Abstract
Cryo-electron microscopy (cryo-EM) enables macromolecular structure determination in vitro and inside cells. In addition to aligning individual particles, accurate registration of sample motion and 3D deformation during exposures are crucial for achieving high resolution reconstructions. Here we describe M, a software tool that establishes a reference-based, multi-particle refinement framework for cryo-EM data and couples a comprehensive spatial deformation model to in silico correction of electron-optical aberrations. M provides a unified optimization framework for both frame-series and tomographic tilt-series data. We show that tilt-series data can provide the same resolution as frame-series on a purified protein specimen, indicating that the alignment step no longer limits the resolution obtainable from tomographic data. In combination with Warp and RELION, M resolves to residue-level a 70S ribosome bound to an antibiotic inside intact bacterial cells. Our work provides a computational tool that facilitates structural biology in cells.
Introduction
Cryo-EM1 is a widely used method for macromolecular structure determination2, 3. Two types of data are commonly analyzed to obtain high-resolution maps. First, samples are prepared at concentrations where individual particles can be distinguished in 2D projections captured in a transmission electron microscope (TEM), and fractionated exposures at constant stage orientation (“frame-series”) are typically acquired. Such data are then subjected to single-particle analysis (SPA). Second, samples containing multiple particles stacked along the projection axis, or samples that capture portions of crowded cellular environments, favor a tomographic approach to distinguish the particles in 3D. Here, the microscope stage is tilted to different angles between sub-exposures (“tilt-series”). Each sub-exposure also comprises a frame-series (“tilt-movie”). Analysis of recurring structures in this data-type has been implemented as sub-tomogram averaging (STA)4–6.
In SPA, many noisy projections of similar particles observed under different orientations are iteratively aligned, classified and averaged to reconstruct 3D maps of the macromolecules’ Coulomb potential7. SPA refinement algorithms assume that each observation shows a single particle in isolation, and can thus be treated independently of other particles8. The same assumption is made in the closely-related STA workflow9–11, where the reference of a single particle is aligned to each sub-tomogram, and surrounding particles are treated as noise.
As samples are irradiated with electrons, beam-induced motion (BIM) leads to changes in particle positions and orientations12. If left uncorrected, these changes decrease the apparent image quality and limit the map resolution. Exposure fractionation into multiple frames captures the particles along their trajectories, allowing for accurate motion registration and the reversal of the detrimental effects of BIM13, 14. Unfortunately, the granularity of the motion model is limited by the low signal per particle. Although each particle’s trajectory is unique, correlations exist on a local scale and can be used to regularize the motion model13, 15. It is thus beneficial to exploit these correlations and treat the contents of a micrograph or tomogram as a multi-particle system embedded in the same physical space rather than isolated particles.
At the data pre-processing stage, the motion model can be fitted based on raw data using reference-free approaches13, 14, 16–20. Frame-series are aligned in 2D, whereas tilt-series are aligned and used to reconstruct tomograms. Extracted particles are fed into SPA or STA pipelines to obtain 3D references. Reference-based alignment can then improve the model accuracy by aligning the raw data to high-resolution reference projections. Such algorithms exist for both frame and tilt-series data6, 15, 21, 22, and improve the accuracy by enforcing local smoothness between particle trajectories on different spatio-temporal scales. However, most implementations remain different for frame and tilt-series data, and are limited to one reference species even in highly heterogeneous datasets. They are further decoupled from other parts of the refinement process, including rotational alignment and contrast transfer function (CTF) fitting, leading to a fragmented workflow and decreased convergence speed, limiting the final map resolution.
Here we present M, a software tool that integrates reference-based refinement of particle motion trajectories with other parts of the structure determination pipeline. We formulate our approach explicitly in a multi-particle framework, which simultaneously optimizes particle poses and hyperparameters describing physically plausible sample deformation within the entire field of view. This allows us to unify the processing of frame and tilt-series, define a set of intuitive regularization constraints such as spatial and temporal resolution, and include any number of particle species at different resolutions. Coupled with a robust approach to CTF correction and with neural network-based map denoising, M achieves higher resolution on several datasets compared to other methods6, 21, 23, 24. We demonstrate how various features of M contribute to these improvements, and achieve the same high resolution for frame and tilt-series data given similar numbers of particles. Most strikingly, we use M to visualize a 70S ribosome bound to an antibiotic in its native cellular context at residue-level resolution from tilt-series data.
Results
Overall design
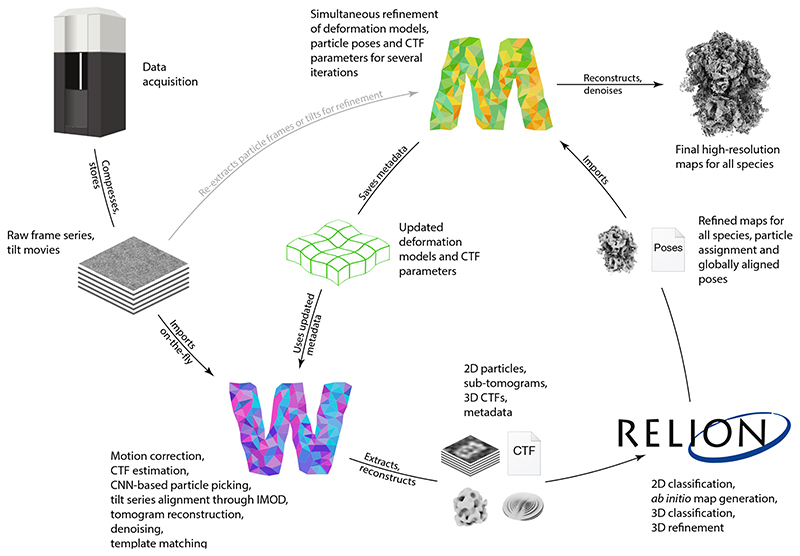
M forms the last part of a cryo-EM data pre-processing and map refinement pipeline – preceded by Warp19 and RELION25, or compatible tools (Fig. 1). Warp performs initial reference-free motion correction and CTF estimation on frame-series or tilt-movies. For tilt-series, Warp, starting with version 1.1.0, calls routines from IMOD26 to perform tilt-series alignment, estimates per-tilt CTF using the tilt angles as constraints, and reconstructs tomographic volumes. Warp then picks particles using a convolutional neural network (CNN) or template matching, and exports them as dose-weighted images or volumes depending on the data type. Particle poses and classes are then determined in RELION27. All classes are imported into M to perform a more accurate, reference-based, multi-particle frame or tilt-series refinement and obtain the final high-resolution maps. Optionally, improved alignments can be applied to re-export particles for further classification in RELION, or to pick additional particles in tomograms.
Figure 1. The Warp–RELION–M pipeline for frame and tilt-series cryo-EM data refinement.
Electron microscopy data are pre-processed on-the-fly in Warp, which then exports particles as images or sub-tomograms. For tilt series, 3D CTF volumes containing the missing wedge and tilt-dependent weighting information are generated for each particle. Particles are imported in RELION, where they can be subjected to a multitude of processing strategies, resulting in 3D reference maps, global particle pose alignments, and class assignments. The particle population encompassing all classes is then imported in M, where reference-based frame or tilt image alignments are performed simultaneously with further refinement of particle poses and CTF parameters. Finally, M produces high-resolution reconstructions that can be used for model building. The improved alignments can be used in Warp to re-export particles for further, more accurate classification in RELION.
M provides a graphical user interface (GUI) that allows users to create, import, export and manage data. Projects are organized as “populations”, which contain “data sources” and “species”. A data source is a set of frame or tilt-series, that stem ideally from the same sample grid and acquisition session. A species is a distinct type of macromolecule, or its compositional and conformational sub-state. The refinement evolution is tracked as a directed graph, parts of which can be stored in different locations while remaining uniquely connected through cryptographic hashes.
Multi-particle system modeling
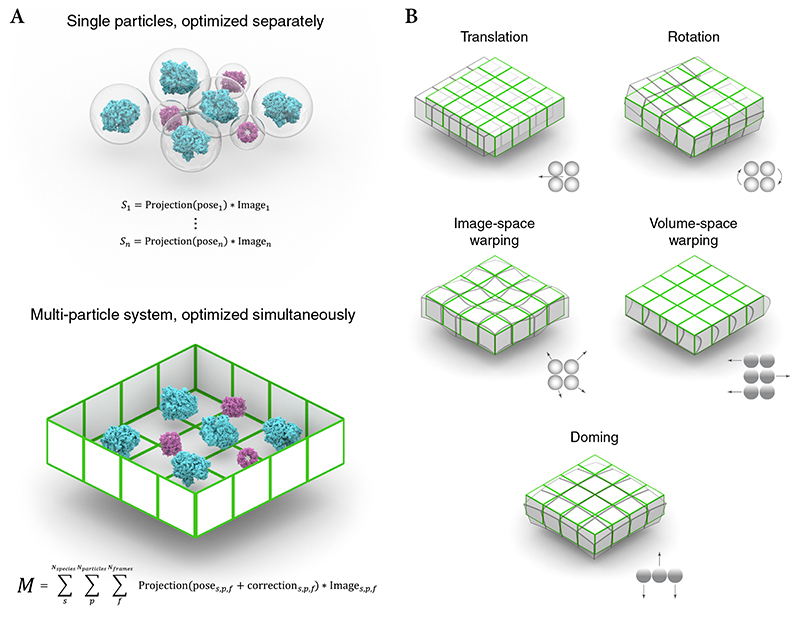
M considers the entire field of view as a physically connected multi-particle system (Fig. 2a). The particles can belong to different species, which can be of varying size, symmetry, and resolution. The particles are subject to the same global transformations including stage translation and rotation, and locally correlated transformations caused by BIM. M performs a reference-based registration of these transformations (Fig. 2b), and reverses them when back-projecting individual particle images to obtain more accurate reconstructions.
Figure 2. Multi-particle system modeling and optimization.
(a) Previous algorithms treat particles as isolated entities and optimize their poses using separate cost functions (top). In M’s multi-particle refinement framework, all particles within the field of view are treated as parts of the same physical volume. Their poses and hyperparameters describing the beam-induced deformation of the volume are optimized simultaneously using a single cost function (bottom).
(b) The multi-particle system deformation model incorporates several modes: Global movement and rotation to account for inaccuracies in stage movement between frames and stage rotation between tilts; image-space warping to model local non-linear deformation in the 2D reference frame of a frame or tilt image; volume-space warping to model the movement of overlapping particles perpendicular to the projection axis (tilt-series only); doming to account for the hypothesized bending of a thin sample along the projection axis (frame-series only).
In frame-series, all transformations occur in the same image reference frame. Their combined effects are parametrized as a pyramid of 3D cubic spline grids (Extended Data Figure 1), to combine fast, global stage motion with slow, local BIM. This model is similar to Warp’s reference-free alignment, but fits more parameters due to the increased signal of high-resolution references. In addition to image-space warping, M can fit doming-like motion12 (Fig. 2b) implemented as parameter grids for defocus and orientation offsets.
For tilt-series, M distinguishes image-space and volume-space effects. Additionally, a coarse model can be fitted for every tilt movie to account for the significant deformation captured in each exposure. Volume-space transformations are resolved in 3D as a function of the accumulated exposure. Because M does not average particle frames or tilts in intermediate steps, per-particle translation and rotation trajectories can be fitted. The temporal resolution of the trajectories can be set for each species depending on the particle’s size and thus the signal available per particle.
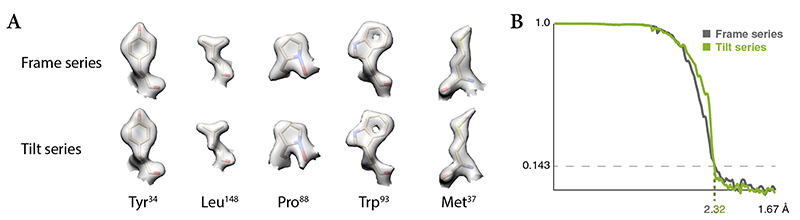
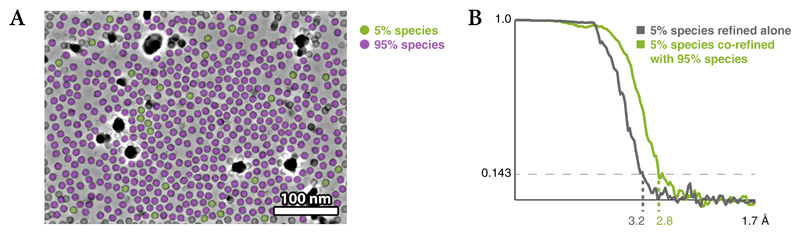
We show the benefit of considering the particles of multiple species in refinement using frame-series of apoferritin (AF-f, Online Methods). We artificially split the apoferritin population in two species comprising 5% or 95% of the particles (Extended Data Figure 2a, Table 1), and assumed no structural similarity between the two species during refinement. Refining the 5% species alone produced a 3.2Å map, while adding the 95% species to the multi-particle system improved the map calculated from the 5% species to 2.8Å (Extended Data Figure 2b).
Correction of electron-optical aberrations
In addition to a geometric deformation model, M fits CTF parameters and higher-order aberrations including beam tilt. For frame-series, defocus is optimized per-particle, similar to cisTEM23 and recent RELION versions24. For tilt-series, defocus is optimized per-tilt, similar to the capability offered in emClarity6. For both data types, astigmatism, anisotropic pixel size and higher-order aberrations are fitted per-series.
