Abstract
Mucosal-associated invariant T (MAIT) cells are innate sensors of viruses and can augment early immune responses and contribute to protection. We hypothesized MAIT cells may have inherent adjuvant activity in vaccine platforms that employ replication-incompetent adenovirus vectors. In mice and humans, ChAdOx1 (chimpanzee adenovirus Ox1) immunization robustly activated MAIT cells. Activation required plasmacytoid dendritic cell (pDC)–derived IFN-α and monocyte-derived IL-18. IFN-α-induced, monocyte-derived TNF was also identified as a key secondary signal. All three cytokines were required in vitro and in vivo. Activation of MAIT cells positively correlated with vaccine-induced T cell responses in human volunteers and MAIT cell-deficient mice displayed impaired CD8+ T cell responses to multiple vaccine-encoded antigens. Thus, MAIT cells contribute to the immunogenicity of adenovirus vectors, with implications for vaccine design.
Mucosal-associated invariant T (MAIT) cells are unconventional T cells that recognize microbe-derived metabolites of vitamin B2 biosynthesis like 5-(2-oxopropylideneamino)-6-ribitylaminouracil (5-OP-RU) (1). However, MAIT cells can also be activated by cytokines, and thereby respond to viruses, which do not synthesize vitamin B2. In vivo, MAIT cells respond to influenza virus to amplify early local immune responses and protect against lethal infection (2–4). We hypothesized that the ability of MAIT cells to augment early immune responses may play a key role in viral vector vaccine immunogenicity. Replication-incompetent adenovirus (Ad) vectors are highly potent vaccine platforms for many human diseases (5). They have been recently licensed for use against Ebola virus (6) and show promise for SARS-CoV-2 infection (7, 8). We sought to determine if such vectors activate MAIT cells and if this impacts vaccine immunogenicity.
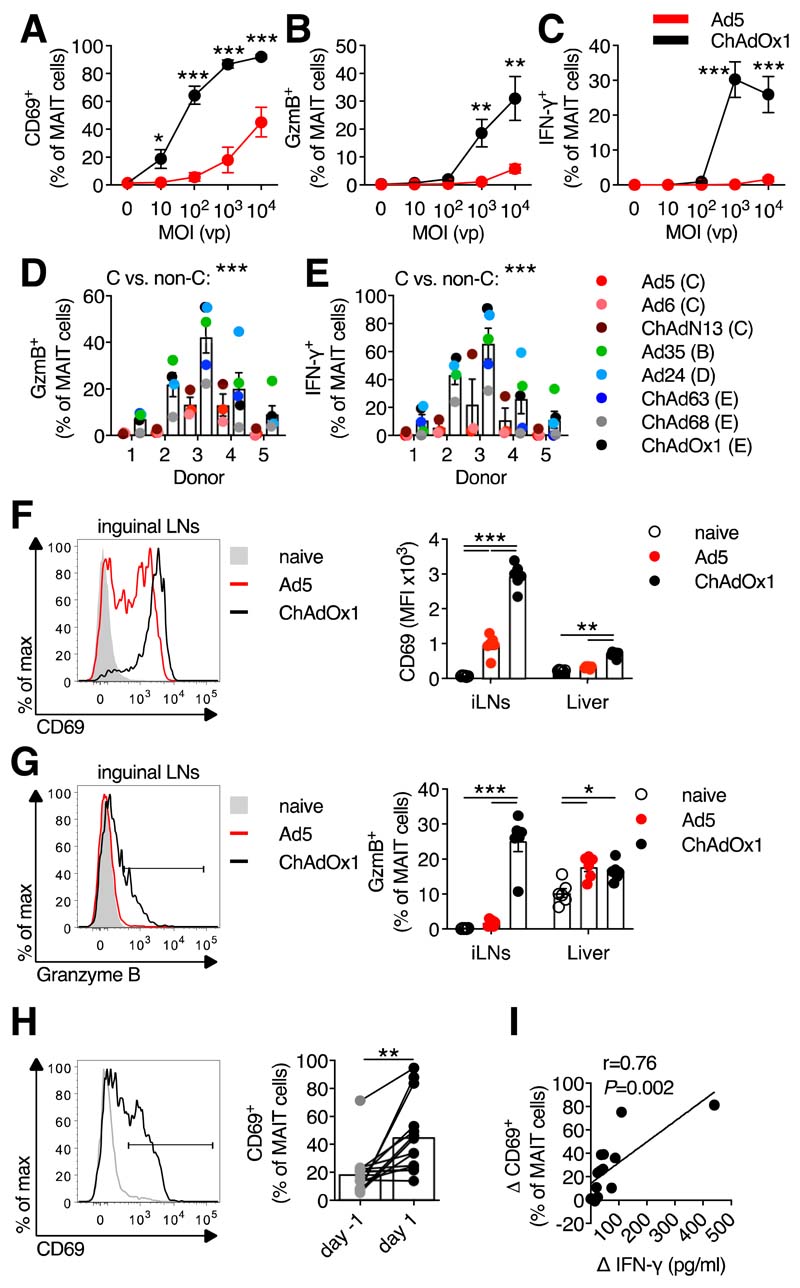
To determine if MAIT cells respond to Ad vectors, we stimulated human peripheral blood mononuclear cells (PBMCs) with Ad5 and chimpanzee adenovirus Ox1 (ChAdOx1), which are leading SARS-CoV-2 candidate vaccines (7, 8). ChAdOx1 induced dose-dependent upregulation of CD69, granzyme B, and interferon (IFN)-γ by MAIT cells (Fig. 1, A to C and fig. S1, A to D), whereas Ad5 only weakly activated MAIT cells. This activation was confirmed using the MR1/5-OP-RU tetramer to identify MAIT cells (fig. S1E).
