Abstract
Purpose
KRAS is the most frequently mutated gene in human cancers. Despite its direct involvement in malignancy and intensive effort, direct inhibition of KRAS via pharmacological inhibitors has been challenging. RNAi induced knockdown using siRNAs against mutant KRAS alleles offers a promising tool for selective therapeutic silencing in KRAS-mutant lung cancers. However, the major bottleneck for clinical translation is the lack of efficient biocompatible siRNA carrier systems.
Methods
Bovine serum albumin (BSA) nanoparticles were prepared by desolvation method to deliver siRNA targeting the KRAS G12S mutation. The BSA nanoparticles were characterized with respect to their size, zeta potential, encapsulation efficiency and nucleic acid release. Nanoparticle uptake, cellular distribution of nucleic acids, cytotoxicity and gene knock down to interfere with cancer hallmarks, uncontrolled proliferation and migration, were evaluated in KRAS G12S mutant A459 cells, a lung adenocarcinoma cell line.
Results
BSA nanoparticles loaded with siRNA resulted in nanoparticles smaller than 200 nm in diameter and negative zeta potentials, displaying optimal characteristics for in vivo application. Encapsulating and protecting the siRNA payload well, the nanoparticles enabled transport to A549 cells in vitro, could evade endosomal entrapment and mediated significant sequence-specific KRAS knockdown, resulting in reduced cell growth of siRNA transfected lung cancer cells.
Conclusions
BSA nanoparticles loaded with mutant specific siRNA are a promising therapeutic approach for KRAS-mutant cancers.
Keywords: BSA, KRAS, lung cancer, siRNA delivery
Introduction
Lung cancer is a major public health problem worldwide (1) and the leading cause of cancer related mortality worldwide. Based on the cell type, lung cancer is categorized into small cell lung cancer (SCLC), which accounts for approximately 15% of the cases, and non-small cell lung cancer (NSCLC), which accounts for the remaining majority of the cases (85%) (2,3). NSCLC is further subclassified into squamous cell carcinoma (25%), large cell carcinoma (10%) and the most prevalent subtype, adenocarcinoma (40%). Kirsten rat sarcoma viral oncogene homolog (KRAS) is the most frequently mutated gene associated with lung cancer, and mutations are found in approximately 27% of lung adenocarcinomas (3–5). KRAS mutations occur predominantly (95%) at codons 12 (>80%) and 13 (4). KRAS, a member of the RAS gene family, encodes a small membrane bound GTPase, which can exist in two different states: the inactive GDP bound state or the active GTP bound state, which further activates downstream signalling cascades that regulate fundamental cell processes (6). KRAS mutations result in constitutively GTP-bound active proteins, thereby activating oncogenic pathways (7). Currently, KRAS mutant lung cancers are treated (8) with conventional chemotherapy regimens based on platinum, most often with limited success (9).
One of the major concerns for drug development is whether KRAS driven tumours retain dependence on KRAS expression. Using an inducible transgenic mouse model of KRAS G12D mutant lung cancer, Roccio et al. showed that transgene withdrawal after formation of visible lung tumours resulted in a dramatic and sustained decrease in tumour volume (10), indicating that mutant KRAS inhibition is critical for successful treatment.
Several strategies, such as farnesyltransferase inhibitors, have been explored as possible inhibitors of KRAS in lung adenocarcinomas. However, direct targeting of RAS proteins has been challenging and has so far not led to a new therapy, mainly because of a lack of specificity. Most often both wild type and mutant KRAS are inhibited resulting in undesirable side effects. RNA interference-based therapy using small interfering RNAs (siRNA) is a promising alternative to small molecule inhibitors since, in principle, can specifically and efficiently inhibit any gene by directly targeting and reducing its mRNA expression. Despite its success in vitro and tremendous therapeutic potential, clinical application of siRNA is limited by its susceptibility to nucleases in serum, reticuloendothelial system clearance and poor cellular uptake due to its large size and strong polarity (11). Therefore, suitable delivery systems are required to protect the sensitive siRNA and mediate its transport to the target cell and subsequent release in the cytoplasm.
In the past decades, apart from viral vectors, various delivery formulations have been developed including cationic lipids, polymers, inorganic nanoparticles and combinations of these groups for siRNA delivery. Cationic lipids and polymers can condense siRNA spontaneously and form nanoscale complexes, showing high gene delivery efficiencies while eluding immune responses, increased risk of mutagenesis and potential genomic integration of viral DNA, often occurring with viral vectors (8). However, these carrier systems often demonstrate high toxicity especially after repeated administration can become immunogenic. An ideal delivery vehicle should be biocompatible and biodegradable, protect the siRNA load from degradation by nucleases during the delivery process, and mediate the intracellular uptake of siRNA (12).
Recently, protein-based nanoparticles have gained importance as delivery devices, since they are not toxic and have low antigenicity, are biodegradable and demonstrate large possibilities for surface modification (13,14). Albumin is the most abundant plasma protein and has been extensively used to prepare nanoparticles. Its half-life in human blood is 19 days on average, thereby improving the plasma area under curve (AUC) value and maintaining high blood ‘drug’ concentrations for a relatively long time. Albumin is also an excellent lyoprotectant and allows immediate redispersion of dried nanoparticles in injectable solutions (15–17) which favours the development of shelf-stable formulations limiting drug release and precipitate formation, which are general problems faced in the development of clinical translatable nanomedicine (17). The most notable example is the enhanced delivery and efficacy of paclitaxel when delivered as an albumin conjugated nanoparticle, Abraxane (18).
Bovine serum albumin (BSA) is an acidic, very stable globular protein (66 kDa) which shows minimal immunogenicity, is non-toxic and biodegradable, making it an ideal candidate as a gene delivery vehicle (15). Due to a high content of charged amino acids (lysine, arginine, aspartate and glutamate), albumin nanoparticles allow electrostatic adsorption of charged molecules. Additionally, albumin nanoparticles represent a promising strategy for targeted drug delivery to tumour cells as they have been shown to accumulate in tumours via specific transendothelial transport processes and through the enhanced permeability and retention mechanism (14,19).
Based on these reports, in this study, we aimed to develop BSA nanoparticles encapsulating an siRNA targeting the G12S mutation of KRAS. We hypothesized that the specific knock down of the constitutively active mutant allele of KRAS would result in sudden cessation of signalling and cancer cell death. The siKRAS G12S-BSA nanoparticles were fully characterized with respect to siRNA protection, gene knockdown and toxicity. We demonstrate that BSA nanoparticles are a non-toxic, efficient siRNA delivery system with optimal properties to preferentially reach lung tumour cells.
Materials And Methods
Synthesis of FITC Labelled BSA
Fluoresceinyl serum albumin was synthesized according to a published protocol (20). In brief, the preparation was performed in a three-step process. In the first step, bovine serum albumin (33.5 mg, 0.5 mmol, Carl Roth) was dissolved in 5 mL Na2CO3-NaHCO3 buffer (pH 9.5, Sigma Aldrich, St. Louis, Missouri, USA) and fluorescein isothiocyanate, FITC (0.6 mg, 1.5 mmol, Sigma Aldrich) dissolved in DMSO was added. The solution was stirred for 5 h at RT in the dark. The fluorescein substituted protein was purified using Vivaspin 6 Centrifugal Concentrator filters (CO 30 kDa) (Sartorius, Gottingen, Germany). The protein concentration was determined using a standard BCA protein assay and the solution was stored in the dark at 4°C.
BSA-Nanoparticle Formation
Three different modifications of previously reported desolvation-crosslinking methods were compared to prepare BSA nanoparticles (21–23). The methods are briefly described, and their differences are highlighted in Table I.