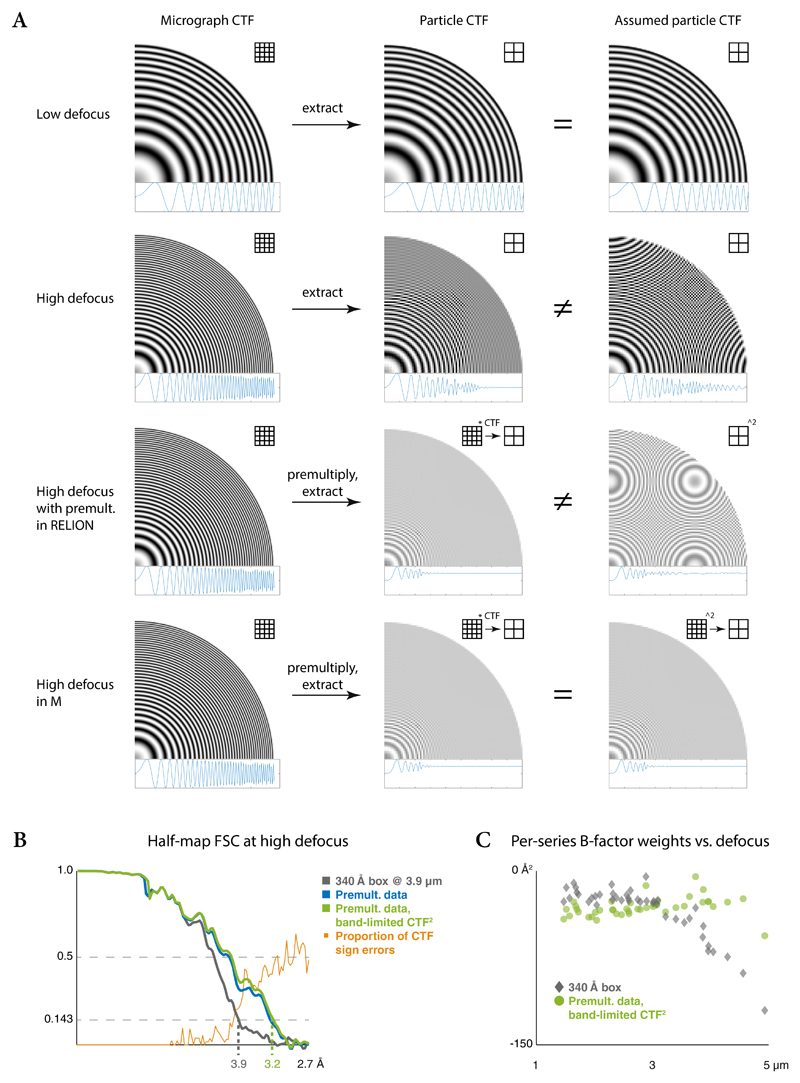
CTF correction at high defocus can introduce artifacts if the chosen particle box size is too small to retain high-resolution Thon rings, leading to their aliasing (Extended Data Figure 3a). M automates the selection of a sufficiently large box size at which the data are pre-multiplied by an aliasing-free CTF. The images are then cropped in real space. To match the underlying CTF of these images, correctly band-limited CTF2 images are constructed in a similar way (Extended Data Figure 3a) to be used for refinement and reconstruction.
We show the benefit of this approach by reconstructing a map from a high-defocus tilt-series of HIV1 virus-like particles (EMPIAR-10164, Table 1). Using a box size twice the particle diameter, the resolution is limited to 3.9Å as the average sign error of the aliased CTF increases (Extended Data Figure 3b). Pre-multiplying the data and CTF at sufficient size and then cropping them improved the resolution to 3.2Å using the same reconstruction box size. Only pre-multiplying the data but using an aliased CTF2 for the Wiener-like reconstruction filter did not decrease the nominal resolution in this case. However, for algorithms that would use aliased models during refinement and classification, we expect these effects to be noticeable. This approach improved the estimated per-tilt-series weighting factors for high-defocus data to the level of low-defocus data for the entire data set (Extended Data Figure 3c).
Optimization procedure
M optimizes all hyperparameters describing geometric deformation, electron-optical aberrations, and particle pose trajectories, simultaneously by applying a gradient-descent optimization using the Limited-memory Broyden–Fletcher–Goldfarb–Shanno (L-BFGS) algorithm28. The target function is the sum of normalized cross-correlations between all extracted particle images contained in a field of view, and reference projections at angles and shifts defined by the particles’ poses and deformation hyperparameters (Fig. 2a).
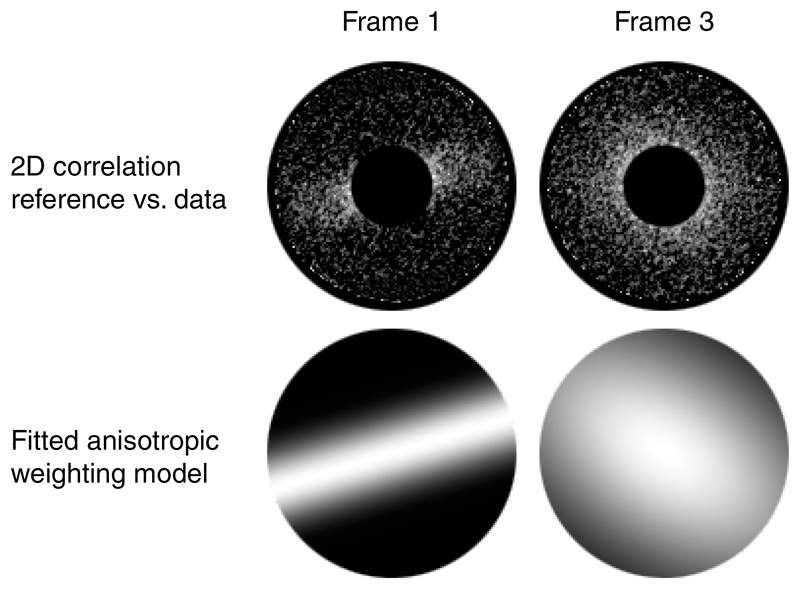
At the end of an optimization iteration, similar to the Fourier Ring Correlation (FRC) approach29, 30, M calculates the per-Fourier component normalized cross-correlation (NCC) between reference projections and image data. This is used to optimize exposure- and tilt-dependent data weighting, and reconstruct half-maps using the updated model, correcting for Ewald sphere curvature31. Because the NCC is resolved in 2D, anisotropic weights can be fitted to make better use of the first frames, which are often affected by strong, unidirectional motion (Extended Data Figure 4).
Map denoising and local resolution
Instead of using a traditional Fourier Shell Correlation (FSC) approach for local resolution estimation32, M trains a CNN-based denoiser using a species’ half-maps to filter them to local resolution for the next refinement iteration (Online Methods). The denoiser applies the noise2noise33 training regime to independently refined34 half-maps obtained at the end of each iteration in M by back-projecting extracted images from the original frames or tilts. Because each half-map is denoised independently, no common artifacts are introduced and amplified over subsequent refinement iterations.
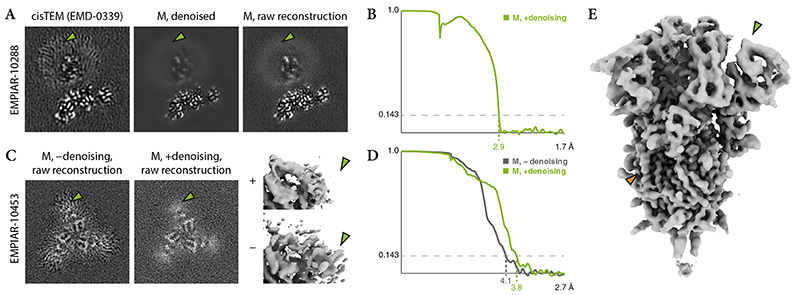
M’s denoising was assessed on the cannabinoid receptor 1-G35 dataset (EMPIAR-10288, Table 1). The original 3.0Å map (EMD-0339) showed overfitting artifacts in the lipid bilayer (Fig. 3a). Processing with Warp, RELION and M led to only slightly improved resolution of 2.9Å (Fig. 3b), while removing the overfitting artifacts in M’s final reconstruction (Fig. 3a).
Figure 3. Effects of deep learning-based denoising of reconstructions during refinement.
(a) 2D XY slices through 3D reconstructions of the cannabinoid receptor 1-G membrane protein35. The original refinement in cisTEM (left) introduced artifacts in the highly disordered lipid region (green arrow). The denoised map (middle) and the raw reconstruction before denoising (right) used in the last refinement iteration in M using 149,308 particles (ca. 15% fewer than in the original study) are devoid of the artifacts because the denoising filtered and downweighed the low-resolution region.
(b) FSC between the half-maps independently refined in M, showing a global resolution of 2.9Å. A value of 3.0Å was reported in the original study, with no FSC curve included with the deposited map.
(c) 2D XY slices and isosurface renderings of the S1 domain in SARS-CoV-2 spike protein36 reconstructions. Refinement in M without denoising introduced visible artifacts (left, bottom–right) in the region (green arrows), which had significantly lower resolution than the rest of the protein. Using denoising, the artifacts were avoided (center, top–right).
(d) FSC between the half-maps refined in M with and without denoising, showing an improvement in global resolution from 4.1Å to 3.8Å when using denoising.
(e) Isosurface rendering of the entire denoised SARS-CoV-2 reconstruction with a global resolution of 3.8Å. Through the denoising process, the more disordered S1 domain (green arrow) was filtered to lower resolution compared to other parts where side chains are visible (orange arrow).
Denoising was also tested on tilt-series of SARS-CoV-2 virions (EMPIAR-10453, Table 1). The S1 domain of the spike protein is conformationally heterogeneous and has significantly lower resolution than the stable parts. Processing with Warp, RELION and M led to a 3.8Å map (Fig. 3c-e), improving over the originally obtained 4.9Å36. Repeating the refinement in M without denoising decreased the global resolution to 4.1Å and generated visible overfitting artifacts in the S1 domain (Fig. 3c,d). This is in line with improvements recently demonstrated using different approaches to local filtering37, 38.
Contribution of different model parameters to map resolution
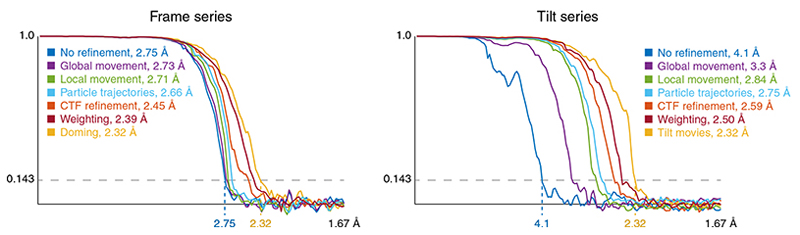
Apoferritin frame and tilt-series data collected from the same grid square under identical conditions (datasets AF-f and AF-t, Online Methods), were used to estimate the contribution of different groups of hyperparameters to map resolution (Fig. 4 and Table 1). For frame-series, particles extracted following reference-free alignment in Warp and refined in RELION (without polishing and CTF refinement) provided a baseline resolution of 2.75Å, which was improved by accumulating the following sets of optimizable parameters in M: Reference-based global motion alignment – 2.73Å; relaxing this constraint to allow local motion alignment – 2.71Å; resolving individual particle pose trajectories – 2.66Å; fitting per-particle defocus and per-frame-series astigmatism and beam tilt – 2.45Å; data-driven anisotropic weight estimation – 2.39Å; resolving doming-like motion – 2.32Å.
Figure 4. Contributions of individual multi-particle system model components to map resolution.
Fourier shell correlation between half-maps for frame-series and tilt-series apoferritin data obtained through extending the set of optimizable parameter groups. Starting with the ‘No refinement’ baseline, in top-down order in the legend, a new group of parameters was added, while keeping the previously added groups, and refinement was performed from scratch. The resolution for each step is given in the legend.
For tilt-series, reference-free tilt movie alignment in Warp, patch tracking-based tilt-series alignment in IMOD, and refinement in RELION provided a baseline resolution of 4.1Å, which was then improved by accumulating the following optimizations in M: Reference-based global tilt image alignment – 3.3Å; relaxing this constraint to allow local image-space warping – 2.84Å; resolving individual particle poses – 2.75Å; fitting per-tilt defocus and astigmatism, and per-tilt-series beam tilt – 2.59Å; data-driven anisotropic weight estimation – 2.50Å; reference-based tilt-movie alignment – 2.32Å. Volume-space warping was not tested because the particles were arranged in a single 2D layer.
We conclude that accurately registering image-space deformation is essential for obtaining high-resolution maps from frame and tilt-series data, whereas modeling other effects leads to smaller improvement that may only become significant in the sub-5Å resolution range. Initial reference-free alignment is less accurate for tilt-series than for frame-series. However, it allows obtaining initial reference maps and particle poses that can be further refined in M. Given similar amounts of particles, M achieved the same resolution with very similar map features (Fig. 5) from either frame or tilt-series data. Thus, collecting data as tilt-series does not incur a resolution penalty. However, because tilt-series are slower to acquire39 and commonly used for crowded, thick samples, we expect maps derived from tilt-series to remain at lower resolution on average.
Figure 5. M achieves similar resolution for frame-series and tilt-series data of an apoferritin sample.
(a) Representative side chain densities observed in the frame-series and tilt-series maps.
(b) Comparison between the global FSC curves for each map.
