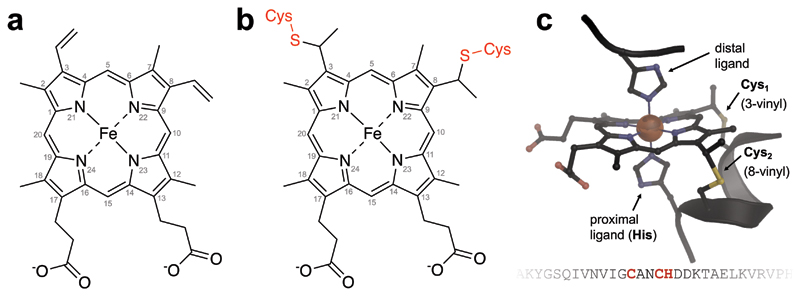
Abstract
The covalent attachment of one or multiple heme cofactors to their protein chain enables cytochromes c to be utilized in electron transfer and redox catalysis in extracytoplasmic environments. A dedicated heme maturation machinery, whose core component is a heme lyase, scans nascent peptides after Sec-dependent translocation for CXnCH binding motifs. Here we report the three-dimensional structure of the heme lyase CcmF from Thermus thermophilus, a 643 aa integral membrane protein. CcmF contains a heme b cofactor at the bottom of a large cavity that opens towards the extracellular side to receive heme groups for cytochrome maturation from the heme chaperone CcmE. A surface groove on CcmF may guide the extended apoprotein to heme attachment at or near a loop containing the functionally essential WXWD motif, situated above the putative cofactor binding pocket. The structure suggests heme delivery from within the membrane, redefining the role of the chaperone CcmE.
Introduction
The tetrapyrrole heme is among the most widespread protein cofactors.1 Its cyclic organic ligand protoporphyrin IX tetradentally coordinates an iron ion in its equatorial plane, promoting an octahedral geometry that leaves a proximal and distal axial position for the interaction with amino acid residues from the peptide chain or – in the case of heme-dependent enzymes – a substrate or cosubstrate (Extended Data Fig. 1a). Heme-containing proteins were named cytochromes for the strong, red color of the Fe-protoporphyrin IX complex and fulfill diverse tasks such as oxygen transport (hemoglobin),2 redox catalysis (catalase,3 nitrite4 and sulfite reductase5), monooxygenation (P450)6 or electron transfer, as in mitochondrial cytochrome c.7 Chemical variations of the cofactor are common and give rise to different classes of cytochromes.8 Among these, cytochromes of type b employ the unmodified heme group that typically binds within a hydrophobic pocket of the protein from where it can dissociate over time. Hemoglobin, myoglobin and the nitrophorins of blood-sucking parasites belong to this class,9,10 and its members are mostly restricted to a single cofactor per peptide chain. In order to avoid the loss of the cofactors in extracytoplasmic environments and allow for a higher cofactor:protein mass ratio,11 evolution has produced a series of systems for the covalent attachment of the cofactor to conserved binding motifs within the protein sequence (CXnCH, most commonly n = 2), defining class c of cytochromes (Extended Data Fig. 1b,c).12,13 To date five such heme maturation systems have been described, all of which contain a conserved heme lyase at their core that performs the remarkable task of scanning any apoprotein chain for the occurrence of a CXnCH motif and subsequently mediating an addition reaction of the cysteine thiols of the motif to the two vinyl side chains of the heme group.14,15 In microorganisms, this maturation process is tightly linked to the Sec-dependent co-translational export of the peptide chain and occurs prior to folding into the final tertiary structure, without any known involvement of further chaperones.14 The resulting cytochromes c can be monoheme proteins, but in organisms such as Geobacter sulfurreducens, whose genome encodes 111 cytochromes c, the largest known member of the family holds no fewer than 36 covalently attached cofactors.16
Heme lyases of all systems operate in diverse cellular contexts and accordingly show high variability. The simplest variation of the theme is exemplified by the human holocytochrome c synthases (HCCS1/2, system III)17 that reside on the inner mitochondrial membrane facing the intermembrane space, where they exclusively cater to the soluble electron carrier cytochrome c and the membrane-integral c 1 heme of respiratory complex III, respectively, and receive both apoprotein and heme from the intermembrane space.18
In contrast, the bacterial and archaeal systems are typically located in the cytoplasmic membrane and are remarkably unspecific in that they are able to attach heme groups to any and all CXnCH motifs in a given peptide chain.19 These heme maturation systems contain additional components, as they must also mediate the translocation of heme through the membrane prior to its attachment to the protein. The most complex of these machineries is system I, the Ccm system (for cytochrome c maturation),20,21 as found in α-, β-proteobacteria, plant mitochondria, deinococci, archaea and some γ-, δ-proteobacteria (Fig. 1a). Here, a set of eight gene products (Fig. 1b) mediates the translocation of heme into the periplasm (CcmABCD), its shuttling to the lyase (CcmE), the reduction of unwanted disulfides in the CXnCH motif (CcmGHI), and finally the covalent attachment to the protein (CcmF). CcmA and CcmB form part of an ABC transporter that associates with the integral membrane protein CcmC, which independently mediates heme translocation,22–24 and the small, monotopic membrane protein CcmD.25 CcmF and CcmHI (which frequently are fused) form a complex in most organisms that scans the protein to locate binding motifs and initiate cofactor attachment.26–28 The heme lyase CcmF is the functional core component of the system. It is a tryptophan-rich protein, containing a functionally essential WXWD motif,29 which is remarkably well conserved in the heme lyases of system II (CcsA) and III (HCCS), as well as in CcmC of system I among others, indicating a common origin and defining the family of ‘heme-handling proteins’ (HHP, Extended Data Fig. 2).30
Fgiure 1. Schematic architecture of heme maturation system I.
