Abstract
Dendritic cells (DC) are professional antigen-presenting cells that orchestrate immune responses. The human DC population comprises two main functionally specialized lineages, whose origins and differentiation pathways remain incompletely defined. Here, we combine two high-dimensional technologies—single-cell messenger RNA sequencing (scmRNAseq) and cytometry by time-of-flight (CyTOF)—to identify human blood CD123+CD33+CD45RA+ DC precursors (pre-DC). Pre-DC share surface markers with plasmacytoid DC (pDC) but have distinct functional properties that were previously attributed to pDC. Tracing the differentiation of DC from the bone marrow to the peripheral blood revealed that the pre-DC compartment contains distinct lineage-committed subpopulations, including one early uncommitted CD123high pre-DC subset and two CD45RA+CD123low lineage-committed subsets exhibiting functional differences. The discovery of multiple committed pre-DC populations opens promising new avenues for the therapeutic exploitation of DC subset-specific targeting.
Dendritic cells (DC) are professional pathogensensing and antigen-presenting cells that are central to the initiation and regulation of immune responses (1). The DC population is classified into two lineages: plasmacytoid DC (pDC) and conventional DC (cDC), the latter comprising cDC1 and cDC2 subpopulations (2,3). Dissecting the origins and differentiation pathways giving rise to DC subpopulations is necessary to understand their homeostasis and role in immune responses and for the development of DC subset-specific therapeutic interventions. Murine DC arise from unique DC-restricted bone-marrow (BM) progenitors known as common DC progenitors (CDP), which differentiate into pDC and DC precursors (pre-DC) and migrate out of the BM into peripheral tissues (4–7). Human equivalents of murine CDP and pre-DC have recently been described (8,9); human pre-DC comprise ~0.001% of peripheral blood mononuclear cells (PBMC) and were identified by their expression of cytokine receptors that mark and drive DC differentiation in mice, including CD117 (c-kit, SCF), CD116 (GMCSF), CD135 (FLT3), and CD123 (IL3-Rα) (9). Previous studies have observed similar receptor expression patterns within human pDC populations, which can differentiate into cDC-like cells when stimulated with interleukin-3 (IL-3) and CD40L (10,11). Therefore, either pDC are precursors of cDC, as proposed (11), or the conventionally defined pDC population is heterogeneous, incorporating an independent pre-DC subpopulation.
To answer this question, we interrogated the blood CD135+HLA-DR+ fraction, which should contain DC and their precursors (12–14), using several integrated, high-dimensional analysis techniques, including single-cell mRNA sequencing (scmRNAseq) and cytometry by time-of-flight (CyTOF). Using these approaches, we identified a novel population of pre-DC within the conventionally defined pDC population. These pre-DC exhibit a unique phenotype and distinct functional properties, which were previously attributed to pDC. Extending our analysis to all DC populations in human blood and BM, we identified the entire DC lineage arising from the BM and revealed the transcriptional priming of pre-DC toward distinct DC subsets. These data offer new insights into DC heterogeneity and ontogeny and highlight unexplored avenues for investigation of the therapeutic potential of DC subset-specific targeting.
Unbiased identification of DC precursors by unsupervised single-cell RNAseq and CyTOF
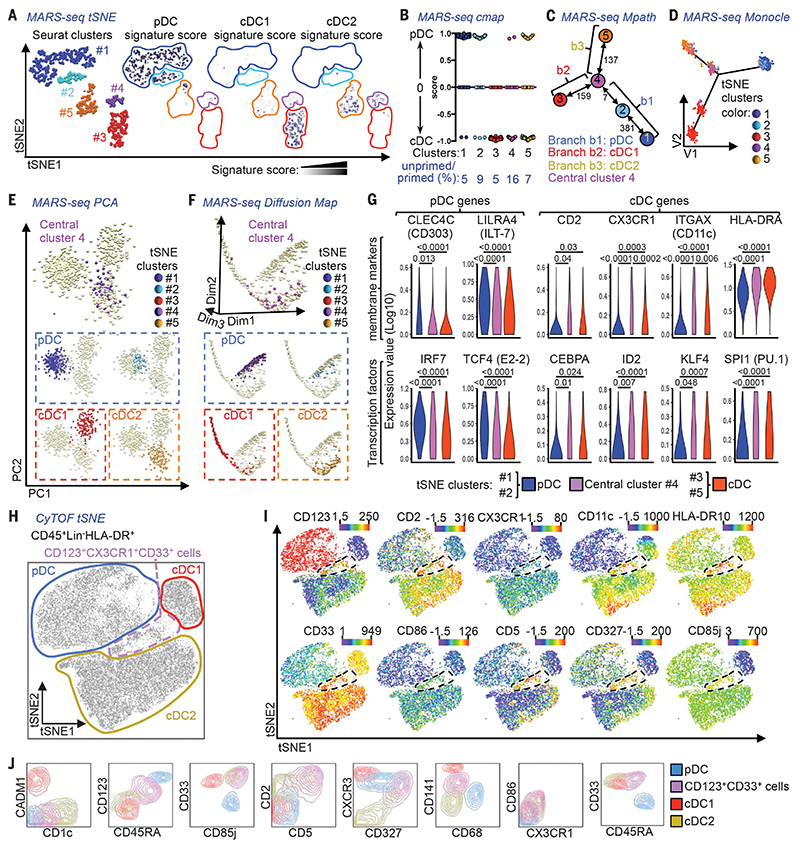
Using PBMC isolated from human blood, we employed massively parallel single-cell mRNA sequencing (MARS-seq) (15) to assess the transcriptional profile of 710 individual cells within the lineage marker (Lin) (CD3/CD14/CD16/CD20/CD34)−, HLA-DR+CD135+ population [Fig. 1, A to G; fig. S1a (16); fig. S1, b to j (17); and table S1 (18)]. The MARS-seq data were processed using nonlinear dimensionality reduction via t-stochastic neighbor embedding (tSNE), which enables unbiased visualization of high-dimensional similarities between cells in the form of a two-dimensional (2D) map (19–21). Density-based spatial clustering of applications with noise (DBSCAN) (22) on the tSNE dimensions identified five distinct clusters of transcriptionally related cells within the selected PBMC population (Fig. 1A and fig. S1g). To define the nature of these clusters, we calculated gene signature scores for pDC, cDC1, and cDC2 [as described in (23) and table S2 (24)] and overlaid the expression of the signatures attributed to each cell onto the tSNE visualization. Clusters #1 and #2 (containing 308 and 72 cells, respectively) were identified as pDC, cluster #3 (containing 160 cells) as cDC1, and cluster #5 (containing 120 cells) as cDC2. Cluster #4 (containing 50 cells) lay in between the cDC1 (#3) and cDC2 (#5) clusters and possessed a weak, mixed pDC/cDC signature (Fig. 1A). We then performed a connectivity map (cmap) analysis (25) to calculate the degree of enrichment of pDC or cDC signature gene transcripts in each individual cell. This approach confirmed the signatures of pDC (#1 and #2) and cDC (#3 and #5) clusters and showed that most cells in cluster #4 expressed a cDC signature (Fig. 1B). The Mpath algorithm (26) was then applied to the five clusters to identify hypothetical developmental relationships based on these transcriptional similarities between cells (Fig. 1C and fig. S2, a and b). Mpath revealed that the five clusters were grouped into three distinct branches, with one central cluster (cluster #4) at the intersection of the three branches (Fig. 1C and fig. S2a). The Mpath edges connecting cluster #4 to cDC1 cluster #3 and cDC2 cluster #5 have a high cell count (159 and 137 cells, respectively), suggesting that the transition from cluster #4 to clusters #3 and #5 is likely valid, and indicating that cluster #4 could contain putative cDC precursors (Fig. 1C). In contrast, the edge connecting cluster #4 and pDC cluster #2 has a cell count of only 7 (Fig. 1C and fig. S2b), which suggests that this connection is very weak. The edge connecting clusters #4 and #2 was retained when Mpath trimmed the weighted neighborhood network (fig. S2b), simply due to the feature of the Mpath algorithm that requires all clusters to be connected (26). To confirm these findings, we tested Monocle (27), principal components analysis (PCA), Wishbone (28), and diffusion map algorithms (29). Monocle and PCA resolved the cells into the same three branches as the original Mpath analysis, with the cells from the tSNE cluster #4 again falling at the intersection (Fig. 1, D and E). Diffusion map and Wishbone analyses indicated that there was a continuum between clusters #3 (cDC1), #4, and #5 (cDC2): Cells from cluster #4 were predominantly found in the DiffMap_dim2low region, and cells from clusters #3 and #5 were progressively drifting away from the DiffMap_dim2low region toward the left and right, respectively. The pDC clusters (#1 and #2) were clearly separated from all other clusters (Fig. 1F and fig. S2c). In support of this observation, cells from these pDC clusters had a higher expression of pDC-specific markers and transcription factors (TF) than the cDC clusters (#3 and #5) and central cluster #4. Conversely, cells in cluster #4 expressed higher levels of markers and TF associated with all cDC lineage than the pDC clusters (Fig. 1G). This phenotype led us to hypothesize that cluster #4 represented a population of putative uncommitted cDC precursors.
Fig. 1. MARS-seq and CyTOF identify rare CD123+CD33+ putative DC precursors (pre-DC).
Lin(CD3/CD14/CD16/CD20/CD34)”HLA-DR+CD135+ sorted PBMC were subjected to MARS-seq. (A) t-stochastic neighbor embedding (tSNE) plot of 710 cells fulfilling all quality criteria, colored by clusters identified by tSNE plus Seurat clustering, or by the relative signature score for pDC, cDC1, and cDC2. (B) Connectivity map (cmap) analysis showing the degree of enrichment for pDC or cDC signature genes in the tSNE/Seurat clusters. (C) Mpath analysis applied to the tSNE/Seurat clusters defining their developmental relationship. Representations of the 710 cells by (D) Monocle, (E) PCA, and (F) Diffusion map, highlighting the tSNE/Seurat clusters identified in (A). (G) Violin plots of tSNE/Seurat pDC clusters, cluster #4, and cDC clusters showing the expression of pDC and cDC signature genes with differential expression between cluster #4 and pDC clusters. Adjusted P values calculated by Kruskal-Wallis test followed by Dunn’s multiple comparisons procedure. (H and I) tSNE plots of CyTOF data from CD45+Lin(CD7/CD14/CD15/CD16/CD19/CD34)−HLA-DR+ PBMC, showing (H) gates defining the CD123+CD33+ cells and DC subsets and (I) relative expression of selected markers. (J) Subsets defined in (H) were overlaid onto 2D-contour plots for phenotypic comparison. The gating strategy before MARS-seq is shown in fig. S1a.
We next employed CyTOF, which simultaneously measures the intensity of expression of up to 38 different molecules at the single-cell level, to further understand the composition of the delineated subpopulations. We designed a panel of 38 labeled antibodies to recognize DC lineage and/or progenitor-associated surface molecules (table S3; Fig. 1, H to J; and fig. S3) and the molecules identified in cluster #4 by MARS-seq, such as CD2, CX3CR1, CD11c, and HLA-DR (Fig. 1I). Using the tSNE algorithm, the CD45+Lin(CD7/CD14/CD15/CD16/CD19/CD34)−HLA-DR+ PBMC fraction (fig. S3a) resolved into three distinct clusters representing cDC1, cDC2, and pDC (Fig. 1H). An intermediate cluster at the intersection of the cDC and pDC clusters that expressed both cDC-associated markers (CD11c/CX3CR1/CD2/CD33/CD141/BTLA) and pDC-associated markers (CD45RA/CD123/CD303) (Fig. 1, I to J, and fig. S3b) corresponded to the MARS-seq cluster #4. The delineation of these clusters was confirmed when applying the phenograph unsupervised clustering algorithm (30) (fig. S3c). The position of the intermediate CD123+CD33+ cell cluster was distinct, and the cells exhibited a high expression of CD5, CD327, and CD85j, together with high levels of HLA-DR and the cDC-associated molecule CD86 (Fig. 1, I to J). Taken together, these characteristics led us to consider whether CD123+CD33+ cells might represent circulating human pre-DC.
