Abstract
Horizontal gene transfer permits rapid dissemination of genetic elements between individuals in bacterial populations. Transmitted DNA sequences may encode favourable traits. However, if acquired DNA has an atypical base composition, it can reduce host fitness. Consequently, bacteria have evolved strategies to minimise the harmful effects of foreign genes. Most notably, xenogeneic silencing proteins bind incoming DNA that has a higher AT-content than the host genome. An enduring question has been to understand why such sequences are deleterious. Here, we show that the toxicity of AT-rich DNA in Escherichia coli frequently results from constitutive transcription initiation within the coding regions of genes. Left unchecked, this causes titration of RNA polymerase and a global downshift in host gene expression. Accordingly, a mutation in RNA polymerase that diminishes the impact of AT-rich DNA on host fitness, reduces transcription from constitutive, but not activator-dependent, promoters.
Introduction
Bacteria obtain DNA from their environment by direct uptake (transformation), the action of viruses (transduction), or the acquisition of transmissible plasmids (conjugation)1. Thus, “horizontal” DNA transfer allows phenotypes to spread through bacterial populations. Transferred traits can be beneficial1. However, acquired sequences with a high AT-content may reduce host fitness2-6. Bacteria have developed mechanisms to diminish the toxicity of AT-rich genes. One approach involves a family of xenogeneic DNA binding proteins7,8. These proteins target DNA that has a higher AT-content than the host genome (“AT-rich” DNA), and silence transcription9–12. However, it is unclear why AT-rich DNA is toxic and why transcriptional silencing is beneficial.
The Histone-like Nucleoid Structuring (H-NS) protein of γ-proteobacteria is the best-characterised xenogeneic silencing protein. To reduce transcription, H-NS oligomerises across AT-rich genes. This process is triggered by nucleation at sequences containing a T:A step14–19. Intriguingly, promoter -10 hexamers (consensus 5’-TATAAT-3’, recognised by the RNA polymerase σ70 subunit) are excellent inducers of H-NS nucleation15,20. Hence, H-NS can exclude RNA polymerase from, or trap RNA polymerase at, promoter DNA8,9,21,22.
A challenge has been to understand why horizontally acquired AT-rich DNA is toxic. As we noted previously, such regions contain a disproportionally high number of promoter-like sequences23–26. Furthermore, we showed that H-NS suppresses transcription primarily from promoters that are intragenic and/or far from gene starts25. Here, we test the hypothesis that transcription from intragenic promoters is a major cause of toxicity in cells lacking H-NS. Working with a sub-set of AT-rich genes, we demonstrate dramatic effects of H-NS on intragenic transcription. When this transcription is disabled, associated fitness costs decrease. Genome-wide, if derepressed, intragenic transcription sequesters RNA polymerase. This causes a global downshift in canonical gene expression. Accordingly, a mutation in RNA polymerase that reduces transcription from constitutive promoters, but not those that are activator-dependent, compensates for loss of H-NS.
Results
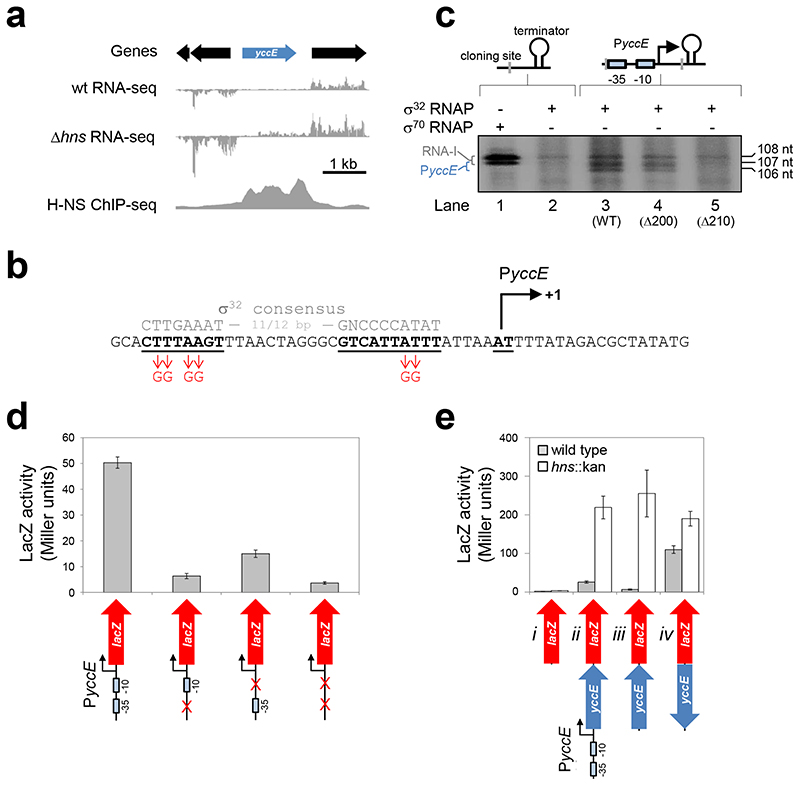
Identification of the canonical yccE promoter
We chose yccE as a model gene to understand xenogeneic silencing. We selected yccE because it is AT-rich (66% A/T), silenced and bound by H-NS27, and contains at least two intragenic promoters25,28. Importantly, yccE can be studied in isolation because the adjacent genes are not H-NS bound (Figure 1a). We sought to understand the source of yccE transcription in cells lacking H-NS. As a starting point, we identified the canonical yccE promoter. We were aided by previous observations that yccE is a target for σ32 (heat shock σ factor)29. Segments of the yccE intergenic region were fused to lacZ in reporter plasmid pRW50 (Figure S1a). Promoter activity located to a 55 base pair (bp) DNA fragment immediately upstream of yccE (Figure S1b). Figure 1b shows the sequence of this DNA fragment, named yccEΔ200. A σ32-dependent promoter sequence is apparent. We monitored transcripts produced from this promoter (PyccE) in vitro. To do this, fragments from the yccE intergenic region were cloned upstream of the λoop terminator in plasmid pSR. Transcripts initiating at PyccE, and terminating at λoop, should be ~106 nucleotides (nt) in length. Coincidentally, the RNA-I transcript, derived from the pSR replication origin, is 108/107 nt long. To avoid confusing transcripts, we first monitored RNAs produced using pSR without a PyccE insert. The data show that σ32 dependent synthesis of the RNA-I transcript is inefficient (Figure 1c, compare lanes 1 and 2). When the entire yccE intergenic region was cloned in pSR σ32 dependent yccE transcripts were identified (Figure 1c, lane 3). The same transcripts were observed upon cloning the truncated yccEA200 fragment (Figure 1c, lane 4). A 10 bp truncation at the 5’ end of yccEA200 abolished transcription (Figure 1c, lane 5). To confirm correct identification of PyccE, DNA recognition elements for σ32 were mutated (detailed in Figure 1b). All mutations greatly reduced promoter activity (Figure 1d).
