Abstract
Mutational signatures are imprints of pathophysiological processes arising through tumorigenesis. We generated isogenic CRISPR-Cas9 knockouts (Δ) of 43 genes in human induced pluripotent stem cells, cultured them in the absence of added DNA damage, and performed whole-genome sequencing of 173 subclones. ΔOGG1, ΔUNG, ΔEXO1, ΔRNF168, ΔMLH1, ΔMSH2, ΔMSH6, ΔPMS1, and ΔPMS2 produced marked mutational signatures indicative of being critical mitigators of endogenous DNA modifications. Detailed analyses revealed mutational mechanistic insights, including how 8-oxo-dG elimination is sequence-context-specific while uracil clearance is sequence-context-independent. Mismatch repair (MMR) deficiency signatures are engendered by oxidative damage (C>A transversions), differential misincorporation by replicative polymerases (T>C and C>T transitions), and we propose a ‘reverse template slippage’ model for T>A transversions. ΔMLH1, ΔMSH6, and ΔMSH2 signatures were similar to each other but distinct from ΔPMS2. Finally, we developed a classifier, MMRDetect, where application to 7,695 WGS cancers showed enhanced detection of MMR-deficient tumors, with implications for responsiveness to immunotherapies.
Keywords: Genomic instability, cancer, cancer genomics, CRISPR-Cas9 systems
Introduction
Somatic mutations arising through endogenous and exogenous processes mark the genome with distinctive patterns, termed mutational signatures1–4. While there have been advancements in the analytical aspects of deriving mutational signatures from human cancers5–7, there is an emerging need for experimental substantiation, elucidating etiologies and mechanisms underpinning these patterns8–11. Cellular models have been used to systematically study mutagenesis arising from exogenous sources of DNA damage8,11. Next, it is essential to experimentally explore genome-wide mutagenic consequences of endogenous sources of DNA damage in the absence of external DNA damaging agents.
Lindahl noted that water and oxygen, essential molecules for living organisms, are some of the most mutagenic elements to DNA12. His seminal work demonstrated that spontaneous DNA lesions occur through endogenous biochemical activities such as hydrolysis and oxidation. Errors at replication are also an enormous potential source of DNA changes. Fortuitously, our cells are equipped with DNA repair pathways that constantly mitigate this endogenous damage13,14. In this work, we combine CRISPR-Cas9-based biallelic knockouts of a selection of DNA replicative/repair genes in human induced Pluripotent Stem Cells (hiPSCs), whole-genome sequencing (WGS), and in-depth analysis of experimentally-generated data to obtain mechanistic insights into mutation formation. It is beyond the scope of this manuscript to study all DNA repair genes. Thus, we have focused on 42 DNA replicative/repair pathway gene knockouts successfully generated through semi-high-throughput methods. We also compared our experimental data with reported cancer-derived signatures.
While there is substantial literature regarding DNA repair pathways and complex protein interactions involved in maintaining genomic integrity15–20, here we focus on directly mapping whole-genome mutational outcomes associated with DNA repair defects, critically, in the absence of applied, external damage. This study, therefore, allows us to identify replicative/repair genes that are fundamentally important to genome maintenance against endogenous DNA damage.
Results
Biallelic knockouts of DNA replicative/repair genes
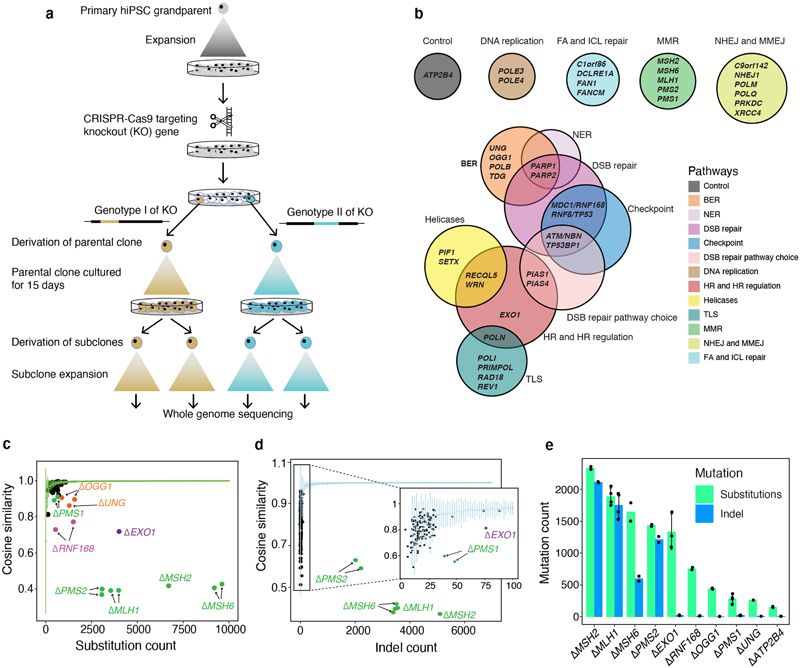
We knocked out (Δ) 42 DNA replicative/repair pathway genes and an unrelated control gene, ATP2B4 (Fig. 1a,b, Supplementary Table 1). A pilot experiment was performed to standardize experimental procedures (Methods, Extended Data Fig. 1). In the full-scale study, two knockout genotypes were generated per gene except for EXO1, MSH2, TDG, MDC1, and REV1, for which only one knockout genotype was obtained. All parental knockout lines were grown for 15 days under normoxic conditions (~20% oxygen). For each genotype, two single-cell subclones were derived for whole-genome sequencing, aiming for four sequenced subclones per edited gene (Fig. 1a). For single-genotype genes, three subclones were derived for ΔEXO1 and ΔMSH2, and four for ΔTDG, ΔMDC1, and ΔREV1.
Figure 1. Mutational consequences of DNA replicative/repair pathway gene knockouts.
(a) Experimental workflow from isolation of gene knockouts to generating subclones for WGS. (b) Forty-three genes were knocked out, including 42 DNA replicative/repair genes and one control gene (ATP2B4). (c) Distinguishing substitution profiles of control subclones and knockout subclones. Green line shows the cosine similarities between bootstrapped profiles of controls against aggregated control substitution profile. X-axis shows the aggregated substitution number of each genotype of a knockout. (d) Distinguishing indel profile of control subclones and knockout subclones. Light blue line shows the cosine similarities between bootstrapped indel profiles of controls against aggregated control indel profile. X-axis shows the aggregated indel number of each genotype of a knockout. (e) De novo mutation number of knockout subclones (n = 2~4, Supplementary Table 2) cultured for 15 days. Bars and error bars represent mean ± SD of subclone observations.
In all, 173 subclones were obtained from 78 genotyped knockouts of 43 genes (Supplementary Table 2). All subclones were sequenced to an average depth of ~25-fold. Short-read sequences were aligned to human reference genome assembly GRCh37/hg19. All classes of somatic mutations were called, subtracting variation of the primary hiPSC parental clone. Rearrangements were too infrequent to decipher specific patterns.
We confirmed that mutational outcomes were neither due to off-target edits nor to the acquisition of new driver mutations (Online Methods). We verified that knockouts were biallelic, validated the protein loss via mass spectrometry, and ensured that subclones in all comparative analyses were single-cell derived (Online Methods).
Mutational consequences of gene knockouts
Under these controlled experimental settings, if simply knocking-out a gene (in the absence of providing additional DNA damage) could produce a signature, then the gene is critical to maintaining genome stability from endogenous DNA damage. It would manifest an increased mutation burden above background and/or altered mutation profile (Extended Data Fig. 2). We found background substitution and indel mutagenesis associated with growing cells in culture occurred at ~150 substitutions and ~10 indels per genome and was comparable across all subclones (Supplementary Table 2, 3).
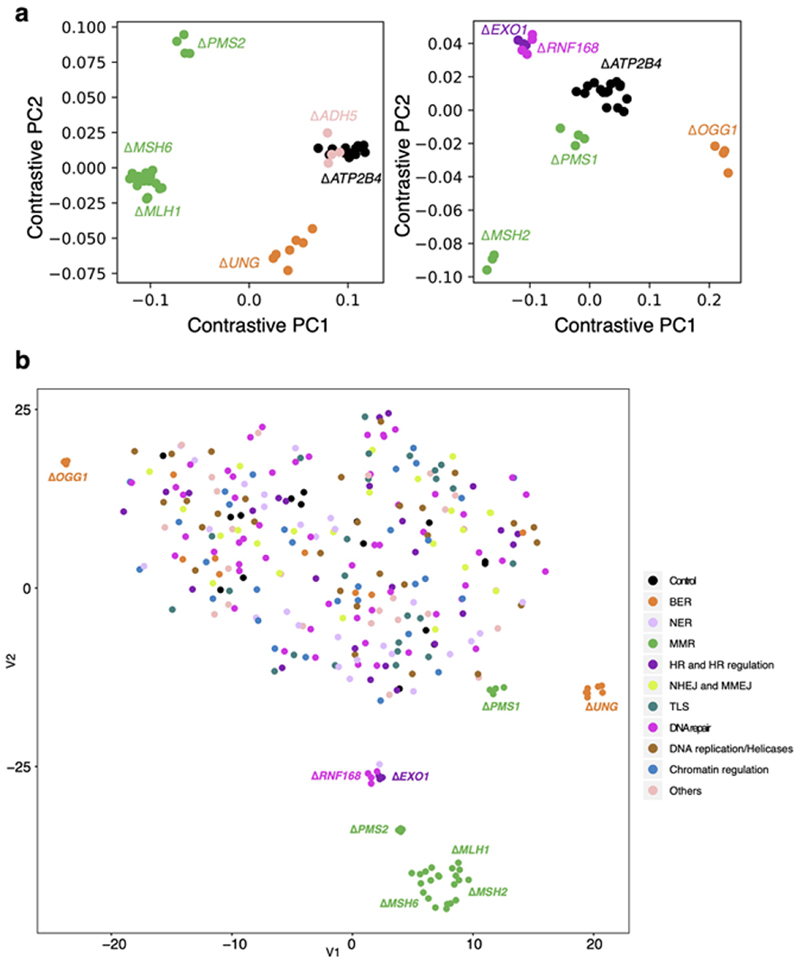
To address potential uncertainty associated with the relatively small number of subclones per knockout and variable mutation counts in each gene knockout (Methods), we generated bootstrapped control samples with variable mutation burdens (50-10,000). We calculated cosine similarities between each bootstrapped sample and the background control (ΔATP2B4) mutational signature (mean and standard deviations). A cosine similarity close to 1.0 indicates that the mutation profile of the bootstrapped sample is near-identical to the control signature. Cosine similarities could thus be considered across a range of mutation burdens (green line in Fig. 1c and light blue line in Fig. 1d). We next calculated cosine similarities between knockout profiles and controls (colored dots in Fig. 1c,d). A knockout experiment that does not fall within the expected distribution of cosine similarities implies a mutation profile distinct from controls, i.e., the gene knockout has a signature. For substitution signatures, two additional dimensionality reduction techniques, namely, contrastive principal component analysis (cPCA)21 and t-Distributed Stochastic Neighbour Embedding (t-SNE)22 were also applied to secure high confidence mutational signatures (Extended Data Fig. 3, Methods). This stringent series of steps would likely dismiss weaker signals and be highly conservative at calling mutational signatures.
We identified nine single substitution, two double substitution, and six indel signatures. Two gene knockouts, ΔOGG1 and ΔUNG, produced only substitution signatures. Five gene knockouts, ΔMSH2, ΔMSH6, ΔMLH1, ΔPMS2, and ΔPMS1, presented substitution and indel signatures. Two gene knockouts, Δ RNF168 and ΔEXO1, had substitution and double substitution signatures. ΔEXO1 also produced an indel signature. The average de novo mutation burden accumulated for these nine knockouts ranged between 250-2,500 for substitutions and 5-2,100 for indels (Fig. 1e). Based on cell proliferation assays, mutation rates for each knockout were calculated and ranged between 6-129 substitutions and 0.39-126 indels per cell division (Supplementary Table 4). In the following sections, we dissect these experimentally-generated signatures, compare them to one another and to cancer-derived mutational signatures to gain insights into the sources of endogenous DNA damage and mutational mechanisms.
Safeguarding the genome from oxidative DNA damage
Oxygen can generate reactive oxygen species (ROS) and oxidative DNA lesions. The commonest is 8-oxo-2’-deoxyguanosine (8-oxo-dG), although over 25 oxidative DNA lesions are known23. 8-oxo-dG is predominantly repaired by Base Excision Repair (BER). A pervasive mutational signature observed in cell-based experiments has been speculated as due to culture-related oxidative damage9,11. It is similar to a mutational signature identified in adrenocortical cancers and neuroblastomas, called RefSig1824 or SBS186. Biallelic loss of MutY DNA-glycosylase gene (MUTYH), which excises adenines inappropriately paired with 8-oxo-dG, has also been reported to generate a hypermutated version of a similar signature25. It is unclear whether other genes responsible for removing oxidative damage would also result in these characteristic patterns.
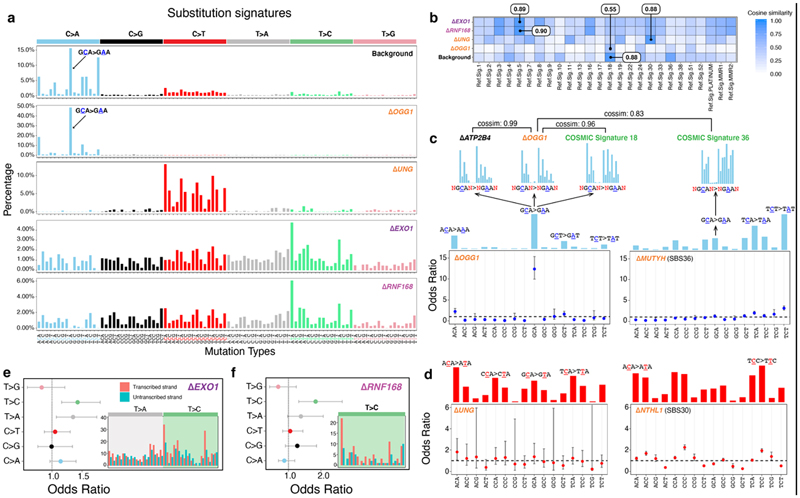
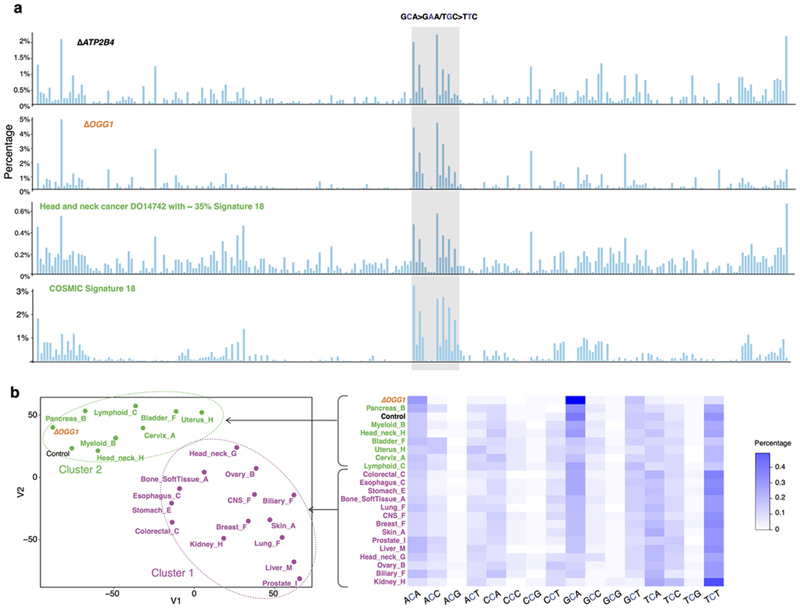
8-oxoguanine glycosylase (OGG1) is responsible for the excision of 8-oxo-dG26. Thus, an ΔOGG1 signature would be an undisputed pattern of 8-oxo-dG-related damage. ΔOGG1 produced a marked G>T/C>A pattern particularly at TGC>TTC/GCA>GAA with additional peaks at TGT>TTT/ACA>AAA, CGA>CTA/GCT>GAT, and AGA>ATA/TCT>TAT (Fig. 2a), similar to the culture-related signature and RefSig18/SBS18 (Fig. 2a,b). This supports the hypothesis that RefSig18/SBS18/culture-related signatures are due to oxidative damage, specifically implicating 8-oxo-dG. We expanded signature channels by considering ±2 bases flanking the mutated base. Higher-resolution assessment of the most dominant peak at TGC>TTC/GCA>GAA in ΔOGG1 showed an almost identical pattern to control samples carrying culture-related signatures and SBS18 (cosine similarity (cossim): > 0.9, Fig. 2c, Extended Data Fig. 4), strengthening the argument that the G>T/C>A transversions observed in cultured cells and SBS18 are indeed caused by 8-oxo-dG-related damage.
