Summary
Regulatory T (Treg) cells are vital for the maintenance of immune homeostasis, while their dysfunction constitutes a cardinal feature of autoimmunity. Under steady state conditions, mitochondrial metabolism is critical for Treg function; however, the metabolic adaptations of Tregsduring autoimmunity are ill defined. Herein, we report that elevated mitochondrial oxidative stress and a robust DNA damage response (DDR) associated with cell death occur in Tregs in individuals with autoimmunity. In an experimental autoimmune encephalitis (EAE) mouse model of autoimmunity, we found a Treg dysfunction recapitulating the features of autoimmune Tregs with a prominent mtROS signature. Scavenging of mtROS in Tregs of EAE mice reversed the DDR and prevented Treg cell death, while attenuating the Th1 and Th17 autoimmune responses. These findings highlight an unrecognized role of mitochondrial oxidative stress in defining Treg cell fate during autoimmunity, which may facilitate the design of novel immunotherapies for diseases with disturbed immune tolerance.
Keywords: autoimmunity, regulatory T cell, metabolism, mitochondrial oxidative stress, DNA damage response, lysosome
Introduction
Regulatory T cell (Treg) deficiency caused by mutations in the Foxp3 gene, the exclusive transcription factor of the Treg cell lineage, results in the life-threatening immune dysfunction polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome, characterized by multi-organ autoimmune inflammation. Furthermore, Foxp3-depleted animals, which lack Treg cells from their periphery, succumb to systemic autoimmunity (Fontenot et al., 2003; Khattri et al., 2003; Sakaguchi et al., 1985). This connection between Treg cell function and disease indicates the vital role Treg cells have in the maintenance of self-tolerance and immune homeostasis (Bennett et al., 2001; Wildin et al., 2001). Consistently, Treg cell dysfunction is a common denominator in autoimmunity. To this end, reduced Treg cell numbers and compromised function have been demonstrated in a wide range of autoimmune diseases, including multiple sclerosis (MS), systemic lupus erythematosus (SLE), type 1 diabetes (T1D), thyroiditis and inflammatory bowel disease (IBD) (Carbone et al., 2014; Dominguez-Villar and Hafler, 2018; Goschl et al., 2019; Grant et al., 2015; Long and Buckner, 2011; Viglietta et al., 2004). Nevertheless, experimental manipulation of Treg cells has profound effects on the incidence, onset and severity of autoimmune manifestations in animal models. As for the clinic, various trials are under way examining the feasibility and efficacy of Treg cell infusion or Treg cell expansion for the treatment of autoimmunity (NCT02691247, NCT02772679, NCT01988506) (Rosenzwajg et al., 2019). Hence, Treg cells have an enormous potential in controlling autoimmune reactions and the re-establishment of self-tolerance, even if widely disturbed. However, despite the extensive phenotypic and functional characterization of Treg cells over the last decade, the molecular mechanisms that underpin the Treg cell dysfunction and contraction during the course of an autoimmune response remain elusive.
Among the many cellular and molecular processes involved in Treg cell fitness, their unique metabolic profile has recently gained much attention. Treg cells under steady state conditions or upon in vitro activation exhibit increased mitochondrial metabolism, with the mitochondrial respiratory chain being vital for their T cell-suppression capacity, stability and survival (Angelin et al., 2017; Galgani et al., 2016; Newton et al., 2016). However, whether autoimmune environments induce differential metabolic rewiring and the metabolic processes that dictate Treg cell survival and function during autoimmunity are completely unknown. Besides the acknowledged role in generation of ATP via oxidative phosphorylation (OXPHOS), mitochondria serve diverse activities in immune cells, vital for cell integrity, proliferation and growth. To this end, production of mitochondrial reactive oxygen species (mtROS), release of cytochrome c and mitochondrial DNA, as well as generation of metabolites can initiate signaling cascades affecting gene expression and cell activation, proliferation and differentiation (Mehta et al., 2017; Mills et al., 2017; Rambold and Pearce, 2018; Weinberg et al., 2015). Importantly, de-regulation of mitochondrial function increases intracellular oxidation and stress, perturbs the functional activities of organelles such as endoplasmic reticulum and lysosomes, induces autophagy and mediates cellular damage and death that could ultimately lead to disturbed homeostasis and pathologies (Mills et al., 2017; Rambold and Pearce, 2018). Therefore, the maintenance of mitochondrial network integrity and activity is indispensable for immune cell homeostasis and function. Overall, delineating the molecular events that dictate the Treg cell metabolic aberrancies in autoimmune diseases represents an unmet clinical need, which holds promise for therapeutic manipulation.
Herein guided by transcriptomic analysis, we reveal a metabolic reprogramming of Treg cells in individuals with autoimmune diseases enriched in mitochondrial oxidative stress, a DNA damage response (DDR) and cell death. Then, using the mouse model of multiple sclerosis, we demonstrate compromised mitochondrial function in Foxp3+ Treg cells and an ensuing increase in mitochondrial oxidative stress, which results in an impaired lysosomal function that promotes a DDR and subsequently cell death. Importantly, scavenging of mtROS either chemically or through targeted expression of the antioxidant enzyme catalase to mitochondria of Foxp3+ Treg cells restrained the DDR, reduced apoptosis and diminished autoimmune responses. Collectively, our findings reveal a metabolic reprogramming of Foxp3+ Treg cells during autoimmunity, which is characterized by mitochondrial oxidative stress and dysfunctional lysosomes instructing a cell death program and thus a breakdown of self-tolerance.
Results
Treg cell metabolic reprogramming in individuals with autoimmune diseases
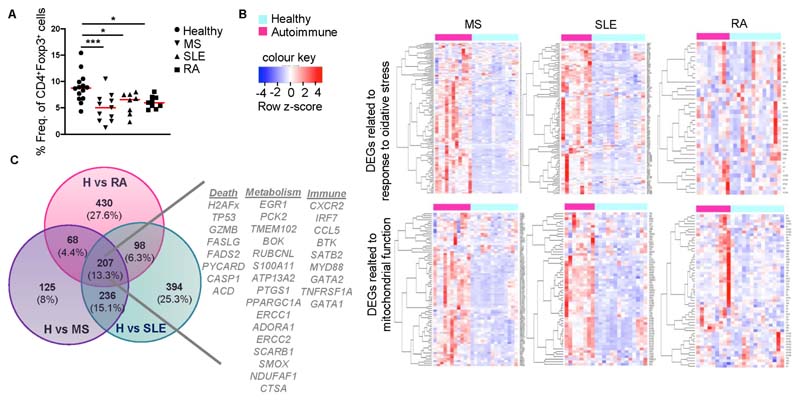
To investigate the metabolic requirements of Treg cells in autoimmune settings we first assessed CD4+CD25+CD127-Foxp3+ Treg cell frequencies in peripheral blood of individuals with multiple sclerosis (MS), rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) (Supplemental Table 1). In line with previous reports (Carbone et al., 2014; Dominguez-Villar and Hafler, 2018; Viglietta et al., 2004), our findings demonstrated a marked reduction of Treg cell frequencies in all three autoimmune conditions compared to healthy individuals (Figure 1A). To dissect the molecular mechanisms underlying Treg cell reduction, isolated CD4+CD25+CD127- Treg cells (Figure S1A, Supplemental Table 1) were subjected to RNA sequencing. Treg cells from individuals with autoimmune diseases (designated hereafter as autoTregs) demonstrated extensive transcriptomic alterations compared to Treg cells from healthy individuals (Figure S1B). Interestingly, gene set enrichment analysis (GSEA), demonstrated a metabolic reprogramming of autoTregs in all groups of autoimmune diseases, highlighted by the presence of oxidative stress, DNA damage response (DDR), mitochondrial dysfunction and cell death related pathways (Figures 1B and S1C). Comparison of these gene sets among the three disease settings revealed 207 (13.3% overlap) common differentially expressed genes (DEGs) in autoTregs (Figure 1C), consisted of a prominent core fingerprint of 33 genes involved in cell death (i.e. TP53, H2AFx, FASLG), metabolism (i.e. TMEM102, ATP13A2, PTGS1) and Treg-mediated immune-related processes (i.e. CXCR2, IRF7, TNFRSF1A) (Figure 1C). These results indicate a robust metabolic reprogramming in Treg cells from individuals with autoimmune diseases, enriched in signatures associated with mitochondrial dysfunction and oxidative stress-induced cell death.
Figure 1. Treg cells isolated from the peripheral blood of individuals with autoimmune diseases are metabolically reprogrammed.
(A) Frequencies of CD4+CD127-CD25+Foxp3+ Treg cells from the peripheral blood of healthy individuals (n = 14) and individuals with autoimmunity: MS (n = 11), SLE (n = 8), RA (n = 9). Results are expressed as mean ± SEM. Statistical significance was obtained by one-way ANOVA. ***P = 0.0008, *P = 0.0242, *P = 0.0196.
(B) Heatmaps of enriched gene sets found during GSEA analysis of healthy vs autoimmune Treg cell samples. Genes from similar enriched processes were pooled and depicted in a single heatmap.
(C) Venn diagram showing the overlap among significant DEGs of Healthy vs MS, Healthy vs RA and Healthy vs SLE comparisons. A signature of 33 selected genes involved in cell death, metabolism and immune related responses is depicted. See also Figure S1 and Supplemental Table 1.
