Abstract
Base editors are RNA-programmable deaminases enabling precise single-base conversions on genomic DNA. However, off-target activity is a concern in the potential use of base editors to treat genetic diseases. Here, we report unbiased analyses of transcriptome-wide and genome-wide off-target modifications effected by cytidine base editors in the liver of mice with phenylketonuria. The intravenous delivery of intein-split cytosine base editors via dual adeno-associated viruses led to the repair of the disease-causing mutation without generating off-target mutations in the RNA and DNA of the hepatocytes. Moreover, the transient expression of a cytidine base editor mRNA and a relevant single-guide RNA intravenously delivered via lipid nanoparticles led to ~21% on-target editing and to the reversal of the disease phenotype, also without detectable transcriptome-wide and genome-wide off-target edits. Our findings support the feasibility of therapeutic cytidine base editing to treat genetic liver diseases.
A large proportion of genetic diseases is caused by single-nucleotide mutations that are potentially targetable by RNA-programmable deaminases, known as base editors (BEs). BEs allow single C·G to T·A or A·T to G·C base pair conversions via uracil and inosine intermediates through single-stranded (ss)DNA-specific cytosine or adenosine deaminases that are fused to a catalytically impaired Cas91,2. These base pair conversions occur independently of double stranded (ds)DNA breaks or homology directed repair (HDR) and allow efficient editing in vivo in post-mitotic tissues. We and others have previously used viral vectors, including Adeno-associated viruses (AAVs), to deliver BEs and correct disease phenotypes in vivo 3–5. More recent ex vivo studies, however, have demonstrated that CBEs can give rise to tens of thousands of sgRNA-independent transcriptome-wide off-target mutations in cell lines6–9, and hundreds of genome-wide off-target mutations in induced pluripotent stem cells and two-cell stage embryos10,11. It has moreover been shown that in vivo overexpression of the most commonly used CBE deaminase, rat APOBEC1 (rAPOBEC1), leads to hyperediting beyond its endogenous target transcript APOB 12–14, causing dysplasia and hepatocellular carcinoma in the mouse liver15. Together, these data suggest that CBE expression could pose considerable risks for application in patients, prompting us to assess off-target effects during in vivocytosine base editing in somatic tissues and to establish a delivery approach where the duration of CBE expression is minimised.
Results
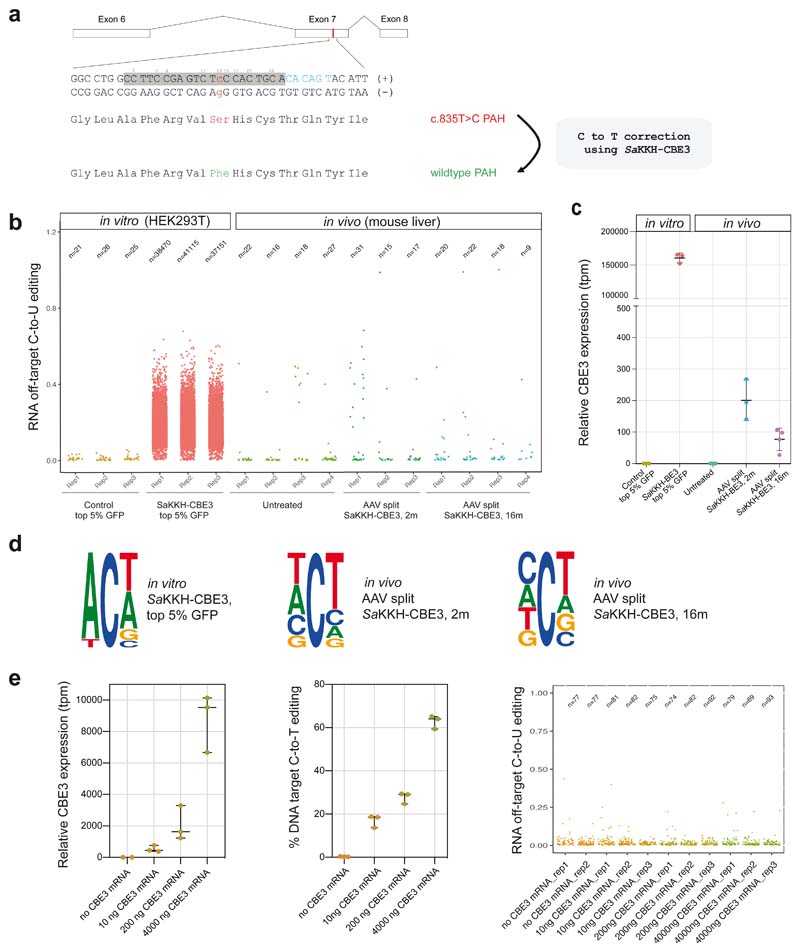
To evaluate off-target deamination of CBEs in vivo we focused on the Pahenu2 mouse model for Phenylketonuria, which can be cured by AAV-mediated delivery of a Staphylococcus aureus (Sa)KKH-CBE3 system into the mouse liver5. In brief, SaKKH-CBE3, consisting of a SaCas9(KKH) nickase variant fused to rAPOBEC1 is systemically introduced into Pahenu2 mice using a dual AAV intein-split system to correct the disease-causing T-to-C mutation in exon 7 (Fig. 1a). We first focused on the assessment of transcriptome-wide off-target deamination using RNA-sequencing (RNA-seq; on average 185 million reads per library). Confirming previous in vitro findings6,9, transfection of an SaKKH-CBE3-expressing plasmid into HEK293T cells resulted in an average of 39 000 C-to-U transitions (Fig. 1b, Suppl. Fig. 1, 3). To determine whether similar rates of transcriptome-wide off-target mutations occur during cytosine base editing in vivo in the liver, SaKKH-CBE3 was systemically introduced via AAV8 into Pahenu2 mice. RNA from the liver was extracted after 8 weeks when peak transgene expression from AAV vectors is reached16, and analysed by RNA-seq. While 23% on-target editing was observed, to our surprise we found no increase in transcriptome-wide C-to-U transitions compared to untreated controls (Fig. 1b, Suppl. Fig 2, 3). Consistent with these results, cytosine edits preferentially occurred within the typical APOBEC consensus motif of ACW (W=A or U) in treated HEK293T cells but not in treated mouse livers (Fig. 1d, Suppl. Fig. 4). Interestingly, when comparing SaKKH-CBE3 expression in different samples, we observed that transcript levels were three orders of magnitude higher in HEK293T cells compared to the liver (Fig. 1c). To investigate, whether excessive CBE overexpression was associated with high off-target editing rates in vitro, we transfected lower doses of SaKKH-CBE3 mRNA and sgRNA into HEK293T and Hepa 1-6 cells containing exon 7 of the Pahenu2 locus. We found that an 18-fold reduction in CBE expression compared to plasmid transfection retained 63% on-target editing, but drastically reduced RNA off-target mutations (Fig. 1b,c,e, Suppl. Fig. 5). These results support our hypothesis that prevalent RNA off-target deamination in HEK293T cells by CBE expression from plasmid DNA is likely associated with extensive rAPOBEC1 overexpression.
