Abstract
To uncover the minimal requirements for morphogen gradient formation, we have engineered a synthetic morphogen in Drosophila wing primordia. We show that an inert protein (GFP) can form a detectable diffusion-based gradient in the presence of surface-associated anti-GFP nanobodies, which modulate the gradient by trapping the ligand and limiting leakage from the tissue. We next fused anti-GFP nanobodies to the receptors of Dpp, a natural morphogen, to render them responsive to extracellular GFP. In the presence of these engineered receptors, GFP could replace Dpp to organize patterning and growth in vivo. Concomitant expression of GPI-anchored non-signaling receptors further improved patterning, to near wild type quality. Theoretical arguments suggest that GPI-anchorage could be important for these receptors to expand the gradient length scale while reducing leakage.
Morphogen transport in epithelia
During development, morphogens provide positional information by forming long range concentration gradients. Despite the importance of morphogens, there is still no consensus on how they spread within tissues (1). The most parsimonious view is that morphogens travel by diffusion (1–3). However, epithelia, monolayered sheets of cells, present a particular challenge for diffusion-based mechanisms since ligand leakage is expected to occur, thus compromising planar gradient formation (4) and possibly affecting the development of other tissues and organs (5). Much of our knowledge about the formation and interpretation of morphogen gradients in epithelia comes from studies of the Bone Morphogenetic Protein (BMP) homolog Decapentaplegic (Dpp) in wing imaginal discs of Drosophila. In these epithelial pouches, Dpp is produced by a stripe of cells and spreads to form a gradient that organizes growth and patterning along the anterior-posterior (A/P) axis (6). It has been suggested that Dpp spreads by planar transcytosis or on specialized filopodia called cytonemes (7, 8). Both mechanisms would ensure planar transport, however, so far, direct functional evidence remains scant. In contrast, there is extensive genetic evidence for the requirement of glypicans in morphogen transport (9–11). It has been suggested that morphogens could piggyback on laterally diffusing glypicans while undergoing dissociation-reassociation cycles for cell-to-cell transfer, thus remaining within the plane of the epithelium. Here we take a forward engineering approach to investigate whether an inert protein can form a diffusion-based gradient in the basolateral space of a developing pseudostratified epithelium and specify positional information.
Extracellular binders reveal a diffusion-based gradient
Synthetic approaches have become a powerful tool to uncover the key features of natural processes (12, 13). To assess the ability of an inert protein to form a gradient in wing imaginal discs of Drosophila, we engineered flies to express, from a localized source, green fluorescent protein appended with a secretion targeting signal (SecGFP) (Fig. 1A). This was achieved by integrating SecGFP-coding sequences into the patched (ptc) locus (Fig. S1), a gene which, like dpp, is expressed along the A/P boundary (Fig.1A). GFP was detectable, albeit weakly, in the expression domain (Fig. 1B, C; no binders). GFP fluorescence was also present uniformly in the peripodial space, an enclosed lumen on the epithelium’s apical side. By contrast, the basolateral space was devoid of detectable GFP, most likely because it is exposed to the larval circulation, which could provide an escape route (see schematic in Fig. S2A). Indeed, GFP can cross the basal lamina to and from the hemolymph (Fig. S2) and leakage could therefore prevent locally expressed GFP from forming a detectable gradient in the basolateral space. Natural morphogens, which form gradients, are known to bind various receptors and extracellular components (11). We therefore asked whether adding GFP-binding species in the extracellular space would reduce leakage and enable the formation of a detectable gradient in SecGFP-expressing wing imaginal discs.
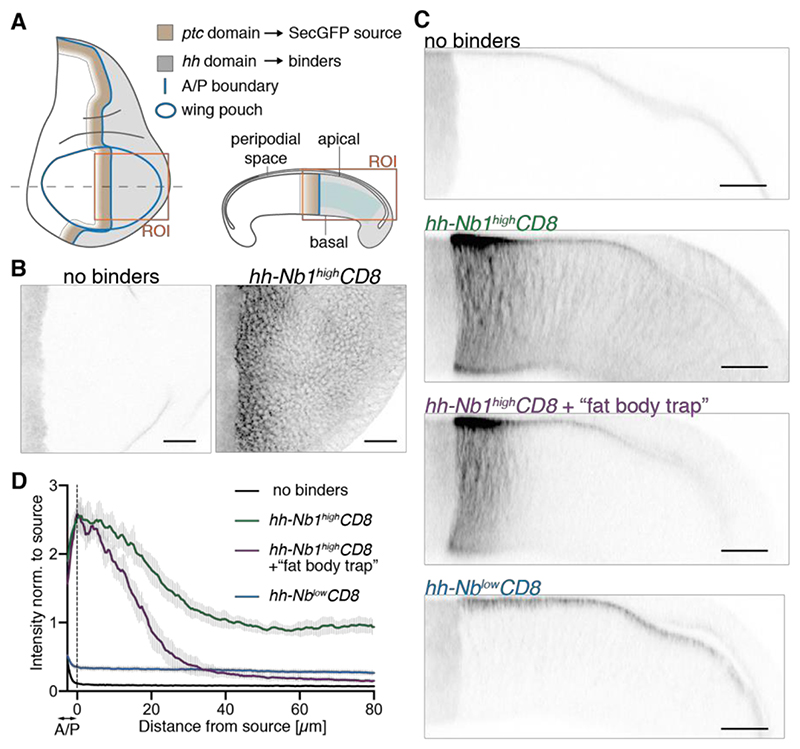
Fig. 1. Establishment of a GFP gradient in a developing epithelium.
