Abstract
Cerebral organoids, or brain organoids, can be generated from a wide array of emerging technologies for modeling brain development and disease. The fact they are cultured in vitro makes them easily accessible both genetically and for live assays such as fluorescence imaging. Here we describe a protocol for reliably generating cerebral organoids of a telencephalic identity and for maintaining longterm viability for later stages of neural development including axon outgrowth and neuronal maturation. The method builds upon earlier cerebral organoid methodology, with modifications of embryoid body size/shape to increase surface area and slice culture to maintain nutrient and oxygen access to the interior regions of the organoid, enabling long-term culture. We also describe approaches for introducing exogenous plasmid constructs and for sparse cell labeling in order to image neuronal axon outgrowth and maturation over time. Together, these methods allow for modeling of later events in cortical development, which are important for neurodevelopmental disease modeling. The protocols described can be easily performed by an experimenter with stem cell culture experience, and take 2-3 months to complete, with long term maturation occurring over several months.
Introduction
Brain organoids are an emerging technology offering a window into human brain development1–3. As such, they are uniquely placed for studies of both basic mechanisms of human neural development and function, as well as pathogenesis of neurodevelopmental and neurological disorders. Their development was originally inspired by the need for more accurate models of human neural tissue for the study of disorders where animal models are suboptimal. Earlier in vitro studies made use of fairly pure preparations of human neurons from directed 2D differentiation of pluripotent stem cells (PSCs). Then, pioneering work from Su-Chun Zhang and James Thomson demonstrated the first PSC-derived neural cultures with tissue-like organization called neural rosettes4. By first generating 3D embryoid bodies followed by a phase of neural induction and finally plating down the aggregates, neuroepithelial cells would grow in a conformation reminiscent of the neural tube. Later, Yoshiki Sasai and colleagues extended these methods and performed similar 3D EB neural inductions but with plating down at an even later stage such that the cultures formed larger cortical-like structures5. These innovative studies formed the foundation for the original cerebral organoid protocol, which involved continued 3D culture (rather than plating down), as well as embedding the organoids in Matrigel (an extracellular-matrix-rich gel derived from EHS mouse sarcoma) and suspension culture with agitation1,6.
In the several years since the initial descriptions of neural organoids, the field has seen an explosion in use of these models and their continued improvement. The advantage of such a large number of research groups moving into this field is the diversity of expertise being applied to furthering this technology. For example, those with expertise in stem cell differentiation have demonstrated great success with direct differentiation of individual brain regions through manipulation of growth factor signaling2,7. Experts in molecular analysis, such as single cell RNA sequencing, have demonstrated remarkable similarities to in vivo developmental trajectories8,9,10,11. Finally, electrophysiological studies have demonstrated a striking ability for brain organoids to develop relatively mature neuronal networks 12,13,14. These more recent studies have highlighted tremendous potential of brain organoids, while at the same time assessing their current capabilities and identifying limitations that still need addressing.
Development of improved cerebral organoid protocols
The original cerebral organoid protocol was exciting, not only for the tissue structure seen, but also the complexity of various brain regions and their association with one another. However, a major limitation was the inability to reliably generate regional identities. Indeed, careful independent assessments revealed a very strong “batch effect” in which organoids made at the same time (and often cultured together in the same bioreactor or dish) were highly similar, but there was huge variability between batches in terms of regional identity formation12,15. This led us to investigate the source of this variability and to identify a very early divergence between batches at the neural induction stage15. Interestingly, there was a correlation between the abundance of non-neural mesodermal and endodermal cells and the quality and reliability of resultant organoids. Thus, we sought to improve the reliable generation of neuroectoderm at an early stage.
Well-organized neuroectoderm tends to form initially around the surface of the early embryoid body15. In order to increase the relative abundance of this surface tissue, we previously experimented with different approaches including simply starting with fewer cells to generate smaller spheres, as well as using fibrous microscaffolds to generate more intricate shapes with higher surface area-to-volume ratios15. Both approaches led to more reliable formation of neural identities, and interestingly generated organoids with primarily a telecephalic identity. This included both dorsal cerebral cortical and choroid plexus identities and ventral ganglionic eminence identities within the same organoid.
Cortical tissue architecture, namely the formation of a dense, radially aligned cortical plate, has been another area of improvement in cerebral organoids in recent years. This cellular organization has been shown to be a prerequisite for formation of the layered grey matter of the cerebral cortex16, and thus influences connectivity of these discrete layer identities. In order to promote cortical plate formation, we previously hypothesized that maintenance of basement membrane may be key as it has been shown in vivo to be necessary for maintenance of the radial glial scaffold,17 which neurons use as a guide to position themselves within the cortical plate. Previous work from the Sasai group had shown that dissolved laminin could maintain basement membrane on the neuroepithelium along the outside18. Similarly, addition of dissolved ECM in the form of Matrigel on cerebral organoids also maintained the basement membrane and led to formation of a distinct cortical plate similar to in vivo 15.
Development of long-term culture at the air-liquid interface
While the 3D conformation of brain organoids allows for remarkable tissue architecture, a major limitation is the lack of nutrients and oxygen inside the core of the organoid. Certain cells, such as radial glia, can survive even quite deep within the organoid, likely due to their different metabolic needs and their elongated shape contacting the exterior. However, as increasing numbers of neurons are produced, they are progressively pushed inside and undergo necrosis due to the lack of nutrients. In order to overcome this limitation, we recently turned to a classic technique of organotypic slice culture19. We reasoned that exposing the interior of the organoid to oxygen and media would result in improvements in cell survival.
Organotypic slice culture at the air-liquid interface (ALI) is a classic method often applied to rodent brain samples. Typically it is used for more short-term studies involving live imaging of cellular behaviors or electrophysiological measurements. However, there are reports of slice cultures being maintained at least six months20 and we found that slice cultures of brain organoids could be maintained over extremely long periods,being limited only by the time point at which one decides to take them for end-point analysis. Indeed, we have maintained such cultures for up to and beyond one year. Under these conditions, neurons within the organoid showed enhanced survival, with an approximately 66% decrease in the presence of TUNEL positive cells. Perhaps more striking though was the degree of morphological maturity, with neurons forming thick tracts similar to the white matter tracts of the brain after 1-2 months of culture at the ALI. We also showed that this morphological maturity translated to functional maturity with the ability to form neural networks and even drive muscle contractions13.
Comparison with alternative methods
Numerous alternative methods now exist for generating neural organoids of various individual brain regions 21. These rely upon the use of small molecules to manipulate growth factor signaling and thus influence cellular identity. These so-called directed differentiation protocols often make use of dual SMAD inhibition22, which was shown in 2D cultures to efficiently promote neuronal identity determination. Further manipulation of signaling pathways involved in axial patterning, such as Hedgehog and Wnt, have been shown to direct formation of specific regional identities within the brain. These protocols are generally quite reproducible due to the powerful inductive abilities of these small molecules. However, one limitation of these approaches is the effect these extrinsic factors have on tissue self-organization. Often, these protocols result in tissues with simpler architecture, and overall smaller neural tube-like buds more reminiscent of neural rosettes23. While complex tissue architecture is not always needed for specific scientific questions, there are many situations where this architecture would be desirable, and so the “guided” approach described here would be beneficial.
Neuronal survival in brain organoids is another issue that has been an area of intense research in the field. Several recent protocols have made use of growth factors, such as BDNF, GDNF and LIF, to promote further maturation and survival12,24,25. While a careful side-by-side analysis has not been done, it seems that application of these growth factors still does not overcome the major problem of necrosis within the organoid. Thus, more focus has been placed recently on attempts to vascularize brain organoids in vitro and in vivo. A few studies have reported that addition of angiogenic cells, such as endothelial or mesodermal precursors, can lead to vascularization in vitro 26,27. While these reports have observed promising primitive endothelial tubes, it remains to be seen whether functional vasculature can be achieved fully in vitro.
In vivo transplantation of brain organoids, with and without cocultured vascular cells, into rodents has also demonstrated the formation of vasculature, in this case with demonstrated blood flow 28. These in vitro and in vivo approaches have demonstrated intriguing improvements in cell survival. However, it is still unclear whether this actually leads to improvements in tissue development (for example, further expansion of a layered cortical plate). Indeed, in most cases, vascular cells appear to invade the neural tissue and, if anything, disrupt tissue architecture rather than support it.
Applications
The reproducible formation of large telencephalic vesicles that develop into layered cortical lobules provides a system to examine processes of brain morphogenesis and disorders that affect these processes, such as microcephaly. The additional improvements in neuronal positioning also enable studies of defects in neuronal migration such as lissencephaly and neuronal heterotopias. In addition, because the protocols described here avoid extensive use of growth factors and small molecules, cell types are generated in accordance with intrinsic developmental programs and timing. For example, LIF has been reported to promote the switch to gliogenesis 29, so application of this factor may artificially speed up radial glial progression through fate decisions. Indeed, without its use we find that healthy, mature astrocytes are not generated in large numbers until 4-5 months, which is remarkably similar to human timing in vivo, but much slower than other protocols using growth factors to speed up maturation, such as BDNF2.
Culture at the ALI provides an easy solution to the necrotic core problem and allows for continued culture in vitro without having to perform challenging transplantations in vivo. This improvement allows neurons within ALI cerebral organoids (ALI-COs) to project their axons throughout the culture, whereas neurons within whole 3D organoids only project along the healthy exterior15. Thus, neuronal projections in ALI-COs exhibit much better organization, showing typical bundling and long-range directionality into the interior, reminiscent of white matter tracts of the brain. Because the tracts exhibit specific projection patterns, they can be used to examine axon guidance, and the response of these human tracts to specific guidance cues. They can also be used to examine defects in long-range connectivity such as in the case of spinal cord injury or degeneration. The improved survival and static culture also allows for electrophysiological studies, such as multielectrode array recordings, over very long time periods, potentially enabling long-term studies of neuronal network maturation.
Limitations
There are still certain limitations that remain to be addressed and will likely be the topic of future studies. In particular, while local patterning seems to produce regions with proper relative positioning, the global macro-scale arrangement is still dissimilar to the brain in vivo. For example, cerebral organoids may exhibit several telencephalic vesicles within a single organoid whereas, in vivo, a single vesicle is formed, which then separates into two hemispheres. Nonetheless, focusing on individual lobules of organoids reveals their remarkable similarity to individual cerebral hemispheres in vivo.
A further limitation that can still pose a problem is a persistent, albeit subtle, batch effect. As the guided approach involving simple conformational changes to the embryoid bodies is a much gentler approach compared with small molecules, the resulting organoids still rely mainly upon self-organization and intrinsic signaling. This means that, while they are predominantly telencephalic in identity, there can be differences in relative abundance of subregions such as dorsal cortex and ganglionic eminences. For example, some batches may even give rise to overabundant choroid plexus with more limited cortical lobules. While usage of small molecules may overcome this variability, we prefer to generate several batches and use careful quality controls to determine identity rather than sacrifice tissue architecture. Finally, some cell lines exhibit biases in identity determination and this must be determined on a line-by-line basis. A universal protocol that works for all PSCs in exactly the same way still remains to be developed, and this limitation even applies to directed differentiation protocols.