Figure 1. Transcriptome-wide C-to-U editing analysis after AAV-mediated SaKKH-CBE3 treatment.
a)The mutant Pahenu2 allele that harbours the disease-causing c.835T>C (p.F263S) mutation (red) on Exon 7 was corrected using SaKKH-CBE3. The sgRNA binds the protospacer (grey) and NNNRRT PAM site (blue). All Cs within the protospacer region are indicated (C2-C20). A Ser to Phe amino acid change at position 263 restores PAH enzyme activity. b) Transcriptome-wide C-to-U editing events are represented as single dots (exact numbers are indicated above, n). Values were derived from 3-4 biolgically independent replicates. in vitro RNA-seq data are from HEK293T cells co-transfected with SaKKH-CBE3, the Pahenu2 -targeting sgRNA, and a GFP-expressing plasmid, sorted for top 5% GFP expression. In vivo RNA-seq data are from mice treated with adeno-associated virus (AAV) to express SaKKH-CBE3 and the Pahenu2 -targeting sgRNA. Control mice were not injected. in vitro samples were analysed 72 hours after transfection. AAV treated mice were analysed 2 months or 16 months after injection of 5 × 1011 vg per AAV vector. c) Relative SaKKH-CBE3 expression levels (rAPOBEC1, tpm=transcripts per million) of samples analysed in b). n = 3 or 4 biologically independent experiments, represented by individual dots. Values represent mean ± s.d. d) Sequence motifs of representative data sets. The motifs were derived from accumulated RNA C-to-U edits of 3 biologically independent replicates per group. in vitro SaKKH-CBE3-treated samples exhibit a typical APOBEC1 ACW motif. Ts should be considered as Us. The motifs depict base probability as letter size. e) SaKKH-CBE3 expression was titrated by transfecting different concentrations of SaKKH-CBE3 mRNA and in vitro transcribed sgRNA into HEK293T cells with exon 7 of Pahenu2 stably integrated. The first panel depicts relative expression of the base editor, the second panel shows corresponding C-to-T edits at the genomic target locus and panel three shows corresponding RNA C-to-U editing events of the samples analysed in panel one and two. Cells were harvested 48 hours after transfection and sorted for co-transfected mCherry expression. n = 3 biologically independent experiments, represented by individual dots. Values represent mean ± s.d. Each dot in panel three represents one C-to-U editing event. Total numbers are indicated above (n). Replicates represent biologically independent replicates.
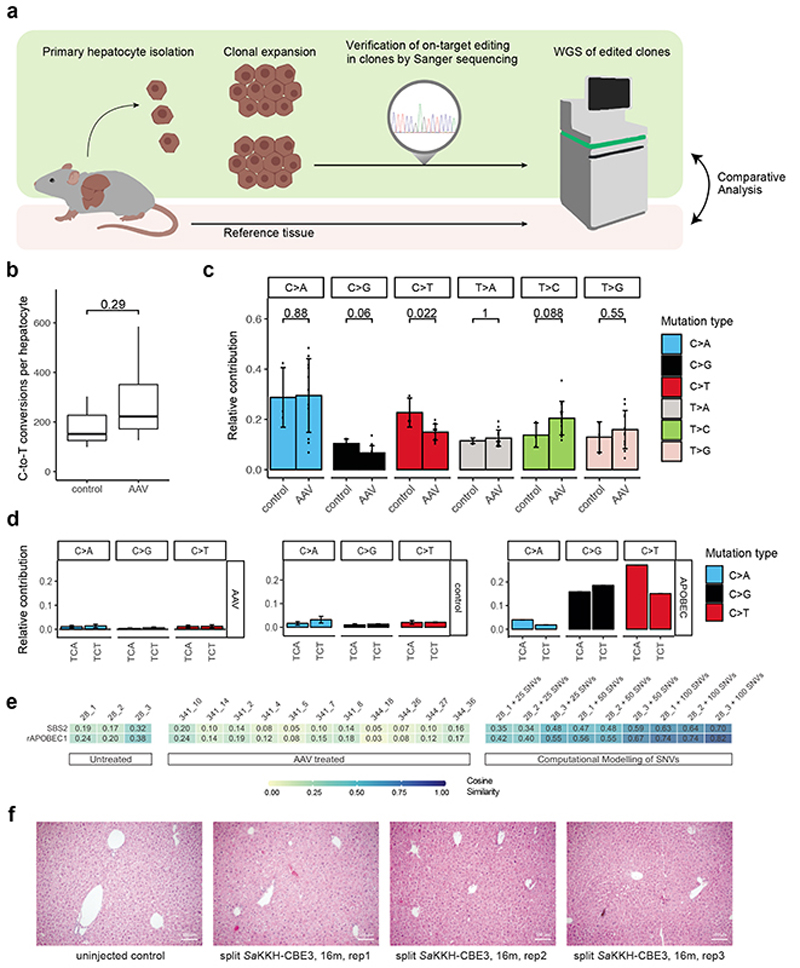
We next assessed whether AAV-mediated delivery of SaKKH-CBE3 into Pahenu2 mice leads to off-target editing on genomic DNA. While we identified no sgRNA-dependent mutations at predicted off-target loci in our previous study5, it remained unclear whether random sgRNA-independent off-target mutations occur during base editing in adult tissues. These mutations are different for each cell and cannot be detected by whole genome sequencing (WGS) of bulk DNA from a pool of cells17. We therefore isolated hepatocytes from treated and untreated control mice, and expanded them clonally as chemically induced liver progenitor (CLiP)18 cells prior to sequencing (Fig. 2a). This allowed us to obtain clonal DNA for WGS without generating excessive noise observed during single cell DNA amplification19,20. We selected three clones from an untreated control animal and 11 clones from AAV treated animals with confirmed editing at the target locus for WGS at 30x coverage (Suppl. Fig. 6). DNA from untreated bulk tissue of the same animal was used to filter out germline variants, and subclonal mutations with a frequency below 20% were dismissed as they likely occurred during in vitro expansion. Analysis of the ratio between synonymous and non-synonymous (dN/dS) mutations showed no shift towards synonymous mutations (Suppl. Fig. 7), suggesting that we did not select against clones with protein-coding mutations. Confirming that WGS reliably detects single base conversions in clonal DNA, we observed C-to-T editing at the target locus in all 11 clones from AAV-treated animals, and identified on average 94% of heterozygous germline SNPs (Suppl. Fig. 6, 8). When we next analysed nucleotide conversion rates in the different groups, we found no significant increase in C-to-T conversions in hepatocyte genomes isolated from AAV treated mice compared to control mice (Fig. 2b). In addition, mutation spectra were similar between treated and untreated clones, but distinct from positive control clones where APOBEC mutations were added in silico (Fig. 2c, Suppl. Fig. 9, 10). Identified C-to-T conversions were moreover not associated with typical APOBEC motifs (Fig. 2d), and cosine similarity analysis showed no enrichment of APOBEC signatures in clones derived from AAV treated mice (Fig. 2e). Taken together, SaKKH-CBE3 expression after AAV-mediated delivery into the liver did not lead to substantial off-target deamination in hepatocytes on RNA and genomic DNA. In line with these results, we found no evidence for malignant transformation in the liver after 16 months of SaKKH-CBE3 expression (Fig. 2f, Suppl. Fig. 11).
