Abstract
The three-dimensional positions of atoms in protein molecules define their structure and the roles they perform in biological processes. The more precisely atomic coordinates are determined, the more chemical information can be derived and the more mechanistic insights into protein function may be inferred. With breakthroughs in electron detection and image processing technology, electron cryo-microscopy (cryo-EM) single-particle analysis has yielded protein structures with increasing levels of detail in recent years1,2. However, obtaining cryo-EM reconstructions with sufficient resolution to visualise individual atoms in proteins has thus far been elusive. Here, we show that using a new electron source, energy filter and camera, we obtained a 1.7 Å resolution cryo-EM reconstruction for a human membrane protein, the β3 GABAA receptor homopentamer3. Such maps allow a detailed understanding of small molecule coordination, visualisation of solvent molecules and alternative conformations for multiple amino acids, as well as unambiguous building of ordered acidic side chains and glycans. Applied to mouse apoferritin, our strategy led to a 1.22 Å resolution reconstruction that, for the first time, offers a genuine atomic resolution view of a protein molecule using single particle cryo-EM. Moreover, the scattering potential from many hydrogen atoms can be visualised in difference maps, allowing a direct analysis of hydrogen bonding networks. The technological advances described here, combined with further approaches to accelerate data acquisition and improve sample quality, provide a route towards routine application of cryo-EM in high-throughput screening of small molecule modulators and structure-based drug discovery.
Multiple factors determine the attainable resolution of reconstructions from single-particle cryo-EM. However, for biological macromolecules, the radiation damage caused by electron interactions with the sample is a fundamental limitation. In order to preserve the molecular structure, damage is restricted by carefully limiting the number of electrons used for imaging4. The resulting counting statistics lead to high levels of noise. The signal-to-noise ratio (SNR) of cryo-EM images drops rapidly with spatial frequency, and at higher spatial frequencies the noise is typically orders of magnitude higher than the signal.
High-resolution reconstructions may still be calculated by averaging over many images of individual particles, provided that their relative orientations can be determined. However, because noise reduction scales with the square root of the number of particles, and because higher SNRs lead to more accurate orientations, acquiring more particles is often less efficient than increasing the SNR in the images. Consequently, although microscope automation5 and faster image processing programs6,7 have allowed reconstructions from larger data sets in recent years, increasing the SNR of the raw data may lead to bigger improvements, as is illustrated by the sudden increase in cryo-EM resolutions with the introduction of more sensitive direct electron cameras in 20138,9.
Here, we report the impact of three technological developments that further increase the SNR of cryo-EM images: a new cold field emission electron gun (CFEG), a new energy filter and the latest generation Falcon direct electron camera (Figure 1a). We demonstrate that combined, these developments lead to a marked increase in the achievable resolution that ultimately enables the visualisation of individual protein atoms in optimised samples.
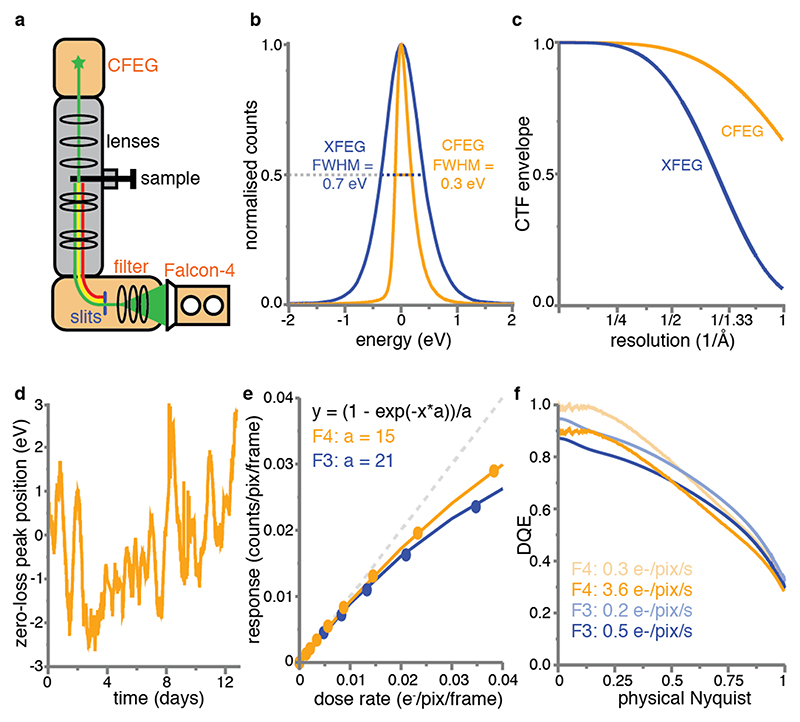
Figure 1. New imaging technologies for cryo-EM.
(a) Schematic overview of an electron cryo-microscope. The new cold field-emission gun (CFEG), energy filter and Falcon-4 camera are highlighted in orange. (b) Energy spread of the XFEG (blue) and the CFEG (orange), with the full width of the curves at half their maximum value (FWHM). (c) Theoretical CTF envelope functions for the XFEG (blue) and the CFEG (orange). (d) The relative position of the zero-loss peak with respect to the centre of the slits in the energy filter over multiple days of operation. (e) Dose response measurements for the Falcon-3 (blue dots) and Falcon-4 (orange dots). The orange and blue lines are the corresponding fits to the data, with the fit parameters indicated in the same colours; the dashed grey line represents the perfect response. Note that the X-axis units are per camera frame, while the Falcon-4 and Falcon-3 read out at operate at 248 and 40 frames per second, respectively. (f) DQE curves for the Falcon-3 at a dose rate of 0.2 (light blue) and 0.5 e-/pixel/s (blue) and the Falcon-4 at a dose rate of 0.3 (light orange) and 3.6 e-/pixel/s (orange).
An electron source optimised for energy spread
The source inside the microscope emits electrons with a range of different wavelengths, or energies. Because not all these electrons can be focused in the same plane due to chromatic aberration in the objective lens, the energy spread of the electrons leads to a blur in the images. The corresponding loss in SNR increases with spatial frequency and is described by an envelope on the contrast transfer function (CTF). Many state-of-the-art electron microscopes are equipped with a field emission gun (FEG) that is operated at a temperature of 1700-1800 K and optimised for brightness (XFEG). In order to optimise the source for its energy spread, a so-called “cold” FEG (CFEG) may be operated at room temperature instead. JEOL have pioneered the use of a CFEG for cryo-EM of biological macromolecules10,11. We used a prototype of a CFEG that was recently developed by Thermo Fisher Scientific (TFS) and is operated at a low beam current. This reduces the brightness of the source from a typical 7.5×107 A/m2srV for a standard TFS XFEG to 5×107 A/m2srV for the CFEG at the beam current used for our experiments. However, compared to an energy spread of 0.7 eV for the XFEG, the new CFEG has an energy spread of 0.3 eV (Figure 1b). As a result, the SNRs at high spatial frequencies increase. Whereas the differences are small for resolutions worse than 2 Å, the use of the CFEG boosts the CTF from 58% to 91% at 1.5Å, and from 26% to 80% at 1.2Å. Since the SNR scales with the square of the CTF, this leads to a 2.5-fold increase in the SNR at 1.5 Å and a 9.5-fold increase at 1.2 Å (Figure 1c, Methods).
A disadvantage of operating the FEG at room temperature is that gas atoms can absorb to the surface of its tip, which reduces the number of electrons emitted. This contamination builds up over time and can be removed by periodically heating the tip. This procedure is called tip flashing and takes less than a minute. By performing tip flashing every 6-10 hours, based on automated monitoring of the beam current in the data acquisition program EPU, we found that the CFEG beam current remains stable over days (Extended Data Figure 1).
A stable energy filter
When electrons pass through the specimen, two types of interactions occur. Elastically scattered electrons maintain their energy and contribute to the signal in the images. Inelastically scattered electrons deposit part of their energy in the sample and contribute noise to the images. Therefore, SNRs can be improved by filtering away electrons that have experienced an energy loss. Gatan has pioneered the use of energy filters with a 90° bending prism for biological cryo-EM and their filters are in widespread use12. JEOL microscopes use a different, Omega-shaped filter design13. We used a new energy filter that TFS developed using the experience that previously led to the energy filter reported by Kahl et al14. The filter is located below the microscope’s projector column and comprising a 90° bending prism with an adjustable energy selecting slit and multiple electromagnetic lenses to correct for aberrations and to enlarge both the energy dispersion and the image. The mechanics of the new filter were designed to minimise the impact of temperature variations on the position of the optical elements, including the energy slit, with respect to the optical axis of the system. The large radius of the bending prism (135 mm compared to 75 mm in Gatan filters) reduces third and fourth order distortions. The new filter is stable over many days of operation: the position of the electrons that have not lost any energy, the so-called zero-loss peak, with respect to the centre of the energy slit changes less than 3 eV in either direction (Figure 1d).