Overview of the procedure
Human PSCs are first dissociated from their substrate and reaggregated as floating embryoid bodies (EBs) (steps 1-9; Figure 1a). We describe here the two approaches for increasing surface area-to-volume of EBs: smaller EBs and fibrous microscaffolds. These modified or engineered EBs are then sequentially taken through a series of media with conditions that are similar to the original cerebral organoid protocol but with some minor improvements in nutrient composition and timing (steps 10-15). Importantly, this still involves embedding in Matrigel droplets and agitation culture on an orbital shaker (steps 16-32). We also describe the process of removing the Matrigel after neuroepithelial buds are well established (steps 33-37), as its presence can impair proper layering. Once neurogenesis is well-established, Matrigel is supplied as a dissolved component within the media (steps 38-39). Upon formation of the cortical plate, organoids are taken for slice culture and kept for long term at the ALI (steps 40-58),. We also describe assays for generating improved cerebral organoids using a commercially optimized kit (Box 1), sparse labelling with Sendai virus (Box 2), and the electroporation of cultures with plasmids encoding transgenes such as GFP (Box 3).
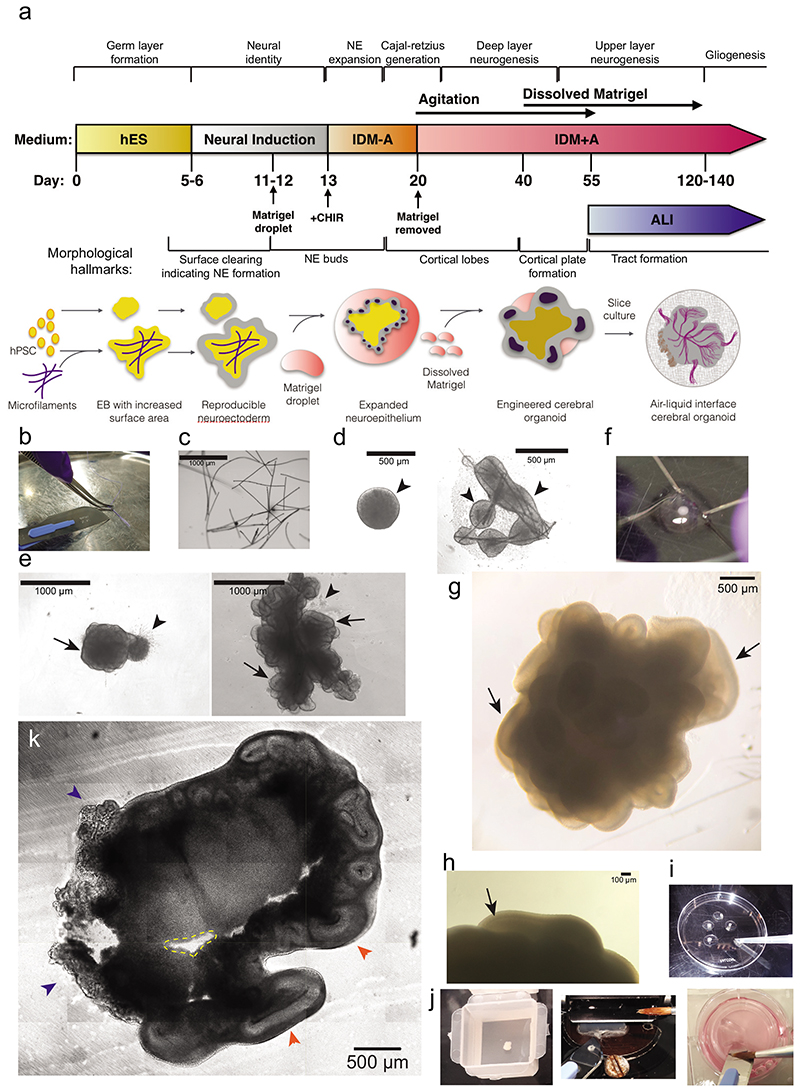
Figure 1. Methodological overview and key steps.
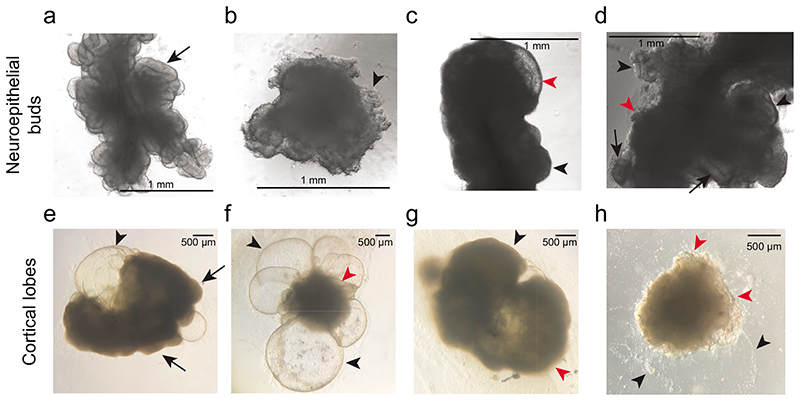
a. Timeline of the protocol for generating improved cerebral organoids and air-liquid interface (ALI) cultures with key developmental stages provided above and morphological hallmarks provided below. NE = neuroepithelium. b. Image to demonstrate microscaffold preparation. The suture is splayed with the blunt side of the scalpel blade, then cut into 1 mm pieces on a sterile tray. c. Image showing rehydrated fibres observed under a phase contrast microscope (scale bar = 1000 μm). d. Images of round EB at day 7 after successful neural induction (left-hand panel), showing a smooth border and brightening edges (arrowheads), and a micropatterned organoid at day 7 (right-hand panel), demonstrating bright smooth edges (arrowheads) indicative of proper neural induction. e. Images of reduced-size organoid made with 3,000 cells at Day 12 observed under phase contrast (left-hand panel) and a micropatterned organoid at Day 13 (right-hand panel). Note that both organoids predominantly consist of neuroepithelial buds (arrows) with sparse disorganized areas of migratory cells (arrowheads). Scale bars = 1000 μm. f. Image to show the process of removing Matrigel droplets by microdissection with syringe needles. One needle is used as a pin to hold the Matrigel droplet while the other is used as a blade to cut or tear off the Matrigel until most is removed. g. Bright-field image at day 50 showing well-formed, large and smooth cortical lobes (arrows) covering the entire organoid. h. Image to show a visible cortical plate under phase contrast on a Day 58 organoid (arrow). Scale bar = 100 μm. i. Image to show process of preparation for slice culture. Organoids are transferred to the lid of a 60 mm dish with a cut 1 ml tip and gently washed with HBSS. j. Image of an organoid just after embedding in 3% low-melt agarose within a peel-A-way mold (left-hand panel). We also provide an image to show the process of section collection with a scalpel and a brush (middle panel); the brush is used to manipulate the section only through contact with the agarose. The tissue section is slid and loaded onto the scalpel blade for transfer, before being deposited onto the cell culture insert by gently pushing the edge of the section off the scalpel blade (right-hand panel). The brush can also be used to apply medium to the scalpel blade to facilitate sliding of the tissue section off the blade. k. Bright-field image of a tissue slice of a 54-day old organoid immediately after sectioning. The ALI-CO displays regions of choroid plexus (blue arrowheads) and large cortical walls organized around what is left of the cortical ventricles (orange arrowheads). Small cavities (dashed yellow line) in the organoid section can be caused by detachment of small regions of necrotic tissue that is particularly brittle. Scale bar = 500 μm.
Box 1.
Generation of improved cerebral organoids using a commercially optimized kit
In our lab, we have also successfully generated advanced cerebral organoids from H9, H1 and IMR-90-4 cell lines using the STEMdiff™ Cerebral Organoid Kit. The kit comprises a set of culture media in a basal medium and supplement format and performs best in our hands when used with small EBs (2000 – 3000 cells) as described in the main procedure. When using the kit, we apply the same techniques as described in the rest of this protocol but with modified timing and the following modifications:
Day 0 – Making embryoid bodies (EB seeding medium). This medium is supplemented with ROCK inhibitor (1:100; final concentration 50 μM) but not bFGF before use. We increase the volume of medium used to 150 μl per EB.
Day 3 – Feeding EBs (EB formation medium). We increase the volume of medium used to 150 μl per EB.
Day 5 – Induction of neruoepithelia (Induction medium). Neural induction can be performed in the same 96-well low-attachment U-bottom plate, by simply changing the media and adding 150 μl of Induction media.
Day 7 – Embedding neuroepithelial tissues and expansion of neuroepithelial buds (Expansion medium). We do not add CHIR when using kit medium.
Day 10 –Growth of cerebral tissue (IDM-A) Transfer to IDM-A and continue as in the main protocol.
Day 14 – Removing Matrigel and further growth of cerebral tissue (IDM+A). From this stage, we can use homemade IDM+A medium interchangeably with the Maturation medium provided in the kit.
Day 30 and later – Growth of cortical plate and maturation of the cerebral tissue (IDM+A+MG)
Due to the slight acceleration of developmental progression of the organoids generated with the kit, cortical plate can be observed as early as day 45-50.
Box 2.
Sparse fluorescence cell labelling using EmGFP Sendai virus *TIMING 30 min
ALI-CO transduction with EmGFP Sendai virus is a simple and efficient way to sparsely label cells for a variety of applications. Addition of viral particles to the medium used for feeding will lead to sparse labeling across the entire surface of the ALI-CO, whereas injection of viral particles into specific regions of the ALI-CO will produce confined and focal GFP fluorescence.
CAUTION Although Sendai virus is a single-stranded pleomorphic RNA virus with no risk of genomic integration, the virus should be handled in a CL1 laminar flow cabinet and you should wear protective clothing. Pipette tips and sharps that have come into contact with the virus should be decontaminated in an RBS50 or Virkon solution prior to appropriate disposal.
Labeling in media
Thaw on ice one aliquot of EmGFP Sendai virus.
-
Add between 12-24 μl of virus to 2-4 ml of SFSC medium.
CRITICAL STEP The virus is typically supplied with a titer between 8.0x107-15.0x107 Cell Infectious Units (CIU)/ml and can be found on the supplier’s website by looking up the LOT number. The total number of cells in each well will vary depending on the size of the ALI-COs and the number of ALI-COs per cell culture insert, making it difficult to precisely estimate the multiplicity of infection (MOI). We find 6 μl volume is sufficient to sparsely label an insert carrying 2 ALI-COs for ~90 days when using a virus with a titer of 8.1x107 CIU/ml.
Aspirate off all medium from two wells containing ALI-CO inserts.
-
Add 1 ml of SFSC medium supplemented with virus to each well.
CRITICAL STEP Adding the virus directly to the medium in the well with the ALI-CO insert can result in uneven labeling across the sample. Premix the virus with fresh medium before feeding.
-
After 3-4 days, check the ALI-COs for fluorescence. Individual cells should be evident by this time point (Figure 2g).
CRITICAL STEP This labeling method is best suited for young ALI-COs (up to ~120 days). As the ALI-COs age, the central areas of the culture are not as easily transduced and labeling is more prevalent along the external regions of the ALI-COs. For mature ALI-COs (over ~120 days), injection of the virus yields better results.
Labeling by injection
Using forceps, transfer an insert of ALI-COs to the lid of a 10 cm dish.
Under a stereomicroscope, use a pair of scissors to open the tip of a pulled borisilicate glass micropipette. Holding the scissors at a ~40-60° angle relative to the long axis of the capillary, cut the tip to attain a ~9-14 mm tip taper length from the shoulder of the pipette.
Thaw on ice one aliquot of EmGFP Sendai virus.
Dip the micropipette tip into the viral particle solution and wait for the micropipette tip to fill up by capillary action.
