Summary
The extensively studied yeast GAL1–10 gene cluster is tightly regulated by environmental sugar availability. Unexpectedly, under repressive conditions the 3′ region of the GAL10 coding sequence is trimethylated by Set1 on histone H3 K4, normally characteristic of 5′ regions of actively transcribed genes. This reflects transcription of a long noncoding RNA (GAL10-ncRNA) that is reciprocal to GAL1 and GAL10 mRNAs and driven by the DNA-binding protein Reb1. Point mutations in predicted Reb1-binding sites abolished Reb1 binding and ncRNA synthesis. The GAL10-ncRNA is transcribed approximately once every 50 min and targeted for degradation by the TRAMP and exosome complexes, resulting in low steady-state levels (approximately one molecule per 14 cells). GAL10-ncRNA transcription recruits the methyltransferase Set2 and histone deacetylation activities in cis, leading to stable changes in chromatin structure. These chromatin modifications act principally through the Rpd3S complex to aid glucose repression of GAL1–10 at physiologically relevant sugar concentrations.
Introduction
Studies of the human and yeast transcriptomes indicate that ncRNAs are much more numerous than initially expected. In metazoans, ncRNAs are implicated in the formation of heterochromatin, X chromosome inactivation, dosage compensation, and allele-specific repression at imprinted loci (reviewed in Munroe and Zhu, 2006; Pauler et al., 2007). However, little is known about the function of the many ncRNA transcripts in euchromatic regions of the genome. The FANTOM3 mouse cDNA sequencing project found many pairs of coding-noncoding transcripts with either coregulation or inverse regulation, suggesting that ncRNAs may play a general role in the regulation of coding transcripts (Katayama et al., 2005). ncRNAs are activated by many of the same factors as coding transcripts; 36% of the predicted binding targets of transcription factors cMyc, Sp1, and p53 occur within or immediately 3′ to annotated genes, suggesting that they activate antisense transcripts to these coding sequences (Cawley et al., 2004).
Widespread transcription of intergenic regions is also predicted in S. cerevisiae from global analyses of RNA pol II density (Steinmetz et al., 2006). ncRNAs derived from intergenic regions and silenced regions of the yeast genome are not normally detected, however, due to the rapid degradation of transcripts by the TRAMP complexes and the nuclear exosome (Houseley et al., 2007; Wyers et al., 2005). Perhaps the best-studied sense-antisense pair is the PHO84 gene, where loss of the nuclear exosome component Rrp6 leads to stabilization of an anti-sense transcript. This results in repression of the sense transcript through recruitment of the histone deacetylase (HDAC) Hda1, which deacetylates the PHO84 promoter (Camblong et al., 2007). In addition, ncRNAs repress Ty1 retrotransposition via histone modifications induced in trans (Berretta et al., 2008). This suggested that ncRNA transcription could contribute to the establishment of chromatin structure within euchromatic regions.
Histone modifications modulate gene expression by controlling the binding of regulatory proteins (reviewed in Ruthenburg et al., 2007). Nucleosomes are generally hyperacetylated at promoters of active genes and hypoacetylated when transcription is repressed. The process of transcription is accompanied by alterations in histone modification patterns, as chromatin is unfolded in front of, and reassembled behind, the transcribing polymerase. Early transcriptional events lead to localized recruitment of the histone methyltransferase Set1, which trimethylates histone H3 lysine 4 (H3 K4me3) over the 5′ end of transcribed coding regions (Ng et al., 2003; Schneider et al., 2004). As RNA polymerase advances through the gene, lysine 36 of H3 becomes increasingly trimethylated (H3 K36me3) by another histone methyltransferase Set2 (reviewed in Lee and Shilatifard, 2007). This latter modification causes the recruitment of the small Rpd3 HDAC complex, via binding of the chromodomain protein Eaf3, leading to deacetylation over transcribed coding regions and preventing spurious internal transcription initiation (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005).
The GAL cluster of S. cerevisiae encodes three genes required for galactose metabolism, GAL1, GAL7, and GAL10, and has been analyzed extensively as a model for regulated gene expression. The GAL cluster has three characterized states depending on the carbon sources available in the medium: induced (in the absence of glucose and the presence of galactose), noninduced (in the absence of glucose and the presence of other carbon sources such as raffinose), and repressed (in the presence of glucose). Such highly regulated nutrient response systems allow S. cerevisiae to thrive on a wide range of carbon sources other than fermenting fruit. Analyses of fermented food products have shown S. cerevisiae to be commonly the most prevalent microbe in cultures derived from cereals (principle sugar maltose) and milk (principle sugar lactose) (reviewed in Jespersen, 2003). Therefore, S. cerevisiae has evolved to metabolize the complex mixtures of sugars present in natural environments.
In this study we demonstrate that binding of the regulatory protein Reb1 to the 3′ end of the GAL10 ORF stimulates transcription of a ncRNA under repressed and noninduced conditions. This causes di- and trimethylation of histone 3 lysine 4 (H3 K4me2, H3 K4me3) within the GAL10 coding region, and H3 K36me3 across the entire GAL1–10 locus, along with decreased H3 acetylation. These chromatin structure modifications lead to increased glucose repression of GAL1–10 in low-sugar media, primarily mediated by H3 K36me3 binding protein Eaf3. We suggest that the GAL10-ncRNA and other ncRNAs modulate gene expression patterns to optimize nutrient utilization.
