Abstract
Alveolata comprises diverse taxa of single-celled eukaryotes, many renowned for their ability to live inside animal cells. Notable examples are apicomplexan parasites and dinoflagellate symbionts, the latter of which power coral reef ecosystems. Although functionally distinct, they evolved from a common, free-living ancestor and must evade their hosts’ immune response for persistence. Both the initial cellular events that gave rise to this intracellular lifestyle and the role of host immune modulation in coral-dinoflagellate endosymbiosis are poorly understood. Here, we use a comparative approach in the cnidarian endosymbiosis model Aiptasia, which re-establishes endosymbiosis with free-living dinoflagellates every generation. We find that uptake of microalgae is largely indiscriminate, but non-symbiotic microalgae are expelled by vomocytosis, while symbionts induce host-cell innate immune suppression and form a LAMP1-positive niche. We demonstrate that exogenous immune stimulation results in symbiont expulsion and conversely, inhibition of canonical toll-like receptor (TLR) signaling enhances infection of host animals. Our findings indicate that symbiosis establishment is dictated by local innate immune suppression, to circumvent expulsion and promote niche formation. This work provides insight into the evolution of the cellular immune response and key steps involved in mediating endosymbiotic interactions.
Introduction
In competition for space and resources, living organisms have evolved effective strategies to exploit diverse environments. Notable examples are the parasitic apicomplexans, such as Plasmodium and Toxoplasma, the causative agents of malaria and toxoplasmosis, and symbiotic dinoflagellates 1. Both belong to the superphylum Alveolata and represent highly successful groups of unicellular, eukaryotic protozoa capable of living inside animal cells. As obligate parasites, apicomplexans infect a wide range of hosts including marine invertebrates, amphibians, reptiles, birds, and mammals, with major impacts on animal and human welfare 2,3. Dinoflagellates are among the most abundant single-celled, marine organisms and include phytoplankton, which are fundamental in marine food webs, as well as parasites that infect both wild and farmed fish and crustaceans 4. Moreover, dinoflagellates play a key role as photosynthetic endosymbionts of reef-building corals and other marine invertebrates 5. As symbionts, they transfer vital nutrients to support nutrition of their animal hosts allowing corals to thrive in nutrient-poor environments 6. The symbiotic association between dinoflagellates and corals is ancient, highly effective, and powers the productivity of entire coral reefs; ecosystems with major ecological and economic implications that are greatly threatened by climate change.
Regardless of the role of the intracellular resident, a unifying feature of all intracellular lifestyles is the ability of the ‘invader’ to live within the constraints of the host immune system. Intracellular apicomplexan parasites and dinoflagellate symbionts are thought to have evolved from a common, free-living photosynthetic ancestor 3,7–9 but continued on distinct trajectories. While intracellular parasites lost photosynthetic capabilities and became strictly dependent on their hosts, dinoflagellate symbionts can switch between an autotrophic, free-living lifestyle and living as intracellular symbionts. The endosymbiotic association between most reef-building corals and their photosynthetic dinoflagellates is established anew in each generation 10,11. Naturally non-symbiotic coral progeny must simultaneously acquire suitable symbionts and reject other microorganisms, especially pathogens, encountered in the environment. Additionally, free-living dinoflagellates must switch their mode of living to accommodate their new intracellular lifestyle. To establish symbiosis, symbionts have to escape the coral innate immune system, establish an intracellular niche, and coordinate cellular functions with their host cell to effectively exchange nutrients. In other words, the initiation of extant coral-dinoflagellate endosymbiosis recapitulates key cell biological steps required to establish an intracellular lifestyle inside an animal cell.
Cnidarians, such as reef-building corals and sea anemones, are relatively simple, evolutionarily ancient animals; yet as sisters to bilaterians, many of the components and pathways of their innate immune system are surprisingly similar to those found in humans 12,13. Accordingly, understanding endosymbiosis establishment in cnidarians may provide key insights into the basic principles of immune evasion governing the transition into an intracellular lifestyle and the molecular pathways that allow animal hosts to distinguish between different microbes encountered in the environment 14,15. It is well established that immune suppression occurs during cnidarian endosymbiosis establishment 12,16–19. Specifically, it was shown that symbionts induce organism-wide immune suppression through NF-κB downregulation, possibly via TGFβ-signaling to promote symbiosis establishment 16,17,20,21. Moreover, heterologous microalgae are thought to activate host innate immunity leading to their subsequent removal reviewed in 12,18,21,22. However, to date it is unknown how suppression of innate immunity contributes to symbiosis establishment and symbiont selection. The prevailing belief is that modulation of the immune response allows symbionts to avoid recognition and/or phagolysosomal digestion by arresting the phagosomal maturation process 12,23–27. Apoptosis has been suggested as an alternative, post-phagocytotic sorting mechanism for incompatible symbionts 25,28. However, neither escaping apoptosis nor digestion has been experimentally shown to play a role in clearance of incompatible microalgae during symbiosis establishment. In fact, no cellular mechanism or molecular pathway of how hosts select symbionts and distinguish between different microbes has been experimentally confirmed, and the role of innate immunity suppression in symbiosis establishment remains unknown.
This limited understanding is largely a consequence of the lack of an experimental system that allows investigation of these questions with cellular resolution. To overcome this, we established larvae of the sea anemone Exaiptasia pallida (previously E. pallida, commonly Aiptasia) as a tractable model 29–31. We exploit the ability of naturally aposymbiotic (symbiont-free) Aiptasia larvae to phagocytose symbionts from the environment with similar specificities as coral larvae in order to dissect symbiosis establishment under controlled conditions. Importantly, due to their small size and transparency they are amenable to high-resolution microscopy and cell biological analyses 19,31–33. Here, we utilize Aiptasia larvae as an experimental system to address the fundamental question of how suppression of host immunity is involved in symbiont selection. We describe a mechanism for the selective maintenance of intracellular symbionts involving cell-specific suppression of the host innate immune system to avoid expulsion from the host cell, the common fate of non-symbiotic microalgae, and not phagolysosomal digestion as commonly thought.
Results
Phagocytosis of microalgae is indiscriminate, but intracellular maintenance is specific
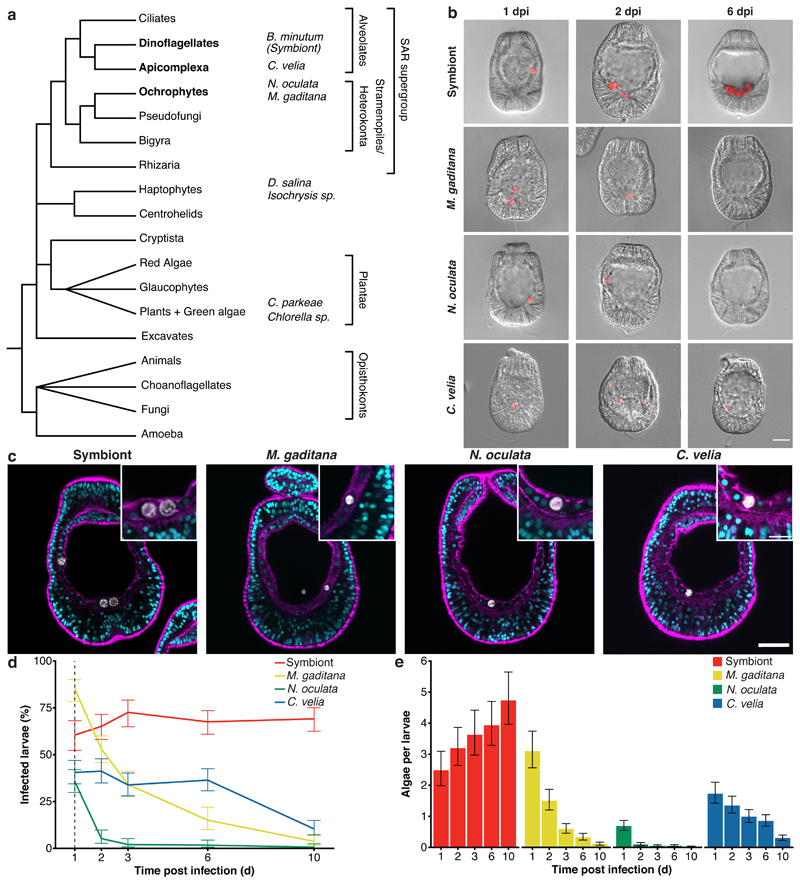
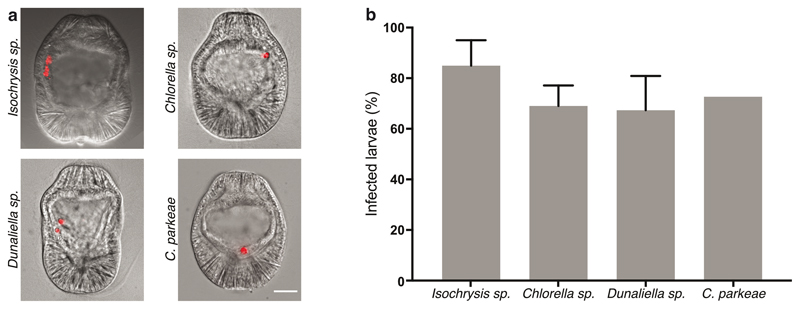
To gain insight into the mechanisms that support intracellular maintenance of symbionts and removal of non-symbiotic microorganisms, we developed a comparative system exploiting the ability of naturally aposymbiotic (symbiont-free) Aiptasia larvae to phagocytose symbionts and other particles from the environment 19,31–33. For comparison to the dinoflagellate symbiont Breviolum minutum (previously Symbiodinium minutum, strain SSB01) 34,35, we first chose Chromera velia. C. velia, the closest known photosynthetic relative of apicomplexan parasites, transiently infects both corals and Aiptasia 22,36. Moreover, we screened six distinct apicomplexan-unrelated microalgae (Fig. 1a) and found that Isochrysis sp., Chlorella sp., Dunaliella salina, Chlamydomonas parkeae, Nannochloropsis oculata, and Microchloropsis gaditana (Fig. 1a) are phagocytosed by the endodermal cells of Aiptasia larvae (Fig. 1b, Extended Data Fig. 1). For further comparative analyses, we chose N. oculata and M. gaditana, neither of which had any previously reported connection to corals or anemones; however, both are known for high lipid contents 37,38 and thus may be used as ‘food’ by Aiptasia larvae. We confirmed that all four microalgae are fully internalized using confocal microscopy (Fig. 1c).
Fig. 1. Aiptasia larvae as a comparative system to dissect symbiont maintenance.
a Phylogenetic tree adapted from 96. The four microalgae used for the comparative analysis belong to the SAR supergroup: B. minutum strain SSB01 (symbiont) belongs to the phylum Dinoflagellata, C. velia belongs to the phylum Apicomplexa, both within Alveolata; while both M. gaditana and N. oculata are members of Ochrophyta. The additional microalgae screened belong to two different phyla: D. salina and Isochrysis sp. - Haptophyta and C. parkeae and Chlorella sp. - Chlorophyta (images in Extended Data Fig. 1). b Aiptasia larvae 1-, 2-, and 6-day(s) post infection (dpi) with symbiont, M. gaditana, N. oculata, and C. velia, as indicated. Larvae were infected at 4-6 days post fertilization (dpf) for 24 hours and were washed into fresh filtered artificial seawater (FASW). Images are merges of DIC (gray) and autofluorescence of microalgae photosynthetic pigments (red), scale bar represents 25 μm (exemplary images from data in d and e). c All microalgae, symbiont, M. gaditana, N. oculata, and C. velia., were taken up in the endodermal tissue. Autofluorescence of microalgal photosynthetic pigment (white), Hoechst-stained nuclei (cyan), phalloidin-stained F-actin (magenta). Scale bar for whole larva represents 25 μm and scale bar for close up represents 10 μm (n=5 with at least 5 larvae imaged each time). d Percentage of Aiptasia larvae infected after exposure to 1.0 x 105 cells ml-1 symbiont, M. gaditana, N. oculata, or C. velia for 24 hours followed by a washout into fresh FASW (indicated by the arrow and dotted line). Error bars indicate mean ± 95 % CI of 5 independent replicates. e Average number of microalgae/larva after exposure to 1.0 x 105 cells ml-1 of symbiont, M. gaditana, N. oculata, or C. velia for 24 hours after which larvae were washed into fresh FASW. Error bars are mean ± 95 % CI of 5 independent replicates with around 50 larvae each.
To compare uptake efficiency and long-term host tolerance between symbiotic microalgae and non-symbiotic microalgae, we quantified the proportion of infected larvae and the number of intracellular microalgae over time. Naturally, infection efficiencies vary between batches, but we found that after 24 hours the proportion of larvae infected with M. gaditana (85.2 %) was significantly higher when compared to N. oculata (35.7 %), while symbionts (B. minutum) and C. velia showed intermediate results (60.5 % and 40.5 %, respectively). The proportion of larvae containing symbionts remained relatively constant, even after removal of microalgae from the environment at 24 hours post infection (hpi) (Fig. 1d, for a statistical analysis see Source Data Fig. 1). In contrast, the proportion of larvae infected with the other microalgae decreased over time. The most rapid reduction occurred with N. oculata, followed by M. gaditana, and a more gradual decrease with C. velia. While the number of symbionts increased due to proliferation within the host (Fig. 1e) 33, the mean microalga cell count per larva for all the other microalgae decreased (Fig. 1e, Source Data Fig. 1). Taken together, our results indicate that an array of microalgae are taken up effectively by Aiptasia larvae, suggesting that decisive symbiont-selection mechanisms occur after uptake, and that host clearance response varies between the distinct microalgae types.
