Abstract
Epithelial cells rapidly adapt their behaviour in response to increasing tissue demands. However, the processes that finely control these cell decisions remain largely unknown. The postnatal period covering the transition between early tissue expansion and the establishment of adult homeostasis provides a convenient model to explore this question. Here, we demonstrate that the onset of homeostasis in the epithelium of the mouse oesophagus is guided by the progressive build-up of mechanical strain at the organ level. Single-cell RNA sequencing and whole-organ stretching experiments revealed that the mechanical stress experienced by the growing oesophagus triggers the emergence of a basal KLF4Bright committed population, which balances cell proliferation and marks the transition towards homeostasis in a YAP dependent manner. Our results point to a simple mechanism whereby mechanical changes experienced at the whole tissue level are integrated with those “sensed” at the cellular level to control epithelial cell fate.
Epithelial tissues possess the remarkable ability to adapt their program of cell fate in response to increasing tissue demands, such as developmental cues and damage1–7. Yet, the mechanisms governing transitions between different modes of cell behaviour remain unclear.
To better understand epithelial cell transitions, we focused on one of the first, and most important, transitions that a specified tissue must face: the establishment of homeostasis. In most tissues, development continues after birth, with a rapid but physiological expansion that encompasses the postnatal growth of the animal, and culminates in the establishment of homeostasis8, 9. At this point, proliferation and differentiation must become balanced to ensure tissue maintenance10. However, adult homeostatic cells retain a degree of plasticity, being able to reactivate developmental features if needed11. Indeed, differentiating or lineage committed cells in a range of tissues, including intestine, lung and skin, have been shown to possess the ability to transition back to a stem cell-like state in response to tissue damage6, 12, 13.
The mouse oesophagus represents an ideal model to study changes in epithelial cell dynamics at the whole tissue level due to its simple architecture10. Although the processes governing adult tissue maintenance have been widely studied, here we aim at shedding light into the unknown mechanisms that initially shape homeostasis, following postnatal development. During embryonic development, the oesophagus originates as a single layer of cuboidal cells that matures into a squamous, multi-layered tissue14. The resulting adult oesophagus is lined by a squamous keratinized epithelium, formed by layers of keratinocytes (Fig. 1a). Proliferation is confined to progenitor cells within the basal layer. Upon commitment to differentiation, basal cells exit the cell cycle and stratify into the suprabasal layers, being ultimately shed at the surface. Quantitative studies have proposed that the homeostatic oesophageal epithelium (OE) is maintained by a single, functionally equivalent, progenitor population that balances the production of proliferating and differentiating cells through stochastic cell fate decisions15.
Growing evidence suggest that cell behaviour is not solely regulated by classically known biochemical cues. Instead, cells are able to alter their behaviour in response to a combination of molecular and mechanical stimuli16–27. This is exemplified during postnatal development, when tissues growing at their own unique pace generate significant levels of mechanical stress at the organ level24, 28. In this context, physical forces influence cell adhesion, shape, as well as niche availability and, in return, feedback on tissue growth, organisation and maturation26, 29.
Here we demonstrate that after birth the developing oesophagus grows at a slower pace than the rest of the body, resulting in the build-up of a physiological tensile strain along its longitudinal axis. Tissue stretching experiments reveal the existence of a defined strain threshold that, once reached, promotes the emergence of a KLF4Brigh committed basal cell population, primed for differentiation, in a YAP dependent fashion. This mechanical transition signals the tilt of progenitor cell fate from a proliferative mode, during tissue expansion, towards the balanced commitment state characteristic of adult homeostasis15. Our results reveal that naturally occurring changes in tissue mechanics orchestrate the establishment of tissue homeostasis.
Results
Oesophageal transition towards homeostasis
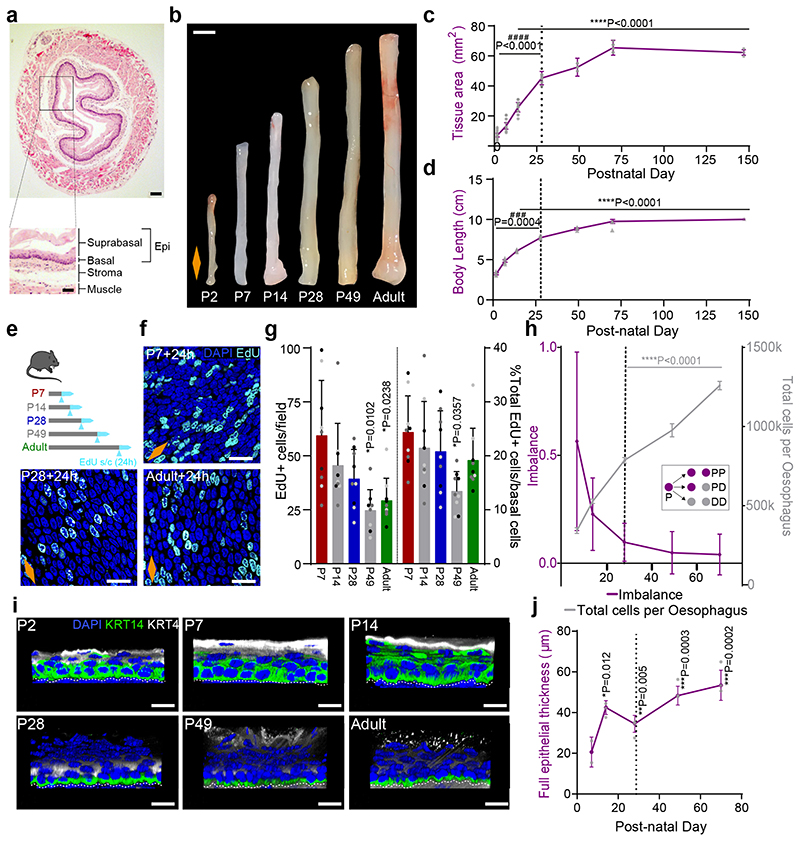
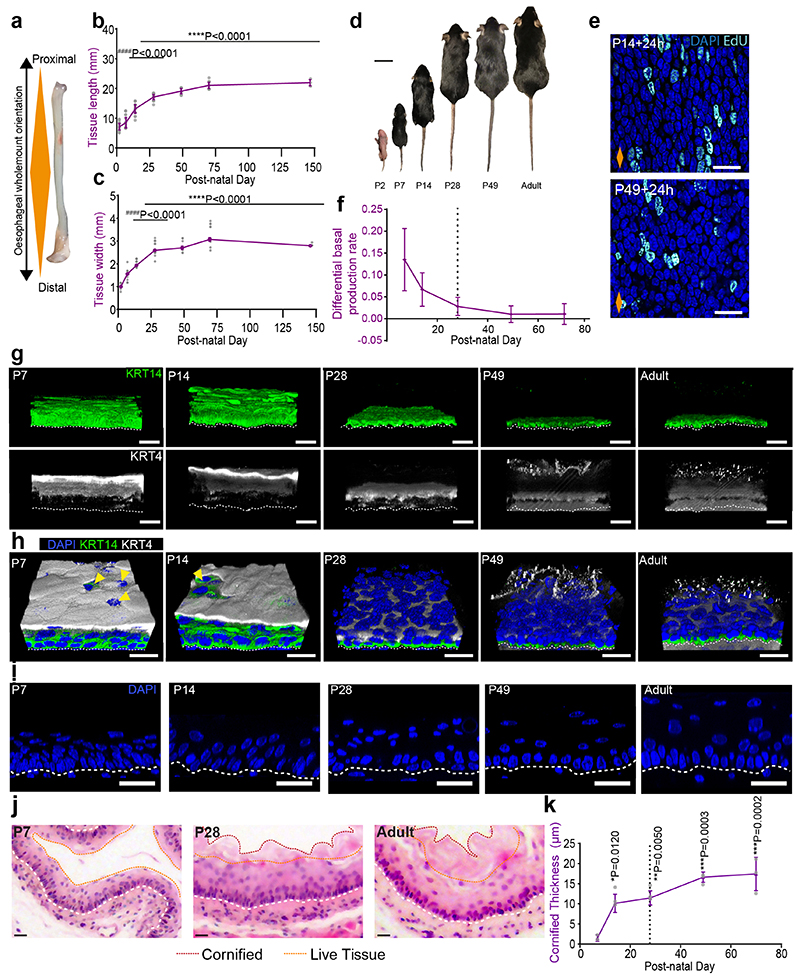
To study changes associated with the establishment of adult homeostasis, we started by measuring tissue dimensions from postnatal day (P)2 to P147 (Extended Data Fig. 1a-c). The oesophagus showed a biphasic mode of growth, encompassing the growth of the animal (Fig. 1b-d and Extended Data Fig. 1b-d). The initial phase of rapid oesophageal expansion slowed down by ~P28, and stabilized at P70, coinciding with the establishment of adult homeostasis15.
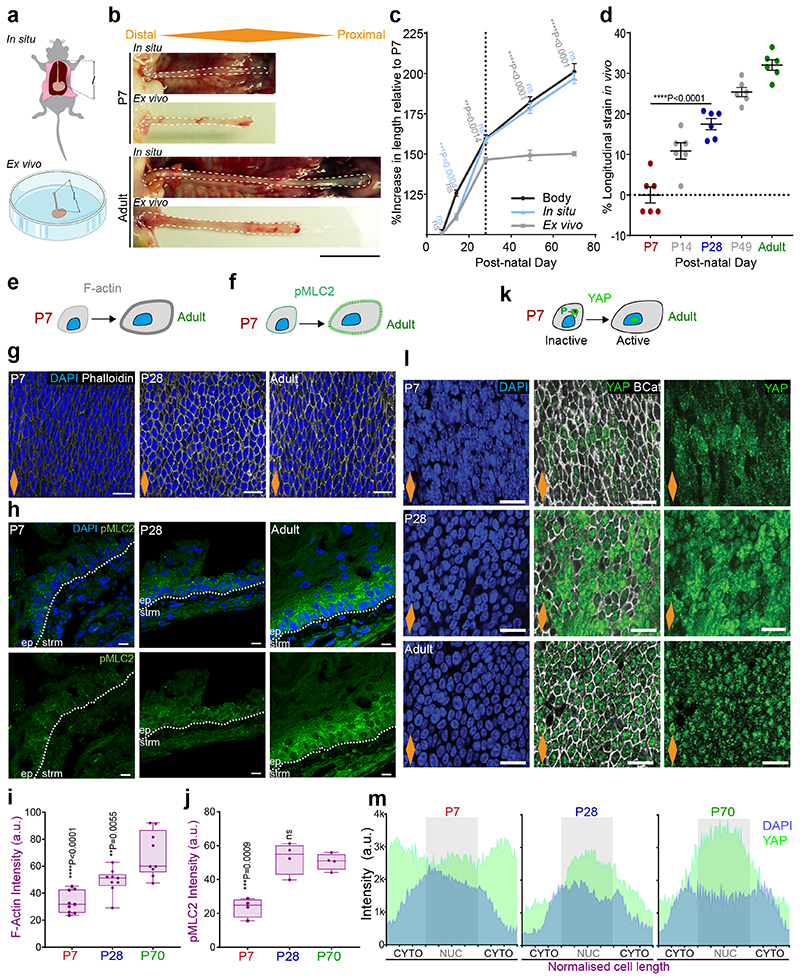
Fig. 1. The postnatal mouse oesophagus as a model of rapid tissue expansion transitioning towards homeostasis.
a, H&E section of adult oesophagus. Inset showing tissue layers. Epi, epithelium.
b, Images of whole oesophagus throughout postnatal development.
c, Tissue area after longitudinally opening the oesophageal tube. n=103 mice; Millimeters, mm.
d, Whole body growth in length excluding tail. n=26 mice; Centimeter, cm.
e, In vivo protocol. Mice were treated with a single EdU injection 24h prior tissue collection at the time points indicated.
f, Basal confocal view of EdU+ cells in typical OE wholemounts from (e).
g, Quantification of EdU+ basal cells per field (left), and relative to the number of DAPI+ basal cells (right) from (e). Presented as mean ± SD. Points show individual measurements, greyscale indicates values from each of 3 mice. n=3 mice.
h, Fraction of cell fate imbalance, showing the degree to which basal cell fate is biased towards duplication over differentiation and loss (left axis; see Methods and model in Fig. 2n). Total cell production throughout postnatal development (right axis; see Methods). Over time OE cells tilt towards balanced cell fate, in line with model of oesophageal homeostasis by Doupe et. al.15 (inset; P, progenitor cell; D, differentiating cell). n=3 mice. See Supplementary Note 1.
i, Representative 3D rendered confocal z-stacks showing side views of OE wholemounts throughout postnatal development.
j, Epithelial thickness, including enucleated terminally differentiated layers. n=3 mice; Micrometre, μm.
All data derived from wild-type C57BL/6J mice, expressed as mean values ± SEM, unless otherwise stated. Data analysis was performed using one-way ANOVA with Tukey’s multiple comparisons test (#p relative to P70; *p relative to P7).
Scale bars. 1a(200 μm), inset(100 μm); 1b(2mm); 1f,i(20μm).
Stainings. Blue, 4’,6-diamidino-2-phenylindole (DAPI); cyan, EdU; green, basal marker KRT14; greyscale, differentiation marker KRT4.
Dashed lines indicate basement membrane. Dotted lines in graphs indicate P28. Orange diamonds depict the longitudinal orientation of the oesophagus where indicated (outlined in Extended Data Fig. 1a). Parts of (e) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided. See also Extended Data Fig. 1.
We next looked at cell proliferation by short-term in vivo EdU incorporation assays. Quantification of EdU+ cells after a 24h chase revealed a reduction in the proliferative activity of progenitors over time (Fig. 1e-g and Extended Data Fig. 1e). The tilt away from progenitor cell duplication was further revealed through an estimate of fate imbalance (Fig. 1h and Extended Data Fig. 1f; Methods and Supplementary Note 1). The degree of fate imbalance is determined as the ratio of the differential change in total basal cell number to the cell division rate. When the fate imbalance is large, progenitor cell divisions are biased towards a symmetric duplicative fate outcome (model in Fig. 1h, inset), while when the fate imbalance is small, cell duplication is approximately balanced by cell differentiation. During the early postnatal period we observed a 60% imbalance towards cell duplication, which drastically falls reaching a near-balance homeostatic-like state between P28 and Adult.
Next, we performed immunostaining for keratin 14 (KRT14) and Keratin 4 (KRT4), which mark the basal and suprabasal layers of the adult OE, respectively. At early time points, both keratins showed an atypical expression pattern, acquiring their characteristic adult profile by P28 (Fig. 1i and Extended Data Fig. 1g,h). The ongoing maturation of the tissue was further illustrated by the progressive keratinization and cornification of the upper-most luminal layers, as well as increasing tissue thickness (Fig. 1i,j and Extended Data Fig. 1h-k).
These results suggest that, unlike the skin, the squamous epithelium of the oesophagus has not reached full maturation and keratinization by birth30. Instead, our data points towards P28 as the approximate age at which the features of the homeostatic OE become established.
