Abstract
The Escherichia coli protein RhlB is an ATP-dependent motor that unfolds structured RNA for destruction by partner ribonucleases. In E. coli, and probably many other related γ-proteobacteria, RhlB associates with the essential endoribonuclease RNase E as part of the multi-enzyme RNA degradosome assembly. The interaction with RNase E boosts RhlB's ATPase activity by an order of magnitude. Here, we examine the origins and implications of this effect. The location of the interaction sites on both RNase E and RhlB are refined and analysed using limited protease digestion, domain cross-linking and homology modelling. These data indicate that RhlB's carboxy-terminal RecA-like domain engages a segment of RNase E that is no greater than 64 residues. The interaction between RhlB and RNase E has two important consequences: first, the interaction itself stimulates the unwinding and ATPase activities of RhlB; second, RhlB gains proximity to two RNA-binding sites on RNase E, with which it cooperates to unwind RNA. Our homology model identifies a pattern of residues in RhlB that may be key for recognition of RNase E and which may communicate the activating effects. Our data also suggest that the association with RNase E may partially repress the RNA-binding activity of RhlB. This repression may in fact permit the interplay of the helicase and adjacent RNA binding segments as part of a process that steers substrates to either processing or destruction, depending on context, within the RNA degradosome assembly.
Keywords: DEAD box helicase, RhlB, ribonuclease, RNase E, RNA degradosome
Introduction
In Escherichia coli the RNA helicase B, RhlB, is a component of the multi-enzyme RNA degradosome (Figure 1(a)). The other main components of the RNA degradosome include the essential endoribonuclease, RNase E, the exo-ribonuclease polynucleotide phosphorylase (PNPase), and the glycolytic enzyme enolase.1,2 On their own RNase E and PNPase have limited activity for structured RNA, but evidence indicates that the RhlB helicase can work in conjunction with those ribonucleases by converting the RNA into suitable substrates through its energy-dependent unwinding activity.
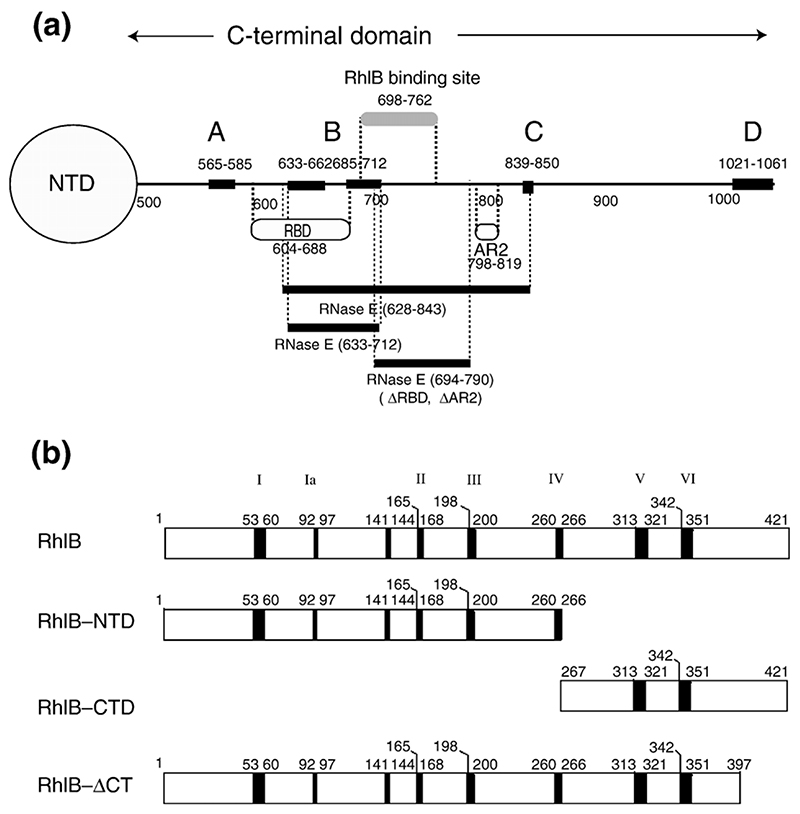
Figure 1. RNase E and RhlB domains and experimental constructions.
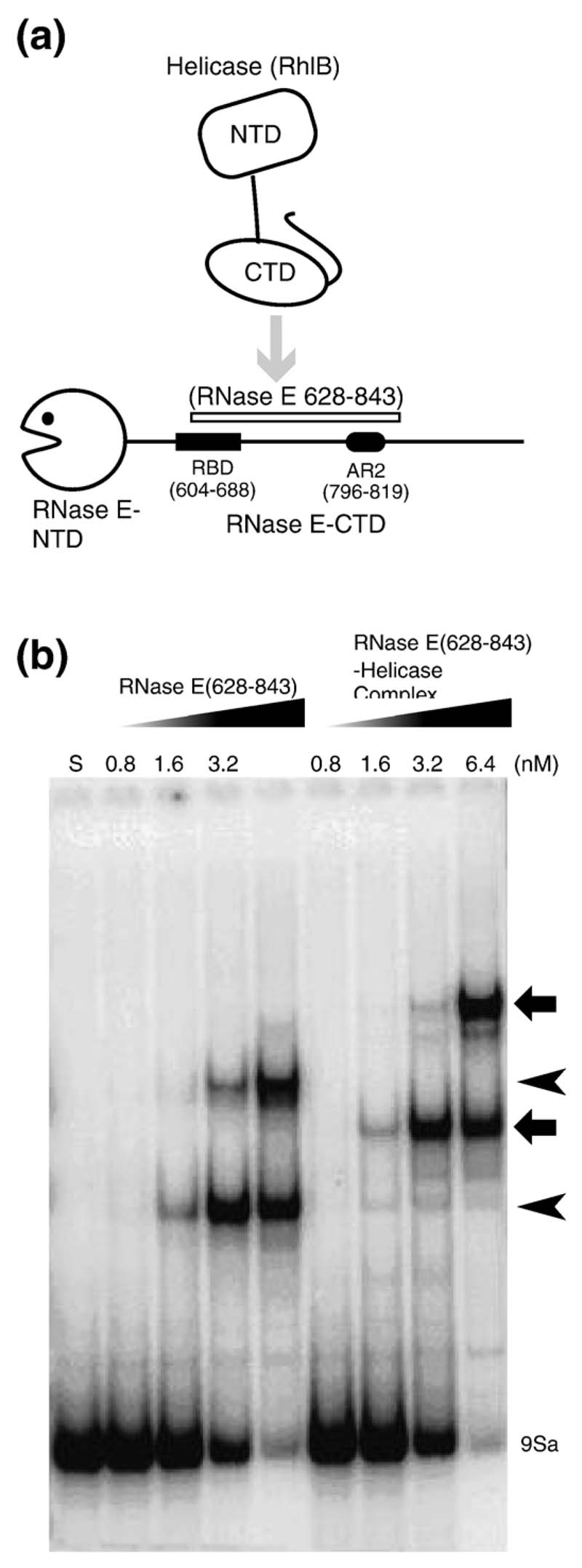
(a) Schematic representation of the recognition elements of the RNase E carboxy-terminal domain, which forms the scaffold of the RNA degradosome. The positions of the recognition sites are indicated along the length of the carboxy-terminal domain (CTD). The regions of structural propensity in the CTD are labelled A to D.12 AR2: a second non-catalytic Arg-rich region that binds RNA independently of the first longer RBD.41 Several segments of RNase E CTD were used in these studies: RNase E (628-843) corresponds to residues 628-843. The RNase E (498-1061) polypeptide is an N-terminal polyhistidine tagged derivative of RNase E in which residues 27-497 of RNase E have been deleted. RNase E (ΔRBD, ΔAR2) is a derivative of RNase E (498-1061) in which the RBD and AR2 have been deleted. NTD is the N-terminal domain, which encompasses the catalytic site. (b) Schematic representation of the RhlB contructs studied here: full-length helicase RhlB, RhlB-CTD, RhlB-NTD, and RhlB-ΔCT. The principal DEAD-box helicase sequence motifs are labelled I, Ia, II, III, IV, Vand VI. Not shown are Ib (follows Ia), Q and GG (which follow III) and QxxR (which follows VI).
RhlB belongs to an extensive helicase family whose members share a structurally conserved ATPase domain containing the amino acid sequence DEAD as part of a Walker sequence motif that identifies residues engaging Mg-ATP.3 Proteins containing “DEAD-box” sequence motifs or its close variants use the free energy of ATP binding and hydrolysis to induce unwinding of nucleic acids, and some can drive the energy-dependent displacement of proteins from RNA.4 The latter activity is perhaps required by the RNA degradosome for manipulating certain RNA substrates.
The universal core of DEAD-box proteins comprises a structural repeat of a fold resembling the single-stranded DNA binding protein, RecA.5 The N-terminal RecA-like domain contains seven key conserved motifs common to the helicase family, (known as I, Ia, Ib, II, III, Q and GG), while the C-terminal domain contains four motifs (IV, V, VI and QxxR). Crystallographic studies of a number of RNA helicases have identified the roles of these sequence motifs in forming an intricate interaction network that supports ATP and RNA binding, catalysis, and inter-domain communication.6–11 Amongst the salient features, the DEAD box (motif II) coordinates a Mg ion that supports the phosphate of the nucleotide and, together with residues in motif V, also binds a water molecule that is likely to attack the terminal γ-phosphate. Motifs Ib and IV make contact to the RNA, and motifs V and VI make contact with the ATP. Motif III positions motifs II and VI, and genetic evidence indicates that it is involved in the unwinding mechanism, so it likely links the substrate translocation with the turnover of ATP. Each of these key motifs is mapped onto the schematic representation of the E. coli RhlB helicase in Figure 1(b).
RhlB has been shown to be a monomer in isolation as well as in the complex with its recognition site on RNase E,12 and it is likely that the enzyme functions as a monomer during the unwinding process. One proposed mechanism for monomeric helicases such as RhlB is the “inchworm” stepping model, in which a successive transfer of the nucleic acid substrate takes place between two nucleic acid binding sites whose affinity alternates during the force-generating step of the ATP-consuming cycle.13 Single molecule measurements of ATPase and helicase activities in the DEAD-box family helicase NS3 support a mechanism involving the interplay of two RNA binding sites within a helicase monomer: a translocator site and a duplex-opener site.14 A 3' singlestranded RNA binding site loads the RNA substrate. The translocator site moves in 11 base-pair steps to recognise the duplex RNA, and the helix-opener site moves in shorter sub-steps of 2-5 base-pairs to bring about the unwinding. An appealing hypothesis holds that the two sites could be located in the two RecA-like domains described above.