The cytochrome c maturation system (Ccm) is a multi-component machinery in the cytoplasmic membrane of most proteobacteria, archaea, deinococcales and plant mitochondria. a, In the Ccm system, the heme cofactor is translocated from its place of biosynthesis in the cytoplasm by the CcmABCD subcomplex centered on the ABC transporter CcmAB, whereby CcmC is required for heme translocation and attachment to the heme chaperone CcmE. CcmD is required for the dissociation of CcmE after loading with cofactor. In parallel, the apocytochrome chain is exported via the Sec system. Its CXXCH heme-binding motifs will oxidize to form disulfides that are reduced by CcmG, and protein and cofactor are then coupled by the heme lyase CcmF. In concert with CcmH it recognizes CXXCH motifs, receives heme from CcmE and attaches a cofactor to each motif in a processive manner from N- to C-terminus of the chain. Only after heme attachment the nascent cytochrome attains its final tertiary structure. b, The Ccm machinery of T. thermophilus is encoded in a single eight-gene operon.
In order to understand the complex interplay of heme transport and binding motif recognition, we have recombinantly produced the heme lyase CcmF from the thermophilic eubacterium Thermus thermophilus (TtCcmF) and have determined its three-dimensional structure by X-ray crystallography. TtCcmF is an integral membrane protein of 643 amino acids (71 kDa) with an extended extracellular domain and a heme group of type b. Its structure provides strong hints towards the sites and modes where heme and protein are bound to afford cofactor attachment.
Results
Overall Structure of CcmF
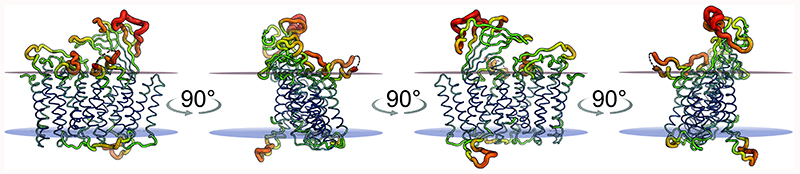
TtCcmF crystallized in the orthorhombic space group P212121, with one monomer in the asymmetric unit. The diffraction data collected from these crystals showed substantial anisotropy, with limiting resolutions of 2.63 Å along the a* axis and 4.90 Å along b* (Supplementary Table 1), in line with a crystal packing characterized by weak protein–protein interactions along the b axis (Supplementary Fig. 1). TtCcmF is a polytopic membrane protein with 15 transmembrane helices (Fig. 2a, Supplementary Fig. 2), at variance with most of the earlier topology predictions that assigned 11-13 transmembrane elements, but correctly derived from a phylogenetic analysis by Ferguson and co-workers.30 The N-terminus of the protein is located on the extracellular (p-) side of the membrane, and the periplasmic face of TtCcmF is dominated by the loop regions connecting helices h2 and h3 (loop 2) as well as h4 and h5 (loop 4) in the N-terminal half of the protein. In addition, the C-terminal part features a large periplasmic domain (residues 508-608) in loop 14, connecting helices h14 and h15 (Fig. 2d). This domain shows an unusual tertiary structure with two antiparallel, four-stranded β-sheets that is similar in topology only to subdomains of obviously functionally unrelated viral Gc glycoproteins of the phenoviridae family (Supplementary Fig. 3). Its increased temperature factors with respect to the membrane-integral part of the heme lyase point towards a degree of structural flexibility that is well in line with substantial conformational changes during its functional cycle (Extended Data Fig. 3). While the recognition and reduction of heme-binding motifs requires CcmF to act in concert with CcmH, the periplasmic face of CcmF with its extended, flexible loop regions likely plays an integral role in this process, by guiding the nascent apocytochrome chain and scanning it for the CXnCH signature motifs (Extended Data Fig. 4).
Fgiure 2. Structure of T. thermophilus CcmF.
a, Cartoon representation of CcmF and its orientation within the membrane in front (left) and side view (right). The accessory heme is shown in ball-and-stick representation and the structure is colored from blue at the N-terminus to red at the C-terminus. Helices are numbered and the periplasmic C-terminal domain is indicated. Lipid and detergent molecules visible in the electron density map are shown as sticks. b, Top view of the heme b binding site from the periplasmic side. Six of the 15 transmembrane helices form a wide channel above the accessory heme that is capped by loop 6, which holds the conserved WXWD motif. c, Side view of helices 6, 7 and 14, with the heme b moiety liganded by residues H259 and H439. The hydrophobic channel is blocked by residues W214 and W251, and loop 6 with the WXWD motif is positioned above a wide vestibule on the periplasmic side. d, The C-terminal periplasmic domain of CcmF and its anchoring transmembrane helices 14 and 15. In the periplasmic domain, eight β-strands form two antiparallel sheets.
