Summary
The exosome plays key roles in RNA maturation and surveillance, but it is unclear how target RNAs are identified. We report the functional characterization of the yeast exosome component Rrp44, a member of the RNase II family. Recombinant Rrp44 and the purified TRAMP polyadenylation complex each specifically recognized lacking a single m1A58 modification, even in the presence of a large excess of total tRNA. This tRNA is otherwise mature and functional in translation in vivo but is presumably subtly misfolded. Complete degradation of the hypomodified tRNA required both Rrp44 and the poly(A) polymerase activity of TRAMP. The intact exosome lacking only the catalytic activity of Rrp44 failed to degrade , showing this to be a specific Rrp44 substrate. Recognition of hypomodified by Rrp44 is genetically separable from its catalytic activity on other substrates, with the mutations mapping to distinct regions of the protein.
Introduction
The nuclear and cytoplasmic forms of the exosome share ten common components, often referred to as the “core” exosome, while the nuclear complex contains an additional 3’ exonuclease Rrp6 (reviewed in Houseley et al. [2006]). Recent studies proposed that Rrp44 is the only catalytically active nuclease in the yeast core exosome (Dziembowski et al., 2007; Liu et al., 2006). The domain organization of Rrp44 resembles that of bacterial RNase II (see Figure 1A) and RNase R (Vincent and Deutscher, 2006), with a cold shock protein RNA-binding domain (CSD), a catalytic RNB domain, and a C-terminal S1 RNA-binding domain. However, Rrp44 has an additional N-terminal PinC domain that has no equivalent in the bacterial enzymes.
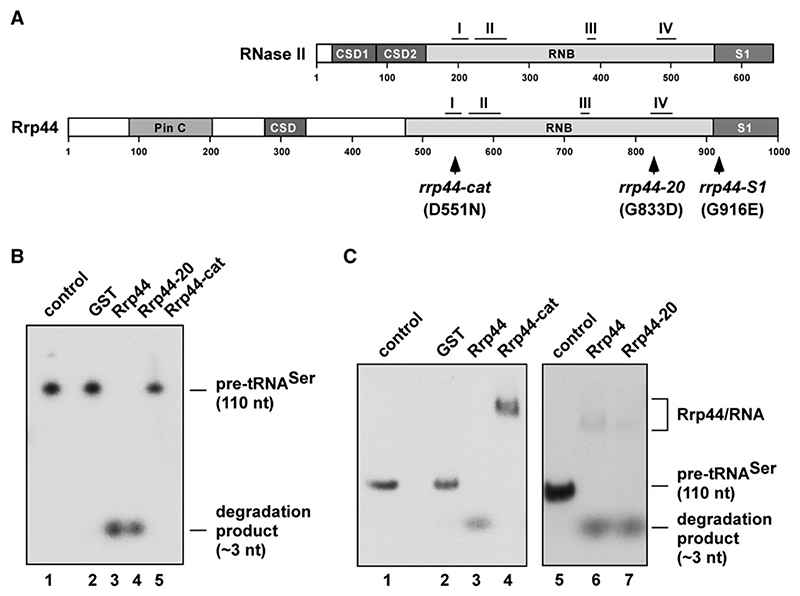
Figure 1. A Point Mutation in the RNB Motif I of Rrp44 Inactivates the Enzyme without Affecting RNA Binding.
(A) Domain structures of E. coli RNase II and S. cerevisiae Rrp44. RNB motifs I-IV (adapted from Frazao et al. [2006]) are shown. Positions of Rrp44 point mutations are indicated.
(B) Exonuclease activity of wild-type GST-Rrp44, GST-Rrp44-20, GST-Rrp44-cat, or free GST on a 5’ end-labeled 110 nt pre-tRNASer transcript. Recombinant protein (100 fmol) and 5 fmol RNA were incubated for 10 min at 37°C. Control: mock-digested substrate without recombinant protein. Reaction products were separated on a 12% polyacrylamide/8 M urea gel and visualized by autoradiography. The full-length RNA and ~3 nt degradation products are indicated on the right.
(C) RNA-binding assay using wild-type GST-Rrp44, GST-Rrp44-cat, GST-Rrp44-20, or free GST and 5’ end-labeled 110 nt pre-tRNASer transcript. Substrate and reaction conditions were as described in (B). After incubation at 37°C, samples were immediately put on ice and then analyzed on a native 4% polyacrylamide gel. The positions of RNA-protein complexes, free full-length RNA, and ~3 nt degradation products are indicated on the right.
