Abstract
Multidimensional single cell-analyses of T cells have fueled the debate about either extensive plasticity or “mixed” priming of T helper cell subsets in vivo. Here, we developed an experimental framework to probe the idea that the site of priming in the systemic immune compartment is a determinant of T helper cell-induced immunopathology in remote organs. By site-specific in vivo labeling of antigen-specific T cells in inguinal (i) or gut draining mesenteric (m) lymph nodes, we show that i-T cells and m-T cells isolated from the inflamed central nervous system in a model of multiple sclerosis are distinct. i-T cells were Cxcr6+ and m-T cells expressed P2rx7. Notably, m-T cells infiltrated white matter while i-T cells were also recruited to grey matter. Therefore, we propose that the definition of T helper cell subsets by their site of priming might guide an advanced understanding of T helper cell biology in health and disease.
Introduction
Organ-specific autoimmune diseases, including multiple sclerosis (MS), are initiated and maintained by the activation of autoreactive T cells in the systemic immune compartment 1. In MS, autoreactive T cells are likely activated through molecular mimics of autoantigens acquired at body surfaces 2. However, it is unclear whether and how the anatomical niche, in which an autoreactive T cell is primed, has an impact on the recruitment, topology, and inflammatory effector function of T cells in remote tissues like the central nervous system (CNS). Particularly in MS, the topology and the type of lesions within the CNS are critical determinants of the clinical phenotype 3, and we hypothesize that lesion development in MS is not random but controlled by immune system intrinsic molecular cues, which in turn might already be imprinted at the site of T cell priming outside the CNS. For instance, the gut microbiome might dictate the susceptibility to and severity of autoimmune diseases 4,5, e.g. by controlling the adjuvanticity in local lymph nodes, thus inducing distinct T helper cell species.
T helper cell subsets have been defined by their signature cytokines IFN-γ (Th1), IL-4 (Th2), and IL-17 (Th17). This concept has been widely accepted (also in T cell mediated autoimmune diseases) because cytokines determine the interaction of T cells with other immune cells, and thus their function in host defense and inflammation 6. While these T cell subsets are well characterized on the molecular level after in vitro differentiation of naive T cells 7, a large overlap between T cell subsets has been observed in vivo and explained by plasticity of T helper cells previously “cleanly” committed to a specific T cell lineage in mice and men 8,9. Alternatively, it is an emerging concept that T cell subsets might be primed as mixed phenotypes from the get-go 10,11.
Here, we label antigen-specific T cells in skin draining inguinal (iLN) and gut draining mesenteric lymph nodes (mLN) in vivo, thus establishing a “provenance mapping system” for the T cells that are later collected in remote non-lymphoid tissues such as in the CNS during the course of experimental autoimmune encephalomyelitis (EAE), a preclinical disease model for MS. We characterize activated T cells defined by their anatomical origin in iLN (i-T cells) or mLN (m-T cells), and provide evidence that the priming site is a fundamental determinant of their commitment to a defined T helper cell lineage. Such imprinting ultimately results in the generation of specific effector functions in the CNS, including white matter vs grey matter infiltration. We show that this concept of “anatomically” defined T helper cell subsets is compatible with some aspects of the extensive molecular characterization of previously defined T helper cell lineages. However, it also extends beyond rigid lineage definitions and instead provides a framework to accommodate and organize the complex “transcriptional continuum” of single T cells in tissues based on their provenance.
Results
Provenance mapping of central nervous system T cells to distinct priming sites
In order to track lymphocytes from their priming sites to remote tissues, a photoconvertible protein tagged to mitochondria (mitoDendra2, mD2) was expressed in T cells using a CD4-Cre driver strain (PhAM T mice). mD2 requires less energy for conversion than the widely used photoconvertible protein Kaede and tagging to mitochondria increases its half-life 12. By irradiation with violet light (405 nm), we were able to photoconvert all T cells in any lymph node from mD2GREEN to mD2RED (Extended Data Fig. 1a-c, including gating strategy). mD2RED T cells could be followed for 5 cell divisions (and about 3 days) before they “lost” the label (Extended Data Fig. 1d, e). First, we examined the population dynamics of mD2RED CD4+ T cells in skin draining inguinal lymph nodes (iLN) in steady state (Fig. 1a). While naive T cells (CD4+CD44intCD62Lhigh) decreased within the mD2RED compartment over time, the relative fraction of antigen-experienced T cells (CD44highFoxp3–) increased – a process that was entirely blocked by FTY720, which prevents trafficking of lymphocytes out of secondary lymphoid tissue (Fig. 1a). Notably, the fraction of Foxp3+ regulatory T (Treg) cells largely prevailed over conventional T cells in lymph node-resident mD2RED T cells in steady state (Fig. 1b). Next, we immunized PhAM T mice subcutaneously with myelin oligodendrocyte glycoprotein peptide 35-55 (MOG(35-55)) in complete Freund’s adjuvant (CFA) according to a standard protocol to induce EAE, which reflects many aspects of the autoimmune T cell response in human MS 13. On day 4 after immunization, T cells in iLN were photoconverted, and their population dynamics were monitored. Again, the fraction of naive T cells decreased and the fraction of antigen-experienced (CD44high) T cells increased within iLN-resident mD2RED T cells in an FTY720-dependent manner (Fig. 1c). In contrast to steady state, the fraction of antigen-experienced effector T cells outnumbered the fraction of Foxp3+ Treg cells in lymph node-resident (mD2RED) T cells of immunized mice (Fig. 1d). In summary, light-induced labeling of T cells allowed for assessing the dynamics of lymph node-resident conventional T cells and Treg cells.
Fig. 1. Provenance mapping of central nervous system T cells to distinct peripheral priming sites.
a, c, Kinetics of mD2REDCD4+ T cell in iLN of FTY720-treated and untreated (no FTY) mice after photoconversion in steady state (a) and on day 4 after EAE induction (c). Representative plot of one PhAM T mouse per time point and one non-photoconverted dark control (bar) from two independent experiments. b, d, Population dynamics of T cell populations 2 days after photoconversion without (left; no FTY) or with FTY (right) in steady state (b) and on day 6 after immunization (d). Red bars represent the frequencies of mD2REDCD4+ T cells of all CD4+ T cells directly (day 0, left) and 2 days after photoconversion (right) in iLN. Representative plot from two independent experiments. e, f, g, Flow cytometric assessment of iLN-labeled (e) and mLN-labeled (f) PhAM T EAE mice 2 days after photoconversion at disease onset in the periphery (e, f) and the CNS (g). Representative plots from n = 15 mice per group for the periphery and the CNS. The site of photoconversion (iLN or mLN) is indicated (lightning symbol). h, Frequencies of regulatory and activated conventional mD2REDCD4+ T cells in PhAM T EAE mice for different tissues. n = 2 mice per group, representative plot from three independent experiments. Data shown as mean. i, Velocity versus Ca2+-indicator ratio (YFP/CFP) of TCRMOG Twitch-2B T cells in iLN and mLN imaged on days 3 or 4 post transfer. Cell numbers and Ca2+ ratios = 24 and 883 for iLN(PBS), 25 and 856 for iLN(MOG), 67 and 2353 for mLN(PBS), and 167 and 5971 for mLN(MOG), respectively. j, Mean T cell fractions with MOG-specific Ca2+ signals in iLN and mLN. n = 3 experiments. Data shown as mean T cell fraction (MOG(CFA) – PBS (CFA)) + s.d.. k, IAb-MOG tetramer binding in mD2RED i-T and m-T cells in the CNS of PhAM T EAE mice. Pool of n = 3 mice per group, representative plot from two independent experiments.
Since the gut microbiome might modulate immunopathology in remote tissues 14, we wanted to compare iLN-derived T cells with T cells primed in gut draining mesenteric lymph nodes (mLN) as to their trafficking to distant sites. We photoconverted all T cells in iLN or mLN in MOG(35-55)/CFA immunized mice at disease onset (score 1, on day 11 to 13 after immunization) and tracked mD2RED T cells at the site of photoconversion and alternative lymph nodes (for experimental design, see Extended Data Fig. 1f). After iLN-labeling, CD44highmD2RED T cells were recovered from iLN but not mLN (Fig. 1e). Similarly, we only detected negligible fractions and absolute numbers of CD44highmD2RED T cells in mLN upon iLN-labeling at earlier time points after immunization (days 6 through 9, Extended Data Fig. 1g). Vice versa, after mLN labeling CD44highmD2RED T cells were found in mLN but not iLN (Fig. 1f). Together, these data indicated that no cross-trafficking occurred between these sites. Notably, we were able to track iLN-labeled and mLN-labeled T cells to the spleen (Extended Data Fig. 1h) and to the CNS (Fig. 1g) without leakiness of the signal (Extended Data Fig. 1i). Next, we interrogated the activation status and the fraction of Foxp3+ Treg cells among iLN-derived mD2RED T cells (i-T cells) and mLN-derived mD2RED T cells (m-T cells) at remote sites. The m-T cell compartment in the CNS comprised higher fractions of Foxp3+ Treg cells, while the m-T cell fraction of CD69+ T cells was smaller than in the i-T cell compartment (Fig. 1h), suggesting that the i-T cell and m-T cell populations in the CNS were distinct. Both CNS-resident i-T cells and m-T cells were CD44high (Fig. 1h). As expected, the CNS mD2GREEN T cell compartment, which was fed by a mixture of all possible source lymph nodes, did not show differences in the fractions of Foxp3+, CD69+, or CD44high T cells in iLN- vs mLN-labeled mice (Extended Data Fig. 1j).
In our model, iLN but not mLN are directly draining the site of antigen inoculation. In order to investigate antigen-specific priming in these distinct LN stations, TCRMOG transgenic T cells (2D2) expressing a Ca2+ sensor 15 were transferred in host mice on day 10 post subcutaneous immunization with MOG(35-55) in CFA followed by two photon-imaging of iLN and mLN three or four days after adoptive transfer. MOG-specific TCR triggering was observed both in iLN and mLN (Fig. 1i, j, Extended Data Fig. 1k, l, Supplementary Videos 1–4), suggesting that as of day 10 after immunization MOG-specific priming occurred both in iLN and mLN. In addition, both i-T cells and m-T cells isolated from the CNS bound IAb-MOG tetramers (Fig. 1k). Thus, MOG-specific i-T cells and m-T cells reached the CNS.
