Abstract
The thiamine-dependent E1o component (EC 1.2.4.2) of the 2-oxoglutarate dehydrogenase complex catalyses a rate-limiting step of the tricarboxylic acid cycle (TCA) of aerobically respiring organisms. We describe the crystal structure of Escherichia coli E1o in its apo and holo forms at 2.6 Å and 3.5 Å resolution, respectively. The structures reveal the characteristic fold that binds thiamine diphosphate and resemble closely the α2ß2 heterotetrameric E1 components of other 2-oxo acid dehydrogenase complexes, except that in E1o, the α and β subunits are fused as a single polypeptide. The extended segment that links the α-like and β-like domains forms a pocket occupied by AMP, which is recognised specifically. Also distinctive to E1o are N-terminal extensions to the core fold, and which may mediate interactions with other components of the 2-oxoglutarate dehydrogenase multienzyme complex. The active site pocket contains a group of three histidine residues and one serine that appear to confer substrate specificity and the capacity to accommodate the TCA metabolite oxaloacetate. Oxaloacetate inhibits E1o activity at physiological concentrations, and we suggest that the inhibition may allow coordinated activity within the TCA cycle. We discuss the implications for metabolic control in facultative anaerobes, and for energy homeostasis of the mammalian brain.
Keywords: thiamine diphosphate, dehydrogenase, E1o subunit, citric acid, metabolic regulation
Introduction
For all eukaryotes, archaea, and most eubacteria, the metabolic “hub” for both catabolic and anabolic processes that provides key metabolic intermediates, and a vital resource of energy, is the citric acid/ tricarboxylic acid (TCA) cycle, also known as the Krebs cycle. The route of metabolites through the cycle is affected by factors such as available carbon source and oxidative state (e.g. aerobic, anaerobic and oxidative stress).1 Control within the cycle is exerted at several points, especially at the reactions that are far from equilibrium, such as those catalysed by isocitrate dehydrogenase2 and the 2-oxoglutarate dehydrogenase (OGDH) multienzyme complex.3 The OGDH complex is largely rate-limiting in the TCA cycle in many species, and is a crucial nexus in the utilization of sugar and amino acids, which are “funnelled” via 2-oxoglutarate and oxidized to succinyl-CoA, releasing CO2 and reducing NAD+ to NADH. The importance of the OGDH complex in highly active tissues is highlighted by its selective inactivation in the pathologies of several neurode-generative disorders, particularly those associated with oxidative stress of mitochondria, such as Alzheimer’s and Parkinson’s diseases.4–6
The OGDH complex is comprised of multiple copies of three types of component enzymes: a 2-oxoglutarate decarboxylase dependent on thiamine diphosphate (E1o, EC 1.2.4.2), a lipoylated succinyl transferase (E2o, EC 2.3.1.6), and a dihydrolipoyl dehydrogenase (E3, EC 1.8.1.4). The quaternary organisation of the OGDH complex ensures that the products of one reaction are efficiently shuttled to the next active site for the subsequent reaction.7 The three-stage catalytic mechanism of the OGDH multienzyme complex (as depicted in Figure 1(a)) is related to that of other 2-oxo acid dehydrogenase complexes (such as pyruvate dehydrogenase, PDH; and branched-chain 2-oxo acid dehydrogenases, BCDH) and extensive investigations into these complexes have also revealed corresponding similarities in their quaternary organisation.8–10
Figure 1.
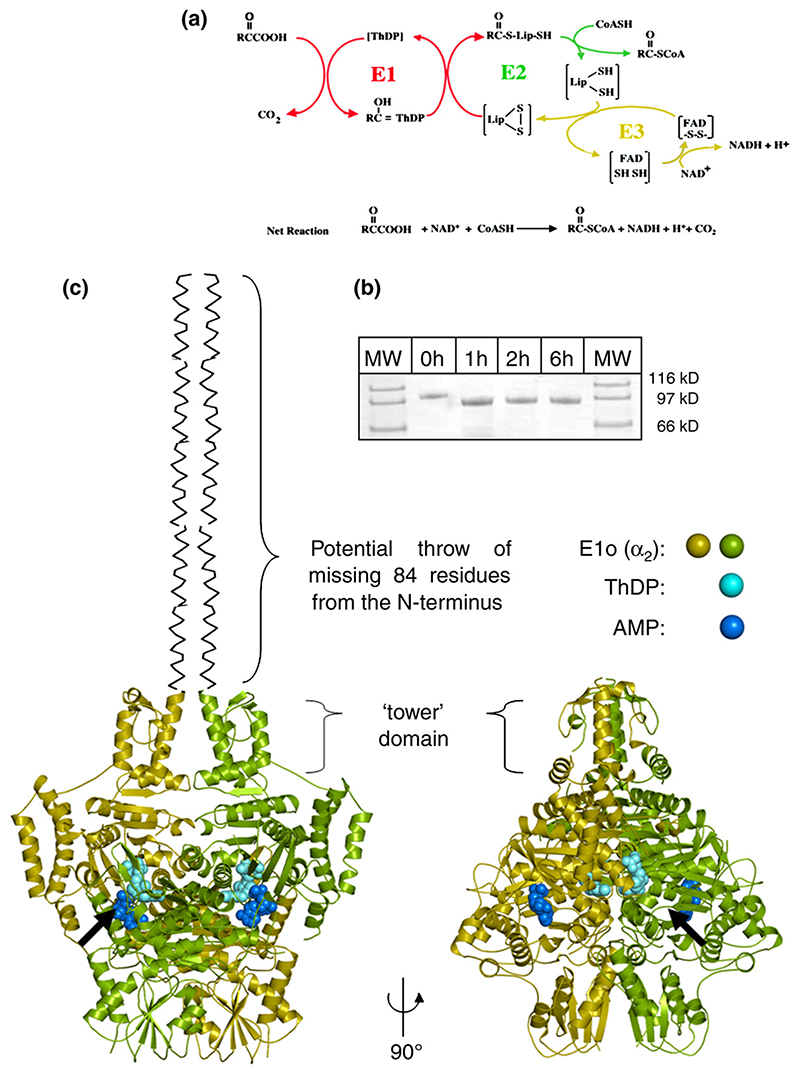
(a) The reaction catalysed by the 2-oxoglutarate dehydrogenase (OGDH) multienzyme assembly. The three component enzymes of the OGDH assembly function in series: E1o is a ThDP-dependent 2-oxoglutarate decarboxylase, E2o a lipoylated succinyltransferase and E3 is a dihydrolipoyl dehydrogenase. The reactions have been colour-coded for each of the three separate enzymes catalysing the three stages. (b) SDS-PAGE showing (1:100 (w/w)) trypsin treatment of E. coli E1o at 0 °C. (c) Two views of E. coli 2-oxoglutarate dehydrogenase E1 component (E1o). E1o is composed of two subunits (green and gold ribbons). Each subunit contains a Mg2+ and ThDP coordinated in the active site (cyan spheres). An AMP is bound in a separate binding pocket (blue spheres). Black arrow indicates the entrance of the active site in the foreground. The 77 N-terminal residues of E1o were removed by limited digestion with trypsin before crystallization. Binding studies indicate this region of sequence is responsible for assembly with the E2o core (Supplementary Data).
In almost all instances, E1 and E3 are recruited into their multi-enzyme complexes through direct interaction with E2, which is an oligomeric enzyme with structurally distinct domains that are linked by flexible regions of polypeptide chain of 28-50 residues that are rich in Ala and Pro. In eukaryotes, however, an additional E2-like protein (referred variously as E3BP or protein X) is required to specifically bind E3. Although the organisation of the three component enzymes is crucial for the activity of the complex, various architectures have evolved for the dehydrogenases, though it is not clear if these differences affect function.7 Thus, the E2 assembles into either a 24-mer with octahedral symmetry or a 60-mer with icosahedral symmetry, depending on the organism. All BCDH, OGDH, and most PDH E2 from Gram-negative bacteria associate to form 24-mers,11 whereas in eukaryotes and Grampositive bacteria, the PDH E2 are icosahedral 60-mers.12 The multiplicity of E2 subunits dictates the upper limit for the number of E1 and E3 that may join the complex, giving rise to ordered clusters of 5×106 to 11 × 106 Da.13 Puzzlingly, fewer E1o bind per E2o in the OGDH complex compared with the other 2-oxo acid dehydrogenase multienzyme complexes14(Supplementary Data).
