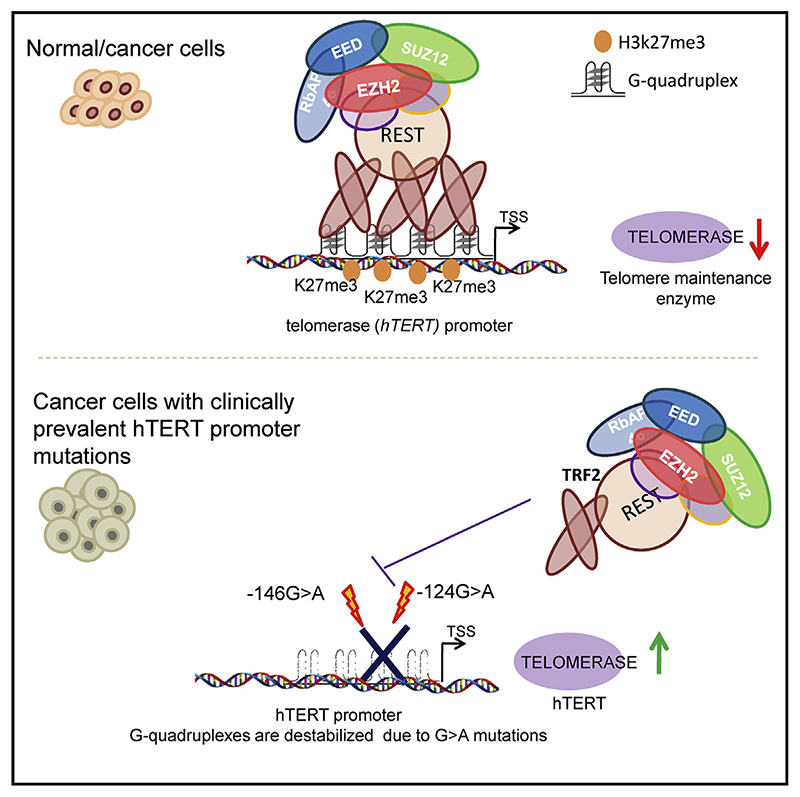
Summary
Human telomerase reverse transcriptase (hTERT) remains suppressed in most normal somatic cells. Resulting erosion of telomeres leads eventually to replicative senescence. Reactivation of hTERT maintains telomeres and triggers progression of >90% of cancers. However, any direct causal link between telomeres and telomerase regulation remains unclear. Here, we show that the telomere-repeat-binding-factor 2 (TRF2) binds hTERT promoter G-quadruplexes and recruits the polycomb-repressor EZH2/PRC2 complex. This is causal for H3K27 trimethylation at the hTERT promoter and represses hTERT in cancer as well as normal cells. Two highly recurrent hTERT promoter mutations found in many cancers, including ~83% glioblastoma multiforme, that are known to destabilize hTERT promoter G-quadruplexes, showed loss of TRF2 binding in patient-derived primary glioblastoma multiforme cells. Ligand-induced G-quadruplex stabilization restored TRF2 binding, H3K27-trimethylation, and hTERT re-suppression. These results uncover a mechanism of hTERT regulation through a telomeric factor, implicating telomere-telomerase molecular links important in neoplastic transformation, aging, and regenerative therapy.
Graphic abstract.
Schematic representation of sample collection and analysis. The figure was created using BioRender.com
Introduction
Telomeres are nucleoprotein complexes that protect chromosome ends. In humans, the reverse transcriptase subunit (hTERT) of telomerase, necessary for telomere synthesis, is repressed in most adult somatic cells (Blackburn et al., 2006; Cech, 2004; Cong et al., 2002; Shay and Wright, 2019). Loss of hTERT repression resulting in increased telomerase activity is instrumental for telomere maintenance, which aids cancer initiation and progression in >90% of human cancers (Artandi and DePinho, 2010; Cao et al., 2002; Pandita et al., 2015; Shay and Bacchetti, 1997). Although these suggest direct molecular links between hTERT regulation and telomeres—possibly through telomeric factors—this remains unclear.
Recent work by us and others show non-telomeric DNA binding of telomeric proteins telomere-repeat-binding-factor 1 (TRF1), telomere-repeat-binding-factor 2 (TRF2), and RAP1 (Martinez et al., 2010; Mukherjee et al., 2018, 2019a; Paeschke et al., 2005; Sarthy et al., 2009; Simonet et al., 2011; Yang et al., 2011). We found ~20,000 TRF2 binding sites genome-wide, a large fraction of which comprised non-duplex structures called G-quadruplexes (Mukherjee et al., 2019a). Binding of TRF2 to DNA secondary structures including DNA and RNA G-quadru-plexes were implicated in multiple studies (Baker et al., 2009; Be-narroch-Popivkeretal.,2016; Biffietal.,2012; Foucheetal.,2006; Islam etal., 2014; Mishraet al., 2016; Pedrosoetal., 2009; Purohit et al., 2018; Rhodes and Lipps, 2015). Furthermore, other telomere-binding proteins like heterochromatin protein 1 (HP1)-alpha, TIN2, and POT1, were found to interact with G-quadruplexes (Baumann and Price, 2010; Chow et al., 2018; Pike et al., 2019; Roach et al., 2020; Smogorzewska et al., 2000; Zaug et al., 2005). Because the hTERT promoter harbors multiple G-quadruplex-forming sequences (Li et al., 2017; Lim et al., 2010; Monsen et al., 2020; Palumbo et al., 2009; Saha et al., 2017; Yu et al., 2012), we asked whether TRF2 associates with the hTERT promoter and if this affects hTERT regulation.
Two somatic hTERT promoter mutations (G > A at the –124th or –146th bp from the translation start site), highly recurrent in multiple cancers including glioblastoma multiforme (GBM) (>80%), melanomas (>70%), hepatocellular (>40%), and urothelial bladder (>50%) carcinomas, induce hTERT reactivation (Horn et al., 2013; Huang et al., 2013; Killela et al., 2013; Liu et al., 2016). Both mutations are within G-quadruplex-forming sequences and destabilized G-quadruplex in solution (Kang et al., 2016). Here, we show these mutations disrupt TRF2 interaction with hTERT promoter G-quadruplexes. As a result, TRF2-induced suppression of hTERT was lost in glioblastoma patient-derived cells, melanoma, and hepatocellular carcinoma cells. In the presence of G-quadruplexes stabilizing ligands, TRF2 binding was regained, re-suppressing telomerase across cells harboring hTERT promoter mutations.
Results
TRF2 directly binds the hTERT promoter and regulates hTERT expression and telomerase activity in cancer and normal cells
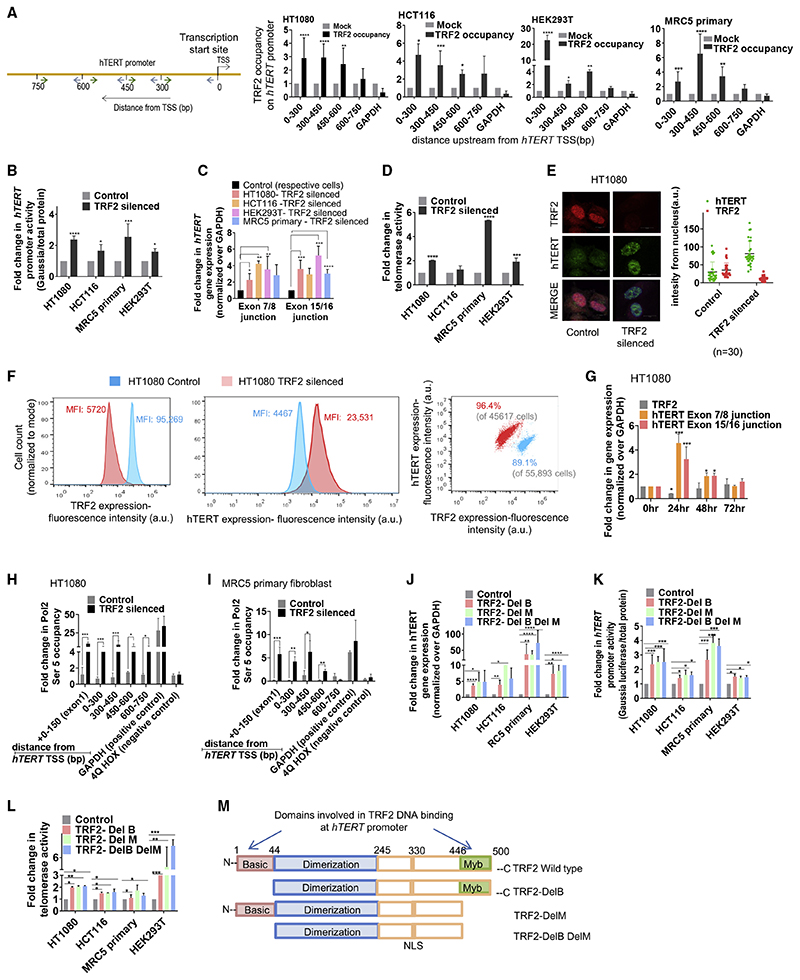
Reads from TRF2 chromatin immunoprecipitation sequencing (ChIP-seq) peaks (recently reported by us) (Mukherjee et al., 2019a) mapped to the hTERT promoter (Figure S1A). TRF2 binding on the hTERT promoter spanned from the transcription start site (TSS) up to ~600 bp upstream in human cancer (fibrosarcoma HT1080 and colon HCT116 cells) as well as normal (MRC5 primary fibroblasts) and immortalized (embryonic kidney HEK293T) cells (Figure 1A). Normal somatic cells are known to have low telomerase activity. We also noted this on comparing hTERT expression and telomerase activity across the four cell types (Figures S1B and S1C). On finding TRF2 occupancy at the hTERT promoter in the non-cancer cell types as well, we studied the role of TRF2 in telomerase regulation in both normal and cancer cells.
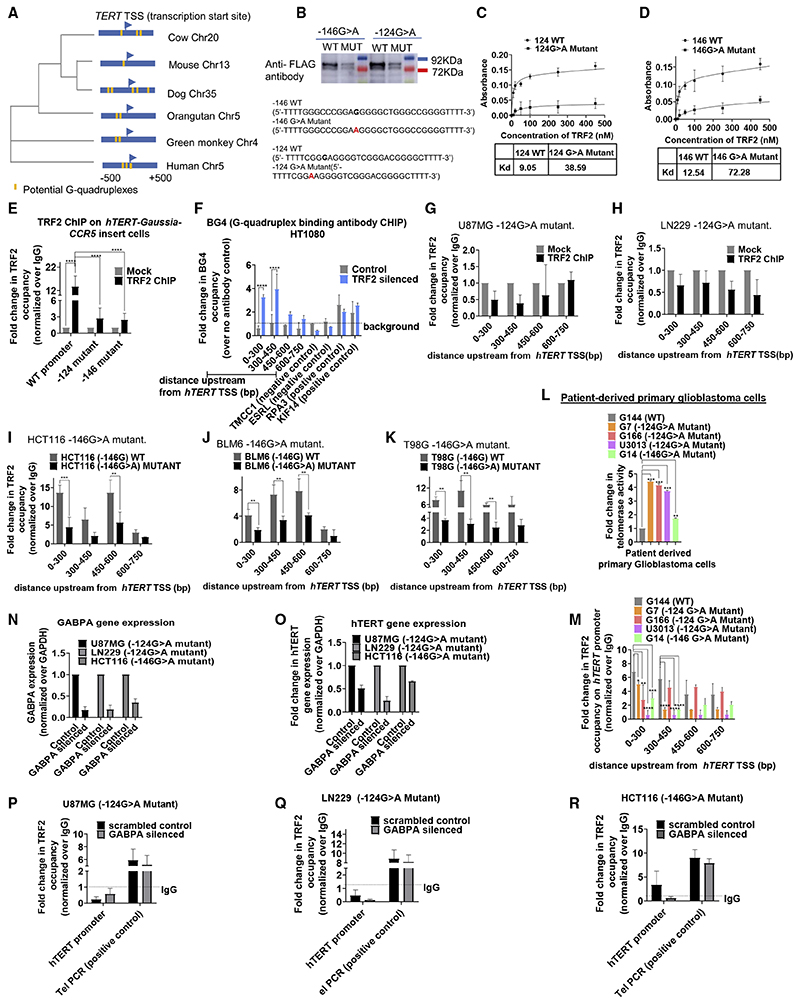
Figure 1. TRF2 binds at the hTERT promoter and transcriptionally represses hTERT .
(A) Scheme showing the hTERT promoter with position of primers designed for ChIP-qPCR (quantitative real-time PCR from ChIP DNA) (indicated by arrows) spanning from 0 to –750 bp of TSS, TRF2 ChIP followed by hTERT promoter-spanning qPCR for TRF2 binding in cancer (HT1080 and HCT116), and MRC5 and HEK293T cells relative to immunoglobulin G (IgG) ChIP (Mock) (see STAR Methods for detail on ChIP DNA qPCR data analysis).
(B) Effect of siRNA-induced TRF2 silencing on hTERT promoter activity in cells 48 h after transfection; +33 to –1,267 bp hTERT promoter cloned upstream of Gaussia luciferase. Cells treated with scrambled siRNA as control.
(C) Effect of TRF2 silencing on hTERT expression; functional (exon 7/8) and full (exon 15/16) transcripts. Fold change normalized over respective cells treated with scrambled siRNA control.
(D) Effect of TRF2 silencing on telomerase activity quantified using telomerase-repeat-amplification-protocol (TRAP) followed by ELISA (see STAR Methods); signal normalized over scrambled treated cells (control). HCT116 cells had relatively high telomerase activity in control than other cells, and the increase following TRF2-silencing was modest.
(E) Immunofluorescence staining of hTERT and TRF2 protein in HT1080 cells. TRF2 and hTERT were stained using Alexa fluor-594 (red signal) and Alexa fluor-498 (green signal), respectively. Quantification of nuclear signal (marked by DAPI, blue) from 30 cells (n = 30) shown in respective right panels.
(F) Flow cytometry using dual staining for hTERT and TRF2 in HT1080 control (scrambled siRNA-treated) and TRF2-silenced cells. Mean intensity of fluorescence (MIF) for hTERT and TRF2 is shown (left and center panel); right panel shows total cell populations monitored: 89.1% of 55,893 cells were analyzed (gated) for control (scrambled treated) cells (with higher TRF2 and relatively low hTERT); 96.4% of 45,617 cells were analyzed for TRF2-silenced cells (lower TRF2 and relatively high hTERT). The cell counts were normalized to respective modes for comparative representation in the left and center panels.
