Abstract
Background
Genetic testing for breast cancer susceptibility is widely used, but for many genes, evidence of an association with breast cancer is weak, underlying risk estimates are imprecise, and reliable subtype-specific risk estimates are lacking.
Methods
We used a panel of 34 putative susceptibility genes to perform sequencing on samples from 60,466 women with breast cancer and 53,461 controls. In separate analyses for protein-truncating variants and rare missense variants in these genes, we estimated odds ratios for breast cancer overall and tumor subtypes. We evaluated missense-variant associations according to domain and classification of pathogenicity.
Results
Protein-truncating variants in 5 genes (ATM, BRCA1, BRCA2, CHEK2, and PALB2) were associated with a risk of breast cancer overall with a P value of less than 0.0001. Protein-truncating variants in 4 other genes (BARD1, RAD51C, RAD51D, and TP53) were associated with a risk of breast cancer overall with a P value of less than 0.05 and a Bayesian false-discovery probability of less than 0.05. For protein-truncating variants in 19 of the remaining 25 genes, the upper limit of the 95% confidence interval of the odds ratio for breast cancer overall was less than 2.0. For protein-truncating variants in ATM and CHEK2, odds ratios were higher for estrogen receptor (ER)-positive disease than for ER-negative disease; for protein-truncating variants in BARD1, BRCA1, BRCA2, PALB2, RAD51C, and RAD51D, odds ratios were higher for ER-negative disease than for ER-positive disease. Rare missense variants (in aggregate) in ATM, CHEK2, and TP53 were associated with a risk of breast cancer overall with a P value of less than 0.001. For BRCA1, BRCA2, and TP53, missense variants (in aggregate) that would be classified as pathogenic according to standard criteria were associated with a risk of breast cancer overall, with the risk being similar to that of protein-truncating variants.
Conclusions
The results of this study define the genes that are most clinically useful for inclusion on panels for the prediction of breast cancer risk, as well as provide estimates of the risks associated with protein-truncating variants, to guide genetic counseling. (Funded by European Union Horizon 2020 programs and others.)
Genetic testing for cancer susceptibility is an established part of medical practice. Until recently, testing was performed mainly in patients with a strong family history of cancer and involved a limited number of genes known to be associated with a high risk of cancer or with specific cancer syndromes. With the advent of affordable sequencing, testing with larger panels of genes has become possible. However, for many genes on such panels, evidence of an association with cancer is often weak and accurate estimates of the cancer risks associated with variants are often not available.1
To better define the set of genes associated with breast cancer risk, we designed a panel consisting of 34 known or suspected breast cancer susceptibility genes, including genes provided on commercial panels. Using this panel, we sequenced germline DNA from more than 60,000 women with breast cancer and more than 53,000 controls participating in studies of the Breast Cancer Association Consortium (BCAC).2 We used these data to estimate the risks of breast cancer overall and tumor subtypes associated with germline protein-truncating variants and rare missense variants in these genes.
Methods
Studies
We included samples from women with breast cancer and unaffected controls participating in 44 BCAC studies (Tables S1 and S2 in the Supplementary Appendix, available with the full text of this article at NEJM.org). All the studies were approved by the relevant ethics review boards and used appropriate consent procedures. Overall, 30 studies did not select patients or controls on the basis of family history (population-based studies); the remaining 14 studies oversampled patients with a family history of breast cancer (family-based studies). After all quality-control steps were taken, 53,461 controls and 60,466 women with an invasive tumor (54,624 [90.3%]), in situ tumor (4187 [6.9%]), or tumor of unknown invasiveness (1655 [2.7%]) were included in the analyses. Of these, 48,826 patients and 50,703 controls were from population-based studies.
Sequence Analysis
We analyzed a panel of 34 known or suspected breast cancer susceptibility genes (Tables S3 and S4), including genes provided on commercial panels. Details regarding library preparation, sequencing, variant calling, quality-control procedures, and variant classification are provided in the Supplementary Methods section in the Supplementary Appendix.
Statistical Analysis
The primary analyses were burden analyses in which odds ratios and 95% confidence intervals for breast cancer associated with the presence of any variant in a given category were estimated by means of logistic regression. The two main categories were protein-truncating variants and rare missense variants (i.e., variants with a population frequency of <0.001). Missense-variant associations were evaluated according to domain and classification of pathogenicity. Analyses were also conducted according to tumor subtype, age, and ancestry (European vs. Asian).
We conducted separate analyses for the population-based studies and the family-based studies. The inclusion of studies that oversampled patients with a family history of breast cancer improves the power to detect association3 but leads to biased risk estimates. Therefore, we report risk estimates, 95% confidence intervals, and P values from the population-based studies but also report the results of association tests from all studies when appropriate. For protein-truncating variants in each gene, evidence of an association is also expressed in terms of Bayesian false-discovery probabilities, reflecting both evidence from the data in this study and evidence from previous research.4 Absolute risks were calculated by combining age-specific estimated odds ratios with population incidences in the United Kingdom in 2016 as a baseline.5,6 Details regarding the statistical analysis are provided in Supplementary Methods.