Structures determined by crystallography are available for each globular component of the OGDH multienzyme complex, with the single exception of E1o.15–18 Moreover, at least one crystal structure is available for all other enzymes that constitute the TCA cycle. We present here the crystal structure and functional analyses of the Escherichia coli 2-oxoglutarate decarboxylase, revealing the molecular details of this last structurally characterised component of the TCA cycle. The structure reveals a homo-dimer in which the protomers have the recurring fold characteristic of thiamine-dependent enzymes; however, the E1o differs by the inclusion of specialized ligandbinding pockets, whose function we explore.
Results
Tertiary and quaternary structure
We were unable to identify crystallization conditions for purified recombinant E. coli E1o (210 kDa) after extensive screening. However, crystals were readily obtained from a 190 kDa trimmed E1o (tE1o) product lacking the first 77 residues from its N terminus that was prepared by treatment with trypsin (Figure 1(b)). tE1o also retained the decarboxylase catalytic activity of the full-length E1o enzyme but failed to assemble with its complementary binding partner, E2o, into an OGDH multienzyme complex19,20(Supplementary Data).This suggests that the N-terminal fragment removed by trypsin plays a role in mediating interactions between E1o and E2o (vide infra).
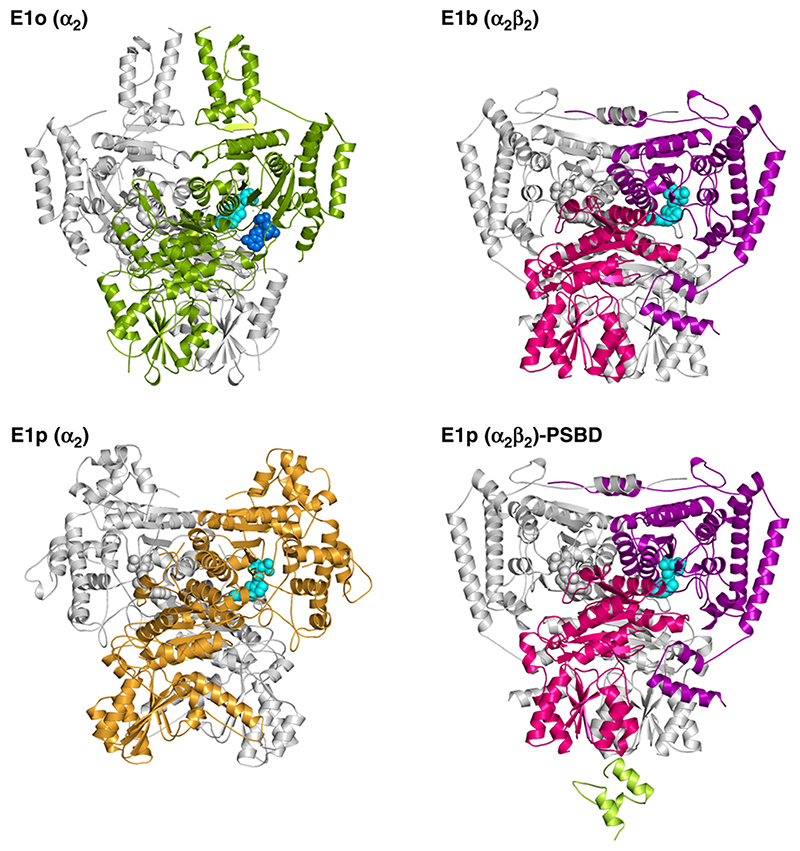
The structure was solved by multiple wavelength anomalous dispersion (MAD) (Table 1). Consistent with its behaviour in solution, tE1o forms a homodimer in the crystal, which is also the content of the crystal asymmetric unit (Figure 1(c)). Despite the absence of any discernible conservation of amino acid sequence between the E1 of different enzymes (identity typically <20%), the core fold of E1o bears a striking resemblance to that of other 2-oxo acid dehydrogenase E1s including pyruvate dehydrogenase (E1p) and branched chain 2-oxo acid dehydrogenases (E1b) (Figure 2). The 2-oxo acid dehydrogenase fold was first described in a related ThDP-dependent enzyme, transketolase, in which the active site is formed at the interface between two subunits, as illustrated for E1o in Figure 2.21 It appears that all OGDH and PDH E1 from Gramnegative bacteria are α2 homodimers, whereas all BCDH and PDH E1 from Gram-positive bacteria and eukaryotic species are α2β2 heterotetramers.7 Comparison of these structures suggests that E1o is in fact more closely related to α2β2 heterotetrameric E1 than other known homodimeric α2 E1. This structural similarity suggests that E1 proteins diverged by gene fusion or fission of the α-like and β-like subunits of a common ancestor. Thus, E1o is the contiguous arrangement of the C-terminal end of the α-like domain with the N-terminal start of the β-like domain. The inter-domain linker tracks along the surface of the E1o. Structure-based sequence alignment22 shows that this adjunct domain is conserved in all E1o, which may reflect its role in binding AMP (described below).
Table 1. Crystallographic data and refinement.
| Holo native | Apo native | Apo Se methionine | |||
|---|---|---|---|---|---|
| Diffraction data | |||||
| Wavelength | Native: 0.9794 Å | Native: 0.97937 Å | Se peak: 0.97955 Å | Se inflection: 0.979700 Å | Se remote: 0.97565 Å |
| Unit cell (Å) | P41212 | P41212 | P41212 | P41212 | P41212 |
| a,b = 143.81 | a,b = 142.49 | a,b = 144.60 | a,b = 144.64 | a,b = 144.60 | |
| c = 254.57 | c = 251.81 | c = 256.93 | c = 256.08 | c = 256.73 | |
| β,γ = 90° | β,γ = 90° | β,γ = 90° | β,γ = 90° | β,γ = 90° | |
| Resolution (Å) | 30.0-3.2 | 40-2.4 | 40-3.0 | 40-3.0 | 40-3.0 |
| ()=Highest resolution shell | (3.28-3.20) | (2.66-2.60) | (3.11-3.00) | (3.11-3.00) | (3.11-3.00) |
| -Rmerge (%) | 30.9 (68.0) | 13.5 (87.3) | 10.0 (24.1) | 9.0 (17.6) | 9.4 (18.1) |
| <l>/<s(l)> | 5.1 (2.5) | 17.2 (3.0) | 16.2 (7.5) | 15.5 (8.2) | 15.5 (8.0) |
| Completeness (%) | 89 (91) | 100 (100) | 100 (100) | 100 (100) | 100 (100) |
| Redundancy | 4.2 (4.3) | 8.7 (8.6) | 7.3 (7.5) | 6.1 (6.1) | 6.6 (6.5) |
| Wilson B Factor (Å2) | 52.3 | 54.9 | |||
| Phasinga | |||||
| Figure of merit | 0.37 | ||||
| Refinement | |||||
| Reflections | 26910 | 76341 | |||
| Atoms: | |||||
| Total | 12,543 | 13,373 | |||
| Protein (non-H) | 12,242 | 12,594 | |||
| Solvent | 1 | 748 | |||
| AMP | 46 | 46 | |||
| ThDP | 52 | 0 | |||
| Oxaloacetate | 18 | 0 | |||
| Mg | 2 | 0 | |||
| Rwork (%) | 23.7 (30.0) | 18.30 (30.6) | |||
| Rfree ^(%) | 30.3 (34.6) | 24.8 (39.8) | |||
| RMSD bonds (Å) | 0.026 | 0.014 | |||
| RMSD angles (°) | 2.40 | 1.18 | |||
| Ramachandran statistics (%) | |||||
| Generously allowed | 0.7 | 0.1 | |||
| Additional allowed | 15.7 | 10.1 | |||
| Most favourable | 83.6 | 89.8 | |||
| Disallowed | 0.1 | None | |||
Anomalous substructure was solved from the MAD data, and figure of merit was estimated by PHASER for SAD data. SAD data were used to calculate phases.
5% of reflections were used for Rfree set.
Figure 2. Domain architecture of the E1o component from the E. coli OGDH.
