Abstract
While central nervous system (CNS) microglia have been studied extensively, surprisingly little is known about macrophages populating the peripheral nervous system (PNS Macs). We performed ontogenic, transcriptomic and spatial characterization of sciatic nerve Macs (snMacs). Using multiple fate-mapping systems, we show that snMacs do not derive from the early embryonic precursors colonizing the CNS, but originate primarily from late embryonic precursors and get replaced by bone marrow-derived Macs over time. Using single-cell profiling, we identified a tissue-specific core signature of snMacs and found two spatially-separated snMacs: Relmα+Mgl1+ snMacs in the epineurium and Relmα- Mgl1- snMacs in the endoneurium. Globally, snMacs lack most core signature genes of microglia, with only the endoneurial subset expressing a restricted number of these genes. Single-cell transcriptomics revealed that in response to injury both snMacs respond differently and that the PNS, in contrast to the CNS, is permissive to prolonged engraftment of monocyte-derived Macs recruited upon injury.
Introduction
The advent of single-cell transcriptomics enables the identification of novel, previously unrecognized cell subsets across tissues and species1. Compared with bulk transcriptomics, single-cell sequencing avoids contaminations from cells of neighboring tissues and prevents related but distinct cells from being studied as a single population. This is particularly relevant for macrophage biology. Macs have been proposed to be one of the most plastic cells of the immune system2. The assumed plasticity of Macs is marked by their capacity to profoundly adapt to their tissue of residence and to change immensely during tissue inflammation or stress. However, contamination with other cells or even engulfment of dying cells could yield contaminated gene expression profiles that overestimate the tissue-specific signature of resident Macs, as we and others have experienced recently3,4. Moreover, monocytes recruited during inflammation will quickly down-regulate monocyte markers and acquire a general macrophage profile, which can also lead to an overestimation of the plasticity of resident Macs during inflammation.
Therefore, there is a clear need to revisit the macrophage profiles at the single-cell level to pinpoint their true identity and fates. Ideally, this should be coupled to both ontogenic and spatial profiling of the distinct macrophage clusters. This is needed to understand which single-cell clusters are merely representing distinct activation states or developmental intermediates of the same cells and which single-cell clusters represent different cell subsets that are found in a different micro-environment in the tissues or are derived from distinct precursors.
Microglia, the resident Macs of the central nervous system (CNS), have been characterized extensively. Early parabiosis and fate mapping studies established that microglia are maintained through self-renewal independently of circulating bone-marrow (BM) derived precursors5,6. They derive exclusively from early embryonic precursors originating from the yolk sac between E7.5 and E9.53,4 in a PU.1, IRF8 and CSF-1R-dependent manner, independent of Myb5,6. Both IL-34, the ligand for CSF-1R, and TGF-β play a prominent role in microglia development, and shape their unique gene signature, hallmarked by genes such as Ccl12, Cd34, Ecscr, Fcrls, Gpr34, Hexb, Kcnd1, Lag3, Mef2c, Nav3, Sall1, Sall3, Sema4d, Siglech, Slc2a5, Sparc, Tmc7, Tmem119 and Trem2 9–11. Elegant fate mapping tools revealed their clonal expansion in steady-state and during pathology, advancing our knowledge on their self-maintenance capacity7. Additionally, also other non-parenchymal macrophage populations at CNS interfaces are starting to be characterized 8–10. Not all border-associated CNS Macs are derived from embryonic precursors. The choroid plexus Macs for example contain both embryonic Macs and Macs derived from BM monocytes 8,10.
In striking contrast, the cellular origin and self-maintenance capacity of Macs in the peripheral nervous system (PNS) remains largely unknown. Neither their gene signature nor ontogeny has been characterized so far. Furthermore, we have at present no idea whether the PNS is permissive to long term BM-derived macrophage engraftment upon injury or whether the macrophage pool reverts to the original resident macrophage population that was present before the injury, as is observed in the CNS9,11,12. To investigate this, we used a model of sciatic nerve crush, which leads to Wallerian degeneration. During Wallerian degeneration, the axons distal from the site of the crush disintegrate and the myelin debris generated is cleared to create a permissive environment for axon regrowth. Macs play an essential role in this process, not only to clear the myelin debris and provide nerve growth factors 13, but also to induce new blood vessel formation via VEGF, which enables Schwann cells to align along these vessels and guide newly outgrowing axons14.
Through a combination of genetic lineage tracing, parabiosis, and shielded irradiation experiments we determined the ontogeny and self-maintenance capacity of the resident Macs in healthy and injured PNS (sciatic nerve). We combined this with bulk- and single-cell RNA sequencing to define their transcriptomic signature. Our study identifies two distinct, spatially separated snMacs populations and indicates that bulk transcriptomics overestimates the plasticity of Macs by pooling resident and recruited Macs. Moreover, we report that snMacs differ strongly from CNS microglia in terms of ontogeny, gene expression profile and response to injury.
Results
Sciatic nerve macrophages possess a unique gene expression profile
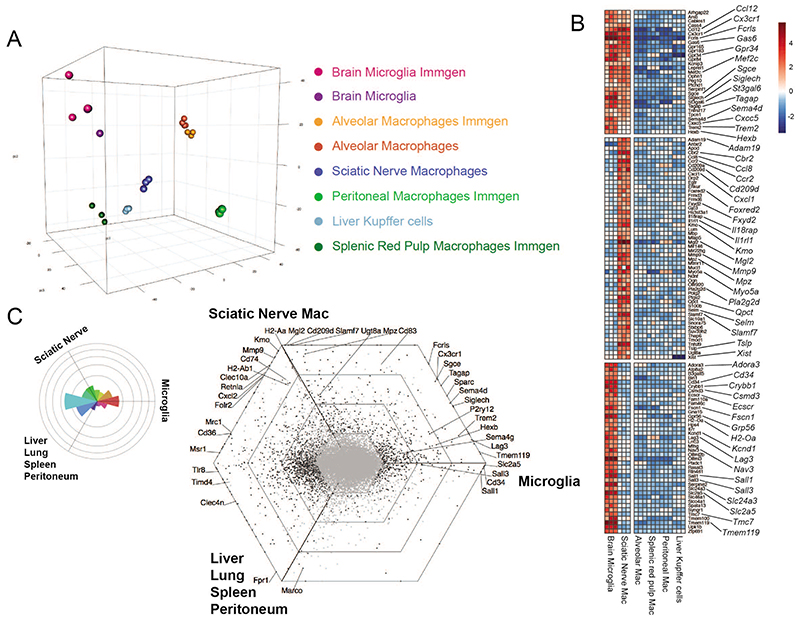
Macrophages are strongly imprinted by their tissue of residence15–17. To compare the gene expression of sciatic nerve macrophages (snMacs) to other tissue resident Macs we performed a micro-array profiling utilizing the same isolation protocols, amplification kits and micro-array chips as the Immgen Consortium (www.immgen.org). To ensure compatibility of the acquired data, we compared the Immgen profiles to our in-house profiling of brain microglia and alveolar macrophages. The transcriptomic profile of liver Kupffer cells was reported previously18,19. Digestion of tissues can induce the expression of immediate early response genes 10,20,21. Since each tissue is digested differently, we decided to remove the immediate early response genes to avoid that these genes would be over-represented in a given macrophage population. The principle component analysis (PCA) of six different tissue-resident Macs confirmed that each macrophage population possesses a distinct gene expression profile and is profoundly adapted to its tissue of residence (Figure 1A). Macs profiled in-house or by Immgen clustered together. Surprisingly, snMacs did not cluster with brain microglia in the PCA (Figure 1A). Brain microglia and snMacs shared some genes, including Ccl12, Cx3crl, Fcrls, Gpr34, Hexb, Mef2c, Sema4d, Siglech and Trem2, but many of those genes were more highly expressed in brain microglia (Figure 1B). Other core brain microglia enriched genes like Cd34, Ecscr, Kcnd1, Lag3, Nav3, Sall1, Sall3, Slc2a5, Sparc, Tmc7 or Tmem119 were completely lacking in snMacs.
Figure 1. Sciatic nerve macrophages possess a unique gene expression profile.
A. PCA analysis of transcriptional profiles of brain microglia, alveolar macrophages, sciatic nerve (SN) macrophages, liver Kupffer cells, peritoneal macrophages or splenic red pulp macrophages. Transcriptional profiles were obtained from cells sorted in-house or from the Immgen consortium (www.immgen.com). snMacs were sorted as CD45+F4/80+CD64+ live cells. B. Heat map showing differentially expressed (DE) genes in brain microglia versus sciatic nerve macrophages, or alveolar macrophages, liver Kupffer cells, peritoneal macrophages or splenic red pulp macrophages. Top panel shows genes shared between brain microglia and snMacs, middle panel and bottom panel show genes enriched in either brain microglia or snMacs. The heatmap displays the relative log2 normalized expression per gene. This was determined by calculating the mean expression value per gene over all cell types in the heatmap and then subtracting this mean from each cell type’s particular gene expression value. C. Rose plot and hexagonal “triwise” plot showing a visualization of DE genes between brain microglia, snMacs and all other macrophage populations analyzed in A and B. Bulk microarray data (A-B-C) were obtained from three independent replicates from three different experiments. The gating strategy for sorting snMacs is depicted in Supplementary Figure 1A.
The snMac profile, however, was enriched for several genes when compared with the other tissue-resident Macs such as Adam19, Cbr2, Cd209d, Foxred2, Fxyd2, Mgl2, Mmp9, ll1rl1, Kmo or Tslp (Figure 1B). A few genes picked up by this bulk transcriptomic profiling of snMacs appear to be typical Schwann cell-related genes (Mpz, Drp2, Ugt8a), which suggests that they could be derived from the phagocytosis of Schwann cells by snMacs. The relatively low similarity between CNS and snMac gene expression could also be visualized in a triwise plot where snMacs were compared to brain microglia or to Macs outside the nervous system (Figure 1C). As visualized by the Rose plot on the left, the majority of the genes were either shared between snMacs and other tissue Macs, or were microglia specific while only few genes were shared between microglia and snMacs. This indicates that snMacs possess a unique gene expression profile that is strikingly different from that of CNS microglia.
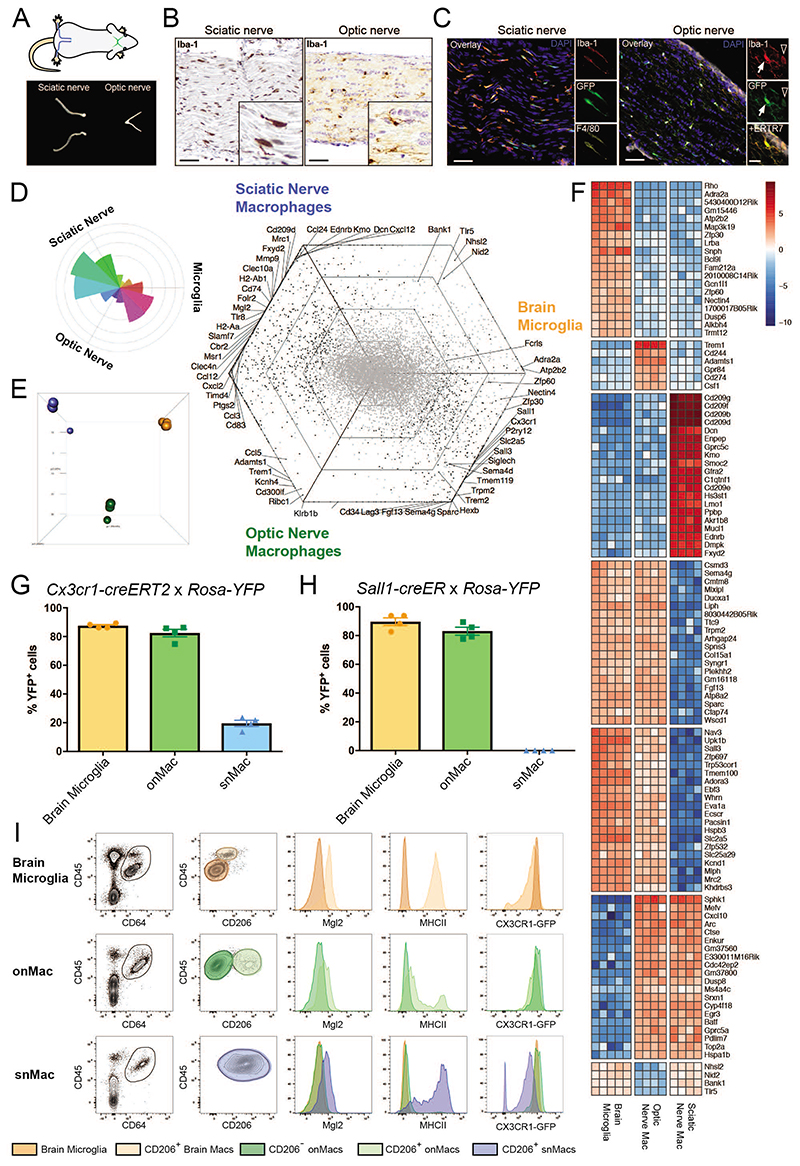
We next decided to compare snMacs and brain microglia to optic nerve Macs (onMacs) by bulk RNA-Sequencing. The cranial nerves are generally considered of PNS origin, but the optic nerve forms one of the exceptions as it arises as a tract of the diencephalon. Iba1+ Cx3cr1-GFP+ Macs could be found both within the sciatic and optic nerve bundles (Figures 2A, B, C). However, transcriptomic analysis revealed that snMacs cluster separately from onMacs (Figures 2D, E). When the major directionalities of genes co-expressed between the three types of nervous system Macs were analyzed in the triwise plot (Figure 2D), it was clear that onMacs cluster between snMacs and brain microglia, sharing genes with each subset, while again only a few genes were found in common between brain microglia and snMacs. Similarly, to what we found before for the bulk transcriptomic analysis of snMacs, the bulk transcriptomic analysis of onMacs revealed the presence of genes known to be expressed by CNS tissue cells and not by Macs themselves such as Adra2a, Atp2b2, Fgf13, Trpm2, Zfp30 or Zfp60 (expressed by brain astrocytes, neurons or endothelial cells respectively) (for single-cell analysis of these transcripts, see http://dropviz.org/)22 (Figures 2D, F). It should also be noted that in the bulk-sequencing of the onMacs, a minor fraction of CD206+ Macs might be present (Figure 2I, gating strategy Supp Figure 1). This confirms that we have to be cautious when analyzing bulk transcriptomic signatures of Macs 20. Nonetheless, it allowed the evaluation of the expression of typical microglia genes in onMacs and snMacs. In general, most genes characteristic for CNS microglia were higher expressed in onMacs compared to snMacs (Figure 2D). Even for the microglia-associated genes we found earlier to be present in PNS (such as Ccl12, Cx3cr1, Fcrls, Gpr34, Hexb, Mef2c, Sema4d, or Siglech - Figures 1 B,C), their expression level in PNS was always lower compared to onMacs or microglia (Figure 2D). Several other microglia core genes (such as Cd34, Ecscr, Kcnd1, Lag3, Nav3, Sall1, Sall3, Slc2a5, Sparc, Tmc7 or Tmem119) were shared between brain microglia and onMacs, but were completely lacking in snMacs (Figures 1C, 2D, F).
