Abstract
Current materials used for in vitro 3D cell culture are often limited by their poor similarity to human tissue, batch-to-batch variability and complexity of composition and manufacture. Here, we present a “blank slate” culture environment based on a self-assembling peptide gel free from matrix motifs. The gel can be customised by incorporating matrix components selected to match the target tissue, with independent control of mechanical properties. Therefore the matrix components are restricted to those specifically added, or those synthesised by encapsulated cells. The flexible 3D culture platform provides full control over biochemical and physical properties, allowing the impact of biochemical composition and tissue mechanics to be separately evaluated in vitro. Here, we demonstrate that the peptide gels support the growth of a range of cells including human induced pluripotent stem cells and human cancer cell lines. Furthermore, we present proof-of-concept that the peptide gels can be used to build disease-relevant models. Controlling the peptide gelator concentration allows peptide gel stiffness to be matched to normal breast (<1 kPa) or breast tumour tissue (>1 kPa), with higher stiffness favouring the viability of breast cancer cells over normal breast cells. In parallel, the peptide gels may be modified with matrix components relevant to human breast, such as collagen I and hyaluronan. The choice and concentration of these additions affect the size, shape and organisation of breast epithelial cell structures formed in co-culture with fibroblasts. This system therefore provides a means of unravelling the individual influences of matrix, mechanical properties and cell-cell interactions in cancer and other diseases.
Keywords: Biomaterials, Cancer, Stem cells, Extracellular matrix, Stiffness
Introduction
In many research areas, but particularly in cancer research and disease modelling, there is an increasing emphasis on the use of biomaterials to grow cells in 3D [1–3]. It is now well-understood that culturing most cells on 2D surfaces results in inferior physiological conditions affecting cell morphology, phenotype and cell-matrix interactions [4–6]. As a result, there is a growing body of literature focussed on the development of biomaterials as biomimetic culture platforms, to produce more tissue-realistic cell behaviour in vitro. It has become clear that there is unlikely to be a one-size-fits-all solution, with 3D in vitro culture environments requiring the same capacity for variability and specificity as provided by natural in vivo matrix microenvironments [1]. Therefore, the major hurdle still to be overcome is the provision of a system that is both highly tunable and reproducible in composition and mechanical properties.
Materials for 3D culture may broadly be separated into natural and artificially derived materials [7]. Natural materials, most notably collagen gels and Matrigel™, are the most established, with a long history of use in tissue culture for applications ranging from cell migration and invasion studies to regenerative medicine [8–10]. Since these materials are commonly based on matrix proteins, cells grown on or within them are able to adhere and grow to form tissue-realistic structures [9,11]. However, the biological origin of natural materials also results in batch-to-batch variability and uncertainty in composition, most particularly for Matrigel™, a basement membrane extract derived from Engelbreth-Holm-Swarm mouse sarcoma [12]. For this reason, and to provide opportunities for adding functionality, there has been a shift in focus towards more highly defined, synthetic alternatives [7].
Apart from a few notable exceptions [13,14], there have thus far been few models designed specifically to allow customisable matrix composition. Similarly, many models have a matrix component (commonly collagen, laminin or hyaluronan) as an essential part of their make-up, making it hard to discriminate endogenous matrix production by encapsulated cells from the 3D matrix itself. Here, we present the optimisation of a self-assembling peptide gel as the basis for a 3D culture platform with user-controlled composition, mechanical properties, and cell-cell interactions. Based on a short, octapeptide gelator [15], the raw materials required for the peptide gels can be produced reliably and rapidly and are widely commercially available. By defining a protocol that enables independent control over mechanical and biochemical properties, we aim to provide a platform technology suitable for studies decoupling the influences of matrix stiffness and composition on cell behaviour. Here, we demonstrate the application of the peptide gel to investigate the role of matrix stiffness and functionalisation on a model of breast cancer.
Results
Controlled gelation produces a fully-defined environment
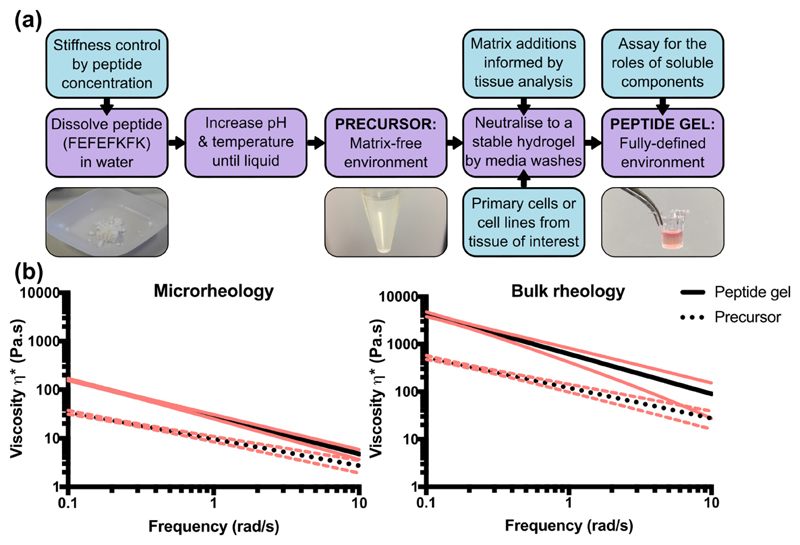
Peptide gel fabrication is primarily a two-stage process, but with multiple degrees of freedom engineered into the design as illustrated in Fig. 1 (a). The first stage is to create a matrix-free precursor by peptide dissolution in water. This precursor contains no organic components other than the octapeptide gelator FEFEFKFK, the concentration of which will determine the stiffness of the final peptide gel. The second stage is to incorporate the cells and matrix components of interest for the desired application, yielding a final peptide gel with user-defined stiffness, matrix composition and cellularity. The peptide itself is commercially available from several suppliers, and importantly we have verified that our fabrication method is effective for peptide preparations obtained from different companies.
Fig. 1.
Schematic illustrating the two-stage process of peptide gel fabrication, along with the degrees of freedom achievable in design. (a) The initial formation of a cell- and matrix-free precursor allows subsequent functionalisation by physically mixing in matrix components of interest. (b) Quantification of the increase in complex viscosity between the precursor and the final 6 mg/mL peptide gel, measured by microrheology and bulk oscillatory rheology. Trendlines of data collected from 5 replicates are shown along with 95% confidence intervals (red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
This two-stage process is crucial as, to ensure homogeneity, the precursor must first be taken to the liquid state by modulation of pH and temperature. Since the precursor is cell- and matrix-free, it can be heated to 80 °C with no detrimental effects. Prior to cell and/or matrix addition, the precursor is cooled to 37 °C. Although the precursor is self-supporting at 37 °C, its viscosity is sufficiently low to allow it to be treated as a liquid. In this way, cells and matrix additions may be stirred into the gel by simple pipetting. Mixing the precursor gel with these components at a 1 in 5 ratio produces a peptide gel with a final concentration of 6 mg/mL FEFEFKFK peptide preparation. Sequential media washes then produce the final peptide gel, which has higher viscosity than the precursor due to complete pH neutralisation.
To quantify this change in viscosity, we employed two measurement methods: bulk oscillatory rheology, and microrheology measurements based on the Brownian motion of micron-sized beads [16,17]. As shown in Fig. 1(b), both measurement methods show approximately an order of magnitude increase in viscosity between the precursor and peptide gel. Interestingly, the absolute values measured by microrheology are an order of magnitude lower than those measured by bulk rheology. Since the viscosity measurements for control samples (100% Matrigel™ for bulk rheology and pure water for microrheology, Supplementary Fig. 1) matched the expected literature values [18,19], this difference appears to reflect a property of the peptide gel itself. A similar difference between bulk and microrheological properties was also observed for 100% Matrigel™ (Supplementary Fig. 1(e)). It is therefore likely that this is a more general property of some hydrogels. It has also recently been reported that the normal force applied to a sample when setting up the bulk oscillatory rheometer can impact the viscoelastic response of the material [20,21]. It is possible that this, along with the impact of material heterogeneity at each length scale [22], could have contributed to the discrepancy between the bulk and microrheology measurements. Although exploring this further was beyond the scope of the current study, we anticipate that microrheology will be invaluable in future studies, investigating local variations in stiffness on the single cell scale. However, since bulk rheology is an established method for materials characterisation, we focussed on this technique for the remainder of the study.
In particular, we report an average value of the storage modulus G' (at 1 rad/s) of G' = 600 ± 90 Pa for the 6 mg/mL peptide gel, and as a means of comparison, an average value of G' = 120 ± 20 Pa for 100% Matrigel™ (mean ± SEM) in agreement with previously reported values [18]. These measurements therefore indicate that, whereas Matrigel™ is acknowledged to provide a 3D culture environment with artificially low stiffness [23], peptide gels may be produced with a final stiffness falling within the same range as many soft tissues in vivo, with the 6 mg/mL condition particularly similar to the stiffness of normal breast [24].
Matrix-free environments support cell viability
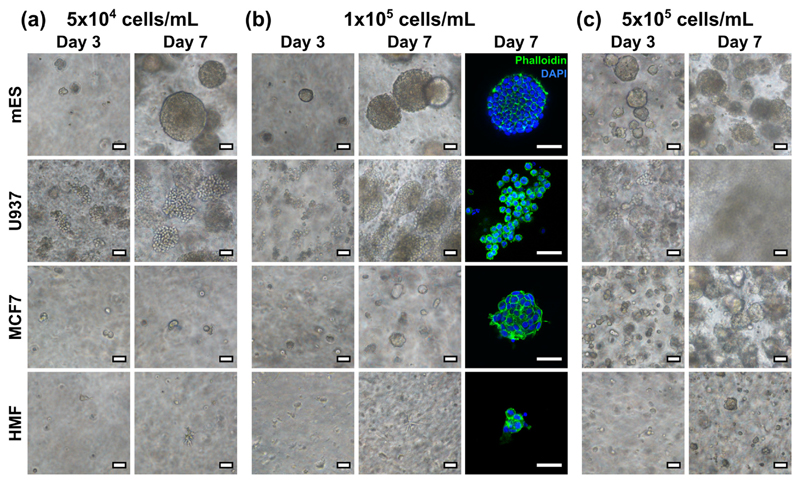
In its simplest form, peptide gel fabrication allows cells to be encapsulated in a matrix-free environment, achieved by mixing the precursor with cells suspended in cell culture medium. To demonstrate this, four cell lines were encapsulated in this way: mouse embryonic stem cells (mES), human leukaemic cells (U937), human breast cancer cells (MCF7) and human mammary fibroblasts (HMF). As shown in Fig. 2, although all cell types showed some ability to grow within the matrix-free gel, the growth characteristics of the cell populations differed dramatically between lines. In particular, although the mES and U937 cells formed large colonies within 7 days, as expected due to their anchorage independence, the adherent MCF7 and HMF cell lines formed smaller colonies at a slower rate.