(a) The homo-dimeric quaternary structure of E. coli E1o (left) compared with the corresponding E1b from the Pseudomonas putida, E1p from E. coli and Bacillus stearothermophilus. The E1p from B. stearothermophilus PDH complex is shown complexed to the PSBD from the E2 component. The ThDP cofactor and (in the E1o) AMP ligand are shown as spheres coloured cyan and blue, respectively.
The dimer of β subunits from E1p and E1b forms the contact surface for recruitment into the enormous 2-oxo acid multienzyme complexes.23 However, in E1o these pseudo-β subunits are splayed apart, which may explain why E1o does not bind to E2o via the latter’s peripheral subunit binding domain (PSBD), instead interacting with the E2o core directly via its own N-terminal domain (Figure 2 and Supplementary Data).9 This domain has a columnar arrangement of four helices formed by residues 85-190 (Figure 1(b)). The intra-protomer helical pairs pack locally as a right-handed coiled-coil. The N-terminal regions from each subunit meet at the top of the inner pair of helices, from where a further 15 residues are disordered in the structure. The full length E1o will have 77 additional residues here that are predicted to have modest helical propensity based on sequence composition. Thus, in the complete protein the helices may extend outwards further from the N terminus. Structure-based sequence alignment shows the conservation of this extension in all known E1o, although its length varies from 71 to 113 amino acids and is longest in eukaryotic species. In contrast, this region is absent in all heterotetrameric E1p and E1b.
The active site
The E1o has the common amino acid sequence motif for binding ThDP; however, the cofactor could not be found in the crystal structure (Table 1). It thus appears that this structure represents the apo form of the enzyme. Although ThDP was not added during purification or crystallization, other E1 that have been crystallized have retained in the active site an endogenous ThDP that originated from the expression host.23
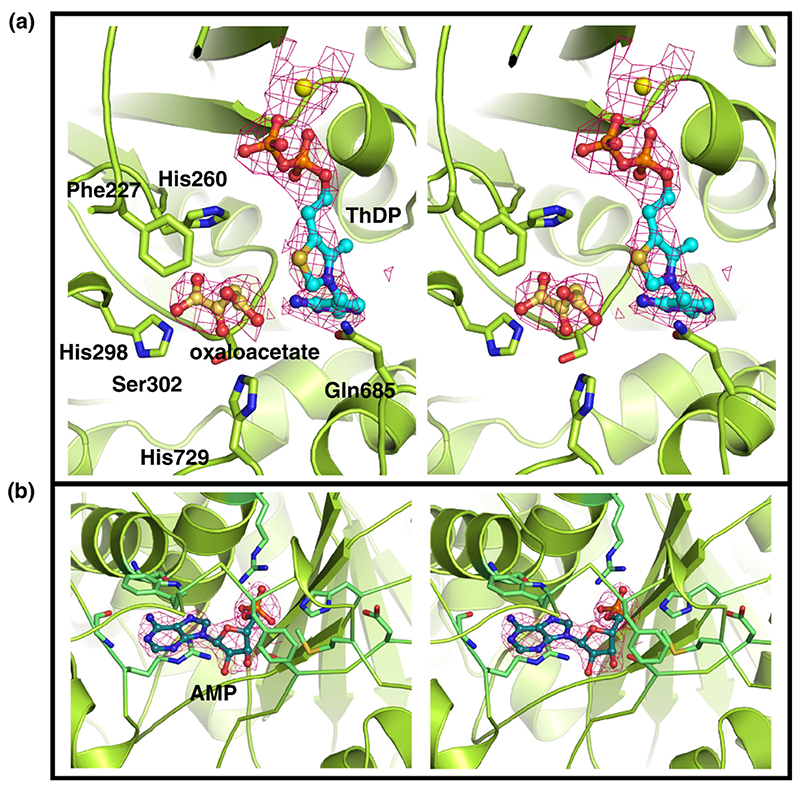
To obtain the structure of the cofactor-bound form of E1o, we attempted soaking the apo crystals; however, these crystals failed to diffract, which may be the consequence of conformational changes caused by the association of ThDP or disruption of crystal contacts. Instead, we co-crystallized apo-E1o in the presence of ThDP and Mg2+, which eventually produced a few small crystals that were multi-layered and only diffracted to modest resolution (3.5 Å). However, the redundancy of these data was high, and the phases for the structure were from the well-resolved apo form. Thus, despite the limited resolution and data quality, it was possible to observe unbiased density in difference maps at the active sites. This density occurs in both subunits related by non-crystallographic symmetry, and matched the shape of ThDP with Mg bound (Figure 3 and Table 1).
Figure 3. Stereo views of the electron density of putative ligands; the map was generated with sigmaA-weighted coefficients and contoured to 1σ.
(a) Active site of E1o showing Mg2+, ThDP and an oxaloacetate molecule bound (yellow sphere, cyan and gold stick format, respectively). This structure represents E1o in an inhibited conformation as oxaloacetate occludes the site where the substrate is expected to bind. The residues mediating the interactions with oxaloacetate are shown in stick format. (b) AMP binding pocket of E1o.
The cofactor adopts the conserved V-conformation and the surrounding residues are likely to be responsible for activation of the ThDP, substrate recognition and catalysis.21 A putative Mg was located at a site near the diphosphate that is consistent with observations of other ThDP enzymes. All other ThDP-dependent enzymes contain an invariant glutamate residue,21 which appears necessary for activation of ThDP. In E1o this function is served by Glu580, which also marks the entrance of a cavity within E1o connecting both active sites through the middle of the protein. The channel is lined by the side-chains of six acidic residues, three from each subunit (Asp368, Glu580 and Glu662). A similar cavity solvated with several entrained water molecules is found in all 20 of the ThDP-dependent enzyme structures available to date; however, the location of these water molecules is not revealed in E1o because of the limited resolution of the holo structure. A recently proposed catalytic mechanism for thiamine-dependent enzymes holds that each active site serves as a long range general acidbase that communicates through a similar acidic channel in E1p, which serves as a proton wire.24 The presence of such a channel in E1o and its acidic composition indicates that a similar mechanism may operate in this enzyme.
The active site loops are disordered in both the apo- and holo-E1o structures (residues 391-407, 458-471). An analogous loop in E1p and E1b is important for catalysis and recognition of the lipoyl domain, and the loop contains a conserved motif with sequence pattern YR-(x)3-H-(x)3-4-D-(x)4-YR-(x)2-DE.23‘25‘26 The equivalent loop of the E. coli E1o is conserved within the E1o family, but its corresponding sequence motif (YRR-x-GHNE-x-D-x-P-(x)3-Q) clearly differs from that found in E1p and E1b. The question remains whether these active site loops perform a similar role to those found in E1p and E1b, or if they have evolved an alternative set of interactions for achieving the same function.
Surrounding the cofactor are three prominent His residues (residues 260, 298 and 729). Given their proximity to the ThDP thiazolium, these residues are likely to define either the substrate-binding pocket or to be of catalytic importance. Unexpectedly, additional density (from difference maps) was identified between these His residues, and serine 321, obstructing the catalytic C2 nucleophile of ThDP. No organic constituents were added to the protein during purification or crystallization, with the exception of citrate and ethylene glycol. However, the extra density does not appear to have the correct shape for either of these molecules, and the separation of the coordinating histidine residues is too small to accommodate citrate. Although the ligand could not be definitively identified, its location suggested to us that the E1o active site can bind the small dicarboxylic acid, oxaloacetate, which is well disposed to form highly favourable salt-bridges with the flanking histidine residues. Given the occludent position of this unidentified ligand, it seems likely that it could inhibit the activity of E1o.