(A) Schematic representation of a wing imaginal disc of Drosophila. SecGFP is expressed under the control of the ptc promoter (brown), while a membrane-tethered anti-GFP nanobody is expressed under the control of the hh promoter (grey). ROI indicates the areas depicted in panels B and C. Blue shading indicates the region used to generate the profiles shown in D. (B) In the absence of binders, SecGFP can be seen at the source but is not detectable in basolateral focal planes. Upon expression of high-affinity binders in the posterior compartment (hh-Nb1highCD8), a gradient is readily seen. (C) Cross-sections of imaginal discs expressing SecGFP in the ptc domain, showing that, in the absence of binders, GFP is detectable in the peripodial space but not in the basolateral space. In the presence of binders (hh-Nb1highCD8), a gradient can be seen in the basolateral space but with a non-zero tail, which is largely abrogated by concomitant activation of UAS-Nb1highCD8 in the fat body (+ “fat body trap”). Only a shallow basolateral gradient is detected when a low-affinity binder is expressed (hh-NblowCD8). (D) Fluorescence intensity profiles derived from preparations like those shown in panel C. The vertical dotted line marks the estimated posterior edge of the source. The numbers of discs analyzed are as follows: No binders, n=10; hh-Nb1highCD8, n=11; hh-Nb1highCD8 + fat body trap, n=7; hh-NblowCD8, n=10. Scale bars: 20 μm
Extracellular GFP-binding proteins are readily engineered by fusing a transmembrane protein (e.g. human CD8) to one of the many characterized anti-GFP nanobodies (14), such
as GBP1 (aka vhhGFP4), which binds GFP with a Kd of 0.23 nM (15, 16) (referred henceworth as Nb1high). DNA encoding this fusion protein (Nb1highCD8) was knocked into the hedgehog (hh) locus so that it be expressed at ‘physiological level’ in a domain (grey shading in Fig. 1A) that abuts the ptc expression domain, where SecGFP is produced. In the presence of both genetic modifications, a gradient of GFP fluorescence was readily detectable (Fig. 1B, C, hh-Nb1highCD8) in both the basolateral and apical regions. Here we focus on the basolateral gradient; a discussion of the apical gradient can be found in Fig. S3. The basolateral GFP profile (green curve in Fig. 1D) differed somewhat from a classic exponential, with a shoulder near the source and a non-zero tail far from the source (length scales and non-zero tail values are listed in Table S1). Since GFP can diffuse in and out of imaginal discs, we considered the possibility that the non-zero tail could arise from GFP that escaped into the hemolymph (GFPhemo). This was tested by trapping GFP in the hemolymph with Nb1highCD8 expressed at the surface of the fat body, a sprawling organ that lines the body cavity (see schematic in Fig. S2A). In the resulting imaginal discs, the GFP profile decayed all the way to background level, showing that the tail indeed originated from the hemolymph (Fig. 1C, D, purple curve). In conclusion, a single extracellular binding species reveals the gradient of an inert protein in vivo but leakage in the hemolymph occurs and interferes with the gradient’s shape, most obviously far from the source, at the tail end of the gradient.
Key parameters of gradient formation
Having established that an inert protein can form a gradient in a developing epithelium, we set out to investigate the importance of the surface binders’ affinity for GFP. To ask if the high affinity of Nb1 for GFP (0.23 nM) is needed for a detectable gradient to form, this parameter was changed by using, as an extracellular binder, LaG3, which binds GFP with a Kd of 25nM (17) (hence referred to as Nblow). In imaginal discs carrying ptc-SecGFP and hh-NblowCD8, GFP fluorescence was above background but not detectably graded (Fig. 1C, D, compare blue and black curves; see Fig. S4 for a discussion of Nb-mediated GFP fluorescence boosting). This indicates that a low-affinity binder can trap extracellular GFP, but also that sufficiently high affinity is needed for a meaningful gradient of surface-associated GFP to form.
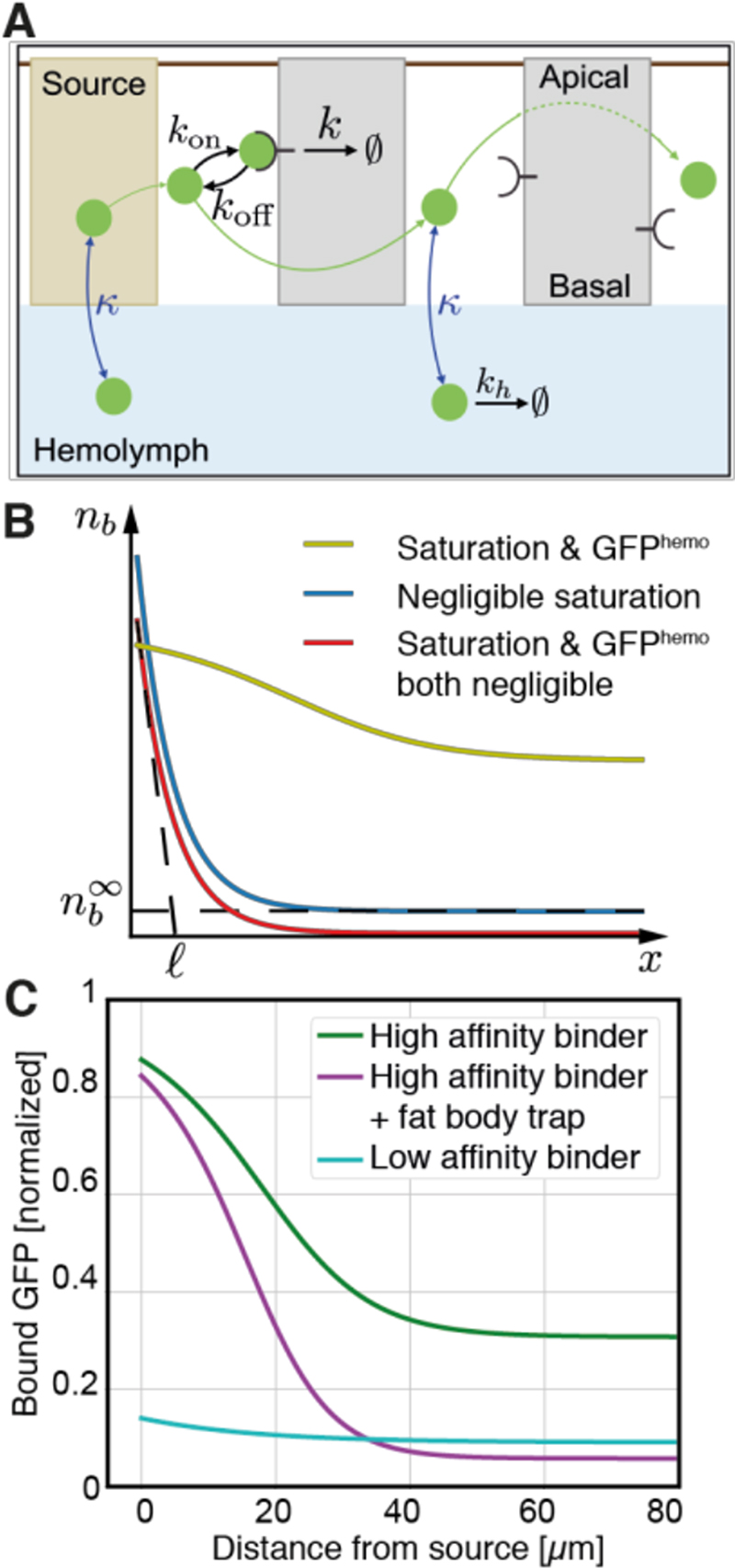