Figure 2. Genome-wide off-target analysis after AAV-mediated SaKKH-CBE3 treatment.
a) Primary hepatocytes were isolated after in vivo base editing and clonally expanded as chemically induced liver progenitor cells (CLiPs). On-target editing was confirmed by Sanger sequencing, and selected clones were analysed by whole genome sequencing (WGS). Called variants were compared to bulk reference tissue to filter out germline variants. b) C·G to T·A editing events were summarized as C-to-T conversions per hepatocyte clone. Clones were derived from AAV-treated (n=11), and untreated control (n=3) mice. Box plots are standard Tukey plots, where the centre line represents the median, the lower and upper hinges represent the first and third quartiles, and whiskers represent + 1.5 the interquartile range. Wilcoxon test was used for comparison. c) Relative contributions of respective single base substitutions observed in the genome of AAV-treated clones (n=11) versus untreated control clones (n=3). C>A includes G>T, C>G includes G>C, C>T includes G>A, T>A includes A>T, T>C includes A>G and T>G includes A>C. Values represent mean ± s.d. Wilcoxon test was used for comparison. d) Relative contributions of APOBEC-relatable sequence motifs of C>A (G>T), C>G (G>C) and C>T (G>A) conversions of AVV-treated hepatocytes (n=11) versus unedited controls (n=3). The typical APOBEC signature, derived from COSMIC data40, is shown in the right panel. Typical APOBEC signatures are absent in base edited hepatocytes after AAV delivery of SaKKH-CBE3. Values represent mean ± s.d. e) Heatmap showing the cosine similarity of mutational signatures of clones to the COSMIC signature SBS240 and a predetermined rat APOBEC1 signature10 as displayed in Suppl. Fig. 10. Heatmap; 1=exact match, 0=no similarity observed. f) Haematoxylin and Eosin staining of representative mouse liver paraffin sections from long-term AAV-treated mice 16 months after AAV split SaKKH-CBE3 (5 × 1011 vg per AAV vector) administration (n=3). 4 individual pictures were taken per animal where 3 animals were analysed per group and whole sections were evaluated by a trained pathologist. Controls sections were derived from uninjected Pahenu2 mice. Scale bar, 100 μm.
Further analysis of the Pahenu2 locus in edited mice revealed that SaKKH-CBE3 expression led to frequent indel formation at the target locus due to simultaneous nicking and base excision repair (BER) on opposite DNA strands21 (Suppl. Fig. 12). In an attempt to reduce indel formation, we exchanged SaKKH-CBE3 with nuclease-dead SaKKH-CBE2 and Gam- SaKKH-CBE4, a fourth-generation base editor, where binding of the bacteriophage Mu-derived Gam protein to dsDNA breaks has been suggested reduce indel formation21. However, although SaKKH-CBE2 and Gam- SaKKH-CBE4 led to fewer indels, SaKKH-CBE3 surpassed these constructs with regard to the number of correctly edited alleles (Suppl. Fig. 13, 14). As we identified no large deletions or chromosomal rearrangements (Suppl. Fig. 15), and small indels at the Pah locus are unlikely to drive malignant transformation, we decided to continue with SaKKH-CBE3 for all subsequent steps in our study.
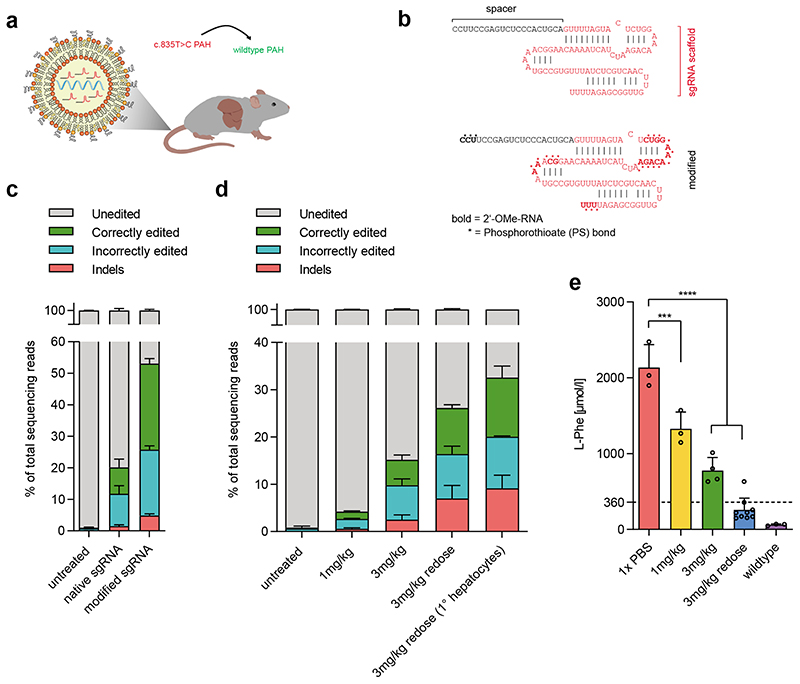
In postmitotic cells, including hepatocytes, AAV genomes can persist over many years22. While this makes AAVs ideal vectors for gene replacement therapies, ‘hit and run’ genome editing only requires transient expression of editing components. To reduce potential off-target effects associated with long-term expression of Cas9 nucleases, previous studies harnessed lipid nanoparticles (LNPs) to deliver short-lived Cas9 mRNA and sgRNA for transient genome editing in the mouse liver23,24. To explore whether a similar approach would be feasible for base editing, we targeted the Pahenu2 mouse with SaKKH-CBE3 mRNA and the respective sgRNA using LNP (Fig. 3a). In order to reduce susceptibility to degradation of the mRNA and sgRNA by nucleases, we used 5-methoxyuridine-modified SaKKH-CBE3 mRNA and introduced chemical modifications to the SaCas9 sgRNA in compliance with structural data of SaCas925 (Fig. 3b). The chemically modified sgRNA allowed more efficient editing compared to the unmodified sgRNA in vitro (Fig. 3c), prompting us to encapsulate SaKKH-CBE3 mRNA and modified sgRNA into LNP formulations optimised for mRNA delivery26. Administration of a single 3 mg/kg dose via the tail vein was well tolerated (Suppl. Fig.16), and resulted in target C-to-T conversion of 10.7% in whole liver lysates, with 5.5% reads supporting restoration of the PAH enzyme (Fig. 3d; Suppl. Fig. 17, 18). After confirming the transient nature of sgRNA and SaKKH-CBE3 mRNA in hepatocytes (Suppl. Fig. 19), we examined how repeated dosing affects editing by administering two doses of 3mg/kg at an interval of one week. On-target C-to-T editing increased to 18.8%, with 10.8% reads supporting restoration of the PAH enzyme (Fig. 3d; Suppl. Fig. 17, 18). Notably, when we analysed isolated hepatocytes instead of whole liver lysates, editing rates increased to 21% (Suppl. Fig. 20). Analysing blood L-Phenylalanine (L-Phe) levels, we found that correction efficiencies after redosing were sufficient to reduce the elevated L-Phe levels of Pahenu2 mice below the therapeutic threshold of 360μmol/l27 (Fig. 3e), leading to a reversion of the Pahenu2 -associated fur colour phenotype (Suppl. Fig. 21).
Figure 3. Correction of the disease locus of the Pahenu2 mouse model via LNP-mediated delivery of base editing components.
a) Concept of in-vivo correction of the mutated Pahenu2 locus by systemic administration of LNP-encapsulated SaKKH-CBE3 mRNA and sgRNA. b) Chemical modifications introduced to the Sa Cas9 sgRNA targeting the c.835T>C mutation. c) Correctly edited reads support restoration of the correct PAH amino acid sequence by C-to-T conversion of the target C13, including any synonymous mutations. Incorrectly edited reads summarize mutations that are not synonymous to the wildtype PAH amino acid sequence, including C-to-T conversions at positions additional to or other than C13, and conversions that are not C-to-T. Indels are shown separately. Values represent mean of three independent biological replicates performed on separate days ± s.d. d) Editing efficiencies after systemic administration of 1 mg/kg, 3 mg/kg or two doses of 3 mg/kg in vivo in whole liver lysates and in isolated primary hepatocytes. Correctly edited reads restore the wildtype PAH amino acid sequence, incorrectly edited reads mean mutations that lead to amino acid sequences nonsynonymous to the wildtype PAH amino acid sequence. Values represent mean of three independent biological replicates performed on separate days ± s.d. e) Blood L-Phe levels from homozygous Pahenu2 and wildtype C57BL6 mice were determined one week after the latest injection. Values (n=3 per treatment group) represent mean ± s.d. A two-way ANOVA with Dunnett’s multiple comparisons test was performed to account for multiple comparisons to a single control (untargeted): ***P < 0.0002, ****P < 0.0001.