Comparison with RELION on atomic-resolution frame-series data
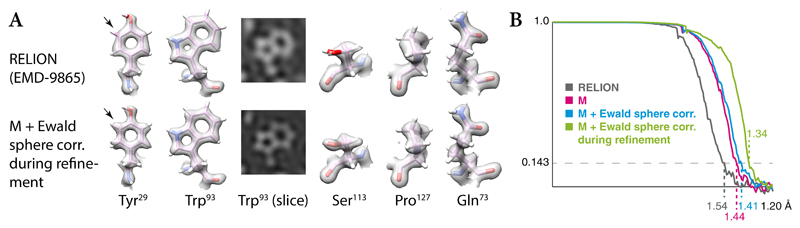
M’s frame-series performance was assessed on apoferritin data previously processed with RELION 3.140 (EMPIAR-10248, Table 1). The data acquired on a JEOL microscope with a cold-field emission gun achieved an atomic resolution of 1.54Å. At this resolution, we were able to assess the effect of Ewald sphere correction with the single side-band algorithm31 (Extended Data Figure 5). Applying it to the reconstruction alone, as done in RELION 3.0, improved the resolution from 1.44 to 1.41Å. Considering the sphere curvature during refinement, improved the resolution to 1.34Å. Coupled with the demonstrated benefits of multi-species refinement and map denoising, this makes M a useful addition to the frame-series SPA pipeline.
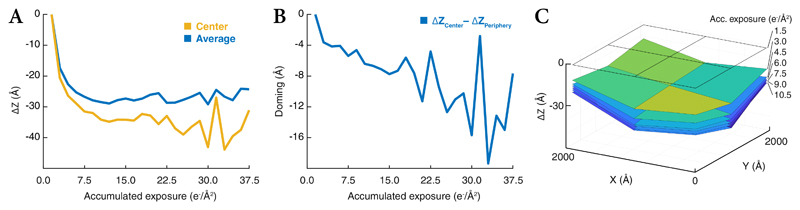
The data’s high resolution also enabled analysis of the sample’s doming behavior, showing that the defocus of the entire field of view changed by over -25Å during the first 7.5e-/Å2 of exposure (Extended Data Figure 6a), corresponding to the sample moving away from the electron source. A more localized, steadily increasing bending of the center relatively to the periphery followed, reaching a difference of -16Å after 37.5e-/Å2 (Extended Data Figure 6b,c).
Comparison with other tools for tilt-series data refinement
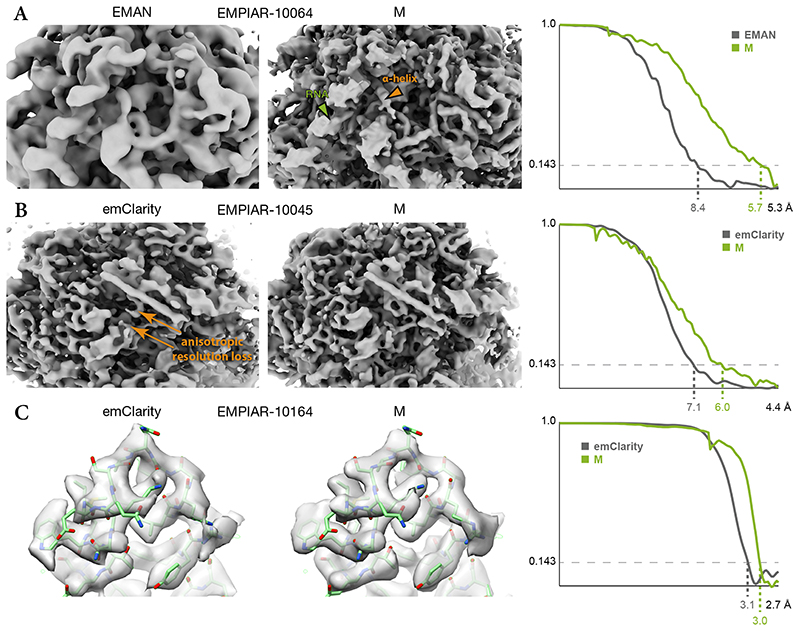
M’s performance on tilt-series was compared with the EMAN221 and emClarity6 packages on data used in the respective publications (Fig. 6 and Table 1). EMAN2 reached a resolution of 8.4Å on an in vitro 80S ribosome sample (EMPIAR-10064), while emClarity reached 8.6Å for the same data6, improving upon a previous 13Å result41. M improved the resolution to 5.7Å and produced a map with secondary structure elements and helical RNA groves (Fig. 6a). We attribute some of this improvement to M’s application of constraints between individual particle tilt images, which is absent in EMAN2.
Figure 6. Comparison of maps obtained from published tilt-series using M or other software.
(a) 80S ribosome data from EMPIAR-10064 were used to benchmark tilt-series processing in EMAN (EMD-0529). M achieved higher resolution, accompanied by visibly better resolved features such as RNA (green arrow) and α-helices (orange arrow).
(b) 80S ribosome data from EMPIAR-10045 were used to benchmark emClarity. The originally published map (EMD-8799, not shown) exhibited strong resolution anisotropy. A recently updated map42 still suffered from resolution anisotropy (“smearing” direction indicated by orange arrows). M achieved higher and more isotropic resolution, aiding the map’s interpretability.
(c) HIV-1 capsid-SP1 data from EMPIAR-10164 were used to benchmark emClarity (EMD-8986). M achieved slightly higher resolution using ca. 30% of the particle number used by emClarity. Doubling the number of particles did not increase the resolution. PDB-5L93 was rigid-body fitted into the maps for visualization.
emClarity reached a resolution of 7.8Å on purified 80S ribosomes6 (EMPIAR-10045), and was later improved to 7.1Å42, surpassing the original 12.9Å result43. M reached 6.0Å, accompanied by improved resolution isotropy and map features (Fig. 6b). We attribute the improved isotropy to M’s denoising-based filtering, whereas emClarity employs an FSC-based approach that may have to be tuned more conservatively to achieve the desired robustness.
emClarity also reached 3.1Å on isolated HIV-1 capsid-SP1 assemblies (EMPIAR-10164), improving upon previous 3.9Å44 and 3.4Å45 results. M reached 3.0Å, accompanied by local improvements in map quality (Fig. 6c). We attribute the slight improvement to M’s more accurate deformation model and simultaneous optimization of all parameters, in contrast to emClarity’s separate steps for full image alignment and particle alignment.
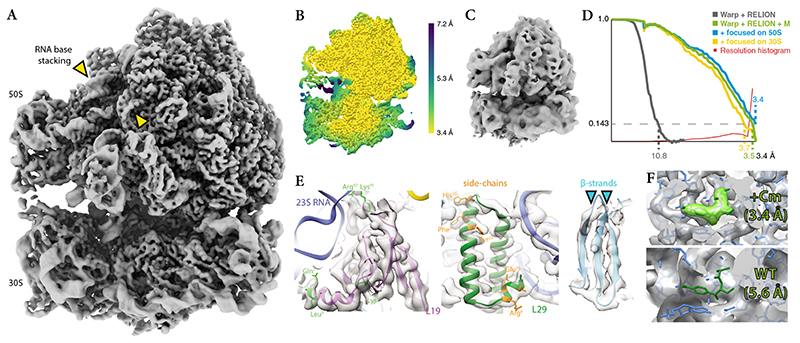
M enables the visualization of an antibiotic bound to 70S ribosomes at 3.5Å in cells
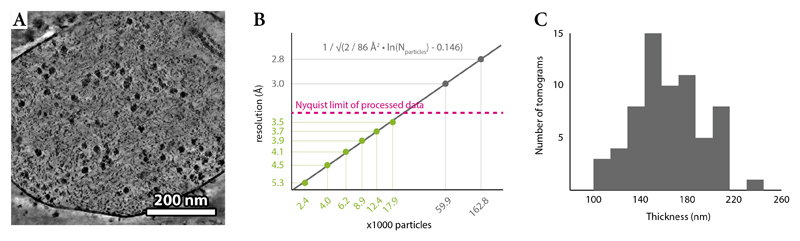
M’s performance on tilt-series data of intact cells was assessed using data of chloramphenicol (Cm)-treated Mycoplasma pneumoniae 46 (Extended Data Figure 7a). M refined the 70S ribosome to 3.5Å (Fig. 7a,d) based on 17,890 particles from 65 tomograms, and a B-factor47 of 86Å2 (Extended Data Figure 7b, Table 1). The large 50S ribosomal subunit dominated the alignment and had a higher average resolution (Fig. 7b,d), with much of its core reaching the 3.4Å Nyquist limit. Independent refinement of the 30S and 50S subunits improved the resolution to 3.7Å and 3.4Å, respectively (Fig. 7d). In contrast, processing these data with Warp and RELION alone led to a 10Å map (Fig. 7c). M’s result constitutes a dramatic increase in structural detail (Fig. 7e); features typical for this resolution range, such as amino-acid side chain stubs and individually resolved β-strands (Fig. 7e), are observed in the map. A rigid body fit of an E. coli 70S ribosome–Cm structure (PDB-4v7t) confirmed the presence of the Cm molecule at its expected binding site (Fig. 7f), marking the first direct visualization of a drug bound to its target inside a cell. The density was absent in a reconstruction from untreated M. pneumoniae 46 (Fig. 7f). Therefore, tilt-series data of an intact, ca. 160nm thick (Extended Data Figure 7c), cellular specimen can lead to residue-level resolution structures of macromolecules in their native biological context.
Figure 7. M. pneumoniae 70S ribosome-antibiotic map at 3.5Å refined with the new Warp–RELION–M pipeline from tilt-series data set of intact cells.
(a) Isosurface representation of the 3.5Å resolution map.
(b) Isosurface of the map colored by local resolution. Despite stalling of the ribosome that is induced by antibiotic binding, residual ratcheting leads to higher resolution in the large 50S subunit, which dominates the alignment, and lower resolution in the small 30S subunit.
(c) Isosurface of a 10.8Å map derived from the same data set using only Warp and RELION.
(d) FSC curves showing the resolution improvement achieved through global and focused refinement in M. The overlaid local resolution histogram shows that a significant portion of the map is resolved close to the data’s Nyquist limit of 3.4Å.
(e) High-resolution features, such as large amino acid side chains (in green and orange) and well-separated β-strands (cyan arrows), are resolved at a level expected for this resolution range.
(f) Atomic model of a Cm-bound 70S ribosome (PDB-4v7t) fitted into the 3.4Å 50S map (top) shows correspondence of map density (light green) to the Cm molecule (dark green). Fitting the same model into a 5.6Å 70S ribosome map of untreated M. pneumoniae cells (EMD-10683, bottom) does not show any density for Cm, providing a negative control.
Discussion
Our results demonstrate that treating cryo-EM frame and tilt-series as multi-particle systems rather than sets of isolated particles, and integrating their reference-based refinement with particle alignment and CTF refinement improves map resolution. The new framework removes previous technical limitations of tilt-series data processing, allowing to achieve resolution at par with state-of-the-art frame-series results, provided similar amounts and quality of data. Because correlation between a 3D reference and a sub-tomogram was shown to be equivalent to the weighted sum of correlations between reference projections and the image series from which the sub-tomogram was reconstructed48, we were able to formulate the refinement process in M identically for frame and tilt-series. Processing of image series rather than pre-reconstructed sub-tomograms can lower the computational complexity48 and enable flexible refinement of tilt angles and offsets.
Although M’s refinement is constrained based on multi-particle assumptions, its forward model and reconstruction algorithms, as well as RELION’s 3D classification, assume isolated particles. While this is rarely an issue for in vitro data, refinement of crowded cellular data stands to benefit from extending these algorithms as well. Future work on reconstruction and classification algorithms within M’s flexible optimization framework may address this shortcoming by modeling the multi-particle system explicitly to achieve higher resolution.
M’s ability to resolve ribosomes in cells at a resolution previously considered exclusive to samples of isolated particles demonstrates that structures can be, in an ideal case, visualized directly inside cells at a resolution suitable to arrive to atomic models. The ribosome is an outlier in terms of size and abundance in cells. Whereas smaller complexes may be refined to similar resolutions in principle (Fig. 6c), the number of such instances may be limited by the scarcity and heterogeneity of many complexes, and the difficulty of localizing them in crowded cellular environments. Concentrating proteins significantly in cells is likely to perturb the organism. The only way to overcome this is to collect more data; although sample preparation and data acquisition are becoming more streamlined, collecting enough particles of a rare protein complex to reach high resolution may prove impractical for an individual research team. To help overcome this limitation, M offers data pooling and distributed processing mechanisms to allow the community to share data and explore their potential. We show that including more particle species in a multi-particle refinement improves the resolution of all species involved. Thus, everyone stands to benefit from having more proteins identified and refined in shared data.
In conclusion, M can be combined with the established programs Warp and RELION into a powerful pipeline for cryo-EM data processing, that includes a comprehensive and transferable tilt-series workflow. This workflow avoids conversion of file formats and conventions between different software packages, enabling non-expert users to achieve state-of-the-art results. It has the potential to achieve residue-level resolution maps of particles inside cells and to capture macromolecular machines in action within their native environment. Together with complementary approaches, it further establishes the foundation for the emerging field of high-resolution structural biology in cells.
Online Methods
Data management
M requires data sources initialized based on a Warp project folder. Beside a list of frame/tilt-series items, it stores the deformation model to be refined. M saves the refined deformation model for each item in the same XML metadata files previously created by Warp. Due to a shared code base, Warp can use the updated model when calculating new frame-series averages or tomographic reconstructions. Multiple data sources of either type can be combined in a single population to facilitate the sharing and pooling of valuable data sets capturing complex cellular environments that can contribute to far more than one project, but do not contain enough data for any single project on their own. To account for minor pixel size miscalibrations between different microscopes, the pixel size can be refined alongside other parameters in M.
A species is initialized from the refinement results of RELION or other compatible software, taking the unfiltered half-maps, a mask, and the particle coordinates and poses (i.e. translations and rotations) as a starting point. The state of a species after each refinement iteration comprises the reconstructed half-maps, the weights of the trained denoising model, various filtered and sharpened maps, a denoised map, and a list of particle coordinates and poses with multiple temporal sampling points if desired. Various map metrics, including global, local, and anisotropic resolution, are calculated. The particles reference their data source items by their data hash to avoid naming conflicts between different data sources.