Table I. Differences in the three methods to prepare BSA nanoparticles.
| 2% BSA solution (ul) | Bulk DNA (ug) | EtOH volume 1 (ml) | 25% aqueous Glutaraldehyde (ul) | Ethanol volume 2 (ml) | |
|---|---|---|---|---|---|
| Method 1 | 500 | 150 | 1.0* | 4 | 1.0 |
| Method 2 | 500 | 150 | 1.0 | 4 | 1.0 |
| Method 3 | 500 | 150 | 1.4 | 4 | 0 |
In Method 1, BSA-DNA mixture was added dropwise over one minute into 1.0 ml EtOH
First, 500 μl BSA solution (in purified water 2% w/v) was incubated with 150 μg bulk DNA (50 μg/μl stock in nanopure water). Subsequently, ethanol (EtOH) was added dropwise (over 1 min) to the suspension, while stirring. Immediately, 4 μl glutaraldehyde (25% aqueous solution) was added. The NP suspension was stirred for 20 h at RT and then 1 ml EtOH was added to the NP suspension to remove excess glutaraldehyde. The nanoparticles were separated by centrifugation at 20,238 g for 30 min. The resulting nanoparticle pellet was resuspended in 500 μl nanopure water by vortexing and ultrasonication.
Method 3 was found to be most suitable and used for subsequent experiments. For BSA nanoparticles encapsulating plasmid DNA (pDNA), 50 μg of pDNA was used, and for siRNA containing nanoparticles, 100 μM (0.1 nmol or 1.3 μg) and 200 μM (0.2 nmol or 2.6 μg) siRNA was encapsulated. FITC-labelled BSA nanoparticles were prepared without DNA.
Characterization of BSA Nanoparticles
To measure hydrodynamic diameters, and polydispersity indices (PDI), a 1:30 dilution of the nanoparticle suspension was prepared in water and 90 μl was added into a disposable micro-cuvette (Malvern Instruments, Malvern, UK). Sizes were determined with a Zetasizer Nano ZS (Malvern Instruments) at 173° backscatter angle running 15 sub runs three times per sample. The measurements were done in triplicates. Results are represented as average hydrodynamic diameter (nm) ± standard deviation.
To measure zeta potentials, nanoparticle suspensions were diluted 1:300 in 10 mM NaCl and transferred to a folded capillary cell (Malvern Instruments) for each sample using the Zetasizer Nano ZS (Malvern Instruments). Zeta potential measurements were performed in triplicates by laser Doppler anemometry (LDA), with each run consisting of 30–50 scans. Results are shown in mV ± standard deviation.
After centrifugation and pelleting the nanoparticles, the supernatant was collected and the amount of free BSA in the supernatant was measured by the standard BCA protein assay. The nanoparticle yield was calculated as following:
where, BSA total is the amount of BSA added, and BSA free is the amount of dissolved BSA remaining after centrifugation as quantified by the BCA assay.
Encapsulation efficiency of the BSA nanoparticles was measured directly and indirectly. For direct measurements, 100 μl of the nanoparticle suspension was pelleted and the pellet was digested by 100 μg/ml proteinase K (Sigma Aldrich) for 5 h at 37°C. DNA was then purified using the GenElute PCR Clean-up Kit (Sigma Aldrich) following manufacturer’s instructions. Indirect encapsulation efficiency was measured by estimating the ‘unencapsulated’ DNA present in the supernatant after centrifugation. DNA was quantified in both cases using the SYBR Gold Assay as previously described (24). Briefly, purified DNA was diluted 1:100, and 30 μl of the dilution was distributed in triplicate wells of a white FluoroNunc 96 in each well of a white FluoroNunc 96-well plate (Thermo Fisher Scientific, Waltham, MA, US) followed by 30 μl of a 4× SYBR® gold solution was added to each well and incubation for 10 min in the dark at RT. The fluorescence of each sample was measured using a FLUOstar Omega (BMG Labtech, Ortenberg, Germany) at an excitation wavelength of 485/20 nm and an emission wavelength of 520/ 20 nm. Samples were quantified against a standard curve of two-fold serially diluted DNA. All measurements were performed in triplicates and the results are displayed as mean value ± standard deviation.
The loading efficiency was calculated as the amount of encapsulated DNA (μg DNA) present per mg of BSA, according to the encapsulation efficiency and BSA nanoparticle yield respectively.
Scanning Electron Microscopy
Surface morphology of the nanoparticles was measured using scanning electron microscopy (SEM) with a FEI, Helios G3 UC, (Thermofisher Scientific, Waltham, MA, USA) as described before (25). Briefly, 5 μl of the nanoparticle suspension was dropped onto filter paper attached to an adhesive carbon tape and mounted on the aluminium stab.
Cell Culture
Human lung adenocarcinoma cell line, A549 (ATCC® CCL-185) was cultured in complete RPMI-1640 (10% FBS, 1%Pen/Strep) at 37°C in 5% CO2. During sub-culturing, cells were washed in 1 × PBS, trypsinized with 0.25% (w/v) trypsin, and sub-cultivated at the ratio of 1:5 to 1:10. The A549 cells used here were mycoplasma free and were regularly tested for contamination. In addition, they are not listed in the database of commonly misidentified cell lines maintained by ICLAC.
Cellular Uptake and Transfection
A549 cells were seeded in a 24-well plate, at a density of 1 × 105 cells per well, the night before the experiment. The uptake of BSA nanoparticles was tested in cells with 50–60% cellular confluency. The medium was removed and FITC-BSA, pre-dispersed in 0.5 ml RPMI-1640, was added at increasing concentrations (100 μg/ml, 150 μg/ml, and 200 μg/ml). Untreated cells were used as blank control, and every concentration was tested in triplicates. After 3 h of incubation at 37°C or 4°C, the cells were washed in 500 μl PBS, and detached from the well plate with 200 μl of 0.05% trypsin solution. The trypsin was neutralized with 400 μl of RPMI-1640, and cells were centrifuged at 450 g for 5 min. After removing the supernatant, the cell pellets were resuspended in 500 μl of FACS buffer (BD Biosciences, Franklin Lakes, New Jersey, USA) and uptake efficiency was assessed by flow cytometry using the Attune™ NxT Acoustic Focusing Cytometer.
Similar experiments were performed using BSA nanoparticles encapsulating YOYO-1 labelled DNA, pDNA or siRNA labelled with Alexa 350. For uptake experiments, BSA nanoparticles were used at a final bulk DNA concentration of 250 ng and 500 ng, for pDNA at 250 ng, 500 ng, 1 μg, 1.5 μg and 2 μg, and for siRNA experiments with 30 nM and 50 nM siRNA, taking into account the loading efficiencies of each formulation.
Agarose Gel Electrophoresis
To evaluate their stability, siRNA loaded nanoparticles were treated with 50 μl FBS, 50 μl of RNase solution (10 μg/ml) or a mixture of both FBS and RNase, at 37°C for 1 h. The nanoparticles were then degraded after proteinase K treatment (100 μg/ml) and centrifuged. The supernatant was loaded onto a 20% polyacrylamide gel (Novex™ TBE Gels, 20%, Invitrogen, Carlsbad, California, USA). The gels were run in 1x Tris Borate EDTA (TBE, Sigma Aldrich) buffer for 1 h at 120 V and stained with a 3x concentrated solution of Gel Green (Biotium, Landing Parkway Fremont, California, USA) for 30 min at RT, while gentle shaking. The gel was imaged with the Chemidoc (Bio-Rad Hercules, California, USA) at 254 nm.
