Abstract
Cancer-associated fibroblasts (CAFs) within the tumour microenvironment perform diverse roles and can modulate therapy responses. CAF functions include remodelling the extracellular matrix (ECM), promoting cancer cell invasion via heterotypic cell-cell contacts, and producing a variety of inflammatory modulating chemokines and cytokines. The inflammatory environment within tumours influences responses to many therapies, including the efficacy of oncolytic viruses, however the role of CAFs in this context remains unclear. Further, little is known about the cell signalling triggered by heterotypic cancer cell - fibroblast contacts nor about what activates fibroblasts to express inflammatory mediators. In this study, we show that direct contact between cancer cells and CAFs triggers the expression of a wide range of inflammatory modulators by fibroblasts. This is initiated following transcytosis of cytoplasm from cancer cells into fibroblasts leading to the activation of STING and IRF3-mediated expression of IFNB1 and other cytokines. IFNB1 then drives interferon-stimulated transcriptional programmes in both cancer cells and stromal fibroblasts and ultimately undermines the efficacy of oncolytic viruses, both in vitro and in vivo. Further, targeting IRF3 solely in stromal fibroblasts restores oncolytic HSV function.
Results and Discussion
The phenotype of cancers is dictated by the interplay of cancer cells with genetic mutations and, in many squamous cell carcinoma, oncogenic HPV infection with non-transformed cells in the tumour microenvironment1. Cancer-Associated Fibroblasts (CAFs) are an important component of the tumour microenvironment and well established functions in remodelling the extracellular matrix and promoting cancer invasion2,3. More recently, an invasion promoting role of direct cancer cell-CAF contact has been reported4. Further, they are increasingly appreciated to have a role in modulating the efficacy of both conventional cytotoxic and targeted cancer therapies5,6. The presence and sub-type of CAF can also be correlated with differing levels of both lymphocytes and myeloid cells within tumours7,8 and ablation of αSMA positive cells increased the efficacy of α-CTLA4 therapy in a mouse pancreatic cancer model9. Interestingly, in this same model the ablation of αSMA positive cells in the absence of therapy leads to more aggressive disease, thereby highlighting the complex interplay between CAFs and tumour phenotype. However, it remains unclear what promotes the immune modulatory effects of CAFs and how this might influence cancer therapies. Oncolytic viral therapies, such as the HSV derivative talimogene laherparepvec (T-VEC)10, aim to exploit differences in cancer cells to enhance viral replication and killing of malignant cells while sparing normal cells11. Further, they can re-awaken immune responses to the tumour12. However, oncolytic viruses only work in a minority of patients and an improved understanding of the determinants of efficacy is needed11. The activation of innate viral defence mechanisms that sense aberrant cytosolic nucleic acids in cancer cells can undermine viral infection13, but little is known about their role in stromal modulation of oncolytic viral therapies. In this work, we explore mechanisms triggering the expression of inflammatory modulators by CAFs and investigate how they modulate oncolytic viral therapy.
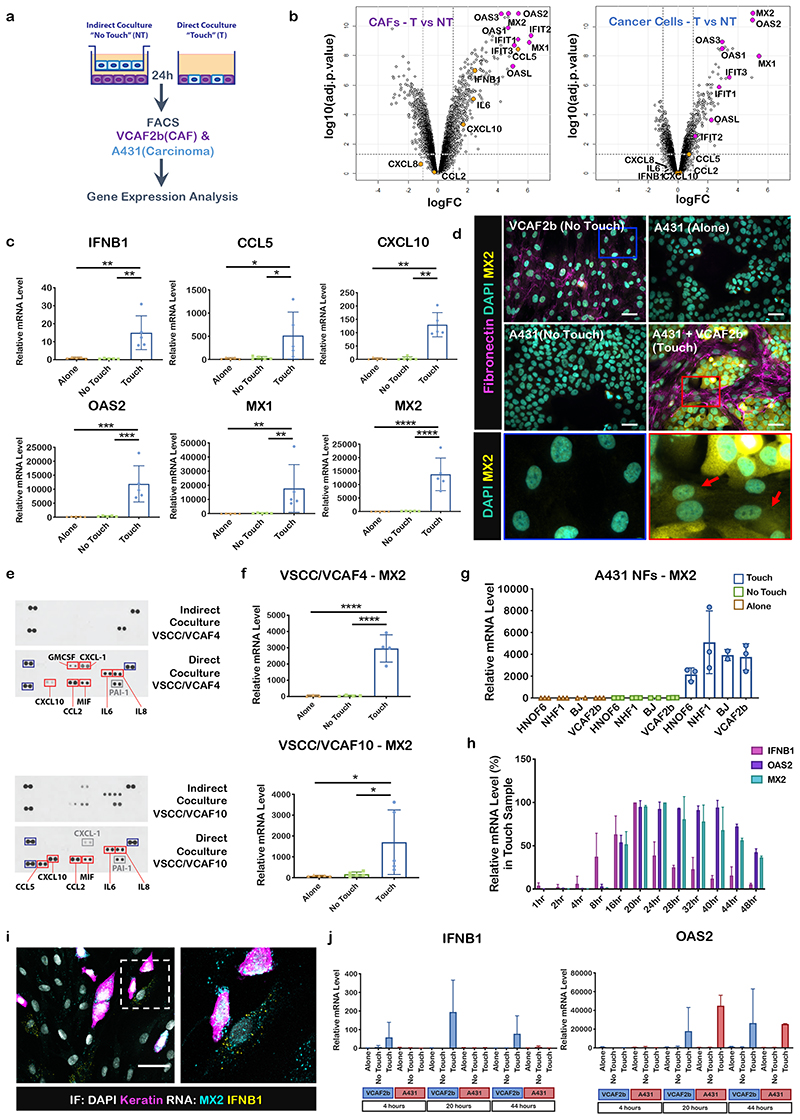
To study the consequences of heterotypic contact between cancer cells and fibroblasts we performed transcriptomic analysis in mono-cultures, direct co-cultures, or indirect co-cultures involving the different cell types separated by a 0.4μm filter (Figure 1a). These analyses revealed a dramatic increase in the expression of numerous inflammatory modulators in CAFs, including chemokines, cytokines, and Interferon-Stimulated Genes (ISGs), in direct co-culture conditions but not when cultured either side of a 0.4μm filter (Figure 1b, Supplementary Figure 1a&b and Supplementary Table 1). QRT-PCR analysis for the soluble factors IFNB1, CCL5, CXCL10, and the ISGs OAS2, MX1, and MX2 confirmed the microarray analysis (Figure 1c). MX2 induction was also readily detectable by immunofluorescence (Figure 1d). We confirmed these data by separating the epithelial and fibroblastic components from two patients with vulval SCC and using cytokine arrays to monitor the levels of a wide range of immune-modulatory molecules (Figure 1e). Unfortunately, IFNB1 was not on this array but we additionally confirmed the up-regulation of the ISG, MX2, in these paired patient cultures (Figure 1f). Co-culture of A431 cancer cells with fibroblasts isolated from normal tissue also promoted a dramatic increase in MX2, indicating that the induction of ISGs is not restricted to CAFs (Figure 1g). Non-transformed HaCAT cells did not trigger ISG expression (Supplementary Figure 1c). A431 cancer cells were not able to induce ISGs when co-cultured with human endothelial umbilical vein derived cells (HUVEC) or macrophages derived from peripheral mononuclear blood cells (PBMCs) (Supplementary Figure 1d&e). Time course analysis indicated that the up-regulation of IFNB1 began 4 hours after contact, whereas the majority of other genes, including the ISGs OAS2 and MX2, were induced later (Figure 1h), with up-regulation of ISGs persisting for 4 days (Supplementary Figure 1f). Intriguingly, in situ RNA analysis indicated that IFNB1 up-regulation was only observed in <5% of fibroblasts (Figure 1i – individual cell culture controls are shown in Supplementary Figure 1g). Furthermore, IFNB1 was specifically up-regulated in fibroblasts in contact with cancer cells when silicone templates were used to recreate defined cancer cell/fibroblast interfaces (Supplementary Figure 1h). This contrasts with the up-regulation of the ISG, MX2, in almost all SCC and CAF cells. QRT-PCR analysis of cancer cells and stromal fibroblasts sorted using FACS after co-culture confirmed the in situ analysis (Figure 1j). Together, these data indicate an amplification loop in which IFNB1 expression is triggered following cell-cell contact in a subset of CAFs and this leading to paracrine signalling inducing ISGs in both cell types.
Figure 1. Cancer cell-CAF contact triggers the expression of inflammatory modulators including IFNB1.
