Abstract
The catalytic cofactor thiamine diphosphate is found in many enzymes of central metabolism and is essential in all extant forms of life. We demonstrate the presence of an oxygen-dependent free radical in the thiamine diphosphate-dependent Escherichia coli2-oxoglutarate dehydrogenase, which is a key component of the tricarboxylic acid (Krebs) cycle. The radical was sufficiently long-lived to be trapped by freezing in liquid nitrogen, and its electronic structure was investigated by electron paramagnetic resonance (EPR) and electron–nuclear double resonance (ENDOR). Taken together, the spectroscopic results revealed a delocalized π radical on the enamine-thiazolium intermediate within the enzyme active site. The radical is generated as an intermediate during substrate turnover by a side reaction with molecular oxygen, resulting in the continuous production of reactive oxygen species under aerobic conditions. This off-pathway reaction may account for metabolic dysfunction associated with several neurodegenerative diseases. The possibility that the on-pathway reaction may proceed via a radical mechanism is discussed.
Introduction
One of the greatest ecological changes of the postbiotic earth was the advent of molecular oxygen. The environmental change presented by this powerful oxidant is likely to have focused evolutionary pressure on metabolism to avoid redox processes that are sensitive to interception by oxygen.1 However, life within an oxygen-rich biosphere evolved ways not only to withstand the hazard posed by the reactivity of O2 but also to harness the vast energy to be derived from oxygen reduction.2
Within the context of a cell, reactions with molecular oxygen can yield toxic byproducts, such as superoxide anions, hydrogen peroxide, peroxynitrite, and hydroxy radicals that are collectively termed “reactive oxygen species” (ROS).3 Despite this potential hazard, it appears that the metabolic advantage conferred by reactions exploiting O2 as the terminal oxidant in respiration has more than outweighed the costs of ROS containment. However, the distinctive enzymes of obligate anaerobes may have retained the earlier metabolism that predated an oxygenrich atmosphere and survived by occupying ecological niches free of molecular oxygen.4
One ancient metabolic chemistry still shared by all forms of life is the use of thiamine diphosphate (ThDP, Figure 1A) as an enzymatic cofactor in many diverse chemical transformations.5 It has been suggested that the oxygen intolerant, ThDP-dependent enzymes of obligate anaerobes could have dominated the use of ThDP before the advent of an oxygen-rich biosphere.6 One salient example is provided by 2-oxoacid-ferredoxin oxidoreductases (EC 1.2.7.1, OFOR), which catalyze the transformation of 2-oxoacids to acyl-coenzyme A by an electrontransfer mechanism that involves a radical intermediate.7,8 A [4Fe-4S] cluster adjacent to ThDP mediates the generation of this radical intermediate.6,8-10 In contrast, aerobic organisms do not have this single globular [4Fe-4S] containing, ThDP-dependent enzyme. Instead, this catalytic role is delegated to three different enzymes that constitute the 2-oxoacid dehydrogenase multienzyme complex. The complex is composed of ThDP-dependent 2-oxoacid decarboxylase (E1), lipoate-dependent acyl transferase (E2) and FAD-dependent dihydrolipoyl dehydrogenase (E3), which convert 2-oxoacids to acyl-CoA, CO2, and reducing equivalents.11 In a remarkable display of supramolecular architecture, these three enzymes are organized into multimeric complexes that specifically channel reaction intermediates between the multiple active sites of the complex.11
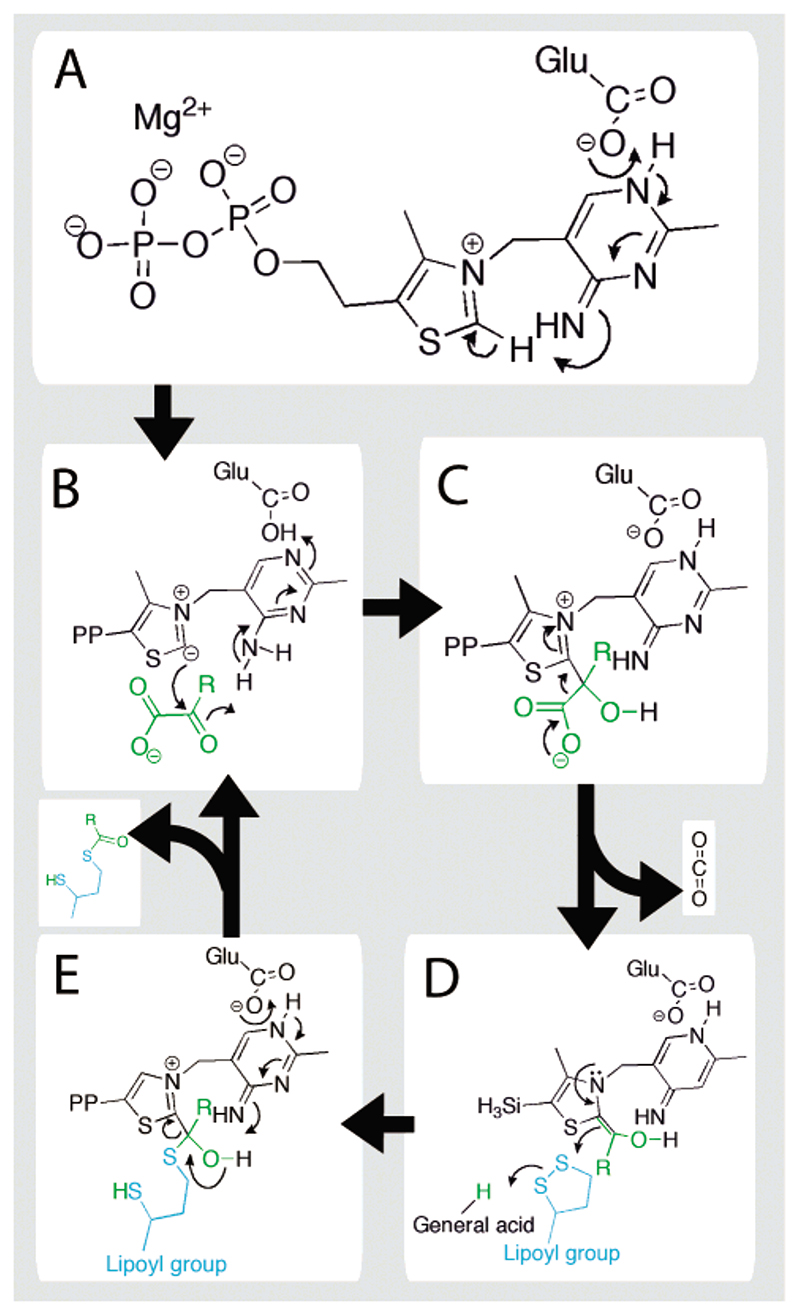
Figure 1.