Figure 1. Activation of human and murine MAIT cells by adenovirus vectors.
(A to C) Human PBMCs (n=9; four experiments) were stimulated with Ad5-GFP or ChAdOx1-GFP (multiplicity of infection (MOI)=0 to 104 vp (viral particles)). MAIT cell CD69 (A), granzyme B (GzmB) (B), and IFN-γ (C) expression was measured after 24 hours. (D and E) Human PBMCs (n=5; two experiments) were stimulated with the indicated vectors (species in parentheses). MAIT cell GzmB (D) or IFN-γ (E) expression were measured after 24 hours. (F and G) C57BL/6J mice (n=6 per group; representative of two experiments) were immunized intramuscularly (i.m.) with 108 IU (infectious units) of Ad5-GFP or ChAdOx1-GFP. Inguinal LN MAIT cell CD69 (F) and GzmB (G) expression was measured after 24 hours. (H and I) Healthy human volunteers (n=14) were immunized with a 5×1010 vp dose of ChAdOx1 MenB.1. (H) MAIT cell CD69 expression 1 day pre- and 1 day post-immunization. (I) Pearson correlation of change in plasma IFN-γ levels following vaccination with the change in MAIT cell CD69 expression. *, P<0.05; **, P<0.01; ***, P<0.001. Unpaired t test (A to C), two-way ANOVA (D and E), one-way ANOVA with Sidak correction for multiple comparisons (F and G), or Wilcoxon rank-sum test (H). Symbols indicate average response (A to C) or individual mice/volunteers (D to I). Mean ± SEM are shown.
Species C-derived Ad vectors have been shown to poorly stimulate innate immune responses as compared to non-species C vectors (9–11). We tested the relative ability of three species C vectors (Ad5, Ad6, and ChAdN13) and five non-species C vectors (Ad24, Ad35, ChAd63, ChAd68, and ChAdOx1) (fig. S1F) to activate MAIT cells. Following stimulation, we observed greater average activation by non-species C compared to species C vectors (Fig. 1, D and E).
We next tested the ability of Ad vectors to activate MAIT cells in vivo. Intramuscular (i.m.) ChAdOx1 immunization of C57BL/6J mice strongly induced upregulation of CD69 and granzyme B by MAIT cells, whereas Ad5 induced significantly weaker activation (Fig. 1, F and G and fig. S2, A to C). We also observed significant upregulation of CD69 on MAIT cells 1 day following immunization of human volunteers with a candidate ChAdOx1 vaccine (Fig. 1H and fig. S3, A to C). Plasma IFN-γ levels markedly increased post-vaccination (fig. S3D), as seen in non-human primates (10). This increase correlated with levels of MAIT cell activation (Fig. 1I).
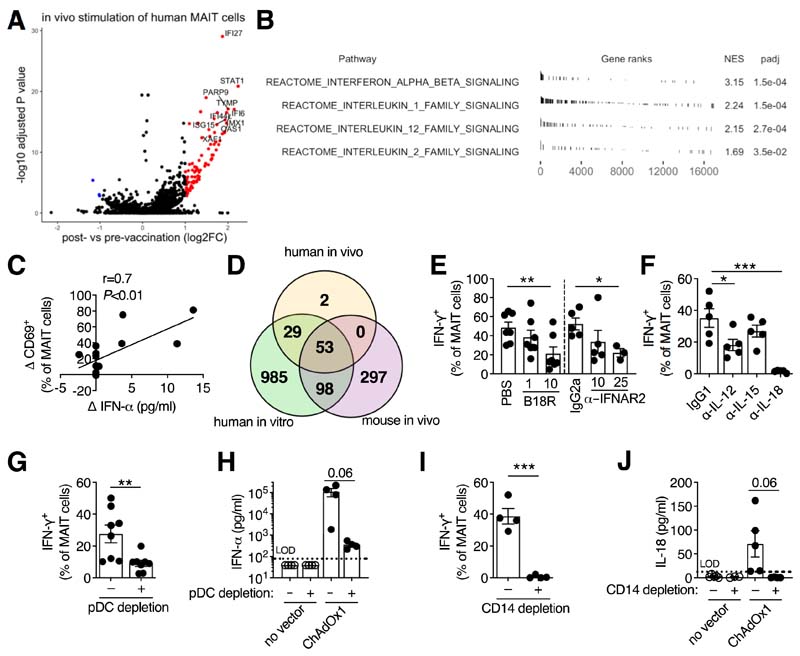
To investigate the pathways involved, RNA sequencing (RNA-Seq) of MAIT cells was performed. Eighty-four genes were significantly upregulated in human MAIT cells following vaccination (Fig. 2A and data S1). GSEA (12) identified the strong induction of type I IFN, interleukin (IL)-1 family, IL-12 family, and IL-2 family signaling pathways (Fig. 2B). Changes in post-vaccination plasma IFN-α or CCL2, an IFN-regulated chemokine (13), strongly correlated with MAIT cell activation (Fig. 2C and fig S3, D and E). Comparison of genes upregulated in MAIT cells following human vaccination, vaccination of mice, or in vitro stimulation showed a high degree of overlap. Ninety-eight percent of vaccine-upregulated genes in humans were upregulated in at least one of the other two conditions and 63% were upregulated in both (Fig. 2D and fig. S4, A and B and data S2 to S4). GSEA on murine MAIT cells and in vitro-stimulated human MAIT cells identified similar enrichment of these cytokine signaling pathways (fig. S4, C and D).