Figure 2. Optic nerve macrophages are closer to brain microglia than snMacs.
A. Scheme of sciatic nerves (blue) and optic nerves (green) and representative pictures of isolated nerves. B. Light microscopy images showing Iba1+ macrophages (brown) in sciatic nerve and optic nerve. Scale bar, 50μm. C. Immunofluorescence microscopy images showing Iba1+F4/80+Cx3cr1gfp+ sciatic nerve macrophages and Iba1+Cx3cr1gfp+ optic nerve macrophages both inside the optic nerve (arrow) and within the surrounding ERTR7+ meninges (arrowhead). Scale bar, 50μm, insets 20μm. D. Rose and hexagon “triwise” plots of genes detected by bulk RNA sequencing in snMacs, onMacs and brain microglia. snMacs were sorted as CD45+F4/80+CD64+ live cells, onMacs and microglia were sorted as CD45LoCD11bhi live cells. E. Principle component analysis (PCA) of the bulk RNA-sequencing transcriptome profiles. Sciatic nerve macrophages in blue, optic nerve macrophages in green and brain microglia in orange. F. Heatmap of the top genes differentially expressed or shared between the brain microglia, optic nerve macrophages or sciatic nerve macrophages. The heatmap displays the relative log2 normalized expression per gene. This was determined by calculating the mean expression value per gene over all cell types in the heatmap and then subtracting this mean from each cell type’s particular gene expression value. G, H. Adult Cx3cr1creER:R26-YFP (G) or Sall1creER:R26-YFP (H), were injected with TAM and YFP expression was analyzed after two weeks by flow cytometry. Each symbol represents one mouse. I. Expression of CD206, CD45, Mgl2, MHCII and CX3CR1-GFP by CD45+CD64+ macrophages measured by flow cytometry. Plots are depicting CD206- brain microglia (orange) and CD206+ brain macrophages (light orange) as well as CD206- (green) and CD206+ (light green) onMacs. Sciatic nerve macrophages (blue). Histograms show the expression levels of MGL2, MHCII and Cx3cr1-GFP in the individual populations. Each histogram comes from four pooled mice. (B, C) Representative data from two independent microscopy or flow cytometry experiments. (D, E, F) Bulk RNA-sequencing data were obtained from four to five independent replicates from at least three different experiments. For snMacs and onMacs, cells from five mice were pooled per replicate. (G, H) Pooled data coming from 2 independent experiments (4 mice in total). (I) Representative data from two independent flow cytometry experiments. The gating strategy for sorting snMacs is depicted in Supplementary Figure 1B.
The absence or lower expression of microglia-associated genes in snMacs is of importance for the functional study of snMacs in vivo. Neither the Cx3cr1-creERT2, nor the Sall1-creER systems, designed to target microglia, resulted in strong labeling of snMacs (four times lower labeling with the Cx3cr1-creERT2 mice, no labeling at all with the Sall1-creER mice), while onMacs were targeted to a similar extent as microglia (Figures 2G, H).
In summary, snMacs express a unique transcriptional profile with only a partial overlap with the CNS microglia core signature.
The sciatic nerve lacks microglia-like macrophages
In addition to the fact that the bulk transcriptomic profile of Macs can be contaminated by surrounding tissue cells, there is also the risk to pool different subtypes of Macs residing in distinct micro-environments within a given tissue. It has recently been demonstrated that the brain harbors border- or CNS-associated Macs (BAMs or CAMs) that express a different transcriptomic and surface receptor profile compared to microglia14,15,23. Microglia can be distinguished from BAMs by lower expression of the MHC class II complex (encoded by H2-Aa, H2-Ab1 and Cd74), CD206 (encoded by Mrc1), and CD45. Mrc1, H2-Aa, H2-Ab1 and Cd74 were enriched in snMacs as compared to other tissue-resident Macs (Figure 1C) or microglia and onMacs (Figure 2D). snMacs also expressed high Mgl2 (Figures 1C, 2D). We therefore evaluated the protein expression of these surface receptors. We distinguished CD45HiCD206Int BAMs from CD45LowCD206Low microglia in the brain as described before8,10. CD45HiCD206Int BAMs were found to express higher levels of Mgl2 and MHCII. The ON harbored Macs with a similar surface expression profile as in the brain: a dominant population of CD45LowCD206Low ON microglia and a minor population resembling CD45HiCD206Int BAMs (Figure 2I). In contrast, all snMacs expressed high levels of CD206, and Mgl2. The MHC class II expression in this population varied from low to high. We further investigated the expression of CX3CR1 in Macs subsets derived from Cx3cr1 GFP/WT mice. snMacs had a lower expression of CX3CR1 as compared to brain microglia and ON microglia (confirming the position of Cx3cr1 on the Triwise plot as being enriched in onMacs and microglia – Figure 2D). Note, that some of the snMacs expressed little or no CX3CR1 (Figure 2I). Altogether, this transcriptomic and proteomic profiling of nervous system-associated Macs indicated that the PNS lacks CNS-like microglia. In contrast, microglia represented the dominant macrophage population in the brain and the optic nerve: these cells express microglia “core” genes such as Cd34, Ecscr, Fscn1, Gpr34, Kcnd1, Nav3, Sall1, Sall3, Slc2a5, Sparc and Tmem119, display a similar CX3CR1-GFPHiCD45LowCD206Low MHCIILowMgl2Low surface expression profile and are both efficiently targeted by Sall1-creERT2 or Cx3cr1- creERT2 mice. Surprisingly, these cells are not present in the sciatic nerve and this prompted us to further characterize the snMacs.
Sciatic nerve macrophages derive predominantly from late embryonic precursors that are slowly replaced by BM-derived monocytes
Microglia in the brain derive from early embryonic precursors that give rise to yolk sac Macs (YS Macs), which seed the brain around E9.57,10,25. Subsequent waves of precursors, including embryonic fetal liver monocytes and hematopoietic stem cell (HSC)-derived monocytes do not engraft in the microglia pool, resulting in the pure early embryonic origin of microglia 23,24. As snMacs do not contain any microglia-like cells, we next sought to unravel the ontogeny of snMacs using multiple fate mapping models.
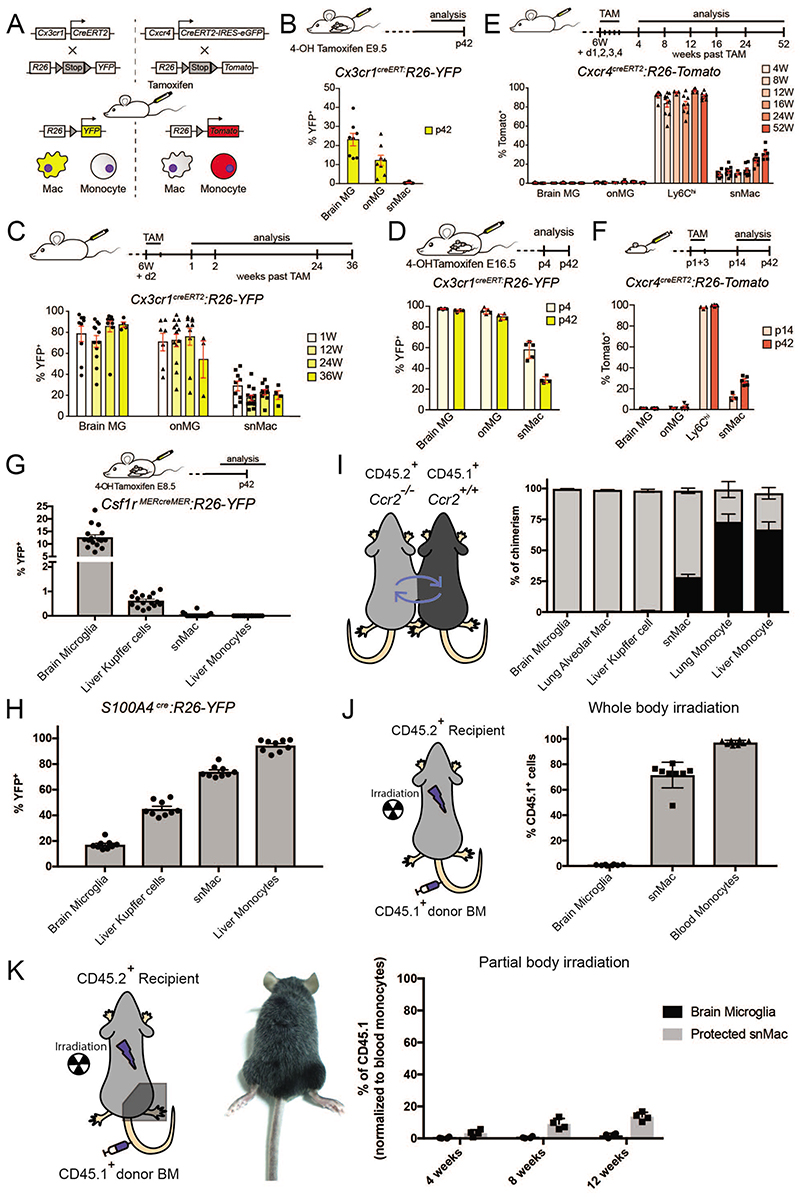
To tag early embryonic YS Macs we treated Cx3cr1-creERT2 × Rosa26-YFP pregnant mice at E9.5 with tamoxifen (TAM)25. In addition, to label HSC-derived circulating monocytes we treated adult Cxcr4-creERT2 × Rosa26-Tomato mice with TAM (Figure 3A). Administration of TAM to Cx3cr1-creERT2 × Rosa26-YFP pregnant mice at E9.5, labeled microglia in the brain and onMacs in adult mice, but surprisingly not snMacs (Figure 3B). Administration of TAM to adult Cx3cr1-creERT2 × Rosa26-YFP mice labeled both microglia in the brain and onMacs and these cells remained highly labeled for up to 36 weeks (Figure 3C). snMacs, were less efficiently labeled but their labeling was grossly maintained for 36 weeks and only dropped slightly, implying that all nervous system tissue-associated Macs possess a relatively strong self-maintenance capacity. Administration of TAM to Cx3cr1-creERT2 × Rosa26-YFP pregnant mice at E16.5 labeled microglia in the brain and ON and this labeling was maintained. In contrast, the snMacs labeling dropped by half in adult mice, suggesting a partial replacement of late embryonic snMacs by BM-derived monocytes in the first weeks of life (Figure 3D).
Figure 3. Ontogeny of sciatic nerve macrophages and brain microglia is completely different.