To formalize the role of extracellular binders and leakage in GFP gradient formation, we devised a diffusion-degradation-leakage mathematical model (Fig. 2A and Supplementary Theory), building on previous work (18, 19). Free GFP was assumed to diffuse in the intercellular space with a diffusion constant D and to bind/unbind at rates k on/k off receptors internalized and degraded at rate k. The flux between hemolymph and the epithelium was assumed to be driven by the concentration difference between them, with a proportionality coefficient k. At steady state, in the posterior compartment (where the receptors are expressed), the concentrations of free (c) and receptor-bound (n b) GFP follow the equations:
| (1) |
| (2) |
where n T refers to the density of receptors at the cell surface, h is the intercellular distance, and c H is the free GFP concentration in the hemolymph. Analytical exploration of the model showed that essential features of the bound GFP gradient profile are recapitulated (Fig. 2B; Fig. S5): (i) close to the source, receptor saturation leads to a shoulder; (ii) further away from the source, the profile decays on a length scale determined by the diffusion constant, the degradation of receptors, and leakage to the hemolymph; (iii) far from the source, the concentration of GFP remains at a constant non-zero value that depends on the hemolymph GFP concentration.
Fig. 2. A diffusion-degradation-leakage model for GFP gradient formation.
(A) Schematic representation of the model for gradient formation. Parameters are described in the main text and Supplementary Theory. (B) Main features of bound GFP profiles predicted by the model using parameters listed in Table S2 (unless specified otherwise). Yellow-green curve: Profile exhibiting receptor saturation near the source, and a non-zero tail due to GFPhemo (ligand production rate j = 0.5nM/s, n T = 100nM. μm). Blue curve: Without saturation, the gradient is an exponential with a non-zero tail and a decay length that depends on diffusion, effective degradation with rate k r and leakage with rate K (j = 3. 10−4nM/s, n T = 3. 104 nM. μm). Red curve: Setting the ligand concentration to zero in the hemolymph abolishes the non-zero tail (as blue curve with c H =0). See section 1.3 of the Supplementary Theory for full parameter definitions. (C) Bound GFP profiles normalized to the total concentration of receptors. The blue and green curves were obtained with the known on and off rates for the low- and high-affinity receptors respectively. The purple curve was obtained by increasing degradation in the hemolymph. Compare to corresponding experimental curves in Fig. 1D.
We then tested whether the model, and its consideration of leakage in particular, could quantitatively account for observed experimental profiles. To derive the concentration c H of GFP in the hemolymph, we surmised that it is set by a balance of input from tissue leakage and loss by degradation in the hemolymph (k H) (Supplementary Theory, where we also discuss the contribution of other larval tissues that produce and degrade the ligand). Parameters were chosen from published data or estimated, with the remaining unknown parameters obtained from a fit to experimental curves, as described in Table S2. Our fitting procedure indicated a substantial leakage rate, K ~1/(13s). Comparison of Figs. 2C and 1D shows that the model provides a suitable framework to rationalize experimental observations. The effect of reducing affinity was recapitulated by setting this parameter to that measured for Nblow (blue curve in Fig. 2C). Being a poor binder, NblowCD8 is unable to trap much GFP at the cell surface, reducing the amplitude of the gradient near the source. Consequently, NblowCD8 takes up and degrades GFP at a relatively slow rate, leading to increased leakage (section 1.5 in Supplementary Theory). Thus, lowering ligand-binder affinity adversely affects the gradient both by reducing gradient amplitude and increasing the concentration of GFP in the hemolymph. The model also replicated the effect of the fat body trap by increasing the hemolymph degradation rate ~20-fold (magenta curve in Fig. 2C).
