Abstract
Mononuclear phagocytes (MNPs) are vital for maintaining intestinal homeostasis but in response to acute microbial stimulation can also trigger immunopathology, accelerating recruitment of Ly6Chi monocytes to the gut. The regulators that control monocyte tissue adaptation in the gut remain poorly understood. Interferon Regulatory Factor 5 (IRF5) is a transcription factor previously shown to play a key role in maintaining the inflammatory phenotype of macrophages. Here we investigate the impact of IRF5 on the MNP system and physiology of the gut at homeostasis and during inflammation. We demonstrate that IRF5 deficiency has a limited impact on colon physiology at steady state but ameliorates immunopathology during Helicobacter hepaticus induced colitis. Inhibition of IRF5 activity in MNPs phenocopies global IRF5 deficiency. Using a combination of bone marrow chimera and single cell RNA-sequencing approaches we examined the intrinsic role of IRF5 in controlling colonic MNP development. We demonstrate that IRF5 promotes differentiation of Ly6Chi monocytes into CD11c+ macrophages and controls the production of anti-microbial and inflammatory mediators by these cells. Thus, we identify IRF5 as a key transcriptional regulator of the colonic MNP system during intestinal inflammation.
Introduction
The term Inflammatory Bowel Disease (IBD) encompasses a group of debilitating inflammatory conditions of the gastrointestinal tract that affects ~0.5-1% of westernised populations (1). The IBDs are associated with high morbidity and burden healthcare systems (2, 3). Conventional IBD therapies are limited by moderate-high rates of adverse events, or patient unresponsiveness, whilst approximately 40% of patients successfully treated with anti-TNFα become refractory to therapy (2). Therefore, there is unmet clinical need for IBD therapies. The aetiology of IBD is unknown, but interplay between host genetics, and environmental factors, and the microbiota contribute to disease pathogenesis (1).
Mononuclear Phagocytes (MNPs), including monocytes, macrophages, and Dendritic Cells (DCs), are present in large numbers in the colonic Lamina Propria (cLP), and carry out diverse, overlapping functions critical to the maintenance of intestinal homeostasis. The dysregulation of the intestinal MNP system leads to infection and inflammation (4–11).
The origins of the intestinal MNP systems has been the topic of considerable debate in recent years, clouded by inconsistent nomenclature and shared surface markers between macrophages and DCs. Intestinal Lamina Propria DCs at the steady state are largely derived from pre-DC precursors, generated in the bone marrow, which are understood differentiate into three major intestinal DC subsets. These subsets comprise an XCR1 positive (Xcr1+SIRPα-CD103+Cd11b-CX3CR1-) population that is analogous to classical dendritic cells (cDC) 1 DCs, and two cDC2-like SIRPα positive (SIRPα+Xcr1- Cd11b+CX3XR1+) subsets which can be further discriminated by CD103 expression(12, 13). In addition, the existence of a discrete population of hybrid macrophage/DC cells within the cDC2 intestinal compartment has been described (14). The ontogeny of intestinal DCs during inflammation is more complicated since some monocyte-derived cells may acquire phenotypic and functional DC hallmarks (15–17).
Intestinal Lamina Propria macrophages have dual origins: from embryonically derived macrophages (CD4+ Tim4+) that self-renew, and monocytes, but in the adult mouse, most of the macrophage turnover is of monocytic origin (18, 19). In mice, the differentiation of monocytes to macrophages in the cLP has been termed the “monocyte waterfall” (19). After entering the cLP, naïve Ly6Chi, MCHII- (P1) monocytes begin maturing by acquiring expression of MHCII (P2) before downregulating Ly6C expression. The pool of MHCII+ cells comprises of Ly6C+/-CX3CR1Int monocyte/macrophage intermediates (P3) and fully mature Ly6C-CX3CR1hiF4/80hiCD64hiMHCIIhi macrophages (P4) (19, 20). During infection, de novo recruited monocytes give rise to CD11c+ intestinal macrophages that are phenotypically pro-inflammatory (21). The blood origin of intestinal macrophage subsets was also confirmed in human studies where two monocyte-derived macrophage populations: CD11c+ with high turnover and CD11c- with slow turnover, were identified at steady state (22). It was suggested that CD11c+ macrophages might be an intermediate between blood monocytes and tissue resident CD11c- macrophages (22).
The regulators that control the transition of monocytes through a number of intermediate differentiation states are largely unknown, but the cytokines, TGFβ and IL10, have been linked to the development of cLP tissue-resident macrophages (23, 24). CX3CR1IL10R- mice exhibited heightened inflammation, which maintained a pro-inflammatory monomacrophage state, preventing their full differentiation and initiating spontaneous colitis (24). Loss of TGFβ-Receptor on macrophages resulted in a minor impairment of macrophage differentiation, defined by transcriptional profiling of monocyte to macrophage transition in the cLP (23).
One candidate intrinsic regulator of the intestinal macrophage signature is Interferon Regulatory Factor 5 (IRF5), which was described to promote an inflammatory macrophage phenotype(25) and has variants that are genetic risk factors for Ulcerative Colitis and Crohn’s Disease (26–28). IRF5 is activated by phosphorylation and ubiquitination events downstream of Pattern Recognition Receptors (PRRs), e.g. NOD2, TLR2, and TLR4, and directly regulates many cytokines associated with IBD (IL-lβ, IL-6, IL-10, IL-12, IL-23, TNF), placing IRF5 as a nexus for the regulation of inflammatory responses (1, 25, 29). To formally examine the role of IRF5 in the establishment of intestinal MNP system, we compared the continuum of cell states of wild type (WT) and IRF5 deficient (Irf5-/-) MNPs at steady state and during Helicobacter hepaticus (Hh) induced intestinal inflammation using a combination of competitive Mixed Bone Marrow Chimaera (MBMC), single cell gene expression analysis (scRNA-seq) and functional validation approaches. Hh infection concomitant with the administration of anti-Interleukin 10 Receptor (αIL10R) antibodies triggers IL-23 dependent intestinal inflammation with robust TH1/TH17 T cell response, which carries many features of human IBD(9, 30–32). In this model, CX3CRint and CD11c+ monocyte/macrophages intermediates drive immunopathology by producing pro-inflammatory cytokines such as IL-23, IL-lβ and TNFα (9, 33). We show that IRF5 promotes the differentiation of monocytes into a bactericidal and inflammatory CD11c+ macrophage phenotype during Hh + αIL10R-induced colitis and is essential for the development of immunopathology in this model.
Results
IRF5 deficiency has limited impact on colon physiology at steady-state
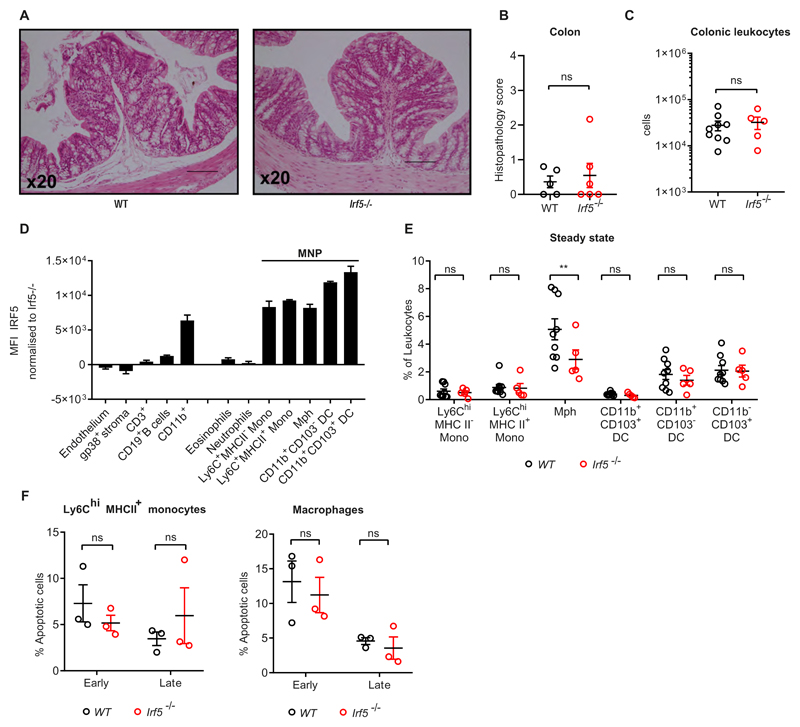
In steady state, we found that the colons (Fig.1A) and caeca (Supplementary Fig. S1A) of WT and Irf5-/- were comparable in morphology. Sections were scored for epithelial hyperplasia, nucleated cell infiltrate, area affected, and submucosal oedema and displayed no obvious signs of inflammation (score < 3) and no morphological differences between WT and Irf5-/- (Fig. 1B and Supplementary Fig S1B). The immune compartment of the cLP was evaluated by flow cytometry and revealed that the number of leukocytes in the colon (live CD45+) were comparable between WT and Irf5-/- (Fig. 1C).
Figure 1. IRF5 deficiency has limited impact on colon physiology at steady-state.
A) Representative H&E sections of colons from WT (left) and Irf5-/- (right) mice at steady state. B) Histopathology scoring of WT (n=6) and Irf5-/- (n=5) colons. C) Number of cLPLs retrieved from steady state WT (n=9) and Irf5-/- (n=5) mice. D) IRF5 expression in the steady state cLP of WT mice (n=3). E) The frequency of intestinal MNPs in the cLP of steady state WT (n=9) and Irf5-/- (n=5) mice. F) Quantification of early phase (Annexin V+ Live/Dead-), and late phase (Annexin V+ Live/Dead+) cell death assessed by Annexin V labelling combined with viability dye staining in WT (n=3) and Irf5-/- (n=3) cLP Ly6Chi MHC II+ (P2) monocytes and macrophages using flow cytometry immediately after cell isolation. B, C, E: Data are pooled from two independent experiments. D, F: data are representative of two independent experiments. B, C) Mann-Whitney U test. E, F) Two-Way ANOVA with Sidak correction. Data presented are mean ± SEM, ns = not significant, ** p ≤ 0.01
Next, we assessed the levels of IRF5 expression in the cells of the colon and demonstrated that non-myeloid, and non-leukocyte populations expressed low levels of IRF5 compared to CD11b+ myeloid cells (Fig.1D). Among myeloid cells, MNPs, i.e. monocytes, macrophages and DCs, expressed the highest levels of IRF5 (Fig.1D). The composition of the cLP myeloid compartment in WT and Irf5-/- was profiled using the gating strategy that included definition of the stages of monocyte differentiation(9, 20) (Supplementary Fig.S1C). Frequencies and absolute numbers of Ly6ChiMHCII- (P1) and Ly6ChiMHCII+ (P2) monocytes and CD11b+ DCs among the infiltrated leukocytes were similar in Irf5-/- animals but a higher frequency of F4/80+ macrophages was observed in WT mice (5.1%) than in Irf5-/- (2.9%) (Fig.1E, Supplementary Fig S1E). IRF5-deficient and WT Ly6Chi MHC II+ monocytes and macrophages were no different in their levels of apoptosis (Fig. 1F). Thus, we hypothesised that IRF5 may promote differentiation of monocytes to macrophages in the cLP.