To increase efficient delivery and reduce off-target editing, genome editing therapies that target genetic liver diseases would benefit from high hepatotropism. LNPs have previously been shown to interact with apolipoprotein E (ApoE), facilitating LDL receptor interaction and subsequent internalization by hepatocytes28,29. Confirming hepatotropism of our LNP formulation, we observed liver-specific mCherry expression after systemic administration of 1 mg/kg mCherry mRNA (Suppl. Fig. 22), and only marginal on-target editing in tissues other than the liver after administration of two doses of 3 mg/kg (Suppl Fig. 23).
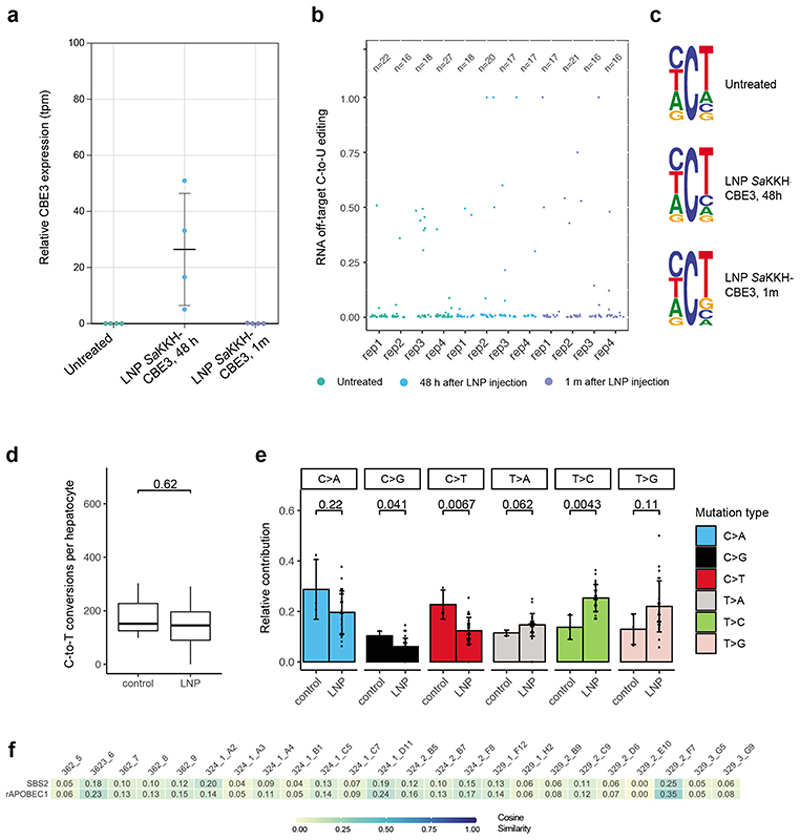
We next analysed whether LNP-mediated base editing in the liver causes off-target deamination on RNA or DNA. Considering that this approach only leads to transient expression of SaKKH-CBE3, we performed RNA-seq analysis 48h after injection of a 3 mg/kg dose. Importantly, we found that CBE expression levels were within the range of endogenous mAPOBEC1, and did not lead to increased C-to-U transitions compared to untreated controls (Fig. 4a,b; Suppl. Fig. 24, 25). Cytosine edits moreover did not preferentially occur within a typical APOBEC consensus motif (Fig. 4c). One month after LNP delivery SaKKH-CBE3 expression was completely abolished, and unsurprisingly again no C-to-U off-target mutations were observed (Fig. 4a, b, c). To next assess whether LNP-mediated delivery of SaKKH-CBE3 led to off-target deamination on genomic DNA, we first analysed 10 computationally predicted off-target loci30 using high-throughput sequencing (>10 000x coverage). Similar to our previous study where CBE was delivered via AAV5, we observed no C-to-T conversions above background at these sites (Suppl. Fig. 26). We then focused on sgRNA-independent off-target mutations, and isolated hepatocytes one month after treatment for clonal expansion. 24 clones with confirmed on-target editing were selected (Suppl. Fig. 6), and analysed by WGS at 30x coverage. We neither observed an increase of C-to-T conversions compared to control clones, nor an enrichment of typical APOBEC motifs (Fig. 4d, e; Suppl. Fig. 27, 28). In line with these results, cosine similarity analyses revealed no enrichment of APOBEC signatures in clones derived from LNP treated mice (Fig. 4f). Taken together, base editing via LNP-mediated delivery of SaKKH-CBE3 mRNA and chemically modified sgRNA enabled correction of disease phenotypes without detectable off-target deamination on the transcriptome and genome.
Figure 4. Transcriptome- and genome-wide off-target analysis after LNP-mediated SaKKH-CBE3 treatment.
a)Relative SaKKH-CBE3 expression levels (quantified by rAPOBEC1 expression, tpm=transcripts per million) of untreated mouse livers (n=4) and edited mouse livers 48 hours or 1 month after LNP treatment (n=4). Shown are biologically independent replicates, where n is indicated as individual dots. Values represent mean ± s.d. b) Jitter plots show transcriptome-wide C-to-U editing events. RNA-seq data from untreated mouse livers and LNP-treated livers analysed 48 hours or 1 month after injection. Each dot represents one editing event, exact values are indicated above (n). c) Sequencing motifs for all RNA C-to-U editing events shown in b). Motifs were derived from cumulated RNA C-to-U edits of 3 independent replicates per sample. Ts should be considered as Us. The motifs depict base probability as letter size. d) C·G to T·A editing events were summarized as C-to-T conversions per hepatocyte clone. 24 clones were derived from LNP-treated mice, and 3 clones from untreated control mice. Box plots are standard Tukey plots, where the centre line represents the median, the lower and upper hinges represent the first and third quartiles, and whiskers represent + 1.5 the interquartile range. Wilcoxon test was used for comparison. e) Relative contributions of single base substitutions observed in the genome of hepatocytes isolated from LNP-treated mice (n=24) versus hepatocytes isolated from control mice (n=3). Their frequency in the genome (y-axis) is shown. C>A includes G>T, C>G includes G>C, C>T includes G>A, T>A includes A>T, T>C includes A>G and T>G includes A>C conversions. Values represent mean ± s.d. Wilcoxon test was used for comparison. f) Heatmap showing the cosine similarity of mutational signatures from each clones to the COSMIC signature SBS240 and the predetermined rat APOBEC1 signature10 as displayed in Suppl. Fig. 10. Heatmap; 1=exact match, 0=no similarity observed.
Discussion
Transient non-viral gene editing is an attractive approach to target monogenetic liver diseases in a clinical setting. LNP-mediated delivery of mRNA and sgRNA is therefore particularly promising and has previously been used for classical Cas9 endonucleases23,24. For base editors, a recent study had already explored LNP-mediated delivery for BEs31. However, they only obtained correction of the targeted mutation in less than 1% of hepatocytes, which is insufficient for therapeutic application for most genetic diseases. In contrast, in this study we acquired editing efficiencies of 21% per haploid genome, exceeding correction efficiencies required to cure a range of monogenic liver diseases, including phenylketonuria32. Interestingly, when transducing SaKKH-CBE3 via AAV vectors, correction of the Pahenu2 locus at rates similar to LNP-mediated delivery was only observed after 8 weeks post injection5. Considering that SaKKH-CBE3 expression levels were higher in AAV-treated mice compared to LNP-treated mice, we speculate that lower editing rates resulted from inefficient reconstitution of the full-length SaKKH-CBE3 by dual intein-split AAV vectors5. Besides the constraint that full-length BEs are too large to be packaged into single viral particles, AAV vectors suffer from several other limitations: (1)Circulating antibodies and capsid-specific memory T-cells from previous exposure to wild-type AAVs are prevalent in humans22. (2) AAVs have been shown to integrate into murine hepatocyte genomes33–35, and although it is controversial whether integration also occurs in human genomes caution is warranted. (3) Long-term expression of bacterial proteins (such as Cas9) often provoke immune responses, which over time may lead to a rejection of cells expressing Cas9 or BEs33.
