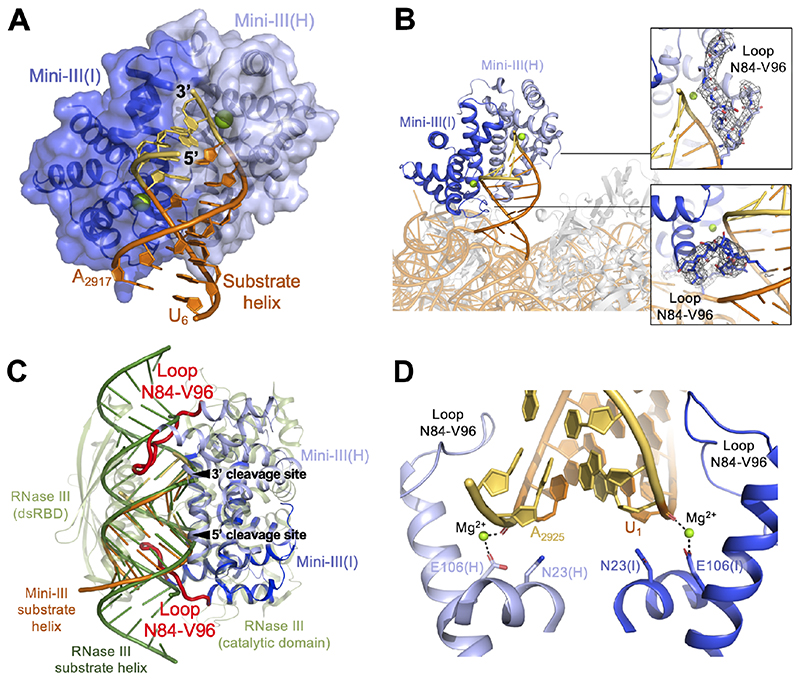
The cryoEM structure of the Mini-III/50S(pre-23S) complex reveals how Mini-III recognises and matures pre-23S rRNA within the 50S subunit. Dimerisation of Mini-III creates a catalytic groove into which the pre-23S rRNA substrate helix fits snugly (Figure 3A). The substrate helix assumes a regular A-form, and each cleavage site is positioned similarly within the symmetrically aligned catalytic pockets of the Mini-III subunits (Figure 3A). The A-form 23S rRNA substrate helix is recognised partly via a loop containing residues N84-V96 (Figure 3B). Mini-III adopts an RNase III-like fold, known to recognise and cleave A-form dsRNA (Gan et al., 2006) (Figure 3C). An alignment of Mini-III with RNase-III bound to a dsRNA substrate reveals that these loops would also facilitate binding of longer dsRNAs previously identified for Mini-III (Głów et al., 2015) (Figure 3C). Each Mini-III subunit coordinates one Mg2+ ion via the conserved residue E106 (Figure 3D) and the phosphodiester backbone of U1 and A2925 at the cleavage sites, thereby positioning the RNA for maturation. Based on other RNase-III like structures, a second Mg2+ ion is expected to bind to conserved residues E106 and D23 (mutated to N here to inactivate catalysis) (Figures S3A, S3B). The positive charge of the metal cofactors stabilises the highly negatively charged active site to facilitate the catalytic assembly (Figure S3C). Taken together, the similarities between RNase III and Mini-III suggest that Mini-III utilises an RNase III-like catalytic mechanism (Gan et al., 2006, 2008) for pre-23S rRNA maturation.
Figure 3. Mini-III binding to the substrate helix of pre-23S rRNA.
(A) Mini-III dimer (light/dark blue) shown as both transparent surface representation and cartoon representation bound to the pre-23S rRNA substrate helix (mature nts in orange, extension nts in yellow). Mg2+ ions (green) are shown in each active site.
(B) Mini-III bound to the 50S(pre-23S) particle, coloured as in Figure 1A, with r-proteins shown in grey. The insert shows the cryoEM density for loop N84-V96 for each Mini-III subunit.
(C) Mini-III/pre-23S rRNA superimposed on Aquifex aeolicus RNase III/dsRNA substrate (PDB 2NUG). Mini-III RNA-binding loops (residues N84-V96) are highlighted in red. Both RNA cleavage sites are indicated with arrows.
(D) Mini-III active site organisation coloured as in (A). Mini-III RNA-binding loops are indicated. Catalytically important residues are shown in stick representation.
Our cryoEM structure further shows interactions between Mini-III, mainly via chain I, and 50S(pre-23S) outside of the substrate helix. Mini-III interacts extensively with the phosphodiester backbone of a stretch of neighbouring nts (U2812 and C2814C2815C2816) in the 23S rRNA, via a region containing conserved residues (chain I: R83, N84, R100) (Figure 4A). The conserved residue, R100(I), also binds to the base of G2916, which base-pairs with C2816 from this rRNA stretch (Figure 4B). Interestingly, A2817, located downstream of this stretch, is flipped out relative to its conformation in mature 50S, to form a π-π stacking interaction with F63(I), thus clamping Mini-III to the pre-23S rRNA (Figure 4A insert). In mature 23S rRNA, A2817 would sterically clash with Q97(I) of Mini-III (Figure 4C). While Mini-III lacks a dsRBD generally found in members of the RNase III family, the extensive network of interactions between Mini-III and the pre-23S rRNA, outside of the substrate helix, eliminates the need for such a dsRBD.
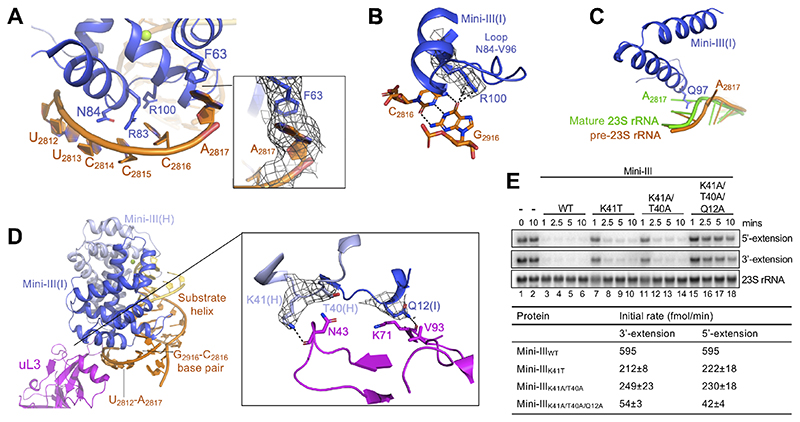
Figure 4. Interactions between Mini-III and 50S(pre-23S).
(A) Mini-III interactions outside of the substrate helix. Conserved residues are shown in stick representation. Insert shows the cryoEM density for the flipped-out A2817 that forms a π-π stacking interaction with F63(I).
(B) CryoEM density for Mini-III residue R100(I) bound to the C2816-G2916 base-pair from pre-23S rRNA, coloured as in Figure 1A.
(C) Superimposition of mature 23S rRNA (green, PDB 5NJT) with Mini-III-bound pre-23S rRNA (orange). Mini-III residue Q97(I) (dark blue, stick representation) sterically clashes with A2817 in mature 23S rRNA, but not in pre-23S rRNA.
(D) Mini-III (dark/light blue), uL3 (pink), and pre-23S rRNA (orange/yellow) interactions. RNA elements shown in (A), (B), and (C) are highlighted. The insert shows cryoEM density for residues from Mini-III (dark/light blue) able to bind to uL3 (pink).