(G) Expression of TRF2 and hTERT(exon7/8 and exon15/16) 24,48, and 72 h following TRF2 siRNA treatment in HT1080 cells. The siRNA complex was removed 6 h after initial transfection.
(H and I) Pol2 (Ser5) occupancy spanning hTERT promoter following TRF2 silencing in HT1080 (H) and MRC5 cells (I). Cells treated with scrambled siRNA as control.
(J–L) Expression of the full-length hTERT transcript (exon 15/16) (J) and hTERT promoter activity (K) and telomerase activity (L) following expression of TRF2 deletion mutants. Results were normalized to untransfected control cells in each case.
(M) Scheme showing the full-length and mutant forms of TRF2 used in the study.
All error bars represent ± SDs from mean. p values calculated by paired/unpaired t test and two-way ANOVA in (G) and (J)–(L) (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001).
hTERT promoter activity (from +33 to –1,267 bp promoterluciferase reporter construct in plasmid) was enhanced on small interfering RNA (siRNA)-mediated TRF2 silencing in all the four cell types (Figure 1B). Silencing of TRF2 upregulated hTERT expression (both the reverse transcriptase [exon 7/8] and the full-length [exon 15/16] transcripts) (Figure 1C) and telomerase activity across cell lines (Figure 1D).
Immunofluorescence (IF), following TRF2 silencing, revealed 2-to 3-fold enhanced hTERT within nucleus in HT1080 (Figure 1E). In fluorescence-activated cell sorting (FACS) on TRF2 silencing, the mean fluorescence intensity (MFI) of hTERT increased by ~5.2-fold relative to control (scrambled treated) cells (Figure 1F). TRF2 silencing experiments were performed using previously published TRF2 siRNA and confirmed using western blots (Figure S1D).
Further, to confirm direct impact of TRF2 on hTERT expression, TRF2 was first depleted using siRNA in HT1080 cells, which gave enhanced hTERT expression (see STAR Methods for details). Thereafter, cells were maintained for 72 h with no further siRNA addition when TRF2 levels gradually increased—concomitant decline in hTERT was evident (Figure 1G). As expected, Pol-II (initiation RNA polymerase phospho-Ser5) occupancy increased significantly at the hTERT promoter, including exon 1 (0 to +150 bp from TSS), following TRF2 silencing in both HT1080 and MRC5 cells (Figures 1H and 1I). Taken together, these results suggested transcriptional control of functional hTERT by TRF2. The antibody used for hTERT was confirmed by FACS and IF in super-telomerase cells that constitutively overexpress telomerase (characterized earlier) (Cristofari and Lingner, 2006) (Figure S1E).
Both MYB and basic domains of TRF2 are necessary for transcription regulation of hTERT
Overexpression of FLAG-tagged TRF2-DelM (lacking C-terminal-Myb [M] domain), TRF2-DelB (lacking N-terminal-basic [B] domain), or TRF2-DelB-DelM (lacking both B and M domains) mutants gave enhanced expression of the hTERT full-transcript (Figure 1J), enhanced promoter (Figure 1K), and telomerase activity (Figure 1L) in HT1080, HCT116, MRC5, and HEK293T cells (scheme comparing TRF2 deletion mutants with wild-type TRF2 shown in Figure 1M; dimerization domain was common in all). ChIP with anti-FLAG antibody following expression of the FLAG-tag-TRF2 deletion mutants in HT1080 cells did not show occupancy of TRF2-DelM, TRF2-DelB, or TRF2-DelB-DelM at the hTERT promoter (Figure S1F). Expression of the deletion mutants was confirmed in each case using anti-FLAG antibody (Figure S1 G).
Further, occupancy of the full-length endogenous TRF2 at the hTERT promoter was significantly reduced following expression of TRF2-DelB, DelM, or DelB-DelM mutants (Figure S1H). The TRF2 antibody recognized the full-length endogenous TRF2 as well as the TRF2-deletion mutants (Figure S1I). Therefore, in the absence of any binding of the TRF2 deletion mutants at the hTERT promoter (as shown above (Figure S1F), the reduced TRF2 ChIP is likely to result from lack of endogenous full-length TRF2 occupancy on expressing TRF2-deletion mutants.
We next checked the effect of the TRF2-deletion mutants on hTERT repression. All the three TRF2-deletion mutants (TRF2-DelB, DelM, or DelB-DelM) induced hTERT promoter activity, whereas full-length TRF2 expression did not affect hTERT promoter activity significantly (Figure S1J). Expression of the FLAG-tagged full-length TRF2 or the respective TRF2 deletion mutants was confirmed using anti-FLAG antibody (Figure S1 K). Together, these findings show that both the N-terminal B domain and the C-terminal M domain of TRF2 are required for hTERT repression.
Epigenetic state of chromatin at the hTERT promoter is TRF2-dependent
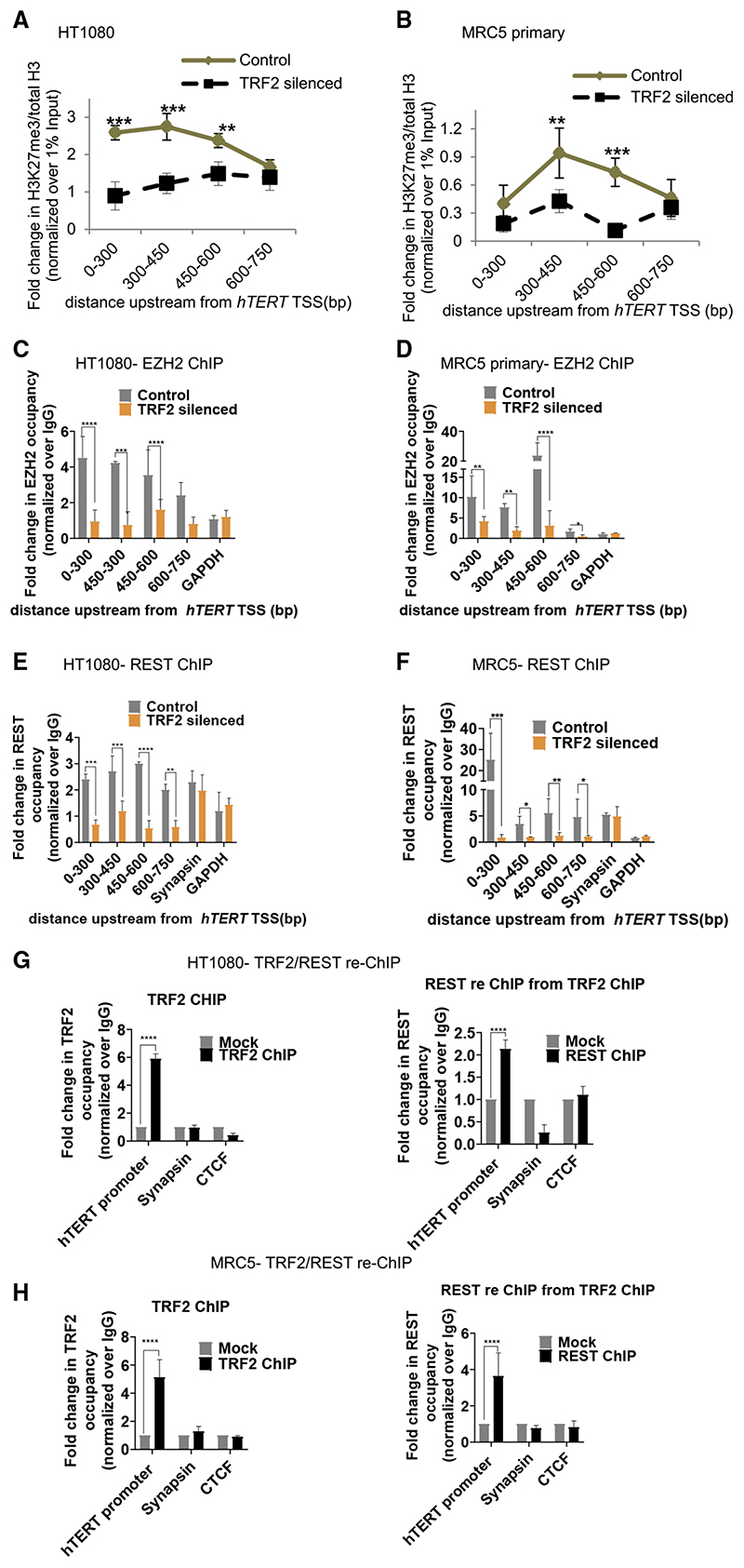
TRF2-mediated promoter histone methylation was observed earlier (Benetti et al., 2008; Mukherjee et al., 2018, 2019a; Ye et al., 2014). Here, we checked histone-activation (H3K4me1 and H3K4me3) and repressor (H3K27me3 and H3K9me3) marks at the hTERT promoter following TRF2 silencing. ChIP-qPCRfor the four histone marks spanning 750 bp upstream of hTERT TSS (as in Figure 1A) showed significant loss of the H3K27me3 repressor in both HT1080 and MRC5 cells (Figures 2A and 2B); H3K4me1, H3K4me3, or H3K9me3 did not change significantly on TRF2 silencing (Figures S2A and S2B).
Figure 2. TRF2 recruits the polycomb repressor complex (PRC2) at the hTERT promoter.
(A and B) Effect of TRF2 silencing on H3K27me3 occupancy (ChIP-qPCR) spanning 0–750 bpofthe hTERT promoterHT1080
(A) and MRC5 (B) cells. Fold change represented as H3K27me3 ChIP over total H3 ChIP, normalized to 1% input in respective cases (see STAR Methods for detail).
(C and D) EZH2 occupancy on the hTERT promoter (spanning 0–750 bp) on silencing TRF2 in HT1080 (C) and MRC5 cells (D). Scrambled siRNA-treated cells as control.
(E and F) REST occupancy on the hTERT promoter on silencing TRF2 in HT1080 (E) and MRC5 (F) cells. Synapsin promoter reported for REST binding was used as control forTRF2-inde-pendent REST occupancy. Scrambled siRNA-treated cells as control.
(G and H) TRF2 ChIP followed by REST re-ChIP: TRF2 ChIP (left panel) and REST re-ChIP (right panel) in HT1080 (G) and MRC5 (H) cells at the hTERT core promoter (+38 to −237 bp). Sya-napsin, where REST binding is independent of TRF2 used as control for TRF2-REST co-binding in TRF2/REST-re-ChIP. GAPDH across replicates was not detectable following reChIP therefore CTCF used as negative control for reChIP experiments. All error bars represent ± SDs from mean values; p values calculated by paired/unpaired t test, for (A)–(F) two-way ANOVA was used (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001).
Recruitment of the polycomb repressor complex (PRC2) at the hTERT promoter induced by TRF2
The EZH2 subunit of the polycomb-repressor-complex-2 (PRC2) catalyzes histone H3K27 trimethylation resulting in gene inactivation (Margueron and Reinberg, 2011; Stern et al., 2017). In both HT1080 and MRC5 cells, TRF2 silencing resulted in loss of EZH2 occupancy at the hTERT promoter (Figures 2C and 2D). However, silencing of EZH2 did not affect TRF2 occupancy (Figure S2C) suggesting TRF2-induced recruitment of EZH2/ PRC2 at the hTERT promoter.
Recruitment of PRC2 by the RE1-silencing-factor (REST) (Dietrich et al., 2012; McGann et al., 2014) and REST-dependent gene silencing has been reported (Bruce et al., 2004). Further, interaction of REST with TRF2 was reported (Mukherjee et al., 2018, 2019a; Zhang et al., 2011). Here, we tested if TRF2 recruited REST to the hTERT promoter. TRF2 silencing, in HT1080 and MRC5 cells, gave reduced REST association at the hTERT promoter (spanning 750 bp) showing TRF2-depen-dent REST occupancy (Figures 2E and 2F). This was supported by intracellular TRF2-REST interaction in HT1080 cells (Hussain et al., 2017) and in MRC5 cells shown here using co-immunoprecipitation (coIP) of REST with anti-TRF2 antibody (Figure S2D).
TRF2 silencing resulted in loss of REST occupancy (Figures 2E and 2F) whereas REST silencing did not reduce TRF2 binding at the hTERT promoter (Figure S2E). Further, REST-reChIP, from the TRF2-ChIP fraction, in HT1080 and MRC5 cells confirmed TRF2-REST association at the hTERT promoter (Figures 2G and 2H). The synapsin promoter reported for REST binding, but not TRF2 (Mukherjee et al., 2018, 2019a), was used as control: REST-reChIP as expected was negative for the synapsin promoter. Reverse of this, i.e., TRF2-reChIP following immunoprecipitation with anti-REST antibody confirmed TRF2-REST co-binding (Figure S2F). TRF2-reChIP was negative for the synapsin promoter as expected.
Overexpression of the TRF2-delB orTRF2-delM deletion mutants resulted in loss of REST occupancy from the hTERT promoter (Figure S2G); whereas REST occupancy at the synapsin promoter (where REST and TRF2 do not interact) remained unaffected. Together, these showed full-length TRF2 binding at the hTERT promoter is required to engage the EZH2/PRC2-REST repressor complex. This is causal for the H3K27me3 modification inducing restrictive chromatin, which suppressed hTERT expression.
CoIP of TRF2 with REST was clear (Figure S2D). As reported earlier (Dietrich et al., 2012), we confirmed coIP of REST with EZH2 (Figure S2H). However, coIP of EZH2 with TRF2 was not evident (data not shown). Therefore, it is likely that TRF2 recruits REST and EZH2 association is through REST, suggesting a multi-protein complex where direct TRF2-EZH2 binding is relatively weak for detection by coIP.