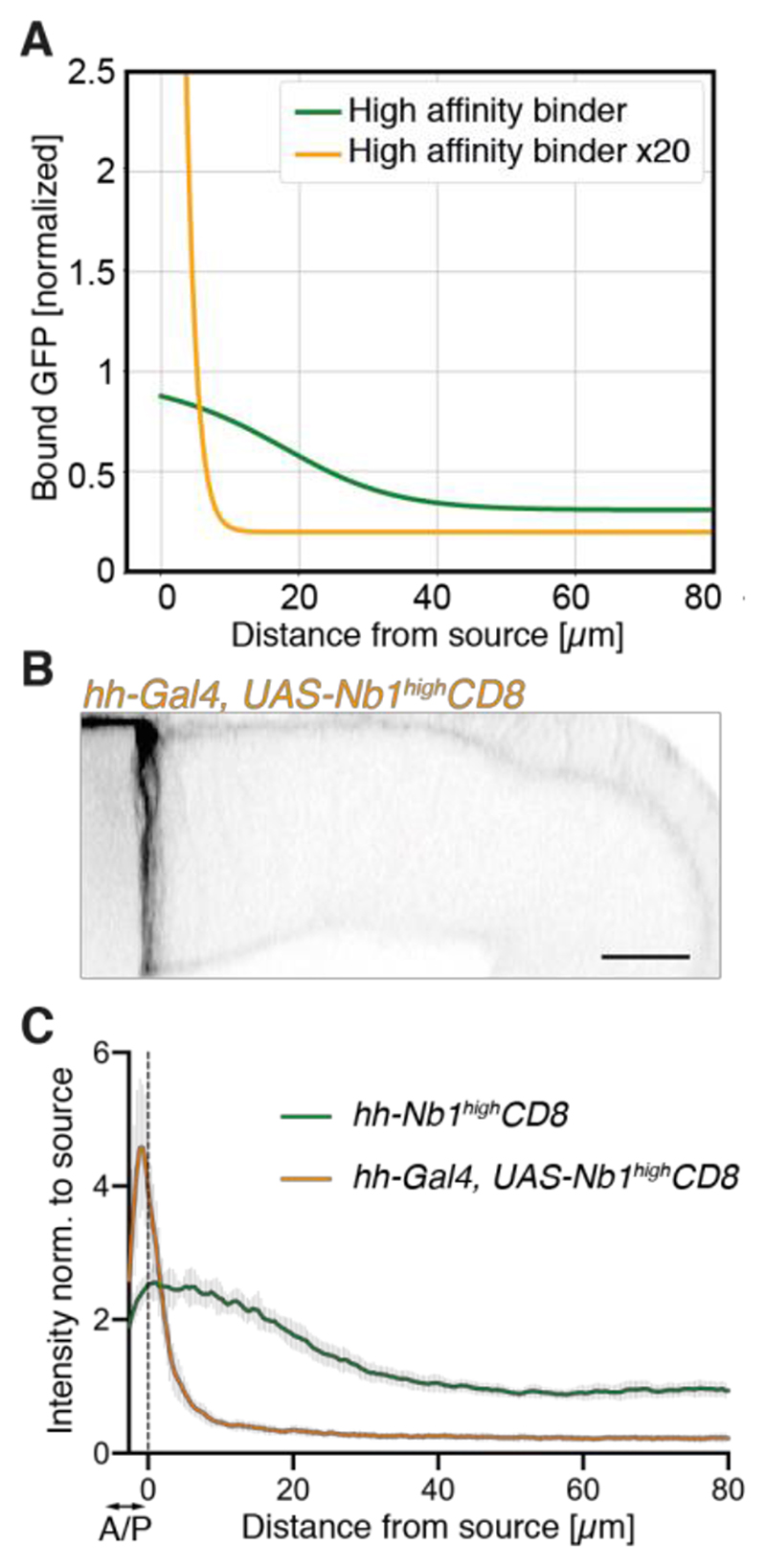
The model could also be used for de novo predictions. With the parameters determined above, it predicted that increasing ligand production at the source should lead to gradient extension as well as flattening near the source because of saturation (Fig. S6A). This was indeed found experimentally in imaginal discs overexpressing SecGFP under the control of ptc-Gal4 (Fig. S6B-D). Also confirmed experimentally was the prediction that increasing receptor expression (achieved by boosting the level of Nb1highCD8 ~20-fold, with hh-Gal4 and UAS-Nb1highCD8; see Fig. S7A for quantification) would lead to a steepening of the gradient and an increase in GFP level near the source, although the latter was not as marked in the experiment as in the model (compare Figs. 3A and 3C). Both model and experiment showed a reduced non-zero tail in this condition, confirming that receptor-mediated internalization contributes to limiting leakage into the hemolymph. Therefore, increasing ligand-receptor avidity could contribute to reducing signaling by GFPhemo flowing back in the tissue, although this could be at the cost of a reduced range.
Fig. 3. Predicted and experimental effect of increasing binder expression.