(E) Top: Northern blot showing removal of 5’-and 3’-extensions from pre-23S rRNA by Mini-IIIWT, Mini-III-K41T, Mini-III-K41A/T40A, and Mini-III-K41A/T40A/Q12A. The first two lanes show pre-23S rRNA at 0 and 10 minutes in the absence of Mini-III protein. The blot is representative of at least 3 independent experiments. Bottom: Initial reaction rates for each mutant. Initial slopes (%/min) were calculated for each curve by regression of the linear portion of the curves shown in Figure S2D, with 100% corresponding to 700 fmol pre-23S rRNA.
Many of the nts outside of the substrate helix that are associated with Mini-III are wedged between Mini-III and the r-protein cofactor uL3 (Figure 4D). As such, uL3 both maintains the Mini-III binding site in the mature 23S rRNA and helps to position the substrate helix for Mini-III cleavage. The cryoEM density supports binding of residues Q12(I), T40(H) and K41(H) from Mini-III to residues N43, K71 and V93 in uL3, suggesting that uL3 also serves as an additional anchor point for Mini-III to the 50S(pre-23S) (Figure 4D insert). Variants of Mini-III with single- or double mutations of residues in the Mini-III/uL3 interface (Mini-IIIK41T and Mini-IIIK41A/T40A) showed a ~2.5-fold decrease in initial reaction rate compared to WT, for the pre-23S rRNA maturation in 50S(pre-23S) particles, whereas a triple mutant (Mini-IIIK41A/T40A/Q12A) exhibited a more severe ~12-fold decrease (Figures 4E and S3D). The mutated residues are located far from the active site of either Mini-III subunit and are not involved in RNA binding. Furthermore, differential scanning fluorimetry (DSF), sizeexclusion chromatography (SEC) and circular dichroism (CD) analysis shows that none of the point mutations causes protein misfolding or aggregation (Figures S3E, S3F and S3G). Thus, binding of Mini-III to uL3, primarily via K41(H) and Q12(I), play a crucial role in cleavage activity, likely by stabilising the Mini-III/50S(pre-23S) interaction.
CryoEM structure of the M5/50S(pre-5S) complex
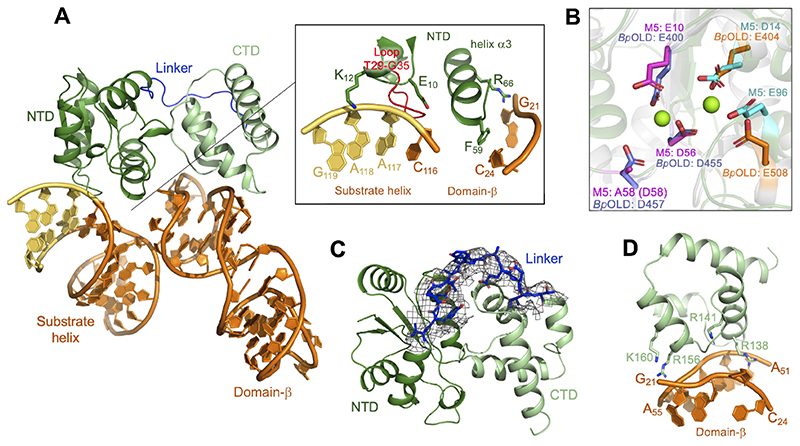
The cryoEM structure of M5/50S(pre-5S) complex reveals an M5 monomer associated with the 50S(pre-5S) subunit (Figure 5A), agreeing with the observation that M5 is a monomer in solution (Figure S4A). M5 binds to two regions of the pre-5S rRNA, i.e. the substrate helix and the arm known as domain-β (Szymanski et al., 2002) (Figure 5A). The NTD, with a classic Toprim fold (Allemand et al., 2005), binds to both of these regions. It binds the backbone of nts C116 to G119 from the 3’-strand of the substrate helix via conserved residues E10 and K12, and a loop (T29-G35) containing conserved residues N30, G31, and A33 (Figures 5A insert and S4B). The NTD helix α3 sits between the two RNA regions and interacts with domain-β via the conserved residue R66 with G21, and residue F59 with C24 though π-π stacking with the base (Figure 5A insert).
Figure 5. M5 binding to the substrate helix of pre-5S rRNA.
(A) M5 (dark/light green) bound to pre-5S rRNA substrate helix (mature nts in orange, extension nts in yellow) and domain-β (in orange). The linker between the M5 NTD and CTD is shown in dark blue. The insert shows the interactions between the M5 NTD and pre-5S rRNA substrate. Conserved residues in close proximity to the RNA substrate are shown in stick representation. The T29-G35 loop is shown in red.
(B) Superimposition of M5 NTD (dark green) from the cryoEM M5/50S(pre-5S) structure with the Toprim domain from Burkholderia pseudomallei OLD (BpOLD) protein (PDB 6NK8) (grey) bound to two Mg2+ ions (green). Acidic residues involved in catalysis are shown in stick representation for both proteins: M5 NTD (magenta), BpOLD (purple) for the conserved Toprim triad coordinating one Mg2+ ion in BpOLD, and M5 NTD (cyan), BpOLD (orange) for acidic residues coordinating a second Mg2+ ion in BpOLD.
(C) M5 NTD (dark green) and CTD (light green) with the linker region (dark blue) represented as sticks. The cryoEM density is shown for the linker.
(D) Conserved M5 CTD (light green) residues located in proximity to the pre-5S rRNA substrate to allow interaction.
Structural comparison of the M5 NTD with the Burkholderia pseudomallei OLD (BpOLD) protein (PDB 6NK8), which cleaves one strand of a DNA duplex using a catalytic Toprim domain (Schiltz et al., 2019), reveals a common spatial organisation of active-site acidic residues (Figure 5B). Like BpOLD, M5 has a strictly conserved Toprim catalytic triad E10/D56/D58 (D58 here mutated to A), and additional conserved, acidic residues (D14, E96), that are positioned for coordination of two Mg2+ ions (Figure 5B). This suggests that M5 cleaves each RNA strand of the dsRNA substrate helix, by a two-metal ion dependent mechanism (Yang, 2008), similarly to BpOLD. Initial binding of M5 to the 3’-cleavage site agrees with previous data suggesting removal of the 3’-extension prior to cleavage of the 5’-strand (Condon et al., 2001), and implies that a remodelling of the complex occurs between cleavages. A long linker (residues N109-T122) connects the NTD to the CTD (Figure 5C). Deuterium exchange experiments suggest that this linker is flexible (Figure S5A) and could allow the domains to move relative to each other, which – in combination with a possible change in pre-5S rRNA substrate conformation after removal of the 3’-extension – would allow the 5’-cleavage reaction to occur.