Crystallographic studies of RNA helicases have illuminated the potential inter-domain movements that are associated with stepping and the interplay of the two proposed binding sites. The structure of the helicase domain of the Drosophila Vasa protein, which has been solved in complex with single-stranded RNA and a non-hydrolyzable ATP analogue, reveals that the RNA is bound at an interfacial cleft formed by the interaction of the two RecA-like domains.9 The complex is more “closed”, with more domain-domain interactions than seen in the “open” structures where the RNA is absent. Interdomain movement is thus associated with the cycle of ATP binding, hydrolysis and RNA unwinding or translocation. The displacement of the two strands during ATP turnover may result from “pushing” the duplex against a wedge-like α-helix located in the N-terminal domain.
Many monomeric DEAD-box helicases appear to function by themselves; however, RhlB, the eukaryotic translation initiation factor eIF4AIII and certain helicases involved in eukaryotic RNA splicing appear to require protein partners for full activity.15,16 The interaction of RhlB and RNase E boosts ATPase activity by at least an order of magnitude,12,17,18 but it is not presently clear how the interaction with RNase E activates RhlB.
Here, we map the site of interaction between RNase E and RhlB. Our results reveal cooperation between RhlB and two binding sites in RNase E that affects unwinding. In the context of the RNA degradosome, the helicase/RNase E cooperation pre-organises the substrate for cleavage by the neighbouring phosphorolytic nuclease PNPase and perhaps the RNase E catalytic domain. We discuss the implications of these findings for the operation of the helicase within the context of the degradosome.
Results
Mapping the site of interaction between RhlB and RNase E
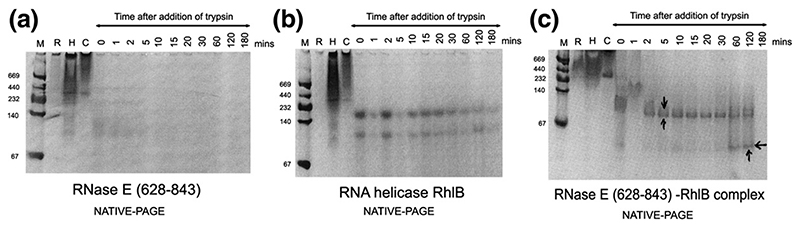
We had found earlier that RhlB forms a stable complex with the segment of RNase E corresponding to residues 628-843 (segments will be referred to as RNase E (628-843), etc.).12,18 The dissociation constant of this interaction lies in the 50 nM range. RNase E (628-843) includes the putative RNA binding segments AR2 and part of RBD (Figure 1(a)). To refine the boundaries for the protein-protein interaction, a protease digestion time course was undertaken. In the absence of the helicase, RNase E (628-843) is sensitive to digestion, probably due to its intrinsically unstructured character,12 whereas RhlB is comparatively more resistant. When the two proteins are mixed to form a complex, and the digest products analysed by denaturing gels, a pattern results that approximates the sum of the digestions of the individual components, although some protected fragments can be discerned. The generation of a protected species becomes more apparent from electrophoretic analysis of the digest products under non-denaturing conditions (Figure 2(a)-(c)), which identifies new bands for the treated complex that are not seen in digests of the individual components (indicated by arrows in Figure 2(c)).
Figure 2. Mapping the site of interaction of RhlB helicase and RNase E.
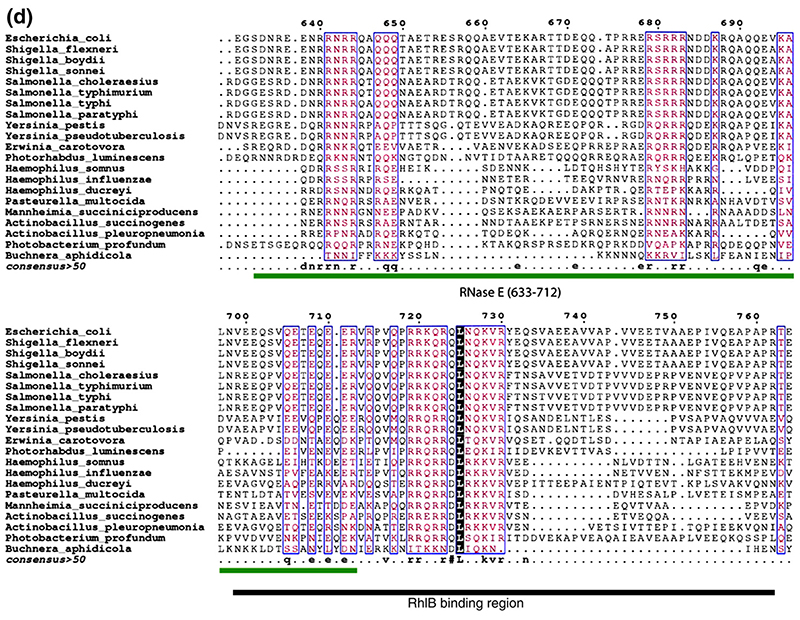
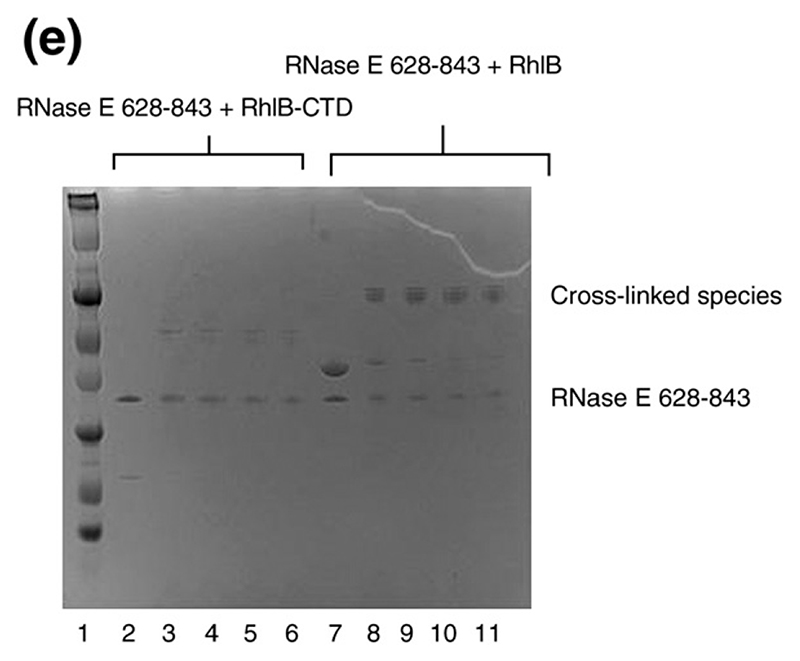
(a)–(c) Limited protease digestion of the RhlB-RNase E (628–843) complex. Lanes M, R, H, and C correspond to molecular weight makers (masses indicated). (d) Conservation of putative RNA binding regions in the vicinity of the helicase binding site in RNase E. The boundary of the RhlB binding site is indicated. Segment B corresponds to a region with predicted propensity to form a four-helix bundle.12 This prediction is consistent with sequence profile library searching38 of RNase E (640–710). (e) Interactions of the carboxy-terminal domain of RhlB (RhlB-CTD) with RNase E. Chemical cross-linking was used to evaluate the interaction of RhlB-CTD with RNase E (628-843). All samples were buffer exchanged into 0.2 M triethanolamine (pH 8.0), and dimethylsuberimidite (DMS) added to a final concentration of 10 mM, and results were analysed by Coomassie-stained SDS/PAGE. Lane 1, molecular mass markers. Lane 2 shows RNase E (628-843) plus RhlB-CTD, and lane (7) RNase E (628-843) plus RhlB before the addition of DMS. Lanes 3 to 6 show RNase E (628-843) and RhlB-CTD at 30, 60, 120 and 180 min, respectively, after the addition of DMS. Lanes 8 to 11 show RNase E (628-843) and RhlB at 30, 60, 120 and 180 min, respectively, after the addition of DMS. Similar results were found using glutaraldehyde as the cross-linking agent (results not shown).
Digest products were analysed by mass spectrometry. Two short regions of RNase E (628-843) are identified, corresponding to residues 696-712 and residues 730-762 (Supplementary Data, Figure S1). We can infer that segment 696-712 is not sufficient by itself to bind RhlB avidly, because we find that RNase E (633-712), which encompasses this segment, does not interact with RhlB by either pull-down experiments or by size exclusion chromatography (results not shown). A 13-mer synthetic peptide corresponding to RNase E residues 730 to 742 failed to interact with RhlB, or to boost its ATPase activity, in the micromolar concentration range, suggesting that this segment is insufficient for supporting either activity. Trypsin prefers to cut at peptide bonds preceding basic residues, so it has a limited resolution for mapping the boundaries of the RhlB binding site on RNase E (628-843); however, similar regions were also protected from proteolysis by the enzymes chymotrypsin, which cuts at hydrophobic residues, and V8 protease, which has preference for acidic residues. Sequencing of the product peptides suggests the N-terminal boundary of the recognition site to be the RNase E residue 698. Taken together, these results indicate that a minimal region for RhlB recognition by RNase E can be narrowed to residues 698-762. This region of RNase E is comparatively well conserved amongst homologues (Figure 2(d)).
The C-terminal tail of the helicase, corresponding to residues 399 to 421, is arginine-rich and is readily liberated by trypsin; however, the digest product still binds RNase E avidly, showing that the RhlB C-terminal tail is not required for protein-protein recognition.12 However, residues elsewhere in the carboxy-terminal domain (CTD) are likely to interact with RNase E, since the fragments of RhlB in the digested complex in Figure 2(a) matched peptides mainly from the C-terminal RecA-like domain. In the following section we present evidence that corroborates that this domain is the site of interaction.