After phagocytosis, non-symbiotic microalgae are cleared by expulsion and not digestion
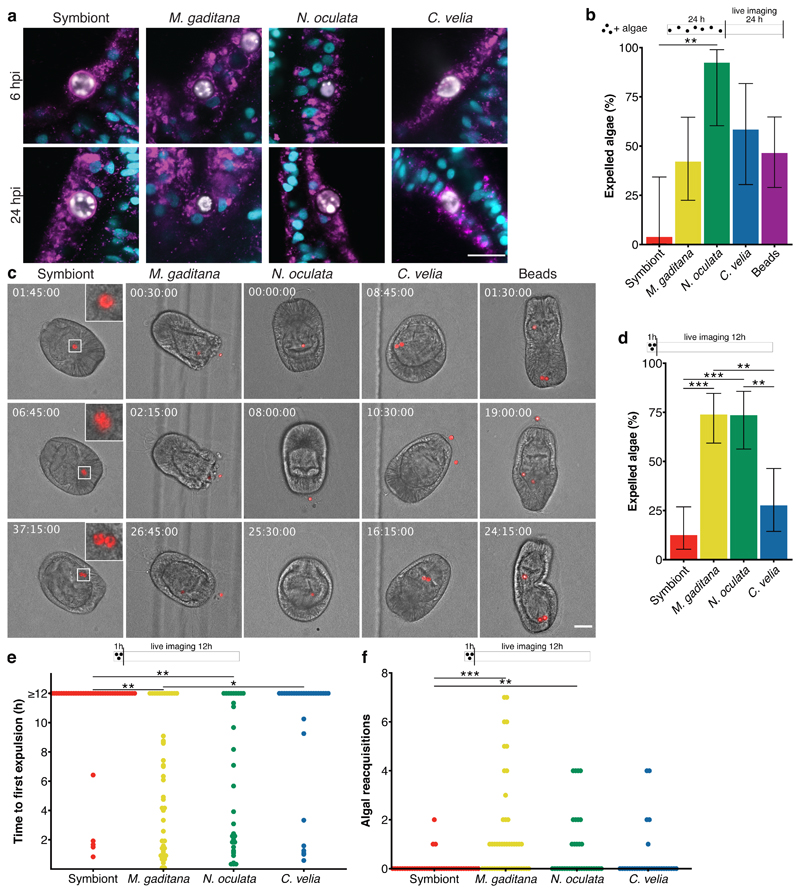
Microalgae, including symbionts, are intracellularized by phagocytosis, an ancient process by which cells internalize large particles from the environment. Originally utilized by single-celled organisms such as amoeba to acquire food, phagocytosis has evolved to become an important part of immunity for killing invading microbes in macrophages of higher metazoans 39,40. Accordingly, we speculated that non-symbiotic microalgae are cleared by intracellular digestion, characterized by the consecutive fusion of the nascent phagosome with endocytic vesicles to ultimately mature into the digestive phagolysosome. Interestingly, N. oculata- and M. gaditana-containing phagosomes appeared to be void of the lysosomal-associated membrane protein 1 (LAMP1), a marker protein associated with late endosomes and lysosomes 41, and was found to be only weakly associated with C. velia-containing phagosomes. In contrast, the symbiosome, the organelle in which the symbiont resides, was already heavily decorated with LAMP1 shortly after symbiont uptake (Fig. 2a, Supplementary Videos 1-3). Thus, it appears that symbionts, and not the non-symbiotic ‘invaders,’ rapidly establish a LAMP1-positive niche allowing intracellular persistence. This then raises the question: how are non-symbiotic microalgae, destined for removal (Fig. 1d), cleared by the host cell after phagocytosis?
Fig. 2. Non-symbiotic microalgae are removed by expulsion.
a Only the symbiont was maintained in a LAMP1-positive symbiosome, as early as 6 hpi, while the other microalgae showed no, or only slight, LAMP1 staining. LAMP1 (magenta); nuclei (cyan); microalgae (white). (n=4 with 30 larvae per n). Scale bar represents 10 μm. b During live imaging of larvae, symbionts were rarely expelled and replicated within the host endoderm. M. gaditana, N. oculata, C. velia, and beads were all expelled at some point during 48 hours of imaging. For the symbiont 13 larvae were imaged with a total of 27 algae; M. gaditana: 7 larvae/19 microalgae; N. oculata: 7 larvae/13 microalgae; C. velia: 8 larvae/12 microalgae; beads: 11 larvae/28 beads. Expulsion was significantly higher in N. oculata-containing larvae (p = 0.0077) than in symbiont-containing larvae. Error bars represent mean ± 95 % CI. c Exemplary images during live imaging. Timestamps indicate time after the start of imaging (hh:mm:ss), which coincides with 24 hours post infection. Stills are DIC and red autofluorescence of microalgae photosynthetic pigments, scale bar represents 30 μm, inserts are zoomed 3-fold. For corresponding Supplementary Videos see Supplementary Videos S6-10. d Live imaging during 12 hours after 1-hour infection reveal low levels of symbiont expulsion but significantly higher levels of expulsion of M. gaditana (p = 0.0000058) and N. oculata (p = 0.000021), also compared to compared to C. velia (p = 0.0014, p = 0.0035, respectively). Microalgae that were not expelled during the time of observation were set to 12 for the quantification. Error bars represent mean ± 95 % CI. e Time to the first expulsion was highly stochastic during this time with significant differences between the symbiont and M. gaditana (p = 0.0025) and N. oculata (p = 0.0052) as well as between M. gaditana and C. velia (p = 0.036). f M. gaditana in particular (p = 0.00010), but also N. oculata (p = 0.0049) were frequently reacquired during the observed period compared to the symbiont. d, e, f: n=4 with ~10 larvae each. Statistics are based on a two-sided generalized linear mixed model of the data with pairwise p-values adjusted with the Tukey method. *P < 0.05, **P < 0.01, ***P < 0.001.
To monitor elimination of non-symbiotic microalgae, we established live imaging to observe their fate over time. After 24 hours of infection, larvae were embedded in low-gelling agarose (LGA) and imaged every 15 minutes for 48 hours. We selected larvae that appeared to have intracellular microalgae, indicated by continuous and synchronous movement of both the microalga and the rotating larva (Supplementary Video 4); this was clearly distinct from the non-synchronous movement of microalgae located in the gastric cavity (* in Supplementary Video 4). Nearly all intracellular symbionts were maintained inside Aiptasia larvae for the entire duration of the observation period (Fig. 2b + c, Supplementary Video 5, Supplementary Table 1), with one exception where at one time-point a pair of symbionts moved asynchronously in the gastric cavity of a larva (Supplementary Video 6). Furthermore, symbionts frequently replicated (11 replication events in 13 larvae) suggesting that the immobilization in LGA does not affect larval or symbiont physiology or symbiosis stability.
Interestingly, most larvae that had phagocytosed M. gaditana cells (4/7), N. oculata cells (7/8), and C. velia cells (6/8) expelled the microalgae (42.1 %, 92.3 %, 58.3 % of total microalgae respectively), while digestion was not observed (Fig. 2b + c, Supplementary Videos 7-9, Supplementary Table 1). After expulsion, the microalgae appeared intact and healthy, and re-acquisition of non-symbiotic microalgae by larvae occurred frequently (Supplementary Table 1). To test whether expulsion was a common response to inert particles in addition to living microalgae, we imaged larvae containing intracellular polystyrene beads of comparable size as the microalgae. We saw that beads were also both expelled from (11/11, 46.4 % of total microalgae) and re-acquired by all larvae (Fig. 2b + c, Supplementary Table 1, Supplementary Video 10). It is also worth noting that in one event, a C. velia cell replicated during imaging (Supplementary Video 9). Together, this indicates that expulsion, and not phagolysosomal digestion, is the prevalent response to the uptake of non-symbiotic particles in Aiptasia larvae. Uptake and subsequent removal appear to occur non-specifically and continuously, as a trial-and-error mechanism to probe for symbionts in the environment.
To test whether differential expulsion dynamics contribute to the differences in uptake efficiency and/or rates of loss between the distinct microalgae (Fig. 1d), we monitored symbionts, M. gaditana, N. oculata, and C. velia by live imaging during the first 12 hours after uptake (after infection for 1 hour). We found that while symbionts and C. velia were expelled relatively rarely (12.5 % and 27.6 %, respectively), M. gaditana and N. oculata were expelled with high frequency (73.9 % and 73.5 %, respectively) (Fig. 2d). The average time until the first expulsion was significantly shorter for M. gaditana (5.6 hours) and N. oculata (5.7 hours) when compared to C. velia (9.7 hours) and symbionts (10.8 hours); however, the expulsion dynamics were highly variable suggesting that it is a stochastic process (Fig. 2e). Microalgae were frequently re-acquired and the average number of re-acquisition events per microalga was particularly high for M. gaditana (Fig. 2f). This suggests that differences in infection between distinct microalgae (Fig. 1d) may be a consequence of variable expulsion and re-acquisition frequencies. For instance, the high initial infection efficiency of M. gaditana may result from its frequent re-acquisition, while after washout, the rapid loss of M. gaditana and N. oculata cells from Aiptasia larvae is likely due to the comparatively quick expulsion.
Expulsion of incompatible microalgae is actin-independent and is regulated by ERK5
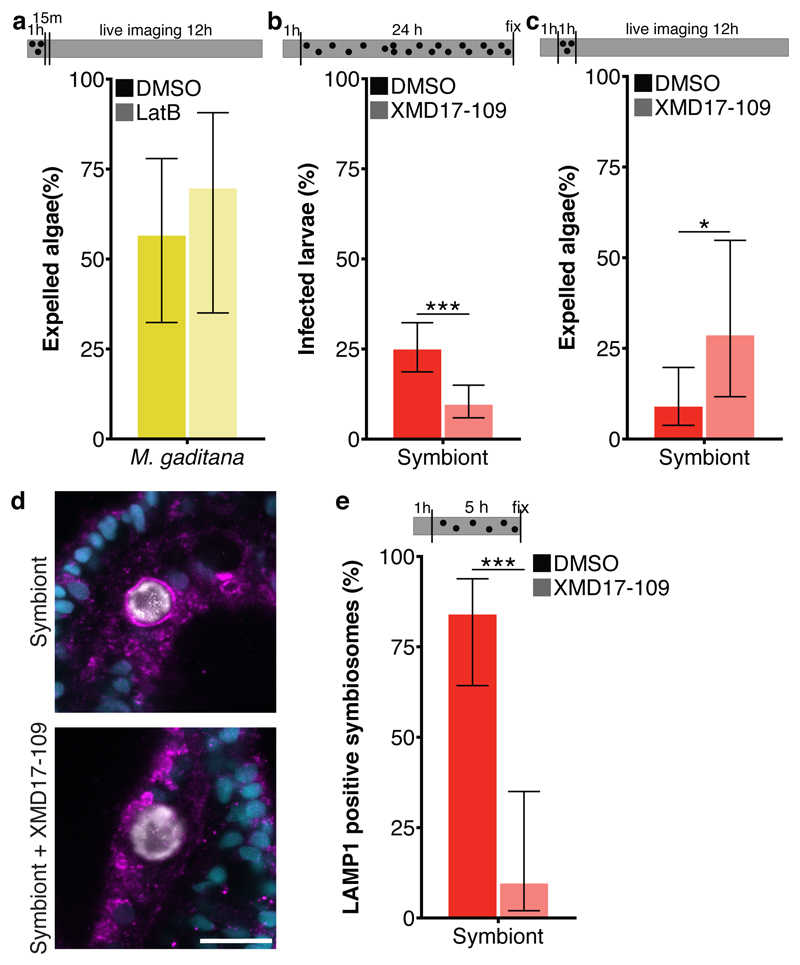
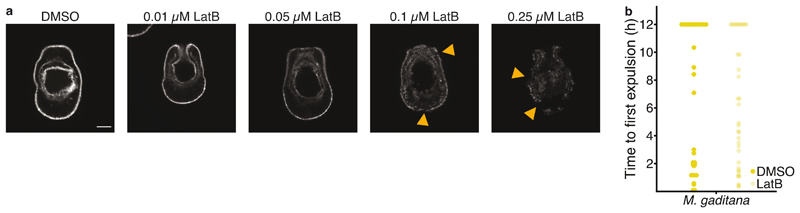
Interestingly, expulsion of microalgae by Aiptasia larvae resembles the non-lytic expulsion of a living organism from phagocytic cells, known as ‘vomocytosis’; both the phagocyte and the expelled microbe remain undamaged during this process 42,43,reviewed in 44. Because the host phagocyte remains intact during this process, immune stimulation may be prevented, facilitating the dissemination of pathogens within the infected organism. To date, vomocytosis has been observed in an array of animal macrophages and amoeba in response to fungal pathogens reviewed in 44. The dynamics and mechanism of vomocytosis are distinct from constitutive exocytosis, the default expulsion mechanism to release phagosomal content in amoeba. While constitutive exocytosis occurs consistently ~80 minutes after phagocytosis and depends on actin and the actin-regulating protein WASH (WASP and SCAR homolog), vomocytosis is stochastic and occurs between 2-12 hours after phagocytosis and is actin-independent 45–47. Indeed, expulsion of microalgae by Aiptasia larvae is a highly stochastic process (Fig. 2e). Moreover, we found that expulsion of M. gaditana was unaffected by inhibition of actin polymerization via Latrunculin B suggesting that vomocytosis, rather than constitutive exocytosis, is at play (Fig. 3a and Extended Data Fig. 2).