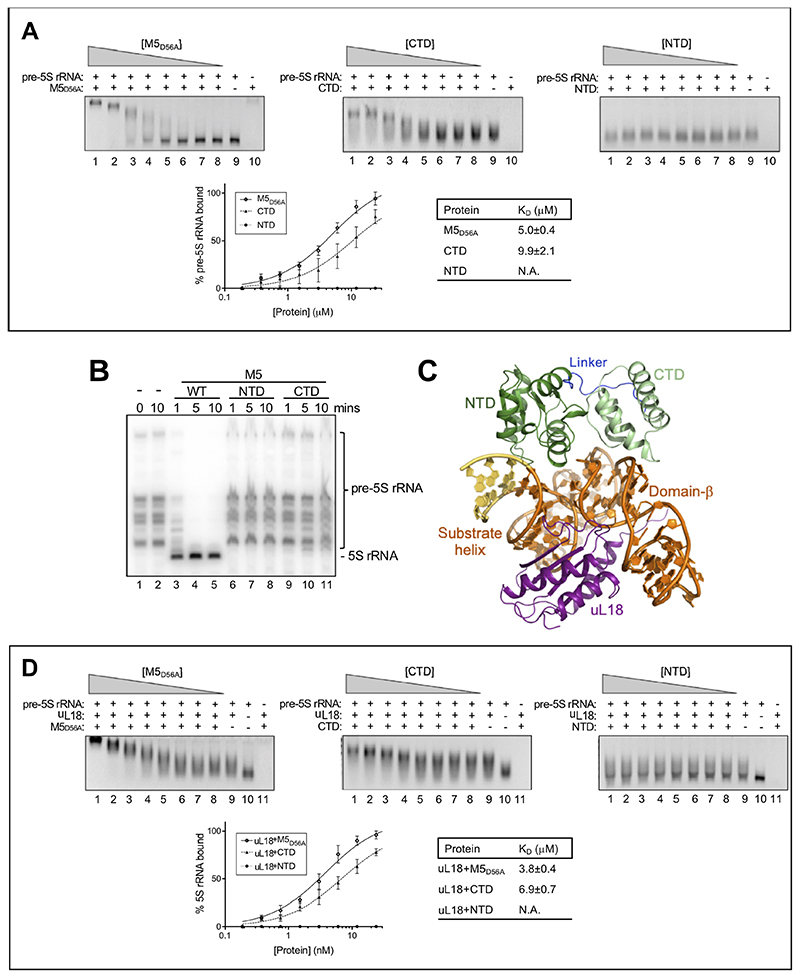
The structure of the CTD shows a bundle of four tightly-coiled a-helices arranged as two pairs of antiparallel elements. A DALI search identified the best structural match as the death domain (DD) (Table S3), but with low Z-scores (< 5.6). The CTD deviates from all known DDs by lacking key structural DD-fold elements (Figure S5B) and by operating in a different biological pathway. The CTD is therefore not a DD, but has a novel fold with an unknown function. In the cryoEM structure, the M5 CTD anchors M5 to the pre-5S rRNA by binding both strands of domain-β. The conserved residues R138, R141, and R156 bind to one strand at nts A51-A55 (Figure 5D), whereas the conserved residue K160 binds the other strand at G21, which is also recognised by the NTD. An electrophoretic mobility-shift assay (EMSA) of K/R-to-E mutant M5 proteins with in vitro transcribed pre-5S rRNA confirmed a crucial role for R156 and K160 in pre-5S rRNA binding (Figure S6A). R156 could further bind the r-protein uL5 (Figure S6B), which is, however, not required for M5 activity in vitro (Stahl et al., 1984). When repeating the EMSA with inactive M5D56A and domain fragments (NTD, CTD), only M5D56A and the CTD shifted the pre-5S rRNA in a concentration-dependent manner (Figure 6A). The NTD is thus not able to bind the pre-5S rRNA in the absence of the CTD. In an activity assay, only M5WT, but neither the CTD nor NTD had RNase activity (Figure 6B). Taken together, these results confirm that the CTD drives pre-5S rRNA binding of M5 via basic residues, and that the structure of the M5 CTD represents a novel RBD fold.
Figure 6. The role of uL18 in M5 substrate-binding and RNase activity.
(A) Top: EMSA in 2.5% native agarose gels for inactive, full-length M5D56A (first panel), M5 CTD (second panel), and M5 NTD (last panel) with in vitro transcribed pre-5S rRNA. Gels show free pre-5S rRNA (bottom band) and M5-bound pre-5S rRNA (top band) for a protein titration (lane 1-8). Lane 9 corresponds to pre-5S rRNA alone, and lane 10 to protein alone. Bottom left: plot of M5-bound pre-5S rRNA (%) as a function of M5 protein concentration. Each point represents the area-under-the-curve from band integration with the area for pre-5S rRNA alone (lane 9) set to 100%. Standard deviations are calculated from integration of bands from three independent experiments. Bottom right: KD-values determined by curve-fitting using a least-squares fit for one site with specific binding.
(B) Northern blot showing processing of pre-5S rRNA in 50S particles by M5WT, M5 NTD, or M5 CTD. Lanes 1 and 2 show the reaction at 0 and 10 minutes without M5 protein, respectively.
(C) uL18 (purple) bound to the pre-5S rRNA substrate helix and domain-β, opposite from M5 (dark/light green, linker in dark blue).
(D) As in (A), but with incubation of each sample of pre-5S rRNA with uL18 at a ratio of 1:1, prior to M5 protein titration. Protein titrations corresponds to lane 1-8, lane 9 to pre-5S rRNA + uL18, lane 10 to pre-5S rRNA alone, and lane 11 to M5 protein + uL18.
In our structure, the cofactor r-protein uL18 lies on the opposite side of the substrate rRNA from M5 (Figure 6C), and – unlike uL3 for Mini-III – does not make any direct contacts with the RNase. Instead, uL18 moulds the rRNA substrate for optimal M5 recognition by bringing together domain-β and the substrate helix (Figure 6C). In an EMSA with the CTD, NTD and M5D56A, addition of uL18 decreased the KD for the substrate for both the CTD and M5D56A by a factor of ~1.5, and did not facilitate binding of the NTD (Figure 6D). This shows that the pre-5S folding by uL18 primarily serves a catalytic purpose by positioning the CTD-bound domain-β in proximity to the substrate helix for cleavage by the NTD, explaining the absolute requirement of uL18 for M5 cleavage activity in vitro (Stahl et al., 1984).
Discussion
Here we present the structure of the maturation RNases M5 and Mini-III bound to B. subtilis 50S(pre-5S) and 50S(pre-23S) particles, respectively, on which they carry out the final maturation of their respective pre-RNA substrate. Currently, only one structure is available that describes the maturation of pre-rRNAs bound in the ribosome, i.e. the exosome bound to the yeast 60S(pre-7S) subunit during 7S-to-5.8S rRNA processing (Schuller et al., 2018), leaving bacterial rRNA maturation entirely unexplored. The data presented here provide the first structural insights into rRNA maturation in bacteria by revealing how these RNases recognise and process dsRNA pre-rRNA substrates. The observed binding modes differ from those found for proteins maturing eukaryotic pre-rRNAs, emphasising the diversity of rRNA maturation mechanisms and indicating that they likely arose through convergent evolution.
Protein titration with microscope visualisation allows capture of low-affinity complexes
The technique deployed here for capturing Mini-III and M5 bound to their respective pre-rRNA substrates, circumvents issues of complex-instability often encountered when working with low-affinity systems such as those relying on enzyme/RNA interactions. By titrating the large ribosomal particle with increasing amounts of the smaller enzyme component and visualising each ribosome:enzyme ratio with a cryo-electron microscope, we were able to obtain two low-affinity complexes at 210x excess of enzyme. This ratio did not disturb the cryoEM image background and thereby did not present a problem for particle picking. We believe that this method can be widely deployed to capture low affinity complexes consisting of a small and large component.
