Abstract
Recurrent herpesvirus infections can lead to numerous forms of diseases ranging from cold sore and genital herpes to encephalitis. Currently, there is an incomplete understanding of the genetic and immunological factors conferring susceptibility to recurrent HSV2 infections in the central nervous system (CNS). Here we describe two adult patients with recurrent HSV2 lymphocytic Mollaret’s meningitis each carrying a rare monoallelic variant in one of the autophagy proteins ATG4A and LC3B2. Importantly, HSV2-activated autophagy was abrogated in patient primary fibroblasts, which also exhibited significantly increased viral replication together with enhanced cell death. HSV2 antigen was captured in autophagosomes of infected cells, and genetic inhibition of autophagy by disruption of autophagy genes, including ATG4A and LC3B2, led to enhanced viral replication and cell death in primary fibroblasts and a neuroblastoma cell line. Activation of autophagy by HSV2 was sensitive to UV irradiation of the virus and inhibited in the presence of acyclovir, highlighting the need for a functional viral DNA genome and viral replication. However, HSV2-induced autophagy was independent of the DNA-activated STING pathway. Finally, reconstitution of wildtype ATG4A and LC3B2 expression using lentiviral gene delivery or electroporation of in vitro transcribed mRNA into patient cells restored virus-induced autophagy and the ability to control HSV2 replication. This study represents the first autophagy defect causing a primary immunodeficiency with increased susceptibility to viral infection described in humans which altogether suggests an important role for autophagy in anti-HSV2 immunity in the CNS.
Introduction
Recurrent lymphocytic meningitis (RLM) is an infectious disease of the meninges, occasionally involving the brain parenchyma. This rare disease, first described by the French neurologist Pierre Mollaret in 1944, and commonly referred to as Mollaret’s meningitis, is characterized by recurrent and self-limited episodes of meningeal irritation with fever and pleocytosis of mononuclear predominance(1, 2). Although self-limiting, patients may be left with long-lasting neurological and neurocognitive sequelae. RLM has been defined by a clinical history of more than three episodes of low grade fever and the meningismus triad; nuchal rigidity, photophobia, and headache; lasting between two to five days, followed by spontaneous resolution (3, 4). The most common reported etiological agent in RLM is herpes simplex virus type 2 (HSV2), accounting for up to 95% of cases(5). Other etiological agents reported in a minority of patients are HSV1 and varicella zoster virus (VZV). Most patients do not report having genital or orolabial lesions at the time of presentation(6). Diagnosis is made by clinical history in conjunction with PCR testing for HSV2 DNA in cerebrospinal fluid (CSF)(3, 4). In clinical practice, it is commonly accepted to treat such patients with intravenous acyclovir during the acute phase, and some patients may benefit from a prophylactic regimen, although clinical evidence for this is lacking(7). Despite the potentially debilitating clinical nature and risk of neurocognitive complications of the disease, the pathophysiology of RLM and factors determining differential susceptibility to develop recurrent HSV2 reactivation and infection in the CNS remain largely unknown.
Genetic inborn errors of immunity, leading to increased susceptibility to HSV1 encephalitis (HSE) infections, have been thoroughly studied and described over the past 15 years. In particular, the Toll-like receptor (TLR)3 pathway has been demonstrated to play a pivotal role in preventing the spread of HSV1 infection from the olfactory bulb or trigeminal ganglion to the brain parenchyma and thus mediate antiviral immunity to life-threatening HSE. Inborn errors in molecules of the TLR3 signaling pathway, including defects in TLR3(8–10), UNC93B(11), TRIF(12), TRAF3(13), TBK1(14), and IRF3(8, 15, 16) were shown to be implicated in predisposing children and adults to HSE. Studies in induced pluripotent stem cell (iPSC)s demonstrated a seminal role of TLR3 signaling pathway in intrinsic antiviral immunity to HSV1 in cortical neurons and oligodendrocytes(17). The above-mentioned immunodeficiencies all lead to impaired production of antiviral cytokines of the type I interferon (IFN) family(18). More recently, IFN-independent antiviral mechanisms in the CNS have been identified through genetic analysis of viral encephalitis patients. These include non-redundant defects in the RNA debranching enzyme DBR1 as well as SNORA31 encoding small nucleolar RNA of the H A/A class, which were identified in children with brain stem encephalitis, and HSE, respectively (19, 20). The mechanistic basis for the antiviral activity of these molecules remain to be fully understood, and suggests that innate immune mechanisms independent of pattern recognition receptors and downstream inflammatory responses are important for innate antiviral defense(21).
Autophagy is an essential and highly conserved cellular homeostatic mechanism promoting maintenance of cellular integrity(22, 23). Autophagy is permanently active at some basal level in all cells and can be further upregulated under several cytopathological conditions, such as nutrient starvation, inflammation, infection, and oxidative stress(23, 24). The hallmark of macroautophagy (referred to as ‘autophagy’ from this point) is sequestration of misfolded proteins, defective organelles and intracellular invaders in autophagic compartments, thereby preventing their aggregation and replication (22). Catabolic degradation biproducts are also used to replenish deficient nutrients in states of starvation or to reduce the infectious particle load, promoting cytosolic sensing of molecular patterns to trigger the innate immune response(22). The sequestrated cargo is enclosed within an elongating double-membraned phagophore, which then develops to become a mature cargo-containing vacuole named the autophagosome. This process is completed by fusion with lysosomal compartments, and lysosomal hydrolytic enzyme-mediated lysis of the cargo(22). Autophagy may be either pro- or antiviral(25–28). As evidence of the importance of autophagy in antiviral defenses, there are numerous examples of viruses that have developed sophisticated immune evasion strategies to inhibit or dampen autophagy pathways in order to replicate and propagate(25, 27). The mechanisms of virus-autophagy interactions include viral degradation(26), increased innate sensing(29), regulation of inflammation (30), promotion of cell survival(25), and enhanced antigen presentation(31). Studies have convincingly implicated autophagy in antiviral activities in mice during HSV1 infection(32, 33). HSV1 has evolutionarily adapted to evade autophagy degradation through the neurovirulence factor ICP34.5, which binds to Beclin-1 and blocks autophagy-mediated degradation of the virus(34).
In this study, we identify rare variants in host genes encoding the autophagy-related proteins ATG4A and LC3B2 in two heterozygous patients with Mollaret’s meningitis. We demonstrate that HSV2 induces autophagy in control primary fibroblasts. Interestingly, patients harboring predicted deleterious genetic variants in ATG4A and MAP1LC3B2 exhibit impaired autophagy induction following HSV2 infection, together with increased viral load and enhanced cell death. To the best of our knowledge, this study represents the first description of a functionally important antiviral role of autophagy in herpesvirus infections in humans and suggests that inborn errors in autophagy may represent a novel class of primary immunodeficiencies.
Results
Clinical and demographic characteristics of patients with HSV2 recurrent lymphocytic meningitis
Here we describe two out of a total of fifteen patients diagnosed with HSV2 RLM, defined by three or more episodes of clinical signs of neuroinfection, CSF pleocytosis, and detection of HSV2 by PCR in the CSF. These two patients were selected based on the identification of rare gene variants in autophagy-encoding genes (see next section). Patient 1 (P1) was a 50-year-old female of Caucasian (Danish) origin. At the time of recruitment, P1 presented to the department of infectious diseases with signs and symptoms of serous meningitis. In her medical history, five previous episodes had been documented with positive identification of HSV2 DNA in the CSF on four occasions and a validated seropositivity to HSV2. Patient 2 (P2) was a 78-year-old female of Caucasian (Danish) origin. P2 presented to the department of infectious diseases with a history of twelve previous episodes of serous meningitis and both positive identification of HSV2 DNA in the CSF and seropositivity to HSV2. Both patients had negative PCR for HSV1 and varicella zoster virus (VZV) in the CSF. Table 1 summarizes the demographics, clinical presentations and CSF findings of P1 and P2. Both patients described long-lasting states of fatigue and headache and slight neurocognitive dysfunction abiding for months following each episode. Persistent short-term memory loss and disorientation constituted their measurable permanent neurological sequelae. Both patients had normal routine clinical immunological evaluation, including immunoglobulins, B and T cell numbers and phenotypes within normal ranges. Neither of them had a medical history with unusual frequency or severity of infections, except for HSV2 RLM.
Table 1. Summary of demographics and main clinical and paraclinical findings in patients (P1 and P2) with recurrent lymphocytic Mollaret’s meningitis.
| ID | Age, Sex | Recurr. (N) | Clinical manifestations | Temp (°C) | CSF1 findings |
|---|---|---|---|---|---|
| P1 | 50, Female | 6 | Nuchal rigidity, confusion, headache, minor cognitive changes | 38.1 | Total cell count, 66/mm3, mononuclear predominance; Protein, 0.67 g/L; Glucose, 2.6 mmol/L; HSV2 positive by PCR |
| P2 | 78, Female | 12 | Nuchal rigidity, headache, Long-term neurocognitive sequelae | 38.4 | Total cell count, 987/mm3, mononuclear predominance, Protein 1.1 g/L; Glucose, 3.5 mmol/L; HSV2 positive by PCR |
CSF, cerebrospinal fluid; HSV, herpes simplex virus; VZV, varicella zoster virus.
Both P1 and P2 were negative for HSV1 and VZV by PCR. P1 was serum IgG negative for HSV1 and IgG positive for HSV2, whereas P2 was serum IgG positive for both HSV1 and HSV2.