i-T cells and m-T cells share expanded TCR families but no public clones
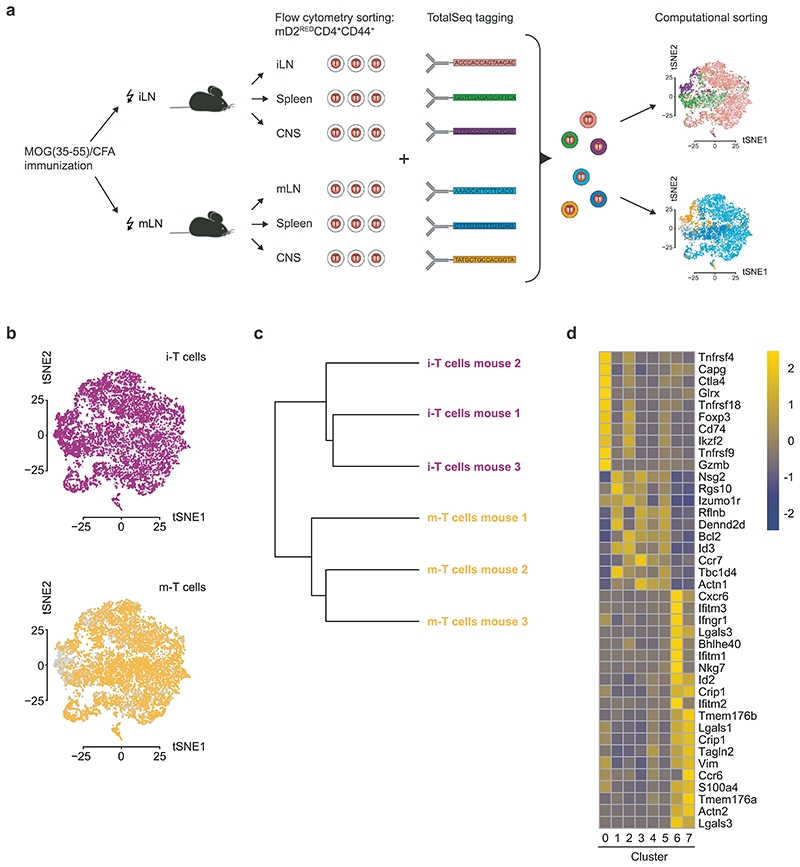
We used single-cell RNA sequencing (scRNAseq) to further characterize i-T cells and m-T cells at their priming site, in the spleen – a transit station on their way to the CNS – and in the CNS. Photoconverted (mD2RED) CD44high i- and m-T cells were sorted, tagged, and analyzed according to an experimental set-up that avoided batch effects (Extended Data Fig. 2a). Unsupervised clustering of the combined i-T cell and m-T cell dataset identified 11 clusters (Fig. 2a). In order to validate our provenance mapping system, we used the TCR as an endogenous “barcoding system”. Single T cells were grouped according to FindMarkers function in the Seurat R package. Clusters 1-5 were combined into a single cluster because none of the “cluster markers” had a log fold change (FC) value higher than 1. For all other clusters, at least 8 of the top markers had a logFC higher than 1. Single T cells in clusters 8 and 9 were excluded from further analysis because they showed high levels of cell cycle genes. Single T cells in cluster 10 were also excluded because TCR gene expression was low. We found expanded TCR clones in i-T cells and m-T cells of all clusters (Fig. 2b). In order to test whether the clones of the “i-stream” and the “m-stream” (Extended Data Fig. 2b) were related across organs, we performed hierarchical clustering of CDR3 sequences. The CDR3 repertoires within each stream (but not across streams e.g. according to organs) clustered together, suggesting that the expanded i-T cell and m-T cell clones were distinct (Extended Data Fig. 2c). More i-T cell than m-T cell clones were related among each other in the phenotypic clusters 6 and 7 corresponding to spleen and CNS. In contrast, cluster 0, which extended across organs, contained more related m-T cell clones than i-T cell clones (Fig. 2c). TRBV families 5, 12, 19, and 20 were overrepresented in the spleen and CNS while TRBV3 was most prominent in cluster 0 (Fig. 2d). Notably, since immune regulatory genes (including Foxp3, Ikzf2, Ctla4, and Gitr (Tnfrsf18)) were overrepresented in cluster 0 as compared to cluster 6 (Extended Data Fig. 2d), distinct TCR-Vβ families appeared to be associated with regulatory and effector functions of T cells, respectively. As i-T cells and m-T cells shared the same TRBV families in the spleen and CNS, we concluded that similar antigens might be recognized by i-T cells and m-T cells, and some of the overrepresented TRBV motifs were consistent with MOG-reactivity 16,17. However, the lack of public clones between i-T cells and m-T cells suggested separate trajectories from the peripheral immune system to the CNS in both streams without “cross-over”.
Fig. 2. Clonal expansion of i-T cells vs m-T cells.
a, Unsupervised clustering t-distributed stochastic neighbour embedding (t-SNE) plot of all single mD2RED CD4+CD44high T cells colored according to the cell cluster (upper plot), i-stream (middle), and m-stream (lower). n = 5 PhAM T EAE mice per group, n = 14621 cells, i-LN n = 4940 cells, i-Spleen n = 1840 cells, i-CNS n = 448 cells, m-LN n = 4888 cells, m-Spleen = 2001 cells, m-CNS n = 504 cells. Clusters 0, 1-5, 6, and 7 are highlighted by grey lines. b, TCR repertoire clonality analysis for each group plotted on a two-layer donut chart. First layer indicates the frequency of singleton (“1”, met once), doubleton (“2”, met twice), and clonotypes that met three or more times (“3+”). The second layer (quantile), displays the abundance of top 20% (Q1), next 20% (Q2) and up to Q5 clonotypes for clonotypes from “3+” set. The letter at the beginning corresponds to the i- or m-stream and the number corresponds to the cluster where those single T cells originated from in the unsupervised clustering. c, Repertoire overlap analysis using hierarchical clustering. Dendrogram shows weighted clonal overlaps for TRBCDR3 sequences among clusters, analyzed using F pairwise similarity metric in VDJtools. Branch length shows the distance between repertoires. For each stream a clonotype tracking heatmap is shown. Each row corresponds to a clonotype that is shared between samples of the same stream. Only top shared most abundant clonotypes are plotted. d, Heatmap representation of TRBV gene families across samples. Sample clustering based on variable segment usage. Weighted variable segment usage profiles were used, hierarchical clustering was performed using Euclidean distance. b-d, n = 3 PhAM T EAE mice per group, n = 8254 cells, i0 n = 723 cells, i1-5 n = 2603 cells, i7 n = 321 cells, i6 n = 411 cells, m0 n = 643 cells, m1-5 n = 3166 cells, m7 n = 240 cells, m6 n = 147 cells.
Transcriptional signatures of i-T cells and m-T cells are different from classical T helper cell signatures
Unsupervised clustering of single cell transcriptomes i- and m-T cells in all organs yielded 10 different clusters. Most strikingly, in the CNS we observed a large preponderance of cluster 9 in i-T cells and of cluster 5 in m-T cells (Fig. 3a, b). To test whether the transcriptomes of i-T cells and m-T cells were distinct, we used the minimum density hyperplane projection 18 to cluster mD2RED T cells within the CNS, using the top 2000 most variable genes, and performed Hartigan’s dip test of multimodality10,19. In isolation, the i-T cell and m-T cell data sets did not show a “dip” (Fig. 3c, left and middle). In contrast, we found a significant “dip” in the density distribution of the combined set of i-T cells and m-T cells in the CNS, suggesting that the distribution of the combined i- and m-T cell population was at least bimodal (Fig. 3c, right). Therefore, i-T cells and m-T cells in the CNS likely constituted distinct T cell subsets, indicating that their site of priming was a determinant of the segregation of their transcriptomes. RNA velocity analysis illustrated a trajectory from the lymph node to the spleen to the CNS within both the i-stream and the m-stream (Fig. 3d), consistent with the TCR sequencing data. In the CNS, neither i-T cell nor m-T cell phenotypes were congruent with classical Th signatures (Fig. 3e, Extended Data Fig. 3a). Accordingly, Th17 cells – either current or historic IL-17 producers as marked by mD2 using an IL-17A-Cre driver in PhAM IL-17 mice – were primed both in iLN and mLN and trafficked to the CNS in both scenarios (Extended Data Fig. 3b). Despite their mixed phenotypes, however, i-T cells showed an overall greater similarity to Th1 cells than m-T cells (Fig. 3e, Extended Data Fig. 3c). “Th2-ness” was evenly distributed among i-T cells and m-T cells. Notably, when using a published pathogenic Th17 signature (which comprises a mixture of Th1 and Th17 features) as a reference 20, i-T cells were enriched in the Th17path signature in the lymph node, the spleen, and the CNS compartment (Fig. 3f), arguing in favor of this signature to be pre-imprinted already in the systemic immune compartment (i.e. in skin draining lymph nodes but not in gut draining lymph nodes).
Fig. 3. Imprinting of T helper cell signatures in i-T cells vs m-T cells.
a, Unsupervised clustering t-distributed stochastic neighbour embedding (t-SNE) plot of all single mD2RED CD4+CD44high T cells colored according to the cell cluster (left) or i- and m-stream (right). n = 2 PhAM T EAE mice per group, n = 4169 cells. b, Cluster distribution of i-stream (top) vs m-stream (bottom) by tissue. c, Two-sided Hartigan’s dip test of multimodality applied to i-T cells in the CNS (left), m-T cells in the CNS (middle), and all mD2RED (combined i- and m-) T cells per CNS (right) without adjustments for multiple comparison. d, RNA velocity analysis using ratios of unspliced-to-spliced transcripts plotted in UMAP space for single T cells from i-stream (left) and m-stream (right). The directional flow of the velocity arrows between cell clusters shows the projection from the observed state to predicted future state. e, T-cell signature scores based on the expression of key genes. Th1 n = 553 cells, at least 3 genes out of Csf1, Cxcr6, Il12rb2, Il18r1, Klrc1, and Hopx; Th2 n = 389 cells, at least 2 genes out of Il4, Tnfsf13b, Batf, Nfil3, and Atf5; Th17 n = 127 cells, at least 4 genes out of Il17f, Il17a, Il21, Il2, Tnfrsf13b, Ptgfrn, Ahr, Irf4, Rora, and Plagl2; Treg n = 373 cells, at least 3 genes out of Ikzf2, Nrp1, Fosb, Tnfsf11, and Irf8. f, GSEA for gene signatures (see text) differentially expressed in m-stream vs i-stream tissues.
The core signatures of i-T cells and m-T cells are distinct
Since i-T cells and m-T cells could not be described as either classical Th1 cells or Th17 cells, we sought to define the private “core signatures” of i-T cells and m-T cells. We used our scRNAseq data set of i-T cells and m-T cells in order to identify genes uniquely upregulated in i-T cells or m-T cells in all organs (lymph nodes, spleen, CNS) (Fig. 4a, b). The i- and m-T cell core signatures comprised 94 genes and 50 genes, respectively (Fig. 4b). Examples of i-T cell associated genes were transcription factors like Rora and Runx2, costimulatory molecules like Icos and Cd2, cytokine receptors like Il1r1 and Il18r1 as well as chemokine receptors and integrins such as Cxcr6 and Itgb1, respectively (Fig. 4c, e, Extended Data Fig. 4a, b). In the m-T cell core signature, we found Malt1, Ahr, as well as Cd9 and P2rx7, and the integrin subunit Itga4 (Fig. 4d, e, Extended Data Fig. 4a, b). Eventually, in order to define transcriptional modules private to i-T cells and m-T cells, respectively, we used the SCENIC algorithm that ranks transcription factors (TFs) according to the expression levels of known target genes 21. This TF activity assessment yielded high scores for RORγt, Runx3, and Crem in i-T cells and for Ahr, Foxo1, and Ezh2 in m-T cells (Extended Data Fig. 4c). Based on the i-T cell and m-T cell core signatures, we speculated that one of the discriminating features of these T cell subsets might be different trafficking properties. As examples of guidance molecules, we investigated putative transactivators of Cxcr6 and Itga4, respectively. TFs controlling Cxcr6 (including Tbx21, Crem, Rorc, and Runx family members) were almost exclusively more active in i-T cells than in m-T cells, consistent with the observation that Cxcr6 was more abundantly expressed in i-T cells than in m-T cells (Extended Data Fig. 4d). In contrast, TFs controlling Itga4 were distinct in i-T cells and m-T cells with Klf2 and Ets family members in the Itga4 regulon of i-T cells and m-T cells, respectively (Extended Data Fig. 4e). In summary, i-T cells and m-T cells acquire a distinct core signature in the systemic immune compartment.
Fig. 4. Core signatures of i-T cells vs m-T cells.
a, Unsupervised clustering t-distributed stochastic neighbour embedding (t-SNE) plot of all single mD2REDCD4+CD44high T cells colored according to the lymph node of origin (top), the spleen (middle), and the CNS (bottom) for the i- and m-stream. n = 2 PhAM T EAE mice per group, n = 4169 cells, i-LN n = 1296 cells, i-Spleen n = 542 cells, i-CNS n = 233 cells, m-LN n = 1404 cells, m-Spleen = 525 cells, m-CNS n = 169 cells. b, Core gene set from differentially expressed genes in all three tissues with P-value < 0.05 for the i-stream (top) vs the m-stream (bottom). Two-sided Wilcoxon rank-sum test. c, d, Average gene expression for the 94 genes in the i-stream core gene set (c) and the 50 genes in the m-stream core gene set (d) per tissue as indicated. e, Flow cytometric assessment of Cxcr6 (top), P2rx7 (middle) vs CD44 and CD49d (Itga4) vs CD29 (Itgb1) (bottom) of mD2REDCD4+CD44high i- and m-T cells in the spleen of PhAM T EAE mice 2 days after photoconversion at disease onset. Representative plots from n = 3 mice per group for Cxcr6, P2rx7 and CD49d/CD29.