Figure 2. Safeguarding the genome from oxidative damage and cytosine deamination.
(a) Substitution signatures of background mutagenesis (from control ΔATP2B4), ΔOGG1,ΔUNG,ΔEXO1 and ΔRNF168. (b) Cosine similarity between mutational signature of gene knockouts and cancer-derived mutational signatures24. (c) Odds ratio of C>A occurring at 16 trinucleotides for ΔOGG1 and ΔMUTYH (SBS36)6. Calculation was corrected for distribution of trinucleotides in the reference genome. Odds ratio less than 1 with 95% confidence interval (CI) < 1 implies that C>A mutations at that particular trinucleotide are less likely to occur. The mutational profiles of C>A at GCA with ±2 flanking bases are shown for ΔATP2B4,ΔOGG1, SBS18 and SBS36. (d) Odds ratio of C>T occurring at all 16 trinucleotides for ΔUNG and ΔNTHL1 (SBS30)6. Transcriptional strand asymmetry of (e) ΔEXO1 signature and (f) ΔRNF168 signature. Dots and error bars in (c-f) represent calculated odds ratio with 95% confidence interval. The insets show the count of T>C/A>G mutations on transcribed and non-transcribed strands.
ΔOGG1 signature is qualitatively analogous to the signature of ΔMUTYH-related adrenocortical cancers25 (recently renamed SBS366), although the latter demonstrates hypermutator phenotypes and has its tallest peak at TCT (Fig. 2c). These similarities are explained by related but distinct roles played by OGG1 and MUTYH in repairing oxidation-related lesions: 8-oxo-dG can pair with C or with A during DNA synthesis. 8-oxo-G/C mismatches are, however, not mutagenic and oxidized guanines are simply excised by OGG127. By contrast, 8-oxo-G/A mismatches are first repaired by MutY-glycosylase, which removes the A, and repair synthesis by pol-β or -λ inserts a C opposite the oxidized base. The resulting 8-oxo-G/C pair is then excised by OGG1 as outlined earlier. This mechanistic relatedness likely explains why mutational signatures of ΔOGG1 and ΔMUTYH are qualitatively alike, if quantitatively dissimilar. Notably, that simple knockouts of OGG1 or MUTYH can result in overt mutational phenotypes suggests that these genes are indispensable for maintaining the genome against endogenous oxidative damage.
Lastly, we examined ΔOGG1 G>T/C>A mutations correcting for frequencies of the 16 trinucleotides in the reference genome and found that ΔOGG1 is depleted of mutations at GG/CC dinucleotides (Fig. 2c). Yet, prior literature reports 5’-G in GG and the first two Gs in GGG are more likely to be oxidized through intraduplex electron transfer reactions28,29. Therefore, one would expect elevated G>T/C>A mutation burdens in GG-rich regions when OGG1 is defunct. Our results may be explained by previous experiments which demonstrate that 8-oxo-dG excision rates by OGG1 are sequence-context dependent30: 8-oxo-dG excision at consecutive 5’-GG s is reported as inefficient compared to 5’-CGC/5’-GCG and 5’-AGC/5’-GCT because OGG1 employs a bend-and-flip strategy to recognize 8-oxo-dG31–33. Stacked adjacent 8-oxo-dGs have an increased kinetic barrier, preventing flipping out and removal of 8-oxo-dG30. While this may explain why OGG1 cannot repair oxidized guanines at GG/CC motifs, it remains unclear how these motifs are repaired as guanine oxidation does occur at such sites. At some GG/CC motifs, we suggest a possibility in the section on mismatch repair genes later.
Maintaining cytosines from deamination to uracil
Deamination involves hydrolytic loss of an amine group. At CpG dinucleotides, deamination of 5-methylcytosine into thymine is a well-studied, universal process34,35,36, with C>T at CpGs (Signature 1) found in many tumor types. Hypermutator phenotypes of C>T at CpGs, however, have been reported in cancers with biallelic loss of methyl-binding domain 4 (MBD4)37. This example underscores a mutational process that is customarily under tight MBD4 regulation, wherein its knockout uncovers the potential magnitude of unrepaired endogenous deamination.
Spontaneous hydrolytic cytosine deamination to uracil occurs more slowly at ~100-500/cell/day 38. Cytosine deamination to uracil is rectified by UNG (uracil-N-glycosylase) via BER39. Uracils that are not removed prior to replication can result in C>T mutations (Fig. 2a). There are signatures associated with enhanced APOBEC-related deamination in many cancers (Signatures 2 and 13). However, the consequence of UNG dysfunction is less clear. The ΔUNG signature comprised mainly C>T transitions. When corrected for reference genome trinucleotide frequencies, no trinucleotide preferences were observed (Fig. 2d), suggesting a general role for UNG activity on all uracils regardless of sequence context. ΔUNG signature is most similar to RefSig 30 (cossim 0.88), previously associated with ΔNTHL1 40. Both UNG and NTHL1 are BER glycosylases that process aberrant pyrimidines, which may explain the similarities between these signatures. However, when corrected for trinucleotide frequencies, ΔNTHL1 signature shows preference for ACC, CCC, and TCC trinucleotides in contrast to ΔUNG, supporting that they are signatures of different aetiologies.
Preserving thymines and adenines from T>C/A>G transitions
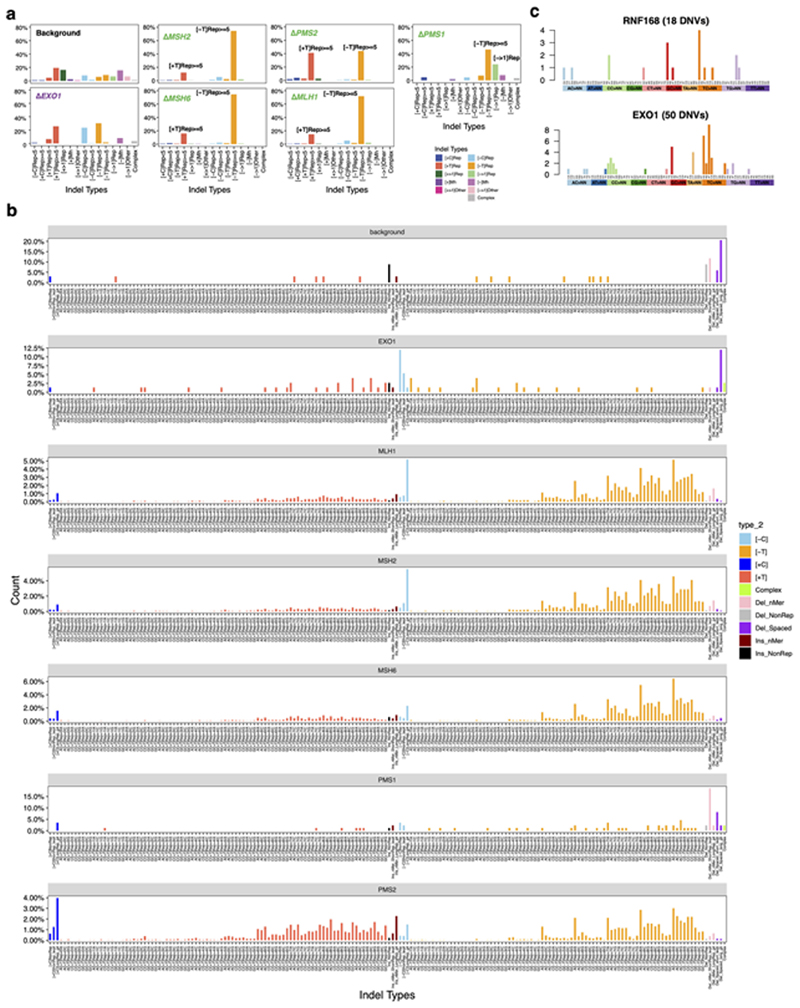
Two genes EXO1 and RNF168, with wide-ranging roles in repair/checkpoint pathways41,42,43 showed mutational signatures. EXO1 encodes a 5’ to 3’ exonuclease with RNase H activity. ΔEXO1 generated substitution, double-substitution, and indel signatures in hiPSC (Fig. 2a and Extended Data Fig. 5), consistent with previous report of ΔEXO1 in HAP1 lines9. In HAP1 cells, ΔEXO1 had stronger C>A components, probably reflecting differences in model systems. ΔEXO1 also produced a double substitution pattern defined by TC>AT, TC>AA, and GC>AA mutations, and an indel signature characterized by 1 bp A/T insertions at long poly[d(A-T)] (>= 5 bp) and 1 bp deletions at short poly[d(A-T)] or poly[d(C-G)] (< 5 bp) (Extended Data Fig. 5).
RNF168 encodes an E3 ubiquitin ligase involved in DNA double-strand break (DSB) repair43 that regulates 53BP1, BRCA1, and RAD18 recruitment to DSBs through ubiquitin-dependent signaling44–46. The substitution signature of ΔRNF168 has two T>C peaks at ATA>ACA and TTA>TCA (Fig. 2a) and shares similarity with ΔEXO1(cossim: 0.94). Double substitution patterns were defined by TC>AA and GC>AA mutations. Indel signature was not observed for ΔRNF168.
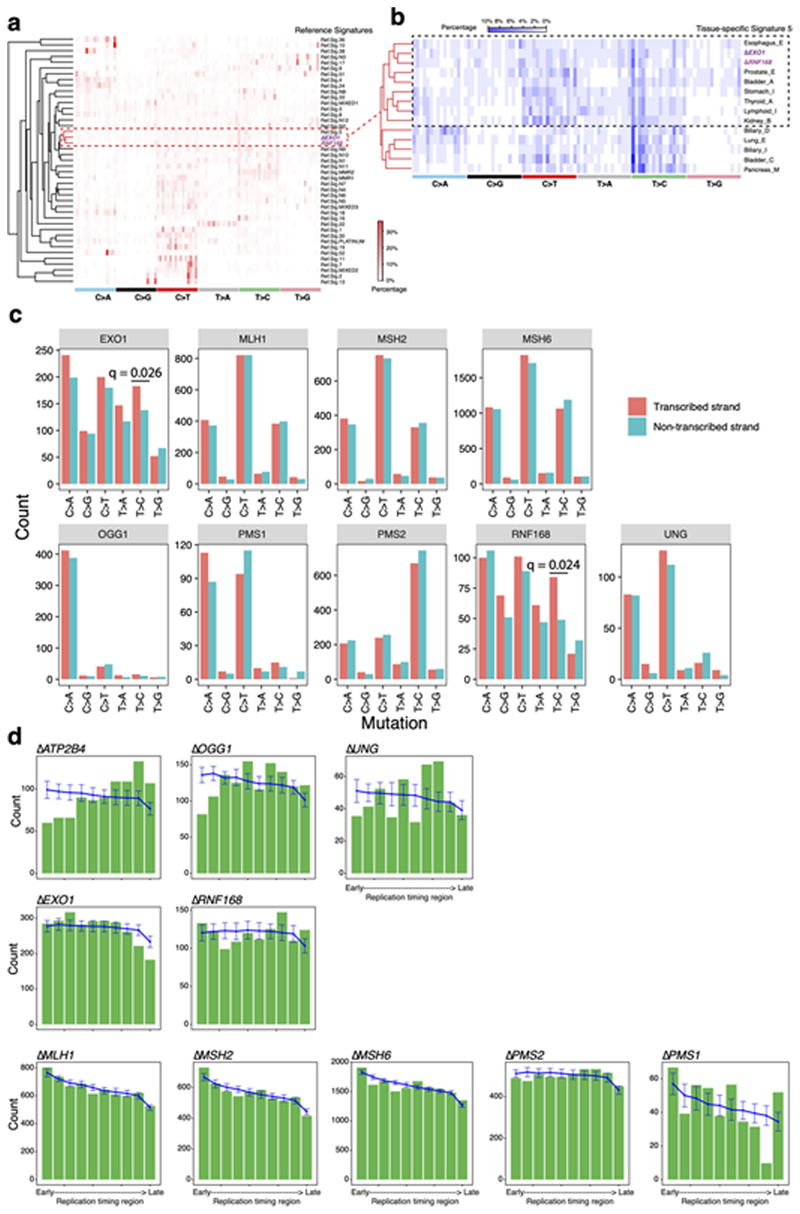
Substitution signatures of ΔEXO1 and ΔRNF168 are most similar to RefSig5 of cancer-derived signatures (Fig. 2b, cossim: 0.89-0.9, Extended Data Fig. 6a,b), defined mainly by T>C/A>G substitutions. Additionally, ΔEXO1 and ΔRNF168 signatures show transcriptional strand bias for T>C/A>G mutations (Fig. 2e,f, Extended Data Fig. 6c), in particular, at ATA and TTA context, with bias for T>C on the transcribed strand (A>G on non-transcribed strand). This is in-keeping with T>C/A>G transcriptional strand asymmetry in Signature 5. The etiology of Signature 5 is currently unknown, although a hypermutator phenotype has been reported in association with ERCC2 loss7. Due to its similarity to ΔEXO1 and ΔRNF168 signatures, the wide-ranging roles played by these proteins and the transcriptional strand bias observed, we speculate that Signature 5 has a complex origin, and may be associated with endogenous DNA damage that are repaired by multiple repair pathway proteins.
