Abstract
Lipoyl-lysine swinging arms are crucial to the reactions catalysed by the 2-oxo acid dehydrogenase multienzyme complexes. A gene encoding a putative lipoate protein ligase (LplA) of Thermoplasma acidophilum was cloned and expressed in Escherichia coli. The recombinant protein, a monomer of molecular mass 29 kDa, was catalytically inactive. Crystal structures in the absence and presence of bound lipoic acid were solved at 2.1 Å resolution. The protein was found to fall into the α/β class and to be structurally homologous to the catalytic domains of class II aminoacyl-tRNA synthases and biotin protein ligase, BirA. Lipoic acid in LplA was bound in the same position as biotin in BirA. The structure of the T. acidophilum LplA and limited proteolysis of E. coli LplA together highlighted some key features of the post-translational modification. A loop comprising residues 71–79 in the T. acidophilum ligase is proposed as interacting with the dithiolane ring of lipoic acid and discriminating against the entry of biotin. A second loop comprising residues 179–193 was disordered in the T. acidophilum structure; tryptic cleavage of the corresponding loop in the E. coli LplA under non-denaturing conditions rendered the enzyme catalytically inactive, emphasizing its importance. The putative LplA of T. acidophilum lacks a C-terminal domain found in its counterparts in E. coli (Gram-negative) or Streptococcus pneumoniae (Grampositive). A gene encoding a protein that appears to have structural homology to the additional domain in the E. coli and S. pneumoniae enzymes was detected alongside the structural gene encoding the putative LplA in the T. acidophilum genome. It is likely that this protein is required to confer activity on the LplA as currently purified, one protein perhaps catalysing the formation of the obligatory lipoyl-AMP intermediate, and the other transferring the lipoyl group from it to the specific lysine residue in the target protein.
Keywords: pyruvate dehydrogenase complex, lipoic acid, limited proteolysis, surface loops, domain
Introduction
Lipoic acid is a crucial prosthetic group in the ubiquitous family of multienzyme complexes that catalyse oxidative decarboxylation of 2-oxo acids and in the glycine cleavage system of plants and bacteria.1–3 In all these instances, a lipoyl group attached in amide linkage to the N6-amino group of a lysine residue in one of the component enzymes acts as a “swinging arm”, helping to shuttle covalently bound reaction intermediates between the active sites of the component enzymes.1,4,5 In the 2-oxo acid dehydrogenase complexes, the lipoic acid is attached to a small 9 kDa protein domain, the so-called lipoyl domain that forms part of the dihydrolipoyl acyltransferase (E2; EC 2.3.1.12) component. In the glycine cleavage system, it is similarly attached to the H-protein, a protein that is structurally homologous to the lipoyl domain.1,6 Indeed, it turns out that the lipoyl group cannot function if detached from the protein, and the unravelling of the underlying protein–protein interactions explains the rate-enhancements and substrate channelling that are prominent features of what are effectively nanomachines catalysing multistep reactions.1,5,7
Two pathways, LplA and LipB, which carry out the lipoylation, a post-translational modification of the target proteins, have been described. The LplA protein activates exogenously derived lipoic acid by reaction with ATP to form lipoyl-AMP before reacting this with a conserved lysine residue in the target protein to form the protein-bound lipoamide group.
The second pathway uses lipoic acid synthesized from octanoate, in turn derived from the fatty acid biosynthetic pathway,8 and this pathway requires the LipB gene product. LipB is capable of transferring either the octanoyl group from octanoyl-acyl carrier protein (octanoyl-ACP) or the lipoyl group from lipoyl-ACP onto an apo lipoyl domain.9 The lipoyl group is synthesized from an octanoyl precursor by the LipA protein, a metalloenzyme that makes use of S-adenosyl-L-methionine (SAM) and iron–sulphur clusters in its reaction mechanism. Although LipA can act on octanoyl-ACP, its preferred substrate appears to be an octanoylated lipoyl-protein.10 The proposed pathway thus is, first, the transfer of an octanoyl group from octanoyl-ACP onto a lipoyl domain, a reaction catalysed by LipB, and secondly, the subsequent conversion of the attached octanoyl group to the lipoyl group, catalysed by LipA.
A similar prosthetic group, biotin, is attached to target proteins by the formation of an amide linkage between the carboxy group of biotin and the N6-amino group of a specific lysine residue, creating the biotinyl-lysine swinging arm that is required for moving one-carbon moieties about in catalysis by the numerous biotin-dependent enzymes.1 This reaction is catalysed by a biotin protein ligase, encoded by the BirA gene. BirA has a two-step reaction mechanism like that of LplA: the generation of biotinyl-AMP from biotin and ATP, followed by the transfer of the biotinyl group to the target protein.11 Moreover, the lysine residue in the target protein is housed and displayed in a biotinyl domain, the structure of which is very similar to that of a lipoyl domain.12,13 Such striking similarities have prompted studies into the selectivity of the post-translational modifications.14–16
These investigations indicate that specific amino acid sequences surrounding the target lysine residue in particular, together with crucial differences in a prominent surface loop, ensure that the lipoyl and biotinyl domains do not suffer aberrant lipoylation/biotinylation at the hands of LplA and BirA in Escherichia coli.14–16 Thus, lipoylation and biotinylation stand in marked contrast to other post-translational modifications such as phosphorylation, where the amino acid sequence surrounding the target site is the principal determinant of kinase specificity and action.
The published structures of E. coli BirA17,18 have helped identify the points of interaction in the protein with substrate and reaction intermediates,19,20 and the defining features of the interaction of BirA with the biotinyl domain have been identified.21–23 The first lipoate protein ligase gene to be cloned and expressed, and the most widely studied, was LplA of E. coli 24 but structural and mechanistic work on lipoylation has lagged behind. Here, we describe the cloning and expression of the gene encoding what was thought to be the lipoate protein ligase of Thermoplasma acidophilum and the determination of its structure at 2.1 Å resolution. This structure, together with the results of limited proteolysis of the E. coli LplA, throws new light on the mechanism, notably the importance of surface loops in and around the active site. It has also revealed that the lipoate protein ligases of Archaea may require a second complementary protein. The T. acidophilum gene encoding this second protein has been tentatively identified.
Results and Discussion
Cloning and expression of the gene encoding T. acidophilum LplA
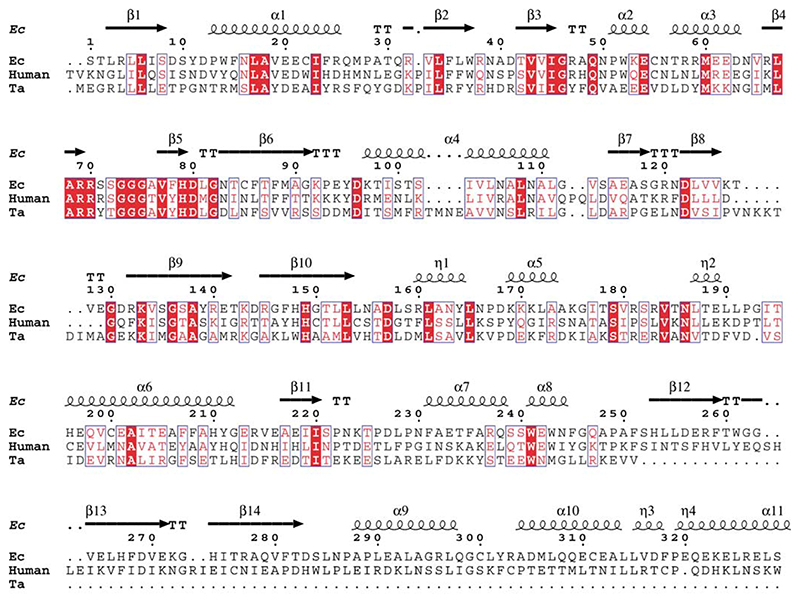
The genomes of two thermophilic organisms, T. acidophilum, and Thermus thermophilus, were searched for putative lipoate-protein ligases in the expectation that a protein from a thermophile might prove readily amenable to crystallization. Using the amino acid sequence of E. coli LplA as a query, a BLASTsearch of the T. acidophilum genome revealed one potential homologue (protein Ta0514). When its inferred primary structure was compared with those published for known lipoate and biotin protein ligases, it was found to contain all the distinguishing sequence signatures of this family25 and was shown to share significant secondary structure homology to E. coli LplA and the human lipoyl transferase (Figure 1). The gene encoding protein Ta0514 was therefore chosen for cloning.