Identification of rare genetic variants in autophagy genes by whole exome sequencing
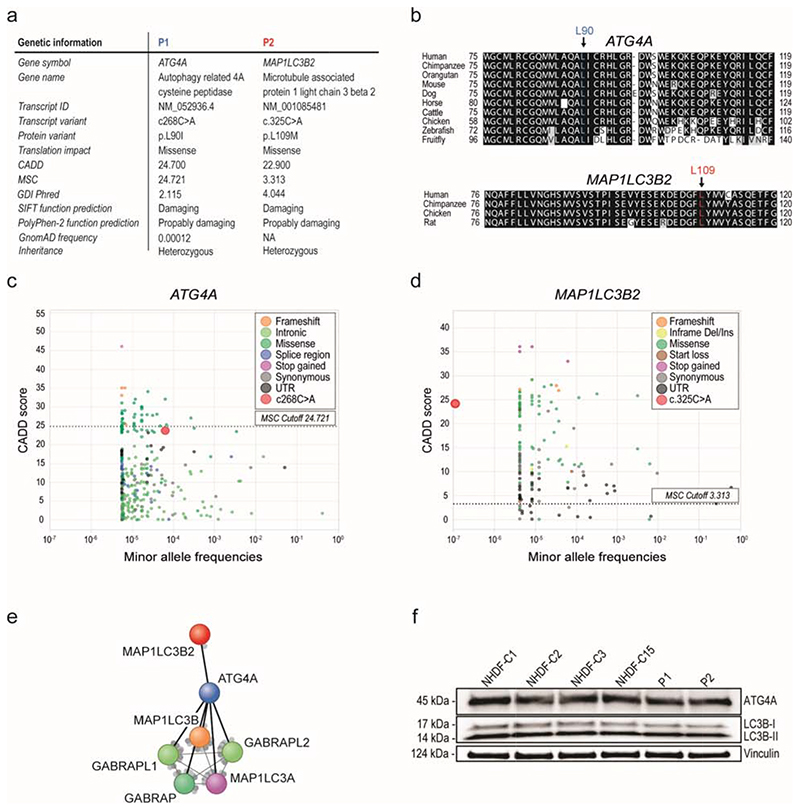
We applied whole exome sequencing (WES) analysis to investigate for genetic predisposition in RLM patients. Since there was no obvious family clustering and neither male predominance described in the literature nor in our cohort of 15 patients with a history of Mollaret’s meningitis, we applied a model of autosomal dominance with incomplete penetrance. Briefly, common variants were kept if they were observed in at less than 0.01% in common variant databases (1000 genomes, ExAC, gnomAD, and NHLBI ESP), and further based on their predicted deleteriousness i.e. to represent probable disease-associated variants according to having a CADD score > 15 and bioinformatic predictions tools including SIFT, PolyPhen2, and GDI. First, WES data were searched for variants shared by several or all of the 15 patients without identifying any common ones. Next through individual analysis, we identified a variant in the gene ATG4A encoding autophagy regulating protease 4A in P1, and a missense variant in MAP1LC3B2 (microtubule-associated protein 1A/1B light chain 3 beta 2) encoding the protein LC3B2 in P2 (Figure 1a), The variants have CADD scores of 24.7 and 22.9 respectively, suggesting a high probability of deleteriousness. Moreover, the identified variants were rare with a minor allele frequency (MAF) of 0.00012 (among these 3 hemizygous) for the ATG4A variant and not previously reported for MAP1LC3B2 in the gnomAD database (Figure 1a). We further sought into gnomAD for the presence of loss-of-function (LoF) alleles in the coding sequences of these genes. For ATG4A only one variant is reported compared to 18 expected based on prediction of variants using depth corrected probability of variants. Furthermore, fewer missense variants are detected (95) than predicted (145). The low loss-of-function observed/expected upper bound fraction, LOEUF score of 0.262 for ATG4A indicates strong selection against predicted loss-of-function (pLoF). For MAP1LC3B2 The LOEUF score is 1.366 indicating no selection against LoF, however only 3 high confidence LoF alleles are reported among Europeans, resulting in 5/100.000, and as such must be considered a rare event. One MAP1LC3B2 variant E25Q with a MAF gnomAD European of 0.01 is present; however this variant is predicted to be benign and only found in one patient. The exomes of P1 and P2 also contained other rare mutations, none of which were likely to be disease-causing based on known functions (Supplementary Table 1).
Figure 1. Identification of rare potentially disease causing variants in the autophagy genes ATG4A and MAP1LC3B2.
(a) Summary of genetic characteristics of the ATG4A and MAP1LC3B2 variants identified in P1 and P2. The variants cause missense mutations in the autophagy proteins ATG4A and LC3B2. (b) Alignment of the peptide sequence around the identified mutations in ATG4A (top) and MAP1LC3B2 (bottom) across species. (c-d) Minor allele frequency (MAF) and combined annotation dependent depletion (CADD) scores of variants reported in the genome aggregation database (gnomAD) database for (c) ATG4A and (d) MAP1LC3B2. The dotted line corresponds to the mutation significance cutoff (MSC). (e) STRING analysis of ATG4A and MAP1LC3B2. (f) Expression levels of ATG4A and LC3B2 in lysates from patient and control primary fibroblasts. GDI, gene damage index.
The c.268C>A variant in ATG4 results in a missense mutation and consequently an amino acid change from Leucine (Leu) to Isoleucine (Ile) within the peptidase domain of the molecule. For MAP1LC3B2 the c.325C>A variant causes an amino acid change from Leucine (Leu) to Methionine (Met) within the C-terminal β-sheet of the molecule (Figure 1a). The L90 residue of ATG4A and L109 residue LC3B2 are highly conserved among species throughout evolution, suggesting their importance in human biology (Figure 1b). The high CADD scores combined with the low allele frequencies in gnomAD suggest a high probability of disease-causing potential of these variants (Figures 1c-d)(35). ATG4A is a cysteine protease located in the cytoplasm and required for autophagy. ATG4A cleaves the C-terminal amino acid of LC3B2 family proteins to conjugate it to phosphatidylethanolamine (PE). MAP1LC3B2 encodes the LC3B2 ubiquitin-like protein involved in formation and maturation of autophagosomes(23). A total of 452 known PID-associated genes were investigated and P1 and P2 were not found to harbor variants predicted to be deleterious in any of those genes. Additionally, we did not identify any other autophagy-related variants in an in-house cohort of twenty previously exome-sequenced HSE patients(10) using the same criteria as the ones applied here. Likewise, we specifically looked for variants in genes previously associated with HSE, i.e. those in the TLR3 pathway as well as DBR1 and SNORA31, in the present Mollaret’s meningitis cohort and did not identify any of these.
Search tool for the retrieval of interacting genes/proteins (STRING) analysis of both protein interactomes showed strong reported interactions between each of the proteins by themselves and within the autophagy pathway. Interestingly, both protein-reported interactions are substantial and robust in experimental, database, co-expression, gene fusion and co-occurrence sources in homo sapiens (Figure 1e). When performing immunoblotting analysis on cell lysates from both patients and four individual control primary fibroblasts, we found that ATG4A and LC3B2 were expressed at similar levels in controls and P1 and P2, respectively (Figure 1f). This demonstrates that the L90I ATG4A and L109M LC3B2 mutations have no or possibly only a minor effect on expression of ATG4A and LC3B2 proteins in P1 and P2.
Impaired autophagy activity in patients harboring predicted deleterious variants in ATG4A and MAP1LC3B2 compared to controls
We next evaluated the capacity for HSV2 to induce autophagy in patient and control cells. LC3B2 is a member of the MAP1LC3 family (36). Following activation of the autophagy pathway, LC3-I synthesis is upregulated to cytosolic abundance. The exposed glycine residue of LC3 is then conjugated to phosphatidylethanolamine (PE) and subsequently integrated in the expanding phagophore and termed LC3-II. Thus, calculation of the ratio between LC3B-II:LC3-I immunoblot bands as well as quantification of LC3 punctate-positive cells in immunofluorescence provides pivotal information on the levels of induced autophagy (37, 38). The autophagy receptor p62/SQSTM1 specifically binds cargo to be sequestrated in autophagosomes, thus allowing p62 foci formation in immunofluorescence microscopy or p62 turnover measured by immunoblot to reflect induction of autophagy and degradation of cargo-bearing autophagosomes, respectively (38). Recently, a new antibody was developed to measure the phosphorylation of endogenous ATG16L1, which is present only in newly-formed autophagosomes, thereby providing information about recent events as opposed to prolonged cellular stress in cell culture(39). In this study, we have combined the three readouts mentioned above to generate an examination and interpretation of the autophagy function in healthy controls and patient cells, as well as in neuroblastoma cell lines.
To determine whether patient cells had defective activation of autophagy in response to HSV2, we initially examined LC3B-I to LC3-II conversion following HSV2 infection in primary fibroblasts. Basal autophagy was observed in mock-infected control cells, and this was further elevated following HSV2 infection and with increasing viral MOI (Figure 2a, Supplementary Figure 1a). Induction of autophagy occurred between 4 to 8h post-infection, as measured by LC3B-I to LC3-II conversion (Supplementary Figure 1b-c). The magnitude of autophagy induction was roughly similar in all four primary control fibroblasts (Supplementary Figure 1d-e). Importantly, virus-induced autophagy was abrogated in patient cells, as measured by LC3-I to LC3-II conversion and ATG16L phosphorylation (p-ATG16L1/ATG16L1) (Figure 2a, Supplementary Figure 1f-h). To corroborate these findings using complementary approaches, we performed co-immunostaining for LC3B and the viral protein ICP5 following HSV2 infection in control and patient primary fibroblasts. An elevated formation of LC3B puncta in HSV2-infected control but not patient cells was detected by immunofluorescent microscopy (Figure 2b, c). Similar observations were made when autophagy was assessed by quantification of p62 foci, which were less abundantly induced in cells from P1 and P2 than in control cells upon HSV2 infection (Figure 2d, e). Interestingly, the autophagy response in HSV2-infected control cell cultures was observed both in cells showing positive and negative staining for viral ICP5 antigen (Figure 2b, d). The impairment of autophagy in patient cell cultures was observed most profoundly in ICP5-negative cells (Supplementary Figure 1i). To directly quantify autophagosome-lysosome fusion events, the Lysotracker dye was used and fluorescence microscopy showed increased HSV-2-induced autophagy/lysosome fusion in controls but not in P1 and P2 (supplementary Figure 1 j,k).
Figure 2. Impaired autophagy in response to HSV2 infection in primary fibroblasts from P1 and P2.
(a) Immunoblot for LC3-I:LC3-II levels in lysates from P1, P2 and control primary fibroblasts isolated 12 h after HSV2 infection (MOI of 1 and 3). (b) Immunofluorescence staining with anti-HSV1 ICP5 (red), anti-LC3B (green), and DAPI (blue) in primary fibroblasts from P1, P2 and control 24 h after infection with HSV2 (MOI 0.1). (Scale bar 20 μm). (c) Quantification of LC3B punctate-positive cells. (d) Immunofluorescence staining with anti-HSV1 ICP5 (red), anti-p62 (green), and DAPI (blue) in primary fibroblasts from P1, P2 and control 24 h after infection with HSV2 (MOI 0.1). (Scale bar 20 μm). (e) Quantification of p62-punctate-positive cells (f) Flow cytometric analysis of autophagy using Cyto-ID autophagosome staining in control, P1 and P2 primary fibroblasts following HSV2 infection (MOI 1, 12 h) or stimulation with the autophagy inducer rapamycin (500 nM). (g) Quantification of data shown in panel f. (j) Quantification of autophagy flux by treating cells with chloroquine (100 μM) together with infection by HSV2 (MOI 1) or treatment with rapamycin (500 nM) for 12h. Data shown are representative of three independent experiments. Statistics was performed by 2Way ANOVA. For the histograms in panel c, d, f, and g, data are shown as mean values +/- st.dev. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001 ns, not significant.