Fig. 3. Symbiosis establishment relies on vomocytosis inhibition.
a Inhibition of actin polymerization with Latrunculin B did not affect expulsion of M. gaditana from infected larvae during 12 hours of live imaging, as would be expected from constitutive exocytosis. (n=3) b ERK5 inhibition with XMD17-109 significantly (p = 0.000013) reduced the fraction of symbiotic larvae when pre-treated for 1 hour and treated during a 24-hour infection (n=6) and c increased (p = 0.013) the percentage of symbionts expelled during 12 hours of live imaging (n=5). d Larvae with normal ERK5 activity were able to form a LAMP1-positive symbiosome, while larvae treated with the ERK5 inhibitor XMD17-109 showed massively reduced accumulation of LAMP1 when pre-treated for 1 hour and treated during a 5-hour infection. LAMP1 (magenta); nuclei (cyan); symbiont (white). Scale bar represents 10 μm. e ERK5 inhibition significantly (p = 0.0000027) reduced the fraction of symbionts with LAMP1 accumulation (n=3). In all graphs, error bars represent mean ± 95 % CI. Statistics are based on a two-sided generalized linear mixed model accounting for repeated measurements. Bars above graphs represent duration of infection (black dots), treatment (grey fill) and live imaging (where applicable). Bars above graphs represent duration of infection (black dots), treatment (grey fill) and live imaging (where applicable). *P < 0.05, **P < 0.01, ***P < 0.001.
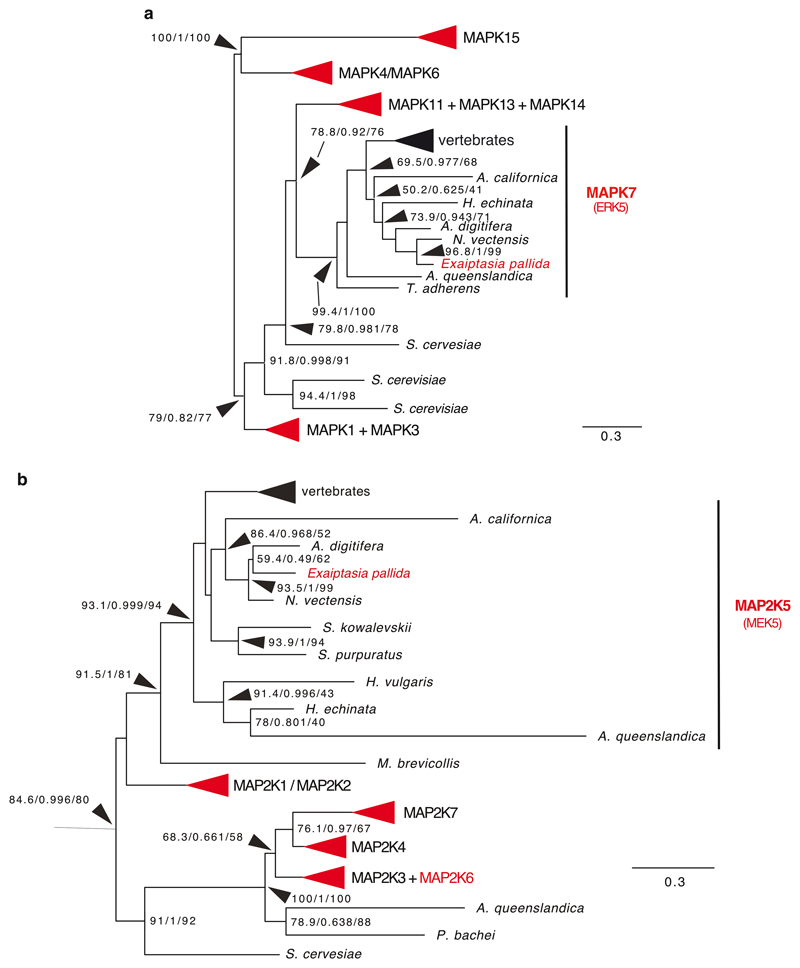
In vertebrate macrophages, vomocytosis is negatively regulated by ERK5 (also known as MAPK 7), as well as the upstream MAPK kinase, MEK5. ERK5 inhibition by XMD17-109 significantly increased rates of vomocytosis 48. Aiptasia contains several MAP kinases and one ERK5 homolog, with a conserved ATP binding site (aa 61-69 in H. sapiens ERK5 – the target of XMD17-109), as well as a clear MEK5 homolog (Extended Data Fig. 3). To examine whether the function of ERK5 as a negative regulator of vomocytosis is conserved in Aiptasia, we incubated Aiptasia larvae with the ERK5 inhibitor shortly before, and during, a 24-hour infection period. We found that the number of larvae infected with symbionts was massively reduced upon ERK5 inhibition (Fig. 3b), due to increased symbiont expulsion (Fig. 3c). Moreover, upon inhibition of ERK5, LAMP1 accumulation did not occur, resembling non-symbiotic microalgae-containing phagosomes (Fig. 3d + e). This further supports that Aiptasia larvae use vomocytosis for removal of incompatible microalgae, a process that symbionts escape to initiate the stable partnership between symbiont and host cell and to promote niche formation.
Modulation of host immunity prevents symbiont ‘vomocytosis’
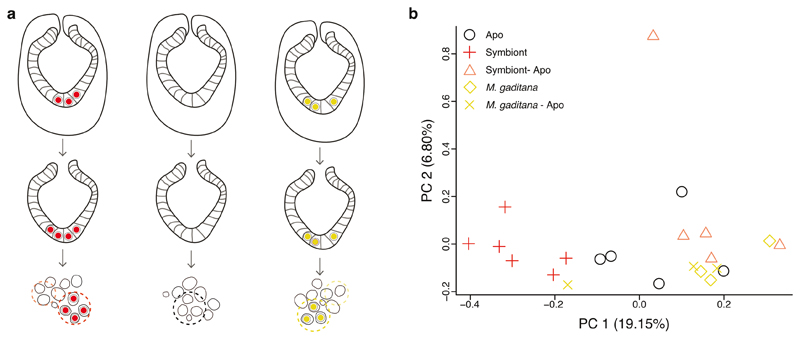
How do symbionts circumvent vomocytosis? Animals have evolved an array of defense mechanisms to protect themselves from invaders, and modulation of host immunity occurs in both parasitic and mutualistic symbioses, including those involving apicomplexan parasites and dinoflagellate symbionts reviewed in 12,49,50. To specifically ask whether host immune modulation is involved in escaping vomocytosis, we first asked how intracellular symbionts or non-symbiotic microalgae affect host immunity at the cellular level by comparing the expression levels of known immune-related genes (based on KEGG 51 annotations for Aiptasia) in cells containing either symbionts or M. gaditana, which are lost rapidly, yet remain intracellular long enough for analysis (Fig. 1d). We infected Aiptasia larvae with symbionts or M. gaditana for 24-48 hours, dissociated infected larvae, and sequenced groups of 8-12 endodermal cells. Specifically, we compared symbiont-containing cells to cells containing M. gaditana, as well as aposymbiotic neighboring cells from symbiont- or M. gaditana-containing endoderms, and aposymbiotic cells from naïve endoderms (Fig. 4a, Extended Data Fig. 4).
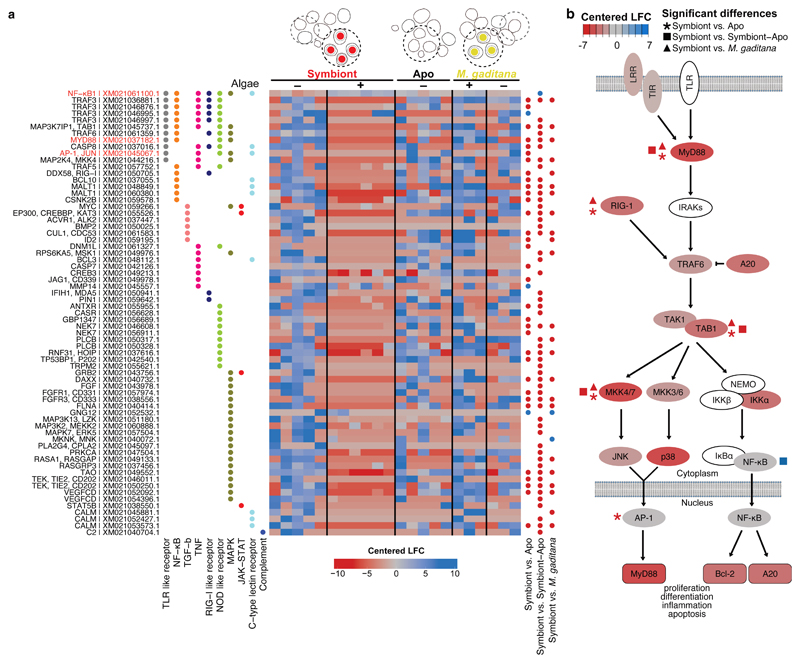
Fig. 4. Local suppression of host innate immunity is a prerequisite for symbiosis establishment.
a Several genes in immunity-related pathways were differentially downregulated in symbiont-containing cells but not in M. gaditana-containing cells or aposymbiotic cells from infected larvae (colors indicated centered log fold change according to Deseq2 with downregulation in red and upregulation in blue). The heatmap shows all differentially regulated genes symbiont vs Apo, symbiont vs symbiont-Apo, symbiont vs M. gaditana within the following KEGG pathways: C-type lectin receptor signaling pathway (ko04625), Complement and coagulation cascades (ko04610), JAK-STAT (ko04630), MAPK (ko04010), NF-κB (ko04064), NOD-like receptor (ko04621), RIG-I-like receptor (ko04622), TGFβ (ko04350), TNF (ko04668), and TLR-like receptor (ko04620). Significantly differentially expressed genes compared between populations of single cells are indicated with blue (upregulated) or red (downregulated) dots. Gene names in red indicate special interest as mentioned throughout the text. KEGG annotation was automated based on homology. b Simplified TLR pathway according to KEGG annotations (genes in white could not be identified in Aiptasia) representing gene expression in symbiotic cells (centered log fold change as in a). Statistically significant changes between symbiotic and aposymbiotic cells are indicated with asterisks, changes between symbiotic and aposymbiotic cells from symbiotic larvae are indicated with squares, and changes between symbiont- and M. gaditana-containing cells with triangles.
We found that the expression of an array of immune genes was significantly lower in symbiont-containing cells compared to all other samples (Fig. 4a). This involved 10 KEGG pathways related to metazoan innate immunity with some genes present in multiple pathways and multiple transcripts annotated as the same gene (e.g. TRAF3) (Fig. 4a). This suggests that suppression of host innate immunity upon intracellular establishment of dinoflagellate symbionts may occur at a much broader scale at the cellular level than previously reported for whole organisms 12. Specifically, the expression of genes associated with NF-κB and AP-1/Jun-related signaling pathways, such as toll-like receptor (TLR) (Fig. 4a) and tumor necrosis factor (TNF) pathways, differed proportionally the most (∼20% of genes from each pathway) (Fig. 4a, Supplementary Table 2). Moreover, genes of the TLR pathways were significantly affected when symbiotic cells are compared to cells across other conditions (cells from aposymbiotic larvae, non-symbiotic cells from symbiotic larvae, or M. gaditana-containing cells) (Fig. 4b).
