Abstract
Cytochrome c 6A is a unique dithio-cytochrome of green algae and plants. It has a very similar core structure to that of bacterial and algal cytochromes c 6 but is unable to fulfill the same function of transferring electrons from cytochrome fto photosystem I. A key feature is that its heme midpoint potential is more than 200 mV below that of cytochrome c 6 despite having His and Met as axial heme-iron ligands. To identify the molecular origins of the difference in potential, the structure of cytochrome c 6 from the cyanobacterium Phormidium laminosum has been determined by X-ray crystallography and compared with the known structure of cytochrome c 6A. One salient difference of the heme pockets is that a highly conserved Gln (Q51) in cytochrome c 6 is replaced by Val (V52) in c 6A. Using protein film voltammetry, we found that swapping these residues raised the c 6A potential by +109 mV and decreased that of c 6 by almost the same extent, -100 mV. X-ray crystallography of the V52Q protein showed that the Gln residue adopts the same configuration relative to the heme as in cytochrome c 6 and we propose that this stereochemistry destabilizes the oxidized form of the heme. Consequently, replacement of Gln by Val was probably a key step in the evolution of cytochrome c 6A from cytochrome c6, inhibiting reduction by the cytochrome b 6 f complex and facilitating establishment of a new function.
Introduction
Cytochromes c are involved in many different biological electron transfer (ET) reactions. The most important functional characteristic of any individual cytochrome (cyt) is its midpoint redox potential (E m). Unfolded mitochondrial cyt c has E m,7 ≈ –150 mV, but in the folded protein this increases dramatically to +260 mV.1 Thus, exclusion of solvent and the structure of the protein environment are of crucial importance in determining the E m of the native protein. In different c-type cyts E m,7 ranges between ~ – 400 mV and +400 mV, but in any homologous group of cyts of similar function the redox potential is tuned to a relatively narrow range of values. Such conservation of E m must depend upon conservation of stereochemistry of the heme pocket, and hence of the amino acid sequence, but a completely satisfactory description has not yet been achieved.
In all Class I cyts c the heme is bound near the protein N-terminus by the motif –CXXCH–, in which the His is one of the axial heme ligands. The redox potential is determined by the relative stabilities of the iron-porphyrin ring system in the reduced form, which is electrically neutral, and the oxidized form that carries a positive charge. The nature of the sixth ligand divides Class I cyts into a low-potential group, in which coordination is with the imidazole side chain of a His residue, and a high-potential group that uses the Sδ atom of a Met. In mitochondrial cyt c an extensive hydrogen bond network, which includes a conserved water molecule (wat166) and several conserved amino acids, is important in determining the redox potential.2,3
Cyt c6 has a relatively simple hydrogen bond network surrounding the heme cavity, and there is no redox dependent conformational change4 as observed with mitochondrial cyt c.3 This family of cyts c falls into two sections, “conventional” cyt c 6 and cyt c 6A, with E m,7 values of ~+340 mV for c 6 and ~+100 mV for c 6A,5 putting them at opposite ends of the redox potential scale of cyts c with His and Met as the axial ligands. Cyt c 6 functions interchangeably with the copper-protein plastocyanin as the electron donor to photosystem I in cyanobacteria and algae, with the relative levels of the two proteins determined by copper availability.6–8 In green plants, plastocyanin is an essential component of the photosynthetic electron transport chain, so the discovery in Arabidopsis thaliana of a gene encoding a cyt c 6-like protein was a surprise.9,10 Amino acid sequence and structure place the corresponding protein in the c 6 family, and the three-dimensional structure of the A. thaliana protein confirms a high structural similarity with cyt c 6 from algal and cyanobacterial sources as well as the presence of His and Met as axial ligands to the heme iron.5,11 The most striking structural difference from algal and cyanobacterial cyt c 6, which was also inferred from sequence analysis, is the presence of a surface exposed 12 residue loop insertion peptide (LIP) containing a disulfide bridge between Cys67 and Cys73. In view of these differences, the novel form of the protein has been named cyt c 6A.
In spite of the similarity between cyt c 6A and conventional cyt c 6, the former is unable to replace the function of plastocyanin. Unlike plastocyanin and cyt c 6, it is only slowly oxidized by photosystem I,12 and its low redox potential makes it unable to oxidize the cyt b6f complex. The natural reductant of cyt c 6A remains unidentified. However, if the redox potential of the heme in cyt c 6A remained similar to that of the ancestral cyt c 6, reduction by the cyt b6f complex would still be possible, even though the role of the newly evolved cyt c 6A was to be reduced by something else. Continued reduction by the cyt b 6 f complex would presumably compromise the new function of cyt c 6A. The evolution of an unusually low redox potential for the heme in cyt c 6A can therefore be seen as an important step in the development of a novel function for the protein. The aim of the present work was to identify the origin(s) of the difference in Em between cyt c 6A and cyt c 6.
We have determined the X-ray structure of a cyt c 6 from the cyanobacterium Phormidium laminosum and compared the heme pocket to that of A. thaliana cyt c 6A. This reveals a number of subtle differences which have been probed by site-directed mutagenesis, protein film voltammetry, and X-ray crystallography. This study has revealed that a single residue, which is not a heme ligand, is responsible for tuning the heme redox potential by ~100 mV. We suggest that mutation of this residue was a crucial step in the evolution of cyt c 6A from cyt c 6.
Materials and Methods
Expression Plasmid for P. laminosum cyt c 6
Recombinant P. laminosum cyt c6 was obtained by expression in Escherichia coli of a synthetic gene containing the coding region of the mature P. laminosum cyt c6 fused to the signal peptide from Anabaena cyt c6 (the sequence of this synthetic gene is reported in the Supporting Information). The synthetic gene was generated by back-translation of the amino acid sequence and the codon usage of E. coli genes that are highly expressed during exponential growth. The product was cloned into pGEMT-easy for expression under the T7-promoter. Correct sequence and direction of insertion were confirmed by sequencing. The resulting expression plasmid was named pPlc6.
Site-Directed Mutagenesis
Mutations were introduced using the Stratagene QuikChange method.13 The mutants V52Q and V52Q-AA of the A. thaliana cytochrome c 6A gene were generated by replacing the codon “GTG” in the wild-type or I17A/G18A cyt c 6A expression plasmids (pAtc6a and pAtc6aAA, respectively)5 by “CAG”. Mutant A31K was generated by replacing the codon “GCG” in plasmid pAtc6a by “AAA”. The mutant Q51V of the P. laminosum cyt c 6 gene was generated by replacing the codon “CAG” in the expression plasmid pPlc6 by “GTG”. Incorporation of the correct mutations and absence of undesired changes were corroborated by sequencing of the mutated constructs.