A) Scheme of experimental set-up of co-culture experiments highlighting ‘touch’ and ‘no-touch’ assays. B) Volcano plots comparing gene expression in indirect (‘no touch’ NT) vs direct (‘touch’ T) co-culture – changes in both fibroblasts (left) and cancer cells (right) are shown. Cytokines are highlighted in orange and Interferon Stimulated Genes (ISGs) in magenta. n=2 biological replicates. Expression and adjuvant-p-values are available in Supplementary Table 1 C) qRT-PCR analysis of subset of cytokine and ISG genes in of the cells on their own (Alone) or in co-cultures (Indirect, Direct). mRNA levels were normalized against two housekeeping genes. Each dot represents a biological replicate (n=5). One-way ANOVA with Dunnet’s multiple comparisons test. IFNB1 ANOVA F=11.45, R square=0.6561 DF=14, p=0.0017 TvsA Mean Diff. 14.43, 95% CI of diff. 5.836 to 23.02, p=0.0028, TvsNT Mean Diff. 14.02, 95% CI of diff. 5.43 to 22.62 p=0.0023. CCL5 ANOVA F=4.592, R square=0.4336 DF=14, p=0.0330, TvsA Mean Diff. 492.4, 95% CI of diff. 27.97 to 956.8 p=0.0381, TvsNT Mean Diff. 481.6, 95% CI of diff. 17.15 to 946 p=0.0424. OAS2 ANOVA F=16.13, R square=0.7289, DF=14, p=0.0004, TvsA Mean Diff. 11687, 95% CI of diff. 5778 to 17595 p=0.0007, TvsNT Mean Diff. 11538, 95% CI of diff. 5629 to 17446 p=0.0007. MX2 ANOVA F=25.62, R square=0.8103, DF=14, p<0.0001 TvsA Mean Diff. 13722, 95% CI of diff. 8196 to 19248 p<0.0001, TvsNT Mean Diff. 13657, 95% CI of diff. 8131 to 19183 p<0.0001. Mann-Whitney test CCL5 TvsA Median Diff 270 p=0.0079, TvsNT Median Diff 266 p=0.0159. MX1 TvsA Median Diff 9895 p=0.0079, TvsNT Median Diff 9826 p=0.0079. D) Micrographs of immunofluorescent (IF) stainings of co-cultures of A431 cells and VCAF2b and cells on their own (Alone and No-Touch). DAPI (green), MX2 (yellow) and fibronectin (magenta). Blue and red boxes depict zoomed images from the No Touch (blue) and Touch (red) conditions. Red arrows highlight MX2 expression in the cytoplasm of VCAF2b cells. Shown representative images from three biological replicates. Scale bar is 30μm. E) Protein cytokine arrays of conditioned medium from direct or indirect co-cultures of the indicated pair of cancer cells and CAFs. Blue boxes are the hotspots showing positive control/orientation makers, red boxes show cytokines differently expressed between co-cultures, grey boxes show cytokines that are at similar levels between the different co-cultures. F) qRT-PCR of the ISG MX2 from two pairs of patient derived cell lines and CAFs. mRNA levels were normalized against two housekeeping genes. Each dot is a biological replicate. One-way ANOVA with Sidak’s multiple comparisons test. VSCC/VCAF4, n=4 ANOVA F=40.68, Rsquare=0.9105, DF=10, p<0.0001. TvsA Mean Diff.2917, SE of diff. 393.6, p<0.0001, TvsNT, Mean Diff.2911, SE of diff. 364.4 p<0.0001. VSCC/VCAF10, n=5 ANOVA F=5.152, Rsquare=0.462, DF=14 p=0.0243, TvsA, Mean Diff.1614, SE of diff. 567.4 p=0.0293, TvsNT, Mean Diff.1538, SE of diff. 567.4 p=0.0375. G) qRT-PCR of MX2 after co-culture of A431 cells with fibroblasts derived from normal tissue and VCAF2b as control. Each dot is a biological replicate (n=3 except for BJ n=2), represented. mRNA levels were normalized against two housekeeping genes. H) qRT-PCR-time course of IFNB1, MX2 and OAS2 of direct co-culture of A431 and VCAF2b, lapsed hours indicate number of hours after A341 cells are added to VCAF2b. n=2 biological replicates. mRNA levels are represented as percentage from the highest value. I) Micrographs of RNAScope combined with IF of A431-VCAF2b direct co-culture. IF: DAPI (grey), Keratin (magenta), RNA probes: MX2 (cyan), IFNB1 (yellow), scale bar is 50μm. Shown representative images from three biological replicates J) qRT-PCR of ISG MX2 and OAS2 of FAC Sorted A431 and VCAF2b cells after co-culture at 4, 20 and 44 hours after A431 addition to VCAF2b cells.n=2 biological replicates. mRNA levels were normalized against two housekeeping genes. For all graphs, represented is mean with standard deviation (SD). Statistics, ns not significant, * p<0.05, ** p<0.005, *** p<0.001, **** p<0.0001
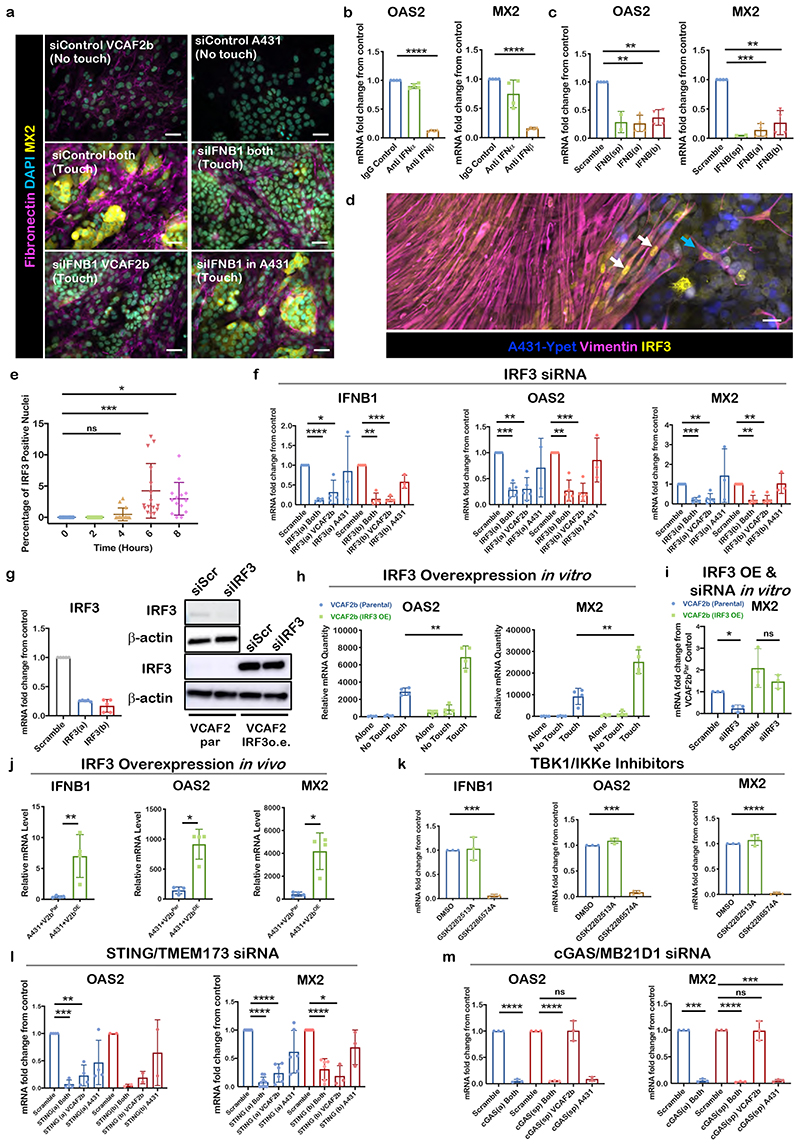
We next sought to determine the pathway driving the expression of ISGs, such as MX2 and OAS2. IFNB1 was the most highly up-regulated interferon in the transcriptomic analysis. Importantly, blockade of IFNB1 using either siRNA or a blocking antibody reduced MX2 and OAS2 expression (Figure 2a-c). Consistent with these data, depletion of the IFNB1 receptors, IFNAR1&2, in both cells of different cancer cell-CAF pairs also reduced MX2 and OAS2 expression (Supplementary Figure 2a&b). IRF9 was also required for MX2 and OAS2 induction, but did not regulate IFNB1 mRNA levels, consistent with its reported role downstream of IFNB114 and IFNAR1&2 (Supplementary Figure 2c). In contrast, IRF3 is one of several transcription factors involved in the regulation of IFNB1 and we noted an increase in the nuclear accumulation of IRF3 in a subset of CAFs 4 hours after initiating heterotypic cell-cell contact (Figure 2d & e). The functional importance of IRF3 was demonstrated by the lack of MX2 protein induction following siRNA-mediated depletion in VCAF2b (Supplementary Figure 2f). Further, mRNA analysis using two independent siRNA demonstrated that IRF3 was required for the induction of IFNB1, as well as the ISGs MX2 and OAS2 (Figure 2f – shown for a second cancer cell-CAF pair in Supplementary Figure 2e). Depletion of IRF3 in CAFs alone prevented the induction of IFNB1 and ISGs (Figure 2f & g and Supplementary Figure 2f – knockdown confirmed in Figure 2g). Furthermore, over-expression of IRF3 led to enhanced induction of ISGs (Figure 2h) and over-expression of IRF3 that cannot be targeted by siRNA restores the induction of MX2 in co-cultures (Figure 2i - over-expression is confirmed in Figure 2g). In vivo experiments involving co-injection of A431 cells with either control or IRF3 over-expressing CAFs demonstrated increased expression of IFNB1, OAS2, and MX2 (Figure 2j), while expression of STING and cGAS was equivalent in the control and IRF3 over-expressing tumours (Supplementary Figure 2h). Supplementary Figure 2g confirms the presence of extensive cancer cell fibroblast interfaces in tumour arising from co-injection of cancer cells and CAFs. These data indicate that IRF3 function in CAFs can drive IFNB1 and ISG expression in vivo. Interestingly, depletion of IRF3 did not affect the pro-invasive function of CAFs and had little effect on the production of fibronectin fibrils (Supplementary Figure 2j-l). These data indicate that regulation of inflammatory modulators and invasion are separable functions in fibroblasts with distinct regulatory mechanisms.
Figure 2. Cytoplasmic DNA mechanisms converge on IRF3 to drive ISG expression following cancer cell-CAF contact.
A) Micrographs of IF staining of different culture conditions after cell-type specific siRNA knockdown (KD) of IFNB1. Fibronectin (magenta), MX2 (yellow), DAPI (cyan). Shown representative images from 2 biological replicates. Scale bar is 50 μm. B) qRT-PCR of ISGs MX2 and OAS2 after co-culture with IFN type-1 specific blocking antibodies. mRNA levels shown were normalized to IgG control. Each dot is a technical replicate from two biological replicates. One sample t test, OAS2 anti-IFNβ vs IgG control Theoretical mean 1, Actual mean 0.1288, Discrepancy -0.8712, 95% CI -0.8884 to -0.8539, t=160.7, df=3, p<0.0001. MX2 anti-IFNβ vs IgG control Theoretical mean 1, Actual mean 0.1606, Discrepancy - 0.8394, 95% CI -0.8652 to -0.8136, t=103.6, df=3 p<0.0001. C) qRT-PCR of ISGs MX2 and OAS2 after siRNA KD of IFNB1 of both cell types using a pool of two separate siRNA sequences, and then each siRNA individually (a, b). Each dot is a biological replicate (n=4). mRNA levels shown were normalized to siRNA scramble control. One sample t test, OAS2 siIFNB1(a) Theoretical mean 1, Actual mean 0.2652, Discrepancy -0.7348, 95% CI -0.9650 to -0.5047, t=10.16, df=3, p=0.0020. siIFNB1(b) Theoretical mean 1, Actual mean 0.3697, Discrepancy -0.6303, 95% CI -0.8516 to -0.4090, t=9.063, df=3, p=0.0028. MX2 siIFNB1(a) Theoretical mean 1, Actual mean 0.1432, Discrepancy -0.8568, 95% CI -1.030 to -0.6838, t=15.76, df=3, p=0.0006. siIFNB1(b) Theoretical mean 1, Actual mean 0.2674, Discrepancy - 0.7326, 95% CI -1.060 to -0.4049, t=7.113, df=3, p=0.0057. D) Micrographs of IF stainings from A431/VCAF2b co-culture with defined borders (silicone inserts used) showing nuclear translocation of IRF3 in VCAF2b upon contact with A431. White arrows highlight nuclear IRF3 signal in VCAF2b in the boundary. Phalloidin (grey), Keratin (magenta), IRF3 (yellow), DAPI (cyan), Shown representative images from two biological replicates. Scale bar is 50μm. E) Quantification of micrographs from A431-VCAF2b co-culture stained with IRF3, over a time course. Hours indicating time passed after addition of A431 cells to VCAF2b. Depicted is the percentage of IRF3 positive nuclei of VCAF2b cells. Each dot indicates a 20x field of view from four different technical replicates. One-way ANOVA with Sidak’s multiple comparisons test. ANOVA, F=8.681, R square=0.3553 df=67, p<0.0001. 