Results
Protein-Truncating Variants
Risk of Breast Cancer Overall
Protein-truncating variants in 5 genes (ATM, BRCA1, BRCA2, CHEK2, and PALB2) were associated with a significant risk of breast cancer overall (P<0.0001) (Table 1 and Figs. 1 and 2). For these genes, odds ratios ranged from 2.10 to 10.57. In CHEK2, the c.1100delC variant accounted for approximately 80% of the protein-truncating variants. The effect size for this variant (odds ratio, 2.66; 95% confidence interval [CI], 2.27 to 3.11; P = 1.1x10-33 [P = 5.3x10-53 in all studies]) did not differ significantly from the effect size for all other protein-truncating variants in CHEK2 (odds ratio, 2.13; 95% CI, 1.60 to 2.84; P = 3.0x10-7 [P=7.4x10-10 in all studies]), with a P value of 0.19 for the difference in the odds ratios.
Table 1. Risk of Breast Cancer Overall Associated with Protein-Truncating Variants in 34 Genes in Population-Based Studies and All Studies.* .
| Gene | Population-Based Studies (48,826 patients and 50,703 controls)† | All Studies (60,466 patients and 53,461 controls)† | Prior Probability‡ | BFDP | |||
|---|---|---|---|---|---|---|---|
| No. of Carriers of Protein Truncating Variants | Odds Ratio (95% CI) | P Value | P Value | ||||
| Women with Breast Cancer | Controls | ||||||
| ABRAXAS1 | 17 | 19 | 0.98 (0.50–1.94) | 0.96 | 0.93 | 0.1 | 0.98 |
| AKT1 | 3 | 6 | 0.47 (0.12–1.93) | 0.29 | 0.14 | 0.1 | 0.94 |
| ATM | 294 | 150 | 2.10 (1.71–2.57) | 9.2×10–13 | 5.5×10–20 | 0.8 | 1.3×10–18 |
| BABAM2 | 7 | 9 | 0.62 (0.23–1.71) | 0.36 | 0.34 | 0.1 | 0.95 |
| BARD1 | 62 | 32 | 2.09 (1.35–3.23) | 0.00098 | 0.00011 | 0.2 | 0.0076 |
| BRCA1 | 515 | 58 | 10.57 (8.02–13.93) | 1.1×10–62 | 3.7×10–65 | 0.99 | 1.5×10–64 |
| BRCA2 | 754 | 135 | 5.85 (4.85–7.06) | 2.2×10–75 | 8.4×10–77 | 0.99 | 3.1×10–76 |
| BRIP1 | 86 | 75 | 1.11 (0.80–1.53) | 0.54 | 0.54 | 0.2 | 0.85 |
| CDH1 | 11 | 12 | 0.86 (0.37–1.98) | 0.72 | 0.58 | 0.2 | 0.94 |
| CHEK2 | 704 | 315 | 2.54 (2.21–2.91) | 3.1×10–39 | 3.2×10–61 | 0.99 | 1.3×10–60 |
| c.1100delC variant | 548 | 245 | 2.66 (2.27–3.11) | 1.1×10–33 | 5.3×10–53 | ||
| Other variants | 156 | 70 | 2.13 (1.60–2.84) | 3.0×10–7 | 7.4×10–10 | ||
| EPCAM | 14 | 19 | 0.73 (0.36–1.49) | 0.39 | 0.13 | 0.1 | 0.95 |
| FANCC | 71 | 65 | 1.26 (0.89–1.79) | 0.20 | 0.20 | 0.1 | 0.87 |
| FANCM | 302 | 300 | 1.06 (0.90–1.26) | 0.48 | 0.28 | 0.1 | 0.96 |
| GEN1 | 31 | 43 | 0.66 (0.41–1.06) | 0.088 | 0.18 | 0.1 | 0.95 |
| MEN1 | 2 | 5 | 0.37 (0.07–1.97) | 0.24 | 0.64 | 0.1 | 0.95 |
| MLH1 | 5 | 9 | 0.58 (0.19–1.77) | 0.34 | 0.55 | 0.1 | 0.95 |
| MRE11 | 48 | 55 | 0.88 (0.59–1.32) | 0.54 | 0.34 | 0.1 | 0.98 |
| MSH2 | 13 | 13 | 1.06 (0.47–2.36) | 0.89 | 0.80 | 0.1 | 0.92 |
| MSH6 | 39 | 23 | 1.96 (1.15–3.33) | 0.013 | 0.021 | 0.1 | 0.55 |
| MUTYH | 232 | 231 | 1.00 (0.83–1.21) | 0.99 | 0.88 | 0.1 | 1.00 |
| NBN | 90 | 103 | 0.90 (0.67–1.20) | 0.48 | 0.65 | 0.2 | 0.95 |
| NF1 | 31 | 17 | 1.76 (0.96–3.21) | 0.068 | 0.011 | 0.2 | 0.25 |
| PALB2 | 274 | 55 | 5.02 (3.73–6.76) | 1.6×10–26 | 1.1×10–32 | 0.99 | 2.9×10–32 |
| PIK3CA | 3 | 12 | 0.21 (0.06–0.75) | 0.016 | 0.19 | 0.1 | 0.94 |
| PMS2 | 40 | 36 | 1.16 (0.73–1.85) | 0.53 | 0.37 | 0.1 | 0.92 |
| PTEN | 14 | 6 | 2.25 (0.85–6.00) | 0.10 | 0.0040 | 0.2 | 0.14 |
| RAD50 | 120 | 121 | 1.08 (0.83–1.40) | 0.57 | 0.45 | 0.1 | 0.95 |
| RAD51C | 54 | 26 | 1.93 (1.20–3.11) | 0.0070 | 0.00026 | 0.3 | 0.0090 |
| RAD51D | 51 | 25 | 1.80 (1.11–2.93) | 0.018 | 0.0018 | 0.3 | 0.044 |
| RECQL | 103 | 120 | 0.84 (0.64–1.10) | 0.21 | 0.89 | 0.1 | 0.95 |
| RINT1 | 32 | 49 | 0.72 (0.46–1.14) | 0.17 | 0.31 | 0.1 | 0.96 |
| STK11 | 6 | 5 | 1.60 (0.48–5.28) | 0.44 | 0.50 | 0.2 | 0.70 |
| TP53 | 7 | 2 | 3.06 (0.63–14.91) | 0.17 | 0.015 | 0.8 | 0.033 |
| XRCC2 | 15 | 18 | 0.96 (0.47–1.93) | 0.90 | 0.81 | 0.1 | 0.98 |
BFDP denotes Bayesian false-discovery probability, and CI confidence interval.