TRF2 association at the hTERT promoter is independent of telomeres
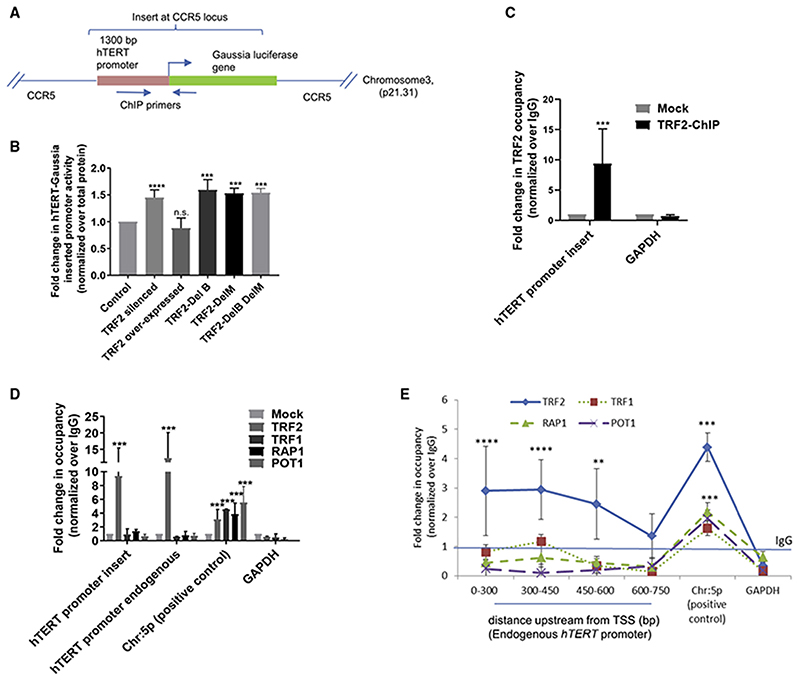
We inserted a hTERT promoter Gaussia luciferase reporter at the CCR5 safe-harbor locus, 46 Mb away from the nearest telomere, by CRISIP/Cas9-mediated editing in HEK293T cells (Figure 3A). Luciferase expression from the reporter was enhanced on silencing TRF2 or on overexpression of the TRF2 deletion mutants, whereas overexpression of full-length TRF2 had no significant effect on hTERT expression (Figure 3B). TRF2 binding at the inserted hTERT promoter was clearly observed (Figure 3C). ChIP-qPCRwas performed using primers specific to the inserted loci (+113 to −196 bp of TSS, indicated in Figure 3A).
Figure 3. TRF2 binding on hTERT promoter is independent of telomeres.
(A) Scheme showing insertion of Gaussia luciferase downstream ofthe hTERT promoter (+33 to –1,267 bp) at CCR5 locus using CRISPR/Cas9-mediated editing in HEK293T cells (see Supplemental information for characterization of cells). Position of primers designed for ChIP-qPCR indicated by arrows.
(B) Effect of TRF2 silencing or expression of full-length TRF2 or TRF2-deletion mutants TRF2-DelB, TRF2-DelM, and TRF2-DelB-DelM on hTERT-promoter Gaussia luciferase activity relative to untreated control cells. Normalized using total protein in each case.
(C) qPCR following TRF2 ChIP at the inserted-hTERT promoter at CCR5 locus using primers shown in scheme above (A); normalized over mock (IgG). GAPDH promoter was used as negative control for TRF2 occupancy.
(D and E) qPCRfollowing ChIPforTRF1, POT1, and RAP1: at the CCR5-locus-inserted-hTERT promoterand the endogenous hTERT promoter (+38to −237 bp) in HEK293T cells(D) and spanningthe endogenous hTERT promoter in HT1080cells (E). Chromosome 5p region 100 kb downstream ofthehTERT locus reported for physical association with telomeres by looping was used as positive control and GAPDH as negative control.
All error bars represent ± SDsfrom mean values; pvalues calculated by paired/unpairedttest; for(B), (D), and (E) two-wayANOVAwas used (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001).
We reasoned interaction with telomeres by looping (Kim et al., 2016) would show presence of the shelterin factors POT1, TRF1, and RAP1, along with TRF2, at the inserted promoter (at the CCR5 locus) in HEK293T cells. In contrast to TRF2, binding of the other shelterins POT1, TRF1, or RAP1 was not found (Figure 3D). However, occupancy of POT1, TRF1, or RAP1 was observed ~100 kb downstream of the hTERT locus reported to engage telomeres (Kim et al., 2016), which we used as positive control. Telomeric binding of POT1, TRF1, RAP1, and TRF2 was confirmed independently using telomere-specific ChIP-PCR followed by dot-blot (Figure S3A; STAR Methods). Therefore, it is unlikely that the inserted hTERT promoter at the CCR5 locus (46 Mb away from telomeres) is bridged to the telomeres through looping interactions.
Next, we checked if TRF2 binding at the endogenous hTERT promoter was from telomeres through looping. Arguing as above, we tested occupancy of POT1, TRF1, and RAP1. Although TRF2 occupancy was clear (Figure 1A), we did not find POT1, TRF1, or RAP1 up to −750 bp of the endogenous hTERT promoter in HT1080 cells (Figure 3E); however, their binding in the region reported to engage telomeres by looping (Kim et al., 2016) was clear. Telomeric binding of POT1/TRF1/ RAP1 was confirmed independently by ChIP-qPCR followed by dot-blot (Figure S3B; STAR Methods). Therefore, TRF2 association at the hTERT promoter was unlikely from telomere looping.
Although recruitment of RAP1 to TRF2 sites was noted earlier (Janouskova et al., 2015; Sarthy et al., 2009), we did not observe this in the case of the hTERT promoter. This is consistent with ChIP-seq showing that TRF2 and RAP1 binding is exclusive in a substantial number of sites suggesting all TRF2 binding sites may not recruit RAP1 (Yang et al., 2011).
TRF2 binding at the hTERT promoter was dependent on G-quadruplex
The hTERT promoter harbors an unusually high number of tandem G-quadruplex-forming sequences (31–35, 41) (Figure S4A). This is evolutionarily conserved, because other vertebrates also have putative G-quadruplexes within 500 bp of TERT TSS (Figure 4A). Interaction of TRF2 with G-quadruplex was reported by others and us (Biffi et al., 2012; Islam et al., 2014; Mishra et al., 2016; Mukherjee et al., 2019a; Pedrosoetal.,2009; Purohit et al., 2018; Rhodes and Lipps, 2015). Here, we selected two hTERT promoter G-quadruplex-forming tracts (Figures S4A and S4B). The two tracts harbored the mutations (−124 bp (G > A) and −146 bp (G > A) (Figure S4A) frequently found in cancers, including GBM (Horn et al., 2013; Huang et al., 2013; Killela et al., 2013; Liu et al., 2016), that destabilized the G-quadruplexes in vitro (Kang et al., 2016) (Figure S4B).
Figure 4. TRF2-induced repression of hTERT is G-quadruplex-dependent.
(A) Phylogenetic tree based on the sequence spanning ±500 bp of the TERT TSS across vertebrates. Presence and position of putative G-quadruplexes (configuration: stem of three Gs and loop size up to 15 bases) in respective organisms is shown in yellow.
(B) Oligonucleotide pull-down from cell lysate of HT1080 cells expressing FLAG-tagged TRF2; 5’-biotin-tagged oligonucleotides from hTERT wild-type (WT) or with mutations (MUT) at the −124th or −146th position were used for pull-down followed by western blot and probed using anti-FLAG antibody. Sequence of respective WT or MUT (base substitution shown in red), with Illi overhangs to minimize steric interactions because of biotin or on ELISA plate (C and D below) given in the bottom panel.
(C and D) ELISA experiments using biotin-tagged hTERT promoter oligonucleotides for WT and the corresponding G > A mutation and increasing concentrations of purified TRF2 protein, WT with −124G > A mutant (C), and WT with corresponding 146G > A mutation (D). Significance for each point was calculated by paired t test, p value across all was p < 0.0001 in both (C) and (D).
(E) qPCR following TRF2 ChIP at the exogenously inserted WT or with −124/−146G > A mutation, hTERT promoter at the CCR5 locus in HEK293T cells relative to IgG (Mock). Scheme of the inserted hTERT promoter with ChIP-qPCR primer positions as in Figure 3A.
(F) qPCR following BG4 ChIP at the hTERT promoter spanning up to 750 bp upstream of TSS: fold-change in BG4 occupancy over experiment using no-antibody control (as per manufacturer’s protocol) shown in TRF2-silenced or scrambled siRNA-treated HT1080 cells (control). Positive and negative controls for BG4 antibody were used as reported earlier.
(G and H) TRF2 ChIP-qPCR spanning 0–750 bp upstream of the hTERT promoter in glioblastoma U87MG (G) and LN229 (H) cell lines with −124G > A promoter mutation relative to IgG ChIP (Mock).
(I–K) TRF2 ChIP-qPCR spanning the hTERT promoter in cancer cell lines with or without the −146G > A promoter mutation: HCT116 cells (I), BLM6 cells (J), or T98G cells (K). Normalized over respective IgG ChIPs (see STAR Methods for details on data analysis). Single base substitutions were made in each case using CRISPR/Cas9-mediated editing.
(Land M) Telomerase activity quantified by ELISA TRAP (see STAR Methods) (L) and TRF2 ChIP-qPCR spanning the hTERT promoter in patient-derived primary glioblastoma cells (M): G144 (wild-type hTERT promoter); G7, G166, U3013 (−124G > A mutant hTERT promoter); and G4 (−146G > A mutant hTERT promoter). (N and O) GABPA (N) and hTERT (O) gene expression following GABPA silencing using qRT-PCR relative to scrambled siRNA control.
(P–R) TRF2 ChIP followed by ChIP-qPCR for TRF2 occupancy at the hTERT mutant promoter following GABPA silencing in U87MG −124G > A mutant (P), LN229 −124G > A mutant (Q), or HCT116 −146G > A(R) mutant cells.
All error bars represent ± SDs from mean values. p values calculated by paired/unpaired t test, for (C)–(F), (L), and (M) two-way ANOVA was used (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001).
To test interaction of TRF2 with hTERT-promoter G-quadru-plex, flag-tagged TRF2 was expressed in HT1080 cells. Lysate from the cells was incubated with biotinylated wild-type or mutant (−124G > A/−146G > A) oligonucleotides (after allowing G-quadruplex formation by the oligonucleotides; see STAR Methods) and pulled down using streptavidin beads. Using anti-flag-antibody we observed enhanced TRF2 interaction with wild-type relative to the mutant oligonucleotides (Figures 4B and S4C). ELISA with recombinant TRF2 showed ~4-to 6fold higher affinity for the hTERT promoter G-quadruplex(es) relative to the respective mutant oligonucleotide(s) that destabilized the G-quadruplex (Figures 4C and 4D).
Next, we used the hTERT promoter Gaussia luciferase reporter at the CCR5 locus (Figure 3A). G > A substitutions were introduced either at the −124th or the −146th positions from translation start site of hTERT. TRF2 occupancy at the inserted hTERT promoter was significantly depleted for both the substitutions relative to the unsubstituted promoter (Figure 4E). As expected, TRF2 occupancy at the endogenous hTERT promoter remained unaltered in these cells (Figure S4D).
Although sequence tracts at the hTERT promoter that form G-quadruplexes in solution were shown, in vivo evidence for G-quadruplex formation has not been reported. We directly tested for in vivo presence of G-quadruplex at the endogenous hTERT promoter by ChIP using the reported G-quadruplex-binding antibody BG4 (Hansel-Hertsch et al., 2018). Surprisingly, BG4 occupancy at the hTERT promoter was not detectable (Figure 4F). This could be because the G-quadruplexes were bound by pro-tein(s) that restrict BG4 binding, as mentioned by authors earlier (Hansel-Hertsch et al., 2018). Therefore, we tested BG4 binding after silencing TRF2. In cells lacking TRF2, occupancy of BG4 on the hTERT promoter was evident (Figure 4F).
The conformation of the hTERT promoter G-quadruplex remained intact in presence of recombinant purified TRF2 (Figure S4E). Ligands that stabilize G-quadruplex formation induced TRF2 binding whereas mutations that destabilized G-quadru-plexes showed reduced/loss in TRF2 occupancy (described below). Further, multiple groups, including earlier work by us, show TRF2 binds/supports G-quadruplex formation (Biffi et al., 2012; Hussain et al., 2017; Mishra et al., 2016; Mukherjee et al., 2019a; Pedroso et al., 2009; Purohit et al., 2018; Rhodes and Lipps, 2015; Traczyk et al., 2020). Therefore, it is unlikely that TRF2 adversely affected G-quadruplex formation at the hTERT promoter excluding BG4 binding. Together, these results support in vivo G-quadruplex-dependent TRF2 binding at the hTERT promoter.
TRF2 occupancy is lost in cancers with hTERT promoter mutations
Two GBM cell lines, U87MG and LN229, both carrying endogenous −124G > A hTERT promoter mutation (Patil et al., 2015), were tested first. TRF2 binding up to −750 bp of the hTERT promoter was not detectable in both cell types (Figures 4G and 4H). Moreover, TRF2 overexpression did not result in TRF2 binding at the hTERT promoter in U87MG or LN229 (Figure S4F).
For the −146G > A hTERT promoter mutation, we tested three pairs of cancer cell lines with/without the mutation (a gift from the Tergaonkar laboratory; characterized earlier) (Akıncılar et al., 2016). In HCT116 colon cancer cells, the −146G > A mutation was introduced resulting in telomerase activation as expected (Akıncılar et al., 2016). In BLM6 melanoma and T98G GBM cells, −146G > A was corrected by A > G substitution, which gave telomerase repression (Akıncılar et al., 2016). In all the three cases, we found TRF2 occupancy was significantly reduced at the hTERT promoter with −146G > A mutation relative to the corresponding cell line without this change (Figures 4I–4K).
Loss/gain of H3K27me3 was TRF2-dependent (Figures 2A and 2B). Here, we checked HCT116 cells as a candidate case: loss of H3K27me3 modification from the hTERT promoter in cells with −146G > A mutation (as expected from loss of TRF2) was clearly observed relative to HCT116 cells without the mutation (Figure S4G). Loss of the H3K27me3 repressor mark in multiple cancer cell types with the hTERT promoter mutations has been reported (Akıncılar et al., 2016; Stern et al., 2017).
Next, we studied primary cells obtained from five grade 4 GBM patients. On sequencing the hTERT promoter (38 bp downstream to 237 bp upstream of ATG), we found three cases with −124G > A (G7, G166, and U3013), one case with −146G > A mutation (G14), and one case with no mutation (G144) (STAR Methods). Telomerase activity, as expected, was several-folds higher in GBM cells with either −124/−146 G > A mutation (G7, G166, U3013, or G14) compared to G144 with no hTERT promoter mutation (Figure 4L). TRF2 occupancy at the hTERT promoter was significantly reduced in G7, G166, U3013, and G14 relative to the G144 primary GBM case (Figure 4 M). Therefore, in different cancer cell types, including primary patient-derived GBM, TRF2 binding at the hTERT promoter was lost on G-quadruplex de-sta-bilization in case of −124/−146 G > A mutations.