Pre-DC exist within the pDC fraction and give rise to cDC
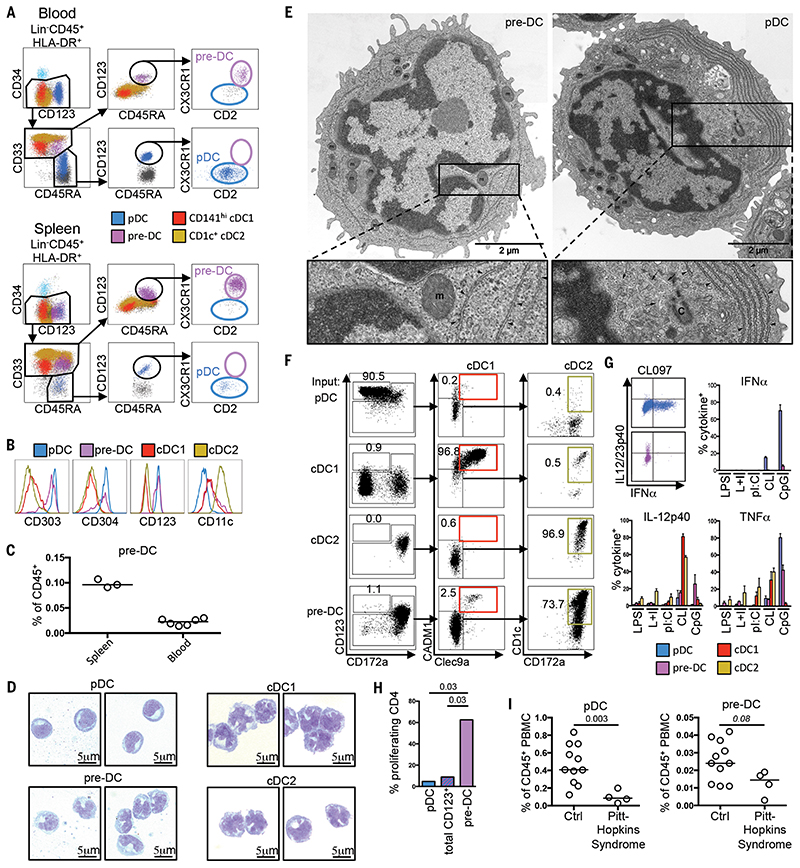
We analyzed the CD123+CD33+ cell cluster within the Lin−HLA-DR+ fraction of the PBMC by flow cytometry. Here, we identified CD123+CD33− pDC, CD45RA+/-CD123− cDC1 and cDC2, and CD33+CD45RA+CD123+ putative pre-DC (Fig. 2A and fig. S4a). The putative pre-DC expressed CX3CR1, CD2, CD303, and CD304, with low CD11c expression, whereas CD123+CD33− pDC exhibited variable CD2 expression (Fig. 2, A and B, and fig. S4, b and c). We then extended our analysis to immune cells from the spleen and identified a similar putative pre-DC population, which was more abundant than in blood and expressed higher levels of CD11c (Fig. 2, A and C, and fig. S4d). Both putative pre-DC populations in the blood and spleen expressed CD135 and intermediate levels of CD141 (fig. S4c). Wright-Giemsa staining of putative pre-DC sorted from the blood revealed an indented nuclear pattern reminiscent of classical cDC, a region of perinuclear clearing, and a basophilic cytoplasm reminiscent of pDC (Fig. 2D). At the ultrastructural level, putative pre-DC and pDC exhibited distinct features, despite their morphological similarities (Fig. 2E and fig. S4e): Putative pre-DC possessed a thinner cytoplasm, homogeneously distributed mitochondria (m), less rough endoplasmic reticulum (RER), an indented nuclear pattern, a large nucleus, and limited cytosol, compared with pDC; pDC contained a smaller nucleus, abundant cytosol, packed mitochondria, and well-developed and polarized cortical RER organized in parallel cisterna alongside numerous stacks of RER membranes, suggesting a developed secretory apparatus, in agreement with previously published data (31).
Fig. 2. Characterization of human pre-DC.
(A) Flow cytometric identification of pre-DC and pDC within PBMC and spleen cell suspensions. (B) Expression of CD303/CD304/CD123/CD11c by blood pre-DC and DC subsets. (C) % pre-DC within spleen (n = 3) and PBMC (n = 6) CD45+ populations. (D) Wright-Giemsa staining of sorted blood pre-DC and DC subsets. (E) Electron micrographs of pre-DC and pDC [RER (arrowheads), centriole (C), and microtubules (small arrows) near RER cisterna are indicated]. (F) DC subsets or pre-DC were cocultured for 5 days with MS-5 feeder cells, FLT3L, GM-CSF, and SCF. Their capacity to differentiate into cDC1 or cDC2 was measured by flow cytometry (n = 3). (G) Intracellular detection of cytokines in DC subsets and pre-DC post-TLR stimulation. TNF-α and IL-12p40 production by pDC and pre-DC, alongside mean % cytokine-positive pre-DC and DC subsets exposed to LPS, LPS+IFN-γ (L+I), polyI:C (pI:C), CL097 (CL), or CpG-ODN2216 (CpG) (n = 4). (H) Proliferation of naïve CD4+ Tcells cultured for 6 days with allogeneic pDC, total CD123+HLA-DR+ cells, or pre-DC (n = 2). (I) Frequency of pDC and pre-DC from control subjects (Ctrl) (n = 11) and Pitt-Hopkins syndrome (PHS) patients (n = 4). P values calculated by Mann-Whitney test. Error bars, mean ± SEM.
We then compared the differentiation capacity of pre-DC to that of cDC and pDC through stromal culture in the presence of FLT3L, GMCSF, and SCF, as previously described (8). After 5 days, the pDC, cDC1, and cDC2 populations remained predominantly in their initial states, whereas the putative pre-DC population had differentiated into cDC1 and cDC2 in the known proportions found in vivo (14,23,32,33) (Fig. 2F, fig. S4f, and fig. S5). Altogether, these data suggest that CD123+CD33+ CD45RA+CX3CR1+CD2+ cells are circulating pre-DC with cDC differentiation potential. Breton and colleagues (9) recently reported a minor population of human pre-DC (highlighted in fig. S6a), which shares a similar phenotype with the Lin− CD123+CD33+CD45RA+ pre-DC defined here (fig. S6, a and b). Our results reveal that the pre-DC population in blood and spleen is markedly larger than the one identified within the minor CD303−CD141−CD117+ fraction considered previously (fig. S6, c and d).
Pre-DC are functionally distinct from pDC
Interferon-α (TNF-α)-secreting pDC can differentiate into cells resembling cDC when cultured with IL-3 and CD40L (10,11) and have been considered DC precursors (11). However, when we used traditional ILT3+ILT1− (10) or CD4+CD11c− (11) pDC gating strategies, we detected a “contaminating” CD123+CD33+CD45RA+ pre-DC subpopulation in both groups (fig. S6, e and f). We questioned, therefore, whether other properties of traditionally classified “pDC populations” might be attributed to pre-DC. TLR7/8 (CL097) or TLR9 (CpG ODN 2216) stimulation of pure pDC cultures resulted in abundant secretion of TNF-α but not IL-12p40, whereas pre-DC readily secreted IL-12p40 but not TNF-α (Fig. 2G and fig. S7). Furthermore, although pDC were previously thought to induce proliferation of naïve CD4+ T cells (10,34), here we found that only the pre-DC subpopulation exhibited this attribute (Fig. 2H). Reports of potent allostimulatory capacity and IL-12p40 production by CD2+ pDC (34) might then be explained by CD2+ pre-DC “contamination” (35) (fig. S8).
Pitt-Hopkins syndrome (PHS) is characterized by abnormal craniofacial and neural development, severe mental retardation, and motor dysfunction and is caused by haplo-insufficiency of 7CF4,which encodes the E2-2 transcription factor—a central regulator of pDC development (36). We confirmed that patients with PHS had a marked reduction in their blood pDC numbers compared with healthy individuals but that they retained a population of pre-DC (Fig. 2I and fig. S9), which likely accounts for the unexpected CD45RA+CD123+CD303lo) cell population reported in these patients (37). Taken together, our data indicate that, although pre-DC and pDC share some phenotypic features, they can be separated by their differential expression of several markers, including CD33, CX3CR1, CD2, CD5, and CD327. pDC are bona fide TNF-α-producing cells, but the reported IL-12 production and CD4+ T cell allostimulatory capacity of pDC can likely be attributed to “contaminating” pre-DC, which can give rise to both cDC1 and cDC2.
Identification and characterization of committed pre-DC subsets
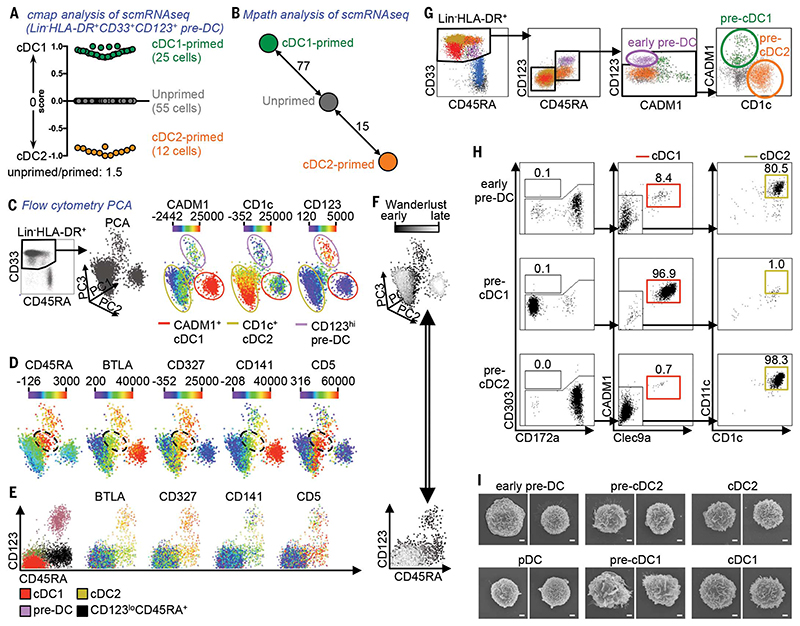
The murine pre-DC population contains both uncommitted and committed pre-cDC1 and pre-cDC2 precursors (7). We asked whether the same was true for human blood pre-DC using microfluidic scmRNAseq [fig. S10a (38); fig. S10, b and c (39); and table S4 (40)]. The additional single-cell gene expression data relative to the MARS-seq strategy used for Fig. 1, A to G (2.5 million reads per cell and an average of 4742 genes detected per cell versus 60,000 reads per cell and an average of 749 genes detected per cell, respectively), were subjected to cmap analysis, which calculated the degree of enrichment for cDC1 or cDC2 signature gene transcripts (23) for each single cell (Fig. 3A). Among the 92 analyzed pre-DC, 25 cells exhibited enrichment for cDC1 gene expression signatures, 12 cells for cDC2 gene expression signatures, and 55 cells showed no transcriptional similarity to either cDC subset. Further Mpath analysis showed that these 55 “unprimed” pre-DC were developmentally related to cDC1-primed and cDC2-primed pre-DC, and thus their patterns of gene expression fell between the cDC1 and cDC2 signature scores by cmap (Fig. 3B and fig. S11). These data suggest that the human pre-DC population contains cells exhibiting transcriptomic priming toward cDC1 and cDC2 lineages, as observed in mice (7).