Endogenous DNA damage managed by mismatch repair (MMR)
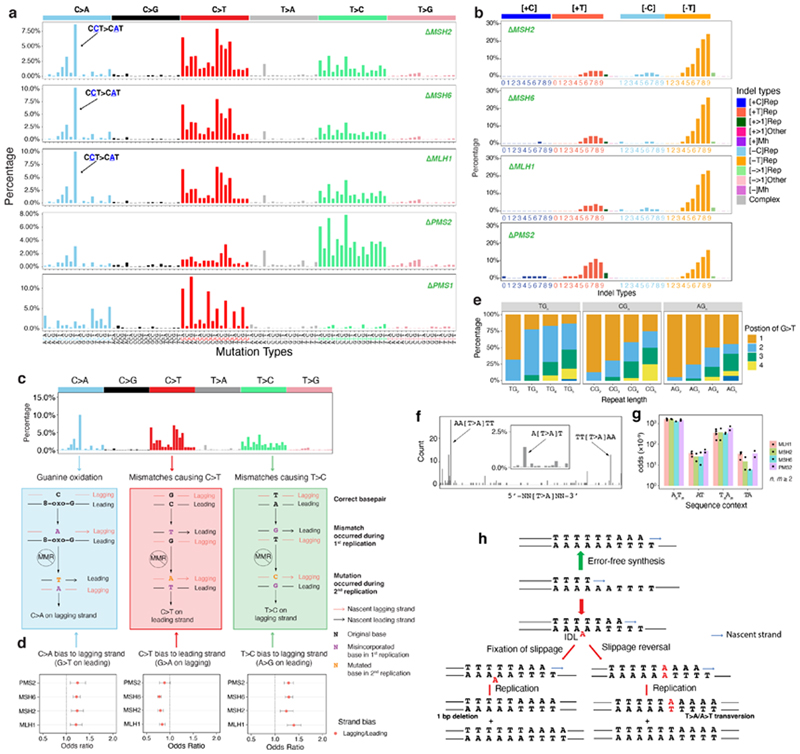
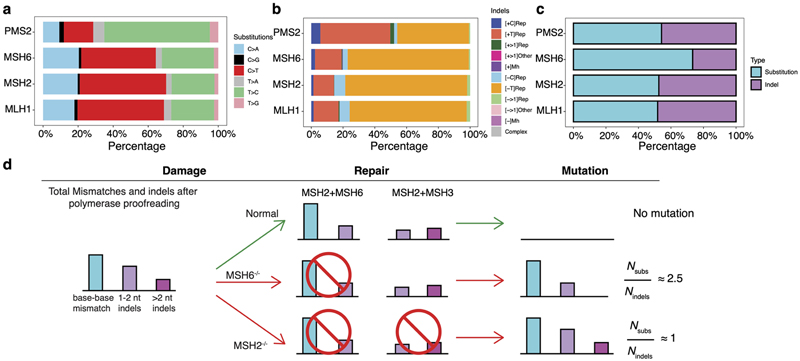
Knockouts of five genes involved in the MMR pathway47–49, MSH2, MSH6, MLH1, PMS2, and PMS1, produced substitution and indel signatures (Fig. 3a,b). ΔMLH1, ΔMSH2, and ΔMSH6 produced qualitatively identical substitution signatures (cossim: 0.99) characterized by a single strong peak at CCT>CAT/AGG>ATG, and multiple peaks of C>T and T>C (Fig. 3a). In contrast, ΔPMS2 generated a signature of predominantly T>C transitions with a predominance at ATA, ATG, and CTG (Fig. 3a). The single peak at CCT>CAT/AGG>ATG remains visible in the ΔPMS2 signature, albeit markedly reduced (10% to 3%). In addition, ΔMSH2, ΔMSH6, and ΔMLH1 generated indel signatures dominated by A/T deletions at long repetitive sequences. In contrast, ΔPMS2 produced similar proportions of A/T insertions and A/T deletions at long repetitive sequences (Fig. 3b, Extended Data Fig. 5a,b). ΔPMS1 generated A/T deletions only at long poly[d(A-T)] (>=5 bp) and long deletions (> 1bp) at repetitive sequences (Extended Data Fig. 5a).
Figure 3. Multiple endogenous sources of DNA damage managed by mismatch repair.
(a) Substitution and (b) indel signatures for five mismatch repair gene knockouts. The indel signature of ΔPMS1 is shown in Extended Data Fig. 5a. (c) Dissection of DNA mismatch repair mutational signatures: C>A mutations believed to be due to oxidative damage of guanine and proposed mechanism of how DNA polymerase errors contribute to mis-incorporated bases that result in C>T and T>C. All other mismatch possibilities and their outcomes are demonstrated in Extended Data Fig. 7. The red and black strands represent lagging and leading strands, respectively. The arrowed strand is the nascent strand. (d) Replicative strand asymmetry observed for mutational signatures generated by four MMR gene knockouts. Dots and error bars represent odds ratio with 95% confidence interval. (e) The relative frequency of occurrence of G>T/C>A in polyG tracts for ΔMSH6. The count and relative frequency of occurrence of G>T/C>A in polyG tracts for ΔMSH2 and ΔMLH1 are shown in Extended Data Fig. 8. (f) T>A mutation frequency is highest at junctions of poly(A)poly(T) or poly(T)poly(A). The inset shows that T>A mutations have a striking peak at ATT. (g) Odds for T>A mutations occurring at poly(A)poly(T) or poly(T)poly(A) are higher than AT sequences flanked by other nucleotides, corrected for sequence context through whole genome. Data are represented as mean ± SEM. N= 2~4, see Supplementary Table 2. (h) Putative ‘reverse template slippage’ model: T>A substitutions at poly(A)poly(T) or poly(T)poly(A) junctions arise due to template strand slippage and subsequent reversal of the slipped template strand. IDL: insertion-deletion loop.
In-depth analysis of these mutational signatures allowed us to determine putative sources of endogenous DNA damage (Fig. 3c) acted upon by MMR.
First, we consistently observed replication strand bias across ΔMLH1, ΔMSH2, ΔMSH6, and ΔPMS2: C>A on the lagging strand (equivalent to G>T leading strand bias), C>T on the leading strand (or G>A lagging) and T>C lagging (or A>G leading) (Fig. 3d). Similar results were previously reported in yeast and human cancers50–52. Under our experimental settings where exogenous DNA damage was not administered, mismatches may be generated by DNA polymerases α, δ or ε during replication. In the absence of MMR, these lesions become permanently etched as mutations. To understand which replicative polymerases could be causing these mutations, we analysed putative progeny of all 12 possible base/base mismatches (Extended Data Fig. 7). T/G mismatches are the most thermodynamically stable and represent the most frequent polymerase error53. Our assessment suggests that the predominance of T>C transitions on the lagging-strand can only be explained by misincorporation of T by lagging strand polymerases, pol-α and/or pol-δ, leading to G/T mismatches (Fig. 3c). Similarly, the observed bias for C>T transitions on the leading strand is likely to be predominantly caused by misincorporation of G on lagging strand by pol-α and/or pol-δ resulting in T/G mismatches (Fig. 3c).
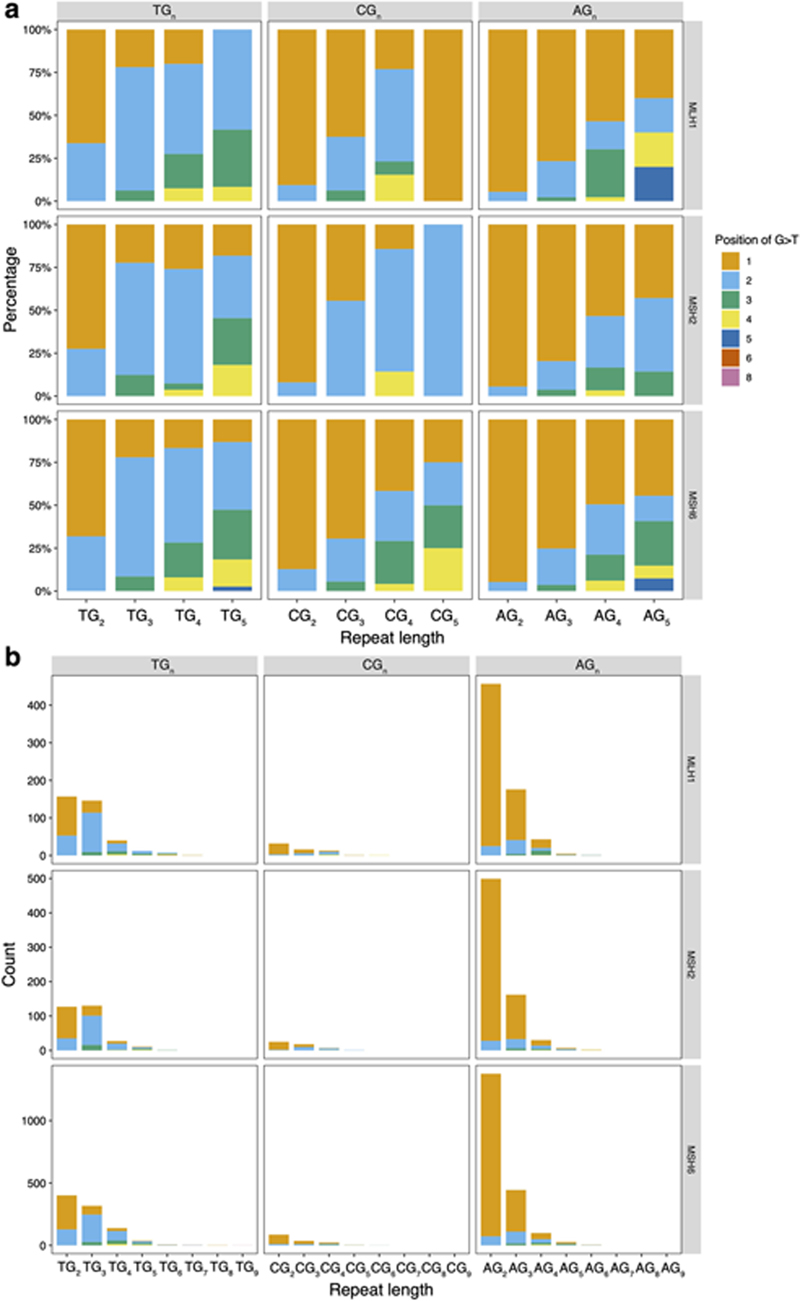
Second, the C>A predominance could be explained by differential processing of 8-oxo-dGs (Fig. 3c). The principal C>A/G>T peak in MMR-deficient cells occurs at CCT>CAT/AGG>ATG followed by CCC>CAT/GGG>GTG and is distinct from the C>A/G>T peaks observed in ΔOGG1. However, we previously showed a depletion of mutations at CC/GG sequence motifs for ΔOGG1. Intriguingly, the experimental data suggest that the 8-oxo-G:A mismatches can be repaired by MMR, preventing C>A/G>T mutations54. Furthermore, G>T/C>A mutations of MMR-deficient cells occurred most frequently at the second G in 5’-TG n(n>=3) in ΔMLH1, ΔMSH2, and ΔMSH6 (Fig. 3e and Extended Data Fig. 8). This is consistent with previous reports55 of the classical imprint of guanine oxidation at polyG tracts where site reactivity in double-stranded 5’-TG1G2G3G4T sequence is reported as G2 > G3 > G1 > G4. These results implicate MMR involvement in repairing 8-oxo-G:A mismatches at GG motifs that perhaps cannot be cleared by OGG1 in BER. As for G>T leading strand bias, studies in yeast have demonstrated that an excess of 8-oxo-dG-associated mutations occurs during leading strand synthesis56. Furthermore, translesion synthesis polymerase η is also more error-prone when bypassing 8-oxo-dG on the leading strand57, which would result in increased 8-oxo-G:A mispairs on the leading strand.
Third, T>A transversions at ATT were strikingly persistent in MMR knockout signatures, although with modest peak size (<3% normalized signature, Fig. 3a). Additional sequence context information revealed that T>A occurred most frequently at AATTT or TTTAA, junctions of poly(A) and poly(T) tracts (Fig. 3f)58,59. Moreover, the length of 5’- and 3’- flanking homopolymers influenced the likelihood of mutation occurrence: T>A transversions were one to two orders of magnitude more likely to occur when flanked by homopolymers of 5’poly(A)/3’poly(T) (AnTm) or 5’poly(T)/3’poly(A) (TnAm), than when there were no flanking homopolymeric tracts (Fig. 3g).
Since polynucleotide repeat tracts predispose to indels due to replication slippage, a known source of mutagenesis in MMR-deficient cells, we hypothesize that T>A transversions observed at abutting poly(A)/poly(T) tracts are the result of ‘reverse template slippage’. In this scenario, the polymerase replicating across a mixed repeat sequence such as AAAAAATTTT, in which the template slipped at one of the As, would incorporate five instead of six Ts opposite the A repeat (red arrow pathway in Fig. 3h). If at this point the template were to revert to its original correct alignment, A/A mismatch would occur, resulting in a T>A transversion. If the slippage remained, this would give rise to a single nucleotide deletion, a characteristic feature of MMR-deficient cells known as microsatellite instability (MSI) (Fig. 3b, indel signatures).
Gene-specific mutational signatures in MMR-deficiency
There are uncertainties regarding which cancer-derived signatures are truly MMR-deficiency signatures. It was suggested that SBS6, SBS14, SBS15, SBS20, SBS21, SBS26, and SBS44 were MMR-deficiency related6. In an independent analytical exercise, only two MMR-associated signatures were identified24, with variations seen in different tissue-types24. An experimental process would help to obtain clarity in this regard8–11.
As described earlier, the substitution and indel patterns of ΔMSH2,ΔMSH6, and ΔMLH1 showed qualitative similarities and were distinct from ΔPMS2 (Fig. 3a,b and Extended Data Fig. 9a,b). While qualitative indel profiles of ΔMSH2, ΔMSH6, and ΔMLH1 were very similar (Fig. 3b), their quantitative burdens were different (Fig. 1e). ΔMLH1 and ΔMSH2 had high indel burdens, whereas ΔMSH6 had half the indel burden (Fig. 1e). Substitution-to-indel ratios showed that ΔMSH2, ΔPMS2, and ΔMLH1 produced similar numbers of substitutions and indels, while ΔMSH6 generated nearly 2.5 times more substitutions than indels (Extended Data Fig. 9c,d). This is in-keeping with known protein interactions: MSH2 and MSH6 form heterodimer MutSα that addresses base-base mismatches and small (1-2 nt) indels48,60. MSH2 can also heterodimerize with MSH3 to form heterodimer MutSβ, which does not recognize base-base mismatches, but can address indels of 1-15 nt61. This functional redundancy in small indel repair between MSH6 and MSH3 explains the smaller number of indels observed in ΔMSH6 (Fig. 1e, Extended Data Fig. 9d) compared to ΔMSH2 cells. This is consistent with the near-identical MSI phenotypes of Msh2-/- and Msh3-/-; Msh6-/-mice62.