Fig. 2.
Bright field and fluorescence images of various cell lines growing in the 6 mg/mL peptide gel, with initial seeding density of (a) 5 × 104, (b) 1 × 105, and (c) 5 × 105 cells/mL. All scale bars 50 μm.
To test the effect of seeding density on cell growth, each cell line was also suspended at three seeding densities: 5 × 104, 1 × 105 and 5 × 105 cells/mL within the peptide gel. At the lowest seeding density, mES and U937 both formed defined colonies with clearly distinguishable boundaries, with mES cells forming a larger number of smaller colonies at high seeding density, and U937 rapidly colonising the entire gel with separate colonies merging together. MCF7 cells also formed approximately spherical colonies at this seeding density, similar in morphology to the mES cell colonies. HMF showed limited proliferation at all seeding densities, forming small rounded clusters rather than the classic elongate morphology typical of fibroblasts [25].
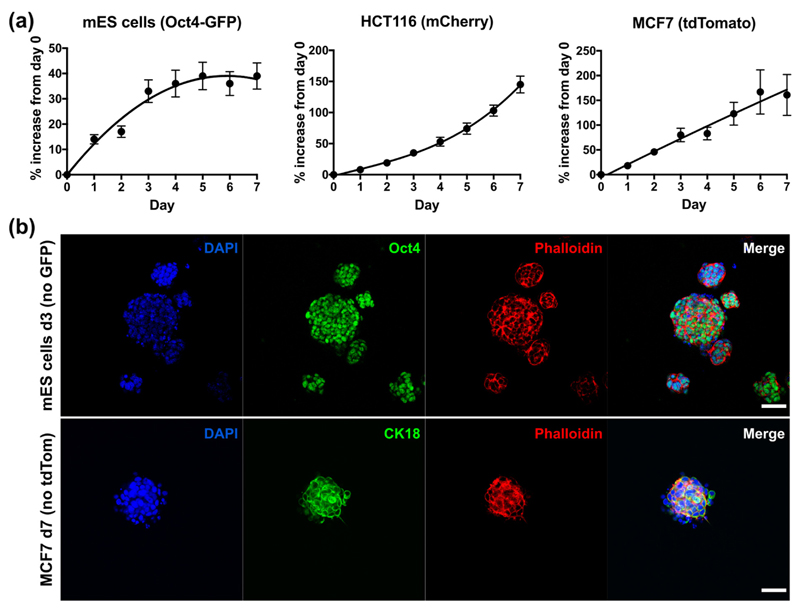
The optical transparency of the peptide gels also allows quantitative read-outs of fluorescence (reporters or constitutively expressed) allowing realtime measurement of viable cell number. Fig. 3(a) shows the increase in fluorescence signal relative to the value at day 0 for three cell lines: mES with an Oct4-GFP reporter, mCherry-HCT116 colorectal cancer cells, and tdTomato-MCF7 breast cancer cells. Each cell line gave a distinct growth profile. The fluorescence values for both HCT116 and MCF7 increased steadily up to day 7, and whilst an approximately linear increase in fluorescence over time was observed for MCF7, the increase for HCT116 became even more pronounced towards day 7. The Oct4-GFP construct in the mES cells is active in pluripotent cells and will switch off as the cells differentiate and Oct4 is down-regulated. Monitoring for GFP over 7 days in 3D culture, we observed an increase in signal due to proliferation (days 0–3), followed by a plateau (days 4–7). This in good agreement with the images in Fig. 2(c), which show the formation of stable colonies by day 3. After this point, the packed clusters of cells stop proliferating as rapidly and start differentiating, as seen by the levelling off of GFP.
Fig. 3.
Methods for assaying cell growth and pluripotency in the 6 mg/mL peptide gels. (a) Real-time measurements of % increase in signal relative to day 0, from fluorescently-tagged cell lines encapsulated in matrix-free gels (seeding density 5 × 105 cells/mL). Graphs show mean ± standard deviation for n = 3 independent experiments. Trendlines are intended as a visual guide only. (b) End-point immunostaining and microscopy of unmodified mES (Oct4 stain) and MCF7 (CK18 stain) cell lines seeded within the matrix-free peptide gels. Scale bar 50 μm.
Quantitative fluorescent read-outs can be used in parallel with end-point fluorescent immunostaining and microscopy, as shown in Fig. 3(b). In this case, peptide gels containing E14 mES (without the Oct4-GFP reporter) and unlabelled MCF7 cell lines were fixed and stained for the pluripotency marker Oct4 and the epithelial marker cytokeratin 18 (CK18) respectively. Cells were seeded at 5 × 105 cells/mL so that, in each case, the gels could be fixed at the time point where the cells gave their maximum fluorescent expression: day 3 for the mES cells, and day 7 for MCF7. This demonstrates that both realtime fluorescence readouts and end-point staining of unlabelled cells can be used to examine cell growth and behaviour within the peptide gels.
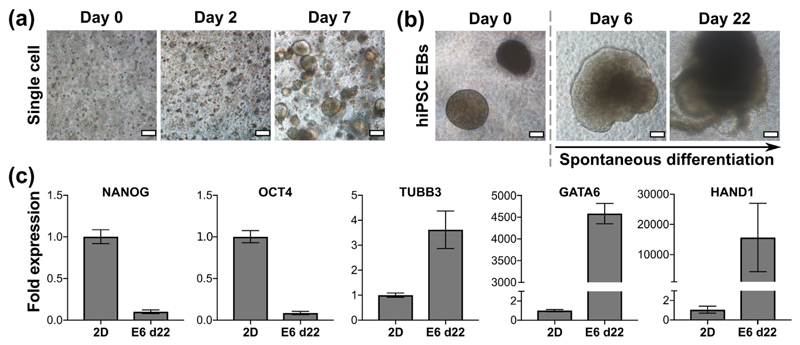
Embryoid bodies spontaneously differentiate in matrix-free peptide gels
Increasingly, researchers and industry are recognising that the extracellular matrix (ECM) plays a key role for in vitro control of stem cell pluripotency and differentiation, similar to its essential role for in vivo development [26–28]. However, human induced pluripotent stem cells (hiPSCs) are notoriously difficult to culture and differentiate reliably into mature functional cell types, often requiring illdefined complex matrices and/or the inclusion of small molecule inhibitors of intracellular signalling. This is a problem in the field as potential therapeutic applications and the need for robust reproducibility in disease models requires batch-to-batch consistency and xeno-free/GMP-compliance of components used to support differentiation. As shown in Fig. 4(a), hiPSCs encapsulated in the peptide gels as single cells formed round, well-defined colonies by day 7 when cultured in E8 medium; a defined, xeno-free formulation. To drive rapid differentiation, hiPSCs were induced to form embryoid body (EB) type clusters using the hanging-drop method. Following encapsulation of these clusters in the peptide gel in E6 medium (xeno-free and lacking the FGF-2/TGFβ required to maintain hiPSC pluripotency) we observed good viability and dramatic changes in cell morphology as expected for differentiating colonies, Fig. 4(b). To validate this, RNA was successfully extracted from the hiPSC EBs encapsulated in peptide gels after 22 days of differentiation and used to perform qRT-PCR for quantification of pluripotency and lineage-specific differentiation, Fig. 4(c). Expression of NANOG and OCT4 (pluripotency) were markedly decreased compared with control hiPSCs grown in E8 in 2D, and three germ layer markers TUBB3 (ectoderm), GATA6 (endoderm) and HAND1 (mesoderm) all increased. This therefore supports the application of the peptide gels as synthetic, fully-defined 3D environments to support hiPSC differentiation with the potential to add functionality to direct differentiation and enhance maturity in lineages of interest.
Fig. 4.
hiPSC growth and differentiation seeded in matrix-free 6 mg/mL peptide gels. (a) hiPSC seeded as a single cell suspension at 1 × 106 cells/mL, (b) embryoid bodies (EBs) formed of 2000 cells per EB, seeded at 8–10 EBs/gel and maintained in E6 medium. Scale bar 100 μm. (c) qPCR results for EBs maintained to day 22 in E6. Fold expression is shown relative to hiPSC grown in 2D on vitronectin to day 3, normalised to RPLPO. OCT4 and NANOG are used as markers of pluripotency, whilst TUBB3, GATA6 and HAND1 are used as markers of ectoderm, endoderm and mesoderm respectively. Graphs show mean ± SEM for n = 2 independent experiments.