A next-generation camera
The relatively large pixel size of Falcon direct-electron cameras, each measuring 14 x 14μm15, permits using a thick epilayer which increases the signal to noise ratio16,17. Noise in the resulting images arises mainly from the variation in height and shape of the charge distribution that is generated by individual incident electrons, or events. This noise may be reduced by a counting algorithm that positions a constant signal distribution at an estimated centre position for each event. But in order to identify all individual events, the number of events that are in close proximity of each other on a single frame should be minimized. With a frame rate of 40 Hz, the Falcon-3 was the first Falcon camera that was fast enough for electron counting, but only when spreading the electron dose over typical exposure times of close to a minute.
The Falcon-4 camera operates at 248 Hz, while an improved counting algorithm estimates event positions with sub-pixel accuracy and its epilayer design optimises the event size versus event signal strength and thereby reduces the number of events that are lost during counting (Figure 1e). As a result, images with typical exposure times of a few seconds can be recorded, while the detective quantum efficiency remains similar to the Falcon-3 camera in counting mode (Figure 1f). In addition, the resulting movies can be written out in a new data format, called electron-event representation (EER)18, which stores each detected electron event as an x,y-position on a four-times oversampled grid and its original time, or frame, of recording. We adapted our image processing software RELION19 (see Methods) to read the new data format, thus allowing motion correction by the Bayesian polishing algorithm20 and reconstructions of movie frames with flexible dose fractionation schemes, up to the original frame rate of the camera.
Application to a human GABAA receptor sample
To evaluate the impact of the technological advances described above, we used a homopentameric β3 GABAA receptor (GABAAR) variant, a ~200kDa human membrane protein bound to the Mb25 megabody to overcome its severe preferential orientation in ice3,21. GABAARs are a large family of pentameric ligand-gated chloride channels that mediate inhibitory neurotransmission and are targeted by a wide range of psychoactive drugs22. Yet, structure-based drug development has been hampered by the difficulty of solving GABAAR structures, currently limited to 2.5-3 Å resolution in the best cases21,23. While the majority of GABAARs are heteromeric, β3 subunits form functional homomeric channels gated by histamine. Their 5-fold symmetry allowed us to collect multiple data sets in a time-efficient manner, as one would need five times more data to reach the same resolution with an asymmetrical sample.
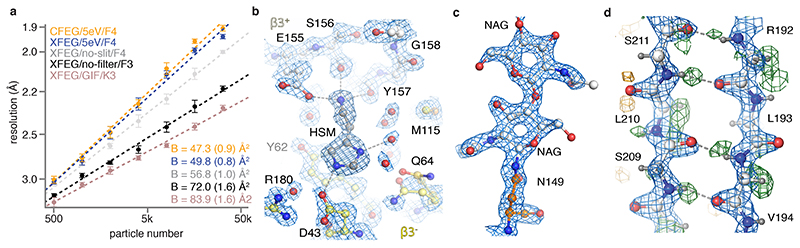
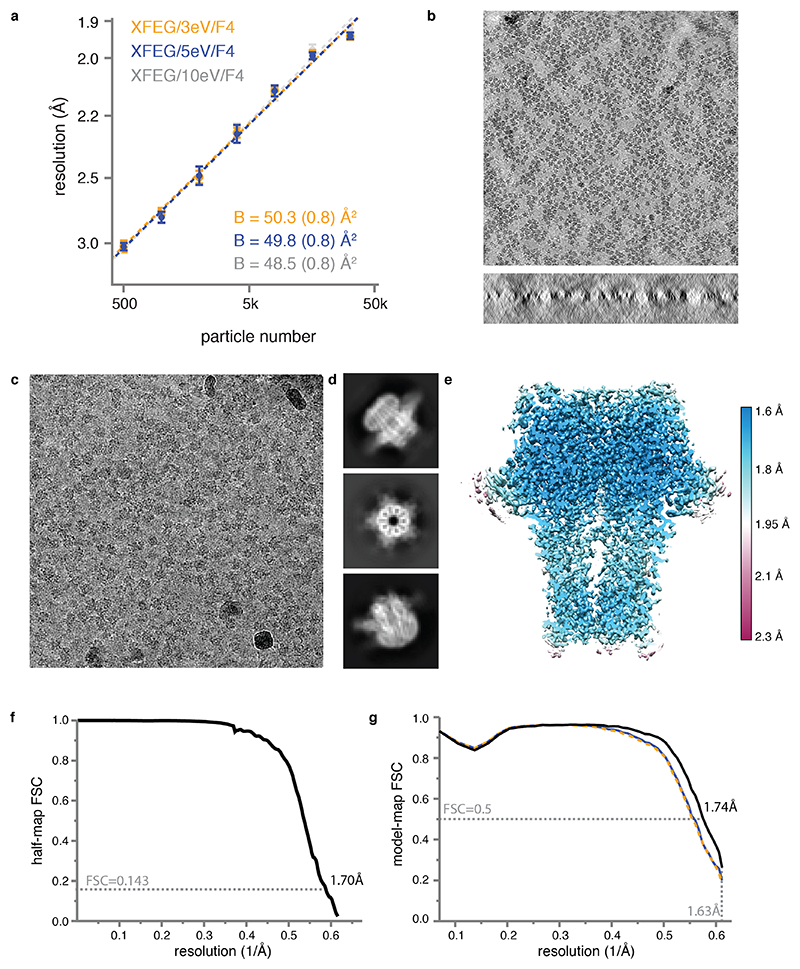
We collected seven data sets from three different GABAAR grids on three different Titan Krios microscopes (Extended Data Table 1; Extended Data Figure 2) and assessed their quality in terms of their estimated B-factors24 (Methods, Figure 2a). To assess the current state-of-the art, we collected two data sets on a first grid on a microscope with an XFEG using either a Gatan K3 camera and a Gatan energy filter (with a slit width of 20 eV), or a bottom-mounted Falcon-3 camera. This resulted in B-factors of 87 and 72 Å2, respectively. We do note that data acquisition on the K3 was three times faster than on the Falcon-3. Therefore, reconstructions at a given target resolution may be obtained within less time on the K3. When we imaged a second grid on a microscope with an XFEG, the new energy filter (with retracted slits) and a Falcon-4 camera, an improvement of 15 Å2 in the B-factor over the Falcon-3 data set was observed. Using the same grid and the same microscope, a further improvement of 7 Å2 in the B-factor was achieved when using the energy filter with a slit width of 5 eV. Varying the energy slit widths to 3 eV and 10 eV did not have a noticeable effect on the B-factors (Extended Data Figure 2a). Imaging a third grid on yet another microscope, with a CFEG, the new energy filter (with a slit width of 5 eV) and a Falcon-4 camera, led to a small improvement for those reconstructions with resolutions better than 2 Å. The latter is expected from the relatively small gains in the CTF envelope at those resolutions when using a CFEG instead of an XFEG (Figure 1c). Finally, we performed tomography on the same GABAAR grid used in the CFEG experiment to determine its ice thickness, which we found to be in the range of 320-370 Å (Extended Data Figure 2b).
Figure 2. GABAAR reconstructions.
