Abstract
Raised albumin-creatinine ratio (ACR) is an indicator of microvascular damage and renal disease. We aimed to identify genetic variants associated with raised ACR and study the implications of carrying multiple ACR-raising alleles with metabolic and vascular related disease. We performed a genome-wide association study of ACR using 437,027 individuals from the UK Biobank in the discovery phase, 54,527 more than previous studies, and followed up our findings in independent studies. We identified 62 independent associations with ACR across 56 loci (P<5x10-8), of which 20 were not previously reported. Pathway analyses and the identification of 20 of the 62 variants (at r 2>0.8) coinciding with signals for at least sixteen related metabolic and vascular traits, suggested multiple pathways leading to raised ACR levels. After excluding variants at the CUBN locus, known to alter ACR via effects on renal absorption, an ACR genetic risk score was associated with a higher risk of hypertension, and less strongly, type 2 diabetes and stroke. For some rare genotype combinations at the CUBN locus, most individuals had ACR levels above the microalbuminuria clinical threshold. Contrary to our hypothesis, individuals carrying more CUBN ACR-raising alleles, and above the clinical threshold, had a higher frequency of vascular disease. The CUBN allele effects on ACR were twice as strong in people with diabetes – a result robust to an optimization-algorithm approach to simulating interactions, validating previously reported gene-diabetes interactions (P≤4x10-5). In conclusion, a variety of genetic mechanisms and traits contribute to variation in ACR.
Introduction
High urinary albumin excretion is a marker of chronic kidney disease (CKD) and a predictor of mortality and cardiovascular events in the general population and in clinical populations, such as individuals with diabetes (1). Moreover, raised albuminuria is believed to be indicative of systemic endothelial microvascular damage (2). Albumin-creatinine ratio (ACR) is an accepted marker of urinary excretion of albumin (3) available in large numbers of individuals through routine clinical testing.
Genetic studies of ACR are important as individuals with raised ACR levels due to genetics affecting tubular reabsorption, for example, may not be at higher risk of vascular disease. Conversely, individuals with low ACR levels due to genotypes that directly alter ACR may be missed in clinical tests for microvascular damage. Genetic studies of ACR may identify new, or clarify the role of known, pathways altering microvascular function or kidney function, or both.
Prior to the availability of data from the UK Biobank, previous genetic association studies had identified only one region of the genome robustly associated with ACR at the cubilin (CUBN) locus (4–6). Associations were mainly represented by a common single-nucleotide polymorphism (SNP) (rs1801239) with a minor allele frequency (MAF) of 10% (5). In addition, van Zuydam et al. showed a putative association between a signal on chromosome 6 (GABRR1) and diabetic microalbuminuria (7). Most recently, a rare SNP (rs141640975) with a MAF of 0.8% was identified as associated with albuminuria through exome sequencing (8). Ahluwalia et al. also identified evidence of association for three genes (HES1, CDC73 and GRM4) after performing gene-based tests. Both Teumer et al. and Ahluwalia et al. provided evidence that the CUBN variant has a 3 to 4-fold larger effect in people with diabetes. Teumer et al. identified two additional loci with stronger effects in individuals with diabetes (RAB38/CTSC and HS6ST1).
Mutations in the CUBN gene cause an autosomal recessive disorder, Imerslund Gräsbeck Syndrome, characterized by Vitamin B12 malabsorption and, in many cases, mild proteinuria (9). Proteinuria in this context is likely to be due to a defect of the cubilin receptor which binds albumin in renal tubuli thus decreasing albumin reabsorption (10). Therefore, the CUBN variant may alter ACR directly without being associated with renal damage.
To understand more about the genetic factors associated with variation in ACR and study the implications of carrying multiple ACR-raising alleles with metabolic and vascular related disease, we performed a genome-wide association study (GWAS) of ACR using 437,027 individuals from the UK Biobank study with subsequent replication using publicly available data from the CDKGen consortium (5) and the EXTEND study. Our study follows those from Haas et al. and Zanetti et al. who identified 46 and 21 genetic associations with ACR at the conventional P<5x10-8 threshold respectively in smaller subsets of UK Biobank participants (N=382,500 and N=218,450 discovery sets respectively) (11, 12). In addition to the identification of 20 novel loci in our larger sample size, we focused our analyses on those very difficult to perform in the context of a GWAS consortium of many smaller studies and not performed by Haas et al. These analyses included testing haplotype effects at the CUBN locus, colocalization of genetic associations, and gene-disease interactions in the UK Biobank.
Results
Clinical characteristics of the 437,027 UK Biobank individuals of European ancestry and with ACR available are presented in Table 1 and Supplementary Figure 1. The characteristics of the EXTEND study individuals are also presented in Table 1.
Table 1. Characteristics of participants from the UK Biobank and EXTEND analysed.
Data presented as mean (±standard deviation) or median [25th -75th percentile] where not otherwise stated.
| UK Biobank | EXTEND | |
|---|---|---|
| N | 437,027 | 5,679 |
| Age (years) | 57.28 (± 8.02) | 54.31 (± 14.83) |
| Sex (%Female - %Male) | 54.18 - 45.82 | 62.85 - 37.15 |
| Height (cm) | 168.7 (± 9.2) | 168.5 (± 9.1) |
| BMI | 27.38 (± 4.74) | 26.52 (± 4.62) |
| ACR (mg/mml) | 1.08 [0.68 - 1.82] | 0.73 [0.42 - 1.34] |
| CAD (%Yes - %No) | 10.47 - 89.53 | 3.79 - 96.21 |
| T2D (%Yes - %No) | 3.17 - 96.83 | 1.04 - 98.96 |
| Systolic BP (mmHg) | 144.2 (± 24.0) | 131.1 (± 19.8) |
| Diastolic BP (mmHg) | 86.3 (± 13.5) | 77.4 (± 10.8) |
ACR = albumin-creatinine ratio; CAD = coronary artery disease; T2D = type 2 diabetes; BP = blood pressure.
GWAS of ACR in UK Biobank identified sixty-two associated variants across fifty-six loci, twenty of which have not been previously associated with ACR
We identified 62 statistically independent SNPs in 56 loci associated with ACR at P<5×10-8 of which 42 reached P<6×10-9, a threshold that we estimate reflects better the 5% type 1 error rate (13). Of the 62 associations, 18 were located in loci not previously reported to be associated with ACR, and 2 were in low linkage disequilibrium (r 2<0.1) with lead SNPs previously reported. Of these 20 associations, 9 reached P<6×10-9 (Table 2) The LD score regression intercept was 1.05 indicating limited inflation due to population stratification. Conditional analysis revealed five loci containing one additional signal and one containing two additional signals (CUBN) (Supplementary Table 1 and Supplementary Figs. 2-3).