Figure 1. Characterisation of the yccE locus.
a) Genomic context of yccE and its promoter. The panel shows yccE, and surrounding genes, alongside data describing H-NS binding (ChIP-seq27) and RNA abundance (standard RNA-seq; this work, done in duplicate). Data are representative.
b) Sequence of the yccEΔ200 DNA fragment containing PyccE. The PyccE -10 and -35 elements are in bold and underlined. A consensus σ32 promoter sequence is in grey for comparison. Mutations made to disrupt sequence elements are in red. Transcription can initiate at adjacent nucleotides. These are labelled (+1) and are highlighted by a bent arrow.
c) Analysis of transcripts generated from PyccE in vitro. The gel image shows transcripts generated by RNA polymerase, associated with either σ70 or σ32, from pSR plasmid DNA templates. The schematic diagrams above the gel image represent the native cloning site of pSR (left hand side) or derivatives containing a PyccE insert (right hand side). The different PyccE containing DNA fragments inserted are indicated below the gel image in parenthesis. PyccE derived transcripts manifest as a 107/106 nucleotide (nt) doublet. The 108/107 nt RNA-I transcript is derived from the plasmid replication origin. The experiment was done three times. Data are representative.
d) Effect of PyccE mutation in vivo. The graph shows LacZ activity data obtained from E. coli JCB387 cells carrying different yccEΔ200 derivatives cloned in pRW50. Assays were done in triplicate and error bars show standard deviation from the mean.
e) Induction of yccE transcription in the absence of H-NS does not require PyccE in vivo. The panel illustrates a series of yccE::lacZ fusions labelled i-iv. Genes are shown as block arrows and PyccE is shown as a bent line arrow. The ß-galactosidase activity was measured in lysates of M182 (grey bars) and M182hns::kan cells (open bars). Assays were done in triplicate and error bars show standard deviation from the mean.
Increased transcription of yccE in Δhns cells does not require PyccE
Convention dictates that PyccE should cause increased yccE transcription in cells lacking H-NS. We considered this unlikely given the low activity, and σ32 dependence, of PyccE. Hence, we generated a series of yccE::lacZ fusions to investigate the contribution of PyccE. The different constructs, labelled i through iv, are illustrated below the graph in Figure 1e. We first measured lacZ expression in wildtype cells (Figure 1e, grey bars). When yccE was in the forward orientation, low-level lacZ expression was apparent. This was abolished upon deletion of PyccE (compare constructs ii and iii). In the reverse orientation, yccE stimulated higher lacZ expression (construct iv). This is consistent with our previous identification of a strong antisense promoter at the 5’ end of yccE (adjacent to lacZ in this assay)28. Remarkably, in the absence of H-NS, lacZ expression increased in all scenarios (Figure 1e, white bars). This transcription must be due to intragenic promoters.
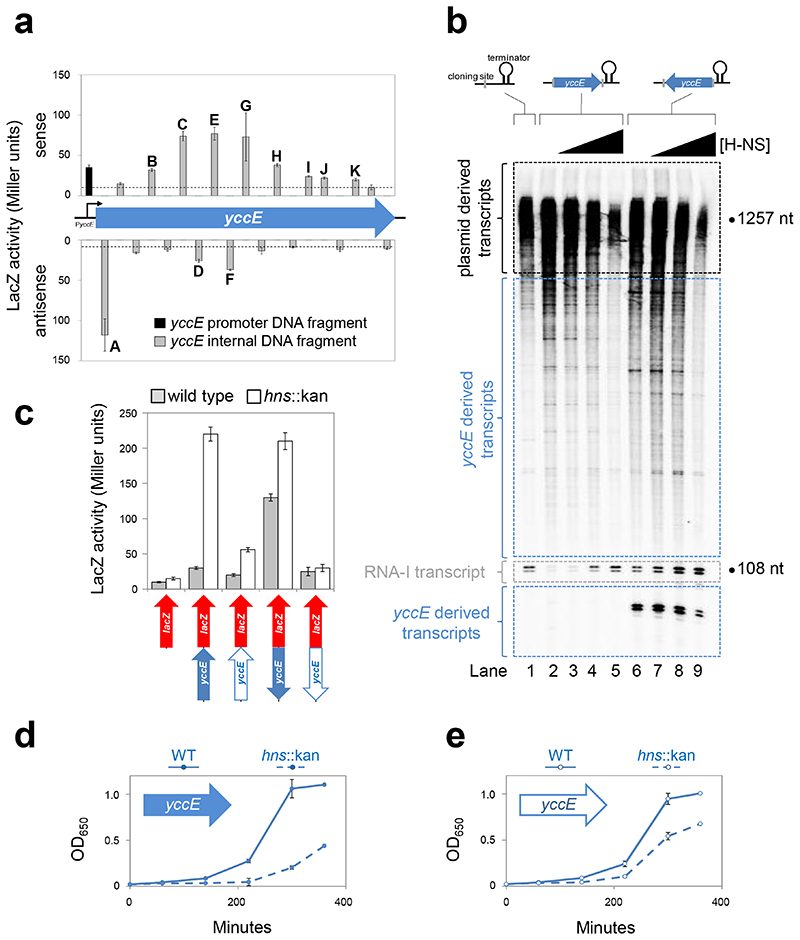
The yccE coding sequence is enriched for promoters
A search identified 21 possible promoters within yccE. To test for function, we isolated promoters on 56 bp DNA fragments and fused them to lacZ (Figure 2a). Note that such sequences are too short to be subject to repression by H-NS25,30. Eleven DNA fragments stimulated β-galactosidase expression twofold or more above background (labelled A-K in Figure 2a). Under the conditions of our experiment, 7 of the 11 intragenic promoters were more efficient at driving transcription than PyccE (Figure 2a, black bar). Note that fragment “A” contains the strong antisense promoter described previously28.