Figure 1. Amino acid sequence alignment of the E. coli (E.c), T. acidophilum (T.a) and human lipoate protein ligases.
The secondary structure elements of the E. coli LplA enzyme are highlighted.
An appropriate DNA fragment was excised from the T. acidophilum genome, cloned into plasmid pET3a and expressed in E. coli BL21 DE3 cells (see Materials and Methods for details). The sequence of the insert was confirmed by automated DNA sequence analysis. The recombinant protein was purified and found to exhibit a single band of apparent molecular mass of ca 30 kDa when submitted to SDS-PAGE. The apparent molecular mass of the protein inferred from gel filtration on a Superdex 75 column was also ca 30 kDa. The N-terminal sequence was found to be MEGRLLL- and the molecular mass, determined by means of electrospray mass spectrometry, was 29,872(±3) Da. The molecular mass calculated from the inferred amino acid sequence is 29,876 Da. All these data are consistent with the protein being a monomer and having the amino acid sequence depicted in Figure 1.
However, when the purified protein was assayed for lipoate protein ligase activity, using an E. coli lipoyl domain or the E2 of the T. acidophilum PDH complex as substrate, it was found to be inactive. At first sight this was puzzling but the probable reason emerged later when the three-dimensional structure was determined (see below).
Structure of T. acidophilum LplA
The structure of the putative T. acidophilum LplA was partially solved at 2.5 Å resolution by means of the single-wavelength anomalous dispersion technique using a selenomethionine-labelled protein crystal (see Materials and Methods for details). The crystallographic data are given in Table 1. This partial structure was then used as a model for molecular replacement in conjunction with datasets of crystals of native T. acidophilum LplA before and after soaking with lipoic acid. This led to good quality structures with and without bound lipoic acid at 2.1 Å (see Tables 2 and 3).
Table 1. Selenomethionine-labelled protein.
| A.Crystallographic data | |
| Resolution (Å) | 34–2.5 |
| Space group | P41 |
| Unit cell parameters | |
| a (Å) | 58.82 |
| b (Å) | 58.82 |
| c (Å) | 82.55 |
| α (deg.) | 90.00 |
| β (deg.) | 90.00 |
| γ (deg.) | 90.00 |
| B. Merging statistics | |
| Figure of merit | 0.74 (0.63) |
| No. of reflections | 9109 |
| Average redundancy | 2.2 (1.7) |
| Mosaicity (deg.) | 1.45 |
| Completeness (%) | 90.8 (60.8) |
| R-merge (linear) | 0.117 (0.226) |
| Chi2 | 4.8 (1.4) |
| C. Refinement statistics | |
| R-factor (%) | 35 |
| R-free (%) | 40 |
Table 2. Native protein.
| A. Crystallographic data | |
| Resolution (Å) | 20–2.1 |
| Space group | P21 |
| Unit cell parameters | |
| a (Å) | |
| b (Å) | 118.35 |
| c (Å) | 105.59 |
| α (deg.) | 90.00 |
| β (deg.) | 93.74 |
| γ (deg.) | 90.00 |
| B. Merging statistics | |
| No. of reflections | 68,337 |
| Average redundancy | 10 |
| Mosaicity (deg.) | 0.68 |
| Completeness (%) | 95.3 (72.3) |
| R-merge (linear) | 0.068 (0.234) |
| Chi2 | 1.04 (0.86) |
| C. Refinement statistics | |
| RMSD bond length (Å) | 0.01 |
| RMSD bond angle (deg.) | 1.2 |
| Residues in disallowed region (%) | 0 |
| Mean B value (Å2) | 42.1 |
| R-factor (%) | 20.8 |
| R-free (%) | 25.2 |
Table 3. Native crystal soaked with lipoic acid.
| A. Crystallographic data | |
| Resolution | 20–2.1 Å |
| Space group | P21 |
| Unit cell parameters | |
| a (Å) | 53.37 |
| b (Å) | 117.87 |
| c (Å) | 105.67 |
| α (deg.) | 90.00 |
| β (deg.) | 93.52 |
| γ (deg.) | 90.00 |
| B. Merging statistics | |
| No. of reflections | 97,539 |
| Average redundancy | 4.0 |
| Mosaicity (deg.) | 0.8 |
| Completeness | 99.7 (99.2) |
| R-merge (linear) | 0.064 (0.206) |
| Chi2 | 1.023 (0.714) |
| C. Refinement statistics | |
| RMSD bond length | 0.008 |
| RMSD bond angle | 1.095 |
| Mean B value (Å2) | 32.5 |
| Residues in disallowed regions (%) | 0 |
| R-factor (%) | 20.7 |
| R-free (%) | 23.9 |
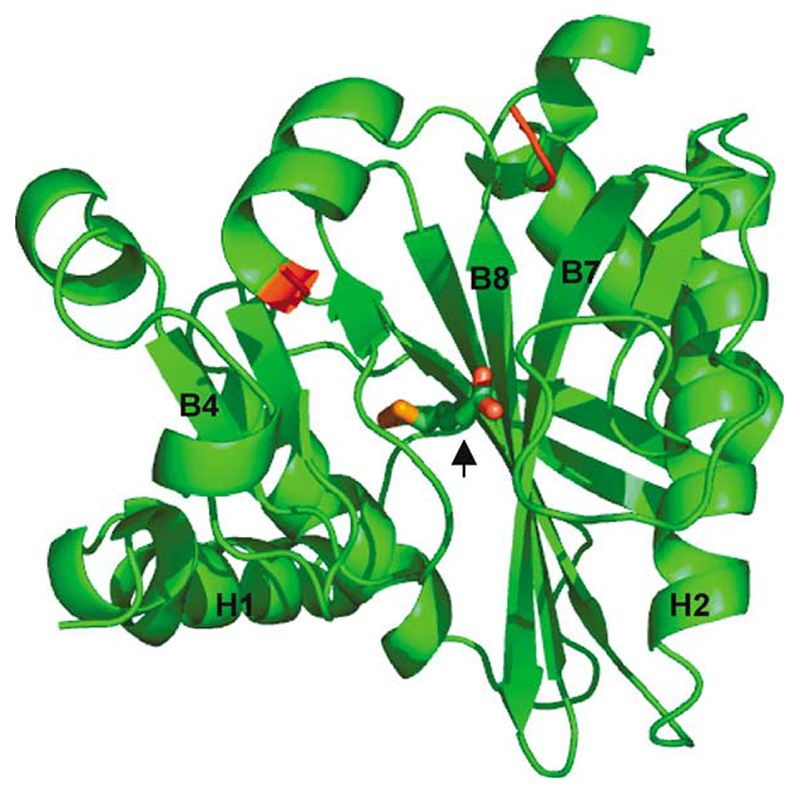
Very little difference was observed between the structures with and without bound lipoic acid and only the structure with substrate present is shown (Figure 2). LplA belongs to the α/β class of proteins, which consist of a central parallel or mixed β-sheet surrounded by α-helices. The structure of the central region is dominated by a wall of seven β-strands of varying length and directionality, two of which are labelled B7 and B8 in Figure 2. On either side of this wall reside a pair of helices. Lipoic acid is accommodated in a pocket formed by the central β-sheet, the helix pair labelled H1 and the three short β-strands labelled B4 (Figure 2) close to a disordered loop situated between amino acid residues 179 and 193. The N-terminal end of this disordered loop occurs at the end of a short helix that emerges from the β-strand labelled B8; the C-terminal end sits between strands B7 and B8. Both ends of the disordered loop are coloured red in Figure 2. Bulky hydrophobic amino acid interactions dominate between the central β-sheet region and the helix pair labelled H2, pointing to a role in conferring stability for this subsection of the protein. Another loop in LplA, comprising residues 71–79, is observed in a position that has an important counterpart in a loop in BirA that only becomes ordered upon binding biotin. This loop is potentially important in selecting lipoic acid as substrate, as discussed below.