To further quantify both the steady state and the increased levels of autophagy in response to various stimuli in patient vs control cells, a Cyto-ID® Autophagy Detection Kit was used and flow cytometry detection was performed. In control cells, HSV2 infection triggered a significant increase in autophagy as demonstrated by the nearly 3-fold increase in Cyto ID staining in infected cells (Figure 2f, g, and Supplementary Figure 1l-m). The capacity to induce autophagy through the mTOR pathway was also assessed in controls cells by using rapamycin, which significantly increased autophagy in control cells compared to mock treated cells (Figure 2f, g). In sharp contrast, neither HSV2 infection nor treatment with rapamycin resulted in any significant induction of autophagy in cells from P1 and P2 (Figure 2f, g). Similarly, induction of autophagy by Earle’s Balanced Salt Solution (EBSS) starvation was reduced in P1 and P2, although present to a certain extent (Figure 3a; Supplementary Figure 2a).
Figure 3. Impaired autophagy Flux in response to infection with HSV2 MS strain and clinical isolates in primary fibroblasts from P1 and P2.
(a) Quantification of flow cytometric analysis of autophagy using Cyto-ID autophagosome staining in three pooled controls, P1 and P2 primary fibroblasts following incubation with EBSS medium for 30 min. (b) Flow cytometric analysis of autophagy using Cyto-ID autophagosome staining in three pooled controls, P1 and P2 primary fibroblasts following HSV2 infection (MOI 1, 12 h) with or without Chloroquine (100 μM). (c) Quantification of autophagy flux by treating cells with chloroquine (100 μM) together with infection by HSV2 (MOI 1) for 12h. (d) Flow cytometric analysis of autophagy using Cyto-ID autophagosome staining in three pooled controls, P1 and P2 primary fibroblasts following infection with clinical isolates of HSV2 at an MOI of 1 for 24 h.
Next, autophagic flux was assessed by the use of chloroquine treatment, measurement of the Cyto-ID signal, and calculation of the Δ mean florescence intensity (MFI). A significant increase in autophagy flux was evident in control primary fibroblasts following HSV2 infection, however, this induction was largely abrogated in P1 and P2 primary fibroblasts (Figure 3b, c; Supplementary Figure 2b). Importantly, the impairment of HSV-2-induced autophagy in P1 and P2 was reproducible using three different clinical isolates of HSV2 (Figure 3d; Supplementary Figure 2 c-e). Collectively, these data demonstrate that patients with inborn variants in ATG4A (P1) and MAP1LC4B2 (P2) have significantly impaired autophagic flux in response to infection by HSV2 strains as well as HSV2 clinical isolates in primary fibroblasts.
Impaired autophagy increases viral burden and cytotoxicity in HSV2-infected patient primary fibroblasts
We next sought to unravel the cellular impact of impaired autophagy in P1 and P2 following HSV2 infection. Cellular morphology of primary fibroblasts from P1, P2 and controls was visualized under wide-field microscope after HSV2 infection. Control primary fibroblasts infected with low MOIs (0.1 and 0.5) showed no cytopathic effects, whereas cell rounding, detachment and various other cytopathic effects were observed at higher MOIs (Figure 4a, upper panel). In contrast, significant cell pathology and cell death was observed at low infection dose (MOI of 0.1 and 0.5) in both P1 and P2 (Figure 4a, middle and lower panels). Next, we measured cell viability of infected cells from P1 and P2 compared to controls using a CellTiter 96® AQueous assay. Whilst pooled control cells showed a mild decrease in cell viability from MOI 0.1-1, both P1 and P2 exhibited significantly reduced cell viability under these conditions (Figure 4b, c). Similar results were obtained when cytotoxicity was evaluated by measurement of LDH release in cell culture supernatants in response to the HSV2 MS strain (Figure 4d) as well as to clinical isolates of HSV2 (Figure 4e).
Figure 4. Defective autophagy in patient primary fibroblasts causes impaired control of viral replication, decreased cell viability and accelerated cytotoxicity during HSV2 infection.
(a) Inverted light microscopy of primary fibroblasts from control and patients following 16 h of infection with HSV2 at an MOI of 0.1, 0.5, and 1. (b, c) Cell viability measurement of control and P1 (b) and P2 (c) following HSV2 infection (MOI 0.1, 16 h) using CellTiter 96 Aqueous One Solution Cell Proliferation Assay.(d) Measurement of LDH levels in supernatants from primary fibroblasts infected with HSV2 (MOI 0.1, 24 h). (e) Measurement of LDH levels in supernatants form primary fibroblasts from controls or P1 and P2 infected with clinical isolates of HSV2 at an MOI of 1 for 24h. (f) Flow cytometric analysis of GFP signal of control, P1 and P2 primary fibroblasts following infection with HSV2 VP26_GFP for 24 h. (g) Quantification of the data in panel f. (h) Plaque assay for HSV2 yield in supernatants from control, P1 and P2 primary fibroblasts following 2 h, 12h, 16 h or 24 h of HSV2 infection with MOI 0.1 and 1. (i) Quantification of immunofluorescence staining with anti-HSV1 ICP5 (red) in primary fibroblasts from P1, P2 and control 24 h after infection with HSV2 (MOI 0.1). Data shown are representative of three independent experiments; For the graphs in panel b-d, and f-h, data are shown as mean values +/- st.dev. Statistics on the data presented in panel b,c,g and h was performed using 2Way ANOVA, except for panel d which, in which Mann-Whitney test was used. * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.000; ns not significant.
To explore the ramifications of altered autophagy on HSV2 replication in patient cells, we first infected control and patient primary fibroblasts with HSV2 VP26-GFP. Control cells infected at a MOI of 1 and 3 exhibited only marginal increase in GFP florescence intensity compared to mock-infected cells (Figure 4f, upper panel, Figure 4g), suggesting low infection levels. In contrast, cells from both P1 and P2 exhibited significant increase in GFP fluorescence intensity following infection (Figure 4f, middle and lower panels, 4g), demonstrating an increased intracellular viral burden. These results were confirmed by determination of viral titers in supernatants from patients and control cells using plaque assay demonstrating significantly enhanced viral replication at 12h, 16h and 24h post- infection (Figure 4h), and by quantification of staining for ICP5 in infected cells (Figure 4i). Collectively, these data demonstrate elevated viral replication and enhanced cell death in patient cells harboring variants in ATG4A and MAP1LC3B2.
Autophagy protects against HSV2 infection in neuroblastoma cell lines
Based on the identification of mutations in autophagy-related genes in RLM patient and the observed impaired antiviral response in patient-derived cells, we wanted to examine whether alteration of autophagy impacted on antiviral activity against HSV2 in an isogenic system. To this end, we used the neuroblastoma cell line SK-N-SH(40). First, we examined whether HSV2 infection stimulated activation of autophagy in these cells, and whether this was affected by targeting of the cardinal autophagy genes ATG5 and ATG6 by CRISPR/Cas9 technology. ATG5 and Beclin1 are both essential for the initiation of autophagy(41). Similar to what was observed in control primary fibroblasts, HSV2 infection stimulated the conversion of LC3-I to LC3-II in SK-N-SH cells (Supplementary Figure 3a), and the formation of autophagosomes (Figure 5a). This was dependent on the essential autophagy-related genes ATG5 and ATG6 (Supplementary Figure 3b-c). The major HSV2 capsid Protein ICP5 was readily detected in p62-positive foci in HSV2-infected cells (Figure 5a), suggesting actual autophagocytic sequestration of the virus. Of note, p62-ICP5 co-localization was observed mainly in cells with low levels of viral antigens, similar to what was observed in primary fibroblasts (Figure 2b, d). Lysotracker staining demonstrated HSV-2-induced autophagosome-lysosome fusion in controls cells but not in cells from P1 and P2 (Supplementary Figure 3d). In ATG5 gRNA and ATG6 gRNA cells, an increased GFP fluorescence intensity, accelerated cell detachment, and altered cellular morphology was observed compared to the WT infected cells (Supplementary Figure 3e-i). Thus, inhibition of autophagy in an isogenic system impairs the control of HSV2. Taking advantage of this system, we next treated the SK-N-SH-derived SH-SY5Y neuroblastoma cell line with gRNAs targeting the specific genes mutated in P1 and P2, ATG4A and MAP1LC3B2, respectively. These autophagy molecules are involved at a later stage of autophagy in the elongation process before fusion of autophagosomes with lysosomes(42). Similar to what was observed in cells depleted for ATG5 or ATG6, genetic alteration of ATG4A (affected in P1) or LC3B2 (affected in P2) expression resulted in significantly increased cytotoxicity, viral ICP5 gene expression, and enhanced production of progeny virus following HSV2 infection (Figure 5b-e). Thus, autophagy is antiviral against HSV2, and ATG4A and LC3B2 are essential for exerting this autophagy-mediated antiviral response in neuronal-derived cell lines.
Figure 5. SH-SY5Y neuroblastoma cell lines deficient in autophagy proteins ATG4A and LC3B2 show increased cell death and decreased autophagy.
(a) Confocal microscopy imaging of the SK-N-SH-derived SH-SY5Y neuroblastoma cells infected with HSV2 at MOI 0.1 for 8 h and stained with anti-p62, anti-ICP5, and DAPI. The 2nd. row is productively infected (virus in the nucleus) and 3rd. row non-productively infected. (b) Inverted light microscopy of Mock or HSV2-infected SH-SY5Y cells not receiving gRNA or gRNA targeting AAVIC1 (empty vector), ATG4A, or LC3B2. The images were acquired 16 and 24 h post infection at MOI 1. (c) Immunoblot analysis of VP5, ATG4A, p62, LC3B, and vinculin (VCL) in lysates from SK-SY5Y cells treated with gRNAs targeting AAV1 (empty vector), ATG4A, or LB3B2 and infected with HSV2 (MOI 1 and 3) for 24 h (d) Measurement of LDH levels in supernatants from cells receiving the same treatment as in panel b. (e) Plaque assay for HSV2 yield in supernatants from the HSV2 infected SH-SY5Y cells from panel b. Data shown are representative of three independent experiments. For the histograms in panel c, d, data are shown as mean values +/- st.dev. For statistics Mann-Whitney was used for data presented in panel d, whereas 2Way ANOVA was used for data presented panel e. *,p<0.05; ****, p<0.0001; ns, not significant.