The −124/−146G > A mutations create novel binding site(s) resulting in binding of the ETS factor GABPA at the hTERT promoter (Akıncılar et al., 2016; Bell et al., 2015; Stern et al., 2017). Therefore, it is possible that GABPA binding excludes/ competes with TRF2 binding at the hTERT promoter with the mu-tation(s). To test this we checked for TRF2 occupancy at the hTERT promoter after silencing GABPA in the hTERT promoter mutant cell lines U87MG, LN229 (−124G > A mutant) and HCT116 (−146G > A mutant as described above) using previously published siRNA against GABPA (Stern et al., 2017). GABPA silencing was confirmed using qRT-PCR (Figure 4N); and hTERT expression was repressed on GABPA silencing as reported earlier (Akıncılar et al., 2016; Stern et al., 2017) (Figure 4O). We found that in absence of GABPA, TRF2 occupancy was not restored (hTERT core promoter +38 to −237 bp) across any of the three cell lines with hTERT promoter mutation(s) (Figures 4P–4R). Therefore, it is unlikely that TRF2 and GABPA compete for binding at the mutant hTERT promoter and further support loss of TRF2 because of de-stabilization of the promoters G-quadruplexes.
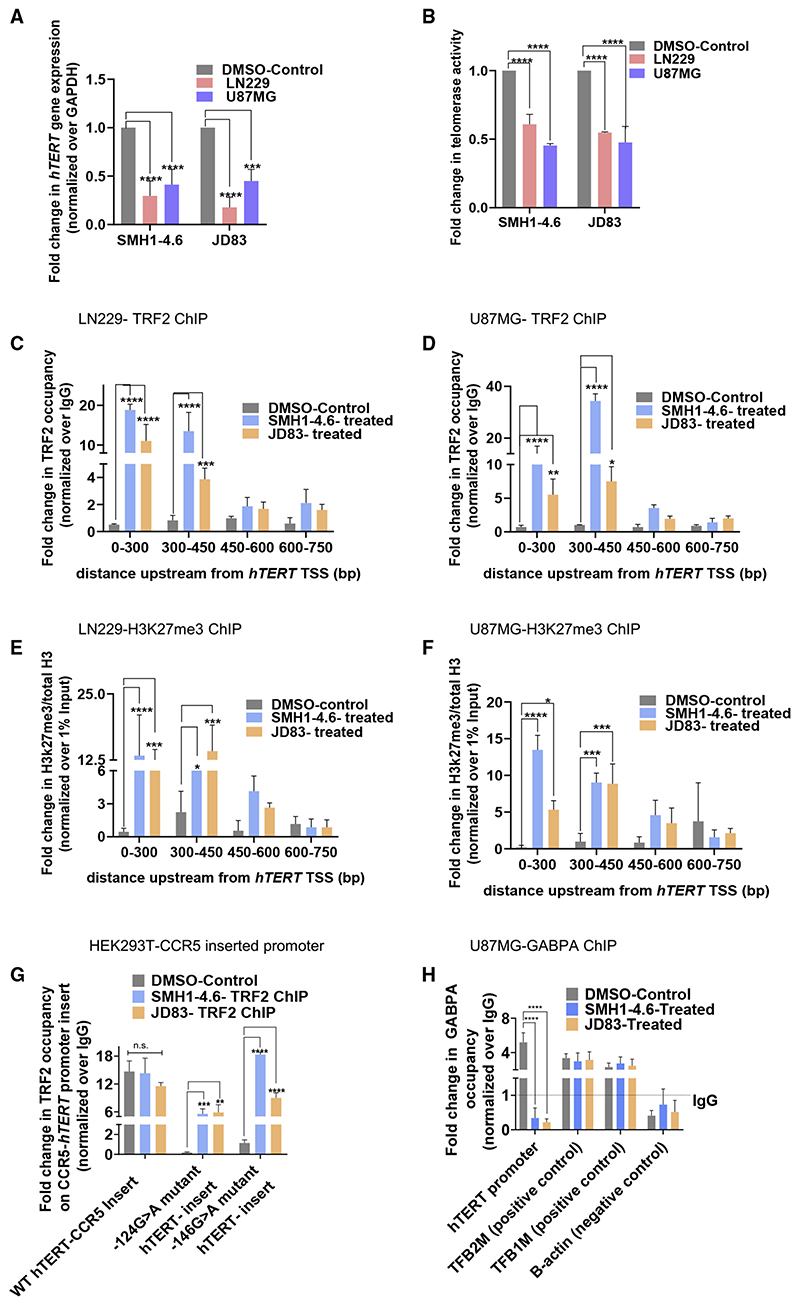
Stabilization of G-quadruplex using ligands reinstates TRF2 binding and re-suppresses activated telomerase
On observing loss of TRF2 binding in cells with hTERT promoter mutations that destabilized G-quadruplex formation, we next tested if stabilization of the G-quadruplex rescued TRF2 binding. Four reported intracellular G-quadruplex binding ligands (Figure S5A; Table S1) were screened using LN229 GBM cells (harboring −124G > A mutation) with reactivated telomerase. Two ligands, SMH1−4.6 and JD83, induced hTERT repression in LN229 cells (Figure 5A). SMH1−4.6 or JD83 treatment gave ~40%–50% repression of hTERT in U87MG cells (Figure 5A) and significant repression of telomerase activity in both U87MG and LN229 cells (Figure 5B). TRF2 expression remained relatively unaltered in presence of the ligands (Figure S5B).
Figure 5. G-quadruplex-binding ligands re-suppress activated hTERT in glioblastoma multiforme with mutations in the hTERT promoter.
(A and B) hTERT expression (A) and telomerase activity (B) in glioblastoma multiforme (GBM) cell lines U87MG and LN229 (with −124G > A hTERT promoter mutation) following treatment with G-quadruplex-binding ligands SMH1−4.6 and JD83 (2.5 μM) or DMSO for 24 h.
(C and D) TRF2 ChIP-qPCR spanning the hTERT promoter following treatment with SMH1−4.6 and JD83 (2.5 μM) or DMSO for 24 h in LN229 (C) or U87MG (D) cells.
(E and F) Fold change in repressor histone mark H3K27me3 by ChIP-qPCR spanning the hTERT promoter following treatment with SMH1−4.6 and JD83 (2.5 μM) or DMSO for 24 h in LN229 (E) or U87MG (F) cells. Fold-change shown with respect to total H3 ChIP; respective ChIPs were normalized to 1% input.
(G) TRF2 ChIP-qPCR at the exogenously inserted CCR5-locus-hTERT promoter with either the wild-type or −124G/−146G > A hTERT promoter mutations in HEK293T cells following treatment with SMH1−4.6 and JD83 (2.5 μM) or DMSO-treated (control) for 24 h. Normalized to IgG ChIP in each case.
(H) GABPA ChIP followed by qPCR at the hTERT core-promoter (+38 to −237 bp); in U87MG (−124G > A mutant) cells following treatment with SMH1−4.6 and JD83 (2.5 μM) or DMSO (control) for 24 h. Normalized to IgG in each case. TFB1M and TFB2M are positive control and B-actin is negative control for GABPA ChIP. All error bars represent ± SDs from mean values. p values calculated by paired/unpaired t test or two-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001).
On treatment with SMH1−4.6 or JD83, TRF2 binding at the hTERT promoter (up to 450 bp from TSS) increased significantly in both LN229 and U87MG cells (Figures 5C and 5D). On observing that the ligands SMH1−4.6 or JD83 restored TRF2 occupancy at the hTERT promoter, we tested their effect on the conformation of the mutant hTERT promoter sequence. In the presence of either SMH1−4.6 or JD83, the mutant oligonucleotides were restored to conformations similar to the wild-type G-quadruplex (Figure S5C). On the other hand, in presence of ligands 260697 or FC4ND08 that did not suppress hTERT expression, the mutant hTERT promoter oligonucleotides did not regain the G-quadruplex conformation (Figure S5D). Unlike SMH1−4.6/JD83, in presence of the ligands 260697 or FC4ND08, TRF2 occupancy at the mutant hTERT promoter was not restored (Figures S5E and S5F). Further, we noted a gain in H3K27me3 repressor modification spanning the hTERT promoter in both cell lines on treatment with SMH1−4.6/JD83 (Figures 5E and 5F), consistent with TRF2-induced gain in H3K27me3 seen earlier (Figures 2A and 2B).
Next, we used the CCR5-locus hTERT promoter Gaussia luciferase reporter cell lines with/without the −124/−146 mutation. Treatment with SMH1−4.6 or JD83 resulted in a significant increase of TRF2 occupancy only at the inserted mutant hTERT promoter—and not at the wild-type hTERT promoter (Figure 5G).
GABPA is known to bind the hTERT mutant promoter (Akıncı-lar et al., 2016; Bell et al., 2015; Stern et al., 2017). Therefore, we next tested the effect of ligand-mediated promoter G-quadruplex stabilization on GABPA occupancy. Following treatment with SMH1−4.6 or JD83 in U87MG (−124G > A) mutant cells, we found significant loss in GABPA occupancy at the hTERT promoter (qPCR with hTERT promoter-spanning primers); GABPA occupancy at the positive controls (TFB1M and TFB2M) genes and negative control (B-actin) gene remained unaltered (Figure 5H). Together, these demonstrate ligand-mediated G-quadruplex stabilization results in recovery of promoter TRF2 occupancy, a gain in histone repressor H3K27me3, loss of GABPA binding, re-suppression of activated hTERT, and telomerase activity in cells with hTERT promoter mutations. These suggest that telomerase reactivation, frequently found in many cancers with −124/−146 hTERT promoter mutations, is likely due to loss of TRF2-induced repression of hTERT.
Discussion
Here, we show TRF2-induced direct recruitment of the EZH2/ PRC2-REST repressor complex at the hTERT promoter. This was causal for repressor histone modifications that maintained non-permissive chromatin at the hTERT promoter resulting in TRF2-mediated repression of hTERT expression and telomerase activity. A hTERT reporter, introduced using CRISPR/Cas9-mediated editing at the CCR5 locus, confirmed TRF2-induced repression of hTERT promoter activity. Using cells with/without specific mutations introduced at the endogenous hTERT promoter (using CRISPR/Cas9-mediated single-base editing) that disrupt the TRF2 binding site, we demonstrate direct TRF2 binding and transcriptional role of TRF2 in hTERT regulation.
TRF2 binding at the hTERT promoter was dependent on the stability of promoter G-quadruplex. Loss of TRF2 binding was observed in cells with hTERT promoter mutation(s) that destabilize G-quadruplex. The resulting decrease in repressor histone H3K27 trimethylation at the hTERT promoter and the enhanced hTERT expression was clear in all the cases we tested.
Depletion of hTERT functional protein was recently reported to induce telomere de-protection (Killedar et al., 2015; Tomlinson et al., 2015). This could also be independent of hTERT catalytic activity and result in growth arrest or apoptosis (Cesare et al., 2013; Perera et al., 2019; Sarthy and Baumann, 2010). G-quad-ruplex binding ligands have been previously reported to inhibit telomerase activity by targeting G-quadruplexes at telomeres (Balasubramanian et al., 2011; Bryan and Baumann, 2011; Gran-otier et al., 2005). Enrichment of the heterochromatin protein HP1-alpha at telomeres was recently noted to affect telomere structure and protection (Chow et al., 2018), possibly through association of HP1-alpha with G-quadruplex formed by telomeric DNA and RNA (transcribed from telomeres called telomeric repeat-containing RNA [TERRA]) (Roach et al., 2020; Smekalova and Baumann, 2013). POT1 independently, or in complex with shelterin proteins TIN2/TPP1, is essential for chromosome-end protection and telomerase processivity through disruption of telomeric G-quadruplexes (Bae and Baumann, 2007; Baumann and Price, 2010; Calvete et al., 2015; Pike et al., 2019; Zaug et al., 2005). Consistent with these findings, we observed growth arrest and cell death in GBM cells, on treatment with G-quadru-plex binding ligands (data not shown). We note this could be through mechanisms, as mentioned above, in addition to hTERT depletion shown here. Further, sustained TRF2 depletion is known to induce telomeric DNA damage resulting in apoptosis and/or arrest in cell growth. In experiments where we silenced TRF2, this was transient, and experiments were conducted within 48 h of silencing. As expected, we did not detect any change in cell viability across cell lines.
Although transcriptional repression of hTERT by TRF2 has not been reported earlier, high TRF2 along with low hTERT levels was observed in CD4-T-lymphocytes and an osteosarcoma-derived cell line (El Maïet al., 2014; Escoffier et al., 2005). Smo-gorzewska et al. (2000), on the other hand, did not find any change in hTERT mRNA or promoter activity on TRF2 induction. In our hands, TRF2 silencing enhanced hTERT expression across cells; however, TRF2 overexpression did not alter hTERT expression significantly. It is therefore likely that chromatin at the hTERT promoter is constitutively repressed (with saturated levels of TRF2 occupancy), which is de-regulated on TRF2 downregulation. This is consistent with the observation showing that across 127 human tissues (NIH Epigenomics Roadmap), the region upstream of hTERT maintains a resting polycomb signature keeping it repressed—disruption of this signature results in telomerase activation (Valentijn et al., 2015).
Constitutive hTERT repression across normal tissue appears consistent with our observations from cells that are not derived from cancer—MRC5 and HEK293T cell lines. On finding TRF2 occupancy at the hTERT promoter in MRC5 and HEK293T cells, we asked whether, and if so how, TRF2 might affect hTERT regulation in non-cancer conditions. Although normal somatic cells are largely known to be devoid of telomerase expression and activity, we found basal expression of hTERT and telomerase activity as has been reported before in normal cells from multiple human tissues (Broccoli et al., 1995; Kyo et al., 1997; Masutomi et al., 2003; Wong et al., 2014). We observed downregulation of TRF2 and resulting loss of repressor histones promoted permissive chromatin changes and enhanced hTERT and telomerase activity. Further, Stern et al. (2017) recently showed EZH2/ PRC2 binding on the hTERT core promoter across cancer cells. Adding to this and to work by others showing epigenetic regulation in cancer (Brien et al., 2016; Mukherjee et al., 2019b), we demonstrate TRF2 binding to be causal for recruitment of EZH2/PRC2 at the hTERT promoter. These results support the role of TRF2 in maintaining the repressor polycomb signature at the hTERT promoter necessary for repressed hTERT and telomerase activity in normal conditions.
Promoter mutation(s) in hTERT was reported to generate site(s) for binding of the ETS transcription factor GABPA (Akıncı-lar et al., 2016; Bell et al., 2015; Li et al., 2015; Stern et al., 2017) leading to telomerase reactivation in cancer cells. We find the following aspects of interest in this context. First, GABPA silencing did not re-instate TRF2 binding (Figures 4P–4R). Second, ligand-induced stabilization of promoter G-quadruplex restored TRF2 occupancy at the mutant hTERT promoter (Figures 5C and 5D). Third, interestingly, GABPA binding at the mutant hTERT promoter was compromised on G-quadruplex stabilization (Figure 5H). Together, therefore, it is likely that GABPA binding (in case of mutations in the hTERT promoter) is facilitated by the permissive chromatin state due to destabilization of the G-quadruple and resultant loss of TRF2 and the PRC2 repressor complex.