The carboxy-terminal domain of RhlB is the site of RNase E recognition
We prepared a fragment of RhlB from the C-terminal RecA-like domain that includes a subset of the conserved helicase motif elements (namely, V and VI), referred to here as RhlB-CTD (Figure 1(b)). This protein was found to bind avidly to RNase E (628-843) by various methods. When a mixture of RNase E (628-843) and RhlB-CTD was applied to size exclusion chromatography (S200), a peak was observed that elutes earlier than the individual components and which contains both proteins (not shown). Cross-linking of RhlB-CTD and RNase E (628-843) with dimethyl suberimidate (DMS) gave a novel band in denaturing electrophoresis, confirmed to contain both proteins by mass spectrometry (Figure 2(e)). No cross-linked species were observed for the controls of RNase E (628-843) and RhlB-CTD alone, indicating that these molecules do not selfassociate, while in control experiments, while enolase was found to cross-linked to itself, as expected for this homo-dimeric enzyme (data not shown). The stoichiometry for RhlB-CTD/RNase E (628-843) complex is 1:1, as judged by SDS/PAGE of the complex from size-exclusion chromatography, from mass of the cross-linked species in Figure 2(e), and from mass estimates by nanoflowelectrospray mass spectrometry (P. Ilag and C. Robinson, personal communication). A 1:1 stoichiometry is consistent with results reported by Khemici et al.,18 who measured the ATPase activity of RhlB at different concentrations of RNase E (628-843) or RNase E (694-790), and with the reported mass of the complex of full-length RhlB and RNase E (628-843).12
RNase E (628–843) and RNase E (694–790) stimulate the RNA unwinding activity of RhlB
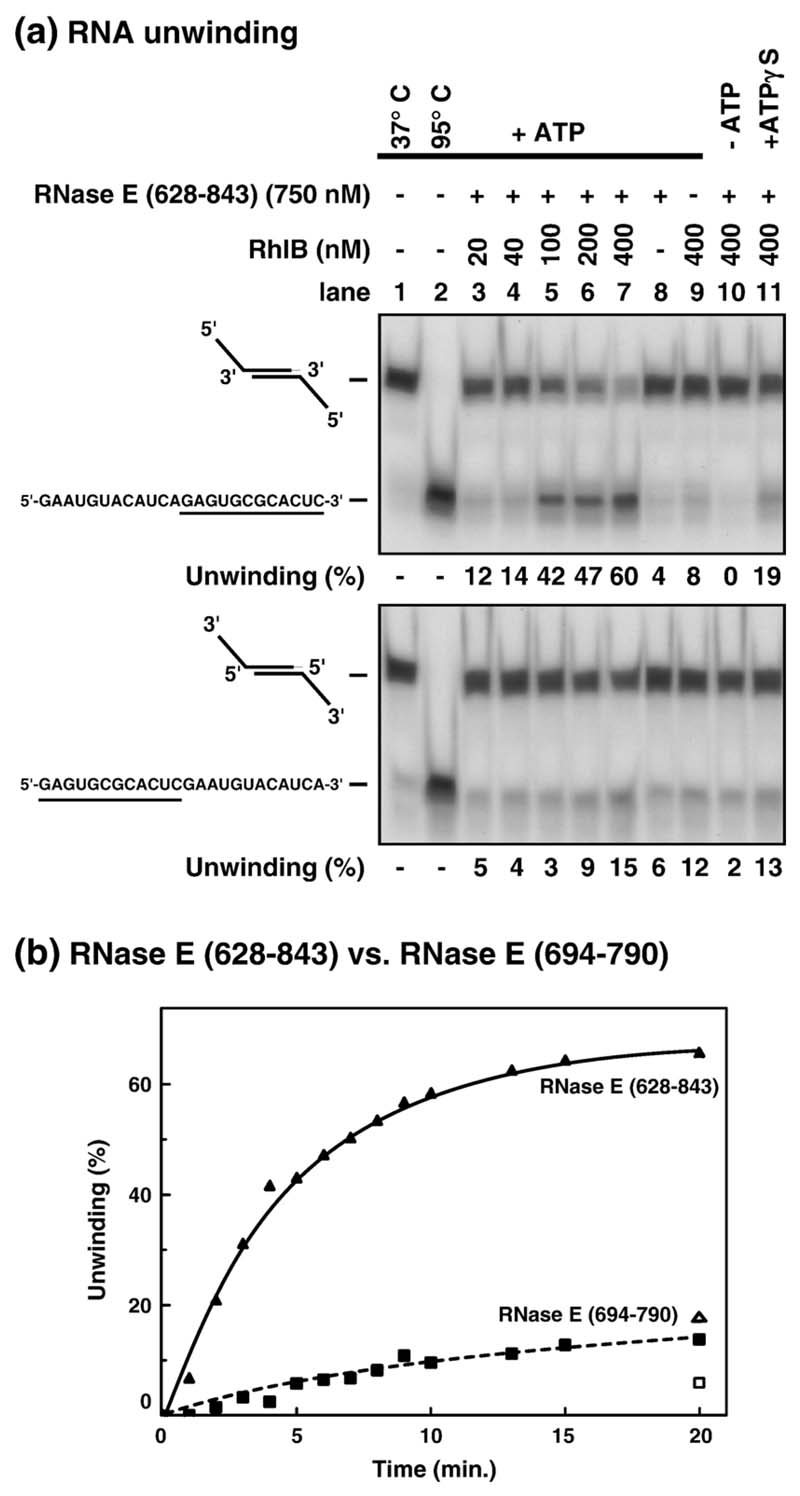
Earlier studies have shown that the ATPase activity of helicase RhlB is boosted by interaction with RNase E. Here, we have explored whether this interaction stimulates RNA unwinding, and whether the RNA-binding segments of RNase E contribute to the unwinding process. We first tested two self-annealing oligomers with identical duplex-forming regions and 5' or 3' single-strand tails that have the same sequence (Figure 3(a), lanes 1 and 2). In the presence of RhlB, saturating concentrations of RNase E (628-843) and ATP, extensive unwinding is observed for the 5' tailed RNA duplex (60% of duplex unwound with 400 nM RhlB, lane 7 upper panel). In the absence of ATP, no unwinding was detected even with the greatest protein concentration tested (lane 10). By itself, RNase E (628-843) lacks significant unwinding activity (lane 8, values ranged from 0 to 5% in different experiments). In contrast, a low but reproducible unwinding occurs when RhlB is incubated alone (8%, lane 9). These controls show that RhlB requires ATP for unwinding and that RNase E (628-843) stimulates its activity. To test the requirement for ATP hydrolysis in unwinding, we used the non-hydrolysable analogue, ATPγS. A reproducible unwinding of the duplex was detected (19%, lane 11) although it is threefold lower than observed with ATP (60%, lane 7). These results show that the unwinding is stimulated by ATP hydrolysis. The limited unwinding associated with the non-hydrolysable analogue could be due to RNA binding as a consequence of the large excess of protein relative to substrate in these assays.
Figure 3. Stimulation of RNA unwinding activity by RNase E (628–843).
(a) RNase E (628-843) and RhlB were incubated as described in Materials and Methods with 5' or 3'-tailed duplex RNA (0.1 nM) for 30 min at 37 °C (upper and lower panels, respectively). Each substrate was formed from a self-annealing synthetic RNA oligonucleotide whose sequence is indicated on the left. The sequence of the single-strand and duplex forming (underlined) region of both substrates is identical. The oligonucleotides were 5'-labelled with γATP. Unwinding was analysed by native PAGE (12%) and autoradiography. The unwinding was quantified using a phosphorimager and is indicated as a percentage under each panel. Lane 1 shows duplex RNA incubated under unwinding conditions but without protein. The percentage of single-stranded RNA in this control (typically 5%) was subtracted as a background in the unwinding reactions. Lanes 2 shows duplex RNAs denatured by heating to 95 °C for 3 min. (b) The stimulation of RhlB unwinding activity by RNase E (694–790) is comparatively less efficient. 400 nM RhlB and 0.1 nM 5'-tailed duplex were incubated with 750 nM RNase E (628–843) (filled triangle) or 660 nM RNase E (694-790) (filled square) at 37 °C with ATP. In the presence of ATPγS, 17% (open triangle) and 4% (open square) of single-stranded substrate is produced after 20 min with RNase E (628–843) and RNase E (694–790), respectively, compared to 66% and 14% with ATP.
The same conditions were used to test the unwinding of the 3'-tailed RNA duplex (Figure 3(a), lower panel). This substrate resists unwinding under conditions where the 5' tailed duplex was 60% unwound (compare lanes 7, upper and lower panels). Since the spontaneous re-annealing of both products is similar (less than 5% in 10 min, data not shown), this result is not due to faster re-annealing of the 3'-tailed substrate. Furthermore, the unwinding of the 3'-tailed substrate is similar with ATP or ATPγS (lower panel, compare lanes 7 and 11), showing that ATP hydrolysis is not required for unwinding of that substrate. We conclude that the RNA helicase activity of RhlB has specificity for duplexes with 5' single-stranded extensions. The 5' singlestrand ends are necessary, since a blunt-ended duplex prepared by treatment with RNase T2 was not unwound in this assay (data not shown).
We tested the capacity of the shorter fragment of RNase E (694-790), which lacks the RNA binding domains, to stimulate RNA unwinding by RhlB. As shown in the time course in Figure 3(b), the RNase E (694-790)/RhlB complex (squares) can unwind the 5'-tailed duplex although it is less active than the RNase E (628-843)/RhlB complex (triangles). In the presence of ATPγS (open symbols), unwinding by RNase E (694-790) is reproducibly three to fourfold lower than observed with ATP. Thus, although less active than the longer RNase E (628-843), RNase E (694-790) stimulates ATPase-dependent unwinding. After correction for the values with ATPγS at 20 min, the hydrolysis-dependent unwinding activity of RNase E (694-790)/RhlB is about 20% compared to RNase E (628-843)/RhlB. This is an overestimation, since the difference in initial rates in Figure 3 is closer to tenfold. The comparison of the activity of these complexes indicates that the RNA-binding domain (RBD) and arginine-rich region 2 of RNase E (AR2) boost RNA unwinding. It was observed earlier that RNase E segments 694-790 and 628-843 were roughly equivalent in stimulating the ATPase activity of the helicase;18 thus the difference in their ability to activate the unwinding by the helicase is likely to be due to the RNA binding segments RBD and AR2 in the RNase E. This suggests that there is a dynamic interplay between the RNA binding segments of the RNase E and the helicase during the unwinding process. We explored this interplay in the following experiments.
RNA recognition by RNase E and RhlB
Using an electrophoresis mobility shift assay, we have shown that RNase E (628-843) binds the 7S precursor of 5S ribosomal RNA (rRNA) avidly and forms two complexes that migrate at different rates.12 Those earlier experiments measured binding by competition with non-specific RNA and suggested that the affinities lie in the nanomolar range. Here, we have measured direct binding, in the absence of competitor, of 9Sa RNA, which is a fragment of the longer 9S precursor of 5S rRNA.19 We again observe two distinct bands with affinities estimated to be in the nanomolar concentration range (Figure 4(b), lanes 1-5). The two bands may arise either from the binding of two 9S rRNA molecules by one RNase E (628–843) molecule, or by the association of two molecules of RNase E (628–843) to the same 9S rRNA molecule. We also observe that the complex of RNase E (628–843) and RhlB gave rise to two species whose mobilities are comparatively retarded (Figure 4(b), lanes 6–9).
Figure 4. The ternary complex of helicase-RNase E and rRNA.
(a) Schematic representation of the domains used in the experiments. (b) Gel shift assay with RNase E (628–843) alone or complexed with RhlB using the 9Sa RNA substrate. Lane S is the lane for the RNA alone. The free RNA is indicated at the bottom of the gel. The concentrations of the proteins (in nM) are shown above each lane. The shifted bands for the RNase E (628–843)-RNA complex are indicated by arrowheads on the right, and the ternary RNase E (628–843)–helicase–RNA complex are indicated by the broad arrows. (c) Example of a double filter RNA binding assay showing a side-by-side measurement using RNase E (628-843) alone or complexed with RhlB. Quantification of the radioactive RNA on the filters and curve fitting were performed as described in Materials and Methods. The binding isotherms shown here were fitted to data from several independent determinations (see Table 1).
RhlB also binds 9Sa rRNA in the nM concentration range by the mobility assay, but the bands are not as well defined as those observed for RNase E (628-843) (data not shown).