Treg cells during autoimmunity display impaired mitochondrial function and enhanced accumulation of damaged mitochondria
To illuminate the cellular processes dictating Treg metabolic reprogramming in autoimmunity we employed the experimental autoimmune encephalitis (EAE) mouse model, a well-characterized animal model that resembles human MS, and analyzed the Treg cell metabolic properties through assessment of oxygen consumption rate (OCR), as an indicator of mitochondrial oxidative phosphorylation (OXPHOS) status. Thus, CD4+Foxp3+ (Tregs) were isolated from the draining lymph nodes (dLNs) of Mog35-55/CFA-immunized Foxp3-gfp KI mice before disease onset (pre-diseased) and subsequently subjected to Seahorse flux analysis. Treg cells from pre-diseased mice exhibited increased OCR compared to Treg cells isolated from non-immunized, naïve animals (Figure 2A). Notably, despite the enhanced OCR, Treg cells exhibited reduced ATP levels, at the pre-disease stage and during established disease, compared to naïve Treg cells (Figure 2B). The suppressive function of Treg cells is highly dependent on mitochondrial metabolism, with extensive engagement of the mitochondrial respiratory chain (Field et al., 2020; Fu et al., 2019; Newton et al., 2016; Weinberg et al., 2019). Thus, we sought to assess Treg cell mitochondrial function in the course of autoimmune responses. To this end, pre-diseased Treg cells demonstrated reduced mitochondrial membrane potential as assessed by TMRE and JC-1 (Figures 2C and S2A), accompanied by decreased expression of cytochrome c, one of the main proteins that control redox signaling in mitochondrial oxidative phosphorylation (OXPHOS) (Figure 2D), while mitochondrial mass and mitochondrial DNA (mtDNA) content were markedly increased compared to naïve Treg cells (Figures 2E and S2B) or to pre-diseased non Foxp3 CD4+ T cells (Figure S2C). Electron microscopy confirmed the enhanced accumulation of damaged mitochondria with severely aberrant mitochondrial morphology, impaired cristae organization, and loss of mitochondrial electron density in pre-diseased Treg cells (Figure 2F), consistent with mitochondrial dysfunction. In support, GSEA analysis demonstrated that Treg cells from individuals with MS were enriched in “mitochondrial depolarization”, with BOK, ABCD1, ALOX12, IFI6, ATP5IF1 being among the top enriched genes and “mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway”, with TMEM102, TP53, GZMB, HSPA1A, CHCHD10 being among the top enriched genes (Figures 2G-H). Overall, these findings indicate that in autoimmunity-associated Treg cells are characterized by compromised mitochondrial function and increased accumulation of damaged mitochondria.
Figure 2. Treg cells during autoimmune responses experience an impaired mitochondrial function.
(A)Seahorse analysis of oxygen consumption rate (OCR) in CD4+CD25+GITR+ Treg cells isolated from dLNs (inguinal) of naïve or immunized mice 9 d following s.c. Mog35-55/CFA injection. For B-F CD4+Foxp3+ Treg cells were isolated from dLNs (inguinal for pre-diseased or cervical for diseased) of naïve, pre-diseased or EAE induced with disease activity score > 3.5, Foxp3gfp.KI mice.
(B)Intracellular ATP values are depicted for naïve (n = 6), pre-diseased (n = 6) and diseased (n = 5) Foxp3+ Treg cells (****P < 0.0001).
(C) Mitochondrial membrane potential measured by flow cytometry using TMRE. Mean fluorescence intensity (MFI) of TMRE is depicted for naïve (n = 9), pre-diseased (n = 10) and diseased Treg cells (n = 8) (*P = 0.0423, *P = 0.05).
(D) MFI of cytochrome c in naïve or pre-diseased Treg cells. One representative experiment of two is depicted (n = 4 mice per group, *P = 0.0492).
(E) Representative immunofluorescence confocal microscopy images for Mitotracker (red) and DAPI (blue) and Mitotracker puncta/cell, (n = 5 mice per group, *P = 0.0111).
(F) Representative images of Foxp3+ Treg cells using transmission electron microscopy are depicted.
(G) Treg cells were isolated as CD4+CD127-CD25+ cells from peripheral blood of healthy individuals (n = 14) and subjects with MS (n = 11). GSEA plot showing the enrichment of “GO Mitochondrial depolarization” (NES 1.42, FDR 0.23) gene set. The top enriched genes are listed to the right of each plot.
Results are presented as mean ± SEM. Statistical significance was obtained by One-way ANOVA or unpaired Student’s t-test. See also Figure S2
Mitochondrial oxidative stress is a hallmark of Treg cells in autoimmune responses
It is well established that diminished mitochondrial function can result in elevated mitochondrial reactive oxygen species (mtROS) production (Sena and Chandel, 2012). Importantly, increased mtROS production was evident in Treg cells of pre-diseased and diseased animals (Figure 3A) along with significantly enhanced DNA oxidation, based on 8-Oxo-2’-deoxyguanosine (8-OHdG) expression (Figure 3B) compared to naïve Treg cells or pre-diseased non Foxp3 CD4 T cells (Figure S2D). Of interest, 8-OHdG foci co-localized with the translocase of outer membrane 20 (TOM20) (Figure 3B), indicating a mitochondrial-oriented oxidative stress response, mirroring the Treg cell transcriptomic analysis from individuals with autoimmune diseases (Figures 1B, C and S1C). In addition, the activity of the mitochondrial antioxidant enzymes manganese superoxide dismutase (Mn-SOD) and catalase, which play an essential role in mtROS scavenging (Sies and Jones, 2020), was significantly lower in pre-diseased compared to naïve Treg cells (Figure 3C). Notably, GSEA analysis of human Treg cells demonstrated an enriched expression of transcripts related to “response to oxidative stress” in individuals with MS, with SOD2, TP53, MT-CO1, ATP13A2, MT-NDs being among the top enriched genes (Figure 3D). These results suggest that compromised mitochondrial function promotes Treg cell mitochondrial oxidative stress during autoimmune responses.
Figure 3. Mitochondrial oxidative stress is a hallmark of Treg cells in autoimmune environments.
For A-C CD4+Foxp3+ Treg cells were isolated from dLNs (inguinal for pre-diseased or cervical for diseased) of naïve, pre-diseased or EAE induced with disease activity score > 3.5 Foxp3gfp.KI mice.
(A) Mean Fluorescence Intenstity (MFI) of mtROS production measured by flow cytometry using mitosox Red in naïve (n = 13), pre-diseased (n = 9) and diseased Treg cells (n = 5) (*P = 0.0191, **P = 0.0075).
(B) Immunofluorescence confocal microscopy for 8-OHDG (red), TOM20 (green) and DAPI (blue) in isolated Treg cells. Representative fields (10 μm scale bar) from three independent experiments are depicted. Pearson correlation analysis for co-localization efficiency is depicted (***P < 0.0001).
(C) SOD2 (**P = 0.0069) and mCAT (*P = 0.0336) enzyme activity measured in isolated mitochondria from naïve (n = 4) or pre-diseased Treg cells (n = 3). Each sample was pooled from n = 4 - 5 mice.
(D) Treg cells were isolated as CD4+CD127-CD25+ cells from peripheral blood of healthy individuals (n = 14) and subjects with MS (n = 11). GSEA plot showing the enrichment of “GO Response to Oxidative Stress” (NES 1.43, FDR 0.23) gene set is depicted. The top enriched genes are listed to the right of each plot.
Results are expressed as mean ± SEM. Statistical significance was obtained by unpaired Student’s t- test (C) or One-way ANOVA (A, B).
Impaired lysosomal function and defective mitophagy occurs in autoimmune Treg cells
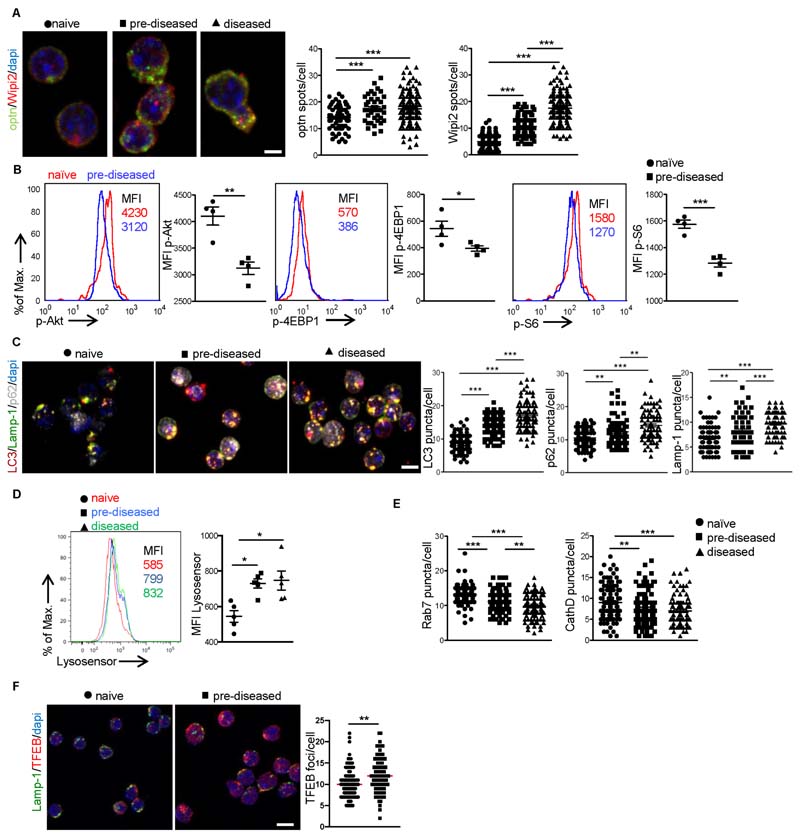
Mitophagy is essential for removal of damaged mitochondria and maintenance of cellular homeostasis (Galluzzi et al., 2017), while persistent mtROS impairs lysosomal function (Baixauli et al., 2015; Demers-Lamarche et al., 2016), which might compromise mitophagy pathway. Thus, we asked whether mitophagy is functional in Treg cells during the autoimmune response. Upon mitochondria depolarization, ubiquitination of the outer membrane and recruitment of mitophagy receptors take place to deliver damaged mitochondria to autophagolysosomes for degradation (Galluzzi et al., 2017; Gkirtzimanaki et al., 2018; Heo et al., 2015; Zachari et al., 2019). Our data show increased expression of mitophagy receptors such as optineurin (Optn) and ubiquinated TANK-binding kinase 1 (TBK1) proteins together with the repeat domain phosphoinositide-interacting protein 2 (Wipi2) in pre-diseased Treg cells (Figure 4A and S3A). Autophagosome formation is initiated upon downregulation of AKT/mTOR pathway (Martina et al., 2012). To this end, the levels of phosphorylated AKT, the mammalian target of rapamycin (mTOR), signaling components S6 and eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) (Figure 4B) were significantly decreased in Treg cells from pre-diseased mice compared to naïve Treg cells, suggesting a proper formation of autophagosome, which was also evident by the increased expression of the autophagosomal protein LC3 (Figure 4C). In support, electron microscopy analysis of Treg cells showed enhanced formation of double membrane structures around destroyed mitochondria (Figure S3B). The final step of mitophagy involves delivery of damaged mitochondria to autophagolysosomes for degradation (Klionsky et al., 2016; Ponpuak et al., 2010). Immunofluorescence microscopy revealed increased co-localization of TOM20 with lysosomal-associated membrane protein 1 (Lamp-1) and phospho serine 65 ubiquitin (pser65-Ub) in Treg cells from pre-diseased or diseased compared to naïve animals (Figure S3C), indicating that damaged mitochondria are efficiently delivered to lysosomes. Surprisingly, expression of the adaptor protein SQSTM1/p62 that targets ubiquitinated proteins for lysosomal degradation (Klionsky et al., 2016; Pankiv et al., 2007; Ponpuak et al., 2010) was markedly increased in pre-diseased compared to naïve Treg cells (Figure 4C). This was not attributed to p62 transcription (Figure S3D), thus suggesting an aberrant lysosomal function in pre-diseased and diseased Treg cells. Investigation of the autoimmune Treg lysosomal function, measuring lysosomal pH using LysoSensor Green (DND-189), a specific lysosomal pH-sensitive probe that accumulates in acidic organelles and its fluorescence increases upon protonation, indicated increased pH in lysosomes of Treg cells isolated from pre-diseased or diseased mice compared to naïve animals (Figure 4D) or to pre-diseased non Foxp3 CD4+ T cells (Figure S3E). In line with this, lysosomal protease Cathepsin D (cathD) activity and the expression of Rab7 GTPase, known to regulate intracellular membrane trafficking of endosomal/lysosomal compartments (Wang et al., 2011), were significantly decreased in pre-diseased or diseased compared to naïve Treg cells (Figure 4E). In addition, expression of the transcription factor EB (TFEB), the master regulator of lysosomal biogenesis (Baixauli et al., 2015), was significantly increased in pre-diseased Treg cells, indicating the presence of compensatory mechanisms operating during lysosomal dysfunction (Figure 4F). Collectively, our findings suggest an impaired mitophagy clearance due to lysosomal dysfunction in Treg cells occurs during autoimmunity.