Figure 2. Activation of MAIT cells by adenovirus vectors requires pDC-derived IFN-α and monocyte-derived IL-18.
(A and B) Gene expression analysis of MAIT cells isolated from the PBMCs of human volunteers 1 day pre- and 1 day post-vaccination with ChAdOx1 MenB.1 (n=14). (A) Volcano plot of differentially expressed genes (log2 FC>1, adjusted P<0.05). The top 10 upregulated genes are annotated. (B) Selected cytokine signaling pathways from the Reactome database enriched by Gene Set Enrichment Analysis. (C) Pearson correlation of change in plasma IFN-α level following vaccination with the change in MAIT cell CD69 expression. (D) Overlap of genes upregulated in MAIT cells from ChAdOx1-vaccinated volunteers, from human PBMCs stimulated with ChAdOx1, and from the draining inguinal LNs of ChAdOx1-vaccinated mice. (E and F) Human PBMCs were stimulated with ChAdOx1-GFP and the following inhibitors were used: vaccinia virus-derived type I interferon antagonist B18R (1 or 10 μg/ml; n=7; three experiments) or anti-IFNAR2 antibody (10 or 25 μg/ml; n=5 or 3; two or one experiments, respectively) (E) or anti-IL-12, anti-IL-15, or anti-IL-18 antibodies (10 μg/ml; n=5; two experiments) (F). MAIT cell IFN-γ expression was measured after 24 hours. (G and H) PBMCs were depleted of CD123+ pDCs or left untreated and stimulated with ChAOx1-GFP. MAIT cell IFN-γ expression (n=8; three experiments) (G) or levels of IFN-α in the cell culture supernatant (n=4; one experiment) (H) were measured after 24 hours. (I and J) PBMCs were depleted of CD14+ monocytes or left untreated and stimulated with ChAdOx1-GFP. MAIT cell IFN-γ expression (n=4; two experiments) (I) or IL-18 levels in the supernatant (n=4; three experiments) (J) were measured after 24 hours. *, P<0.05; **, P<0.01; ***, P<0.001. Repeated-measures one-way ANOVA with Dunnett Correction (E and F), or unpaired t test (G to J). Symbols indicate individual donors. Mean ± SEM are shown.
In vitro, inhibition of type I IFN signaling reduced MAIT cell IFN-γ production by >50% (Fig. 2E). Blockade of IL-18 (an IL-1 family member) or IL-12 also reduced MAIT cell activation. By contrast, blockade of IL-15 (an IL-2 family member) had no effect (Fig. 2F and fig. S5A). MAIT cell activation by Ad vectors was independent of TCR signaling (fig. S5B) (2, 3).
To understand the cellular origins of these critical cytokines, we examined the cell populations transduced by Ad5 and ChAdOx1. Monocytes or conventional dendritic cells (cDCs) were the major transduced population by both vectors (>80% of GFP+ cells) (fig. S5, C to F). ChAdOx1 also efficiently transduced CD123+ plasmacytoid dendritic cells (pDCs), whereas Ad5 did not (fig. S5F) (11). Importantly, depletion of CD123+ pDCs resulted in a significant (67%) reduction in IFN-γ production by MAIT cells (Fig. 2G) and reduced IFN-α levels by >99% following ChAdOx1 stimulation (Fig. 2H).
Depletion of CD14+ monocytes significantly reduced MAIT cell activation after ChAdOx1 stimulation (Fig. 2I and fig. S5G) and abrogated the secretion of IL-18 (Fig. 2J). The Cathepsin B–NLRP3 inflammasome pathway (14) was the source of IL-18 in response to ChAdOx1 (fig. S6). Thus, pDC-derived IFN-α and monocyte-derived IL-18 play critical roles in activating MAIT cells in response to Ad vectors. Ad5 induced negligible amounts of IFN-α (fig. S7, A and B) (10, 11). Despite transducing monocytes, Ad5 did not induce IL-18 or IL-12p70 (fig. S7, C and D). By sharp contrast, ChAdOx1 induced robust production of IFN-α and IL-18.