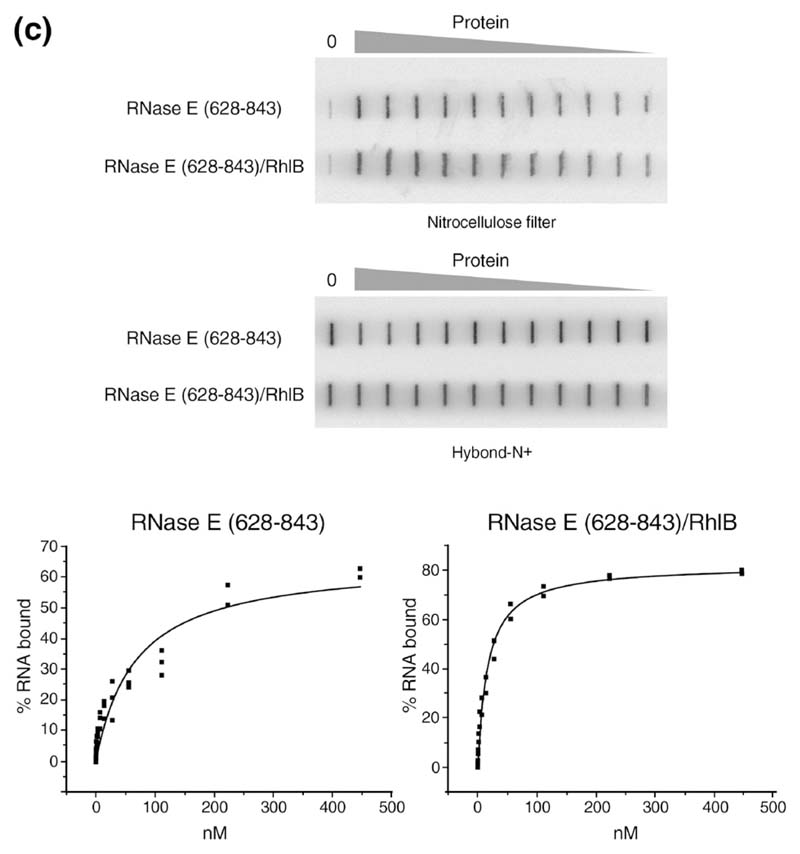
We used a double-membrane filter binding assay to measure the binding of RhlB. The method includes a photo-cross-linking step to capture the protein-RNA complex. This yielded more consistent and reproducible binding profiles, but as the method involves an irreversible step, it must be taken as an approximation to the equilibrium binding. From the binding data, we measured an apparent K D value of 48 nM for RhlB alone (Table 1). A truncated derivative of RhlB lacking an arginine-rich C-terminal tail (RhlB-Δ CT) binds RNA more weakly than the full length RhlB (193 nM; Table 1), indicating a role of this basic tail in RNA association. The individual domains of RhlB were tested for binding, and while RhlB-CTD forms a complex, RhlB-amino terminal domain (NTD) did not measurably bind the RNA in the range of concentrations tested (not shown). Thus, the CTD is likely to be the primary site of RNA interaction in the absence of ATP.
Table 1. Apparent K D determined by filter binding.
| Protein | K D (nM) |
|---|---|
| RNase E (628–843) | 58±11 |
| RhlB | 48±13 |
| RhlB ΔCT | 193±54 |
| RNase E (628–843)–RhlB | 25±6 |
| RNase E (628–843)–RhlB ΔCT | 60±18 |
Dissociation constants were determined assuming simple one site binding. Each value is the average of at least two independent measurements. RhlB ΔCT lack the C-terminal basic region and corresponds to residues 1–397.
We also measured by filter binding the affinity of the complexes of RNase E (628-843)/RhlB (Figure 4(c)) and RNase E (628-843)/RhlB-ΔCT. Their apparent K D values were 25 nM and 60 nM, respectively (Table 1). In comparison, the KD value of RNase E (628-843) alone was 58 nM (Figure 4(c) and Table 1). The reduced affinity of the complex containing RhlB-ΔCT compared with the complex containing the fulllength helicase is consistent with the reduced affinity of the isolated RhlB-ΔCT. Note, however, that theaffinity of the individual components in the complexes of RNase E (628-843)/RhlB and RNase E (628-843)/RhlB-ΔCT is not additive. That is, if the components fully cooperate in RNA binding, the apparent KD of the complex should be much lower than measured here. This observation suggests that the helicase and the RNase E segment only partially cooperate to bind RNA in the absence of ATP.
Degradation of REP-containing mRNA by mini-degradosomes
Previous in vitro work showed that RhlB in the degradosome facilitates the degradation of structured RNA by PNPase.1,20 We examined whether the RBD and AR2 RNA-binding regions of RNase E participate in this RNA-degrading reaction. Using the procedures described by Khemici et al.,18 we reconstituted “mini-degradosomes” comprising the C-terminal portion of RNase E (residues 497 to 1061) with PNPase and helicase.
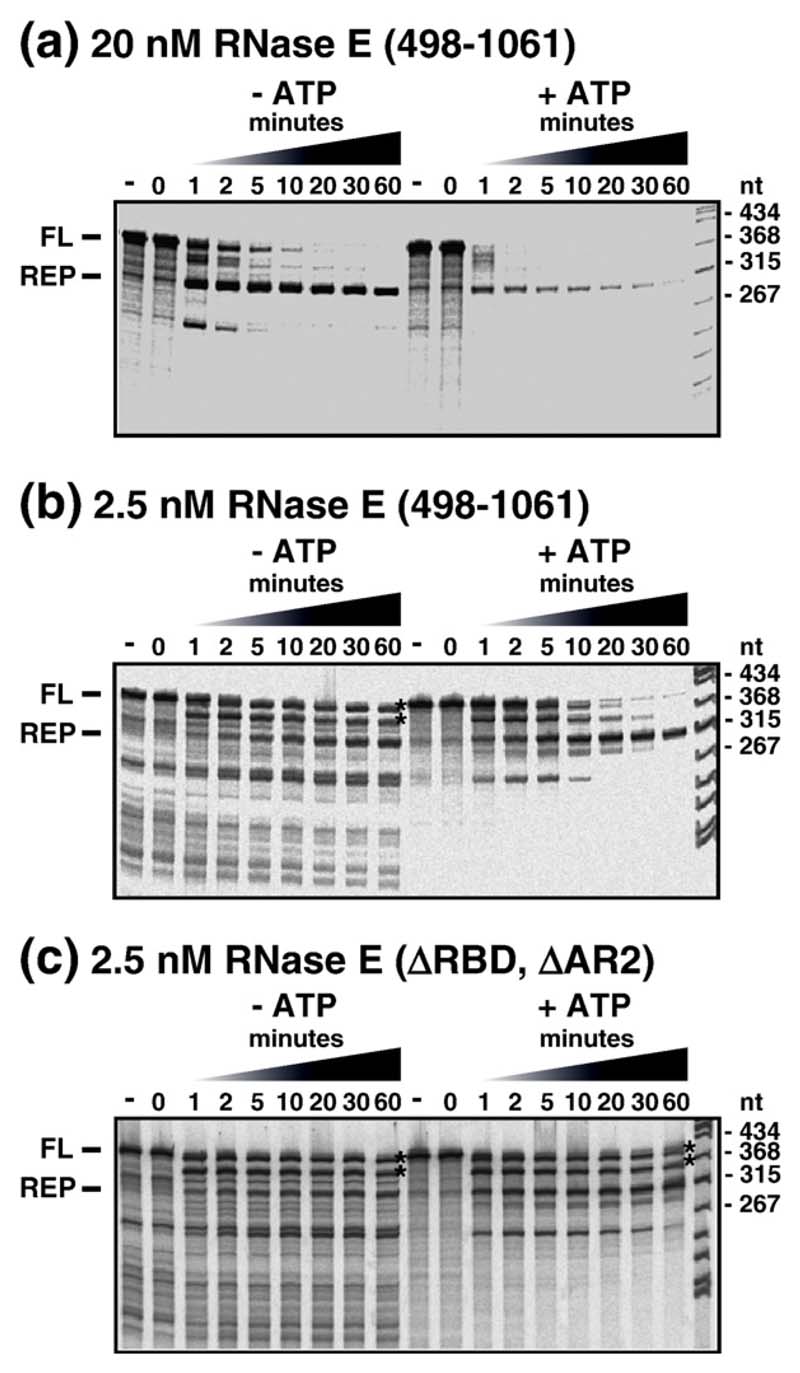
Figure 5 shows a series of RNA degradation experiments using mini-degradosomes and a small mRNA substrate derived from the malEF intergenic region, containing two repeated extragenic palindromic (REP) sequences, that when transcribed into RNA, fold into an extended RNA stem loop structure.21 The upper panel shows the wild-type control with intact RNase E (497-1061). In the absence of ATP, purified PNPase by itself or as part of the mini-degradosome pauses at the base of this stem loop producing the decay intermediate (REP) indicated to the left of the upper panel. Note that, in the time course to the left, most of the full length (FL) substrate has been attacked within 5 min, while substantial amounts of the REP intermediate accumulate and persist even after 60 min. Nevertheless, in a comparable reaction, but with ATP, the degradation of the full-length substrate is accelerated; there are lower levels of the REP intermediate, and most of the intermediate is degraded within 20 min. These results indicate that RhlB in the mini-degrado-some facilitates degradation of the REP structure by PNPase in a reaction that requires ATP hydrolysis, in agreement with earlier studies.1,20
Figure 5.
Helicase-facilitated RNA degradation in the mini-degradosome. Mini-degradosomes were employed in the degradation of an mRNA fragment derived from the intergenic region of the malEF operon containing two repetitive extragenic palindrome (REP) sequences (see the text). The panels show autoradiograms in which the products of digestion in a time course were separated by denaturing gel electrophoreses (6% (w/v) acrylamide, 29:1, 8 M urea). To the left of each panel, FL indicates the full-length substrate and REP, the intermediate in which degradation arrests a few nucleotides 3' of the REP structure. In each panel, two time courses are presented: in the absence of ATP (left) and in the presence of ATP (right). Mini-degradosomes were assembled from RhlB, PNPase and RNase E (498–1061) or its derivatives. (a) Degradation reaction using a mini-degradosome assembled from intact RNase E (498–1061). The concentration of the complex, based on PNPase content, was 20 nM in a reaction containing 20 nM substrate. (b and c) Degradation reactions with 2.5 nM complex and 20 nM substrate in which the conditions had been altered to reduce the activity of PNPase (see the text).
Mini-degradosomes were prepared in which the RBD, AR2 or both have been deleted from the RNase E (497-1061) polypeptide. The reconstitution of these derivatives with RhlB and PNPase was comparable to intact RNase E (497-1061) (data not shown). Thus, as expected, deletion of the RNA binding regions did not affect assembly of the complex. For each mutant, under the conditions employed in the experiment in the upper panel of Figure 5, the pattern of degradation in the absence or presence of ATP was comparable to the intact RNase E (497-1061) control (data not shown).