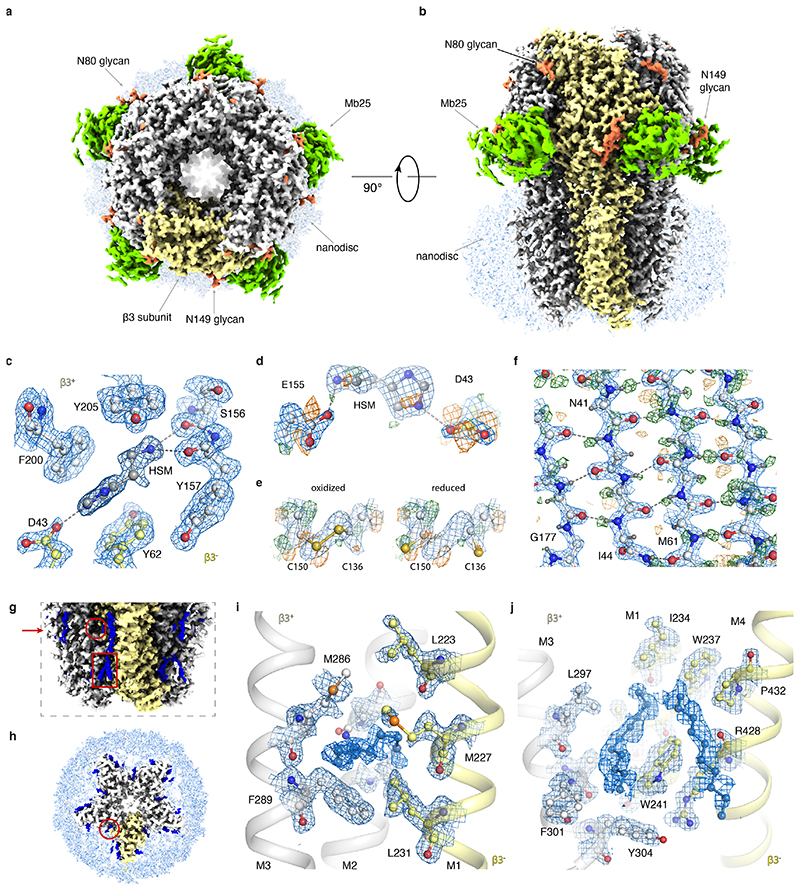
(a) B-factor plots for four data sets using: the new CFEG, the new energy filter with a slit width of 5 eV and a Falcon-4 camera (orange); an XFEG, the new energy filter with a slit width of 5 eV and a Falcon-4 (blue); an XFEG, the new energy filter with the slits retracted and a Falcon-4 (grey); an XFEG, no energy filter and a (bottom-mounted) Falcon-3 detector (black); and an XFEG, a Gatan Imaging Filter (GIF) and a K3 detector (brown). B-factors estimated from the slope of fitted straight lines are shown in the same colours. The numbers in parentheses and the error bars represent the estimated and the sample standard deviations from seven-fold random resampling, respectively. (b) Overview of map quality at the agonist binding pocket, illustrating HSM coordination and multiple water molecules (red spheres). (c) The N-acetyl glucosamine moieties attached to Asn149. (d) Difference map (green positive; orange negative) visualises hydrogen atoms in the hydrogen bonding network between β-strands.
By combining the data sets with the new filter on both the XFEG and the CFEG, we obtained a 1.7 Å resolution reconstruction from 371,693 particles (Figure 2b-d, Extended Data Figure 2c-g, Extended Data Figure 3, Extended Data Table 2). The resulting map provides unprecedented levels of detail for a protein structure, other than apoferritin, solved by single-particle cryo-EM. The N-linked glycan on Asn149 shows density of exceptional quality; alternative side chain conformations are visible for many amino acids; lipid molecules are visible in the transmembrane region of the receptor; and in the most ordered regions hydrogen-bonding networks can be visualised directly in difference maps calculated in REFMAC25 with a refined model from which the hydrogen atoms were omitted. The agonist pocket is occupied by a histamine (HSM) molecule, which binds in a similar manner as observed for benzamidine3 and the neurotransmitter GABA23. However, for the first time, it is now possible to visualize the coordinating carboxyl groups of Glu155 and Asp43, alongside a network of ordered water molecules that surround the ligand to fill the agonist pocket. A movie of the dose-dependent evolution of the cryo-EM density indicates that the structure of the agonist pocket may be affected by radiation damage (Supplementary Information Video 1). Similarly, the disulfide bond between Cys136 and Cys150, a signature of the Cys-loop receptor family to which GABAAR belong, appears to be partially reduced during the electron exposure (Supplementary Information Video 2).
Atomic resolution structure of apoferritin
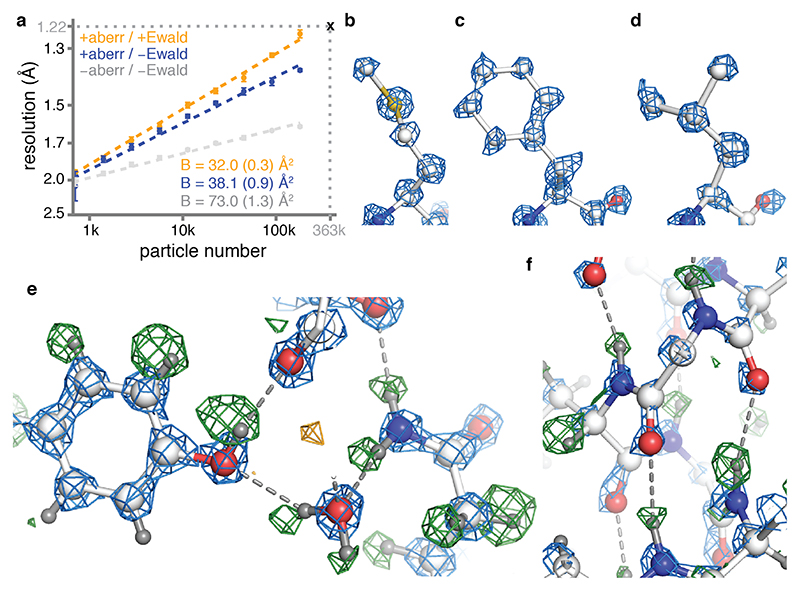
To explore the potential of the new technologies to yield even higher resolution reconstructions, we tested a mouse apoferritin sample. Apoferritin is a popular cryo-EM benchmark because its molecular stability and its 24-fold symmetry allow high-resolution reconstructions to be calculated from relatively few particles. Using JEOL’s CFEG and Omega energy filter with a K2 camera, the same mouse apoferritin sample, but a different grid, resulted in the best apoferritin map reported thus far: 1.53 Å resolution from 120,295 particles, with a B-factor of 46 Å2 11. We previously also reported a reconstruction of human apo-ferritin with a resolution of 1.65 Å from 426,450 particles recorded on a Titan Krios microscope with an XFEG, a Gatan energy filter and a Gatan K2 camera19. The B-factor for that data set was 66 Å2.
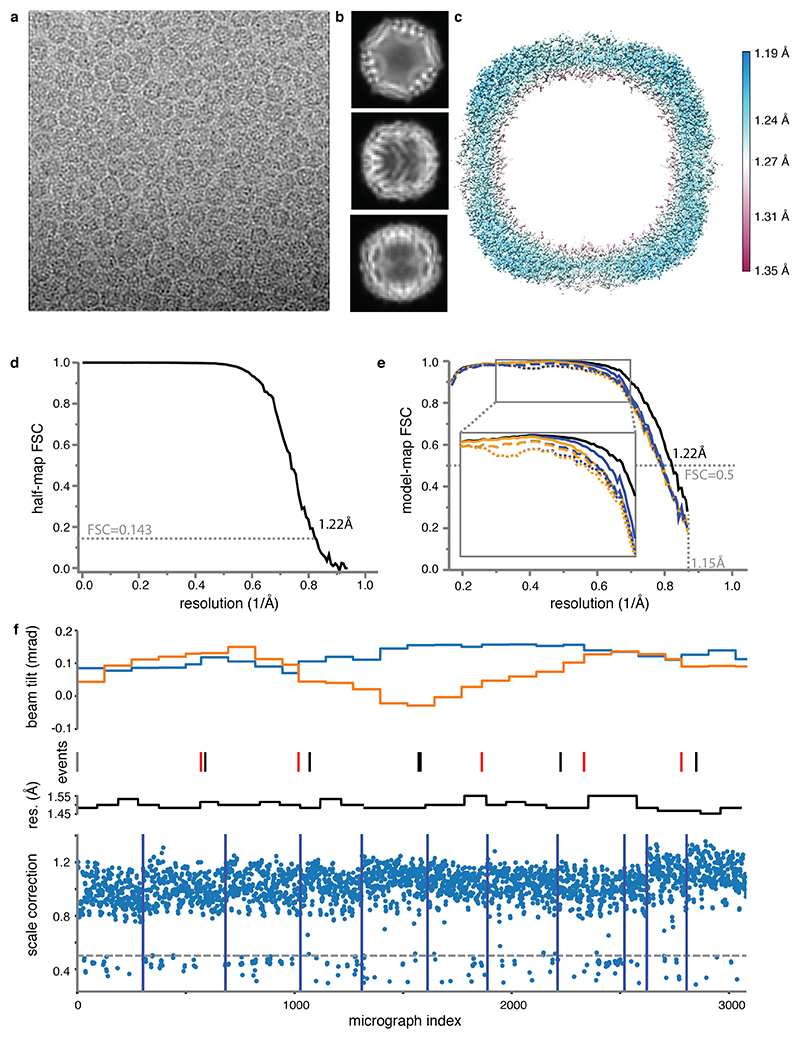
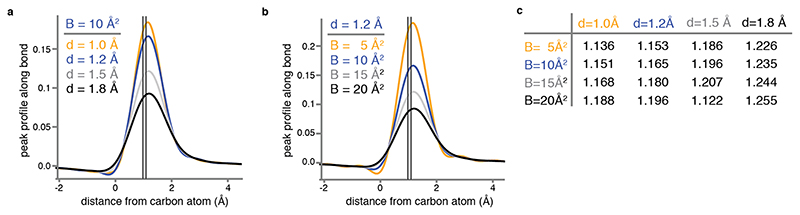
We imaged mouse apoferritin on the CFEG microscope, using the Falcon-4 camera and a slit width of 10 eV in the new energy filter. From 3,370 movies, we obtained a reconstruction with a resolution of 1.22 Å from 363,126 particles (Extended Data Figure 4, Supplementary Information Video 3). Corrections during image processing, for third and fourth-order optical aberrations26, as well as for the curvature of the Ewald sphere19,27, were crucial in obtaining a B-factor of 32 Å2 (Figure 3a). After sharpening of the map, most atoms are resolved as individual blobs of density; C=O bonds and some of the C-N bonds in the peptide planes are less well separated (Figure 3b-d). Many hydrogen atoms, including those from individual water molecules, are resolved in a REFMAC difference map, more clearly than in the GABAA receptor case and allowing a direct analysis of hydrogen bonding networks (Figure 3e-f). Detailed analysis of the hydrogen positions revealed that the density peaks in the difference map were consistently further away from their bonded atom than the hydrogen atoms at its expected position. This result is explained by the observation that electron imaging visualises electrostatic potential, which depends both on the positions of nuclei and electrons (see Methods, Extended Data Figure 5).