Earlier, we reported TRF2 silencing resulted in altered H3K4me3 and H3K4me2 histone marks at the p21 promoter. This was due to the reduced binding of TRF2-mediated REST/ co-REST and LSD1 repressor complex (Mukherjee et al., 2018). In the case of the hTERT promoter, however, we observed significant change in the H3K27Me3 mark through TRF2-dependent engagement of the REST/PRC2 complex, whereas LSD1 occupancy remained unaffected (Figure S5G). The variation in the histone marks at the two promoters is likely due to the difference in the histone modifying complexes that TRF2 recruits at the two promoters. In addition, consistent with this, promoter-specific histone alterations on TRF2 silencing was found in multiple promoters studied by us earlier (Mukherjee et al., 2018, 2019a).
Further, the hTERT promoter was shown to coexist in multiple transient folded/unfolded states (Yu et al., 2012). This is consistent with our results suggesting that, in the presence of stabilizing ligands, the folded G-quadruplex form is promoted. This folded state is likely to result in enhanced TRF2 binding and/or disrupt the GABPA binding sites. In addition, because of reports showing hTERT G-quadruplex stabilization mask Sp1 and CTCF binding to the hTERT promoter (Li et al., 2017; Palumbo et al., 2009), the possibility of G-quadruplex-dependent mechanisms have been discussed (Kim et al., 2016; Li and Tergaonkar, 2016; Rhodes and Lipps, 2015). However, the role of hTERT promoter G-quadruplex-TRF2 interaction in directly determining the fate of telomerase regulation has not been reported earlier.
Consistent with our earlier results in case of p21 and PCGF3 promoters (Hussain et al., 2017; Purohit et al., 2018), we observed that both the N-terminal (B) and C-terminal (M) domains of TRF2 are important for binding at the hTERT promoter. The M domain was reported to be necessary for double-strand DNA binding at the telomeres (van Steensel et al., 1998). The B domain of TRF2 was reported in binding DNA secondary struc-tures—e.g., in t-loop stabilization, double-single strand DNA junctions (Benarroch-Popivker et al., 2016; Fouche et al., 2006; Schmutz et al., 2017), and the G-quadruplex (Hussain et al., 2017; Mukherjee et al., 2019a; Pedroso et al., 2009; Purohit et al., 2018). It is possible, therefore, that binding of TRF2 outside telomeres, and particularly at promoters with DNA secondary structures, involves both M and B domains of full-length TRF2.
Moreover, DNA binding by TRF2 was shown to involve the homodimeric form of TRF2 (Choi et al., 2011; Court et al., 2005; van Steensel et al., 1998). Therefore, because in both TRF2-delB and TRF2-delM, where the dimerization domain is intact, the mutants would dimerize with the endogenous full-length TRF2. These interactions are likely to sequester endogenous TRF2 resulting in loss of TRF2 binding at the hTERT promoter. Similar observations were made for the loss of endogenous TRF2 from telomeres in presence of the TRF2-delM mutant (van Steensel et al., 1998). These resports also support our observations that the mutant forms not only do not bind to the hTERT promoter (Figure S1F) but also inhibit binding of the full-length endogenous TRF2 to the hTERT promoter (Figure S1 H).
With the growing understanding of the function of telomerase in telomere protection in addition to telomere synthesis (Cesare et al., 2013; Perera et al., 2019; Sarthy and Baumann, 2010), mechanisms of telomerase regulation shown here, implicating potential crosstalk with telomeres, might be significant. Recently, we showed non-telomeric TRF2 binding to be telomere-dependent (Mukherjee et al., 2018, 2019a). Together, findings here might be relevant in further understanding how telomerase is regulated, and in turn how telomeres are managed/maintained during telomere-dependent physiological processes like cellular senescence, DNA damage response, cancer, and aging (Abbas and Dutta, 2009; Arnoult and Karl-seder, 2015). The case of telomeres in cancer is particularly relevant. Although further work will be required to test this, based on our findings here, it is possible that establishment of a telomeretelomerase crosstalk through TRF2 (telomeric along with non-te-lomeric binding at the hTERT promoter) is key to how telomeres are managed in cancer cells. In addition, relatively enhanced levels of telomerase and long telomeres are crucial for mainte-nance/survival of pluripotent stem cells (Aguado et al., 2017; Vinayagamurthy et al., 2020; Zou et al., 2017) It is possible, therefore, that TRF2-mediated hTERT regulation, linked to telomeres, is of significance in pluripotency.
Kim et al. (2016) showed looping of the 5p chromosome telomere to a region ~1.2 Mb away and 100 kb downstream of hTERT. This loop further engaged the hTERT promoter, and as a consequence, authors concluded telomere-bound TRF2 was physically associated to the hTERT promoter. However, direct TRF2-dependent hTERT transcription was not studied. We show direct binding of TRF2 to the hTERT promoter that is independent of telomere looping. First, we reasoned telomere-association would show binding of other telomeric factors like TRF1, RAP1, and POT1 at the hTERT promoter along with TRF2—this was not the case (Figure 3E). Second, considering the likelihood of telomere looping to diminish with physical distance, we inserted an exogenous hTERT promoter-reporter ~46 Mb away from the nearest telomere. Here, again, TRF2 binding was clear whereas other telomere-bound factors were absent (Figures 3C and 3D). This is consistent with earlier work showing non-telo-meric TRF2 binding throughout the genome (Mukherjee et al., 2018,2019a). Therefore, it is likely that both telomere-dependent (Kim et al., 2016) and telomere-independent mechanisms of TRF2 interaction regulate hTERT. Further work will be required to understand in what contexts these mechanisms work, particularly with respect to aging (telomere shortening).
In conclusion, results showing TRF2-induced re-suppression of hTERT using small molecule ligands in GBM and other cancers offer potential therapeutic opportunity. We show mechanisms that maintain hTERT in a repressed state in normal cells. Deregulation of which induced hTERT reactivation in cancer cells. Together, these results suggest direct molecular links between telomeres and telomerase that might be critical in advancing the understanding of cell-intrinsic functions including neoplastic transformation, aging, and pluripotency/differentiation.
Star★Methods
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TRF2 rabbit polyclonal | Novus | Cat#NB110−57130; RRID:AB_844199 |
| TRF2 mouse monoclonal | Abcam | Cat#4A794 ab13579; RRID:AB_300474 |
| hTERT rabbit monoclonal Y182 | Abcam | Cat#ab32020; RRID:AB_778296 |
| REST rabbit polyclonal | Millipore | Cat#17−641; RRID:AB_11212617 |
| Histone H3 rabbit polyclonal | Abcam | Cat# ab1791; RRID:AB_302613 |
| H3K4me1 rabbit polyclonal | Abcam | Cat# ab8895; RRID:AB_306847 |
| H3K4me3 mouse monoclonal | Abcam | Cat# ab1012; RRID:AB_442796 |
| H3K27me3 mouse monoclonal | Abcam | Cat# ab6002; RRID:AB_305237 |
| H3K9me3 rabbit polyclonal | Abcam | Cat# ab176916; RRID:AB_2797591 |
| BG4 G4 specific antibody monoclonal | Millipore | Cat# MABE917; RRID:AB_2750936 |
| EZH2 rabbit polyclonal | Cell Signaling Technologies | Cat#4905; RRID:AB_2278249 |
| GAPDH mouse monoclonal | Santacruz | Cat#6C5 SC−32233; RRID:AB_627679 |
| TRF1 mouse monoclonal | Novus | Cat# NB110−68281; RRID:AB_1111093 |
| RAP1 mouse monoclonal | Santa Cruz | Cat#4C8/1 SC−53434; RRID:AB_630189 |
| POT1 mouse monoclonal | Santacruz | Cat#M1P1H5 SC−81711; RRID:AB_1128696 |
| Monoclonal anti-FLAG M2 Mouse antibody | Sigma | Cat# F1804; RRID:AB_262044 |
| LSD1 primary anti-rabbit antibody | Cell Signaling Technologies | Cat#2139; RRID:AB_2070135 |
| anti-Rabbit-HRP | Cell Signaling Technologies | Cat#7074S; RRID:AB_2099233 |
| anti-Mouse-HRP | Cell Signaling Technologies | Cat#7076S; RRID:AB_330924 |
| anti-rabbit Alexa Fluor® 488 | Molecular Probes, Life Technologies | Cat#A11034; RRID:AB_2576217 |
| anti-mouse Alexa Fluor® 594 | Molecular Probes, Life Technologies | Cat#A11062; RRID:AB_1500656 |
| Critical commercial assays | ||
| Qubit dsDNA HS Assay Kit | Invitrogen | Cat#Q32851 |
| Magnetic Dyna-beads (protein G/A) for immunoprecipitation | Invitrogen | Cat# 10009D |
| FUGENE HD | Promega | Cat# E2311 |
| Amersham Rapid-Hyb buffer | GE healthcare | Cat#RPN1635 |
| TeloTAGGG Telomerase PCR ELISA kit | Sigma Millipore Merck | Cat#11854666910 |
| Experimental models: Cell lines | ||
| HT1080 | ATCC | Cat#ATCC-CCL−121 |
| MRC5 | ATCC | Cat#ATCC® CCL−171 |
| HEK293T | ATCC | Cat#ATCC® CRL−3216 |
| HCT116 | Gift form Prof.Vinay Tergaonkar laboratory | N/A |
| U87MG | Gift from Prof. Ellora Sen laboratory | N/A |
| LN229 | Gift from Prof. Ellora Sen laboratory | N/A |
| Oligonucleotides | ||
| hTERT (+38 to −237) FP-CCAGGCCGGGCTCCCAGTGGAT | This paper | N/A |
| hTERT (+38 to −237) RP-GGCTTCCCACGTGCGCAGCAGGA | This paper | N/A |
| ChIP-q-PCR Primers used for hTERT-gaussia inserted at the CCR5 locus FP GACCGCGCTTCCCACGTGGCGGAG | This paper | N/A |
| ChIP-q-PCR Primers used for hTERT-gaussia inserted at the CCR5 locus RP GCCTCGGCCACAGCGATGCAGATCAG | This paper | N/A |
| Tel PCRFP-CGGTTTGTTTGGGTTTG GGTTTGGGTTTGGGTTTGGGTT | This paper | N/A |
| Tel PCRRP-GGCTTGCCTTACCCT TACCCTTACCCTTACCCTTACCCT | This paper | N/A |
| gRNA sequence GGAGAGCTTGGCTCTGTTGGGGG | This paper | Sigma (custom synthesis) |
| TRF2 siRNA 5’GGCUGGAGUGCAGAAAUAU3’ | This paper | N/A |
| TRF2 siRNA 5’CUGGGCUGCCAUUUCUAAA3’ | This paper | N/A |
| TRF2 siRNA 5’GCUGCUGUCAUUAUUUGUA3’ | This paper | N/A |
| For other oligonucleotides, refer to supplemental information | N/A | N/A |
| Recombinant DNA | ||
| AY10_pS. Donor.R5.TS | Addgene | Cat#100292 |
| hTERT promoter driven Gaussia Luciferase insert construct | Genecopoeia | Cat# HPRM25711-PG04 |
| pX459 v2.0 | Gift from Feng Zhang lab | N/A |
| TRF2 overexpression pCMV6 plasmid | Origene | Cat#PS100001 |
| AY10_pS. Donor.R5.TS | Addgene | Cat#100292 |
| Software and algorithms | ||
| Quadbase 2 server for G-quadruplex detection | Dhapola and Chowdhury, 2016 | http://quadbase.igib.res.in/TetraPlexFinder |
| MUSCLE for sequence alignment | Edgar, 2004 | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| Prism 8 Graphpad Prism | https://www.graphpad.com/scientific-software/prism/ | N/A |
| G-quadruplex binding ligands | ||
| 260697 | Mergny et al., 2002; gift from Jean Francois Riou’s laboratory | N/A |
| FC4ND08 | Collie et al., 2012; gift from Jean Francois Riou’s laboratory | N/A |
| SMH1−4.6 | Hampel et al., 2010; gift from Jean Francois Riou’s laboratory | N/A |
| JD83 | Dash et al., 2008; gift from Jean Francois Riou’s laboratory | N/A |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shantanu Chowdhury (shantanuc@igib.in).
Materials availability
Reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Experimental Model And Subject Details
Source and maintenance details of all cancer, immortalized and primary cell lines used in the study
Cancer cell lines
HT1080 fibrosarcoma cell line was purchased from ATCC (ATCC-CCL−121) and cultured in Modified Eagle’s medium (MEM) supplemented with 10% Fetal Bovine Serum (FBS). HCT116 colorectal cancer cell line was a Gift from Prof. Vinay Tergaonkar’s lab, these were cultured in Dulbecco’s Modified Eagle’s Medium-High Glucose (DMEM-HG) supplemented with 10% FBS with 1X Anti-Anti. U87MG and LN229 Glioblastoma cell lines were a kind gift from Prof. Ellora Sen from NBRC, these were both cultured in DMEM-HG with 1X Glutamax, 1X Anti-Anti (GIBCO) and 10% FBS. All cell lines were maintained at 37 Degree Celsius, with 5% CO2 and 95% humidity.
hTERT promoter mutant isogenic cell lines
Isogneic pairs of HCT116, T98G and BLM cells with their respective hTERT promoter mutant cell lines were a kind gift form Prof. Vinay Tergaonkar’s lab. These were all maintained in Dulbecco’s Modified Eagle’s Medium-High Glucose (DMEM-HG) supplemented with 10% FBS with 1X Anti-Anti. All cell lines were maintained at 37 Degree Celsius, with 5% CO2 and 95% humidity.
Primary and Normal immortalized cell lines
MRC5 Primary fibroblasts were obtained from ATCC (ATCC® CCL−171) and cultured in Modified Eagle’s medium (MEM) supplemented with 10% Fetal Bovine Serum (FBS) and 1x Glutamax with 1X Anti-Anti. HEK293T (human embryonic kidney normal immortalized cell line) was also obtained from ATCC (ATCC® CRL−3216) and cultured in Dulbecco’s Modified Eagle’s Medium-High Glucose (DMEM-HG) supplemented with 10% FBS. All cell lines were maintained at 37 Degree Celsius, with 5% CO2 and 95% humidity.