IRF5 deficiency protects against intestinal inflammation
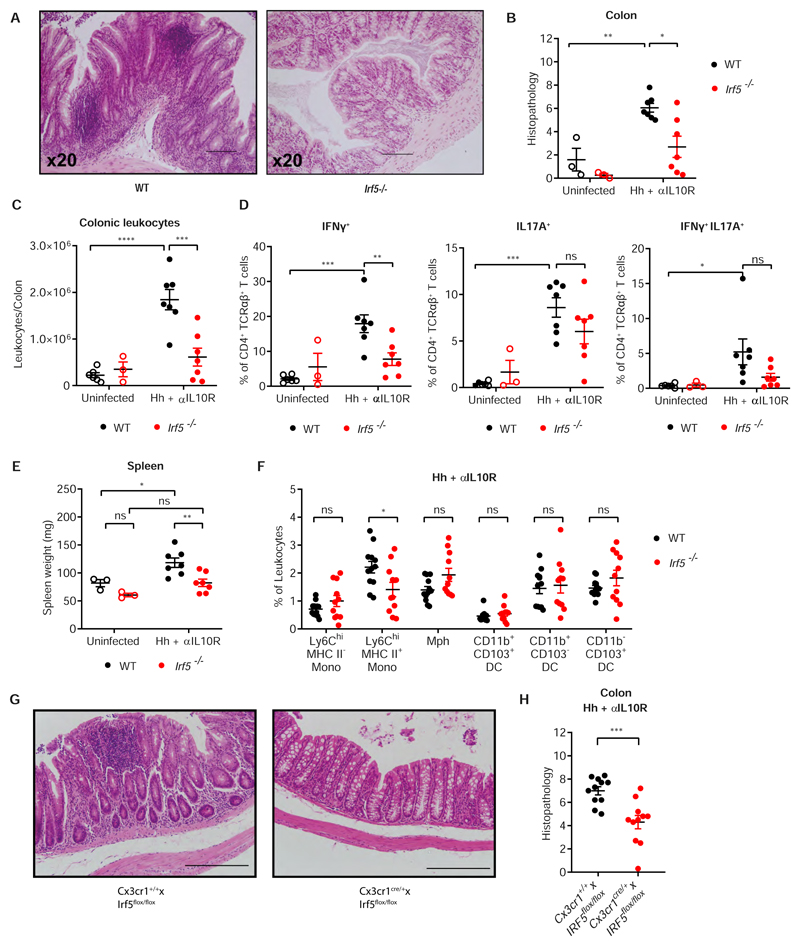
Next, we evaluated the effect of IRF5 deficiency on the pathogenesis of intestinal inflammation. WT and Irf5-/- mice were subjected to Hh + αIL10R colitis for 21 days and inflammatory indices were analysed upon sacrifice. Morphological analysis (Fig 2A, Supplementary Fig. S2A) and histological scoring indicated that both colons (Fig. 2B) and caeca (Supplementary Fig. S2B) were protected from colitis by IRF5-deficiency. The leukocyte infiltrate to the cLP was significantly reduced in Irf5-/- mice (Fig. 2C), consistent with the reduced levels of inflammation in the colon and caecum of Irf5-/- animals. Next, we profiled TH1 and TH17 lymphocyte responses that are involved in the pathogenesis of colitis (34). Irf5-/- mice displayed a significantly reduced TH1 effector response as quantified by emergence of IFNγ+ CD4+ T cells, and a non-significant reduction in the number of IL-17a+ TH17 cells and double positive IFNγ+/IL-17a+ cells (Fig. 2D).
Figure 2. IRF5 deficiency protects against intestinal inflammation.
A) Representative H&E sections of colons from WT (left) and Irf5-/- (right) mice at d21 Hh + αILI0R. B) Histopathology scoring of WT and Irf5-/- colons. C) Number of cLPLs retrieved from steady state and d21 Hh + αILI0R WT and Irf5-/- mice. D) frequencies of IFNγ-, IL17A- and IFNγ/IL17-producing CD4+ T cells in WT and Irf5-/- mice following 4 hours of culture with PMA/Ionomycin and brefeldin assessed by intracellular flow cytometry. E) Spleen weights of WT and Irf5-/- mice at steady state and d21 Hh + αILI0R. B,E: Data are representative of two independent experiments, (ss n=3, Hh + αILI0R n=7). Two-Way ANOVA with Tukey correction. C,D: Data are representative of two independent experiments, (WT ss n=6, Irf5-/- ss n = 3 Hh + αIL10R n=7). Two-Way ANOVA with Tukey correction. F) The frequency of intestinal MNPs in the cLP at d21 Hh + αIL10R WT (n=12) and Irf5-/- (n=11) mice. G) Representative H&E sections of colons from CX3CR1IRF5+ (left) and CX3CR1IRF5- (right) mice at d21 Hh + αIL10R. H) Histopathology scoring of colons from CX3CR1IRF5+ (n=11) and CX3CR1IRF5- (n=11) mice at d21 Hh + αIL10R. F,H: Data are pooled from two independent experiments. F) Two-Way ANOVA with Sidak correction, H) Unpaired t-test. Data presented are mean ± SEM, ns = not significant, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p < 0.0001
Hh + αIL10R colitis in WT mice led to splenomegaly, but not in Irf5-/- mice (Fig. 2E), indicating that they were also protected from systemic aspects of disease. Despite the altered immune response, Hh presence in the caecal faeces were unaffected in Irf5-/- compared to WT, quantified by detection of the Hh Cytolethal Distending Toxin B (cdtB) gene (Supplementary Fig. S2C), ruling out differential bacterial colonisation in WT and IRF5-deficient animals.
Myeloid cells make up a significant part of the leukocyte pool at the peak of inflammation in the colon(33). Ly6Chi monocytes are rapidly recruited to the gut in response to inflammatory signals, with Ly6ChiMHCII+ inflammatory monocytes becoming the predominant cells that carry out inflammatory effector functions (9, 15, 16, 20, 24, 35, 36). Indeed, we observed an increase in the frequency of Ly6ChiMHCII+ inflammatory monocytes (0.9% to 2.2%) at the peak of Hh + αIL10R induced inflammation, while the frequency of F4/80+ macrophages diminished (Figs 2F and 1E; Supplementary Fig. S2D). The frequencies of the DC populations remained unaffected by ongoing inflammation (Figs 2F and 1E). IRF5 deficiency significantly attenuated the predominance of Ly6ChiMHCII+ inflammatory monocytes (Fig. 2F), approximating the monocyte-macrophage waterfall observed at the steady state (Fig. 1F). At the peak of inflammation all MNP populations in Irf5-/- animals were smaller in absolute numbers than those of their wildtype counterparts (Supplementary Fig S2E). Finally, to confirm that IRF5 activity in MNPs is a major contributor into the immunopathology of intestinal inflammation, we subjected Cx3cr1crexIRF5flox/flox animals, which are deficient in IRF5 specifically in their MNP compartment (Supplementary Fig S2E), to the Hh + αIL10R colitis model. Histological scoring indicated that both the colons (Fig. 2G, H) and caeca (Supplementary Fig S2F, E) of these animals were protected from colitis by IRF5-deficiency in MNPs.
These data demonstrate that IRF5 plays a critical role in the pathogenesis of intestinal inflammation via the MNP system.
IRF5 has limited effect on monocyte development in the bone marrow and blood
At homeostasis, a higher frequency of fully differentiated F4/80+ macrophages was observed in WT compared to Irf5-/- mice (Fig 1), suggesting that IRF5 may play role in monocyte differentiation. However, in Hh + aILI0R induced colitis, the different inflammatory environments between WT and Irf5-/- animals obscured this effect (Fig 2). To compare the differentiation competence of WT and Irf5-/- monocytes in a shared environment, we performed mixed bone marrow chimera experiments. The lethally irradiated mice were reconstituted with 50:50 WT:Irf5-/- bone marrow mix and the efficiency of reconstitution in the bone marrow, of blood monocytes, and of the cLP MNP compartment were investigated. We observed no difference in reconstitution of long term (LT)- or short term (ST)-haematopoietic stem cells (HSCs), myeloid progenitors (common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMP) and megakaryocyte-erythrocyte progenitors (MEP) or Ly6Chi mature monocyte population in the bone marrow (Supplementary Fig S3A, B). IRF5 expression assessed by intracellular staining using flow cytometry was negligible in LT-HSCs, ST-HSCs, and CMPs but detectable in GMPs and MEPs. Ly6Chi monocytes express the highest levels of IRF5 among the tested progenitor and mature cell populations (Supplementary Fig. S3C). The reconstitution of Ly6Chi monocytes in the blood was not affected by IRF5 deficiency, but more Ly6Clo monocytes appeared to be derived from WT progenitors (Supplementary Fig. S3D).