Fig. 3. Identification of committed human pre-DC subsets.
(A and B) Single-cell mRNA sequencing (scmRNAseq) of 92 Lin(CD3/14/16/19/20)”HLA-DR+CD33+CD123+ cells (sort gating strategy in fig. S8a). (A) Connectivity map (cmap) enrichment score of cells (cDC1-versus cDC2-specific signatures). (B) Mpath analysis showing the developmental relationship between “unprimed,” cDC1-primed, and cDC2-primed cells defined in (A). (C) Lin”HLA-DR+CD33+ PBMC analyzed by flow cytometry and visualized as 3D-PCA of three cell clusters (pre-DC, cDC1, and cDC2) and the relative expression of CADM1, CD1c, and CD123.
(D) Relative expression of CD45RA, BTLA, CD327, CD141, and CD5 in the same 3D-PCA plot. The dashed black circles indicate the intermediate CD45RA+ population. (E) CD45RA/CD123 dot plots showing overlaid cell subsets defined in the 3D-PCA plot (left panel) with the relative expression of BTLA, CD327, CD141, and CD5. (F) Overlay of the Wanderlust dimension [progression from early (dark) to late (clear) events is shown] onto the 3D-PCA and CD45RA/CD123 dot plots. (G) Gating strategy starting from live CD45+Lin(CD3/14/16/19/20)”CD34”HLA-DR+ PBMC to define pre-DC subsets among CD33+CD45RA+ cDC. (H) Pre-DC subsets were cocultured for 5 days with MS-5 feeder cells, FLT3L, GM-CSF, and SCF (n = 3). Their capacity to differentiate into Clec9A+CADM1+ cDC1 (red) or CD1c+CD11c+ cDC2 (beige) was analyzed by flow cytometry. (I) Scanning electron microscopy of pre-DC and DC subsets. Scale bar, 1 μm.
We next asked whether we could identify this heterogeneity within the pre-DC population by flow cytometry, using either pre-DC-specific markers (CD45RA, CD327, and CD5) or markers expressed more intensely by pre-DC compared with cDC2 (BTLA and CD141). 3D-PCA analysis of the Lin−HLA-DR+CD33+ population (containing both differentiated cDC and pre-DC) identified three major cell clusters: CADM1+ cDC1, CD1c+ cDC2, and CD123+ pre-DC (Fig. 3C and fig. S12a). Interestingly, although cells located at the intersection of these three clusters (Fig. 3D) expressed lower levels of CD123 than pre-DC but higher levels than differentiated cDC (Fig. 3C), they also expressed high levels of pre-DC markers (Fig. 3D and fig. S12a). We reasoned that these CD45RA+ CD123lo cells might be committed pre-DC that are differentiating into either cDC1 or cDC2 (Fig. 3E). The Wanderlust algorithm (41), which orders cells into a constructed trajectory according to their maturity, confirmed the developmental relationship between pre-DC (early events), CD45RA+CD123lo cells (intermediate events), and mature cDC (late events) (Fig. 3F). Flow cytometry of PBMC identified CD123+CADM1−CD1c− putative uncommitted pre-DC, alongside putative CADM1+CD1c− pre-cDC1 and CADM1−CD1c+ pre-cDC2 within the remaining CD45RA+ cells (Fig. 3G and fig. S12b). These three populations were also present, and more abundant, in the spleen (fig. S12c). Importantly, in vitro culture of pre-DC subsets sorted from PBMC did not give rise to any CD303+ cells (which would be either undifferentiated pre-DC or differentiated pDC), whereas early pre-DC gave rise to both cDC subsets, and pre-cDC1 and pre-cDC2 differentiated exclusively into cDC1 and cDC2 subsets, respectively (Fig. 3H, fig. S12d, and fig. S13).
Scanning electron microscopy confirmed that early pre-DC are larger and rougher in appearance than pDC and that committed pre-DC subsets closely resemble their mature cDC counterparts (Fig. 3I and fig. S14a). Phenotyping of blood pre-DC by flow cytometry (fig. S14b) identified patterns of transitional marker expression throughout the development of early pre-DC toward pre-cDC1/2 and differentiated cDC1/2. Specifically, CD45RO and CD33 were acquired in parallel with the loss ofCD45RA; CD5, CD123, CD304, and CD327were expressed abundantly by early pre-DC, intermediately by pre-cDC1 and pre-cDC2, and rarely if at all by mature cDC and pDC; FceRI and CD1c were acquired as early pre-DC commit toward the cDC2 lineage, concurrent with the loss of BTLA and CD319 expression; early pre-DC had an intermediate expression of CD141 that dropped along cDC2 differentiation but was increasingly expressed during commitment toward cDC1, with a few pre-cDC1 already starting to express Clec9A; and IRF8 and IRF4—transcription factors regulating cDC lineage development (2,3)—were expressed by early pre-DC and pre-cDC1, whereas pre-cDC2 maintained only IRF4 expression (fig. S14c).
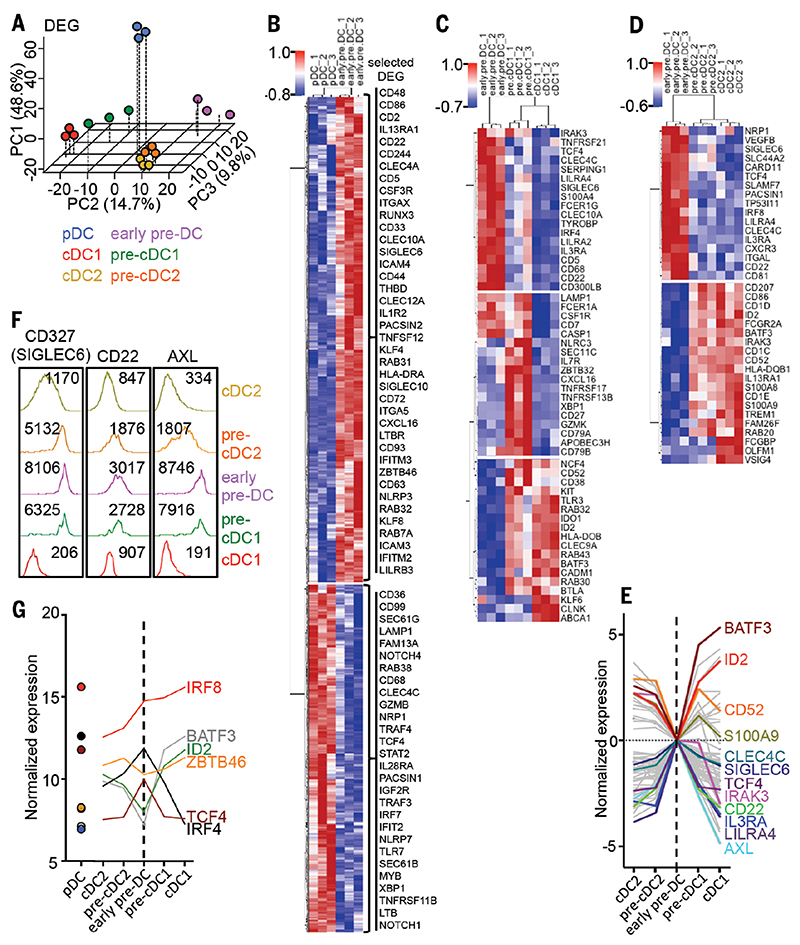
We next sorted pre-DC and DC subsets from blood and performed microarray analyses to define their entire transcriptome. 3D-PCA analysis of the microarray data showed that pDC were clearly separated from other pre-DC and DC subsets along the horizontal PC1 axis (Fig. 4A and fig. S15). The combination of the PC2 and PC3 axes indicated that pre-cDC1 occupied a position between early pre-DC and cDC1 and, although cDC2 and pre-cDC2 exhibited similar transcriptomes, pre-cDC2 were positioned between cDC2 and early pre-DC along the PC3 axis (Fig. 4A). Hierarchical clustering of differentially expressed genes (DEG) confirmed the similarities between committed pre-DC and their corresponding mature subset (fig. S16). The greatest number of DEG was between early pre-DC and pDC (1249 genes), among which CD86, CD2, CD22, CD5, ITGAX (CD11c), CD33, CLEC10A, SIGLEC6 (CD327), THBD, CLEC12A, KLF4, and ZBTB46 were more highly expressed by early pre-DC, whereas pDC showed higher expression of CD68, CLEC4C, TCF4, PACSIN1, IRF7, and TLR7 (Fig. 4B). An evolution in the gene expression pattern was evident from early pre-DC to pre-cDC1 and then cDC1 (Fig. 4C), whereas pre-cDC2 were similar to cDC2 (Fig. 4D and fig. S16). The union of DEG comparing pre-cDC1versus early pre-DC and cDC1 versus pre-cDC1 has 62 genes in common with the union of DEG from comparing pre-cDC2 versus early pre-DC and cDC2 versus pre-cDC2. These 62 common genes include the transcription factors BATF3, ID2, and TCF4 (E2-2), and the pre-DC markers CLEC4C (CD303), SIGLEC6 (CD327), and IL3RA (CD123) (Fig. 4E, fig. S17, and table S5). The progressive reduction in transcript abundance of SIGLEC6 (CD327), CD22, and AXL during early pre-DC to cDC differentiation was also mirrored at the protein level (Fig. 4F). Key transcription factors involved in the differentiation and/or maturation of DC subsets showed a progressive change in their expression along the differentiation path from pre-DC to mature cDC (Fig. 4G). Finally, pathway analyses revealed that pre-DC exhibited an enrichment of cDC functions relative to pDC and were maintained in a relatively immature state compared with mature cDC (fig. S18).
Fig. 4. DC and pre-DC subset gene expression analysis.
(A) Microarray data from sorted DC and pre-DC subsets (shown in Fig. 3) were analyzed by 3D PCA using differentially expressed genes (DEG). For each PCA dimension (principal component, PC), the variance explained by each component is indicated. (B to D) Heat maps of DEG between (B) early pre-DC/pDC, (C) early pre-DC/pre-cDC1/cDC1, and (D) early pre-DC/pre-cDC2/cDC2. (E) Expression profiles of 62 common genes identified from DEG analysis comparisons along the lineage progression from early pre-DC to mature cDC for cDC1 and cDC2, respectively. The profiles were plotted with the log2 foldchange values (versus early pre-DC). (F) Expression level of CD327 (SIGLEC6), CD22, and AXL proteins by DC and pre-DC subsets evaluated by flow cytometry. The mean fluorescence intensities are indicated. (G) Expression profile of selected transcription factors.