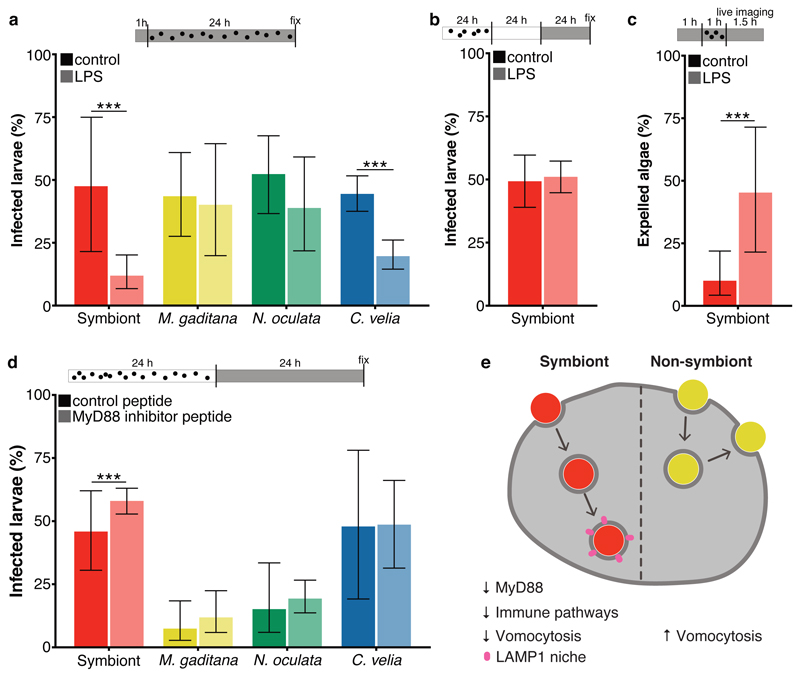
TLR signaling plays a fundamental role in animal innate immunity to recognize and eliminate invasive microbes, including the apicomplexan parasites from the genera Plasmodium and Toxoplasma 49,50. TLRs function as pattern recognition receptors (PRRs) and consist of an extracellular recognition domain constructed of tandem copies of a leucine-rich repeat (LRR) motif, a transmembrane domain, and a cytoplasmic signaling domain (TIR domain) 52. To date, canonical TLRs that have both extracellular LRR domains and intracellular TIR domains have not been identified in Aiptasia; however, two proteins containing only TIR-domains have been identified, along with two proteins containing only LRR domains 29, and it is known that the functional TLR of Hydra is similarly split into two proteins (hyLRR + hyTRR) 53. To test whether modulation of TLR-signaling affects the initiation of the interaction between the host and microalgae, we challenged aposymbiotic Aiptasia larvae with lipopolysaccharides (LPS), a ligand for mammalian TLR, which is commonly used to elicit an immune response, for 1 hour before and during infection with microalgae (Fig. 5a). LPS treatment massively decreased the proportion of symbiont-infected larvae to 25 % of the untreated control (Fig. 5a), reflecting previous observations in Aiptasia adult anemones 16. We observed a similar effect for C. velia, but no significant effects for N. oculata and M. gaditana infected larvae (Fig. 5a). Surprisingly, once symbionts were intracellularly integrated for more than 24 hours, LPS treatment did not affect symbiosis stability (Fig. 5b). Together, this suggests that for both the dinoflagellate symbiont and the apicomplexan-related ‘invader,’ activation of host immunity pathways conflicts with the initiation of the symbiotic interaction; however, once symbionts are stably integrated into the host cell, symbiosis stability is not compromised by immune activation. This is likely due to the fact that a large proportion of host immune signaling is transcriptionally repressed by that time (Fig. 4a).
Fig. 5. Immune stimulation enhances expulsion of symbionts during initial interaction.
a Percentage of symbiont-infected Aiptasia larvae was significantly (p = 0.0000000024) reduced when larvae were pre-treated for 1 hour with LPS, followed by a 24-hour exposure to microalgae and LPS. Percentage of C. velia-infected larvae was also reduced (p = 0.00000000024), but to a lesser degree; there was no effect observed for M. gaditana- or N. oculata-infected larvae. (n=5) b LPS exposure for 24 hours after established symbiosis (24 hours infection + 24 hours incubation without symbionts) did not influence symbiont maintenance. (n=5) c LPS treatment induced expulsion. LPS exposure for 1 hour before and during 1 hour of infection, significantly (p = 0.00038) enhanced the number of larvae with expulsion events as observed during 1.5 hours of live imaging. For the LPS treatment n=3 with total 42 microalgae; for the control n=4 with total of 50 microalgae. d MyD88 inhibition enhanced maintenance of symbionts in larvae (p = 0.0000053), while maintenance of other microalgae was unaffected. For the symbionts n=10; for M. gaditana n=3; for N. oculata n=13; and for C. velia n=3. e Model of microalgae uptake in Aiptasia endodermal cells. While uptake of symbionts (red) leads to downregulation of immunity genes until a functional LAMP1-positive symbiosome is formed, non-symbiotic microalgae (yellow) do not elicit a strong transcriptomic response in the host cells and are subsequently expelled by vomocytosis. All graphs show mean ± 95 % CI with statistics based on a two-sided generalized linear mixed model accounting for repeated measurements. Bars above graphs represent duration of infection (black dots), treatment (grey fill) and live imaging (where applicable). *P < 0.05, **P < 0.01, ***P < 0.001.
To directly test if immune activation via LPS treatment induces symbiont expulsion, we used live imaging to monitor the fate of phagocytosed symbiont cells following LPS exposure. We found that in the presence of LPS, symbionts were expelled more frequently (Fig. 5c). Thus, immune stimulation by LPS counteracts the robustness of symbiosis establishment due to an increase in expulsion events of newly phagocytosed symbionts. In contrast, LPS treatment did not significantly reduce the infection efficiency for N. oculata and M. gaditana microalgae, likely because the removal via vomocytotsis is stochastic and naturally occurs so quickly within the first 12-24 hours that artificial immune stimulation by LPS may not have an additional effect (Fig. 1d and Fig. 5a). Taken together, this suggests that vomocytosis is an immune response in Aiptasia larvae that is used for protection against ‘invaders.’ However, compatible symbionts suppress vomocytosis, a key step to begin an endosymbiotic lifestyle with their hosts.
TLR signaling is modulated via MyD88 during intracellular establishment of symbionts
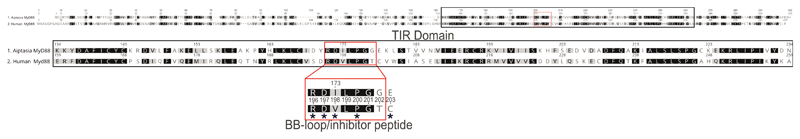
In mammals, most TLR-mediated activation of the immune system depends on the critical adapter molecule myeloid differentiation primary-response gene 88 (MyD88). During Plasmodium and Toxoplasma infection, MyD88 plays a key role in mounting an immune response in the host. For example, MyD88 is involved in innate sensing of Toxoplasma gondii infection, and MyD88 knockout mice are more susceptible to infection reviewed in 50. Interestingly, expression levels of Aiptasia MyD88 were significantly lower in cells that phagocytosed symbionts when compared to neighboring aposymbiotic cells, M. gaditana-containing cells, or aposymbiotic cells from naïve larvae (Fig. 4a, b). To directly test whether decreased MyD88 activity affects retention of symbionts or non-symbiotic microalgae, we infected Aiptasia larvae for 24 hours, and then inhibited the activity of MyD88 using an inhibitory peptide that specifically interferes with human MyD88 homodimer formation 54. The peptide mimics the solvent exposed BB-loop within the MyD88 TIR domain, a stretch of 7 amino acids, which is highly conserved between human and Aiptasia MyD88 (Extended Data Fig. 5). Indeed, we found that addition of the inhibitory peptide significantly increased the number of larvae that contained symbionts compared to the control; however, no such effect was observed for M. gaditana, N. oculata, or C. velia (Fig. 5d). This suggests that inhibition of MyD88 facilitates symbiont persistence within the first 24-48 hours of infection but is not sufficient on its own to allow persistence of non-symbiotic microalgae.
Discussion
A model to explain selective maintenance of intracellular dinoflagellate symbionts
The selection of compatible symbionts from the environment is a key step during the establishment of cnidarian-dinoflagellate endosymbiosis. Here, we observe the fate of phagocytosed compatible symbionts and incompatible microalgae in Aiptasia larvae in real-time, and we further investigate these observations with gene expression analysis, chemical perturbations, and cell biology approaches. Our findings lead us to propose a model in which aposymbiotic Aiptasia larvae constitutively acquire and release diverse microalgae from and to the environment. By default, phagocytosed, non-symbiotic microalgae are removed by vomocytosis. Acquisition of suitable symbionts leads to suppression of innate immunity pathways in the host cell, likely by suppressing the highly conserved TLR signaling pathway by targeting MyD88. This suggests that local modulation of innate immunity in the host cell is crucial for symbionts to escape expulsion by vomocytosis and promote intracellular LAMP1-niche formation (Fig. 5e). Thus, we define ‘escaping vomocytosis’ as a critical step during symbiosis establishment that requires suppression of host immunity, allowing us to directly link host immune modulation with a biological function.
Towards a molecular understanding of symbiont selection and niche formation
Experimental evidence suggests that one symbiont selection criterion is cell size: smaller microalgae are taken up more efficiently than larger ones. Thus, the physical properties of the symbiont play a role in symbiosis establishment 19,55. Moreover, specific recognition mechanisms between symbiont and host cells have been implied to promote the initial interaction 25,56,57, but cnidarian hosts also nonspecifically take up inert beads and incompatible microalgae. Both fail to establish a long-term partnership, likely due to their inability to manipulate the host immune system to promote a stable symbiotic interaction 18,22,55,56. At the mechanistic level, it is commonly thought that symbionts employ similar tactics as pathogenic microorganisms to accommodate long-term intracellular residency 16,20,25,58,59. Some pathogens, for example, modulate the TGFβ pathway during infection to promote their own tolerance 60,61. Similarly, the TGFβ pathway has been proposed to be involved in cnidarian-dinoflagellate endosymbiosis; specifically, the addition of an anti-human TGFβ antibody impairs symbiont uptake in Aiptasia anemones and coral larvae 16,20. It remains unclear, however, whether modulation of the TGFβ pathway occurs in vivo during symbiosis establishment and if so whether this is specific to symbiont-containing cells or rather an organismal-wide effect. Here, we uncover a different means for manipulating the immune response to regulate symbiosis. Our data suggest that transcriptional repression of MyD88, a conserved cytoplasmic adapter that integrates virtually all TLRs in mammals 50, may promote symbiosis establishment in Aiptasia larvae. While MyD88 activity has been shown to impair infection with the apicomplexan parasites Plasmodium and Toxoplasma in mammals 49,50, its role in endosymbiosis establishment was unclear until now. Analogous to counteracting apicomplexan infection in mammals, MyD88 activity may impair the initiation of cnidarian-dinoflagellate symbiosis, a principle that appears to be common for immune challenge in mutualistic and parasitic intracellular alveolates.
Along those lines, it is also worth noting that various pathogens including Listeria monocytogenes, Salmonella enterica, and Cryptococcus neoformans have been shown to reside in pathogen-containing vacuoles (PCV) that are decorated with LAMP1 62–66 similar to what we observe here for the intracellular symbionts in Aiptasia larvae, revealing another commonality between endosymbionts and certain intracellular pathogens (Fig. 2a). These types of PCVs are thought to resemble ‘modified lysosomes’ that provide an intracellular niche for replication without being acidic or digestive, two common features of classical lysosomes 41. Currently, the intracellular niche of dinoflagellates in the cnidarian host is not well described; however, in this context, we make three interesting discoveries: 1. We find that LAMP1-niche formation is impaired upon inhibition of ERK5, a negative regulator of vomocytosis (Fig. 3d+e), 2. non-specific immune stimulation via LPS increases symbiont expulsion within the first 24 hours of symbiosis establishment (Fig. 5a+c), but 3. LPS treatment has no effect once symbionts have been integrated into the host cells for 24 hours (Fig.5b). This suggests that activation of host immunity pathways conflicts with the initiation of the symbiotic interaction; however, once symbionts are stably integrated within the LAMP1-niche, symbiosis stability is not compromised by immune activation, likely due to the fact that a large proportion of host immune signaling is transcriptionally repressed by that time (Fig. 4a).
In contrast to previous organismal-wide analyses 17,19,21, we find that expression of the NF-κB transcription factor itself was not downregulated in symbiotic cells. In fact, compared to aposymbiotic cells from symbiotic larvae, NF-κB was even upregulated (Fig. 4a). This is in accordance with a host cell-specific response directly linked to symbiont phagocytosis and suggests that transcriptional downregulation of NF-κB itself is not required for symbiosis establishment at the individual host cell level but may play a role at the organismal level or once the symbiotic association becomes more mature. However, we observe significant transcriptional repression of multiple effectors of the second branch of TLR signaling, which is mediated by the transcription factor AP-1, including one transcript identified as AP-1, in symbiont-containing cells (Fig. 4b). This shows that host immune suppression occurs at a much broader scale at the cellular level than previously reported for whole organisms 12, a distinct response that likely reflects the immediate impact of symbiont phagocytosis by host cells.
Establishing endosymbiosis in cnidarian cells relies on escaping expulsion, not digestion
We found that symbiont uptake induces immune suppression in order to bypass clearance by vomocytosis, the fate of non-symbiotic phagocytosed particles, and not to escape phagolysosomal digestion. Phagocytosis is an ancient process used by unicellular organisms to acquire and digest food 67. Moreover, microbial killing by professional macrophages in higher metazoans is thought to derive from phagotrophy 39,67. Thus, being a primitive and highly conserved mechanism, it seemed likely that intracellular digestion would be used by cnidarians to eliminate incompatible invaders. In fact, symbiont digestion has been implicated in maintenance of stable dinoflagellate numbers in cnidarian-dinoflagellate symbiosis and in coral bleaching68,69. Accordingly, symbionts halting phagosomal maturation and thus avoiding intracellular digestion has been implied to play a role in symbiosis establishment 24,26,70. On the other hand, symbiont expulsion has been described as a key mechanism by which intact symbiotic microalgae are lost from host tissue during heat-induced bleaching 71–75. Additionally, it was suggested that symbiont expulsion is involved in regulating symbiont density in the host 76. However, the proportion of healthy symbiont cells, debris, and other microorganisms, as well as the life stage of the symbiont, differs between the expelled material of different species examined 77. Here, we uncover a vital role for expulsion in defining partner specificity during larval stages in the endosymbiotic relationship between cnidarians and their dinoflagellate symbionts. While non-pathogenic microalgae are removed by expulsion, selective intracellular persistence of symbionts requires avoidance of expulsion by vomocytosis, a phenomenon preceded by host immune suppression. This raises the interesting question of whether the same mechanisms are at play in bleaching, regulating symbiont density, and symbiosis establishment. For example, while our data suggest that Aiptasia larvae probe their environment using a trial-and-error mechanism, it is possible that the constitutive acquisition and release of incompatible microalgae that we observe in aposymbiotic larvae only occurs until a compatible symbiont strain has colonized the host. The immune suppression associated with the uptake may slow or halt this indiscriminate process to favor symbiont replication and dispersal rather than continuous de novo acquisition. Accordingly, even in partially bleached hosts, the remaining symbionts may suppress uptake of new microalgae from the environment to re-colonize the host by distributing descendants of the original symbiont strain. This could explain why anemones, and even entire coral colonies, are often stably inhabited by only a single Symbiodiniaceae type, or a few different ones at most 78–81. Alternatively, differences may exist between larval and adult life stages, larvae being more promiscuous and effective in phagocytosing microalgae when compared to their adult counterparts 36.