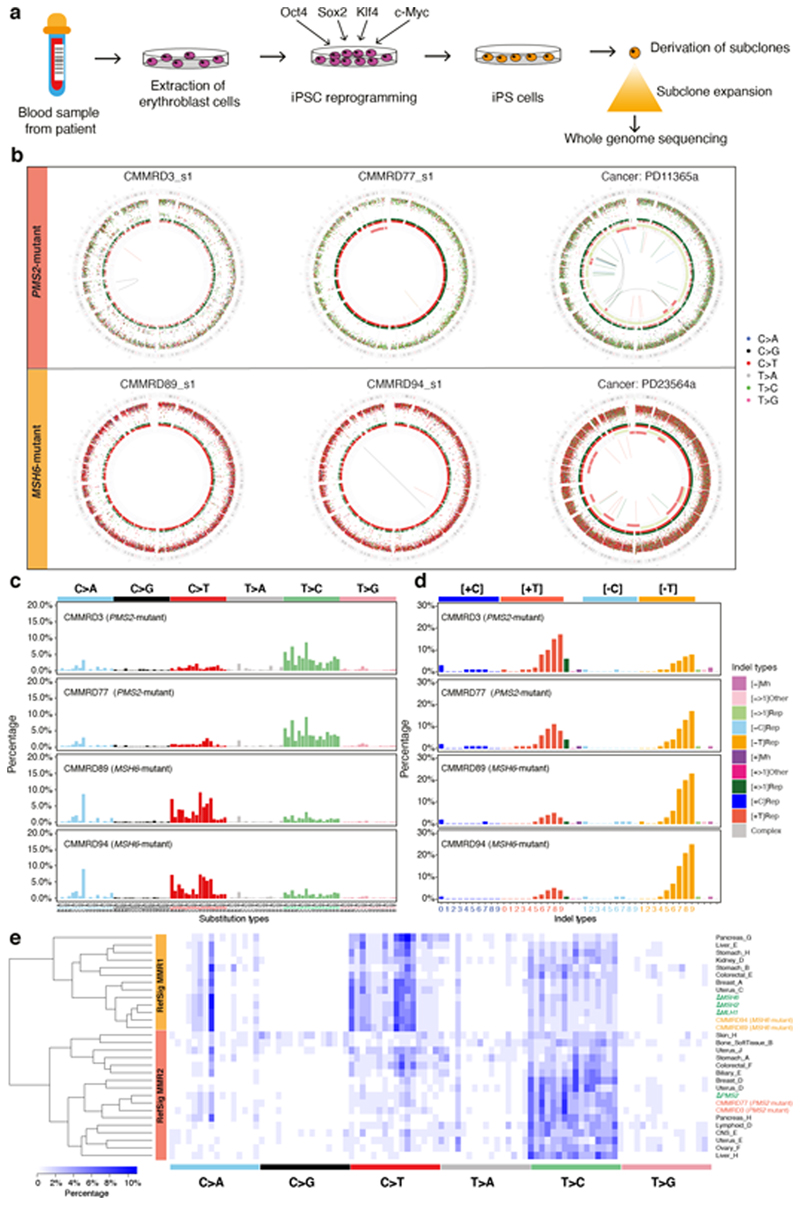
Thus, there are clear qualitative differences between substitution and indel profiles of ΔMSH2, ΔMSH6, and ΔMLH1 from ΔPMS2. To validate these MMR-gene-specific knock-out signatures, we interrogated genomic profiles of normal cells derived from patients with inherited autosomal recessive defects in MMR genes resulting in Constitutional Mismatch Repair Deficiency (CMMRD) – a severe, hereditary cancer predisposition syndrome characterized by increased risk of early-onset (often pediatric) malignancies and cutaneous café-au-lait macules63,64. hiPSCs were generated from erythroblasts derived from four CMMRD patients (one PMS2 homozygote, one PMS2 heterozygote and two MSH6 homozygotes) and one healthy control65. hiPSC clones obtained were genotyped65, and expression arrays and cellomics-based immunohistochemistry were performed to ensure pluripotent stem cells were generated (Methods). Parental clones were grown for mutation accumulation, single-cell subclones were derived and whole-genome sequenced (Fig. 4a).
Figure 4. Gene-specific features of signatures of mismatch repair (MMR) deficiency are recapitulated in other model systems.
(a) Experimental workflow including generation of hiPSCs from patients with Constitutional Mismatch Repair Deficiency (CMMRD), subcloning of hiPSCs and whole-genome sequencing. (b) Genome plots of MMR knockouts demonstrate consistent gene-specificity regardless of model system, e.g., cancer (in vivo) and CMMRD patient-derived hiPSCs (in vitro). Top: whole genome plots of two iPSC subclones from two PMS2 mutated CMMRD patients and a breast tumor with PMS2 deficiency. Bottom: genome plots of two iPSC subclones derived from two MSH6 mutant CMMRD patients and a breast tumor with MSH2/MSH6 deficiency. Genome plots show somatic mutations including substitutions (outermost, dots represent six mutation types: C>A, blue; C>G, black; C>T, red; T>A, grey; T>C, green; T>G, pink), indels (the second outer circle, colour bars represent five types of indels: complex, grey; insertion, green; deletion other, red; repeat-mediated deletion, light red; microhomology-mediated deletion, dark red) and rearrangements (innermost, lines representing different types of rearrangements: tandem duplications, green; deletions, orange; inversions, blue; translocations, grey). (c) 96-channel substitution profiles. (d) 45-channel indel profiles. (e) Hierarchical clustering of cancer-derived tissue-specific MMR signature and MMR knockout signatures. 96-bar plots of ΔPMS2-related tissue-specific signatures can be viewed here:
https://signal.mutationalsignatures.com/explore/cancer/consensusSubstitutionSignatures/6
Mutational signatures seen in CMMRD hiPSCs were virtually identical to those of the CRISPR-Cas9 knockouts (Fig. 4b-d). The ΔPMS2 CMMRD patterns carried the same propensity for T>C mutations, small contribution of C>T mutations and single C>A/G>T at CCT/AGG peak as ΔPMS2, and the MSH6 CMMRD patterns carried the excess of C>T mutations with pronounced C>A/G>T at CCT/AGG similar to ΔMLH1, ΔMSH2 and ΔMSH6 clones (Fig. 4c). Indel propensities were also reflected in patient-derived cells (Fig. 4d). Accordingly, gene-specificity of signatures generated in the experimental knockout system is well-recapitulated in independent patient-derived stem cell models.
Furthermore, gene-specific MMR signatures were seen in the International Cancer Genome Consortium (ICGC) cohort of >2,500 primary WGS cancers24. Biallelic MSH2/MSH6/MLH1 mutant tumors carried the same signature (RefSig MMR1) as ΔMSH2/ΔMSH6/ΔMLH1 clones (Fig. 4e). We also identified biallelic PMS2 mutants in several cancers, including breast and ovarian cancers with mutation patterns (RefSig MMR2) that were indistinguishable from experimentally-generated ΔPMS2 signatures (Fig. 4e).
A mutational-signature-based classifier of MMR-deficiency
Algorithms to classify MMR-deficient tumors developed using massively-parallel sequencing data depend on detecting elevated tumor mutational burden (TMB) or microsatellite instability (MSI)66–71. New knowledge from our experimental data and awareness of tissue-specific signature variation (Fig. 4e) led us to derive an MMR-deficiency classifier.
We obtained WGS data on 336 colorectal cancers from patients recruited via the National Health Service-based UK 100,000 Genomes Project (UK100kGP) run by Genomics England (GEL). Critically, these samples had accompanying immunohistochemistry (IHC) testing of MMR-deficiency status based on protein staining of MSH2, MSH6, MLH1, and PMS2, and 79 out of 336 cases were identified as MMR-deficient (~24%). This cohort of 336 samples was randomly assigned into a training set (180 MMR-proficient and 56 MMR-deficient samples) or a test set (77 MMR-proficient and 23 MMR-deficient samples). We developed a logistic regression classifier, called MMRDetect, using new mutational-signatures-based parameters derived from the experimental insights gained from this study: 1) the exposure of MMR-deficient substitution signatures (E MMRD); 2) the cosine similarity between substitution profile of the tumor and that of MMR knockouts (S sub); 3) the mutation burden of indels in repetitive regions (N repindel) and 4) the cosine similarity between repeat-mediated deletion profile of the tumor and that of MMR knockouts (S repdel) (further details in Methods, Extended Data Fig. 10, Supplementary Table 5,6). Ten-fold cross-validation was conducted in the training set (Extended Data Fig. 10f). As a comparator, we applied another widely-used MSI classifier, MSIseq66 to the same cohort of 336 colorectal cancers.
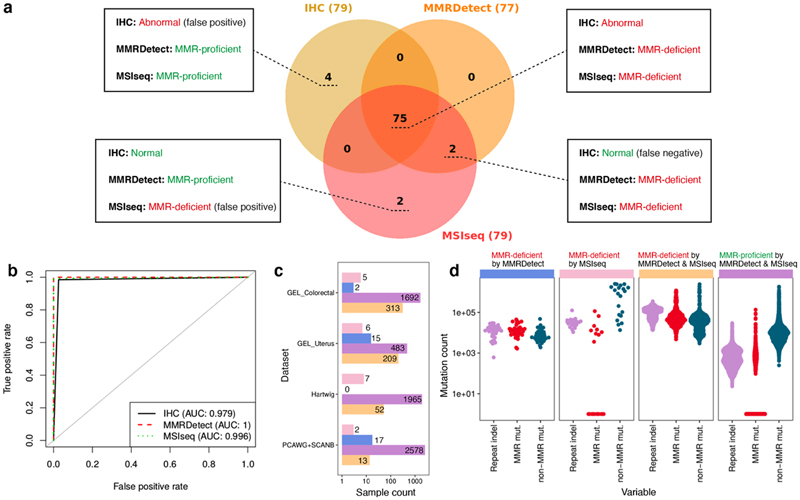
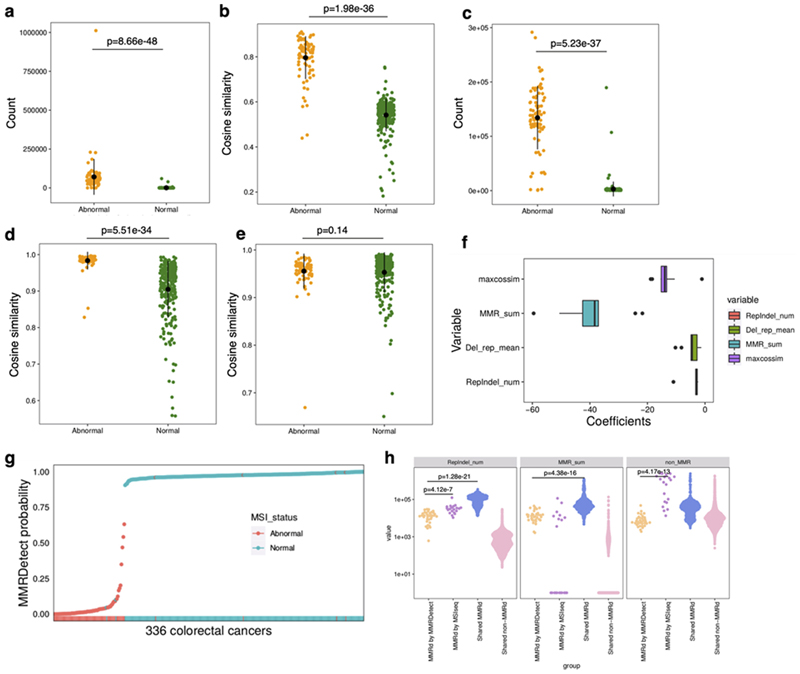
Samples with MMRDetect-calculated probability < 0.7 are defined as MMR-deficient by MMRDetect (Extended Data Fig. 10g). In all, 75 of 336 samples were concordantly determined as MMR-deficient by MMRDetect, MSIseq, and IHC (Fig. 5a, Supplementary Table 5). Eight samples had discordant statuses, including four samples with MMR-deficiency only by IHC, two samples by MSIseq and MMRDetect, and two samples uniquely called by MSIseq. To understand these discordances, we sought driver mutations. Among these eight samples, two samples missed by IHC had confirmed loss-of-function mutations in MMR genes. Additionally, two cases uniquely called by MSIseq were misclassified, and were, in fact, POLE mutated and not MMR-deficient (Fig. 5a, Supplementary Table 5). While receiver operating characteristic (ROC) curves generated by these three methods showed excellent performance across the board, MMRDetect had the highest AUC of 1 in this dataset (Fig. 5b).
Figure 5. Mutational signature-based mismatch repair (MMR) deficiency classifier, MMRDetect.
(a) Concordance of three MMR-deficiency detection methods - immunohistochemistry (IHC) staining, MSIseq and MMRDetect - on 336 colorectal cancers is illustrated in the Venn diagram. IHC staining, MSIseq and MMRDetect identified 79, 79 and 77 MMR-deficient samples, respectively. Details of the eight samples with discordant outcomes from the three methods are provided in Supplementary Table 5. Four samples classified as MMR-proficient by MMRDetect and MSIseq have abnormal IHC staining (shown in dark yellow). However, no functional mutations in MMR genes were found. Two samples classified as MMR-proficient by MMRDetect and IHC staining were identified as MMR-deficient by MSIseq (shown in pink) and did not have MMR gene mutations but had POLE mutations and signatures instead. Two samples classified as MMR-deficient by MMRDetect and MSIseq have normal IHC staining (shown in orange). Both have mutations in MMR genes. (b) Receiver operating characteristic (ROC) curves of IHC staining, MMRDetect and MSIseq classification. (c) Concordance between MSIseq and MMRDetect on 2,012 GEL colorectal cancers, 713 GEL uterine cancers, 2,024 Hartwig metastatic cancers and 2,610 cancers from PCAWG & SCANB projects. The bars show the numbers of samples that were identified as MMR deficient by only MSIseq (pink), only MMRDetect (blue), both (yellow) and none (purple). (d) The distribution of three variables amongst samples that were discordantly (blue, pink) and concordantly (yellow and purple) detected by MSIseq and MMRDetect: the number of repeat-mediated indels, number of mutations associated with MMRD signatures and non-MMRD mutations. Numbers of MMR-deficient samples determined by MMRDetect only (blue), MSIseq only (pink), both (yellow) and none (purple) are 34, 20, 587 and 6,718, respectively.
We next compared MMRDetect and MSIseq on another 2,012 colorectal and 713 uterine samples from UK100kGP, 2,610 published WGS primary cancers72–74 and 2,024 WGS metastatic cancers 75 (Supplementary Table 7-10, Methods). There was a very high concordance between MMRDetect and MSISeq for classifying tumors (0.97 to 0.997 (Fig. 5c)). To understand the discrepancies between the two algorithms, we compared variables that were used by the two classifiers (Fig. 5d) and found that samples uniquely identified as MMR-deficient by MSIseq had a significantly higher number of repeat-mediated indels (N repindel) and non-MMR-deficiency signatures (E non-MMRD) than the ones identified as MMR-deficient by only MMRDetect (p < 0.001, two-sided Mann-Whitney test, Extended Data Fig. 10h). This indicated a higher likelihood of misclassifying samples with high indel loads caused by non-MMR-deficient mutational processes (i.e., false positives) for MSIseq, a known generic problem reported for NGS indel-based classifiers76. Many of these samples showed signatures associated with proofreading POLE mutations. This demonstrates that MMRDetect has improved specificity over MSIseq.
Of note, samples identified as MMR-deficient by only MMRDetect had significantly lower numbers of repeat-mediated indels (N repindel) and MMR-related substitution signatures (E MMRD) than samples concordantly identified as MMR-deficient by both MSIseq and MMRDetect (p < 0.001, two-sided Mann-Whitney test, Extended Data Fig. 10h), suggesting that MMRDetect has improved sensitivity for MMR-deficient cancers with lower overall MMR-related mutation counts (E MMRD). Indeed, of 15 bona fide MMR-deficient breast cancers, a tumor-type that is not as proliferative as colon/uterine cancer and has lower mutation numbers in general, MMRDetect identified 13 cases (87%), whilst MSIseq identified five (~33%) of 15 samples, as the remaining ten samples had lower repeat-mediated indel loads (2,885-18,863, Supplementary Table 10). The two cases missed by MMRDetect had very low levels of MMR-related signatures and were complicated by high levels of APOBEC-related mutagenesis. Thus, MMRDetect has enhanced sensitivity, particularly at detecting MMR-deficient samples with lower mutation burdens (Fig. 5d), although it could miss cases where MMR-deficiency is present at a very low level. We note that the current version of MMRDetect classifier has been trained on highly-proliferative colorectal cancers. More sequencing data are required to improve MMRDetect’s detection sensitivity in other tumor types.