Comparison of 50S particles with pre-rRNA and mature 50S particles
A comparison of the 50S(pre-23S) and 50S(pre-5S) particles with mature 50S in the assembled, mature 70S particle from B. subtilis (PDB 6HA1, (Crowe-McAuliffe et al., 2018)) reveals no significant structural differences between these particles apart from in the studied pre-rRNA/RNase regions, indicating the processing occurs on otherwise mature subunits. Furthermore, the 5’ and 3’ extensions are located at regions distal to the 30S particle in the 70S ribosome, and therefore do not sterically block subunit association, in agreement with the observation that cells with 50S(pre-5S) or 50S(pre-23S) subunits are viable. Indeed, we have previously shown that 5S precursors accumulate in 70S ribosomes and polysomes in strains lacking RNase M5 (Condon et al., 2001) and that the 23S rRNA is almost entirely in the precursor state in cells lacking Mini-III and depleted for RNase J1 (Redko and Condon, 2010), showing that both pre-5S rRNA and pre-23S rRNA are tolerated in assembled, functional ribosomes. This is consistent with previous data (Bunner et al., 2010) suggesting that processing step occurs very late in ribosome assembly and can be used as a quality control indicator that assembly has occurred correctly.
Mini-III and M5 use different strategies to recognise their ds rRNA substrate
The two key RNases characterised here, Mini-III and M5, both have dsRNA substrates, but differ in their rRNA substrate recognition pattern and in their oligomeric state, and as a result, have different dsRNA cleavage patterns. Dimeric Mini-III recognises both strands of the dsRNA substrate, and utilises each subunit to cleave one strand of the dsRNA helix with a mechanism likely resembling that of RNase III. Interestingly, Mini-III lacks a dsRBD found in all other classes of the RNase-III family of proteins. Here, we identify interactions between Mini-III and the r-protein uL3 that anchors the RNase to the pre-23S substrate in the pre-50S particles, which compensates for the absent dsRBD/substrate interactions and further explains why RNase III is unable to cleave at this site (Redko et al., 2008). Of note, no significant differences were observed between the crystal structure of B. subtilis Mini-III (4OUN) and the cryoEM structure of the same protein, showing that the interaction with uL3 unlikely have any allosteric effect on the enzyme. Monomeric M5 is anchored to the pre-5S rRNA substrate within the 50S particle via its CTD with a novel RBD fold, which in turn positions the catalytic NTD appropriately for rRNA substrate helix cleavage. In our structure, the NTD solely recognises the 3’-strand, suggesting that it cleaves the substrate one strand at a time in a 3’ to 5’ order. Previous studies showed that the nature of the nucleotide immediately 5’ to the 5’-strand cleavage site (G-1) is important for activity (Stahl et al., 1980). Initial removal of the 3’-strand would open up the otherwise base-paired 5’-strand to base-recognition of G-1 by the M5 NTD. Thus, rearrangements of the rRNA substrate subsequent to 3’-strand cleavage to accommodate the 5’-strand in the active site, could well explain this base dependency. The M5 NTD contains a long flexible loop (residues 80-99) covering a number of basic residues that could be involved in binding and positioning of the 5’-strand subsequent to removal of the 3’-strand.
Mini-III and M5 use a two-metal ion dependent cleavage mechanism
Despite the differences in recognition and cleavage pattern, both enzymes utilise a similar two-metal dependent cleavage mechanism (Yang, 2008), involving a number of acidic aspartate and glutamate residues to coordinate Mg2+ ions. This two-metal ion catalysis is shared by all members of the RNase III family whereas one or two-metal ion dependent cleavage mechanisms are used by members of the Toprim family. All Toprim members have the conserved E and DxD motif that critically bind one Mg2+ ion, but additional active site components vary between Toprim family members based on specific functional requirements. Topoisomerases and gyrases contain a catalytic tyrosine that facilitates catalysis by covalently binding the DNA backbone at the cleavage site (Sissi and Palumbo, 2009; Yang, 2010). DnaG primases have extra acidic residues that coordinate multiple metals needed for nucleotide binding and polymerase activity (Keck et al., 2000; Podobnik et al., 2000; Rymer et al., 2012). M5 matures the pre-5S rRNA without such a tyrosine residue (Allemand et al., 2005). Instead additional acidic residues, similar to those found in the recently characterised Toprim domaincontaining protein BpOLD, could bind a second Mg2+ ion near the scissile bond, suggesting that M5 and BpOLD proteins share a similar mechanism for substrate cleavage using two Mg2+ ions.
R-proteins serve a role in quality control and ribosome assembly
In addition to metal-ions, both Mini-III and M5 rely on r-proteins, uL3 and uL18, respectively, for activity. Both r-protein cofactors shape and maintain the structures of the rRNA substrates for RNase recognition and catalysis, and uL3 additionally anchors Mini-III to the 50S particle to compensate for a lack of an RBD in Mini-III. As such, the RNases only operate at full efficiency in the presence of their r-protein cofactors when their rRNA substrate is correctly folded, tightly coupling the final rRNA maturation steps to the quality control of ribosome assembly.
The necessity for co-factors for RNase activity is mostly observed for proteins carrying out rRNA processing and can be appreciated in the context of quality control. Like M5 and Mini-III, the maturation of the 3’-end of 16S by YbeY in E. coli may involve r-protein S11 (Vercruysse et al., 2016), which could play a role in structuring the rRNA for RNase-recognition or in directly recruiting the RNase itself. Failure to cleave the 3’-end of 16S rRNA by the YbeY homolog in B. subtilis (YqfG) results in degradation of the ribosome by the exoribonuclease RNase R, demonstrating the importance of this quality control step in ribosome assembly (Baumgardt et al., 2018). In contrast to the stimulatory roles investigated here, r-proteins may also play activity-reducing roles to link processing to quality control of ribosome assembly. The 5’-3’ exoribonuclease activity of B. subtilis RNase J1, required for maturation of the 5’-end of 16S rRNA, is thought to be blocked by as yet unidentified r-proteins from progressing beyond the mature 5’ end and degrading the 16S rRNA completely (Mathy et al., 2007). The activity of RNases involved in non-rRNA maturation reactions can also be modulated by protein co-factors, e.g. RNase III is modulated by YmdB (Kim et al., 2008), RNase E by RraA/B (Gao et al., 2006; Lee et al., 2003) and RNase Y by the Y-complex (Deloughery et al., 2016; DeLoughery et al., 2018). However, in contrast to the rRNA maturating RNases, the amplitude of the modulation effects on these non-rRNA maturation RNases is often lower, suggesting that their purpose is to fine tune enzyme activity or specificity according to cellular needs, rather than to act in ‘all-or-nothing’ pathways, typified by those related to quality control.
Mini-III and M5 could be engineered to serve other functions
Double-strand RNases of the RNase III family serve as a front-line defence against virus attack, through RNA silencing (Dicer and Drosha) (Bernstein et al., 2001; Zeng et al., 2005), and in CRISPR phage defence mechanisms, through the processing of crRNAs by RNase III (Deltcheva et al., 2011). Both Mini-III and M5 recognise dsRNA differently from RNase III. These RNases could be recruited for similar defence functions in bacteria, or could be engineered to carry out these functions in the future. Mini-III could further serve as a novel dsRNA-cleavage counterpart to currently available dsDNA-cleavage restriction enzymes. For this function, sequence specificity is essential, and previous work showed that B. subtilis Mini-III cleaves long dsRNA, independent of uL3, in a sequence-dependent and specific manner (Głów et al., 2015). Our understanding of their RNA recognition mode clears the path to engineering of these enzymes for other biological applications.