HSV2-induced autophagy is dependent on viral replication but independent of the STING pathway
Next, we examined the molecular mechanism, whereby HSV2 induces autophagy. HSV1-induced autophagy in macrophages is dependent on STING(43), which is also involved in anti-HSV2 defense in mouse models(44). In addition, it has recently been reported that cyclic-GMP-AMP (cGAMP), the STING agonist produced by the DNA sensor cGAMP synthase (cGAS), induces anti-HSV1 activity partly dependent on autophagy (45). We first observed that cGAMP treatment activated autophagy and the IFN pathway in both control and patient-derived primary fibroblasts (Figure 6a; Supplementary Figure 4a,b). Therefore, STING depletion from primary fibroblasts, using the CRISPR/Cas9 technology, led to a drastic reduction in STING levels and ablated inflammatory and autophagy responses to the STING agonist cGAMP (Figure 6a). Importantly, however, HSV2-induced autophagy and viral replication was not affected by the lack of STING (Figure 6b-d). In addition, the neuron-like cell line SK-N-SH, in which HSV2 activates autophagy, has very low to now detectable levels of either cGAS or STING (Supplementary Fig 4c), and does not respond to cGAMP treatment as denoted by the absence of IFIT1 mRNA induction (Supplementary Figure 4d), further arguing against a major role of the cGAS-STING pathway in HSV2-induced autophagy in fibroblasts and neuronal cells. Since HSV1-induced autophagy has been described to be STING dependent, we next examined autophagy responses to HSV1 (Supplementary Figure 4e)(43, 45, 46). Compared to HSV2, we found that two HSV1 strains triggered a weaker autophagy response (Supplementary Figure 4e), which exhibited partial STING dependence (Supplementary Figure 4f,g). Interestingly, we also observed that although HSV2 infection of primary fibroblasts activated autophagy, it did not trigger expression of ISGs in fibroblasts, neither at 24 h of infection, at early time points (2h and 8h) nor at high MOIs (Figure 6e, f; Supplementary Figure 4h. These data suggest that the STING pathway is not at play in HSV2-activated autophagy, and that the IFN system does not have a critical role in anti-HSV2 activity in some cell types. The requirement for ULK1 in the HSV2-induced autophagy response was evaluated by CRISPR/Cas9-mediated depletion of ULK1 in fibroblasts. Knocking-out ULK1 from cells neither affected LC3-I to LC3-II conversion nor viral replication (supplementary Figure 4 h,i), implying that HSV2 activates autophagy in fibroblasts largely independently of ULK1. Finally, to gain knowledge on the viral requirements for activation of autophagy, HSV2 was treated with UV light prior to infection. UV treatment totally abolished HSV2-induced autophagy (Figure 6g,h), suggesting that a functional viral genome is essential for evoking this cellular response. Further, pre-treatment of control fibroblasts with acyclovir, a blocker of viral DNA replication, before HSV2 infection fully inhibited induction of autophagy (Figure 6i,j, Supplementary Figure 4k). Collectively, these data demonstrate that HSV2 activates autophagy through a mechanism involving viral DNA replication and acting independently of STING and ULK1, suggesting a late stage viral product or event, or a more general form of cellular stress.
Figure 6. HSV2-induced autophagy in primary fibroblasts relies on viral replication but acts independently of STING.
(a-d) Primary fibroblasts were treated with gRNAs targeting STING using two different guide RNAs (STING #1 and STING #2) or AAVS1 (empty vector). Cells were (a) stimulated with cGAMP extracellularly (100 μg/mL) for 3h, or (b, c) infected with HSV2 MOI 0.1 and 1 for 24 h. Whole-cell extracts were analyzed for (a-c) STING signaling events and autophagy by immunoblotting and (d) HSV2 viral titers in the supernatants were assessed by plaque assay. (e) Control primary fibroblasts were either infected with HSV2 (MOI 1) for 24h or stimulated with IFNβ1 (100 UI/mL) for 3h. Whole-cell extracts were subjected to immunoblotting and assessed for LC3-I to LC3-II conversion. (f) Control and patient cells were infected with HSV2 (MOI 1) for 24h, and RNA was isolated for analysis of immune-related-genes by Nanostring. (g) Primary fibroblasts were infected with live or UV-inactivated HSV2 (MOI 0.1 and 1) for 24h. Whole-cell extracts were subjected to immunoblotting and assessed for autophagy induction and (h) LC3-I:LC3-II ratios were quantified (i) Primary fibroblasts were pre-treated with acyclovir (4.5 μM) for 6 h before infection with HSV2 (MOI 1) and whole-cell extracts were subjected to immunoblotting and assessed for autophagy induction. (j) Quantification of LC3-I:LC3-II ratios. Statistics used was 2Way ANOVA.
Expression of WT ATG4A and MAP1LC3B2 rescues HSV2-induced autophagy and reduces viral load
Finally, we investigated whether re-introduction of WT ATG4A and MAP1LC3B2 into primary fibroblasts from P1 and P2, respectively, would rescue HSV2-induced autophagy and control of HSV2 infection. P1 and P2 primary fibroblasts were electroporated with mRNA encoding WT ATG4A, L90I ATG4A, WT LC3B2, or L109M LC3B2 as illustrated (Figure 7a, b). The electroporated primary fibroblasts were infected with HSV2 at a MOI of 1 for 24 hours. Following electroporation of WT ATG4, but not the mutant L90I ATG4A, into P1 primary fibroblasts the cells regained the ability to induce autophagosomes as measured by Cyto-ID (Figure 7a). Similarly, reintroduction of WT LC3B2, but not mutant L109M LC3B2, into cells from P2 enabled the cells to activate autophagy in response to HSV2 infection, (Figures 7b). Next, virus yield in supernatants from the transduced cells was quantified by plaque assay. In agreement with the reconstituted autophagy response following transduction with WT alleles, the cells from P1 and P2 regained the ability to control viral replication (Figure 7c, d), and to limit virus-induced cell death (Figure 7e, f) to a level comparable to control cells. To confirm these data, we next performed reconstitution of patient fibroblasts by an alternative approach, namely lentiviral gene transduction. This reproduced the above findings on restoration of autophagy responses, including autophagy flux, (Supplementary Figure 5a-d), viral control (Supplementary Figure 5e,f) and cell survival (Supplementary Figure 5g,h) after expression of WT, but not mutant versions, of ATG4A and LC3B2 in fibroblasts from P1 and P2, respectively. To further explore the mechanism whereby the ATG4A and LC3B2 variants may impact on autophagy, we next examined the impact of expressing the mutant autophagy proteins in control primary fibroblasts depleted of endogenous gene expression. These data demonstrated abolished HSV2-induced autophagy in LC3B2-depleted cells which was restored by expression of LCB3B2 WT but not by the L109M LC3B2 mutant (Figure 7g, h). Moreover, cells regained the ability to control HSV2 replication upon introduction of WT LC3B2 (Figure 7i). Despite several attempts using different guide or combination of guide RNAs, we were not successful in depleting control fibroblasts for ATG4A and were therefore not able to perform the reconstitution experiments. Finally, expression of increasing amounts of ATG4A L90I and LC3B2 L109M did not inhibit HSV2-induced autophagy in WT fibroblasts, thus arguing against a dominant negative effect of the autophagy variants, further suggesting haploinsufficiency as the genetic mechanism (Figure 7j-m). Altogether, these data demonstrate that reconstitution of WT ATG4A and WT MAP1LC3B2 into P1 and P2 primary fibroblasts, respectively, rescues the autophagy machinery allowing patient cells to re-gain antiviral properties and the ability to control viral infection.
Figure 7. Reconstitution of WT ATG4A and LC3B2 in P1 and P2 primary fibroblasts, respectively, rescues autophagy and the antiviral response following HSV2 infection.
(a) Flow cytometric analysis (CytoID) of HSV2-activated autophagy P1 primary fibroblasts electroporated with L90I ATG4 or wt ATG4A with quantification of ATG4 expression. (b) Flow cytometric analysis (CytoID) of HSV2-activated autophagy in P2 primary fibroblasts electroporated with L109M LC3B2 or wt LCB3B with quantification by immunoblotting of LC3B2 protein expression. (c, d) Plaque assay for HSV2 replication in supernatants from P1 (c) and P2 (d) fibroblasts electroporated with the relevant mutant and wt alleles of ATG4A and LC3B2. (e, f) Cytotoxicity measured as LDH release from untreated and HSV2-infected (MOI 1, 24h) primary fibroblasts from P1 (e) and P2 (f) electroporated with the relevant mutant and wt alleles of ATG4A and LC3B2. (g,h) Wt fibroblasts were depleted for endogenous LCB32 by CRISPR/Cas9 technology or electroporated with the guide AAVS1 as control, followed by expression of either WT LC3B2 or L109M LC3B2 by electroporation, and (g) measurement of LC3-I and LCB3-II levels in cells after xh and h) quantification of HSV2-induced autophagy in the modified cells by CytoID. (i) Plaque assay showing HSV2 replication in the supernatants of cells from (g,h). (j-m) Expression of increasing amounts of L90I ATG4A and L109M LC3B2 mutants in WT fibroblasts by electroporation and (j.k) measurement of protein levels and (l,m) HSV2-induced autophagy in the cells from (j,k) by CytoID. The experiments shown were performed at least twice. For the histograms in panel a, b,c,d,h and i, data are shown as mean values +/- st.dev. 1Way ANOVA was used for the data presented in panel a,b,c,d,h, whereas Mann-Whitney was used for panel i * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001; ns, not significant.
Discussion
In the present work, we describe two adult patients with recurrent HSV2 Mollaret meningitis carrying missense mutations and functional defects in the autophagy-related genes ATG4A and MAP1LC3B2. These data suggest that autophagy plays an essential role in anti-HSV2 immunity, either through inability to maintain HSV2 latency in sensory ganglia or through increased productive viral replication in the CNS in cells of the meninges and subarachnoid space. We demonstrate that primary fibroblasts from these patients exhibit impaired autophagy in response to HSV2, as well as to rapamycin and EBSS starvation, albeit in the setting of preserved basal autophagy. Moreover, we show that patient primary fibroblasts have significantly increased viral replication together with enhanced infection-induced cell death. Using a neuron-like cell line, we found that autophagy also plays a major role in antiviral activity and cell survival during HSV2 infection. CRISPR/Cas9 knock-down of the DNA sensor adaptor molecule STING showed that autophagy and antiviral activity induced by HSV2 is STING independent, which was in contrast to autophagy induction by HSV1. Moreover, induction of autophagy by HSV2 was independent on ULK1 but sensitive to UV irradiation of the virus. Autophagy induction was also inhibited in the presence of acyclovir, revealing the need for a functional viral genome and viral replication. This suggests that autophagy is induced as a response to cellular stress inflicted by active viral replication and accumulation of virus or viral components in the cell. Finally, we show that reconstitution of patient primary fibroblasts with WT ATG4A and MAP1LC3B2 alleles either by mRNA electroporation or lentiviral transduction lead to restoration of HSV2-induced autophagy and re-establishment of antiviral defenses, thus providing strong evidence for the disease-causing role of the identified variants. Finally, expression of the mutant ATG4A and LC3B variants in WT fibroblasts did not reconstitute cells and also did not exert a dominant negative effect, thus suggesting haploinsufficiency as the mechanism. From a physiological and clinical perspective, this might be the most likely scenario, given that complete autophagy defect is lethal in mice within 24h of birth and likely would be incompatible with life in humans (47). Altogether, based on these findings, we suggest an important role of autophagy in controlling antiviral immunity to HSV2 in the CNS in humans.