Protein Expression and Purification
Wild-type and mutant A. thaliana cyt c 6A were expressed and purified as described in ref 5. Expression of wild-type and mutant P. laminosum cyt c 6 differed as follows: E. coli BL21(DE3) was used instead of GM119, and expression cultures were induced with 100 mg/L IPTG after overnight growth at 30 °C and 170 rpm. Purification was analogous to the procedure used for A. thaliana cyt c 6A.
Crystallization and X-ray Data Collection
Crystals of P. lami- nosum cyt c 6 and A. thaliana cyt c 6A V52Q-AA variant were grown by the hanging drop vapor diffusion method with 1 μL of protein solution mixed with 1 μL of reservoir solution. For P. laminosum cyt c 6 the reservoir solution contained 2.5 M NaCl, 100 mM imidazole, 200 mM Zn(OAc)2, pH 6.0. For A. thaliana cyt c 6A V52Q-AA variant, 1.26 M ammonium sulfate, 100 mM acetate pH 4.5, and 200 mM NaCl were used as precipitant. Prior to data collection, protein crystals were immersed in their respective precipitant solution containing 20% v/v glycerol, followed by rapid freezing in liquid nitrogen. X-ray diffraction data were collected on beamline ID 29-1 at the ESRF Grenoble, France. Data were indexed, integrated, and scaled with Denzo and Scalepack.14
Structure Determination and Refinement
Molecular replacement was used to solve both structures. The program PHASER was employed15 for P. laminosum cyt c 6, using the structure of M. braunii cyt c 6 (PDB entry 1ctj) as a search model.16 Data collected at the Feedge were useful to corroborate the position of the Fe atom in the maps and for modeling using anomalous Fourier synthesis. For the V52Q-AA variant the program MOLREP17 was used in conjunction with the oxidized wild-type-AA cyt c 6A structure (PDB code 2ce0) as the search model.5 A solution was obtained with an R-factor of 36.2% and a correlation coefficient of 65.0%. For both structures several rounds of rigid body and restrained refinement were carried out with REFMAC518 followed by automatic building of waters in ARP/WARP.19 Iterative cycles of model building with Coot20 followed by restrained refinement resulted in final models with the statistics reported in Table 1. The coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 2v08 (r2v08sf) for P. laminosum cyt c 6 and 2v07 (r2v07sf) for A. thaliana V52Q-AA cyt c 6A. Structure figures were prepared using the program Pymol.21
Table 1.
Crystallographic Data Processing and Refinement Statistics for P. laminosum Cytochrome c 6 and the V52Q-AA Variant of A. thaliana Cytochrome c 6A
| Pl cyt c 6 | V52Q-AA At cyt c 6A | |
|---|---|---|
| Data Collection and Processing | ||
| space group | P63 | P3221 |
| unit cell parameters (Å) | a = b = 57.4, | a = b = 57.8, |
| c = 89.5 | c = 65.1 | |
| no. of measured reflections | 36 142 | 16 890 |
| no. of unique reflections | 11 288 | 16 451 |
| -R merge (%)a | 9.6(23.1)b | 8.6 (26.4) |
| average I/σ(I) | 12.3 (4.8) | 19.2(5.6) |
| multiplicity | 3.2 (3.1) | 7.8 (4.8) |
| completeness (%) | 99.7(100.0) | 98.9 (97.5) |
| Refinement Statistics | ||
| resolution range (Å) | 25.59–2.0 | 50.1–1.60 |
| (2.05–2.00) | (1.64–1.60) | |
| R-factor (%) | 18.4(22.8) | 19.8(18.1) |
| R free-factor (%) | 25.5 (30.5) | 23.6 (22.6) |
| Quality of Model | ||
| rmsd bond lengths (Å) | 0.019 | 0.013 |
| rmsd bond angles (deg) | 1.779 | 1.356 |
| Ramachandran Plot Quality | ||
| % in most favored region | 84.9 | 84.8 |
| % in additional allowed region | 15.1 | 15.2 |
| Model | ||
| atoms (amino acids) | 1217(chain A3–86, chain B 3–86) | 929 (95) |
| ligands | 2 hemes | heme |
| water molecules | 116 | 115 |
| B-factor (Å2) | 26.8 | 19.5 |
Rmerge = Σi |I i – ⟨I⟩|/Σ ⟨I⟩ where ⟨I⟩ is the mean intensity of Nreflections with intensities I i and common indices h, k, and l (scalepack output). b Values of reflections recorded in the highest resolution shell are shown in parentheses.
Protein Film Voltammetry
Reduction potentials were measured by protein film voltammetry as described previously.22 All potentials are reported relative to the standard hydrogen electrode.
Results
Crystal Structure of Reduced P. laminosum cyt c6 and Comparison of the Heme Pocket with A. thaliana cyt c 6A
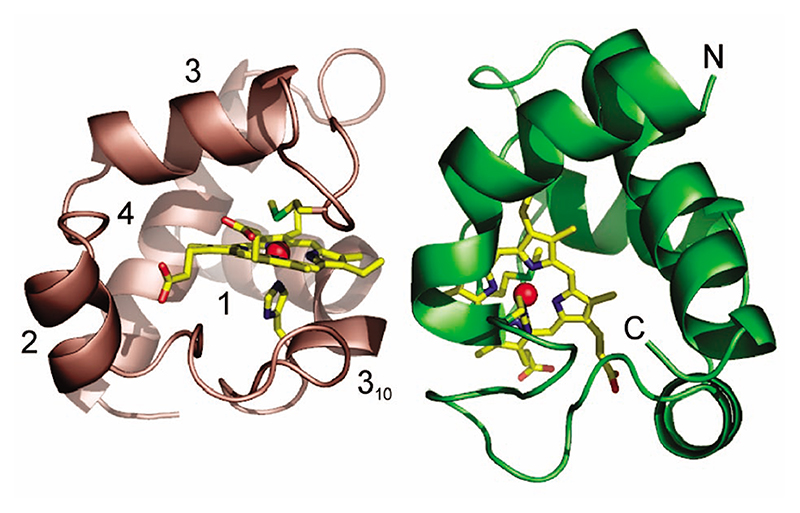
Reduced crystals of P. laminosum cyt c 6 contain two protein molecules in the asymmetric unit (chains A and B: residues 3–86) with the Cα-atoms of the two chains superimposing with a root-mean-square deviation (rmsd) of 0.09 Å. Statistics for data collection and refinement are summarized in Table 1. The two molecules form a dimer with an interface which is small (294 Å2), poorly packed, and largely hydrophobic (78% apolar atoms). An Fe–Fe distance of 16.2 Å and nonplanar orientation of the two hemes suggest optimization for fast intermolecular ET. A high interface planarity is also calculated indicative of weak interactions within the homodimer interface.23,24 The overall fold of each monomer is homologous to other cyts c 6 (Figure 1), and the backbone can be superimposed on that of cyt c 6A (excluding the LIP) with an rmsd of 1.3 Å. The heme iron is coordinated via the Nϵ1 of H19 and the Sδ of M59, with the latter having a trans orientation along the Cβ–Cγ bond, with R stereochemistry at the pro-chiral sulfur ligand. No hydrogen bond interaction to the Sδ atom of the Met ligand is observed.