To further investigate an impact of IRF5 deficiency on the composition and phenotype of monocytes in the blood in non-inflammatory conditions, we conducted single cell RNA-sequencing (scRNA-seq) analysis of CX3CR1+ WT and Irf5-/- cells from five mixed bone marrow chimeras. We identified five subpopulations of cells (Supplementary Fig. 4A) that included discrete sets of Ly6c2 hi, Cd36 hi, Cd74 hi and Cd74 hi/Cd209 hi cells similar to those reported previously(37), (Supplementary Fig. 4C). As would be expected for immature monocytes, the set of Ly6c2 hi cells (Cluster I) also showed high expression of Sell and Ccr2 (Supplementary Fig 4D). Two clusters of Ly6clo Cd36+ve cells (Clusters II & III) were also positive for the transcription factors Cebpb and Nr4a1 which are known to regulate the transition from Ly6chi to Ly6clo monocytes(37) (Supplementary Fig 4D). Of these, the largest (Cluster II) was distinguished by high expression of Itgal, while the smaller (Cluster III) showed high expression of Apoe (Supplementary Fig 4C). The remaining two clusters (IV and V) both expressed Cd74 and Ccr2, with the smallest cluster (V) also showing expression of Cd209a, Ciita, Batf3 and H2-Dmb1 suggestive of a monocyte-derived DC (moDC) precursor phenotype(38) (Supplementary Fig 4D). Overall, we found broadly similar proportions of WT and Irf5-/- in the different clusters, although, consistent with the FACS data (Supplementary Fig S3D), the knockout did show a small decrease in Cd36hi (47.2% Irf5-/- vs 55.7% WT) together with a concomitant increase in Ly6c2hi (32.6% Irf5-/- vs 26.1% WT) cell frequency relative to WT (Supplementary Fig 4B). The transcriptional phenotypes of the Irf5-/- and WT cells were highly similar within each of the clusters. In total only 8 genes (including Irf5) were found to be significantly differentially expressed (|fc| > 1.5, BH adjusted p value < 0.05, Wilcoxon tests), with nearly all of the differences being identified in the putative moDC precursor population (Supplementary Fig 6A). Together, these data indicate that although IRF5 is unlikely to have a global impact on monocyte development and phenotypes in the bone marrow and blood, it may help to promote the transition of blood monocytes from Ly6C hi to Ly6C lo/Cd36 hi and play a role in shaping the development of Cd74 hi/Cd209 hi moDCs.
IRF5 has subtle effect on CD11c+ intestinal macrophages at steady state
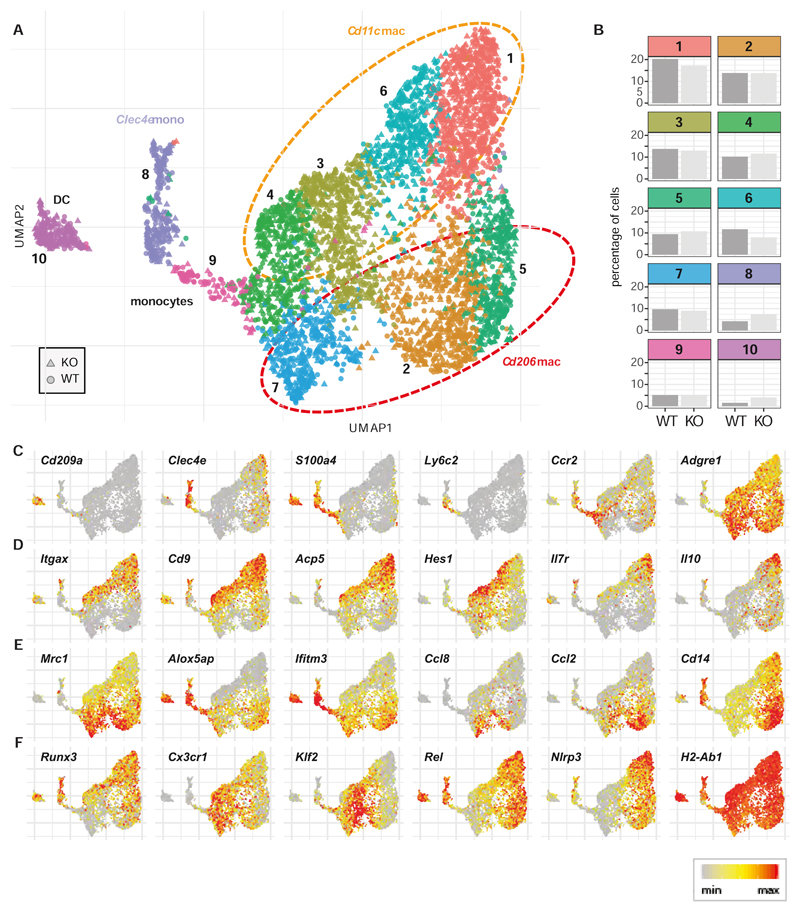
Under homeostatic conditions Ly6Chi monocytes continuously extravasate into the colon where they give rise to heterogenous populations of macrophages (13). Because a change in this process may affect susceptibility to colitis we investigated whether IRF5 can act in a cell-intrinsic fashion to regulate the composition of MNP pools in the steadystate cLP. While the reconstitution of Ly6Chi monocytes in the blood was not affected by IRF5 deficiency, more Ly6Chi monocytes in the cLP were derived from WT progenitors (Supplementary Fig. S3E). This finding is consistent with the previously reported more efficient recruitment of donor WT than Irf5-/- monocytes into the tissue in mixed bone marrow chimera (39). Next, we performed scRNA-seq analysis of Cx3cr1+ve MNP extracted from the cLP of the steady state WT/Irf5 -/- mixed bone marrow chimera. We identified ten distinct subpopulations (Fig 3A and Supplementary Fig 5) that comprised Cd209a +ve dendritic cells (cluster 10), Clec4a+ and Ly6c2hi monocytes (clusters 8 and 9) and seven Adgre1+ve (encoding for F4/80) macrophage populations (clusters 1-7) (Fig 3A). As expected, the Mϕ clusters represented the majority of MNPs in uninfected mixed bone marrow chimera, consistent with the FACS-based analysis (Fig 1E and Fig S6C). The Mϕ broadly split into two compartments that were distinguished by expression of Itgax (Cd11c) and Mrc1 (Cd206) (orange and red ellipses Fig 3A). The Cd11c Mϕ also showed expression Cd9 and Acp5 and comprised separate populations of Hes1 +ve and Il7r +ve/Il10 +ve cells that may represent epithelial associated and resident tolerogenic Mϕ populations respectively (33, 40) (Fig 3D). Macrophages with high expression of Mrc1 also showed higher expression of Alox5ap and the anti-inflammatory molecule Ifitm3 (41) (Fig 3E). Subpopulations of the Mrc1 Mϕ were characterised by high expression of key monocyte chemo-attractants Ccl8 (42) and Ccl2 (which encode the ligand for CCR2). Orthogonal to groupings by Cd11c vs Mrc1 status the Mϕ showed differences in the expression of Runx3 and Cx3cr1 which are associated with mature macrophages (Fig 3F). While surface expression of CX3CR1 protein is known to be associated with maturity, Cx3cr1 gene expression was higher in Runx3-ve cells suggesting that transcription of this gene is down-regulated as Mϕ mature. Additionally, both the Cd11c and Mrc1 Mϕ populations exhibited apparent differences in activation state being split between expression of Klf2, which is known to inhibit the pro-inflammatory activation of immune cells(43) and expression of genes associated with Mϕ activation such as the key NF-κB target gene Rel and the Nlrp3 inflammasome (Fig 3F). No Timd4 (Tim-4) expression was detected in any of the macrophage clusters (data not shown), indicating that, as expected, the monocyte-independent resident macrophage population was not represented amongst the donor-derived cells(18). Overall the distribution of WT and Irf5-/--cells between the clusters was similar (Fig 3B) although there were fewer Irf5 -/- (25.2%) than WT (32.1%) cells in the activated Cd11c Mϕ (clusters 1 and 6). In contrast, there was an increase in the frequency of DCs (4.0% vs 1.6%) and Clec4e +ve monocytes (cluster 8; 7.4% vs 4.3%) amongst the Irf5-/- cells. Across the clusters only 34 genes were significantly affected (|fc| > 1.5, BH adjusted p value < 0.05, Wilcoxon tests) by the lack of IRF5, with the majority of differences (n=23) being observed in the DC cluster (Supplementary Fig 6B). However, amongst the Cd11c +ve macrophage clusters 1, 3, and 6 we did note a consistent down-regulation of Ccl4 (also known as Macrophage inflammatory protein-1β, Mip-lβ) in the Irf5-/- cells (Supplementary Fig 6B).
Figure 3. IRF5 has subtle effect on CD11c+ intestinal macrophages at steady state.
WT and Irf5-/- CD45+CD11b+SiglecF-Ly6G-CX3CR1+ cells were sorted from the colons of five mixed bone marrow chimera animals and subjected to scRNA-Seq analysis. A) Graph based clustering(58) of equal numbers of WT and Irf5-/- cells (n=4780 total) identified nine clusters of MNPs and one cluster of dendritic cells. B) The bar plots show the percentages of WT and Irf5-/- cells that were found in each cluster. Panels C) – F) show the expression of cell type markers C), genes expressed in Cd11c +ve macrophages D), genes expressed in Mrc1 +ve macrophages and genes with associated with macrophage differentiation and activation E).
Altogether absence of the IRF5 had a subtle effect on the composition and phenotype of the steady state colonic lamina propria MNP compartment that was suggestive of a role for IRF5 in controlling CD11c+ MNP development.