Primary Glioblastoma cell lines
Primary glioma patient-derived cell lines G144 (51yr old male) and G166 (74yr old female) were as reported (Pollard et al., 2009). U3013 cell line (derived from 78yr old female) (Savary et al., 2013; Xie et al., 2015). G14 and G7 cells were first reported in Fael Al-Mayhani et al. (2009) and Pollard et al. (2009); however no patient information was available in the earlier reports or could be retrieved by us. G144, G166, G14 and G7 were kind gift from Dr. Steve Pollard, Edinburgh. These cells were cultured by Dr. Deo Prakash Pandey’s laboratory as described in Pollard et al., 2009. U3013, was a gift from Prof. Lene Uhrnbom and these were cultured according to protocol stated in Savary et al. (2013) and Xie et al. (2015). All cell lines were maintained at 37°C, with 5% CO2 and 95% humidity.
Confirmation of the hTERT promoter mutations in patient-derived glioblastoma cells
Bases corresponding to −146 or −124 mutation are marked in green (wild-type) or red (in case of mutation) below.
G144
CCTCGCCGTTGGGAGCAATGCTGCCCGTGGGAGCCCAGGGGACCCGGGCACCCGTCCTGCCCCTTCACCTTCCAGCTCCG CCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTCCCGGGTCCCCGGCCCAGCCCCCTCCGGGCCCTCCCAGCC CCTCCCCTTCCTTTCCGCGGCCCCGCCCTCTCCTCGCGGCGCGAGTTTCAGGCAGCGCTGCGTCCTGCTGCGCACGTGG-GAAGCCCCT
No mutations (WT)
G7
GAACTAATAGAGATGTAAGCGTGTTGAAGCGATGGCGGAGGGACTAGGGGACCCGGGCACCCGTCCTGCCCCTTCACCTTC-CAGCTCCGCCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTCCCGGGTCCCCGGCCCAGCCCCTTCCGGGCCCTCC-CAGCCCCTCCCCTTCCTTTCCGCGGCCCCGCCCTCTCCTCGCGGCGCGAGTTTCAGGCAGCGCTGCGTCCTGCTGCGCA CGTGGGAAGCC
−124 C > T/G > A mutation
G166
GAACTAATAGAGATGTAAGCGTGTTGAAGCGATGGCGGAGGGACTAGGGGACCCGGGCACCCGTCCTGCCCCTTCACCTTC-CAGCTCCGCCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTCCCGGGTCCCCGGCCCAGCCCCTTCCGGGCCCTCC-CAGCCCCTCCCCTTCCTTTCCGCGGCCCCGCCCTCTCCTCGCGGCGCGAGTTTCAGGCAGCGCTGCGTCCTGCTGCGCAC GTGGGAAGCC
−124 C > T/G > A mutation
G14
CCGGGGAAAGAACAGGACGCGCTCCCACGTGGCGGAGGGACTGGGGACCCGGGCACCCGTCCTGCCCCTTCACCTTCCA GCTCCGCCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTTCCGGGTCCCCGGCCCAGCCCCCTCCGGGCCCTCC-CAGCCCCTCCCCTTCCTTTCCGCGGCCCCGCCCTCTCCTCGCGGCGCGAGTTTCAGGCAGCGCTGCGTCCTGCTGCGCA CGTGGGAAGCACATTTTATGATAATTTTGACGTTACTTATCATTTTTATACTGCATACATCCTAAGGACAAAAAACAAAAT-TAAATCTGATTATCAACCTTTTCCGCGACAATATTTTTACAC
−146C>T/G>A mutation
U3013
CTCCTTTACAGCCGGACGCGCTTCCACGTGGCGGAGGGACTGGGGACCCGGGCACCCGTCCTGCCCCTTCACCTTCCAGC TCCGCCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTCCCGGGTCCCCGGCCCAGCCCCTTCCGGGCCCTCC-CAGCCCCTCCCCTTCCTTTCCGCGGCCCCGCCCTCTCCTCGCGGCGCGAGTTTCAGGCAGCGCTGCGTCCTGCTGCG-CACGTGGGAAGCCA
−124 C > T/G > A mutation
CRISPR-mediated insertion of hTERT promoter-driven gaussia luciferase in to the CCR5 locus
The hTERT promoter driven Gaussia Luciferase insert construct was obtained from a Genecopoeia promoter reporter clone. The insert sequence was cloned into AY10_pS. Donor.R5.TS. The hTERT promoter donor vector with mutation at −124 position was generated using Quikchange SDM kit (Agilent) according to the manufacturers’ protocol. For cleavage at CCR5 locus a reported gRNA sequence (5’-GGAGAGCTTGGCTCTGTTGGGGG−3’) was cloned into the pX459 v2.0, a gift from Feng Zhang that co-expresses cas9 protein and the gRNA. The gRNA cloned pX459 and the donor vector were co-transfected using FUGENE HD transfection agent according to the manufacturers’ protocol. Starting from 36 hours post transfection, the cells were treated with 2ug/ml puromycin for 3 days for selecting cells that have taken up pX459 plasmid. After growing for 5 days after selection, the cells were seeded into 96 well plates after dilution for clonal selection. 40 clones were screened using gaussia luciferase activity and PCR to find out the positive clones.
Confirmation of CCR5-hTERT-Gaussia insert by sequencing and culture conditions
The inserted regions was PCR amplified with primers aligning to the CCR5 locus region flanking the TERT-Gaussia on both sides and sequenced.
In case of mutations, the inserted promoter was amplified (with primers flanking the CCR5 locus, as mentioned above) and confirmed by sequencing.
The highlighted bases in the following sequences correspond to −146 and −124 respectively from start of sequence (5’ to 3’ in the anti-sense strand). Green designates WT and purple designates mutated bases.
Post confirmation these cells were maintained in Dulbecco’s Modified Eagle’s Medium-High Glucose (DMEM-HG) supplemented with 10% FBS with addition of 1X anti-anti to cells post single cell seeding of CRISPR mutated pooled cell population.
CCR5 TERT -Gaussia insert WT
CGCCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTCCCGGGTCCCCG GCCCAGCCCCCTCCGGGCCCTCCCAGCCC CTCCCCTTCCTTTCCGCGGCC
CCR5 TERT -Gaussia insert −124 C > T/G > A mutation
CGCCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTCCCGGGTCCCCG GCCCAGCCCCTTCCGGGCCCTCCCAGCCCCTCCCCTTCCTTTCCGCGGCC
CCR5 TERT – Gaussia insert −146 C > T/G > A mutation
CGCCTCCTCCGCGCGGACCCCGCCCCGTCCCGACCCCTTCCGGGTCCCCG GCCCAGCCCCCTCCGGGCCCTCCCAGCCCCTCCCCTTCCTTTCCGCGGCC
Methods Details
Analysis of TERT promoter sequence across vertebrates for G-quadruplex
G-quadruplex sequences were identified within ± 500 bp TERT TSS in various mammalian clades using reported Quadbase 2 server which detects G-quadruplexes from sequence (Dhapola and Chowdhury, 2016). Sequence homology and conservation scores were determined using neighbor joining cluster generation algorithm in the publicly available multiple sequence alignment tool MUSCLE (Edgar, 2004).
ChIP (chromatin immunoprecipitation)
ChIP assays were performed as per protocol previously reported in Mukherjee et al. (2018). ChIP assays were performed using relevant primary antibody. IgG was used for isotype control in all ChIP experiments. Three million cells were harvested and fixed for each were fixed with ~1% formaldehyde for 10 min and lysed. Chromatin was sheared to an average size of ~200−300 bp using Biorupter (Diagenode). 10% of sonicated fraction was processed as input using phenol-chloroform and ethanol precipitation. ChIP was performed using 3 μg of the respective antibody incubated overnight at 4°C. Immune complexes were collected using herring sperm DNA-saturated Magnetic Dyna-beads (protein G/A) and washed extensively using a series of low salt, high salt and LiCl Buffers. The Dynabeads were then resuspended in TE (Tris-EDTA pH 8.1) buffer and incubated with proteinase K at 55°C for 1hr. Then, phenolchloroform-isoamyl alcohol was utilized to extract DNA from the proteinase K treated fraction. DNA was precipitated by centrifugation after incubating overnight at −20°C with isopropanol with glycogen and 3M sodium acetate. The precipitated pellet was washes with freshly prepared 70% ethanol and resuspended in nuclease free water. ChIP DNA was further validated by q-PCR method.
Analysis of ChIP experiments-
ForTRF2, REST and EZH2, ChIP-q-PCR was performed with equal amount of DNA (quantified by qubit HS DNA kit) from each ChIP and its respective m([0−9]+)ock (IgG). Thereafter respective mean Ct values were used for calculating fold change over IgG (mock).
For histone ChIP assays, ChIP-qPCR was performed with equal amount of DNA from each histone ChIP and its respective total H3 ChIP. This was normalized to 1% input. The input normalized mean Ct values for ChIP and total H3 were used for calculating fold change over total H3.
In case of BG4 ChIP, since the antibody was produced in E. coli, a no antibody mock control was used. Similar to the other ChIP assays, ChIP-qPCR was performed with equal amount of ChIP DNA and mock immunoprecipitation before calculating fold change over mock.
Tel PCR
Tel PCR was used as a positive control for TRF2 ChIP, as TRF2 is a bonafide Telomere binding protein. Following, Chromatin immunoprecipitation using Anti-TRF2 antibody, we performed a Real-Time PCR with equal amounts of DNA template form ChIP and IgG fractions, using telomere specific primers (O’Callaghan et al., 2008) at an annealing standardized at an annealing temperature of 52.5°C. The ChIP fraction readout was normalized over IgG to confirm if there was substantial TRF2 binding at the telomeres over background.
LSD1 chromatiniImmunoprecipitation
ChIP (Chromatin Immunoprecipitation) was performed using ChIP protocol same as above. LSD1 primary antibody (2139-CST antirabbit) was used −3 μl per pull down along with respective Mock(IgG) immunoprecipitation.
Re-ChIP
For Re-ChIPof TRF2 immunoprecipitated fraction with REST, the above stated ChIP protocol was followed with a starting harvest of 6 million cells with pull-down of TRF2 mouse monoclonal antibody using protein-G-dynabeads. For Re-ChIP, half the pull-down fraction was resuspended in TE buffer with 10mM DTT after the salt buffer washes and incubated for 30 mins at RT. Following this the fraction was centrifuged at 10K rpm at 4°C for 10 mins and supernatant was used as lysate for REST ChIP using REST rabbit monoclonal antibody and pull down using protein A Dynabeads.
Immunoprecipitation of proteins
Six million cells were collected and washed in cold 1X PBS and lysed using RIPA (Sigma) with 1x mammalian Protease inhibitor Cocktail as per manufacturers’ protocol. For immunoprecipitation experiments 1 mg of protein was incubated for 4 hours at 4°C with primary antibody in ratio recommended by manufacturer for immunoprecipitation. The pull-down was performed using Catch and Release co-immunoprecipitation kit (Millipore) as per manufacturer’s protocol.
Immunofluorescence microscopy
Adherent cells were seeded on coverslips and allowed to reach a confluency of ~70%. Cells were fixed using freshly prepared 4% Paraformaldehyde by incubating for 10 min at RT. Cells were permeabilised with 0.5% Triton X−100 (10 min at RT) and treated with blocking solution (5% BSA in PBS) for 2 hr at RT. All the above stated were followed by three washes with ice cold PBS for 5 mins each. Post-blocking, cells treated with relevant antibodies as follows: anti-TRF2 antibody mouse (1:200), anti-hTERT antibody rabbit (1:200) and incubated overnight at 4°C in a humid chamber. Post-incubation, cells were washed alternately with PBS and PBST three times and probed with secondary Ab (rabbit Alexa Fluor® 488(1:1000) / mouse Alexa Fluor® 594 (1:1000)) for 2 hr at RT. Cells were washed again alternately with PBS and PBST three times and mounted with Prolong® Gold anti-fade reagent with DAPI. Images were taken on Leica TCS-SP8 confocal microscope. LEICA LAS-AF software was used to calculate TRF2 and hTERT signal intensity (a.u.) post (region of interest) ROI definitions for nuclear signal.
Immuno-flow cytometry
3 million cells for each condition were fixed using 4% formaldehyde for 10 mins at RT followed by 3 ice cold PBS washes for 5 mins each. Cells were permeabilised using 90% Methanol (pre-chilled) for 5 mins and followed by three ice cold PBS washes for 5 mins each. Dilution of primary antibodies were made (hTERT rabbit and TRF2 mouse) in 1% BSA (in PBS) in 1:250 ratio by volume. Cells were incubated with primary antibody cocktail for 2 hr at RT. Three ice cold PBS washes to cells (10 mins each) were given and secondary antibodies-rabbit Alexa Fluor® 488(1:1000) / mouse Alexa Fluor® 594(1:1000) in 1% BSA (in PBS) were added. Cells were incubated at RT for 1hr and given three ice cold PBS washes (10 mins each). Cells were resuspended in 0.5 mL of PBS and scored for Fluorescence intensity in an Acuuri c6 flow cytometer in the FL1 (488 nm) and FL3 (594 nm) channels. The FCS files were analyzed using Flow-Jo (version 10) software.
Transfections and TRF2 silencing
Cells were transfected in a 1:3 complex of FUGENE HD and DNA/RNA using protocols previously described forTRF2 WT and mutant mammalian expression plasmids and TRF2 siRNA pool was used forTRF2 silencing at 150pMol concentration keeping cells treated with same concentration of scrambled RNA as control; as described in Mukherjee et al. (2018, 2019a). Cells were incubated with transfection complex for 12 hours in media post which a media change was given. Cells were given fresh media changes every 24 hours. All experiments were performed following transient TRF2 silencing for 48hrs.
GABPA silencing
We transfected cells with siRNA against GABPA as published previously by Stern et al. (2015). A pool of four siRNAs used was at a concentration of 100pM, transfected using fugene. Following transfection we confirmed silencing using RT-PCR with primers to assess GABPA expression levels.
TRF2 silencing recovery experiment
Post Transfection with the TRF2 silencing siRNA-FUGENE complex, the transfection complex was removed from the HT1080 cells post 6hrs and fresh media change was given. One well was kept un-transfected; this was collected as 0hrtime point. Transfected cells were collected post 18hrs after first media change; this was 24hr time point. A media change was given to the remaining cells at this stage and 24hr later the 48hr time point cells were collected. A final media change was given to the cells in the remaining well which were collect post next 24 hr as the 72hr time point. All the collected cells were stored in Tirzol and RNA isolation, cDNA synthesis and RT PCR for TRF2 and hTERT gene expressions were performed together.