Figure 4. Treg cells exhibit incomplete mitophagy during autoimmune responses.
CD4 Foxp3 Treg cells were isolated from dLNs of naïve, pre-diseased or diseased Foxp3gfpKI mice.
(A) Immunofluorescence confocal microscopy for Optn (red), Wipi2 (green) and DAPI (blue) (10 pm scale bar). Fields from one representative experiment of three are depicted. Optn (***P < 0.0001) and Wipi2 (*** P < 0.0001) puncta/cell are depicted.
(B) MFI of p-Akt (**P = 0.0032), p-4EBP1 (*P = 0.0487) and p-S6 (***P = 0.0005) (n = 4 mice per group).
(C) Immunofluorescence confocal microscopy for LC3 (red), Lamp-1 (green), p62 (silver white) and DAPI (blue) (10 pm scale bar). Fields from one representative experiment of three are shown. LC3 (***P < 0.0001), Lamp-1 (**P = 0.0032, ***P = 0.0002, ***P < 0.0001) and p62 (**P = 0.0020, **P = 0.0026, ***P < 0.0001) puncta/cell are depicted.
(D) MFI of Lysosensor Green DND (n = 5 mice per group, *P = 0.0166, *P = 0.0290).
(E) Rab7 (**P = 0.0021, ***P < 0.0001) and CathD (**P = 0.0086, ***P = 0.0005) puncta/cell are depicted.
(F) Immunofluorescence confocal microscopy for TFEB (red), Lamp-1 (green) and DAPI (blue) (10 pm scale bar). Fields from one representative experiment of three are depicted. TFEB (**P = 0.011) puncta/cell are depicted. Results are expressed as mean ± SEM. Statistical significance was obtained by One-way ANOVA or unpaired Student’s t-test. See also Figure S3.
Incomplete mitophagy aggravates Treg cell mtROS and promotes apoptotic cell death
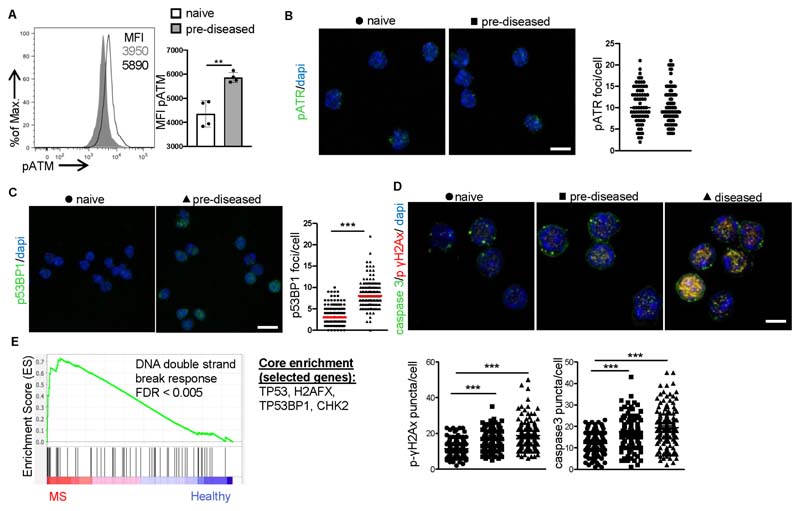
Lysosomal malfunction exacerbates mitochondrial oxidative stress (Jinn et al., 2017; Yu et al., 2018), which in turn initiates a DDR (Corrado and Campello, 2016) (Gkikas et al., 2018; Guerra-Castellano et al., 2018) that impacts cell fate. We therefore assessed the DDR in naïve and pre-diseased Treg cells and observed a marked increase in the levels of phosphorylated Ataxia-telangiectasia mutated (ATM), a protein kinase involved in the regulation of the cellular response to DNA double-strand breaks (DSBs) (Shiloh and Ziv, 2013), in pre-diseased Treg cells compared to naïve ones (Figure 5A) or to pre-diseased non Foxp3 CD4+ T cells (Figures S4A). However, there was no difference in the number of accumulated phosphorylated Ataxia telangiectasia and Rad3 related (ATR) protein foci (Figure 5B), which represents the response to persistent single-stranded DNA. Moreover, pre-diseased Treg cells displayed increased accumulation of p53-binding protein 1 (p53BP1) foci (Figure 5C), a central adapter of the DDR (Cuella-Martin et al., 2016); hence, suggesting the presence of double-strand DNA breaks. Importantly, pre-diseased and diseased Treg cells demonstrated elevated levels of phospho(Ser139)-histone γH2AX (phospho-γH2AX), a hallmark protein denoting double-strand DNA breaks (Figure 5D), together with abundance of caspase-3 (Figure 5D) and displayed at least two-fold higher programmed cell death compared to naïve Treg cells (Figure S4A) or to pre-diseased non Foxp3 CD4+ T cells (Figure S4C), and decreased frequencies (Figure S4D). In accordance, GSEA analysis of human Treg cells, demonstrated an enriched expression of transcripts related to “DNA double strand break response” in individuals with MS, with TP53, H2AFX, TP53BP1, CHK2 being among the top enriched genes (Figure 5E). Collectively, these findings demonstrate that Treg cells exhibit an enhanced DDR, culminating in increased cell death during the course of an autoimmune response.
Figure 5. Increased DDR and cell death are Treg cell checkpoints in autoimmunity.
For A-D Treg cells were isolated from Foxp3gfp.KI mice.
(A) MFI of pATM (**P = 0.0021), (n = 4 mice per group). One representative experiment of three is depicted.
(B) Immunofluorescence confocal microscopy for pATR (green) and dapi (blue) (10 pm scale bar) (n = 4 mice per group). Fields from one representative experiment of two are depicted. pATR puncta/cell are depicted.
(C) Immunofluorescence confocal microscopy for p53BP1 (green) and dapi (blue) (10 μm scale bar) (n = 4 mice per group). Fields from one representative experiment of two are depicted. p53BP1 puncta/cell (***P < 0.0001).
(D) Immunofluorescence confocal microscopy for pH2Ax (red), caspase 3 (green) and DAPI (blue). Representative fields from three independent experiments (10 μm scale bar). Caspase 3 (***P < 0.0001) and pH2AX (***P < 0.0001) puncta/cell are depicted.
(E) Treg cells were isolated as CD4+CD127-CD25+ cells from peripheral blood of healthy individuals (n = 14) and subjects with MS (n = 11). GSEA plot showing the enrichment of “GO DNA double strand break response” (NES 1.78, FDR < 0.005) gene set is depicted. The top enriched genes are listed to the right of the plot.
Results are expressed as mean ± SEM. Statistical significance was obtained by One-way ANOVA or unpaired Student’s t-test. See also Figure S4.
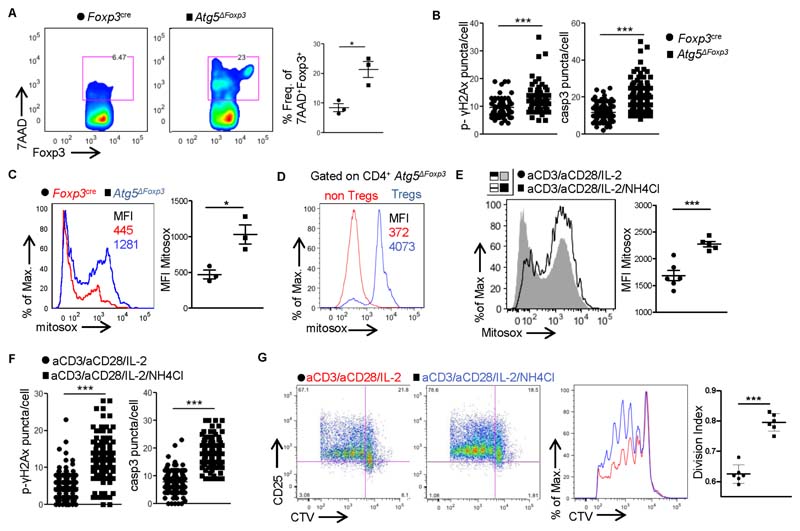
To mechanistically link the lysosomal-mediated mitophagy defects with the increased oxidative stress and DDR in pre-diseased Tregs we specifically ablated mitophagy in Treg cells by crossing mice carrying a loxP-flanked Atg5 allele (Atg5 fl/fl) (Hara et al., 2006), an essential gene for autophagosome formation, with mice expressing a YFP-Cre fusion from the Foxp3 locus (Foxp3 Cre) (Rubtsov et al., 2008), to generate Foxp3 Cre Atg5 fl/fl (denoted as Atg5ΔFoxp3) mice. In previous reports Atg5ΔFoxp3 mice succumbed at 10-12 weeks of age due to fatal autoimmunity (Wei et al., 2016); however, the molecular mechanism underlying the Treg cell defects remained elusive. Flow cytometric analysis revealed significantly higher cell death in Foxp3+ Treg cell compartment of Atg5ΔFoxp3 mice (Figures 6A and S5A), accompanied by enhanced DNA damage and caspase 3 expression (Figure 6B). Notably, mitophagy-deficient Treg cells, but not non-Treg CD4+ T cells, were characterized by excessive mtROS (Figures 6C, D) suggesting a vicious cycle via which mitophagy defects fuel the mitochondria oxidative stress response. This was further supported upon lysosomal inhibition of in vitro activated Treg cells treated with ammonium chloride, which increases the pH in intracellular vacuoles. This treatment markedly induced mtROS (Figure 6E), increased DNA breaks and caspase 3 expression (Figure 6F), followed by Treg apoptotic cell death (Figure S5B) and impaired Treg cell suppressive activity (Figure 6G). Taken together, these findings suggest that impaired lysosomal function increases mitochondrial oxidative stress and reduces survival of Treg cells.