0hvs4h Mean diff - 0.4756, 95% CI -2.966 to 2.015, p=0.9538, 0hvs6h Mean diff -4.244,95% CI -6.703 to - 1.785, p=0.0002, 0hvs8h Mean diff -2.961,95% CI -5.487 to -0.4354, p=0.0164. F) qRT-PCR of, IFNB1, OAS2 and MX2 in direct A431/VCAF2b co-cultures after cell-type specific siRNA KD of IRF3 using two different sequences. Each dot is a biological replicate. mRNA levels shown were normalized to siRNA scramble control. One sample t test, IFNB1 siIRF3(a) Both Theoretical mean 1, Actual mean 0.1207, Discrepancy -0.8793, 95% CI -0.9368 to -0.8218, t=48.65, df=3, p<0.0001. siIRF3(a) VCAF2b Theoretical mean 1, Actual mean 0.3201, Discrepancy -0.6799, 95% CI -1.159 to -0.2011, t=4.519, df=3, p=0.0203. siIRF3(b) Both : Theoretical mean 1, Actual mean 0.1486, Discrepancy -0.8514, 95% CI -1.086 to -0.6171, t=11.56, df=3, p=0.0014. siIRF3(b) VCAF2b Theoretical mean 1, Actual mean 0.1443, Discrepancy -0.8557, 95% CI -0.9640 to -0.7475, t=25.16, df=3, p<0.001. OAS2 siIRF3(a) Both, Theoretical mean 1, Actual mean 0.2840, Discrepancy -0.7160, 95% CI -0.8788 to - 0.5532, t=12.21, df=4, p=0.0003. siIRF3(a) VCAF2b, Theoretical mean 1, Actual mean 0.3033, Discrepancy -0.6967, 95% CI -0.9663 to -0.4271, t=7.176, df=4, p=0.0020. siIRF3(b) Both, Theoretical mean 1, Actual mean 0.2698, Discrepancy -0.7302, 95% CI -0.9827 to - 0.4777, t=8.028, df=4, p=0.0013, siIRF3(b) VCAF2b p=0.0006, siIRF3(b) A431 Theoretical mean 1, Actual mean 0.3201, discrepancy -0.6799, 95% CI -1.159 to -0.2011, t=4.519, df=3, p=0.6231. MX2 siIRF3(a) Both p<0.0001, siIRF3(a) VCAF2b p=0.0028, siIRF3(a) A431 p=0.6327. siIRF3(b) Both p=0.0014, siIRF3(b) VCAF2b, Theoretical mean 1, Actual mean 0.2370, Discrepancy -0.7630, 95% CI -0.9808 to -0.5452, t=9.728, df=4. p=0.0012. G) IRF3 siRNA Knockdown efficiency shown through both qRT-PCR (left) and Western blot (right). qRT-PCR of IRF3 expression in VCAF2b cells after treatment with either scrambled control or two different IRF3 siRNA (a - red, b - blue). Each dot is a biological replicate (n=4) and mRNA levels shown were normalized to siRNA scramble control. Top panel of western blot shows IRF3 expression in VCAF2b. Bottom panels show IRF3 expression in VCAF2b parental (VCAF2b par) and lentiviral transfected VCAF2b with siRNA resistant IRF3 (VACF2b o.e.) (bottom). To avoid saturating the signal for the over-expressed IRF3, the lower panel is a shorter exposure and hence the endogenous protein is not easily visible. Representative blot of two biological replicates. IRF3 (50-55kDa) and loading control Antibeta actin (43kDa). H) qRT-PCR of OAS2 and MX2 in direct, indirect co-cultures and cells on their own using A431 and VCAF2 parental (VCAF2b par) and VCAF2b transfected with IRF3 construct (VCAF2b IRF3 OE). mRNA levels were normalized against two housekeeping genes. Each dot represents a biological replicate (n=4). Unpaired t test, Touch parental vs Touch IRF3 OE OAS2, Mean difference -680.8, SE of diff 680.8, T=5.87, df=6, p=0.0010. MX2, Mean difference -16016, SE of diff 3308, T=4.842, df=6, p=0.0028 I) qRT-PCR of MX2 of direct co-cultures of A431 with VCAF2b par and VCAF2b OE after transfection with IRF3 siRNA. Each dot is a biological replicate (n=3). mRNA levels shown were normalized to siRNA scramble control. One sample t test, VCAF2b par siIRF3, Theoretical mean 1, Actual mean 0.2351, Discrepancy -0.7649, 95% CI -1.161 to -0.3686, t=8.305, df=2, p=0.0142. Unpaired t test VCAF2b OE Scramble vs siIRF3, mean difference - 0.6156, 95% CI -2.128 to 0.8966, t=1.13, df=4. p=0.3215 J) qRT-PCR of IFNB1, OAS2 and MX2 from subcutaneous tumours four days after injection of A431-VCA2b par or A431-VCAF2b OE in the flank of Balb/c nude mice. Each dot represents a sample from an individual tumour (n=4). mRNA levels were normalized against two housekeeping genes. Unpaired t test IFNB1, Mean difference 6.578, 95%CI of diff. 2.33 to 10.83, T=3.789, df=6, p=0.0091. Mann Whitney test, OAS2, diff btw medians 877.7, p=0.0286 MX2, diff btw medians 4430 p=0.0286 K) qRT-PCR of IFNB1, OAS2 and MX2 in direct co-cultures after treatment with DMSO, structural control agent (GSK2282513A) or TBK/IKKε inhibitor (GSK2286574A). Each dot is a biological replicate (n=3). mRNA levels shown were normalized to DMSO control. One sample t test for GSK2286574A IFNB1 Theoretical mean 1, Actual mean 0.06098, Discrepancy -0.939, 95% CI -1.009 to -0.8688, t=57.53, df=2, p=0.0003, OAS2 Theoretical mean 1, Actual mean 0.08724, Discrepancy -0.9128, 95% CI - 0.9914 to -0.8341, t=49.93, df=2, p=0.0004. MX2 Theoretical mean 1, Actual mean 0.02398, Discrepancy -0.976, 95% CI -1.024 to -0.9282, t=87.82, df=2, p=0.0001. L) qRT-PCR of OAS2 and MX2 after siRNA KD of two different TMEM173/STING sequences in both cell types and cell-type specific. Each dot is a biological replicate. mRNA levels shown were normalized to siRNA scramble control. One sample t test OAS2 STING(a) Both, Theoretical mean 1, Actual mean 0.07346, Discrepancy -0.9265, 95% CI -1.046 to -0.8073, t=24.72, df=3, p<0.0001, STING(a) VCAF2b, Theoretical mean 1, Actual mean 0.2311, Discrepancy - 0.7689, 95% CI -1.074 to -0.4636, t=8.014, df=3, p=0.0041. STING(a) A431, Theoretical mean 1, Actual mean 0.4696, Discrepancy -0.5304, 95% CI - -1.177 to 0.1164, t=2.61, df=3 p=0.0797. MX2 STING(a) Both, Theoretical mean 1, Actual mean 0.07983, Discrepancy - 0.9202, 95% CI -0.9891 to -0.8513, t=30.80, df=8, p<0.0001. STING(a) VCAF2b, Theoretical mean 1, Actual mean 0.2416, Discrepancy -0.7584, 95% CI -0.9282 to -0.5886, t=11.48, df=5, p<0.0001. STING(a) A431, Theoretical mean 1, Actual mean 0.6166, Discrepancy - 0.3834, 95% CI -0.7782 to 0.01149, t=2.496, df=5, p=0.0548. STING(b) Both, Theoretical mean 1, Actual mean 0.3092, Discrepancy -0.6908, 95% CI -0.8864 to -0.4953, t=9.083, df=5, p=0.0003, STING(b) VCAF2b, Theoretical mean 1, Actual mean 0.1898, Discrepancy - 0.8102, 95% CI -1.262 to -0.3582, t=7.712, df=2, p=0.0164. STING(b) A431, Theoretical mean 1, Actual mean 0.6952, Discrepancy -0.3048, 95% CI - -1.066 to 0.4562, t=1.723, df=5, p=0.2270. M) qRT-PCR of OAS2 and MX2 in direct A431/VCAF2b co-cultures after cell-type specific siRNA KD of cGAS/MB21D1. Each dot is a biological replicate (n=3). mRNA levels shown were normalized to siRNA scramble control. OAS2 cGAS(a) Both, Theoretical mean 1, Actual mean 0.05284, Discrepancy -0.9472, 95% CI -1.021 to -0.8731, t=55.06, df=2, p=0.0003. cGAS(sp) Both, Theoretical mean 1, Actual mean 0.05240, Discrepancy -0.9476, 95% CI -0.9661 to -0.9291, t=220.7, df=2, p<0.0001. cGAS(sp) VCAF2b, Theoretical mean 1, Actual mean 1.007, Discrepancy 0.006660, 95% CI -0.4635 to 0.4769, t=0.06095, df=2, p=0.9569. MX2 cGAS(a) Both, Theoretical mean 1, Actual mean 0.05284, Discrepancy -0.9472, 95% CI -1.021 to -0.8731, t=55.06, df=2, p=0.0003. cGAS(sp) Both Theoretical mean 1, Actual mean 0.02928, Discrepancy -0.9707, 95% CI - 0.9944 to -0.9470, t=176.2, df=2, p<0.0001. cGAS(sp) VCAF2b, Theoretical mean 1, Actual mean 0.9911, Discrepancy -0.008935, 95% CI -0.4588 to 0.4409, t=0.08546, df=2, p=0.9397. cGAS(sp) A431, Theoretical mean 1, Actual mean 0.05162, Discrepancy - 0.9484, 95% CI -1.006 to -0.8906, t=70.57, df=2, p=0.0002. For all graphs, represented is mean with SD. Statistics: ns not significant, * p<0.05, ** p<0.005, *** p<0.001, **** p<0.0001.
The data outlined above establish IRF3 expression in CAFs as a key factor regulating the expression of inflammatory modulators in co-culture experiments. Analysis of human HNSCC biopsies and adjacent non-malignant tissue confirmed the presence of nuclear IRF3 in elongated stromal cells adjacent to invading tumour strands in the majority of samples (Supplementary Figure 2m). Conventional immunohistochemical methods preclude the simultaneous analysis of more than 2 or 3 variables, therefore we interrogated single cell RNA sequencing analysis of HNSCC15. Interestingly, high levels of IRF3 or the functionally related IRF7 were found in >20% of stromal fibroblasts (Supplementary Figure 2i shows that IRF7 also plays a role in IFNB1 expression in CAFs). Further, these IRF3/7+ve fibroblasts exhibited a highly significant up-regulation of ISGs compared to IRF3/7 -ve fibroblasts (Supp Figure 2n). These data provide support for IRF3/7 activation in fibroblasts being linked to ISG expression in patients. IRF3/7 +ve fibroblasts showed similar expression of CAF markers such as ACTA2 and FAP as IRF3/7 -ve fibroblasts (Supplementary Figure 2o), and RNA expression of these markers was also similar among CAFs that triggered different ISGs levels in co-culture (Supplementary Table 2), suggesting that IRF3 activation is not associated specifically with either ACTA2+ve or FAP+ve CAF sub-types16. Inhibition of two kinases capable of phosphorylating IRF3, TBK1 and IKKε, also prevented the induction of IFNB1, MX2, and OAS2 (Figure 2k). Numerous upstream regulatory mechanisms have been documented for TBK1 and IKKε, including TLR3, RIG-I, MDA5, DDX41, ZBP1 (also known as DAI) and cGAS/STING. We therefore depleted these molecules and tested the induction of ISGs following cancer cell – fibroblast contact. siRNA targeting STING and ZBP1 combined with DDX41 reduced the levels of ISGs following contact (Figure 2l and Supplementary Figure 2p), whereas there was little effect when the other molecules were targeted. Cell type specific siRNA-mediated depletion indicated that STING was exclusively required in CAFs (Figure 2l). In contrast, cGAS, which generates the STING activator cGAMP, was required in the cancer cells suggesting that it may be sensing DNA damage in the cancer cells (Figure 2m). Interestingly, ZBP1 and DDX41 co-depletion in CAFs alone reduced the induction of ISGs, suggesting that the sensing of aberrant nucleotides can also occur in stromal fibroblasts. Together, these data suggest parallel mechanisms of IRF3 activation with aberrant nucleotides being sensed by ZBP1&DDX41 in fibroblasts and by cGAS in cancer cells, both of which converge on STING and IRF3 in CAFs. Interestingly, A431 cells alone do not respond to cGAMP (Supplementary Figure 3a). This is probably due to inactivation of STING/IRF3 pathway, which is a common feature of many cancers 17,18. Consistent with this, we observed much lower levels of STING mRNA in A431 cancer cells compared to VCAF (Supplementary Figure 3b), which could explain why A431 in mono-culture do not trigger ISG expression despite ongoing genomic stress.