An accessory heme cofactor
The transmembrane helices of CcmF group into three segments (Fig. 2a), with helices h1–h4 forming a compact four-helix bundle at the apex of the protein that terminates with the extended loop 4. Helices h5–h10 and h11–h15 then frame a large central channel that spans the entire width of the lipid bilayer but is blocked on the intracellular side by a bound heme (Fig. 2b, Extended Data Fig. 5). This cofactor is non-covalently bound to the protein (heme b), but is held through axial coordination of the iron ion by residues H259 in helix h7 and H493 in helix h14 (Fig. 2c). The presence of a non-exchanging heme b in CcmF was reported previously,31 and this cofactor was shown not to represent the substrate heme to be attached to an apoprotein heme-binding motif, but rather to play a role as a redox-active accessory heme, with a midpoint potential determined for E. coli CcmF to –147 ± 2 mV at pH 7.32 In vitro, this heme group was reduced by quinols,31 and while the addition of a cysteine thiol to a double bond of a heme vinyl side chain is not a net redox process, the accessory heme in CcmF might serve to reduce a second (substrate) heme that is delivered by CcmE. This substrate heme is covalently attached to CcmE via a histidine, and reducing the metal to the Fe2+ state was suggested to facilitate the breaking of this bond in preparation for the nucleophilic attack of a cysteine thiol.33 It remains to be elucidated whether this is a concerted process and whether the electron is transferred back to the accessory heme after the reaction or leaves CcmF on the substrate heme.
Binding cavity for a substrate heme group
The structure of CcmF only contains the accessory cofactor, but one of its most prominent features is a wide channel right above this heme that is partly capped on the extracellular side by loop 6, connecting helices h6 and h7. This loop region contains the hallmark WXWD motif and in it, both tryptophan residues face the lumen of the channel (Fig. 2c). The aspartate that forms part of this motif, D241, is oriented away from the channel, but plays an obvious role by capping the positive end of helix h7 through H-bond formation with the amide NH groups of V243 and E244. Such capping residues are known to stabilize α-helices,34 and the conservation of D241 in the general heme-handling WXWD motif points towards a crucial role for helix h7 that may well have to undergo conformational changes within CcmF during uptake and alignment of the substrate heme (vide infra). In the structural model, the cavity between the WXWD-containing loop 6 and the accessory heme group is occupied by the hydrophobic tails of a detergent molecule (dodecyl maltoside) and a phospholipid (PE), whose polar head groups reside at the membrane/water interface and thus help to define the position of the protein within the lipid bilayer, in line with a prediction obtained from the orientation of proteins in membranes (OPM) server (Fig. 2a).35 The protein surface with respect to the position of the membrane reveals a substantial incision in the otherwise compact arrangement of transmembrane helices (Fig. 3a) where helices h6, h8 and h14 create a vestibule that opens laterally, forming a gate from the central channel to the outer leaflet of the membrane between helices h11 and h13. This cleft is of a size and shape to allow for the insertion of a second heme cofactor – the substrate heme – that in system I cytochrome c maturation is delivered by the CcmE protein (Fig. 3b).
Fgiure 3. Heme binding to CcmF and structural clues to protein function.
a, A channel crossing the entire heme lyase consists of two vestibules near the cytoplasmic and periplasmic faces of the membrane, respectively, separated by a narrow, hydrophobic constriction. Each vestibule is of sufficient size to accommodate a heme cofactor, but only the cytoplasmic vestibule contains the accessory heme b moiety of CcmF. The periplasmic vestibule is almost completely open to the outer leaflet of the membrane. Helices h10 and h11 are partially omitted for clarity. b, Van-der-Waals surface representation of CcmF, colored from –5 k B T to +5 k B T. The two orientations shown highlight the opening of the periplasmic vestibule and the surface groove presumed to guide the apocytochrome chain. c, Molecular docking of a second heme group in the periplasmic vestibule, in an orientation as suggested by the buoy model for heme delivery. d, View from within the membrane onto the docked substrate heme in the periplasmic vestibule.
Molecular docking of an additional heme group in this position yielded a very good fit (Fig. 3c,d), and in fact also led to a displacement of the side chains of residues W238 and W240 within the WXWD motif, such as to sandwich the porphyrin moiety of the substrate heme, in accordance with the general concept of ‘heme handling’ by proteins containing this particular motif (Extended Data Fig. 6).30 Notably, H172 and H301 that were suggested to axially ligate the substrate heme group when bound to CcmF (Supplementary Fig. 4) are not in direct proximity to the postulated binding site of the substrate heme in this structure.29 Residue H301 is located on a disordered loop connecting helices h8 and h9 and may reach the bound heme upon changing its conformation, while H172 within helix h5 faces towards the vestibule, but resides at a distance of about 17 Å from the substrate heme iron. It is tempting to speculate on a sequence of events where the heme-handling WXWD motif receives the substrate heme from CcmE as in our docking model, followed by a conformational change that pulls the substrate heme into the core of CcmF to be liganded by H172 and H301 prior to attachment to the apoprotein chain. However, the further elaboration of the dynamic action of the heme lyase will require more direct structural evidence.