Results
H3 K4 Is Methylated over the 3′ Coding Region of GAL10 under Repressive Conditions
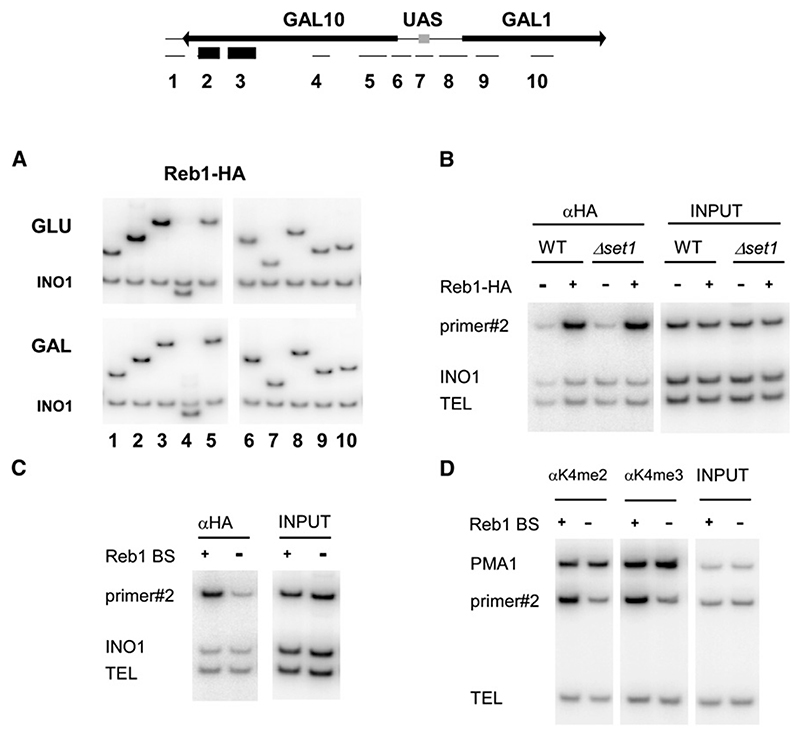
Methylation of histone H3 K4 is closely linked to transcriptional activity of RNA polymerase II (RNA pol II). H3 K4me3 is generally restricted to the 5′ end of transcribed genes, where it correlates with the stable recruitment of the methyltransferase Set1 (Ng et al., 2003). In contrast, H3 K4me2 can spread into the body of genes in higher eukaryotes and appears even more widely distributed in yeast (Pokholok et al., 2005; Santos-Rosa et al., 2002; Schneider et al., 2004). However, we observed an unusual pattern of H3 K4me3 over the GAL1–10 cluster (Figure 1A).
Figure 1. H3 K4 Methylation Pattern at the GAL1–GAL10 Locus.
(A) Schematic of the GAL locus showing the location of primer sets used for ChIP analysis.
(B) ChIP analysis of H3 K4 methylation and HA-Set1 binding. Cells were grown to log phase in rich media containing 2% galactose (GAL) or 2% glucose (GLU), and chromatin was precipitated with antibodies to H3 K4me2, H3 K4me3, and the HA-epitope. PCR was performed with primer sets shown in (A) and a telomeric primer set (TEL) as a loading control.
(C) Quantification of ChIP data as fold enrichment of signal at the GAL locus over telomeric signal normalized to the input.
Chromatin immunoprecipitation (ChIP) was performed across the GAL1–10 locus on cells grown in inducing medium (2% galactose [GAL]) or repressive medium (2% glucose [GLU]) using antibodies specific to H3 K4me2, H3 K4me3, and HA-tagged Set1. As expected, K4me3 was present over the 5′ end of both GAL1 and GAL10 in galactose and correlated with the presence of HA-Set1 (Figures 1B and 1C, left panels). In cells grown on glucose medium, the peaks of K4me2, K4me3, and HA-Set1 binding over the 5′ ends of GAL1 and GAL10 disappeared, consistent with repression of the GAL1–10 promoter (Figures 1B and 1C, right panels). However, a prominent peak of K4me2 and K4me3 appeared over the 3′ coding region of GAL10. The same peak was seen in cells grown in noninducing raffinose medium (data not shown). This peak of H3 K4me is not due to increased histone density over the GAL10-3′ region, as demonstrated by ChIP using an antibody against the C terminus of histone H3 (see Figure S1 available online). Moreover, both K4me2 and K4me3 at this site were abolished in a set1Δ strain (Figure S2). However, stable binding of HA-Set1 was not observed over the GAL10-3′ region, despite the normally tight correlation between Set1 binding to the 5′ end of transcribed genes and the presence of K4me3 (Ng et al., 2003). We conclude that Set1 methylates H3 K4 over the 3′ region of GAL10 in the absence of induction of the GAL1–10 promoter.
The Transcription Regulator Reb1 Is Required for H3 K4 Methylation in the 3′ Region of GAL10
The presence of H3 K4me2 and H3 K4me3 would not be expected in transcriptionally repressed genes (Ng et al., 2003; Pokholok et al., 2005). We therefore suspected that a DNA binding protein was driving transcription from this region, a hypothesis supported by the presence of a DNase I hypersensitive site in the 3′ region of the GAL10 ORF (Proffitt, 1985).
Inspection of the DNA sequence identified one perfect consensus binding site for Reb1 (Reb1 BS) and three closely matching sites. Reb1 is a myb-related protein known to be involved in transcription initiation of RNA pol I and II genes and in termination of RNA pol I transcription (Ju et al., 1990; Morrow et al., 1990). ChIP analyses using a genomic Reb1-HA fusion revealed that a peak of Reb1-HA binding was present over the 3′ region of GAL10 in cells grown in glucose (Figure 2A, GLU) or raffinose medium (data not shown) but was absent in galactose (Figure 2A, GAL). These data correlate well with the presence of the observed H3 K4me peaks, suggesting a causal relation between Reb1 binding and H3 K4 methylation. Extending the analysis to adjacent regions, a second peak of Reb1-HA binding was observed immediately downstream of GAL7, corresponding to the position of a further putative Reb1 BS (Figure S3).
Figure 2. Reb1 Binding to GAL10 in Glucose Causes H3 K4 Methylation.
(A) ChIP analysis of Reb1-HA binding at the GAL locus in 2% glucose (GLU) and 2% galactose (GAL) medium, performed as in Figure 1. Primers are shown in schematic (top). A PCR primer set to the INO1 coding region was used as internal loading control.
(B) ChIP analysis of Reb1-HA binding at the GAL locus in wild-type and set1Δ cells grown in glucose media, performed as in Figure 1.
(C) Reb1-HA binding in the presence and absence of Reb1-binding sites (Reb1 BS+/−) is shown by ChIP from cells grown in glucose media. Primer #2 was used to amplify the GAL locus. Primer sets to the INO1 coding region (INO1) and a telomeric DNA (TEL) were used as loading controls.
(D) ChIP analysis of H3 K4 methylation at the GAL locus in wild-type and Reb1 BSΔ mutant grown in glucose media. ChIP analysis was performed as in Figure 1; primer sets to PMA1 and TEL act as positive and negative controls for H3 K4 methylation.
Reb1 contains SANT domains, which were previously reported to mediate the binding of c-myb to histone H3 (Mo et al., 2005). It was therefore conceivable that H3 K4me is necessary for the binding of Reb1 to chromatin. However, ChIP analyses demonstrated that Reb1-HA was bound to the 3′ region of GAL10 in both the wild-type and in set1Δ cells that lack H3 K4 methylation (Figure 2B). Conversely, H3 K4 methylation could be a consequence of Reb1 binding to the GAL10-3′ region. Deletion of REB1 is lethal, so we abolished its binding to GAL10 by introducing point mutations into all four putative Reb1-binding sites (Reb1 BSΔ mutant). The resulting strain carrying a mutated GAL10 grew indistinguishably from the wild-type on standard 2% galactose media. These mutations reduced both the Reb1-HA and K4me2 and -me3 ChIP signals at this locus to background levels (Figures 2C and 2D). We conclude that Reb1 binding in the GAL10 coding region is required for H3 K4 methylation close to the Reb1-binding site.