Figure 2. H-NS represses intragenic yccE transcription and associated fitness costs.
a) Identification of intragenic yccE promoters. The data are β-galactosidase activities driven by short intragenic yccE DNA fragments in strain JCB387. The bars align with the location of the DNA fragment relative to yccE and show sense (upper) and antisense (lower) transcription. Bars labelled “a”-“k” have at least 2-fold over background activity (empty pRW50; shown by dashed line). The black bar represents the canonical promoter PyccE. Note that, two DNA fragments resisted cloning. Hence, 19 of the 21 potential promoters were tested. Assays were done in triplicate. Error bars show standard deviation from the mean.
b) Transcription can initiate at multiple sites within yccE in vitro. Gel image showing RNA generated in vitro separated by denaturing gel electrophoresis. DNA templates, with the yccE gene cloned upstream of the λoop terminator in plasmid pSR, are illustrated above the gel. Transcripts generated by RNA polymerase (400 nM) with empty pSR plasmid (lane 1) are highlighted by a black dashed line. Transcripts initiating within yccE in the forward (lanes 2-5) or reverse (lanes 6-9) orientation are highlighted by a blue dashed line. The control RNA-I transcripts are highlighted by a grey dashed line. H-NS was added at concentrations of 0.8, 1.5, or 3.0 μM. The experiment, done three times, is representative.
c) Mutation of promoters within yccE prevents induction in Δhns cells. The lower illustrations show yccE (blue arrows) cloned upstream of lacZ (red arrow). A solid blue arrow represents wild type yccE whereas open arrows indicate yccE with mutated intragenic promoter -10 elements. For each lacZ fusion, β-galactosidase activity was measured in lysates of M182 (grey bars) and M182hns::kan cells (open bars). Assays were done in triplicate and error bars show standard deviation from the mean.
d,e) The fitness cost of yccE is reduced when intragenic promoters are mutated. The figure illustrates changes in culture OD650 following inoculation of LB medium. The inoculum was M182 (solid line) or M182hns::kan (dashed line) transformed with the pSR plasmid carrying d) wild type yccE or e) yccE with internal promoter -10 elements mutated. Cells were grown at 37°C. The experiment was done in triplicate. Error bars show standard deviation from the mean.
Transcription initiation within yccE is repressed by H-NS in vitro
In the context of the full yccE gene, intragenic promoters should be repressed by H-NS. We tested this in vitro using the pSR system. As noted above, with empty pSR, the 108/107 nt RNA-I transcript is produced. In addition, larger transcripts are generated from genes native to the plasmid. The complete set of transcripts produced from pSR is shown in Lane 1 of Figure 2b. In the figure, large (>1000 nt) pSR derived transcripts are highlighted by a black box and RNA-I is highlighted by a grey box. We introduced yccE into pSR, in either the forward or reverse orientation, upstream of the λoop terminator. Since yccE is 1257 bp in length, most intragenic transcripts should be separable from pSR derived transcripts. As expected, numerous transcripts between 100 and 1000 nt in length were detected for both yccE containing plasmids (compare Lanes 1, 2 and 6 in Figure 2b). For the plasmid containing yccE in the reverse orientation an additional small transcript was detected (highlighted by lower blue box in Figure 2b). This was expected, the transcript generated from the strong antisense promoter at the 5’ end of yccE, has a size of 90 nt in this assay. Regardless of yccE orientation, addition of H-NS inhibited synthesis of most yccE derived transcripts (Lanes 3-5 and 6-9). Synthesis of the RNA-I transcript was enhanced by H-NS suggesting that RNA polymerase is titrated by promoters within yccE.
Intragenic promoters are the source of increased yccE transcription in cells lacking H-NS
To confirm that H-NS repressed intragenic yccE promoters in vivo, we made derivatives of our yccE::lacZ fusions. The derivatives carry mutations in intragenic -10 elements. Schematics are below the graph in Figure 2c. The open arrows show yccE lacking internal promoters. Full details are in Figure S2. In the absence of H-NS, the ability of mutated yccE alleles to stimulate lacZ expression was reduced (Figure 2c). Thus, both biochemical and genetic inspection show silencing of intragenic transcription by H-NS. We refer to this as “pseudo-regulation” that occurs independently of, and may be mistaken for, the control of mRNA synthesis. Thus, supposed gene regulatory effects of H-NS may often be due to intragenic promoters.
The fitness cost of yccE is a consequence of intragenic transcription
We predicted a link between intragenic transcription and reduced fitness associated with loss of H-NS. Hence, E. coli M182, and the hns::kan variant, were transformed with pSR carrying yccE with or without internal promoters. Importantly, yccE mRNA cannot be expressed in this scenario; no upstream promoter is present. We monitored cultures inoculated with these strains (Figure 2d,e). In all cases, cells lacking hns had reduced fitness compared to the parent. However, this fitness defect was smaller for the yccE derivative lacking internal promoters (Figure 2e). Complete elimination of the fitness defect was not expected; AT-rich genes present on the E. coli chromosome also contribute.