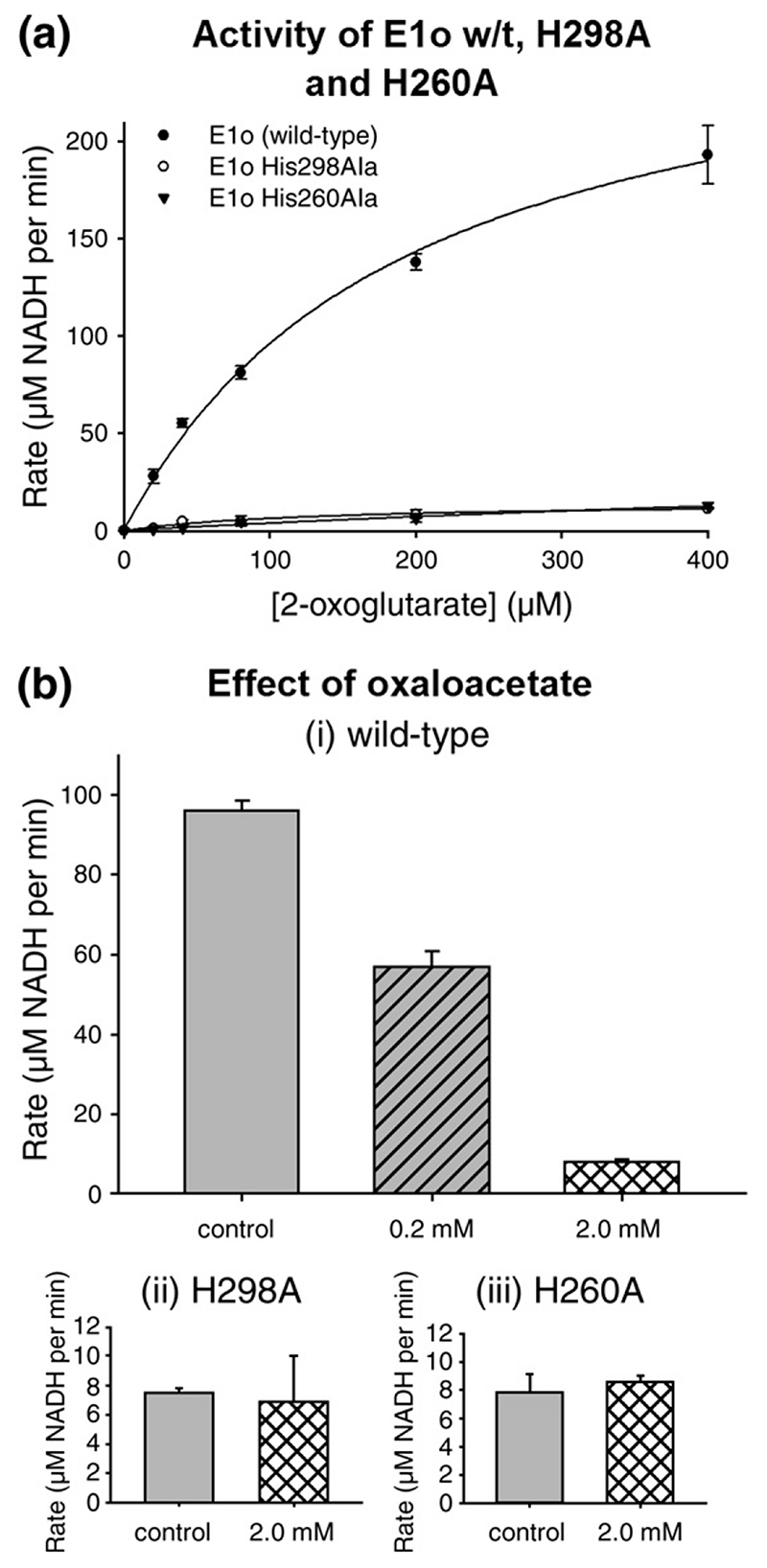
Hypothesising that the pocket can avidly bind oxaloacetate via the active site histidine residues, we reasoned this ligand would block access for 2-oxoglutarate and thus inhibit the enzyme. To test this hypothesis, we examined the effects of oxaloacetate on the turnover of 2-oxoglutarate using assays for the full OGDH complex, which measures the generation of NADH from NAD+. A significant level of inhibition was seen at physiological concentrations of oxaloacetate (Figure 4(b) and Supplementary Data) and this inhibition was lifted by raising the concentration of substrate, suggesting that oxaloacetate is an inhibitor of E1o, consistent with the electron density for this ligand in the E1o-ThDP crystal structure.27‘28 We compared the inhibition by oxaloacetate with other intermediates of the Krebs cycle, such as citrate, and the cryoprotectant ethylene glycol, but found that neither had a significant effect (Supplementary Data).
Figure 4. Effects of Krebs metabolites on OGDH complex activity monitoring the NADH product of the reaction.
(a) Substrate-dependence of the rate of reaction of E. coli OGDH complex with E1o wild-type and mutants in which active site His residues (H298 and H260) are replaced with Ala. (b) The rate of the OGDH complex containing wild-type E1o or active site mutant of E1o. The assay was performed with 400 μM 2-oxoglutarate in the presence of varying concentrations of oxaloacetate.
To test the importance of the residues potentially coordinating the oxaloacetate, we substituted Ala for His260 in one construct and Ala for His298 in another. Both of these E1o mutants had dramatically reduced catalytic rates (Figure 4(a)), which is expected as both His are well positioned to facilitate recognition of the distal carboxylate of 2-oxoglutarate. In comparison to the wild-type E1o, however, neither mutant was inhibited by oxaloacetate (Figure 4(b)), consistent with their auxiliary role in binding oxaloacetate. Since oxaloacetate is an intermediate of the Krebs cycle, its potential capacity to inhibit 2-OGDH could conceivably function to affect a cross-regulation within the cycle.
E1o contains an AMP binding site
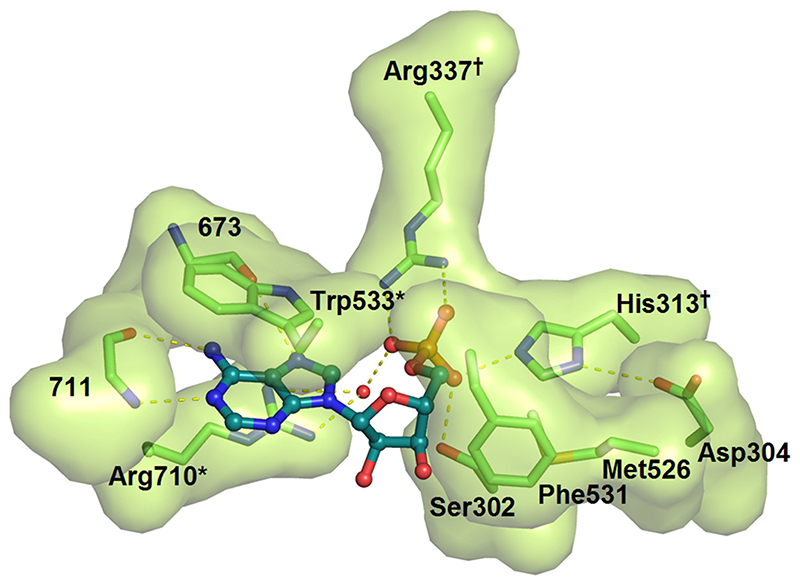
Unexpectedly, a ligand was discovered in a unique binding pocket situated away from the active site and that lies under a flexible linker connecting the α and β-like sub-domains of E1o. The electron density fitted adenosine monophosphate (AMP) unambiguously (Figure 3). The pattern of hydrogen bonds that the protein makes with the adenine ring discriminates between this base and other purines such as inosine and guanine. The pocket was occupied in both subunits of the apo and ThDP-bound forms of the enzyme. This suggests that the AMP and ThDP-binding sites are not in communication. The phosphate group of the AMP forms hydrogen bonds with Arg337, Arg710 and His313, and its purine ring stacks against the indole ring of Trp533 (Figure 5). This nucleotide is situated more than 20 Å from the active site, and its binding site shares no known homology to other well studied nucleotide-binding motifs, such as the Rossmann fold.29 We tested whether AMP or ATP might have an effect on the activity of E. coli E1o as measured by the decarboxylation30 and the full-complex assays.31 No significant inhibitory or activating effect was observed for either nucleotide (results not shown). Other adenine derivatives were also tested, such as cyclic AMP and ADP, but these had no significant effect on catalysis. Furthermore, two double mutants of E1o were designed to disrupt interactions with the adenine ring (Arg710Ala, Trp533Ala) or phosphate (Arg337Ala, His313Ala). The first and second double mutants are expected to ablate the aromatic stacking of the ring and the hydrogen bonding interactions, respectively. Neither of the mutants showed an effect on catalytic activity with in vitro enzyme assays (data not shown).
Figure 5.