The sample sizes reflect totals after quality control. Analyses for genes other than BRCA1 and BRCA2 excluded carriers of protein-truncating variants in BRCA1 and BRCA2; therefore, sample sizes were slightly lower in those analyses (47,522 patients and 50,475 controls in population-based studies, and 58,728 patients and 52,976 controls in all studies).
Details regarding the prior probabilities are provided in the Supplementary Methods section in the Supplementary Appendix.
Figure 1. Frequency of Protein-Truncating Variants in 34 Genes in Population-Based Studies.
Shown are percentages of women with breast cancer and controls who were carriers of protein-truncating variants in 34 genes. The genes are listed in order of increasing estimated odds ratios for breast cancer overall.
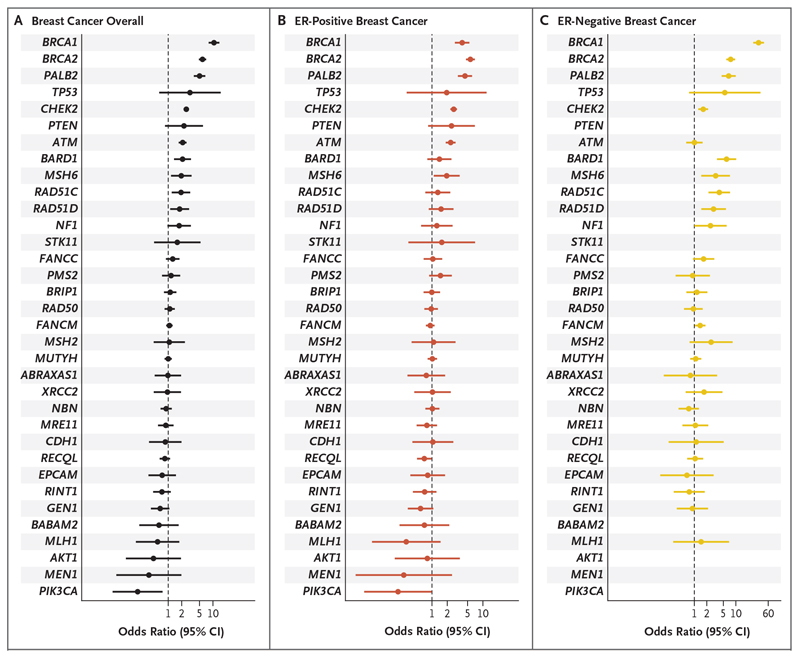
Figure 2. Risk of Breast Cancer Overall and Tumor Subtypes Associated with Protein-Truncating Variants in 34 Genes in Population-Based Studies.
Shown are odds ratios and 95% confidence intervals (CIs) for breast cancer overall (Panel A), estrogen receptor (ER)-positive breast cancer (Panel B), and ER-negative breast cancer (Panel C) associated with protein-truncating variants in 34 genes. The genes are listed in order of decreasing estimated odds ratios for breast cancer overall.
There was more modest evidence of an association with breast cancer overall for proteintruncating variants in 7 other genes: BARD1 (odds ratio, 2.09; 95% CI, 1.35 to 3.23; P = 0.00098 [P = 0.00011 in all studies]), RAD51C (odds ratio, 1.93; 95% CI, 1.20 to 3.11; P=0.0070 [P = 0.00026 in all studies]), RAD51D (odds ratio, 1.80; 95% CI, 1.11 to 2.93; P = 0.018 [P = 0.0018 in all studies]), PTEN (odds ratio, 2.25; 95% CI, 0.85 to 6.00; P=0.10 [P=0.0040 in all studies]), NF1 (odds ratio, 1.76; 95% CI, 0.96 to 3.21; P=0.068 [P=0.011 in all studies]), TP53 (odds ratio, 3.06; 95% CI, 0.63 to 14.91; P=0.17 [P=0.015 in all studies]), and MSH6 (odds ratio, 1.96; 95% CI, 1.15 to 3.33; P = 0.013 [P = 0.021 in all studies]) (Table 1). For 4 of these genes (BARD1, RAD51C, RAD51D, and TP53), the Bayesian false-discovery probability was less than 0.05 (Table 1).
For the 12 genes that had evidence of an association with breast cancer overall, the effect size for protein-truncating variants did not differ significantly between European women and Asian women (Table S7). Of the remaining 22 genes, all but 3 (AKT1, MSH2, and STK11) had an upper limit of the 95% confidence interval of the odds ratio of less than 2.0 in the population-based studies.