Catalytic mechanism of the ThDP-dependent E1o component of the OGDH multienzyme complex. The substrate, 2-oxoglutarate, is shown in green, where R is –C2H4−CO2 −. The dithiolane ring of lipoate is shown in blue.
The reaction starts at the E1 subunit and, as depicted in Figure 1, involves decarboxylation followed by reductive acylation of a lipoyl group.
ThDP is first activated by the abstraction of a proton from the C2 carbon via the amino pyrimidine group (Figure 1A) and onto an invariant Glu, giving rise to a highly reactive carbanion.12–14 The reaction proceeds by nucleophilic attack on the R-carbon of the substrate, 2-oxoglutarate (Figure 1B). This intermediate eliminates CO2 (Figure 1C) and forms the meta-stable enamine-ThDP intermediate15,16 (also referred to as hydroxy-carboxyethylidene-ThDP) (see Figure 1D). This electronrich intermediate reductively acylates the lipoyl group, which requires a general acid thought to be a conserved His within the active site of E1.17–19 The catalytic cycle is completed with the aid of a general base provided by the aminopyrimidine ring of ThDP (see Figure 1E). The acyl-lipoyl product of the reaction catalyzed by E1 is covalently retained within the dehydrogenase complex and is channelled to the active sites of E2 and E3 yielding acyl-CoA and reducing equivalents, respectively.11
Two key examples of the 2-oxoacid dehydrogenases are pyruvate dehydrogenase (PDH) and 2-oxoglutarate dehydrogenase (OGDH), which, in eukaryotes, are both located in mitochondria. In humans, both are inactivated under oxidative stress and conditions associated with neurodegenerative disease,20 and there is evidence that peroxide is generated by OGDH within neurons.21,22 In addition, several earlier studies have found that other ThDP-dependent enzymes are susceptible to para-catalytic inactivation in the presence of oxidants.23–25 We hypothesized that these biological and chemical properties may be related. To probe and characterize the capacity of 2-oxoacid dehydrogenases to form ROS, we have prepared each of the recombinant E. coli OGDH subunits [E1o (EC 1.2.4.2), E2o (EC 2.3.1.61), and E3 (EC 1.8.1.4)]. In testing the ROS-generating activity of these enzymes, we made the surprising discovery that the ThDP-dependent E1o from OGDH generates hydrogen peroxide, with the concomitant formation of a radical on the ThDP cofactor. Here we discuss the formation of this radical and the implications of its existence for enzyme function.
Results
Fluorimetry
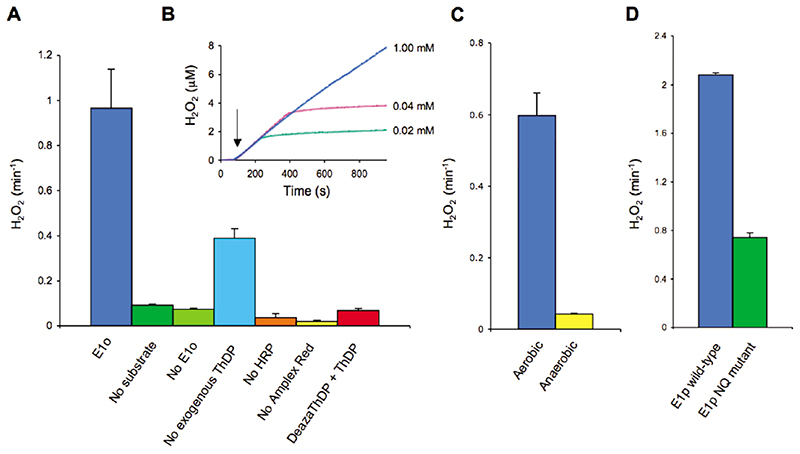
Using a fluorometric assay, we first examined whether the E1o component of the OGDH multienzyme assembly contributes to the generation of reactive oxygen species through the formation of peroxide (Figure 2). E1o produced peroxide at a significant rate in the presence of its substrate, 2-oxoglutarate, but not significantly in its absence (Figure 2A). Peroxide generation required the addition of ThDP, and it was inhibited in the presence of a ThDP analogue, 3-deazaThDP, which is a potent inhibitor of 2-oxoglutarate turnover.26 When substrate was depleted, peroxide generation ceased, suggesting that peroxide formation requires substrate turnover (Figure 2B). Moreover, the reaction produced far more than 1 equiv of peroxide per active site, also indicating that the peroxide was generated catalytically (data not shown); however, we cannot exclude the possibility that a proportion of the enzymes are also para-catalytically inactivated.24 Varying the substrate concentration within physiological range (0.02–2 mM) did not significantly alter the rate of peroxide production (Figure 2B). The generation of peroxide was almost entirely abolished in the presence of catalase, corroborating the specificity of the assay. Finally, to confirm that molecular oxygen is involved in the E1o-catalyzed production of peroxide, we repeated the assay under anaerobic conditions. With less than 20 ppm O2, peroxide generation was barely detectable (Figure 2C).