The AMP binding pocket highlighting key residues that engage the ligand. The interacting residues are shown with a solvent-accessible surface. The adenine ring is base-stacked against the indole ring of Trp533 and the aliphatic portion of Arg710. Hydrogen bonds from the back-bone of residue 711 and 673 specify the adenine form of the purine ring. The phosphate of AMP forms a bidentate salt-bridge with Arg337, a water-mediated H-bond with Arg710, and H-bonds with Ser302 and His313. Two double Ala mutants were engineered to disrupt interactions with the adenine ring (*) and the phosphate (†) of AMP.
Given that our in vitro analysis did not reveal a function, we examined the effects of the above mutations on organism fitness in order to infer the importance of the AMP-pocket for the overall E. coli metabolism in vivo (see Supplementary Data). We used a strain of E. coli in which the gene encoding E1o was deleted (ΔsucA). These ΔsucA cells did not grow on minimal media containing acetate as the sole carbon source, which is expected, since acetate may only support viability when metabolised through the Krebs cycle, which in turn is dependent on the catalytic activity of E1o. ΔsucA cells were transformed with plasmids encoding the E1o gene harbouring mutations in the AMP binding pocket, but these constructs had only modest effects on growth rates in acetate compared with wild-type enzyme (Supplementary Data). Thus, the AMP binding pocket is probably not essential for viability under these conditions.
Discussion
An AMP binding site in E1o
Our structural data reveal unambiguous electron density for AMP, which is sequestered under a loop that connects two structural domains. Earlier studies had reported that ATP and AMP/ADP have either modest or no detectable allosteric effects on prokaryotic and eukaryotic E1o.32–35 Cellular levels of Ca2+ and ratios of ATP/AMP or ADP have been shown to modulate the activity of OGDH in vitro, though a molecular basis for this control has not yet been discovered. The residues that contact the AMP are well conserved in homologous E1o from other bacterial species and in some plant species such as Arabidopsis thaliana. However, it is apparent that AMP binding is specific for E1o and would not be expected to bind the paralogous E1p/E1b, in which a two-residue insertion fills the binding pocket. Additionally, many of the interactions of E1o with AMP are supported by the extended loop connecting the α-like and β-like domains, a feature which appears to be conserved in all E1o (Figures 2 and 6). It seems paradoxical that a prevalent ligand binding site has little if any effect on organism fitness. Double mutations to severely perturb the AMP binding site had no effect on the catalytic activity of E1o, suggesting that it is unlikely that the AMP has a purely structural role. However, it is not inconceivable that AMP binding to this site is an artefact of purification and instead some other, perhaps much larger AMP containing molecules, such as RNA, may bind at this site. Indeed, interactions between core metabolic enzymes and RNA have been seen with isocitrate dehydrogenase, which regulates translation by interactions with AMP and mRNA via its active site and a HEAT domain, respectively.36 It is also a possibility that the modest effects of the nucleotides as allosteric modulators might become more pronounced under certain stress conditions, such as exposure to strong oxidants or under starvation. Thus, further in-depth studies are needed to uncover the role, if any, of this curious ligand pocket.
Quaternary organization
Our observations from proteinase digestion demonstrate a role for the N terminus of E1o in mediating interactions with the core domain of E2o. It appears likely that this mode of interaction via the N terminus is conserved in other, if not all, E1o and is similar to that identified in another homodimeric 2-oxo acid decarboxylase, namely the E1p of Azotobacter vinelandii.37 Cryo-electron microscopy of E1o-E2o sub-complexes show that E1o is displaced at a distance of at least several nanometers from the E2o core,10 which seems at odds with the biochemical finding that E1o binds directly to the E2o core.9 However, the discovery of the N-terminal extension from the E1o may explain how E1o is able to maintain a gap between its globular domain and the OGDH core (Figure 1(b)). The feature of an annular gap between E2 and the peripheral E1 and E3 components has been identified and extensively studied in pyruvate dehydrogenase complexes.13,38 Thus, the OGDH complex has evolved an alternative protein apparatus to the pyruvate dehydrogenase complex that nonetheless achieves the same architectural outcome; apparently necessary in all 2-oxo acid dehydrogenase multienzyme complexes.
Regulating the TCA cycle
We observed additional electron density in the proximity of the ThDP cofactor in the structure of holo E1o, which fitted oxaloacetate. However, the resolution of the holo E1o structure is insufficient to identify the ligand definitively. Nevertheless, the engagement of this ligand in the active site suggests it might act as an inhibitor. The effects of oxaloacetate on E1o activity were therefore tested, and we found that at physiological concentrations of oxaloacetate, E1o activity is markedly reduced (Figure 4).
To explore the conservation of this putative inhibitory site, we aligned sequences of E1o, which revealed that the coordinating three histidine residues and one serine (in E. coli E1o, His260, His729, His298 and Ser321) form an invariant motif present in all E1o. It is therefore predicted that E1o universally bind this ligand. In contrast, E1p sequences do not adhere to this consensus. However, several E1b, including the human protein, contain the residues necessary to coordinate such a ligand. It is of note that the Thermus thermophilus E1b, for which a crystal structure is available,39 also has this motif. A recent report comments on the presence of electron density at the active site of this enzyme, but does not identify the ligand.40 Coincidentally, E1o and this structure were obtained by crystallization in a citrate buffer. We inspected the deposited coordinates and structure factors for the T. thermophilus E1b (Protein Data Bank code: 1umb) and find that the density is similar to that seen in our E. coli E1o structure, and also occurs at the corresponding position of the conserved motif. This finding supports the suggestion that the motif predisposes the enzyme to bind an inhibitor. However, further investigation is required to verify if oxaloacetate binds in the active site of T. thermophilus and human E1b as we predict.
Earlier studies had shown that oxaloacetate competes with 2-oxoglutarate for binding for binding with bacterial E1o.35,41These extensive kinetic analyses of the Azotobacter vinelandii OGDH, a close homologue of the E. coli OGDH, suggest that oxaloacetate occludes the active site of E1o. Our findings, that oxaloacetate is potentially present in the structure and acts as an inhibitor in vitro, fit well with these earlier reports. As oxaloacetate is an intermediate of the Krebs cycle, its capacity to inhibit the E1o of OGDH at physiologically relevant concentrations might account for regulatory “cross-talk” within the cycle.