Figure 1. Three-dimensional structure of the homodimer of reduced P. laminosum cyt c 6.
Chains A and B are colored green and salmon, respectively. The heme and axial ligands are in stick representation with the Fe as a red sphere. Each monomer is composed of four α-helices, three β-turns, and one γ-inverse turn. Helix 1 is distorted at C15 with residues 15–18 part of a 310 helix fragment. The N and C terminal helices are indicated for chain A.
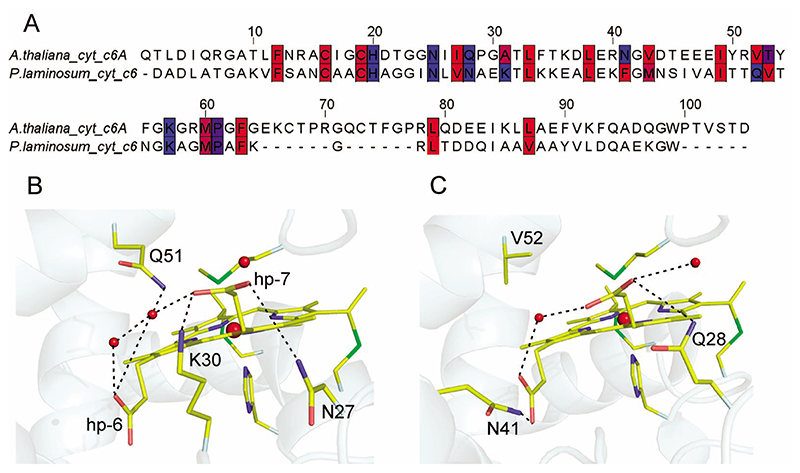
In both P. laminosum cyt c 6 and A. thaliana cyt c 6A the heme environment is predominantly hydrophobic, although differences in residue properties occur at postions 31, 41, 52, and 53 (A. thaliana numbering, Figure 2A) which may influence the heme E m value. A water molecule, interconnecting the heme propionates via hydrogen bonds, is observed in both structures (Figure 2). This water is structurally conserved in all cyt c 6 structures, but there is no evidence to suggest that it moves upon change of oxidation state in cyt c 6A or cyt c 6.4,5 A structural rearrangement involving a water molecule contributes to stabilizing the positively charged oxidized heme in the mitochondrial cyts c.2,3 The most extensive polar interactions are in the heme pocket of P. laminosum cyt c 6 (Figure 2 and Table S1 Supporting Information) which is dominated by the Q51 side chain. This is fixed into position by a hydrogen bond with the conserved water molecule and pinions the side chain so that its Nϵ2 atom is in van der Waals contact (3.0 Å) with a methine carbon atom of the porphyrin ring (Figure 2B).
Figure 2. Structural comparison of the heme binding sites.
(A) Sequence alignment between A. thaliana cyt c 6A and P. laminosum cyt c 6 (sequence identity 34%). Residues whose side-chain atoms are within 5 Å of a heme atom are colored red to blue according to their hydrophobicity (blue = most polar, red = least polar),45 using the JalView alignment editor.46 (B and C) The heme binding pockets of reduced P. laminosum cyt c 6 and reduced A. thaliana cyt c 6A, respectively. Water molecules are shown as small red spheres with the predominant polar interactions indicated by dashed lines; the larger spheres are the heme iron. The heme propionates are assigned as hp. A summary of the respective heme cavity polar interactions is given in Table S1.
The absence of the Gln residue is the predominant feature distinguishing the heme pocket of cyt c 6A from cyts c 6 (Figure 2B,C). No electrostatic interaction involving an electropositive amino acid side chain and a heme propionate is present in cyt c 6A, whereas a hydrogen bond between the heme propionate-6 and the side chain Nζ of K30 is present in P. laminosum cyt c 6 (Figure 2B, Table S1 Supporting Information). An electropositive side chain in this position is highly conserved within the cyt c 6 family but is an Ala (A31 in A. thaliana) or Ser in all known cyt c 6A sequences. The presence of a strongly electron withdrawing group interacting with a heme propionate would be expected to affect the heme E m 25,26 by electrostatic destabilization of the positive charge on the oxidized cyt and by a downward shift of the propionate pK in the oxidized form. Based on these differences, single-site variants were constructed in P. laminosum cyt c 6 (Q51V-c 6) and A. thaliana cyt c 6A (V52Q-c 6A and A31K-c 6A) to explore the individual contributions of these molecular substitutions on heme E m in the c 6 family.
