Abstract
Supervised learning algorithms trained on medical images will often fail to generalize across changes in acquisition parameters. Recent work in domain adaptation addresses this challenge and successfully leverages labeled data in a source domain to perform well on an unlabeled target domain. Inspired by recent work in semi-supervised learning we introduce a novel method to adapt from one source domain to n target domains (as long as there is paired data covering all domains). Our multi-domain adaptation method utilises a consistency loss combined with adversarial learning. We provide results on white matter lesion hyperintensity segmentation from brain MRIs using the MICCAI 2017 challenge data as the source domain and two target domains. The proposed method significantly outperforms other domain adaptation baselines.
Keywords: Domain adaptation, Adversarial learning, Brain MR
1. Introduction
In medical imaging, fully automated tools using deep learning techniques are increasing in popularity for numerous clinical tasks, including image segmentation, image classification and instance counting [8]. Among these tools, deep learning frameworks exhibit excellent performance (often described as ‘superhuman’) when applied on images drawn from the same distribution (scanner type, parameters, patient pool etc.) as the one used in training the model. However, the performance may deteriorate drastically when the algorithm is applied in previously unseen domains. This performance gap is a critical barrier to the safe implementation and widespread adoption of these techniques in clinical practice.
The process of adapting a model from a ‘source’ domain to a target domain is called ‘domain adaptation’. Successful methods have included:
Training with a small number of labeled examples from the target domain. While this solution is theoretically straightforward, its practical use is limited as it requires additional labelling on the target domain.
Embedding the imaging data in a latent space. This latent space is learnt so as to ignore domain-specific features (e.g. contrast), while retaining domaininvariant features (pathology). Adversarial approaches have been proposed to address this angle in the context of lesion segmentation [5]and heart structure segmentation between MR and CT [2]. In both cases, the adversarial training is used to make the latent space as uninformative as possible about the domain the images come from.
Semi-supervised methods use a model trained on a small number of labeled examples to provide pseudo-labels for unlabeled data, which is then trained on. The model-fitting and updating semi-supervised labels can be seen as a form of expectation maximisation and has been used in medical imaging [1].
Enforcing output robustness to input perturbation. Recent methods have exploited the property that the distribution of predictions should be invariant to small perturbations on the input data. This observation can be expressed as , where is an augmented/perturbed version of x. The enforcement of this property has the additional advantage of limiting the unwanted behaviour of drastic output change for minimal input perturbations, which can be seen as improving robustness. For instance Perone et al. [9] proposed a teacher-student framework ensuring consistency between the outputs when passing to the student an augmented version of the unlabeled input of the teacher, that is similarly augmented afterwards.
Methods 2 to 4 fall under the purview of ‘Unsupervised Domain Adaptation’ (UDA), as does the presented work. In general, UDA does not rely on labeled training examples from the desired target domain. This is especially desirable in medical imaging, where labelling is time-consuming and highly variable, and the ‘domain’ depends on scanner manufacturer, acquisition protocol and reconstruction strategy. The augmentations required to create the perturbed input data can either be generic (geometric or contrast operations) or application-specific. In the context of medical imaging, the latter includes physics-based image augmentation, synthetic bias field addition or registration-based approaches [14]. These methods lean on domain-specific knowledge to generate plausible transformations.
We propose a UDA pipeline applied to the segmentation of white matter hyperintensities (WMH) which introduces a paired consistency (PC) loss which guides the adaptation. The proposed (PC) method enforces the output consistency between the results obtained on two separate acquisitions per subject: an in-plane and a volumetric FLAIR sequence. We aim to (1) segment WMH lesions on completely unlabeled examples and (2) to make these predictions similar between the in-plane and volumetric cases. In other words, we regularize the fitting by explicitly promoting similarity in the labels generated by each FLAIR acquisition. This adaptation method was supplemented with an adversarial loss in order to prevent the model from getting stuck in bad local minima. After an overview of the proposed approach and its variants in Sect. 2, we present the experiments which show that our proposed method leverages the unlabeled data to produce more consistent lesion segmentations across all domains.