Manually poke the ALI-CO with the micropipette.
-
After penetrating into the tissue, gently tap the glass micropipette to allow release of the virus solution.
CRITICAL STEP Alternatively, insert the micropipette into the dropper unit that comes supplied with the capillaries to eject the virus solution.
After 3-4 days, check the ALI-COs for fluorescence.
Box 3.
Electroporation of plasmid DNA into organoid ventricles *TIMING 2-4 h
Sometime between days ~40-55, organoids should display large lobes with fluid-filled cavities into which constructs can be injected and electroporated to interrogate a variety of biological processes from early neurogenesis onwards. Here we describe injection and electroporation of a farnesyl-GFP Sleeping Beauty transposon expression construct in combination with the SB100X transposase for stable labeling of radial glial progenitors and their progeny. However, the same approach can be taken to deliver any plasmid construct relevant for the specific research question.
Injection and electroporation
Assemble the micropipetting apparatus and prewarm IDM+A+MG as well as a small volume (2-3 ml) of OptiMEM.
Under a stereomicroscope, use scissors to cut the tip of a pulled borosilicate glass micropipette. Holding the scissors at a ~40-60° angle relative to long axis of the capillary, cut the tip to attain a tip taper length between ~9-14 mm from the shoulder of the pipette.
-
Store the open micropipette in a separate 10 cm cell culture dish with a Blu Tac or plasticine clay holder and put the lid on to prevent clogging of the micropipette by dust particles.
CRITICAL STEP For maximal accuracy and best results, especially in the case of young organoids with relatively small ventricles, it is essential to produce micropipettes with tip taper above 9 mm. Holding the scissors at an angle when opening the micropipette ensures that the tip is sharp and can easily penetrate into the cortical lobes with minimal tissue disruption. Furthermore, a tip with such features will not fill or release its contents unless negative or positive pressure is applied, respectively.
Prepare the DNA solution for injection by combining the fGFP and the SB100X constructs to a final concentration of 80 ng/μl and 240 ng/μl, respectively, in PBS. Prepare 5 μl of DNA solution per organoid to be injected. To the total volume, add Fast Green to a final concentration of 0.05%. This will help to monitor filling of the individual ventricles and visualization of the injected lobes.
-
Place a stereomicroscope in a horizontal flow cabinet workstation along with a small square of parafilm, a P10 pipette, a box of 10 μl tips and the DNA solution.
CRITICAL STEP If a horizontal flow cabinet is not available, the procedure can be performed on a bench. In order to minimise the risk of contamination, organoids should be fed with fresh medium supplemented with Antibiotic-Antimycotic (1x) the day after electroporation.
For an electroporation dish with a chamber of 5 mm interelectrode gap, program the square electroporator to deliver 5 pulses of 1 ms duration and 40-80 V amplitude with 1 s inter-pulse interval.
Select an organoid with large, well-developed and clearly visible lobes. In a laminar flow cabinet, transfer it to the electroporation dish using a cut P1000 tip.
-
Replace the medium used for transfer with prewarmed OptiMEM.
CRITICAL STEP To best visualize the organoid lobes during electroporation, we recommend adjusting the volume of OptiMEM in the chamber to minimize the meniscus, which can cause strong reflection and glare and make it difficult to visualise the organoid morphology.
Cover the electroporation dish with a lid and transfer it to the stereomicroscope stage.
Pipette 5 μl of DNA solution on the parafilm square to form a droplet.
Insert the open micropipette into the holder at one end of the pipetting apparatus and a clean 200 μl filter tip in the mouth piece at the other end.
Test the micropipette by dipping it into PBS and blowing into the system – bubbles should form in the PBS.
-
Aspirate the DNA solution into the micropipette.
CRITICAL STEP We recommend loading the micropipette by applying small bursts of suction and checking that the filter tip does not become blocked with saliva. If a clog forms, this can produce a vacuum that causes the DNA solution to spray along the silicone tubing.
-
Gently poke into the lobes of the organoid with the micropipette tip and immediately start filling the lobes by blowing into the mouthpipetting apparatus.
CRITICAL STEP A slow, gentle and steady outflow of DNA will prevent the lobes from bursting.
Probe all lobes of the organoid or, alternatively, target only a subset of lobes that are of interest, then position the organoid with lobes oriented toward the positive electrode so that plasmid DNA will enter the ventricular surface upon electric field application.
-
Once filling is completed, clamp the electroporator leads to the chamber electrodes, tap the omega (Ω) icon to measure the preload resistance and run the protocol – bubbles should form on the medium surface.
CRITICAL STEP The organoid should be oriented relative to the electrical field in such a way as to maximize the number of cortical lobes targeted.
-
After electroporation, return the organoid to a new cell culture dish with warm medium. Wash the micropipette tip multiple times by aspirating and ejecting PBS from a dish.
CRITICAL STEP Multiple washes (at least 2-3) are required to prevent debris from clogging the capillary
After 2-4 days, positive GFP fluorescence should become visible and ALI-COs can be prepped from the electroporated organoids so that a variety of biological processes can be studied over time by fluorescence live imaging (Figure 2 h,i).
Experimental design
There are several aspects to consider before starting the experiment, including decisions regarding the specifics of protocol implementation, timing (with regard to assaying particular developmental events), and proper controls.
Protocol implementation
Before implementing this protocol, there are several decisions to consider. The first is whether to take the smaller EB or the microscaffold approach. The former is technically easier as it does not require preparation of microfibers. However, because the EBs are smaller, they generate fewer telencephalic lobules, which may be a limitation depending on downstream applications (for example, quantitative measurement or electroporation). Second, the protocol can be used with either homemade media or ready-made media available as a kit from Stem Cell Technologies. We find that both options produce similar results. However, for reasons that are unclear to us, the use of kit media requires adjustments to the timing of the protocol (see Box 1 for more detail), as it seems the stem cells progress faster through EB and neural induction stages (based on morphology). This adjusted timeline is in line with the timing suggested by the manufacturer.
We also describe the use of the small molecule CHIR99021, a GSK3β inhibitor and activator of Wnt signaling, to promote expansion of the telencephalic neuroepithelium. This is optional as the organoids will still form the proper tissue identity and architecture without it. However, its use promotes larger lobules that are easier to manipulate, particularly for injections of virus or electroporation of plasmid constructs. Importantly, CHIR99021 application is performed well after identity determination (day 13), thus minimizing any effects on neural induction or regional identity.
Finally, the generation of ALI-COs from later organoids is entirely optional, and organoids can instead be maintained indefinitely in 3D suspension culture. However, as previously demonstrated13, if left as floating aggregates, organoids exhibit progressive necrosis in the core with resultant neuronal death.
Quality control
Throughout the protocol, there are various stages at which you can assess the quality and success of the protocol. We find that morphological hallmarks are particularly predictive of proper identity and can indicate whether the protocol is proceeding successfully. Performing a series of quality control steps throughout the protocol should result in highly comparable organoids (within and across batches) that also exhibit proper tissue architecture and function. Throughout the protocol, we point to key morphological features that should be present. However, before even beginning this protocol, the cell line to be used should be checked for pluripotency and its morphology should also be checked, as any indication of differentiation will usually lead to suboptimal organoids.
Another consideration is which controls to use. This should always be considered before beginning the experiment to be sure that the proper comparisons are made. Whenever possible, organoids from the same batch (made at the same time and treated identically) should be compared with each other. Then multiple batches should be handled as similarly as possible, in order to compare the effects of the treatment across batches.
Comparing experimental conditions within a batch is not always possible; for example, when working with genetically modified cell lines or patient cell lines. In this instance, the phenotype of interest should first be assayed in controls from different batches to get a sense of its inherent variability. Each read-out will have a different range of variability, which should be assessed in a pilot study for each one individually. Comparisons can then be done on experimental conditions after proper power calculation to determine the number of batches needed. This is often tedious to assess but necessary for any meaningful statistical comparison (as it the case in more well-established fields, such as mouse genetics).
Developmental timing
Finally, an important consideration is the stage at which to collect and assay organoids. We have performed extensive characterization of various stages of organoid development and find that it recapitulates in vivo timelines remarkably well. Much of this characterization is published, but we also provide a timeline of the key stages of developmental events to help guide experimental design (Figure 1a). When used in combination with morphological characterization, this should help you to judge the stage of development and enable accurate downstream assessment.
Materials
Reagents
Cells
H9 (WA09, RRID:CVCL_9773) & H1 (WA01, RRID:CVCL_9771) human ES cells, feeder-independent (Wisconsin International Stem Cell Bank, Wicell Research Institute) ! CAUTION Use of human tissues and human stem cells must adhere to institutional and funding body regulations as well as relevant ethical guidelines.
-
Reprogrammed feeder-independent iPS cells (Wisconsin International Stem Cell Bank, Wicell
Research Institute, iPS (IMR-90)-4 cells (RRID:CVCL_C437)
Growth media and supplements
StemFlex medium (Thermo Fisher Scientific, cat. no. A3349401)
• DMEM-F12 (Thermo Fisher Scientific, cat. no. 11330057)
DMEM, high glucose, GlutaMAX Supplement, pyruvate (Thermo Fisher Scientific, cat. no. 31966047)
Knockout Serum Replacement (KOSR; Thermo Fisher Scientific, cat. no. 10828028)
hESC-quality fetal bovine serum (FBS; Thermo Fisher Scientific, cat. no. 10439024)
Fetal bovine serum (FBS; Thermo Fisher Scientific, cat. no. 10270106)
Glutamax (Thermo Fisher Scientific, cat. no. 35050038)
MEM-Non-essential amino acids (MEM-NEAA; Merck, cat. no. M7145)
2-Mercaptoethanol (Merck, cat. no. 8057400005)
bFGF (FGF2; Peprotech, cat. no. 100-18B) *CRITICAL We have not tested bFGF from other vendors for this protocol.