Committed pre-DC subsets are functional
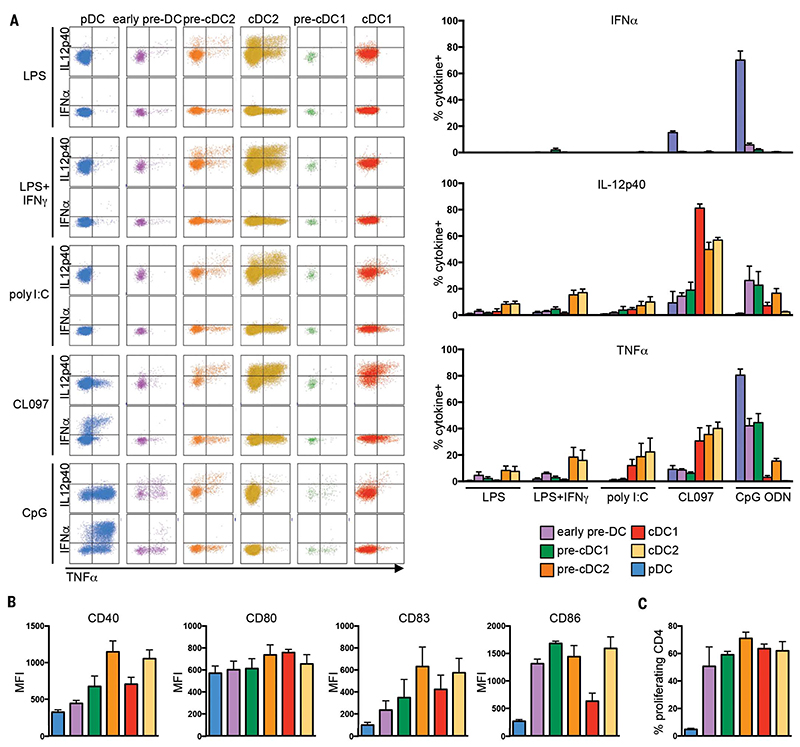
We then asked to what extent the functional specializations of DC (1,42) were acquired at the precursor level by stimulating PBMC with Toll-like receptor (TLR) agonists and measuring their cytokine production (Fig. 5A). Pre-DC produced significantly more tumor necrosis factor-α (TNFα) and IL-12p40 when exposed to CpG ODN 2216 (TLR9 agonist) than to either lipopolysaccharide (LPS) (TLR4 agonist) or polyI:C (TLR3 agonist) (P = 0.03, Mann-Whitney test). We confirmed that pDC were uniquely capable of robust TNF-α production in response to CL097 and CpG ODN 2216. CpG ODN 2216 stimulation also triggered IL-12p40 and TNFa production by early pre-DC, pre-cDC1, and to a lesser extent pre-cDC2. Although TLR9 transcripts were detected only in early pre-DC (fig. S19a), these data indicate that, contrary to differentiated cDC1 and cDC2, pre-cDC1 and pre-cDC2 do express functional TLR9. Interestingly, although pre-cDC2 resembled cDC2 at the gene expression level, their responsiveness to TLR ligands was intermediate between that of early pre-DC and cDC2. Pre-DC subsets also expressed T cell costimulatory molecules (Fig. 5B) and induced proliferation and polarization of naïve CD4+ T cells to a similar level as did mature cDC (Fig. 5C and fig. S19b).
Fig. 5. Functional analysis of DC and pre-DC subsets.
(A) Frequency of cytokine production by pre-DC and DC subsets upon TLR stimulation was measured by intracellular flow cytometry. Dot plots (left panel) show TNF-α, IL-12p40, and TNFa production by pDC, early pre-DC, pre-DC2, cDC2, pre-DC1, and cDC1. Bar charts (right panel) show the mean relative numbers of pre-DC and DC subset cells producing TNF-α+, IL-12p40+, or TNF-α+ in response to LPS, LPS+IFN-γ (L+I), pI:C, CL097 (CL), or CpG ODN2216 (CpG) (n = 4). (B) Expression level [represented as mean fluorescence intensity (MFI)] of costimulatory molecules (CD40, CD80, CD83, and CD86) by blood pre-DC and DC subsets (n = 4). (C) Proliferation of naïve CD4+ Tcells after 6 days of culture with allogenic pre-DC and DC subsets (n = 3). P values calculated by Mann-Whitney test. Error bars, mean ± SEM.
Unsupervised mapping of DC ontogeny
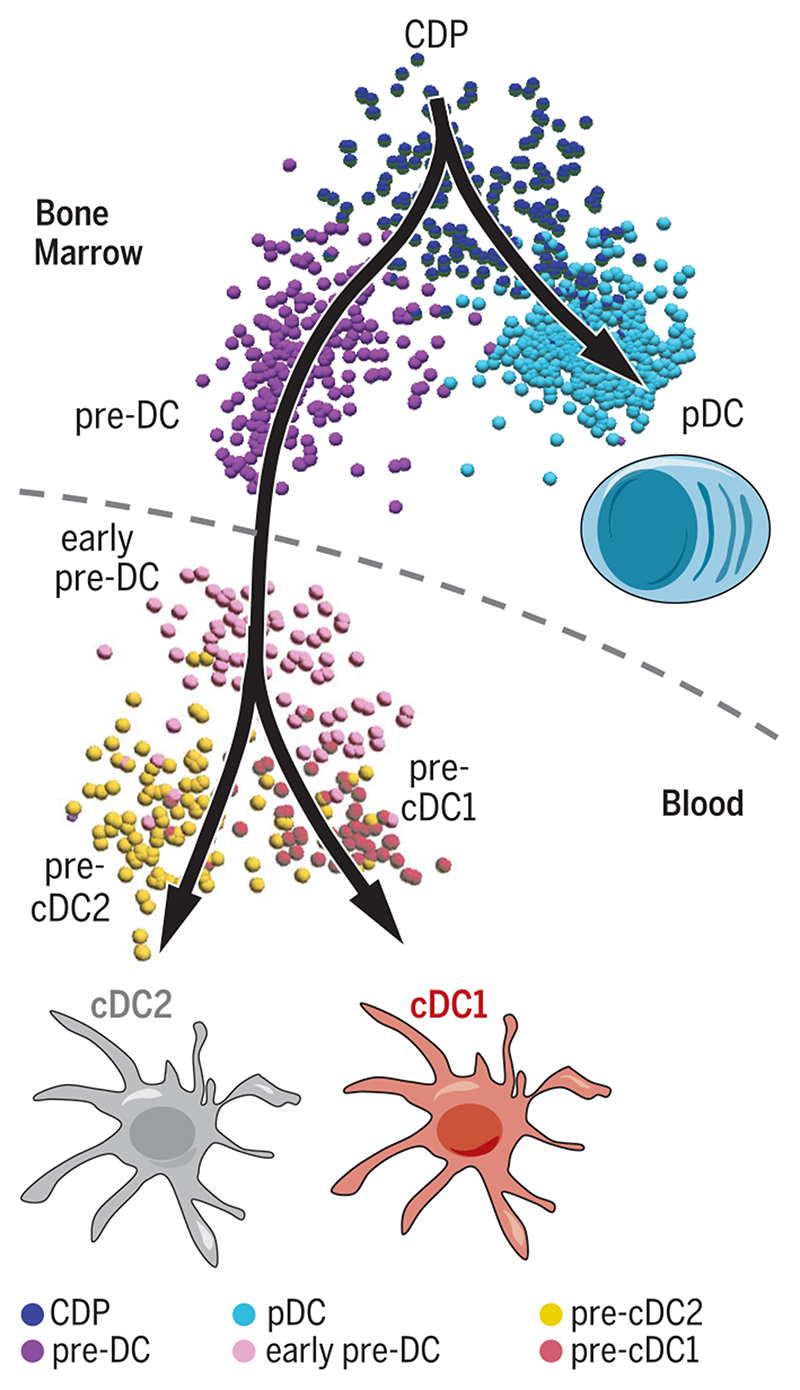
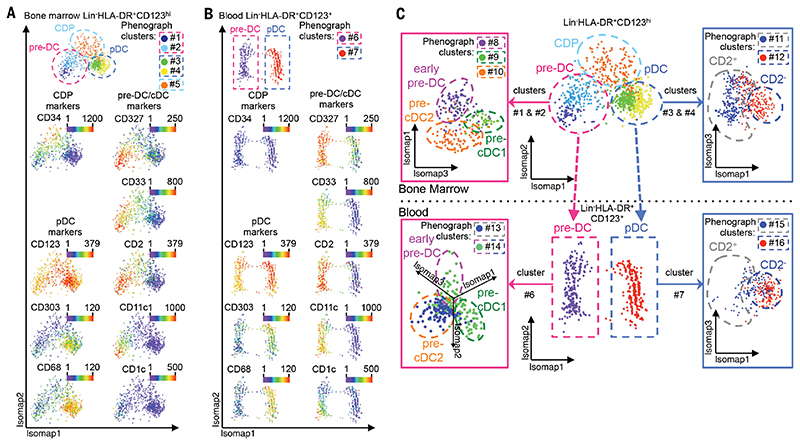
To understand the relatedness of the cell subsets, we performed an unsupervised Isomap analysis (43) of human BM cells, obtained from CyTOF analysis, for nonlinear dimensionality reduction (Fig. 6A and fig. S20a). This analysis focused on the Lin−CD123hi fraction and identified CD123hiCD34+ CDP (phenograph cluster #5), from which branched CD34−CD123+CD327+CD33+ pre-DC (clusters #1 and #2) and CD34−CD123+CD303+CD68+ pDC (clusters #3 and #4), which both progressively acquired their respective phenotypes. Cells in the pre-DC branch increasingly expressed CD2, CD11c, CD116, and, at a later stage, CD1c. Isomap analysis of Lin−CD123+ cells in the peripheral blood identified two parallel lineages, corresponding to pre-DC and pDC, in which a CDP population was not detected (Fig. 6B). Isomap and phenograph analysis of pre-DC extracted from the Isomap analysis of Fig. 6A (BM, clusters #1 and #2) and Fig. 6B (blood, cluster #6) revealed the three distinct pre-DC subsets (Fig. 6C) as defined by their unique marker expression patterns (fig. S20, b and c).
Fig. 6. Unsupervised mapping of DC ontogeny using CyTOF.
CyTOF data from bone marrow (BM) and PBMC were analyzed using Isomap dimensionality reduction to compare overall phenotypic relatedness of cell populations and were automatically subdivided into clusters using the phenograph algorithm. (A and B) Isomap plots showing the expression level of common DC progenitor (CDP), pDC, pre-DC, and cDC-specific markers within (A) BM and (B) blood Lin(CD3/CD7/CD14/CD15/CD19/CD34)−HLA-DR+CD123+ cells. (C) Phenotypic association between Lin-HLA-DR+CD123hl BM and CD123+ PBMC, showing progression from CDP toward pDC or pre-DC in the BM and the clear separation of pDC and pre-DC in the blood. Cells within the pre-DC phenograph clusters (clusters #1 and #2 in the BM and #6 in the blood) and cells within the pDC phenograph clusters (clusters #3 and #4 in the BM and #7 in the blood) were further analyzed by Isomap to define pre-DC subsets (left panels and fig. S20, c and d) and heterogeneity among pDC (right panels and fig. S20, d and e).In summary, we traced the developmental stages of DC from the BM to the peripheral blood through CyTOF, showing that the CDP population in the BM bifurcates into two pathways, developing into either pre-DC or pDC in the blood (Fig. 6, A to C). This pre-DC population is heterogeneous and exists as distinct subsets detectable in both the blood and BM (Fig. 6C and fig. S20, b and c). Furthermore, we uncovered an intriguing heterogeneity in blood and BM pDC that warrants further investigation (Fig. 6C and fig. S20, d and e).
In summary,we traced the developmental stages of DC fromthe BM to the peripheral blood through CyTOF, showing that the CDP population in the BM bifurcates into two pathways, developing into either pre-DC or pDC in the blood (Fig. 6, A to C). This pre-DC population is heterogeneous and exists as distinct subsets detectable in both the blood and BM (Fig. 6C and fig. S20, b and c). Furthermore, we uncovered an intriguing heterogeneity in blood and BM pDC that warrants further investigation (Fig. 6C and fig. S20, d and e).
Discussion
Using unsupervised scmRNAseq and CyTOF analyses, we have unraveled the complexity of the human DC lineage at the single-cell level, revealing a continuous process of differentiation that starts in the BM with CDP and diverges at the point of emergence of pre-DC and pDC potentials, culminating in maturation of both lineages in the blood. A previous study using traditional surface marker-based approaches had suggested the presence of a minor pre-DC population in PBMC (9), but the combination of high-dimensional techniques and unbiased analyses employed here shows that this minor population had been markedly underestimated: We reveal a population of pre-DC that overlaps with that observed by Breton and colleagues (9) within the CD117+CD303−CD141− fraction of PBMC but accounts for >10-fold the number of cells in peripheral blood than was originally estimated and is considerably more diverse (fig. S6c).