Figure 3. Apo-ferritin reconstruction.
(a) B-factor plots for reconstructions using: high-order aberration and Ewald sphere correction (orange); high-order aberration correction only (blue); and no correction (grey). B-factors estimated from the slope of fitted straight lines are shown in the same colours. The numbers in parentheses and the error bars represent the estimated and the sample standard deviations from seven-fold random resampling, respectively. (b) Density for M100 in the 1.22 Å map is shown in blue. (c) Density for F51. (d) Density for L175. (e) Hydrogen bonding network around Y32 and water-302 is visible in the difference map (green positive; orange negative) (f) the α-helix hydrogen bonding network involving residues 21NRQIN25, illustrated as in (e).
Finally, we calculated reconstructions for each of the 434 camera frames that were acquired per exposure and were able to follow the dose-dependent evolution of the cryo-EM density with an unprecedented resolution of 0.1 electrons (e-) per Å2 (Supplementary Information Video 4). Following the initial dose of 1.5 e-/Å2, the resolution of the single-frame reconstructions extends to 1.7 Å and remains better than 2 Å up to an accumulated dose of 11.5 e-/Å2. However, the first few reconstructions are of relatively poor quality, probably because initial beam-induced motions are not modelled adequately. As radiation damage is minimal in the earliest frames, further improvements in beam-induced motion correction, possibly by considering particle rotations and height changes28, may thus extend attainable resolution even further.
Outlook
The three developments in electron microscopy hardware described in this paper provide a step-change in the achievable resolution by single-particle cryo-EM. Using apoferritin, we show that these developments allow structure determination of a protein to true atomic resolution, as per the Sheldrick criterion29,30. The improved energy spread of the CFEG plays a crucial role in extending resolutions to better than 1.5 Å, which is in agreement with similar observations made with the JEOL CFEG10. There are two alternatives to the CFEG strategy to improve the CTF envelope function at atomic resolution. Firstly, one could use objective lenses with a lower chromatic aberration, which are common in material science, although their narrow gap polepieces would limit the amount by which the sample may be tilted. Secondly, one could reduce the energy spread with a monochromator31. The disadvantage of this strategy would be a considerably more complicated experimental setup.
Our results on the GABAAR illustrate how the new technology can improve cryo-EM structures at lower resolutions, beyond the highly stable test samples that apoferritin represents. Although variations between microscopes and grids can complicate direct comparisons of B-factors, our results on GABAAR suggest that the Falcon-4 camera in combination with the new filter provide the best image quality (Figure 2a). At this stage, it is unclear whether the removal of inelastically scattered electrons from our thin samples alone would be enough to account for this improvement, or whether other effects, e.g. increased amplitude contrast32, play a role too. In the future, it will also be of interest to explore whether, instead of removing them, inelastically scattered electrons could still be used to add to the signal in a chromatic aberration corrected microscope33,34.
The increased SNR of cryo-EM images enabled by the technology described here will expand the applicability of both single-particle analysis and electron tomography to more difficult samples, including membrane proteins in lipid bilayers, small proteins and structurally heterogeneous macromolecular complexes. Moreover, higher resolution reconstructions will allow improved visualization of hydrogen bonding networks, ordered waters and alternative side chain conformations, which will further our understanding of protein function and facilitate cryo-EM fragment-based drug discovery.
Methods
Characterisation of the new microscopy hardware
The energy spread of the field emission gun was measured using the new energy filter. Post-slit multi-poles of the filter were set to spectroscopy mode in order to form an enlarged image of the spectrum on the camera. The intensity of the incoming beam was carefully reduced in order not to saturate or damage the camera. The energy dispersion at the camera was 8 meV/pixel. The exposure time was 31 ms. The energy spread as full width at half the maximum value (FWHM) of ΔE=0.71 for XFEG, and ΔE=0.28 for CFEG (Figure 1b) were used to calculate CTF envelope function, Δf, using the following equation31:
using Cc=2.7mm; ΔV/V=0.02ppm and ΔI/I=0.1ppm. Note that the latter two contributions account for less than 1% drop in the CTF envelope.
The stability of the energy filter was assessed by repeatedly measuring the position of the zero-loss peak with respect to the energy-selecting slit. To this end, we mechanically moved one slit edge to the optical axis and fine-tuned an offset on the high tension supply such that the slit edge blocked half of the beam intensity. The resulting offset of the high tension was read out as the shift in the zero loss position. Measurements, each taking approximately 40 seconds, were repeated continuously for 14 days.
Dose response curves for the Falcon-3 and Falcon-4 cameras were measured using the flu-screen and fixed magnification steps. Before each experiment, the flu-screen was carefully calibrated with a Faraday cup in its linear range (0.4-5.0 nA at 300 kV). The read-out from the flu-screen was interpreted as the expected electron dose and plotted against the counted number of electrons on the camera.
DQE measurements were performed as described35 at 300 keV on a Titan Krios G2 microscope at the LMB for the Falcon-3 and on a Titan Krios G3i microscope at the Materials Science Department of the University of Cambridge for the Falcon-4. For calibration of the flu-screen on the microscope with the Falcon-4, a Faraday cage and a Keithley 6485 picoammeter (kindly provided by Gerard van Hoften, Thermo Fisher Scientific) were used. For the microscope with the Falcon-3, the drift tube of a Gatan energy filter (GIF; BioQuantum 686) was used as a Faraday cup and the current was measured using a Deben 087-001 SEM Probe current meter. Because the beam currents used in counting mode imaging are too low to be accurately measured, a defined beam with a measurable current was set at a low magnification and the desired electron flux on the camera was obtained by increasing the magnification. The relative magnification change was measured by using a higher current to fully illuminate the flu-screen at the low magnification and noting the change in current upon going to the higher magnification.
To account for the non-linearity that is introduced by coincidence loss, DQEs were first measured at a low dose rate (0.19 e-/pixel/s for the Falcon-3; 0.32 e- /pixel/s for the Falcon-4) and then extrapolated to a typical dose rate (0.5 e- /pixel/s for the Falcon-3; 3.6 e-/pixel/s for the Falcon-4) using the measured response versus dose-rate curves (Figure 1e). Modulation transfer functions (MTFs) were measured using the knife-edge method from a straight part of a shadow of the pointer on the camera. The actual area of the images used was chosen carefully by looking at the difference between the observed image and that predicted from the inferred MTF, to ensure that the edge in the selected area was straight and free of defects. In the case of the Falcon-4, the only part of the pointer edge that was suitable was at a 15 degrees angle with the pixel array of the camera, the effect of which was accounted for when calculating the MTF. Even at the lowest dose rates used, there was still a small decrease in the power spectra at low-spatial frequency due to coincidence loss. To avoid overestimating the resulting DQE at low spatial frequency, the noise power spectra were extrapolated from a Gaussian fit of the power between 0.2 and 0.8 of Nyquist. The amplitudes and widths of fitted Gaussians to the noise power spectra were observed to be in excellent agreement with those expected from the number of counted electrons per frame and estimates from the point spread function of images of individual incident electrons.