Bovine Serum Albumin (BSA; Thermo Fisher Scientific, cat. no. BP1605-100)
Sterile water
Heparin (Sigma-Aldrich, cat. no. H3149)
ROCK inhibitor Y27632 (Merck cat. no. 688000-5)
N2 supplement (Thermo Fisher Scientific cat. no. 17502048)
B27-vit. A supplement (Thermo Fisher Scientific cat. no. 12587010)
B27+vit. A supplement (Thermo Fisher Scientific cat. no. 17504044)
Penicillin-Streptomycin (Thermo Fisher Scientific, cat. no. 15140122)
Neurobasal medium (Thermo Fisher Scientific, cat. no. 21103049)
Insulin solution (Merck cat. no. I9278-5ML)
Amphotericin B (Merck, cat. no. A2942-20ML)
CHIR99021 (Tocris, cat. no. 4423)
Dimethyl Sulphoxide (DMSO, Thermo Fisher Scientific, cat. no. BP231-100)
Ascorbic Acid (Merck, cat. no. A4544)
Sodium bicarbonate (Merck, cat. no. S5761-500)
D(+)-Glucose Anyhidrous (Formedium, cat no. 50-99-7)
Antibiotic-Antimycotic (100x) (Thermo Fisher Scientific, cat no. 15240062)
STEMdiff Cerebral Organoid Kit (Stem Cell Technologis, cat no. 08570)
Enzymes and other reagents
Growth factor reduced Matrigel (Corning, cat. no. 356230)
EDTA 0.7mM
Sterile D-PBS without calcium and magnesium, pH 7.2
Matrigel (Corning, cat. no. 356234)
Spray bottle containing 70% Ethanol
Accutase (Sigma-Aldrich, cat. no. A6964)
Trypan blue (Thermo Fisher Scientific, cat. no. 15250061)
Vicryl polyglactin coated suture, violet (Ethicon, cat. no. W9567)
Opti-MEM Reduced Serum Medium (Thermo Fisher Scientific, cat no. 31985062)
Virkon tablets (Antec International, cat. no. 14030061)
Cleaner RBS 50 non-foam solution (Medline, cat. no. RBS50)
Fast Green FCF (Sigma-Aldrich, cat.no. F7252)
HBSS with calcium, magnesium and no phenol red (Thermo Fisher Scientific, cat. no. 14025092)
Low-melt agarose (Sigma-Aldrich, cat. no. A9414)
CytoTune EmGFP Sendai Fluorescence Reporter (Thermo Fisher Scientific, cat. no. A16519)
pT2-CAG-fGFP plasmid (Addgene #108714)
pCAG-SB100X plasmid (Addgene #127909)
Equipment
CO2 incubators (Eppendorf, cat. no. 6731000022)
Biological Safety Cabinet (Walker class II)
Horizontal Laminar Flow Workstation
BTX Gemini X2 HT Electroporation System (BTX, cat. no. 452008)
Petri Dish Platinum Electrode for Tissues Chamber Kit, 5 mm gap (BTX, 45-0505)
Leica VT1000 S Vibrating blade microtome
Millicell Cell Culture Insert, 30 mm diameter, hydrophilic PTFE, 0.4 μm (Merck Millipore, cat. no. PICM0RG50)
Disposable Scalpel Sterile No 22 (Swann-Morton, cat. no. 0508)
Round art paintbrush No 5
Rectangular ice pan, 4-9 L capacity
Rectangular black rigid plastic backdrop, 27 x 40 cm size
Peel-A-way embedding molds (Sigma-Aldrich, cat. no. E6032)
Single edge razor blades (Fisherbrand, cat. no. 12-640)
Multipette/Repeater (X)stream (Eppendorf, cat. no. 4986 000.025)
Eppendorf Combitips advanced Biopur 10ml orange single wrap (Eppendorf, cat. no. E0030089677)
SAMCO stainless steel scissors (S Murray, cat. no. N828/01)
Aspirator tube assemblies for calibrated microcapillary pipettes (Sigma-Aldrich, cat. no. A5177-5EA)
Silicone tubing 2mm-id 3mm-od 1m length (Fisher Brand, cat. no. PCST/2X4)
Select Pipette Filter 0.2 μm (Scientific Laboratory Supplies Ltd, cat. no. PIP6043)
Capillary Tubes Glass 50μl volume 100mm length (Drummond Scientific, cat. no. 1-000-0500)
QIAGEN Plasmid Plus Maxi Kit (25) (Qiagen Ltd, cat. no. 12963)
Loctite Super Glue (Loctite, cat. no. 411001)
Blu Tack (Bostik, cat. no. 801103)
6-well tissue culture dishes (Corning, cat. no. 3516)
Sterile filter pipette tips (1 ml, 200 μl and 10μl; Starlab, cat. no. S1122-1830-C, S1120-8810-C, S1120-3810-C)
Sterile microcentrifuge tubes (1.5 ml size; Sarstedt, cat. no. 72.692)
Sterile microcentrifuge tubes (0.5 ml size; Sterstedt, cat. no. 72.699)
Stericup 0.2 μm filter unit (500 ml, Merck, cat. no. SCGP00525)
96-well U-bottom ultra low attachment plates (Corning, cat. no. 7007)
15 ml conical tube (Sarstedt, cat. no. 62.553.041)
50 ml conical tube (Sarstedt, cat. no. 62.547.004)
24-well ultra low attachment plates (Corning, cat. no. 3473)
Parafilm (Bemis, cat. no. PM996)
100 mm tissue culture dish (Corning, cat. no. 353003)
10 ml syringe (Becton Dickinson, cat. no. 302188)
50 mm tissue culture dish (Sterilin, cat. no. 124)
35 mm Easy-Grip Tissue Culture Dishes (Corning, cat. no. 353001)
Orbital shaker (Infors Celltron)
Pipette controller (Jencons Scientific, cat. no. 613-4442)
5, 10 and 25 ml serological pipettes (Sarstedt, cat. no. 86.1253.001, 86.1254.001, GreinerBio-One cat. no. 760160)
Sterilized scissors
37°C and 40 °C water bath with heat transfer beads (Grant instruments cat. no.SAP18 and Lab armor)
Stereomicroscope (Leica MZ6)
P-2000 micropipette puller (Sutter Instrument)
Inverted tissue culture microscope (EVOS M5000, Thermo Scientific, cat. no. AMF5000)
Inverted point-scanning confocal microscope with heated incubation chamber for live imaging
Automated cell counter (BioRad, TC10)
Benchtop centrifuge (Eppendorf, 5810)
Sterile standard forceps (Fine Science Tools, cat. no. 11000)
U-base 96-well tissue culture plate (Corning, cat. no. 3799)
Disposable scalpel No. 22 (Swann Morton, cat. no. 0501)
Sterile stainless steel tray
Sterile scissors
Sterile empty 10μl tip rack
Crushed ice
Reagent Setup
Human pluripotent stem cell lines
Culture feeder-independent human pluripotent stem cells (hPSC) using standard procedures in 5% CO2 incubator at 37°C. In brief, hPSC are maintained in StemFlex medium and cultured on Matrigel coated plates. 0.5 mg growth factor reduced (GFR) Matrigel is dissolved in 6 ml cold DMEM-F12 and 1ml is used to coat each well of an entire 6-well plate. hPSC are passaged using 0.7 mM EDTA in sterile D-PBS without calcium and magnesium.
Growth factors and other additives
Prepare a 10% solution of BSA by dissolving 1 g in 10 ml sterile PBS and store at -20°C for 1-2 years. Reconstitute 50 μg bFGF in 5 ml sterile 0.1% BSA in PBS to obtain a 10 μg ml-1 solution. Make 25 μl aliquots and store at -20°C for up to 6 months (avoid repeated freezing and thawing and do not thaw aliquot at temperatures higher than room temperature). Reconstitute heparin in sterile PBS to a final stock concentration of 1 mg ml-1 and store at 2-8°C for up to 2 years. Reconstitute 5 mg ROCK inhibitor Y27632 in 2.96 ml sterile water to obtain a final concentration of 5 mM. Make 150 μl aliquots and store at -20°C for up to one year. If using CHIR99021, then reconstitute in DMSO to the final concentration of 3mM, aliquot and store at -20°C for up to one year. Dissolve 352 mg of ascorbic acid in 50 ml DMEM/F12 for 40 mM stock, sterile filter, and store at -20°C as 5 ml aliquots for up to one year. Aliquot and store N2 and B27 supplements at -20°C for up to one year.
Reagents for vibratome sectioning and ALI culture
Prepare 250 ml of a 50% (w/v) Glucose solution in PBS by mixing 125 g of anhydrous glucose and ~170 ml of PBS, and heating the solution on a hotplate with stirring. Once the glucose has fully dissolved, filter-sterilize through a 0.2 μm filter unit. The sterile solution can be stored at 2-8 °C for several months. In case a precipitate forms, re-dissolve the glucose by incubating the solution at 37 °C before making Serum-free slice culture (SFSC) medium. Dissolve 1.2 g of low-melt agarose powder in 40 ml of HBSS with Ca2+ and Mg2+ to obtain a 3% (w/v) low-melt agarose solution. The solution can be stored at room temperature for a maximum of one month. Thaw 100 ml of Antibiotic-Antimycotic (100x) solution overnight at 2-8 °C. This solution is extremely heat labile and, following a second freeze-thaw cycle, it should be used only once and any remaining solution discarded. For this reason, one-shot aliquots should be prepared according to the daily SFSC medium requirement. Aliquoting should be performed in a laminar flow cabinet, the Antibiotic-Antimycotic solution should be kept cold throughout and the aliquots should be stored at -20 °C. The CytoTune®-EmGFP Sendai Fluorescence Reporter is supplied as 100 μl of viral particle solution. Thaw this solution on ice, mix by gently pipetting up and down, and prepare 12-24 μl aliquots (depending on the number of cell culture inserts transduced each time) in pre-chilled 500 μl microcentrifuge tubes and store at -80 °C. Typically 12 μl of virus is sufficient for 1-2 inserts and 24 μl for 2-4 inserts. Plasmids for electroporation are prepped using QIAGEN Plasmid Plus Kits or alternatively QIAGEN EndoFree Plasmid Kits. Purification is carried out as detailed in the kit’s manual and elution is performed in sterile Milli-Q water (typically 100-250 μl, to achieve plasmid concentrations between 2-4 μg/μl). Dissolve Fast Green FCF dye in distilled water to a final concentration of 10% (w/v).
Aliquoting Matrigel
Thaw Matrigel on ice at 4°C for >4h or overnight. Pre-cool 1 ml pipette tips, and 10 microcentrifuge tubes at -20°C for 10-15 min. In a sterile hood, pipette Matrigel up and down to mix using cold pipette tips and transfer 1ml to each tube. Store aliquots at -20°C for up to one year. Avoid repeated freezing and thawing.
CRITICAL Matrigel will polymerise at room temperature so it is important all materials that come in contact with the solution are kept cold and aliquotting is done quickly to minimize time at room temperature.
hES media
For approx. 500 ml medium, combine 400 ml DMEM-F12, 100 ml KOSR, 15 ml ES-quality FBS, 5 ml Glutamax, 5 ml MEM-NEAA, and 3.5 μl 2-Mercaptoethanol. Sterile filter using a vacuum driven 0.2 μm Stericup filter unit. hES media can be stored for up to two weeks at 2-8°C. Add bFGF to a final concentration of 4 ng ml-1 for low bFGF hES media. *CRITICAL To avoid loss of activity of bFGF over time through repeated warming of the media, add bFGF immediately before use only to the volume needed.
Neural induction media
For approx. 100 ml of medium, combine 97 ml of DMEM-F12 with 1 ml N2 supplement, 1 ml Glutamax supplement and 1 ml MEM-NEAA. Add Heparin for the final concentration 1 μg ml-1 and filter using a vacuum driven 0.2 μm filter unit. Store at 2-8°C for up to two weeks.
Cerebral organoid improved differentiation media without vitamin A (IDM-A)
For approx. 250 ml medium, combine 125 ml DMEM-F12, 125 ml Neurobasal, 1.25 ml N2 supplement, 62.5 μl Insulin, 2.5 ml Glutamax supplement, 1.25 ml MEM-NEAA and 2.5 ml penicillin-streptomycin and 5 ml B27 supplement without vitamin A. Prepare a 1:100 dilution of 2-Mercaptoethanol in DMEM-F12 and add 87.5 μl of this to the media. Filter the media using a vacuum driven 0.2 μm filter unit and store at 2-8°C for up to two weeks. For IDM-A with CHIR, add 1:1000 CHIR99021 stock solution for the working concentration of 3 μM.