IRF5 promotes generation of CD11c+ macrophages in inflamed colon
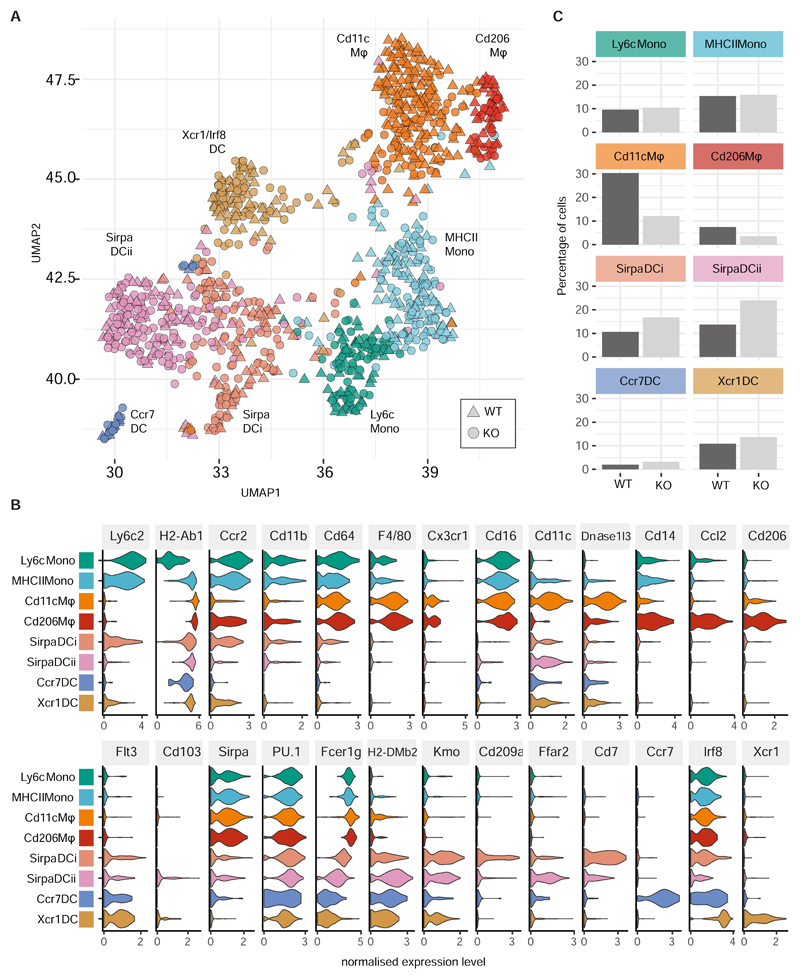
Next we subjected CX3CR1+ WT and Irf5-/- MNPs isolated from the inflamed cLP of three Hh + αIL10R MBMCs to scRNA-seq analysis (Supplementary Fig. S7A). Examination of the top cluster markers genes (Supplementary Fig. S7B) revealed the existence of two groups of monocytes, two clusters of macrophages and four clusters of dendritic cells (Fig. 4A). The monocyte clusters comprised a set of Ly6c2-high immature monocytes (“Ly6c Mono”) and a group of mature monocytes (“MHCII Mono”) that expressed MHCII genes such as H2-Ab1 and the pro-inflammatory cytokine Il1b (Fig. 4B). The two macrophage clusters were clearly demarcated by expression of Adgre1 (F4/80), Cd81 and Cx3cr1. The largest cluster of Cd11c Mϕ was characterised by the high expression level of Itgax (Cd11c) and known cLP macrophage markers, such as MHC glycoproteins (H2-M2), complement molecules (C1qa,b,c), tetraspanins (Cd63, Cd72, Cd81), oxidative stress response (Hebp1) and anti-microbial molecules (Lyz2, Acp5, Dnase1l3) (Fig. 4B and Supplementary Fig. S7B). The second cluster of Cd206 Mϕ lacked Itgax expression but was defined by high expression of Mrc1 (Cd206), chemokines (e.g. Ccl2, Ccl3, Ccl4, Ccl7, Ccl8, Ccl12, Cxcl2), scavenger, phagocytic and immunoactivating receptors (Cd36, Fcgr4, Clec4b1) and anti-viral molecules (Ch25h, Gbp2b) (Fig. 4B and Supplementary Fig. S7B). The remaining four clusters of cells lacked Cd64 expression and showed expression of established dendritic cell markers such as Flt3, Cd11c, and the Ciita- dependent DC-specific MHCII genes H2-DMb2 and H2-Oa(44). “Sirpa DC i” and “Sirpa DC ii” clusters displayed a DC2-like profile being marked by expression of Sirpa, Kmo, Cd209a and Cd7 (Fig. 4A & Supplementary Fig S7B). The cells in these clusters also strongly expressed PU.1, but were distinguished by low Flt3 expression suggesting that they may be moDC (38, 45) (Fig 5). The remaining two DC clusters comprised a set of Xcr1 high Irf8 high Sirpa low cells (“Xcr1 DC”) that are likely to correspond to conventional cDC1 cells and a small group of migratory Ccr7 positive DCs (“Ccr7 DC”). In comparison to that observed in uninfected animals, the MNP population structure at the peak of Hh + αIL10R induced inflammation (Fig 4A,C) showed a marked increase in the numbers of Ly6c2 hi MhcII + inflammatory monocytes, a larger and more heterogeneous DC population along with a diminished frequency of macrophages, consistent with above analysis (Fig 2F and Supplementary Fig. S7C).
Figure 4. IRF5 promotes generation of CD11c+ macrophages in inflamed colon.
WT and Irf5-/- CD45+CD11b+SiglecF-Ly6G-CX3CR1+ cells were sorted from the colons of three mixed bone marrow chimera animals at d21 of Hh + αIL10R colitis and subjected to droplet-based single cell transcriptomic analysis. A) Graph based clustering(58) of equal numbers of WT and Irf5-/- cells (n=1106 total) identified four clusters of MNPs and four clusters of dendritic cells. B) The violin plots show the expression levels (x axes) of selected known cLP MNP and DC sub-population markers in each of the identified clusters (y axes). C) The bar plots show the percentages of WT and Irf5-/- cells that were found in each cluster.
Figure 5. IRF5 defines an inflammatory MNP signature during colitis.
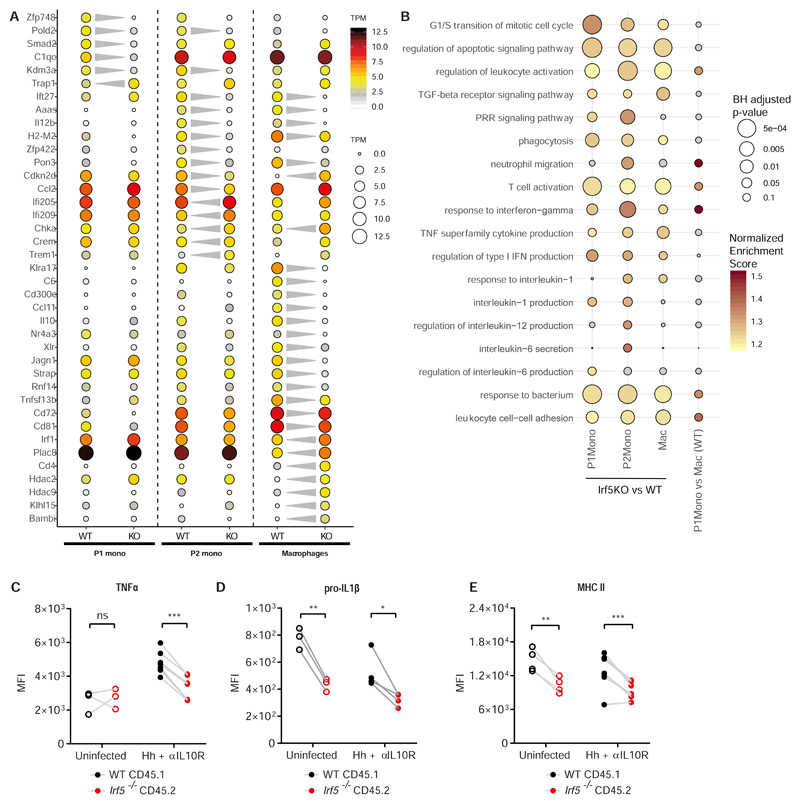
WT and Irf5-/- P1 monocytes, P2 monocytes and macrophages were sorted from three mixed bone marrow chimera animals at d21 Hh + αIL10R. A) The dot plot shows the expression (mean TPM, n=3 biological replicates) of selected genes found to be differentially expressed between WT and Irf5-/- cells at one or more stages of the monocyte waterfall. The significant changes (| fc | > 2, BH adjusted p < 0.05) are indicated by the grey triangles. B) Selected GO Biological process categories that showed a significant enrichment (coloured dots, GSEA analysis, BH adjusted p < 0.1) in at least one of the three Irf5 KO vs WT small bulk RNA-seq comparisons. C-E) Intracellular or extracellular flow cytometry labelling was used to quantify the expression of C) TNFα, D) pro-IL1β, E) MHC II on WT vs Irf5-/- macrophages in mixed bone marrow chimera uninfected (n=3), and at d21 Hh + αIL10R colitis (n=4). Two-Way ANOVA with Sidak Correction. Data from one representative experiment presented are mean ± SEM, ns = not significant, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001
When clusters were split by genotype, it was found that monocyte clusters had similar numbers of WT and Irf5-/- cells, whereas macrophage clusters, especially Cd11c Mϕ, had higher numbers of WT cells, while the Sirpa DC i and ii clusters contained higher numbers of Irf5-/- cells (Fig. 4C), findings which were confirmed by FACS-based analysis (Supplementary Fig S7C). The decrease in the frequency of Irf5-/- Cd11c Mϕ relative to WT was more striking during the peak of inflammation (Fig 4C) than at steady state (Fig 3B), suggesting that the propensity of IRF5 to promote generation of CD11c+ macrophages is accentuated by the inflammatory environment. Together with the fact that CD11c+ monocyte/macrophages can drive immunopathology(9) this observation provided a possible explanation for the pathogenic role of IRF5 in intestinal inflammation (Fig. 2).
IRF5 defines an inflammatory MNP signature during colitis
To investigate the effect of IRF5 on the transcriptional phenotype of intestinal monocytes and macrophages, we conducted small bulk RNA-seq analysis of WT and Irf5-/- Ly6ChiMHCII- (P1) monocytes, Ly6ChiMHCII+ (P2) monocytes and Ly6C-MHCII+F4/80+ macrophages (n=100 cells/sample) from each of the inflamed colons of three Hh + αIL10R mixed bone marrow chimera animals. First, we identified the genes that showed significant variation between the WT monocyte and macrophage samples. Hierarchical clustering of these genes revealed that the transcriptome of P2 monocytes overlaps with the profiles of both P1 monocytes and macrophages in line with the concept that they represent a transitional state of monocyte to macrophage differentiation (Supplementary Fig. S8A). We also detected high expression levels for genes previously shown to be associated with mature intestinal macrophages, such as MHC molecules (H2-M2), tetraspanins (Cd72, Cd81), complement molecules (C1qa, C1qb, C1qc), chemokines (Ccl5, Ccl8) and phagocytic and immunoactivating receptors (Fcgr4, Fcer1g, Cd300e) in the macrophage populations(23). Next, we identified genes that were significantly (BH adjusted p < 0.05, fold change| > 2) regulated by IRF5 in each of the P1 (n=607 genes), P2 (n=761 genes) and macrophage (n=977 genes) compartments (Supplementary Fig. S8B). Amongst the differentially expressed genes, Ly6ChiMHCII- Irf5-/- P1 monocytes showed significantly lower levels of Smad2 and Kdm3a which respectively transduce and positively regulate TGF-B and Jak2/Stat3 signalling, pathways of known importance for monocyte maturation (Fig. 5A). In line with this observation IRF5 deficient macrophages failed to down-regulate genes highly expressed in P1 and P2 monocytes including Plac8, Cdkn2d (P19ink4d) and Irf1 and also showed significantly lower expression of the MHC class II molecule H2-M2 (Fig. 5A). At the same time, in macrophages, loss of IRF5 reduced expression of the key pro-inflammatory cytokines (Il-12b, Ccl11, Tnfsf13b/BAFF), expression of the immunoactivating receptor Cd300e, tetraspanins (Cd81 and Cd72) as well as IL-10. A significant reduction in the expression of the key pro-inflammatory cytokine Il-12b was also observed in the P2 monocytes. These changes were accompanied by up and down-regulation of the epigenetic regulators Hdac2 and Hdac9 in IRF5 deficient macrophages. At the pathway level, geneset enrichment analysis of Gene Ontology (GO) Biological Process categories revealed that IRF5 broadly modulated inflammatory pathways including “leukocyte activation”, “response to interferon-gamma”, “response to bacterium” and “regulation of T-cell activation” in both monocytes (P1 & P2), and macrophages (BH adjusted p-value < 0.1, Fig. 5B). Genes regulated by IRF5 in the P2 monocyte compartment displayed a significant enrichment of genes involved in “regulation of interleukin-12 production”, “interleukin 6-secretion”, “interleukin-1 production”, while genes associated with “response to interleukin-1” and “TNF superfamily cytokine production” were also affected in macrophages (Fig. 5B). These pathways, and specifically production of IL-23, IL-1, and TNF, have been previously associated with colitis development and/or IBD (9, 29, 33). In independent experiments, using flow cytometry, we confirmed that colonic WT macrophages in the MBMCs produced higher levels of cytokines TNFα and IL-1β cytokines than Irf5-/- cells (Fig. 5C, D). The surface expression of MHCII was higher on WT macrophages relative to Irf5-/- (Fig. 5E).