GABAA receptor production, purification and nanodisc reconstitution
Human GABAA receptor β3-subunit with a truncated M3-M4 loop was transiently expressed in HEK293S GnTI- cells as previously described3. The sample used for XFEG datasets collected on Falcon4 contained a point mutation, K279T, thought to stabilise the receptor and a C-terminal 1D4 tag for purification. The sample used for the CFEG dataset, as well as the Falcon3 and K3 datasets, contained the wild-type sequence and an N-terminal streptavidin-binding peptide (SBP) tag for purification. Subsequent comparison of the resulting cryo-EM maps revealed that the K279T mutation had no structural impact, hence the Falcon4 datasets could be merged during data processing. Frozen cell pellets were resuspended in 50 mM HEPES pH 7.6, 300 mM NaCl, 1 mM histamine supplemented with 1% (v/v) mammalian protease inhibitor cocktail (Sigma-Aldrich) and solubilised with 1% (w/v) lauryl maltose neopentyl glycol (LMNG) and 0.1% (w/v) cholesteryl hemisuccinate Tris salt (CHS). No CHS was added to the sample that was used for the CFEG dataset. Receptors were purified by 1D4 and SBP affinity chromatography, respectively. Nanodisc reconstitution was performed as previously described23. Briefly, after the binding step, while the receptor was still bound to the affinity beads, an excess of bovine brain extract (BBE; type I, Folch fraction I, Sigma-Aldrich, 20 mg/mL stock) and phosphatidylcholine (POPC, Avanti, 10 mg/mL) mixture (15:85, v/v) was added to the samples and incubated for 30 min. Receptors were then reconstituted into membrane scaffold protein 2N2 (MSP2N2) lipid nanodiscs by incubating with an excess of MSP2N2 (0.6 mg/mL final concentration) and bio-beads for 2 h. The resin was then washed extensively and the GABAAR samples were eluted with a buffer containing 12.5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.6, 75 mM NaCl, 1 mM histamine and 2mM 1D4 peptide or 2.5 mM biotin, respectively.
Cryo-EM grid preparation
Purified GABAAR was incubated with ~1 μM Megabody-2521 and 3.5 μl sample was applied to glow discharged 300 mesh 1.2/1.3 and 2/2 UltraAuFoil gold grids (Quantifoil) for 30 s then blotted for 5.5 s prior to plunge-freezing with liquid ethane cooled by liquid nitrogen. Grid vitrification was performed using a Leica plunger (Leica Microsystems; XFEG datasets) or Vitrobot Mark IV (Thermo Fisher Scientific; CFEG datasets) at 100% humidity and 14 °C.
A frozen aliquot of 7 mg/ml mouse apoferritin in 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM dithiothreitol (DTT) and 5% trehalose, which we received from the Kikkawa Lab at Tokyo University, was thawed at room temperature and cleared by centrifugation at 10,000g for 10 min. The supernatant was diluted to 5 mg/ml with 20 mM HEPES pH 7.5 150 mM NaCl, and 3 μl of the diluted sample was applied onto glow-discharged R1.2/1.3 300 mesh UltrAuFoil gold grids (Quantifoil) for 30 s and then blotted for 5 s before plunge-freezing the grids into liquid ethane cooled by liquid nitrogen. Plunge-freezing was performed using a Vitrobot Mark IV (Thermo Fisher Scientific) at 100% humidity and 4 °C.
Cryo-EM data acquisition
All cryo-EM data were collected on Falcon or K3 cameras in electron counting mode using Titan Krios microscopes (Thermo Fisher Scientific) operating at 300 kV. Before data acquisition, two-fold astigmatism was corrected and beam tilt was adjusted to the coma-free axis using the autoCTF program (Thermo Fisher Scientific). All datasets were acquired automatically using EPU software (Thermo Fisher Scientific). Detailed data acquisition parameters for all data sets are given in Extended Data Table 1.
For GABAAR, two data sets were acquired on a Titan Krios at the Department of Biochemistry, University of Cambridge, UK. This microscope is equipped with an XFEG and a bottom-mounted (BM) Falcon-3 camera and a K3 camera behind the Gatan energy filter. All other data sets were acquired at the Thermo Fisher Scientific RnD division in Eindhoven, The Netherlands. Four GABAAR data sets were collected on a Titan Krios equipped with an XFEG and a prototype Falcon-4 camera that was mounted behind the new energy filter. These datasets were collected consecutively over a period of 5 days from the same grid to study the effect of varying energy slit widths. To keep the ice thickness similar between these experiments, all the exposure areas were selected at the start using a constant ice filter within EPU software. One GABAAR data set was acquired on a microscope with a CFEG and a prototype Falcon-4 camera that was mounted behind the new energy filter. The slit width of the energy filter was set to 5 eV. For this data set, 5573 movies were acquired using a 100 μm objective aperture and 3160 movies were collected without an objective aperture. The CFEG was automatically flashed every 5 hours using the EPU software.
For the apoferritin data set, we used the same microscope with the CFEG, the new filter and the prototype Falcon-4 camera. No objective aperture was used and the slit width of the energy filter was set to 10 eV. Flashing of the CFEG had no noticeable effect on data quality (Extended Data Figure 4f).
Data acquisition was not optimised in terms of speed and the prototype Falcon-4 version did not write movies at the highest possible rate. Typically, two movies were collected per hole, with physical stage movements in between each hole. The resulting data acquisition rate of ~95 movies per hour (including Dewar filling times and CFEG flashing). The 3,370 apoferritin movies were collected within 36 hours; GABAAR datasets allowing ~2 Å reconstructions in ~9 hours. Fringe free imaging (FFI) and aberration-free image shift (AFIS), which allow acquisition of multiple images per hole and multiple holes per stage position, would allow considerably higher throughputs.
The EER movie format
Electron event representation (EER) is a movie format that is introduced with the Falcon-4 camera18. Whereas in the existing MRC image format groups of subsequent camera frames, or dose fractions, are summed and represented as images, EER stores the location and time of recording of each detected electron event. The EER format records the event coordinates in a four times oversampled, that is a 16k×16k, grid. The time resolution is the original frame rate of the camera, so that dose fractionation during data acquisition is no longer needed. Instead, beam-induced sample motions or stage drift can be corrected without a loss in temporal resolution. The electron event stream is stored on disk after compression with a run-length encoding algorithm. For GABAAR, data sets were acquired at a dose rate of 4.7 electrons per pixel per second, at a pixel size of 0.727 Å and a total dose of 18.1 electrons per pixel. These settings yielded EER movies consisting of 1113 frames and with an average size of 485 MB. For apoferritin, data were acquired at a dose rate of 4.5 electrons per pixel per second, at a pixel size of 0.457 Å and a total dose of 40 electrons per Å2, resulting in movies of 434 frames occupying on average 160 MB.
To read EER movies, we modified the publicly available RELION-3.1-beta. Inside RELION, the events from the 16k×16k grid are positioned as binary pixels in a 8k×8k grid, which is then Fourier cropped into a 4k×4k grid. Using the modified version, EER movies can be used for frame alignment using RELION’s implementation of the MotionCor2 algorithm36 and for per-particle beam-induced motion correction using its Bayesian polishing program20. For both programs, the user needs to specify a dose fractionation rate.
Ice-thickness measurement
The ice thickness was measured on a GABAAR grid using tomography. Tilt series were collected on three different grid-squares with a 10eV slit on a Falcon4 camera. Data was acquired covering an angular range from -45° to +45° with 3° angular increments recorded automatically using the dose-symmetric tilting scheme under low-dose conditions using TEM Tomography software (TFS). Each tilt series was collected with a nominal defocus value of 5 μm in electron counting mode using a dose of 4 e-/Å2 per tilt and a calibrated pixel size of 1.898 Å. Tomograms were reconstructed with patch tracking and ice thickness measured with IMOD software37.
Apoferritin image processing
A total of 3370 movies in EER format were motion corrected with RELION’s implementation of the MotionCor2 algorithm36. For this purpose, original hardware movie frames were dose fractionated into groups of 14 frames, corresponding to an accumulated dose of 1.3 e-/Å2 per fraction. CTF estimation was performed with CTFFIND-4.1.1338 using the sums of power spectra from combined fractions corresponding to an accumulated dose of 4 e-/Å2. Micrographs whose estimated resolution from CTFFIND was worse than 5 Å were removed, leaving 3080 micrographs for further processing.