The TRAMP4 complex is a major nuclear cofactor for the exosome (LaCava et al., 2005; Vanacova et al., 2005) and comprises the poly(A) polymerase Trf4, the putative ATP-dependent RNA helicase Mtr4, and either Air1 or Air2, which are zinc-knuckle putative RNA-binding proteins that appear to be functionally redundant. The TRAMP/exosome system functions in the surveillance of many nuclear RNA species. In yeast strains lacking the activity of the tRNA 1-methyladenosine58 methyltransferase (trm6-504), several tRNAs fail to be methylated, including the initiator tRNAMet. This tRNA has a unique structure, which is particularly sensitive to loss of methylation (Basavappa and Sigler, 1991). The hypomethylated is stable and functional for translation at 25°C; however, transfer to 37° C leads to degradation of the precursor pre- (Kadaba et al., 2006). Hypomodified pre- was stabilized by mutations in the exosome components Rrp44 (rrp44-20) or Rrp6 (rrp6D) or in the TRAMP complex (mtr4-20, trf4-20), but it was unclear which of these factors specifically recognizes and degrades the defective pre-tRNA (Kadaba et al., 2004).
Results
Mutations in Rrp44 Alter RNA Binding and Exonuclease Activities In Vitro
The rrp44-20 mutation was identified as a suppressor of the temperature-sensitive lethal mutation trm6-504 (Kadaba et al., 2004; Zuo et al., 2006). The genomic location of the rrp44-20 allele was identified by linkage analysis. Sequencing of the mutant allele revealed a single nucleotide substitution G2498 → A resulting in a Gly833 → Asp substitution in Rrp44, within domain IV of the RNB motif (G833D; see Figure 1A).
Analysis of RNase II in E. coli indicated that RNA binding and catalysis involve distinct regions of the protein (Frazao et al., 2006; Zuo et al., 2006). This was confirmed by the isolation of a point mutation (D209N) in RNB motif I (see Figure 1A), which inactivates the exonuclease activity of RNase II without affecting RNA binding (Amblar and Arraiano, 2005). Sequence comparisons predicted that an Asp551 → Asn mutation (D551N: rrp44-cat) should abolish the catalytic activity of Rrp44 (Figure 1A) (Dziembowski et al., 2007).
Wild-type Rrp44, Rrp44-20, and Rrp44-cat were expressed in E. coli as GST fusions and purified on glutathione Sepharose (see Figure S1 in the Supplemental Data available with this article online). These were tested in exonuclease and RNA-binding assays using in vitro-transcribed, 5’ end-labeled pre-tRNASer as a fully unmodified substrate (Figures 1B and 1C). In contrast to previous analyses, the assays were performed at low magnesium concentrations (0.25 mM) similar to the reported intracellular concentration of eukaryotic cells (0.1-1 mM) (Romani and Scarpa, 1992).
In the in vitro exonuclease experiment shown in Figure 1B, ~12 ng (100 fmol) protein and ~5 fmol RNA were incubated for 10 min at 37°C. The reaction products were separated on a denaturing 12% polyacrylamide/8 M urea gel and visualized by autoradiography. Both Rrp44 and Rrp44-20 degraded pre-tRNASer in an apparently processive manner, leaving a short oligonucleotide (~3 nt) product (Figure 1B). In contrast, Rrp44-cat failed to detectably degrade pre-tRNASer (Figure 1B) and was also inactive on a37 nt pBS transcript, poly(A30), poly(U30), and (Figure 2, Figure S2, and data not shown). This confirmed that Asp551 is part of the catalytic site and that the activity seen for wild-type Rrp44 is not due to a contaminating E. coli exonuclease.
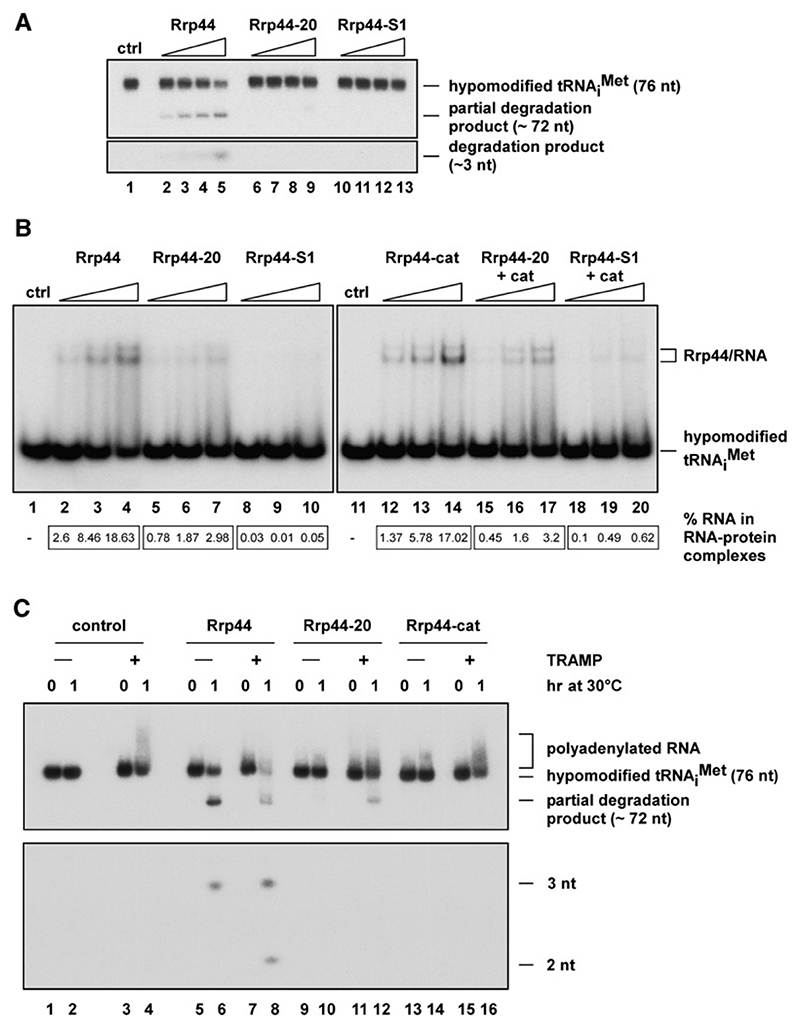
Figure 2. Recognition and Degradation of Native, Hypomodified by Rrp44 and TRAMP.