Discussion
In accompanying the cofactor from the translocase subcomplex CcmABCD to the lyase CcmF, the role of the CcmE protein was described as a heme chaperone.36 It receives heme from CcmC that itself contains a WXWD motif and mediates an anti-Markovnikov addition reaction of the β-carbon of the 3-vinyl group of heme to a conserved histidine (H118 in T. thermophilus CcmE) of the chaperone. NMR structures of cofactor-free CcmE showed a compact β-barrel fold,37,38 implying that the heme group, when bound, must be largely exposed. A conserved tyrosine (Y122 in T. thermophilus CcmE) was identified as an axial ligand to heme and interpreted such that the hydrophobic cofactor might lay flat on the outside of CcmE, shielded from the hydrophilic environment of the periplasm by the C-terminus of the protein while in transit from CcmC to CcmF.33 The structure of CcmF now provides new insights that suggest a more straightforward mechanism of heme shuttling. It also explains the unusual configuration of CcmE as a ‘chaperone’ that does not seem to shield its cargo at all. As CcmF does not function as a transporter, the substrate heme group was suggested to reach the lyase from the periplasmic side. However, the large lateral gate in the integral membrane protein and the appropriate size for the internal cavity in CcmF to hold a bound substrate heme (Fig. 3c, Extended Data Fig. 6) opens the possibility to insert the cofactor from within the membrane, without ever requiring its complete extraction from the hydrophobic phase. The enzyme ferrochelatase (HemH) that finalizes heme biosynthesis with the insertion of Fe2+, is located in the cytoplasm in some bacteria, but associated with the cytoplasmic membrane in others,39,40 so that direct delivery of the cofactor into the membrane is possible.
A rationale for the apparent complexity of heme attachment thus is that while the porphyrin ring – given that the Fe ion is shielded by suitable ligands – will readily partition into a lipid bilayer due to its hydrophobicity, the charged propionate side chains will prefer the polar head group region, as generally seen in the known structures of membrane-integral cytochromes b (Extended Data Fig. 7). This facilitates lateral heme movement, but not a flip-flop rearrangement between inner and outer leaflet. In cofactor-loaded CcmE, however, heme is attached via the 3-vinyl group that in this orientation would point towards the center of the membrane. In the Ccm system, the membrane protein CcmC is strictly required for loading CcmE with heme,41 while CcmD is needed for the release of holo-CcmE from CcmC.42 The ABC transporter CcmAB is not indispensable for heme maturation and does not transport heme from the cytoplasm to the periplasm.22,24 Its role remains under debate, and it was speculated that its ATP-dependent conformational changes might provide mechanical force for the action of CcmCDE. Alternatively, it might either serve as a heme flippase acting to enrich the cofactor in the outer leaflet of the membrane or help to rotate the heme group such that attachment of the 3-vinyl to His118 of CcmE becomes feasible. Either way, the CcmC-mediated attachment to CcmE assures a location of the cofactor in the outer leaflet of the lipid bilayer, providing direct access to the lateral gate of CcmF (Fig. 4). In this model the chaperone acts like a buoy, retaining heme at the outer leaflet and assisting its insertion into CcmF in the correct orientation for the highly stereospecific cofactor attachment. CcmE also directly interacts with CcmH(I), even in its heme-free state,43 and its free diffusion between isolated subcomplexes was questioned previously, raising the possibility of a functional maturase supercomplex consisting of all parts of the Ccm machinery.44 A protein machinery of this size and complexity would require substantial intrinsic flexibility, in particular for the periplasmic domains of CcmF, CcmE and CcmH(I), but would provide a most efficient arrangement with distinct components responsible for heme delivery, apoprotein recognition and cofactor attachment. Experimental evidence for the functionality of physiologically relevant maturase supercomplexes remains to be provided, and the definition of further conformational states will be a challenging yet rewarding target for future structural analyses. Interestingly, the heme lyase invariably attaches the second cysteine of the heme-binding CXnCH motif to the 8-vinyl group of the heme cofactor first, implying that the release from CcmE follows only after this step, so that the heme group always remains covalently bound to either the chaperone or the cytochrome chain.45 A ternary complex was reported, where the apoprotein chain is linked to CcmH(I) via a disulfide bond and to heme via a thioether to the 8-vinyl, while the 3-vinyl remains attached to CcmE.43
Fgiure 4. A buoy model for heme delivery to CcmF.
Considering that the hydrophobic porphyrin moiety will partition into the membrane such that its hydrophilic propionate sidechains remain at the membrane surface, the ABC transporter CcmAB might act as a heme flippase translocating the cofactor from the inner to the outer leaflet of the cytoplasmic membrane. Based on the hypothesis that heme-handling proteins containing the WXWD motif work by orienting the cofactor by rotation, CcmF then accepts the cofactor into its hydrophobic binding pocket and arranges it such that its vinyl groups are accessible for an attack by the cysteine thiols of the heme-binding motifs of the apocytochrome chain.