Figure 6. Defective lysosomal function and mitophagy in Treg cells enhance mitochondrial oxidative stress and DDR, promoting cell death.
For A-D Treg cells were isolated from peripheral LNs or spleen (when indicated) of naïve Foxp3Cre or Atg5AFoxp3 n = 3 mice per group. One representative experiment of three is depicted.
(A) Flow cytometric analysis and frequencies of 7AAD+CD4+Foxp3+ Treg cells (*P = 0.0132).
(B) pH2Ax (*** P < 0.0001) and caspase 3 (***P < 0.0001) puncta/cell.
(C) MFI of Mitosox (*P = 0.0191).
(D) MFI of Mitosox gated on CD4+Foxp3- T cells or CD4+Foxp3+ Treg cells is depicted. For E-G CD4+Foxp3+ Treg cells isolated from LNs of naïve Foxp3gfp K\ mice were in vitro activated with aCD3/aCD28 beads and IL-2 in the presence or absence of ammonium chloride for 16 hrs.
(E) MFI of Mitosox in control (n = 6) or ammonium chloride treated activated Treg cells (n = 5) (***P = 0.0007). One representative experiment of three is depicted.
(F) pH2Ax (***P < 0.0001) and caspase 3 (***P < 0.0001) puncta/cell.
(G) CD4+Foxp3+ Treg cells isolated from LNs of naïve Foxp3gp.KI mice were in vitro cultured with IL-2 in the presence or absence of ammonium chloride for 16 hrs. Treg cells were then washed and co-cultured with cell trace violet (CTV) labeled CD4+Foxp3- responder T cells in the presence of aCD3/aCD28 beads. Suppressive activity of Treg cells was measured 96h later. Flow cytometric analysis of CTV dilution and CD25 expression. Division index of CD4+Foxp3- cells (n = 6 mice per group ***P < 0.0001) is depicted.
Results are presented as mean ± SEM. Statistical significance was obtained by unpaired Student’s t-test. See also Figure S5.
Scavenging of mtROS inhibits Treg cell death and ameliorates autoimmunity
To address the role of mitochondrial oxidative stress in the induction of Treg cell death, we treated activated Treg cells with carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a mitochondrial uncoupler, and MitoTEMPO, a mitochondria-specific superoxide scavenger. Scavenging of mtROS following mitochondrial depolarization resulted in inhibition of Treg cell death in vitro (Figure S6A). In addition, in vivo treatment of pre-diseased animals with mitoTEMPO reduced mtROS production (Figure S6B) and diminished Treg cell DNA damage and apoptosis (Figures 7A and S6C). This was associated with restoration of lysosomal function as evidenced by the increased p62 degradation, reduced lysosomal pH and elevated CathD and Rab7 protein expression (Figures 7B and S6D-E). Notably, therapeutic mtROS scavenging significantly attenuated the clinical score of EAE (Figure 7C) and alleviated the mononuclear cell infiltration into the spinal cords (Figure 7C). These results were accompanied by reduced CD4+ T cell frequencies (Figure S6F), dampened Th1 (IFNγ) and Th17 (IL-17) cell infiltration (Figure 7D) and increased Treg cell accumulation into spinal cords of MitoTEMPO-treated mice (Figure 6E). In support, scavenging of mtROS (Figure S6G) in young Mog35-55/CFA-immunized Atg5AFoxp3 mice (denoted hereafter as pre-diseased/MT Atg5ΔFoxp3), restrained the DDR (Figure S6H) and lowered the Treg cell death (Figure S6I), thereby suppressing the autoimmune responses as indicated by the reduced antigenspecific IFNγ and IL-17 cytokine levels in supernatants of autoantigen re-stimulated dLN cells (Figure 7E).
Figure 7. Scavenging of mtROS in Treg cells attenuates the DDR response, reverses Treg cell death and ameliorates Th1 and Th17 autoimmune responses.
For A-D CD4+Foxp3+ Treg cells were isolated from dLNs of naïve or pre-diseased Foxp3gfp.KI mice treated with or without MT.
(A) Caspase-3 and pH2AX (***P < 0.0001) puncta/cell measured by immunofluorescence confocal microscopy, in Treg cells isolated from dLNs of untreated (n = 4) or MT treated (n = 4) pre-diseased mice. One representative experiment of three.
(B) Lamp-1 (*P = 0.0223, ***P < 0.0001), p62 (**P = 0.0029, ***P < 0.0001), Rab7 (**P = 0.0017, ***P = 0.0005, ***P < 0.0001) and CathD (***P < 0.0001) puncta/cell measured by IF are depicted (n = 4 mice per group). One representative experiment of three.
(C) Mean clinical score (*P = 0.0152, *P = 0.0160) and EAE severity (*P = 0.0160) from diseased mice treated with (n = 6) or without MT (n = 4). Representative H&E sections and score from spinal cords of control diseased (clinical score 4) and diseased mice treated with mitotempo (clinical score 1.5) at 14d post immunization (p.i.) (n = 8 mice per group, ***P = 0.0004). One representative experiment of two is depicted.
(D) Flow cytometric analysis and frequencies of CD4+IL-17+ Th17 cells (*P = 0.0456) and CD4+IFNγ+ Th1 cells (*P = 0.0151) in spinal cords of diseased (clinical score 4) and diseased mice treated with MT (clinical score 1.5) at 14d post-immunization (p.i.). One representative experiment on two is depicted.
(E) dLN cells were isolated from (7-9 weeks old) Mog35-55/CFA immunized Atg5AFoxp3 (denoted as pre-diseased Atg5AFoxp3) mice treated or not with MT, 9d p.i. (n = 8 mice per group) and cultured in the presence or absence of Mog35-55 peptide for 48hrs. IFNγ (**P = 0.036) and IL-17 (*P = 0.0101) measured by ELISA in the supernatants.
For F-I CD4+Foxp3+ Treg cells were isolated from dLNs of Mog35-55/CFA immunized mCATFoxp3 mice, over-expressing the antioxidant enzyme mCAT in their Treg cell compartment, 9d p.i. (n = 4 mice per group). One representative experiment of two is depicted.
(F) Immunofluorescence confocal microscopy for 8-OHDG (red), TOM20 (green) and DAPI (blue). Representative fields (10 μm scale bar). Pearson correlation analysis for colocalization efficiency of 8-OHDG and TOM20 (****p < 0.0001).
(G) Immunofluorescence confocal microscopy for pH2Ax (red), caspase 3 (green) and DAPI (blue). pH2AX (***P < 0.0001) puncta/cell.
(H) Frequency of 7AAD+CD4+Foxp3+ Treg cells (*P = 0.0452).
(I) dLN cells were isolated and cultured in the presence or absence of Mog35-55 peptide for 48hrs. IFNγ (**P = 0.0041) and IL-17 (*P = 0.0163) measured by ELISA in the supernatants (n = 6 mice per group).
Results are presented as mean ± SEM. Statistical significance was obtained by One-way ANOVA or unpaired Student’s t-test. See also Figure S6.
To provide direct evidence for the mitochondrial oxidative stress-mediated Treg cell DDR and death, the antioxidant enzyme catalase (CAT) was over-expressed and specifically targeted to mitochondria (mCAT) of Treg cells. In particular, mice that harbor a mitochondrial-targeted human CAT gene (Dai et al., 2009), directed by the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter/enhancer regions in a BAC transgene (containing a loxP-flanked STOP cassette that prevents transcription of the downstream mCAT gene) were crossed with Foxp3cre mice to generate mCATFoxp3 animals. Notably, pre-diseased Treg cells in mCATFoxp3 mice had lower mitochondrial DNA oxidization, compared to Foxp3cre littermates (Figure 7F) confirming that mCAT overexpression in Treg cells was sufficient to restrain mitochondrial oxidative stress in an autoimmune setting. Importantly, Treg cell specific scavenging of mtROS led to a marked decrease of the DDR, as evident by the accumulation of pH2Ax foci (Figure 7G), and the lower Treg cell death (Figure 7H). Additionally, mCATFoxp3 mice exhibited a marked reduction of IFNγ and IL-17 levels in recall assays of dLN cells isolated from autoimmune mice (Figure 7I). Together, these findings reveal that mitochondrial oxidative stress dictates the Treg cell fate in the course of an autoimmune response and scavenging of mtROS maintains the Treg cell fitness, which is essential for prevention of autoimmunity.
Discussion
Treg cells are instrumental in the maintenance of self-tolerance, while their dysfunction leads to the development of autoimmune diseases. In steady state, Treg cells exhibit a unique metabolic profile, relying mostly on mitochondria metabolism to meet their energy demands, characterized by high levels of fatty acid oxidation and low to modest glycolysis, with mitochondrial respiratory chain being vital for their T cell-suppressive capacity, stability and survival (Galgani et al., 2016; He et al., 2017; Newton et al., 2016). Our data reveal that in the course of an autoimmune response, Treg cells exhibit an elevated mitochondrial oxidative stress response that results in attenuation of lysosomal function, induction of a DDR and ultimately culminating in cell death. Of interest, Treg cells isolated from the periphery of individuals with autoimmune diseases demonstrated an intense metabolic reprogramming recapitulating the aforementioned findings.