Table 2. GWAS summary statistics in the UK Biobank for the 20 SNPs located in loci not previously reported or in low linkage disequilibrium (r 2<0.1) with lead SNPs previously reported.
Comparisons with publicly available data from the CKDGen consortium and lookups in the EXTEND study can be found in Supplementary Tables 2 and 3, respectively.
| Nearest Gene | SNP | Chr | Position (b37) | EA/OA | EA Freq | Beta | SE | P |
|---|---|---|---|---|---|---|---|---|
| MST01 | rs35202981 | 1 | 155578042 | G/A | 0.139 | 0.017 | 0.003 | 1.4E-08 |
| EDEM3 | rs78444298 | 1 | 184672098 | G/A | 0.980 | 0.045 | 0.007 | 1.4E-09 |
| GPD2 | rs111688960 | 2 | 157599687 | A/G | 0.013 | 0.051 | 0.009 | 1.2E-08 |
| FAT1 | rs62342738 | 4 | 187656129 | C/G | 0.181 | 0.015 | 0.003 | 5.5E-09 |
| C5orf56 | rs11242113 | 5 | 131777234 | A/G | 0.188 | 0.016 | 0.003 | 1.6E-09 |
| KCNK5 | rs1544935 | 6 | 39124448 | G/T | 0.216 | 0.017 | 0.002 | 2.2E-11 |
| VEGFA | rs3734692 | 6 | 43817791 | T/A | 0.310 | 0.016 | 0.002 | 1.8E-13 |
| AUTS2 | rs35692677 | 7 | 69902654 | G/A | 0.813 | 0.015 | 0.003 | 1.1E-08 |
| ZBTB1 | rs11990607 | 8 | 81363534 | A/G | 0.835 | 0.015 | 0.003 | 2.5E-08 |
| MLLT10 | rs6482189 | 10 | 21889138 | G/A | 0.683 | 0.013 | 0.002 | 1.6E-09 |
| CYP26A1 | rs2068888 | 10 | 94839642 | G/A | 0.550 | 0.012 | 0.002 | 1.0E-09 |
| SBF2 | rs11042685 | 11 | 10262551 | C/T | 0.493 | 0.011 | 0.002 | 2.1E-08 |
| NUMA1 | rs7115200 | 11 | 71752160 | G/T | 0.440 | 0.012 | 0.002 | 1.4E-09 |
| OAF | rs12790943 | 11 | 120058623 | T/C | 0.421 | 0.011 | 0.002 | 3.0E-08 |
| NAV3 | rs10860332 | 12 | 78748014 | A/G | 0.414 | 0.011 | 0.002 | 4.4E-08 |
| DLEU1/BCMS | rs3116613 | 13 | 51143055 | G/T | 0.211 | 0.014 | 0.002 | 3.9E-08 |
| WDR81 | rs550628400 | 17 | 1639795 | G/A | 0.006 | 0.075 | 0.013 | 2.1E-08 |
| CYP2A7 | rs79600176 | 19 | 41392490 | T/C | 0.978 | 0.038 | 0.007 | 3.6E-08 |
| CCDC97 | rs56254331 | 19 | 41826020 | A/C | 0.831 | 0.017 | 0.003 | 7.9E-10 |
| ZBTB46 | rs11697610 | 20 | 62379531 | G/A | 0.387 | 0.012 | 0.002 | 1.9E-08 |
SNP = single nucleotide polymorphism; b37 = build 37; EA/OA = effect allele / other allele; Freq = Frequency; SE = standard error; P = P-value.
Genetic variants validate in independent data
We used summary statistics from the CKDGen consortium meta-analysis of 54,451 individuals (5) to check for directional consistency of the ACR associated variants. Of the 62 variants, 47 were available in the summary statistics (including proxy variants at r 2>0.8). The effect estimates of 39/47 SNPs showed directional consistency (Pbinomiai<0.0001) (Supplementary Table 2). The variants not previously identified also showed strong evidence of validation - 15 of the 20 were present in the CKDGen consortium (or their proxies), and 11 of these were directionally consistent. In addition, 8/15 analyzed by the CKDGen consortium reaching P<6×10-9 in our analysis, 7/8 effect estimates were directionally consistent. Of the 47 SNPs, the maximum variance explained by a single SNP in UK Biobank was 0.03%. We note the power to detect an association at P=5×10-8 explaining 0.03% of the variance in 54.451 individuals was 8%, and 76% at P=8×10-4 after Bonferroni correction for 62 tests (0.05/62). In the EXTEND study of 5,679 individuals with measures of ACR, the overall variance explained by all 62 ACR-associated SNPs was 0.61% of the inverse-normalized trait (Supplementary Table 3).
High genetic risk for ACR increases risk of microalbuminuria
We next assessed the risk of being above the clinical threshold of 3 mg/mmol for microalbuminuria using a genetic risk score (GRS) for ACR. A one unit increase in the ACR GRS was equivalent to 0.07 mg/mmol increase in ACR. In the UK Biobank, a one unit increase in the GRS for ACR was associated with a higher risk for being over the clinical threshold and this effect was stronger in men (odds-ratio (OR)=1.124, 95%CI: 1.116-1.133, P=4.0x10-202) than women (OR=1.059, 95%CI: 1.053-1.065, P=3.4×10-85; P interaction=9.3x10-34). To avoid inflated effect estimates due “winner’s curse” we repeated this analysis in the independent EXTEND study. A one unit higher genetic risk score for ACR was associated with an overall OR=1.062 (95%CI: 1.015 – 1.110, P=0.009). When restricting the analysis to CUBN raising alleles, we showed that the 19.9% of UK Biobank individuals carrying at least one CUBN raising allele had an OR=1.153 (95%CI: 1.12-21.185, P=1.0x10-24) for being above the clinical threshold compared to those carrying no ACR-raising alleles at the locus.