Recent reports have shown that BEs can cause substantial sgRNA-independent off-target deamination on RNA and DNA in in vitro cultured cells and two-cell stage embryos6–9,10,11. Contrary to these findings, we observed no increase in C-to-U/T transitions and no enrichment of APOBEC signatures following in vivo SaKKH-BE3 delivery. We argue that the lack of off-target deamination is a result of moderate CBE expression below the levels of housekeeping genes such as GAPDH. Supporting this hypothesis, we found that ex vivo in HEK293T cells RNA off-target effects were critically dependent on excessive CBE overexpression.
Notably, analysis of genome wide off-target deamination on a single cell level required clonal expansion of isolated hepatocytes prior to WGS to obtain sufficient amounts of genomic DNA. Thus, we could only analyse a relatively small number of hepatocytes, and potentially missed a subpopulation with increased off-target rates. Considering that protein-damaging mutations could limit the potential of hepatocytes to dedifferentiate into CLIPs and expand ex vivo, it is moreover feasible that we selected against hepatocytes that accumulated a large number of mutations, thereby underestimating the true off-target rates. However, arguing against the latter concern, we were able to detect hundreds of naturally occurring somatic SNVs per hepatocyte without observing a shift towards synonymous (silent) mutations. Finally, it is worth noting that the number of somatic SNVs and in vitro mutations that occur within the first CLIP cell division set the detection limit for CBE-induced mutations. In silico introduction of APOBEC mutations to untreated control clones, nevertheless, revealed that in our experimental setup less than 50 mutations would have been robustly detected by Cosine similarity analysis (Fig. 4d). Thus, in the context of our previous work, where we showed that genomes of human liver cells naturally accumulate thousands of mutations over a lifetime20, our results suggest that in vivo base editing in the liver might be well tolerated in clinical applications, in particular when CBE expression is temporally limited.
Prior to application in humans, additional safety studies would be required. These might include base-editing studies in animal models with sensitized genetic backgrounds highly vulnerable to malignant transformation, and studies in large animal models that investigate potential immune responses to CBEs. As our results indicate that CBE expression levels play a crucial role for off-target generation, it would furthermore be important to investigate whether the chosen delivery method leads to excessive overexpression in a subset of transfected cells. In addition, it would be interesting to test new CBE variants that were optimized to reduce Cas9-independent deamination9,36–39. These variants might better tolerate variations in CBE expression levels, and thus further increase safety of in vivo base editing approaches.
In summary, we developed a non-viral and transient base editing approach to treat monogenic liver diseases without substantial off-target effects. Several aspects of LNP-mediated RNA delivery, such as its synthetic nature and the high degree of scalability of all components, support the potential of this approach for therapeutic use.
Methods
AAV vector production
All pseudotyped AAV2/8 vectors were produced by the Viral Vector Facility of the Neuroscience Center Zurich. AAV vectors were ultracentrifuged and diafiltered. Physical titers (vector genomes ml–1) were determined using a Qubit 3.0 Fluorometer. Identity of the packaged genomes of each AAV vector was confirmed by Sanger DNA-sequencing.
Cell culture transfection protocol and genomic DNA preparation
HEK293T (ATCC CRL-3216) cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) plus GlutaMax (Thermo Fisher), supplemented with 10% (v/v) fetal bovine serum (FBS) and 1× penicillin-streptomycin (Thermo Fisher Scientific) at 37°C and 5% CO2. Cells were maintained at confluency below 90% and seeded on 48-well cell culture plates (Greiner). 12-16h after seeding, at approximately 70% confluency, cells were transfected using 1.5μl of Lipofectamine 2000 (Thermo Fisher Scientific) and 500 ng base editor mRNA and 50 ng sgRNA. Cells were incubated for 3 days and genomic DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s protocol. For FACS experiments, SaKKH-CBE3 (Addgene #85170) was co-transfected with a control plasmid expressing GFP in HEK293T at 70-80% confluency in 6-well plates (1.5ug BE plasmid, 0.5ug GFP-expressing plasmid, 0.5ug sgRNA-expressing plasmid). An intein-split BE was co-transfected in HEK293T at 70-80% confluency in 6-well plates (N-terminal and C-terminal constructs in a 1:1 ratio, at 1.25ug plasmid each) or the C-terminal part only as RFP control (1.25ug plasmid)5. Different amounts of SaKKH-CBE3 mRNA (4000ng, 200ng, 10ng), together with 400ng mCherry mRNA and 2000ng sgRNA were transfected into HEK293T and Hepa 1-6 cells in a 6-well format and harvested after 48 hours for CBE titration experiments.
RNA synthesis
Chemically modified sgRNAs were synthesized by Axolabs (Kulmbach, Germany). The product purity is > 90%. Full length SaKKH-CBE3 mRNA was transcribed by TriLink Biotech, fully substituted with 5-Methyl-C and Pseudo-U and capped.
Formulation of LNPs containing base editor mRNA and sgRNA
LNP were formulated as described previously26. In brief, LNP consisted of 1,2-distearoyl-sn-glycero-3-phosphocholine, cholesterol, a PEG-lipid, and an ionizable cationic lipid with branched tail structure as described in ‘US 2016/0376224 A1’. The lipid components in an ethanol solution were rapidly mixed with an aqueous solution (pH 4.0) containing SaKKH-CBE3 mRNA and gRNA (1:1 weight ratio) through an in-line mixer at a mRNA:lipid ratio as previously described26. The resulting LNP formulation was dialyzed overnight against 1 x PBS, 0.2 μm sterile-filtered, and stored at −80 °C at a concentration of 1 μg/μl total RNA. LNP had an average hydrodynamic diameter of 67-71 nm with a polydispersity index of 0.02-0.06 as determined by dynamic light scattering (Malvern NanoZS Zetasizer) and a mode size of 67-75 nm as determined by nanoparticle tracking analysis (Malvern Panalytical NanoSight NS300). Encapsulation efficiencies of SaKKH-CBE3 mRNA and sgRNA in the LNP were both at 96% measured by the Quant-iT Ribogreen Assay (Life Technologies).
Animal studies
Animal experiments were performed in accordance with protocols approved by the Kantonales Veterinäramt Zürich. Pahenu2 mice were housed in a pathogen-free animal facility at the Institute of Molecular Health Sciences at ETH Zurich and kept in a temperature-and humidity-controlled room on a 12 h light-dark cycle. Mice were fasted for 3-4 h before blood was collected from the tail vein for L-Phe determination but were otherwise fed a standard laboratory chow. Mice were genotyped at weaning. Wild type C57BL/6 mice were used as controls for physiological blood L-Phe levels. Homozygous Pahenu2 were injected with 1-6 mg/kg RNA. The control group was injected with 1 x PBS. Injection volumes were 120-150 μl.
Histology
Tissues were fixed using 4% Paraformaldehyde (PFA). Tissues were dehydrated with before paraffinization. Paraffin blocks were cut into 5-μm thick sections, deparaffinized with xylene, and rehydrated. Sections were HE-stained and examined for histopathological changes.
Cytokine analysis
Blood was collected 4 h after injection of either 1x PBS or 3 mg/kg LNPs and allowed to clot at room temperature for 1-2 h, centrifuged at 1000 × g for 10 minutes. Serum cytokine analysis was done with cytolab, an analytical service.