Vomocytosis as an ancient mechanism of innate immunity
Currently, vomocytosis is thought to function as an escape route for pathogens from specialized immune cells, possibly in an effort to promote dissemination 48. This concept is largely based on the experimental evidence for fungal pathogens from the genus Cryptococcus that are vomocytosed from cultured macrophages and zebrafish 42,46,62. Specifically, it was thought that fungal pathogens had evolved the capacity for vomocytosis in response to selective pressure exerted during the interaction with predatory amoeba 82. It is unclear whether vomocytosis is initiated by the host or the ‘invader,’ but the inability of heat-killed C. neoformans cells to undergo vomocytosis and its dependency on the fungal polysaccharide capsule suggest that vomocytosis is triggered by the pathogen 44,46,62. Interestingly, sporadic reports about other pathogens that appear to use ‘vomocytosis-like’ mechanisms are increasing 44,83–85. Here, we extend the observation of vomocytosis to a non-pathogenic interaction occurring between Cnidaria and diverse microalgae and inert beads. We find that Aiptasia larvae indiscriminately phagocytose microalgae from the environment and, only after uptake, select for suitable symbionts by eliminating the incompatible microalgae or indigestible particles via vomocytosis, suggesting that the process is likely triggered by the host. This is distinct from amoeba which release only live fungal cells via vomocytosis but rely on WASH-mediated constitutive exocytosis for expulsion of inert beads and heat-killed fungal cells 45. Mechanistically, vomocytosis involves the prevention of LAMP1 accumulation in cnidarian larvae, which is in line with observations for vomocytosed C. neoformans cells in animal macrophages this study, 86.
To date, constitutive exocytosis has not been assessed in cnidarian larvae; however, we find that inhibition of actin polymerization, which halts constitutive exocytosis, does not alter the frequency or dynamics of the expulsion of M. gaditana cells (Fig. 3a) suggesting that, at least for large particles (≥ 3 μm), vomocytosis is the predominant mechanism at play in these organisms. Vomocytosis of indigestible material, non-symbiotic microalgae, or even pathogenic microbes may be an effective clearing mechanism for these small, motile organisms, which are in constant contact with the environment. Similarly, constitutive exocytosis and vomocytosis are efficient clearance mechanisms for free-living, unicellular amoebae, while the phagocytic cells of multicellular organisms rely on the retention and digestion of the phagocytosed material to avoid infecting neighboring cells or tissue. Aiptasia larvae represent an interesting intermediate between these two - they are multicellular organisms with structured tissues, however both the ectoderm and endoderm are in constant contact with the environment, therefore the tight regulation of vomocytosis that occurs in macrophages is not only unnecessary in Aiptasia larvae but detrimental for successful symbiosis establishment. Moreover, our data suggest that symbionts circumvent this default clearing by inducing localized host immune suppression. Therefore, vomocytosis may not be specific to certain pathogens but rather an evolutionarily ancient defense mechanism to fight ‘invaders’ that was co-opted by symbiotic cnidarians to selectively promote maintenance of intracellular symbionts. More broadly, we propose that vomocytosis is an ancient clearing mechanism of animal cells, and that the ability to escape vomocytosis was key for the evolution of an intracellular lifestyle for alveolates.
Conclusion
Cnidarians, such as corals and anemones, are evolutionarily ancient animals with simple body plans, yet possess cells with complex immune capacities providing a powerful experimental system to dissect the basis for the evolution of intracellular lifestyles and partner specificity. Uncovering the cell biology of symbiosis establishment using comparative, experimental approaches in a dinoflagellate-cnidarian model will be key to understanding the origin of mutualistic and parasitic intracellular lifestyles of alveolates within animal cells.
Methods
Live Organism Culture and Maintenance
Microalgae maintenance
For infection experiments of Aiptasia larvae, we used Breviolum minutum clade B (SSB01, symbiont) 35, Microchloropsis gaditana CCMP526 (NCMA, Bigelow Laboratory for Ocean Sciences, Maine, USA), Nannochloropsis oculata, and Chromera velia (NORCCA K-1276, NIVA, Oslo, Denmark). All cultures were grown in cell culture flasks in 0.22 μm filter-sterilized 1x Diago IMK medium (Wako Pure Chemicals, Osaka, Japan) on a 12-h light:12-h dark (12L:12D) cycle under 20–25 μ mol m−2 s−1 of photosynthetically active radiation (PAR) as measured with an Apogee PAR quantum meter (MQ-200; Apogee, Logan, USA). Sterile stock cultures of B. minutum (SSB01, symbiont), N. oculata, and C. velia were grown at 26 °C and M. gaditana at 18 °C. All microalgae (including M. gaditana) cultures to be used for infections were kept at 26 °C for 1-2 weeks post splitting before infection.
Aiptasia spawning and larval culture conditions
Aiptasia clonal lines F003 and CC7 (Carolina Biological Supply Company #162865; Burlington, USA) were induced to spawn following the previously described protocol 30. Aiptasia larvae were maintained in glass beakers in filter-sterilized artificial seawater (FASW) at 26 °C and exposed to a 12L:12D cycle.
Infection assay
At least three biological replicates (i.e. distinct spawning events) of naturally aposymbiotic Aiptasia larvae were collected and diluted to a concentration of 300-500 larvae per milliliter of FASW in glass beakers. Between 4- and 8-days post fertilization (dpf), the larvae were infected with 1.0 x 105 microalgae ml -1 of the respective microalgae types. Beakers were kept at 26 °C and exposed to a 12L:12D cycle. After a 24-hour infection, the larvae were washed to remove the microalgae and fresh FASW was added.
Quantification of infection efficiency
Infected larvae were fixed at 1-, 2-, 3-, 6-, and 10-day(s) post infection (dpi) using a 4% formaldehyde solution (#F1635, Sigma-Aldrich, Munich, Germany) for 30 minutes at room temperature (RT), followed by two washes in 0.1% Triton X-100 (PBS-Triton) (#3051, Carl Roth GmbH, Karlsruhe, Germany), and mounted in 87% glycerol (#G5516, Sigma-Aldrich) in PBS with the addition of 2.5 mg ml -1 1,4-Diazabicyclo[2.2.2]octane (DABCO) (#D27802, Sigma-Aldrich). At least 50 larvae per replicate per microalgal type were counted (Nikon Plan Fluor 20x objective). Data recording was performed in Microsoft Excel v16.16.6. Representative differential interference contrast (DIC) and epi-fluorescent images of the microalgal autofluorescence were taken. Microscopic analysis was conducted with a Nikon Eclipse Ti inverted microscope using a Nikon Plan Fluor 40x air objective. Images were processed with Fiji software 87.
Imaging and staining procedures in Aiptasia larvae
Fluorescent staining for f-actin
Infected larvae were fixed 24 hpi using 4% formaldehyde solution for 30 minutes at RT, followed by one wash in 0.05% Tween20 (#P7949, Sigma-Aldrich) in PBS (PBS-Tween20) for 5 minutes. For permeabilization, larvae were rotated in 1.5 ml Eppendorf tubes at 0.25 rpm in a solution of 1% PBS-Triton and 20% DMSO (#67-68-5, FisherScientific, Schwerte, Germany) for 1 hour at RT. For blocking, the permeabilization solution was exchanged with 5% normal goat serum (#005-000-121, Jackson ImmunoResearch Laboratories, Inc., Ely, UK) in 0.05% PBS-Tween20. Larvae remained in the blocking buffer for 30 minutes at RT while rotating. After two washes in 0.05% PBS-Tween20, larvae were incubated in Phalloidin Atto 565 (#94072, Sigma-Aldrich) diluted 1:200 in 0.05% PBS-Tween20 on a rotator and protected from light. Larvae were washed three times in 0.05% PBS-Tween20 before incubation in 10 μg ml-1 Hoechst (#B2883, Sigma-Aldrich) diluted in Tris-buffered saline, pH 7.4; 0.1% Triton X-100; 2% bovine serum albumin (#A7906, Sigma-Aldrich); 0.1% sodium azide (#S2002, Sigma-Aldrich) for 30 minutes on a rotator at RT in the dark. Larvae were washed three times for 5 minutes with 0.05% PBS-Tween20 and washed step-wise into glycerol from 30% to 50% to 100% and finally mounted. Confocal microscopic analysis was carried out with a Leica TCS SP8 stand using 63x glycerol immersion objective (NA 1.30), Leica LAS X software and Fiji software 87. Hoechst, Atto 565, and symbiont autofluorescence were excited with 405, 561, and 633 nm laser lines, respectively. Fluorescence emission was detected at 410-501 nm for Hoechst, 542-641 nm for Phalloidin Atto 565, and 645-741 for symbiont autofluorescence.
Live imaging
For live imaging of infected larvae, chambers were prepared by placing two 2.5 mm x 5 mm strips of non-toxic double-sided tape (TES5338, Tesa, Norderstedt, Germany) at the periphery of 35 mm μ-Dishes (# 81166, Ibidi, Gräfelfing, Germany). 1.5% low gelling agarose (LGA) (#A4018, Sigma-Aldrich) was dissolved in FASW by heating to 80 °C until liquid. The liquid LGA was cooled to and kept at 37 °C until use. 300 larvae in a volume of 1 ml were placed in a 1.5 ml Eppendorf centrifuge tube and quickly vortexed using a Sprout mini centrifuge (#552021, Biozym Scientific GmbH, Hessisch Oldendorf, Germany) to pellet larvae. The larvae and 1.5% LGA at 37 °C were mixed for a final LGA concentration of 1.14% LGA. The mixture was pipetted into the center of the prepared imaging chamber and a glass coverslip was pressed on top of the droplet to adhere on either end to the double-sided tape. The Ibidi plate was filled with 2 ml of FASW. Microscopic analysis was conducted with a Nikon Eclipse Ti inverted microscope using a Nikon Plan Fluor 20x air objective, with images taken every 5 or 15 minutes (as indicated) in DIC and TexasRed channels. Images were processed with Fiji software 87.
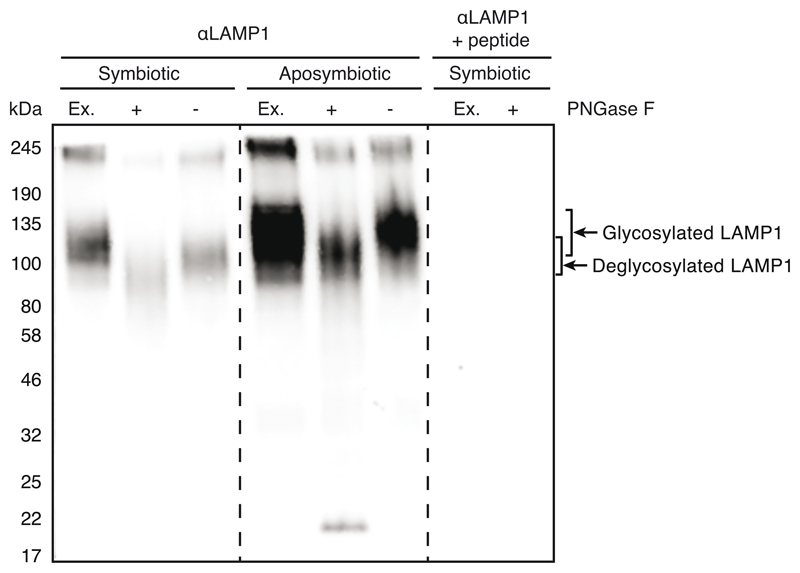
Aiptasia-specific anti-LAMP1 antibody purification
An antibody against the Aiptasia LAMP1 homolog (LOC110235349) was raised against the peptide IIGRRKSQRGYEKV (KXJ16564.1) coupled to the adjuvant keyhole limpet hemocyanin in rabbits (BioScience GmbH). The antibody was affinity purified from the third bleed using the synthetic peptide coupled to N-hydroxysuccinimide esters (NHS)-activated sepharose (17090601, GE Health Care Life Sciences) according to manufacturer’s protocols.