Risk of Tumor Subtypes
Of the 12 genes that had evidence of an association with breast cancer overall, 2 had a stronger association with estrogen receptor (ER)-positive breast cancer than with ER-negative breast cancer: ATM (odds ratio, 2.33 [95% CI, 1.87 to 2.91] for ER-positive disease vs. 1.01 [95% CI, 0.64 to 1.59] for ER-negative disease; P=0.00055 for the difference) and CHEK2 (odds ratio, 2.67 [95% CI, 2.30 to 3.11] for ER-positive disease vs. 1.64 [95% CI, 1.25 to 2.16] for ER-negative disease; P=3.6x10-5 for the difference) (Fig. 2 and Table S8). CHEK2 also had evidence of an association with ER-negative, non-triple-negative breast cancer (odds ratio, 2.53; 95% CI, 1.75 to 3.67) but not with triple-negative breast cancer (odds ratio, 1.06; 95% CI, 0.63 to 1.76) (Table S9).
In contrast, for BARD1, BRCA1, BRCA2, PALB2, RAD51C, and RAD51D, odds ratios were higher for ER-negative breast cancer than for ER-positive breast cancer (P<0.05 for all genes). Of these genes, 3 had a stronger association with triplenegative breast cancer than with ER-negative, non-triple-negative breast cancer: BARD1 (P=0.044), BRCA1 (P = 9.6x10 %, and BRCA2 (P = 7.8x10 (Table S8).
Among the genes that had no evidence of an association with breast cancer overall, FANCM had some evidence of an association with ER-negative breast cancer (P=0.0050 in all studies) and FANCC had evidence of an association with triplenegative breast cancer (P=0.0021 in all studies) (Table S9).
For BRCA1, BRCA2, and PALB2, odds ratios were higher for invasive tumors than for in situ tumors. In contrast, for ATM and CHEK2, odds ratios for invasive tumors and in situ tumors were similar (Table S10).
Age Effects and Absolute Risk
In the population-based studies, odds ratios decreased significantly with increasing age for 6 genes: BRCA1, BRCA2, CHEK2, PALB2, PTEN, andTP53 (P<0.01 for all genes) (Tables S11 and S12). Estimated absolute risks of breast cancer were derived by combining age-specific estimated odds ratios with population incidences in the United Kingdom (Fig. 3). For carriers of protein-truncating variants in BRCA1, BRCA2, and PALB2, the estimated absolute risks by 80 years of age exceeded the 30% threshold for high risk, as defined by National Institute for Health and Care Excellence guidelines.7 For carriers of protein-truncating variants in ATM, BARD1, CHEK2, RAD51C, and RAD51D, the estimated absolute risks by 80 years of age were within the 17 to 30% range for moderate risk.
Figure 3. Estimated Absolute Risk of Breast Cancer Associated with ProteinTruncating Variants in 8 Genes.
Shown are absolute risks of breast cancer through 80 years of age associated with protein-truncating variants in 8 genes that had significant evidence of an association with breast cancer overall, on the basis of estimated odds ratios from population-based studies. The absolute risk was not calculated for TP53 because of the wide 95% confidence interval for the odds ratio and the known association with a substantial risk of childhood cancer. Baseline absolute risks were derived from population incidences in the United Kingdom in 2016.6 The I bars indicate 95% confidence intervals.
Rare Missense Variants
There was evidence of an association with breast cancer overall for rare missense variants in 6 genes: CHEK2 (odds ratio, 1.42; 95% CI, 1.28 to 1.58; P=2.5x10-11 [P=2.9x10-18 in all studies]), ATM (odds ratio, 1.06; 95% CI, 1.00 to 1.13; P=0.051 [P = 0.0010 in all studies]), TP53 (odds ratio, 1.10; 95% CI, 0.91 to 1.31; P = 0.32 [P = 0.00080 in all studies]), BRCA1 (odds ratio, 1.11; 95% CI, 1.02 to 1.20; P = 0.010 [P = 0.027 in all studies]), CDH1 (odds ratio, 1.10; 95% CI, 0.98 to 1.23; P = 0.096 [P = 0.042 in all studies]), and RECQL (odds ratio, 1.12; 95% CI, 1.00 to 1.26; P = 0.047 [P=0.036 in all studies]) (Table 2). Of these 6 genes, 2 had a stronger association with ER-positive breast cancer than with ER-negative breast cancer (CHEK2 [P = 9.1x10 and CDH1 [P = 0.012]) and 1 had a stronger association with ER-negative breast cancer than with ER-positive breast cancer (BRCA1 [P = 0.01]). Odds ratios decreased with increasing age for BRCA1 (P = 0.0026), CHEK2 (P = 0.00022), and TP53 (P = 0.00023).