Figure 2. Structure of the LplA of T. acidophilium with lipoic acid bound at the active site.
The red colouring denotes the boundaries of the disordered region that comprises residues 179–193. The lipoic acid is depicted in sticks and designated with an arrowhead. The helices of interest are numbered H1 and H2; the β-strands of interest are numbered B4, B7 and B8.
Lipoic acid binding
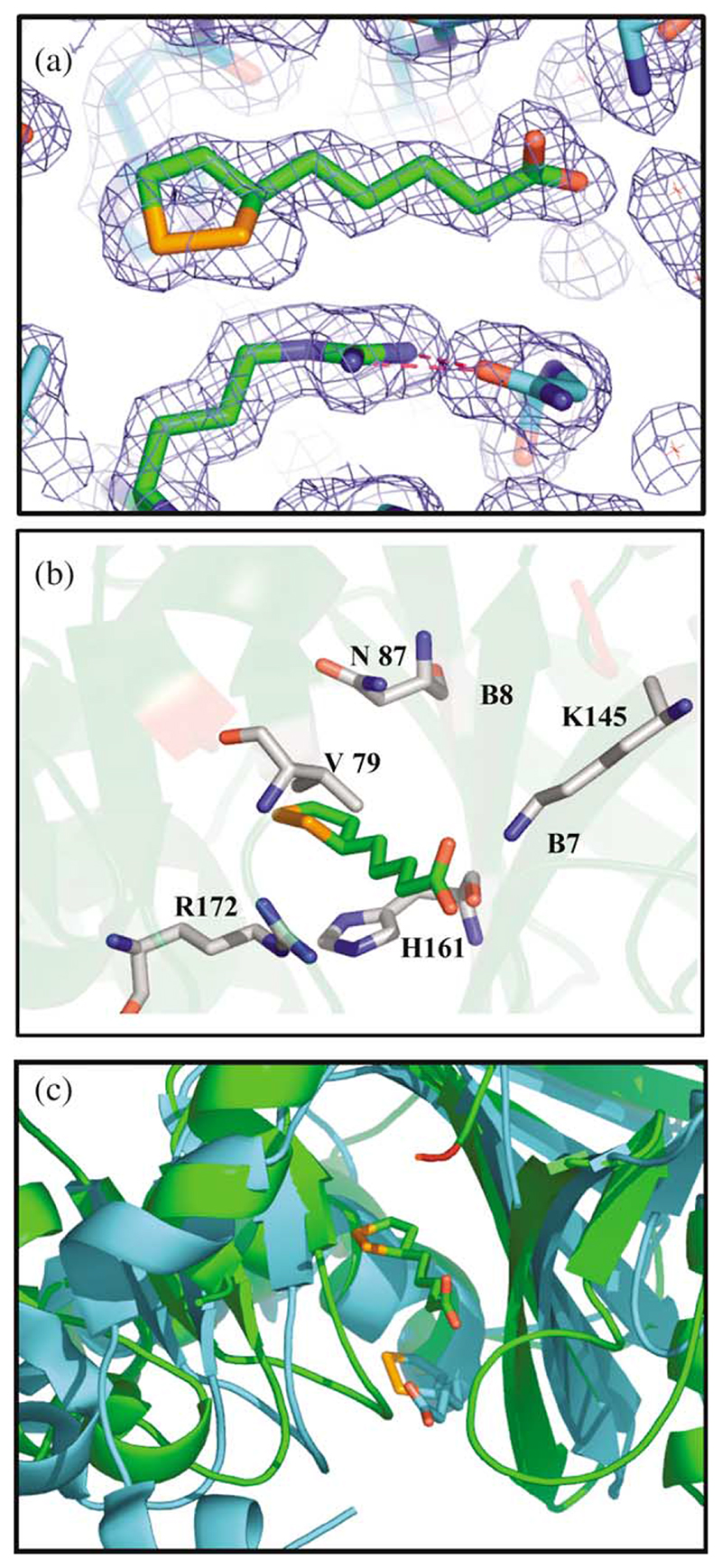
The lipoic acid-binding pocket was observed to occur in the vicinity of the switch point region which defines the locale of the active site of α/β proteins.26 The electron density from the crystal that was soaked in lipoic acid unambiguously showed the substrate bound in high occupancy (Figure 3(a)). A racemic mixture of lipoic acid was used in the crystallization trials, but it appears (not surprisingly) that the structure is enriched for one isomer. The electron density appears to fit best with the S-form but we would wish to test this biochemically before making a definitive assignment. The R-enantiomer of lipoic acid is required for the E. coli 2-oxo acid dehydrogenase complexes,1 so it is important to be fully satisfied with the evidence before making a categorical statement about the archaeal enzyme. The temperature factors for the ligand are very similar to those of the contacting side-chains from the protein, which indicates the occupancy to be approaching 100%. The switch point region where lipoic acid binds can be identified by the presence of the crevice between strands B7 and B8 in Figure 3(b). Interestingly, Lys145, which is conserved in both prokaryotic and eukaryotic lipoate protein ligases and in all biotin protein ligases, sits at the very top of β-strand B7 (Figure 3(b)). Here, it makes a direct interaction with the carboxy group of the lipoic acid which rests along the central β-sheet motif perpendicular to the direction of the strands. All the amino acids that lie below lipoic acid in this sheet are either small and hydrophobic or have their sidechains pointing directly away from the ligand, thereby creating a suitable environment of hydrophobic interactions for the long aliphatic chain of the lipoic acid. Residues that may play a more specific role in lipoic acid binding are Val79, Ile46, Arg72, Asn87 and His161 (Figure 3(b)). All these amino acid residues in the binding site are highly conserved in lipoate protein ligases.
Figure 3. Lipoic acid binding in the LplA of T. acidophilum.
(a) 2Fo—Fc map of electron density from a crystal of the native LplA soaked with R,S-lipoic acid. Lipoic acid can be seen forming interactions with Arg72 which itself forms a hydrogen bond with the carbonyl oxygen of Gly77. This interaction is shown by the broken red lines. The bond distances to the carbonyl oxygen are 3.06 and 2.86 Å. (b) Amino acid residues in the lipoic acid binding site that are highly conserved in LplAs. The residue numbers are shown in black using T. acidophilum LplA numbering. (c) Comparison of the proposed lipoic acid-binding sites in the T. acidophilum and E. coli lipoate protein ligases. The T. acidophilum protein and lipoic acid are coloured green whereas the E. coli protein and lipoic acid are coloured blue.
Since the completion of this work the X-ray crystallographic structure of E. coli LplA in the presence of lipoic acid at 2.4 Å resolution has been reported27 and deposited in the Protein Data Bank (1X2H). A comparison of the proposed lipoic acid-binding sites between the E. coli and T. acidophilum enzymes surprisingly shows the lipoic acid to be bound in different areas (Figure 3(c)). The significance of this difference remains to be seen.