When small bulk RNA-seq data were compared to scRNA-Seq gene expression data, a good correspondence between the genes expressed in the Ly6c and MHCII monocyte clusters and P1 and P2 samples respectively was observed (Supplementary Fig S8C). Both the Cd11c and Cd206 macrophage clusters showed similarities to the small-bulk macrophage sample (Supplementary Fig S8C).
IRF5 promotes monocyte to Cd11c macrophage differentiation during intestinal inflammation
Given the reduction in Irf5-/- macrophages numbers that we saw in the mixed bone marrow chimera at the peak of Hh + αlL10R induced colitis (Fig 4C and Supplementary Fig S7C) we examined an apparent role for IRF5 in controlling monocyte differentiation in more detail. First, we performed a global comparison of genes regulated by IRF5 in macrophages with those that were associated with differentiation from P1 monocytes to macrophages in the inflamed cLP. Overall, this analysis revealed a significant positive correlation between genes up-regulated during macrophage differentiation and those positively regulated by IRF5 in macrophages (Spearman’s’ rho: 0.45, p = 5.5 x 10-110) (Supplementary Fig. S9). Close examination of the scatter plot, however, also revealed a large number of genes that were up-regulated in macrophages independent of the presence of IRF5 including C1qc, Ptgs1 (Cox1) and Mmp13 (green dots, Supplementary Fig. S9). These data support a cell-intrinsic role for IRF5 in regulating myeloid cell differentiation and phenotype during intestinal inflammation.
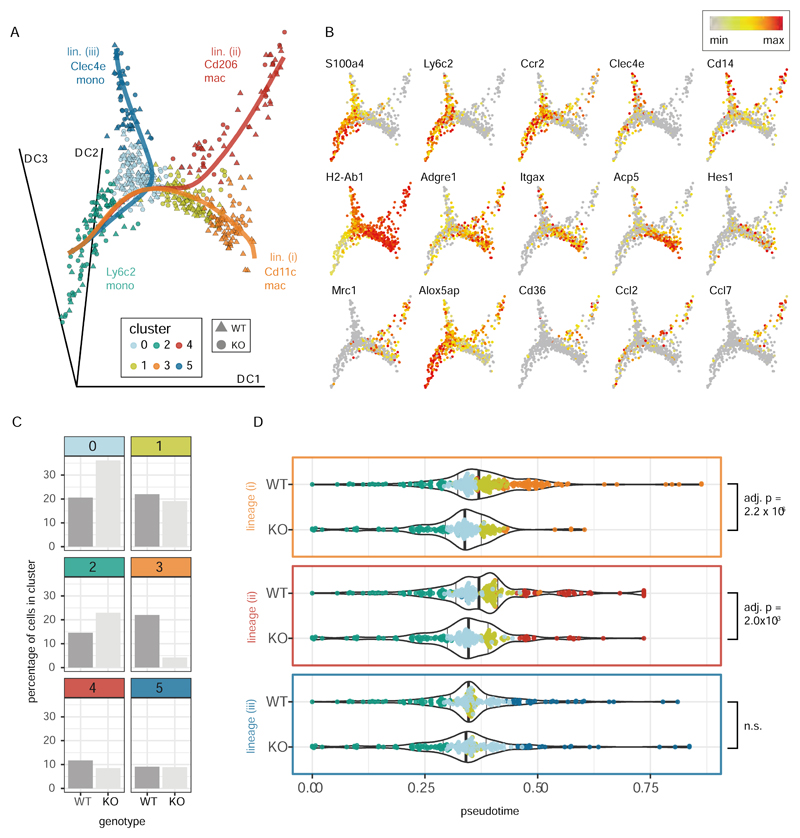
To dissect the role of IRF5 in controlling MNP differentiation in the inflamed cLP in more detail we applied the Slingshot pseudo-time algorithm(46) to our scRNA-Seq data. After exclusion of the dendritic cells, higher-resolution analysis identified 6 clusters of monocytes and macrophages (Supplementary Fig. S10) that fell into three predicted lineages (with Ly6c2 monocytes assumed to represent the ‘root’ state) (Fig. 6A and 6B). These represented the differentiation of (i) Cd11c (Itgax) macrophages that also expressed Acp5, (ii) Cd206 (Mrc1) macrophages that were also positive for Cd36, Ccl2 and Ccl7 and (iii) a small population of Clec4e expressing mature (MHCIIhi) monocytes that resembled those found in the steady state data (see Fig 3). Most notably, the Irf5 -/- cells were under-represented (4.3% vs 22% of WT cells) in the terminal cluster of the Cd11c lineage (Fig 6C, cluster 3) with progression of Irf5-/- cells in pseudotime along this lineage being significantly different to that of the wildtype cells (Fig 6D, Bonferroni adjusted p = 2.2x10-6). The pseudotime distribution of Irf5-/- cells along the Cd206 lineage was also significantly altered (Fig 6D, Bonferroni adjusted p=0.003) but this was associated with only a slight reduction in the number of Irf5-/- Cd206 macrophages (Fig 6C, cluster 4, 8.5% vs 11.7% of WT cells). In contrast, a similar percentage of Irf5-/- and WT cells were found in the Clec4e monocyte cluster (Fig 6C, cluster 5) and there was no difference between the progression of Irf5-/- and WT cells along this lineage (Fig 6D).
Figure 6. IRF5 promotes monocyte to Cd11c macrophage differentiation during intestinal inflammation.
WT and Irf5-/- monocytes and macrophages from the inflamed intestine (Fig 4) were re-clustered at higher resolution (Supplementary Fig S10) and subject to pseudotime analysis. A) Embedding of the cells in the first three dimensions of a diffusion map shows the three differentiation trajectories (solid lines) identified by the Slingshot pseudotime algorithm (with the Ly6c2 monocytes assumed to represent the root state) B) Expression of selected cell type marker genes and genes associated with Cd11c (Itgax) an Mrc1 (Cd206) macrophages. C) The bar plots show the percentages of WT and Irf5-/- cells that were found in each cluster. D) The violin plots show the progression of the WT and Irf5-/- (KO) cells through pseudotime along the three identified trajectories (as shown in A)). Differences in the distribution of cells in pseudotime between the genotypes were assessed with a KS tests (p-values adjusted using the Bonferroni correction). The position of the cells in pseudotime is shown on top of the violin plots (cells colored by cluster as in A)). The position of the 50th quantiles is indicated by the bold vertical lines.
The reduction of Irf5-/- cells in the macrophage clusters was paralleled by an increase in the number of Irf5-/- cells in the immature Ly6c2 monocyte cluster (cluster 2, 23.0% vs 14.6% WT cells) and differentiating monocyte cluster (cluster 0, 36% vs 20.6% of WT cells) (Fig 6C). These data suggest that IRF5 may promote the acquisition of CD11c expression by Ly6ChiMHCII+ monocytes and their differentiation to CD11c+ macrophages.
CD11c+ macrophages occupy a distinct colonic niche
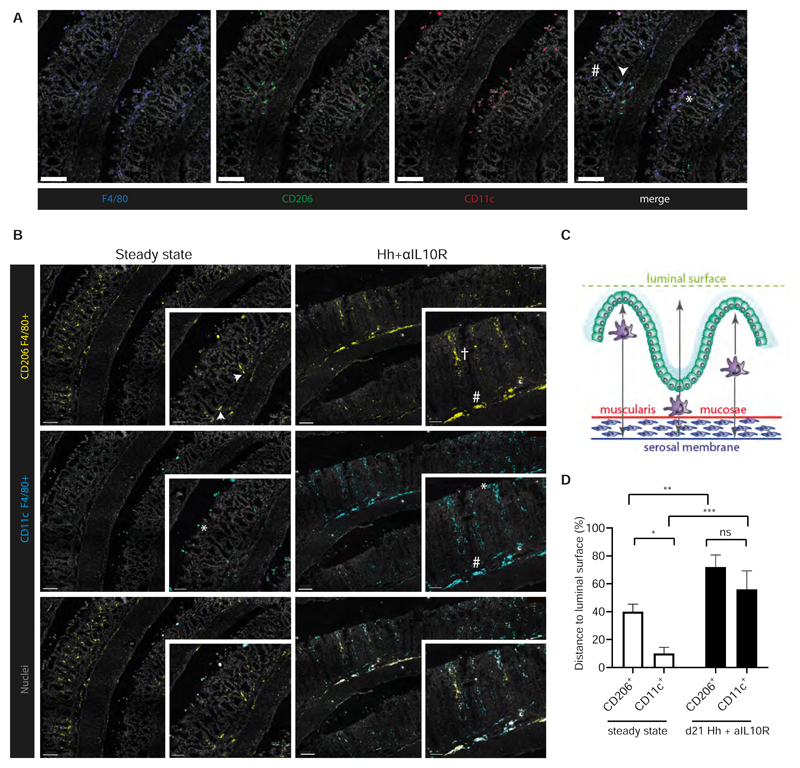
We assessed the localisation of CD206+ and CD11c+ macrophage subsets in the colon, by performing labelling of colonic tissue sections with antibodies against CD11c, CD206 and F4/80 and subsequent analysis by confocal microscopy (Fig 7A). This analysis revealed that the two macrophage subsets localized at distinct sites at steady state. CD206+/F4/80+ cells (CD206+ macrophages) dominated the colonic macrophage pool and were located within the lamina propria, with some found to be residing at the base of intestinal crypts (Fig 7B). Macrophages at the base of crypts are believed to be involved in response to the mucosal barrier damage via secretion of CCL8 (42) and other chemokines and may transmit regenerative signals to neighbouring colonic epithelial progenitors (47). Indeed, chemokines (Ccl2, Ccl7, Cxcl2) were distinctively expressed in Cd206 Mϕs during Hh + αIL10R-induced colitis (Supplementary Fig S11A).