Table 2. Risk of Breast Cancer Overall Associated with Rare Missense Variants in 34 Genes in Population-Based Studies and All Studies.
| Gene | Population-Based Studies (48,826 patients and 50,703 controls)* | All Studies (60,466 patients and 53,461 controls)* | |||
|---|---|---|---|---|---|
| No. of Carriers of Rare Missense Variants | Odds Ratio (95% CI) | P Value | P Value | ||
| Women with Breast Cancer | Controls | ||||
| ABRAXAS1 | 233 | 242 | 1.04 (0.86–1.25) | 0.70 | 0.40 |
| AKT1 | 142 | 156 | 0.96 (0.76–1.21) | 0.72 | 0.63 |
| ATM | 2411 | 2471 | 1.06 (1.00–1.13) | 0.051 | 0.0010 |
| BABAM2 | 167 | 170 | 1.01 (0.81–1.26) | 0.91 | 0.63 |
| BARDI | 591 | 616 | 1.00 (0.89–1.12) | 0.94 | 0.41 |
| BRCA1 | 1393 | 1300 | 1.11 (1.02–1.20) | 0.010 | 0.027 |
| BRCA2 | 2831 | 3038 | 0.98 (0.93–1.04) | 0.50 | 0.58 |
| BRIP1 | 868 | 961 | 0.95 (0.86–1.04) | 0.25 | 0.54 |
| CDH1 | 682 | 668 | 1.10 (0.98–1.23) | 0.096 | 0.042 |
| CHEK2 | 895 | 697 | 1.42 (1.28–1.58) | 2.5×10–11 | 2.9×10–18 |
| EPCAM | 290 | 328 | 0.97 (0.82–1.14) | 0.69 | 0.43 |
| FANCC | 597 | 620 | 0.95 (0.85–1.07) | 0.42 | 0.80 |
| FANCM | 1434 | 1566 | 0.95 (0.88–1.02) | 0.17 | 0.85 |
| GEN1 | 701 | 707 | 1.05 (0.94–1.17) | 0.38 | 0.25 |
| MEN1 | 109 | 130 | 0.86 (0.66–1.12) | 0.25 | 0.81 |
| MLH1 | 677 | 711 | 1.02 (0.91–1.13) | 0.78 | 0.68 |
| MRE11 | 552 | 611 | 0.94 (0.84–1.06) | 0.33 | 0.93 |
| MSH2 | 908 | 1024 | 0.92 (0.84–1.01) | 0.093 | 0.12 |
| MSH6 | 1088 | 1155 | 1.00 (0.92–1.09) | 0.98 | 0.74 |
| MUTYH | 659 | 702 | 1.00 (0.90–1.12) | 1.00 | 0.58 |
| NBN | 665 | 725 | 0.95 (0.85–1.06) | 0.37 | 0.71 |
| NF1 | 816 | 899 | 0.94 (0.85–1.03) | 0.19 | 0.53 |
| PALB2 | 805 | 892 | 0.96 (0.87–1.06) | 0.39 | 1.00 |
| PIK3CA | 170 | 205 | 0.83 (0.67–1.02) | 0.080 | 0.33 |
| PMS2 | 934 | 963 | 0.95 (0.87–1.05) | 0.31 | 0.62 |
| PTEN | 68 | 70 | 1.08 (0.76–1.53) | 0.65 | 0.48 |
| RAD50 | 1046 | 1089 | 0.99 (0.91–1.08) | 0.83 | 0.44 |
| RAD51C | 196 | 206 | 0.93 (0.76–1.14) | 0.49 | 0.60 |
| RAD51D | 224 | 212 | 1.05 (0.86–1.27) | 0.64 | 0.57 |
| RECQL | 656 | 627 | 1.12 (1.00–1.26) | 0.047 | 0.036 |
| RINT1 | 732 | 762 | 1.01 (0.91–1.12) | 0.89 | 0.18 |
| STK11 | 114 | 139 | 0.83 (0.64–1.07) | 0.15 | 0.16 |
| TP53 | 257 | 244 | 1.10 (0.91–1.31) | 0.32 | 0.00080 |
| XRCC2 | 207 | 213 | 1.03 (0.84–1.25) | 0.80 | 0.53 |
The sample sizes reflect totals after quality control. The analysis for each gene excluded carriers of protein-truncating variants in that gene.
For missense variants (in aggregate) in BRCA1, the risk of breast cancer overall differed according to domain (P=3.0x10-6); there was an increased risk associated with variants located in the regions encoding the RING domain (P= 1.0x10-6) and the BRCT1 domain (P=0.00020) (Tables S13 and S14 and Fig. S1A). For missense variants in CHEK2, the odds ratios did not vary according to location (P = 0.52), such that the risk associated with variants within domains (P=4.2x10-9) was similar to the risk associated with variants outside domains (P = 0.001) (Tables S13 and S15 and Fig. S1B). For ATM, there was an increased risk associated with variants in the FRAP-ATM-TRRAP (FAT) domain (P=0.00019 in all studies) and the protein kinase domains (P = 0.00092 in all studies) (Tables S13 and S16 and Fig. S1C). There was no evidence of a risk associated with missense variants in specific domains in BRCA2 (P = 0.27) or PALB2 (P = 0.48) (Tables S13, S17, and S18 and Figs. S1D and S1E).
We specifically examined rare missense variants in BRCA1, BRCA2, and TP53 that would be classified as pathogenic according to clinical guidelines. There was clear evidence of an association with breast cancer overall for pathogenic variants (in aggregate) in each gene (odds ratio, 16.11 [95% CI, 5.83 to 44.50] in BRCA1, 5.68 [95% CI, 2.62 to 12.29] in BRCA2, and 2.91 [95% CI, 1.71 to 4.98] in TP53) but not for other missense variants (in aggregate) in each gene (odds ratio, 1.06 [95% CI, 0.98 to 1.15] in BRCA1, 0.97 [95% CI, 0.92 to 1.02] in BRCA2, and 0.94 [95% CI, 0.77 to 1.14] in TP53) (Table S19 and Figs. S2A, S2B, and S2C).