KLF4 marks the emergence of an early committed population
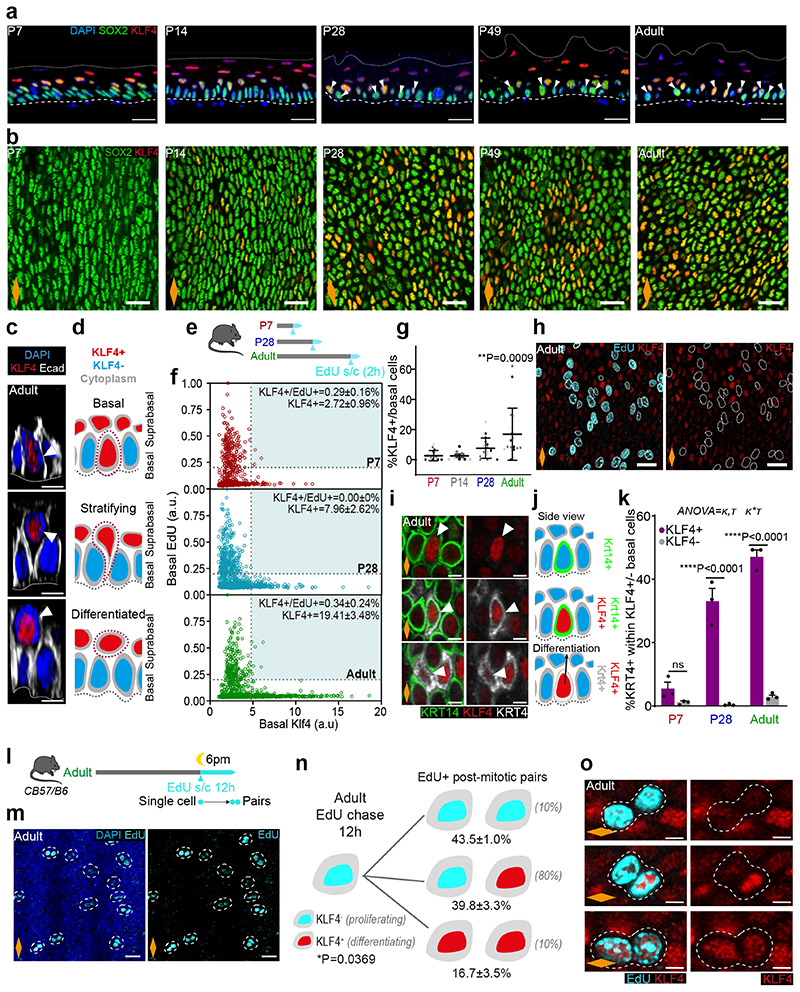
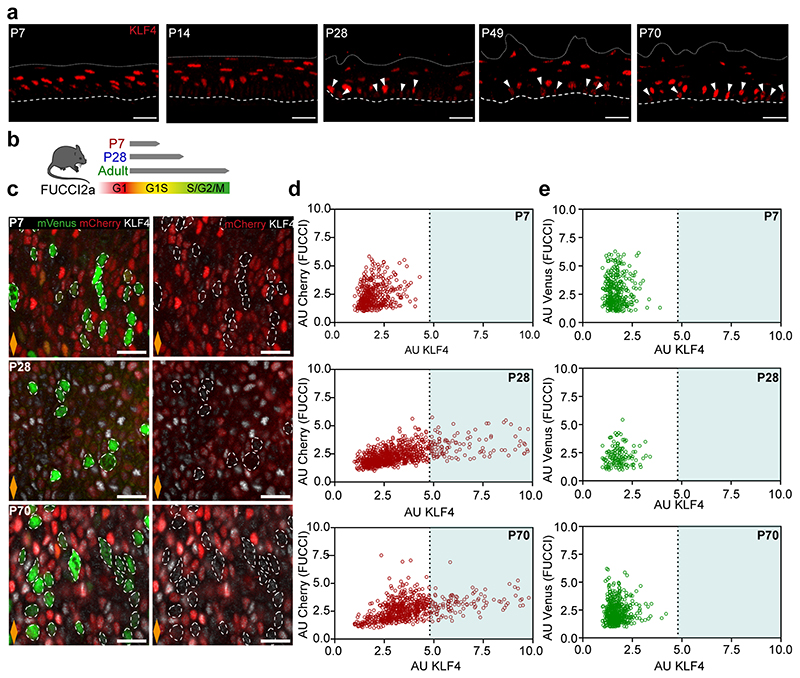
To further understand the transition towards homeostasis, we analysed the expression pattern of key transcription factors associated with cell fate in oesophageal wholemounts by 3D confocal imaging. These included SOX2, which marks basal progenitor cells in the OE31, 32, and KLF4, a well-characterised transcription factor marking suprabasal differentiated cells33–37. Similarly to KRT14 and KRT4 (Fig. 1i), SOX2 and KLF4 expression at early time points was not restricted to their typical basal and suprabasal location in adults, showing an immature pattern of expression. By P28, SOX2 and KLF4 acquired their characteristic adult distribution, reflecting the maturation of the tissue (Fig. 2a).
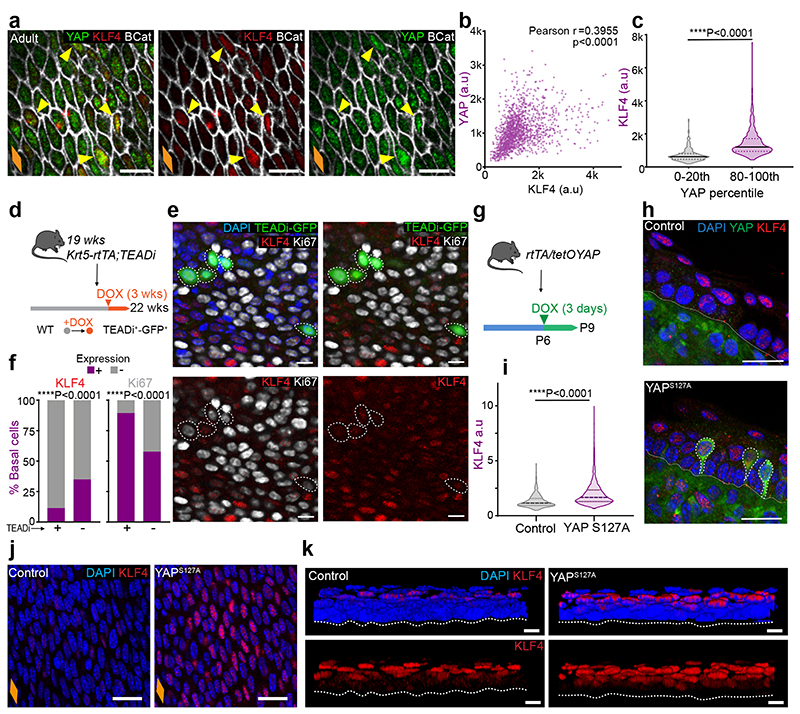
Fig. 2. KLF4 marks the emergence of an early committed population in the OE.
a, Confocal z-stack side views of typical OE wholemounts. White arrow, basal KLF4+ cells; Dashed lines, basement membrane; Dotted lines, upper limit of the OE.
b, Representative basal views of OE wholemounts from (a).
c, 3D confocal side views of basal insets from adult OE show KLF4+ cells detaching from basement membrane (dotted line) and transitioning from basal to suprabasal layers. Basement membrane (dotted line).
d, Illustration of transitioning KLF4+ (red) cells as shown in (c).
e, In vivo protocol. 2h EdU chase at indicated time points.
f, Basal EdU and KLF4 intensity quantification (a.u.) from (e) (see Methods). n=3 mice.
g, Percentage of KLF4+ basal cells expressed as mean values ± SD. One-way ANOVA with Tukey’s multiple comparisons test (*p, relative to P7). Points show individual measurements, greyscale indicates values from different mice, n=3 mice.
h, Basal views of adult OE from (e). Dotted lines, EdU+ basal cells.
i, Basal views of adult OE wholemounts showing colocalization of KLF4 with basal (KRT14, green) and suprabasal (KRT4, greyscale) markers. White arrows, cell of interest.
j, Side view illustration depicting KLF4+ (red), KRT14 (green) and KRT4 (grey) basal colocalization combinations as shown in (i).
k, Percentage of KRT4+ cells within KLF4+/- basal population (mean ± SEM, Two-way ANOVA with Tukey’s multiple comparisons test, ns, not significant, n=3 mice).
l, In vivo protocol. Adult mice were administered a single dose of EdU at 6pm, and sampled after 12 hours to identify post-mitotic EdU+ basal pairs.
m, Representative basal views of adult OE from (l). Dotted lines, EdU+ pairs.
n, Schematic showing quantitative data on KLF4 expression in EdU+ pairs from (l). One-way ANOVA, with Tukey’s multiple comparisons test; n=3 mice. Estimates from oesophageal homeostasis model by Doupe et. al.15 shown between parentheses. See Supplementary Note 1.
o, Basal confocal views of typical adult OE wholemounts from (m). Dotted lines, representative pairs.
Scale bars. 2a,b,h, and m(20 μm); 2c,i, and o(5 μm).
Orange diamonds, longitudinal orientation of the oesophagus (Extended Data Fig. 1a). Parts of (e, l) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided. See also Extended Data Fig. 2.
Interestingly, we noticed a progressive increase in the percentage of basal cells expressing high levels of KLF4 (KLF4 bright and dim cells will be referred to as KLF4+/-; Methods). KLF4+ cells were initially rare, representing ~2-3% of the basal population. This increased to ~9% by P28, reaching ~19% in the adult OE (Fig. 2a-g and Extended Data Fig. 2a). This results in a “salt and pepper pattern” in the basal progenitor compartment, with SOX2+ cells interspersed with double SOX2+/KLF4+ cells (Fig. 2a,b). The temporal changes in basal KLF4 expression, coinciding with the transition window at P28, identify KLF4 as a basal signature marking the transition towards adult homeostasis.
Since KLF4 is a transcription factor characteristic of differentiated cells, we reasoned that the previously unreported basal KLF4+ population identified here could represent a pool of early committed cells transitioning towards differentiation in OE (Fig. 2c,d). Consistent with this notion, previous work has shown how targeted deletion of Klf4 in the mouse oesophagus results in increased proliferation in the basal layer33. To explore this hypothesis, we initially investigated the proliferative activity of the KLF4+ basal subpopulation. Short-term EdU incorporation assays (Fig. 2e-h), as well as quantitative imaging analysis of wholemounts obtained from cell cycle reporter mouse line Fucci2a (R26Fucci2aR; Extended Data Fig. 2b-e), reveal that the KLF4+ population identified in the basal layer of the OE is in a non-proliferative resting state. We next evaluated the expression profile of KRT4 and KRT14 in the KLF4+/- basal populations. We noted that, although KLF4+ basal cells could be found expressing any potential combination of these keratins (Fig. 2i,j), the adult basal KLF4+ population was enriched in KRT4, revealing a bias towards differentiation (Fig. 2k). Taken together, the data support the committed state of the KLF4+ basal population. The emergence of this committed population by P28 inevitably slows down the rapid tissue expansion that takes place immediately after birth, favouring the balanced proliferation/differentiation typical of adult homeostasis.
We next explored a debated question in the field, whether basal progenitors initiate commitment toward differentiation from the moment of division, or whether cell fate outcome is assigned later, following a refractory period in the basal compartment15, 38–42. For this, we quantified the expression of KLF4 in post-mitotic EdU basal pairs, as marked by a 12h EdU chase43 (Fig. 2l,m). Our results revealed KLF4 levels being reliably detected in EdU+ sister cells just hours after their physical post-mitotic separation, supporting the notion that fate can be assigned from the time of division (Fig. 2n,o; Supplementary Note 1).
Collectively, we conclude that the emergence of KLF4 in the basal cell compartment defines an early committed population, marking the transition towards homeostasis. This data provides further insight into the temporal resolution of cell fate commitment, revealing that commitment towards differentiation can be made from the division stage.
Single-cell RNA sequencing captures onset of homeostasis
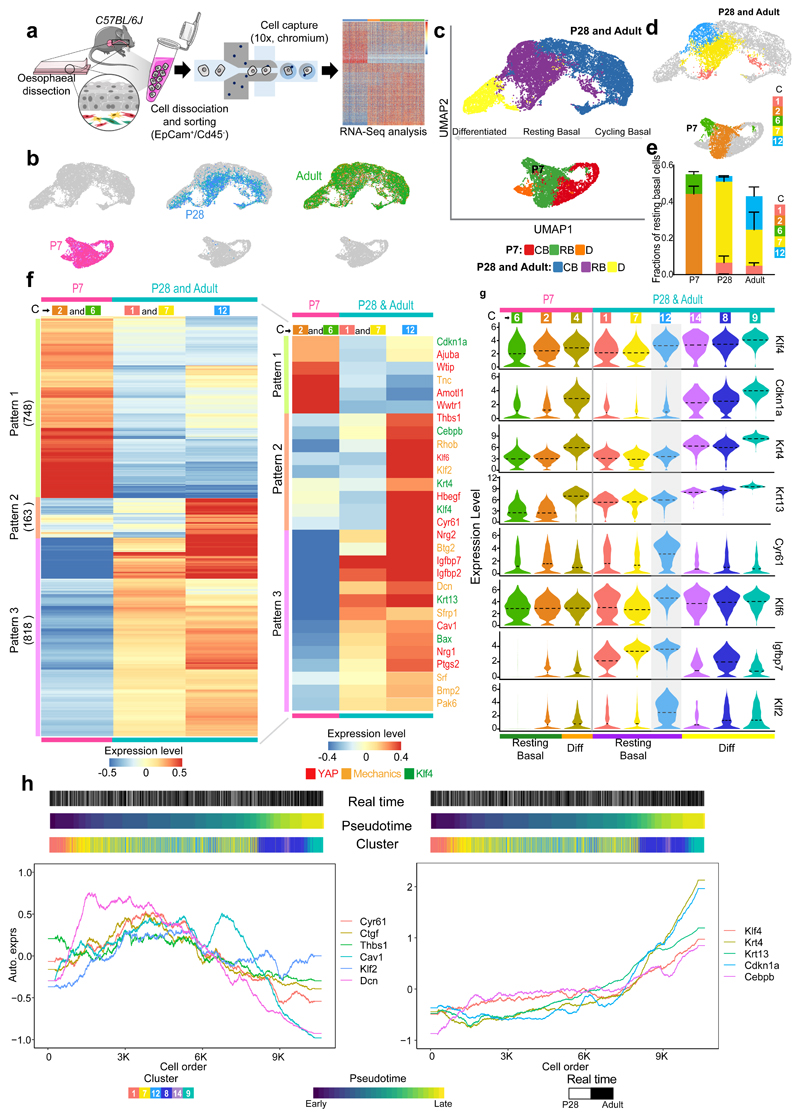
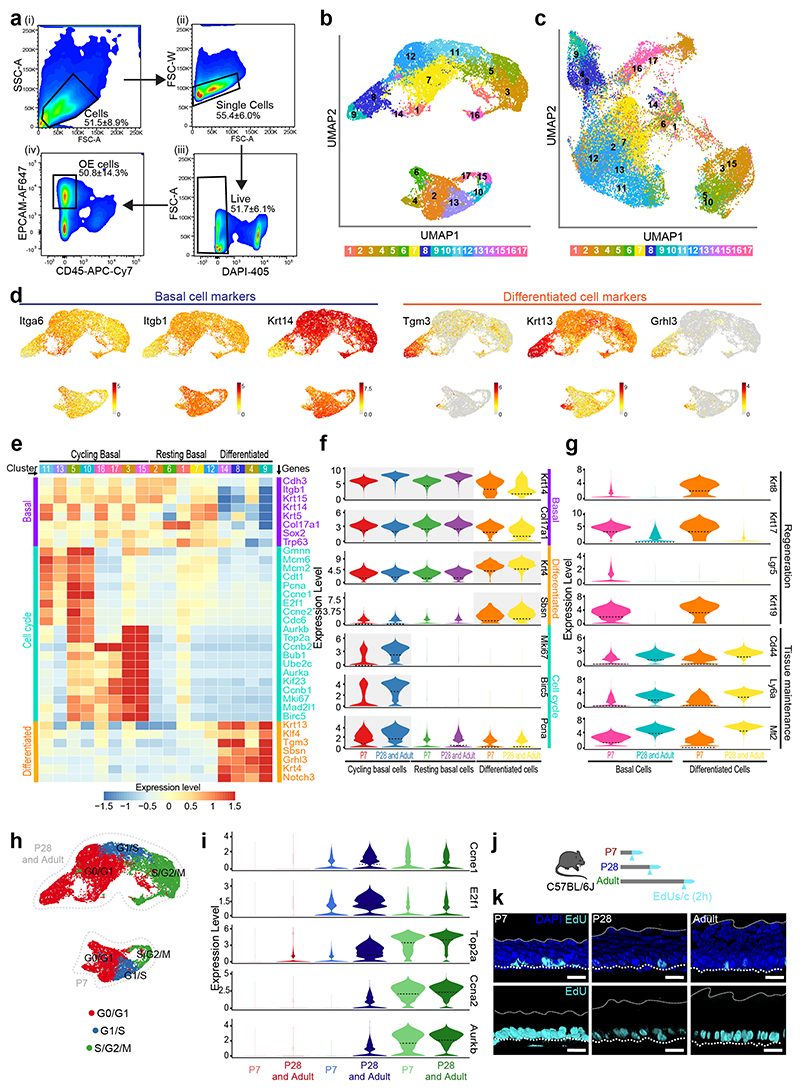
To explore the mechanisms regulating the emergence of the KLF4+ basal population and the transition to homeostasis, we made use of large-scale transcriptomics at single cell resolution, focusing on three critical time points spanning the postnatal transition: P7, P28 and Adult. Single-cell RNA sequencing (scRNA-seq) was performed on the viable epithelial cell fraction (EpCam+/Cd45-) isolated from the esophagus of C57BL/6 wild-type mice (Fig. 3a and Extended Data Fig. 3a and Supplementary Table 1). The UMAP distribution of the single-cell data revealed a noticeable separation of P7 cells, while P28 and Adult cluster closely together (Fig. 3b). This confirms that by P28, although not fully mature, the OE closely resembles the adult homeostatic tissue at the transcriptional level. Hence, P28 and Adult samples were grouped when exploring gene expression patterns, unless otherwise stated.
Fig. 3. Single-cell transcriptional profiling defines transition towards homeostasis.
a, Schematic of single-cell RNA-seq data generation from OE sorted cells (EpCam+/Cd45-) using 10X Genomics platform. Cells were isolated form 15 mice (P7), 12 mice (P28), and 9 (adults); 3 libraries (x10, Chromium) for P7 and P28, and 6 libraries for Adult.
b, Cell distribution in the dimension reduction space UMAP at different postnatal time points.
c, UMAPs representing annotated cell types. CB, cycling basal; RB, resting basal; D, differentiated.
d, UMAPs representing resting basal cell clusters from Extended Data Fig. 3b.
e, Fraction of resting basal cell clusters per time point. Fraction, expressed as mean values ± SEM. n=3 for P7 and P28 and n=6 for Adult.
f, Heatmap representing expression of individual genes belonging to distinctive patterns of gene expression in resting basal cells as defined in (c) and (d), number of genes provided in brackets. For expression values, log2-transformed normalized UMIs were scaled and averaged across all cells belonging to each group of resting basal cell cluster(s). Scale bar denotes expression range. Inset shows the expression of selected genes related to KLF4, YAP and response to mechanical stimuli. The colours of gene names denotes the relevancy to KLF4, YAP and response to mechanical stimuli.
g, Violin plots showing the expression distribution from resting basal through to differentiated clusters of selected genes in (f) related to KLF4 and tissue mechanics. The expression is log2-transformed normalized UMIs. Dotted lines, mean.
h, Expression of relevant genes along the pseudotime trajectory from resting basal to differentiated cells for P28 and Adult. Left panel, YAP target genes (Cyr61, Ctgf, Thbs1) and genes associated with a response to mechanical stimuli (Cav1, Klf2, Dcn). Right panel, depicts KLF4 target genes (Krt4, Krt13, Cdnk1a, Cebpb). Gene expression is represented as auto-scaled, log2-transformed normalized UMIs smoothed using a rolling mean along its trajectory with a window size of 5% of cells. The three bars on the top denote the arrangement of cells according to real time points, pseudotime and clusters in Extended Data Fig. 3b, respectively.