Figure 2. Peroxide generation by ThDP-dependent enzymes assayed with horseradish peroxidase and Amplex Red. The assay was performed measuring fluorescence (at 590 nm) or absorbance (at 560 nm) of the resorufin.
(A) Bar graph showing rate of peroxide generation from E. coli E1o, detected by the HRP-Amplex Red coupled assay. Peroxide generation is dependent on substrate (2-oxoglutarate), the enzyme E1o, and ThDP and may be inhibited by the addition of 3-deazathiamine diphosphate. (B) Trace of peroxide dependent fluorescence of Amplex Red for E1o with varying substrate concentration. Arrow indicates the addition of 2-oxoglutarate. It is apparent that peroxide generation is proportional with but not stoichiometric to substrate turnover. (C) Peroxide generation by E1o in an aerobic and anaerobic (<20 ppm O2) atmosphere determined by absorbance of the resorufin product. (D) Bar graph showing the rate of peroxide generation by B. stearothermophilus E1p and a catalytically impaired mutant form of this protein (E1p D180N,E183Q) in which two acidic residues within the channel have been replaced with their amide equivalents.27 Error bars correspond to 1 standard deviation.
We also examined the activity of the ThDP-dependent E1 subunit (E1p) of Bacillus stearothermophilus pyruvate dehydrogenase and found that it too generated peroxide after its substrate, pyruvate, was added to the reaction (Figure 2D). We have earlier described mutations in an acidic tunnel within E1p that profoundly inhibit pyruvate turnover.27 The peroxide-generating activity of one of these mutants (D18ON,E183Q (E1p NQ)) is significantly reduced (Figure 2D), supporting the view that peroxide generation is associated with catalytic turnover.
We also tested pure recombinant E2o and E3 of the E. coli OGDH complex to determine whether they contribute to peroxide generation, as suggested in earlier studies of its mammalian homologue.21 However, the enzymatic peroxide assay used here was not suitable for investigating E2o or E3 since one of the substrates, dihydrolipoamide, covalently bound to the E2o chain, interferes with the assay (Supporting Information). This observation suggests that caution should be used in interpreting earlier findings of peroxide generation by the full PDH and OGDH complexes. However, we cannot rule out that the E2 and E3, as well as E1, could contribute to ROS generation.
EPR Spectroscopy
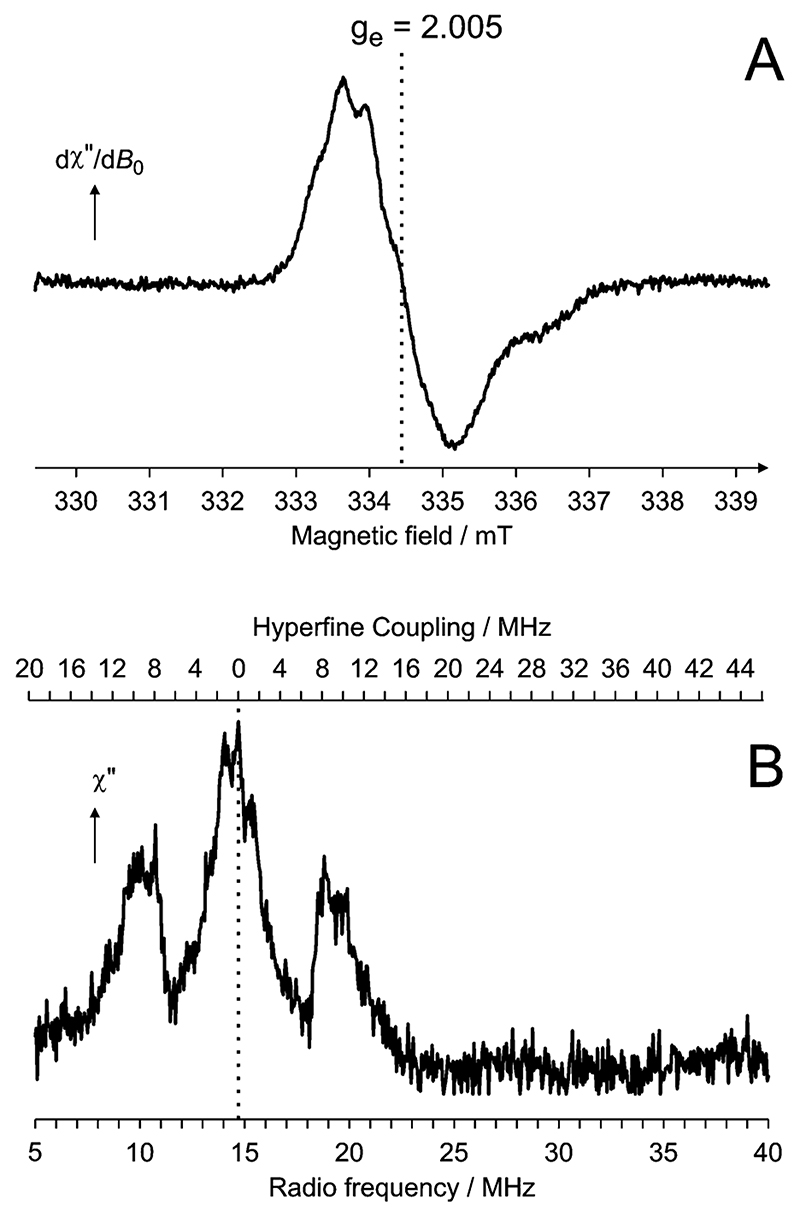
To further establish the origin of hydrogen peroxide generation by E1o we investigated if there is a radical intermediate present during catalytic turnover. We flash-froze samples to “trap” any intermediates generated during the reaction and then probed using continuous-wave electron
paramagnetic resonance (cw-EPR) spectroscopy. Pure E1o prepared with excess ThDP, but in the absence of substrate, had no paramagnetic signal when measured by cw-EPR. In contrast, samples with an excess of substrate and flash-frozen before substrate could be fully depleted exhibited a cw-EPR signal showing that the turnover of substrate involves the formation of a radical (Figure 3A). The g-factor of 2.005 and 1.5 mT peak-to-peak line width of the radical strongly suggest the presence of an unpaired π electron within an organic center of the enzyme, as opposed to a σ radical which one would expect to produce a far broader resonance.28 These spectroscopic features and the partially resolved hyperfine coupling (hfc) pattern of the organic radical in E1o are similar to but distinct from those of the radicals in 2-oxoglutarate/ferredoxin oxidoreductase8 and PFOR.7,9 The same EPR signal was also observed in the full E1-E2-E3 OGDH assembly under aerobic conditions and in the presence of substrate (results not shown).