Star methods text
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
- RESOURCE AVAILABILITY
- Lead Contact
- Materials Availability
- Data and Code Availability
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
- B. subtilis strain constructions and isolation of 50S ribosomal subunits
- METHODS DETAILS
- Cloning, expression and purification of M5, uL18 and Mini-III
- RNase activity assays
- CryoEM sample preparation and data acquisition
- CryoEM image processing
- Model building and refinement of cryoEM structures
- Crystallisation, data collection, and X-ray structure determination
- Hydrogen deuterium exchange – mass spectrometry (HDX-MS)
- Differential scanning fluorimetry (DSF)
- Circular dichroism (CD)
- In vitro transcription of pre-5S rRNAs
- Determination of KD for the M5 interaction with pre-5S rRNA and uL18
QUANTIFICATION AND STATISTICAL ANALYSIS
Resource Availability
Lead Contact
Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact, Carine Tisné (carine.tisne@cnrs.fr).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Experimental Model And Subject Details
B. subtilis strain constructions and isolation of 50S ribosomal subunits
The construction of the B. subtilis strain CCB038 (ΔrnmV) has been described previously (Condon et al., 2001). Strain CCB1262 (ΔmrnC ΔrnjA) was built by transforming strain SSB1044 (ΔmrnC/yazC) (Redko et al., 2008) with chromosomal DNA from strain CCB434 (ΔrnjA) (Figaro et al., 2013). 50S subunits carrying pre-rRNAs were isolated from 200 mL cultures of CCB038 and CCB1262 grown in 2xYT medium. Cells were pelleted, washed in buffer A (20 mM Tris-HCl pH 7.5, 200 mM NH4Cl, 6 mM β-mercaptoethanol) supplemented with 10 mM MgCl2. Pellets were resuspended in 1.7 mL buffer A containing 10 mM MgCl2, and lysed by French Press (Glen Mills; 20000 Psi). The lysate was clarified by centrifugation, and layered on top of a 38 mL 10-30% sucrose gradient in buffer A containing 3 mM MgCl2 (4°C), prepared in ultracentrifuge tubes (Seton), with a Biocomp Gradient Master 108 (Serlabo) gradient maker. Gradients were centrifuged 16 hrs at 22,100 rpm in a Beckmann SW28 rotor at 4°C. Fractions were collected, and those containing 50S subunits with pre-rRNA were pooled, concentrated, and resuspended in 20-40 μL of a buffer containing 20 mM Tris-HCl pH 7.4, 100 mM NaCl, 100 mM KCl, 6 mM β-mercaptoethanol and 10 mM MgCl2.
Methods Details
Cloning, expression and purification of M5, uL18 and Mini-III
DNA fragments encoding Geobacillus stearothermophilus M5 (residues 1-187) or separate domains (NTD: residues 1-113, CTD: 105-187), and G. stearothermophilus uL18, were subcloned into the pET28a vector modified to incorporate an N-terminal His6-tag followed by a Tobacco Etch Virus protease (TEV) cleavage site. B. subtilis Mini-III (residues 1-143) was cloned as described (Redko et al., 2008). Mutations were generated using the QuikChange Site-Directed Mutagenesis Kit (Agilent), and verified by sequencing. Plasmids were transformed into E. coli BL21(DE3) CodonPlus RIL competent cells, cultured in Lysogenic Broth at 37°C, and protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3h growth at 37°C. Cell pellet from 2 L culture was lysed by sonication in a buffer containing 20 mM Tris pH 8.0, 1 M NaCl, 5% glycerol, 1 mM DTT, 20 mM imidazole, and clarified by centrifugation. The clarified lysate was applied to a 5 mL HisTrap HP column (GE Healthcare) and eluted with a gradient of 20-500 mM imidazole. M5 proteins were buffer exchanged into 50 mM HEPES pH 7.5, 200 mM NaCl, 5% glycerol, 1 mM DTT and applied to a 5 mL HiTrap Heparin HP column (GE Healthcare). Protein was eluted with a gradient of 200-1000 mM NaCl, treated with TEV for 12h at room temperature and applied to a 5 mL HisTrap HP column (GE Healthcare) to remove the His6-tag. Mini-III protein from HisTrap HP purification was buffer exchanged into 50 mM HEPES pH 7.5, 50 mM NaCl, 5% glycerol, 1 mM DTT, applied to a 6 mL Resource S column (GE Healthcare), and eluted with a gradient of 50-1000 mM NaCl. Purified proteins were buffer exchanged into 20 mM Tris pH 7.4, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 1 mM CaCl2 before storage. The oligomeric state of each protein was evaluated by analytical size exclusion chromatography (SEC) against molecular weight standards (BioRad), or by coupling SEC with multi-angle light scattering analysis to either a Superdex 75 or 200 10/300 GL (GE Healthcare) 24 mL column.
RNase activity assays
Assays were performed in 10 μL reactions containing 0.8 μg (0.7 pmol) 50S subunits with pre-rRNA prepared from CCB038 or CCB1260 cells and 25 ng (1.2 pmol) M5 protein or 10 ng (0.3 pmol dimers) Mini-III protein in buffer A containing 10 mM MgCl2 at 37°C. Mini-III reactions were stopped at times indicated by adding 10 μL of running dye (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), applied to 1% agarose gels without prior denaturation and capillary transferred to hybond-N (GE Healthcare) membranes. M5 samples were phenol extracted, precipitated with 0.1 volumes of 20 mg/mL glycogen, 0.1 volumes 10 mM LiCl, and 3 volumes of 95% EtOH, resuspended in 10 μL H2O, and added 10 μL running dye. Samples were denatured at 95°C for 5 mins before migration on 5% polyacrylamide/7M urea gels, and electro-transferred (300 mA) to hybond-N (GE Healthcare) membranes. Membranes were UV-cross-linked and probed with oligo HP246 (5’-ATC GGC GCT GAA GAG CTT AAC TTC C-3’) complementary to the mature 5S rRNA sequence as described previously (Condon et al., 2001), or oligos CC2563 (5’-CCG TTA AAA AGA ATC ACT ATG TGA TAT CTT G-3’) and CC2564 (5’-GAT AAC AGA TGT GAC ATC ATT CAA AAA TAT G-3’), complementary to pre-23S rRNA 5’- and 3’-precursor sequences, respectively. Membranes with Mini-III samples were stripped and re-probed for the mature 23S rRNA sequence, using oligo CC257 (5’-ATA TGA GCT CCA TCG GCT CCT AGT GCC AAG GCA TC-3’), to serve as loading control. Initial reaction rates (fmol/min) were derived by linear regression in GraphPad Prism of time-points from 0-1 min for wild-type protein or 0-2.5 min for mutant proteins in a plot of remaining precursor (%) over time (min), where 100% corresponds to 700 fmol.
CryoEM sample preparation and data acquisition
Samples for cryoEM analysis were prepared in a buffer of 20 mM Tris pH 7.4, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 1 mM CaCl2, by mixing 50S(pre-5S) subunits with RNase M5 at ratios ranging from 1:1 to 1:1000. G. stearothermophilus M5 was utilised as it binds B. subtilis pre-5S rRNA with a similar KD as B. subtilis M5, but proved less prone to aggregation. A final 50S(pre-23S) to RNase ratio of 1:210 was used, giving rise to final concentrations of 0.47 μM B. subtilis 50S(pre-23S) to 100 μM B. subtilis Mini-IIID23N protein, and 1 μM B. subtilis 50S(pre-5S) to 210 μM G. stearothermophilus M5D58A protein. 3 μL of each mixture was applied to glow-discharged UltrAuFoil Au 300 1.2/1.3 grids, blotted at 100 % humidity, 4° C for 3 s at a blot force of 5 for M5/50S(pre-5S) and 0 for Mini-III/50S(pre-23S), then plunge frozen into liquid ethane using a Vitrobot (FEI). Images were recorded on a FEI Talos Artica microscope with a Falcon III direct electron detector at 200 kV operating in counting mode. Data collection parameters are summarised in Table S1.