Variations in the Heme Reduction Potentials
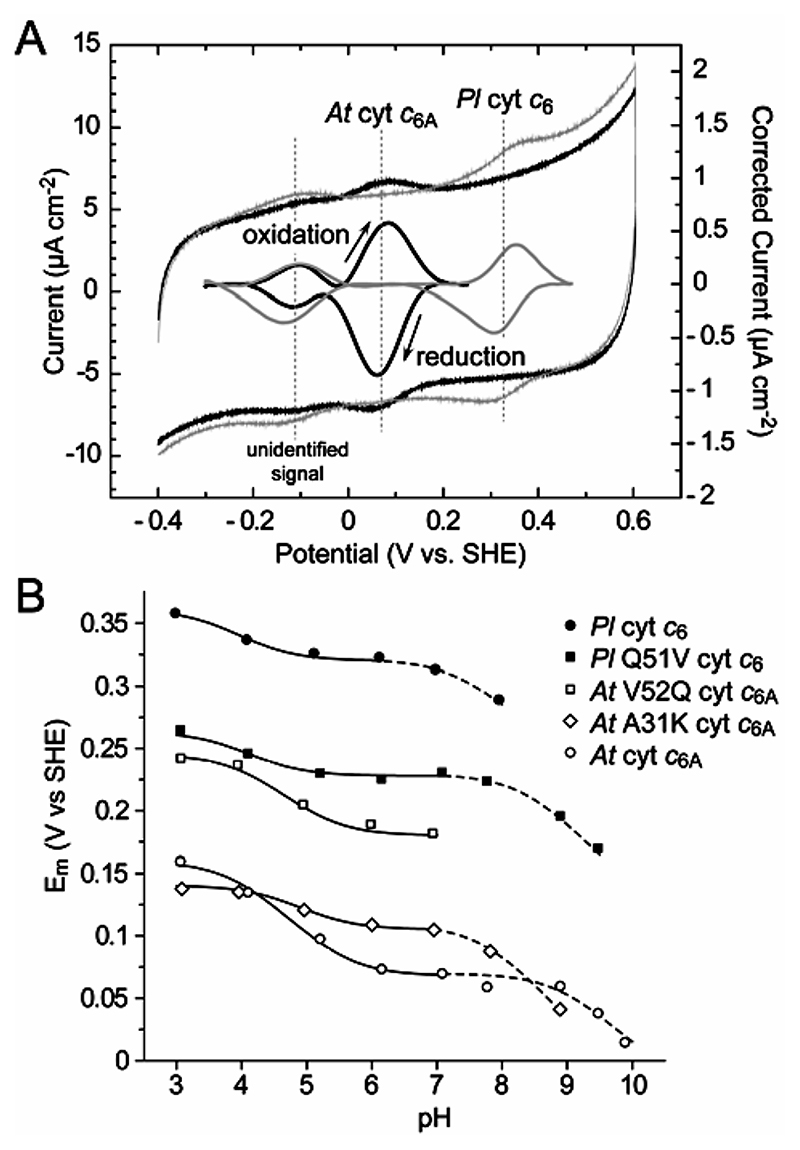
Typical cyclic voltammograms recorded for P. laminosum cyt c 6 and A. thaliana cyt c 6A are shown in Figure 3A. The small peak-to-peak separations observed (30–50 mV) confirm that the proteins were adsorbed onto the pyrolytic graphite edge (PGE) electrode surface, and the E m values are taken from the average of the reductive and oxidative peak potentials. Table 2 reports the pH-independent E m values for each protein, observed at close to neutral pH. The potential for A. thaliana cyt c 6A is ~250 mV lower than that of P. laminosum cyt c 6 which in thermodynamic terms corresponds to ~6 kcal/mol in the free energy of oxidation of the ferrous heme (ΔG°). For V52Q-c 6A an increase in Em of 109 mV is observed (Table 2), and an almost corresponding decrease is observed in the converse mutation (Q51V) in P. laminosum cyt c 6 (Table 2). Thus the Gln to Val mutation has accounted for almost half the difference in redox potential between cyt c 6 and cyt c 6A. Interestingly, residue 51/ 52 (Gln/Val), which lies close to the porphyrin ring, is responsible for the 2 nm bathochromic shift of the α-band in cyt c 6A (Table 2). Insertion of an electropositive residue in the vicinity of the heme propionate-6 in cyt c 6A (A31K) raises the E m of the heme (Table 2). However, this increase is only a third of that observed for the Val to Gln mutation in cyt c 6A (Table 2).
Figure 3.
(A) Cyclic voltammograms for cyt c 6A and cyt c 6 adsorbed on a PGE electrode, A. thaliana cyt c 6A (black) and P. laminosum cyt c 6 (gray), with their background corrected signals. The high potential redox couple is characteristic for protein-bound heme, whereas the low potential redox couple is from an unidentified contaminant (which is not visible on SDS gels and, hence, constitutes a small percentage of protein present but appears to react better with the electrode). Conditions: pH 7.1, scan rate 50 mV s-1, 25 °C. (B) pH dependence of the midpoint potentials (E m) at 25 °C. The data are fitted using the Nernst equation, E m = E acid – RT/F In[(1 + K ox/a H+)/(1 + Kred/a H+)], where E acid is the pH-independent value at low pH for pK ox1 and pK red1 (solid lines) and the pH-independent value at neutral pH for pK ox2 (dashed lines).
Table 2. Reduction Potentials, pK Values, and Spectral Data for Wild-Type and Mutant Proteins.
| protein | E m(mV)a neutral pH | ΔE m (vs WT) | pK ox1 | pK red1 | pK ox2 | α-band max (nm) |
|---|---|---|---|---|---|---|
| A. thaliana cyt c 6A | ||||||
| wild-type | +71 | - | 3.9 | 5.4 | 9.0 | 555.4 |
| V52Q | + 180 | + 109 | 4.1 | 5.2 | n.d. | 553.1 |
| A31K | + 105 | +34 | 4.6 | 5.2 | 7.8 | 555.4 |
| AA | +83 | +12 | 3.7 | 4.9 | 8.8 | 555.4 |
| AA-V52Q | + 168 | +97 | 3.6 | 5.1 | 8.5 | 553.1 |
| P. laminosum cyt c 6 | ||||||
| wild-type | +325 | - | 3.6 | 4.3 | 7.4 | 553.1 |
| Q51V | +225 | -100 | 4.0 | 4.6 | 8.3 | 555.5 |
a E m, the pH-independent reduction potential at approximately neutral pH. pK values were determined by fitting the data in Figure 3B (see text). Errors are ±5 mV for E m and ±0.05 for the pK values.
In each case variation of E m with pH displays three phases (Figure 3B). The physiologically significant pH range is probably 5 to 7, mostly in the pH independent region. The “low-pH” transition has been characterized using the Nernst equation for a single protonation event27 using two pK values, pK ox1 and pK red1, referring to the oxidized and reduced states, respectively (Table 2). pK ox1 and pK red1 probably refer to one of the heme propionates, most likely the less-solvent-exposed propionate-7, in analogy to other c-type cyts.28,29 The pK 1 values for V52Q-c 6A and Q51V-c 6 are similar to those of their respective wild-type proteins, but the separation between pKox1 and pKred1 is considerably larger for cyt c 6A (1.5) than c 6 (0.7), indicating increased interaction between the sites of electronation and protonation. The introduction of the positive charge with the A31K-c 6A mutant might be expected to lower pK ox1, whereas a small increase was actually observed (Table 2).
The “alkaline transition” described by pK ox2 occurs at a variable pH (Table 2) and has been modeled similarly by a coupled deprotonation event. The variants of each species show significant variations in their pK ox2 values, though the Q51V mutation shifts the value of P. laminosum cyt c 6 toward that of A. thaliana cyt c 6A, and the A31K-c 6A mutation shifts the value of A. thaliana cyt c 6A toward that of P. laminosum cyt c 6.