Discussion
In standardized experiments performed in a diploid, non-transformed human stem cell model, biallelic gene knockouts that produce mutational signatures in the absence of administered DNA damage are indicative of genes that are important at maintaining the genome from intrinsic DNA damage sources (Fig. 6). We find substitution, double substitution and/or indel signatures of nine genes: ΔOGG1, ΔUNG,ΔEXO1,ΔRNF168,ΔMLH1,ΔMSH2,ΔMSH6,ΔPMS2, and ΔPMS1, suggesting that these proteins are critical guardians of the genome in non-transformed cells. Many gene knockouts did not show mutational signatures under these conditions. This does not mean that they are not important DNA repair proteins. There may be redundancy, or the gene may be crucial to the orchestration of DNA repair, even if not imperative at directly preventing mutagenesis. It is also possible that some knockouts have very low rates of mutagenesis such that statistically distinct signatures cannot be distinguished from background mutagenesis within our experimental time frame. For genes involved in double-strand-break (DSB) repair, hiPSCs may not be permissive for surviving DSBs to report signatures. Other genes may require alternative forms of endogenous DNA damage that manifest in vivo but notin vitro, for example, aldehydes, tissue-specific products of cellular metabolism, and pathophysiological processes such as replication stress. Likewise, for genes in the nucleotide excision repair pathway, bulky DNA adducts, whether exogenous (e.g., ultraviolet damage) or endogenous (e.g., cyclopurines and by-products of lipid peroxidation) may be a pre-requisite before these compromised genes reveal associated signatures. Experimental modifications such as addition of DNA damaging agents or using alternative cellular models (e.g., cancer lines or permissive cellular models of specific tissue-types), could amplify signal, but they could also modify mutational outcomes. That must be taken into consideration when interpreting data. Also, not all genes have been successfully knocked out in this endeavor and could have similarly important roles in directly preventing mutagenesis.
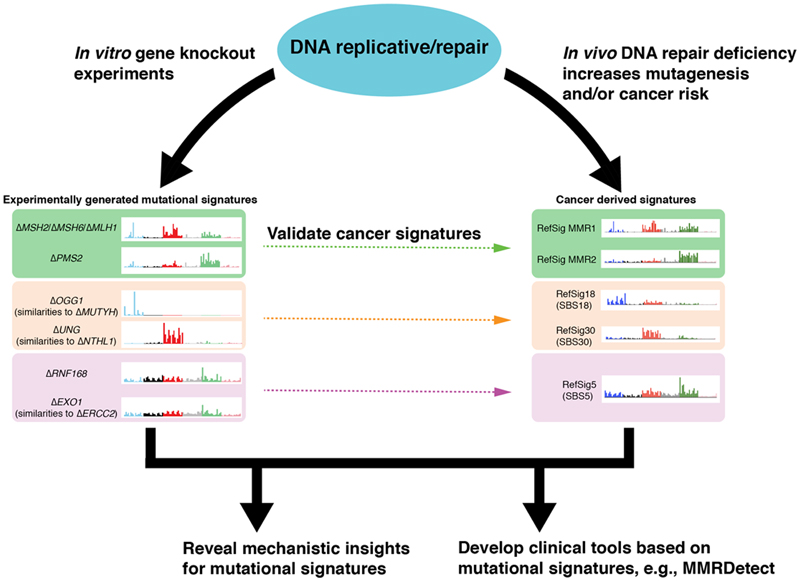
Figure 6. Impact of experimental validation of cancer-derived mutational signatures on biological understanding and development of clinical applications.
Some genes (often involved in DNA repair pathways) which are important guardians against endogenous DNA damage under non-malignant circumstances, have been identified in this work. They help to validate and to understand the etiologies of cancer-derived mutational signatures. The biological insights help to drive the development of new genomic clinical tools to detect these abnormalities with greater accuracy and sensitivity across tumor types.
Detailed dissection of experimental signatures revealed interesting mutational insights, including how OGG1 and MMR sanitize oxidized guanines at specific sequence motifs. By contrast, UNG maintains all cytosines from hydrolytic deamination, irrespective of sequence context. Exhaustive assessment of DNA mismatches and their putative outcomes also uncovered precise polymerase errors that are repaired by MMR, including misincorporation of T resulting in T>C transitions and misincorporation of G resulting G>A/C>T transitions by lagging strand polymerases. We also observe a T>A substitution pattern at abutting poly(A) and poly(T) tracts and postulate a mechanism called reverse template slippage.
While it is known that 8-oxo-dGs can result in G>T mutations, our work demonstrates that the etiology of the culture-related signature and cancer-derived Signature 18 is mainly 8-oxo-dG. We highlight the importance of functional EXO1 and RNF168 in preventing Signature 5, a relatively ubiquitous signature characterized by T>C/A>G transitions. We define gene-specificities of MMR deficiency signatures, prove that these are robust in normal stem cells derived from patients with CMMRD, and identify gene-specific signatures in human cancers.
Finally, unlike signatures of environmental mutagens that are historic, signatures of repair pathway defects are likely to be on-going. They could serve as biomarkers in precision medicine13,14,18 (Fig. 6) to identify pathway defects where selective therapeutic strategies are available. Our experiments led to the development of a more sensitive and specific assay to detect MMR deficiency, MMRDetect. Current TMB-based assays may have reduced sensitivity to detect MMR deficiency in tissues that do not have high proliferative rates. They may also falsely call MMR-proficient cases as MMR-deficient because single components were used for measurement (e.g., indel or substitution burdens only). High mutational burdens can be due to different biological processes77. Consequently, assays based on burden alone are unlikely to be adequately specific. As a community, we are at the early stages of seeking experimental validation of mutational signatures. However, we hope that our approach, which leans on experimental data, provides a template for improving biological understanding of how mutational patterns arise, and this, in turn, could help propose improved tools for tumor characterization going forward.
Methods
Cell lines and culture
The human iPSC line used in this study is previously described11. The line was derived at the Wellcome Sanger Institute (Hinxton, UK). The use of this cell line model was approved by Proportionate Review Sub-committee of the National Research Ethics (NRES) Committee North West - Liverpool Central under the project “Exploring the biological processes underlying mutational signatures identified in induced pluripotent stem cell lines (iPSCs) that have been genetically modified or exposed to mutagens” (ref: 14.NW.0129). It is a long-standing iPSC line that is diploid and does not have any known driver mutations. It does carry a balanced translocation between chromosomes 6 and 8. It grows stably in culture and does not acquire a vast number of karyotypic abnormalities. This is confirmed through mutational and copy number assessment of the WGS data reviewed of all subclones.
Cell culture reagents were obtained from Stem Cell Technologies unless otherwise indicated. Cells were routinely maintained on Vitronectin XF-coated plates (10-15 ug/mL) in TeSR-E8 medium. The medium was changed daily, and cells were passaged every 4–8 days depending on the confluence of the plates using Gentle Cell Dissociation Reagent.
All cell lines were grown at 37°C, with 20% oxygen and 5% carbon dioxide in a humidified incubator, except for the pilot study in which the iPSCs knockouts were also grown under hypoxic condition (3% oxygen) as one of the experimental conditions. Cells were cultivated as monolayers in their respective growth medium and passaged every 3-4 days to maintain sub-confluence during the mutation accumulation step. All cell lines were tested negative for mycoplasma contamination using MycoAlertTM Mycoplasma Detection Kit and LookOut® Mycoplasma PCR Detection Kit according to the manufacturers’ protocol.
CMMRD patient sample collection
Four CMMRD patients were recruited under the auspices of the Insignia project. This included two PMS2-mutant patients and two MSH6-mutant patients. Supplementary Table 11 shows the genotypes of these four patients. A healthy donor was recruited as control. Ethical approval for the generation of hiPSCs from patients and healthy control was received for the Insignia project under the title “Exploring the biological processes underlying mutational signatures identified in patients with inherited disorders and in patients exposed to mutagens”, with reference number 13/EE/0302, from the East of England Cambridgeshire and Hertfordshire Research Ethics Committee.
Generation of DNA repair gene knockouts in human iPSCs
Biallelic DNA repair gene knockouts in human iPSCs were performed by the High Throughput Gene Editing team of Cellular Operations at the Sanger Institute, Hinxton, UK. These knockouts were generated based on the principles of CRISPR/Cas9-mediated HRD and NHEJ as described previously 78.
Generation of donor plasmids for precise gene targeting via HDR
All knockouts were generated using an established protocol that was found to minimize potential off-target effects 78. Briefly, the intermediate targeting vectors were generated for each gene using GIBSON assembly of the four fragments: pUC19 vector, 5’ homology arm, R1-pheS/zeo-R2 cassette and 3’ homology arm. Gene-specific homology arms were amplified by PCR from the iPSC gDNA and were either gel-purified or column-purified (QIAquick, QIAGEN). pUC19 vector and R1-pheS/zeo-R2 cassette were prepared as gel-purified blunt fragments (EcoRV digested). Fragments were assembled via GIBSON assembly reactions (Gibson Assembly Master Mix, NEB, E2611) according to the manufacturer’s instructions. Assembly reaction mix was transformed into NEB 5-alpha competent cells and clones resistant to carbenicillin (50 μg/mL) and zeocin (10 μg/mL) were analyzed by Sanger sequencing to select for correctly-assembled constructs. Sequence-verified intermediate targeting vectors were converted into donor plasmids via a Gateway exchange reaction. LR Clonase II Plus enzyme mix (Invitrogen, 12538120) was used to perform a two-way reaction exchanging only the R1-pheSzeo-R2 cassette with the pL1-EF1αPuro-L2 cassette as previously described 79. The latter was generated by cloning synthetic DNA fragments of the EF1α promoter and puromycin resistance cassette into one of pL1/L2 vector 79. Following Gateway reaction and selection on yeast extract glucose (YEG) + carbenicillin agar (50 μg/mL) plates, correct donor plasmids were verified by capillary sequencing across all junctions.
Guide RNA design & cloning
For every gene knockout, two separate gRNAs targeting within the same critical exon of a gene were also selected. The gRNAs were selected using the WGE CRISPR tool 80 based on their off-target scores. Selected gRNAs were suitably positioned to ensure DNA cleavage within the exonic region, excluding any sequence within the homology arms of the targeting vector. To generate individual gene targeting plasmids, gene-specific forward and reverse oligos were annealed and cloned into BsaI site of either U6_BsaI_gRNA (kindly provided by Sebastian Gerety, unpublished). The gRNA sequences are listed in Supplementary Table 12. All the oligos were synthesized by Integrated DNA Technologies (IDT)
Delivery of KO-targeting plasmids, donor templates and Cas9, selection and genotyping
Human iPSCs were dissociated to single cells and nucleofected with Cas9-coding plasmid (hCas9, Addgene 41815), sgRNA plasmid and donor plasmid on Amaxa 4D-Nucleofactor program CA-137 (Lonza). Following nucleofection, cells were selected for up to 11 days with 0.25 μg/mL puromycin. Edited cells were expanded to ~70% confluency before subcloning. Approximately 1000 cells were subcloned onto 10 cm tissue culture dishes precoated with SyntheMAX substrate (Corning) at a concentration of 5 μg/cm2to allow colony formation for 8-10 days until colonies are approximately 1-2 mm in diameter. Individual colonies were picked into U-bottom 96-well plates using a dissection microscope and a p20 pipette, grown to confluence and then replica plated. Once confluent, the replica plates were either frozen as single cells in 96-well vials or the wells were lysed for genotyping.
To genotype individual clones from a 96-well replica plate, cells were lysed and used for PCR amplification with LongAmp Taq DNA Polymerase (NEB, M0323). Insertion of the cassette into the correct locus was confirmed by visualizing on 1% E-gel (Invitrogen, G700801) PCR products generated by gene-specific (GF1 and GR1) and cassette specific primers (ER: TGATATCGTGGTATCGTTATGCGCCT and PF: CATGTCTGGATCCGGGGGTACCGCGTCGAG) for both 5’ and 3’ ends. We also confirmed single integration of the cassette by performing a qPCR copy number assay. To check the CRISPR site on the non-targeted allele, PCR products were generated from across the locus, using the same 5’ and the 3’ gene-specific genotyping primers. The PCR products were treated with exonuclease I and alkaline phosphatase (NEB, M0293; M0371) and Sanger sequenced to verify successful knockouts. Sequence reads and their traces were analysed and visualised on a laboratory information management system (LIMS)-2. For each targeted gene, two independently-derived clones with different specific mutations were isolated and studied further.
Generation of iPSCs from Constitutional Mismatch Repair Deficiency (CMMRD) Patients
Peripheral blood mononuclear cells (PBMCs) isolation, erythroblast expansion, and IPSC derivation were done by the Cellular Generation and Phenotyping facility at the Wellcome Sanger Institute, Hinxton, according to Agu et al 201565. Briefly, whole blood samples collected from consented CMMRD patients were diluted with PBS, and PBMCs were separated using standard Ficoll Paque density gradient centrifugation method. Following the PBMC separation, samples were cultured in media favoring expansion into erythroblasts for 9 days. Reprogramming of erythroblasts enriched fractions was done using non-integrating CytoTune-iPS Sendai Reprogramming kit (Invitrogen) based on the manufacturer’s recommendations. The kit contains three Sendai virus-based reprogramming vectors encoding the four Yamanaka factors, Oct3/4, Sox2, Klf4, and c-Myc. Successful reprogramming was confirmed via genotyping array and expression array.
Proteomics analysis
Cell pellets were dissolved in 150 μL buffer containing 1% sodium deoxycholate (SDC), 100mM triethylammonium bicarbonate (TEAB), 10% isopropanol, 50mM NaCl and Halt protease and phosphatase inhibitor cocktail (100X) (Thermo, #78442) using pulsed probe sonication followed by boiling at 90 °C for 5 min. Aliquots containing 50 μg of total protein, measured with the Coomassie Plus Bradford Protein Assay (Pierce), were reduced with 5 mM tris-2-carboxyethyl phosphine (TCEP) for 1 h at 60 °C and alkylated with 10 mM Iodoacetamide (IAA) for 30 min in dark. Proteins were then digested with 75 ng/μL trypsin (Pierce) overnight. The tryptic digests from the ATP2B4, EXO1, OGG1, PMS1, PMS2, RNF168 and UNG knock-out clones as well as three biological replicates of the parental cell line were labelled with the TMTpro 16plex reagents (Thermo) according to manufacturer’s instructions. The digests from MLH1, MSH2, MSH6 clones were subjected to label-free single-shot analysis. The TMTpro labelled peptides were fractionated with offline high-pH Reversed-Phase (RP) chromatography (XBridge C18, 2.1 x 150 mm, 3.5 μm, Waters) on a Dionex Ultimate 3000 HPLC system with 1% gradient. Mobile phase A was 0.1% ammonium hydroxide and mobile phase B was acetonitrile, 0.1% ammonium hydroxide. LC-MS analysis was performed on the Dionex Ultimate 3000 system coupled with the Orbitrap Lumos Mass Spectrometer (Thermo Scientific). Selected TMTpro peptide fractions were loaded to the Acclaim PepMap 100, 100 μm × 2 cm C18, 5 μm, 100? trapping column and were analyzed with the EASY-Spray C18 capillary column (75 μm × 50 cm, 2 μm). Mobile phase A was 0.1% formic acid and mobile phase B was 80% acetonitrile, 0.1% formic acid. The TMTpro peptide fractions were analyzed with a 90 min gradient from 5%-38% B. MS spectral were acquired with mass resolution of 120 k and precursors were isolated for CID fragmentation with collision energy 35%. MS3 quantification was obtained with HCD fragmentation of the top 5 most abundant CID fragments isolated with Synchronous Precursor Selection (SPS) and collision energy 55% at 50k resolution. For the label-free experiments, peptides were analyzed with a 240 min gradient and HCD fragmentation with collision energy 35% and ion trap detection. Database search was performed in Proteome Discoverer 2.4 (Thermo Scientific) using the SequestHT search engine with precursor mass tolerance 20 ppm and fragment ion mass tolerance 0.5 Da. TMTpro at N-terminus/K (for the labelled samples only) and Carbamidomethyl at C were defined as static modifications. Dynamic modifications included oxidation of M and Deamidation of N/Q. The Percolator node was used for peptide confidence estimation and peptides were filtered for q-value < 0.01. All spectra were searched against reviewed UniProt human protein entries. Only unique peptides were used for quantification. The results of proteomics analysis are provided in Supplementary Table 13.