In modified reaction conditions, we employed the same salt and buffer conditions as in the RNA unwinding experiments in Figure 4, and we lowered the concentration of mini-degradosome from 20 nM to 2.5 nM and the concentration of inorganic phosphate from 10 mM to 1 mM. Under these conditions, the overall rate of degradation was significantly slower (Figure 5, middle panel). In the absence of ATP, a variety of decay intermediates persist for up to 60 min and the REP intermediate only slowly accumulates. This pattern suggests a loss of processivity and the accumulation of intermediates that are poor substrates for re-binding to PNPase. The asterisks after the 60 min lane indicate the positions of transient intermediates in degradation by PNPase that have been mapped previously.1,21 They correspond to RNA with short inverted repeats at the 3' end that can fold into modestly stable stem loops. Note that in the presence of ATP, after 20 min of incubation, most of the transient intermediates have disappeared and the REP intermediate, which has accumulated to significant levels, is essentially resistant to further degradation under these conditions. In comparable reactions with mini-degradosomes assembled from the RNase E (497-1061) lacking the RDB or AR2 domains, the pattern of degradation was comparable to wild-type, showing the deletion of either the RBD or AR2 did not affect this reaction (data not shown). However, with the mini-degradosome in which both domains were deleted, the transient intermediates persist even after 60 min in the presence of ATP (Figure 5, lower panel, asterisks). Thus, the deletion of both the RBD and AR2 impairs the capacity of RhlB to facilitate PNPase-mediated decay, at least under the limiting conditions used in these reactions. This observation suggests that the RNA-binding regions of RNase E cooperate with the helicase to process the RNA substrate.
Prediction of the recognition surface in RhlB, and the possible origins of ATPase activation
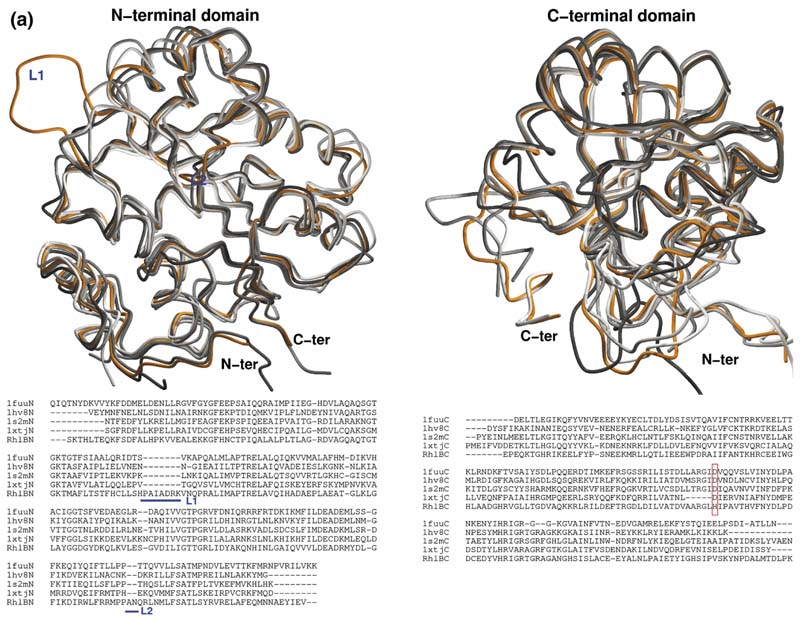
We prepared a model of the E. coli RhlB based on the crystal structures of its homologues. An overlay of the individual domains of the RhlB model with crystal structures of the four available DEAD box helicases reveals the location of insertions and deletions in loops of RhlB for both NTD and CTDs on what is otherwise a structurally similar core (Figure 6(a)). Calculations for potential ligand sites and protein partners predicted patterns involving surface sites corresponding to the conserved regions of the DEAD box helicase family (segments I to VI in Figure 1(b)) and are seen to be sites for recognition of ATP, RNA or domain-domain interaction in the cocrystal structure of the Drosophila Vasa DEAD-box helicase with single-stranded RNA (Figure 6(b)). Thus, we expect RhlB to engage the RNA and ATP and to make domain-domain interactions in a very similar fashion to the Drosophila Vasa enzyme, and we used that structure as a scaffold to generate a model of the complex of RhlB with RNA and ATP. However, this model reveals the potentially important substitution of a histidine for asparate at position 320 in box V of RhlB and its closest bacterial homologues (Figure 6(a), lower panel, highlighted by the red box). H320 could in principle be responsible for the activation of the helicase by the RNase E (694–768).
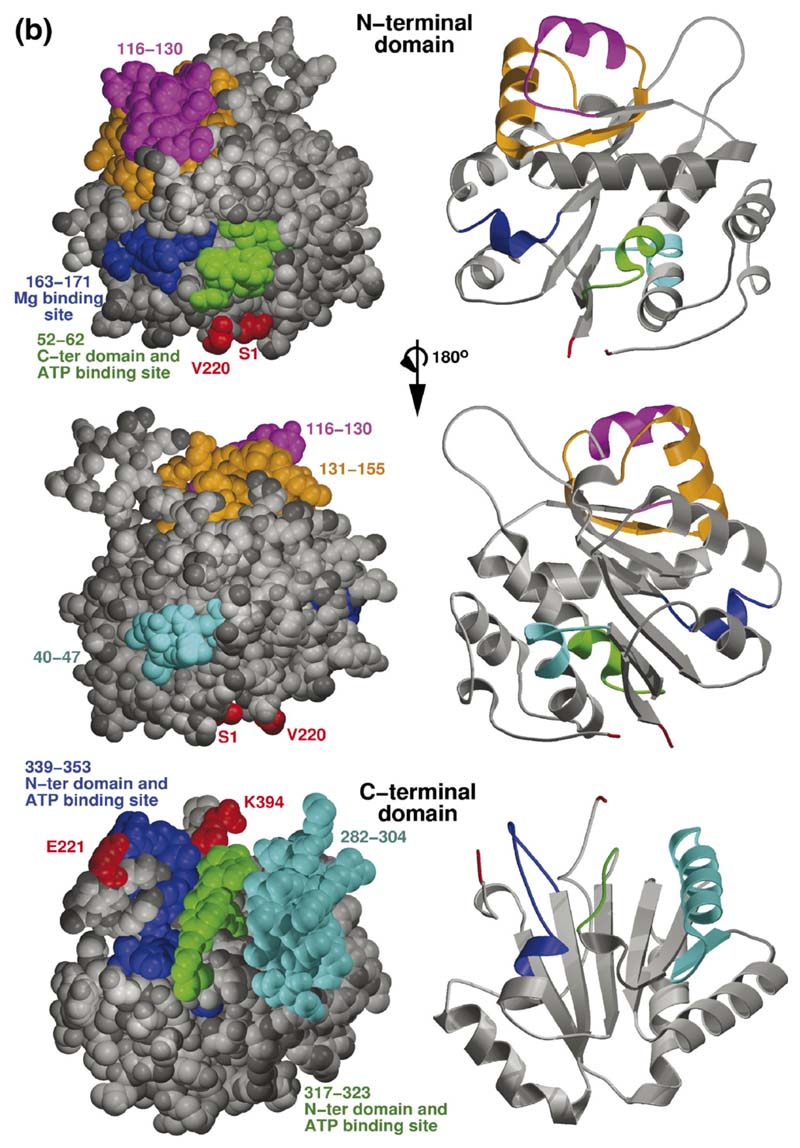
Figure 6. Homology model of RhlB and the predicted interaction site with RNase E.
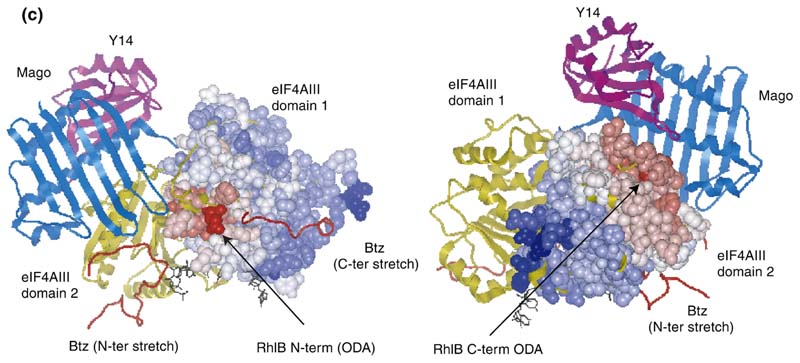
(a) Overlay of the independent domains with the templates (grey) and predicted models (orange) for the NTD and CTD of RhlB, indicating the differences in the loops. The inset shows the structure-based sequence alignment of the structural homologues, with the inserted loops indicated. The boxed region shows a conserved difference between RhlB and its homologues in motif V (see Supplementary Data, Figure S2 for sequence alignment of RhlB proteins from other Gram-negative bacteria in which the RNase E has a conserved potential helicase binding site). (b) Functional patterns on the surface of RhlB predicted by the program FPSPD.40 Three functional patterns are predicted on the surface of the C-terminal domain and five on the surface of the N-terminal domain. Pattern 52-62 is predicted to be involved in interactions with the C-terminal domain and ATP, and pattern 163-171 is most likely involved in interactions with the magnesium ion. Patterns 40-47,116-130 and 131-155 are probably not involved in domain interactions and may be involved in other interactions. Residues in red correspond to the N and C termini for each domain. (c) Comparison of the optimal docking area (ODA) identified on RhlB with the protein-protein interacting region on eIF4AIII in the exon junction complex (pdb code 2JOS).11 The left panel shows an overlay of the RhlB-NTD with eIF4AIII-domain 1 (RMSD 2.3 Å) and the right panel shows the overlay of RhlB-CTD with eIF4AIII-domain 2 (RMSD 3.3 Å). RhlB residues are shown as CPK spheres here. The ODA identified on both domains of RhlB is shown in red.
The surface of the RhlB domains were analysed with an algorithm that evaluates propensity for forming protein-protein interactions. The method, known as the optimal docking area (ODA), calculates the effective desolvation energy of burying a surface patch from the solvent, based on atomic solvation parameters optimized for protein-protein docking.22,23 While the ODA values found should be interpreted with caution, as some artefacts from modelling (for instance, over-exposed side-chains) could affect the calculations, they nonetheless can be a good indicator of the surface that is more likely to be interacting with other proteins. In particular, we note that the main ODA regions in the RhlB-CTD lie on a surface that has an exposed ridge of a β-sheet, corresponding to residues 230-235 (Figure 6(c)). This surface could form a sheet-like hydrogen bonding interaction with the recognition segment of RNase E, and the encircling ring of negatively charged residues could make electrostatic complementarity to the conserved basic residues of our identified helicase binding site in RNase E (see sequence alignment, Supplementary Data, Figure S2).