Comparison of T. acidophilum LplA with E. coli BirA implicates loop regions in substrate recognition and catalysis
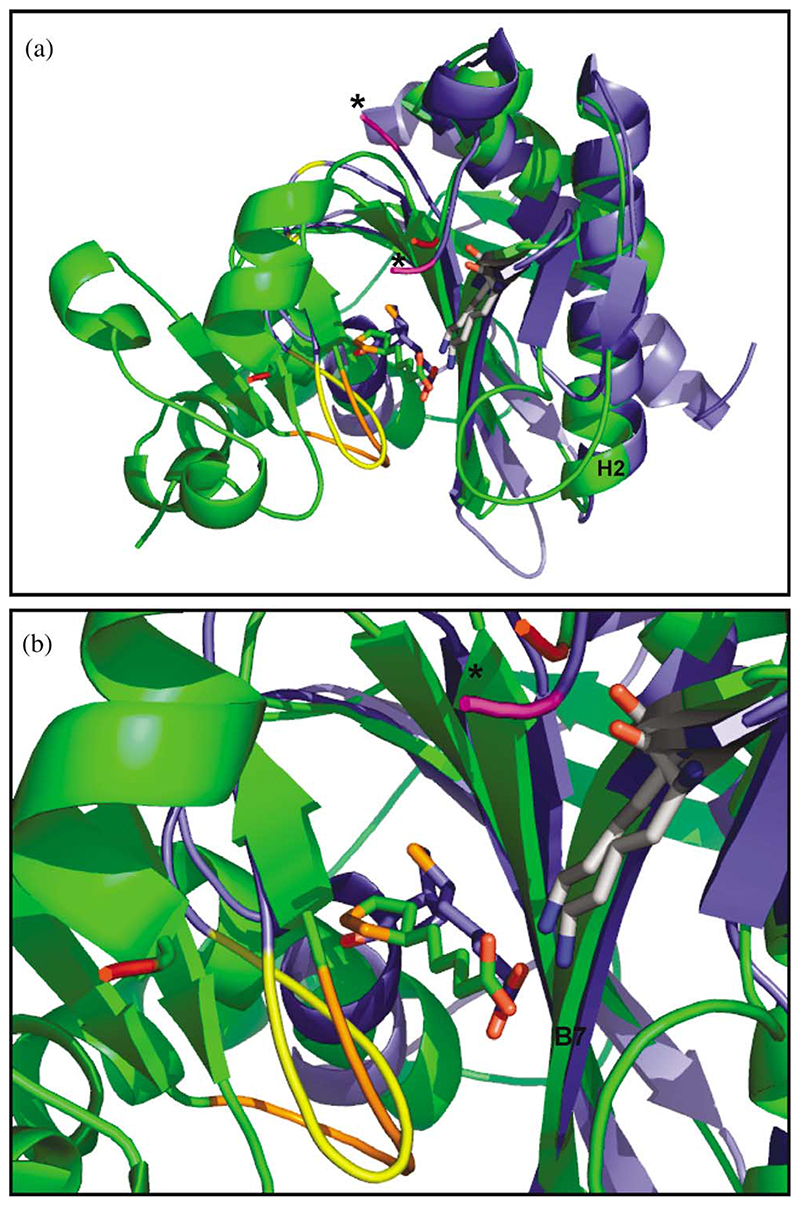
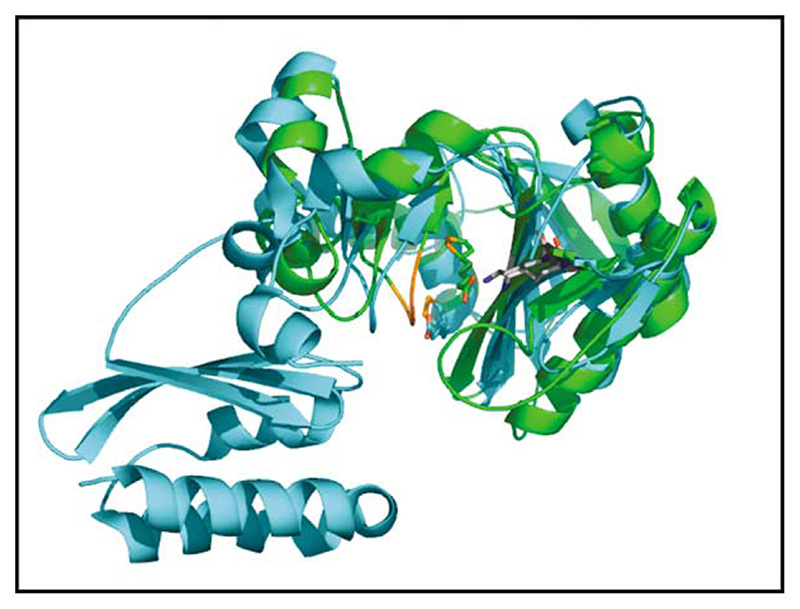
Fatty acid synthases, amino-acyl tRNA synthases, biotin protein ligases and LplAs all share a common reaction mechanism involving an acyl-adenylate intermediate. Structural relationships have long been postulated to exist between different members of this group.28–30 In particular, a clear similarity has been demonstrated between the active sites of BirA and class II aminoacyl-tRNA synthases.31 When the cofactor-binding sites of the E. coli BirA18 and T. acidophilum LplA are superimposed, it can be seen that a lysine residue (Lys145 in T. acidophilum numbering) that is conserved in the primary structures of all biotin and lipoate protein ligases is located in exactly the same position and orientation in the two structures, thus underlining its importance (Figure 4(a) and (b)). Within the two enzymes, lipoic acid and biotin occupy almost identical positions (Figure 4(b)), but different from the lipoic acid-binding site in the E. coli LplA described elsewhere.27 There is little in the way of obvious steric hindrance that would prevent biotin from binding in the lipoic acid-binding site of LplA, which raises interesting questions about the origins of specificity. It may be that many slightly unfavourable interactions within the pocket cumulatively discriminate against biotin, but it may be that biotin is somehow prevented from entering the LplA active site altogether.
Figure 4. (a) The catalytic domain of E. coli BirA with biotin bound (all in blue) superimposed on the LplA of T. acidophilum with lipoic acid bound (all in green).
The boundaries of a disordered loop region in the BirA are indicated in purple colouring and marked by black asterisks. The boundaries of an equivalent disordered loop in the T. acidophilum LplA are indicated in red; the C-terminal end of the LplA loop lies close in space to the C-terminal end of the BirA loop (asterisked), whereas the N-terminal end is located at the end of an a-helix on the other side of the lipoic acid-binding site. A region in BirA that becomes ordered upon biotin binding is coloured yellow and the loop in the T. acidophilum LplA that contains Gly77 is coloured orange. (b) A higher magnification view of (a) showing the close alignment of the lipoic acid and biotin in their respective binding sites plus the almost identical positions of the nearby loop regions and conserved lysine residue near the switch point region in BirA and LplA. The colour coding is the same as in (a).
In this regard, the loops that connect α-helices and β-sheets within secondary structure motifs are often important in enzyme catalysis and may provide a clue to substrate specificity in lipoate protein ligases. In the apo form of E. coli BirA, there are four unstructured loops. Two of these loops have been directly implicated in biotin and/or ATP binding32,33. The loop comprising residues 110–128 (coloured yellow in Figure 4(a) and (b)) becomes ordered upon biotin binding and is thought to interact with the biotinyl-AMP intermediate. When Arg118 within this loop is replaced by glycine, BirA loses its specificity and biotinylates not only other proteins but itself as well.20 The explanation proposed is that this loop plays a part in sequestering the biotinyl-AMP in the active site; replacement of Arg118 allows the reactive biotinyl-AMP to escape more readily and react with any nearby exposed lysine residues that it encounters. In the T. acidophilum LplA, a loop encompassing residues 71–79 occurs in a similar place in the structure (coloured orange in Figure 4(a) and (b)) that could play an analogous role.
This loop contains amino acids that are conserved in all lipoate protein ligases, and previous studies have implicated this region in lipoic acid binding in E. coli LplA.34,35 A G76S mutation in the loop (corresponding to Gly77 in the T. acidophilum protein) prevents E. coli LplA from using seleno-lipoic acid (an analogue of lipoic acid in which the two sulphur atoms of the dithiolane ring are each replaced with selenium) as substrate, while only partly decreasing its ability to use lipoic acid. This could be taken as evidence that the loop interacts with the dithiolane ring of lipoic acid as it enters the active site and that the wild-type loop cannot distinguish between lipoic and selenolipoic acid. However, it is easy to imagine that it could act to reject biotin, which contains a much more bulky and chemically different ring structure in place of the normal dithiolane ring of lipoic acid. In addition, it can be seen from the T. acidophilum structure that the carbonyl oxygen of Gly77 forms an interesting bifurcated hydrogen bond with Arg72, also a conserved amino acid within this loop, and clearly situated in the active site of the T. acidophilum LplA. By partaking in this hydrogen bond, the arginine forms an attractive surface for lipoic acid to interact with (Figure 3(a)). Thus, the G76S mutation in E. coli LplA could have caused a slight reorganization of the corresponding Arg72 side-chain, which in turn could have contributed to preventing selenolipoic acid from entering the lipoic acid binding pocket. Whatever the precise explanation, this loop plainly has an influential effect on selecting the lipoic acid substrate for binding.