Proliferation assay
Cells were seeded at 5,500 per well on 96-w plates. Measurements were taken at 24 h intervals post-seeding over a period of 5 days according to manufacturer’s instructions. Briefly, plates were removed from the incubator and allowed to equilibrate at room temperature for 30 minutes, and equal volume of CellTiter-Glo reagent (Promega) was added directly to the wells. Plates were incubated at room temperature for 2 minutes on a shaker and left to equilibrate for 10 minutes at 22°C before luminescence was measured on PHERAstar FS microplate reader. Luminescence readings were normalized and presented as relative luminescence units (RLU) to time point 0 (t0). Supplementary Table 14 shows the statistics of 6 replicates for each time point per indicated knockout lines. Doubling time was calculated based on replicate-averaged readings on the linear portion of the proliferation curve (exponential phase) using formula:
Genomic DNA extraction and WGS
Samples were quantified with Biotium Accuclear Ultra high sensitivity dsDNA Quantitative kit using Mosquito LV liquid platform, Bravo WS and BMG FLUOstar Omega plate reader and cherrypicked to 500ng/120µl using Tecan liquid handling platform. Cherrypicked plates were sheared to 450bp using a Covaris LE220 instrument. Post-sheared samples were purified using Agencourt AMPure XP SPRI beads on Agilent Bravo WS. Libraries were constructed (ER, A-tailing and ligation) using ‘Agilent Sureselect kit’ on an Agilent Bravo WS automation system. KapaHiFi Hot start mix and IDT 96 iPCR tag barcodes were used for PCR set-up on Agilent Bravo WS automation system. PCR cycles include 6 standard cycles: 1) Incubate 95C 5 mins; 2) Incubate 98C 30 secs; 3) Incubate 65C 30 secs; 4) Incubate 72C 1 min; 5) Cycle from 2, 5 more times; 6) Incubate 72C 10 mins. Post PCR plate was purified using Agencourt AMPure XP SPRI beads on Beckman BioMek NX96 liquid handling platform. Libraries were quantified with Biotium Accuclear Ultra high sensitivity dsDNA Quantitative kit using Mosquito LV liquid handling platform, Bravo WS and BMG FLUOstar Omega plate reader, then pooled in equimolar amounts on a Beckman BioMek NX-8 liquid handling platform and finally normalized to 2.8 nM ready for cluster generation on a c-BOT and loading on requested Illumina sequencing platform. Pooled samples were loaded on the X10 using 150 PE run length, sequenced to ~25X coverage. The details of sequence coverage for all clones and subclones are provided in Supplementary Table 2.
Alignment and somatic variant-calling
Short reads were aligned to human reference genome GRCh37/hg19 assembly using the BWA-MEM algorithm81. Three algorithms, CaVEMan (http://cancerit.github.io/CaVEMan/)82, Pindel (http://cancerit.github.io/cgpPindel)83 and BRASS (https://github.com/cancerit/BRASS) were used to call somatic substitutions, indels and rearrangements in all subclones, respectively.
Assurance of knockout state using WGS data
First, we examined whether there were CRISPR-Cas9 off-target effects by seeking relevant mutations in other DNA repair genes besides the genes of interest. We also searched for potential off-target sites based on gRNA target sequences using COSMID84 and confirmed that there were no off-target hits in knockouts that generated mutational signatures (Supplementary Table 15). We confirmed chromosome copy number in all subclones remained stable and unchanged from their parent. Second, we confirmed that there are frameshift indels near the gRNA targeted sequence in the genes of interest for all knockout subclones. One UNG knockout was found to be heterozygous and was excluded in the downstream analysis. Third, we checked mislabelled samples by examining the shared mutations between subclones. Subclones originally derived from the same parental knockout clone would share some mutations, in contrast to subclones from different knockouts. Consequently, one ΔPRKDC, one ΔTP53 and two ΔNBN subclones were removed from downstream analysis. Fourth, variant allele fraction (VAF) distribution for each knockout subclone was examined. VAF>=0.4 was used as a cut-off for determination of whether the subclone was derived from a single-cell. When contrasting mutation burden between subclones, we only selected subclones that were derived from single-cells, cultured for 15 days. Shared mutations among subclones were removed to obtain de novo somatic mutations accumulated after knocking out the gene of interest. Supplementary Table 2 summarizes the number of de novo mutations (substitutions and indels) for all subclones.
Determination of gene knockout-associated mutational signatures
An intrinsic background mutagenesis exists in normal cells grown in culture. Knocking out a DNA repair gene that is involved in repairing endogenous DNA damage may result in increased unrepaired DNA damage and thereby result in mutation accumulation with subsequent rounds of replication. Whole-genome sequencing of these knockouts can detect the mutations that occur as a result of being a specified knockout. If mutation burden and mutational profile of a knockout is significantly different from the control subclones which have only the background mutagenesis, it is most likely that there is gene knockout-associated mutagenesis. Based on this principle, our approach to identify gene knockout-associated mutational signature involved three steps: 1) we determined the background mutational signature; 2) we determined the difference between the mutational profile of knockout and background mutation profiles. 3) we removed the background mutation profile from mutation profile of the knockout subclone.
Substitution profiles were described according to the classical convention of 96 channels: the product of 6 types of substitution multiplied by 4 types of 5’ base (A,C,G,T) and 4 types of 3’ base (A,C,G,T). Indel profiles were described by type (insertion, deletion, complex), size (1-bp or longer) and flanking sequence (repeat-mediated, microhomology-mediated or other) of the indel. Here, we used two sets of indel channels. Set one contains 15 channels: 1bp C/T insertion at short repetitive sequence (<5 bp), 1bp C/T insertion at long repetitive sequence (>=5 bp), long insertions (> 1bp) at repetitive sequences, microhomology-mediated insertions, 1bp C/T deletions at short repetitive sequence (<5 bp), 1bp C/T deletions at long repetitive sequence (>=5 bp), long deletions (> 1bp) at repetitive sequences, microhomology-mediated deletions, other deletion and complex indels (Extended Data Fig. 5a). Set two contains 45 channels, in which the 1 bp C/T indels at repetitive sequences are further expanded according to the exact length of the repetitive sequences (Fig. 3b). Indel channel set one was applied to all knockout subclones, whilst channel set two was only applied to four MMR gene knockouts (ΔMLH1, ΔPMS2, ΔMSH2, ΔMSH6) to obtain a higher resolution of mutational signatures of MMR gene knockouts.
Identifying background signatures
The mutational profile of control subclones were used to determine background mutagenesis. Aggregated substitution profiles of all control subclones (ΔATP2B4) were used as the background substitution mutational signature. Aggregated indel profiles of all subclones containing <= 8 indels were used as the background indel mutational signature.
Distinguishing mutational profiles of control and gene-edited subclone profiles
Signal-to-noise ratio affects mutational signature detection. In this study, ‘noise’ is largely background mutagenesis. The averaged mutation burden caused by the background mutagenesis in control cells for substitution and indels are around 150 and 10, with standard deviation of 10 and 1.4, respectively. ‘Signal’ represents the elevated mutation burden caused by gene knockouts. The averaged mutation burden in knockouts range from 63 to 2360 for substitution, and 0 to 2122 for indels after 15 days in culture, as shown in Supplementary Table 2.
The costs associated with whole genome sequencing is prohibitive, thus we have 2-4 subclones per knockout. The intrinsic fluctuation of detected mutation burden in each sample and the limited subclone numbers impose a greater uncertainty in mutational signature detection. Thus, to distinguish high-confidence mutational signatures from noise, we employed three different methods.
First, we evaluated the similarity of mutational profile between control and each gene knockout. According to the mutational profile of control subclones, , for a given number of mutations N (0 < N < 10000), one could generate L bootstrapped samples:
| (1) |
where . One can calculate the cosine similarities (sl) between bootstrapped control samples (ml) and experimentally-obtained control profile (P control) to obtain a distribution of cosine similarities P(S):
| (2) |
We can then calculate the cosine similarity (S knockout) between control profile (pcontrol) and knockout profile (pknockout). As shown in Figs. 1c and 1d, when the mutation count is low, the bootstrapped samples are less similar to the actual control profile than the bootstrapped samples with higher mutation count. Comparing S knockout and P(S) at a given mutation number, N knockout, one could identify which gene knockouts having distinct mutational profiles from the control (p value of S knockout is less than 0.01 in P(S)).
Second, we used contrastive principal component analysis (cPCA)21, which efficiently identified directions that were enriched in the knockouts relative to the background through eliminating confounding variations present in both (Extended Data Fig. 3a), to recognize gene knockout-specific patterns from background signature.
Third, we used t-Distributed stochastic neighbor embedding (t-SNE)22, which is a visualization technique for viewing pairwise similarity data resulting from nonlinear dimensionality reduction based on probability distributions. In t-SNE implementation, mutational profiles that are similar to each other were plotted nearby each other, whereas profiles that are dissimilar are plotted distantly in a 2D space (Extended Data Fig. 3b).
Subtraction of the background mutational signature from knockout mutation profile
The experiment-associated mutational signature can then be obtained by subtracting the background mutational signature from the mutational profile of treated subclones through quantile analysis. First, one can generate a set of bootstrap samples of each treated subclone in order to determine the distribution of mutation number for each channel. According to the distribution, the upper and lower boundaries (e.g., 99% CI) for each channel can be identified. Then, based on the background mutational signature and averaged mutation burden (as initial value), one can construct bootstrapped background profiles, and subtract it from the centroid of bootstrap subclone samples. Due to data noise, some channels may have negative values, in which case, the negative values are set to zero. Occasionally, the number of mutations in a few channels will fall outside the lower boundary after removing the background profile. To avoid negative values, the background mutation pattern is maintained but burden is scaled down through an automated iterative process.
Topography analysis of signatures
Strand bias
Reference information of replicative strands and replication-timing regions were obtained from Repli-seq data of the ENCODE project (https://www.encodeproject.org/)85. The transcriptional strand coordinates were inferred from the known footprints and transcriptional direction of protein coding genes. First, for a given mutational signature, one could calculate the ‘expected’ ratio of mutations between transcribed and non-transcribed strand, or between lagging and leading strands, according to the distribution of trinucleotide sequence context in these regions. Second, the ‘observed’ ratio of mutations between different strands can be identified through mapping mutations to the genomic coordinates of all gene footprints (for transcription) or leading/lagging regions (for replication). Third, all mutations were orientated towards pyrimidines as the mutated base (as this has become the convention in the field). This helped denote which strand the mutation was on. Fourth, the level of asymmetry between different strands was measured by calculating the odds ratio of mutations occurring on one strand (e.g., transcribed or leading strand) vs. on the other strand (e.g., non-transcribed or lagging strand). IntersectBed86 was used to identify mutations overlapping certain genomic features.
MMRDetect algorithm
We trained a mismatch repair (MMR) deficiency logistic regression-based classifier, called MMRDetect, based on mutational signatures obtained from the experimental work. We obtained mutation data from 336 WGS colorectal cancers with accompanying immunohistochemistry (IHC) staining of the four MMR proteins (MSH2, MSH6, MLH1 and PMS2) from UK100,000 Genomes Project (UK100kGP). Within this cohort of 336 colorectal cancers, there were 79 (24%) cancers with abnormal IHC staining indicative of MMR deficiency. 336 cancers were randomly divided into a training set and a test set by using the R function sample(). The training set had 180 MMR-proficient and 56 MMR-deficient samples. The test data set had 77 MMR-proficient and 23 MMR-deficient samples (Supplementary Table 5). Based on the experimental data, we investigated four potential predictor variables in MMRDetect (Extended Data Fig. 10):
-
1)
The sum of exposures of MMR mutational signatures. We fitted tissue-specific substitution signatures to each tumor using an R package (signature.tools.lib) published by Degasperi et al 24.
-
2)
The maximum cosine similarities between the substitution profiles of cancer samples and those of MMR gene knockouts. For each cancer sample, we calculated the cosine similarity between the substitution profile and substitution signatures of the four MMR gene knockouts. The maximum value was used in fitting the model.
-
3)
The number of repeat-mediated indels. We examined the sequence context of each indel. Only the indels occurring at repetitive regions were used.
-
4)
The cosine similarities between the profiles of repeat-mediated deletions of cancer samples and those of MMR gene knockouts. For each cancer sample, we calculated the cosine similarity between the repeat-mediated deletion profile and those of the four MMR gene knockouts. The mean value was used for fitting the model.
The values of different variables were transformed to between 0 and 1 using formula x’ = x/max (x) for comparability. Supplementary Table 5 shows calculated parameters of 336 tumors for MSIseq and MMRDetect. The logistic regression algorithm (function glm()) provided in R package glmnet was employed as the framework of MMRDetect. Supplementary Table 6 provides the weight (coefficients) of the four variables obtained from training the model using the training data set. A ten-fold cross validation was performed for the training data to evaluate the stability of the weights (Extended Data Fig. 10f).
Additional four datasets were used to compare the performance of MMRDetect and MSIseq:
Data analysis
All statistical analysis were performed in R87. P values were calculated using two-sided Mann-Whitney test, wilcox.test() in R. The enrichment of mutations on specific trinucleotide sequences was assessed by calculating the odds ratio (OR) between observed ratio and expected ratio. The 95% confidence interval (CI) was calculated to estimate the precision of the OR. All plots were generated by ggplot288.