Cerebral organoid improved differentiation media with vitamin A (IDM+A)
For approx. 500 ml medium, combine 250 ml DMEM-F12, 250 ml Neurobasal, 2.5 ml N2 supplement, 125 μl Insulin, 5 ml Glutamax supplement, 2.5 ml MEM-NEAA, 5 ml penicillin-streptomycin, 5 ml ascorbic acid solution, 3.5 μl neat 2-Mercaptoethanol, 10 ml B27 supplement with vitamin A and 500 mg sodium bicarbonate. Filter the media using a vacuum driven 0.2 μm filter unit and store at 2-8°C for up to two weeks. Directly before use, add 1:500 Amphothericin B. For IDM+A with dissolved Matrigel, combine 50 ml of cold IDM+A with 1ml of Matrigel.
Serum-supplemented slice culture (SSSC) medium
For approximately 500 ml of medium, combine 445 ml of high glucose DMEM supplemented with GlutaMAX, 50 ml of FBS and 5 ml of 50% (w/v) glucose solution. Sterile filter using a Stericup 0.2 μm filter unit, prepare 40 ml aliquots in 50 ml conical tubes and store at -20 °C for up to 6 months. Prior to use, supplement the required volume of SSSC medium with Antibiotic-Antimycotic (100x) solution to 1x final concentration.
Serum-free slice culture (SFSC) medium
For approximately 500 ml of media, combine 480 ml of Neurobasal medium with 10 ml of B27 supplement, 5 ml of 50% glucose solution and 5 ml of GlutaMAX. Following sterilization by filtration through a 0.2 μm Stericup filter unit, the medium can be stored at 2-8 °C for up to one month. Prior to feeding, supplement the required volume of SFSC medium with Antibiotic-Antimycotic (100x) solution to 1x final concentration.
Equipment Setup
Orbital shaker
Clean the orbital shaker by spraying with 70% v/v ethanol and place it on a shelf in the tissue culture incubator. Make sure the shelf above is positioned with enough room for tissue culture dishes to be placed on the shaker. Move the power cable to the side so that it can exit the incubator without hanging in front of tissue culture dishes and tape to hold in place to ensure that the incubator door closes easily. Set the orbital shaker to 57 rpm. This speed is ideal for the described shaker with an orbit diameter of 25 mm, and a dish size of 5-6 mm diameter with 5 ml of media.
Vibrating microtome and sectioning equipment
Install a vibrating microtome, also known as vibratome, in a laminar flow hood and sterilize by UV light irradiation. After each use, clean the stage chamber, specimen plate and blade holder with 70% ethanol (v/v) and apply UV treatment. The set-up should be used exclusively for live samples and should not come into contact with fixatives. Due to the delicate nature of organoid samples, the vibrating microtome should allow for a wide range of amplitude, frequency and speed settings. It is usually necessary to adjust these parameters from organoid to organoid to preserve tissue integrity and obtain optimal results. All our ALI-CO specimens are prepared on a Leica VT1000 S vibrating blade microtome system. If a laminar flow hood is not available, vibratome sectioning can be performed under non-sterile conditions, although this is not recommended. The antibiotics and antimycotics in the SSSC and SFSC media should prevent (or strongly limit) contamination, thus allowing long-term culture despite potential exposure to contaminants. Prior to use, paintbrushes, peel-A-way molds and razor blades should be sterilised by UV-treatment.
Micropipetting tubing apparatus assembly
Take an aspirator tube assembly unit for calibrated microcapillary pipettes, replace the micropipette holder with a 0.2 μm select pipette filter, attach a ~30 cm piece of silicone tubing to the other side of the filter and at the free end of the silicone tubing insert the micropipette holder. Prior to use, insert a sterile 200 μl filter tip in the mouth piece.
Borosilicate glass micropipette pulling
Pull borosilicate glass micropipettes using a micropipette puller. We use a P2000 micropipette puller (Sutter Instrument) with the following settings: heat – 550, filament – 5, velocity – 25, delay – 150, pull – 150. Exact settings may need to be adjusted due to instrument-to-instrument variability. Store the micropipettes in a 10 cm cell culture dish with a bit of Blu Tac or plasticine to hold them in place and protect the tip.
Procedure
Preparation of fibrous microscaffolds *TIMING 0.5-1 h
CRITICAL Perform all steps in a cell culture hood to avoid contamination
-
1.
Remove vicryl-polyglactin-coated suture from protective wrapping and place the free end on a sterile tray. With the blunt side of a scalpel, splay about 1 cm of the suture (Figure 1b). Cut into 1 mm (approx.) pieces. Cut a total of 5 cm along the suture length and gather in one place on the tray.
-
2.
Pipette 5 ml hES media into a 15 ml tube.
-
3.
Aspirate about 500 μl of hES media with a cut p1000 tip and pipette the microfibers up and down to fully immerse.
CRITICAL STEP Cut a p1000 pipette tip with sterile scissors to obtain an opening of 3-5 mm in diameter to allow the fibres to enter the pipette tip.
-
4.
Transfer media with microfibers to a 15 ml conical tube.
-
5.
Repeat until most of the fibres have been collected. Avoid any large clumps that might have formed.
-
6.
With a cut p200 tip, transfer 150 μl of the microfiber suspension into a well of a regular 96-well round bottom plate.
CRITICAL STEP Cut a p200 pipette tip with sterile scissors to obtain an opening of 2-3 mm in diameter to allow the fibres to enter the tip.
-
7.
Observe the fibers under a microscope at low magnification (Figure 1c). Estimate the number of microfibers in the well. For a 1x solution, you need 5-10 fibers per well. If, for example, there are 30 fibres in the well, the fiber suspension is therefore approximately 6x.
? TROUBLESHOOTING
Generation of embryoid bodies *TIMING 1-2 h
-
8.
When hESC or iPSC colonies grown in one well of a 6-well plate have reached about 80% confluency, the cells are ready for generation of embryoid bodies (EBs). Typically, one well will yield approximately 96 EBs. If making organoids with scaffolds, follow option A. Alternatively, if starting with smaller EBs (with fewer cells), follow option B.
-
CRITICAL STEP The quality of the stem cell colonies is critical to the success of cerebral tissue formation. The colonies should have compact morphology with no differentiation and display optimal features of pluripotency.
-
Using a fibrous microscaffold
Prepare 1x fibre suspension by diluting the concentrated fibre suspension in low bFGF hES media with ROCK inhibitor (1:100, final concentration 50 μM). Prepare 150 ul of 1x fibre suspension per EB. When handling fibres, use a cut p200 or p1000 tip
Prepare an additional 2 ml of low bFGF hES media with ROCK inhibitor without fibres
Remove the culture media and wash the cells with 1 ml D-PBS without calcium and magnesium. Add 500 μl Accutase solution per well of a 6-well plate and place the cells back in the incubator for 4-6 min.
Tap the dish vigorously to detach the cells
Spray the well with 1 ml of low bFGF hES media with ROCK inhibitor and pipette up and down up to 5 times to achieve a single cell suspension. Transfer to a 15 ml conical tube
Centrifuge cells at 270g for 5 min. Aspirate the supernatant without disturbing the pellet and re-suspend in 1 ml low bFGF hES media with ROCK inhibitor
Count live cells by taking a small aliquot and adding an equal volume of Trypan blue to stain dead cells and count using a hemocytometer or an automated cell counter. Use the average of two replicates to calculate the live cell number
Add the required volume of the cell suspension to the 1x fibre suspension from Step 1 to obtain 18,000 live cells/150 μl
With a cut p200 tip, plate 150 μl in each well of a low attachment 96-well U-bottom plate. Agitate the tube between wells to ensure even distribution of cells and fibres. When seeding one whole or multiple 96-well plates, transfer the cell suspension and fibre solution to a reservoir and use a multichannel pipette with cut pipette tips for seeding
-
Starting with smaller EBs (decreased diameter) containing fewer cells
Prepare 150 μl low bFGF hES media with 1:100 ROCK inhibitor per EB.
Prepare an additional 2ml of low bFGF hES media with ROCK inhibitor
Remove the culture media and wash the cells with 1 ml D-PBS without calcium and magnesium. Add 500 ul Accutase solution per well of a 6-well plate and place the cells back in the incubator for 4-6 min
Tap the dish vigorously to detach the cells
Add 1 ml of low bFGF hES media with ROCK inhibitor to the well and pipette up and down up to 5 times to achieve a single cell suspension. Transfer to a 15 ml conical tube
Centrifuge cells at 270 g for 5 min. Aspirate the supernatant without disturbing the pellet and re-suspend in 1 ml low bFGF hES media with ROCK inhibitor
Count live cells by taking a small aliquot and adding an equal volume of Trypan blue to stain dead cells and count using a hemocytometer or automated cell counter. Use the average of two replicates to calculate the live cell number
Add the required volume of media from Step 1 to obtain 2,000 live cells/150μl.
With a p200, plate 150 μl in each well of a low attachment 96-well U-bottom plate. Agitate the tube between wells to ensure even distribution of cells.
-
-
9.
Once cells have been seeded into the 96-well plate, place the plate in the incubator and try not to disturb it for the first 24 hours. The day of generation of the EBs is Day 0.
Feeding EBs and initiation of germ layer differentiation *TIMING 5-6 d
-
10.
Observe the plate under a tissue culture microscope 24 hours after seeding. By now, a single EB should have formed in each well. The presence of some dead cells around the EB and at the bottom of the well will not disturb development of the EB at the center.
? TROUBLESHOOTING
-
11.
Feed the EBs on Day 3 with hES without bFGF or ROCK inhibitor. Carefully aspirate approximately half of the medium without disturbing the EB at the bottom of the well. Top up with 150 μl of fresh media.
Induction of primitive neuroepithelia *TIMING 5-6 d
-
12.
When the EBs are bright at the periphery and with smooth edges (typically day 5-6) (Figure 1d), transfer the EBs to a low attachment 24-well plate containing 500 μl Neural induction medium. Gently pipette up individual EBs into a cut p200 pipette tip and place 1 EB in each well. Be careful not to disrupt the EB and try to minimize the volume of medium carried over with the tissue.
-
CRITICAL STEP Use a cut p200 tip to produce an opening of about 2-3 mm when transferring EBs. Do not attempt to scoop the EB out of the well with a spatula or other tools as this will damage the EB.
? TROUBLESHOOTING
-
13.
After 48 h, carefully aspirate about half of the spent media without disturbing the EB and top up with 500 μl Neural induction medium.
-
14.
After another 48 hrs, observe the EBs under a tissue culture microscope. The EBs should have become brighter around the outside, indicating neuroectodermal differentiation.
-
15.
After 4-5 days in Neural induction media, the primitive neuroepithelia should begin to show radial organization of a pseudostratified epithelium.
CRITICAL STEP Healthy cell aggregates should have smooth edges. The neuroepithelium develops on the outer surface and appears translucent and radially organised6. We occasionally observe outgrowths or buds of translucent tissue that is not radially organized but that does not usually interfere with organoid formation.
Embedding neuroepithelial tissues in Matrigel droplets *TIMING 1-2 h
-
16.
When the aggregates display radially-organized optically-translucent epithelia, embed them in Matrigel droplets (usually on day 11).
CRITICAL STEP Embed the tissues in Matrigel soon after the neuroepithelium becomes evident since late transfer can impact later morphology of cerebral tissues.
-
17.
Thaw Matrigel on ice at 4°C for 2 h to overnight.
-
18.
In a sterile hood, prepare a dimpled Parafilm sheet for supporting Matrigel droplets. Place a 5x5 cm square of Parafilm on an empty tip rack for size p10 tips, paper side up. Press down into the Parafilm over each hole to create small indentations. We found that using a 1.5 ml microcentrifuge tube to apply pressure produces good results. Although Parafilm cannot be sterilized, we found that cleaning with 70% ethanol prevents culture contamination Alternatively, UV treatment can be employed.