Accumulating evidence proposes mitochondrial dynamics to imprint on Treg cell fate and raises the possibility that defective mitochondrial function could be the driving force for Treg cell defects during autoimmunity. In support of this notion, ablation of mitochondrial respiratory chain complex III in Foxp3+ Treg cells led to fatal autoimmunity similar to the phenotype of scurfy mice (Weinberg et al., 2019). Further, Treg cells from mice deficient in mitochondrial ND6 gene, which impairs the complex I of the electron transport chain, lost their suppressive capacity in vitro (Angelin et al., 2017). In addition, specific ablation of the metabolic sensor LKB1 in Treg cells resulted in impaired OXPHOS, diminished ATP levels, compromised numbers of Treg cells and development of autoimmunity (He et al., 2017; Yang et al., 2017). Moreover, mitochondrial transcription factor A (Tfam), essential for mitochondrial respiration, mitochondrial DNA, transcription and packaging, has been recently shown to be essential for Treg cell maintenance in non-lymphoid tissues in the steady state and in tumors (Fu et al., 2019). In a similar fashion, fatty acid binding protein 5 (FABP5) (Field et al., 2020) and CD36 (Wang et al., 2020) expression by Treg cells were required for maintenance of their mitochondrial fitness and suppressive function in the tumor microenvironment. Our results extend those findings and demonstrate that mitochondrial dysfunction is a hallmark of Treg cells in an autoimmune environment with diminished intracellular ATP levels, decreased mitochondrial membrane potential, dampened cytochrome c expression and increased mtROS production, all hallmarks of the mitochondrial oxidative stress response.
Damaged mitochondria are cleared through mitophagy, a specialized form of autophagy, targeting mitochondria to lysosomes for degradation (Galluzzi et al., 2017; Xu et al., 2019). Malfunctioning mitochondria have been implicated in various abnormalities of the immune system. Specifically, accumulation of damaged mitochondria, due to defects in mitophagy, triggers increased mtROS production, elevated cytoplasmic calcium levels, and intracellular debris, such as mtDNA release to the cytosol, which can all act as danger signals for the immune system (Corrado and Campello, 2016; Guerra-Castellano et al., 2018; Zhang et al., 2014; Zhou et al., 2011). In line with this, we recently demonstrated that monocytes from individuals with systemic lupus erythematosus, exhibited an impaired mitophagic degradation leading to mtDNA accumulation, which instructs inflammatory cascade (Gkirtzimanaki et al., 2018). Although, the role of autophagy in homeostasis and function of Treg cells in the steady state is established (Wei et al., 2016), whether autophagy and mitophagy pathways are deregulated in Treg cells during autoimmune responses remain elusive. Our results address these needs and demonstrate that both mitophagy and autophagosome formation initiate normally in autoimmune Treg cells whereas lysosomal degradation is defective impairing therefore completion of mitophagy.
A crosstalk between mitochondria and lysosomes is well established, with lysosomal dysfunctions to induce mitochondria defects and vice versa. To this end, deletion of the mitochondrial protein AIF, OPA1, or PINK1, as well as chemical inhibition of the electron transport chain, impaired lysosomal activity, a process dependent on ROS (Demers-Lamarche et al., 2016). Moreover, deletion of mitochondrial transcription factor A (Tfam) in CD4+ T lymphocytes, impaired lysosome function, disrupted autophagy and enhanced p62 accumulation, promoting inflammatory responses (Baixauli et al., 2015). On the other hand, lysosomal dysfunction is also capable of inducing mitochondria defects and metabolic de-regulation. In this line, deletion of TRAF3IP3 specifically in Treg cells restricted mTORC1 signaling by recruiting the serine-threonine phosphatase catalytic subunit (PP2Ac) to lysosomes, leading to impaired Treg cell function and exacerbation of inflammatory disorders (Yu et al., 2018). Silencing ATP13A2, a gene encoding lysosomal P-type ATPs, induces mitochondrial fragmentation (Ramonet et al., 2012), increased mitochondrial mass, associated with defective mitochondria (Gusdon et al., 2012) as well as reduced mitochondrial respiratory rate and membrane potential (Grunewald et al., 2012). Depletion of the lysosomal K+ channel TMEM175 (Jinn et al., 2017) in SHSY5Y cells reduced lysosomal degradation impairing mitochondrial respiration and ATP production. Likewise, mutation of GBA, the gene encoding the lysosomal enzyme glucosylceramidase beta/β-glucocerebrosidase, triggered mitochondrial dysfunction by blocking mitophagy (Li et al., 2013). In this line, our results reveal a ying-yang effect between mitochondria and lysosomal function imprinted by mtROS in Treg cells during autoimmunity. Specifically, mitophagy-deficient autoimmune Treg cells exhibited increased mtROS, an enhanced DDR and cell death, while mtROS scavenging restored lysosomal degradation and inhibited Treg cell death. In support, in a recent paper, autophagy defects increased mtROS in CD4+ T cells from young individuals, a process linked to Th17-mediated inflammation (Bharath, 2020). The precise mechanisms and mediators influencing this ying-yang effect in Treg cells during autoimmunity remain to be investigated. A possible scenario could be a defect of the mitochondrial antioxidant defense in Treg cells. Indeed, our results showed decreased expression of manganese superoxide dismutase (Mn-SOD) and catalase in autoimmune Treg cells, which holds true for autoTregs from individuals with autoimmune diseases (MS). Another possibility might be deficiencies in lysosomal enzymes as depicted in individuals developing lysosomal storage disorders that are prone to developing autoimmune diseases (Rigante et al., 2017). In support, pathway analysis revealed lysosomal genes to be differentially expressed in Treg cells from individuals with autoimmune diseases (i.e. in MS: LAMP3, LAMP5, ATP13A2, VPS33B, BLOC1S2, GLMP, HPS4, LAMTOR1, LAMTOR2, LAMTOR5, ACP2, RILP, in SLE: LAMTOR2, LAMP5, ACP2, BLOC1S3, GLMP, LAMP3, ATP13A2 and in RA: ATP13A2, CTNS, LAMTOR2, LAMP5, RILP, GLMP). Taking into consideration that many susceptibility genes are shared across multiple autoimmune disorders, the Treg cell aberrancies described here could be a cell-intrinsic phenomenon, which influences the predisposition to autoimmunity. Alternatively, common autoimmune inflammatory mediators could instruct the Treg cell defects leading to cell death and disturbance of self-tolerance. Future studies should address these important issues to shed light into the pathogenesis of autoimmune diseases.
The therapeutic potential of ex-vivo expanded autologous Treg cells either alone (NCT02691247) or in combination with IL-2 administration (NCT02772679) is evaluated in individuals with T1D. In addition, administration of low doses of IL-2 promoting the induction of Tregs was found beneficial in several autoimmune diseases including RA, SLE, psoriasis and Crohn’s disease (Rosenzwajg et al., 2019) and a respective clinical trial (NCT01988506) in 14 autoimmune and auto-inflammatory diseases is expected to launch results in 2021. Our results shed light to a previously unrecognized mechanism of Treg dysfunction during autoimmunity, highlighting mitochondrial oxidative stress as important determinant of Treg cell fate. Thus, the design of novel therapies using mitochondria-specific anti-oxidants arises as a promising road to treat autoimmune diseases (Mendiola et al., 2020). Indeed, preclinical studies using mtROS scavengers yielded promising results (Lehmann et al., 1994; Mao et al., 2013; Stanislaus et al., 2005), while antioxidant compounds such as mitoQ are currently in clinical trial for MS (mitoQ for MS fatigue NCT03166800). Overall, understanding the mechanisms of Treg defects in the course of an autoimmune response may pave the way for novel therapeutic interventions not only in autoimmunity but also in other inflammatory settings characterized by disturbed peripheral tolerance.
Limitations of study
The mechanistic findings of our study were obtained in a mouse model of induced autoimmunity. It will be of interest to examine whether similar metabolic aberrancies operate in Treg cells in spontaneous autoimmune models. Furthermore, an unanswered question is which are the upstream mechanisms that drive Treg dysfunction during autoimmunity. It would be of interest to delineate whether the metabolic defects in Treg cells are cell-intrinsic and may contribute to susceptibility of autoimmunity or whether influences of the autoimmune inflammatory environment imprint on Treg cells to engage the mitochondrial oxidative stress response.
Star Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Panayotis Verginis (pverginis@bioacademy.gr)
Materials Availability
The mouse line generated in this study (mCatFoxp3 mice) is available from the lead contact upon request.
Experimental Model and Subject Details
Human subjects
Peripheral blood samples were obtained from individuals diagnosed with autoimmune diseases or healthy (n = 14) individuals. Subjects with RA (n = 9) were diagnosed according to the 1987 American College of Rheumatology (ACR) criteria (Arnett et al., 1988). Individuals with SLE (n = 8) met the 1999 American College of Rheumatology revised criteria (Aringer et al., 2019). Subjects with Relapsing Remitting MS (RRMS, n = 11) were diagnosed based on the revised McDonald criteria (Thompson et al., 2018). At the time of sampling all individuals with SLE had moderate to severe disease activity (SLEDAI > 7), subjects with RA had moderate disease activity (DAS > 4.2) according to the disease activity score based on the 28 joint counts (Prevoo et al., 1995), while subjects with MS were all active or highly active. Subjects with autoimmune diseases and healthy individuals were recruited through the Rheumatology and Clinical Immunology Department, 4th Clinical Pathology, Attikon University Hospital (Athens, Greece) and the Neurological Clinic of Athens Naval Hospital. The Clinical Research Ethics Board of all hospitals approved this study. Informed consent was obtained from all individuals prior to sample collection. All subjects were emitted treatment for at least 24h prior to blood drawing. Clinical and demographic characteristics are summarized in Supplemental Table 1.
Animal Studies
All mice used were in C57BL/6 background. C57BL/6J, Rag1–/– and mCAT mice were obtained from the Jackson Laboratory. Foxp3-gfp KI (Fontenot et al., 2005) and Foxp3YFP/cre (Rubtsov et al., 2008) mice were provided by Alexander Rudensky (Department of Immunology, Memorial Sloan-Kettering Cancer Center, New York, New York, USA). Atg5ΔFoxp3 mice were generated by crossing Alg5fl/fl mice (Hara et al., 2006) (kindly given from Dr. Noboru Mizushima (RIKEN, Saitama prefecture, Japan) and Foxp3YFP mCATFoxp3 mice were generated by crossing mCAT mice and Foxp3YFP/cre Mice were housed 6 per cage in a temperature (21-23°C) and humidity controlled colony room, maintained on a 12hr light/dark cycle (07:00 to 19:00 light on), with standard food (4RF21, Mucedola Srl, Italy) and water provided ad libitum and environmental enrichments. All mice in the animal facility were screened regularly by using a health-monitoring program, in accordance to the Federation of European Laboratory Animal Science Association (FELASA) and were free of pathogens (Mahler Convenor et al., 2014). During all experiments mice were daily monitored. All mice used in the experiments were female 8-10 weeks old and each experiment was repeated three times. The total number of mice analyzed for each experiment is detailed in the figure legends. Littermates of the same genotype were randomly allocated to experimental groups. All procedures were in accordance to institutional guidelines and were approved by the Directorate of Agriculture and Veterinary Policy, Region of Attika, Greece (Athens, Greece, protocol 1474).