A. Scheme of the TAM induced recombination in either macrophages or monocytes in the Cx3cr1CreERT2:R26-YFP and Cxcr4CreERT2: R26- Tomato reporter mouse lines. B. YFP expression in yolk sac derived macrophages was induced by 4-OH TAM injection in pregnant Cx3cr1CreERT2:R26-YFP mice at embryonic day (E)9.5. The percentage of YFP+ brain microglia (MG), onMacs and sciatic nerve macrophages (snMac) was quantified by flow cytometry at postnatal day (p)42. C. YFP expression was induced by TAM injection in six weeks (6W) old Cx3cr1CreERT2:R26-YFP mice. The percentage of YFP+ brain MG, onMacs, and snMacs was quantified by flow cytometry. D. YFP expression in embryonic macrophages was induced by 4-OH TAM injection in pregnant Cx3cr1CreERT2:R26-YFP mice at E16.5. The percentage of YFP+ brain MG, onMacs and snMacs was quantified by flow cytometry. E. F. Tomato expression was induced by injecting TAM at 6W of age (E) or at postnatal days (p)1+3 (F) in Cxcr4CreERT2:R26-Tomato mice. The percentage of Tomato+ brain MG, onMacs, Ly6Chi blood monocytes (Ly6Chi) and snMacs was quantified by flow cytometry. G. YFP expression in yolk sac derived macrophages was induced by 4-OH TAM injection in pregnant Csf1rMERcreMER:R26-YFP mice at E8.5. The percentage of YFP+ brain MG, ON MG, Ly6Chi and snMacs was quantified by flow cytometry at p42. H. The percentage of YFP+ brain MG, Kupffer cells, snMacs and liver monocytes was quantified by flow cytometry in adult S100A4Cre:R26-YFP mice. I. Parabiotic mice were generated by suturing together CD45.1+ WT and CD45.2+CCR2-/- mice. The percentage of cells of CD45.1+ WT donor origin was determined in the macrophages and monocytes from indicated tissues from the CD45.2+CCR2-/- parabionts. Data represent two independent experiments involving four independent parabionts. J. K. CD45.2+ recipient mice underwent full-body irradiation (J) or partial-body irradiation with one leg protected by a lead shield (K). Mice were reconstituted with CD45.1+ donor BM and percentage of CD45.1+ brain MG, snMacs and blood monocytes was quantified by flow cytometry. All data are shown as mean ±SEM. Symbols represent individual animals. (B) Pooled data coming from 3 independent experiments (8 mice in total). (C) Pooled data from 3 independent experiments (at least 9 mice in total), except for week 36 (2 independent experiments with at least 3 mice). (D) Data from 2 pregnant females. Offspring divided over 2 time points (p4 (5 mice), p42 (4 mice). (E) Pooled data from 2 independent experiments (with at least 6 mice), except for week 12 (1 experiment with 4 mice). (F) P14 time point: 1 experiment with 3 mice; p42: data pooled from 2 independent experiments with 6 mice in total. (G) Pooled data from 2 independent experiments with 16 mice in total. (H) Pooled data from 2 independent experiments with 9 mice in total. (I) Data represent two independent experiments involving four independent parabionts. (J) Pooled data of two independent experiments with 8 mice in total. (K) One experiment shown of two independent experiments with 4 mice per experiment.
Administration of TAM in adult Cxcr4-creERT2 × Rosa26-Tomato mice efficiently labeled circulating monocytes (Figure 3E). In contrast, brain and ON microglia did not show any labeling, confirming that circulating monocytes do not engraft in the brain or ON microglia pool during adulthood. In contrast, snMacs showed low labeling and this increased very slowly with time in adult mice, confirming their strong self-maintenance capacity. Administration of TAM in neonatal Cxcr4-creERT2 × Rosa26-Tomato mice also efficiently labeled circulating monocytes without labeling brain or ON microglia, while the labeling of snMacs doubled between the second and the sixth week of life (Figure 3F). To further substantiate these findings we utilized additional fate-mapping tools. Treating Csf1r-Mer-CRE-Mer × Rosa26-YFP pregnant mice with TAM at E8.5, labeled microglia but not snMacs, demonstrating by two independent fate-mapping models that snMacs do not derive from early YS Macs (Figures 3G). We next tested the S100a4-CRE × Rosa26-YFP reporter mice. In these mice, YS Macs are lowly labeled (around 10 %), late embryonic fetal liver monocytes show an intermediate labeling (around 40 %), while adult circulating HSC-derived monocytes are highly labeled (around 90 %) 26. Microglia showed low YFP labeling, fitting with a YS Mac origin, while snMacs displayed YFP labeling that was around 65 % (Figure 3H). This indicates that snMacs do not derive entirely from circulating monocytes, but could potentially represent a mixture of cells derived from intermediately labeled late embryonic fetal liver monocytes and highly labeled adult circulating monocytes. To verify the contribution of adult circulating monocytes we generated parabiotic mice (Figure 3I). CD45.1+Ccr2+/+ mice were surgically attached to CD45.2+Ccr2-/- mice. As monocytes require CCR2 for their exit of the BM27, monocytes in CD45.2+Ccr2-/- recipient mice are predominantly derived from the CD45.1+Ccr2+/+ parabiotic partner. While microglia, alveolar macrophages and Kupffer cells remained of almost pure host origin, about 20 % of the snMacs were derived from circulating precursors originating from the parabiotic partner after 8 weeks (Figure 3I). To further substantiate these findings we generated BM chimeras. A first group of CD45.2+ mice underwent whole body irradiation followed by injection of CD45.1+ BM. In these mice, microglia were largely radio-resistant and remained of pure CD45.2+ background. snMacs were radiosensitive, as most of these cells were replaced by CD45.1+ BM derived cells (Figure 3J). This whole body irradiation experiment suggested a much higher rate of replacement by BM monocytes as compared to the results utilizing the Cxcr4-creERT2 or the parabiosis experiments. As this could be due to an artificial irradiation-induced myeloid engraftment, as we have described before for microglia 28, we also performed partial irradiation chimeras in which the right hind leg, and thus the sciatic nerve, was shielded from irradiation (Figure 3K). The snMacs that were shielded from irradiation still contained some cells derived from the CD45.1+ BM cells but the increase in this chimerism level as a function of time was very slow with still less than 20 % of the snMacs deriving from CD45.1+ BM cells 12 weeks after the BM transfer. Altogether, these experiments indicate that, in contrast to CNS microglia, snMacs are not derived from early embryonic precursors, but are primarily derived from late embryonic precursors that get replaced gradually by circulating monocytes after birth. In the first four to six weeks of life this replacement is relatively fast with about half of the embryonic population being replaced by circulating monocytes. In adult life, the replacement by circulating monocytes is much slower with a half-life of more than six months (Figures 3C, 3E, 3K). snMacs are therefore not only different from CNS microglia in terms of transcriptomic identity and surface receptor profile, but also in their ontogeny.
Macrophage gene expression assessed by bulk sequencing strongly fluctuates upon sciatic nerve injury
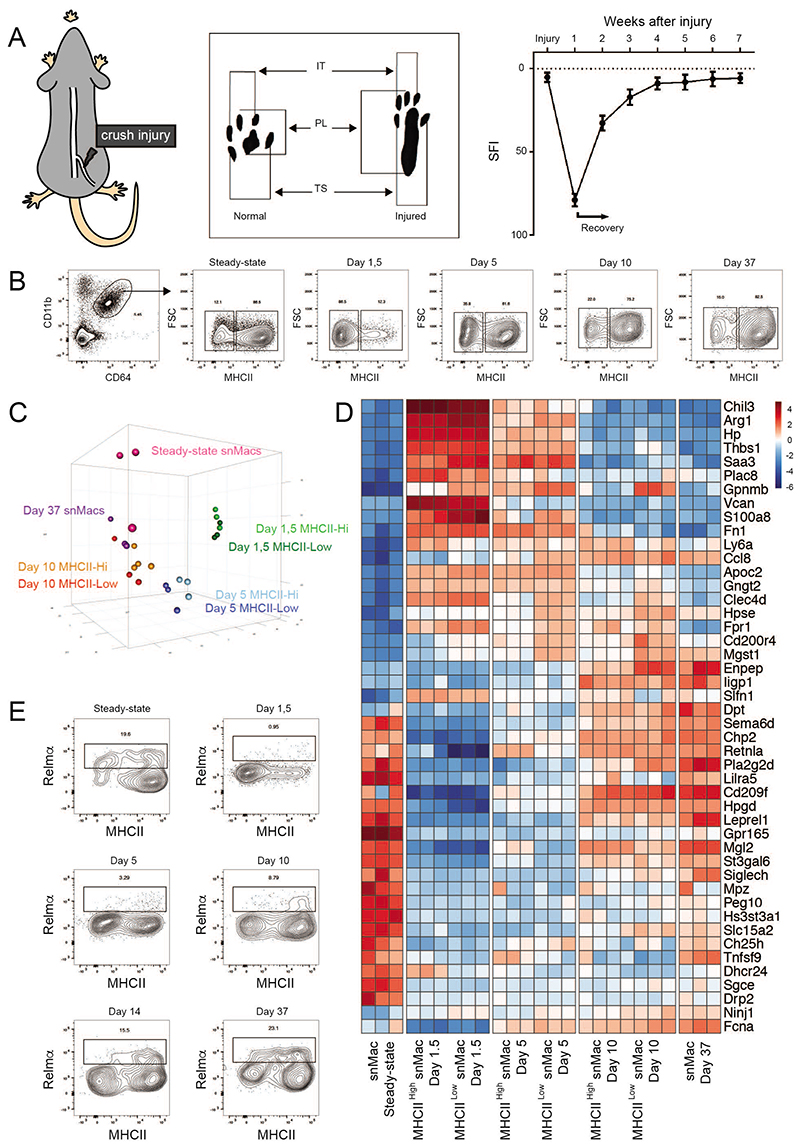
To assess the effect of peripheral nerve injury on snMac activation, we adopted a model of sciatic nerve crush. In this nerve injury model, mice suffer from a maximal injury score one week post injury (as reflected by the sciatic nerve functional index (SFI), based on foot print analysis), but then start recovering thanks to the regenerative capacity of the PNS (Figure 4A). We isolated the nerve part distal from the injury, which is the part of the nerve that will disintegrate completely during the process of Wallerian degeneration. We first profiled Macs by flow cytometry at multiple time-points after a sciatic nerve crush (Figures 4A, B), and this revealed a marked drop in MHC class II expression in snMacs upon injury and a restoration of MHC class II levels upon recovery. As MHC class II has been proposed to be characteristic of classical “M1” macrophage activation29, this would fit into a theoretical model where snMacs acquire a reparative alternative “M2” activation during nerve injury as previously proposed 30,31. To characterize macrophage activation during nerve injury, we performed bulk RNA-sequencing at multiple time-points after sciatic nerve crush (Figure 4C). We used MHCII as a distinguishing marker to sort MHCIILow and MHCIIHi Macs, following the proposed “M1/M2” assumption that the presence or absence of MHCII could define two different activation states of Macs. At day 1.5, 5 and 10 post crush, we obtained enough cells for both MHCIIHi and MHCIILow subsets for RNA sequencing. For steady-state and snMacs from day 37 post crush almost all Macs were MHCIIHi and the cells were sorted as one population. PCA analysis revealed that day 1,5 and day 5 Macs differed greatly from steady-state snMacs, but that in the following days the gene expression profile of the snMacs gradually returned to that observed under homeostatic conditions. In addition, MHCIIHi and MHCIILow cells of the various time-points strongly clustered together indicating that high or low MHC class II expression does not correlate with a different macrophage activation state. In fact, further analysis of the differentially expressed genes exposed the severe limitations of applying the in vitro M1/M2 concept to in vivo Macs. Relmα (encoded by Retnla), Mgl2, Ym1 (encoded by Chil3) and Arginase (encoded by Arg1) are all considered prototypical “M2” markers 32, but these were not always co-expressed and their expression profiles did not correlate with higher or lower MHCII expression (Figure 4D). Mgl2 and Retnla were already highly expressed in steady-state snMacs and dropped upon injury, while Arg1 and Chil3 were strongly up-regulated during injury and dropped again upon recovery. Strikingly, many of the genes that represent the “core” snMac signature (Figure 1B) were strongly down-regulated upon injury (Figure 4D). This suggests that snMacs lose their tissue identity and acquire a distinct activation state upon nerve injury. To investigate this further, we examined the protein expression of Relmα+. While this confirmed a drop of the Relmα+ expression on snMacs upon injury, it also revealed that Relmα is in fact only expressed by a small fraction of steady-state snMacs, suggesting a previously unappreciated macrophage heterogeneity in the sciatic nerve which could severely endanger our conclusions made by bulk RNA sequencing. Thus, to allow for this heterogeneity, we continued to examine the snMacs using single-cell RNA sequencing.
Figure 4. Peripheral nerve injury strongly affects snMacs gene expression.
A. Graphical representation showing the crush injury model (left panel) and an example of a footprint diagram (middle panel), which is analysed at different time points upon the crush injury and results in a typical SFI (sciatic functional index) score (right panel). B. Gating strategy to identify MHCIIlow and MHCIIhi macrophages that were sorted at different time points upon crush injury. C. PCA analysis showing clustering of the different transcriptional profiles obtained from microarray analysis of different sorted macrophage populations at different time points upon injury. D. Heat map analysis showing DE genes between the MHCIIlow and MHCIIhi populations isolated at different time points upon analysis. The heatmap displays the relative log2 normalized expression per gene. This was determined by calculating the mean expression value per gene over all cell types in the heatmap and then subtracting this mean from each cell type’s particular gene expression value. E. Flow cytometry analysis showing Relmα expression in SN MHCIIlow and MHCIIhi populations at different time points upon injury. The replicate bulk transcriptomic profiles (C,D) of each individual time-point were acquired during 3 independent experiments. The flow cytometry plots in (B) are representative plots of at least 3 mice per time-point performed in 3 independent experiments. (E) Flow cytometry plots are representative for at least two independent experiments, except for d10 (1 experiment with 3 mice).