Control of matrix composition
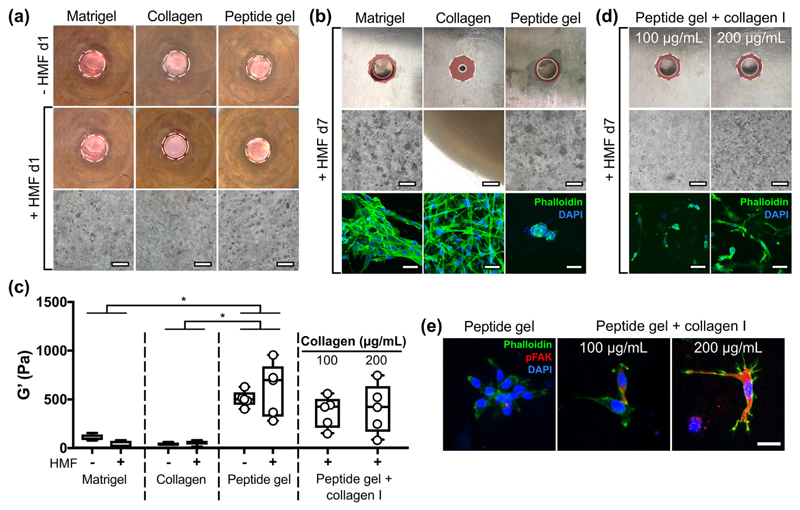
The matrix-free environment of the peptide gels prevents attachment and spreading of fibroblasts (as seen in Fig. 2), in contrast to their typical adherent culture morphology. To examine this, HMF were encapsulated in 100% Matrigel™, rat tail collagen I or the peptide gel. Fig. 5(a) shows that, after only 24 h, the presence of matrix influences cell behaviour. Relative to the cell free condition, a small degree of contraction is evident in the presence of cells for the collagen gel, but not for the other materials. The fibroblasts display a typical elongated morphology in the collagen gel by day 1, and by day 7 (Fig. 5(b)) this is also evident in 100% Matrigel™, but not in the peptide gel. As shown in Fig. 5(b), by day 7 the collagen gel has become a dense mass as contraction continues. Matrigel™ has also undergone a modest degree of contraction at day 7, with no contraction observed in the matrix-free peptide gel. Phalloidin staining of F-actin revealed that the HMF formed a regular network of elongated cells in collagen, dense clusters of elongated cells in Matrigel™, and sparse clusters of rounded cells in the peptide gel. This provides additional evidence for the lack of cell-attachment motifs within the non-functionalised gel, making them effectively inert to adhesive cell types.
Fig. 5.
Human mammary fibroblast (HMF) elongation and contraction in peptide gels is influenced by matrix modifications. (a) Macroscopic contraction and HMF morphology at day 1 in 100% Matrigel™, 1.5 mg/mL rat tail collagen I gel and unmodified 6 mg/mL peptide gel, all seeded at 5 × 105 cells/mL; (b) contraction and HMF morphology with Phalloidin staining at day 7; (c) box plots (n = 5) of bulk oscillatory rheology measurements of each condition at day 1, with and without encapsulated HMF (individual Matrigel™ and collagen data points not included for clarity), along with results from peptide gels with increasing collagen I modification; (d) contraction and HMF morphology at day 7 in peptide gels with increasing collagen I modification. (e) pFAK staining of modified and unmodified peptide gels at day 7. Bright field images have been sharpened (using Fiji software) to facilitate comparison of fibroblast morphology. All macroscopic images are 3 mm across, and contraction is shown relative to the cell free condition at day 1: original images can be found in Supplementary Fig. 2. Scale bar 100 μm (bright field images), 50 μm (Phalloidin images), 25 μm (pFAK images). *indicates p < 0.05 (two-way ANOVA with Tukey post-hoc).
Bulk oscillatory rheology was used to measure the mechanical properties of each material, and to assess the impact of encapsulated cells (Fig. 4(c) and Supplementary Fig. 2). All measurements were taken at day 1 after seeding, since cell-matrix interactions were observed at this time point in the absence of dramatic changes to the macroscopic material properties. The presence of cells does not influence gel stiffness at day 1, however, the peptide gel displays significantly higher stiffness (G') than either Matrigel™ or collagen (Fig. 5(c)). The lack of interaction between cells and the peptide gel could therefore be influenced by stiffness or matrix composition. To test this, fibroblasts were seeded into peptide gels containing collagen I at 100 or 200 μg/mL. Although no significant change in stiffness or viscoelasticity (Supplementary Fig. 2) was observed relative to the unmodified peptide gel, doubling collagen concentration resulted in a greater degree of cell elongation, as well as modest gel contraction (Fig. 5(c) and (d)). This suggests that interaction between HMFs and the peptide gel is determined primarily by matrix composition rather than stiffness. To validate these results, we examined the distribution of phosphorylated focal adhesion kinase (pFAK) within the encapsulated HMF. The increasing intensity of pFAK staining with increased collagen concentration validates the observed differences in HMF morphology, and demonstrates how simple functionalisation can alter behaviour of encapsulated cells by controlling their cell-matrix interaction.
Distinguishing exogenous and endogenous matrix
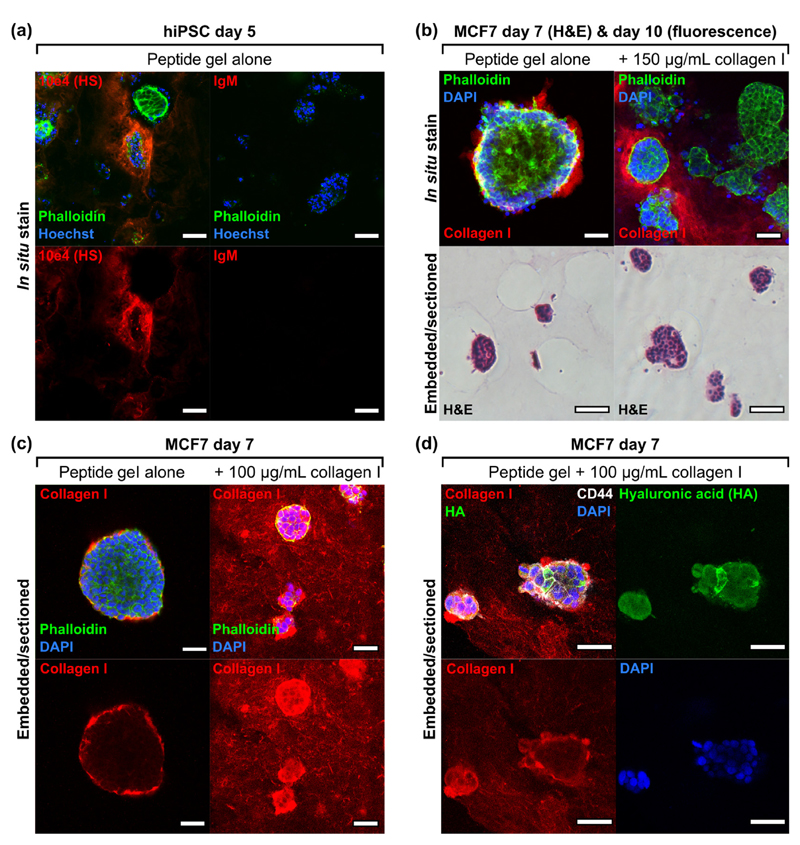
The ability to discriminate and visualise matrix functionalisation (exogenous) as opposed to cell-deposited matrix (endogenous) is crucial for application of the peptide gels to the study of cell-matrix interactions. Immunostaining and microscopy can be used to detect heparan sulphate deposition by hiPSC (Fig. 6(a)) and collagen I deposition by MCF7 (Fig. 6(b), upper panel) seeded in matrix-free peptide gels, highlighting predominantly cell-associated but non-uniform distribution in the 3D cultures.
Fig. 6.
Exogeneous and endogenous matrix may be distinguished using immunostaining in 6 mg/mL peptide gels. (a) Heparan sulphate (HS) deposition by human induced pluripotent stem cells (1 × 106 cells/mL), (b) collagen I localisation on culture with MCF7 in unmodified peptide gel or with collagen I modification, with corresponding H&E (5 × 105 cells/mL), (c) improved collagen I localisation on embedding and sectioning (seeding density reduced to 1 × 105 cells/mL for 100 μg/mL collagen to avoid overconfluence at day 7 in this condition), (d) co-stain for exogenous collagen I and endogenous HA (biotinylated hyaluronic acid binding protein, bHABP, detected using TRITC-streptavidin) in peptide gel modified with collagen I (1 × 105 cells/mL). Negative control images can be found in Supplementary Fig. 4. Scale bar 100 μm for fluorescence images in panel (b), otherwise all scale bars are 50 μm.
When we applied the same technique to visualise collagen I in a modified peptide gel, again confocal microscopy revealed a positive collagen I signal, although this showed some spatial heterogeneity as displayed in the single z-slice in Fig. 6(b), upper right panel. It was not possible to distinguish using this technique whether this heterogeneity was a true reflection of the collagen I localisation within the gel, or a limitation of the in situ (whole gel) staining and imaging method. To clarify this, gels were embedded in agar blocks to allow sectioning. As a first test, the agar blocks were paraffin-embedded to allow microtomy and haematoxylin and eosin (H&E) staining (lower panel, Fig. 6(b)). This method was successful in revealing the cross-sectional structure of cell clusters, whilst preserving gel integrity. To facilitate immunostaining of collagen I throughout the gel, the agar blocks were thick-sectioned at 500 μm using a vibratome, rather than paraffin-embedding. This alternative approach meant that the resulting hydrated sections could be immunostained directly, avoiding the rehydration and antigen retrieval stages necessary following paraffin-embedding (see Supplementary Fig. 3). As shown in Fig. 6(c), this method was successful for immunostaining both endogenous and exogenous collagen I. Multiple matrix components could be visualised clearly in vibratome-generated sections (Fig. 6(d)), where the same MCF7 structures grown in peptide gel with collagen I additions showed endogenously deposited hyaluronic acid (HA). We observed a clear distinction between the relatively homogeneous distribution of collagen I (exogenous matrix) and the cell-localised deposition of HA (endogenous matrix). Increased intensity of collagen I staining was observed around cell clusters, likely to be the result of both endogenous collagen I production (see Fig. 6(c)) and localised interactions of the cells with the exogenous collagen. Again, the peptide gel model provides the opportunity to access and probe these events, assaying changes in matrix composition and organisation driven by reciprocal interaction with encapsulated cells.