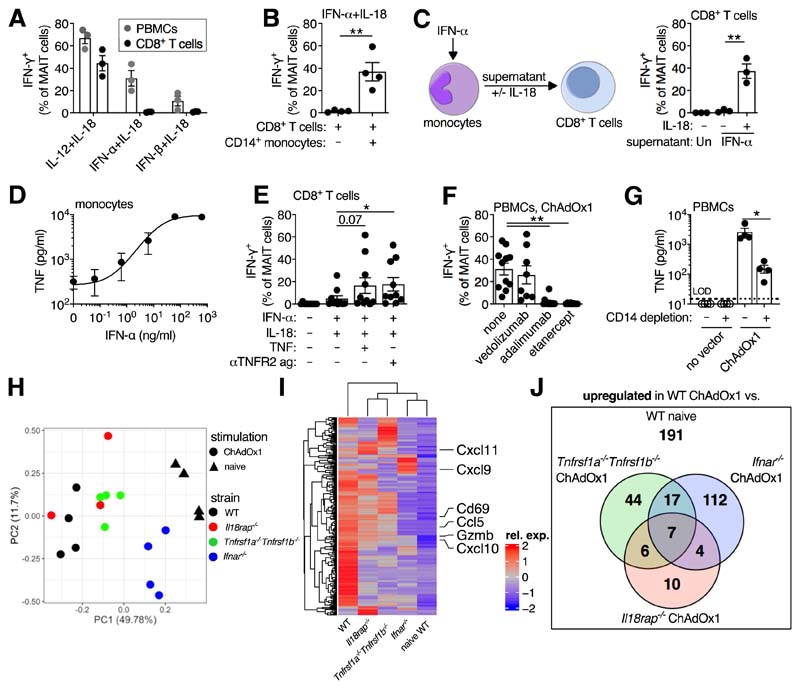
Notably, although IFN-α/β + IL-18 induced production of IFN-γ by MAIT cells in PBMC culture, this was not seen using isolated CD8+ T cells (~75% of human MAIT cells express CD8 (15)) (Fig. 3A), despite the induction of CD69 (fig. S8A). Depletion of monocytes reduced MAIT cell IFN-γ production following IFN-α + IL-18 stimulation (fig. S8B). The addition of monocytes rescued the response (Fig. 3B). Since conditioned supernatant, or provision of PBMCs across a transwell significantly rescued MAIT cell IFN-γ production (Fig. 3C and fig. S8C), this suggested the presence of a soluble, monocyte-derived, IFN-α-dependent signal. Unexpectedly, IFN-α-treated monocytes secreted TNF (Fig. 3D and fig. S8D). Additionally, TNFR2 signaling pathways were strongly induced in stimulated MAIT cells (fig. S8, E to G). Thus, we investigated if TNF was the IFN-α-dependent intermediary signal. Addition of TNF or an anti-TNFR2 agonist to isolated CD8+ T cells stimulated with IFN-α + IL-18 enhanced MAIT cell IFN-γ production by >300% (Fig. 3E and fig. S8H). Addition of an anti-TNF antibody (adalimumab) inhibited MAIT cell IFN-γ production in response to IFN-α + IL-18 stimulation, or conditioned supernatant (fig. S8, I and J). TNF blockade using either adalimumab or recombinant TNFR2–Fc fusion protein (etanercept), but not a control antibody (vedolizumab), inhibited IFN-γ production by MAIT cells in response to ChAdOx1 (Fig. 3F). Depletion of monocytes reduced ChAdOx1-induced TNF production by 94% (Fig. 3G). Ad5 induced minimal TNF (fig. S8K), consistent with the poor ability to stimulate IFN-α (fig. S7A).
Figure 3. IFN-α acts directly and indirectly through the induction of TNF to activate MAIT cells.