To enable multiple users to collaborate and pool their results, M tracks precisely the chain of refinements and other operations on data. After each refinement iteration, a “commit” is generated to save the new state. Similar to version-control systems like Git49, the commit’s hash is based on the exact state of the system committed. The hash of each data source item is calculated from the raw data, the refined deformation and imaging models, and the hashes of all species used for their refinement. The hash of each species is calculated based on the half-maps, the weights of the denoising model, the particle coordinates and poses, and the hashes of all data source items contributing information. The hashes can be used to verify a graph representing all steps that led to a particular state of a data source or species. Similar to the “pull request” mechanism in Git, species can be added to a population taking into account potential physical collisions with existing particles. This enables the maintenance of a centralized population repository from which multiple users can obtain pre-aligned data sources, identify new particle species or reclassify existing particles into more states, and contribute the results back to the repository.
Deformation model
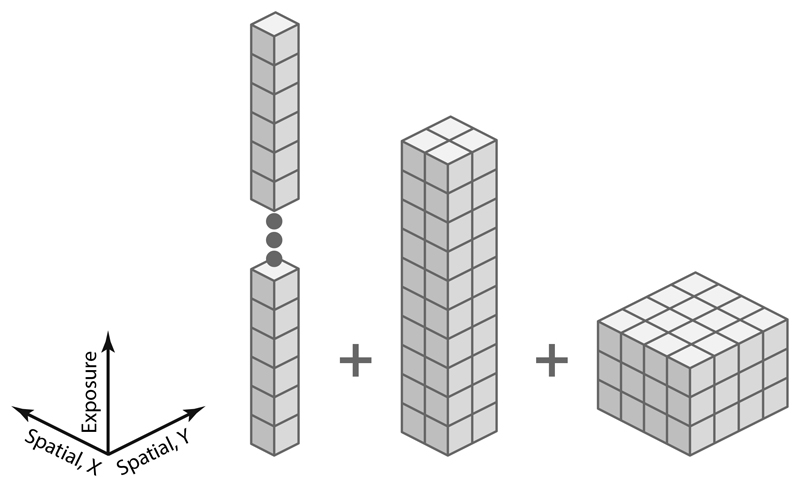
For frame-series data, deformation of the multi-particle system is modeled in the XY plane only, with a pyramid (Extended Data Figure 1) of cubic spline grids19 GF,j(δ,i) (where j is the index within the pyramid, δ is the spatial interpolation coordinate, and i is the temporal interpolation coordinate) going from high temporal/low spatial to low temporal/high spatial resolution. This accounts for the fast-changing, global stage movement, and the slowly-developing, local BIM. Furthermore, translation and rotation of individual particles as a function of exposure can be modeled with 2–3 control points depending on the particle size and overall exposure.
The model for tilt-series data is more complex, owing to the higher potential for perturbations in the system between individual tilt exposures. As the mechanical rotation of the microscope stage and the estimated orientation of the tilt axis are imperfect, the assumed stage orientation can be randomly off in every tilt. M thus refines an independent set of stage rotation angle corrections ωi for every tilt i. These corrections only affect the particle orientations to avoid redundancy, as the induced changes in the projected particle positions can be fully modeled by a deformation grid that must already be employed for other purposes.
Similarly, stage translation varies randomly between individual tilts. BIM patterns can be very different across adjacent tilt images as additional exposures are taken for focusing and tracking in-between. Particle positions can further deviate due to other imaging artifacts, such as wrongly calibrated magnification anisotropy50. M employs an “image warp” grid of cubic splines GTI with a spatial resolution of 3–5 control points in X and Y and per-tilt temporal resolution to model these geometric displacements in image space collectively. Furthermore, in vitro and in situ sample types for which tilt-series are commonly used contain multiple overlapping layers of particles. Some deformations of densely filled volumes, such as shearing, or bending in the Z dimension when viewed at a high tilt angle, cannot be modeled accurately by XY translations in image space. M employs an additional “volume warp” grid GTV, implemented as a 4D grid of control points with quadrilinear interpolation between them that is anchored in volume space rather than image space. Hence it rotates with the sample and can model slow, continuous deformation that affects the particles’ projected positions in image space. As with frame-series data, per-particle translation and rotation as a function of exposure is also modeled for tilt-series.
Finally, a single tilt image exposure is usually fractionated in multiple frames, making it a tilt movie. At 1–3 e-/Å2, the exposure in a single tilt movie is usually short, but still requires additional modeling to compensate motion. M parametrizes the XY translation as a combination of a grid with no spatial and per-frame temporal resolution, and a grid with a spatial resolution of 3x3 and a temporal resolution of 3. Stage and particle orientations are assumed to remain constant throughout a tilt movie, as the biggest beam-induced changes have been shown to occur in the very beginning of each of the short exposures20. Overall, the number of parameters for tilt-series is larger than for frame-series, requiring a higher particle density to achieve equivalent accuracy.
Imaging model
The ability to model imaging conditions such as defocus, astigmatism, magnification or higher-order aberrations is equally important for obtaining high-resolution reconstructions. Frame- and tilt-series offer different advantages for refining some of these parameters.
For particles in frame-series data, the Z coordinate and thus the relative offset from the global defocus of the micrograph is unknown. Although local defocus estimation based on amplitude spectrum fitting has been shown to increase resolution19, reference-based refinement of per-particle defocus can lead to a further increase in resolution24. M refines per-particle defocus and a per-series astigmatism for frame-series, assuming constant values throughout the series.
Tilt-series provide accurate Z coordinates for all particles. However, the initial amplitude spectra-based global defocus estimates for each tilt have lower accuracy due to very short exposures, and cannot be assumed to remain constant throughout the series due to stage movement and refocusing. Furthermore, these estimates can be biased by contrast-rich objects that are not the particles of interest, such as a carbon film below or above the particles, or the platinum coating layer for FIB-thinned samples51. The astigmatism can also change between tilts due to fluctuating electron optics. M refines per-tilt defocus and astigmatism for tilt-series, and calculates per-particle tilt CTFs based on these values and the Z coordinate of a particle’s position transformed according to the fitted stage orientation. Particles in tilt-series can potentially have more accurate defocus values because the number of parameters that can be fitted scales with the number of tilts or particles for tilt- or frame-series, respectively. In many cases the number of tilts will be significantly lower than the number of particles.
In both frame- and tilt-series, M also models per-series anisotropic magnification and higher-order optical aberrations. Refinement of a global set of Zernike polynomials representing the aberrations based on a 2D phase residual image calculated from all particles in a data set has been shown to improve the resolution significantly for slightly misaligned microscopes52. Within individual tilt-series, beam tilt can vary as it is applied to compensate stage misalignments during tracking. Unfortunately, the signal in individual tilts is insufficient for accurate beam tilt estimation, and such an option is not implemented in M.
Optimization procedure
M seeks to maximize the following target function M, which is essentially a weighted, normalized cross-correlation between all particle images and the corresponding reference projections:
where s is a particle species, p is a particle of that species, and i is the index of a frame or tilt in a series; ⋆ denotes the dot product between two complex vectors, where the complex numbers are treated as pairs of scalars; |…| denotes the L2 norm; W is the anisotropic exposure- and tilt angle-dependent amplitude weighting of frame or tilt i; P is a projection operator in Fourier space sampling a central slice of the volume of species s at orientation Θ, taking into account the anisotropic scaling τ, bent to account for the Ewald sphere curvature determined by the species’ diameter; ▪ denotes scalar multiplication; T is the complex-valued beam tilt compensation; FT denotes the discrete Fourier transform; CTF is the real-valued CTF taking into account the defocus at position Λ and the astigmatism in frame or tilt i; AS is the real-valued, rotational average over the amplitude spectra of all particle images of all species extracted from tilt i or the average of all aligned frames, used for spectrum whitening, scaled and cropped to the respective species size and resolution; I is the FT of a particle image extracted from frame or tilt i at position δ, cropped to the respective species resolution; D is a soft circular mask with particle diameter d.
Similar target functions in previous literature used P · CTF to model the contents of I 23, 25. However, in M’s implementation I is pre-multiplied by CTF to avoid CTF aliasing despite using small particle windows. This change does not affect the numerator part of M due to the associativity of complex number multiplication; its impact on the denominator part of M does not affect the achieved resolution in any way. It also avoids the additional memory footprint of storing pre-calculated CTFs, or the computational overhead of calculating them on-the-fly.
M can consider the Ewald sphere curvature during refinement if this is made necessary by a large species and/or high resolution53. In this case 2 copies of CTF · I are prepared using the single side-band algorithm31: CTFP · I and CTFQ · I. To calculate the cost function, one is correlated with a bent central slice P, and the other with a central slice bent in the opposite direction. The resulting cost functions MP and MQ are then added. As with previous implementations24, the absolute handedness for the correction must be provided by the user.
For frame-series, the position and orientation of particle p in frame i are calculated as:
where λ is the value of the refined particle position trajectory interpolated at the accumulated exposure of frame i; GOF is a deformation grid pyramid produced by Warp’s original reference-free alignment that is not altered in M refinement; GF is a deformation grid pyramid that is refined in M; Z is the refined defocus value of particle p that is added as the Z coordinate to its position; θ is the value of the refined particle orientation trajectory interpolated at the accumulated exposure of frame i.
For tilt-series, the position and orientation of particle p in tilt i are calculated as:
where R and RXYZ construct a rotation matrix based on a set of Euler and XYZ angles, respectively, and R −1 calculates a set of Euler angles based on a rotation matrix; CV is the center of the volume in which the multi-particle system is anchored, and Ci is the center of the full tilt image; Zi is the refined defocus value of tilt i that is added to the Z coordinate of the transformed particle position; Ω is the stage orientation determined in the initial, reference-free tilt-series alignment that is not altered in M refinement; · denotes matrix multiplication here.
For frames in tilt movie i, the position of particle p in frame k is calculated as:
where GOF is the deformation grid pyramid produced by Warp’s original reference-free alignment of the tilt movie that is not altered in M refinement; GTF is a deformation grid pyramid for the tilt movie that is refined in M.
Due to the very large number of parameters, M employs L-BFGS28 to perform almost all of the optimization. Only the initial defocus search is done exhaustively over a limited range to avoid getting trapped in a local optimum because of the quickly oscillating nature of the CTF. Every L-BFGS search iteration requires the calculation of a partial derivative of the target function with respect to each optimizable parameter. Reevaluating M twice per parameter to compute the gradient with the central differences numerical scheme would be very computationally expensive. Like Warp, M takes a computational shortcut for most of the parameters.
Before optimization starts, M calculates the partial derivatives of the X and Y components of all Λp,i with respect to all warping grid parameters and all control points of a particle’s position trajectory that affect them. Similarly, the partial derivatives of the individual Euler angle components of all Θp,i with respect to all stage angle correction parameters and all control points of a particle’s orientation trajectory are calculated. As each parameter influences only a small fraction of particle frames or tilts, most of the derivatives are 0. They are excluded from the precalculated lists to avoid unnecessary computation. Then, during optimization, once per search iteration, the partial derivative of for each particle frame or tilt is calculated with respect to X, Y and the Euler angles. This amounts to evaluating M 10 times. A useful approximation for the derivative for each parameter n can then be calculated as follows:
where α∈{x,y,ϕ,ϑ,ψ}, i. e. one of the translation axes or Euler angles; ‖ denotes the concatenation of two tuples; (…)α denotes the selection of component α from a tuple.
The deformation parameters make up the bulk of all parameters. Parameters such as absolute magnification and beam tilt do not benefit from the same shortcut and their derivatives must be calculated independently with the central differences scheme. The CTF-related parameters are few, but the calculation of their derivatives is especially expensive because it requires the particles to be reextracted at an aliasing-free size, pre-multiplied by the altered CTF, and cropped to refinement size – all involving expensive FT steps. M calculates the values of M by adding up the results from small batches of particles. This allows the cost of the first FT at aliasing-free size to be amortized over all optimizable CTF parameters, as its result is reused for all subsequent calculations. The gradients for all per-particle or per-tilt defocus and astigmatism parameters can all be calculated in the same pass as each of them affects only one particle or tilt.
If defocus is to be optimized, an iterative grid search can be executed before the L-BFGS optimization starts. The search runs for 5 iterations. For the first iteration, a range of ±300 nm around the current values is sampled in 10 nm steps. For each subsequent iteration, the search step is halved, and a range of ± the new search step around the 2 best values for each particle or tilt from the previous iteration is sampled.
Memory footprint considerations
Traditional SPA refinement treats every particle as an isolated entity, thus requiring no more than one particle to be held in memory at any given time if parallelization is not considered. A multi-particle approach, however, needs to rapidly evaluate the state of the entire multi-particle system during refinement. The particle frame-/tilt-series need to be stored in memory because re-extracting and reprocessing them for every evaluation would be too inefficient. While an in vitro sample usually contains a single layer of proteins with up to 1000–2000 particles in a field of view, a densely packed in situ volume has the potential to contribute tens of thousands of particles to refinement if enough species can be identified. The image size is selected to be twice the particle diameter to account for signal delocalization and interpolation artifacts, leading to significant overlap even in the single-layer case. At high refinement resolution, the memory requirements of all extracted particle frame-/tilt-series in a system can vastly exceed those of the original data, rising to tens or even hundreds of gigabytes.
Although M uses GPUs for acceleration wherever possible, currently available consumer-level cards offer up to 12 GB, which would be insufficient in many cases. Therefore, the extracted particle frame-/tilt-series are held in “pinned” (i.e. page-locked) CPU memory where they can be transparently accessed by the GPU. Despite the low bandwidth of CPU–GPU memory transfers, the GPU does not experience a significant performance penalty when correlating them to reference projections. This is because the particle data accesses are sequential and highly coalesced, whereas the creation of reference projections on-the-fly accesses the GPU memory randomly, creating significant overhead. As faster CPU–GPU interfaces are being developed, the penalty should become more negligible in the future.