-
19.
Press a grid of 4x4 dimples (16 in total). Pick up the sheet with sterile tweezers, shake off excess ethanol and trim off the edges with sterile scissors. Place the square of Parafilm into a 60 mm tissue culture dish and, if any ethanol is left on the sheet, allow to air dry.
CRITICAL STEP A grid of 4x4 will fit in a 60 mm dish, but not a larger piece of parafilm. We recommend working with a maximum of 16 droplets at a time to prevent drying out of the tissues and premature polymerisation of Matrigel.
-
20.
Using a cut p200 tip, transfer individual organoids into each Parafilm dimple.
CRITICAL STEP Cut a p200 pipette tip with sterile scissors to obtain an opening of 2-3 mm in diameter. Be sure the opening is not too small as this will disrupt the EB. Do not attempt to move the EBs using tools such as a spatula as this will damage the tissues.
-
21.
Once all tissues have been transferred, remove excess media from around each aggregate by aspirating with an uncut p200 tip.
CRITICAL STEP Point the tip opening away from the tissue to prevent it from entering the tip, which will result in damage.
-
22.
Immediately add about 30 μl of Matrigel to each aggregate. We find that pipetting up a larger volume of Matrigel and then slowly releasing dropwise into each dimple (rather than pipetting 30 μl at a time) helps to prevent formation of bubbles. The exact volume of Matrigel is not crucial.
CRITICAL STEP Add the Matrigel quickly to prevent drying out of the tissues.
-
23.
Using a p10 tip, position each aggregate in the center of its droplet.
CRITICAL STEP This must be done immediately after adding the droplet before Matrigel starts to polymerise.
-
24.
Place the 60 mm dish containing droplets on Parafilm in the 37°C incubator and allow the Matrigel to polymerise for 20-30 min.
-
25.
Add 5 ml of Neural Induction medium to the 60 mm dish.
-
26.
To detach residual Matrigel droplets (some might have detached by themselves already), turn the Parafilm sheet over using sterile forceps and agitate either the dish or the sheet until the droplets fall off. Wash any persistent droplets off the Parafilm sheet with a gentle stream of media ejected from a p1000 pipette.
-
27.
Return the dish to the 37°C incubator.
-
28.
After 1-2 days, observe the embedded tissues under the microscope. Radially organized neuroepithelial buds with small central lumens should begin to be visible (Figure 1e).
Expansion of the neuroepithelial buds *TIMING 3 d
-
29.
2 days after Matrigel embedding (usually on day 13), aspirate most of the spent media from the 60 mm dish. Be careful not to disrupt the tissues.
-
30.
(OPTIONAL) Replace the media with 5 ml of IDM-A with 3 μM CHIR99021 and return to the 37°C incubator for 3 days (see Experimental Design section).
-
31.
On day 16, when large neuroepithelial buds should be readily apparent with limited non-neural outgrowth, replace the media with fresh IDM-A (without CHIR99021).
? TROUBLESHOOTING
Growth of cerebral tissue *TIMING 22d
-
32.
On day 18, refresh the medium and transfer the 60 mm dish with organoids to an orbital shaker installed in the 37°C incubator.
CRITICAL STEP The optimal speed for the orbital shaker used in this protocol and for the model of orbital shaker listed is 57 rpm. We find that an incorrect shaker speed disrupts cortical tissue organization.
-
33.
On day 20, transfer half of the organoids from the 60cm dish to a fresh dish using a cut p1000 tip and remove the spent medium.
CRITICAL STEP Cut a p1000 pipette tip with sterile scissors to obtain an opening of 3-5 mm in diameter. Be sure the opening is not too small as this will disrupt the EB. Do not attempt to move the EBs using tools such as a spatula as this will damage the tissues.
-
34.
With two syringe needles, carefully remove most of the Matrigel from around each organoid. Use one needle as a pin to hold the Matrigel droplet and the side of the opening of the other one as a blade to cut or tear off the Matrigel (Figure 1f) This step can be performed in a sterile dissection cabinet or against a dark background in a cell culture hood. Alternatively, Matrigel can be broken off by gently pipetting the organoids up and down in a 5 ml serological pipette 3-5 times.
CRITICAL STEP We found that removing Matrigel improves the architecture of tissues to be analysed by microscopy.
CRITICAL STEP Removing Matrigel by pipetting into a serological pipette is less precise and can result in tissue damage if performed with too much force.
-
35.
Carefully remove the Matrigel debris from the dish by aspirating with an uncut p1000 tip.
-
36.
Add 5 ml IDM-A and return to the orbital shaker at 37°C.
-
37.
Refresh the media every 3-4 days.
Growth of cortical plate and maturation of the cerebral tissue *TIMING 10-20 d
-
38.
On day 40, prepare IDM+A supplemented with 1:50 dissolved Matrigel (IDM+A+MG). Thaw Matrigel on ice at 4°C for 2 h to overnight and add to cold IDM+A before warming to room temperature.
CRITICAL STEP Matrigel needs to be added to cold medium; otherwise, it will polymerise and precipitate.
CRITICAL STEP IDM+A+MG must be used on the same day as it was prepared. If the medium is cooled down and then re-warmed, the Matrigel polymerises and precipitates.
CRITICAL STEP Addition of dissolved Matrigel is required for cortical plate formation.
-
39.
Change the media every 3-4 days and monitor organoids at each media change by examining under a transmission or dissecting microscope. Sometime between days 50-60, the lobes should become quite large (Figure 1g) and the cortical plate should form (seen as a darker line adjacent to lighter margin on the outline of a cortical bud under a phase contrast microscope (Figure 1h)).
-
CRITICAL STEP If the culture medium turns yellow before the next media change, divide organoids from each 60 mm dish into two dishes. Use a cut p1000 to handle the organoids.
? TROUBLESHOOTING
ALI-CO preparation *TIMING 3-8 h
CRITICAL Only organoids with obvious lobes and visible cortical plates should proceed to air-liquid interface culture.
-
40.
Heat up the 3% low-melt agarose solution in a microwave and mix at regular intervals to prevent formation of clumps. Once the agarose has melted completely, place in a 40 °C water bath with heat transfer beads to equilibrate for at least one hour.
-
41.
Prepare all equipment necessary for the embedding procedure in a laminar flow cabinet or, if this is not available, on a clean lab bench.
-
42.
Approximately 1-1.5 h after placing the agarose at 40 °C, cut a 1 ml tip at an angle to produce an opening of 2-3 mm and transfer the selected organoids as individual droplets onto the lid of a 10 cm dish (Figure 1i). As the lid has shallower edges than the plate, it is more suitable for this procedure. After transfer, aspirate the medium from the droplet using a P1000 pipette, taking care not to damage the tissue.
-
43.
By pipetting up and down with a cut 1 ml tip, wash the individual organoids in ice-cold HBSS with Ca2+ and Mg2+, taking care to remove any leftover Matrigel attached to the surface of the organoid.
-
CRITICAL STEP It is essential to remove all Matrigel from the surface of the organoid as this can cause the slice to detach from the agarose during sectioning, thus compromising the quality of the preparation.
? TROUBLESHOOTING
? TROUBLESHOOTING
-
44.
Agarose embedding. Remove the HBSS used for the wash and fill the required number of peel-A-way molds with 5-8 ml of 3% low-melt agarose solution.One after the other, process the organoids as follows: using a cut 1 ml tip, apply a drop of agarose on the organoid, pipette the organoid up and down to dilute any remaining HBSS and then transfer it to the mold. By this point, the agarose in the mold has become more viscous so although the organoid sinks to the bottom, it should not come into contact with the plastic bottom of the mold, stopping a few millimeters above it (Figure 1j).
-
CRITICAL STEP Low melt agarose tends to solidify quickly at room temperature so it’s important to process the organoids quickly and in small batches (max 5 organoids at a time). The ideal number of organoids is 1-2 per mold, but we have successfully processed up to 4 organoids in one mold. The rate-limiting step in the preparation of ALI-COs is sectioning, and samples should be processed on a vibratome within approximately 1.5-2 h of embedding.
? TROUBLESHOOTING
-
45.
Incubate the molds containing the embedded organoids on ice for a minimum of 20 min. A good indicator of fully hardened agarose ready for sectioning is the formation of water condensation on the surface of the agarose block.
-
46.
During the incubation of the blocks on ice, prepare the cell culture inserts for organoid slice collection. Place the required number of cell culture inserts in a 6-well tissue culture dish – one organoid typically yields between 4 to 12 slices and we do not recommend exceeding 3 slices per insert; 2 to 4 inserts should be sufficient for one organoid.
-
47.
Apply 1 ml of SSSC medium to the side of the wells to bathe the cell culture inserts from below.
CRITICAL STEP The hydrophobicity of the cell culture insert membranes can vary depending on the lot, leading to formation of air bubbles under the insert. If this occurs, simply wash the bottom side of the insert by pipetting, or agitate side to side in the media.
-
48.
Remove the peel-A-way mold without damaging the agarose block and cut off excess agarose around the organoid using a razor blade.
-
CRITICAL STEP Do not remove too much agarose as this could damage the organoids during sectioning. On the other hand, having a large amount of agarose around the organoid can intensify the vibration of the slice during sectioning, leading to detachment of tissue from the surrounding agarose. It is therefore essential to minimize the size of the block, ensuring that the tissue is surrounded evenly by agarose. For a square block, there is no preference for its orientation relative to the sectioning direction; however, for a rectangular block, sectioning should be performed along the direction of the short side of the rectangle in order to minimize vibration.
? TROUBLESHOOTING
-
49.
Glue the sample to the specimen disk using superglue and allow it to dry for a few minutes.
-
50.
Secure the specimen disk to the buffer tray and fill the buffer chamber with ice-cold HBSS with Ca2+ and Mg2+. To maintain a cool temperature, exchange the HBSS regularly or, alternatively, fill the cooling bath with crushed ice.
-
51.
Secure the blade to the knife holder, adjust the clearance, fix the knife holder to the vibrating microtome and proceed to sectioning.
-
CRITICAL STEP The sectioning thickness should be set to 300 μm. Ideal frequency and speed settings can vary from instrument to instrument and should be determined empirically. On a Leica VT1000S system, we typically section at frequencies of 30-60 Hz and speeds of 0.15-0.30 mm/s. It is essential to adjust these parameters to preserve tissue integrity and ensure that the tissue slice does not detach from the supporting agarose and its integrity is entirely preserved. If necrosis has already started to develop in the organoid core, dead tissue may detach during sectioning, leaving a large cavity in the centre. This should not negatively affect the preparation, provided the live tissue remains intact, but it should be avoided. Early necrosis in the core can occur if the organoids are particularly large due to accelerated growth or as a result of fusion of multiple organoids. The age at which the organoids are sectioned should be adjusted accordingly. Typically, we perform sectioning between days 45-60.
? TROUBLESHOOTING
-
52.
Pick up the section using a disposable scalpel and a brush (Figure 1j).
-
CRITICAL STEP The brush should only be used to manipulate the agarose as direct contact with the tissue would compromise the section. Alternatively, a second scalpel can be used in place of a brush for collection of the section. In fact, while the tissue sticks to the bristles of the brush, it does not stick to the scalpel blade. This collection method is particularly useful if the tissue has detached from the surrounding agarose.
? TROUBLESHOOTING
-
53.
Lay down the section on a cell culture insert by gently pushing the agarose edge with a brush or a scalpel (Figure 1j).