Previous studies have identified primary immunodeficiencies in the TLR3 signaling pathway predisposing to HSE(8, 9, 11, 14). These studies consolidated the notion that cell intrinsic IFN production in neurons and oligodendrocytes is pivotal in host defense against HSV1 infection in the CNS. Cells from the two patients described here did not exhibit defective virus-induced IFN responses. The two patients presented here were identified to harbor rare genetic variants predicted to be damaging in the two key genes of the autophagy machinery, ATG4A and MAP1LC3B2. Cross-checking for the presence of autophagy-associated variants in our in-house cohorts of patients suffering from HSE (N=25) or from severe influenza infection (N= 30) did not yield any positive results with the defined search criteria. There are no previous studies to suggest a genetic etiology of Mollaret’s meningitis. First, this has never been investigated genetically. Second, there are no reports of multiplex families, possibly because the inheritance may show incomplete penetrance, some defects may be de novo mutations, and also because HSV2 infection, mostly acquired in adolescence or early adulthood, precludes the disease in children. A major finding of this present work is the novel role of autophagy in antiviral defense against HSV2 infection and identification of a cellular pathway, which, if deficient, may predispose individuals to severe viral CNS infection and primary immunodeficiency. To our knowledge, this is the first published study on a cohort of Mollaret meningitis patients applying WES analysis with demonstration of the cellular phenotypes in relevant patient-derived materials showing an autophagy defect and its cellular and clinical consequences.
In the context of HSV CNS infection, it has been reported that eIF2α phosphorylation by PKR following HSV1 CNS infection induces autophagy, leading to HSV1 clearance(48). Moreover, HSV1 has evolved to evade autophagy through the neurovirulence factors ICP34.5 and US11. ICP34.5 binds to the autophagy protein Beclin-1 and interferes with formation of the autophagosome(34, 48). ICP34.5 was also shown to inhibit TBK1 signaling and by this mechanism, to inhibit autophagy(49). HSV1 tegument protein US11 interacts with PKR and inhibit eIF2α phosphorylation. The fact that HSV1 has developed immune evasion strategies to avoid autophagy proposes an essential importance of autophagy during herpes simplex CNS infections. The extent to which these two HSV genes, which have orthologues in HSV2 contribute to evasion of autophagy remains to be determined. The effect of autophagy in HSV infection appears to be highly cell type specific. Iwasaki and colleagues described a neuron-specific role of autophagy and suggested that post mitotic cells, such as primary neurons may utilize autophagy rather than classical IFN-based antiviral mechanisms in order to avoid the pathological and detrimental effect of IFN towards neuronal cells in the CNS(50). HSV has been demonstrated to induce autophagy during infection, as for example described for mouse primary trigeminal neurons, in which HSV was demonstrated to trigger the formation of p62-mediated autophagosomes(51). Studies in human fibroblasts and mouse DCs have concluded that autophagy can be triggered by HSV dsDNA in a manner dependent on the adaptor STING but independent of viral replication(43, 52). STING-mediated activation of autophagy was proven to occur through a mechanism dependent on WIPI2 and ATG5, but independent of the ULK and VPS34–Beclin kinase complexes(45, 53). Here we show that the autophagy response important for anti-HSV2 defense in primary fibroblasts was activated in a manner independent of cGAS and STING, and further independent of ULK1, but required the expression of early and late genes as well as the ability for the virus to replicate. This was based on the observation that activation of autophagy was abrogated by cross-linking of the viral genome by UV irradiation, and treatment with acyclovir, respectively. These mechanistic findings indicate that HSV2 activates autophagy through a mechanism whereby cells sense processes requiring replicating virus and a late stage viral product or event, possibly involving a form of cellular stress. In this context, it is interesting that immunofluorescence microscopy of both primary fibroblasts and neuroblastoma cell lines revealed abundant HSV2-induced autophagy also in cells not expressing the viral late gene ICP5, which opens up the possibility of a degree of paracrine signaling involved in the virus-induced stress response.
The interaction between autophagy and antiviral IFN and cytokine responses is complex, as autophagy may exert both stimulatory and inhibitory effects. As an example of the latter, triggering of cGAS-STING signaling by HSV1 has been shown to trigger negative regulatory activities on the type I IFN response, through GAS and STING(30, 54). In many cases, autophagy serves a role in promoting cell survival and limiting pathology. This is evidenced by the finding that murine gro29 cells with high basal autophagy tightly restrict HSV replication(55). Importantly, HSV1 induced autophagy in fibroblasts has a cytoprotective effect and occurs in a TBK1-dependent manner (56). Recently Vance and associates reported on the finding that HSV1-induced autophagy in mice is mediated through STING and TBK1 but independently of IRF3, thus suggesting IFN-independent but STING-dependent antiviral responses exerted through autophagy (46). Taken together, autophagy may represent a major antiviral mechanism in the CNS instead of the classical antiviral effector mechanisms, particularly type 1 IFN, described in other cell types and tissues. This hypothesis is further supported by the finding of IFN-responses to HSV2 infection of primary fibroblasts and neuroblastoma-like cells being rather weak, possibly suggesting a major role of autophagy rather than IFN in antiviral defenses in the CNS, which might serve to protect the brain against type 1 IFN-mediated immunopathology. In line with this we recently proposed that a large panel of constitutive innate immune mechanisms, play important roles in controlling infections and limiting activation of pattern recognition receptors (21).
Finally, since Mollaret’s meningitis occurs during reactivation of HSV2 from latency in sensory neural ganglia, the identified autophagy defect in these two patients may be directly caused by failure to maintain viral latency. Alpha herpesvirus latency is maintained by multiple mechanisms, including tight control of viral replication by viral genome circularization in an episomal manner, and viral genome packaging in histones, which maintains minimal synthesis of viral proteins to avoid lytic reactivation(57). Importantly, among the oncogenic gammaherpesviruses, Epstein-Bar virus (EBV), Rhesus macaque rhadinovirus (RRV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), it was reported that autophagy contributes to establishment and maintenance of latency in lymphoid tissue(58). The prevalence of Mollaret’s meningitis is unknown but is much lower than the incidence of a single episode of HSV2 meningitis. Since 70-80 % of an adult population is seropositive for HSV2 and harbors the virus latently in sensory ganglia, the proportion of HSV2 infected individuals who repeatedly fails to control viral latency and develop Mollaret’s meningitis is extremely low. However, since our experimental set-up represents a lytic HSV2 infection of fibroblasts and neuronal-like cells, rather than a precise reflection of herpesvirus latency in sensory neurons, this study does not allow us to distinguish between a failure to maintain viral latency versus unrestricted viral replication in the CNS once re-activated in patients with autophagy defects and Mollaret’s meningitis.
In summary, our study demonstrates a causal relationship between the clinical phenotype in two Mollaret’s meningitis patients, the genetic variants identified, and the functional in vitro studies. In these patients, we identified missense mutations in the genes encoding the autophagy proteins ATG4A and LC3B2, leading to reduced viral induction of autophagy, impaired control of HSV2 replication, correlating with increased cell death. Our findings may thus suggest a role for autophagy in antiviral defense in the CNS in humans. From a biological and evolutionary perspective, the use of autophagy in antiviral defense in the CNS may enable elimination of invading pathogens in a non-destructive manner in an anatomical location where inflammatory activity, including production of excessive type I IFN, can be pathological and cause irreversible damage. Overall, the present study increases insight and knowledge on infection immunology in the CNS and identifies a novel category of primary immunodeficiencies, in which defects in autophagy pathways may predispose to severe viral infection in the CNS in humans. Finally, these findings indicate that therapies stimulating cellular autophagy pathways may be beneficial in the treatment and prophylaxis of HSV2 recurrent lymphocytic meningitis.
Materials and Methods
Collection of patient material
The two patients (P1 and P2) included in this study are part of a larger cohort study on RLM patients, including 15 patients. For collection of primary dermal fibroblasts, skin biopsies from patients were taken. Following aseptic technique and local anaesthesia with subdermal lidocaine injection, a clean 4-mm punch biopsy was obtained and stored in 10 mL supplemented RPMI 1640. Under microscope guidance, the punch biopsies were segmented to approximately 0.2 mm sections and cultured in DMEM high glucose media supplemented with 20% heat inactivated fetal calf serum (FCS) and 100 IU/mL penicillin and 100 ug/mL streptomycin (BioWest, France). These conditions favor fibroblasts growth over keratinocytes. After two passages, the keratinocyte population had regressed with fibroblasts overgrowth and expansion. Primary dermal fibroblasts from passages two and three were counted and cryopreserved in 10% DMSO in FBS for 25 h, then stored for long-term in a liquid nitrogen tank. Upon experimental use, cryopreserved primary fibroblasts were promptly thawed in RT DMEM high glucose complemented with 10% FCS and antibiotics(59). The primary fibroblasts were counted, seeded, and incubated overnight at 37°C in 5% CO2 before infection. Control primary fibroblasts from healthy individuals were a kind gift from the Department of Immunodermatology and Experimental Allergology, Hannover School of Medicine, Germany. Normal human derived fibroblasts (NHDF)s were purchased (C12302, Promocell, Heidelberg).
Generation of ATG5 and ATG6/Beclin1 knockout in SK-N-SH neuroblastoma cell lines
The neuroblastoma cell line SK-N-SH was maintained in 10% FCS-supplemented DMEM. The ATG5 sgRNA and ATG6 sgRNA SK-N-SH neuroblastoma cell lines were generated by cloning annealed sgRNA sequences into lentiCRISPRv2 puro plasmid (Addgene plasmid #98290). The cell lines were generated as a pool by transducing lentiviral vector encoding Cas9 and the corresponding sgRNA followed by selection with puromycin to remove non-transduced cells. We designed two sets of sgRNAs for each gene. The sgRNAs targeting ATG5 were 1-hatg5-sense: 5’-CACCGATCACAAGCAACTCTGGAT-3’, 1-hatg5-antisense: 5’-AAACATCCAGAGTTGCTTGTGATC-3’ and 2-hatg5-sense: 5’-CACCGCCTTAGATGGACAGTGCAGA-3’, 2-hatg5-antisense: 5’-AAAC TCTGCACTGTCCATCTAAGGC-3’. The sgRNAs targeting ATG6 were 1-hatg6-sense: 5’-CACC GAAACTCGTGTCCAGTTTCAG-3’, 1-hatg6-antisense: 5’-AAACCTGAAACTGGACACGAGTTTC-3’ and 2-hatg5-sense: 5’-CACCGGACACGAGTTTCAAGATCC-3’, 2-hatg6-antisense: 5’-AAACGGATCTTGAAACTCGTGTCC-3’. Genomic DNA was harvested following PCR amplification of the region of interest. The following primers were used for PCR amplification and sequencing of ATG5: Forward hatg5 5’-CTCCATTCCACATCTTTGCC-3’, Reverse hatg5 5’-CTTAGAGTAATGGCATTTCC-3’. The following primers were used for PCR amplification and sequencing of ATG6: Forward hatg6 5’-GAGGTGAAGAGCATCGGGGG-3’, Reverse hatg6 5’-CCATCACGCCCGGCTAATTT-3’. All sequences were analyzed using TIDE version 2.0.1(60). The efficiency of gene targeting on whole pools of cells was determined by Western blotting.