Individuals with a genetic risk score for high ACR have a higher risk of hypertension
We next tested the combined role of ACR-associated variants in five diseases related to vascular dysfunction in the UK Biobank: hypertension, type 2 diabetes (T2D), coronary artery disease (CAD), chronic kidney disease (CKD), and stroke. We reasoned this would reveal whether individuals with a high ACR genetic risk score were at high risk of vascular-related disease. The genetic risk score of raised ACR was associated with higher risk of hypertension [OR=1.013, 95%CI: 1.010-1.016, P=1.6x10-16], but much weaker or no evidence of association with stroke [OR=1.011, 95%CI: 1.001-1.022, P=0.027], T2D [OR=1.008, 95%CI: 1.000-1.017, P=0.045], CAD [OR=1.004, 95%CI: 0.999-1.010, P=0.13] and CKD [OR=0.996, 95%CI: 0.982-1.010, P=0.59]. These results were not strongly influenced by the large effect of SNPs in CUBN - we obtained similar results when using a GRS excluding the three variants in CUBN (Supplementary Table 4). We had no direct measure of vascular disease and therefore we were unable to establish if the association between the ACR genetic risk score and hypertension was directly due to vascular dysfunction, but they imply that a higher genetic risk score for ACR is not benign.
Analyses of pathways and variants previously associated with vascular and metabolic traits implicate multiple mechanisms
We next examined the 62 ACR-associated variants for pleiotropic effects. We observed these variants to likely influence ACR through a wide variety of mechanisms, many of which are known to be causally related to, or strongly associated with, vascular diseases and related traits. Previous GWAS had identified 20 of the ACR associated variants (or those in strong LD (r2 >0.8)) as associated with a trait related to metabolic, inflammatory or vascular disease at genome-wide significance levels (P<5x10-8). These associations were with a wide variety of traits, including blood lipid profiles, fasting glucose, blood pressure, and type 2 diabetes (Supplementary Table 5). Two variants represented known type 2 diabetes signals (a variant in the ARL15 locus and a variant in the SNX17 locus that is in LD with highly pleiotropic variants at the GCKR locus (r2>0.85)), three represented known coronary artery disease signals (a variant near KCKN5, one near TRIB1 and one in the intron of CCDC97) and one represented a known blood pressure signal (a variant in HOTTIP). These variants are likely examples of variants with pleiotropic effects that affect ACR through additional mechanisms. For seven of the 20 ACR associated variants in strong linkage disequilibrium with known signals for other traits, data was available to perform a co-localization analysis. All seven showed a high probability (>0.7) that variants associated with ACR represented the same signal as that previously reported – including with those for blood lipid profiles, fasting glucose, blood pressure, and type 2 diabetes (Supplementary Table 6). Using MAGMA (14) we identified an enrichment of genes at associated loci involved in pathways related to lipid metabolism, genital and digestive tract development at P<0.05 after adjustment for multiple testing (Supplementary Table 7). We did not observe evidence of tissue enrichment for genes in the associated loci (Supplementary Figure 4).
We observed three signals at the previously reported CUBN locus
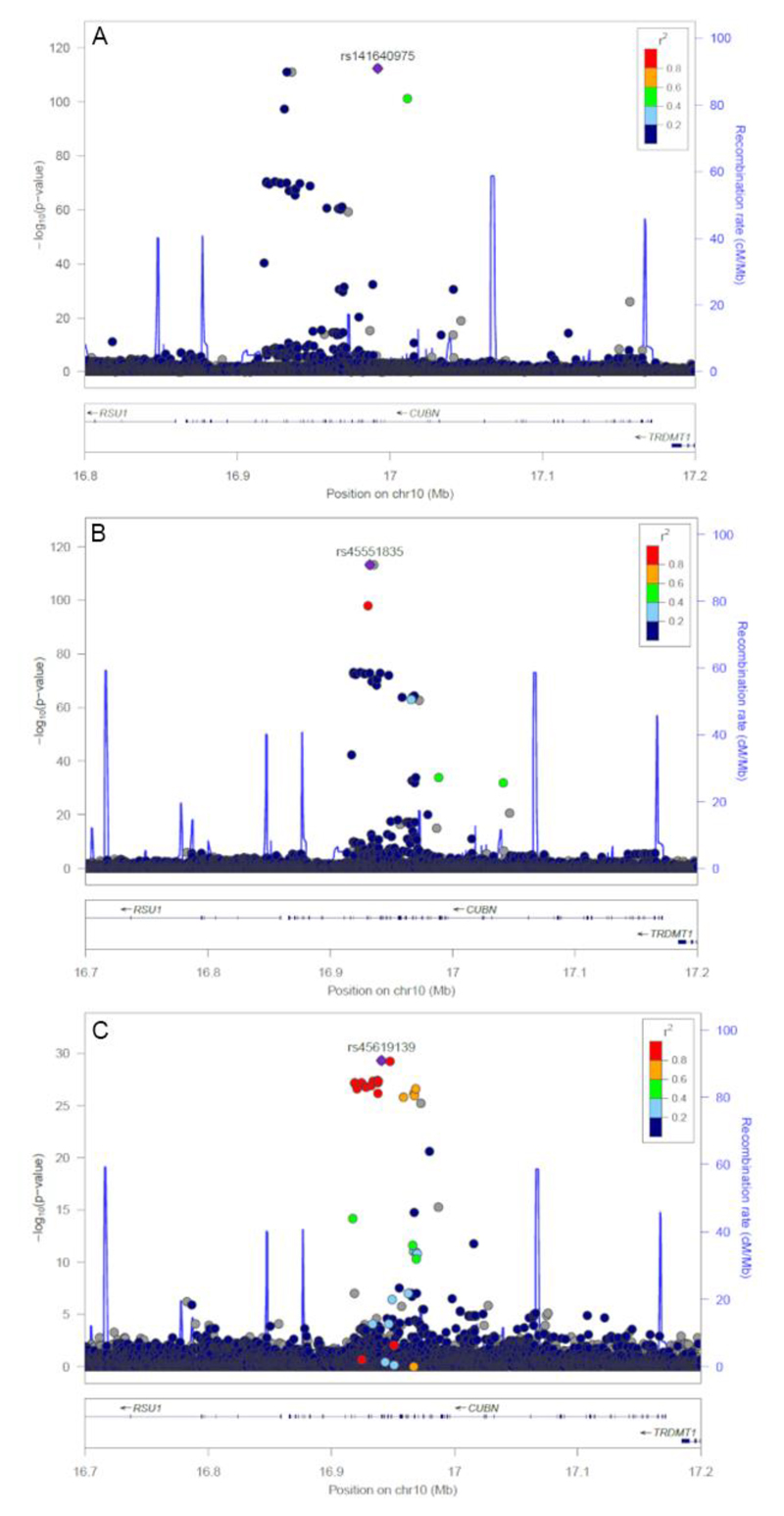
We identified three independent SNPs associated with ACR at the CUBN locus (rs45619139, rs45551835 and rs141640975). These variants had common (rs45619139; minor allele frequency (MAF) 10.1%), low (rs45551835; MAF 1.4%) and rare (rs141640975; MAF 0.3%) allele frequencies and were weakly correlated (low linkage disequilibrium). The minor alleles at the common, low frequency and rare variants were associated with 0.06, 0.19 and 0.46 standard deviation higher ACR, respectively (Table 3 and Figure 1). The common variant previously reported was rs1801239, but this association was abolished after the adjustment for stronger lead SNP rs45619139 in the UK Biobank (r 2=0.78). The low frequency (rs45551835) and rare (rs141640975) variants both alter the amino acid sequence of CUBN (g.16932384G>A, p.Ala2914Val) and (g.16992011G>A, p.Ala1690Val) respectively, and have previously been associated with variation in ACR.