C, Cluster. Parts of (a) were drawn by using and/or adapting diagrams from Servier Medical Art.
See also Extended Data Fig. 3,4.
The combined dataset underwent unsupervised clustering, and annotation using signatures of known lineage and cell cycle markers (Fig. 3c and Extended Data Fig. 3b-i). We identified three different populations for all three time points: i) actively cycling basal progenitors, defined by increased expression of both basal and cell cycle markers; ii) resting/committed basal cells, with enriched basal signature expression and reduced levels of cycling markers; and iii) differentiated cells, marked by increased expression of genes associated with differentiation, as well as reduced levels of both basal and cell cycle marker genes (Fig. 3c and Extended Data Fig. 3j,k; Methods). Integration analysis of the unsupervised clusters confirmed the equivalence of the assigned populations between time points (Extended Data Fig. 3c, Methods). We concluded that, consistent with the current model15, the basal layer of the OE is formed by a mixture of actively cycling and resting cells.
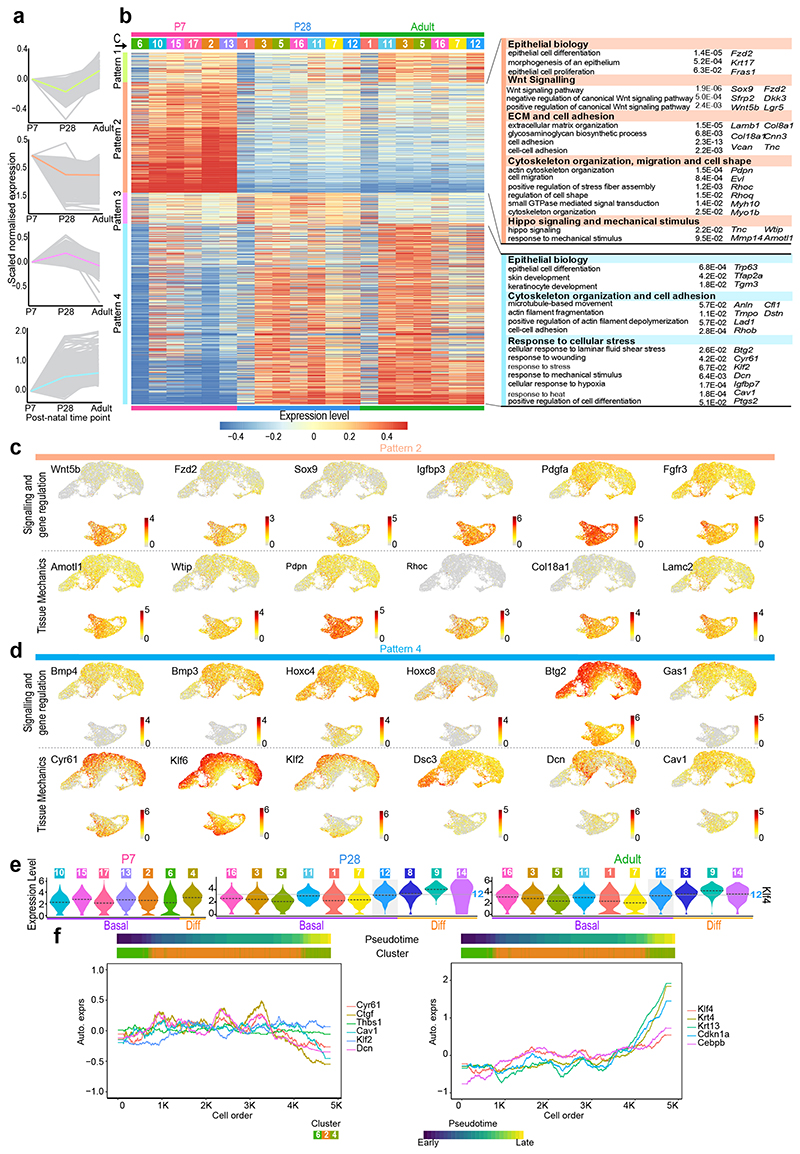
To shed light on the regulatory processes governing the epithelial transition towards homeostasis, we focused our analysis on the basal cell compartment, where progenitor cells reside. We identified four major patterns defining changes in gene expression over time (Extended Data Fig. 4a,b and Supplementary Table 2; Methods). Gene Ontology (GO) enrichment analysis of the pattern genes revealed a particularly strong WNT signature at P7 (Wnt5b, Fzd2, Sox9, Sfrp2, Lgr5, Dkk3, and Wnt11) (Extended Data Fig. 4b,c). Other signalling genes, with notable differences at P7, included increased expression of IGF related genes (Igfbp3, Igfbp4, Igf2, and Igf1r), as well as Pdgfa, Fgfr2, and Fgfr3 (Extended Data Fig. 4b,c and Supplementary Table 3), hinting at an active crosstalk between epithelial and mesenchymal cells at early postnatal stages44, 45. In contrast, later time points were indicative of a maturation process; drifting towards BMP-driven regulation (Bmp3, Bmp4, Btg2, Hoxc8, Hoxc4) (Extended Data Fig. 4b,d)46–48, and showing changes in genes related to protein biogenesis and metabolism (Supplementary Table 3)49, 50. Consistent with a transition away from the developmental state of the early postnatal oesophagus51–53, P28 and Adult cells showed an enrichment in genes involved in tissue maintenance and homeostasis, including Cd44, Ly6a, and Mt2 54. This differs from the P7 epithelium, which presented a marked expression of genes directly associated with epithelial development, wound healing and cancer (Krt8, Krt17, Krt19 and Lgr5)14, 55, 56 (Extended Data Fig. 3g). These transcriptional profiles reflect the fact that, from P28 onwards, basal cells favour maturation, leaving behind the proliferative state that governs tissue expansion shortly after birth51–53.
A subset of related GO terms were found to change at both early and later time points. We interpreted these shared changes as an indication of an active switch, suggesting their relevance during this developmental transition. These included GO terms associated with extracellular matrix (ECM) and cytoskeleton organisation, cell adhesion, and response to mechanical stimulus (Extended Data Fig. 4b-d), all critical for the mechanical stability and behaviour of epithelial cells57, 58. Indeed, the transcriptional signature of basal cells showed a marked transition from active cytoskeletal remodelling at P7 (Tnc, Pdpn, Rhoc, Evl, Vcan, Mmp14) to an increased cell adhesion at later time points (Dcn, Lad1, Cfl1, Dstn, Dsc3, Rhob). To explore whether these processes were related to the emergence of the KLF4+ committed population in the basal layer (Fig. 2), we started by assessing Klf4 expression across all clusters and time points. As predicted from tissue observations, Klf4 expression was higher in differentiated clusters, as well as in a subset of the resting basal population that becomes progressively established from P28 onward, defined by cluster (C) 12 (Extended Data Fig. 4e). In order to investigate what makes C12 distinctive, we further compared the transcriptional profile of this Klf4 high cluster with the remaining resting basal clusters (C2 and C6 for P7; C1 and C7 for P28 and Adult; Fig. 3d-g and Supplementary Table 4). We identified 3 patterns of differential gene expression in the resting basal compartment: i) Pattern 1, genes elevated at P7; ii) Pattern 2, genes highly expressed in Klf4 high C12; and iii) Pattern 3, genes progressively increasing over time (Fig. 3f). This analysis showed that Klf4 and Klf4 associated genes, including downstream targets Krt4, Krt13, Cdkn1a and Cebpb 59–61, presented a higher expression level at later time points (P28 and Adult; Fig. 3f,g). Additionally, we were able to confirm that the Klf4 signature seen in C12 is a feature of an emerging committed basal population, which becomes more marked as cells fulfil their differentiation program (C14, C8, C9; Fig. 3g). The Klf4 expression profile, therefore, strengthens histological observations suggesting that KLF4+ basal cells represent a committed population established at the onset of homeostasis.
Another feature defining resting basal cells at later time points was the differential expression of genes associated with a response to mechanical stimuli and YAP mechanosensing (Fig. 3f)62–65. Interestingly, these genes were enriched at later time points (P28 and Adult), and more particularly in the cluster containing the Klf4 high population (C12; Fig. 3f,g). More specifically, P28 and Adult samples were marked by an increase in Cyr61 expression, a direct downstream target of YAP in mouse keratinocytes, as well as other genes modulated by YAP, including, among others, Thbs1, Nrg1, Nrg2, Klf6, Hbegf, Ptgs2, Igfbp2 and Igfbp7 (Fig. 3f,g)66–71. Cav1, a key cellular mechanoregulator also known to positively control YAP in response to substrate stiffness, showed increased expression at P28 and Adult66, 72, 73. Additionally, the expression of Klf2, a shear stress-responsive transcription factor known to be critical for heart valve development, was also significantly increased at later time points74, 75(Fig. 3f,g). Accordingly, Amotl1, Wtip, and Ajuba, which have been proposed to act as negative regulators of the mechanosensing pathway Hippo76, 77, showed increased expression at P7 compared to later time points (Fig. 3f).
Given the concurrence of genes associated with KLF4 and tissue mechanics, we then focused on the dynamic pattern of gene expression to unveil any clues suggesting potential co-regulation. For this, we performed pseudotime analysis, inferring the differentiation path within the resting basal population at early (P7; Extended Data Fig. 4f) and later time points (P28 and Adult; Fig. 3h; Methods). Next, we examined gene expression along the pseudotime trajectory. At later time points, YAP associated genes, including upstream regulators (Cav1) and direct targets (Cyr61, Ctgf and Thbs1), presented a distinctive pattern of expression that peaks in C12, decreasing thereafter as differentiation becomes established (C14, C8 and C9; (Fig. 3g,h). Other mechanics related genes, such as Dcn and Klf2, presented a similar trend, with Dcn showing a much wider peak of expression that spans C7 and C12. Klf4 expression was also higher in C12 but, as anticipated for a differentiation marker, its expression increased progressively throughout the differentiation trajectory (C14, C8, C9; Fig. 3g,h). Accordingly, KLF4 downstream targets (Krt4, Krt13, Cdkn1a, and Cebpb) showed a subsequent increase in the late phases of the pseudotime trajectory, representing the most differentiated stages (Fig. 3g,h). These results point to a link between a response to mechanical stimuli and the expression of KLF4 in the basal layer of the oesophageal epithelium at late time points; an similar association could not be made at P7 (Extended Data Fig. 4f).
Our scRNA-seq analysis highlights a clear shift in cell state through postnatal development, from being highly morphogenic at P7, to a more mature state at P28 and Adult. Moreover, the uncovered transcriptional signature suggests a potential link between changes in tissue mechanics and a transition towards commitment in basal progenitor cells.
Differential growth leads to build-up in mechanical strain
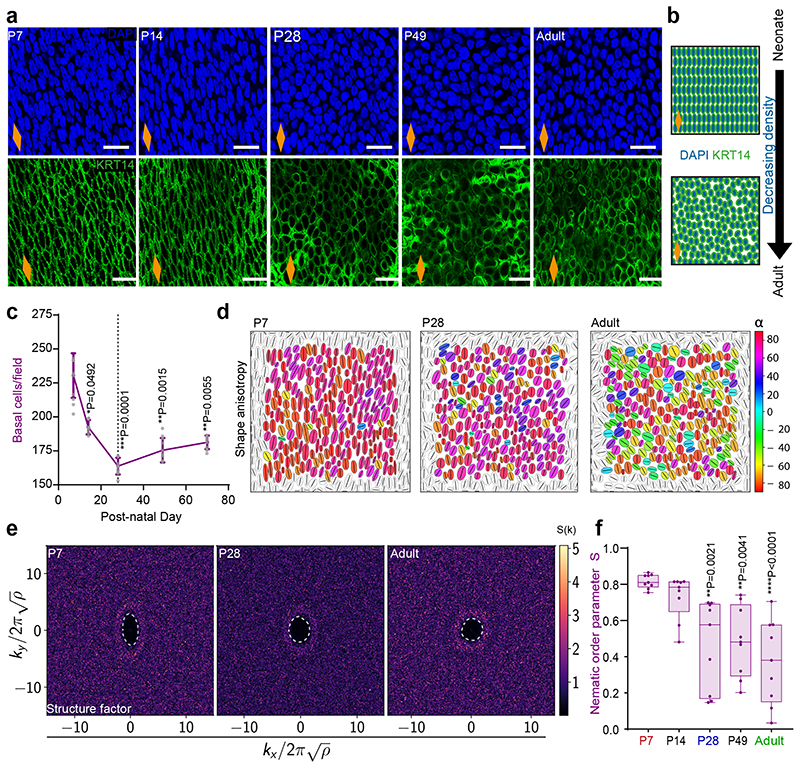
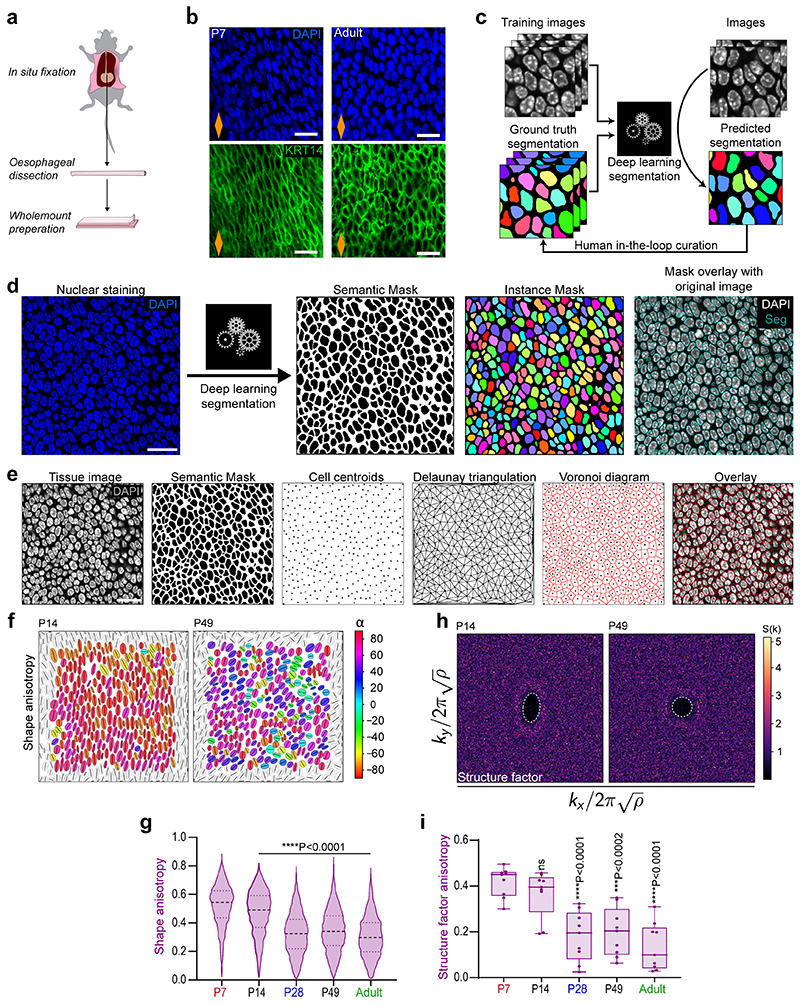
The biomechanical scRNA-seq signatures, together with considerable changes in basal cell morphology and density (Fig. 4a-c and Extended Data Fig. 5a,b), prompted us to look deeper into the architecture of the basal OE. We developed a deep learning image segmentation analysis pipeline (Extended Data Fig. 5c-e) to accurately quantify postnatal changes in cell shape, orientation and spatial organisation. Our analysis shows that the anisotropic pattern and long-range orientational alignment observed in basal cells at early time points, becomes less apparent from P28 onwards (Fig. 4d-f and Extended Data Fig. 5f-i), reflecting temporal changes in the physical properties of the tissue (Supplementary Note 1).
Fig. 4. Homeostatic transition coincides with tissue spatial reorganisation.
a, Typical basal views of confocal z-stacks showing changes in cell shape, alignment and density in OE wholemounts. Blue, DAPI; green, KRT14; scale bar 20 μm.
b, Schematic exemplifying changes seen in (a).
c, Quantification of basal cell density. Expressed as mean values ± SEM; n=3 mice.
d, Cell shape anisotropy tensor represented as an ellipse calculated from the nuclear centroid position of each basal cell. Long axis of each ellipse is proportional to the dominant eigen value of the tensor. Orientation is colour-coded. Results from representative experiment are shown; n=3 mice.
e, Bidimensional structure factor quantifying basal cell spatial organisation. Changes in the dashed white outline (from ellipse to circle) depict a transition from anisotropic to isotropic spatial cell distribution over time; n=3 mice.
f, Nematic order parameter indicative of the orientational order of cells in the tissue. Box-and-whisker plots: Box plots show median and quartiles; and whiskers, minima/maxima. n=3 mice
Nuclei at the edge of the image frame were discarded in (d) and (e) to avoid confounding effects from partially captured cells.