Generation of ATG4A and LC3B2 knockout in SH-SY5Y neuroblastoma cell lines
SH-SY5Y cells were cultured and maintained in DMEM-High glucose medium (Sigma Aldrich) supplemented with 10% heat inactivated FCS and 10000 IU/mL penicillin and streptomycin (Sigma Aldrich) and 2mM L-glutamine (Sigma Aldrich). ATG4A and LC3B2 knock-outs cell lines were generated as a pool using Amaxa 4D-Nucleofector protocol for SH-SY5Y cells (Lonza Colonge,GmbH) following manufacture’s instruction. Briefly, cells in the logarithmic growth phase were detached using (TrypLE Express, Thermo scientific) and 7 x 105 cells were subjected to nucleofection (4D-Nucleofector, Lonza) with a complex of 24 μg of Cas9 Nuclease V3 protein (IDT technology) + 13 μg of SgRNA (Synthego, USA.). After nucleofection cells were re-suspended in prewarmed medium and incubated for 48 hrs. After 48 h, the knock-out were validated by western blot and used for the experiments. Sequences of the synthetic guide RNA used are as follows; ATG4A -UGGGGAUGUAUGCUACGCUG; LC3B – UUCAAGCAGCGCCGCACCUU.
Whole exome sequencing (WES) and bioinformatics
Genomic DNA was isolated from whole blood from each patient as previously described(16, 61, 62). WES was performed employing Kapa HTP Library preparation and Nimblegen SeqCap EZ MedExome Plus kit, analysis on Illumina Nextseq 550 system, SNP calling relative to hg19 with BWA, PCR and optical duplicates were identified and marked. The alignment was recalibrated using GATK. Single nucleotide polymorphisms were called employing HaplotypeCaller from the GATK package. Variant call files (VCF) files were uploaded to Ingenuity Variant Analysis and filtered according to confidence (call quality at least 30.0, read depth at least 5.0, allele fraction at least 25.0). We then used different variant scoring systems, including combined annotation dependent depletion (CADD), mutation significance cut-off (MSC), gene damage index (GDI), SIFT, and PolyPhen-2 to identify variants of interest. The exomes were analyzed for deleterious variants using combined annotation dependent depletion (CADD) score >15 and a frequency below 0.01% in the general population based on frequencies reported to the Genome Aggregation Database (gnomAD), ExAC, and NHLBI ESP. Finally, identified variants were manually checked by inspecting BAM files using IGV
Sequence alignment
The following ATG4A sequences were obtained from UniProt (http://www.uniprot.org/): Homo sapiens isoform 1 Q8WYN0, Chimpanzee G2HGQ3, Orangutan Q5R699, Mouse Q8C9S8, Dog E2RLY1, Bovin Q6PZ05, Horse F7BWS2, Chicken Q5ZIW7, Zebrafish Q4VBK1, Fruitfly M9PBM3. The following LC3B2 sequences were obtained from UniProt (http://www.uniprot.org/): Homo sapiens A6NCE7, Chimpanzee A0A2J8ME52, Chicken Q5ZLF8, Rat A0A0G2K9Q7. Sequences were aligned using Clustal Omega CLUSTAL O(1.2.4) multiple sequence alignment(63). BoxShade 3.21 was used for converting the alignment file to a EPS.
PopViz
The PopViz plot was generated by use of PopViz webserver and subsequently modified in Adobe Illustrator SC6(64).
STRING analysis
Protein interactome was obtained based on reported evidence from experimental, database, co-expression, gene fusion and co-occurrence interaction sources in homo sapiens. Minimum required interaction score was set to highest confidence (0.900). All nodes shown (in red) are query proteins and first shell of interactors; second shell and proteins of unknown 3D structure were excluded(65, 66).
HSV1 and HSV2 infection
All HSV2 infections were performed using HSV2 MS strain unless mentioned otherwise. Other HSV strains used were the laboratory HSV2 strain 333, and the laboratory HSV1 strains KOS and 17+. The clinical HSV2 isolates used were two from Swedish patients (90.036 and 90.263) and one from a patient from Uganda (BA327UR). Patient primary fibroblasts, SK-N-SH, and SH-SY5Y neuroblastoma cell lines were infected by addition of virus at an MOI of 0.1-3 (as indicated in each figure) diluted in serum-free DMEM onto the 85% confluent cell monolayer. Virus-infected cell cultures were left for 60 min at room temperature (RT) with gentle tilting to promote viral particles adsorption. Supernatant was removed, and cells were washed twice in DMEM and left to incubate at 37°C in 10% FCS-supplemented DMEM.
NanoString nCounter
Primary fibroblasts from P1, P2 and three controls were infected with HSV2 at MOI 1 for 18h. Following infection, cells were lysed and total RNA was purified using High Pure RNA Isolation kit (Roche, Switzerland). Approximately 100 ng of total RNA from both untreated and virus treated cells was subjected to nCounter™ SPRINT (NanoString Technologies, USA) analysis according to the manufacturer’s instructions using the nCounter® Human Immunology v2 Panel (579 target genes and 15 reference genes). The raw data were background subtracted (Threshold count value of 25) and normalized using the geometric mean of the positive controls according to manufacturer’s recommendation. Then another normalization was performed using the geometric mean of the six most stable reference genes (ALAS1, EEF1G, G6PD, OAZ1, GUSB and RPL19). The heatmaps with accompanying hierarchical cluster analyses were generated using the Advanced Analysis Module of the nSolver™ software.
Cyto-ID Flow Cytometry
Primary fibroblasts from P1 and P2 and controls were seeded at a density of 1.8x105 cells in 12 well plates and infected as described above. Rapamycin was used at a concentration of 500 nM as a positive control for autophagy induction. Cells were incubated for 30 min with Eagle balanced salt solution (EBSS) (Sigma-Aldrich, Saint-Louis, Missouri, USA). At 12h post infection (hpi) or treatment, cells were washed twice with PBS supplemented with 5% FBS and stained with Cyto-ID Green Detection Dye for 30 minutes at 37°C according to the manufacturer’s instructions (ENZ-KIT175-0200, eNZO Life Sciences, NY, USA). Cells were trypsinized and analyzed using BD FACSVerse flow cytometer. Data were analyzed using FlowJo 10.5.3. Using mean fluorescence intensity (MFI) of the Cyto-ID signal, autophagy flux was calculated, based on values from infected and uninfected cells in the presence or absence of chloroquine (CQ, 100 μM), a lysosomotropic molecule preventing lysosomal acidification leading to inhibition of autophagosome fusion was measured. Under these conditions, stained autophagosomes accumulate over time and the total change in cellular Cyto-ID signal can be calculated [(ΔMFI Cyto-ID = MFI Cyto-ID (+CQ) - MFI Cyto-ID (-CQ)] for each treatment and assessment of the autophagy flux can be made(67–69).
Immunofluorescence microscopy
Cells were grown at 50% confluency on 13 mm diameter coverslips and were fixed with 100% methanol on ice, before washing them with 1X phosphate buffered saline (PBS). Blocking was done in 10%-FBS. PBS and cells were permeabilized with 0.1% triton. Staining was done in a moist chamber. Images were taken with 63X oil immersion lens on widefield fluorescence microscope (Zeiss Axio Observer). Antibodies used for immunofluorescence were anti-LC3B (1:500 dilution, Abcam), anti-SQSTM1/p62 (8025, Cell Signaling 1:500), anti-ICP5 (1:500 dilution, Abcam), Alexa Fluor-488 and -594 conjugated secondary antibodies (1:750 and 1:1000 dilutions respectively, Life Technologies). Coverslips were mounted to the slides using DAPI-containing ProLong Diamond antifade mounting media (Thermo Fisher, Waltham, MA). Lysotracker staining was performed following the instructions of the manufacturer (ThermoFisher, Waltham, MA).
Cell viability assay
Primary fibroblasts were seeded at a concentration of 2x104 cells/mL on a 96-well plate in DMEM and infected at MOI 0.1, 1, and 3. Cells were left incubating for 24h, after which all samples were treated with CellTiter 96® AQueous One Solution Reagent (Promega, Wisconsin, USA) according to the manufacturer’s instructions. Absorption at 490 nm was read using a 96-well plate reader.
LDH Cytotoxicity assay
Cells were seeded in a 96-well plate as described above. Once cells achieved 50% confluence, they were infected with HSV2 MS strain at an MOI of 0.1 for 16 h or 24 h, after which supernatants were harvested. Percentage of cytotoxicity was calculated as according to manufacturer instructions (catalog # 88954, Thermo Fisher Scientific, Massachusetts, USA).
Flow cytometry on cells infected with HSV2 GFP
Primary fibroblasts were seeded in a 12-well plate at a density of 1.8x105 and infected with HSV2 (333 strain) containing a UL27-eGFP tag at MOI of 1 and 3 as described above. At 16 hpi cells were washed twice with PBS complemented with 10% FBS, trypsinized and the GFP signal was measured using BD FACSVerse flow cytometer. Data were analyzed using FlowJo 10.5.3.
Plaque Assay
1.8x105 primary fibroblasts were seeded in a 12-well plate overnight. Infection was carried out as describe above. At 16 and 24 hpi, supernatants were collected and cryopreserved until use. Near confluent Vero cells seeded in 6-well plates were infected with the serially-diluted supernatants. After 1h supernatants were removed and DMEM containing 2% FBS, 1% pencillin and 1% streptomycin with human Beriglobin (CSL Behring) was added. Plates were incubated at 37°C in a humidified atmosphere at 5% CO2 for 48 h. These cells were washed and fixed with ice-cold methanol and stained with 0.1% (w/v) crystal violet and 2% (v/v) ethanol in H2O. Plaques were counted using a binocular microscope and the pfu/mL was calculated.