Noncoding RNAs in the GAL Cluster
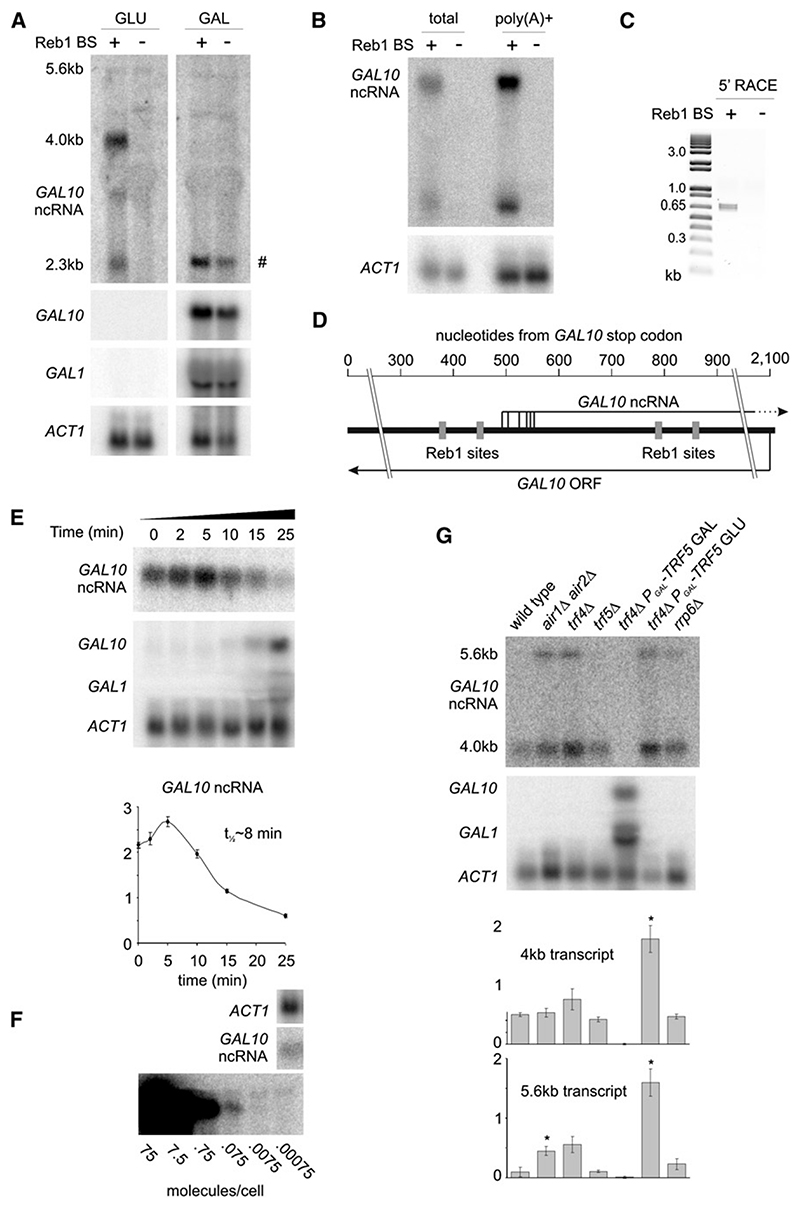
H3 K4me3 is generally associated with sites of transcription initiation, and Reb1 was reported to act as a transcription activator for RNA pol II (Remacle and Holmberg, 1992). To identify Reb1-dependent transcripts, northern blots of RNA from wild-type and Reb1 BSΔ cells grown in glucose and galactose were probed with strand-specific probes to the region of the H3 K4me3 peak (Figure 3A). A major transcript of 4 kb and a weaker 2.3 kb transcript, both running antisense to the GAL10 gene, were observed in the wild-type strain in glucose but not in galactose medium, consistent with the observed H3 K4 methylation. These transcripts were not detected in the Reb1 BSΔ strain in either sugar. A faint signal from a transcript of 5.6 kb was also observed. This may be related to the additional peak of Reb1-HA binding observed at the 3′ end of GAL7 (Figure S3). Oligo-dT selection revealed that the ncRNAs are polyadenylated (Figure 3B), and cap-dependent 5′ RACE showed that they are also capped (Figure 3C). Sequencing of cloned 5′ RACE products identified transcription start sites over a 60 bp range between the second and third Reb1-binding sites (Figure 3D).
Figure 3. A Cryptic Transcript Is Produced from the 3′ End of GAL10 .
(A) Northern analysis of RNA from wild-type and Reb1 BSΔ mutant cells grown in rich media containing 2% glucose or 2% galactose. Probes are GAL10 sense, GAL10 antisense, GAL1, ACT1. Bands marked # in galactose samples are GAL10 mRNA, detected since small amounts of antisense RNA probe are produced by template strand switching (“turnaround transcription”) at the end of the DNA substrate during in vitro transcription.
(B) Total and oligo(dT) affinity selected RNA from wild-type and Reb1 BSΔ mutant cells hybridized with GAL10 sense and ACT1 probes.
(C) Cap-dependent 5′ RACE analysis of total RNA performed using a GeneRacer kit with primer GAL10F2. Products were cloned and sequenced to determine transcriptional start site.
(D) Schematic of GAL10 ncRNA 5′ ends as determined by 5′ RACE.
(E) Quantification of transcript half-life after addition of 2% galactose to cells growing exponentially in raffinose medium. RNA was probed for GAL10 antisense, GAL10, GAL1, and ACT1. Quantification represents data from three cultures; y axis is in arbitrary units; error bars indicate ±1 standard error.
(F) GAL10 antisense signal from 20 mg FT4 wild-type RNA compared to signal from in vitro transcribed truncated GAL10 antisense RNA in 20 mg of wild-type RNA. Panels showing GAL10 antisense and the in vitro transcribed RNA derive from the same exposure of the same blot, and no differential processing has been applied.
(G) Northern analysis of GAL10 antisense in TRAMP mutants. Cells were grown to mid-log phase at 25° in rich media containing 2% glucose, except the trf4Δ GAL-trf5 strain,which was grown on 2% galactose prior to a 24 hr shift to 2% glucose. RNA analysis as in (E). *p < 0.05 for Student’s t test of wild-type versus mutant.
The major 4 kb ncRNA transcript produced in glucose medium (herein referred to as the GAL10-ncRNA) therefore has its promoter at the 3′ end of GAL10, runs antisense through the GAL10 ORF, across the bidirectional GAL1–10 promoter and extends through GAL1 in the sense orientation to its 3′ end (see schematic in Figure 4A). The presence of a 5′ cap and a poly(A) tail strongly suggest that the GAL10-ncRNA is produced by RNA pol II.