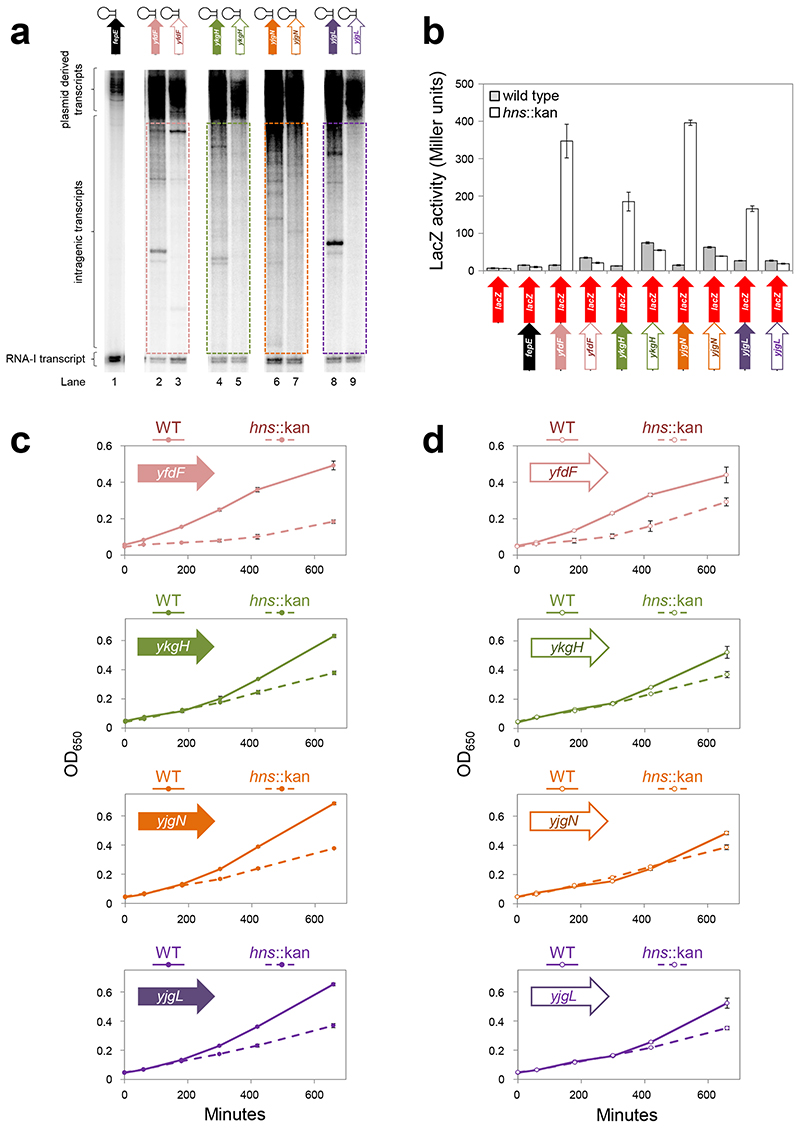
Repression of intragenic transcription by H-NS reduces the fitness cost of many AT-rich genes
We identified other solitary genes targeted by H-NS: yfdF, ykgH, yjgN and yjgL. We cloned these, and derivatives lacking intragenic promoters, upstream of the λoop terminator in pSR. The fepE gene, which has an AT-content of 55%, was included as a control. The constructs are illustrated above the gel image in Figure 3a. Full gene sequences are in Figure S3. Figure 3a shows results of in vitro transcription experiments. Whilst RNA polymerase did not initiate transcription within fepE (Lane 1), intragenic transcription was observed for yfdF, ykgH, yjgN and yjgL (Lanes 2, 4, 6 and 8). Mutation of intragenic promoters reduced this transcription (Lanes 3, 5, 7 and 9). The coding regions described above were also cloned upstream of lacZ in pRW50 (Figure 3b). Expression of lacZ was measured in M182 or the hns::kan derivative (Figure 3b). Upon deletion of hns, expression of lacZ downstream of yfdF, ykgH, yjgN or yjgL, but not fepE, increased (solid arrows). In contrast, no such increase occurred when intragenic promoters were mutated (open arrows). These analyses are consistent with silencing of intragenic promoters by H-NS at all loci tested. Hence, we measured the fitness cost of multicopy yfdF, ykgH, yjgN and yjgL in cells with and without H-NS. In all cases, the fitness deficit between wild-type and hns::kan cells decreased upon mutation of intragenic promoters (Figure 3c,d). Furthermore, changes in fitness and lacZ expression were significantly correlated (Figure S4a).
Figure 3. H-NS represses intragenic transcription and associated fitness costs at many loci.
a) Transcription initiation within the coding regions of H-NS target genes in vitro. An in vitro transcription assay using different AT-rich genes, cloned upstream of the λoop terminator in plasmid pSR, as a template. The different DNA constructs are illustrated above the gel. The cloned genes have an AT-content of 65% (yfdF), 63% (ykgH), 63% (yjgN) and 68% (yjgL). For each cloned gene a solid arrow represents the wild type DNA sequence whereas an open arrow is a derivative where intragenic promoter -10 elements have point mutations. Note that the fepA gene was used as a control and has an AT-content of 55%. The positions of transcripts generated by RNA polymerase (400 nM) from the fepE control (lane 1) or the other cloned genes (lanes 2-9) are labelled. The in vitro transcription assays were run on three separate occasions. Data are representative.
b) Increased transcription in cells lacking H-NS frequently requires intragenic promoters in vivo. The panel illustrates a series of DNA constructs where different gene coding regions have been cloned upstream of lacZ (red arrow). For each cloned gene a solid arrow represents the wild type DNA sequence whereas an open arrow is a derivative where intragenic promoter -10 elements have point mutations. The cloned genes have an AT-content of 65% (yfdF), 63% (ykgH), 63% (yjgN) and 68% (yjgL). Note that the fepA gene is used as a control and has an AT-content of 55%. For each lacZ fusion β-galactosidase activity was measured in lysates of M182 (grey bars) and M182Δhns (open bars) cells. Assays were done in triplicate and error bars show standard deviation from the mean.
c,d) The toxicity of many AT-rich genes is a consequence of spurious intragenic transcription. The figure illustrates growth of M182 (solid line) or M182hns::kan (dashed line) cells transformed with the pSR plasmid carrying different AT-rich genes. Panel c) shows wild type gene derivatives and
d) shows derivatives with internal promoter -10 elements mutated (open arrows). Experiments were done using M9 minimal media at 30°C. The experiment was done in triplicate and error bars show standard deviation from the mean.