Independent control of matrix stiffness
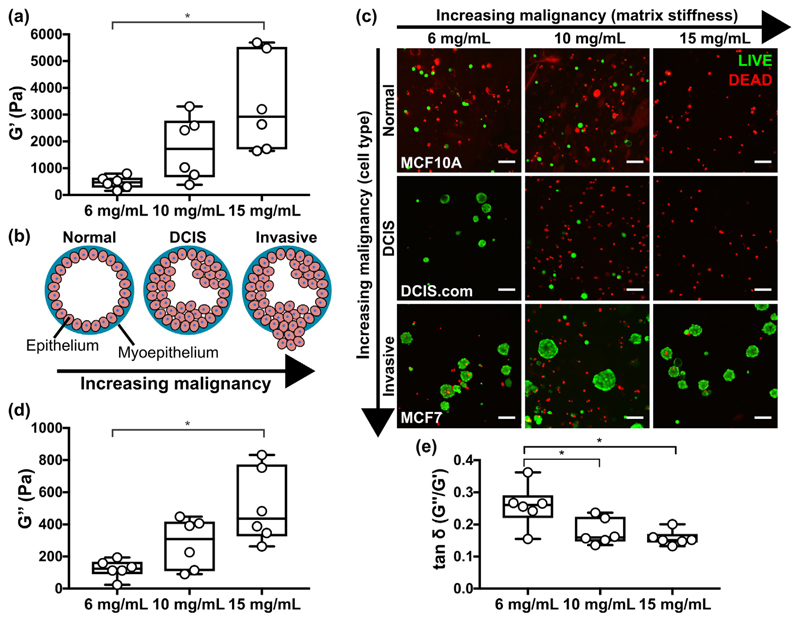
A benefit of the peptide gels is that their mechanical properties may be altered with no change in matrix composition, simply by controlling the amount of peptide preparation added to the precursor. We used this approach to create peptide gels with G' values spanning an order of magnitude, from 500 Pa to 5 kPa (Fig. 7(a)). Importantly, this range is relevant to human tissue: increasing the peptide gel concentration from 6 to 15 mg/mL yields a change in G' representative of the increasing tissue stiffness associated with tumourigenesis in breast cancer, Fig. 7(b) [29]. This raised the interesting question of how human breast cell lines representative of different stages of malignancy would respond to culture under this range of stiffness conditions. Fig. 7(c) shows the results of a cell viability stain following 7 days culture of three cell lines in each peptide gel concentration: MCF10A (non-tumourigenic, normal breast), MCF10DCIS.com (ductal carcinoma in situ, pre-invasive) and MCF7 (invasive breast cancer). Interestingly, the more malignant the cell type, the greater its viability in the peptide gels, with only MCF7 forming stable clusters across all conditions. Limited DCIS.com acinar growth was also seen in the lowest peptide gel stiffness. Otherwise, no acini were observed, and very few viable MCF10A or DCIS.com were detected in the highest stiffness 15 mg/mL gels. Matched quantification of gel stiffness and cell viability within a representative experiment (Supplementary Fig. 5) confirmed that changes in stiffness with peptide concentration were constant between the cell lines, and therefore that the changes in viability are cell-type specific responses to their mechanical environment. In addition to the effect of matrix stiffness, it is also of note that the viscoelastic response of the gels, as quantified by the loss modulus G“ and the G“/G' ratio tan δ (Fig. 7(d, e)), also changes with peptide concentration. In particular, tan δ shows a significant decrease in magnitude with increasing peptide concentration. This indicates a decreasing relative contribution from the viscous (energy loss) material response: a factor recently identified as crucial for determining the extent of matrix remodelling [30]. Even in the absence of relevant matrix proteins and glycosaminoglycans, the peptide gels may therefore be used to probe the effect of mechanical environment on cell behaviour and relative survival, as demonstrated using cell models of breast cancer progression.
Fig. 7.
Increasing peptide gel concentration produces corresponding increase in stiffness. (a) Box plots (n = 6) showing G' results of increasing peptide gel concentrations characterised using bulk oscillatory rheology, (b) schematic of the increase in malignancy in breast cancer associated with increasing matrix stiffness, (c) LIVE/DEAD staining of breast epithelial cell lines MCF10A, DCIS.com and MCF7 in peptide gels of increasing stiffness. Box plots (n = 6) showing (d) G“ and (e) tan δ measurements correspond to the G' values shown in (a). Scale bar 100 μm. *indicates p < 0.05 (one-way ANOVA with Tukey post-hoc).
Enhancing cell viability using multicellular models
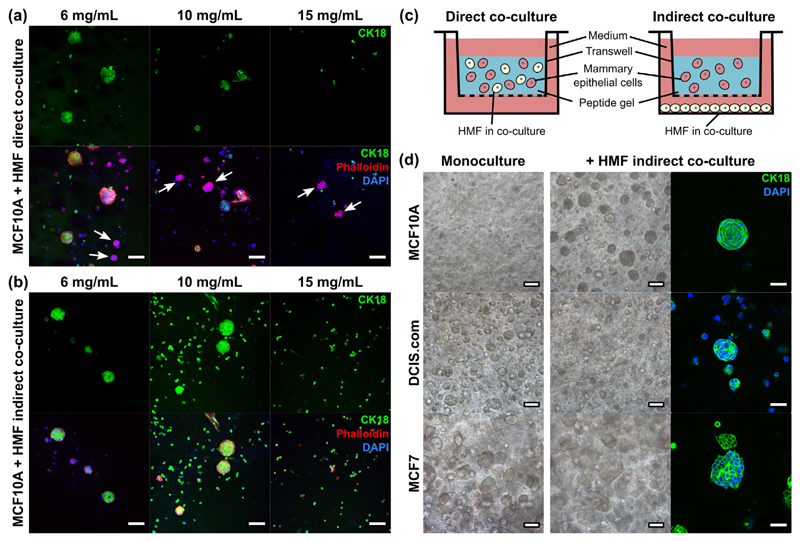
Another degree of freedom in peptide gel design is the ability to add more than one cell type in co-culture. This may be achieved by encapsulating multiple cell types within the same gel (Fig. 8(a)) or alternatively by culturing the peptide gel in a transwell insert, with a second cell type at the base of the well (Fig. 8(b)). This approach allows cell-cell interactions to be studied when the cells are in direct or indirect co-culture, as illustrated in Fig. 8(c). Co-culturing MCF10A breast epithelial cells with HMFs allows the MCF10A to form clusters in both 6 mg/mL and 10 mg/mL peptide gels, either in (a) direct or (b) indirect co-culture - in contrast to their lack of viability in stiffer gels in monoculture (see Fig. 7(c)). A CK18 (epithelial cell specific) co-stain with Phalloidin was used to distinguish between MCF10A and HMF clusters, revealing that, in the direct co-culture conditions, HMF formed small, distinct clusters in all peptide gel concentrations (Supplementary Movie 1). Interestingly, the ability of HMFs to support MCF10A viability does not require direct contact, with MCF10A acini observed in indirect co-culture in the 6 mg/mL and 10 mg/mL gels and single cells/small clusters in 15 mg/mL gels. The lack of Phalloidin+/CK18− cells verifies that no HMFs were able to penetrate the transwell filter and migrate into the gel during indirect co-culture. Comparing the impact of indirect co-culture with HMFs between MCF10A, DCIS.com and MCF7s, the normal breast cell line demonstrated the most marked difference in growth with stromal cell conditioning of the gel enabling the MCF10As to form tight cell clusters. Co-culture in non-functionalised gels had less impact on the morphology or growth of encapsulated DCIS.com or MCF7s (Fig. 8(d)).
Fig. 8.
Co-culture with HMF supports MCF10A viability. (a) Direct co-culture of both MCF10A and HMF within the gel, white arrows indicate CK18− clusters, i.e. HMF, (b) MCF10A within the gel with HMF indirect co-culture, (c) schematic illustrating the set-up of each co-culture variant, (d) indirect co-culture of the three breast epithelial cell lines MCF10A, DCIS.com and MCF7 (6 mg/mL peptide gels). Each cell type was seeded at 5 × 105 cells/mL, shown at day 7. Scale bar 50 μm for CK18 stain in panel (d), otherwise all scale bars are 100 μm.
Matrix additions support MCF10A 3D culture
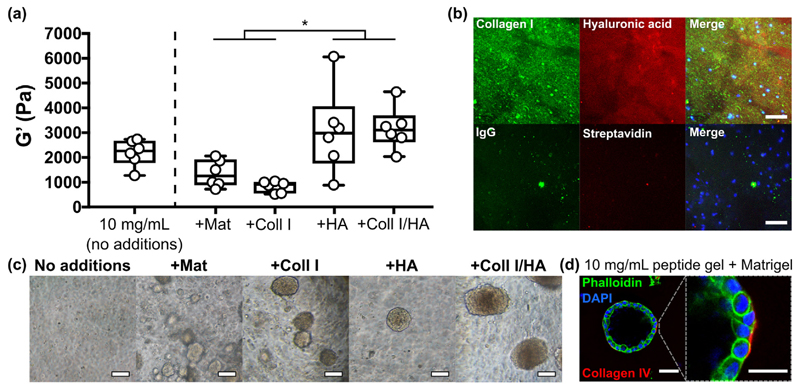
Building on our initial observation that adding ECM components to the peptide gel altered stromal cell growth and viability (Fig. 5), we investigated the effect of varying matrix composition on MCF10A cell behaviour. Rheological characterisation of 10 mg/mL peptide gels (Fig. 9 (a) and Supplementary Fig. 6) showed that whereas additions of 100 μg/mL collagen I or 20% Matrigel™ produced stiffnesses closer to that of normal breast tissue (G' = 800 ± 200 and 1400 ± 500 Pa respectively, mean ± SEM), the addition of 804 kDa HA significantly increased peptide gel stiffness in either the presence or absence of collagen I (G' = 3200 ± 400 and 3000 ± 700 Pa respectively). Interestingly, peptide gels with HA modifications alone showed behaviour closer to an elastic solid than the other modified conditions, as characterised by a significant decrease in tan δ (Supplementary Fig. 6(b)). Immunostaining (or use of an HA binding peptide) enables visualisation of the matrix components added to the gel, demonstrating that both components are homogeneously distributed throughout the gel (Fig. 9(b)).
Fig. 9.