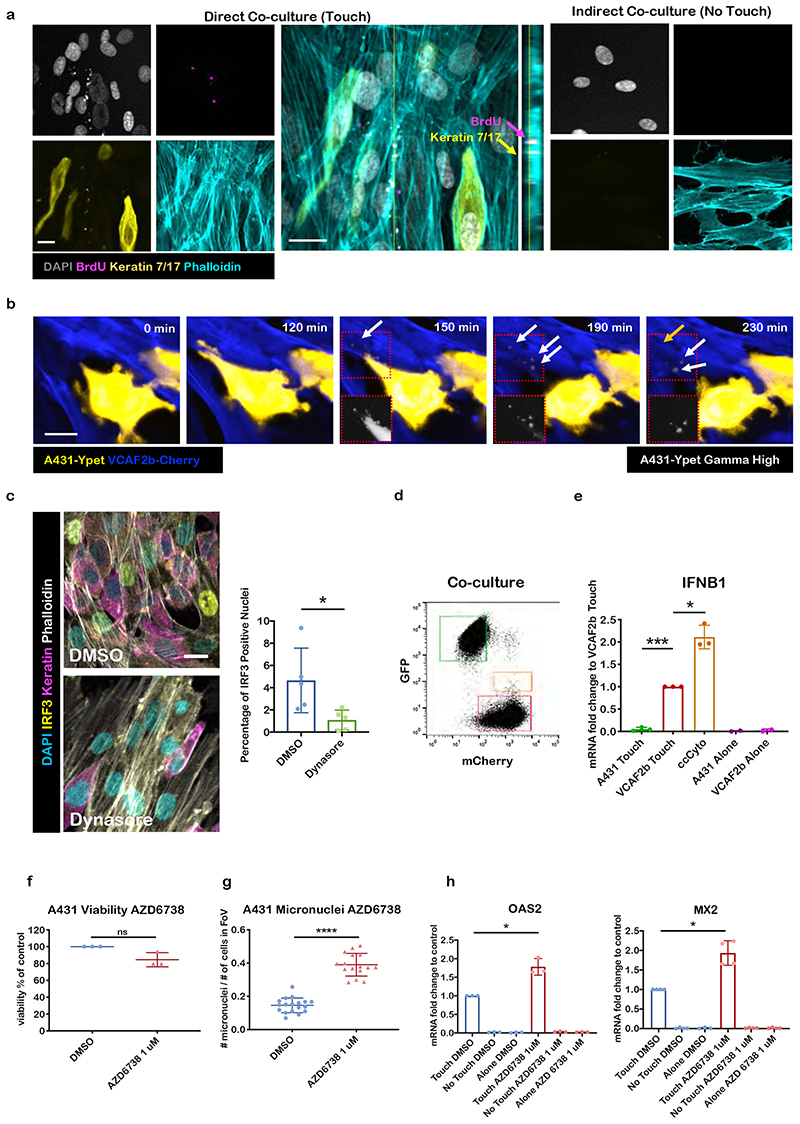
The data above indicate that activation of a cytoplasmic DNA sensing signalling axis is responsible for the activation of IFNB1 in CAFs and subsequent ISG expression in both cancer cells and fibroblasts. However, we were puzzled how direct contact between the two cell types would trigger a cytoplasmic DNA response and the requirement for cGAS and STING in different cell types. We considered whether gap junctions might enable the transfer of the STING agonist cGAMP from cancer cells to CAFs19; however, we were unable to observe dye transfer between cancer cells and CAFs, even after 24hrs (Supplementary Figure 3c), which would indicate functional gap junctions. The transfer of dye between CAFs after 2hrs confirmed the functionality of the assay. Further, pharmacological inhibition of gap junctions did not reduce ISG induction (Supplementary Figure 3d). Our previous 3D electron microscopy of the interface between cancer cells and CAFs revealed extensive membrane protrusions and complex intertwining of plasma membranes 4. This led us to speculate that there might be some cytoplasm transfer between the two cell types. Immunofluorescence of co-cultures demonstrated that epithelial keratins and cancer cell DNA that had been labelled with BrdU prior to co-culture could be found in puncta within stromal fibroblasts (Figure 3a). Live imaging of cells in co-culture demonstrated that CAFs could internalise cytoplasmic material from membrane ruffles of cancer cells that there were in contact with (Figure 3b). The fluorescent signal from the vesicles internalised in the CAFs would abruptly disappear, possibly suggest release of the contents into the CAF cytoplasm (Figure 3b – transcytosed vesicles marked with white arrows and disappearance of vesicle marked with orange arrow, see also Suppl. Movie 1). Cytoplasmic transfer of material into fibroblasts was not restricted to cancer cells; but also observed with non-transformed cells but did not lead to ISG expression, presumably due to the low levels of genomic stress in non-transformed cells (Supplementary Figure 3 e-g). Consistent with other reports, we also observed transfer of material from cancer cells to macrophages and endothelial cells and, in the reverse direction, from CAFs to cancer cells (Supplementary Figure 3h). The transfer of cytoplasmic material between cells is a dynamin-dependent process termed transcytosis20. Blockade of dynamin function using the inhibitor dynasore prevented the nuclear accumulation of IRF3 (Figure 3c), thereby suggesting a link between transcytosis and IRF3 activation. In addition, depletion of E-cadherin, which forms heterotypic adhesions with N-cadherin in CAFs, partially reduced the induction of IGSs (Supplementary Figure 3i). Flow cytometry enabled us to purify the small fraction of CAFs that contained fluorescent material from cancer cells (Figure 3d). Strikingly, CAFs containing mCherry fluorescence derived from cancer cells had significantly higher expression of IFNB1 (Figure 3e). These data indicate that transfer of material from cancer cells to fibroblasts is linked to IFNB1 induction.
Figure 3. CAFs use transcytosis to sense genomic stress in cancer cells.
A) Confocal micrographs of IF staining of a direct and indirect A431/VCAF2b co-cultures. BrdU (magenta), keratin (yellow), Phalloidin (cyan), DAPI (grey). Larger middle panel shows XY and YZ projections showing keratin and BrdU inside VCAF2b. Shown representative images from five biological replicates. Scale bar is 20μm. B) Still images of a time-lapse showing the transcytosis of material from an A431-yPET cell (yellow) into a mCherryLifeAct expressing CAF (blue). White arrows show transcytosed material and orange arrow highlights disappearance of a vesicle. Shown representative images from two biological replicates. Scale bar is 20μm. C) Micrographs of A431-VCAF2b co-culture after 6h treatment with DMSO(control) or dynasore 50μM, Phalloidin (grey), Keratin (magenta), IRF3 (yellow), DAPI (cyan), Shown representative images from 2 biological replicates. Scale bar is 20μm (left). Quantification of percentage of IRF3 positive nuclei. Each dot represents one field of view (FoV). Two biological replicates. Unpaired t-test t=2.627 df=8, Mean Diff. -3.573, 95% CI of diff. -6.71 to -0.4363, p=0.0303. D) Sorting strategy after 20h co-culture of A431-yPet and VCAF2b-mCherry. Green square=A431-Ypet Touch (66.3%), orange square=ccCyto (mCherry positive fibroblasts that have shifted in the GFP channel, 0.055%), red square VCAF2b-mCh Touch (32.6%). E) qRT-PCR of IFNB1 of different FACS sorted A431/VCAF2b populations after co-culture. mRNA levels were normalized to VCAF2b Touch. Each dot is a biological replicate (n=3, except n=2 for A431 Alone and VCAF2b Alone) one sample t test A431 Touch, Theoretical mean 1, Actual mean 0.05032, Discrepancy -0.9497, 95% CI -1.053 to -0.8463, t=39.54, df=2, p=0.0006. ccCyto Theoretical mean 1, Actual mean 2.111, Discrepancy 1.111, 95% CI 0.4526 to 1.769, t=7.261, df=2, p=0.0184. F) Viability of A431 cells after 48h of AZD6738 treatment compared with control. Each dot is a biological replicate (n=3). Wilcoxon Rank test 1μM, Theoretical median 100, Actual median 79.71, Discrepancy -20.29 p=0.25 G) Number of micronuclei normalized to the number of cells within a FoV after 48h treatment of AZD6738 1μM or DMSO. Each dot represents one FoV, results from three biological replicates. Unpaired t-test, t=12.54 df=32, Mean Diff. 0.2442, 95% CI of diff. 0.2045 to 0.2839 p<0.0001. H) qRT-PCR of OAS2 and MX2 in A431-VCAF2b 20h co-cultures, after 48h treatment of A431 with either DMSO or AZD6738 1μM. Each dot is a biological replicate (n=3 for OAS2, n=4 for MX2). mRNA levels shown were normalized to DMSO control. One sample t test. OAS2, Theoretical mean 1, Actual mean 1.782, Discrepancy 0.7818, 95% CI 0.2263 to 1.337, t=6.056, df=2, p=0.0262. MX2 Theoretical mean 1, Actual mean 1.934, Discrepancy 0.9343, 95% CI 0.4331 to 1.435, t=5.933, df=3, p=0.0096. For all graphs, represented is mean with SD. Statistics: ns not significant, * p<0.05, ** p<0.005, *** p<0.001, **** p<0.0001.
The data described above suggest that fibroblasts may utilise transcytosis to ‘sense’ genomic damage in neighbouring cancer cells. To test this, we induced additional DNA damage in A431 cells using an ATR inhibitor, AZD6738, prior to co-culturing with CAFs. This increased the level of micronuclei in A431 cells without compromising viability (Figure 3f & g and Supplementary Figure 3j). Further, pre-treatment with AZD6738 boosted the induction of ISGs, thereby confirming that fibroblasts sense genomic stress and the associated formation of micronuclei in cancer cells via transcytosis of material from the cytoplasm of cancer cells (Figure 3h). Lastly, we generated A431 cells with varying intrinsic abilities to produce cytoplasmic DNA. Single cell cloning enabled us to isolate A431 sub-lines that exhibited different levels of BrdU+ve cytoplasmic DNA (Supplementary Figure 3k shows clones with differences in cytoplasmic replicated DNA). Sub-clones that could not generate cytoplasm DNA did not trigger ISG expression when co-cultured with fibroblasts (Supplementary Figure 3l) even though they transferred cytoplasm with the same efficiency (Supplementary Figure 3m) - we termed these A431 cells ‘non-inducers’. Together these data establish that CAFs can sense genomic stress in cancer cells that they come into contact with provided that the cancer cells generate cGAMP, there is effective transcytosis, and sufficient IRF3 in the CAFs.