Redox Potential and Structure of the Oxidized V52Q-AA Variant of A. thaliana cyt c6A
The double Ala variant, I17A-G18A (AA), was used to study the structure of the V52Q-c 6A mutation. The reason for using the double AA mutation is that it crystallizes with a simpler unit cell and higher crystallographic order than the wild-type protein.5,11 The double AA mutations lie in the – CXXCH– heme binding motif and have minimal effect on the structure of cyt c 6A compared to the wild-type protein.5,11 Two Ala residues are found in the corresponding positions in P. laminosum cyt c 6 (Figure 2A) and many other cyts c 6. The effect on E m for the AA substitutions in cyt c 6A compared to the wild-type protein is small, with a slight increase in E m being observed (Table 2). The V52Q-AA-c 6A variant shows a large increase in E m over the AA variant, as observed between the wild-type protein and the V52Q-c 6A variant. The effect is a little smaller in the AA background than in the wild type (97 mV compared with 109 mV). The AA-c 6A and V52Q-AA-c 6A variants also display small shifts in the pK values (Table 2).
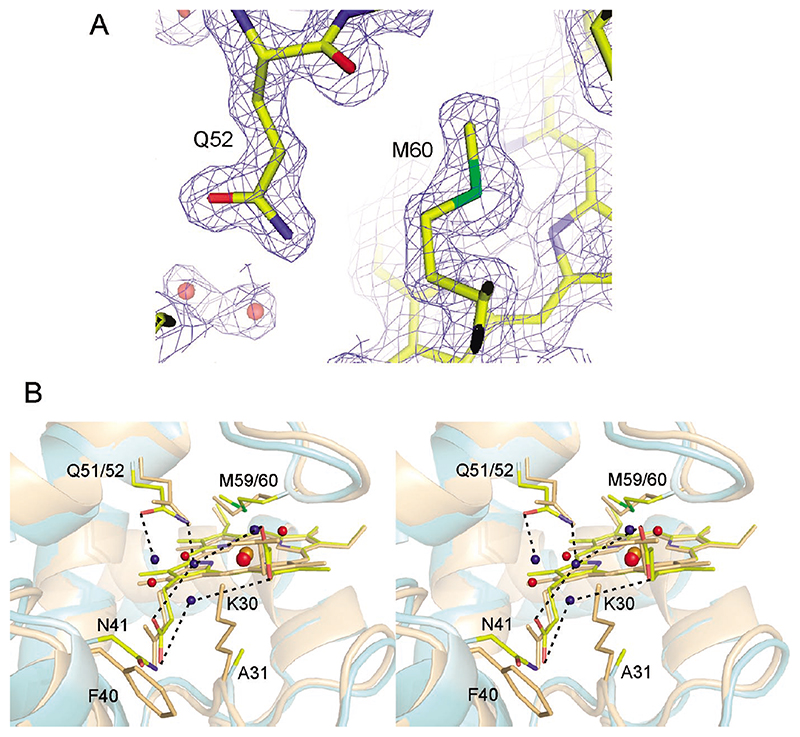
The structure of the oxidized V52Q-AA-c 6A variant was refined at 1.6 Å resolution; statistics for data collection and refinement are summarized in Table 1 and electron density shown in Figure 4A. Unambiguous electron density for a Gln residue was observed in difference maps of the V52Q-AA-c 6A, when using the AA-c 6A structure as the search model.5 Figure 4B compares the structures of the heme cavities in V52Q-AA-c 6A and P. laminosum cyt c 6. The side-chain orientation of Q52 is similar to the “naturally’ occurring Gln residue in P. laminosum cyt c 6 with the Nϵ2 atom hydrogen bonding with the structurally conserved bridging propionate water. This bridging water, a conserved presence throughout the c 6 structural family, facilitates orientation of the Gln side-chain amide group to within van der Waals contact (3.2 Å) of a methine carbon of the porphyrin ring. Additional small structural differences between V52Q-AA-c 6A and the AA-c 6A are confined to the periphery of the heme pocket. In V52Q-AA-c 6A the heme propionate-6 carboxyl groups are perpendicular instead of parallel to the heme plane producing small changes in length of hydrogen bonds from the atom O2D (Figure 4B and Table S1). This structural change also results in another water molecule bridging the two heme propionates (Figure 4B and Table S1). At the distal side of the heme, the axial heme ligand, M60, rearranges from a trans to a cis conformation along the Cβ-Cγbond retaining the same stereochemistry at the chiral sulfur (Figure 4A), but shifting slightly the main-chain atoms of M60 and P61.
Figure 4.
(A) Part of a σ-weighted electron density map contoured at 1.0 σ for the oxidized V52Q-AA-c 6A variant structure of A. thaliana. The Gln52 and Met60 referred to in the text are indicated. (B) Stereoview of the heme regions of an overlay of reduced P. laminosum cyt c 6 (light orange) and the oxidized V52Q-AA-c 6A variant of A. thaliana (pale cyan). Water molecules are shown as small spheres: red for P. laminosum cyt c 6 and blue for the V52Q-AA A. thaliana cyt c 6A variant. The iron atoms are shown as larger spheres: orange for P. laminosum and red for A. thaliana. Hydrogen bonding interactions are depicted by dashed lines; certain amino acids discussed in the text are labeled and shown as yellow sticks for A. thaliana or light orange sticks for P. laminosum.
Calculations of the Solvent Exposure of the Hemes in cyts c6 and c 6A
Differences in the solvent exposure of the heme in cyts c 6 and c 6A may contribute to the large difference in reduction potential (see Table 3). The total exposure of the heme is dominated by that of propionate-6. However, the propionic acid side chain is not part of the conjugated ring system, and its main effect on E m is the result of electrostatic stabilization of the ferric heme state;28,30 in all cases studied the propionate remains mainly charged in the pH range 6–7. Alternatively, calculating the solvent exposure for the iron-porphyrin ring system and atoms within one α-bond of the ring, as suggested by Tezcan et al.,1 may prove more meaningful (see Table 3). The smaller areas of exposure result largely from the methyl-3 carbon atom. Although the cyts c 6 mostly show lower exposures than cyt c 6A, by both criteria the spread of values is wide and the number of samples for cyt c 6A small. The V52Q-AA-c 6A substitution increased the total heme exposure but had an insignificant effect on the porphyrin ring exposure, perhaps because of the slight readjustment in the position of the main chain discussed above.
Table 3.