(A)Exonuclease activity of wild-type GST-Rrp44, GST-Rrp44-20, or GST-Rrp44-S1 on a 5’ end-labeled, native, hypomodified substrate. Recombinant protein (3, 6, 12, or 60 fmol) was incubated with 5 fmol RNA for 90 min at 30°C in the presence of 0.25 mM MgCl2. Reaction products were separated on a 12% polyacrylamide/8 M urea gel and visualized by autoradiography. Fulllength and partially degraded species are indicated on the right.
(B)RNA-binding assay using wild-type or mutant GST-Rrp44 proteins and 5’ endlabeled, native hypomodified . Recombinant protein (3, 6, or 12 fmol) was incubated with 5 fmol RNA for 10 min at 30°C in the presence of 0.25 mM MgCl2. Reactions were then analyzed as described in Figure 1C. The lower panel shows Phosphor Imager quantification of the same experiment.
(C)Combined exonuclease/polyadenylation assay using 2 ml TAP-purified TRAMP complex, 200 fmol GST-Rrp44 fusion protein (wild-type, 44-20, or cat), and 50 fmol 5’ endlabeled, native hypomodified substrate. Reactions were performed at 30°C with 0.5mM MgCl2 and 1 mMATP for the times indicated. Products were separated on a 12% polyacrylamide/8 M urea gel and visualized by autoradiography. Full-length , polyadenylated RNA, and degraded RNA species are indicated.
To confirm that the impaired nuclease activity of Rrp44-cat does not result from reduced substrate binding, we performed RNA band shift assays (Figure 1C). Reaction conditions were as above, except that the samples were put on ice after incubation at 37°C and separated on a precooled, native 4% polyacrylamide gel. Incubation of pre-tRNASer with Rrp44 or Rrp44-20 generated degradation products with greater gel mobility than the free substrate (Figure 1C, lanes 6 and 7). In addition, a low level of slower-migrating RNA was detected. We interpret this as showing that they bind unmodified pre-tRNASer but then rapidly degrade the RNA. In contrast, Rrp44-cat generated only bands with slowed migration (Figure 1C, lane 4), which represent stable RNA-protein complexes. The wild-type protein also formed stable RNA-protein complexes when incubated in the presence of 10 mM EDTA to inhibit nuclease activity (data not shown).
We conclude that the D551N mutation in Rrp44-cat abolishes the exonucleolytic activity of Rrp44 without affecting its ability to bind RNA, consistent with the presence of functionally separable domains that are required for either RNA binding or catalysis, as previously reported for E. coli RNase II (Frazao et al., 2006; Zuo et al., 2006). However, Rrp44-20 was apparently unaffected in RNA binding or catalysis on a fully unmodified pre-tRNASer substrate.
To characterize the putative defect in Rrp44-20 activity on a more biologically relevant substrate, we purified and 5’ end labeled the native, hypomodified from a trm6 Δ yeast strain (Anderson et al., 2000) (Figure 2A). To facilitate analyses of RNA binding, we also generated a point mutation (Glyg16 → Glu; Rrp44-S1) in the S1 RNA-binding domain, which is predicted to disrupt its activity. This was expressed in E. coli as a GST fusion (Figure S1).
On the native, hypomodified substrate, wild-type Rrp44 generated a partial degradation product (Figure 2A). The gel mobility of this would correspond to removal of the 4 nt 3’ overhang comprising the CCA tail and the 3’ encoded “discriminator” base. In addition, a 3 nt product was detected (lower panel). We predict that this results from complete, Rrp44-mediated degradation of tRNA fragments present in the input. In contrast, Rrp44-20 and Rrp44-S1 showed no detectable nuclease activity (Figure 2A).