Figure 3. A thiamine radical in the E1o subunit of E. coli 2-oxoglutarate dehydrogenase. (A) cw-EPR spectrum. (B) Pulsed ENDOR spectrum.
In PFOR, a radical arises by single electron transfer from a ThDP-derived intermediate to an Fe-S cluster during the normal course of the enzyme-catalyzed reaction. Thus by analogy, it appears likely that the radical species in the E1o enzyme is a ThDP-derived intermediate. However, the E1o does not contain an Fe-S cluster, which we confirmed recently by solving the X-ray crystal structure of the protein to 2.4 Å resolution.29 Thus an alternative electron acceptor must be considered to account for the formation of the radical. Since peroxide generation by E1o is oxygen-dependent, we reasoned that the single electron acceptor that gives rise to the thiamine radical is molecular oxygen. Consistent with the oxygen-dependence of hydrogen peroxide generation by E1o, samples of E1o with substrate and prepared under anaerobic conditions (<20 ppm O2) showed only a very weak cw-EPR signal (data not shown).
To further characterize this radical, pulsed electron–nuclear double resonance (ENDOR) spectroscopy experiments were performed, Figure 3B. The broad lines are indicative of a protein bound radical containing conformationally restrained protons that interact in nonequivalent ways with the unpaired electron. These geometric restraints of the intermediate are likely to be important aspects of substrate recognition and of active site organization to favor catalysis.30
The most significant features of the ENDOR spectrum are the hyperfine couplings (hfc’s) observed in the frequency range 6–16 MHz and are due to protons interacting strongly with the unpaired electron. This broad feature in the ENDOR of E1o is similar to that observed for PFOR which extends over the range 12–20 MHz.10 The hfc’s are most likely to derive from protons on the aliphatic side group that is at an α position to the radical center (R group in Figure 1D), as suggested for the radical species in PFOR.9,10 However, the substrates for PFOR and E1o are different, and while the intermediate in PFOR contains an immobilized methyl group10 with three protons, in E1o, a methylene with two protons occupies this position. This chemical difference is likely to account for the main features distinguishing the ENDOR spectra of PFOR and E1o. Nevertheless, both the EPR and ENDOR spectra of E1o are consistent with a π radical delocalized over the thiazolium ring of the enamine-ThDP intermediate.10
In order to model the electronic structures of possible thiamine-derived radicals the coordinates of an enamine-ThDP intermediate were taken from a high-resolution crystal structure of an enzyme related to E1o, pyruvate oxidase.31 The heavy atoms remained fixed, while protons were added as appropriate and energy optimized using Density Functional Theory (DFT). In pyruvate oxidase, this intermediate has three protons at C2β, while, in E1o, the different substrate results in the replacement of one of these protons by an aliphatic chain. The hfc’s of the remaining two (methylene) protons at C2β are affected by the orientation of the aliphatic side group. Since the geometry of the latter is not known we have truncated the R group with a proton for the calculations.
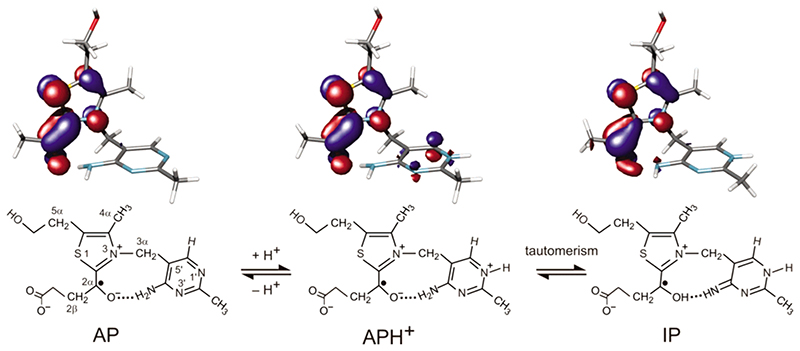
The enamine-ThDP radical intermediate may potentially exist in three forms that differ in the protonation states of N1', N4', and O2β (Figure 4). The 4'-aminopyrimidine (AP) may be converted into 4'-aminopyrimidinium (APH+) by the addition of a proton at N1',32 which can undergo a tautomerism by the transfer of a proton from the 4'-amino group to O2β, forming the 1 ‘,4'-iminopyrimidinium (IP) intermediate.
Figure 4.
Structures of three potential thiamine-derived radicals and representations of their singly occupied molecular orbitals. The molecular orbitals are presented at a contour level of 0.05 e/au.3 The blue and red areas denote regions of opposite sign of the wave function.