The transcriptomes of i-T cells vs m-T cells are controlled by different transcription factors
In order to directly assess the transcriptional landscape of i-T cells and m-T cells, we photoconverted T cells in iLN or mLN in immunized mice and sorted i-T cells or m-T cells from the spleen two days later to subject them to ATACseq. When we restricted the RNA core signatures of i-T cells and m-T cells to those loci that also exhibited a homologously differentially open chromatin, the i-T cell signature was reduced to 47 genes, and the m-T cell signature to 18 genes (Fig. 5a, b and Supplementary Table 1). Salient examples of loci with differentially open chromatin that were also differentially transcribed included Cxcr6 in i-T cells and Itga4 in m-T cells (Fig. 5c, Extended Data Fig. 5a). GO pathway analysis of the “ATACseq-validated” i- and m-T cell core signatures revealed “IL-1 receptor activity” and “purine metabolism” as distinctive between i- and m-T cells, respectively, while trafficking and adhesion pathways were detected to be active in both subsets (Fig. 5d). Differential open chromatin regions (OCR) in the i- and m-T cell core signature were analyzed for transcription factor activity 22,23. When ranked by transcription factor activity score, transcription factors associated with general activation of T cells, including Tcf1 (encoded by Tcf7), Maf, Irf4, Prdm1, and Foxo1, appeared to be more active in m-T cells than i-T cells while Crem and JunB, but also T-bet and the ROR transcription factors, which have been associated with specific effector T cell commitment, were more operational in i-T cells than m-T cells (Fig. 5e). Accordingly, Crem, Fosl2, and T-bet binding motifs were among the top motifs in differential OCR i-signature genes. Conversely, Irf and Ets transcription factor motifs were preferentially found in differential OCR m-signature genes (Extended Data Figs. 5b, c), largely corroborating our SCENIC analysis. Taken together, the ATACseq data supported the concept that i-T cells and m-T cells constitute distinct T helper cell subsets that may be controlled by specific transcriptional networks.
Fig. 5. Transcriptional modules of i-T cells vs m-T cells.
a, Normalized ATAC signal displaying all peaks for i- and m-T cells from spleen (grey). Dashed lines represent the border of fold enrichment between i- and m-T cells > 2; peaks with < 5-fold enrichment over background signal were discarded. Peaks associated to genes overexpressed in CNS i-stream are shown in purple; peaks associated to genes overexpressed in CNS m-stream are shown in orange. Some relevant genes are labeled: Ahr, Itga4, Nt5e, Adk, Malt1, and P2rx7 (m-stream); Ifngr1, Itgb1, Icos, Cxcr6, Il1r1, Satb1, and Cd7 (i-stream). Pool of n = 2 PhAM T EAE mice per group. b, Average gene expression of genes (n = 65, 47 i-stream up and 18 m-stream up) that are both in the core gene set of differentially expressed genes and contain differentially open chromatin. c, Genome browser view of some of the signature genes mentioned in (a) which are also associated to differentially open chromatin regions. Displayed tracks correspond to ATACseq for i-T cells (purple) and m-T cells (orange). In all regions, differential peaks are highlighted with arrows. d, List of the top 20 enriched pathways found for i-stream (left) and m-stream (right) based on genes associated to differential open chromatin regions which were also differentially expressed (core signature genes). Two-tailed Fisher’s exact test with Benjamini-Hochberg FDR correction. e, Transcription factor activity score as assessed with Regulatory Genomics Toolbox in i-signature genes with differentially open chromatin regions as compared to m-T cells and in m-signature genes with differentially open chromatin regions as compared to i-T cells, ranked by transcription factor activity score. Y-axis represents the differences in the transcription factor dynamics between the two conditions, and x-axis shows the position of transcription factors in the ranking. Transcription factors active in the m- and i-stream are positioned on the left and right side of the plot, respectively. Some relevant transcription factors are highlighted.
The anatomical lymph node niche dictates the signature of i-T cells and m-T cells independently of the mode of immunization
Next, we asked whether the distinct i- and m-T cell signatures were determined by the antigen dose or route of antigen delivery that might create differential T cell priming milieus in iLN and mLN. In order to address this question, we selected Cxcr6 and Itgb1 as well as P2rx7 and Itga4 that segregated with i-T cells and m-T cells, respectively, and tested their expression on the protein level in splenic CD44high mD2RED i- or m-T cells two days after photoconversion of iLN or mLN of PhAM T mice subjected to various immunization regimens. First, subcutaneous injection of MOG(35-55) at remote sites (neck) did not essentially alter the pattern of i- and m-T cell marker expression (Fig. 6a). Second, subcutaneous immunization with another antigen (OVA(323-339)) in CFA also led to the preferential expression of Cxcr6 and Itga4lowItgb1high in i-T cells and P2rx7 and Itga4highItgb1low in m-T cells (Fig. 6a). When we modified our subcutaneous immunization protocol by using no adjuvant (IFA) or alternative adjuvants (LPS or peptidoglycan), we consistently observed that i-T cells had an Itga4lowItgb1high phenotype while m-T cells were Itga4highItgb1low irrespective of the adjuvant (Extended Data Fig. 6a). Non-immunogenic or moderately immunogenic s.c. immunizations blunted the differential expressions of P2rx7 in m-T cells and Cxcr6 in i-T cells (Extended Data Fig. 6a), suggesting that the “bedrock” T cell signature imprinting capacity of iLN and mLN can be modified (but is not abolished) by different adjuvants. Importantly, we also tested our i-T cell and m-T cell markers in TCRMOG transgenic PhAM T mice (2D2 x Pham T) after i.v. administration of MOG(35-55), which would not bias the antigen dose in favor of iLN. Two days after i.v. injection of MOG peptide into 2D2 x Pham T mice, iLN or mLN were photoconverted followed by the analysis of i-T cells and m-T cells in the spleen after another two days. Similar to the subcutaneous immunization protocols, we did not observe cross-trafficking between iLN and mLN (Extended Data Fig. 6b). Again, antigen specific CD44high i-T cells had an Itga4lowItgb1high phenotype and expressed more Cxcr6 while m-T cells were Itga4highItgb1low and expressed more P2rx7 than their counterparts derived from the alternative lymph node station (Fig. 6a). In an attempt to definitely prove that the i- and m-T cell signatures were linked to the priming of T cells in iLN or mLN, we bred the PhAM T provenance reporter into the double transgenic TCRMOG and BCRMOG background (2D2 x TH). 2D2 x TH PhAM T mice develop spontaneous EAE at around 5 weeks of age (in the absence of any immunization) 24,25. At 5 weeks, iLN or mLN of 2D2 x TH PhAM T mice were photoconverted and i-T cells and m-T cells were tested in the spleen. Again, we found robust Itga4lowItgb1high and Itga4highItgb1low phenotypes in i-T cells and m-T cells, respectively. P2rx7 was higher in m-T cells while Cxcr6 expression was marginally elevated in i-T cells as compared to m-T cells in the spontaneous EAE model (Fig. 6a). When we analyzed i-T cells and m-T cells isolated from lymph nodes and spleen of 2D2 x TH PhAM T mice by RNAseq, we identified major i- and m-core signature genes segregating with i-T cells and m-T cells isolated from spontaneous EAE mice (Fig. 6b), and GSEA confirmed the enrichment of i-signature genes in i-T cells as well as of m-signature genes in m-T cells isolated from lymph nodes and spleens of 2D2 x TH PhAM T mice (Fig. 6c, d). In summary, these data support the idea that the specific ecosystem of iLN and mLN is a major determinant for the imprinting of distinct transcriptomes in T cells during antigen specific priming.
Fig. 6. The i- and m-signatures of antigen specific T cells are robust under various priming conditions.
a, Flow cytometric assessment of mD2REDCD4+CD44high T cells in the spleen of iLN-labeled (purple) and mLN-labeled (orange) Pham T mice, 2D2 Pham T mice, and 2D2 x TH Pham T mice for Cxcr6 (top), P2rx7 (middle), and CD49d (Itga4) vs CD29 (Itgb1) (bottom). Pham T mice were subjected to either MOG(35-55)/CFA or OVA(323-339)/CFA s.c. immunization at the base of tail or MOG(35-55)/CFA s.c. immunization at the neck (between the shoulder blades) plus pertussis toxin i.v. on day 0 and day 2 after immunization (as indicated) and analyzed 2 days after photoconversion on day 11 after immunization. 2D2 Pham T mice were immunized with 40 μg MOG(35-55) i.v., photoconverted on day 2, and analyzed on day 4 after immunization. 2D2 x TH Pham T mice were photoconverted at disease onset (approximately at 5 weeks of age) and analyzed 3 days after photoconversion. Representative plots from n = 3 Pham T mice per group and immunization condition, n = 4 2D2 Pham T mice and 2D2 x TH Pham T mice per group. Numbers indicate mean fluorescent intensities. b, Gene expression heatmap of all differentially expressed genes in the bulk sequencing data (up and down regulated) in i-T cells and m-T cells isolated from the spleen of 2D2 x TH Pham T mice. Differentially expressed genes (p-value < 0.05) were detected using the Seurat “FindMarkers” function, based on two-sided Wilcoxon rank-sum test and adjusted by the Bonferroni method. Art2b, Itga4, Itgb1, Ifitm2, Cd7, Rarg and Stx11 position is indicated. c, GSEA of i- and m-T cell core signatures (see Supplementary Table 1) in bulk RNAseq data of i-T cells vs m-T cells isolated from lymph nodes (top row) and spleen (bottom row) of 2D2 x TH Pham T mice. d, Waterfall plot of genes expressed in i-T cells (left) versus m-T cells (right) from the spleen of 2D2 x TH Pham T mice, ranked by fold-change. Significant genes at p-value < 0.05 are colored. Two-sided Wilcoxon rank-sum test adjusted by the Bonferroni method. Some genes are indicated by name.
The CNS infiltration patterns of i-T cells vs m-T cells are distinct
To test whether the phenotypes of i-T cells and m-T cells resulted in distinct effector functions in the CNS, we photoconverted T cells in the iLN or mLN in MOG(35-55)/CFA-immunized mice, and analyzed the infiltration of i-T cells and m-T cells in the spinal cord and brain stem by histology. While the amount of i-T cells and m-T cells was in the same range in the brain stem, m-T cells were virtually absent in the spinal cord, which was readily targeted by i-T cells (Fig. 7a, b). Since both groups of mice were immunized according to an identical protocol (and then differentially labeled either in iLN or mLN), the disease severity and phenotype was similar (Extended Data Fig. 7a). While at least one third of i-T cells were negative for α4 integrin (Itga4, CD49d) expression, essentially all m-T cells expressed the α4 integrin subunit in the CNS (Fig. 7c). Conversely, Cxcr6 was strongly expressed in the majority of CNS i-T cells but not m-T cells (Fig. 7c). In order to link this observation with the priming of autoreactive T cells in iLN or mLN, we adoptively transferred MOG-specific i-T cells or m-T cells isolated from the spleen of 2D2 x Pham T mice into Rag1 –/– secondary host animals. First, we noted that the i- vs m-phenotypes of transferred T cells were stable according to the differential expression of Itga4 (Extended Data Fig. 7b). When we analyzed the host animals two days into clinical disease, T cell infiltrates in the brain stem were similar irrespective of whether the host mice had received i-T cells or m-T cells (Fig. 7d (i, ii)). Yet white matter inflammatory foci in the brain stem were more prominently found in m-T cell recipients (Fig. 7d (ii, arrow heads)). In addition, in the cerebellum, only m-T cells but not i-T cells infiltrated into the white matter (Fig. 7d (ii, arrows and iv (vs iii)). In the spinal cord, the infiltration pattern was scattered comprising white matter and grey matter in mice that had received i-T cells. In contrast – similar to the cerebellum –, m-T cell recipients exhibited primarily white matter infiltrates in the spinal cord (Fig. 7d (v, vi and vii, viii, quantified in Fig. 7e)). In order to test whether the differential expression of Cxcr6 was necessary to produce the differential infiltration pattern observed in i- vs m-T cells, we used Cxcr6 gfp/gfp mice (functional Cxcr6 –/–). Upon immunization with MOG(35-55)/CFA, the disease severity in Cxcr6 –/– mice was comparable to wild type control mice (Extended Data Fig. 7c). However, T cell infiltrates in the spinal cord of Cxcr6 –/– mice were essentially restricted to white matter areas while the T cell infiltrates of wild type mice extended to the grey matter as well (Fig. 7f, quantified in Fig. 7g). Strikingly, in the cerebellum, T cell infiltrates in Cxcr6 –/– mice were exclusively observed in the white matter (Fig. 7h), suggesting that Cxcr6 expression contributed to the targeting of autoreactive T cells to grey matter. Therefore, the differential expression of crucial guidance molecules between i-T cells and m-T cells might be responsible for their differential targeting to specific CNS areas.