All data derived from wild-type C57BL/6J mice. Data analysis was performed using one-way ANOVA with Tukey’s multiple comparisons test (*p, relative to P7; ns, not significant).
Dashed line in graphs indicates P28. Orange diamonds depict the longitudinal orientation of the oesophagus where indicated (outlined in Extended Data Fig. 1a).
Source data are provided. See also Supplementary Note 1 and Extended Data Fig. 5.
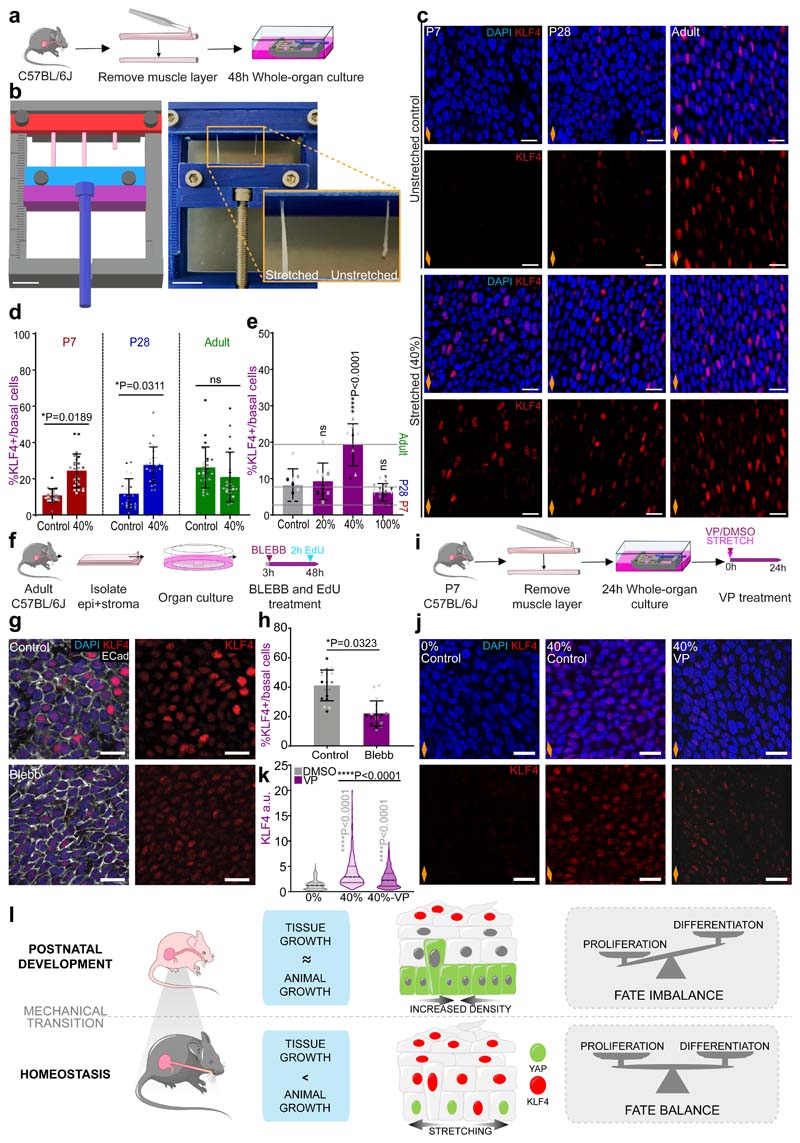
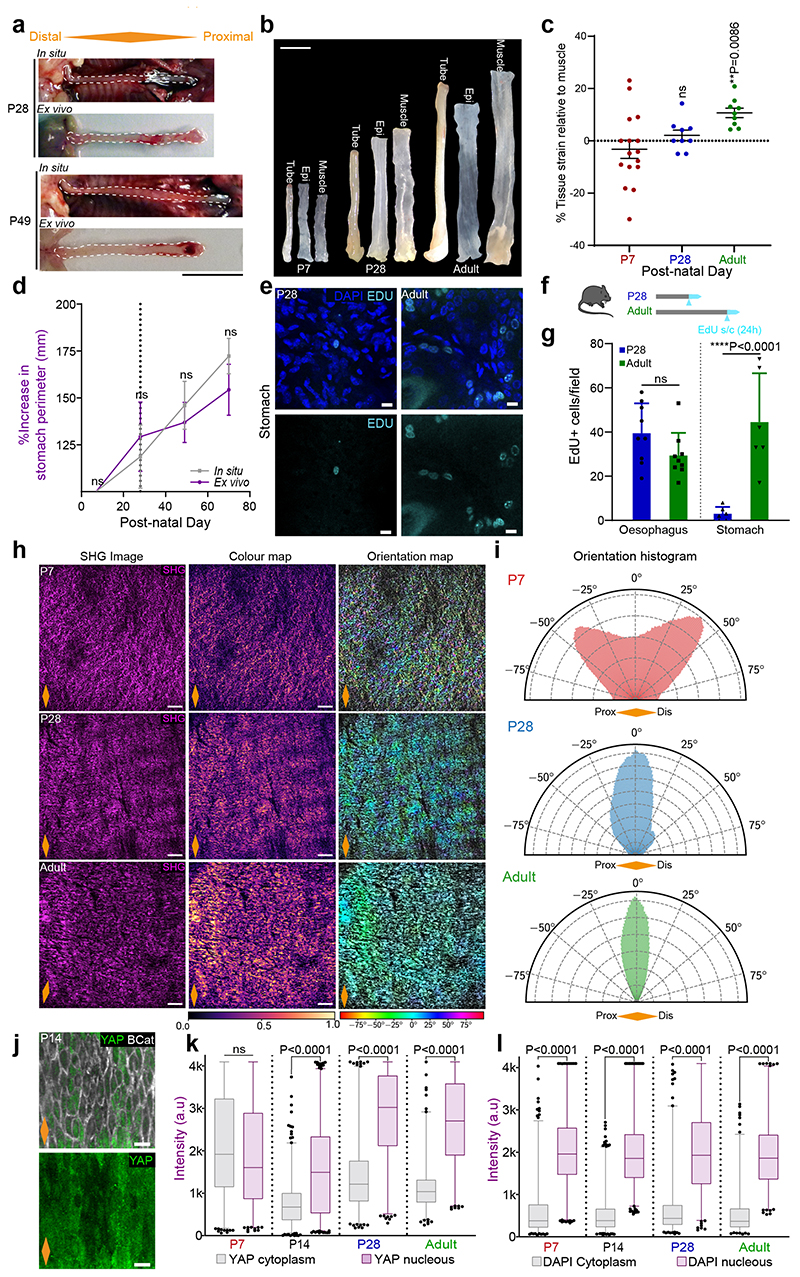
Our data, so far, hints to a possible link between changes in tissue mechanics and the transition towards homeostasis. We speculated that a potential source of mechanical stress may originate from growth anisotropy between the OE, neighbouring tissues and/or the whole body of the animal. To explore this scenario, we measured the length of the oesophagus post-mortem (in situ), within the animal, and after tissue dissection (ex vivo) at different postnatal time points (Fig. 5a,b and Extended Data Fig. 6a). The increase in length relative to P7 (in situ and ex vivo) was compared to that of the body length (Fig. 5c). Interestingly, we observed a discrepancy in oesophageal length before and after dissection, with the ex vivo oesophagus becoming significantly shorter from P28 to Adult. We established that the differential growth of the developing oesophagus, under normal physiological conditions, results in the build-up of a progressive strain, estimated to be between 20% and 40% from P28 to adult (Fig. 5d and Extended Data Fig. 6b,c; Supplementary Note 1). This growth pattern was not observed in the stomach, which showed a continuous increase in size and proliferative activity during the final postnatal stages (Extended Data Fig. 6d-g). Interestingly, this mechanical transition coincided with an active remodelling of the ECM, as evidenced by temporal changes in second harmonic generation (Extended Data Fig. 6h,i; Supplementary Note 1). We, therefore, conclude that the OE undergoes a mechanically-mediated transition, from being densely packed at earlier time points, when rapid proliferation promotes cell crowding (Fig. 4c), to becoming more loosely packed and stretched upon homeostasis (Fig. 5c,d).
Fig. 5. Differential growth generates longitudinal tissue strain sensed at the cellular level.
a, Method for oesophageal length measurements in situ and ex vivo (l).
b, In situ and immediate ex vivo images of oesophageal tubes captured in P7 and adult mice. Dashed lines delineate oesophageal tube.
c, Percentage increase in oesophageal length (in situ and ex vivo) compared to body length (excluding tail) throughout postnatal development (P7 set to 100). Dashed line indicates P28. Data expressed as mean ± SEM. Each time point was analysed using two tailed unpaired t-test (ns, not significant; grey and blue indicate difference between body and ex vivo, body and in vivo, respectively.); n=6 mice.
d, Longitudinal tissue strain in vivo represented as percentage. Data expressed as mean ± SEM; n=6 mice.
e and f, Schematic representing changes in F-actin (grey) and pMLC2 (green) levels in OE cells throughout postnatal development.
g, Basal views of typical OE wholemount showing Phalloidin staining for F-actin at indicated time points. Blue, DAPI; greyscale, Phalloidin.
h, Representative side views of tissue sections showing pMLC2 staining at indicated time points. Blue, DAPI; green, pMLC2; Dotted lines, basement membrane; ep, epithelium; strm, stroma.
i and j, Quantification of basal F-actin and pMLC2 staining, respectively (see Methods). Box plots show median and quartiles; and whiskers, minima/maxima; (i) n=3 animals, (j) n=4 mice.
k, Schematic representation of changes in YAP (green) localization during postnatal development.
l, Basal view of representative OE wholemounts showing progressive translocation of YAP to the nucleus as tissue matures. Blue, DAPI; green, YAP; greyscale, B-Catenin (BCat).
m, Quantification of basal nuclear and cytoplasmic YAP staining (see Methods). A total of 60 cells per time point were measured from 3 animals. Localization pattern shows the average intensity for one representative animal per time point. Full quantification in Extended Data Fig. 6 j, k.
Scale bars. 5b(1 cm); 5g,l(20 μm); 5h (10 μm).
Data analysis for d, i, and j was performed using one-way ANOVA with Tukey’s multiple comparisons test (*p; relative to P7; ns, not significant).
Orange diamonds, longitudinal orientation of the oesophagus (Extended Data Fig. 1a). Parts of (a) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided. See also Supplementary Note 1 and Extended Data Fig. 6.
To test whether the observed changes in the mechanical properties of the oesophagus stimulated a switch in tension and cytoskeleton remodelling of epithelial cells, we labelled the OE for F-actin and phosphorylated myosin light chain II (pMLC2), a marker of cortical actomyosin contractility. Both presented a similar profile, increasing their signal as tissue matures (Fig. 5e-j; Supplementary Note 1). We next interrogated the cytoplasmic-to-nuclear translocation of the transcription factor YAP, an effector of the mechanosensing pathway Hippo78. YAP nuclear localization is associated with increased space availability, cell spreading, and subsequent cytoskeletal reorganisation. As predicted by the increase in tensile strain observed in the OE, YAP was found to translocate from its cytoplasmic localization at P7 to the nucleus in adult animals, with an intermediate state at P28 reflecting a mechanosensing transition (Fig. 5k-m and Extended Data Fig. 6j-l). This data is indicative of basal progenitors sensing the physiological changes in strain and space availability experienced by the tissue.
We conclude that the differential growth of the oesophagus results in the build-up of a longitudinal tensile strain as the OE transitions towards homeostasis. Changes in the cellular response to mechanical stimulus, initially evidenced at the transcriptional level, were further reinforced by the cytoplasmic to nuclear translocation of the Hippo effector YAP.
YAP promotes basal commitment through KLF4
The experiments above suggest a temporal correlation between YAP and KLF4 expression patterns, with both transcription factors becoming localized to the nucleus of basal cells as the tissue transitions towards homeostasis (Fig. 2 and Fig. 5l,m). To explore the existence of a potential link between the two, we used three different approaches. First, we co-stained and correlated the levels of both proteins in the nucleus of adult basal cells (Fig. 6a-c). Second, we looked at KLF4 expression upon interfering with YAP signalling in adult mice. For this, we used an inducible mouse line (Krt5-rtTA;TEADi-GFP79), where genetic induction results in the blockage of YAP- and TAZ-induced TEAD activity in the basal cell compartment, detected by the TEADi-GFP reporter. (Fig. 6d-f). Third, we investigated the impact of YAP overactivation in the early postnatal OE (P6-P8), when basal cells are still negative for KLF4 (Fig. 2a,b,g), using the YAPS127A Doxycycline (Dox) inducible mouse strain R26-rtTA;tetO-YAPS127A (rtTA/tetOYAP). (Fig. 6g-k). Collectively, these experiments showed a correlation between YAP and KLF4 expression, and demonstrated that YAP nuclear activity promotes KLF4 expression in the basal cell compartment, balancing progenitor cell behaviour through commitment (Supplementary Note 1).
Fig. 6. YAP nuclear localization promotes increased levels of basal commitment through KLF4.
a, Confocal basal views of typical adult OE from C57BL/6J WT mice. Yellow arrows indicate colocalization of KLF4 (high) and YAP (high) staining.
b, Correlation plot for KLF4 and YAP intensities in the basal layer the adult OE (a). n=3 mice. (r, Pearson correlation coefficient)
c, Quantification of KLF4 intensity (a.u) within the lowest and highest YAP expressing cells (0-20th and 80-100th percentile respectively). n=296 cells per percentile from 3 mice.
d, In vivo protocol. Adult Krt5-rtTA;TEADi mice were doxycycline (DOX) treated for 3 weeks to block YAP and TAZ-induced TEAD activity in basal cells. TEADi expressing cells are marked by GFP expression (TEADi+).
e, Representative basal views of OE wholemounts from (d). TEADi expressing cells are marked by GFP expression. Dotted circles indicate GFP+ cells.
f, Percentage of KLF4+/- and Ki67+/- cells in TEADi+/- basal cells from (d, as marked by GFP). A total of 800 TEADi+/-basal cells were quantified from 72 fields out of 2 mice. Analysed by one sided Chi-squared test.
g, In vivo protocol. P6 rtTA/tetOYAP pups were doxycycline treated for 3 days to induce an active form of YAP (S127A), and culled at P9.
h, Staining in OCT embedded OE cryosections (10 μm thick) of YAP activated mice from (g). Nuclear KLF4 expression colocalizes with YAP+ cells. Dotted white lines, YAP+ cells.
i, KLF4 intensity quantification (a.u) in control and YAP overexpressing mice as shown in (j). n=1362-1779 cells from 3 animals.
j, Representative basal views of OE wholemounts from (g).
k, Representative 3D rendered side views of OE wholemounts from (g), showing expanded KLF4 suprabasal compartment in rtTA/tetOYAP mice compared to controls.
Scale bars. 6a,e and k(10 μm), 6h,j(20μm).
Stainings. Blue, DAPI. 6A, 6H, 6J, 6K (green, YAP; red, KLF4; greyscale, BCat). 6A (green, TEADi; red, KLF4; greyscale, Ki67)
Dotted line, basement membrane (h, k). Violin plots (c, i) show median (solid line) and quartiles (dotted lines). Data from (c) and (i) were analysed using two-tailed unpaired t test.
Orange diamonds depict the longitudinal orientation of the oesophagus where indicated (outlined in Extended Data Fig. 1a). Parts of (d, g) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided. See also Supplementary Note 1.