Method Details
Human cell isolation from peripheral blood
Heparinized blood (20ml) was collected from healthy subjects and individuals with autoimmune diseases. PBMCs were isolated on Histopaque-1077 (Sigma) density gradient. Briefly, blood was diluted 1:1 with PBS and carefully layered over Histopaque medium. Tubes were centrifuged at 1800 rpm for 30 min with no brake at room temperature. PBMCs layer was collected and cells were washed with PBS.
In vivo immunization protocol
Mice were immunized by subcutaneous (s.c) injections at the base of the tail, with 100 μg Mog35-55 (Genemed synthesis) emulsified (1:1) in complete Freund’s adjuvant (CFA, Sigma Aldrich) and sacrificed 9d post-injection, inguinal LNs were isolated. For EAE induction mice were immunized with 200 μg Mog35-55 emulsified (1:1) in IFA, plus 4 μg/ml Mycobacterium (BD Difco™). Emulsion was injected s.c over three sites (i.e. 50 μl per site): one along the midline of the back between the shoulders, and two on either side of the midline on the lower back. Additionally, mice received intraperitoneally (i.p.) 300 ng pertussis toxin (Sigma Aldrich), on day 0 and day 2. Mice were monitored daily for clinical signs of disease as described (Alissafi et al., 2017; Alissafi et al., 2015). Mice were sacrificed when the disease scored 4 and their cervical LNs and spinal cord were isolated. For scavenging of mitochondrial ROS mice received i.p. injections of 30 μg mitoTEMPO (Sigma-Aldrich) every other day.
Flow cytometry and cell sorting
For analysis and isolation of Treg cells, single-cell suspensions from human peripheral blood mononuclear cells (PBMCs) or mouse dLNs and spinal cord were stained with conjugated antibodies against mouse: CD4, CD25, GITR, cytochrome c, pH2A.x (ser139), 7AAD (Biolegend), pAkt (S473), pS6 (S235/236), p4E-BP1 (T36/T45), phospho ATM (ser1981) TMRE, Mitosox, Mitotracker Red CMX ROS, Lysosensor (Molecular Probes) and JC-1 (1:100, eBiosciences), and against human: CD4, CD25, CD127 (Biolegend). For Foxp3 intracellular staining, cells were fixed and stained using the Foxp3 Staining Set (eBioscience) according to the manufacturer’s instructions. For intracellular cytokine staining, dLN cells were incubated with 50 ng/ml PMA (Sigma-Aldrich), 2 μg/ml ionomycin (Sigma-Aldrich), and Golgi plug (1/1000; Becton Dickinson Biosciences) for 6 hours at 37°C, and stained for IFN-γ and IL-17 (Biolegend) using the BD Cytofix/Cytoperm Plus Fixation/Permeabilization kit (Becton Dickinson Biosciences). For caspases staining, FITC-VAD-FMK (eBioscience) in complete medium was incubated with the cell suspensions for 1 h at 37C and 5% CO2, washed, and stained for the extracellular markers, according to the vendors instructions. For intracellular phospho protein staining, cells were permeabilized with the intracellular Fixation & Permeabilization buffer set (eBioscience) according to the manufacturer’s instructions and stained with antibody against phospho proteins. For isolation of Foxp3+ Treg cells, spleen and LNs isolated from Foxp3gfp.KI mice, were enriched for CD4+ T cells (Miltenyi microbeads). CD4+ Foxp3+ or Foxp3- cells were sorted on a FACS ARIA III (BD Biosciences). Cells were sorted on a FACS ARIA III (BD Biosciences) and the BD FACSDIVA v8.0.1 software (BD Biosciences). Cell purity was above 95%. Analysis was performed with FlowJo software.
ATP Detection Assay
Cells were sorted and 1x105 cells/well were plated in a 96well-plate. ATP was measured according to the manufacturer instructions (ATPlite Luminescence Assay System, Perkin Elmer).
Measurement of Oxygen Consumption Rate (OCR) by Seahorse analysis
dLN cells were stained with anti-GITR-APC, anti-CD25-PE and anti-CD4-PB and Treg cells were sorted as CD4+ CD25+ GITR+. After sorting cells were kept on ice until plating into 96-well Seahorse plates, which were previously coated with CellTAK according to manufacturer’s instructions. 100,000 cells were plated in 50μl XF Seahorse medium (Agilent, Santa Clara, CA, USA) (pH 7.4) supplemented with 2mM glutamine, 10mM glucose and 2mM sodium pyruvate. Plates were centrifuged at 200g without break and cells were then incubated for 30 min at 37°C without CO2. Afterwards, 130μl of the same XF medium was added to each well. OCR was measured at basal conditions and after sequential stimulation of the cells by 1μm Oligomycin, 1μM FCCP and 0.5μM Rotenone/Antimycin (all included in the Mitostress kit, Agilent, Waldbronn, Germany) in a Seahorse XFe96 Analyzer (Agilent) using the Wave Software (Agilent).
Quantitative PCR analysis
Cells were lysed in Buffer RA1 (Macherey-Nagel) and RNA was extracted using a NucleoSpin® RNA isolation kit according to the manufacturer’s instructions. First-strand cDNA synthesis was performed using PrimeScript reverse transcriptase (Takara). qPCR was carried out using the Kapa Sybr® Fast Universal kit (Kapa Biosystems). Relative expression of target genes was calculated by comparing them to the expression of the housekeeping gene Hprt.
RNA sequencing pipeline
Total RNA was extracted as described by manufacturer (NucleoSpin® RNA XS) and mRNA libraries were generated using the Illumina TruSeq Sample Preparation kit v2. Single-end 75-bp or 100-bp mRNA sequencing was performed on Illumina NextSeq 500. Raw fastq sequencing reads were aligned against the human reference genome sequence (GRCh38/hg38) using the STAR aligner v2.6 (Dobin et al., 2013), discarding all non-uniquely aligned reads. Gene quantification was performed using HTSeq (Anders et al., 2015) with the “union” mode counting reads matching exons of the genocde v.29 annotation.
Further processing was performed with the R Bioconductor package edgeR v.3.26.8 (Robinson et al., 2010). Reads were normalized for intra- and inter-sample variances using the functions “calcNormFactors” and “estimateDisp”. Differential gene expression analysis was performed with the functions “glmQLFit” and “glmQLFTest”, reporting p-value, false-discovery rate (FDR) and log2 fold-changes between any possible pair-wise comparison and gene in R (Team, 2018). Genes with p-value ≤ 0.05 and fold change|FC| > 1.5 were considered statistically significantly differentially expressed (DEGs). Heatmaps were created in R with an in-house developed script which is based on ggplot package.
Enrichment analysis
Pathway and gene ontology (GO) enrichment analysis was performed using Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) in order to reveal enriched signatures in our gene sets based on the Molecular Signatures Database (MSigDB) v7.0. Gene sets were ranked by taking the -log10 transform of the p-value multiplied by the FC. Significantly upregulated genes were at the top and significantly downregulated genes were at the bottom of the ranked list. GSEA pre-ranked analysis was then performed using the default settings. Enrichment was considered significant by the GSEA software for FDR (q-value) < 0.25.
Histological Analysis and Immunofluorescence
H&E sections of spinal cord were performed as described (Alissafi et al., 2017; Alissafi et al., 2015). For autophagy immunofluorescence (Alissafi et al., 2017; Alissafi et al., 2018), Treg cells were seeded in coverslips pretreated with poly lysine, fixed with 4% PFA for 15 min in room temperature (RT) followed by 10 min of fixation with ice cold methanol in -20° C, washed twice with PBS and ice cold methanol. Cells were permeabilized by using 0.1% saponin (Sigma-Aldrich) and stained with mouse anti-LC3 antibody (1:20, nanoTools), rat anti-Lamp-1 (1:400, Santa Cruz Biotechnology) and rabbit anti-p62 (1:500, MBL). For mitophagy, lysosomal function and DNA damage immunofluorescence cells were seeded in coverslips pretreated with poly lysine, fixed with 4% PFA for 15 min in room temperature followed by 10 min of fixation with ice cold methanol in -20° C, washed twice with PBS and ice cold methanol. Cells were permeabilized by using 0.5% Triton-X-100 (Sigma-Aldrigh) for 5 min in RT and stained with rabbit anti-phospho-ubiquitin (Ser65) (1:200, Millipore) and mouse anti-Tom20 (1:100, Calbiochem) or mouse anti-WIPI2 (1:200, Biorad), rabbit anti-pTBK (ser172) (1:50, Cell Signaling Technologies), mouse anti-mono and poly UB (1:100, Enzo), rabbit anti-Optineurin (1:100 C-terminal, Cayman), or mouse anti-Rab7 (1:2000, Abcam), and goat anti-cathepsin D (1:100, Santa Cruz), or mouse anti-phospho-Histone H2A.X (ser139)(1:200, Millipore) and rabbit anti-Caspase-3 (1:150, Cell Signaling Technologies), or mouse anti 8-OHDG DNA/RNA damage antibody (1:50, Novus Biologicals) and rabbit anti-Tom20 (1:200, CST), or rabbit phospho ATR (1:100, GeneTex), or rabbit p53BP1 (1:200, Novus Biologicals) or rabbit TFEB (1:100 Thermo Fischer Scientific), followed by incubation with Alexa fluor® 555 anti-mouse IgG (1:500, Molecular Probes), Alexa fluor® 647 anti-rabbit IgG (1:200, Molecular Probes), Alexa fluor® 488 anti-rat IgG (1:250, Molecular Probes). For visualization of the nuclei Dapi (Sigma-Aldrich) was used. Samples were coverslipped with moviol and visualized using inverted confocal live cell imaging system Leica SP5. Puncta/cell were calculated using a macro developed in Fiji software. Colocalization of puncta was calculated using cross-correlation analysis with velocity software (Costes et al., 2004).