Fig. 7. Distinct functional phenotypes of i-T cells vs m-T cells in the CNS.
a, b, Histology of mD2GR(EEN) and mD2RED T cells in the lumbar spinal cord (SC(L)) of PhAM T EAE mice 2 days after photoconversion at disease onset (a) and quantification in SC(L) and brain stem (BST) (b). Scale bars 500 μm (left) and 20 μm (magnified; middle and right). Ten sections per mouse from n = 5 mice for i-T cells and n = 3 mice for m-T cells. Unpaired two-tailed t-test. ****P=0.000033. Data shown as mean + s.e.m.. c, Flow cytometric assessment of Cxcr6 and Itga4 vs CD44 in mD2REDCD4+ i- and m-T cells in the CNS of PhAM T EAE mice 2 days after photoconversion at disease onset. Representative plots from three mice per group. d, e, Histology of transferred TCRMOG i-T cells and m-T cells in BST and cerebellum (i-iv) and SC(L) (v-viii), comprising white matter (WM) and grey matter (GM) of recipient Rag1 –/– mice. Scale bars 500 μm (i, ii, v, vi), 50 μm (iii, iv), and 100 μm (vii, viii) (d). Quantification of GM frequencies of i- and m- T cells per SC(L) transsection (e). n = 3 mice per group. Unpaired two-tailed t-test. *P=0.046. Data shown as mean + s.d.. f, g, Immunostaining (f) and quantification of GM frequencies (g) of CD4+ T cells in SC(L) of wild-type (WT) and Cxcr6 –/– mice. WT n = 7 mice, Cxcr6 –/– n = 5 mice. Unpaired two-tailed t-test. **P=0.0055. FM, FluoroMyelin Red. Data shown as mean + s.d.. Scale bars 500 μm. h, Immunostaining of CD4+ T cells in the cerebellum of WT and Cxcr6 –/– mice. n = 3 mice per group. Scale bars 100 μm. i, Unsupervised clustering t-SNE plot of cerebrospinal fluid (CSF) single CD4+ T cells from untreated MS patients, representations of clusters (left) and TCR clonality (right). T cell clones were considered as expanded when found > 2. j, Unsupervised clustering t-SNE plot, colored according to Cxcr6 gene expression. n = 6 CSF samples, n = 14338, cluster0 = 6119, cluster1 = 4821, cluster2 = 2403, cluster3 = 995 cells.
Representation of i-T cell and m-T cell core signatures in the CSF of MS patients
Finally, in an effort to translate our provenance-determined T cell signatures into human T cells, we performed scRNAseq on CSF cells of untreated patients with relapsing remitting MS (Supplementary Table 2). Unsupervised clustering of CSF CD4+ T cells identified 4 clusters, with expanded T cells accumulating in clusters 2 and 0 (Fig. 7i, Supplementary Table 3). Next, we projected established T helper cell signatures, including Th1, Th2, Th17, and Treg cell signatures, on the transcriptomes of single CSF CD4+ T cells. However, except for Treg cells (that localized to cluster 3), Th signatures segregated only insufficiently with CSF T cell clusters (Extended Data Fig. 7d). In contrast, CXCR6 and other molecules comprised in the i-T cell signature (including IL2RB, IL18RAP, SYTL2, and CD2) were mainly expressed in cluster 2 T cells (Fig. 7j, Extended Data Fig. 7e) while m-signature gene expression (e.g. NT5E, DST, and AIG1) was overrepresented in T cells mapping to clusters 0, 1, and 3. ITGA4 positive cells were more widely distributed in human CSF CD4+ T cells (Extended Data Fig. 7e). These data suggested that “provenance-defined” T cell subsets might be identified in human CSF T cells as well with some markers being universal across species.
Discussion
In this study, we introduce the concept of provenance-defined T cell signatures that are maintained in remote tissues. Since i-T cells and m-T cells did not cross-traffic to alternative lymph node stations in relevant amounts, it was likely that the specific (anatomically and functionally defined) niches in iLN and mLN were determinants of the distinct transcriptomes and effector phenotypes of i-T cells and m-T cells. In fact, transcription factors like Crem and Ahr that were associated with i-T cells and m-T cells, respectively, are known to be activated by environmental cues 26,27. While transcription factor activities were different in i-T cells vs m-T cells, potential non-redundant transcriptional modules of i-T cells and m-T cells remain to be defined.
Priming site-specific imprinting of T cell features has been described, in particular in the context of tissue-selective lymphocyte trafficking 28. For instance, lamina propria-derived dendritic cells (DCs) exposed to vitamin A induce CCR9 and α4β7 integrin expression in T cells, thus promoting the capacity of those T cells to home back to the gut 29. Conversely, DCs in skin draining lymph nodes suppress gut homing molecules and induce CCR10 as well as cutaneous lymphocyte antigen (CLA) in T cells to enable them to home to the skin 30,31. Moreover, a series of more recent papers suggest that “precursors” of tissue-resident effector or regulatory T cell subsets are found in the spleen 14,32. Here, we propose that the differential imprinting of T cell phenotypes in different lymph node stations is co-opted by autoreactive T cells that then aberrantly home to remote target tissues and exert distinct effector functions according to adjuvant cues they received at their original priming site. The microbiome at body surfaces might provide some of these adjuvant cues. While the significance of the skin microbiome in MS pathology has not been explored to the same extent as the gut microbiome 33, our study might provide a universal framework for the translation of this “environmental information” into immunopathology in remote target tissues.
Neither i-T cells nor m-T cells fully overlapped with Th1 or Th17 cells, respectively, but constituted distinct T cell species. IL-17-producing cells were induced in both iLN and mLN and reached the CNS. Yet, their transcriptomes and functional phenotypes were distinct. For instance, while m-T cells were primarily recruited to white matter, i-T cells were licensed to infiltrate both grey and white matter. Cxcr6 expression was necessary for this behavior, and Cxcr6 was expressed in i-T cells but much less in m-T cells. Recently, Cxcr6 has been described as a marker of tissue residency in human CSF and brain T cells 34–36, even though white vs grey matter distribution has not been analyzed. Cxcr6 is a constituent of the core signature of CD4+ tissue resident memory T (TRM) cells 37. Conversely, while essentially lacking Cxcr6, m-T cells expressed higher levels of P2rx7 than i-T cells. In line with the preferential expression of P2rx7 in m-T cells, retinoic acid has been reported as inducer of P2rx7 in intestinal T cells with diverse functional consequences 38,39. In CD8+ T cells, sensing of extracellular ATP through P2rx7 is associated with longevity and enhanced metabolic fitness, and could represent an alternative means to promote tissue residency by inducing the expression of TGF-βRII 40,41.
Finally, besides the identification of meaningful functional markers on effector T cells, our approach of classifying T helper cells according to “provenance”-guided traits might provide a more realistic T helper cell classification than the traditional cytokine-defined T helper cell categorization in particular in humans where a multifunctional and heterogenic cytokine expression by T cells blurs the classification of T helper cells into the Th1, Th2, or Th17 lineage 11. In fact, ex vivo isolated human memory T cells can be best described according to a graded effectorness-model rather than T helper cell subsets by scRNAseq analysis 42. Our concept of provenance-defined T helper cell subsets embraces the continuity in TCR signaling strength and cytokine production of ex vivo isolated effector T cells but still defines distinct T helper cell traits that might be exploited for prognostic and therapeutic patient stratification in autoimmune diseases, and perhaps any kind of adaptive immunity.
Materials and Methods
Human data
Patients were prospectively recruited in the LMU neuroimmunology outpatient department for diagnostic procedures, including CSF sampling. Inclusion criteria for the additional sequencing analysis was a diagnosis of relapsing-remitting multiple sclerosis according to the revised McDonald criteria 43 and no current treatment with any disease modifying drug or glucocorticosteroids. The study was approved by the local ethics committee of the LMU Munich (Project-No 163-16 and 18-419) and written informed consent was granted by all participants included in the study.
Animals
C57BL/6J wild-type mice (#000664), Phamfloxed reporter mice (#018385) 44, CD4-Cre mice (#022071) 45, IL-17A-Cre mice (#016879) 46, TCRMOG mice (also known as 2D2, #006912) 47, and Rag1 –/– mice (#002216) 48 were obtained from The Jackson Laboratory. BCRMOG mice (also known as TH) 49 were kindly provided by H. Wekerle (Max-Planck-Institut für Neurobiologie, Martinsried), Cxcr6 gfp/gfp mice (#005693) 50 were kindly provided by P. Knolle (Klinikum rechts der Isar, Technical University of Munich, Germany). To generate mice with cell type-specific excision of loxP-flanked cassettes, mice were bred with CD4-Cre mice to create PhAM T mice or with IL17A-Cre to create PhAM IL17 mice. TH mice were bred with 2D2 mice and PhAM T mice to generate 2D2 x TH PhAM T mice that develop spontaneous EAE 24,25. All mouse strains were on C57BL/6J background. Animals were kept in a specific pathogen-free facility at the Technical University of Munich or at the Ludwig Maximilian University of Munich with a dark/light cycle of 12 hours, a temperature of 20-24°C, and a humidity of 45-60%. All experimental protocols were approved by the standing committee for experimentation with laboratory animals of the Bavarian state authorities and were carried out in accordance with the corresponding guidelines (TVA AZ: ROB-55.2-2532.Vet_02-13-29, ROB-55.2-2532.Vet_02-17-69, ROB-55.2-2532.Vet_02-17-234, ROB-55.2-2532.Vet_02-14-95, ROB-55.2-2532.Vet_03-18-53). 8-12-week-old age- and sex-matched mice were used for all experiments, except for spontaneous EAE experiments where 5-8 week-old age- and sex-matched 2D2 x TH PhAM T mice were used.
Induction of EAE
EAE was induced by subcutaneous immunization in the base of tail or in the neck (between the shoulder blades where indicated) with 200 μg of MOG(35-55) peptide (MEVGWYRSPFSRVVHLYRNGK; Auspep) in complete Freund’s adjuvant (CFA) containing 500 μg Mycobacterium tuberculosis H37Ra (BD Biosciences, #231141) per mouse plus intravenous (i.v.) or intraperitoneal (i.p.) injection of 200 ng pertussis toxin (Sigma-Aldrich, #P7208-50UG) on days 0 and 2 after immunization. Disease progression and severity were assessed as described before 51. The onset of disease was typically between 11 and 13 days and the peak of disease was typically between 15 and 20 days post immunization.