AT-rich genes titrate RNA polymerase and cause a global downshift in housekeeping transcription
RNA polymerase levels are limited in E. coli 31. Consequently, multiple copies of a strong promoter can hinder growth and titrate transcription of the lac operon (Figure S4b). Interestingly, RNA polymerase levels do not increase in cells lacking H-NS (Figure S5). Therefore, competition for the enzyme must increase; more promoters compete for a limited supply of RNA polymerase. Logically, migration of RNA polymerase to spurious promoters should cause a global downshift in canonical transcription. However, despite many studies of the H-NS controlled transcriptome, a universal downshift has never been reported. We reasoned that this might result from data normalisation approaches used previously. Briefly, transcriptome analysis compares RNA levels in two strains. Comparison requires a point of reference believed to be consistent between the strains. For example, it may be assumed that housekeeping genes are similarly transcribed or that averaged transcription across all genes will be equivalent. Problematically, these approaches cannot differentiate between technical variation and genuine shifts in global transcription. To circumvent this problem, spiked-in RNA standards can be used34. We adapted this tactic to quantify global effects of H-NS on transcription. Briefly, we grew E. coli MG1655, and the hns::kan variant, to mid-log phase. We then counted colony-forming units for each E. coli culture. The same was done for a single culture of Salmonella Typhimurium. Accordingly, we were able to mix a defined number of clonal S. Typhimurium cells with each E. coli strain. The cell mixtures were subject to transcriptome analysis. For identically grown S. Typhimurium cells, differences in RNA abundance must result from processing variation. Hence, at the end of the procedure, transcripts mapping uniquely to S. Typhimurium were used to normalise the data. Figure 4a shows a post-normalisation plot of read depth for each E. coli gene in each strain. The diagonal blue line shows the expected position of data points if transcription is unchanged between strains. There is a downshift in transcription of genes not bound by H-NS (black). Conversely, H-NS bound genes (red) are transcribed more frequently. This behaviour is exemplified in Figure 4b.
Figure 4. Most transcription is uniformly downregulated in cells lacking H-NS.
a,b) Most transcription is uniformly downregulated in cells lacking H-NS. a) the plot illustrates changes in global transcription caused by loss of H-NS. Data points represent H-NS bound (red) and unbound (black) genes. Genes with unaltered transcription should fall on the diagonal blue line. Data are from duplicate RNA-seq experiments. b) The basally expressed fad genes whose transcription is reduced in the absence of H-NS. Genes bound by H-NS are in red and other genes are black. Graphs show H-NS binding27 and RNA abundance (RNA-seq with spiked in control; this work). Data are representative.
c) RNA polymerase is redistributed in cells lacking H-NS. RNA polymerase distribution in wild type (top) and hns::kan cells (middle). Each heat map shows the average position of DNA-bound (i.e. transcribing or interacting with a promoter) RNA polymerase molecules within the cell. The bottom panelshows the average position of Ter as determined by visualising a TetR-mYpet fusion bound at an array of Ter proximal tetO sequences. Each distribution was generated from 100 cells between 3.5 and 4.5 μm in length. Each square is 1/624 of total cell area.
d) The σ70 G424D mutation hinders constitutive but not activator dependent NM501 promoter activity in vitro. KMnO4 footprinting reactions analysed on a denaturing polyacrylamide gel. Bands are indicative of open complex near the transcription start site (+1). RNA polymerase added at concentrations of 200, 250, 300 or 350 nM and CRP was 1.0 μM. The experiment, done three times, is representative.
e,f) The σ70 G424D mutation hinders constitutive but not activator dependent promoter activity in vivo. Different promoter DNA fragments were cloned upstream of lacZ (red arrow). For each promoter the location of key DNA sequence elements is represented by a box. In each case, the box is coloured according to the relationship between the DNA sequence and the consensus sequence for that element: perfect (dark blue), imperfect (pale blue) or completely absent (white). For each lacZ fusion β-galactosidase activity was measured in lysates of JCB387rpoD::kan carrying pVRσ (white bars) or pVRσG424D (black bars). The experiment was done in triplicate. Error bars show standard deviation from the mean.
RNA polymerase titration can be visualised directly
Genes bound by H-NS are overrepresented near the chromosome replication terminus (Ter)35,36. Conversely, most RNA polymerase binds near the origin of replication (Ori)37. These loci occupy distinct intracellular territories; Ter typically frequents mid-cell whilst Ori migrates to the poles37–39. Consequently, it should be possible to visualise titration of RNA polymerase at the cellular level. Previously, we used super resolution microscopy to track individual RNA polymerase molecules in live E. coli 40. Here, we repeated this analysis to examine the effect of deleting hns. In wild-type cells, RNA polymerase clusters near the quarter cell positions (Figure 4c, top). However, in cells lacking H-NS, RNA polymerase is redistributed and mid-cell is occupied (Figure 4c, middle). Consistent with unaltered RNA polymerase abundance, occupancy of mid-cell corresponds to reduced RNA polymerase abundance elsewhere (Figure 4c).
RNA polymerase mutation can compensate for loss of H-NS by favouring regulated transcription
An aspartic acid substitution in the RNA polymerase σ70 subunit (G424D in E. coli) can compensate for loss of H-NS41. This side chain could clash with the promoter -10 element during transcription initiation41. According to our model, promoters within genes are constitutive; they rely solely on their DNA sequence to bind RNA polymerase. Conversely, canonical promoters are regulated; a complex array of transcriptional activators stabilise RNA polymerase binding and DNA unwinding42. We reasoned that transcriptional defects, due to the G424D mutation, might be more pronounced at constitutive promoters. To test this, we purified RNA polymerase containing σ70 or the G424D derivative. We then investigated the ability of the RNA polymerase derivatives to stimulate unwinding of a promoter -10 element using KMnO4 footprinting. The semi-synthetic NM501 promoter was used because it has constitutive activity, by virtue of a consensus -10 element, but can be upregulated by the transcriptional activator CRP43. The G424D mutation completely abolished DNA opening by RNA polymerase in the absence of CRP (Figure 4d, compare lanes 2-5 and 6-9). In contrast, when CRP was present, the σ70 G424D mutation had little effect (compare lanes 10-13 and 14-17). Similarly, in vivo, the σ70 G424D mutation caused transcriptional defects at 7 of 8 constitutive promoters (Figure 4e). However, σ70 G424D functioned as well as, or better than, wild type RNA polymerase at activator dependent promoters (Figure 4f).
Discussion
The AT-rich genes examined here impose a fitness cost due to intragenic promoters. This phenomenon is likely to be widespread in bacteria; functional homologues of H-NS are apparent in diverse species8. Furthermore, the DNA binding properties of RNA polymerase are highly conserved42. We suggest that misappropriation of cellular resources underlies the hns phenotype. Redeployment of the finite RNA polymerase pool causes uniform suppression of canonical transcription. Whilst this general effect is likely to be pervasive, we do not exclude organism-specific complications. For example, in Pseudomonas aeruginosa, loss of H-NS-like proteins causes prophage induction and cell death44,45. Interestingly, linear H-NS filaments do not pose a barrier to transit of RNA polymerase46. Accordingly, transcription of an mRNA, and silencing of intragenic promoters, could occur simultaneously. For example, in this work, PyccE was active both in isolation and upstream of H-NS bound yccE. We speculate that gene silencing by H-NS may have evolved to discriminate between canonical and spurious RNA synthesis.