Changes in tissue mechanics influence basal KLF4 expression
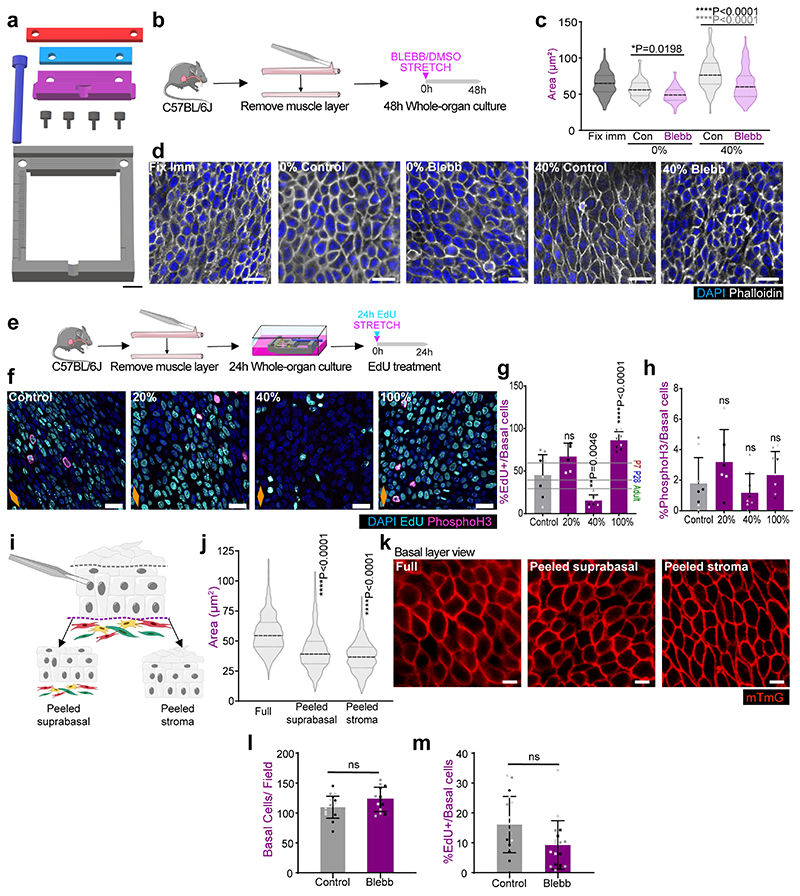
Having identified an increase in tensile strain throughout postnatal development, we next interrogated whether the mechanical stress imposed at the whole organ level was sensed at the cellular level. To answer this question, we used a whole-organ in vitro approach utilising a 3D printed stretching device80 (Fig. 7a,b and Extended Data Fig. 7a) that allows for controlled uniaxial tissue stretching (Methods). Adult OE were stretched along their longitudinal axis at a static strain of 40%, representing the physiological levels measured in the adult oesophagus (Fig. 5d and Extended Data Fig. 6c; Methods). As a result, there was an increase in basal cell area (Extended Data Fig. 7b,c) that could be rescued by blebbistatin (BLEBB) treatment, which inhibits myosin II motor activity disrupting tissue tension and cell junction stability (Extended Data Fig. 7b-d; 40% stretch). Accordingly, BLEBB lead to a marked cellular disorganisation. These results suggest that basal cells bear part of the mechanical load experienced upon whole-organ stretching (Supplementary Note 1).
Fig. 7. Changes in tissue mechanics influence basal KLF4 expression.
a, In vitro protocol. Oesophageal tubes were exposed to a 40% stretch using 3D-printed stretcher and kept in vitro as whole-organ cultures for 48h.
b, 3D model of stretching device (left), in use (right).
c, Confocal basal views of stretched and unstreched control samples after 48h.
d, Basal quantification of KLF4+ cells expressed as percentage of DAPI+ basal cells from (c). P7 and P28 n=5 mice, Adult n=6 mice.
e, Threshold response. Basal quantification of KLF4+ cells in P7 whole-organ cultures after exposing to 20%, 40% or 100% longitudinal stretch, or unstreched control conditions for 24h (as per a). n=3 mice.
f, In vitro protocol. Adult epithelial-stromal composites were kept in vitro and treated with 25μM blebbistatin (BLEBB) for 48h and EdU for the final 2h.
g, Confocal basal views after a 48 hour BLEBB treatment in vitro (f).
h, Percentage of KLF4+ basal cells relative to DAPI+ basal cells from (f). n=3 mice.
i, In vitro protocol. Oesophagi were exposed to 40% stretch and treated with DMSO/Verteporfin (VP; 1 μM) for 24 hours.
j, Confocal basal views after 24 hour VP treatment in vitro (i).
k, KLF4 intensity quantification (a.u) in OE basal cells from (i). A total of n=794-1398 cells from 3 animals per condition.
l, Suggested model. The physiological stretching experienced by the postnatal oesophagus promotes the expression of KLF4 in the basal progenitor cell compartment in a YAP dependent manner. The onset of basal KLF4 expression marks the emergence of an early committed population, which balances proliferation, and defines the transition towards adult homeostasis.
Scale bars. 7b(1cm), inset (5mm) c,g,j(20μm).
Stainings. Blue, DAPI; red, KLF4; greyscale, Ecad.
Data expressed as mean±SD. Analysis for (e) and (k) one-way ANOVA with Tukey’s multiple comparisons test (ns, not significant). (e) relative to Control. Analysis for (d) and (g) was performed using two-tailed unpaired t test (ns, not significant). Individual points show individual measurements, greyscale indicates values from each of 3 mice.
Orange diamonds, longitudinal orientation of the oesophagus (Extended Data Fig. 1a). Parts of (a, f, i, l) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided. See also Extended Data Fig. 7.
We next investigated the relevance of mechanical stress in driving the transition to homeostasis. Oesophageal samples from P7, P28 and Adult animals were stretched to adult physiological levels (40%). The expression of KLF4 was analysed as a proxy to identify changes in cell commitment in the basal layer. Reassuringly, the unstretched in vitro controls exhibited a similar KLF4 expression profile to that observed in vivo (Fig. 7c, top panels versus Fig. 2b). Moreover, KLF4 expression remained unchanged in adult stretched samples, confirming that the strain was within physiological range (Fig. 7c,d). P7 and P28 samples, instead, showed a significant increase in KLF4 basal levels that reached control adult values upon stretching (Fig. 7c,d). This data suggests that physiological levels of tensile stress promote KLF4 expression in the basal compartment. Further experiments exposing P7 samples to a range of tensile strains, revealed that changes in basal KLF4 expression respond to a strain threshold between 20% and 40% (Fig. 7e), matching the levels found at the transition between P28 and Adult (Fig. 5d and Extended Data Fig. 6c; Methods). Accordingly, the proliferative activity of the oesophagus at P7 was reduced at 40% strain (Extended Data Fig. 7e-h). Contrary to physiological levels of strain, stretching by 100% promoted an increase in proliferative activity, in line with previous studies on overstretched tissues81. These results support the notion of a biomechanical switch operating during the homeostatic transition of the OE that dictates a shift in cell behaviour.
Remarkably, KLF4 levels in adult unstretched controls closely resembled those of its in vivo counterpart (Fig. 7c). This suggests that, while an increase in strain is sufficient to activate KLF4 expression in basal cells, it does not seem necessary to maintain it in the adult, where suprabasal keratinization provides the means to retain tissue architecture to a certain extent (Extended Data Fig. 1j,k and Extended Data Fig.7i-k)82, 83.
Supporting the idea that basal KLF4 expression responds to changes in tissue mechanics, in vitro treatment with BLEBB to lower tissue tension (Extended Data Fig. 7l), by disrupting actomyosin contractility, led to a reduction in KLF4 expression in the adult OE (Fig. 7f-h), although no impact on cell proliferation was observed (Extended Data Fig. 7l,m). Further mechanistic insight was achieved by treating stretched P7 oesophagi with Verteporfin, an established pharmacological inhibitor of YAP84, 85. Verteporfin rescued the strain-induced KLF4 response (Fig. 7i-k), demonstrating that the mechanical regulation of KLF4 expression is mediated by YAP.
Taken together, our results demonstrate that the longitudinal tensile strain, generated by the anisotropic growth of the oesophagus during postnatal development, must reach a critical threshold in order to affect cell behaviour. Once this threshold is reached, basal KLF4 expression increases modulated via YAP. This results in progenitor cells tilting their balance towards commitment, which ensures the establishment of tissue homeostasis (Fig. 7l).
Discussion
Here we report that the physiological strain emerging during the normal postnatal development of the oesophagus controls its maturation, temporally guided by a mechanical threshold. In particular, we show that the differential growth of the oesophagus relative to the entire body generates a level of mechanical stretch that influences epithelial cell fate and promotes the transition towards homeostasis. Mechanistically, our data indicates that the physiological stretching of the oesophagus results in the nuclear re-localization of YAP mechanosensor, which in turn promotes the emergence of a KLF4+ committed population in the basal progenitor compartment. The onset of basal cell commitment towards differentiation balances proliferation, and ultimately defines the transition to adult homeostasis.
Tissue morphogenesis has long been known to be orchestrated by finely balanced gradients of morphogens. More recent studies in a growing number of developmental systems are starting to uncover the impact that diverse types of mechanical forces have on major organs in the body24, 29, 81, 86, 87. From the shear stress dictating the development of the heart and lungs in mouse and zebrafish embryos18, 74, 75, to the mechanical stress guiding the patterning of the intestine and skin follicles in the developing chick and mouse19–21, 88. However, there is surprisingly limited knowledge on the mechanisms regulating how developmental processes come to an end, safeguarding adult tissue homeostasis from abnormal growth and disease. Our findings in the postnatal mouse oesophagus reveal that this transition is linked to the mechanical changes experienced by the tissue. The rapid expansion of the oesophagus after birth is progressively restricted as the OE matures and keratinizes. When that occurs, the oesophagus no longer grows at the same rate as the rest of the body, resulting in the progressive stretching of the OE. This is marked by a transcriptional switch, showing an upregulation in the expression of genes known to regulate the response to mechanical stimuli (Klf2, Cav1, Dcn)72, 74, 75, including YAP associated genes (Cyr61, Thbs1, Igfbp2, Klf6, among others)66, 67, 69, 71. Interestingly, this transcriptional signature is a feature of a particular committed cell population that emerges at P28 in the basal progenitor compartment, and is marked by high levels of KLF4 expression. The presence of a differentiating population in the basal layer promotes a more balanced progenitor cell behaviour, signalling the transition towards adult homeostasis15. Ex vivo stretching experiments reveal that the build-up in tissue strain experienced by the oesophagus during its postnatal development is responsible for the emergence of this KLF4+ basal committed population. Remarkably, we demonstrate the existence of a defined strain threshold that triggers the tilt in basal cell fate towards commitment, offering a means by which the temporal regulation of this process may be achieved. Genetic mouse models provide further mechanistic insights revealing that KLF4 expression is regulated in a YAP dependent manner. Our results establish a direct link between differential growth, tissue stretching and the acquisition of a committed basal population that defines the transition towards homeostasis. This supports the idea that changes in tissue mechanics can regulate epithelial behaviour in response to specific tissue needs.
Several in vitro and in vivo studies have shown that activation of typical mechanosensors such as YAP/TAZ and Piezo1 triggers an epithelial proliferative response66, 89–94. Additional work in the mouse intestinal epithelium indicates that YAP can also regulate cell differentiation, specifically through its interaction with KLF495. Our observations suggest a model in which changes in cell behaviour are regulated by integrating the physiological levels of mechanical stress happening at the organ level, with those sensed by local neighbouring cells in a YAP dependent fashion. This provides a simple solution to orchestrate the maintenance of a whole tissue at the cellular level (model in Fig. 7l).
Our study offers a new perspective in understanding the mechanisms governing epithelial cell transitions. These observations are likely to have wider implications, and open exciting new questions as to whether cell behaviour is regulated similarly in other mechanically active tissues96–100. Future studies should elucidate whether regulatory processes controlled through differential growth are a broader phenomenon operating in other tissue models, as well as their translational relevance in regeneration and cancer6, 87, 101.
Methods
Mice strains and allele induction
All animal experiments were approved by the local ethical review committees. Work was approved by the University of Cambridge, and conducted according to Home Office project license PPL70/8866 at the Wellcome Trust-Medical Research Council Cambridge Stem Cell Institute, Cambridge University. Additional mouse work was conducted according to approved protocols LCMB-034 and 19-329 from the NIH-Intramural Animal Care and Use Committee (ACUC) of the National Cancer Institute (NIH), or under permit number 2017-15-0201-01381 reviewed and approved by The National Animal Ethics Committee in Denmark.
Unless otherwise specified, C57BL/6J mice wild type (wt) (Charles River, UK; strain code, 632) were used. Genetically engineered mouse lines used include: FUCCI2a (R26Fucci2aR, kindly provided by Ian J. Jackson102); tetO-YAP S127A (kindly provided by Jonas Larsson103); Rosa26-rtTAM2 (stock #006965, Jackson Laboratory); Krt5-rtTA;TEADi (kindly provided by Ramiro Iglesias-Bartolome79); and Rosa26-mT/mG (stock # 007676, Jackson Laboratory104). Animals cohorts were used at exact postnatal days (P) indicated, the adult cohort constitutes mice between 10-14 weeks of age.
All strains were maintained in a C57BL/6 background. All experiments comprised male and female mice, with no gender specific differences observed. Only male mice were used in single-cell RNA sequencing (scRNA-seq) experiments, to avoid confounding effects due to oestrous cycle. All animals were housed between 19-23°C, 45-65% humidity and a day/night light cycle of 12 hours on, 12 hours off.
Tissue dimensions
Oesophageal dimensions were measured from P2-Adult. Wt mice were culled and oesophagi were excised, taking as reference two anatomical landmarks: the cricoid cartilage and the opening of stomach fundus. Oesophageal tubes were longitudinally opened and flattened, to measure length and width under a dissecting microscope, and tissue area was calculated. Tissue area averaged from n = 3-22 per time point.
For in situ vs ex vivo comparison, we performed an immediate in situ post-mortem tissue measurement, followed by a subsequent measurement after dissection (ex vivo) (Fig. 5a). Wt mice were culled at indicated time points and organs surrounding the oesophagus and stomach were removed for clear view. In situ images were taken, from which the length (oesophagus) and perimeter outline (stomach) were measured. This was repeated immediately after tissue dissection. Measurements were expressed as a percentage relative to their value at P7.
Histology
Haematoxylin and Eosin (H&E) staining was performed by the Histology Core Service at Cambridge Stem Cell Institute and imaged using EvosXL Core Cell Imaging System.
Tissue preparation and Immunofluorescence
For in situ fixation of the oesophagus (Extended Data Fig. 5a,b), wt mice were culled and organs surrounding the oesophagus and stomach were removed. The thoracic cavity was filled with 4% paraformaldehyde (Alfa Aesar; 043368) in PBS (4% PFA) for 10 minutes (min) for fixation. The oesophagus was then excised and processed for immunostaining as indicated below.
For oesophageal wholemounting (WM), oesophagi were excised, opened longitudinally, the muscle layer removed, and the tissue flattened under a dissecting microscope. The epithelial-stromal composite was then fixed in 4% PFA, 30min. Samples were incubated for 30min in permeabilization buffer (PB; 0.5% Bovine serum albumin (VWR International; 126575-10), 0.25% Fish skin gelatin (Sigma; G7765), 1% Triton X-100 (Fisher Scientific Ltd; 10102913) in PBS), and blocked for 2 hours (h) in PB containing 10% Donkey Serum (PBDS; Scientific Laboratory Supplies; D9663). Primary antibodies (Supplementary Table 5) were incubated in PBDS for 3 days (d) at 4°C followed by 4 washes over 24h with 0.2% Tween-20 (Promega UK Ltd; H5151) in PBS. Secondary antibodies were incubated overnight at 4°C in PBDS and washed 4 times over 4h with 0.2% Tween-20 in PBS. Cell nuclei were stained with 1μg/ml DAPI (Simga; D9542) in PBS at 4°C before mounting. Wholemounts (WM) were cleared in 1.52 Rapiclear mounting media (SUNJin Lab; RC152001). Organ culture samples were stained following the same procedure.
WMs of the oesophageal epithelium (OE) were stained with specific antibodies. These were prepared by incubating oesophagi in 5mM EDTA (Life Technologies; 15575020) in PBS for 2.5h at 37°C following muscle layer removal. The epithelium was peeled from the stroma and fixed as above. Staining was performed using low Triton PB (pB; 0.5% Bovine serum albumin, 0.25% Fish skin gelatine, 0.5% Triton X-100 in PBS) and the following altered timings: permeabilization in pB, 10min; blocking in pBDS, 1h; primary antibodies in pBDS, overnight at 4°C; 0.2% Tween-20 in PBS washes, over 2h; secondary antibodies in pBDS, 3h; washes as with primary; cell nuclei staining as above.
For staining of 10μm cryosections, fixed oesophagi were embedded in optimal cutting temperature compound (OCT; Fisher Scientific Ltd; 12678646) for cryosectioning onto glass slides. Sections were permeabilized in pB for 10min, blocked in pBDS for 30min, incubated overnight with primary antibodies in pB at 4°C, followed by a 1h incubation with secondary antibodies in pB. Sections were washed 4 times for 5min in 0.2% Tween-20 in PBS between incubations.
Where indicated, EdU (5-Ethynyl-2’-deoxyuridine) incorporation was detected using Click-iT imaging kits according to the manufacturer’s instructions (Invitrogen).
EdU tracing
For in vivo tracing, EdU (Life Technologies; A10044) was administered 10ul/g body-weight at 0.1mg/ml via subcutaneous (s/c) injection. Tissues were collected 2 and 24h after injection. In organ cultures, EdU was added to the culture media at a 10μM final concentration, and incubated for 2h at 37°C and 5% CO2. EdU+ cells were quantified in WMs from 3-9 confocal z-stacks per animal.
For tracking EdU+ post-mitotic cell pairs in adult mice, EdU was administered at 18:00 (s/c), when the proportion of S-phase cells is low in the circadian cycle, making clonal density labelling feasible, and chased for 12h43, 105. KLF4 expression profile was determined in 225 EdU pairs from 3 different animals.
Basal cell density
Basal cell density in oesophageal WMs was calculated by quantifying the number of DAPI+ basal cells per field in 3-6 images per animal.
Total cell production and imbalance
First, we estimated the total number of basal cells in the whole oesophagus over time, by multiplying basal cell density by total tissue area at each time point. Total basal cell production rate, Ḃ, was then estimated as the average change in basal cell number, B, divided by the time step.
The degree of fate imbalance, i.e. the degree to which basal cell fate is biased towards duplication over differentiation and cell loss, is estimated as (1/λ)Ḃ/B, where λ is the cell division rate. The division rate is determined from the results of short-term (2h) EdU incorporation assay (Fig. 2e) and normalized by the reported division rate of basal cells in the homeostatic adult tissue15.