CryoEM image processing
397 movies were collected for the 50S(pre-23S)/Mini-III assembly and 794 movies (one dataset of 409 movies and one dataset of 385 movies) for the 50S(pre-5S)/M5 assembly. Data processing was performed using Relion-3.0 (Zivanov et al., 2018) and CryoSparc-2.8 (Punjani et al., 2017), and is summarised in Figure S1B (50S(pre-5S)-M5) and S2A (50S(pre-23S)/Mini-III). With Relion-3.0, a particle subset was manually selected after motion correction and ctf estimation to calculate reference-free 2D class averages, which were then used as templates for automated particle picking. 2D-classification and several rounds of 3Dclassification were used to iteratively clean up the particle sets. 3D auto-refinement resulted in isotropic maps at near atomic resolution. After per-particle motion correction and radiationdamage weighting, the polished particles were subjected to per particle ctf refinement in Relion-3.0. Particles were then transferred to CryoSparc and subjected to heterogenous refinement and/or non-uniform refinement in CryoSparc-2.8, followed by global sharpening, local resolution estimation and local filtering. The final resolutions of the maps were estimated by 0.143 cut-off of the global Fourier shell correlation (FSC) and by a 0.5 cut-off for the local resolution estimation. Data processing parameters for all specimens are summarised in Table S1.
Model building and refinement of cryoEM structures
Models were docked to the density in Chimera-1.11.2 (Pettersen et al., 2004), and 5’- and 3’-extensions were modelled manually in Coot-0-8-9-1 (Emsley et al., 2010) from the CCP4 (Ballard et al., 2014) of programs. After model building both structures were subjected to density weighted molecular dynamics fitting with Namdinator (Kidmose et al., 2019) followed by geometrical optimisation with ISOLDE (Croll, 2018) and cycles of real-space refinement in Phenix (Afonine et al., 2012) against the locally filtered cryo-EM maps, and manual fitting in Coot. Model vs. map statistics are reported in Table S1.
Crystallisation, data collection, and X-ray structure determination
Extensive attempts to crystallise M5 protein were unsuccessful. Instead, crystals of M5 CTD and M5 NTD were grown by vapour diffusion at 20°C from sitting drops composed of 200 nL of protein (4 mg/ml) and 200 nL of reservoir solution containing 0.1 M sodium cacodylate pH 6.5, 0.2 ammonium sulphate and 30% PEG 8000 for the CTD, and drops of 50 nL of protein (26.6 mg/ml) and 100 nL of reservoir solution of 0.1 M HEPES pH 7.5, 1.4 M sodium citrate tribasic dihydrate for the NTD. Crystals were cryo-protected with reservoir solution supplemented with 20% (v/v) glycerol, and flash-cooled in liquid nitrogen. Diffraction data were collected to 1.3 Å for the NTD and to 1.5 Å for the CTD. Phases for the CTD were determined by ab initio molecular replacement (MR) of α-helical segments in ARCIMBOLDO (Rodríguez et al., 2012). This procedure generated an initial model covering the entire domain. Phases for the NTD were determined by MR using a homology model generated with iTasser (Yang and Zhang, 2015) covering the entire domain. Further modelling and refinement was carried out using Refmac5 (Murshudov et al., 1997) and COOT (Emsley et al., 2010). The asymmetric unit (ASU) contained two protein molecules of the CTD, and one molecule for the NTD. Residues W117-D186 were assigned for the CTD, and the NTD covered residues M1-E113, plus an N-terminal GGS sequence from the vector. For the CTD, the two molecules in the asymmetric unit align with an RMSD of 0.6 Å for the Cα atoms, with the main differences attributed to residues at the chain termini or segments involved in crystal contacts. Final statistics are reported in Table S2.
Hydrogen deuterium exchange – mass spectrometry (HDX-MS)
A 16 μM M5D56A sample in 20 mM Tris pH 7.4, 200 mM NaCl, 2 mM MgCl2, 1 mM β-mercaptoethanol, 5% glycerol was incubated at 20°C for five deuteration time points (0.5, 2, 10, 30 and 60 minutes for deuterated experiments) in 20 mM Tris, 200 mM NaCl, 2 mM MgCl2, 1 mM β-mercaptoethanol with D2O pD 7.4. Exchange reactions were stopped by addition of 1:1 (v/v) quench buffer (2M guanidine-HCl, 100 mM Glycine pH 2.35) at 1°C over 0.5 minutes. Quenched samples were injected with a CTC PAL robot (Leap Technologies, Zwingen, Switzerland) into an Acquity UPLC system with HDX technology (Waters, Manchester, UK) and digested through a pepsin-immobilized cartridge in 0.1% aqueous formic acid solution at a flow rate of 200 μL/min, and next trapped and desalted on a UPLC pre-column (ACQUITY UPLC BEH C18 VanGuard pre-column, 2.1 mm I.D. x 5 mm, 1.7 μM particle diameter, Waters) for 3 minutes. Digested peptides were separated on a UPLC reversed-phase column (ACQUITY UPLC BEH C18, 1.0 mm I.D. x 100 mm, 1.7 μM particle diameter, Waters) at 0.1°C with a gradient elution of solvent A (0.1% formic acid aqueous) and solvent B (acetonitrile with 0.1% formic acid) [2% -40% B (7 min), 40% -85% B (0.5 min), and 85% B (1 min)] at a flow rate of 40 μL/min. MS/MS analyses were acquired on a Synapt G2SI HDMS (Waters, Manchester, UK) in positive polarity and resolution mode initially calibrated, and using a lock-mass correction with glufibrinogen peptide. Data were acquired in MSE acquisition mode with the following parameters: capillary voltage, 3 kV; sampling cone voltage, 40 V; source temperature, 80°C; desolvation gas, 150°C and 600 L/h; acquisition range, acquisition range, 50-2000 m/z; scan time, 0.3 s; trap collision energy, 15 to 40 eV. MSE data were processed using Waters ProteinLynx Global Server 2.5.3 and filtered with DynamX 3.0 as follows: the experiment was carried out as three analytical replicates, where only peptides identified in all replicates between 5 and 30 residues were kept, with a minimum fragment per amino acid of 0.2 and a minimum intensity at 103. Deuterium uptakes for all identified peptides were manually validated. Deuterium uptakes were not corrected for back-exchange, represented as relative.
Differential scanning fluorimetry (DSF)
DSF was performed in a 96-well plate using a CFX96 Touch real-time PCR detection system (Bio-Rad) with excitation and emission filters of 450-490 and 515-530 nm, respectively. Each well consisted of 2 μL protein in a buffer of 20 mM HEPES pH 7.5, 150 mM NaCl to a final concentration of 5 μM, 2 μL of SYPRO Orange diluted 5000-fold in buffer from the manufacturer’s stock (Invitrogen). Fluorescence intensities were measured from 25 to 95°C with a ramp rate of 1°C/min. Tm-values were determined by curve-fitting using GraphPad Prism v.5.01 software of three technical replicates like previously described (Niesen et al., 2007).
Circular dichroism (CD)
CD spectra were measured (Aviv215 spectropolarimeter, Aviv Biomedical) with protein samples at 1 mg.mL-1 in a buffer of 50 mM HEPES pH7.5, 230 mM NaCl, 10 mM MgCl2, 1 mM TCEP. Far-UV CD spectra were recorded between 200 and 260 nm using a cylindrical cell with a 0.02 cm path length and an averaging time of 1 s per step. The scan was repeated consecutively three times and merged to produce an averaged spectrum. This spectrum was corrected using buffer baselines measured under the same conditions and normalised to the molar peptide bond concentration and path length as Delta Epsilon coefficient per residue. Secondary structure estimations were derived from the normalised spectra using BeStSel (Micsonai et al., 2018).