Gaussia-luciferase assay
Minimal promoter region of TERT (~1300 bp starting from 48 bp downstream of Transcription start site) procured from Genecopoeia-HPRM25711-PG04 (pEZX-PG04.1 vector). Gaussia luciferase kit from Promega was used for detecting secreted Gaussia luciferase signal as per manufacturer’s protocol.
Real time PCR for mRNA expression
Total RNA was isolated using TRIzol® Reagent (Invitrogen, Life Technologies) according to manufacturer’s instructions. RNA was quantified and used for cDNA preparation using Applied Biosciences kit. A relative transcript expression level for genes was measured by quantitative real-time PCR using a SYBR Green based method. Average fold change was calculated by difference in threshold cycles (Ct) between test and control samples. GAPDH gene was used as housekeeping control for normalizing the cDNA concentration of each sample. Each experiment was performed in biological triplicates; where each time control and test conditions were set up separately and each sample was run in q-PCR thrice as technical replicates. Following this the technical replicates were averaged. We calculated the fold change of test over respective control conditions and designated the test condition as “1” for plotting; p values in all cases were calculated taking into conversation the variability of both the control and test readings.
Dot blot analysis
For dot blot analysis, Genomic/ ChIP DNA was denatured at 95°C and dot blotted on N+ hybond membrane (Amersham) in pre -wetted in 2X SSC buffer. The DNA was UV cross-linked. Membranes were pre-hybridized in Rapid-Hyb buffer (Amersham) for 30 min at 37°C. Following this, hybridization with a 24-bp radio-labeled telomeric probe (AATCCC)4 was performed for 4 hr at 37°C and membranes washed with 2XSSC and 0.2XSSC + 0.1% SDS twice at hybridization temperature before exposing overnight on phosphoimager imaging plate. All data were scanned using Bio-Rad Personal Molecular Imager. Data was processed and quantified using ImageJ image analysis software.
Western blotting
For western blot analysis, protein lysates were prepared by resuspending cell pellets in passive lysis buffer/RIPA with 1x mammalian Protease inhibitor Cocktail. Protein was separated using 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon FL, Millipore). After blocking the membrane was incubated with primary antibodies-anti-TRF2 antibody (Novus Biological), anti-TERT antibody (Abcam), anti-REST(Millipore), anti-EZH2(CST) and anti-GAPDH antibody (Santa-cruz). Secondary antibodies, anti-mouse and anti-rabbit HRP conjugates were from CST. The blot was finally developed by using Millipore HRPchem-iluminescence detection kit and images in a GE chemiluminescence imager.
G-quadruplex preparation
5 μM of oligonucleotide in each case was diluted in 10 mM Tris HCl ph7.5 and 140 mM KCl and denatured by heating to 95°C for 5 min and slowly cooled to 25° C for overnight.
Circular dichroism
The circular dichroism (CD) spectra were recorded on a Jasco−810 Spectropolarimeter equipped with a Peltier temperature controller. Experiments were carried out using a 1 mm path-length cuvette over a wavelength range of 200−330 nm. The G-quadruplex formation was induced as described above, and the G-quadruplexes formed were used as such for monitoring the CD spectra. The CD spectra reported are representations of three averaged scans taken at 25°C and are baseline corrected for signal contributions due to the buffer.
In the CD experiments performed with the ligand and TRF2 purified protein. First, the CD spectra ofthe 5uM solution (in quadruplex buffer) of mutant hTERT promoter oligo (G-quadruplex) was recorded. This was followed by taking a blank reading for buffer+ protein/ ligand. Meanwhile, the hTERT promoter mutant oligos were incubated with 5 times higher molar concentration of ligand or protein. The ligand/protein and the oligos were mixed well and then allowed to be incubated for 20min after which their CD spectra were recorded again.
Elisa
5’-biotinylated oligonucleotides were induced to form G-quadruplex as described above. The 5 uM G-quadruplex stock solution was diluted to a final concentration of 50 pM in 1X Tris-buffer saline, 0.1% Tween 20 (TBST) buffer; 20 ul of this was loaded into each well of a 384 well streptavidin coated pre-blocked plate from Thermo Scientific (Pierce) and incubated at 37°C on shaker for 2 hours to allow streptavidin and biotin binding. Following this the excess unbound oligonucleotides was washed off using 1XTBST buffer X3 washes.
TRF2 protein was diluted in 1X Phosphate buffer saline, 0.1% Tween 20 (PBST) buffer and 20ul of dissolved protein was incubated with bound quadruplex (in the wells) in increasing concentration across wells for 2 hours on shaker at 4°C. Unbound protein was washed off with 1X PBST buffer X3 washes. Anti-TRF2 antibody (Novus) was used at 1:1000 dilution (20 μl per well) and incubated for 1 hr at room temperature on shaker. Wells were washed three times with 1X PBST. Alkaline phosphatase conjugated Anti-IgG antibody (Sigma) was used at 1:1000 dilution (20 μl/well) and incubated for 45 minutes at room temperature on shakerand then wells were washed once with 1X PBST and twice with 1X PBS. 20 μl BCIP/NBT substrate was added into each well and absorbance was recorded at 610 nm wavelength for 1hour with 10 min interval on TECAN multimode reader. GraphPad Prism7 was used for analysis.
Telomerase activity using ELISA TRAP
One million cells were lysed using CHAPS lysis buffer provided in the kit, we add RNase inhibitor and protease inhibitor to avoid and protein degradation. Post lysate preparation protein concentration is estimated. Following this all protein samples were diluted to 0.5ug/ul concentration so a uniform volume of 2ul could be picked to add 1ug of lysate for TRAP assay as guided in the protocol. As a precaution we performed a second round of protein estimation here to confirm our dilutions were to an exact 0.5ug/ul concentration. Next telomerase repeat amplification protocol (TRAP) (Kim et al., 1994) PCR was performed using PCR master mix provided in the kit. The primers were AG(GGTTAG)7 sequence repeats 3’biotnylated P1-TS primers. This allows telomerase to add the substrate TTAGGG six-mer substrate on the P1-TS primers, substrate primers a (TTAGGG)6 repetitive sequences-P2. P3 is dioxygenin tagged used to amplify the telomeric sequence to a specific number of cycles. Post PCR as per the protocol conditions, the product is estimated using ELISA performed on streptavidin coated plate using anti-dig-Pod antibody using ROCHE TeloTAGGG Telomerase PCR ELISA kit. The method is well reported (Giri et al., 2010; Shi et al., 2014).
Oligonucleotide-pulldown assay
Total cell lysate of > 2000ug concentration was isolated using RIPA buffer (without SDS) with 1X mPIC. Lysate was pre-cleared for cellular biotin by adding 60ul of Dynabeads MyOne Streptavidin C1 (cat no65001) beads per sample and rotating at 4-degree celsius for 2 hours. Streptavidin beads were then removed using a magnetic stand and the lysate was divided into two equal parts. To one the wild-type biotinylated oligonucleotide was added, while to the other mutant biotinylated oligonucleotide was added, amounting to 50 pmoles in each. The lysate was incubated on rotor with oligonucleotides for 16hrs at 4°C. Thereafter the protein and DNA were cross-linked for 15min in UV crosslinker. Thereafter 100ul of Streptavidin beads were added to each tube post twice washing of beads in 1XPBST. Beads were incubated with cross-liked lysate for 2 hours. Post this, beads were separated on magnetic stand and washed twice in 1X wash buffer (20 mM Tris+10mM NaCl+ Tween 0.1%). Lastly the bound protein was eluted using Elution buffer (1MTris HCl pH6.8+10% SDS+ Bitoin 25mM). The beads were re-suspended in 50ul of elution buffer and heated at 95°C for 5 min, the buffer was then stored in fresh tube, the process was repeated with 50ul of elution buffer. Of this total eluted protein, 60ul was run on SDS-PAGE gel after adding 6X protein loading dye, as in a normal western blot protocol.
TRF2 ChIP-seq coverage on TERT promoter
Sorted Alignment files (BAM) for TRF2 ChIP-seq (Mukherjee et al., 2019a) was visualized using the publicly available software IGVfor Coverage of reads on hTERT promoter with Transcription start site (TSS) defined using transcript variant NM_198253 (RefSeq).
Quantification And Statistical Analysis
All experiments were performed in three biological replicates. Based on the triplicate readings all error bars represent ± standard deviations from mean values; p values calculated. For a comparison of two datasets (control and TRF2 silenced conditions) we compared using paired/unpaired t test. Further multiple comparisons were performed using two-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001). All the calculations were performed using Prism Graphpad, which was also used to plot all datasets.
Supplementary Material
Highlights.
Non-telomeric TRF2 suppresses re-activated human telomerase in glioblastoma cells
PRC2 recruitment depends on the TRF2 hTERT-G-quadruplex interaction
Clinically deleterious hTERT promoter mutations disrupt G4-TRF2 association
G4 stabilization reinstates TRF2-induced hTERT repression in patient glioblastoma
In brief.
Sharma et al. show transcriptional repression of hTERT by non-telomeric TRF2. TRF2 binds hTERT promoter G-quadruplex and recruits the REST/PRC2-complex. High-frequency clinical mutations associated with cancer destabilize hTERT promoter Gquadruplex, compromise TRF2 binding, and result in hTERT re-activation. Stabilization of G-quadruplex reinstates TRF2 binding and hTERT re-suppression in patient-derived glioblastoma cells.
Acknowledgments
We thank Vinay Tergaonkar for the gift of cell lines with or without hTERT promoter mutations, Ellora Sen for GBM cell lines, and Lene Uhrbom and Smitha Sreedheran for U3013 GBM cells. We acknowledge Peter Maillietforsynthesis of G-quadruplex binding ligands used in this study. We are grateful to Balam Singh for laboratory management, Venkatesh Prasad for suggestions on immunofluorescence experiments, and Dr. Debayan Ganguly, Yasmeen Khan, and Dipanjali Sinha for helping with the CD experiments. We thank all members of the S.C. laboratory for their inputs/suggestions and the IGIB HPC and Core Imaging Facility. Research fellowships to S.S., A.K.M., S.S.R., A.S., M.K. (CSIR), S.B. (DBT), and M.V. (DST) are acknowledged. S.L. acknowledges Helse Sør-Øst, Norway and the Medical Student Research Program, University of Oslo, Oslo, Norway for fellowships. This work was supported by research grants from Wellcome Trust/DBT India Alliance Fellowship (IA/S/18/2/504021) to S.C. Support from Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology (DBT) to S.C. and from Helse Sør-Øst, Norway and Medical Student Research Program, University of Oslo, Oslo, Norway to D.P. is also acknowledged.
Footnotes
Author Contributions
S.S. did the TRF2 ChIP experiments and hTERT expression-related experiments, hTERT promoter activity assays, telomerase activity assays, immunofluorescence microscopy, ligand treatment experiments and downstream assays, histone ChIP, BG4 ChIP, oligonucleotide pull-down assays, ELISA, western blots for TRF2 silencing, and immunoprecipitation for REST and EZH2, ChIP for REST and EZH2, Re-CHIP for REST, FLAG ChIP for delB delM, TRF2 ChIP with DelB DelM overexpression, GABPA silencing experiments, GABPA ChIP followed by ligand overexpression, CD with ligand and CD with TRF2, data curation, TERT sequence homology across species, figures, and manuscript writing. A.K.M. did the histone ChIP assays, flow-cytometry, ChIP for shelterin components (TRF2, TRF1, RAP1, POT1), dot blots, ChIP and Re-CHIP for REST, TERT expression analysis, EZH2 ChIP, FLAG ChIP for delB delM, REST ChIP with DelB DelM overexpression, ChIP-seq reanalysis, data curation, figures, and manuscript writing. S.S.R. did the generation of CCR5-TERTGaussia cell lines and characterization. S.B. did the TRF2 protein purification and TRF2 ChIP assays. S.L. and G.N. did the TRF2 ChIP and telomerase activity in patient-derived cell lines. D.P. did the TRF2 ChIP in patient-derived cell lines and telomerase activity and provided suggestions in manuscript. M.V. did the circular dichroism experiments. A.S. did the TRF2 protein purification. M.K. did the image acquisition for immunofluorescence experiments. S.C provided the conceptualization of the idea, design/plan of experiments, supervision, grants for funding, figures, and manuscript writing.
Declaration of Interests
S.C. is a scientific advisor for Rejuvas Biotech India Pvt. Ltd.
Inclusion and Diversity
The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Data and code availability
This study did not generate datasets/code.