Figure 7. CD11c+ macrophages occupy a distinct colonic niche.
A) Representative images of immunofluorescent labelling of colonic sections at steady state. Individual channels visualise the distribution of F4/80+ (blue), CD206+ (green) and CD11c+ (red) cells within the structure of the colon. CD206+ macrophages (white arrow) and CD11c+ macrophages (*) as well as single-positive F4/80+ cells (#) can be detected in the merged image. Cell nuclei are labelled with Sytoxblue (grey). Scalebars represent 50 μM. B) Localisation of double-positive CD11c+F4/80+ (cyan) and CD206+F4/80 (yellow) cells, in steady state (n=6) and colitic mouse colons (n=6) by immunehistofluorescence. Separate channels based on overlap of staining were created. Cell nuclei were labelled with Sytoxblue (grey). A minimum of 5 sections per mouse were evaluated. Macrophages at the base of the crypts (white arrow), at the tips of the villi (*), interspersed within the villi (†); at the Muscularis mucosae membrane (#). Scalebars represent 100 μM in the overview images and 50μM in the enlargement. C) Schematic depiction of image quantification analysis. Localisation of macrophages is assessed by their minimal distance (black arrow) to the tip of the villi (artificial luminal surface depicted in green), muscularis mycosae (red) and serosal membranes (blue). D) Quantification of minimal distance of CD206+ F4/80+ and CD11c+ F4/80+ cells in steady state and d21 Hh + αIL1 OR to the luminal surface. Minimal distance is presented as a percentage of the distance to the tip of the villi to the total distance between the luminal surface and serosal membrane. Two-way Anova with Tukey’s multiple comparisons test. Data presented as mean ± SEM, ns p > 0.05, * p < 0.05, ** p < 0.01, *** p <0.001.
CD11c+ F4/80+ cells (CD11c+ macrophages) were more sparsely located and mainly found at the luminal surface (Fig 7B). Thus, CD11c+ macrophages may represent a primed macrophage phenotype, ready to respond to microbial encroachment (21). In fact, many genes specifically expressed in Cd11c Mϕs during Hh + αIL10R-induced colitis belonged to an anti-microbial defence programme (Dnase1l3, Acp5, Mmp14 etc) and protein recycling (Ctsa, Ctsh, Ctsz) (Supplementary Fig S11A). We quantified the distance of the macrophage subsets in relation to the luminal surface and the serosal membrane (Fig 7C). At steady state, CD11c+ macrophages were found to be significantly closer to the luminal surface than CD206+ macrophages, but this proximity was lost during inflammation (Fig 7D), when CD206+ and CD11c+ macrophages started to intersperse throughout the cLP and at the muscularis mucosae membrane (Fig 7B).
IRF5 controls the phenotype of CD11c+ macrophages
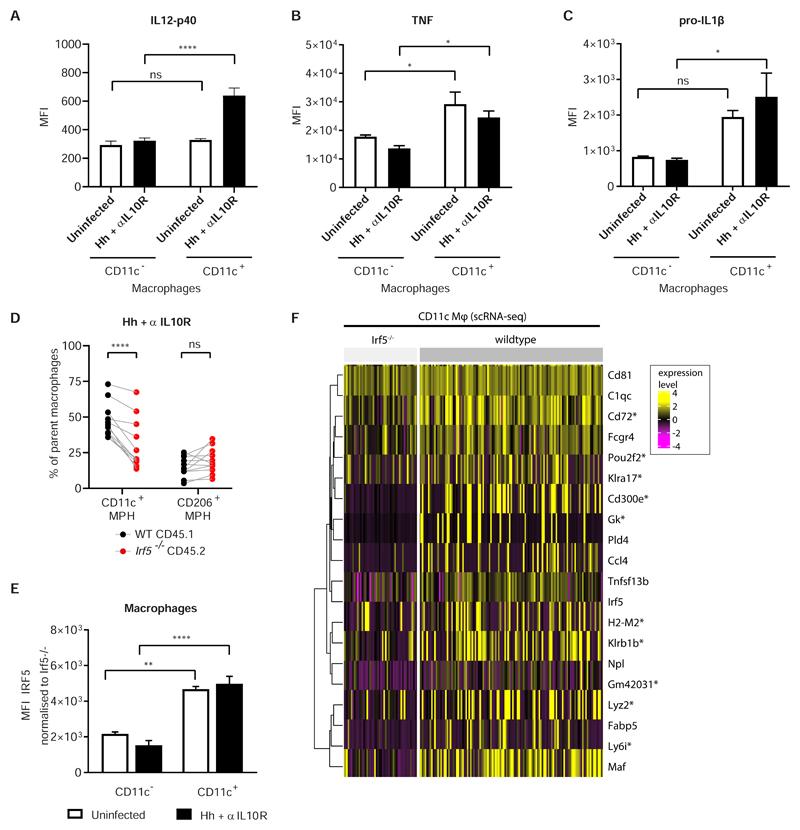
Previously, CD11c+F4/80+ monocytes and macrophages were shown to be critical effector cells in the development of Hh + αIL10R-induced experimental colitis via the production of IL-23 (9). Here we confirmed that CD11c+F4/80+ macrophages (Supplementary Fig. S11B) produced higher levels of IL-12p40, a subunit of IL-23 (Fig. 8A), as well as other inflammatory cytokines, such as TNF and IL-1ß, than CD11c- macrophages (Fig. 8B,C). In the setting of the mixed bone marrow chimera, we confirmed that IRF5 promoted the development of CD11c+ macrophages both at the peak of Hh + αlL1OR induced colitis (Fig. 8D) and in steady state (Supplementary Fig. S11C). In keeping with this observation, the expression of IRF5 protein was higher in CD11c+ macrophages and Ly6ChiMHCII+ monocytes, compared to their CD11c- counterparts (Fig 8E and Supplementary Fig 11D).
Figure 8. IRF5 controls phenotype of CD11c+ macrophages.
A, B, C) Comparison of IL12p40, TNF and IL-1β inflammatory cytokine expression in CD11c+ vs CD11c- cLP macrophages assessed by intracellular flow cytometry, in uninfected MBMC (n=3) and d21 Hh + αIL10R (n=4). One experiment. Two-Way ANOVA with Tukey Correction. Data presented are mean ± SEM, ns = not significant, * p ≤ 0.05, **** p < 0.0001. D) The frequency of parent WT and Irf5-/- macrophages expressing CD11c or CD206 at d21 Hh + αIL10R colitis. Two-Way ANOVA with Sidak Correction. Data presented are mean ± SEM from two independent experiments, ns = not significant, **** p < 0.0001. E) IRF5 expression in CD11c+ vs CD11c- macrophages in mixed bone marrow chimera assessed by intracellular flow cytometry. One representative experiment, uninfected n=3, Hh + αIL10R n=4. F) Heatmap of expression of selected genes in WT and Irf5-/- Cd11c macrophages from the inflamed cLP of the mixed bone marrow chimeras (see Fig 5). All of the genes shown were found to be significantly differentially expressed between the WT and Irf5-/- cells of this cluster (Wilcoxon tests, BH adjusted p < 0.05). *’s denote significant differential expression between the genotypes in the macrophage small-bulk RNA-seq data.
At the peak of Hh + αlL10R induced colitis IRF5 positively regulated a cassette of genes that defined cLP macrophage phenotypes, such as MHC molecules (H2-M2), tetraspanins (Cd72, Cd81), complement molecules (C1q), chemokines (Ccl4), acid phosphatase 5 (Acp5) and phagocytic and immunoactivating receptors (Fcgr4, Fcer1g, Cd300e) in the Cd11c Mϕ population. A number of killer cell lectin-like receptor family members (Klrb1b, Klra2, Klra17), not previously associated with macrophage function were also affected by the lack of IRF5 in this compartment (Fig 8F). Together our data show that IRF5 promotes the differentiation and inflammatory phenotype of Cd11c+ macrophages during Hh + αIL10R induced colitis.
Discussion
Using a model of mononuclear phagocyte development in the gut and a combination of mixed bone marrow chimera approaches and single cell analysis of gene expression, we have demonstrated the importance of IRF5 in promoting the generation of macrophages in the cLP. We found that it dictates an inflammatory CD11c+F4/80+ macrophage phenotype in inflammation, and controls the immunopathology of Hh + αIL10R-induced colitis.
Our results revealed a cell-intrinsic role for IRF5 in the control of a wide-range of genes and biological pathways related to monocyte differentiation, leukocyte activation, response to bacterium, pattern recognition receptor signalling pathway and regulation of T-cell activation in inflamed intestine (Fig 5). Indeed, mice with a global or MNP-specific loss of IRF5 were protected from Hh + αIL10R colitis (Fig. 2). In comparison, IRF5 had a much more limited impact on gene expression at steady state intestine (Figs 1,3) and we observed no morphological differences in cLP between WT and Irf5-/- at steady state (Fig 1), consistent with the recently published report (48).
The most consistent function of IRF5 identified in this study is its ability to promote a pro-inflammatory monocyte and macrophage state, which is positive for CD11c. Cd11c+ macrophages were found at the luminal surface at homeostasis and throughout the cLP in inflammation (Fig 7). They transcribe anti-microbial molecules, such as cathepsins, and are efficient producers of inflammatory cytokines, such as TNF and IL-1β, that support pathogenic T cell responses in the intestine(34, 49, 50) (Fig 8 and Supplementary Fig S8). CD11c+ macrophages produce high quantities of IL-23 in the early stages of Helicobacter hepaticus induced colitis and are essential for triggering intestinal immunopathology (9, 15). They are also essential producers of IL-1b and IL-23 in Citrobacter rodentium induced colitis (21). CD11c+ intestinal macrophages were marked by high level of IRF5 (Fig. 8). IRF5 deficiency ameliorated the accumulation of CD11c+ macrophages in cLP (Fig. 8D, Supplementary Fig S11B). With the recently established link between CD11c+ macrophages and IRF5 in the development of atherosclerotic lesions (51), our data here support the notion that IRF5 may guide monocyte differentiation towards inflammatory CD11c+ macrophages in a variety of tissues and pathologies. While we only found a subtle effect for IRF5 on monocyte development in the bone marrow and subset conversion in the blood at steady state (Supplementary Fig S4), it is possible that it has a larger effect in these compartments during inflammation.