Discussion
This large study, in which we evaluated coding variation in putative breast cancer susceptibility genes, involved more than 60,000 patients and 53,000 controls, permitting the calculation of risk estimates that are more precise than those obtained previously. We found strong evidence of an association with breast cancer risk (Bayesian false-discovery probability, <0.05) for proteintruncating variants in 9 genes, with a P value of less than 0.0001 for 5 genes (ATM, BRCA1, BRCA2, CHEK2, and PALB2) and a P value of less than 0.05 for the other 4 genes (BARD1, RAD51C, RAD51D, and TP53). We found that, for protein-truncating variants in most of these genes, the odds ratio differed according to breast cancer subtype. Protein-truncating variants in ATM and CHEK2 were more strongly associated with ER-positive disease than with ER-negative disease, a finding consistent with earlier observations,5,8 whereas proteintruncating variants in BARD1, BRCA1, BRCA2, PALB2, RAD51C, and RAD51D were more strongly associated with ER-negative disease than with ER-positive disease. None of the other 25 genes in the panel had a Bayesian false-discovery probability of less than 0.10. Of note, 19 genes had an upper limit of the 95% confidence interval of the odds ratio of less than 2.0, with 2.0 representing a proposed threshold for “pathogenic, moderate risk alleles”9; we therefore conclude that these genes are not informative for the prediction of breast cancer risk. We confirmed that missense variants in BRCA1, BRCA2, and TP53 that would be classified as pathogenic according to clinical guidelines are indeed associated with clinically significant risks. We also found that rare missense variants in CHEK2 overall, as well as variants in specific domains in ATM, are associated with moderate risk.
Our results are broadly consistent with the results of an analysis by Lee et al.,10 in which the Clinical Genome Resource (ClinGen) clinical validity framework was used (Table S20). Of the 10 genes that were regarded as having definitive evidence of an association with breast cancer risk in that analysis,10 7 genes had variants associated with breast cancer risk in our analysis; deleterious variants in the remaining 3 genes (CDH1, PTEN, and STK11) are very rare and confer a predisposition to specific cancer syndromes. Of the 18 genes that were regarded as having moderate, limited, or disputed evidence of an association with breast cancer risk in that analysis, 2 genes (RAD51C and RAD51D) had protein-truncating variants associated with breast cancer risk in our analysis; 13 of the remaining genes had an upper limit of the 95% confidence interval of the odds ratio of less than 2.0 (and thus were classified as not conferring moderate or high risk according to our analysis), and the other 3 genes (MSH2, MSH6, and NF1) could not be classified. An association between breast cancer risk and variants in RAD51C and RAD51D is highly plausible, given that the functions of these genes are related to one another, that both genes had a stronger association with ER-negative breast cancer than with ER-positive breast cancer, and that both genes have evidence of an association with ovarian cancer.11
Association analyses of rare variants are susceptible to bias. Inclusion of studies that over-sampled patients with a family history of breast cancer improves the power to detect an association but leads to an upward bias in estimated odds ratios. The expected larger effect size in the familybased studies (as compared with the populationbased studies) was seen for protein-truncating variants in all the genes with evidence of an association with breast cancer, except BRCA1 and BRCA2, and was seen for missense variants in ATM, CHEK2, and TP53 (Tables S21 and S22). Protein-truncating variants in some of these genes are also associated with other types of cancer. Moreover, testing for cancer susceptibility genes in cancer genetics clinics can lead to a downward bias, because carriers identified through previous testing may be excluded. We observed a depletion of carriers of protein-truncating variants in BRCA1 and BRCA2 among women with breast cancer in the family-based studies. Therefore, we regard the estimates from the population-based studies to be the most reliable.
With regard to rare missense variants, the clearest evidence of an association with increased breast cancer risk was for CHEK2. For rare missense variants (in aggregate) in CHEK2, the odds ratio was approximately 1.4. The association was independent of location, which suggests that a large proportion of missense variants in CHEK2 confer risk (albeit below the threshold suggested for “pathogenic, moderate risk alleles”9). We also found some evidence of an association with risk for rare missense variants (in aggregate) in ATM, BRCA1, CDH1, and TP53, with odds ratios of approximately 1.1 (P<0.05) for all genes. Furthermore, we found evidence of an association with risk for missense variants in BRCA1, BRCA2, and TP53 that would be classified as pathogenic according to standard criteria but not for other missense variants in these genes; the odds ratios for the pathogenic variants (in aggregate) were similar to odds ratios for protein-truncating variants in these genes. For ATM, evidence of an association with risk appeared to be restricted to variants located in the regions encoding the phosphatidylinositol 3-kinase and 4-kinase and FAT domains, a finding consistent with previous observations.12
The absolute risk estimates for protein-truncating variants in previously established risk genes were broadly consistent with previous estimates, except estimates for BRCA2 and TP53, which were lower than risks reported in previous family-based studies.13-15 However, the estimate for TP53 in this study was based on data from only seven carriers and had a wide confidence interval. The low estimate may reflect the significant early mortality among carriers of protein-truncating variants in TP53; alternatively, it is possible that the earlier family-based studies had overestimated the risk.16 The analysis for TP53 is further complicated by the possibility of unrecognized somatic mutations in the blood due to age-related clonal hematopoiesis.17-20 The fact that the estimate for BRCA2 was lower than the estimate for BRCA1 may reflect the older age distribution in this study, as compared with previous studies, and perhaps a disproportionately large effect of genetic modifiers on risk among BRCA2 carriers, as compared with BRCA1 carriers.