Using RELION’s template-based algorithm, 428,590 particles were picked and subjected to 2D classification. After selection of good classes, 380,236 particles were subjected to standard 3D auto-refinement to give an initial reconstruction with an estimated resolution of 2.3 Å. Subsequently, three runs of CTF refinement26 were performed: first refining magnification anisotropy; then refining optical aberrations (up to the 4th order); and finally refining per-particle defocus and per-micrograph astigmatism. A 3D auto-refinement with the refined values yielded an estimated resolution of 1.7 Å. Bayesian polishing to optimise per-particle beam-induced motion tracks, followed by another round of auto-refinement resulted in a 1.43 Å map. CTF refinement was then repeated for optical aberration correction, magnification anisotropy, per-particle defocus and per-micrograph astigmatism. To compensate for changes in the tilt of the electron beam during the data acquisition session, separate optics groups were created for particles belonging to consecutive groups of 125 micrographs, or whenever the CFEG was flashed. At this point, 201 micrographs with refined scale factors (rlnGroupScaleCorrection) with values below 0.5 were also removed, resulting in 363,126 particles in the final set (Extended Data Figure 4f). Following a last round of auto-refinement, these particles were subjected to a second round of Bayesian polishing with a large particle extraction box (spanning 320 Å), such that high-resolution signals that are delocalised by the CTF were still contained in the images. At this point, seven hardware frames were grouped into each dose fraction to be able to capture more rapid motions. Since Ewald sphere curvature was predicted to limit the resolution of a particle with a diameter of 120 Å to approximately 1.3 Å39, the particles were then used for reconstruction with Ewald sphere correction27. The estimated resolution of the final map, following standard post-processing procedures in RELION, was 1.22 Å.
GABAA receptor image processing
GABAAR data sets were processed following the same strategy as for apoferritin, with the following exceptions. All movies, except for the ones acquired on the microscope with a CFEG, were recorded in MRC format. For the CFEG data, hardware movie frames were initially dose fractionated into groups of 24 frames, corresponding to an accumulated dose of 0.86 e-/Å2 per fraction. For the second round of Bayesian polishing, a dose fractionation of 12 hardware frames was used. Particle picking was performed using an in-house generated Linux port of the of BoxNet2D neural network in Warp40. Prior to the last auto-refinement, 3D classification into three classes without alignment was performed to select the particles contributing to the best reconstruction. The first part of the CFEG data was acquired with a 100 μm objective aperture. Charging of the aperture probably led to third and fourth order optical aberrations, which were estimated during CTF refinement in optics groups of 160 micrographs, or whenever the CFEG was flashed or liquid nitrogen was refilled. The second part of the CFEG data was acquired without an objective aperture. For these images, no significant higher-order optical aberrations were observed and creating multiple optics groups was deemed unnecessary. When merging the energy-filtered CFEG and XFEG data sets, refinement of per-micrograph scale factors and per-particle normalisation corrections were unstable, and these were switched off using the --no_norm and --no_scale arguments. The mask used for resolution estimation during post-processing only contained density for GABAAR; all density corresponding to the megabodies was removed.
B-factor estimation
For each GABAAR dataset, a random subset of 32,000 particles was selected from the final set of particles after the second round of Bayesian polishing. This subset was subjected to 3D auto-refinement and the resulting orientations were used to calculate reconstructions for each of the two random halves used in the auto-refinement. Similar half-set reconstructions were also calculated for random subsets of 16,000, 8,000, 4,000, 2,000, 1,000 and 500 particles. This procedure was repeated seven times. For each subset and repeat, resolution estimation was again performed using standard post-processing functionality in RELION. The inverse square of the resulting estimated resolutions were then plotted against the natural logarithm of the number of particles in the subset, and B-factors were calculated from the slope of a straight line fitted through all points in the plot. For apoferritin, the same procedure was used, starting from a subset of 180,000 particles and halving the subset size eight times.
Atomic modelling
The atomic models for GABAAR and apoferritin were derived from PDB-4cof and PDB-6s61, respectively. Both models were manually adjusted in COOT41. Difference maps were used to rebuild the model, add alternative conformations and waters. Real-space refinement was performed using PHENIX, version 1.16.354942 for GABAAR. Both models were refined with anisotropic atomic B-factors in reciprocal space using REFMAC, version 5.8.025825. For cross-validation, the atomic models refined against the full map was perturbed by random shifts of the atoms of up to 0.3 Å and the perturbed model was refined in REFMAC against one of the two half-maps. FSC curves for the resulting model against that half-map (FSCwork) and against the other half-map (FSCfree) are shown with blue and dashed orange lines, respectively, in Extended Data Figures 2 and 4. Molprobity score, clash score, rotamer and Ramachandran analyses were performed using MOLPROBITY43.
For refinement and difference map calculation, maps were first scaled globally using Gaussian scaling: i.e. where Fo is the Fourier transform of the post-processed map and Fc is the Fourier transform of a map calculated from a model without hydrogens. Next, each resolution shell was scaled by finding a scale factor D and sigma per shell with a maximum likelihood method: . Finally, difference maps were calculated as Fo - DFc. Since Fo is FSC weighted, the resulting map will be also FSC weighted. FSC is calculated from half maps. Structural figures were prepared using PyMOL (Schrödinger) and UCSF ChimeraX44.
Hydrogen position calculations
X-ray scattering and electron scattering factors are related through the Mott-Bethe formula, which is the solution of Poisson equations in Fourier space under certain conditions:
Where ; m is the mass of an electron; e is the charge of an electron, h is the Planck constant; and ϵ 0 is the dielectric constant of the vacuum; Z is the nuclear charge; fX(s) is the X-ray scattering factor; fe(s) is the electron scattering factor; and s=1/d is the resolution. Adding the position-independent, isotropic atomic B-factor gives:
This formula is valid if the centre of electron density coincides with the position of the nucleus. If this is not the case, the formula should be modified according to the vector Δx between the centre of electron density and the nuclear position (assuming that the centre of electron density is at the origin):
Note that, in general, the form factor for electron scattering becomes a complex quantity. For all non-hydrogen atoms, it is safe to assume that the centre of electron density and the nuclear position coincide.
For hydrogen atoms, the centre of electron density and the nuclear position do not coincide and we are interested in the peak height for the density of electrostatic potential along the bond between a hydrogen and its parent atom. The below ignores the fact that electron densities will be non-spherical, as they are elongated along the bond, thus adding additional complexity to Mott-Bethe formula.
The densities for the proton and the electron of a hydrogen atom at a resolution smax and with a given B value is given by:
,
To calculate the density at a position x we need to calculate:
Even if both densities corresponding to the proton and the electron are spherically symmetric, the combined density will not be spherical. Extended Data Figure 5 shows density profiles and their maxima for a hydrogen bonded to a carbon atom for different resolutions and B values. These profiles were calculated with the assumption that the electron-carbon distance is 0.98 Å and the proton-carbon distance is 1.09 Å.
Extended Data
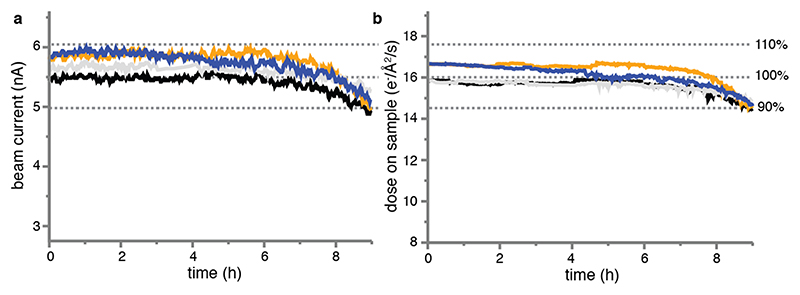
Extended Data Figure 1. Characteristics of the new cryo-EM technology.
(a) Four consecutive measurements of the CFEG beam current over a period of nine hours. The FEG tip was flashed just before the start of each measurement. (b) The dose at the sample as measured for the same four experiments in (a).
Extended Data Figure 2. Cryo-EM for GABAAR.
(a) B-factor plots for three data sets using an X-FEG, the new energy filter with a slit width of 3eV (orange), 5eV (blue) and 10 eV (grey) and a Falcon 4 camera. (b) Two orthogonal views of an electron tomogram for ice thickness measurement. Scale bar is 50 nm. (c) Representative electron micrograph from the CFEG dataset. Scale bar is 30 nm. (d) A selection of 2D class average images. (e) Local resolution map. (f) Fourier Shell Correlation (FSC) between the two independently refined half-maps. (g) FSC between the model and the map calculated for the model refined against the full reconstruction (black); the model refined in the first half-map against that half-map (FSCwork; blue); and the model refined in the first half-map against the second half-map (FSCtest; dashed orange). Atomic models were refined including spatial frequencies up to 1.63 Å.