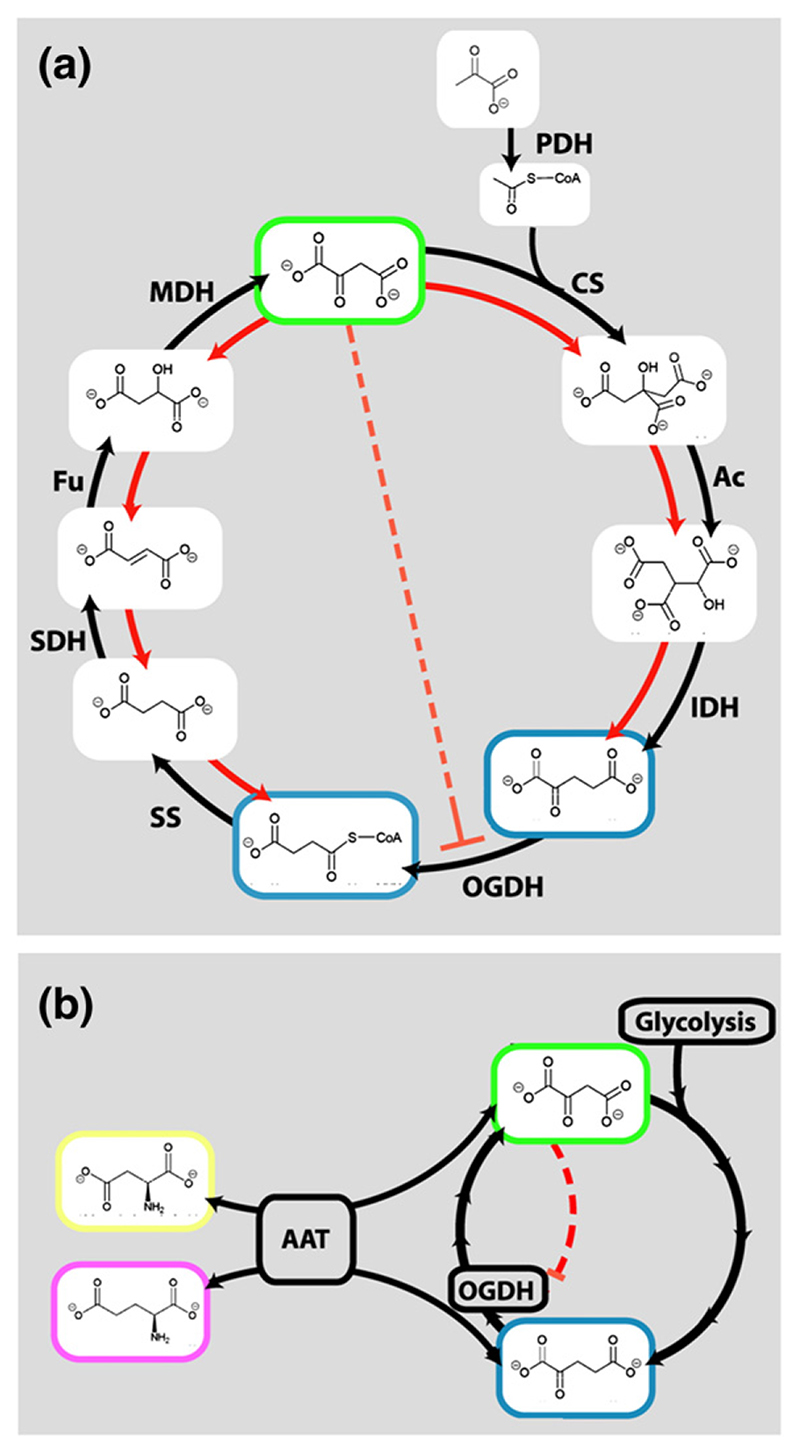
If oxaloacetate, a key intermediate in the Krebs cycle, interacts in vivo with E1o, which is the rate limiting, up-stream Krebs cycle enzyme, then this might provide a mechanism for achieving metabolic regulation. In facultative anaerobes, regulation of precisely this nature is required during the switch from aerobic respiration via the Krebs cycle to anaerobic growth in the presence of an abundant glycolytic carbon source.42 Evidence for such a switch has been found not only in enteric facultative anaerobic bacteria, such as E. coli, and also in eukaryotes, such as Saccharomyces cerevisiae 43 (Figure 6(a)). In these organisms, the concentration of oxaloacetate may serve as an indicator of glucose levels.42 Thus, it can be envisaged that the inhibitory effects of oxaloacetate on E1o activity may be responsible for feedback within the cycle to steer metabolism to aerobic or anaerobic routes in accordance with metabolic demands.
Figure 6. An “oxaloacetate switch”.
(a) Proposed mechanism of regulating the TCA cycle in facultative anaerobes, such as E. coli. Under aerobic conditions (black arrows) pyruvate is metabolised via PDH, citrate synthase (CS), aconitase (Ac), isocitrate dehydrogenase (IDH), OGDH, succinyl-CoA synthetase (SS), succinate dehydrogenase (SDH), fumarase (Fu) and malate dehydrogenase (MDH). In the presence of high concentration of glucose (or other glycolytic substrates), the concentration of oxaloacetate is raised,52 and MDH, Fu, SDH and SS function in reverse, allowing anaerobic fermentation (red arrows). It is likely this pathway is aided by oxaloacetate-dependent inhibition of E1o. (b) Proposed regulation of 2-oxoglutarate (OG) flux by the oxaloacetate (green outline) in the mammalian brain. When oxaloacetate is in excess, OGDH is inhibited, allowing diversion of OG to generate the excitatory neurotransmitter, glutamate (magenta outline). Transamination between glutamate-oxaloacetate/aspartate-2-oxoglutarate is catalysed by amino acid aminotransferase. Isotope labelling studies suggest the amine for this product may be donated by aspartate (yellow outline) to leucine (to produce a substrate for the BCDH).44
Another context in which the oxaloacetate switch may function is in the mammalian brain, as suggested by the conservation of the putative oxaloacetate-binding motif in human E1o. As well as a proportionately high metabolic demand, neurones and glial cells must balance the utilization of 2-oxoglutarate, for energy metabolism via the Krebs cycle and transamination of this intermediate to produce the major excitatory neurotransmitter, glutamate and inhibitory neurotransmitter, γ-aminobutyric acid (GABA) (Figure 6(b)).44 If too much 2-oxoglutarate is directed to the generation of glutamate, then oxaloacetate will become depleted and the Krebs cycle can no longer support the energy demands of the cell. Glutamate levels are also balanced by aspartate, which is produced by the transamination of oxaloacetate in neurones and the glial cells that provide metabolic support for neurones.45 The ability of E1o to sense and respond to the levels of oxaloacetate suggests a mechanism that may enable the concerted regulation of both the energy and the signalling demands of mitochondria in the brain.
Metabolic pathways that are cyclical with multiple, branching, entrance and exit points, such as the Krebs cycle represent a regulatory challenge in order to prevent the loss of intermediates as well as adapting to changing metabolic needs.1 The proposed mechanism, in which oxaloacetate modulates the Krebs cycle via E1o, offers one possible explanation for how that cycle can be regulated over its multiple branch points.
Experimental Procedures
Protein over-expression and purification
Mutant and wild-type E.coli E1o genes were cloned into a pET11c vector and over-expressed in E. coli BL21(DE3) using induction with 0.5 mM IPTG.20 Cells were harvested 3 h post-induction, lysed, and lysates were enriched for E1o by ammonium sulphate precipitation (20-50% saturation); precipitated proteins were dissolved in 20 mM potassium phosphate (pH 7.0) and loaded onto an anion exchange column with HiLoad Q-Sepharose (Amersham) and eluted with a linear gradient using the same buffer containing 1 M NaCl. Enriched fractions were pooled, concentrated with a 100 kDa MWCO centrifugal filter and fractionated by Superdex S200 size exclusion chromatography (Amersham). Purified proteins were analysed by electrospray ionisation-time of flight mass spectrometry.
Mutagenesis
E. coli sucA was cloned from genomic DNA by PCR and ligated into a pET11c expression vector. Expression and purification are described elsewhere.20 Site-directed mutagenesis of sucA was carried out using a QuikChange® PCR Site-Directed Mutagenesis kit (Stratagene). PCR products were transformed in E. coli DH5α from which engineered plasmids were prepared and analysed for the desired mutation by DNA sequencing. Mutant plasmids were expressed and purified as the wild-type.20
Enzymatic assays
For analysis of the E1o activity, OGDH assemblies were prepared by mixing purified mutant or wild-type E1o enzymes with wild-type E2o and E3 enzymes in vitro. The activity of the complex was monitored by measuring the formation of NADH at 340 nm. The reaction mixtures contained 60 pmol E1o, 240 pmol E2o, 180 pmol E3, 100 mM Tris-HCl (pH 7.4), 1 mM ThDP, 2 mM MgCl2 and 5 mM NAD+. In mixtures containing oxaloacetate, the metabolite was added to a concentration of 0.2 mM or 2 mM. Reactions were started by adding 2-oxoglutarate and lithium co-enzyme A to a final concentration of 400 μM and 1 mM, respectively. Enzyme activity was immediately followed by measuring the rate of increase in absorbance at 340 nm using a temperature-controlled spectrophotometer at 35 °C.
Crystallization of truncated E. coli E1o
Extensive crystallization trials were performed using pure, full-length E1o. Conditions were found that required several months for crystals to appear, but these were not easily reproduced. A strategy of limited proteolysis was tested to identity a “trimmed” E1o product more suited to crystallization. Trypsin was used to produce a single cleaved product (tE1o), which lacked 66 residues from its N terminus. Ten mg/ml of tE1o in 20 mM potassium phosphate pH 7 was used in subsequent crystal trials and produced 1 mm×0.05 mm×0.05 mm crystals under several conditions within 24 h. Crystallization was performed by sitting drop using 1 μl of tE1o and 1 μl of precipitating agent from a reservoir of 12% (w/v) polyethylene glycol 4000, 50 mM sodium citrate pH 5.6.