Statistics and reproducibility
No statistical method was used to predetermine sample size. We produced two genotypes for each gene, and two subclones for each genotype. Four subclones were obtained for most of genes, except for ΔEXO1 and ΔMSH2. Off-target gene knockouts and/or mislabelled samples could cause erroneous results by reporting a signature incorrectly, thus, to reduce the likelihood of errors, we excluded these samples. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. This was however a systematic experimental study performed with identical conditions across all knockouts and thus all sequencing data generated afterwards was agnostic and fully comparable to one another.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
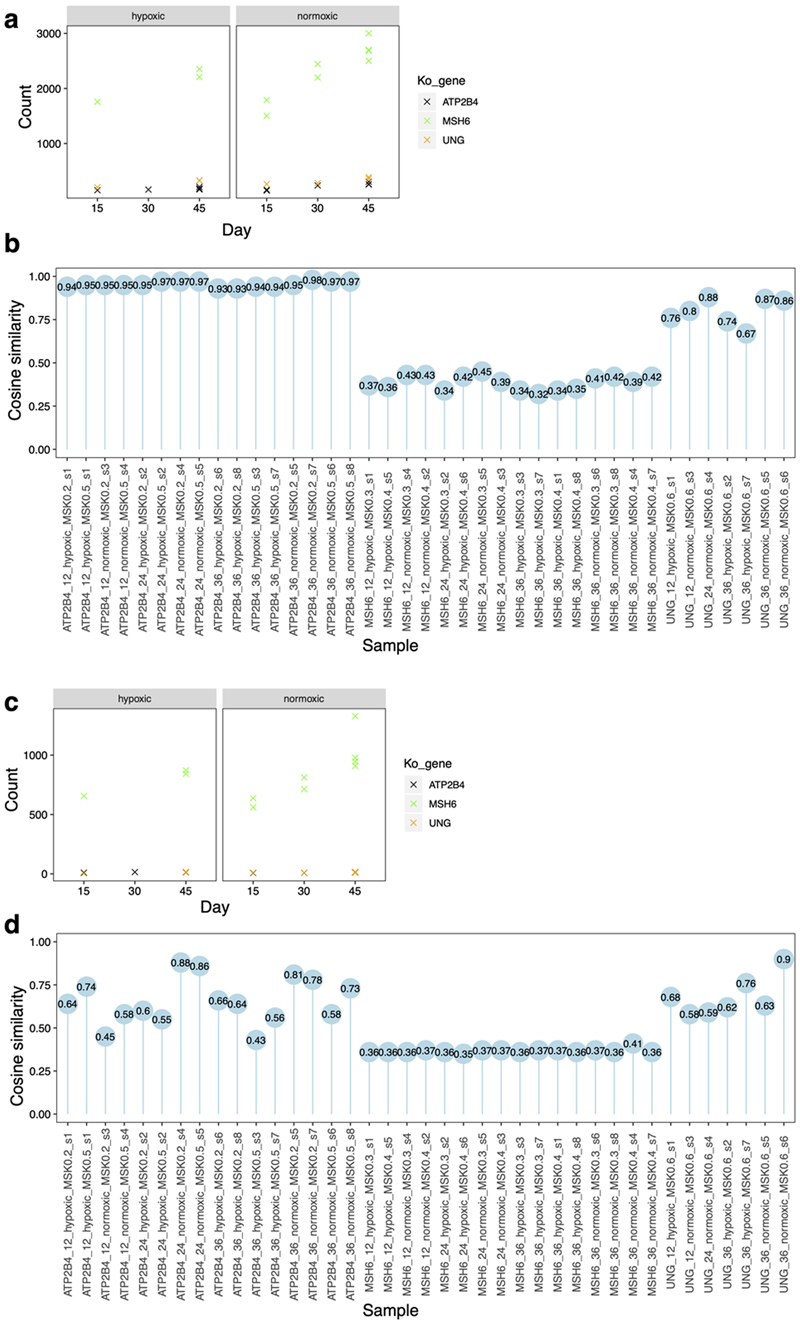
Extended Data Fig. 1. Results of pilot study.
Three genes were selected for knockout (&#Δ): MSH6, UNG and ATP2B4 (negative control). Two genotypes per gene were obtained and grown in culture to gauge reproducibility of signatures between different genotypes of a gene-knockout. These lines were cultured under normoxic (20%) and hypoxic (3%) states, for defined culture times of ~15, 30 or 45 days. Two single-cell subclones were derived for whole genome sequencing for each parental line (equivalent to four subclones per gene edit). One of the UNG genotypes appeared to be heterozygous, which was excluded in downstream analysis. (a) Substitution burden for knockouts of ATP2B4, UNG and MSH6 under hypoxic and normoxic conditions as well as different culturing time. (b) The cosine similarities between the mutational profile of each subclone and background signature of culture. (c) Indel burden for knockouts of ATP2B4, UNG and MSH6 under hypoxic and normoxic conditions as well as different culturing time. (d) The cosine similarities between the mutational profile of each subclone with background signature of culture. Overall, the differences between normoxic and hypoxic conditions were not marked, although normoxic conditions produced slightly more mutations. Time in culture made only a marginal, non-linear difference to burden of mutagenesis. Given the results of the pilot, weighing up the costs and risks associated with prolonged culture time (risk of infection, risk of selection, marked increase in cost of experimental reagents) with the minimal return in terms of mutation number, and also intending to minimize transitions between hypoxic to normoxic conditions while handling cell cultures, we opted to proceed with the full-scale study under normoxic conditions and for 15 days for the rest of study.
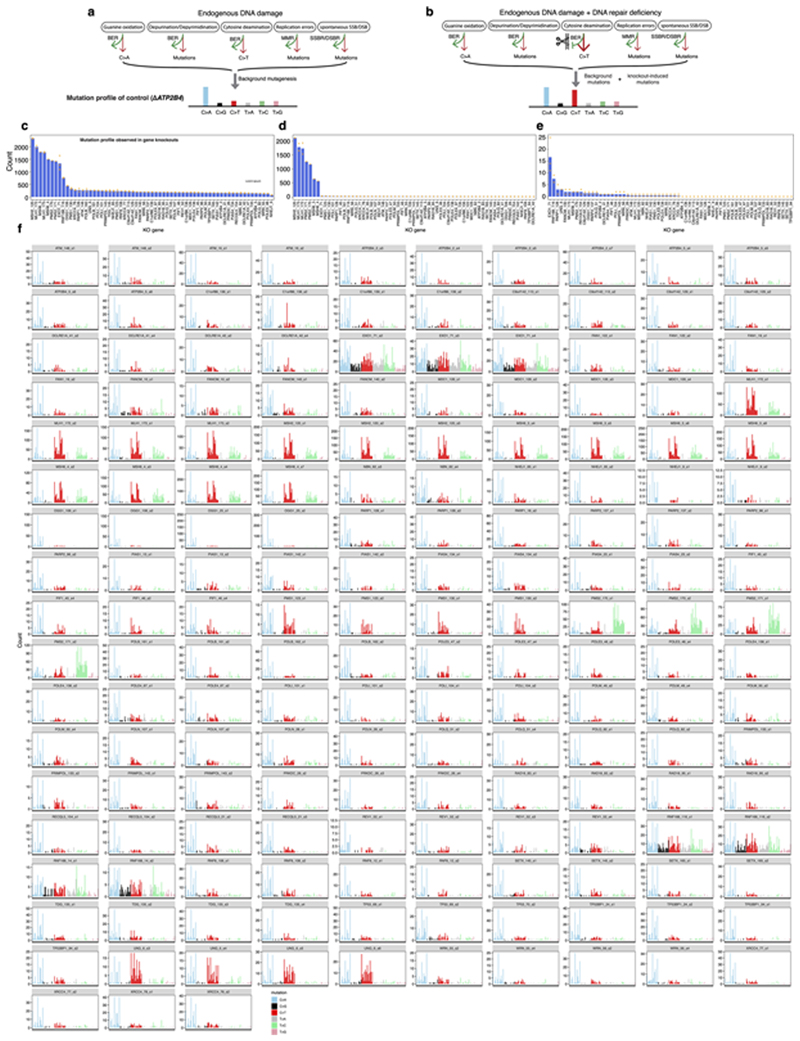
Extended Data Fig. 2. Detecting mutational consequences of knockouts in the absence of added external DNA damage.
(a)(b) Schematic illustration of potential components of background signature (a) and Possible mutational consequences of the DNA repair gene knockouts for proteins that are critical mitigators of mutagenesis (b). (c)-(e) Mutation burden of whole-genome-sequenced subclones of gene knockouts. (c) Substitution, (d) indel and (e) double substitution. Bars represent the mean. Individual data points are shown in orange dots. In all comparative analyses, all gene knockouts were cultured for 15 days and only daughter subclones that were fully clonal (i.e., clearly derived from a single cell) were included. N = 2~4, which is the number of clonal knockout subclones cultured under normoxic condition for 15 days (see Supplementary Table 2). (f) 96-channel substitution mutation profiles of 173 gene knockout subclones.
Extended Data Fig. 3. Results of contrastive principal component analysis and t-SNE.
(a) Contrastive principal component analysis (cPCA) was employed to discriminate knockout profiles from control profiles (&#ΔATP2B4). Each figure contains six different genes. Nine gene knockouts separate from the controls. Using this method, &#ΔADH5 did not separate clearly from &#ΔATP2B4, indicative of either having no signature or a weak signature. Dot colours indicate the repair/replicative pathway that each gene is involved: in black - control; green - MMR; orange – BER; dark purple – HR and HR regulation; light purple - checkpoint. Each dot represents a subclone. The number of subclones for each gene knockout (N = 2~4) can be found in Supplementary Table 2. (b) The t-SNE algorithm was applied to discriminate the mutational profiles of gene knockouts from those of control knockouts. Gene knockouts that produce mutational signatures separate clearly from control subclones and other knockouts which do not have signatures. Subclones of the gene knockouts which produce signatures are clustered together, indicating consistency between subclones.
Extended Data Fig. 4. Oxidative damage-associated mutational signatures.
(a) Relative mutation frequency of G>T/C>A in 256 possible channels which take two adjacent bases 5’ and 3’ of each mutated base (4×4×4×4=256) for &#ΔATP2B4, &#ΔOGG1, a head and neck cancer with strong Signature 18 and COSMIC Signature 18. (b) Left: tSNE plot of tissue-specific mutational signature 18. Two groups are featured with predominant peaks at TGC>TTC/GCA>GAA (highlighted in green) and AGA>ATA/TCT>TAT (highlighted in purple), respectively. Right: heatmap of 21 tissue-specific mutational signatures at C>A. We compared experimental signatures to previously published cancer-derived signatures, focusing on 21 tissue-specific variations of Signature 18. Interestingly, we found two distinct groups of Signature 18. Signatures of &#Δ OGG1, cellular models and signatures derived from head and neck tumors, pancreas, myeloid, bladder, uterus, cervix, lymphoid tumors were most similar to each other, with the predominant G>T/C>A peak at TGC>TTC/GCA>GAA. By contrast, an alternative version of this signature with a predominant G>T/C>A peak at AGA>ATA/TCT>TAT was noted in colorectal, esophagus, stomach, bone, lung, CNS, breast, skin, prostate, liver, head and neck tumors (Signature Head_neck_G), ovary, biliary and kidney cancers. Indeed, there are many types of oxidative species which could fluctuate between tissues, variably affecting trinucleotides resulting in the variation observed in Signature 18.
Extended Data Fig. 5. Indel signatures and double substitution signatures.
(a) 15-channel Indel signatures. (b) 186-channel Indel signatures. (c) Aggregated double substitution profile of &#ΔRNF168 and &#ΔEXO1.
Extended Data Fig. 6. Similarities between &#ΔEXO1, &#ΔRNF168 signatures and Signature 5 and results of analysis on transcriptional strand bias and distribution of mutations on replication timing domains.
(a) Hierarchical clustering of cancer-derived reference signatures with &#ΔEXO1 and &#ΔRNF168 signatures. (b) Hierarchical clustering of tissue-specific signature 5 with &#ΔEXO1 and &#ΔRNF168 signatures. (c) Transcriptional strand bias in 9 gene knockouts. Pearson's Chi-Squared test (chisq.test()) was used to calculate the p-value. P-value was corrected using p.adjust(). Unlike mutational signatures of environmental mutagens, we do not observe striking transcriptional strand bias in signatures generated by DNA repair gene knockouts, except for T>C generated by &#ΔEXO1 and &#ΔRNF168. Since transcriptional strand bias is largely induced by NER repairing DNA bulky adducts, lack of it indicates that most of the endogenous DNA damage is not particularly bulky or DNA-deforming. (d) Distribution of mutation density across replication timing domains (separated into deciles) for signatures associated with different gene knockouts. Green bars indicate observed distribution. Blue lines indicate expected distribution with correction of trinucleotide density of each domain. Bars and error bars represent mean ± SD of bootstrapping replicates (n=100).
Extended Data Fig. 7. Putative outcomes of all possible base-base mismatches.
Outcomes from 12 possible base-base mismatches. The red and black strands represent lagging and leading strands, respectively. The arrowed strand is the nascent strand. The highlighted pathways are the ones that generate C>A (blue), C>T (red) and T>C mutations (green) in the &#ΔMSH2 mutational signature.
Extended Data Fig. 8. Distribution of G>T/C>A mutations in polyG tracts of &#ΔMSH2, &#ΔMSH6 and &#ΔMLH1.
(a) Relative frequency of occurrence of G>T/C>A in polyG tracts. (b) Occurrence of G>T/C>A in polyG tracts.
Extended Data Fig. 9. Gene-specific mutational signatures in MMR-deficiency.
Proportion of different mutation types of substitution (a) and indel (b) signatures for 4 MMR gene knockouts. (c) The ratio of substitution and indel burden. (d) Schematic interpretation of the relative mutation burdens of &#ΔMSH2 and &#ΔMSH6.
Extended Data Fig. 10. Development of MMRDetect.
(a)-(e) Distribution of the five parameters across IHC-determined MMR gene abnormal (orange) and MMR gene normal (green) samples. black dots and error bars represent mean ± SD of the paramenters. N Abnormal = 79 samples (yellow); N Normal = 257 samples (green). (a) Exposure of MMR signatures. (b) Cosine similarity between the substitution profile of cancer samples and that of MMR gene knockouts. (c) Number of indels in repetitive regions. (d) Cosine similarity between the profile of repeat-mediated deletions of cancer sample and that of knockout generated indel signatures, (e) the cosine similarity between the profile of repeat-mediated insertion of cancer sample and that of knockout generated indel signatures. P-values were calculated through two-sided Mann-Whitney test. (f) Distribution of coefficients from 10-fold cross validation using training data set. Box plots denote median (horizontal line) and 25th to 75th percentiles (boxes). The lower and upper whiskers extend to 1.5× the inter-quartile range. N = 10 iterations. (g) MMRDetect-calculated probabilities for 336 colorectal cancers. With cut-off of 0.7, 77 out of 336 were predicted to be MMR-deficient samples (probability < 0.7). Colour bars represent the MSI status determined by IHC staining: red – abnormal; blue – normal. Four samples with abnormal IHC staining have probabilities > 0.7, whilst 2 samples with normal IHC staining have probabilities < 0.7. The 4 samples were revealed to be false positive cases and the 2 samples were false negative ones for IHC staining through validation using MSIseq and seeking coding mutations in MMR genes. (h) Distribution of the mutation number of repeat-mediated indels, MMR-deficiency signatures and non-MMR-deficiency signatures across four groups of samples: MMR-deficient samples determined by only MMRDetect (yellow), MMR-deficient samples determined by only MSIseq (purple), MMR-deficient samples determined by both MMRDetect and MSIseq (blue) and non-MMR-deficient samples determined by both MMRDetect and MSIseq (pink). P-values were calculated through two-sided Mann-Whitney test. Numbers of MMR-deficient samples determined by MMRDetect only (blue), MSIseq only (pink), both (yellow) and none (purple) are 34, 20, 587 and 6718, respectively.