Still, memory requirements can become too high even for CPU memory. To reduce the footprint, M exploits the varying information content of frames/tilts over the course of a series. As sample damage from radiation is accounted for by applying a Gaussian (“B-factor”) weighting function in Fourier space14, 24, the contribution of higher-frequency components becomes negligible at high exposure. M crops extracted particle images in Fourier space to a resolution that corresponds to the weighting function value falling below 0.25, resulting in considerable space savings once high resolution is reached. Assuming an increase in the weighting B-factor of 4 Å 2 per 1 e-/Å2 of accumulated exposure, the maximum useful frequency at exposure d is , and the image size m scales with a factor of min(1,fmax/frefine). Thus, the upper bound for memory consumption in case of low refinement resolution and/or low overall exposure is O(m 2 d), while the lower bound is Ω(m 2ln(d)) in case of high refinement resolution and/or high overall exposure.
Avoiding CTF aliasing
Cryo-EM data of thin biological specimens are usually acquired at defocus to achieve phase contrast. In the absence of a phase plate device, and often in the case of in situ tomography, defocus values can exceed 4 μm to enable better visual interpretation of the raw data. Higher defocus results in stronger delocalization of the signal in real space, as reflected by faster oscillations of the CTF in Fourier space. As the CTF oscillates between -1 and 1, combining signals with different defoci would result in an average value of 0 at higher spatial frequencies. Thus, a phase shift of π must be applied to frequency components modulated by negative CTF values prior to averaging. Furthermore, it is desirable to compute the reconstruction as a weighted average, using the CTF for the weighting. Multiplying the FT of a particle image by the corresponding real-valued CTF achieves both goals.
Current SPA packages advise the user to select the particle box size as 1.5–2 the particle diameter to account for Fourier-space interpolation artifacts, not considering the image defocus. When an image is cropped around a particle, the Fourier-space modulation pattern becomes band-limited to the new window size. If CTF oscillations are too fast to be resolved, the band-limited values for the amplitudes of the corresponding frequency components will converge to 0. Even worse, the analytical 2D CTF model used in refinement and reconstruction is not band-limited, and contains solely aliasing artifacts past the Fourier-space Nyquist frequency instead of converging to 0. This can put a hard limit on the achievable resolution for small particles and those acquired at high defocus that is independent of the actual data quality.
This problem can be mitigated by selecting a box size large enough to avoid CTF aliasing54 at the highest defocus value in a data set. However, the required size m can exceed 1000 px at high resolution or defocus, significantly slowing down refinement algorithms whose complexity and memory footprint are O(m 2) and O(m 3), respectively. This increase can be entirely avoided by pre-multiplying particle images by the CTF at an aliasing-free size, and cropping them to a smaller size for refinement or reconstruction. As the modulation pattern is CTF2 after pre-multiplication, the band-limited oscillations will converge to 0.5 instead of 0. The 2D CTF model used in refinement and reconstruction must be similarly band-limited to match the data. As M operates on all particles of an entire frame-/tilt-series at a time and extracts the particle images on-the-fly, such considerations are made automatically for the currently needed resolution.
The minimum box size needed for CTF correction at a given resolution is dictated by the maximum oscillation rate of the CTF within the available spatial frequency range. This is not necessarily the oscillation rate at the highest spatial frequency as φ is not a monotonic function: A combination of low underfocus and high Cs will cause the oscillations to slow down significantly and accelerate again at higher spatial frequencies. The oscillation rate can be calculated as the first derivative of φ. In practice, it is easier to evaluate dφ/dk numerically within the relevant range of spatial frequencies to find its maximum absolute value. To fully resolve the oscillation, one period must be rasterized onto at least 2 pixels, i.e. the window size must be chosen such that max(dφ/dk) = 2π/2px. While this guarantees a fully resolved CTF in 1D, a CTF rasterized on a Cartesian 2D grid has an anisotropic sampling rate. At its lowest, i.e. along the diagonals, it requires the sampling rate of the 1D case.
Before particle extraction, the size padding factor at which the images will be pre-multiplied by the CTF has to be determined, taking into consideration the maximum defocus value expected in a frame-/tilt-series, and the expected maximum resolution. During refinement, the latter is set to the refinement resolution. For the final reconstruction, it is set to 1.25x the current global resolution. Particles are extracted using the calculated minimum box size (or twice the particle diameter in case that value is larger), and pre-multiplied by the CTF in Fourier space. Then the inverse FT (IFT) is applied, the particles are cropped to the refinement or reconstruction size in real space, and transformed back to Fourier space for refinement. The band-limited CTF2 model is prepared by simulating the function at the same aliasing-free size in Fourier space, cropping its IFT in real space, and taking the real components of the result’s FT.
Data-driven weighting
To account for radiation damage as a function of accumulated exposure, or increasing sample thickness as a function of the stage orientation, several heuristics and empirical approaches have been proposed14, 24, 43. By default, M adopts the heuristic introduced in RELION 1.443. The B-factor is increased by 4 Å2 per 1 e-/Å2 of exposure, and each tilt is weighted as cosϑ. Once high resolution is reached, the weights can be estimated empirically using a reference correlation-based approach similar to the one introduced in RELION 3.024.
In a departure from RELION’s scheme, the normalized correlation (NC) is calculated between particle images and reference projections at the end of a refinement iteration are not combined across the entire data set. It is kept as a 2D image to enable the fitting of anisotropic weights rather than averaging rotationally. The correlation data can then be recombined in different ways to calculate different kinds of weights. Furthermore, because M supports the refinement of multiple species with different resolution, the per-species correlation vectors for each frame or tilt need to be combined. This is done by weighting each one by the FSC calculated between the half-maps of the respective species. This produces a set of vectors NCd,i,k, where d is the series, i is the frame or tilt, and, optionally, k is the tilt movie frame.
The procedure then iteratively calculates () as:
and optimizes the weighting parameters to minimize the following cost function:
where · denotes scalar multiplication; G is an anisotropic 2D Gaussian B-factor weighting function; B is a vector describing the B-factor along the X and Y axes, and their rotation; W is a scalar weight; is the weighted average of all particle CTFs in one frame or tilt. The B-factors in each group are constrained such that the highest value in a group is set to 0.
In this default formulation, the weighting scheme allows to assign separate weights not only to individual frames/tilts, but also to weight the contribution of an entire series. For data with high particle density this scheme can be extended to assign different weights to frames/tilts of each individual series. Anisotropic B-factors improve the weighting of frames with significant intra-frame motion (Extended Data Figure 4). Combined with per-series, per-frame weighting, such granularity allows to rescue more information from the first few frames of an exposure if parts of them are less affected by BIM.
Map reconstruction
Previous refinement packages took two different approaches to map reconstruction from frame- and tilt-series data. For frame-series, weighted averages were prepared either directly from the initial, reference-free alignments, or based on a “polishing” procedure24. These 2D averages were then weighted based on a 2D CTF model and a spectral signal-to-noise ratio (SSNR) term25, and back-projected to obtain the reconstruction. For tilt-series, the algorithms operated on intermediate per-particle 3D reconstructions (‘sub-tomograms’) with fixed translational and rotational offsets between individual tilt images. These 3D sub-tomograms were then weighted based on a 3D CTF model43 and a spectral signal-to-noise (SSNR) term, and back-projected to obtain the reconstruction.
M seeks to unify the handling of both types of data and uses the original, non-interpolated 2D data at every step, including reconstruction. For tilt-series, this approach avoids any artifacts from intermediate interpolation and reconstruction steps. For frame-series, the requirement for identical orientation of all particle frames no longer exists as they are not averaged in 2D, enabling the modeling of particle orientation as a function of exposure. Only for individual tilt movie frames a shortcut is taken to save memory and computation, and they are pre-averaged in 2D using the approach described for Warp19 after a separate multi-particle refinement of the respective tilt movie.
Thus, for the reconstruction, individual particle frames or tilts are weighted by an exposure-dependent function to account for radiation damage, and an aliasing-free 2D CTF model (see previous section) that incorporates the exact defocus and astigmatism values for that position and frame/tilt. The weighted data are then back-projected through Fourier space summation, accounting for Ewald sphere curvature. The reconstruction is finalized by dividing the summed data component by the summed weights component25.
Map denoising
Reconstructions of biological specimens derived from cryo-EM data rarely have homogeneous resolution throughout all parts of the macromolecule. Using a map filtered to its global resolution for particle alignment can have detrimental effects. Poorly resolved regions, such as floppy protein domains or the lipid bilayer around transmembrane domains, will make the alignment worse by adding noise to reference projections below the refinement resolution. In the case of fully independent half-maps34, the noise patterns that the particles will be aligned against are independent, and amplifying them over several iterations only has the potential of making the resolution worse. In the case of refinement with merged half-maps23, where overfitting is avoided by limiting the refinement resolution, the poorly resolved regions may be well below that limit, leading to a common, overfitted noise pattern in both half-maps.
Past attempts at filtering maps based on local resolution estimates for refinement55, 56 applied FSC-based approaches32 to estimate the local resolution and performed the filtering in the Fourier domain. As only one set of estimates can be made based on one pair of half-maps, any spurious patterns in the estimated values will be introduced into both half-maps when the filtering is performed. The locality and accuracy of the estimates depends on the window size32. A smaller window increases locality at the expense of accuracy. Once introduced, the noise pattern can become amplified over multiple iterations, leading to overestimated local resolution and phantom features that can be misinterpreted. More advanced regularization schemes have been proposed37, 38 since to deal with this problem.
M implements a new approach to map filtering that uses neural network-based denoising. The recently proposed noise2noise training principle33 allows the training of differentiable denoiser models without a noise-free ground truth, using only two independently noisy observations. It has been successfully applied to micrograph19 and tomogram19, 57 denoising. The implementation in M utilizes gold-standard34 half-map reconstructions, which represent another obvious case of two independently noisy observations of the same signal, and are interchangeably used as input and target in training. The reconstructions are obtained at the end of each refinement iteration in M by back-projecting extracted images from the original frames or tilts, using the particle half-sets carried over from RELION at the beginning of the workflow. We find that a denoiser trained on one pair of half-maps not only matches closely the result of conventional global resolution filtering when applied to maps with homogeneous resolution, but also provides locally smooth, artifact-free local resolution filtering. As such models can train on and denoise sets of micrographs or tomograms with different defocus values and thus different noise models, they can also recognize and adapt to different noise levels within the same reconstruction. In another important departure from FSC-based methods, the denoising step is applied to the half-maps independently and the denoiser sees only one of them at a time. Thus, even if some spurious pattern is introduced as part of the denoising, it is independent between the half-maps.
The neural network architecture, implemented in TensorFlow 1.10, is identical to the one used for tomogram denoising in Warp. A separate denoising model is maintained for every species, and trained only on the respective pair of half-maps. The model is initialized with random values and trained for 800 iterations upon the creation of a new species. It is later retrained for another 800 iterations after every refinement. Spectrum whitening is applied to the maps before training to restore high-frequency amplitudes23, similar to B-factor-based sharpening47. During training, 643 px volumes are extracted from both maps at the same random position and orientation, and presented to the network as input and output in mini-batches of 3. The random orientations make sure the network learns the noise model rather than merely learning the average map. The learning rate for the Adam optimizer is exponentially decreased from 10-3 to 10-5 throughout the training. For the denoising of each half-map, the map is partitioned in 643 px windows overlapping by 24 px, denoised, and the results from each window are inserted into the output volume. Regardless of regions with above-average resolution being potentially present, the refinement resolution is set conservatively to the global map resolution. In addition to the two half-maps for refinement, a denoised average map is also prepared by applying the same denoising model to the average of the spectrum-whitened half-maps.
Assessment of map denoising
Frame-series data were downloaded for the EMPIAR-10288 entry (Fig. 3a,b). Frame alignment and local CTF estimation were performed in Warp with a spatial resolution of 5x5. 1,033,994 particles were picked with a retrained BoxNet model in Warp and exported at 1.5 Å/px. 2D classification, 3D classification and refinement were performed in RELION using EMD-0339 as the initial reference. 149,328 particles corresponding to the best 3D class were imported in M. The particle poses were given a temporal resolution of 2, the deformation grid resolution was set to 2x2, and refinement of all parameters was performed for 5 iterations (Table 1). Data-driven weight estimation was performed to assign unique weights to every frame index.
Pre-aligned tilt movies were downloaded for the EMPIAR-10453 entry (Fig. 3c,d). Gold fiducials were picked with BoxNet in Warp, and fiducial-based tilt-series alignment was performed in IMOD. Tilt-series CTF estimation and reconstruction of full tomograms at 12 Å/px was performed in Warp. A binary classifier based on a 3D CNN (in development, not part of Warp and M) was trained using 5 manually segmented tomograms to segment the SARS-CoV-2 virions. Another 3D CNN-based binary classifier was trained on manually picked spike protein positions in 7 tomograms. Automatically picked spike protein positions were cross-referenced with the segmented virions to remove particles further away than 200 Å, obtaining 38,742 particles. Sub-tomograms were reconstructed at 5 Å/px for refinement in RELION. After ab initio map generation, 3D refinement was performed, reaching the 10 Å Nyquist limit. The results were imported in M, where a 1x1x41 image warping grid and particle poses were optimized for 2 iterations. Sub-tomograms were reconstructed at 5 Å/px using the improved alignments, and subjected to classification into 4 classes in RELION. 22,998 particles from 2 classes showing the spike trimer were imported in M, where a 3x2x41 image warping grid, CTF, and particle poses were optimized for 4 iterations with C3 symmetry (Table 1). For the comparison, the refinement procedure was modified to omit the denoising step. Refinement was then restarted at 10 Å and performed for 5 iterations using the same settings.