MCF10A growth and morphology is dictated by matrix additions. (a) Box plots (n = 6) showing bulk oscillatory rheology of 10 mg/mL peptide gels with matrix modifications, (b) immunostaining of collagen I and HA distribution in the modified 10 mg/mL peptide gels seeded with MCF10A (DAPI, blue, scale bar 50 μm), (c) acinar morphology in modified 10 mg/mL gels at day 14 (scale bar 100 μm), (d) single cell acinus formed in 20% Matrigel™ at day 14 (scale bar 50 μm (left), 25 μm (right)). *indicates p < 0.05 (one-way ANOVA). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In contrast to the lack of growth observed in the unmodified peptide gel, MCF10A 3D growth and the formation of cell clusters was supported in all modified conditions (Fig. 9(c)). However, only the 20% Matrigel™ condition was able to promote formation of the classic polarised single-cell layer acini typically observed in 100% Matrigel™ (Supplementary Fig. 7), with peripheral collagen IV deposition (indicative of a nascent basement membrane) by day 14 (Fig. 9(d)). This highlights that additional or alternative influences, beyond functionalisation with collagen I and HA, are required for this complex cell behaviour, in agreement with previously published studies [31,32].
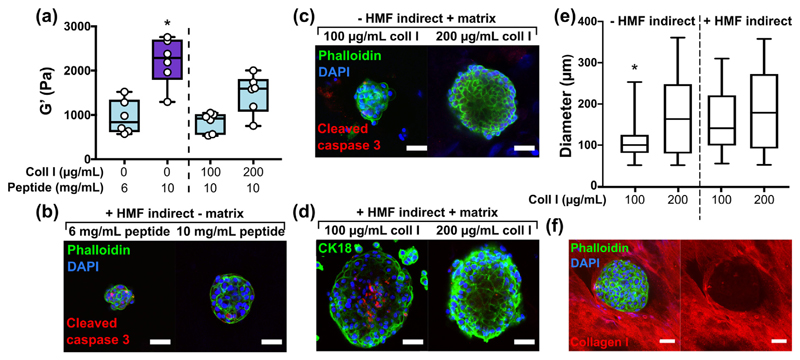
We next investigated how independent control of matrix stiffness, composition and co-culture conditioning could alter MCF10A organisation. Functionalisation with collagen I significantly decreased the stiffness of a 10 mg/mL peptide gel (Fig. 10(a) and Supplementary Fig. 6). Therefore, a 6 mg/mL peptide gel with equal stiffness to the collagen-containing conditions was included as a matrix-free control. After 14 days of culture, immunostaining revealed that neither indirect co-culture with HMFs (Fig. 10(b)) nor the presence of collagen I (Fig. 10(c)) was sufficient to produce organised acinar structure, indicated here by the lack of focused cleaved caspase 3 staining in the core of MCF10A clusters.However, where applied in combination, collagen I and HMF co-culture produced large acini, with evidence in the 100 μg/mL collagen I condition of organised cleaved caspase 3 staining, an early stage of lumen formation (Fig. 10(d)). Quantification of acinar diameter (Fig. 10(e)) revealed that HMF co-culture produced a significant increase in acinar size in peptide gels containing 100 μg/mL collagen I. A significant increase in diameter was also observed on increasing collagen concentration from 100 to 200 μg/mL, although the acini formed in the 200 μg/mL condition were dense with no central lumen, as shown in Fig. 10(b).
Fig. 10.
Combined control of matrix additions and co-culture influences MCF10A organisation. (a) Box plots (n = 6) showing bulk oscillatory rheology results of peptide gels with and without matrix modifications, *indicates p < 0.05 relative to all other conditions (one-way ANOVA), (b) acinar morphology on indirect co-culture with HMF, (c) acinar morphology in MCF10A monoculture in 10 mg/mL peptide gels with matrix additions, (d) acinar morphology on indirect co-culture with HMF in 10 mg/mL peptide gels with matrix additions, (e) quantification of acinar diameter for each condition, *indicates p < 0.05 relative to all other conditions (Kruskal-Wallis, >25 acini across 2 independent experiments per condition), (f) immunostaining of an acinus surrounded by collagen I matrix in a 10 mg/mL peptide gel modified with 200 μg/mL collagen I. Scale bar 50 μm.
Interestingly, although it is clear that additional or alternative matrix functionalisation is required to promote further acini maturation, the MCF10As were able to organise and re-engineer their surrounding matrix, indicating reciprocal interactions between the encapsulated cells and their local environment. After 14 days MCF10A culture in a peptide gel containing 200 μg/mL collagen I, Fig. 10(f), the epithelial cells appear to distort the collagen, with collagen excluded entirely from the cell cluster and surrounding the acinus at its periphery.
Experimental procedures
Cell line maintenance
The human mammary fibroblast cell line HMFU19 (a gift from Professor Mike O'Hare, Ludwig Institute, London, UK), leukemia cell line U937 (DSMZ GmBH) and colorectal cancer line mCherry-HCT116 (a gift from Prof. Anna Grabowska, University of Nottingham) were cultured in HMF cell culture medium: RPMI-1640 with 10% fetal bovine serum (FBS) and 1% l-glutamine. The murine embryonic stem cell line E14TG2a and Oct4-GFP reporter line (both gifts from Prof. Austin Smith, University of Cambridge, UK) were maintained on tissue culture flasks coated with 0.1% gelatin (G1890 Sigma), in knockout Dulbecco's modified Eagle medium (DMEM) with 10% FBS (HyClone), 1% Non Essential Amino Acids, 1% l-glutamine, 0.1% β mercaptoethanol, and leukemia inhibitory factor (LIF, ESG1107 Millipore). The breast cancer cell line MCF7 and the modified tdTomato-MCF7 (a gift from Prof. Anna Grabowska, University of Nottingham) were maintained in high glucose DMEM with 10% FBS and 1% l-glutamine. The breast cell line MCF10DCIS.com (Asterand) was cultured in advanced DMEM with 5% horse serum and 1% L-Glutamine. MCF10A were maintained in DMEM/F12 (D8062 Sigma) with 5% horse serum, 1% L-Glutamine, 10 μg/mL insulin (I9278 Sigma), 0.5 mg/mL hydrocortisone (50237 Tocris), 20 ng/mL epidermal growth factor (ABC016 Source Biosciences) and 100 ng/mL cholera toxin (C8052 Sigma). All cell lines were obtained from ATCC unless specified.
hiPSC culture and EB formation
For human induced pluripotent stem cell experiments (hiPSC), REBL-PAT (non-disease) cells were used as established and characterised previously [49]. hiPSC were maintained in Essential 8 medium (E8) on tissue culture flasks coated with recombinant vitronectin peptide (VTN-N) following manufacturer's instructions. For hiPSC passage, cells incubated with TrypLE Express were collected in E8 supplemented with 10 μM Y-27632 ROCK inhibitor (72304, Stem Cell Technologies, UK). The hanging drop method used to generate embryoid bodies (EBs) from hiPSCs was adapted from [50]. hiPSCs were harvested 48 h after seeding and resuspended in E8 with 10 μM Y-27632 and 4 mg/mL polyvinyl alcohol (Sigma, UK). 20 μL droplets containing 2000 cells/droplet were pipetted onto the lid of a 10 cm petri dish containing 10 mL PBS to maintain hydration. The EBs were formed for 24 h at 37 °C, then collected in DMEM. EBs were allowed to sediment at the bottom of a 15 ml falcon tube for 10–15 min at 37 °C and were subsequently cultured in peptide gels maintained in Essential 6 medium (E6) to allow spontaneous differentiation.
All cell lines were maintained in antibiotic-free conditions, at 37 °C and 5% CO2 in a humidified atmosphere. All media components were obtained from Gibco, UK unless specified.
Precursor formation
A commercially available peptide preparation in powder form was used as the source of the octapeptide gelator (FEFEFKFK, Phe-Glu-Phe-Glu-Phe-Lys-Phe-Lys). As part of this study we used peptide sourced from Cambridge Research Biochemicals (batch 32597) although we also verified the fabrication method using a second peptide source (Pepceuticals, UK). To form each precursor, a mass of between 7.5 and 18.75 mg peptide preparation was dissolved in 800 μL sterile water (W3500 Sigma), using a 3 min vortex step followed by centrifugation (3 min at 1000 rpm) and a 2 h incubation at 80 °C. After incubation, 0.5 M NaOH (S2770 Sigma) was added incrementally to the gels until optically clear. Gels were vortexed, buffered by addition of 100 μL 10× PBS (70011 Gibco), and incubated at 80 °C overnight. The resulting precursors could be stored at 4 °C until required.
Peptide gel formation
Prior to peptide gel formation, each precursor was heated at 80 °C until liquid to ensure homogeneity, before transferring to a 37 °C water bath. Peptide gel formation was then induced by pH neutralisation on addition of cell culture medium. A final volume of 1.25 mL was obtained from each preparation, by adding 250 μL of cell culture medium to a precursor volume of 1 mL. The end concentration of peptide preparation therefore ranged between 6 and 15 mg/mL. Medium was thoroughly mixed with the precursor by gentle (reverse) pipetting, before plating at 100 μL per well into a 96-well plate, or at 200 μL per well into a hanging insert within a 24-well plate (MCRP24H48 Millipore). The wells were then flooded with cell culture medium and incubated at 37 °C and 5% CO2 in a humidified atmosphere. Sequential media changes (at least two) over the next 24 h ensured complete neutralisation and therefore gelation.
For cell encapsulation, the 250 μL volume of cell culture medium was prepared as a cell suspension at 5× the intended final seeding density, to allow for the dilution factor on mixing with the precursor. Trypsin-EDTA (0.25%), or TrypLE Express in the case of REBL-PAT, was used to detach all adherent cell lines from 2D culture at sub-confluence. Cells were resuspended in 250 μL cell culture medium at a density between 2.5 × 105 and 5 × 106 cells/mL, giving final seeding densities in the peptide gel between 5 × 104 and 1 × 106 cells/mL.