Table 3. Independent signals identified at the CUBN locus on chromosome 10.
Associations in the CUBN locus with ACR in UK Biobank whole cohort. Univariable results when each SNP is entered individually and multivariable results when all three SNPs are entered in the same regression model.
| SNP | Position (b37) | Effect/Other Allele | Frequency Effect Allele | Model | Beta | SE | P | |
|---|---|---|---|---|---|---|---|---|
| CUBN | rs141640975 | 10:16992011 | A/G | 0.003 | Univariable Multivariable |
0.463 0.470 |
0.022 0.022 |
4E-99 3E-102 |
| rs45551835 | 10:16932384 | A/G | 0.014 | Univariable Multivariable |
0.188 0.156 |
0.009 0.010 |
2E-90 1E-56 |
|
| rs45619139 | 10:16940846 | G/C | 0.101 | Univariable Multivariable |
0.059 0.040 |
0.004 0.004 |
1E-57 7E-25 |
SNP = single nucleotide polymorphism; b37 = build 37; SE = standard error; P = P-value
Figure 1.
Significance of SNP associations for three independent signals at the CUBN locus. A) association of SNPs from the initial GWAS analysis, B) strength of SNP associations after the first round of conditional analysis, and C) strength of SNP associations after the second round of conditional analysis.
Haplotype analysis suggests alleles at the CUBN locus have additive effects on ACR
To test the effects of carrying more than one of the three CUBN variants, and whether in cis (one copy of the CUBN gene carrying 2 or 3 minor alleles) or trans (both copies of the CUBN gene carrying at least one minor allele), we estimated the haplotypes formed by the three SNPs. The correlation between the three SNPs was low. However, the D’ measure of linkage disequilibrium was high, suggesting a limited number of recombination events have occurred between the three variants: D’=1 between rs141640975 and rs45551835 and between rs141640975 and rs45619139, and D’ 0.92 between rs45551835 and rs45619139. The variants formed five out of a maximum of eight haplotypes (Table 4). As expected from its very low frequency, the minor allele (A) at the rare variant rs141640975 occurred on only one haplotype – together with the common alleles at the two other variants (G-C-A; frequency=0.3%) and had the largest effect (0.48 SD). All four potential haplotypes formed by the common (rs45619139) and low frequency (rs45551835) CUBN variants were detected – indicating a recombination event must have occurred between them (or, less likely, a second identical mutation). However, the ACR-raising allele (A) at the low frequency variant occurred much more frequently on the same haplotype with the ACR-raising allele at the common variant (G) (A-G-G; frequency=1.3%), rather than with the ACR-lowering allele (C) (A-C-G; frequency=0.1%; Table 4). The A-G-G haplotype was associated with 0.2 SDs higher ACR compared to the commonest haplotype, formed by the three common alleles (G-C-G). This effect was larger than that of the two haplotypes carrying only one of the ACR raising alleles for the low-frequency or common variant (A-C-G and G-G-G), consistent with an additive effect of the two alleles, suggesting that the presence of the two alleles in cis on the same haplotype did not change their effects (Table 4).
Table 4. Haplotype associations with ACR based on the 3 SNPs in the CUBN locus.
Effect sizes are given in standard deviations of inverse-normalized ACR and are relative to the baseline haplotype group formed by the three common alleles of the three SNPs. The χ2 test statistics and P-values for each haplotype correspond to the significance of the association when compared against all other haplotypes pooled. Alleles are ordered across haplotypes based on genomic position and represent 1) the low-frequency variant rs45551835, 2) the common variant rs45619139, and 3) the rare variant rs141640975.
| Haplotype | Frequency | Additive Effect | 95% CI | ▫2 | P |
|---|---|---|---|---|---|
| G-C-G (000) | 0.895 | REF | REF | 352.7 | 1E-78 |
| G-G-G (010) | 0.086 | 0.039 | 0.031, 0.047 | 72.6 | 2E-17 |
| A-G-G (110) | 0.013 | 0.201 | 0.181, 0.221 | 370.1 | 2E-82 |
| G-C-A (001) | 0.003 | 0.482 | 0.437, 0.527 | 421.9 | 2E-94 |
| A-C-G (100) | 0.001 | 0.138 | 0.065, 0.211 | 15.2 | 1E-04 |
95% CI = ninety-five percent confidence interval; χ2= chi-squared test statistic; P = P-value
Carriers of ACR-raising CUBN alleles have higher disease frequency
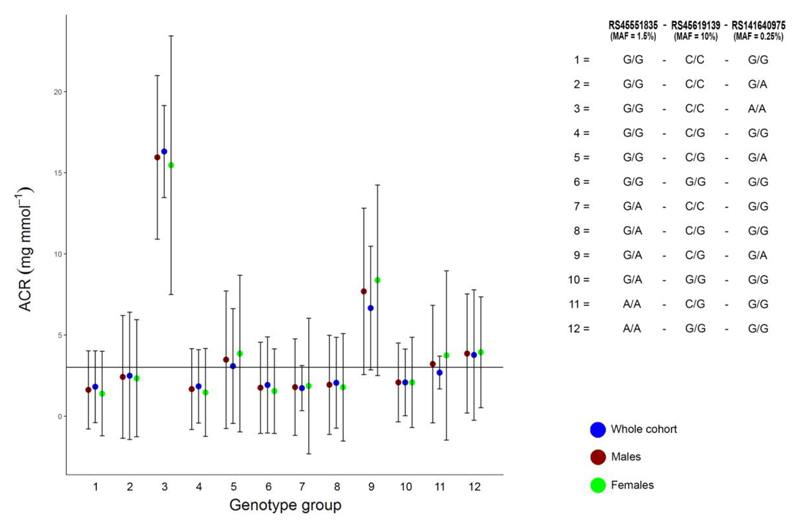
We next classified UK Biobank individuals into 12 groups based on genotype combinations of the three CUBN variants (Supplementary Table 8 and Figure 2). We tested the hypothesis that people with clinically classifiably microalbuminuria partly due to their CUBN genetics would have a lower frequency of vascular related diseases compared to those without ACR raising alleles at the CUBN locus. We noted that for some very rare CUBN genotype combinations the majority of individuals would be classified as having microalbuminuria. For example, of the 25 individuals heterozygous at each of the three CUBN SNPs (<0.01% of the UK Biobank study), 21 (84%) had an ACR value that would classify them as having microalbuminuria (P<0.001 Fisher’s exact test, compared to individuals carrying no ACR raising alleles at the CUBN locus) (Figure 2).