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado T, Gutiérrez FJ, Aix E, Schneider RP, Giovinazzo G, Blasco MA, Flores I. Telomere Length Defines the Cardiomyocyte Differentiation Potency of Mouse Induced Pluripotent Stem Cells. Stem Cells. 2017;35:362–373. doi: 10.1002/stem.2497. [DOI] [PubMed] [Google Scholar]
- Ak?nc?lar SC, Khattar E, Boon PLS, Unal B, Fullwood MJ, Ter-gaonkar V. Long-Range Chromatin Interactions Drive Mutant TERT Promoter Activation. Cancer Discov. 2016;6:1276–1291. doi: 10.1158/2159-8290.CD-16-0177. [DOI] [PubMed] [Google Scholar]
- Arnoult N, Karlseder J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat Struct Mol Biol. 2015;22:859–866. doi: 10.1038/nsmb.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomol-ogous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Baker AM, Fu Q, Hayward W, Lindsay SM, Fletcher TM. The Myb/SANT domain of the telomere-binding protein TRF2 alters chromatin structure. Nucleic Acids Res. 2009;37:5019–5031. doi: 10.1093/nar/gkp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadru-plexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584:3779–3784. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RJA, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, Bauwens S, Djerbi N, Latrick CM, Fraisier V, Pei B, et al. TRF2-Mediated Control ofTelomere DNATopology asaMechanism forChro-mosome-End Protection. Mol Cell. 2016;61:274–286. doi: 10.1016/j.molcel.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Schoeftner S, Muñoz P, Blasco MA. RoleofTRF2 in the assembly of telomeric chromatin. Cell Cycle. 2008;7:3461–3468. doi: 10.4161/cc.7.21.7013. [DOI] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, Balasubramanian S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J Am Chem Soc. 2012;134:11974–11976. doi: 10.1021/ja305734x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- Brien GL, Valerio DG, Armstrong SA. Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs. Cancer Cell. 2016;29:464–476. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Baumann P. G-quadruplexes: from guanine gels to chemotherapeutics. Mol Biotechnol. 2011;49:198–208. doi: 10.1007/s12033-011-9395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Her-nandez B, Fernandez V, Dominguez F, Salas C, Romero-Laorden N, Garcia-Donas J, et al. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat Commun. 2015;6:8383. doi: 10.1038/ncomms9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li H, Deb S, Liu J-P. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene. 2002;21:3130–3138. doi: 10.1038/sj.onc.1205419. [DOI] [PubMed] [Google Scholar]
- Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage. Mol Cell. 2013;51:141–155. doi: 10.1016/j.molcel.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Farrell AS, Lakamp AS, Ouellette MM. Characterization of the DNA binding specificity of Shelterin complexes. Nucleic Acids Res. 2011;39:9206–9223. doi: 10.1093/nar/gkr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow TT, Shi X, Wei J-H, Guan J, Stadler G, Huang B, Blackburn EH. Local enrichment of HP1alpha at telomeres alters their structure and regulation of telomere protection. Nat Commun. 2018;9:3583. doi: 10.1038/s41467-018-05840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie GW, Promontorio R, Hampel SM, Micco M, Neidle S, Parkinson GN. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J Am Chem Soc. 2012;134:2723–2731. doi: 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- Cong Y-S, Wright WE, Shay JW. Human telomerase and its regulation. MicroBiol Mol Biol Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash J, Shirude PS, Balasubramanian S. G-quadruplex recognition by bis-indole carboxamides. Chem CommuN (Camb) 2008;26:3055–3057. doi: 10.1039/b806042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhapola P, Chowdhury S. QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 2016;44(W1):W277–83. doi: 10.1093/nar/gkw425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N, Rappsilber J, Södersten E, Hansen K. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012;8:e1002494. doi: 10.1371/journal.pgen.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Maï M, Wagner K-D, Michiels J-F, Ambrosetti D, Borderie A, Destree S, Renault V, Djerbi N, Giraud-Panis M-J, Gilson E, Wagner N. The Telomeric Protein TRF2 Regulates Angiogenesis by Binding and Activating the PDGFRß Promoter. Cell Rep. 2014;9:1047–1060. doi: 10.1016/j.celrep.2014.09.038. [DOI] [PubMed] [Google Scholar]
- Escoffier E, Rezza A, Roborel de Climens A, Belleville A, Gazzolo L, Gilson E, Duc Dodon M. A balanced transcription between telomerase and the telomeric DNA-binding proteins TRF1, TRF2 and Pot1 in resting, activated, HTLV-1-transformed and Tax-expressing human T lymphocytes. Retrovirology. 2005;2:77. doi: 10.1186/1742-4690-2-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fael Al-Mayhani TM, Ball SLR, Zhao J-W, Fawcett J, Ichimura K, Collins PV, Watts C. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J NeuroSci Methods. 2009;176:192–199. doi: 10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Fouché N, Cesare AJ, Willcox S, Ozgür S, Compton SA, Griffith JD. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- Giri S, Nieber K, Acikgöz A, Pavlica S, Keller M, Bader A. Molecular and Cellular Biochemistry. Springer; 2010. Telomerase activity and hepatic functions of rat embryonic liver progenitor cell in nanoscaffold-coated model bioreactor; pp. 137–149. [DOI] [PubMed] [Google Scholar]
- Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, Mergny JL, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel SM, Sidibe A, Gunaratnam M, Riou JF, Neidle S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg Med Chem Lett. 2010;20:6459–6463. doi: 10.1016/j.bmcl.2010.09.066. [DOI] [PubMed] [Google Scholar]
- Hansel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13:551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Saha D, Purohit G, Kar A, Kishore Mukherjee A, Sharma S, Sengupta S, Dhapola P, Maji B, Vedagopuram S, et al. Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci Rep. 2017;7:11541. doi: 10.1038/s41598-017-11177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MA, Thomas SD, Murty VV, Sedoris KJ, Miller DM. c-Myc quadruplex-forming sequence Pu-27 induces extensive damage in both telomeric and nontelomeric regions of DNA. J Biol Chem. 2014;289:8521–8531. doi: 10.1074/jbc.M113.505073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoušková E, Necasova I, Pavloušková J, Zimmermann M, Hluchý M, Marini V, Novakova M, Hofr C. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res. 2015;43:2691–2700. doi: 10.1093/nar/gkv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H-J, Cui Y, Yin H, Scheid A, Hendricks WPD, Schmidt J, Sekulic A, Kong D, Trent JM, Gokhale V, et al. A Pharmacological Chaperone Molecule Induces Cancer Cell Death by Restoring Tertiary DNA Structures in Mutant hTERT Promoters. J Am Chem Soc. 2016;138:13673–13692. doi: 10.1021/jacs.6b07598. [DOI] [PubMed] [Google Scholar]
- Killedar A, Stutz MD, Sobinoff AP, Tomlinson CG, Bryan TM, Beesley J, Chenevix-Trench G, Reddel RR, Pickett HA. A Common Cancer Risk-Associated Allele inthe hTERT Locus Encodes a Dominant Negative Inhibitor of Telomerase. PLoS Genet. 2015;11:e1005286. doi: 10.1371/journal.pgen.1005286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;200:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kim W, Ludlow AT, Min J, Robin JD, Stadler G, Mender I, Lai T-P, Zhang N, Wright WE, Shay JW. Regulation of the Human Telomerase Gene TERT by Telomere Position Effect-Over Long Distances (TPE-OLD): Implications for Aging and Cancer. PLoS Biol. 2016;14:e2000016. doi: 10.1371/journal.pbio.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyo S, Takakura M, Kohama T, Inoue M. Telomerase activity in human endometrium. Cancer Res. 1997;57:610–614. [PubMed] [Google Scholar]
- Li Y, Tergaonkar V. Telomerase reactivation in cancers: Mechanisms that govern transcriptional activation of the wild-type vs. mutant TERT promoters. Transcription. 2016;7:44–49. doi: 10.1080/21541264.2016.1160173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou Q-L, Sun W, Chandrasekharan P, Cheng HS, Ying Z, Lakshmanan M, Raju A, Tenen DG, Cheng S-Y, et al. Non-ca-nonicalNF-kB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat Cell Biol. 2015;17:1327–1338. doi: 10.1038/ncb3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P-T, Wang Z-F, Chu I-T, Kuan Y-M, Li M-H, Huang M-C, Chiang P-C, Chang T-C, Chen C-T. Expression of the human telomerase reverse transcriptase gene is modulated by quadruplex formation in its first exon due to DNA methylation. J Biol Chem. 2017;292:20859–20870. doi: 10.1074/jbc.M117.808022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KW, Lacroix L, Yue DJE, Lim JKC, Lim JMW, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J Am Chem Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- Liu T, Yuan X, Xu D. Cancer-SpecificTelomerase ReverseTranscriptase (TERT) Promoter Mutations: Biological and Clinical Implications. Genes (Basel) 2016;7:38. doi: 10.3390/genes7070038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gómez-López G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- McGann JC, Oyer JA, Garg S, Yao H, Liu J, Feng X, Liao L, Yates JR, 3rd, Mandel G. Polycomb- and REST-associated histonedeacetylases are independent pathways toward a mature neuronal phenotype. eLife. 2014;3:e04235. doi: 10.7554/eLife.04235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny J-L, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tawani A, Mishra A, Kumar A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO. The hTERT core promoterforms three parallel G-quadruplexes. Nucleic Acids Res. 2020;48:5720–5734. doi: 10.1093/nar/gkaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee AK, Sharma S, Sengupta S, Saha D, Kumar P, Hussain T, Srivastava V, Roy SD, Shay JW, Chowdhury S. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 2018;14:e1007782. doi: 10.1371/journal.pgen.1007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee AK, Sharma S, Bagri S, Kutum R, Kumar P, Hussain A, Singh P, Saha D, Kar A, Dash D, Chowdhury S. Telomere repeat-binding factor 2 binds extensively to extra-telomeric G-quadruplexes and regulates the epigenetic status of several gene promoters. J Biol Chem. 2019a;294:17709–17722. doi: 10.1074/jbc.RA119.008687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee AK, Sharma S, Chowdhury S. Non-duplex G-Quadruplex Structures Emerge as Mediators of Epigenetic Modifications. Trends Genet. 2019b;35:129–144. doi: 10.1016/j.tig.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan N, Dhillon V, Thomas P, Fenech M. A quantitative real-time PCR method for absolute telomere length. Biotechniques. 2008;44:807–809. doi: 10.2144/000112761. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interac-tive ligands. J Am Chem Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandita RK, Chow TT, Udayakumar D, Bain AL, Cubeddu L, Hunt CR, Shi W, Horikoshi N, Zhao Y, Wright WE, et al. Single-strand DNA-binding protein SSB1 facilitates TERT recruitment to telomeres and maintains telomere G-overhangs. Cancer Res. 2015;75:858–869. doi: 10.1158/0008-5472.CAN-14-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V, Pal J, Somasundaram K. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget. 2015;6:43452–43471. doi: 10.18632/oncotarget.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso IM, Hayward W, Fletcher TM. The effect of the TRF2 N-terminal and TRFH regions on telomeric G-quadruplex structures. Nucleic Acids Res. 2009;37:1541–1554. doi: 10.1093/nar/gkn1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera ON, Sobinoff AP, Teber ET, Harman A, Maritz MF, Yang SF, Pickett HA, Cesare AJ, Arthur JW, MacKenzie KL, Bryan TM. Telomerase promotes formation of a telomere protective complex in cancer cells. Sci Adv. 2019;5:eaav4409. doi: 10.1126/sciadv.aav4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike AM, Strong MA, Ouyang JPT, Greider CW. TIN2 Functions with TPP1/POT1 To Stimulate Telomerase Processivity. Mol Cell Biol. 2019;39:e00593-18. doi: 10.1128/MCB.00593-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Purohit G, Mukherjee AK, Sharma S, Chowdhury S. Extrate-lomeric Binding of the Telomere Binding Protein TRF2 at the PCGF3 Promoter Is G-Quadruplex Motif-Dependent. Biochemistry. 2018;57:2317–2324. doi: 10.1021/acs.biochem.8b00019. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach RJ, Garaví M, González C, Jameson GB, Filichev VV, Hale TK. Heterochromatin protein 1a interacts with parallel RNA and DNA G-quadruplexes. Nucleic Acids Res. 2020;48:682–693. doi: 10.1093/nar/gkz1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Singh A, Hussain T, Srivastava V, Sengupta S, Kar A, Dhapola P, Dhople V, Ummanni R, Chowdhury S. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J Biol Chem. 2017;292:15205–15215. doi: 10.1074/jbc.M117.792077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy JF, Baumann P. Apollo-taking the lead in telomere protection. Mol Cell. 2010;39:489–491. doi: 10.1016/j.molcel.2010.08.018. [DOI] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary K, Caglayan D, Caja L, Tzavlaki K, Bin Nayeem S, Bergström T, Jiang Y, Uhrbom L, Forsberg-Nilsson K, Westermark B, et al. Snail depletes the tumorigenic potential of glioblastoma. Oncogene. 2013;32:5409–5420. doi: 10.1038/onc.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz I, Timashev L, Xie W, Patel DJ, de Lange T. TRF2 binds branched DNA to safeguard telomere integrity. Nat Struct Mol Biol. 2017;24:734–742. doi: 10.1038/nsmb.3451. [DOI] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20:299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- Shi Y-A, Zhao Q, Zhang L-H, Du W, Wang X-Y, He X, Wu S, Li Y-L. Knockdown of hTERT by siRNA inhibits cervical cancer cell growth in vitro and in vivo. Int J Oncol. 2014;45:1216–1224. doi: 10.3892/ijo.2014.2493. [DOI] [PubMed] [Google Scholar]
- Simonet T, Zaragosi L-E, Philippe C, Lebrigand K, Schouteden C, Augereau A, Bauwens S, Ye J, Santagostino M, Giulotto E, et al. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 2011;21:1028–1038. doi: 10.1038/cr.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smekalova E, Baumann P. TERRA -a calling card for telomerase. Mol Cell. 2013;51:703–704. doi: 10.1016/j.molcel.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. Mutation of the TERT promoter, switch to active chromatin, and mono-allelic TERT expression in multiple cancers. Genes Dev. 2015;29:2219–2224. doi: 10.1101/gad.269498.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Paucek RD, Huang FW, Ghandi M, Nwumeh R, Costello JC, Cech TR. Allele-Specific DNA Methylation and Its Interplay with Repressive Histone Marks at Promoter-Mutant TERT Genes. Cell Rep. 2017;21:3700–3707. doi: 10.1016/j.celrep.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson CG, Moye AL, Holien JK, Parker MW, Cohen SB, Bryan TM. Two-step mechanism involving active-site conformational changes regulates human telomerase DNA binding. BioChem J. 2015;465:347–357. doi: 10.1042/BJ20140922. [DOI] [PubMed] [Google Scholar]
- Traczyk A, Liew CW, Gill DJ, Rhodes D. Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1. Nucleic Acids Res. 2020;48:4562–4571. doi: 10.1093/nar/gkaa171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volck-mann R, van Noesel MM, George RE, Tytgat GAM, Molenaar JJ, Versteeg R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Vinayagamurthy S, Ganguly A, Chowdhury S. Extra-telomeric impact of telomeres: Emerging molecularconnections in pluripotencyorstem-ness. J Biol Chem. 2020;295:10245–10254. doi: 10.1074/jbc.REV119.009710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MS, Wright WE, Shay JW. Alternative splicing regulation of telomerase: a new paradigm? Trends Genet. 2014;30:430–438. doi: 10.1016/j.tig.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Bergström T, Jiang Y, Johansson P, Marinescu VD, Lindberg N, Segerman A, Wicher G, Niklasson M, Baskaran S, et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine. 2015;2:1351–1363. doi: 10.1016/j.ebiom.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Renault VM, Jamet K, Gilson E. Transcriptional outcome of telomere signalling. Nat Rev Genet. 2014;15:491–503. doi: 10.1038/nrg3743. [DOI] [PubMed] [Google Scholar]
- Yu Z, Gaerig V, Cui Y, Kang H, Gokhale V, Zhao Y, Hurley LH, Mao H. Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J Am Chem Soc. 2012;134:5157–5164. doi: 10.1021/ja210399h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Casaday-Potts R, Precht P, Jiang H, Liu Y, Pazin MJ, Mattson MP. Nontelomeric splice variant of telomere repeat-binding factor 2 maintains neuronal traits by sequestering repressor element 1-silencing transcription factor. Proc Natl Acad Sci USA. 2011;108:16434–16439. doi: 10.1073/pnas.1106906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Tong HJ, Li M, Tan KS, Cao T. Telomere length is regulated by FGF-2 in human embryonic stem cells and affects the life span of its differentiated progenies. Biogerontology. 2017;18:69–84. doi: 10.1007/s10522-016-9662-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets/code.