To determine whether the Rrp44-20 mutation specifically impaired recognition of , we performed RNA gel mobility assays (Figure 2B). Rrp44-20 showed reduced binding to the hypomodified compared to the wild-type or Rrp44-cat protein (quantified in lower panel of Figure 2B). The reduction in binding of Rrp44-20 to hypomodified was modest but was reproducibly observed in six independent experiments. In contrast, RNA binding was greatly reduced but not abolished in the Rrp44-S1 mutant. To analyze RNA binding in the absence of substrate degradation, the rrp44-20 and rrp44-S1 mutations were combined with the catalytic (D551N) mutation to generate Rrp44-20 + cat and Rrp44-S1 + cat. This abolished formation of degradation products (data not shown) but did not clearly alter the relative abilities of the wild-type, Rrp44-20, and Rrp44-S1 proteins to bind hypomodified (Figure 2B).
These results suggested that the suppression phenotype of rrp44-20 might result from impaired binding to the misfolded pre-. However, subsequent analyses revealed that Rrp44-20 also showed reduced binding to other substrates tested, poly(A30) (Figure S2B), and the 37 nt pBS transcript (Figure S2D). In contrast to the reduced band shift, the catalytic activity of Rrp44-20 was not clearly impaired on the 37 nt transcript (Figure S2C) and was only mildly impaired on the poly(A30) substrate (Figure S2A). Rrp44-S1 showed greatly reduced catalytic activity on both substrates.
Comparing Rrp44 binding and nuclease activity on various RNA substrates, we suggest that wild-type Rrp44 can bind and completely degrade unstructured substrates but can only partially degrade the hypomodified, but otherwise mature, (Figure 2). This indicates that Rrp44 can remove the presumably unstructured 3’ overhang in the hypomodified on its own, but it needs cofactors to completely degrade this nearnative tRNA, possibly to assist in opening the stable structure of the RNA.
The Rrp44-20 protein showed marked defects in stable RNA binding on some, but not all, substrates. This reduced binding has only limited effects on the ability of the protein to degrade unstructured unmodified pre-tRNASer, poly(A30), or 37 nt pBS substrates but completely abolished the partial 3’ degradation of hypomodified seen for the wild-type protein (Figure 2A). From these results we conclude that the suppression phenotype of rrp44-20 is probably not due to reduced overall binding of the hypomodified but might result from impaired positioning of the unstructured 3’ overhang for catalysis. This indicates that a single exosome subunit, Rrp44, is directly involved in the recognition of its cognate substrate.
TRAMP Stimulates the Degradation of Hypomodified by Rrp44
The identification of mutations in the TRAMP complex as suppressors of the trm6-504 phenotype (Kadaba et al., 2004) suggested that TRAMP could be a cofactor for Rrp44-mediated degradation of .
To test this hypothesis in vitro, we incubated native, 5’ end-labeled hypomodified with recombinant GST-tagged Rrp44 and/or TAP-purified TRAMP complex, in the presence of 1 mM ATP (Figure 2C and Figure S3). The RNA substrate was polyadenylated by the TRAMP complex in the absence of Rrp44 (Figure 2C, lane 4), indicating that TRAMP can recognize hypomodified , consistent with previous results (Vanacova et al., 2005). Addition of the TRAMP complex stimulated the activity of Rrp44 (Figure 2C, lanes 6 and 8), enhancing both the fraction of the tRNA that was degraded (upper panel) and the yield of the 2 nt product of complete digestion (lower panel). TRAMP did not stimulate the activity of Rrp44-cat (lane 16). The activity of Rrp44-20 was slightly stimulated by TRAMP (Figure 2C, compare lanes 10 and 12), but degradation was much less robust than with the wild-type protein. The stimulation of Rrp44-mediated degradation of by TRAMP required the presence of ATP in the reaction (Figure S3) and was inhibited by mutation of highly conserved residues (Asp236,238 → Ala) in the active site of Trf4 (DADA mutation). This mutation was previously shown to abolish poly(A) polymerase activity (Vanacova et al., 2005).
These data indicate that the TRAMP complex is a required cofactor in the complete, Rrp44-mediated degradation of a highly structured, native tRNA that contains all modifications but one.
Rrp44 and TAP-Purified TRAMP Complex Specifically and Independently Recognize Hypomodified in Total tRNA
We next addressed whether tRNA degradation by recombinant Rrp44 and TRAMP is specific for the misfolded, hypomodified . Wild-type and mutant Rrp44 proteins and/or TAP-purified TRAMP complex were incubated with total transfer RNAs isolated from either wild-type or trm6-504 mutant yeast strains. Northern hybridization was used to analyze individual tRNAs (Figure 3A).