Additional immunization regimens
Mice were immunized subcutaneously in the base of tail with 200 μg of OVA(323-339) (ISQAVHAAHAEINEAGR; peptides&elephants) in CFA containing 500 μg Mycobacterium tuberculosis H37Ra (BD Biosciences, #231141) or 200 μg of MOG(35-55) in either incomplete Freund’s adjuvant (IFA), IFA plus 50 μg lipopolysaccharide (LPS, E. coli O111:B4; Sigma-Aldrich, #L4391-1MG), or IFA plus 50 μg peptidoglycan (PGN, S. aureus; Sigma-Aldrich, #77140-10MG) per mouse plus i.v. or i.p. injection of 200 ng pertussis toxin (Sigma-Aldrich, #P7208-50UG) on days 0 and 2 after immunization.
Oral treatment with FTY720
Mice were treated with 2 μg/ml FTY720 (Fingolimod, Novartis, #L04AA27) orally over the drinking water ad libitum. Treatment in experiments with naive animals was started 5 days and in experiments with EAE animals 1 day before lymph node irradiation, and was continued until the end of the experiment. To ensure a sufficient dose of FTY720 in EAE animals, these animals received one shot of 1 μg/g bodyweight FTY720 i.p. at the start of treatment.
Preparation of single cell suspensions and T cell cultures
Lymph nodes were passed through a 30-μm cell strainer (Sysmex, #04-004-2326) and spleens were passed through a 70-μm cell strainer (Greiner Bio-One, #542070), followed by gravity centrifugation (400 rcf, 4 °C, 10 min). Spleen samples underwent erythrocyte lysis with BD Pharm Lyse (BD Biosciences, #555899) or ACK solution (NH4Cl 8.024 mg/l (Merck, #1011451000), KHCO3 1.001 mg/l (Merck, #1048540500), EDTA.Na2·2H2O 3.722 mg/l (Merck, #1084181000)).
In retroviral transduction experiments, B cells were depleted from the cell suspensions using the Dynabeads B220 depletion kit (Thermo Fisher, #11441D). The remaining T cells were cultured in the presence of 0.5 μg/ml anti-CD3 (500A2, Invitrogen, #16-0033-86) and anti-CD28 (37.51, Invitrogen, #16-0281-86) stimulating antibodies in complete DMEM (Sigma-Aldrich, #D5671) (supplemented with 2 mM L-Glutamine (Sigma-Aldrich, #G7513), 100 μg/ml Penicillin/Streptomycin (Sigma-Aldrich, #4333), 36 mg/l Asparagine (Sigma-Aldrich, #A4284), 1 mM Sodium-Pyruvate (Sigma-Aldrich, #8636), 10 ml/l Non-essential amino acids (Sigma-Aldrich, #M7145), 4 μl/l β-Mercaptoethanol (Merck KGaA, #805740) and 10% fetal calf serum (Sigma-Aldrich, #S0615)).
Preparation of mononuclear cells from the central nervous system (CNS)
Mice were perfused through the left cardiac ventricle with ice-cold phosphate-buffered saline (PBS). The brain was dissected and the spinal cord was flushed out with PBS by hydrostatic pressure. CNS tissue was cut into pieces and digested with 1 mg/ml collagenase D (Roche Diagnostics, #11088866001) and 40 μg/ml DNase I (Roche Diagnostics, #11088866001) at 37 °C for 30 min. Mononuclear cells were isolated by passing the CNS tissue through a 100-μm cell strainer (Greiner Bio-One, #542000). The cells were pelleted by gravity centrifugation (400 rcf., 4 °C, 10 min), followed by a Percoll gradient (70%/37%, GE Healthcare, #17-0891-01) centrifugation (640 rcf, 20 °C, 22 min). The cells were removed from the interphase, washed, and resuspended in FACS buffer (PBS with 2% FCS) for further analysis.
Antibodies and flow cytometry
Cell suspensions from lymphoid organs were stained with the following fluorochrome-conjugated antibodies in this study: APC anti-mouse CD3e (145-2C11, BD Biosciences, #553066, 1:100), PerCP-eF710 anti-mouse CD4 (RM4-5, eBioscience, #46-0042-82, 1:800), APC anti-mouse CD4 (RM4-5, BD Biosciences, #553051, 1:1000), BV786 anti-mouse CD4 (RM4-5, BD Biosciences, #563727, 1:800), APC-Cy7 anti-mouse CD8a (53-6.7, Biolegend, #100714, 1:200), BV786 anti-mouse CD29 (HM β1-1, BD Biosciences, #740899, 1:100), APC-R700 anti-mouse CD44 (IM7, BD Biosciences, #565480, 1:200), BV421 anti-mouse CD44 (IM7, BD Biosciences, # 563970, 1:200), APC anti-mouse CD49d (R1-2, Biolegends, #103622, 1:100), APC anti-mouse CD62L (MEL-14, BD Biosciences, #553152, 1:800), BV785 anti-mouse CD62L (MEL-14, Biolegend, #104440, 1:800), AF647 anti-mouse CD186 (CXCR6, SA051D1, Biolegend, #151115, 1:100), BV421 anti-mouse CD186 (Cxcr6, SA051D1, Biolegend, #151109, 1:100), BV421 anti-mouse TCR γ/δ (GL3, Biolegend, #118119, 1:100), and APC anti-mouse P2rx7 (1F11, Biolegend, #148706, 1:100). For dead cell exclusion, LIVE/DEAD fixable Near-IR stain kit (Invitrogen, #L34976, 1:500) was used and to avoid Fc-receptor binding, mouse Fc Block (BD Biosciences, #553142, 1:100) was used. For cell proliferation analysis, CellTrace violet cell proliferation kit (Thermo Fisher, #C34557) was used.
For cell hashing in combination with 10x Chromium Single Cell 3’ Solution, TotalSeq-A anti-mouse Hashtag 1 to 12 (M1/42; 39-F11, Biolegend, #155801, #155803, #155805, #155807, #155809, #155811, #155813, #155815, #155817, #155819, #155821, #155823; 1:100 for all) were used and for cell hashing in combination with 10x Chromium Single Cell 5’ Solution, TotalSeq-C anti-mouse Hashtag 1 to 9 (M1/42; 39-F11, Biolegend, #155861, #155863, #155865, #155867,#155869, #155871, #155873, #155875, #155877; 1:100 for all) were used.
For profiling of protein levels at a single cell level in combination 10x Chromium Single Cell 5’ Solution, TotalSeq-C anti-human CD4 (RPA-T4, Biolegend, #300567), TotalSeq-C anti-human CD8 (RPA-T8, Biolegend, #301071), and TotalSeq-C Mouse IgG1, κ isotype Ctrl Antibody (MOPC-21, Biolegend, #400187) were used. All antibodies were diluted 1:100.
In order to perform intracellular staining on mD2RED reporter cells, a two-step fixation protocol was used to conserve the reporter’s fluorescent signal. After surface staining, the cells were fixed with 1% PFA for 15 min on ice and Fix/Perm from the Foxp3 staining kit (eBioscience, #00-5523-00) for 30 min on ice, followed by permeabilization and staining of eF450 anti-mouse Foxp3 (FKJ-16s, eBioscience, #48-5773, 1:100). For I-Ab OVA 323-339 (MBL, #TS-M710-2) and I-Ab MOG 35-55 (MBL, #TS-M704-2) tetramer stainings, cells were treated with 0.7 units/ml of neuraminidase (Sigma-Aldrich, #N-2133) for 30 min at 37 °C and 5% CO2, followed by treatment with 10 nM of Dasatinib (Selleckchem, #S1021) for 30 min at 37 °C and 5% CO2, and subsequent tetramer staining for 2 h at room temperature.
Flow cytometric analysis was performed on a CytoFLEX flow cytometer (Beckman Coulter) with CytExpert (v.2.3.1.22) software or a FACS Aria III (BD Biosciences) with BD FACSDIVA (v.8.0.1) software, and flow cytometric data were analyzed using FlowJo (v10.5.1) software (Tree Star).
Fluorescence-activated cell sorting analysis of T cells
For single-cell RNA sequencing experiments, single cell suspensions of lymph nodes, spleen, and CNS from iLN- or mLN-labeled PhAM T EAE mice (2 days after photoconversion at disease onset) were incubated with LIVE/DEAD fixable Near-IR and mouse Fc Block in PBS for 30 min on ice. Cells were washed with PBS and incubated with antibodies against surface markers and cell hashing antibodies for 30 min on ice. Cells were sorted on live mD2redCD4+CD44highCD8α– into PBS with 2% BSA on a FACS Aria III machine (BD Biosciences). Positive mD2Red fluorescence was judged against a non-photoconverted PhAM T EAE animal.
For adoptive transfer experiments of photoconverted animals, single cell suspensions of spleens from iLN- or mLN-labeled PhAM T x 2D2 EAE mice (2 days after photoconversion/day 8 after EAE induction) were treated as above except for LIVE/DEAD fixable Near-IR and cell hashing antibody treatment.
Retroviral transduction of T cells
HEK293T cells were cultured in complete DMEM (see above). HEK293T cells were purchased from ATCC (#CRL-3216) and authenticated prior to receipt by the commercial vendor using the STR method but not tested for mycoplasma after receipt. For the generation of retrovirus harboring Twitch-2B DNA, HEK293T cells were transfected using the polyethylenimine (PEI) (Polysciences, #24765) transfection method with 6 μg of pMSCV-Δneo-Twitch-2B and 3.5 μg of pCL-Eco plasmid. The culture medium was refreshed at 24 h after PEI transfection. The virus-containing supernatant was harvested after 48 h and 72 h and viral particles were concentrated using Amicon® Ultra Filters (100 kDa cut-off, Merck, #UFC910024). After two days of culture, T cells were resuspended at a concentration of 4 x 106 cells/ml in complete DMEM with the concentrated retroviral supernatant at a dilution of 1:10, supplemented with 8 μg/ml polybrene (Sigma-Aldrich, #TR-1003-G) and 10 ng/ml IL-2 (PeproTech, #212-12). The T cells were spin-infected in 12 well plates with 0.5 ml per well (2000 rcf., room temperature, 90 min).
Adoptive transfer of T cells
For all adoptive transfer experiments with photoconverted T cells, 7,000 - 12,000 splenic i-T cells or m-T cells (mD2redCD4+CD44high cells) from PhAM T x 2D2 mice at day 8 after EAE induction were sorted on a FACS Aria III machine (BD Biosciences), washed twice with PBS and transferred into Rag1 –/– EAE-matched recipients (immunized at the same time as the donor mice). The cells of one mouse were transferred into one donor.
For all adoptive transfer experiments with intravital lymph node imaging, 20-30 x 106 Twitch-2B transduced T cells were transferred into C57BL/6 wild-type mice one day after retroviral transduction and on day 10 post immunization.
Surgical exposure of lymph nodes for photoconversion and intravital imaging
Animals were sedated with a cocktail of medetomidine, midazolam and fentanyl (MMF) and the fur was removed with depilatory cream (Veet, #07768282). For intravital lymph node imaging experiments, the mice were tracheotomized after anesthesia and ventilated with 1.5 % isoflurane (CP Pharma, #1415) throughout the surgery and imaging procedure. Body temperature was regulated by a custom made heat mat.
In photoconversion experiments of T cells, the inguinal lymph node was irradiated directly through the skin with a 405nm diode laser (LuxX 405-60, Soliton, #OMI.115.15001) for 20 min or the mesenteric lymph node was irradiated after surgical exposure. Surgical exposure of the mesenteric lymph node was achieved by making a small incision in the skin and the abdominal wall. The caecum was pulled out to expose the mesenteric lymph nodes and bedded on a dampened compress (Fink & Walter GmbH, #731042), which was constantly moisturized with 0.9% sodium chloride solution (Fresenius Kabi Deutschland GmbH, # 6605514). The largest and closest lymph node to the caecum was then irradiated for 10 min. The abdominal wall and the skin were sutured after T cell photoconversion and the anesthesia was antagonized with a cocktail of atipamezole, flumazenil, and naloxone (AFN). Postoperative pain treatment was carried out with buprenorphine. The mice were sacrificed for analysis 2-3 days after T cell photoconversion.