Figure 2.
ACR by CUBN genotype group. ACR mean values and standard deviations by genotype group based on SNPs in the CUBN locus. Solid black line is the clinical threshold for microalbuminuria.
We next selected all UK Biobank individuals above the clinical threshold for microalbuminuria to quantify the extent to which those with at least one ACR-raising allele at CUBN would have a lower frequency of vascular related disease compared to those without a CUBN raising allele. Of the 40,491/437,027 individuals above the clinical threshold for microalbuminuria, 31,592 carried no ACR-raising alleles at the CUBN locus. Contrary to our hypothesis that CUBN ACR-raising alleles are benign for vascular related diseases, people above the clinical threshold and carrying ACR-raising alleles at the CUBN locus had a higher frequency of CAD (carriers 11.2% vs non-carriers 10.2%, P=0.005), T2D (carriers 6.7% vs non-carriers 5.7%, chi-squared P<0.001) and stroke (carriers 3.4% vs non-carriers 2.9%, chi-squared P=0.006, Supplementary Table 8).
Additional ACR-diabetes interaction effects at the CUBN locus observed
We performed interaction analyses to test whether the 62 lead variants had stronger effects in individuals with diabetes as previously observed for the CUBN locus (5, 8). We replicated the statistical interactions between the common and rare CUBN signals and diabetes status. In people with diabetes, each copy of the minor allele for the common variant rs45619139 was associated with a 0.12 SD (95%CI: 0.08-0.15) effect on ACR (inverse-normalized). The observed effect in people without diabetes was 0.06 SD (95%CI: 0.05-0.06) (P interaction=1×10-5) (Table 5 and Supplementary Table 9). The minor alleles at the low frequency and rare CUBN variants also showed evidence of statistical interaction with diabetes status, with larger effects on ACR in people with diabetes (rs45551835 P interaction=6× 10-7; rs141640975 P interaction=4× 10-5). These types of interaction are prone to statistical artefacts, especially when dichotomising groups of people (15). We recently developed a negative control method based on an computational optimization algorithm to assess the likelihood of such artefacts (16) (url: https://github.com/drarwood/gags) - after 1,000 repeated interaction analyses of groups of individuals sampled to have the same means and standard deviations of ACR as individuals with and without a diagnosis of diabetes, we observed an empirical P-value ≤0.02 (19 of 1000 tests were more significant than the observed interaction at most). This suggests that the interaction was unlikely to be a statistical artefact of the differences in the two underlying distributions of ACR (Table 5 and Supplementary Figure 5). After accounting for multiple testing, we found no evidence of interaction for the remaining 59 index variants outside of the CUBN locus with main effects on ACR (Supplementary Table 10).
Table 5. Associations of the three statistically independent SNPs at the CUBN locus within individuals with diabetes and without diabetes.
The interaction term with diabetes is presented. Empirical P refers to the control experiment that involved the repeated sampling of two groups (x1,000) matched on the distributions of ACR observed in individuals with- and without diabetes prior to interaction analysis of the dummy group variable.
| Individuals with diabetes | Individuals without diabetes | Interaction Term | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Effect Allele | Beta | SE | P | Beta | SE | P | Beta | SE | P | Empirical P |
| rs45551835 | A | 0.358 | 0.044 | 3.8E-16 | 0.179 | 0.010 | 1.4E-78 | 0.213 | 0.043 | 6E-07 | 0.001 |
| rs45619139 | G | 0.115 | 0.017 | 4.7E-11 | 0.055 | 0.004 | 3.7E-49 | 0.075 | 0.017 | 1E-05 | 0.002 |
| rs141640975 | A | 0.781 | 0.107 | 2.6E-13 | 0.449 | 0.022 | 1.3E-89 | 0.426 | 0.104 | 4E-05 | 0.020 |
SNP = single nucleotide polymorphism; SE = standard error; P = P-value
When combining variants into a weighted GRS (excluding CUBN variants), we observed evidence of interaction with diabetes status, with the GRS having larger effects in people with diabetes (P interaction =2×10-6). After random sampling (described above) we observed an empirical P interaction=0.11, suggesting the interaction may not be specific to diabetes (Supplementary Figure 5). This T2D interaction may be a feature of metabolic disease in general because we also identified evidence of interaction between the ACR GRS and other diseases - all with stronger effects in cases than controls: hypertension (GRS β cases=0.036, 95%CI: 0.034-0.038; GRS β controls=0.025, 95%CI: 0.023-0.027; P interaction =2×10-18), CKD (GRS β cases=0.043, 95%CI: 0.030-0.056; GRS β controls=0.031, 95%CI: 0.030-0.033; P interaction 4×10-05), CAD (GRS β cases=0.034, 95%CI: 0.030-0.039; GRS β controls=0.032, 95%CI: 0.031-0.034; P interaction=3×10-02), and stroke (GRS β cases=0.039, 95%CI: 0.030-0.048; GRS β controls=0.032, 95%CI: 0.031-0.034; P interaction=2×10-02). In contrast to a previous study (5), we found no evidence of a gene-diabetes interaction at the HS6ST1 locus (Pinteraction 0.76) or RAB38/CTSC locus (P interaction 0. 06).
Discussion
We performed a GWAS of ACR in 437,027 individuals from the UK Biobank and identified 62 SNPs in 56 loci associated with increased ACR. Of these, we identified 20 associations not previously known, of which 9 reached a stricter significance threshold of P<6×10-9. Prior to the availability of the UK Biobank data, only one of these 56 loci (CUBN) was known to be associated with variation in ACR in individuals of European ancestry (5, 6, 8). Recent analysis, of smaller sets of unrelated individuals from the UK Biobank, identified 46 and 22 variants associated with ACR. The 62 variants we identified include 41/46 loci reported by the recent Haas et al. analysis (11). The five detected by Haas et al. but not us, despite our larger sample size, fell below the GWAS threshold of P<5x10-8 (P-values ranged between 1.5x10-5 and 4.9x10-8). This difference may reflect sampling error between our analyses. A more recent analyses of the UK Biobank data by Zenetti et al. (12) identified 19 associations with urinary ACR, but this analysis identified 22 variants after splitting the data into a discovery and replication dataset. These three analyses of the same data show the value in different groups analysing large genetic datasets with different approaches.