The second major population of macrophages detected in our analyses was marked by the expression of CD206 and predominantly located at the base of crypts at steady state (Fig. 7). These macrophages expressed phagocytic receptors, the scavenger receptor Cd36, which is critical for lysosomal lipolysis (52), the anti-inflammatory gene Ifitm3 (41), Alox5ap involved in leukotriene biosynthetic pathway, and a milieu of chemokines (Ccl2, Ccl7, Cxcl2 etc) (Figs 3, 4 and Supplementary Fig. S11). These may represent resident macrophages involved in the clearance of senescent epithelial and apoptotic cells, sensing and regulating response to mucosal damage and possibly contributing to epithelial renewal. The Cd206 macrophages appeared largely unaffected by IRF5 deficiency (Fig. 3) and unlikely to be major contributors to the Hh-induced pathology(33). Pseudo-time analysis of our scRNA-seq data indicate that the Cd206 and Cd11c macrophages broadly represent alternative macrophage differentiation trajectories during intestinal inflammation (Fig. 6). Together with the distinct distribution of these two macrophage populations in the cLP (Fig. 7) and their unequal dependence on IRF5 (Fig. 8) these data suggest that they may emerge independently in specific environmental niches. In fact, our steady state single-cell data (Fig 3) suggest that there is substantial heterogeneity within both the Cd206 and Cd11c macrophage populations (the Cd206 populations can be readily split, for example, by Cd14 status) and further imaging and lineage tracing studies are needed to resolve the niches, origins and functions of these subsets. In both our steady state and inflamed single cell datasets we noted the presence of a small mature (MHCII hi), activated (Rel +ve, Nlrp3 +ve) Clec4e hi monocyte (F4/80 -ve) population that appeared unaffected by absence of IRF5 (Fig. 3 and Fig. 6). Given the known roles of Clec4e (Mincle), a C-type lectin receptor (CLR), these cells may play important roles in host defence and tissue repair in the intestine (53).
The Cd206 macrophages in the cLP transcribed high levels of CCL2 (Fig. 4), a critical chemokine for accumulation of monocytes in the cLP (20). Consistent with previously published analysis (39), we observed more efficient recruitment of donor WT than donor Irf5-/- monocytes to cLP in the mixed bone marrow chimera animals, highlighting another mechanism by which IRF5 could modulate inflammation i.e. via controlling a pathogenic positive-feedback loop of inflammatory monocyte recruitment.
Finally, our data suggest that in an inflammatory environment, IRF5 specifically promotes key aspects of macrophage differentiation whilst repressing DC transition (Fig. 4). The observed changes in expression of the histone deacetylases Hdac2 and Hdac9 (Fig 5) may be consistent with a role for IRF5 in controlling of the epigenetic state of these cells. This process may be due to loss of competition for IRF binding sites, and engagement of an IRF4-dependent differentiation program(54, 55). IRF4 and IRF5 were shown to compete for binding to Myeloid Differentiation primary response 88 (MyD88) and activation following TLR4 ligation(56). IRF4 is a key regulator of intestinal CD11b+ DC subsets and a critical transcription factor in the DC fate of monocytes in in vitro bone marrow cultures(55, 57). Thus, in the absence of IRF5, IRF4 may be able to dominate the fate choice of monocytes, explaining the increased predisposition to DC fate in Irf5-/-.
Although intestinal DCs are believed to be largely derived of FLT3L-dependent progenitors (13), several studies have provided evidence that Sirpa CD11b+ DCs are replenished by monocytes in the inflamed cLP (16, 17). It is intriguing that MHCII+Cd209+ blood monocytes, previously identified as precursors of moDCs (38), showed the highest number of genes affected by the lack of IRF5, while all other monocyte populations remained largely unaffected (Supplementary Fig S4, S6A). This may reflect the more advanced differentiated state of MHCII+Cd209+ monocytes, but more functional characterisation of the populations are needed.
In summary, the data presented here reveals that IRF5 controls the MNP system in the colon, is a critical driver of intestinal inflammation and promotes monocyte differentiation towards bactericidal and inflammatory CD11c+ macrophages.
Materials and Methods
Study Design
The purpose of this study was to understand the intrinsic role of IRF5 in directing macrophage polarisation and intestinal inflammation. Flow cytometry, bulk- and single cell-RNA-sequencing, and immunofluorescence labelling of intestinal tissue sections were used to analyse the leukocyte milieu in the colons of wild type or Irf5-/- or mixed bone marrow chimeric mice. Mice were aged between 8-16 weeks at the commencement of experiments. Experimental sample sizes were not predetermined. Helicobacter hepaticus infections were ended upon mouse sacrifice at d21 post-infection. In general, experiments were performed at least twice unless indicated otherwise. Data were not excluded from analysis except for QC failures in RNAseq analysis detailed in Material and Methods (Supplementary materials). Histopathology assessment was conducted in a blinded manner independently by two researchers. Experimenters were not blinded to intervention groups for flow cytometry analysis.
Supplementary Material
One sentence summary.
The transcription factor IRF5 promotes macrophage differentiation in Helicobacter-driven intestinal inflammation in mice
Acknowledgements
We are grateful to S. Teichmann (Wellcome Sanger Institute) for help in establishing Smart-Seq2 protocol and generating preliminary data that inspired our subsequent single cell analysis. We thank the High-Throughput Genomics Group (Wellcome Trust Centre for Human Genetics) for the generation of the sequencing data; C. Pearson for assistance with the generation of mixed bone marrow chimaeras; J. Webber for assistance with cell sorting, and the Kennedy Institute Histopathology Team for sectioning and staining of mouse colons.
Funding
This work was supported by the Kennedy Trust for Rheumatology Research (ALC, MGV, DB, SNS), the MRC CGAT programme (SNS), the Novo Nordisk Foundation (Tripartite Immunometabolism Consortium - grant NNF15CC0018486 to IAU), and the Wellcome Trust (Investigator Award 095688/Z/11/Z to FP and 209422/Z/17/Z to IAU).
Footnotes
Author contributions:
ALC performed all experiments, except as noted below; MGV and SNS performed all computational analyses; DLB conducted immunofluorescence microscopy; MA generated scRNA-seq libraries; ICA provided advice and assisted with H.h infections and phenotype analysis; IAU, SNS and FP devised and directed the study. ALC, IAU, SNS wrote the manuscript.
Competing financial interests:
FP received research funding or consultancy fees from GSK, Genentech, Roche and UCB. Other authors declare that they have no competing interests.
Data availability
Next generation sequencing datasets are available via the Gene Expression Omnibus (GEO) via accession codes GSE129354 (GM-DMDM data) and GSE129258 (MBMC small bulk and single-cell data).
References
- 1.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 2.Bryant RV, Brain O, Travis SPL. Conventional drug therapy for inflammatory bowel disease. Scandinavian Journal of Gastroenterology. 2015;50:90–112. doi: 10.3109/00365521.2014.968864. [DOI] [PubMed] [Google Scholar]
- 3.Bravatà I, Fiorino G, Allocca M, Repici A, Danese S. New targeted therapies such as anti-adhesion molecules, anti-IL-12/23 and anti-Janus kinases are looking toward a more effective treatment of inflammatory bowel disease. Scandinavian Journal of Gastroenterology. 2015;50:113–120. doi: 10.3109/00365521.2014.993700. [DOI] [PubMed] [Google Scholar]
- 4.Meuret G, Bitzi A, Hammer B. Macrophage turnover in Crohn’s disease and ulcerative colitis. Gastroenterology. 1978;74:501–503. [PubMed] [Google Scholar]
- 5.Bune AJ, Hayman AR, Evans MJ, Cox TM. Mice lacking tartrate-resistant acid phosphatase (Acp 5) have disordered macrophage inflammatory responses and reduced clearance of the pathogen, Staphylococcus aureus. Immunology. 2001;102:103–113. doi: 10.1046/j.1365-2567.2001.01145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, et al. CX3CR1-Mediated Dendritic Cell Access to the Intestinal Lumen and Bacterial Clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 7.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103(+), but not CX3CR1(+), antigen sampling cells migrate in lymph and serve classical dendritic cell functions. The Journal of Experimental Medicine. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosin-Roger J, Ortiz-Masia D, Calatayud S, Hernandez C, Esplugues JV, Barrachina MD. The activation of Wnt signaling by a STAT6-dependent macrophage phenotype promotes mucosal repair in murine IBD. Mucosal Immunol. 2016;9:986–998. doi: 10.1038/mi.2015.123. [DOI] [PubMed] [Google Scholar]
- 9.Arnold IC, Mathisen S, Schulthess J, Danne C, Hegazy AN, Powrie F. CD11 c+ monocyte/macrophages promote chronic Helicobacter hepaticus-induced intestinal inflammation through the production of IL-23. Mucosal Immunol. 2016;9:352–363. doi: 10.1038/mi.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang MH, Sougawa N, Tanaka T, Hirata T, Hiroi T, Tohya K, Guo Z, Umemoto E, Ebisuno Y, Yang B-G, Seoh J-Y, et al. CCR7 Is Critically Important for Migration of Dendritic Cells in Intestinal Lamina Propria to Mesenteric Lymph Nodes. The Journal of Immunology. 2006;176:803. doi: 10.4049/jimmunol.176.2.803. [DOI] [PubMed] [Google Scholar]
- 11.Worthington JJ, Czajkowska BI, Melton AC, Travis MA. Intestinal Dendritic Cells Specialize to Activate Transforming Growth Factor-β and Induce Foxp3(+) Regulatory T Cells via Integrin αvβ8. Gastroenterology. 2011;141:1802–1812. doi: 10.1053/j.gastro.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bain CC, Montgomery J, Scott CL, Kel JM, Girard-Madoux MJH, Martens L, Zangerle-Murray TFP, Ober-Blöbaum J, Lindenbergh-Kortleve D, Samsom JN, Henri S, et al. TGFβR signalling controls CD103+CD11b+ dendritic cell development in the intestine. Nature Communications. 2017;8:620. doi: 10.1038/s41467-017-00658-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joeris T, Muller-Luda K, Agace WW, Mowat AM. Diversity and functions of intestinal mononuclear phagocytes. Mucosal Immunol. 2017;10:845–864. doi: 10.1038/mi.2017.22. [DOI] [PubMed] [Google Scholar]