Materials and Methods
Strains, plasmids and general methods
E. coli JCB387 Δnir Δlac and MG1655 have been described previously47,48. M182hns::kan and KF26hns::kan were constructed by P1 transduction of hns::kan from MG1655 into the respective parent strains. The MG1655 hns::kan strain was provided by Ding Jin. M182rpoD::kan was generated using gene doctoring according to the protocol of Lee et al49 using the plasmids and oligonucleotides listed in Table S1. Note that, prior to gene doctoring, M182 strains were transformed with plasmid pVRσ that encodes rpoD 50. Quickchange mutagenesis was used to introduce the G424D mutation into pVRσ encoded rpoD (Table S1). Fortuitously, chromosomal and plasmid encoded σ70 were produced at indistinguishable levels (Figure S6). Sample sizes for all experiments were selected to ensure reproducibility in line with our previous work.
DNA fragments and gene expression assays
Promoter::lacZ fusions were made by cloning DNA fragments upstream of lacZ in the low copy number plasmid pRW5051. The nested deletions in the yccE intergenic region were generated by PCR and oligonucleotides shown in Table S1. The various yccE, yfdF, ykgH, yjgN and yjgL alleles were synthesised by Invitrogen and some contain silent mutations to remove EcoRI or HindIII restriction sites to facilitate cloning (Figures S2a and S3). Oligonucleotides used to amplify the different alleles for cloning into pSR and pRW50 are shown in Table S1. The 56 bp intragenic yccE fragments were generated with overlapping oligonucleotides (Table S1). The resulting single stranded overhangs were filled with DNA polymerase before cloning. β-galactosidase assays were done using the protocol of Miller52. All assay values are the mean of three biological replicates and the error bars show standard deviation from the mean. Experiments were done at least twice. Cells were grown aerobically at 37°C, to mid-log phase, in LB media.
Bioinformatic analysis of genes and design of new coding regions
Our stringent search criteria selected putative σ70 dependent promoters as described previously25. Thus, sequences were selected that matched the motifs 5’-TAnAAT-3’, 5’-TATnAT-3’ or 5’-TATAnT-3’. The relaxed search selected the sequence 5’-TAnnnT-3’. To inactivate promoter -10 elements the initial 5’-TA-3’ was replaced with 5’-GG-3’
Proteins, KMnO4 footprinting and in vitro transcription assays
H-NS and RNA polymerase were prepared as described previously53. DNA fragments for KMnO4 footprinting experiments were derived from Qiagen maxi-preparations of plasmid pSR. Thus, promoter DNA fragments were excised from pSR by sequential digestion with HindIII and then AatII. After digestion fragments were labelled at the HindIII end using [γ-32P]-ATP and polynucleotide kinase. Footprints were done as described by Grainger et al.54. The in vitro transcription experiments were done as described by Savery et al.55 using the system of Kolb et al.56. A Qiagen maxiprep kit was used to purify supercoiled pSR plasmid carrying the different promoter inserts. This template (16 pg ml-1) was pre-incubated with purified H-NS in buffer containing 20 mM Tris pH 7.9, 5 mM MgCl2, 500 μM DTT, 50 mM KCl, 100 μg ml-1 BSA, 200 μM ATP, 200 μM GTP, 200 μM CTP, 10 μM UTP with 5 μCi [α-32P]-UTP. The reaction was started by adding purified E. coli RNA polymerase. Labelled RNA products were analysed on a denaturing polyacrylamide gel. All in vitro assays were repeated at least three times in their entirety.
Growth assays
Cells lacking H-NS rapidly acquire compensatory mutations41. Consequently, reproducible changes in growth were only obtained when precautions were taken to minimise this phenomenon. The primary precaution was to reduce the number of division cycles that strains passed though during experimental setup. Thus, M182 and the hns::kan derivative were taken directly from long-term -80°C storage and used immediately to inoculate LB medium. After incubation for several hours at 37°C cells were harvested and competency was induced using ice cold CaCl2. The cells were then transformed with desired plasmids and transformants were isolated on selective agar plates. A colony from each plate was suspended in LB medium and aliquots of this were used immediately to inoculate fresh media so that growth could be monitored. Cells were grown either in LB medium at 37°C or in M9 minimal medium at 30°C. Values shown are from three biological replicates and the experiments were done on two separate occasions.
Western blotting
To determine relative protein levels in different strains cells were grown in LB media at 37°C. Aliquots of the culture were harvested at indicated time points and CFUs determined. Following this quantification the same number of cells from each aliquot were resuspended in SDS-PAGE gel loading buffer. After heating to 90°C for two minutes we separated proteins present in each lysate using SDS-PAGE. The proteins were then transfered to a Hybond-ECL nitrocellulose membrane using an Invitrogen XCell II Blot Module. Mouse anti-sera against the σ70 and β subunits of RNA polymerase (Neoclone) and H-NS (a gift from Jay Hinton) was used to detect the relevant proteins. Primary antibody binding was probed with horseradish peroxidise-linked rabbit anti-mouse antisera (Sigma-Aldrich A9044). The experiments were done on two separate occasions.
Standard RNA-seq
RNA-seq experiments were done in duplicate. E. coli MG1655 and E. coli MG1655 Δhns were grown in LB medium at 30°C to an OD600 of 0.5-0.7. RNA was harvested, RNA-seq libraries were constructed and libraries were sequenced as described previously57.
RNA-seq with a spiked in control
Transcriptome analysis experiments shown were performed in duplicate using E. coli MG1655 and E. coli MG1655 Δhns. We also ran the same experiment, again in duplicate, using E. coli M182 and the hns::kan derivative. The results were near identical. For each replicate three mid-log phase bacterial cultures were prepared. The three cultures were E. coli MG1655, E. coli MG1655 Δhns or Salmonella Typhimurium 14028s in LB medium. Before harvesting RNA the number of colony forming units (CFUs) per unit volume was determined for each culture by plating dilutions of each culture on nutrient agar and counting bacterial colonies. Aliquots of the two E. coli cultures were mixed S. Typhimurium cells. The volume of Salmonella cells was normalised to the CFUs for the corresponding E. coli culture. This step is crucial because it allows processing artefacts, due to differences in lysis efficiency, RNA recovery or cDNA synthesis, to be removed. Thus, in the final transcriptome analysis, the number of sequencing reads corresponding to the S. Typhimurium genome should be identical for each sample; they were derived from the same number of clonal S. Typhimurium cells. Hence, differences can only result from downstream processing. RNA was harvested, and RNA-seq libraries were prepared, as described previously57. RNA-seq libraries were sequenced on am Illumina Hi-Seq 2000 instrument (University at Buffalo, SUNY, Buffalo, NY, USA).