A higher GRS for ACR was associated with higher risk of being above the clinical threshold for ACR. Like the Haas et al. study, we showed that people with a high ACR genetic risk score were at higher risk of hypertension compared to those with a lower ACR, but there was no association with other diseases, including diabetes.
Our analysis of individual variants suggested that a higher genetic ACR results from a wide range of pathogenic mechanisms. Twenty of 62 ACR-associated SNPs have been previously associated at genome-wide significance with metabolic, inflammatory and vascular diseases and related traits. We identified over 200 separate ACR-SNP-second trait association entries in the NHGRI GWAS catalog (17), although over half included highly pleiotropic GCKR variants in LD with the our signal in SNX17 (18). With the exception of the SNX17 signal near GCKR, there were notably few that were known blood pressure, diabetes or CAD signals, and of these, they were not the most strongly associated variants for these traits suggesting that the variants identified as associated with ACR are likely pleiotropic. The only T2D variant apart from that near GCKR associated with ACR is that in ARL15, where the ACR raising allele is associated with higher risk of T2D, but with a much lower odds ratio (1.06 [95%CI: 1.04-1.09)] (19) than many other known T2D variants. The ACR raising allele at ARL15 is also associated with apparently paradoxical effects on body composition (20) and poorer kidney function as measured by eGFR (6). That 20 of the 62 ACR-associated signals were previously known signals for a wide variety of traits, together with our pathway analyses implicating lipid metabolism and gut and genital development suggests a wide variety of mechanisms involved in normal variation in ACR. In addition, we note that 19/20 variant-ACR associations we present as novel are available in a recent meta-analysis of eGFR by Wuttke et al. (21). Of these, 16/19 effect estimates are directionally consistent with eGFR, with 10/16 at P<0.05 and 4/16 at P<5x10-8
Compared to previous studies, we performed a more extensive analysis of the previously reported CUBN locus (4, 5). We showed that the low frequency ACR-raising allele at rs45551835 most often occurs in cis with the more common ACR-raising allele at rs45619139 but this haplotype was associated with ACR consistent with the additive effects of the two alleles.
The association with hypertension indicates that disease processes underlie a high ACR GRS, rather than benign effects on kidney reabsorption and means that it would be inappropriate to tailor the clinical threshold to a person’s GRS for ACR. This is likely to be the case even for people carrying ACR-raising genotypes at the CUBN locus. These alleles might have been expected to be benign on disease risk, but for individuals above the microalbuminuria threshold our results provide evidence of higher frequencies of type 2 diabetes, coronary heart disease, and stroke among CUBN allele carriers compared to non-carriers.
The availability of individual level data from a large study also allowed us to test for genediabetes status interactions more extensively than before. Teumer et al. (2016) and Ahluwalia et al. (2019) showed that the ACR raising minor-allele in CUBN had a 3 to 4-fold effect in people with diabetes compared to controls. Teumer et al. also reported larger effects in individuals with diabetes for variants in the HS6ST1 and RAB38/CTSC loci. We tested these reported interactions and performed negative control experiments to control for the different distributions of ACR between people with and without diabetes (15). We replicated the previously reported interactions for the common and rare signal at the CUBN locus but did not find evidence of gene-diabetes interactions or main effects at the HS6ST1 or RAB38/CTSC loci.
Our analysis had a number of strengths, including the availability of individual level data from a single large-study that provided homogeneous measures of ACR. Access to individual level data facilitated analyses that would otherwise be difficult to perform, including haplotype analysis, disease prevalence among rare genotype groups, and interaction analysis with follow-up negative control experiments.
The main limitations of our study include the cross-sectional nature of the associations with disease prevalence. We are presently limited in our ability to evaluate the impact of ACR on disease outcomes prospectively in the UK Biobank within individuals who do not report having kidney disease. Second, the rare and low-frequency nature of the genetic variants with the largest effects mean replication of these signals will be difficult in many studies given relatively small sample sizes. Third, we calculated ACR in individuals with albumin levels below assay detection limits. Albumin levels were set to study-specific limits of detection – an approach previously used by other GWAS analyses of ACR (5). Finally, further work needs to be undertaken in other ethnicities to determine whether the findings in our study replicate in other ancestries.
In conclusion, we have performed one of the largest genetic association studies on ACR and have gained further insight into the biological causes and clinical implications of raised ACR.
Materials and methods
Study individuals
We used 437,027 UK Biobank individuals that had measures of ACR and were classified as European ancestry through principal components analyses and a k-means clustering approach using the first 4 principal components and 1000 Genomes Project samples as reference ancestral centers.
Albumin-creatinine ratio in UK Biobank
Measures of ACR were derived using urinary levels of albumin and creatinine. Albumin was measured in the UK Biobank samples using immuno-turbidimetric analysis method (Randox Bioscience, UK) while creatinine was measured using enzymatic analysis method (Beckman Coulter, UK). If albumin was <6.7 mg/L (the assay detection level in UK Biobank) then albumin was set at 6.7 mg/L prior to the calculation of the ratio, an approach consistent with previous studies (5).
Albumin-creatinine ratio in EXTEND
Albumin and creatinine were measured in samples using immuno-turbidimetric and enzymatic methods, respectively. If albumin was <2.9 mg/L (the assay detection level) then albumin was set at 2.9 mg/L prior to the calculation of the ratio.
Genome-wide association analysis
Genetic associations for inverse-normalized ACR were tested using a linear-mixed model approach, implemented in BOLT-LMM (22). Models were adjusted for baseline age, sex, study center, and genotyping array. Imputed genotypes from the Haplotype Reference Consortium (HRC) from the UK Biobank were used(23). Variants with imputation quality (INFO) <0.3 or minor allele frequency (MAF) <0.1% were excluded. After quality control, 12,082,474 variants for association analysis remained. Lead SNPs were defined as those with the smallest P-value. Locus boundaries were defined using a ±0.5 Mb distance from the lead SNP. Conditional analysis was performed by subsequently adding all lead SNPs for each locus as covariates.
Lookups of associations using summary statistics from the CKDGen consortium
We downloaded summary statistics from the latest meta-analysis of ACR performed by CKDGen consortium to enable a comparison of directions of estimated effects for SNPs associated with ACR in the UK Biobank (5). We used proxies with r2>0.8 within 500kb if unavailable in CKDGen.