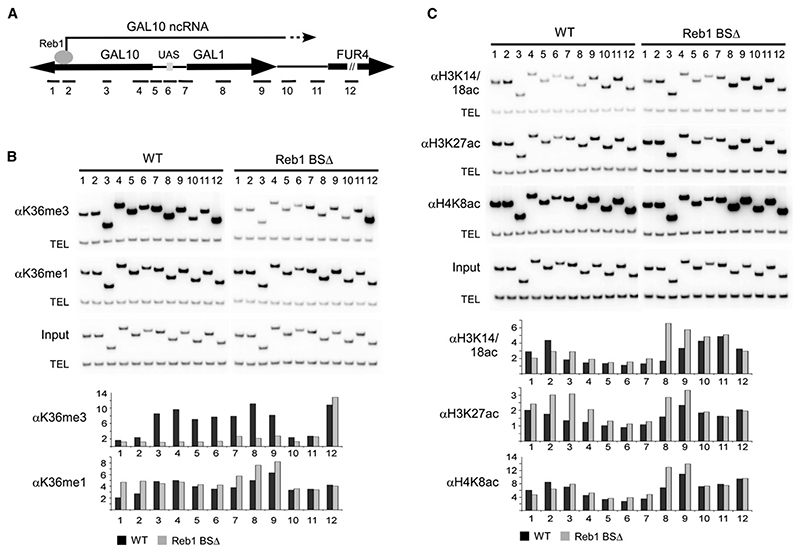
Figure 4. Histone Modifications Induced by the GAL10 ncRNA.
(A) Schematic of the GAL locus showing the location of primer sets used for ChIP analysis.
(B) ChIP analysis of H3 K36 methylation in the presence and absence of the GAL10 ncRNA (Reb1 BS+/−). Cells were grown to log phase in rich media containing 2% raffinose, and chromatin was precipitated with antibodies against H3 K36me3 and H3 K36me1. PCR was performed with primer sets shown on the schematic diagram for the GAL locus and a telomeric primer set (TEL) as a loading control.
(C) ChIP analysis of histone acetylation in the presence and absence of the GAL10 ncRNA (Reb1 BS+/−). Cells were grown to log phase in rich media containing 2% raffinose, and chromatin was precipitated with antibodies against acetylated H3 K14/18, H3 K27, and H4 K8. PCR was performed with primer sets shown on schematic diagram for the GAL locus and a telomeric primer set (TEL) as a loading control.
Graphs in (B) and (C) show quantification of ChIP data as fold enrichment of signal at the GAL locus over telomeric signal normalized to the input. Data shown is the average of two independent experiments.
The half-life of the GAL10-ncRNA transcript is ~8 min after addition of 2% galactose to cells growing in 2% raffinose (Figure 3E), suggesting that it is relatively stable. The steadystate level was quantified at ~0.07 molecules per cell, or one transcript in 14 cells, by comparison of hybridization intensity to in vitro transcribed RNA (Figure 3F). The ncRNA was detected at similar levels in cells grown in glucose and raffinose, and in three different wild-type strain backgrounds (data not shown).
Many ncRNAs in S. cerevisiae are targeted by the TRAMP4/5 surveillance complexes for rapid degradation immediately after transcription (Wyers et al., 2005). Loss of both poly(A) polymerases of the TRAMP complexes, Trf4 and Trf5, is lethal, so the level of the GAL ncRNA was assessed in a conditional trf4Δ PGAL-TRF5 strain 24 hr after transfer to glucose medium. This strain showed a 3.5-fold increase in the abundance of the GAL10-ncRNA relative to wildtype (Figure 3G), with a larger increase in the level of the 5.6 kb ncRNA transcript. The abundance of the 5.6 kb transcript was also strongly increased by simultaneous loss of both the Air1 and Air2 components of the TRAMP complexes, which show functionally redundancy (Figure 3G). We conclude that the GAL10-ncRNA is targeted for degradation by the TRAMP complexes. Most transcripts are rapidly degraded in the nucleus, but the fraction that escapes nuclear surveillance is relatively stable. Based on the ncRNA half-life and steady-state levels, and compensating for the degraded fraction, we calculate that one transcript is produced per cell every 50 min. Consistent with such a low transcription rate, RNA pol II recruitment to GAL10 was not detected by ChIP (data not shown).
Expression of the GAL10-ncRNA Modifies Chromatin
Transcription by RNA pol II leads to H3 K36me over coding regions (reviewed in Lee and Shilatifard, 2007). We investigated whether ncRNA transcription is associated with H3 K36me3 over the repressed GAL1–GAL10 genes. ChIP was performed using antibodies specific to H3 K36me1 or K36me3, and the precipitated DNA was analyzed by PCR with primers scanning the GAL1–GAL10 region (Figure 4A). In glucose medium, high levels of H3 K36me3 were observed over GAL10, GAL1, and the GAL1–10 promoter region in the strain with the intact Reb1-binding sites (Figure 4B, wild-type). In contrast, only background levels were seen in the Reb1 BSΔ strain (Figure 4B). H3 K36me1 was also elevated over the entire region relative to the internal loading control, but this was independent of the Reb1-binding sites in GAL10. We cannot exclude the possibility that the 5.6 kb ncRNA transcript is involved in maintaining this modification. These results show that cryptic ncRNA transcription can direct H3 K36me3 formation over repressed genes and intergenic regions, which otherwise lack this histone modification.
H3 K36me3 is generally associated with subsequent histone deacetylation, so we assessed whether this is the case for GAL1–GAL10. Cells were grown in raffinose in order to avoid overlapping histone deacetylation induced by Mig1/Tup1/Hda1-mediated glucose repression. ChIP was performed with antibodies against the acetylated forms of H3 K14/18, H3 K27, and H4 K8, and the chromosomal region encompassing GAL1 and GAL10 was analyzed by PCR in the wild-type and Reb1 BSΔ strains (Figure 4C). The wild-type strain showed reduced H3 K27 acetylation (H3 K27ac) over both the GAL1 and GAL10 coding regions relative to the Reb1 BSΔ strain, whereas H3 K14/18ac was clearly decreased only over the GAL1 coding region, and histone H4 K8ac was marginally altered. We conclude that Reb1-driven ncRNA transcription causes reduced acetylation of specific histone residues over the GAL1 and GAL10 coding regions.