Peptide gel formation with matrix modifications
Modified peptide gels were created using the method above, by incorporating matrix additions into the 250 μL volume added to the precursor. For collagen I additions, rat tail collagen I (A10483 Gibco) was neutralised directly before use with 1 M NaOH according to manufacturer instructions, and diluted with sterile water and 10× PBS to a concentration of 0.5–1.5 mg/mL. For hyaluronic acid (HA) additions, streptococcal HA polymer with molecular weight 804 kDa (HA804 Iduron) was reconstituted in PBS at 0.5 mg/mL and sterilised using a 0.2 μm syringe filter. Corning Matrigel™ (354234 Fisher Scientific) was used for the 20% Matrigel™ condition. All matrix preparations were kept on ice. Modified peptide gels were created by preparing a 250 μL volume containing each matrix component at 5× the desired final concentration (diluted with cell culture medium if necessary), and mixing with 1 mL precursor as described above. Cells were incorporated into this 250 μL volume at 5× the desired final seeding density as described.
Matrigel™ and collagen gels
Neutralised rat tail collagen I was prepared at 1.5 mg/mL as described above, and plated at 200 μL per well into a 24-well plate hanging insert. Corning Matrigel™ was plated in the same way. All solutions were kept on ice during use. To seed cells into these gels, a cell pellet was prepared and suspended in either the neutralised collagen solution or in pure Matrigel™, giving a final seeding density of 5 × 105 cells/mL. Matrigel™ and collagen gels were incubated at 37 °C and 5% CO2 in a humidified atmosphere for 30 min to allow gelation, before flooding the wells with cell culture medium.
Bulk oscillatory rheology
Peptide gel samples were prepared for bulk rheology as described above, by plating at 200 μL per well into 24-well plate hanging inserts, and incubating overnight as described above. At day 1 after seeding, samples were removed from the inserts with a scalpel and mounted onto a Physica MCR 301 rheometer (Anton Paar) with Peltier plate set to 37 °C. The linear viscoelastic region was determined for each sample condition by carrying out an amplitude sweep from 0.1 to 100% strain at 1 rad/s. Following this, a constant strain of 0.5% was used to obtain frequency sweeps from 0.1 to 100 rad/s, as well as 5 min time sweeps at a constant frequency of 1 rad/s. The same tests were carried out on the precursor samples, which could be pipetted directly onto the rheometer plate. All tests were carried out using an 8 mm diameter parallel plate set-up with a spacing of 1 mm.
Microrheology
The microrheological properties of both precursor and peptide gel were tested by measuring the Brownian motion of 2 μm diameter polystyrene beads (19814 Polysciences) embedded into the samples. For the precursor, beads were suspended at a final concentration of 2 × 105 beads/mL by incorporating them with the addition of 10× PBS. This relatively low bead concentration was chosen to avoid clustering and to ensure that only one bead was present in the field of view for the duration of the experiment. After a standard overnight incubation at 80 °C, precursor samples were equilibrated for 1 h at 37 °C and 5% CO2 in a humidified atmosphere prior to transfer into an 8-well coverslip (80821 IBIDI) at 200 μL/well for testing. For the peptide gel, beads were added to the gel by suspension into the 250 μL of cell culture medium used for neutralisation, at a final concentration of 2 × 105 beads/mL. Peptide gels were seeded into the 8-well coverslips and incubated at 37 °C and 5% CO2 in a humidified atmosphere overnight. The beads were imaged in wide-field transmission with a 100× oil-immersion objective lens (numerical aperture = 1.3) using an inverted microscope (Eclipse Ti-S, Nikon) and a CMOS camera (Optimos, QImaging). The trajectories of 5 individual beads per condition were tracked using a centre of mass algorithm. For each bead at least 50,000 frames were recorded at a frame rate of 600 frames per second (exposure time 1500 μs), this high frame rate was achieved by imaging a small region of interest. For some measurements, the number of frames was limited by the bead diffusing out of the field of view (for such gels the adoption of a relatively low laser power for optically trapping the bead of interest could be considered in future studies). In-house LabVIEW programs (LabVIEW, 2013, National Instruments, USA) were used for (i) bead tracking, (ii) trajectory conversion to mean squared displacement, and (iii) for extracting the complex viscosity of the gel immediately surrounding the beads [51]. The experimental set-up was verified by taking control measurements of the viscosity of water (Supplementary Fig. 1). For these experiments, a 1064 nm continuous wave laser (Ventus 1064, Laser Quantum Ltd., UK) operating at spatial Gaussian mode (TEM00) was used to optically trap beads suspended in water with a laser power <5 mW. 500,000 frames were recorded at a frame rate of 600 fps.
Live cell imaging and detection
Fluorescence signal from peptide gels containing fluorescently labelled cells was detected using a Fluostar Omega Plate Reader (BMG LabTech). An Eclipse TI-S microscope (Nikon) was used for bright field imaging during culture. For quantification of cell cluster diameter, all regions containing cell clusters were imaged and Fiji software was used for manual diameter measurement [52]. For LIVE/DEAD staining, peptide gels were washed with PBS, removed from their hanging inserts, and incubated for 15 min in a solution of 40 μM Ethidium homodimer and 20 μM calcein AM (L3224 Fisher) in PBS. A Leica TCS SPE laser scanning confocal microscope was used for acquisition of fluorescence images.
Immunofluorescence staining
After washing in PBS, and removal from hanging inserts if necessary, peptide gels were incubated for 1 h in paraformaldehyde (Polysciences) diluted to 4% (v/v) in PBS. Samples were washed in PBS in preparation for immunofluorescence staining. Samples were incubated in blocking buffer, consisting of 0.1% Triton X-100 and 0.5% bovine serum albumin (Sigma) for 1 h, and incubated overnight at 4 °C with a solution of primary antibody in blocking buffer: Oct4 (83932 Cell Signalling Technologies (CST), 1:400), CK18 (53981582 Thermofisher, 1:50–1:100), rabbit IgG (PP64, Chemicon International, 1:1000), pFAK (Tyr397, 44-624G Thermofisher, 1:100), collagen I (ab34710 AbCam, 1:100–1:500), collagen IV (ab6311 AbCam, 1:200), cleaved caspase 3 (9661 CST, 1:400) or CD44 (3570 CST, 1:400). After further washes in blocking buffer, samples were incubated overnight at 4 °C with a solution of secondary antibody in blocking buffer (a21042/a21050/a11010/a11029/a11034 Invitrogen, 1:400). For HA staining, biotinylated hyaluronic acid binding protein (bHABP, AMS.HKD-BC41 AMSBio, 1:100) was added with the primary antibody, and TRITC-streptavidin (Stratech) with the secondary antibody. Samples were incubated in a 300 nM DAPI solution (D3571 Invitrogen) for 1 h at room temperature prior to imaging. For heparan sulphate staining (10e4, 370255-1 AMSBio, 1:100), 10% goat serum (Sigma) in PBS was substituted as blocking buffer, with Hoechst substituted in place of DAPI counterstain. Where Phalloidin was used for F-Actin staining (F432/R415, Thermofisher, 1:1000), this was added either alone or with the secondary antibody, as a solution in blocking buffer as described above.
Embedding and sectioning
Peptide gels were embedded in a 2–4% solution of agar (SLS) in distilled water, and set for several hours at 4 °C prior to further processing. Agar blocks were sectioned using a Leica Vibratome at a thickness of 500 μm. Agar slices were stored in PBS, and were stained using the same immunofluorescence techniques described above. Alternatively, the agar blocks were transferred to a tissue processor and set in a paraffin block. 10 μm slices were sectioned onto SuperFrost slides (Thermo Scientific, UK) using a microtome. After drying at 37 °C, slides were dewaxed in xylene and rehydrated in an ethanol series to allow staining with Haematoxylin and Eosin (3 min each). Slides were washed in running water at each stage, and incubated for 20 s in acid alcohol and 1 min in Scott's tap water between stains. Slides were dehydrated, cleared in xylene, and coverslipped using DPX mounting medium (Thermo Scientific, UK).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted and purified from cells within peptide gels using the Nucleospin RNA kit (Machery Nagel) according to the manufacturer's instructions. 700 μg of RNA was reverse-transcribed to cDNA using SuperScript™ IIIReverse Transcriptase, following the manufacturer's instructions. The GoTaq qPCR Master Mix (Promega) was used to run the PCR reaction on a LightCycler® 480 (Roche). Human RPLPO and HSP90AB1 were used as reference genes for normalisation and hiPSCs cultured in 2D were used to calculate relative expression using the ΔΔCt method. Primers used are listed in Supplementary Table 1.
Statistics
Prism v.7.0d and SPSS v.24 (IBM) were used for statistical analysis. One-way or two-way ANOVA with Tukey HSD post-hoc test were used as appropriate where measurements were normally distributed, verified by the Shapiro-Wilk test. In the one case where the data were not normally distributed (the acini diameter measurements in Fig. 10), a non-parametric Kruskal-Wallis test was used with Bonferroni correction for multiple comparisons. Statistical significance for all tests was declared at p < 0.05.
Discussion
An increasing awareness of the importance of matrix components in regulating cell behaviour has necessitated the continued improvement of in vitro 3D models of human tissue. As recently highlighted in a high-profile technology feature, the ECM governs a surprising number of cellular functions, which must be adequately modelled in vitro to better understand development, differentiation and the progression to disease [1]. A major hurdle to this has been unpicking the multitude of influences exerted by the matrix on neighbouring cells. The self-assembled peptide gel presented here allows the independent control of two critical factors: matrix composition and bulk stiffness. This in turn facilitates the application of customised (“bespoke”) gels to mimic multiple matrix microenvironments tailored for specific applications.
Self-assembling peptide gels are intermediates between natural and synthetic materials; chemically synthesised, but formed from natural building blocks, with a biomimetic, fibrillar nanostructure [23,33]. Here, the octapeptide gelator sequence FEFEFKFK was chosen due to the high biocompatibility demonstrated by gelation at pH 7 at 37 °C [15]. Previous studies have explored the application of similar gels for regenerative medicine applications [15,34] however, to our knowledge they have not yet been used to create fully-defined matrices for cell encapsulation. By exploiting the pH-dependent viscosity of the gels, we could physically incorporate cells and/or matrix components into the precursor, and these remained homogeneously distributed on final gelation.