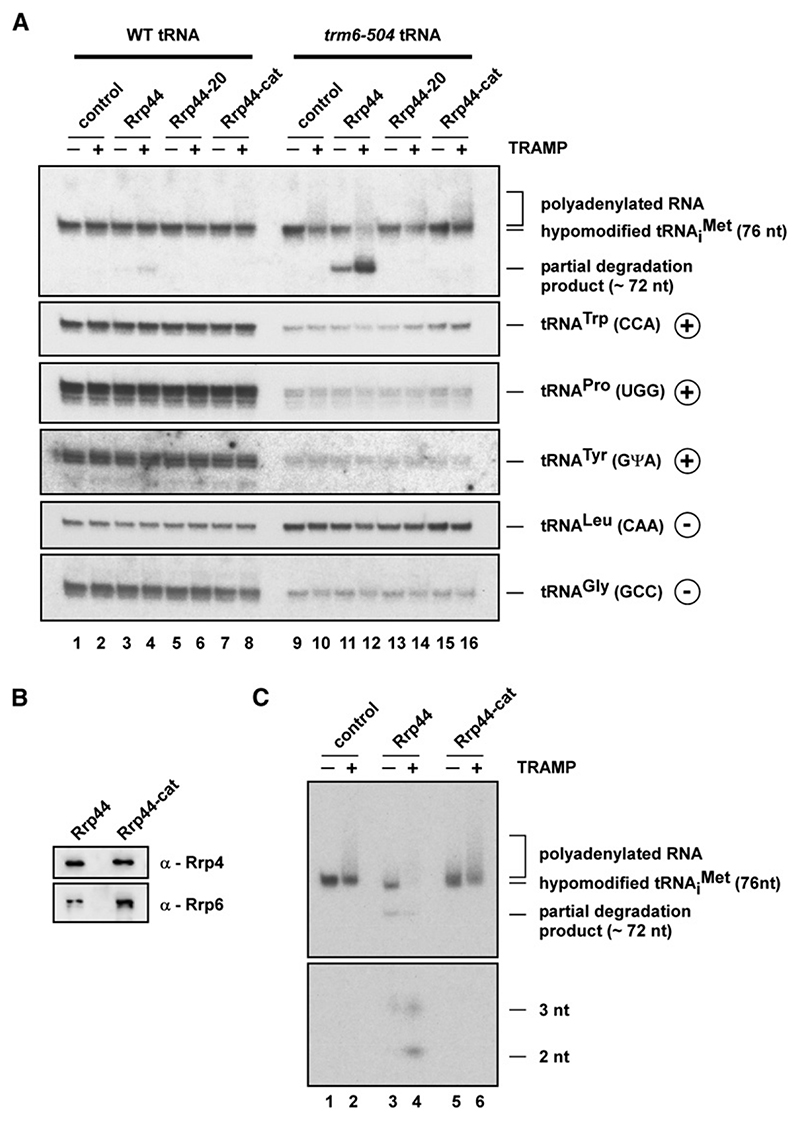
Figure 3. Rrp44 and TRAMP Independently Recognize Hypomodified in Total tRNA.
(A)Combined exonuclease/polyadenylation assay using 0.5 ml TAP-purified TRAMP complex, 100 fmol GST-Rrp44 fusion proteins (wild-type, 44-20, or cat), and total tRNAs purified from wild-type (WT) or trm6-504 yeast strains. In order to achieve equal levels in these tRNA preparations, the reaction was performed with 120 ng wild-type tRNAs and 40 ng trm6-504 tRNAs. After a 10 min incubation at 30°C, the reaction products were separated on a 12% polyacrylamide/8 M urea gel and analyzed by northern hybridization using oligo probes specific for or other tRNAs either containing (+) or lacking (−) the m1A58 modification.
(B)Western analysis of TAP-purified exosome from GAL::rrp44 strains transformed with a plasmid expressing szz-tagged wild-type Rrp44 or the mutant Rrp44-cat protein. Strains were grown in glucose medium at 30°C. Exosomes were purified on IgG Sepharose, eluted by TEV cleavage, and analyzed by western blotting with anti-Rrp4 and anti-Rrp6 antibodies.
(C)Combined exonuclease/polyadenylation assay using 2.5 μl TAP-purified yeast exosome, 0.5 μl TRAMP complex, and 2.5 fmol 5’ end-labeled, native, hypomodified substrate in the presence of 1 mM ATP. Reactions were incubated for 90 min at 30°C at 0.75 mM MgCl2 and analyzed as described in Figure 2C.
Wild-type Rrp44 on its own degraded the hypomodified from the trm6-504 strain to an ~72 nt product but showed much lower activity with the fully modified tRNA from the wild-type strain (Figure 3A, lanes 11 and 3) and no detectable degradation of othertRNAs. These included three species that also undergo Trm6-mediated m1A modification (indicated with ⨁ in Figure 3A). TAP-purified TRAMP independently recognized and polyadenylated from the trm6-504 strain (Figure 3A, lane 10), but not the fully modified (Figure 3A, lane 2) or other tRNAs tested. Addition of the TRAMP complex strongly enhanced the degradation activity of Rrp44 (Figure 3A, lanes 11 and 12) but did not stimulate Rrp44-20 or Rrp44-cat (Figure 3A, lanes 13-16).
These data demonstrate that both Rrp44 and TRAMP specifically and independently recognize the misfolded hypomodified in vitro, even in the presence of a large excess of other tRNA species. However, TRAMP plus Rrp44 was not sufficient to efficiently degrade the hypomodified to small products in the presence of other tRNAs. This result is in contrast to the experiment shown in Figure 2A, where a single, 5’ end-labeled tRNA substrate was used. This suggests that the intact nuclear exosome complex might be needed for efficient and complete degradation of pre- in vivo.