(A) Human PBMCs or purified CD8+ T cells (n=3; one experiment) were stimulated with the indicated cytokines (50 ng/ml). MAIT cell IFN-γ expression was measured after 24 hours. (B) Purified CD8+ T cells ± CD14+ monocytes (n=4; one experiment) were stimulated with IFN-α + IL-18 (50 ng/ml). MAIT cell IFN-γ expression was measured after 24 hours. (C) Purified monocytes (n=3; one experiment) were stimulated with IFN-α (50 ng/ml), or left untreated. After 24 hours, supernatants were transferred ± IL-18 (50 ng/ml) to autologous purified CD8+ T cells. MAIT cell IFN-γ expression was measured after 24 hours. (D) TNF production by IFN-α-treated CD14-purified monocytes was measured after 24 hours (n=3; one experiment). (E) Purified CD8+ T cells (n=10; four experiments) were stimulated with IFN-α and IL-18 ± TNF (50 ng/ml) or anti-TNFR2 agonist antibody (2.5 μg/ml). MAIT cell IFN-γ expression was measured after 24 hours. (F) PBMCs were stimulated with ChAdOx1 and the following inhibitors were added: vedolizumab (anti-α4β7 integrin antibody, n=8; two experiments), adalimumab (anti-TNF antibody, n=11; three experiments), or etanercept (TNFR2-Fc fusion protein, n=8; two experiments) (10 μg/ml). MAIT cell IFN-γ expression was measured after 24 hours. (G) PBMCs ± CD14-depletion were stimulated with ChAdOx1. Concentration of TNF in the supernatant was measured after 24 hours (n=4; one experiment). (H to J) C57BL/6J (n=4), Il18rap−/− (n=3), Tnfrsf1a−/−Tnfrsf1b−/− (n=4), or Ifnar−/− (n=4) mice were immunized intramuscularly with 108 IU of ChAdOx1-GFP. Naive C57BL/6J mice (n=4) were used as a control. After 24 hours, MAIT cells were isolated from the inguinal LNs and sorted for RNA sequencing (one experiment). (H) Principal component analysis. (I) Heatmap of the upregulated differentially expressed genes (log2 FC>1, adjusted P<0.05) between MAIT cells from ChAdOx1-immunized and naive C57BL/6J mice, with all other groups shown for comparison. (J) Overlap of the genes upregulated (log2 FC>1, adjusted P<0.05) in MAIT cells from ChAdOx1-immunized and naïve C57BL/6J mice, and the genes upregulated in MAIT cells from ChAdOx1-immunized C57BL/6J mice as compared to each of the ChAdOx1-immunized knockout strains. *, P<0.05; **, P<0.01. Unpaired t test (B, C and G), repeated-measures one-way ANOVA with Dunnett Correction (E and F). Symbols indicate individual donors. Mean ± SEM are shown.
These data suggest a model in which pDC-derived IFN-α acts directly and indirectly via induction of TNF by monocytes (with IL-18) to activate MAIT cells in response to ChAdOx1 (fig. S9). To test this model in vivo, wild-type (WT) C57BL/6J, Il18rap−/−, Ifnar−/−, and Tnfrsf1a−/−Tnfrsf1b−/− mice were immunized with ChAdOx1. MAIT cells from these animals were then analyzed by RNA-seq (fig. S10, A and B and data S5 to 8). Principal component analysis identified a strong gradient of activation, where MAIT cells from Ifnar−/− mice were most similar to those from naïve animals and MAIT cells from Tnfrsf1a−/−Tnfrsf1b−/− and Il18rap−/− mice had intermediate transcriptional profiles (Fig. 3, H and I). The effector genes Cd69 (and at the protein level), Cxcl10, Cxcl11, Ccl5, and Gzmb were all regulated along this gradient (fig. S10, C and D). Other genes (like Cxcl9) were only regulated by TNF signaling (fig. S10D). In total, 51% of the genes induced by vaccination were regulated by ≥1 of these cytokine pathways, and 11% were co-regulated by ≥2 of these pathways (Fig. 3J and data S9). Thus, TNF, IL-18, and especially type I IFN play a critical role in vivo in Ad vector-induced MAIT cell activation.