In vitro assays
1x106 sorted CD4+Foxp3gfp+ Treg cells were labeled with the division-tracking dye CellTrace violet (CTV, Invitrogen) according to the manufacturer’s protocol, then co-cultured with beads coated with monoclonal antibody (mAb) to the invariant signaling protein CD3 plus mAb to CD28, at a ratio of 1 bead per 4 cells (Invitrogen), in the presence of IL-2 (5.000 U/ml) and treated either with 2.5 μM CCCP (Tocris Bioscience) or with Dimethyl Sulfoxide (DMSO) control. After 96 h, cells were stained (7-AAD) and subjected to FACS. For lysosomal inhibition 2x105 sorted CD4+Foxp3gfp+ Treg cells were plated in 96-well round-bottom plates and cultured with beads coated with monoclonal antibody (mAb) to the invariant signaling protein CD3 plus mAb to CD28, at a ratio of 1 bead per 4 cells (Invitrogen) and IL-2 (5.000 U/ml) in the presence or absence of the lysososmal inhibitor ammonium chloride (NH4Cl, 15 mM, Sigma-Aldrich) for 24 hours. For Treg suppression assay with lysosomal inhibition, 2x105 CD4+Foxp3gfp+ Treg cells were plated in 96-well round-bottom plates and cultured with 300 U/ml IL-2 in the presence or absence of 15 mM ammonium chloride for 20 hrs. Tregs were then collected, washed and cultured with 105 CD4+Foxp3- CTV labelled responder T cells in the presence of beads coated with monoclonal antibody (mAb) to the invariant signaling protein CD3 plus mAb to CD28 for 96h (ratio responderT/Treg/beads 4/1/2). For dLN cells restimulation, 2x105 cells were cultured in 96 round bottom plates in the presence or absence of 30 μg/ml Mog35-55 peptide. Supernatants were collected 48hrs and IFNγ or IL-17 was measured using ELISA kits (R&D Biosystems) according to the manufacturer’s instructions. For all experiments, cells were cultured in RPMI-1640 supplemented with 100 U/ml penicillin and streptomycin, 10% heat inactivated fetal bovine serum (FBS) (all Gibco) and 50 μM 2-Mercaptoethanol (sigma).
Measurement of mitochondrial anti-oxidant enzymes activity
For measurement of mitochondrial catalase and MnSOD activity, 5x106 CD4+Foxp3+ Treg cells were sorted from dLNs and pooled from 5 Foxp3 gfp mice. Cells were centrifuged at 1800 rpm for 20 min at 4°C and pellet was homognized with sonication (3 rounds of 3s, 5s, 10s, 0.25 amplitude) in 300 ul PBS. Cell homogenates were then centrifuged at 1500 x g for 15 min at 4°C. Supernatants were then collected and centrifuged at 10.000 x g for 15 min at 4°C. Pellets containing the mitochondrial fraction were resuspended in 50 ul PBS. To determine MnSOD (SOD2) activity samples were treated with 2 mM potassium cyanide. MnSOD activity was measured in fresh samples utilizibg the Superoxide Dismutase (SOD) Colorimetric Activity Kit (Invitrogen) following the manufacturers instructions. For measurement of mitochondrial catalase activity, the colorimetric catalase activity kit (abcam) was used following the manufacturers instructions.
Electron Microscopy
For conventional electron microscopy, the cells were pelleted at 800g for 5min. Pellets were fixed with 3% paraformaldehyde – 2% glutaraldehyde made up in 0.1M Phosphate Buffer (PB) for 1h at RT. After subsequent buffer washes, pellets were embedded in 4% low-melting agarose in 0.1M PB. Following solidification small cubes were cut and post-fixed with 1% osmium tetroxide for 1h on ice. After washing with buffer, the samples were dehydrated in a graded ethanol series, and embedded in Epon/Araldite resin mixture. Ultrathin sections (65nm) were cut with a Diatome diamond knife at a thickness of 65nm on a Leica EM UC7 ultramicrotome (Leica Microsystems, Vienna, Austria), were then mounted onto 300 mesh copper grids and stained with uranyl acetate and lead citrate. Sections were examined with a Philips 420 transmission electron microscope and photographed with a Megaview G2 CCD camera.
Quantification and Statistical Analysis
Statistical analysis was performed using unpaired Student’s t-test or One way ANOVA using Tukey post test in GraphPad Prism v5 software. Data are presented as means ± S.E.M. P value < 0.05 was considered as indicative of statistical significance. All P values and n are reported in the figure legends. The investigators were not blinded to the identities of the samples. Compared samples were collected and analyzed under the same conditions. G*power analysis was performed (with 90% power and 0.001 type 1 error) to calculate the appropriate sample size. No data were excluded. All data showed normal distribution.
Supplementary Material
Acknowledgments
We thank Aikaterini Hatzioannou, for assisting with experiments and cell sorting, Giannis Vatsellas for assisting with RNAseq and for providing technical advice, Anastasia Apostolidou for cell sorting, Nikos Malissovas for providing technical assistant, Antigoni Pieta for assisting with patient sample collection, Evi Goulielmaki for providing information for DDR IF, Agapaki Anna for assisting with histology, Stamatis Pagakis and Eleni Rigana for providing assistance with confocal microscopy and quantification of confocal images, Pavlos Alexakis for assisting with mice. This work was supported by grants from the Greek General Secretariat of Research and Technology (Aristeia II 3468 to P.V.). T.C. was supported by the Deutsche Forschungsgemeinschaft (SFB-TRR 127 and SFB 1181). D.B. was supported by the ERC under the European Union’s Horizon 2020 research and innovation program (grant agreement No 742390). T.A. is supported by IKY Fellowships of Excellence for Postgraduate Studies in Greece, Siemens Program. D.B., T.A. and P.V. were supported by the European Union’s Horizon 2020 research and innovation program under grant agreement No 733100.
Footnotes
Author contributions
AT designed the study, performed experiments, analyzed data, generated figures, and wrote the manuscript. KL and LM performed experiments and analyzed data. KI performed electron microscopy experiments. MM assisted with immunohistochemical analysis. LJH and AI performed Seahorse experiments. FA performed bioinformatic analysis of RNAseq data. DT performed clinical evaluation of individuals with MS and provided human specimens. KNT, GGA and CT provided materials, assisted in designing experiments and critically edited the manuscript. BD performed clinical evaluation of individuals with RA and SLE, provided human specimens and materials, wrote and critically edited the manuscript. PV designed and supervised the study, performed data analysis, and wrote the manuscript.
Declaration of Interests. The authors declare no competing interests.
Data and Code Availability
The data that support the findings of this study are available from the lead contact (pverginis@bioacademy.gr) upon reasonable request. The RNAseq data have been deposited in the European Genome-Phenome Archive, which is hosted by the EBI, under accession number EGAS00001004470.
References
- Alissafi T, Banos A, Boon L, Sparwasser T, Ghigo A, Wing K, Vassilopoulos D, Boumpas D, Chavakis T, Cadwell K, Verginis P. Tregs restrain dendritic cell autophagy to ameliorate autoimmunity. J Clin Invest. 2017;727:2789–2804. doi: 10.1172/JCI92079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alissafi T, Hatzioannou A, Ioannou M, Sparwasser T, Grun JR, Grutzkau A, Verginis P. De novo-induced self-antigen-specific Foxp3+ regulatory T cells impair the accumulation of inflammatory dendritic cells in draining lymph nodes. J Immunol. 2015;194:5812–5824. doi: 10.4049/jimmunol.1500111. [DOI] [PubMed] [Google Scholar]
- Alissafi T, Hatzioannou A, Mintzas K, Barouni RM, Banos A, Sormendi S, Polyzos A, Xilouri M, Wielockx B, Gogas H, Verginis P. Autophagy orchestrates the regulatory program of tumor-associated myeloid-derived suppressor cells. J Clin Invest. 2018;128:3840–3852. doi: 10.1172/JCI120888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelin A, Gil-de-Gomez L, Dahiya S, Jiao J, Guo L, Levine MH, Wang Z, Quinn WJ, 3rd, Kopinski PK, Wang L, et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017;25:1282–1293.:e1287. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aringer M, Costenbader K, Daikh D, Brinks R, Mosca M, Ramsey-Goldman R, Smolen JS, Wofsy D, Boumpas DT, Kamen DL, et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019;71:1400–1412. doi: 10.1002/art.40930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Baixauli F, Acin-Perez R, Villarroya-Beltri C, Mazzeo C, Nunez-Andrade N, Gabande-Rodriguez E, Ledesma MD, Blazquez A, Martin MA, Falcon-Perez JM, et al. Mitochondrial Respiration Controls Lysosomal Function during Inflammatory T Cell Responses. Cell Metab. 2015;22:485–498. doi: 10.1016/j.cmet.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- Bharath LP. Metformin Enhances Autophagy and Normalizes Mitochondrial Function to Alleviate Aging-Associated Inflammation. Cell Metabolism. 2020 doi: 10.1016/j.cmet.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone F, De Rosa V, Carrieri PB, Montella S, Bruzzese D, Porcellini A, Procaccini C, La Cava A, Matarese G. Regulatory T cell proliferative potential is impaired in human autoimmune disease. Nat Med. 2014;20:69–74. doi: 10.1038/nm.3411. [DOI] [PubMed] [Google Scholar]