In vitro transcription of pre-5S rRNAs
Single stranded DNA template for G. stearothermophilus pre-5S rRNA (5’-AmTmT GCC TAG CAG CGA CCT ACT CAC ATC GGC GCT GGA GGG CTT AAC TTC CGT GTT CGG GAT GGG AAC GGG TGT TTC CCC TCC GCT ATC ACC ACT AGG CAA TCC C TAT AGT GAG TCG TAT TA-3’) (Eurogentec) was used for in vitro transcription with a T7 RNA polymerase primer (5’-TAA TAC GAC TCA CTA TAG-3’). RNAs were transcribed in a 5 mL reaction with 0.2 μM DNA template in a buffer of 20 mM Tris-HCl pH 8.0, 0.5 mM spermidine, 2.5 mM DTT, 0.005% Triton X-100, supplemented with 5 mM of each rNTP, 5.6 mM GMP, 49.7 mM MgCl2 and T7 RNA polymerase. The reaction was incubated for 4h at 37°C, and stopped with 100 mM EDTA. The reaction mixture was purified on an 8 mL MonoQ 10%100 GL anion exchange column (GE Healthcare) in a buffer of 25 mM sodium phosphate pH 6.5, 50 mM NaCl, and eluted in a similar buffer with a 50 mM to 1 M NaCl gradient. The RNA was buffer exchanged into 1.7 M (NH4)2SO4 and applied to an 8 mL Phenyl superose HR 10/10 column (GE Healthcare), and eluted in H2O with a 1.7 M to 0 M (NH4)2SO4 gradient.
Determination of KD for the M5 interaction with pre-5S rRNA and uL18
The EMSA was performed in a 20 μL reaction mixture with 3 μM pre-5S rRNAs, 3 μM uL18 and M5 proteins at 0.375-48 μM, in a buffer of 20 mM Tris-HCl pH 7.4, 100 mM NaCl, 100 mM KCl, 10 mM MgCl2, 1 mM CaCl2. For mixtures with one M5 concentration, protein was added to 24 μM. Reaction mixtures were incubated at room temperature for 30 minutes, mixed with 2 μL 30% glycerol, and applied to a 4°C 2.5% agarose gel with in-cast ethidium bromide, and electrophoresed in 4°C TB buffer (45 mM Tris-base pH 8.3, 45 mM boric acid) at room temperature for 10 min at a constant voltage of 20 V/cm. Bands were visualised using the Bio-Rad ChemiDoc Imaging System, and quantified using ImageJ (Schneider et al., 2012). KD-values were determined by curve-fitting using GraphPad Prism v.5.01 software (Niesen et al., 2007) from three independent experiments as a least-squares fit for one site with specific binding from a plot of protein concentration against percentage of bound pre-5S rRNA.
Quantification And Statistical Analysis
See Methods Details for details on image processing and model building and refinements.
All p values for the ribosome profiling data were calculated using t tests based on custom R or Python scripts. Statistical analysis of the remaining datasets was performed with Microsoft Excel. Typically, data were averaged, standard deviations calculated, and statistical significance was assessed using the T.Test function assuming two-tailed distribution and unequal variance.
Supplementary Material
Acknowledgments
All grids were prepared and cryo-EM data collected at the BIOCEM facility, Department of Biochemistry, University of Cambridge. We thank Dr Dimitri Y. Chirgadze, Dr Steve Hardwick and Mr Lee Cooper for assistance with data collection at the Cryo-EM Facility. We thank the staff at the Synchrotron Soleil beamline Proxima-1 (PX1) for support during data collection, Christophe Velours from the Institute de Biologie Integrative de la Cellule at the I2BC platform for assistance with SEC-MALS and Bruno Baron from the PFT Biophysique Moléculaire of C2RT at Institut Pasteur for CD experiments. This work was supported by funds from the IDEX USPC, CNRS (UMR8261, UMR7178), Paris University, University of Strasbourg (Unistra), the Agence Nationale de la Recherche (Project ARNr-QC, ANR-15-CE11-0009), the Labex Dynamo (ANR-11-LABX-0011), the French Proteomic Infrastructure (ProFI; ANR-10-INBS-08-03) and the Edmond de Rothschild foundation. BFL and TD are supported by a Welcome Trust Investigator Award and an AstraZeneca studentship (TD). MB was supported by a PhD fellowship from the Région Alsace. The BIOCEM facility is funded by the Welcome Trust (206171/Z/17/Z; 202905/Z/16/Z), Department of Biochemistry, Department of Chemistry, School of Biological Sciences, School of Clinical Medicine, and University of Cambridge. SC and MB thank GIS IBiSA, Région Alsace, Communauté Urbaine de Strasbourg, FEDER and the IdeX program of the University of Strasbourg for financial support for purchase of HDX-MS and native MS instruments.
Footnotes
Author contributions
CC and CT conceived the project. SO, MC, LG and AT prepared the samples for CryoEM analysis; SO, TD and BFL performed CryoEM data collection, processing, and refinement; SO, MC, CD, PB and CT performed the structural study of M5; MB and SC performed mass spectrometry analysis. SO, CC and CT analysed the data and prepared the manuscript with input from all authors. All authors discussed the data analysis, critically reviewed the manuscript, and approved the final version.
Declaration of interests
The authors declare no competing interests.
Data and code Availability
All software and algorithms are listed in the Key Resources Table.
The cryoEM maps generated in this study have been deposited in the Protein data Bank (PDB and the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-10535 (B. subtilis Mini-IIID23N / B. subtilis 50S(pre-23S)), and EMD-10543 (G. stearothermophilus M5D58A / B. subtilis 50S(pre-5S)). The atomic coordinates have been deposited in the PDB under the accession numbers 6TNN (B. subtilis Mini-IIID23N / B. subtilis 50S(pre-23S)), 6TPQ (G. stearothermophilus M5D58A / B. subtilis 50S(pre-5S)), 6TG6 (G. stearothermophilus M5 NTD), and 6TGJ (G. stearothermophilus M5 CTD).