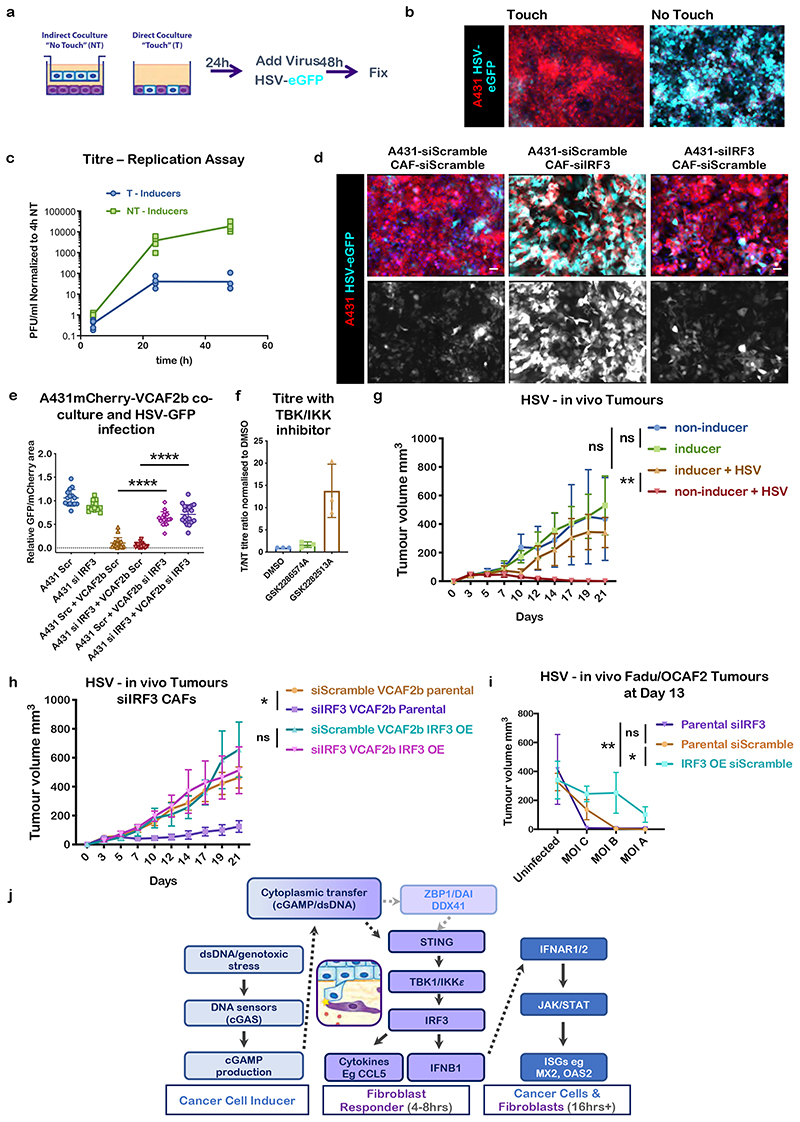
A major function of the interferon stimulated gene response is to provide protection against viral infection, with gene products such as MX1 and OAS interfering with viral replication21. Therefore, we were interested to tests if the induction of ISGs downstream of IRF3 in CAFs would affect oncolytic viral therapy (Figure 4a). We used HSV171622 a GFP-expressing HSV derivative similar to the only currently licenced oncolytic virus, T-VEC. Supplementary Figure 4a shows that ‘non-inducer’ A431 cells incapable of triggering IRF3 signalling in stromal fibroblasts were efficiently infected with HSV, whereas those that could trigger IRF3 in fibroblasts remained largely uninfected when co-cultured with CAFs. Importantly, the protection of ‘inducer’ A431 cells against viral infection depended upon direct cell-cell contact. HSV was highly effective at infecting A431 cells capable of triggering IRF3 in neighbouring fibroblasts when separated from CAFs by a 0.4μm filter, as assessed by HSV-eGFP+ve cells; but this susceptibility to infection was almost completely abrogated in direct co-culture conditions (Figure 4b). Similar results were obtained with two other classes of virus, Vaccinia and Reovirus (Supplementary Figure 4c&d) and with a different pair of cancer cell-CAF lines (FaDu-OCAF1 Supp. Figure 4e). HSV viral titre analysis demonstrated almost a thousand-fold lower viral titre 48 hours after infection of direct co-cultures compared to indirect co-cultures (Figure 4c). Calculation of replication rates demonstrated that the there was a similar reduction in the rate of virus replication between 24 and 48 hours (Supplementary Figure 4b). Even more crucially, if we blocked the ability of CAF to respond to cytoplasmic DNA by depleting IRF3 then we restored the efficacy of HSV infection (Figure 4d & e). TBK1/IKKε inhibition also promoted viral infection rates (Figure 4f). Co-culture with either macrophages or endothelial cells was unable to provide protection against viral infection, which is consistent with the lack of ISG induction in macrophage and endothelial cell co-cultures (Supp. Figure 4f&g, Supplementary Figure 1d&e). These data establish that contact mediated activation of the IRF3 pathway in CAFs protects cancer cells from oncolytic viruses.
Figure 4. Contact mediated IRF3 activation undermines viral therapy.
A) Schematic cartoon of experimental design for oncolytic virus treatment of cancer cell-CAF co-cultures. B) Micrographs of direct (touch, T) or Indirect (non-touch, NT) co-cultures after 48h infection with HSV-eGFP (Multiplicity of Infection (MOI) =1). HSV-eGFP (green), A431-mCherry (red), scale bar is 50μm. Images are representative of three biological replicates C) HSV viral replication assay comparing A431 Inducers in direct and indirect co-culture with VCAF2b, values normalized to 4h NT (indirect co-culture). Each dot is a technical replicate, data from three biological replicates. Line connects means at 4-24-48h. D). Micrographs of A431-VCAF2b direct co-cultures after 48h of HSV infection. Cells were previously incubated with scramble or IRF3 siRNA for 24h. HSV-eGFP (green), A431-mCherry (red), DAPI (blue). Scale bar is 50 μm. Representative images from three biological replicates E) Quantification of D); HSV-eGFP infection in A431/VCAF2b co-cultures after cell-type specific siRNA KD of IRF3. Each dot represents a FoV, data from three biological replicates. Represented mean and SD. One-way ANOVA with Tukey’s multiple comparisons test. ANOVA F=149, R square=0.8816, DF=105 p<0.0001. A431Scramble+VCAF2b Scramble vs A431Scramble+VCAF2bsiIRF3 Mean difference -0.5251, SE of diff 0.04559, p<0.0001. A431siIRF3+VCAF2b Scramble vs A431siIRF3+VCAF2bsiIRF3, Mean difference -0.6602, SE of diff 0.04559, p<0.0001. F) HSV-eGFP titre ratio in T/NT co-cultures after treatment with DMSO, structural control agent (GSK2282513A) or TBK/IKKε inhibitor (GSK2286574A). Each dot is a biological replicate (n=3). Represented mean and SD. G) Growth curves of tumour volume after subcutaneous (s.c.) injection of VCAF2b+A431 inducers or non-inducers co-cultures in the flank of ICRF nu/nu mice with or without addition of HSV-eGFP virus (MOI= 0.005) upon injection. Data from two independent experiments, Inducer (uninfected) n=9, Non-Inducer (uninfected) n=9. Inducer+HSV n=6, Non-Inducer+HSV n=6. Statistical test at day 14, One-way ANOVA with Sidak’s multiple comparisons, ANOVA F=8.353, R square=0.4908, DF=29, p=0.0005. Non-Ind vs Ind Mean difference -0.0, SE of diff 0.04559, p=0.6400, Ind vs Ind+HSV p=0.1901, Non-Ind vs Non-Ind+HSV p=0.0019. Represented mean with SD H) Growth curves of tumour volume after s.c. injection of VCAF2b+A431 inducers co-cultures in the flank ICRF nu/nu mice treated with HSV-eGFP virus (MOI= 0.005) upon injection. VCAF2b cells were incubated with Scramble or IRF3 siRNA for 24h before co-culture. For VCAF2b parental groups n=12 mice from four biological replicates. For VCAF2b IRF3 oe n=6 mice from 2 biological replicates. Statistical test at day 14, One-way ANOVA with Sidak’s multiple comparisons, ANOVA p=0.0173. Parental siScramble vs siIRF3 p=0.0219. IRF3 oe siScramble vs siIRF3 p=0.5852. Represented mean with SD I) Dose-response graph showing tumour volume at day 13 after s.c. co-injection of HSV treated FaDu-OCAF2 co-culture in the flank of BalbC nude mice. OCAF2 parental and IRF3 oe cells were previously incubated with Scramble or IRF3 siRNA for 24h before co-culture. MOI, A= 0.005, B = 0.00158 and C= 0.0005. n=36 mice from one biological replicate. Statistics Two-way ANOVA with Tukey’s multiple comparisons, interaction p=ns, MOI effect p<0.001 (n=9 per condition), IRF3 expression effect p= 0.0055 (n=12 per condition) Parental siIRF3 vs siScramble p=0.9789, Parental siIRF3 vs IRF3 oe siScramble p=0.0098, parental siScramble vs IRF3 oe siScramble p=0.0155. J) Schematic overview of current understanding of signalling pathways.
Finally, we tested whether activation of the IRF3 pathway in CAFs would affect oncolytic virus efficacy in vivo. We compared the effect of the modified oncolytic HSV-eGFP virus on tumours formed using either A431 that effectively activate IRF3/IFNB1 in CAFs or ‘non-inducer’ A431 that are unable to trigger this pathway, possibly as a result of lower levels of genomic stress (described in Supplementary Figure 3k and confirmed in Supplementary Figure 4l). In both cases, the A431 cells were admixed with VCAF2b prior to injection. Figure 4g shows that the tumours grew at equivalent rates regardless of whether the IRF3/IFNB1 pathway was triggered; however, tumours that were unable to trigger this pathway were much more sensitive to oncolytic virus and were entirely eliminated after three weeks. We further demonstrated the importance of stromal IRF3 by depleting the gene in stromal fibroblast prior to injection into mice. Figure 4h shows that previously virus insensitive tumours lacking IRF3 only in the fibroblast compartment became highly sensitive to HSV infection. This effect was rescued if the VCAF2b expressed an siRNA resistant form of IRF3, thereby confirming the specificity of the siRNA approach (Figure 4h). We additionally explored a second tumour model based on FaDu SCC cells and OCAF2. This model exhibited greater ‘baseline’ sensitivity to HSV and, therefore, represented an ideal model to test whether increased IRF3 function in CAFs could confer resistance to viral therapy. Figure 4i shows the effect HSV across a range of MOIs. At all MOI tested, tumours containing IRF3 over-expressing OCAF2 showed resistance to HSV. Furthermore, at lower MOI depletion of IRF3 increased the sensitivity of FaDU/OCAF2 tumours to HSV infection. Uninfected tumours grew at equivalent rate regardless of IRF3 levels in OCAF2 (Supplementary Figure 4 i-k).
Taken together, this analysis sheds light on several aspects of tumour biology with broader implications for anti-viral defences in non-transformed tissue. We provide a mechanism to explain the up-regulation of various cytokines, in particular IFNB1, in stromal fibroblasts. It is increasingly apparent that stromal fibroblasts influence the inflammatory environment in tumours7,8 and their ability to activate STING in response to genomic damage in neighbouring cancer cells may underpin some of these observations. The relationship between interferon function and oncolytic viruses is complex23; whilst the anti-viral interferon response can support the immune-mediated component of oncolytic virotherapy12,24,25, our data illustrate how contact between CAF and tumour cells may restrict the early stages of tumour cell infection and immunogenic cell killing, thus compromising eventual therapy. In the longer term, it will be interesting to determine the role of touch-mediated triggering of fibroblast STING/IRF3 signalling on tumour immunity. Activation of interferon stimulated genes has been linked to both favourable and unfavourable outcomes following immunotherapy. These discrepancies could be explained by either differences in acute vs chronic interferon signalling, with the latter leading to loss of T-cell functionality, or differences in which cells are expressing ISGs26. We observe strong similarity between ‘touch-induced’ genes and signatures linked to ineffective radio- and immuno-therapy27-29 (Supplementary Figure 4m). This is of particular interest given ongoing efforts to combine radiotherapy and immunotherapy in the treatment of mucosal SCC, such as head and neck SCC. STING activation can trigger senescence in fibroblasts and, although we find no evidence for cell cycle exit in our transcriptomic analysis (Supplementary Figure 2k: TOP2A +ve marker for proliferation and CDKN2A +ve marker for senescence) and do not observe the morphological changes associated with senescence in CAFs with nuclear IRF3, it is possible that the mechanism we elucidate could push CAFs towards senescence over longer time-frames30. This could then explain why senescent fibroblasts are found in some tumours31.
The evolutionary origin of the touch-dependent mechanism we describe is likely to lie in responding to physical tissue damage. Keratinocytes and fibroblasts are normally separated by a basement membrane and hence not in direct contact, but only exchange soluble factors in a similar manner to our ‘no-touch’ assays. Upon damage that disrupts the basement membrane direct contact becomes possible and a touch-mediated mechanism for triggering cytokine and chemokine expression would constitute a means of sensing structural damage to tissue and prophylactically alerting defences against any bacteria or viruses that may accompany the structural damage. Our observation of transcytosis between non-transformed keratinocytes and fibroblasts support the notion that this process is not specific to cancer (Supplementary Figure 3g). In the future, it will be interesting to explore the mechanistic details of the transcytosis process and release of cGAMP. Our data indicate that this depends on dynamin and partially on heterotypic adherens junctions (Figure 3c and Supplementary Figure 3i). The ability of fibroblasts to sense aberrant cytoplasmic nucleotides in neighbouring cells may also act as a back-up mechanism for counteracting viral infections. It is well established that many viruses are able to disable STING/IRF3-dependent signalling in infected cells32, but transcytosis of cGAMP signals to adjacent cells may represent an evolutionary response to the viral gene products that target STING/IRF3. It is interesting to note that the development of the oncolytic HSV virus deliberately targeted the ICP34.5 gene product that inactivates IRF333. The rationale for this was that it is linked to the virulence of the virus, which would constitute an undesirable side-effect, and the frequent inactivation of STING/IRF3 in cancer cells would make targeting this axis unnecessary.