The Exosome Purified from a Strain Expressing Only Catalytically Inactive Rrp44 Does Not Degrade Hypomodified
To determine whether Rrp44 is the only exosome component capable of degrading hypomodified , the exosome was purified from the GAL::rrp44 strain grown on glucose medium and complemented by plasmids expressing either Rrp44-szz or Rrp44-cat-szz. Complexes were purified on IgG Sepharose and eluted by TEV protease cleavage. Recovery of intact exosome complexes was confirmed by western blotting of the core component Rrp4 and the nuclear-specific component Rrp6 (Figure 3B) and by Coomassie staining (data not shown). The yield of Rrp4 was very similar for wild-type and mutant Rrp44, but ~2-fold more Rrp6 was recovered with tagged Rrp44-cat.
5’ end-labeled hypomodified was incubated with equivalent amounts of exosome copurified with Rrp44-szz or Rrp44-cat-szz, in the absence or presence of TAP-purified TRAMP. The degradation pattern generated by the wild-type exosome closely resembled that of recombinant Rrp44. The exosome on its own partially degraded the hypomodified (Figure 3C, lane 3), while the addition of the TRAMP complex stimulated the exonuclease activity and led to a complete degradation of the substrate (Figure 3C, lane 4). In contrast, the exosome copurified with Rrp44-cat-szz showed no exonuclease activity, while addition of the TRAMP complex led to polyadenylation of the RNA.
From these results we conclude that Rrp44 is the only exosome subunit that can recognize and, in the presence of the TRAMP complex, completely degrade the hypomodified .
Discussion
All ten core components of the yeast exosome are essential for viability, and conditional depletion of any of these proteins leads to very similar phenotypes on RNA metabolism. This has made it difficult to determine whether individual substrates are directed to, or targeted by, specific components of the complex. Here, we report that yeast Rrp44 plays a direct role in the specific recognition and degradation of defective .
How Is the Hypomodified Distinguished from the Fully Modified RNA?
The initiator shows structural details that differ from other tRNAs, and methylation of position A58 plays a uniquely important role in maintaining the correct tertiary structure (Basavappa and Sigler, 1991). The hypomethy-lated can be specifically and independently recognized by both the TRAMP complex and Rrp44, even in the presence of a large excess of other tRNA species (see also Vanacova et al. [2005]). It seems unlikely that the specific change in base chemistry induced by the loss of adenosine methylation is the determinant for recognition. We envisage that Rrp44 and the TRAMP complex, and probably other tRNA surveillance systems (Alexandrov et al., 2006), are unable to bind correctly folded and modified tRNAs but can bind tRNAs that are misfolded. This would allow recognition of any defects in tRNA maturation that lead to altered structures. Conceptually, this might resemble the ability of protein chaper-ones to recognize and bind any protein that shows generic features of “misfolding” regardless of the specific protein sequence.
Relating Rrp44 Structure and Function
A single point mutation in the predicted catalytic site of Rrp44 in RNB motif I abolished nuclease activity without affecting RNA binding. This implies that Rrp44, like E. coli RNase II (Amblar and Arraiano, 2005), contains functionally separable domains that are primarily responsible for RNA binding or catalysis.
Two distinct regions of RNase II, the “anchor” and “catalytic” regions, establish contacts with the RNA and act synergistically to provide processive degradation (see Figure 4A) (Frazao et al., 2006). The catalytic site lies deep within a cavity and is accessible only to single-stranded RNA, which is held in place by a “clamp” formed by two conserved aromatic residues (Tyr253 and Phe358). When the RNA is shortened to less than five nucleotides, binding to the clamp is lost and an oligonucleotide (4 nt) product is released.