The calculated orbitals of the three structures suggest that the unpaired electron is largely restricted within the thiazolium moiety in a π-type radical (Figure 4), as was predicted for the radical in PFOR.10 However, the predicted hfc’s do show significant variations, Table 1. Whereas AP and APH+ radical models have proton hfc’s in the same range as those observed in the ENDOR spectrum, the very large hfc’s predicted for the protons bound at C2β and O2β of IP are absent from the ENDOR spectrum. Furthermore, the large energy difference (105 kJ mol–1) suggests that APH+ is thermodynamically favored over IP.
Table 1. Isotropic hfc’s of Possible Structures of the Enamine-ThDP Radical; All Values Are in MHz.
| structure | |||
|---|---|---|---|
| position | AP | APH+ | IPH |
| N(1')H | − | −1.2 | −0.3 |
| C(2'α)¾ | 0.0 | −0.4 | −0.1 |
| N(4α)H(1) | −0.3 | 0.4 | −0.4 |
| N(4α)H(2) | 0.5 | 3.3 | − |
| C(6')H | −0.4 | −7.1 | −0.6 |
| C(3α)H(1) | 1.8 | 3.4 | 1.4 |
| C(3α)H(2) | 7.8 | 6.1 | 7.3 |
| C(4α)H3 | −2.9 | −1.2 | 0.7 |
| C(5α)H(1) | 9.1 | 7.4 | 9.6 |
| C(5α)H(2) | 0.3 | 0.1 | 0.3 |
| C(2,α)H(1) | −0.3 | −0.4 | 16.6 |
| C(2,β)H(2) | 13.3 | 11.7 | 62.4 |
| C(2,β)H(3) | 10.6 | 8.5 | 33.3 |
| O(2,β)H(3) | − | − | 76.2 |
Discussion
Taken together, our evidence suggests that the normal enamine-ThDP intermediate is converted by molecular oxygen into a species harboring a radical, as summarized in the left panel of Figure 5. As the reacting oxygen is reduced by one electron, it forms a superoxide anion. A second oxidation step would be expected to produce hydrogen peroxide and the acyl-ThDP product. Alternatively, the superoxide may dismutate to produce peroxide, allowing a second molecule of oxygen to receive the second electron. The acyl-ThDP product may then be hydrolyzed spontaneously to produce succinate and complete the catalytic cycle.33
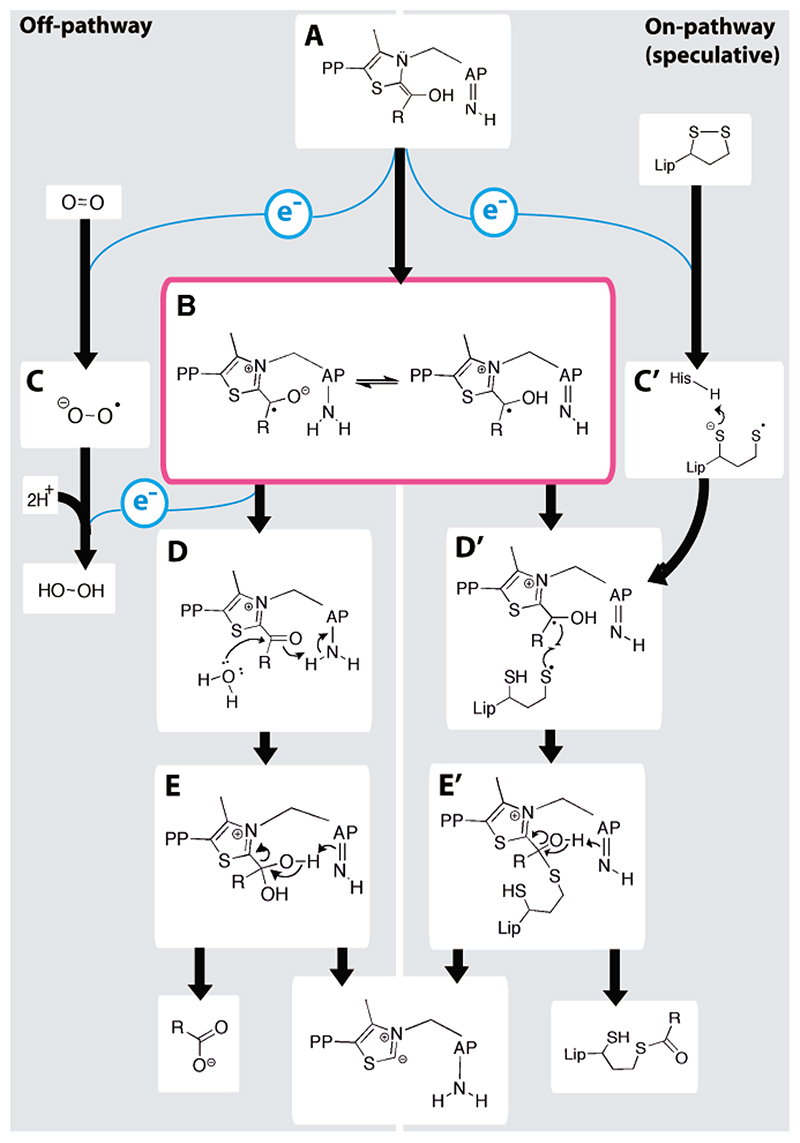
Figure 5. A mechanism involving radical intermediates for the thiamine-dependent 2-oxo acid dehydrogenases. Two competing routes are presented on the left and right for the off- and on-pathway, respectively.