Single-cell RNA sequencing reveals two major subsets of steady-state snMacs residing in distinct locations
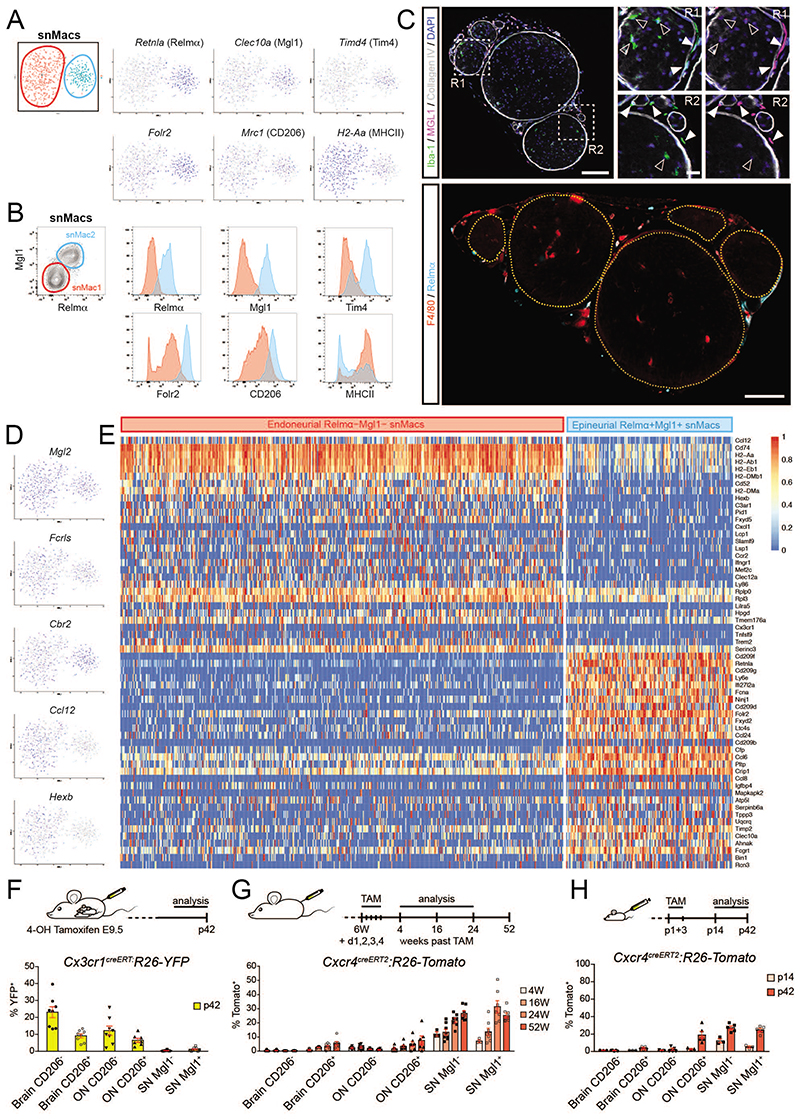
To better tackle snMac heterogeneity, we first performed single-cell RNA sequencing (scRNA-seq) on steady-state snMacs. This readily revealed the presence of two subsets of snMacs: a bigger snMac1 cluster and a smaller snMac2 cluster (Figure 5A). The smaller blue snMac2 cluster expressed higher levels of Retnla (coding for Relma), Clec10a (coding for Mgl1), Folr2, Timd4 (coding for TIM4) and Mrc1 (coding for CD206) (Figure 5A). Retnla, Clec10a and Folr2 were homogeneously expressed by almost all Macs in the blue cluster and were therefore unlikely to represent contaminations from the occasional uptake of surrounding tissue cells. We therefore verified their surface expression by flow cytometry (Figure 5B). This provided surface markers to distinguish Relmα+Mgl1+ snMacs2 from Relmα- Mgl1- snMacs1. These surface markers were then used to localize Relmα+Mgl1+ snMacs2 and Relmα Mgl1 snMacs1 by confocal microscopy. Strikingly, Relmα+Mgl1+ Iba-1+ snMacs2 were found to reside in the epineurium, the connective tissues surrounding nerve fascicles, while Relmα- Mgl1- Iba-1+ snMacs1 were found inside the endoneurium of the sciatic nerve (Figure 5C), indicating the presence of distinct anatomical macrophage niches in the sciatic nerve17.
Figure 5. Single-cell sequencing reveals the presence of two macrophage subsets in the sciatic nerve.
snMacs were sorted for single-cell RNA sequencing as F4/80+CD64+ cells. A. tSNE plots of about 600 snMacs revealing the presence of two snMac subsets (red and blue) with distinct gene expression patterns. B. Representative flow cytometry of snMacs confirming the identification of two snMac subsets at the protein level. snMac1 (red) were Relmα+Mgl1+, whereas snMac2 (blue) were Relmα-Mgl1-. Other markers are shown in histograms. Representative plots are shown from 2 independent experiments. C. Confocal microscopy of sciatic nerve vibratome cross sections from C547BL/6 mice. Top: Location of Iba-1+Mgl1- snMac1 (filled arrowheads) and Iba-1+Mgl+ snMac2 (open arrowheads) is depicted. The perineurium and basal membranes of blood vessels were stained with Collagen IV. Nuclei were counterstained with DAPI. Images are representative of at least five individual mice. Bottom: Nerve fascicle boundaries are marked by dashed yellow delineations. Location of F4/80+Relmα- snMac1 and F4/80+Relmα+ snMac2 is depicted. Scale bar: overview 100μm, insets 20μm. D. E Single-cell tSNE plot showing the expression profile of individual genes (D) and heatmap of the differentially expressed genes between Relmα+Mgl1+ snMacs and Relmα-Mgl1- snMacs (E). The heatmap shows the scaled log2 normalised expression values. The log2 normalised expression values were scaled using the ‘scale_quantile’ function of the SCORPIUS package with default parameters (v1.0). F. YFP expression in yolk sac derived macrophages was induced by 4-OH TAM injection in pregnant Cx3cr1CreERT2:R26-YFP mice at 9.5. The percentage of YFP positive CD206- brain microglia, CD206+ brain Macs, CD206- onMacs, CD206+ ON Macs, Mgl1- snMac1 and Mgl1+ snMac2 was quantified by flow cytometry at postnatal day (p)42. G, H. Tomato expression was induced by injecting TAM at 6W of age (G) or at postnatal days (p)1+3 (H) in Cxcr4CreERT2:R26-Tomato mice. The percentage of Tomato+ CD206- brain microglia, CD206+ brain Macs, CD206- ON microglia, CD206+ ON macrophages, Mgl1- snMac1 and Mgl1+ snMac2 was quantified by flow cytometry. All data are shown as mean ±SEM. Symbols represent individual animals. (F) Data pooled from 3 independent experiments with in total 8 mice. (G) Data from 2 independent experiments with at least 7 mice, except for week 4 (1 experiment with 3 mice). (H) P14 time point: 1 experiment with 3 mice; p42: data pooled from 2 independent experiments with 6 mice in total.
We further compared the transcriptomic identity of epineurial Relmα+Mgl1+ snMacs and endoneurial Relmα- Mgl1- snMacs. First, we evaluated the expression of the “core” genes earlier found via bulk transcriptomic profiling to be enriched in snMacs as compared to other tissue-resident Macs (Figure 1B, Supplementary Figure 2A). This revealed that most genes defined as snMac genes by bulk sequencing appeared more highly expressed in either epineurial Relmα+ Mgl1+ snMacs (e.g. Cc18, Cd209a, Cd209d, Fxyd2, Tslp and Mmp9) or in endoneurial Relmα Mgl1 snMacs (e.g. Ccr2, Cxcl1, ll1rl1, Selm, Pla2g2d, Qpct, Tnfsf9, Xist). A few genes were highly expressed in both snMac populations (e.g. Cbr2, Fcrls, Gas6 and Mgl2)(Supplementary Figure 1A and Figure 5D). Interestingly, the endoneurial Relmα-Mgl1- snMacs expressed many CNS microglia-associated genes like Ccl12, Cx3cr1, Gpr34, Gpr183, Hexb, Mef2c, St3gal6, Tagap and Trem2 (Supplementary Figure 2A and Figure 5D-E). This implies that the shared CNS microglia signature picked up in the bulk analysis between brain microglia and snMacs was coming almost entirely from the endoneurial Relmα- Mgl1- Iba-1+ snMacs that reside within the nerve fascicles, in close contact with neurons and Schwann cells. This was also associated to a stronger Cx3cr1GFP signal in endoneurial Macs in the Cx3cr1 GFP/+ reporter mice (Supplementary Figures 2B-E).
Next, we directly compared the epineurial Relmα+Mgl1+ snMacs with the endoneurial Relmα- Mgl1- snMacs (Figure 5E). In addition to the microglia-associated genes discussed above, endoneurial Relmα- Mgl1- snMacs also expressed many genes not found in CNS microglia like Ccr2, Cd74, H2-Aa, H2-Ab1, H2-Eb1, H2-DMa, H2-DMb1, Fxyd5, Slamf7, Lsp1, Lilra5 and Tfnsf9. Unexpectedly, we also found that the immediate early response genes10,20, such as Atf3, Fos, Jun or ler2 were more strongly expressed by the endoneurial Relmα- Mgl1- snMacs as compared to epineurial Relmα+Mgl1+ snMacs (Supplementary Figure 3). This is surprising as both cells were present in the same sciatic nerve and therefore digested and processed together. This could suggest that endoneurial Relmα- Mgl1- snMacs are intrinsically more responsive and will more easily express immediate early response genes. Alternatively, the endoneurial Macs could also be subjected to more severe digestion stress than epineurial Macs, as the endoneurium could represent a tighter tissue and this increased mechanical stress could induce a stronger expression of immediate early response genes. The epineurial Relmα+Mgl1+ snMacs subset expressed higher levels of Cc18, Cd209a, Cd209d, Fxyd2, Tslp and Mmp9 (Supplementary Figure 3).
We have also compared the snMacs to CNS BAMs and microglia10 by combining both sc-RNA-seq datasets in one tSNE plot (Supplementary Figure 4). This illustrates that microglia are the most distinctive macrophages with unique expression of Sall1, Slc2a5, Cd34 and Sparc. Conversely, snMacs and BAMs display higher levels of Pf4 and Mgl2 as compared to microglia. Nonetheless, snMacs did not completely overlap with BAMs, as shown by stronger expression of Gpr65 in BAMs but not in snMacs, the stronger expression of Retnla and Cd209d in epineurial snMacs, the stronger expression of Lilra5 in endoneurial snMacs and choroid plexus BAMs (CP-BAMs) but not other BAMs, or the lower expression of Ly6e in endoneurial snMacs, for example.
The epineurial Relmα+Mgl1+ snMacs also displayed a higher expression of Lyve1 as compared to endoneurial Relmα-Mgl1- snMacs (Supplementary Figure 5). It was recently demonstrated that the lung interstitium contains two subsets of Macs: Lyve1HiMHCIILow Macs that are often associated with blood vessels and Lyve1LowMHCIIHi Macs that are in close interaction with nerve bundles33. We therefore stained for CD31+ blood vessels and LYVE1. Epineurial Relmα+Mgl1+ snMacs indeed expressed higher levels of Lyve1 (Supplementary Figure 5A, B). Moreover, these cells we often found in association with CD31+CollagenIV+ blood vessels (Supplementary Figure 5C, D), as is observed in the lungs.
Having identified these distinct populations, we next revisited the cellular origin of the different snMac populations. Confirming our previous analysis, we found no significant contribution from early embryonic YS Macs to either snMac population based on the absence of labeling in the adult offspring from pregnant Cx3cr1-creERT2 × Rosa26-YFP mice treated with TAM at E9.5 (Figure 5F) and equivalent low-grade engraftment by circulating monocytes in both snMac populations based on the small but steadily increasing labeling over time in adult Cxcr4-creERT2 × Rosa26-Tomato adult mice treated with TAM (Figure 5G). Both populations showed a stronger increase in labeling in the first week of life when Cxcr4-creERT2 × Rosa26-Tomato neonatal mice were treated with TAM (Figure 5H), underlining that the increased engraftment of monocytes in the snMac pool in the neonatal period applies to both snMac populations. Altogether, this indicates that epineurial Relmα+Mgl1+ snMacs and endoneurial Relmα- Mgl1- snMacs are both primarily derived from late embryonic precursors and both get slowly replaced by circulating monocytes after birth.
In summary, single-cell transcriptomic analysis, flow cytometry and confocal microscopy revealed the presence of two major snMac populations in the steady-state sciatic nerve. The differential localization of both subsets implies that only Relmα- Mgl1- snMacs are in close contact with nerve axons (and Schwann cells) inside the endoneurium and it is this population that expresses some of the CNS microglia associated genes (e.g. Ccl12, Cx3cr1, Gpr34, Gpr183, Hexb, Mef2c, St3gal6, Tagap, Trem2), while epineurial Relmα+Mgl1+ snMacs are not in contact with neurons and do not express these nervous-system-associated genes. This prompted us to evaluate the effect of crush injury on the distinct snMac populations using sc-RNA-seq.