Acquisition of apoferritin benchmark data
To compare the resolution achievable with frame and tilt-series data and assess individual algorithms implemented in M, we acquired two data sets of human heavy-chain apoferritin: AF-f (frame-series) and AF-t (tilt-series). To make sure that any observed differences came from data type and processing strategies rather than local variance in sample quality, neighboring holes within the same grid square were used for both data sets.
GST-tagged apoferritin was overexpressed in E. coli, captured on Gluthatione-sepharose beads after cell lysis, cleaved off the resin by TEV protease and purified to homogeneity by size exclusion chromatography in 50 mM Tris-HCl pH 7.5, 100 mM NaCl and 0.5 mM TCEP.
3 μl of apoferritin at 3.8 mg/ml were applied to freshly glow discharged R 1.2/1.3 holey carbon grids (Quantifoil) at 4˚C and 100% relative humidity followed by plunge-freezing in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). The sample concentration resulted in a dense, single-layered hole coverage. Data were collected on a Titan Krios TEM (Thermo Fisher Scientific) operated at 300 kV and a magnification resulting in a calibrated pixel size of 0.834 Å. The energy filter (Gatan) was operated in zero loss mode with a slit width of 20 eV. The K3 direct electron detector (Gatan) was operated in counting mode with a freshly acquired reference for gain correction. The exposure rate was adjusted to 20 e-/px/s. SerialEM58 was used for frame and tilt-series acquisition.
Positions for both data sets were selected to be distributed evenly over the same grid area to maximize the similarity in ice thickness and particle density. For AF-f, 150 frame-series were collected with a total series exposure of 32 e-/Å2, fractionated in 40 frames. For AF-t, 135 tilt-series ranging from -40 to +40 degrees were collected in a grouped dose-symmetric scheme59 with a group size of 2 and in 2 degree steps. Each tilt was exposed to 2.7 e-/Å2, fractionated in 3 frames.
Comparison between frame and tilt-series performance
Using data set AF-f, frame-series alignment and local CTF estimation were performed in Warp with a spatial resolution of 8x5, owing to the rectangular format of the K3 chip. 22,122 particles were picked with a retrained BoxNet model in Warp and exported at full resolution in 512 px boxes. Global 3D refinement with octahedral symmetry was performed in RELION 3.0. The results were imported in M. The particle poses were given a temporal resolution of 3, the deformation grid resolution was set to 6x4, and refinement of all parameters was performed for 5 iterations (Table 1). Data-driven weight estimation was performed to assign unique weights to every series and frame index.
Using data set AF-t, tilt movie frame alignment was performed in Warp using a model without spatial resolution. Initial tilt-series alignment was performed in IMOD using patch tracking on 6x binned images with default settings. Tilt-series CTF estimation was performed in Warp. 18,991 particles were picked using Warp’s 3D template matching in full tomograms reconstructed at 10 Å/px. Sub-tomograms and 3D CTF volumes were exported at 2 Å/px using 140 px boxes. Global 3D refinement with octahedral symmetry was performed in RELION 3.0. The results were imported in M. The particle poses were given a temporal resolution of 3, the image warp grid resolution was set to 6x4x41, and refinement of all parameters was performed for 5 iterations, including tilt movie frame alignment in the last 2 iterations (Table 1). Data-driven weight estimation was performed to assign unique weights to every series and tilt index.
Assessment of multi-species refinement
Particles from each frame-series of the AF-f data set were split in 5% and 95% sub-populations, resulting in species with 3,710 and 70,497 particles, respectively. Frame alignments and particle poses previously obtained from Warp and RELION were reused. In the first scenario, the 5% species was refined alone. In the second scenario, the 5% species was co-refined with the 95% species. Both species were assumed to be structurally independent and did not contribute particles to each other’s reconstructions. For both tested scenarios, a 6x4 starting grid for the deformation was used, the resolution of all species was set to 4.0 Å and only one refinement iteration was performed in M to avoid possible benefits from the higher resolution the 95% species would reach after the first iteration.
Comparison with RELION on atomic-resolution frame-series data
Frame-series data were downloaded for the EMPIAR-10248 entry and pre-processed in Warp. 109,437 particles were exported at 0.6 Å/px using 466 px boxes and refined in RELION. The resulting particle poses and half-maps were imported in M and refined for 5 iterations starting with a resolution of 3.0 Å in the first iteration. A starting grid of 4x4 was used for the deformation model, and the number of frames was truncated to 25. All CTF-related parameters were refined, including doming, per-series beam tilt and a 3x3 grid model for local astigmatism (Table 1). For the last 2 iterations, anisotropic per-series, per-frame B-factor weights were estimated. The final iteration was completed in ca. 24 hours, using 4 GeForce 2080 Ti GPUs. The original mask deposited with EMD-9865 was used to estimate the final resolution.
To analyze the doming behavior, fitted doming model parameters were averaged across the data set. Because doming was fitted after per-particle defocus, which was dominated by frames 3–4 due to weighting, the values were normalized by subtracting those of frame 1 from all. As a larger, planar inclination spanning the field of view was observed in the fits in addition to the more local bending of the center relative to the periphery, a plane was fitted into each frame’s values and subtracted from them before quantifying the doming.
Comparison with other tools for tilt-series data refinement
Tilt-series data were downloaded for the EMPIAR-10064 entry. Initial tilt-series alignment was performed in IMOD using manually picked gold fiducials on 4x binned images with default settings. Tilt-series CTF estimation was performed in Warp. 3,566 particles were picked using Warp’s 3D template matching in full tomograms reconstructed at 10 Å/px. Sub-tomograms and 3D CTF volumes were exported at 5.0 Å/px. Global 3D refinement reached a resolution of 13 Å. The results were imported in M. The particle poses were given a temporal resolution of 3, the image warp and volume warp grid resolutions were set to 8x8x41 and 4x4x2x20, respectively, and refinement of all parameters was performed for 5 iterations (Table 1). Data-driven anisotropic weight estimation was performed to assign unique weights to every series and tilt index.
The processing of EMPIAR-10045 tilt-series was performed in exactly the same way as descried in the previous paragraph for EMPIAR-10064, using 3,058 particles (Table 1).
Tilt-series movie data were downloaded for the EMPIAR-10164 entry. Tilt movie frame alignment was performed in Warp using a model without spatial resolution. Initial tilt-series alignment was performed in IMOD using gold fiducials automatically picked in Warp, on 6x binned images with default settings. Tilt-series CTF estimation was performed in Warp. 130,658 particles were picked using Warp’s 3D template matching with a template derived from EMD-3782 in full tomograms reconstructed at 10 Å/px. Sub-tomograms and 3D CTF volumes were exported at 5 Å/px using 56 px boxes. Global 3D refinement with C6 symmetry was performed in RELION 3.0, and reached the 10 Å Nyquist limit. The results were imported in M. The particle poses were given a temporal resolution of 3, the image warp and volume warp grid resolutions were set to 8x8x41 and 3x3x3x20, and refinement of all parameters was performed for 5 iterations, including tilt movie frame alignment in the last 2 iterations (Table 1). Data-driven anisotropic weight estimation was performed to assign unique weights to every series, tilt index and tilt frame index.
Acquisition and refinement of M. pneumoniae in situ tilt-series data
Data previously used in another study46 were re-analyzed with the release version of M. As described there, Mycoplasma pneumoniae strain M129 (ATCC 29342) cells were grown on 200 mesh gold grids coated with a holey carbon support (R 2/1, Quantifoil). Cells were cultivated at 37 °C in modified Hayflick medium: 14.7 g/L Difco PPLO (Becton Dickinson, USA), 20% (v/v) Gibco horse serum (New Zealand origin, Life Technologies, USA), 100 mM HEPES-Na (pH 7.4), 1% (w/w) glucose, 0.002% (w/w) phenol red and 1,000 U/mL freshly dissolved penicillin G. Chloramphenicol (Cm; Sigma-Aldrich, USA) was added 15 minutes prior to vitrification, at a final concentration of 0.5 mg/ml. Grids were quickly washed with PBS buffer containing 10 nm protein A-conjugated gold beads (Aurion, Netherlands), blotted from the back side for 2 seconds, and plunged into mixed liquid ethane/propane at liquid N2 temperature with a manual plunger (Max Planck Institute of Biochemistry, Germany). The cryo-EM grids were stored in a sealed box in liquid N2 before usage.
Tilt-series data were collected on a Titan Krios TEM operated at 300 kV (Thermo Fisher Scientific) equipped with a field-emission gun, a Gatan K2 Summit direct detector and a Quantum post-column energy filter (Gatan). Images were recorded in exposure-fractionation, counting mode using SerialEM 3.7.2. Tilt-series were acquired with a dose-symmetric scheme using dedicated scripts59 with the following settings: TEM in nano-probe mode, magnification 81,000 with a calibrated pixel size of 1.7 Å, energy filter in zero loss mode, defocus range 1.5 to 3.5 μm, tilt range -60° to 60° with 3° tilt increment and constant exposure per tilt, total exposure of 120 e-/Å2. In total, 65 tilt-series were collected from Cm-treated cells.
Raw tilt movies were processed in Warp. De novo tilt-series alignment was performed in IMOD using gold fiducials picked automatically with Warp’s BoxNet, and the results were imported in Warp, where the tilt-series CTFs were estimated. Using full tomograms reconstructed at 10 Å/px, two tomograms were denoised using Warp’s Noise2Map tool to pick the ribosome particles manually. Using these coordinates, sub-tomograms were exported from Warp to RELION to obtain an initial reference. This reference was used to perform template matching in Warp at 10 Å/px. In addition, a binary classifier based on a 3D CNN was trained on the 2 manually picked tomograms to remove false positives (membranes, carbon hole edges etc.) from the template matching results. 24,202 particles were obtained this way. Sub-tomograms for all particles were exported from Warp to RELION and aligned against the previously refined low-resolution reference. No classification was performed. The results were imported in M. There, global movement and rotation, a 5x5x41 image-space warping grid, a 8x8x2x10 volume-space warping grid, as well as particle pose trajectories with 3 temporal sampling points were refined over 5 iterations (Table 1). Starting with iteration 3, CTF parameters were also refined. At the beginning of iteration 4, reference-based tilt movie alignment was performed, resulting in a 3.7 Å map. Using the improved alignments, sub-tomograms were reconstructed at 3 Å/px. Classification into 5 classes was performed in RELION. 17,890 particles from the 2 best classes were imported in M and refined for another iteration using the same settings to obtain a 3.5 Å map. The final iteration was completed in ca. 6 hours, using 4 GeForce 2080 Ti GPUs. Afterwards, focused refinements were performed in M using masks limited to the 30S and 50S subunits, optimizing only image warping and particle poses.
To calculate the Rosenthal–Henderson47 plot, deformation, weighting and CTF parameters from the last iteration of 70S refinement were kept. The number of particles was reduced by excluding entire tilt-series from the data set, thus keeping the average particle density per series constant. Resolution was reset to 10 Å at the beginning of each subset’s refinement, and only the particle pose trajectories were optimized for 3 iterations.
Extended Data
Extended Data Figure 1.
A pyramid results from a combination of several grids to model the in-plane motion occurring in a frame-series with 40 frames as a function of position and dose. Each cubical cell represents a sampling point. The top grid has full temporal (per-frame exposure) and no spatial resolution to model fast, global motion (left, 1x1x40, shown truncated). For subsequent grids, temporal resolution is reduced by a factor of 4 and spatial resolution is doubled to model slower, local motion (center, 2x2x10; right 4x4x3). The spatial resolution of the first grid can be set higher if there is enough particle signal to fit.
Extended Data Figure 2.
Apoferritin frame-series were refined using a small 5% sub-population of the particles alone, and together with another 95% sub-population that improved the accuracy of the multi-particle system hyperparameters, but did not contribute particles to the 5% half-maps.
(a) Exemplary micrograph (n = 150 micrographs collected from the same sample) showing the distribution of the 2 sub-populations within a frame-series.
(b) FSC curves between the half-maps of the 5% population in both scenarios, showing the benefit of multi-species refinement.
Extended Data Figure 3.
High-resolution information is delocalized at high defocus. Choosing an insufficiently large particle box size results in loss of that information. In Fourier space, this results in CTF oscillations becoming too fast to be resolved at the sampling rate provided by the small box, averaging to 0. M chooses the box size automatically for each frame- or tilt-series’ defocus, pre-multiplies the data and simulated CTF by the CTF to eliminate the oscillations and localize the signal, and then crops the data to the desired map size. This avoids the pitfall of losing map resolution due to an inappropriately chosen box size.
(a) Visualization of the delocalization and aliasing effects in Fourier space as 2D and rotationally averaged 1D CTFs; grids depict sampling rate. At low defocus (row 1), all signal is localized within the box and no aliasing is seen in the simulated CTF used for the image formation model during refinement. At high defocus (row 2), high-resolution signal is delocalized outside the small particle box. Once the particle is extracted, the fast CTF oscillations are averaged to 0 and high-resolution information is lost. At the same time, the simulated CTF is filled with aliasing artifacts because it is not low-pass filtered in the same way. If the particle data are pre-multiplied by the CTF at a box size large enough to contain all signal and resolve all CTF oscillations (row 3), as can be done optionally in RELION, all particle signal is contained in the box after cropping it to a smaller size, and the CTF averages to 0.5. However, the simulated CTF2 does not match this and contains aliasing artifacts. M applies the pre-multiplication to both particle data and simulated CTF in a larger box before cropping (row 4) to avoid the mismatch.
(b) FSC between the half-maps reconstructed from HIV1 virus-like particles of a single high-defocus (3.9μm) tilt-series in an insufficiently large box. Using data extracted without pre-multiplication, as is currently common, limits the resolution to 3.9Å (grey). Pre-multiplying both particle data and CTF in a larger box, as automated in M, improves the result to 3.2Å (green). Pre-multiplying only particle data is only slightly worse here (blue), but would likely lead to noticeably worse results in RELION as the aliased CTF2 would be used in the image formation during refinement. The FSC curves diverge as the proportion of CTF sign errors (orange) increases.