2. Methods
The proposed training strategy for domain adaptation occurs in two phases. In the first phase the network is trained only on labeled data until convergence. During the second phase of the training, the paired unlabeled data is presented in addition to the labeled data and a consistency term is added to the loss function. This consistency term is inspired from the loss proposed by Xie et al. [13] that aims at minimizing the Kullback-Leibler divergence between the output probability distribution y when conditioned on the unlabeled input x from the set U or its augmented countrapart drawn from .
| (1) |
We adapt this method to the segmentation task by using the dice loss [7] instead of the KL divergence. In the following, we denote as yl the labeled ground truth, ŷl the prediction over labeled images, ŷu the prediction over unlabeled input and ŷû the prediction over its augmented/paired counterpart. The losses used in our framework are thus expressed as follows:
| (2) |
We trained networks using ℒtot as specified in (2) and denote them as PC. These networks fθ(h|x) produce a feature representation h from which y is calculated.
Preventing Trivial Solutions
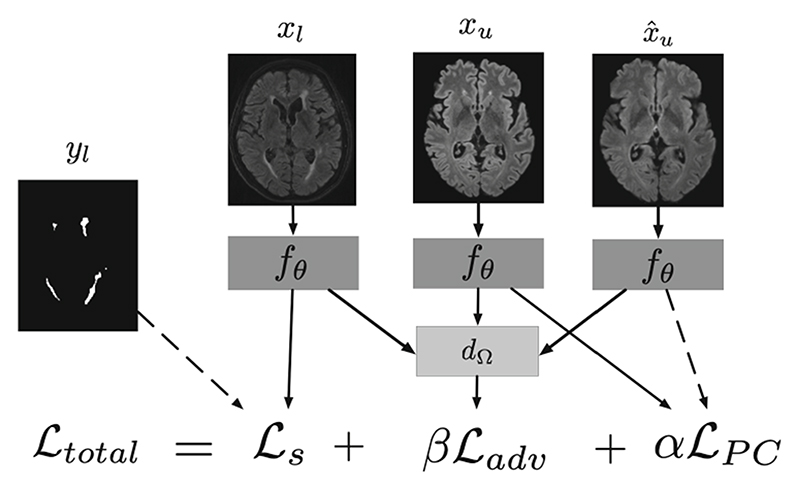
Early in our experiments, we encountered a specific degenerate solution: our network was able to produce one solution for source images (a good lesion mask) while producing a trivial result on the target domain (in this case, a mask of the foreground). This meant that there was good agreement between in-plane and volumetric FLAIRs because they simply segmented foreground—ignoring the lesions altogether. This means that the network was identifying the domain of the images and using this to inform its solution: undesirable behaviour. We introduced an additional adversarial term to avoid these ‘solutions’. Inspired by the domain adversarial literature (methods 2 in Sect. 1) we propose an adversarial loss to minimize the amount of information about domain contained in h. We introduce a discriminator dΩ which takes h as input and outputs a domain prediction . The adversarial loss, ℒadv is given by the cross-entropy, where n is the number of domains, is the multi-class cross entropy loss, d is a one-hot encoded vector of the domain label and is the model’s domain prediction as in [11]. We use a gradient reversal layer as in [5] in order to minimize Ltot whilst maximizing Ladv. Figure 1 presents the diagram of the proposed method with the combination of different losses, where β controls the strength with which the model is adapting its features whereas α controls the weights the consistency effect.
Fig. 1.
Diagram of proposed method. At training time, xu, xl and yl are supplied to the network. xu is an image from the unlabeled target domain and x̂u is the result of applying some augmentation function to xu. A labeled image, xl, is passed through the network, fθ before combining with a label yl to form the segmentation loss, ℒs. The image representations are fed to a domain discriminator d Ω which attempts to maximise the cross-entropy between predicted domain and actual domain, ℒadv. Finally, similarity is promoted between the network predictions on xu and using ℒPC.
Augmentation
In [13] the authors suggest various properties of augmented samples necessary for performing Unsupervised Data Augmentation. Samples should be realistic, valid (meaning they should not alter the underlying label), smooth, diverse and make use of targeted inductive biases (domain knowledge). In the absence of sufficiently realistic augmentation functions we use paired scans which are considered as augmented samples. However, taking them as they are makes for a discrete augmentation function with discontinuous jumps. In order to encourage continuity we used a large range of augmentations on the paired data, including generic geometric transformations and MR specific non-geometric transformations. Geometric augmentations were sampled independently and combined as one affine transform, using random rotations (all axis ranging from –10 to 10°), random shears ([0.5, 0.5]) and random scaling ([0.75,1.5]). For the non-geometric augmentations we applied k-space motion artefact augmentation as described in [12] and bias field augmentation as implemented in [3]. We measure how useful these additional augmentations are in our experiments.