GAL10-ncRNA Expression Enhances Glucose Repression
Previously described ncRNAs function in either the activation or repression of transcription (Hongay et al., 2006; Uhler et al., 2007); however, loss of the GAL10-ncRNA did not change steady-state levels of GAL1 or GAL10 mRNA or their induction rate in standard 2% (20 g l–1) galactose medium (Figures S4A and S4B). Such high galactose concentrations will rarely be encountered by S. cerevisiae in the wild, and we therefore suspected that the ncRNA might modulate responses at lower galactose levels. Galactose induction increases between 0.1 g l–1 and 1.0 g l–1 (Li et al., 2000), and across this range the level of the GAL10-ncRNA decreased, suggesting competition with expression of GAL1–10 (Figure S5A). We selected the lowest concentration that gave clear GAL1 induction (0.1g l–1) for further analysis, which is physiologically relevant for S. cerevisiae, as it falls within the range of galactose concentrations found in grapes (0.05-0.16 g kg–1).
A number of feedback mechanisms operate during galactose induction that may be altered by mutation of the Gal10 protein sequence, despite the Reb1 BSΔ mutant having no discernable growth phenotype on high galactose. To avoid secondary effects from mutation of Gal10, we constructed a second Reb1-binding site mutant (Reb1 BSΔ-silent), which did not change the GAL10 coding sequence (Figure S6A), and confirmed that this mutant lacked GAL10-ncRNA expression by northern blot (Figure 5A) and that H3 K36me3 over GAL1–10 was reduced (data not shown). As before, this mutant strain showed no difference in GAL induction at high galactose concentrations (Figure S6B). This silent Reb1 mutant strain was used for all further experiments.
Figure 5. The Effect of GAL10 ncRNA Expression on Glucose Repression.
(A) GAL induction in response to low-galactose/glucose mixtures. Cells were grown to OD ~0.4 in 2% raffinose, and then glucose and galactose were added to the indicated concentration. Cells were harvested after 3 hr, and RNA was probed for GAL1, GAL10, and ACT1, and for the GAL10 ncRNA. The 18S rRNA signal is from ethidium staining. Quantification of GAL10 mRNA levels was determined relative to ACT1 signal. The increasing level of 18S and ACT1 with increasing glucose concentrations is due to the elevated growth rate.
(B) Quantification of GAL induction. Cells were grown to OD ~0.4 in 2% raffinose, and then glucose and galactose were added to the indicated concentration. Cells were harvested after 2 hr, and RNA was probed for GAL1 and GAL10. Data were normalized to 18S ribosomal RNA levels determined by ethidium staining. Quantification represents data from six cultures; y axis is in arbitrary units; error bars indicate ±1 standard error.
(C) Cells were grown overnight to OD ~0.2 in rich 2% raffinose media; no dilution or manipulation of the cultures was performed prior to the experiment. Cultures were induced with 0.1 g l–1 galactose/0.2 g l–1 glucose, and samples were taken at the indicated time points. RNA analysis as in (B).
(D) Quantification of time course data from (C). Data are from three cultures; y axis is in arbitrary units; error bars indicate ±1 standard error. * p < 0.05 for Student’s t test of wild-type versus mutant.
Induction experiments with wild-type and Reb1 BSΔ-silent using 0.1 g l–1 galactose as the sole carbon source revealed only small differences with poor reproducibility (Figure 5A, lanes 1 and 2). We therefore added increasing concentrations of glucose to reveal potential effects of the ncRNA on glucose repression. A clear difference in the level of induction after 3 hr was observed with the addition of 0.1–0.4 g l–1 glucose, while addition of 0.8 g l–1 glucose repressed induction below detectable levels (Figure 5A). Quantification of induction with 0.1 g l–1 galactose for 2 hr in the presence of 0.2 g l–1 glucose confirmed a very reproducible difference in mRNA levels (Figure 5B). In three experiments, each with two independent Reb1 BSΔ-silent clones compared to wild-type, mRNA levels were lower in the wild-type than in the mutant (p < 0.0005 for GAL10 mRNA and p < 0.01 for GAL1 mRNA). Across an induction time course (Figures 5C and 5D), this difference was reproducible up to 4–5 hr but became more variable at 5–7 hr, which coincides with the slowed growth at OD > 0.8.
The addition of small quantities of glucose both reduced induction rate and introduced a delay in induction (compare wild-type induction with no glucose [Figure S5B] with induction in the presence of glucose [Figure 5D]). This explains the difference in wild-type GAL induction between 0 g l–1 and 0.2 g l–1 glucose at 3 hr (Figure 5A, lanes 1 and 5) and 2 hr (Figure 5B, lanes 1 and 3).
The effect of the GAL10-ncRNA on histone acetylation and H3 K36me3 over the GAL1–10 region under these growth conditions is similar to that observed in 2% raffinose, indicating that the same histone modifiers are recruited (Figure S7). Furthermore, H3 K4me3 was increased in the Reb1 BSΔ mutant over GAL1 in 0.1 g l–1 galactose + 0.2 g l–1 glucose, presumably reflecting the increased GAL1 transcription relative to the WT under these growth conditions.
From these data we conclude that the GAL10-ncRNA acts to enhance glucose repression of GAL1–10 induction at low environmental sugar concentrations.
GAL10-ncRNA Enhances Glucose Repression In Cis Primarily through Eaf3
Characterized ncRNAs in budding yeast can act both in cis (Hongay et al., 2006) and in trans (Berretta et al., 2008). If the ncRNA acts in trans on GAL1–10 induction, ncRNA from the wild-type allele in a heterozygous Reb1 BSΔ-silent diploid strain should suppress increased GAL induction from the mutant allele, but this will not occur if the ncRNA acts in cis. GAL1–10 induction analysis at the 3 hr time point in these strains revealed that the GAL10-ncRNA does not repress induction from a mutant allele in trans. A highly significant difference in induction was observed between a homozygous wild-type diploid and a heterozygous Reb1 BSΔ-silent diploid (Figure 6A).
Figure 6. Mechanistic Analysis of GAL10 ncRNA’s Role in Glucose Repression.
(A) GAL induction in haploid and diploid strains. Analysis of three haploid parental strains (FT4, Reb1 BSΔ-silent, and BY4742) and two diploid strains (FT4 x BY4742 and Reb1 BSΔ-silent x BY4742), grown overnight to OD -0.2 in rich 2% raffinose media and induced with 0.1 g l–1 galactose/0.2 g l–1 glucose. Cells were harvested after 3 hr, and RNA was analyzed and quantified as in Figure 5B. Quantification represents data from four cultures of each strain.
(B) Allele-specific ChIP analysis of H3 K36 trimethylation and H3 K27 acetylation at the 5′ end of GAL10 ncRNA in cells grown in 2% raffinose. One replicate of FT4 and Reb1 BSΔ-silent and three replicates each for BY4742 and the two diploid strains were performed. PCR product normalized to loading control (Tel) and input DNA are shown for each allele in each strain; error bars indicate ±1 standard deviation.