Microscopy
Mouse tissue was imaged using a Zeiss Apotome. Imaging conditions and intensity scales were matched for all images. Images were analysed using Fiji ImageJ software (v1.51n)37.
Amplification and high-throughput sequencing of genomic DNA samples
Genomic DNA from mouse tissues was isolated using the DNeasy Blood and Tissue kit or the RNeasy Mini Kit (Qiagen). Subsequent PCR reactions generated amplicons for HTS and were performed using NEB Next High-Fidelity 2x PCR Master Mix. In brief, 500 ng genomic DNA was amplified in 26 cycles for the first PCR in a 20 μl reaction. The PCR product was purified using Agencourt AMPure XP beads (Beckman Coulter), and amplified with primers containing sequencing adaptors. The products were gel purified and quantified using the Qubit 3.0 fluorometer with the dsDNA HS assay kit (Thermo Fisher Scientific). Samples were sequenced on an Illumina Miseq.
RT-qPCR for RNA fold change over time
Hepatocytes where then harvested at different time points and RNA isolated using the RNeasy (Qiagen) kit according to the manufacturer’s instructions. cDNA was generated using GoScript reverse transcriptase (Promega). qPCR to determine RNA fold-change over time was determined using a StepOnePlus system (Thermo Fisher)
HTS data analysis
Sequencing reads were demultiplexed using Miseq Reporter (Illumina), and analysed using a custom script. In short, reads were merged with PEAR v0.9.838 and mapped to the Ensembl mouse genome v38.90 using BWA MEM39. Base editing frequency was quantified in R using CrispRVariants v1.7.5540 and Biostrings v2.46.0641. Scripts are available at https://github.com/HLindsay/Villiger_deaminase.
Tissue cryosections
Mice were euthanized with CO2 and perfused through the inferior vena cava with PBS, followed by freshly prepared PLP buffer containing 75 mM L-Lysine (Sigma-Aldrich), 30.4 mM Na2HPO4, 7.1 mM NaH2PO4 (Sigma-Aldrich), NaIO4 (Sigma Aldrich) and 1% PFA. Tissues were isolated and fixed in PLP buffer overnight at 4°C, followed by 3 washing steps with buffer containing 81 mM Na2HPO4 and 19 mM NaH2PO4 at pH 7.4. Tissues were transferred to a 30% sucrose solution for 6 h at 4°C and embedded in OCT compound in cryomolds (Tissue-Tek). Frozen tissues were sectioned at 10 μm at -20 °C, and mounted directly on SuperFrost Plus slides (Thermo Fisher Scientific). Cryosections were counterstained with DAPI (Thermo Fisher Scientific) and mounted in Vectashied mounting medium (Vector Labs). Two frozen sections were analysed per mouse per tissue.
Unidirectional enrichment of target site for the detection of structural variants and large deletions
Genomic DNA from treated mice was sheared using Covaris E220 ultrasonicator to a fragment size of 350 bp. Ends were repaired and Illumina Y-adapters annealed using KAPA HTP Library Preparation kit (Kapa Biosystems). Unbiased detection of structural variants and large deletions was facilitated by unidirectional amplification using 3’ and 5’ Enrichment primers (5’-GGAGTTCAGACGTGTGCTCTTCCGATCTccgtcctgttgctggcttac or 5’-GGAGTTCAGACGTGTGCTCTTCCGATCTTGAGCATCCATTGTGGTTGG) combined with an adapter specific primer. Amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter). Sequencing was performed on a 300 cycle flow cell in paired end mode on an Illumina Miseq platform. Reads were aligned to the mouse reference genome (Mus_musculus, Ensembl, GRCm38.p5) using BWA-MEM39 (0.7.17). samblaster42 (0.1.24) was used to exclude PCR duplicates and to add Matetag (--excludeDups --addMateTags --maxSplitCount 2 --minNonOverlap 20). Discordant paired-end alignments and the split-read alignments were extracted and sorted using samtools43 (1.3.1). Structural variant analysis was performed using LUMPY44 (0.2.13) and DELLY45 (0.7.6).
Statistical analyses
A priori power calculations to determine sample sizes for animal experiments were performed using the R ‘pwr’ package. Statistical analyses were performed using GraphPad Prism 6.01 for Windows. Sample sizes and statistical tests are listed in corresponding figure legends. In brief, the Dunnett’s test was used to compare multiple variables to a single control for blood L-Phe levels. Group averages are presented as mean ± s.d.
RNA-seq experiments and data analysis
RNA library preparation was performed using the TruSeq Stranded Total RNA kit (Illumina) with ribosomal RNA (rRNA) deletion. RNA-seq libraries were sequenced on an Illumina NovaSeq machine at the Functional Genomics Center in Zurich (FGCZ), achieving an average of 185 Million paired-end (PE) reads per library.
Quality control, pre-processing, alignment of RNA-seq reads
Quality of Illumina PE RNA-seq reads was evaluated using FastQC v0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Using FastqScreen version 0.11.1 (https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/), potential sample contaminations (genomic DNA, rRNA, Mycoplasma etc) were screened against a custom database including UniVec (https://www.ncbi.nlm.nih.gov/tools/vecscreen/univec/), refseq mRNA sequences, selected genome sequences (human, mouse, arabidopsis, bacteria, virus, phix, lambda, mycoplasma) (https://www.ncbi.nlm.nih.gov/refseq/), and SILVA rRNA sequences (https://www.arb-silva.de/). Illumina PE reads were pre-processed using Trimmomatic version 0.36 to trim off sequencing adaptors and low quality ends (average quality lower than 20 within a 4 nt window). Flexbar version 3.0.3 was used to remove the first 6 bases of each read, which showed priming bias introduced by the library preparation protocol46. PE RNA-seq reads were generated with different read length (2X51 and 2X151). After adapter and quality trimming, if the read length was longer than 50 nt, only the first 50 nt were kept for downstream STAR mapping and variant calling. Quality controlled reads (average quality 20 and above, read length 20 and above) were aligned to the reference genomes (mouse reference genome: GRCm38.p5, Ensembl release 91; human reference genome: GRCh38.p10, Ensembl release 91) using STAR version 2.7.0e with 2-passes mode. PCR-duplicates were marked using Picard version 2.9.0. Read alignments were comprehensively evaluated in terms of different aspects of RNA-seq experiments, such as sequence quality, gDNA and rRNA contamination, GC/PCR/sequence bias, sequencing depth, strand specificity, coverage uniformity and read distribution over the genome annotation, using R scripts in ezRun (https://github.com/uzh/ezRun/) developed at FGCZ.
RNA sequence variant calling and filtering
Variant calling from RNA-seq reads was performed according to GATK Best Practices (https://gatkforums.broadinstitute.org/gatk/discussion/3891/calling-variants-in-rnaseq). In details, GATK (version 4.1.2.0) tool SplitNCigarReads was applied to post-processed the read alignments. Afterwards variants were called using HaplotypeCaller (GATK version 4.1.2.0) on PCR-deduplicated, post-processed aligned reads. Variant loci in base-editor overexpression experiments were filtered to exclude sites without high-confidence reference genotype calls in the control experiment. For a given SNV, the read coverage in the control experiment should be >90th percentile of the read coverage across all SNVs in the corresponding overexpression experiment. Only loci having at least 99% of reads containing the reference allele in the control experiment were kept.
Quantification of gene expression
Transcript expression was calculated using kallisto (version 0.44.0).