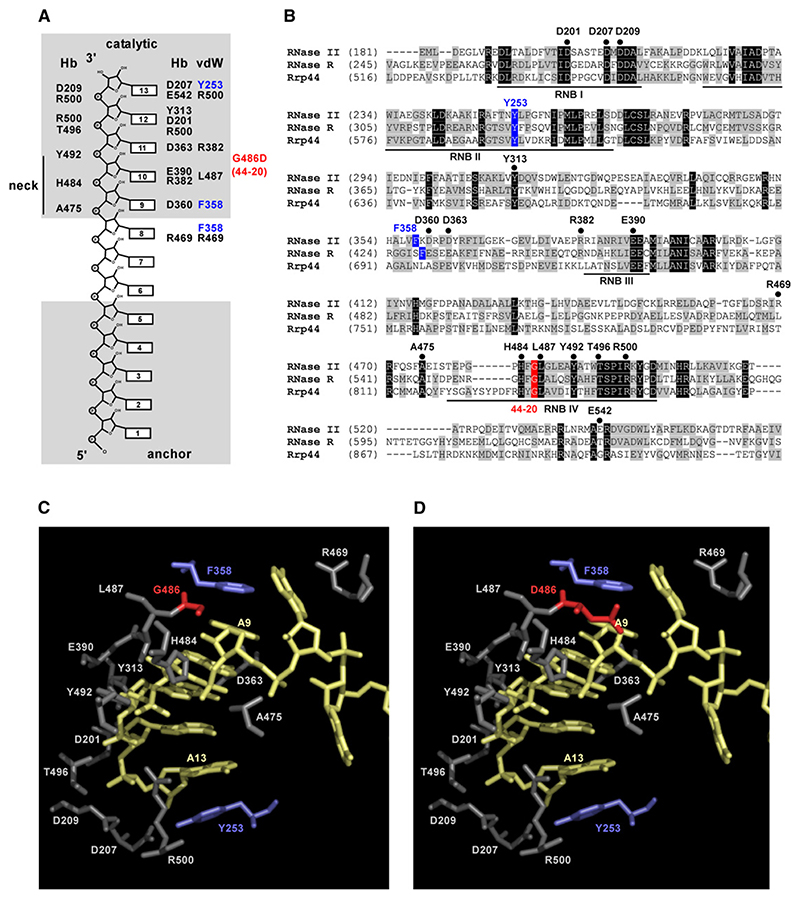
Figure 4. The Rrp44-20 Suppressor Mutation Is Predicted to Affect Specific Interactions between the Rrp44 Catalytic Domain and the 3’ End of the Substrate.
(A)Atomic interactions between RNA and protein residues in the RNA-bound complex of E. coli RNase II (modified from Frazao et al. [2006]). Hb, hydrogen bonds; vdW, van der Waals interactions. The two aromatic residues forming the RNA clamp in RNase II are highlighted in blue. The RNase II residue (G486) corresponding to the rrp44-20 suppressor mutation (G833D) is marked in red.
(B)Alignment of the RNB domains of E. coli RNase II and RNase R and S. cerevisiae Rrp44. Motifs I-IV are underlined. Protein residues involved in RNA interactions are indicated as in (A).
(C)Structural model of the RNA-bound complex of E. coli RNase II (PDB accession number 2IX1) created with PyMol. Protein residues involved in RNA interactions and the position of Gly486 are shown. The enzyme-bound poly(A) RNA is depicted in yellow.
(D)Modeling of Asp486, the mutation present in rrp44-20, onto the structure of RNA-bound E. coli RNase II. The G486D amino acid replacement was performed in a backbone-dependent manner using PyMol.
Sequence alignments suggest that Rrp44 differs from RNase II in the region of the RNA clamp (Tyr253 and Phe358, marked blue in Figure 4). Tyr253 is part of the highly conserved RNB motif II, and equivalent residues are present in RNase II, RNase R, and Rrp44. In contrast, we could find no counterpart for Phe358 in Rrp44. We hypothesize that these differences in the clamp region may be related to differences in substrate recognition.
The activity of RNase II is blocked by secondary structure (Amblar et al., 2006), whereas RNase R can degrade double-stranded RNAs with single-stranded 3’ overhangs of at least 7 nt (Vincent and Deutscher, 2006). Rrp44 was able to remove the unstructured 4 nt of the 3’ CCA tail from the hypomodified , whereas further degradation also required the TRAMP complex. We speculate that recognition of by Rrp44 is permitted by the predicted wider RNA clamp, which might enable it to recognize and bind specific double-stranded RNAs with a short 3’ overhang. Nuclease activity on such a substrate would, however, leave the double-stranded region of the RNA in the catalytic cavity, which is likely to prevent unwinding of the substrate. Addition of the TRAMP complex allowed complete degradation of the by Rrp44, and this stimulation required ATP and the poly(A) polymerase activity of Trf4 (see Figure S3). We propose that following the specific recognition and removal of the 3’ overhang by Rrp44, Trf4 adds a poly(A) tail to the substrate. The bound RNA might then “back-up,” allowing rebinding of the substrate with the longer single-stranded tail along the entire RNA-binding groove. Degradation would then resume with the double-stranded RNA structure being opened by Mtr4, possibly acting in conjunction with the S1 and CSD RNA-binding domains of Rrp44, leading to processive degradation of the remainder of the substrate. A mutation in the S1 domain (G916E) also reduced but did not completely abolish RNA binding and greatly inhibited activity on all substrates tested.
Analysis of rrp44-20 revealed that degradation of the hypomodified was inhibited by a single amino acid substitution (Gly833 → Asp). In E. coli RNase II the counterpart of Rrp44 Gly833 (Gly486) is located in the “neck” region of the substrate-binding site of the catalytic domain (Figures 4A and 4C), which contains many amino acids conserved between RNase R and Rrp44. The neck forms specific hydrogen bonds (Hb) and van der Waals interactions (vdW) with the 3’ end of the substrate (see Figures 4A and 4C) (Frazao et al., 2006). Introduction of a negatively charged side chain (Gly486 → Asp) might reduce the affinity for RNA near the catalytic site (Figure 4D), while not affecting substrate binding by the anchor domain. Consistent with this, Rrp44-20 showed reduced binding to the hypomodified , as well as poly(A30) and a 37 nt transcript from the pBS polylinker. However, loss of degradation was specific for . The less-structured RNA substrates may more readily enter the catalytic cavity of Rrp44, making specific interactions between the RNA and the neck of the catalytic region dispensable for catalysis. These observations imply that specific recognition and/or correct positioning of the 3’ end of the hypomodified takes place in the catalytic domain of Rrp44.