State (A) shows the canonical enamine-ThDP intermediate that is common to all ThDP-dependent enzymes (AP is the pyrimidine group of ThDP). A single electron is transferred to either oxygen or a lipoyl group to form (B) the ThDP radical and (C) superoxide or (C') the thiyl radical of the dihydrolipoyl group. The ThDP radical continues in the off-pathway reaction by giving a second electron to either superoxide or oxygen, forming a second superoxide or hydrogen peroxide, respectively, and (D) an acyl-ThDP intermediate, which is hydrolyzed to produce succinate and regenerate the active ThDP carbanion. In the on-pathway, (D') the thiyl lipoyl and enamine-ThDP radical terminate, giving (E') the tetrahedral intermediate, and so the reaction may produce a reductively acylated lipoyl group.
The OFOR enzymes7 and pyruvate oxidase31 both give rise to a similar enamine-ThDP radical intermediate by single electron transfers to a [4Fe–4S] cluster and a flavin, respectively. Thus, the enamine-ThDP intermediate itself possesses an innate propensity for radical chemistry,33 and, in the case of E1o, this allows oxygen to be a suitable single electron acceptor.
Given the tendency of the enamine-ThDP intermediate to undergo single electron transfers, as in E1o to molecular oxygen, the question arises whether a radical step occurs in the “on-pathway” reaction of 2-oxoacid dehydrogenases (Figure 5). An on-pathway mechanism can be envisaged in an electronically similar way to the off-pathway reaction, in which the dioxygen is replaced with the disulfide of lipoate. Similar electron transfers between cofactors and reactive sulfurs, followed by the formation of a cofactor-sulfur adduct, have been observed in other systems.34,35 Favoring this possibility, we have found that the reduced form of the lipoyl group (dihydrolipoamide) readily transfers single electrons, demonstrated by its ability to reduce cytochrome c 6 by one electron (Supporting Information). Although direct evidence of the on-pathway mechanism is lacking, the sequential transfer of two electrons via a radical intermediate may be the preferred route by which the electronegative enamine-ThDP intermediate of E1o not only may reduce
the electron-rich disulfide center of the lipoate on E2o but also provides a mechanism for directing the formation of the lipoyl adduct (Figure 5, right).36
The detection of peroxide generation by both E1o and E1p suggests that the off-pathway reaction with oxygen might occur in the E1 components of 2-oxoacid dehydrogenases generally. This potentially toxic side reaction of E1-bound intermediates may help to explain why these proteins and their reaction intermediates are not free to diffuse but are retained within large assemblies containing E2 and E3.11 Restriction of all the enzymatic components and intermediates in close proximity may confer a protective advantage by kinetically favoring the normal reaction as envisaged in the “hot potato hypothesis”.37
In the presence of excess substrate, the off-pathway reaction may be suppressed (Figure 5). However, if one of the downstream substrates of 2-oxoglutarate is limiting, the off-pathway reaction involving oxygen may compete with the on-pathway reaction. This balancing act is perhaps nowhere more carefully controlled than in the human brain, where neurons are sustained by an unusually high metabolic rate and are particularly susceptible to oxidative stress. Any limit of O2 supply or imbalance (e.g., glutamate excitotoxicity) is quickly reflected in oxidative stress and cellular damage. For example, the 2-oxoacid dehydrogenases have been found to be inactivated early in the pathogenesis of several chronic neurodegenerative diseases20,39 and in ischemia-reperfusion injury.39,21 The discovery that the ThDP-dependent components of the PDH and OGDH assemblies undergo radical side reactions with oxygen further implicates these complexes in such disorders. Indeed, since the enamine-ThDP radical intermediate is common to all thiamine-dependent enzymes studied so far,5 it will be interesting to discover if other ThDP-dependent enzymes are also susceptible to side reactions with oxygen.
Materials and Methods
Preparation of Dehydrogenase Subunits and Assembly
Wild type E. coli E1o, E2o, and E3 were each cloned into a pET11c vector and overexpressed in E. coli BL21(DE3) using induction with 0.5 mM IPTG.40 Cells overexpressing E1o, E2o and E3 were expressed and purified separately. Cells were harvested 3 h postinduction and lysed, and lysates were enriched for E1o/E2o/E3 by ammonium sulfate precipitation and redissolved in 20 mM potassium phosphate pH 7.0. The samples were further purified by eluting from an anion exchange HiLoad Q-Sepharose (Amersham) with a linear gradient of the same buffer containing 1 M NaCl. Enriched fractions were pooled, concentrated with a 30 kDa MWCO centrifugal filter, and fractionated by Superdex S200 size exclusion chromatography (Amersham). Purified proteins were analyzed by electrospray ionization–time-of-flight mass spectrometry.
To evaluate the proper assembly of the reconstituted complex the activity of the full 2-oxoglutarate dehydrogenase multienzyme complex was measured by the formation of NADH at 340 nm. The reaction mixture contained 450 pmol of E1o, 900 pmol of E2o, 450 pmol of E3, 100 mM potassium phosphate pH 7.0, 1 mM ThDP, 1 mM MgCl2, 0.26 mM cysteine hydrochloride, 2.5 mM NAD+. The reaction was started by adding 2-oxoglutarate and lithium co-enzyme A to a final concentration of 1 mM each. Enzyme activity was immediately followed by measuring the rate of increase in absorbance at 340 nm using a temperature-controlled spectrophotometer at 30 °C.
Wild type and the acidic tunnel mutant of Bacillus stearothermo-philus E1p were prepared as described by Frank et al.27
Measuring Generation of Hydrogen Peroxide
Peroxide was detected by the horseradish peroxidase (HRP)-Amplex Red fluorescent dye coupled assay.41 The assay includes horseradish peroxidase to specifically oxidize Amplex Red in the presence of peroxide, producing the fluorescent product, resorufin. Each 1 mL sample contained 0.5 μM recombinant E. coli E1o, 2 mM thiamine diphosphate, 2 mM MgCl2, 50 mM Tris-HCl buffer pH 7.4. Samples were incubated for 2 h at 37 °C before the start of each experiment to allow for the binding of cofactors. Fluorescence was followed at 37 °C in a quartz cuvette using a Philips Fluorimeter with excitation at 540 nm and emission recorded at 590 nm. 2 U·mL-1 horseradish peroxidase and 2 mM Amplex Red reagent were added. After 5 min, substrate, 1 mM 2-oxoglutarate, was added and the change in fluorescence was measured. For samples with Bacillus stearothermophilus E1p, the same assay conditions were used, except that pyruvate was used as the substrate. When comparing aerobic and anaerobic samples, the change in absorbance at 560 nm was measured inside and outside an anaerobic (<20 ppm O2) glovebox (Belle Technology, Portesham, UK) at room temperature.