-
54.
During sectioning, keep the plate with the sections on ice.
-
55.
After sectioning, incubate the slices for 1-2h at 37 °C
-
56.
Observe the sections to check their morphology. Sections should be flat against the insert and lobes should still be readily apparent, with clearer ventricular zones and darker intermediate zones and cortical plate (Figure 1k). Replace the SSSC medium with 1ml of SFSC medium.
-
57.
For daily feeding, aspirate ~850 μl of medium with a P1000 pipette and discard the tip. Repeat this step for all wells with ALI-CO cultures.
-
58.
Add 850 μl of SFSC medium to each well using the dispense function of a repeater pipette. Use individually wrapped sterile combitips and do not let the tip come into contact with any surface during feeding.
-
CRITICAL STEP Changing pipette tips when removing medium from different wells and using sterile combitips will prevent the spread of potential contamination across multiple inserts. Do not dispense the medium on the slice but along the wall of the well so as to bathe the slice from below and maintain air above the slice. Do not exceed ~1 ml of total medium in the well as it would permeate through the insert and submerge the slice.
PAUSE POINT ALI-COs can be maintained indefinitely as slices, as long as they are fed every day. Usually, the agarose will remain, and will not pose a problem. On the contrary, it seems to provide a degree of structural support, particularly for the outgrowth of escaping tracts. Therefore, it is not necessary to remove the attached agarose.
? TROUBLESHOOTING
Timing
Steps 1 – 7, preparation of fibrous microscaffolds: 0.5-1 h
Steps 8 – 9, generation of embryoid bodies: 1-2 h
Steps 10 – 11, feeding EBs and initiation of germ layer differentiation: 5-6 d
Steps 12 – 15, induction of primitive neuroepithelia: 5-6 d
Steps 16 – 28, embedding neuroepithelial tissues in Matrigel droplets: 1-2 h
Steps 29 – 31, expansion of the neuroepithelial buds: 3 d
Steps 32 – 37, growth of cerebral tissue: 22 d
Steps 38 – 39, growth of cortical plate and maturation of the cerebral tissue: 10-20 d
Steps 40 – 58, ALI-CO preparation: 3-8 h
Anticipated Results
This protocol outlines the generation and growth of mature cerebral organoids with a primarily telencephalic identity. These methods result in tissues with better morphology and reproducibility than unpatterned cerebral organoids with more mixed identities. However, they are more limited to telencephalon and therefore do not typically generate other brain regions (i.e. retina, midbrain, etc.)15. The air-liquid interface culture can be applied to any neural organoid to allow modelling and visualization of later stages of neurogenesis, astrogliogenesis, axon guidance and the establishment of neuronal networks. This approach allows for extended culture over periods of months or even years with enhanced overall tissue health and cell survival, in particular neuronal survival.
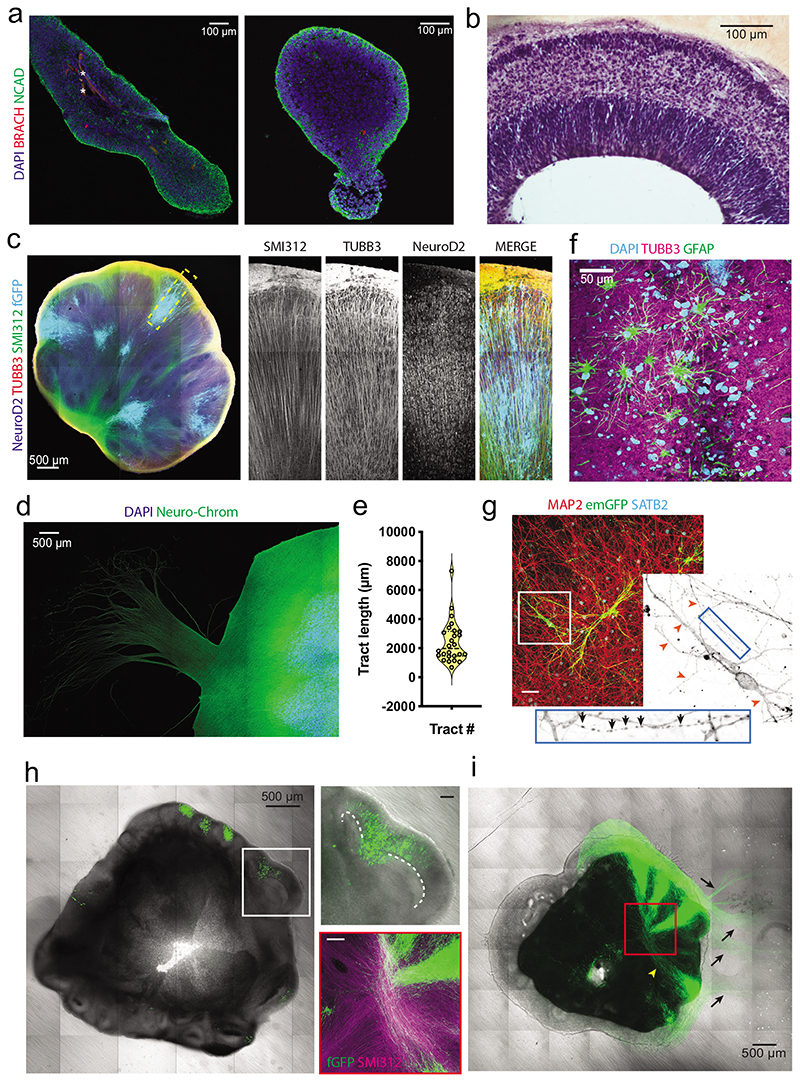
The use of a microscaffold or reduced spheroid size to increase surface area-to-volume ratio is a very effective way to improve neural induction efficiency without the use of small molecules (Figure 2a). However, this protocol should also be compatible with small molecules, such as dual-SMAD inhibitors, which may be necessary for certain cell lines to achieve efficient neural identity induction. Neural induction should be evident based on the degree of clearing of EBs. Darker EBs that lack smooth borders are generally a sign of nonneural identity or cell death, both of which will scatter light and make the EBs appear darker. Due to its epithelial nature, proper neuroectoderm will exhibit an ordered arrangement that scatters light less, allowing more of the light to pass through, and will thus appear more translucent.
Figure 2. Expected results upon histological and immunohistochemical analysis.
a. Image of a micropatterned organoid at Day 11 (left-hand panel) and a non-micropatterned organoid made with 3,000 cells at Day 10 stained for germ layer markers N-cadherin (N-cad) for neural ectoderm and Brachyury (Brach) for mesoderm (right-hand panel). Both organoids show primarily neural differentiation. Autofluorescent fibres are marked with an asterisk. Scale bars = 100 μm. b. Image to show radial arrangement of cell nuclei in the cortical plate as visible by hematoxylin and eosin (H&E) staining of a section of a Day 69 organoid. Scale bar = 100 μm. c. Immunofluorescence image of an 88-day-old H9 ALI-CO (60+28 days at the ALI) electroporated with fGFP on day 58. The section was stained for the neuronal nuclear marker NeuroD2, the neuronal cytoplasmic marker TUBB3 and the axonal marker SMI312. Large SMI312+/TUBB3+ axon bundles are seen to project between different lobules of the ALI-CO, and the lobules display radial organization as shown by the combination of markers used. A magnified view of the boxed area in the far-left-hand image is shown in the other images, showing radial organization of neurons (NeuroD2+) within lobules to form an ordered array of cell bodies (fGFP+), axons (SMI312+) and more widely neuronal processes (TUBB3+), as well as a merged image showing all of these processes at once. Scale bar = 500 μm. d. Immunofluorescence image of an 88-day-old H9 ALI-CO (54+34 days at the ALI), showing a large escaping tract positive for the panaxonal marker Neuro-Chrom but devoid of cell bodies (DAPI-). Scale bar = 500 μm. e. Violin plot of tract length quantification performed on a total of 26 fGFP+ tracts from 6 ALI-COs aged between 69-75 days total derived from H1 and H9 hESCs. The solid line corresponds to the median (M=1978.5 μm) and the dashed lines to the first (Q1=1472.7 μm) and third (Q3= 3126.5 μm) quartiles. f. Immunofluorescence image of a 168-day-old H9 ALI-CO (68+100 days at the ALI), showing a dense TUBB3+ neuropil populated by GFAP+ astrocytes with characteristic star-shape. Scale bar = 50 μm g. Immunofluorescence image of a 280-day-old H9 ALI-CO (45+235 days at the ALI) sparsely labeled with EmGFP Sendai virus and stained for the dendritic marker MAP2 and the nuclear neuron upper-layer marker SatB2 (left-hand panel). The image reveals a thick and healthy neuropil with individual EmGFP+ neurons displaying mature morphology. The right-hand panel is a magnified view of the area highlighted by a white box in the left-hand panel. The blue box shows a further magnification of an axon with varicosities (black arrows). Scale bars 50 μm. h. Overlay of bright-field and fluorescence images of an fGFP electroporated 60-day-old ALI-CO immediately after sectioning. The top-right panel, which is a magnified view of the area highlighted by a white box in the left-hand panel, shows individual electroporated cortical lobes. The dashed white line marks the apical surface. Scale bars = 500 μm (left-hand panel) and 100 μm (right-hand panels). i. Overlay of bright-field and fluorescence images of the ALI-CO slice shown in Figure 2h after 28 days at the ALI (60+28 days total). Over time, the ALI-CO has developed thick internal (yellow arrowhead) and external (black arrows) fGFP+ tracts. The red box marks the magnified region stained for panaxonal marker SMI312+ in combination with the electroporated fGFP+, showing their presence in a specific tract within the interior. Scale bars = 500 μm.
If neural identity is properly attained, the EBs should generate large neural tube-like buds upon Matrigel embedding. These buds should appear bright, with smooth borders, and should be several cell layers thick from the internal lumen to the surface (Figure 3a). If neural identity is not properly attained, many other cell types and structures can be observed. Sometimes, these can also appear to be epithelial, but such nonneural epithelial buds will be very small (Figure 3b) and fail to exhibit the extensive pseudostratification seen in the neuroepithelium. Some neural crest outgrowth is often seen, which is evident as migratory fibroblast-type cells. This is generally not a problem, but too much outgrowth can make it difficult to observe development of telencephalic lobes. In general, suboptimal neuroepithelial buds (Figure 3b-d) indicate incomplete or absent neural differentiation and these organoids should not be cultured further. Over the course of several weeks, the buds will become large lobes containing fluid-filled ventricles (Figure 1g), and the use of dissolved Matrigel will enable formation of a dense cortical plate along the perimeter of these lobes (Figure 1h, 2b). However, some organoids will start out looking promising at the neuroepithelial bud stage but later develop abnormally. For example, cysts sometimes develop and, although occasional cysts can be acceptable (Figure 3e), an overabundance can impact adjacent telencephalic tissue development (Figure 3f). Furthermore, cortical lobes should continue to exhibit a smooth border and should protrude from the main mass. Often, the ventricular zone and even intermediate zone, cortical plate, and marginal zone are visible under transmitted light (Figure 1h). The absence of these morphological hallmarks (Figure 3g-h) suggests suboptimal development and we prefer to remove these as a quality control measure.