3. Experiments and Results
Data
In this work we focus on white matter hyperintensity segmentation. The data comes from two separate studies. As a source domain we use the White Matter Hyperintensity challenge data presented in MICCAI 2017 [6]. The other dataset was used as target domain and comes from a sub-study within the Pharmacokinetic and Clinical Observations in PeoPle over fiftY (POPPY) [4]. In this study two different FLAIR sequences were acquired during the same MR session for all 72 subjects on a Philips 3T scanner. The in-plane FLAIR was an axial acquisition with 3 mm slice thickness and 1 mm2 planar resolution (Repetition time (TR) 8000 ms, Inversion time (TI) 2400 ms and echo time (TE) 125 ms) while the volumetric FLAIR was of resolution 1.04 × 1.04 × 0.56 mm3 (TR = 8000ms TI = 1650 ms TE = 282 ms). Both images were rigidly coregistered to the 1 mm3 T1 sequence acquired during the session. All individuals were male with mean age of 59.1 ± 6.9 yrs, including HIV-positive subjects and population-matched controls.
Implementation Details and Training
The MICCAI Challenge dataset was split with a train:validation:test assignment of 40:10:10 subjects. For the POPPY dataset, the split was 38:15:20.
Training was done using 2d axial slices of size 256 × 256 with inference carried out by concatenating the predictions across all slices to form a 3d volume. The segmentation network uses the U-Net architecture [10] with depth of 4 and a maximum number of filters of 256 at the deepest layer, with ReLU as the activation function. Initial training on the MICCAI dataset only was performed using the Adam optimizer with an initial learning rate 10-3 and a learning rate decay schedule decaying with γ = 0.1 (γ is a multiplicative factor of learning rate decay) at epoch 300 and 350. The validation set is used for early stopping, thus the baseline model takes the network configuration at the epoch where it showed the highest accuracy on the validation set. All adaptation models and adversarial models were initialized with the weights of this trained baseline model.
The choice of α parameter balancing the segmentation and the consistency loss in the domain adaptation runs proved to be important. Generally, high values of a led to degenerate solutions, where predictions on the target dataset were no longer capturing lesions. Since scheduling a slowly increasing a did not help, a was fixed at 0.2 in all experiments.
In case of an adversarial setting, empirical assessment of the best choice of architecture for the discriminator led to the following choice: four 2D convolutional layers with a kernel size of 3 × 3 and a stride of 2 followed by batch normalisation and leaky ReLU activation. The number of output channels is 4 to begin with and doubles at each layer to a total of 32. Finally, there are three fully connected layers with output sizes of 64, 32, and 2 with relu activations and dropout applied (p = 0.5).
Points of Comparison
In order to assess the relevance of the proposed paired consistency, we compared the proposed PC with adversarial setting and augmentation (PC+Adv+Aug) to the version without adversarial setting (PC+Aug) and the simplest version removing also the augmentation (PC). In addition, we trained classical UDA methods with a mean-teacher framework (MT) as well as the adversarial setting without PC with (Adv+Aug) and without augmentation (Adv). Finally we compared to the baseline U-Net model trained only on the MICCAI dataset with (Baseline+Aug) and without (Baseline) augmentation.
For the final results table checkpoints were chosen for each of the experiments by looking at the performance across the validation set.
Reported Metrics
As a first metric of consistency, we compute the Dice score overlap between the two volumes. However, high dice agreement may arise without predicting lesions, for instance with the segmentation of foreground or of another anatomical structure. Such degenerate solutions can indeed occur as the consistency term in the loss can be minimized for any consistent prediction between volumes.
As there are no lesion segmentations for the POPPY dataset, we use the known association between age and white matter hyperintensity load reported for this dataset [4] as surrogate evaluation that the segmented elements are lesions. The effect size is a useful metric for determining whether the lesion loads predicted by the various models agree with the reported literature. For the eight compared models, the effect size ranged from 1.2-fold to 1.5-fold increase in lesion load normalized by total intracranial volume per decade. This compares well with the reported effect size on the POPPY dataset of 1.4-fold with a 95th confidence interval of [1.0; 2.0]. Predictions from in-plane POPPY and volumetric POPPY were compared using the dice overlap, the 95th percentile Hausdorff distance measured in mm (HD95), the recall (or sensitivity), the ratio of difference in volume between the two predictions (VD) as was used in [6].