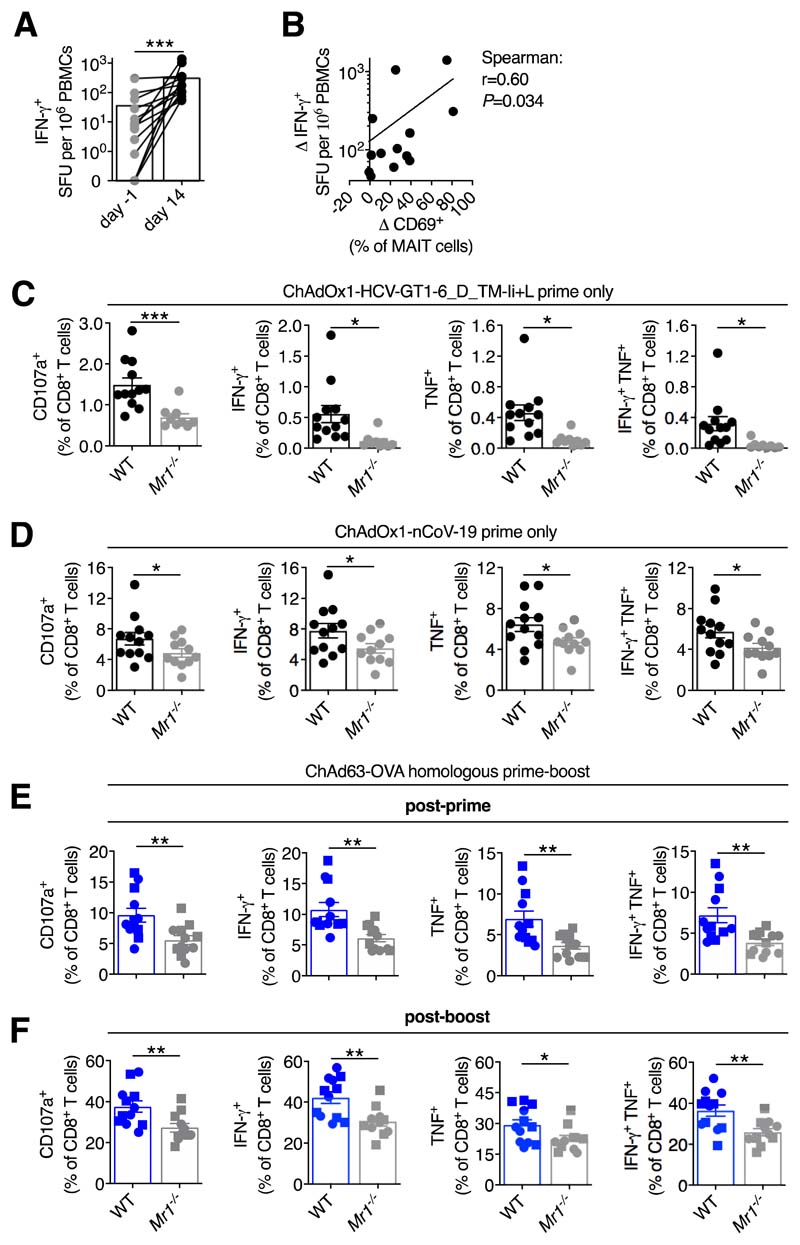
Human volunteers showed a significant increase in IFN-γ-producing T cells following ChAdOx1 boosting immunization (Fig. 4A). The degree of expansion positively correlated with MAIT cell activation (Fig. 4B). To determine if this was a causal relationship, WT and Mr1−/− mice, which lack MAIT cells (16), were used (fig. S11, A to C). Following vaccination with ChAdOx1 expressing an optimized HCV antigen (17), Mr1−/− mice had significantly reduced frequencies of HCV-specific CD8+ T cells compared with WT mice (Fig. 4C; fig. S11D). No significant defect in HCV-specific CD4+ T cells was observed (fig. S11E). We also observed defects in the CD8+ T cell responses of Mr1−/− mice vaccinated with the candidate SARS-CoV-2 vaccine, ChAdOx1-nCoV-19 (Fig. 4D and fig. S11F) (7). WT and Mr1−/− mice were then given a homologous ChAd63-OVA prime-boost immunization (fig. S11G) (18). Mr1−/− mice displayed reduced OVA-specific CD8+ T cell responses after both priming and boosting (Fig. 4, E and F). Differences in the microbiome (19) or general immunodeficiency of Mr1−/− mice did not explain these differences in immunogenicity (fig. S12).
Figure 4. MAIT cell deficiency impacts on T cell responses following ChAdOx1 or ChAd63 immunization.
(A) Frequency of IFN-γ-producing PBMCs measured by peptide ELISPOT in ChAdOx1 MenB.1 vaccinated volunteers pre-boost (n=14) or day 14 post-boost (n=13). (B) Spearman rank correlation analysis of the change in MAIT cell CD69 expression from pre-boost to day 1 post-boost versus the increase in IFN-γ-producing PBMCs from pre-boost to day 14 post-boost. (C) C57BL/6J (n=12) or Mr1−/− (n=9) mice were immunized intramuscularly (i.m.) with 108 IU of ChAdOx1-HCV-GT1-6_D_TM-Ii+L (two experiments). On day 16, HCV-specific CD107a+, IFN-γ +, TNF+, or IFN-γ +TNF+ CD8+ T cell responses were measured. (D) C57BL/6J (n=12) or Mr1−/− (n=11) mice were immunized i.m. with 108 IU of ChAdOx1-nCoV-19 (two experiments). On day 13, SARS-CoV-2 spike-specific CD107a+, IFN-γ +, TNF+, or IFN-γ +TNF+ CD8+ T cell responses were measured. (E and F) C57BL/6J (n=12) or Mr1−/− (n=12, n=11 post-boost) were primed i.m. with 107 IU of ChAd63-OVA and boosted intravenously on day 28 with 108 IU (squares) or 109 IU (circles) of ChAd63-OVA. SIINFEKL-specific CD107a+, IFN-γ +, TNF+, or IFN-γ +TNF+ CD8+ T cell responses were measured either 3 weeks post-prime (E) or 3 weeks post-boost (F). *, P<0.05; **, P<0.01; ***, P<0.001. Wilcoxon rank-sum test (A), or two-way ANOVA (C to F). Symbols indicate individual volunteers/mice. Mean ± SEM are shown.
In summary, MAIT cells can sense the diversity of the Ad vector-induced innate immune activation landscape (e.g. IFN-α, TNF, IL-18), integrating these signals to augment vaccine-induced CD8+ T cell immunity. The blend of signals required to maximally trigger MAIT cells described here includes a critical pathway via type I IFN-dependent TNF release and relies on cross-talk between two distinct populations of transduced cells, and varies between adenovirus serotypes. Our data, coupled with studies in the lung (4, 20, 21), support a model that places MAIT cells in a critical bridging position between innate and adaptive immunity. The mechanism by which MAIT cell activation promotes antigen-specific CD8+ T cell responses remains to be defined. However, local production of chemokine CXC10 represents a promising candidate as it can promote CD8+ T cell priming (22).