Recent work in mice found uncommitted and subset-committed pre-DC subsets in the BM (7,44). Here, we similarly identified three functionally and phenotypically distinct pre-DC populations in human PBMC, spleen, and BM: uncommitted pre-DC and two populations of subset-committed pre-DC (figs. S21 and S22). In line with the concept of continuous differentiation from the BM to the periphery, the proportion of uncommitted cells was higher in the pre-DC population in the BM than in the blood. Altogether, these findings support a two-step model of DC development whereby a central transcriptomic subset-specific program is imprinted on DC precursors from the CDP stage onward, conferring a core subset identity irrespective of the final tissue destination; in the second step of the process, peripheral tissue-dependent programming occurs to ensure site-specific functionality and adaptation (7,44). Future studies will be required to reveal the molecular events underlying DC subset lineage priming and the tissue-specific cues that regulate their peripheral programming and to design strategies that specifically target DC subsets at the precursor level. In addition, how the proportions of uncommitted pre-DC versus committed pre-DC are modified in acute and chronic inflammatory settings warrants further investigation.
An important aspect of unbiased analyses is that cells are not excluded from consideration on the basis of preconceptions concerning their surface phenotype. We found that pre-DC express most of the markers that classically defined pDC, such as CD123, CD303, and CD304. Thus, any strategy relying on these markers to identify and isolate pDC will have inadvertently included CD123+CD33+ pre-DC as well. Although this calls us to urgently reconsider some aspects of pDC population biology, it may also explain earlier findings, including that pDC cultures possess cDC potential and acquire cDC-like morphology (10,11), as recently observed in murine BM pDC (45); pDC mediate Th1 immunity through production of TNF-α and IL-12 (10,46–50); pDC exhibit naïve T cell allostimulatory capacity (34,48); and pDC express costimulatory molecules and exhibit antigen-presentation/cross-presentation capabilities at the expense of TNF-α secretion (46,51). These observations could be attributed to the undetected pre-DC in the pDC populations described by these studies, andindeed it has been speculated that the IL-12 production observed in these early studies might be due to the presence of contaminating CD11c+ cDC (52). We directly addressed this possibility by separating CX3CR1+CD33+CD123+CD303+CD304+ pre-DC from CX3CR1−CD33−CD123+CD303+CD304+ “pure” pDC and showing that pDC could not polarize or induce proliferation of naïve CD4 T cells, whereas pre-DC had this capacity, and that pDC were unable to produce IL-12, unlike pre-DC, but were the only cells capable of strongly producing TNF-α in response to TRL7/8/9 agonists, as initially described (53). Thus, it is of paramount importance that pre-DC be excluded from pDC populations in future studies, particularly when using commercial pDC isolation kits. Finally, if pDC are stripped of all their cDC properties, it raises the question as to whether they truly belong to the DC lineage or rather are a distinct type of innate IFN-I-producing lymphoid cell. It also remains to be shown whether the BM CD34+CD123hi CDP population is also a mixture of independent bona fide cDC progenitors and pDC progenitors.
Despite their classification as precursors, human pre-DC appear functional in their own right, being equipped with some T cell costimulatory molecules and with a strong capacity for naïve T cell allostimulation and cytokine secretion in response to TLR stimulation (Figs. 2 and 5 and figs. S7 and S19). Pre-DC produced low levels of TNF-α in response to CpG ODN 2216 exposure and secreted IL-12 and TNF-α in response to various TLR ligands. Hence, it is reasonable to propose that pre-DC have the potential to contribute to both homeostasis and various pathological processes, particularly inflammatory and autoimmune diseases, where dysregulation of their differentiation continuum or their arrested development could render them a potent source of inflammatory DC ready for rapid recruitment and mobilization.
Beyond the identification of pre-DC, our data revealed previously unappreciated transcriptional and phenotypic heterogeneity within the circulating mature DC populations. This was particularly clear in the case of cDC2 and pDC, which were grouped into multiple Mpath clusters in the single-cell RNAseq analysis and showed marked dispersion in the tSNE analysis of the CyTOF data with phenotypic heterogeneity. Isomap analysis of the CyTOF data also revealed another level of pDC heterogeneity by illustrating the progressive phenotypic transition from CDP into CD2+ pDC in the BM, involving intermediate cells that could be pre-pDC. Whether a circulating pre-pDC population exists remains to be concluded. Finally, defining the mechanisms that direct the differentiation of uncommitted pre-DC into cDC1 or cDC2 or that maintain these cells in their initial uncommitted state in health and disease could lead to the development of new therapeutic strategies to modulate this differentiation process.
Materials and methods
Blood, bone marrow, and spleen samples
Human samples were obtained in accordance with a favorable ethical opinion from Singapore SingHealth and National Health Care Group Research Ethics Committees. Written informed consent was obtained from all donors according to the procedures approved by the National University of Singapore Institutional Review Board and SingHealth Centralised Institutional Review Board. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Paque (GE Healthcare) density gradient centrifugation of apheresis residue samples obtained from volunteer donors through the Health Sciences Authorities (HSA, Singapore). Blood samples were obtained from 4 patients with molecularly confirmed Pitt-Hopkins syndrome (PHS), who all showed the classical phenotype (54). Spleen tissue was obtained from patients with tumors in the pancreas who underwent distal pancreatomy (Singapore General Hospital, Singapore). Spleen tissue was processed as previously described (23). Bone marrow mononuclear cells were purchased from Lonza.
Generation of single-cell transcriptomes using MARS-seq
MARS-Seq using the Biomek FXP system (Beckman Coulter) as previously described (15)was performed for scmRNAseq of the DC compartment of the human peripheral blood. In brief, lineage marker (Lin)(CD3/14/16/19/20/34)−CD45+CD135+HLA-DR+ CD123+CD33+ single cells were sorted into individual wells of 384-well plates filled with 2 μl lysis buffer (Triton 0.2% (Sigma Aldrich) in molecular biology grade H2O (Sigma Aldrich), supplemented with 0.4 U/μl protein-based RNase inhibitor (Takara Bio Inc.), and barcoded using 400 nM IDT. Details regarding the barcoding procedure with poly-T primers were previously described (15). Samples were pre-incubated for 3 min at 80°C and reverse transcriptase mix consisting of 10 mM DTT (Invitrogen), 4 mM dNTPs (NEB), 2.5 U/μl SuperScript III Reverse Transcriptase (Invitrogen) in 50 mM Tris-HCl (pH 8.3; Sigma), 75 mM KCl (Sigma), 3 mM MgCl2 (Sigma), ERCC RNA Spike-In mix (Life Technologies), at a dilution of 1:80 × 107 per cell was added to each well. The mRNA was reverse-transcribed to cDNA with one cycle of 2 min at 42°C, 50 min at 50°C, and 5 min at 85°C. Excess primers were digested with ExoI (NEB) at 37°C for 30 min then 10 min at 80°C, followed by cleanup using SPRIselect beads at a 1.2x ratio (Beckman Coulter). Samples were pooled and second strands were synthesized using a Second Strand Synthesis kit (NEB) for 2.5 hours at 16°C, followed by a cleanup using SPRIselect beads at a 1.4x ratio (Beckman Coulter). Samples were linearly amplified by T7-promoter guided in vitro transcription using the T7 High Yield RNA polymerase IVT kit (NEB) at 37°C for 12 hours. DNA templates were digested with Turbo DNase I (Ambion) for 15 min at 37°C, followed by a cleanup with SPRIselect beads at a 1.2x ratio (Beckman Coulter). The RNA was then fragmented in Zn2+ RNA Fragmentation Solution (Ambion) for 1.5 min at 70°C, followed by cleanup with SPRIselect beads at a 2.0 ratio (Beckman Coulter). Barcoded ssDNA adapters [IDT; details of barcode, see (15)] were then ligated to the fragmented RNAs in 9.5% DMSO (Sigma Aldrich), 1 mM ATP, 20% PEG8000 and 1 U/μl T4 RNA ligase I (NEB) solution in 50 mM Tris HCl pH7.5 (Sigma Aldrich), 10 mM MgCl2 and 1 mM DTT for 2 hours at 22°C. A second reverse transcription reaction was then performed using Affinity Script Reverse Transcription buffer, 10 mM DTT, 4 mM dNTP, 2.5 U/μl Affinity Script Reverse Transcriptase (Agilent) for one cycle of 2 min at 42°C, 45 min at 50°C, and 5 min at 85°C, followed by a cleanup on SPRIselect beads at a 1.5x ratio (Beckman Coulter). The final libraries were generated by subsequent nested PCR reactions using 0.5 μM of each Illumina primer [IDT; details of primers, see (15)] and KAPA HiFi HotStart Ready Mix (Kapa Biosystems) for 15 cycles according to manufacturer’s protocol, followed by a final cleanup with SPRIselect beads at a 0.7x ratio (Beckman Coulter). The quality and quantity of the resulting libraries was assessed using an Agilent 2200 TapeStation instrument (Agilent), and libraries were subjected to next generation sequencing using an Illumina HiSeq1500 instrument [PE no index; read1:61 reads (3 reads random nucleotides, 4 reads pool barcode, 53 reads sequence), read2:13 reads (6 reads cell barcode, 6 reads unique molecular identifier)].
Preprocessing, quality assessment, and control of MARS-seq single-cell transcriptome data
Cell specific tags and Unique Molecular Identifiers (UMIs) were extracted (2,496 cells sequenced) from sequenced data-pool barcodes. Sequencing reads with ambiguous plate and/or cell-specific tags, UMI sequences of low quality (Phred < 27), or reads that mapped to E. coli were eliminated using Bowtie1 sequence analysis software (55), with parameters “-M1 -t-best-chunkmbs 64 -strata.” Fastq files were demultiplexed using the fastx_ barcode_splitter from fastx_toolkit, and R1 reads (with trimming of pooled barcode sequences) were mapped to the human hg38 + ERCC pseudo genome assembly using Bowtie “-m 1 -t-best-chunkmbs 64 -strata.” Valid reads were then counted using UMIs if they mapped to the exonbased gene model derived from the BiomaRt HG38 data mining tool provided by Ensembl (56). A gene expression matrix was then generated containing the number of UMIs for every cell and gene. Additionally, UMIs and cell barcode errors were corrected and filtered as previously described (15).
Normalization and filtering of MARS-seq single-cell transcriptome data
In order to account for differences in total molecule counts per cell, we performed a down-sampling normalization as suggested by several studies (15,57). Here, we randomly down-sampled every cell to a molecule count of 1,050 unique molecules per cell (threshold details discussed below). Cells with molecule counts <1,050 were excluded from the analysis [table S1 (18)]. Additionally, cells with a ratio of mitochondrial versus endogenous genes exceeding 0.2, and cells with <90 unique genes, were removed from the analysis. Prior to Seurat analysis (58), expression tables were filtered to exclude mitochondrial and ribosomal genes to remove noise. The validation of down sampling threshold for normalization of MARS-seq single-cell transcriptome data are detailed in the supplementary methods.
Analysis of MARS-seq single-cell transcriptome data
Analysis of the normalized and filtered singlecell gene expression data (8,657 genes across 710 single-cell transcriptomes used in the final expression table) was achieved using Mpath (26), PCA, tSNE, connectivity map (cmap) (25) and several functions of the Seurat single-cell analysis package. cmap analysis was performed using DEG between pDC and cDC derived from the Gene Expression Omnibus data series GSE35457 (23). For individual cells, cmap generated enrichment scores that quantified the degree of enrichment (or “closeness”) to the given gene signatures. The enrichment scores were scaled and assigned positive or negative values to indicate enrichment for pDC or cDC signature genes, respectively. A permutation test (n = 1,000) between gene signatures was performed on each enrichment score to determine statistical significance. For the tSNE/Seurat analysis, a Seurat filter was used to include genes that were detected in at least one cell (molecule count = 1), and excluded cells with <90 unique genes. To infer the structure of the single-cell gene expression data, a PCA was performed on the highly variable genes determined as genes exceeding the dispersion threshold of 0.75. The first two principle components were used to perform a tSNE that was combined with a DBSCAN clustering algorithm (22) to identify cells with similar expression profiles. DBSCAN was performed by setting 10 as the minimum number of reachable points and 4.1 as the reachable epsilon neighborhood parameter; the latter was determined using a KNN plot integrated in the DBSCAN R package (59) (https://cran.r-project.org/web/packages/dbscan/). The clustering did not change when using the default minimal number of reachable points.