In intravital imaging experiments, the blood vessels were visualized by i.v. infusion of Texas Red-conjugated dextran (50 μg, 70 kDa, Invitrogen, #11580226) and electrocardiograms and physiological parameters were monitored throughout the procedure. The mesenteric lymph node was surgically exposed as described above and was fixed between a dampened compress and a glass cover slip using tissue adhesive (3M Animal Care Products, #70200742529) on a heated custom-made stage. To expose the inguinal lymph nodes, the skin was cut open 2 cm along the medial abdomen and the incision was extended to the knee of the mouse. Then the skin was retracted towards the mouse’s back until the inguinal lymph node was exposed. The skin was fixed with a needle, with the lymph node laying on a dampened compress on a heated custom-made stage. Connective and adipose tissue were cleared from the lymph node. A cover slip was fixed on the skin using tissue adhesive surrounding the lymph node.
Imaging of Ca2+ signaling with intravital two-photon microscopy
On day 3 or 4 post adoptive transfer, i.e. day 13 or 14 post immunization, the mesenteric or inguinal lymph nodes of recipient mice were prepared for intravital imaging as described above. Time-lapse two-photon laser-scanning microscopy was performed using a SP2 confocal microscope (Leica) equipped with a 10 W Millenia/Tsunami laser (Newport Spectra Physics) and the Leica LCS (v.2.6.1.5173) software. Excitation wavelength was tuned to 835 nm and routed through a water immersion objective (25x, NA 0.95, Leica). With a zoom of 2x, areas of 240 x 240 μm size were scanned, and 30-50 μm z-stacks were acquired with 3-4 μm z-step. The acquisition rate was 25.219 s interval time with images line-averaged twice. Fluorescent signals were detected using non-descanned photomultiplier tube detectors (Hamamatsu) equipped with 475/50 nm, 537/26 nm, and 630/69 nm band-pass filters (Semrock).
Two-photon image processing
For the processing of the images a Gaussian blur filter was used before Z-projecting the stacks with maximum intensity to obtain two-dimensional videos. Ratiometric pseudocolor images were generated by dividing the cpVenusCD (FRET) by the mCerulean3 (CFP) channel and changing to a fire lookup table. To analyze the Ca2+ ratios, the cell shapes were manually outlined in each time frame in the 2D maximum projection to create a region of interest (ROI) for every cell to be tracked. The bleed-through of CFP into the FRET channel was found to be 44%, hence the FRET signal was corrected to YFP = FRET – 0.44 x CFP. The average signal intensities of all pixels in each ROI were defined to calculate the Ca2+ ratio at every given time point. To define Ca2+ signals, the threshold was set to a FRET ratio of 0.69, which was above the maximum level spontaneously reached by 98% of MOG-specific T cells in the mLN of mice immunized with PBS/CFA. Further, according to previous studies 15, elevated Ca2+ concentrations were stratified by duration: signals lasting shorter than 2 minutes were considered as short-term signaling, whereas Ca2+ levels above threshold lasting for more than 2 minutes were referred to as long-term Ca2+ signaling. Motility parameters were determined from the ROI coordinates over time using ImageJ (v.1.49k).
Immunohistochemistry
Mice were perfused intracardially with ice cold PBS (30 ml) and 4% PFA (15 ml). Intact CNS tissue was dissected and immunohistochemical analysis was performed on 10 μm thick cryosections. For antigen retrieval, the tissue sections were incubated with 10 mM citrate buffer (pH 6.0) for 20 minutes at 70 °C. Afterwards the samples were treated with blocking solution (antibody diluent, Agilent Technologies, #S080983-2) and incubated with anti-CD4 (EPR6855, Abcam, #ab183685, 1:100) at 4 °C for 16-18 h. After incubation with primary antibody, the tissue sections were washed thoroughly with PBST (PBS + 0.05% Tween-20) and incubated with anti–rabbit AF647 (Abcam, #ab150087, 1:500) at 37 °C for 1 h. The sections were washed and stained with FluoroMyelin Red (Invitrogen, # F34652, 1:50) and mounted with ProLong Gold antifade mounting media (Life Technologies, #P36965). The images were acquired at 20x magnification with a Nikon Eclipse Ti2 microscope and NIS Elements AR (v.5.20.00) software or at 20x and 60x magnifications with a Leica SP8 confocal microscope and the Leica Applications Suite X (v.3.5.6.21594) software.
Single-cell RNA sequencing
Two mouse single-cell RNA experiments were performed. In the first experiment, scRNAseq and Cell Hashing libraries were prepared using the 10x Chromium Single Cell 3’ Solution (Chromium Single Cell 3’ v3, 10x Genomics, #1000092), combined with cell hashing as per established protocols 52. In the second experiment, scRNAseq, scTCRseq and Cell Hashing libraries were prepared using the 10x Chromium Single Cell 5’ Solution (Chromium Next GEM Single Cell VDJ v1.1 with feature barcoding technology for cell surface protein, 10x Genomics, #1000167, #1000020, and #1000071). In brief, after cells have been sorted, they were centrifugated and the supernatant was carefully removed. Cells were resuspended in the mastermix plus 37.8 μl of water before 70 μl of the cell suspension were transferred to the chip. Integrity of the pellet was checked under the microscope to ensure that all cells were loaded onto the chip. From here on, 10x experiments were performed according to the manufacturer’s instructions. QC was performed with a Bioanalyzer 2100 (Agilent Technologies, #5067-4626) as recommended in the protocol, and libraries were quantified with the Qubit dsDNA hs kit (Life Technologies, #Q32851). All steps were performed using RPT filter tips (Starlab, #S1183-1710, #SS1180-8710, #S1182-1730) and LoBind tubes (Eppendorf, #EP0030108051, #EP0030108078, #EP0030124359). Libraries were sequenced on an Illumina HiSeq 2500 using read lengths of 28 bp read 1, 8 bp i7 index, 91 bp read 2.
Fresh human CSF samples were processed within one hour after collection. CSF samples (3-6 ml) were centrifuged at 300 g for 10 minutes. The pellet was then transferred to a 2-ml tube, and stained with TotalSeq-C anti-human CD4, CD8A and mouse IgG1 isotype control (Biolegend, #300567, #301071, #400187; 0.5 μg of each) according to the cell surface labeling protocol available from 10x Genomics with the difference that all centrifugations were done at 300 g for 10 minutes. The human CSF scRNAseq and scTCRseq libraries were prepared using the 10x Chromium Single Cell 5’ Solution (Chromium Next GEM Single Cell VDJ v1.1, 10x Genomics with feature barcoding technology for cell surface protein, #1000167, #1000020 and #1000005) as per the manufacturer’s protocol. CSF cells were loaded in total (with a maximum target cell number of 10,000). Libraries were sequenced on an Illumina NovaSeq6000 S4 using read lengths of 150 bp read 1, 8 bp i7 index, 150 bp read 2.
Single-cell sequencing data processing
Cell Ranger software (10x Genomics, v.3.1.0) was used to demultiplex samples, process raw data, align reads to either the mouse mm10 or the human GRCh38 reference genome and summarize unique molecular identifier (UMI) counts. Filtered gene-barcode and hashing-barcode matrices that contained only barcodes with UMI counts that passed the threshold for cell detection were used for further analysis. Cell hashing analysis of libraries generated using the Chromium Single Cell 3’ v2 kit was performed using the CITE-seq-Count software version 1.4.3 (DOI 10.5281/zenodo.2590196). Then, we processed the filtered UMI count matrices using R (v.3.6.1) and the R package Seurat (v.3.1.0) 53,54. Cell hashing raw counts were normalized using centered log ratio (CLR) transformation, where counts were divided by the geometric mean of an HTO across cells and log-transformed. As QC steps we filtered out: doublets originating from two cells from different samples (one single barcoded cell was positive for two HTO) and cells where no HTO was detected. We further discarded cells where the number of detected genes was <200 or >5,000 and/or 10% of the counts belonged to mitochondrial genes. After QC, only raw gene counts in high-quality singlets were submitted to: log-normalization; identification of high variable genes by using the MeanVarPlot method; scaling; and regression against the number of UMIs and mitochondrial RNA content per cell.
Two clustering analyses were performed on the mouse scRNAseq data sets. For single cell gene expression analysis, only cells analyzed in the 10x Chromium Single Cell 3’ experiment were used for the first clustering analysis and identification of genes differentially expressed in each cluster-organ-stream combination. Additionally, for TCR analysis, both 10x 3’ and 5’ datasets were merged. After merging, integration was performed to remove batch effects and the integrated assay was used for principal component analysis and unsupervised clustering. After marker identification for each cluster, only cells where a TCR clonotype was detected, were used for upstream analysis of TCR repertoire. Single-cell TCR annotation was performed using the 10x cellranger vdj pipeline. CDR3 information of the clonotype detected in each single-cell and frequency for each clonotype was added to the Seurat meta.data and the combined dataset was used to generate the input files required for the VDJtools software to perform comparative post-analysis of immune CDR3 repertoires. We used velocyto 55 to determine in which direction cells were changing in the cross-tissue trajectories both in the i- and m-stream. For the analysis we used the R package velocyto.R and Seurat wrapper package provided by Seurat.
For the human CSF scRNAseq data, only single cells, to which a TCR was assigned and in which expression of CD4 but not CD8 was detected, were subjected to clustering analysis.
Hartigan’s dip test
An automatic classifier called Minimum Density Hyperplane 18 was used to cluster cells based on the top 2000 most variables genes for several subsets of the 3’ single-cell RNA dataset with the R package PPCI (v.0.1.5). Briefly, the dimensionality reduction merges clusters with contiguous regions and projects the data on two dimensions. The pairwise distances between all cells were calculated for those dimensions and the multimodality of those distances were tested using Hartigan’s Dip Test Statistic for Unimodality 19 with the R package diptest (v.0.75-7).
RNAseq
Total RNA was isolated from sorted i-T cells and m-T cells recovered from iLN and mLN, respectively, as well as from splenic i-T cells and m-T cells (mD2redCD4+CD44high cells) of 2D2 x TH Pham T mice using the RNAeasy Plus micro kit (Qiagen, #74034). Quality and integrity of total RNA was controlled on a Bioanalyzer 2100 (Agilent Technologies). Library preparation for bulk-sequencing of poly(A)-RNA was done as described previously 56. Briefly, barcoded cDNA of each sample was generated with a Maxima RT polymerase (Thermo Fisher, #EP0742) using oligo-dT primer containing barcodes, unique molecular identifiers (UMIs) and an adaptor. Ends of the cDNAs were extended by a template switch oligo (TSO) and full-length cDNA was amplified with primers binding to the TSO-site and the adaptor. NEB UltraII FS kit was used to fragment cDNA. After end repair and A-tailing a TruSeq adapter was ligated and 3’-end-fragments were finally amplified using primers with Illumina P5 and P7 overhangs. In comparison to previous descriptions 56, the P5 and P7 sites were exchanged to allow sequencing of the cDNA in read1 and barcodes and UMIs in read2 to achieve a better cluster recognition. The library was sequenced on a NextSeq 500 (Illumina) with 67 cycles for the cDNA in read1 and 16 cycles for the barcodes and UMIs in read2. Data was processed using the published Drop-seq pipeline (v1.0) to generate sample- and gene-wise UMI tables 57. Reference genome (GRCm38) was used for alignment. Transcript and gene definitions were used according to GENCODE version M25.
ATACseq
ATACseq was performed as described by Corces and colleagues 58 with minor modifications for limited cell number. As many cells as possible (viability > 90%) were sorted and pre-washed with ice-cold ATAC Resuspension Buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2), then pelleted (500 rcf., 4°C, 10 min) and re-suspended in the corresponding volume of ATAC lysis plus transposition mixture (minimal volume 10 μl), containing 5 μl of 2x Tagment DNA Buffer, 0.5 μl of Tn5 Transposase (both from Illumina Tagment DNA Enzyme and Buffer Kit), 0.95 μl of water, 3.3 μl of PBS, 0.05 μl of 2% digitonin (Promega), 0.1 μl of 10% Tween-20, and 0.1 μl of 10% NP-40. Lysis and transposition were performed together to avoid cell loss.