Western blot
Primary fibroblasts were seeded in complemented DMEM, incubated overnight and treated with the different drugs and/or infected with HSV2 as described above. Western Blot procedure was performed as previously described(70). In brief, cells were washed twice with ice-cold PBS and lysed in ice-cold Pierce RIPA lysis buffer (Thermo Scientific) supplemented with 10 mM NaF, 1x complete protease cocktail inhibitor (Roche) and 5 IU/mL benzonaze (Sigma Aldrich). Protein concentration was determined using a BCA protein assay kit (Thermo Scientific). Whole-cell lysates were denatured for 3 min at 95°C in presence of 1x XT Sample Buffer (BioRad) and 1x XT reducing agent (BioRad), and 10-20 μg of reduced samples were separated by SDS-PAGE on 4-20% Criterion TGX precast gradient gels (BioRad). Each gel was run initially for 15 min at 70V and 45 min at 120V. Transfer onto PVDF membranes (BioRad) was done using a Trans-Blot Turbo Transfer system for 7 min. Membranes were blocked for 1h with 5% skim-milk (Sigma Aldrich) at room temperature in PBS supplemented with 0.05% Tween-20 (PBST). Membranes were fractionated in smaller pieces and probed overnight at 4°C with any of the following specific primary antibodies in PBST: anti- LC3B2 (3868, Cell Signaling 1:1000; 192890 Abcam 1:1000), anti-SQSTM1/p62 (8025, Cell Signaling 1:1000); anti-ATG16l 1:1000 (AB188642 Abcam, Cambridge, UK); anti-p-ATG16L 1:1000 (AB195242 Abcam, Cambridge UK), anti-TBK1/NAK (3013, Cell Signaling 1:1000), anti-phospho-TBK1/NAK (5483, Cell Signaling 1:1000), anti-IRF3 (11904, Cell Signaling 1:1000), anti-phospho-IRF3 (4947, Cell Signaling 1:500), anti-STING (13647, Cell Signaling 1:1000), anti-phospho-STING (40818, Cell Signaling); anti-IFIT1 (14769, Cell Signaling 1:1000), anti-ISG15 (2758, Cell Signaling 1:1000), anti-HSV1-2 VP5 (ab6508, AbCam 1:1000), anti-ULK1 (8054S Cell Signaling technology, Denver, MA) and anti-Vinculin (18799, Cell Signaling 1:1000) used as loading control. After three washes in PBST, secondary antibodies, peroxidase-conjugated F(ab)2 donkey anti-mouse IgG (H+L) (1:10000) or peroxidase-conjugated F(ab)2 donkey anti-rabbit IgG (H+L) (1:10000) (Jackson ImmunoResearch) were added to the membrane in PBST 1% skimmed milk for 1h at room temperature. All membranes were washed three times and developed using either the SuperSignal West Pico PLUS chemiluminescent substrate or the SuperSignal West Femto maximum sensitivity substrate (ThermoScientific) and a Chemidoc Imaging system (Biorad). Specific bands were quantified by densitometry using the Image J software.
Genome editing in primary fibroblasts using CRISPR/Cas9 strategy
The cell lines were generated as a pool by resuspension of primary fibroblasts (1x105 cells) in 15 μL of Opti-MEM and incubated at room temperature for 25 min with a 5 uL Opti-MEM solution containing 6 ug of Cas9 Nuclease V3 protein (IDT) and 3.2 ug of modified synthetic guide RNA (Synthego). The different guide RNA sequences were designed using Synthego’s website and were as follows: AAVS1 (GGGGCCACUAGGGACAGGAU); STING guide 1 (g1) (CAGUCCUCCAGUAGCUGCCC); STING guide 2 (g2) (ACAGGUACCGGGGCAGCUAC). LC3B2 (UUCAAGCAGCGCCGCACCUU); ATG4A guide 1 (UGGGGAUGUAUGCUACGCUG); ATG4A guide 2 (AGUAAUCUGAUCUUCAUACU); ATG4A guide 3 (AUAUCCAGAUACAGAUGAGC); ATG4A guide 4 (UUAGGAUGUUCAUUCGCUGU); ATG4A guide 5 (UAGGAUGUUCAUUCGCUGUG) ULK1 guide 1 (g1) (GUCCUCGCUCAGCGUGCGCA); ULK1 guide 2 (g2) (AGCAGAUCGCGGGCGCCAUG). After incubation, the 20 μL of solution containing cells, Cas9 protein and guide RNAs were transferred to a strip electroporation cassette and subsequently subjected to electroporation using a 4D Nucleofection machine (Lonza). Primary Cell P3 and pulsing code CM 138 were used to achieve maximal efficiency. After electroporation, 100 uL of pre-warmed medium was added to each well. Cells were then transferred to regular tissue culture plates and expanded for at least 3-4 days before further use.
Lentiviral vector production and transduction of primary fibroblasts
To create the pCCL/PGK-ATG4A and pCCL/PGK-MAP1LC3B2 plasmid vectors, ATG4 isoform A and MAP1LC3B2 isoform B2 cDNA were commercially acquired (PlasmID: HsCD00418846 and HsCD00431847, respectively) and inserted into a BamHI -and XhoI-digested pCCL/PGK-eGFP using NEBuilder HiFi DNA Assembly (New England Biolabs, Ipswich, MA) according to manufacturer’s instructions(71). Subsequently, the ATG4A 268C>A (L90I))and MAP1LC3B2 325C>A (L109M) patient variants were introduced by overlapping PCR amplification of ATG4A and MAP1LC3B2, respectively, and subsequent insertion into pCCL/PGK-eGFP as described above, to create pCCL/PGK-ATG4A(268C>A) and pCCL/MAP1LC3B2(325C>A). Lentiviral vectors were produced by standard calcium phosphate transfection of the packaging plasmids pMD2.G, pRSV-Rev and pMDIg/pRRE together with either pCCL/PGK-eGFP, pCCL/PGK-ATG4A, pCCL/PGK-MAP1LC3B2 pCCL/PGK-ATG4A(268C>A) or pCCL/MAP1LC3B2(325C>A) into HEK293T cells. 48 h after transfection, virus-containing supernatants were harvested by filtering through 0.45-μm filters and finally polybrene was added to a final concentration of 8 μg/ml. For transduction of patient and control primary fibroblasts, filtered supernatants were added to fibroblasts seeded at 2-3 × 105 cells/flask in T25 culture flasks one day prior to transduction.
Statistical analysis
Experiments were performed in experimental triplicates and repeated two or three times. Statistics were calculated using nonparametric Mann-Whitney test or 1-way or 2-way ANOVA, depending on whether or not the data were normally distributed. (GraphPad Prism 8). Tests were two-sided and p-values < 0.05 were considered significant.
Ethics
The patients and healthy controls were included following oral and written consent and in accordance with The Helsinki Declaration and national ethics guidelines and after approval from the Danish National Committee on Health Ethics (# 1-10-72-275-15), the Data Protection Agency, and Institutional Review Board.
Supplementary Material
Summary.
Autophagy defects predispose to recurrent HSV2 infection in the central nervous system and reveals an important role for autophagy in antiviral immunity in humans
Acknowledgments
We wish to thank the patients participating in this study. In addition, we wish to thank technician Kirsten Stadel Pedersen for excellent technical assistance and Dr. Beatrice Dyring Andersen for performing skin biopsies on the two patients.
Funding
The PhD scholarship to ASH was funded by the European Union under the Horizon 2020 research and innovation program (H2020) and Marie Skłodowska-Curie Actions–Innovative Training Networks Programme MSCA-ITN GA 675278 EDGE (Training Network providing cutting-EDGE knowlEDGE on Herpes Virology and Immunology. DO salary was supported by a Weiman fond Associate Professor Fellowship. THM was funded by The Independent Research Fund Denmark (4004-00047B), Aarhus University Research Fund (AUFF-E-2015-FLS-66), and The Lundbeck Foundation (R268-2016-3927). SRP was supported by grants from The European Research Council (786602), The Lundbeck Foundation (R198-2015-171, R268-2016-3927), and The Novo Nordisk Foundation (NNF18OC0030274).