Data normalisation and transcriptome analysis
Our normalisation procedure is based on the addition of a proportional number of S. Typhimurium cells to each sample of E. coli cells immediately before harvesting RNA. All reads were first mapped to the E. coli MG1655 genome using CLC Genomics Workbench (Version 8.0; default parameters except that a perfect match was required). Unmapped reads were mapped to the S. Typhimurium 14028s genome (same parameters as above). The number of mapped S. Typhimurium reads, for each E. coli sample, was used to determine a correction factor for each sample. For example, if Sample A has twice as many S. Typhimurium reads as Sample B, the correction factor for Sample A will be twice that for Sample B. Having calculated a correction factor for each sample, we remapped all sequence reads to the S. Typhimurium 14028s genome using CLC Genomics Workbench (same parameters as above). Unmapped reads were then mapped to the E. coli MG1655 genome using CLC Genomics Workbench (same parameters as above). Total read coverage per gene was calculated using a custom Python script. These values were normalised to the length of the gene, and further normalised using the correction factor (described above). Raw data are available from the ArrayExpress database using accession number E-MTAB-4751.
Super resolution microscopy of RNA polymerase
Cell preparation, microscopy, and analysis were performed as described previously40,58. In brief, we used an endogenous fusion of photoactivatable fluorescent protein PAmCherry, with the β’ subunit of RNA polymerase, encoded by E. coli strain KF2658. Glycerol stocks of KF26, and the hns::kan derivative, were used to inoculate fresh Rich Defined Media (RDM, Teknova). When the culture attained an OD650 value of ~0.2 cells were collected by centrifugation and resuspended. Cells in 1 μl of the suspension were placed on an RDM agarose pad and imaged for 300000 frames at 15 ms exposure on a custom built single-molecule TIRF microscope. For each strain, 9 fields of view were imaged. The experiment was done on 3 separate occasions. Molecules were imaged by photoactivating and localising fluorophores, and joining localisations over multiple frames to obtain trajectories of individual molecules. To measure RNA polymerase mobility, we calculated an apparent diffusion coefficient from the mean squared displacement of trajectories of individual molecules, and used a threshold to distinguish transcriptionally active and promoter bound molecules from the rest of the population. Cell outlines were determined from the brightfield image, and the average intracellular location of RNA polymerase was established from a 2D histogram of the mean trajectory positions relative to the cell outline for all DNA-bound molecules in at least 100 cells. Ter positioning was determined using similar analysis of strain PZ111 carrying a tetO array inserted at Ter, and expressing a TetR-mYpet fusion. The strain was constructed by P1 transduction of tetR- mYPet kan, into an AB1157 strain with an array of 240 tetO sequences 50 kb clockwise of dif (ter3)59.
Acknowledgements
This work was funded by Leverhulme Trust project grant (RPG-2013-147) and Wellcome Trust Career Development Fellowship (WT085092MA) awarded to D.C.G. Support for J.T.W. was a National Institutes of Health Director’s New Innovator Award (1DP2OD007188). A.N.K. and M.S. were supported by BBSRC grant (BB/N018656/1; to ANK and MS), and a Wellcome Trust Investigatorship (110164/Z/15/Z; to ANK). We thank Jay Hinton for the gift of anti-H-NS anti-sera.
Footnotes
Contributions
D.C.G. and J.T.W. designed the study and wrote the manuscript. L.E.L., G.B., S.S.S., A.M.S., R.P.B., and M.S. generated the data and prepared it for publication. M.S. and A.N.K. provided new analytical tools and critically discussed the manuscript with D.C.G. and J.T.W. All authors contributed to data analysis and interpretation.
Data availability
The data that support these findings are available from the corresponding author upon request. The raw data for RNA-seq experiments are available from the ArrayExpress database using accession number E-MTAB-4751. Original gel images are shown in Figure S7.
References
- 1.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 2.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 3.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popa O, Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol. 2011;145:615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Raghavan R, Kelkar YD, Ochman H. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci. 2012;109:14504–14507. doi: 10.1073/pnas.1205683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evol. 2013;8:489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–61. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 8.Singh K, Milstein JN, Navarre WW. Xenogeneic Silencing and Its Impact on Bacterial Genomes. Annu Rev Microbiol. 2016;70:199–213. doi: 10.1146/annurev-micro-102215-095301. [DOI] [PubMed] [Google Scholar]
- 9.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 10.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;28:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smits WK, Grossman AD. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 2010;6:e1001207. doi: 10.1371/journal.pgen.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon BR, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman CJ. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid. 2014;75:1–11. doi: 10.1016/j.plasmid.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol. 2007;14:441–418. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 15.Gordon BR, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, Navarre WW, Xia B, Liu J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci. 2011;108:10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–73. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–44. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landick R, Wade JT, Grainger DC. H-NS and RNA polymerase: a love-hate relationship? Curr Opin Microbiol. 2015;24:53–59. doi: 10.1016/j.mib.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Winardhi RS, Yan J, Kenney LJ. H-NS Regulates Gene Expression and Compacts the Nucleoid: Insights from Single-Molecule Experiments. Biophys J. 2015;109:1321–1329. doi: 10.1016/j.bpj.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. J Biol Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- 23.Huang Q, Cheng X, Cheung MK, Kiselev SS, Ozoline ON, Kwan HS. High-density transcriptional initiation signals underline genomic islands in bacteria. PLoS One. 2012;7:e33759. doi: 10.1371/journal.pone.0033759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh SS, Grainger DC. H-NS can facilitate specific DNA-binding by RNA polymerase in AT-rich gene regulatory regions. PLoS Genet. 2013;9:e1003589. doi: 10.1371/journal.pgen.1003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. Widespread suppression of intragenic transcription initiation by H NS. Genes Dev. 2014;28:214–219. doi: 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam KN, Charles TC. Strong spurious transcription likely contributes to DNA insert bias in typical metagenomic clone libraries. Microbiome. 