(C) Analysis of GAL gene induction in histone deactylase mutants. Cell growth, induction, and analysis as in (A). GAL10 ncRNA is visualized with probe to GAL1/GAL10. Quantification represents data from three cultures of each strain; error bars indicate ±1 standard deviation.
(D) Analysis of GAL gene induction in HDAC mutants combined with Reb1 BSΔ. Cell growth, induction, and analysis as in (C). Quantification represents data from seven cultures in two experiments (WT, sir2Δ, and eaf3Δ strains) and from three cultures in one experiment (hda1Δ strains); error bars indicate ±1 standard deviation.
Histone modification analyses were performed in the same heterozygous diploids containing the wild-type and the Reb1 BSΔ-silent allele, along with the parental haploids, using allelespecific PCR primers to the 5′ end of the GAL10-ncRNA (Figure S8A). Consistent with GAL10-ncRNA being cis-acting, H3 K36me3 occurred only over the WT allele (Figure 6B). The two-fold difference in H3 K27 acetylation between wild-type and mutant alleles was maintained in the heterozygous diploid, showing that histone deacetylation also occurs in cis (Figures 6B and S8B).
To identify HDACs acting at the GAL1–10 cluster in WT cells, deletion mutants of nine of the ten known budding yeast HDACs were tested for GAL1–10 induction with 0.1 g l–1 galactose + 0.2 g l–1 glucose (Figure 6C). hos1Δ, hst1Δ, hst2Δ, hst3Δ, and hst4Δ caused detectable but not significant increases in GAL1–10 induction, as confirmed by the Student’s t test. Only hda1Δ and sir2Δ significantly affected GAL1–10 expression under these conditions and were chosen for further investigation (Figure 6C). The remaining HDAC, Rpd3, could not be tested in this assay as it conferred a mild slow growth phenotype that interfered with the interpretation of induction data. Nevertheless, ChIP analyses in raffinose showed that deletion of RPD3 led to increased H3 K27 acetylation. Neither H3 K9 nor H3 K14/K18 acetylation were affected, likely due to redundancy with other HDACs (Figure S9). These data suggested that the GAL10-ncRNA acts through recruitment of Hda1, Sir2, or Rpd3.
If one HDAC were directly recruited by the GAL10-ncRNA, its absence should cause increased GAL1–10 induction—but this should not be additive with the Reb1 BSΔ mutation. To test this possibility, HDA1 and SIR2 were deleted in WT and Reb1 BSΔ-silent strain backgrounds, and GAL1–10 induction was tested as described above. Since rpd3Δ could not be analyzed by GAL induction, we instead deleted EAF3, a subunit of the Rpd3S complex, which did not cause any detectable growth phenotype.
The hda1Δ deletion increased GAL1–10 induction in WT cells but had a much greater effect in the Reb1 BSΔ-silent strain (Figure 6D), showing that Hda1 acts synergistically with the GAL10-ncRNA. ChIP analysis of wild-type and hda1Δ cells growing in 2% raffinose also revealed a synergistic effect between Hda1 and the GAL10-ncRNA on H3 K14/18 acetylation (Figure S9). Since the effects of the GAL10-ncRNA are seen in the absence of Hda1, this cannot be a major downstream target of the ncRNA. Hda1 was previously shown to aid glucose repression through its interaction with the Tup1/Ssn6 corepressor (Wu et al., 2001). Recruitment of this complex to GAL1–10 by the glucose repressors Mig1 and Nrg1 may explain the observed increase in H3 K14/18ac and GAL1–10 derepression in hda1Δ cells.
The sir2Δ deletion increased GAL1–10 induction in the WT and the Reb1 BSΔ-silent strains to similar levels. However, loss of Sir2 also reproducibly reduced expression of the GAL10-ncRNA (Figures 6C and 6D). This suggests that depression of GAL1–10 in sir2Δ partly reflects the loss of the ncRNA. Derepression of GAL1–10 in the sir2Δ strain was, however, stronger than that in the Reb1-BSΔ-silent mutants, indicating that the effects of sir2Δ are not solely due to loss of GAL10-ncRNA synthesis. The basis of the requirement for Sir2 in production of the GAL10-ncRNA is currently unclear but does not simply reflect interference from the increased GAL1–10 transcription (Figures 6C and 6D compare GAL10-ncRNA levels in sir2Δ and hda1Δ, which show similar GAL10 mRNA). Sir2 activity is NAD+ dependent, and these data may be related to recently reported effects of the cellular NAD/NADP ratio on GAL1–10 expression (Kumar et al., 2008).
The eaf3Δ mutation greatly reduced the difference between the wild-type and the Reb1 BSΔ strain (on average across seven experiments) (Figure 6D), indicating that Eaf3 is required for the effects of the GAL10-ncRNA on GAL1–10 expression. Over other transcribed regions, Eaf3 binds to H3 K36me3 and recruits the Rpd3S HDAC complex, and subsequent deacetylation then acts to inhibit spurious transcription initiation. We conclude that the GAL10-ncRNA acts in cis through a similar mechanism, with transcription-coupled deposition of H3 K36me3 leading to histone deacetylation by Rpd3S and increased glucose repression of GAL1–10 under low-glucose conditions.
Discussion
Reb1 Drives ncRNA Transcription
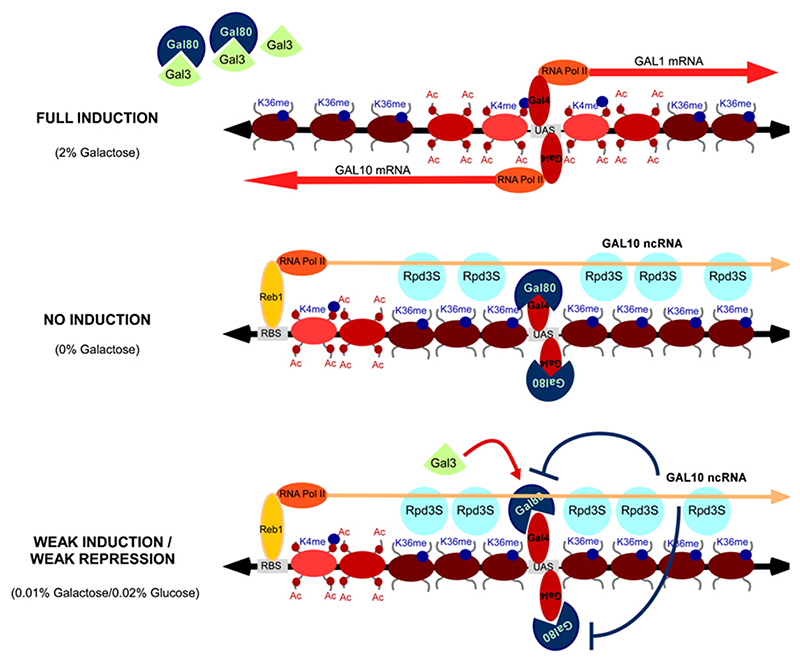
Recent studies have suggested that noncoding transcripts are so common in budding yeast that almost the entire genome is transcribed by RNA pol II (Steinmetz et al., 2006), making it an excellent system for their functional analysis. We here show that binding of the transcription factor Reb1 within the GAL10 coding region drives the expression of an antisense transcript covering almost the entire GAL1–10 locus under noninduced or repressing conditions (Figure 7).