Comparison of Heme Solvent Accessibility between A.thaliana Cyt c 6A and Cyt c 6 for Which Structures Have Been Determined with X-ray Crystallography
| solvent exposure Å2a | ||||
|---|---|---|---|---|
| Pdb code | E m,7(mV) | total heme | porphyrin ringe | |
| Cytochrome c 6A | ||||
| Arabidopsis thaliana wild-type | 2dge11 | 71b,d/94c,5 | 70.4 | 15.9 |
| I17A/G18A oxidized | 2ce05 | 83b·d/89c·5 | 78.9 | 21.6 |
| I17A/G18A reduced | 2ce15 | 79.6 | 20.6 | |
| I17A/G18A/V52Q oxidized | 2v07d | 168b·d | 97.2 | 19.8 |
| Cytochrome c 6 | ||||
| Arthrospira maxima | 1f1f32 | 314c,47 | 62.3 | 17.9 |
| Cladophora glomerata | 1ls948 | 355 b,48 | 60.9 | 11.2 |
| Chlamydomonas reinhardtii | 1cyi/1cyj31 | 370c,49 | 70.0 | 9.4 |
| Monoraphidium braunii | 1ctj16 | 358c,50 | 60.7 | 7.4 |
| Phormidium laminosum | 2v08d | 325b,d | 57.7 | 14.7 |
| Porphyra yezoensis | 1gdv51 | 52.1 | 10.1 | |
| Scenedesmus obliquus | ||||
| oxidized | 1c6o4 | 73.6 | 8.8 | |
| reduced | 1c6r4 | 67.4 | 12.0 | |
a Calculated with NACCESS.52 b Obtained by CV. c Potentiometric titration. d Present work. e Cβ, O1, and O2 atoms were excluded from the calculation.
Discussion
As part of the present work we have determined the structure of reduced cyt c 6 from the cyanobacterium P. laminosum which reveals a homodimer in the asymmetric unit. The overall fold and heme architecture are very similar to other cyts c 6 for which structures are known. Homodimers in the solid and solution state have been reported for other cyts c 6.4,31,32 However in the solid state none has interface properties resembling a transient ET complex as is observed here for P. laminosum cyt c 6. The P. laminosum cyt c 6 monomer was used to search for the molecular origins of the difference in heme redox potential displayed between cyt c 6 and cyt c 6A. At physiological pH the latter has an Em of +71 mV measured using PFV and is the lowest for a naturally occurring cyt c with His and Met as heme axial ligands. This potential is more in line with bis-His cyts where the replacement of the axial Met with a His residue results in a downward shift in potential as observed in the semisynthetic bis-His cyt c.33 Thus the low heme midpoint potential of cyt c 6A suggests that factors other than the nature of the axial ligand are responsible.
Residue 51/52 is a major regulator of Em between cyt c 6 and cyt c 6A
Removal of the polar Gln and replacement with the hydrophobic Val, as found “naturally” in many cyts c 6A, would a priori have been predicted to increase the heme redox potential, as the positive charge on the oxidized heme is destabilized by the increased hydrophobicity. However, the effect is the opposite. In all known cyt c 6 structures and in the V52Q-AA-c 6A variant, the conserved Gln side chain is hydrogen bonded to a structurally conserved water molecule. This water locks the side chain into an orientation in which the amide group is pointing toward the porphyrin ring and within van der Waals distance of a methine carbon on the porphyrin ring. We suggest that this positioning of the Gln side chain is responsible for ~100 mV of heme redox potential in the cyts c 6, with the conserved water having an important structural role.
One possibility is that the amide group of the Gln and the porphyrin ring form an unusual aromatic hydrogen bond (one proton from the amide group (the donor) points toward a ring carbon atom (the acceptor)). Aromatic hydrogen bonds are estimated to have half the strength of conventional hydrogen bonds.34 Such an interaction could perturb the electron spin density of the porphyrin ring and so influence the redox potential. Amide/aromatic H-bonds have a larger interplanar angle between the ring perpendicular and the donor than the more common amide/aromatic stacking interactions, a separation of less than 3.8 Å between the donor (the sp2 nitrogen) and acceptor, and an N–H···C angle greater than 120°.35 In both P. laminosum cyt c 6 and the V52Q-AA-c 6A structures, the sp2 hybridized nitrogen of the Gln side chain is less than 3.5 Å from the ring carbon of the aromatic porphyrin, with a N–H···C of ~105°. Thus the interaction does not quite fulfill all the criteria for an amide/aromatic hydrogen bond, but the van der Waals contact does suggest that it is a key determinant of the physical properties of the porphyrin. This is consistent with the spectral shift caused by the presence of the Gln residue (Table 2) which is a clear indication that it interacts with the π-electron system of the porphyrin ring.36
A second possibility is a simple dipolar interaction with the polar Gln side chain destabilizing the positively charged oxidized heme. A similar electrostatic interaction has been reported in mitochrondrial cyts c involving a side chain dipole of an Asn and a water molecule.37 Upon substitution of the Asn for an Ile the water molecule in the heme pocket is lost together with the side chain dipole. This results in a smaller destabilization of the oxidized state and a 55 mV decrease in E m.37
Other Contributors to the Differences in Em between cyt c 6 and cyt c 6A
Although the Gln/Val substitution results in a significant change in reduction potential, it does not account for all of the difference (~ 250 mV) between cyt c 6 and cyt c 6A. Other factors which may be important are the electron spin-density distribution in the porphyrin ring system, the Fe–Sδ- (Met) bond strength, further electrostatic interactions, and heme solvent exposure. In V52Q-AA-c 6A a change in the side chain orientation of M60 from trans to cis is observed (Figure 4A) with the stereochemistry of the sulfur atom retained. This change in orientation of the Met ligand will influence the electronic structure of the porphyrin which in turn could have an influence on the E m.38 However, from extensive work by Walker and co-workers the axial ligand orientation is expected to have only a small effect on the heme E m in cyts c.39
Previous structural analysis suggested the electrostatic interaction between an electropositive amino acid side chain and a heme propionate was the key determinant of the decreased reduction potential of cyt c 6A.11 Here, study of the A31K-c 6A variant showed that introduction of a positively charged residue does indeed raise the heme’s redox potential, but by only 34 mV (Table 2). This substitution could therefore be considered to “fine-tune” the reduction potential, as it does not account for a major part of the difference in potential between cyts c 6 and c 6A. Likewise mutation of the corresponding residue in a cyt c 6, K29 to a His resulted in a downward shift in redox potential of 65 mV.40 Together residues 31 and 51/52 contribute ~ 150 mV of heme redox potential (ΔG° ≈ 3.5 kcal/mol) which equates to ~50% of the known accessible redox space sampled in Class I cyts c and 60% of the difference between the proteins studied here. Other residues, identified above, which could contribute to a further lowering of redox potential are N41 and T53. The side chain of N41 is within hydrogen-bonding distance of heme propionate-7, but cyt c 6 from several species also has an Asn in this position. T53 would be expected to have some effect on heme potential, but this would probably be smaller than that of a Gln at position 51/52 because the side chain Oγis not in van der Waals contact with the porphyrin ring and the side chain dipole is smaller than that of Gln. Moreover, cyt c 6A of Chlamydomonas has Ile instead of Thr in this position.41
Differences in the solvent exposure of the porphyrin rings may, in comparison to the results of Tezcan et al.,1 contribute up to 100 mV to the reduction potential. Although examination of space filling models of the known crystal structures suggests a larger gap adjacent to the heme methyl-3 carbon atom in cyt c 6A compared with c 6, we have been unable to find a single residue that is conserved in one group and not in the other and could be responsible for the change in solvent exposure of the heme.