Cytotoxicity
For cell viability assessment via MTS assay, 3000 A549 cells were seeded per well in phenol red free RPMI-1640 medium (Sigma-Aldrich) in a 96-well plate 24 h prior to the experiment. Cells were transfected with 100, 150 and 200 μg of empty BSA nanoparticles and incubated for 48 h or with 200 μg of BSA nanoparticles for 24, 48, 72 and 96 h at 37°C, 5%CO2. Untreated cells were correspondingly incubated as blank controls. For assessment of cell viability after siRNA mediated knock down, 5000 A549 cells were seeded per well in phenol red free RPMI-1640 medium in a 96 well plate. Cells were then treated with non-specific, scrambled siCtrl, siKRAS (esiRNA, Sigma Aldrich) or siKRAS G12S (Eurofins Genomics, Ebersberg, Germany) at a final siRNA concentration of 30 nM. Cells were incubated for 48 h at 37°C, 5% CO2. Subsequently, 20 μl of CellTiter 96® AQueous One Solution (Promega, Madison, USA) was added to each well and incubated for 4 h at 37°C, 5% CO2 before absorption measurements were performed at 490 nm using a FLUOstar Omega (BMG Labtech, Ortenberg, Germany).
Apoptosis Assay
For apoptosis assessment via Annexin V and Propidium iodide staining, 50,000 A549 cells were seeded per well in a 24 well plate in RPMI-1640 complete medium 24 h prior to the experiment. Cells were then treated with non-specific, scrambled siCtrl, siKRAS (esiRNA, Sigma Aldrich) or siKRAS G12S (Eurofins Genomics, Ebersberg, Germany) at a final siRNA concentration of 30 nM. Cells were incubated for 48 h at 37°C, 5% CO2. Subsequently, cells were washed two times in cold PBS and resuspended in Annexin V Binding Buffer at a concentration of 1 × 106 cells/ml, and 100 μl of cell suspension was taken from the suspension above to be treated with 5 μl of Annexin V-FITC (BD Biosciences) and 10 μl Propidium iodide (BD Biosciences). The cell suspension was incubated for 15 min in the dark, following which 400 μl of Annexin V Binding buffer was added to each tube before measurement using the Attune™ NxT Acoustic Focusing Cytometer.
Migration Assay
For migration assays, Boyden chambers (VWR, 8 μm pore size) placed in 24 well plates were used. In the lower compartment, 500 μl of complete RPMI-1640 medium was added and A549 cells transfected 24 h before with 30 nM siRNA were added to the upper compartment (20,000 cells in 300 μl serum free medium RPMI-1640). The plates were incubated at 37°C, 5% CO2 for 16 h, after which the cells were washed with PBS and fixed with 4% PFA. Cells were removed from the upper membrane surface with a cotton tip and washed 2 times with PBS. Cells were then stained with 0.1% crystal violet (Sigma Aldrich) prepared in 20% ethanol and quantified by counting three fields (20x magnification) per chamber.
Western Blot
Total protein extract was isolated from A549 cells transfected with siCtrl, siKRAS and siKRAS G12S 48 h after transfection using RIPA Buffer (Sigma Aldrich) supplemented with 1x each protease and PhosSTOP phosphatase inhibitor cocktails (Roche, Basel, Switzerland). The samples were analysed by Western blotting following standard protocols (26) and using antibodies specific for KRAS (sc-30, Santa Cruz Biotechnology, Dallas, Texas, United States), pAKT (Ser 473) (sc-7985, Santa Cruz), pERK1/2 (#9102, Cell Signaling Technology, Danvers, Massachusetts, United States) and Histone H3 (sc-517,567, Santa Cruz).
Cellular Distribution
For confocal images, 20,000 A549 cells were seeded in 8 well μ-slides (ibidi, Martinsried, Germany) and transfected with 250 μg ofYOYO-1 labelled DNA encapsulated in BSA nanoparticles. After 24 h of incubation, cells were harvested and washed with PBS. Wells were stained with 100 μM LysoTracker Red DND-99 (Invitrogen) in pre-warmed cell culture medium for 1 h at 37°C, 5% CO2. After washing, cells were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min and washed again. DAPI was added to appropriate wells at a final concentration of 1 μg/ml in PBS and incubated for 20 min. All cells were washed again and mounted using FluorSave reagent (Merck Millipore, Billerica, USA) prior to analysis with a SP8 inverted scanning confocal microscope (Leica Camera, Wetzlar, Germany).
Statistical Analysis
All results are depicted as mean value ± standard deviation (SD) and experiments were performed in triplicates and. Oneway ANOVA was performed in GraphPad Prism (GraphPad Software, La Jolla, USA).
Results
Nanoparticle preparation and characterization
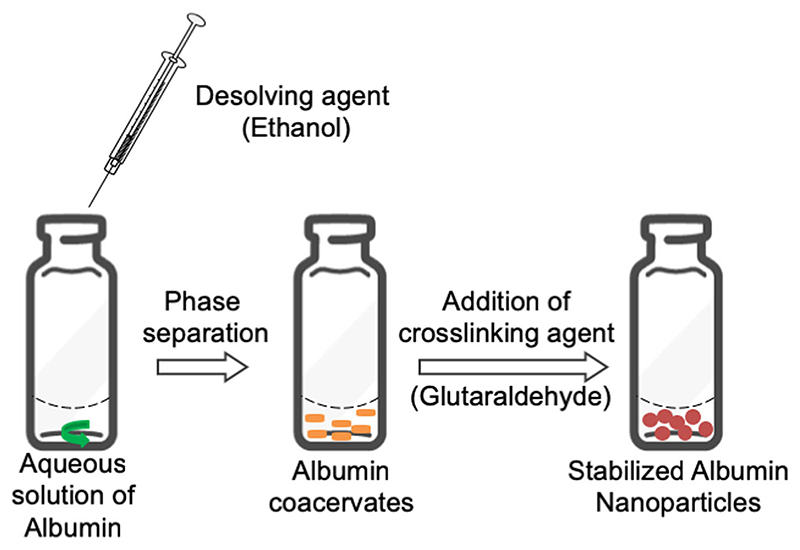
BSA nanoparticles were prepared by desolvation of an aqueous BSA solution under addition of ethanol and stabilized by the addition of glutaraldehyde (Fig. 1). Three previously published protocols were compared, and the influence of different variables on the manufacturing process was analysed. Keeping the concentration of BSA constant at 2% w/v, the amount of crosslinker, glutaraldehyde, and the desolvating agent, ethanol, was investigated with respect to their amount and/or order of addition to the aqueous BSA solution. Langer et al. previously demonstrated that the rate of addition of ethanol influences particle size (22). Therefore, we used a constant rate of ethanol addition at 1 ml/min for all three methods.
Fig. 1. Schematic representation of the desolvation method used to prepare BSA nanoparticles.
Hydrodynamic diameter, polydispersity index (PDI) and zeta potential of all prepared nanoparticles were determined by dynamic light scattering and laser Doppler anemometry, respectively. All three methods compared here resulted in nanoparticles of optimal size (below 200 nm in diameter) and a polydispersity index (PDI) below 0.15 (Table II). It was observed that the addition of more desolvating agent before purification resulted in slightly bigger particles and did not improve the particle size. Therefore, it was concluded that the third method resulted in optimized nanoparticles with regard to size and PDI.
Table II. Characterization of the BSA Nanoparticles obtained using three different methods.
| Method | Diameter (nm) | PDI | Zeta Potential (mV) | % | Loading efficiency (μg DNA/mg BSA) | |
|---|---|---|---|---|---|---|
| BSA NP Yield | DNA Encapsulation | |||||
| 1 | 149 ± 2.51 | 0.13 ± 0.01 | −14.1 ±0.75 | 81.10 ± 2.35 | 63 ± 2.12 | 11.66 ± 1.87 |
| 2 | 147 ± 3.03 | 0.07 ± 0.005 | −13.8 ± 0.54 | 75.80 ± 1.86 | 61 ± 3.53 | 12.07 ± 2.65 |
| 3 | 132 ± 1.9 | 0.05±0.01 | −20.5 ± 1.2 | 95.13 ± 1.65 | 85 ± 1.27 | 13.40 ± 1.33 |
The surface charge of nanoparticles affects their stability and blood circulation as it influences particle aggregation, opsonization and consequent uptake by the reticuloendothelial system (27). The particles obtained were negative, with zeta potential for all methods between –10 and −20 mV, both in case of empty or bulk DNA-loaded particles.