However, we find that the activation of STING and IRF3 in stromal fibroblasts and the subsequent induction of ISGs in both fibroblasts and cancer cells fundamentally undermines the efficacy of oncolytic viruses. This knowledge may help to identify patients that are less likely to respond to oncolytic viruses. Further we demonstrate that pharmacological targeting of TBK and IKKε is a possible way to restore viral killing.
Materials and Methods
Cell culture
We used VCAF2b, a patient derived vulvar cancer associated fibroblast (CAF) cell line. Two other patient derived SCC/CAF pairs were also used: VSCC4/VCAF4 and VSCC10/VCAF10. VCAF2b, VSCC4, VSCC10, VCAF4 and VCAF10 were derived from tumours obtained under REC 15/EE/0151 OCAF1 is a CAF cell line derived from an oral cavity tumour, HNOF6 are fibroblasts derived from non-involved tissue of an oral carcinoma (obtained under ethical approval REC: 06/Q0403/125). Isolation of cell lines from fresh tumour tissues was performed as previously reported elsewhere2. Briefly, we receive material fresh from the operating theatre. It is then cultured at the interface between cell culture plastic and a collagen/Matrigel mix with media containing serum, hydrocortisone, insulin, transferrin, and selenium. After a week or two, fibroblasts will have invaded from the tumour piece onto the culture dish. Differential adhesion and lack of supporting growth factors for keratinocytes is used to favour the growth of CAFs. Different culture conditions are used to favour the expansion of cancer cells (this includes fibroblast feeder layers). Once the CAFs are sufficiently expanded (T175 flask) then cells are frozen, STR tested (to ensure that they are unique and not the result of inadvertent cross-contamination), and stained for markers. Primary fibroblasts are immortalized by transfecting a hTERT plasmid using lentivirus. A431 (SCC line ECACC), FaDu (pharynx line ATCC), HaCat (Human skin keratinocytes), HUVEC (Human umbilical vein endhothelial cells), BJ (foreskin fibroblasts immortalized with h-TERT), NHF1-hTERT (normal human fibroblasts immortalized with h-TERT) cell lines were obtained from the Crick central Cell Services facility. Routine screening for Mycoplasma was performed for all cell lines. All epithelial cells were cultured in DMEM (ThermoFisher #41966052) containing 10% Foetal Bovine Serum (PAA Labs), 1% antibiotics (Pen/Strep) (Invitrogen #15140122). Fibroblasts were cultured with the addition of 1% Insulin-transferrin-Selenium (Invitrogen #41400045) and kept at 37˚C and 5% CO2. Fibroblasts were not allowed to reach >90% confluency. New flasks were used after every passage. HUVEC Cells were cultured in EBMTM-2 Basal Medium (Lonza CC-3156) and EGMTM-2 SingleQuotsTM Supplements (Lonza CC-4176) Cell lines that are not commercially obtainable are available from the authors upon reasonable request.
Cell Co-cultures
Mono-cultures (alone), direct co-cultures (touch), or indirect co-cultures (no touch) separated by a 0.4μm filter (Sarsteadt #83.3932.040), were performed using different combinations of epithelial cell lines, fibroblasts, endothelial cells and macrophages. Co-cultures were performed using a 1:1 ratio between cancer cells and fibroblasts, typically 5x104 cells of each cell type in a 24-well plate. Cells were co-cultured together for 20h unless otherwise specified. All co-cultures were done in DMEM (ThermoFisher #41966052) containing 10% Foetal Bovine Serum (PAA Labs), 1% antibiotics (Pen/Strep) (Invitrogen #15140122) and 1% Insulin-transferrin-Selenium (Invitrogen #41400045. For the experiments in which a defined cancer cell-CAF interface was needed 5x103 VCAF2b were seeded in one chamber of a 2-well silicone insert (Ibidi Cat.No:80209). After 5 days 2x104 A431 cells were seeded in the opposite chamber. 24h later the insert was removed. When the cells had established a defined interface (usually around 36h later) cells were fixed using 4% paraformaldehyde and stained as explained below.
Transfection of small interfering RNA
Cells were plated into 6-well plates (Falcon, Thermo Fisher) at ~70% confluence, siRNA was diluted into serum free media [Opti-Mem] (Thermo Fisher, Cat No: 31985047) and mixed with 2μL/well DharmaFECT[2] transfection reagent (Dharmacon #T-2002-03), for a final siRNA concentration of 100nM and incubated at room temperature for 20 minutes to allow formation of complexes. All siRNAs were purchased from Dharmacon or Sigma, sequences are shown in Table 2. Cells were incubated in siRNA containing media for 24 hours before trypsinizing and plating for further experiments.
Table 2. Catalogue number and sequence of all siRNAs used in experiments.
| Gene | Catalogue Number | Sequence |
|---|---|---|
| IRF3 | D-006875-04-0005 D-006875-05-0005 | 5’-GGGAAGAGUGGGAGUUCGA-3’ 5’-CCAAGAGGCUCGUGAUGGU-3’ |
| IFNB1 | D-019656-02-0005 D-019656-04-0005 | 5’-CAACAAGUGUCUCCUCCAA-3’ 5’-UGGCUAAUGUCUAUCAUCA-3’ |
| IRF7 | D-011810-04 D-011810-05 | 5’-GAGAGUGGCUCCUUGGAGA-3’ 5’-GACAUCGAGUGCUUCCUUA-3’ |
| IRF9 | SASI_Hs01_00025478 SASI_Hs01_00025479 | 5’-CACAGAAUCUUAUCACAGU-3’ 5’-GCAGAGACUUGGUCAGGUA-3’ |
| IFNAR1 | SASI_Hs01_00121376 | 5’-CGUACAAGCAUCUGAUGGA-3’ |
| IFNAR2 | SASI_Hs01_00208506 | 5’-GAAGCAUAAACCCGAAAUA-3’ |
| STING | D-024333-01 D-024333-02 D-024333-03 D-024333-04 | 5’-GCACCUGUGUCCUGGAGUA-3’ 5’-GGUCAUAUUACAUCGGAUA-3’ 5’-GCAUCAAGGAUCGGGUUUA-3’ 5’-ACAUUCGCUUCCUGGAUAA-3’ |
| cGAS | D-015607-01-0002 D-015607-02-0002 D-015607-03-0002 | 5’-GAAGAAACAUGGCGGCUAU-3’ 5’-GAAGAGAAAUGUUGCAGGA-3’ 5’-GUAAGGAAUUUCUGACAAA-3’ |
| ZBP1 | D-014650-01-0002 D-014650-02-0002 D-014650-03-0002 | 5’-GCAAAGUCAGCCUCAAUUA-3’ 5’-GGACACGGGAACAUCAUUA-3’ 5’-GGACCCAACAGCUGGAUUU-3’ |
| LRRFIP1 | D-019842-01-0002 D-019842-02-0002 D-019842-03-0002 | 5’-GCACAAUACUGAGAAAGAA-3’ 5’-GUAGAAACCCUUAAAGAUG-3’ 5’-GGUAAAGUCUACUGACAGA-3’ |
| IFI16 | D-020004-01-0002 D-020004-02-0002 D-020004-03-0002 | 5’-GGAGUAAGGUGUCCGAGGA-3’ 5’-GAUUAGAAGUGCCAGCGUA-3’ 5’-GGACCAGCCCUAUCAAGAA-3’ |
| DDX41 | D-010394-01-0002 D-010394-02-0002 D-010394-03-0002 | 5’-GCAAAGAGUCUGCCAAGGA-3’ 5’-UCAAAGCGCUGCUGCUAGA-3’ 5’-CAAGGGAGCUGCGGAGGAA-3’ |
| CDH1 | D-003877-02 D-003877-07 D-003877-06 D-003877-05 | 5’-GGAGAGCGGUGGUCAAAGAUU-3’ 5’-GCAGUACAUUCUACACGUAUU-3’ 5’-GAGAACGCAUUGCCACAUAUU-3’ 5’-ACCAGAACCUCGAACUAUAUU-3’ |
Generation of siRNA-resistant IRF3 overexpressing fibroblasts
Fibroblasts were infected using a lentiviral plasmid (PCSII-IRES-BLASTI) (Fucci Lab) containing IRF3 (Human V5-IRF3-pcDNA, Addgene #32713) comprising of 3 mutations from position 1121 – 1141 (shown below). These mutations were specific to the IRF3 siRNA targeting region and altered the nucleotide sequence without changing the amino acid sequence. Cells were selected using 5μg/mL blasticidin.
| IRF3 siRNA target sequence | ACCaagaggCTCgtgatgGTC |
| IRF3 mutations | ACGaagaggCTGgtgatgGTG |
Drug treatments
For TBK1/IKKε inhibition, cells were plated for mono-culture, direct or indirect co-culture in media containing either DMSO or drugs. TBK1/IKKε inhibitor (250nM) compounds were kindly provided by GlaxoSmithKline (GSK); GSK2 (GSK2286574A) and GSK3 control (GSK2282513A). The same method was used for perturbing IRF3 nuclear location using Dynasore (50μM) (Selleckchem #S8047). Cells were fixed after 6 hours before immunofluorescent staining. Again, the same method was used for Gap junction inhibition, using Tonabersat 40 μM (Sigma SML1354) and Enoxolone 100 μM (Abcam ab142579). Cells were cultured for 20 hours before harvesting in RNA Protect (Qiagen #76526) for gene expression analyses. AZD6738 (Selleckchem #S7693) was used at 1μM to treat the cancer cells for 48h, after that cells were trypsinized and plated in direct or indirect co-culture with fibroblasts or on their own for 20 hours before harvesting in RNA Protect (Qiagen #76526) for gene expression analyses. For micronuclei analysis, after treatment cells were fixed with 4% paraformaldehyde and stained with DAPI 1:1000 (Sigma D9542) for imaging in Zeiss LSM510 microscope. Images were analysed using ImageJ.
Type-1 IFN blocking antibodies
For blocking antibody treatment cells were plated for mono-culture, direct or indirect co-culture in media containing either anti IFN alpha (PBL #31110-1) or beta (PBL #31401-1) at 1000 neutralising units/ml or IgG isotope control. Cells were cultured for 20 hours before harvesting in RNA Protect (Qiagen #76526) for gene expression analyses.
In vivo experiments
The Francis Crick’s Institute Animal Welfare and Ethical Review Body and UK Home Office authority provided by Project License 70/8380 approved all animal model procedures. A431 inducer or non-inducer cell lines were co-cultured with VCAF2b in 1:1 ratio for 24h before tumour induction. Tumours were induced by injecting ICRF nu/nu or Balb/c nu/nu mice with 4x106 co-cultured cells in PBS/30% matrigel (BD biosciences #356235) either with (at Multiplicity of Injection (MOI) of 0.005, 0.00158, or 0.0005) or without HSV-eGFP oncolytic virus. Tumours were measured three times a week. Mice were culled when tumours reached more than 12mm in mean tumour diameter, or the tumour ulcerated significantly.
Oncolytic Virus
Cancer cells and fibroblasts were cultured in direct, indirect co-culture or alone for 24 hours prior to addition of oncolytic virus for 18h (vaccinia) or 48 hours (HSV, Reo) (MOIs varied depending on cell type and oncolytic virus, MOIs are indicated in the relevant experimental figures). Subsequently cells were fixed in 4% paraformaldehyde (PFA) for 10 minutes. We used a Zeiss LSM 510 system to acquire epifluorescence micrographs. For siRNA experiment cells were incubated for 24h with the specific siRNA before trypsinization and plating in the different co-culture conditions. HSV1716-eGFP, Vaccinia-RFP (GL-ONC1) and Reovirus were kind gifts from Sorrento Therapeutics, Genelux and Oncolytics Biotech Inc. respectively.