Of course, this entire complexity could be circumvented by simply translocating the accessory heme group, bound in CcmF with its propionate moieties facing the cytoplasm, to the outer leaflet and the WXWD motif situated right above its binding site. This would alleviate the need for heme flipping and subsequent rotation and – most strikingly – this seems to be exactly what is realized in the cytochrome c synthases of system II, CcsBA.46 In fact, the HHP family members CcsBA and CcmF(HI) share key structural features, but CcsBA unites the functionalities of heme translocation, peptide scanning and heme attachment in a single machinery. The heme lyase CcmF seems to have evolved from this integrated device, and why it has differentiated into the complex multi-part Ccm system with its heme translocator and chaperone/shuttle components remains an open question within this important pathway of posttranslational maturation of cofactor-containing metalloenzymes.
Online Methods
Cloning, production, and purification of recombinant proteins
The gene encoding for TtCcmF was amplified via PCR from the genomic DNA of Thermus thermophilus HB27 and inserted into a modified pGEX-3 plasmid by Gibson cloning.47 The GST-tag of the pGEX-3 vector was replaced by a StrepTag(II) sequence (5’-AGCTGGAGCCACCCGCAGTTCGAAAAA-3’) followed by a linker (5’-GGCGCC-3’) and a TEV cleavage site (5’-GAGAACCTGTACTTCCAATCA-3’), resulting in N-terminally Strep-tagged CcmF. All constructs were verified by DNA sequencing (Eurofins GATC).
TtCcmF was produced in E. coli C43 cells grown in LB medium supplemented with 100 μg·mL−1 ampicillin. A 100 mL pre-culture was inoculated from single colonies and was incubated at 37 °C and 180 rpm until an OD600 of ~0.6 was reached. The pre-culture was used to inoculate 9 l of main culture which was incubated at 37 °C and 180 rpm to an OD600 of ~0.9. The culture was induced with 100 mM IPTG (isopropyl-β-D-thiogalactopyranoside) and further incubated at 20 °C and 180 rpm overnight.
Cell growth for selenomethionine-derivatized TtCcmF was done in M9 minimal media supplemented with 0.004 % (w/v) arginine, asparagine, glutamine, glutamate, histidine, isoleucine, leucine, phenylalanine, serine, threonine, tryptophan, tyrosine, valine, 0.2 % (w/v) glucose, 2 mM MgSO4, 100 μM CaCl2 and 0.00005 % (w/v) thiamine at 37 °C at 180 rpm until an OD600 of ~0.9 was reached. The culture was supplemented with threonine, lysine, phenylalanine (100 mg each per L of culture), with leucine, isoleucine and valine (50 mg each per L of culture) and L-(+)-selenomethionine (60 mg per L of culture) and incubated for 15 min. Afterwards, protein production was induced with 1 mM IPTG and FeSO4 (6.25 μg·mL−1).
Cells were harvested at 5,000×g, resuspended in lysis buffer (50 mM Tris/HCl pH 7.5, 100 mM NaCl) and disrupted in a microfluidizer (Microfluidics). The lysed cells were centrifuged at 15,000×g for 30 min, and subsequently at 300,000×g for 2 h. The resulting membrane pellet was resuspended in lysis buffer and stored at –80 °C until further use.
For purification the membrane pellet was thawed and solubilized for 1 h in lysis buffer supplemented with 1 % (w/v) DDM (n-dodecyl-β-D-maltopyranoside). The suspension was centrifuged at 100,000×g for 30 min and the supernatant was loaded onto a Strep-tactin superflow column equilibrated with loading buffer (50 mM Tris/HCl pH 7.5, 100 mM NaCl, 0.02 % (w/v) DDM). The column was washed subsequently with loading buffer and TtCcmF was eluted with elution buffer (50 mM Tris/HCl pH 7.5, 500 mM NaCl, 0.02 % (w/v) D12M, 5 mM D-desthiobiotin). Protein containing fractions were pooled and concentrated in VivaSpin concentrators (Sartorius, 100 kDa cut-off). The concentrated protein was further purified via size exclusion chromatography (Superdex S200 16/60, GE Healthcare) in SEC buffer (20 mM Tris/HCl pH 7.5, 100 mM NaCl, 0.02 % (w/v) DDM) and concentrated in VivaSpin concentrators (Sartorius, 100 kDa molecular weight cut-off). Protein samples were either used directly for crystallization or stored at –80 °C until use.
Crystallization
Initial crystallization was set up with an OryxNano crystallization robot (Douglas Instruments) with initial screens prepared internally in the group. Initial hits were refined by systematic variation of crystallization conditions. Crystals were obtained at 20 °C within 3-7 days in 0.6 μL drops containing 5 mg·mL−1 protein mixed with the reservoir in a 1:1 ratio. The best crystals of TtCcmF appeared in conditions with 18-19 % (v/v) low molecular weight polyethylene glycols (PEG 400, PEG 550 MME, PEG 600, PEG 1000), 0.1 M BisTris pH 6.5, 150 mM MgCl2 and 4 % (v/v) trifluorethanol and were plate shaped with dimensions of 200×60×10 μm3. Crystals were harvested from their drops in nylon loops, briefly placed in reservoir drops mixed with 10 % (v/v) 2R,3R-butane diol and vitrified in liquid nitrogen.