(A) Predicted GFP profile following a 20-fold increase in binder expression (orange). Compare to green curve, which is reproduced from Fig. 2C. Note the steep gradient and the lower non-zero tail. Bound-GFP concentrations are normalized to the lower value of total receptor concentration. (B) Cross-section of a ptc-SecGFP imaginal disc overexpressing the high-affinity binder (hh- Gal4, UAS-Nb1highCD8). (C) GFP profiles in hh-Gal4, UAS-Nb1highCD8 (orange curve, n=8) and hh-Nb1highCD8 discs (green curve from Fig. 1D). Scale bars: 20 μm
Engineering a GFP-dependent signaling gradient in vivo
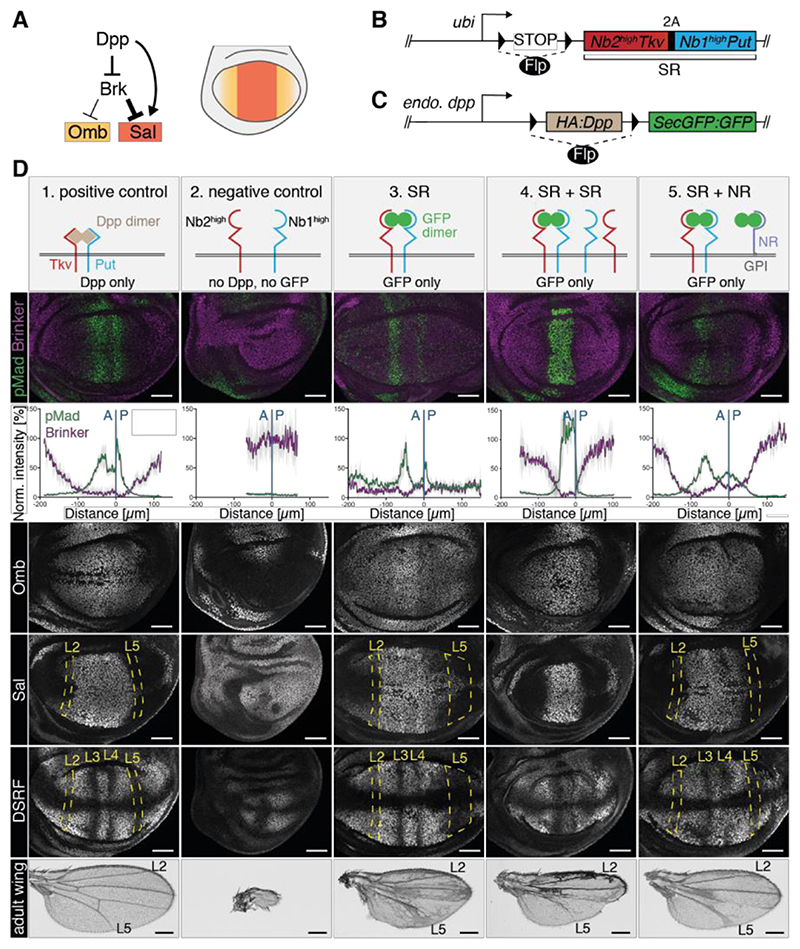
So far, we have identified minimal conditions for an inert protein to form a gradient along the plane of a developing epithelium. We next asked if this gradient could provide positional information. The best characterized morphogen in wing imaginal discs is Dpp, which promotes growth and specifies the position of veins along the A/P axis. As a first step towards asking if GFP could substitute for Dpp in vivo, we engineered the Dpp receptors Thickveins (Tkv) and Punt (Put) to render them responsive to GFP. Normally, Dpp dimers bind to two pairs of Tkv and Put, leading to phosphorylation of Mad (20) and transcriptional repression of brinker (brk) (21). The resulting inverse Brk gradient in turn controls the nested expression of target genes such as spalt (sal) and optomotor-blind (omb) (22, 23) (Fig. 4A). We reasoned that GFP dimers might initiate the same signaling cascade if Tkv and Put were fused to anti-GFP nanobodies (GBP1 = Nb1high, and GBP6, hereafter called Nb2high) that recognize non-overlapping epitopes (16, 24) (Fig. S8A). We created plasmids to express Nb2highTkv and Nb1highPut and co-transfected them in S2 cells. Addition of GFP dimers (or monomers) to the culture medium led to accumulation of phospho-Mad (pMad), suggesting that the chimeric receptors can be activated by GFP (Fig. S8B), although we cannot be sure that the signaling kinetics normally achieved by the natural ligand were entirely recapitulated.
Fig. 4. Rescue of growth and patterning by GFP.
(A) Target genes of Dpp signaling in the pouch, which give rise to the wing. (B) Schematic representation of SR, the transgene for conditional expression of engineered receptors. (C) Dpp locus engineered to allow Flp-mediated replacement of an essential region by sequences encoding secreted GFP dimers. Throughout this study, rn-Gal4 and UAS-Flp were used to inactivate Dpp and/or trigger SR expression specifically in the pouch. (D) Phenotypes of wing imaginal discs and adult wings of various genotypes (columns). The positive control (column 1) shows imaginal discs and wings from a dpp-[>Dpp>SecGFP:GFP] homozygous larva. Flp is absent and Dpp is therefore expressed as in wild type discs. For the negative control (column 2), we used larvae homozygous for a different conditional allele (dpp-[>Dpp>]) (25) and carrying the SR transgene. Here, Flp expression inactivates Dpp in the pouch without triggering the GFP:GFP production, while at the same time activating expression of the engineered receptors. The resulting phenotypes recapitulated those of classical Dpp mutants (e.g. brk derepression and growth impairment). Abrogation of Dpp activity shows that the engineered receptors do not trigger signaling in the absence of GFP. If, in combination with the SR transgene, the dpp-[>Dpp>SecGFP:GFP] allele is used (column 3, SR), signaling activity (e.g. pMad immunoreactivity near the source) and growth are restored, albeit imperfectly. Note the occasional spots of pMad throughout the pouch, the expanded zone of brk repression, the fuzzy boundary of sal expression in the posterior compartment, and the disrupted vein pattern. Adding a second SR transgene (column 4, same genotype as in column 3 with one additional SR transgene, SR+SR) led to enhanced pMad at the source and a narrowing of the signaling gradient (relative to SR alone). Addition of non-signaling receptors (column 5, same genotype as in column 3 with addition of dally-NblowGPI, SR+NR) extended the signaling gradient (relative to SR alone). Note the absence of background pMad far from the source and the wild type-like expression of target genes. Note however that vein L4 was often disrupted and vein L5 was slightly broadened in the distal part. Scale bar wing discs: 50 μm. Scale bar adult wings: 0.25 mm.