Electron Paramagnetic Resonance Spectroscopy
Components were mixed at the following concentrations at room temperature for 30-45 min to allow enzyme to bind cofactors: 200 μM E1o, 50 mM Tris-HCl buffer pH 7.4, 4 mM ThDP, 4 mM MgCl2, 4 mM CaCl2. The mixture was then transferred into 3 mm inner diameter EPR tubes, and the reactions were started at room temperature with the addition of 2-oxoglutarate to a 2 mM final concentration. After 10-15 s, the samples were then flash-frozen in liquid nitrogen.
X-band (9 GHz) continuous-wave EPR spectra were recorded at 80 K using a pulsed EPR spectrometer (Bruker Elexsys E580) equipped with an EPR resonator (4122SHQE) cooled by a helium cryostat (Oxford ESR 910). Conditions used were as follows: microwave frequency, 9.39 GHz; microwave power, 20 mW; modulation frequency, 100 kHz; modulation amplitude, 0.2 mT; temperature, 80 K.
Pulsed ENDOR spectra were recorded at 80 K with the same spectrometer and an ENDOR accessory (Bruker E560 D-P) including a radio frequency amplifier (250A; Amplifier Research, Souderton, PA) and a dielectric-ring ENDOR resonator (Bruker EN4118X-MD-4-W1), which was immersed in a helium-gas flow cryostat (CF935; Oxford Instruments, Oxford, U.K.). For Davies-type ENDOR, a microwave pulse-sequence π-t-π/2-τ–π using 64- and 128-ns π/2-pulses and π-pulses, respectively, and an RF pulse of 10 μs duration and starting 1 μs after the first microwave pulse was used. The separation times t
and τ between the microwave pulses were selected to be 13 μs and 500 ns, respectively. The entire pulse pattern was repeated with a frequency of 200 Hz so as to avoid saturation effects caused by long relaxation times. The ENDOR spectrum was recorded at a magnetic field position corresponding to the center of the cw-EPR signal (Figure 3A). ENDOR signals were detected in two spectral regions between 0 and 30 MHz and are arranged in pairs (symmetrically distributed around the free proton Larmor frequency, νH = 14.75 MHz).
For doublet-state radicals, two ENDOR lines are expected per group of magnetically equivalent nuclei. These are separated by the orientation-dependent hfc constant, A, that quantifies the interaction of the nuclear magnetic moment with the electron magnetic moment. In the weak-coupling case, when |A| < 2|ν n| (ν n = g n β,B 0/h is the Larmor precessional frequency of the nucleus at the respective magnetic field B 0, gn and βn are the nuclear g-value and the Bohr magneton, respectively), and the resonance frequencies are ν ± ENDOR = |ν n ± (A/ 2)|.
Computations
DFT was performed at the unrestricted B3LYP/6-31G(d,p) level of theory, as implemented in program package Gaussian 03.42 Isotropic hfc’s were calculated for the optimized structure at the same level of theory. Graphical representation of molecular orbitals was achieved using the Molden program package,43 followed by rendering with POV-Ray.
Supplementary Material
Acknowledgment
This work was supported by the Wellcome Trust. Access to the UCL Research Computing Services for DFT calculations is gratefully acknowledged. We thank Finian Leeper for providing 3-deazathiamine diphosphate, Paul Skelton for help with mass spectrometry, and Torsten Reda for help with the anaerobic glovebox. We are grateful to Simon Lewis, Richard Cammack, Hal Dixon, Peter Leadlay, Martin Brand, Jonathan Worrall, Derek Bendall, and Finian Leeper for helpful discussions.