Single-cell RNA sequencing reveals that the distinct snMacs subsets respond differentially to injury
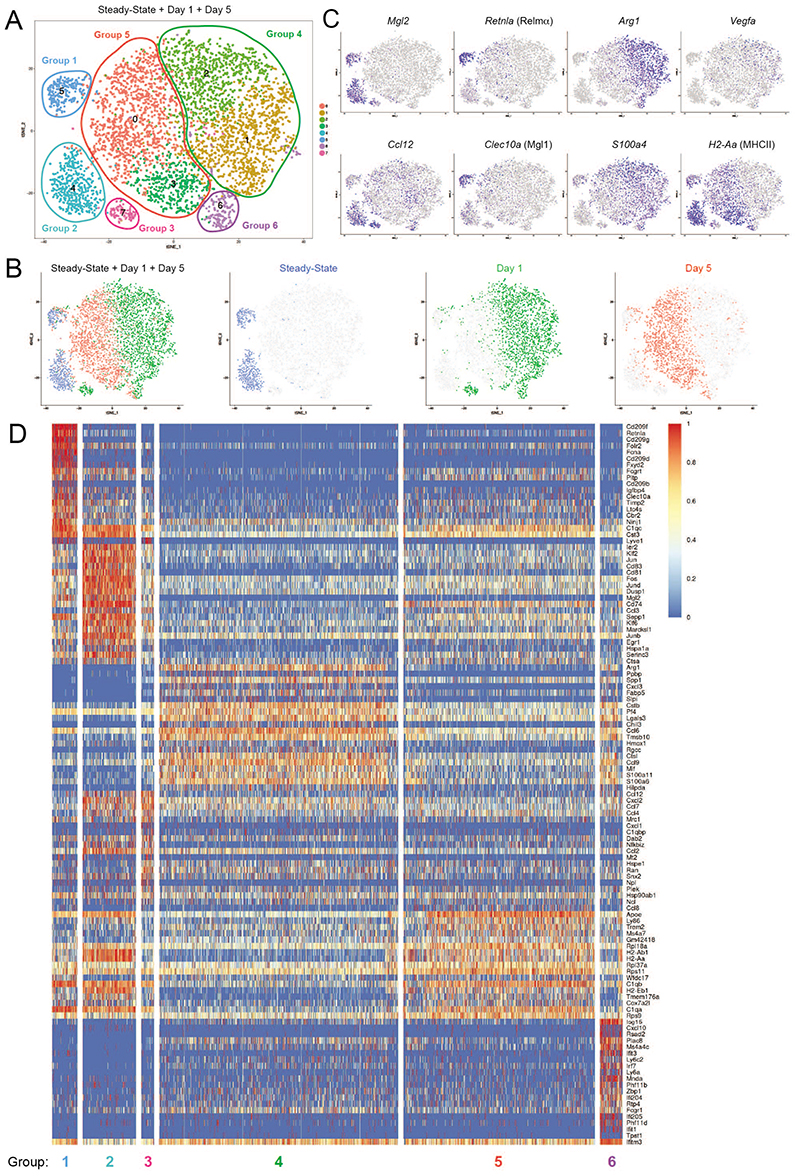
We next performed single-cell sequencing on day 1 and day 5 post crush to compare the changes in the gene expression profile measured by bulk RNA-sequencing and scRNA-seq analysis. This allowed us to investigate whether the changes in gene expression profile measured by bulk RNA-sequencing were occurring in both subsets of snMacs and to gauge the presence of recruited monocyte-derived Macs within the pool of snMacs. For this analysis we kept the early response genes in the heatmaps, as we reasoned the expression of some of these genes (like chemokines) could be influenced by the injury. The analysis revealed the presence of multiple small tSNE clusters that were fused into six main groups of cells (Figure 6A). We next mapped the cells coming from the distinct timepoints post injury on the tSNE plot (Figure 6B). Steady-state Macs clustered in group 1 and group 2 (Figure 6B). snMacs retrieved at day 1 post crush clustered mainly in group 4, but also in group 1, group 3 and group 6. snMacs retrieved at day 5 post crush clustered mainly in group 5, but also in group 1, group 2 and group 6. We next looked at the genes identified in the two snMac subsets in the steady-state (Figure 5). Group 1 represented epineurial Relmα+Mgl1+ resident snMacs as revealed by their high Retnla, high Clec10a, high Cbr2, high Mgl2 and low Ccl12 expression (Figure 6C-D) and contained cells from the steady-state, day 1 and day 5 post injury (Figure 6B-D). Group 2 represented endoneurial Relmα- Mgl1- resident snMacs as revealed by their low Retnla, low Clec10a, and high Cbr2, high Mgl2 and high Ccl12 expression (Figure 6C-D), but only contained cells from the steady-state and from day 5 (Figure 6B). Group 3 was only present at day 1 and represented endoneurial Relmα- Mgl1- resident snMacs (low Retnla, low Clec10a, high Cbr2, high Mgl2, high Ccl12). Endoneurial Relmα- Mgl1- snMacs thus clustered separately on day 1 post crush injury and analysis of the DE genes between group 2 and group 3 revealed that Relmα- Mgl1- snMacs had been temporarily activated by the crush injury, as shown by their increased expression of the monocyte-macrophage chemo-attractants Ccl6, Ccl7, Ccl8 and Ccl12. IPA analysis of the differentially expressed genes between group 3 and group 2 revealed “granulocyte adhesion and diapedesis” as one of the top 5 canonical pathways. Conversely, we found no differentially expressed genes among the epineurial Relmα+Mgl1+ resident snMacs between steady-state, day 1 and day 5, suggesting that as these cells are outside the endoneurium they might not sense the damage to the neurons or the Schwann cell activation. Group 4 represented a major population that was not present in the steady-state sciatic nerve, but was present at day 1 post injury and expressed high levels of S100a4, suggesting that this group represented recently recruited monocyte-derived Macs. These cells represented 90 % of all Macs present at day 1 post injury. Group 5 was present at day 5 only and again represented 90 % of snMacs, suggesting that these were the monocyte-derived Macs recruited upon injury that had further adapted to the SN environment. Group 6 represented a minor subset of Macs not present during the steady-state that appeared at day 1 post injury and largely maintained its gene expression profile at day 5.
Figure 6. Epineurial and endoneurial snMacs respond differently to peripheral nerve injury.
A. tSNE analysis of sc-RNA-Seq data reveals the presence of 6 major groups during nerve injury. B. tSNE plot as in A) with an overlay of different colors corresponding to cells isolated at different days post sciatic nerve injury. Steadystate macrophages cluster in Group 1 and Group 2. snMacs retrieved at day 1 postcrush cluster mainly in Group 3 and Group 4, but also (partially) in Group 1 and Group 6. snMacs retrieved at day 5 post-crush cluster mainly in Group 5 and partially in Group 6. C. tSNE plots showing expression of individual genes in the different clusters. D. Heat map showing the top differentially genes in each of the individual groups of the tSNE plot shown in (A). The heatmap shows the scaled log2 normalised expression values. The log2 normalised expression values were scaled using the ‘scale_quantile’ function of the SCORPIUS package with default parameters (v1.0).
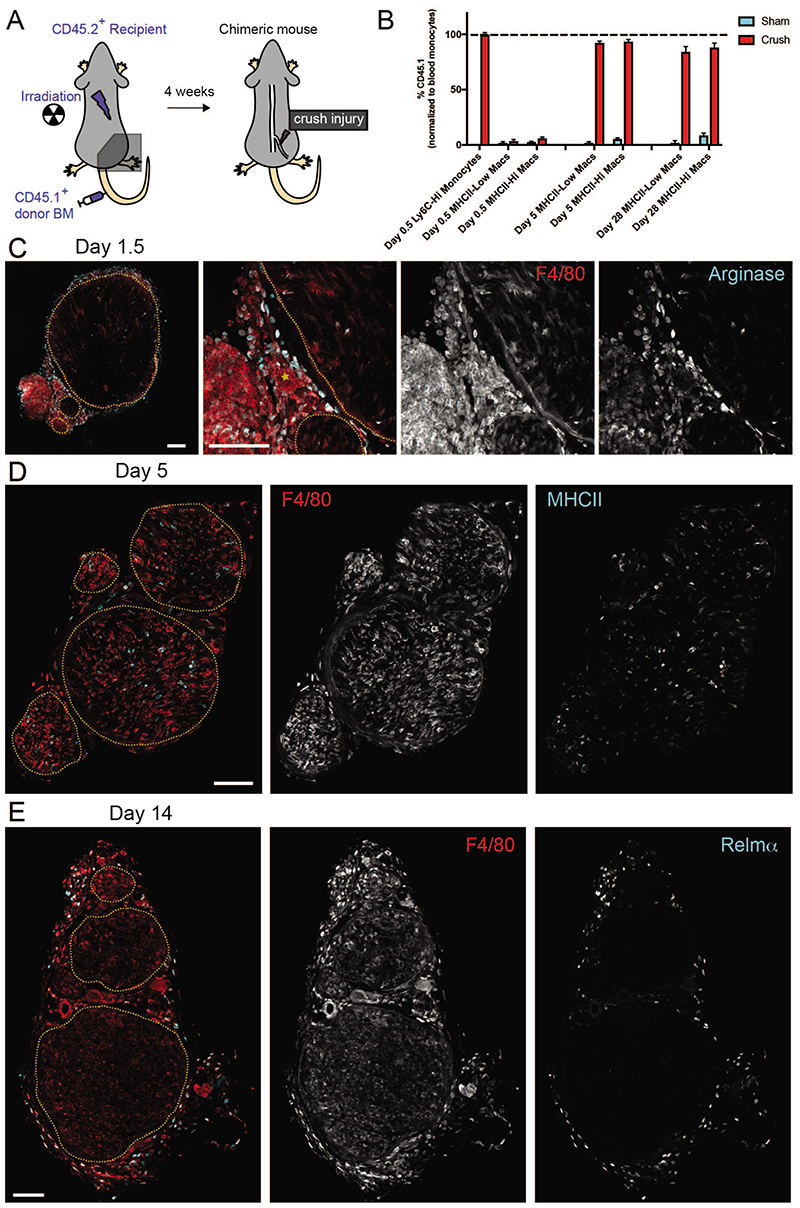
IPA analysis revealed that monocyte-derived Macs recruited at day 1 turned on functions associated with enhanced glucose metabolism (“glycolysis”, “sucrose degradation” and “granulocyte adhesion and diapedesis”), while on day 5 pathways associated with “antigen presentation” became prominent which fits well with the upregulation of the MHC class II level from day 1 to day 5 by flow cytometry (Figure 4B). Interestingly, the predominant signature associated with group 6, a small subset of monocyte-derived recruited Macs that was shared between day 1 and day 5 was “type I IFN signaling”. All in all, this annotation suggested that the resident snMacs would represent only 5 to 10 % of the total snMac population upon injury. To validate this by fate-mapping we partially irradiated mice, keeping their right hind leg (and sciatic nerve) protected from irradiation and reconstituted these mice with CD45.1 BM (Figure 7A). At day 0.5 we could still identify resident Macs based on the higher CD64 and lower Ly6C expression (Supplementary Figure 6A). Using the chimeras we could confirm that these Ly6CLowCD64Hi cells were still resident Macs of host origin, while the Ly6CHi monocytes were derived from the BM (Figure 7B). However, during the subsequent days the recruited monocytes differentiated into Macs following a process we have previously dubbed the “mono-mac waterfall”34,35, during which monocytes lose Ly6C expression and gain CD64 expression making it impossible to distinguish recruited macrophages from resident macrophages based on these markers. Interestingly, recruited Macs seemed to develop into both MHCIIHi and MHCIILow Macs. This was confirmed in the chimeric mice as at day 5 post injury, more than 90 % of the MHCIIHi and MHCIILow snMacs were derived from circulating BM-derived precursors (Figure 7B). This confirmed that in the sc-RNA-seq data only Groups 1, 2 and 3 represent resident snMac populations, while the remaining cells represent the dominant population of recruited monocyte-derived Macs. The strong recruitment of monocyte-derived Macs was also confirmed by the global expansion of the number of macrophages (Supplementary Figure 6B). Staining for Relmα by confocal microscopy confirmed that resident Relmα+Mgl1+ snMacs remained in the epineurium during the injury (Figure 7 and Supplementary Figure 7) and were therefore not in close contact with the injured axons, likely explaining why the gene expression profile of these cells was not affected by the crush injury. The increase in number of Macs observed by flow cytometry was also confirmed by confocal microscopy (Supplementary Figure 7).
Figure 7. The sciatic nerve is permissive to long-term monocyte engraftment after peripheral nerve injury.
A. Schematic representation of the partial-body irradiation experiment in which the right leg and its sciatic nerve were protected from irradiation. B. CD45.2+ recipient mice underwent partial-body irradiation with one leg protected by a lead shield. Mice were reconstituted with CD45.1+ donor BM and 4 weeks later the nerve that was protected from irradiation underwent a crush injury. The percentage of CD45.1+ snMacs and blood monocytes was quantified by flow cytometry at the indicated time-point post injury. All data are shown as mean ±SEM. Data represent 4 mice per time-point from two independent experiments. C. Arginase positive macrophages are present both in- and outside the nerve fascicles at 1.5 days after nerve crush. Overview image (left) and higher zoom (3 rightmost) images are shown. A blood vessel is marked by a yellow asterisk. D. MHCIIHi and MHCIILow macrophages are present both in and outside the nerve fascicles at 5 days after nerve crush. E. Relmα+ macrophages are excluded from the nerve fascicles and reside mostly in the perineurium and epineurium connective tissues at 14 days after nerve crush. In (C), (D) and (E), nerve fascicle boundaries are marked by yellow dashed delineations and F4/80 is used as a pan macrophage marker. Images are representative of nerves of 2 (C) or 3-5 mice. Scale bar = 100μm.