Previously, decoupling of matrix stiffness and composition to detect their respective influence on cell behaviour has often been conducted in the presence of a complex matrix environment. For instance, ribose-mediated collagen cross-linking was used to increase the stiffness of a collagen-rBM (reconstituted basement membrane) composite, demonstrating that ErbB2 signalling was necessary to promote mammary epithelial invasion in stiff matrices [9]. In an alternative approach, rBM was combined with alginate gels to determine the interplay between matrix stiffness and rBM concentration, with greater epithelial cell malignancy observed in stiffer matrices only when rBM concentration was held constant [18]. A similarly elegant approach combined collagen with methacrylated gelatin, allowing independent control of collagen concentration and matrix stiffness [35]. Using this system, the authors discovered that MDA MB 231 breast cancer cell invasion was best supported by matrices with low stiffness but high collagen concentration. By using a non matrix-derived self-assembling peptide gel as a starting point, the method described here is distinct from these approaches in that it not only allows independent control of matrix and mechanical properties, but also allows the inclusion of selected matrix components, specific for the application. This flexibility is permitted by the two-stage fabrication method; the first stage creates a matrix-free precursor to define the stiffness, the second stage defines the composition.
We were able to demonstrate the increase in viscosity during gelation of the matrix-free gel using two distinct measurement techniques: bulk oscillatory rheology, and microrheological measurements based on the Brownian motion of micron sized beads embedded in the gel. Indeed, despite the absolute measurement values obtained from the two methods differing by approximately an order of magnitude (likely due to the different length scales explored by the two techniques), the relative increase in viscosity on gelation was found to be very similar between the two experimental procedures. Nonetheless, bulk rheology is a well-established experimental method, allowing us to compare the viscoelastic properties of the peptide gel with those of other established 3D cell culture platforms. Whereas naturally derived gels, such as Matrigel™/rBM and collagen tend to be far less stiff than the tissues they are used to mimic [23], we are able to control the peptide gel storage modulus in the range 500–5000 Pa, which covers a wide range of in vivo tissue stiffnesses, such as brain and breast [23,29,36]. It is important to note, however, that bulk rheology is not well-suited to measuring cell-induced changes in gel mechanical properties during culture. This is because it cannot measure local material changes at the length scale of a single cell. Therefore, we are developing a novel microrheology method, combined with the ability to optically trap a bead in the case of low modulus gels [37–39], to measure the gels' mechanical properties at a cell-scale; particularly at the cell-matrix interface, as cells re-engineer their microenvironment.
In good agreement with our current understanding of cell-matrix interactions, anchorage- independent cells proliferate within unmodified peptide gels, with the lack of matrix attachment motifs effectively enabling the cells to form structures similar to those seen in suspension culture. Adhesive cell types, such as fibroblasts, additionally require matrix components, such as collagen I, to achieve their characteristic morphology. Importantly, the gel formulation method presented here allows for biochemical functionalisation, whilst also providing control conditions with matched peptide gel stiffness. Although cell adhesion motifs have classically been considered necessary for interactions between matrix stiffness and cell behaviour [40], we have also shown a clear link between stiffness and cell response in the absence of cell binding sequences. It has recently been demonstrated that cells encapsulated in 3D materials rapidly synthesise their own matrix, with initial matrix stiffness implicated as a key factor determining the extent of this early matrix deposition [30,41]. Importantly, the peptide gels allow independent assessment of the effects of biochemical functionalisation and of the mechanical environment initially presented to encapsulated cells.
In trying to create artificial culture environments where the control of biochemical and physical properties is required, researchers often combine naturally derived and synthetic elements. This can be seen in the chemical modification of natural materials, e.g. hyaluronic acid with thiol modifications to allow cross-linking, or in the incorporation of biological components into synthetically produced hydrogels [23] as well as the current study. A particularly successful approach is that taken by the Lutolf group, functionalising synthetic polyethylene glycol (PEG) hydrogels with laminin-111 to produce highly complex tissue models, including the human intestine [13]. Matrix remodelling by encapsulated cells can be regulated and reported by the inclusion of matrix metalloproteinase (MMP) cleavable cross-links or reporters [23]. Within the peptide gels, encapsulated cells appear to readily organise and remodel their surrounding matrix without the need to engineer in specific cleavage sites. The ability to image both endogenous and exogenous matrix within the fully-synthetic system is a significant advantage here and will enable monitoring of matrix reorganisation as cells grow, differentiate and migrate. Another feature of importance to cell biologists is the optical transparency of the peptide gel that enables simple assessment of cell growth in real-time by microscopy. The ability to use automated plate readers to read-out endogenous fluorescence as well as end-point analysis by immunostaining and fluorescent microscopy is likely to be useful for high-throughput analyses and applications such as toxicity screening or biomarker identification.
The ability to investigate the impact of stromal cells in co-culture with epithelial cells is particularly valuable when studying the microenvironmental control of cancer initiation, growth and metastasis. Bidirectional cross-talk is thought to promote cancer progression, with exosomal-mediated signalling between neighbouring cells likely to play an important role [42–44]. Stromal cells can additionally alter the hormone-dependence of nearby epithelia: for instance estrogen treatment of uterine epithelial cells increased their proliferation only when in culture with stromal cells [45]. For some effects, direct cell contact appears to be unnecessary, with the exposure of epithelial cancer cells to stromal-conditioned media sufficient to alter their sensitivity to chemotherapy and radiation [46]. In the current study, we were able to demonstrate differences in the behaviour of encapsulated cells when in the presence of direct or indirect stromal cell co-culture, with the flexibility of the culture system providing a useful test environment in which to study the regulation of cancer cells by their microenvironment.
As demonstrated in multiple studies, a benefit of using short peptides to create hydrogels for cell encapsulation is the ability to simply and cheaply engineer in covalently immobilised peptidic functional motifs e.g. matrix-derived cell adhesion sequences (RGD, IKVAV etc.) A less reductionist approach can also be achieved by functionalisation with complex sequences, combining cell attachment and proteolytic motifs [47]. These approaches have clear benefits for mechanistic investigations and additionally highlight the potential for using peptide gels to move to fully synthetic xeno-free, matrix-inspired 3D culture. In the current study, we chose to incorporate full-length proteins and glycans to enable the visualisation of cell-mediated changes in matrix organisation, detailing cell contraction and immunocytochemical imaging of matrix organisation and synthesis. Importantly, by using “naked” matrix free gels, we can also detail matrix deposition by encapsulated cells. This is particularly valuable when studying glycans such as heparan sulphate where the conservation of structure between species makes it impossible to differentiate glycosaminoglycans deposited by encapsulated (human) cells from those present in complex animal-derived matrices (e.g. Matrigel™). Cell-deposited matrices have themselves been used for 3D culture [5] and the study of differential ECM deposition under different conditions, such as stromal activation in the presence of cancer cells [48] is increasingly studied to identify potential targets for novel therapeutic strategies. The use of indirect stromal cell co-culture to effectively condition the peptide gels will hopefully prove useful in these studies.
Conclusions
In summary, we present the optimisation of a well-established, simple and relatively inexpensive peptide gel for the study of cell-matrix interactions in a wide variety of cell types. By eliminating or significantly reducing the need for animal-derived components e.g. Matrigel™, this synthetic gel also helps researchers move away from the batch-to-batch variability associated with their use, and addresses the need to replace, refine and reduce the use of animals in research. The cell encapsulation protocol has been specifically designed to ensure that reliable, reproducible 3D culture is achievable within a standard cell culture laboratory setting with independent control of the biochemical and mechanical influences of the matrix microenvironment. In this study, as well as demonstrating broad applicability across multiple adhesive and non-adhesive cell types, we have demonstrated how the peptide gel can be applied to unpick the role of extracellular regulation on the behaviour of cell lines used to model the progression from normal breast to breast cancer. We hope the peptide gels will be of interest to the matrix biology community, with the optimised protocol and commercially available precursors ensuring that the technology is approachable for any cell culture laboratory.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.matbio.2019.06.009.
Acknowledgements
The research described was funded by the National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs) grant NC/N001583/1 (J.A., G.F., C.L.R.M.), and supported by the EPSRC/BBSRC/MRC (Engineering and Physical Sciences Research Council/Biotechnology and Biological Sciences Research Council/Medical Research Council) joint grants EP/R035067/1, EP/R035156/1 and EP/R035563/1 (M.T., A.J.W., C.L.R.M.), MRC grant MR/P003214/1 (K.P.A.), EPSRC/MRC CDT Regenerative Medicine and Faculty of Medical and Health Sciences (J.R.J., A.T., A.J.W., C.L.R. M.), EPSRC grant EP/N006615/1 (J.L.T., C.L.R. M.) and Swedish Research Council grant 2015-06532 (S.P.G.). We thank Robin Ketteler and Alexander Agrotis for technical help and the MRC for support of the LMCB High-Content Biology Laboratory at University College London (MC_U12266B, MR/M02492X/1). We thank Anna Grabowska and Pam Collier (University of Nottingham) for their kind gift of the labelled HCT116-mCherry and MCF7-td Tomato cell lines and for advice on real-time cell assays, and Tony Day (University of Manchester) for helpful advice and discussions.
Footnotes
CRediT authorship contribution statement
J.C. Ashworth:Conceptualization, Formal analysis, Investigation.J.L. Thompson:Investigation.J.R. James:Investigation.C.E. Slater:Investigation.S. Pijuan-Galitó:Supervision.K. Lis-Slimak:Investigation.R.J. Holley:Conceptualization.K.A. Meade: Conceptualization.A. Thompson:Supervision.K.P. Arkill:Methodology, Supervision.M. Tassieri:Formal analysis.A.J. Wright:Conceptualization, Formal analysis, Funding acquisition.G. Farnie:Conceptualization, Funding acquisition, Writing - review & editing.C.L.R. Merry:Conceptualization, Funding acquisition, Writing - review & editing.