Selenomethionine incorporated E1o (E1oSeMet) was prepared using a modification of the methionine suppression method. An overnight culture of cells in 250 ml LB (with 100 μg/ml of ampicillin) was harvested by centrifugation and used to inoculate 250 ml of minimal media with glucose as a carbon source, which was incubated at 37 °C until the A600 nm reached 0.8. Inhibition of methionine synthesis was triggered by the addition of 100 mg each of amino acids (Lys, Thr, Phe, Leu, Ile, Val, and SeMet). After 15 min, the temperature was lowered to 20 °C and expression was induced with 0.5 mM IPTG for 3 h. Purification and crystallization were as described for the native tE1o, except that 10 mM dithiothreitol (DTT) was included in the precipitation reservoir. Incorporation of selenomethionine was assayed by mass spectrometry showing at least 22 of the 24 methionine residues in each subunit were replaced with selenomethionine.
Co-crystals of tE1o in complex with the cofactors, thiamine diphosphate (ThDP) and Mg2+ were prepared by purifying tE1o as described for the native protein, but in the presence of 5 mM ThDP and 2 mM Mg2+. After the final step of purification tE1o-Mg-ThDP samples contained 9 mg/ml of tE1o-Mg-ThDP in 5 mM ThDP, 20 mM potassium phosphate pH 7. tE1o-Mg-ThDP co-crystals were grown using 1 μl of sample and 1 μl of precipitation agent (8% polyethylene glycol, 50 mM sodium citrate (pH5.6)).
Data collection and processing
tE1o, tE1o-Mg-ThDP and tE1oSeMet crystals were cryoprotected using 22.5% ethylene glycol, 12% polyethylene glycol 4000, 50 mM sodium citrate (pH 5.6.) and flash-frozen in liquid nitrogen before X-ray data collection at 100 K on beamlines ID14.4, 23.1, and 29, at the European Synchrotron Radiation Facility, Grenoble. Data from a single native tE1o crystal, a single tE1o-Mg-ThDP crystal, and three multi-wavelength anomalous dispersion (MAD) datasets from one tE1oSeMet crystal were collected to 2.4 Å, 3.5 Å and 3.5 Å resolution, respectively. tE1o and MAD data were auto-indexed and scaled with HKL2000,46 and the tE1o-Mg-ThDP dataset was processed with MOSFLM and SCALA.47 In all cases, the space group is P41212, and the asymmetric unit of the cell fit one homodimer of tE1o chains with a solvent content of 59%. The crystals of the holo form were of small size and poor quality and multi-layered, so that reflections for the holo data set were of poor quality, resulting in high values of R-merge. However, the redundancy was high, and the phases from the apo were sufficiently good quality to see unbiased density in difference maps. For refinement of the holo structure, data were used at 3.5 Å.
Phase determination and structure refinement
The MAD data were further scaled, merged, and truncated to 5 Å with XPREP, and 32 of the 48 selenium sites were identified after >200 rounds of searching the Patterson difference map using ShelxD.48 However, refinement and density modification of these sites failed to yield an interpretable electron density map. These sites were analysed with PROFESSS,47 which identified a dyad symmetry axis in the sub-structure, consistent with the non-crystallographic symmetry. The selenium atoms were refined against the peak data (to 3.0 Å resolution) and the remaining eight Se atoms were found using PHASER.49 The phases generated by these refined selenium sites were used for non-crystallographic symmetry averaging in DM.47 An electron density map using these phases and coefficients produced an easily interpretable electron density map. Arp/wArp47 was then used to automatically build 80% of the protein model into the experimental electron density. The model was then manually fitted in Coot50 and refined in REFMAC.51 N-terminal sequence analysis show that tE1o contained residues YSST, corresponding to residue 76. Except for several surface exposed loops (in chain A: 119–121, 133, 146–148, 191, 224–225, 287–288, 392–406, 459–469; and in chain B: 116–118, 223–225, 287–290, 392–406, 459–473, 658–659), residues 85–933 from both chains of the homodimeric tE1o were accounted for in the electron density. Electron density fitting the dimensions of an adenosine monophosphate ligand were identified within a highly selective binding pocket in the tE1o. Both the electron density and the geometry of the interactions at the site are consistent with adenine (rather than guanine or hypoxanthine). The pocket was identified in both sites related by non-crystallographic dyad symmetry. After the model was refined against the high-resolution native tE1o data, the model included 750 water molecules.
The tE1o-Mg-ThDP structure was solved by molecular replacement using the apo-tE1o homodimer as a search model in MOLREP.47 The solution was rigid body and positional refined, with tight NCS restraints using REFMAC to 3.5 Å. A sigma-A weighted map and difference map was examined in Coot, revealing positive electron density matching the shape of thiamine diphosphate and a divalent cation, assumed to be Mg2+. Adjacent to this was electron density corresponding to another ligand, which was consistent with oxaloacetate. The model was manually fitted using Coot and refined in REFMAC5. The atomic geometry of the models was validated using PROCHECK.47
Accession numbers
Atomic coordinates and structure factor amplitudes have been deposited with the RCSB Protein Data Bank under accession codes 2jgd and r2jgdsf.
Supplementary Material
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2007.01.080
Acknowledgements
This work was supported by the Wellcome Trust. We thank Chris Titman, David Christianson, Hal Dixon and Philip Oliver for advice and helpful discussions; the staff of the GenoBase resource at the Nara Institute of Science and Technology for kindly providing bacterial strains; the staff of ESRF for use of facilities, and Randy Read and Airlie McKay for help with their program PHASER.
Abbreviations
- OGDH
2-oxoglutarate dehydrogenase
- PDH
pyruvate dehydrogenase
- TCA
tricarboxylic acid cycle
- MAD
multi-wavelength anomalous dispersion
- BCDH
branched-chain 2-oxo acid dehydrogenase.
References
- 1.Srere PA. 17th Fritz Lipmann Lecture. Wanderings (wonderings) in metabolism. Biol Chem Hoppe Seyler. 1993;374:833–842. [PubMed] [Google Scholar]
- 2.Anderson SL, Minard KI, McAlister-Henn L. Allosteric inhibition of NAD+-specific isocitrate dehydrogenase by a mitochondrial mRNA. Biochemistry. 2000;39:5623–5629. doi: 10.1021/bi000272e. [DOI] [PubMed] [Google Scholar]
- 3.Hansford RG, Zorov D. Role of mitochondrial calcium transport in the control of substrate oxidation. Mol Cell Biochem. 1998;184:359–369. [PubMed] [Google Scholar]
- 4.Gibson GE, Park LC, Sheu KF, Blass JP, Calingasan NY. The alpha-ketoglutarate dehydrogenase complex in neurodegeneration. Neurochem Int. 2000;36:97–112. doi: 10.1016/s0197-0186(99)00114-x. [DOI] [PubMed] [Google Scholar]
- 5.Tretter L, Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. J Neurosci. 2004;24:7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 8.Packman LC, Perham RN. Limited proteolysis and sequence analysis of the 2-oxo acid dehydrogenase complexes from Escherichia coli Cleavage sites and domains in the dihydrolipoamide acyltransferase components. Biochem J. 1987;242:531–538. doi: 10.1042/bj2420531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Packman LC, Perham RN. Chain folding in the dihydrolipoyl acyltransferase components of the 2-oxo-acid dehydrogenase complexes from Escherichia coli Identification of a segment involved in binding the E3 subunit. FEBS Letters. 1986;206:193–198. doi: 10.1016/0014-5793(86)80979-6. [DOI] [PubMed] [Google Scholar]
- 10.Wagenknecht T, Grassucci R, Schaak D. Cryoelectron microscopy of frozen-hydrated alphaketoacid dehydrogenase complexes from Escherichia coli . J Biol Chem. 1990;265:22402–22408. [PubMed] [Google Scholar]
- 11.Mattevi A, Obmolova G, Kalk KH, Westphal AH, de Kok A, Hol WG. Refined crystal structure of the catalytic domain of dihydrolipoyl transacetylase (E2p) from Azotobacter vinelandii at 2.6 Å resolution. J Mol Biol. 1993;230:1183–1199. doi: 10.1006/jmbi.1993.1235. [DOI] [PubMed] [Google Scholar]