A second loop in E. coli BirA, comprising residues 212–234, may also have a role in substrate binding. Although this loop does not become ordered in the co-crystal with biotin,18 biochemical evidence has shown that it becomes less sensitive to proteolysis after biotin or biotin-AMP is bound31,36, the degree of protection being greater in the presence of the adenylate intermediate. The terminal amino acid residues of this loop are coloured purple and marked by asterisks in Figure 4(a). Interestingly, this loop coincides well with the disordered loop (residues 179–193) in the T. acidophilum LplA structure. As shown in the superimposed structures in Figure 4(a), the N-terminal end of the BirA loop maps close to the N-terminal end of a short helix in T. acidophilum LplA and at the C-terminal end of that helix begins the N-terminal region of the disordered loop in the T. acidophilum protein (the N and C-terminal ends of the T. acidophilum LplA loop are coloured red). However, at the C-terminal end of the loops in the two proteins, the superimposition is almost perfect, both sitting directly in the crevice of the conserved, switch point region. They are thus well placed to play a key role in ligand binding and/or catalysis. Interestingly, comparison of the T. acidophilum and E. coli LplA sequences shows that this disordered loop matches to a position that is extremely trypsin-sensitive in E. coli LplA.30 In light of this, a study of the limited proteolysis of E. coli LplA was undertaken to see if any further parallels could be drawn between the T. acidophilum and E. coli proteins.
Limited proteolysis of E. coli LplA
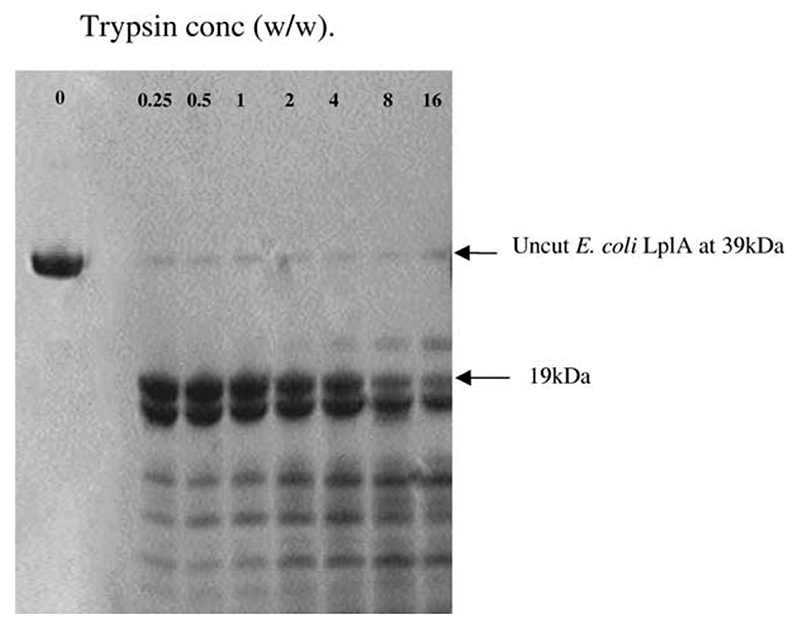
In the earlier study, E. coli LplA was subjected to limited proteolysis at 30 °C using concentrations of 0.1% and 0.5% (w/w) trypsin or chymotrypsin.30 Four proteinase-resistant fragments with molecular masses of around 20 kDa were identified, all arising from the C-terminal part of the protein. The cleavage sites were found to lie between amino acid residues 178 and 190, a region that corresponds with the disordered loop (residues 179–193) in the T. acidophilum structure. No proteinase-resistant fragments from the N-terminal part of the chain were observed.
In the present study, E. coli LplA was exposed to increasing concentrations of trypsin from 0.25% to 16% (w/w) overnight at 0 °C. The products were subjected to SDS-PAGE (Figure 5). It can be seen that exposure to trypsin under these conditions generated two predominant bands of apparent molecular mass 17–19 kDa. Both fragments appeared to be resistant to the action of trypsin (totally stable until trypsin reached 4% (w/w) and still present until it reached 16%). Both tryptic fragments were characterized by means of N-terminal sequence analysis and mass spectrometry. The smaller fragment had a molecular mass of 17,653(±7) Da and an N-terminal sequence SRVTNL-, whereas the larger fragment had a mass of 19,039(±10) Da and the same N-terminal sequence (STLRL-) as the intact LplA. Encouragingly, these molecular masses and N-terminal sequences tally exactly and enable us to depict the effect of trypsin on the E. coli LplA protein as in Figure 6. The N and C-terminal fragments are separated by an 11 amino acid residue segment, evidently an exposed loop and susceptible to trypsin, as observed previously.30 Given that Green et al.30 did not observe it after carrying out the digestion with trypsin at 30 °C, we infer that the N-terminal fragment must be less stable than the C-terminal fragment.
Figure 5. Limited proteolysis of E. coli LplA with trypsin.
The protein was treated with varying concentrations of trypsin at pH 7.5 and 0 °C and the products were subjected to SDS-PAGE. The left-hand lane shows LplA with no trypsin added, the other seven lanes represent the effects of trypsin at the individual concentrations shown.
Figure 6. The trypsin cleavage sites in E. coli LplA mapped onto the amino acid sequence.
The loop that was excised by trypsin is shown in red. The dotted box indicates a sequence at the N-terminal end of the lower molecular mass fragment that was protected from excision by the binding of lipoic acid.
After exposure to trypsin, the cleaved E. coli LplA was found to behave during purification essentially as the wild-type enzyme; in particular it emerged on gel filtration in the same position as the native enzyme, with an apparent molecular mass of around 39 kDa. However, it was found to be catalytically inactive, no longer able to lipoylate an E. coli lipoyl domain in the presence of lipoic acid and ATP. It would appear that the loss of activity can safely be attributed to the cleavage of the loop region, highlighting its importance, directly or indirectly, in the catalytic mechanism.
If lipoic acid and magnesium ions were added before the E. coli LplA was treated with trypsin, the pattern of digestion was altered somewhat. The larger (N-terminal) fragment remained unchanged, whereas the lower molecular mass (C-terminal) fragment became extended by six amino acid residues, its N-terminal sequence beginning GITSVR-(Figure 6). The previous site of cleavage, at the arginine residue of GITSVR-, was clearly no longer accessible to the proteinase. It is obvious that the structural environment of the loop region must have been changed significantly for this to have happened. The C-terminal regions of these analogous disordered loops in T. acidophilum LplA and E. coli BirA sit directly in the switch-point region.
Taken together, these results suggest that the loop region plays an important part in the mechanism of both biotinylation and lipoylation.
The structure suggests two proteins may be needed for lipoylation in T. acidophilum
The results described above indicate that E. coli LplA and the putative T. acidophilum LplA share a catalytically important flexible loop that is located close to the lipoic acid-binding site. As discussed above, the X-ray crystallographic structure of E. coli LplA at 2.4 Å has been reported.27 A superimposition (Figure 7) shows that overall the two structures are very similar, apart from the position of the proposed lipoic acid-binding sites, as described above, and also the fact that the E. coli LplA contains a large additional segment beyond the C-terminal end of the T. acidophilum protein. When we map our limited proteolysis results onto the structure of the E. coli enzyme, it confirms that the flexible loop separates two globular structural domains, which can be identified further with the larger N-terminal fragment and smaller C-terminal fragment that we obtained by carrying out the limited proteolysis at low temperature (Figure 6). It is apparent from the crystal structure listed in the database for a putative lipoate protein ligase from Streptococcus pneumoniae (Protein Data Bank accession code: 1QVZ), that it resembles the E. coli LplA with two comparable domains. S. pneumoniae is Gram-positive, whereas E. coli is Gram-negative, but both are eubacteria.