LAMP1 antibody validation and deglycosylation assay
Two aposymbiotic or symbiotic adult Aiptasia were homogenized in 50 mM TrisHCl pH 7.5, 200 mM NaCl, and 1% NP-40 with 2X Halt Protease Inhibitor Cocktail (78430, Thermo Fisher Scientific) and then sonicated on ice (Sonifier 250, Branson Ultrasonics) with two rounds of 25 pulses at duty cycle 40%, output control 1.8. The homogenate was centrifuged at maximum speed at 4°C for 10 minutes and supernatant was transferred to a new tube. Deglycosylation assay using PNGase F (New England BioLabs Inc., P0704S) was performed according to manufacturer’s protocol with the exception of incubating the reaction for 3 hours at 37°C followed by overnight at RT. 0.5 mg/ml of LAMP1 antibody was pre-adsorbed with 1 mg/ml of LAMP1 peptide in 4% milk in 0.1% PBT overnight at 4°C. Untreated and treated extracts were diluted 1:1 in 5X loading dye and heated to 100°C or 60°C, respectively, for 5 minutes. Samples were loaded into a 4-20% precast gel (Bio-Rad Laboratories Inc., 4561095), which was run at 90 V for 15 minutes then at 200 V for 1 hour at room temperature in 1X SDS running buffer. The proteins were transferred onto a nitrocellulose membrane at 0.37 A for 1 hour and 15 minutes at RT in 1X transfer buffer (100 ml methanol, 100 ml 10X transfer buffer, 800 ml water). The membrane was blocked for 1 hour at room temperature in 4% milk in 0.1% Triton X-100 in PBS. The blot was divided in two and incubated in either LAMP1 antibody diluted 1:2000 in blocking buffer or pre-adsorbed LAMP1 overnight at 4°C. The following day the blots were washed in 0.1% Triton X-100 in PBS 3 x 15 minutes at RT. The secondary antibody, goat-α-rabbit-HRP (Jackson ImmunoResearch), was added at a dilution of 1:5000 for 1 hour at RT, protected from light, followed by 3 x 15 minutes washes in either 0.1% Triton X-100 in PBS and one final wash in 1X PBS. The blot was developed using 1:1 ECL (GERPN2232, Sigma-Aldrich) and imaging on ECL Imager (ChemoCam, Intas).
Immunofluorescence staining (LAMP1)
Larvae were fixed for 45 minutes in 4% formaldehyde at RT, followed by three washes in 0.2% PBS-Triton and one wash in PBS. Larvae were then permeabilized in PBS-Triton for 1.5 hours at RT, followed by blocking in 5% normal goat serum and 1% BSA in PBS-Triton (blocking buffer) for 1 hour. Primary antibody (rabbit-α-LAMP1) (see above, validation in Extended Data Fig. 6) was diluted 1:100 in blocking buffer and incubated overnight at 4°C. After three washes in PBS-Triton, the secondary antibody (goat-α-rabbit AlexaFluor 488, #ab150089, abcam, Berlin, Germany) was diluted 1:500 in blocking buffer and incubated for 1.5 hours at RT. Larvae were washed two times in PBS-Triton, followed by a 15-minute incubation with 10 μg ml-1 Hoechst protected from light at RT, and two final washes in PBS-Triton and one in PBS before mounting in 87% glycerol. Larvae were imaged on a Leica TCS SP8 confocal laser scanning microscope using a 63x glycerol immersion lens (NA 1.30) and Leica LAS X software. Hoechst, Alexa 488, and symbiont autofluorescence were excited with 405, 496, and 633 nm laser lines, respectively. Fluorescence emission was detected at 410-501 nm for Hoechst, 501-541 nm goat-α-rabbit AlexaFluor 488, and 645-741 for symbiont autofluorescence.
Transcriptome
Sample collection and sequencing
Larvae (300 ml-1) were infected with either 105 ml-1 symbionts, 105 ml-1 M. gaditana, or left uninfected at 6-7 days post fertilization (dpf) for 24 to 48 hours. Per replicate 3-5 larvae were transferred to 5 ml of Calcium- and Magnesium-free artificial seawater (CMF-SW, doi:10.1101/pdb.rec12053) and incubated for 5 min. They were then incubated in 70 μl of Pronase (0.5 % in CMF-SW, 10165921001, Sigma-Aldrich) and sodium thioglycolate (1 % in CMF-SW, T0632, Sigma-Aldrich) for approximately 2 min, after pipetting up and down 3-5 times to remove the ectodermal cells from the larvae. The endoderm was transferred to 70 μl of FAWS, and residual ectodermal cells were mechanically removed, before transfer to fresh FAWS. Cells were separated using tweezers, with the whole procedure not taking longer than 30 min. Pools of 7-20 cells, either symbiotic, aposymbiotic from symbiotic animals, aposymbiotic from aposymbiotic animals, M. gaditana-containing cells, or microalgae free cells from M. gaditana-containing larvae were picked using a microcapillary needle (Science Products GB100T-8P) pulled to an opening diameter of 8-12 μm diameter (Micropipette Puller P-97, Sutter Instrument). Capillaries contained 4.3 μl of lysis buffer (0.2 % TritonX-100, 1U/μl Protector RNase inhibitor {3335399001, Sigma-Aldrich}, 1.25 μM oligo- dT30VN, and 2.5 mM dNTP mix) and cells were flushed out of the capillary with lysis buffer, and then flash frozen. Sequencing libraries were prepared by RNA reverse transcription and pre-amplification of cDNA of 21 PCR cycles as described in Picelli et al. 88. cDNA libraries were prepared for Illumina sequencing and sequenced on NextSeq500 (Illumina) with 75 bp paired-end reads. Reads have been deposited on NCBI sequence read archive (SRA) with following accession numbers: cells from aposymbiotic larvae (SRX7119772-7119776), symbiotic cells (SRX7119782-7119787), aposymbiotic cells from symbiotic larvae (SRX7119777-7119781), combined in the SRA project SRP229372, M. gaditana-containing cells (SRX7229078-7229080), and microalgae-free cells from M. gaditana-containing larvae (SRX7229075-7229077) combined in the SRA project SRP233508.
Computational methods
Paired reads were mapped to the Aiptasia genome version GCF_001417965.1 using HISAT2 version 2.1.0 at default settings, except –X 2000 --no-discordant --no-unal --no-mixed. Transcripts were quantified in Trinity v2.5.1 using salmon v0.10.2 at default settings. Principal component analysis was conducted using a perl script supplied with Trinity for all samples. Differential expression was analyzed using DEseq2 89 (log2-fold change ≥ 1, adjusted p-value ≤ 0.05) in R! v3.5.2 90. Graphing of results was restricted to pathways involved in innate immunity as found in KEGG 51, using the R! package “ComplexHeatmap” 91 in KNIME 92. Mean corrected log fold expressions from Deseq2 were used in “pathview” 93 in R! as basis for the shown pathway (Fig. 4b). The custom KNIME workflow used for processing and analyzing the data is available at https://doi.org/10.24433/CO.0872345.v1.
Exogenous immune perturbations
LPS/ ERK5 inhibitor treatment
Aiptasia larvae were washed and diluted to a concentration of 300-500 larvae ml-1 and were then incubated with 20 μg μl-1 LPS (lipopolysaccharides from Escherichia coli O127:B8, Sigma-Aldrich), 1 μM XMD17-109 (0.1% DMSO {Dimethylsulfoxid, Carl-Roth}), 0.1 % DMSO, or without for 1 hour. Microalgae cultures were then added to a final concentration of 1 x 105 microalgae ml-1 of the respective microalgal types and incubated at 26 °C and exposed to a 12L:12D cycle. After a 24-hour infection, the larvae were fixed in 4% formaldehyde for 30 minutes, washed in PBS, and mounted in 100% glycerol for counting. Infection status was quantified in at least 100 larvae per infection using a Nikon Eclipse Ti epifluorescent microscope using a Nikon Plan Fluor 20x objective, utilizing microalgal autofluorescence. Data recording was performed in Microsoft Excel v16.16.6.
LPS post-infection treatment
Washed and diluted Aiptasia larvae (300-500 ml-1) were mixed with B. minutum (1.0 x 105 microalgae ml-1) and left for infection at 26 °C with a 12L:12D cycle for 24 hours. They were then washed/filtered to remove excess microalgae and incubated further for 24 hours. Half of the infected larvae were exposed to 20 μg ml-1 of LPS for 24 hours, before fixation with 4% formaldehyde. They were then washed in PBS and mounted in glycerol. Infection status was assessed in at least 100 larvae per infection using a Nikon Eclipse Ti epifluorescent microscope using a Nikon Plan Fluor 20x objective, utilizing microalgae autofluorescence. Data recording was performed in Microsoft Excel v16.16.6.
Live imaging of early infection with LPS / ERK5 inhibitor treatment
Aiptasia larvae (300-500) were either incubated with 20 μg ml-1 LPS, 1 μM XMD17-109 (0.1% DMSO), 0.1 % DMSO, or without for 1 hour, before infection with B. minutum (1.0 x 105 microalgae ml-1) for 1 hour. They were then mounted in μ-Dishes as described previously with LPS, XMD17-109, DMSO or without in both agar and surrounding FASW) and then imaged on a Nikon Eclipse Ti inverted microscope using a Nikon Plan Fluor 20x air objective, with images taken every 5 min in DIC and TexasRed channels. Images were processed with Fiji software 87. For each independent experiment (n=3 (LPS) or n=4(ERK5)), between 6 and 14 larvae with a total of 7-24 microalgae were observed.
MyD88 inhibitor peptide treatment
6 dpf larvae were infected with 1.0 x 105 cells ml-1 of B. minutum (SSB01, symbiont) or N. oculata for 24 hours. Infected larvae were washed with FASW to remove microalgae from water and 10 units ml-1 penicillin and 10 μg ml-1 streptomycin (P4333 Sigma-Aldrich) were added. Infected larvae were incubated for 24 hours at 26 °C under a 12L:12D cycle with either 50 μM of MyD88 inhibitor peptide or 50 μM control peptide (NBP2-29328, Novus Biologicals, Centennial, CO, USA). Larvae were fixed in 4% formaldehyde and infection efficiency was quantified as previously described.
LAMP1 assessment after ERK5 inhibitor treatment
Aiptasia larvae were exposed to 1 μM ERK5 inhibitor (XMD17-109) or not for 1 hour prior to and during infection with 1.0 x 105 cells ml-1 of B. minutum (SSB01, symbiont) for 5 hours. Larvae were then stained for LAMP1 as described above. Symbionts with strong LAMP1 staining were assessed on a Leica TCS SP8 confocal laser scanning microscope using a 63x glycerol immersion lens (NA 1.30) and Leica LAS X software. For control, the LAMP1 staining was evaluated in 20 symbiotic larvae; however, due to low infection numbers after XMD17-109 treatment, only 8, 4, and 7 larvae could be evaluated in the respective biological replicates.
Live imaging of early infection with Latrunculin B treatment
To determine the optimal concentration of latrunculin B (LatB), Aiptasia larvae were incubated with 0.01, 0.05, 0.1 or 0.25 μM LatB for 6 hours, fixed and stained with Phalloidin to visualize f-actin as described above. Actin levels were reduced at 0.05 μM LatB when compared to the control, but the larvae still appeared healthy and intact, in contrast to what was observed at higher concentrations (Extended Data Fig. 2a) Therefore, we used 0.05 μM LatB for the live imaging. Briefly, Aiptasia larvae (300-500) were infected with M. gaditana (1.0 x 105 microalgae ml-1) for 1 hour followed by a 15-minute incubation with either 0.05 μM (LatB) in DMSO or with DMSO as a control. Larvae were mounted in μ-Dishes in both LGA and FASW as described above, supplemented with LatB or DMSO and imaged on a Nikon Eclipse Ti inverted microscope using a Nikon Plan Fluor 20x air objective. Images were taken every 5 minutes in DIC and the TexasRed channel. Images were processed with Fiji software 87. For each independent experiment (n=3), between 8 and 10 larvae with a total of 8 to 16 microalgae were observed.
ERK5 / MEK5 phylogeny
To resolve the evolutionary history and classification of Aiptasia MAPKs and MAP2Ks we first used well-defined ‘bait’ sequences (human MAPK7 and MAP2K5) and reciprocal BLAST searches to identify and collate the MAPK and MAP2K repertoire in Aiptasia. Aiptasia sequences were then used to retrieve other MAPK and MAP2K sequences of animals including key cnidarian species from public databases using BLASTP. H. echinata sequences were downloaded from https://research.nhgri.nih.gov/hydractinia/download/ and manually curated and translated. Resulting protein sequences were deduplicated and then aligned using ClustalW (GONNET, goc: 3, gec: 1.8). Then, automated trimming was performed using trimAI (using standard parameters; http://trimal.cgenomics.org/, 94). Best-fitting amino acid substitution models were determined using ModelFinder (-m MF -msub nuclear -nt AUTO) within FigTree and Maximum likelihood trees generated. Trees were finalized using FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) and Adobe Illustrator CC 2018 and Affinity Designer v1.9.1. Original tree files with accession numbers are provided (Supplementary Files 1+2).
Statistical notes
Complete statistical analysis, including raw data, R-script, results and repeat numbers, can be found under https://doi.org/10.24433/CO.0872345.v1. For analysis a two-sided generalized linear mixed model was used in R v4.0.3 90 using the gam function from the mgcv-package 95. Treatment of larvae within one well in a repeat was taken into account with the random Intercept “s(Well_ID, bs=“re”)”. For all experiments that were performed side-by-side the random intercept “s(Repeat.Date.,bs=‘re’)” was used to account for repeat measurements. Additionally, for all graphs in Fig 1 and Fig 2 multiple comparisons were performed with the emmeans package, using the default Tukey correction methods of p-values.