Among European women, approximately 6.8% of the patients and 2.0% of the controls had protein-truncating variants in any of the 9 genes associated with breast cancer risk; in addition, 2.2% of the patients and 1.4% of the controls had missense variants in CHEK2. The frequency of protein-truncating variants among Asian women (4.4% of the patients and 1.3% of the controls) was lower than the frequency among European women; this finding is attributable to the much lower frequency of the c.1100delC variant in CHEK2 among Asian women.
The absolute risk estimates place proteintruncating variants in BRCA1, BRCA2, and PALB2 in the high-risk category and place proteintruncating variants in ATM, BARD1, CHEK2, RAD51C, and RAD51D in the moderate-risk category. These results may guide screening, as well as prevention with risk-reducing surgery or medication, in accordance with national guidelines. However, because breast cancer risk is influenced by other genetic and lifestyle factors, in addition to family history, the incorporation of this information into risk models would be required to give appropriate estimates.21
Despite the size of this study, the evidence of an association with breast cancer risk for several of the genes we analyzed (e.g., FANCM, MSH6, and NF1) remains equivocal, and even for the genes that had a clear asso ciation with risk, the confidence intervals for the risk estimates are wide. Incorporation of pedigree data and combined analyses with other studies may improve the precision of these estimates. In the meantime, these results may help to guide the clinical reporting of results generated by multigene-panel testing and the counseling of women who are undergoing genetic testing for breast cancer susceptibility.
Supplementary Material
Footnotes
Supported by the European Union Horizon 2020 research and innovation programs BRIDGES (grant number, 634935) and B-CAST (633784), the Wellcome Trust (v203477/Z/16/Z), and Cancer Research UK (C1287/A16563). Details regarding funding of specific studies are provided in the Supplementary Appendix.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Requests for individual-level data used in these analyses should be made through the BCAC Data Access Co-ordinating Committee (bcac@medschl.cam.ac.uk).
Contributor Information
Breast Cancer Association Consortium:
Leila Dorling, Sara Carvalho, Jamie Allen, Anna Gonzalez-Neira, Craig Luccarini, Cecilia Wahlström, Karen A. Pooley, Michael T. Parsons, Cristina Fortuno, Qin Wang, Manjeet K. Bolla, Joe Dennis, Renske Keeman, M. Rosario Alonso, Nuria Älvarez, Belen Herraez, Victoria Fernandez, Rocio Nunez-Torres, Ana Osorio, Jeanette Valcich, Minerva Li, Therese Törngren, Patricia A. Harrington, Caroline Baynes, Don M. Conroy, Brennan Decker, Laura Fachal, Nasim Mavaddat, Thomas Ahearn, Kristiina Aittomäki, Natalia N. Antonenkova, Norbert Arnold, Patrick Arveux, Margreet G.E.M. Ausems, Päivi Auvinen, Heiko Becher, Matthias W. Beckmann, Sabine Behrens, Marina Bermisheva, Katarzyna Bialkowska, Carl Blomqvist, Natalia V. Bogdanova, Nadja Bogdanova-Markov, Stig E. Bojesen, Bernardo Bonanni, Anne-Lise Borresen-Dale, Hiltrud Brauch, Michael Bremer, Ignacio Briceno, Thomas Brüning, Barbara Burwinkel, David A. Cameron, Nicola J. Camp, Archie Campbell, Angel Carracedo, Jose E. Castelao, Melissa H. Cessna, Stephen J. Chanock, Hans Christiansen, J. Margriet Collée, Emilie Cordina-Duverger, Sten Cornelissen, Kamila Czene, Thilo Dörk, Arif B. Ekici, Christoph Engel, Mikael Eriksson, Peter A. Fasching, Jonine Figueroa, Henrik Flyger, Asta Försti, Marike Gabrielson, Manuela Gago-Dominguez, Vassilios Georgoulias, Fabian Gil, Graham G. Giles, Gord Glendon, Encarna B. Gómez Garcia, Grethe I. Grenaker Alnæs, Pascal Guénel, Andreas Hadjisavvas, Lothar Haeberle, Eric Hahnen, Per Hall, Ute Hamann, Elaine F. Harkness, Jaana M. Hartikainen, Mikael Hartman, Wei He, Bernadette A.M. Heemskerk-Gerritsen, Peter Hillemanns, Frans B.L. Hogervorst, Antoinette Hollestelle, Weang Kee Ho, Maartje J. Hooning, Anthony Howell, Keith Humphreys, Faiza Idris, Anna Jakubowska, Audrey Jung, Pooja Middha Kapoor, Michael J. Kerin, Elza Khusnutdinova, Sung-Won Kim, Yon-Dschun Ko, Veli-Matti Kosma, Vessela N. Kristensen, Kyriacos Kyriacou, Inge M.M. Lakeman, Jong Won Lee, Min Hyuk Lee, Jingmei Li, Annika Lindblom, Wing-Yee Lo, Maria A. Loizidou, Artitaya Lophatananon, Jan Lubinski, Robert J. MacInnis, Michael J. Madsen, Arto Mannermaa, Mehdi Manoochehri, Siranoush Manoukian, Sara Margolin, Maria Elena Martinez, Tabea Maurer, Dimitrios Mavroudis, Catriona McLean, Alfons Meindl, Arjen R. Mensenkamp, Kyriaki Michailidou, Nicola Miller, Nur Aishah Mohd Taib, Kenneth Muir, Anna Marie Mulligan, Heli Nevanlinna, William G. Newman, Bprge G. Nordestgaard, Pei-Sze Ng, Jan C. Oosterwijk, Sue K. Park, Tjoung-Won Park-Simon, Jose