Figure 7. Superimposition of the structures of T. acidophilum LplA (green) and E. coli LplA (blue), highlighting the extra domain in the E. coli enzyme.
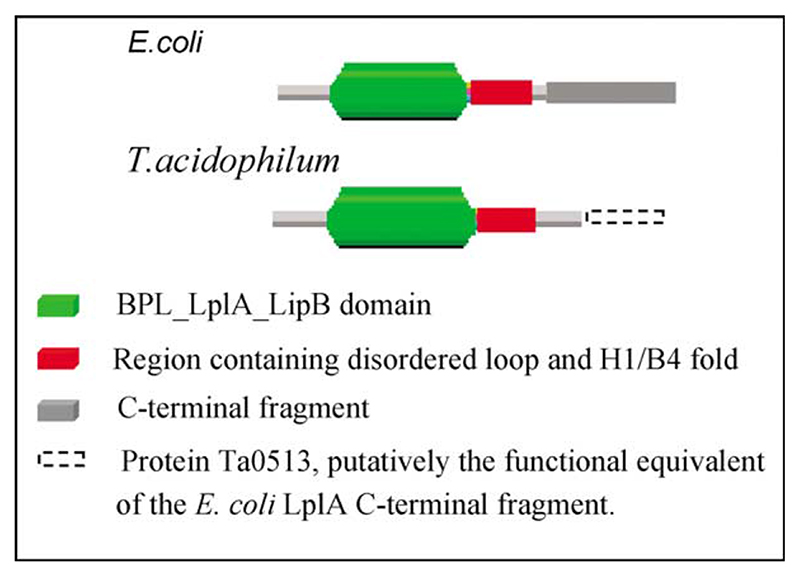
Using the program Pfam37 to analyze the domain organization of E. coli LplA, the N-terminal part was identified as containing the BPL_LplA_LipB domain i.e. the domain that all biotinylating and lipoylating enzymes possess (Figure 8). Even though the C-terminal domain of the E. coli LplA protein is distinct, and structurally stable, as judged by limited proteolysis, Pfam revealed no matching domain in its database. Assuming the N-terminal domain contains the machinery to bind lipoic acid, it is tempting to hypothesize a role for the C-terminal domain in recognising the lipoyl domain and/or transferring the lipoyl group onto it from the lipoyl-AMP intermediate, though of course the situation may not be as clear-cut as this. In E. coli BirA, a C-terminal domain is present that has been demonstrated to contribute to the interaction with both ATP and the biotinyl domain.21 This domain is distinct from the central biotinbinding domain that is depicted in Figure 4(a) and could be envisaged perhaps as analogous to the C-terminal domain of E. coli LplA.
Figure 8. Domain structure of lipoate protein ligases predicted by analysis using the program Pfam.
Inspection of the sequence alignments in Figure 1 and the three-dimensional structure in Figure 2, however, indicates that no obvious comparison can be drawn for the putative T. acidophilum LplA. When the same Pfam analysis was carried out on the T. acidophilum LplA structure, the majority of the protein was found to be accounted for in terms of the BPL_LipA_LipB domain, leaving only a much smaller C-terminal fragment remaining (Figure 8). This pattern of differences is repeated throughout other bacterial and archaeal lipoate protein ligases; C-terminal fragments of length 172 to 193 amino acid residues are observed in the eubacterial enzymes whereas in their archaeal counterparts the C-terminal segment is significantly smaller, ranging in size from 87 to 107 amino acid residues. This decrease in size could perhaps be explained by the widely observed compaction that can accompany the greater stability of the archaeal enzymes but, set against this, no corresponding compaction of the BPL_LplA_LipB domain was observed when crossing from bacterial to archaeal sources. The most likely explanation is that something is missing from the C-terminal part of the archaeal enzymes rather than it is merely reduced in size. This raises the intriguing possibility that in T. acidophilum two separate proteins are required to carry out the two-step lipoylation reaction; indeed it may be recalled that the putative T. acidophilum ligase that we describe here is catalytically inactive (see above) even though the crystal structure clearly shows it to have the BPL_LplA_LipB fold, to resemble closely the same domain in S. pneumoniae and E. coli LplAs, and to bind lipoic acid in what is obviously an appropriate active site (Figure 3(a) and (b)).
The level of sequence similarity between the C-terminal regions of bacterial lipoate protein ligases is low. It is not surprising, therefore, that using these sequences to search the T. acidophilum genome for possible homologues produced mixed results. However one hit, a 10 kDa protein, Ta0153, that was retrieved by BLAST searching using C-terminal sequences from Bacillus subtilis and Treponema denticola LplAs, proved of more than usual interest, given that it is located in the genome immediately next to Ta0154, representing the T. acidophilum lipoate protein ligase described here. Indeed the last A of the Ta0153 termination codon (TGA) is also the A of the initiation codon (ATG) of Ta0154 i.e. TGATG. When the T. acidophilum Ta0513 sequence was analysed in the program Fugue,38 which uses structure-based environmental matches to seek structural homologues, the C-terminal domain of the S. pneumoniae LplA structure was matched with a high degree of confidence, suggesting that these proteins are functionally equivalent. The potential importance of this common domain is further supported by the observation that the sequences of the C-terminal domains of LplA from a wide variety of bacteria were likewise predicted to have significant structural similarity to the C-terminal domain of S. pneumoniae LplA. The way is now open to test experimentally whether lipoylation in Archaea requires two enzymes to catalyse the two-part reaction. It is worth noting that the need for two proteins as part of the LplA system in mammalian cells has been reported earlier. Curiously, the mammalian LplAs, though they contain both the N and C-terminal domains that are present in the E. coli/S. pneumoniae LplAs (as judged by Pfam/ Fugue analysis) and exhibit some 30% sequence identity with E. coli LplA, appear unable to catalyse the activation of lipoic acid to lipoyl-AMP and only the transfer of the lipoyl group to the apo-lipoyl domain.39,40 Thus, the parallel is not exact and there is clearly much left to discover about the enzymes that catalyse this important post-translational modification.
Since our paper was submitted for publication, the structure of the T. acidophilum LplA has been reported by others.41 The two structures are essentially identical; the Kim et al. structure41 complements ours in that it has the lipoyl-AMP in place, and its location is in accord with our proposals. However, these authors do not indicate whether their enzyme was catalytically active in lipoylating a target protein, they do not identify loop regions as playing a substantial part in the catalytic mechanism, nor do they comment on the potential need for another protein to fulfil the role of the terminal domain that is missing from the T. acidophilum enzyme in comparison with the LplA of E. coli or S. pneumoniae.
Materials and Methods
Plasmid construction, gene expression and protein purification
Genomic DNA of T. acidophilum DSM 1728 was a kind gift from Peter Zwickl, Max-Planck-Institut für Bio-chemie, Martinsried, Germany. Standard protocols for molecular biology were used as described elsewhere.42 A fragment of DNA encoding the putative lipoate protein ligase was amplified from this template by means of PCR, with the N-terminal primer 5’-TTTCATATGGAAGG CAGGCTTCTTTT and the C-terminal primer, 5’-TTTGGATCCCTATACGACCTCTTTCCTC, thereby generating NdeI and BamHI restriction cleavage sites, respectively. The amplified fragment was cloned into plasmid pET 3a and expressed in E. coli BL21(DE3) cells grown in LB medium, which were induced at an A600 of 0.7 with a final concentration of 0.45 mM IPTG for 4 h. The cells were collected by centrifugation and lysed in 20 mM Tris-HCl (pH 7.5), 20% (v/v) glycerol, 1 mM EDTA, 1 mM DTT using a French press. The soluble fraction, which contained the desired protein, was separated by centrifugation (20,000g, 30 min). The protein was purified using ion-exchange chromatography on a Q Sepharose HP column, equilibrated in 10 mM potassium phosphate buffer (pH 7.0), containing 1 mM DTT (buffer A) and eluted with a gradient of 0%–100% buffer B (buffer A + 200mM NaCl) over ten column volumes. Fractions containing T. acidophilum ligase were pooled and dialysed against buffer A overnight. Fractionation was then carried out on a ceramic hydroxyapatite (type I) column pre-equilibrated in buffer A and eluted using a gradient of buffer A to buffer B (200 mM KH2PO4 (pH 7.0), 1 mM DTT) over ten column volumes. Pooled fractions from this step were concentrated to 1 ml and subjected to gelfiltration chromatography on a Superdex 75 column in 10 mM Tris–HCl (pH 7.5) buffer, containing 1 mM DTT, 150 mM NaCl. All steps were carried out at 4 °C and the purified protein was stored at −80 °C. Throughout the procedure, the purification was monitored by submitting samples to SDS-PAGE and the protein was judged to be > 95% pure on Coomassie-stained gels.