The results, gathered in Table 1, reporting median and interquartile range are ordered according to the average significance ranking, follows the guidelines of the MICCAI Decathlon challenge 20181.
Table 1.
Performance of different methods on the target (POPPY) and the source domain (MICCAI 2017 WMH Challenge). We report the dice between our models’ predictions and the ground truth annotations in the source domain as well as the HD95. The evaluation on target domains is done with the Dice, the HD95, the volume difference (VD) and the recall. A significative rank measure is calculated across all metrics. Results are reported with the format median (IQR) in percentages for all metrics except the HD95 in mm. Best results are in bold andunderlined when significantly better than all others (p < 0.05 paired Wilcoxon tests).
| POPPY | MICCAI | ||||||
|---|---|---|---|---|---|---|---|
| Dice | HD | VD | Recall | Dice | HD | Rank | |
| PC+Adv+Aug | 54.5 (10.6) | 32.7 (9.8) | 15.2 (22.8) | 52.4 (14.4) | 81.4 (9.6) | 28.5 (8.6) | 2.5 |
| PC+Aug | 53.2 (15.1) | 39.2 (15.5) | 25.4 (15.6) | 43.5 (12.5) | 81.6 (15.5) | 18.6 (4.8) | 3.3 |
| PC | 50.7 (17.0) | 35.1 (11.9) | 16.6 (21.4) | 43.6 (11.0) | 81.4 (22.6) | 17.2 (3.6) | 3.4 |
| MT | 48.6 (12.3) | 33.6 (14.8) | 33.7 (19.0) | 40.9 (5.0) | 80.0 (18.2) | 20.0 (7.3) | 4.3 |
| Baseline+Aug | 42.8 (14.6) | 34.9 (11.1) | 39.3 (22.3) | 33.5 (12.6) | 80.6 (14.8) | 17.8 (4.9) | 4.9 |
| Baseline | 43.0 (16.2) | 33.3 (15.1) | 40.3 (24.8) | 33.3 (14.8) | 81.1 (16.9) | 17.5 (3.3) | 5.6 |
| Adv | 41.8 (15.4) | 32.6 (6.1) | 25.2 (24.0) | 33.5 (12.7) | 82.5 (12.0) | 17.6 (5.2) | 5.7 |
| Adv+Aug | 41.4 (16.4) | 36.6 (9.0) | 38.0 (16.0) | 33.6 (13.9) | 81.9 (11.1) | 19.7 (11.0) | 6.3 |
4. Discussion
In this work, we presented a novel method of performing unsupervised multidomain adaptation. A pretrained model from one domain is retrained on paired unlabeled data from two target domains, encouraging consistent predictions. The proposed approach was evaluated against existing UDA strategies including representation learning approaches using domain adversarial training [5], and the ‘Mean Teacher’ algorithm for unsupervised domain adaptation [13] as well as an unsupervised baseline for WMH segmentation. Overall, our method was able to produce more consistent predictions across two target domains while retaining similar performance on its original training domain. More specifically, adaptation techniques optimizing pairwise consistency not only outperformed baseline models not benefitting from any adaptation but also adversarial strategies. Furthermore, it appeared that the PC method while closest to the mean teacher algorithm, outperformed this approach potentially thanks to differences in the optimisation strategies. Understanding the reasons for these differences also reported by [13] could be an interesting avenue of future investigation. Regarding the adversarial results, the observed inferior performance suggests that depending on the adaptation problem, the learning of a latent space invariant to domain (as enforced in the adversarial approach) may cause an information loss detrimental to the segmentation task. Additionally, the effects of data augmentation (which normally impacts performance positively) did not provide any benefit in the pure adversarial setting. Specific investigation of the effect of each type of augmentation would be needed to better understand this behaviour. While a pure adversarial setting proved ineffective, best performance across all models was obtained when combining it with our proposed PC strategy as it promoted a good label distribution in our target images. Future work will focus on removing the need for paired data by finding sufficiently realistic augmentation functions.
In conclusion, PC is a promising method to adapt automated image segmentation tools to different scanner manufacturers, MR sequences and other confounds. This adaptation is critical to the clinical translation of these tools notably in the context of scanner upgrades and multicentre trials.