It is striking that the activation of MAIT cells is tightly linked to the immunogenicity of adenovirus vectors. This technology has emerged as a potent platform for T cell immunogenicity in clinical trials for HIV (23), and as vaccines for emerging viruses such as Ebola (6) and SARS-CoV-2 (7, 8). This knowledge can be harnessed to improve the design of these vaccines against major pathogens and cancers.
Supplementary Material
One-sentence summary.
Maximal immunogenicity of candidate adenovirus vaccine vectors requires the activation of MAIT cells.
Acknowledgements
We thank H Ferry and L. Hardy for assistance with cell sorting; S. Slevin, C.-P. Hackstein, and C. Willberg for critical discussions; M. Salio and V. Cerundolo for the Mr1−/− mice; J. Rehwinkel for the Ifnar−/− mice; M. Esposito, H. Al-Mossawi, L. Ni Lee, and T. Donnison for reagents; the NIH Tetramer Facility for the MR1 tetramers; and all of the volunteers for sample donation and participation in the trial.
Funding
N.M.P. is supported by an Oxford-UCB Postdoctoral Fellowship. A.A. is supported by a Wellcome Clinical Training Fellowship [216417/Z/19/Z]. L.C.G. is supported by a Wellcome PhD Studentship [109028/Z/15/Z]. M.E.B.F. is supported by an Oxford-Celgene Doctoral Fellowship. S.B.M. and T.S.C.H. are supported by the Wellcome [211050/Z/18/Z and 211050/Z/18/A]. E.B. is supported by the Medical Research Council (STOP-HCV and MR/R014485/1), an NIHR Senior Fellowship, the NIHR Biomedical Research Centre (Oxford), and the UKRI/NIHR through the UK Coronavirus Immunology Consortium (UK-CIC). C.S.R. is supported by the NIHR Biomedical Research Centre and is a Jenner Institute Investigator. A.J.P. is supported by the NIHR Oxford Biomedical Research Centre and is an NIHR Senior Investigator. P.K. is supported by the Wellcome [WT109965MA], the NIHR Biomedical Research Centre (Oxford), the UKRI/NIHR through the UK Coronavirus Immunology Consortium (UK-CIC), and an NIHR Senior Fellowship. The ChAdOx1 MenB.1 clinical trial is funded by the Medical Research Council DPFS (MRM0076931). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Footnotes
Author contributions:
N.M.P. and P.K. designed the project. N.M.P., A.J.S., C.D., T.S.C.H., E.B., C.S.R., A.J.P., and P.K. designed the experiments. N.M.P., A.A., L.C.G., A.J.S., C.D., C.H., L.S.R., M.E.B.F., M.U., H.S., and S.B.M. performed the experiments. S.C., B.O., M.R., F.T., T.L., S.C., A.F., E.B., C.S.R., and A.J.P. provided samples and reagents. All authors contributed to the writing and editing of the manuscript.
Competing interests:
C.D., C.S.R., and A.J.P. are named inventors on a patent application in the field of meningococcal vaccines. A.J.P. waives his rights under any patent. P.K. is a named inventor on a patent application in the field of cancer vaccines.
Data Availability
All gene expression data are deposited under GSE158835. All data are available in the manuscript or the supplementary materials.
References and Notes
- 1.Provine NM, Klenerman P. MAIT Cells in Health and Disease. Annu Rev Immunol. 2020;38:203–228. doi: 10.1146/annurev-immunol-080719-015428. [DOI] [PubMed] [Google Scholar]
- 2.Loh L, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc Natl Acad Sci USA. 2016;113:10133–10138. doi: 10.1073/pnas.1610750113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Wilgenburg B, et al. MAIT cells are activated during human viral infections. Nature Communications. 2016;7:11653–11. doi: 10.1038/ncomms11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Wilgenburg B, et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nature Communications. 2018;9:4706. doi: 10.1038/s41467-018-07207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitelli A, et al. Chimpanzee adenoviral vectors as vaccines - challenges to move the technology into the fast lane. Expert Rev Vaccines. 2017;16:1241–1252. doi: 10.1080/14760584.2017.1394842. [DOI] [PubMed] [Google Scholar]
- 6.European Medicines Agency. New vaccine for prevention of Ebola virus disease recommended for approval in the European Union. 2020:1–2. https://www.ema.europa.eu/en/documents/press-release/new-vaccine-prevention-ebola-virus-disease-recommended-approval-european-union_en.pdf.