Reactions were incubated for 30 min at 37°C in a thermomixer shaking at 1000 rpm and DNA was purified using the Qiagen PCR clean-up MinElute kit (Qiagen, #28006) and eluted in 10 μl of elution buffer (10 mM Tris pH 8.0). The transposed DNA was subsequently amplified in 50 μl reactions with custom primers as described by Buenrostro and colleagues 59. After 4 cycles of amplification, libraries were monitored with qPCR: 5 μl PCR sample in a 15 μl reaction with the same primers. qPCR output was monitored for the ΔRN; 0.25 ΔRN cycle number was used to estimate the number of additional cycles of the PCR reaction needed for the remaining PCR samples. Amplified libraries were purified with the Qiagen PCR clean-up MinElute kit and size selected for fragments with less than 600 bp, using the Agencourt AMPure XP beads (Beckman Coulter, #A63881). Libraries were quality controlled by Qubit and Agilent DNA Bioanalyzer analysis and then deeply sequenced on a HiSeq 1500 system, according to the standard Illumina protocol for 50 bp single-end reads.
Quantification and statistical analysis
Statistical evaluations of cell frequency measurements, cell numbers, mRNA amounts, and protein levels were performed with the unpaired Student’s t test when two populations were compared. Two-tailed p values < 0.05 were considered significant. Multiple comparisons were performed with two-way ANOVA followed by post hoc multiple comparisons tests as indicated in the legends to the figures. EAE scores between groups were analyzed as disease burden per individual day with one-way-ANOVA and post hoc test as indicated. Survival curve was calculated using Kaplan-Meier analysis and the p values were analyzed with log-rank test (Mantel-Cox). A p value of p < 0.05 was considered significant.
Motility parameters for Ca2+ imaging were calculated from the cell coordinates over time using ImageJ (v.1.49k). Immunohistochemistry images were processed using NIS elements AR (v.5.20.00) software. Calculations and the generation of graphs were performed using Graph Pad Prism v7.05 (Graph-Pad software).
ATACseq reads were aligned to the mouse genome mm10 using Bowtie (v.1.1.2) 60 with options “-q -n 2 --best --chunkmbs 2000 -p 32 -m 1 -S.”. Duplicated reads were removed using picard MarkDuplicates, and then ATAC peaks over input background were identified using HOMER (v.4.9) 61 findPeaks.pl with option “-style factor”. Differential ATAC peaks were determined based on fold change > 2 and genomic feature annotation of found peaks was performed using HOMER (v.4.9) annotatePeaks.pl.
Gene expression from the scRNAseq 3’ dataset was averaged and normalized in reads per million. Gene Set Enrichment Analysis (GSEA, v.4.0.3 Broad Institute) software was used to test enrichment of gene set signatures. All default options were used, except for the permutation type that was set to “gene_set”. Normalized Enrichment Score (NES) represents the degree to which a gene set is overrepresented accounting for the gene set size, using a weighted Kolmogorov–Smirnov-like statistic originally described in 62. False Discovery Rate (FDR) is the estimated probability that a gene set is a false positive. Gene sets in Fig. 3f were obtained using published data from GEO accession number GSE39820 20 and GSE56021 63. Differential gene expression was performed using GEO2R (p-value < 0.05, maximum 200 genes, Benjamini & Hochberg correction) for each condition. Gene sets in Fig. 6c were obtained from the core signatures (Supplementary Table 1).
Pathway analysis based on differential signature genes associated to ATAC peaks was performed using the R package l2p (v.0.0.3), with following databases: GO, KEGG, PANTH and REACTOME. Transcription factor motif prediction was done using the R package HOMER (v.4.9) findMotifsGenome.pl with the HOCOMOCO database v11 64. Transcription factors in the differential signature genes associated with ATAC peaks were further explored using tools from the python library Regulatory Genomics Toolbox.
Transcriptional modules were created using the R package SCENIC (v1.1.2-2) and post-processed using the R package scFunction (v.0.0.0.9000) from the scRNAseq mouse 3’ dataset. All genes controlled by a transciption factors (TF) form a regulon. A regulon specificity score (RSS) is calculated using Jensen-Shannon divergence and allows to order regulons by differential activity in the inguinal and mesenteric origin.
Extended Data
Extended Data Fig. 1. Site-specific labeling of T cells by photoconversion in inguinal and mesenteric lymph nodes in vivo.
a, Schematic of photoconversion of T cells in the iLN and mLN of PhAM T mice. b, Flow cytometric gating strategy for mD2GREEN or mD2RED CD4+ T cells. The DUMP channel comprised LIVE/DEAD™-Near-IR and CD8α-APC-Cy7. c, Flow cytometric assessment in the indicated LN of iLN-labeled (top) and mLN-labeled (bottom) PhAM T mice immediately after photoconversion (photo) or without photoconversion (dark). Representative plots of n = 3 mice per group. d, e, Flow cytometric proliferation readout of in vitro-activated PhAM T CD4+ T cells 4 days after photoconversion (d) and mD2RED signal over time (e). Representative plots from two independent experiments. f, Experimental design of iLN (top) or mLN (bottom) irradiation in MOG(35-55)/CFA immunized Pham T mice. g, Flow cytometric assessment of mD2REDCD44high frequencies (top) and absolute numbers (bottom) in different tissues in iLN-labeled PhAM T EAE mice 2 days after photoconversion at indicated time points. Representative plots from two mice per time point. h, i, Flow cytometric assessment in the spleen of iLN-labeled (left) and mLN-labeled (right) PhAM T EAE mice 2 days after photoconversion at disease onset (h) and of the CNS in non-photoconverted (dark) PhAM T EAE mice (i). Representative plots from spleen n = 15 mice per group, CNS n = 7 mice. j, Frequencies of regulatory and activated conventional mD2GREENCD4+ T cells in PhAM T EAE mice for different tissues. n = 2 mice per group, representative plot from three independent experiments for Foxp3 and CD44 and two for CD69. k, Frequency and duration of Ca2+ signaling in iLN (top) or mLN (bottom) of mice immunized on days 3 or 4 post transfer, i.e. days 13 or 14 post immunization. Left panels: individual Ca2+ signaling durations of TCRMOG Twitch-2B T cells. The dotted line indicates the cut-off (2 minutes) to distinguish between short- and long-term Ca2+ signaling. Cumulative results from iLN(PBS) and mLN(PBS) n = 2, iLN(MOG) and mLN(MOG) n = 3 mice. Right panels: fractions of T cells presenting short- and long-term Ca2+ signaling or no Ca2+ spikes. Data shown as mean. l, Representative images of TCRMOG Twitch-2B T cells from intravital time-lapse two-photon microscopy for Ca2+ imaging depicted by a fluorescence overlay of T cells (left) and a pseudocolor Ca2+ ratio image (right) from iLN(PBS) and mLN(PBS) n = 2, iLN(MOG) and mLN(MOG) n = 3 mice (see Supplementary Videos 1–4). Scale bars 25 μm.
Extended Data Fig. 2. TCR repertoire analysis of i-T cells and m-T cells.
a, Schematic of single cell sequencing of sorted iLN- or mLN-derived photoconverted T cells from MOG(35-55)/CFA immunized PhAM T mice in combination with TotalSeq hashtag barcoding antibodies. b, Unsupervised clustering t-distributed stochastic neighbour embedding (t-SNE) plot of all single mD2RED CD4+CD44high T cells analyzed. In the upper plot only single T cells from i-stream are colored (purple). In the lower plot only single T cells from m-stream are colored (orange). n = 5 PhAM T EAE mice per group, n = 14621 cells, i-T cells n = 7228 cells, m-T cells n = 7393 cells. c, Repertoire overlap analysis using hierarchical clustering. Dendrogram shows weighted clonal overlaps for TRB-CDR3 sequences among mice, analyzed using F pairwise similarity metric in VDJtools. Branch length shows the distance between repertoires. n = 3 PhAM T EAE mice per group. d, Average gene expression of all single mD2RED CD4+CD44high T cells analyzed for the TOP10 differentially expressed genes of cluster 0 to 7.
Extended Data Fig. 3. Single cell transcriptome analysis in i-T cells and m-T cells.
a, Average gene expression of all single mD2RED CD4+CD44high T cells analyzed from i- and m-stream in spleen and CNS grouped into T cell subsets based on key signature genes (see Fig. 3). b, Flow cytometric assessment in iLN-labeled (left) and mLN-labeled (right) PhAM IL17 EAE mice 2 days after photoconversion at disease onset. LN (top row), spleen (middle row), and CNS (bottom row). Representative plots of n = 3 mice per group. c, Unsupervised clustering t-SNE plot, colored according to i- and m-stream cell group and key gene marker (Tbx21, Rorc, Ccr6, Csf2, Ifng, Il17a, Il10) expression. n = 2 PhAM T EAE mice per group, n = 4169 cells.
Extended Data Fig. 4. Core signatures and transcription factor regulons of i-T cells and m-T cells.
a, b, Gene expression (violin plots) of i-stream signature genes Cxcr6 and Itgb1 and m-stream signature genes P2rx7 and Itga4 in the spleen (a) and CNS (b). c-e, Transcriptional module analysis of scRNAseq data from Fig. 4 based on SCENIC algorithm in i- and m-T cells. All genes controlled by a given transcription factor build a regulon. The regulon specificity score (RSS) was calculated using Jensen-Shannon divergence. c, Genome-wide regulons with RSS > 0.2 in at least one condition are displayed for i-T cells and m-T cells (i-stream and m-stream across all organs). d, e, Regulons that control Cxcr6 (d) and Itga4 (e) with no RSS threshold requirement.
Extended Data Fig. 5. ATACseq of i-T cells and m-T cells isolated from the spleen.
a, Genome browser view of key gene markers (Tbx21, Rorc, Ifng, Csf2, Il21, and Ccr6). Displayed tracks correspond to ATACseq for i-T cells (purple) and m-T cells (orange). In all regions, differential peaks (if existing) are highlighted with arrows. b, c, Ranked list of the top transcription factor motifs predicted by HOMER based on cumulative hypergeometric distribution testing for differential ATAC peaks associated to signature genes for splenic i-T cells (b) and m-T cells (c). For i-T cells 48 transcription factor sequences out of 154 are shown and for m-T cells, 16 sequences out of 30 are shown.
Extended Data Fig. 6. Robust i-T cell and m-T cell signatures in various immunization protocols.
a, Flow cytometric assessment of mD2REDCD4+CD44high T cells in the spleen of iLN-labeled (purple) and mLN-labeled (orange) Pham T mice, for Cxcr6 (top), P2rx7 (middle), and CD49d (Itga4) vs CD29 (Itgb1) (bottom). Pham T mice were subjected to different s.c. immunization regimens at the base of tail plus pertussis toxin i.v. on day 0 and day 2 after immunization (as indicated) and analyzed 2 days after photoconversion on day 11 after immunization. Representative plots from n= 3 Pham T mice per group (iLN- and mLN-labeled) and immunization condition. Numbers indicate mean fluorescent intensities. b, i-T cells and m-T cells do not cross-traffic after i.v. immunization. 2D2 Pham T mice were injected with 40 μg MOG(35-55) i.v., photoconverted at iLN (left) or mLN (right) on day 2, and analyzed for the fraction of CD44high mD2RED T cells in iLN and mLN on day 4 after injection. Representative plots from n = 4 2D2 Pham T mice per group.