Evolution of Heme Redox Potential in cyt c 6A and Its Biological Implications
The original function of cyt c 6 in chloroplasts was to transfer electrons from the cyt b 6 f complex to photosystem I (and in photosynthetic bacteria it may also have served as an electron donor to cyt c oxidase). Oxidation of the cyt b 6 f complex requires a high redox potential. Attempts to modify the redox potential of cyts by site-directed mutagenesis have usually led to a lowering of potential. Lett and Guillemette found only one mutant of yeast iso-1-cyt c in which the potential was raised,25 and McClendon and co-workers found that the potential of wild-type cyt b562 lay at the upper end of a range of potentials of mutant forms.42,43 The same authors suggested that many cyts (and possibly many other redox proteins) have evolved to maximize the redox potential by stabilizing their reduced state relative to the oxidized state, consistent with cyt c 6 accepting electrons from the cyt b 6 f complex. By contrast, the evolution of cyt c 6A has followed a path of increasing stabilization of the oxidized state relative to the reduced, leading to an unusually low E m for a cyt c with His-Met ligation.
The function of cyt c 6A remains unknown, but it is not involved in photosynthetic electron transfer beween the cyt b 6 f complex and photosystem I.12,44 Given the high degree of conservation of residues forming the heme pocket of cyt c 6A, it is likely that the low redox potential is a universal feature of the cyt in higher plants. Assuming that oxidation and reduction of the heme are an essential feature of the function of cyt c 6A, the lowering of midpoint potential of cyt c 6 by ~100 mV by the Gln to Val substitution is an example of how a single amino acid change could, in principle, alter the properties of the protein to abolish its original function and perform a new one more effectively. Presumably substitution of the Gln would have been preceded by a duplication of the cyt c6 gene, to allow retention of a “conventional” cyt c 6 for photosynthetic electron transfer, as in Chlamydomonas.41 One form of the protein acquired a new function, perhaps associated with insertion of the cysteine-containing stretch into the LIP. Substitution of the Gln ensured that reduction by the cyt b6f complex could not interfere with the new function, and this was probably followed by additional tuning of cyt c 6A to lower its midpoint potential further, resulting in the protein seen today.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust, BBSRC, the Medical Research Council, the Nuffield Foundation, and the Newton Trust. We thank the staff of the European Synchrotron Radiation Facility (ESRF) for support with data collection.
The abbreviations used are as follows
- cyt c
cytochrome c
- PGE
pyrolytic graphite edge
- LIP
loop insertion peptide
- IPTG
Isopropyl-D-thiogalactoside
- Em
midpoint potential
- ET
electron transfer
References
- (1).Tezcan FA, Winkler JR, Gray HB. J Am Chem Soc. 1998;120:13383–13388. [Google Scholar]
- (2).Berghuis AM, Brayer GD. J Mol Biol. 1992;223:959–976. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- (3).Berghuis AM, Guillemette JG, McLendon G, Sherman F, Smith M, Brayer GD. J Mol Biol. 1994;236:786–799. doi: 10.1006/jmbi.1994.1189. [DOI] [PubMed] [Google Scholar]
- (4).Schnackenberg J, Than ME, Mann K, Wiegand G, Huber R, Reuter W. J Mol Biol. 1999;290:1019–1030. doi: 10.1006/jmbi.1999.2944. [DOI] [PubMed] [Google Scholar]
- (5).Marcaida MJ, Schlarb-Ridley BG, Worrall JAR, Wastl J, Evans TJ, Bendall DS, Luisi BF, Howe CJ. J Mol Biol. 2006;360:968–977. doi: 10.1016/j.jmb.2006.05.065. [DOI] [PubMed] [Google Scholar]
- (6).Ho KK, Krogmann DW. Biochim Biophys Acta. 1984;766:310–316. [Google Scholar]
- (7).Merchant S, Bogorad L. Mol Cell Biol. 1986;6:462–469. doi: 10.1128/mcb.6.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wood PM. Eur J Biochem. 1978;87:9–19. doi: 10.1111/j.1432-1033.1978.tb12346.x. [DOI] [PubMed] [Google Scholar]
- (9).Gupta R, He Z, Luan S. Nature. 2002;417:567–571. doi: 10.1038/417567a. [DOI] [PubMed] [Google Scholar]
- (10).Wastl J, Bendall DS, Howe CJ. Trends Plant Sci. 2002;7:244–245. doi: 10.1016/s1360-1385(02)02280-x. [DOI] [PubMed] [Google Scholar]
- (11).Chida H, Yokoyama T, Kawai F, Nakazawa A, Akazaki H, Takayama Y, Hirano T, Suruga K, Satoh T, Yamada S, Kawachi R, et al. FEBS Lett. 2006;580:3763–3768. doi: 10.1016/j.febslet.2006.05.067. [DOI] [PubMed] [Google Scholar]
- (12).Molina-Heredia FP, Wastl J, Navarro JA, Bendall DS, Hervas M, Howe CJ, De La Rosa MA. Nature. 2003;424:33–34. doi: 10.1038/424033b. [DOI] [PubMed] [Google Scholar]
- (13).Braman J, Papworth C, Greener A. Methods Mol Biol. 1996;57:3144. doi: 10.1385/0-89603-332-5:31. [DOI] [PubMed] [Google Scholar]
- (14).Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- (15).McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr, Sect D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- (16).Frazao C, Soares CM, Carrondo MA, Pohl E, Dauter Z, Wilson KS, Hervas M, Navarro JA, De la Rosa MA, Sheldrick GM. Structure. 1995;3:1159–1169. doi: 10.1016/s0969-2126(01)00252-0. [DOI] [PubMed] [Google Scholar]