I.A. Perez, Paolo Peterlongo, David J. Porteous, Karolina Prajzendanc, Darya Prokofyeva, Paolo Radice, Muhammad U. Rashid, Valerie Rhenius, Matti A. Rookus, Thomas Rüdiger, Emmanouil Saloustros, Elinor J. Sawyer, Rita K. Schmutzler, Andreas Schneeweiss, Peter Schürmann, Mitul Shah, Christof Sohn, Melissa C. Southey, Harald Surowy, Maija Suvanto, Somchai Thanasitthichai, Ian Tomlinson, Diana Torres, Thérèse Truong, Maria Tzardi, Yana Valova, Christi J. van Asperen, Rob M. Van Dam, Ans M.W. van den Ouweland, Lizet E. van der Kolk, Elke M. van Veen, Camilla Wendt, Justin A. Williams, Xiaohong R. Yang, Sook-Yee Yoon, M. Pilar Zamora, D. Gareth Evans, Miguel de la Hoya, Jacques Simard, Antonis C. Antoniou, Äke Borg, Irene L. Andru-lis, Jenny Chang-Claude, Montserrat García-Closas, Georgia Chenevix-Trench, Roger L. Milne, Paul D.P. Pharoah, Marjanka K. Schmidt, Amanda B. Spurdle, Maaike P.G. Vreeswijk, Javier Benitez, Alison M. Dunning, Anders Kvist, Soo H. Teo, Peter Devilee, and Douglas F. Easton
References
- 1.Easton DF, Pharoah PDP, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243–57. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.University of Cambridge and Cancer Research UK. Breast Cancer Association Consortium. 2020 http://bcac.ccge.medschl.cam.ac.uk.
- 3.Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: implications for design of association studies. Genet Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- 4.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–27. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt MK, Hogervorst F, van Hien R, et al. Age- and tumor subtype-specific breast cancer risk estimates for CHEK2*1100delC carriers. J Clin Oncol. 2016;34:2750–60. doi: 10.1200/JCO.2016.66.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Research UK. Breast cancer incidence (invasive) statistics. 2020 https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/breast-cancer/incidence-invasive.
- 7.National Institute for Health and Care Excellence. Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer. 2019 Nov 20; https://www.nice.org.uk/guidance/cg164. [PubMed]
- 8.Renault A-L, Mebirouk N, Fuhrmann L, et al. Morphology and genomic hallmarks of breast tumours developed by ATM deleterious variant carriers. Breast Cancer Res. 2018;20:28. doi: 10.1186/s13058-018-0951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spurdle AB, Greville-Heygate S, Antoniou AC, et al. Towards controlled terminology for reporting germline cancer susceptibility variants: an ENIGMA report. J Med Genet. 2019;56:347–57. doi: 10.1136/jmedgenet-2018-105872. [DOI] [PubMed] [Google Scholar]
- 10.Lee K, Seifert BA, Shimelis H, et al. Clinical validity assessment of genes frequently tested on hereditary breast and ovarian cancer susceptibility sequencing panels. Genet Med. 2019;21:1497–506. doi: 10.1038/s41436-018-0361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song H, Dicks E, Ramus SJ, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol. 2015;33:2901–7. doi: 10.1200/JCO.2015.61.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavtigian SV, Oefner PJ, Babikyan D, et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet. 2009;85:427–46. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antoniou A, Pharoah PDP, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–16. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 15.Hwang SJ, Lozano G, Amos CI, Strong LC. Germline p53 mutations in a cohort with childhood sarcoma: sex differences in cancer risk. Am J Hum Genet. 2003;72:975–83. doi: 10.1086/374567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortuno C, James PA, Spurdle AB. Current review of TP53 pathogenic germline variants in breast cancer patients outside Li-Fraumeni syndrome. Hum Mutat. 2018;39:1764–73. doi: 10.1002/humu.23656. [DOI] [PubMed] [Google Scholar]
- 17.Watson CJ, Papula AL, Poon GYP, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367:1449–54. doi: 10.1126/science.aay9333. [DOI] [PubMed] [Google Scholar]
- 18.Boettcher S, Miller PG, Sharma R, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science. 2019;365:599–604. doi: 10.1126/science.aax3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffee B, Cox HC, Kidd J, et al. Detection of somatic variants in peripheral blood lymphocytes using a next generation sequencing multigene pan cancer panel. Cancer Genet. 2017;211:5–8. doi: 10.1016/j.cancergen.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Coffee B, Cox HC, Bernhisel R, et al. A substantial proportion of apparently heterozygous TP53 pathogenic variants detected with a next-generation sequencing hereditary pan-cancer panel are acquired somatically. Hum Mutat. 2020;41:203–11. doi: 10.1002/humu.23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21:1708–18. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.