References
- [1].Madhusoodanan J. Matrix mimics shape. Nature. 2019;566:563–565. doi: 10.1038/d41586-019-00681-1. [DOI] [PubMed] [Google Scholar]
- [2].Cruz-Acuña R, García AJ. Synthetic hydrogels mimicking basement membrane matrices to promote cell-matrix interactions. Matrix Biol. 2017;57–58:324–333. doi: 10.1016/j.matbio.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].David L, Dulong V, Le Cerf D, Chauzy C, Norris V, Delpech B, Lamacz M, Vannier J-P. Reticulated hyaluronan hydrogels: a model for examining cancer cell invasion in 3D. Matrix Biol. 2004;23:183–193. doi: 10.1016/j.matbio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- [4].Baker BM, Chen CS. Deconstructing the third dimension-how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125(13):3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- [6].Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15(6):753–762. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Caliari SR, Burdick JA. A practical guide to hydrogels for cell culture. Nat Methods. 2016;13(5):405–414. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Buxton PG, Bitar M, Gellynck K, Parkar M, Brown RA, Young AM, Knowles JC, Nazhat SN. Dense collagen matrix accelerates osteogenic differentiation and rescues the apoptotic response to MMP inhibition. Bone. 2008;43(2):377–385. doi: 10.1016/j.bone.2008.03.028. [DOI] [PubMed] [Google Scholar]
- [9].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20(8):931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/S1046-20230300032-X. [DOI] [PubMed] [Google Scholar]
- [12].Hansen KC, Kiemele L, Maller O, O'Brien J, Shankar A, Fornetti J, Schedin P. An in-solution ultrasonication-assisted digestion method for improved extracellular matrix proteome coverage. Mol Cell Proteomics. 2009;8(7):1648–1657. doi: 10.1074/mcp.M900039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Designer matrices for intestinal stem cell and organoid culture. Nat Publ Group. 2016;539(7630):560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- [14].Gjorevski N, Lutolf MP. Synthesis and characterization of well-defined hydrogel matrices and their application to intestinal stem cell and organoid culture. Nat Protoc. 2017;12(11):2263–2274. doi: 10.1038/nprot.2017.095. [DOI] [PubMed] [Google Scholar]
- [15].Mujeeb A, Miller AF, Saiani A, Gough JE. Self-assembled octapeptide scaffolds for in vitro chondrocyte culture. Acta Biomater. 2013;9(1):4609–4617. doi: 10.1016/j.actbio.2012.08.044. [DOI] [PubMed] [Google Scholar]
- [16].Rizzi L. Microrheology of biological specimens. Encyclopedia of Analytical Chemistry. 2006:1–24. doi: 10.1002/9780470027318.a9419. [DOI] [Google Scholar]
- [17].Del Giudice F, Tassieri M, Oelschlaeger C, Shen AQ. When microrheology, bulk rheology, and microfluidics meet: broadband rheology of hydroxyethyl cellulose water solutions. Macromolecules. 2017;50(7):2951–2963. doi: 10.1021/acs.macromol.6b02727. [DOI] [Google Scholar]
- [18].Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13(June):1–35. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- [19].Lee MP, Padgett MJ, Phillips D, Gibson GM, Tassieri M. Dynamic stereo microscopy for studying particle sedimentation. Opt Express. 2014;22(4):4671. doi: 10.1364/oe.22.004671. [DOI] [PubMed] [Google Scholar]
- [20].Jaspers M, Dennison M, Mabesoone MF, MacKintosh FC, Rowan AE, Kouwer PH. Ultra-responsive soft matter from strain-stiffening hydrogels. Nat Commun. 5 doi: 10.1038/ncomms6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Broedersz CP, Kasza KE, Jawerth LM, Münster S, Weitz DA, MacKintosh FC. Measurement of nonlinear rheology of cross-linked biopolymer gels. Soft Matter. 2010;6(17):4120–4127. doi: 10.1039/C0SM00285B. [DOI] [Google Scholar]
- [22].Megone W, Roohpour N, Gautrot JE. Impact of surface adhesion and sample heterogeneity on the multiscale mechanical characterisation of soft biomaterials. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-24671-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shan J, Chi Q, Wang H, Huang Q, Yang L, Yu G, Zou X. Mechanosensing of cells in 3D gel matrices based on natural and synthetic materials. 2014 doi: 10.1002/cbin.10325. [DOI] [PubMed] [Google Scholar]
- [24].Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9(2):108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ashworth JC, Mehr M, Buxton PG, Best SM, Cameron RE. Optimising collagen scaffold architecture for enhanced periodontal ligament fibroblast migration. J Mater Sci Mater Med. 29 doi: 10.1007/s10856-018-6175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hagbard L, Cameron K, August P, Penton C, Parmar M, Hay DC, Kallur T. Developing defined substrates for stem cell culture and differentiation. Philos Trans R Soc, B. 373 doi: 10.1098/rstb.2017.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lin X, Wei G, Shi Z, Dryer L, Esko JD, Wells DE, Matzuk MM. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224(2):299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- [28].Smyth N, Meyer M, Paulsson M, Edgar D, Murray P, Vatansever HS, Frie C. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J Cell Biol. 2002;144(1):151–160. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol. 2015;7(10):1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Loebel C, Mauck RL, Burdick JA. Local nascent protein deposition and remodelling guide mesenchymal stromal cell mechanosensing and fate in three-dimensional hydrogels. Nat Mater. doi: 10.1038/s41563-019-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Muschler J, Streuli CH. Cell-matrix interactions in mammary gland development and breast cancer. Cold Spring Harb Perspect Biol. 2011;2:a003202. doi: 10.1101/cshperspect.a003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, Bundred NJ. Novel cell culture technique for primary ductal carcinoma in situ: role of notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99(8):616–627. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- [33].Saiani A, Mohammed A, Frielinghaus H, Collins R, Hodson N, Kielty CM, Sherratt MJ, Miller AF. Self-assembly and gelation properties of α-helix versus β-sheet forming peptides. Soft Matter. 2008;5(1):193–202. doi: 10.1039/B811288F. [DOI] [Google Scholar]
- [34].Wan S, Borland S, Richardson SM, Merry CL, Saiani A, Gough JE. Self-assembling peptide hydrogel for intervertebral disc tissue engineering. Acta Biomater. 2016;46:29–40. doi: 10.1016/j.actbio.2016.09.033. [DOI] [PubMed] [Google Scholar]
- [35].Berger AJ, Linsmeier KM, Kreeger PK, Masters KS. Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin-methacrylate and collagen. Biomaterials. 2017;141:125–135. doi: 10.1016/j.biomaterials.2017.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Budday S, Sommer G, Haybaeck J, Steinmann P, Holzapfel GA, Kuhl E. Rheological characterization of human brain tissue. Acta Biomater. 2017;60:315–329. doi: 10.1016/j.actbio.2017.06.024. [DOI] [PubMed] [Google Scholar]
- [37].Tassieri M. Linear microrheology with optical tweezers of living cells'is not an option’! Soft Matter. 2015;11(29):5792–5798. doi: 10.1039/c5sm01133g. [DOI] [PubMed] [Google Scholar]
- [38].Tassieri M. Microrheology with optical tweezers: peaks and troughs. Curr Opin Colloid Interface Sci. (Accepted for Publication) [Google Scholar]
- [39].Watts F, Tan LE, Wilson CG, Girkin JM, Tassieri M, Wright AJ. Investigating the micro-rheology of the vitreous humor using an optically trapped local probe. J Opt. 16 doi: 10.1088/2040-8978/16/1/015301. [DOI] [Google Scholar]
- [40].Harunaga JS, Yamada KM. Cell-matrix adhesions in 3D. Matrix Biol. 2011;30:363–368. doi: 10.1016/j.matbio.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ferreira SA, Motwani MS, Faull PA, Seymour AJ, Yu TT, Enayati M, Taheem DK, Salzlechner C, Haghighi T, Kania EM, Oommen OP, et al. Bi-directional cell-pericellular matrix interactions direct stem cell fate. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-06183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fu H, Yang H, Zhang X, Xu W. The emerging roles of exosomes in tumor-stroma interaction. J Cancer Res Clin Oncol. 2016;142:1897–1907. doi: 10.1007/s00432-016-2145-0. [DOI] [PubMed] [Google Scholar]
- [43].Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70(23):9621–9630. doi: 10.1158/0008-5472.can-10-1722. [DOI] [PubMed] [Google Scholar]
- [44].Yeung V, Webber JP, Dunlop EA, Morgan H, Hutton J, Gurney M, Jones E, Falcon-Perez J, Tabi Z, Errington R, Clayton A. Rab35-dependent extracellular nanovesicles are required for induction of tumour supporting stroma. Nanoscale. 2018;10(18):8547–8559. doi: 10.1039/c8nr02417k. [DOI] [PubMed] [Google Scholar]
- [45].Astrahantseff KN, Morris JE. Estradiol-17 β stimulates proliferation of uterine epithelial cells cultured with stromal cells but not cultured separately. In Vitro Cell Dev Biol Anim. 1994;30(11):769–776. doi: 10.1007/BF02631300. [DOI] [PubMed] [Google Scholar]
- [46].Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, Ji B, Evans DB, Logsdon CD. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68(3):918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhu J. Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering. Biomaterials. 2010;31(17):4639–4656. doi: 10.1016/j.biomaterials.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gioiella F, Urciuolo F, Imparato G, Brancato V, Netti PA. An engineered breast cancer model on a chip to replicate ECM-activation in vitro during tumor progression. Adv Healthc Mater. 2016;5(23):3074–3084. doi: 10.1002/adhm.201600772. [DOI] [PubMed] [Google Scholar]
- [49].Mosqueira D, Mannhardt I, Bhagwan JR, Lis-Slimak K, Katili P, Scott E, Hassan M, Prondzynski M, Harmer SC, Tinker A, Smith JGW, et al. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardio-myopathy. Eur Heart J. 2018;39(43):3879–3892. doi: 10.1093/eurheartj/ehy249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lin Y, Chen J. StemBook. Cambridge (MA): Harvard Stem Cell Institute; 2014. Embryoid body formation from human pluripotent stem cells in chemically defined E8 media. [Internet] [DOI] [PubMed] [Google Scholar]
- [51].Yao A, Tassieri M, Padgett M, Cooper J. Microrheology with optical tweezers. Lab Chip. 2009;9(17):2568–2575. doi: 10.1039/b907992k. [DOI] [PubMed] [Google Scholar]
- [52].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.