A selenomethionine derivative of the protein was generated by gene expression for 6–8 h according to a protocol developed by Dr Maryn Symmons (see web site†) which is based on an earlier method.43 The selenomethionine-labelled protein was purified by means of the same procedure as that developed for the wild-type enzyme, except that all buffers were extensively degassed and contained 5 mM DTT to prevent oxidation of the selenomethionine. The incorporation of selenomethionine into T. acidophilum LplA was confirmed as 100% by means of mass spectrometry.
E. coli LplA was purified as described elsewher14 using ion-exchange chromatography on Q Sepharose-HP and gel-filtration on Superdex 75, except that after the ion-exchange step the partly purified protein sample was dialysed for 4 h against 20 m Tris–HCl (pH 7.5) containing 1 mM DTT, 1 mM EDTA and 10% glycerol and subjected to heparin Sepharose chromatography using exactly the same buffers and running conditions as described14 for the ion-exchange step. The material was then passed down the Superdex 75 column to give > 95% pure protein in 10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM DTT.
Protein chemical techniques
Proteins were characterized by means of SDS-PAGE, automated N-terminal sequencing (Protein and Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge) and electrospray mass spectrometry using a VG BioQ quadrapole mass spectrometer with myoglobin as a calibration standard, all as described elsewhere.14
Enzyme assays
The ability of T. acidophilum and E. coli lipoate protein ligases to catalyse lipoylation was determined by means of an assay described previously.14 Briefly, R,S-lipoic acid (200 μM), ATP (1.5 mM) and MgCl2 (1.5 mM) were incubated with 250 μg of either E. coli lipoyl domain or the E2 of T. acidophilum pyruvate dehydrogenase complex (a kind gift from Professor M. J. Danson, University of Bath) for 12 h at 20 °C in the dark. Reactions were initiated by adding 5 μg of the relevant ligase, all in a final volume of 250 μl. Evidence of lipoylation was checked for by the increase of mass as judged by mass spectrometry or, in the case of the E. coli lipoyl domain, by the change in electrophoretic mobility on non-denaturing PAGE that accompanies lipoylation.14
Limited proteolysis of E. coli LplA
A sample (1 ml) of E. coli LplA (1 mg/ml in 20mM Tris–HCl (pH 7.5), 1 mM DTT, 150 mM NaCl) was digested at 0 °C with varying concentrations of trypsin in 20 mM Tris–HCl (pH 7.5). In time-course experiments, aliquots of 20 μl were taken at given times, mixed with 1 μl of 50 mM PMSF to inhibit the trypsin, and stored at −80 °C. When studying the effect of substrate binding, lipoic acid and magnesium (both at final concentrations of 2 mM) were pre-incubated with LplA for 30 min at 20 °C, then left at 0 °C for 10 min before addition of trypsin. The digestion products were analysed by SDS-PAGE using Mes running buffer, as gels run in Mops buffer were unable to resolve the two fragments generated by tryptic cleavage of the E. coli LplA.
Crystallization of T. acidophilum lipoyl ligase, data collection and model building
The protein solution as effluent from the Superdex 75 gel filtration column in 10 mM Tris–HCl (pH 7.5) buffer, containing 1 mM DTT, 150 mM NaCl was concentrated to 10 mg/ml and centrifuged at 20,000g for 30 min at 4°C. Hampton screens I and II were employed using the hanging-drop, vapour-diffusion method. Protein solution (2 μl) was mixed with precipitant (1 μl) and the tray left at 20 °C. After two weeks crystals were observed in 20% PEG 8000, 0.1 M sodium cacodylate buffer (pH 6.5) containing 0.2 M magnesium acetate. Switching to Mes buffer and fine screening around this condition resulted in good quality reproducible crystals that diffracted to 2.9 Å in-house using 25% glycerol as a cryo-protectant.
Selenomethionine-labelled protein was subjected to the same fine crystal screen at a protein concentration of 10 mg/ml. The crystals that formed in this case, however, were not reproducible, were in a different crystal form from the native protein, and showed very mosaic diffraction patterns. One crystal was harvested that diffracted to 2.8 Å in-house though the diffraction pattern was of generally poorer quality than that of the native protein crystal. A single-wavelength anomalous dispersion (SAD) data set was colleted to 2.5 Å resolution at the National Synchrotron Light Source in Brookhaven, USA on beam line X9A using 1 s exposure and 1° oscillation slicing. The data were indexed using DENZO and SCALEPACK.44 The data were processed in a P41 space group and from the SCALEPACK output (see Table 1) it can be seen to be of rather poor quality. The average Chi squared value for all reflections was 4.8, mosaicity was 1.4 and overall completeness was 90%. A model of the T. acidophilum LplA was prepared by Dr T.A. Terwiliger using the PHENIX suite.45 Owing to the poor quality of the crystal, the model is not perfect (see Table 1) with an R-free of 0.4 and 147 out of 262 residues placed. However, this partial structure was used as a starting model for molecular replacement using the good quality native crystal data, which allowed the model to be built completely.
Synchrotron data were collected from a crystal of the native protein and a native protein crystal soaked with a racemic (R,S) mixture of lipoic acid, at ESRF, Grenoble on beam line ID29. Both these crystals belong to space group P21, and the data extend to 2.1 Å in both instances, with good quality overall (Tables 2 and 3 and Figures 3(a) and 9). Molecular replacement was performed with the programme PHASER46 using the selenomethionine partial structure as a model. Building of the model for the resulting solution was carried out using COOT.47 The solution was refined using Refmac48 and ArpWarp49 on the CCP4 graphical user interface.
Figure 9. Quality of the electron density map from the crystal structure of T. acidophilum LplA.
A β-strand with the sequence KLWHAA (residues 158–163) is illustrated.
Protein Data Bank accession codes
The coordinates have been submitted to the RCSB PDB and have accession codes 2C71 and 2C8M.
Acknowledgements
E.M. thanks the Biotechnology and Biological Sciences Research Council and the Department of Biochemistry, University of Cambridge for a Research Studentship. This work was supported by the wellcome trust. We are grateful to John Lester for automated DNA sequencing, Mike Weldon for N-terminal sequence analysis of proteins, and Charles Hill for DNA primer synthesis (all Department of Biochemistry, University of Cambridge). We thank Dr Bob Sweet, Dr Herb Klei, Dr Tom Terwiliger and all the skilled staff at the RapiData 2004 crystallography course for their help.