(c) Relation between tilt-series defocus and associated contribution of high-resolution information to the reconstruction. For the larger dataset, not pre-multiplying the data results in a strong correlation, where high-defocus data is down-weighted to contribute less (grey). The correlation disappears when pre-multiplication is applied, so more tilt-series contribute high-resolution information (green).
Extended Data Figure 4.
Normalized 2D cross-correlation between reference projections and data, averaged over all particles in a single frame is shown for the 1st and 3rd frame of the same exposure. Values in the low-frequency region are excluded to reduce the value range. The fitted B-factor is highly anisotropic for the 1st frame because of intra-frame motion: 0Å2 and -62Å2 along X and Y, respectively. For the 3rd frame, the fit is much more isotropic due to lack of intra-frame motion, but some high-resolution information is lost to radiation damage: -8Å2 and -10Å2 along X and Y, respectively.
Extended Data Figure 5.
Atomic-resolution data of apoferritin previously refined with RELION 3.1 to 1.54Å (EMD-9865) were processed with M to achieve a resolution of 1.34Å.
(a) Examples of side-chain densities produced by RELION (top) and M (bottom), showing cases of improved atomic features such as one of the hydrogens in Tyr29 (black arrow).
(b) FSC between the half-maps produced by RELION (grey) and M (green), showing a general improvement in resolution through M.
Extended Data Figure 6.
Doming models describing per-frame, spatially resolved (3x3 points) defocus offsets fitted during the refinement of atomic-resolution data of apoferritin (EMPIAR-10248) were averaged across the dataset, showing significant changes in the CTF during exposure.
(a) Defocus change plotted against the accumulated exposure show a fast change in both the central point and the average of the entire field of view’s 3x3 points at the beginning of the exposure. After the first 7.5e-/Å2 of exposure, the average change stabilizes, while the central point continues to decrease in defocus.
(b) When corrected for global inclination, the difference between the central and peripheral defocus change indicates a steady increase in doming within the field of view as a function of accumulated exposure.
(c) Surface rendering of the spatially resolved defocus change for the first 7 frames shows an inclination of the entire field of view, as well as a more localized dent in the center. The observed change in the CTF can also be caused by electrostatic lensing effects due to sample charging, and further experiments are necessary to investigate the exact nature of doming.
Extended Data Figure 7.
(a) 2D XY slice through an exemplary denoised tomogram (n = 65 tilt-series collected from the same sample. Each tilt-series captures a single cell).
(b) Resolution plotted against the number of particles shows that 5Å can be obtained with less than 3000 large, asymmetric particles in cells. Extrapolation beyond the Nyquist limit of the data (magenta line) is speculative, but indicates that 3Å could be surpassed with less than 100,000 particles, given data with higher magnification.
(c) Histogram of manually measured cell thickness values from 65 tomograms.
ED_Table1. Refinement parameters for all datasets.
| Series type | Tilt range | Particles picked | Particles classified | Image warp | Volume warp | Tilt movie alignment | Per-particle trajectory samples | CTF refinement | Weight fitting | Higher-order aberrations | Doming | Iterations | Symmetry | Resolution | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EMPIAR-10288 | F | 1,033,994 | 149,328 | 2×2×40 | 2 | ✓ | ✓ | 5 | C1 | 2.9 Å | |||||
| EMPIAR-10453 | T | ±60 | 38,742 | 22,998 | 3×2×41 | — | ✓ | 1 | ✓ | ✓ | 4 | C3 | 3.8 Å | ||
| AF-t | T | ±40 | 18,991 | 6×4×41 | — | ✓ | 3 | ✓ | ✓ | ✓ | ✓ | 5 | O | 2.3 Å | |
| AF-f | F | 22,122 | 6×4×40 | 3 | ✓ | ✓ | ✓ | ✓ | 5 | O | 2.3 Å | ||||
| EMPIAR-10248 | F | 109,437 | 4×4×25 | 2 | ✓ | ✓ | ✓ | ✓ | 5 | O | 1.34 Å | ||||
| EMPIAR-10064 | T | ±40 | 3,566 | 8×8×41 | 4×4×2×20 | 3 | ✓ | ✓ | 5 | C1 | 5.7 Å | ||||
| EMPIAR-10045 | T | ±60 | 3,058 | 8×8×41 | 4×4×2×20 | 3 | ✓ | ✓ | 5 | C1 | 6.0 Å | ||||
| EMPIAR-10164 | T | ±60 | 130,658 | 8×8×41 | 3×3×20 | ✓ | 3 | ✓ | ✓ | 5 | C6 | 3.0 Å | |||
| M. pneumoniae | T | ±60 | 24,202 | 17,890 | 5×5×41 | 8×8×2×10 | ✓ | 3 | ✓ | ✓ | 6 | C1 | 3.5 Å |
Supplementary Material
A pyramid results from a combination of several grids to model the in-plane motion occurring in a frame-series with 40 frames as a function of position and dose. Each cubical cell represents a sampling point. The top grid has full temporal (per-frame exposure) and no spatial resolution to model fast, global motion (left, 1x1x40, shown truncated). For subsequent grids, temporal resolution is reduced by a factor of 4 and spatial resolution is doubled to model slower, local motion (center, 2x2x10; right 4x4x3). The spatial resolution of the first grid can be set higher if there is enough particle signal to fit.
Apoferritin frame-series were refined using a small 5% sub-population of the particles alone, and together with another 95% sub-population that improved the accuracy of the multi-particle system hyperparameters, but did not contribute particles to the 5% half-maps.
(a) Exemplary micrograph (n = 150 micrographs collected from the same sample) showing the distribution of the 2 sub-populations within a frame-series.
(b) FSC curves between the half-maps of the 5% population in both scenarios, showing the benefit of multi-species refinement.
High-resolution information is delocalized at high defocus. Choosing an insufficiently large particle box size results in loss of that information. In Fourier space, this results in CTF oscillations becoming too fast to be resolved at the sampling rate provided by the small box, averaging to 0. M chooses the box size automatically for each frame- or tilt-series’ defocus, pre-multiplies the data and simulated CTF by the CTF to eliminate the oscillations and localize the signal, and then crops the data to the desired map size. This avoids the pitfall of losing map resolution due to an inappropriately chosen box size.
(a) Visualization of the delocalization and aliasing effects in Fourier space as 2D and rotationally averaged 1D CTFs; grids depict sampling rate. At low defocus (row 1), all signal is localized within the box and no aliasing is seen in the simulated CTF used for the image formation model during refinement. At high defocus (row 2), high-resolution signal is delocalized outside the small particle box. Once the particle is extracted, the fast CTF oscillations are averaged to 0 and high-resolution information is lost. At the same time, the simulated CTF is filled with aliasing artifacts because it is not low-pass filtered in the same way. If the particle data are pre-multiplied by the CTF at a box size large enough to contain all signal and resolve all CTF oscillations (row 3), as can be done optionally in RELION, all particle signal is contained in the box after cropping it to a smaller size, and the CTF averages to 0.5. However, the simulated CTF2 does not match this and contains aliasing artifacts. M applies the pre-multiplication to both particle data and simulated CTF in a larger box before cropping (row 4) to avoid the mismatch.
(b) FSC between the half-maps reconstructed from HIV1 virus-like particles of a single high-defocus (3.9μm) tilt-series in an insufficiently large box. Using data extracted without pre-multiplication, as is currently common, limits the resolution to 3.9Å (grey). Pre-multiplying both particle data and CTF in a larger box, as automated in M, improves the result to 3.2Å (green). Pre-multiplying only particle data is only slightly worse here (blue), but would likely lead to noticeably worse results in RELION as the aliased CTF2 would be used in the image formation during refinement. The FSC curves diverge as the proportion of CTF sign errors (orange) increases.
(c) Relation between tilt-series defocus and associated contribution of high-resolution information to the reconstruction. For the larger dataset, not pre-multiplying the data results in a strong correlation, where high-defocus data is down-weighted to contribute less (grey). The correlation disappears when pre-multiplication is applied, so more tilt-series contribute high-resolution information (green).
Normalized 2D cross-correlation between reference projections and data, averaged over all particles in a single frame is shown for the 1st and 3rd frame of the same exposure. Values in the low-frequency region are excluded to reduce the value range. The fitted B-factor is highly anisotropic for the 1st frame because of intra-frame motion: 0Å2 and -62Å2 along X and Y, respectively. For the 3rd frame, the fit is much more isotropic due to lack of intra-frame motion, but some high-resolution information is lost to radiation damage: -8Å2 and -10Å2 along X and Y, respectively.
Atomic-resolution data of apoferritin previously refined with RELION 3.1 to 1.54Å (EMD-9865) were processed with M to achieve a resolution of 1.34Å.
(a) Examples of side-chain densities produced by RELION (top) and M (bottom), showing cases of improved atomic features such as one of the hydrogens in Tyr29 (black arrow).
(b) FSC between the half-maps produced by RELION (grey) and M (green), showing a general improvement in resolution through M.
Doming models describing per-frame, spatially resolved (3x3 points) defocus offsets fitted during the refinement of atomic-resolution data of apoferritin (EMPIAR-10248) were averaged across the dataset, showing significant changes in the CTF during exposure.
(a) Defocus change plotted against the accumulated exposure show a fast change in both the central point and the average of the entire field of view’s 3x3 points at the beginning of the exposure. After the first 7.5e-/Å2 of exposure, the average change stabilizes, while the central point continues to decrease in defocus.
(b) When corrected for global inclination, the difference between the central and peripheral defocus change indicates a steady increase in doming within the field of view as a function of accumulated exposure.
(c) Surface rendering of the spatially resolved defocus change for the first 7 frames shows an inclination of the entire field of view, as well as a more localized dent in the center. The observed change in the CTF can also be caused by electrostatic lensing effects due to sample charging, and further experiments are necessary to investigate the exact nature of doming.
(a) 2D XY slice through an exemplary denoised tomogram (n = 65 tilt-series collected from the same sample. Each tilt-series captures a single cell).
(b) Resolution plotted against the number of particles shows that 5Å can be obtained with less than 3000 large, asymmetric particles in cells. Extrapolation beyond the Nyquist limit of the data (magenta line) is speculative, but indicates that 3Å could be surpassed with less than 100,000 particles, given data with higher magnification.
(c) Histogram of manually measured cell thickness values from 65 tomograms.
Acknowledgements
The human H-chain apoferritin plasmid and purification protocol were kindly provided by L. Fairall, C. Savva and the Protex facility of the University of Leicester. We thank Thomas Hoffmann and the EMBL IT support. We thank B. Engel, whose generous sharing of in situ data enabled the development of algorithms that later became M. P.C. was supported by the Deutsche Forschungsgemeinschaft within SFB860 and SPP1935, Germany’s Excellence Strategy (EXC 2067/1-390729940), the European Research Council (ERC) Advanced Investigator Grant TRANSREGULON (693023), and the Volkswagen Foundation. J.M. was supported by the EMBL and ERC starting grant 3DCellPhase- (760067).
Footnotes
Author contributions
D.T. designed M’s architecture and algorithms, and carried out all implementation and application. L.X. and J.M. provided tilt-series data of Cm-treated M. pneumoniae, assisted in testing M and interpretation of the maps. D.T. and L.X. solved the Cm-bound ribosome structure. C.D. collected apoferritin frame and tilt-series, and analysed the frame-series data using RELION. P.C. provided scientific environment, funding and additional interpretations and implications. D.T., P.C. and J.M. wrote the manuscript with input from all authors.
Competing financial interests
The authors declare no competing financial or other interests.
Data availability
Maps were deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers 11603 (AF-t), 11611 (AF-f), 11652 (EMPIAR-10248), 11653 (EMPIAR-10045), 11654 (EMPIAR-10064), 11655 (EMPIAR-10164), 11656 (EMPIAR-10288), 11651 (EMPIAR-10453), 11650, 11998, 11999 (Cm-treated M. pneumoniae 70S ribosome, 30S, and 50S subunits, respectively). Raw data for AF-t and AF-f were deposited in the EMPIAR database under accession number 10491, and raw data of Cm-treated M. pneumoniae under accession number 10499. Data from previous studies reanalyzed here were obtained from EMPIAR (10045, 10064, 10164, 10248, 10288, 10453). Maps from previous studies used for comparisons were obtained from EMDB (0339, 0529, 3782, 8986, 9865, 10683). Atomic models from previous studies used for comparisons were obtained from PDB (4V7T, 5L93).
Code availability
M and Warp binaries, source code, and user guide are available as Supplementary Software. Updated versions can be found at https://github.com/cramerlab/warp and https://warpem.com.