The yield of nanoparticle formation following the three methods was calculated. It was found that BSA nanoparticle yield using method 3 was 95.13%, while that of method 1 and 2 was 81.1% and 75.80%, respectively, indicating that the third method was also the most effective with respect to nanoparticle yield.
Encapsulation efficiency and release
The encapsulation efficiency of the three methods as measured by bulk DNA content of the nanoparticles was analysed using standard SYBR® Gold assays. Methods 1 and 2 both showed approximately 60% encapsulation of DNA while method 3 more efficiently packaged 85% of bulk DNA in the same amount of BSA. Based on the size, zeta potential, nanoparticle yield and encapsulation efficiency (Table II), method 3 was considered most suitable and used for further analysis.
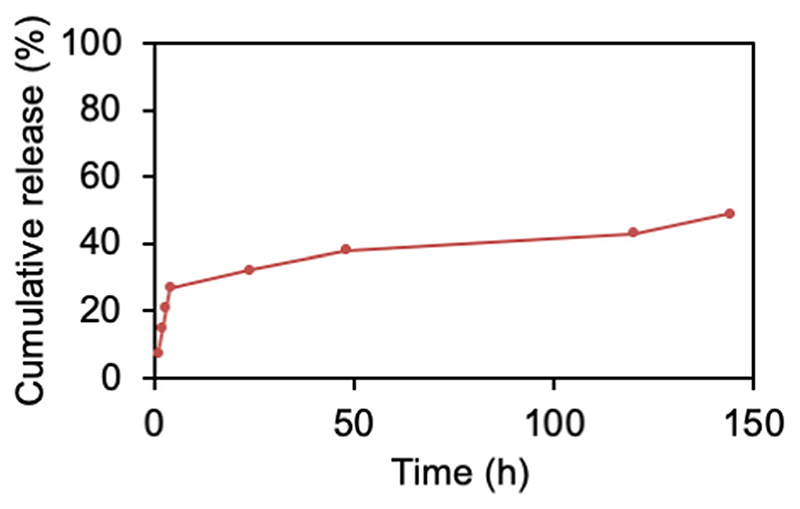
In vitro release of DNA from BSA nanoparticles was evaluated by dispersing freshly prepared nanoparticles in 1 ml PBS followed by shaking at 300 rpm at 37°C. The suspension was then centrifuged at 20,000 g for 10 min at predetermined time intervals, and the DNA released in the supernatant was evaluated by SYBR Gold assay. As shown in Fig. 2, the release of DNA from the BSA nanoparticles follows a biphasic curve with an initial fast release phase, wherein 30% of the DNA was released within the first 6 h followed by a slow and sustained release phase, during which another 18% of the DNA was released. However, after 1 week, the BSA particles showed aggregation with an average size of 400 nm.
Fig. 2. DNA release profile of bulk DNA loaded nanoparticles prepared following Method 3.
Released DNA (%, Y axis) was measured by SYBR Gold assay and plotted against time (hours, X-axis). Data points indicate mean ±SD, n = 3.
Uptake of fluorescently labelled BSA nanoparticles
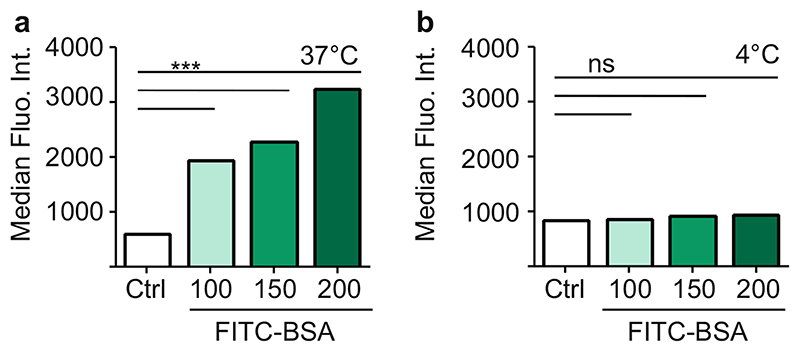
The uptake of FITC-labelled BSA particles in vitro was evaluated by flow cytometry. Lung cancer cells (A549) were treated with increasing concentrations of FITC-BSA nanoparticles, 100, 150 and 200 μg/ml dispersed in medium. Figure 3 shows the median fluorescence intensity (MFI) measured in A549 cells 4 h after treatment as compared to unlabelled BSA treated cells (Control, Ctrl). We observed a rapid uptake of FITC-BSA nanoparticles and a concentration dependent increase in cells grown at 37°C, as shown in Fig. 3. To distinguish between passive adsorption on the outer cell membrane and active uptake mechanisms, fluorescence association was compared at 4°C and 37°C. At 37°C, a rapid and concentration dependent increase was observed in cellular fluorescence while there was only minimal increase in fluorescence at 4°C despite increased particle concentration. No significant increase compared to the control cells was observed, indicating that BSA particles do not heavily adsorb to A549 cells. The fluorescence observed at 37°C was significantly higher (p <0.001 for all tested concentrations) indicating that an energy-based uptake mechanism based on endocytosis can be assumed and that BSA based nanoparticles are efficiently taken up in A549 cells.
Fig. 3. Cellular uptake of FITC-BSA nanoparticles in A549 cells at (a) 37°C and (b) 4°C measured by flow cytometry represented as median fluorescence intensity (MFI, Y axis) Data points indicate mean ± SD, n = 3; One-way ANOVA, ***, p < 0.001, n.s., not significant).
Cytotoxicity
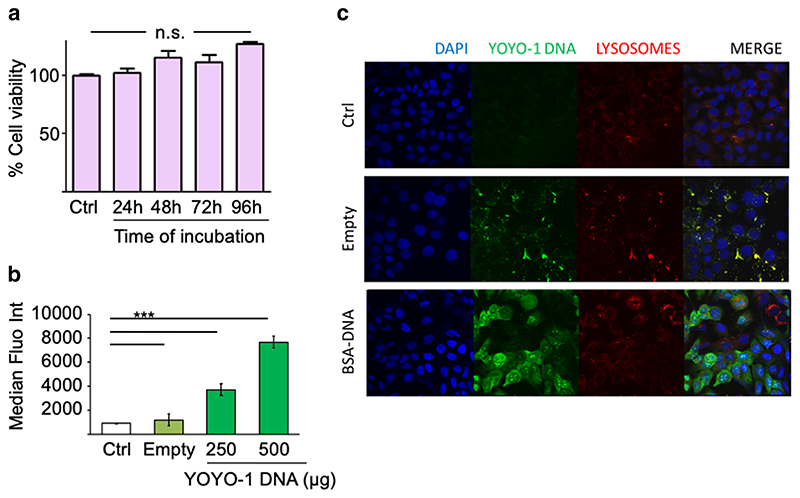
One critical requirement of gene delivery vectors is compatibility with living cells. Therefore, we evaluated the cytotoxicity of BSA nanoparticles over 4 days. A549 cells were incubated with bulk DNA containing BSA nanoparticles at increasing amounts of BSA (100, 150 and 200 μg) for 48 h and 200 μg BSA for up to 96 h, and cell viability was evaluated by MTS assay. As shown in Supplementary Fig. 1 and Fig. 4a, cell viability remained above 80% over the entire incubation period of 96 h, indicating that BSA nanoparticles do not strongly/significantly affect cell proliferation of A549 cells.
Fig. 4. BSA nanoparticles can safely and efficiently deliver DNA to A549 cells.