- 14.Sheng J, Chen Q, Soncin I, Ng SL, Karjalainen K, Ruedl C. A Discrete Subset of Monocyte-Derived Cells among Typical Conventional Type 2 Dendritic Cells Can Efficiently Cross-Present. Cell Reports. 2017;21:1203–1214. doi: 10.1016/j.celrep.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 15.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy Ansuman T, Friedlander G, Mack M, Shpigel N, Boneca Ivo G, Murphy Kenneth M, Shakhar G, et al. Ly6Chi Monocytes in the Inflamed Colon Give Rise to Proinflammatory Effector Cells and Migratory Antigen-Presenting Cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 16.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. The Journal of Experimental Medicine. 2012;209:139. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber HA, Loschko J, Karssemeijer RA, Escolano A, Meredith MM, Mucida D, Guermonprez P, Nussenzweig MC. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. The Journal of Experimental Medicine. 2013;210:2025–2039. doi: 10.1084/jem.20130903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw TN, Houston SA, Wemyss K, Bridgeman HM, Barbera TA, Zangerle-Murray T, Strangward P, Ridley AJL, Wang P, Tamoutounour S, Allen JE, et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. The Journal of Experimental Medicine. 2018 doi: 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AM. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014;15:929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, Guilliams M, Malissen B, Agace WW, Mowat AM. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo S-U, Kuffa P, Kitamoto S, Nagao-Kitamoto H, Rousseau J, Kim Y-G, Núñez G, Kamada N. Intestinal macrophages arising from CCR2+ monocytes control pathogen infection by activating innate lymphoid cells. Nature Communications. 2015;6:8010. doi: 10.1038/ncomms9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bujko A, Atlasy N, Landsverk OJB, Richter L, Yaqub S, Horneland R, Øyen O, Aandahl EM, Aabakken L, Stunnenberg HG, Bækkevold ES, et al. Transcriptional and functional profiling defines human small intestinal macrophage subsets. The Journal of Experimental Medicine. 2018;215:441. doi: 10.1084/jem.20170057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schridde A, Bain CC, Mayer JU, Montgomery J, Pollet E, Denecke B, Milling SWF, Jenkins SJ, Dalod M, Henri S, Malissen B, et al. Tissue-specific differentiation of colonic macrophages requires TGF[beta] receptor-mediated signaling. Mucosal Immunol. 2017 doi: 10.1038/mi.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zigmond E, Bernshtein B, Friedlander Catherine G, Walker R, Yona S, Kim K-W, Brenner O, Krauthgamer R, Varol C, Müller W, Jung S. Macrophage-Restricted Interleukin-10 Receptor Deficiency, but Not IL-10 Deficiency, Causes Severe Spontaneous Colitis. Immunity. 2014;40:720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, Udalova IA. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 26.Balasa A, Gathungu G, Kisfali P, Smith EOB, Cho JH, Melegh B, Kellermayer R. Assessment of DNA methylation at the interferon regulatory factor 5 (IRF5) promoter region in inflammatory bowel diseases. International Journal of Colorectal Disease. 2010;25:553–556. doi: 10.1007/s00384-010-0874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dideberg V, Kristjansdottir G, Milani L, Libioulle C, Sigurdsson S, Louis E, Wiman A-C, Vermeire S, Rutgeerts P, Belaiche J, Franchimont D, et al. An insertion-deletion polymorphism in the Interferon Regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Human Molecular Genetics. 2007;16:3008–3016. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 28.Gathungu G, Zhang CK, Zhang W, Cho JH. A two-marker haplotype in the IRF5 gene is associated with inflammatory bowel disease in a North American cohort. Genes And Immunity. 2012;13:351. doi: 10.1038/gene.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 30.Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, Cua DJ, Powrie F, Cheever AW, Maloy KJ, Sher A. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. The Journal of Experimental Medicine. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus Triggers Colitis in Specific-Pathogen-Free Interleukin-10 (IL-10)-Deficient Mice through an IL-12-and Gamma Interferon-Dependent Mechanism. Infection and Immunity. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullberg MC, Jankovic D, Gorelick PL, Caspar P, Letterio JJ, Cheever AW, Sher A. Bacteria-triggered CD4(+) T Regulatory Cells Suppress Helicobacter hepaticus-induced Colitis. The Journal of Experimental Medicine. 2002;196:505–515. doi: 10.1084/jem.20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bain CC, Oliphant CJ, Thomson CA, Kullberg MC, Mowat AM. Proinflammatory Role of Monocyte-Derived CX3CR1int Macrophages in Helicobacter hepaticus-Induced Colitis. Infection and Immunity. 2018;86 doi: 10.1128/IAI.00579-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahern PP, Izcue A, Maloy KJ, Powrie F. The interleukin-23 axis in intestinal inflammation. Immunological Reviews. 2008;226:147–159. doi: 10.1111/j.1600-065X.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- 35.Farache J, Koren I, Milo I, Gurevich I, Kim K-W, Zigmond E, Furtado GC, Lira SA, Shakhar G. Luminal Bacteria Recruit CD103(+) Dendritic Cells into the Intestinal Epithelium to Sample Bacterial Antigens for Presentation. Immunity. 2013;38:581–595. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science. 2014;343 doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mildner A, Schönheit J, Giladi A, David E, Lara-Astiaso D, Lorenzo-Vivas E, Paul F, Chappell-Maor L, Priller J, Leutz A, Amit I, et al. Genomic Characterization of Murine Monocytes Reveals C/EBP[beta]; Transcription Factor Dependence of Ly6C-Cells. Immunity. 2017;46:849–862.:e847. doi: 10.1016/j.immuni.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Menezes S, Melandri D, Anselmi G, Perchet T, Loschko J, Dubrot J, Patel R, Gautier EL, Hugues S, Longhi MP, Henry JY, et al. The Heterogeneity of Ly6Chi Monocytes Controls Their Differentiation into iNOS+ Macrophages or Monocyte-Derived Dendritic Cells. Immunity. 2016;45:1205–1218. doi: 10.1016/j.immuni.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Feng D, Bi X, Stone RC, Barnes BJ. Monocytes from Irf5(-/-) mice have an intrinsic defect in their response to pristane-induced lupus. Journal of immunology (Baltimore, Md. : 1950) 2012;189:3741–3750. doi: 10.4049/jimmunol.1201162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung GA, Cool T, Valencia CH, Worthington A, Beaudin AE, Forsberg EC. The lymphoid-associated interleukin 7 receptor (IL7R) regulates tissueresident macrophage development. Development. 2019;146 doi: 10.1242/dev.176180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alteber Z, Sharbi-Yunger A, Pevsner-Fischer M, Blat D, Roitman L, Tzehoval E, Elinav E, Eisenbach L. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol Cell Biol. 2018;96:284–297. doi: 10.1111/imcb.12000. [DOI] [PubMed] [Google Scholar]
- 42.Asano K, Takahashi N, Ushiki M, Monya M, Aihara F, Kuboki E, Moriyama S, Iida M, Kitamura H, Qiu CH, Watanabe T, et al. Intestinal CD169(+) macrophages initiate mucosal inflammation by secreting CCL8 that recruits inflammatory monocytes. Nat Commun. 2015;6:7802. doi: 10.1038/ncomms8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jha P, Das H. KLF2 in Regulation of NF-kappaB-Mediated Immune Cell Function and Inflammation. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18112383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson DA, Grajales-Reyes GE, Satpathy AT, Vasquez Hueichucura CE, Murphy TL, Murphy KM. Revisiting the specificity of the MHC class II transactivator CIITA in classical murine dendritic cells in vivo. European Journal of Immunology. 2017;47:1317–1323. doi: 10.1002/eji.201747050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richter L, Landsverk OJB, Atlasy N, Bujko A, Yaqub S, Horneland R, Øyen O, Aandahl EM, Lundin KEA, Stunnenberg HG, Bækkevold ES, et al. Transcriptional profiling reveals monocyte-related macrophages phenotypically resembling DC in human intestine. Mucosal Immunology. 2018;11:1512–1523. doi: 10.1038/s41385-018-0060-1. [DOI] [PubMed] [Google Scholar]
- 46.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMc Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey SP, Yan J, Turner JR, Abraham C. Reducing IRF5 expression attenuates colitis in mice, but impairs the clearance of intestinal pathogens. Mucosal Immunol. 2019;12:874–887. doi: 10.1038/s41385-019-0165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghoreschi K, Laurence A, Yang X-P, Tato CM, McGeachy MJ, Konkel J, Ramos HL, Wei L, Davidson T, Bouladoux N, Grainger J, et al. Generation of Pathogenic Th17 Cells in the Absence of TGF-β Signaling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, Maloy KJ. IL-1β mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4+ Th17 cells. The Journal of Experimental Medicine. 2012;209:1595. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seneviratne AN, Edsfeldt AO, Cole JE, Kassiteridi C, Swart M, Park I, Green P, Khoyratty TE, Saliba DG, Goddard ME, Sansom SN, et al. Interferon Regulatory Factor 5 Controls Necrotic Core Formation in Atherosclerotic Lesions by Impairing Efferocytosis. Circulation. 2017 doi: 10.1161/CIRCULATIONAHA.117.027844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, Yan C, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu X, Nagata M, Yamasaki S. Mincle: 20 years of a versatile sensor of insults. Int Immunol. 2018;30:233–239. doi: 10.1093/intimm/dxy028. [DOI] [PubMed] [Google Scholar]
- 54.Xu D, Meyer F, Ehlers E, Blasnitz L, Zhang L. Interferon Regulatory Factor 4 (IRF-4) Targets IRF-5 to Regulate Epstein-Barr Virus Transformation. Journal of Biological Chemistry. 2011;286:18261–18267. doi: 10.1074/jbc.M110.210542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goudot C, Coillard A, Villani A-C, Gueguen P, Cros A, Sarkizova S, Tang-Huau T-L, Bohec M, Baulande S, Hacohen N, Amigorena S, et al. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity. 2017;47:582–596.:e586. doi: 10.1016/j.immuni.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 56.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. Negative regulation of Toll-like-receptor signaling by IRF-4. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Persson Emma K, Uronen-Hansson H, Semmrich M, Rivollier A, Hägerbrand K, Marsal J, Gudjonsson S, Håkansson U, Reizis B, Kotarsky William K, Agace W. IRF4 Transcription-Factor-Dependent CD103+CD11b+ Dendritic Cells Drive Mucosal T Helper 17 Cell Differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 58.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36:411. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next generation sequencing datasets are available via the Gene Expression Omnibus (GEO) via accession codes GSE129354 (GM-DMDM data) and GSE129258 (MBMC small bulk and single-cell data).