Finally, we note the occurrence of another putative region of interaction is an exposed coil in the vicinity of residues 377-383. The proposed role of this site in RhlB-RNase E interaction is in agreement with functional data. Results from E. coli two-hybrid studies showed that RhlB (residues 194-421) binds the RNase E while RhlB (residues 194-368) does not.24 These results, taken together with our observations that the tail (397-421) does not affect RNase E interaction, indicate that at least part of the binding must be contributed directly or indirectly by residues 368 to 397, which encompasses our identified potential interaction site. Residues in this coil are co-conserved with H320 amongst RhlB homologues, which we noted is the distinctive component of the motif V element that distinguishes RhlB from other helicases that do no require partners for activity. The co-variation suggests a link between RNase E binding on the one hand and the activation of the catalytic site, on the other, although the pathway of communication between these spatially distinct sites is not presently clear.
The crystal structure of the eukaryotic DEAD-box helicase eukaryotic initiation factor 4AIII (eIF4AIII) has been reported in complex with partner proteins of the exon junction complex.10,11 The surface of eIF4AIII that engages the partner proteins coincides with the predicted RhlB-RNase E interaction site (Figure 6(c)).
Discussion
RhlB is an important functional component of the RNA degradosome, and it is activated by the proteinprotein interactions with the carboxy-terminal portion of the ribonuclease RNase E. In this study, the molecular recognition and activation of RhlB by RNase E has been explored. Limited proteolysis locates the RhlB binding site to a region within residues 698-762 of RNase E. Conversely, a portion of the C-terminal RecA-like domain of RhlB has been identified here as being the key recognition site for RNase E using a variety of methods. The comparable stability of the complexes formed between RNase E (628-834) with full length RhlB and its isolated C-terminal domain implicates that domain as being the principle and probably the exclusive region of interaction between RNase E and RhlB.
Based on crystal structures of several homologues, a predicted model of RhlB was prepared (Figure 6). We have identified a site on the surface of the RhlB-CTD that may recognize the cognate RNase E peptide (indicated in Figure 6(c)). This predicted site of interaction is in agreement with the data from E. coli two-hybrid studies.20 The predicted interaction site uses a similar surface to that of eIF4AIII with its partner proteins in the exon junction complex (Figure 6(c)). In particular, sequence analysis highlights Y383 on this exposed surface that is likely to be one of the determinants for interaction with the RNase E. The identified surface includes an exposed ridge of a beta-sheet, which can in principle form an extended beta-sheet with a partner protein (for instance, as seen in the interaction of the TonB protein with its receptor, the TonB box25). Indeed, the helicase-recognition site in RNase E (698-762) has predicted propensity for forming beta-strands, one of which could be the target for the helicase. That segment also has predicted segments of alpha-helix, and the secondary structural elements may condense into a mixed alpha/beta-fold upon association with the helicase.
RhlB-CTD has been proposed to interact with polynucleotide phosphorylase to generate a compact RNA-degrading assembly that is in effect a subassembly of the RNA degradosome.24,26 The identified potential binding surface on RhlB-CTD might be involved in mediating this interaction, but further experiments are required to test if the binding of PNPase and RNase E might be exclusive, so that they compete for the same surface, or if they may share the site.
A model of the complex of RhlB with RNA and ATP was also generated using the structure of the Drosophila Vasa protein.9 This model indicates that the bacterial RhlB and the eukaryotic Vasa will share many details of recognition, with the single exception of residue H320 in box V of RhlB and its closest homologues. We propose that H320 could be partially responsible for the activation of the helicase by the RNase E (694-768) segment. The residue at this position is aspartate in the E. coli paralogues, SrmB and RhlE, which is striking because both these helicases have intrinsic ATPase activity by themselves and are not allosterically activated by their interactions with RNase E (at sites different from where RhlB binds).18
The surface of RhlB, which we propose to interact with RNase E, lies at a distance from the helicase's catalytic site (Figure 6(c)). Thus, it is not immediately clear how the interaction with RNase E would boost the ATPase activity of the RhlB. However, it seems possible that activation results indirectly through an allosteric effect on the RhlB-CTD that changes its RNA binding properties or the presentation of a conserved arginine residue in the CTD that could interact with the nucleotide engaged in the NTD, as occurs in the hexameric T7 helicase through subunit-subunit interactions.27
Both the helicase and the RNase E segment bind RNA, and the experiments in this study explored whether the proteins might associate with the nucleic acid cooperatively. In isolation, helicase and RNase E (628-834) will bind each other and RNA with apparent dissociation constants in the 50 nM range (Table 1). The truncated RhlB lacking an arginine-rich tail (RhlB-ΔCT) showed fourfold weaker RNA binding compared to full length RhlB, indicating that this tail contributes significantly to the energy of RNA binding. One surprising finding is that the affinity of the RhlB-RNase E (628-834) complex for structured RNA is not much greater than the isolated components. We would have expected their binding energies are additive, in the absence of positive or negative cooperativity. Taken together, the results suggest that the RNA binding activity of one component, most likely the helicase, is partially repressed in the protein-protein equilibrium complex; thus, the interaction is likely to represent negative cooperativity, with important mechanistic consequences.
Our results suggest that the RBD and AR2 RNA binding regions of RNase E facilitate RNA unwinding by RhlB. We observe a relatively higher unwinding rate for the RNase E (628-843)-RhlB complex versus the smaller RNase E (694-790)-RhlB that lacks both the RBD and AR2 domains. Since both RNase E (628-843) and RNase E (694-790) bind to RhlB with roughly the same affinity and stimulate the RNA-dependent ATPase activity to the same extent,18 the greater unwinding activity of the former is most probably attributable to the participation of the RNA-binding domains.
We explored the linkage between RNA unwinding by the RNase E-RhlB complex and phosphorolytic degradation of the substrate (Figure 5). Because the ribonuclease destroys the unwound, single-stranded product, the net unwinding process has proportionally less of the competing reverse reaction in which the single-strands re-anneal to regenerate the duplex substrate. Thus, RNA-unwinding is effectively coupled with RNA-degradation, and experiments could be performed with equimolar complex and substrate, or even with substrate excess. This contrasts with conditions used for many studies of other helicases, where helicase and ATP must be in great excess to drive the equilibrium in order to detect the unwinding products. Using this unwinding assay in the context of the mini-degradosome, we find a marked effect of ATP on the turnover of messenger RNA. Notably, this activity is optimal when the RNA binding domains of the RNase E are present, which suggests that they must work cooperatively with the helicase to unwind the substrate.
Studies of a number of DEAD-box helicases reveal that they require a single-stranded loading strand for unwinding activity and often recognise a defined polarity of the polynucleotide. While some helicases require a 5' single-stranded overhang, others require a 3' overhang, while yet others can unwind irrespective of the 5' or 3' orientation of the single strand.28–31 The specificity of RhlB for the 5'-tailed RNA duplex (Figure 4) implies 5'-to-3' directionality for RNA unwinding. In contrast, PNPase degrades RNA in the reverse direction, 3'-to-5', and the opposing polarities of helicase and PNPase might seem incompatible. However, if PNPase pauses on a 3' single strand downstream of a stable RNA duplex, RhlB could bind to the 5' single strand upstream of the duplex, and RhlB could then unwind one or a few base-pairs to expose a fresh 3' single-stranded segment for 3' to 5' degradation by PNPase. Since most mRNA substrates will contain a mixture of single and double-stranded regions, PNPase can act alone on the single stranded regions. This is consistent with a large body of literature that concludes that PNPase by itself can efficiently degrade unstructured polynucleotides in a processive reaction.32 Since most RNAs would not be expected to have extensive base-paired regions, the unwinding of one or a few base-pairs by RhlB should be sufficient to disrupt most structures. Thus, RhlB would be acting “locally”, since its specificity would be for RNA duplexes with 5' single-stranded extensions that are bound to PNPase.
It may also be considered that interaction with especially well folded RNA serves to check the fidelity for the fold of structured RNA substrates that will be processed by the RNase E, such as precursors of ribosomal RNA, tRNA and tmRNA. If they are not properly folded, they might be unfolded during the interplay of RNA binding by the helicase and the AR2 and RBD segments, and thus continue on to a fate of destruction. We speculate that properly folded substrates may withstand the opposing forces and thus be released by a translocase effect of the helicase, after appropriate processing by the catalytic domain of the RNase E internally, or trimming at the 3' end by the activity of PNPase.
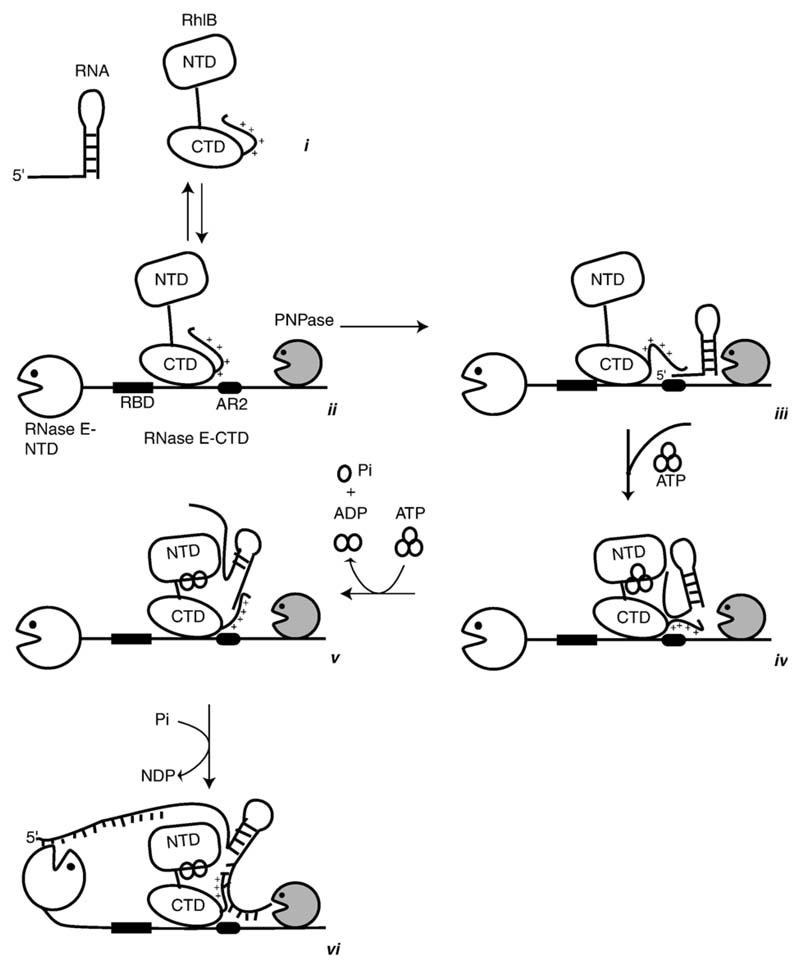
We interpret our findings in a speculative model for the sequential process of RNA unwinding and degradation or processing in the RNA degradosome, as shown in the idealised schematic in Figure 7. The model is based in part on our hypothesis that the interactions with RNase E suppress the RNA binding activity of RhlB by negative cooperativity, but enhance its ATPase and unwinding activities by positive cooperativity. In the first stages of the proposed process, RNA is bound at either the RBD or AR2 sites of RNase E, perhaps depending on the fold of the substrate. In this resting state, when the ATP is absent, the basic C-terminal tail of the helicase can interact with the RNA. With ATP binding, the helicase becomes activated perhaps by allostery, so that now the surfaces of the NTD and CTD of the helicase can recruit the substrate. With ATP hydrolysis, the NTD and CTD rock apart, causing the duplex to unwind. The new single-stranded 5' region re-engages somewhere on the surface of the helicase. With each successive cycle of ATP binding and hydrolysis, the substrate translocates. The unfolded RNA is now a substrate for the 3'-to-5' phosphorolytic activity of the PNPase. In the context of the degradosome, the exposed single-stranded regions may now become accessible for internal cleavage by the endonucleolytic activity of the RNase E catalytic domain.33
Figure 7.