Supplementary Material
Acknowledgments
We thank the Wellcome Sanger Institute Cellular Genetics and Phenotyping facility for assistance, the CASM IT team, Julia Foreman and Grace Ping for assistance in carrying out and completing this project. We thank the COMSIG Consortium spearheaded by Professor Steve Jackson. In Cambridge, this work was funded by Cancer Research UK (CRUK) Advanced Clinician Scientist Award (C60100/A23916), Dr Josef Steiner Cancer Research Award 2019, Medical Research Council (MRC) Grant-in-Aid to the MRC Cancer unit, CRUK Pioneer Award, Wellcome Strategic Award (WT101126), and Wellcome Sanger Institute faculty funding and supported by the NIHR Cambridge BRC and UK Regenerative Medicine Platform (MR/R015724/1). The work of T.I.R and J.S.C was funded by the CRUK Centre grant with reference number C309/A25144.
Support for the MMRDetect classifier was enabled by access to data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care) funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
This publication and the underlying research are facilitated by data that were generated by the Hartwig Medical Foundation (HMF) and the Center for Personalized Cancer Treatment (CPCT) in the Netherlands, and the International Cancer Genome Consortium.
Footnotes
Author Contributions Statement
Conceptualization: SNZ, BS; Writing: SNZ, XZ, GK, JJ, BS; Clinical sample collections: LS, GB, VPA, DR, SNZ; Laboratory work: GK, KU, TIR, CAA, WB, CG; Data curation/Formal analysis: XZ, GK, ASN, AD, TIR, JSC, SNZ; Administration: RH, WB, JY.
Competing Interests Statement
SNZ holds patents on clinical algorithms of mutational signatures and during the completion of this project, served advisory roles for Astra Zeneca, Artios Pharma Ltd and Scottish Genome Project.
Contributor Information
Genomics England Research Consortium:
John C. Ambrose, Prabhu Arumugam, Emma L. Baple, Marta Bleda, Freya Boardman-Pretty, Jeanne M. Boissiere, Christopher R. Boustred, Helen Brittain, Mark J. Caulfield, Georgia C. Chan, Clare E. H. Craig, Louise C. Daugherty, Anna de Burca, Andrew Devereau, Greg Elgar, Rebecca E. Foulger, Tom Fowler, Pedro Furió-Tarí, Adam Giess, Joanne M. Hackett, Dina Halai, Angela Hamblin, Shirley Henderson, James E. Holman, Tim J. P. Hubbard, Kristina ibáñez, Rob Jackson, Louise J. Jones, Dalia Kasperaviciute, Melis Kayikci, Athanasios Kousathanas, Lea Lahnstein, Kay Lawson, Sarah E. A. Leigh, Ivonne U. S. Leong, Javier F. Lopez, Fiona Maleady-Crowe, Joanne Mason, Ellen M. McDonagh, Loukas Moutsianas, Michael Mueller, Nirupa Murugaesu, Anna C. Need, Pter O’Donovan, Chris A. Odhams, Andrea Orioli, Christine Patch, Mariana Buongermino Pereira, Daniel Perez-Gil, Dimitris Polychronopoulos, John Pullinger, Tahrima Rahim, Augusto Rendon, Pablo Riesgo-Ferreiro, Tim Rogers, Mina Ryten, Kevin Savage, Kushmita Sawant, Richard H. Scott, Afshan Siddiq, Alexander Sieghart, Damian Smedley, Katherine R. Smith, Samuel C. Smith, Alona Sosinsky, William Spooner, Helen E. Stevens, Alexander Stuckey, Razvan Sultana, Mélanie Tanguy, Ellen R. A. Thomas, Simon R. Thompson, Carolyn Tregidgo, Arianna Tucci, Emma Walsh, Sarah A. Watters, Matthew J. Welland, Eleanor Williams, Katarzyna Witkowska, Suzanne M. Wood, and Magdalena Zarowiecki
Data availability
Raw sequence files are deposited at the European Genome-phenome Archive with accession numbers EGAS00001000800 and EGAS00001000874. Mutation calls have been deposited at Mendeley: http://dx.doi.org/10.17632/ymn3ykkmyx.3. hiPSCs can be obtained directly from the authors. The curated data is available for general browsing from our reference Mutational Signature website, SIGNAL (https://signal.mutationalsignatures.com). The age information of the human patient samples is not publicly available as this information could compromise privacy and lead to identification of the individuals.
Publicly available genomic datasets reanalyzed here to compare the performance of MMRDetect and MSIseq are available at European Genome-phenome Archive (EGAS0000100117872), http://dcc.icgc.org/pcawg/ 73, https://data.mendeley.com/datasets/2mn4ctdpxp/1 74, https://resources.hartwigmedicalfoundation.nl/ 75 and the Genomics England Research Environment (main programme v8) via https://re.extge.co.uk/ovd/.
Source data are available for this study. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
Code can be obtained here: https://github.com/Nik-Zainal-Group/COMSIG_KO.git
References
- 1.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haradhvala NJ, et al. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nature Communications. 2018;9:1746. doi: 10.1038/s41467-018-04002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov LB, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nature Genetics. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nik-Zainal S, et al. The genome as a record of environmental exposure. Mutagenesis. 2015;30:763–770. doi: 10.1093/mutage/gev073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou X, et al. Validating the concept of mutational signatures with isogenic cell models. Nature Communications. 2018;9:1744. doi: 10.1038/s41467-018-04052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen S, et al. 5-Fluorouracil treatment induces characteristic T>G mutations in human cancer. Nature Communications. 2019;10:4571. doi: 10.1038/s41467-019-12594-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucab JE, et al. A Compendium of Mutational Signatures of Environmental Agents. Cell. 2019;177:821–836.e16. doi: 10.1016/j.cell.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–8. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 13.Mardis ER. The Impact of Next-Generation Sequencing on Cancer Genomics: From Discovery to Clinic. Cold Spring Harbor Perspectives in Medicine. 2019;9 doi: 10.1101/cshperspect.a036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nature Reviews Clinical Oncology. 2018;15:353–365. doi: 10.1038/s41571-018-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunkel TA, Erie DA. DNA MISMATCH REPAIR. Annual Review of Biochemistry. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 17.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA Repair Genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 19.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends in Cell Biology. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 21.Abid A, Zhang MJ, Bagaria VK, Zou J. Exploring patterns enriched in a dataset with contrastive principal component analysis. Nature Communications. 2018;9:2134. doi: 10.1038/s41467-018-04608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Maaten L, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 23.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Degasperi A, et al. A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies. Nature Cancer. 2020;1:249–263. doi: 10.1038/s43018-020-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilati C, et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. The Journal of Pathology. 2017;242:10–15. doi: 10.1002/path.4880. [DOI] [PubMed] [Google Scholar]
- 26.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:8010–5. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruner SD, Norman DPG, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 28.Lee YA, Durandin A, Dedon PC, Geacintov NE, Shafirovich V. Oxidation of guanine in G, GG, and GGG sequence contexts by aromatic pyrenyl radical cations and carbonate radical anions: relationship between kinetics and distribution of alkali-labile lesions. The journal of physical chemistry B. 2008;112:1834–1844. doi: 10.1021/jp076777x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugiyama H, Saito I. Theoretical Studies of GG-Specific Photocleavage of DNA via Electron Transfer: Significant Lowering of Ionization Potential and 5‘-Localization of HOMO of Stacked GG Bases in B-Form DNA. Journal of the American Chemical Society. 1996;118:7063–7068. [Google Scholar]
- 30.Allgayer J, Kitsera N, von der Lippen C, Epe B, Khobta A. Modulation of base excision repair of 8-oxoguanine by the nucleotide sequence. Nucleic Acids Research. 2013;41:8559–8571. doi: 10.1093/nar/gkt620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee A, Verdine GL. A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex. Proceedings of the National Academy of Sciences. 2006;103:15020–15025. doi: 10.1073/pnas.0603644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman JI, Stivers JT. Detection of Damaged DNA Bases by DNA Glycosylase Enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat Res. 1999;437:11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 35.Shen JC, Rideout WM, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–6. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waters TR, Swann PF. Thymine-DNA glycosylase and G to A transition mutations at CpG sites. Mutat Res. 2000;462:137–47. doi: 10.1016/s1383-5742(00)00031-4. [DOI] [PubMed] [Google Scholar]
- 37.Sanders MA, et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood. 2018;132:1526–1534. doi: 10.1182/blood-2018-05-852566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes DE, Lindahl T. Repair and Genetic Consequences of Endogenous DNA Base Damage in Mammalian Cells. Annual Review of Genetics. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 39.Mol CD, et al. Crystal structure and mutational analysis of human uracil-DNA glycosylase: Structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 40.Grolleman JE, et al. Mutational Signature Analysis Reveals NTHL1 Deficiency to Cause a Multi-tumor Phenotype. Cancer Cell. 2019;35:256–266.e5. doi: 10.1016/j.ccell.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Genschel J, Modrich P. Mechanism of 5′-Directed Excision in Human Mismatch Repair. Molecular Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 42.Bolderson E, et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Research. 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattiroli F, et al. RNF168 biquitinates K13-15 on H2A/H2AX to Drive DNA Damage Signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Bohgaki M, et al. RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks. Proceedings of the National Academy of Sciences. 2013;110:20982. doi: 10.1073/pnas.1320302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 46.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 47.Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSβbound to unpaired DNA loops. Nat Struct Mol Biol. 2012;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palombo F, et al. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 49.Warren JJ, et al. Structure of the Human MutSα \DNA\ Lesion Recognition Complex. Molecular Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 50.Andrianova MA, Bazykin GA, Nikolaev SI, Seplyarskiy VB. Human mismatch repair system balances mutation rates between strands by removing more mismatches from the lagging strand. Genome research. 2017;27:1336–1343. doi: 10.1101/gr.219915.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lujan SA, et al. Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity. PLOS Genetics. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morganella S, et al. The topography of mutational processes in breast cancer genomes. Nat Commun. 2016;7 doi: 10.1038/ncomms11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aboul-ela F, Koh D, Tinoco I, Jr, Martin FH. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T) Nucleic acids research. 1985;13:4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazurek A, Berardini M, Fishel R. Activation of Human MutS Homologs by 8-Oxo-guanine DNA Damage. Journal of Biological Chemistry. 2002;277:8260–8266. doi: 10.1074/jbc.M111269200. [DOI] [PubMed] [Google Scholar]
- 55.Morikawa M, et al. Analysis of guanine oxidation products in double-stranded DNA and proposed guanine oxidation pathways in single-stranded, double-stranded or quadruplex DNA. Biomolecules. 2014;4:140–159. doi: 10.3390/biom4010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pavlov YI, Newlon CS, Kunkel TA. Yeast Origins Establish a Strand Bias for Replicational Mutagenesis. Molecular Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- 57.Mudrak SV, Welz-Voegele C, Jinks-Robertson S. The Polymerase η Translesion Synthesis DNA Polymerase Acts Independently of the Mismatch Repair System To Limit Mutagenesis Caused by 7,8-Dihydro-8-Oxoguanine in Yeast. Molecular and Cellular Biology. 2009;29:5316. doi: 10.1128/MCB.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meier B, et al. Mutational signatures of DNA mismatch repair deficiency in C. elegans and human cancers. Genome Research. 2018;28:666–675. doi: 10.1101/gr.226845.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lang GI, Parsons L, Gammie AE. Mutation Rates, Spectra, and Genome-Wide Distribution of Spontaneous Mutations in Mismatch Repair Deficient Yeast. G3: Genes, Genomes, Genetics. 2013;3:1453. doi: 10.1534/g3.113.006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 61.Palombo F, et al. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Current Biology. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 62.Wind Nd, et al. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nature Genetics. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- 63.Poulogiannis G, Frayling IM, Arends MJ. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology. 2010;56:167–179. doi: 10.1111/j.1365-2559.2009.03392.x. [DOI] [PubMed] [Google Scholar]
- 64.Heinen CD. Mismatch repair defects and Lynch syndrome: The role of the basic scientist in the battle against cancer. DNA Repair. 2016;38:127–134. doi: 10.1016/j.dnarep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agu Chukwuma A, et al. Successful Generation of Human Induced Pluripotent Stem Cell Lines from Blood Samples Held at Room Temperature for up to 48 hr. Stem Cell Reports. 2015;5:660–671. doi: 10.1016/j.stemcr.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ni Huang M, et al. MSIseq: Software for Assessing Microsatellite Instability from Catalogs of Somatic Mutations. Scientific Reports. 2015;5:13321. doi: 10.1038/srep13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niu B, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2013;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C, Liang C. MSIpred: a python package for tumor microsatellite instability classification from tumor mutation annotation data using a support vector machine. Scientific Reports. 2018;8:17546. doi: 10.1038/s41598-018-35682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cortes-Ciriano I, Lee S, Park W-Y, Kim T-M, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nature Communications. 2017;8:15180. doi: 10.1038/ncomms15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite Instability Detection by Next Generation Sequencing. Clinical Chemistry. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 71.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nature Medicine. 2016;22:1342. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 72.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell PJ, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staaf J, et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nature Medicine. 2019;25:1526–1533. doi: 10.1038/s41591-019-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Priestley P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujimoto A, et al. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Research. 2020;30:334–346. doi: 10.1101/gr.255026.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Campbell BB, et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell. 2017;171:1042–1056.e10. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bressan RB, et al. Efficient CRISPR/Cas9-assisted gene targeting enables rapid and precise genetic manipulation of mammalian neural stem cells. Development. 2017;144:635. doi: 10.1242/dev.140855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tate PH, Skarnes WC. Bi-allelic gene targeting in mouse embryonic stem cells. Methods. 2011;53:331–8. doi: 10.1016/j.ymeth.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hodgkins A, et al. WGE: a CRISPR database for genome engineering. Bioinformatics. 2015;31:3078–80. doi: 10.1093/bioinformatics/btv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013:1303–3997. [Google Scholar]
- 82.Jones D, et al. cgpCaVEManWrapper: Simple execution of CaVEMan in order to detect somatic single nucleotide variants in NGS data. Current protocols in bioinformatics. 2016;56 doi: 10.1002/cpbi.20. 15.10.1-15.10.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raine KM, et al. cgpPindel: Identifying Somatically Acquired Insertion and Deletion Events from Paired End Sequencing. Current protocols in bioinformatics. 2015;52 doi: 10.1002/0471250953.bi1507s52. 15.7.1-15.7.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Molecular therapy. Nucleic acids. 2014;3:e214–e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.The Encode Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. [Google Scholar]
- 88.Wickham H. ggplot2: elegant graphics for data analysis. Springer New York: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence files are deposited at the European Genome-phenome Archive with accession numbers EGAS00001000800 and EGAS00001000874. Mutation calls have been deposited at Mendeley: http://dx.doi.org/10.17632/ymn3ykkmyx.3. hiPSCs can be obtained directly from the authors. The curated data is available for general browsing from our reference Mutational Signature website, SIGNAL (https://signal.mutationalsignatures.com). The age information of the human patient samples is not publicly available as this information could compromise privacy and lead to identification of the individuals.
Publicly available genomic datasets reanalyzed here to compare the performance of MMRDetect and MSIseq are available at European Genome-phenome Archive (EGAS0000100117872), http://dcc.icgc.org/pcawg/ 73, https://data.mendeley.com/datasets/2mn4ctdpxp/1 74, https://resources.hartwigmedicalfoundation.nl/ 75 and the Genomics England Research Environment (main programme v8) via https://re.extge.co.uk/ovd/.
Source data are available for this study. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code can be obtained here: https://github.com/Nik-Zainal-Group/COMSIG_KO.git