Primary Hepatocyte Isolation and clonal expansion
Primary hepatocytes were isolated using a two-step perfusion method. Briefly, after pre-perfusion with HANKS’s Buffer (HBSS, 0.5 mM EDTA, 25 mM HEPES) via inserting the cannula through the superior vena cava and cutting the portal vein, the liver was perfused at low speed for approximately 10 min with Digestion Buffer (low Glucose DMEM, 1mM HEPES) with freshly added 32 μg/mL Liberase TM (Roche). The digestion was stopped using Isolation Buffer (low glucose DMEM, 10% FBS) and cells were separated from the matrix by gently pushing with a cell scraper. The cell suspension was filtered through a 100 μm filter before 2x low speed centrifugation at 50 x g for 2 min. Hepatocytes were expanded as chemically induced liver progenitors (CLiPs)18. First, cells were plated at low density (450 – 900 cells / ml) on Matrigel coated plates (16 ul Matrigel, Corning Life Sciences, per ml SHM medium). Full SHM Medium (SHM with freshly added YAC) was changed every other day until colonies were big enough to be picked. For picking, the plate was incubated shortly with TryplE (Thermo Fisher Scientific) until the colony edges started to detach. TryplE was inhibited by adding medium and colonies were picked into Matrigel coated 96-well plates. Picked clones were expanded and upon confluency divided for on-target sequencing and further expansion. On-target sequencing was performed upon direct lysis of the cell pellet and PCR amplification for 29 cycles on the diluted lysate using GoTaq G2 HotStart Green Master Mix (Promega). PCR amplification was performed using the following forward (FW) and reverse (Rev) primers: FW: cgacatccctcagtaatgcca, Rev: gcagtggatcatggggacca. Upon confirmation of a unique amplicon, the PCR product was purified using Ampure XP beads and then sequenced using an in sequence primer: acatgacccaaagcagtagg using the Sanger method (Microsynth).
Whole genome sequencing and data analysis
Upon confirmation of on-target editing, DNA was harvested using QIAamp DNA Mini Kit (Qiagen) or Quick DNA Microprep kit (Zymo Research) according to the manufacturer’s instructions. DNA concentrations were determined using Qubit dsDNA HS kit (Invitrogen). WGS was performed at a mean coverage of 30x using an Illumina Novaseq.
Read alignment, variant calling and variant filtering
Sequence reads were mapped against mouse reference genome GRCm38 by using Burrows-Wheeler Aligner v0.7.5 mapping tool39 with settings ‘bwa mem -c 100 -M’. Sequence reads were marked for duplicates by using Sambamba v0.4.732 and realigned per donor by using Genome Analysis Toolkit (GATK) IndelRealigner v2.7.2. Raw variants were multisample-called by using the GATK HaplotypeCaller v3.4-4647 and GATK-Queue v3.4-46 with default settings and additional option ‘EMIT_ALL_CONFIDENT_SITES’. The quality of variant and reference positions was evaluated by using GATK VariantFiltration v3.4-46 with options ‘-snpFilterName LowQualityDepth -snpFilterExpression “QD < 2.0” -snpFilterName MappingQuality -snpFilterExpression “MQ < 40.0” -snpFilterName StrandBias -snpFilterExpression “FS > 60.0” -snpFilterName HaplotypeScoreHigh -snpFilterExpression “HaplotypeScore > 13.0” -snpFilterName MQRankSumLow -snpFilterExpression “MQRankSum < −12.5” -snpFilterName ReadPosRankSumLow-snpFilterExpression “ReadPosRankSum < −8.0” -cluster 3 -window 35’. Full pipeline description and settings also available at: https://github.com/UMCUGenetics/IAP.
To obtain high-quality somatic mutation catalogs, we applied postprocessing filters as described20. Briefly, we considered variants at autosomal chromosomes without any evidence from a paired control sample (day 0 isolation for mouse 324 and 329; gDNA isolated from tail for mouse 341, 344 and the control); passed by VariantFiltration with a GATK phred-scaled quality score ≥100 for base substitutions and ≥250 for indels; a base coverage of at least 20X in the clonal and paired control sample; mapping quality (MQ) of ≥60; no overlap with single nucleotide polymorphisms (SNPs) in the Single Nucleotide Polymorphism Database v142. We additionally filtered base substitutions with a GATK genotype score (GQ) lower than 99 or 10 in clonal or paired control sample, respectively. For indels, we filtered variants with a GQ score lower than 99 in both clonal and paired control sample and filtered indels that were present within 100 bp of a called variant in the control sample. In addition, for both SNVs and INDELs, we only considered variants with a variant allele frequency of 0.2 or higher in the clones to exclude in vitro accumulated mutations19,20. The scripts are available at https://github.com/ToolsVanBox/SNVFI and https://github.com/ToolsVanBox/INDELFI.
Due to the karyotypically unstable nature of the cells and for the fair comparison of the number of mutations in the later analysis, only the mutations from the regions considered as diploid (1.5 < ratio < 2.5 from the Control-FREEC48 output when the samples were treated as diploid) and callable were included. The absolute number of mutations were corrected for the lengths of the accounted genomic regions.
Mutational profile and signature analysis
The number of 6-substitution types (C>A, C>G, C>T, T>A, T>C and T>G) or 96-trinucleotide mutation types (6 substitution types with 5’-and 3’-flanking bases) were reported and the frequencies of the 96-trinucleotide mutations were plotted for every mouse using an in-house developed R package49. For the normalized absolute number and relative amount of 6-substitution types, the samples were classified based on the injected chemicals; for each group, the mean and the standard deviation were calculated and plotted.
To illustrate the potential APOBEC activity in the samples, COSMIC signature 2 and 13 were selected as a positive control since these signatures have been associated with APOBEC activity36,50. The 96-trinuclueotide frequencies were pooled from the two signatures and normalized so that the frequencies add up to 1. As the C>N substitutions characterizes the APOBEC signature, the contributions of the C>N substitutions were selected for the visualization.
The 96-nt rat APOBEC signature was deduced based on the experimentally determined C>T frequencies10 under the assumption that other substitutions do not contribute to the rat APOBEC signature. For the three control mouse samples, the 96-nt mutational profile was constructed and normalized by the total number of SNVs and multiplied by the median number of SNVs (428 SNVs) to make them comparable between the samples. To mimic the APOBEC activity on the mutational profile, the additional number of SNVs were distributed over the 96-nt mutational patterns according to the determined rat APOBEC signature, for 10, 25, 50 and 100 SNVs. Any decimal values were rounded, and summed to the profiles of the controls. Here, addition of every 10 APOBEC SNV’s were visualized using MutationalPatterns package49 in R. For all the samples and the APOBEC-signature-added controls, cosine similarity with the human SBS236 or rat APOBEC signature was calculated using MutationalPatterns49 in R.
To calculate the variant detection sensitivity of our method, we identified germline variants and counted how many of them were found in the clones. In order to exclude potential artefacts in our data, the direct output from the IAP pipeline were further filtered with the following criteria: located in diploid and CALLABLE region, passed by VariantFiltration with a GATK phred-scaled quality score ≥100, GATK genotype score (GQ) equals 99, base coverage of at least 20X in all the clones and the bulk samples, not overlaps with the variants in our blacklists (available upon request), and present as a heterozygous variant (VAF ≥ 0.3) in the three bulk samples. Our filtering resulted with 86 heterozygous variants, and any position with < VAF 0.3 were counted as absent in the clones.
For the determination of non-synonymous to synonymous (dN/dS ratio) mutations for all the detected single base substitutions, the position of the mutations, reference base and alternative base was extracted from the VCF files for AAV-treated and LNP treated mice separately. The global maximum-likelihood estimates and the confidence intervals for both mice group were calculated using dNdScv package51 and plotted using ggplot252 in R.