Based on these encouraging results with cultured cells, we created a transgene that expresses, in a Flippase (Flp)-dependent manner, both engineered receptors under the control of the ubiquitin (ubi) promoter (Fig. 4B). This transgene, ubi-[>STOP>Nb2highTkv 2A
Nb1highPut], where > indicates Flp recombination targets, will henceforth be referred to as SR (for signaling receptors). We also developed a dpp allele that can be inactivated, while at the same time made to express secreted GFP dimers upon Flp expression (dpp-[>Dpp>SecGFP:GFP], Fig. 4C, validated in Fig. S9). First, we used the previously described dpp-[>Dpp>]allele (25) to confirm that inactivation of Dpp throughout the wing primordium with rotund-Gal4 (rn-Gal4) and UAS-Flp abrogated growth and patterning, even in the presence of GFP-responsive receptors (Fig. 4D, column 2). Crucially with dpp-[>Dpp>SecGFP:GFP], which produces GFP upon Dpp inactivation, recognizably patterned wings developed (Fig. 4D; column 3). Note that no GFP gradient was detectable in this genetic background, perhaps because of rapid internalization and degradation of GFP by the signaling receptors. The rescuing activity of secreted GFP dimers was further assessed in imaginal discs by staining for various markers of Dpp signaling. pMad immunoreactivity was unexpectedly low in the GFP-producing cells (see Fig. S10). Most relevant to this study, however, signaling activity was graded on either side of the source, including in the posterior compartment, which relies entirely on ligand diffusion (26). In addition, pMad immunoreactivity was also present in a salt and pepper manner throughout the whole pouch, as if residual signaling activity persisted far from the source. In agreement, brk was repressed over a wider range than in control discs. Both sal and omb were expressed in GFP-rescued discs, although in a range that did not recapitulate the wild type situation; in the posterior compartment, the Sal domain boundary was fuzzy whereas the domain of Omb was too broad. Normally Sal and Omb ensure the patterned downregulation of DSRF, which is required for vein fate specification (27). However, in the ‘rescue’ condition, an oversized domain of DSRF downregulation could be seen in the posterior compartment, and a corresponding sprawling vein L5 in surviving adult wings. Ectopic vein material was also seen throughout the wings, probably a result of global ectopic Dpp signaling. Despite these limitations, the above results suggest that a GFP gradient can stimulate growth and provide substantial patterning information through engineered receptors.
Signaling activity far from the GFP source - e.g in the form of ectopic pMad – suggests the presence of GFP dimers throughout the disc, perhaps as a result of re-entry from the hemolymph. During our initial analysis of the GFP gradient, we found that leakage could be reduced by increasing the level of extracellular binders (Fig. 3B,C). We therefore asked if a similar strategy could be used to reduce non-zero tail signaling in rescued discs. Indeed, with two copies of the transgene expressing engineered GFP-responsive receptors (SR + SR), background pMad immunoreactivity was largely abrogated (Fig. 4D, column 4). The target genes omb and sal were still expressed in a nested fashion, however, the width of these domains, as well as that of the zone of brk repression were narrower than in the wild type (compare with Fig. 4D, column 1). This suggests that a two-fold increase in receptor expression had the beneficial effect of reducing the adverse effect of leakage on signaling activity far from the source, but at the expense of a reduced range. These results can be understood qualitatively in the context of our gradient model (see section 1.3.2 in Supplementary Theory): at low receptor density, receptor activation is too low to trigger target gene activation; at intermediate receptor density, leakage can lead to a high ligand concentration in the hemolymph, triggering signaling and target gene activation far from the source (Fig S5F-G); at higher receptor density, hemolymph concentration drops but the gradient scale shortens because of increased degradation in the tissue. It appears therefore that long-range GFP gradients with low residual signaling far from the source may only be achievable within a narrow parameter regime (Fig. S5F-G).
Beneficial effects of GPI-anchored non-signaling receptors
The above analysis suggests that, by solely modulating the expression of signaling receptors, it is difficult to reduce leakage without shortening the gradient. Natural morphogens bind not only to signaling receptors but also to non-signaling extracellular proteins such as glypicans, GPI-anchored heparan sulfate proteoglycans (11). We therefore set out to investigate whether low-affinity, GPI-anchored, extracellular binders (non-signaling receptors, NR) would improve the performance of the signaling gradient. As a first step, we created a DNA fragment encoding Nblow (Kd: 25 nM, chosen to mimic the affinity of BMP for heparin (28)), tethered to the extracellular face of the plasma membrane by a GPI anchor, NblowGPI). This fragment was expressed from a UAS transgene with rn-Gal4, which concomitantly triggered expression of Flp to inactivate Dpp and initiated expression of SecGFP and the engineered receptors. In the resulting imaginal discs, the signaling activity of SR was suppressed (Fig. S11), perhaps because excess NblowGPI prevented SecGFP from accessing the signaling receptors. To achieve a more reasonable expression level we inserted the NblowGPI-encoding fragment in the dally locus, one of the two glypican-encoding genes of Drosophila. This allele (dally-NblowGPI) was then combined with all the previously described genetic elements needed for a GFP signaling gradient to form. Addition of this non-signaling receptor extended the pMad gradient while narrowing the domain of brk repression, an indication of reduced GFPhemo signaling far from the source (compare columns 3 and 5 in Fig. 4D). In fact, with this combination of signaling and non-signaling receptors, target gene expression (controlled entirely by GFP dimers) was comparable to that in wildtype imaginal discs, and the resulting wings were remarkably well patterned, proportioned and consistently sized (Fig. S12).