(a) Cell viability (% Y-axis) as measured by MTS assay in A549 cells treated with 200 μg of BSA nanoparticles for 24, 48, 72 and 96 h (Time, X-axis). (b) Transfection of A549 cells with BSA nanoparticles loaded with a YOYO-1 labelled DNA measured by flow cytometry as median fluorescence intensity (MFI, Y-axis). μg DNA is DNA amount used for transfection, taking into account encapsulation efficiency for each preparation. (c) Confocal images of A549 cells 24 h after transfecting with BSA nanoparticles loaded with YOYO-1 labelled DNA (green) and staining with DAPI (blue, depicting the cell nuclei) and LysoTracker Red DND-99(red, representing the lysosomes). Empty nanoparticles exhibit auto fluorescence on excitation at 488 nm. Ctrl, untreated cells. Data points indicate mean ± SD, n = 3; One-way ANOVA, ***, p < 0.001; ns, not significant.
Cellular delivery and endosomal escape of DNA
Cellular uptake of YOYO-1 labelled bulk DNA loaded BSA nanoparticles was investigated by flow cytometry. As shown in Fig. 4b, the MFI of A549 cells treated for 24 h with nanoparticles containing 500 ng YOYO-1 labelled DNA was significantly higher than after treatment with nanoparticles containing 250 ng DNA.
To confirm whether the DNA cargo is released from the carrier and can escape the endosomal compartment, cellular distribution of the YOYO-1 labelled DNA was performed via confocal microscopy. Empty BSA particles exhibit measurable autofluorescence if excited at 488 nm. Lysosomes were stained using Lysotracker-Red. Figure 4c shows punctate distribution of the empty BSA particles, mostly colocalizing with the lysosomes. Interestingly, YOYO-1 labelled DNA is found homogenously spread within the cells, as well as in the nuclei.
BSA nanoparticles facilitate gene expression in vitro
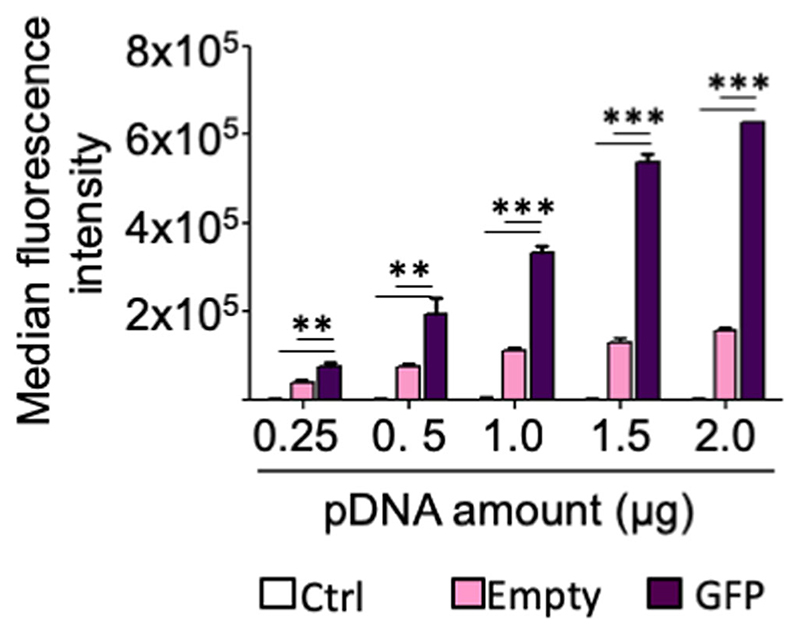
Although uptake and cellular localization of YOYO-1 DNA confirmed efficient internalization of the DNA, its functionality remained to be evaluated. To this extent, BSA nanoparticles were loaded with a plasmid DNA expressing GFP under the CMV promoter. A549 cells were transfected with BSA nanoparticles containing increasing amounts of pDNA, and after 48 h, GFP levels were measured by flow cytometry. Figure 5 confirms successful transfection efficiency of the plasmid DNA loaded BSA nanoparticles in comparison to just autofluorescence of the empty particles. Even at plasmid doses as low as 250 ng, significantly increased GFP expression was measured. These results support the use of BSA nanoparticles as efficient nucleic acid carriers for potential therapeutic lung cancer approaches.
Fig. 5. Transfection of A549 cells with BSA nanoparticles loaded with a GFP expressing plasmid measured by flow cytometry as median fluorescence intensity (MFI) ofGFP (Y-axis).
Ctrl, untreated cells. μg pDNA is pDNAamount used for transfection, taking into account encapsulation efficiency for each preparation. Data points indicate mean ± SD, n = 3; One-way ANOVA, ***, p < 0.001; ns, not significant.
siRNA loaded nanoparticles
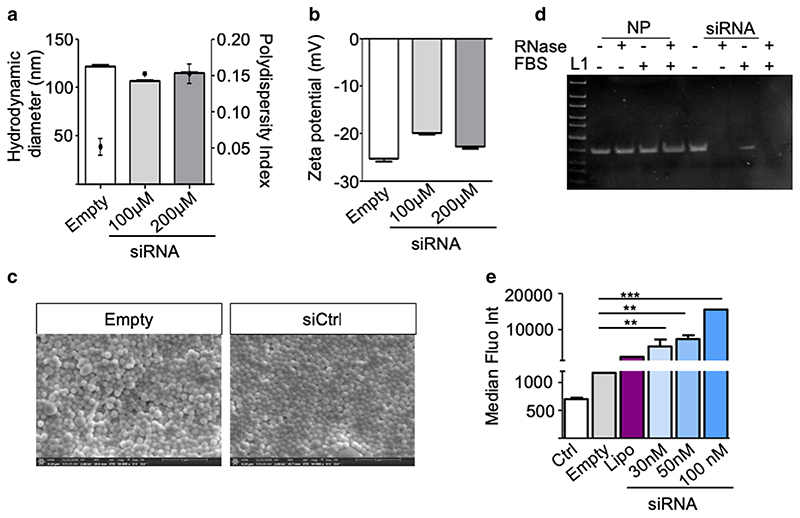
In an approach to achieve therapeutic gene silencing, we also prepared siRNA loaded BSA nanoparticles. As shown in Fig. 6a, b, the siRNA loaded BSA nanoparticles had an average size of 100 nm, with PDI below 0.2 and a zeta potential around -25 mV. Moreover, BSA nanoparticles were able to efficiently condense siRNA, with an encapsulation efficiency of 87.14 ± 0.95% (Table III). Scanning electron microscopy (Fig. 6c) images revealed structures with spherical shape and confirmed the homogenous dispersity of the nanoparticles.
Fig. 6. Characterization of siRNA loaded BSA nanoparticles.
(a) Hydrodynamic diameters (left y-axis, bars) and polydispersity indices (PDI, right y-axis, dots) (b) zeta potentials of the nanoparticles (c) Scanning electron microscope (SEM) micrographs of empty (left) and siRNA loaded (right) BSA nanoparticles. (d) Electrophoretic mobility of naked siRNA (siRNA), siRNA loaded BSA nanoparticles (–) following incubation with FBS (+ FBS) or RNase (+ RNase) or both (+ +). L, ultra-low range DNA ladder. (e) Cellular uptake of BSA nanoparticles loaded with Alexa 350 labelled siRNA in A549 cells measured by flow cytometry and represented as median fluorescence intensity (MFI, Y axis). Data points indicate mean ± SD, n = 3.