The study of the cellular origin of Macs during the repair process also revealed that one month after the injury, Macs were still predominantly derived from recruited circulating precursors and did not revert to the original resident macrophage population (Figure 7B). This is in strong contrast to what is observed for CNS microglia9,11,12, again confirming that the PNS macrophage niche is permissive for circulating monocytes to engraft.
Having experimentally confirmed the annotation of the tSNE clusters, we compared the differentially expressed genes obtained by bulk RNA sequencing (Figure 4D) and by scRNA-seq (Figure 6D). While the bulk sequencing suggested that resident Macs down-regulated Retnla and Mgl1 and acquired Arg1 and Chil3, the scRNA-seq experiment uncovered that Retnla and Clec10a expression is maintained in epineurial snMacs (tSNE group 1), while Arg1 and Chil3 are mainly expressed by recruited monocyte-derived Macs (tSNE group 4).
Interestingly, localization of the arginase-expressing monocyte-derived Macs indicated that F4/80Hi Macs already expressed arginase before entering the endoneurial compartment (Figure 7C). Relmα+ macrophages clearly localize outside the nerve fascicle (Figure 7E) and remain there throughout the course of the crush injury (Supplementary Figure 7). Note also that HLA-Aa Hi and HLA-Aa Low cells were identified in each of the tSNE Groups and immunofluorescence confirmed that MHCIIHi and MHCIILow Macs were found inside and outside the endoneurium (Figure 7D), explaining why MHCIIHi and MHCIILow cells did not differ in the bulk RNA-sequencing (Figure 4D).
Altogether, this suggests that epineurial Relmα+Mgl1+ resident snMacs do not significantly react to the nerve injury, while endoneurial Relmα- Mgl1- resident snMacs react by producing monocyte chemoattractants. This in turn recruits Macs that rapidly acquire Arginase, Chil3 and Vegf production and are the predominant macrophage population during the repair process.
Discussion
In this study we first examined the ontogeny and bulk transcriptomic signature of snMacs compared with brain microglia, onMacs, liver Kupffer cells, lung alveolar Macs, spleen red-pulp Macs and peritoneal Macs. We could show that both brain and onMacs were derived from early embryonic precursors that were efficiently tagged at E8.5 and E9.5 in two independent TAM-inducible Csf1r-MERcreMER and Cx3cr1-creERT2 mouse models. In addition, we found that these CNS Macs displayed little or no engraftment of circulating monocytes using Cxcr4-creERT2 mice. This confirms a recent study reporting that most onMacs are long-lived Macs36. In addition, brain microglia and onMacs share an important CNS microglia “core” gene signature, including expression of Ccl12, Cd34, Ecscr, Kcnd1, Fcrls, Hexb, Lag3, Nav3, Mef2c, Sall1, Sall3, Sema4d, Siglech, Slc2a5, Sparc, Tmc7, Tmem119 and Trem2. This corroborates recent reports showing that CNS microglia display a relatively conserved gene expression profile across different anatomical CNS regions 37. The main onMac population is thus to be considered as CNS microglia based on their ontogeny and transcriptomic identity.
Contrary to this, we found that snMacs differ in many ways from CNS microglia. First, these cells do not derive from early embryonic precursors as shown by the minimal labeling of these Macs using tamoxifen-based fate-mapping at E8.5 and E9.5 in Csf1r-MERcreMER and Cx3cr1-creERT2 mouse models, respectively. Instead, they derive predominantly from late embryonic precursors that are slowly replaced by BM-derived precursors after birth. This was shown by a multitude of techniques, including Cxcr4-creERT2 and Sl00a4-cre fatemapping, parabiosis and partially irradiated BM-chimeras. Secondly, snMacs express only a minor part of the CNS microglia “core” genes, including Ccl12, Cx3cr1, Gpr34, Gpr183, Hexb, Mef2c, St3gal6, Tagap and Trem2, but completely lack the expression of Adora3, Crybb1, Csmd3, Fscn1, Ecscr, Gpr56, Kcnd1, Lag3, Nav3, Sall1, Sall3, Slc2a5, Sparc, Tmc7 and Tmem119. This list of genes is particularly striking because monocytes that gain access to the emptied microglia-niche in the brain following complete depletion of resident embryonic-derived microglia also fail to acquire these genes even after spending months in the brain38–40. Moreover, these genes are controlled by one central microglia-associated transcription factor: Sall141. Knock-down of Sall1 in CNS microglia was also associated with the increased expression of Adam19, Clec12a, Cd74, H2-Aa, H2-Ab1, Lilra5, Lilrb4, Ms4a7, Myo5a and Tmem176a by microglia. This set of genes is also highly expressed by snMacs as compared with other tissue-resident Macs (Figure 1 and41). In addition, these genes are also more highly expressed in monocyte-derived microglia-like cells in the CNS 38,39. This links the lack of a large part of the microglia core signature in snMacs to their distinct ontogeny and that fact that these cell do not derive from early embryonic precursors.
We have recently proposed the macrophage niche hypothesis in which we postulated the existence of a restricted number of macrophage niches per tissue that would confer tissue identity and self-maintenance capacity to Macs17. This hypothesis fits well with our data in the liver and the lungs where monocyte-derived Macs acquire almost all genes expressed by their embryonic counterparts and can self-maintain for months even in presence of embryonic Macs 18,42,43. The CNS niche forms an exception: monocyte-derived microglia-like cells lack the central microglia-associated Sall1 gene module and fail to self-maintain in the CNS as long as some embryonic-derived microglia remain to outcompete them. This implies that the CNS niche is unique in that it provides an exclusive competitive advantage to early embryonic YS Macs. This could be closely linked to the unique capacity of these early embryonic YS Macs to acquire the transcriptional repressor Sall1, as Sall1 protects embryonic microglia from excessive activation, which may give them a competitive advantage. We show here that the PNS niche does not provide such a competitive advantage to early embryonic YS Macs and is in fact completely permissive to the prolonged engraftment of BM monocyte-derived cells.
The bulk PNS core macrophage signature also contained many genes uniquely expressed by snMacs as compared with CNS microglia or other tissue-resident Macs. However, most of the genes in the core PNS Mac signature defined by bulk RNA sequencing could be assigned to either Relmα+Mgl1+ snMacs located in the epineurium or Relmα- Mgl1- snMacs inside the endoneurium. Mgl2, a C-type lectin recognizing N-acetylgalactosamine, and Cbr2, a carbonyl reductase, formed interesting exceptions as these were enriched in the PNS as compared with other tissues and were shared between the two PNS Mac subsets. Interestingly, Mgl2 and Cbr2 were also highly expressed by nerve-associated interstitial Macs in the lungs 33. Epineurial Relmα+Mgl1+ snMacs displayed high expression of Cc18, Ccl24, Cd209 genes, Clec10a (coding for Mgl1), Fcna, Folr2, Fxyd2 and Retnla (coding for Relmα). Many of these genes have been associated with the alternative “M2” activation of Macs. However, the binary in vitro-based “M1/M2” classification of Macs does not hold at all for in vivo Macs44 and this is also the case for snMacs. First, epineurial snMacs already express the so-called “M2 markers” Relmα and Mgl1 in the steady-state. Moreover, the other prototypical “M2” genes Arginase and Chil3 are acquired mainly by the recruited monocytes and not by the resident Relmα+Mgl1+ epineurial snMacs during nerve injury, showing that the typical core cassette of “M2” genes is not expressed by one and the same cell type in vivo.
While it is clear that the “M1/M2 framework” is not applicable in vivo, it will be interesting to investigate what micro-environmental signals imprint the expression of Relmα and Mgl1 in the Macs in the epineurium in steady-state and which cells produce these signals. As these genes are inducible by IL-4 and IPA proposes IL-4 as one of the most likely upstream regulators of the gene expression profile of Relmα+Mgl1+ snMacs, there may be an unexpected cellular source for IL4 in the micro-environment of the epineurial snMacs. The presence of TH2 cells or ILC2 cells in the epineurium has to our knowledge not been reported, but it would be worthwhile to investigate. Epineurial Relmα+Mgl1+ snMacs also express Tslp and as Tslp is also associated with type 2 responses, this could also influence macrophage activation.
The second subset of snMacs that was identified in steady-state were Relmα- Mgl1- snMacs located inside the endoneurium expressing higher levels of Ccr2, ll1rl1, Cxcll, Selm ( also known as Selenom and coding for Selenoprotein M), Pla2g2d, Qpct, Tnfsf9, Xist and genes of the MHC class II complex, H2-Aa, H2-Ab1 and Cd74. The expression of ll1rl1, encoding the T1/ST2 receptor for IL33, is particularly interesting as we found that Schwann cells, which reside in the endoneurium, express high levels of Il33 in steady-state and further increase their expression of Il33 upon injury)(data not shown). The expression of Ccr2 is consistent with the continuous engraftment of circulating monocytes in the snMac pool. We found both MHCIIHi and MHCIILow snMacs in the nerve fascicle and outside the endoneurium, but the majority of endoneurial Relmα- Mgl1- snMacs were of the MHCIIHi profile.
We found that PNS Macs subsets react differently to damage in a model of nerve crush injury. While the epineurial snMacs did not show any differentially expressed gene upon injury, endoneurial snMacs, which are in direct contact with the damaged axons and dedifferentiating Schwann cells, rapidly responded to the injury. They immediately started expressing monocyte recruiting chemokines, coinciding with a massive influx of circulating monocytes that swiftly outnumbered the resident Macs. The profile of activated endoneurial Relmα- Mgl1- snMacs did not resemble the profile of “disease-associated microglia” (DAMs) as these cells do not express the typical DAM-associated genes like Ank, Axl, Clec7a, Cox6a2, Cst1, Cst7, Fabp3, Fabp5, Igf1, Itgax, Rab7b45 and if anything seemed to down-regulate Apoe and Trem2 expression. This again underlines the important differences between CNS microglia and snMacs.
At first we had profiled the Macs during injury by bulk RNA-sequencing but this turned out to be misleading. Not only did this mix up two distinct resident Mac subsets, it also gave the false impression of a drop in genes expressed by the Macs in the epineurium due to the dilution of resident Macs by recruited monocyte-derived Macs (drop in Cd209f, Fcna or Retnla for example), while in fact the transcriptomic identity of the resident epineurial snMacs was not affected by the injury at all (Cd209f, Fcna and Retnla expression remained equally high in these cells). This highlights that relying on bulk RNA sequencing can lead to misinterpretation of the data as here we would have likely speculated on potential signals explaining the down-regulation of Retnla and a potential loss of “M2” identity during the first days of peripheral nerve injury.
All monocytes entering the injured nerve expressed Vegf, recently shown to be essential for sciatic nerve regeneration14. In addition, they also expressed very high levels of Arginase, while blood monocytes typically express low levels of Arg. This could be due to a local type 2 skewing nerve environment, which would rapidly upregulate Arg1 expression upon entering the nerve, consistent with our previous findings that the cytokine IL-13 is induced within the first few hours upon nerve injury46. Alternatively, it is also possible that peripheral nerve injury would trigger a rapid response in the CNS leading to a general systemic immunosuppression that may reprogram circulating monocytes, such as has been proposed in conditions of spinal cord injury or stroke (central nervous system injury-induced immune deficiency syndrome or CIDS)47. Thus, this represents an interesting avenue for future investigations.
Altogether, we describe the presence of two anatomically distinct subsets of Macs in the peripheral nerve, located in the endoneurium and epineurium, respectively. In contrast to CNS microglia these Macs derive from late embryonic precursors and get slowly replaced by BM-Macs and therefore completely lack the cardinal Sall1-module of CNS microglia. PNS crush injury does not greatly affect epineurial Macs, but does lead to the activation of endoneurial Macs that increase their production of monocyte-attracting chemokines. Monocytes recruited to the injured nerve rapidly develop into macrophages and express high levels of Arg1, Chil3 and Vegf. Although this remains to be demonstrated in vivo, our data would therefore suggest that the main Mac subset driving the nerve repair during this model of Wallerian degeneration is the recruited monocyte-derived Macs. These cells are numerically dominant and express the highest level of Vegf, which has been proposed to be essential for the generation of new blood vessels, which in turn enable the outgrowth of the axons14. After the injury these recruited Macs adopt the resident PNS Mac signature and remain in the PNS. This underlines that in stark contrast to the CNS, the PNS niche is permissive to BM-derived monocyte-derived Macs. The striking differences with CNS microglia also imply that the tools used to study CNS microglia will not be useful to study snMacs. Thus, we hope that the single-cell transcriptomic, spatial and ontogenic characterization of snMacs described here will pave the way towards the generation of novel PNS Macs specific tools to study the functional role of these cells specifically in vivo.
Material & Methods
Mice
C57BL/6 mice purchased from Charles River Laboratories were used as WT mice. Cx3cr1CreER mice25, Sall1CreER mice48, Cxcr4CreER-IRES-GFP (mice generated by Ralf Stumm, manuscript entitled “Fate mapping reveals a Cxcr4-dependent immune response of monocytes to experimental stroke” currently under revision at Nature Neuroscience), Rosa26-YFP mice49 and Rosa26-Tomato mice50 were bred in-house on a C57BL/6 background under specific pathogen-free conditions. Cx3cr1 CreER and Sall1CreER mice were crossed to Rosa26-YFP and Cxcr4CreER-IRES-GFP were crossed to Rosa26-Tomato mice. CD45.1+ and CD45.2+ mice were purchased from Harlan. Csf1rMERcreMER:R26-YFP and S100A4cre:R26-YFP have been described before29 and were kindly provided for this study by Florent Ginhoux. All animal experiments were approved by local administration in Germany (Regierungspräsidium Freiburg) or Belgium (University of Antwerp and University of Ghent) and were performed in accordance to the respective national, federal and institutional regulations and the guidelines of the Federation of European Laboratory Animal Science Associations (FELASA).