Data collection and structure determination
X-Ray diffraction data were collected at 100 K on beamline X06SA at the Swiss Light Source (Paul Scherrer Institut, Switzerland) equipped with an Eiger 16M detector (Dectris). Datasets for phasing were collected at λ=0.9797 Å using a low-exposure procedure with the beam transmission set to 6 % on a selenomethionine-derivatized crystal, while diffraction data from native crystals were collected at λ=1.0000 Å. Data were processed with XDS,48 autoPROC,49 and StarAniso. For phasing, seven datasets of a single selenomethionine-derivatized crystal were processed separately and scaled and merged in Aimless. Heavy atom site search, phasing, density modification and initial structure building were performed in AutoSol50 from the Phenix suite.51 The initially obtained protein model was of poor quality and completeness and was manually built in COOT 52 and refined in BUSTER 53 in iterative cycles. The sequence register was fixed with the help of selenium sites obtained from the selenomethionine-derivatized datasets. Final refinement was carried out against two merged data sets obtained from the same crystal, with limiting resolutions of 2.7 Å and 3.1 Å along a * and c * axes and 4.9 Å along axis b *, as evaluated with StarAniso (Global Phasing Ltd.). The final steps of model refinement were carried out with phenix.refine,51 and model validation was done with Molprobity.54 Data collection and refinement statistics are listed in Supplementary Table 1.
Extended Data
Extended Data Fig. 1. The heme cofactor and c-type cytochromes.
a, Fe-protoporphyrin IX, the widely used tetrapyrrole cofactor heme, with IUPAC numbering of the carbon atoms in the aromatic ligand. Two vinyl side chains are located at position 3 and 8, and the negatively charged propionate side chains at positions 13 and 17 are relevant for the translocation of the cofactor across a lipid bilayer. b, in cytochromes of type c, the heme cofactor is covalently linked to two cysteine residues of the protein chain via thioether bonds. This linkage is catalyzed by heme lyases and allows for a high cofactor/protein ratio. c, heme cofactors are bound to signature CXXCH motifs in the protein sequence, with the two cysteine residues of the motif forming the thioether linkages, and the subsequent histidine acting as a proximal axial ligand to the iron ion of the cofactor. Figure made from cytochrome c nitrite reductase (PDB ID 1FS7).1
Extended Data Fig. 2. The tryptophan-rich signature motif (WXWD).
In all classes of heme lyases, a tryptophan-rich motif is suggested to be directly involved in the handling of the heme cofactor. It is found in human cytochrome c synthase (HCCS), as well as in the lyase CcsA of system II and the components CcmC and CcmF of system I.
Extended Data Fig. 3. B-factor distribution in TtCcmF.
Elevated B-factors provide a measure of structural flexibility within the structure of CcmF. The cytoplasmic face of the protein shows high B-factors only in the loop connecting helices h8 and h14 near the C-terminus. In contrast, the periplasmic face of the heme lyase features multiple regions with increased flexibility, notably including the periplasmic domain in the loop connection helices h14 and h15.
Extended Data Fig. 4. Surface representations of TtCcmF.
a, Stereo representation of a low-resolution molecular surface of CcmF and its orientation within the membrane. The membrane is represented by a red disc for the periplasmic boundary and a blue disc for the cytoplasmic boundary. b, Three different views of the surface of CcmF with bound lipids in stick representation, highlighting the extensive periplasmic protrusion that is made up predominantly by the C-terminal periplasmic domain.
Extended Data Fig. 5. Environment of the accessory heme group in CcmF.
The stereo figure shows the b-type heme group liganded by residues H259 in helix h7 and H493 in helix h14. The open space above the accessory heme is the vestibule suggested to accommodate the substrate heme group for attachment to an apocytochrome chain. The refined 2Fo–Fc electron density map is contoured at the 1 σ level.
Extended Data Fig. 6. Docking model for a substrate heme group in CcmF.
The stereo figure details the docking result for a second b-type heme cofactor as a substrate heme within the cavity located above the accessory heme, seen from the opening of this vestibule towards the inner leaflet of the membrane. In order to accommodate the substrate heme, the two tryptophane residues of the WXWD motif in loop 6, W238 and W240, had to relocate, interacting with the bound heme moiety via π-stacking interactions.
Extended Data Fig. 7. Orientation of b-type heme groups in membrane proteins.
Membrane-integral heme groups are most commonly canonical Fe-protoporphyrin IX (heme b). Even bound within the protein matrix, they consistently orient with their propionate sidechains towards the hydrophilic surface of the membrane, revealing this to be a preferred orientation in either leaflet of the membrane. Panels show the orientation of protein complexes in the membrane above a detail of the orientation of the membrane-integral heme b groups. In the respiratory complexes cytochrome bc 1 (a) from oxidative phosphorylation and cytochrome b 6 f (b) from oxygenic photosynthesis, the low- and high-potential heme b moieties have both propionates facing the hydrophilic phase. c, in TtCcmF, the single heme b cofactor is located at the boundary of membrane and cytoplasm, with an approximate 40° rotation with respect to a and b, but in a highly similar arrangement as the heme group in the cytoplasmic leaflet of formate dehydrogenase (d) and nitrate reductase (e).
Supplementary Material
Acknowledgements
The authors thank Flore Kersten and Susana Andrade for support and helpful discussions. This work was supported by the European Research Council (grant no. 310656) and Deutsche Forschungsgemeinschaft (CRC 1381, project ID 403222702, and RTG 2202, project ID 46710898). We thank the beam line staff at the Swiss Light Source, Villigen, Switzerland, for excellent assistance with data collection.