Extended Data Figure 3. GABAAR reconstruction details.
(a) The GABAAR cryo-EM map viewed from the extracellular space (top view). The density of one subunit of the homopentamer is highlighted in yellow; glycans are orange; the nanobody domain of Mb25 green; and the lipid nanodisc light blue. (b) As in b), but viewed parallel to the plasma membrane space (side view). (c) Stacking of histamine with aromatic residues in the ligand-binding pocket. Water molecules removed for clarity. (d) Radiation damage in the ligand-binding pocket illustrated by difference maps of Asp43 and Glu155 carboxyl groups. (e) Radiation damage causes partial reduction of the disulfide bond between Cys136 and Cys150. Left panel: oxidized state, right panel: reduced state. Only Cα, Cβ and sulphur atoms depicted for clarity. (f) Difference map revealing the hydrogen bonding network between β-strands. (g) A close-up view of the lipids (blue) surrounding the transmembrane region of the receptor. Compared to (a-b), the contour level is decreased. Red arrow indicates section level as depicted in panel (h). The general anaesthetics pocket and the neurosteroid modulation site are indicated by the red circle and rectangle, respectively. (h) Top-view of a transverse section through the GABAAR transmembrane region shows semi-ordered lipids surrounding individual subunits. (i) Close-up view of the general anaesthetics pocket. (j) Close-up view of the neurosteroid modulation site. Lipids in (i) and (j) could not be identified unambiguously and therefore are modelled as aliphatic chains.
Extended Data Figure 4. Cryo-EM for apoferritin.
(a) Representative electron micrograph. Scale bar is 20 nm. (b) 2D class average images. (c) Local resolution map. (d) Fourier Shell Correlation (FSC) between the two independently refined half-maps. (e) FSC between the model and the map as calculated for the model refined against the full reconstruction against that map (black); the model refined in the first half-map against that half-map (FSCwork; blue); and the model refined in the first half-map against the second half-map (FSCtest; dashed orange). FSCwork and FSCtest curves are shown for three different models: with hydrogens and anisotropic B-factors (solid lines); with hydrogens and isotropic B-factors (dashed lines); and without hydrogens and with isotropic B-factors (dotted lines). Including hydrogens and using anisotropic B-factors gives a better fit to the data at medium resolution, but lead to a small amount of overfitting at high resolution. Atomic models were refined including spatial frequencies up to 1.15 Å. (f) The beam tilt in X (blue) and Y (orange); the resolution (res.) for subsets of 5,000 consecutive particles (black); and RELION’s rlnGroupScaleCorrection values for individual micrographs (blue dots) during the data acquisition experiment. Events like CFEG flashing (red), liquid nitrogen filling (black) and moving the grid to another grid square (dark blue) are indicated with vertical lines. The cutoff in scale correction values used to discard part of the micrographs is indicated with a dashed grey line.
Extended Data Figure 5. Electrostatic potential of hydrogen atoms.
(a) Calculated profile (see Methods) of the electrostatic scattering potential along the bond between a carbon and a hydrogen atom, for a B-value of 10 Å2 and resolutions (d) of 1.0 Å (orange), 1.2 Å (blue), 1.5 Å (grey) and 1.8 Å (black). The two vertical lines indicate the electron-carbon distance of 0.98 Å and the proton-carbon distance of 1.09 Å. (b) As in (a), but for a resolution of 1.2 Å and B-values of 5 Å2, 10 Å2, 15 Å2 and 20 Å2. (c) Distances from the carbon atom (in Å) at which the calculated profile of the electrostatic scattering potential along the bond between a carbon and a hydrogen atom is at its maximum, for different resolutions (d) and B-values.
Extended Data Table 1. Cryo-EM data collection parameters.
| Apoferritin | GABAA-β3 | GABAA-β3 | GABAA-β3 | GABAA-β3 | GABAA-β3 | GABAA-β3 | GABAA-β3 | |
|---|---|---|---|---|---|---|---|---|
| Data collection and processing | ||||||||
| Electron Gun | CFEG | CFEG | XFEG | XFEG | XFEG | XFEG | XFEG | XFEG |
| Detector | Falcon4 | Falcon4 | Falcon4 | Falcon4 | Falcon4 | Falcon4 | BM-Falcon3 | K3 |
| Magnification | 270k | 165k | 165k | 165k | 165k | 165k | 96k | 130K |
| Energy filter slit width (eV) | 10 | 5 | open | 10 | 5 | 3 | NA | 20 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 |
| Flux on detector (e/pix/sec) | 4.5 | 4.7 | 2.7 | 2.8 | 2.8 | 2.8 | 0.7 | 14.9 |
| Electron exposure on sample (e–/Å2) | 40 | 40 | 35 | 35 | 35 | 35 | 37 | 40 |
| Target defocus range (μm) | 0.3-0.9 | 0.3-1.1 | 0.9-1.8 | 0.3-1.1 | 0.3-1.1 | 0.3-1.1 | 0.5-1.5 | 0.3-1.5 |
| Observed defocus range (95 th percentile in μm) | 0.27-0.97 | 0.30-1.11 | 0.80-1.83 | 0.31-0.95 | 0.32-0.87 | 0.34-1.00 | 0.58-1.51 | 0.48-1.46 |
| Calibrated pixel size (Å) | 0.457 | 0.727 | 0.735 | 0.735 | 0.735 | 0.735 | 0.827 | 0.65482 |
| Symmetry imposed | O | C5 | C5 | C5 | C5 | C5 | C5 | C5 |
| Number of collected movies | 3370 | 8733 | 1244 | 1268 | 1556 | 1952 | 1501 | 1507 |
| Initial particle images (no.) | 428590 | 1105069 | 192053 | 199867 | 242059 | 264675 | 412913 | 215957 |
| Final particle images (no.) | 363126 | 233576 | 33166 | 42780 | 45009 | 50328 | 129810 | 46441 |
| Map resolution at FSC=0.143 (Å) | 1.22 | 1.73 | 1.97 | 1.89 | 1.87 | 1.88 | 1.97 | 2.32 |
Extended Data Table 2. Refinement and validation statistics.
| Refinement | Apoferritin (EMD-11638) (PDB 7A4M) (EMPIAR-10424) | GABAA-β3 (EMD-11657) (PDB 7A5V) (EMPIAR-10500) |
|---|---|---|
| Map resolution range refined against (Å) | 1.22 - 136.5 | 1.6 - 291.1 |
| Map sharpening B factor (Å2) | 2.3 | 23 |
| Model composition | ||
| Non-hydrogen atoms Protein residues Waters Ions Glycans HSM Lipids |
40512 37584 2880 48 - - - |
21144 19239 1020 10 515 40 320 |
| Isotropic B factors (Å2) | ||
| Protein Waters Ions Glycans HSM Lipids |
25.3 39 33.4 - - |
46 40.8 50.5 73.2 23.2 105.4 |
| R.m.s. deviations | ||
| Bond lengths (Å) Bond angles (°) |
0.0155 1.739 |
0.009 1.285 |
| Validation | ||
| MolProbity score Clashscore Poor rotamers (%) |
1.69 4.96 2.94 |
1.17 2.69 0 |
| Ramachandran plot | ||
| Favored (%) Allowed (%) Disallowed (%) |
97.65 2.35 0.00 |
97.33 2.67 0.00 |
Supplementary Material
Acknowledgements
We thank Mike van Beers, Marcel Veerhoek, Roland Jonkers for maintaining the Titan Krios microscopes; Walter van Dijk, Bram van de Kerkhof, Stan Konings and Gijs van Duinen for advice on optics and microscope alignments; Bart van Knippenberg, Andreas Voigt and Fanis Grollios for support with EPU software; Haruaki Yanagisawa from the Kikkawa lab for providing the apoferritin sample; Paul Miller for the GABAA expression vectors; Adrian Koh, Toby Darling and Jake Grimmett for support with computing; Garbi Lezcano Singla, Erik Franken for support with the EER format; Gerald van Hoften and Gerard Hosmar for support with the Falcon-4 camera and Eirini Ioannou for logistics support. This work was supported by the EM facilities at the MRC-LMB, the Biochemistry Department of Cambridge University and Thermo Fisher Scientific. The Cryo-EM Facility at Department of Biochemistry is funded by the Wellcome Trust (206171/Z/17/Z; 202905/Z/16/Z) and the University of Cambridge. We acknowledge funding from the UK Medical Research Council (MC_UP_A025_1012 to G.M., MR/L009609/1 and MC_UP_1201/15 to A.R.A. and MC_UP_A025_1013 to S.H.W.S.); the Japan Society for the Promotion of Science (Overseas Research Fellowship to T.N.); MRC-LMB, Cambridge Trust and School of Clinical Medicine, University of Cambridge (LMB Cambridge Scholarship and Cambridge MB/PhD fellowship to A.S.); the European Commission (Marie Skłodowska-Curie Actions H2020-MSCA-IF-2015/709054 to L.M. and H2020-MSCA-IF-2017/793653 to P.M.G.E.B.); EMBO (long term fellowship 300-2015 to L.M.); Cancer Research UK (T.M., grants C20724/A14414 and C20724/A26752 to Christian Siebold, University of Oxford); Boehringer Ingelheim Fonds (PhD Fellowship to J.M.); Research Foundation - Flanders (FWO, PhD Fellowship to T.U.).