2015;3:22. doi: 10.1186/s40168-015-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli . Nucleic Acids Res. 2011;39:2073–91. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chintakayala K, Singh SS, Rossiter AE, Shahapure R, Dame RT, Grainger DC. E. coli Fis protein insulates the cbpA gene from uncontrolled transcription. PLoS Genet. 2013;9:e1003152. doi: 10.1371/journal.pgen.1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wade JT, Castro Roa D, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Extensive functional overlap between sigma factors in Escherichia coli . Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 30.Haycocks JR, Sharma P, Stringer AM, Wade JT, Grainger DC. The molecular basis for control of ETEC enterotoxin expression in response to environment and host. PLoS Pathog. 2015;11:e1004605. doi: 10.1371/journal.ppat.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piper SE, Mitchell JE, Lee DJ, Busby SJ. A global view of Escherichia coli Rsd protein and its interactions. Mol Biosyst. 2009;5:1943–1947. doi: 10.1039/B904955j. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasan R, Scolari VF, Lagomarsino MC, Seshasayee AS. The genome-scale interplay amongst xenogene silencing, stress response and chromosome architecture in Escherichia coli . Nucleic Acids Res. 2015;43:295–308. doi: 10.1093/nar/gku1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–53. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 34.Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarei M, Sclavi B, Cosentino Lagomarsino M. Gene silencing and large-scale domain structure of the E. coli genome. Mol Biosyst. 2013;9:758–767. doi: 10.1039/c3mb25364c. [DOI] [PubMed] [Google Scholar]
- 37.Dame RT, Kalmykowa OJ, Grainger DC. Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in Gram negative bacteria. PLoS Genet. 2011;7:e1002123. doi: 10.1371/journal.pgen.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Junier I, Boccard F, Espéli O. Polymer modeling of the E. coli genome reveals the involvement of locus positioning and macrodomain structuring for the control of chromosome conformation and segregation. Nucleic Acids Res. 2014;42:1461–1473. doi: 10.1093/nar/gkt1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youngren B, Nielsen HJ, Jun S, Austin S. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 2014;28:71–84. doi: 10.1101/gad.231050.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stracy M, Lesterlin C, Garza de Leon F, Uphoff S, Zawadzki P, Kapanidis AN. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc Natl Acad Sci USA. 2015;112:E4390-9. doi: 10.1073/pnas.1507592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali SS, Soo J, Rao C, Leung AS, Ngai DH, Ensminger AW, Navarre WW. Silencing by H-NS potentiated the evolution of Salmonella . PLoS Pathog. 2014;10:e1004500. doi: 10.1371/journal.ppat.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DJ, Minchin SD, Busby SJ. Activating transcription in bacteria. Annu Rev Microbiol. 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 43.Miroslavova NS, Busby SJ. Investigations of the modular structure of bacterial promoters. Biochem Soc Symp. 2006;73:1–10. doi: 10.1042/bss0730001. [DOI] [PubMed] [Google Scholar]
- 44.Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci USA. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Wally H, Miller SJ, Lu CD. The Multifaceted Proteins MvaT and MvaU, Members of the H-NS Family, Control Arginine Metabolism, Pyocyanin Synthesis, and Prophage Activation in Pseudomonas aeruginosa PAO1. J Bacteriol. 2009;191:6211–6218. doi: 10.1128/JB.00888-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotlajich MV, Hron DR, Boudreau BA, Sun Z, Lyubchenko YL, Landick R. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. Elife. 2015;4:e04970. doi: 10.7554/eLife.04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page L, Griffiths L, Cole JA. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch Microbiol. 1990;154:349–354. doi: 10.1007/BF00276530. [DOI] [PubMed] [Google Scholar]
- 48.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605-12. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee DJ, Bingle LE, Heurlier K, Pallen MJ, Penn CW, Busby SJ, Hobman JL. Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains. BMC Microbiol. 2009;9:252. doi: 10.1186/1471-2180-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodius VA, Busby SJ. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase sigma70 subunit: application of suppression genetics. J Mol Biol. 2000;299:311–324. doi: 10.1006/jmbi.2000.3737. [DOI] [PubMed] [Google Scholar]
- 51.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini NR. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol Lett. 1992;74:271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- 52.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 53.Grainger DC, Goldberg MD, Lee DJ, Busby SJ. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol Microbiol. 2008;68:1366–1377. doi: 10.1111/j.1365-2958.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 54.Grainger DC, Belyaeva TA, Lee DJ, Hyde EI, Busby SJ. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with the C-terminal domain of the RNA polymerase alpha subunit. Mol Microbiol. 2004;51:1311–1320. doi: 10.1111/j.1365-2958.2003.03930.x. [DOI] [PubMed] [Google Scholar]
- 55.Savery NJ, Lloyd GS, Kainz M, Gaal T, Ross W, Ebright RH, Gourse RL, Busby SJ, et al. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolb A, Kotlarz D, Kusano S, Ishihama A. Selectivity of the Escherichia coli RNA polymerase E sigma 38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 1995;23:819–826. doi: 10.1093/nar/23.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stringer AM, Currenti S, Bonocora RP, Baranowski C, Petrone BL, Palumbo MJ, Reilly AA, Zhang Z, Erill I, Wade JT. Genome-scale analyses of Escherichia coli and Salmonella enterica AraC reveal noncanonical targets and an expanded core regulon. J Bacteriol. 2014;196:660–671. doi: 10.1128/JB.01007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endesfelder U, Finan K, Holden SJ, Cook PR, Kapanidis AN, Heilemann M. Multiscale spatial organization of RNA polymerase in Escherichia coli . Biophys J. 2013;105:172–181. doi: 10.1016/j.bpj.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zawadzki P, Stracy M, Ginda K, Zawadzka K, Lesterlin C, Kapanidis AN, Sherratt DJ. The Localization and Action of Topoisomerase IV in Escherichia coli Chromosome Segregation Is Coordinated by the SMC Complex, MukBEF. Cell Rep. 2015;13:2587–2596. doi: 10.1016/j.celrep.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support these findings are available from the corresponding author upon request. The raw data for RNA-seq experiments are available from the ArrayExpress database using accession number E-MTAB-4751. Original gel images are shown in Figure S7.