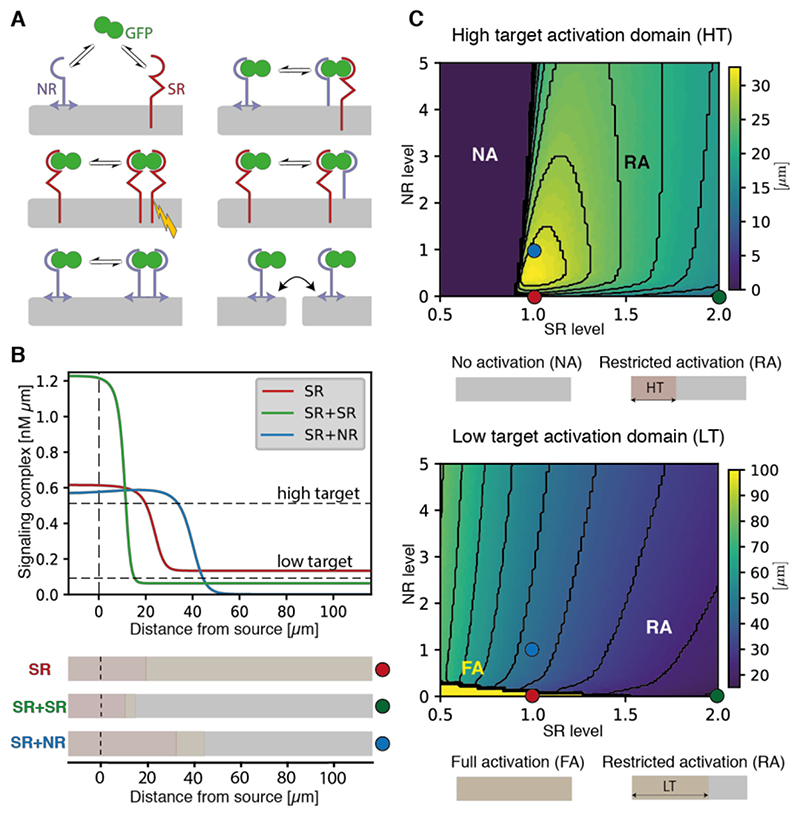
To rationalize how non-signaling receptors could improve the signaling gradient’s characteristics, we devised a formal description of the relevant molecular interactions (Fig. 5A, Fig. S13C, Supplementary Theory, and Table S3). In this framework, GFP dimers bind signaling and non-signaling receptors and transit from one configuration to another. Signaling receptors typically undergo rapid endocytosis upon binding to their ligands. By contrast, GPI-anchored binders could have a longer lifetime (29), allowing them to handover ligand to signaling receptors (30). Simulations showed however that addition of membrane tethered non-receptors does not extend the signaling gradient, although they can alter its shape near the source (Supplementary Theory and Fig. S13D). We next considered the relevance of GPI anchors’ labile nature (31). Locally expressed GFP-GPI spreads within wing imaginal discs (32, 33) suggesting that GPI-anchored proteins can detach from cells and possibly reinsert themselves nearby, a process we call hopping (Fig. 5A). Simulations introducing a tissue-scale effective diffusion constant D r = 0.1μm2/s for GFP-NblowGPI (representing lateral diffusion in the cell membrane and intercellular hopping) can explain the extension of the signaling gradient by Nblow-GPI (Fig. 5B). In fact, in the absence of NR diffusion (or with a low diffusion constant), NRs can only shorten the gradient since non-diffusing NRs provide an additional route for ligand degradation without contributing to ligand spread (Fig. S13D-E and Supplementary Theory). At high NR concentration, competition for the ligand with SR inhibits signaling (Fig. 5C), as observed experimentally (Fig. S11). For intermediate values of NR concentration however, NR diffusion enables the gradient range to increase, while avoiding uniform activation of low target genes by leaked ligand.
Fig. 5. Modeling the effect of GPI-anchored non-signaling receptors on gradient length scale.
(A) Schematic representation of the molecular interactions considered by our model, including GFP dimer handover and NR hopping. Signal transduction (yellow lightning bolt) is activated by GFP-bound signaling receptor dimers (SR). See Supplementary Theory for details. (B) Predicted profiles of signaling complexes in three conditions: a reference case with signaling receptors only (SR; red), doubling SR levels (SR + SR; green), and adding non-signaling receptors (SR + NR; blue). As observed experimentally, doubling SR leads to a steeper gradient, while adding NR reduces GFPhemo signaling and extends the gradient, due to non-signaling receptor effective diffusion. For illustration, arbitrary thresholds are chosen to indicate the position where high- and low-level target genes would be activated (see Supplementary Tables 2 and 3 for parameter values). (C) Width of the high (top) and low (bottom) target activation domain (arbitrary threshold shown in panel B), as a function of normalized levels of SR and NR. Warmer colors indicate a wider target activation domain. Colored dots show parameter combinations used in panel B. Top: For the normalized SR value of 1, increasing NR initially lengthens the high target domain, while further increase shortens it by preventing access of GFP to SR (as observed experimentally, Fig. S11). Bottom: For the normalized SR value of 1 and in the absence of NR, GFPhemo signaling dominates and low target gene is activated throughout (bright yellow region). Increasing SR or NR production both lead to a reduction in the low target domain size.
Conclusion
We have shown that, in the presence of extracellular binders, GFP can form a gradient in an epithelium. Because GFP is inert in wing imaginal discs, it is unlikely to spread by a specialized transport mechanism such as planar transcytosis. The low off rate of Nb1high (k off= 1.7 × 10-4 s-1) also limits the contribution of ligands passing from one receptor to another. We therefore suggest that the GFP gradient forms by free diffusion even though the readily detectable gradient is largely made up of bound GFP (see Fig. 1C and Fig. S5H-K). In the presence of engineered GFP-responsive receptors (SR), diffusing GFP can act as a morphogen. One limitation of free diffusion is that it allows leakage into the circulation, a potential threat to positional information. As we have shown, signaling from leaked GFP could be reduced by increasing the level of signaling receptors. However, this was at the expense of a reduced range. Leakage can be reduced without concomitant decrease in gradient range by adding GPI-anchored non-signaling receptor (NR). We suggest that, by virtue of their labile association with cell membranes GPI-anchored non-signaling receptor could undergo tissue-level diffusion and thus extend the gradient. Although this hypothesis remains to be demonstrated experimentally, our results so far show that, a combination of free and NR-assisted diffusion suffices to emulate the range and activity of a natural morphogen.
Supplementary Material
One Sentence Summary.
GFP can from a gradient in vivo and, with engineered GFP-responsive receptors, replace a morphogen that coordinates patterning and growth.