Extended Data Fig. 7. Distinct functional phenotypes of i-T cells and m-T cells in the CNS compartment both in mice and humans.
a, EAE progression in iLN-labeled and mLN-labeled PhAM T EAE mice. Photoconversion at disease onset and analysis 2 days later. iLN-labeled n = 5 mice, mLN-labeled n = 3 mice, representative plot from five independent experiments. Data shown as mean ± s.d. b, Flow cytometric assessment of CD49d (Itga4) vs CD44 in transferred TCRMOG mD2REDCD4+CD44high i- and m-T cells isolated from the spleen of secondary recipient Rag1 –/– mice (approximately 20 days after transfer). n = 3 mice per group. c, EAE progression in wild-type (WT) and Cxcr6 –/– mice. WT n = 5 mice, Cxcr6 –/– n = 6 mice, representative plot from three independent experiments. Data shown as mean ± s.d. d, Unsupervised clustering t-distributed stochastic neighbour embedding (t-SNE) plot of cerebrospinal fluid (CSF) single CD4+ T cells isolated from untreated MS patients, colored according to T helper cell and Treg cell signature gene expression. Th1 (CSF1, CXCR6, IL12B2, IL18R1, KLRC1, HOPX), Th2 (IL4, TNFSF13B, BATF, NFIL3, ATF5), Th17 (IL17A, IL21, IL2, TNFRSF13B, PTGFRN, AHR, IRF4, RORA, PLAGL2), and Treg (FOXP3, IKZF2, NRP1, FOSB, TNFSF11, IRF8). e, Gene marker expression of a selection of i-stream core signature genes (IL2RB, IL18RAP, SYTL2) (top row), and m-stream core signature genes (NT5E, DST, AIG1) (bottom row). d, e, n = 6 CSF samples from untreated MS patients, n = 14339 cells.
Supplementary Material
Acknowledgements
We would like to thank Veronika Husterer and Monika Pfaller for skillful technical assistance, as well as Anja Domadenik, Vladyslav Kavaka, and Monika Hammel for help with library preparation. This study was supported by funds from the European Research Council (ERC, CoG 647215 to TK), from the Deutsche Forschungsgemeinschaft (SFB1054-B06 (TK), SFB1054-B05 (DB, VB, SG), SFB1371-P04 (DB, KS, SJ), GA 2913/1-1 (CG), DO 420/7-1 (KD), EXC 2145 (SyNergy) ID 390857198 (to TK and KD), TRR128-A07 and TRR128-A12 (TK), and TRR274-A01 (TK), TRR274-B03 (TM), TRR274-C02 (TM), and TRR274-Z02 (TK, TM)), from the Gemeinnützige Hertie-Stiftung (Hertie Network of Excellence in Clinical Neuroscience to CG and TK), and from the Langmatz-Foundation.
Footnotes
Author contributions
MH designed experiments, performed most of the experiments, analyzed data, and drafted the manuscript. EB performed the human scRNAseq experiments and analyzed the mouse and human scRNAseq experiments. RK performed histology and analyzed data. GL analyzed the mouse scRNAseq and bulk RNAseq experiments. HDM performed and analyzed the ATACseq experiments. ST, SK, AM, CS, LA, BK, GG, AMA, LAG, and SF performed experiments and analyzed data. IB performed and analyzed the Ca2+ imaging experiments. RÖ performed the mouse bulk RNAseq experiments and analyzed data. SG, SJ, and KS performed the mouse scRNAseq experiments and analyzed data. CG analyzed the human scRNA experiments. VRB, RR, TKü, NK, BH, DHB, TM, and KD designed experiments and analyzed data. TK conceptualized and directed the study, supervised the experiments, analyzed data, and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Data availability
NGS raw data and processed gene expression data have been deposited into the GEO repository under the accession number GSE156718 (scRNAseq mouse), GSE172003 (scRNAseq human), GSE172513 (ATACseq mouse) and GSE171122 (bulk RNAseq mouse). Clinical data for patient samples can be found in Supplementary Table 2. All other data generated or analyzed during this study are included in the published article or are available from the corresponding author upon reasonable request.
Code availability
No custom code or algorithms were used in this study.
References
- 1.Bielekova B, et al. Encephalitogenic potential of the myelin basic protein peptide (amino acids 83-99) in multiple sclerosis: results of a phase II clinical trial with an altered peptide ligand. Nat Med. 2000;6:1167–1175. doi: 10.1038/80516. [DOI] [PubMed] [Google Scholar]
- 2.Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 3.Roosendaal SD, Barkhof F. Imaging phenotypes in multiple sclerosis. Neuroimaging Clin N Am. 2015;25:83–96. doi: 10.1016/j.nic.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 5.Kadowaki A, Quintana FJ. The Gut-CNS Axis in Multiple Sclerosis. Trends Neurosci. 2020;43:622–634. doi: 10.1016/j.tins.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 7.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 8.Zhou L, Chong MMW, Littman DR. Plasticity of CD4+ T cell lineage differentiation. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Sallusto F. Heterogeneity of Human CD4(+) T Cells Against Microbes. Annu Rev Immunol. 2016;34:317–334. doi: 10.1146/annurev-immunol-032414-112056. [DOI] [PubMed] [Google Scholar]
- 10.Kiner E, et al. Gut CD4+ T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat Immunol. 2021;22:216–228. doi: 10.1038/s41590-020-00836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becattini S, et al. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science (New York, NY) 2015;347:400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 12.Kremers G-J, Hazelwood KL, Murphy CS, Davidson MW, Piston DW. Photoconversion in orange and red fluorescent proteins. Nat Methods. 2009;6:355–358. doi: 10.1038/nmeth.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold R, Linington C, Lassmann H. Understanding pathogenesis and therapy of multiple sclerosis via animal models: 70 years of merits and culprits in experimental autoimmune encephalomyelitis research. Brain. 2006;129:1953–1971. doi: 10.1093/brain/awl075. [DOI] [PubMed] [Google Scholar]
- 14.Wu H-J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyratsous NI, et al. Visualizing context-dependent calcium signaling in encephalitogenic T cells in vivo by two-photon microscopy. Proc Natl Acad Sci USA. 2017;114:E6381–E6389. doi: 10.1073/pnas.1701806114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alli R, Nguyen P, Geiger TL. Retrogenic modeling of experimental allergic encephalomyelitis associates T cell frequency but not TCR functional affinity with pathogenicity. J Immunol. 2008;181:136–145. doi: 10.4049/jimmunol.181.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen P, et al. Discrete TCR repertoires and CDR3 features distinguish effector and Foxp3+ regulatory T lymphocytes in myelin oligodendrocyte glycoprotein-induced experimental allergic encephalomyelitis. J Immunol. 2010;185:3895–3904. doi: 10.4049/jimmunol.1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlidis NG, Hofmeyr DP, Tasoulis SK. Minimum density hyperplanes. Journal of Machine Learning Research. 2018;17:1–33. [Google Scholar]
- 19.Hartigan JA, Hartigan PM. The dip test of unimodality. Tha annals of statistics. 1985;13:70–84. [Google Scholar]
- 20.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, et al. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019;20:45–21. doi: 10.1186/s13059-019-1642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettelli E, Baeten D, Jäger A, Sobel RA, Kuchroo VK. Myelin oligodendrocyte glycoprotein-specific T and B cells cooperate to induce a Devic-like disease in mice. J Clin Invest. 2006;116:2393–2402. doi: 10.1172/JCI28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnamoorthy G, Lassmann H, Wekerle H, Holz A. Spontaneous opticospinal encephalomyelitis in a double-transgenic mouse model of autoimmune T cell/B cell cooperation. J Clin Invest. 2006;116:2385–2392. doi: 10.1172/JCI28330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foulkes NS, Borjigin J, Snyder SH, Sassone-Corsi P. Rhythmic transcription: the molecular basis of circadian melatonin synthesis. Trends Neurosci. 1997;20:487–492. doi: 10.1016/s0166-2236(97)01109-0. [DOI] [PubMed] [Google Scholar]
- 27.Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol. 2019;72:625. doi: 10.1038/s41577-019-0125-8. [DOI] [PubMed] [Google Scholar]
- 28.Sigmundsdottir H, Butcher EC. Environmental cues, dendritic cells and the programming of tissue-selective lymphocyte trafficking. Nat Immunol. 2008;9:981–987. doi: 10.1038/ni.f.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwata M, et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Sigmundsdottir H, et al. DCs metabolize sunlight-induced vitamin D3 to ‘program’ T cell attraction to the epidermal chemokine CCL27. Nat Immunol. 2007;8:285–293. doi: 10.1038/ni1433. [DOI] [PubMed] [Google Scholar]
- 31.Biedermann T, et al. IL-12 instructs skin homing of human Th2 cells. J Immunol. 2006;177:3763–3770. doi: 10.4049/jimmunol.177.6.3763. [DOI] [PubMed] [Google Scholar]
- 32.Delacher M, et al. Precursors for Nonlymphoid-Tissue Treg Cells Reside in Secondary Lymphoid Organs and Are Programmed by the Transcription Factor BATF. Immunity. 2020;52:295–312.:e11. doi: 10.1016/j.immuni.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akdis CA. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat Rev Immunol. 2021 doi: 10.1038/s41577-021-00538-7. [DOI] [PubMed] [Google Scholar]
- 34.Beltrán E, et al. Early adaptive immune activation detected in monozygotic twins with prodromal multiple sclerosis. J Clin Invest. 2019 doi: 10.1172/JCI128475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smolders J, et al. Tissue-resident memory T cells populate the human brain. Nature communications. 2018;9:4593–14. doi: 10.1038/s41467-018-07053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herich S, et al. Human CCR5high effector memory cells perform CNS parenchymal immune surveillance via GZMK-mediated transendothelial diapedesis. Brain. 2019 doi: 10.1093/brain/awz301. [DOI] [PubMed] [Google Scholar]
- 37.Kumar BV, et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto-Hill S, Friesen L, Kim M, Kim CH. Contraction of intestinal effector T cells by retinoic acid-induced purinergic receptor P2X7. Mucosal Immunol. 2017;10:912–923. doi: 10.1038/mi.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rissiek B, Haag F, Boyer O, Koch-Nolte F, Adriouch S. P2X7 on Mouse T Cells: One Channel, Many Functions. Front Immun. 2015;6:204. doi: 10.3389/fimmu.2015.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borges da Silva H, et al. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature. 2018;559:264–268. doi: 10.1038/s41586-018-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borges da Silva H, et al. Sensing of ATP via the Purinergic Receptor P2RX7 Promotes CD8+ Trm Cell Generation by Enhancing Their Sensitivity to the Cytokine TGF-β. Immunity. 2020;53:158–171.:e6. doi: 10.1016/j.immuni.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cano-Gamez E, et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nature communications. 2020;11:1801–15. doi: 10.1038/s41467-020-15543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson AJ, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17:162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- 44.Pham AH, McCaffery JM, Chan DC. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 2012;50:833–843. doi: 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 46.Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bettelli E, et al. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197:1073–1081. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 49.Litzenburger T, et al. B lymphocytes producing demyelinating autoantibodies: development and function in gene-targeted transgenic mice. J Exp Med. 1998;188:169–180. doi: 10.1084/jem.188.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unutmaz D, et al. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J Immunol. 2000;165:3284–3292. doi: 10.4049/jimmunol.165.6.3284. [DOI] [PubMed] [Google Scholar]
- 51.Korn T, et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat Med. 2007;13:423–431. doi: 10.1038/nm1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoeckius M, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. The impact of amplification on differential expression analyses by RNA-seq. Sci Rep. 2016;6:25533. doi: 10.1038/srep25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11, Unit 11.7-11.7.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinemann C, et al. IL-27 and IL-12 oppose pro-inflammatory IL-23 in CD4(+) T cells by inducing Blimp1. Nature communications. 2014;5:3770. doi: 10.1038/ncomms4770. [DOI] [PubMed] [Google Scholar]
- 64.Kulakovskiy IV, et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018;46:D252–D259. doi: 10.1093/nar/gkx1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS raw data and processed gene expression data have been deposited into the GEO repository under the accession number GSE156718 (scRNAseq mouse), GSE172003 (scRNAseq human), GSE172513 (ATACseq mouse) and GSE171122 (bulk RNAseq mouse). Clinical data for patient samples can be found in Supplementary Table 2. All other data generated or analyzed during this study are included in the published article or are available from the corresponding author upon reasonable request.
No custom code or algorithms were used in this study.