Table III. Encapsulation efficiency of siRNA loaded BSA Nanoparticles.
| siRNA starting amount (ng) | siRNA encapsulated (ng) | % siRNA encapsulation | Loading efficiency (ng siRNA/mg BSA) |
|---|---|---|---|
| 1000 | 878.14 | 87.8 | 175.6 |
| 2000 | 1721.04 | 86.05 | 344.2 |
| 4000 | 3502.83 | 87.57 | 700.5 |
One critical requirement for non-viral gene delivery vehicles is the ability to protect the nucleic acid cargo against enzymatic degradation in the biological environment, which is especially relevant to the labile siRNA. To evaluate their stability, siRNA loaded nanoparticles were treated with FBS, an environment known to be rich in RNases, and in RNase solution (10 μg/ml) at 37°C for 1 h. The nanoparticles were then degraded by proteinase K treatment. The integrity ofthe released siRNA was confirmed with agarose gel electrophoresis. Figure 6d shows similar size bands for untreated control siRNA, and siRNA released from untreated particles, as well as from particles incubated with RNase, FBS or a mixture of RNase and FBS. Based on the results, it is concluded that the siRNA was intact after FBS and RNase treatment of the particles but not after treatment of the free siRNA. This observation also led to the hypothesis that by encapsulation in BSA, enhanced siRNA stability was achieved for in vivo applications.
Next, cellular uptake of Alexa Fluor 350-labelled siRNA (AF350-siRNA) was evaluated by flow cytometry. Since empty BSA nanoparticles show auto fluorescence at 488 nm, Alexa Fluor 350 was used to label the siRNA. Figure 6e shows the MFI of A549 cells 24 h after transfection with nanoparticles composed of AF350-siRNA at a final concentration of 30, 50 and 100 nM, calculated according to encapsulation efficiency. Untreated cells (Ctrl) and empty BSA nanoparticles were used as negative controls, and lipofectamine 2000 (Lipo), a commonly used transfection reagent, as positive control. As observed with YOYO-1 labelled DNA and pDNA, high cellular uptake was observed, even as compared to Lipofectamine. These results confirm the suitability of BSA nanoparticles to deliver siRNA to lung cancer cells.
BSA nanoparticles targeting mutant KRAS
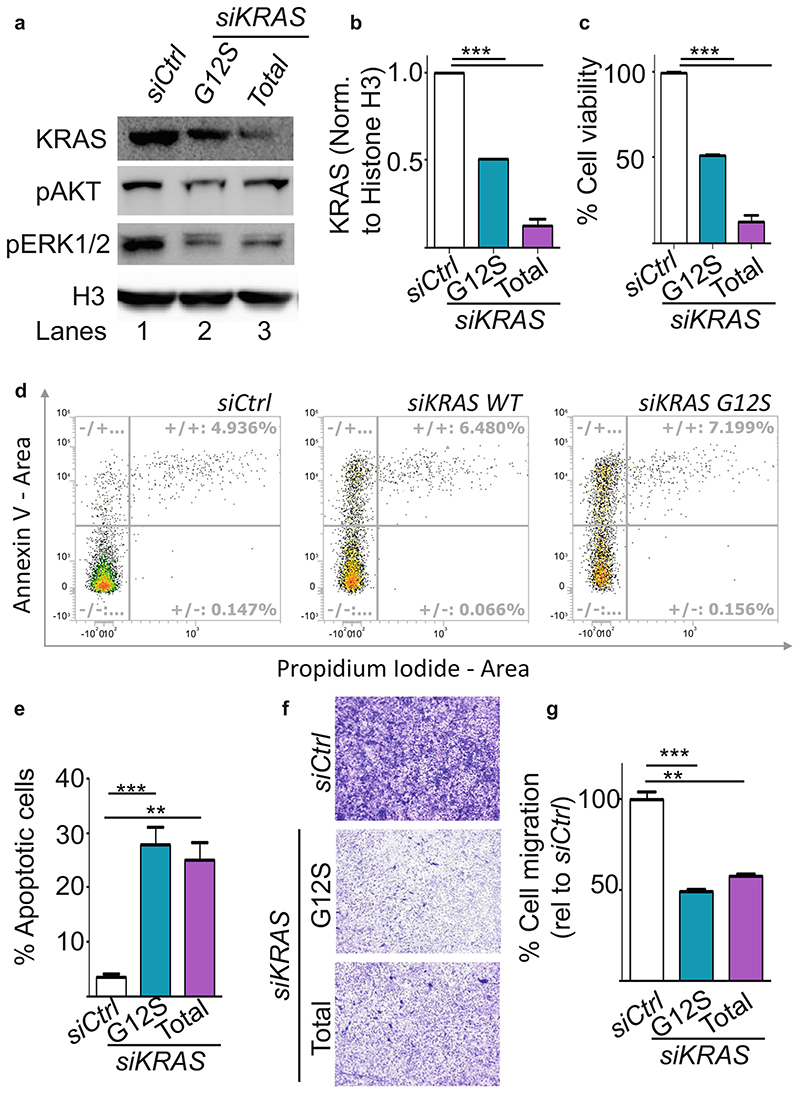
In order to use the BSA nanoparticles therapeutically as carriers for anti-tumour nucleic acid delivery, we then prepared BSA nanoparticles loaded with siRNA specific to a mutant allele of KRAS, i.e. KRAS G12S. As control, we used a siRNA pool against the wild type KRAS allele. We transfected A549 cells, containing the KRAS G12S mutant allele, with BSA particles containing the siRNAs, scrambled control siRNA (Ctrl), siRNA against KRAS WT or KRAS G12S, at a final concentration of 50 nM. siRNA mediated loss of function of KRAS was evaluated by western blot (Fig. 7a) using an antibody specific for KRAS. It was observed that 50 nM of the siRNA pool against KRAS WT resulted in almost 90% loss of KRAS. While, using the mutant specific siRNA led to a 45% loss in KRAS, as measured by western blot using an antibody against WT KRAS. Both siRNAs could reduce the levels of pAKT (ser 473) and pERK1/2, the downstream effectors of the signalling pathways. Interestingly, the allele specific knockdown of KRAS G12S resulted in a stronger inhibition of signalling as compared to the loss of WT KRAS, confirming that mutation specific inhibition of KRAS is essential.
Fig. 7. Mutation specific knock of KRAS after transfection with siRNA loaded BSA nanoparticles.
(a) Protein extracts from A549 cells transfected with BSA nanoparticles loaded with 30 nM of scrambled control siRNA (siCtrl, Lane I), siRNA against KRAS GI2S (siKRAS GI2S, Lane 2) or siKRAS against both WT and mutant allele (siKRAS Total, Lane 3) were analysed by western blot using the indicated antibodies. Histone H3 (H3) was used as loading control. (b) Quantification of the KRAS knockdown observed in A. (c) Cell viablilty (% Y-axis) as measured by MTS assay in A549 cells transfected with siCtrl, siKRAS GI2S and siKRAS total. (d) Dot plot showing the percentage of Annexin V- or propidium iodide positive A549 cells after transfection with siCtrl, siKRAS GI2S or siKRAS Total. x and y axes denote Propidium iodide and Annexin V signals respectively, (e) Quantitation of Annexin V-positive cells represented as percentage of apoptotic cells. (f) A549 were subjected to transwell migration assays 24 h aftertransfection as indicated. The crystal violet dye staining images of the membranes are shown. (g) Percentage of migrated cells was quantified by counting three fields (20x magnification) per chamber and compared with controls. Magnification, X20X. Data points indicate mean ± SD, n = 3. One-way ANOVA, **, p < 0.01, ***, p < 0.001.