- 12.Izard T, Aevarsson A, Allen MD, Westphal AH, Perham RN, de Kok A, Hol WG. Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc Natl Acad Sci USA. 1999;96:1240–1245. doi: 10.1073/pnas.96.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne JL, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu X, et al. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 2002;21:5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettit FH, Hamilton L, Munk P, Namihira G, Eley MH, Willms CR, Reed LJ. Alpha-keto acid dehydrogenase complexes. XIX. Subunit structure of the Escherichia coli alphaketoglutarate dehydrogenase complex. J Biol Chem. 1973;248:5282–5290. [PubMed] [Google Scholar]
- 15.Ricaud PM, Howard MJ, Roberts EL, Broadhurst RW, Perham RN. Three-dimensional structure of the lipoyl domain from the dihydrolipoyl succinyltransferase component of the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli . J Mol Biol. 1996;264:179–190. doi: 10.1006/jmbi.1996.0632. [DOI] [PubMed] [Google Scholar]
- 16.Knapp JE, Mitchell DT, Yazdi MA, Ernst SR, Reed LJ, Hackert ML. Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J Mol Biol. 1998;280:655–668. doi: 10.1006/jmbi.1998.1924. [DOI] [PubMed] [Google Scholar]
- 17.Robien MA, Clore GM, Omichinski JG, Perham RN, Appella E, Sakaguchi K, Gronenborn AM. Three-dimensional solution structure of the E3-binding domain of the dihydrolipoamide succinyltransferase core from the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli . Biochemistry. 1992;31:3463–3471. doi: 10.1021/bi00128a021. [DOI] [PubMed] [Google Scholar]
- 18.Mande SS, Sarfaty S, Allen MD, Perham RN, Hol WG. Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure. 1996;4:277–286. doi: 10.1016/s0969-2126(96)00032-9. [DOI] [PubMed] [Google Scholar]
- 19.Frank RAW. Structural enzymology of 2-oxo acid dehydrogenases: symmetry and multiplicity. PhD thesis, University of Cambridge; 2004. [Google Scholar]
- 20.Ricaud P. Structure and interactions of the lipoyl domain of the 2-OGDH complex. PhD thesis, Cambridge University; 1998. [Google Scholar]
- 21.Lindqvist Y, Schneider G, Ermler U, Sundstrom M. Three-dimensional structure of transketolase, a thiamine diphosphate dependent enzyme, at 2.5 Å resolution. EMBO J. 1992;11:2373–2379. doi: 10.1002/j.1460-2075.1992.tb05301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 23.Frank RA, Pratap JV, Pei XY, Perham RN, Luisi BF. The molecular origins of specificity in the assembly of a multienzyme complex. Structure (Camb) 2005;13:1119–1130. doi: 10.1016/j.str.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 24.Frank RA, Titman CM, Pratap JV, Luisi BF, Perham RN. A molecular switch and proton wire synchronize the active sites in thiamine enzymes. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]
- 25.Hawes JW, Schnepf RJ, Jenkins AE, Shimomura Y, Popov KM, Harris RA. Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase (BCKDH) in catalysis and phosphorylation site recognition by BCKDH kinase. J Biol Chem. 1995;270:31071–31076. doi: 10.1074/jbc.270.52.31071. [DOI] [PubMed] [Google Scholar]
- 26.Fries M, Chauhan HJ, Domingo GJ, Jung HI, Perham RN. Site-directed mutagenesis of a loop at the active site of E1 α2β2 of the pyruvate dehydrogenase complex. A possible common sequence motif. Eur J Biochem. 2003;270:861–870. doi: 10.1046/j.1432-1033.2003.03444.x. [DOI] [PubMed] [Google Scholar]
- 27.Barron JT, Kopp SJ, Tow J, Parrillo JE. Fatty acid, tricarboxylic acid cycle metabolites, and energy metabolism in vascular smooth muscle. Am J Physiol. 1994;267:H764–H769. doi: 10.1152/ajpheart.1994.267.2.H764. [DOI] [PubMed] [Google Scholar]
- 28.Cortassa S, Aon MA, Marban E, Winslow RL, O’Rourke B. An integrated model of cardiac mitochondrial energy metabolism and calcium dynamics. Biophys J. 2003;84:2734–2755. doi: 10.1016/S0006-3495(03)75079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossman MG. Evolution of glycolytic enzymes. Phil Trans Roy Soc ser B. 1981;293:191–203. doi: 10.1098/rstb.1981.0072. [DOI] [PubMed] [Google Scholar]
- 30.Khailova LS, Bernkhardt R, Khiubner G. Study of the kinetic mechanism of the pyruvate-2,6-dichlorophenolindophenol reductase activity of muscle pyruvate dehydrogenase. Biokhimiia. 1977;42:113–117. [PubMed] [Google Scholar]
- 31.Danson MJ, Fersht AR, Perham RN. Rapid intramolecular coupling of active sites in the pyruvate dehydrogenase complex of Escherichia coli: mechanism for rate enhancement in a multimeric structure. Proc Natl Acad Sci USA. 1978;75:5386–5390. doi: 10.1073/pnas.75.11.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornfeld S, Benziman M, Milner Y. Alphaketoglutarate dehydrogenase complex of Acetobacter xylinum Purification and regulatory properties. J Biol Chem. 1977;252:2940–2947. [PubMed] [Google Scholar]
- 33.Parker MG, Weitzman PD. The purification and regulatory properties of alpha-oxoglutarate dehydrogenase from Acineobacter iwoffi . Biochem J. 1973;135:215–223. doi: 10.1042/bj1350215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heckert LL, Butler MH, Reimers JM, Albe KR, Wright BE. Purification and characterization of the 2-oxoglutarate dehydrogenase complex from Dictyostelium discoideum . J Gen Microbiol. 1989;135:155–161. doi: 10.1099/00221287-135-1-155. [DOI] [PubMed] [Google Scholar]
- 35.Meixner-Monori B, Kubicek CP, Habison A, Kubicek-Pranz EM, Rohr M. Presence and regulation of the alpha-ketoglutarate dehydrogenase multienzyme complex in the filamentous fungus Aspergillus niger . J Bacteriol. 1985;161:265–271. doi: 10.1128/jb.161.1.265-271.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson SL, Lin AP, McAlister-Henn L. Analysis of interactions with mitochondrial mRNA using mutant forms of yeast NAD(+)- specific isocitrate dehydrogenase. Biochemistry. 2005;44:16776–16784. doi: 10.1021/bi0515568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hengeveld AF, de Kok A. Identification of the E2-binding residues in the N-terminal domain of E1 of a prokaryotic pyruvate dehydrogenase complex. FEBS Letters. 2002;522:173–176. doi: 10.1016/s0014-5793(02)02931-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhou ZH, McCarthy DB, O’Connor CM, Reed LJ, Stoops JK. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc Natl Acad Sci USA. 2001;98:14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai T, Nakagawa N, Maoka N, Masui R, Kuramitsu S, Kamiya N. Ligand-induced conformational changes and a reaction intermediate in branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8, as revealed by X-ray crystallography. J Mol Biol. 2004;337:1011–1033. doi: 10.1016/j.jmb.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Machius M, Wynn RM, Chuang JL, Li J, Kluger R, Yu D, et al. A versatile conformational switch regulates reactivity in human branched-chain alpha-ketoacid dehydrogenase. Structure. 2006;14:287–298. doi: 10.1016/j.str.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Bunik V, Westphal AH, de Kok A. Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii evidence for the formation of a precatalytic complex with 2-oxoglutarate. Eur J Biochem. 2000;267:3583–3591. doi: 10.1046/j.1432-1327.2000.01387.x. [DOI] [PubMed] [Google Scholar]
- 42.Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Frick O, Wittmann C. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13C flux analysis. Microb Cell Fact. 2005;4:30. doi: 10.1186/1475-2859-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magistretti PJ. Brain energy metabolism. In: Squire R, editor. Fundamental Neuroscience. 2nd. Elselvier; New York: 2003. pp. 339–360. [Google Scholar]
- 45.Magistretti PJ, Pellerin L, Rothman DL, Shulman RG. Energy on demand. Science. 1999;283:496–497. doi: 10.1126/science.283.5401.496. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Collaborative computational project, no 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallog sect D. 1999;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallog sect D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, Storoni LC, Read RJ. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallog sect D. 2004;60:1220–1228. doi: 10.1107/S0907444904009990. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallog sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallog sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Neidhardt FC, Curtiss R, 3rd, Ingraham JL, Lin ECC, Brooks Low K, Magasanik B, et al. In: Escherichia coli and Salmonella typhimurium. Neidhardt FC, editor. ASM Press; Washington, DC: 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.