References
- (1).Falkowski PG. Science. 2006;311:1724–1725. doi: 10.1126/science.1125937. [DOI] [PubMed] [Google Scholar]
- (2).Raymond J, Segre D. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- (3).Halliwell B, Gutteridge J. Free radicals in biology and medicine. Oxford University Press; Oxford: 1998. [Google Scholar]
- (4).Martin W, Muller M. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- (5).Frank RA, Leeper FJ, Luisi BF. Cell Mol Life Sci. 2007;64:892–905. doi: 10.1007/s00018-007-6423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chabriere E, Vernede X, Guigliarelli B, Charon MH, Hatchikian EC, Fontecilla-Camps JC. Science. 2001;294:2559–2563. doi: 10.1126/science.1066198. [DOI] [PubMed] [Google Scholar]
- (7).Ragsdale SW. Chem Rev. 2003;103:2333–2346. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
- (8).Cammack R, Kerscher L, Oesterhelt D. FEBS Lett. 1980;118:271–273. [Google Scholar]
- (9).Bouchev VF, Furdui CM, Menon S, Muthukumaran RB, Ragsdale SW, McCracken J. J Am Chem Soc. 1999;121:3724–3729. [Google Scholar]
- (10).Mansoorabadi SO, Seravalli J, Furdui C, Krymov V, Gerfen GJ, Begley TP, Melnick J, Ragsdale SW, Reed GH. Biochemistry. 2006;45:7122–7131. doi: 10.1021/bi0602516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Perham RN. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- (12).Breslow R. J Am Chem Soc. 1957;79:1762–1762. [Google Scholar]
- (13).Kern D, Kern G, Neef H, Tittmann K, Killenberg-Jabs M, Wikner C, Schneider G, Hubner G. Science. 1997;275:67–70. doi: 10.1126/science.275.5296.67. [DOI] [PubMed] [Google Scholar]
- (14).Muller YA, Lindqvist Y, Furey W, Schulz GE, Jordan F, Schneider G. Structure. 1993;1:95–103. doi: 10.1016/0969-2126(93)90025-c. [DOI] [PubMed] [Google Scholar]
- (15).Schellenberger A. Biochim Biophys Acta. 1998;1385:177–186. doi: 10.1016/s0167-4838(98)00067-3. [DOI] [PubMed] [Google Scholar]
- (16).Jordan F. Nat Prod Rep. 2003;20:184–201. doi: 10.1039/b111348h. [DOI] [PubMed] [Google Scholar]
- (17).Jones DD, Stott KM, Reche PA, Perham RN. J Mol Biol. 2001;305:49–60. doi: 10.1006/jmbi.2000.4257. [DOI] [PubMed] [Google Scholar]
- (18).Nemeria N, Arjunan P, Brunskill A, Sheibani F, Wei W, Yan Y, Zhang S, Jordan F, Furey W. Biochemistry. 2002;41:15459–15467. doi: 10.1021/bi0205909. [DOI] [PubMed] [Google Scholar]
- (19).Fries M, Jung HI, Perham RN. Biochemistry. 2003;42:6996–7002. doi: 10.1021/bi027397z. [DOI] [PubMed] [Google Scholar]
- (20).Gibson GE, Park LC, Sheu KF, Blass JP, Calingasan NY. Neuro Chem Int. 2000;36:97–112. doi: 10.1016/s0197-0186(99)00114-x. [DOI] [PubMed] [Google Scholar]
- (21).Tretter L, Adam-Vizi V. J Neurosci. 2004;24:7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. J Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Khailova LS, Bernkhardt R, Khiubner G. Biokhimiia. 1977;42:113–117. [PubMed] [Google Scholar]
- (24).Sumegi B, Alkonyi I. Arch BioChem Biophys. 1983;223:417–424. doi: 10.1016/0003-9861(83)90605-7. [DOI] [PubMed] [Google Scholar]
- (25).Tse MT, Schloss JV. Biochemistry. 1993;32:10398–10403. doi: 10.1021/bi00090a015. [DOI] [PubMed] [Google Scholar]
- (26).Mann S, Perez Melero C, Hawksley D, Leeper FJ. Org BioMol Chem. 2004;2:1732–1741. doi: 10.1039/b403619k. [DOI] [PubMed] [Google Scholar]
- (27).Frank RA, Titman CM, Pratap JV, Luisi BF, Perham RN. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]
- (28).Carrington A, McLachlan AD. Introduction to Magnetic Resonance. Harper International; New York: 1969. [Google Scholar]
- (29).Frank RA, Price AJ, Northrop FD, Perham RN, Luisi BF. J Mol Biol. 2007;368:639–651. doi: 10.1016/j.jmb.2007.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Mesecar AD, Stoddard BL, Koshland DE., Jr Science. 1997;277:202–206. doi: 10.1126/science.277.5323.202. [DOI] [PubMed] [Google Scholar]
- (31).Wille G, Meyer D, Steinmetz A, Hinze E, Golbik R, Tittmann K. Nat Chem Biol. 2006;2:324–328. doi: 10.1038/nchembio788. [DOI] [PubMed] [Google Scholar]
- (32).Nemeria N, Chakraborty S, Baykal A, Korotchkina LG, Patel MS, Jordan F. Proc Natl Acad Sci USA. 2007;104:78–82. doi: 10.1073/pnas.0609973104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Flournoy DS, Frey PA. Biochemistry. 1986;25:6036–6043. doi: 10.1021/bi00368a030. [DOI] [PubMed] [Google Scholar]
- (34).Kay CWM, Schleicher E, Kuppig A, Hofner H, Rudiger W, Schleicher M, Fischer M, Bacher A, Weber S, Richter G. J Biol Chem. 2003;278:10973–10982. doi: 10.1074/jbc.M205509200. [DOI] [PubMed] [Google Scholar]
- (35).Kolberg M, Strand KR, Gaff P, Andersson KK. Biochim Biophys Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- (36).Pan K, Jordan F. Biochemistry. 1998;37:1357–1364. doi: 10.1021/bi971835y. [DOI] [PubMed] [Google Scholar]
- (37).Perham RN. Philos Trans R Soc London, Ser B. 1975;272:123–136. doi: 10.1098/rstb.1975.0075. [DOI] [PubMed] [Google Scholar]
- (38).Bunik VI, Schloss JV, Pinto JT, Gibson GE, Cooper AJ. NeuroChem Res. 2007;32:871–891. doi: 10.1007/s11064-006-9239-z. [DOI] [PubMed] [Google Scholar]
- (39).Lucas DT, Szweda LI. Proc Natl Acad Sci USA. 1999;96:6689–6693. doi: 10.1073/pnas.96.12.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ricaud P., Ph.D . Thesis. Cambridge University; 1998. [Google Scholar]
- (41).Mohanty JG, Jaffe JS, Schulman ES, Raible DG. J Immunol Methods. 1997;202:133–141. doi: 10.1016/s0022-1759(96)00244-x. [DOI] [PubMed] [Google Scholar]
- (42).Frisch MJ, et al. Gaussian 03. Gaussian, Inc; Pittsburgh, PA: 2003. revision B.04 ed. [Google Scholar]
- (43).Schaftenaar G, Noordik JH. J Comput-Aided Mol Des. 2000;14:123–134. doi: 10.1023/a:1008193805436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.