Variation explained and validation of GRS in the EXTEND study
We used the Exeter 10,000 (EXTEND) study to calculate the variance explained by discovered genetic associations in an independent cohort. EXTEND is a population-based study in the South West of England. Genotyping was performed using the Illumina Infinium Global Screening Array. Imputation of genotypes was performed using the Haplotype Reference Consortium (HRC) imputation reference panel (24). Analyses were based on 5,679 individuals with genotype data and measures of ACR. Association analyses were carried out using RVTEST, adjusting for relatedness and ancestry through a genomic relationship matrix (25).
Genetic risk score derivation for raised ACR
A weighted ACR genetic risk score (GRS) was calculated for each participant using the index variants identified from the UK Biobank analysis. Dosages were re-coded to ACR-increasing alleles prior to weighting using the respective effect sizes observed in the UK Biobank. A weighted score was subsequently calculated as the sum of the weighted dosages (Equation 1) prior to re-scaling to reflect the number of ACR increasing alleles (Equation 2).
| (1) |
| (2) |
Testing the ACR genetic risk score against risk of clinically defined microalbuminuria
We tested whether a higher ACR genetic risk score was associated with being above the 3 mg/mmol clinical threshold (NICE, https://www.nice.org.uk/guidance/cg182) for microalbuminuria using logistic regression.
Testing whether a high ACR genetic risk score is associated with diabetes and vascular related disease
We used logistic regression to test the combined role of ACR-associated variants in five diseases related to vascular dysfunction and diabetes in the UK Biobank - hypertension, type 2 diabetes (T2D), coronary heart disease (CAD), chronic kidney disease (CKD) and stroke. Disease definitions were derived using a combination of questionnaire data, hospital episode statistics and interviews. Hypertension was defined as a systolic blood pressure of >140 mmHg, or a diastolic blood pressure of >90 mmHg, or the report of blood pressure medication usage using the baseline UK Biobank questionnaire. T2D was defined through self-report of diabetes using the UK Biobank questionnaire at baseline, >35 years of age, and without reporting of insulin use within the first year of diagnosis. We excluded individuals reporting diabetes diagnosed less than one year prior to baseline data collection (N = 1,757) to exclude those who may be on insulin treatment within the first year of diagnosis and therefore could have other forms of diabetes. Incident cases (relative to UK Biobank baseline visit) of T2D were included using Hospital Episode Statistics (HES) data (from ICD10 code: E11.*). In addition, having any form of diabetes was defined for individuals who reported being informed of having the disease by their doctor (UK Biobank data field 2443). CAD was defined from HES and self-reported data from the UK Biobank questionnaire at baseline. Reporting of angina and/or myocardial infarction was used as criteria. CKD was defined using relevant primary and secondary ICD9 (580-629) and ICD10 codes (N00 to N99) available from HES data. Stroke was defined using codes 1583 (ischaemic stroke), 1081 (stroke), 1086 (subarachnoid haemorrhage) and 1491 (brain haemorrhage) from clinic nurses’ codes for non-cancer illness (UK Biobank data field 20002).
Investigating the overlap of loci associated with ACR with other traits from previous genome-wide association studies
We downloaded association statistics from the NHGRI-EBI GWAS Catalog (17). We looked up lead SNPs for ACR and proxies (r2 > 0.8) against SNPs catalogued with P- value<5x10-8.
Colocalization analysis of ACR-associated SNPs associated with other traits
We performed colocalization analysis to determine the likelihood of shared causal variants at associated loci that overlap for other traits in the GWAS catalog. We used summary statistics for SNPs 500Kb either-side of the lead ACR-associated variant using publicly available genome-wide association study data (16, 26–32), aligning the effects to the ACR effect alleles. We used the coloc.abf function to estimate the probability of each locus sharing a causal variant.
Gene-set and tissue enrichment analyses
Gene-set analyses and tissue expression analyses were performed using MAGMA (14) as implemented in the online Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA) tool (33).
Haplotype estimation and testing of associated SNPs in the CUBN locus
Estimation and testing of haplotypes were performed using UNPHASED (version 3.1.7) (34). Genotypes were converted to best-guess genotypes (0, 1, or 2) prior to haplotype estimation. Effect estimates were made relative to the reference haplotype comprising of the common alleles. This analysis was performed in a subset of 367,882 unrelated UK Biobank individuals (<3rd degree relatives).
Interaction analyses
We performed interaction analyses for both novel and previously reported (5) SNPs to test for differences in effect sizes between diabetes cases and controls. These analyses were performed in the unrelated subset of 367,882 UK Biobank individuals. Of these, we classified 17,671 as having some form of diabetes. We compared effect sizes in diabetes cases versus controls by fitting the multiplicative interaction model and testing if not equal to zero - specifically:
| (3) |
Negative control experiment to test specificity of interactions
We tested whether putative interactions were specific to the interacting condition (e.g. diabetes), or an artefact of the highly skewed distribution. Using a computational optimization (genetic) algorithm (url: https://github.com/drarwood/gags), we repeatedly sampled individuals from the UK Biobank to derive groups matched to the ACR distributions of diabetes cases and controls but randomized to diabetes status. We repeated this random sampling 1,000 times and compared the results to the observed interaction (15).
Supplementary Material
Aknowledgments
This research has been conducted using the UK Biobank Resource under application numbers 9072 and 10171. The authors would like to acknowledge the use of the University of Exeter High-Performance Computing (HPC) facility in carrying out this work.
Funding
This paper presents independent research supported by the NIHR Exeter Clinical Research. The views expressed in this publication are those of the author(s) and not necessarily those of the NIHR Exeter Clinical Research Facility, the NHS, the NIHR or the Department of Health in England. A.R.W. and T.M.F. are supported by the European Research Council grant SZ-245 50371-GLUCOSEGENES-FP7-IDEAS-ERC. R.B. is funded by the Wellcome Trust and Royal Society grant 104150/Z/14/Z. J.T. is funded by a Diabetes Research and Wellness Foundation Fellowship. S.E.J. is funded by the Medical Research Council (grant MR/M005070/1). A.M. and M.N.W. is supported by the Wellcome Trust Institutional Strategic Support Award (WT097835MF). We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z) for the generation of the array data for validation.