To annotate the clusters, we used the gene signatures of blood pDC, cDC1 and cDC2 derived from the Gene Expression Omnibus data series GSE35457 (23) [table S2 (24), data processing described below] to calculate the signature gene expression scores of cell type-specific gene signatures, and then overlaid these signature scores onto the tSNE plots. Raw expression data of CD141+ (cDC1), CD1c+ (cDC2) DCs and pDC samples from blood of up to four donors (I, II, V and VI) was imported into Partek Genomics Suite software, version 6.6 Copyright; 2017 (PGS), where they were further processed. Data were quantile-normalized and log2-transformed, and a batch-correction was performed for the donor using PGS. Differential probe expression was calculated from the normalized data (ANOVA, Fold-Change ≥ 2 and FDR-adj. P value < 0.05) for the three comparisons of every cell type against the remaining cell types. The three lists of differentially expressed (DE) probes were intersected and only exclusively expressed probes were used for the cell-type specific gene signatures. The probes were then reduced to single genes, by keeping the probe for a corresponding gene with the highest mean expression across the dataset. Resulting gene signatures for blood pDCs, CD1c+ and CD141+ DCs contained 725, 457 and 368 genes, respectively. The signature gene expression score was calculated as the mean expression of all signature genes in a cluster. In order to avoid bias due to outliers, we calculated the trimmed mean (trim = 0.08).
Monocle analysis was performed using the latest prepublished version of Monocle v.2.1.0 (27). The data were loaded into a monocle object and then log-transformed. Ordering of the genes was determined by dispersion analysis if they had an average expression of ≥0.5 and at least a dispersion of two times the dispersion fit. The dimensionality reduction was performed using the reduceDimension command with parameters max_components = 2, reduction_method = “DDRTree” and norm_ method = “log.” The trajectory was then built using the plot_cell_trajectory command with standard parameters.
Wishbone analysis (28) was performed using the Python toolkit downloaded from https://github.com/ManuSetty/wishbone. MARS-seq data were loaded using the wishbone.wb.SCData. from_csv function with the parameters data_ type = ‘sc-seq’ and normalize = True. Wishbone was then performed using wb.run_wishbone function with parameter start_cell = “run1_ CATG_AAGACA”, components_list = [1, 2, 3, 4], num_waypoints = 150, branch = True. Start_cell was randomly selected from the central cluster #4. Diffusion map analysis was performed using the scdata.run_diffusion_map function with default parameters (29). Wishbone revealed three trajectories giving rise to pDC, cDC1 and cDC2 respectively. Along each trajectory, the respective signature gene shows increasing expression (fig. S2c). Although Wishbone results might be interpreted to suggest that cDC2 are early cells and differentiate into pDC and cDC1 on two separate branches, this is simply because Wishbone allows a maximum of two branches and assumes all cells fall on continuous trajectories. Nevertheless, it is able to delineate the three trajectories that are in concordance with Mpath, monocle, and diffusion map analysis.
C1 single-cell mRNA sequencing
Lin(CD3/14/16/19/20)−HLA-DR+CD33+CD123+cells at 300 cells/μl were loaded onto two 5-10 μm C1 Single-Cell Auto Prep integrated fluidic circuits (Fluidigm) and cell capture was performed according to the manufacturer’s instructions. Individual capture sites were inspected under a light microscope to confirm the presence of single, live cells. Empty capture wells and wells containing multiple cells or cell debris were discarded for quality control. A SMARTer Ultra Low RNA kit (Clontech) and Advantage 2 PCR Kit (Clontech) was used for cDNA generation. An ArrayControl RNA Spots and Spikes kit (with spike numbers 1, 4 and 7) (Ambion) was used to monitor technical variability, and the dilutions used were as recommended by the manufacturer. The concentration of cDNA for each single cell was determined by Quant-iT PicoGreen dsDNA Reagent, and the correct size and profile was confirmed using DNA High Sensitivity Reagent Kit and DNA Extended Range LabChip (Perkin Elmer). Multiplex sequencing libraries were generated using the Nextera XT DNA Library Preparation Kit and the Nextera XT Index Kit (Illumina). Libraries were pooled and subjected to an indexed PE sequencing run of 2 × 51 cycles on an Illumina HiSeq 2000 (Illumina) at an average depth of 2.5-million row reads per cell.
C1 single-cell analysis
Raw reads were aligned to the human reference genome GRCh38 from GENCODE (60) using RSEM program version 1.2.19 with default parameters (61). Gene expression values in transcripts per million were calculated using the RSEM program and the human GENCODE annotation version 22. Quality control and outlier cell detection was performed using the SINGuLAR (Fluidigm) analysis toolset. cmap analysis was performed using cDC1 versus cDC2 DEG identified from Gene Expression Omnibus data series GSE35457 (23), and the enrichment scores were obtained. Similar to the gene set enrichment analyses, cmap was used to identify associations of transcriptomic profiles with cell-type characteristic gene signatures.
Mpath analysis of MARS or C1 single-cell mRNA sequencing data
Developmental trajectories were defined using the Mpath algorithm (26), which constructs multibranching cell lineages and reorders individual cells along the branches. In the analysis of the MARS-seq single-cell transcriptomic data, we first used the Seurat R package to identify five clusters: For each cluster, Mpath calculated the centroid and used it as a landmark to represent a canonical cellular state; subsequently, for each single cell, Mpath calculated its Euclidean distance to all the landmarks, and identified the two nearest landmarks. Each individual cell was thus assigned to the neighborhood of its two nearest landmarks. For every pair of landmarks, Mpath then counted the number of cells that were assigned to the neighborhood, and used the determined cell counts to estimate the possibility of the transition between landmarks to be true. A high cell count implied a high possibility that the transition was valid. Mpath then constructed a weighted neighborhood network whereby nodes represented landmarks, edges represented a putative transition between landmarks, and numbers allocated to the edges represented the cell-count support for the transition. Given that single-cell transcriptomic data tend to be noisy, edges with low cell-count support were considered likely artifacts. Mpath therefore removed the edges with low cell count support. n represents cell count on the edge, and -n represents the distance between nodes. By applying a minimum spanning tree algorithm, Mpath fíndsthe shortest path that connectsall nodeswith the minimum sum of distance. Consequently, the resulting trimmed network is the one that connects all landmarks with the minimum number of edges and the maximum total number of cells on the edges. Mpath was then used to project the individual cells onto the edge connecting its two nearest landmarks, and assigned a pseudo-time ordering to the cells according to the location of their projection points on the edge. In the analysis of the C1 single-cell transcriptome data, we first used the cmap analysis to identify cDC1-primed, unprimed, and cDC2-primed clusters, and then used Mpath to construct the lineage between these three clusters. The Mpath analysis was carried out in an unsupervised manner without prior knowledge of the starting cells or number of branches. This method can be used for situations of nonbranching networks, bifurcations, and multibranching networks with three or more branches.
Mass cytometry staining, barcoding, acquisition, and data analysis
For mass cytometry, preconjugated or purified antibodies were obtained from Invitrogen, Fluidigm (preconjugated antibodies), Biolegend, eBioscience, Becton Dickinson, or R&D Systems as listed in table S3. For some markers, fluorophore- or biotin-conjugated antibodies were used as primary antibodies, followed by secondary labeling with anti-fluorophore metal-conjugated antibodies (such as the anti-FITC clone FIT-22) or metal-conjugated streptavidin, produced as previously described (19). Briefly, 3 × 106 cells per well in a U-bottom 96 well plate (BD Falcon, Cat# 3077) were washed once with 200 μL FACS buffer (4% FBS, 2mM EDTA, 0.05% Azide in 1X PBS), then stained with 100 μL 200 μM cisplatin (Sigma-Aldrich, Cat# 479306-1G) for 5 min on ice to exclude dead cells. Cells were then incubated with anti-CADM1-biotin and anti-CD19-FITC primary antibodies in a 50 μL reaction for 30 min on ice. Cells were washed twice with FACS buffer and incubated with 50 μL heavy-metal isotopeconjugated secondary mAb cocktail for 30 min on ice. Cells were then washed twice with FACS buffer and once with PBS before fixation with 200 μL 2% paraformaldehyde (PFA; Electron Microscopy Sciences, Cat# 15710) in PBS overnight or longer. Following fixation, the cells were pelleted and resuspended in 200 uL 1X permeabilization buffer (Biolegend, Cat# 421002) for 5 min at room temperature to enable intracellular labeling. Cells were then incubated with metal-conjugated anti-CD68 in a 50 μL reaction for 30 min on ice. Finally, the cells were washed once with permeabilization buffer and then with PBS before barcoding.
Bromoacetamidobenzyl-EDTA (BABE)-linked metal barcodes were prepared by dissolving BABE (Dojindo, Cat# B437) in 100 mM HEPES buffer (Gibco, Cat# 15630) to a final concentration of 2 mM. Isotopically purified PdCl2 (Trace Sciences Inc.) was then added to the 2 mM BABE solution to a final concentration of 0.5 mM. Similarly, DOTA-maleimide (DM)-linked metal barcodes were prepared by dissolving DM (Macrocyclics, Cat# B-272) in L buffer (MAXPAR, Cat# PN00008) to a final concentration of 1 mM. RhCl3 (Sigma) and isotopically purified LnCl3 was then added to the DM solution at 0.5 mM final concentration. Six metal barcodes were used: BABE-Pd-102, BABE-Pd-104, BABE-Pd-106, BABE-Pd-108, BABE-Pd-110 and DM-Ln-113.
All BABE and DM-metal solution mixtures were immediately snap-frozen in liquid nitrogen and stored at -80°C. A unique dual combination of barcodes was chosen to stain each tissue sample. Barcode Pd-102 was used at 1:4000 dilution, Pd-104 at 1:2000, Pd-106 and Pd-108 at 1:1000, Pd-110 and Ln-113 at 1:500. Cells were incubated with 100 μL barcode in PBS for 30 min on ice, washed in permeabilization buffer and then incubated in FACS buffer for 10 min on ice. Cells were then pelleted and resuspended in 100 μL nucleic acid Ir-Intercalator (MAXPAR, Cat# 201192B) in 2% PFA/PBS (1:2000), at room temperature. After 20 min, cells were washed twice with FACS buffer and twice with water before a final resuspension in water. In each set, the cells were pooled from all tissue types, counted, and diluted to 0.5 × 106 cells/mL. EQ Four Element Calibration Beads (DVS Science, Fluidigm) were added at a 1% concentration prior to acquisition. Cell data were acquired and analyzed using a CyTOF Mass cytometer (Fluidigm).
The CyTOF data were exported in a conventional flow-cytometry file (.fcs) format and normalized using previously described software (62). Events with zero values were randomly assigned a value between 0 and -1 using a custom R script employed in a previous version of mass cytometry software (63). Cells for each barcode were deconvolved using the Boolean gating algorithm within FlowJo. The CD45+Lin (CD7/CD14/CD15/CD16/CD19/CD34)−HLA-DR+ population of PBMC were gated using FlowJo and exported as a .fcs file. Marker expression values were transformed using the logicle transformation function (64). Random subsampling without replacement was performed to select 20,000 cell events.The transformedvalues of subsampled cell events were then subjected to t-distributed Stochastic Neighbor Embedding (tSNE) dimension reduction (21) using the markers listed in table S3, and the Rtsne function in the Rtsne R package with default parameters. Similarly, isometric feature mapping (Isomap) (43) dimension reduction was performed using vegdist, spantree, and isomap functions in the vegan R package (65).