Footnotes
Author contributions
E.d.J., J.K., M.B., J.M., and P.T. contributed to microscope hardware and software developments; G.McM. performed DQE analysis; D.K., A.S. and S.M. prepared cryo-EM grids, T.U. produced megabody, A.K., L.Y., E.V.P, S.W.H and D.Y.C. acquired cryo-EM data; T.N., A.K, A.S., L.Y., S.M., P.M.G.E.B., I.T.G., L.M., J.M. and S.H.W.S analysed cryo-EM data; T.N., A.R.A., T.M. and G.M. prepared atomic models; T.N., A.K., A.R.A. and S.H.W.S. conceived the project and designed the experiments; G.M., A.R.A. and S.H.W.S. wrote the manuscript, with input from all authors.
Competing interest statement
A.K., S.M., L.Y., D.K., E.V.P., E.d.J., J.K., M.B., J.M., and P.T are employees of Thermo Fisher Scientific.
Data availability
Atomic coordinates for the GABAAR construct and apoferritin have been deposited in the Protein Data Bank with accession codes 7A5V and 7A4M, and the cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-11657 and EMD-11638, respectively. Raw movies of all datasets have been deposited in the Electron Microscopy Public Image Archive with accession codes EMPIAR-10500 for GABAAR and EMPIAR-10424 for apoferritin.
Code availability
Both the implementation of reading EER movies into RELION and the EER format itself are distributed as free, open-source software, with downloads available from https://github.com/3dem/relion and https://github.com/fei-company/EerReaderLib. In-house generated Linux port of the BoxNet2D neural network in Warp is available from https://gist.github.com/biochem-fan/76ff5934495585da56ab2c0af4fe2563.
References
- 1.Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyumkis D. Challenges and opportunities in cryo-EM single-particle analysis. J Biol Chem. 2019;294:5181–5197. doi: 10.1074/jbc.REV118.005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glaeser RM. Specimen Behavior in the Electron Beam. Methods Enzymol. 2016;579:19–50. doi: 10.1016/bs.mie.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5 doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 8.McMullan G, Faruqi AR, Clare D, Henderson R. Comparison of optimal performance at 300keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy. 2014;147C:156–163. doi: 10.1016/j.ultramic.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kühlbrandt W. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 10.Hamaguchi T, et al. A new cryo-EM system for single particle analysis. J Struct Biol. 2019;207:40–48. doi: 10.1016/j.jsb.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Kato T, et al. CryoTEM with a Cold Field Emission Gun That Moves Structural Biology into a New Stage. Microsc Microanal. 2019;25:998–999. [Google Scholar]
- 12.Gubbens A, et al. The GIF Quantum, a next generation post-column imaging energy filter. Ultramicroscopy. 2010;110:962–970. [Google Scholar]
- 13.Tanaka . Journal of Microscopy. Wiley Online Library; 1999. A new 200 kV Ω-filter electron microscope. [DOI] [PubMed] [Google Scholar]
- 14.Kahl F, et al. Chapter Three - Test and characterization of a new post-column imaging energy filter. In: Hawkes PW, Hÿtch M, editors. Advances in Imaging and Electron Physics. Vol. 212. Elsevier; 2019. pp. 35–70. [Google Scholar]
- 15.Kuijper M, et al. FEI’s direct electron detector developments: Embarking on a revolution in cryo-TEM. J Struct Biol. 2015;192:179–187. doi: 10.1016/j.jsb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 16.McMullan G, Faruqi AR, Henderson R. Direct Electron Detectors. Methods Enzymol. 2016;579:1–17. doi: 10.1016/bs.mie.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 17.Ruskin RS, Yu Z, Grigorieff N. Quantitative characterization of electron detectors for transmission electron microscopy. J Struct Biol. 2013;184:385–393. doi: 10.1016/j.jsb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, et al. Electron Event Representation (EER) data enables efficient cryoEM file storage with full preservation of spatial and temporal resolution. 2020 doi: 10.1101/2020.04.28.066795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zivanov J, Nakane T, Scheres SHW. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchański T, et al. Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM. 2019 doi: 10.1101/812230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieghart W, Savić MM. International Union of Basic and Clinical Pharmacology. CVI: GABAA Receptor Subtype- and Function-selective Ligands: Key Issues in Translation to Humans. Pharmacol Rev. 2018;70:836–878. doi: 10.1124/pr.117.014449. [DOI] [PubMed] [Google Scholar]
- 23.Masiulis S, et al. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature. 2019;565:454–459. doi: 10.1038/s41586-018-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Nicholls RA, Tykac M, Kovalevskiy O, Murshudov GN. Current approaches for the fitting and refinement of atomic models into cryo-EM maps using CCP-EM. Acta Crystallogr Sect Struct Biol. 2018;74:492–505. doi: 10.1107/S2059798318007313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zivanov J, Nakane T, Scheres SHW. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo CJ, Henderson R. Ewald sphere correction using a single side-band image processing algorithm. Ultramicroscopy. 2018;187:26–33. doi: 10.1016/j.ultramic.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tegunov D, Xue L, Dienemann C, Cramer P, Mahamid J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.7 Å inside cells. 2020 doi: 10.1101/2020.06.05.136341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheldrick G. Phase Annealing in Shelx-90 - Direct Methods for Larger Structures. Acta Crystallogr Sect A. 1990;46:467–473. [Google Scholar]
- 30.Wlodawer A, Dauter Z. ‘Atomic resolution’: a badly abused term in structural biology. Acta Crystallogr Sect Struct Biol. 2017;73:379–380. doi: 10.1107/S205979831700225X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiemeijer PC, Bischoff M, Freitag B, Kisielowski C. Using a monochromator to improve the resolution in TEM to below 0.5Å. Part I: Creating highly coherent monochromated illumination. Ultramicroscopy. 2012;114:72–81. doi: 10.1016/j.ultramic.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Langmore JP, Smith MF. Quantitative energy-filtered electron microscopy of biological molecules in ice. Ultramicroscopy. 1992;46:349–373. doi: 10.1016/0304-3991(92)90024-e. [DOI] [PubMed] [Google Scholar]
- 33.Rose HH. Future trends in aberration-corrected electron microscopy. Philos Transact A Math Phys Eng Sci. 2009;367:3809–3823. doi: 10.1098/rsta.2009.0062. [DOI] [PubMed] [Google Scholar]
- 34.Haider M, Hartel P, Müller H, Uhlemann S, Zach J. Current and future aberration correctors for the improvement of resolution in electron microscopy. Philos Transact A Math Phys Eng Sci. 2009;367:3665–3682. doi: 10.1098/rsta.2009.0121. [DOI] [PubMed] [Google Scholar]
- 35.McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastronarde DN, Held SR. Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeRosier DJ. Correction of high-resolution data for curvature of the Ewald sphere. Ultramicroscopy. 2000;81:83–98. doi: 10.1016/s0304-3991(99)00120-5. [DOI] [PubMed] [Google Scholar]
- 40.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casañal A, Lohkamp B, Emsley P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci Publ Protein Soc. 2020;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr Sect Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams CJ, et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci Publ Protein Soc. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci Publ Protein Soc. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates for the GABAAR construct and apoferritin have been deposited in the Protein Data Bank with accession codes 7A5V and 7A4M, and the cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-11657 and EMD-11638, respectively. Raw movies of all datasets have been deposited in the Electron Microscopy Public Image Archive with accession codes EMPIAR-10500 for GABAAR and EMPIAR-10424 for apoferritin.
Both the implementation of reading EER movies into RELION and the EER format itself are distributed as free, open-source software, with downloads available from https://github.com/3dem/relion and https://github.com/fei-company/EerReaderLib. In-house generated Linux port of the BoxNet2D neural network in Warp is available from https://gist.github.com/biochem-fan/76ff5934495585da56ab2c0af4fe2563.