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand F, Mathy N, Brechemier-Baey D, Condon C. The 5S rRNA maturase, ribonuclease M5, is a Toprim domain family member. Nucleic Acids Res. 2005;33:4368–4376. doi: 10.1093/nar/gki752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard C, Keegan R, Krissinel E, Lebedev A, Uski V, Waterman D, Wojdyr M. CCP4: a resource for macromolecular crystallography. Acta Crystallogr Sect A Found Adv. 2014;70:C1723-C1723 [Google Scholar]
- Baumgardt K, Gilet L, Figaro S, Condon C. The essential nature of YqfG, a YbeY homologue required for 3’ maturation of Bacillus subtilis 16S ribosomal RNA is suppressed by deletion of RNase R. Nucleic Acids Res. 2018;46:8605–8615. doi: 10.1093/nar/gky488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckert B, Abdelshahid M, Schäfer H, Steinchen W, Arenz S, Berninghausen O, Beckmann R, Bange G, Turgay K, Wilson DN. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 2017;36:2061–2072. doi: 10.15252/embj.201696189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bunner AE, Nord S, Wikström PM, Williamson JR. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J Mol Biol. 2010;398:1–7. doi: 10.1016/j.jmb.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet-D’Orval B, Batista M, Bouvier M, Quentin Y, Fichant G, Marchfelder A, Maier LK. Insights into RNA-processing pathways and associated RNA-degrading enzymes in Archaea. FEMS MicroBiol Rev. 2018;42:579–613. doi: 10.1093/femsre/fuy016. [DOI] [PubMed] [Google Scholar]
- Condon C. In: Encyclopedia of Microbiology. Schmidt, RNA Processing, editor. Elsevier; 2009. pp. 395–408. [Google Scholar]
- Condon C, Brechemier-Baey D, Beltchev B, Grunberg-Manago M, Putzer H. Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: Mature 5S rRNA is dispensable for ribosome function. Rna. 2001;7:242–253. doi: 10.1017/s1355838201002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll TI. ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect D Struct Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe-McAuliffe C, Graf M, Huter P, Takada H, Abdelshahid M, Nováče k, Murina V, Atkinson GC, Hauryliuk V, Wilson DN. Structural basis for antibiotic resistance mediated by the Bacillus subtilis ABCF ATPase VmlR. Proc Natl Acad Sci U S A. 2018;115:8978–8983. doi: 10.1073/pnas.1808535115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System, Version 1.8. Schrödinger LLC; 2014. http://www.pyMolorg. [Google Scholar]
- Deloughery A, Dengler V, Chai Y, Losick R. Biofilm formation by Bacillus subtilis requires an endoribonuclease-containing multisubunit complex that controls mRNA levels for the matrix gene repressor SinR. Mol Microbiol. 2016;99:425–437. doi: 10.1111/mmi.13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoughery A, Lalanne JB, Losick R, Li GW. Maturation of polycistronic mRNAs by the endoribonuclease RNase Y and its associated Y-complex in Bacillus subtilis. Proc Natl Acad Sci U S A. 2018;115:E5585–E5594. doi: 10.1073/pnas.1803283115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J Bacteriol. 2013;195:2340–2348. doi: 10.1128/JB.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Miller OL. Transcription mapping of the escherichia coli chromosome by electron microscopy. J Bacteriol. 1989;171:4207–4216. doi: 10.1128/jb.171.8.4207-4216.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol Microbiol. 2008;67:143–154. doi: 10.1111/j.1365-2958.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, Moore CJ, Cohen SN, Georgiou G. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol Microbiol. 2006;61:394–406. doi: 10.1111/j.1365-2958.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- Głów D, Pianka D, Sulej AA, Kozłowski ŁP, Czarnecka J, Chojnowski G, Skowronek KJ, Bujnicki JM. NAR Breakthrough Article: Sequence-specific cleavage of dsRNA by Mini-III RNase. Nucleic Acids Res. 2015;43:2864–2873. doi: 10.1093/nar/gkv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6:225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- Kidmose RT, Juhl J, Nissen P, Boesen T, Karlsen JL, Pedersen BP. Namdinator -Automatic molecular dynamics flexible fitting of structural models into cryo EM and crystallography experimental maps. IUCr J. 2019;6:526–531. doi: 10.1107/S2052252519007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Manasherob R, Cohen SN. YmdB: A stress-responsive ribonuclease-binding regulator of E. coli RNase III activity. Genes Dev. 2008;22:3497–3508. doi: 10.1101/gad.1729508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhan X, Gao J, Qiu J, Feng Y, Meganathan R, Cohen SN, Georgiou G. RraA: A protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell. 2003;114:623–634. [PubMed] [Google Scholar]
- Li N, Chen Y, Guo Q, Zhang Y, Yuan Y, Ma C, Deng H, Lei J, Gao N. Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res. 2013;41:7073–7083. doi: 10.1093/nar/gkt423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. Polyadenylation of stable RNA precursors in vivo. Proc Natl Acad Sci U S A. 1998;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 5’-to-3’ Exoribonuclease Activity in Bacteria: Role of RNase J1 in rRNA Maturation and 5’ Stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Micsonai A, Wien F, Bulyáki É, Kun J, Moussong É, Lee YH, Goto Y, Réfrégiers M, Kardos J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46:W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Nomura M. Assembly of bacterial ribosomes. Fed Proc. 1972;31:18–20. [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera -A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Podobnik M, McInerney P, O’Donnell M, Kuriyan J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J Mol Biol. 2000;300:353–362. doi: 10.1006/jmbi.2000.3844. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Redko Y, Condon C. Ribosomal protein L3 bound to 23S precursor rRNA stimulates its maturation by Mini-III ribonuclease. Mol Microbiol. 2009;71:1145–1154. doi: 10.1111/j.1365-2958.2008.06591.x. [DOI] [PubMed] [Google Scholar]
- Redko Y, Condon C. Maturation of 23S rRNA in Bacillus subtilis in the absence of mini-III. J Bacteriol. 2010;192:356–359. doi: 10.1128/JB.01096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redko Y, Bechhofer DH, Condon C. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol Microbiol. 2008;68:1096–1106. doi: 10.1111/j.1365-2958.2008.06207.x. [DOI] [PubMed] [Google Scholar]
- Rodríg uez, Sammito M, Meindl K, De Ilarduya ĨM, Potratz M, Sheldrick GM, Usón I. Practical structure solution with ARCIMBOLDO. Acta Crystallogr Sect D Biol Crystallogr. 2012;68:336–343. doi: 10.1107/S0907444911056071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymer RU, Solorio FA, Tehranchi AK, Chu C, Corn JE, Keck JL, Wang JD, Berger JM. Binding mechanism of metalśNTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure. 2012;20:1478–1489. doi: 10.1016/j.str.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz CJ, Lee A, Partlow EA, Hosford CJ, Chappie JS. Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage. Nucleic Acids Res. 2019;47:9448–9463. doi: 10.1093/nar/gkz703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller JM, Falk S, Fromm L, Hurt E, Conti E. Structure of the nuclear exosome captured on a maturing preribosome. Science. 2018;360:219–222. doi: 10.1126/science.aar5428. [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. Mechanism And Regulation Of Bacterial Ribosomal RNA Processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- Stahl DA, Meyhack B, Pace NR. Recognition of local nucleotide conformation in contrast to sequence by a rRNA processing endonuclease. Proc Natl Acad Sci U S A. 1980;77:5644–5648. doi: 10.1073/pnas.77.10.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DA, Pace B, Marsh T, Pace NR. The ribonucleoprotein substrate for a ribosomal RNA-processing nuclease. J Biol Chem. 1984;259:11448–11453. [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S ribosomal RNA database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse M, Köhrer C, Shen Y, Proulx S, Ghosal A, Davies BW, RajBhandary UL, Walker GC. Identification of YbeY-protein interactions involved in 16S rRNA maturation and stress regulation in Escherichia coli. MBio. 2016;7 doi: 10.1128/mBio.01785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. An equivalent metal ion in one-and two-metal-ion catalysis. Nat Struct Mol Biol. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Topoisomerases and site-specific recombinases: Similarities in structure and mechanism. Crit Rev Biochem Mol Biol. 2010;45:520–534. doi: 10.3109/10409238.2010.513375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software and algorithms are listed in the Key Resources Table.
The cryoEM maps generated in this study have been deposited in the Protein data Bank (PDB and the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-10535 (B. subtilis Mini-IIID23N / B. subtilis 50S(pre-23S)), and EMD-10543 (G. stearothermophilus M5D58A / B. subtilis 50S(pre-5S)). The atomic coordinates have been deposited in the PDB under the accession numbers 6TNN (B. subtilis Mini-IIID23N / B. subtilis 50S(pre-23S)), 6TPQ (G. stearothermophilus M5D58A / B. subtilis 50S(pre-5S)), 6TG6 (G. stearothermophilus M5 NTD), and 6TGJ (G. stearothermophilus M5 CTD).