The vegdist function was run with method = “euclidean.” The spantree function was run with default parameters. The Isomap function was run with ndim equal to the number of original dimensions of input data, and k = 5. Phenograph clustering (30) was performed using the markers listed in table S3 before dimension reduction, and with the number of nearest neighbors equal to 30. The results obtained from the tSNE, Isomap and Phenograph analyses were incorporated as additional parameters in the .fcs files, which were then loaded into FlowJo to generate heat plots of marker expression on the reduced dimensions. The above analyses were performed using the cytofkit R package which provides a wrapper of existing state-of-the-art methods for cytometry data analysis (66).
Human cell flow cytometry: Labeling, staining, analysis, and cell sorting
All antibodies used for fluorescence-activated cell sorting (FACS) and flow cytometry were mouse anti-human monoclonal antibodies (mAbs), except for chicken anti-human CADM1 IgY primary mAb. The mAbs and secondary reagents used for flow cytometry are listed in table S6. Briefly, 5 × 106 cells per tube were washed and incubated with Live/Dead blue dye (Invitrogen) for 30 min at 4 C in phosphate buffered saline (PBS) and then incubated in 5% heat-inactivated fetal calf serum (FCS) for 15 min at 4 °C (Sigma Aldrich). The appropriate antibodies diluted in PBS with 2% fetal calf serum (FCS) and 2 mM EDTA were added to the cells and incubated for 30 min at 4 °C, and then washed and detected with the secondary reagents. For intra-cytoplasmic or intra-nuclear labeling or staining, cells were fixed and perme-abilized with BD Cytofix/Cytoperm (BD Biosciences) or with eBioscience FoxP3/Transcription Factor Staining Buffer Set (eBioscience/Affimetrix), respectively according to the manufacturer’s instructions. Flow cytometry was performed using a BD LSRII or a BD FACSFortessa (BD Biosciences) and the data analyzed using BD FACSDiva 6.0 (BD Biosciences) or FlowJo v.10 (Tree Star Inc.). For isolation of precursor dendritic cells (pre-DC), PBMC were first depleted of T cells, monocytes and B cells with anti-CD3, anti-CD14 and anti-CD20 microbeads (Miltenyi Biotec) using an AutoMACS Pro Separator (Miltenyi Biotec) according to the manufacturer’s instructions. FACS was performed using a BD FACSAriaII or BD FACSAriaIII (BD Biosciences). Wanderlust analysis (41) of flow cytometry data was performed using the CYT tool downloaded from https://www.c2b2.columbia.edu/danapeerlab/html/cyt-download.html. As Wanderlust requires users to specify a starting cell, we selected one cell at random from the CD45RA+ CD123+ population.
Cytospin and scanning electron microscopy
Cytospins were prepared from purified cells and stained with the Hema 3 system according to the manufacturer’s protocol (Fisher Diagnostics). Images were analyzed at 100X magnification with an Olympus BX43 upright microscope (Olympus). Scanning electron microscopy was performed as previously described (23).
Dendritic cell (DC) differentiation coculture assay on MS-5 stromal cells
MS-5 stromal cells were maintained and passaged as previously described (8). MS-5 cells were seeded in 96-well round-bottom plates (Corning) at 3,000 cells per well in complete alpha-Minimum Essential Media (a-MEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Serana) and 1% penicillin/streptomycin (Nacalai Tesque). A total of5,000 sorted purified cells were added 18-24 hours later, in medium containing 200 ng/mL Flt3L (Miltenyi Biotec), 20 ng/mL SCF (Miltenyi Biotec), and 20 ng/mL GM-CSF (Miltenyi Biotec), and cultured for up to 5 days. The cells were then resuspended in their wells by physical dissociation and filtered through a cell strainer into a polystyrene FACS tube.
Intracellular cytokine detection following stimulation with TLR ligands
A total of 5 × 106 PBMC were cultured in Roswell Park Memorial Institute (RPMI)-1640 Glutmax media (Life Technologies) supplemented with 10% FBS, 1% penicillin/streptomycin and stimulated with either lipopolysaccharide (LPS, 100 ng/mL; InvivoGen), LPS (100 ng/mL) + interferon gamma (IFNγ, 1,000 U/mL; R&D Systems), Flagellin (100 ng/mL, Invivogen), polyI:C (10 μg/mL; InvivoGen), Imidazoquinoline (CL097; Invivogen) or CpG oligodeoxynucleotides 2216 (ODN, 5 μM; InvivoGen) for 2 hours, after which 10 μg/ml Brefeldin A solution (eBioscience) was added and the cells were again stimulated for an additional 4 hours. After the 6 hours stimulation, the cells were labeled with cytokine-specific antibodies and analyzed by flow cytometry, as described above.
Mixed lymphocyte reaction
Naïve T cells were isolated from PBMC using Naïve Pan T-Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer’s instructions, and labeled with 0.2 μM carboxyfluorescein succinimidyl ester (CFSE) (Life Technologies) for 5 min at 37°C. A total of5,000 cells from sorted DC subsets were cocultured with 100,000 CFSE-labeled naïve T cells for 7 days in Iscove’s Modified Dulbecco’s Medium (IMDM; Life Technologies) supplemented with 10% KnockOut Serum Replacement (Life Technologies). On day 7, the T cells were stimulated with 10 μg/ml phorbol myristate acetate (InvivoGen) and 500 μg/ml ionomycin (Sigma Aldrich) for 1 hour at 37°C. 10 μg/ml Brefeldin A solution was added for 4 hours, after which the cells were labeled with cytokine-specific antibodies and analyzed by flow cytometry, as described above.
Electron microscopy
Sorted cells were seeded on poly-lysine-coated coverslips for 1 hour at 37°C. The cells were then fixed in 2% glutaraldehyde in 0.1 M cacoldylate buffer, pH 7.4 for 1 hour, post fixed for 1 hour with 2% buffered osmium tetroxide, then dehydrated in a graded series of ethanol solutions, before embedding in epoxy resin. Images were acquired with a Quemesa (SIS) digital camera mounted on a Tecnai 12 transmission electron microscope (FEI Company) operated at 80 kV.
Microarray analysis
Total RNA was isolated from FACS-sorted blood pre-DC and DC subsets using a RNeasy Micro kit (Qiagen). Total RNA integrity was assessed using an Agilent Bioanalyzer (Agilent) and the RNA Integrity Number (RIN) was calculated. All RNA samples had a RIN ≥7.1. Biotinylated cRNA was prepared using an Epicentre TargetAmp 2-Round Biotin-aRNA Amplification Kit 3.0 according to the manufacturer’s instructions, using 500 pg of total RNA starting material. Hybridization of the cRNA was performed on an Illumina Human-HT12 Version 4 chip set (Illumina). Microrarray data were exported from GenomeStudio (Illumina) without background subtraction. Probes with detection P values > 0.05 were considered as not being detected in the sample, and were filtered out. Expression values for the remaining probes were log2 transformed and quantile normalized. For DEG analysis, comparison of one cell subset with another was carried out using the limma R software package (67) with samples paired by donor identifiers. DEG were selected with Benjamini-Hochberg multiple testing (68) corrected P value < 0.05. In this way, limma was used to select up and down-regulated signature genes for each of the cell subsets in the pre-DC data by comparing one subset with all other subsets pooled as a group. Expression profiles shown in Fig. 4E were from 62 common genes identified from the union of DEG from comparing pre-cDC1 versus early pre-DC and cDC1 versus pre-cDC1, and the union of DEG from comparing pre-cDC2 versus early pre-DC and cDC2 versus pre-cDC2 [table S5 (69) and fig. S17(70)].
Luminex drop array assay on sorted and stimulated pre-DC and DC populations
A total of 2,000 cells per well of sorted pre-DC and DC subsets were seeded in V-bottom 96 well plates and then incubated for 18 hours in 50 μL complete RPMI-1640 Glutmax media (Life Technologies) supplemented with 10% FBS and 1% penicillin/streptomycin, and stimulated with either LPS, LPS + IFNγ, Flagellin, polyI:C, Imidazoquinoline or CpG oligodeoxynucleotides (ODN) 2216. Cells were then pelleted and 30 μL supernatant was collected. A Luminex Drop Array was performed using 5 μL of the supernatant. Human G-CSF, GM-CSF, TNF-α2, IL-10, IL-12p40, IL-12p70, IL-15, IL-1RA, IL-1a, IL-1b, IL-6, IL-7, IL-8, MIP-1b, TNF-a, and TNF-ß were tested by multiplexing (EMD Millipore) with DropArray-bead plates (Curiox) according to the manufacturer’s instructions. Acquisition was performed using xPONENT 4.0 (Luminex) acquisition software, and data analysis was performed using Bio-Plex Manager 6.1.1 (Bio-Rad).
Statistical analyses
The Mann-Whitney test was used to compare data derived from patients with Pitt-Hopkins Syndrome and controls and the intracellular detection of IL-12p40 and TNF-α in pre-DC stimulated with LPS or poly I:C versus CpG ODN 2216. The Kruskal-Wallis test, followed by the Dunn’s multiple comparison test, was used to compare the expression level of individual genes in single cells in the MARS-seq single-cell RNAseq dataset. Differences were defined as statistically significant when adjusted P < 0.05. All statistical tests were performed using GraphPad Prism 6.00 for Windows (GraphPad Software). Correlation coefficients were calculated as Pearson’s correlation coefficient.
Supplementary Material
Introduction
Dendritic cells (DC) are professional antigen-presenting cells that orchestrate immune responses. The human DC population comprises multiple subsets, including plasmacytoid DC (pDC) and two functionally specialized lineages of conventional DC (cDC1 and cDC2), whose origins and differentiation pathways remain incompletely defined.
Rationale
As DC are essential regulators of the immune response in health and disease, potential intervention strategies aiming at manipulation of these cells will require in-depth insights of their origins, the mechanisms that govern their homeostasis, and their functional properties. Here, we employed two unbiased high-dimensional technologies to characterize the human DC lineage from bone marrow to blood.
Results
We isolated the DC-containing population (Lineage−HLA−DR+CD135+ cells) from human blood and defined the transcriptomes of 710 individual cells using massively parallel single-cell mRNA sequencing. By combining complementary bioinformatic approaches, we identified a small cluster of cells within this population as putative DC precursors (pre-DC). We then confirmed this finding using cytometry by time-of-flight (CyTOF) to simultaneously measure the expression of a panel of 38 different proteins at the single-cell level on Lineage−HLA−DR+ cells and found that pre-DC possessed a CD123+CD33+CD45RA+ phenotype. We confirmed the precursor potential of pre-DC by establishing their potential to differentiate in vitro into cDC1 and cDC2, but not pDC, in the known proportions found in vivo.
Interestingly, pre-DC also express classical pDC markers, including CD123, CD303, and CD304. Thus, any previous studies using these markers to identify or isolate pDC will have inadvertently included CD123+CD33+ pre-DC. We provide here new markers that can be used to identify unambiguously pre-DC from pDC, including CD33, CX3CR1, CD2, CD5, and CD327. When CD123+CD33+pre-DC andCD123+CD33− pDC were isolated separately, we ob served that pre-DC have unique functional properties that were previously attributed to pDC. Although pDC remain bona fide interferon-α-producing cells, their reported interleukin-12 (IL-12) production and CD4 T cell allostimulatory capacity can likely be attributed to “contaminating” pre-DC.
Human DC emerge from BM CDP, diverge at the point of emergence of pre-DC and pDC potential, and culminate in maturation of both lineages in the blood.