Abbreviations used
- ACP
acyl carrier protein
- IPTG
isopropyl-β-D-thiogalactopyranoside
- DTT
dithiothreitol
- PDB
Protein Data Bank
Footnotes
References
- 1.Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 2.de Kok A, Hengeveld AF, Martin A, Westphal AH. The pyruvate dehydrogenase multienzyme complex from Gram-negative bacteria. Bio-chim BioPhys Acta. 1998;1385:353–366. doi: 10.1016/s0167-4838(98)00079-x. [DOI] [PubMed] [Google Scholar]
- 3.Motokawa Y, Fujiwara K, Okamura-Ikeda K. in Health and Disease. In: Packer L, Cadenas E, editors. Biothiols in Health and Disease. Marcel Dekker; New York: 1995. pp. 389–407. [Google Scholar]
- 4.Reed LJ, Hackert ML. Structure-function relationships in dihydrolipoamide acyltransferases. J Biol Chem. 1990;265:8971–8974. [PubMed] [Google Scholar]
- 5.Perham RN. Domains, motifs and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional enzyme. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Addad C, Pares S, Sieker L, Neuburger M, Douce R. The lipoate arm in the glycine decarboxylase complex is not freely swinging. Nature Struct Biol. 1995;2:63–68. doi: 10.1038/nsb0195-63. [DOI] [PubMed] [Google Scholar]
- 7.Milne JLS, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu X, et al. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 2002;21:5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brody S, Oh C, Hoja U, Schweizer E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae . FEBS Letters. 1997;408:217–220. doi: 10.1016/s0014-5793(97)00428-6. [DOI] [PubMed] [Google Scholar]
- 9.Jordan SW, Cronan JE., Jr The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein transferase. J Bacteriol. 2003;185:1582–1589. doi: 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem Biol. 2003;10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Chapman-Smith A, Cronan JE., Jr The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends BioChem Sci. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- 12.Athappilly FK, Hendrickson WA. Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure. 1995;3:1407–1419. doi: 10.1016/s0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
- 13.Roberts EL, Shu N, Howard MJ, Broadhurst RW, Chapman-Smith A, Wallace JC, et al. Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry. 1999;38:5045–5053. doi: 10.1021/bi982466o. [DOI] [PubMed] [Google Scholar]
- 14.Reche P, Li YL, Fuller C, Eichhorn K, Perham RN. Selectivity of post-translational modification in biotinylated proteins: the carboxy carrier protein of the acetyl-CoA carboxylase of Escherichia coli . BioChem J. 1998;329:589–596. doi: 10.1042/bj3290589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DD, Horne HJ, Reche PA, Perham RN. Structural determinants of post-translational modification of the pyruvate dehydrogenase multienzyme complex of Escherichia coli . J Mol Biol. 2000;295:289–306. doi: 10.1006/jmbi.1999.3335. [DOI] [PubMed] [Google Scholar]
- 16.Reche P, Perham RN. Structure and selectivity in post-translational modification: attaching the biotinyl-lysine and lipoyl-lysine swinging arms in multifunctional enzymes. EMBO J. 1999;18:2673–2682. doi: 10.1093/emboj/18.10.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzyme synthase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver LH, Kwon K, Beckett D, Matthews BW. Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc Natl Acad Sci USA. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon K, Streaker ED, Beckett D. Binding specificity and the ligand dissociation process in the E. coli biotin holoenzyme synthase. Protein Sci. 2002;11:558–570. doi: 10.1110/ps.33502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi-Rhee E, Schulman H, Cronan JE. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2004;13:3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman-Smith A, Mulhern TD, Whelan F, Cronan JE, Jr, Wallace JC. The C-terminal domain of biotin protein ligase from E. coli is required for catalytic activity. Protein Sci. 2001;10:2608–2617. doi: 10.1110/ps.22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver LH, Kwon K, Beckett D, Matthews BW. Competing protein:protein interactions are proposed to control the biological switch of the E. coli biotin repressor. Protein Sci. 2001;10:2618–2622. doi: 10.1110/ps.32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reche PA, Howard MJ, Broadhurst RW, Perham RN. Heteronuclear NMR studies of specificity of the post-translational modification of biotinyl domains by biotin protein ligases. FEBS Letters. 2000;479:93–98. doi: 10.1016/s0014-5793(00)01829-9. [DOI] [PubMed] [Google Scholar]
- 24.Morris TW, Reed KE, Cronan JE. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli: molecular cloning and characterization of the LplA gene and gene product. J Biol Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- 25.Reche PA. Lipoylating and biotinylating enzymes contain a homologous catalytic module. Protein Sci. 2000;9:1922–1929. doi: 10.1110/ps.9.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branden C. Relation between structure and function of alpha/beta proteins. Quart Rev Biophys. 1980;13:317–338. doi: 10.1017/s0033583500001712. [DOI] [PubMed] [Google Scholar]
- 27.Fujiwara K, Toma S, Okamura-Ikeda K, Motokawa Y, Nakagawa A, Taniguchi H. Crystal structure of lipoate-protein ligase A from Escherichia coli determination of the lipoic acid-binding site. J Biol Chem. 2005;280:33645–33651. doi: 10.1074/jbc.M505010200. [DOI] [PubMed] [Google Scholar]
- 28.Berg P. Studies on the enzymatic utilization of amino acyladenylates; the formation of adenosine triphosphate. J Biol Chem. 1958;233:601–607. [PubMed] [Google Scholar]
- 29.Eisenberg MA. Biotin: biogenesis, transport, and their regulation. Advan EnzyMol Relat Areas Mol Biol. 1973;38:317–372. doi: 10.1002/9780470122839.ch7. [DOI] [PubMed] [Google Scholar]
- 30.Green DE, Morris TW, Green J, Cronan JE, Jr, Guest JR. Purification and properties of the lipoate protein ligase of Escherichia coli . BioChem J. 1995;309:853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artymiuk PJ, Rice DW, Poirrette AR, Willet P. A tale of two synthases. Nature Struct Biol. 1994;1:758–760. doi: 10.1038/nsb1194-758. [DOI] [PubMed] [Google Scholar]
- 32.Streaker ED, Beckett D. Ligand-linked structural changes in the Escherichia coli biotin repressor: the significance of surface loops for binding and allostery. J Mol Biol. 1999;292:619–632. doi: 10.1006/jmbi.1999.3086. [DOI] [PubMed] [Google Scholar]
- 33.Kwon K, Streaker ED, Ruparelia S, Beckett D. Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J Mol Biol. 2000;304:821–833. doi: 10.1006/jmbi.2000.4249. [DOI] [PubMed] [Google Scholar]
- 34.Reed KE, Morris TW, Cronan JE., Jr Mutants of Escherichia coli K-12 that are resistant to a selenium analog of lipoic acid identify unknown genes in lipoate metabolism. Proc Natl Acad Sci USA. 1994;91:3720–3724. doi: 10.1073/pnas.91.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris TW, Reed KE, Cronan JE., Jr Lipoic acid metabolism in Escherichia coli the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Nenortas E, Beckett D. Evidence for distinct ligand-bound conformational states of the multifunctional Escherichia coli repressor of biotin biosynthesis. Biochemistry. 1995;34:16624–16631. doi: 10.1021/bi00051a010. [DOI] [PubMed] [Google Scholar]
- 37.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucl Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 39.Fujiwara K, Okamura-Ikeda K, Motokawa Y. Purification and characterization of lipoyl-AMP:N epsilon-lysine lipoyltransferase from bovine liver mitochondria. J Biol Chem. 1994;269:16605–16609. [PubMed] [Google Scholar]
- 40.Fujiwara K, Suzuki M, Okumachi Y, Okamura-Ikeda K, Fujiwara T, Takahashi E, Motokawa Y. Molecular cloning, structural characterization and chromosomal localization of human lipoyltransferase gene. Eur J Biochem. 1999;260:761–767. doi: 10.1046/j.1432-1327.1999.00204.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim DJ, Kim KH, Lee HH, Lee SJ, Ha JY, Yoon HJ, Suh SW. Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains. J Biol Chem. 2005;280:38081–38089. doi: 10.1074/jbc.M507284200. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 43.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallog sect D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallog sect D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallog sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallog sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Perrakis A, Morris R, Lamzin VS. Huber R, editor. Automated protein model building combined with iterative structure refinement. Nature Struct Biol. 1999;6:458–463. doi: 10.1038/8263. Received 16 August 2005. received in revised form 3 November 2005; accepted 15 November 2005) Available online 5 December 2005. [DOI] [PubMed] [Google Scholar]