Extended Data
Extended Data Fig. 1. Uptake of microalgae is indiscriminate.
a Additional microalgae screened: Isochrysis sp., Chlorella sp., D. salina, and C. parkeae. Images are DIC and red autofluorescence of microalgae photosynthetic pigments. Scale bar indicates 25 μm. b Aiptasia larvae were infected at 4-6 days post fertilization (dpf) for 24 hours and were washed into fresh FASW. Error bars represent SEM. n=3 for all except C. parkeae with n= 1
Extended Data Fig. 2. Inhibition of actin polymerization does not affect expulsion of non-symbiotic microalgae.
a Analysis of the effects of different concentrations of LatrunculinB (LatB) on Aiptasia larvae to determine a suitable concentration for live imaging analysis. Larvae were incubated for 6 hours in LatB, washed, fixed and the f-actin levels were assessed by fluorescence microscopy. 0.01 μM does not affect the overall levels or distribution of actin. In contrast, LatB concentrations >0.1 μM substantially decreased actin levels and impacted the integrity of Aiptasia larvae (see arrowheads pointing to “holes” within the organisms). Accordingly, an intermediate concentration of 0.05 μM LatB which substantially reduced f-actin levels without compromising larval integrity was used for live imaging in Fig. 3a). b Inhibition of actin polymerization with Latrunculin B did not affect the time to expulsion of M. gaditana from infected Aiptasia larvae.
Extended Data Fig. 3. ERK5 and MEK5 homologues in Aiptasia.
Phylogenetic analysis of ERK5 and MEK5 from Aiptasia. a + b are collapsed trees of Aiptasia MAPK (A) or MAP2K (B) in comparison to several other cnidarian and vertebrate species. Red arrowheads or writing indicate presence of an Aiptasia homolog. Both Aiptasia ERK5 and MEK5 cluster within ERK5 (MAPK7) or MEK5 (MAP2K5), respectively. Full tree can be accessed through Supplementary Files 1 and 2.
Extended Data Fig. 4. Cell-specific characterization for transcriptomic analysis.
a Schematic of Aiptasia larvae used for cell-specific sequencing. Ectodermal cells were removed resulting in only endodermal cells that were dissociated and selected for based on contents: aposymbiotic cells from symbiotic larvae (Symbiont-Apo), symbiotic cells from symbiotic larvae (Symbiont (red)), aposymbiotic cells from aposymbiotic larvae (Apo), cells containing M. gaditana from larvae infected with M. gaditana (M. gaditana (yellow)), and aposymbiotic cells from larvae infected with M. gaditana (M. gaditana-Apo). b Principal Component Analysis (PCA) plot of host gene expression in different conditions.
Extended Data Fig. 5. Amino acid sequence similarity between human and Aiptasia MyD88.
Human MyD88 homo-dimerizes to trigger a downstream signaling cascade leading to immune activation. It consists of three domains, the death domain (DD), the interdomain (ID) and the C-terminal TIR domain 97. The human TIR domain is key for homo-dimerization with other TIR domains from MyD88 or other TIR domain containing proteins. Three distinct regions contributing to homo-dimerization have been identified by crystallography, NMR and mammalian two-hybrid analysis 98. However, the so-called BB-loop within the TIR domain, a solvent-exposed stretch of 7 residues (RDLVPGT) is particularly critical for homodimerization in human MyD88. Accordingly, cell-permeable peptides mimicking the 7 residues of the BB-loop of human MyD88 interfere with homo-dimerization 99,55,100. The TIR domains (black box/upper alignment) of mammals and Aiptasia are well conserved (50% sequence identity). Moreover, the BB-loop (red box) is almost identical and key residues (*) are conserved. Identical amino acids have black background, similar aa have gray background and aa with white background are not similar according to blosume62 scoring.
Extended Data Fig. 6. N-deglycosylation of LAMP1 in Aiptasia homogenate.
Verification of α-LAMP1 antibody used in Figs. 2a, 3d + e by Western blot. LAMP1 has been observed to run at a higher than predicted weight (38 kDa) because it is heavily glycosylated 101. Deglycosylation of homogenates of symbiotic and aposymbiotic adult Aiptasia CC7 using PNGase F resulted in a shift to a lower molecular weight. As control, extracts were detected with LAMP1 antibody pre-adsorbed with the peptide used for raising the antibody.
Supplementary Material
Acknowledgements
We thank Dinko Pavlinic and Vladimir Benes (Genecore Facility, EMBL Heidelberg) for assistance with the SmartSeq2 protocol and sequencing library preparation; David Ibberson (Deepseqlab, Heidelberg University) for assistance with the SmartSeq2 protocol; Carsten Rippe for access to the BioAnalyzer; Liz Hambleton for help with antibody purification; Bruno Gideon Bergheim for initiating live imaging of Aiptasia larvae; Moritz Mercker (Bionum) for advice on statistical analysis, Friedrich Frischknecht, Thomas Gilmore, Thomas Holstein, and Steffen Lemke for advice; Robin May for advice and comments on the manuscript.
Funding was provided by the Deutsche Forschungsgemeinschaft (DFG) (Emmy Noether Program Grant GU 1128/3–1) and the H2020 European Research Council (ERC Consolidator Grant 724715) to AG, a scholarship to SR by the CellNetworks Excellence Cluster (Heidelberg University) Postdoctoral Program, and a Ph.D. scholarship within the Graduate School “Evolutionary Novelty & Adaptation by the Baden-WÜrttemberg Landesgraduiertenforderung Program to PAV.
Footnotes
Author Contributions
Conceptualization, M.R.J., S.R. and A.G.; Methodology, M.R.J., S.R., P.A.V., I.M. and A.G.; Software, P.A.V. and S.G.G.; Formal Analysis, M.R.J., S.R., Investigation, M.R.J., S.R. and I.M.; Data Interpretation, M.R.J., S.R., I.M. and A.G.; Resources, A.G.; Data Curating, P.A.V. and S.G.G.; Writing - Original Draft, M.R.J., S.R. and A.G., Writing–Review & Editing, M.R.J., S.R., S.G.G., P.A.V. and A.G.; Visualization, M.R.J., S.R.; Supervision, A.G., Project Administration, M.R.J., S.R., and A.G.; Funding Acquisition, A.G.
Declaration of Interests
The authors declare no competing interest.
Data availability
RNA sequencing data: raw reads can be accessed at NCBI sequence read archive (SRA) with following accession numbers: cells from aposymbiotic larvae (SRX7119772-7119776), symbiotic cells (SRX7119782-7119787), aposymbiotic cells from symbiotic larvae (SRX7119777-7119781), combined in the SRA project SRP229372, M. gaditana-containing cells (SRX7229078-7229080), and microalgae-free cells from M. gaditana-containing larvae (SRX7229075-7229077), combined in the SRA project SRP233508.
Code availability
The Knime 92 workflow used for analysis as well as other raw data and R scripts for analysis can be found under https://doi.org/10.24433/CO.0872345.v1
References
- 1.Saldarriaga JF, “Max” Taylor FJR, Cavalier-Smith T, Menden-Deuer S, Keeling PJ. Molecular data and the evolutionary history of dinoflagellates. Eur J Protistol. 2004;40:85–111. [Google Scholar]
- 2.Seeber F, Steinfelder S. Recent advances in understanding apicomplexan parasites. F1000Research. 2016;5:1369. doi: 10.12688/f1000research.7924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong WK, del Campo J, Mathur V, Vermeij MJA, Keeling PJ. A widespread coral-infecting apicomplexan with chlorophyll biosynthesis genes. Nature. 2019;568:103–107. doi: 10.1038/s41586-019-1072-z. [DOI] [PubMed] [Google Scholar]
- 4.de Vargas C, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605-1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 5.Baker AC. Flexibility and Specificity in Coral-Algal Symbiosis: Diversity, Ecology, and Biogeography of Symbiodinium. Annu Rev Ecol Evol Syst. 2003;34:661–689. [Google Scholar]
- 6.Yellowlees D, Rees TAV, Leggat W. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 2008;31:679–694. doi: 10.1111/j.1365-3040.2008.01802.x. [DOI] [PubMed] [Google Scholar]
- 7.Janouškovec J, et al. Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles. eLife. 2019;8:e49662. doi: 10.7554/eLife.49662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janouškovec J, et al. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci. 2015;112:10200–10207. doi: 10.1073/pnas.1423790112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janouškovec J, Horák A, Oborník M, Lukeš J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baird AH, Guest JR, Willis BL. Systematic and Biogeographical Patterns in the Reproductive Biology of Scleractinian Corals. Annu Rev Ecol Evol Syst. 2009;40:551–571. [Google Scholar]
- 11.Schwarz JA, Krupp DA, Weis VM. Late Larval Development and Onset of Symbiosis in the Scleractinian Coral Fungia scutaria. Biol Bull. 1999;196:70–79. doi: 10.2307/1543169. [DOI] [PubMed] [Google Scholar]
- 12.Mansfield KM, Gilmore TD. Innate immunity and cnidarian-Symbiodiniaceae mutualism. Dev Comp Immunol. 2018;90:199–209. doi: 10.1016/j.dci.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 13.Poole AZ, Weis VM. TIR-domain-containing protein repertoire of nine anthozoan species reveals coral–specific expansions and uncharacterized proteins. Dev Comp Immunol. 2014;46:480–488. doi: 10.1016/j.dci.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Buchmann K. Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front Immunol. 2014;5 doi: 10.3389/fimmu.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh J, et al. Invertebrate immune diversity. Dev Comp Immunol. 2011;35:959–974. doi: 10.1016/j.dci.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Detournay O, Schnitzler CE, Poole A, Weis VM. Regulation of cnidarian–dinoflagellate mutualisms: Evidence that activation of a host TGFβ innate immune pathway promotes tolerance of the symbiont. Dev Comp Immunol. 2012;38:525–537. doi: 10.1016/j.dci.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Mansfield KM, et al. Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci Rep. 2017;7:16025. doi: 10.1038/s41598-017-16168-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews JL, et al. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci. 2017;10:201710733-13199. doi: 10.1073/pnas.1710733114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfowicz I, et al. Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci Rep. 2016;6:32366. doi: 10.1038/srep32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berthelier J, et al. Implication of the host TGFβ pathway in the onset of symbiosis between larvae of the coral Fungia scutaria and the dinoflagellate Symbiodinium sp. (clade C1f) Coral Reefs. 2017;36:1263–1268. [Google Scholar]
- 21.Mansfield KM, et al. Varied effects of algal symbionts on transcription factor NF-κB in a sea anemone and a coral: possible roles in symbiosis and thermotolerance. bioRxiv. 2019 doi: 10.1101/640177. [DOI] [Google Scholar]
- 22.Mohamed AR, et al. Deciphering the nature of the coral-Chromera association. ISME J. 2018;12:776–790. doi: 10.1038/s41396-017-0005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M-C, et al. ApRab11, a cnidarian homologue of the recycling regulatory protein Rab11, is involved in the establishment and maintenance of the <i>Aiptasia-<\i>Symbiodinium endosymbiosis. Biochem Biophys Res Commun. 2005;338:1607–1616. doi: 10.1016/j.bbrc.2005.10.133. [DOI] [PubMed] [Google Scholar]
- 24.Chen M-C, Cheng Y-M, Sung P-J, Kuo C-E, Fang L-S. Molecular identification of Rab7 (ApRab7) in Aiptasia pulchella and its exclusion from phagosomes harboring zooxanthellae. Biochem Biophys Res Commun. 2003;308:586–595. doi: 10.1016/s0006-291x(03)01428-1. [DOI] [PubMed] [Google Scholar]
- 25.Davy SK, Allemand D, Weis VM. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev MMBR. 2012;76:229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohamed AR, et al. The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: the symbiosome as an arrested early phagosome. Mol Ecol. 2016;25:3127–3141. doi: 10.1111/mec.13659. [DOI] [PubMed] [Google Scholar]
- 27.Voolstra CR, et al. The host transcriptome remains unaltered during the establishment of coral-algal symbioses. Mol Ecol. 2009;18:1823–1833. doi: 10.1111/j.1365-294X.2009.04167.x. [DOI] [PubMed] [Google Scholar]
- 28.Dunn SR, Weis VM. Apoptosis as a post-phagocytic winnowing mechanism in a coral-dinoflagellate mutualism. Environ Microbiol. 2009;11:268–276. doi: 10.1111/j.1462-2920.2008.01774.x. [DOI] [PubMed] [Google Scholar]
- 29.Baumgarten S, et al. The genome of Aiptasia a sea anemone model for coral symbiosis. Proc Natl Acad Sci. 2015;112:11893–11898. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grawunder D, et al. Induction of Gametogenesis in the Cnidarian Endosymbiosis Model Aiptasia sp. Sci Rep. 2015;5:15677. doi: 10.1038/srep15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hambleton EA, et al. Sterol transfer by atypical cholesterol-binding NPC2 proteins in coral-algal symbiosis. eLife. 2019;8:e43923. doi: 10.7554/eLife.43923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bucher M, Wolfowicz I, Voss PA, Hambleton EA, Guse A. Development and Symbiosis Establishment in the Cnidarian Endosymbiosis Model Aiptasia sp. Sci Rep. 2016;6:19867. doi: 10.1038/srep19867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hambleton EA, Guse A, Pringle JR. Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. J Exp Biol. 2014;217:1613–1619. doi: 10.1242/jeb.095679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaJeunesse TC, et al. Systematic Revision of Symbiodiniaceae Highlights the Antiquity and Diversity of Coral Endosymbionts. Curr Biol. 2018;28:2570–2580.:e6. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Xiang T, Hambleton EA, DeNofrio JC, Pringle JR, Grossman AR. Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity1. J Phycol. 2013;49:447–458. doi: 10.1111/jpy.12055. [DOI] [PubMed] [Google Scholar]