To understand the functional benefit of this KRAS knock down, we subsequently performed an MTS assay 48 h after transfecting A549 cells with 50 nM siRNA. Figure 7c shows 33% cell viability in siKRAS G12S treated cells and 25% cell viability in siKRAS treated cells. Furthermore, we measured the apoptotic cells by Annexin V/Propidium iodide double staining using flow cytometry. Staining with Annexin V was used in conjunction with the vital dye, propidium iodide for identification of early and late apoptotic cells. Viable cells with intact membranes exclude propidium iodide, whereas the membranes of dead and damaged cells are permeable to it. Therefore, viable cells are double negative, while cells that are in early apoptosis are Annexin V positive and propidium iodide negative, and cells that are in late apoptosis or already dead are positive for both stains. As seen in Fig. 7d, e, the population of normal cells decreased while the population of apoptotic cells significantly increased (p < 0.05) in cells transfected with siRNA against KRAS compared with the Ctrl transfected cells. Interestingly, cells transfected with siKRAS G12S showed a significant increase of early apoptotic cells as well as 1% more late apoptotic cells as compared to cells transfected with siRNA against total KRAS underlining the benefits of G12S-specific knock down.
We further studied the effects of KRAS knock-down on another hallmark of cancer, i.e. cell migration using a transwell assay. The migratory ability of A549 cells transfected with siKRAS G12S and siKRAS decreased by 53 ± 0.7% and 47 ± 2.1% respectively (Fig. 7f, g) as compared to siCtrl transfected cells. These results confirm the therapeutic benefit of the KRAS G12S mutant specific siRNA mediated knock down.
Discussion
siRNAs are an attractive new class of therapeutics, especially when used against undruggable targets for the treatment of cancer and other diseases. Despite the potential of siRNAs in therapy, its clinical application still faces many challenges, including rapid degradation and poor cellular uptake of siRNA as well as off-target effects. Design of siRNAs and use of nanocarriers as delivery vehicles offer significant opportunities to overcome these challenges.
In the present study, BSA nanoparticles were used as the delivery vehicle for siRNA. Albumin nanoparticles represent a promising strategy for drug delivery to tumour cells. The anatomical differences between normal and tumour cells, such as high vascular density, extensive production of vascular mediators, rapid and defective angiogenesis and large pores in the tumour vasculature, allow higher vascular permeability in tumour tissues. Such leaky vasculature permits the extravasation of circulating nanoparticles within the tumour interstitium which then accumulate due to impaired lymph drainage (28). This passive targeting is known as the Enhanced Permeability and Retention (EPR) effect, which currently is discussed very controversially in the literature (29,30) and may not have any impact in clinical lung cancer treatment.
Additional to the EPR effect, the enhanced uptake of BSA nanoparticles in solid tumours is mediated by binding of albumin to albumin binding proteins, such as the membrane associated 60 kDa glycoprotein, gp60, and the secreted protein, acidic and rich in cysteine, SPARC (14,25,31). As albumin binds to gp60 receptor, it initiates the endothelial transcytosis. Gp60 binds to CAVEOLIN-1, and initiates the formation of caveolae, which transport albumin across the extravascular space. SPARC is an extracellular glycoprotein (32,33) which binds to albumin with high affinity (33), and is upregulated in tumour tissue, thereby resulting in the accumulation of albumin, also in form of nanoparticles, in the tumour interstititium.
In this report, BSA nanoparticles were prepared by a desolvation-crosslinking method. Three different methods for BSA nanoparticle formation were evaluated, wherein the amount of crosslinker, glutaraldehyde, and the desolvating agent, ethanol, was investigated. As seen in Fig. 1, all three methods resulted in nanoparticles smaller than 200 nm, presenting optimal characteristics for in vivo use, which is well in accordance with previous reports (21–23). Interestingly, higher effective concentration of glutaraldehyde (Method 3) favours the formation of more condensed particles. This reduced particle size resulted in a significant increase in surface area of the nanoparticle, allowing more surface adsorption of the negatively charged nucleic acids, confirmed by the high encapsulation efficiency and resulted in a slightly decreased zeta potential. The degree of crosslinking also influences the release kinetics of the encapsulated nucleic acid. We observed a cumulative release of 48% of the encapsulated DNA over 6 days in PBS. The initial fast release phase can be attributed to the immediate desorption of surface adsorbed DNA, while the slow but steady release phase can be due to the release of tightly entrapped DNA. These results are in accordance with those reported previously (21), wherein high crosslinking of BSA resulted in sustained gene delivery over one week while low crosslinking resulted in 100% DNA release within 2 days. However, since the BSA nanoparticles started to aggregate after 6 days at 37°C, we were unable to estimate the time required for complete DNA release. High and efficient DNA release is also exemplified by the uptake of YOYO-1 labelled DNA packaged in BSA nanoparticles. Our results show a significant increase in fluorescence using 250 ng of YOYO-1 labelled DNA, as compared to control, within 24 h, indicating rapid and efficient internalization of the BSA nanoparticles and the release of the DNA.
In addition to optimal release of the nucleic acid cargo, the other rate-determining step is intracellular trafficking and endosomal release. BSA nanoparticles loaded with YOYO-1 labelled DNA revealed almost no colocalization with lysosomes and nuclear transport of the DNA, correlating with previous reports (21,34). Using caveolae and clathrin transport inhibitors, Mo et al. have shown that both caveolae and clathrin are involved in the endocytosis of albumin particles (21). Since no lysosomal colocalization can be seen with YOYO-1 labelled DNA, the particles could have entered the cell via clathrin-dependent endocytotic pathways, subsequently reaching the late endosomes and lysosomes from where they may rapidly escape. However, the mechanism for the escape of the particles from endo/lysosome or other intracellular vesicles remains to be the scope of future work.
Since siRNAs are especially sensitive to nucleases, an ideal nanocarrier should be able to protect the labile siRNA cargo against enzymatic degradation in the biological environment. Gel electrophoresis after treatment with RNases and FBS shows intact siRNA released from the nanoparticles, indicating that the nanoparticle encapsulation enhances siRNA stability, making it resistant to nuclease degradation. These results confirm the hypothesis that siRNAs encapsulated within BSA nanoparticles can be used as robust vehicles for siRNA transport to tumour cells.
To achieve therapeutically relevant gene silencing, we used an siRNA against a KRAS mutant, KRAS G12S. The G12S mutation results in an amino acid substitution at position 12 in KRAS, from a glycine (G) to a serine (S). KRAS mutations are always present as heterozygous mutations, indicating that only one mutant allele will be present in the cells. Poloxamer – human IgG hybrid nanoparticles were used to deliver siKRAS G12S in vivo and were found to regress lung tumours in xenograft models, suggesting it as a promising prospect for treatment of NSCLC (35). Similar observations were also observed using siRNA against KRAS G12D mutant tumours in a mouse model for pancreatic cancer (36), confirming that mutated KRAS is indeed an ideal target for RNAi based therapies. In this report, 45% loss of KRAS was obtained after transfection with the mutant specific siRNA and 90% loss using the total KRAS siRNA pool. Both approaches resulted in increased apoptosis, decreased proliferation and migratory abilities as compared to siCtrl transfected cells. The siRNA pool targets the KRAS mRNA at multiple locations resulting in a higher, but non-specific knockdown affecting both, WT and mutant KRAS alleles. Notably, despite the stronger knock down using siKRAS total, the functional benefit obtained was not as pronounced as compared to siKRAS G12S. KRAS mediated downstream signalling occurs via the PI3K-AKT pathway or the RAF-ERK1/2 pathway. We observed a stronger reduction in both pAKT and pERK1/ 2 in the siKRAS G12S transfected cells as compared to siKRAS transfected cells. These results support the theory of oncogene addiction, wherein cancer cells are ‘addicted’ to extreme signalling levels and sudden cessation of signalling results in apoptosis (37).
Conclusions
In summary, BSA nanoparticles encapsulating siRNA against a point mutation in KRAS were developed. These particles showed an optimal size and zeta potential, high encapsulation efficiency. BSA nanoparticles exhibited minimal toxicity and could protect the siRNA against RNases resulting in high knock down efficiencies in vitro. The BSA nanoparticles overcome several barriers associated with gene delivery vehicles for RNAi based therapeutics and will be evaluated in vivo in ongoing research.