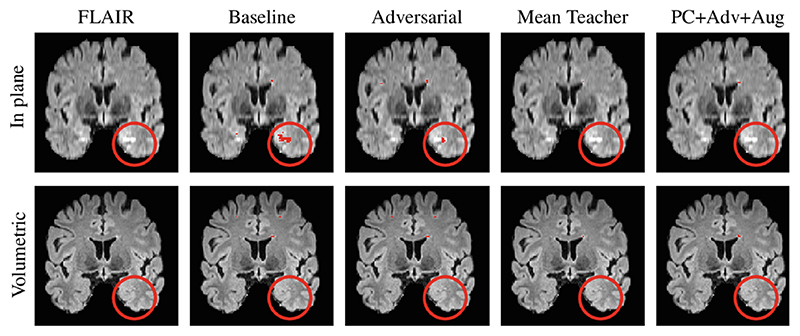
Fig. 2.
Qualitative results on a single slice from a single subject in the POPPY dataset. The top row shows a slice from the in-plane FLAIR acquisition whilst the bottom row shows a slice from the volumetric FLAIR acquisition. Each column shows a model’s predictions on that row’s image. This slice is used to highlight an example of an artefact (shown in the red circle) introduced by the in-plane acquisition. The baseline method introduces a false positive in this region whilst the domain adaptation methods perform better at ignoring it. Our approach shows the best in-plane to volumetric agreement.
Acknowledgements
We gratefully acknowledge the support of NVIDIA Corporation with the donation of one Titan Xp. This project has received funding from the EU H2020 under the Marie Sklodowska-Curie grant agreement No 721820, Wellcome Flagship Programme (WT213038/Z/18/Z) and Wellcome EPSRC CME (WT203148/Z/16/Z). Carole H. Sudre is supported by AS-JF-17-011 Alzheimer’s Society Junior Fellowship.
Footnotes
References
- 1.Bai W, et al. Semi-supervised learning for network-based cardiac MR image segmentation. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, editors. MICCAI 2017. Vol. 10434. Springer; Cham: 2017. pp. 253–260. (LNCS). [DOI] [Google Scholar]
- 2.Dou Q, et al. Unsupervised cross-modality domain adaptation of convnets for biomedical image segmentations with adversarial loss; Proceedings of the 27th International Joint Conference on Artificial Intelligence; 2018. pp. 691–697. [Google Scholar]
- 3.Gibson E, et al. NiftyNet: a deep-learning platform for medical imaging. arXiv. 2017 doi: 10.1016/j.cmpb.2018.01.025. preprint arXiv:1709.03485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haddow LJ, et al. Magnetic resonance imaging of cerebral small vessel disease in men living with HIV and HIV-negative men aged 50 and above. AIDS Res Hum Retroviruses. 2019;35(5):453–460. doi: 10.1089/aid.2018.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamnitsas K, et al. Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In: Niethammer M, et al., editors. IPMI 2017. Vol. 10265. Springer; Cham: 2017. pp. 597–609. (LNCS). [DOI] [Google Scholar]
- 6.Kuijf HJ, et al. Standardized assessment of automatic segmentation of white matter hyperintensities; results of the WMH segmentation challenge. IEEE Trans Med Imaging. 2019;99:1. doi: 10.1109/TMI.2019.2905770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milletari F, et al. V-net: fully convolutional neural networks for volumetric medical image segmentation; 3DV; 2016. pp. 565–571. [Google Scholar]
- 8.Miotto R, et al. Deep learning for healthcare: review, opportunities and challenges. Briefings bioinform. 2017;19(6):1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perone CS, et al. Unsupervised domain adaptation for medical imaging segmentation with self-ensembling. NeuroImage. 2019;194:1–11. doi: 10.1016/j.neuroimage.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. MICCAI 2015. Vol. 9351. Springer; Cham: 2015. pp. 234–241. (LNCS). [DOI] [Google Scholar]
- 11.Schoenauer-Sebag A, et al. Multi-domain adversarial learning. arXiv. 2019 preprint arXiv:1903.09239. [Google Scholar]
- 12.Shaw R, et al. MRI k-space motion artefact augmentation: model robustness and task-specific uncertainty; MIDL; 2019. pp. 427–436. [Google Scholar]
- 13.Xie Q, et al. Unsupervised data augmentation. 2019 arXiv:1904.12848. [Google Scholar]
- 14.Zhao A, et al. Data augmentation using learned transforms for one-shot medical image segmentation. CoRR abs/1902.09383. 2019 http://arxiv.org/abs/1902.09383. [Google Scholar]