- Corrado M, Campello S. Autophagy inhibition and mitochondrial remodeling join forces to amplify apoptosis in activation-induced cell death. Autophagy. 2016;12:2496–2497. doi: 10.1080/15548627.2016.1226738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuella-Martin R, Oliveira C, Lockstone HE, Snellenberg S, Grolmusova N, Chapman JR. 53BP1 Integrates DNA Repair and p53-Dependent Cell Fate Decisions via Distinct Mechanisms. Mol Cell. 2016;64:51–64. doi: 10.1016/j.molcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DF, Santana LF, Vermulst M, Tomazela DM, Emond MJ, MacCoss MJ, Gollahon K, Martin GM, Loeb LA, Ladiges WC, Rabinovitch PS. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers-Lamarche J, Guillebaud G, Tlili M, Todkar K, Belanger N, Grondin M, Nguyen AP, Michel J, Germain M. Loss of Mitochondrial Function Impairs Lysosomes. J Biol Chem. 2016;291:10263–10276. doi: 10.1074/jbc.M115.695825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Villar M, Hafler DA. Regulatory T cells in autoimmune disease. Nat Immunol. 2018;19:665–673. doi: 10.1038/s41590-018-0120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CS, Baixauli F, Kyle RL, Puleston DJ, Cameron AM, Sanin DE, Hippen KL, Loschi M, Thangavelu G, Corrado M, et al. Mitochondrial Integrity Regulated by Lipid Metabolism Is a Cell-Intrinsic Checkpoint for Treg Suppressive Function. Cell Metab. 2020;31:422–437.:e425. doi: 10.1016/j.cmet.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Fu Z, Ye J, Dean JW, Bostick JW, Weinberg SE, Xiong L, Oliff KN, Chen ZE, Avram D, Chandel NS, Zhou L. Requirement of Mitochondrial Transcription Factor A in Tissue-Resident Regulatory T Cell Maintenance and Function. Cell Rep. 2019;28:159–171.:e154. doi: 10.1016/j.celrep.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgani M, De Rosa V, La Cava A, Matarese G. Role of Metabolism in the Immunobiology of Regulatory T Cells. J Immunol. 2016;197:2567–2575. doi: 10.4049/jimmunol.1600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikas I, Palikaras K, Tavernarakis N. The Role of Mitophagy in Innate Immunity. Front Immunol. 2018;9:1283. doi: 10.3389/fimmu.2018.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkirtzimanaki K, Kabrani E, Nikoleri D, Polyzos A, Blanas A, Sidiropoulos P, Makrigiannakis A, Bertsias G, Boumpas DT, Verginis P. IFNalpha Impairs Autophagic Degradation of mtDNA Promoting Autoreactivity of SLE Monocytes in a STING-Dependent Fashion. Cell Rep. 2018;25:921–933.:e925. doi: 10.1016/j.celrep.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goschl L, Scheinecker C, Bonelli M. Treg cells in autoimmunity: from identification to Treg-based therapies. Semin Immunopathol. 2019;41:301–314. doi: 10.1007/s00281-019-00741-8. [DOI] [PubMed] [Google Scholar]
- Grant CR, Liberal R, Mieli-Vergani G, Vergani D, Longhi MS. Regulatory T-cells in autoimmune diseases: challenges, controversies and--yet--unanswered questions. Autoimmun Rev. 2015;14:105–116. doi: 10.1016/j.autrev.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Grünewald A, Arns B, Seibler P, Rakovic A, Munchau A, Ramirez A, Sue CM, Klein C. ATP13A2 mutations impair mitochondrial function in fibroblasts from patients with Kufor-Rakeb syndrome. Neurobiol Aging. 2012;33:1843 e1841–1847. doi: 10.1016/j.neurobiolaging.2011.12.035. [DOI] [PubMed] [Google Scholar]
- Guerra-Castellano A, Diaz-Quintana A, Perez-Mejias G, Elena-Real CA, Gonzalez-Arzola K, Garcia-Maurino SM, De la Rosa MA, Diaz-Moreno I. Oxidative stress is tightly regulated by cytochrome c phosphorylation and respirasome factors in mitochondria. Proc Natl Acad Sci U S A. 2018;115:7955–7960. doi: 10.1073/pnas.1806833115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusdon AM, Zhu J, Van Houten B, Chu CT. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiol Dis. 2012;45:962–972. doi: 10.1016/j.nbd.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- He N, Fan W, Henriquez B, Yu RT, Atkins AR, Liddle C, Zheng Y, Downes M, Evans RM. Metabolic control of regulatory T cell (Treg) survival and function by Lkb1. Proc Natl Acad Sci U S A. 2017;114:12542–12547. doi: 10.1073/pnas.1715363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn S, Drolet RE, Cramer PE, Wong AH, Toolan DM, Gretzula CA, Voleti B, Vassileva G, Disa J, Tadin-Strapps M, Stone DJ. TMEM175 deficiency impairs lysosomal and mitochondrial function and increases alpha-synuclein aggregation. Proc Natl Acad Sci U S A. 2017;114:2389–2394. doi: 10.1073/pnas.1616332114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann D, Karussis D, Misrachi-Koll R, Shezen E, Ovadia H, Abramsky O. Oral administration of the oxidant-scavenger N-acetyl-L-cysteine inhibits acute experimental autoimmune encephalomyelitis. J Neuroimmunol. 1994;50:35–42. doi: 10.1016/0165-5728(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol. 2013;6:19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SA, Buckner JH. CD4+FOXP3+ T regulatory cells in human autoimmunity: more than a numbers game. J Immunol. 2011;187:2061–2066. doi: 10.4049/jimmunol.1003224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler Convenor M, Berard M, Feinstein R, Gallagher A, Illgen-Wilcke B, Pritchett-Corning K, Raspa M. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab Anim. 2014;48:178–192. doi: 10.1177/0023677213516312. [DOI] [PubMed] [Google Scholar]
- Mao P, Manczak M, Shirendeb UP, Reddy PH. MitoQ, a mitochondria-targeted antioxidant, delays disease progression and alleviates pathogenesis in an experimental autoimmune encephalomyelitis mouse model of multiple sclerosis. Biochim Biophys Acta. 2013;1832:2322–2331. doi: 10.1016/j.bbadis.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MM, Weinberg SE, Chandel NS. Mitochondrial control of immunity: beyond ATP. Nat Rev Immunol. 2017;17:608–620. doi: 10.1038/nri.2017.66. [DOI] [PubMed] [Google Scholar]
- Mendiola AS, Ryu JK, Bardehle S, Meyer-Franke A, Ang KK, Wilson C, Baeten KM, Hanspers K, Merlini M, Thomas S, et al. Transcriptional profiling and therapeutic targeting of oxidative stress in neuroinflammation. Nat Immunol. 2020 doi: 10.1038/s41590-020-0654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Kelly B, O’Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol. 2017;18:488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- Newton R, Priyadharshini B, Turka LA. Immunometabolism of regulatory T cells. Nat Immunol. 2016;17:618–625. doi: 10.1038/ni.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HWt, Kyei GB, Johansen T, Vergne I, Deretic V. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevoo ML, van ‘t Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–48. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- Rambold AS, Pearce EL. Mitochondrial Dynamics at the Interface of Immune Cell Metabolism and Function. Trends Immunol. 2018;39:6–18. doi: 10.1016/j.it.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Ramonet D, Podhajska A, Stafa K, Sonnay S, Trancikova A, Tsika E, Pletnikova O, Troncoso JC, Glauser L, Moore DJ. PARK9-associated ATP13A2 localizes to intracellular acidic vesicles and regulates cation homeostasis and neuronal integrity. Hum Mol Genet. 2012;21:1725–1743. doi: 10.1093/hmg/ddr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigante D, Cipolla C, Basile U, Gulli F, Savastano MC. Overview of immune abnormalities in lysosomal storage disorders. Immunol Lett. 2017;188:79–85. doi: 10.1016/j.imlet.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzwajg M, Lorenzon R, Cacoub P, Pham HP, Pitoiset F, El Soufi KCRI, Bernard C, Aractingi S, Banneville B, et al. Immunological and clinical effects of low-dose interleukin-2 across 11 autoimmune diseases in a single, open clinical trial. Ann Rheum Dis. 2019;78:209–217. doi: 10.1136/annrheumdis-2018-214229. [DOI] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Fukuma K, Kuribayashi K, Masuda T. Organ-specific autoimmune diseases induced in mice by elimination of T cell subset. I. Evidence for the active participation of T cells in natural self-tolerance; deficit of a T cell subset as a possible cause of autoimmune disease. J Exp Med. 1985;161:72–87. doi: 10.1084/jem.161.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Sies H, Jones DP. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat Rev Mol Cell Biol. 2020 doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- Stanislaus R, Gilg AG, Singh AK, Singh I. N-acetyl-L-cysteine ameliorates the inflammatory disease process in experimental autoimmune encephalomyelitis in Lewis rats. J Autoimmune Dis. 2005;2:4. doi: 10.1186/1740-2557-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R. C. R: A language and envinonment for statistical computing. R Foundation for Statistical Computing; 2018. [Google Scholar]
- Thompson AJ, Banwell BL, Barkhof F, Carroll WM, Coetzee T, Comi G, Correale J, Fazekas F, Filippi M, Freedman MS, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17:162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Viglietta V, Baecher-Allan C, Weiner HL, Hafler DA. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. J Exp Med. 2004;199:971–979. doi: 10.1084/jem.20031579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, Mohmood SR, Fernandez-Garcia J, Tsai CH, Schulze I, et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol. 2020;21:298–308. doi: 10.1038/s41590-019-0589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ming Z, Xiaochun W, Hong W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal. 2011;23:516–521. doi: 10.1016/j.cellsig.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, Wu C, Vogel P, Neale G, Green DR, Chi H. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol. 2016;17:277–285. doi: 10.1038/ni.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martinez-Reyes I, Gao P, Helmin KA, Abdala-Valencia H, Sena LA, et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Xu Y, Shen J, Ran Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy. 2019:1–15. doi: 10.1080/15548627.2019.1603547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Blanco DB, Neale G, Vogel P, Avila J, Clish CB, Wu C, Shrestha S, Rankin S, Long L, et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature. 2017;548:602–606. doi: 10.1038/nature23665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Teng XL, Wang F, Zheng Y, Qu G, Zhou Y, Hu Z, Wu Z, Chang Y, Chen L, et al. Metabolic control of regulatory T cell stability and function by TRAF3IP3 at the lysosome. J Exp Med. 2018;275:2463–2476. doi: 10.1084/jem.20180397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachari M, Gudmundsson SR, Li Z, Manifava M, Shah R, Smith M, Stronge J, Karanasios E, Piunti C, Kishi-Itakura C, et al. Selective Autophagy of Mitochondria on a Ubiquitin-Endoplasmic-Reticulum Platform. Dev Cell. 2019;50:627–643.:e625. doi: 10.1016/j.devcel.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Liu Z, Liu J, Ren JX, Sun TS. Mitochondrial DNA induces inflammation and increases TLR9/NF-kappaB expression in lung tissue. Int J Mol Med. 2014;33:817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the lead contact (pverginis@bioacademy.gr) upon reasonable request. The RNAseq data have been deposited in the European Genome-Phenome Archive, which is hosted by the EBI, under accession number EGAS00001004470.