Tamoxifen treatments
To induce expression of EYFP or tdTomato in young animals (Cx3cr1creER:Rosa26-EYFP or Cxcr4creER-IRES-GFP:Rosa26-tdTomato mice, respectively), 0.1 mg Tamoxifen (TAM, T5648, Sigma-Aldrich) was dissolved in 5 μl corn oil (Sigma-Aldrich, Taufkirchen, Germany) and was administered by i.p. injection at postnatal (p) day p1 and p3. To induce reporter gene expression in adult mice, Cxcr4creER-IRES-GFP:Rosa26-tdTomato mice were i.p. injected with 1 mg TAM in 100μl corn oil for five consecutive days starting at an age of six weeks. Adult Cx3cr1creER:Rosa26-EYFP and Sall1creER:Rosa26-EYFP mice were s.c. injected with 4mg TAM in 200μl corn oil twice on day 0 and day 2 of the treatment. To induce expression of EYFP in yolk sac derived macrophages, 75 mg/kg 4-HydroxyTAM (H7904, Merck) and 37.5 mg/kg Progesterone (P0130, Merck) dissolved in corn oil were given by i.p. injection to pregnant females carrying Cx3cr1creER:Rosa26-EYFP embryos at embryonic day (E)9.5 or E16.5. At E19.5, pups were delivered by caesarean section and given to foster mothers.
Generation of parabiotic mice
Parabiosis experiments were performed at the University of Marseille according to French and European guidelines as described previously51. Shortly, CD45.1+ Ccr2+/+ mice were sutured together with CD45.2+ Ccr2-/- mice and subsequently kept under Bactrim 8 weeks before analysis.
Irradiation
For irradiation experiments, ketamine/xylazine anesthetized CD45.2+ recipient mice were irradiated with a single dose of 10Gy, and subsequently intravenously injected with bone marrow cells derived from femur and tibia bones of wild type C57/BL6 donor CD45.1+ mice. During partial body irradiation procedure, the right hind leg of mice was protected using lead shielding.
Induction of peripheral nerve injury
Nerve crush experiments of the Nervus ischiadicus (sciatic nerve) were conducted in 6- to 8-week-old C57BL/6 mice (either sex was used) as previously described (Ydens et al., 2016 NLR paper). Briefly, mice were anesthetized with a single intraperitoneal injection of ketamine (Ketalar; Pfizer; 150 mg/kg) and xylazine (Rompun; Bayer; 10 mg/kg). An incision was made at the right thigh, and gluteal and hamstring muscles were carefully separated to expose the sciatic nerve. The sciatic nerve was crushed, and the wound was closed by sutures. The contralateral (control) side was left untouched. For analgesia, bupronorphinum (Temgesic; Schering-Slough; 0.1 mg/kg) was injected after surgery. For all experiments described the part distal to the nerve injury was used for analysis.
Flow cytometry
For isolation of microglia, the brain was dissected, homogenized with a tissue grinder potter in HBSS containing 15 mM HEPES buffer and 0.54 % glucose and filtered through a 70μm mesh. After centrifugation (220g for 5 min at 4°C), the pellet was resuspended in 37% Percoll followed by centrifugation for 30 min, 800g at 4°C without brake. Myelin was removed from the top layer and the cell pellet was washed once with 1X PBS followed by antibody staining.
In part of the experiments, sciatic nerves or optic nerves were collected in HBSS containing 0.5 mg/ml Collagenase (Sigma-Aldrich, 11088866001) D, 0.5 mg/ml Dispase II (Sigma-Aldrich, D4693) supplemented with 15 mM HEPES and 0.54% D-(+)-Glucose. The nerves were cut into small pieces, digested for 45 min at 37°C and single cells were collected in FACS buffer (2% fetal calf serum and 0.01M EDTA in PBS). Fc receptors were blocked with an anti-CD16/CD32 antibody (1:250, BD Bioscience, clone 2.4G2) prior to incubation with the following antibodies: anti-CD45 (1:200, ThermoFisher, clone 30-F11), anti-CD11c (1:300, ThermoFisher, clone N418), anti-F4/80 (1:200, ThermoFisher, clone BM8) and anti-CD3e (1:300, ThermoFisher, clone eBio500A2), anti-CD11b (1:300, BioLegend, B1/70), anti-CD64 (1:200, BioLegend, clone X54-5/7.1), anti-Mgl1 (1:200, BioLegend, clone LOM-8.7), anti-Ly6C (1:200, BD Bioscience, clone AL-21), anti-Ly6G (1:300, BD Bioscience, clone 1A8), anti-SiglecF (1:200, BD Bioscience, E50-2440), anti-MHC-II (1:300, BD Biosciences, clone M5/114.15.2) and anti-Lyvel (1:200, ThermoFisher, clone ALY7). DAPI was added for dead cell exclusion. Cells were acquired on a BD LSRFortessa™ cell analyzer (BD Biosciences, Heidelberg, Germany).
In part of the experiments, sciatic nerves were dissected and mechanical and enzymatic dissociated for 30 min in an RPMI solution containing DNase (0.01 U/μl; Roche) and Liberase TM (0.02 mg/ml; Roche). The cell suspension was filtered over a 100-μm nylon mesh. Antibodies used were from BD Biosciences or Biolegend: allophycocyanin-Cy7-conjugated anti-MHC Class II (1:1500, I-A/I-E), allophycocyanin-conjugated anti-CD64 (1:300), PE-conjugated anti-Siglec-F (1:1000) and anti-Ly6G (1:1000), PE-Cy5-conjugated anti-CD3 (1:300) and anti-CD19 (1:300), PE-Cy7-conjugated anti-CD11c (1:800), FITC-conjugated anti-Ly6C (1:1000, AL21), Pacific Blue-conjugated anti-CD11b (1:800, M1/70), Alexa 700-conjugated anti-CD45.2 (1:300), APC-conjugated anti-CD45.1 (1:300), biotin-conjugated Relmα (1:100). The LIVE/DEAD fixable Aqua dead cell stain kit (Life Technologies) was used to distinguish live from dead cells. Multiparameter analysis was performed on a FACS LSRII system (BD Biosciences) and analyzed with FlowJo software (Tree Star, Inc.). Doublets were excluded from all analysis. For intracellular markers, cells, stained for extracellular markers, were fixed and permeabilized using the FoxP3 kit according to manufacturer’s protocol and incubated with antibodies against the intracellular markers.
Cell sorting
For cell sorting, sciatic nerves and optic nerves from 8-10 week old female C57BL/6 mice were prepared for flow cytometry as described and sorted on a MoFlo Astrios EQ cell sorter (Beckman Coulter) or on a FACSAria (BD Biosciences).
Histology, immunofluorescence and microscopy
In part of the experiments, sciatic nerves were fixed in 4% PFA (Electron Microscopy Sciences, RT15710), washed in PBS and embedded in 40°C ultra-low melting point agarose (Invitrogen 16520-050). Next, solidified agarose blocks were trimmed and mounted using cyanoacrylate adhesive, in order to cut 50 μm cross-sections on a vibratome (Microm International HM650V). Immunofluorescence staining procedures were performed with the sections floating in solution, employing Pelco Prep-Eze wellplate inserts (Ted Pella 36172) to retain the sections during solution exchanges. After 3 initial washes in PBS, sections were blocked and permeabilized in normal goat serum (1:500, Dako Belgium NV X0907) diluted in PBT buffer (PBS + 0.5% BSA + 0.02% Triton-X-100) for 2h at room temperature. Samples were incubated in primary antibodies over-night at 4°C, washed 4x for 5 min and 1x for 30 min in PBT and incubated with secondary antibodies for 3 h at room temperature or over-night at 4°C. After 3 washes of 5 min in PBT and 1 wash of 30 min in PBS, sections were stained with Hoechst 33342 (1:20.000 in PBS, Life Technologies H3570) for 1 h at room temperature, washed 3x for 5 min and 1x for 30 min in PBS and mounted on standard microscopy slides in anti-fade mounting medium (Dako Belgium NV S302380-2). The following antibody combinations and concentrations were used: Rat anti-F4/80 (1:100 - 1:200, AbD Serotec/BioRad MCA497A, clone Cl:A3-1) detected with Goat anti-Rat-AlexaFluor594 (1:500, Life Technologies A11006); Rabbit polyclonal anti-Relm-α (1:200, Peprotech 500-P214Bt) detected with Donkey anti-Rabbit-AlexaFluor647 (1:500, Life Technologies A31573) or Goat anti-Rabbit-AlexaFluor488 (1:500, Life Technologies A11008), Goat polyclonal anti-Arginase (1:100, Santa Cruz (N-20): sc-18351) detected with Donkey anti-Goat-AlexaFluor488 (1:500, Life Technologies A11055), Rat anti-MHCII-FITC (1:100, Biolegend clone M5/114.15.2) detected with Goat polyclonal anti-Fluorescein/OregonGreen-AlexaFluor488 (1:150, Invitrogen A-11096 AB_221558). Images were acquired on a Zeiss LSM700 laser scanning confocal microscope using either a Plan-Neofluar 10x/0.30 or a Plan-Apochromat 20x/0.8 objective. Sequential scanning was used to eliminate cross-talk of the fluorescence channels. Stitching of image tiles and merging of z-stacks with maximum intensity projections was done with either the Zeiss Zen software or the Fiji distribution of ImageJ51,52.
In part of the experiments, dissected sciatic nerves and optic nerves were fixed in 4% PFA in PBS and washed in 1x PBS. To prepare vibratome cross-sections, PFA fixated sciatic nerves were embedded in low-melting agarose (Lonza 50101). Solidified agarose blocks were trimmed and 400μm thick cross-sections were cut with a Leica VT1000 S vibratome. Immunofluorescence stainings were performed with sections floating in buffer. Sections were blocked in PBT buffer (PBS + 5% normal donkey serum + 0.5% Triton X-100) over-night at 4°C. Primary antibodies were incubated for 48h at 4°C in PBT, sections were washed 3x 30 min in PBT and incubated with secondary antibodies for 24h at 4°C. After washing 2x 30 min with PBT and 1x 30 min with PBS, sections were mounted on SuperFrost Plus slides (R. Langenbrinck GmbH) in Mowiol (Merck Millipore) mounting medium. Nuclei were counterstained with DAPI. For cryosections, PFA fixated sciatic nerves and optic nerves were dehydrated in 30 % sucrose in PBS and embedded in Tissue-Tek® O.C.T. compound (Sakura Finetek Germany GmbH). 20μm thick longitudinal sections were obtained using a Leica CM 1900 UV crytostat and collected on SuperFrost Plus slides (R. Langenbrinck GmbH). Sections were blocked in PBS + 5% normal donkey serum + 0.01% Triton X-100 buffer for 1h at room temperature. Primary antibodies were incubated over night at 4°C in PBS + 5% normal donkey serum + 0.01% Triton X-100 followed by 3x 5min washing with PBS + 0.1% Triton X-100. Secondary antibodies were incubated for 2h at room temperature in PBS + 5% normal donkey serum + 0.01% Triton X-100 followed by washing 2x 5min with PBS and mounting in Mowiol mounting medium. Nuclei were counterstained with DAPI. The following primary antibodies were used: Rabbit polyclonal anti-Iba-1 (1:500, Abcam ab178846), Rat anti-CD301a(Mgl1) (1:200, BioLegend 145602, clone LOM-8.7), Rabbit polyclonal anti-Lyve1 (1:500, Abcam ab14917) and Goat polyclonal anti-Collagen IV (1:500, Merck AB769). Following secondary antibodies were used: Donkey anti-Goat-AlexaFluor405 (1:500, Abcam ab175664), Donkey anti-Rat-AlexaFluor568 (1:500, Abcam, ab175475) and Donkey anti-Rabbit-AlexaFluor647 (1:500, ThermoFisher, A31573). Fluorescence Images were taken using an Olympus BX-61 conventional fluorescence microscope equipped with a camera for fluorescence images (Olympus XM10IR). Confocal images were acquired on an Olympus FV 1000MPE laser scanning microscope using a Plan-Apochromat 20x/1.0 (Olympus XLUMPLFLN-W) objective.
For paraffin sections, sciatic nerves and optic nerves were fixed in 4% PFA in PBS. The tissues were then embedded in paraffin and 4μm thick longitudinal sections were cut with a Microm HM360 microtome (ThermoFisher Scientific). Slides were stained with Rabbit polyclonal anti-Iba-1 (1:500, WAKO 019-19741). Bright-field microscopy Images were taken using an Olympus BX-61 microscope equipped with a camera for bright-field images (Olympus XC10).