- 7.Folegatti PM, et al. Articles Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. The Lancet. 2020:1–13. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu F-C, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. The Lancet. 2020:1–10. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinn KM, et al. Antigen expression determines adenoviral vaccine potency independent of IFN and STING signaling. J Clin Invest. 2015;125:1129–1146. doi: 10.1172/JCI78280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teigler JE, Iampietro MJ, Barouch DH. Vaccination with adenovirus serotypes 35, 26, and 48 elicits higher levels of innate cytokine responses than adenovirus serotype 5 in rhesus monkeys. J Virol. 2012;86:9590–9598. doi: 10.1128/JVI.00740-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson MJ, et al. Type I IFN induced by adenovirus serotypes 28 and 35 has multiple effects on T cell immunogenicity. J Immunol. 2012;188:6109–6118. doi: 10.4049/jimmunol.1103717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samarajiwa SA, Forster S, Auchettl K, Hertzog PJ. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 2008;37:D852–D857. doi: 10.1093/nar/gkn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature Immunology. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner LC, Klenerman P, Provine NM. Insights Into Mucosal-Associated Invariant T Cell Biology From Studies of Invariant Natural Killer T Cells. Front Immunol. 2018;9:911–25. doi: 10.3389/fimmu.2018.01478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 17.Donnison T, et al. Viral vectored hepatitis C virus vaccines generate pan-genotypic T cell responses to conserved subdominant epitopes. Vaccine. 2020;38:5036–5048. doi: 10.1016/j.vaccine.2020.05.042. [DOI] [PubMed] [Google Scholar]
- 18.Gola A, et al. Prime and target immunization protects against liver-stage malaria in mice. Sci Transl Med. 2018;10:eaap9128. doi: 10.1126/scitranslmed.aap9128. [DOI] [PubMed] [Google Scholar]
- 19.Varelias A, et al. Recipient mucosal-associated invariant T cells control GVHD within the colon. J Clin Invest. 2018;128:1919–1936. doi: 10.1172/JCI91646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meierovics A, Yankelevich W-JC, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci USA. 2013;110:E3119-28. doi: 10.1073/pnas.1302799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meierovics AI, Cowley SC. MAIT cells promote inflammatory monocyte differentiation into dendritic cells during pulmonary intracellular infection. J Exp Med. 2016;59 doi: 10.1084/jem.20160637. jem.20160637-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peperzak V, et al. CD8+ T cells produce the chemokine CXCL10 in response to CD27/CD70 costimulation to promote generation of the CD8+ effector T cell pool. J Immunol. 2013;191:3025–3036. doi: 10.4049/jimmunol.1202222. [DOI] [PubMed] [Google Scholar]
- 23.Barouch DH, et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19) Lancet. 2018;392:232–243. doi: 10.1016/S0140-6736(18)31364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dicks MDJ, et al. A Novel Chimpanzee Adenovirus Vector with Low Human Seroprevalence: Improved Systems for Vector Derivation and Comparative Immunogenicity. PLoS ONE. 2012;7:e40385. doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antrobus RD, et al. Clinical Assessment of a Novel Recombinant Simian Adenovirus ChAdOx1 as a Vectored Vaccine Expressing Conserved Influenza A Antigens. Mol Ther. 2013;22:668–674. doi: 10.1038/mt.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colloca S, et al. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med. 2012;4:115ra2. doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provine NM, et al. Unique and Common Features of Innate-Like Human Vδ2+ γδT Cells and Mucosal-Associated Invariant T Cells. Front Immunol. 2018;9:756. doi: 10.3389/fimmu.2018.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Delft A, et al. The generation of a simian adenoviral vectored HCV vaccine encoding genetically conserved gene segments to target multiple HCV genotypes. Vaccine. 2018;36:313–321. doi: 10.1016/j.vaccine.2017.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Provine NM, et al. Immediate Dysfunction of Vaccine-Elicited CD8+ T Cells Primed in the Absence of CD4+ T Cells. J Immunol. 2016;197:1809–1822. doi: 10.4049/jimmunol.1600591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Provine NM, et al. Longitudinal Requirement for CD4+ T Cell Help for Adenovirus Vector-Elicited CD8+ T Cell Responses. J Immunol. 2014;192:5214–5225. doi: 10.4049/jimmunol.1302806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ussher JE, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. 2014;44:195–203. doi: 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata M, et al. Novel epoxysuccinyl peptides. Selective inhibitors of cathepsin B, in vitro. FEBS Lett. 1991;280:307–310. doi: 10.1016/0014-5793(91)80318-w. [DOI] [PubMed] [Google Scholar]
- 35.Coll RC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muñoz-Planillo R, et al. K+ Efflux Is the Common Trigger of NLRP3 Inflammasome Activation by Bacterial Toxins and Particulate Matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Calvo M, et al. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998;273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 38.Graham SP, et al. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. bioRxiv. 2020;18:313–11. doi: 10.1038/s41541-020-00221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnitz RA, Imam S, Yates K, Haining WN. Isolation of RNA and the synthesis and amplification of cDNA from antigen-specific T cells for genome-wide expression analysis. Methods Mol Biol. 2013;979:161–173. doi: 10.1007/978-1-62703-290-2_13. [DOI] [PubMed] [Google Scholar]
- 40.Picelli S, et al. Full-length RNA-seq from single cells usingSmart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 41.Cribbs AP, et al. CGAT-core: a python framework for building scalable, reproducible computational biology workflows. F1000Res. 2019;8:377–13. [Google Scholar]
- 42.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019:060012 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All gene expression data are deposited under GSE158835. All data are available in the manuscript or the supplementary materials.