Microarray
VCAF2b and A431 with stably expressing fluorescent proteins were FACS sorted 24 hours after seeding them in mono-culture, direct or indirect co-culture. The cells were directly sorted into 500μl RLT buffer containing 1% β-mercaptoethanol (Sigma #M6250) and RNA was extracted using RNAeasy microkit (Qiagen #74034). n=2 biological replicates Gene expression data were analysed using Bioconductor 2.7 (http://bioconductor.org), running on R 2.12.1. Normalised probe set expression measures were calculated using log2 transformation and quantile normalisation using the ‘Lumi’ package (Du p et al (2008). To identify significant differences in gene expression in different groups, moderated Student’s t-tests were performed using empirical Bayes statistics in the ‘Limma’ package, and the resulting P-values were adjusted for multiple testing using the false discovery rate (FDR) Benjamini and Hochberg method (Smyth, 2004); probe sets with adjusted P-value FDR q<0.05 were called differentially expressed. Gene expression data were analysed using Bioconductor 2.7 (http://bioconductor.org), running on R 2.12.1. Normalised probe set expression measures were calculated using log2 transformation and quantile normalisation using the ‘Lumi’ package3. To identify significant differences in gene expression in different groups, moderated Student’s t-tests were performed using empirical Bayes statistics in the ‘Limma’ package, and the resulting P-values were adjusted for multiple testing using the false discovery rate (FDR) Benjamini and Hochberg method4; probe sets with adjusted P-value FDR q<0.05 were called differentially expressed. Microarray data is available at GEO under record GSE121058.
Single-cell RNA seq
Data analysis GSE1033225 was downloaded and single cell RNA normalised expression was analysed to determine gene expression of cells categorised as fibroblasts by the authors.
Gene set enrichment analysis (GSEA)
Gene set enrichment Analysis, GSEA, (version 2.2.3) 6 was carried out between VCAF_CO and VCAF_SOL groups using gene sets extracted from Twyman-Saint Victor7 et al. All parameters were kept as default. Gene signatures with FDR q-value equal or less than 0.05 were considered statistically significant.
Analysis of human CAF RNA sequencing data
Sequencing data was obtained and analysed previously in our group as explained in Park et al8. This data is deposited at the NCBI Gene Expression Omnibus under GSE121536. We extracted the transcripts-per-million values for the expression of the desired genes.
Invasion Assay
The cancer cell/CAF organotypic co-culture assay set up was adapted from a previously described assay1. In brief, a mixture of collagen 1 (BD Bioscience #354249, 4mg/mL) and Matrigel (BD Bioscience #356235, 2mg/mL) was set at 37°C for 1h. Subsequently fluorescently labelled, siRNA treated VCAF2b and A431 cells were added on top of the collagen I/Matrigel mix at a concentration of 5x105 cells each in DMEM/10%FBS/1%ITS/PenStrep. 24 hours later the gel was mounted on a metal bridge and fed from below with DMEM/10%FBS/1%ITS/PenStrep. After 6 days, the co-culture was fixed with 4%PFA/0.25% glutaraldehyde, the invasion depth was imaged with confocal microscopy and analysed using ImageJ.
Cytokine array
Cancer cells (A431, VSCC4, VSCC10) and VCAFs (VCAF2b, VCAF4, VCAF10) were grown in 6-well plates in direct or in-direct co-cultures for 24h. Conditioned media was collected, filtered using a 0.4μm low protein binding PVDF Miltex syringe-driven filter (Millipore #SLHV033RS) to remove cell debris and subsequently used on cytokine arrays (R&D systems #ARY005) according to the manufacturer’s instructions.
Calcein transfer (Parachute) Assays
A431 or VCAF2b were seeded to create a confluent monolayer 24h before adding Calcein loaded mCherry expressing A431 or VCAF2b. Calcein loading was accomplished by adding 1uM Calcein AM (Invitrogen #C1429) in HBSS onto the cells for 20 minutes at 37°C. Subsequently, cells were trysinized and added to the monolayer cultures for at least 2h. Imaging was performed on live cells with DRAQ5 as a nuclear marker added just before imaging.
5-Bromo-2’-deoxyuridine (BrdU) treatment
A431 cancer cells were cultured in media containing 100μM BrdU (Santa Cruz #40024) for 48 hours before trypsinizing and plating with VCAF2b cells for subsequent co-culture experiments.
Crash Assay
2 well culture-inserts (Thistle #80209) were adhered to glass bottom plates (MatTek #p24-1.0-13-f) coated with 40μg/ml rat tail collagen (ThermoFisher #354249) in PBS. VCAF2b fibroblasts were plated in 1 well of the insert 3 days before plating A431 cancer cells in the adjacent well. 24 hours later the insert was removed and cells were left for ~20 hours to collide before fixing with 4% PFA for further experiments involving RNA Scope and or immunofluorescence.
IRF3 Time Course
Fibroblasts were seeded on MatTek plates as above for 24 hours before A431 cancer cells were plated on top. Cells were then fixed in 4% PFA at 2, 4, 6 and 8 hours before staining for immunofluorescence. Primary antibody used; IRF3 (Cell Signaling, D614C).
Immunofluorescence
Cells in culture Cells were seeded on MatTek plates as above and fixed in 4% PFA before carrying out immunofluorescence. Cells were permeabilized (0.25% Triton-X in PBS), blocked (5% BSA in PBS with 0.05% Tween-20) and incubated overnight in 4˚C in primary antibodies diluted in 1% BSA in PBS with 0.05% Tween-20 (PBST). Primary antibodies include; monoclonal rat anti-BrdU clone BU1/75, 1:1000 (Bio-Rad #OBT0030CX) and monoclonal mouse anti-keratin 7/17 clone CAM5.2, 1:200 (in-house), polyclonal rabbit anti-MX2, 1:50 (Novus Biologicals, #NBP1-81018), polyclonal rabbit anti-Fibronectin, 1:400 (Sigma #F3648), monoclonal mouse anti-Vimentin clone LN-6, 1:200 (Sigma #V2258), mouse monoclonal anti-dsDNA clone AE-2, 1:200 (Merck Millipore, #MAB1293), mouse monoclonal anti-phospho histone H2A.X ser 139 clone JBW301, 1:500 (Merk Millopore #05-636), reovirus–σ3 4F2 (DSHB, University of California). 4F2 (reovirus) was deposited to the DSHB by Dermody, T.S. (DSHB Hybridoma Product 4F2 (reovirus)). Cells were then washed before incubating in secondary antibody diluted in 1% BSA in PBST for 1 hour. Cell were washed before adding DAPI 1:1000 (Sigma D9542) and Phalloidin (dilution 1:500, Sigma, 68825) and imaged using Zeiss LSM780 or Zeiss LSM510 microscope. Images were analysed using ImageJ.
Patient material
Patient material was obtained from the Royal Marsden Hospital after informed consent through the project CCR2924 (Approved by the London/Chelsea NHS REC with code REC: 06/Q0403/125). All material was handled according to relevant ethical regulations by the Human Tissue Act. After excision the tumours were flash-frozen. Briefly, fresh frozen sections were fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X-100 and stained with anti-IRF3 antibody (dilution 1:100 Cell Signaling, D614C). Phalloidin (dilution 1:500, Sigma, 68825) and DAPI (dilution 1:1000, Sigma D9542) were added as counterstaining. Sections were mounted using MOWIOL reagent and imaged using a Zeiss LSM 780 microscope.
RNA scope
Cells were plated on chambered glass slides (LAB-TEK, Sigma #S6690-1PAK) coated with 40μg/ml rat tail collagen (ThermoFisher #354249) in PBS and fixed in 4% PFA. RNA scope was carried out in compliance with manufacturer’s instructions (Advanced Cell Diagnostics). Probes against IFNβ (C3) and MX2 (C1) were used and cells were mounted using ProLong™ Diamond Antifade Mountant (Invitrogen #P36965). Immunofluorescence for keratin7/17 or fibronectin was carried out after RNA Scope and slides were re-mounted, and imaged using Zeiss LSM780 or Zeiss LSM510 microscope.
RNA isolation and qRT-PCR
Cells were harvested using RNAprotect cell reagent (Qiagen #76526) and tumour samples were retained in RNA Later (Qiagen #76106) and kept frozen until RNA extraction using either Qiagen micro/mini-kit (Qiagen #74034/74104) or MagMAX™-96 magnetic bead-based purification kits (ThermoFisher #AM1830). The cDNA was prepared using M-MuLV reverse transcriptase (Promega # m3682), and qPCR was performed using Platinum SYBR green qRT-PCR SuperMix-UDG with Rox (ThermoFisher #11744100), using the QuantStudio 7 Real-Time PCR systems (Applied Biosystems). RNA levels were normalised using at least two housekeeping genes using the ΔΔCt method. For experiments in which we used a treatment control group we then normalised to to the appropriate control (i.e. IgG for antibody treatments, scramble siRNA for the silencing experiments, and DMSO for the drug treatment experiments).
Fluorescence-activated cell sorting (FACS)
Cells were sorted using a flow cytometry cell sorter MoFlo XDP. Fluorescently labelled A431-Ypet and VCAF2b-mCherry cells were co-cultured for 20 hours. The cells were then trypsinized and re-suspended in 3%BSA in PBS + 1mM EDTA in preparation for sort. Cells were separated into 3 populations; A431-Ypet, VCAF2b-mCherry, and VCAF2b-mCheryy cells containing A431-Ypet material (ccCyto) using 488nm laser with collection filter 530/40 for Ypet and 561nm laser with collection filter 610/20 for mCherry. Gates were designed based on negative and single colour controls. All cell populations were purity tested and data was analysed using FlowJo software.
Western blotting
For western blot, cells were lysed in buffer containing 50mM Tris HCL pH 8, 150mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 1% SDS and 1mM EDTA (all purchased from Sigma). Just before use a protease inhibitor cocktail tablet (Roche #11873580001) was added to the buffer. Cells were grown in a 6-cm dish and washed with PBS before adding the lysate buffer. Cells were then mechanically lysed using a cell scraper, transferred into eppendorfs and incubated on ice for 15 minutes. The lysates were Sonicated between 10 and 14 microns (amplitude) for 10 seconds. Samples were centrifuged at 4˚C for 5 minutes before measuring protein levels. Total protein was quantified using the Bicinchoninic Acid (BCA) method in accordance with manufacturer’s instructions (ThermoFisher #23225). Following protein quantification, 20μg of sample was loaded on a 4-12% gradient Bis-Tris gel (Invitrogen #4561085) and transferred to a PVDF membrane (Invitrogen #1704156) for botting. The membrane was blocked for 1 hour in a mix of 5% BSA/Milk in TBST and then incubated overnight at 4˚C with IRF3 (D6I4C) antibody (dilution 1:1000, Cell Signalling #11904S) or anti-β-actin (AC-15) antibody (dilution 1:100 000, Sigma #A1978). The following day, the membrane was washed before adding the HRP-conjugated secondary (Dilution 1:25 000, ThermoFisher), and incubating for 1 hour at room temperature. The membrane was washed again before developing with Luminata Western HRP substrate kit (Millipore #WBLUR0100) and imaged.
Statistical Analysis
Statistical analysis was performed using Prism software (Graphpad Software). Mean values and standard deviation are shown in graphs. P-values were obtained from Mann-Whitney t test for comparing two datasets and ANOVA followed by Dunnett’s multiple-comparison post hoc test for multiple datasets when sample size was large enough to confirm normality using Shapiro-Wilk normality test. For the QRT-PCR normalized data, one sample t-test referenced to an hypothetical value was used. Level of significance is indicated with * (p < 0.05), ** (p < 0.01), *** (p < 0.001), **** (p < 0.0001). All statistic tests performed were two-tailed.