Tissue thickness
Tissue thickness was measured using side views of rendered confocal oesophageal WM images. 18 individual measurements were made for each of 3 animals.
Imaging, analysis and segmentation
Confocal images were acquired using either inverted Leica SP5 with standard laser configuration, SP8-X with white light laser or upright SP8-X with standard laser configuration with LAS X 3.5.5.19976 Leica software. Typical confocal settings used included: 40X objective, optimal pinhole, scan speed 400Hz, line average 3, optimal step size, and resolutions of 512x512 or 1024x1024 pixels. For quantification, images where acquired with a digital zoom of 2X or 3X.
Images were reconstructed from optical sections using Volocity 5.3.3 software (PerkinElmer) and ImageJ 1.51W. KLF4, EdU, Fucci2a (mCherry and mVenus), as well as YAP intensity measurements (Fig. 6b,c) were performed on Volocity 5.3.3 using semi-automated nuclear segmentation. For the analysis of KLF4 bright cells (KLF4+), the threshold was set to include nuclei above the 80th percentile of KLF4 intensity (Fig. 2b,f,g and Extended Data Fig. 2c-e). This was visually confirmed to be over the background level of expression for nuclear KLF4 staining. EdU was set as >0.2 as detected in representative confocal images. F actin (Phalloidin labelling) and phosphorylated myosin light chain II (pMLC2) were quantified using image segmentation protocol described in Supplementary Note 1.
YAP localization was assessed by measuring the fluorescent intensity of YAP immunolabeling and DAPI staining across the major cell axis of each individual cell. Nuclear and cytoplasmic YAP intensities were assigned per pixel based on DAPI intensity. Those within the lower 20th percentile of DAPI intensity were considered cytoplasmic, while those within the top 20th percentile of DAPI intensity were measured as nuclear (Fig. 5m and Extended Data Fig. 6j-l). A total of 240 cells were analysed from 3 separate animals at four different time points (P7, P14, P28 and Adult; 60 cells per time point).
Collagen fibres were imaged using second harmonic generation (SHG), alongside NucRed647 (Life Technologies; R37106) in WMs. Imaging was performed using an inverted Leica TCS SP5 acousto-optic beam splitter (AOBS) multiphoton laser scanning microscope equipped with a Modelocked Ti:Sapphire Laser (Chameleon Ultra II, Coherent Inc., 680-1080nm tuning range, 80-MHz repetition rate, 140-fs pulse width) that allows for SHG imaging. For image acquisition the excitation laser (Ti:Sapphire) was tuned to 900nm, producing emission of SHG signal at 450nm. Typical settings included: 40X objective, pinhole 600μm, scan speed 400Hz, line average 2, 0.15μm step size, and resolution 1024x1024 pixels. 3 images per animal were captured, and the collagen fibre orientation was assessed using pipeline outlined in Supplementary Note 1.
For single cell image segmentation of WMs, single 2μm z-slices of the basal layer, acquired using a Leica SP5 confocal microscope system, were used. For single cell image segmentation of cryosections, single 2μm z-slices of tissue cross-sections were acquired with the criteria outlined above. A minimum of 3 images were captured per animal. Analysis performed via pipeline described in Supplementary Note 1.
Note that images from in situ oesophagi (fixed post-mortem undissected) were compared with oesophagi fixed immediately after dissection to confirm that basal cell shape was not affected by tissue harvesting (Fig. 4a versus Extended Data Fig. 5a,b).
RNA sequencing
Single cell and RNA isolation
Epithelial-stromal composites were obtained as indicated above. For each sample, oesophagi were pooled from different animals (C57BL/6 wild-type mice) to enable the capture of sufficient cell numbers (P7, 5 mice; P28, 4 mice; Adult, 3 mice; 3 independent samples for P7 and P28, 6 independent samples for adult).
Following initial preparation, WMs were cut into 3mm2 pieces, and incubated in 0.5mg/ml Dispase (Sigma; D4818) in PBS containing 1U/μl RNAse Inhibitor (Life Technologies; AM2696) at 37°C for 10min. The epithelium was then peeled away from the stroma and thoroughly minced before placing back into Dispase for a 5min incubation at 37°C. EDTA was then added to a final concentration of 5mM, and suspension diluted 1/5 by adding FACS staining buffer (SB; 2% heat-inactivated Foetal bovine serum (Life Technologies; 26140079), 25mM HEPES (Life Technologies; 15630056), 1mM EDTA in PBS) in order to reduce Dispase activity. Samples were then filtered through 30μm cell strainer and centrifuged at 300xg for 10min at 4°C, and resuspended in SB. Staining with primary antibodies or isotype controls was performed for 15min at 4°C, followed by a final wash in FACS buffer (FACS staining buffer without EDTA) at 300xg for 5min at 4°C. The final cell suspension was placed in FACS buffer containing 1U/μl RNAse Inhibitor.
The following immunoglobulins were used (Supplementary Table 5)for sorting: 20μg/ml Ep-Cam-AF647 to stain epithelial cells, and 8μg/ml CD45-APC-Cy7 to exclude immune contamination. Isotype controls were used at same concentration as specific antibodies: AF647 Rat IgG2a and APC-Cy7 Mouse IgG2a..
Work was carried out using RNAse free and/or PCR grade sterile reagents and plastic ware wherever possible.
Viable epithelial (Ep-Cam+/CD45-/DAPI-) cells were sorted on a BD FACSAria™ III cell sorter with BS FACSDiva 8.0.1 software, utilizing DAPI incorporation (4μg/ml) to exclude dead cells. Approximately 5000 cells per sample followed the 10X Chromium pipeline for scRNA-seq. FlowJo v10.6.2 software was used to generate sorting plots (Extended Data Fig. 3a).
Library preparation and sequencing of RNA from single cells
scRNA-seq libraries were generated and sequenced using 10X Genomics kits (Single Cell 3′ v3) at the CRUK-CI Genomics Core Facility of the CRUK Cambridge Institute. Libraries were generated in two different batches/dates: Batch 1, 3 biological replicates for P7 and 3 for Adult were used for 6 libraries; Batch 2, 3 biological replicates for P28 and 3 for Adult were used for 6 libraries. Presence of Adult samples in both batches provided common samples to control batch effect. The cells for each biological replicate were loaded into one 10X Chromium microfluidics chip channel to package them into one library. 12 libraries in total were sequenced on Illumina NovaSeq 2 SP and 2 S2 flow cells (Supplementary Table 1).
Single-cell RNA-seq analysis
Data processing
The raw sequencing data from the 10X Genomics platform were processed using CellRanger (v3.0.2) (Supplementary Table 1). CellRanger aligned reads, filtered empty dropouts and counted unique molecular identifiers (UMIs) to generate a count matrix. We used Ensembl GRCm38/mm10 (release 92) as the reference genome for read alignment. To filter out low quality cells, basic QC metrics were calculated using R package scater (1.12.2)106 and cells with <1251 genes were removed. Cells with mitochondrial proportions >15% and genes expressed in <3 cells were also discarded from analysis. Genes expressed in less than 3 cells were removed. Read counts were normalized by a deconvolution method using the R package scran (v1.12.1)107. Cells expressing fibroblast marker genes including Col1a2, Col1a1, Fn1, and Pdgfra were considered as non-epithelial contaminants. To filter out non-epithelial cells, the normalized read count values for the genes above were averaged and the cells with averaged normalized values for the four genes > 99 percentile (=0.4301828) of the distribution were filtered out. As a result, a total of 26,482 cells from 3-6 samples per time point were selected for further analysis (For basic statistics and QC metrics see Supplementary Table 1).
Dimension reduction and data visualization
PCA combined with technical noise modeling was applied to the normalized data for dimension reduction, implemented by the denoise_PCA function in the R package scran. The data were then projected using two-dimensional Uniform Manifold Approximation and Projection (UMAP) or t-Distributed Stochastic Neighbour Embedding (t-SNE) with default parameter setting using the R package scater. The biological replicates for each time point overlapped well with each other, confirming negligible batch effects between samples and conditions, therefore no batch effect correction was necessary. R package Seurat (v3.0.2)108 was used to visualize cells in the dimension reduction space.
Data clustering and cluster annotation
Next, a louvain community detection method was used for clustering109. First, a shared nearest-neighbour graph was constructed using k=20 nearest-neighbours of each cell (buildSNNGraph function in the R package scran). In this graph, two cells are connected by an edge if they share nearest-neighbours, with the edge weight determined by the highest average rank of the shared neighbours. Then the Walktrap method from the R package igraph (v1.2.4.1) (with steps = 4 as the default option) was used to identify densely connected communities that were considered to be cell clusters.
Cell clusters were annotated based on differentially expressed genes (DEGs) and known marker genes for cell types. If a few neighbouring clusters in the dimension reduction spaces shared key expression patterns, they were merged into one cell type manually. First, based on basal cell markers such as Krt14, Itgb1, and Itga6, and differentiation markers such as Tgm3, Krt13, and Grhl3, all clusters were classified into basal cells (clusters 1-3, 5-7, 10-13, 15-17) and differentiated cells (clusters 4, 8, 9, 14) (Extended Data Fig. 3b,d-f). The predominant expression of basal progenitor markers throughout the entire dataset (including Krt14, Itgb1 and Itga6) reflects an enrichment of basal cells under the experimental conditions used (Extended Data Fig. 3d,e). Accordingly, increased expression of differentiation markers (such as Krt13, Krt4, Tgm3, Grhl3, and Sbsn) was restricted to a few clusters (Extended Data Fig. 3d,e).
Then we classified all cells into different cell cycle phases based on cell cycle genes: clusters 1, 2, 4, 6, 7, 8, 9, 12, and 14 for G0/G1; clusters 11 and 13 for G1/S; clusters 3, 5, 10, 15, 16, and 17 for S/G2/M (Extended Data Fig. 3h,i). As a result, all cells were annotated as one of three cell types as follows: clusters 3, 5, 10, 11, 13, 15, 16, and 17 for basal cycling cells; clusters 1, 2, 6, 7 and 12 for resting basal cells (RBC); clusters 4, 8, 9, and 14 for differentiated cells (Extended Data Fig. 3d-i). Of note, although suprabasal cells at early time points do not proliferate (Extended Data Fig. 3j,k), they tend to express key markers of basal identity (Fig. 1i and Extended Data Fig. 3d-f,i). For this reason, clusters containing RBCs at P7 likely represent a combination of both basal and basal-like suprabasal cells.
To confirm that the annotated three cell types matched well between different time points, we integrated the data for P7, P28, and Adult using a standard workflow of Seurat (v3.0.2) (Extended Data Fig. 3c). Specifically, we normalised the data for each time point separately using SCTransform and then integrated them by applying the following built-in functions sequentially with default parameters: SelectIntegrationFeatures (nfeatures = 3000), PrepSCTIntegration, FindIntegrationAnchors, and IntegrateData. For cluster annotation, we visualized gene expression in the dimension reduction and in violin plots using built-in functions of Seurat (v3.0.2).
Classifying gene expression for basal cells into distinct dynamic patterns along the time course and Gene Ontology analysis of the different expression patterns
After annotating cell types, we focused on molecular change for basal cells over time. To this end, we first identified DEGs for each time point using the function ‘findMarkers’ of scran (v.1.12.1) by defining them as the genes having FDR<0.05 and absolute value of log2(fold-change)>(95 percentile of all log2(fold-change)) in the three comparisons of P7 vs. P28, P28 vs. Adult, P7 vs. Adult. The total number of DEGs was 1,738. We then scaled the normalized expression value of DEGs for all cells and calculated the average of the scaled expression value across the cells for each time point. Then scaled log2(fold-change) for each time was calculated by subtracting the averaged scaled value at P7 from those at all time points (thus, scaled log2(fold-change) at P7 = 0).
The scaled log2(fold-change) profiles for DEGs (1,738 genes) were clustered based k-means clustering (k=10) using pheatmap (1.0.12) and the resulting clusters were grouped into 4 major patterns: Pattern 1, 148 genes; Pattern 2, 545 genes; Pattern 3, 153 genes; Pattern 4, 892 genes (Extended Data Fig. 4a,b and Supplementary Table 2). The genes in each major pattern was used for Gene Ontology (GO) analysis by DAVID (https://david.ncifcrf.gov/)110. The resulting GO terms were manually curated and selected based on experimental observations and biological knowledge (Extended Data Fig. 4b and Supplementary Table 3).
We then examined the transcriptional heterogeneity in RBCs over time. First, we calculated the proportion of RBC clusters at each time point (Fig. 3d,e); clusters 2 and 6 at P7, clusters 1, 7 and 12 at P28 and Adult. This was marked by the emergence of cluster 12 over time, which represents a subset of RBCs expressing higher levels of Klf4. To understand temporal changes in RBCs from P7 through P28 to Adult at molecular level, we then characterised gene expression changes from cluster 2 over cluster 7 to cluster 12. To this end, we calculated marker genes for the RBC clusters (2, 7, and 12) using the function findMarkers as described above: marker genes for each cluster were defined as the genes having FDR<0.05 and log2(fold-change)>(97.5 percentile of all log2(fold-change)) in the comparison of each cluster with the rest of the RBC clusters. The total number of all marker genes was 1,729. Next, based on our observation in Fig. 3c-e, we grouped clusters of RBCs into three groups to represent each time point as follows: clusters 2 and 6 to represent P7; clusters 1 and 7 for P28/Adult; cluster 12 for P28/Adult. For the 1,729 marker genes, their normalised expression value was scaled among RBCs and then averaged across the cells of each group. The average expression for the three groups were clustered based on k-means clustering (k=6) using using pheatmap (1.0.12) and the resulting 6 clusters were grouped into 3 major patterns (Fig. 3f): Pattern 1, 748 genes; Pattern 2, 163 genes; Pattern 3, 818 genes. Then we performed GO analysis using DAVID for each of the 3 major patterns to understand underlying biological processes (Supplementary Table 4). As a result, we found that enriched biological processes are very similar between basal cells (Extended Data Fig. 4b) and RBCs only (Supplementary Table 4). Of note, well-known Klf4 and Yap related genes, amongst other genes associated with response to mechanical stimuli belonged to the 3 major patterns from the RBC clusters, showing dynamic expression across the clusters (Fig. 3f).
We then explored a potential regulatory relationship between genes denoting a response to mechanical stimuli, including YAP targets, and Klf4 related genes in RBCs. We examined the temporal order of expression of YAP and Klf4 target genes at single cell level from RBC state and along the differentiation trajectory. Thus, we performed pseudotime analysis using the R package monocle (v2.12.0) for RBCs and differentiated cells for P7 and P28/Adult separately to project the expression profile of relevant genes along differentiation. To this end, we first identified DEGs for RBCs and differentiated cells at P7 and P28/Adult separately by applying the criteria of FDR<0.05 and absolute value of log2(fold-change)>(95 percentile of all absolute value of log2(fold-change)). As a result, the number of DEGs was 1,018 for P7 and P28/Adult, respectively. Based on the DEGs, dimension reduction was performed by the DDRTree algorithm implemented in the R package monocle. Next, the pseudotime order was calculated using the orderCells function of monocle with default parameters. We then displayed the expression profiles for relevant mechanics and Klf4 related genes along the pseudotime for P7 and P28/Adult, separately (Fig. 3h and Extended Data Fig. 4f); for each gene, auto-scaled gene expression was plotted using a rolling mean along its trajectory with a window size of 5% of cells.
Whole oesophageal organ culture and tissue stretching
3D printed stretcher was developed from prototype in80 using Fusion360 Autodesk and Ultimaker Cura software and printed using Ultimaker3 3D printer in ABS (biocompatible plastic). A millimetric scale was included in printing of device to guide stretch. Once printed and sterilized, a 10cm adjustable screw was inserted into the lower clamp to allow for controlled tissue stretching (Fig. 7b). The system was then placed in a sterile plastic container with lid, allowing for the adjustable screw to be accessible from the outside of the container.
OE samples were collected at time points indicated and the muscle layer removed whilst retaining the tubular structure. Samples were then placed into the upper and lower clamps under no tension. Control samples were clamped at one end (top clamp, Fig. 7b; inset) and cultured under no longitudinal tension. Stretched samples were tightened into clamps using M3 nuts and screws. The millimetric scale and adjustable screw were used to apply a 40% stretch in tissue length. Whole-organ cultures were kept for 24/48h at 37°C, 5% CO2 in minimal medium containing 1 part DMEM (4.5 g/L D-Glucose, Pyruvate, L-Glutamine; Invitrogen 11971-025): 1 part DMEM/F12, 5μg/ml insulin, 1.8x10-4M adenine, 5% foetal calf serum, 5% Penicillin-Streptomycin and 5μg/ml Apo-Transferrine.
In vitro stretched tissues were treated with pharmacological compounds to inhibit myosin II motor activity (25μM Blebbistatin-48h) or YAP activity (1μM Verteporfin-24h), respectively. Vehicle (DMSO) was added to control samples.
Tissue response to different levels of tensile strain was assessed in P7 samples subjected to increasing levels of longitudinal stretching (20%, 40% and 100%) for 24h. For EdU incorporation assays, EdU was added to media at a final concentration of 10μM for 1h, 24h prior to collection.