- 36.Cumbo VR, et al. Chromera velia is endosymbiotic in larvae of the reef corals Acropora digitifera and A. tenuis . Protist. 2013;164:237–244. doi: 10.1016/j.protis.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Budisa A, et al. Marine microagae Microchoropsis gaditana and Pseudochoris wilhelmii cultivated in oil refinery wastewater - A perspective on remediation and biodiesel production. Fresenius Environ Bull. 2019;28:10. [Google Scholar]
- 38.Ma X-N, Chen T-P, Yang B, Liu J, Chen F. Lipid Production from Nannochloropsis. Mar Drugs. 2016;14:61. doi: 10.3390/md14040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boulais J, et al. Molecular characterization of the evolution of phagosomes. Mol Syst Biol. 2010;6:423. doi: 10.1038/msb.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flannagan RS, Jaumouillé V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol. 2012;7:61–98. doi: 10.1146/annurev-pathol-011811-132445. [DOI] [PubMed] [Google Scholar]
- 41.Luzio JP, Hackmann Y, Dieckmann NMG, Griffiths GM. The Biogenesis of Lysosomes and Lysosome-Related Organelles. Cold Spring Harb Perspect Biol. 2014;6:a016840-a016840. doi: 10.1101/cshperspect.a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bojarczuk A, et al. Cryptococcus neoformans Intracellular Proliferation and Capsule Size Determines Early Macrophage Control of Infection. Sci Rep. 2016;6:21489. doi: 10.1038/srep21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith LM, May RC. Mechanisms of microbial escape from phagocyte killing. Biochem Soc Trans. 2013;41:475–490. doi: 10.1042/BST20130014. [DOI] [PubMed] [Google Scholar]
- 44.Seoane PI, May RC. Vomocytosis: What we know so far. Cell Microbiol. 2020;22 doi: 10.1111/cmi.13145. [DOI] [PubMed] [Google Scholar]
- 45.Watkins RA, et al. Cryptococcus neoformans escape from Dictyostelium amoeba by both WASH-mediated constitutive exocytosis and vomocytosis. Front Cell Infect Microbiol. 2018;8:108. doi: 10.3389/fcimb.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma H, Croudace JE, Lammas DA, May RC. Expulsion of Live Pathogenic Yeast by Macrophages. Curr Biol. 2006;16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 47.Johnston SA, May RC. The Human Fungal Pathogen <i>Cryptococcus neoformans</> Escapes Macrophages by a Phagosome Emptying Mechanism That Is Inhibited by Arp2/3 Complex-Mediated Actin Polymerisation. PLoS Pathog. 2010;6:e1001041. doi: 10.1371/journal.ppat.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilbert AS, et al. Vomocytosis of live pathogens from macrophages is regulated by the atypical MAP kinase ERK5. Sci Adv. 2017;3:e1700898. doi: 10.1126/sciadv.1700898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gazzinelli RT, Mendonça-Neto R, Lilue J, Howard J, Sher A. Innate Resistance against Toxoplasma gondii: An Evolutionary Tale of Mice, Cats, and Men. Cell Host Microbe. 2014;15:132–138. doi: 10.1016/j.chom.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh D, Stumhofer JS. Do you see what I see: Recognition of protozoan parasites by Toll-like receptors. Curr Immunol Rev. 2013;9:129–140. doi: 10.2174/1573395509666131203225929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Botos I, Segal DM, Davies DR. The Structural Biology of Toll-like Receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosch TCG, et al. Uncovering the evolutionary history of innate immunity: The simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Loiarro M, et al. Pivotal Advance: Inhibition of MyD88 dimerization and recruitment of IRAK1 and IRAK4 by a novel peptidomimetic compound. J Leukoc Biol. 2007;82:801–810. doi: 10.1189/jlb.1206746. [DOI] [PubMed] [Google Scholar]
- 55.Biquand E, et al. Acceptable symbiont cell size differs among cnidarian species and may limit symbiont diversity. ISME J. 2017;11:1702–1712. doi: 10.1038/ismej.2017.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neubauer EF, Poole AZ, Weis VM, Davy SK. The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ. 2016;4:e2692. doi: 10.7717/peerj.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neubauer E-F, et al. A diverse host thrombospondin-type-1 repeat protein repertoire promotes symbiont colonization during establishment of cnidarian-dinoflagellate symbiosis. eLife. 2017;6:961. doi: 10.7554/eLife.24494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarz JA, et al. Coral Life History and Symbiosis: functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genomics. 2008;9:97. doi: 10.1186/1471-2164-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 2008;23:369–376. doi: 10.1016/j.tree.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 60.Ndungu FM, Urban BC, Marsh K, Langhorne J. Regulation of immune response by Plasmodium-infected red blood cells. Parasite Immunol. 2005;27:373–384. doi: 10.1111/j.1365-3024.2005.00771.x. [DOI] [PubMed] [Google Scholar]
- 61.Waghabi MC, Keramidas M, Feige J-J, Araujo-Jorge TC, Bailly S. Activation of transforming growth factor β by Trypanosoma cruzi . Cell Microbiol. 2005;7:511–517. doi: 10.1111/j.1462-5822.2004.00481.x. [DOI] [PubMed] [Google Scholar]
- 62.Alvarez M, Casadevall A. Phagosome Extrusion and Host-Cell Survival after Cryptococcus neoformans Phagocytosis by Macrophages. Curr Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 63.Birmingham CL, et al. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 64.Levitz SM, et al. Cryptococcus neoformans Resides in an Acidic Phagolysosome of Human Macrophages. Infecction Immun. 1999;67:6. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liss V, et al. Salmonella enterica Remodels the Host Cell Endosomal System for Efficient Intravacuolar Nutrition. Cell Host Microbe. 2017;21:390–402. doi: 10.1016/j.chom.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 66.Madan R, Rastogi R, Parashuraman S, Mukhopadhyay A. Salmonella acquires lysosome-associated membrane protein 1 (LAMP1) on phagosomes from Golgi via SipC protein-mediated recruitment of host Syntaxin6. J Biol Chem. 2012;287:5574–5587. doi: 10.1074/jbc.M111.286120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hartenstein V, Martinez P. Phagocytosis in cellular defense and nutrition: a food-centered approach to the evolution of macrophages. Cell Tissue Res. 2019;377:527–547. doi: 10.1007/s00441-019-03096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Downs CA, et al. Symbiophagy as a cellular mechanism for coral bleaching. Autophagy. 2009;5:211–216. doi: 10.4161/auto.5.2.7405. [DOI] [PubMed] [Google Scholar]
- 69.Titlyanov E, et al. Degradation of zooxanthellae and regulation of their density in hermatypic corals. Mar Ecol Prog Ser. 1996;139:167–178. [Google Scholar]
- 70.Chen M-C, Cheng Y-M, Hong M-C, Fang L-S. Molecular cloning of Rab5 (ApRab5) in Aiptasia pulchella and its retention in phagosomes harboring live zooxanthellae. Biochem Biophys Res Commun. 2004;324:1024–1033. doi: 10.1016/j.bbrc.2004.09.151. [DOI] [PubMed] [Google Scholar]
- 71.McCloskey LR, Cove TG, Verde EA. Symbiont expulsion from the anemone Anthopleura elegantissima (Brandt) (Cnidaria; Anthozoa) J Exp Mar Biol Ecol. 1996;195:173–186. [Google Scholar]
- 72.Hoegh-Guldberg O, McCloskey LR, Muscatine L. Expulsion of zooxanthellae by symbiotic cnidarians from the Red Sea. Coral Reefs. 1987;5:201–204. [Google Scholar]
- 73.Hoegh-Guldberg O, Smith GJ. The effect of sudden changes in temperature, light and salinity on the population density and export of zooxanthellae from the reef corals Stylophora pistillata Esper and Seriatopora hystrix Dana. J Exp Mar Biol Ecol. 1989;129:279–303. [Google Scholar]
- 74.Bieri T, Onishi M, Xiang T, Grossman AR, Pringle JR. Relative Contributions of Various Cellular Mechanisms to Loss of Algae during Cnidarian Bleaching. PLoS One. 2016;11:e0152693. doi: 10.1371/journal.pone.0152693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Treuren W, et al. Live imaging of Aiptasia larvae, a model system for coral and anemone bleaching, using a simple microfluidic device. Sci Rep. 2019;9:9275. doi: 10.1038/s41598-019-45167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baghdasarian G, Muscatine L. Preferential expulsion of dividing algal cells as a mechanism for regulating algal-cnidarian symbiosis. Biol Bull. 2000;199:278–286. doi: 10.2307/1543184. [DOI] [PubMed] [Google Scholar]
- 77.Steele R. The Significance of Zooxanthella-Containing Pellets Extruded by Sea Anemones. Bull Mar Sci. 1977;27:591–594(4). [Google Scholar]
- 78.Mieog JC, van Oppen MJH, Cantin NE, Stam WT, Olsen JL. Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs. 2007;26:449–457. [Google Scholar]
- 79.Thornhill DJ, LaJeunesse TC, Kemp DW, Fitt WK, Schmidt GW. Multi-year, seasonal genotypic surveys of coral-algal symbioses reveal prevalent stability or post-bleaching reversion. Mar Biol. 2006;148:711–722. [Google Scholar]
- 80.LaJeunesse TC, et al. Specificity and stability in high latitude eastern Pacific coral-algal symbioses. Limnol Oceanogr. 2008;53:719–727. [Google Scholar]
- 81.Silverstein RN, Correa AMS, Baker AC. Specificity is rarely absolute in coral–algal symbiosis: implications for coral response to climate change. Proc R Soc B Biol Sci. 2012;279:2609–2618. doi: 10.1098/rspb.2012.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chrisman CJ, Alvarez M, Casadevall A. Phagocytosis of Cryptococcus neoformans by, and Nonlytic Exocytosis from, Acanthamoeba castellanii . Appl Environ Microbiol. 2010;76:6056–6062. doi: 10.1128/AEM.00812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by Tubercular Mycobacteria Is Spread by Nonlytic Ejection from Their Amoeba Hosts. Science. 2009;323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miao Y, Wu J, Abraham SN. Ubiquitination of Innate Immune Regulator TRAF3 Orchestrates Expulsion of Intracellular Bacteria by Exocyst Complex. Immunity. 2016;45:94–105. doi: 10.1016/j.immuni.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Song J, et al. TLR4-mediated expulsion of bacteria from infected bladder epithelial cells. Proc Natl Acad Sci. 2009;106:14966–14971. doi: 10.1073/pnas.0900527106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith LM, Dixon EF, May RC. The fungal pathogen Cryptococcus neoformans manipulates macrophage phagosome maturation. Cell Microbiol. 2015;17:702–713. doi: 10.1111/cmi.12394. [DOI] [PubMed] [Google Scholar]
- 87.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 89.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.R Core Team. R: A language and environment for statistical computing. 2018 [Google Scholar]
- 91.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma Oxf Engl. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 92.Berthold MR, et al. Studies in classification, data analysis, and knowledge organization (GfKL 2007) Springer; 2007. KNIME: the Konstanz Information Miner. [Google Scholar]
- 93.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wood SN. Generalized Additive Models: An Introduction with R. Chapman and Hall/CRC; 2017. [DOI] [Google Scholar]
- 96.Keeling PJ, Burki F. Progress towards the Tree of Eukaryotes. Curr Biol. 2019;29:R808–R817. doi: 10.1016/j.cub.2019.07.031. [DOI] [PubMed] [Google Scholar]
- 97.Hardiman G, Rock FL, Balasubramanian S, Kastelein RA, Bazan JF. Molecular characterization and modular analysis of human MyD88. Oncogene. 1996;13:2467–2475. [PubMed] [Google Scholar]
- 98.Vyncke L, et al. Reconstructing the TIR Side of the Myddosome: a Paradigm for TIR-TIR Interactions. Structure. 2016;24:437–447. doi: 10.1016/j.str.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 99.Loiarro M, et al. Peptide-mediated Interference of TIR Domain Dimerization in MyD88 Inhibits Interleukin-1-dependent Activation of NF-κB. J Biol Chem. 2005;280:15809–15814. doi: 10.1074/jbc.C400613200. [DOI] [PubMed] [Google Scholar]
- 100.Loiarro M, Ruggiero V, Sette C. Targeting TLR/IL-1R signalling in human diseases. Mediators Inflamm. 2010;2010:674363. doi: 10.1155/2010/674363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Winchester BG. Lysosomal membrane proteins. Eur J Paediatr Neurol. 2001;5:11–19. doi: 10.1053/ejpn.2000.0428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data: raw reads can be accessed at NCBI sequence read archive (SRA) with following accession numbers: cells from aposymbiotic larvae (SRX7119772-7119776), symbiotic cells (SRX7119782-7119787), aposymbiotic cells from symbiotic larvae (SRX7119777-7119781), combined in the SRA project SRP229372, M. gaditana-containing cells (SRX7229078-7229080), and microalgae-free cells from M. gaditana-containing larvae (SRX7229075-7229077), combined in the SRA project SRP233508.
The Knime 92 workflow used for analysis as well as other raw data and R scripts for analysis can be found under https://doi.org/10.24433/CO.0872345.v1