Figure 3. Examples of morphological hallmarks and suboptimal morphologies.
a-d. Representative images of acceptable and suboptimal organoids based on the morphology of neuroepithelial buds. a. An optimal organoid 3 days after Matrigel embedding with excellent neuroepithelial bud morphology demonstrated by their brightness, smooth borders and apicobasal thickness (arrow). b. A suboptimal organoid 2 days after Matrigel embedding, displaying no evidence of proper neuroepithelial bud formation, instead forming very thin and convoluted epithelia (arrowhead), likely of a nonneural identity. c. A suboptimal organoid 5 days after Matrigel embedding, displaying suboptimal buds that are dark and lack a visible lumen (black arrowhead). A cyst can also be seen (red arrowhead), which should not be present at this early time point and further indicates improper identity. d. A suboptimal organoid 5 days after Matrigel embedding, displaying some healthy neuroepithelial buds (arrows) but an abundance of nonneural thin epithelia (red arrowheads) and other dark buds lacking a smooth border (black arrowhead). e-h. Representative images of acceptable and suboptimal organoids based on cortical lobe morphology. e. An acceptable organoid at day 55 with numerous cortical lobules (arrows). Although a large cyst is present (arrowhead), the adjacent cortical tissue appears well developed and healthy. f. A suboptimal organoid at day 55, lacking visible cortical lobes and displaying numerous cysts (black arrowheads) and central disorganized tissue (red arrowhead). g. A suboptimal organoid at day 55, displaying regions with a smooth border but that fail to form protruding lobules (black arrowhead). The rest of the tissue also appears disorganized and lacks smooth borders (red arrowhead). h. A suboptimal organoid at day 40, displaying convoluted thin epithelia (red arrowheads) and numerous migrating cells (black arrowheads).
It is important to keep in mind that, although the reliability of telencephalic organoids with good morphology is greatly improved with this protocol, there can still be more subtle variability in terms of timing or maturation, particularly between different batches of organoids made on different days. There is also variability between cell lines, and the protocol may need to be optimized for different cell lines, particularly with respect to timing. The protocol timing described here works well for the three cell lines described. However, the user should empirically test new cell lines and adjust timing based upon key morphological features described in this protocol.
When preparing ALI-COs, the exact day the samples should be sectioned can vary depending on the batch and the individual organoid. While early sectioning is advisable to preserve the population of deep-layer neurons, late sectioning will favor the upper-layer neuron population. Immediately after sectioning, organoids will display ventricles comprising radially aligned progenitors and what remains of the fluid-filled ventricles. These structures appear as more translucent areas (Figure 1k). Overlying these progenitor zones is a layer of neurons and basal progenitors that appears as a darker cell layer. Because the ALI-COs described here are derived from telencephalic organoids, it is not uncommon to observe various regions of telencephalic identity, in addition to dorsal cortex, such as choroid plexus. It is possible to monitor the maturation of ALI-COs by brightfield microscopy. Over the course of weeks, the clear areas corresponding to the progenitor zones will progressively shrink and translucent internal tracts will develop, as well as escaping tracts that project into the agarose.
After approximately one month in culture, immunofluorescence analysis of high-quality preparations from organoids with a cortical plate reveals extensive growth of large bundles, reminiscent of CNS tracts, projecting into the internal regions of the tissue (Figure 2c). The cortical plate also still remains and has thickened, forming an ordered lattice with radial alignment. It is also possible to observe large, robust escaping bundles positive for the pan-neuronal marker Neuro-Chrom and mostly lacking cell bodies (Figure 2d). These tracts can become quite long, extending over several mm over the course of 2-3 weeks (Figure 2e). As this approach allows for prolonged maintenance of complex tissue (we have successfully cultured ALI-COs for periods of over one year), it can be used to study other cells types that arise later in development, such as astrocytes (Figure 2f), within a healthy cellular milieu.
For studies of cell morphology, we present two alternative EmGFP Sendai transduction methods to sparsely label cells in ALI-COs. Using a very basic set of tools, this method can be used to efficiently label neurons and, in combination with immunostaining, it enables the study of their morphology, highlighting details such as dendrites, dendritic spines and even single axons and their boutons (Figure 2g). Furthermore, because of the high signal-to-noise ratio, this methodology is well suited for segmentation studies where high contrast is required.
ALI-CO maturation and processes such as progenitor cell division, neuronal migration and axon guidance can be studied in detail by injection and electroporation of a construct expressing farnesy-lGFP (fGFP) into the ventricles of organoids prior to sectioning. Farnesyl-GFP is incorporated into the plasma membrane of cells and, as such, it aids in the visualization of cell processes, such as radial glia endfeet, axons and growthcones. Within the first week of electroporation, we are able to monitor the behaviour of progenitor cells within cortical lobes, processes such as interkinetic nuclear migration (IKNM) and progenitor divisions (Figure 2h). Over time, robust fGFP+ tracts can be seen connecting different lobes of the organoid and projecting away from the slice (Figure 2i). Immunofluorescence staining for the axonal marker SMI312 on the same sample reveals that fGFP+ tracts are only a small subset of a large and complex network of tracts that develop over time (Figure 2i).
Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 7 | Fibers are not straight and are curly Fibers are clumped |
Fibers have been shaved too much Fibers have not been separated well enough |
Try a more upright angle of the scalpel blade in order to cut individual fibers off without shaving them or dragging the blade along the fibre Use a narrower angle in order to separate individual fibers while cutting Press down on the suture to splay the individual fibers before cutting |
| 10 | Cells are not coating the fibers and are clumped in the middle Microscaffold EBs are very round with fibers inside the sphere |
Fibers are too long Fibers are too short |
Make sure to cut fibers about 1mm in length Same as above |
| 12 | EBs are very dark | Lack of neuroectoderm EBs are too large |
Check the quality of starting PSCs. PSCs grown in StemFlex or E8 tend to be more efficient at neural induction. Try the commercially optimized kit. Small molecules (dual SMAD inhibitors) may be needed. Check the accuracy of the cell counting |
| 31 | A lot of outgrowth of non-neural identities | Failed neural induction Too much neural crest |
Same as above Try again, it may be an issue with the batch Try the commercially optimized kit |
| 39 | Buds remain small and underdeveloped Lack of formation of a cortical plate |
Shaker speed is not optimal Tissue culture vessel or volume is not optimal Shaker speed or vessel is not optimal Matrigel is not fully dissolved |
Be sure to adjust shaker speed depending on orbit diameter Use a 60mm dish with the shaker speed at 57rpm. A smaller or larger dish will result in non-optimal centrifugal forces Same as above Be sure to add Matrigel to cold media just before feeding to avoid Matrigel precipitation |
| 51 | The organoid tissue detaches from the agarose matrix during sectioning | Leftover Matrigel or media around the organoid can cause the tissue to detach | Remove all matrigel from the surface of the organoid prior to embedding Rinse the organoid in HBSS and incubate in warm agarose for a few minutes before placing in embedding mold and cooling |
| 51 | The organoid is damaged during sectioning | Sectioning speed and/or vibration frequency may be too high Necrosis has already developed in the core The organoid is too large |
Decrease the sectioning speed and the frequency of vibration Section at earlier stages Section at earlier stages and do not let the organoids fuse |
| 52 | Difficulties handling sections and maintaining tissue integrity | Too much or too little agarose left around the organoid The bristles of the brush come into contact with the tissue |
Cut the agarose around the organoid leaving ~2-mm space all around the organoid Orient the block on the specimen plate so as to minimize the travel distance of the blade Only manipulate the agarose surrounding the organoid section Use two scalpels to manipulate the sections |
| 58 | Poor ALI-CO health after sectioning Contamination has spread across multiple inserts |
Too slow organoid transfer to agarose Too long incubation on ice The antibiotic-antimycotic solution used became spoiled The same tip was used to remove spent medium from multiple wells The repeated pipette combitip came into contact with a contaminated surface |
Embed fewer organoids at one time and work quickly Embed fewer organoids at one time After thawing an aliquot do not refreeze and discard Thaw the antibiotic solution at 4 °C overnight For each well, use a new 1 ml tip to remove the medium Do not touch the side of the wells containing the ALI-COs or any other potentially non-sterile surface with the combitip |
| Box 3,step 13 | Problems loading or expelling the micropipette | Bubbles in the micropipette that interfere with the vacuum | Use a 10 ml syringe to push the air bubbles out of the micropipette. Wash with PBS and ensure there are no further bubbles. |
Acknowledgements
The authors would like to thank members of the Lancaster lab for helpful discussions, especially I. Kelava, as well as Z. Neuburger for photos of the ALI-CO procedure. We also thank the LMB light microscopy facility for assistance with imaging. Work in the Lancaster lab is supported by the Medical Research Council (MC_UP_1201/9) and the European Research Council (ERC STG 757710).
Footnotes
Declaration of competing financial interests
M.A.L. is an inventor on two cerebral organoid patents with licensing agreements with third parties, including Stem Cell Technologies.
Data availability statement
All data generated or analysed during the development of this protocol are included in the figures.
References
- 1.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paşca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadoshima T, et al. Self-organization of axial polarity, inside-out layer pattern, and speciesspecific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S-C, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 5.Eiraku M, et al. Self-Organized Formation of Polarized Cortical Tissues from ESCs and Its Active Manipulation by Extrinsic Signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nature Protocols. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cederquist GY, et al. Specification of positional identity in forebrain organoids. Nat Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. PubMed - NCBI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camp JG, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhaduri A, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka Y, Cakir B, Xiang Y, Sullivan GJ, Park I-H. Synthetic Analyses of Single-Cell Transcriptomes from Multiple Brain Organoids and Fetal Brain. Cell Rep. 2020;30:1682–1689.:e3. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velasco S, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quadrato G, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giandomenico SL, et al. Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22:669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trujillo CA, et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell. 2019;25:558–569.:e7. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones EG, Rakic P. Radial Columns in Cortical Architecture: It Is the Composition That Counts. Cereb Cortex. 2010;20:2261–2264. doi: 10.1093/cercor/bhq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halfter W, Dong S, Yip Y-P, Willem M, Mayer U. A Critical Function of the Pial Basement Membrane in Cortical Histogenesis. J Neurosci. 2002;22:6029–6040. doi: 10.1523/JNEUROSCI.22-14-06029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasu M, et al. Robust Formation and Maintenance of Continuous Stratified Cortical Neuroepithelium by Laminin-Containing Matrix in Mouse ES Cell Culture. PLOS ONE. 2012;7:e53024. doi: 10.1371/journal.pone.0053024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daza RAM, Englund C, Hevner RF. Organotypic slice culture of embryonic brain tissue. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4914. pdb.prot4914. [DOI] [PubMed] [Google Scholar]
- 20.Croft CL, Noble W. Preparation of organotypic brain slice cultures for the study of Alzheimer’s disease. F1000Res. 2018;7:592. doi: 10.12688/f1000research.14500.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paşca SP. Assembling human brain organoids. Science. 2019;363:126–127. doi: 10.1126/science.aau5729. PubMed - NCBI. [DOI] [PubMed] [Google Scholar]
- 22.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelava I, Lancaster MA. Stem Cell Models of Human Brain Development. Cell Stem Cell. 2016;18:736–748. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Jo J, et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe M, et al. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. PubMed - NCBI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cakir B, et al. Engineering of human brain organoids with a functional vascular-like system. Nature Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pham MT, et al. Generation of human vascularized brain organoids. NeuroReport. 2018;29:588–593. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansour AA, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. PubMed - NCBI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller FD, Gauthier AS. Timing Is Everything: Making Neurons versus Glia in the Developing Cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during the development of this protocol are included in the figures.