RNA sequencing
Total RNA was extracted from FACS sorted cells using Picopure RNA extraction kit (Life Technologies) according to manufacturer’s protocol. The SMARTer Ultra Low Input RNA Kit for Sequencing v4 (Clontech Laboratories, Inc., Mountain View, CA, USA) was used to generate first strand cDNA from 500 to 750 pg total-RNA. Double stranded cDNA was amplified by LD PCR (11 cycles) and purified via magnetic bead clean-up. Library preparation was carried out as described in the Illumina Nextera XT Sample Preparation Guide (Illumina, Inc., San Diego, CA, USA). 150 pg of input cDNA were tagmented (tagged and fragmented) by the Nextera XT transposome. The products were purified and amplified via a limited-cycle PCR program to generate multiplexed sequencing libraries. For the PCR step 1:5 dilutions of index 1 (i7) and index 2 (i5) primers were used. The libraries were quantified using the KAPA SYBR FAST ABI Prism Library Quantification Kit (Kapa Biosystems, Inc., Woburn, MA, USA). Equimolar amounts of each library were pooled, and the pools were used for cluster generation on the cBot with the Illumina TruSeq SR Cluster Kit v3. The sequencing run was performed on a HiSeq 1000 instrument using the indexed, 50 cycles single-read (SR) protocol and the TruSeq SBS v3 Reagents according to the Illumina HiSeq 1000 System User Guide. Image analysis and base calling resulted in .bcl files, which were converted into .fastq files with the CASAVA1.8.2 software. Library preparation and RNaseq were performed at the Genomics Core Facility “KFB - Center of Excellence for Fluorescent Bioanalytics” (University of Regensburg, Regensburg, Germany; www.kfb-regensburg.de).
Microarray analysis
Cell populations were isolated from the sciatic nerve by sorting as described above. RNA was obtained using the RNEasy Plus Micro Kit (QIAGEN) following manufacturer’s instructions and RNA integrity was assessed using a Bioanalyser 2100 (Agilent). Using the Ambion WT Expression Kit, per sample, an amount of 50 ng of total RNA per sample spiked with bacterial poly-A RNA positive controls (Affymetrix) was converted to double stranded cDNA in a reverse transcription reaction. Samples were fragmented and labeled with biotin in a terminal labeling reaction according to the Affymetrix WT Terminal Labeling Kit. A mixture of fragmented biotinylated cDNA and hybridisation controls (Affymetrix) was hybridised on Affymetrix GeneChip Mouse Gene 1.0 ST Arrays. The chips were washed and stained in the GeneChip® fluidics station 450 (Affymetrix) according to the manufacturer’s procedures and scanned using a GeneChip® scanner 3000 (Affymetrix). Samples were subsequently analyzed using R/Bioconductor. All samples passed quality control, and the Robust Multi-array Average (RMA) procedure was used to normalize data within arrays (probeset summarization, background correction and log2-transformation) and between arrays (quantile normalization). Only probesets that mapped uniquely to one gene were kept, and for each gene, the probeset with the highest expression level was kept. To create the heatmaps, we transformed the log2 normalized values by calculating the mean per gene over all cell types in the heatmap and subsequently subtracting this mean from the cell type expression value for each particular gene. This results in a positive value for the cell types where the gene is expressed more than the mean and a negative value for the cell types where the gene is expressed less than the mean.
Single-cell sequencing
Sorting and RNA isolation
Macrophages were sorted as CD64+F4/80+CD45+ Live cells from the SN. Cells were sorted into PBS 0.04% BSA, spun down and resuspended in PBS/0.04%BSA at an estimated final concentration of 1000 cells/μl. Cellular suspensions were loaded on a GemCode Single-Cell Instrument (10x Genomics, Pleasanton, CA, USA) to generate single-cell Gel Bead-in-EMulsion (GEMs). Single-cell RNA-Seq libraries were prepared using GemCode Single-Cell 3’ V1 Gel Bead and Library Kit (10x Genomics) according to the manufacturer’s instructions. Briefly, GEM-RT was performed in 96-deep well reaction module: 55°C for 45min, 85°C for 5 min; end at 4°C. After RT, GEMs were broken down and the cDNA was cleaned up with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific, 37002D) and SPRIselect Reagent Kit (SPRI; Beckman Coulter; B23318). cDNA was amplified with 96-Deep Well Reaction Module: 98 °C for 3 min; cycled 12 times : 98°C for 15s, 67°C for 20 s, and 72°C for 1 min; 72°C for 1 min; end at 4°C. Amplified cDNA product was cleaned up with SPRIselect Reagent Kit prior to enzymatic fragmentation. Indexed sequencing libraries were generated using the reagents in the GemCode Single-Cell 3’ Library Kit with the following intermediates: (1) end repair; (2) A-tailing; (3) adapter ligation; (4) post-ligation SPRIselect cleanup and (5) sample index PCR. Pre-fragmentation and post-sample index PCR samples were analysed using the Agilent 2100 Bioanalyzer.
RNA sequencing analysis
Sequencing libraries were loaded on an Illumina NextSeq500 with sequencing settings following recommendations of 10X Genomics (26/8/0/98 - 2.1pM loading concentration). Sequencing was performed at the VIB Nucleomics Core (VIB, Leuven). The demultiplexing of the raw data was done by the 10x’s CellRanger software (version 2.0.0 (KCs, AMs) or version 2.0.2 (Microglia, splenic macs, colonic macs); cellranger mkfastq which wraps Illumina’s bcl2fastq). The reads obtained from the demultiplexing were used as the input for ‘cellranger count’ (10x’s CellRanger software) which align the reads to the mouse reference genome (mm10) using STAR and collapses to unique molecular identifier (UMI) counts. The result is a large digital expression matrix with cell barcodes as rows and gene identities as columns. The aggregation of the different samples was done using ‘cellranger aggr’ (10x’s CellRanger software).
Preprocessing data
Preprocessing of the data was done by the scran and scater R package according to workflow proposed by the Marioni lab (Lun et al., 2016). Outlier cells were identified based on 3 metrics (library size, number of expressed genes and mitochondrial proportion) and cells were tagged as outliers when they were 3 median absolute deviation (MADs) away from the median value of each metric across all cells. Low-abundance genes were removed using the ‘calcAverage’ function and the proposed workflow. The raw counts were normalised and log2 transformed by first calculating “size factors” that represent the extent to which counts should be scaled in each library. Detecting highly variable genes, finding clusters and creating tSNE plots was done using the Seurat pipeline. Marker genes per identified subpopulation were found using the findMarker function of the Seurat pipeline. Additional low quality (low UMI counts, high % of mitochondrial genes) were removed from the analysis.
Supplementary Material
Acknowledgements
We would like to thank Gert Van Isterdael from the Flow Cytometry Core Facility for assistance. This study benefitted from data assembled by the Immgen Consortium. We would also like to thank VIB TechWatch and the VIB Single Cell Accelerator program for supporting the 10x Genomics experiments. YS’s lab is supported by the FWO and YS is a Marylou Ingram Scholar. CLS is supported by FWO grants. MG’s lab is supported by grants from the FWO, an ERC Consolidator grant and a Ghent University GOA grant. M.P. is supported by the Sobek Foundation, the Ernst-Jung Foundation, the German Research Foundation (SFB 992, SFB1160, DFG SFB/TRR167, Reinhart-Koselleck-Grant) and the Ministry of Science, Research and Arts, Baden-Wuerttemberg (Sonderlinie “Neuroinflammation”) and by the BMBF-funded competence network of multiple sclerosis (KKNMS). His research is supported by the German Research Foundation (DFG) under Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project ID 390939984).
FG is an EMBO YIP awardee and is supported by Singapore Immunology Network (SIgN) core funding as well as a Singapore National Research Foundation Senior Investigatorship (NRFI) NRF2016NRF-NRFI001-02. The study was supported by the University of Antwerp and the Fund for Scientific research (FWO-Flanders). EY was supported by an FWO-SB doctoral fellowship. SJ is a recipient of an ERC Consolidator Grant, several FWO program grants and an EOS grant (G0G7318N).
Abbreviations
- BM
bone marrow
- BAMs
border-associated macrophages
- CNS
central nervous system
- CAMs
CNS-associated macrophages
- DAMs
disease-associated microglia
- HSC
hematopoietic stem cell
- Macs
macrophages
- snMacs
sciatic nerve macrophages
- PNS
peripheral nervous system
- PCA
Principle component analysis
- SN
sciatic nerve
- SFI
sciatic nerve functional index
- sc-RNA-seq
single-cell RNA sequencing
- TAM
tamoxifen
References
- 1.Giladi A, Amit I. Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell. 2018;172:14–21. doi: 10.1016/j.cell.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mass E, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz C, et al. A Lineage of Myeloid Cells Independent of Myb and Hematopoietic Stem Cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 6.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 7.Tay TL, et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat Neurosci. 2017:1–14. doi: 10.1038/nn.4547. [DOI] [PubMed] [Google Scholar]
- 8.Goldmann T, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. 2016 doi: 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordão MJC, et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363:eaat7554. doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 10.Van Hove H, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci. 2019;13:206. doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 11.Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FMV. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci. 2011;14:1142–1149. doi: 10.1038/nn.2887. [DOI] [PubMed] [Google Scholar]
- 12.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FMV. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 13.Chen P, Piao X, Bonaldo P. Role of macrophages in Wallerian degeneration and axonal regeneration after peripheral nerve injury. Acta Neuropathol. 2015;130:605–618. doi: 10.1007/s00401-015-1482-4. [DOI] [PubMed] [Google Scholar]
- 14.Cattin A-L, et al. Macrophage-Induced Blood Vessels Guide Schwann Cell-Mediated Regeneration of Peripheral Nerves. Cell. 2015;162:1127–1139. doi: 10.1016/j.cell.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavin Y, et al. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guilliams M, Scott CL. PERSPECTIVES. Nat Rev Immunol. 2017:1–10. doi: 10.1038/nri.2017.42. [DOI] [Google Scholar]
- 18.Scott CL, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nature Communications. 2016;7:10321. doi: 10.1038/ncomms10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott CL, et al. The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity. 2018;49:312–325.:e5. doi: 10.1016/j.immuni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haimon Z, et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Vol. 19. Nature Publishing Group; 2018. pp. 636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Brink SC, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 22.Saunders A, et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell. 2018;174:1015–1030.:e16. doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginhoux F, Prinz M. Origin of microglia: current concepts and past controversies. Cold Spring Harbor Perspectives in Biology. 2015;7:a020537. doi: 10.1101/cshperspect.a020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prinz M, Erny D, Hagemeyer N. Ontogeny and homeostasis of CNS myeloid cells. Vol. 18. Nature Publishing Group; 2017. pp. 385–392. [DOI] [PubMed] [Google Scholar]
- 25.Yona S, et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoeffel G, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Vol. 7. Nature Publishing Group; 2006. pp. 311–317. [DOI] [PubMed] [Google Scholar]
- 28.Mildner A, et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 29.Movahedi K, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Research. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 30.Nadeau S, et al. Functional recovery after peripheral nerve injury is dependent on the pro-inflammatory cytokines IL-1β and TNF: implications for neuropathic pain. J Neurosci. 2011;31:12533–12542. doi: 10.1523/JNEUROSCI.2840-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shechter R, et al. Recruitment of Beneficial M2 Macrophages to Injured Spinal Cord Is Orchestrated by Remote Brain Choroid Plexus. Immunity. 2013;38:555–569. doi: 10.1016/j.immuni.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jablonski KA, et al. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE. 2015;10:e0145342. doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakarov S, et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 2019;363:eaau0964. doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- 34.Tamoutounour S, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 35.Tamoutounour S, et al. Origins and Functional Specialization of Macrophages and of Conventional and Monocyte-Derived Dendritic Cells in Mouse Skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 36.O’Koren EG, et al. Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity. 2019;50:723–737.:e7. doi: 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuda T, et al. Spatial and temporal heterogeneity of mouse and human microglia at singlecell resolution. Nature. 2019:1–23. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 38.Shemer A, et al. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nature Communications. 2018:1–16. doi: 10.1038/s41467-018-07548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett FC, et al. A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron. 2018;98:1170–1183.:e8. doi: 10.1016/j.neuron.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cronk JC, et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. Journal of Experimental Medicine. 2018;215:1627–1647. doi: 10.1084/jem.20180247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buttgereit A, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol. 2016;17:1397.:EP —1406. doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
- 42.van de Laar L, et al. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity. 2016 doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 43.Bonnardel J, et al. Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity. 2019;51:638–654.:e9. doi: 10.1016/j.immuni.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray PJ. Macrophage Polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 45.Keren-Shaul H, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169:1276–1290.:e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 46.Ydens E, et al. Acute injury in the peripheral nervous system triggers an alternative macrophage response. J Neuroinflammation. 2012;9:1–17. doi: 10.1186/1742-2094-9-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meisel C, Schwab JM, Prass K, Meisel A, Dirnagl U. Central nervous system injury-induced immune deficiency syndrome. Nat Rev Neurosci. 2005;6:775–786. doi: 10.1038/nrn1765. [DOI] [PubMed] [Google Scholar]
- 48.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoue S, et al. A mouse line expressing Sall1-driven inducible Cre recombinase in the kidney mesenchyme. Genesis. 2010;48:207–212. doi: 10.1002/dvg.20603. [DOI] [PubMed] [Google Scholar]
- 50.Srinivas, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jun 28;9(7):676–82. doi: 10.1038/nmeth.2019. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin J, et al. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015 Jul-Aug;82(7-8):518–29. doi: 10.1002/mrd.22489. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.