Footnotes
Authorship statement
THM conceived the idea; ASH, THM, CTB and SRP developed the idea; CTB, MH, and ASH cared for patients and collected patient material; KE provided clinical isolates of HSV2; ASH, DO, KAS, SML, NSBA, MMT, MRF, CB, LIM, FR, LK, DDK, DVDH, SD, IP, and SR performed experiments and analyzed data with THM, SRP, VSS, SM, JK, LSK, and JGM; MC assisted in bioinformatical analysis; ASH and THM wrote the first draft of the manuscript, all authors read, commented and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.M P. La meningite endothelio-leucocytaire multirecurrente benigne. Syndrome nouveau ou maladie nouvelle. Documents clinique. Rev Neurol (Paris) 1944;76:57–76. [Google Scholar]
- 2.Min Z, Baddley JW. Mollaret’s meningitis. Lancet Infect Dis. 2014;14:1022. doi: 10.1016/S1473-3099(14)70874-6. [DOI] [PubMed] [Google Scholar]
- 3.Shalabi M, Whitley RJ. Recurrent benign lymphocytic meningitis. Clin Infect Dis. 2006;43:1194–1197. doi: 10.1086/508281. [DOI] [PubMed] [Google Scholar]
- 4.Tedder DG, Ashley R, Tyler KL, Levin MJ. Herpes simplex virus infection as a cause of benign recurrent lymphocytic meningitis. Ann Intern Med. 1994;121:334–338. doi: 10.7326/0003-4819-121-5-199409010-00004. [DOI] [PubMed] [Google Scholar]
- 5.Wright WF, Palisoc K, Baghli S. Mollaret meningitis. J Neurol Sci. 2019;396:148–149. doi: 10.1016/j.jns.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 6.Schlesinger Y, Tebas P, Gaudreault-Keener M, Buller RS, Storch GA. Herpes simplex virus type 2 meningitis in the absence of genital lesions: improved recognition with use of the polymerase chain reaction. Clin Infect Dis. 1995;20:842–848. doi: 10.1093/clinids/20.4.842. [DOI] [PubMed] [Google Scholar]
- 7.Bergstrom T, Alestig K. Treatment of primary and recurrent herpes simplex virus type 2 induced meningitis with acyclovir. Scand J Infect Dis. 1990;22:239–240. doi: 10.3109/00365549009037909. [DOI] [PubMed] [Google Scholar]
- 8.Zhang SY, Jouanguy E, Sancho-Shimizu V, von Bernuth H, Yang K, Abel L, Picard C, Puel A, Casanova JL. Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol Rev. 2007;220:225–236. doi: 10.1111/j.1600-065X.2007.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, Segal D, Sancho-Shimizu V, Lorenzo L, Puel A, Picard C, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 10.Mork N, Kofod-Olsen E, Sorensen KB, Bach E, Orntoft TF, Ostergaard L, Paludan SR, Christiansen M, Mogensen TH. Mutations in the TLR3 signaling pathway and beyond in adult patients with herpes simplex encephalitis. Genes Immun. 2015;16:552–566. doi: 10.1038/gene.2015.46. [DOI] [PubMed] [Google Scholar]
- 11.Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, Alcais A, Picard C, Mahfoufi N, Nicolas N, Lorenzo L, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 12.Sancho-Shimizu V, Perez de Diego R, Lorenzo L, Halwani R, Alangari A, Israelsson E, Fabrega S, Cardon A, Maluenda J, Tatematsu M, Mahvelati F, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez de Diego R, Sancho-Shimizu V, Lorenzo L, Puel A, Plancoulaine S, Picard C, Herman M, Cardon A, Durandy A, Bustamante J, Vallabhapurapu S, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33:400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman M, Ciancanelli M, Ou YH, Lorenzo L, Klaudel-Dreszler M, Pauwels E, Sancho-Shimizu V, Perez de Diego R, Abhyankar A, Israelsson E, Guo Y, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouanguy E, Zhang SY, Chapgier A, Sancho-Shimizu V, Puel A, Picard C, Boisson-Dupuis S, Abel L, Casanova JL. Human primary immunodeficiencies of type I interferons. Biochimie. 2007;89:878–883. doi: 10.1016/j.biochi.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Andersen LL, Mork N, Reinert LS, Kofod-Olsen E, Narita R, Jorgensen SE, Skipper KA, Honing K, Gad HH, Ostergaard L, Orntoft TF, et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med. 2015;212:1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmer B, Ewaleifoh O, Harschnitz O, Lee YS, Peneau C, McAlpine JL, Liu B, Tchieu J, Steinbeck JA, Lafaille F, Volpi S, et al. Human iPSC-derived trigeminal neurons lack constitutive TLR3-dependent immunity that protects cortical neurons from HSV-1 infection. Proc Natl Acad Sci U S A. 2018;115:E8775–E8782. doi: 10.1073/pnas.1809853115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mogensen TH. IRF and STAT Transcription Factors - From Basic Biology to Roles in Infection, Protective Immunity, and Primary Immunodeficiencies. Front Immunol. 2018;9:3047. doi: 10.3389/fimmu.2018.03047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang SY, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, Mandel H, Garcia P, Ciancanelli MJ, Biran A, Lafaille FG, et al. Inborn Errors of RNA Lariat Metabolism in Humans with Brainstem Viral Infection. Cell. 2018;172:952–965.:e918. doi: 10.1016/j.cell.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafaille FG, Harschnitz O, Lee YS, Zhang P, Hasek ML, Kerner G, Itan Y, Ewaleifoh O, Rapaport F, Carlile TM, Carter-Timofte ME, et al. Human SNORA31 variations impair cortical neuron-intrinsic immunity to HSV-1 and underlie herpes simplex encephalitis. Nat Med. 2019;25:1873–1884. doi: 10.1038/s41591-019-0672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paludan SR, Pradeu T, Masters SL, Mogensen TH. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat Rev Immunol. 2020 doi: 10.1038/s41577-020-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243–251. doi: 10.1080/15548627.2017.1402992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmad L, Mostowy S, Sancho-Shimizu V. Autophagy-Virus Interplay: From Cell Biology to Human Disease. Front Cell Dev Biol. 2018;6:155. doi: 10.3389/fcell.2018.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong X, Levine B. Autophagy and viruses: adversaries or allies? J Innate Immun. 2013;5:480–493. doi: 10.1159/000346388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 30.Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, Reinert L, Cai Y, Jensen SB, Skouboe MK, Nyengaard JR, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37 doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, Desjardins M. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. 2007;81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connell D, Liang C. Autophagy interaction with herpes simplex virus type-1 infection. Autophagy. 2016;12:451–459. doi: 10.1080/15548627.2016.1139262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, Scott E, Shah I, Stenson PD, Gleeson J, Cooper DN, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13:109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006;273:2553–2562. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian W, Alsaadi R, Guo Z, Kalinina A, Carrier M, Tremblay ME, Lacoste B, Lagace D, Russell RC. An antibody for analysis of autophagy induction. Nat Methods. 2020;17:232–239. doi: 10.1038/s41592-019-0661-y. [DOI] [PubMed] [Google Scholar]
- 40.Biedler JL, Helson L, Spengler BA. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 1973;33:2643–2652. [PubMed] [Google Scholar]
- 41.Gallagher LE, Chan EY. Early signalling events of autophagy. Essays Biochem. 2013;55:1–15. doi: 10.1042/bse0550001. [DOI] [PubMed] [Google Scholar]
- 42.Kaminskyy V, Zhivotovsky B. Proteases in autophagy. Biochim Biophys Acta. 2012;1824:44–50. doi: 10.1016/j.bbapap.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iversen MB, Reinert LS, Thomsen MK, Bagdonaite I, Nandakumar R, Cheshenko N, Prabakaran T, Vakhrushev SY, Krzyzowska M, Kratholm SK, Ruiz-Perez F, et al. An innate antiviral pathway acting before interferons at epithelial surfaces. Nat Immunol. 2016;17:150–158. doi: 10.1038/ni.3319. [DOI] [PubMed] [Google Scholar]
- 45.Gui X, Yang H, Li T, Tan X, Shi P, Li M, Du F, Chen ZJ. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashiro LH, Wilson SC, Morrison HM, Karalis V, Chung JJ, Chen KJ, Bateup HS, Szpara ML, Lee AY, Cox JS, Vance RE. Interferon-independent STING signaling promotes resistance to HSV-1 in vivo. Nat Commun. 2020;11:3382. doi: 10.1038/s41467-020-17156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 48.Lussignol M, Queval C, Bernet-Camard MF, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J Virol. 2013;87:859–871. doi: 10.1128/JVI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma Y, Jin H, Valyi-Nagy T, Cao Y, Yan Z, He B. Inhibition of TANK binding kinase 1 by herpes simplex virus 1 facilitates productive infection. J Virol. 2012;86:2188–2196. doi: 10.1128/JVI.05376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 2012;12:334–345. doi: 10.1016/j.chom.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katzenell S, Leib DA. Herpes Simplex Virus and Interferon Signaling Induce Novel Autophagic Clusters in Sensory Neurons. J Virol. 2016;90:4706–4719. doi: 10.1128/JVI.02908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J Virol. 2011;85:4212–4221. doi: 10.1128/JVI.02435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu D, Wu H, Wang C, Li Y, Tian H, Siraj S, Sehgal SA, Wang X, Wang J, Shang Y, Jiang Z, et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 2019;26:1735–1749. doi: 10.1038/s41418-018-0251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, Oh BH, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Sage V, Banfield BW. Dysregulation of autophagy in murine fibroblasts resistant to HSV-1 infection. PLoS One. 2012;7:e42636. doi: 10.1371/journal.pone.0042636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmad L, Mashbat B, Leung C, Brookes C, Hamad S, Krokowski S, Shenoy AR, Lorenzo L, Levin M, O’Hare P, Zhang SY, et al. Human TANK-binding kinase 1 is required for early autophagy induction upon herpes simplex virus 1 infection. J Allergy Clin Immunol. 2019;143:765–769.:e767. doi: 10.1016/j.jaci.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Grinde B. Herpesviruses: latency and reactivation - viral strategies and host response. J Oral Microbiol. 2013;5 doi: 10.3402/jom.v5i0.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lussignol M, Esclatine A. Herpesvirus and Autophagy: “All Right, Everybody Be Cool, This Is a Robbery!”. Viruses. 2017;9 doi: 10.3390/v9120372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vangipuram M, Ting D, Kim S, Diaz R, Schule B. Skin punch biopsy explant culture for derivation of primary human fibroblasts. J Vis Exp. 2013:e3779. doi: 10.3791/3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen SE, Christiansen M, Ryo LB, Gad HH, Gjedsted J, Staeheli P, Mikkelsen JG, Storgaard M, Hartmann R, Mogensen TH. Defective RNA sensing by RIG-I in severe influenza virus infection. Clin Exp Immunol. 2018;192:366–376. doi: 10.1111/cei.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nissen SK, Christiansen M, Helleberg M, Kjaer K, Jorgensen SE, Gerstoft J, Katzenstein TL, Benfield T, Kronborg G, Larsen CS, Laursen A, et al. Whole Exome Sequencing of HIV-1 long-term non-progressors identifies rare variants in genes encoding innate immune sensors and signaling molecules. Sci Rep. 2018;8:15253. doi: 10.1038/s41598-018-33481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P, Bigio B, Rapaport F, Zhang SY, Casanova JL, Abel L, Boisson B, Itan Y. PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics. 2018;34:4307–4309. doi: 10.1093/bioinformatics/bty536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirouac DC, Saez-Rodriguez J, Swantek J, Burke JM, Lauffenburger DA, Sorger PK. Creating and analyzing pathway and protein interaction compendia for modelling signal transduction networks. BMC Syst Biol. 2012;6:29. doi: 10.1186/1752-0509-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Budida R, Stankov MV, Dohner K, Buch A, Panayotova-Dimitrova D, Tappe KA, Pohlmann A, Sodeik B, Behrens GMN. Herpes simplex virus 1 interferes with autophagy of murine dendritic cells and impairs their ability to stimulate CD8(+) T lymphocytes. Eur J Immunol. 2017;47:1819–1834. doi: 10.1002/eji.201646908. [DOI] [PubMed] [Google Scholar]
- 68.Chan LLY, Shen D, Wilkinson AR, Patton W, Lai N, Chan E, Kuksin D, Lin B, Qiu J. A novel image-based cytometry method for autophagy detection in living cells. Cancer Res. 2014;74 doi: 10.4161/auto.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo S, Liang Y, Murphy SF, Huang A, Shen H, Kelly DF, Sobrado P, Sheng Z. A rapid and high content assay that measures cyto-ID-stained autophagic compartments and estimates autophagy flux with potential clinical applications. Autophagy. 2015;11:560–572. doi: 10.1080/15548627.2015.1017181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olagnier D, Brandtoft AM, Gunderstofte C, Villadsen NL, Krapp C, Thielke AL, Laustsen A, Peri S, Hansen AL, Bonefeld L, Thyrsted J, et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat Commun. 2018;9:3506. doi: 10.1038/s41467-018-05861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jakobsen M, Stenderup K, Rosada C, Moldt B, Kamp S, Dam TN, Jensen TG, Mikkelsen JG. Amelioration of psoriasis by anti-TNF-alpha RNAi in the xenograft transplantation model. Mol Ther. 2009;17:1743–1753. doi: 10.1038/mt.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.