- (17).Vagin A, Teplyakov A. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- (18).Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr, Sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- (19).Perrakis A, Sixma TK, Wilson KS, Lamzin VS. Acta Crystallogr, Sect D. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- (20).Emsley P, Cowtan K. Acta Crystallogr, Sect D. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (21).DeLano WL. The Pymol Molecular Graphics Systems. San Carlos, CA, U.S.A: 2002. [Google Scholar]
- (22).Leggate EJ, Hirst J. Biochemistry. 2005;44:7048–7058. doi: 10.1021/bi050189x. [DOI] [PubMed] [Google Scholar]
- (23).Nooren IM, Thornton JM. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- (24).Sato K, Crowley PB, Dennison C. J Biol Chem. 2005;280:19281–19288. doi: 10.1074/jbc.M500842200. [DOI] [PubMed] [Google Scholar]
- (25).Lett CM, Guillemette JG. Biochem J. 2002;362:281–287. doi: 10.1042/0264-6021:3620281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lo TP, Komar-Panicucci S, Sherman F, McLendon G, Brayer GD. Biochemistry. 1995;34:5259–5268. doi: 10.1021/bi00015a041. [DOI] [PubMed] [Google Scholar]
- (27).Clark WM. Oxidation-Reduction Potentials of Organic Systems. Bailliere, Trindall and Cox; London: 1960. [Google Scholar]
- (28).Takayama SJ, Mikami S, Terui N, Mita H, Hasegawa J, Sambongi Y, Yamamoto Y. Biochemistry. 2005;44:5488–5494. doi: 10.1021/bi047498s. [DOI] [PubMed] [Google Scholar]
- (29).Ye T, Kaur R, Wen X, Bren KL, Elliott SJ. Inorg Chem. 2005;44:8999–9006. doi: 10.1021/ic051003l. [DOI] [PubMed] [Google Scholar]
- (30).Mao J, Hauser K, Gunner MR. Biochemistry. 2003;42:9829–9840. doi: 10.1021/bi027288k. [DOI] [PubMed] [Google Scholar]
- (31).Kerfeld CA, Anwar HP, Interrante R, Merchant S, Yeates TO. J Mol Biol. 1995;250:627–647. doi: 10.1006/jmbi.1995.0404. [DOI] [PubMed] [Google Scholar]
- (32).Sawaya MR, Krogmann DW, Serag A, Ho KK, Yeates TO, Kerfeld CA. Biochemistry. 2001;40:9215–9225. doi: 10.1021/bi002679p. [DOI] [PubMed] [Google Scholar]
- (33).Raphael AL, Gray HB. Proteins. 1989;6:338–340. doi: 10.1002/prot.340060316. [DOI] [PubMed] [Google Scholar]
- (34).Levitt M, Perutz MF. J Mol Biol. 1988;201:751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- (35).Mitchell JB, Nandi CL, McDonald IK, Thornton JM, Price SL. J Mol Biol. 1994;239:315–331. doi: 10.1006/jmbi.1994.1370. [DOI] [PubMed] [Google Scholar]
- (36).Ponamarev MV, Schlarb BG, Howe CJ, Carrell CJ, Smith JL, Bendall DS, Cramer WA. Biochemistry. 2000;39:5971–5976. doi: 10.1021/bi9928997. [DOI] [PubMed] [Google Scholar]
- (37).Langen R, Brayer GD, Berghuis AM, McLendon G, Sherman F, Warshel A. J Mol Biol. 1992;224:589–600. doi: 10.1016/0022-2836(92)90546-v. [DOI] [PubMed] [Google Scholar]
- (38).Shokhirev NV, Walker FA. J Biol Inorg Chem. 1998;3:581–594. [Google Scholar]
- (39).Walker FA, Huyuh BH, Scheidt WR, Osvath SR. J Am Chem Soc. 1986;108:5298–5297. [Google Scholar]
- (40).De la Cerda B, Diaz-Quintana A, Navarro JA, Hervas M, De la Rosa MA. J Biol Chem. 1999;274:13292–13297. doi: 10.1074/jbc.274.19.13292. [DOI] [PubMed] [Google Scholar]
- (41).Wastl J, Purton S, Bendall DS, Howe CJ. Trends Plant Sci. 2004;9:474–476. doi: 10.1016/j.tplants.2004.08.005. [DOI] [PubMed] [Google Scholar]
- (42).Springs SL, Bass SE, Bowman G, Nodelman I, Schutt CE, McLendon GL. Biochemistry. 2002;41:4321–4328. doi: 10.1021/bi012066s. [DOI] [PubMed] [Google Scholar]
- (43).Springs SL, Bass SE, McLendon GL. Biochemistry. 2000;39:6075–6082. doi: 10.1021/bi0001675. [DOI] [PubMed] [Google Scholar]
- (44).Weigel M, Varotto C, Pesaresi P, Finazzi G, Rappaport F, Salamini F, Leister D. J Biol Chem. 2003;278:31286–31289. doi: 10.1074/jbc.M302876200. [DOI] [PubMed] [Google Scholar]
- (45).Kyte J, Doolittle RF. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- (46).Clamp M, Cuff J, Searle SM, Barton GJ. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- (47).Cho YS, Wang QJ, Krogmann D, Whitmarsh J. Biochim Biophys Acta. 1999;1413:92–97. doi: 10.1016/s0005-2736(99)00124-8. [DOI] [PubMed] [Google Scholar]
- (48).Dikiy A, Carpentier W, Vandenberghe I, Borsari M, Safarov N, Dikaya E, Van Beeumen J, Ciurli S. Biochemistry. 2002;41:14689–14699. doi: 10.1021/bi026473v. [DOI] [PubMed] [Google Scholar]
- (49).Gorman DS, Levine RP. Plant Physiol. 1966;41:1637–1642. doi: 10.1104/pp.41.10.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Campos AP, Aguiar AP, Hervas M, Regalla M, Navarro JA, Ortega JM, Xavier AV, De La Rosa MA, Teixeira M. Eur J Biochem. 1993;216:329–41. doi: 10.1111/j.1432-1033.1993.tb18150.x. [DOI] [PubMed] [Google Scholar]
- (51).Yamada S, Park SY, Shimizu H, Koshizuka Y, Kadokura K, Satoh T, Suruga K, Ogawa M, Isogai Y, Nishio T, Shiro Y, et al. Acta Crystallogr, Sect D. 2000;56:1577–1582. doi: 10.1107/s090744490001461x. [DOI] [PubMed] [Google Scholar]
- (52).Hubbard SJ, Thornton JM. NACCESS, Computer program. Department of Biochemistry and Molecular Biology, University College; London: 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.