In understanding RNA turnover, a major limitation has been the lack of in vitro systems that faithfully recapitulate in vivo surveillance and degradation mechanisms. The results reported here should facilitate more detailed analyses of RNA surveillance.
Experimental Procedures
Expression and Purification of Recombinant Proteins
Point mutations in the expression construct for GST-Rrp44 (GST-Dis3sc) (Noguchi et al., 1996) were created using the QuikChange kit (Stratagene) and oligos Rrp44-D551N-F/R (Rrp44-cat), Rrp44-G833D-F/R (Rrp44-20), or Rrp44-G916E-F/R (S1) (see Table S1). E. coli strain BL21(DE3)pLysS (Stratagene) transformed with plasmids encoding GST-Rrp44 or GST alone (pGEX-4T1) was grown at 23°C to an OD600 of 0.4, treated with IPTG at 1 mM, and incubated at 23°C to an OD600 of 1. Cell lysis and purification were performed in buffer NB (20 mM Tris/HCl [pH 7.6], 150 mM NaCl, 5 mM MgCl2, 8.7% glycerol, and 0.1% Nonidet P40) supplemented with complete EDTA-free protease inhibitor (Roche). GST fusion proteins were affinity purified on glutathione-Sepharose beads and eluted with 20 mM glutathione, which was subsequently removed by buffer exchange on NAP columns (GE Healthcare).
Exosome and TRAMP Purification
One-step purifications of exosome (at 150 mM NaCl) and TRAMP complex (at 120 mM NaCl) on IgG Sepharose columns followed by TEV elution were performed as described (LaCava et al., 2005). For exosome isolation, GAL::rrp44 strains transformed with plasmids encoding szz-tagged Rrp44 were grown in SD —His/—Ura medium to an OD600 of 1.3 at 30°C. For the isolation of TRAMP complexes, an rrp6D strain transformed with a plasmid expressing zz-tagged Trf4 (Vanacova et al., 2005) was grown in SD –Leu medium to an OD600 of 1 at 25°C. Each preparation used 2 liters of culture, producing 200-300 μl of the final TEV fraction. Enzyme concentrations were normalized by immunoblotting using anti-Rrp4 or anti-Rrp6 (Mitchell et al., 2003) antibodies.
In Vitro Assays
Exonuclease and RNA-binding assays were performed in 10 mM Tris/ HCl (pH 7.6), 75 mM NaCl, 2 mM DTT, 100 μg/ml BSA, 0.8 U/μl RNasin, 4.5% glycerol, 0.05% Nonidet P40, and 0.25-0.75 mM MgCl2. In combined exonuclease/polyadenylation assays, ATP was added to 1 mM. 5’ end-labeled substrates were poly(A30) and poly(U30) (Dharmacon), in vitro-transcribed pre-tRNASer, a 37 nt RNA derived from the pBS polylinker (Mitchell et al., 1997), and purified, native hypomodified . In vitro transcription, 5’ end labeling, and gel purification of the labeled RNAs were performed as described (LaCava et al., 2005). Total tRNA was isolated from wild-type and trm6-504 strains, and hypomodified was purified from strain yJA146 (gcd10DhcIMT4) as described (Anderson et al., 2000).
Supplementary Material
Acknowledgments
Wethank W. Kellerand J. Houseley for yeast strains and reagents and J. Houseley, S. Granneman, K. Kotovic, N. Watkins, and M. Kos for critical reading of the manuscript. This work was supported by the Wellcome Trust and a long-term HFSP fellowship to C.S.
References
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Amblar M, Arraiano CM. A single mutation in Escherichia coli ribonuclease II inactivates the enzyme without affecting RNA binding. FEBS J. 2005;272:363–374. doi: 10.1111/j.1742-4658.2004.04477.x. [DOI] [PubMed] [Google Scholar]
- Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J Mol Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa R, Sigler PB. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;74:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 30 processing of stable RNAs. Mol Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Hayashi N, Azuma Y, Seki T, Nakamura M, Nakashima N, Yanagida M, He X, Mueller U, Sazer S, Nishimoto T. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 1996;15:5595–5605. [PMC free article] [PubMed] [Google Scholar]
- Romani A, Scarpa A. Regulation of cell magnesium. Arch Biochem Biophys. 1992;298:1–12. doi: 10.1016/0003-9861(92)90086-c. [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Vincent HA, Zhang J, Wang Y, Deutscher MP, Malhotra A. Structural basis for processivity and single-strand specificity of RNase II. Mol Cell. 2006;24:149–156. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.