Footnotes
Author Contributions A.B. and O.E. designed the experiments, A.B. and L.I. produced protein and generated crystals, A.B. solved the crystal structure, A.B. and L.Z. built and refined the crystal structure, A.B. and O.E. analysed data and wrote the manuscript.
Author Information The authors declare no competing financial interests.
Data Availability
The structural model and structure factors for TtCcmF have been deposited with the Protein Data Bank at www.pdb.org under the accession number 6ZMQ.
References
- 1.Anson ML, Mirsky AE. The Heme Compounds in Nature and Biological Oxidations. Science. 1928;68:647–648. doi: 10.1126/science.68.1774.647. [DOI] [PubMed] [Google Scholar]
- 2.Pauling L, Coryell CD. The Magnetic Properties and Structure of Hemoglobin, Oxyhemoglobin and Carbonmonoxyhemoglobin. Proc Natl Acad Sci U S A. 1936;22:210–216. doi: 10.1073/pnas.22.4.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fita I, Rossmann MG. The Active Center of Catalase. J Mol Biol. 1985;185:21–37. doi: 10.1016/0022-2836(85)90180-9. [DOI] [PubMed] [Google Scholar]
- 4.Einsle O, et al. Structure of cytochrome c nitrite reductase. Nature. 1999;400:476–480. doi: 10.1038/22802. [DOI] [PubMed] [Google Scholar]
- 5.Hermann B, Kern M, La Pietra L, Simon J, Einsle O. The octaheme MccA is a heme c-copper sulfite reductase. Nature. 2015;520:706–709. doi: 10.1038/nature14109. [DOI] [PubMed] [Google Scholar]
- 6.Schlichting I, et al. The catalytic pathway of cytochrome P450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 7.Margoliash E, Barlow GH, Byers V. Differential Binding Properties of Cytochrome c: Possible Relevance for Mitochondrial Ion Transport. Nature. 1970;228:723. doi: 10.1038/228723a0. [DOI] [PubMed] [Google Scholar]
- 8.Poulos TL. Heme Enzyme Structure and Function. Chem Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weichsel A, Andersen JF, Roberts SA, Montfort WR. Nitric oxide binding to nitrophorin 4 induces complete distal pocket burial. Nat Struct Biol. 2000;7:551–554. doi: 10.1038/76769. [DOI] [PubMed] [Google Scholar]
- 10.Perutz MF, Kendrew JC, Watson HC. Structure and Function of Hemeoglobin .2. Some Relations between Polypeptide Chain Configuration and Amino Acid Sequence. J Mol Biol. 1965;13:669. [Google Scholar]
- 11.Barker PD, Ferguson SJ. Still a puzzle: why is heme covalently attached in c-type cytochromes? Structure. 1999;7:R281–290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- 12.Moore GR, Pettigrew GW. Cytochromes c Evolutionary, structural and physicochemical aspects. Springer-Verlag; 1990. [Google Scholar]
- 13.Keilin D. On Cytochrome, a Respiratory Pigment, Common to Animals, Yeast, and Higher Plants. Proc R Soc London, B. 1925;98 [Google Scholar]
- 14.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- 16.Methé BA, et al. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 17.Babbitt SE, Sutherland MC, Francisco BS, Mendez DL, Kranz RG. Mitochondrial cytochrome c biogenesis: no longer an enigma. Trends Biochem Sci. 2015;40:446–455. doi: 10.1016/j.tibs.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babbitt SE, et al. Mechanisms of Mitochondrial Holocytochrome c Synthase and the Key Roles Played by Cysteines and Histidine of the Heme Attachment Site, Cys-XX-Cys-His. J Biol Chem. 2014;289:28795–28807. doi: 10.1074/jbc.M114.593509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens JM, Uchida T, Daltrop O, Ferguson SJ. Covalent cofactor attachment to proteins: cytochrome c biogenesis. Biochem Soc Trans. 2005;33:792–795. doi: 10.1042/BST0330792. [DOI] [PubMed] [Google Scholar]
- 20.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thöny-Meyer L. Cytochrome c maturation: a complex pathway for a simple task? Biochem Soc Trans. 2002;30:633–638. doi: 10.1042/bst0300633. [DOI] [PubMed] [Google Scholar]
- 22.Schulz H, Fabianek RA, Pellicioli EC, Hennecke H, Thöny-Meyer L. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc Natl Acad Sci U S A. 1999;96:6462–6467. doi: 10.1073/pnas.96.11.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baysse C, et al. Co-ordination of iron acquisition, iron porphyrin chelation and iron-protoporphyrin export via the cytochrome c biogenesis protein CcmC in Pseudomonas fluorescens. Microbiol SGM. 2003;149:3543–3552. doi: 10.1099/mic.0.26566-0. [DOI] [PubMed] [Google Scholar]
- 24.Page MD, Ferguson SJ. Mutational analysis of the Paracoccus denitrificans c-type cytochrome biosynthetic genes ccmABCDG: disruption of ccmC has distinct effects suggesting a role for CcmC independent of CcmAB. Microbiol-Uk. 1999;145:3047–3057. doi: 10.1099/00221287-145-11-3047. [DOI] [PubMed] [Google Scholar]