Acknowledgments
We thank C. Alexandre for generating vg-Gal4, dally-attP and advice on genome engineering and the Crick Fly Facility for DNA injections. We also acknowledge the technical and intellectual contributions of S. Crossman and I. McGough. J. Briscoe provided comments on the manuscript. We are also grateful to A. Lander for pointing out the possible relevance GPI’s loose anchorage to membranes. We thank H. Ashe, G. Pflugfelder and A. Salzberg for the generous gift of antibodies. The Developmental Studies Hybridoma Bank also provided antibodies. Drosophila stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This work was supported by core funding from the Francis Crick Institute (FC001204 to JPV and FC001317 to GS) and a Wellcome Trust Investigator Award to JPV (206341/Z/17/Z). KS was the recipient of a PhD studentship from the Wellcome Trust (109054/Z/15/Z). This project was conceived by KS, JPV and GS. KS designed and performed all the experiments. The results were analyzed by all authors. The model was conceived by GS, MDG and LC, and numerical simulations were performed by MDG and LC. The main text was written by KS, JPV and GS with comments from MDG and LC. The authors declare no competing or financial interests. All data are described in the main text or supplementary materials. All materials are available upon request. A link to the computer code can be found in Supplementary Theory.
References
- 1.Müller P, Rogers KW, Yu SR, Brand M, Schier AF. Morphogen transport. Development. 2013;140:1621–1638. doi: 10.1242/dev.083519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou S, et al. Free extracellular diffusion creates the Dpp morphogen gradient of the Drosophila wing disc. Curr Biol. 2012;22:668–675. doi: 10.1016/j.cub.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu SR, et al. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461:533–536. doi: 10.1038/nature08391. [DOI] [PubMed] [Google Scholar]
- 4.Lander AD, Nie Q, Vargas B, Wan FY. Size-normalized Robustness of Dpp Gradient in Drosophila Wing Imaginal Disc. J Mech Mater Struct. 2011;6:321–350. doi: 10.2140/jomms.2011.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornberg TB, Guha A. Understanding morphogen gradients: a problem of dispersion and containment. Curr Opin Genet Dev. 2007;17:264–271. doi: 10.1016/j.gde.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 7.Entchev EV, Schwabedissen A, González-Gaitán M. Gradient formation of the TGF-beta homolog Dpp. Cell. 2000;103:981–991. doi: 10.1016/s0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 8.Roy S, Huang H, Liu S, Kornberg TB. Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein. Science. 2014;343:1244624. doi: 10.1126/science.1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Häcker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 10.Nakato H, Li JP. Functions of Heparan Sulfate Proteoglycans in Development: Insights From Drosophila Models. Int Rev Cell Mol Biol. 2016;325:275–293. doi: 10.1016/bs.ircmb.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009;1:a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P, et al. Morphogen gradient reconstitution reveals Hedgehog pathway design principles. Science. 2018;360:543–548. doi: 10.1126/science.aao0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toda S, et al. Engineering Synthetic Morphogen Systems that Can Program Multicellular Patterning. Science. 2020 doi: 10.1126/science.abc0033. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmansa S, Alborelli I, Bieli D, Caussinus E, Affolter M. A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. Elife. 2017;6 doi: 10.7554/eLife.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothbauer U, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 16.Kirchhofer A, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct & Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 17.Fridy PC, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander AD, Nie Q, Wan FYM. Do morphogen gradients arise by diffusion? Dev Cell. 2002;2:785–796. doi: 10.1016/s1534-5807(02)00179-x. [DOI] [PubMed] [Google Scholar]
- 19.Bollenbach T, Kruse K, Pantazis P, Gonzalez-Gaitan M, Julicher F. Morphogen transport in epithelia. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;75:011901. doi: 10.1103/PhysRevE.75.011901. [DOI] [PubMed] [Google Scholar]
- 20.Newfeld SJ, et al. Mothers against dpp participates in a DDP/TGF-beta responsive serine-threonine kinase signal transduction cascade. Development. 1997;124:3167–3176. doi: 10.1242/dev.124.16.3167. [DOI] [PubMed] [Google Scholar]
- 21.Müller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 22.Barrio R, De Celis JF. Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway. Proc Natl Acad Sci U S A. 2004;101:6021–6026. doi: 10.1073/pnas.0401590101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivasankaran R, Vigano MA, Muller B, Affolter M, Basler K. Direct transcriptional control of the Dpp target omb by the DNA binding protein Brinker. EMBO J. 2000;19:6162–6172. doi: 10.1093/emboj/19.22.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang JC, et al. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell. 2013;154:928–939. doi: 10.1016/j.cell.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosch PS, Ziukaite R, Alexandre C, Basler K, Vincent J-P. Dpp controls growth and patterning in Drosophila wing precursors through distinct modes of action. Elife. 2017;6 doi: 10.7554/eLife.22546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans CJ, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Celis JF. Pattern formation in the Drosophila wing: The development of the veins. Bioessays. 2003;25:443–451. doi: 10.1002/bies.10258. [DOI] [PubMed] [Google Scholar]
- 28.Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur J Biochem. 1996;237:295–302. doi: 10.1111/j.1432-1033.1996.0295n.x. [DOI] [PubMed] [Google Scholar]
- 29.Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- 30.Schlessinger J, Lax I, Lemmon M. Regulation of growth factor activation by proteoglycans: what is the role of the low affinity receptors? Cell. 1995;83:357–360. doi: 10.1016/0092-8674(95)90112-4. [DOI] [PubMed] [Google Scholar]
- 31.Müller GA. The release of glycosylphosphatidylinositol-anchored proteins from the cell surface. Arch Biochem Biophys. 2018;656:1–18. doi: 10.1016/j.abb.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Greco V, Hannus M, Eaton S. Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell. 2001;106:633–645. doi: 10.1016/s0092-8674(01)00484-6. [DOI] [PubMed] [Google Scholar]
- 33.Tempesta C, Hijazi A, Moussian B, Roch F. Boudin trafficking reveals the dynamic internalisation of specific septate junction components in Drosophila. PLoS ONE. 2017;12:e0185897. doi: 10.1371/journal.pone.0185897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.