Blebbistatin treatment of adult oesophageal organ culture
Epithelial-stroma composites were cut into ~3x4mm pieces and placed onto transparent ThinCert™ inserts (Greiner Bio-One Ltd; Cat# 657641), stromal side to the membrane. Tissues were dried for 5min at 37°C, and cultured in minimal medium at 37°C, 5% CO2 for 3h to ensure adequate tissue adhesion to the membrane. 25μM Blebbistatin was then added to the media. Control explants were treated with DMSO vehicle alone. Samples were collected after 45h.
For EdU incorporation assays, EdU was added and detected as above.
Changes in basal cell area upon tissue composite separation
Oesophagi from Rosa26-mT/mG mice were dissected and the upper most keratinized layers of the epithelium were mechanically separated with fine forceps. Alternatively, stromal layers were removed by incubating oesophagi in 5mM EDTA in PBS for 2.5h, 37°C.
Basal cells were immediately imaged by confocal microscopy. Endogenous expression of the tdTomato reporter in cell membranes allowed for imaging without fixation and immunolabeling. Cell area was measured using the manual ROI segmentation feature of the Volocity software and compared to that of basal cells from control unprocessed tissue samples.
Statistics and Reproducibility
A minimum of 3 independent mice/tissue cultures were used in all cases, with the exception of TEADi experiments (see Fig. 6d-f) due limitations associated to the COVID-19 pandemic. The number of biological replicates and animals is indicated in figure legends (“n” in legends refers to number of independent replicates per time point and/or condition). All replicates showed consistent results.
For image analysis, a minimum of 3 independent samples were inspected in all cases, capturing at least 3 replicate images each. All figures show representative images of a minimum of 3 mice/tissue cultures. Data are expressed as median values ± SEM, unless otherwise indicated.
Differences between groups were assessed by using two-tailed unpaired t-test, one-way, two-way analysis of variance (ANOVA) or Chi-squared analysis as indicated in figure legends. ANOVA based analysis was followed by Tukey’s test for multiple comparisons. The correlation between YAP and KLF4 expression levels in immunostained WMs was calculated by computing the Pearson correlation coefficient (r). All tests were two-sided. Exact p values are indicated in relevant figures up to four decimal places. Statistical differences between groups were assessed utilizing GraphPad Prism software. No statistical method was used to predetermine sample size. Experiments were performed without randomization or blinding.
Extended Data
Extended Data Fig. 1. Postnatal characterisation. Related to Fig. 1.
a, Diagram illustrating longitudinal oesophageal orientation from proximal to distal as marked by orange diamond. b, and c, Oesophageal tissue growth in length and width over time, respectively. Data expressed as mean ± SEM and analysed using one-way ANOVA with Tukey’s multiple comparisons test (n = 103 mice; #p relative to P70; *p relative to P7). d, Images showing animal body growth throughout postnatal development. e, Representative images showing EdU+ basal cells 24 hours post-labelling in P14 and P49 from Fig. 1e-g. f, Graphical representation of differential basal cell production rate throughout postnatal development. See Methods; data expressed as mean ± SEM; n=3 mice. g, 3D rendered z-stacks showing split confocal channels from Fig. 1i. h, Typical 3D rendered confocal z-stacks showing tilted side views from Fig. 1i. Yellow arrows indicate immature epithelial barrier. i, Representative side views of confocal z-stacks showing the thickening of the oesophageal epithelium (OE). j, Representative H&E sections of the oesophagus showing increasing cornification, as delimited by dotted lines. k, Quantification of the cornified thickness. n=3 mice; Micrometer, µm. Data expressed as mean ± SEM and analysed using one-way ANOVA with Tukey’s multiple comparisons test (#p relative to P70; *p relative to P7).
Scale bars. S1d(2 cm); S1e,g-j(20 µm). Stainings. Blue, DAPI; cyan, EdU; green, KRT14; greyscale, KRT4.
All data derived from wild-type C57BL/6J mice.
Dashed lines indicate basement membrane. Dotted lines in graphs indicate P28. Orange diamonds depict longitudinal orientation of the oesophagus where indicated.
Source data are provided.
Extended Data Fig. 2. KLF4 basal cell profile in FUCCI2a mice. Related to Fig. 2.
a, Representative confocal z-stacks showing side views of OE wholemounts from Fig. 2a. Dashed lines indicate basement membrane; dotted lines mark the upper limit of the OE; white arrows indicate basal KLF4+ cells. Red, KLF4. b, In vivo protocol. Oesophagi from FUCCI2a mice were collected at time points indicated. Schematic indicating expression pattern of fluorescent proteins in FUCCI2a mouse model. c, Confocal images showing basal views of typical FUCCI2a OE wholemounts in (b). Orange diamonds indicate longitudinal orientation of the oesophagus. White dashed lines indicate mVenus+ cells; green, mVenus; red, mCherry. d and e, Correlation between KLF4 protein expression and reporter fluorescent proteins mCherry (d)/mVenus (e) in the basal layer from (b) and (c). n=3 mice; Scale bars 20 µm. Parts of (b) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided.
Extended Data Fig. 3. Single-cell RNA sequencing annotation. Related to Fig. 3.
a, Flow cytometry gating strategy for isolation of OE cells. Oesophageal cell suspensions (i) were gated to sort the single (ii) viable (iii) population, enriched for epithelial cells (iv; EpCam+/CD45-). Cells were isolated from 15 mice (P7), 12 mice (P28), and 9 (adults). Representative plots from adult sample are shown. b, UMAP representing cell clusters based on louvain clustering. c, UMAP representing the distribution of cell clusters in (b) after integrating data from different time points (see Methods). d, UMAP showing expression of representative makers for basal (left panel) and differentiated cells (right panel) in OE. e, Heatmap showing expression of representative marker genes for basal cells, cell cycle, and differentiation for the 17 clusters shown in (cluster number from b in upper bar). Expression values were log2-transformed normalized UMIs followed by scaling and averaging across cells in the same clusters. f, Violin plots showing expression of representative epidermal (basal vs. differentiated) and cell cycle markers at different postnatal stages split by annotated cell cohorts in Fig. 3c. g, Violin plots showing expression of genes associated with regeneration and homeostasis for basal and differentiated cell types at distinct postnatal stages. Basal cells include both cycling and resting cells. h, UMAP showing spatial distribution of distinct cell cycle phases, annotated using cell cycle analysis by R package scran (v 1.12.1) combined with manual curation based on genes in (e). i, Violin plots showing expression of representative cell cycle genes at postnatal stages split by cell cycle cohorts identified in (h). Colour scheme for cell cycle phases as in (h). j, In vivo protocol. Mice were treated with EdU 2 hours prior culling at indicated time points. k, Typical 3D rendered confocal side views showing basal EdU population (see Methods). Dashed white lines, basement membrane. Dotted white lines, upper OE limit. Blue, DAPI; cyan, EdU; scale bar 10 µm. For violin plots in (f), (g) and (i), expression level means log2-transformed normalized UMIs, dotted lines indicate the median of the distribution. Colour bars of UMAPs in (d) denotes expression range. Parts of (j) were drawn by using and/or adapting diagrams from Servier Medical Art.
Extended Data Fig. 4. Single-cell RNA sequencing expression profile. Related to Fig. 3.
a, Distinctive patterns (Pt) of gene expression in basal cells as defined in (Fig. 3c). Grey, relative expression profiles of individual genes belonging to each pattern. Solid coloured lines, median values at each time point. To calculate the relative expression profiles, log2-transformed normalized UMIs were scaled and averaged across all basal cells at each time point and adjusted compared to the value at P7. b, Heatmap representing expression of individual genes belonging to the 4 patterns in (a). For expression values, log2-transformed normalised UMIs were scaled and averaged across all basal cells for each cluster and time point. The table on the right shows selected GO terms for major Pt2 and Pt4, corresponding p-values and representative genes. Closely related GO terms are grouped together. See Supplementary Table 3 for GO analysis result for all 4 expression patterns. c and d, UMAPs showing expression of genes related to key biological processes from Gene Ontology analysis for Patterns 2 (c) and 4 (d) in (b). e, Violin plots showing expression of Klf4 for cells in individual clusters at P7 (left), P28 (middle) and Adult (right). f, Expression of relevant genes along the pseudotime trajectory from basal resting to differentiated cells for P7. Left panel, YAP target genes (Cyr61, Ctgf, Thbs1) and genes associated with a response to mechanical stimuli (Cav1, Klf2, Dcn). Right panel, depicts KLF4 target genes (Krt4, Krt13, Cdnk1a, Cebpb). For violin plots in (e), expression level means log2-transformed normalized UMIs and dotted lines indicate the median of the distribution. Colour bars of UMAPs in (c) and (d) indicate log2-transformed normalized UMIs. Gene expression in (f) is represented as auto-scaled, log2-transformed normalized UMIs smoothed using a rolling mean along its trajectory with a window size of 5% of cells. Two bars on the top denotes the arrangement of cells according to pseudotime and clusters in Extended Data Fig. 3b, respectively.
Extended Data Fig. 5. Deep Learning based segmentation. Related to Fig. 4.
a, Protocol for in situ fixation of the oesophagus, and b, Images from in situ oesophagi compared with oesophagi fixed immediately after dissection confirm that basal cell shape was not affected by tissue harvesting (Supplementary to Fig. 4a). Blue, DAPI; green, KRT14. Scale bar, 20 µm. c, Schematic depicting deep learning based segmentation principle. Manually or semi-automatically annotated “ground truth” images were used to train a U-Net convolutional neural network. Training of the network was assessed on validation images and iteratively optimized until the achievement of satisfactory automated segmentation. d, Schematic of pipeline utilised for segmentation of single z-slice confocal images of OE basal layer. Nuclear segmentation was based on DAPI staining (blue). Mask overlay shows the match between the binary mask and the original fluorescence image. Scale bar, 20 µm. e, Schematic describing the computation of Voronoi diagrams of the tissue. Single z-slice confocal images of the OE basal layer are segmented as described in (d). Cell centroids are computed using the binary mask. Delaunay triangulation of cells was performed using cell centroids coordinates. Voronoi diagrams are calculated as the dual of Delaunay triangulation of cells in the tissue and overlaid onto the original fluorescence image. Scale bar, 20 µm. f, Cell shape anisotropy tensor at P14 and P49 (supplementary to Fig. 4d). n=3 mice. Orientation is colour-coded. Results from a representative experiment are shown; n=3 mice. g, Violin plots showing the distribution in cell shape anisotropy throughout postnatal development. n=2052-2594 cells from 3 animals per time point. Black dashed line, median. One-way ANOVA with Tukey’s multiple comparisons test (*p relative to P7). h, Bidimensional structure factor at P14 and P49 (supplementary to Fig. 4e). Changes in the dashed white outline (from ellipse to circle) depict a transition from anisotropic to isotropic spatial distribution over time; n=3 mice. i, Structure factor shape anisotropy distribution as shown in (h) and Fig. 4e. Box plots show median and quartiles; and whiskers, minima/maxima. Individual measurements from n=3 mice; (ns, not significant; *p relative to P7).
All data derived from wild-type C57BL/6J mice. Parts of (a) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided.
Extended Data Fig. 6. Oesophageal tissue strain and second harmonic generation. Related to Fig. 5.
a, In situ and immediate ex vivo images of oesophageal tubes at P28 and P49 (supplementary to Fig. 5b). White dashed lines delineate oesophageal tube; scale bar 1 cm. b, Representative images showing the size of combined and separate oesophageal layers; Full oesophageal tube (Tube), epithelial composite (Epi) and muscle layer (Muscle); scale bar 5 mm. c, Longitudinal tissue strain relative to muscle, represented as percentage. Data expressed as mean ± SEM. One-way ANOVA with Tukey’s multiple comparisons test (P7 n=16, P28 and Adult n=9; *p relative to P7; ns, not significant). d, Measure of the stomach perimeter over time. Mean values ± SEM; n=3 mice; Millimetre, mm. e, Basal confocal view of EdU+ cells in wholemounts of typical squamous stomach epithelium from (f). f, In vivo protocol. Mice were treated with a single EdU injection 24h prior stomach collection at the time points indicated. g, Quantification of EdU+ basal cells per field from (f). Presented as mean ± SD; n = 3- h, Representative views of stroma underlying OE basement membrane using second harmonic generation (SHG). Left panels, collagen in magenta. Middle panels, colour map of SHG signal intensity. Right panels, colour-coded local orientation map of SHG signal. Scale bar 100 µm. i, Representative histograms depicting orientation distribution of collagen fibres in (h). n=3 mice. j, Basal view of representative OE wholemounts at P14. Green, YAP; greyscale, B-Catenin (BCat). Scale bar 10 µm. k and l, Quantification of basal nuclear and cytoplasmic staining of YAP and DAPI, respectively (see Methods; supplementary to Fig. 5m). A total of 20 cells for 3 different animals were measured.
All data derived from wild-type C57BL/6J mice. Data analysis for c and d was performed using one- and two-way ANOVA, respectively, with Tukey’s multiple comparisons test (n=3-9; ns, not significant; (c) relative to P7). g, k and l was performed using two-tailed unpaired t test (ns, not significant). Box plots show median and quartiles; whiskers, 0.1 and 0.99 percentiles.
Orange diamonds depict longitudinal orientation of the oesophagus where indicated. Parts of (f) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided.
Extended Data Fig. 7. Changes in tissue mechanics at cellular level. Related to Fig. 7.
a, Model of parts required for 3D printed stretcher. Scale bar, 1 cm. b, In vitro protocol. Oesophagi were exposed to 40% stretch using stretcher (Fig. 7a) and treated with/without 25 µM blebbistatin (BLEBB) for 48 hours. c, Individual basal cell areas. Data analysis was performed using Two-way ANOVA with Tukey’s multiple comparisons test (n=3 mice; ns, not significant; black indicates significance between control vs. BLEBB conditions; grey indicates statistical differences between stretching conditions). d, Confocal basal views of typical organ cultures after 48 hour BLEBB treatment in vitro from (b). Scale bar, 20 µm. e, In vitro protocol. Oesophagi stretched at the indicated levels, treated with EdU for 1 hour, and kept in vitro as whole-organ cultures for a 24 hours. f, Confocal basal views showing EdU and phosphohistone H3 (PhosphoH3) staining in cultures from (e). Scale bar, 20 µm. g and h, Basal quantification of EdU+ and PhosphoH3 cells expressed as percentage of DAPI+ cells from (e). Mean ± SD One-way ANOVA with Tukey’s multiple comparisons test (n=3 mice; *p relative to Control; ns, not significant;). i, In vivo protocol. Oesophageal wholemounts from Rosa26-mT/mG mice were mechanically peeled, separating cornified suprabasal layers or underlying stroma (see Methods). j, Individual area of basal cells. Data analysis was performed using One-way ANOVA with Tukey’s multiple comparisons test (n=3 mice; *p relative to full tissue). k, Confocal basal views of typical OE wholemounts after mechanical separation. Red, mTmG; scale bar, 10 µm. l, Basal cell density, expressed in number of cells per field after BLEBB treatment in Fig. 7e. Mean ± SD; n=3 mice. Unpaired t test (ns, not significant). m, Basal quantification of EdU+ cells expressed as percentage of DAPI+ cells after BLEBB treatment in Fig. 7e. Mean ± SD; n=3 mice. Unpaired t test (ns, not significant).
Data derived from wild-type C57BL/6J mice, unless otherwise stated. Individual points show individual measurements, greyscale indicates values from each of 3 mice. Parts of (b, e) were drawn by using and/or adapting diagrams from Servier Medical Art.
Source data are provided.
Supplementary Material
Acknowledgments
We thank members of the Alcolea lab, Simons lab and Bartomeu Colom (Sanger Institute) for comments and suggestions; the staff of the University Biomedical Services, Gurdon Institute; Peter Humphreys in imaging core facilities at JCBC; Alex Sossick, Richard Butler and Nicola Lawrence at the Gurdon Institute Imaging Facility; CRUK-CI Genomics Core Facility at the CRUK Cambridge Institute; Jonas Larsson (tetO-YAP S127A); Ian J. Jackson (Fucci2a); NIHR Cambridge BRC Cell Phenotyping Hub for FACS support; the Jeffrey Cheah Biomedical Centre (JCBC) core facilities. We thank T. Savin, E. Hannezo, B. Ladoux, G. Charras, S. Hénon, N. Harmand, G. Duclos and J-B. Lugagne for their insight on deep learning based image analysis and/or tissue mechanics. This work was mainly supported by funding from the Wellcome Trust and The Royal Society (105942/Z/14/Z to M.P.A), and a core support grant from the Wellcome Trust (203151/Z/16/Z) and MRC (MC_PC_17230) to the Wellcome-Medical Research Council Cambridge Stem Cell Institute. J.M was supported by a CRUK Cambridge Cancer Centre PhD fellowship. A.H was supported by a Wellcome Trust Junior Interdisciplinary Research Fellowship (098357/Z/12/Z) and a Herchel Smith Postdoctoral Research Fellowship. The work also received support from the Novo Nordisk Foundation (NNF18CC0033666 to K.K; NNF17OC0028730 to K.B.J and NNF17CC0027852 to K.B.J); Human Frontier Science Program (LT000092/2016-L) and Basic Science Research Program (NRF-2014R1A6A3A01005675) to S.H; National Institutes of Health (ZIA BC 011763 to R.I.B); Wellcome PhD stutentship (220088/Z/20/Z to F.J.E); C.V. was supported by the Engineering and Physical Sciences Research Council (1506089). The Royal Society and European Research Council (‘CellFateTech’, 772798 to K.J.C). B.D.S. acknowledges funding from the Wellcome Trust (098357/Z/12/Z and 219478/Z/19/Z) and the Royal Society in the form of an E.P. Abraham Research Professorship (RP\R1\180165). A.H, S.H and B.D.S acknowledge the support of the core funding to the Wellcome Trust/CRUK Gurdon Institute (203144/Z/16/Z & C6946/A24843).