Fluorescence-activated cell sorting and RNA extraction
Cells were incubated for 72 h post-transfection. Before sorting, cells were washed with phosphate-buffered saline (PBS) and incubated with TrypLE (Gibco) until they detached. After two washing steps with PBS, cells were resuspended in FACS Buffer (PBS with 2% FBS and 2 mM EDTA) and filtered through 35 μm nylon mesh cell strainer snap caps (Stemcell technologies). Flow cytometry was done on a FACSAria III sorter (BD Biosciences) using FACSDiva version 8.0.1 (BD Biosciences). Gating is shown in Suppl. Fig. 29, 30. RNA from sorted cells was extracted using Qiagen RNA Quick and easy kit according to the manufacturer’s instructions.
Supplementary Material
Acknowledgements
We thank S. Iyer and JK. Joung for sharing technical details about variant filtering for transcriptome off target identification, H. Vatandaslar, S. Pantasis for their support with primary hepatocyte isolations and microscopy analysis, the members of the FGCZ for whole genome and RNA sequencing, H.M. Grisch-Chan for her help with L-Phe analysis, M. Tanner and N. Rimann for their support with animal work. D. Lenggenhager for the evaluation of histological samples for pathologies. This work was supported by the Swiss National Science Foundation grant #310030_185293 (to GS), the Swiss National Science Foundation Sinergia grant #180257 (to BT), the Swiss National Science Foundation grant #205 321_169 612 (to JH), the Novartis Foundation for Medical-Biological Research #19A004 (to DW), and by the PHRT grant #528 (to GS, MS, BT).
Footnotes
Author contributions
L.V. and T.R. designed the study, performed experiments, analysed data, and wrote the manuscript. D.W. contributed to the design of the study, and edited the manuscript. P.J.C.L. and Y.K.T. developed, prepared, and characterized lipid nanoparticles, contributed to the design of the study, and edited the manuscript. M.B.B. assisted with LNP formulation. C.B. and J.H. assisted with the design of sgRNA modifications. F.R. conducted in vitro transfection experiments. S.J. conducted HTS data analysis. W.Q. and H.R. analysed RNA-seq data. R.O. and R.v.B. performed WGS analysis. M.S. and B.T. provided reagents and conceptual advice. G.S. designed and supervised the research and wrote the manuscript. All authors approved the final version.
Competing interests
P.J.C.L., M.B.B. and Y.K.T. are employees of Acuitas Therapeutics.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-01X-XXXX-X.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes form the corresponding authors on reasonable request. High-throughput data is publicly available (accession number for the GEO datasets for HTS and RNA-seq: GSE148349; accession number for the SRA dataset for WGS: PRJEB38134).
Code availability
The scripts used to quantify on-target and off-target editing are available at https://github.com/HLindsay/Villiger_deaminase.
References
- 1.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533 doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaudelli NM, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chadwick AC, Wang X, Musunuru K. In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arterioscler Thromb Vasc Biol. 2017 doi: 10.1161/ATVBAHA.117.309881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu S-M, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 5.Villiger L, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 6.Grünewald J, et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019 doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grünewald J, Zhou R, Iyer S, Lareau CA, Garcia SP. CRISPR adenine and cytosine base editors with reduced RNA off-target activities. 2019:1–25. doi: 10.1038/s41587-019-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rees HA, Wilson C, Doman JL, Liu DR. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci Adv. 2019 doi: 10.1126/sciadv.aax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou C, et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019 doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 10.Zuo E, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science (80-) 2019;126:21. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGrath E, et al. Targeting specificity of APOBEC-based cytosine base editor in human iPSCs determined by whole genome sequencing. Nat Commun. 2019;10:1–9. doi: 10.1038/s41467-019-13342-8. 2019 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamanaka S, Poksay KS, Driscoll DM, Innerarity TL. Hyperediting of multiple cytidines of apolipoprotein B mRNA by APOBEC-1 requires auxiliary protein(s) but not a mooring sequence motif. J Biol Chem. 1996;271:11506–11510. doi: 10.1074/jbc.271.19.11506. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol. 2011;18:230–238. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sowden M, Hamm JK, Smith HC. Overexpression of APOBEC-1 results in mooring sequence-dependent promiscuous RNA editing. J Biol Chem. 1996;271:3011–3017. doi: 10.1074/jbc.271.6.3011. [DOI] [PubMed] [Google Scholar]
- 15.Yamanaka S, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 17.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012 doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 18.Katsuda T, et al. Conversion of Terminally Committed Hepatocytes to Culturable Bipotent Progenitor Cells with Regenerative Capacity. 2017 doi: 10.1016/j.stem.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Jager M, et al. Measuring mutation accumulation in single human adult stem cells by whole-genome sequencing of organoid cultures. 13:59–78. doi: 10.1038/nprot.2017.111. [DOI] [PubMed] [Google Scholar]
- 20.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. 2016 doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komor AC, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017 doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nathwani AC, et al. Adenovirus-Associated Virus Vector–Mediated Gene Transfer in Hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn JD, et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22 doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Yin H, et al. structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimasu H, et al. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway A, et al. Non-viral Delivery of Zinc Finger Nuclease mRNA Enables Highly Efficient In Vivo Genome Editing of Multiple Therapeutic Gene Targets. Mol Ther. 2019;27:866–877. doi: 10.1016/j.ymthe.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell JJ, Trakadis YJ, Scriver CR. Phenylalanine hydroxylase deficiency. Genet Med. 2011;13:697–707. doi: 10.1097/GIM.0b013e3182141b48. [DOI] [PubMed] [Google Scholar]
- 28.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mui BL, et al. Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of siRNA Lipid Nanoparticles. Mol Ther - Nucleic Acids. 2013;2:1–8. doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016 doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song C-Q, et al. Adenine base editing in an adult mouse model of tyrosinaemia. 2019 doi: 10.1038/s41551-019-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okano Y, et al. Molecular Basis of Phenotypic Heterogeneity in Phenylketonuria. N Engl J Med. 1991;324:1232–1238. doi: 10.1056/NEJM199105023241802. [DOI] [PubMed] [Google Scholar]
- 33.Nelson CE, et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kay MA. Nature Reviews Genetics. Vol. 12. Nature Publishing Group; 2011. State-of-the-art gene-based therapies: The road ahead; pp. 316–328. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser J. How safe is a popular gene therapy vector? Science. 2020 doi: 10.1126/science.367.6474.131. [DOI] [PubMed] [Google Scholar]
- 36.Yu Y, et al. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-15887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YB, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grünewald J, et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nature Biotechnology. 2019 doi: 10.1038/s41587-019-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doman JL, Raguram A, Newby GA, Liu DR. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol. 2020:1–9. doi: 10.1038/s41587-020-0414-6. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013;00:1–3. [Google Scholar]
- 44.Lindsay H, et al. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat Biotechnol. 2016;34:701–702. doi: 10.1038/nbt.3628. [DOI] [PubMed] [Google Scholar]
- 45.Pagès H, Aboyoun P, G R, D S. Biostrings: Efficient manipulation of biological strings [Google Scholar]
- 46.Faust GG, Hall IM. SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, et al. The Sequence Alignment / Map (SAM) Format and SAMtools 1000 Genome Project Data Processing Subgroup. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014 doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rausch T, et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. 2010;38:1–7. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Depristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–501. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boeva V, et al. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: Comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:1–11. doi: 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petljak M, et al. Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell. 2019;176:1282–1294.e20. doi: 10.1016/j.cell.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017 doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wickham H. ggplot2: elegant graphics for data analysis. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2016 doi: 10.1007/978-3-319-24277-4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes form the corresponding authors on reasonable request. High-throughput data is publicly available (accession number for the GEO datasets for HTS and RNA-seq: GSE148349; accession number for the SRA dataset for WGS: PRJEB38134).
The scripts used to quantify on-target and off-target editing are available at https://github.com/HLindsay/Villiger_deaminase.