Supplementary Material
Table 1. STR Profiles of cultured fibroblasts and patient derived cancer cell lines.
| Cell Line | OCAF1 | OCAF2 | VCAF2B | VCAF4 | VSCC4 | VCAF10 | VSCC10 | A431 (Ind) | A431 (NonInd) |
|---|---|---|---|---|---|---|---|---|---|
| FGA | 19,20 | 22,26 | 23,26 | 20,25 | 20,25 | 20,25 | 25,25 | 20,20 | 20,20 |
| TPOX | 8,11 | 8,8 | 8,8 | 11,11 | 11,11 | 11,11 | 11,11 | 11,11 | 11,11 |
| D8S1179 | 13,14 | 14,15 | 8,15 | 15,15 | 15,15 | 15,17 | 15,17 | 13,13 | 13,13 |
| vWA | 14,17 | 15,17 | 14,15 | 16,17 | 16,17 | 14,14 | 14,14 | 15,17 | 15,17 |
| AMEL | X,Y | X,X | X,X | X,X | X,X | X,X | X,X | X,X | X,X |
| Penta D | 13,14 | 9,13 | 11,12 | 10,11 | 10,11 | 11,11 | 11,11 | 9,11 | 11,11 |
| CSF1PO | 11,12 | 11,12 | 10,10 | 10,13 | 10,10 | 11,11 | 11,11 | 11,12 | 11,12 |
| D16S539 | 11,13 | 11,12 | 12,13 | 8,9 | 8,9 | 10,12 | 12,12 | 12,14 | 12,14 |
| D7S820 | 11,12 | 10,11 | 9,12 | 10,10 | 10,10 | 10,11 | 10,11 | 10,10 | 10,10 |
| D13S317 | 9,13 | 11,12 | 12,12 | 8,12 | 8,12 | 10,10 | 10,10 | 13,13 | 9,13 |
| D5S818 | 11,11 | 12,12 | 9,9 | 12,13 | 13,13 | 11,11 | 11,11 | 12,13 | 12,13 |
| Penta E | 12,12 | 11,18 | 7,11 | 12,16 | 12,16 | 13,14 | 13,14 | 12,13 | 13,13 |
| D18S51 | 14,16 | 12,16 | 12,12 | 14,20 | 14,20 | 14,16 | 16,16 | 13,17 | 13,17 |
| D21S11 | 28,31 | 27,28 | 27,31 | 29,30 | 29,30 | 29,33.2 | 29,33.2 | 28,28 | 28,28 |
| TH01 | 9.3,9.3 | 6,6 | 7,9 | 6,9.3 | 6,9.3 | 9,9 | 9,9 | 9,9 | 9,9 |
| D3S1358 | 15,19 | 15,15 | 14,16 | 16,17 | 17,17 | 15,17 | 17,17 | 14,14 | 14,14 |
| Comments | Unique profile | Unique profile | Unique profile | Profile compares with VSCC4 | Profile compares with VCAF4 | Profile compares with VSCC10 | Profile compares with VCAF10 | Profile compares with A431 (NonInd) | Profile compares with A431 (Ind) |
Table 3. Primers used to amplify genes upregulated upon cancer cell-fibroblast contact.
| Gene | Sequence | |
|---|---|---|
| IFNB1 | Forward | 5’-GCGACACTGTTCGTGTTGTC-3’ |
| Reverse | 3’-ATAGATGGTCAATGCGGCGT-5’ | |
| CCL5 | Forward | 5’-AGCCCTCGCTGTCATCCT-3’ |
| Reverse | 3’-CACTTGGCGGTTCTTTCG-5’ | |
| CXCL10 | Forward | 5’-CCTGCAAGCCAATTTTGTCCACGTGTT-3’ |
| Reverse | 3’-AGCACTGCATCGATTTTGCTCCCCTC-5’ | |
| OAS2 | Forward | 5’-CCGTTGGTGTTGGCATCTTC-3’ |
| Reverse | 3’-GCATTGTCGGCACTTTCCAA-5’ | |
| MX1 | Forward | 5’-CACACCGTGACGGATATGGT-3’ |
| Reverse | 3’-TTTGGACTTGGCGGTTCTGT-5’ | |
| MX2 | Forward | 5’-GCCCTTAGCATGCTCCAGAA-3’ |
| Reverse | 3’-ATCGTGCTCTGAACAGTTTGG-5’ | |
| TMEM173/STING | Forward | 5’-ATATACAGCCGCTGGCTCAC-3’ |
| Reverse | 3’-GATATCTGCGGCTGATCCTG-5’ | |
| cGAS/MB21D1 | Forward | 5’-CAGCCGCGATGATATCTCCA-3’ |
| Reverse | 3’-GCAGAAATCTTCACGTGCTCATA-5’ | |
| IRF3 | Forward | 5’-TAAACGCAACCCTTCTTTGC-3’ |
| Reverse | 3’-GATGCACAGCAGGAGGATTT-5’ | |
Table 4. Primers used to amplify housekeeping genes in which to normalise gene expression to.
| House Keeping Gene | Sequence | |
|---|---|---|
| GAPDH | Forward | 5’-CAAAAGGGTCATCATCTCTGC-3’ |
| Reverse | 3’-AGTTGTCATGGATGACCTTGG-5’ | |
| ACTIN | Forward | 5’-GGACTTCGAGCAAGAGATGG-3’ |
| Reverse | 3’-AGCACTGTGTTGGCGTACAG-5’ | |
| B2M | Forward | 5’-CGTGGCCTTAGCTGTGC-3’ |
| Reverse | 3’-AATGTCGGATGGATGAAACC-5’ | |
Acknowledgements
We thank Probir Chakravarty for patient assistance with Bioinformatics, Shane Foo and Ramin Sadri for technical assistance, Joan Massague for discussion, laboratory colleagues for support and advice. We thank Andy Blanchard at GSK (Stevenage UK) for provision of compounds.
Funding
E.N.A, E.M. A.R., S.D., S.H. and E.S. are supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001144), the UK Medical Research Council (FC001144) and the Wellcome Trust (FC001144). E.N.A. was additionally supported by the Wellcome Trust (096084/B/11/Z). A.R. by the Spanish Society for Medical Oncology (Beca Fundación SEOM), E.M. by a BBSCR-GSK CASE Fellowship. K.J.H, A.M. and D.M. are supported by The Royal Marsden/The Institute of Cancer Research National Institute of Health Research Biomedical Research Center, and a Cancer Research UK grant (A23275).
Footnotes
Author Contributions
Conceptualization and Design: E.S., E.N.A., E.M., A.R., & K.J.H, Development of Methodology: S.D., E.N.A., E.M., & A.R., Acquisition of Data: E.N.A., S.D., E.M., S.H., T.K., D. M., & A.R.; Analysis and Interpretation of Data: E.N.A, E.S., A.R., E.M., K.J.H., & A.M., Writing & Editing, E.N.A, A.R., E.M., E.S., K.J.H., & A.M.
Competing interest declaration
E.S. and E.M received research funds from GSK through a BBSCR-GSK CASE Fellowship. The authors declare that there is no other competing interest.
Data availability statement
Microarray data have been deposited to GEO database under record GSE121058 and a read-only access token is available to reviewers upon request this embargo will be lifted after publication. Publicly accessed databases used are accessible at the NCBI Gene Expression Omnibus under records GSE103322 and GSE121536 as indicated in the methods section. Other data that support the findings are available upon reasonable request from the corresponding authors.
References
- 1.Barros MR, et al. Activities of stromal and immune cells in HPV-related cancers. J Exp Clin Cancer Res. 2018;37:137. doi: 10.1186/s13046-018-0802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 3.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 4.Labernadie A, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol. 2017;19:224–237. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirata E, Sahai E. Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harb Perspect Med. 2017:a026781. doi: 10.1101/cshperspect.a026781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su S, et al. CD10 + GPR77 + Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 2018;172:841–856.:e16. doi: 10.1016/j.cell.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Costa A, et al. Fibroblast heterogeneity and immunosuppressive environment in Human breast cancer. Cancer Cell. 2018:1–17. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Givel AM, et al. MiR200-regulated CXCL12β promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Özdemir BC, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andtbacka RHI, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 11.Pol J, et al. Trial Watch—Oncolytic viruses and cancer therapy. Oncoimmunology. 2015;5:e1117740. doi: 10.1080/2162402X.2015.1117740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribas A, et al. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell. 2017;170:1109–1119.:e10. doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type i interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 15.Puram SV, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171:1611–1624.:e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Öhlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017:jem.20162024. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaston J, et al. Intracellular STING inactivation sensitizes breast cancer cells to genotoxic agents. Oncotarget. 2016;7 doi: 10.18632/oncotarget.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Q, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533:493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong SM, et al. Co-regulation of transcellular and paracellular leak across microvascular endothelium by dynamin and Rac. Am J Pathol. 2012;180:1308–1323. doi: 10.1016/j.ajpath.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Verhelst J, Hulpiau P, Saelens X. Mx Proteins: Antiviral Gatekeepers That Restrain the Uninvited. Microbiol Mol Biol Rev. 2013;77:551–566. doi: 10.1128/MMBR.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackie RM, Stewart B, Brown SM. Intralesional injection of herpes simplex virus 1716 in metastatic melanoma Controlled oxygen therapy and carbon dioxide retention during exacerbations of chronic obstructive pulmonary disease. Lancet. 2001;357:525–526. doi: 10.1016/S0140-6736(00)04049-6. [DOI] [PubMed] [Google Scholar]
- 23.Ilkow CS, et al. Reciprocal cellular cross-talk within the tumor microenvironment promotes oncolytic virus activity. Nat Med. 2015;21:530–536. doi: 10.1038/nm.3848. [DOI] [PubMed] [Google Scholar]
- 24.Wongthida P, et al. Type III IFN interleukin-28 mediates the antitumor efficacy of oncolytic virus VSV in immune-competent mouse models of cancer. Cancer Res. 2010;70:4539–4549. doi: 10.1158/0008-5472.CAN-09-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamarin D, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. TGF- β upregulates CD70 expression and induces exhaustion of effector memory T Targeted depletion of an MDSC subset unmasks pancreatic ductal. Sci Transl Med. 2014;6:226ra32. doi: 10.1126/scitranslmed.3008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benci JL, et al. Opposing Functions of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell. 2019;178:933–948.:e914. doi: 10.1016/j.cell.2019.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benci JL, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. 2016;167:1540–1554.:e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayers M, et al. IFN- γ – related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Twyman-Saint Victor C, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015 doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glück S, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol. 2017;19:1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassona Y, Cirillo N, Heesom K, Parkinson EK, Prime SS. Senescent cancer-associated fibroblasts secrete active MMP-2 that promotes keratinocyte dis-cohesion and invasion. Br J Cancer. 2014;111:1230–1237. doi: 10.1038/bjc.2014.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manivanh R, Mehrbach J, Knipe DM, Leib A. Role of Herpes Simplex Virus 1 γ34.5 in the Regulation of IRF3 Signaling. 2017;91:1–13. doi: 10.1128/JVI.01156-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calvo F, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 36.Smyth G. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:1. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Material and Methods section
- 1.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 2.Calvo F, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 4.Smyth G. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:1. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 5.Puram SV, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171:1611–1624.:e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twyman-Saint Victor C, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015 doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park D, et al. Extracellular matrix anisotropy is determined by TFAP2C-dependent regulation of cell collisions. Nat Mater. 2019 doi: 10.1038/s41563-019-0504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data have been deposited to GEO database under record GSE121058 and a read-only access token is available to reviewers upon request this embargo will be lifted after publication. Publicly accessed databases used are accessible at the NCBI Gene Expression Omnibus under records GSE103322 and GSE121536 as indicated in the methods section. Other data that support the findings are available upon reasonable request from the corresponding authors.