Footnotes
Author Contributions Statement
Experiments were designed, validated and conducted by J.M, F.J.E, R.I.B and M.P.A, with J.M, F.J.E working under the supervision of M.P.A; K.K and S.U carried out experiments under the supervision of K.B.J; A.H developed the deep learning based image segmentation and other computational image analysis pipelines to quantify cell fluorescence levels, cell shape, tissue spatial organisation and SHG images. S.H guided the experimental design for single-cell RNA sequencing and performed analysis of the data. A.H and S.H were both under the supervision of B.D.S; C.V designed and provided expertise on the 3D printed stretcher device with input from K.J.C; A.H, R.I.B and K.J.C provided insights and technical expertise on tissue mechanics and stretching experiments; K.B.J and B.D.S supervised parts of the study and provided expertise in the epithelial stem cell field; J.M and M.P.A conceived and coordinated the project, supervised experiments and wrote the manuscript with input from A.H and B.D.S. Review & Editing of final manuscript by all authors. Funding acquisition by A.H, R.I.B, K.J.C, K.B.J, B.D.S, and M.P.A.
Competing Interests Statement The authors declare no competing interests.
Data Availability
Single-cell RNA sequencing (scRNA-seq) data that support the findings of this study have been deposited in ArrayExpress under accession code EMTAB-8662. Source data have been provided. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code Availability
The statistical analysis of the single-cell RNA sequencing data was performed using R codes developed for this study. Image segmentation and computational image analysis was performed using Python codes developed and/or adapted for this study. All codes for the computational analysis above are available at https://github.com/BenSimonsLab/McGinn_Nat-Cell-Biol_2021.
References
- 1.Varga J, Greten FR. Cell plasticity in epithelial homeostasis and tumorigenesis. Nat Cell Biol. 2017;19:1133–1141. doi: 10.1038/ncb3611. [DOI] [PubMed] [Google Scholar]
- 2.Tata PR, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 5.Ge Y, et al. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell. 2017;169:636–650.:e614. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yui S, et al. YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell. 2018;22:35–49.:e37. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Que J, Garman KS, Souza RF, Spechler SJ. Pathogenesis and Cells of Origin of Barrett’s Esophagus. Gastroenterology. 2019;157:349–364.:e341. doi: 10.1053/j.gastro.2019.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiu J, et al. Tracing the origin of adult intestinal stem cells. Nature. 2019;570:107–111. doi: 10.1038/s41586-019-1212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen MS, et al. Tracing the cellular dynamics of sebaceous gland development in normal and perturbed states. Nat Cell Biol. 2019;21:924–932. doi: 10.1038/s41556-019-0362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcolea MP, Jones PH. Tracking cells in their native habitat: lineage tracing in epithelial neoplasia. Nat Rev Cancer. 2013;13:161–171. doi: 10.1038/nrc3460. [DOI] [PubMed] [Google Scholar]
- 11.Blanpain C, Fuchs E. Stem cell plasticity Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344:1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tata PR, et al. Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity. Dev Cell. 2018;44:679–693.:e675. doi: 10.1016/j.devcel.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donati G, et al. Wounding induces dedifferentiation of epidermal Gata6(+) cells and acquisition of stem cell properties. Nat Cell Biol. 2017;19:603–613. doi: 10.1038/ncb3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosekrans SL, Baan B, Muncan V, van den Brink GR. Esophageal development and epithelial homeostasis. Am J Physiol Gastrointest Liver Physiol. 2015;309:G216–228. doi: 10.1152/ajpgi.00088.2015. [DOI] [PubMed] [Google Scholar]
- 15.Doupe DP, et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammoto T, et al. Mechanochemical control of mesenchymal condensation and embryonic tooth organ formation. Dev Cell. 2011;21:758–769. doi: 10.1016/j.devcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barriga EH, Franze K, Charras G, Mayor R. Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo. Nature. 2018;554:523–527. doi: 10.1038/nature25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, et al. The Strength of Mechanical Forces Determines the Differentiation of Alveolar Epithelial Cells. Dev Cell. 2018;44:297–312.:e295. doi: 10.1016/j.devcel.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Shyer AE, et al. Villification: how the gut gets its villi. Science. 2013;342:212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shyer AE, Huycke TR, Lee C, Mahadevan L, Tabin CJ. Bending gradients: how the intestinal stem cell gets its home. Cell. 2015;161:569–580. doi: 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shyer AE, et al. Emergent cellular self-organization and mechanosensation initiate follicle pattern in the avian skin. Science. 2017;357:811–815. doi: 10.1126/science.aai7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heckel E, et al. Oscillatory Flow Modulates Mechanosensitive klf2a Expression through trpv4 and trpp2 during Heart Valve Development. Curr Biol. 2015;25:1354–1361. doi: 10.1016/j.cub.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 23.Vermot J, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7:e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilmour D, Rembold M, Leptin M. From morphogen to morphogenesis and back. Nature. 2017;541:311–320. doi: 10.1038/nature21348. [DOI] [PubMed] [Google Scholar]
- 25.Segel M, et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature. 2019;573:130–134. doi: 10.1038/s41586-019-1484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biggs LC, Kim CS, Miroshnikova YA, Wickstrom SA. Mechanical Forces in the Skin: Roles in Tissue Architecture, Stability, and Function. J Invest Dermatol. 2019 doi: 10.1016/j.jid.2019.06.137. [DOI] [PubMed] [Google Scholar]
- 27.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LeGoff L, Lecuit T. Mechanical Forces and Growth in Animal Tissues. Cold Spring Harb Perspect Biol. 2015;8:a019232. doi: 10.1101/cshperspect.a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petridou NI, Spiro Z, Heisenberg CP. Multiscale force sensing in development. Nat Cell Biol. 2017;19:581–588. doi: 10.1038/ncb3524. [DOI] [PubMed] [Google Scholar]
- 30.Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- 31.Liu K, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeWard AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tetreault MP, et al. Esophageal squamous cell dysplasia and delayed differentiation with deletion of kruppel-like factor 4 in murine esophagus. Gastroenterology. 2010;139:171–181.:e179. doi: 10.1053/j.gastro.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc Natl Acad Sci U S A. 2006;103:18668–18673. doi: 10.1073/pnas.0608658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 36.Fortunel NO, et al. KLF4 inhibition promotes the expansion of keratinocyte precursors from adult human skin and of embryonic-stem-cell-derived keratinocytes. Nat Biomed Eng. 2019;3:985–997. doi: 10.1038/s41551-019-0464-6. [DOI] [PubMed] [Google Scholar]
- 37.Kypriotou M, Huber M, Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ’fused genes’ family. Exp Dermatol. 2012;21:643–649. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 38.Mesa KR, et al. Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell Stem Cell. 2018;23:677–686.:e674. doi: 10.1016/j.stem.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rompolas P, et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science. 2016;352:1471–1474. doi: 10.1126/science.aaf7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulukian A, Fuchs E. Spindle orientation and epidermal morphogenesis. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130016. doi: 10.1098/rstb.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 42.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 43.Alcolea MP, et al. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat Cell Biol. 2014;16:615–622. doi: 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Driskell RR, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crisera CA, Maldonado TS, Longaker MT, Gittes GK. Defective fibroblast growth factor signaling allows for nonbranching growth of the respiratory-derived fistula tract in esophageal atresia with tracheoesophageal fistula. J Pediatr Surg. 2000;35:1421–1425. doi: 10.1053/jpsu.2000.16404. [DOI] [PubMed] [Google Scholar]
- 46.Botchkarev VA, Sharov AA. BMP signaling in the control of skin development and hair follicle growth. Differentiation. 2004;72:512–526. doi: 10.1111/j.1432-0436.2004.07209005.x. [DOI] [PubMed] [Google Scholar]
- 47.Park S, et al. B-cell translocation gene 2 (Btg2) regulates vertebral patterning by modulating bone morphogenetic protein/smad signaling. Mol Cell Biol. 2004;24:10256–10262. doi: 10.1128/MCB.24.23.10256-10262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Cao X. BMP signaling and skeletogenesis. Ann N Y Acad Sci. 2006;1068:26–40. doi: 10.1196/annals.1346.006. [DOI] [PubMed] [Google Scholar]
- 49.Blanco S, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koria P, Andreadis ST. Epidermal morphogenesis: the transcriptional program of human keratinocytes during stratification. J Invest Dermatol. 2006;126:1834–1841. doi: 10.1038/sj.jid.5700325. [DOI] [PubMed] [Google Scholar]
- 51.Sadagurski M, et al. Insulin-like growth factor 1 receptor signaling regulates skin development and inhibits skin keratinocyte differentiation. Mol Cell Biol. 2006;26:2675–2687. doi: 10.1128/MCB.26.7.2675-2687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim X, et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang C, et al. Wnt/beta-catenin signaling is critical for dedifferentiation of aged epidermal cells in vivo and in vitro. Aging Cell. 2012;11:14–23. doi: 10.1111/j.1474-9726.2011.00753.x. [DOI] [PubMed] [Google Scholar]
- 54.Joost S, et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst. 2016;3:221–237.:e229. doi: 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haake AR, Cooklis M. Incomplete differentiation of fetal keratinocytes in the skin equivalent leads to the default pathway of apoptosis. Exp Cell Res. 1997;231:83–95. doi: 10.1006/excr.1996.3441. [DOI] [PubMed] [Google Scholar]
- 56.Ge Y, Fuchs E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet. 2018;19:311–325. doi: 10.1038/nrg.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Philippeos C, et al. Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J Invest Dermatol. 2018;138:811–825. doi: 10.1016/j.jid.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nystrom A, Bruckner-Tuderman L. Matrix molecules and skin biology. Semin Cell Dev Biol. 2019;89:136–146. doi: 10.1016/j.semcdb.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 59.Ghaleb AM, Yang VW. Kruppel-like factor 4 (KLF4): What we currently know. Gene. 2017;611:27–37. doi: 10.1016/j.gene.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 61.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morgan JT, Raghunathan VK, Chang YR, Murphy CJ, Russell P. Wnt inhibition induces persistent increases in intrinsic stiffness of human trabecular meshwork cells. Exp Eye Res. 2015;132:174–178. doi: 10.1016/j.exer.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mendez MG, Janmey PA. Transcription factor regulation by mechanical stress. Int J Biochem Cell Biol. 2012;44:728–732. doi: 10.1016/j.biocel.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morse EM, et al. PAK6 targets to cell-cell adhesions through its N-terminus in a Cdc42-dependent manner to drive epithelial colony escape. J Cell Sci. 2016;129:380–393. doi: 10.1242/jcs.177493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kopf J, Petersen A, Duda GN, Knaus P. BMP2 and mechanical loading cooperatively regulate immediate early signalling events in the BMP pathway. BMC Biol. 2012;10:37. doi: 10.1186/1741-7007-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci U S A. 2011;108:2270–2275. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maglic D, et al. YAP-TEAD signaling promotes basal cell carcinoma development via a c-JUN/AP1 axis. EMBO J. 2018;37 doi: 10.15252/embj.201798642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schutte U, et al. Hippo signaling mediates proliferation, invasiveness, and metastatic potential of clear cell renal cell carcinoma. Transl Oncol. 2014;7:309–321. doi: 10.1016/j.tranon.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen J, et al. Hippo component YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breast cancer. J Exp Clin Cancer Res. 2018;37:175. doi: 10.1186/s13046-018-0850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He C, et al. YAP forms autocrine loops with the ERBB pathway to regulate ovarian cancer initiation and progression. Oncogene. 2015;34:6040–6054. doi: 10.1038/onc.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xin M, et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreno-Vicente R, et al. Caveolin-1 Modulates Mechanotransduction Responses to Substrate Stiffness through Actin-Dependent Control of YAP. Cell Rep. 2018;25:1622–1635.:e1626. doi: 10.1016/j.celrep.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Echarri A, Del Pozo MA. Caveolae-mechanosensitive membrane invaginations linked to actin filaments. J Cell Sci. 2015;128:2747–2758. doi: 10.1242/jcs.153940. [DOI] [PubMed] [Google Scholar]
- 74.Steed E, et al. klf2a couples mechanotransduction and zebrafish valve morphogenesis through fibronectin synthesis. Nat Commun. 2016;7:11646. doi: 10.1038/ncomms11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goddard LM, et al. Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis. Dev Cell. 2017;43:274–289.:e275. doi: 10.1016/j.devcel.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Das Thakur M, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Misra JR, Irvine KD. The Hippo Signaling Network and Its Biological Functions. Annu Rev Genet. 2018;52:65–87. doi: 10.1146/annurev-genet-120417-031621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan Y, et al. YAP1/TAZ-TEAD transcriptional networks maintain skin homeostasis by regulating cell proliferation and limiting KLF4 activity. Nat Commun. 2020;11:1472. doi: 10.1038/s41467-020-15301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verstreken CM, Labouesse C, Agley CC, Chalut KJ. Embryonic stem cells become mechanoresponsive upon exit from ground state of pluripotency. Open Biol. 2019;9:180203. doi: 10.1098/rsob.180203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aragona M, et al. Mechanisms of stretch-mediated skin expansion at single-cell resolution. Nature. 2020;584:268–273. doi: 10.1038/s41586-020-2555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 83.Hatzfeld M, Keil R, Magin TM. Desmosomes and Intermediate Filaments: Their Consequences for Tissue Mechanics. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a029157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang L, et al. Unbalanced YAP-SOX9 circuit drives stemness and malignant progression in esophageal squamous cell carcinoma. Oncogene. 2019;38:2042–2055. doi: 10.1038/s41388-018-0476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nowlan NC, Francis-West P, Nelson C. Mechanics of development. Philos Trans R Soc Lond B Biol Sci. 2018;373 doi: 10.1098/rstb.2017.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fiore VF, et al. Mechanics of a multilayer epithelium instruct tumour architecture and function. Nature. 2020;585:433–439. doi: 10.1038/s41586-020-2695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huycke TR, et al. Genetic and Mechanical Regulation of Intestinal Smooth Muscle Development. Cell. 2019;179:90–105.:e121. doi: 10.1016/j.cell.2019.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 90.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 91.Benham-Pyle BW, Pruitt BL, Nelson WJ. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science. 2015;348:1024–1027. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gudipaty SA, et al. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature. 2017;543:118–121. doi: 10.1038/nature21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elbediwy A, et al. Integrin signalling regulates YAP and TAZ to control skin homeostasis. Development. 2016;143:1674–1687. doi: 10.1242/dev.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schlegelmilch K, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Imajo M, Ebisuya M, Nishida E. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat Cell Biol. 2015;17:7–19. doi: 10.1038/ncb3084. [DOI] [PubMed] [Google Scholar]
- 96.Gokhin DS, Ward SR, Bremner SN, Lieber RL. Quantitative analysis of neonatal skeletal muscle functional improvement in the mouse. J Exp Biol. 2008;211:837–843. doi: 10.1242/jeb.014340. [DOI] [PubMed] [Google Scholar]
- 97.Domenga V, et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chao CM, Moiseenko A, Zimmer KP, Bellusci S. Alveologenesis: key cellular players and fibroblast growth factor 10 signaling. Mol Cell Pediatr. 2016;3:17. doi: 10.1186/s40348-016-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dekoninck S, et al. Defining the Design Principles of Skin Epidermis Postnatal Growth. Cell. 2020;181:604–620.:e622. doi: 10.1016/j.cell.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paszek MJ, et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mort RL, et al. Fucci2a: a bicistronic cell cycle reporter that allows Cre mediated tissue specific expression in mice. Cell Cycle. 2014;13:2681–2696. doi: 10.4161/15384101.2015.945381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jansson L, Larsson J. Normal hematopoietic stem cell function in mice with enforced expression of the Hippo signaling effector YAP1. PLoS One. 2012;7:e32013. doi: 10.1371/journal.pone.0032013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 105.Burns ER, Scheving LE, Fawcett DF, Gibbs WM, Galatzan RE. Circadian influence on the frequency of labeled mitoses method in the stratified squamous epithelium of the mouse esophagus and tongue. Anat Rec. 1976;184:265–273. doi: 10.1002/ar.1091840302. [DOI] [PubMed] [Google Scholar]
- 106.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Subelj L, Bajec M. Unfolding communities in large complex networks: combining defensive and offensive label propagation for core extraction. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:036103. doi: 10.1103/PhysRevE.83.036103. [DOI] [PubMed] [Google Scholar]
- 110.Huang da, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA sequencing (scRNA-seq) data that support the findings of this study have been deposited in ArrayExpress under accession code EMTAB-8662. Source data have been provided. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
The statistical analysis of the single-cell RNA sequencing data was performed using R codes developed for this study. Image segmentation and computational image analysis was performed using Python codes developed and/or adapted for this study. All codes for the computational analysis above are available at https://github.com/BenSimonsLab/McGinn_Nat-Cell-Biol_2021.