Supplementary Material
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11095-019-2665-9) contains supplementary material, which is available to authorized users.
Acknowledgments and Disclosures
This work was in part supported by the Deutsche Gesellschaft für Muskelkranke e.V. (Grant Number 2017-Me7/1) and ERC Starting Grant ERC-2014-StG – 637,830 “Novel Asthma Therapy” to Olivia Merkel as well as the LMU Excellent Nachwuchsförderungsfonds to Aditi Mehta.
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–94. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dogan S, Shen R, Ang DC, Johnson ML, D’Angelo SP, Paik PK, et al. Molecular epidemiology of EGFR and KRAS mutations in 3, 026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–77. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ElOsta B, Behera M, Kim S, Berry LD, Sica G, Pillai RN, et al. Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: the lung Cancer mutation consortium experience. J Thorac Oncol. 2019 doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3(6):459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 7.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–8. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 8.Merkel OM, Zheng M, Debus H, Kissel T. Pulmonary gene delivery using polymeric nonviral vectors. Bioconjug Chem. 2012;23(1):3–20. doi: 10.1021/bc200296q. [DOI] [PubMed] [Google Scholar]
- 9.Shepherd FA, Domerg C, Hainaut P, Janne PA, Pignon JP, Graziano S, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31(17):2173–81. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464(7287):436–40. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kandil R, Merkel M. Therapeutic delivery of RNA effectors: diseases affecting the respiratory system. Pharmazie. 2016;71(1):21–6. [PubMed] [Google Scholar]
- 12.Merkel OM, Kissel T. Nonviral pulmonary delivery of siRNA. Acc Chem Res. 2012;45(7):961–70. doi: 10.1021/ar200110p. [DOI] [PubMed] [Google Scholar]
- 13.Tarhini M, Benlyamani I, Hamdani S, Agusti G, Fessi H, Greige-Gerges H, et al. Protein-based nanoparticle preparation via nanoprecipitation method. Materials (Basel) 2018;11(3) doi: 10.3390/ma11030394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elzoghby AO, Samy WM, Elgindy NA. Protein-based nanocarriers as promising drug and gene delivery systems. J Control Release. 2012;161(1):38–49. doi: 10.1016/j.jconrel.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 15.Elzoghby AO, Samy WM, Elgindy NA. Albumin-based nanoparticles as potential controlled release drug delivery systems. J Control Release. 2012;157(2):168–82. doi: 10.1016/j.jconrel.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 16.Gong J, Chen M, Zheng Y, Wang S, Wang Y. Polymeric micelles drug delivery system in oncology. J Control Release. 2012;159(3):312–23. doi: 10.1016/j.jconrel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Tong R, Yala L, Fan TM, Cheng J. The formulation of aptamer-coated paclitaxel-polylactide nanoconjugates and their targeting to cancer cells. Biomaterials. 2010;31(11):3043–53. doi: 10.1016/j.biomaterials.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kratz F, Beyer U. Serum proteins as drug carriers of anticancer agents: a review. DrugDeliv. 1998;5(4):281–99. doi: 10.3109/10717549809065759. [DOI] [PubMed] [Google Scholar]
- 19.Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release. 2008;132(3):171–83. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Monsigny M, Roche AC, Midoux P. Uptake of neoglycoproteins via membrane lectin(s) of L1210 cells evidenced by quantitative flow cytofluorometry and drug targeting. Biol Cell. 1984;51(2):187–96. doi: 10.1111/j.1768-322x.1984.tb00298.x. [DOI] [PubMed] [Google Scholar]
- 21.Mo Y, Barnett ME, Takemoto D, Davidson H, Kompella UB. Human serum albumin nanoparticles for efficient delivery of Cu, Zn superoxide dismutase gene. Mol Vis. 2007;13:746–57. [PMC free article] [PubMed] [Google Scholar]
- 22.Langer K, Balthasar S, Vogel V, Dinauer N, vonBriesen H, Schubert D. Optimization of the preparation process for human serum albumin (HSA) nanoparticles. Int J Pharm. 2003;257(1–2):169–80. doi: 10.1016/s0378-5173(03)00134-0. [DOI] [PubMed] [Google Scholar]
- 23.Wartlick H, Spankuch-Schmitt B, Strebhardt K, Kreuter J, Langer K. Tumour cell delivery of antisense oligonuclceotides by human serum albumin nanoparticles. J Control Release. 2004;96(3):483–95. doi: 10.1016/j.jconrel.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 24.Feldmann DP, Cheng Y, Kandil R, Xie Y, Mohammadi M, Harz H, et al. In vitro and in vivo delivery of siRNA via VIPER polymer system to lung cells. J Control Release. 2018;276:50–8. doi: 10.1016/j.jconrel.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andima M, Costabile G, Isert L, Ndakala AJ, Derese S, Merkel OM. Evaluation of beta-Sitosterol loaded PLGA and PEG-PLA nanoparticles for effective treatment of breast Cancer: preparation, physicochemical characterization, and antitumor activity. Pharmaceutics. 2018;10(4) doi: 10.3390/pharmaceutics10040232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta A, Cordero J, Dobersch S, Romero-Olmedo AJ, Savai R, Bodner J, et al. Non-invasive lung cancer diagnosis by detection of GATA6 and NKX2-1 isoforms in exhaled breath condensate. EMBO Mol Med. 2016;8(12):1380–9. doi: 10.15252/emmm.201606382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wacker M. Nanocarriers for intravenous injection-the long hard road to the market. IntJ Pharm. 2013;457(1):50–62. doi: 10.1016/j.ijpharm.2013.08.079. [DOI] [PubMed] [Google Scholar]
- 28.Jhaveri AM, Torchilin VP. Multifunctional polymeric micelles for delivery of drugs and siRNA. Front Pharmacol. 2014;5:77. doi: 10.3389/fphar.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols JW, Bae YH. EPR: evidence and fallacy. J Control Release. 2014;190:451–64. doi: 10.1016/j.jconrel.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 30.Danhier F. To exploit the tumor microenvironment: since the EPR effect fails in the clinic, what is the future of nanomedicine? J Control Release. 2016;244(Pt A):108–21. doi: 10.1016/j.jconrel.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Park K. Albumin: a versatile carrier for drug delivery. J Control Release. 2012;157(1):3. doi: 10.1016/j.jconrel.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Gradishar WJ. Albumin-bound paclitaxel: a next-generation taxane. Expert Opin Pharmacother. 2006;7(8):1041–53. doi: 10.1517/14656566.7.8.1041. [DOI] [PubMed] [Google Scholar]
- 33.Fu Q, Sun J, Zhang W, Sui X, Yan Z, He Z. Nanoparticle albuminbound (NAB) technology is a promising method for anti-cancer drug delivery. Recent Pat Anticancer Drug Discov. 2009;4(3):262–72. doi: 10.2174/157489209789206869. [DOI] [PubMed] [Google Scholar]
- 34.Langiu M, Dadparvar M, Kreuter J, Ruonala MO. Human serum albumin-based nanoparticle-mediated in vitro gene delivery. PLoS One. 2014;9(9):e107603. doi: 10.1371/journal.pone.0107603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perepelyuk M, Shoyele O, Birbe R, Thangavel C, Liu Y, Den RB, et al. siRNA-encapsulated hybrid nanoparticles target mutant K-ras and inhibit metastatic tumor burden in a mouse model of lung Cancer. Mol Ther Nucleic Acids. 2017;6:259–68. doi: 10.1016/j.omtn.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ZordeKhvalevsky E, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci USA. 2013;110(51):20723–8. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torti D, Trusolino L. Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: promises and perils. EMBO Mol Med. 2011;3(11):623–36. doi: 10.1002/emmm.201100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.