Figure 7. Model for GAL10 ncRNA-Induced Histone Modifications over the GAL Cluster.
Full induction: GAL1–10 are transcribed. Reb1 binding to the GAL10 3 is absent, and no ncRNA is produced. No induction: GAL1–10 are not transcribed. Reb1 binds within the GAL10 coding region and induces GAL10 ncRNA transcription. This results in high levels of K36me3 and increased histone deacetylation over the GAL1–10 cluster. Weak induction/weak repression: GAL10 ncRNA-induced chromatin modifications repress GAL1–10 transcription in condition of concomitant weak induction and repression.
Reb1 is an essential protein initially identified as a Pol I transcription termination and initiation factor (Morrow et al., 1990) and subsequently shown to stimulate RNA pol II transcription at many housekeeping genes (Graham and Chambers, 1994; Packham et al., 1996; Remacle and Holmberg, 1992; Schuller et al., 1994; Yagi et al., 1994). The role of Reb1 in driving ncRNA transcription is unlikely to be restricted to the GAL locus. The consensus Reb1-binding site (CGGGTAA) can be identified close to the initiation site of the PHO84 antisense transcript, and further inter-genic Reb1 binding targets were detected in a whole-genome study using ChIP-on-chip (Harbison et al., 2004). BLAST searches reveal 215 perfect matches to the Reb1-binding sequence within coding regions in S. cerevisiae (see Figure S5). In addition, Reb1 acts in the rDNA and as an insulator at subtelomeric Y′ elements (Fourel et al., 2001; Fourel et al., 1999; Kulkens et al., 1992). Recent studies have identified ncRNAs in both the rDNA and telomeric regions with transcriptional start sites in close proximity to Reb1-binding sites (Houseley et al., 2007; Li et al., 2006; Vasiljeva et al., 2008). We speculate that Reb1 may also act in these regions through the induction of ncRNAs and concomitant long-range changes in chromatin structure.
Transcription of the ncRNA Alters the Chromatin Structure In Cis
ncRNAs can act in cis, in which case their function is limited to the genomic region of their production, or in trans, targeting chromosomal loci with sequence similarity to the ncRNA itself. A role for the GAL10-ncRNA as a diffusible, trans-acting factor seemed unlikely, due to its low steady-state level of ~0.07 copies per cell. This was confirmed by analyses in a heterozygous diploid, which showed that only the chromosome expressing the ncRNA was subject to its effects.
Transcription of the GAL10-ncRNA was estimated to occur only once every 50 min on average. Despite its apparently infrequent expression, GAL10-ncRNA transcription profoundly altered the chromatin structure over the GAL1 and GAL10 genes (Figure 7). By specifically mutating the Reb1-binding sites, we were able to investigate histone modifications directly caused by the transcript. As with other RNA pol II transcribed regions, nucleosomes over the 5′ end of the GAL10-ncRNA were di- and trimethylated at H3 K4 by the methyltransferase Set1. Stable binding of Set1 was not detected by ChIP, probably as a result of the low frequency of ncRNA transcription.
Most previously characterized repressive ncRNAs in yeast such as SRG1, which regulates SER3, act via a transcriptional interference mechanism, whereby high-level transcription through the promoter region of a coding gene interferes with transcription factor binding (Martens et al., 2004). The low transcription frequency of the GAL10 ncRNA indicates that this mechanism is unlikely to make a substantial contribution to the observed GAL regulation, and a mechanism involving persistent histone modification was therefore sought.
Over transcribed protein-coding regions, RNA pol II elongation induces H3 K36me3, which is important for repression of spurious transcription initiation within transcribed regions (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). We detected H3 K36me3 over the entire region covered by the ncRNA transcript in the wild-type, but not in the Reb1-BSΔ mutant strain. H3 K36me3 has been shown to recruit the Rpd3S HDAC complex, leading to histone deacetylation (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). Indeed, histone acetylation was increased at all analyzed lysine residues in the Reb1-BSΔ mutant strain. Analysis of acetylation levels further confirmed a role of Rpd3 in deacetylating lysine residues over the GAL1–10 region (Figure S9). Therefore, the rarely transcribed GAL10-ncRNA induces histone modification patterns otherwise associated only with strongly transcribed coding regions.
The differences in histone acetylation induced by the ncRNA were more marked over the GAL1 and GAL10 ORFs than over the actual GAL1–10 promoter. This suggests that the alterations in GAL induction either result from changes in promoter clearance or transcription elongation, or are due to effects at a distance, such as perturbation of the formation of gene loops or other active chromatin conformations.
Over protein-coding genes, the Rpd3S HDAC complex is recruited by the H3 K36me3-binding protein Eaf3 (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). Deletion of EAF3 caused derepression GAL1–10 in low-glucose conditions, which was not further increased by the Reb1 BSΔ-silent mutation. This indicates that the effects of the GAL10-ncRNA on GAL1–10 expression require the activity of Eaf3, consistent with recruitment of Rpd3S. The yeast genome encodes nine other putative HDACs, and all of these were also tested for effects on GAL1–10 induction. Of these, loss of Hda1 showed synergistic enhancement of GAL1–10 expression under the low-glucose conditions, indicating that it functions in parallel with the ncRNA/Rpd3S pathway. Loss of Sir2 resulted in the reduction of the GAL10-ncRNA, via a currently unknown mechanism, and this was accompanied by derepression of GAL1–10. No other HDAC mutation suppressed the effects of the loss of the GAL10-ncRNA, consistent with the model that this acts predominately through Eaf3/Rpd3S.