The pre-DC compartment further differentiates into functionally and phenotypically distinct lineage-committed subpopulations, including one early uncommitted CD123+ pre-DC subset (early pre-DC), which give rise to both cDC1 and cDC2 through corresponding CD45RA+CD123b pre-cDC1 and pre-cDC2 lineage-committed subsets, respectively.
We then asked whether the pre-DC population contained both uncommitted and committed pre-cDC1 and pre-cDC2 precursors, as recently shown in mice. Using microfluidic single-cell mRNA sequencing (scmRNAseq), we showed that the human pre-DC population contains cells exhibiting transcriptomic priming toward cDC1 and cDC2 lineages. Flow cytometry and in vitro DC differentiation experiments further identified CD123+CADM1−CD1c− putative uncommitted pre-DC, alongside CADM1+CD1c− pre-cDC1 and CADM1−CD1c+ pre-cDC2. Finally, we found that pre-DC subsets expressed T cell costimulatory molecules and induced comparable proliferation and polarization of naïve CD4 T cells as adult DC. However, exposure to the Toll-like receptor 9 (TLR9) ligand CpG triggered IL-12p40 and tumor necrosis factor-a production by early pre-DC, pre-cDC1, and pre-cDC2, in contrast to differentiated cDC1 and cDC2, which do not express TLR9.
Conclusion
Using unsupervised scmRNAseq and CyTOF analyses, we have unraveled the complexity of the human DC lineage at the single-cell level, revealing a continuous process of differentiation that starts in the bone marrow (BM) with common DC progenitors (CDP), diverges at the point of emergence of pre-DC and pDC potential, and culminates in maturation of both lineages in the blood and spleen. The pre-DC compartment contains functionally and phenotypically distinct lineage-committed subpopulations, including one early uncommitted CD123+ pre-DC subset and two CD45RA+CD123lo lineage-committed subsets. The discovery of multiple committed pre-DC populations with unique capabilities opens promising new avenues for the therapeutic exploitation of DC subset-specific targeting.
Dendritic cells (DC) are professional antigen-presenting cells that orchestrate immune responses. The human DC population comprises two main functionally specialized lineages, whose origins and differentiation pathways remain incompletely defined. Here, we combine two high-dimensional technologies—single-cell messenger RNA sequencing (scmRNAseq) and cytometry by time-of-flight (CyTOF)—to identify human blood CD123+CD33+CD45RA+ DC precursors (pre-DC). Pre-DC share surface markers with plasmacytoid DC (pDC) but have distinct functional properties that were previously attributed to pDC. Tracing the differentiation of DC from the bone marrow to the peripheral blood revealed that the pre-DC compartment contains distinct lineage-committed subpopulations, including one early uncommitted CD123high pre-DC subset and two CD45RA+CD123low lineage-committed subsets exhibiting functional differences. The discovery of multiple committed pre-DC populations opens promising new avenues for the therapeutic exploitation of DC subset-specific targeting.
Acknowledgments
We thank L. Robinson of Insight Editing London for critical review and editing of the manuscript; P. Y. J. Ai from the SingHealth Flow Cytometry Core Platform; and M. L. Ng, S. H. Tan, and T. B. Lu from the Electron Microscope Unit of NUS for their assistance. This work was supported by Singapore Immunology Network core funding (F.G. and E.W.N.); Agency for Science, Technology and Research (A*STAR), Singapore; the A*STAR Graduate Scholarship (P.S.); the Wellcome Trust (WT 107931/Z/15/Z) (M.H.); and the National Research Foundation Singapore under its cooperative basic research grant new investigator grant (NMRC/BNIG/2026/2014) and administered by the Singapore Ministry of Health’s National Medical Research Council (C.-A.D). This work was supported by the French National Research Agency through the “Investments for the Future” program (France-BioImaging, ANR-10-INSB-04), LABEX DCBIOL (ANR-10-IDEX-0001-02 PSL and ANR-11-LABX-0043), and grants from Agence Nationale de Recherche contre le SIDA et les hépatites virales (ANRS) (P.B., N.R., M.J., and E.G.-M.). J.L.S., M.B., and A.S. are members of the Excellence Cluster ImmunoSensation. J.L.S. is funded by Sonderforschungsbereich 645 and 704. A.S. is funded by an Emmy-Noether fellowship (SCHL 2116/1-1) of the German Research Foundation and a Young Investigator Award of the Biomedical Research Council Singapore. F.G. and P.S. are inventors on patent application 10201607246S held by A*STAR, which covers the methods for the identification, targeting, and isolation of human dendritic cell (DC) precursors (“pre-DC”) and their use as biomarkers of inflammatory diseases. The MARS-seq, microfluidic scmRNAseq, and microarray data sets are deposited in the Gene Expression Omnibus under accession numbers GSE98052, GSE98011, and GSE80171, respectively.
References and Notes
- 1.Schlitzer A, McGovern N, Ginhoux F. Dendritic cells and monocyte-derived cells: Two complementary and integrated functional systems. Semin Cell Dev Biol. 2015;41:9–22. doi: 10.1016/Jsemcdb.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guilliams M, et al. Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginhoux F, et al. The origin and development of nonlymphoid tissue CD103+DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onai N, et al. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 2013;38:943–957. doi: 10.1016/Jimmuni.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Schlitzer A, et al. Identification of cDC1-and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 2015;16:718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med. 2015;212:385–399. doi: 10.1084/jem.20141442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breton G, et al. Circulating precursors of human CD1c+and CD141+dendritic cells. J Exp Med. 2015;212:401–413. doi: 10.1084/jem.20141441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cella M, et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 11.Grouard G, et al. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J Exp Med. 1997;185:1101–1112. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 13.Griffin JD, et al. Differential expression of HLA-DR antigens in subsets of human CFU-GM. Blood. 1985;66:788–795. [PubMed] [Google Scholar]
- 14.Dzioneketal A. BDCA-2, BDCA-3, and BDCA-4: Three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol165.11.6037. [DOI] [PubMed] [Google Scholar]
- 15.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorting strategy from total Lin−HLA-DR+CD135+cells for MARS-seq [Google Scholar]
- 17.Workflow and quality control of MARS-seq single-cell data analysis [Google Scholar]
- 18.Number of detected genes per cell in the total DC MARS-seq experiment [Google Scholar]
- 19.Becher B, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 20.Amir AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Maaten L. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2625. [Google Scholar]
- 22.Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. 1996;1996:226–231. [Google Scholar]
- 23.Haniffa M, et al. Human tissues contain CD141hicrosspresenting dendritic cells with functional homology to mouse CD103+nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/Jimmuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DC subset signature genes derived from Gene Expression Omnibus data series GSE35457 and used for MARS-seq and C1 data analyses [Google Scholar]
- 25.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Schlitzer A, Chakarov S, Ginhoux F, Poidinger M. Mpath maps multi-branching single-cell trajectories revealing progenitor cell progression during development. Nat Commun. 2016;7:11988. doi: 10.1038/ncomms11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setty M, et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coifman RR, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: Multiscale methods. Proc Natl Acad Sci USA. 2005;102:7432–7437. doi: 10.1073/pnas.0500896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine JH, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/Jcell2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadaka C, Marloie-Provost M-A, Soumelis V, Benaroch P. Developmental regulation of MHC II expression and transport in human plasmacytoid-derived dendritic cells. Blood. 2009;113:2127–2135. doi: 10.1182/blood-2008-10-178152. [DOI] [PubMed] [Google Scholar]
- 32.Jongbloed SL, et al. Human CD141+(BDCA-3)+dendritic cells (DCs) represent a unique myeloid DC subset that crosspresents necrotic cell antigens. J Exp Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald KPA, et al. Characterization of human blood dendritic cell subsets. Blood. 2002;100:4512–4520. doi: 10.1182/blood-2001-11-0097. [DOI] [PubMed] [Google Scholar]
- 34.Matsui T, et al. CD2 distinguishes two subsets of human plasmacytoid dendritic cells with distinct phenotype and functions. J. Immunol. 2009;182:6815–6823. doi: 10.4049/jimmunol0802008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H, et al. Human BDCA2+CD123+CD56+dendritic cells (DCs) related to blastic plasmacytoid dendritic cell neoplasm represent a unique myeloid DC subset. Protein Cell. 2015;6:297–306. doi: 10.1007/s13238-015-0140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: Recent progress and open questions. Annu Rev Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/Jcell2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorting strategy of Lin−HLA-DR+CD33+CD45RA+CD1clo/-CD2+CADM1lo/-CD123+pre-DC for C1 scmRNAseq [Google Scholar]
- 39.Workflow and quality control of C1 scmRNAseq data analysis [Google Scholar]
- 40.Number of expressed genes detected per cell in the pre-DC C1 scmRNAseq experiment [Google Scholar]
- 41.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/Jcell2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 44.Grajales-Reyes GE, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8a+conventional DC clonogenic progenitor. Nat Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlitzer A, et al. Identification of CCR9−murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood. 2011;117:6562–6570. doi: 10.1182/blood-2010-12-326678. [DOI] [PubMed] [Google Scholar]
- 46.Krug A, et al. Toll-like receptor expression reveals CpG DNA as a unique microbial stimulus for plasmacytoid dendritic cells which synergizes with CD40 ligand to induce high amounts of IL-12. Eur J Immunol. 2001;31:3026–3037. doi: 10.1002/1521-4141(2001010)31:10<3026::AID-IMMU3026>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 47.Dzionek A, et al. Plasmacytoid dendritic cells: From specific surface markers to specific cellular functions. Hum Immunol. 2002;63:1133–1148. doi: 10.1016/S0198-8859(02)00752-8. [DOI] [PubMed] [Google Scholar]
- 48.Cella M, Facchetti F, Lanzavecchia A, Colonna M. Plasmacytoid dendritic cells activated by influenza virus and CD40L drive a potent TH1 polarization. Nat Immunol. 2000;1:305–310. doi: 10.1038/79747. [DOI] [PubMed] [Google Scholar]
- 49.Ito T, et al. Plasmacytoid dendritic cells prime IL-10-producing T regulatory cells by inducible costimulator ligand. J Exp Med. 2007;204:105–115. doi: 10.1084/jem.20061660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fonteneau J-F, et al. Activation of influenza virus-specific CD4+and CD8+T cells: A new role for plasmacytoid dendritic cells in adaptive immunity. Blood. 2003;101:3520–3526. doi: 10.1182/blood-2002-10-3063. [DOI] [PubMed] [Google Scholar]
- 51.Hoeffel G, et al. Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity. 2007;27:481–492. doi: 10.1016/Jimmuni.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y-J. IPC: Professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annuRevimmunol23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 53.Siegal FP, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 54.de Winter Cf, et al. Phenotype and natural history in 101 individuals with Pitt-Hopkins syndrome through an internet questionnaire system. J Orphanet Rare Dis. 2016;11:37. doi: 10.1186/s13023-016-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grün D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 58.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahsler M, Piekenbrock M. dbscan: Density Based Clustering of Applications with Noise (DBSCAN) and Related Algorithms. R package version 1.0-0. 2017 https://CRAnR-projectorg/package=dbscan. [Google Scholar]
- 60.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83A:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/Jimmuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69A:541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 65.Oksanen J, et al. vegan: Community Ecology Package. R package version 2.4-2. 2017 https://CRAnR-projectorg/package=vegan. [Google Scholar]
- 66.Chen H, et al. Cytofkit: A bioconductor package for an integrated mass cytometry data analysis pipeline. PLOS Comput Biol. 2016;12:e1005112. doi: 10.1371/journalpcbi.1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 68.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Stat Soc B. 1995;57:289–300. [Google Scholar]
- 69.Lists of genes identified from the microarray DEG analysis comparisons along the lineage progression from early pre-DC to mature cDC, for cDC1 and cDC2, respectively, and the list of the 62 common genes [Google Scholar]
- 70.Venn diagram comparison of the two lists of DEG and identification of the 62 common genes [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.