A model for the sequential steps in the unwinding and translation mechanism of the cooperating helicase-ribonuclease assembly. The helicase is envisaged as a two-domain structure with a flexible inter-domain linker and positively charged C-terminal tail. The isolated helicase interacts with RNA (i), but may interact only weakly while engaged to RNase E (ii). In the cartoon, PNPase is also engaged with RNase E (ii). RNA may bind to RNase E at RBD or AR2 (as indicated here by interaction with the stem-loop RNA structure, (iii) or both. Cycles of ATP binding and hydrolysis may expose the surfaces of the RecA-like domains of RhlB to interact with the RNA and stabilise its interaction during unwinding and 5' to 3' translocation (iv) and (v). The exposed terminal regions of the RNA are subject to phosphorolytic cleavage by the adjacent PNPase or hydrolytic attack internally by the catalytic domain of RNase E (vi). See the text for further narrative.
Materials and Methods
Expression and purification of RhlB, RNase E (628–843) and RNase E (694–790)
RhlB and RNase E (628-843) were expressed and purified as described by Callaghan et al.12 For the helicase unwinding assays, extracts of overexpressed RNase E (628-843) and RNase E (694-790) were prepared as described by Vanzo et al.17 The proteins from high speed supernatants, prepared by successive centrifugations (15 min at 10,000g then 1 h at 260,000g), were precipitated with ammonium sulphate (40% saturation for RNase E (628-843) and 60% for RNase E (694-790)).19 The proteins were further purified from the resuspended pellet by exclusion chromatography on Sephacryl S-200 (Pharmacia). Peak fractions were pooled and dialysed against buffer A containing 50 mM NaCl. The RNase E (628-843) polypeptide was applied to S-Sepharose (Pharmacia) and eluted with 450 mM NaCl. The RNase E (694-790) polypeptide was purified on a Heparin-agarose colomn (Pharmacia) and eluted with 150 mM NaCl. Both proteins were judged to be more than 90% pure by silver stained SDS/PAGE and were quantified by protein assay (BioRad) with a bovine serum albumin standard.
Cloning, expression and purification of RhlB-CTD, RhlB-NTD and truncated RhlB
DNA fragments encoding the RhlB NTD (1-226) and CTD (230-421) were amplified from pRHLB1 using FailSafe (Epicentre), with primers adding upstream NdeI and downstream BamHI restriction sites, and cloned into pCRII-TOPO by the Topo TA method (Invitrogen). The recombinant plasmids were digested with NdeI and BamHI and the RhlB reading frame containing fragments purified and ligated into NdeI/BamHI digested pET20b (Novagen). Vectors for T7 RNA polymerase-dependent overproduction34 of the RhlB NTD (pGA41) and CTD (pGA43), lacking the pET20b encoded N-terminal pelB leader and C-terminal His-Tag, were confirmed by DNA sequencing. BL21 DE3 was transformed by electroporation, and was used to prepare cultures in YT and 2xYT broth supplemented with carbenicillin at 0.1 mg/ml. Expression of the RhlB-CTD was induced at A600 nm of 0.6 with 1 mM IPTG, and the culture was incubated at 37 °C for a further 3 h. Cells were harvested by centrifugation at 4000g for 15 minutes at 4 °C, and the pellet resuspended in cell lysis buffer (50 mM bis-Tris-HCl (pH 7.0), 0.1 mM EDTA, 1 mg/ml of lysozyme, 3 mM DTT). DNase I was added and the cells were ruptured with an emulsifier. The cell debris was removed by centrifugation at 14,000g for 30 min at 4 °C, and the supernatant was dialysed against buffer H (50 mM Hepes (pH 6.5) with 100 mM NaCl) at 4 °C using a membrane with a pore size of 7 kDa. The dialysed supernatant was applied to a HiTrap SP HP column equilibrated with buffer H containing 100 mM NaCl, and the chromatogram was developed with a linear gradient of buffer H from 100 mM NaCl to 1 M NaCl. Samples were analysed by SDS/PAGE, and fractions enriched in RhlB-CTD were re-applied to the HiTrap SP HP column and resolved by using a shallower salt gradient with buffer H from 100 mM NaCl to 600 mM NaCl. Fractions containing the purest RhlB-CTD were pooled, aliquoted and stored at -20 °C. The purified sample was evaluated by ten cycles of N-terminal sequencing (PNAC facility, Department of Biochemistry) to confirm the identity of the protein. N-terminal sequencing gave the expected N-terminal sequence of MRIKEELFYP, confirming the identity of the protein.
For the RhlB-NTD, expression was carried out as described above with clones transformed with plasmid DNA pGA41 with RhlB-NTD (1-266) in pET20b. The soluble fraction after cell lysis predominantly contained the RhlB-NTD protein and was used for the RNA binding experiments. Truncated helicase RhlB (residues 1-397) was cloned into pET11 vector and transformed in E. coli BL21(DE3) cells. The expression protocol was the same as for the full length RhlB described above. Truncated helicase was precipitated from the cell lysate by adding NH4SO4 to 60% saturation. The sample was continuously stirred at 4 °C for 1 h and then spun at 30,000g for 40 min. The pellet was resuspended in buffer M (20 mM Mes (pH 6.5), 300 mM NaCl, 2 mM DTT). The protein was desalted into buffer B (20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 2 mM DTT, 10% (v/v) glycerol) and loaded on a Mono Q column for further purification. A linear gradient from 0% to 25% of elution buffer (buffer B+ 2 M NaCl) was used to elute the protein.
Size exclusion chromatography
A 1:1 stoichiometric complex of RNase E with RhlB-CTD was formed by mixing the purified recombinant components, and 200 μl of the complex at 1 mg/ml concentration was injected onto Superdex HiLoad 16/60 S75 and S200 analytical gel filtration columns pre-equilibrated with 50 mM Tris-HCl (pH 7.5), 100 mM NaCl buffer. Control experiments with the individual proteins were also carried out.
Limited proteolysis
Proteolytic reactions were initiated by the addition of sequencing grade trypsin (final concentration of 1 μg/ml; Roche), corresponding to 1/1000 of the target protein concentration (1 mg/ml) in 5 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM CaCl2,10 mM MgCl2 and 1 mM DTT. Samples were collected at regular intervals and either mixed 1 : 1 with Tris-glycine native sample buffer and stored at -20 °C until native-PAGE analysis or the reactions were quenched by addition of 1 : 1 of reducing and denaturing sample buffer (NuPAGE), followed by incubation at 95 °C for three minutes and frozen until SDS/PAGE analysis. The resolved limited digest products were analysed by in-gel protease digestion and MALDI fingerprinting in the PNAC facility, Department of Biochemistry, Cambridge, UK.
Chemical cross-linking of RhlB-CTD and RNase E (628–843)
RhlB-CTD and RNase E (628-843) were diluted to a concentration of approximately 0.5 mg/ml. DMS at 10 mM concentration was added to each of the samples (RNase E (628-843) alone, RhlB-CTD only, and RhlB-CTD and RNase E (628-843)), and time points taken at 30 min, 60 min, 120 min and 180 min after addition of DMS. The various samples were mixed with 6× loading buffer (80 mM Tris-HCl (pH 6.5), 100 mM DTT, 2% (w/v) SDS, 15% glycerol) and boiled for 3 min to quench the crosslinking reaction. Samples were analysed by SDS/PAGE. The experiment was repeated with DMS concentrations of 20 mM and 30 mM, and with enolase with DMS at a concentration of 10 mM. For glutaraldehyde cross-linking, 10 μl of 1% (v/v) glutaraldehyde was added to 90 μl of sample and time points taken as before. For the crosslinking in triethanolamine, samples of RNase E (628-843), RhlB-CTD, enolase and full length RhlB were buffer exchanged into 0.2 M triethanolamine (pH 8.0) using BioRad Bio-Spin 6 columns. The samples were then diluted to a protein concentration of approximately 0.5 mg/ml using 0.2 M triethanolamine (pH 8.0).
RNA binding assays by electrophoretic mobility shift
Frozen protein stocks (except full length mutant RNase E and degradosome sub-complexes) were thawed on ice and diluted 40-fold with dilution buffer (50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10% (v/v) glycerol, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 2 μg/ml of aprotinin, 0.8 μg/ml of pepstatin A and 0.8 μg/ml of leupeptin) which was pre-warmed to 30 °C. Proteins were incubated in ice for 5 min and then clarified by centrifugation at 14,000g for 20 min at 4 °C. Triton X-100 was added to a final concentration of 0.2% to the supernatant, and frozen at -20 °C in 50 μl aliquots. Protein concentrations were determined by running a range of protein dilutions along with standards of bovine serum albumin (100-1200 ng) on an 8% SDS/PAGE gel. Gels were run at 8 mA for around 16 h. Gels were stained with SYPRO orange (Interchim) and imaged using a Molecular Dynamics FluorImager.