These data support the model shown in Figure 7. Reb1-driven transcription of the GAL10-ncRNA leads to K36 trimethylation of histone H3. This recruits the Rpd3S complex via the H3 K36me3-binding protein Eaf3, leading to increased deacetylation across the GAL1–10 locus and increased repression of the divergent GAL1–10 promoter. Moreover, this mechanism is probably not restricted to GAL1–10. Preliminary analysis of the remaining core gene of the GAL operon, GAL2, revealed a peak of H3 K4me3 at the 3′ end of the ORF and a corresponding ncRNA that runs antisense to the entire GAL2 gene (data not shown).
The phenotypic consequences of GAL10-ncRNA expression were not manifested during growth in standard laboratory media. However, clear differences in GAL1–10 expression were detected in glucose-galactose mixtures, which may represent more physiological conditions. In fact, most natural substrates will contain complex mixtures of nutrients, many present at low levels. This suggested that antisense ncRNAs might participate in the regulation of many metabolic genes and would be detectable as peaks of H3 K4me3 at the 3′ end of the open reading frames. Examination of published ChIP-on-chip data (Kirmizis et al., 2007) revealed peaks of H3 K4me3 at the 3′ end of ORFs of many glucose-repressed metabolic genes, including HXT[8, 13, 15, 16, and 17], MPH2, MPH3, JEN1, ICL2, and ICL1. This strongly suggests that ncRNAs participate in the regulation of multiple catabolic genes, acting collectively to optimize gene expression in response to the complex and rapidly changing mix of nutrients available to yeast populations growing on natural substrates.
Experimental Procedures
Strains and Media
Strains were constructed by standard methods and are listed in Table S1; details are given in the Supplemental Data. For ChIP analysis, colonies from YPD (2% glucose) plates were pregrown overnight in YPD (2% glucose), YPR (2% raffinose), or YPG (2% galactose), diluted into the same media to OD600 0.25 and grown to OD600 1. For RNA analysis, cells were grown at 30° to OD600 0.2–0.6 in rich media (1% yeast extract, 2% peptone, 2% sugar), except for in Figure S4B, where synthetic media was used (0.5% NH4SO4, 0.17% yeast nitrogen base, 1 × amino acids, 2% sugar). For highly reproducible longer (3 hr) GAL inductions, cells were grown overnight to OD 0.2–0.5 in YPR and diluted to precisely OD 0.2 prior to induction at 15 s intervals with glucose/galactose mixtures.
ChIP
ChIP and PCR conditions were essentially as described in Suka et al. (2001). Primer sequences are indicated in Table S3. Additional washings (3×10 min) with lysis buffer previous to sonication were used with αHA antibody and initial 140 mM NaCl concentration was adjusted to 275 mM for incubation with αHA antibody. Antibodies used are described in Table S4.
RNA Methods
Cells were fixed by addition of ethanol to 70% prior to RNA extraction, and northern blotting and probing were performed as described (Houseley et al., 2007). Details of hybridization conditions are given in the Supplemental Data.
Supplementary Material
The Supplemental Data include Supplemental Experimental Procedures, five tables, and nine figures and can be found with this article online at http://www.cell.com/molecular-cell/supplemental/S1097-2765(08)00810-1.
Acknowledgments
We thank Kevin Struhl for providing the FT4 WT and HA-Set1 strains. J.H. was funded by the Leverhulme Trust, D.T. was funded by The Wellcome Trust, L.R. and M.G. were funded by NIH GM23674, and M.V. was funded by NIH GM23674 and The Wellcome Trust MV37766. We thank the members of the Grunstein, Tollervey, and Vogelauer labs for helpful discussions.
References
- Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, Simmen KC, Muller K, Li R, Mermod N, Gilson E. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2001;2:124–132. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IR, Chambers A. A Reb1p-binding site is required for efficient activation of the yeast RAP1 gene, but multiple binding sites for Rap1p are not essential. Mol Microbiol. 1994;12:931–940. doi: 10.1111/j.1365-2958.1994.tb01081.x. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen L. Occurrence and taxonomic characteristics of strains of Saccharomyces cerevisiae predominant in African indigenous fermented foods and beverages. FEM Yeast Res. 2003;3:191–200. doi: 10.1016/S1567-1356(02)00185-X. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Ju QD, Morrow BE, Warner JR. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990;10:5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T, van der Sande CA, Dekker AF, van Heerikhuizen H, Planta RJ. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992;11:4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PR, Yu Y, Sternglanz R, Johnston SA, Joshua-Tor L. NADP regulates the yeast GAL induction system. Science (New York, NY. 2008;319:1090–1092. doi: 10.1126/science.1151903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Li J, Wang S, VanDusen WJ, Schultz LD, George HA, Herber WK, Chae HJ, Bentley WE, Rao G. Green fluorescent protein in Saccharomyces cerevisiae: real-time studies of the GAL1 promoter. Biotechnol Bioeng. 2000;70:187–196. doi: 10.1002/1097-0290(20001020)70:2<187::aid-bit8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Li C, Mueller JE, Bryk M. Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol Biol Cell. 2006;17:3848–3859. doi: 10.1091/mbc.E06-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Mo X, Kowenz-Leutz E, Laumonnier Y, Xu H, Leutz A. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 2005;19:2447–2457. doi: 10.1101/gad.355405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow BE, Ju Q, Warner JR. Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem. 1990;265:20778–20783. [PubMed] [Google Scholar]
- Munroe SH, Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell Mol Life Sci. 2006;63:2102–2118. doi: 10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Packham EA, Graham IR, Chambers A. The multifunctional transcription factors Abf1p, Rap1p and Reb1p are required for full transcriptional activation of the chromosomal PGK gene in Saccharomyces cerevisiae. Mol Gen Genet. 1996;250:348–356. doi: 10.1007/BF02174393. [DOI] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 2007;23:284–292. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genomewide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Proffitt JH. DNase I-hypersensitive sites in the galactose gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:1522–1524. doi: 10.1128/mcb.5.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle JE, Holmberg S. A REB1-binding site is required for GCN4-independent ILV1 basal level transcription and can be functionally replaced by an ABF1-binding site. Mol Cell Biol. 1992;12:5516–5526. doi: 10.1128/mcb.12.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schuller HJ, Schutz A, Knab S, Hoffmann B, Schweizer E. Importance of general regulatory factors Rap1p, Abf1p and Reb1p for the activation of yeast fatty acid synthase genes FAS1 and FAS2. Eur J Biochem. 1994;225:213–222. doi: 10.1111/j.1432-1033.1994.00213.x. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specifc antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M. TUP1 utilizes histone H3/H2B-specifc HDA1 deacetylase to repress gene activity in yeast. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Yagi S, Yagi K, Fukuoka J, Suzuki M. The UAS of the yeast GAPDH promoter consists of multiple general functional elements including RAP1 and GRF2 binding sites. J Vet Med Sci. 1994;56:235–244. doi: 10.1292/jvms.56.235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.