- 25.Richard-Fogal CL, Frawley ER, Kranz RG. Topology and function of CcmD in cytochrome c maturation. J Bacteriol. 2008;190:3489–3493. doi: 10.1128/JB.00146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders C, et al. The Cytochrome c Maturation Components CcmF, CcmH, and CcmI Form a Membrane-integral Multisubunit Heme Ligation Complex. J Biol Chem. 2008;283:29715–29722. doi: 10.1074/jbc.M805413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahuja U, Rozhkova A, Glockshuber R, Thöny-Meyer L, Einsle O. Helix swapping leads to dimerization of the N-terminal domain of the c-type cytochrome maturation protein CcmH from Escherichia coli. FEBS Lett. 2008;582:2779–2786. doi: 10.1016/j.febslet.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Fabianek RA, Hofer T, Thöny-Meyer L. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch Microbiol. 1999;171:92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- 29.San Francisco B, Sutherland MC, Kranz RG. The CcmFH complex is the system I holocytochrome c synthetase: engineering cytochrome c maturation independent of CcmABCDE. Mol Microbiol. 2014;91:996–1008. doi: 10.1111/mmi.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Harvat EM, Stevens JM, Ferguson SJ, Saier MH. Evolutionary origins of members of a superfamily of integral membrane cytochrome c biogenesis proteins. Bba-Biomembranes. 2007;1768:2164–2181. doi: 10.1016/j.bbamem.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 31.Richard-Fogal CL, et al. A conserved heme redox and trafficking pathway for cofactor attachment. EMBO J. 2009;28:2349–2359. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.San Francisco B, Bretsnyder EC, Rodgers KR, Kranz RG. Heme Ligand Identification and Redox Properties of the Cytochrome c Synthetase, CcmF. Biochemistry. 2011;50:10974–10985. doi: 10.1021/bi201508t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.San Francisco B, Kranz RG. Interaction of HoloCcmE with CcmF in Heme Trafficking and Cytochrome c Biosynthesis. J Mol Biol. 2014;426:570–585. doi: 10.1016/j.jmb.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoemaker KR, Kim PS, York EJ, Stewart JM, Baldwin RL. Tests of the Helix Dipole Model for Stabilization of α-Helices. Nature. 1987;326:563–567. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]
- 35.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulz H, Hennecke H, Thöny-Meyer L. Prototype of a heme chaperone essential for cytochrome c maturation. Science. 1998;281:1197–1200. doi: 10.1126/science.281.5380.1197. [DOI] [PubMed] [Google Scholar]
- 37.Enggist E, Thöny-Meyer L, Guntert P, Pervushin K. NMR structure of the heme chaperone CcmE reveals a novel functional motif. Structure. 2002;10:1551–1557. doi: 10.1016/s0969-2126(02)00885-7. [DOI] [PubMed] [Google Scholar]
- 38.Arnesano F, et al. Solution structure and characterization of the heme chaperone CcmE. Biochemistry. 2002;41:13587–13594. doi: 10.1021/bi026362w. [DOI] [PubMed] [Google Scholar]
- 39.Ferreira GC, et al. Structure and Function of Ferrochelatase. J Bioenerg Biomembr. 1995;27:221–229. doi: 10.1007/BF02110037. [DOI] [PubMed] [Google Scholar]
- 40.Dailey TA, Dailey HA. Identification of [2Fe-2S] clusters in microbial ferrochelatases. J Bacteriol. 2002;184:2460–2464. doi: 10.1128/JB.184.9.2460-2464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richard-Fogal C, Kranz RG. The CcmC:Heme:CcmE Complex in Heme Trafficking and Cytochrome c Biosynthesis. J Mol Biol. 2010;401:350–362. doi: 10.1016/j.jmb.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahuja U, Thöny-Meyer L. CcmD is involved in complex formation between CcmC and the heme chaperone CcmE during cytochrome c maturation. J Biol Chem. 2005;280:236–243. doi: 10.1074/jbc.M410912200. [DOI] [PubMed] [Google Scholar]
- 43.Verissimo AF, Mohtar MA, Daldal F. The Heme Chaperone ApoCcmE Forms a Ternary Complex with CcmI and Apocytochrome c. J Biol Chem. 2013;288:6272–6283. doi: 10.1074/jbc.M112.440024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verissimo AF, Daldal F. Cytochrome c biogenesis System I: An intricate process catalyzed by a maturase supercomplex? BBA-Bioenergetics. 2014;1837:989–998. doi: 10.1016/j.bbabio.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verissimo AF, et al. The thioreduction component CcmG confers efficiency and the heme ligation component CcmH ensures stereo-specificity during cytochrome c maturation. J Biol Chem. 2017;292:13154–13167. doi: 10.1074/jbc.M117.794586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci U S A. 2009;106:10201–10206. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.BUSTER v. 2.10.3. Global Phasing Ltd; Cambridge, United Kingdom: 2010. [Google Scholar]
- 54.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structural model and structure factors for TtCcmF have been deposited with the Protein Data Bank at www.pdb.org under the accession number 6ZMQ.