Radiolabelled RNA was prepared from DNA templates by in vitro transcription using labeled UTP or CTP. All reagents used were from Promega. In vitro transcription reaction was setup with 5.5 μl of diethyl pyrocarbonate-treated water, 10 μl of 5× transcription buffer, 5 μl of 0.1 M DTT, 1.5 μl of 40 units/μl Rnasin,10 μl of 5×NTP mix (2.5 mM ATP, CTP, GTP), 5 μl of 0.5 mM rUTP, 10 μl of [33P]UTP (10 μCi/μl) 2 μl of 20 ng/μl of T7 PCR template, 1 μl of T7 RNA polymerase (80units/μl). The reaction mix was incubated at 37 °C for 1 h. One μl of T7 RNA polymerase was added again to the reaction and incubated at 37 °C for 1 h. Six μl of 10×RQ1 DNase buffer and 4 μl of 1unit/μl of RQ1 DNase was added and incubated for 15 min at 37 °C. Finally 40 μl of diethyl pyrocarbonate-treated water was added and extraction was done using phenol/chloroform/isoamyl alcohol followed by purification with a G-50 column (Pharmacia). Protein aliquots after initial dilution were thawed in ice and spun at 14,000g for 20 min and the supernatant was used to make various dilutions. The reaction buffer was 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10% glycerol, 0.1% (v/v) Genapol, 1 mM EDTA, 1 mM DTT and 40 units of RNasin. Reactions were set up with 0.4 nM RNA in each reaction. Reactions were set up in ice and then incubated at 30 °C for 10 min before loading the gel. For the acrylamide gel electrophoresis, all gel plates, combs, spacers and gel boxes were washed extensively with detergent or bleach and then soaked in 0.1 M NaOH and 1 mM EDTA for 20 min and finally rinsed with MilliQ water. Samples were loaded on a 5% (w/v) native polyacrylamide gel under tension (200 V). The running buffer used was 0.5× TBE (10 mM Tris-borate, 1 mM EDTA (pH7.0)) with 0.1% Triton X-100. Gels were ran in the cold room at 200 V for 2-4 h and then dried on a gel dryer.
RNA-binding assays by filter binding
We used the double-filter binding procedure as described by Amblar et al.35 Radiolabelled RNA was prepared as described in the previous section. Briefly, 0.5 fmol of 33P-labelled RNA was incubated with serial dilutions of protein in 60 μl of binding buffer (100 mM KCl, 20 mM Tris-HCl (pH 8), 2 mM DTT, 10 mM EDTA, 10% glycerol) for 15 min at room temperature. Reactions were chilled on ice and subjected to 254 nm UV crosslinking for ten cycles of 1 min exposures and 1 min pauses. The reactions were filtered at room temperature; the filters were separated, dried and quantified by phosphorimaging. In this method, the first filter (nitrocellulose) binds protein and the second filter binds free RNA (Hybond N+). These data were fitted for a simple single binding site model using Origin software.
RNA helicase assay
Self-annealing RNA oligonucleotides were synthesized with an Applied Biosystems automated synthesizer. The sequence of the oligonucleotide for the 5' single strand end substrate is 5'-GAAUGUACAUCA GAGUGCGCACUC-3'. The oligonucleotide used for the 3' single strand ends substrate is 5'-GAGUGCGCACUCGAAUGUACAUCA-3' (the underlined nucleotides form the duplex). The oligonucleotides were phosphorylated with T4 polynucleotide kinase (Biolabs) or 3'-labelled with pCp using T4 RNA ligase (Biolabs) and purified on a Sephadex G-25 column (Pharmacia). Specific activity of the two substrates was the same, ~4×106 cpm/pmol for 5'-labelling and ~ 2×106 cpm/pmol for 3'-labelling. The labelled oligonucleotides were further purified by electrophoresis on a 12% DNA sequencing gel (0.5×TBE). Oligonucleotides were eluted from gel with an elution buffer (500 mM ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, 0.1% SDS). After a brief centrifugation to remove gel fragments, the oligonucleotides were precipitated with 2.5 volumes of 100% ethanol and the pellets resuspended in 50 μl of hybridization buffer (20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 1 mM EDTA). The oligonucleotides were heated at 90 °C for 3 min then hybridized overnight at room temperature (more than 90% of the RNA forms duplexes).
The unwinding activity of RhlB was assayed in vitro by measuring the conversion of the 12-mer RNA duplexes to monomers. The 12 bp RNA helices have a predicted ΔG value of -22.2 kcal/mol (37 °C) and Tm of 48 °C (0.1 nM).36 Purified proteins were incubated with 6 fmol (0.1 nM) of duplex in a 50 μl reaction containing 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 60 mM KCl, 40 mg of bovine serum albumin (BSA), 100 ng of yeast tRNA, 50 units of RNasin and when indicated 2 mM ATP or ATPγS. After incubation at 37 °C, reactions were stopped by the addition of 50 μl of 20% glycerol, 0.2% SDS, 4 mM EDTA and analyzed directly by electrophoresis on a native 0.5×TBE-12% polyacrylamide gel (29:1). The gel was dried and RNA visualized by autoradiography. The ratio of single-stranded products to duplex substrates was quantified using a Fuji Bas 1000 Phosphorimager.
Reconstitution of mini-degradosomes and RNA degradation assays
Mini-degradosomes were prepared as described by Khemici et al.18 Digestion assays of the 33P-labelled 375 nt malE-malF intergenic region RNA (REP RNA) were performed as described,1 with minor changes. KCl was removed from the reaction buffer; 5 mM DTT and 40 units of RNasin were added. Degradosome reconstitutes containing 200 ng PNPase were diluted in elution buffer with protease inhibitors and 2 μl was added to 38 μl reactions at 30 °C. Four μl aliquots were treated with 0.25 mg/ml proteinase K, then diluted into formamide/urea/dye mix. The radioactive RNA was visualized with a Phosphorimager (Fuji). For a typical RNase E (497-1061) reconstitution, reactions contained roughly 60 ng of RNase E (497-1061), 200 ng of PNPase (20 nM) and 120 ng of RhlB.
Homology modelling and surface calculations
The closest structural homologues were identified by sequence profile library searching against the structural homologue database HOMSTRAD,37 using the program FUGUE.38 A three dimensional structure was prepared using the DEAD-box proteins from Methanococcus janna-shii (Protein Data Bank (pdb) code 1HV8) and Sacchar-omyces cerevisiae (pdb:1S2 M), the initiation factor 4A from Saccharomyces cerevisiae (pdb code 1FUU) and the human UAP 56 (pdb code 1XTJ) as structural templates with the program MODELLER.39 The model of the complex with single-stranded RNA and ATP was based on the Drosophila Vasa structure (pdb code 2DB3). The functional signature search was carried out using the program FPSPD.40 The optimal docking area (ODA) computations used the procedure of Fernandez-Recio et al.22,23 Secondary structure predictions were made using JPRED†.
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2006.12.014
Acknowledgements
We thank Louise Matheson and Ola Zaid for help with the protease digestion experiments, Simon Donhauser for help preparing the domains of RhlB and Ping Ling for technical assistance. We thank John Lester for DNA sequencing, Len Packman for peptide analyses by mass spectrometry and Mike Weldon for peptide amino acid sequencing. We thank Nick Crump and Jonathan Worrall for helpful discussions. The Wellcome Trust (to B.F.L.), the CNRS (to A.J.C.) and the National Science Fondation (grant no. MCB-0344286 to C.M.B.) supported this work with additional funding from the Fundamental Microbiology Program of the MENRT (to A.J.C.) and the Franco-British Alliance Program (to A.J.C. and B.F.L.). V.C. was supported by a Nehru Cambridge Scholarship.
Abbreviations used
- AR2
arginine-rich region 2 of RNase E (EC 3.1.26), corresponding to residues 798 to 819
- DMS
dimethylsuberimidate
- ODA
optimal docking area
- PNPase
polynucleotide phosphorylase (EC 2.7.7.8)
- RBD
RNA-binding domain of RNase E, corresponding to residues 604 to 688
- REP
repetitive extragenic palindrome
- RhlB
RNA helicase B (EC 3.6.1)
- RhlB-CTD
carboxy terminal domain of RhlB helicase, corresponding to residues 267 to 421
- RhlB-NTD
amino terminal domain of RhlB helicase, corresponding to residues 1 to 266
- RhlB-ΔCT
derivative of RhlB lacking the arginine-rich carboxy-terminal residues 398 to 421
- RNase E (628–843)
a segment of ribonuclease RNase E corresponding to residues 628 to 843
- RNase E (498–1061)
segment of RNase E containing residues 1–26 and 498 to 1061
- RNase E (ΔRBD, ΔAR2)
is a derivative of RNase E (498–1061) in which the RBD and AR2 have been deleted
Footnotes
References
- 1.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 2.Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. Proteins associated with the RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DexH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 5.Caruthers JM, McKay DB. Helicase structure and mechanisms. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 6.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii . Proc Natl Acad Sci USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci USA. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEADbox protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 10.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 12.Callaghan AJ, Aurriko JP, Grossmann JG, Kühnel K, Poljak L, et al. Studies of the RNA degradosome-organising domain of the Escherichia coli RNase E. J Mol Biol. 2004;340:965–979. doi: 10.1016/j.jmb.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 13.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 14.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers GW, Jr, Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucl Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 16.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 17.Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, et al. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol Microbiol. 2004;54:1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- 19.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 20.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3’ exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaren RS, Newbury SF, Dance GS, Causton HC, Higgins CF. mRNA degradation by processive 3’-5’ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- 22.Fernandez-Recio J, Totrov M, Abagyan R. Identification of protein-protein interaction sites from docking energy landscapes. J Mol Biol. 2004;335:843–865. doi: 10.1016/j.jmb.2003.10.069. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Recio J, Totrov M, Skorodumov C, Abagyan R. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins: Struct Funct Genet. 2005;58:134–143. doi: 10.1002/prot.20285. [DOI] [PubMed] [Google Scholar]
- 24.Liou G-G, Chang H-Y, Lin C-S, Lin-Chao S. DEAD box RHlB helicase physically associates with exoribonuclease PNPase to degrade doublestranded RNA independent of the degradosome-assembling region of RNase E. J Biol Chem. 2002;277:41157–41162. doi: 10.1074/jbc.M206618200. [DOI] [PubMed] [Google Scholar]
- 25.Shultis DD, Purdy MD, Banchis CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 26.Lin PH, Lin-Chao S. RhlB helicase rather than enolase is the beta-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)-exoribonuclease complex. Proc Natl Acad Sci USA. 2005;102:16590–16595. doi: 10.1073/pnas.0500994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 28.Rogers GWJ, Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 29.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nature Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277:12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- 31.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 32.Littauer UZ, Soreq H. Polynucleotide Phosphorylase. 3rd edit. Academic Press; New York: 1982. [Google Scholar]
- 33.Khemici V, Poljak L, Toesca I, Carpousis AJ. Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNase E. Proc Natl Acad Sci USA. 2005;102:6913–6918. doi: 10.1073/pnas.0501129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 35.Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J Mol Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 36.Xia T, Santa Lucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 37.deBakker PIW, Bateman A, Burke DF, Miguel RN, Mizuguchi K, Shi J, et al. HOMSTRAD: adding sequence information to structure-based alignments of homologous protein families. Bioinformatics. 2001;17:748–749. doi: 10.1093/bioinformatics/17.8.748. [DOI] [PubMed] [Google Scholar]
- 38.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 39.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 40.Miguel RN. Sequence patterns derived from the automated prediction of functional residues in structurally-aligned homologous protein families. Bioinformatics. 2004;20:2380–2389. doi: 10.1093/bioinformatics/bth255. [DOI] [PubMed] [Google Scholar]
- 41.Leroy A, Vanzo NF, Sousa S, Dreyfus M, Carpousis AJ. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol Microbiol. 2002;45:1231–1243. doi: 10.1046/j.1365-2958.2002.03104.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.