Abbreviations
- ACR
Albumin-creatinine ratio
- CAD
Coronary artery disease
- CI
Confidence interval
- CKD
Chronic kidney disease
- EXTEND
Exeter 10,000
- GRS
Genetic risk score
- GWAS
Genome-wide association study
- HES
Hospital Episode Statistic
- HRC
Haplotype Reference Consortium
- ICD
International Classification of Diseases
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- OR
Odds Ratio
- SD
Standard Deviation
- SNP
Single nucleotide polymorphism
- T2D
Type 2 diabetes
Footnotes
Conflict of Interest Statement
None declared.
Author Contributions. F.C., T.M.F. and A.R.W designed the study. R.N.B., F.C., Y.J., S. E.J., J.T. and A.R.W. performed data processing and analyses. A.T.H. and A.C.S. assisted in the provision of study-specific data prior to statistical analysis. R.N.B., F.C., T.M.F., A.M., A.C.S., J.T., M.N.W. and A.R.W. were involved in the preparation of the manuscript draft. A.R.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Data availability
Summary results for the GWAS analyses performed will be available from http://www.t2diabetesgenes.org/data/ on publication of this manuscript.
References
- 1.Gerstein HC, Mann JF, Yi Q, Zinman B, Dinneen SF, Hoogwerf B, Halle JP, Young J, Rashkow A, Joyce C, et al. Albuminuria and risk of cardiovascular events, death, and heart failure in diabetic and nondiabetic individuals. JAMA. 2001;286:421–426. doi: 10.1001/jama.286.4.421. [DOI] [PubMed] [Google Scholar]
- 2.Salmon AH, Satchell SC. Endothelial glycocalyx dysfunction in disease: albuminuria and increased microvascular permeability. J Pathol. 2012;226:562–574. doi: 10.1002/path.3964. [DOI] [PubMed] [Google Scholar]
- 3.Wu HY, Peng YS, Chiang CK, Huang JW, Hung KY, Wu KD, Tu YK, Chien KL. Diagnostic performance of random urine samples using albumin concentration vs ratio of albumin to creatinine for microalbuminuria screening in patients with diabetes mellitus: a systematic review and meta-analysis. JAMA Intern Med. 2014;174:1108–1115. doi: 10.1001/jamainternmed.2014.1363. [DOI] [PubMed] [Google Scholar]
- 4.Boger CA, Chen MH, Tin A, Olden M, Kottgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, et al. CUBN is a gene locus for albuminuria. J Am Soc Nephrol. 2011;22:555–570. doi: 10.1681/ASN.2010060598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teumer A, Tin A, Sorice R, Gorski M, Yeo NC, Chu AY, Li M, Li Y, Mijatovic V, Ko YA, et al. Genome-wide Association Studies Identify Genetic Loci Associated With Albuminuria in Diabetes. Diabetes. 2016;65:803–817. doi: 10.2337/db15-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattaro C. Genome-wide association studies of albuminuria: towards genetic stratification in diabetes? J Nephrol. 2018;31:475–487. doi: 10.1007/s40620-017-0437-3. [DOI] [PubMed] [Google Scholar]
- 7.van Zuydam NR, Ahlqvist E, Sandholm N, Deshmukh H, Rayner NW, Abdalla M, Ladenvall C, Ziemek D, Fauman E, Robertson NR, et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects With Type 2 Diabetes. Diabetes. 2018;67:1414–1427. doi: 10.2337/db17-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahluwalia TS, Schulz CA, Waage J, Skaaby T, Sandholm N, van Zuydam N, Charmet R, Bork-Jensen J, Almgren P, Thuesen BH, et al. A novel rare CUBN variant and three additional genes identified in Europeans with and without diabetes: results from an exome-wide association study of albuminuria. Diabetologia. 2019;62:292–305. doi: 10.1007/s00125-018-4783-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grasbeck R. Imerslund-Grasbeck syndrome (selective vitamin B(12) malabsorption with proteinuria) Orphanet J Rare Dis. 2006;1:17. doi: 10.1186/1750-1172-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wahlstedt-Froberg V, Pettersson T, Aminoff M, Dugue B, Grasbeck R. Proteinuria in cubilin-deficient patients with selective vitamin B12 malabsorption. Pediatr Nephrol. 2003;18:417–421. doi: 10.1007/s00467-003-1128-y. [DOI] [PubMed] [Google Scholar]
- 11.Haas ME, Aragam KG, Emdin CA, Bick AG, International Consortium for Blood, P. Hemani G, Davey Smith G, Kathiresan S. Genetic Association of Albuminuria with Cardiometabolic Disease and Blood Pressure. Am J Hum Genet. 2018;103:461–473. doi: 10.1016/j.ajhg.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanetti D, Rao A, Gustafsson S, Assimes TL, Montgomery SB, Ingelsson E. Identification of 22 novel loci associated with urinary biomarkers of albumin, sodium, and potassium excretion. Kidney Int. 2019;95:1197–1208. doi: 10.1016/j.kint.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, Jeffries AR, Dashti HS, Hillsdon M, Ruth KS, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10:343. doi: 10.1038/s41467-018-08259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyrrell J, Wood AR, Ames RM, Yaghootkar H, Beaumont RN, Jones SE, Tuke MA, Ruth KS, Freathy RM, Davey Smith G, et al. Gene-obesogenic environment interactions in the UK Biobank study. Int J Epidemiol. 2017;46:559–575. doi: 10.1093/ije/dyw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, Nolte IM, Cocca M, Taliun D, Gomez F, et al. 1000 Genomes-based metaanalysis identifies 10 novel loci for kidney function. Sci Rep. 2017;7:45040. doi: 10.1038/srep45040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxena R, Diabetes GeneticsInitiative of Broad Institute of H. Mit LU, Novartis Institutes of BioMedical R. Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 19.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ried JS, Jeff MJ, Chu AY, Bragg-Gresham JL, van Dongen J, Huffman JE, Ahluwalia TS, Cadby G, Eklund N, Eriksson J, et al. A principal component meta-analysis on multiple anthropometric traits identifies novel loci for body shape. Nat Commun. 2016;7:13357. doi: 10.1038/ncomms13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, Tin A, Wang L, Chu AY, Hoppmann A, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51:957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–1426. doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao W, Rasheed A, Tikkanen E, Lee JJ, Butterworth AS, Howson JMM, Assimes TL, Chowdhury R, Orho-Melander M, Damrauer S, et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat Genet. 2017;49:1450–1457. doi: 10.1038/ng.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surakka I, Horikoshi M, Magi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O’Reilly PF, Cabrera CP, Warren HR, Rose LM, et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertension. 2017;70:e4–e19. doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary results for the GWAS analyses performed will be available from http://www.t2diabetesgenes.org/data/ on publication of this manuscript.