References
- 1.Dubochet J, Lepault J, Freeman R, Berriman JA, Homo JC. Electron microscopy of frozen water and aqueous solutions. J Microsc. 1982;128:219–237. [Google Scholar]
- 2.Danev R, Yanagisawa H, Kikkawa M. Cryo-Electron Microscopy Methodology: Current Aspects and Future Directions. Trends in biochemical sciences. 2019;44:837–848. doi: 10.1016/j.tibs.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Lyumkis D. Challenges and opportunities in cryo-EM single-particle analysis. J Biol Chem. 2019;294:5181–5197. doi: 10.1074/jbc.REV118.005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bharat TA, Scheres SH. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nature protocols. 2016;11:2054–2065. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castaño-Díez D, Kudryashev M, Arheit M, Stahlberg H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. Journal of structural biology. 2012;178 doi: 10.1016/j.jsb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Himes BA, Zhang P. emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nat Methods. 2018;15:955–961. doi: 10.1038/s41592-018-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank J, Goldfarb W, Eisenberg D, Baker TS. Reconstruction of glutamine synthetase using computer averaging. Ultramicroscopy. 1978;3:283–290. doi: 10.1016/s0304-3991(78)80038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank J. Single-Particle Reconstruction of Biological Molecules–Story in a Sample (Nobel Lecture) Angewandte Chemie International Edition. 2018;57:10826–10841. doi: 10.1002/anie.201802770. [DOI] [PubMed] [Google Scholar]
- 9.Knauer V, Hegerl R, Hoppe W. Three-dimensional reconstruction and averaging of 30 S ribosomal subunits of Escherichia coli from electron micrographs. Journal of molecular biology. 1983;163:409–430. doi: 10.1016/0022-2836(83)90066-9. [DOI] [PubMed] [Google Scholar]
- 10.Oettl H, Hegerl R, Hoppe W. Three-dimensional reconstruction and averaging of 50 S ribosomal subunits of Escherichia coli from electron micrographs. Journal of molecular biology. 1983;163:431–450. doi: 10.1016/0022-2836(83)90067-0. [DOI] [PubMed] [Google Scholar]
- 11.Leigh KE, et al. Subtomogram averaging from cryo-electron tomograms. Methods Cell Biol. 2019;152:217–259. doi: 10.1016/bs.mcb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Brilot AF, et al. Beam-induced motion of vitrified specimen on holey carbon film. Journal of structural biology. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartesaghi A, Lecumberry F, Sapiro G, Subramaniam S. Protein Secondary Structure Determination by Constrained Single-Particle Cryo-Electron Tomography. Structure (London, England : 1993) 2012;20:2003–2013. doi: 10.1016/j.str.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mastronarde DN. In: Electron Tomography. Frank J, editor. SpringerLink; 2006. pp. 163–185. [Google Scholar]
- 17.Lawrence A, Bouwer J, Perkins G, Ellisman M. Transform-based backprojection for volume reconstruction of large format electron microscope tilt-series. Journal of structural biology. 2006;154 doi: 10.1016/j.jsb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez J-J, Li S, Agard DA. Consideration of sample motion in cryo-tomography based on alignment residual interpolation. Journal of structural biology. 2019;205:1–6. doi: 10.1016/j.jsb.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, et al. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat Methods. 2019;16:1161–1168. doi: 10.1038/s41592-019-0591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Ren G. IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure. PloS one. 2012;7 doi: 10.1371/journal.pone.0030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant T, Rohou A, Grigorieff N. cisTEM, user-friendly software for single-particle image processing. eLife. 2018;7:e35383. doi: 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of structural biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 27.Scheres SH. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Nocedal J. Updating quasi-Newton matrices with limited storage. Mathematics of Computation. 1980;35:773–773. [Google Scholar]
- 29.Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 30.van Heel MKW, Schutter W, van Bruggen EFJ. Arthropod hemocyanin studies by image analysis. In: Wood EJ, editor. Life Chemistry Reports (2nd ed), The Structure and Function of Invertebrate Respiratory Proteins; EMBO Workshop; Leeds. 1982. pp. 69–73. [Google Scholar]
- 31.Russo C, Henderson R. Ewald Sphere Correction Using a Single Side-Band Image Processing Algorithm. Ultramicroscopy. 2018;187:26–33. doi: 10.1016/j.ultramic.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. Journal of structural biology. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehtinen J, et al. Noise2Noise: Learning Image Restoration without Clean Data; Proceedings of the 35th International Conference on Machine Learning; 2018. pp. 2965–2974. [Google Scholar]
- 34.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishna Kumar K, et al. Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell. 2019;176:448–458.:e412. doi: 10.1016/j.cell.2018.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turoňová B, et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020 doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramlaul K, Palmer CM, Nakane T, Aylett CHS. Mitigating Local Over-fitting During Single Particle Reconstruction with SIDESPLITTER. Journal of structural biology. 2020;211 doi: 10.1016/j.jsb.2020.107545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Punjani A, Zhang H, Fleet DJ. Non-uniform refinement: Adaptive regularization improves single particle cryo-EM reconstruction. Nat Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 39.Eisenstein F, Danev R, Pilhofer M. Improved applicability and robustness of fast cryo-electron tomography data acquisition. Journal of structural biology. 2019;208:107–114. doi: 10.1016/j.jsb.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato T, et al. CryoTEM with a Cold Field Emission Gun That Moves Structural Biology into a New Stage. Microscopy and Microanalysis. 2019;25:998–999. [Google Scholar]
- 41.Khoshouei M, Pfeffer S, Baumeister W, Forster F, Danev R. Subtomogram analysis using the Volta phase plate. Journal of structural biology. 2017;197:94–101. doi: 10.1016/j.jsb.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Himes BA. emClarity Wiki, change log, “2018-Nov-14” entry. 2020 https://github.com/bHimes/emClarity/wiki.
- 43.Bharat TA, Russo CJ, Lowe J, Passmore LA, Scheres SH. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure (London, England : 1993) 2015;23:1743–1753. doi: 10.1016/j.str.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schur FK, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science New York NY. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 45.Turonova B, Schur FKM, Wan W, Briggs JAG. Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4A. Journal of structural biology. 2017;199:187–195. doi: 10.1016/j.jsb.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Reilly FJ, et al. In-cell architecture of an actively transcribing-translating expressome. Science (New York, NY) 2020;369:554–557. doi: 10.1126/science.abb3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of molecular biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Sánchez RM, Mester R, Kudryashev M. In: Image Analysis. Felsberg M, Forssén P-E, Sintorn I-M, Unger J, editors. Springer International Publishing; Cham: 2019. pp. 415–426. [Google Scholar]
- 49.Git – free and open source distributed version control system. 2020 https://git-scm.com.
- 50.Grant T, Grigorieff N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. Journal of structural biology. 2015;192:204–208. doi: 10.1016/j.jsb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahamid J, et al. Visualizing the Molecular Sociology at the HeLa Cell Nuclear Periphery. Science (New York, NY) 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 52.Zivanov J, Nakane T, Scheres SHW. Estimation of High-Order Aberrations and Anisotropic Magnification From cryo-EM Data Sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeRosier D. Correction of High-Resolution Data for Curvature of the Ewald Sphere. Ultramicroscopy. 2000;81:83–98. doi: 10.1016/s0304-3991(99)00120-5. [DOI] [PubMed] [Google Scholar]
- 54.Penczek PA, et al. CTER–Rapid estimation of CTF parameters with error assessment. Ultramicroscopy. 2014;140:9–19. doi: 10.1016/j.ultramic.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ludtke SJ. Single Particle Refinement and Variability Analysis in EMAN2.1. Methods Enzymol. 2016;579:159–189. doi: 10.1016/bs.mie.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schilbach S, et al. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature. 2017;551:204–209. doi: 10.1038/nature24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchholz T-O, Jordan M, Pigino G, Jug F. Cryo-CARE: Content-Aware Image Restoration for Cryo-Transmission Electron Microscopy Data; 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); Venice, Italy. 2019. pp. 502–506. [Google Scholar]
- 58.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 59.Hagen WJH, Wan W, Briggs JAG. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. Journal of structural biology. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A pyramid results from a combination of several grids to model the in-plane motion occurring in a frame-series with 40 frames as a function of position and dose. Each cubical cell represents a sampling point. The top grid has full temporal (per-frame exposure) and no spatial resolution to model fast, global motion (left, 1x1x40, shown truncated). For subsequent grids, temporal resolution is reduced by a factor of 4 and spatial resolution is doubled to model slower, local motion (center, 2x2x10; right 4x4x3). The spatial resolution of the first grid can be set higher if there is enough particle signal to fit.
Apoferritin frame-series were refined using a small 5% sub-population of the particles alone, and together with another 95% sub-population that improved the accuracy of the multi-particle system hyperparameters, but did not contribute particles to the 5% half-maps.
(a) Exemplary micrograph (n = 150 micrographs collected from the same sample) showing the distribution of the 2 sub-populations within a frame-series.
(b) FSC curves between the half-maps of the 5% population in both scenarios, showing the benefit of multi-species refinement.
High-resolution information is delocalized at high defocus. Choosing an insufficiently large particle box size results in loss of that information. In Fourier space, this results in CTF oscillations becoming too fast to be resolved at the sampling rate provided by the small box, averaging to 0. M chooses the box size automatically for each frame- or tilt-series’ defocus, pre-multiplies the data and simulated CTF by the CTF to eliminate the oscillations and localize the signal, and then crops the data to the desired map size. This avoids the pitfall of losing map resolution due to an inappropriately chosen box size.
(a) Visualization of the delocalization and aliasing effects in Fourier space as 2D and rotationally averaged 1D CTFs; grids depict sampling rate. At low defocus (row 1), all signal is localized within the box and no aliasing is seen in the simulated CTF used for the image formation model during refinement. At high defocus (row 2), high-resolution signal is delocalized outside the small particle box. Once the particle is extracted, the fast CTF oscillations are averaged to 0 and high-resolution information is lost. At the same time, the simulated CTF is filled with aliasing artifacts because it is not low-pass filtered in the same way. If the particle data are pre-multiplied by the CTF at a box size large enough to contain all signal and resolve all CTF oscillations (row 3), as can be done optionally in RELION, all particle signal is contained in the box after cropping it to a smaller size, and the CTF averages to 0.5. However, the simulated CTF2 does not match this and contains aliasing artifacts. M applies the pre-multiplication to both particle data and simulated CTF in a larger box before cropping (row 4) to avoid the mismatch.
(b) FSC between the half-maps reconstructed from HIV1 virus-like particles of a single high-defocus (3.9μm) tilt-series in an insufficiently large box. Using data extracted without pre-multiplication, as is currently common, limits the resolution to 3.9Å (grey). Pre-multiplying both particle data and CTF in a larger box, as automated in M, improves the result to 3.2Å (green). Pre-multiplying only particle data is only slightly worse here (blue), but would likely lead to noticeably worse results in RELION as the aliased CTF2 would be used in the image formation during refinement. The FSC curves diverge as the proportion of CTF sign errors (orange) increases.
(c) Relation between tilt-series defocus and associated contribution of high-resolution information to the reconstruction. For the larger dataset, not pre-multiplying the data results in a strong correlation, where high-defocus data is down-weighted to contribute less (grey). The correlation disappears when pre-multiplication is applied, so more tilt-series contribute high-resolution information (green).
Normalized 2D cross-correlation between reference projections and data, averaged over all particles in a single frame is shown for the 1st and 3rd frame of the same exposure. Values in the low-frequency region are excluded to reduce the value range. The fitted B-factor is highly anisotropic for the 1st frame because of intra-frame motion: 0Å2 and -62Å2 along X and Y, respectively. For the 3rd frame, the fit is much more isotropic due to lack of intra-frame motion, but some high-resolution information is lost to radiation damage: -8Å2 and -10Å2 along X and Y, respectively.
Atomic-resolution data of apoferritin previously refined with RELION 3.1 to 1.54Å (EMD-9865) were processed with M to achieve a resolution of 1.34Å.
(a) Examples of side-chain densities produced by RELION (top) and M (bottom), showing cases of improved atomic features such as one of the hydrogens in Tyr29 (black arrow).
(b) FSC between the half-maps produced by RELION (grey) and M (green), showing a general improvement in resolution through M.
Doming models describing per-frame, spatially resolved (3x3 points) defocus offsets fitted during the refinement of atomic-resolution data of apoferritin (EMPIAR-10248) were averaged across the dataset, showing significant changes in the CTF during exposure.
(a) Defocus change plotted against the accumulated exposure show a fast change in both the central point and the average of the entire field of view’s 3x3 points at the beginning of the exposure. After the first 7.5e-/Å2 of exposure, the average change stabilizes, while the central point continues to decrease in defocus.
(b) When corrected for global inclination, the difference between the central and peripheral defocus change indicates a steady increase in doming within the field of view as a function of accumulated exposure.
(c) Surface rendering of the spatially resolved defocus change for the first 7 frames shows an inclination of the entire field of view, as well as a more localized dent in the center. The observed change in the CTF can also be caused by electrostatic lensing effects due to sample charging, and further experiments are necessary to investigate the exact nature of doming.
(a) 2D XY slice through an exemplary denoised tomogram (n = 65 tilt-series collected from the same sample. Each tilt-series captures a single cell).
(b) Resolution plotted against the number of particles shows that 5Å can be obtained with less than 3000 large, asymmetric particles in cells. Extrapolation beyond the Nyquist limit of the data (magenta line) is speculative, but indicates that 3Å could be surpassed with less than 100,000 particles, given data with higher magnification.
(c) Histogram of manually measured cell thickness values from 65 tomograms.
Data Availability Statement
Maps were deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers 11603 (AF-t), 11611 (AF-f), 11652 (EMPIAR-10248), 11653 (EMPIAR-10045), 11654 (EMPIAR-10064), 11655 (EMPIAR-10164), 11656 (EMPIAR-10288), 11651 (EMPIAR-10453), 11650, 11998, 11999 (Cm-treated M. pneumoniae 70S ribosome, 30S, and 50S subunits, respectively). Raw data for AF-t and AF-f were deposited in the EMPIAR database under accession number 10491, and raw data of Cm-treated M. pneumoniae under accession number 10499. Data from previous studies reanalyzed here were obtained from EMPIAR (10045, 10064, 10164, 10248, 10288, 10453). Maps from previous studies used for comparisons were obtained from EMDB (0339, 0529, 3782, 8986, 9865, 10683). Atomic models from previous studies used for comparisons were obtained from PDB (4V7T, 5L93).
M and Warp binaries, source code, and user guide are available as Supplementary Software. Updated versions can be found at https://github.com/cramerlab/warp and https://warpem.com.